 Open Access
Open Access
ARTICLE
Convolutional Neural Network-Based Classification of Multiple Retinal Diseases Using Fundus Images
Lahore College for Women University, Lahore, 54000, Pakistan
* Corresponding Author: Saima Farhan. Email:
Intelligent Automation & Soft Computing 2023, 36(3), 2607-2622. https://doi.org/10.32604/iasc.2023.034041
Received 05 July 2022; Accepted 22 September 2022; Issue published 15 March 2023
Abstract
Use of deep learning algorithms for the investigation and analysis of medical images has emerged as a powerful technique. The increase in retinal diseases is alarming as it may lead to permanent blindness if left untreated. Automation of the diagnosis process of retinal diseases not only assists ophthalmologists in correct decision-making but saves time also. Several researchers have worked on automated retinal disease classification but restricted either to hand-crafted feature selection or binary classification. This paper presents a deep learning-based approach for the automated classification of multiple retinal diseases using fundus images. For this research, the data has been collected and combined from three distinct sources. The images are preprocessed for enhancing the details. Six layers of the convolutional neural network (CNN) are used for the automated feature extraction and classification of 20 retinal diseases. It is observed that the results are reliant on the number of classes. For binary classification (healthy vs. unhealthy), up to 100% accuracy has been achieved. When 16 classes are used (treating stages of a disease as a single class), 93.3% accuracy, 92% sensitivity and 93% specificity have been obtained respectively. For 20 classes (treating stages of the disease as separate classes), the accuracy, sensitivity and specificity have dropped to 92.4%, 92% and 92% respectively.Keywords
The retina is a thin layer of tissue located at the back wall of a human eye and contributes to vision formation. It comprises millions of light-sensitive nerve cells, optic nerve and macula. Any retinal disorder can interrupt its function, leading to gradual vision loss or blindness. The disorders can occur in the macula, optic nerve, or retinal vessels. With the increase in life expectancy of the population, the number of patients suffering from chorioretinal diseases has also increased [1]. It is observed that 300 million people are visually impaired and 45 million are blind all over the world, with more than 90% in developing countries [2]. These figures are growing day by day and it is estimated that the elder population, especially over 75 years of age, have a high prevalence of visual impairments [1]. Diagnosis and treatment at an early stage are crucial to overcome this problem, especially for the high-risk group. Ophthalmic photography is for the most part and ordinarily utilized by the ophthalmologist to contemplate the status of a patient’s eye and diagnose the disease, if any. Ophthalmic photographs are taken using special eye imaging equipment. These include optical coherence tomography (OCT), color fundus retinal photography, fluorescein angiography, indocyanine green angiography, corneal topography and slit-lamp photography.
With the advancements in technology, computer systems can be used to automate the diagnosis process of eye diseases. This helps in replacing the manual analysis of ophthalmic images by the ophthalmologists. Time and effort taken in manual diagnosis can, therefore, be saved and utilized for the treatment of the disease. Since the rise of machine learning techniques, computers are capable of training themselves from environment and perform appropriate actions according to circumstances just as a human brain would do. Several researchers suggested various methods of machine learning to classify eye diseases. They proposed the use of artificial neural networks (ANN), support vector machines (SVM), Naive Bayes, K-nearest neighbors (KNN), in addition to many more, to classify different retinal diseases [3–5].
Although machine learning techniques have outperformed several manual and traditional approaches yet they have some limitations. The major issue with these methods is their support of hand-crafted features. The selection of features may vary and affect results each time. Moreover, they are expensive and become a bottleneck for traditional machine learning methods. Deep learning algorithms overcome the limitations posed by traditional machine learning algorithms. It is a subset of machine learning known as “deep neural network” because of the association of multiple layers of neural network [6]. Researchers moved to automated feature extraction using a deep learning approach instead of hand-crafted feature selection as traditional machine learning does.
Different researchers came up with deep learning approaches for feature extraction and selection. They worked on Deep Belief Networks (DBN), CNN and pre-trained transfer learning models like inception V3 and AlexNet for the classification of retinal diseases. They contributed to the automation of Age-related Macular Degeneration (AMD) diagnosis [7,8], automated detection of Diabetic Retinopathy (DR) [9,10], glaucoma and cataract automatic identification [11,12]. They also contributed in extracting various retinal parts of the fundus images to find irregularity if any e.g., segmentation of retinal blood vessels [13,14], classification of arterioles and venules [15], and detection of papilledema [16]. But still, gates to the multi-category classification of multiple retinal diseases are open.
This paper presents a deep learning approach to classify fundus retinal images for multi-category data after performing a few preprocessing steps on the images. The deep learning approach proposed here does not require the extraction and selection of hand-crafted features. The paper is organized as follows: details of the dataset used by this study, the preprocessing steps and the structure of the proposed deep learning model are presented in the Materials and Methods section. Outcomes are organized in the Results section and the findings of this research compared with the literature are presented in the Discussion section. The paper ends with a conclusion.
Insights of the dataset, data preprocessing operations and the CNN model proposed in this study are presented in this section.
Publicly accessible retinal images datasets are smaller in size and are insufficient for multiple retinal disease classification. These datasets either contain merely a couple of retinal diseases or just stages of a particular single disease. Deep learning models demand a huge measure of information to train them. This research, therefore, consolidated data by taking retinal images from three distinct resources including project of STARE (Structured Analysis of the Retina) (http://cecas.clemson.edu/∼ahoover/stare/), dataset provided by Kaggle team for competition purpose (https://www.kaggle.com/linchundan/fundusimage1000) and High-Resolution Fundus (HRF) image database [17]. The STARE dataset contains 400 images of different retinal diseases. The dataset is highly imbalanced as some diseases have hardly one or two instances, whereas a few are not even labelled. Excluding such images, the rest are used in this research. The Kaggle dataset comprises 1000 images belonging to 37 categories of retinal diseases. A subset of diseases from this dataset has been selected for the proposed model. The HRF image dataset contains 45 images and is uniquely intended for vessel segmentation. The entire dataset has been incorporated into the present research for the identification of retinal diseases.
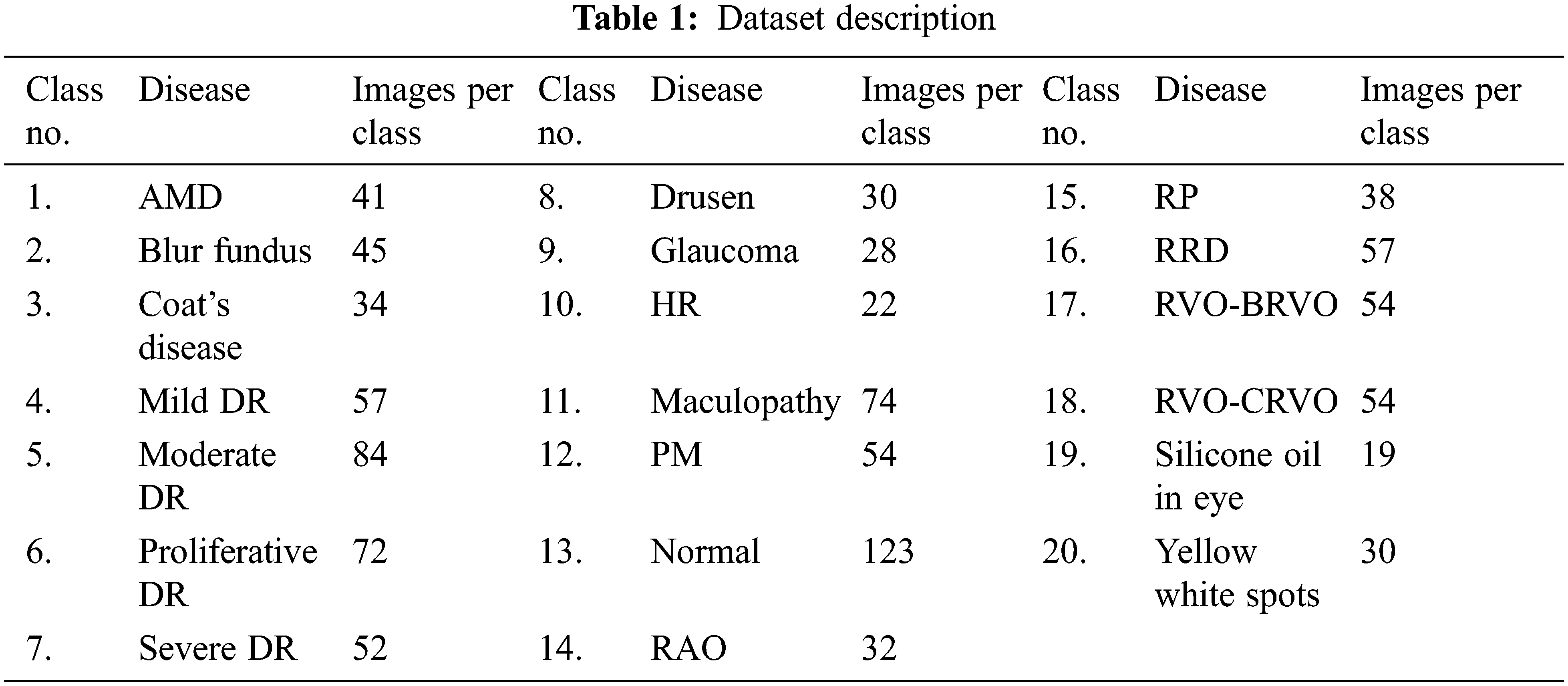
Retinal diseases covered in this research are AMD, Blur Fundus, Coat’s disease, Mild DR, Moderate DR, Proliferative DR (PDR), Severe DR, Drusen, Glaucoma, Hypertensive Retinopathy (HR), Maculopathy, Pathologic Myopia (PM), Retinal Artery Occlusion (RAO), Retinitis Pigmentosa (RP), Rhegmatogenous Retinal Detachment (RRD), Retinal Vein Occlusion (RVO)–(i) Branch Retinal Vein Occlusion (BRVO) and (ii) Central Retinal Vein Occlusion (CRVO), Silicone Oil in eye and Yellow White spots. The proposed CNN model is capable of distinguishing among the diseases as well as their stages. A normal control class is also included for distinction in addition to 19 case classes. Retinal diseases included in this research and the number of instances against each disease/stage are presented in Table 1.
Fundus images in the dataset vary in size as these have been collected from different sources. The first step in preprocessing is resizing and scaling of the images. Images with higher resolution generate more parameters while going through CNN layers, requiring more space to reside in memory. Moreover, the learning model must handle all those parameters resulting in higher computational costs. Generally, a large set of parameters and a small number of training instances may lead to overfitting. ImageNet pre-trained models use images with a resolution of 224 × 224. Therefore, all images in the dataset are resized to a resolution of 224 × 224 to save memory and computational cost.
The second task is the selection of green channel from RGB fundus images. Processing images with three channels demand more computational cost. Conversion is possible either by splitting channels or by grayscale conversion. In fundus images, the vascular plane of the retina, macula, optic nerve and blood vessels must be visible for the analysis and diagnosis of the disease. Green channel of an image provides more details that can be useful in the diagnosis process. The red, green and blue channels as well as grayscale images are presented in Fig. 1.

Figure 1: RGB image, red, blue, green channels and gray scale image
Most retinal diseases affect blood vessels which are extremely thin in nature. Sharpening operation enhances these vessels by convolving the image
It is observed that the macula is always dull and the optic disc is bright in fundus images. Contrast adjustment rectifies the illumination effect on vessels, macula and optic disc. Contrast Limited Adaptive Histogram Equalization (CLAHE) has been performed to enhance the low-contrast regions by clipping limit to avoid noise.
A deep learning model to classify multi-retinal diseases, comprising various CNN layers, is presented in this paper. The flow of preprocessing steps and the deep learning model are depicted in Fig. 2.

Figure 2: Flow diagram of proposed work
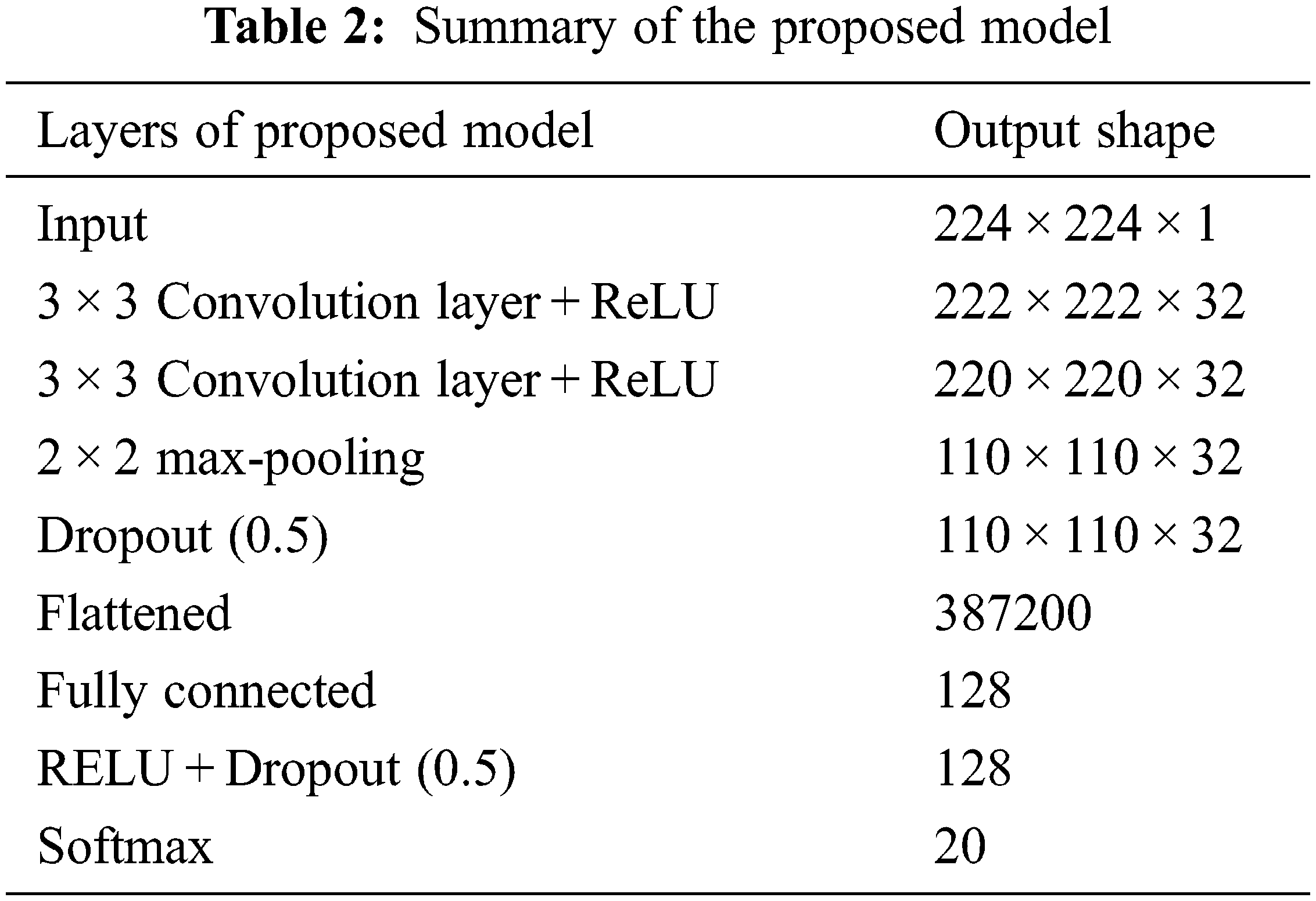
The presented deep learning model is constituted using different CNN layers. CNN is a well-known deep learning model, especially for the classification of images. Various researchers have presented its variations by changing the number and arrangement of CNN layers. However, its fundamental layers i.e., convolutional layers, activation functions, pooling layers and fully connected layers remain almost the same [18]. Convolutional layers are always meant to be the primary layers of the network that act as feature identifier. All other layers are responsible for dimensionality reduction and to overcome the overfitting problem. Activation layers offer non-linearity to the system to speed up computations. Pooling layers perform downsampling and the fully connected layer selects the strongly correlated features belonging to a particular class. The proposed model uses a pair of convolutional layers alongside Rectifier Linear Unit (ReLU) activation functions, dropout layers, fully connected layer and softmax layer. Each layer utilizes the outputs of its predecessor layer, as shown in Eq. (2).
In each convolutional layer, input images are convolved with 32 filters of size 3 × 3 to extract features from them as expressed in Eq. (3).
ReLU activation function has been applied after each convolutional layer to activate the function depending on the values of x. ReLU activation function f(x) is shown in Eq. (4).
The task of the pooling layer is to reduce the resolution of the images. Max pooling algorithm has been applied since it reserves a maximum value in a pooling region with a stride size of 2 for downsampling. Dropout layer is added to the model with dropout rate of 0.5 to avoid overfitting [19]. All these layers worked as automatic feature learning and extraction. Following it, the outcomes have been transformed to a single dimension (flattened) and a fully connected layer is stacked on the previous layers which serves as a classification layer. This layer has a large number of parameters because of connections to the previous layer’s neurons and may result in overfitting. To avoid this problem, ReLU and dropout layers are added. The last layer is softmax, which capitalizes on the softmax function by providing an end-to-end trainable model. The overall arrangement of layers with their output shape is presented in Table 2.
The proposed model has been implemented with 10-fold cross-validation on 1000 images belonging to 20 categories. Once a model is structured and implemented, its correctness and reliability can be assessed using various evaluation metrics. Accuracy is one such metric that reflects the correctly classified instances. However, the retinal fundus imaging dataset used in this research has imbalanced classes; therefore, model evaluation based only on accuracy is not enough. Two evaluation metrics i.e., sensitivity and specificity are also considered. Sensitivity measures the strength of the model for correctly classifying true diseased subjects, whereas, specificity measures the competence of the model for correctly identifying those subjects that do not belong to a particular class.
This study has three assessment dimensions. Initially, all images are classified into binary categories after preprocessing. Unfortunately, the model has not learned features appropriately for normal vs. abnormal subjects because of the imbalanced nature of the dataset (123 vs. 877). For this reason, binary classification has been performed over a single category vs. a single category. The results are presented as accuracy, sensitivity and specificity as shown in Tables 3, 4 and 5 respectively.
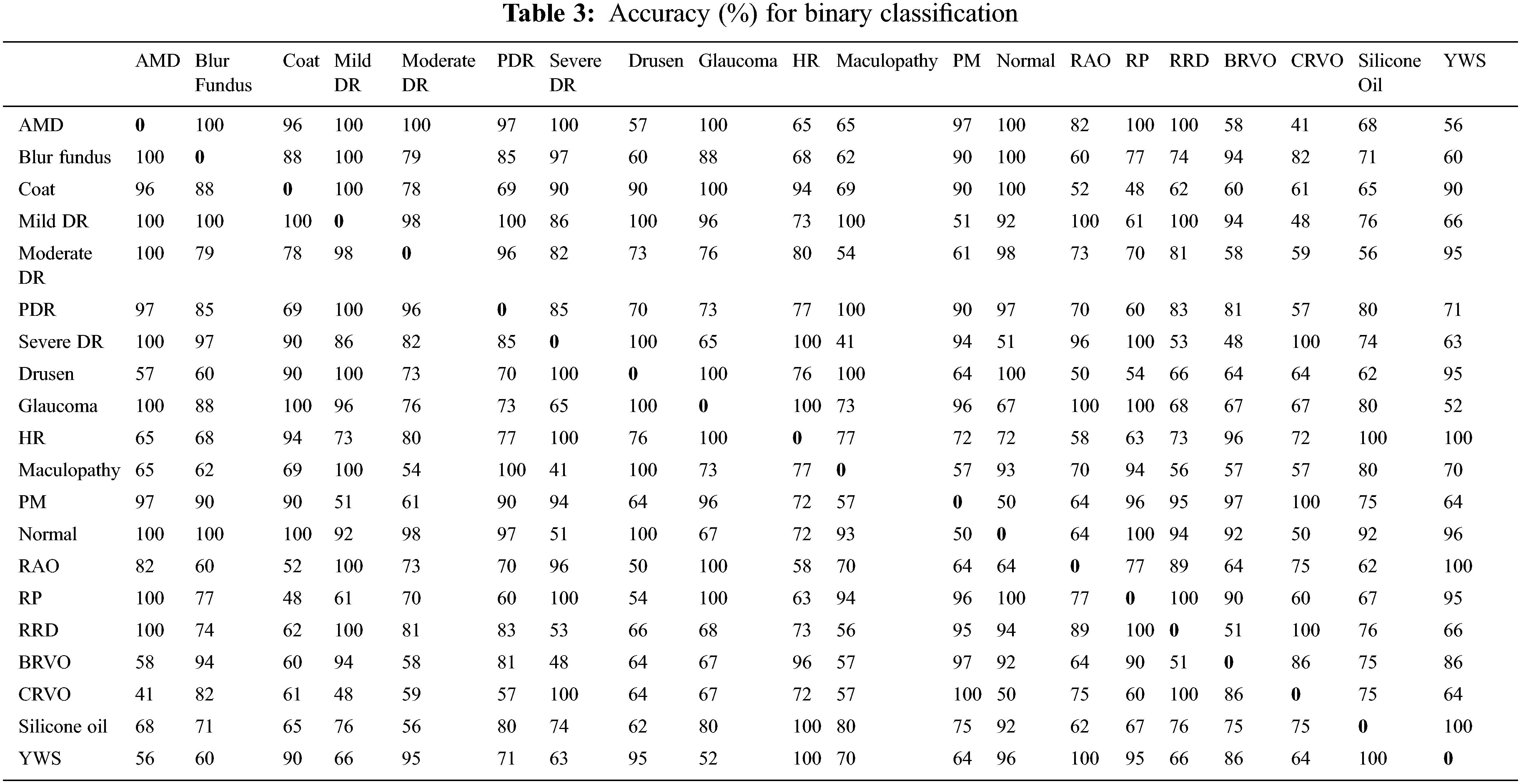
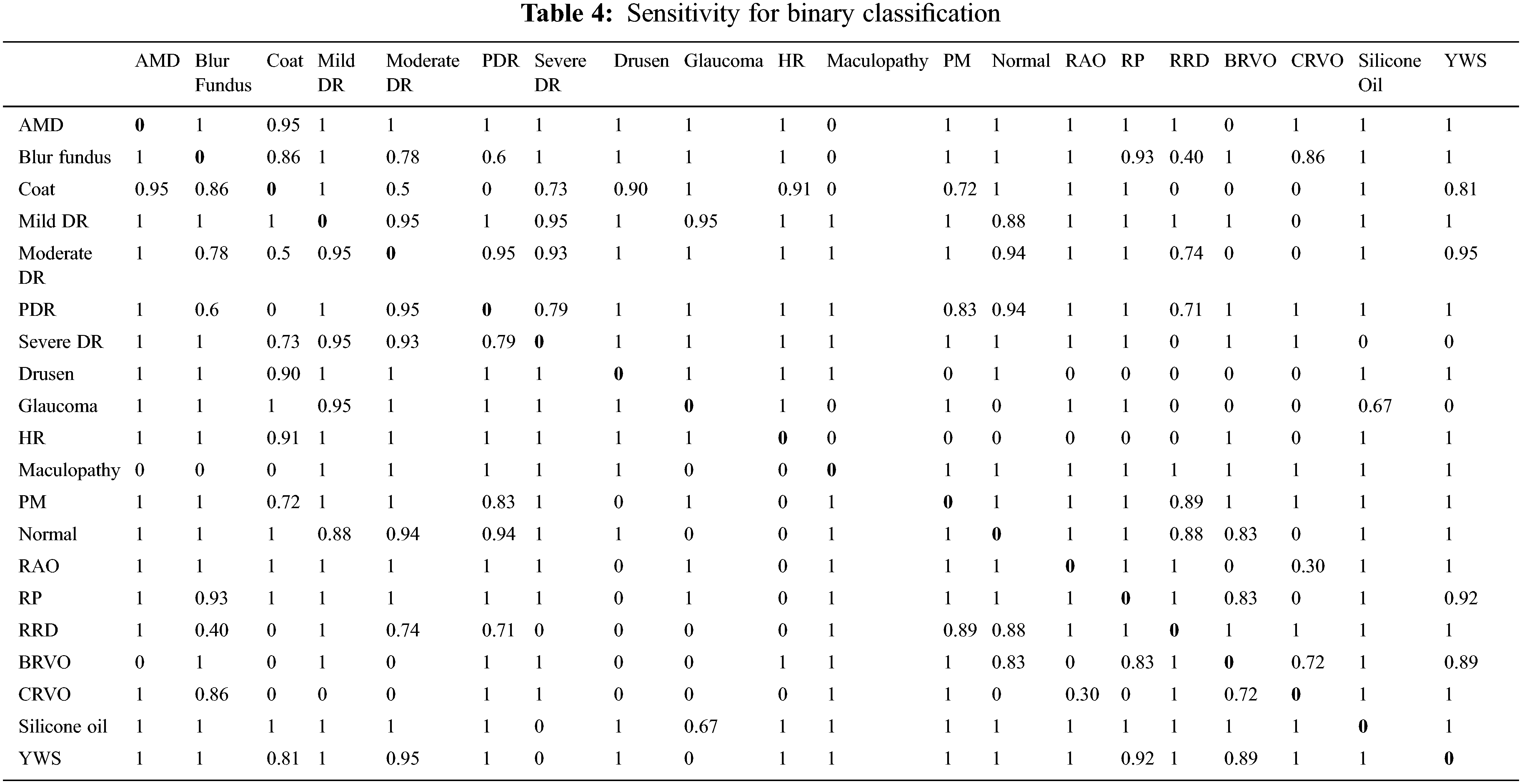
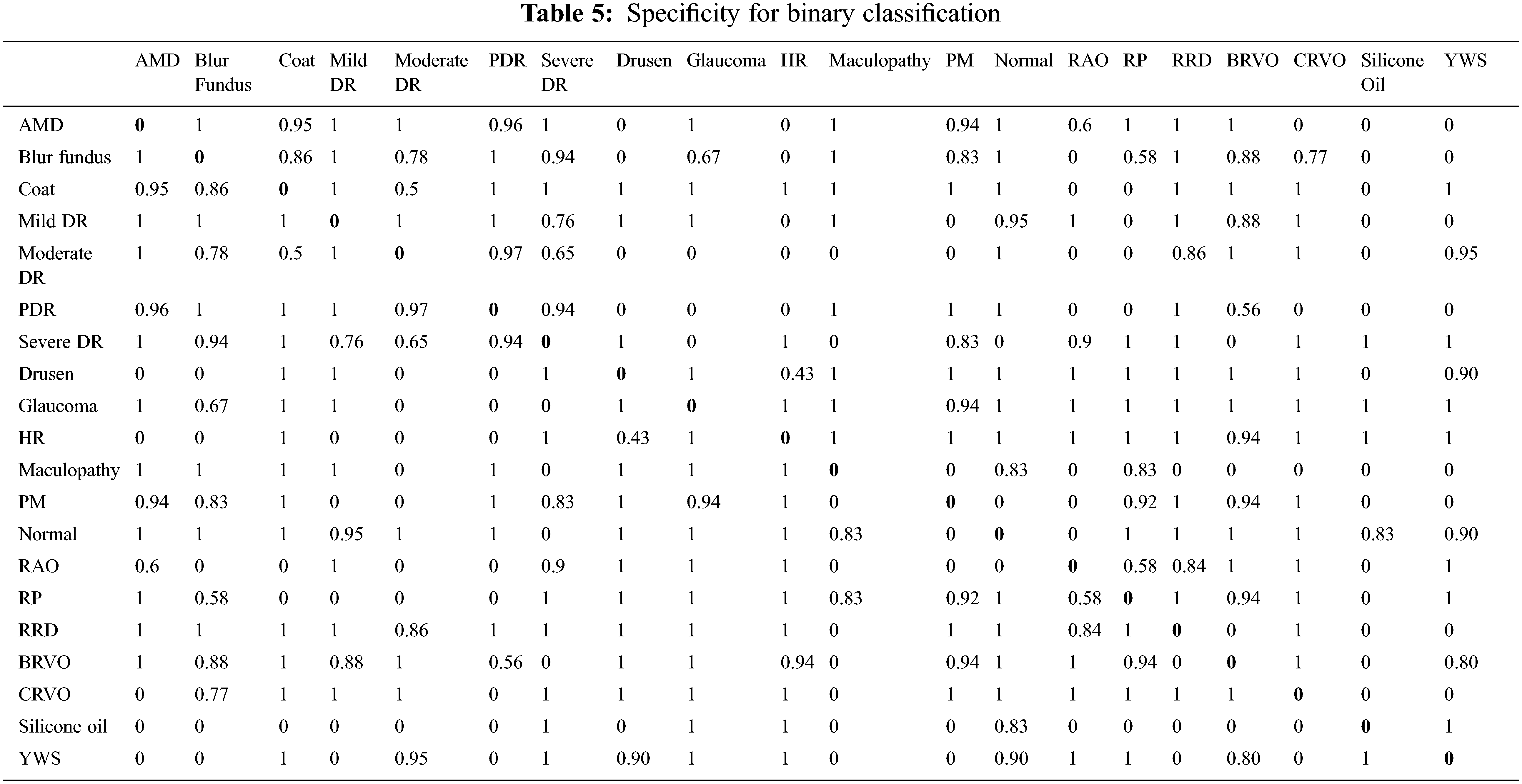
Disease vs. disease classification is insignificant for ophthalmologists. Normal vs. disease classification is more logical and practical. Emphasis has been given to normal vs. disease classification and the obtained results are shown in Table 6. It has been observed that in most cases, it earns up to 100% results, but for a few cases model could not learn due to data biases. The solution for the imbalanced class problem is data augmentation. Since binary classification is not the primary focus of our research, therefore, data augmentation technique is not connected here and left as future work.
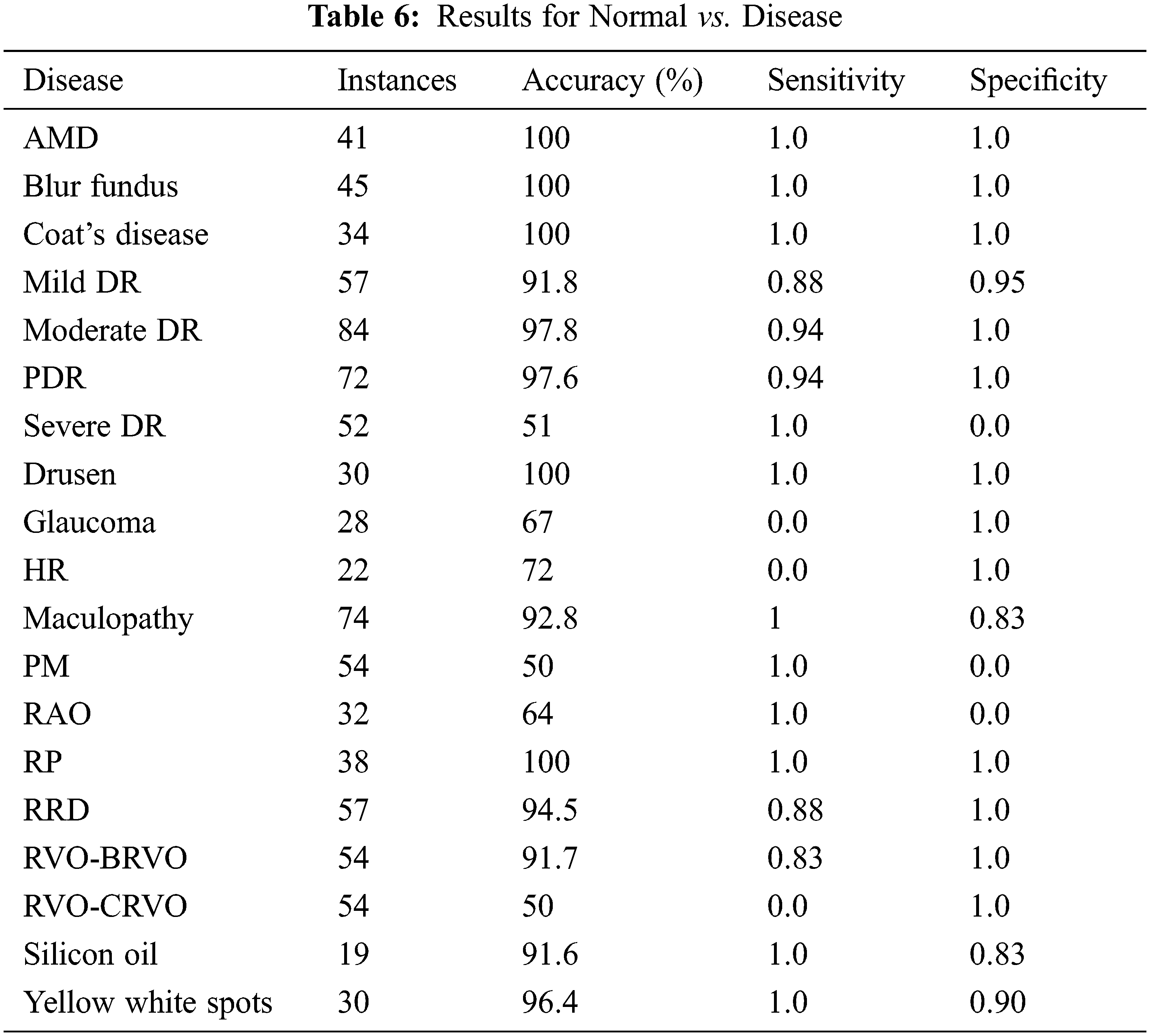
The second dimension of the study is concerned with multi-categories classification and analysis of its results. The total number of classes considered here is 16 (15 disease classes and one normal control class). The diseases consist of AMD, blur fundus, coat’s disease, DR, Drusen, glaucoma, HR, maculopathy, PM, RAO, RP, RRD, RVO, silicon oil in eye and yellow-white spots. It is necessary to mention here that sub-classes/stages of various diseases have been collectively taken as a single class. The proposed model has been tested with original as well as preprocessed images. It is observed that classification without preprocessing turned out to be unsatisfactory. Improvement in classification results can be witnessed with an enhancement of original images. The presented accuracy is 93.3% with preprocessing and 43.5% without preprocessing. The achieved sensitivity and specificity with preprocessing are 0.92 and 0.93 respectively. The results are presented in Table 7 and Receiver Operating Characteristic (ROC) curves for all the classes are shown in Fig. 3a.
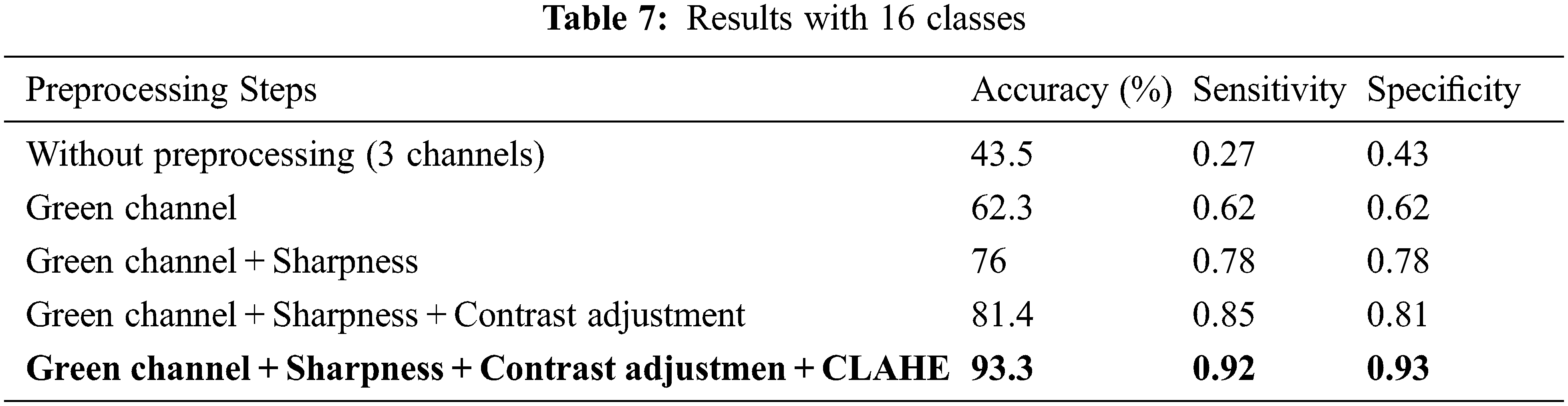

Figure 3: ROC curve for (a) 16 classes, (b) 20 classes
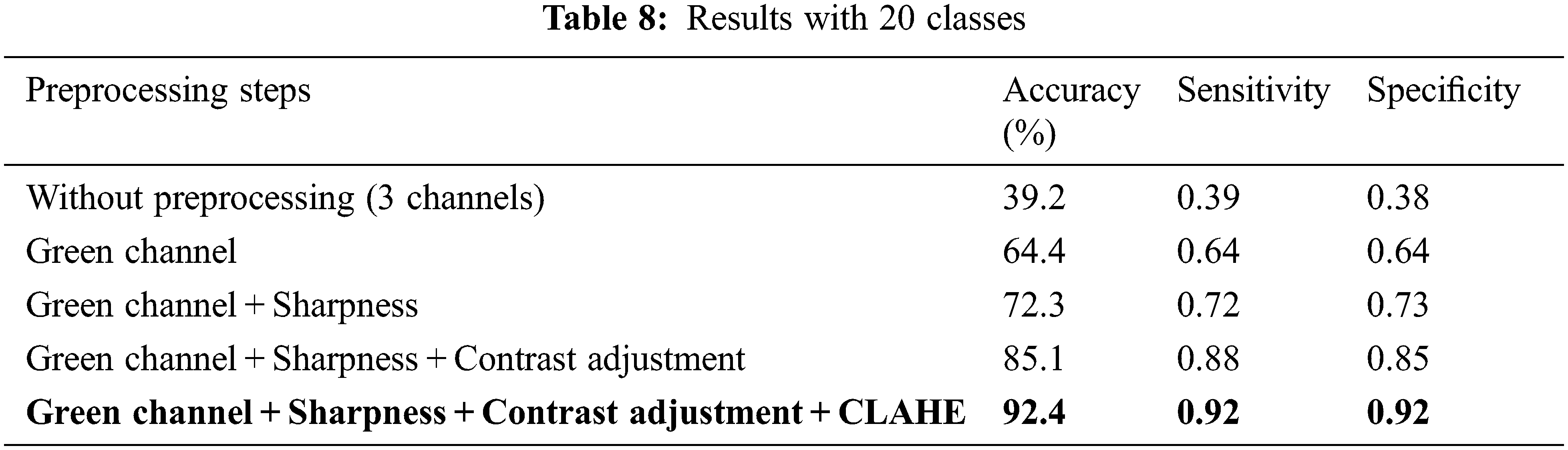
The third dimension of this study is the classification of fundus images by treating all stages of various diseases as separate classes. Here, altogether 20 classes are considered (19 disease classes along with their stages as separate classes and one normal control class). The diseases with substages/classes include DR (mild, moderate, severe and proliferative) and RVO (BRVO and CRVO). The results have very slightly dropped due to the addition of sub-stages of diseases as compared to previous results with 16 classes. The results including accuracy, sensitivity and specificity are presented in Table 8. Accuracy for preprocessed images has turned out to be 92.4%, whereas without preprocessing it was 39.2%. Sensitivity and Specificity with preprocessing are 0.92. The ROC curves for all 20 classes are shown in Fig. 3b.
It is observed that most of the past works are lacking in either of 3 things: (i) the dataset size is small (ii) research is conducted for binary classification only and/or (iii) an engineered set of features is used. This study copes with these lacking by bringing a comparatively large dataset into play and introducing an automated approach for the identification of 19 distinct retinal diseases using CNN which does end-to-end classification and drops the step of hand-crafted feature extraction and selection. The preprocessing operations applied in this study comprise green channel selection, image sharpening, contrast adjustment, and CLAHE to enhance the image features required by the model. It has been observed that with the proposed preprocessing steps, the outcomes of the model have improved.
A lot of research has been done on the classification of various retinal diseases using traditional machine learning algorithms [20–24] with less work on deep learning. The traditional methods suffer from hand-crafted feature extraction which introduces bias. Most importantly, the existing researches cover binary classification or classification of just a couple of retinal diseases presenting fair results as compared to our research which covers a wide range of retinal diseases showing excellent results.
Binary classification of diabetic retinopathy (healthy vs. unhealthy) using five layers of CNN for feature extraction and decision tree for classification is performed with fivefold cross-validation, achieving 97% accuracy, 94% sensitivity and 98% specificity [25]. The fundus images are brightness adjusted and resized to 512 × 512. Another research regarding diabetic retinopathy classification using the data augmentation technique on 1000 images collected from Kaggle Community achieved 94.5% accuracy [26]. Again the research is focused on binary classification using CNN. The model consists of four blocks each containing two CNN layers with varying filter sizes followed by max-pooling layers and two fully connected layers. Data augmentation is performed by applying rotation, flipping, shearing, rescaling and translation on the original three-channel images to enlarge the dataset. The preprocessing steps of our proposed approach are well-suited to improve results rather than augmentation techniques for data expansion. A CNN architecture with six layers for feature extraction and a softmax layer for classification of arteriole and venule from retinal fundus images resulted in 82.26% accuracy, 82% sensitivity and specificity [15]. The research involves various preprocessing steps to perform data augmentation, morphological thinning and segmentation of the fundus images before classification.
It is tricky to tackle multiple classes for classification. Some key points must be considered for classifying data consisting of a large number of categories [27]. The size of a dataset matters a great deal because convolutional layers demand a huge measure of contribution to extract features. In contrast to the previous studies, the dataset in the proposed research is built up from three distinct resources to provide a large and more diverse range of fundus images for training and testing. The constructed dataset comprises a total of 1000 images that have been gathered from publicly available resources. The details of the dataset and resources used in this research are provided in the materials and methods section. One of the reasons for the improved results of this research work is that the convolutional layers can extract a variety of features from a wide range of input as compared to previous works. Secondly, more computation sometimes affects performance. At times, a common or widely used CNN architecture may not be able to extract features for multiple classes due to computational expensiveness. Therefore, this research focuses on preprocessing to highlight features and utilizes a deep learning model with two convolutional layers to extract features. Next, the density or resolution of input images must be critically chosen. The images with greater density produce a large number of parameters that require more memory space and make the model computationally expensive. Consequently, it is recommended to resize input images to a resolution of 224 × 224 or 200 × 200. The renowned pre-trained models like VGG, ResNet and AlexNet, etc. also get images with a resolution of 224 × 224. This exploration has been executed on a 64-bit CPU with 4 GB RAM. The input images have been resized to a resolution of 224 × 224 as one of its preprocessing steps to make it consume less memory and remain computationally inexpensive.
There is a lot of valuable work by a significant number of researchers on the classification of retinal diseases using a deep learning approach using different retinal imaging modalities. Some of the researchers constructed their deep learning models, while others utilized pre-trained models like VGG Net, AlexNet and Inception V3, etc. Karri presented a paper on classification using the pre-trained model GoogleNet with three classes i.e., Diabetic macular edema, dry AMD and normal controls using OCT imaging modality with an accuracy of 94% [28]. Using slit lamp imaging modality, Gao contributed to the automated feature learning of cataract disease with remarkable results [29]. DBN for feature extraction, General Regression Neural Network (GRNN) for selecting robust features and reducing dimensionality, and SVM for classification of fundus AMD, diabetic and healthy subjects have been proposed by showing up to 96.73% accuracy, 0.79 sensitivity and 0.97 specificity [30].
Most of the researchers worked for binary classification with less focus on multi-category classification of retinal diseases. A few studies exist that cover multiple retinal diseases or the classification of multiple stages of a single disease but the results are not much satisfactory. J. Y. Choi presented a pilot study employing a small database to classify retinal images consisting of ten classes [31]. STARE database, which is also a part of this research, has been investigated with 279 images by applying data augmentation with translation and rotation, brightness enhancement and additive Gaussian noise to expand the dataset. It utilized a pre-trained model VGG-19 for feature extraction and random forest for classification. Classification of nine retinal diseases is performed involving Background DR, PDR, Dry AMD, Wet AMD, RVO, RAO, HR, coat’s disease and retinitis with an accuracy of 30.5%. All of the specified nine diseases are covered in the proposed research. The worthwhile findings of the research are that with the increase of categories, the performance of the model degrades. It can be observed that for multi-category classification the results are not satisfactory as compared to the proposed approach.
The prediction of fundus AMD with 13 classes (nine AREDS stages, three late AMD stages and one for unlabeled images) is conducted by extracting features using CNN and performing classification using an ensemble of random forest [8]. The images are processed for illumination correction and color balance and are resized to 512 × 512 pixels. The research resulted in an accuracy of 63.3%. The study is particularly for AMD detection which appears at macula only, therefore, it adapted illumination correction and color balance. The target of our proposed research is the classification of various diseases which appear in the macula, optic nerves and vessels. So, the normalization of input images has been done by contrast adjustment and CLAHE and achieved 92.4% accuracy.
Another study is conducted for the identification of five retinal diseases using a dataset of 157 instances of STARE database. The dataset is preprocessed with an upgraded CLAHE filter and data augmentation is applied. The study achieved 100% results [32]. Although several kinds of research are conducted for the retinal disease diagnosis based on deep learning strategies using fundus photographs [33–35] but all these researches cover a small range of images. In contrast, this paper presents a study with a large scope. It makes use of a large dataset collected from different sources and works on a vast variety of retinal diseases.
Regardless of the high accuracies as reported in Table 9, the advantages of the proposed approach overweight others. Our approach targeted classification of a wide range of retinal diseases i.e., 20, validated over ten folds cross-validation. The results of the proposed work have been evaluated in three dimensions i.e., (i) binary classification producing an accuracy of up to 100%, (ii) multi-class classification with 16 classes (stages of diseases taken as the same class) producing 93.3% accuracy and (iii) multi-class classification with 20 classes (stages of diseases as separate classes) producing 92.4% accuracy. This research is different from existing ones in that it not only deals with major retinal diseases but also produces excellent results. Feature engineering through CNN results in robust features to be the candidate for differentiating multiple diseases.
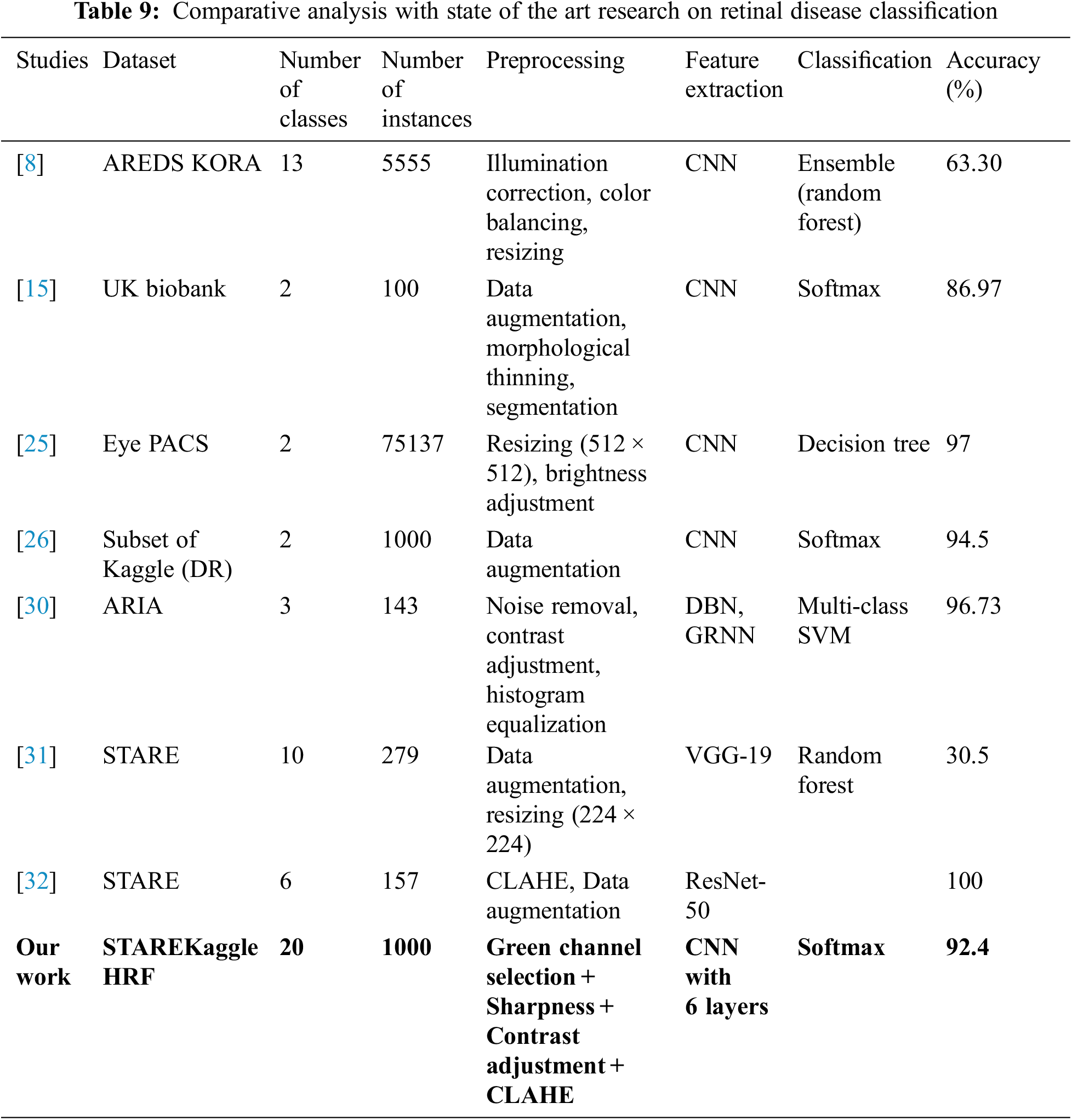
A comprehensive comparison of state-of-the-art research with our proposed approach is presented in Table 9. The comparison focuses on the dataset used, size of the dataset, number of classes in the dataset, preprocessing operations, the technique used for feature extraction, classification models used and results achieved by the approach.
This paper has proposed a deep learning model for the automated identification of multiple retinal diseases using fundus imaging modality. Six layers of the convolutional neural network have been used for feature extraction and a softmax layer for classification after pursuing some image processing techniques as preprocessing steps. The paper comes up with auspicious results. For binary classification, we achieved up to 100% accuracy. With 16 classes the obtained accuracy is 93.3%, whereas, for 20 classes, it turned out to be 92.4%. The research concludes that as the number of classes increases, results deteriorate. Compared to existing studies, it has been observed that proposed preprocessing steps along with the CNN, result in promising outcomes. In the future, it is planned to solve the imbalanced classes issue using the data augmentation technique and to incorporate Principal Component Analysis (PCA) to speed up computations. Moreover, the proposed model is planned to be implemented in the clinical environment to assist ophthalmologists.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. R. Klein and B. E. Klein, “The prevalence of age-related eye diseases and visual impairment in aging: Current estimates,” Investigative Ophthalmology & Visual Science, vol. 54, no. 14, pp. ORSF5–ORSF13, 2013. [Google Scholar]
2. J. Muecke, D. I. Sia, H. Newland, R. J. Casson and D. Selva, “Perspective on ophthalmic support in countries of the developing world,” Clinical & Experimental Ophthalmology, vol. 41, no. 3, pp. 263–271, 2013. [Google Scholar]
3. U. R. Acharya, C. M. Lim, E. Y. K. Ng, C. Chee and T. Tamura, “Computer-based detection of diabetes retinopathy stages using digital fundus images,” Proceedings of the Institution of Mechanical Engineers, Part H: Journal of Engineering in Medicine, vol. 223, no. 5, pp. 545–553, 2009. [Google Scholar] [PubMed]
4. J. Nayak, R. Acharya, P. S. Bhat, N. Shetty and T. -C. Lim, “Automated diagnosis of glaucoma using digital fundus images,” Journal of Medical Systems, vol. 33, no.5, pp. 337–346, 2009. [Google Scholar] [PubMed]
5. Z. Qiao, Q. Zhang, Y. Dong and J. -J. Yang, “Application of SVM based on genetic algorithm in classification of cataract fundus images,” in 2017 IEEE Int. Conf. on Imaging Systems and Techniques (IST), Beijing, BJ, China, pp. 1–5, 2017. [Google Scholar]
6. Y. LeCun, Y. Bengio and G. Hinton, “Deep learning,” Nature, vol. 521, no.7553, pp. 436–444, 2015. [Google Scholar] [PubMed]
7. P. Burlina, K. D. Pacheco, N. Joshi, D. E. Freund and N. M. Bressler, “Comparing humans and deep learning performance for grading AMD: A study in using universal deep features and transfer learning for automated AMD analysis,” Computers in Biology and Medicine, vol. 82, pp. 80–86, 2017. [Google Scholar] [PubMed]
8. F. Grassmann, J. Mengelkamp, C. Brandl, S. Harsch, M. E. Zimmermann et al., “A deep learning algorithm for prediction of age-related eye disease study severity scale for age-related macular degeneration from color fundus photography,” Ophthalmology, vol. 125, no. 9, pp. 1410–1420, 2018. [Google Scholar] [PubMed]
9. V. Gulshan, L. Peng, M. Coram, M. C. Stumpe, D. Wu et al., “Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs,” Jama, vol. 316, no. 22, pp. 2402–2410, 2016. [Google Scholar] [PubMed]
10. L. Zhou, Y. Zhao, J. Yang, Q. Yu and X. Xu, “Deep multiple instance learning for automatic detection of diabetic retinopathy in retinal images,” IET Image Processing, vol. 12, no. 4, pp. 563–571, 2018. [Google Scholar]
11. M. Christopher, A. Belghith, C. Bowd, J. A. Proudfoot, M. H. Goldbaum et al., “Performance of deep learning architectures and transfer learning for detecting glaucomatous optic neuropathy in fundus photographs,” Scientific Reports, vol. 8, no. 1, pp. 1–13, 2018. [Google Scholar]
12. Y. Dong, Q. Zhang, Z. Qiao and J. -J. Yang, “Classification of cataract fundus image based on deep learning,” in 2017 IEEE Int. Conf. on Imaging Systems and Techniques (IST), Beijing, BJ, China, pp. 1–5, 2017. [Google Scholar]
13. T. J. Jebaseeli, C. A. D. Durai and J. D. Peter, “Retinal blood vessel segmentation from diabetic retinopathy images using tandem PCNN model and deep learning based SVM,” Optik, vol. 199, pp. 163328, 2019. [Google Scholar]
14. H. Boudegga, Y. Elloumi, M. Akil, M. H. Bedoui, R. Kachouri et al., “Fast and efficient retinal blood vessel segmentation method based on deep learning network,” Computerized Medical Imaging and Graphics, vol. 90, pp. 101902, 2021. [Google Scholar] [PubMed]
15. R. Welikala, P. Foster, P. Whincup, A. R. Rudnicka, C. G. Owen et al., “Automated arteriole and venule classification using deep learning for retinal images from the UK biobank cohort,” Computers in Biology and Medicine, vol. 90, pp. 23–32, 2017. [Google Scholar] [PubMed]
16. T. Saba, S. Akbar, H. Kolivand and S. A. Bahaj, “Automatic detection of papilledema through fundus retinal images using deep learning,” Microscopy Research and Technique, vol. 84, no. 12, pp. 3066–3077, 2021. [Google Scholar] [PubMed]
17. A. Budai, J. Odstrcilik, R. Kolar, J. Hornegger, J. Jan et al., “A public database for the evaluation of fundus image segmentation algorithms,” Investigative Ophthalmology & Visual Science, vol. 52, no. 14, pp. 1345–1345, 2011. [Google Scholar]
18. K. O’Shea and R. Nash, “An introduction to convolutional neural networks,” arXiv Preprint arXiv:1511.08458, 2015. [Google Scholar]
19. N. Srivastava, G. Hinton, A. Krizhevsky, I. Sutskever and R. Salakhutdinov, “Dropout: A simple way to prevent neural networks from overfitting,” The Journal of Machine Learning Research, vol. 15, no. 1, pp. 1929–1958, 2014. [Google Scholar]
20. M. S. Ramya, “Diabetic retinopathy analysis using machine learning,” Turkish Journal of Computer and Mathematics Education (TURCOMAT), vol. 12, no. 9, pp. 2930–2937, 2021. [Google Scholar]
21. R. G. Ramani, L. Balasubramanian and S. G. Jacob, “ROC analysis of classifiers in automatic detection of diabetic retinopathy using shape features of fundus images,” in 2013 Int. Conf. on Advances in Computing, Communications and Informatics (ICACCI), Mysore, MYQ, India, pp. 66–72, 2013. [Google Scholar]
22. K. Narasimhan, V. Neha and K. Vijayarekha, “An efficient automated system for detection of diabetic retinopathy from fundus images using support vector machine and Bayesian classifiers,” in 2012 Int. Conf. on Computing, Electronics and Electrical Technologies (ICCEET), Nagercoil, NGL, India, pp. 964–969, 2012. [Google Scholar]
23. S. Agarwal, K. Acharjya, S. K. Sharma and S. Pandita, “Automatic computer aided diagnosis for early diabetic retinopathy detection and monitoring: A comprehensive review,” in 2016 Online Int. Conf. on Green Engineering and Technologies (IC-GET), Coimbatore, CBE, India, pp. 1–7, 2016. [Google Scholar]
24. S. Roychowdhury, D. D. Koozekanani and K. K. Parhi, “DREAM: Diabetic retinopathy analysis using machine learning,” IEEE Journal of Biomedical and Health Informatics, vol. 18, no. 5, pp. 1717–1728, 2013. [Google Scholar]
25. R. Gargeya and T. Leng, “Automated identification of diabetic retinopathy using deep learning,” Ophthalmology, vol. 124, no. 7, pp. 962–969, 2017. [Google Scholar] [PubMed]
26. K. Xu, D. Feng and H. Mi, “Deep convolutional neural network-based early automated detection of diabetic retinopathy using fundus image,” Molecules, vol. 22, no. 12, pp. 2054, 2017. [Google Scholar] [PubMed]
27. J. Deng, A. C. Berg, K. Li and L. Fei-Fei, “What does classifying more than 10,000 image categories tell us?” in European Conf. on Computer Vision, Berlin, Heidelberg: Springer, pp. 71–84, 2010. [Google Scholar]
28. S. P. K. Karri, D. Chakraborty and J. Chatterjee, “Transfer learning based classification of optical coherence tomography images with diabetic macular edema and dry age-related macular degeneration,” Biomedical Optics Express, vol. 8, no. 2, pp. 579–592, 2017. [Google Scholar] [PubMed]
29. X. Gao, S. Lin and T. Y. Wong, “Automatic feature learning to grade nuclear cataracts based on deep learning,” IEEE Transactions on Biomedical Engineering, vol. 62, no. 11, pp. 2693–2701, 2015. [Google Scholar] [PubMed]
30. R. Arunkumar and P. Karthigaikumar, “Multi-retinal disease classification by reduced deep learning features,” Neural Computing and Applications, vol. 28, no. 2, pp. 329–334, 2017. [Google Scholar]
31. J. Y. Choi, T. K. Yoo, J. G. Seo, J. Kwak, T. T. Um et al., “Multi-categorical deep learning neural network to classify retinal images: A pilot study employing small database,” PloS One, vol. 12, no. 11, pp. e0187336, 2017. [Google Scholar] [PubMed]
32. S. S. M. Muhammad, T. S. Tan, M. A. Ansari, W. H. W. Hitam and J. S. Y. Sia, “Retinal disease identification using upgraded CLAHE filter and transfer convolution neural network,” ICT Express, vol. 8, no. 1, pp. 142–150, 2022. [Google Scholar]
33. B. Goutam, M. F. Hashmi, Z. W. Geem and N. D. Bokde, “A comprehensive review of deep learning strategies in retinal disease diagnosis using fundus images,” IEEE Access, vol. 10, pp. 57796–57823, 2022. [Google Scholar]
34. F. Saeed, M. Hussain and H. A. Aboalsamh, “Automatic diabetic retinopathy diagnosis using adaptive fine-tuned convolutional neural network,” IEEE Access, vol. 9, pp. 14344–41359, 2021. [Google Scholar]
35. K. M. Kim, T. Y. Heo, A. Kim, J. Kim, K. J. Han et al., “Development of a fundus image-based deep learning diagnostic tool for various retinal diseases,” Journal of Personalized Medicine, vol. 11, no. 5, pp. 321, 2021. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools