 Open Access
Open Access
ARTICLE
Identification and Transcriptional Regulation of CAMTA Genes in Liriodendron chinense
State Key Laboratory of Tree Genetics and Breeding, College of Life Sciences, Nanjing Forestry University, Nanjing, 210037, China
* Corresponding Author: Liming Yang. Email:
Phyton-International Journal of Experimental Botany 2024, 93(3), 413-425. https://doi.org/10.32604/phyton.2024.047739
Received 15 November 2023; Accepted 17 January 2024; Issue published 28 March 2024
Abstract
This study explores CAMTA genes in the rare and endangered Chinese plant species, Liriodendron chinense. Despite the completion of whole-genome sequencing, the roles of CAMTA genes in calcium regulation and stress responses in this species remain largely unexplored. Within the L. chinense genome, we identified two CAMTA genes, Lchi09764 and Lchi222536, characterized by four functional domains: CG-1, TIG, ANK repeats, and IQ motifs. Our analyses, including phylogenetic investigations, cis-regulatory element analyses, and chromosomal location studies, aim to elucidate the defining features of CAMTA genes in L. chinense. Applying Weighted Gene Co-Expression Network Analysis (WGCNA), we explored the impact of CAMTA genes on different organs and their regulation under abiotic stress conditions. The identification of significant gene modules and the prediction of promoter binding sites revealed co-expressed genes associated with CAMTA transcription factors. In summary, this study provides initial insights into CAMTA genes in L. chinense, laying the groundwork for future research on their evolution and biological roles. This knowledge enhancement contributes to a better understanding of plant responses to environmental stress—an essential aspect of plant biology.Keywords
Supplementary Material
Supplementary Material FileIn plants, calcium ions (Ca2+) are essential minerals that form an integral part of the cell wall and act as secondary messengers within the cells [1]. Ca2+ is essential for controlling a number of physiological functions, such as detecting outside stimuli and preserving the membrane potential across cell membranes [2]. A collection of Ca2+-dependent sensors and modulators are necessary for the Ca2+-mediated signal transduction. Calmodulins (CaMs), calcineurin B-like proteins (CBLs), calmodulin-like proteins (CMLs), calcium-dependent protein kinases (CDPKs), and calcineurin-binding transcription activators (CAMTAs) are some examples of these sensors [3,4].
CAMTAs are transcription factors that are widely distributed in multicellular organisms and have the ability to bind to CaM. Numerous anchor protein repeat sequences that facilitate protein-protein interactions, a Ca2+-dependent CaM-binding domain (CaMBD), a TIG domain involved in non-specific DNA binding, a CG-1 DNA-binding domain at the N-terminus, and varying numbers of non-Ca-dependent CaM-binding motifs are some of the distinctive structural domains that define them [5]. The roles of the CAMTA genes are varied. They have the ability to precisely identify and bind to DNA cis-elements found in downstream genes’ promoter regions, controlling the expression of those genes [6]. For instance, EDS1 (Enhanced Disease Susceptibility 1) serves as an immune regulatory factor upstream in the CAMTA signaling pathway. In Arabidopsis thaliana, CAMTA1 can specifically bind to the CG box in the promoter region of the EDS1 gene, thereby suppressing its expression [7]. Moreover, A. thaliana’s CAMTA3 can bind to the CM2 sequence (CCGCGT) in the CBF2 gene’s promoter region, activating CBF2 as well as other genes [8]. Moreover, CAMTAs are involved in maintaining the pH within vacuoles [9], regulating auxin transport [10], regulating cell division and growth, reacting to outside signal inputs, and taking part in disease resistance [11], modulating fruit development [12] and maturation, and adapting to environmental stressors [13].
For instance, in A. thaliana, AtCAMTA1 and AtCAMTA5 are expressed during pollen development [14]. Wheat’s CAMTAs play a role in signal transduction against rust pathogen invasion [15]. In Brassica campestris, BcCAMTAs exhibit significantly increased expression during cold stress but are suppressed during drought, heavy metal, and nitrate stress [16]. In Populus trichocarpa, under short-term mannitol stress, a response is characterized by the upregulation of all 7 PtCAMTA genes in both roots and leaves. However, during prolonged treatment, the responses of PtCAMTA3, PtCAMTA6, and PtCAMTA7 are relatively subdued [17].
Typically found in the southern Yangtze River region of China, L. chinense is a widely dispersed, valuable timber and decorative tree species. L. chinense’s genome sequencing offers the chance to identify the CAMTA genes, perform bioinformatics analysis, and investigate the transcriptional regulation of these genes. With the help of this work, the biological roles of the CAMTA genes in L. chinense will hopefully be better understood.
2.1 The Identification of CAMTA Genes in L. chinense
The genomic and chromosome annotation data for L. chinense were obtained from the genome database through downloading (https://www.ncbi.nlm.nih.gov/datasets/genome/GCA_003013855.2/).
The CAMTA gene and protein sequences from the A. thaliana genome database were downloaded from https://www.arabidopsis.org. The CAMTA gene and protein sequences from the Oryza sativa genome database were obtained from http://rapdb.dna.affrc.go.jp.
The genome sequences from L. chinense genome database were downloaded from ftp://ftp.cngb.org. Hidden Markov Model (HMM) files were acquired from the Pfam database (http://pfam.xfam.org/). The TIG domain (PF01833), ANK repeat (PF00023), and IQ motif (PF00612) in the Pfam database were the sources of Hidden Markov Model (HMM) files, which were selected using the E-value <10−5 criterion. Protein sequences of CAMTA genes from A. thaliana and O. sativa were utilized in the construction of a Hidden Markov Model.The software for protein domain alignment, HMMER 3.0 (http://hmmer.janelia.org/) was utilized to identify all potential CAMTA genes members in the L. chinense genome database. We constructed a CAMTA genes database for L. chinense using the gene sequences from O. sativa and A. thaliana. The sequences were aligned using the BLAST 2.9.0 + tool, and alignment results were obtained. We’ll use the conventional E-value cutoff of 10. These BLAST results were then cross-referenced with the pfam database to acquire a more comprehensive set of data. The last phase resulted in the definitive dataset for L. chinense’s CAMTA genes. We discovered where the results of the pfam alignment and the BLAST alignment intersected.
2.2 Physicochemical Property Analysis and Phylogenetic Tree Analysis
Basic information about the L. chinense CAMTA genes, including protein molecular weight, isoelectric point, and tertiary structure, was analyzed using the ExPASy database (http://www.expasy.org/). Conservation motifs within the L. chinense CAMTA genes were identified using the MEME online tool (https://meme-suite.org/meme/tools/meme), and motif diagrams were generated. A. comosus, Carica papaya, Solanum lycopersicum, Citrus sinensis, Theobroma cacao, Nicotiana sylvestris, P. trichocarpa, Brassica napus, Malus domestica, Nicotiana tomentosiformis, Dorcoceras hygrometricum, A. thaliana, Chlorella variabilis, O. sativa CAMTA sequences were downloaded from the website http://planttfdb.gao-lab.org/. Constructing the phylogenetic tree using the maximum likelihood method was performed with MEGA11 software.
Using the Tbtools toolkit, gene architecture, conserved domains, conserved protein motifs, and chromosomal positions were visualized and examined. Predict cis-acting elements using the PlantCARE software (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/), visualization processing through TBtools version 0.674.
2.3 Transcriptome Data Analysis and qPCR Validation
After being obtained from NCBI, raw transcriptome data (PRJNA559687 [18], PRJNA679089, PRJNA679101 [19]) were obtained from NCBI and underwent quality control using fastp. To do differential gene analysis, the R (version 4.2.2) package Desq2 was used. A padj value less than 0.01 and an absolute |log2foldchange| more than two were used to identify differentially expressed genes for additional research.Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment were predicted using Eggnog-Mapper (http://eggnog-mapper.embl.de/). The GO enrichment analysis database was sourced from http://purl.obolibrary.org/obo/go/go-basic.obo, while the KEGG enrichment analysis database was obtained from https://www.yuque.com/cjchen/hirv8i/ghaxyl. Visualization of the analysis results was conducted using TBtools. Upstream 2000 bp sequences were selected as the promoter and transcription factor binding sites were predicted within the promoter region using https://jaspar.genereg.net/ [20].
The comprehensive repertoire of transcription factors for A. thaliana was obtained from http://planttfdb.gao-lab.org/ and utilized for comparative analysis with the complete transcription factor genome of L. chinense. Genes demonstrating a Pearson correlation coefficient exceeding 0.6 and a p-value below 0.05 were selected as those co-expressed with the CAMTA genes.
Furthermore, R version 4.2.2 was used to create a heatmap based on Weighted Gene Co-expression Network Analysis (WGCNA), and Cytoscape v3.10.1 was used to show the co-expression network. Venn diagrams were created via the web-based tool available at http://www.interactivenn.net/. The font colors of the generated figures were manually adjusted using Adobe Illustrator version 28.0. Finally, a heatmap depicting the expression levels of the CAMTA gene in L. chinense was created using R version 4.2.2.
L. chinense seedlings underwent low-temperature treatment for 0, 1, 3, 6, 12, 24, and 72 h. To ensure the accuracy and reliability of the results, quantitative RT-PCR (qRT-PCR) validation was performed. The 2−ΔΔCt method was employed to calculate the relative expression levels of the CAMTA genes. The internal reference gene selected for normalization was the 18S gene of L. chinense. All qRT-PCR primers were designed using Primer 5.0. The primer design details are provided in Supplementary Table 1.
3.1 Identification of the CAMTA Genes in L. chinense
Two CAMTA genes have been successfully identified in the genome of L. chinense. These genes possess distinct structural domains and exhibit specific biochemical properties, including mild acidity, variability in stability, and hydrophilicity. Designated as Lchi09764 and Lchi22536, these genes possess four essential functional domains: the CG-1 domain, TIG domain, ANK repeat structure, and IQ motif. They are located on chromosomes Chr4 and Chr3, respectively (Supplementary Tables 2, 3). In the L. chinense CAMTA gene, a total of 5 motifs were identified and designated as motif 1, motif 2, motif 3, motif 4, and motif 5 (Supplementary Fig. 1).
3.2 Evolutionary Tree Analysis
An evolutionary tree was constructed using a set of plant species, including L. chinense and A. comosus, C. papaya, S. lycopersicum, C. sinensis, T. cacao, N. sylvestris, P. trichocarpa, B. napus, M. domestica, N. tomentosiformis, D. hygrometricum, A. thaliana, C. variabilis, O. sativa (Fig. 1). Evolutionary tree analysis reveals that the CAMTA genes in L. chinense are relatively closely related to O. sativa and C. papaya.
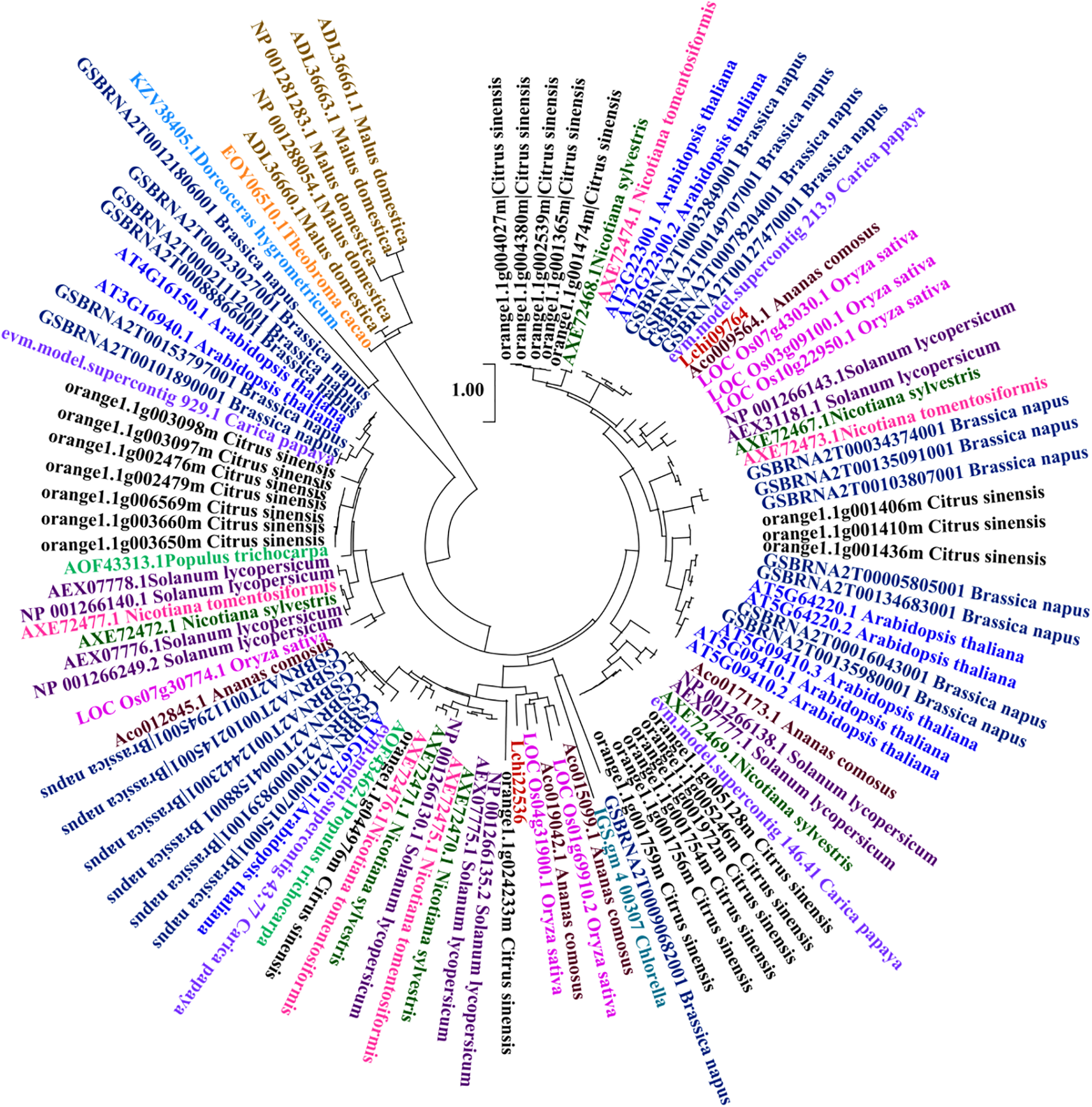
Figure 1: Analysis of the evolutionary tree of L. chinense CAMTA genes. Different font colors represent different species
3.3 Identification of the CAMTA Genes in L. chinense
L. chinense CAMTA transcription factor can modulate gene expression by binding to its cis-element during stress response (Supplementary Fig. 2, Supplementary Table 4). The promoter region, which is a sequence approximately 2000 bp upstream of the translation start site, was submitted to PlantCARE to identify cis-elements.
3.4.1 Differential Gene Analysis under Abiotic Stress
According to transcriptome study, there are three main types of stress that cause the expression of CAMTA genes: heat, drought, and cold. Nonetheless, it was noted that these genes’ expression profiles differed depending on the type of stress. Create a Venn diagram that shows the number of genes that are differentially expressed in response to various abiotic stimuli, as shown in Supplementary Fig. 3. The heatmap of genes co-expressed with L. chinense CAMTA genes that are differently expressed is displayed in Supplementary Fig. 4A. The number of differentially expressed genes in L. chinense that are upregulated and downregulated in response to different abiotic stress treatments is displayed in a bar chart in Supplementary Fig. 4B. Based on Pearson correlation coefficient-based co-expression network analysis, it can be inferred that the L. chinense CAMTA genes may co-regulate abiotic stress responses alongside transcription factors such as BES1, C3H, ERF, GATA, LSD, MYB-related, NAC, SBP, WRKY, and YABBY (Fig. 2).
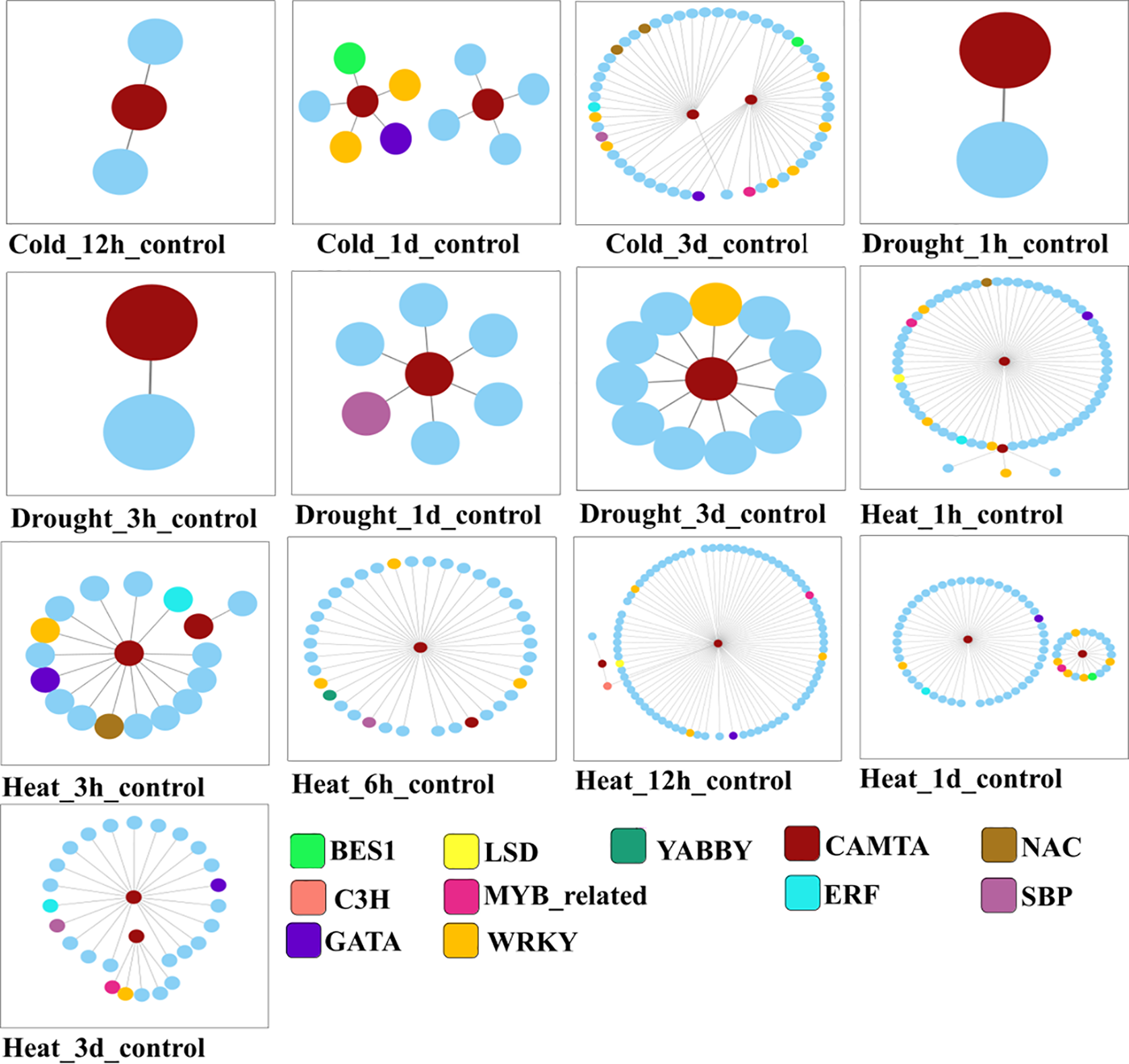
Figure 2: L. chinense CAMTA genes and co-expression dynamics under abiotic stress. Under each abiotic stress treatment, the nodes reflect differentially expressed genes of L. chinense, and the various colors correspond to different matching transcription factors. Co-expression is indicated by the connecting lines. The association between genes with variable expression is shown by the lines
3.4.2 Enrichment Analysis of Differentially Expressed Genes Co-Expressed with the L. chinense CAMTA Genes
We performed GO and KEGG enrichment analyses on differentially expressed genes responsive to abiotic stress and co-expressed with L. chinense CAMTA genes (Fig. 3). These differentially expressed genes that co-expressed the CAMTA genes were shown to be enriched in terms of Molecular Function, Cellular Component, and Biological Process, with the highest level of enrichment observed in the GO enrichment analysis. This implies that a number of elements of plant growth and development are regulated by the L. chinense CAMTA genes. This gene set’s involvement in several biological processes, such as glycosylation, RNA biology, cytoskeleton organization, chromosomal structure and function, and gene expression, was shown by the KEGG enrichment analysis.
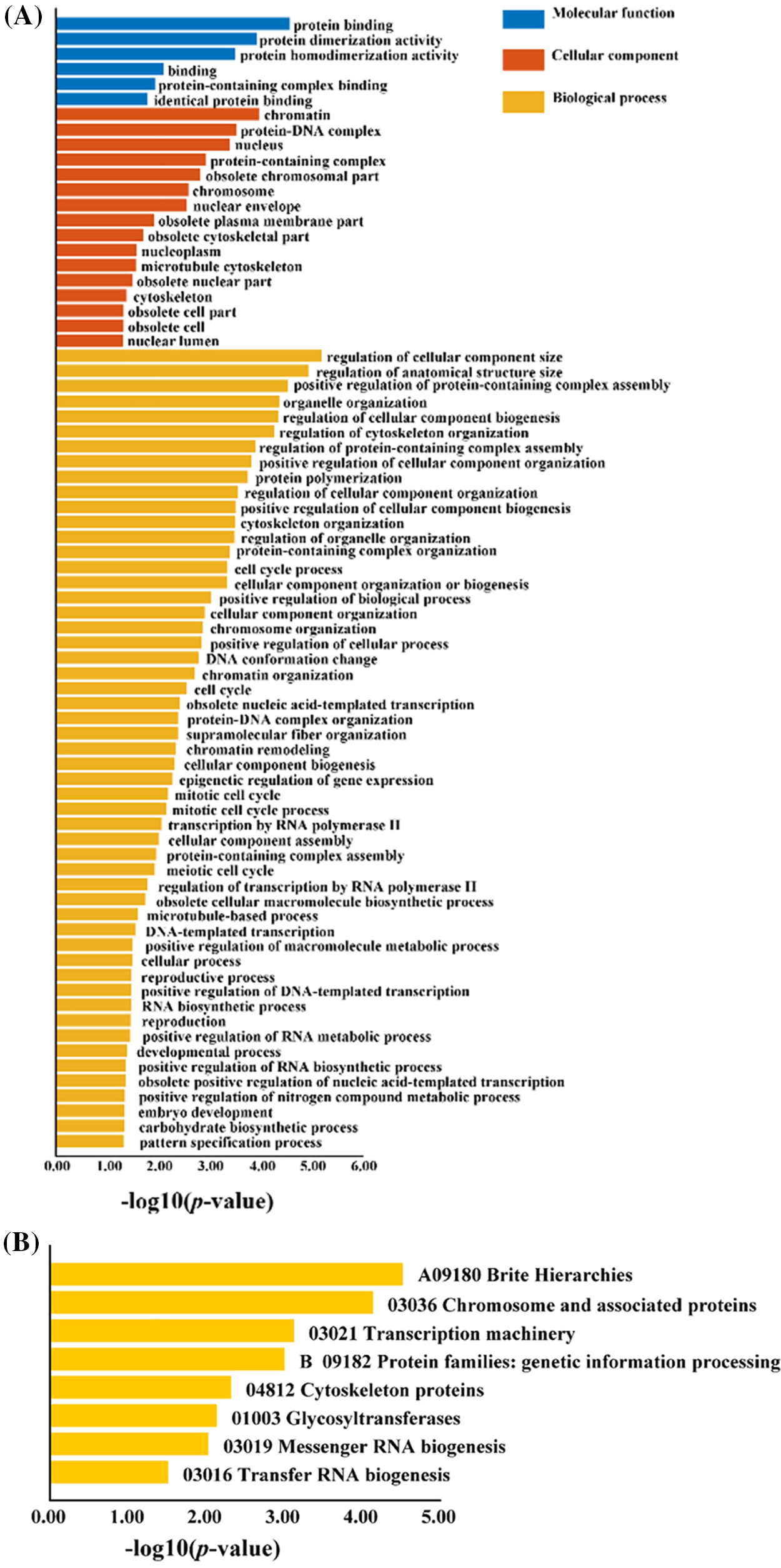
Figure 3: GO enrichment and KEGG enrichment analysis of differentially expressed genes co-expressed with the L. chinense CAMTA genes
3.4.3 WGCNA Analysis under Abiotic Stress Conditions
WGCNA was performed on the differentially expressed genes of L. chinense under abiotic stress, revealing a total of 4642 genes with altered expression patterns (Supplementary Fig. 5). Originally, only genes with variances larger than the fourth percentile were kept in the gene selection process. We then used the Pearson correlation coefficient to do a sample clustering study and produced a dendrogram that illustrates the sample clustering tree. (Supplementary Fig. 5A). To construct an unscaled network, a soft threshold of 16 was chosen (Supplementary Fig. 5B), leading to the generation of an adjacency matrix and the creation of a topological overlap matrix (Supplementary Fig. 6A). Six unique gene modules were successfully found by WGCNA; these gene modules are distinguished by genes that have similar expression patterns (Supplementary Fig. 6B). This approach greatly enhances our comprehension of the interrelationships and expression profiles of these genes.
Additionally, we chose modules whose sample characteristics had correlation coefficients higher than 0.5. The brown, green, blue, and yellow modules made up these modules. Interestingly, we found differentially expressed genes that co-expressed with CAMTA genes within the blue module. Thus, we put up the theory that the blue module’s genes and the CAMTA genes in L. chinense may work together to coordinate the plant’s reaction to a 6-h heat stress. Genes co-expressed with CAMTA under non-biological stress are found in the blue module. We used Cytoscape to create a network graph (Fig. 4). A diagram displaying the genes found in the blue module. This thorough investigation may reveal important relationships between genes and stress reactions, expanding our knowledge in this area. The prediction of transcription factor binding sites in the upstream promoter regions of genes in the blue module reveals that Lchi10447 and Lchi18769 are not only co-expressed with the L. chinense CAMTA transcription factor but also have CAMTA transcription factor binding sites in their upstream regions (Supplementary Tables 5, 6, Supplementary Fig. 7).
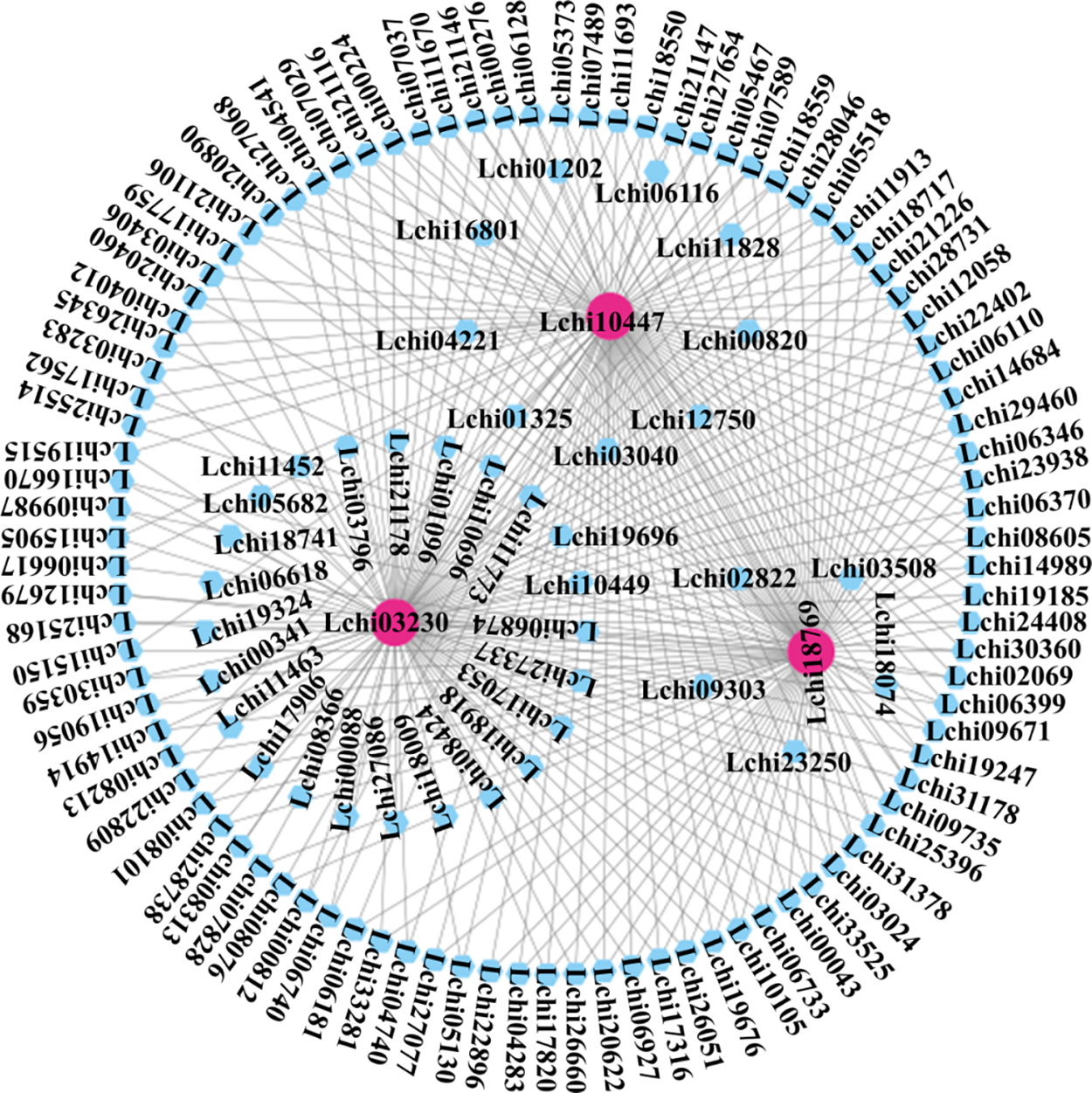
Figure 4: Gene co-expression network plot within the blue module of WGCNA. Differentially expressed genes under abiotic stress conditions are represented by the nodes in the network. Genes that co-express with the L. chinense CAMTA genes are shown by pink nodes. The correlation between pairs of differentially expressed genes is shown by the connecting lines
3.4.4 Differential Gene Expression Analysis for Different Organs Using WGCNA
We carried out a WGCNA on a set of 4943 differential genes, obtained from various organs including bract, leaf, petal, pistil, shoot, sepal, and stamen, in order to further investigate the regulatory roles of the CAMTA genes in the development of different organs in L. chinense (Supplementary Fig. 8).
First, we kept genes whose variances were higher than the fourth quartile criteria in order to undertake gene selection. The Pearson correlation coefficient was then used to perform sample clustering analysis, which led to the creation of a sample clustering tree (Supplementary Fig. 8A). To establish an unscaled network, we selected a soft threshold of 18 (Supplementary Fig. 8B). Following this step, we generated an adjacency matrix and constructed a topological overlap matrix, as illustrated in the (Supplementary Fig. 9A). WGCNA successfully identified six gene modules, each comprising genes with analogous expression patterns, thereby enhancing our comprehension of the correlations and expression profiles among these genes (Supplementary Fig. 9B).
To begin gene selection, we first retained genes whose variances above the fourth quartile threshold. Following sample clustering analysis using the Pearson correlation coefficient, a sample clustering tree was produced. Significantly elevated correlation coefficients (Supplementary Fig. 10). And an analysis of motifs in the upstream promoter sequences was conducted (Supplementary Fig. 11). There are 161 genes altogether in the yellow module. This strategy has great potential to further our knowledge of how the CAMTA genes in L. chinense regulate relevant genes’ transcriptional expression throughout leaf development.
The upstream 2000 bp sequences of genes in the yellow module were extracted for further analysis. Motif analysis was conducted, and the prediction of transcription factor binding sites was performed using JASPAR (Supplementary Tables 7, 8). The presence of CAMTA binding sites within the promoter regions of Lchi14980 and Lchi01238 has been identified.
To further validate the transcriptional regulation by the CAMTA genes in the L. chinense, we conducted qRT-PCR validation following different time points of cold stress treatment (1, 3, 6, 12, 24 h, 3 d). The qRT-PCR results revealed a distinct expression pattern for Lchi09764 and Lchi22536. These genes initially exhibited an upregulation in response to cold stress, followed by a subsequent decrease, and then a subsequent increase as the duration of cold stress treatment extended (Supplementary Fig. 12).
We may now go on to the CAMTA genes, which actively contribute to the regulation of these processes, after talking about calcium signals and their function in plants’ responses to stressors. In particular, the CAMTA gene family members display a range of actions in response to several biotic and abiotic stressors, such as temperature and light [21], salinity [22], hormones [23], and pathogens [24]. This family of transcription factors plays a crucial role in orchestrating plant growth and development under challenging environmental conditions. Researchers have identified this transcription factor in various species, including seven members in Populus [17], eight in Phaseolus vulgaris [25], fifteen in wheat [26], and ten in Heimia myrtifolia [27]. The CAMTA genes exhibit slight variations across different species. In our study, we identified two CAMTA genes in L. chinense, and these identified genes may exhibit distinct functionalities.
Since the CAMTA gene is involved in both the control of abiotic stress responses and plant growth and development, it serves a dual role in various plant species. For example, in A. thaliana, AtCAMTA3 expression levels are downregulated during fruit development but significantly upregulated during fruit ripening. AtCAMTA5 is essential for the formation of pollen [28], whereas AtCAMTA6 is a key regulator of seed germination [29]. In tea plants, CsCAMTAs play a crucial role in regulating the aging process and flowering period [30]. In tomatoes, there is a high differential expression during both fruit development and ripening processes [31]. In Musa acuminata, all MuCAMTA genes exhibit upregulation under drought stress [32]. Our research highlights the critical function that the CAMTA genes perform in L. chinense. In addition to components associated with the regulation of diurnal cycles and light responsiveness, this gene also contains cis-acting elements relevant to meristem expression. Notably, under non-biotic stress circumstances, the CAMTA genes in L. chinense co-express with differently expressed genes, providing more evidence for their potential involvement in regulating plant growth and development. In academic discourse, these discoveries offer significant new information for a fuller understanding of plants’ capacity for environmental adaptation. The effects of low temperature stress are substantial in the field of plant biology because they have a major influence on various processes related to growth and development.
Consequently, there is a need to establish intricate regulatory networks that confer resilience and adaptability that enable plants to withstand and adapt to the challenges posed by cold conditions [33]. In A. thaliana, the dehydration-responsive element (DRE) and C-repeat motif (C-REPEAT) are crucial elements for the expression of cold-induced genes. The DRE-binding DREB1/CBF transcription factors play a major regulatory role in cold responses, and their absence results in reduced cold tolerance in plants [34]. In A. thaliana cold stress, CAMTA3 and CAMTA5 induce DREB1B/C via temperature/Ca2+, and RVE4/8 activates DREB1A/C expression during daylight, slightly activating DREB1B regardless of temperature. RVE4/8 uses EE/EE-like sequences, while CAMTA3 and CAMTA5 use CGCG-box/CGCG-like sequences [34]. CAMTA transcription factors may collaborate with other transcription factors such as NAC, MYB, bZIP, CBF, and WRKY during cold stress [35]. In A. thaliana, the mutation of CAMTA3 leads to a significant reduction in CBF2 and other cold-induced genes under cold conditions [36]. In tea plants, the expression levels of CsCAMTA1/3/4/6 are higher in cold-tolerant varieties than in cold-sensitive ones, indicating their crucial role throughout the entire cold acclimation process [30]. In the context of the current research, understanding the regulatory mechanisms of the CAMTA genes in cold stress becomes crucial for the L. chinense. This study validated the response of the L. chinense CAMTA genes to cold stress at different time points through qRT-PCR. For high-temperature and drought conditions, a more in-depth analysis may be required in future studies.
The CAMTA genes are crucial for immunological responses because they actively regulate calcium ion levels and stress reactions [37]. In A. thaliana, the CAMTA3 mutant demonstrates constitutive immunity under low-temperature conditions, whereas the functionally acquired CAMTA3-3D mutant exhibits a markedly diminished disease resistance [38]. In tomatoes, upon infection with Botrytis cinerea, all CAMTA genes manifest an upregulated expression pattern in leaf tissues [39]. Unfortunately, despite our thorough bioinformatics investigation of the CAMTA genes in L. chinense, no immune-related functions were found. This discovery deviates from the immune-related roles found in other species’ CAMTA genes. This discrepancy could be caused by variables peculiar to a particular species or by special CAMTA gene regulation mechanisms in L. chinense immune responses. Further research is necessary to confirm these findings with greater rigor, explore the specific function of the CAMTA genes in immune control in L. chinense, and develop a more thorough knowledge of their significance in plant biology. It is noteworthy that the CAMTA genes is intricately linked with hormone regulation [33]. In Cucumis sativus, a majority of CAMTA genes possess diverse hormone-responsive elements and stress-related motifs, exhibiting varying expression patterns across different tissues and inducibility by hormones such as IAA and ABA [40]. Similarly, in tea plants, CAMTA genes harbor cis-acting elements related to hormone regulation [41]. The promoter regions of CitCAMTAs in Citrus sinensis contain cis-acting elements responsive to growth hormones, including Auxin Response Elements (AuxR-core) and TGA elements [42]. In Linum usitatissimum, the promoter regions of LuCAMTAs contain Abscisic Acid Response Elements (ABRE) and Salicylic Acid Response Elements (TCA elements) [43]. In this study, putative cis-acting elements associated with hormone responses within the L. chinense CAMTA genes have been predicted.
However, it is crucial to stress that further experimental validation is required for these first investigations. More research is necessary to confirm the precise involvement of the CAMTA genes in various biological processes, particularly in the areas of hormone control, stress response, and physiological and biochemical regulatory mechanisms.
This study found two CAMTA genes in L. chinense (Lchi09764 and Lchi22536). Bioinformatics analyses covered gene structure, evolution, organ regulation, and responses to abiotic stress. WGCNA analysis highlighted key co-regulated genes in organ development and abiotic stress, with Lchi10447 and Lchi18769 identified as pivotal co-expressed with CAMTA. Additionally, WGCNA emphasized the importance of Lchi14980 and Lchi01238 in coordinating organ development with CAMTA, linked to the yellow module related to leaf growth. These findings deepen our understanding of L. chinense CAMTA in regulating abiotic stress responses and organ growth, validated by qPCR under cold stress indicating significant regulatory roles for CAMTA genes.
Acknowledgement: None.
Funding Statement: This research was funded by the National Natural Science Foundation of China (No. 31971682) and the Research Startup Fund for High-Level and High-Educated Talents of Nanjing Forestry University.
Author Contributions: K.H. conducted a literature review and drafted the manuscript. Y.R. and J.C. examined and edited the manuscript. T.M. provided assistance with image modifications, data analysis, and language polishing. L.Y. secured funding and supervised the progress of manuscript.
Availability of Data and Materials: The datasets analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.047739.
References
1. Khan F, Goher F, Paulsmeyer M, Hu CG, Zhang JZ. Calcium (Ca2+) sensors and MYC2 are crucial players during jasmonates-mediated abiotic stress tolerance in plants. Plant Biol. 2023;25(7):1025–34. doi:10.1111/plb.13560. [Google Scholar] [PubMed] [CrossRef]
2. Shi X, Bao J, Lu X, Ma L, Zhao Y, Lan S, et al. The mechanism of Ca2+ signal transduction in plants responding to abiotic stresses. Environ Exp Bot. 2023;216:105514. doi:10.1016/j.envexpbot.2023.105514. [Google Scholar] [CrossRef]
3. Ren H, Zhang Y, Zhong M, Hussian J, Tang Y, Liu S, et al. Calcium signaling-mediated transcriptional reprogramming during abiotic stress response in plants. Theor Appl Genet. 2023;136(10):1–22. doi:10.1007/s00122-023-04455-2. [Google Scholar] [PubMed] [CrossRef]
4. Zeng H, Zhu Q, Yuan P, Yan Y, Yi K, Du L. Calmodulin and calmodulin-like protein-mediated plant responses to biotic stresses. Plant, Cell Environ. 2023;46(12):3680–703. doi:10.1111/pce.14686. [Google Scholar] [PubMed] [CrossRef]
5. Liu C, Tang D. Comprehensive identification and expression analysis of CAMTA gene family in phyllostachys edulis under abiotic stress. PeerJ. 2023;11:e15358. doi:10.7717/peerj.15358. [Google Scholar] [PubMed] [CrossRef]
6. Yang F, Dong F, Hu F, Liu Y, Chai J, Zhao H, et al. Genome-wide identification and expression analysis of the calmodulin-binding transcriptionactivator (CAMTA) gene family in wheat (Triticum aestivum L.). BMC Genetics. 2020;21(1):1–10. doi:10.1186/s12863-020-00916-5. [Google Scholar] [CrossRef]
7. Zhang L, Du L, Shen C, Yang Y, Poovaiah B. Regulation of plant immunity through ubiquitin-mediated modulation of Ca2+–calmodulin-At SR1/CAMTA 3 signaling. Plant J. 2014;78(2):269–81. doi:10.1111/tpj.12473. [Google Scholar] [PubMed] [CrossRef]
8. Carlow C. Analysis of the vitis C-repeat binding factor (CBF) genes and their potential roles in both the CBF and stomatal development pathways. University of Guelph; 2016. [Google Scholar]
9. Dong F, Yang F, Liu Y, Jia W, He X, Chai J, et al. Calmodulin-binding transcription activator (CAMTA)/factors in plants. In: Calcium transport elements in plants. Netherlands: Elsevier; 2021. p. 249–66. doi:10.1016/B978-0-12-821792-4.00017-5. [Google Scholar] [CrossRef]
10. Iqbal Z, Iqbal MS, Sangpong L, Khaksar G, Sirikantaramas S, Buaboocha T. Comprehensive genome-wide analysis of calmodulin-binding transcription activator (CAMTA) in Durio zibethinus and identification of fruit ripening-associated DzCAMTAs. BMC Genom. 2021;22(1):1–22. doi:10.1186/s12864-021-08022-1. [Google Scholar] [PubMed] [CrossRef]
11. Chang Y, Bai Y, Wei Y, Shi H. CAMTA3 negatively regulates disease resistance through modulating immune response and extensive transcriptional reprogramming in cassava. Tree Physiol. 2020;40(11):1520–33. doi:10.1093/treephys/tpaa093. [Google Scholar] [PubMed] [CrossRef]
12. Yu J, Song B, Gu K, Cao B, Zhao K, Wu J, et al. Genome-wide identification and expression analysis of CAMTA gene family implies PbrCAMTA2 involved in fruit softening in pear. Hortic. 2023;9(4):467. doi:10.3390/horticulturae9040467. [Google Scholar] [CrossRef]
13. Meenakshi, Kumar A, Kumar V, Dubey AK, Narayan S, Sawant SV, et al. CAMTA transcription factor enhances salinity and drought tolerance in chickpea (Cicer arietinum L.). Plant Cell, Tissue Organ Cult (PCTOC). 2022;148(2):319–30. doi:10.1007/s11240-021-02191-3. [Google Scholar] [CrossRef]
14. Zhu X, Wang B, Wei X, Du X. Characterization of the CqCAMTA gene family reveals the role of CqCAMTA03 in drought tolerance. BMC Plant Biol. 2022;22(1):428. doi:10.1186/s12870-022-03817-0. [Google Scholar] [PubMed] [CrossRef]
15. Li M, Yang Z, Liu J, Chang C. Wheat susceptibility genes TaCAMTA2 and TaCAMTA3 negatively regulate post-penetration resistance against Blumeria graminis forma specialis tritici. Int J Mol Sci. 2023;24(12):10224. doi:10.3390/ijms241210224. [Google Scholar] [PubMed] [CrossRef]
16. Hu R, Wang Z, Wu P, Tang J, Hou X. Identification and abiotic stress analysis of calmodulin-binding transcription activator/signal responsive genes in non-heading Chinese cabbage (Brassica campestris ssp. chinensis Makino). Plant Omics. 2015;8(2):141–7. doi:10.3316/INFORMIT.065797151928435. [Google Scholar] [CrossRef]
17. Wei M, Xu X, Li C. Identification and expression of CAMTA genes in Populus trichocarpa under biotic and abiotic stress. Sci Rep. 2017;7(1):17910. doi:10.1038/s41598-017-18219-8. [Google Scholar] [PubMed] [CrossRef]
18. Zong Y, Hao Z, Tu Z, Shen Y, Zhang C, Wen S, et al. Genome-wide survey and identification of AP2/ERF genes involved in shoot and leaf development in Liriodendron chinense. BMC Genom. 2021;22(1):1–17. doi:10.1186/s12864-021-08119-7. [Google Scholar] [PubMed] [CrossRef]
19. Wu W, Zhu S, Zhu L, Wang D, Liu Y, Liu S, et al. Characterization of the Liriodendron Chinense MYB gene family and its role in abiotic stress response. Front Plant Sci. 2021;12:641280. doi:10.3389/fpls.2021.641280. [Google Scholar] [PubMed] [CrossRef]
20. Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, Van Der Lee R, et al. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;46(D1):D260–6. doi:10.1093/nar/gkx11260. [Google Scholar] [CrossRef]
21. Zhang H, Zhu J, Gong Z, Zhu JK. Abiotic stress responses in plants. Nat Rev Genet. 2022;23(2):104–19. doi:10.1038/s41576-021-00413-0. [Google Scholar] [PubMed] [CrossRef]
22. Kumar A, Kumar V, Dubey A, Narayan S, Sawant S, Pande V, et al. CAMTA1 transcription factor regulates salinity and drought tolerance in chickpea (Cicer arietinum L.). 2021. doi:10.21203/rs.3.rs-840327/v1. [Google Scholar] [CrossRef]
23. Yang Y, Sun T, Xu L, Pi E, Wang S, Wang H, et al. Genome-wide identification of CAMTA gene family members in Medicago truncatula and their expression during root nodule symbiosis and hormone treatments. Front Plant Sci. 2015;6:459. doi:10.3389/fpls.2015.00459. [Google Scholar] [PubMed] [CrossRef]
24. Kim YS, An C, Park S, Gilmour SJ, Wang L, Renna L, et al. CAMTA-mediated regulation of salicylic acid immunity pathway genes in Arabidopsis exposed to low temperature and pathogen infection. The Plant Cell. 2017;29(10):2465–77. doi:10.1105/tpc.16.00865. [Google Scholar] [PubMed] [CrossRef]
25. Büyük İ., İlhan E, Şener D, Özsoy AU, Aras S. Genome-wide identification of CAMTA gene family members in Phaseolus vulgaris L. and their expression profiling during salt stress. Mol Biol Rep. 2019;46:2721–32. doi:10.1007/s11033-019-04716-8. [Google Scholar] [PubMed] [CrossRef]
26. Yang F, Dong FS, Hu FH, Liu YW, Chai JF, Zhao H, et al. Genome-wide identification and expression analysis of the calmodulin-binding transcription activator (CAMTA) gene family in wheat (Triticum aestivum L.). BMC Genet. 2020;21(1):1–10. doi:10.1186/s12863-020-00916-5. [Google Scholar] [PubMed] [CrossRef]
27. Yang L, Zhao Y, Zhang G, Shang L, Wang Q, Hong S, et al. Identification of CAMTA gene family in Heimia myrtifolia and expression analysis under drought stress. Plants. 2022;11(22):3031. doi:10.3390/plants11223031. [Google Scholar] [PubMed] [CrossRef]
28. Mitsuda N, Isono T, Sato MH. Arabidopsis CAMTA family proteins enhance V-PPase expression in pollen. Plant Cell Physiol. 2003;44(10):975–81. doi:10.1093/pcp/pcg137. [Google Scholar] [PubMed] [CrossRef]
29. Shkolnik D, Finkler A, Pasmanik-Chor M, Fromm H. Calmodulin-binding transcription activator 6: a key regulator of Na+ homeostasis during germination. Plant Physiol. 2019;180(2):1101–18. doi:10.1104/pp.19.00119. [Google Scholar] [PubMed] [CrossRef]
30. Li B, He S, Zheng Y, Wang Y, Lang X, Wang H, et al. Genome-wide identification and expression analysis of the calmodulin-binding transcription activator (CAMTA) family genes in tea plant. BMC Genom. 2022;23(1):667. doi:10.1186/s12864-022-08894-x. [Google Scholar] [PubMed] [CrossRef]
31. Yang T, Peng H, Whitaker BD, Conway WS. Characterization of a calcium/calmodulin-regulated SR/CAMTA gene family during tomato fruit development and ripening. BMC Plant Biol. 2012;12(1):1–13. doi:10.1186/1471-2229-12-19. [Google Scholar] [PubMed] [CrossRef]
32. Meer L, Mumtaz S, Labbo AM, Khan MJ, Sadiq I. Genome-wide identification and expression analysis of calmodulin-binding transcription activator genes in banana under drought stress. Sci Hortic. 2019;244:10–4. doi:10.1016/j.scienta.2018.09.022. [Google Scholar] [CrossRef]
33. Kidokoro S, Konoura I, Soma F, Suzuki T, Miyakawa T, Tanokura M, et al. Clock-regulated coactivators selectively control gene expression in response to different temperature stress conditions in Arabidopsis. Proc Natl Acad Sci. 2023;120(16):e2216183120. doi:10.1073/pnas.2216183120. [Google Scholar] [PubMed] [CrossRef]
34. Kidokoro S, Shinozaki K, Yamaguchi-Shinozaki K. Transcriptional regulatory network of plant cold-stress responses. Trends Plant Sci. 2022;27(9):922–35. doi:10.1016/j.tplants.2022.01.008. [Google Scholar] [PubMed] [CrossRef]
35. Xiao P, Feng JW, Zhu XT, Gao J. Evolution analyses of CAMTA transcription factor in plants and its enhancing effect on cold-tolerance. Front Plant Sci. 2021;12:758187. doi:10.3389/fpls.2021.758187. [Google Scholar] [PubMed] [CrossRef]
36. Doherty CJ, van Buskirk HA, Myers SJ, Thomashow MF. Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. The Plant Cell. 2009;21(3):972–84. doi:10.1105/tpc.108.063958. [Google Scholar] [PubMed] [CrossRef]
37. Matsumura M, Nomoto M, Itaya T, Aratani Y, Iwamoto M, Matsuura T, et al. Mechanosensory trichome cells evoke a mechanical stimuli-induced immune response in Arabidopsis thaliana. Nat Commun. 2022;13(1):1216. doi:10.1038/s41467-022-28813-8. [Google Scholar] [PubMed] [CrossRef]
38. Huang J, Sun Y, Orduna AR, Jetter R, Li X. The mediator kinase module serves as a positive regulator of salicylic acid accumulation and systemic acquired resistance. Plant J. 2019;98(5):842–52. doi:10.1111/tpj.14278. [Google Scholar] [PubMed] [CrossRef]
39. Li X, Huang L, Zhang Y, Ouyang Z, Hong Y, Zhang H, et al. Tomato S.R./CAMTA transcription factors SlSR1 and SlSR3L negatively regulate disease resistance response and SlSR1L positively modulates drought stress tolerance. BMC Plant Biol. 2014;14(1):1–19. doi:10.1186/s12870-014-0286-3. [Google Scholar] [PubMed] [CrossRef]
40. Gao R, Luo Y, Yun F, Wu X, Wang P, Liao W. Genome-wide identification, expression profile, and alternative splicing analysis of CAMTA family genes in cucumber (Cucumis sativus L.). Agron. 2021;11(9):1827. doi:10.3390/agronomy11091827. [Google Scholar] [CrossRef]
41. Zhou Q, Zhao M, Xing F, Mao G, Wang Y, Dai Y, et al. Identification and expression analysis of CAMTA genes in tea plant reveal their complex regulatory role in stress responses. Front Plant Sci. 2022;13:910768. doi:10.3389/fpls.2022.910768. [Google Scholar] [PubMed] [CrossRef]
42. Zhang J, Pan X, Ge T, Yi S, Lv Q, Zheng Y, et al. Genome-wide identification of citrus CAMTA genes and their expression analysis under stress and hormone treatments. J Hortic Sci Biotechnol. 2019;94(3):331–40. doi:10.1080/14620316.2018.1504631. [Google Scholar] [CrossRef]
43. Ali E, Raza MA, Cai M, Hussain N, Shahzad AN, Hussain M, et al. Calmodulin-binding transcription activator (CAMTA) genes family: genome-wide survey and phylogenetic analysis in flax (Linum usitatissimum). PLoS One. 2020;15(7):e0236454. doi:10.1371/journal.pone.0236454. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools