 Open Access
Open Access
ARTICLE
Genome-Wide Identification and Expression Analysis of Calmodulin-Like Proteins in Tobacco
College of Agronomy and Biotechnology, Southwest University, Chongqing, 400715, China
* Corresponding Author: Liquan Zhu. Email:
(This article belongs to the Special Issue: Plant Genetic Diversity and Evolution)
Phyton-International Journal of Experimental Botany 2025, 94(1), 157-179. https://doi.org/10.32604/phyton.2025.060566
Received 04 November 2024; Accepted 05 December 2024; Issue published 24 January 2025
Abstract
Calmodulin-like (CMLs) proteins are critical in calcium signaling and essential for plant growth, development, and stress responses. In many species, the CMLs families have been identified and described. However, the characterization and expression profiling of CMLs genes in tobacco is retrievable. In this study, a comprehensive whole-genome identification and analysis, and 75 NtCML genes were identified in tobacco, each containing two to four EF-hand domains. Most NtCML proteins exhibited conserved gene structures and motifs. Notably, most NtCML proteins were intron-less and distributed across 18 chromosomes. Two pairs of tandemly duplicated genes and seven pairs of segmentally duplicated genes were identified within the tobacco genome. Furthermore, 22 pairs of orthologous CMLs genes were discovered between Arabidopsis and tobacco. Cis-acting element analysis revealed that elements associated with hormones, stress responses, and plant growth and development were found in the promoter regions. Expression analysis indicated that some NtCML genes displayed tissue-specific expression patterns. Specifically, NtCML12, NtCML18, NtCML27, and NtCML28 showed significant upregulation during cold acclimation treatment. These results indicate that tobacco CMLs act as Ca2+ signal transducers, regulating plant growth and abiotic stress responses.Keywords
Calcium ions (Ca2+) are essential for plants, serving as structural componentd of cell walls and membranes, as well as vital secondary messengers. Plant growth, development, and adaptability to biotic and abiotic stresses are significantly influenced by calcium ions [1]. Different abiotic and biotic stresses, including hormones, temperature, drought, salt, disease, and so on, can change variations in the level of cytoplasmic calcium ions and affect the movements of calcium ions in plant cells [1,2]. Calcium-binding proteins can sense and interpret intracellular calcium concentrations before undergoing conformational changes and interacting with downstream signaling partners, supporting the plant in responding to environmental stresses [3,4]. Calcium-binding proteins act as calcium sensors, and there are four kinds of calcium ion sensors of plants: calmodulins (CaMs), calmodulin-like proteins (CMLs), Ca2+-dependent protein kinases (CDPKs) and calcineurin B-like proteins (CBLs), which contain one or more EF-hand motifs [5–8]. They commonly contain elongation factor hand (EF-hand) motifs. The EF-hand, characterized by a helix-loop-helix structure, can bind calcium ions and subsequently undergo a conformational change. This alteration allows it to interact with downstream proteins and adjust its catalytic activity, which includes gene regulation, protein interactions, protein phosphorylation, and metabolic changes [9–11]. Plant CaMs have four EF-hand domains, whereas CMLs have one to six and no extra functional domains. CMLs typically have 16%–75% of their amino acid composition in common with CaMs [12].
The CMLs genes in Arabidopsis thaliana and rice currently exhibit 50 and 32 genes, respectively [12,13]. Since the completion of various plant genome sequencing projects, the CMLs gene family has been identified in numerous other plant species, including tomato [14], cucumber [10], apple [4], grape [11], chrysanthemum [15], barley [16]. Previous research has shown that the genes of CMLs play a significant role in plant development, growth, and stress responses [17]. CML39, CML24, and CML25 are crucial calcium ion sensors in plant growth and development [18–20]. CML9, CML8, and CML41 are involved in plant immune response [21–23]. CML9, CML20, CML24, CML37, CML38, and CML39 participate in plant salt stress response [24–27]. MtCML42 regulates flowering time and cold tolerance by gradually increasing MtFTa expression and decreasing MtABI5 [28]. SlCML39 is a significant negative regulator for high-temperature tolerance [29]. Overexpression of ShCML44 demonstrated increased resistance to salt, drought, and cold stress [30]. CMLs genes are involved in multiple physiological functions.
Tobacco is a significant cash crop and model organism worldwide. Although several NtCMLs have been reported in various publications, a comprehensive examination of tobacco CMLs has yet to be conducted. In this study, 75 NtCMLs genes were identified in the tobacco genome. Comprehensive analyses including gene structure, chromosomal distribution, gene duplications, motifs or domains, cis-acting elements, evolutionary relationships, organ-based gene expression profiles, and under cold acclimation conditions. The findings might provide important insights into physiological and molecular research on the NtCMLs genes.
2.1 Identification of NtCMLs Genes and Sequence Analysis
The published CMLs gene sequences in Arabidopsis and rice were obtained from the TAIR database (https://www.arabidopsis.org) (accessed on 04 December 2024) and the TIGR database (http://rice.plantbiology.msu.edu) to identify members of the CMLs gene family in the tobacco genome. Then, 32 OsCMLs and 50 AtCMLs proteins were used as query sequences to perform BLASTP search (E-value < e − 5), and the redundant and repetitive sequences were removed manually. Meanwhile, the HMMER (PF13499) was utilized as a keyword in the databases above to conduct searches. The NCBI Conserved Domain Database (http://www.ncbi.nlm.nih.gov/cdd) (accessed on 04 December 2024), InterProScan (http://www.ebi.ac.uk/Tools/pfa/iprscan5/) (accessed on 04 December 2024), and SMART (http://smart.embl-heidelberg.de/) (accessed on 04 December 2024) were used to predict the structural domains of EF-hands, eliminating protein sequences that lack EF-hands or contain other functional domains. Additionally, AtCaM2 was used to guarantee the NtCMLs by acting as usual CaMs and ensuring that the amino acid identity was less than 80%. The nucleotide and predicted amino acid sequences of the discovered genes, which were named NtCML1 through NtCML75, were used for additional investigation.
ExPASyProtParam (http://web.expasy.org/protparm/) (accessed on 04 December 2024) was used to estimate the physicochemical properties of NtCMLs, such as the number of amino acids, theoretical point (pI), grand average of hydropathicity (GRAVY), and aliphatic index. SMART was used to forecast the number of EF-hands (https://smart.embl.de/smart/set_mode.cgi?NORMAL=1) (accessed on 04 December 2024). Cell-PLoc 2.0 (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) (accessed on 04 December 2024) was used to predict subcellular localization.
2.2 Gene Structure and Conserved Motif Analysis
MEME (http://meme-suite.org/index.html) (accessed on 04 December 2024) was used to study the conserved domains, and ten motifs were chosen. The exon and intron structures of NtCMLs were ascertained using the Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/) (accessed on 04 December 2024).
Using 1000 bootstrap replicates and the neighbor-joining method in MEGA7, the phylogenetic tree was constructed for evolutionary analysis. The NtCML proteins were classified based on the evolutionary relationships of 50 AtCML proteins. Evolutionary tree landscaping can be achieved using the website ChiPlot (https://www.chiplot.online/) (accessed on 04 December 2024).
2.4 Cis-Acting Elements Analysis
PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html) (accessed on 04 December 2024) was used to examine the 2000 bp upstream sequences of NtCMLs genes to identify the cis-acting elements in the promoter region of NtCMLs genes. The Simple BioSequence Viewer from TBtools is used to visualize.
2.5 Chromosome Localisation and Collinearity Analysis
The online website mg2c (http://mg2c.iask.in/mg2c_v2.1/) (accessed on 04 December 2024) was utilized to visualize the chromosomal position. TBtools software was used to examine the homologous relationship of NtCML genes. The Multiple Collinearity Scan toolbox (MCScanX) examined gene duplication occurrences.
2.6 Tissue Specifc Expression Analysis
Tobacco K326 tissue data from 19 tissues was downloaded from the EMBL-EBI website. The corresponding gene expression profile data were obtained by comparing the sample numbers of NtCMLs gene family members. The data was plotted using the Heat Map tool in the software TBtools.
2.7 Cold Acclimation Stress Treatments
The tobacco cultivar ‘Yunyan87’ was sown in soil and allowed to grow at room temperature (26°C) until the plants had six or seven leaves. The plants were then divided into two groups. The P seedlings were grown in an artificial climate chamber and were treated with cold acclimation therapy for three days at a temperature of 12°C (night/day). The other group of seedlings that were not cold-acclimated is known as N. The N and P seedling groups were treated normally for 7 days and recorded as NCT and PCT. NCT and PCT seedlings were put through cold treatment (8°C) for three days and recorded as NCL and PCL. Each treatment involved the collection of three biological samples, which were immediately frozen in liquid nitrogen and stored at −80°C until they could be examined further.
2.8 RNA Extraction and Gene Expression Analysis Using Real-Time PCR
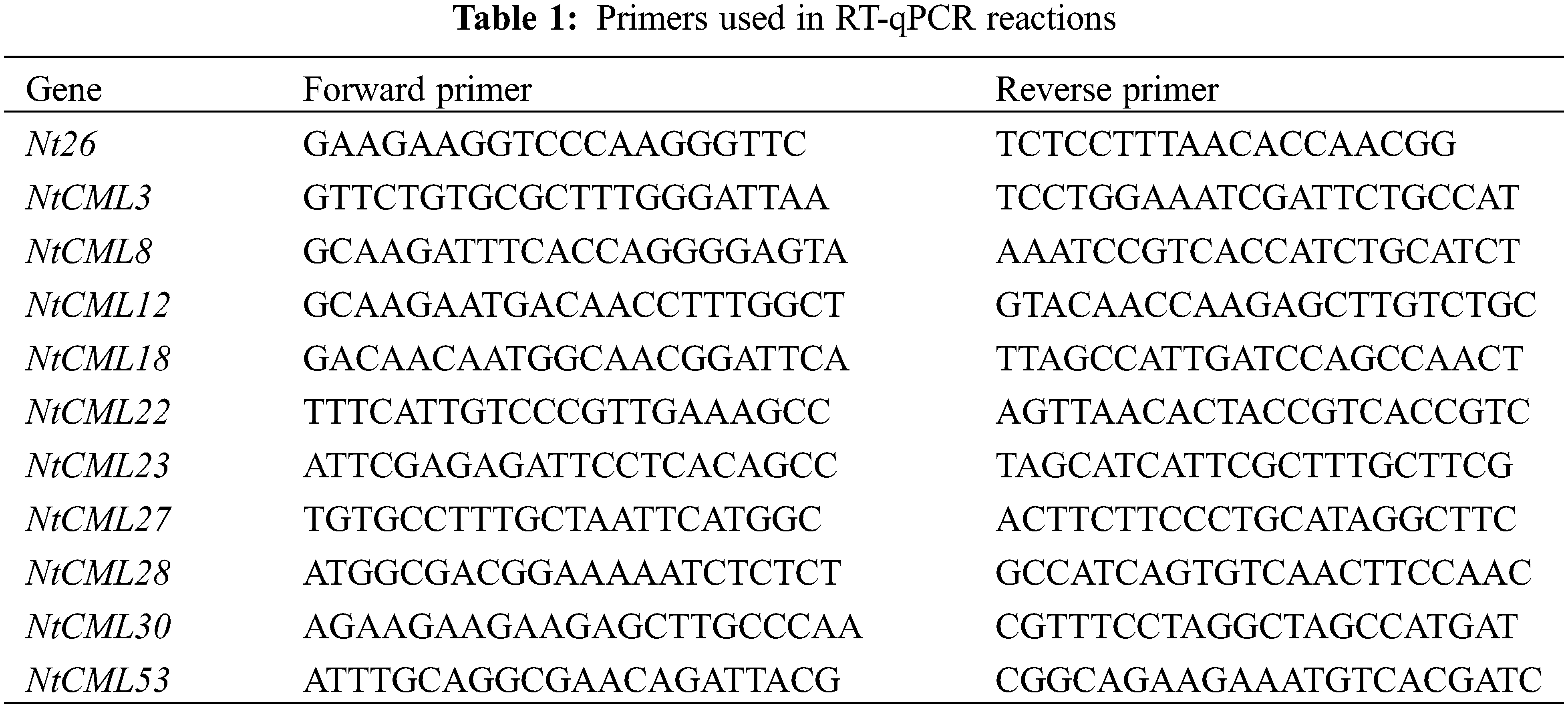
The Spin Column Plant Total RNA Purification Kit (Sangon Biotech, Shanghai, China) was used after the frozen samples had been thoroughly ground to powder in liquid nitrogen. 1% agarose gel electrophoresis was used to assess the quality of the RNA. The All-In-One 5X RT Master Mix (ABM, Shanghai, China) is then used for the cDNA synthesis. The qPrimerDB qPCR Primer Database online resource was utilized to design the primers used in the RT-qPCR. Table 1 lists the particular primers used for qRT-PCR.Quantitative reverse transcription-polymerase chain reaction (qRT-PCR) was carried out in real-time utilizing a CFX-1000 Real-Time System (BioRad) and SYBR Premix Ex Taq II (TaKaRa, China). Three duplicates of the real-time PCR were conducted. We used the N, NCT, and NCL treatments as the baseline to calculate P, PCT, and PCL expression. The N, NCT, and NCL treatments were utilized as a baseline to calculate P, PCT, and PCL expression. The e 2−ΔΔCt technique was used to analyze relative expression [31]. The data shows the average of three biological replicates.
2.9 Protein Interaction Network Prediction
A network of 75 NtCMLs protein sequences was analyzed using the STRING online server (https://cn.string-db.org/) (accessed on 04 December 2024).
SPSS26.0 data processing software is utilized to perform variance analysis and significance experiments on experimental data. Origin2021 software is employed to perform mapping analysis on experimental data.
3.1 Identification and Characterization of NtCMLs Family Members in Tobacco
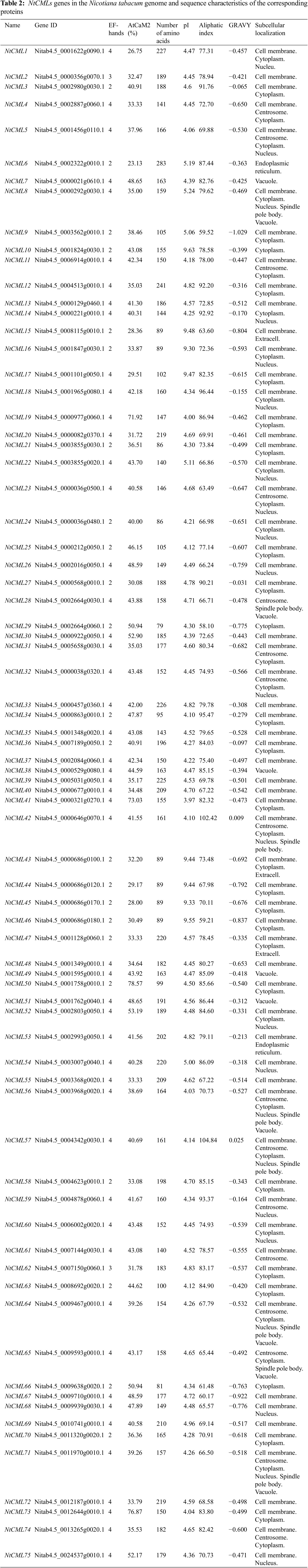
A total of 75 NtCMLs genes were retrieved from the genome of Tobacco and were named in the order of their chromosomal locations (NtCML1 to NtCML40 and NtCML41 to NtCML75) (Table 2). The results were confirmed by analyzing the deduced peptides using Pfam, InterProScan, and SMART databases. Physicochemical property analysis showed that the number of amino acids in the NtCMLs proteins ranged from 79 AA (NtCML39) to 283 AA (NtCML6) and the pI ranged from 3.97 (NtCML41) to 9.63 (NtCML10). The Aliphatic index ranged from 58.10 (NtCML29) to 104.84 (NtCML57). The NtCMLs shared 23%~78% identity with AtCaM2. Most NtCMLs proteins contained two to four EF-hand domains. The GRAVY values of most NtCMLs proteins were negative, indicating that NtCMLs proteins in cucumber are hydrophilic. Subcellular localization demonstrates that most NtCMLs are found in the cell membrane and cytoplasm.
3.2 Gene Structure and Conserved Motif Analysis of the NtCMLs in Tobacco
Motifs are important in identifying Transcription Factor Binding Sites, which helps understand the mechanisms that regulate gene expression [32]. The MEME tool was employed to locate conserved motifs for a more thorough examination of the NtCML proteins (Fig. 1A). Motif 1 and motif 2 were present in all 75 NtCMLs family members. Motifs 3, 4, 5, 6, and 8 are most commonly observed in the N-terminus and motifs 1, 2, and 7 are present in the C-terminus. Some paralogous proteins contained different motifs, such as NtCML14 and NtCML54, NtCML12 and NtCML31, NtCML6 and NtCML9, NtCML41 and NtCML50, while NtCML18 and NtCML59, NtCML38 and NtCML49, NtCML48 and NtCML74, NtCML11 and NtCML37, NtCML40 and NtCML55 had the same motif. Motif 9 is exclusively in NtCML3 and NtCML27.
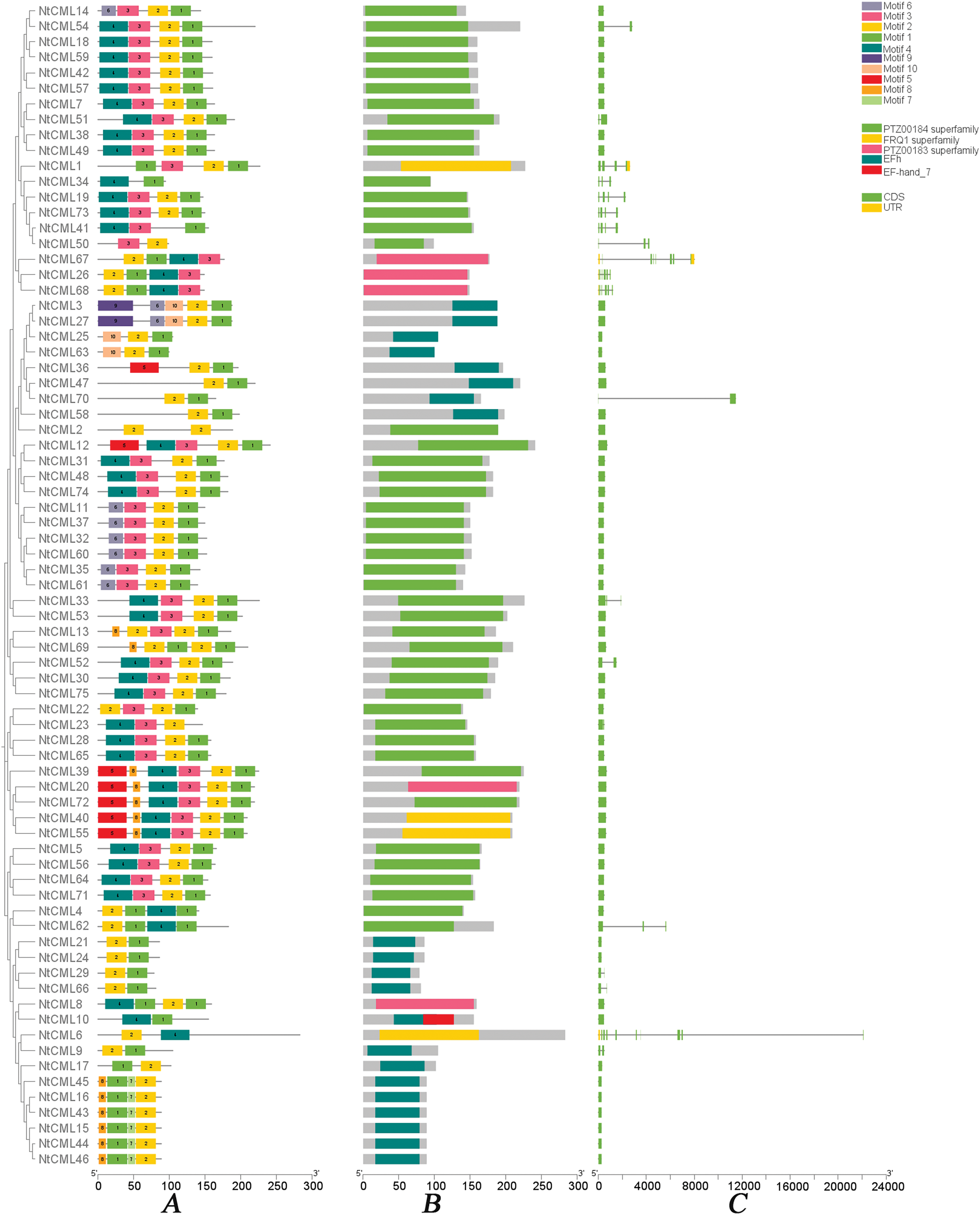
Figure 1: (A) Motif structure of NtCMLs; (B) Domain of NtCMLs; (C) The gene structure of NtCMLs
Most NtCMLs belong to the PTZ00184 superfamily, which is also known as the EF-hand protein superfamily (Fig. 1B), Introns can increase transcript levels, and exons as enhancers are crucial in protein synthesis [33,34]. We analyzed the exon-intron structure of NtCMLs genes to describe their conservation and differences (Fig. 1C). According to the findings, NtCMLs contain between one and eight exons, and the most of their members lack introns.
3.3 Phylogenetic Analysis of NtCMLs Proteins
To further comprehend the links between these compounds, phylogenetic analysis was performed using the recovered tobacco protein sequences, we constructed the phylogenetic tree with 125 NtCMLs protein sequences, including 75 sequences from tobacco (NtCMLs) and 50 from Arabidopsis (AtCMLs). These NtCMLs protein were classified into Five subgroups (Group I–Group V) (Fig. 2). The smallest subgroup was Group I which consisted of 4 CMLs (3 AtCMLs and 1 NtCMLs). The largest subgroup was Group V which consisted of 49 CMLs (19 AtCMLs and 30 NtCMLs). Group II included 18 CMLs members (3 AtCMLs and 15 NtCMLs). The Group III included 17 CML members (7 AtCMLs and 10 NtCMLs). The Group IV included 37 CMLs members (18 AtCMLs and 19 NtCMLs). Furthermore, the majority of NtCML was homologous to Arabidopsis. The findings suggest that CMLs are conserved across all plant species.
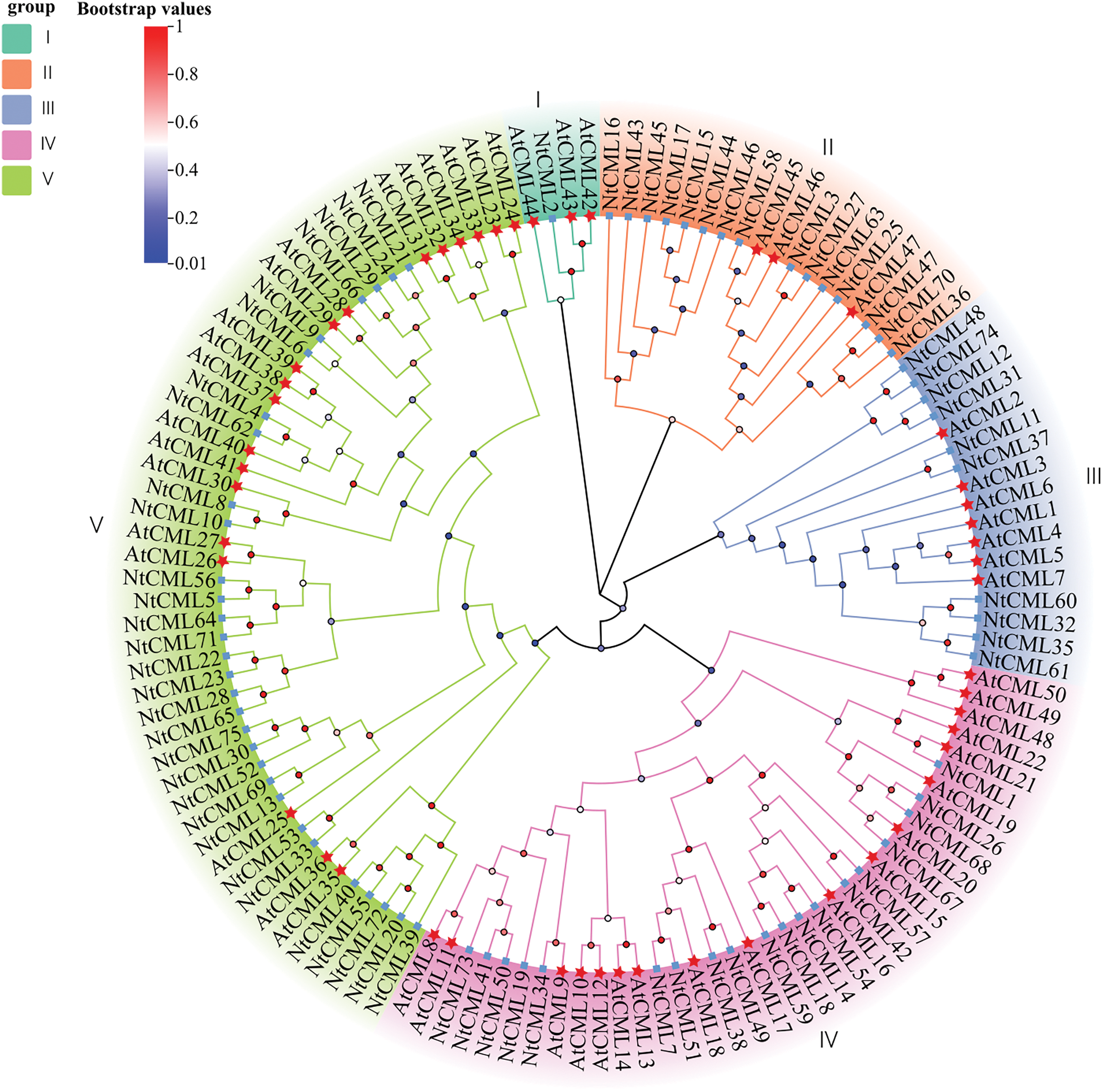
Figure 2: Phylogenetic tree of NtCMLs proteins in tobacco and Arabidopsis
3.4 Cis-Acting Elements Analysis in Promoter Regions of NtCMLs Genes
Gene expression is significantly influenced by parallel trans-regulatory factors and cis-elements. To better understand transcriptional and functional control of the NtCMLs genes, the extraction of 2000-bp upstream sequences of the NtCMLs coding areas was performed by analyzing cis-acting elements (Fig. 3). The primary cis-acting factors linked to plant growth and development, biotic and abiotic stressors, light, and hormones (Fig. 4). Among the elements that respond to plant hormones are abscisic acid (ABRE), methyl jasmonate (CGTCA-motif, TGACG-motif), ethylene (ERE), and salicylic acid (TCA). Abiotic and biotic stressors such as low temperature (LTR), drought (MBS), anaerobic (ARE), and W-box are associated with the element type. Other components of the stress response have also been identified, such as the light element G-box and the trauma response element WUN-motif. The cis-element analysis highlighted the regulatory complexity of the NtCMLs gene family, suggesting that the most of NtCMLs genes are essential for plant development and growth, as well as hormone and stress responses.
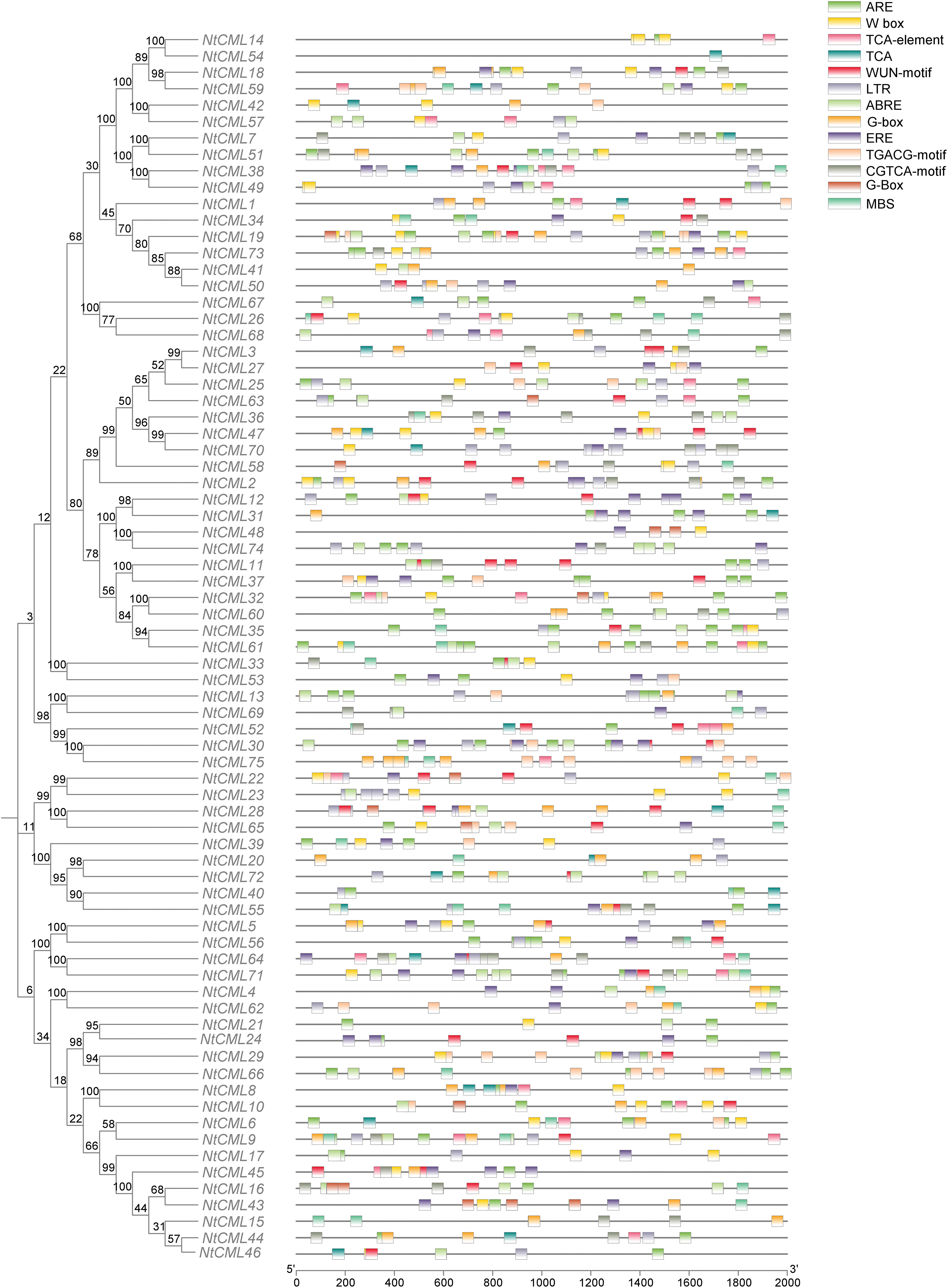
Figure 3: Cis-acting elements analysis of NtCMLs
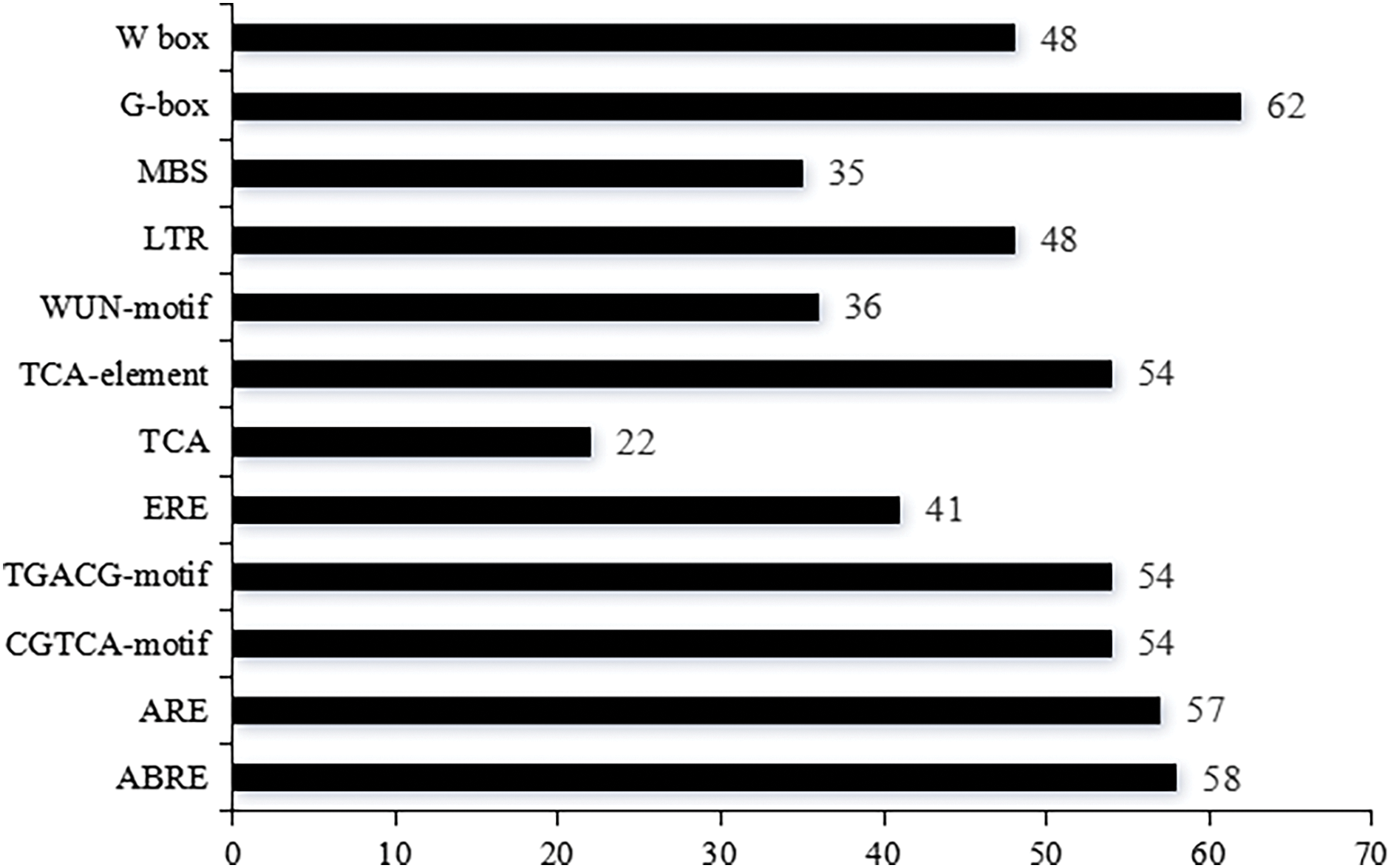
Figure 4: Number of NtCMLs genes containing cis-acting elements
3.5 Chromosomal Location and Synteny Analysis of NtCMLs Genes
The protein sequences of the discovered NtCMLs were used to determine their chromosomal locations. The findings revealed that tobacco has 18 chromosomes, which contain 40 NtCMLs genes (Fig. 5). Particularly, chromosome 17 has the most genes, followed by chromosome 12, which contains five NtCML genes, and chromosome 4 contains four NtCML genes. Ten Chromosomes contain only one NtCMLs genes (Chromosomes 1, 2, 5, 6, 8, 9, 13, 14, 15, 24). The results demonstrated that NtCMLs were randomly distributed across different chromosomes.
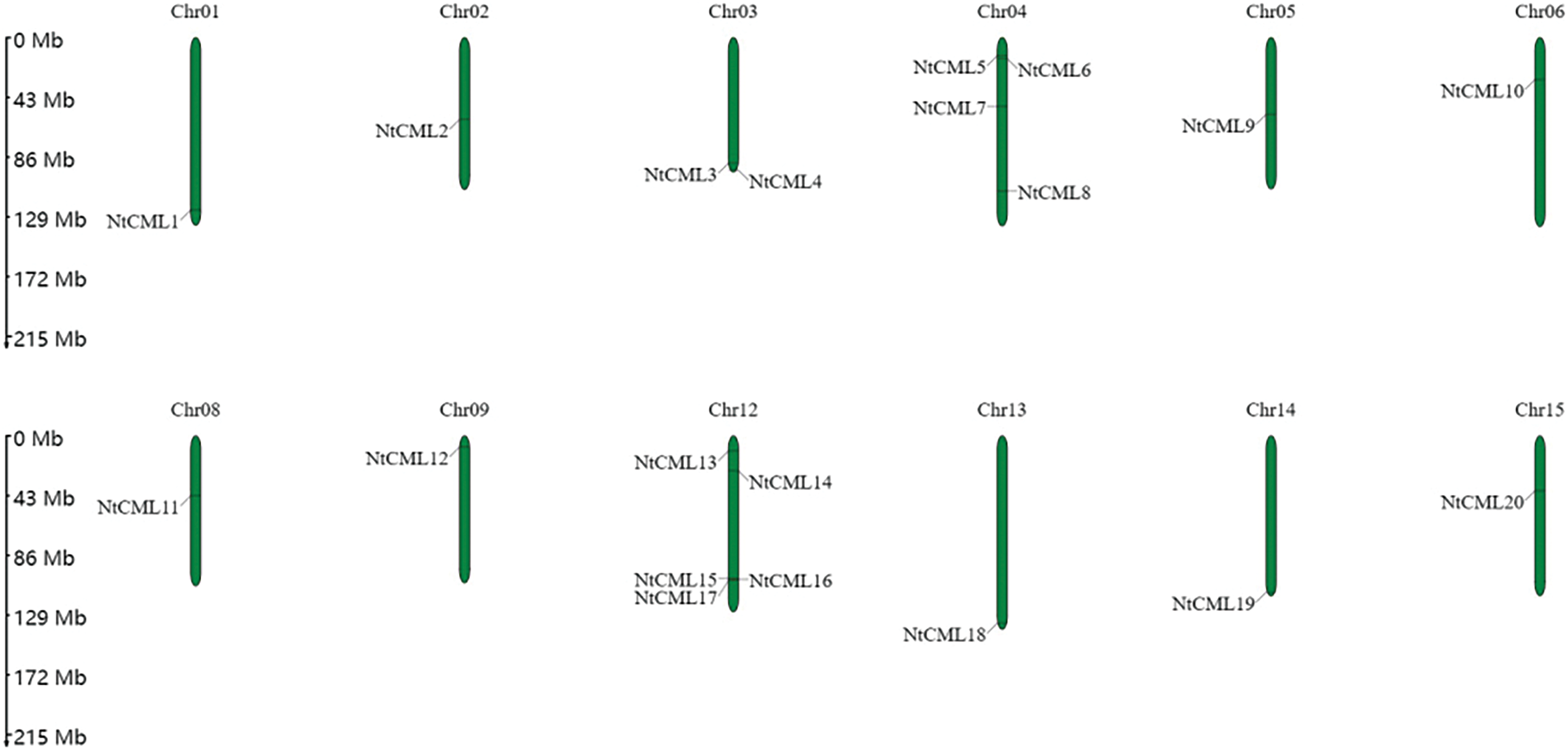
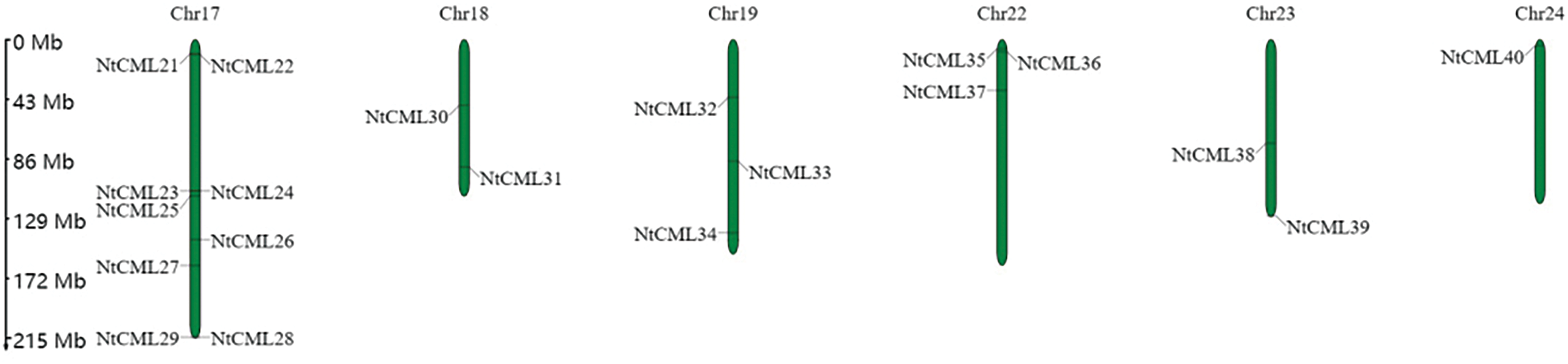
Figure 5: Chromosome distribution of NtCMLs genes in Nicotiana tabacum
To determine the NtCMLs gene duplication of the NtCMLs genes, the segmental duplication events in the NtCMLs gene family were conducted. 12 NtCMLs genes were predicted to be segmentally duplicated on chromosomes 3, 8, 12, 15, 17, 19 and 22. The tobacco genome include seven segmental duplicated gene pairs and two tandemly duplicated gene pairs (Fig. 6). These duplicate genes are probably caused by intra- or inter-chromosomal segmental duplication of other genes.
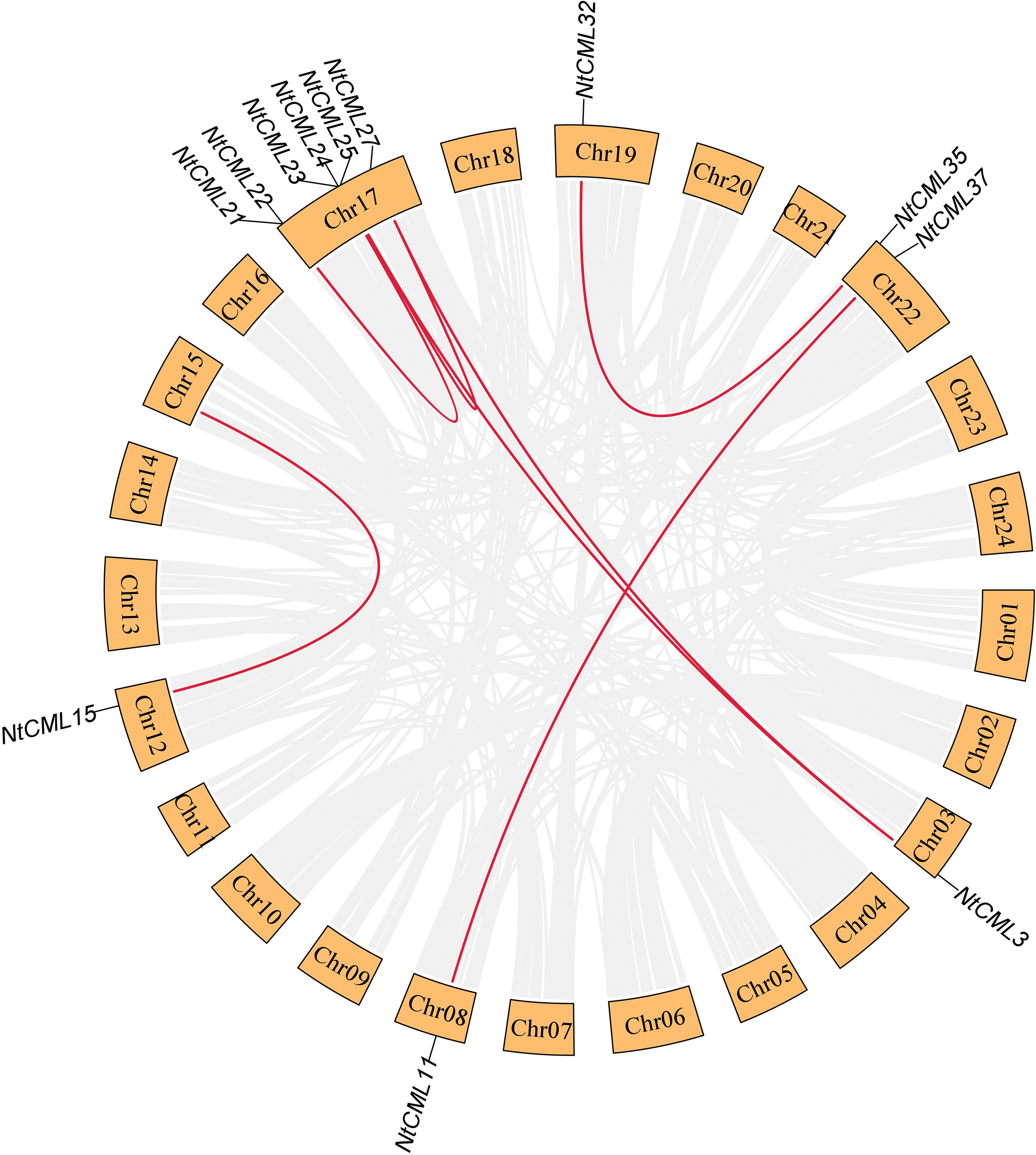
Figure 6: Interchromosomal relationshipsf of NtCMLs in Nicotiana tabacum
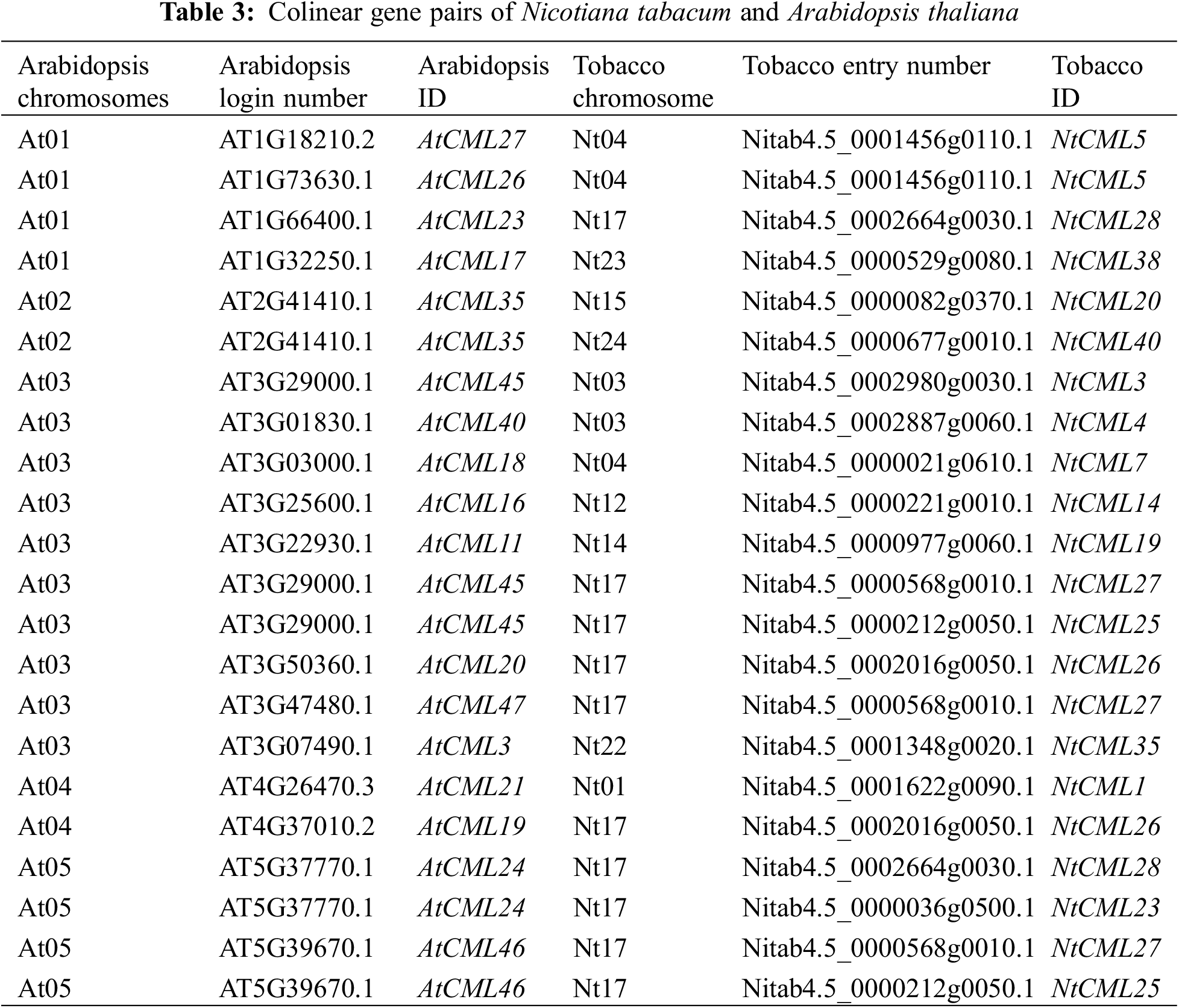
A comparative analysis of the genomes of tobacco and Arabidopsis was conducted to gain a better understanding of the evolution of NtCMLs genes (Fig. 7). 22 pairs of orthologous genes were present between tobacco and Arabidopsis, there was one-to-many or many-to-one collinearity between the NtCMLs and the AtCMLs (Table 3). For instance, four genes (NtCML5, MtCML28, NtCML25, NtCML26) had two homologous genes in Arabidopsis, while one gene (NtCML27) had three orthologous genes. The existence of ancient gene pairs during the divergence of Arabidopsis and tobacco has been demonstrated, and their functions may be similar.
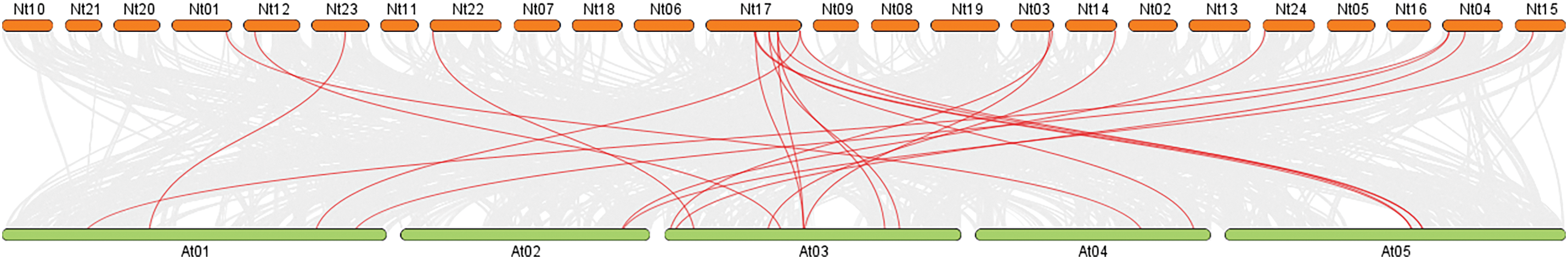
Figure 7: Synteny analysis of CMLs in Nicotiana tabacum and Arabidopsis
3.6 Spatio-Temporal Expression Patterns of NtCMLs Genes in Different Tissues
To determine the possible roles of NtCMLs, the public transcription data for several tobacco K326 tissues, including seed, shoot, flower, stem, and root was downloaded. The different expression patterns in most of the 19 tissues and developmental stages were analyzed (Fig. 8). Some NtCMLs genes exhibited a tissue-specific expression pattern. Eight NtCMLs genes (NtCML19, NtCML41, NtCML50, NtCML73, NtCML6, NtCML10, NtCML38, NtCML49) were highly expressed in the seed, NtCML21, NtCML24, NtCML66, NtCML8, and NtCML29 were highly expressed in flowers, three NtCMLs genes (NtCML70, NtCML36, NtCML47) were highly expressed in young shoot. Furthermore, some NtCMLs genes showed similar expression patterns, including NtCML75 and NtCML30, NtCML41 and NtCML50, NtCML62 and NtCML4, NtCML54 and NtCML14. The findings suggested that these genes may have similar functions in plant growth and development.
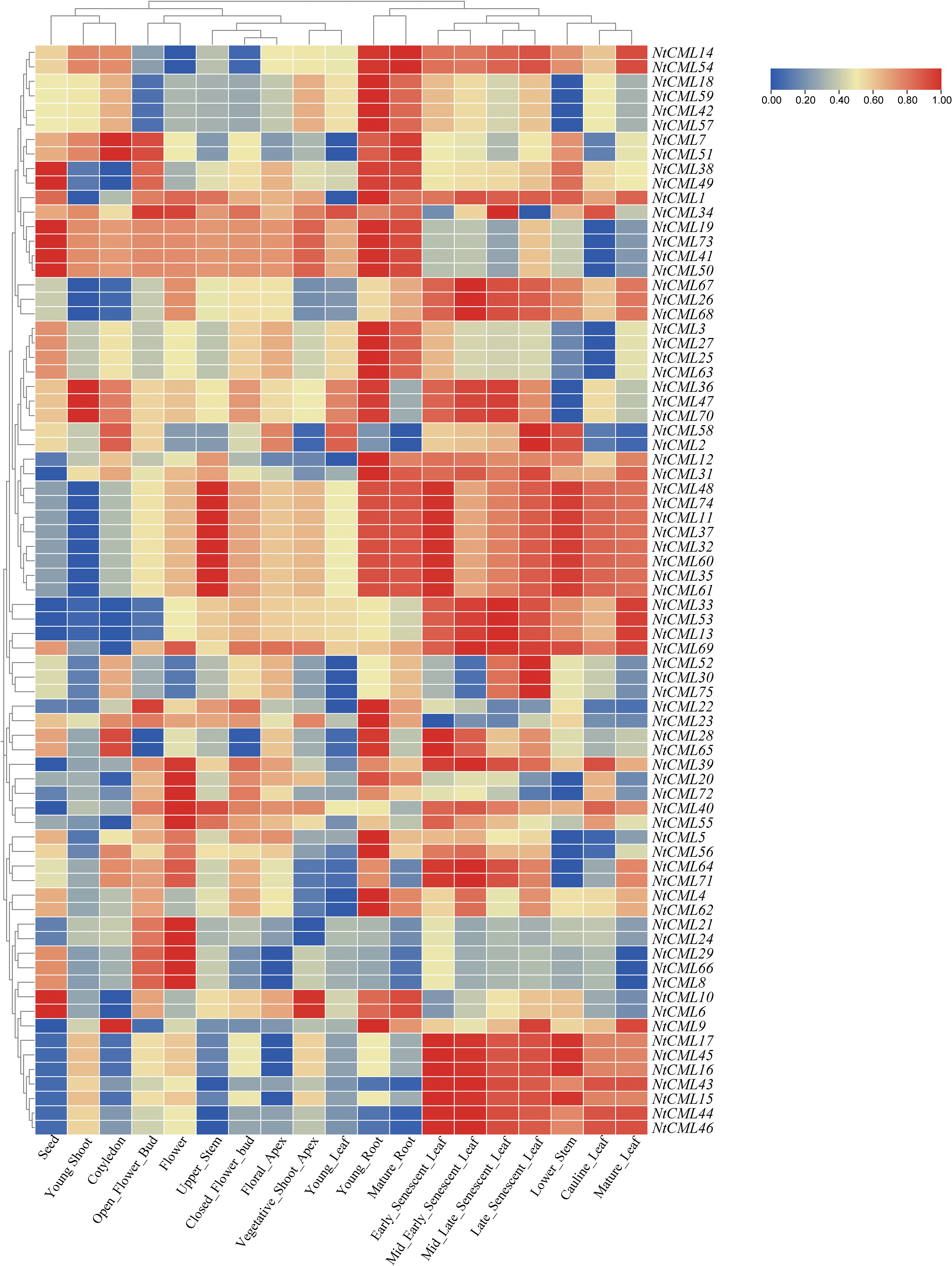
Figure 8: The relative expression patterns of NtCMLs genes in different tissues
3.7 Expression of the Selected 10 NtCMLs Genes in Response to Cold Acclimation
Ten NtCMLs genes were examined for their expression in response to cold acclimation (Fig. 9). The transcript levels of NtCML8, NtCML18, NtCML12, NtCML23, NtCML27, and NtCML28 were significantly upregulated following exposure to cold conditions. In contrast, transcripts for NtCML22 and NtCML53 showed a significant reduction. However, no significant changes were observed in the expression of NtCML30 and NtCML3 under cold stress when compared with the other genes. After 7 days of cold acclimation, we found that NtCML3, NtCML12, NtCML18, NtCML27, and NtCML28 were still significantly upregulated, while NtCML8 and NtCML30 were significantly down-regulated. However, NtCML22, NtCML23, and NtCML53 had no significant change. However, when exposed to low-temperature stress once more, NtCML3, NtCML12, NtCML22, NtCML18, NtCML23, NtCML27, NtCML28, NtCML30, and NtCML53 significantly upregulated. These findings imply that the NtCMLs gene has distinct functions in the cold acclimation recovery process.
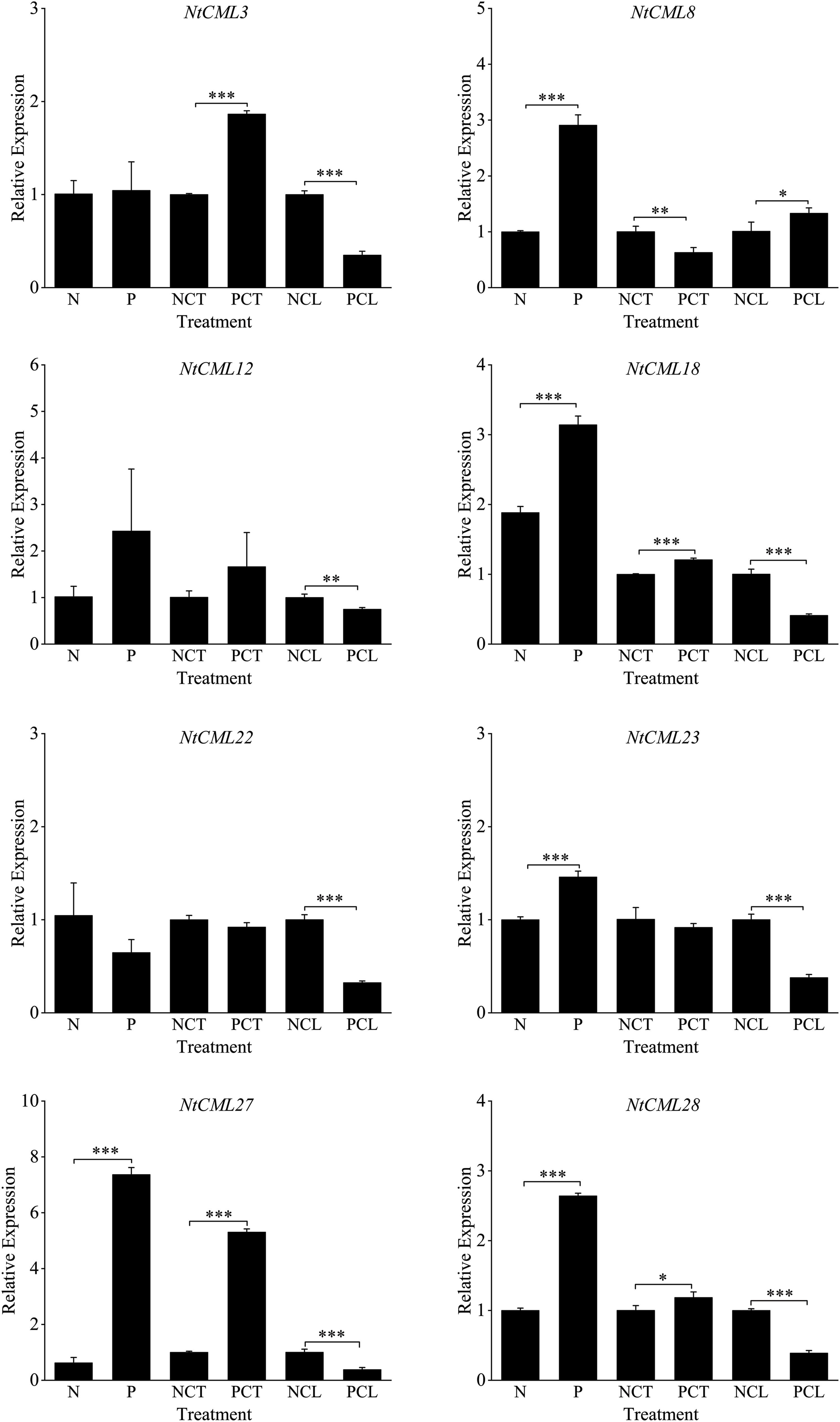
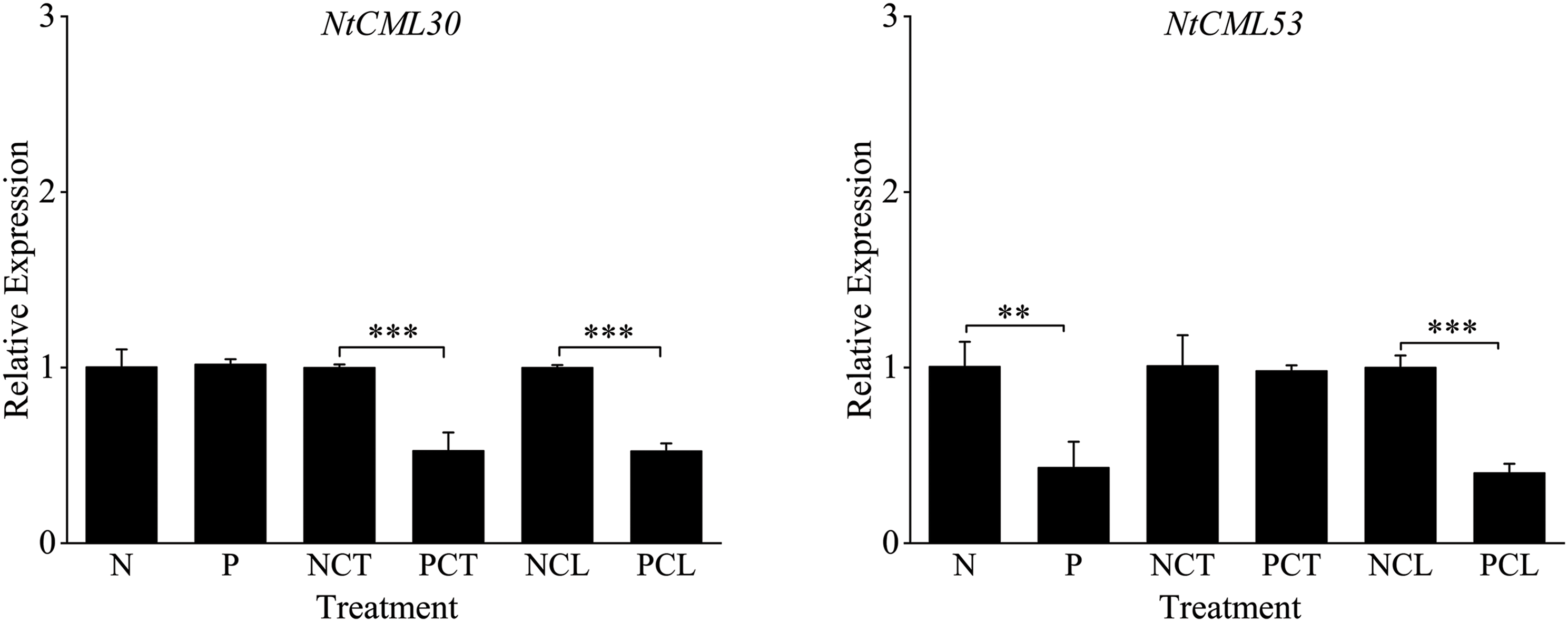
Figure 9: Levels of NtCMLs family members’ relative expression during cold adaptation. Each sample underwent three separate experiments. *, **, *** indicates significant difference at 0.05, 0.01.0.001
3.8 Protein Interaction Network Prediction
In this study, 75 NtCMLs proteins were analyzed using STRING to predict the protein interaction network in tobacco (Fig. 10). 13 NtCMLs proteins were identified as participating in the interaction network, and four CMLs proteins exhibited correlations with more than four other CMLs. Notably, CML46 was associated with ten NtCMLs proteins. The protein-protein associations suggested that some NtCMLs proteins are likely co-expressed based on findings from Arabidopsis research. NtCML4, NtCML59, and NtCML2 demonstrated a close protein interaction and exhibited potential co-expression and co-occurrence patterns. The analysis of the protein interaction network indicated that NtCMLs regulate downstream gene expression through interactions with other proteins, thereby providing a valuable resource for further research.
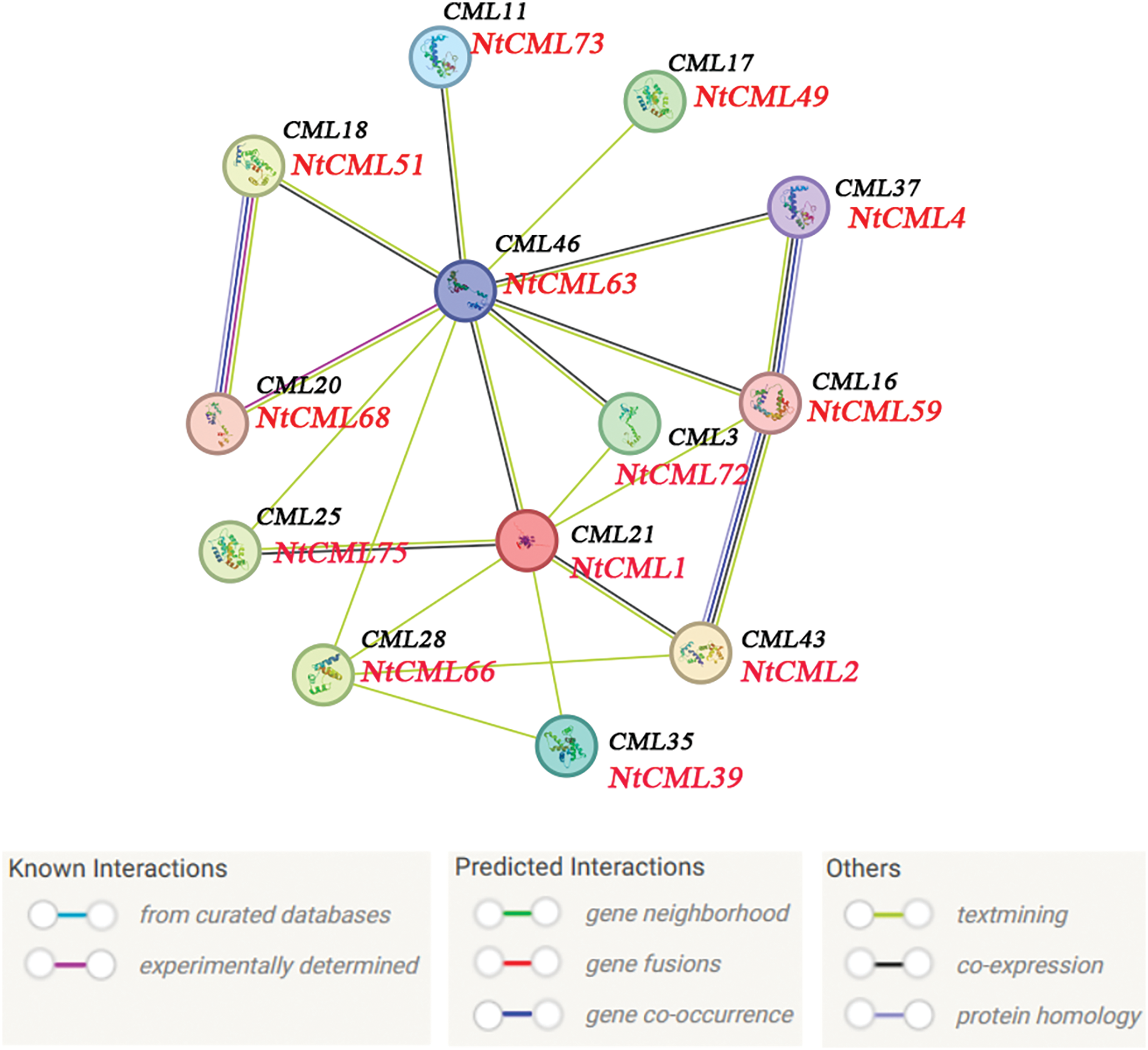
Figure 10: Protein interaction network of CMLs proteins. The homologous genes from tobacc and Arabidopsis are in red and black, respectively
Numerous aspects of plant growth and development, including stress responses, are significantly influenced by calcium ions (Ca2+). Calmodulin-like proteins (CMLs), which act as calcium ion sensors, play a crucial role in cellular signaling networks by regulating a wide range of targets [1]. Using bioinformatics techniques, 75 NtCMLs genes were found in the tobacco genome in this study. In comparison to Arabidopsis thaliana (50 CMLs) [12], rice (32 CMLs) [13] and tomato (52 CMLs) [14], tobacco possesses a greater number of CMLs genes, This discrepancy may be attributed to the allotetraploid nature of tobacco. A comprehensive bioinformatics analysis revealed significant variations in amino acid number, pI, aliphatic index, and instability index between different NtCMLs proteins. The majority of NtCMLs (85%) were acidic, consistent with the performance of CMLs family members in cucumber [10], apple [4], and tomato [14]. Previous studies have demonstrated that CaMs and CMLs exhibit high preservation properties. In the Arabidopsis [12], cucumber [10], and papaya [35], the CMLs shared the 16.1%~74.5%, 24%~77%, and 22.4%~88.1% identity with AtCaM2. In this study, We decided to use 16%~80% amino acid similarity as the selection criterion.
Gene location analysis showed that the 40 NtCMLs genes were not evenly dispersed across the tobacco’s 18 chromosomes, with the highest concentration being on chromosome 17. Calcium ion binding may be affected by the number of CMLs proteins that have conserved structural domains of the EF hands [10]. In Arabidopsis, which usually has 2~6 EF-hands structural domains [12], while tobacco possesses 2~4, which is consistent with cabbage [36] and common beans [37]. Previous studies have demonstrated that the majority of CMLs lack introns [38]. In tobacco, most NtCMLs genes lack introns. This instance aligned with grape [11], cucumber [10], and cabbage [36], proving that the gene structure of CMLs has been preserved through plant evolution. The conserved motif results demonstrated that both motif 1 and motif 2 were present in all members of the NtCMLs family. The diverse NtCMLs were indicated by the different conserved motifs.
A phylogenetic evolutionary tree analysis of tobacco and Arabidopsis reveals that the 125 CMLs proteins are divided into five groups, with at least one tobacco and Arabidopsis CMLs protein in each group, while four subgroups are present in Solanum pennellii [39] and seven are present in cucumber [10]. Additionally, the analysis indicates that many NtCMLs proteins are homologous to Arabidopsis. Gene duplication is an essential process for organisms to acquire new genes, which leads to genetic novelty, and has resulted in numerous new gene functions that have greatly advanced biological evolution [40]. Seven segmental duplicated gene pairs and two tandemly duplicated gene pairs were found in the tobacco genome during our investigation, while two tandemly duplicated gene pairs and three segmentally duplicated gene pairs in the cucumber genome [10]. 22 pairs of orthologous genes were present between tobacco and Arabidopsis, while five collinear gene pairs between cucumber and Arabidopsis [10]. It suggests that plants are highly conserved during evolution.
Gene expression regulates the growth, development, and adaptation of plants. This process is dependent on promoters, which are the cis-acting regions that initiate transcription [41]. This research revealed that the promoters of CMLs in tobacco were enriched with cis-acting elements that were linked to biological stress responses and plant hormones. Some specific cis-elements (ABRE, CGTCA-motif, LTR, MBS, ERE, and WUN-motif, among others) were in the promoter regions of NtCMLs genes. Similar to the reports of Medicago truncatula [2], apple [4], and Chrysanthemum seticuspe [15]. Previous studies have demonstrated that some CMLs genes are involved in hormonal or abiotic stress responses. CML9, CML24, showed response to ABA and salt stresses [19,21]. In this study, NtCML21, NtCML24 and CML9, CML24 belong to the same subfroup, and NtCML21, NtCML24 include ABA cis-acting elements, indicating that they could be involved in ABA response. CML37, CML38, CML39, MtCML40, and MpCML40 showed extensive responses to salt stress [27,28,31], CML39 is involved in the growth and development of seeds [18]. These previous studies have demonstrated that NtCMLs genes are crucial for plant growth, development, and adaptation to adverse conditions.
Examining the spatiotemporal differential expression patterns of NtCMLs in various organs may help to better understand their potential roles in tobacco growth and development. In this study, the NtCMLs gene family displayed a wide range of expression patterns in diverse tissues and organs during different phases of development. CML23 and CML24 are connected with flower development, our findings indicate that NtCML21, NtCML24, and NtCML66 are substantially upregulated in flowers and belong to the same subgroup, implying that they may be involved in flowering and fruit growth [26,41]. NtCML59, NtCML57, and NtCML42 genes were highly expressed in roots, which were presumed to be involved in the process of root growth and development. In conclusion, the spatiotemporal expression pattern of 75 NtCMLs genes demonstrated a tissue specificity.
The physiological and biochemical functions of plants can be regulated by short-term exposure to non-lethal cold temperatures, which can enhance their resistance to cold stress. This phenomenon is known as cold acclimation (CA) [42]. Plants have developed the CA mechanism to reduce the negative effects of cold stress, which is recognized as a common process that enables many temperate species to develop cold tolerance and resistance to freezing [43]. During CA treatment, elevated expression levels of calmodulin phosphatase b-like proteins (CBLs), calmodulin-interacting protein kinases (CIPKs), calmodulin-like proteins (CMLs), calcium-dependent protein kinases (CDPKs) were observed [44]. The results of the study indicate that NtCML12, NtCML18, NtCML27, and NtCML28 were significantly upregulated during the CA treatment. Conversely, the majority of NtCMLs genes were significantly down-regulated following re-exposure to low-temperature stress, indicating that the plant may have accumulated certain substances during the recovery period to enhance its cold tolerance. These findings make it easier to investigate the role of NtCML genes during CA in tobacco. In conclusion, NtCMLs are connected to cold stress, and earlier research has demonstrated that some genes in plants like Medicago truncatula [28] and Solanum lycopersicum [14] are similarly impacted by cold stress. During this investigation, it was discovered that NtCML27 experienced a significant upregulation in response to cold stress and was chosen as a candidate gene to be confirmed in the next phase.
Acknowledgement: We are grateful to the Chiplot and Tbtools software creators. We also want to express our gratitude to the entire crew for their assistance during the trial.
Funding Statement: This study was supported by College of Agronomy and Biotechnology, Southwest University (4412200577).
Author Contributions: Study conception and design: Mengjie Xu, Anbin Wang; data collection: Mengjie Xu, Anbin Wang, Tonghong Zuo; analysis and interpretation of data: Mengjie Xu, Anbin Wang, Tonghong Zuo, Hecui Zhang, Zhihao Hu; draft manuscript preparation: Mengjie Xu; revised the manuscript: Liquan Zhu, Hecui Zhang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are available from the corresponding author, L. Q. Z., upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Perochon A, Aldon D, Galaud JP, Ranty B. Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie. 2011;93(12):2048–53. doi:10.1016/j.biochi.2011.07.012. [Google Scholar] [PubMed] [CrossRef]
2. Sun Q, Yu S, Guo Z. Calmodulin-Like (CML) gene family in medicago truncatula: genome-wide identification, characterization and expression analysis. Int J Mol Sci. 2020;21(19):7142. doi:10.3390/ijms21197142. [Google Scholar] [PubMed] [CrossRef]
3. Wang C, Luan S. Calcium homeostasis and signaling in plant immunity. Curr Opin Plant Biol. 2023;77:102485. [Google Scholar] [PubMed]
4. Li C, Meng D, Zhang J, Cheng L. Genome-wide identification and expression analysis of calmodulin and calmodulin-like genes in apple (Malus × domestica). Plant Physiol Biochem. 2019;139:600–12. doi:10.1016/j.plaphy.2019.04.014. [Google Scholar] [PubMed] [CrossRef]
5. Ma X, Li QH, Yu YN, Qiao YM, Haq SU, Gong ZH. The CBL-CIPK pathway in plant response to stress signals. Int J Mol Sci. 2020;21(16):5668. doi:10.3390/ijms21165668. [Google Scholar] [PubMed] [CrossRef]
6. Virdi AS, Singh S, Singh P. Abiotic stress responses in plants: roles of calmodulin-regulated proteins. Front Plant Sci. 2015;6:809. [Google Scholar] [PubMed]
7. Tong T, Li Q, Jiang W, Chen G, Xue D, Deng F, et al. Molecular evolution of calcium signaling and transport in plant adaptation to abiotic stress. Int J Mol Sci. 2021;22(22):12308. doi:10.3390/ijms222212308. [Google Scholar] [PubMed] [CrossRef]
8. Aldon D, Mbengue M, Mazars C, Galaud JP. Calcium signalling in plant biotic interactions. Int J Mol Sci. 2018;19(3):665. doi:10.3390/ijms19030665. [Google Scholar] [PubMed] [CrossRef]
9. Jessica LG, Michael PW, Hans JV. Structures and metal-ion-binding properties of the Ca2+-binding helix-loop–helix EF-hand motifs. Biochem J. 2007;405(2):199–221. doi:10.1042/BJ20070255. [Google Scholar] [CrossRef]
10. Liu Y, Yin F, Liao L, Shuai L. Genome-wide identification and expression analysis of calmodulin-like proteins in cucumber. PeerJ. 2023;11:14637. doi:10.7717/peerj.14637. [Google Scholar] [CrossRef]
11. Vandelle E, Vannozzi A, Wong D, Danzi D, Digby AM, Dal Santo S, et al. Identification, characterization, and expression analysis of calmodulin and calmodulin-like genes in grapevine (Vitis vinifera) reveal likely roles in stress responses. Plant Physiol Biochem. 2018;129:221–37. doi:10.1016/j.plaphy.2018.06.003. [Google Scholar] [CrossRef]
12. McCormack E, Braam J. Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 2003;59:585–98. [Google Scholar]
13. Boonburapong B, Buaboocha T. Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 2007;7:4. doi:10.1186/1471-2229-7-4. [Google Scholar] [PubMed] [CrossRef]
14. Munir S, Khan MR, Song J, Munir S, Zhang Y, Ye Z, et al. Genome-wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiol Biochem. 2016;102:167–79. doi:10.1016/j.plaphy.2016.02.020. [Google Scholar] [CrossRef]
15. Fu M, Wu C, Li X, Ding X, Guo F. Genome-wide identification and expression analysis of CsCaM/CML gene family in response to low-temperature and salt stresses in Chrysanthemum seticuspe. Plants. 2022;11(13):1760. doi:10.3390/plants11131760. [Google Scholar] [PubMed] [CrossRef]
16. Cai K, Kuang L, Yue W, Xie S, Xia X, Zhang G, et al. Calmodulin and calmodulin-like gene family in barley: identification, characterization and expression analyses. Front Plant Sci. 2022;13:964888. doi:10.3389/fpls.2022.964888. [Google Scholar] [CrossRef]
17. Zeng H, Xu L, Singh A, Wang H, Du L. Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front Plant Sci. 2015;6:600. [Google Scholar] [PubMed]
18. Midhat U, Ting MKY, Teresinski HJ, Snedden WA. The calmodulin-like protein, CML39, is involved in regulating seed development, germination, and fruit development in Arabidopsis. Plant Mol Biol. 2018;96:375–92. doi:10.1007/s11103-018-0703-3. [Google Scholar] [PubMed] [CrossRef]
19. Yang X, Wang SS, Wang M, Qiao Z, Bao CC, Zhang W. Arabidopsis thaliana calmodulin-like protein CML24 regulates pollen tube growth by modulating the actin cytoskeleton and controlling the cytosolic Ca2+ concentration. Plant Mol Biol. 2014;86:225–36. doi:10.1007/s11103-014-0220-y. [Google Scholar] [PubMed] [CrossRef]
20. Wang SS, Diao WZ, Yang X, Qiao Z, Wang M, Biswa RA, et al. Arabidopsis thaliana CML25 mediates the Ca2+ regulation of K+ transmembrane trafficking during pollen germination and tube elongation. Plant Cell Environ. 2015;38(11):2372–86. doi:10.1111/pce.v38.11. [Google Scholar] [CrossRef]
21. Leba LJ, Cheval C, Ortiz-Martin I, Ranty B, Beuzon CR, Galaud JP, et al. CML9, an Arabidopsis calmodulin-like protein,contributes to plant innate immunity through a flagellin-dependentsignalling pathway. Plant J. 2012;71(6):976–89. doi:10.1111/tpj.2012.71.issue-6. [Google Scholar] [PubMed] [CrossRef]
22. Xu B, Cheval C, Laohavisit A, Hocking B, Chiasson D, Tjelvar SG, et al. Calmodulin-like protein regulates plasmodesmal closure during bacterial immune responses. New Phytol. 2017;215(1):77–84. doi:10.1111/nph.2017.215.issue-1. [Google Scholar] [PubMed] [CrossRef]
23. Zhu XY, Robe E, Jomat L, Aldon D, Mazars C. CML8, an Arabidopsis calmodulin-like protein, plays a role in Pseudomonas syringae plant immunity. Plant Cell Physiol. 2017;58(2):307–19. [Google Scholar] [PubMed]
24. Magnan F, Ranty B, Charpenteau M, Sotta B, Galaud JP, Aldon D. Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 2008;56(4):575–89. doi:10.1111/tpj.2008.56.issue-4. [Google Scholar] [PubMed] [CrossRef]
25. Wu X, Qiao Z, Liu H, Acharya BR, Li C, Zhang W. CML20, an Arabidopsis Calmodulin-like protein, negatively regulates guard cell ABA signaling and drought stress tolerance. Front Plant Sci. 2017;8:824. doi:10.3389/fpls.2017.00824. [Google Scholar] [PubMed] [CrossRef]
26. Delk NA, Johnson KA, Chowdhury NI, Braam J. CML24, regulated in expression by diverse stimuli, encodes a potential Ca2+ sensor that functions in responses to abscisic acid, daylength, and ion stress. Plant Physiol. 2005;139:240–53. doi:10.1104/pp.105.062612. [Google Scholar] [PubMed] [CrossRef]
27. Vanderbeld B, Snedden WA. Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol Biol. 2007;64:683–97. doi:10.1007/s11103-007-9189-0. [Google Scholar] [CrossRef]
28. Sun Q, Huang R, Zhu H, Sun Y, Guo Z. A novel Medicago truncatula calmodulin-like protein (MtCML42) regulates cold tolerance and flowering time. Plant J. 2021;108:1069–82. doi:10.1111/tpj.v108.4. [Google Scholar] [PubMed] [CrossRef]
29. Ding H, Qian Y, Fang Y, Ji Y, Sheng J, Ge C. Characteristics of SlCML39, a tomato calmodulin-like gene, and its negative role in high temperature tolerance of arabidopsis thaliana during germination and seedling growth. Int J Mol Sci. 2021;22:11479. doi:10.3390/ijms222111479. [Google Scholar] [PubMed] [CrossRef]
30. Munir S, Liu H, Xing Y, Hussain S, Ouyang B, Zhang Y, et al. Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci Rep. 2016;6:317. [Google Scholar]
31. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–8. doi:10.1006/meth.2001.1262. [Google Scholar] [PubMed] [CrossRef]
32. Hashim FA, Mabrouk MS, Al-Atabany W. Review of different sequence motif finding algorithms. Avicenna J Med Biotechnol. 2019;11:130–48. [Google Scholar] [PubMed]
33. Willson J. Exons as enhancers. Nat Rev Genet. 2020;21:68–9. doi:10.1038/s41576-019-0207-2. [Google Scholar] [PubMed] [CrossRef]
34. Shaul O. How introns enhance gene expression. Int J Biochem Cell Biol. 2017;9:145–55. [Google Scholar]
35. Ding XC, Zhang LP, Hao YW, Xiao SL, Wu ZX, Weixin C, et al. Genome-wide identification and expression analyses of the calmodulin and calmodulin-like proteins reveal their involvement in stress response and fruitripening in papaya. Postharvest Biol Technol. 2018;143:3–27. [Google Scholar]
36. Nie S, Zhang M, Zhang L. Genome-wide identification and expression analysis of calmodulin-like (CML) genes in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genom. 2017;18(1):842. doi:10.1186/s12864-017-4240-2. [Google Scholar] [CrossRef]
37. Zhao H, Gao Y, Du Y, Du J, Han Y. Genome-wide analysis of the CML gene family and its response to melatonin in common bean (Phaseolus vulgaris L.). Sci Rep. 2023;13:1196. doi:10.1038/s41598-023-28445-y. [Google Scholar] [CrossRef]
38. Mohanta TK, Kumar P, Bae H. Genomics and evolutionary aspect of calcium signaling event in calmodulin and calmodulin-like proteins in plants. BMC Plant Biol. 2017;17:38. doi:10.1186/s12870-017-0989-3. [Google Scholar] [PubMed] [CrossRef]
39. Shi J, Du X. Identification, characterization and expression analysis of calmodulin and calmodulin-like proteins in Solanum pennellii. Sci Rep. 2020;10:7474. doi:10.1038/s41598-020-64178-y. [Google Scholar] [CrossRef]
40. Clark JW. Genome evolution in plants and the origins of innovation. New Phytol. 2023;240:2204–9. doi:10.1111/nph.19242. [Google Scholar] [PubMed] [CrossRef]
41. Tsai YC, Delk NA, Chowdhury NI, Braam J. Arabidopsis potential calcium sensors regulate nitric oxide levels and the transition to flowering. Plant Signal Behav. 2007;2:446–54. doi:10.4161/psb.2.6.4695. [Google Scholar] [PubMed] [CrossRef]
42. Mithra SVA, Kulkarni K, Srinivasan R. Plant promoters: characterization and applications in transgenic technology. In: Abdin M, Kiran U, Kamaluddin A, editors. Plant biotechnology: principles and applications. Singapore: Springer; 2017. p. 117–72. [Google Scholar]
43. Xu C, Wang Y, Yang H, Tang Y, Liu B, Hu X, et al. Cold acclimation alleviates photosynthetic inhibition and oxidative damage induced by cold stress in citrus seedlings. Plant Signal Behav. 2023;18(1):2285169. doi:10.1080/15592324.2023.2285169. [Google Scholar] [PubMed] [CrossRef]
44. Xu J, Chen Z, Wang F, Jia W, Xu Z. Combined transcriptomic and metabolomic analyses uncover rearranged gene expression and metabolite metabolism in tobacco during cold acclimation. Sci Rep. 2020;10:5242. doi:10.1038/s41598-020-62111-x. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools