 Open Access
Open Access
ARTICLE
TCD5 Enhances the Photosynthesis Capacity, Increases the Panicle Number and the Yield in Rice
1 Zhejiang Provincial Key Laboratory of Biometrology and Inspection & Quarantine, College of Life Sciences, China Jiliang University, Hangzhou, 310018, China
2 CAS Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai, 200032, China
3 Institute of Virology and Biotechnology, Zhejiang Academy of Agricultural Sciences, Hangzhou, 310021, China
4 Agrotechnical Extension Station in Tiantai County, Taizhou, 317200, China
5 College of Life Sciences, Shanghai Normal University, Shanghai, 200234, China
* Corresponding Authors: Yanjun Dong. Email: ; Sheng Teng. Email:
(This article belongs to the Special Issue: High-Yield Rice Physiology & Genetics)
Phyton-International Journal of Experimental Botany 2023, 92(9), 2649-2663. https://doi.org/10.32604/phyton.2023.030710
Received 19 April 2023; Accepted 19 May 2023; Issue published 28 July 2023
Abstract
Improvement of photosynthetic efficiency is a major approach to increase crop yield potential. Previously, we cloned a gene encoding the chloroplast-located putative monooxygenase TCD5, which is essential in plastid development under low temperature in rice (Oryza sativa L.). In this study, the effects of TCD5 on the photosynthesis and the yields were investigated in rice. Two sets of genetic materials with three levels of TCD5 expression, including tcd5 mutant or TCD5 RNAi transgenic lines and TCD5 over-expression transgenic lines in Jiahua1 and Nipponbare backgrounds, were used in the field trails of multi-locations and multi-years. TCD5 positively affected the panicle number and the yield at dosage. Compared with the wild-types, the panicle numbers were 12.4%–14.6% less in tcd5 mutant and 8.3%–38.6% less in TCD5 RNAi lines, but 26.2%–61.8% more in TCD5 over-expression lines. The grain yields per plant were 9.1%–18.4% less in tcd5 mutant and 14.3%–56.7% less in TCD5 RNAi lines, but 6.9%–56.5% more in TCD5 over-expression lines. The measurements of net photosynthetic rate indicated that mutation or knock down of TCD5 decreased the net photosynthetic rate by 10.4% and 15.6%, respectively, while increasing it by 8.9% and 8.7% in the TCD5 over-expression lines in Jiahua1 and Nipponbare backgrounds, respectively. Accordingly, the measurements of chlorophyll fluorescence parameters showed that the electron transport rate and quantum yield decreased in tcd5 mutant or TCD5 RNAi lines but increased in TCD5 over-expression lines, both in Jiahua1 and Nipponbare backgrounds. IP-MS screening revealed that TCD5 interacts with 29 chloroplast proteins involved in chlorophyll synthesis, photo-reactions of the photosynthesis, carbon assimilation and metabolism, energy metabolism, redox balance, protein synthesis and transportation. Two TCD5 interacted proteins, D1 and FBA were effective targets for improving photosynthesis. These results suggest a potentially new strategy for increasing rice yield by enhancing photosynthesis.Keywords
Rice is the staple food for half of the world’s population, providing 20% of the world’s dietary energy supply and up to 70% in Southeast Asia. Therefore, it is closely linked to global food security [1]. Furthermore, rice is considered an important strategic commodity due to its close connection with employment, economic growth, social stability even regional peace. The demand for increasing rice production has always existed in order to meet the needs of a growthing world population [2]. Great progress, including three qualitative leaps for increasing rice yield has been obtained since the 1950s [3]. In the 1950s, the use of the semi-dwarf 1 (sd1) gene produced the first green revolution in rice [3,4]. In the 1970s, the utilization of the heterosis to develop hybrid rice made the second breakthrough in rice yield [3]. In the 1990s, the utilization of ideal plant architecture, combined with the heterosis between subspecies, led to the third breakthrough [3,5]. The focus of improvement over the past 70 years has mainly been on morphology, which is related to source-sink partition and canopy configuration [3–5]. However, per unit yield has effectively plateaued in China, Japan, Korea, and Indonesia [6], which implies that this strategy had reached its limit.
Improving physiological characteristics, such as photosynthetic efficiency, is another potential option for increasing yield [7,8]. Some convincing evidence has been approved that crop yield can be improved by enhancing photosynthesis [9–11]. Transgenic tobacco increased the protein levels of sedoheptulose bisphosphatase (SBPase), fructose-1, 6-bisphosphate aldolase (FBA), and cyanobacterial putative-inorganic carbon transporter B (ictB), which in turn increased the rate of photosynthetic carbon assimilation, leaf area, and biomass yield [12]. Over-expression of violaxanthin de-epoxidase (VDE), zeaxanthin epoxidase (ZEP), and PsbS affects the rate of NPQ adjustment, increases the speed of recovery from photoprotection, and results in increased leaf carbon dioxide uptake and plant dry matter productivity in tobacco [13], as well as soybean yield [14]. Engineering photorespiratory bypasses by multigene transformation in Arabidopsis, tobacco, and rice substantially increased the photosynthetic efficiency and the product [15–17]. The nuclear origin supplementation of the Photosystem II protein D1 driven by a heat-responsive promoter significantly enhanced net CO2 assimilation rates with increases in biomass and grain yield [18]. Recently, the over-expression of OsDREB1C and maize transcription factor mEmBP-1 has been shown to increase photosynthesis efficiency, biomass, and yield [19,20]. Knocking out the transcription factor NEGATIVE REGULATOR OF PHOTOSYNTHESIS 1 also increased leaf photosynthesis and biomass production in the field of rice [21]. Progress has as well been made in exploring excellent alleles in natural variations of photosynthesis in rice. The partially functional allele of the Narrow Leaf 1 gene from a high-yield variety Takanari increased the photosynthesis rate [22,23]. This elite allele optimized the balance between leaf photosynthesis and plant architecture to increase the rice yield [22].
Previously, we cloned a thermal-dependent chlorophyll-deficient mutant, tcd5, which encodes a putative monooxygenase located in the chloroplast [24]. In this study, we found that TCD5 increased photosynthetic efficiency, panicle number, and grain yield in rice. Furthermore, we discovered that TCD5 interacts with proteins involved in chlorophyll synthesis, photo-reaction of photosynthesis, carbon assimilation and metabolism, energy metabolism, redox balance, protein synthesis, and transportation, which could contribute to the effect of TCD5 on photosynthesis.
Two sets of genetic materials were used in this study in Jiahua1 and Nipponbare background (Oryza sativa L.). The transgenic lines with CaMV35S driven TCD5 fused with MYC tag in tcd5 mutant (TCD5-M-6 and TCD5-M-3) and the TCD5 RNAi in Nipponbare (TCD5-RNAi-J-3 and TCD5-RNAi-J-4) were generated as described [24]. The construct of CaMV35S driven TCD5 was also transferred into Nipponbare by Agrobacterium method used previously [24]. The homozygous T2 generation seeds were collected from the positively screened T0 and T1 generation plants. Two independent TCD5 over-expression lines (TCD5-OE-J-6 and TCD5-OE-J-7) were used for further experiments.
The field trails were performed at Shanghai and Hainan, China from 2020 to 2022. The plants of wild-type Jiahua1, tcd5 mutant, TCD5-M-6 and TCD5-M-3 were cultivated on the paddy field from December 2021 to April 2022 at Hainan and from May to October 2022 at Shanghai. The plants of wild-type Nipponbare, TCD5-RNAi-J-3, TCD5-RNAi-J-4, TCD5-OE-J-6, TCD5-OE-J-7 were cultivated on the paddy field from May to October in 2020 and 2022 respectively at Shanghai. We planted seedlings in 15 rows with a distance of 20 cm; each row contained 10 seedlings with a distance of 20 cm. For net photosynthetic and chlorophyll fluorescence parameters measurement, the plants from the field were moved into the environmentally controlled phytotron (28°C day and 25°C night, 1200 µmol photons m–2 s–1 photosynthetic photon flux density, 16 h : 8 h light : dark photoperiod, and ~75% relative humidity). The plants at early tillering stage were taken from the field with soil into 6 liter pots and moved in phytotron for recovering for 7 days.
2.3 Measurement of Net Photosynthetic CO2 Uptake Rate
A LI6400 Portable Photosynthesis System (LiCor, Lincoln, NE, USA) was used to measure the leaf gas exchange of the second leaf of the plants at tillering stage. Measurements were performed between 10:00 and 13:30. The measurements were finished within 2 days for one set materials. Net photosynthetic CO2 uptake rate of each line was measured more than twenty duplicates.
2.4 Measurement of Chlorophyll Fluorescence Parameters
Chlorophyll fluorescence parameters were measured using the second leaf of the plants at 25°C with a pulse amplitude modulated fluorimeter (DUAL-PAM101, Walz) [25]. The quantum yield and the photosynthetic electron transfer rates were measured and calculated following the literature [26–28].
Grain yield and its components (panicle number per plant, spikelet number per panicle, filled grain number per panicle and 1000-grain weight) were measured with 30 plants randomly selected from the inside of the plots per line.
The Student’s t-test was used to analysis the significance of the difference between the wild-type and other materials using Microsoft Excel software (Microsoft Inc., USA). The p-values are shown in the figures (*p < 0.05, **p < 0.01).
2.7 RNA Isolation, Reverse Transcription and Quantitative PCR
Total RNA was extracted from leaves of the plants at the tillering stage using the TRIzol Reagent (15596026, Invitrogen) according to the manufacturer’s instructions. The first-strand cDNA was synthesized by using First Strand cDNA Synthesis Kit (Toyobo, Japan). qRT-PCR was conducted in Stratagene Mx3000P (Agilent Technologies) with the SYBR Green Mix (TaKaRa, Japan) with specific primers as described [24]. The PCR cycling conditions were 95°C for 10 min, followed by 40 cycles of denaturation at 95°C for 15 s, annealing and elongation at 60°C for 60 s. The 2−∆∆CT method was used to analyze the target genes’ relative expression levels. Three biological replicates were used per sample and the rice ACTIN1 (LOC_Os03g50885) was used as an internal control for normalization [29].
IP-MS was performed as previously described [30] with minor modification. Briefly, proteins were isolated from the rice leaves of Jiahua1 and TCD5-M-6. The MYC antibody was diluted 30 times in the Co-IP buffer. Approximately 1 mg of the Jiahua1 and TCD5-M-6 protein was incubated with 300 μl of diluted MYC antibody, respectively. A 40-μL aliquot of 50% washed protein G-agarose beads (Santa Cruz Biotechnology) was added to the antigen–antibody complex. The solubilized proteins were immunoprecipitated with anti-MYC protein G-agarose beads. After washing, the proteins remaining on the beads were re-solubilized in 20 μL of SDS-loading buffer and then separated via SDS-PAGE. The stained gel lanes were cut into slices. LC-MS/MS were used to analyze the tryptic peptides, and the MASCOT software was used to compare the MS data with the Uniprot protein database. The chloroplast proteins were identified according to the annotation of Uniprot.
3.1 Expression of the TCD5 Gene in Two Sets of Rice Varieties at Tillering Stage
The TCD5 expression levels in Jiahua1 wild-type and tcd5 mutant together with two independent transgenic lines with CaMV35S driven TCD5 in tcd5 mutant were measured at early tillering stage. The TCD5 expression level of tcd5 mutant is about 12.6% of Jiahua1 wild-type. The TCD5 expression level in two transgenic lines TCD5-M-6 and TCD5-M-3 were 10.17 and 6.69 times of Jiahua1 wild-type, respectively (Fig. 1A).
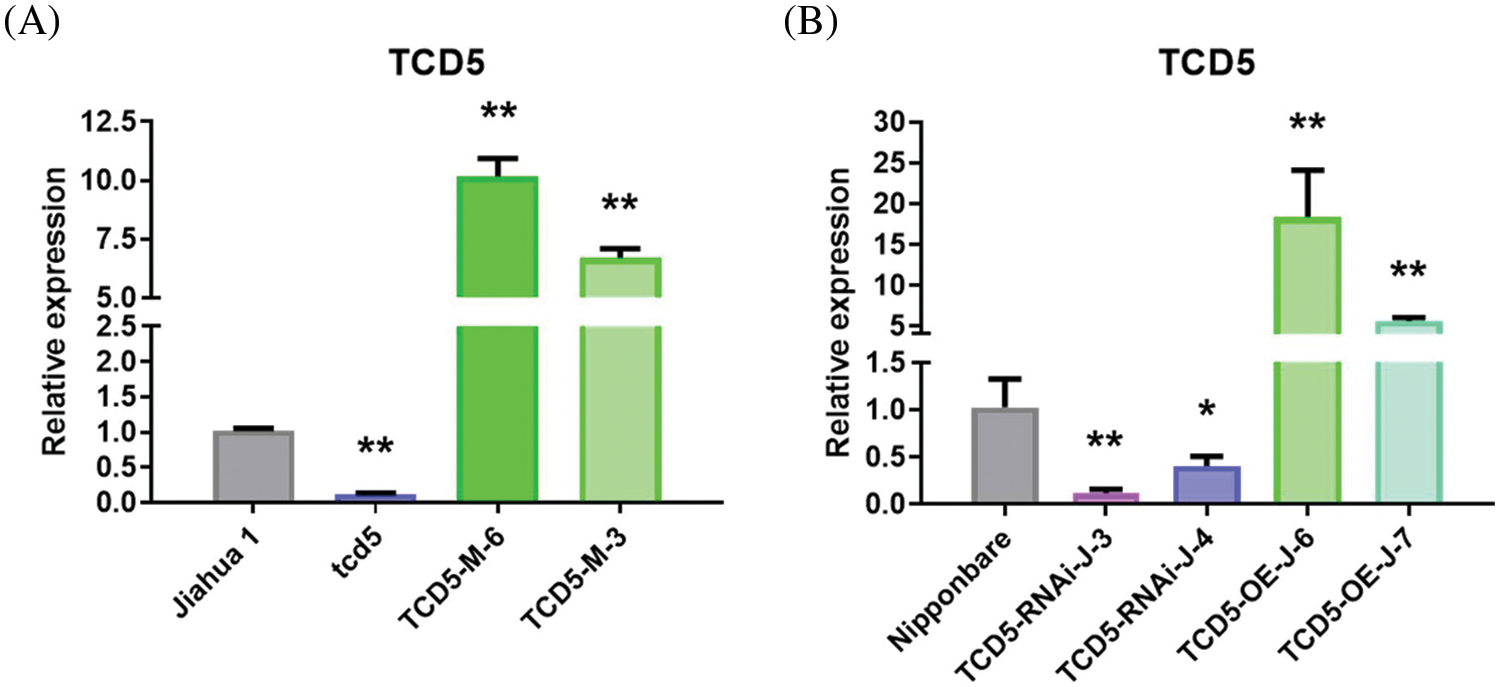
Figure 1: The expression levels of TCD5 in WT, tcd5 mutant or TCD5 RNAi lines, and TCD5 over-expression lines in Jiahua1 and Nipponbare background. (A) The expression levels of TCD5 in Jiahua1, tcd5 mutant and CaMV35S driven TCD5 in tcd5 mutant. (B) The expression levels of TCD5 in Nipponbare, TCD5 RNAi lines and over-expression line. The values are shown as means ± SD of triplicate experiments. Asterisks indicate significant difference between wild-type and other genetic materials, as determined by Student’s t-test (*p < 0.05, **p < 0.01)
The TCD5 expression levels in Nipponbare wild-type, two independent TCD5 RNAi lines and two independent TCD5 over-expression transgenic lines were also measured at tillering stage. The TCD5 expression level in TCD5 RNAi lines TCD5-RNAi-J-3 and TCD5-RNAi-J-4 are about 11.8% and 40.5% respectively of the Nipponbare wild-type. The TCD5 expression level in two transgenic lines TCD5-OE-J-6 and TCD5-OE-J-7 were 18.33 and 5.59 times respectively of that of Nipponbare wild-type (Fig. 1B).
3.2 TCD5 Increases the Panicle Number and the Yield
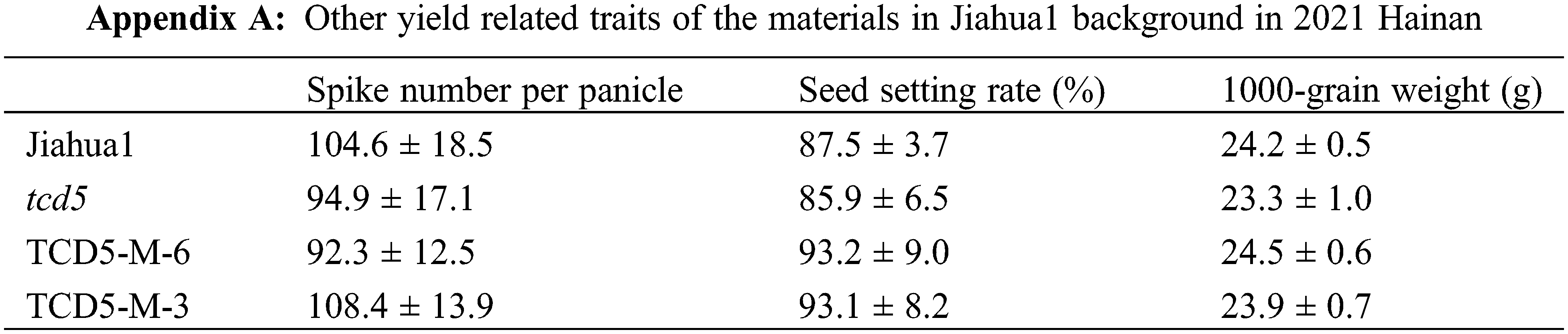
The materials in Jiahua1 background, including Jiahua1 wild-type, tcd5 mutant and two TCD5 over-expression lines (TCD5-M-6 and TCD5-M-3) were planted in Hainan 2021 and Shanghai 2022, respectively. In 2021, Hainan, the panicle number of tcd5 mutant was reduced 12.4% compared with that of Jiahua1. Two TCD5 over-expression lines, TCD5-M-6 and TCD5-M-3 increased 46.4% and 38.2% respectively compared with that of Jiahua1 wild-type (Fig. 2A). The yield of tcd5 mutant was reduced 18.4% compared with that of Jiahua1. In two TCD5 over-expression lines, TCD5-M-6 and TCD5-M-3 the grain yields increased 46.2% and 29.7% respectively compared with that of Jiahua1 wild-type (Fig. 2B). In 2022, Shanghai, the panicle number of tcd5 mutant was reduced 14.6% compared with that of Jiahua1, two TCD5 over-expression lines, TCD5-M-6 and TCD5-M-3 increased 43.1% and 26.2% respectively compared with that of Jiahua1 wild-type (Fig. 2C). In 2022, Shanghai, the yield of tcd5 mutant was reduced 14.6% compared with that of Jiahua1, the yields of two TCD5 over-expression lines, TCD5-M-6 and TCD5-M-3 increased 18.9% and 6.9% respectively compared with that of Jiahua1 wild-type (Fig. 2D). Other yield traits such as spike number per panicle, seed setting rate and1000-grain weight were not affected by TCD5 (Appendixes A--B).
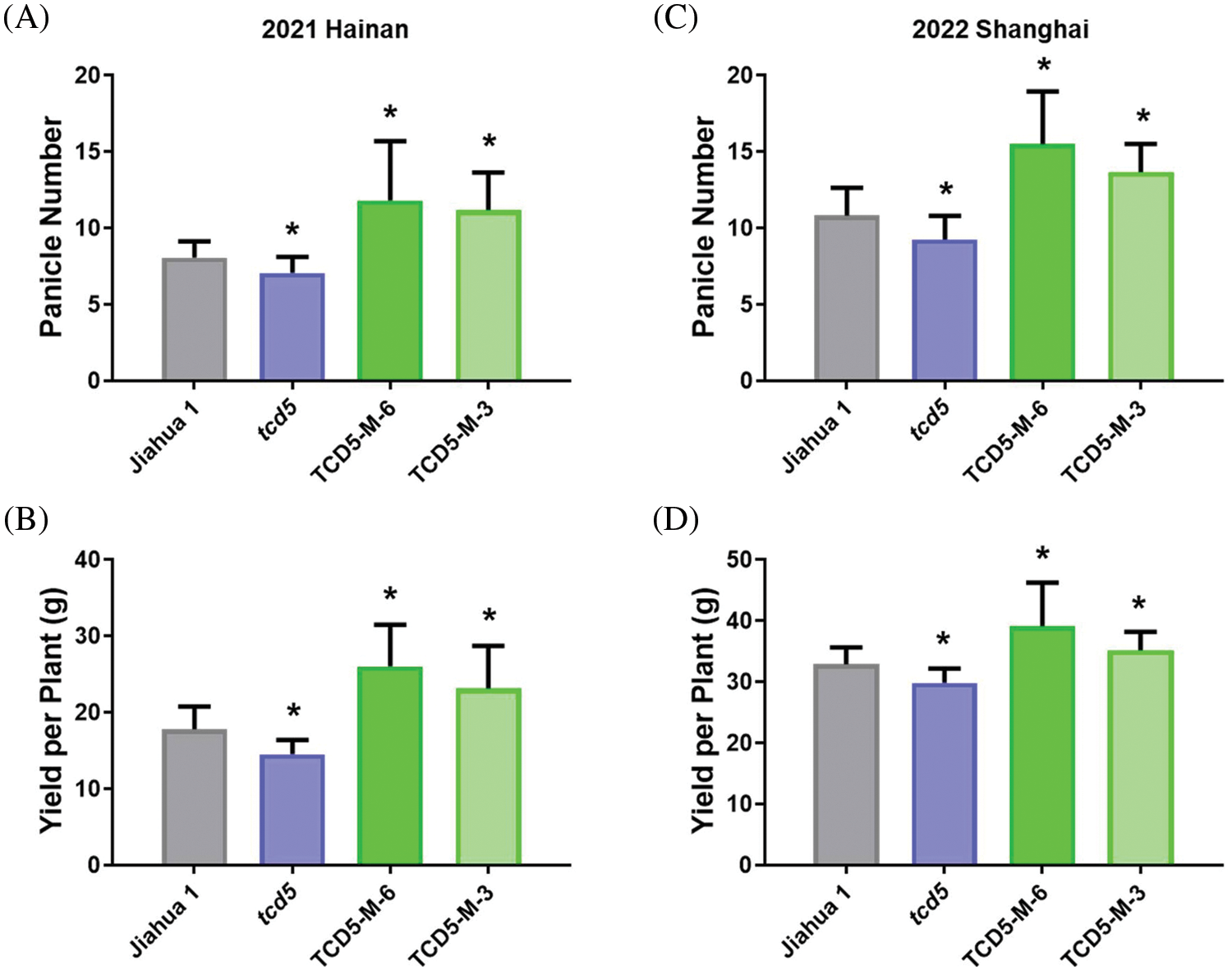
Figure 2: The panicle number and the yield of materials in Jiahua1 background. (A) Panicle numbers of Jiahua1, tcd5 mutant and two TCD5 over-expression lines at Hainan in 2021. (B) Yields of Jiahua1, tcd5 mutant and two TCD5 over-expression lines at Hainan in 2021. (C) Panicle numbers of Jiahua1, tcd5 mutant and two TCD5 over-expression lines at Shanghai in 2022. (D) Yields of Jiahua1, tcd5 mutant and two TCD5 over-expression lines at Shanghai in 2022. The values are shown as means ± SD of 30 plants. Asterisks indicate significant difference between wild-type and other genetic materials, as determined by Student’s t-test (*p < 0.05)
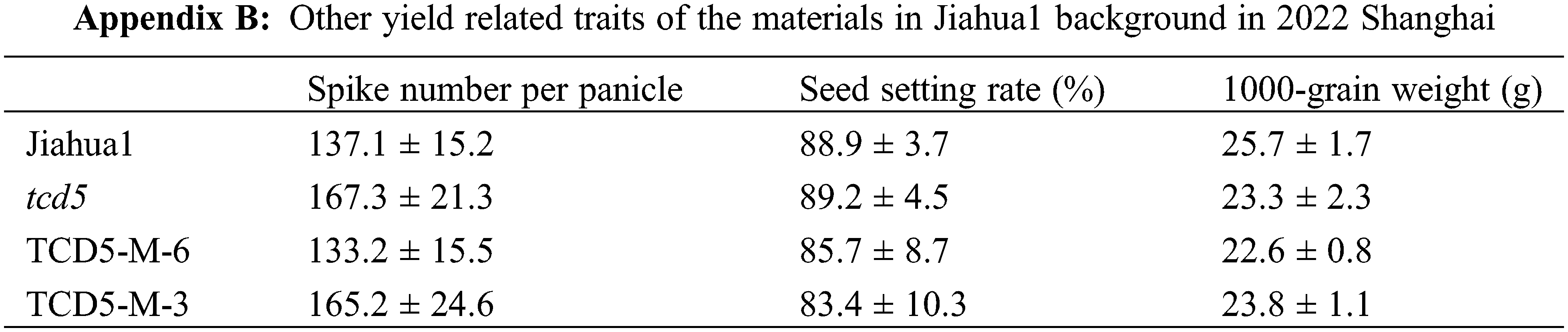
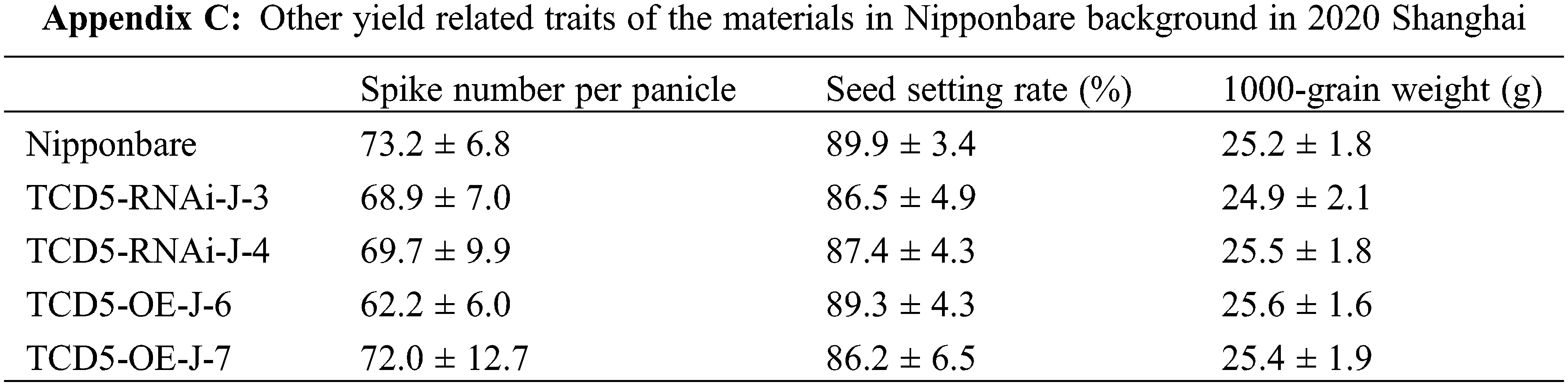
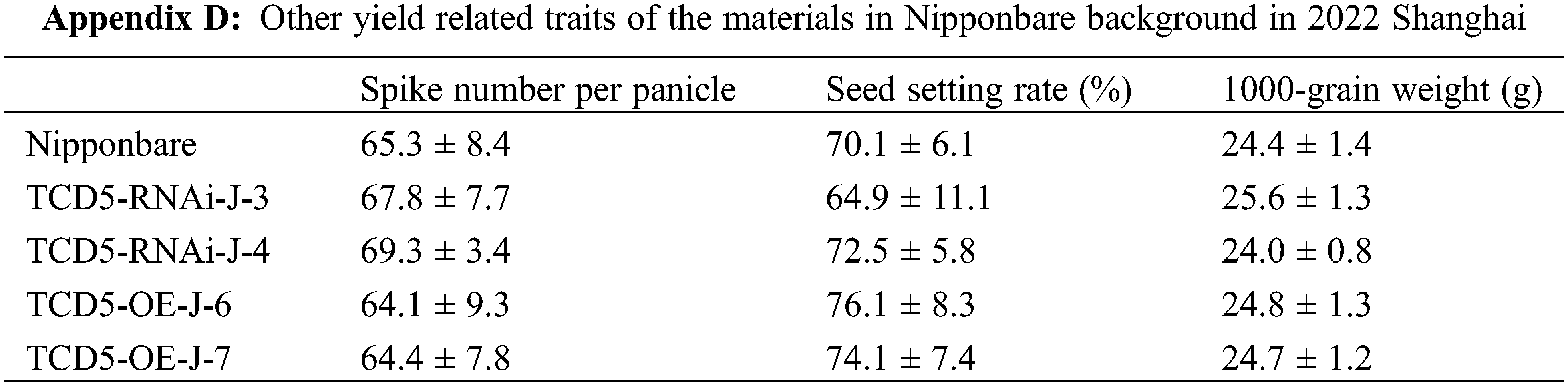
The materials in Nipponbare background, including Nipponbare wild-type, two TCD5 RNAi lines (TCD5-RNAi-J-3 and TCD5-RNAi-J-4) and two TCD5 over-expression lines (TCD5-OE-J-6 and TCD5-OE-J-7) were planted in Shanghai in 2020 to 2022, respectively. In 2020, Shanghai, the panicle number of the two TCD5 RNAi lines, TCD5-RNAi-J-3 and TCD5-RNAi-J-4, were less 30.6% and 38.6% compared with that of Nipponbare (Fig. 3A), and the grain yield reduced 56.7% and 38.7%, respectively (Fig. 3B). The panicle number the two TCD5 over-expression lines, TCD5-OE-J-6 and TCD5-OE-J-7, increased 55.9% and 61.9% respectively compared with Nipponbare wild-type (Fig. 3A). The grain yield per plant of TCD5-RNAi-J-3 and TCD5-RNAi-J-4 were significantly 56.7% and 38.7% respectively lower than that Nipponbare wild-type. The grain yields per plant of TCD5-OE-J-6 and TCD5-OE-J-7 are significantly 53.0% and 56.5% respectively higher than that Nipponbare wild-type (Fig. 3B). In 2022 Shanghai, the panicle number of the two TCD5 RNAi lines, TCD5-RNAi-J-3 and TCD5-RNAi-J-4, were less 11.0% and 8.3% compared with that of Nipponbare (Fig. 3C), and the grain yield reduced 14.9% and 18.9%, respectively (Fig. 3D). The panicle number of the two TCD5 over-expression lines, TCD5-OE-J-6 and TCD5-OE-J-7, increased 37.8% and 43.3% respectively more panicles than Nipponbare wild-type (Fig. 3C) the grain yield per plant of TCD5-OE-J-6 and TCD5-OE-J-7, were significantly 56.7% and 38.7% respectively lower than that Nipponbare wild-type. The grain yields per plant of TCD5-OE-J-6 and TCD5-OE-J-7 are significantly 27.4% and 18.3% respectively higher than that Nipponbare wild-type (Fig. 3D). These results indicated that TCD5 increases the panicle number and the yield. Other yield traits such as spike number per panicle, seed setting rate and 1000-grain weight were not affected by TCD5 (Appendixes C--D).
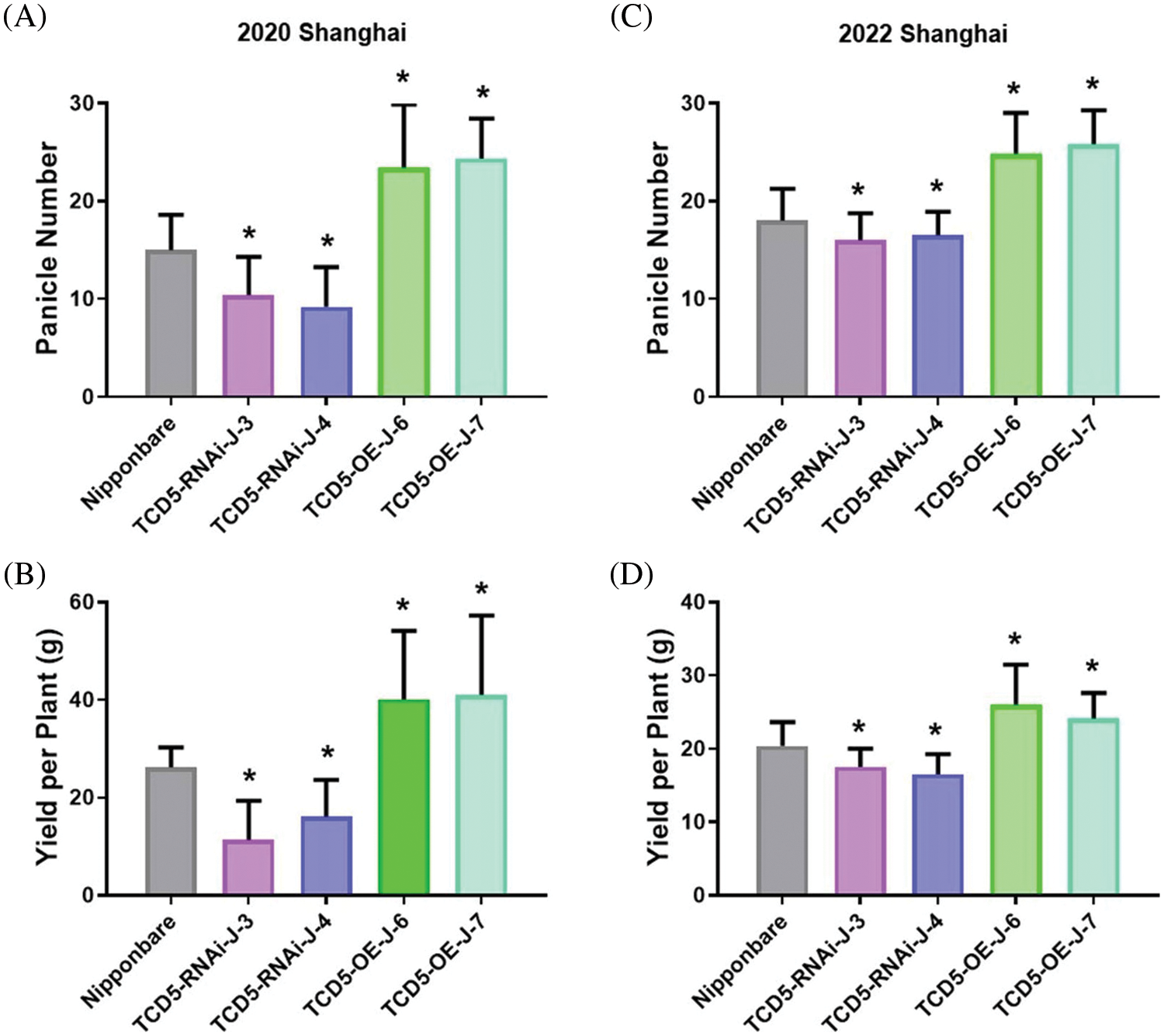
Figure 3: The panicle number and the yield of the materials in Nipponbare background. (A) Panicle numbers of Nipponbare, two TCD5 RNAi lines and two TCD5 over-expression lines at Shanghai in 2020. (B) Yields of Nipponbare, two TCD5 RNAi lines and two TCD5 over-expression lines at Shanghai in 2020. (C) Panicle numbers of Nipponbare, two TCD5 RNAi lines and two TCD5 over-expression lines at Shanghai in 2022. (D) Yields of Nipponbare, two TCD5 RNAi lines and two TCD5 over-expression lines at Shanghai in 2022. The values are shown as means ± SD of 30 plants. Asterisks indicate significant difference between wild-type and other genetic materials, as determined by Student’s t-test (*p < 0.05)
3.3 TCD5 Enhances the Photosynthesis Performance at Early Tillering Stage
The net photosynthetic rate of the youngest fully expanded leaves at tillering stage was measured in the genetic materials in Jiahua1 background (Jiahua1, tcd5 and TCD5-M-6) and Nipponbare (Nipponbare, TCE5-RNAi-J-3 and TCD5-OE-J-7). The net photosynthetic rate of tcd5 was 10.4% lower than that of Jiahua1, whereas that of TCD5-M-6 was 8.9% higher than that of Jiahua1 (Fig. 4A). In the materials in Nipponbare background, the net photosynthetic rate of TCD5-RNAi-J-3 was 15.6% lower than that of Nipponbare, whereas that of TCD5-M-6 was 8.7% higher than that of Nipponbare (Fig. 4B).
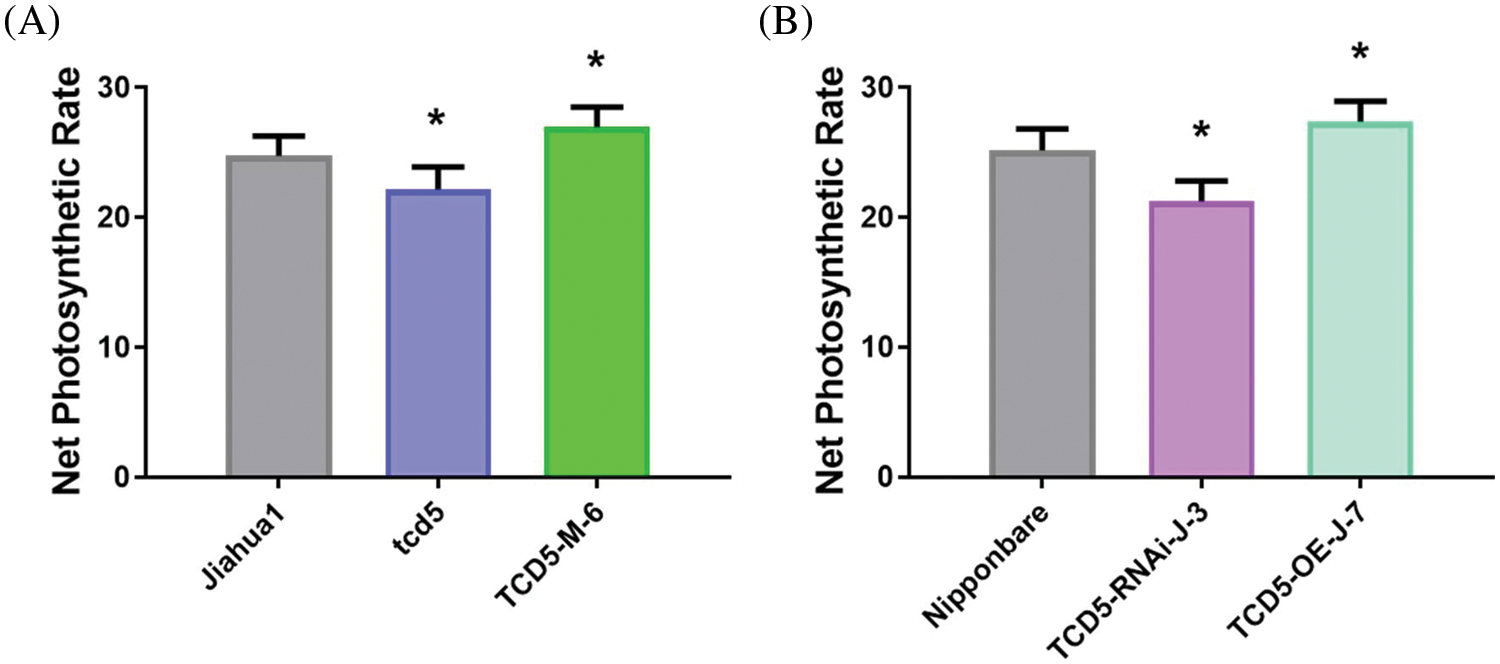
Figure 4: The net photosynthetic rate of WT, mutant or RNAi lines, and over-expression lines in Jiahua1 and Nipponbare background at tillering stage. The values are shown as means ± SD of twenty repeats. Asterisks indicate significant difference between wild-type and other genetic materials, as determined by Student’s t-test (*p < 0.05)
3.4 TCD5 Increased Electron Transport Rate and the Quantum Yield
The electron transport rate and the quantum yield were measured in plants at tillering stage by using the youngest fully expanded leaves. The electron transport rate of Jiahua1 wild-type is significantly higher (p < 0.05) than that of tcd5 mutant but lower (p < 0.05) than that of TCD5 over-expression line (Fig. 5A). The electron transport rate of Nipponbare wild-type is higher (p < 0.05) than that of TCD5 RNAi line but lower (p < 0.05) than that of TCD5 over-expression line (Fig. 5B). Similar results were also observed in the quantum yield. The quantum yield of Jiahua1 wild-type is higher than that of tcd5 mutant but lower than that of TCD5 over-expression line (Fig. 5C). The quantum yield of Nipponbare wild-type is higher than that of TCD5 RNAi line but lower than that of TCD5 over-expression line (Fig. 5D). These results indicated that TCD5 enhanced the electron transport rate and the quantum yield at tillering stage.
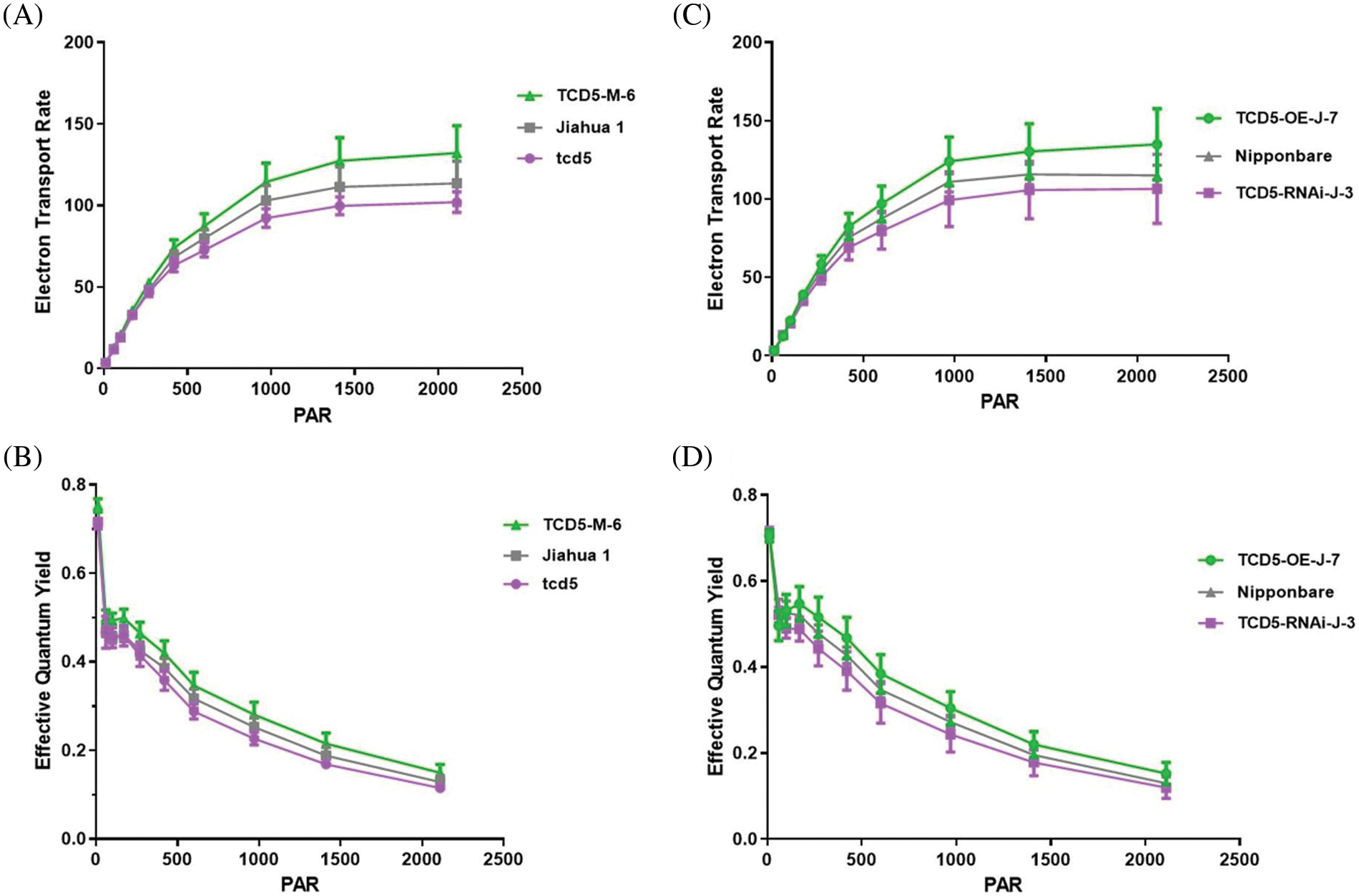
Figure 5: The electron transport rate and the quantum yield of WT, tcd5 mutant or TCD5 RNAi lines, and TCD5 over-expression lines in Jiahua1 and Nipponbare background. The values are shown as means ± SD of twenty repeats
3.5 D1 Were Identified by IP-MS Screening of TCD5 Interact Protein
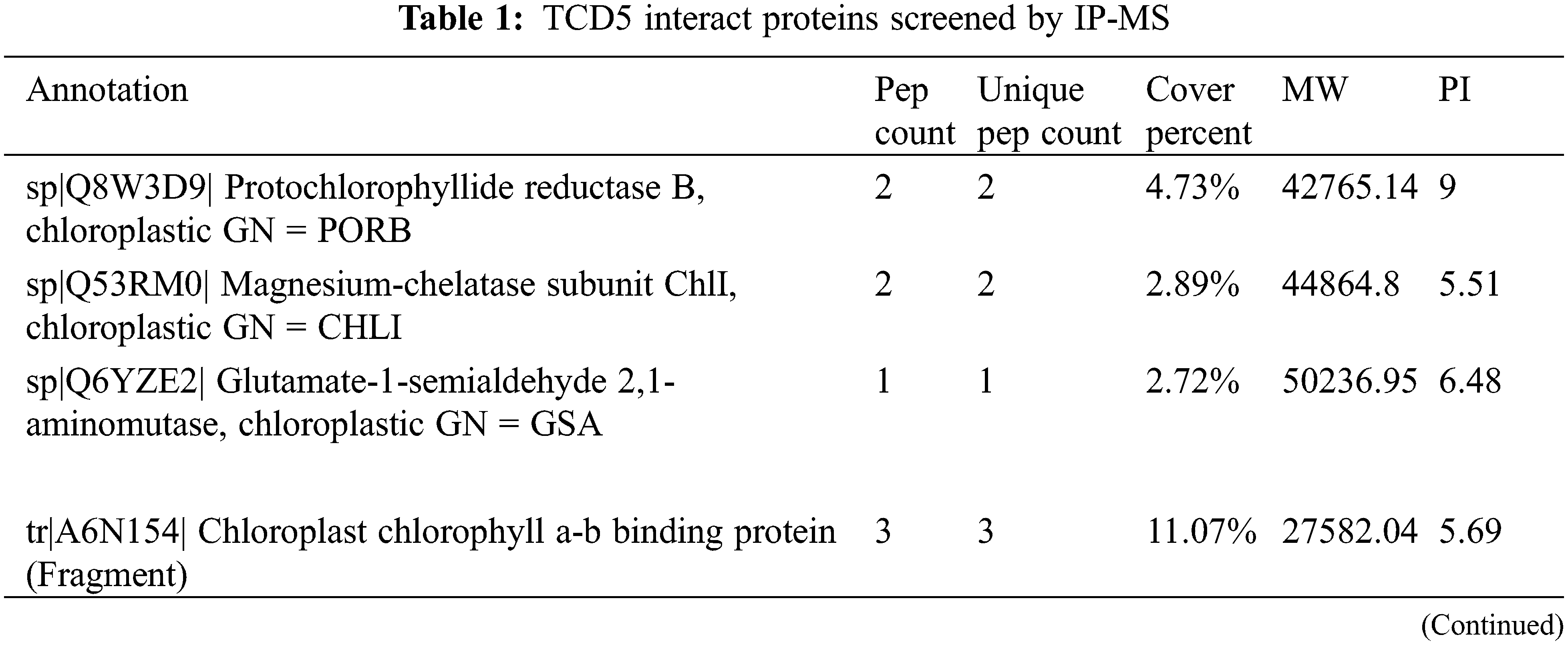
A total of 29 no-redundant putative chloroplast or photosystem-related proteins were identified by using IP-MS screening (Table 1). Among these proteins, Protochlorophyllide reductase B (PorB), Magnesium-chelatase subunit ChlI (ChlI) and Glutamate-1-semialdehyde 2, 1-aminomutase (Gsa) are involved in chlorophyll synthesis; chlorophyll a-b binding protein (CAB), Photosystem I P700 chlorophyll a apoprotein A1 (psaA), Photosystem I iron-sulfur center (psaC), and Photosystem II protein D1 (psbA) are involved in photo-reaction of the photosynthesis; Ribulose bisphosphate carboxylase/oxygenase activase (RCA), Ribulose bisphosphate carboxylase small chain (RBCS), Fructose-bisphosphate aldolase (FBA) and Ribulose-phosphate 3-epimerase (Rpe) are involved in carbon assimilation and metabolism; ATP synthase subunits (aptA, atpB, atpE and atpF) and Chaperone protein ClpC1 (ClpC1) a subunit of ATPase are involved in energy metabolism; Ferredoxin--NADP reductase (Fenr1) and 2-Cys peroxiredoxin BAS1 (Bas1) are involved in redox balance; 30S ribosomal protein S2, S3, S4 and S12, 50S ribosomal protein L5, L18 and L33 together with Translation factor GUF1 homolog are involved in protein synthesis; Chloroplast inner envelope protein is involved in protein transportation.
Since the transformation of maize genes involved in C4 photosynthesis, such as PEPC, etc., into rice in the 1990s [31], more and more efforts have been invested in improving the photosynthesis efficiency in rice and other crops [32]. Several workable strategies to improve crop photosynthesis have been identified and proved based on systems modeling and genetic manipulations. These strategies include 1. Optimize the carbon assimilation and metabolism pathway, including increasing the protein levels of SBPase, FBA, and ictB to optimize the Calvin-Benson cycle [12,33] and engineering photorespiratory bypasses [15–17], 2. Optimize the light reaction pathway, including increasing the speed of recovery from photoprotection by over-expression of VDE, ZEP, and PsbS [13,14], supplementation of the nuclear original D1 protein [18], 3. Over-expression of transcription factors positively regulating the photosynthesis genes [20] or knocking out the transcription factor negatively regulating the photosynthesis genes [21]. Furthermore, progress has been made in exploring the excellent alleles for high photosynthesis in natural variations in rice. The partially functional allele of the Narrow Leaf 1 gene was cloned as an increasing photosynthesis gene [22]. A further study indicated that this elite Narrow Leaf 1 allele balances the leaf photosynthesis and the plant architecture and resulting in higher rice yield [23]. Recently, it has been found that over-expression of the Chlorophyllide-a oxygenase 1 (OsCAO1) gene increases the chlorophyll b content, the photosynthetic rate, and the grain yield [34]. In this study, we found that TCD5 increase photosynthesis efficiency, the panicle number, and the grain yield in a dosage manner based on the results of field experiments in multiple year/environments using the genetic materials of three expression levels in two backgrounds.
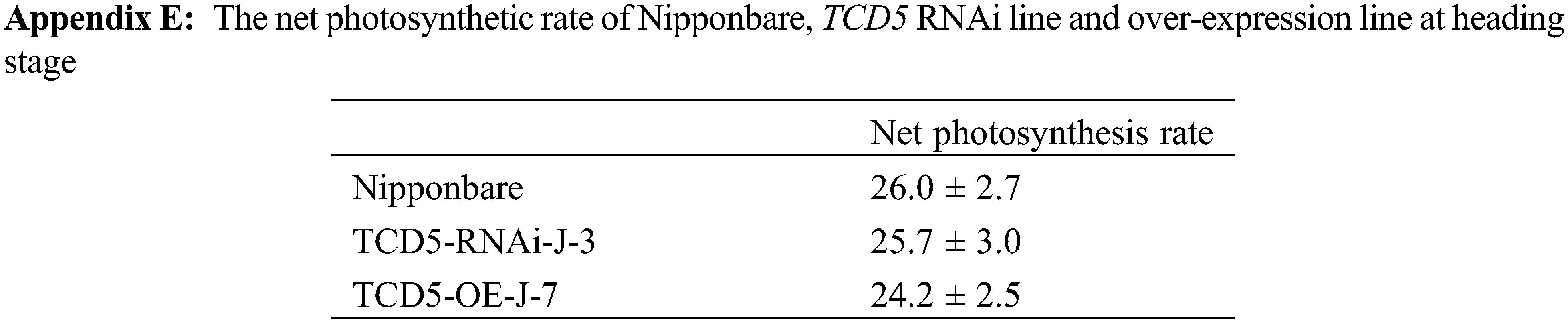
Enhancement of photosynthesis has been reported to result in an increase in tiller number in rice [17–21], which is consistent with our findings. It has recently been reported that sucrose, a photosynthetic product, inhibits the response to strigolactone and promotes tillering in rice [35]. These results elucidated the molecular mechanism of the association between photosynthesis and tiller number in rice. It is somehow surprising that only the panicle number was affected in this study, compared to the reported results [17–21]. We found that there is no significant difference in the net photosynthetic rate between Nipponbare, TCD5 RNAi line, and TCD5 over-expression line at the heading stage (Appendix E), implying that the effects of TCD5 on photosynthesis were expressed specifically at the vegetative stage.
TCD5 was identified by three different laboratories due to the albino leaf phenotype mutant caused by defective chloroplast development under low temperature at the seedling or tillering stage. It was named TCD5, CSV1, and TSV, respectively [24,36,37]. TCD5/CSV1/TSV is conserved among land plants, green algae, and cyanobacteria [36]. TCD5/CSV1/TSV highly expresses in young leaves and immature spikes and is induced by low temperatures [24,36,37]. TCD5/CSV1/TSV is essential for the activity of plastid-encoded RNA polymerase (PEP) and the expression of genes participating in chlorophyll synthesis under restrictively low temperatures [24,37]. The orthologue protein of TCD5/CSV1/TSV in Arabidopsis putatively functions in photosynthetic chain electron flow, and the mutant presented a pale green and growth-defective phenotype. However the development of the chloroplast was unaffected [38]. In this study, we found that TCD5/CSV1/TSV affected the electronic transport rate and the quantum yield under normal growth conditions, which could explain the effect of TCD5/CSV1/TSV on the photosynthesis efficiency.
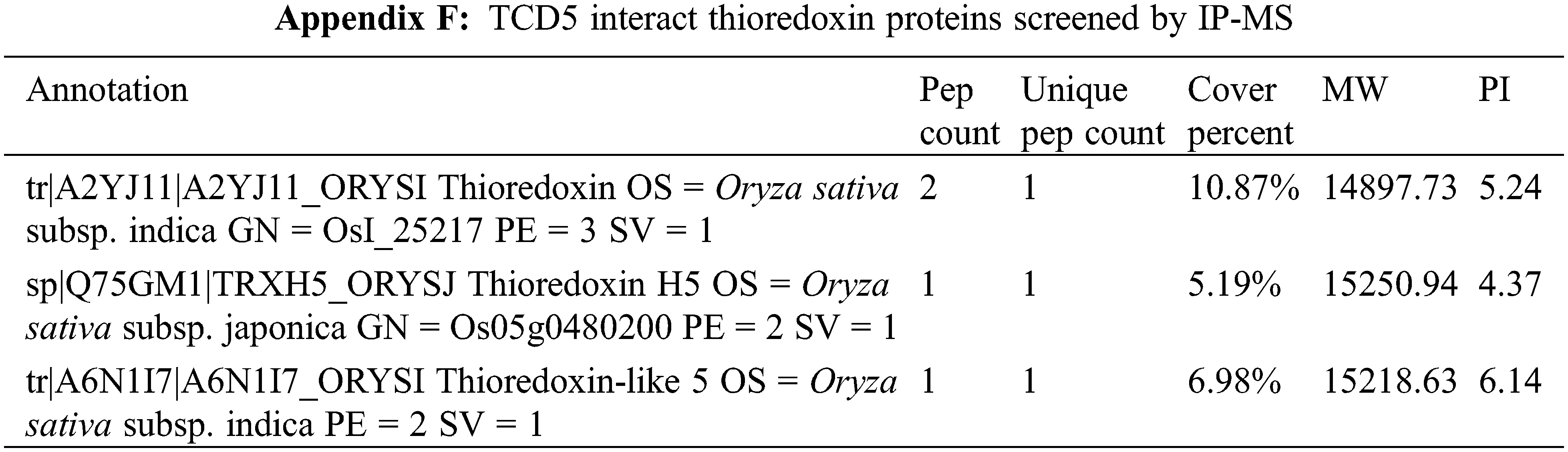
TCD5/CSV1/TSV was reported to interact with OsTrxZ, a thioredoxin Z protein [37]. In this study, three thioredoxin proteins were identified as interactors of TCD5/CSV1/TSV (Appendix F), although OsTrxZ was not included. Interestingly, the targets of photosynthetic modification D1 and FBA [12,18] were identified as TCD5/CSV1/TSV interact proteins. These two proteins may be more stable through the interaction with TCD5/CSV1/TSV, which is similar to OsTrxZ [37]. These may explain why TCD5/CSV1/TSV increases the electron transport rate and the quantum yield. Further exploration will be conducted on the contribution of TCD5/CSV1/TSV interacting with other proteins to enhance photosynthesis.
Acknowledgement: We thank the Professor Genyun Chen for the help of the leaf gas exchange measurement and Professor Hualin Mi for the help of chlorophyll fluorescence parameters measurement.
Funding Statement: This work was funded by the National Natural Science Foundation of China (Grand No. U19A2025) and Scientific Research Foundation of China Jiliang University.
Author Contributions: The authors confirm contribution to the paper as follows: Sheng Teng designed research and wrote the manuscript. Jing Yang performed the most experiments and edited the manuscript. Yufeng Wang prepared for and identified all materials. Zhanghua Hu and Xiaoping Chen performed field experiments and trait evaluation. Yanjun Dong provided critical analyses and edited the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. GRiSP (2013). Rice almanac: Source book for one of the most important economic activities on earth, 4th editionPhilippine: International Rice Research Institute. [Google Scholar]
2. Yadev, S., Kumar, V. (2018). Feeding the world while caring for the planet. Direct Seeded Rice Consortium, 1(2), 1–18. [Google Scholar]
3. Qian, Q., Guo, L., Smith, S. M., Li, J. (2016). Breeding high-yield superior quality hybrid super rice by rational design. National Science Review, 3(3), 283–294. [Google Scholar]
4. Peng, Y., Hu, Y., Qian, Q., Ren, D. (2021). Progress and prospect of breeding utilization of Green Revolution gene SD 1 in rice. Agriculture, 11(7), 611. [Google Scholar]
5. Cheng, S. H., Cao, L. Y., Zhuang, J. Y., Chen, S. G., Zhan, X. D. et al. (2007). Super hybrid rice breeding in China: Achievements and prospects. Journal of Integrative Plant Biology, 49(6), 805–810. [Google Scholar]
6. Grassini, P., Eskridge, K. M., Cassman, K. G. (2013). Distinguishing between yield advances and yield plateaus in historical crop production trends. Nature Communication, 4(1), 2918. [Google Scholar]
7. Zhu, X. G., Long, S. P., Ort, D. R. (2010). Improving photosynthetic efficiency for greater yield. Annual Review of Plant Biology, 61(1), 235–261. [Google Scholar] [PubMed]
8. Long, S. P., Marshall-Colon, A., Zhu, X. G. (2015). Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell, 161(1), 56–66. [Google Scholar] [PubMed]
9. Theeuwen, T. P. J. M., Logie, L. L., Harbinson, J., Aarts, M. G. M. (2022). Genetics as a key to improving crop photosynthesis. Journal of Experimental Botany, 73(10), 3122–3137. [Google Scholar] [PubMed]
10. Kromdijk, J., McCormick, A. J. (2022). Genetic variation in photosynthesis: Many variants make light work. Journal of Experimental Botany, 73(10), 3053–3056. [Google Scholar] [PubMed]
11. Leister, D. (2023). Enhancing the light reactions of photosynthesis: Strategies, controversies, and perspectives. Molecular Plant, 16(1), 4–22. [Google Scholar] [PubMed]
12. Simkin, A. J., McAusland, L., Headland, L. R., Lawson, T., Raines, C. A. (2015). Multigene manipulation of photosynthetic carbon assimilation increases CO2 fixation and biomass yield in tobacco. Journal of Experimental Botany, 66(13), 4075–4090. [Google Scholar] [PubMed]
13. Kromdijk, J., Głowacka, K., Leonelli, L., Gabilly, S. T., Iwai, M. et al. (2016). Improving photosynthesis and crop productivity by accelerating recovery from photoprotection. Science, 354(6314), 857–861. [Google Scholar] [PubMed]
14. de Souza, A. P., Burgess, S. J., Doran, L., Hansen, J., Manukyan, L. et al. (2022). Soybean photosynthesis and crop yield are improved by accelerating recovery from photoprotection. Science, 377(6608), 851–854. [Google Scholar] [PubMed]
15. Kebeish, R., Niessen, M., Thiruveedhi, K., Bari, R., Hirsch, H. J. et al. (2007). Chloroplastic photorespiratory bypass increases photosynthesis and biomass production in Arabidopsis thaliana. Nature Biotechnology, 25(5), 593–599. [Google Scholar] [PubMed]
16. South, P. F., Cavanagh, A. P., Liu, H. W., Ort, D. R. (2019). Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science, 363, eaat9077. [Google Scholar] [PubMed]
17. Shen, B. R., Wang, L. M., Lin, X. L., Yao, Z., Xu, H. W. et al. (2019). Engineering a new chloroplastic photorespiratory bypass to increase photosynthetic efficiency and productivity in rice. Molecular Plant, 12(2), 199–214. [Google Scholar] [PubMed]
18. Chen, J. H., Chen, S. T., He, N. Y., Wang, Q. L., Zhao, Y. et al. (2020). Nuclear-encoded synthesis of the D1 subunit of photosystem II increases photosynthetic efficiency and crop yield. Nature Plants, 6(5), 570–580. [Google Scholar] [PubMed]
19. Wei, S., Li, X., Lu, Z., Zhang, H., Ye, X. et al. (2022). A transcriptional regulator that boosts grain yields and shortens the growth duration of rice. Science, 377(6604), eabi8455. [Google Scholar] [PubMed]
20. Perveen, S., Qu, M., Chen, F., Essemine, J., Khan, N. et al. (2020). Overexpression of maize transcription factor mEmBP-1 increases photosynthesis, biomass, and yield in rice. Journal of Experimental Botany, 71(16), 4944–4957. [Google Scholar] [PubMed]
21. Chen, F., Zheng, G., Qu, M., Wang, Y., Lyu, M. J. A. et al. (2021). Knocking out NEGATIVE REGULATOR OF PHOTOSYNTHESIS 1 increases rice leaf photosynthesis and biomass production in the field. Journal of Experimental Botany, 72(5), 1836–1849. [Google Scholar] [PubMed]
22. Takai, T., Adachi, S., Taguchi-Shiobara, F., Sanoh-Arai, Y., Iwasawa, N. et al. (2013). A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Scientific Reports, 3(1), 2149. [Google Scholar] [PubMed]
23. Ouyang, X., Zhong, X., Chang, S., Qian, Q., Zhang, Y. et al. (2022). Partially functional NARROW LEAF1 balances leaf photosynthesis and plant architecture for greater rice yield. Plant Physiology, 189(2), 772–789. [Google Scholar] [PubMed]
24. Wang, Y., Zhang, J., Shi, X., Peng, Y., Li, P. et al. (2016). Temperature-sensitive albino gene TCD5, encoding a monooxygenase, affects chloroplast development at low temperatures. Journal of Experimental Botany, 67(17), 5187–5202. [Google Scholar] [PubMed]
25. Schreiber, U., Klughammer, C. (2008). Non-photochemical fluorescence quenching and quantum yields in PS I and PS II: Analysis of heat-induced limitations using Maxi-Imaging-PAM and Dual-PAM-100. PAM Application Notes, 1, 15–18. [Google Scholar]
26. Klughammer, C., Schreiber, U. (2008). Complementary PSII quantum yields calculated from simple fluorescence parameters measured by PAM fluorometry and the saturation pulse method. PAM Application Notes, 1, 27–35. [Google Scholar]
27. Kramer, D. M., Johnson, G., Kiirats, O., Edwards, G. E. (2004). New fluorescence parameters for the determination of QA redox state and excitation energy fluxes. Photosynthesis Research, 79, 209–218. [Google Scholar] [PubMed]
28. Yamori, W., Kondo, E., Sugiura, D., Terashima, I., Suzuki, Y. et al. (2016). Enhanced leaf photosynthesis as a target to increase grain yield: Insights from transgenic rice lines with variable Rieske FeS protein content in the cytochrome b6/f complex. Plant, Cell & Environment, 39, 80–87. [Google Scholar]
29. Livak, K. J., Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods, 25, 402–408. [Google Scholar] [PubMed]
30. Hu, X., Yang, Y., Gong, F., Zhang, D., Zhang, L. et al. (2015). Protein sHSP26 improves chloroplast performance under heat stress by interacting with specific chloroplast proteins in maize (Zea mays). Journal of Proteomics, 115, 81–92. [Google Scholar] [PubMed]
31. Ku, M. S., Agarie, S., Nomura, M., Fukayama, H., Tsuchida, H. et al. (1999). High-level expression of maize phosphoenolpyruvate carboxylase in transgenic rice plants. Nature Biotechnology, 17(1), 76–80. [Google Scholar] [PubMed]
32. Yeh, S. Y., Lin, H. H., Chang, Y. M., Chang, Y. L., Chang, C. K. et al. (2022). Maize Golden2-like transcription factors boost rice chloroplast development, photosynthesis, and grain yield. Plant Physiology, 188(1), 442–459. [Google Scholar] [PubMed]
33. Zhu, X. G., de Sturler, E., Long, S. P. (2007). Optimizing the distribution of resources between enzymes of carbon metabolism can dramatically increase photosynthetic rate: A numerical simulation using an evolutionary algorithm. Plant Physiology, 145(2), 513–526. [Google Scholar] [PubMed]
34. Hu, P., Ma, J., Kang, S., Li, S., Wu, X. et al. (2023). Chlorophyllide-a oxygenase 1 (OsCAO1) over-expression affects rice photosynthetic rate and grain yield. Rice Science, 30(2), 87–91. [Google Scholar]
35. Patil, S. B., Barbier, F. F., Zhao, J., Zafar, S. A., Uzair, M. et al. (2022). Sucrose promotes D53 accumulation and tillering in rice. New Phytologist, 234(1), 122–136. [Google Scholar] [PubMed]
36. Morita, R., Nakagawa, M., Takehisa, H., Hayashi, Y., Ichida, H. et al. (2016). Heavy-ion beam mutagenesis identified an essential gene for chloroplast development under cold stress conditions during both early growth and tillering stages in rice. Bioscience, Biotechnology, and Biochemistry, 81, 1–12. [Google Scholar]
37. Sun, J., Zheng, T., Yu, J., Wu, T., Wang, X. et al. (2017). TSV, a putative plastidic oxidoreductase, protects rice chloroplasts from cold stress during development by interacting with plastidic thioredoxin Z. New Phytologist, 215(1), 240–255. [Google Scholar] [PubMed]
38. Vlad, D., Rappaport, F., Simon, M., Loudet, O. (2010). Gene transposition causing natural variation for growth in Arabidopsis thaliana. PLoS Genetics, 6(5), e1000945. [Google Scholar] [PubMed]
Appendixes
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools