 Open Access
Open Access
ARTICLE
Physiological Dynamics and Transcriptomic Analysis of Cut Roses ‘Carola’ Treated with KNO
College of Agriculture & Biology, Zhongkai University of Agriculture and Engineering, Guangzhou, 510225, China
* Corresponding Author: Shenggen He. Email:
Phyton-International Journal of Experimental Botany 2023, 92(12), 3267-3290. https://doi.org/10.32604/phyton.2023.045453
Received 27 August 2023; Accepted 08 October 2023; Issue published 28 December 2023
Abstract
The consumption of cut roses (Rosa hybrida) has always ranked first in the world. However, it is vulnerable to rapid petal and leaf wilting due to leaf stomatal water loss, which seriously affects its ornamental quality and economic value. Stomatal movement, a key in plant physiological processes, is influenced by potassium and nitrate. Advancing comprehension of its physiological and molecular mechanism holds promise for preserving the freshness of cut roses. This study observed the impacts of different concentrations of KNO3 vase treatments on stomatal opening and water loss in cut rose ‘Carola’ leaves, as well as their transcriptional responses to KNO3. Water loss rates were influenced by KNO3 concentrations, with the 25 and 75 mmol/L treatments exhibiting the highest water loss rates. The stomatal aperture reached its widest value when treated with 75 mmol/L KNO3. Transcriptional sequencing analysis was performed to identify differentially expressed genes (DEGs) of which 5456 were up-regulated, and 6607 were down-regulated associated with photosynthesis, starch and sucrose metabolism, metabolic pathways, plant-pathogen interaction, plant hormone signal transduction, and related pathways. 246 DEGs were selected related to response to KNO3 treatment, of which gene ontology (GO) enrichment were nitrate and terpenoid metabolism, ion transport, and response to stimuli. Further heatmap analysis revealed that several genes related to nitrate transport a metabolism, K+ transport, vacuoles, and aquaporin were in close association with the response to KNO3 treatment. Weighted gene co-expression network analysis (WGCNA) revealed that hub genes, including LAX2, TSJT1, and SCPL34 were identified in turquoise, black, and darkgreen module. Transcription factors such as NAC021, CDF3, ERF053, ETR2, and ARF6 exhibited regulatory roles in the response to KNO3 treatment under light conditions. These findings provide valuable insights into the physiological and molecular mechanisms underlying the response of cut rose leaves to KNO3 treatment.
Keywords
Supplementary Material
Supplementary Material FileNomenclature
| DEGs | Differentially expressed genes |
| GO | Gene ontology |
| PCA | Principal component analysis |
| WGCNA | Weighted gene co-expression network analysis |
| KEGG | Kyoto encyclopedia of genes and genomes |
| SE | Standard error |
| TPK | Tandem-pore K+ channel |
| ABA | Abscisic Acid |
Cut rose (Rosa hybrida), reigns as the pinnacle among the ‘Top Four Cut Flowers’ worldwide and is highly cherished, and adored for its beauty and grace. Nonetheless, the post-harvest period poses a significant challenge for cut roses due to their heightened vulnerability to disrupted water metabolism, resulting in consequential wilting of petals and leaves and a pronounced deterioration in overall quality. Stomata, serving as vital conduits for gas exchange and moisture regulation between plants and their environment, assume a critical role in fundamental physiological processes encompassing photosynthesis, transpiration, and stress responses [1–3]. The intricate regulation of stomatal aperture, predominantly governed by the coordinated actions of guard cells, is primarily dependent upon their expansion and contraction. Intriguingly, a myriad of influential factors capable of modulating stomatal movements exists, including environmental stimuli (e.g., light intensity, carbon dioxide levels), cellular osmolytes (including potassium ions, calcium ions, nitrates, and various sugar compounds), as well as plant hormones (e.g., abscisic acid) [4,5].
Potassium (K) plays a pivotal role in maintaining the charge balance of inorganic and organic anions, regulating membrane potential, stabilizing pH homeostasis, and modulating cell osmotic pressure. Several families of potassium channels have been identified, comprising the Shaker family, TPK (Tandem-Pore K+ channel) family, Kir-like family, and various other potassium channels [6]. The Shaker family represents the earliest identified group, including AKT1 (Arabidopsis K+ transporter 1), KAT1 (K+ channel in Arabidopsis thaliana 1), KAT2, AKT2, SKOR (stelar K+ outward rectifier), and GORK (guard cell outward rectifying K+ channel) [6–8]. Contrary to the Shaker family, the TPK family and Kir-like family do not rely on voltage regulation. In Arabidopsis, the TPK family presently encompasses five members, namely TPK1 to TPK5, while KCO3 (K+ channel, Ca2+-activated, outward rectifying 3) belongs to the Kir-like family. Homologous genes associated with these channels have been identified in diverse plants, such as rice, rubber, tobacco, and poplar. Additionally, the repertoire of potassium channels includes the CNGC (cyclic nucleotide-gated channels) family and others [6–10]. Moreover, three types of potassium ion transporters, namely KUP/HAK/KT (K+ uptake permeases/high-affinity K+/K+ transporter), CPA (cation proton antiporters), and HKT (high-affinity K+ transporter), are primarily involved in potassium transportation, aiding plants in K+ acquisition from potassium-deficient soil environments [11–13]. Recent research has provided insights into the functional mechanisms and regulatory principles of potassium channels in stomatal movement. The phosphorylation event hyperpolarizes the plasma membrane, induces the opening of potassium ion channels, triggers potassium ion influx, and concurrently facilitates the influx of related anions, resulting in osmotic water uptake and subsequent stomatal opening. Interestingly, red light may enhance the phosphorylation effect of blue light during this process [14–16]. Recently, the cryo-electron microscopy structure of the inward-rectifying potassium ion channel KAT1 in Arabidopsis guard cells was elucidated, revealing its distinctive “non-domain-swapped” topology and the presence of a Thr-Thr-Gly-Tyr-Gly potassium ion selectivity filter [17].
Nitrogen (N) is a vital macronutrient that profoundly impacts plant growth and development by serving as a crucial component of essential molecules such as amino acids, chlorophyll, nucleic acids, and various secondary metabolites. To ensure efficient uptake, transport, and response to nitrogen, plants have evolved sophisticated systems. The dominant form of nitrogen that plants absorb are ammonium (NH4+) and nitrate (NO3−). The application of Rhizophagus irregularis to Lycium arbarum, in the presence of adequate NO3− and NH4+ supply, led to significant enhancements in L. barbarum biomass, and mycorrhizal plants exhibited superior performance specifically when NO3− was supplied [18]. NO3− is the primary nitrogen source and also functions as a signaling molecule, regulating crucial aspects of plant physiology, including root architecture, shoot growth, flowering time, stomatal regulation, and seed dormancy [19]. The transporters/channels responsible for NO3− uptake can be classified into four families: NPF/NRT1, NRT2, CLC, and SLAC/SLAH [20]. In Arabidopsis, the NPF family comprises 53 genes that transport not only NO3− but also peptides, amino acids, nitrite, glucosinolates, auxin, abscisic acid (ABA), and gibberellins [21–25]. NRT2 forms a heterotetramer with AtNAR2.1/NRT3 to facilitate NO3− transport, except for NRT2.7, which localizes to the vacuolar membrane, while other members of the NRT2 family are plasma membrane-localized inward high-affinity transporters that interact with AtNAR2.1/NRT3 [26]. In addition to its role as a nutrient, NO3− acts as a signaling molecule, triggering a cascade of physiological and developmental responses in plants. The NO3− transporter NPF6.3/NRT1.1 is induced by short-term NO3− exposure and undergoes feedback regulation by high NO3− concentrations, stimulating lateral root development through auxin transport [24,27]. Under high NO3− levels, NPF6.3/NRT1.1 functions as a low-affinity transporter, whereas under low NO3− conditions, it transitions into a high-affinity transporter mode upon phosphorylation by the CIPK23 protein kinase, and NPF6.3/NRT1.1 activates phosphatase C, leading to elevated intracellular Ca2+ levels, which regulate the expression of NRT2.1 and TGA1 genes via Ca2+ signaling [27–29]. NO3− signaling and its downstream physiological responses are closely intertwined with plant hormones. Nitrogen availability influences the biosynthesis and transport of auxin, ethylene, cytokinins and ABA [30–33]. Several transcription factors, including NLP7, NLP6, TGA1, TGA4, ANR, Bzip1, LBD37, LBD38, TCP20, NAC4, and SPL9, have been identified as important regulators in response to NO3− treatment [34]. Notably, TGA1 and TGA4 exhibit peak expression levels one to two hours after NO3− treatment, and their binding to the promoter regions of target genes, as validated by ChIP with NRT2.1 and NRT2.2, confirms their regulatory role [35]. TCP20, on the other hand, binds to the promoters of type-A Arabidopsis response regulators (ARRs) ARR5/7 during NO3− treatment, leading to their upregulation in stems, potentially linking nitrogen and cytokinin signaling in cell division [36,37]. Recent studies by Xu et al. have demonstrated that increasing potassium supply under high nitrogen conditions upregulates nitrogen assimilation-related genes, promoting nitrogen uptake by roots and its transport to aboveground plant parts [38]. This approach significantly enhances the activity of nitrogen metabolism-related enzymes (NR, GS, and GOGAT) in leaves and roots, counteracting the inhibition of nitrogen assimilation in apple roots and leaves under high nitrogen conditions [39]. Moreover, potassium ions facilitate nitrogen allocation within leaves, improving photosynthetic nitrogen use efficiency and facilitating nitrogen uptake and assimilation [38,40].
The N-K balance significantly influences plant growth and stress responses. Physiological studies have revealed the close connection between NO3− and K+ uptake and transport [41–43]. However, the molecular mechanisms governing this balance still require extensive investigation. NRT1.5, originally identified as a pH-dependent bidirectional NO3− transporter, plays a role in regulating K+ distribution and NO3− loading into the xylem in plants [44–46]. NRT1.1 mediates the interaction between NO3− and K+ in Arabidopsis. NRT1.1 expression patterns in the root coordinate with K+ channels/transporters to enhance potassium uptake and root-to-shoot distribution, contingent upon sufficient NO3− supply [47]. Liu et al. elucidated the functional unit formed by the NO3− channel SLAH3 and the K+ channels GORK and SKOR, which coordinate to regulate membrane potential by modulating the nitrogen-potassium balance [48].
To date, there is a significant knowledge gap regarding the response and regulatory mechanisms associated with K+ and NO3− in cut roses. The understanding of K+ and NO3− channels/transporters’ involvement in stomatal closure and their intricate regulatory pathways in cut roses is severely limited, necessitating comprehensive and systematic research. In this study, we focused on the utilization of the cut rose ‘Carola’ as the experimental subject. Initially, we examined the impact of KNO3 treatment on the dynamics of stomatal opening of leaves and water loss in cut roses. Subsequent to this, we conducted transcriptome sequencing and analysis on the KNO3-treated cut rose, with the aim of unraveling the interconnectedness among K+ channels, nitrate transport and metabolism, stomatal opening, water loss, and the underlying physiological and molecular mechanisms governing their regulation. This study is expected to provide a solid theoretical foundation for understanding the regulation of stomatal movement and the mitigation of post-harvest water loss in cut roses and other analogous floral species through the utilization of KNO3.
2.1 Treatments of Cut Roses with Different Concentrations of KNO3
Freshly harvested cut roses (‘Carola’) were obtained from a rose plantation in Huadu District, Guangzhou, China (longitude 113°28′12″E, latitude 23°26′24″N), with a commercial maturity level (cultivated for 2 years). The flower stems were placed in plastic buckets filled with tap water and transported to the postharvest experiment at the Biotechnology Research Institute of Zhongkai University of Agriculture and Engineering within 1 h. Cut roses of uniform developmental stage and stem thickness, free from pests and diseases were selected for the experiments. The stems, with a length of 35 cm and retaining the first and second leaves, were trimmed. The stems were initially placed in conical flasks containing deionized water and subjected to three hours of dark treatment. Subsequently, the cut roses were individually transferred to separate conical flasks containing different concentrations of KNO3 treatments: A (control, CK), B (25 mmol/L), C (50 mmol/L), D (75 mmol/L), and E (100 mmol/L), under a light intensity of 100 μmol m–2 s–1, temperature of 20°C ± 2°C, and humidity of 60% ± 5% (Fig. 1).
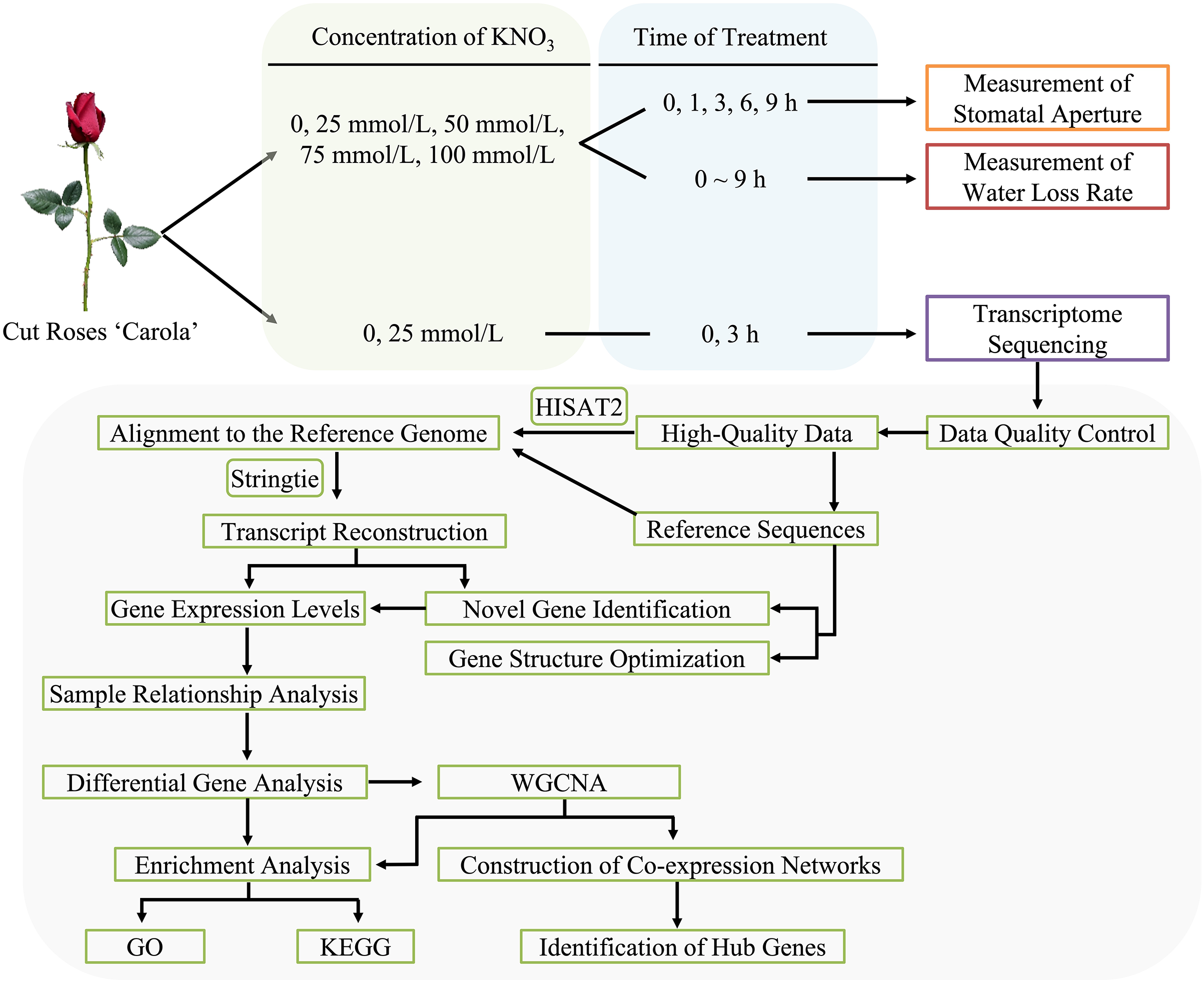
Figure 1: Flowchart of methodology for cut roses ‘Carola’ treated with KNO3
2.2 Measurement of Water Loss Rate and Stomatal Aperture in Cut Roses
The water loss rate of the cut roses within a 9-h light period was continuously monitored using an automated device for precise water loss detection. Data were collected every 5 min, with three biological replicates selected for each treatment.
Four biological replicates, exhibiting consistent growth conditions, were chosen for each treatment. Samples were collected at 0, 1, 3, 6, and 9 h of light exposure. For each stem, a 0.25 cm2 leaf sample was excised using a surgical blade. Two random fields of view from the lower epidermis of rose petals were captured using an optical microscope, resulting in a total of 8 fields of view for statistical analysis. Subsequently, 40 random stomata were selected, and their apertures were quantified using Image-win64 software, enabling the calculation of the mean aperture value (Fig. 1).
2.3 Transcriptome Sequencing and Data Analysis
To induce stomatal closure, the cut roses were placed in darkness for 3 h. Following the dark treatment, the cut roses were individually placed in conical flasks containing deionized water, and 25 mmol/L KNO3 solutions, respectively. Sampling time points of 0, 1, and 3 h were selected under a light intensity of 100 μmol m–2 s–1 (0 h denoted the time point after 3 h of darkness, coinciding with the initiation of white light exposure). Leaf samples were taken from the second leaf of the roses using a surgical blade, immediately flash-frozen in liquid nitrogen, and subsequently stored in a −80°C ultra-low temperature freezer. Each treatment consisted of three biological replicates, with each biological replicate comprising one cut rose. The experimental conditions were maintained at a temperature of 20°C ± 2°C and relative humidity of 60% ± 10%.
RNA extraction, library construction, quality control, and transcriptome sequencing were outsourced to Guangzhou Genedenovo Biotechnology Co., Ltd., Principal component analysis was performed using R based on gene expression levels. Low-quality data filtering was conducted following the approach described by Vidal et al. [43]. The functional annotation of differentially expressed genes (DEGs) was performed by aligning the NCBI rose genome. Alignment analysis based on the reference genome was conducted using HISAT2 software [44]. Differential expression analysis and weighted gene co-expression network analysis (WGCNA) analysis were performed using the OmicSmart transcriptome analysis platform (Fig. 1).
2.4 Quantitative Real-Time (qRT)-PCR
Two differentially expressed genes were selected for analysis, and primer sequences were designed via NCBI (https://www.ncbi.nlm.nih.gov/tools/primer-blast), with the housekeeping gene GAPDH serving as the reference gene. The reaction mixture was prepared using 2 × Real Star SYBR Mixture (with ROX II) reagent (YEASEN, Shanghai, China) as follows: 10 μL of SYBR Mixture (with ROX II), 1 μL of cDNA template, 1 μL of each primer (10 μmol/μL), and 8 μL of nuclease-free water. Real-time PCR reactions were conducted using the CFX Connect Real-Time PCR Detection System (Bio-Rad, USA) under the following conditions: 95°C for 5 min; 95°C for 10 s, 60°C for 30 s, for 40 cycles. Relative quantitative analysis was performed using the 2−ΔΔCT method.
2.5 Data Statistics and Analysis
All data were presented as mean ± standard error (SE). Significance testing was carried out using the least significant difference method (LSD) (p < 0.05). Data analysis was performed using SPSS 26.0 (International Business Machines Corporation, USA) and Sigmaplot 14.0 (Systat Software Inc., USA).
3.1 Effects of KNO3 Treatments on Water Loss
The water loss rate in cut roses ‘Carola’ exhibited distinct responses to various concentrations of KNO3 treatments. During the initial 60 min, the 25 and 75 mmol/L KNO3 treatments exhibited the highest rates of water loss, surpassing the control (CK) treatment, while the 50 and 100 mmol/L KNO3 treatments resulted in the lowest rates. Subsequently, the water loss rates gradually increased in the KNO3 treatments, whereas the control showed a gradual decline. After 150 min, all KNO3 treatments showed higher water loss rates compared to the control, with the 25 and 75 mmol/L KNO3 treatments displaying the highest rates, significantly exceeding the CK treatment (Fig. 2A). Furthermore, the cumulative water loss after 300 min was notably higher in the 25 and 75 mmol/L KNO3 treatments than in the other treatments, including the control (Fig. 2B).
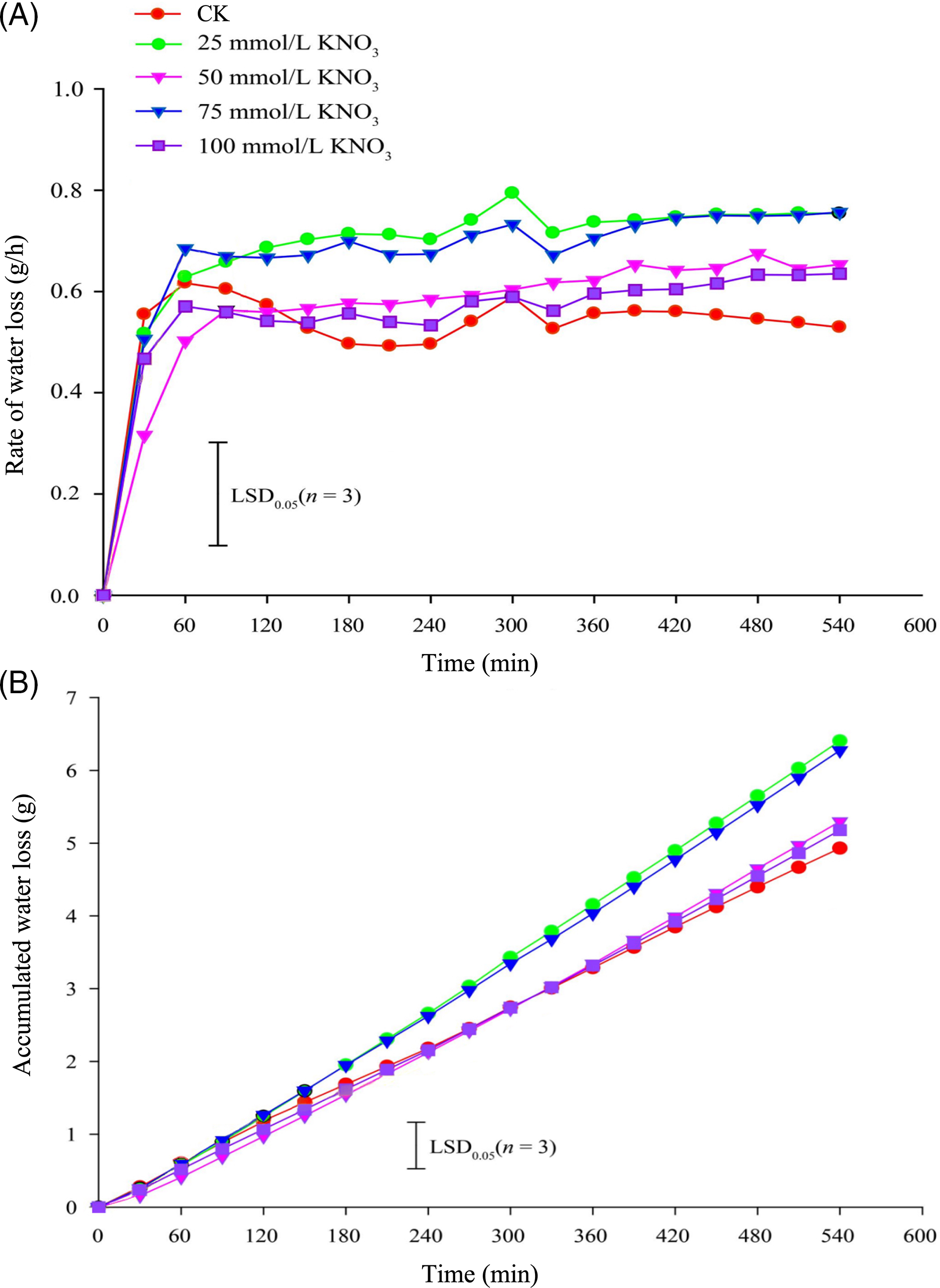
Figure 2: Effects of different concentrations of KNO3 treatments on the water loss rate (A) and cumulative water loss (B) in cut roses ‘Carola’
3.2 Effects of KNO3 Treatments on Stomatal Opening
As depicted in Figs. 3A and 3B, the stomatal apertures of cut roses leaves exhibited distinctive characteristics under the control and various concentration of KNO3 treatments. Specifically, the 75 mmol/L KNO3 treatment consistently demonstrated the widest stomatal aperture under light conditions, significantly exceeding the control. Furtherore, the stomatal apertures of the 25, 50, and 100 mmol/L KNO3 treatments exhibited a rapid increase within the 0 to 3-h time frame, with values noticeably surpassing those of the control by the 3-h mark (Fig. 3).
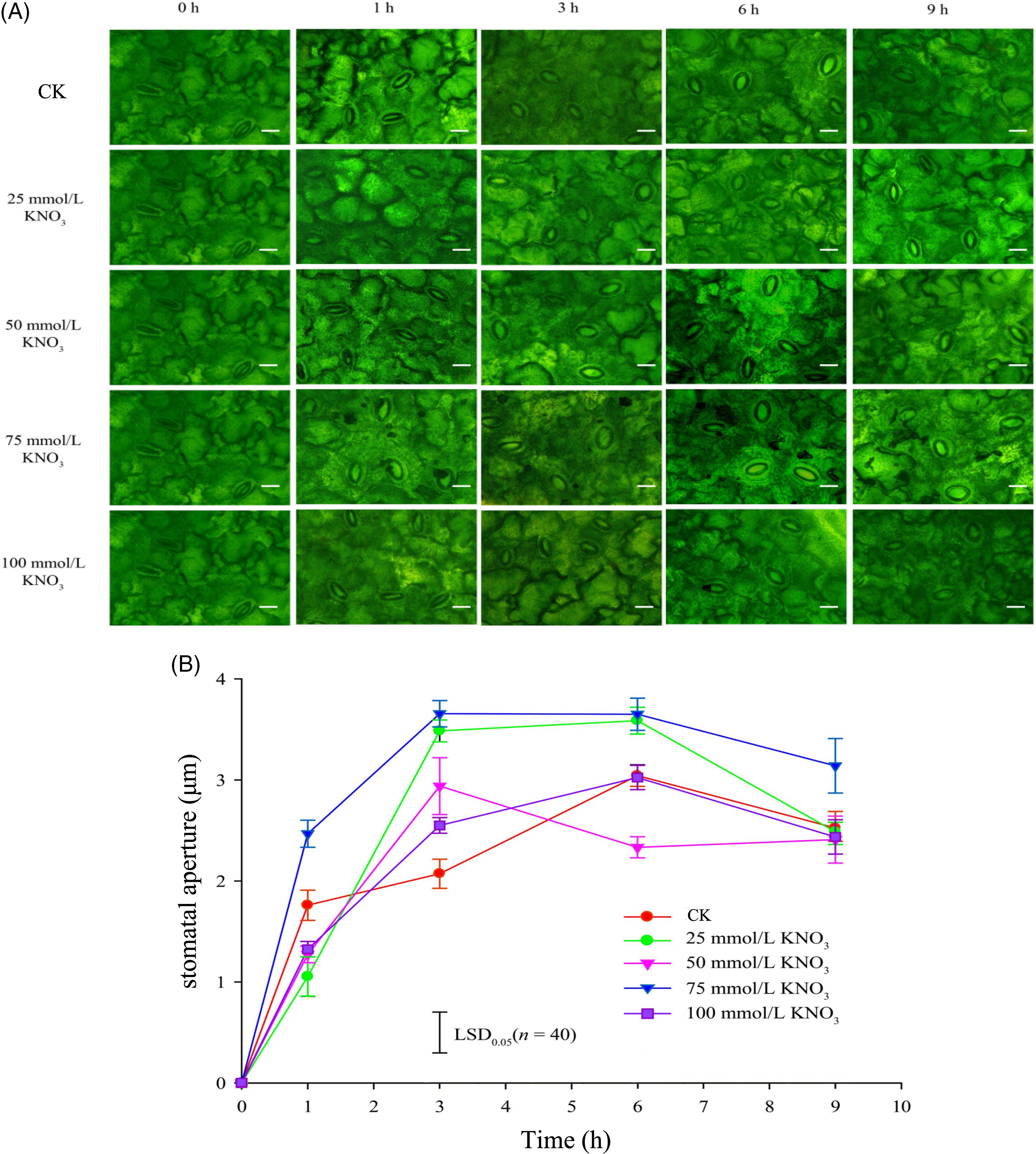
Figure 3: Effects of different concentrations of KNO3 treatments on the opening state of stomata (A) and the stomatal aperture (B) in the lower epidermis of cut rose ‘Carola’ leaves (Scale bar = 10 μm)
3.3 Transcriptional Sequencing and Sample Relationship Analysis
Total RNA from ‘Carola’, a cultivar of cut roses, was extracted and subjected to agarose gel electrophoresis. The ensuing electrophoretic pattern exhibited distinct bands, underscoring the RNA’s suitability for transcriptome sequencing (Fig. S1). Each sample yielded a surplus of 5.4 billion effective reads, with GC content spanning 45.78% to 46.65%. Notably, the Q30 percentage surpassed 91% across all samples, affirming the robustness of the sequencing data underpinning this investigation (Table S1).
Utilizing the expression levels of known genes in each sample, we conducted Principal Component Analysis (PCA), calculated Pearson correlation coefficients, performed sample relationship clustering, and conducted inter-sample correlation analysis. The findings demonstrated high reproducibility among the samples in this transcriptional sequencing study, validating their suitability for subsequent analysis (Fig. 4A).
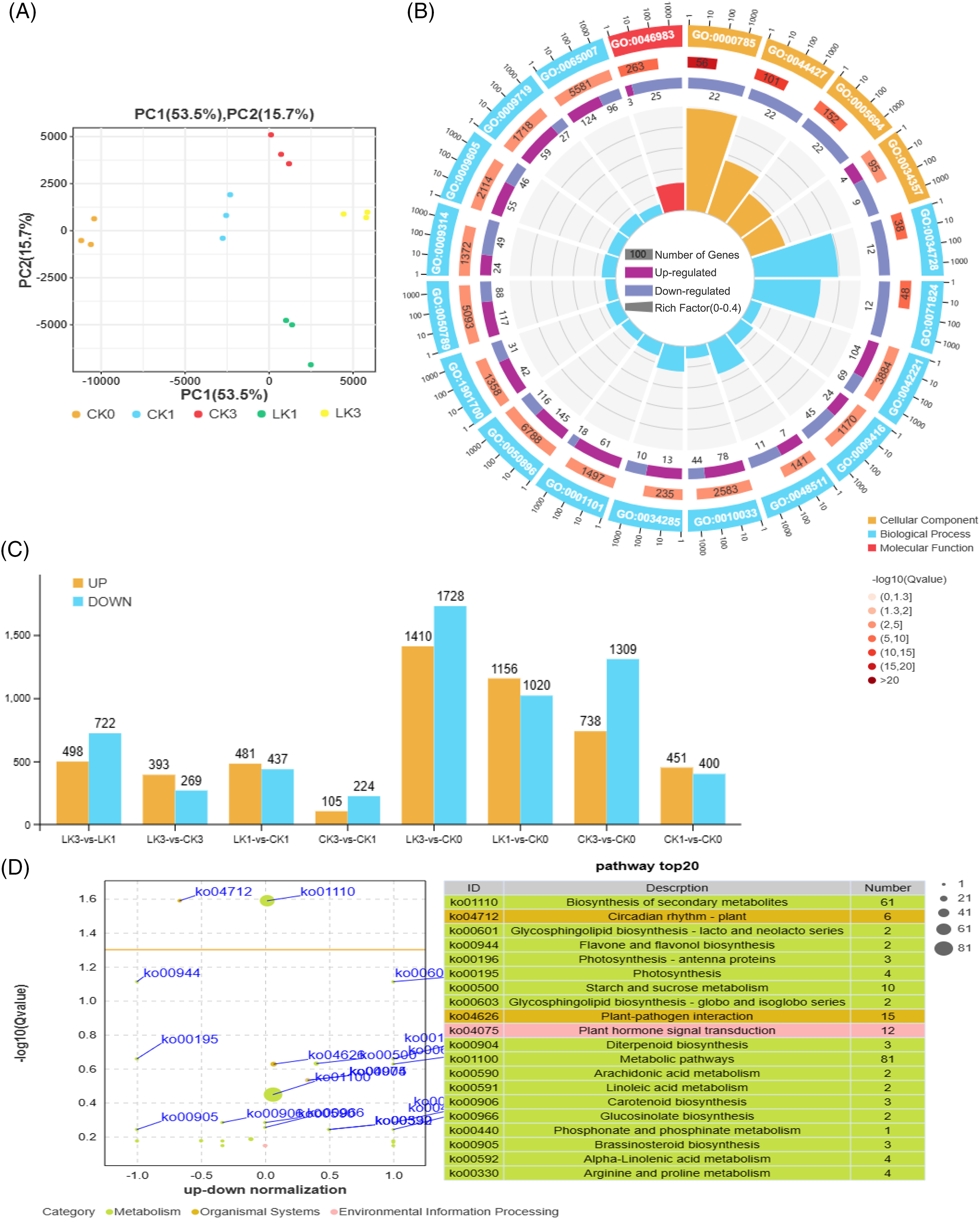
Figure 4: Sample relationship and enrichment analysis of differentially expressed genes (DEGs) in cut rose ‘Carola’ leaves. (A) PCA of samples treated with 25 mmol/L KNO3 and the control; (B) The up-and down-genes between the two treatments; (C) Gene ontology enrichment analysis of the DEGs; (D) KEGG enrichment analysis of the DEGs
A total of 5456 DEGs were up-regulated, while 6607 DEGs were down-regulated (Fig. 4B). Gene ontology (GO) enrichment analysis indicated that the top 20 enriched terms in the cellular component category were chromosome-related (GO:0000785, GO:0044427, GO:0005694), as well as photosynthetic membrane (GO:0034357). In terms of biological process, the major enriched terms were related to biological regulation in response to chemical and light stimuli (GO:0042221, GO:0009416, GO:0050896, etc.). The molecular function category showed enrichment for protein dimerization activity (GO:0046983), suggesting that cut roses exhibit stress responses and initiate gene expression and translation under potassium nitrate treatment in light conditions (Fig. 4C).
Correspondingly, the Kyoto encyclopedia of genes and genomes (KEGG) enrichment analysis revealed that the top 20 enriched pathways included biosynthesis of secondary metabolites, photosynthesis, starch and sucrose metabolism, plant-pathogen interaction, and plant hormone signal transduction (Fig. 4D).
3.4 Functional Annotation and Enrichment Analysis of DEGs
Differential analysis identified 918 genes with significant expression differences between the 25 mmol/L KNO3 treatment and the control after 1 h, along with 662 genes after 3 h of treatment (Fig. 5A). Employing the R programming language, Venn diagrams and UpSetR plots were generated, revealing 246 genes that exhibited differential expression at both 1 and 3 h, underscoring their close association with the response to KNO3 treatment (Fig. 5A).
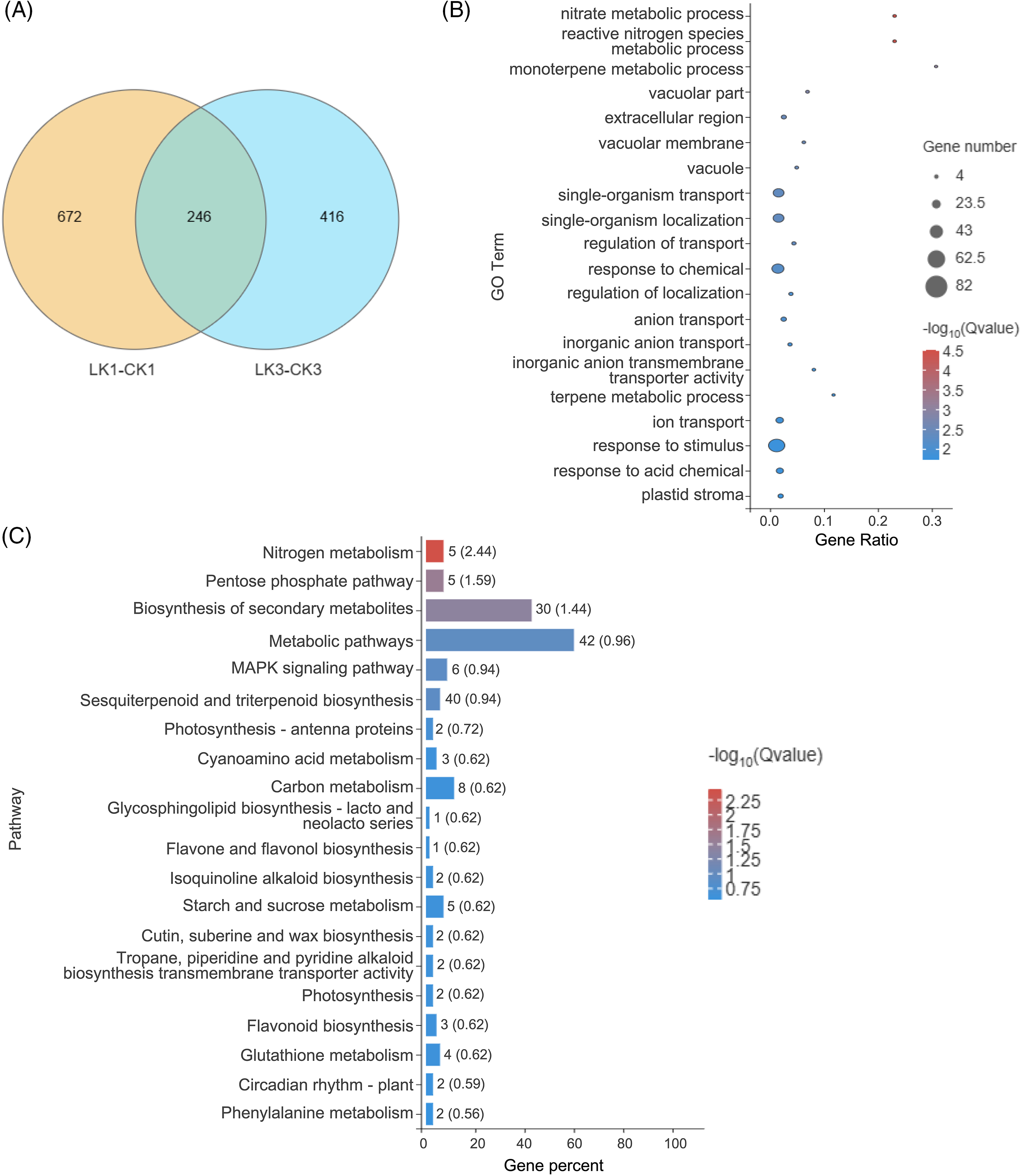
Figure 5: Sample relationship and enrichment analysis of DEGs in cut rose ‘Carola’ leaves. (A) Venn diagram analysis of samples treated with 25 mmol/L KNO3 for 1 and 3 h; (B) GO enrichment analysis of DEGs selected from the Venn diagram; (C) KEGG enrichment analysis of DEGs selected from the Venn diagram
Based on the expression levels of these differentially expressed genes across various samples, GO functional annotation was performed. The top 20 enriched processes encompassed nitrate metabolic processes, terpenoid metabolic processes, vacuolar components (e.g., vacuolar membranes), ion transport, and response to stimuli. These results indicate transcriptional changes in these processes in response to KNO3 treatment, potentially influencing associated physiological processes (Fig. 5B).
Furthermore, the differentially expressed genes were mapped to the KEGG database, revealing highly enriched pathways such as nitrogen metabolism, the pentose phosphate pathway, secondary metabolite biosynthesis, metabolic pathways, and the MAPK (mitogen-activated protein kinase) signaling pathway (Fig. 5C). This suggests the involvement of certain genes in these pathways in the response of cut rose leaves to KNO3 treatment.
3.5 Expression Pattern Analysis of DEGs
Heatmap analysis of expression levels for genes related to NO3− transport and metabolism revealed three classes of differentially expressed genes (Fig. 6A). Class I genes, including nitrate regulatory genes NRG2, APSR, and nitrate transporter NAR2.1, exhibited relatively high expression levels after 1 h of KNO3 treatment, indicating their early response to KNO3 treatment. Class II genes, such as nitrate transporters NRT3.1, NRT2.5, and nitrate reductase NIA1, showed higher expression levels after 1 h of KNO3 treatment, suggesting nitrate transport into cells and associated redox reactions. Class III genes, including nitrate regulatory gene REL2 and nitrate transporter NRT2.7, exhibited decreased expression levels after KNO3 treatment, indicating their suppression during KNO3 treatment (Fig. 6A).
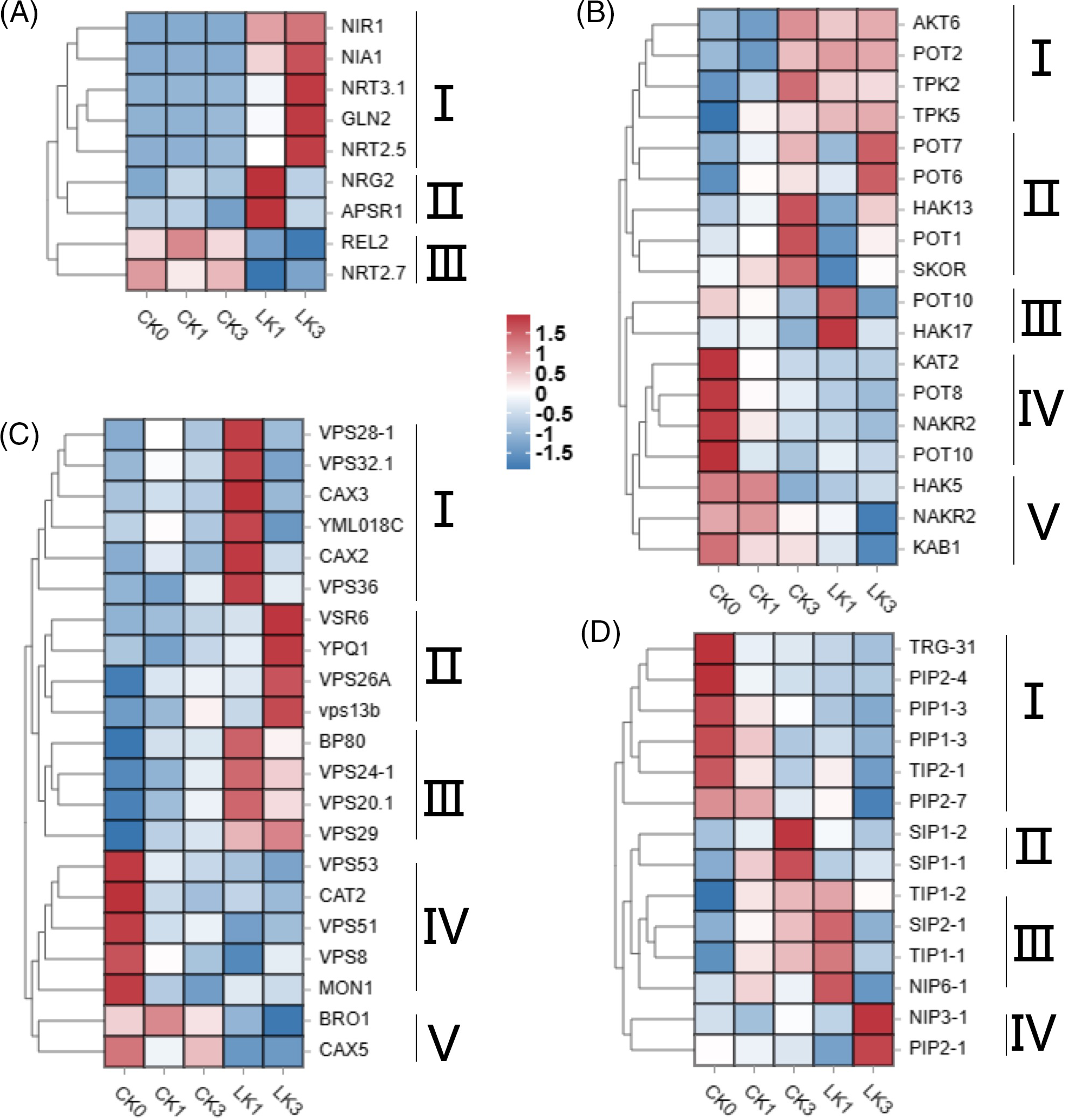
Figure 6: Heatmap of expression levels for differentially expressed genes in cut rose ‘Carola’ leaves. DEGs related to NO3- transport and metabolism (A), K+ channels and transporters (B), vacuoles-associated proteins (C), and aquaporins (D). CK0, CK1 and CK3 correspond to the control group of cut roses for 0, 1 and 3 h, whereas LK1 and LK3 specifically represent samples treated with 25 mmol/L KNO3 for 1 and 3 h
Differential gene heatmap analysis identified five classes of K+ channel and transporter genes (Fig. 6B). Class I genes displayed relatively low expression levels at 0 and 1 h in the control group but exhibited increased expression levels at 3 h, as well as after 1 and 3 h of KNO3 treatment. Class II genes exhibited relatively low expression levels at 0 and 1 h, with increased expression at 3 h. Among them, POT7 (potassium transporter 7) and POT6 displayed higher expression levels after 3 h of KNO3 treatment compared to the control, while HAK13 (high-affinity K+ transporter 13), POT1, and SKOR (stellar K+ outward rectifier) exhibited lower expression levels after 3 h of KNO3 treatment compared to the control. Class III genes, represented by POT10 and HAK17, showed higher expression levels after 1 h of KNO3 treatment compared to other groups. Class IV genes exhibited higher expression levels without light exposure and KNO3 treatment, but their expression decreased under light exposure. Class V genes displayed lower expression levels during KNO3 treatment compared to the control (Fig. 6B).
Five classes of differentially expressed genes related to vacuoles were identified. Class I genes exhibited relatively high expression levels after 1 h of KNO3 treatment, including vacuolar sorting-associated proteins (VPS28-1, VPS32.1, VPS36), vacuolar cation/proton exchangers (CAX3, CAX2), and an unidentified vacuolar membrane protein YML018C. Class II genes showed relatively high expression levels after 3 h of KNO3 treatment, including vacuolar sorting receptors (VSR6), vacuolar sorting-associated proteins (VPS26A and VPS13b), and an amino acid transporter (YPQ1). Class III genes exhibited higher expression levels after KNO3 treatment compared to the control, including vacuolar sorting receptors (BP80) and vacuolar sorting-associated proteins (VPS24-1, VPS20.1, VPS29). Class IV genes displayed higher expression levels without light exposure and KNO3 treatment, with decreased expression under light exposure. Class V genes showed lower expression levels during KNO3 treatment compared to the control (Fig. 6C).
Four classes of aquaporin-related genes were identified, and their expression patterns varied among different treatments. Class IV genes, NIP3-1 (nodulin 26-like intrinsic protein 3–1) and PIP2-1 (plasma membrane intrinsic protein 2–1), displayed higher expression levels after 3 h of KNO3 treatment, indicating their close association with the response to potassium nitrate treatment (Fig. 6D).
3.6 Weighted Gene Co-Expression Network Analysis
By employing WGCNA, we explored the correlation between gene expression levels and their response to potassium nitrate treatment in cut roses. Related genes and their expression patterns were identified, which were then classified into modules based on their similarity in expression. To construct the gene clustering tree, the expression correlation coefficients among genes and determined a power value of 7 was computed (Fig. 7A). The resulting clustering relationships facilitated the partitioning the genes into different modules, and the modules with similar characteristic expression patterns were subsequently merged based on the similarity of their module eigengenes (Fig. 7B). Our network comprised a total of 4860 genes, divided into 18 modules. The turquoise module contained the highest number of genes (1050), while the darkturquoise module had the fewest (58) (Figs. 7A and 7B). Notebly, the turquoise module exhibited decreased gene expression under light exposure, suggesting a potential association between these genes and the response of cut roses to light. Additionally, the lightgreen and greenyellow modules exhibited decreased expression upon KNO3 treatment, while the midnightblue module showed increased expression, indicating their involvement in the response to potassium nitrate treatment. Moreover, the darkturquoise, darkgreen, darked, and yellow modules exhibited relatively higher expression levels after 1 h of potassium nitrate treatment compared to other groups. Similarly, the black module showed relatively higher expression levels after 3 h of potassium nitrate treatment (Fig. 7C). These modules provide valuable insights into subsequent analysis of the gene expression dynamics associated with KNO3 treatment in cut roses.
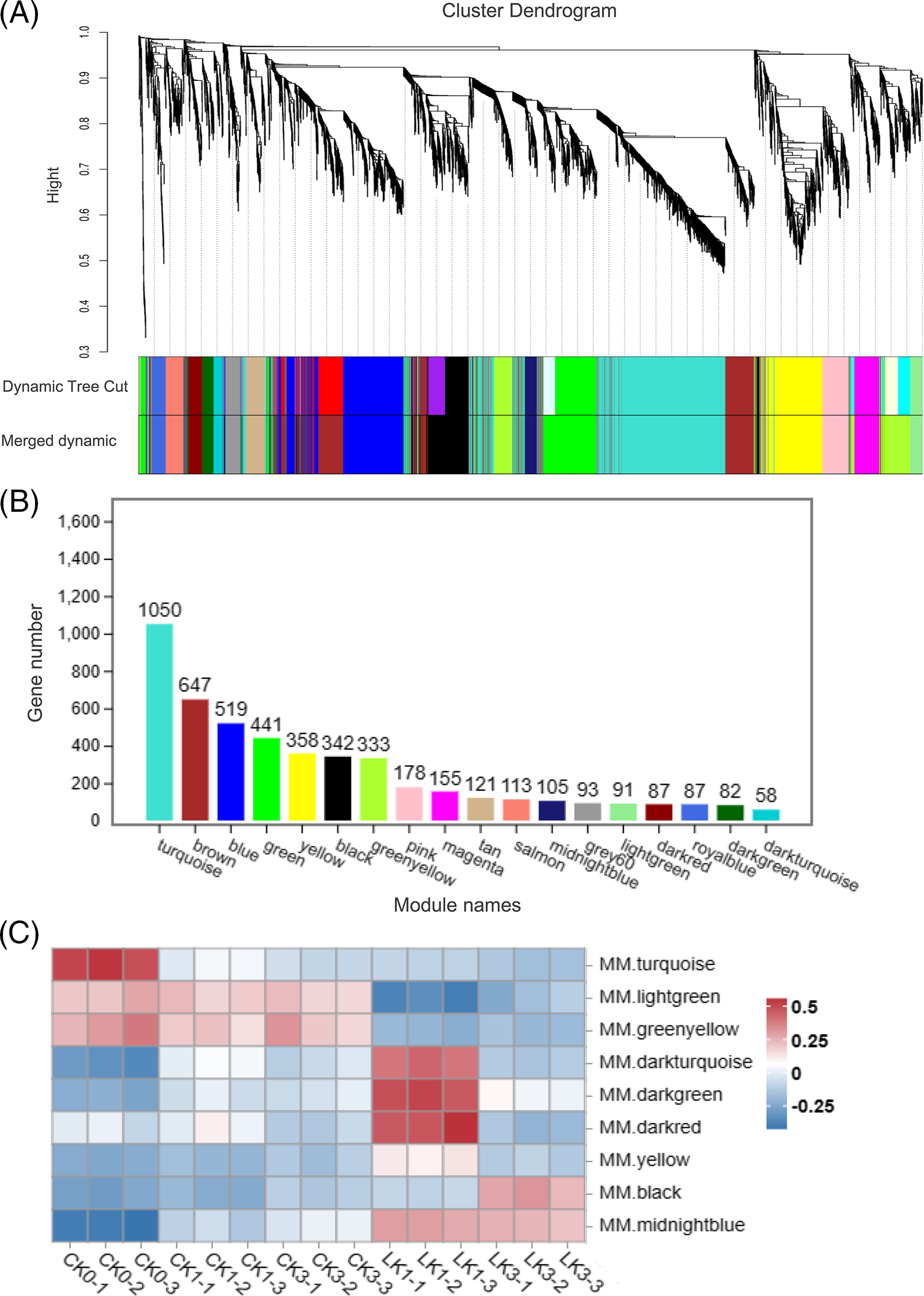
Figure 7: WGCNA of 15 samples of cut rose ‘Carola’ leaves. (A) Hierarchical clustering tree illustrating the co-expression modules identified by WGCNA; (B) Number of genes within each module; (C) Heatmap representing the expression patterns of several modules across the samples
GO analysis revealed that the turquoise module was significantly enriched in processes such as nucleic acid binding transcription factor activity, phosphatidylethanolamine metabolic process, second-messenger-mediated signaling, and chemical homeostasis (Fig. 8A). Moreover, the turquoise module showed a high enrichment level in several KEGG pathways, including Plant hormone signal transduction, Photosynthesis-antenna proteins, and Plant-pathogen interaction (Fig. 8C).
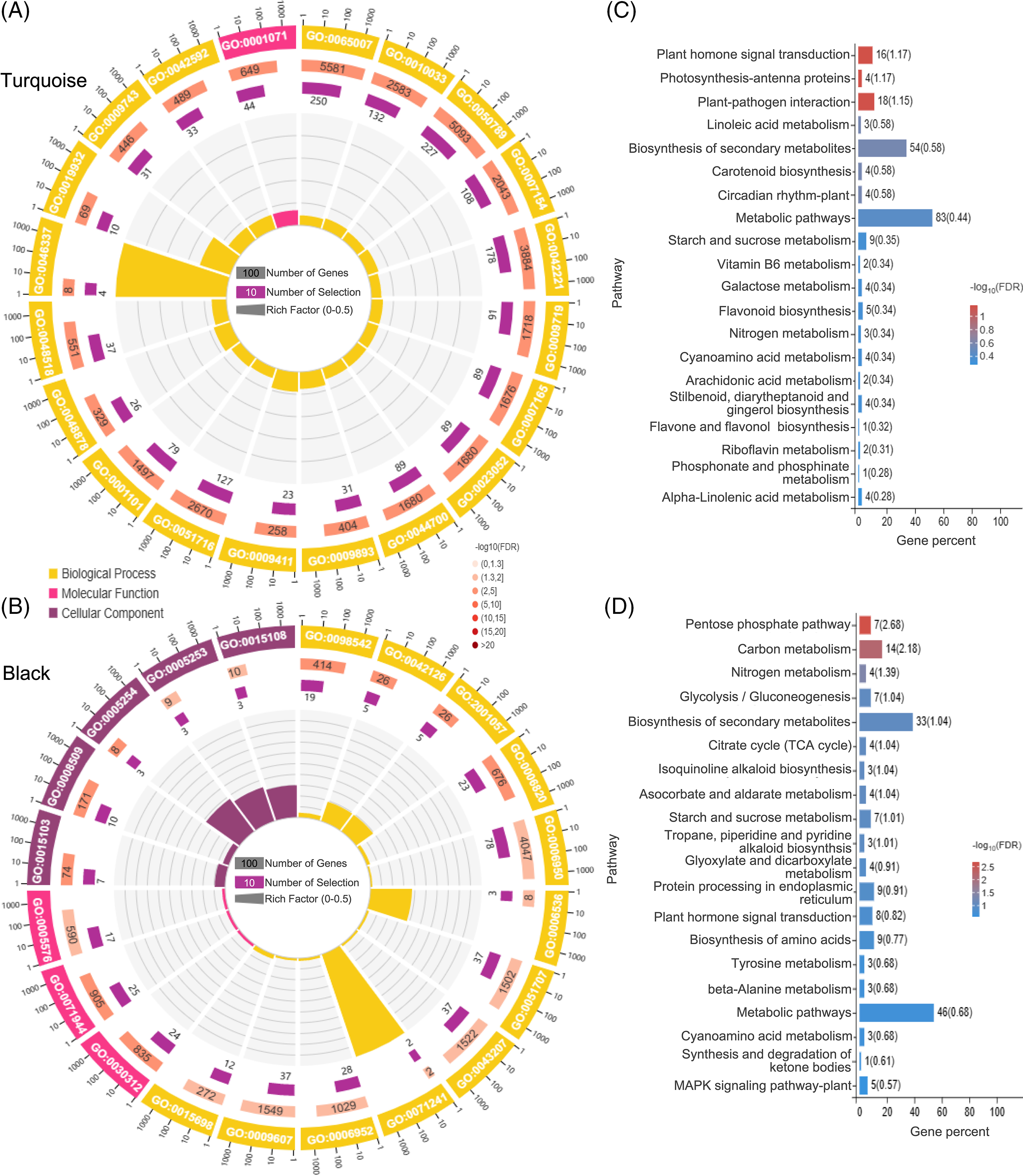
Figure 8: GO and KEGG enrichment analysis. GO enrichment analysis of the top 20 genes in the turquoise module (A) and black module (B); KEGG enrichment analysis of the top 20 genes in the turquoise module (C) and black module (D)
GO analysis of the black module demonstrated a significant enrichment in processes such as anion channel activity, chloride transmembrane transporter activity, cellular response to inorganic substance, glutamate metabolic process, and nitrate metabolic process (Fig. 8B). Furthermore, the black module exhibited a strong enrichment in several KEGG pathways, including the Pentose phosphate pathway, Carbon metabolism, Ascorbate and aldarate metabolism, and Nitrogen metabolism (Fig. 8D).
3.7 Identification of Hub Genes and Construction of Co-Expression Networks
To identify hub genes within each module, we selected the top 100 gene pairs with the highest correlation coefficients (weight values) and constructed a network. Based on our screening, the turquoise module contained 45 genes, the black module contained 83 genes, and the darkgreen module contained 40 genes. According to the NR database annotation, TSJT1 was identified as the hub gene in the turquoise module, known as a stem-specific protein (Fig. 9A); LAX2 (ncbi_112193093) was identified as the hub gene in the black module, characterized as an auxin transporter-like protein involved in signal transduction (Fig. 9B); SCPL34 (ncbi_112197638) was identified as the hub gene in the darkgreen module, classified as a serine carboxypeptidase-like gene (Fig. 9C). Furthermore, several transcription factors such as BEH2, NAC021, CDF3, ethylene-responsive transcription factor (ERF053), ethylene receptor 2 (ETR2), and auxin response factor (ARF6) displayed high connectivity in the turquoise, black, and darkgreen modules, indicating their involvement in the regulatory processes associated with the response of cut roses to potassium nitrate treatment under light conditions (Fig. 9).
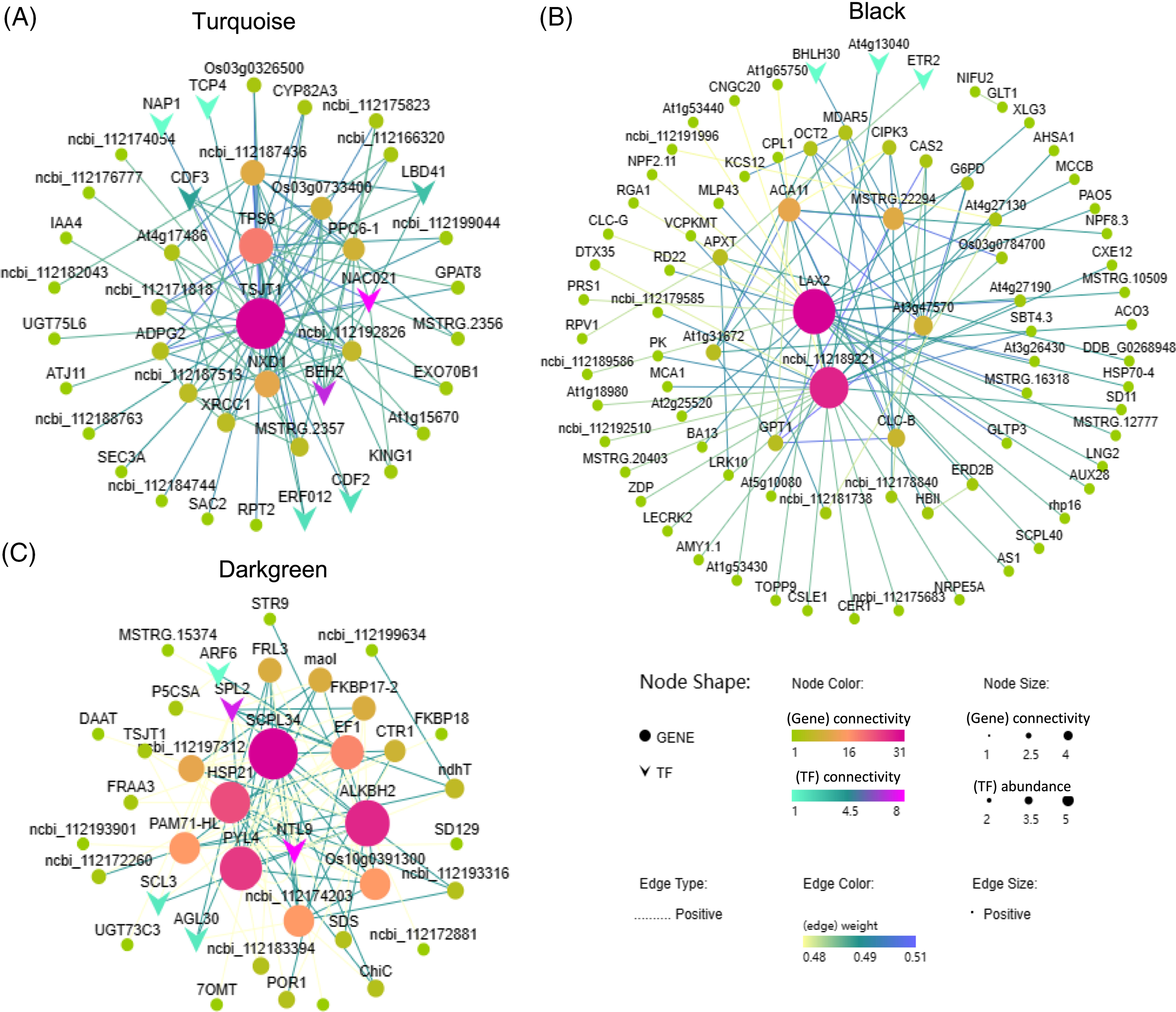
Figure 9: Co-expression network analysis of the turquoise, black, and darkgreen modules. Nodes with darker colors and larger sizes represent higher abundance and stronger connectivity. The color and thickness of the lines between nodes indicate the strength of gene regulatory relationships, with darker colors and thicker lines indicating stronger regulation between genes. (A) Turquoise module; (B) Black module; (C) Darkgreen module
3.8 Validation via qRT-PCR Analysis
To verify the precision and dependability of the RNA-Seq dataset, we employed qRT-PCR for the evaluation of the expression profiles of two selected DEGs, NIA1 and NIR1 (Table S2). The expression patterns of DEGs assessed through q-PCR exhibited a noteworthy degree of concordance, underscoring the robust reliability of the RNA-Seq data produced within the scope of this investigation (Fig. S2).
4.1 Stomatal Opening Observation and Water Loss Rate Analysis of Cut Rose Leaves in Response to KNO3
Previous studies conducted by Huang et al. have provided evidence that cut roses possess the highest stomatal density on their lower epidermis, while petals, sepals, and stems have only scattered stomata [49]. This finding suggests that, like most cut flowers, water loss through stomata is the primary pathway for leaf desiccation in cut roses. In our study, we observed the changes in water loss and stomatal opening on the lower leaf surface of ‘Karola’ cut roses under light conditions after treatment with different concentrations of KNO3. Our results clearly indicate a close relationship between leaf stomata and water loss in the cut roses. We observed varying degrees of differences in the effects of different concentrations of KNO3 treatment on stomatal opening on the hypodermis (Fig. 3), as well as the rate (Fig. 2A) and cumulative amount (Fig. 2B) of water loss in cut roses. Notably, the treatment with 75 mmol/L KNO3 exhibited the most prominent promotion of stomatal opening. In conclusion, our findings demonstrate that different concentrations of KNO3 treatment significantly regulate stomatal opening and water loss in cut roses leaves. Specifically, varying concentrations of KNO3 treatment can promote stomatal opening on the hypodermis (Fig. 3) to different extents and accelerate the water loss rate (Fig. 2) in the flower stems.
4.2 Correlation of NO3− and K+ Transport in Cut Roses under KNO3 Treatment
Numerous key ion channel families are widely present in terrestrial plants; however, moss plants lack ion channels specifically expressed in guard cells, highlighting the essential role of ion osmotic pressure in driving stomatal movement during the evolution of land plants [50]. The activation of nitrate reductase through illumination explained the inverse relationship between light intensity and nitrate content, as it reduces nitrate accumulation [51]. Recent studies have identified the involvement of ion channels or transporters associated with potassium, calcium, and NO3− transport on the vacuolar membrane of guard cells, regulating the opening and closing of plant stomata [52–55]. Aforementioned research demonstrated that the utilization of potassium fertilizer not only impacts rice yield formation but also has consequences for the absorption and utilization of nitrogen within the rice [56]. Upon absorption, NO3− is reduced to NO2−, NH4+, and glutamine by NR, NiR, and glutamine synthetase (GS), respectively, and subsequently metabolized into various amino acids and nitrogenous compounds through glutamate synthase (GOGAT) [34]. Hence, the activity of nitrogen metabolism enzymes is crucial for NO3− absorption and assimilation. In this study, transcriptome GO and KEGG analysis demonstrated the highest enrichment in nitrate metabolism and nitrogen metabolism in cut roses treated with 25 mmol/L KNO3 (Fig. 5). Differentially expressed genes related to nitrate, such as ferredoxin--nitrite reductase (NIR1), nitrate reductase (NIA1), and glutamine synthetase leaf isozyme (GLN2), exhibited expression levels 4 to 46 times higher after treatment with 25 mmol/L KNO3 compared to the control (Fig. 6A). The expression levels of nitrate transporter genes, including NRT2.5 and NRT3.1, increased by 14 and 18.24 times, respectively, after KNO3 treatment compared to the control (Fig. 6A). These findings suggest that following NO3- absorption and assimilation, these genes may significantly contribute to the increased stomatal aperture and water loss rate in cut roses treated with KNO3. Under potassium deficiency, the expression levels of nitrate transporters in Arabidopsis roots were significantly downregulated but rapidly recovered upon potassium supplementation. Potassium activation is essential for nitrogen assimilation-related enzymes and facilitates the transport of NO3− from roots to shoots. Nitrogen levels impact potassium uptake, and excessive nitrogen supply reduces potassium utilization efficiency. Potassium plays a significant role in enhancing NO3− uptake, serving as a crucial factor for nitrogen uptake and assimilation. Notably, the assimilation rate of inorganic nitrogen is higher under light conditions compared to dark conditions. The distribution of nitrogen in plants influences nitrogen uptake and assimilation, and increased potassium supply under high nitrogen conditions enhances the rate of nitrogen distribution to the leaves. In this study, the 25 mmol/L KNO3 treatment resulted in a higher K+ concentration, which partially promoted NO3− absorption.
Stomatal movement is tightly regulated by the turgor pressure of guard cells, with the intracellular concentration of K+ playing a pivotal role in modulating turgor pressure. Unlike other cell types, mature guard cells lack plasmodesmata, necessitating the passage of all inorganic ions across the cell membrane. In this context, potassium ion channels assume a critical role [52,57]. Remarkably, even minor fluctuations in K+ concentration can exert profound effects on membrane charge, triggering a cascade of signaling events associated with K+ acquisition, salinity response, plant immunity, and programmed cell death [58–60]. Within the scope of this study, gene ontology analysis revealed significant enrichment in the category of ion transport (Fig. 5B). Differentially expressed genes implicated in potassium transport exhibited varying degrees of expression variation across different treatments and time points (Fig. 6B). The impact of varying potassium supply levels on potassium ion channels and transporters was evident, highlighting the regulatory role of potassium absorption governed by feedback mechanisms based on the plant’s nutritional status [61]. Accumulation of potassium in the roots leads to a reduced rate of potassium uptake, while under conditions of low potassium levels, the activation and enhancement of potassium ion channels and transporters occur [62]. High salt concentrations in water negatively impact fruit growth, physiology, production, and post-harvest quality by reducing water availability, causing toxicity through Cl− and Na+, and creating nutrient imbalances due to ion competition, including deficiencies of essential nutrients (Ca2+, Mg2+, K+, and NO3−) [63]. K+ and NO3− represent the most abundant cations and anions in plant tissues, with their absorption rates exhibiting a positive correlation and mutual promotion to enhance charge balance. This phenomenon can be attributed to the improvements in charge balance during nutrient uptake and long-distance transport processes, as well as the activation of enzymes involved in nitrate assimilation induced by potassium ions [64]. Excessive nitrogen adversely affects potassium uptake and utilization efficiency, as evidenced by the downregulation of gene expression related to MdAKT1, MdHAK1, and MdPT5 under conditions of high nitrogen stress [38]. Nitrogen transporters, including NRT1.1 and NRT1.5, participate in K+ transport, and intricate physiological and molecular mechanisms interconnect nitrogen and potassium uptake [47,65]. A recent investigation has unveiled that the NO3− channel SLAH3 collaborates synergistically with the K+ channels GORK and SKOR, exerting an influence over nitrogen-potassium homeostasis and the modulation of membrane potential within the realm of plant biology [48]. Within the context of this study, treatment with 25 mmol/L KNO3 resulted in elevated potassium ion concentration, thereby facilitating K+ uptake. However, this process is subject to regulation by the internal potassium status of the plant, and the high concentration of NO3− negatively modulates potassium uptake, ultimately leading to a complex expression pattern of different potassium ion channels and transporters.
4.3 WGCNA Analysis of Cut Rose Leaves Response to Light
Plants can respond to various signals, such as blue light, osmotic stress, sugars, and plant hormones, by activating H+-ATPase [66–68]. The activated H+-ATPase drives K+ uptake by activating inward-rectifying K+ (K+ in) channels, while anions like NO3− and malate enter guard cells, resulting in K+ accumulation and subsequent stomatal opening [1,69,70]. The turquoise module genes exhibited the highest expression levels in darkness (Fig. 7C), which decreased under light conditions. GO analysis revealed enrichment in nucleic acid binding transcription factor activity, phosphatidylethanolamine metabolic process, second-messenger-mediated signaling, and chemical homeostasis. This suggests that processes such as H+-ATPase undergo phosphorylation, signal transmission via second messengers, enhanced transcription factor activity, and regulation of ion and chemical substance homeostasis, ultimately controlling physiological processes like stomatal opening (Fig. 8A). Furthermore, the highly enriched KEGG pathways included plant hormones and photosynthetic antenna proteins, indicating the involvement of photosynthetic antenna proteins in light signal perception and the participation of plant hormones in subsequent signal transduction and physiological responses (Fig. 8C).
4.4 Roles of Plant Hormones in Cut Roses under KNO3 Treatments
Previous studies have provided evidence for the activation of the isopentenyltransferase (IPT) gene by NO3−, facilitating the essential steps of cytokinin synthesis crucial for cell division [71]. Increased NO3− supply induces the biosynthesis of ethylene by activating the 1-aminocyclopropane-1-carboxylate (ACC) synthase (ACS) and ACC oxidase (ACO) genes [31]. Nitrogen influences primary root growth by modulating auxin concentrations, while it affects lateral root growth through the regulation of the AFB3 (an auxin receptor gene) and NPF6.3 [27]. Under NO3− treatment, the transcription levels of the auxin signaling F-box3 (AFB3) receptor gene are enhanced, consequently inducing NAC gene expression. The aminotransferase gene DNR1 (Dull Nitrogen Response1) in rice was identified as a regulator of nitrate uptake and assimilation through modulation of auxin homeostasis. This regulation is achieved by a 520-bp deletion in the DNR1 promoter, leading to decreased transcript levels, elevated auxin content, and enhanced expression of OsARFs, which play a crucial role in activating genes associated with nitrate metabolism [72]. Simultaneously, microRNA 393 (miRNA393) negatively modulates this signaling pathway [34,73]. At 3 h of KNO3 treatment, the black module exhibits relatively higher expression levels (Fig. 6C), with the hub gene identified as an auxin transporter-like protein. GO analysis reveals enrichment in anion channel activity, glutamate metabolic process, nitrate metabolic process, and the KEGG pathway nitrogen metabolism. These findings suggested that after 3 h of KNO3 treatment, the influx of NO3− activates nitrogen metabolism processes, with the involvement of plant hormones such as auxin, promoting stomatal opening and water loss in cut roses (Figs. 8B, 8D).
4.5 Vacuoles Dynamics of Expression in Response to KNO3 Treatment in Cut Roses
During stomatal opening, vacuoles occupy approximately 90% of the guard cell volume, and a significant portion of ions entering the guard cells through the plasma membrane is stored in vacuoles. Conversely, stomatal closure in Arabidopsis guard cells is accompanied by a reduction in cell volume and approximately 20% decrease in vacuole volume [74]. The relationship between vacuole morphology and ion transport represents a dynamic equilibrium. The vacuolar K+ pool is dynamic and serves as a storage reservoir that replenishes ions during abundance or waste, maintaining cytoplasmic concentrations under starvation conditions [75,76]. In the GO analysis of this study, the vacuole-related category and vacuolar membrane showed significant enrichment (Fig. 5B). Differential expression of genes associated with vacuolar protein sorting (VPS) and cationic amino acid transporter (CAX) exhibited notable expression differences in response to the treatment with 25 mmol/L KNO3, suggesting the involvement of vacuole-related genes in regulating the response of cut roses to KNO3 treatment.
Treatment with KNO3 enhances stomatal opening and accelerates water loss in cut roses. Transcriptome analysis revealed distinct gene expression regulation following KNO3 treatment. The findings suggest that under light conditions and with 25 mmol/L KNO3 treatment, a multitude of processes and factors participate in the regulation of stomatal opening and water loss in cut roses. A model describing the response of cut roses to KNO3 signaling under light conditions has been proposed (Fig. 10). Photosynthetic antenna proteins might help detect light signals, while phosphorylation of H+-ATPase plays a role in proton pumping. Signaling relied on second messengers, activating transcription factors and influencing gene expression. Additionally, nitrate transporter-related genes were upregulated for nutrient uptake. Ion channels ensure chemical equilibrium, and active nitrogen metabolism processes were crucial. Plant hormones, such as auxin, potentially played a regulatory role in modulating this process. Vesicle-related gene modulation led to guard cell vacuole fusion, increasing turgor pressure. Collectively, these processes governed stomatal opening and water loss in cut roses. Potassium and nitrogen play pivotal roles in plant growth, yet they also exhibit a nuanced N-K homeostasis. By modulating the channels of potassium and nitrate, it becomes possible to effectively inhibit stomatal aperture. Consequently, the outcomes of this study hold significant implications as a crucial reference in the development of preservatives for cut flowers.
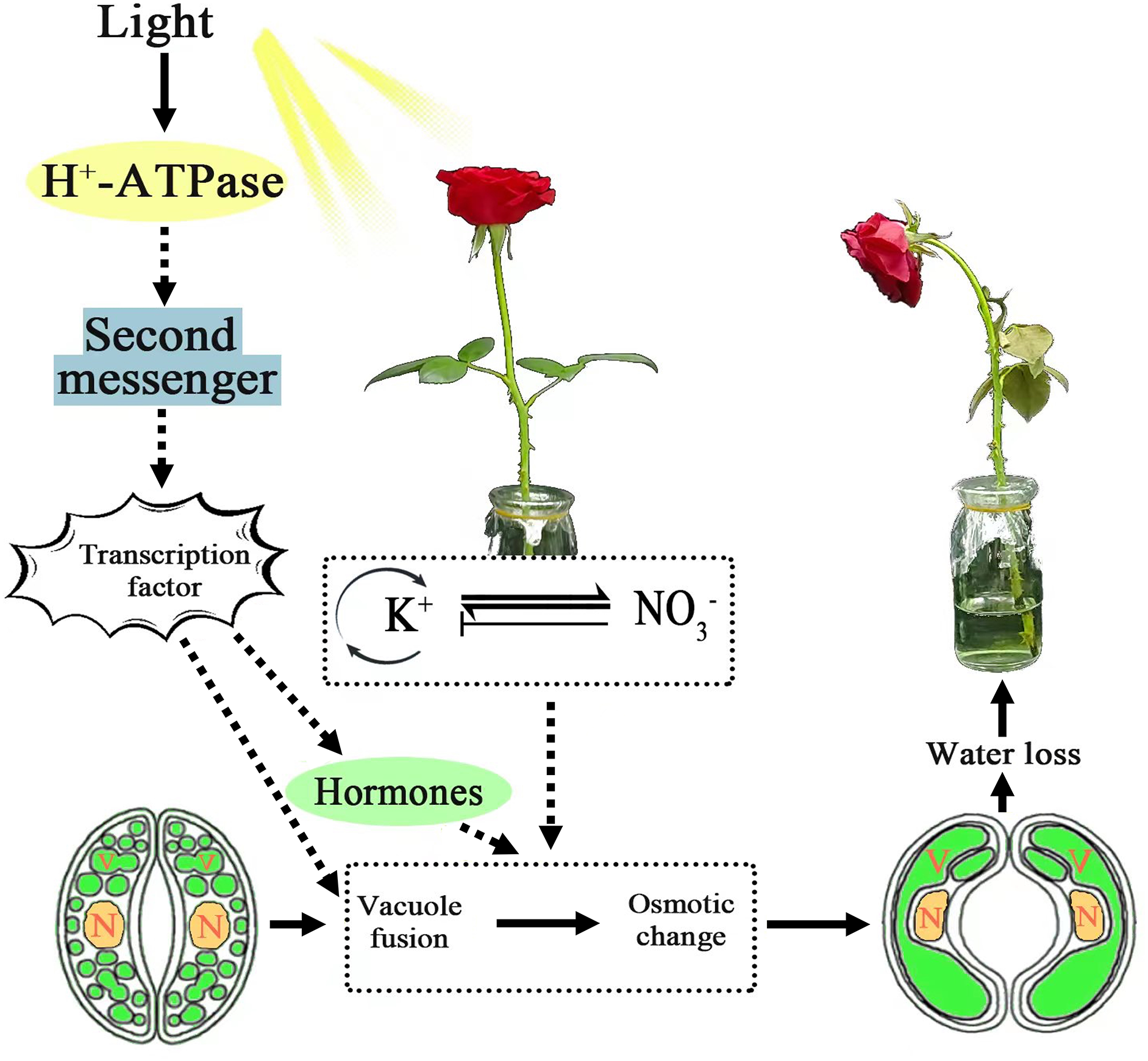
Figure 10: Model of signal transduction pathway of the cut roses in response to KNO3 under light. Solid arrows represent the impact process of integrating the findings of this study into the previous research, while dashed arrows indicate the potential inference process based on the results of this study
Acknowledgement: Not applicable.
Funding Statement: This research was supported by the National Natural Science Foundation of China (Nos. 32002069 and 31972439), the Basic and Applied Research Project of Guangdong Province (2020A1515110961).
Author Contributions: The authors confirm contribution to the paper as follows: Study conception and design: Shenggen He and Hongmei Li; data collection: Yuheng Wu, Songmei Liu, Dongli Cai, Huiling Liang; analysis and interpretation of results: Songmei Liu, Yuheng Wu, Changchun Ye; draft manuscript preparation: Songmei Liu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The raw data supporting the findings of this study are available from the authors upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflict of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2023.045453.
References
1. Shimazaki, K., Doi, M., Assmann, S. M., Kinoshita, T. (2007). Light regulation of stomatal movement. Annual Review of Plant Biology, 58, 219–247. https://doi.org/10.1146/annurev.arplant.57.032905.105434 [Google Scholar] [PubMed] [CrossRef]
2. Kuromori, T., Seo, M., Shinozaki, K. (2018). ABA transport and plant water stress responses. Trends in Plant Science, 23(6), 513–522. https://doi.org/10.1016/j.tplants.2018.04.001 [Google Scholar] [PubMed] [CrossRef]
3. Ye, W., Ando, E., Rhaman, M. S., Tahjib-Ul-Arif, M., Okuma, E. et al. (2020). Inhibition of light-induced stomatal opening by allyl isothiocyanate does not require guard cell cytosolic Ca2+ signaling. Journal of Experimental Botany, 71(10), 2922–2932. https://doi.org/10.1093/jxb/eraa073 [Google Scholar] [PubMed] [CrossRef]
4. Johansson, K. S. L., El-Soda, M., Pagel, E., Meyer, R. C., Toldsepp, K. et al. (2020). Genetic controls of short- and long-term stomatal CO2 responses in Arabidopsis thaliana. Annals of Botany, 126(1), 179–190. https://doi.org/10.1093/aob/mcaa065 [Google Scholar] [PubMed] [CrossRef]
5. Kostaki, K. I., Coupel-Ledru, A., Bonnell, V. C., Gustavsson, M., Sun, P. et al. (2020). Guard cells integrate light and temperature signals to control stomatal aperture. Plant Physiology, 182(3), 1404–1419. https://doi.org/10.1104/pp.19.01528 [Google Scholar] [PubMed] [CrossRef]
6. Dreyer, I., Uozumi, N. (2011). Potassium channels in plant cells. FEBS Journal, 278(22), 4293–4303. https://doi.org/10.1111/j.1742-4658.2011.08371.x [Google Scholar] [PubMed] [CrossRef]
7. Hirsch, R. E. L. B., Spalding, E. P., Sussman, M. R. (1998). A role for the AKT1 potassium channel in plant nutrition. Science, 280(5365), 467–473. [Google Scholar]
8. Ooi, A., Lemtiri-Chlieh, F., Wong, A., Gehring, C. (2017). Direct modulation of the guard cell outward-rectifying potassium channel (GORK) by abscisic acid. Molecular Plant, 10(11), 1469–1472. https://doi.org/10.1016/j.molp.2017.08.010 [Google Scholar] [PubMed] [CrossRef]
9. Gobert, A. I. S., Voelker, C., Czempinski, K., Maathuis, F. J. M. (2007). The two-pore channel TPK1 gene encodes the vacuolar K+ conductance and plays a role in K+ homeostasis. Proceedings of the National Academy of Sciences of the United States of America, 104(25), 10726–10731. https://doi.org/10.1073/pnas.0702595104 [Google Scholar] [PubMed] [CrossRef]
10. Misra, B. B., Reichman, S. M., Chen, S. (2019). The guard cell ionome: Understanding the role of ions in guard cell functions. Progress in Biophysics & Molecular Biology, 146, 50–62. https://doi.org/10.1016/j.pbiomolbio.2018.11.007 [Google Scholar] [PubMed] [CrossRef]
11. Hamamoto, S., Horie, T., Hauser, F., Deinlein, U., Schroeder, J. I. et al. (2015). HKT transporters mediate salt stress resistance in plants: From structure and function to the field. Current Opinion in Biotechnology, 32, 113–120. https://doi.org/10.1016/j.copbio.2014.11.025 [Google Scholar] [PubMed] [CrossRef]
12. Li, W., Xu, G., Alli, A., Yu, L. (2018). Plant HAK/KUP/KT K+ transporters: Function and regulation. Seminars in Cell & Developmental Biology, 74, 133–141. https://doi.org/10.1016/j.semcdb.2017.07.009 [Google Scholar] [PubMed] [CrossRef]
13. Sze, H., Chanroj, S. (2018). Plant endomembrane dynamics: Studies of K+/H+ antiporters provide insights on the effects of pH and ion homeostasis. Plant Physiology, 177(3), 875–895. https://doi.org/10.1104/pp.18.00142 [Google Scholar] [PubMed] [CrossRef]
14. Inoue, S. I., Kinoshita, T. (2017). Blue light regulation of stomatal opening and the plasma membrane H+-ATPase. Plant Physiology, 174(2), 531–538. https://doi.org/10.1104/pp.17.00166 [Google Scholar] [PubMed] [CrossRef]
15. Ando, E., Kinoshita, T. (2018). Red light-induced phosphorylation of plasma membrane H+-ATPase in stomatal guard cells. Plant Physiology, 178(2), 838–849. https://doi.org/10.1104/pp.18.00544 [Google Scholar] [PubMed] [CrossRef]
16. Matthews, J. S. A., Vialet-Chabrand, S., Lawson, T. (2020). Role of blue and red light in stomatal dynamic behaviour. Journal of Experimental Botany, 71(7), 2253–2269. https://doi.org/10.1093/jxb/erz563 [Google Scholar] [PubMed] [CrossRef]
17. Li, S., Yang, F., Sun, D., Zhang, Y., Zhang, M. et al. (2020). Cryo-EM structure of the hyperpolarization-activated inwardly rectifying potassium channel KAT1 from Arabidopsis. Cell Research, 30(11), 1049–1052. https://doi.org/10.1038/s41422-020-00407-3 [Google Scholar] [PubMed] [CrossRef]
18. Gong, M., Zhang, Q., Cheng, K., Zhang, H. (2023). Symbiosis of arbuscular mycorrhizal fungi and Lycium barbarum L. prefers NO3− over NH4+. Horticulturae, 9(6), 637. https://doi.org/10.3390/horticulturae9060637 [Google Scholar] [CrossRef]
19. Raddatz, N., Morales de Los Rios, L., Lindahl, M., Quintero, F. J., Pardo, J. M. (2020). Coordinated transport of nitrate, potassium, and sodium. Frontiers in Plant Science, 11, 247. https://doi.org/10.3389/fpls.2020.00247 [Google Scholar] [PubMed] [CrossRef]
20. Krapp, A., David, L. C., Chardin, C., Girin, T., Marmagne, A. et al. (2014). Nitrate transport and signalling in Arabidopsis. Journal of Experimental Botany, 65(3), 789–798. https://doi.org/10.1093/jxb/eru001 [Google Scholar] [PubMed] [CrossRef]
21. Leran, S., Varala, K., Boyer, J. C., Chiurazzi, M., Crawford, N. et al. (2014). A unified nomenclature of nitrate transporter 1/PEPTIDE transporter family members in plants. Trends in Plant Science, 19(1), 5–9. https://doi.org/10.1016/j.tplants.2013.08.008 [Google Scholar] [PubMed] [CrossRef]
22. Komarova, N. Y., Thor, K., Gubler, A., Meier, S., Dietrich, D. et al. (2008). AtPTR1 and AtPTR5 transport dipeptides in planta. Plant Physiology, 148(2), 856–869. https://doi.org/10.1104/pp.108.123844 [Google Scholar] [PubMed] [CrossRef]
23. Nour-Eldin, H. H., Andersen, T. G., Burow, M., Madsen, S. R., Jorgensen, M. E. et al. (2012). NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature, 488, 531–534. https://doi.org/10.1038/nature11285 [Google Scholar] [PubMed] [CrossRef]
24. Krouk, G., Lacombe, B., Bielach, A., Perrine-Walker, F., Malinska, K. et al. (2010). Nitrate-regulated auxin transport by NRT1.1 defines a mechanism for nutrient sensing in plants. Developmental Cell, 18(6), 927–937. https://doi.org/10.1016/j.devcel.2010.05.008 [Google Scholar] [PubMed] [CrossRef]
25. Tal, I., Zhang, Y., Jorgensen, M. E., Pisanty, O., Barbosa, I. C. et al. (2016). The Arabidopsis NPF3 protein is a GA transporter. Nature Communications, 7(1), 11486. https://doi.org/10.1038/ncomms11486 [Google Scholar] [PubMed] [CrossRef]
26. Chopin, F., Orsel, M., Dorbe, M. F., Chardon, F., Truong, H. N. et al. (2007). The Arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell, 19(5), 1590–1602. https://doi.org/10.1105/tpc.107.050542 [Google Scholar] [PubMed] [CrossRef]
27. Bouguyon, E., Brun, F., Meynard, D., Kubes, M., Pervent, M. et al. (2015). Multiple mechanisms of nitrate sensing by Arabidopsis nitrate transceptor NRT1.1. Nature Plants, 1, 15015. https://doi.org/10.1038/nplants.2015.15 [Google Scholar] [PubMed] [CrossRef]
28. Liu K. H., Huang C. Y., Tsay Y. F. (1999). CHL1 is a dual-affinity nitrate transporter of Arabidopsis involved in multiple phases of nitrate uptake. Plant Cell, 11(5), 865–874. https://doi.org/10.1105/tpc.11.5.865 [Google Scholar] [PubMed] [CrossRef]
29. Ho, C. H., Lin, S. H., Hu, H. C., Tsay, Y. F. (2009). CHL1 functions as a nitrate sensor in plants. Cell, 138(6), 1184–1194. https://doi.org/10.1016/j.cell.2009.07.004 [Google Scholar] [PubMed] [CrossRef]
30. Ma, W., Li, J., Qu, B., He, X., Zhao, X. et al. (2014). Auxin biosynthetic gene TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant Journal, 78(1), 70–79. https://doi.org/10.1111/tpj.12448 [Google Scholar] [PubMed] [CrossRef]
31. Tian, Q. Y., Sun, P., Zhang, W. H. (2009). Ethylene is involved in nitrate-dependent root growth and branching in Arabidopsis thaliana. New Phytologist, 184(4), 918–931. https://doi.org/10.1111/j.1469-8137.2009.03004.x [Google Scholar] [PubMed] [CrossRef]
32. Rahayu, Y. S., Walch-Liu, P., Neumann, G., Romheld, V., von Wiren, N.et al. (2005). Root-derived cytokinins as long-distance signals for NO3− induced stimulation of leaf growth. Journal of Experimental Botany, 56(414), 1143–1152. https://doi.org/10.1093/jxb/eri107 [Google Scholar] [PubMed] [CrossRef]
33. Ondzighi-Assoume, C. A., Chakraborty, S., Harris, J. M. (2016). Environmental nitrate stimulates abscisic acid accumulation in Arabidopsis root tips by releasing it from inactive stores. Plant Cell, 28(3), 729–745. https://doi.org/10.1105/tpc.15.00946 [Google Scholar] [PubMed] [CrossRef]
34. José, A., O’Brien, A. V., Bouguyon, E., Krouk, G., Gojon, A. et al. (2016). Nitrate transport, sensing and responses in plants. Molecular Plant, 9(6), 837–856. https://doi.org/10.1016/j.molp.2016.05.004 [Google Scholar] [PubMed] [CrossRef]
35. Alvarez, J. M., Riveras, E., Vidal, E. A., Gras, D. E., Contreras-Lopez, O. et al. (2014). Systems approach identifies TGA1 and TGA4 transcription factors as important regulatory components of the nitrate response of Arabidopsis thaliana roots. Plant Journal, 80(1), 1–13. https://doi.org/10.1111/tpj.12618 [Google Scholar] [PubMed] [CrossRef]
36. Wang, R., Guan, P., Chen, M., Xing, X., Zhang, Y. et al. (2010). Multiple regulatory elements in the Arabidopsis NIA1 promoter act synergistically to form a nitrate enhancer. Plant Physiology, 154(1), 423–432. https://doi.org/10.1104/pp.110.162586 [Google Scholar] [PubMed] [CrossRef]
37. Ruffel, S., Poitout, A., Krouk, G., Coruzzi, G. M., Lacombe, B. (2016). Long-distance nitrate signaling displays cytokinin dependent and independent branches. Journal of Integrative Plant Biology, 58(3), 226–229. https://doi.org/10.1111/jipb.12453 [Google Scholar] [PubMed] [CrossRef]
38. Xu, X., Liu, G., Liu, J., Lyu, M., Wang, F. et al. (2023). Potassium alleviated high nitrogen-induced apple growth inhibition by regulating photosynthetic nitrogen allocation and enhancing nitrogen utilization capacity. Horticultural Plant Journal, 15, 185. https://doi.org/10.1016/j.hpj.2023.04.003 [Google Scholar] [CrossRef]
39. Zhou, J., Sun, H., Wei, J., Li, P. (2021). Physiologic and transcriptomic insights into the high alkali response of Dunaliella salina. Phyton-International Journal of Experimental Botany, 90(5), 1401–1414. https://doi.org/10.32604/phyton.2021.016514 [Google Scholar] [CrossRef]
40. Hou, W., Trankner, M., Lu, J., Yan, J., Huang, S. et al. (2019). Interactive effects of nitrogen and potassium on photosynthesis and photosynthetic nitrogen allocation of rice leaves. BMC Plant Biology, 19(1), 302. https://doi.org/10.1186/s12870-019-1894-8 [Google Scholar] [PubMed] [CrossRef]
41. Blevins D. G., Barnett N. M., Frost W. B. (1978). Role of potassium and malate in nitrate uptake and translocation by wheat seedlings. Plant Physiology, 62(5), 784–788. https://doi.org/10.1104/pp.62.5.784 [Google Scholar] [PubMed] [CrossRef]
42. Zhang, F., Niu, J., Zhang, W., Chen, X., Li, C. et al. (2010). Potassium nutrition of crops under varied regimes of nitrogen supply. Plant and Soil, 335, 21–34. https://doi.org/10.1007/s11104-010-0323-4 [Google Scholar] [CrossRef]
43. Vidal, E. A., Alvarez, J. M., Araus, V., Riveras, E., Brooks, M. D. et al. (2020). Nitrate in 2020: Thirty years from transport to signaling networks. Plant Cell, 32(7), 2094–2119. https://doi.org/10.1105/tpc.19.00748 [Google Scholar] [PubMed] [CrossRef]
44. Li, H., Yu, M., Du, X. Q., Wang, Z. F., Wu, W. H. et al. (2017). NRT1.5/NPF7.3 functions as a proton-coupled H+/K+ antiporter for K+ loading into the xylem in Arabidopsis. Plant Cell, 29(8), 2016–2026. https://doi.org/10.1105/tpc.16.00972 [Google Scholar] [PubMed] [CrossRef]
45. Lin, S. H., Kuo, H. F., Canivenc, G., Lin, C. S., Lepetit, M. et al. (2008). Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell, 20(9), 2514–2528. https://doi.org/10.1105/tpc.108.060244 [Google Scholar] [PubMed] [CrossRef]
46. Du, X. Q., Wang, F. L., Li, H., Jing, S., Yu, M. et al. (2019). The transcription factor MYB59 regulates K+/NO3− translocation in the Arabidopsis response to low K+ stress. Plant Cell, 31(3), 699–714. https://doi.org/10.1105/tpc.18.00674 [Google Scholar] [PubMed] [CrossRef]
47. Fang, X. Z., Liu, X. X., Zhu, Y. X., Ye, J. Y., Jin, C. W. (2020). The K+ and NO3− interaction mediated by NITRATE TRANSPORTER1.1 ensures better plant growth under K+-limiting conditions. Plant Physiology, 184(4), 1900–1916. https://doi.org/10.1104/pp.20.01229 [Google Scholar] [PubMed] [CrossRef]
48. Liu, B., Feng, C., Fang, X., Ma, Z., Xiao, C. et al. (2023). The anion channel SLAH3 interacts with potassium channels to regulate nitrogen-potassium homeostasis and the membrane potential in Arabidopsis. Plant Cell, 35(4), 1259–1280. https://doi.org/10.1093/plcell/koad014 [Google Scholar] [PubMed] [CrossRef]
49. Huang, X., Lin, S., He, S., Lin, X., Liu, J. et al. (2018). Characterization of stomata on floral organs and scapes of cut ‘Real’ gerberas and their involvement in postharvest water loss. Postharvest Biology and Technology, 142, 39–45. https://doi.org/10.1016/j.postharvbio.2018.04.001 [Google Scholar] [CrossRef]
50. Sussmilch, F. C., Roelfsema M. R. G., Hedrich, R. (2019). On the origins of osmotically driven stomatal movements. New Phytologist, 222(1), 84–90. https://doi.org/10.1111/nph.15593 [Google Scholar] [PubMed] [CrossRef]
51. El Haddaji, H., Akodad, M., Skalli, A., Moumen, A., Bellahcen, S. et al. (2023). Effects of light-emitting diodes (LEDs) on growth, nitrates and osmoprotectant content in microgreens of aromatic and medicinal Plants. Horticulturae, 9(4), 494. https://doi.org/10.3390/horticulturae9040494 [Google Scholar] [CrossRef]
52. Jezek, M., Blatt, M. R. (2017). The membrane transport system of the guard cell and its integration for stomatal dynamics. Plant Physiology, 174(2), 487–519. https://doi.org/10.1104/pp.16.01949 [Google Scholar] [PubMed] [CrossRef]
53. Bassil, E., Zhang, S., Gong, H., Tajima, H., Blumwald, E. (2019). Cation specificity of vacuolar NHX-type cation/H+ antiporters. Plant Physiology, 179(2), 616–629. https://doi.org/10.1104/pp.18.01103 [Google Scholar] [PubMed] [CrossRef]
54. Jaslan, D., Dreyer, I., Lu, J., O’Malley, R., Dindas, J. et al. (2019). Voltage-dependent gating of SV channel TPC1 confers vacuole excitability. Nature Communications, 10(1), 2659. https://doi.org/10.1038/s41467-019-10599-x [Google Scholar] [PubMed] [CrossRef]
55. Liu, X., Cheng, J., Jiang, F., Liang, M., Han, J. et al. (2020). The tonoplast intrinsic protein gene KvTIP3 is responsive to different abiotic stresses in Kosteletzkya virginica. Biomed Research International, 2020, 2895795. https://doi.org/10.1155/2020/2895795 [Google Scholar] [PubMed] [CrossRef]
56. Zhang, T., He, X., Chen, B., He, L., Tang, X. (2021). Effects of different potassium (K) fertilizer rates on yield formation and lodging of rice. Phyton-International Journal of Experimental Botany, 90(3), 815–826. https://doi.org/10.32604/phyton.2021.014168 [Google Scholar] [CrossRef]
57. Wille, A. C., Lucas, W. J. (1984). Ultrastructural and histochemical studies on guard cells. Planta, 160(2), 129–142. https://doi.org/10.1007/BF00392861 [Google Scholar] [PubMed] [CrossRef]
58. Rubio, F., Fon, M., Rodenas, R., Nieves-Cordones, M., Aleman, F. et al. (2014). A low K+ signal is required for functional high-affinity K+ uptake through HAK5 transporters. Physiologia Plantarum, 152(3), 558–570. https://doi.org/10.1111/ppl.12205 [Google Scholar] [PubMed] [CrossRef]
59. Shabala, S. (2017). Signalling by potassium another second messenger to add to the list? Journal of Experimental Botany, 68(15), 4003–4007. https://doi.org/10.1093/jxb/erx238 [Google Scholar] [PubMed] [CrossRef]
60. Demidchik, V., Cuin, T. A., Svistunenko, D., Smith, S. J., Miller, A. J. et al. (2010). Arabidopsis root K+-efflux conductance activated by hydroxyl radicals: Single-channel properties, genetic basis and involvement in stress-induced cell death. Journal of Cell Science, 123(9), 1468–1479. https://doi.org/10.1242/jcs.064352 [Google Scholar] [PubMed] [CrossRef]
61. Wang, Y., Wu, W. H. (2013). Potassium transport and signaling in higher plants. Annual Review of Plant Biology, 64, 451–476. https://doi.org/10.1146/annurev-arplant-050312-120153 [Google Scholar] [PubMed] [CrossRef]
62. Li, C., Liang, B., Chang, C., Wei, Z., Zhou, S. et al. (2016). Exogenous melatonin improved potassium content in Malus under different stress conditions. Journal of Pineal Reseach, 61(2), 218–229. https://doi.org/10.1111/jpi.12342 [Google Scholar] [PubMed] [CrossRef]
63. Silva Filho, A. M. D., Gheyi, H. R., Melo, A. S. D., Silva, A. A. R. D., Bonou, S. I. et al. (2023). Production and quality of west Indian cherry (Malpighia emarginata D. C.) under salt stress and NPK combinations. Horticulturae, 9(6), 649. https://doi.org/10.3390/horticulturae9060649 [Google Scholar] [CrossRef]
64. Coskun, D., Britto, D. T., Kronzucker, H. J. (2017). The nitrogen-potassium intersection: Membranes, metabolism, and mechanism. Plant Cell and Environment, 40(10), 2029–2041. https://doi.org/10.1111/pce.12671 [Google Scholar] [PubMed] [CrossRef]
65. Chen, H., Zhang, Q., Wang, X., Zhang, J., Ismail, A. M. et al. (2021). Nitrogen form-mediated ethylene signal regulates root-to-shoot K+ translocation via NRT1.5. Plant Cell and Environment, 44(12), 3576–3588. https://doi.org/10.1111/pce.14182 [Google Scholar] [PubMed] [CrossRef]
66. Loubna Kerkeb, K. V., Donaire, J. P., Rodríguez-Rosales, M. P. (2002). Enhanced H+/ATP coupling ratio of H+-ATPase and increased 14-3-3 protein content in plasma membrane of tomato cells upon osmotic shock. Physiologia Plantarum, 116(1), 5–41. https://doi.org/10.1034/j.1399-3054.2002.1160105.x [Google Scholar] [PubMed] [CrossRef]
67. Niittyla, T., Fuglsang, A. T., Palmgren, M. G., Frommer, W. B., Schulze, W. X. (2007). Temporal analysis of sucrose-induced phosphorylation changes in plasma membrane proteins of Arabidopsis. Molecular & Cellular Proteomics, 6(10), 1711–1726. https://doi.org/10.1074/mcp.M700164-MCP200 [Google Scholar] [PubMed] [CrossRef]
68. Takahashi, K., Hayashi, K., Kinoshita, T. (2012). Auxin activates the plasma membrane H+-ATPase by phosphorylation during hypocotyl elongation in Arabidopsis. Plant Physiology, 159(2), 632–641. https://doi.org/10.1104/pp.112.196428 [Google Scholar] [PubMed] [CrossRef]
69. Kim, T. H., Bohmer, M., Hu, H., Nishimura, N., Schroeder, J. I. (2010). Guard cell signal transduction network: Advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annual Review of Plant Biology, 61(1), 561–591. https://doi.org/10.1146/annurev-arplant-042809-112226 [Google Scholar] [PubMed] [CrossRef]
70. Qu, X., Cao, B., Kang, J., Wang, X., Han, X. et al. (2019). Fine-tuning stomatal movement through small signaling peptides. Frontiers in Plant Science, 10, 69. https://doi.org/10.3389/fpls.2019.00069 [Google Scholar] [PubMed] [CrossRef]
71. Wang, R., Tischner, R., Gutierrez, R. A., Hoffman, M., Xing, X. et al. (2004). Genomic analysis of the nitrate response using a nitrate reductase-null mutant of Arabidopsis. Plant Physiology, 136(1), 2512–2522. https://doi.org/10.1104/pp.104.044610 [Google Scholar] [PubMed] [CrossRef]
72. Zhang, S., Zhu, L., Shen, C., Ji, Z., Zhang, H. et al. (2021). Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell, 33(3), 566–580. https://doi.org/10.1093/plcell/koaa037 [Google Scholar] [PubMed] [CrossRef]
73. Elena, A., Vidala, V. A., Lub, C., Parryc, G., Greenb, P. J. et al. (2010). Nitrate-responsive miR393/AFB3 regulatory module controls root system architecture in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 107(9), 4477–4482. https://doi.org/10.1073/pnas.0909571107 [Google Scholar] [PubMed] [CrossRef]
74. Eisenach, C., de Angeli, A. (2017). Ion transport at the vacuole during stomatal movements. Plant Physiology, 174(2), 520–530. https://doi.org/10.1104/pp.17.00130 [Google Scholar] [PubMed] [CrossRef]
75. Martinoia, E., Meyer, S., de Angeli, A., Nagy, R. (2012). Vacuolar transporters in their physiological context. Annual Review of Plant Biology, 63, 183–213. https://doi.org/10.1146/annurev-arplant-042811-105608 [Google Scholar] [PubMed] [CrossRef]
76. Izhar Ahmad, F. J. M. M. (2014). Cellular and tissue distribution of potassium: Physiological relevance, mechanisms and regulation. Journal of Plant Physiology, 171(9), 708–714. https://doi.org/10.1016/j.jplph.2013.10.016 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools