 Open Access
Open Access
REVIEW
Biological roles and molecular mechanism of circular RNAs in epithelial-mesenchymal transition of gastrointestinal malignancies
1 School of Basic Medical Sciences, Health Science Center, Ningbo University, Ningbo, 315211, China
2 Department of Gastroenterology, The First Affiliated Hospital of Ningbo University, Ningbo, 315020, China
* Corresponding Authors: YONGFU SHAO. Email: ; GUOLIANG YE. Email:
,
(This article belongs to the Special Issue: Novel Biomarkers and Treatment Strategies in Solid Tumor Diagnosis, Progression, and Prognosis)
Oncology Research 2025, 33(3), 549-566. https://doi.org/10.32604/or.2024.051589
Received 09 March 2024; Accepted 13 June 2024; Issue published 28 February 2025
Abstract
Circular RNAs (circRNAs) are formed by splicing of precursor RNAs and covalently linked at the 5′ and 3′ ends. Dysregulated circRNAs are closely related to the epithelial-mesenchymal transition (EMT) of gastrointestinal malignancies. CircRNAs, including circRNA_0008717, circGOT1, circ-DOCK5, circVPS33B, circPVT1, circMET, circ-OXCT1, circ_67835, circRTN4, circ_0087502, circFNDC38, circ_PTEN1, circPGPEP1, and circ-E-Cad are involved in the EMT process of gastrointestinal malignancies through a variety of mechanisms, such as regulating EMT-inducing transcription factors, signaling pathways, and tumor microenvironments. Gastrointestinal (GI) malignancies are common malignant tumors worldwide, and the heterogeneity and easy metastasis of gastrointestinal malignancies limit the effectiveness of medical treatments. Therefore, investigating the molecular mechanisms involved in the pathogenesis of gastrointestinal malignancies is essential for clinical treatment. This article summarizes the biological roles and molecular mechanism of circRNAs in EMT of gastrointestinal malignancies, providing a theoretical basis for applying EMT-related circRNAs in targeted therapy.Graphic Abstract
Keywords
Gastrointestinal (GI) malignancies are common malignant tumors found worldwide, that seriously endanger human health. According to 2020 global cancer statistics, gastrointestinal malignancies include gastric (GC), colorectal (CRC), esophageal (ESCC), hepatic (HCC), gallbladder, and pancreatic cancer. The incidence rate of these malignancies accounts for more than 50% of all cancers, and the mortality rate exceeds 35%, causing huge economic and social burdens worldwide [1]. Due to the insidious onset of gastrointestinal malignancies, nonspecific clinical manifestations, and rapid progression of the disease, most patients are already in the progressive stage at the time of diagnosis, and the best surgical window is missed. Additionally, the heterogeneity of gastrointestinal malignancies and their tendency to metastasize limit the effectiveness of neoadjuvant radiotherapy, molecular targeted therapy, immunotherapy, and other medical treatments for patients with clinically advanced tumors. Therefore, further understanding of the molecular mechanisms underlying the pathogenesis of gastrointestinal malignancies is of great significance for clinical treatment.
Epithelial-mesenchymal transition (EMT) is a reversible process in which polarized epithelial cells lose the attachment polarity of the basement membrane and the ability of intercellular tight and adhesion junctions in response to a few factors and are converted to mesenchymal cells with infiltrative and migratory abilities. EMT is strongly associated with tumor infiltration and migration and plays an important role in the metastasis of gastrointestinal malignancies [2]. Cancer cells with epithelioid morphology undergo remodeling and detach from the basement membrane with the programmed activation of EMT, transforming into mesenchymal cell morphology, with increased infiltration and migration capacity and spread of cancer cells to the periphery, promoting cancer metastasis [3]. During the EMT process, cadherin is an important molecular marker of EMT and plays an important biological effect. E-cadherin is a key protein mediating intercellular adhesion junctions, which inhibits metastasis by preventing β-cadherin from entering the nucleus of the cell and hindering the action of DNA-binding proteins [4]. When the activation of the EMT process is initiated, the expression of cellular E-cadherin substantially decreases, while the expression of protein markers representing the morphology of mesenchyme, such as N-cadherin and Vimentin, substantially increases, and with the changes in the levels of these proteins, the mobility of the cells and the degradability of the basement membrane consequently increase, ultimately facilitating the spread of the cancer cells to the surrounding stroma [5].
The molecular mechanisms of EMT regulation in gastrointestinal malignancies are complex and regulated by multiple molecules and regulatory networks. In addition to the typical regulation of EMT, epigenetic modification, and post-translational regulation, non-coding RNAs, such as microRNAs (miRNAs) and long non-coding RNAs (lncRNAs), are important regulatory molecules. Additionally, circular RNAs (circRNAs) are important members of the non-coding RNA family and affect the invasion and metastasis of gastrointestinal malignancies by regulating the EMT of cells [6]. In this review, we described the regulatory role and molecular mechanism of circRNAs on EMT in gastrointestinal malignancies from the aspects of EMT-inducing transcription factors (TFs), EMT-related signaling pathways, and tumor microenvironments (TMEs) for a better understanding of the metastasis mechanism in gastrointestinal malignancies, as well as new perspectives for prevention and treatment strategies.
Biological roles of circular RNAs (circRNAs) in GI malignancies
CircRNAs are a class of closed-loop RNA molecules formed by alternative splicing of precursor mRNAs and covalently linked during the 5′ and 3′ ends, which are widely found in eukaryotic cells [7]. Unlike most eukaryotic mature mRNA molecules, circRNAs do not have the typical 5′-terminal m7GTP cap structure and 3′-terminal poly(A) tail, are not easily degraded by ribonuclease (RNase), and have good stability [6]. In addition, circRNAs are relatively tissue, sequence, and disease specific and are closely associated with cancers.
In the field of gastrointestinal malignancies, circRNAs are extensively involved in the regulation of gene expression at the epigenetic, transcriptional, and post-transcriptional levels through interactions with RNAs or proteins, which affects tumorigenesis and development, and is closely related to cancer invasion and metastasis, as well as to the prognosis of patients, and plays an important role in the evolution of gastrointestinal malignancies [7,8]. More and more molecular functions of circRNAs have been explored:
(1) Acting as protein scaffolds to facilitate or inhibit the assembly of protein complexes (Fig. 1A) [9]. For example, circ-Foxo3 regulate the formation of ternary complexes with proteins cyclin-dependent kinase 2 (CDK2) and cyclin-dependent kinase inhibitor 1 (or p21), thereby repressing cell cycle progression [10].
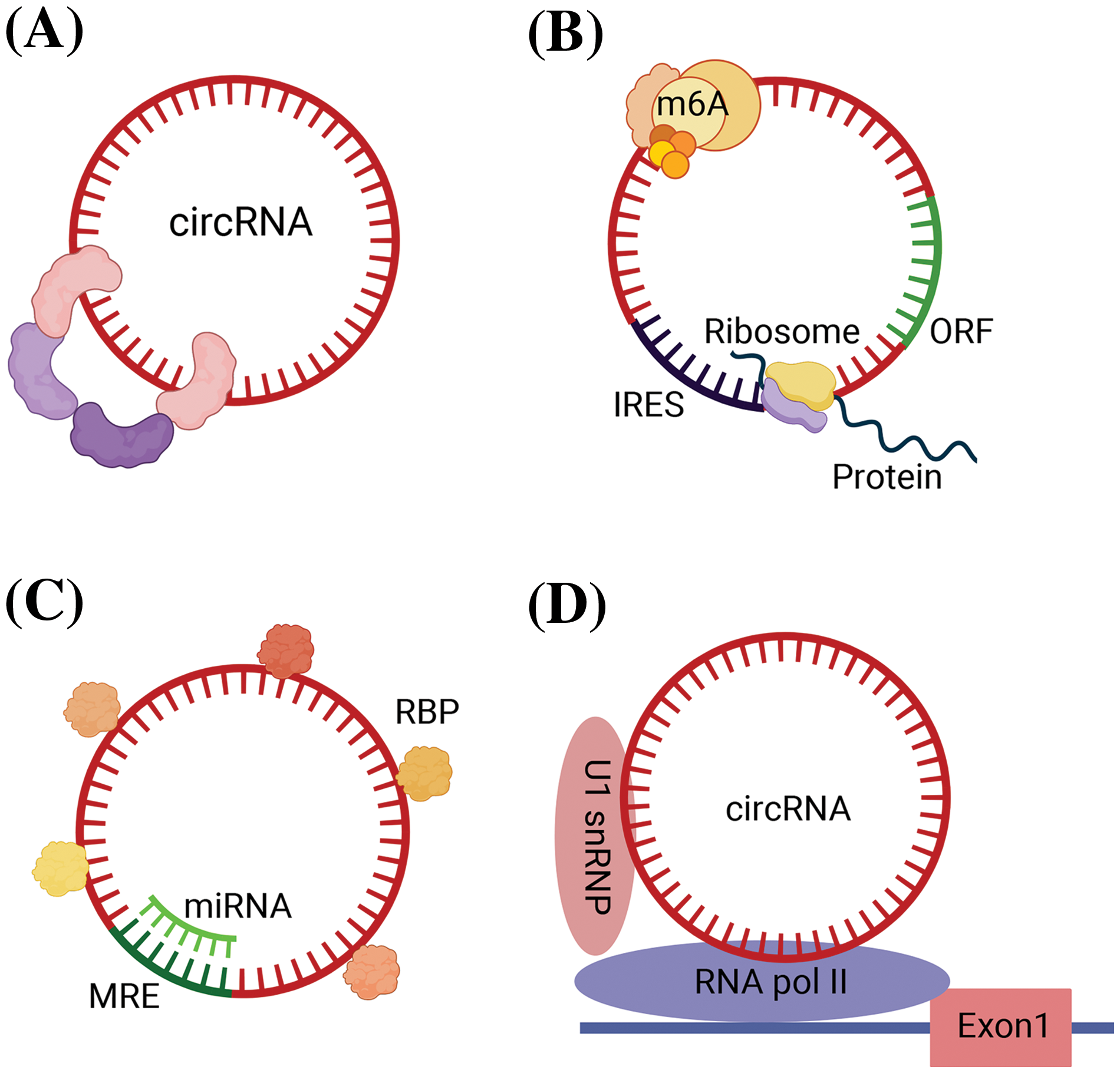
Figure 1: Biological roles of circRNAs in GI malignancies. (A) Function as protein scaffolds to facilitate or inhibit the assembly of protein complexes. (B) Translating into proteins due to IRES and ORF sequence and methylation modification of circRNAs. (C) Function as miRNA sponge and combining with RBP. (D) Regulating transcription. circRNA, circular RNA; GI, gastrointestinal; miRNA, micro RNA; RBP, RNA-binding proteins; IRES, internal ribosome entry site; ORF, open reading frame; MRE, miRNA response elements; m6A, N6-methyladenosine; Exon, exoniensis; U1 snRNP, U1 small nuclear ribonucleoprotein.
(2) Serving as templates for translation, producing functional peptides or proteins in certain contexts. CircRNAs are enriched in methylation-modified adenines, and this methylation modification play a similar role to that of ribosomal entry site sequences, which enables circRNAs to have the ability to be translated into polypeptides and proteins, and play corresponding tumor suppressor or oncogenic roles [11]. Several circRNAs are also capable of directly guiding protein translation due to the presence of internal ribosome entry site (IRES) and open reading frame (ORF) sequences (Fig. 1B). For example, circ-E-Cad encodes a protein named C-E-Cad that promotes the proliferation and migration of gastric cancer via the TGF-β/Smad/C-E-Cad/PI3K/AKT pathway [12,13].
(3) Sponging or binding RNA-binding proteins. CircRNAs are structurally equipped with miRNA response elements (MREs) and can act as miRNA sponges to regulate the expression of downstream target genes by adsorbing specific miRNAs and preventing the binding of miRNAs to downstream mRNA targets in a base-complementary pairing manner (Fig. 1C) [14]. For example, CDR1as contains more than 70 miR-7 binding sites, which can adsorb a large amount of miR-7 by exerting the function of miR-7 sponge, reducing the negative regulation of miR-7, thus indirectly regulating the expression of miRNA downstream target genes to inhibit tumor progression [15].
In addition to MRE, some circRNAs also have one or more RNA binding protein (RBP) binding sites, which can act as protein sponges to bind proteins directly or associate with proteins indirectly under the mediation of RNAs. The RNA-protein complex regulates the interaction between RNA and RBP and participates in variable splicing of RNAs, affecting protein functions and post-transcriptional gene expression (Fig. 1C). For example, circ_0088300 was proved to upregulate the RNA binding protein BOLL, promoting gastric cancer growth and EMT [16].
(4) CircRNAs modulate transcription or epigenetic activities. Some circRNAs can localize to the nucleus, which can bind to Pol II, and binds to U1 small nuclear ribonucleoproteins (snRNPs), regulating the transcriptional activity of the host (Fig. 1D) [17]. For example, the 113-aa protein (p113) encoded by hsa_circ_30402 interacts with Zuotin-related factor 1 (ZRF1) and bromodomain protein 4 (BRD4) to form a transcriptional regulatory complex inducing lipid metabolic repro-gramming and mitochondrial complex I activity, which enhances the oncogenic effects of neuroblastoma cells [18].
Molecular Mechanism of circRNAs in EMT of GI Malignancies
CircRNAs involvement in EMT regulation through EMT-TFs
Epithelial to mesenchymal transition-inducing EMT-TFs are important components of EMT, and a variety of circRNAs can participate in the regulation of EMT in gastrointestinal malignancies by activating EMT-TFs [2]. These EMT-TFs include Snail (Snai1), Slug (Snail2), Twist, and Zeb, all of which can inhibit E-cadherin expression by recognizing E-box sequences, leading to cell adhesion detachment, loss of epithelial cell polarity, and conversion to a mesenchymal phenotype. Although different EMT-TFs widely induce the classical EMT process, they can respond to microenvironmental stimulation in a specific manner and act as molecular switches for EMT (Fig. 2) [19].
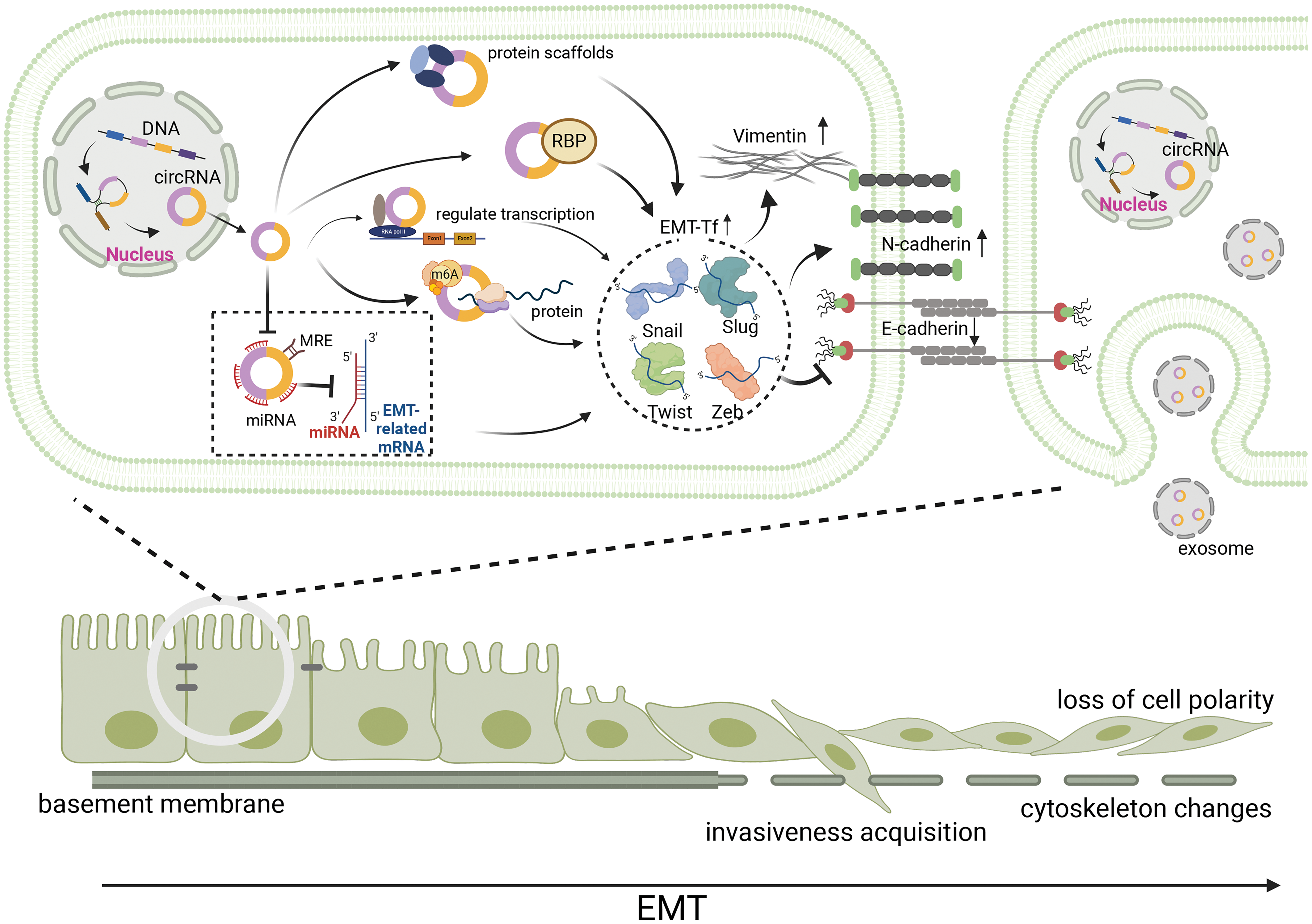
Figure 2: During EMT, circRNAs affect EMT-induced transcription factors, including Snail, Slug, Twist, and Zeb in a variety of ways, and further affected EMT-related molecules, such as E-cadherin, N-cadherin and Vimentin. Hence, epithelial cancer cells with reduced adhesion to each other loss cell polarity, acquire invasivness and become mesenchymal cell. Meanwhile, the basement membrane disrupted and the adhesion between the basement membrane and cells reduced, leading to cytoskeleton changes. circRNA, circular RNA; EMT, epithelial-mesenchymal transition; miRNA, micro RNA; RBP, RNA-binding proteins; m6A, N6-methyladenosine.
CircRNAs regulate snail in EMT
Snail (Snail1) is an EMT-inducing TF with a zinc-finger structure that can bind to the E-box sequence in the proximal promoter region of the E-cadherin gene and represses gene expression. Multiple signaling pathways can participate in EMT initiation and progression by synergistically activating Snail [20]. CircMET (circRNA_0082002), which is abnormally highly expressed in hepatocellular carcinoma tissues, can promote EMT progression by targeting downstream Snail overexpression by binding to miR-30-5p [21]. Snail can induce hepatic cell carcinogenesis through the circMET/miR-30-5p/Snail/DPP4 axis and serve as a TF for DPP4 to induce local immune suppression and participate in the occurrence and development of HCC [21]. In contrast, circFNDC3B was found to be mainly localized in the cytoplasm of intestinal cells, with reduced expression in colon cancer cells and tissues, negatively correlated with colon cancer EMT [22]. Pan et al. [22] demonstrated that circFNDC38 inhibited the process of EMT by encoding a novel protein, circFNDC3B-218aa, which regulates the expression of the Snail and ultimately inhibits cancer cell invasion, migration, and metastasis by decreasing the expression level of mesenchymal markers. Additionally, the low expression of circ-transportin3 (TNPO3) in GC tissues is closely related to the degree of differentiation of GC, weakening its role in stabilizing MYC mRNA by acting as a protein bait for insulin-like growth factor 2-binding protein 3 (IGF2BP3), which finally inhibits the proliferation and metastasis of GC by decreasing the expression of Snail through the MYC-SNAIL axis [23]. However, hsa_circ_0023642, highly expressed in GC tissues, not only acts as a molecular sponge for miR-223-3p, but also regulates the expression of Snail-associated E-cadherin, N-cadherin and Vimentin involved in the process of EMT, promoting cell proliferation, invasion, and migration, and showing a highly positive correlation with the malignant progression of GC [24,25]. Notably, another study found that miR-223-3p targeted SORBS1 to regulate the levels of E-cadherin, N-cadherin, and Vimentin to accelerate the progression of EMT in GC [26].
The zinc finger TF, Slug (also known as Snail2), is another member of the Snail superfamily, which is highly homologous to Snail [27]. Unlike Snail1, which exists as a monomer, Slug is mostly polymeric in conformation, and both can bind specific DNA sequences according to their relative concentrations and cellular environments to inhibit the expression of E-cadherin, which plays an important role in the process of cancer EMT [28]. Slug exogenous overexpression induces effects independent of endogenous Snail expression, which can act as an independent TF to regulate EMT induction [28]. CircRNA-0008717, whose expression was considerably upregulated in ESCC, was positively correlated with EMT and promoted the increase of Slug expression through miR-203 sponging, which ultimately promotes ESCC cell proliferation, migration, and invasion by affecting the levels of E-cadherin and Vimentin [29]. Wong et al. [30] demonstrated that the overexpression of circRTN4 in pancreatic ductal adenocarcinoma regulates miR-497-5p to promote oncogenic lncRNA HOTTIP expression, blocks ubiquitination of EMT driver RAB11FIP1, inhibits cellular RAB11FIP1 degradation, and ultimately promotes EMT, cancer growth, and liver metastasis by altering the levels of TFs, such as Slug. Additionally, bioinformatics analysis revealed that the abnormal high expression of hsa_circ_0001020 in GC could be involved in GC development through the potential p53 signaling pathway [31]. Notably, p53, a key molecule tumor suppressor in the p53 signaling pathway, was in turn confirmed to interact with MDM2 to form the p53-MDM2-Slug complex, which promotes MDM2-mediated Slug degradation and ultimately inhibits cancer invasion and metastasis [32]. The above study suggests a potential novel mechanism by which hsa_circ_0001020 regulates Slug via the p53 signaling pathway to participate in EMT, invasion and metastasis of GC.
CircRNAs regulate twist in EMT
Twist1 and Twist2, which belong to the basic helix-loop-helix (bHLH) TF family, are important EMT-TFs [33]. Unlike other bHLH TFs, Twist1 and Twist2 can regulate E-cadherin and Vimentin expression by binding to the E-box enhancer, which directly or indirectly affects the EMT process [34]. Meng et al. [35] found that Twist1 could directly bind to the Cul2 promoter and selectively promote the expression of Cul2 circRNA (circ-10720) in metastatic hepatocellular carcinoma, which was closely associated with tumor malignance and poor prognosis. Further luciferase assay revealed that miR-490-5p is the main adsorbing mRNA mediating circ_10720 upregulation, which promotes the expression of Vimentin and thus induces EMT [35]. Chen et al. [36] confirmed that circ_67835 was highly expressed in hepatocellular carcinoma cell lines and tissues, which could be used as a prognostic indicator of overall survival in patients, was also found to be the miR-1236-3p sponge to alleviate the inhibitory effect of miR-1236-3p on Twist2. Silencing circ_67835 promotes E-cadherin expression to inhibit the occurrence of EMT, suggesting that circ_67835 could be involved in the EMT of HCC through the miR-1236-3p/Twist2 axis to promote cancer cell proliferation and metastasis, which provides a new explanation for circRNAs-mediated EMT regulation through Twist related markers [36].
The Zeb family comprises Zeb1 and Zeb2 (also known as Smad-interacting protein 1, SIP1), which have partially overlapped genes, both binding to the E-box sequence [37]. Similar to Snail and Twist, Zeb is one of the TFs downstream of the transforming growth factor β (TGF-β) signaling pathway, and TGF-β is associated with EMT progression by inhibiting the expression of the epithelial marker E-cadherin, which in turn regulates the mRNA levels of Zeb1 and Zeb2 [38]. Zeb2 is substantially associated with the progression, malignancy, and prognosis of gastrointestinal malignancies [39,40], and Zeb2 can affect the EMT process of cancers through the Wnt/β-Catenin pathway [41]. CircUBAP2 was upregulated in pancreatic cancer tissues, which enhances the expression of the key downstream gene, Zeb1, through the inhibition of hsa-miR-494 [42]. Meanwhile, miR-494 can target SDC1 to mediate both mRNA and protein expression of E-cadherin and Vimentin, suppressing EMT, metastasis and invasiveness of pancreatic adenocarcinoma cells [43]. Regulatory T cells (Tregs) can inhibit other immune cells from generating anti-tumor immune responses, which can promote cancer cell growth when they undergo activation [44]. Zhao et al. [42] found that Treg markers, such as FOXP3, CCR8, and signal transducer and activator of transcription (STAT)5B were positively correlated with the expression level of Zeb1, suggesting that circUBAP2 can influence the EMT of pancreatic cancer cells through the regulation of miR-494 and target Zeb1 to affect the activity of Tregs for cancer progression. Zhu et al. [45] revealed that circLONP2, highly expressed in the esophagus, can upregulate the expression level of Zeb1 through miR-27b-3p sponging, as well as affect EMT-related proteins to promote the proliferation, migration, and EMT of ESCC cells. Similarly, as a circRNA abnormally highly expressed in ESCC, circ-ZDHHC5 can promote the expression of Zeb1 by binding miR-217, accelerating cell proliferation, migration and invasion [46]. MiR-217 also proved to be a molecular sponge for circRNA_100367 and can target Snail to reduce E-cadherin expression in radiation-resistant ESCC cells with high EMT ability, suggesting that circ-ZDHHC5 has the potential to regulate Zeb1 in the EMT process in ESCC cells [47]. There are limited studies regarding the influence of circRNAs on EMT in gastrointestinal cancers through Zeb, which still need to be further explored.
CircRNAs involvement in EMT regulation through signaling pathways
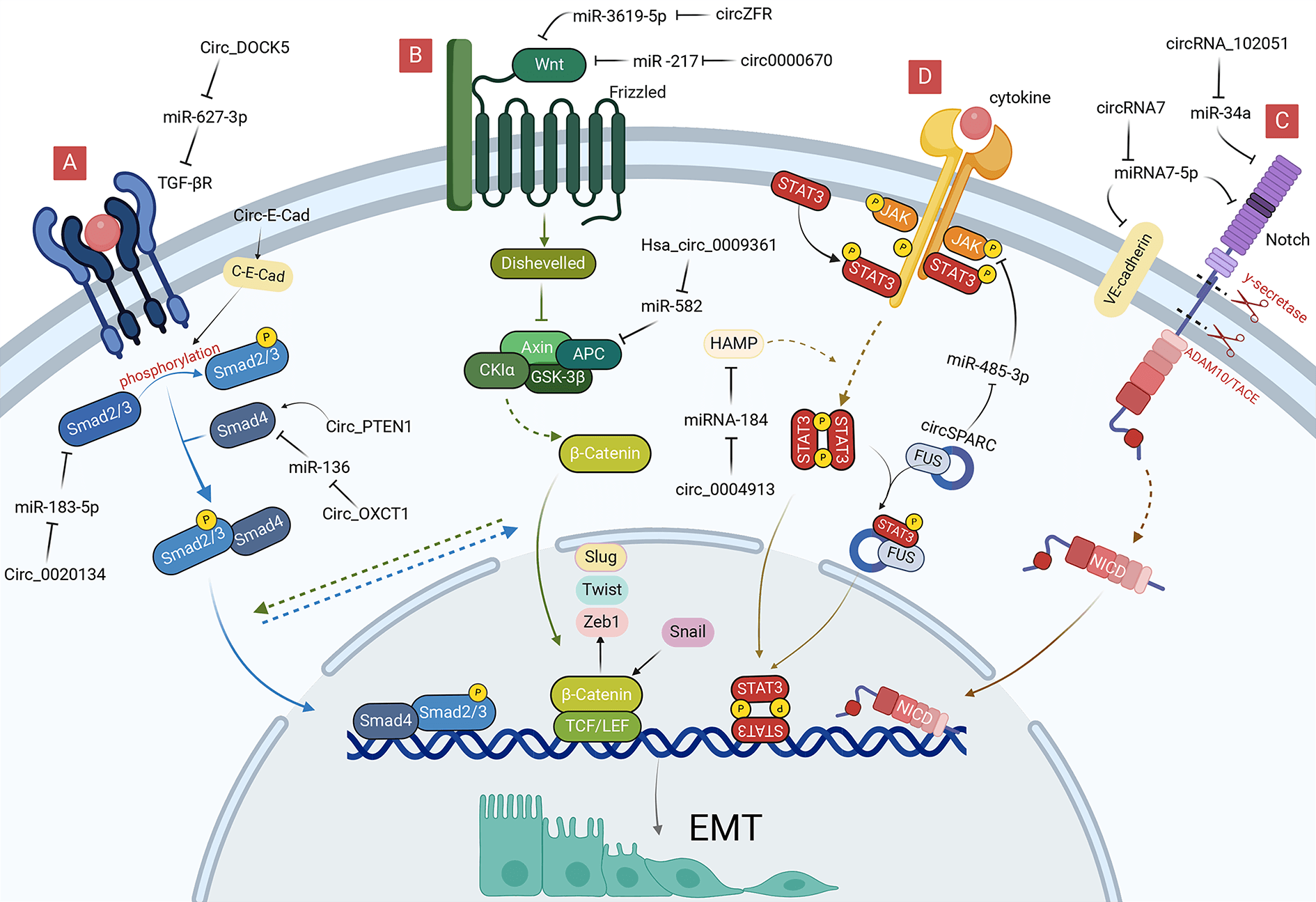
In addition to promoting EMT by regulating EMT-TFs, circRNAs in gastrointestinal malignancies can also induce EMT by regulating signaling pathways, such as TGF-β, Wnt, and Notch. In different contexts, these signaling pathways can be activated by the corresponding signaling molecules, such as TGF-β, Wnt, Notch, and STAT.
TGF-β/Smad pathway-related circRNAs
TGF-β/Smad signaling pathway is the most classical TGF-β-mediated signaling process, considerably promoting the proliferation and metastasis of cancer cells by promoting the proliferation of mesenchymal derived cells, inhibiting the proliferation of epithelial cells and inducing EMT (Fig. 3A) [48–50]. TGFβ RI and TGFβ RII are the main receptors involved in the TGF-β/Smad pathway, which have serine/threonine protein kinase activity and regulate the transcriptional expression of downstream target genes by phosphorylating the Smad protein family. Among them, Smad4 is a key molecule in the TGF-β/Smad pathway, which can bind to phosphorylated Smad2 and Smad3 to form the Smad complex, affecting EMT-TFs and exerting a tumor-suppressive effect [51]. Inactivation, deletion or mutation of Smad4 exists in a variety of malignant tumors, and mutation of TGF-β RII has been demonstrated in gastrointestinal cancer, hepatocellular carcinoma, and gallbladder carcinoma cells, which weaken the negative regulatory mechanism of cancer cells, leading to the development of malignant cancers [52,53].
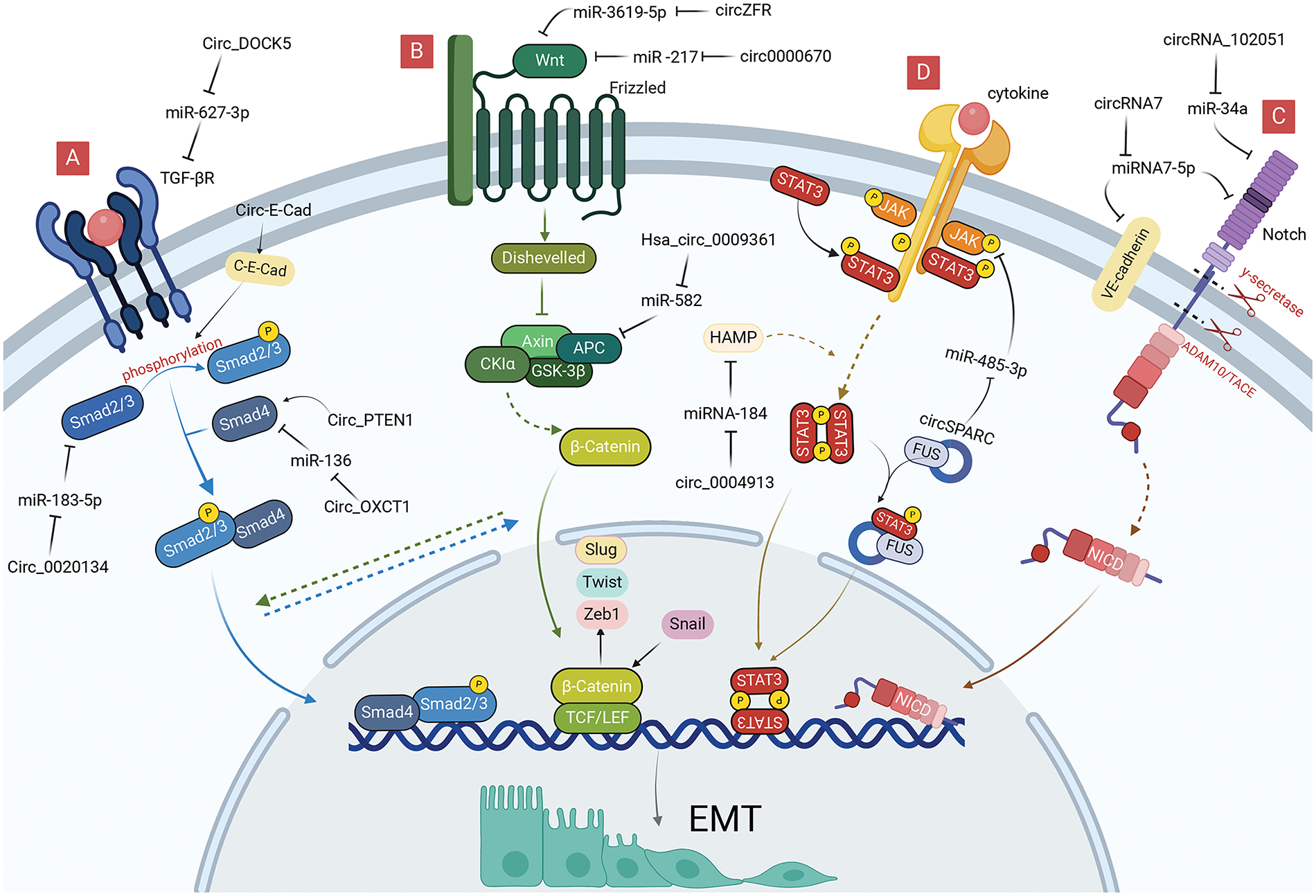
Figure 3: Role of circRNAs in EMT regulation via EMT-signaling pathways in GI cancer cells. (A) CircRNAs can regulate TGF-β/Smad signaling pathway to reduce EMT. (B) CircRNAs can regulate Wnt signaling pathway to reduce EMT. (C) CircRNAs can regulate Notch signaling pathway to reduce EMT. (D) CircRNAs can regulate JAK/STAT signaling pathway to reduce EMT. circRNA, circular RNA; EMT, epithelial-mesenchymal transition; GI, gastrointestinal; miRNA, microRNA.
Inactivation of Smad4 promotes the deterioration and metastasis of gastric malignancies [54]. Zheng et al. [55] found that circ_PTEN1 can bind to the MH2 structural domain of Smad4 to prevent its degradation, which can allow low expression of N-cadherin and Vimentin, which suppresses EMT in colonic epithelial cells by TGF-β. Liu et al. [56] proved that downregulation of circ-OXCT1 could inhibit Smad4 expression by direct binding to miR-136, ultimately modulating the expression of E-cadherin, N-cadherin, and Vimentin through the TGF-β/Smad signaling pathway to promote cell migration, invasion, and EMT processes. CircRNA_0004872 considerably promoted downstream Smad4 gene transcript expression, which inhibited invasion and metastasis of GC cells by interacting with miR-224 [57]. Smad4 could also affect the expression level of circRNA in GC tissues through the Smad4/ADAR1/circRNA_0004872/miR-224/Smad4 feedback loop [57]. Therefore, circRNA_0004872 offers novel potential for targeting TGF-β signaling to prevent colon carcinogenesis.
The Smad2 and Smad3 are mediators of the TGF-β signaling pathway that can be mutated in a variety of cancers [58]. Little is known about the selective activation of Smad2 and Smad3. CircPVT1 serves as a sponge for miR-423-5p in GC, relieving the miR-423-5p-mediated repression of Smad3. Decreased circPVT1 inhibits GC cell EMT by increasing Smad3-related E-cadherin and downregulating Vimentin, Snai1, Twist1, and Zeb1 [59]. Additionally, circ-E-Cad could induce GC proliferation and migration by affecting the phosphorylation of Smad2 and Smad3 by encoding the circ-E-Cad protein. Moreover, circ-E-Cad overexpression upregulated Snail, Slug, and Vimentin expression, which facilitated the EMT of GC cells [13]. Moreover, silencing hsa_circ_0020134 decreases the levels of TGF-β1 and Smad2/3 protein expression in CRC, which inhibits EMT by modulating Slug, Snail, and mesenchymal markers [60].
Mutations in either the TGF-β receptor or Smad4 can abrogate signaling pathways downstream of the TGF-β receptor, similarly, can promote the cellular EMT process induced by TGF-β [61]. Meng et al. [62] discovered that circ-DOCK5, weakly expressed in ESCC, is modulated by Zeb1 and Zeb1-repressed RNA binding proteins, which can act as miR-627-3p molecular sponges, weakening the inhibitory effect of miR-627-3p on the TGFBR2, and ultimately exerting a suppressive effect on cellular EMT and metastasis. Chen et al. [63] found that circ_0087502, highly expressed in pancreatic cancer, could act as a miR-1179 molecular sponge, weakening the inhibitory effect of miR-1179 on the key target gene, TGFBR2, ultimately exerting a role in promoting cell proliferation, migration, and invasion. Yin et al. [64] discovered that circRNA_102610 could cause up-regulation of N-cadherin, Vimentin by targeting miR-103a-3p in patients with Crohn’s disease and exert EMT-promoting effects in intestinal epithelial cells. Further western blotting assays suggest that circRNA_102610 can stimulate the increase of Smad4 expression, which contradicts the inhibitory effect of Smad4 protein on EMT, requiring consideration regarding its association with an aberrant signaling pathway downstream of the Smad4 protein [64].
The Wnt pathway is triggered by the binding of different Wnt ligands to the Frizzled family of receptors on the cell surface, including the classical Wnt, non-classical Wnt-Ca2+, and the Wnt-PCP pathway [65]. In contrast to the classical Wnt pathway, which needs to be mediated by β-catenin, the non-classical Wnt pathway can exert signaling via other molecules acting on the cell, such as protein kinase C and WNT5A [66,67].
The classical Wnt pathway regulates cell differentiation, proliferation, and metastasis by activating the Wnt ligand and Frizzled receptor to allow β-catenin to enter the nucleus where they function as TFs (Fig. 3B) [68]. The Wnt pathway regulates the expression of EMT-TFs by activating β-catenin to bind directly to the corresponding promoters of Slug, Zeb1, and Twist [69]. Conversely, Snail can also enhance the transcriptional activity of β-catenin, a positive feedback effect that makes Wnt signaling more readily available for cellular responses, thus promoting the use of Wnt signaling in cancer cell EMT [70].
Wnt signaling in hepatocellular carcinoma can exert an influence on c-Myc expression by regulating β-catenin, a process that is not only associated with the activation of glycolysis, but also increases the proliferation of HCC and the progression of EMT [71]. Liang et al. found that exosome circ0000670 induced by cigarettes could participate in the Wnt/β-catenin signaling pathway by positively regulating the expression levels of β-catenin and c-Myc using bioinformatics analysis [72]. Nevertheless, silencing circ0000670 resulted in a substantial decrease in the expression of EMT marker proteins and mRNAs, indicating that circ0000670 could modulate the EMT process by impacting the Wnt/β-catenin pathway [72]. Zhou et al. [73] revealed that silencing hsa_circ_0001666 in CRC inhibited cell growth and metastasis, suppressed procalcitonin 10 (PCDH10) expression by directly sponging to miR-576-5p, which reduced β-catenin expression affecting the Wnt pathway, and ultimately regulated cellular EMT by modulating key EMT proteins, such as Snail and Vimentin. Coincidentally in HCC, circUSP10 considerably upregulated and its overexpression accelerated HCC cell proliferation, migration, invasion, and EMT by sponging oncogenic miR-211-5p to regulate TCF12 expression [74]. Moreover, TCF12 is positively correlated with β-catenin in the Wnt signaling pathway, indicating that circUSP10 could regulate EMT through the Wnt pathway [75]. Tan et al. [76] discovered that downregulation of circZFR could inhibit the activity of the Wnt/β-catenin signaling pathway using the luciferase reporter and suppress the proliferation and HCC EMT by regulating the miR-3619-5p/CTNNB1 axis. Liu et al. [47] observed that radiation-resistant ESCC cells have higher EMT expression than regular ESCC cells, and silencing circRNA_100367 reduces cellular β-catenin expression and inhibits the progress of EMT. Moreover, circRNA_100367 attenuates the radio-resistance of ESCC cells via the miR-217/Wnt3 pathway. APC is a CRC anti-oncogene, and the APC protein can form a complex with β-catenin leading to the degradation of the latter when Wnt signaling is abnormal. Without the APC protein, excess β-catenin would accumulate in the nucleus, activating Wnt-targeted genes [77]. Hsa_circ_0009361 is markedly downregulated in CRC tissues, which is associated with poor prognosis, and inhibits progression, migration, invasion, and EMT of CRC cells through the miR-582/APC/β-catenin axis [78].
Notch pathway-related circRNAs
The Notch signaling pathway is widespread in multicellular organisms and generates active Notch fragments (Notch intracellular domain, Notch-ICD) by activating Notch ligands and receptors between neighboring cells, which enter the nucleus, binds to effector proteins and transcriptional promoters, and leads to the expression of target genes (Fig. 3C) [79]. Notch, a key oncogene or cancer suppressor gene, can regulate gene expression in a variety of ways at the transcriptional level [79], and abnormal Notch signaling is often associated with tumorigenesis [80,81]. Notch molecules were found to be significantly elevated in tumor-infiltrating regions and accompanied by the expression of EMT-TFs, such as Snail; thus the Notch signaling pathway plays an important role in regulating cancer EMT.
Some circRNAs regulate Notch transcription. CircAPLP2 is substantially upregulated in CRC. Bioinformatics analysis and luciferase reporter assays have confirmed that miR-101-3p directly interacts with circAPLP2 and targets Notch1 [82]. MiR-101-3p can also reduce the resistance of colon cancer cells to radiation and EMT progression by regulating the expression of E-cadherin and Vimentin, which provided new possibilities for circRNAs to affect EMT through the Notch pathway and a new therapeutic target for colorectal malignant tumors [83]. Notably, bromodomain PHD-finger TF (BPTF) is an attractive target for certain cancers, considerably correlating with the expression levels of EMT markers (Vimentin and E-cadherin), which can promote EMT progression in CRC [84]. Chen et al. [85] illustrated that the substantially upregulated hsa_circRNA_102051 is involved in the tumorigenesis of CRC by activating as a miR-203a sponge, which alleviates the inhibitory effect of miR-203a on Notch1 and BPTF and enhances the stemness of the cancer cells, indicating that hsa_circRNA_102051 may regulate EMT through the miR-203a/Notch1 axis. Bao et al. [86] reported that the expression level of circRNA7 in hepatocellular carcinoma was considerably lower than that in a healthy liver. Vascular endothelial cadherin (VE-cadherin) is one of the key molecules maintaining the adhesion of vascular endothelial cells, promoting EMT and cancer metastasis [87]. Notably, circRNA7 can negatively regulate the expression of VE-cadherin and Notch4 by directly targeting their 3′-untranslated region (UTR) via miRNA7-5p, which ultimately enhances EMT and promotes HCC metastasis [86]. Notably, circRNAs are associated with hepatitis B virus (HBV) infection [88]; circ_00059686 can act as a miR-129-5p sponge to inhibit the progression of HBV-associated HCC through the Notch1 pathway [89]. Currently, there is limited research related to circRNAs regulation of EMT in gastrointestinal malignancies through the Notch pathway, requiring further experimental investigation.
JAK/STAT pathway-related circRNAs
JAK kinases are a class of non-receptor tyrosine kinases that can be activated by cytokine signals and regulate a variety of biological activities in vivo using STAT as a substrate [90]. Its downstream STAT3 is a key signal involved in promoting the survival of cancer cells, and activated STAT3 enters the nucleus to enhance the transcription of downstream EMT-related genes, thereby enhancing the migration, invasion, and chemical resistance of cancer cells (Fig. 3D) [91].
To date, only a few circRNAs exert oncogenic or oncostatic effects through the JAK/STAT3/EMT axis. HAMP, a gene encoding ferredoxin, can exert cancer suppressor effects in the proliferation and migration of hepatocellular carcinoma cells through the STAT3 pathway [92]. Wu et al. [93] demonstrated a substantial decrease in circ_0004913 and HAMP in HCC and identified their target correlation with miRNA-184. Furthermore, circ_0004913 not only inhibits the JAK2/STAT3 signaling pathway through the miR-184/HAMP axis, but also suppresses EMT by reducing the expression of Snail and Vimentin [93]. CircSPARC expression is markedly upregulated in CRC tissues and promotes cell proliferation and migration, which promotes the expression of the JAK2 mRNA by sponging miR-485-3p. This circRNA could bind to the RNA-binding protein, FUS, to stimulate the translocation of phosphorylated STAT3 (p-STAT3) with RNA-binding proteins of the cell nucleus [94]. There is a positive correlation between p-STAT3 and EMT marker expression in CRC [95]. Moreover, miR-485-3p promotes EMT by enhancing the expression of Vimentin, N-cadherin, and Snail [96]. CircSPARC could therefore act as a ceRNA affecting the JAK2/STAT3 pathway to regulate the cellular EMT progression. The abnormally high expression of hsa_circ_0000117 in GC tissues and cells was negatively correlated with miR-337-3p and regulated the downstream target gene, STAT3, to enhance GC cell carcinogenesis [97]. Further bioinformatics analysis revealed that miR-337-3p could act as a molecular sponge to influence the proliferation and EMT of GC [98].
CircRNAs involvement in EMT regulation in the TME
The TME refers to the internal and external environment where cancer cells survive, and oncogenic alterations are induced systemically and locally through endocrine and paracrine secretion [99]. In addition to malignant tumor cells, the TME contains numerous cells, including vascular endothelial, stromal, intrinsic immune (tumor-associated macrophages (TAMs) and natural killer (NK) cells), and acquired immune cells (T and B lymphocytes), whose metabolites, with cytokines included, not only serve as a source of energy supply, but also mediate various cellular messaging [100,101]. The interactions between tumor cells and the TME, mediated by exosomes, directly determine the degree of tumor malignancy [102]. As a stable exosome, circRNAs are able to influence different tumorigenic pathways in the TME, such as angiogenesis and immunosuppression, which ultimately induces EMT in cancer cells [103].
CircRNAs inhibits T cell activation
As a major component of adaptive immunity, T cells are usually involved in immune surveillance and immune editing in cancer, and the exosomal circRNAs-induced inhibitory mechanism of T cell activation can lead to immune escape and promote cancer metastasis (Fig. 4A) [104]. Zhang et al. [105] confirmed the overexpression of circPGPEP1 and NFAT5 in CRC and their target relationship with miR-515-5p. CircPGPEP1 not only increases proliferation and migration by regulating NFAT5, but also promotes the expression of Snail and N-cadherin, which are target genes of the EMT. Moreover, silencing promotes the proliferation of CD8+ and CD4+ cells and participate in the immune escape of CRC cells, indicating that circRNAs could promote EMT by promoting T-cell dysfunction and weakening the immunoreactivity of cancer cells. A similar manifestation was found in HCC, where exosome circCCAR1 expression was upregulated in hepatocellular carcinoma cells, which not only reduced the levels of perforin and granzyme B proteins in CD8+ T cells to promote their apoptosis, but also reduced the secretion of cytokines on their surface, causing dysfunction of CD8+T cells [106]. CircCCAR1 can act as a molecular sponge of miR-127-5p targeting Wilms’ tumor 1-associating protein (WTAP) molecules to promote liver cancer cell growth and metastasis [106]. MiR-127-5p overexpression in hepatocellular carcinoma cells enhances EMT progression by accelerating the activation of the SHC3/ERK signaling pathway to upregulate the expression of Vimentin and N-cadherin, suggesting that circRNAs could promote EMT by accelerating T-cell apoptosis [107]. Paradoxically, in the activated state of effector T cells, pancreatic ductal epithelial cells have substantially reduced E-cadherin expression, and markedly decreased expression of Vimentin and Zeb1, which are manifested in the form of spindle-shaped mesenchymal morphology cells, ultimately facilitating EMT [108].
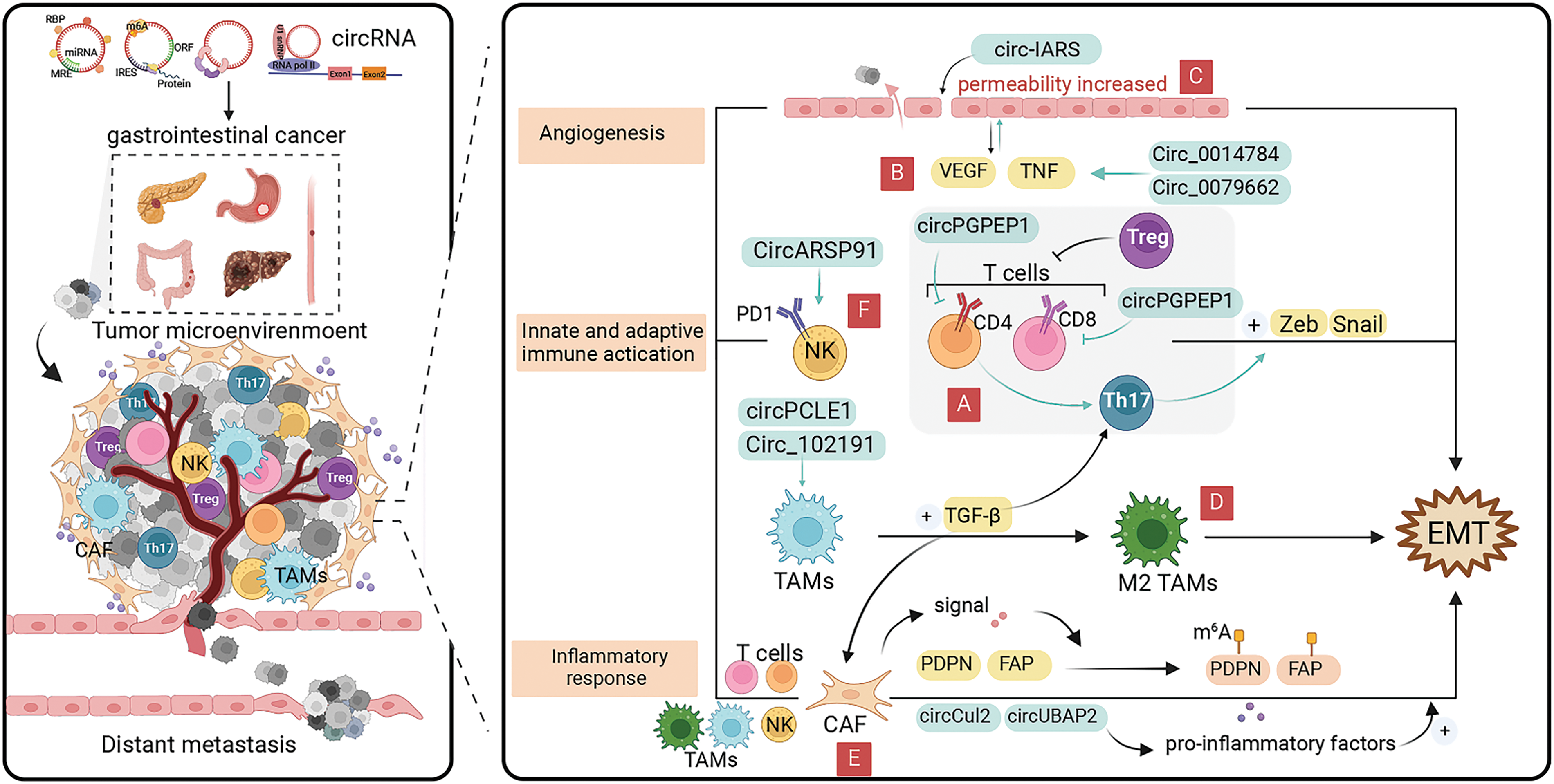
Figure 4: Role of circRNAs in EMT regulation via the tumor microenvironment in GI cancer cells. (A) CircRNAs can affect the EMT process by inhibiting T cell activation. (B) CircRNAs can affect cancer angiogenesis by interacting with the pro-angiogenic factors, such as vascular endothelial growth factor (VEGF) and TNF-α ereby inducing cancer EMT progression. (C) CircRNAs can promote cancer cell proliferation and EMT occurrence by regulating endothelial cell permeability. (D) CircRNAs can promote the conversion of tumor-associated macrophages to M2 phenotype for cancer progression. (E) CircRNAs can induce cancer EMT by regulating cancer-associated fibroblasts which secrete pro-inflammatory factors. (F) CircRNAs can affect the EMT process by modulating NK cell-mediated cytotoxicity. circRNA, circular RNA; EMT, epithelial-mesenchymal transition; GI, gastrointestinal; miRNA, micro RNA; NK, natural killer.
CircRNAs-induced angiogenesis in cancers
Angiogenesis within cancers is a complex process that is co-regulated by multiple factors and plays an important role in cancer growth and metastasis [109]. Cancers can not only obtain sufficient nutrients and remove metabolic wastes through angiogenesis, but also produce a variety of vascular growth factors to further promote the expansion of the vascular network [110]. The major pro-angiogenic factors in the TME include vascular endothelial growth factor (VEGF), platelet-derived growth factor, tumor necrosis factor α (TNF-α), and interleukin (IL)-8 [111]. In the TME, exosomal circRNAs can affect cancer angiogenesis by interacting with the above vascular growth factors, thereby inducing cancer EMT progression (Fig. 4B).
EMT-induced, VEGF-A-mediated angiogenesis has been identified as a connecting mechanism between cancer stem cells and initiation [112]. Liu et al. [113] found that hsa_circ_0014784 not only promotes angiogenic differentiation of vascular endothelial cells via sponge adsorption of miR-214-3p, but also induces the target gene, YAP1, to promote pancreatic cancer cell proliferation, invasion, and EMT. Circ-BANP, which was abnormally highly expressed in hepatocellular carcinoma cells, could promote the expression level of target gene, TLR4, through sponge adsorption of miR-let-7f-5p, and further regulate the expression of VEGF-A and VEGF-2 through the STAT3 signaling pathway to affect the cancer angiogenesis [114]. Additionally, circ-BANP could weaken the inhibition of miR-let-7f-5p on Vimentin and N-cadherin to promote the occurrence of EMT, and accelerate the proliferation and migration of HCC cells [114]. HUR is a post-transcriptional regulator that can bind to a variety of mRNAs to exert different biological functions and play a promotional role in tumorigenesis and development [115]. Upregulated expression of hsa_circ_0000936 in GC tissues and serum could sponge adsorb miR-582-3p to alleviate the inhibitory effect on HUR and enhance the stability of VEGF mRNA, which ultimately leads to the rapid progression of GC in vitro and in vivo [116]. Moreover, silencing miR-582-3p could reduce Vimentin expression by promoting E-cadherin expression through the Wnt/β-catenin pathway and inhibit cancer cell EMT [117].
Exosomal circRNAs also regulate endothelial cell permeability promoting EMT in cancer cells (Fig. 4C) [118]. EMT-capable cancer cells tend to regulate endothelial cell permeability and facilitate cancer cell endocytosis during the pre-invasive phase, leading to distant cell metastasis [119,120]. Tight junction protein, Zo-1, is the site of intercellular junctions, as one of the EMT markers, its expression level is negatively correlated with the occurrence of EMT [120]. Li et al. [121] found that circ-IARS, which was substantially upregulated in plasma exosomes from pancreatic cancer tissues and patients with metastatic cancer, not only enhances endothelial single-molecule cell permeability and promotes cancer cell invasion and metastasis, but also directly binds to miR-122, promoting pancreatic cancer cell EMT by inhibiting downstream ZO-1 expression. The carcinomatous bile-derived exosome circ_CCAC1 can promote cancer progression through sponge adsorption of miR-514a-5p and be translocated to endothelial monolayers, disrupting the integrity of the endothelial barrier and inducing angiogenesis [122]. Silencing miR-514a-5p can reduce E-cadherin expression, promote Vimentin and N-cadherin expression, and induce the EMT process in cancer cells. Therefore, circ_CCAC1 can regulate the EMT occurrence in cancer cells by affecting endothelial cell permeability [123]. A growing number of researchers have focused on the role of circRNAs in angiogenesis and EMT, and in-depth examination of the specific cellular and molecular mechanisms of exosomal circRNAs is expected to lead to the development of new therapies targeting exosomal circRNAs for anti-tumor EMT in the future.
CircRNAs regulates macrophage polarization
TAMs are derived from bone marrow monocytes which are the most prevalent immune cells in the TME [124]. Mediated by exosomal circRNAs, TAMs can lose their killing ability and switch from M1 to M2 phenotype [125]. M2 phenotype TAMs can induce EMT development by interactions with multiple cell types and secretion of TGF-β, thus promoting cancer progression (Fig. 4D). Yi et al. [126] found that circPCLE1 can affect EMT by promoting the upregulation of M1 macrophage markers (TNF-α and IL-6), and decreasing the expression of M2 macrophage markers (IL-10 and MRC1) to induce TAM polarization [126]. Yao et al. [127] demonstrated that tumor-derived circRNA_102191 is a competing endogenous RNA of XPR1 that absorbs miR-493-3p, ultimately promoting the proliferation, migration, and invasion of GC cells by promoting the polarization of M2-type macrophages, as well as enhanced EMT by boosting the level of Vimentin and suppressing the level of E-cadherin. Fatty acid synthase (FASN) is a multifunctional peptidase that is highly expressed in many cancers, supporting cancer cell growth and proliferation and correlating with aggressive capacity [128]. In pancreatic cancer, high expression of circ_0018909 can promote EMT by inducing polarization of M2-type macrophages and regulating miR-545-3p to promote FASN expression, which affects cancer growth, metastasis, and apoptosis [129]. Additionally, circ_0018909 also can induce the polarization of dormant macrophages (M0-type) into M2-type macrophages, ultimately affecting EMT by regulating E-cadherin, N-cadherin, and Vimentin [129].
CircRNAs regulates cancer-associated fibroblasts (CAFs)
CAFs are one of the major components of the TME, which not only remodel the extracellular matrix, but also promote cancer metastasis through various paracrine signaling pathways (Fig. 4E) [130]. Interleukin-6 (IL6) and interleukin-11 (IL-11) belong to the IL6 family of pro-inflammatory factors which can mediate inflammation-driven tumorigenesis. CAFs secretes IL6 and IL11 in response to a variety of stimuli including IL-1β and TGF-β, inducing EMT in tumor cells [131–134]. CircCul2 is specifically expressed in CAFs and can contribute to the production of IL-6 by regulating miR-203a-3p to activate the STAT3 signaling pathway to promote proliferation and metastasis of pancreatic cancer [135]. The upregulated miR-203a-3p has been implicated in inhibiting pancreatic cancer proliferation, EMT, and apoptosis by regulating Slug. Additionally, circCul2 could modulate the CAF-induced EMT process [136]. Cui et al. found that hsa_circ_0006646 and hsa_circ_0061395 could alleviate miRNA-targeted inhibition of IL-11 by competitive binding of miRNAs and promote EMT process in esophageal cancer [137].
CXCL11 is a member of the chemotactic cytokine superfamily that normally recruits selective T cells to mediate inflammatory responses [134]. In hepatocellular carcinoma, the level of CXCL11 secreted by CAFs was considerably elevated compared with that of normal fibroblasts, and CXCL11 stimulated the upregulation of circUBAP2 expression, which further indirectly affected the levels of IL-17 and IL-1β by inhibiting miR-4756 expression [134]. IL-1β, which is considered as a regulator of the TGF-β/Smad pathway, induces the TGF-β-related EMT [138]. Additionally, IL-17a (often referred to as IL-17, is a highly versatile cellular pro-inflammatory cytokine) reduces E-cadherin expression and promotes Vimentin and N-cadherin expression, inducing EMT via the AKT pathway, suggesting the possibility of CAFs involvement in EMT and invasive metastasis of HCC by regulating IL-17 and IL-1β through circUBAP2 [139].
In addition to secreting cytokines, signals released by CAFs can alter the methylation of certain genes, such as E-cadherin, fibroblast activation protein (FAP), and flat foot protein (PDPN) to affect cancer growth and metastasis [140–142]. PDPN and FAP regulate EMT-related proteins in a variety of gastrointestinal cancers [140,142]. CircITGB6, markedly upregulated in metastatic cancer samples, could be induced by TGF-β, and was closely associated with poor prognosis of patients with colon cancer [143]. CircITGB6 enhance the mRNA stability of PDPN by directly binding to IGF2BP3, thereby promoting EMT progression in colon cancer cells by regulating the expression of E-cadherin, N-cadherin, and Vimentin [143].
CircRNAs regulates NK cell-mediated cytotoxicity
NK cells are bone marrow-derived cytotoxic lymphoid stem cells with the ability to self-regulate, rapidly recognize and kill tumor cells, as well as monitor tumor cell metastasis [144]. Exosomal circRNAs are NK cell-derived and involved in the immune response against cancers [145], and some prognostic genes considerably associated with EMT markers were also negatively correlated with NK cell infiltration (Fig. 4F) [146].
Hsa_circ_0048674 is substantially upregulated in HCC and exhibits sponge adsorption of miR-223-3p to regulate PDL1 expression for hepatoma cell growth, migration, and invasion [147]. Additionally, PDL1 can mitigate the effects caused by circRNAs through the PDL1/circRNA_0048674/miR-223-3p/PDL1 feedback pathway [147]. MiR-223-3p inhibits EMT occurrence in hepatocellular carcinoma cells by affecting E-cadherin and Vimentin expression through downregulation of the expression of target gene, FAT1 [148]. Silencing hsa_circ_0048674 also can promote NK cell-mediated cytotoxicity, indicating that hsa_circ_0048674 could participate in EMT process by accelerating NK cell exhaustion [147]. Ma et al. [149] found that circARSP91 could promote the expression of UL16-binding protein 1 (ULBP1) in hepatocellular carcinoma cells at both the mRNA and protein levels, affecting NK cell activation and reinforcing the sensitivity of hepatocellular carcinoma cells to NK cytotoxicity. As a major member of the NKG2D ligand family, ULBP1 plays an important role in NK cell-mediated immune response, which is expressed at a substantially lower level in the mesenchymal phenotype circulating cancer cells than that in the epithelial phenotype, suggesting that circARSP91 may be involved in EMT by altering cytotoxicity of NK cells on hepatocellular carcinoma cells [150]. Intercellular adhesion molecule 1 (ICAM-1) is expressed in concert with EMT-related proteins [151]. High expression of hsa_circ_0007456 in hepatocellular carcinoma cells considerably enhanced susceptibility to NK cells and modulated the expression of downstream target gene, ICAM-1, by directly combining with miR-6852-3p, closely related to EMT regulation [152].
Cancer is also known as a metabolic disease, where cancer cells need access to sufficient nutrients to maintain viability and biosynthesis. Among them, glucose and glutamine are two major nutrients. Cancer cells consume substantially more glucose compared with healthy cells [153,154]. Unlike healthy cells, cancer cells tend to use glycolysis as the main energy source in both aerobic and anaerobic environments, which is known as the “Warburg effect” [155]. Glycolysis can provide sufficient energy for cancer invasion, EMT, and metastasis (Fig. 5A). CircGOT1, whose expression is upregulated in esophageal squamous cell cancers, is able to promote downstream GOT1 expression through direct binding to miR-606, which ultimately affects E-cadherin, N-cadherin, and Vimentin expression for cellular EMT [156]. Additionally, silencing of circGOT1 reduces the consumption of glucose, inhibits acid-lactic acid and ATP synthesis, thus impacting on the process of glycolysis [156]. Uncoincidentally, circVPS33B disrupts oncogenic miR-873-5p in GC cells. Silencing circVPS33B inhibited the uptake of glucose and synthesis of lactic acid, which affected the Warburg effect, as well as promoted the EMT and migration of invasive GC cells by targeting HNRNPK [157].
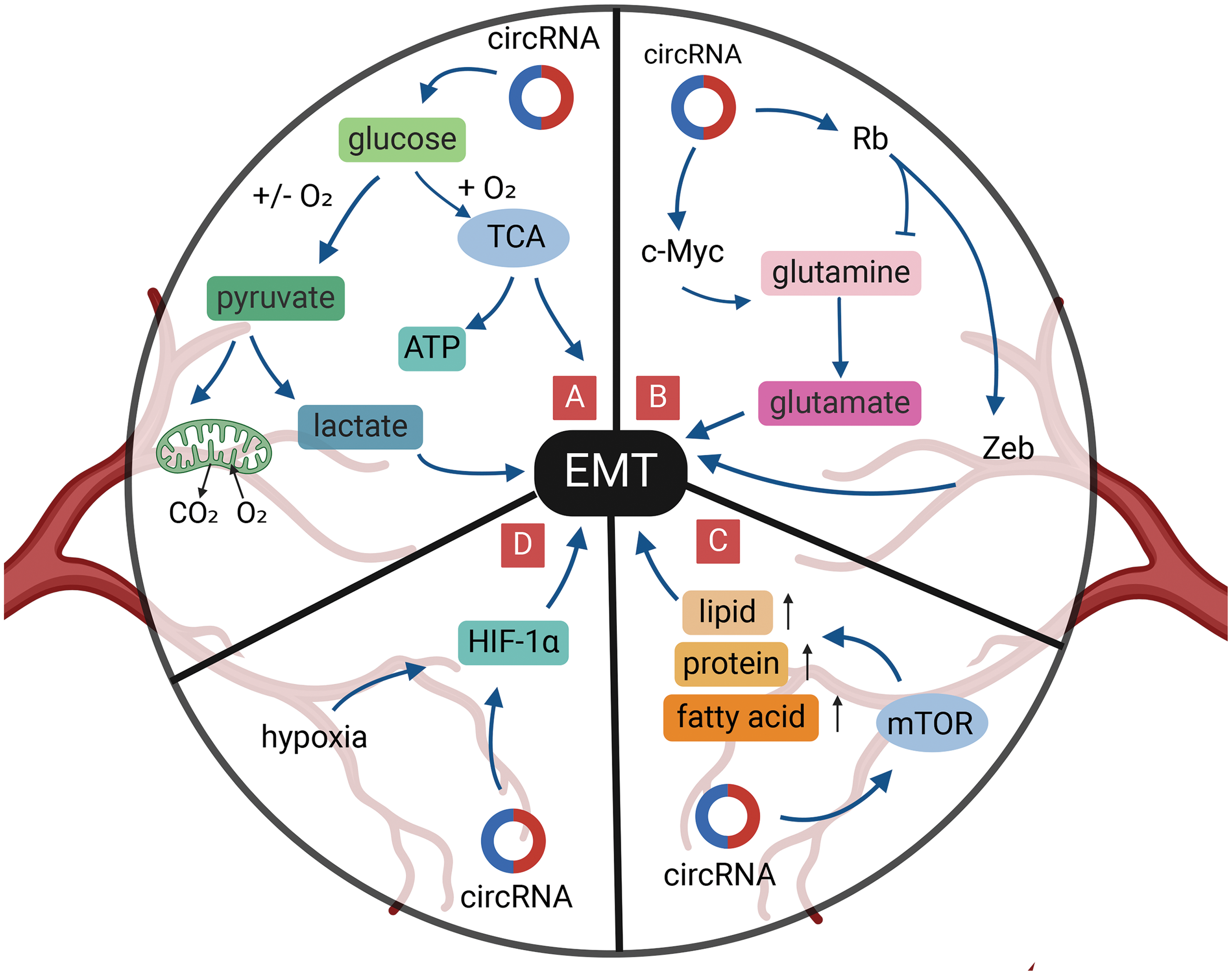
Figure 5: Role of circRNAs in EMT regulation via the tumor metabolism in GI cancer cells. (A) CircRNAs can regulate glucose metabolism to reduce EMT. (B) CircRNAs can regulate glutamine metabolism to reduce EMT. (C) CircRNAs can activate mTOR signal to reduce EMT. (D) CircRNAs can reduce EMT under hypoxia. circRNA, circular RNA; EMT, epithelial-mesenchymal transition; GI, gastrointestinal; miRNA, micro RNA; mTOR, mammalian target of rapamycin; TCA, tricarboxylic acid cycle; ATP, adenosine triphosphate.
The generation of ATP in cancer cell mitochondria involves two key metabolic pathways: the tricarboxylic acid cycle (TCA cycle) and the electron transport chain (ETC). Succinate Dehydrogenase (SDH), as the sole mitochondrial inner membrane protein in the TCA cycle, acts as a bridge between these two important metabolic pathways. SDH catalyzes the oxidation of succinate to fumarate and couples electrons to ubiquinone in the respiratory chain. Inactivation of SDH in tumors leads to the accumulation of succinate, which can promote tumor EMT [158]. It was found that circSDHAF2 translates a heterodimer of SDHAF2, an essential subunit of SDH, with a modified fifth helix, impairing the activation of SDH complex. Silencing circSDHAF2 leads to defects in the SDH complex, which inhibits tumor growth, suggesting the possibility that circSDHAF2 is involved in the tricarboxylic acid cycle to promote EMT [159].
The high demand for glutamine is also seen in cancer cells [160,161] (Fig. 5B). In ESCC cells, highly expressed circ_0001273 regulates SLC1A5 by target binding to miR-622, promoting esophageal cancer cell proliferation, migration, and EMT, and is positively correlated with glutamine consumption [162]. The TF, c-Myc, is the main driver of glutamine utilization by cancer cells, and circ_0005529 increased the expression levels of c-Myc and N-cadherin by binding to miR-527, which promoted GC cell proliferation, migration, and EMT development [163,164]. In contrast, Rb tumor suppressor protein plays a negative regulatory role in glutamine uptake [165]. Moreover, dephosphorylation of the Rb gene also exerts an inhibitory effect on the development of EMT in cancer cells through the Zeb gene [166]. CircRNA_100782, downregulated in GC, is involved in gastric carcinoma development by interacting with miR-574-3p to promote the expression of the oncogene, Rb [167]. Notably, overexpressed miR-574-3p can directly bind to the 3′-UTR of ZEB1 and impinge on the expression of E-cadherin and Vimentin, ultimately promoting EMT progression in GC cells [168]. The above study indicates the possibility that circRNA_100782 regulates glutamate-ammonia metabolism and impacts EMT in GC cells by affecting the Rb and ZEB1 gene.
Mammalian target of rapamycin (mTOR) signaling can also be activated in cancer cells and alters the expression activity of some key metabolic enzymes to regulate cancer metabolism, including ribosome biosynthesis and protein, nucleotide, fatty acid, and lipid synthesis, while mTOR signaling is involved in tumor EMT (Fig. 5C) [169,170]. CircNRIP1 acts as a molecular sponge of miR-149-5p, which can promote cellular EMT through the AKT1/mTOR pathway and affect the proliferation, migration, and invasion of GC cells [171]. Downregulation of circNRIP1 also reduces the lactate content and glucose uptake capacity of GC cells and affects the energy metabolism of cancer cells [171].
Hypoxia is one of the characteristics of the TME and is closely related to patient prognosis. Cancer tissues adapt to environmental changes by activating hypoxia inducible factor (HIF), which in turn promotes the regulation of metabolism (Fig. 5D) [172]. Deletion of HIF-1α inhibits cell proliferation, migration, and EMT, induces G0/G1 cell-cycle arrest, and promotes apoptosis [173]. Liu et al. [174] revealed that circDNMT1 can promote cell proliferation and inhibit apoptosis of GC cells by adsorbing miR-576-3p and promoting the upregulation of HIF-1α expression. Silencing of HIF-1α in GC cells inhibits the level of N-cadherin expression and ultimately hinders EMT in GC cells [175]. Thus, circDNNT1 has the potential to play a role in promoting cellular EMT through the miR-576-3p/HIF-1α axis. Furthermore, circZNF91, circRNA_100859, circDNMT1, and circPRDM4 can affect the “miRNA sponge” to regulate HIF-1α in hypoxia-induced malignant behaviors including pancreatic, colon, and hepatocellular carcinomas [176–179].
Recently, the establishment of the mechanism of circRNAs action in gastrointestinal malignancies has made substantial progress. These studies indicate that circRNAs are key molecules in the process of cancer invasion and metastasis, which can affect EMT and participate in the invasion and metastasis of gastrointestinal malignancies by regulating EMT-TFs, the TME, and signaling pathways, such as Wnt, TGF-β, JAK/STAT, and Notch.
However, some prominent limitations and challenges exist. First, the mechanism of EMT is very complex, involving intracellular and extracellular signaling pathways. The study of whether there are interactions between signaling pathways is still in the early stages, and most of them draw correlation conclusions. In the future, the mechanism of different pathways in EMT needs to be further explored. Second, EMT involves numerous regulatory factors, and understanding the key inducers corresponding to different types of gastrointestinal malignancies may play a decisive role in targeted therapy. Third, although a few EMT databases exist to browse the basic features of EMT-related genes in cancers, there is a lack of EMT databases specifically for circRNAs; new databases should be created and clinical data for diseases corresponding to specific circRNAs associated with EMT should be improved. Fourth, the understanding of the role of EMT-related circRNAs in cancer is incomplete, and studies of these circRNAs affecting EMT are still at the stage of a relatively single-functional mechanism, lacking relevance. Fifth, circRNAs that can act as EMT markers have not yet been identified; newer circRNAs need to be identified and marker correlation studies need to be carried out in the future using high-throughput RNA sequencing technology. Sixth, current studies on circRNAs regulating EMT are mostly focused on common gastrointestinal malignancies, rarely in some diseases such as neuroendocrine gastroeteropancreatic (GEP) cancers [180]. The relationship between circRNAs and EMT in rare tumors of the gastrointestinal system needs more attention. Seventh, the regulation of EMT by circRNAs is primarily focused on the molecular sponge mechanism currently. Only a small number of circRNAs have been identified in gastrointestinal malignancies to participate in the EMT process through translating proteins. Future researches in the field of EMT need to be more in-depth on the side of multiple mechanisms of circRNAs such as translation, transcription, and protein scaffolding. Eighth, the current clinical translational research of circRNAs is insufficient. Most studies on the EMT mechanism of circRNAs remain at the theoretical level, and little is known about the potential clinical applications of EMT-related circRNAs in gastrointestinal malignancies [181]. In the future, we need to increase translational research and design several small molecule targeted drugs targeting circRNAs that regulate EMT by altering their levels to reverse the EMT process.
In summary, circRNAs affect the development of EMT in gastrointestinal malignancies through multiple regulatory mechanisms. Thus far, circRNAs are expected to be ideal biomarkers for gene therapy and prognostic assessment of gastrointestinal malignancies. However, cancer EMT is a multifactorial and multi-stage, complex process. Therefore, the study of circRNAs associated with EMT in gastrointestinal malignancies still faces many challenges.
Acknowledgement: None.
Funding Statement: This study was supported by grants from the Key Scientific and Technological Projects of Ningbo (No. 2021Z133), Ningbo Top Medical and Health Research Program (No. 2023020612), National Natural Science Foundation of China (No. 81702367), the Medical and Health Research Project of Zhejiang Province (No. 2024KY319) and the Youth Medical Backbone Talents Training Program of Ningbo.
Author Contributions: Ziyi Fang and Yongfu Shao made substantial contributions to conception and design of this manuscript. Ziyi Fang was involved in drafting the manuscript and revising it critically for important intellectual content. Jianing Yan and Meng Hu drew the figure in this manuscript. Guoliang Ye and Yongfu Shao reviewed and revised the final manuscript. All authors contributed to the figure discussions and approved the final manuscript submitted.
Availability of Data and Materials: The original data in this study can be obtained from the corresponding author upon request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, et al. Cancer statistics for the year 2020: an overview. Int J Cancer. 2021. doi:10.1002/ijc.33588. Online ahead of print. [Google Scholar] [PubMed] [CrossRef]
2. Ashrafizadeh M, Dai J, Torabian P, Nabavi N, Aref AR, Aljabali AAA, et al. Circular RNAs in EMT-driven metastasis regulation: modulation of cancer cell plasticity, tumorigenesis and therapy resistance. Cell Mol Life Sci. 2024;81(1):214. doi:10.1007/s00018-024-05236-w. [Google Scholar] [PubMed] [CrossRef]
3. Xiao X, Miao X, Duan S, Liu S, Cao Q, Wu R, et al. Single-cell enzymatic screening for epithelial mesenchymal transition with an ultrasensitive superwetting droplet-array microchip. Small Methods. 2023;7(7):e2300096. doi:10.1002/smtd.202300096. [Google Scholar] [PubMed] [CrossRef]
4. Wong SHM, Fang CM, Chuah LH, Leong CO, Ngai SC. E-cadherin: its dysregulation in carcinogenesis and clinical implications. Crit Rev Oncol Hematol. 2018;121:11–22. doi:10.1016/j.critrevonc.2017.11.010. [Google Scholar] [PubMed] [CrossRef]
5. Zhang X, Liu G, Kang Y, Dong Z, Qian Q, Ma X. N-cadherin expression is associated with acquisition of EMT phenotype and with enhanced invasion in erlotinib-resistant lung cancer cell lines. PLoS One. 2013;8(3):e57692. doi:10.1371/journal.pone.0057692. [Google Scholar] [PubMed] [CrossRef]
6. Shao Y, Chen L, Lu R, Zhang X, Xiao B, Ye G, et al. Decreased expression of hsa_circ_0001895 in human gastric cancer and its clinical significances. Tumour Biol. 2017;39(4):1010428317699125. doi:10.1177/1010428317699125. [Google Scholar] [PubMed] [CrossRef]
7. Tao X, Shao Y, Yan J, Yang L, Ye Q, Wang Q, et al. Biological roles and potential clinical values of circular RNAs in gastrointestinal malignancies. Cancer Biol Med. 2021;18(2):437–57. doi:10.20892/j.issn.2095-3941.2020.0348 Online ahead of print. [Google Scholar] [PubMed] [CrossRef]
8. Luo B, Tang CM, Chen JS. CircRNA and gastrointestinal cancer. J Cell Biochem. 2019;120(7):10956–63. doi:10.1002/jcb.28610. [Google Scholar] [PubMed] [CrossRef]
9. Du WW, Fang L, Yang W, Wu N, Awan FM, Yang Z, et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017;24(2):357–70. doi:10.1038/cdd.2016.133. [Google Scholar] [PubMed] [CrossRef]
10. Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44(6):2846–58. doi:10.1093/nar/gkw027. [Google Scholar] [PubMed] [CrossRef]
11. Chen RX, Chen X, Xia LP, Zhang JX, Pan ZZ, Ma XD, et al. N(6)-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10(1):4695. doi:10.1038/s41467-019-12651-2. [Google Scholar] [PubMed] [CrossRef]
12. Fan X, Yang Y, Chen C, Wang Z. Pervasive translation of circular RNAs driven by short IRES-like elements. Nat Commun. 2022;13(1):3751. doi:10.1038/s41467-022-31327-y. [Google Scholar] [PubMed] [CrossRef]
13. Li F, Tang H, Zhao S, Gao X, Yang L, Xu J. Circ-E-Cad encodes a protein that promotes the proliferation and migration of gastric cancer via the TGF-β/Smad/C-E-Cad/PI3K/AKT pathway. Mol Carcinog. 2023;62(3):360–8. doi:10.1002/mc.23491. [Google Scholar] [PubMed] [CrossRef]
14. Guo XY, Sun F, Chen JN, Wang YQ, Pan Q, Fan JG. CircRNA_0046366 inhibits hepatocellular steatosis by normalization of PPAR signaling. World J Gastroenterol. 2018;24(3):323–37. doi:10.3748/wjg.v24.i3.323. [Google Scholar] [PubMed] [CrossRef]
15. Hansen TB, Kjems J, Damgaard CK. Circular RNA and miR-7 in cancer. Cancer Res. 2013;73(18):5609–12. doi:10.1158/0008-5472.CAN-13-1568. [Google Scholar] [PubMed] [CrossRef]
16. Chu S, Fei B, Yu M. Molecular mechanism of Circ_0088300-BOLL interaction regulating mitochondrial metabolic reprogramming and involved in gastric cancer growth and metastasis. J Proteome Res. 2023;22(12):3793–810. doi:10.1021/acs.jproteome.3c00476. [Google Scholar] [PubMed] [CrossRef]
17. Song Z, Lin J, Su R, Ji Y, Jia R, Li S, et al. eIF3j inhibits translation of a subset of circular RNAs in eukaryotic cells. Nucleic Acids Res. 2022;50(20):11529–49. doi:10.1093/nar/gkac980. [Google Scholar] [PubMed] [CrossRef]
18. Yang F, Hu A, Guo Y, Wang J, Li D, Wang X, et al. p113 isoform encoded by CUX1 circular RNA drives tumor progression via facilitating ZRF1/BRD4 transactivation. Mol Cancer. 2021;20(1):123. doi:10.1186/s12943-021-01421-8. [Google Scholar] [PubMed] [CrossRef]
19. Marconi GD, Fonticoli L, Rajan TS, Pierdomenico SD, Trubiani O, Pizzicannella J, et al. Epithelial-mesenchymal transition (EMTthe type-2 EMT in wound healing, tissue regeneration and organ fibrosis. Cells. 2021;10(7):1587. doi:10.3390/cells10071587. [Google Scholar] [PubMed] [CrossRef]
20. Lin Y, Dong C, Zhou BP. Epigenetic regulation of EMT: the Snail story. Curr Pharm Des. 2014;20(11):1698–705. doi:10.2174/13816128113199990512. [Google Scholar] [PubMed] [CrossRef]
21. Huang XY, Zhang PF, Wei CY, Peng R, Lu JC, Gao C, et al. Circular RNA circMET drives immunosuppression and anti-PD1 therapy resistance in hepatocellular carcinoma via the miR-30-5p/snail/DPP4 axis. Mol Cancer. 2020;19(1):92. doi:10.1186/s12943-020-01213-6. [Google Scholar] [PubMed] [CrossRef]
22. Pan Z, Cai J, Lin J, Zhou H, Peng J, Liang J, et al. A novel protein encoded by circFNDC3B inhibits tumor progression and EMT through regulating Snail in colon cancer. Mol Cancer. 2020;19(1):71. doi:10.1186/s12943-020-01179-5. [Google Scholar] [PubMed] [CrossRef]
23. Yu T, Ran L, Zhao H, Yin P, Li W, Lin J, et al. Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol Ther Nucleic Acids. 2021;26:649–64. doi:10.1016/j.omtn.2021.08.029. [Google Scholar] [PubMed] [CrossRef]
24. Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22(8):2297–303. doi:10.26355/eurrev_201804_14818. [Google Scholar] [PubMed] [CrossRef]
25. Zhang Y, Xia L, Wu J, Xu X, Li G. Hsa_circ_0023642 promotes proliferation, invasion, and migration of gastric cancer by sponging microRNA-223. J Clin Lab Anal. 2020;34(10):e23428. doi:10.1002/jcla.23428. [Google Scholar] [PubMed] [CrossRef]
26. Jin X, Qiu X, Huang Y, Zhang H, Chen K. miR-223-3p carried by cancer-associated fibroblast microvesicles targets SORBS1 to modulate the progression of gastric cancer. Cancer Cell Int. 2022;22(1):96. doi:10.1186/s12935-022-02513-1. [Google Scholar] [PubMed] [CrossRef]
27. Brabletz S, Schuhwerk H, Brabletz T, Stemmler MP. Dynamic EMT: a multi-tool for tumor progression. EMBO J. 2021;40(18):e108647. doi:10.15252/embj.2021108647. [Google Scholar] [PubMed] [CrossRef]
28. Bolós V, Peinado H, Pérez-Moreno MA, Fraga MF, Esteller M, Cano A. The transcription factor Slug represses E-cadherin expression and induces epithelial to mesenchymal transitions: a comparison with Snail and E47 repressors. J Cell Sci. 2003;116(3):499–511. doi:10.1242/jcs.00224. [Google Scholar] [PubMed] [CrossRef]
29. Wang T, Wang J, Ren W, Chen S, Cheng YF, Zhang XM. CircRNA-0008717 promotes cell proliferation, migration, and invasion by regulating miR-203/Slug in esophageal cancer cells. Ann Transl Med. 2020;8(16):999. doi:10.21037/atm-20-5205. [Google Scholar] [PubMed] [CrossRef]
30. Wong CH, Lou UK, Fung FK, Tong JHM, Zhang CH, To KF, et al. CircRTN4 promotes pancreatic cancer progression through a novel CircRNA-miRNA-lncRNA pathway and stabilizing epithelial-mesenchymal transition protein. Mol Cancer. 2022;21(1):10. doi:10.1186/s12943-021-01481-w. [Google Scholar] [PubMed] [CrossRef]
31. Yan J, Shao Y, Lu H, Ye Q, Ye G, Guo J. Hsa_circ_0001020 serves as a potential biomarker for gastric cancer screening and prognosis. Dig Dis Sci. 2022;67(8):3753–62. doi:10.1007/s10620-021-07211-y. [Google Scholar] [PubMed] [CrossRef]
32. Wang SP, Wang WL, Chang YL, Wu CT, Chao YC, Kao SH, et al. p53 controls cancer cell invasion by inducing the MDM2-mediated degradation of Slug. Nat Cell Biol. 2009;11(6):694–704. doi:10.1038/ncb1875. [Google Scholar] [PubMed] [CrossRef]
33. Kang Y, Massagué J. Epithelial-mesenchymal transitions: twist in development and metastasis. Cell. 2004;118(3):277–9. doi:10.1016/j.cell.2004.07.011. [Google Scholar] [PubMed] [CrossRef]
34. Sato R, Semba T, Saya H, Arima Y. Concise review: stem cells and epithelial-mesenchymal transition in cancer: biological implications and therapeutic targets. Stem Cells. 2016;34(8):1997–2007. doi:10.1002/stem.2406. [Google Scholar] [PubMed] [CrossRef]
35. Meng J, Chen S, Han JX, Qian B, Wang XR, Zhong WL, et al. Twist1 regulates vimentin through Cul2 Circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78(15):4150–62. doi:10.1158/0008-5472.Can-17-3009. [Google Scholar] [PubMed] [CrossRef]
36. Chen J, Qi Z. The elevated Circ_0067835 could accelerate cell proliferation and metastasis via miR-1236-3p/Twist2 axis in hepatocellular carcinoma. Biomed Res Int. 2022;2022:2825172. doi:10.1155/2022/2825172. [Google Scholar] [PubMed] [CrossRef]
37. Park MK, Lee H, Lee CH. Post-translational modification of ZEB family members in cancer progression. Int J Mol Sci. 2022;23(23):15127. doi:10.3390/ijms232315127. [Google Scholar] [PubMed] [CrossRef]
38. Shirakihara T, Saitoh M, Miyazono K. Differential regulation of epithelial and mesenchymal markers by δEF1 proteins in epithelial mesenchymal transition induced by TGF-β. Mol Biol Cell. 2007;18(9):3533–44. doi:10.1091/mbc.e07-03-0249. [Google Scholar] [PubMed] [CrossRef]
39. Zhang Y, Lin S, Chen Y, Yang F, Liu S. LDH-apromotes epithelial-mesenchymal transition by upregulating ZEB2 in intestinal-type gastric cancer. Onco Targets Ther. 2018;11:2363–73. doi:10.2147/OTT. [Google Scholar] [CrossRef]
40. Xiao Q, Gan Y, Li Y, Fan L, Liu J, Lu P, et al. MEF2A transcriptionally upregulates the expression of ZEB2 and CTNNB1 in colorectal cancer to promote tumor progression. Oncogene. 2021;40(19):3364–77. doi:10.1038/s41388-021-01774-w. [Google Scholar] [PubMed] [CrossRef]
41. Yang S, Li X, Shen W, Hu H, Li C, Han G. MicroRNA-140 represses esophageal cancer progression via targeting ZEB2 to regulate Wnt/β-catenin pathway. J Surg Res. 2021;257:267–77. doi:10.1016/j.jss.2020.07.074. [Google Scholar] [PubMed] [CrossRef]
42. Zhao R, Ni J, Lu S, Jiang S, You L, Liu H, et al. CircUBAP2-mediated competing endogenous RNA network modulates tumorigenesis in pancreatic adenocarcinoma. Aging. 2019;11(19):8484–501. doi:10.18632/aging.102334. [Google Scholar] [PubMed] [CrossRef]
43. Yang Y, Tao X, Li CB, Wang CM. MicroRNA-494 acts as a tumor suppressor in pancreatic cancer, inhibiting epithelial-mesenchymal transition, migration and invasion by binding to SDC1. Int J Oncol. 2018;53(3):1204–14. doi:10.3892/ijo.2018.4445. [Google Scholar] [PubMed] [CrossRef]
44. Tanaka A, Sakaguchi S. Regulatory T cells in cancer immunotherapy. Cell Res. 2017;27(1):109–18. doi:10.1038/cr.2016.151. [Google Scholar] [PubMed] [CrossRef]
45. Zhu C, Bi W, Li H, Wang W. CircLONP2 accelerates esophageal squamous cell carcinoma progression via direct MiR-27b-3p-ZEB1 axis. Front Oncol. 2022;12:822839. doi:10.3389/fonc.2022.822839. [Google Scholar] [PubMed] [CrossRef]
46. Wang Q, Yang L, Fan Y, Tang W, Sun H, Xu Z, et al. Circ-ZDHHC5 accelerates esophageal squamous cell carcinoma progression in vitro via miR-217/ZEB1 axis. Front Cell Dev Biol. 2020;8:570305. doi:10.3389/fcell.2020.570305. [Google Scholar] [PubMed] [CrossRef]
47. Liu J, Xue N, Guo Y, Niu K, Gao L, Zhang S, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging. 2019;11(24):12412–27. doi:10.18632/aging.102580. [Google Scholar] [PubMed] [CrossRef]
48. Tang R, Wang YC, Mei X, Shi N, Sun C, Ran R, et al. LncRNA GAS5 attenuates fibroblast activation through inhibiting Smad3 signaling. Am J Physiol Cell Physiol. 2020;319(1):105–15. doi:10.1152/ajpcell.00059.2020. [Google Scholar] [PubMed] [CrossRef]
49. Chen CL, Huang FW, Huang SS, Huang JS. IGFBP-3 and TGF-β inhibit growth in epithelial cells by stimulating type V TGF-β receptor (TβR-V)-mediated tumor suppressor signaling. FASEB Bioadv. 2021;3(9):709–29. doi:10.1096/fba.2021-00016. [Google Scholar] [PubMed] [CrossRef]
50. Bertrand-Chapel A, Caligaris C, Fenouil T, Savary C, Aires S, Martel S, et al. SMAD2/3 mediate oncogenic effects of TGF-β in the absence of SMAD4. Commun Biol. 2022;5(1):1068. doi:10.1038/s42003-022-03994-6. [Google Scholar] [PubMed] [CrossRef]
51. Kim HS, Woo DK, Bae SI, Kim YI, Kim WH. Microsatellite instability in the adenoma-carcinoma sequence of the stomach. Lab Invest. 2000;80(1):57–64. doi:10.1038/labinvest.3780008. [Google Scholar] [PubMed] [CrossRef]
52. Shin KH, Park YJ, Park JG. Mutational analysis of the transforming growth factor beta receptor type II gene in hereditary nonpolyposis colorectal cancer and early-onset colorectal cancer patients. Clin Cancer Res. 2000;6(2):536–40. [Google Scholar] [PubMed]
53. Kloss CC, Lee J, Zhang A, Chen F, Melenhorst JJ, Lacey SF, et al. Dominant-negative TGF-β receptor enhances PSMA-targeted human CAR T cell proliferation and augments prostate cancer eradication. Mol Ther. 2018;26(7):1855–66. doi:10.1016/j.ymthe.2018.05.003. [Google Scholar] [PubMed] [CrossRef]
54. An HW, Seok SH, Kwon JW, Choudhury AD, Oh JS, Voon DC, et al. The loss of epithelial Smad4 drives immune evasion via CXCL1 while displaying vulnerability to combinatorial immunotherapy in gastric cancer. Cell Rep. 2022;41(13):111878. doi:10.1016/j.celrep.2022.111878. [Google Scholar] [PubMed] [CrossRef]
55. Zheng L, Liang H, Zhang Q, Shen Z, Sun Y, Zhao X, et al. circPTEN1, a circular RNA generated from PTEN, suppresses cancer progression through inhibition of TGF-β/Smad signaling. Mol Cancer. 2022;21(1):41. doi:10.1186/s12943-022-01495-y. [Google Scholar] [PubMed] [CrossRef]
56. Liu J, Dai X, Guo X, Cheng A, Mac SM, Wang Z. Circ-OXCT1 suppresses gastric cancer EMT and metastasis by attenuating TGF-β pathway through the Circ-OXCT1/miR-136/SMAD4 Axis. Onco Targets Ther. 2020;13:3987–98. doi:10.2147/OTT.S239789. [Google Scholar] [PubMed] [CrossRef]
57. Ma C, Wang X, Yang F, Zang Y, Liu J, Wang X, et al. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020;19(1):157. doi:10.1186/s12943-020-01268-5. [Google Scholar] [PubMed] [CrossRef]
58. Brown KA, Pietenpol JA, Moses HL. A tale of two proteins: differential roles and regulation of Smad2 and Smad3 in TGF-beta signaling. J Cell Biochem. 2007;101(1):9–33. doi:10.1002/jcb.21255. [Google Scholar] [PubMed] [CrossRef]
59. Li H, Xue S, Zhang X, Li F, Bei S, Feng L. CircRNA PVT1 modulated cell migration and invasion through epithelial-mesenchymal transition (EMT) mediation in gastric cancer through miR-423-5p/Smad3 pathway. Regen Ther. 2022;21:25–33. doi:10.1016/j.reth.2022.02.003. [Google Scholar] [PubMed] [CrossRef]
60. Yu JH, Tan JN, Zhong GY, Zhong L, Hou D, Ma S, et al. Hsa_circ_0020134 promotes liver metastasis of colorectal cancer through the miR-183-5p-PFN2-TGF-β/Smad axis. Transl Oncol. 2024;39:101823. doi:10.1016/j.tranon.2023.101823. [Google Scholar] [PubMed] [CrossRef]
61. Wang F, Xia X, Yang C, Shen J, Mai J, Kim HC, et al. SMAD4 gene mutation renders pancreatic cancer resistance to radiotherapy through promotion of autophagy. Clin Cancer Res. 2018;24(13):3176–85. doi:10.1158/1078-0432.CCR-17-3435. [Google Scholar] [PubMed] [CrossRef]
62. Meng L, Zheng Y, Liu S, Ju Y, Ren S, Sang Y, et al. ZEB1 represses biogenesis of circ-DOCK5 to facilitate metastasis in esophageal squamous cell carcinoma via a positive feedback loop with TGF-β. Cancer Lett. 2021;519:117–29. doi:10.1016/j.canlet.2021.06.026. [Google Scholar] [PubMed] [CrossRef]
63. Chen M, Liu X, Lu J, Teng H, Yu C, Liu Y, et al. Dysregulation of the circ_0087502/miR-1179/TGFBR2 pathway supports gemcitabine resistance in pancreatic cancer. Cancer Biol Ther. 2023;24(1):2258566. doi:10.1080/15384047.2023.2258566. [Google Scholar] [PubMed] [CrossRef]
64. Yin J, Ye YL, Hu T, Xu LJ, Zhang LP, Ji RN, et al. Hsa_circRNA_102610 upregulation in Crohn’s disease promotes transforming growth factor-β1-induced epithelial-mesenchymal transition via sponging of hsa-miR-130a-3p. World J Gastroenterol. 2020;26(22):3034–55. doi:10.3748/wjg.v26.i22.3034. [Google Scholar] [PubMed] [CrossRef]
65. Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. 2022;7(1):3. doi:10.1038/s41392-021-00762-6. [Google Scholar] [PubMed] [CrossRef]
66. Luna-Ulloa LB, Hernández-Maqueda JG, Castañeda-Patlán MC, Robles-Flores M. Protein kinase C in Wnt signaling: implications in cancer initiation and progression. IUBMB Life. 2011;63(10):915–21. doi:10.1002/iub.559. [Google Scholar] [PubMed] [CrossRef]
67. Chirumbolo S, Bjørklund G. Can Wnt5a and Wnt non-canonical pathways really mediate adipocyte de-differentiation in a tumour microenvironment? Eur J Cancer. 2016;64:96–100. doi:10.1016/j.ejca.2016.05.026. [Google Scholar] [PubMed] [CrossRef]
68. Guo Q, Xu J, Huang Z, Yao Q, Chen F, Liu H, et al. ADMA mediates gastric cancer cell migration and invasion via Wnt/β-catenin signaling pathway. Clin Transl Oncol. 2021;23(2):325–34. doi:10.1007/s12094-020-02422-7. [Google Scholar] [PubMed] [CrossRef]
69. Sánchez-Tilló E, Lázaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, et al. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene. 2010;29(24):3490–500. doi:10.1038/onc.2010.102. [Google Scholar] [PubMed] [CrossRef]
70. Stemmer V, de Craene B, Berx G, Behrens J. Snail promotes Wnt target gene expression and interacts with β-catenin. Oncogene. 2008;27(37):5075–80. doi:10.1038/onc.2008.140. [Google Scholar] [PubMed] [CrossRef]
71. Jiang Y, Han Q, Zhao H, Zhang J. Promotion of epithelial-mesenchymal transformation by hepatocellular carcinoma-educated macrophages through Wnt2b/β-catenin/c-Myc signaling and reprogramming glycolysis. J Exp Clin Cancer Res. 2021;40(1):13. doi:10.1186/s13046-020-01808-3. [Google Scholar] [PubMed] [CrossRef]
72. Liang Z, Fang S, Zhang Y, Zhang X, Xu Y, Qian H, et al. Cigarette smoke-induced gastric cancer cell exosomes affected the fate of surrounding normal cells via the Circ0000670/Wnt/β-catenin axis. Toxics. 2023;11(5). doi:10.3390/toxics11050465. [Google Scholar] [PubMed] [CrossRef]
73. Zhou J, Wang L, Sun Q, Chen R, Zhang C, Yang P, et al. Hsa_circ_0001666 suppresses the progression of colorectal cancer through the miR-576-5p/PCDH10 axis. Clin Transl Med. 2021;11(11):e565. doi:10.1002/ctm2.565. [Google Scholar] [PubMed] [CrossRef]
74. Chen X, Xu Y, Zhou Z, Zhao P, Zhou Z, Wang F, et al. CircUSP10 promotes liver cancer progression by regulating miR-211-5p/TCF12/EMT signaling pathway. Heliyon. 2023;9(10):e20649. doi:10.1016/j.heliyon.2023.e20649. [Google Scholar] [PubMed] [CrossRef]
75. Zhang Y, Yu R, Li Q, Li Y, Xuan T, Cao S, et al. SNHG1/miR-556-5p/TCF12 feedback loop enhances the tumorigenesis of meningioma through Wnt signaling pathway. J Cell Biochem. 2020;121(2):1880–9. doi:10.1002/jcb.29423. [Google Scholar] [PubMed] [CrossRef]
76. Tan A, Li Q, Chen L. CircZFR promotes hepatocellular carcinoma progression through regulating miR-3619-5p/CTNNB1 axis and activating Wnt/β-catenin pathway. Arch Biochem Biophys. 2019;661:196–202. doi:10.1016/j.abb.2018.11.020. [Google Scholar] [PubMed] [CrossRef]
77. Ranes M, Zaleska M, Sakalas S, Knight R, Guettler S. Reconstitution of the destruction complex defines roles of AXIN polymers and APC in β-catenin capture, phosphorylation, and ubiquitylation. Mol Cell. 2021;81(16):3246–61.e3211. doi:10.1016/j.molcel.2021.07.013. [Google Scholar] [PubMed] [CrossRef]
78. Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W, et al. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci. 2019;133(10):1197–213. doi:10.1042/CS20190286. [Google Scholar] [PubMed] [CrossRef]
79. Hanna GJ, Stathis A, Lopez-Miranda E, Racca F, Quon D, Leyvraz S, et al. A phase I study of the pan-notch inhibitor CB-103 for patients with advanced adenoid cystic carcinoma and other tumors. Cancer Res Commun. 2023;3(9):1853–61. doi:10.1158/2767-9764.CRC-23-0333. [Google Scholar] [PubMed] [CrossRef]
80. Maraver A, Fernandez-Marcos PJ, Cash TP, Mendez-Pertuz M, Dueñas M, Maietta P, et al. NOTCH pathway inactivation promotes bladder cancer progression. J Clin Invest. 2015;125(2):824–30. doi:10.1172/JCI78185. [Google Scholar] [PubMed] [CrossRef]
81. Zanotti S, Canalis E. Notch signaling and the skeleton. Endocr Rev. 2016;37(3):223–53. doi:10.1210/er.2016-1002. [Google Scholar] [PubMed] [CrossRef]
82. Wu HB, Huang SS, Lu CG, Tian SD, Chen M. CircAPLP2 regulates the proliferation and metastasis of colorectal cancer by targeting miR-101-3p to activate the Notch signalling pathway. Am J Transl Res. 2020;12(6):2554–69. [Google Scholar] [PubMed]
83. Guo J, Ding Y, Yang H, Guo H, Zhou X, Chen X. Aberrant expression of lncRNA MALAT1 modulates radioresistance in colorectal cancer in vitro via miR-101-3p sponging. Exp Mol Pathol. 2020;115:104448. doi:10.1016/j.yexmp.2020.104448. [Google Scholar] [PubMed] [CrossRef]
84. Xiao S, Liu L, Lu X, Long J, Zhou X, Fang M. The prognostic significance of bromodomain PHD-finger transcription factor in colorectal carcinoma and association with vimentin and E-cadherin. J Cancer Res Clin Oncol. 2015;141(8):1465–74. doi:10.1007/s00432-015-1937-y. [Google Scholar] [PubMed] [CrossRef]
85. Chen Z, Cheng H, Zhang J, Jiang D, Chen G, Yan S, et al. Hsa_circRNA_102051 regulates colorectal cancer proliferation and metastasis by mediating Notch pathway. Cancer Cell Int. 2023;23(1):230. doi:10.1186/s12935-023-03026-1. [Google Scholar] [PubMed] [CrossRef]
86. Bao S, Jin S, Wang C, Tu P, Hu K, Lu J. Androgen receptor suppresses vasculogenic mimicry in hepatocellular carcinoma via circRNA7/miRNA7-5p/VE-cadherin/Notch4 signalling. J Cell Mol Med. 2020;24(23):14110–20. doi:10.1111/jcmm.16022. [Google Scholar] [PubMed] [CrossRef]
87. Shuai Q, Cao L, Qin Z, Zhang Y, Gu Z, Yang J. VE-cadherin fusion protein substrate enhanced the vasculogenic mimicry capability of hepatocellular carcinoma cells. J Mater Chem B. 2020;8(8):1699–712. doi:10.1039/C9TB02790D. [Google Scholar] [PubMed] [CrossRef]
88. Zhang S, Jiang C, Jiang L, Chen H, Huang J, Gao X, et al. Construction of a diagnostic model for hepatitis B-related hepatocellular carcinoma using machine learning and artificial neural networks and revealing the correlation by immunoassay. Tumour Virus Res. 2023;16:200271. doi:10.1016/j.tvr.2023.200271. [Google Scholar] [PubMed] [CrossRef]
89. Fu L, Chen Q, Yao T, Li T, Ying S, Hu Y, et al. Hsa_circ_0005986 inhibits carcinogenesis by acting as a miR-129-5p sponge and is used as a novel biomarker for hepatocellular carcinoma. Oncotarget. 2017;8(27):43878–88. doi:10.18632/oncotarget.16709. [Google Scholar] [PubMed] [CrossRef]
90. Leonard WJ, O’Shea JJ. Jaks and STATs: biological implications. Annu Rev Immunol. 1998;16:293–322. doi:10.1146/annurev.immunol.16.1.293. [Google Scholar] [PubMed] [CrossRef]
91. Hong D, Kurzrock R, Kim Y, Woessner R, Younes A, Nemunaitis J, et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci Transl Med. 2015;7(314):314ra185. doi:10.1126/scitranslmed.aac5272. [Google Scholar] [PubMed] [CrossRef]
92. Shen Y, Li X, Su Y, Badshah SA, Zhang B, Xue Y, et al. HAMP downregulation contributes to aggressive hepatocellular carcinoma via mechanism mediated by Cyclin4-dependent kinase-1/STAT3 pathway. Diagnostics. 2019;9(2):48. doi:10.3390/diagnostics9020048. [Google Scholar] [PubMed] [CrossRef]
93. Wu M, Sun T, Xing L. Circ_0004913 inhibits cell growth, metastasis, and glycolysis by absorbing miR-184 to regulate HAMP in hepatocellular carcinoma. Cancer Biother Radiopharm. 2023;38(10):708–19. doi:10.1089/cbr.2020.3779. [Google Scholar] [PubMed] [CrossRef]
94. Wang J, Zhang Y, Song H, Yin H, Jiang T, Xu Y, et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol Cancer. 2021;20(1):81. doi:10.1186/s12943-021-01375-x. [Google Scholar] [PubMed] [CrossRef]
95. Gao S, Hu J, Wu X, Liang Z. PMA treated THP-1-derived-IL-6 promotes EMT of SW48 through STAT3/ERK-dependent activation of Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2018;108:618–24. doi:10.1016/j.biopha.2018.09.067. [Google Scholar] [PubMed] [CrossRef]
96. Wang H, Wang N, Zheng X, Wu L, Fan C, Li X, et al. Circular RNA hsa_circ_0009172 suppresses gastric cancer by regulation of microRNA-485-3p-mediated NTRK3. Cancer Gene Ther. 2021;28(12):1312–24. doi:10.1038/s41417-020-00280-7. [Google Scholar] [PubMed] [CrossRef]
97. Gao Q, Liu Q, Chen H. Circular RNA hsa_circ_0000117 accelerates the proliferation and invasion of gastric cancer cells by regulating the microRNA-337-3p/signal transducer and activator of transcription 3 axis. Bioengineered. 2021;12(1):1381–90. doi:10.1080/21655979.2021.1918992. [Google Scholar] [PubMed] [CrossRef]
98. Lu RD, Huang Z, Lu JM, Zhang HQ, Shao YF. Effect of Circular RNA hsa_circ_0067582 on the proliferation and invasion ability of gastric cancer cells. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2022;44(1):81–90 (In Chinese). doi:10.3881/j.issn.1000-503X.14113. [Google Scholar] [PubMed] [CrossRef]
99. Zhang Q, Wang W, Zhou Q, Chen C, Yuan W, Liu J, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14. doi:10.1186/s12943-019-1125-9. [Google Scholar] [PubMed] [CrossRef]
100. Bejarano L, Jordāo MJC, Joyce JA. Therapeutic targeting of the tumor microenvironment. Cancer Discov. 2021;11(4):933–59. doi:10.1158/2159-8290.CD-20-1808. [Google Scholar] [PubMed] [CrossRef]
101. Xiao J, Huang K, Lin H, Xia Z, Zhang J, Li D, et al. Mogroside II(E) inhibits digestive enzymes via suppression of interleukin 9/interleukin 9 receptor signalling in acute pancreatitis. Front Pharmacol. 2020;11:859. doi:10.3389/fphar.2020.00859. [Google Scholar] [PubMed] [CrossRef]
102. Wang JJ, Lei KF, Han F. Tumor microenvironment: recent advances in various cancer treatments. Eur Rev Med Pharmacol Sci. 2018;22(12):3855–64. doi:10.26355/eurrev_201806_15270. [Google Scholar] [PubMed] [CrossRef]
103. Wu Z, Yu X, Zhang S, He Y, Guo W. Mechanism underlying circRNA dysregulation in the TME of digestive system cancer. Front Immunol. 2022;13:951561. doi:10.3389/fimmu.2022.951561. [Google Scholar] [PubMed] [CrossRef]
104. Lu Q, Wang X, Zhu J, Fei X, Chen H, Li C. Hypoxic tumor-derived exosomal Circ0048117 facilitates M2 macrophage polarization acting as miR-140 sponge in esophageal squamous cell carcinoma. Onco Targets Ther. 2020;13:11883–97. doi:10.2147/OTT.S284192. [Google Scholar] [PubMed] [CrossRef]
105. Zhang C, Zhang C, Liu X, Sun W, Liu H. Circular RNA PGPEP1 induces colorectal cancer malignancy and immune escape. Cell Cycle. 2023;22(14–16):1743–58. doi:10.1080/15384101.2023.2225923. [Google Scholar] [PubMed] [CrossRef]
106. Hu Z, Chen G, Zhao Y, Gao H, Li L, Yin Y, et al. Exosome-derived circCCAR1 promotes CD8 + T-cell dysfunction and anti-PD1 resistance in hepatocellular carcinoma. Mol Cancer. 2023;22(1):55. doi:10.1186/s12943-023-01759-1. [Google Scholar] [PubMed] [CrossRef]
107. Kong XX, Yang X, Jiang WJ, Zhu M, Kong LB. The long non-coding RNA AC006329.1 facilitates hepatocellular carcinoma progression and metastasis by regulating miR-127-5p/SHC3/ERK axis. J Hepatocell Carcinoma. 2023;10:1085–103. doi:10.2147/JHC.S415309. [Google Scholar] [PubMed] [CrossRef]
108. Goebel L, Grage-Griebenow E, Gorys A, Helm O, Genrich G, Lenk L, et al. CD4+ T cells potently induce epithelial-mesenchymal-transition in premalignant and malignant pancreatic ductal epithelial cells-novel implications of CD4+ T cells in pancreatic cancer development. Oncoimmunology. 2015;4(4):e1000083. doi:10.1080/2162402X.2014.1000083. [Google Scholar] [PubMed] [CrossRef]
109. Nyberg P, Salo T, Kalluri R. Tumor microenvironment and angiogenesis. Front Biosci. 2008;13:6537–53. doi:10.2741/3173. [Google Scholar] [PubMed] [CrossRef]
110. Li D, Zhang Z, Xia C, Niu C, Zhou W. Non-coding RNAs in glioma microenvironment and angiogenesis. Front Mol Neurosci. 2021;14:763610. doi:10.3389/fnmol.2021.763610. [Google Scholar] [PubMed] [CrossRef]
111. Viallard C, Larrivée B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409–26. doi:10.1007/s10456-017-9562-9. [Google Scholar] [PubMed] [CrossRef]
112. Fantozzi A, Gruber DC, Pisarsky L, Heck C, Kunita A, Yilmaz M, et al. VEGF-mediated angiogenesis links EMT-induced cancer stemness to tumor initiation. Cancer Res. 2014;74(5):1566–75. doi:10.1158/0008-5472.CAN-13-1641. [Google Scholar] [PubMed] [CrossRef]
113. Liu B, Gong Y, Jiang Q, Wu S, Han B, Chen F, et al. Hsa_circ_0014784-induced YAP1 promoted the progression of pancreatic cancer by sponging miR-214-3p. Cell Cycle. 2023;22(13):1583–96. doi:10.1080/15384101.2023.2222519. [Google Scholar] [PubMed] [CrossRef]
114. Li G, Kong J, Dong S, Niu H, Wu S, Sun W. Circular BANP knockdown inhibits the malignant progression of residual hepatocellular carcinoma after insufficient radiofrequency ablation. Chin Med J. 2022;135(13):1578–87. doi:10.1097/cm9.0000000000001822. [Google Scholar] [PubMed] [CrossRef]
115. Majumder M, Chakraborty P, Mohan S, Mehrotra S, Palanisamy V. HuR as a molecular target for cancer therapeutics and immune-related disorders. Adv Drug Deliv Rev. 2022;188:114442. doi:10.1016/j.addr.2022.114442. [Google Scholar] [PubMed] [CrossRef]
116. Xie M, Yu T, Jing X, Ma L, Fan Y, Yang F, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020;19(1):112. doi:10.1186/s12943-020-01208-3. [Google Scholar] [PubMed] [CrossRef]
117. Yang M, Ju L, Li C, Cheng H, Li N, Zhang Q, et al. MiR-582-3p participates in the regulation of biological behaviors of A549 cells by ambient PM2.5 exposure. Environ Sci Pollut Res Int. 2022;29(9):13624–34. doi:10.1007/s11356-021-16801-2. [Google Scholar] [PubMed] [CrossRef]
118. Dou R, Liu K, Yang C, Zheng J, Shi D, Lin X, et al. EMT-cancer cells-derived exosomal miR-27b-3p promotes circulating tumour cells-mediated metastasis by modulating vascular permeability in colorectal cancer. Clin Transl Med. 2021;11(12):e595. doi:10.1002/ctm2.595. [Google Scholar] [PubMed] [CrossRef]
119. Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol Cancer. 2019;18(1):116. doi:10.1186/s12943-019-1041-z. [Google Scholar] [PubMed] [CrossRef]
120. Pastushenko I, Brisebarre A, Sifrim A, Fioramonti M, Revenco T, Boumahdi S, et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556(7702):463–8. doi:10.1038/s41586-018-0040-3. [Google Scholar] [PubMed] [CrossRef]
121. Li J, Li Z, Jiang P, Peng M, Zhang X, Chen K, et al. Circular RNA IARS (circ-IARS) secreted by pancreatic cancer cells and located within exosomes regulates endothelial monolayer permeability to promote tumor metastasis. J Exp Clin Cancer Res. 2018;37(1):177. doi:10.1186/s13046-018-0822-3. [Google Scholar] [PubMed] [CrossRef]
122. Xu Y, Leng K, Yao Y, Kang P, Liao G, Han Y, et al. A circular RNA, cholangiocarcinoma-associated circular RNA 1, contributes to cholangiocarcinoma progression, induces angiogenesis, and disrupts vascular endothelial barriers. Hepatology. 2021;73(4):1419–35. doi:10.1002/hep.31493. [Google Scholar] [PubMed] [CrossRef]
123. Li W, Yang FQ, Sun CM, Huang JH, Zhang HM, Li X, et al. circPRRC2A promotes angiogenesis and metastasis through epithelial-mesenchymal transition and upregulates TRPM3 in renal cell carcinoma. Theranostics. 2020;10(10):4395–409. doi:10.7150/thno.43239. [Google Scholar] [PubMed] [CrossRef]
124. Boutilier AJ, Elsawa SF. Macrophage Polarization states in the tumor microenvironment. Int J Mol Sci. 2021;22(13):6995. doi:10.3390/ijms22136995. [Google Scholar] [PubMed] [CrossRef]
125. Wang H, Yung MMH, Ngan HYS, Chan KKL, Chan DW. The impact of the tumor microenvironment on macrophage polarization in cancer metastatic progression. Int J Mol Sci. 2021;22(12):6560. doi:10.3390/ijms22126560. [Google Scholar] [PubMed] [CrossRef]
126. Yi B, Dai K, Yan Z, Yin Z. Circular RNA PLCE1 promotes epithelial mesenchymal transformation, glycolysis in colorectal cancer and M2 polarization of tumor-associated macrophages. Bioengineered. 2022;13(3):6243–56. doi:10.1080/21655979.2021.2003929. [Google Scholar] [PubMed] [CrossRef]
127. Yao M, Mao X, Zhang Z, Xi Y, Gan H, Cui F, et al. Tumor-derived CircRNA_102191 promotes gastric cancer and facilitates M2 macrophage polarization. Cell Cycle. 2023;22(18):2003–17. doi:10.1080/15384101.2023.2271341. [Google Scholar] [PubMed] [CrossRef]
128. Fhu CW, Ali A. Fatty acid synthase: an emerging target in cancer. Molecules. 2020;25(17):3935. doi:10.3390/molecules25173935. [Google Scholar] [PubMed] [CrossRef]
129. Song Y, Wang J, Xu J, Gao Y, Xu Z. Circ_0018909 knockdown inhibits the development of pancreatic cancer via the miR-545-3p/FASN axis and reduces macrophage polarization to M2. J Biochem Mol Toxicol. 2023;37(4):e23293. doi:10.1002/jbt.23293. [Google Scholar] [PubMed] [CrossRef]
130. Ren Q, Zhang P, Lin H, Feng Y, Chi H, Zhang X, et al. A novel signature predicts prognosis and immunotherapy in lung adenocarcinoma based on cancer-associated fibroblasts. Front Immunol. 2023;14:1201573. doi:10.3389/fimmu.2023.1201573. [Google Scholar] [PubMed] [CrossRef]
131. Caja L, Dituri F, Mancarella S, Caballero-Diaz D, Moustakas A, Giannelli G, et al. TGF-β and the tissue microenvironment: relevance in fibrosis and cancer. Int J Mol Sci. 2018;19(5):1294. doi:10.3390/ijms19051294. [Google Scholar] [PubMed] [CrossRef]
132. Grasset EM, Bertero T, Bozec A, Friard J, Bourget I, Pisano S, et al. Matrix stiffening and EGFR cooperate to promote the collective invasion of cancer cells. Cancer Res. 2018;78(18):5229–42. doi:10.1158/0008-5472.Can-18-0601. [Google Scholar] [PubMed] [CrossRef]
133. Lee YE, Go GY, Koh EY, Yoon HN, Seo M, Hong SM, et al. Synergistic therapeutic combination with a CAF inhibitor enhances CAR-NK-mediated cytotoxicity via reduction of CAF-released IL-6. J Immunother Cancer. 2023;11(2):e006130. doi:10.1136/jitc-2022-006130. [Google Scholar] [PubMed] [CrossRef]
134. Liu G, Sun J, Yang ZF, Zhou C, Zhou PY, Guan RY, et al. Cancer-associated fibroblast-derived CXCL11 modulates hepatocellular carcinoma cell migration and tumor metastasis through the circUBAP2/miR-4756/IFIT1/3 axis. Cell Death Dis. 2021;12(3):260. doi:10.1038/s41419-021-03545-7. [Google Scholar] [PubMed] [CrossRef]
135. Zheng S, Hu C, Lin H, Li G, Xia R, Zhang X, et al. circCUL2 induces an inflammatory CAF phenotype in pancreatic ductal adenocarcinoma via the activation of the MyD88-dependent NF-κB signaling pathway. J Exp Clin Cancer Res. 2022;41(1):71. doi:10.1186/s13046-021-02237-6. [Google Scholar] [PubMed] [CrossRef]
136. An N, Zheng B. MiR-203a-3p inhibits pancreatic cancer cell proliferation, EMT, and apoptosis by regulating SLUG. Technol Cancer Res Treat. 2020;19:1533033819898729. doi:10.1177/1533033819898729. [Google Scholar] [PubMed] [CrossRef]
137. Cui Y, Wu Y, Zhu Y, Liu W, Huang L, Hong Z, et al. The possible molecular mechanism underlying the involvement of the variable shear factor QKI in the epithelial-mesenchymal transformation of oesophageal cancer. PLoS One. 2023;18(7):e0288403. doi:10.1371/journal.pone.0288403. [Google Scholar] [PubMed] [CrossRef]
138. Zhang LL, Huang S, Ma XX, Zhang WY, Wang D, Jin SY, et al. Angiotensin(1-7) attenuated angiotensin II-induced hepatocyte EMT by inhibiting NOX-derived H2O2-activated NLRP3 inflammasome/IL-1β/Smad circuit. Free Radic Biol Med. 2016;97:531–43. doi:10.1016/j.freeradbiomed.2016.07.014. [Google Scholar] [PubMed] [CrossRef]
139. Xu QG, Yu J, Guo XG, Hou GJ, Yuan SX, Yang Y, et al. IL-17A promotes the invasion-metastasis cascade via the AKT pathway in hepatocellular carcinoma. Mol Oncol. 2018;12(6):936–52. doi:10.1002/1878-0261.12306. [Google Scholar] [PubMed] [CrossRef]
140. Zou B, Liu X, Zhang B, Gong Y, Cai C, Li P, et al. The expression of FAP in hepatocellular carcinoma cells is induced by hypoxia and correlates with poor clinical outcomes. J Cancer. 2018;9(18):3278–86. doi:10.7150/jca.25775. [Google Scholar] [PubMed] [CrossRef]
141. Wen S, Hou Y, Fu L, Xi L, Yang D, Zhao M, et al. Cancer-associated fibroblast (CAF)-derived IL32 promotes breast cancer cell invasion and metastasis via integrin β3-p38 MAPK signalling. Cancer Lett. 2019;442:320–32. doi:10.1016/j.canlet.2018.10.015. [Google Scholar] [PubMed] [CrossRef]
142. Watanabe N, Kidokoro M, Tanaka M, Inoue S, Tsuji T, Akatuska H, et al. Podoplanin is indispensable for cell motility and platelet-induced epithelial-to-mesenchymal transition-related gene expression in esophagus squamous carcinoma TE11A cells. Cancer Cell Int. 2020;20:263. doi:10.1186/s12935-020-01328-2. [Google Scholar] [PubMed] [CrossRef]
143. Li K, Guo J, Ming Y, Chen S, Zhang T, Ma H, et al. A circular RNA activated by TGFβ promotes tumor metastasis through enhancing IGF2BP3-mediated PDPN mRNA stability. Nat Commun. 2023;14(1):6876. doi:10.1038/s41467-023-42571-1. [Google Scholar] [PubMed] [CrossRef]
144. Terrén I, Orrantia A, Vitallé J, Zenarruzabeitia O, Borrego F. NK cell metabolism and tumor microenvironment. Front Immunol. 2019;10:2278. doi:10.3389/fimmu.2019.02278. [Google Scholar] [PubMed] [CrossRef]
145. Chin DS, Lim CSY, Nordin F, Arifin N, Jun TG. Antibody-dependent cell-mediated cytotoxicity through natural killer (NK) cells: unlocking NK cells for future immunotherapy. Curr Pharm Biotechnol. 2022;23(4):552–78. doi:10.2174/1389201022666210820093608. [Google Scholar] [PubMed] [CrossRef]
146. Kang N, Xie X, Zhou X, Wang Y, Chen S, Qi R, et al. Identification and validation of EMT-immune-related prognostic biomarkers CDKN2A, CMTM8 and ILK in colon cancer. BMC Gastroenterol. 2022;22(1):190. doi:10.1186/s12876-022-02257-2. [Google Scholar] [PubMed] [CrossRef]
147. Li S, Chen Z, Zhou R, Wang S, Wang W, Liu D, et al. Hsa_circ_0048674 facilitates hepatocellular carcinoma progression and natural killer cell exhaustion depending on the regulation of miR-223-3p/PDL1. Histol Histopathol. 2022;37(12):1185–99. doi:10.14670/hh-18-440. [Google Scholar] [PubMed] [CrossRef]
148. Xu J, Wang B, Liu ZT, Lai MC, Zhang ML, Zheng SS. miR-223-3p regulating the occurrence and development of liver cancer cells by targeting FAT1 gene. Math Biosci Eng. 2019;17(2):1534–47. doi:10.3934/mbe.2020079. [Google Scholar] [PubMed] [CrossRef]
149. Ma Y, Zhang C, Zhang B, Yu H, Yu Q. circRNA of AR-suppressed PABPC1 91 bp enhances the cytotoxicity of natural killer cells against hepatocellular carcinoma via upregulating UL16 binding protein 1. Oncol Lett. 2019;17(1):388–97. doi:10.3892/ol.2018.9606. [Google Scholar] [PubMed] [CrossRef]
150. Hu B, Tian X, Li Y, Liu Y, Yang T, Han Z, et al. Epithelial-mesenchymal transition may be involved in the immune evasion of circulating gastric tumor cells via downregulation of ULBP1. Cancer Med. 2020;9(8):2686–97. doi:10.1002/cam4.2871. [Google Scholar] [PubMed] [CrossRef]
151. Mao L, Yang J, Yue J, Chen Y, Zhou H, Fan D, et al. Decorin deficiency promotes epithelial-mesenchymal transition and colon cancer metastasis. Matrix Biol. 2021;95:1–14. doi:10.1016/j.matbio.2020.10.001. [Google Scholar] [PubMed] [CrossRef]
152. Shi M, Li ZY, Zhang LM, Wu XY, Xiang SH, Wang YG, et al. Hsa_circ_0007456 regulates the natural killer cell-mediated cytotoxicity toward hepatocellular carcinoma via the miR-6852-3p/ICAM-1 axis. Cell Death Dis. 2021;12(1):94. doi:10.1038/s41419-020-03334-8. [Google Scholar] [PubMed] [CrossRef]
153. Dunphy MPS, Harding JJ, Venneti S, Zhang H, Burnazi EM, Bromberg J, et al. In vivo PET assay of tumor glutamine flux and metabolism: in-human trial of 18F-(2S,4R)-4-fluoroglutamine. Radiology. 2018;287(2):667–75. doi:10.1148/radiol.2017162610. [Google Scholar] [PubMed] [CrossRef]
154. Yang C, Peng P, Li L, Shao M, Zhao J, Wang L, et al. High expression of GFAT1 predicts poor prognosis in patients with pancreatic cancer. Sci Rep. 2016;6:39044. doi:10.1038/srep39044. [Google Scholar] [PubMed] [CrossRef]
155. Liberti MV, Locasale JW. The warburg effect: how does it benefit cancer cells? Trends Biochem Sci. 2016;41(3):211–8. doi:10.1016/j.tibs.2015.12.001. [Google Scholar] [PubMed] [CrossRef]
156. Zhou S, Guo Z, Lv X, Zhang X. CircGOT1 promotes cell proliferation, mobility, and glycolysis-mediated cisplatin resistance via inhibiting its host gene GOT1 in esophageal squamous cell cancer. Cell Cycle. 2022;21(3):247–60. doi:10.1080/15384101.2021.2015671. [Google Scholar] [PubMed] [CrossRef]
157. Lu Y, Cheng J, Cai W, Zhuo H, Wu G, Cai J. Inhibition of circRNA circVPS33B reduces warburg effect and tumor growth through regulating the miR-873-5p/HNRNPK axis in infiltrative gastric cancer. Onco Targets Ther. 2021;14:3095–108. doi:10.2147/OTT.S292575. [Google Scholar] [PubMed] [CrossRef]
158. Wu JY, Huang TW, Hsieh YT, Wang YF, Yen CC, Lee GL, et al. Cancer-derived succinate promotes macrophage polarization and cancer metastasis via succinate receptor. Mol Cell. 2020;77(2):213–27. doi:10.1016/j.molcel.2019.10.023. [Google Scholar] [PubMed] [CrossRef]
159. Lin HH, Chang CY, Huang YR, Shen CH, Wu YC, Chang KL, et al. Exon junction complex mediates the cap-independent translation of circular RNA. Mol Cancer Res. 2023;21(11):1220–33. doi:10.1158/1541-7786.MCR-22-0877. [Google Scholar] [PubMed] [CrossRef]
160. Pavlova NN, Thompson CB. The emerging hallmarks of cancer metabolism. Cell Metab. 2016;23(1):27–47. doi:10.1016/j.cmet.2015.12.006. [Google Scholar] [PubMed] [CrossRef]
161. Zhang P, Pei S, Wu L, Xia Z, Wang Q, Huang X, et al. Integrating multiple machine learning methods to construct glutamine metabolism-related signatures in lung adenocarcinoma. Front Endocrinol. 2023;14:1196372. doi:10.3389/fendo.2023.1196372. [Google Scholar] [PubMed] [CrossRef]
162. Wu B, Chen Y, Chen Y, Xie X, Liang H, Peng F, et al. Circ_0001273 downregulation inhibits the growth, migration and glutamine metabolism of esophageal cancer cells via targeting the miR-622/SLC1A5 signaling axis. Thorac Cancer. 2022;13(12):1795–805. doi:10.1111/1759-7714.14458. [Google Scholar] [PubMed] [CrossRef]
163. Zhang X, Yang H, Jia Y, Xu Z, Zhang L, Sun M, et al. circRNA_0005529 facilitates growth and metastasis of gastric cancer via regulating miR-527/Sp1 axis. BMC Mol Cell Biol. 2021;22(1):6. doi:10.1186/s12860-020-00340-8. [Google Scholar] [PubMed] [CrossRef]
164. Tu R, Chen Z, Bao Q, Liu H, Qing G. Crosstalk between oncogenic MYC and noncoding RNAs in cancer. Semin Cancer Biol. 2021;75:62–71. doi:10.1016/j.semcancer.2020.10.014. [Google Scholar] [PubMed] [CrossRef]
165. Suresh Babu V, Dudeja G, Sa D, Bisht A, Shetty R, Heymans S, et al. Lack of retinoblastoma protein shifts tumor metabolism from glycolysis to OXPHOS and allows the use of alternate fuels. Cells. 2022;11(20):3182. doi:10.3390/cells11203182. [Google Scholar] [PubMed] [CrossRef]
166. Egger JV, Lane MV, Antonucci LA, Dedi B, Krucher NA. Dephosphorylation of the Retinoblastoma protein (Rb) inhibits cancer cell EMT via Zeb. Cancer Biol Ther. 2016;17(11):1197–205. doi:10.1080/15384047.2016.1235668. [Google Scholar] [PubMed] [CrossRef]
167. Xin D, Xin Z. CircRNA_100782 promotes roliferation and metastasis of gastric cancer by downregulating tumor suppressor gene Rb by adsorbing miR-574-3p in a sponge form. Eur Rev Med Pharmacol Sci. 2020;24(17):8845–54. doi:10.26355/eurrev_202009_22824. [Google Scholar] [PubMed] [CrossRef]
168. Wang M, Zhang R, Zhang S, Xu R, Yang Q. MicroRNA-574-3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting ZEB1 in human gastric carcinoma cells. Gene. 2019;700(49):110–19. doi:10.1016/j.gene.2019.03.043. [Google Scholar] [PubMed] [CrossRef]
169. Cao C, Wang Y, Wu X, Li Z, Guo J, Sun W. The roles and mechanisms of circular RNAs related to mTOR in cancers. J Clin Lab Anal. 2022;36(12):e24783. doi:10.1002/jcla.24783. [Google Scholar] [PubMed] [CrossRef]
170. Li Z, Zhou H, Xia Z, Xia T, Du G, Franziska SD, et al. HMGA1 augments palbociclib efficacy via PI3K/mTOR signaling in intrahepatic cholangiocarcinoma. Biomark Res. 2023;11(1):33. doi:10.1186/s40364-023-00473-w. [Google Scholar] [PubMed] [CrossRef]
171. Zhang X, Wang S, Wang H, Cao J, Huang X, Chen Z, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. doi:10.1186/s12943-018-0935-5. [Google Scholar] [PubMed] [CrossRef]
172. Ghafouri-Fard S, Hussen BM, Shoorei H, Abak A, Poornajaf Y, Taheri M, et al. Interactions between non-coding RNAs and HIF-1α in the context of cancer. Eur J Pharmacol. 2023;943:175535. doi:10.1016/j.ejphar.2023.175535. [Google Scholar] [PubMed] [CrossRef]
173. Tang K, Toyozumi T, Murakami K, Sakata H, Kano M, Endo S, et al. HIF-1α stimulates the progression of oesophageal squamous cell carcinoma by activating the Wnt/β-catenin signalling pathway. Br J Cancer. 2022;127(3):474–87. doi:10.1038/s41416-022-01825-3. [Google Scholar] [PubMed] [CrossRef]
174. Liu A, Xu J. Circ_03955 promotes pancreatic cancer tumorigenesis and Warburg effect by targeting the miR-3662/HIF-1α axis. Clin Transl Oncol. 2021;23(9):1905–14. doi:10.1007/s12094-021-02599-5. [Google Scholar] [PubMed] [CrossRef]
175. Huang YN, Xu YY, Ma Q, Li MQ, Guo JX, Wang X, et al. Dextran sulfate effects EMT of human gastric cancer cells by reducing HIF-1α/TGF-β. J Cancer. 2021;12(11):3367–77. doi:10.7150/jca.55550. [Google Scholar] [PubMed] [CrossRef]
176. Chen ZQ, Zuo XL, Cai J, Zhang Y, Han GY, Zhang L, et al. Hypoxia-associated circPRDM4 promotes immune escape via HIF-1α regulation of PD-L1 in hepatocellular carcinoma. Exp Hematol Oncol. 2023;12(1):17. doi:10.1186/s40164-023-00378-2. [Google Scholar] [PubMed] [CrossRef]
177. Li H, Cao B, Zhao R, Li T, Xu X, Cui H, et al. circDNMT1 promotes malignant progression of gastric cancer through targeting miR-576-3p/Hypoxia inducible factor-1 alpha axis. Front Oncol. 2022;12:817192. doi:10.3389/fonc.2022.817192. [Google Scholar] [PubMed] [CrossRef]
178. Zeng Z, Zhao Y, Chen Q, Zhu S, Niu Y, Ye Z, et al. Hypoxic exosomal HIF-1α-stabilizing circZNF91 promotes chemoresistance of normoxic pancreatic cancer cells via enhancing glycolysis. Oncogene. 2021;40(36):5505–17. doi:10.1038/s41388-021-01960-w. [Google Scholar] [PubMed] [CrossRef]
179. Zhou P, Xie W, Huang HL, Huang RQ, Tian C, Zhu HB, et al. circRNA_100859 functions as an oncogene in colon cancer by sponging the miR-217-HIF-1α pathway. Aging. 2020;12(13):13338–53. doi:10.18632/aging.103438. [Google Scholar] [PubMed] [CrossRef]
180. Campolo F, Sesti F, Feola T, Puliani G, Faggiano A, Tarsitano MG, et al. Platelet-derived circRNAs signature in patients with gastroenteropancreatic neuroendocrine tumors. J Transl Med. 2023;21(1):548. doi:10.1186/s12967-023-04417-8. [Google Scholar] [PubMed] [CrossRef]
181. Huang Y, Shao Y, Yu X, Chen C, Guo J, Ye G. Global progress and future prospects of early gastric cancer screening. J Cancer. 2024;15(10):3045–64. doi:10.7150/jca.95311. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools