 Open Access
Open Access
ARTICLE
DDG1 and G Protein α Subunit RGA1 Interaction Regulates Plant Height and Senescence in Rice (Oryza sativa)
1 Jiangsu Key Laboratory of Eco-Agricultural Biotechnology around Hongze Lake, Regional Cooperative Innovation Center for Modern Agriculture and Environmental Protection, Huaiyin Normal University, Huai’an, 223300, China
2 School of Life Sciences, Huaiyin Normal University, Huai’an, 223300, China
3 Crop Research and Development Center, Huaiyin Institute of Agricultural Sciences of Xuhuai Region in Jiangsu, Huai’an, 223001, China
4 Institute of Bast Fiber Crops, Chinese Academy of Agricultural Sciences, Changsha, 410205, China
* Corresponding Author: Xi Liu. Email:
# These three authors contributed equally to this work
Phyton-International Journal of Experimental Botany 2023, 92(7), 2051-2064. https://doi.org/10.32604/phyton.2023.028196
Received 04 December 2022; Accepted 31 March 2023; Issue published 29 May 2023
Abstract
Many studies have already shown that dwarfism and moderate delayed leaf senescence positively impact rice yield, but the underlying molecular mechanism of dwarfism and leaf senescence remains largely unknown. Here, using map-based cloning, we identified an allele of DEP2, DDG1, which controls plant height and leaf senescence in rice. The ddg1 mutant displayed dwarfism, short panicles, and delayed leaf senescence. Compared with the wild-type, ddg1 was insensitive to exogenous gibberellins (GA) and brassinolide (BR). DDG1 is expressed in various organs, especially in stems and panicles. Yeast two-hybrid assay, bimolecular fluorescent complementation and luciferase complementation image assay showed that DDG1 interacts with the α-subunit of the heterotrimeric G protein. Disruption of RGA1 resulted in dwarfism, short panicles, and darker-green leaves. Furthermore, we found that ddg1 and the RGA1 mutant was more sensitive to salt treatment, suggesting that DDG1 and RGA1 are involved in regulating salt stress response in rice. Our results show that DDG1/DEP2 regulates plant height and leaf senescence through interacting with RGA1.Keywords
Rice feed over two thirds of the world’s population, and the ‘Green Revolution’ gene sd1 has made a significant contribution increases in rice yields [1]. However, the reduction of plant height continues to be the main goal for increasing lodging resistance in rice breeding [2]. So far, many quantitative trait loci (QTLs) and genes that regulate plant height have been isolated and cloned, such as D61, MIT1, D35, and D6 [3–6]. Among these QTLs and genes, a considerable number of genes are involved with plant hormones, especially GA (Gibberellin) and BR (Brassinosteroid). The ‘Green Revolution’ gene sd1 encodes gibberellin 20 oxidase 2 (GA20ox2), which converts GA12/GA53 to the bioactive GA precursors GA9/GA21 [7,8]. In addition, BR biosynthesis genes such as D2, BRD2, and DWARF4 and signal pathway genes such as D61, DLT, and GSK2 regulate rice plant height by controlling cell proliferation and cell expansion [9,10]. However, sd1 is still the dwarf gene most used in rice breeding, and it is urgent to clone a new dwarf gene.
Senescence is the final stage of plant development, and premature leaf senescence negatively impacts rice yield stability [11,12]. Leaf senescence is a highly regulated process by both intrinsic regulators as well as environmental factors [13]. Many leaf senescence-related mutant genes have been isolated in various crops, including OsNAP, OsNaPRT1, OsSRLK, and OsDOS [12,14–16]. The OsNAP gene encodes an NAC (NAM, ATAF, and CUC2) transcriptional activator, and knock-down of OsNAP delays leaf senescence and increases yield [12]. OsNAP regulates the expression of chlorophyll degradation-related gene stay-green (SGR) and senescence-associated gene Osh36 [12]. Similarly, OsNaPRT1 affects the nicotinamide content and regulates the expression of senescence related genes [15]. However, plant leaf senescence is a very complex growth process, and the underlying molecular mechanism of leaf senescence remains largely unknown.
Here, we reported a new dwarfism and delayed leaf senescence mutant in rice, dwarfism and darker green leaf 1 (ddg1). Map-based cloning showed that DDG1 is a new allele of DEP2, which encodes a plant-specific protein without any known functional domain. We also found that DDG1 was expressed in various rice tissues and the DDG1 protein was conserved in plants. In addition, we observed DDG1’s interaction with RGA1 through yeast two-hybrid (Y2H) analysis, bimolecular fluorescent complementation (BiFC) and luciferase complementation image (LCI) assays. Finally, we found that DDG1 positively regulated salt stress response in rice. Our results provide new insight into DDG1/DEP2’s regulation rice growth and development.
2.1 Plant Materials and Cultivation
The ddg1 mutant that exhibited dwarfism and delayed leaf senescence was isolated from the mutant library of the japonica rice variety Kitaake by 1% ethyl methanesulfonate (EMS) in 2017. For EMS treatment, we soaked dry seeds of Kitaake in water for 24 h, added 1% EMS to soak for 10 h, and soaked in water for 24 h, then sowed seeds in the field. The ddg1 mutant was crossed with the indica variety 93–11 to develop the gene mapping population. 521 extreme individuals selected from the F2 population were used to map ddg1 for dwarfism and delayed leaf senescence. The RGA1KO mutant used in this study was kindly provided by Professor Yidan Ouyang and Doctor Shengyuan Sun [17]. All the plants were grown in paddy fields in Huai’an (Jiangsu Province) and Lingshui (Hainan Province), China.
For salt treatment, we germinated seeds of both wild-type (WT, Kitaake) and ddg1 in water for 48 h at 37°C in darkness, and then transferred them to a Yoshida rice nutrient salt mixture (Coolaber, NSP1040, Beijing, China) for 2 weeks, exchanging fresh solution every 3 days. Next, the 2-week-old seedlings were transferred to a hydroponic culture solution supplemented with 150 mM NaCl for 7 d. Finally, the rice seedlings were grown in climate chambers under 14/10 h light/dark and 30°C/25°C day/night conditions.
2.2 Scanning Electron Microscopy (SEM)
At the heading stage, the first internodes of the WT and the ddg1 mutant were collected and fixed with 2.5% glutaraldehyde, and the processed samples were observed with a scanning electron microscope (JSM-840, JEOL).
Chlorophyll was extracted from leaves of wild-type and ddg1 at maturity. 0.1 g fresh leaves was cut and soaked in anhydrous ethanol for 48 h in darkness. The absorbance values at 649, and 665-nm wavelength were measured using a spectrophotometer. Chlorophyll contents were determined according to as previously described method [18].
For GA treatment, we grew seeds of wild-type and ddg1 in hydroponic media with different concentrations of GA for two weeks, and the seedling height of the WT and ddg1 were measured. For BR sensitivity test, leaves with partial leaf blades and sheathes were cut from the 2-week-old seedlings of the wild type and ddg1. Leaves were put into a solution with different concentrations of 24-Epibrassinolide (E1641, Sigma) or control solution, and then incubated in the light for 3 d. Finally, lamina joint angles were measured using IMAGEJ software.
For map-based cloning of DDG1, we chose 521 recessive individual plants showing dwarfism and delayed leaf senescence similar to the phenotype of the ddg1 mutant from an F2 population derived from a cross between ddg1(♀) and the indica variety 93-11(♂). At heading stage, the mutant anthers were emasculated and subjected to the 9311 pollen. Simple sequence repeat (SSR) markers and the developed insertion/deletion (InDel) markers with Primer Premier 5 on chromosome 7 were used for fine mapping. Next, we amplified the corresponding fragments from ddg1 and the WT, and compared sequences using the BioXM 2.6 software.
To verify whether the Os07g0616000 mutation was responsible for the phenotype of ddg1, For constructing the Os07g0616000 complementary vector, the full-length coding sequence (CDS) was amplified and inserted into the p1390Ubi vector. Approximately 2 kb promoter sequence of Os07g0616000 was amplified and substituted with ubiquitin promoter region of p1390Ubi vector. The recombinant was transformed into the calluses from the ddg1 seeds by agrobacterium-infection methods. T2 generation transgenic plants were used for phenotype analysis. The primers used in the map-based cloning and complementary vector construction were listed in Table S1.
2.6 RNA Extraction and qRT-PCR
Total RNAs were extracted using the TRIzol reagent (Invitrogen) from various tissues of the WT and ddg1, and cDNAs were synthesized using a HiScript® II 1st strand cDNA synthesis kit (#R211; Vazyme, Nanjing, China). qRT-PCR experiments were performed using a real-time PCR detection system (BIO-RAD, CFX96) with three biological repetitions using the rice Ubiquitin (LOC_Os03g13170) gene as the internal control. The primers used for qRT-PCR were given in Table S1.
2.7 Yeast Two-Hybrid (Y2H) Assay
The full-length CDS of DDG1 and RGA1 (primers listed in Table S1) were amplified and inserted into the pGBKT7 vector and the pGADT7 vector, respectively. Different combinations of plasmids were also transformed into Saccharomyces cerevisiae strain AH109 on the SD/-Trp/-Leu/-His/-Ade medium. The specific method of yeast transformation can be found in the manufacturer’s instructions, which we followed carefully (http://www.clontech.com/).
2.8 Bimolecular Fluorescent Complementation (BiFC)
For BiFC assays, we inserted the full length CDS of DDG1 and RGA1 (lacking stop codon) into the p2YN and p2YC at PacI and SpeI sites, respectively [19]. We next transformed the DDG1-nYFP and RGA1-cYFP plasmids or corresponding empty vectors into Agrobacterium strain EHA105, and then infiltrated this into leaves of N. benthamiana for 48 h. Fluorescences were then observed by laser confocal microscopy (Zeiss). The primers used for BiFC were listed in Table S1.
2.9 Luciferase Complementation Image (LCI) Assay
For LCI assays, the full-length CDS of DDG1 and RGA1 were amplified and fused to the pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vector, respectively. These recombinants were co-transformed into N. benthamiana leaves by Agrobacterium infection. Empty pCAMBIA 1300-nLUC and pCAMBIA1300-cLUC vectors were used as negative controls. Primers used for LCI assays were listed in Table S1.
3.1 Characterization of the ddg1 Mutant
We isolated a dwarf and dark-green leaves mutant, ddg1, from an ethyl methanesulfonate (EMS)-derived mutagenesis population in a japonica rice variety, Kitaake. Compared with WT, ddg1 exhibited dwarfism and short panicles at maturity (Figs. 1A–1C). Each internode of ddg1 was almost shortened (Fig. 1B). The seeds of ddg1 were small and round (Fig. 1D).The plant height and panicle length of the ddg1 mutant decreased by 30% and 27.8% (Figs. 1E, 1F). We observed no difference in the tiller number between the wild type and ddg1 (Fig. 1G). However, ddg1 showed delayed leaf senescence and had higher chlorophyll content than that of the WT at maturity (Figs. 2A and 2B). Similarly, two senescence-associated genes, SGR and Osh36, were significantly repressed in ddg1 (Figs. 2C and 2D). These results suggest that DDG1 regulates plant height and senescence.
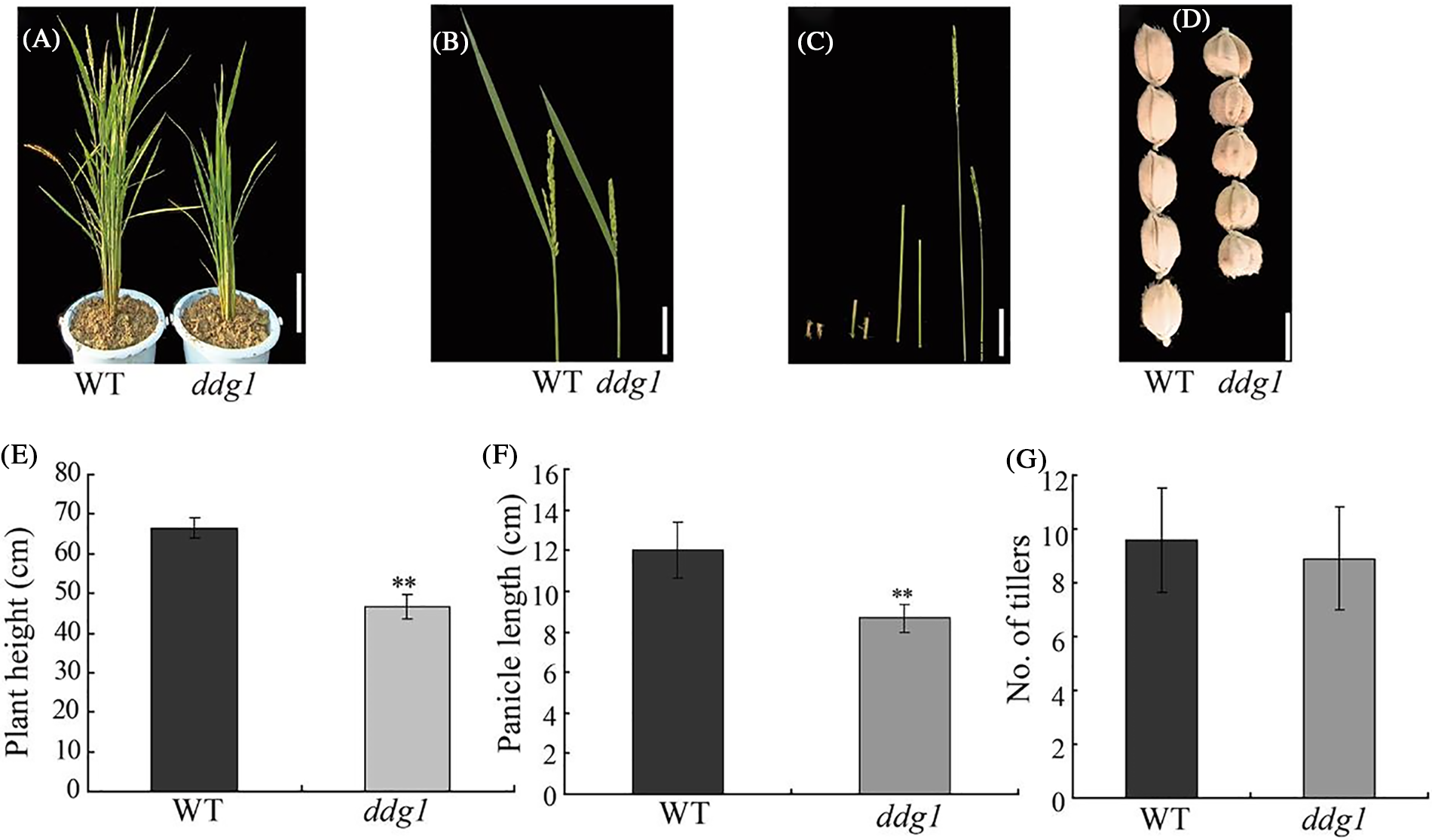
Figure 1: Plant height and panicle length were reduced in the ddg1 mutant plants. (A) Phenotypes of the wild-type (WT) and the ddg1 mutant at the heading stage. Bar = 10 cm. (B) Panicle morphology of WT and ddg1. Bar = 5 cm. (C) The internodes of the wild-type (left) and the ddg1 mutant (right). Bar = 2 cm. (D) Grain morphology of WT and ddg1. Bar = 2 mm. (E) Plant height of the wild-type and the ddg1 mutant. (F) Panicle length of WT and ddg1. (G) Tiller number of WT and ddg1. Data are shown as mean ± SD (n = 20). ** p < 0.01 by Student’s t-test analysis
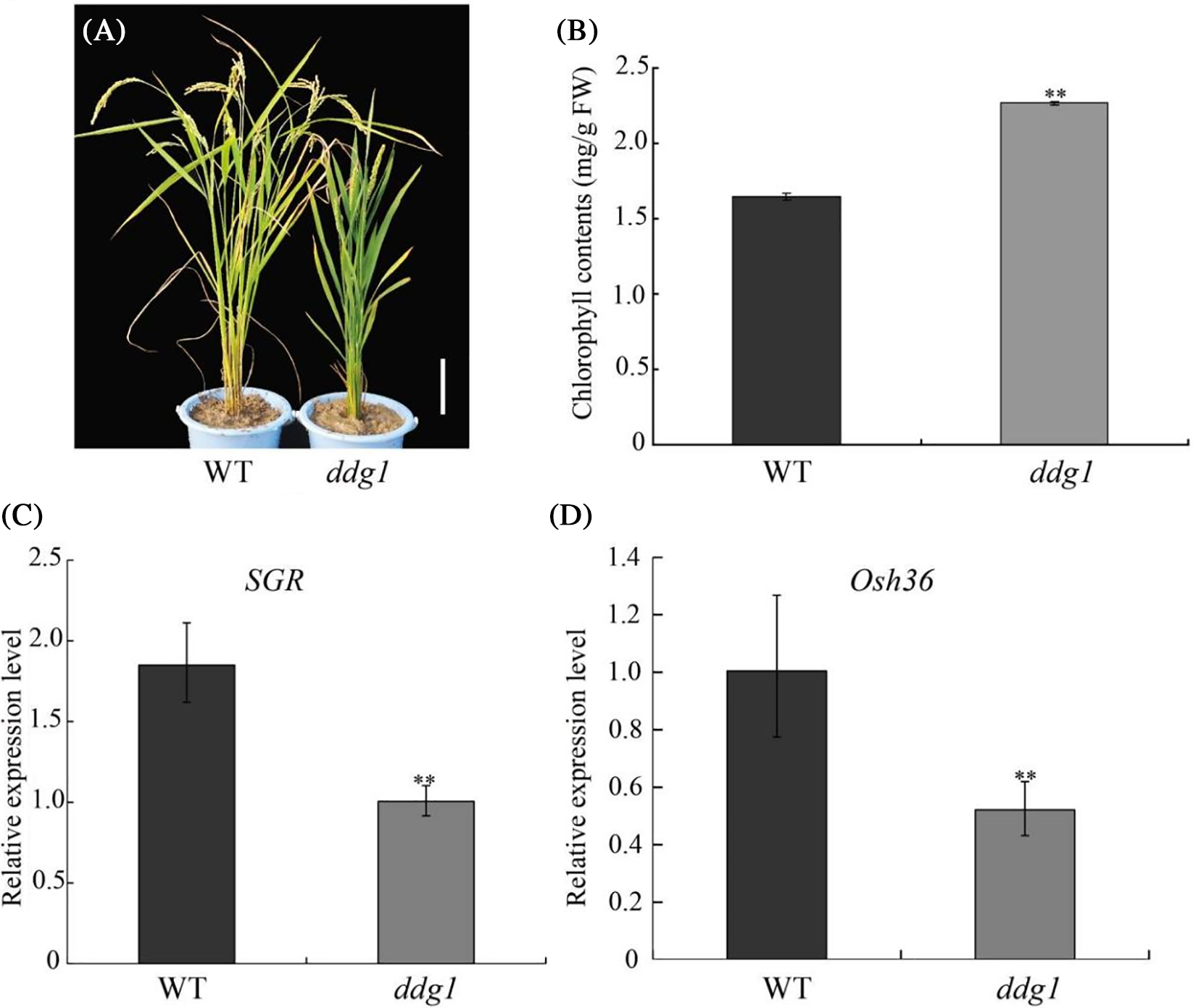
Figure 2: DDG1 negatively affects leaf senescence. (A) Phenotypes of the wild type (WT) and ddg1 at maturity. Bar = 10 cm. (B) Chlorophyll contents of the wild type and ddg1 at maturity. (C and D) Expression analysis of SGR and Osh36 in the wild type and ddg1 by qRT-PCR. Data are shown as means ± SD (n = 10). Bar = 10 cm. ** represents a significant difference at p < 0.01
3.2 Altered Cell Proliferation in ddg1
Normal cell proliferation and cell expansion are responsible for plant organ size. Hence, in order to examine the dwarfism of ddg1 in more detail, we examined the first internodes of the WT and ddg1 at the heading stage using histological analysis. Our SEM observations showed that cell length and width were similar between the wild type and the ddg1 mutant (Fig. S1), which suggests that the DDG1 mutation does not influence cell expansion.
3.3 Decreased Sensitive to GA and BR in ddg1
Both GA and BR are essential for plant growth and developmental processes, including stem development, grain size, senescence, and seed germination [10]. In order to verify whether DDG1 is involved in the GA and BR signaling pathways, we tested the sensitivity of ddg1 to GA and 24-epibrassinolide. First, we cultured the WT and ddg1 seedlings with different concentrations of GA and measured the seedlings’ heights after 2 weeks of growth. As shown in Figs. 3A and 3B, the average seedling height of the wild type and ddg1 increased significantly with an increase in GA concentration, but the rate of increase of seedling height for the ddg1 mutant was significantly lower than that of the wild type, indicating that ddg1 was less sensitive to GA. Secondly, for BR sensitivity test, we used 0, 0.1, or 1 μM of 24-epiBL to treat excised leaf segments of the 2-week-old WT and ddg1 mutant plants and then measured the lamina joint angle. Compared with the wild type, the rate of increase in the lamina joint angle in ddg1 was remarkably reduced (Figs. 3C–3F). The above results indicate strongly that DDG1 has a direct and critical involvement in the GA and BR signaling pathways.
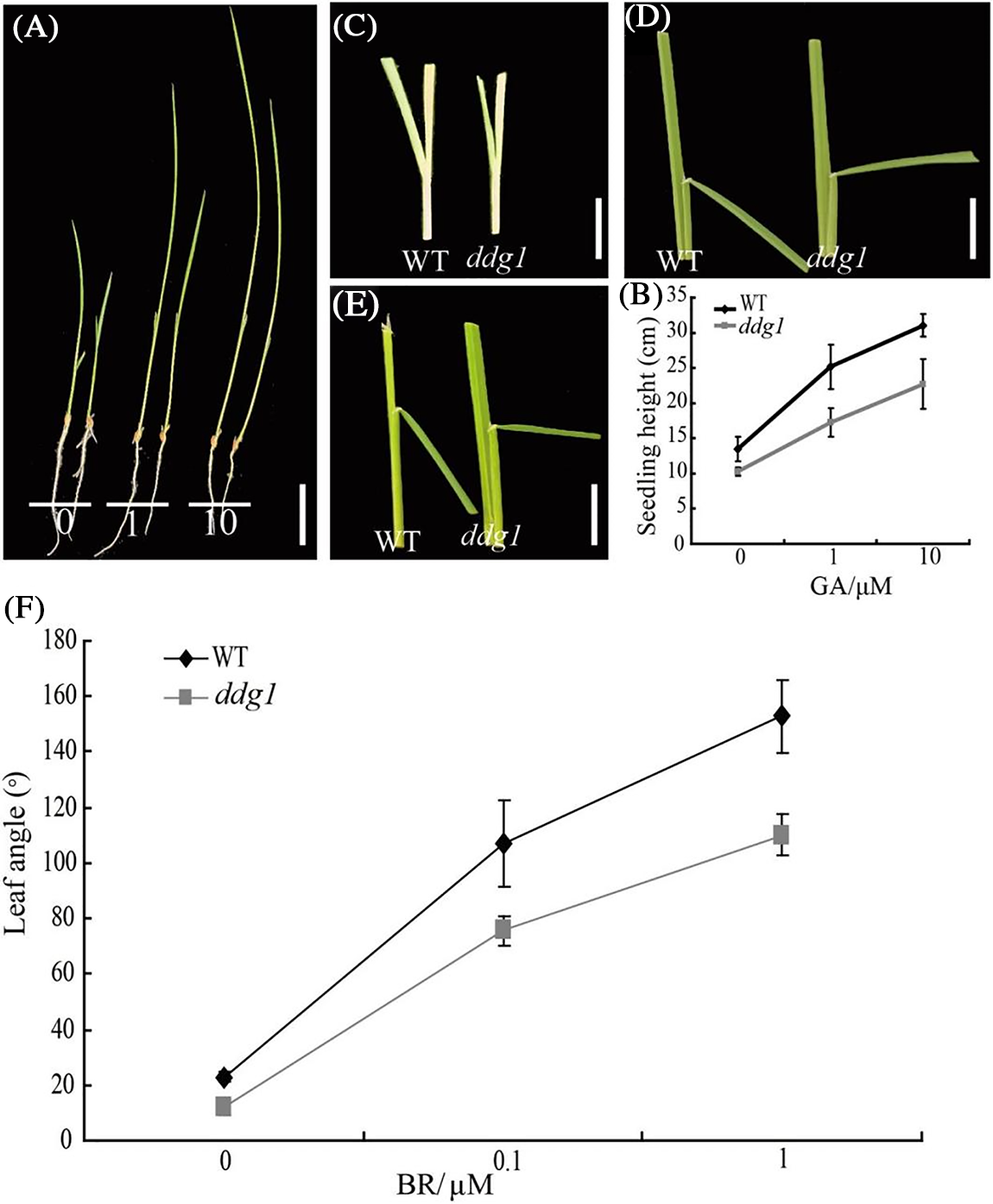
Figure 3: ddg1 is less sensitive to GA and BR. (A) Phenotypes of the wild type (left) and ddg1 (right) with GA treatment. Bar = 5 cm. (B) Seedling height of the wild type and ddg1 after GA treatment. (C–F) Response to 0 (C), 0.1 (D), or 1 (E) μM of 24-epiBL of the lamina joint of the wild type and ddg1. Data represent the means of results from ten seedlings in each sample
To isolate the DDG1 gene responsible for the ddg1 phenotype, we crossed ddg1 with indica var 93–11 to construct the mapping population. Using 521 extreme individuals selected from the F2 population, we mapped DDG1 to a 140-kb region on rice chromosome 7 (Fig. 4A). We found that there were 18 genes in the 140-kb region (http://rice.plantbiology.msu.edu/) (Fig. 4A), and among these 18 genes, one was the DEP2 (Os07g0616000) gene, which regulates panicle architecture and grain size [20,21]. We then sequenced the promoter and coding region of DEP2 between the wild type and ddg1 via Sanger sequencing [22] and found that there was a one-nucleotide deletion of DEP2 in ddg1, compared with the wild type (Fig. 4B). One-nucleotide deletion of DEP2 in ddg1 led to premature termination of the DEP2 protein translation (Fig. S2). Next, in order to study further whether the DEP2 mutation was responsible for the defects in ddg1, we conducted a genetic complementation test. Approximately 2 kb promoter and full-length CDS of DEP2 were transformed into the calluses from the ddg1 mutant and found that the positive complementary plants were similar to the wild type (Fig. 4C), compared to the negative control plants (Fig. S3). Importantly, we found the specific mutant DDG1 gene to be DEP2/Os07g0616000.
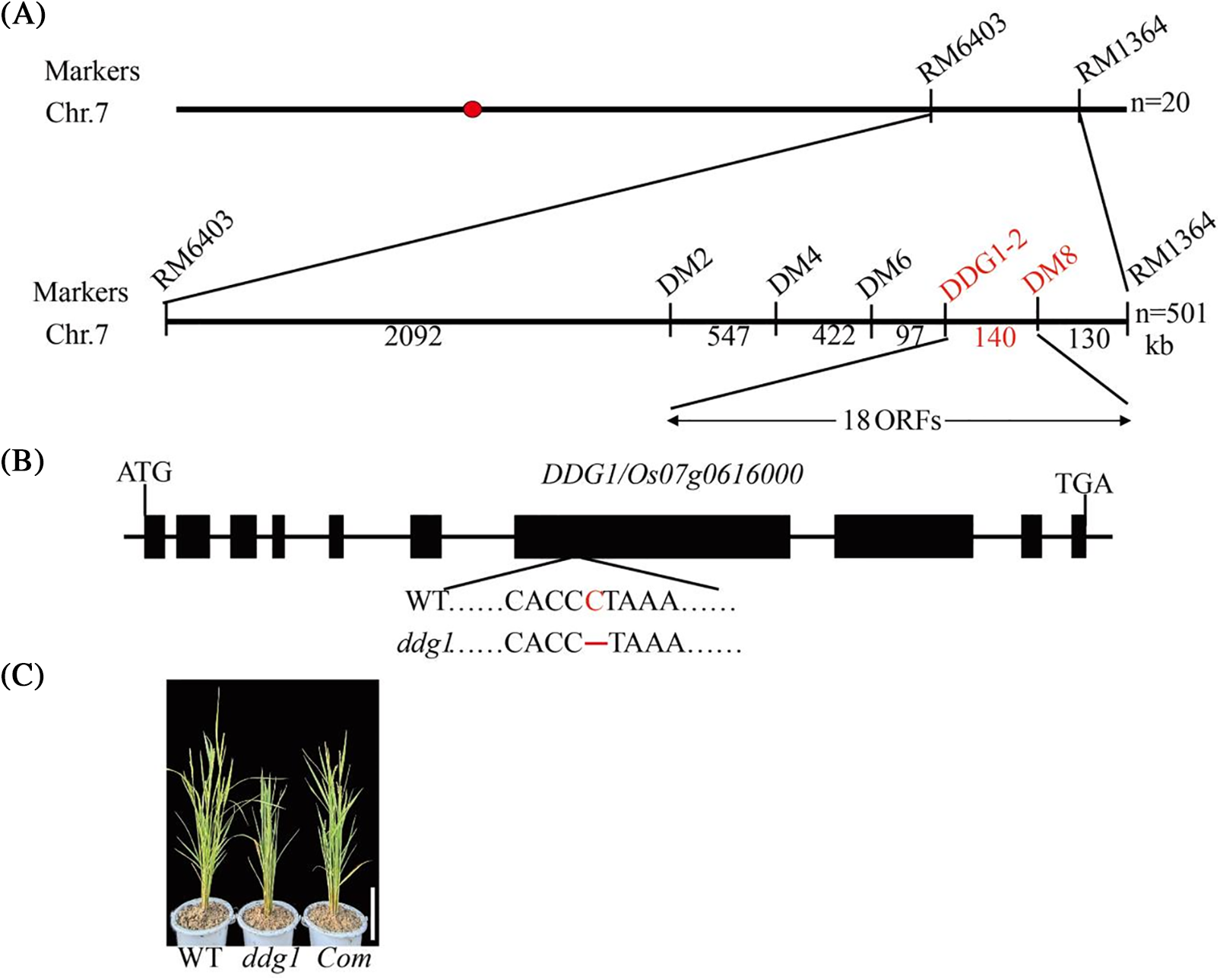
Figure 4: Map-based cloning of DDG1. (A) DDG1 was fine-mapped in the 140-kb interval between InDel markers DDG1-2 and DM8 on chromosome 7. (B) DDG1 gene structure. Black boxes represent exons, and a mutation site is shown in red. (C) Complementation of the ddg1 mutant with the DDG1/Os07g0616000 gene. Bar = 10 cm
3.5 Expression and Evolutionary Analysis of DDG1
To examine the expression pattern of DDG1, we first learned that DDG1 was expressed in both vegetative and reproductive tissues by using the RiceXPro database (Fig. S4; https://ricexpro.dna.affrc.go.jp/). In addition, we performed qRT-PCR to analysis the expression pattern of DDG1. As shown in Fig. 5A, DDG1 was expressed in roots, stem, leaves, panicles, and seeds. Interestingly, we observed that the expression level of DDG1 was significantly repressed in ddg1 (Fig. 5B).
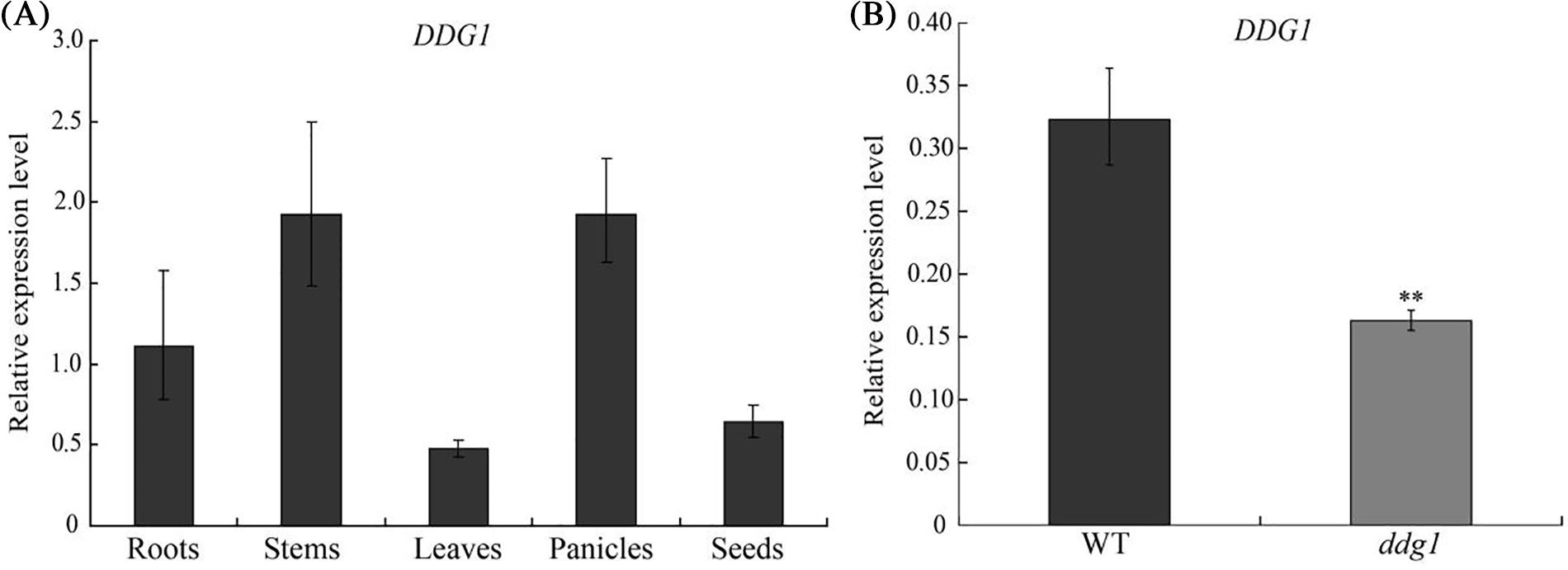
Figure 5: Expression pattern of DDG1. (A) Expression of DDG1 in roots, leaves, panicles, stems and seeds of wild-type plants. (B) Expression level of DDG1 in ddg1 and the wild type leaves. ** represents a significant difference at p < 0.01
DDG1 encodes a plant-specific protein without any known functional domain and that has 10 exons. To analyze the evolutionary relationship of the DDG1 protein, we performed a BLAST search in order to identify DDG1 homologs and found that DDG1 showed high amino acid sequence similarity to proteins in other species, including Oryza brachyantha (88%), Brachypodium distachyon (73%), Triticum dicoccoides (73%), and Panicum virgatum (68%; Fig. S5).
3.6 An Interaction between DDG1 and RGA1
Since the DDG1 protein has no known functional domain, we performed Y2H screening to identify its interacting proteins. To accomplish this, we fused the full-length CDS of DDG1 to the pGBKT7 vector (DDG1-BD) and used it as bait to screen the interacting proteins against a rice leaf yeast library of 2-week-old Kitaake, and we were able to identify one candidate segment belonging to part of the RGA1 gene. After this result, we fused the full-length CDS of RGA1 to the pGADT7 vector (RGA1-AD). Next, DDG1-BD and RGA1-AD were co-transformed into the yeast cells, which grew well on our quadruple dropout media (Fig. 6A; Fig. S6). We then performed bimolecular fluorescent complementation assays to detect the interaction. As shown in Fig. 6B, the fluorescence signal was observed when the fusion constructs p2YN-DDG1 and p2YC-RGA1 were present, but not in the corresponding control. Using a LCI assay, we detected obvious LUC activity signal when DDG1-nLUC was co-expressed with RGA1-cLUC in N. benthamiana (Fig. 6C). Thus, we conclude that DDG1 interacts with RGA1. In addition, compared with the wild type, knock-out of RGA1 showed dwarfism, short panicles, and darker-green leaves, which were similar to the phenotype of ddg1 (Fig. 7).
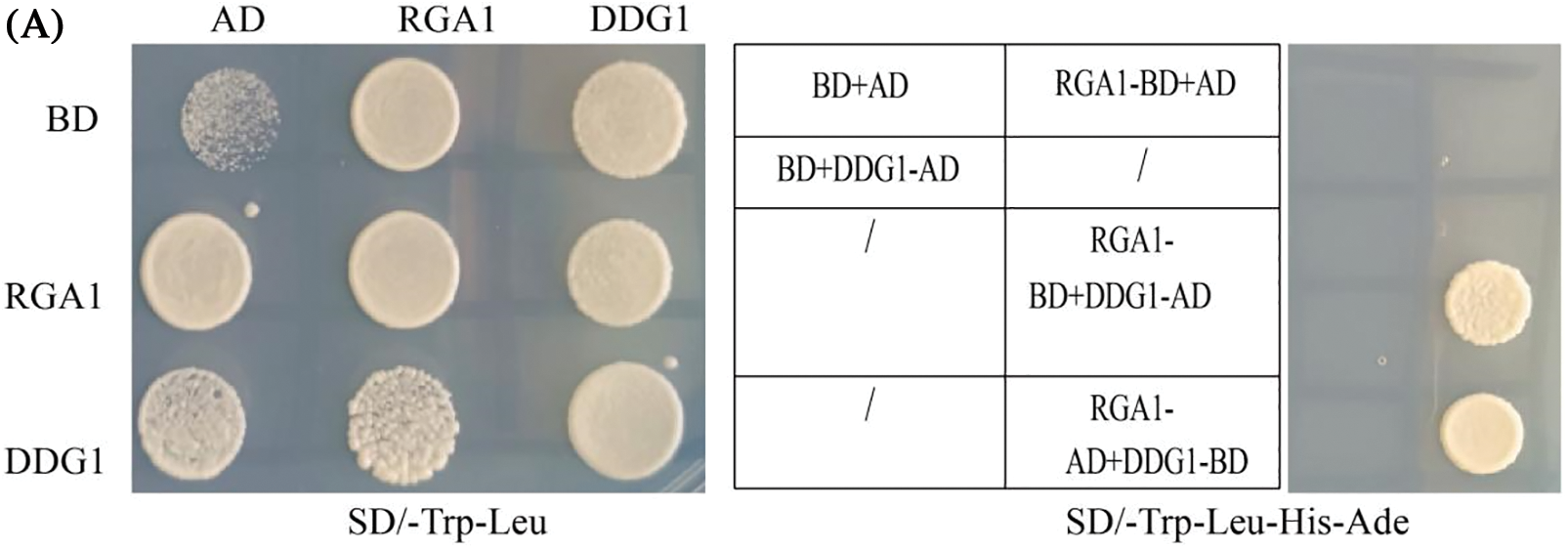
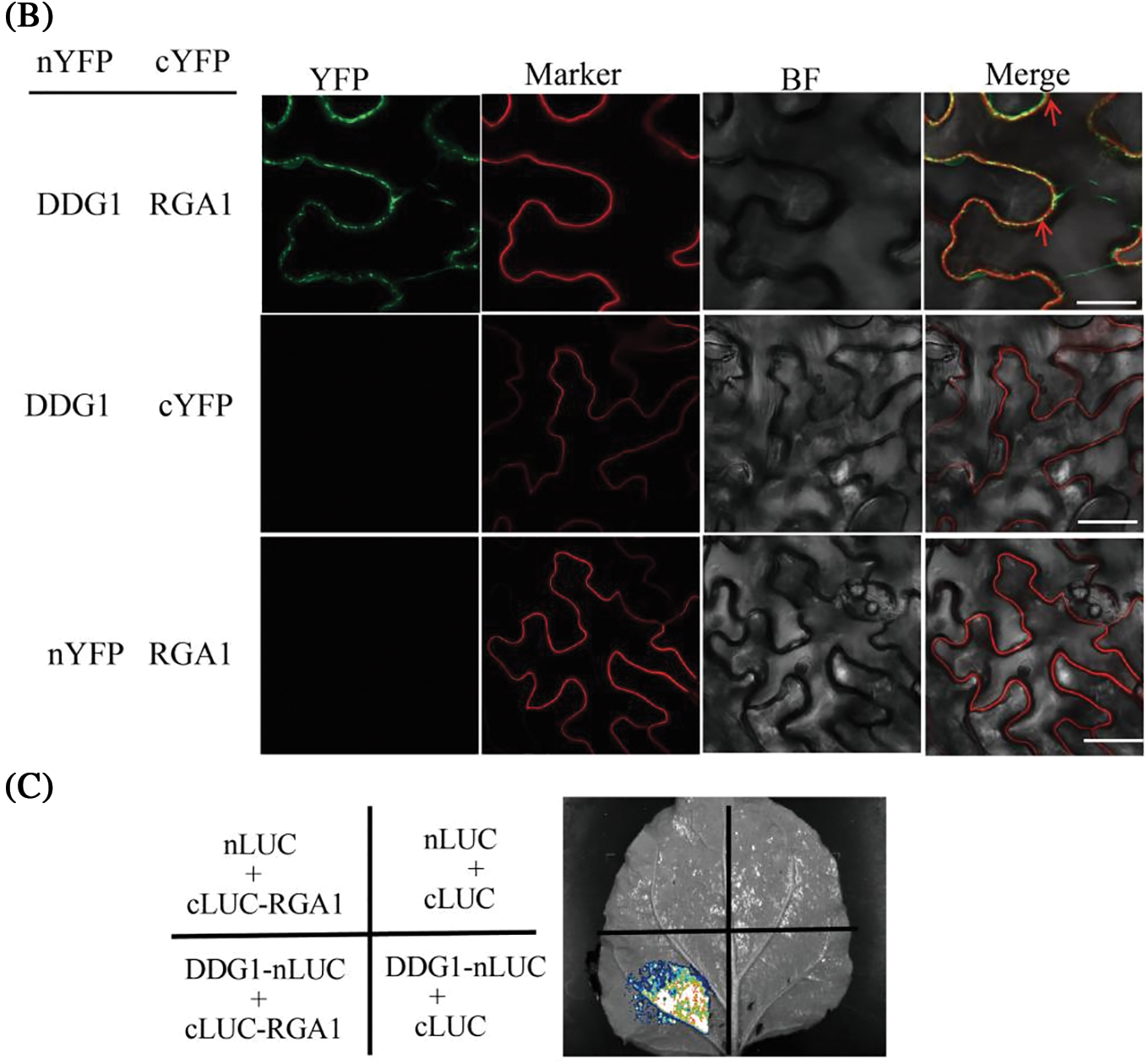
Figure 6: DDG1 interacts with the Gα protein RGA1. (A) Interaction of DDG1 and RGA1 using yeast twohybrid assay. AD: GAL4 activation domain; BD: GAL4 binding domain. (B) BiFC assay. Bar = 10 μm. Plasma membrane protein OsCAMP1-mCherry was used as a marker. (C) LCI assays. LUC activity was detected 72 h after injection
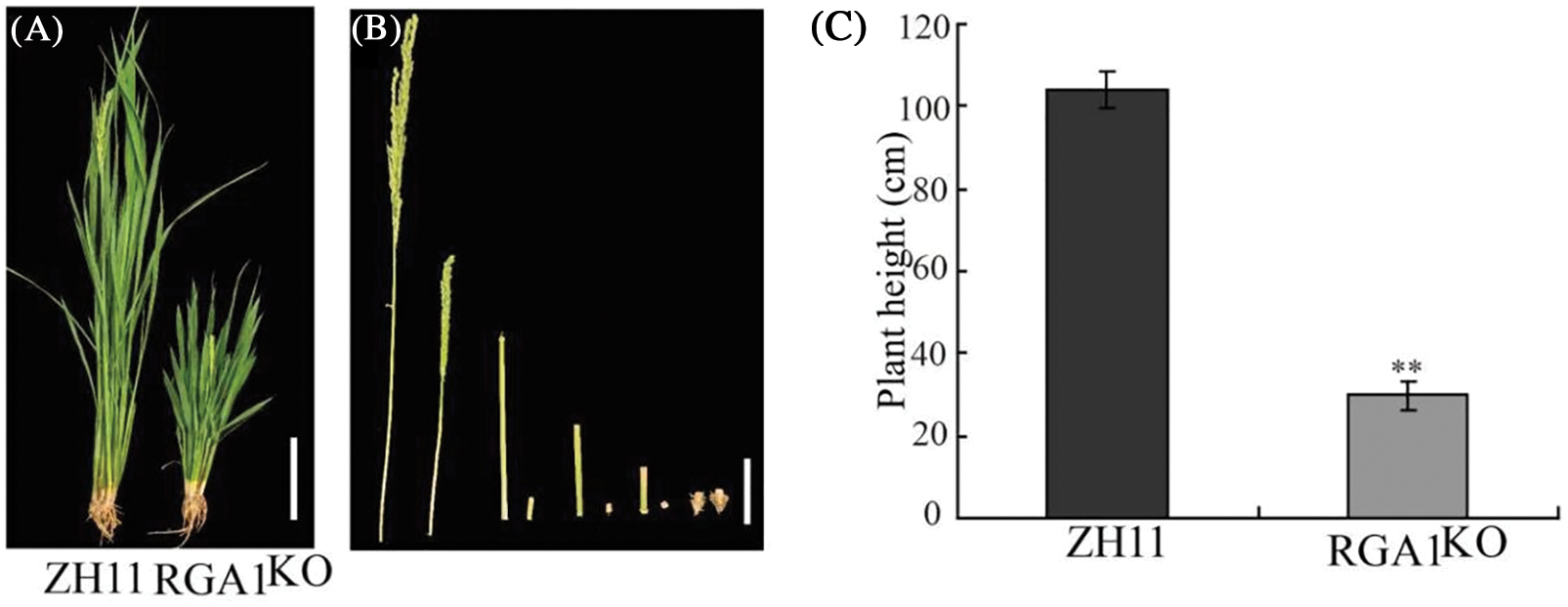
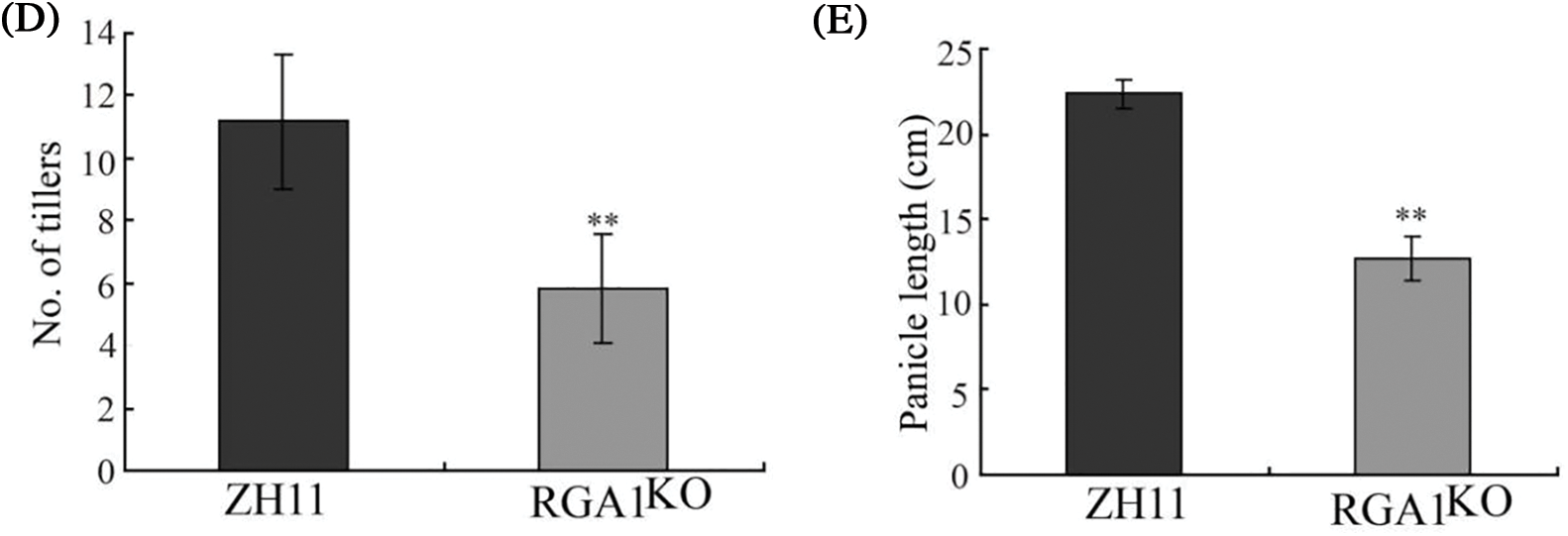
Figure 7: The RGA1KO mutant showed dwarfism and short panicles. (A) Phenotypes of Zhonghua 11 (ZH11) and the RGA1KO mutant at the heading stage. Bar = 10 cm. (B) The internodes of ZH11 (left) and the mutant (right). Bar = 2 cm. (C–E) Plant height, tiller number, and panicle morphology of ZH11 and the mutant. Data are shown as mean ± SD (n = 20). ** p < 0.01 by Student’s t-test analysis
3.7 Decreased Tolerance of ddg1 to Salt Stress Response
Microarray analysis showed that 889 up-regulated and 841 down-regulated genes related-salt stress between control and the RGA1 mutant d1, indicating that RGA1 may be involved in the regulation of salt stress in rice [23]. Therefore, we speculated that DDG1 might also be involved in the regulation of salt stress response in rice. To test this hypothesis, we treated the ddg1 mutant and wild type seedlings with 150 mM NaCl. Compared with the wild type, ddg1 seedlings were more sensitive to salt stress, and the survival frequencies of were approximately 22% and 62% under salt stress conditions in ddg1 and WT plants, respectively (Figs. 8A–8C). Consistent with the salt sensitivity of ddg1, knock-out of RGA1 resulted in a lower survival rate than that of wild-type plants (Fig. S7). Compared with ZH11, the expression level of RGA1 in knock-out of RGA1 plants was decreased (Fig. S8). These results showed that DDG1 and RGA1 both play important roles in rice salt tolerance.
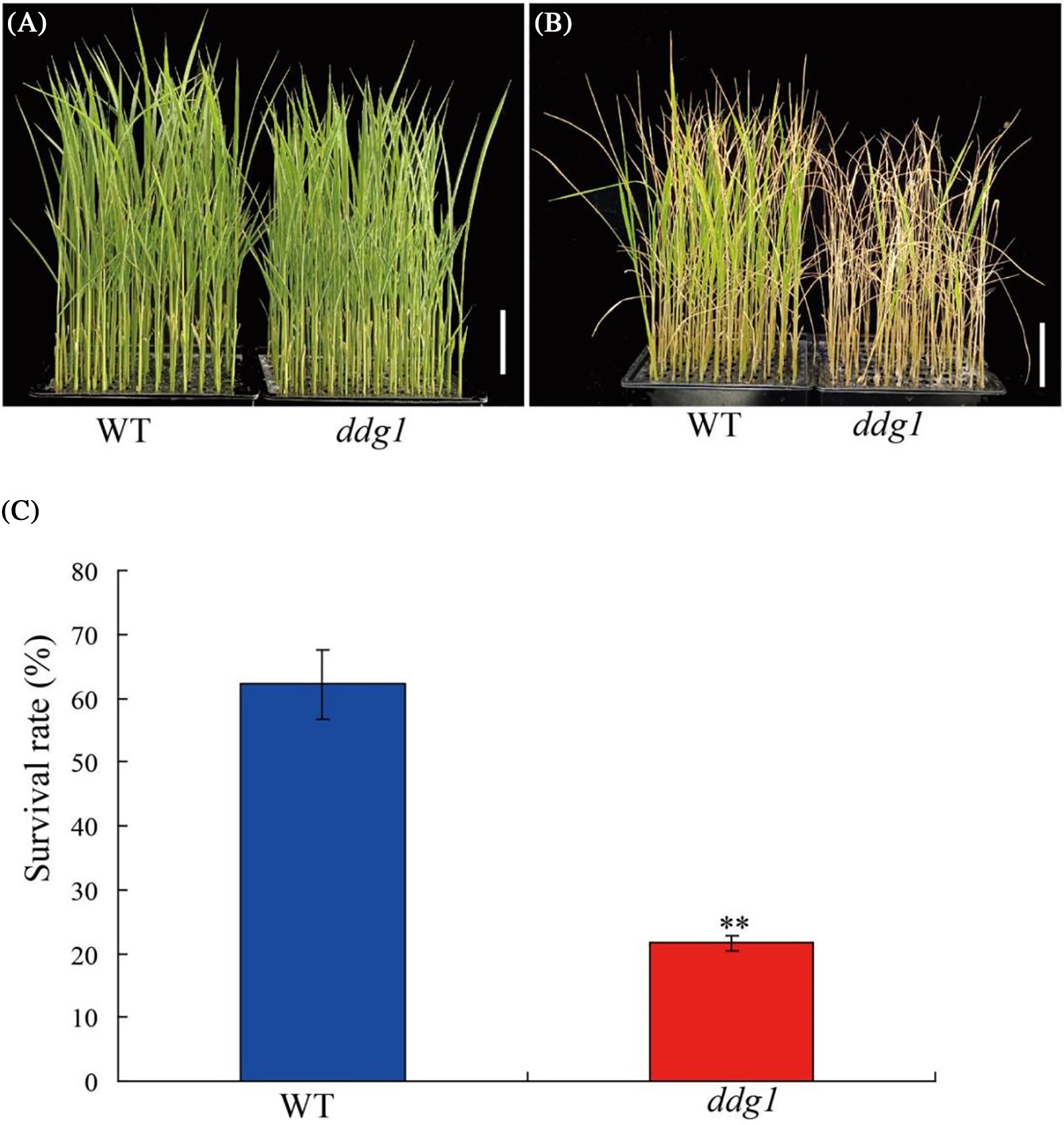
Figure 8: The ddg1 mutant was sensitive to salt stress at the seedling stage. (A and B) Phenotypes of the wild type (WT) and ddg1 before or after 150 mM NaCl treatment. Bar = 5 cm. (C) Survival rates of the WT and ddg1 after salt stress. Three biological repeats were performed with 96 individual plants per repeat. ** indicates a significant difference between WT and ddg1 at p < 0.01
Dwarfism and moderate delayed leaf senescence are essential for increasing rice yield [12]. To date, many genes related to plant height and leaf senescence have already been cloned, but few can be directly applied to rice breeding. In our study, we cloned a novel allele of DEP2, DDG1, which controls plant height and leaf senescence in rice. The DEP2/EP2 allele was first reported to be involved in the regulation of rice panicle erectness [21,24], and it encodes a novel, unknown plant-specific protein and is highly expressed in young panicles. In addition, another allele SRS1, has already been isolated as a regulator of grain size in rice [20]. The srs1 mutant exhibited small and round seeds, and the reduction of cell length and cell number in srs1 have been classified as its phenotype [20]. Along similar lines, our study showed that the reduction of cell number in stems leads to the dwarfism of ddg1. These results indicate that DEP2/EP2/SRS1/DDG1 all regulate cell expansion and cell proliferation. Recently, a cleistogamy gene cl7(t), which is an allele of DEP2, was identified by map-based cloning [25]. cl7(t) showed cleistogamy and closed spikelets, which might be used for the selection of ecologically safe genetically modified crops. Five DEP2 alleles have pleiotropic effects on rice growth and development.
Heterotrimeric G proteins play an important role in plant growth and developmental processes, including hormone responses, plant height, nitrogen-use efficiency, and seed size [26–28]. In Arabidopsis thaliana, there are one G protein α subunit (GPA1), one G protein β subunit (AGB1), and over three G protein γ subunits (AGG1, AGG2 and AGG3), but rice has one Gα gene (RGA1), one Gβ gene (RGB1) and five Gγ homologous genes (RGG1, RGG2, GS3/TT2, qPE9-1/DEP1 and GGC2) [29–31]. In addition, abnormal GPA1, AGB1 and AGG3 can lead to defects in Arabidopsis growth and development [32,33]. In rice, seven G protein subunits are well characterized and are mainly involved in regulating plant architecture, grain size, and stress responses [27,30,31,34]. RGA1/D1 was the first reported G protein in rice [35,36], and the RGA1 mutant d1 has a dwarf, darker-green leaves, and small-seed phenotype and is less sensitive to 24-epiBL and GA [37]. In our study, the phenotype of ddg1 was very similar to that of rga1/d1. Compared with the wild type plants, ddg1 was less sensitive to GA xand BR treatments, similar to rga1/d1. Furthermore, we found that DDG1 interacts with RGA1 from the results of our Y2H, BiFC and LCI assays. The interaction between DEP2 and RGA1 may lead to disruption of the function of the G protein complex, which is our next research focus.
In addition to its function that regulates plant height and senescence, DDG1 seems to play a vital role in controlling salt stress response due to the observed salt-stress-sensitive phenotype of the ddg1 mutant. The RGA1KO mutant was slightly more sensitive to salt stress than the wild type. Based on the DDG1-RGA1-mediated pathway having dual roles in regulating plant growth and salt stress response, it may be possible to manipulate these components to enhance rice yield under stress conditions. Conceivably, DDG1 has multiple biological functions and some unknown interacting proteins of DDG1 possibly still need to be identified in the future.
In conclusion, we isolated a new allele of DEP2, DDG1, which controls plant height and leaf senescence in rice. Y2H, BiFC and LCI assays showed that DDG1 interacted with RGA1. Compared with the wild type, ddg1 was more sensitive to salt stress. In future work, we intend to focus on the interaction proteins of DDG1 to clarify its mechanism of regulating rice’s salt stress response.
Acknowledgement: We thank Professor Yidan Ouyang and Doctor Shengyuan Sun for providing the RGA1KO mutant. The authors thank AiMi Academic Services (www.aimieditor.com) for the English language editing and review services.
Funding Statement: This work was supported by the Program for Huaishang Talents, Huai’an Academy of Agricultural Sciences Initiation and Development of Scientific Research Fund for High-Level Introduced Talents (0062019016B), Jiangsu Collaborative Innovation Center of Regional Modern Agriculture & Environmental Protection (HSXT30133), the Jiangsu Qinglan Project, the Hunan Province Natural Science Fund (2019JJ50714) and the Student Innovation Program of Jinagsu Province (202110323084Y, 202210323029Z).
Author Contributions: Xi Liu and Di Wang designed the project. Chuxuan Zhao, Gen Pan, Xiaonan Ji, Su Gao,Tanxiao Du, Yating Feng, and Wenjing Chen performed the experiments, data analysis and visualization. Xi Liu and Di Wang wrote and revised the manuscript.
Availability of Data and Materials: Sequence data from this article can be found in the GenBank/EMBL data libraries under the following accession numbers: DDG1/DEP2 (Os07g0616000); RGA1 (Os05g0333200); SGR (Os09g0532000); Osh36 (Os05g0475400).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Terao, T., Hirose, T. (2015). Control of grain protein contents through SEMIDWARF1 mutant alleles: Sd1 increases the grain protein content in Dee-geo-woo-gen but not in Reimei. Molecular Genetics and Genomics, 290(3), 939–954. [Google Scholar] [PubMed]
2. Ishimaru, K., Togawa, E., Ookawa, T., Kashiwagi, T., Madoka, Y. et al. (2008). New target for rice lodging resistance and its effect in a typhoon. Planta, 227(3), 601–609. [Google Scholar] [PubMed]
3. Sentoku, N., Sato, Y., Kurata, N., Ito, Y., Kitano, H. et al. (1999). Regional expression of the rice KN1-type homeobox gene family during embryo, shoot, and flower development. Plant Cell, 11(9), 1651–1664. [Google Scholar] [PubMed]
4. Yamamuro, C., Ihara, Y., Wu, X., Noguchi, T., Fujioka, S. et al. (2000). Loss of function of a rice brassinosteroid insensitive1 homolog prevents internode elongation and bending of the lamina joint. Plant Cell, 12(9), 1591–1606. [Google Scholar] [PubMed]
5. Itoh, H., Tatsumi, T., Sakamoto, T., Otomo, K., Toyomasu, T. et al. (2004). A rice semi-dwarf gene, Tan-Ginbozu (D35encodes the gibberellin biosynthesis enzyme, ent-kaurene oxidase. Plant Molecluar Biology, 54(4), 533–547. [Google Scholar] [PubMed]
6. Liu, X., Hu, Q., Yan, J., Sun, K., Liang, Y. et al. (2020). ζ-Carotene isomerase suppresses tillering in rice through the coordinated biosynthesis of strigolactone and abscisic acid. Molecualr Plant, 13(12), 1784–1801. [Google Scholar] [PubMed]
7. Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A. et al. (2002). Green revolution: A mutant gibberellin-synthesis gene in rice. Nature, 416(6882), 701–702. [Google Scholar] [PubMed]
8. Spielmeyer, W., Ellis, M. H., Chandler, P. M. (2002). Semidwarf (sd-1“green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proceedings of the National Academy of Sciences of the United States of America, 99(13), 9043–9048. [Google Scholar] [PubMed]
9. Belkhadir, Y., Jaillais, Y. (2015). The molecular circuitry of brassinosteroid signaling. New Phytologist, 206(2), 522–540. [Google Scholar] [PubMed]
10. Saini, S., Sharma, I., Pati, P. K. (2015). Versatile roles of brassinosteroid in plants in the context of its homoeostasis, signaling and crosstalks. Frontiers in Plant Science, 6, 950. [Google Scholar] [PubMed]
11. Morita, R., Sato, Y., Masuda, Y., Nishimura, M., Kusaba, M. (2009). Defect in non-yellow coloring 3, an α/β hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant Journal, 59(6), 940–952. [Google Scholar]
12. Liang, C., Wang, Y., Zhu, Y., Tang, J., Hu, B. et al. (2014). OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proceedings of the National Academy of Sciences of the United States of America, 111(27), 10013–10018. [Google Scholar] [PubMed]
13. Lim, P. O., Woo, H. R., Nam, H. G. (2013). Molecular genetics of leaf senescence in Arabidopsis. Trends in Plant Science, 8(6), 272–278. [Google Scholar]
14. Kong, Z., Li, M., Yang, W., Xu, W., Xue, Y. (2006). A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiology, 141(4), 1376–1388. [Google Scholar] [PubMed]
15. Wu, L., Ren, D., Hu, S., Li, G., Dong, G. et al. (2016). Down-regulation of a nicotinate phosphoribosyltransferase gene, OsNaPRT1, leads to withered leaf tips. Plant Physiology, 171(2), 1085–1098. [Google Scholar] [PubMed]
16. Shin, N. H., Trang, D. T., Hong, W. J., Kang, K., Chuluuntsetseg, J. et al. (2019). Rice senescence-induced receptor-like kinase (OsSRLK) is involved in phytohormone-mediated chlorophyll degradation. International Journal of Molecular Sciences, 21(1), 260. [Google Scholar] [PubMed]
17. Sun, S. Y., Wang, L., Mao, H., Shao, L., Li, X. et al. (2018). A G-protein pathway determines grain size in rice. Nature Communications, 9(1), 851. [Google Scholar] [PubMed]
18. Liu, X., Xu, Z. Y., Yang, Y. R., Cao, P. H., Cheng, H. et al. (2021). Plastid caseinolytic protease OsClpR1 regulates chloroplast development and chloroplast RNA editing in rice. Rice, 14(1), 45. [Google Scholar] [PubMed]
19. Kudla, J., Bock, R. (2016). Lighting the way to protein-protein interactions: Recommendations on best practices for bimolecular fluorescence complementation analyses. Plant Cell, 28(5), 1002–1008. [Google Scholar] [PubMed]
20. Abe, Y., Mieda, K., Ando, T., Kono, I., Yano, M. et al. (2010). The SMALL AND ROUND SEED1 (SRS1/DEP2) gene is involved in the regulation of seed size in rice. Genes & Genetic Systems, 85(5), 327–339. [Google Scholar]
21. Li, F., Liu, W., Tang, J., Chen, J., Tong, H. et al. (2010). Rice DENSE AND ERECT PANICLE 2 is essential for determining panicle outgrowth and elongation. Cell Research, 20(7), 838–849. [Google Scholar] [PubMed]
22. Sanger, F., Air, G. M., Barrell, B. G., Brown, N. L., Coulson, A. R. et al. (1977). Nucleotide sequence of bacteriophage φX174 DNA. Nature, 265(5596), 687–695. [Google Scholar] [PubMed]
23. Jangam, A. P., Pathak, R. R., Raghuram, N. (2016). Microarray analysis of rice d1 (RGA1) mutant reveals the potential role of G-protein alpha subunit in regulating multiple abiotic stresses such as drought, salinity, heat, and cold. Frontiers in Plant Science, 7, 11. [Google Scholar] [PubMed]
24. Zhu, K., Tang, D., Yan, C., Chi, Z., Yu, H. et al. (2010). Erect panicle2 encodes a novel protein that regulates panicle erectness in indica rice. Genetics, 184(2), 343–350. [Google Scholar] [PubMed]
25. Ni, D. H., Li, J., Duan, Y. B., Yang, Y. C., Wei, P. C. et al. (2014). Identification and utilization of cleistogamy gene cl7(t) in rice (Oryza sativa L.). Journal of Experimental Botany, 65(8), 2107–2117. [Google Scholar] [PubMed]
26. Jin, X., Wang, R. S., Zhu, M., Jeon, B. W., Albert, R. et al. (2013). Abscisic acid-responsive guard cell metabolomes of Arabidopsis wild-type and gpa1 G-protein mutants. Plant Cell, 25(12), 4789–4811. [Google Scholar] [PubMed]
27. Sun, H., Qian, Q., Wu, K., Luo, J., Wang, S. et al. (2014). Heterotrimeric G proteins regulate nitrogen-use efficiency in rice. Nature Genetics, 46(6), 652–656. [Google Scholar] [PubMed]
28. Miao, J., Yang, Z., Zhang, D., Wang, Y., Xu, M. et al. (2019). Mutation of RGG2, which encodes a type B heterotrimeric G protein γ subunit, increases grain size and yield production in rice. Plant Biotechnology Journal, 17(3), 650–664. [Google Scholar] [PubMed]
29. Temple, B. R., Jones, A. M. (2007). The plant heterotrimeric G-protein complex. Annual Review of Plant Biology, 58, 249–266. [Google Scholar] [PubMed]
30. Tao, Y., Miao, J., Wang, J., Li, W., Xu, Y. et al. (2020). RGG1, involved in the cytokinin regulatory pathway, controls grain size in rice. Rice, 13, 76. [Google Scholar] [PubMed]
31. Kan, Y., Mu, X. R., Zhang, H., Gao, J., Shan, J. X. et al. (2022). TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nature Plants, 8(1), 53–67. [Google Scholar] [PubMed]
32. Ullah, H., Chen, J. G., Young, J. C., Im, K. H., Sussman, M. R. et al. (2001). Modulation of cell proliferation by heterotrimeric G protein in Arabidopsis. Science, 292(5524), 2066–2069. [Google Scholar] [PubMed]
33. Li, S., Liu, Y., Zheng, L., Chen, L., Li, N. et al. (2012). The plant-specific G protein γ subunit AGG3 influences organ size and shape in Arabidopsis thaliana. New Phytologist, 194(3), 690–703. [Google Scholar] [PubMed]
34. Swain, D. M., Sahoo, R. K., Srivastava, V. K., Tripathy, B. C., Tuteja, R. et al. (2017). Function of heterotrimeric G-protein γ subunit RGG1 in providing salinity stress tolerance in rice by elevating detoxification of ROS. Planta, 245(2), 367–383. [Google Scholar] [PubMed]
35. Seo, H. S., Kim, H. Y., Jeong, J. Y., Lee, S. Y., Cho, M. J. et al. (1995). Molecular cloning and characterization of RGA1 encoding a G protein α subunit from rice (Oryza sativa L. IR-36). Plant Molecluar Biology, 27(6), 1119–1131. [Google Scholar] [PubMed]
36. Ashikari, M., Wu, J. Z., Yano, M., Sasaki, T., Yoshimura, A. (1999). Rice gibberellin-insensitive dwarf mutant gene Dwarf 1 encodes the α-subunit of GTP-binding protein. Proceedings of the National Academy of Sciences of the United States of America, 96(18), 10284–10289. [Google Scholar] [PubMed]
37. Wang, L., Xu, Y. Y., Ma, Q. B., Li, D., Xu, Z. H. et al. (2006). Heterotrimeric G protein α subunit is involved in rice brassinosteroid response. Cell Research, 16(12), 916–922. [Google Scholar] [PubMed]
Appendix
Supplementary Figure S1: Scanning electron microscope observation of the wild type and ddg1 stems
Supplementary Figure S2: Amino acid sequence alignment of DDG1 and ddg1
Supplementary Figure S3: The phenotype of transgenic negative plant
Supplementary Figure S4: Expression pattern of DDG1 based on the RiceXPro (https://ricexpro.dna.affrc.go.jp/Zapping/)
Supplementary Figure S5: Amino acid sequence alignment of DDG1 and its homologous proteins
Supplementary Figure S6: DDG1 interacts with RGA1 in yeast cell
Supplementary Figure S7: The RGA1 mutant was sensitive to salt stress in rice
Supplementary Figure S8: The expression level of RGA1 in the mutant
Supplementary Table S1: Primers used in this study
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools