 Open Access
Open Access
ARTICLE
Bioinformatics and Functional Analysis of High Oleic Acid-Related Gene GmSAM22 in Soybean [Glycine max (L.) Merr.]
1 College of Agronomy, Jilin Agricultural University, Changchun, 1300118, China
2 College of Plant Protection, Agricultural University, Changchun, 1300118, China
* Corresponding Author: Piwu Wang. Email:
Phyton-International Journal of Experimental Botany 2023, 92(2), 501-519. https://doi.org/10.32604/phyton.2022.023479
Received 28 April 2022; Accepted 16 June 2022; Issue published 12 October 2022
Abstract
High yield, high quality, stable yield, adaptability to growth period, and modern mechanization are the basic requirements for crops in the 21st century. Soybean oleic acid is a natural unsaturated fatty acid with strong antioxidant properties and stability. Known as a safe fatty acid, it has the ability to successfully prevent cardiovascular and cerebrovascular disorders. Improving the fatty acid composition of soybean seeds, can not only speed up the breeding process of high-quality high-oil and high-oleic soybeans, but also have important significance in human health, and provide the possibility for the development of soybean oil as a new energy source. Hence, the aim of this study was to analyze the high oleic acid elated gene GmSAM22 in soybean. In this research the soybean oleic acid-related gene GmSAM22 was screened out by Genome-wide association analysis, a 662 bp fragment was acquired by specific PCR amplification, and the pMD18T cloning vector was linked by the use of a seamless cloning technique. Bioinformatics analysis of the signal peptide prediction, subcellular localization, protein hydrophobicity, transmembrane region analysis, a phosphorylation site, protein secondary and tertiary structure and protein interaction analysis of the protein encoded by the SAM22 gene was carried out. The plasmid of the gene editing vector is pBK041. The overexpression vector was transformed from pCAMBIA3301 as the base vector, and overexpression vector were designed. Positive plants were obtained by genetic transformation by the pollen tube channel method. Fluorescence quantitative PCR was performed on the T2 generation plants to detect the relative expression levels in different tissues. Southern Blot was used to detect the presence of hybridization signal. Screening genes BAR, 35S, and NOS in plants were identified by conventional PCR. 10 seeds with high and low oleic acid content were chosen for quantitative PCR identification, and finally, the concentration and morphology of soybean fatty acids were identified by near-far infrared spectroscopy. On 10 seeds with an upper and lower oleic acid content, a quantitative fluorescence analysis was done. In Southern blot hybridization, the SAM22 gene was integrated into the recipient soybean plant in hands of a sole copy. Fluorescence quantitative PCR appeared that the average relative expression of the SAM22 gene in roots, stems, leaves, and seeds was 1.70, 1.67, 3.83, and 4.41, respectively. Positive expression seeds had a 4.77% increase in oleic acid content. The level of oleic acid in the altered seeds was reduced by 4.13% when compared to CK, and it was discovered that the GmSAM22 gene could be a regulatory and secondary gene that promotes the conversion of stearic acid to oleic acid in soybean. There has not been a discussion of gene cloning or functional verification. The cloning and genetic transformation of the soybean SAM22 gene can effectively increase the content of oleic acid, which lays a foundation for the study of soybean with high oleic acid.Keywords
Soybean [Glycine max (L.) Merr.] is one of the major oil crops in the world. Soybean accounts for about 60% of the world’s oil crop production [1]. Soybean seeds are rich in nutrients, of which the protein content is about it is 40%, and the fat content is about 18%–23% [2]. Oils and fats exist in plants in the form of Triacylglycerols (TAGs), and their quality depends on the composition and content of their fatty acids. Soybean oil mainly contains 5 fatty acids, namely stearic acid (18:0), palmitic acid (16:0), oleic acid (18:1), linolenic acid (18:0), linoleic acid (18:2), wherein oleic acid, linoleic acid, and linolenic acid are unsaturated fatty acids, and stearic acid and palmitic acid are saturated fatty acids (SFA). The nutritional value of oils and fats mainly depends on the length and saturation of the fatty acid carbon chain. Soybean oil quality is determined by the proportion of saturated to unsaturated fatty acids. Eating soybean oil with a high ratio of oleic acid to linoleic acid (O/L) can help prevent cardiovascular disease and contribute to human health. Excessive intake of saturated fatty acids can lead to elevated blood cholesterol and low-density lipoprotein (LDL) levels, cause coronary atherosclerosis and increase the risk of cardiovascular disease. Compared with saturated fatty acids, oleic acid, linolenic acid, and linoleic acid have one or more unsaturated bonds in their structure. Polyunsaturated fatty acids (linoleic acid and linolenic acid) have poor stability, are easily oxidized to peroxides, and are not easy to store. They can reduce the content of harmful LDL and beneficial high-density lipoprotein (HDL) at the same time. Excessive intake increases the risk of cardiovascular disease [3]. Oleic acid is a monounsaturated fatty acid, which contains an unsaturated bond in structure and is relatively stable in nature. It can reduce the content of LDL and maintain the content of HDL, thereby reducing atherosclerosis. Like sclerosis, the role of preventing cardiovascular disease is beneficial to human health, known as safe fatty acids [4].
Plant fatty acid production is mostly in the endoplasmic reticulum and cytoplasm [5]. Sucrose is the main carbon source for fatty acid synthesis, important intermediate metabolite pyruvate is produced through the EMP (Embden-Meyerhof-Parnas) pathway, and the process of glycolysis occurs in the cytoplasm. Acetyl-COA is then produced by pyruvate dehydrogenase as a precursor for the fatty acid synthesis and transported to the corresponding location in the cytoplasm. Acetyl-COA carboxylase catalyzes the formation of Malonyl-COA fatty acid synthase, and Malonyl-COA is the continuous cycle reaction’s substrate. Under the catalysis of fatty acid synthase complex, Malonyl-COA carries out the polymerization reaction of acyl chain by adding 2C polymerized acyl chains each time until saturated fatty acids of 16 to 18C are generated. Oleic acid (18:1Δ9,12) is catalyzed by a 12-fatty acid dehydrogenase to produce linoleic acid, and linoleic acid is catalyzed by a 15-fatty acid dehydrogenase to produce linolenic acid (18:3Δ9,12,15) [6]. The precursor of oleic acid is stearic acid, which is found in soybean fatty acids, and the stearic acid-ACP-desaturase (SACPD) gene family regulates oleic acid biosynthesis. SACPD is a key player in metabolic control [7], and the increase of oleic acid content can be achieved by the positive overexpression of the SACPD gene family, stearic acid (18:0) catalyzes the conversion to oleic acid (18:1) npr1-5 defects in (ssi2) mutants and ASCPD genes encoding salicylic acid-insensitive inhibitors have been identified and validated in various plant genomes [8,9]. The fatty acid dehydrogenase (FAD2) gene family regulates oleic acid, which is produced upstream of linoleic acid biosynthesis. Inhibiting the expression of the FAD2 gene family can accomplish the role of increasing the content of oleic acid. Soybean oleic acid content is affected by genetic additive effects and has epistatic and dominant effects [10].
The SAM22 gene related to oleic acid was screened from 260 spring soybeans by whole-genome sequencing association analysis (GWAS). The determination of high oleic acid was carried out after planting 260 soybeans for drying. 14 soybeans with significant differences in oleic acid were selected for expression detection of candidate genes. In 2018, 20 candidate genes related to oleic acid content were identified, and 8 candidate genes related to oleic acid were detected in 2019 [11]. The gene in this experiment is one of the candidate genes, the gene Glyma was 17G030400, which is located on chromosome 17 of soybean. The gene contribution rate is 15%. And the gene overexpression vector pCAMBIA3301-SAM22 gene and gene editing vector pBGK041-SAM22 gene were inherited by pollen tube pathway method. After transformation, T2 generation seeds were obtained, and routine PCR detection, Fluorescence quantitative PCR, Southern blotting hybridization, and fluorescence quantitative PCR detection were carried out, and the near-infrared spectrometer was used to determine the quality and characteristics of T2 seeds. To explore the effects and changes of SAM22 gene function on soybean quality.
2.1 Experimental Material and Recombinant Vectors
Experiment resources from the Biotechnology Center of Jilin Agricultural University were employed in this study, including Soybean germplasm resource JN28, base vector overexpressing pCAMBIA-3301 and CRISPR-Cas9.0 editing vector pBGK041. Agrobacterium EHA105 cell and Escherichia coliDH5 cell to amplify the recombinant plasmid.
The experiment sources and reagents were listed in Table 1, primers and sequencing services used were provided by Kumei Biotechnology Company, China.
Soybean JN38 was grown indoors, and the young leaves at the later stage of the three-leaf stage of soybean were taken. The soybean genome was extracted using a DNA extraction kit (DP305). A nucleic acid and protein detector was used to determine the purity and quality of the extracted genome. As a template for subsequent experiments, the extracted DNA genome was stored at –20°C for future experiments.
Specific primer sequences for the fatty acid regulation-related gene SAM22 were designed using Primer 5.0 software. Design specific upstream and downstream primers SAM22-A/SAM22-As (Table 2). The PCR system is 25 μL, DNA 2 μL, primerI1 μL, primerII1 μL, ddH2O 7.5 μL, mix mixture 12.5 μL. Pre-denaturation at 95°C, denaturation at 95°C, annealing at 57°C, extension at 57°C, and extension at 72°C for 5 min, 35 s, 40 s, 30 s, and 8 min, respectively, were the PCR reaction conditions. 30 cycles of denaturation, annealing and extension, the position of the band and the target DNA were determined using 0.8% agarose gel electrophoresis, and the target DNA fragment was recovered using a gel purification kit (19101ES50). Seamless connection to pMD18-T was done. The constructed GmSAM22 cloning vector was transformed into Escherichia coli competent cells (DH5α) by heat shock method, and the plate coating method was applied in LB medium for 37°C and 15 h at constant temperature incubation.
2.5 Target Gene Sequence Analysis
Research and analysis of target gene sequences were achieved through different bioinformatics software and online analysis. SignaIP analyzes protein signal, PSORTII analyzes protein subcellular localization, TMHMM, NetPhos, SOPMA, and Swiss-Model are used to analyze protein hydrophobicity and hydrophilicity, protein transmembrane, a protein phosphorylation site, secondary and tertiary structures of proteins. SignaIP analyzes protein signals.
2.6 Overexpression Vector Construction and Identification
The pCAMBIA-3301 vector was linearized with BglII (SV0132), BsteII (SV0132) enzymes. pMD18T-SAM22 was used as a template for PCR-specific amplification. The linearized target segment was purified using a DNA purification kit (K0512) after electrophoresis on a 1.2% agarose gel and the linearized vector large fragment and target fragment were connected to pCAMBIA3301 by seamless assembly cloning kit (D7010S). The cells were transformed into E. coli competent cells (DH5α) by heat shock method, and were coated in LB medium (containing kanamycin in the medium) by plate coating method at 37°C and cultured in a constant temperature incubator for 15 h. After covering the plate with LB solid medium, a single colony was selected and a double enzyme was digested to find whether the gene SAM22 had been resoundingly connected to the overexpression vector.
2.7 Target Information for CRISPR/Cas9 Gene-Editing Vectors
CRISPR/Cas9 gene editing’s target gene, GmSAM22, was knocked out by single-gene and dual-target. Two targets (Fig. 1), the sense strand target sequence is GGGATATAGCTACAGCGTAGTGG, and the antisense strand target sequence is GACCAGCAGCCAATTTGGAGTGG. The gene-editing vector’s basic skeleton is pBGK041, and the vector’s structure is simplified (Fig. 2). The soybean U6 promoter, which is highly efficient and stable, is used. On the vector, there is a Bar gene screening site to see if the editing vector is successfully connected. Cas9.0 was used to encode the two SG sequences, which can efficiently perform directing target fragment DNA double-strand cleavage while minimizing off-target efficiency. Two sets of oligo sequences UP1:TGTGTGGGGATATAGCTACAGCGTAG, Low1:AAACCTACGCTGTAGCTATATCCCCA; UP2:GGGTTGGACCAGCAGCCAATTTGGAG, Low2:AAACCTCCAAATTGGCTGCTGGTCCA.

Figure 1: CRISPR/Cas9 editing vector target
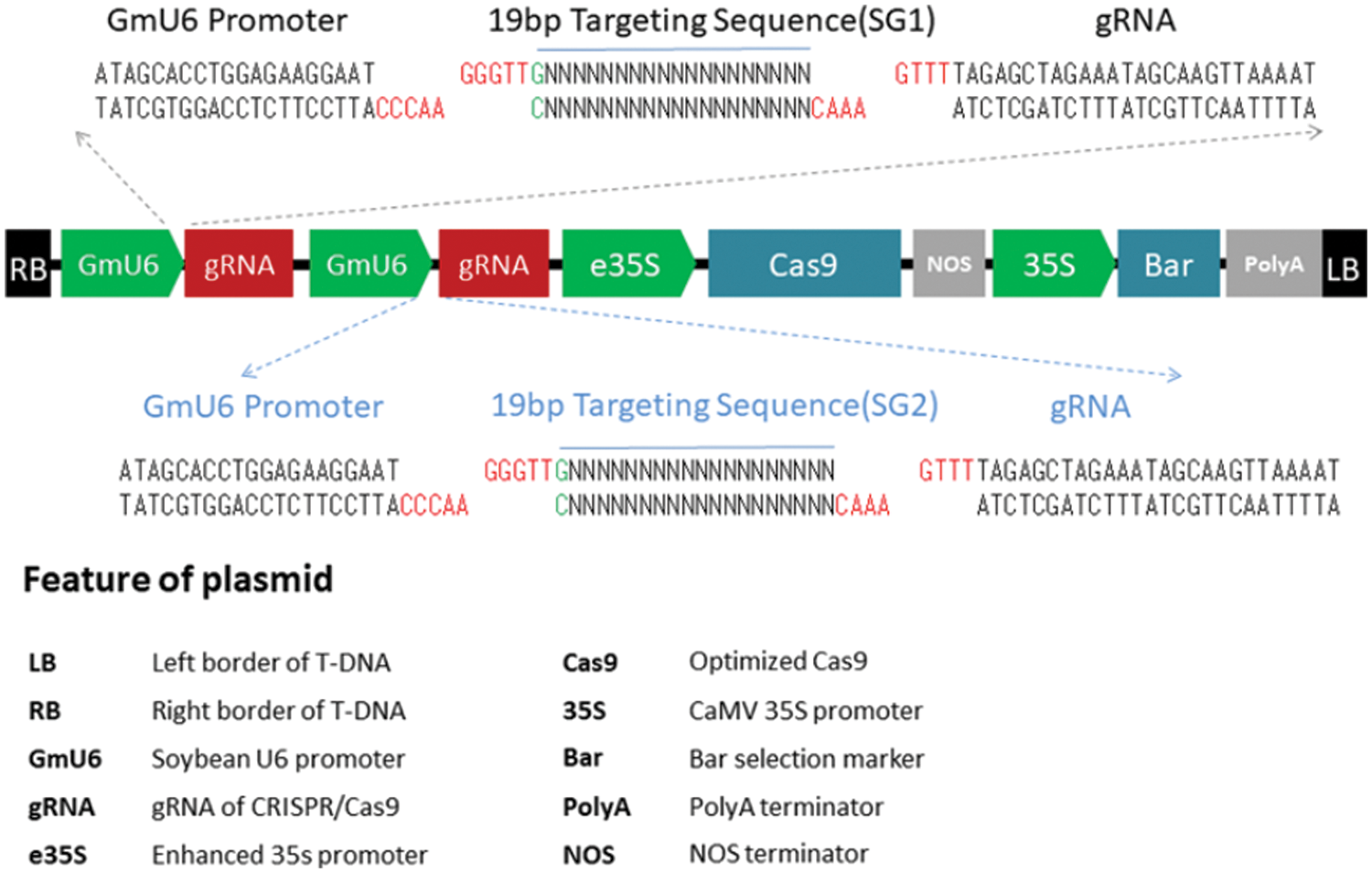
Figure 2: The pBGK041 diagram’s structure
3.1 Sequence Analysis of Soybean SAM22 Gene
In NCBI, Blast analysis was conducted, and the homology between soybean nucleotide sequence and GmSAM22 was as high as 100% (Fig. 3), the target gene sequence was this gene. The ORF sequence was 476 bp long, encoding 161 amino acids, while the complete length of cDNA was 662 bp. The protein has a molecular weight of 16.71 kilodaltons and a predicted isoelectric point of 4.80.
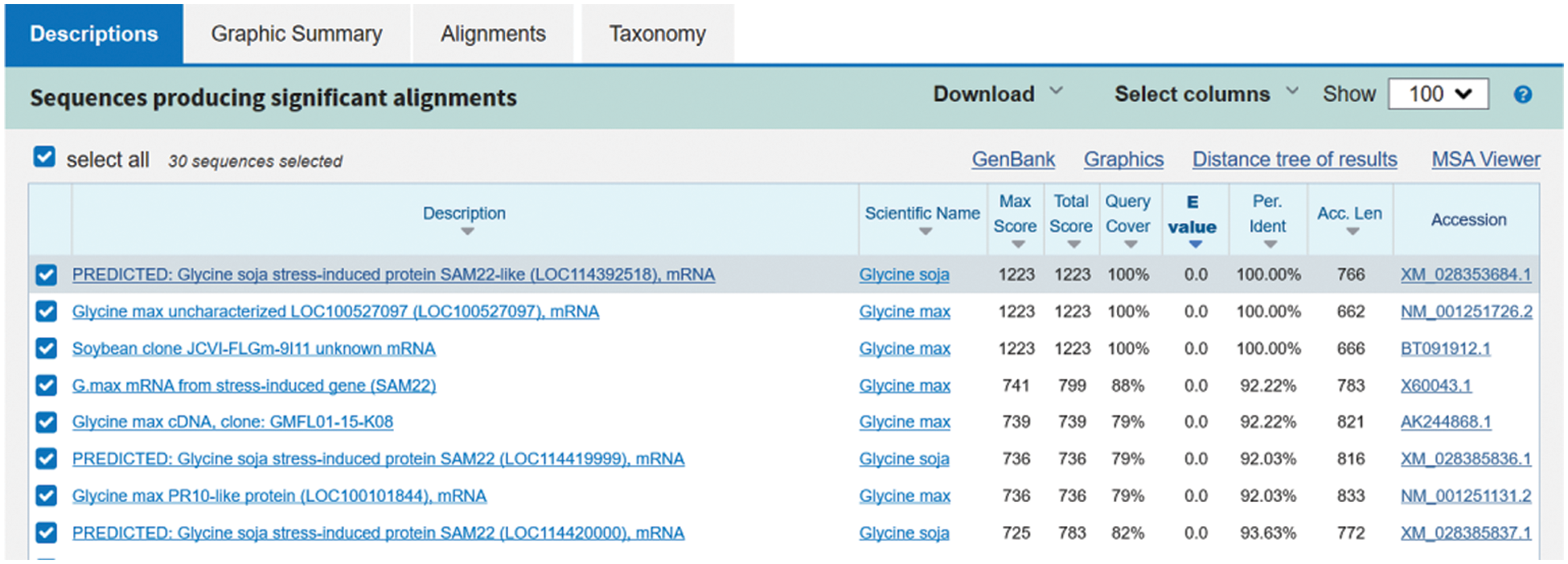
Figure 3: Blast result of SAM22 target gene
3.2 Analysis of the Functional Domain of the Gene SMA22
A functional domain is a unit of a gene-encoded protein that can exert an independent function. The CDD online tool in NCBI was used to perform a functional analysis of the soybean SAM22 amino acid sequence. The results showed that soybean SAM22 has a functional domain, and the peptide segment 76-520 has high homology with members of the SRPBCC family. There are 27 substrate binding sites in this domain (Fig. 4). A deep hydrophobic ligand-binding site exists in the SRPBCC domain of Bet V1 (the primary pollen allergen of birch) and related proteins, and members of this family bind a variety of ligands. Brassinosteroids, cytokinins, flavonoids, and fatty acids can all be bound by Bet V1. They play a role in the development of fruits and flowers, as well as pathogen defense responses.

Figure 4: Analysis of functional domains of soybean SAM22
3.3 The Signal Peptide of the Soybean SAM22-Encoded Protein Was Predicted and Its Subcellular Localization Was Investigated
Protein signal peptides were predicted using Signalp online analysis (Fig. 5). The SAM22 gene’s encoding protein is a non-secreted protein. The results of subcellular localization analysis of the GmSAM22 gene by PSORT II software are shown in (Fig. 6): 47.8% in the cytoplasm and 30.4% in the nucleus were found, demonstrating that the gene is most active in the cytoplasm and nucleus, with mitochondria as a second. It is found on the Golgi apparatus and belongs to a non-secreted protein, as can be seen from the side.
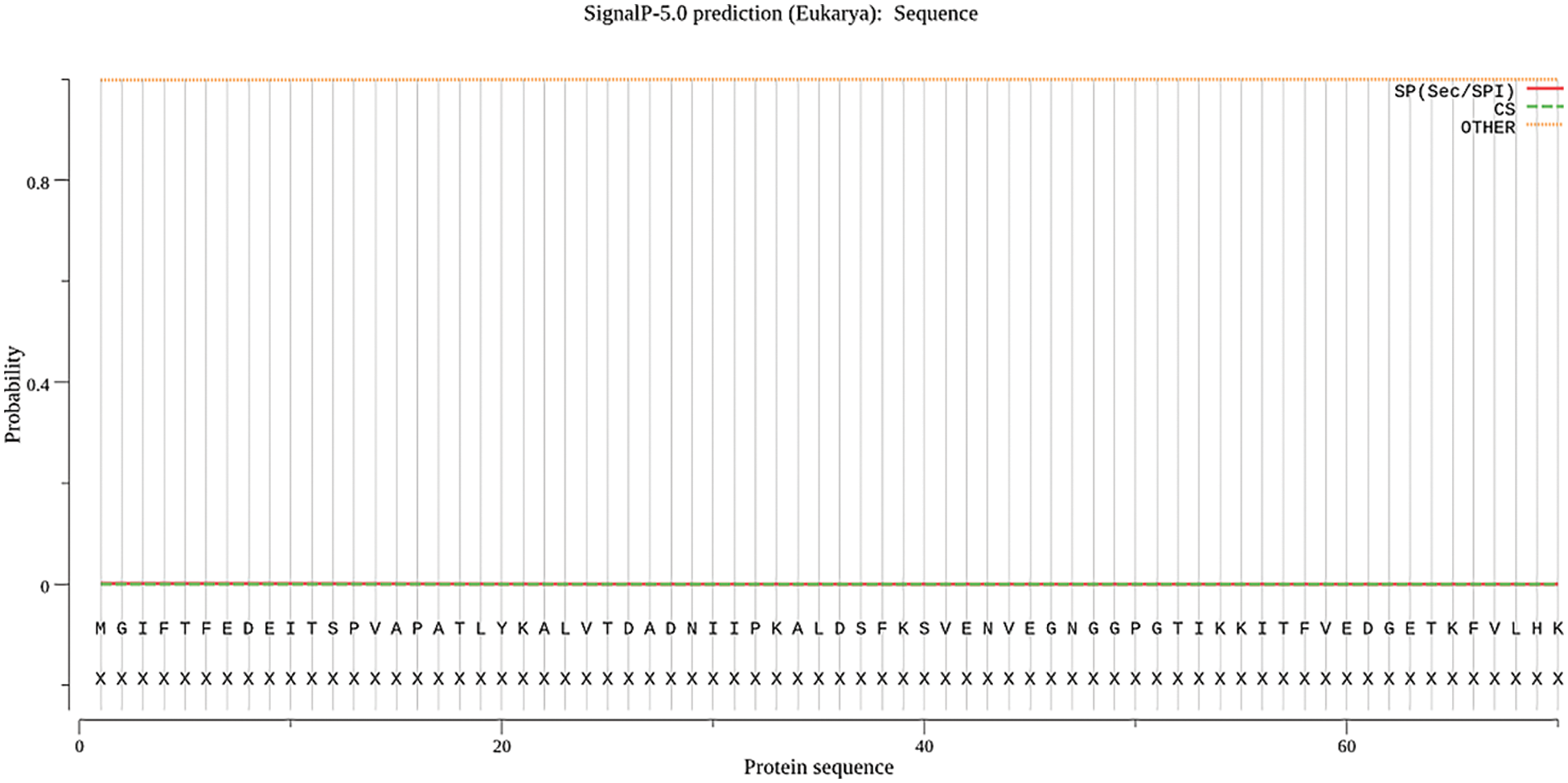
Figure 5: SAM22 gene signal peptide prediction results
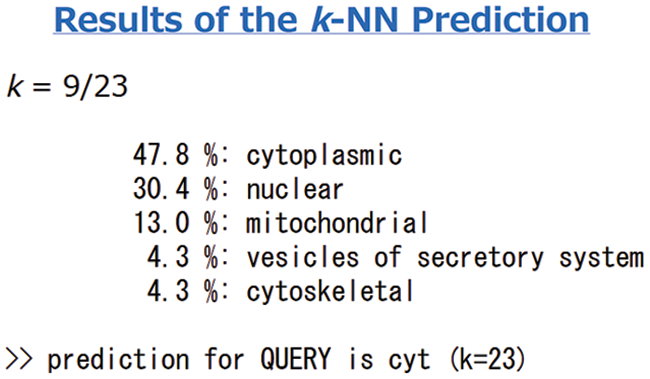
Figure 6: Soybean SAM22 gene protein subcellular localization
3.4 Hydrophilicity and Transmembrane Domain Analysis of Soybean SAM22-Encoded Protein
The following are the results of a ProtScale online analysis of the hydrophilicity and hydrophobicity of soybean SAM22-encoded protein, using Hphob/Kyte and Doolittle as prediction standards (Fig. 7). There was a great deal of low crest value, including the amino acids at positions 119–131, which appear to be clearly hydrophilic. Many of the peaks were rather high, and the amino acids at positions 86–89 and 144–151 exhibited evident hydrophobicity regions, but the amino acid at position 16 has the biggest value at 1.433, indicating that it was the most hydrophobic. Hydrophilic amino acids make up 59.07% of the polypeptide chain, while hydrophobic amino acids make up 40.93%. So the protein was a soluble protein.
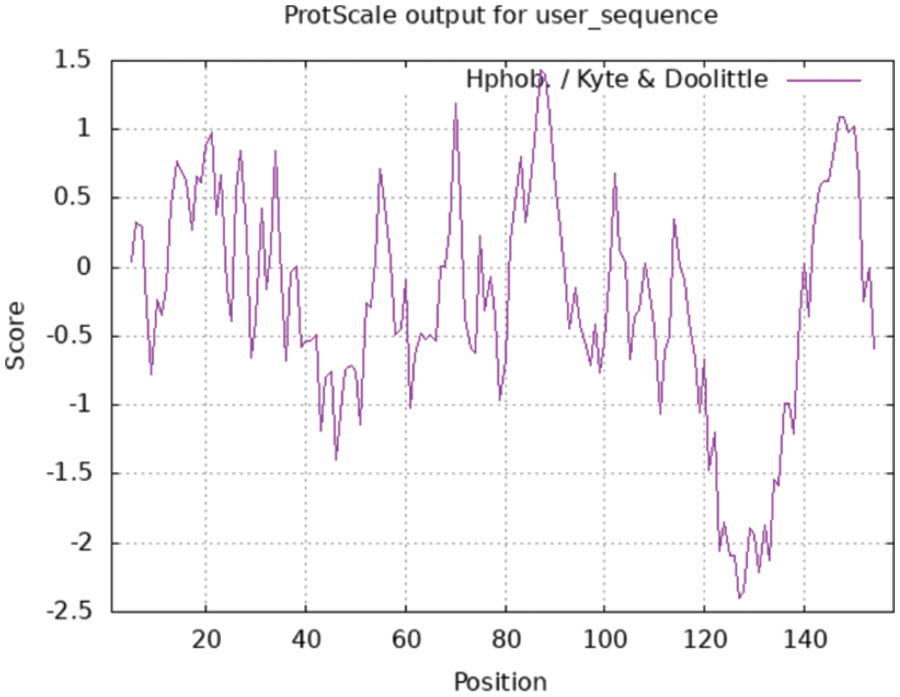
Figure 7: The hydrophilicity and hydrophobicity of the SAM22 protein
The findings of analyzing the protein transmembrane region online with TMHMM Server 2.0 are shown in Fig. 8. The GmSAM22 protein has a transmembrane shape. It is a non-transmembrane protein that does not move around inside cells.
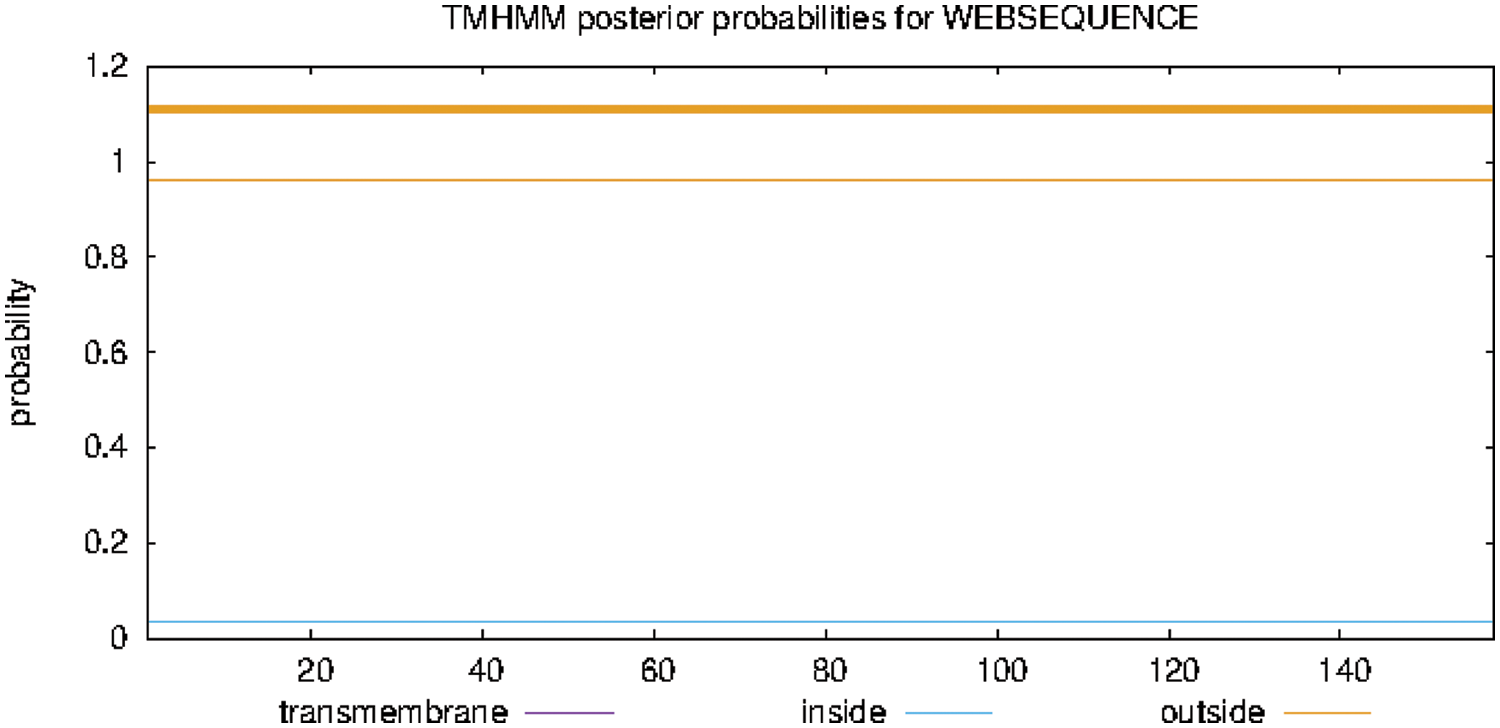
Figure 8: SAM22 gene encoding protein transmembrane region prediction
3.5 Prediction of Phosphorylation Sites GmSAM22 Protein
The following are the results of NetPhos research of SAM22-encoded protein phosphorylation site analysis (Fig. 9): Six serine phosphorylation sites, six threonine phosphorylation sites, and one tyrosine phosphorylation site are among the 13 possible phosphorylation sites on the GmSAM22 protein.
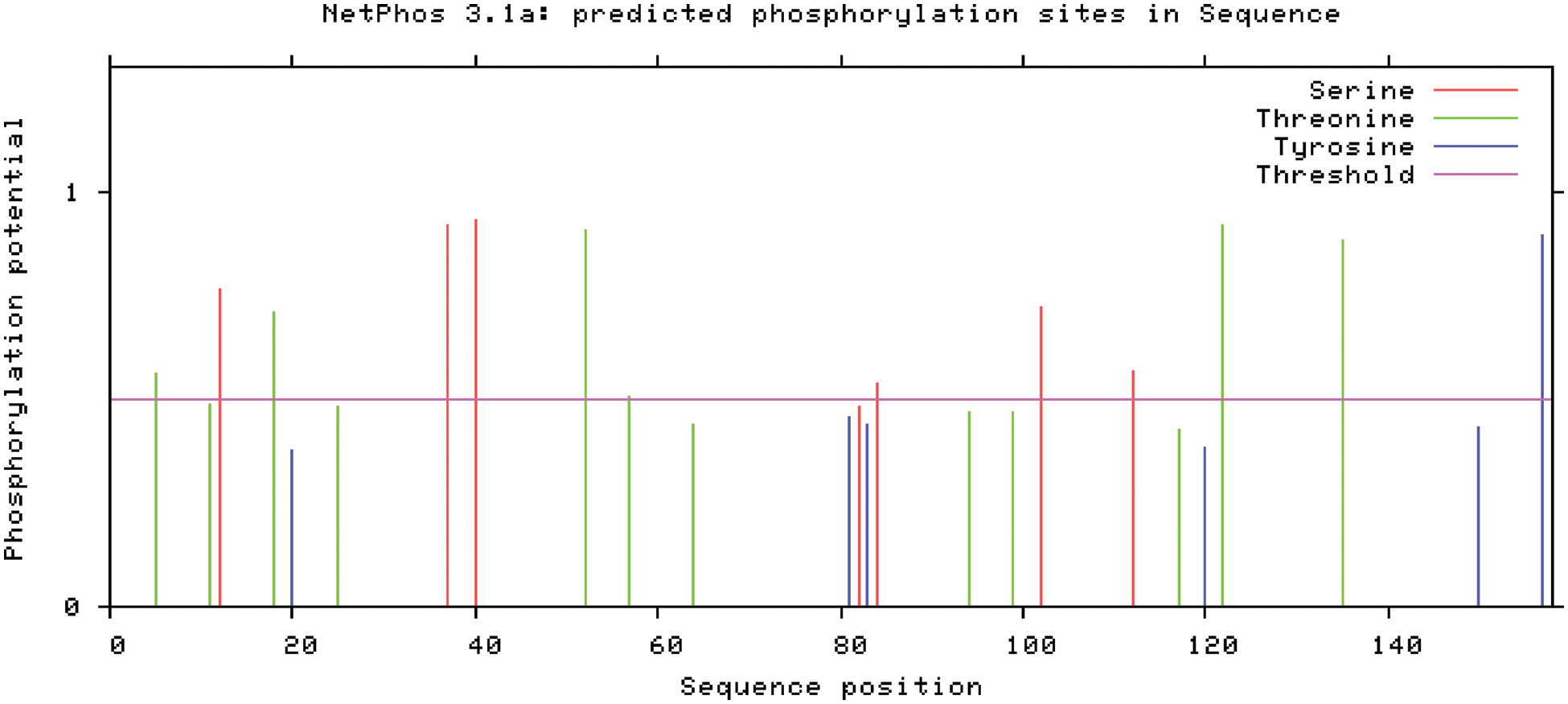
Figure 9: Prediction of phosphorylation of the protein encoded by the soybean SAM22 gene
3.6 Analysis of the Protein Encoded SAM22 Gene in Soybean
The findings of a SOPMA online analysis of the secondary structure of GmSAM22’s amino acid sequence are displayed in Fig. 10: GmSAM22 protein is made up of four parts: α-helix (36.71%), extended chain (25.32%), β-turn (10.76%), and random coil (27.22%). GmSAM22 protein has the highest fraction of α-helical structure, followed by the random coil and extended chain, and β-turn has the lowest.
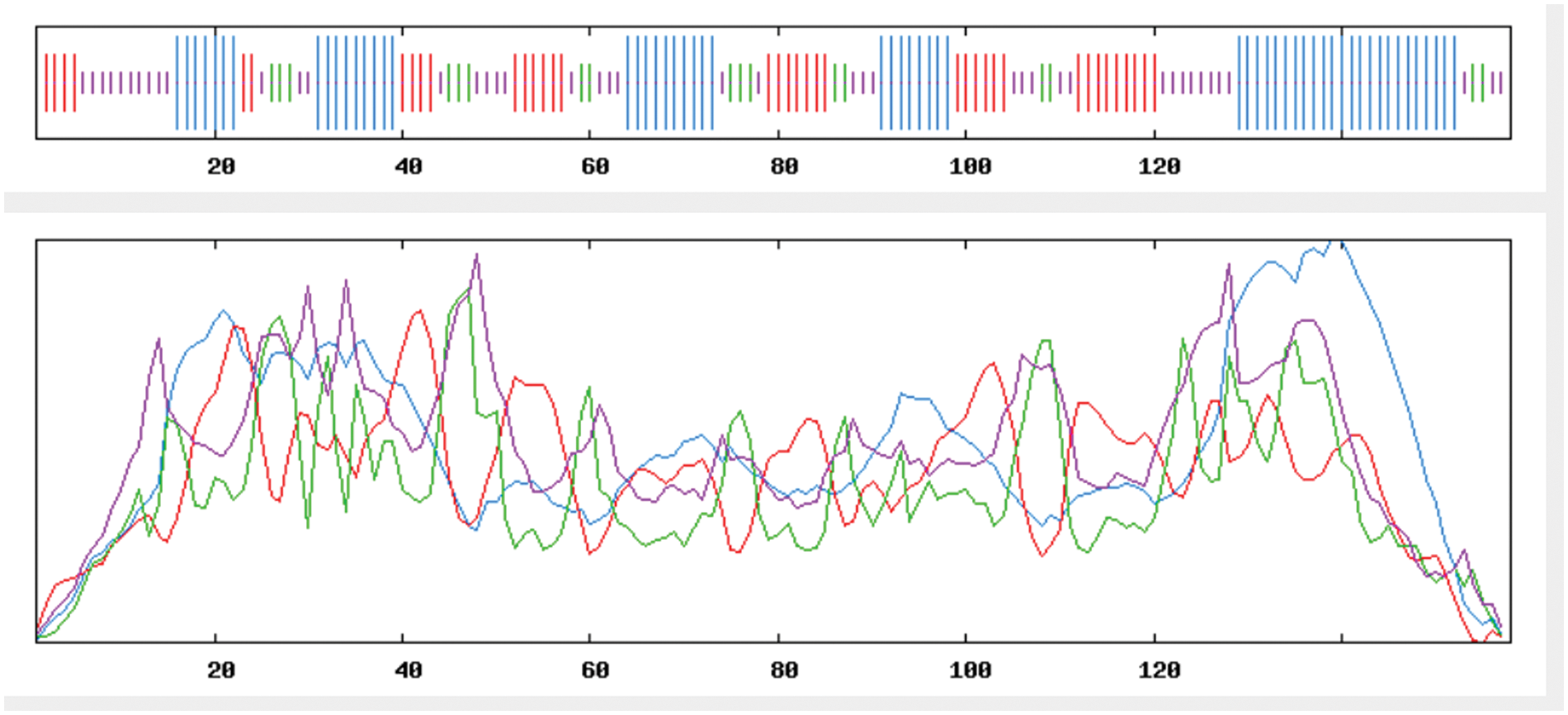
Figure 10: The predicted secondary structure of the SAM22 protein
The tertiary structure of SAM22’s amino acid sequence was predicted and studied using SWISS-MODEL. As shown in Fig. 11, the tertiary structure was consistent with the secondary structure, and both consist of four parts. The three-dimensional structural database contains information on the protein encoded by GmSAM22. The degree of homology with Arabidopsis is 87.52%.
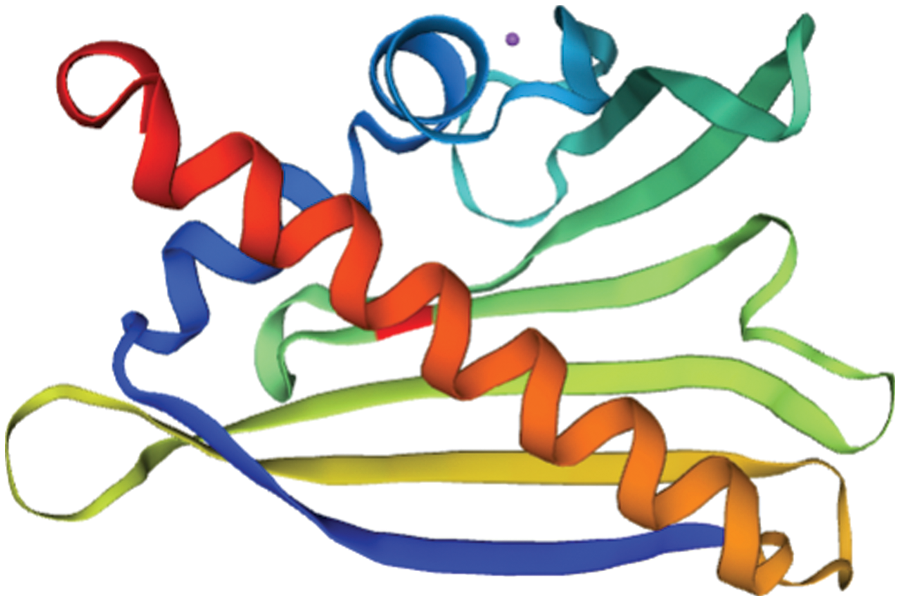
Figure 11: The predicted tertiary structure of the SAM22 protein
3.7 Soybean SAM22 Gene Protein Interaction Analysis
On the GmSAM22 gene, STRING online analysis performed protein interaction analysis, and the results (Fig. 12) showed: SAM22 protein mainly interacts with germ-like protein 20, protein precursor of Bsp domain, peroxidase P7, polyphenol oxidase (chloroplast), Cupin family protein precursor, possible voltage-gated potassium channel subunit β, Kunitz trypsin inhibitor family protein precursor, α-amylase/subtilisin inhibitor, etc. GmSAM22 protein may serve an important role and function in the production of soybean oleic acid, according to a protein interaction study.
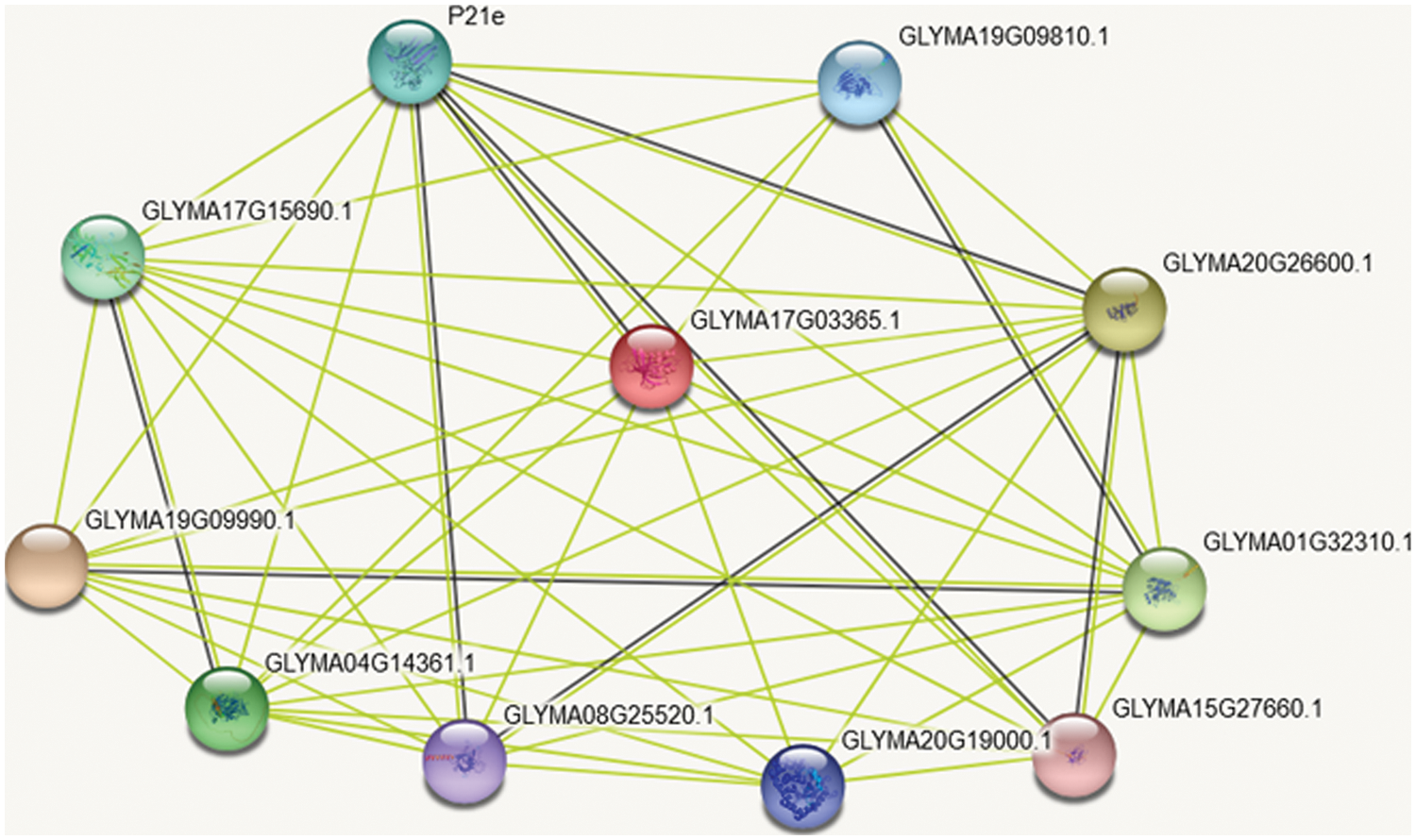
Figure 12: SAM22 protein interaction analysis
3.8 Construction of Overexpression Vector pCAMBIA-3301-SAM22
PCR primers were designed using Primer 5.0 software to specifically amplify the nucleotide sequence of the target gene GmSAM22. The target band was 662 bp (Fig. 13), which was consistent with the predicted fragment size when electrophoresis was performed on a 0.8% agarose gel at 220 V and 110 mA. This region is thought to be the target gene fragment at this time. The pCAMBIA-3301 overexpression vector was provided by the Plant Biotechnology Center for this study. The SAM22 gene vector’s structural diagram is shown below (Fig. 14). The restriction enzyme cleavage site was designed by CE design (Version 3.012) software and DNA man (Version 2.0) software is BglII and BsteII. The cloned target gene was seamlessly cloned and ligated with the pCAMBIA-3301 linearized sizable segment, transformed into Escherichia coli competent cell, and plated on LB and kanamycin medium using a seamless cloning method. For sequencing, it was transported to Shanghai Biosequencing Company. The sequencing results were compared using DNAman, as shown in (Fig. 15). The sequencing results were analyzed by Chromas software, and it was found that the target gene sequencing results had no set of peaks, no spurious peaks, and a good peak graph. Double-enzyme digestion verified whether the ligation was successful. The size of the target fragment was 662 bp, and the large vector fragment was about 9500 bp (Fig. 16), suggesting that the GmSAM22 overexpression vector for soybeans has been successfully constructed.
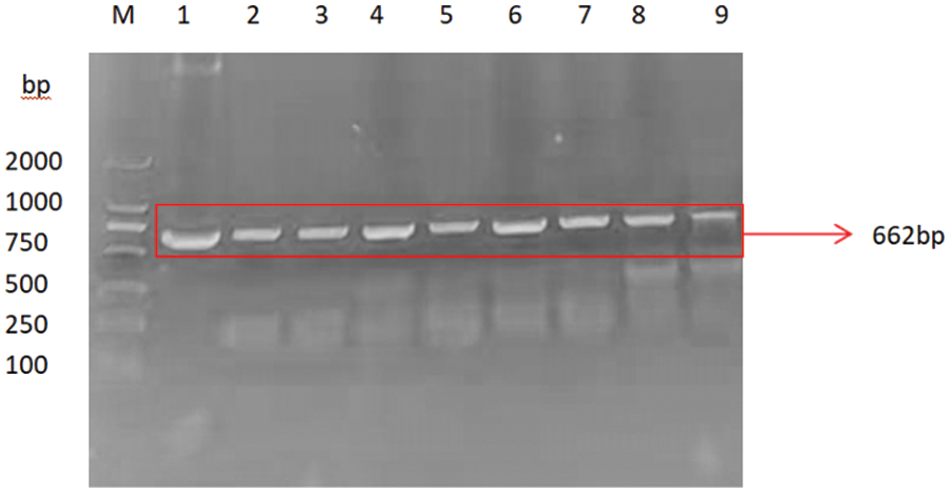
Figure 13: SAM22 gene-specific amplified fragment
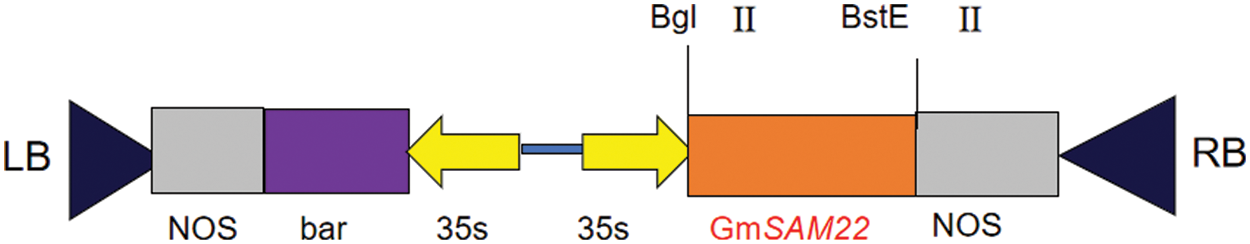
Figure 14: Backbone of SAM22 gene overexpression vector

Figure 15: SAM22 gene sequencing alignment results by DNAman
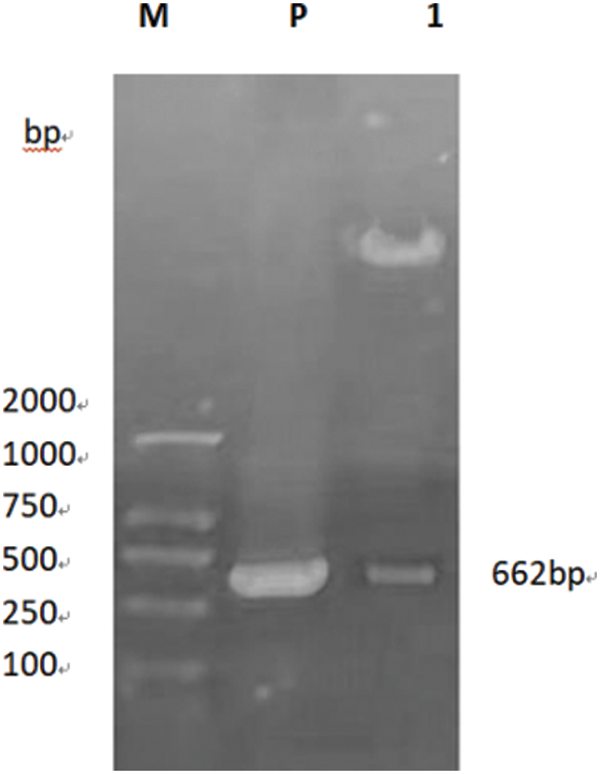
Figure 16: The overexpression vector was double-restricted digested for confirmation
Notes: M: DNA marker; P: Plasmid; 1: Enzyme digestion product.
3.9 Transformation Recipient Soybean
10 positive plants of T1 generation and 18 positive lines of T2 generation were obtained by genetic transformation of pollen tube channel method.
3.10 Transgenic Positive Plants Are Identified Using Conventional PCR Methods
3.10.1 Plants of the T2 Generation are Positively Identified
The promoter gene (35S) and terminator gene (NOS) of the glufosinate-ammonium gene (BAR) were amplified by specific PCR using the soybean genome as the DNA template, and the resulting fragments were 662 in size, 552, 500, and 198 bp (Fig. 17), it can be initially determined that the target gene is integrated into the genome of T2 plants.
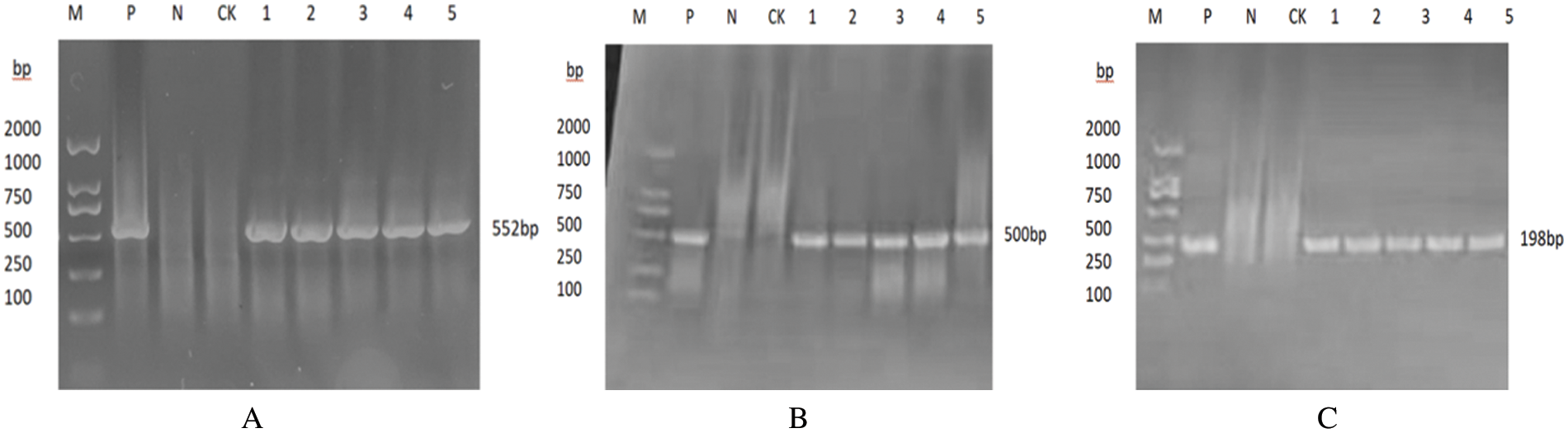
Figure 17: Plants of the T2 generation have been identified by molecular analysis
Notes: Bar, 35 s, and NOS genes are represented by A, B, and C, respectively; M: DNA Marker; P: Plasmid; N: H2O; CK: Untransformed plant. 1-5 plants that were transformed
3.11 Transgenic T2 Plants Were Identified by Southern Blot Hybridization
On the T2 soybeans, plant DNA was extracted by DNA extraction kit (DP305), DNA was digested with enzyme BglII, and the Bar gene was used as a molecular probe for southern blot hybridization. Southern Blot hybridization was used to identify the transformed and untransformed plants at the DNA molecular level. The results show that a hybridization signal occurred in the converted plants’ lanes, but no hybridization signal band appeared in the untransformed plants’ lanes. As shown (Fig. 18) that the SAM22 gene was integrated into the recipient soybean plants in the form of a single genetically transformed scallop, which was consistent with the results of conventional detection PCR. Hybridization bands appeared in the lanes in a bunch of locations, showing that the gene was successfully integrated into the soybean genome. The sites were not the same, hybridized at different positions in the DNA genome
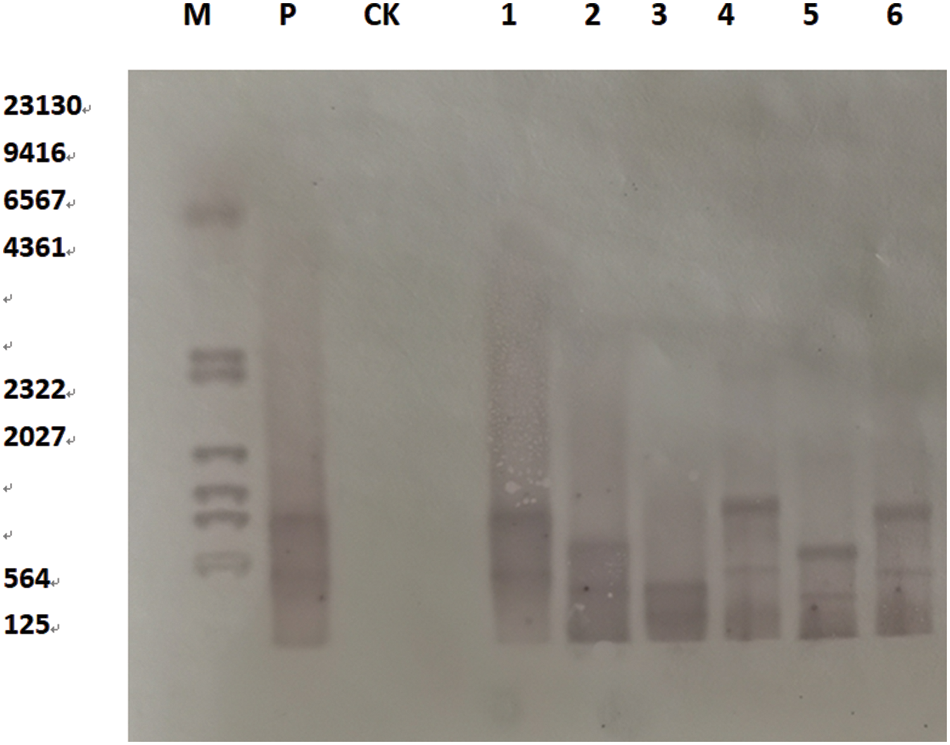
Figure 18: Southern Blot hybridization of T2 transgenic plants
Notes: SAM22 gene; M: Southern dedicated marker; P: Positive control; CK: Untransformed plants; 1–6: Transformed plants.
3.12 T2 Generation Soybean Fluorescence Quantitative PCR Analysis
The mRNAs of plant roots, stems, leaves, and seeds were harvested after the seeds were planted, and the cDNA was reverse transcribed using a fluorescent quantitative reverse transcription kit (B639277). The transgenic lines were identified by conventional PCR detection and Southern blot hybridization, and the soybean cDNA was used as a specific amplification template. Considering β-actin is a highly reliable protein in soybeans, it is chosen as an internal reference gene. Each group of experiments was repeated three times to ensure the experiment’s accuracy. This gene (Fig. 19) showed 1.47, 1.88, and 1.75 relative expression levels on roots, 1.62, 1.73, and 1.67 relative expression levels on stems, 2.60, 2.41, and 3.88 relative expression levels on leaves, and 5.18, 4.17, 3.89 relative expression levels on seeds. The results revealed that seeds had the highest relative expression of the SAM22 gene, followed by leaves and stems. The SAM22 gene was expressed in roots, stems, leaves, and seeds, and the relative expression levels were different, as shown in the above data.
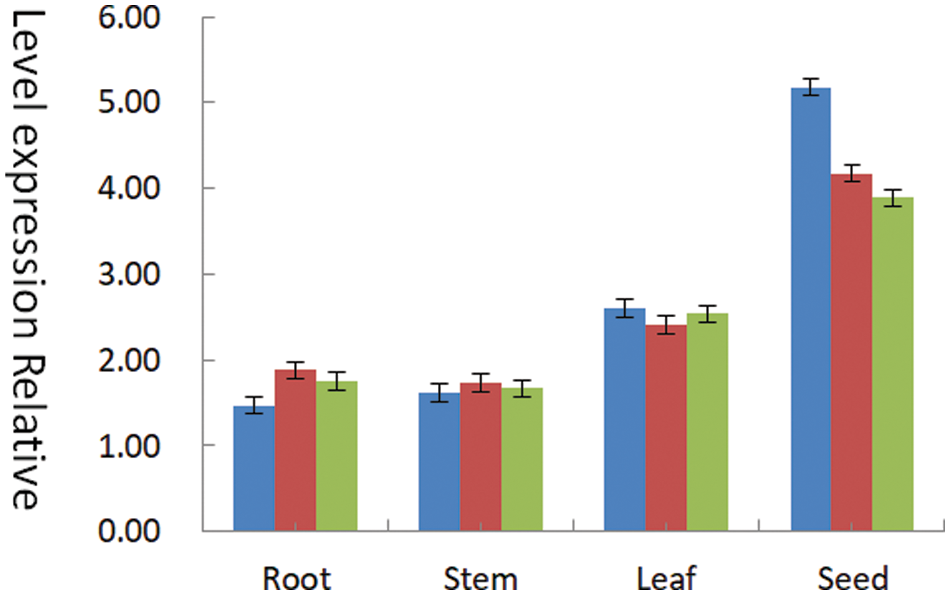
Figure 19: T2 generation identification by fluorescence quantitative PCR SAM22 gene expression in roots, stems, leaves, and seeds
Notes: Green, red, and blue represent three repetitions.
3.13 SAM22 Gene Fluorescence Quantitative PCR in High and Low Oleic Soybeans
Ten soybeans with higher oleic acid and 10 soybeans with lower oleic acid were analyzed in a genome-wide association study (GWAS) of 260 spring soybean germplasm resources. The soybean SAM22 gene is an endogenous gene. To compare the expression of the gene in high and low oleic soybeans, fluorescence quantitative PCR was used on ten high-oleic soybeans and ten low-oleic soybeans. The gene expression level of high oleic soybean (Fig. 20A) was generally higher than 1.50, while the gene expression level of low oleic soybean (Fig. 20B) was generally around 1.0. The relative expression level of this gene was higher in high oleic soybean, and lower in low oleic soybean. This gene may function as a negative regulator in the fatty acid metabolic process of converting oleic acid to linoleic acid.
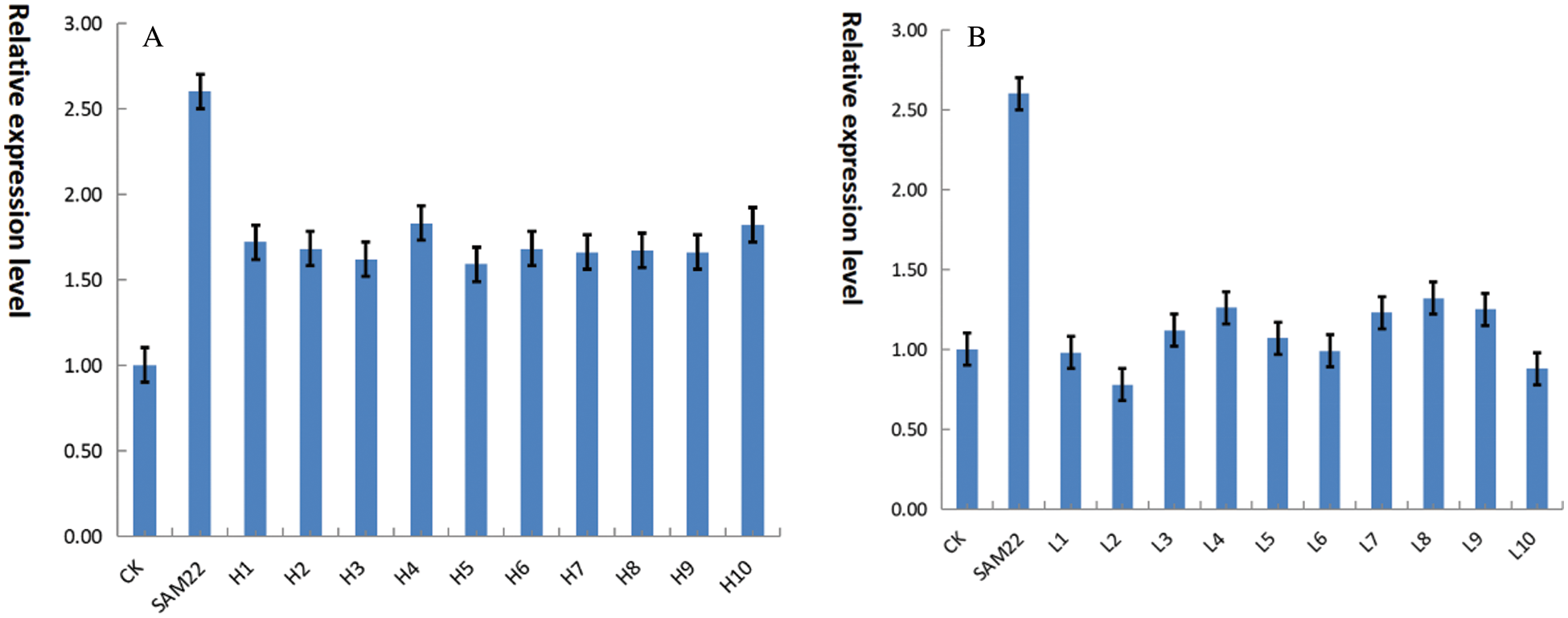
Figure 20: A, real-time PCR of the GmSAM22 gene and high oleic acid germplasm; B, GmSAM22 gene low oleic acid germplasm and real-time quantitative PCR
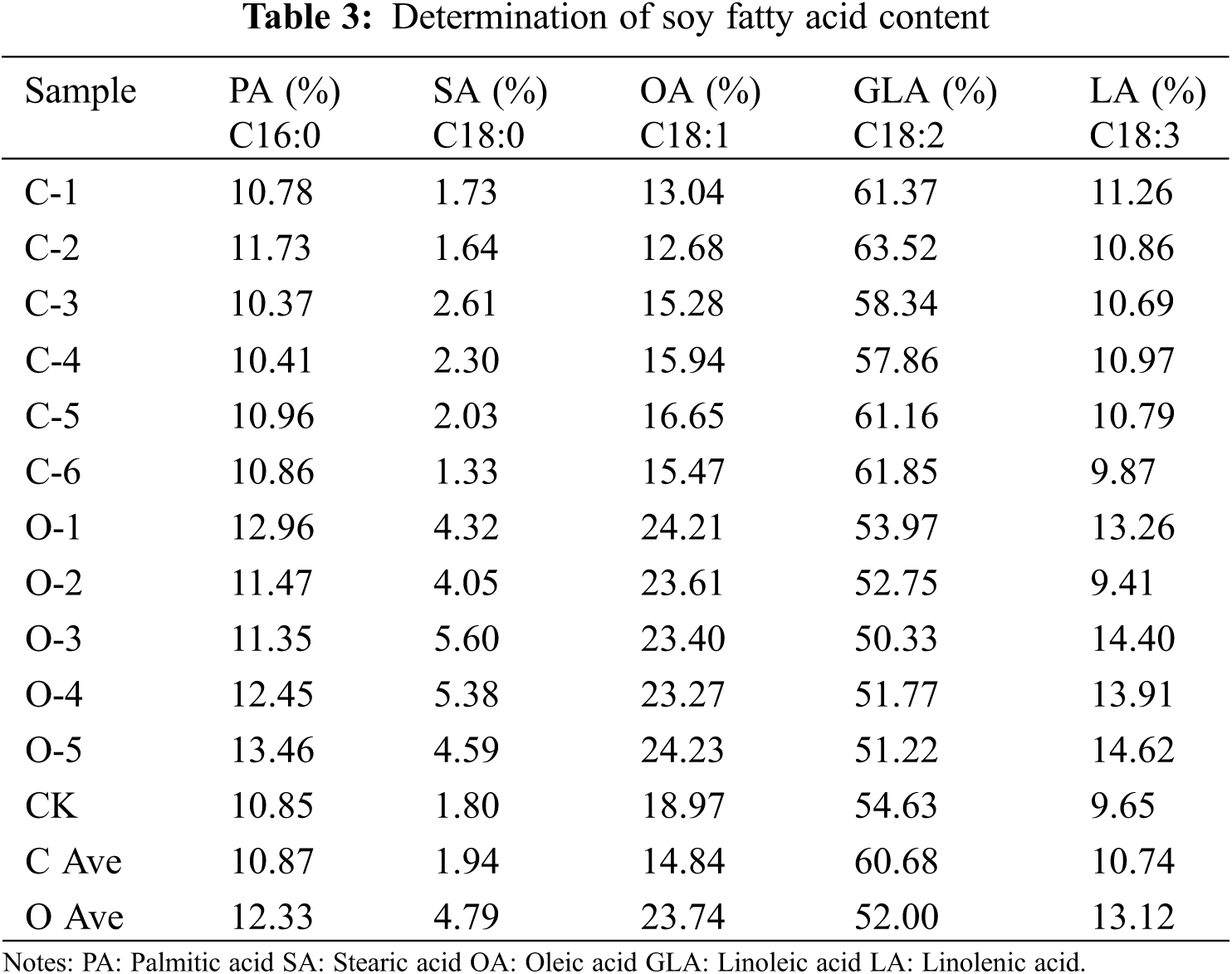
3.14 Analysis of Fatty Acid Content in Transgenic SAM22 Soybean
Fatty acids in soybean T2 seeds were identified and analysed by infrared spectroscopy. The seeds of the overexpressed plants and the seeds of the CRISPR/Cas9 gene-edited plants were taken separately. Linoleic, Oleic, stearic, palmitic, and linolenic acids were measured (Table 3). There was no substantial change in palmitic acid content, according to the results. Between the overexpression vector seeds and CK, the stearic acid content increased, whereas, between the gene-editing vector seeds and CK, the stearic acid content decreased. The effect of overexpression vector seeds on oleic acid content increased by 4.77%, and the effect of gene editing vector seeds on oleic acid content decreased by 4.13% compared with CK. The change in linolenic acid content was not clear. The concentration of soybean linoleic acid decreased by 2.63% in the overexpression vector but increased by 6.05% in the gene-editing vector. The GmSAM22 gene plays a part in the production of linoleic acid. This gene may work as a repressor in the FAD2 family.
3.15 Morphological Identification of SAM22 Gene
Three plants that were found to be positive were chosen at random for comparison with JN28CK and photographed as three replicates of the data (Fig. 21). The traits of 100-grain weight (g), plant height (cm), number of pods (pieces), and branches (pieces) were investigated for the transfected SMA22 gene single plant and the gene-edited single plant, respectively. The results showed that the gene was overexpressed. The average plant height is 102 cm, the number of pods is 108, the number of branches is 5, and the 100-grain weight is 19.66 g. The average plant height of the gene-edited plants was 92 cm, the number of pods was 87, the number of branches was 2, and the 100-grain weight was 17.02 g. It can be seen that the overexpression vector plants grew better, and the gene-editing vector and JN28CK were no discernible differences phenotypically.

Figure 21: Comparison of agronomic traits between soybean overexpressed and CRISPR/Cas9 vector and Jinong 28CK
Notes: A plant with overexpression vector; C is CRISPR/Cas9 vector plant; B and D are 28CK.
The FAD2 gene was first discovered to be cloned from Arabidopsis thaliana. It was also discovered to have a key role in the FA desaturation pathway [12]. In the plant endoplasmic reticulum, oleoyl phosphatidylcholine desaturase (FAD2) is an enzyme that catalyzes the formation of double bonds on the twelfth carbon of fatty acid chains and it is a crucial enzyme that catalyzes the first step in polyunsaturated fatty acid synthesis [13]. At present, when studying the FAD2 gene at home and abroad, the function of the FAD2 gene is often studied by constructing a gene overexpression vector and inhibiting the expression vector into plant cells. Breeding high oleic acid varieties provides resources. Tobacco seed oil, as the raw material for diesel production, contains a high linoleic acid concentration and is susceptible to oxidative degradation. Increasing the content of oleic acid by inhibiting the expression of FAD2 can improve the production performance [14]. Tian et al. [15] used a CRISPR/Cas9 vector to stop the FAD2-2 gene from being expressed in tobacco. The results showed that the content of oleic acid was increased by 68%, and the content of linoleic acid was decreased by 65%, which provided new germplasm for high oleic acid breeding. Huang et al. [16] reported that there were four copies of BnaFAD2 in allotetraploid Brassica napus. All copies of BnaFAD2 were mutated using CRISPR/Cas9 technology. The content of oleic acid and linolenic acid decreased. High oleic soybeans have improved oxidative stability with significant benefits for food and industrial processing. Darr et al. [17] found that natural mutations in the two genes FAD2-1A and FAD2-1B can increase soybean oleic acid content. To test the effects of these allele mutations on seed yield, oleic acid, and protein concentrations, the use of molecular marker method constructed 48 BC3F2:4 line populations, each line was divided into 4 genotype groups, and 3 replicates were set at 6 sites in Tennessee, and 12 lines in each genotype group were evaluated and compared. There was no significant difference in seed output between the high oleic acid genotype group and the other groups, according to the findings. The double mutation FAD2-1A and FAD2-1B groups produced higher oleic acid content than single mutation, and higher total oil and protein content than other groups. The analysis showed that the mutation can effectively increase the content of soybean oleic acid without affecting the yield. Xu et al. [18] constructed an RNAi vector to interfere with the expression of the peanut AhFAD2 gene and transferred the vector into FH1 and HY23 varieties, respectively. The transgenic progeny were analyzed and found to have no significant differences in agronomic traits when compared to the control group. Fluorescence quantitative analysis showed that the expression level of the AhFAD2 gene in the developing seeds of the transgenic lines was lower. The oleic acid content of the transgenic lines was increased by 36.4% and 15.09% in FH1 and HY23, respectively, according to gas chromatography results, the linoleic acid content decreased by 29.8% and 16.2%, respectively, which proved that the method can effectively increase the oleic acid content and provide germplasm resources for high oleic acid flowering seeds.
Gene in this experiment was a candidate gene screened from Genome-wide association analysis (GWAS), with a gene contribution rate of 17% [19]. Experiments show that the positive expression of this gene can promote the biosynthesis of oleic acid. It is speculated that this gene may be a gene that is positively regulated in the biosynthesis of oleic acid. In the FAD2 family, it may operate as a suppressor inhibiting the expression of the FAD2 gene family.
In 1979, Zhou et al. established the pollen tube channel method and proposed the molecular basis of distant hybridization—the demonstration of DNA fragment hybridization, which provided a solid foundation for the establishment of the genetic transformation method of introducing foreign genes through pollen tubes [19,20]. The principle is to use the pollen tube channel formed by the plant after the completion of pollination and inject the DNA extract containing the exogenous gene into it, and finally, the exogenous gene will be integrated into the recipient genome, and then become the carrier of the exogenous gene as the plant grows and develops. The advantage of this method is that it is simple in technology, does not require sophisticated equipment, does not rely on manual tissue culture, and is convenient for most breeders to implement; its disadvantage is that it is greatly restricted by flowering period and environmental factors. At the same time, it is limited by different plant varieties, especially small flower crops, which require certain experience and skills, and the transformation efficiency is low. In recent years, through the genetic engineering breeding of soybean in the pollen tube channel method, Wu et al. [21] introduced the expression vector containing the insect-resistant gene Cry V into soybean by the pollen tube channel method, and successfully obtained the transgenic insect-resistant soybean after additional generation testing, the conversion rate was 1.8%. Wang et al. [22] constructed the pBI121 vector containing the gus gene and transferred it into the soybean variety Liaodou 14 through the pollen tube method. After testing, it was proved that the gus gene could be inherited stably, and the transformation rate was 6.6%. Rong [23] constructed an expression vector containing the HSP17.4 heat shock protein gene, using the Bar gene as a selection marker, and transferred it into soybean Jinong 18 by pollen tube channel method to obtain high-temperature resistant plants, and the transformation rate was 5.83%. In order to improve the transformation efficiency, Wang et al. [24] invented an improved pollen tube channel transformation method, which solved the problem of the low transformation rate of the pollen tube channel method, improved the transformation efficiency, and provided a good foundation for future soybean genetic transformation. In this experiment, the pollen tube channel method was used for genetic transformation, and the pollen tube channel transformation technology of Wang et al. [24] was adopted, and the transformation rate was significantly improved.
According to a bioinformatics study, the protein encoded by the SAM22 gene is a non-secreted protein that primarily functions in the nucleus and cytoplasm, is a soluble protein that does not travel in cells, and includes 13 potential phosphorylation sites. The helix and extended chain are the most prominent. The protein interacts with an array of functions in the protein interaction analysis, so it’s possible that the encoded protein is related to oleic acid. To obtain positive soybean seeds, the overexpression vector pCAMBIA3301-SAM22 and the editing vector pBGK041 were constructed. The plasmids were injected into soybean pollen tubes through the pollen tube channel method. On T2 plants, Southern blot hybridization demonstrated that the transformed plants were integrated into the genome in a single genetically transformed scallop. And seeds had the highest gene expression, according to real-time quantitative PCR. Fatty acids in soybean T2 seeds were identified and analysed by infrared spectroscopy, according to the results. Between the overexpression seeds and CK, the stearic acid content increased, whereas, between the gene-editing seeds and CK, the stearic acid content decreased. The effect of overexpression vector seeds on oleic acid content increased by 4.77%, and the effect of gene editing vector seeds on oleic acid content decreased by 4.13% compared with CK. Overexpression of this gene in seeds can significantly increase the content of soybean oleic acid, which belongs to a positive regulatory gene. Meanwhile, overexpression plants have relatively good morphological identification.
Authors Contribution: Shuo Qu is the executive of the experimental design and experimental research of this study; Shuo Qu and Cai Qi completed the data analysis and the writing of the first draft of the paper; Huimin Cui, Lamboro Abraham, Yaolei Jiao, Guilong Ma, participated in the experimental design, analyzed data, edit the final manuscript; Piwu Wang managed and guided the whole project. All authors have read and agreed to the final text.
Funding Statement: This research was funded by the National Major Special Project for Breeding New Varieties of Genetically Modified Organisms (2016ZX08004-004). National Natural Science Foundation of China (31771817).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Zan, G. M., Zhang, L., Zhang, Y. R., Sheng, Y. H., Zhou, K. et al. (2021). Establishment of a gas chromatographic detection method for fatty acid components in soybean grain. China Agricultural Science Bulletin, 37(9), 118–124 (in Chinese). DOI 10.11924/j.issn.1000-6850.casb2020-0250. [Google Scholar] [CrossRef]
2. Wang, F. J., Zhang, J., Ji, F. D., Li, D. W., Lu, F. (2008). Study on enzymatic hydrolysis and fermentation of defatted soybean mea. China Brewing, 2008, 11, 6–10 (in Chinese). DOI 10.3969/j.issn.0254-5071.2008.11.002. [Google Scholar] [CrossRef]
3. Li, S. J., Cronan, J. E. (1992). The gene encoding the biotin carboxylase subunit of Escherichia coli acetyl-CoA carboxylase. Journal of Biological Chemistry, 267(2), 855–863. DOI 10.1016/S0021-9258(18)48362-7. [Google Scholar] [CrossRef]
4. Byfield, G. E., Xue, H., Upchurch, R. G. (2006). Two genes from soybean encoding soluble Δ9 stearoyl-ACP desaturases. Crop Science, 46(2), 840–846. DOI 10.2135/cropsci2005.06-0172. [Google Scholar] [CrossRef]
5. Zhang, Y. H., Zhang, Y. J., Su, P., Zhang, T., Zhou, R. M. et al. (2019). Metabolic pathways of oils and breeding strategies to increase the oil content of oil crops. Molecular Plant Breeding, 17(9), 3084–3089. [Google Scholar]
6. Topfer, R., Martini, N., Schell, J. (1995). Modification of plant lipid synthesis. Science, 268(5211), 681–686. DOI 10.1126/science.268.5211.681. [Google Scholar] [CrossRef]
7. Zhang, J. T., Li, J. H., Hernan, G. R. (2014). A stearoyl-acyl carrier protein desaturase, NbSACPD-C, is critical for ovule development in Nicotiana benthamiana. Plant Journal, 80(3), 489–502. DOI 10.1111/tpj.12649. [Google Scholar] [CrossRef]
8. Jiang, C. J., Shimono, M., Maeda, S. (2009). Suppression of the rice fatty-acid desaturase gene OsSSI2 enhances resistance to blast and leaf blight diseases in rice. Molecular Plant Microbe Interactions, 22(7), 820–829. DOI 10.1094/MPMI-22-7-0820. [Google Scholar] [CrossRef]
9. Aardra, K. H., John, S. K., Edward, W. (2007). The Arabidopsis stearoyl-acyl carrier protein-desaturase family and the contribution of leaf isoforms to oleic acid synthesis. Plant Molecular Biology, 63(2), 257–271. [Google Scholar]
10. Hou, Z. H., Wu, Y., Cheng, Q., Dong, L. D., Lu, S. J. et al. (2019). Using CRISPR/Cas9 technology to create soybean high oleic acid mutant lines. Acta Agronomica Sinica, 45(6), 839–847 (in Chinese). DOI 10.3724/SP.J.1006.2019.84157. [Google Scholar] [CrossRef]
11. Liu, X., Qin, D., Piersanti, A., Zhang, Q., Miceli, C. et al. (2020). Genome-wide association study identifies candidate genes related to oleic acid content in soybean seeds. BMC Plant Biology, 20(1), 399. DOI 10.1186/s12870-020-02607-w. [Google Scholar] [CrossRef]
12. Nishiuchi, T., Nishimura, M., Arondel, V., Iba, K. (1994). Genomic nucleotide sequence of a gene encoding a microsomal omega-3 fatty acid desaturase from Arabidopsis thaliana. Plant Physiology, 105(2), 767–768. DOI 10.1104/pp.105.2.767. [Google Scholar] [CrossRef]
13. Xue, Y., Yin, N., Chen, B., Liao, F., Win, A. N. et al. (2017). Molecular cloning and expression analysis of two FAD2 genes from chia (Salvia hispanica). Acta Physiologiae Plantarum, 39(4), 95. [Google Scholar]
14. Wu, P. Z., Zhang, S., Zhang, L., Chen, Y. P., Li, M. R. et al. (2013). Functional characterization of two microsomal fatty acid desaturases from Jatropha curcas L. Journal of Plant Physiol, 170(15), 1360–1366. DOI 10.1016/j.jplph.2013.04.019. [Google Scholar] [CrossRef]
15. Tian, Y. S., Chen, K., Li, X., Zheng, Y. P., Chen, F. (2020). Design of high-oleic tobacco (Nicotiana tabacum L.) seed oil by CRISPR-Cas9-mediated knockout of NtFAD2-2. BMC Plant Biology, 20(1), 1–9. DOI 10.1186/1471-2229-9-1. [Google Scholar] [CrossRef]
16. Huang, H. B., Cui, T. T., Zhang, L. L., Yang, Q. Y., Yang, Y. et al. (2020). Modifications of fatty acid profile through targeted mutation at BnaFAD2 gene with CRISPR/Cas9-mediated gene editing in Brassica napus. Theoretical and Applied Genetics, 133(8), 2401–2411. DOI 10.1007/s00122-020-03607-y. [Google Scholar] [CrossRef]
17. Darr, L., Cunicelli, M., Bhandari, H., Feng, C., Li, Z. L. et al. (2020). Field performance of high oleic soybeans with mutant FAD2-1A and FAD2-1B genes in Tennessee. Journal of the American Oil Chemists’ Society, 97(1), 49–56. [Google Scholar]
18. Xu, P. L., Tang, G. Y., Bi, Y. P., Liu, Z. J., Shan, L. (2018). Analysis of transgenic offspring with suppressed expression of AhFAD2 gene in peanut. Chinese Journal of Biological Engineering, 34(9), 1469–1477 (in Chinese). DOI 10.13345/j.cjb.170508. [Google Scholar] [CrossRef]
19. Ni, W. C., Guo, S. D., Jia, S. R. (2020). Genetic transformation of cotton mediated by pollen tube pathway. China Agricultural Science and Technology Herald, 2(2), 27–32 (in Chinese). DOI 10.3969/j.issn.1008-0864.2000.02.007. [Google Scholar] [CrossRef]
20. He, H., Gao, S., Han, D., Wu, N., Qu, J. (2020). Study on transformation of soybean with insect-resistant gene Cry1Iem. Journal of Jilin Agricultural University, 39(6), 665–669 (in Chinese). DOI 10.13327/j.jjlau.2017.3174. [Google Scholar] [CrossRef]
21. Wu, X. X., Liu, W. T., Liu, Q., Li, J., Ma, Y. et al. (2010). Study on transfer of insect resistance gene (cryV) into soybean by pollen tube channel method. Soybean Science, 29(4), 565–568 (in Chinese). DOI 1000-9841(2010)04-0565-04. [Google Scholar]
22. Wang, L. H., Liu, M., Su, Q., Gao, X. R., An, L. J. (2004). Molecular token of transferred soybean via pollen-tube pathway. Molecular Plant Breeding, 2(2), 193–196 (in Chinese). DOI 10.3969/j.issn.1672-416X.2004.02.005. [Google Scholar] [CrossRef]
23. Rong, J. (2017). Construction of plant expression vector of soybean heat shock protein gene HSP17.4 and analysis of heat resistance function (Ph.D. Thesis). Jilin Agricultural University, China. [Google Scholar]
24. Qu, S., Jiao, Y. L., Abraham, L., Wang, P. (2021). Correlation analysis of new soybean [Glycine max (L.) Merr] gene Gm15G117700 with oleic acid. Phyton-International Journal of Experimental Botany, 90(4), 1177–1192. DOI 10.32604/phyton.2021.015206. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools