 Open Access
Open Access
ARTICLE
Transcriptome-Wide Identification and Functional Analysis of PgSQE08-01 Gene in Ginsenoside Biosynthesis in Panax ginseng C. A. Mey.
1 College of Life Science, Jilin Agricultural University, Changchun, 130118, China
2 Jilin Engineering Research Center Ginseng Genetic Resources Development and Utilization, Changchun, 130118, China
3 Jilin Academy of Vegetable and Flower Sciences, Changchun, 130033, China
* Corresponding Authors: Mingzhu Zhao. Email: ; Kangyu Wang. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Plant Secondary Metabolism and Functional Biology)
Phyton-International Journal of Experimental Botany 2024, 93(2), 313-327. https://doi.org/10.32604/phyton.2024.047938
Received 23 November 2023; Accepted 08 January 2024; Issue published 27 February 2024
Abstract
Panax ginseng C. A. Mey. is an important plant species used in traditional Chinese medicine, whose primary active ingredient is a ginsenoside. Ginsenoside biosynthesis is not only regulated by transcription factors but also controlled by a variety of structural genes. Nonetheless, the molecular mechanism underlying ginsenoside biosynthesis has always been a topic in the discussion of ginseng secondary metabolites. Squalene epoxidase (SQE) is a key enzyme in the mevalonic acid pathway, which affects the biosynthesis of secondary metabolites such as terpenoid. Using ginseng transcriptome, expression, and ginsenoside content databases, this study employed bioinformatic methods to systematically analyze the genes encoding SQE in ginseng. We first selected six PgSQE candidates that were closely involved in ginsenoside biosynthesis and then identified PgSQE08-01 to be highly associated with ginsenoside biosynthesis. Next, we constructed the overexpression vector pCAMBIA3301-PgSQE08-01 and the RNAi vector pART27-PgSQE08-01 and transformed ginseng adventitious roots using Agrobacterium rhizogenes, to obtain positive hairy-root clones. Thereafter, quantitative reverse transcription-polymerase chain reaction and high-performance liquid chromatography were used to determine the expression of relevant genes and ginsenoside content, respectively. Then, we focused on the function of PgSQE08-01 gene, which was noted to be involved in ginsenoside biosynthesis. Thus, these findings not only provided a molecular basis for the identification of important functional genes in ginseng but also enriched genetic resources for the biosynthesis of ginsenosides using synthetic biology.Keywords
Supplementary Material
Supplementary Material FileGinseng (Panax ginseng C. A. Mey.), a member of the family Araliaceae, is a valuable medicinal plant of traditional Chinese medicine [1]. This plant species prefers shade, and high temperatures (above 32°C) result in damage to various extents. With a long history of application around the world, its importance is self-evident. It exhibits high economic and medicinal value as it helps in tonifying center-supplementing qi-boosting, generating body fluids, soothing muscles, activating blood cells, and preventing aging [2,3]. Despite its high utility, several issues are associated with the plant species, such as long cultivation time, continuous degradation of varieties, high requirements of environmental conditions and soil, rust disease, and heavy metal pollution [4,5], leading to altered yield and quality that cannot support market demand [6]. At present, there is a serious shortage in the supply of ginsenoside, the main active ingredient of ginseng [7]. Thus, to meet the market demand and protect the limited wild ginseng resources, the application of biosynthetic technology, secondary metabolomics, and molecular methods to innovate ginseng germplasm resources and regulate ginsenoside biosynthesis have become increasingly mainstream to provide a solution to decreasing ginseng quality and yield [8].
Squalene epoxidase (SQE; also referred to as squalene monooxygenase) is a cell membrane-bound enzyme attached to the endoplasmic reticulum that is ubiquitously found in the microsomes of plant, human, and other eukaryotic cells, and functions in the cell membrane, in conjunction with molecular oxygen to perform its catalytic role [9]. SQE plays a vital role in the mevalonic acid (MVA) pathway, in synergy with mevalonate kinase (MVK) and squalene synthase (SS) [10]. It also acts as a precursor and affects the biosynthetic efficiency of triterpenoid saponins. Thus, in plant synthetic biology, the study of the molecular regulatory mechanisms of key enzymes in the ginsenoside biosynthetic pathway has become a hot research topic.
SQE genes have been studied in a variety of plants, such as Arabidopsis thaliana [11], Gynostemma pentaphyllum [12], Panax notoginseng [13], Eleutherococcus senticosus [14], Tripterygium wilfordii [15], Siraitia grosvenorii [16], and Glycyrrhiza glabra [17]. Although sequence analysis, gene cloning, and vector construction have been performed in most plant species, functional studies have been conducted on extremely few plant species, suggesting that SQE genes influence the synthesis of secondary metabolites. SQE, one of the most important regulatory enzymes involved in ginsenoside biosynthesis, regulates the biosynthesis and efficiency of other secondary metabolites together with SS and MVK [18–22]. Several studies have shown that SQE genes regulate the synthesis of triterpenoids, such as saponins and sterols, in ginseng, and others have concluded that SQE activity is positively correlated with ginsenoside content [23]. In recent years, cell factories and synthetic biology have been used to improve the biosynthesis of secondary metabolites in important medicinal plants and provide an important technological means to increase the content of effective medicinal ingredients in traditional Chinese medicinal plants [24]. Although the key enzymes involved in the ginsenoside metabolic pathway have been identified, the mechanism underlying ginsenoside biosynthesis needs further investigation.
Therefore, based on the ginseng transcriptome database established in a previous study, this study screened SQE genes in ginseng and analyzed their functions, evolution, and expression levels. Thereafter, we focused on the SQE gene related to ginsenoside biosynthesis (PgSQE08-01). Using gene overexpression and RNA interference (RNAi), the expression level of PgSQE08-01 gene was determined. Additionally, we analyzed the expression of other key enzymes involved in ginsenoside biosynthesis and the content of saponins in the positive lines. Thus, this study improves the understanding of the role of important functional genes in ginseng using molecular methods and enriches genetic resources for the biosynthesis of ginsenosides using synthetic biology.
2.1 Plant Material and Data Resources
We used two of our previously established databases (high-throughput mRNA sequence databases of various tissues and varieties of ginseng), along with other related databases, in this study. The plant materials used in this study (tap and fibrous roots for total RNA extraction) and adventitious roots for transformation experiments, as well as vectors, were sourced from our laboratory.
2.2 Mining of the Genes Encoding SQE in Ginseng
To determine the sequence of the genes encoding SQE in ginseng, we performed multiple comparisons using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). We retrieved the sequences of genes encoding SQE from NCBI and compared them with the sequences available in the Chinese Ginseng Genome Database [25], the Korean Ginseng Genome Database [26], and a previously established ginseng transcriptome database [27]. Then, we compared the preliminary SQE gene sequences obtained from the former two databases with the latter database and obtained the corresponding sequences in the transcriptome database. Thereafter, we merged the resulting sequences with the aligned SQE gene sequences retrieved from NCBI and the transcriptome database and removed duplicate sequences to obtain the preliminary candidate gene identifier. The cutoff values were set at 1.0E-06. We first used HMMER version 3.0 [28] to build a profile hidden Markov model and search the ginseng proteome database and then used the resulting sequences as query sequences to search against the transcriptome database. Then, the results were merged and de-duplicated, after which we retained all the sequences containing a complete reading frame using the NCBI online query. Using Pfam online software (http://pfam.sanger.ac.uk/), SMART online software (http://smart.embl.de/), and CDD online software (http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), we obtained the final set of SQE gene sequences, which were then subjected to the further bioinformatic analyses.
2.3 Phylogenetic Analysis of PgSQE Genes
A phylogenetic tree was constructed using the maximum likelihood method on MEGA-X [29], with a bootstrap of 2000. Evolview version 3.0 (https://www.evolgenius.info/evolview) [30] was used to edit the phylogenetic tree.
2.4 Gene Ontology (GO) Functional Annotation and Enrichment Analysis of PgSQE Genes
Blast2GO version 5.0 [31] was used to perform GO functional annotation of PgSQE gene sequences, and the sequences were subjected to enrichment analysis based on the annotation results.
2.5 Expression Pattern Analysis of PgSQE Genes
Using Perl scripts, we determined the expression levels of the selected sequences from the Jilin ginseng expression database. TBtools version 1.130 [32] was used to construct heatmaps based on age, tissue type, and cultivars.
2.6 Correlation Analysis between PgSQE Genes Expression and Saponin Content
Perl scripts were used to estimate PgSQE genes expression, and SPSS version 23.0 was used to calculate the Pearson’s correlation coefficient between PgSQE genes expression and saponin content, based on total and individual saponin contents. Then, interaction network analysis was performed between PgSQE genes and the genes encoding known key enzymes of the ginsenoside biosynthetic pathway. By setting stringent p-values, a PgSQE gene was selected (closely related to the genes encoding known key enzymes) as the candidate gene for further experiments.
2.7 PgSQE08-01 Gene Cloning and Vector Construction
The open reading frame (ORF) of PgSQE08-01, containing conserved features, was obtained from NCBI. The upstream promoter and downstream terminator sequences were used to design domain-specific primers and the expression vector, with the XbaI site at the upstream 5′ end and the SmaI site at the downstream 5′ end, respectively. The primers are as follows: 5′-GCTCTAGAAAGGCATTGTAGAGAGAGAGACG-3′ (forward primer); 5′-TCCCCCGGGCTAATCTTGGACCACTGATGAATG-3′ (reverse primer).
RNA was extracted from a four-year-old ginseng plant and reverse-transcribed into cDNA, which was used as the template for polymerase chain reaction (PCR) amplification of the target gene. The target gene was inserted into the XbaI and SmaI restriction sites of the T vector using the pGM-T cloning ligation kit. After digestion with XmaI, the target fragment was inserted into the pCAMBIA3301 vector to construct the pCAMBIA3301-PgSQE08-01 expression vector. The sense and antisense interference fragments were first inserted into the pHANNIBAL vector, and then the region containing the RNAi vector was excised and inserted into the NotI restriction site of vector pART27 to construct the pART27-PgSQE08-01 RNAi vector.
2.8 Genetic Transformation of Explants Derived from Ginseng Adventitious Roots
The adventitious roots of ginseng were cut into 0.8–1.0 cm-long sections and placed on plates containing solid B5 medium. Then, 7–9 small sections of adventitious roots were transferred and evenly placed on culture plates containing solid B5 medium and cultured in the dark for 14–21 d.
The activated bacteria were inoculated in liquid Luria-Bertani medium containing kanamycin. Thereafter, the bacterial culture was incubated at 28°C and 170 rpm until the optical density (at 600 nm; OD600) of the culture reached 0.4–0.6 using SpectraMax M2 (SpectraMax M2, Molecular Devices Company, Silicon Valley, CA, USA). The bacteria were collected and resuspended in half Murashige and Skoog (MS) medium, containing 200 μM AS until the OD600 of the culture reached 0.4–0.6. Then, the 2–3-week-cultured adventitious roots were excised, placed on solid MS medium, and pre-cultured for 48 h under light conditions. The infected explants were then transferred to half MS medium (solid) containing and co-cultured for 48 h in the dark. Finally, the ginseng hairy roots were collected for subsequent experiments.
2.9 PCR-Based Identification of Transformed Ginseng Hairy Roots and Extraction of Ginsenosides
After successful ligation of the overexpression vector, primers were designed using the sequence of the recombinant plasmid linked to the target gene, and transformation was verified using PCR.
The positively transformed hairy roots were dried in an oven at 37°C for 72 h, and 1 g of the plant material was ground into powder using a mortar and pestle. The powder was wrapped in filter paper and immersed in a 50-mL Erlenmeyer flask containing 20 mL of methanol solution. The flask was sealed with a sealing film to prevent methanol from evaporating. After 12 h, it was ultrasonically extracted for 30 min, and then the solution and the filter-paper bag immersed in it were transferred to a Soxhlet extractor. Subsequently, 80 mL of methanol solution was added to the extractor, and the solution was incubated in a water bath at 90°C for 30 h. The solution was then transferred to a rotary evaporator for drying the extract, which was re-dissolved in 20 mL of distilled water.
2.10 Saponin Purification and Determination of Its Content
We added 15 mL of ethyl acetate to the extract, mixed it well, and kept it still for layering. The lower layer of solution was separated using a separatory funnel. This step was repeated three times. Then, 20 mL of water-saturated n-butanol was added to the obtained solution, which was again kept still for layering. Meanwhile, the upper layer of the solution was also separated using a separatory funnel, and this step was repeated three times. The collected solution was dried in a rotary evaporator, and saponins on the wall of the bottle were re-dissolved in 10 mL of chromatographic methanol. The solution was then filtered through a 0.22-μM organic phase filter membrane and sealed for high-performance liquid chromatography (HPLC; Waters e2695 Separations Module, Waters Listed Company, Milford, MA, USA). The chromatographic conditions were referred to the published article [33].
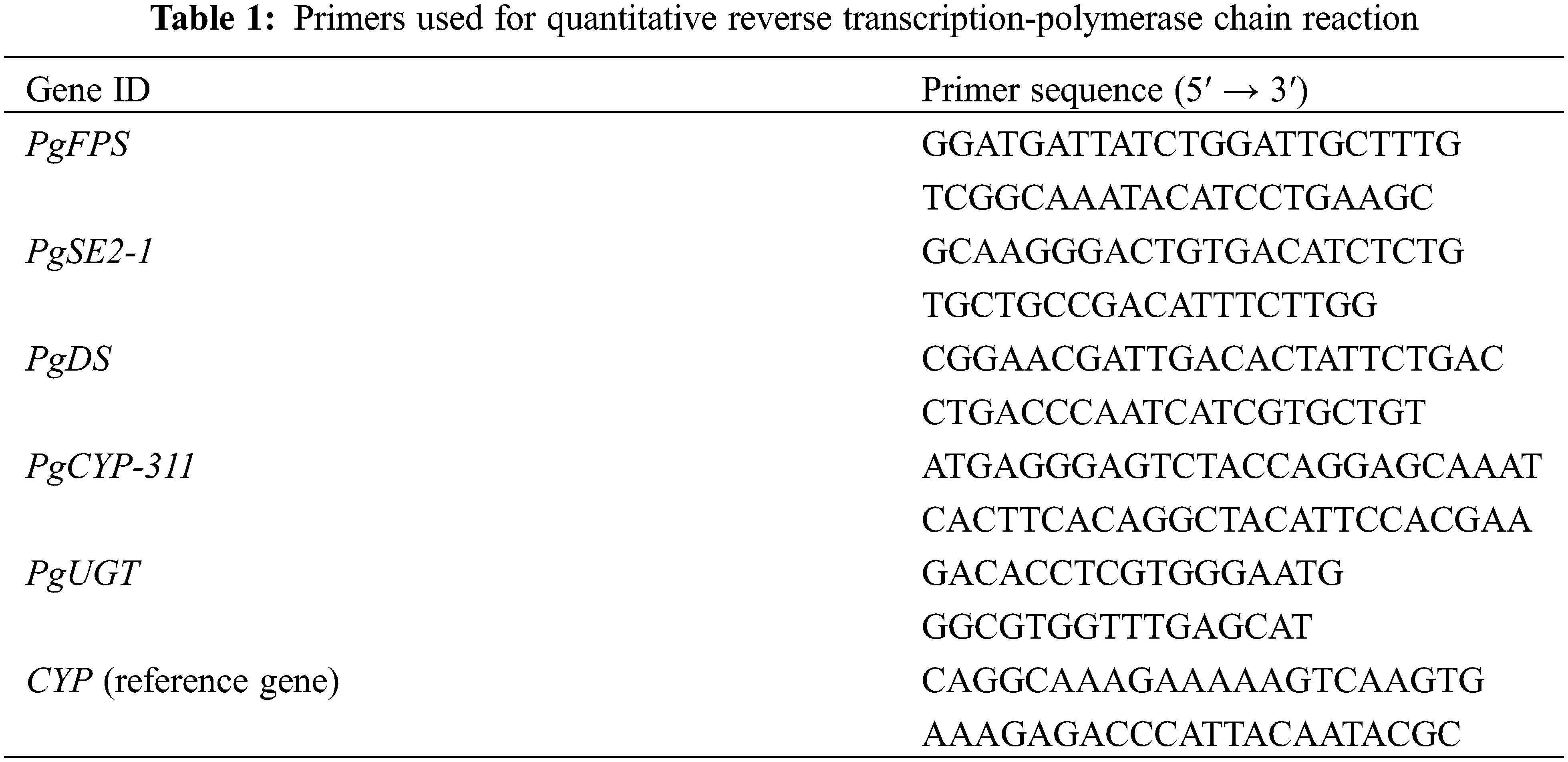
The hairy roots of PgSQE08-01-overexpressing plants were employed to detect the expression levels of five key-enzyme genes (FPS [34], SE2-1 [35], DS [36], CYP-311 [37], and UGT [38]) using quantitative reverse transcription-polymerase chain reaction (qRT-PCR), and the 2−ΔΔCT method was used for estimating the expression levels [39]. All primers for qRT-PCR are shown in Table 1.
3.1 Identification of Genes Encoding SQE in Ginseng
Based on the gene sequences of SQE from all species and the hidden Markov model, we identified several SQE gene sequences in ginseng, with all 20 transcripts exhibiting complete ORFs and conserved domains. We named the 20 PgSQE genes as follows. Each gene name included Pg (short for P. ginseng), the key enzyme name (SQE), and a number depicting the order of naming the gene (Table S1).
3.2 Phylogenetic Analysis of PgSQE Gene Sequences
Using the neighbor-joining method, we constructed a phylogenetic tree with 20 SQE transcripts containing conserved domains and complete ORFs (Fig. 1). Different gene sequences exhibited different aggregation relationships. For example, PgSQE08-01, PgSQE08-02, PgSQE08-03, and PgSQE08-04 (comp65427-c0-seq1, comp65427-c0-seq2, comp65427-c0-seq3, and comp65427-c0-seq4) formed one branch, indicating that they may have similar functions. The transcripts with different gene identifiers aggregated in different branches of the phylogenetic tree, indicating that they may be functionally different.
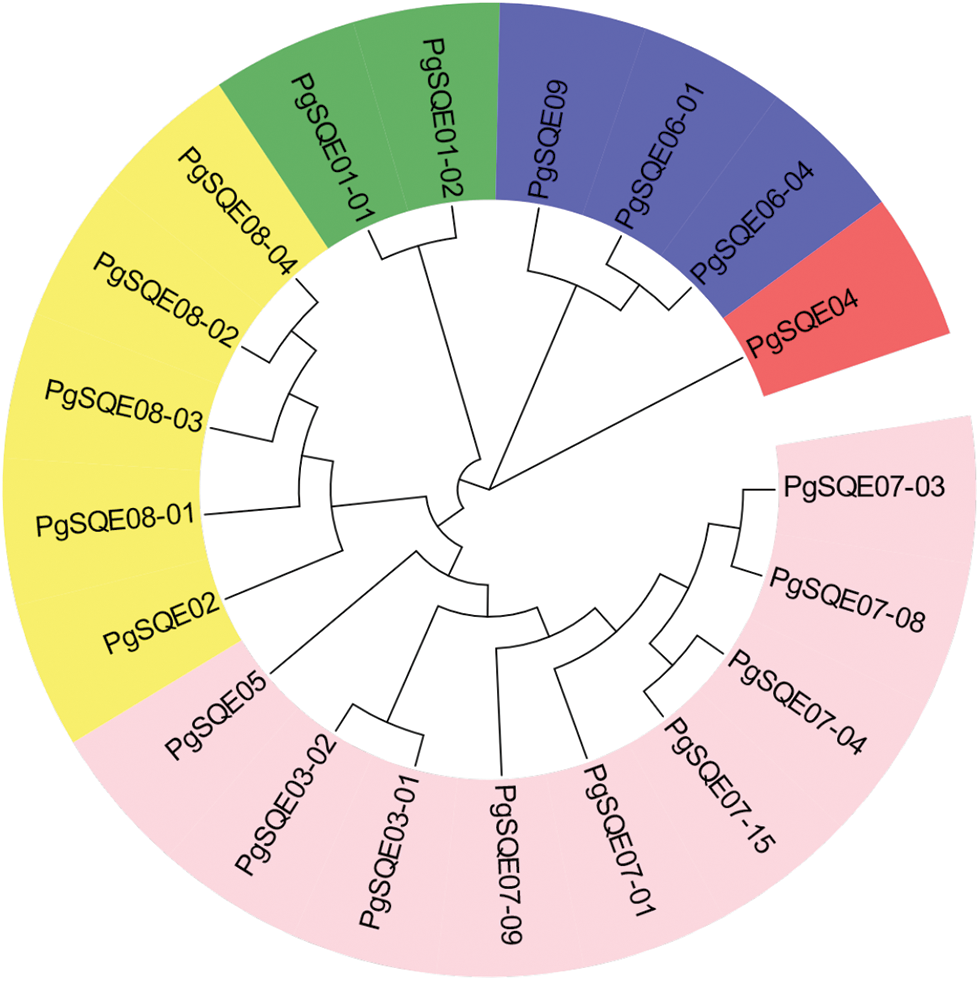
Figure 1: Phylogenetic tree of the 20 PgSQE gene sequences. Different colors represent different clades
3.3 GO Functional Annotation Analysis of PgSQE Gene Sequences
We used Blast2GO to perform a basic functional analysis of the 20 PgSQE gene sequences. All 20 sequences were annotated into three GO function categories, namely biological processes (BP), molecular functions (MF), and cellular components (CC). Among them, the number of genes annotated to MF was the highest (20 genes; 100%), followed by BP (17 genes; 85%) and CC (15 genes; 75%) (Fig. 2A). At level 2, the 20 PgSQE genes were divided into seven functional terms (Fig. 2B), of which we estimated the number of transcripts annotated to three terms. Five PgSQE gene sequences were annotated to all seven terms (25%), whereas two PgSQE gene sequences (10%) were annotated to two of the seven functional terms.
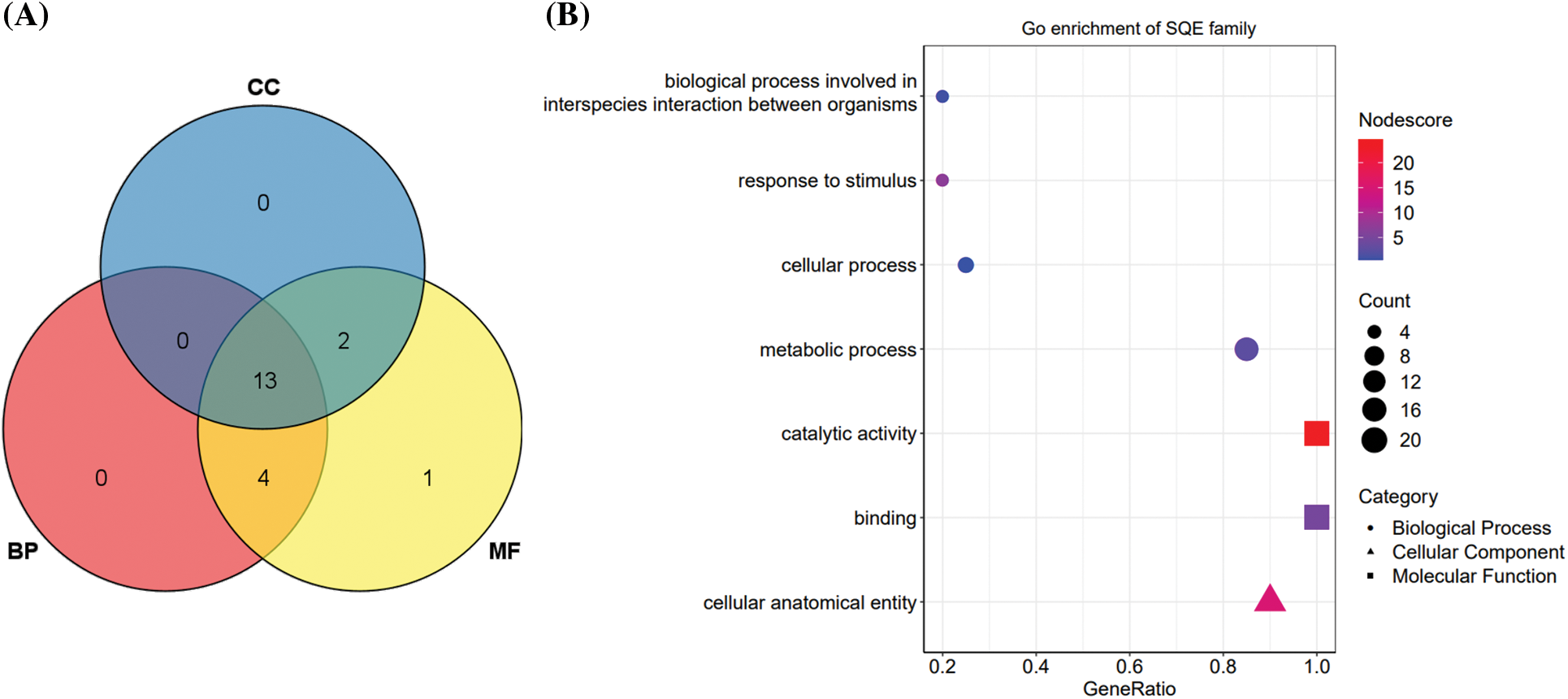
Figure 2: GO functional annotation and enrichment analyses of PgSQE gene sequences. (A) Venn diagram of PgSQE gene transcripts categorized into biological process (BP), molecular function (MF), and cellular component (CC) functional terms. (B) PgSQE transcripts were divided into seven functional categories at level 2, including one CC functional category (triangles), two MF functional categories (squares), and four BP functional categories (circles)
3.4 Expression Patterns of PgSQE Genes
The expression patterns of PgSQE genes are shown in Fig. 3 and Table S2. We found that the expression patterns of PgSQE genes varied with age, tissue type, and cultivar. Similarly, the expression patterns of different PgSQE genes varied in age-, tissue-, and cultivar-matched plants, indicating that the expression levels of PgSQE genes were affected by age, tissue type, cultivar, and external growth environment.
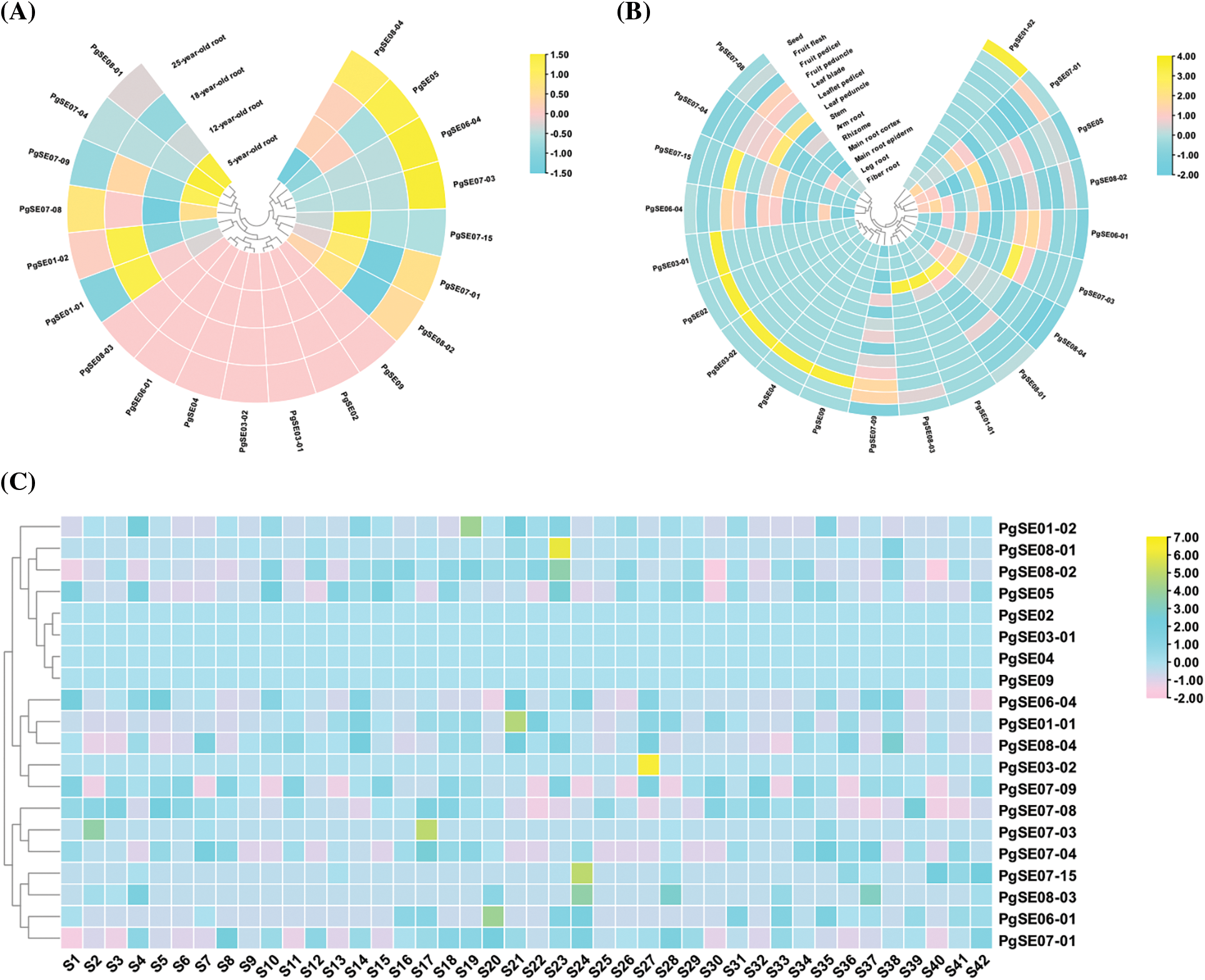
Figure 3: Heatmaps of PgSQE genes. (A) The expression pattern of PgSQE genes in the roots of plants at four different ages. (B) The expression pattern of PgSQE genes in 14 tissues of four-year-old ginseng plants. (C) The expression pattern of PgSQE genes in the roots of four-year-old plants of 42 cultivars. S1–S42 represent 42 ginseng cultivars
3.5 Correlation between Saponin Synthesis and PgSQE Genes
We determined the correlation coefficient between PgSQE genes and saponin content in ginseng. The results showed that, of the 20 PgSQE genes, six genes were associated with saponin synthesis and significantly correlated with multiple monomeric saponins. PgSQE08-01 was correlated with Rf, Rg2, Rc, Re, Rb1, Rb2, Rb3, and Rd. We also estimated the correlation between the genes encoding key enzymes in the ginsenoside biosynthetic pathway and PgSQE genes, which verified that PgSQE08-01 was involved in ginsenoside biosynthesis (Table S3). PgSQE08-01 was significantly correlated with FPS-22, CYP-311, UGT, SE2-1, and DS of the ginsenoside biosynthetic pathway, indicating that they may cooperatively affect the synthesis of saponins.
3.6 PgSQE08-01 Gene Cloning and Vector Construction
The length of the ORF of PgSQE08-01 was 1626 bp. The presence of the sequence of PgSQE08-01 in the pCAMBIA3301-PgSQE08-01 overexpression vector was verified using restriction enzyme digestion (Fig. 4A). The sense and antisense fragments of PgSQE08-01 were ligated into the pHANNIBAL vector, which was transformed into Escherichia coli DH5α competent cells. The construction of recombinant vector pHANNIBAL-PgSQE08-01 was verified by restriction enzyme digestion (Fig. 4B). The pART27-PgSQE08-01 vector was constructed by the digestion of recombinant plasmid pHANNIBAL-PgSQE08-01 and the pART27 vector and verified by enzyme digestion (Fig. 4C). In this way, we successfully constructed PgSQE08-01-overexpression and RNAi vectors.
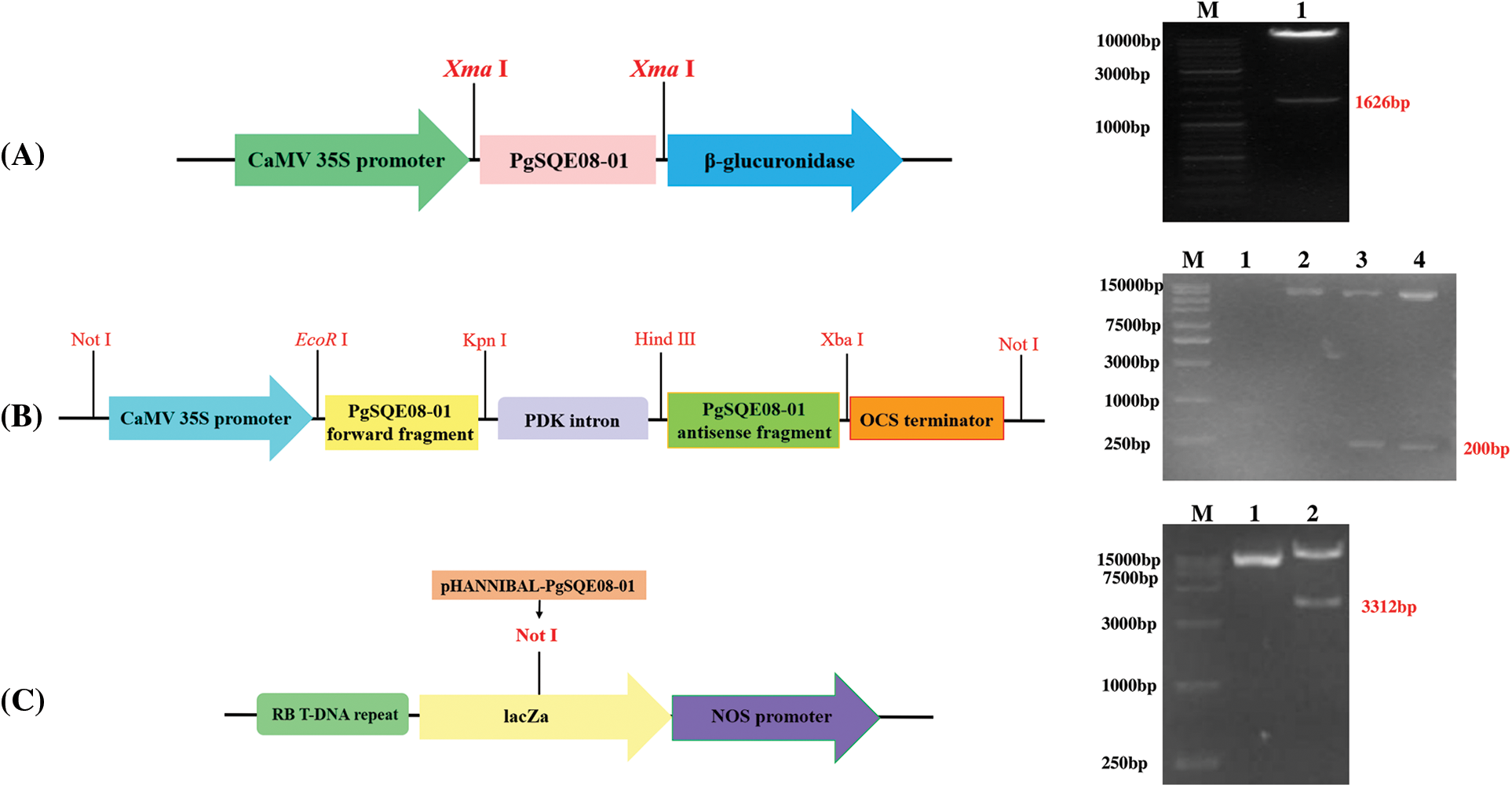
Figure 4: Vector construction for ginseng with PgSQE08-01 gene. (A) Construction of overexpression vector. (B) Construction of intermediate vectors. (C) Construction of the final vector
3.7 Genetic Transformation and Identification of Positively Transformed Adventitious Roots
After transforming the explants with A. rhizogenes A4 and culturing them in the dark for 4–8 weeks, we observed hairy roots growing on the explants. When the hairy roots were 1–2 cm long, we transferred them to a new medium for subculturing. Then, well-developed hairy roots were transferred from the subculture to half MS medium (liquid) for multiplication (Figs. 5A–5I).
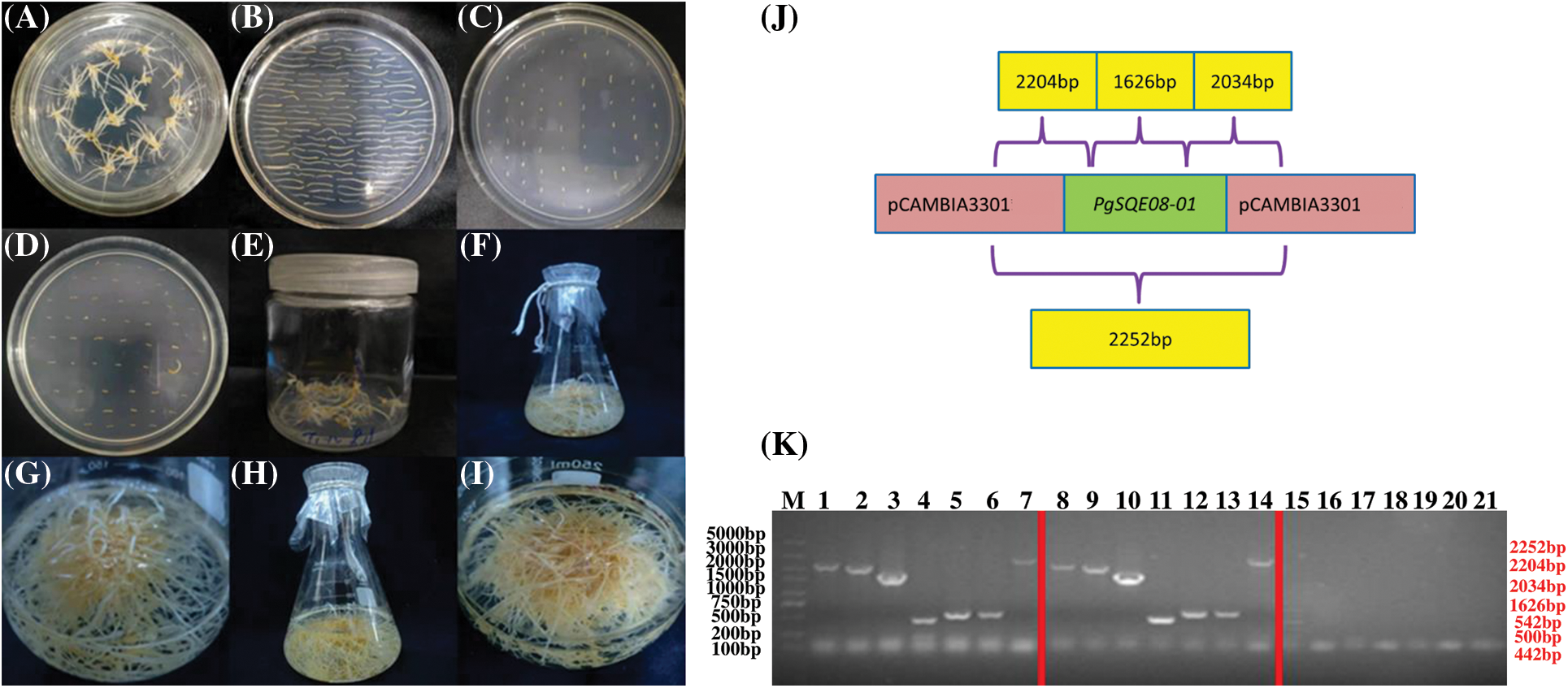
Figure 5: Transformation of hairy roots to overexpress PgSQE08-01. (A) Adventitious roots. (B) Precultured explants. (C) Co-cultured explants. (D) An A4-infected explant containing PgSQE08-01. (E) Induced hairy roots. (F–I) Positively transformed hairy roots. (J) Primer designing for the detection of PgSQE08-01 in positively transformed hairy roots. (K) Agarose gel showing the components of the overexpression vector; M: DL 5000 marker; 1–7, 8–14: T-DNA left region, T-DNA right region, target gene (1626 bp), Bar, GFP, and rol-C; 15–21: negative control (water)
To verify whether the gene of interest was overexpressed in the hairy roots, we extracted DNA from the hairy root for PCR-based detection. The primers were designed as shown in Fig. 5J. The PCR results revealed the following structure.
From left to right: T-DNA left region (2204 bp), T-DNA right region (2034 bp), PgSQE08-01 (1626 bp); Bar (442 bp), GFP (542 bp); rol-C (500 bp), and pCAMBIA3301 vector (2252 bp).
With wildtype hairy roots serving as the negative control and water as the blank, we performed agarose gel electrophoresis to determine the length of the components of the vector. The size of the bands on the gel was consistent with the estimated results, suggesting that PgSQE08-01 was integrated into the genome of the transformed hairy roots (Fig. 5K).
We used the abovementioned approach to generate RNAi vectors with PgSQE08-01 (Figs. 6A–6E), which was followed by PCR validation. The components of the vector are as follows.
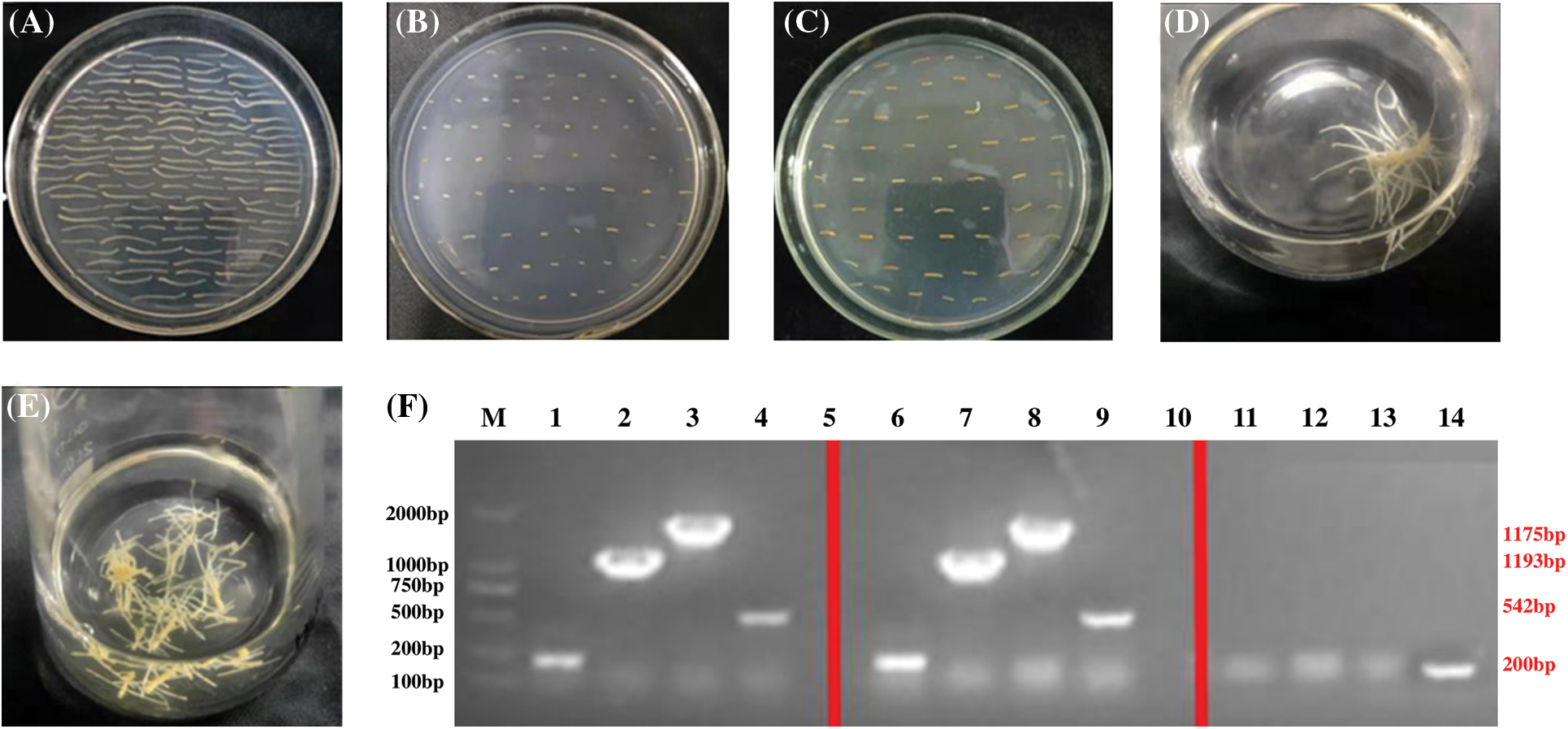
Figure 6: Transformation of hairy roots to silence PgSQE08-01. (A) Precultured explants. (B) Co-cultured explants. (C) An A4-infected explant containing PgSQE08-01. (D and E) Positively transformed hairy roots. (F) Agarose gel showing the components of the RNAi vector; M: DL 5000 marker; 1–4, 6–9: antisense fragment, T-DNA left region, T-DNA right region, and rol-C; 11–14: negative control (water)
From left to right: antisense fragments of the RNAi vector (200 bp), left region of T-DNA (1193 bp), right region of T-DNA (1775 bp), and rol-C (500 bp).
The size of the bands obtained upon electrophoresis was consistent with the calculated results, verifying PgSQE08-01 silencing in the genome of transformed hairy roots (Fig. 6F).
3.8 Expression Levels of the Genes Encoding Five Key Enzymes and Ginsenoside Content in PgSQE08-01-Overexpressing Hairy Roots
To determine the effects of the gene of interest on saponin content, we extracted saponins from hairy roots and estimated their content. We used HPLC to detect monomer saponins in two positively transformed hairy root samples (SQE-8 and SQE-15). The results showed that in the SQE-15 clone, the content of monomer saponins Rb1, Rb2, and Re increased 1.77, 1.94, and 1.71 times, respectively. In the SQE-8 clone, the content of monomer saponin Rb3 and Rd increased 1.65 and 3.08 times, respectively, whereas the content of Rg1 was significantly reduced to 0.64 times. Furthermore, the total glycoside content in SQE-8 and SQE-15 increased by 1.78 and 2.13 times, respectively (Fig. 7).
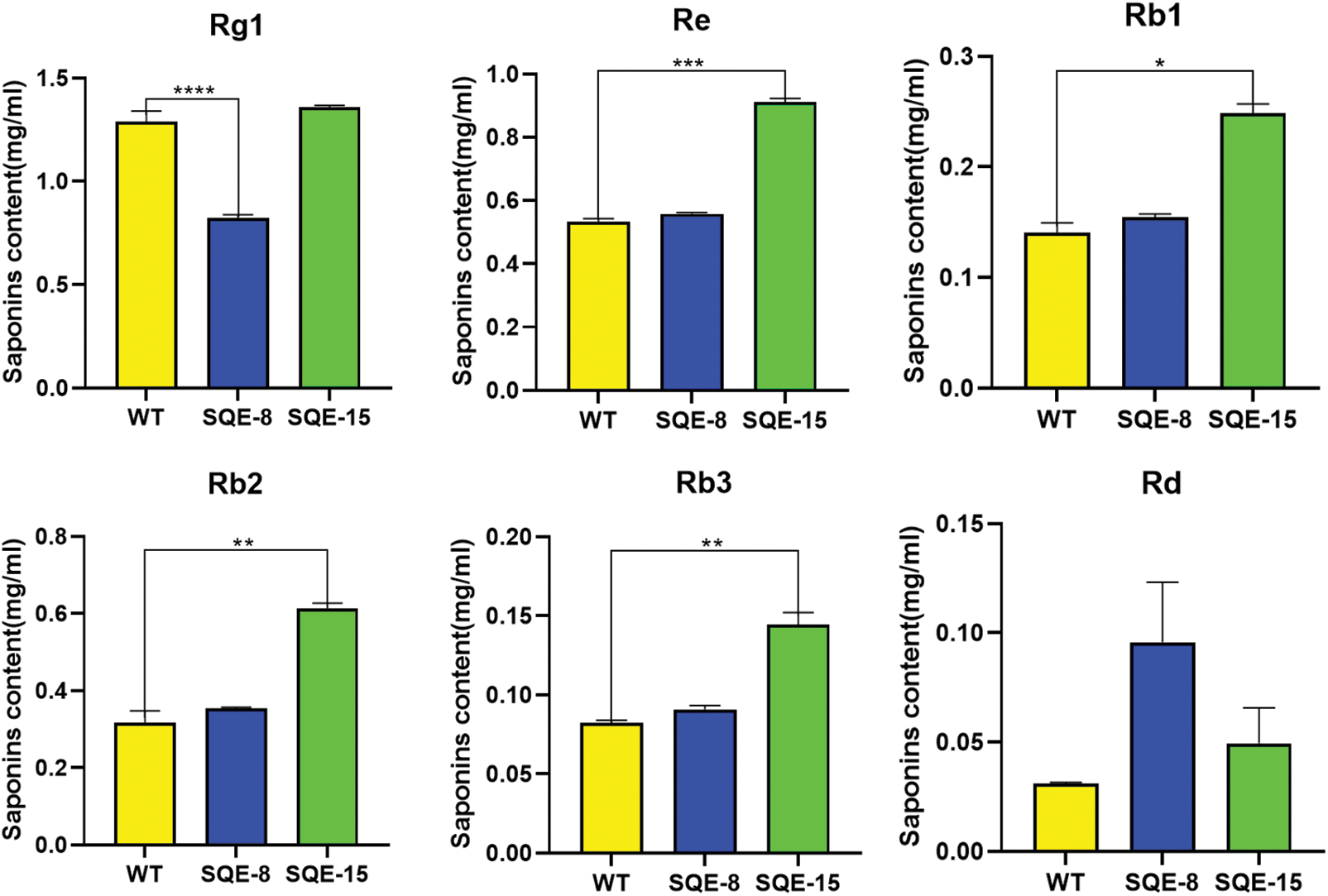
Figure 7: The content of six ginsenosides in positively transformed hairy roots. The X-axis indicates the number of positively transformed hairy roots. WT represents the control. SQE-8 and SQE-15 clones exhibited positively transformed hairy roots. The Y-axis indicated relative ginsenoside content. “*” indicates significant difference at p ≤ 0.05, “**” indicates significant difference at p ≤ 0.01, “***” indicates significant difference at p ≤ 0.001, and “****” indicates significant difference at p ≤ 0.0001, respectively
We further selected FPS, SE2-1, DS, CYP-311 (CYP716A52v2_3), and UGT (UGT71A27_2) of the ginsenoside biosynthetic pathway to study their expression levels in PgSQE08-01-transformed hairy roots. In PgSQE08-01-overexpressing hairy roots, the expression levels of the five genes increased significantly (Fig. 8). This indicated that the overexpression of PgSQE08-01 activated other key enzymes of the saponin biosynthetic pathway, thereby enhancing saponin content and affecting ginsenoside biosynthesis. Thus, it can be concluded the PgSQE08-01 exhibited a significant positive correlation with ginsenoside biosynthesis. Unfortunately, the growth of hairy roots transformed with the RNAi vector was not enough to detect saponin content and gene expression. Thus, further investigations are necessary to understand the molecular mechanism underlying the role of PgSQE08-01 in saponin biosynthesis in ginseng.
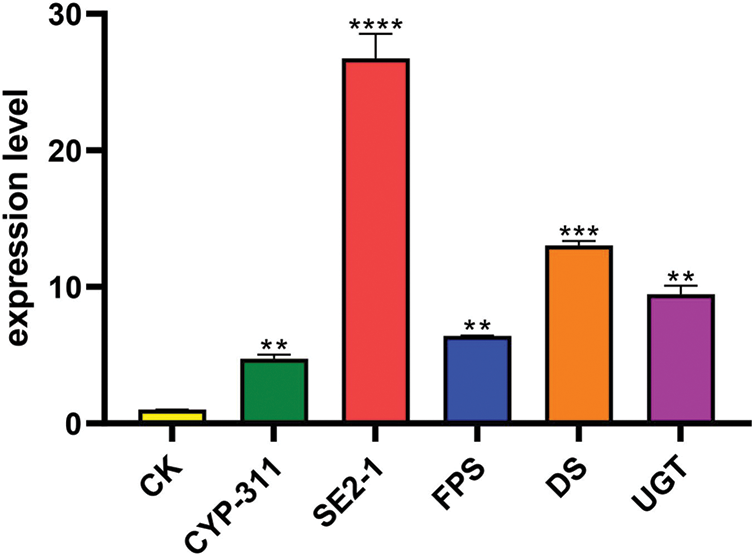
Figure 8: The expression levels of the genes encoding five key enzymes in PgSQE08-01-overexpressing hairy roots. The X-axis indicates the genes encoding five key enzymes. The Y-axis indicates relative expression levels. CK served as the control. The five key enzymes involved in ginsenoside biosynthesis that were used in this study were FPS, SE2-1, DS, CYP-311, and UGT genes. “**” indicates significant difference at p ≤ 0.01, “***” indicates significant difference at p ≤ 0.001, and “****” indicates significant difference at p ≤ 0.0001, respectively
SQE, a key enzyme that functions at the end of the saponin synthesis pathway [40], catalyzes the production of epoxidized squalene from squalene to alter the synthesis efficiency of various terpenoids with squalene as the precursor [3,6]. Therefore, SQE is one of the key enzymes in the terpenoid synthesis pathway [41–43].
In 2020, Han [23] found that overexpression of PgSQE1 increased the contents of Rg1, Re, Rf, Rc, Rb1, Rb2, Rd, and sterols, and the relative expression levels of dammaranediol synthase and cycloenol synthase. These findings are partially consistent with the results of this study. In this study, the content of Rb1, Rb2, and Re significantly increased in the hairy roots of SQE-15, whereas the content of Rb3 and Rd significantly increased in the hairy roots of SQE-8. Moreover, the overexpression of PgSQEO8-01 resulted in an increase in the relative expression of DS, which ultimately increased the content of glycol-type ginsenosides.
The increased content of Rd and Re in SQE-8 and SQE-15 and the decreased content of Rg1 in SQE-8 could not help elucidate the mechanism by which PgSQE08-01 altered the content of triol-type ginsenosides in ginseng. However, qRT-PCR results showed that the overexpression of PgSQE08-01 increased the relative expression of CYP311. Therefore, we hypothesized that the overexpression of PgSQE08-01 gene increased the content of triol ginsenosides; nonetheless, the synthesis mechanism is still unclear.
In 2018, Dong et al. [44] found that the co-expression of SQE and triterpene cyclase promoted the production of triterpenes in plants, indicating that the synergy among the genes encoding key enzymes affected the synthesis of triterpenoids. Accordingly, we proposed two hypotheses to explain the results obtained in the present study. Under the first hypothesis, the overexpression of PgSQE08-01 gene catalyzed the synthesis efficiency of epoxidized squalene, promoting the expression of DS and ultimately increasing the content of triol-type saponins and total saponins. The second hypothesis suggested that the co-expression of PgSQE08-01 and DS increased the content of protopanaxdiol-type ginsenosides and enhanced the expression of P450 or the synergy of P450 with PgSQE08-01 gene and DS (or a gene encoding one of the key enzymes), which ultimately increased the content protopanaxtriol-type ginsenosides and total saponins. However, further studies are required to identify the key enzyme that, or whose co-expression, plays a role in the synthesis of triol-type saponins.
The content of phytosterols is an important indicator of secondary metabolites, and thus understanding sterol biosynthetic pathways has also been a topic for discussion in recent years [2–4]. Though the present study did not detect plant sterols, based on previous literature and the data in this study, it can be inferred that the overexpression of PgSQE08-01 would increase the relative expression of LS and ultimately affect the content of lupeol. Accordingly, we hypothesized that the overexpression of PgSQE08-01 may enhance lupeol content in ginseng. In addition, the overexpression of PgSQE08-01 would increase the expression of β-AS, and thus we hypothesized that it may increase the content of aromatic resins [45,46]. We also hypothesized that the overexpression of PgSQE08-01 resulted in a relative increase in CAS expression, which may increase the content of cycloartenol [47]. According to previous studies and the data in this study, several sterol biosynthetic pathways can be involved in this process. However, the synthesis mechanism is unclear. Thus, multiple sterol biosynthetic pathways and factors affecting the content of sterols need to be further investigated.
In this study, we identified PgSQE08-01 as the gene encoding SQE and determined its effects on the ginsenoside biosynthetic pathway. We transformed the adventitious roots of ginseng with PgSQE08-01-overexpression and RNAi vectors and used positively transformed ginseng hairy roots to determine the expression of genes encoding five key enzymes in ginsenoside biosynthesis and the content of six ginsenosides. We thus investigated the function of PgSQE08-01 gene in ginsenoside biosynthesis and its evolution and expression level in Panax ginseng, which provides a theoretical basis for improving the understanding of important functional genes in the plant species and also adds to the genetic resources for ginsenoside biosynthesis using synthetic biology.
Acknowledgement: Not applicable.
Funding Statement: This work was supported by an award from the Department of Science and Technology of Jilin Province (20210402043GH and 20210204063YY).
Author Contributions: Study conception and design: Yi Wang and Meiping Zhang; analysis and interpretation of results: Yang Jiang and Yu Zhang; draft manuscript preparation: Lei Zhu, Lihe Hou, Mingzhu Zhao and Kangyu Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.047938.
References
1. Kim N, Jayakodi M, Lee S, Choi B, Jang W, Lee J. Genome-and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol J. 2018;16(11):1904–17. [Google Scholar] [PubMed]
2. Li Z, Ji G. Ginseng and obesity. J Ginseng Res. 2018;42:1–8. [Google Scholar] [PubMed]
3. Li X, Liu J, Zuo T, Hu Y, Li Z, Wang H. Advances and challenges in ginseng research from 2011 to 2020: the phytochemistry, quality control, metabolism, and biosynthesis. Nat Prod Rep. 2022;39(4):875–909. [Google Scholar] [PubMed]
4. Zhuravlev Y, Koren O, Reunova G, Muzarok T, Gorpenchenko T, Kats I. Panax ginseng natural populations: Their past, current state and perspectives. Acta Pharmacol Sin. 2008;29(9):1127–36. [Google Scholar] [PubMed]
5. Kim Y, Joo S, Shi J, Hu C, Quan S, Hu J. Metabolic dynamics and physiological adaptation of Panax ginseng during development. Plant Cell Rep. 2018;37(3):393–410. [Google Scholar] [PubMed]
6. Li Y, Wang J, Li L, Song W, Li M, Hua X, et al. Natural products of pentacyclic triterpenoids: from discovery to heterologous biosynthesis. Nat Prod Rep. 2023;40(8):1303–53. [Google Scholar] [PubMed]
7. Yue C, Zhou X, Zhong J. Protopanaxadiol 6-hydroxylase and its role in regulating the ginsenoside heterogeneity in Panax notoginseng cells. Biotechnol Bioeng. 2008;100(5):933–40. [Google Scholar] [PubMed]
8. Wang T, Guo R, Zhou G, Zhou X, Kou Z, Sui F, et al. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: A review. J Ethnopharmacol. 2016;188:234–58. [Google Scholar] [PubMed]
9. Phillips D, Rasbery J, Bartel B, Matsuda S. Biosynthetic diversity in plant triterpene cyclization. Curr Opin Plant Biol. 2006;9:305–14. [Google Scholar] [PubMed]
10. Kim Y, Zhang D, Yang D. Biosynthesis and biotechnological production of ginsenosides. Biotechnol Adv. 2015;33:717–35. [Google Scholar] [PubMed]
11. Rasbery J, Shan H, LeClair RJ, Norman M, Matsuda SPT, Bartel B. Arabidopsis thaliana squalene epoxidase 1 is essential for root and seed development. J Biol Chem. 2007;282:17002–13. [Google Scholar] [PubMed]
12. Guo H, Li R, Liu S, Zhao N, Han S et al. Molecular characterization, expression, and regulation of Gynostemma pentaphyllum squalene epoxidase gene 1. Plant Physiol Biochem. 2016;109:230–39. [Google Scholar] [PubMed]
13. Xia P, Zheng Y, Liang Z. Structure and location studies on key enzymes in saponins biosynthesis of Panax notoginseng. Int J Mol Sci. 2019;20(24):6121. [Google Scholar] [PubMed]
14. Lu H, Liu J, Zhang H, Yin T, Gao S. Ri-mediated transformation of glycyrrhiza uralensis with a squalene synthase gene (GuSQS1) for production of glycyrrhizin. Plant Mol Biol Rep. 2008;26:1–11. [Google Scholar]
15. Liu Y, Zhou J, Hu T, Lu Y, Gao L, Tu L, et al. Identification and functional characterization of squalene epoxidases and oxidosqualene cyclases from Tripterygium wilfordii. Plant Cell Rep. 2020;39(3):409–18. [Google Scholar] [PubMed]
16. Thakur K, Partap M, Kumar P, Sharma R, Warghat A. Understandings of bioactive composition, molecular regulation, and biotechnological interventions in the development and usage of specialized metabolites as health-promoting substances in Siraitia grosvenorii (Swingle) C. Jeffrey. J Food Compos Anal. 2023;116:105070. [Google Scholar]
17. Manzoor MM, Goyal P, Pandotra P, Dar MS, Dar MJ, Misra P, et al. Transcriptome-wide identification of squalene epoxidase genes from Glycyrrhiza glabra L.: expression analysis and heterologous expression of GgSQE1 suggest important role in terpenoid biosynthesis. Protoplasma. 2021;258(5):991–1007. [Google Scholar] [PubMed]
18. Manzoni M, Rollini M. Biosynthesis and biotechnological production of statins by filamentous fungi and application of these cholesterol-lowering drugs. Appl Microbiol Biot. 2002;58(5):555–64. [Google Scholar]
19. Song Y, Guan Z, Merkerk R. Production of squalene in bacillus subtilisby squalene synthase screening and metabolic engineering. J Agric Food Chem. 2020;73(8):305–12. [Google Scholar]
20. Shcherban A, Stasyuk A. Polymorphism of the squalene synthase gene (SQS) in different species of amaranth (Amaranthus L.). Russ J Genet. 2020;56(3):298–306. [Google Scholar]
21. Fu J, Liu G, Yang M. Isolation and functional analysis of squalene synthase gene in tea plant Camellia sinensis. Plant Physiol Bioch. 2019;142:53–8. [Google Scholar]
22. Song Y, Guan Z, Merkerk R. Production of squalene in bacillus subtilis by squalene synthase screening and metabolic engineering. J Agric Food Chem. 2020;68(15):4447–55. [Google Scholar] [PubMed]
23. Han J, Jo H, Choi Y. Overexpression of the squalene epoxidase gene (PgSE1) resulted in enhanced production of ginsenosides and phytosterols in transgenic ginseng. Plant Biotechnol Rep. 2020;14(6):673–82. [Google Scholar]
24. Nam H, Murukarthick J, Sang C. Genome and evolution of the shade-requiring medicinal herb Panax ginseng. Plant Biotechnol. 2018;16(11):1904–17. [Google Scholar]
25. Xu J, Chu Y, Liao B, Xiao S, Yin Q, Bai R, et al. Panax ginseng genome examination for ginsenoside biosynthesis. Gigascience. 2017;6(11):1–15. [Google Scholar]
26. Jayakodi M, Choi B, Lee S. Ginseng genome database: an open-access platform for genomics of Panax ginseng. BMC Plant Biol. 2018;18:62. [Google Scholar] [PubMed]
27. Wang K, Jiang S, Sun C, Lin Y, Yin R, Wang Y, et al. The spatial and temporal transcriptomic landscapes of Ginseng, Panax ginseng CA Meyer. Sci Rep. 2015;5:18283. [Google Scholar] [PubMed]
28. Simon C, Aurélien L, Sean R. HMMER web server: 2018 update. Nucleic Acids Res. 2018;46:W200–4. [Google Scholar]
29. Kumar S, Stecher G, Li M. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018;35(6):1547–9. [Google Scholar] [PubMed]
30. Subramanian B, Gao S, Lercher M, Hu S, Chen W. Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019;47:W270–5. [Google Scholar] [PubMed]
31. Stefan G, Juan MG, Javier T, Tim DW, Shivashankar HN, María JN, et al. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2018;36:3420–35. [Google Scholar]
32. Chen C, Chen H, Zhang Y, Thomas H, Frank M, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13:1194–202. [Google Scholar] [PubMed]
33. Liu C, Zhao M, Ma H, Zhang Y, Liu Q, Liu S, et al. The NAC transcription factor PgNAC41-2 gene involved in the regulation of ginsenoside biosynthesis in Panax ginseng. Int J Mol Sci. 2023;24(15):11946. [Google Scholar] [PubMed]
34. Kim O, Bang K, Jung S, Kim Y, Hyun D, Kim S, et al. Molecular characterization of ginseng farnesyl diphosphate synthase gene and its up-regulation by methyl jasmonate. Biol Plant. 2010;54:47–53. [Google Scholar]
35. Han J, In J, Kwon Y, Choi Y. Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panax ginseng. Phytochemistry. 2010;71:36–46. [Google Scholar] [PubMed]
36. Han J, Kwon Y, Yang D, Jung Y, Choi Y. Expression and RNA interference-induced silencing of the dammarenediol synthase gene in Panax ginseng. Plant Cell Physiol. 2006;47:1653–62. [Google Scholar] [PubMed]
37. Han J, Hwang H, Choi S, Kim H, Choi Y. Cytochrome P450 CYP716A53v2 catalyzes the formation of protopanaxatriol from protopanaxadiol during ginsenoside biosynthesis in Panax ginseng. Plant Cell Physiol. 2012;53:1535–45. [Google Scholar] [PubMed]
38. Jung S, Kim W, Park S, Jeong J, Park M, Lim S, et al. Two ginseng UDP-glycosyltransferases synthesize ginsenoside Rg3 and Rd. Plant Cell Physiol. 2014;55:2177–88. [Google Scholar] [PubMed]
39. Wang K, Zhang Z, Li S, Hu J, Liu T, Jiang Y, et al. Transcriptome-wide analysis for ginsenoside Rb3 synthesis-related genes and study on the expression of methyl jasmonate treatment in Panax ginseng. Life. 2021;11(5):387. [Google Scholar] [PubMed]
40. Li Z, Jiang Y, Guengerich F. Engineering cytochrome P450 enzyme systems for biomedical and biotechnological applications. J Biol Chem. 2019;295(3):758–8763. [Google Scholar]
41. Yan Y, Zhang S, Zhang J. Effect and mechanism of endophytic bacteria on growth and secondary metabolite synthesis in Salvia miltiorrhiza hairy roots. Acta Physiol Plant. 2014;36(5):12–8. [Google Scholar]
42. Liu W, Zhu D, Liu D. Influence of P deficiency on major secondary metabolism in flavonoids synthesis pathway of chrysanthemum morifolium ramat. J Plant Nutr. 2015;38(6):523–8. [Google Scholar]
43. Frapolli M, Pothier JF, Défago G. Evolutionary history of synthesi pathway genes for phloroglucinol and cyanide antimicrobials in plant-associated fluorescent pseudomonads. Mol Phylogenet Evol. 2012;63(3):877–90. [Google Scholar] [PubMed]
44. Dong L, Jacob P, Jean-Etienne B. Co-expression of squalene epoxidases with triterpene cyclases boosts production of triterpenoids in plants and yeast. Metab Eng. 2018;49:1–12. [Google Scholar] [PubMed]
45. Zhao C, Xu T, Liang Y, Zhao S, Ren L, Wang Q, et al. Functional analysis of β-amyrin synthase gene in ginsenoside biosynthesis by RNA interference. Plant Cell Rep. 2015;34(8):1307–15. [Google Scholar] [PubMed]
46. Zhao M, Lin Y, Wang Y, Li X, Han Y, Wang K, et al. Transcriptome analysis identifies strong candidate genes for ginsenoside biosynthesis and reveals its underlying molecular mechanism in Panax ginseng C.A. Meyer. Sci Rep. 2019;9:615. [Google Scholar] [PubMed]
47. Jin M, Lee W, Kim O. Two cycloartenol synthases for phytosterol biosynthesis in polygala tenuifolia willd. Int J Mol Sci. 2017;18(11):2426. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools