 Open Access
Open Access
ARTICLE
Development and Assessment of Reference Genes for qPCR of Malus transitoria under Drought Stress
1 Qinghai Academy of Agriculture and Forestry, Qinghai University, Xining, 810016, China
2 Laboratory for Research and Utilization of Germplasm Resources in Qinghai Tibet Plateau, Xining, 810016, China
3 Key Laboratory of Tree Genetics and Breeding of Qinghai Plateau, National Forestry and Grassland Administration, Xining, 810016, China
* Corresponding Author: Defang Zhang. Email:
Phyton-International Journal of Experimental Botany 2025, 94(3), 911-927. https://doi.org/10.32604/phyton.2025.061770
Received 03 December 2024; Accepted 24 January 2025; Issue published 31 March 2025
Abstract
The use of a stable reference gene is fundamental for achieving reliable quantitative qRT-PCR (qPCR) results. Developing and evaluating the stability of reference genes is necessary for studying the molecular mechanisms of M. transitoria in response to drought stress. In this study, 18 candidate reference genes were selected from transcriptome sequencing data of M. transitoria according to their FPKM values under different drought stress degrees. Cluster-23533.34641 was identified as the most stable reference gene for M. transitoria under drought stress based on qPCR results and combined analysis of Genorm, NormFinder, BestKeeper, and Delta Ct algorithms. The reference genes identified in this research offer improved accuracy for quantifying target gene expression in both M. transitoria and Malus species under drought stress. This study could provide insights into the drought stress-related functional gene or factor in M. transitoria, even in Malus species.Keywords
Malus transitoria, a native Malus species to China, is mostly located in higher elevations regions of arid and semi-arid regions of China, such as Qinghai-Tibetan Plateau, Shanxi, Sichuan, Gansu, and Inner Mongolia [1,2].Based on its drought resistance performance, M. transitoria has been used as a tree species for afforestation and ecology restoration in arid areas, especially in the Qinghai-Tibetan Plateau. Our previous research showed, with climate change, the distribution area of M. transitoria would constantly migrate toward the north, and some populations have lost genetic diversity and showed a trend of shrinking in size [3,4], therefore, it is necessary to study the mechanism and find the key gene related M. transitoria cope with drought stress.
As a key factor in the study of gene function, the stability of the reference gene is influenced by development stage, stress degree, tissue types, and experimental conditions. Therefore, developing and evaluating reliable reference genes is a prerequisite in research on plant development and gene function.
In general, housekeeping genes are consistently expressed across all tissues and play a vital role in sustaining the essential cellular functions of plants, being used as a reference gene. In most of the fruit trees, Ubiquitin extension (UBQ) [5], Actin isoform B (ACTB), 18S rRNA [6], GAPDH, EF1, ACT [7–9], UBQ-1 [7], and others have been used as reference gene.
In the closely related species of M. transitoria, there are some reports about the validation of reference genes. For example, in apples, EF-1α and 18S r RNA were used as reference genes for research of the fruit quality during the fruit coloring period [10], UBQ was used as the reference gene in the different development stages of different genotype apples, and ACTB and UBQ were used in the different tissue [5].WD40, ACT, and GAPDH were identified as the most reliable reference genes for apple peel [9], the suitability of phosphatase 2A (PP2A), ribosomal protein L2 (RPL2), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as reference genes were assessed across various apple tissue types and under diverse biotic stress conditions., but the transitional housekeeping gene, 18S ribosomal RNA (18S), β-tubulin (TUB) and ubiquitin (UBQ) were evaluated as the least stable genes [11].The MDH, SAND, THFS, TMp1, and WD40 were evaluated as the best reference genes to accurately normalize gene expression levels in various tissues of apple [12]. In various tissues and organs of loquat, RPL4, RPL18, HIS3, and TUA3 were identified as the most reliable reference genes [13]. In Herbaceous peonies, EF-1α demonstrated considerable stability as an internal reference gene when exposed to drought stress.[14]. In Kiwifruit, GAPDH and ACT1 are the most suitable reference genes during fruit development [15]. In other species, such as Oryza sativa L. under salt stress, EIF1A and TIP are the most suitable internal reference genes [16]. In Brassica rapa L., EF1-α, UBC, and ACT as the most suitable reference genes [17]. In Kobreisa littledalei, ACTIN and GAPDH as the most suitable reference genes [18]. In Luffa cylindrica, EF-1α was the overall most stable and suitable reference gene [19].
Besides these traditional housekeeping genes, more and more reference genes were developed and assessed with sequencing and bio-information technology zooming. Five potential candidate reference genes were developed and evaluated under various abiotic or biotic stresses in apple roots. Based on the transcriptome sequencing, four potential genes that could be used in post-harvest and other apple gene expression experiments were developed and evaluated [20]. Combining mRNA-seq with qPCR, three potential reference genes are proposed that can be used as reference genes for qPCR in Malus x domestica (apple) [21]. HISTONE 1 (MdH1) was evaluated as the most stable gene in experiments encompassing of apple post-harvest period based on the Transcriptional Profiling [22].
However, previous studies have shown that some housekeeping genes are expressed deferentially at different stages of plant development or under different stresses and conditions [23,24], which suggested that randomly selected reference genes may be inappropriate, leading to deviations or even incorrect results, and further affecting the accuracy of the conclusions [25]. As a result, choosing reference genes with stable expression based on the specific experimental needs is essential for obtaining reliable results.
At present, there are no reports about specific reference genes that have been developed for M. transitoria, especially for stressed samples. To correctly evaluate the expression of a gene that relates to drought resistance in M. transitoria, in this study, we developed and assessed the reference genes based on the transcriptome of drought-stressed samples. The stability of these candidate genes was assessed using four algorithms based on the result of qPCR amplification. These assessed internal reference genes could provide an important basis for improving the accuracy of qPCR in analyzing gene expression under drought stress in M.transitoria. This study could lay a solid foundation for future gene function studies of M. transitoria under drought stress conditions, and further afford useful tools in studying the mechanisms of tolerance in this species.
2.1 Plant Materials Collection
The experiment began on 9 July 2023, at the Forestry Experimental Base of Qinghai University in Qinghai Province. Five treatment groups were set up, with 1.5 kg of soil in the pots in each group. At the beginning, 200 mL of water was simultaneously watered in the 5 treatment groups, for the CK, keep the soil surface moist throughout the entire experimental process, for T1, watering 200 mL every three days, 200 mL every six days for T2, 200 mL every nine days for T3 and 200 mL every twelve days for T4. On the second day after the third round of watering in each treatment group, three biological replicates were taken from each treatment group. The samples were placed in liquid nitrogen and can only be used for subsequent experiments. The phenotype under drought stress is shown in Fig. 1.

Figure 1: The phenotype under drought stress
2.2 Total RNA Extraction and Transcriptome Sequencing
Total RNA extraction was carried out on the recommended protocol of the TaKaRa MiniBEST Plant RNA Extraction Kit (TaKaRa, Dalian, China). The integrity of RNA was evaluated through a combination of 1.0% agarose gel electrophoresis (Biosharp, Hefei, China) and the Nano 6000 Assay Kit, utilizing the Bioanalyzer 2100 system (Agilent Technologies, Santa Clara, CA, USA). Additionally, the RNA concentration and purity were assessed using the Nanodrop One (Thermo Fisher Scientific, Inc., USA).
Sequencing libraries, which were constructed with the NEBNext ® Ultra TM RNA Library Prep Kit for Illumina ® (Illumina, San Diego, CA, USA) and assessed with a Qubit2.0 Fluorometer (Thermo Fisher Scientific, Waltham, MA, USA) and an Agilent 2100 bioanalyzer (Agilent Technologies, CA, USA), were sequenced in the Illumina Novaseq platform (Illumina, USA) according to the recommended protocol.
2.3 Transcriptome Data Processing and Gene Expression Levels Analysis
The raw data was processed to guarantee the reliability and quality of subsequent analyses. Reads containing adapters, undetermined nucleotide bases (denoted as N), or exhibiting low quality—characterized by having over 50% of bases with a Qphred score ≤5—were excluded. This process led to the retrieval of high-quality, clean reads. These clean reads were subsequently assembled using the Trinity software [26]. The annotations were performed based on Nr, Nt, Pfam, KOG/COG, Swiss-prot, KEGG and GO databases.
2.4 Candidate Reference Gene Screening and Primer Design
The FPKM value, which was used for estimating gene expression levels of each sequenced gene, was counted with FeatureCountsv 1.5.0-p3. According to the FPKM value, candidate genes were screened based on the CV and SD value of the FPKM of each gene in different stress levels, The specified requirements are: a coefficient of variation (CV) less than or equal to 0.06, and a standard deviation (SD) not exceeding 4.0. The primer of the candidate gene was designed with Primer 3 software (http://frodo.wi.mit.edu/primer3/, accessed on 18 August 2024) based on the transcriptome sequence of each candidate gene. For each candidate gene, two distinct primer pairs were designed to facilitate PCR amplification.
2.5 First Strand cDNA Synthesis and the Specificity Validation Based on qPCR
The cDNA synthesis was performed using the FastKing gDNA Dispelling RT SuperMix (TIANGEN, China), employing less than 50 ng of total RNA in a final reaction volume of 20 μL. According to the recommended thermocycling conditions, the condition is as follows: 42°C for 15 min, and then 95°C for 3 min.
All of the screened candidate genes were selected to combine PCR amplification with qPCR. The conditions of PCR amplification and qPCR were shown in our previous research about Lycium [27].
2.6 Data Analysis and Stability Assessment
The stability of candidate genes was assessed based on the Genorm, NormFinder, BestKeeper, and Delta Ct from qPCR.
The NormFinder (https://www.moma.dk/software/normfinder) (accessed on 18 August 2024, Anders D. Andersen, Aarhus University, Denmark) analyzes the stability by assessing the evaluation of stability among potential internal reference genes involves ranking them according to their stability scores. The gene exhibiting the lowest stability value is considered the most suitable reference gene [28].
The Genorm (https://genorm.cmgg.be/) (accessed on 18 August 2024, Ghent University, Ghent, Belgium) assesses the stability by the Average Expression Stability Value (M), the smaller M values indicate a more stable gene expression level, while higher values imply poorer stability [24].
The BestKeeper (http://www.Gene-quantification.de/best-keeper.html) (accessed on 18 August 2024, Biometrics, Germany) assesses the stability by analyzing the standard deviation (SD), coefficient of variation (CV), and Pearson correlation coefficient (r) of the Ct value obtained from qPCR were evaluated. Candidate reference genes demonstrated higher r values and lower SD values were considered more stable (threshold set as less than 1), smaller coefficients of variation (CV) are generally considered to reflect greater stability, whereas lower CV values suggest diminished stability [29,30].
In Delta Ct methods, the stability of candidate genes was evaluated by comparing the relative expression levels of all genes, the SD of Delta Ct value represents stability, and the smaller SD indicates more stability of the candidate gene [23]. The expression level result of each gene was drawn with Origin 2021, the Venn diagram was drawn on the website (https://jvenn.toulouse.inrae.fr/app/example.html, accessed on 18 August 2024).
3.1 Sequencing Result of All Samples
A total of 96.1 G of raw base pairs were gathered from 15 samples, consisting of three control samples and 12 drought-stressed samples. Following quality control, 313,136,969 clean reads were generated. The average Q20 and Q30 values were recorded at 97.45% and 93.09%, respectively (Table A1). All raw data have been deposited in the National Center for Biotechnology Information (NCBI) Sequence Read Archive (SRA) database under the accession number PRJNA1172450.
3.2 Expression Profiles of Candidate Reference Genes
A total of 31 potential reference genes were identified through transcriptome sequencing based on FPKM values. The expression data for these genes across various developmental stages are presented in Table 1. The average FPKM values showed significant variation among the different candidate genes. The coefficient of variation (CV) ranged from 0.03 to 0.06, while the standard deviation (SD) varied between 0.17 and 3.77 (see Table 1).
3.3 Amplification Specificity and Efficiency
Eighteen genes were identified that could generate distinct and clear bands, without primer dimers. Specifically amplified were selected after 1.5% agarose gel electrophoresis detection, and then used for subsequent qPCR tests (Table 2). All 18 candidate genes exhibited a distinct peak in the melting curve, demonstrating the amplification specificity of all primers (Fig. 2A–R). Most of the candidate genes showed a slight change in Ct value except for a few genes (Fig. 3A–R and Fig. A1A–1R).
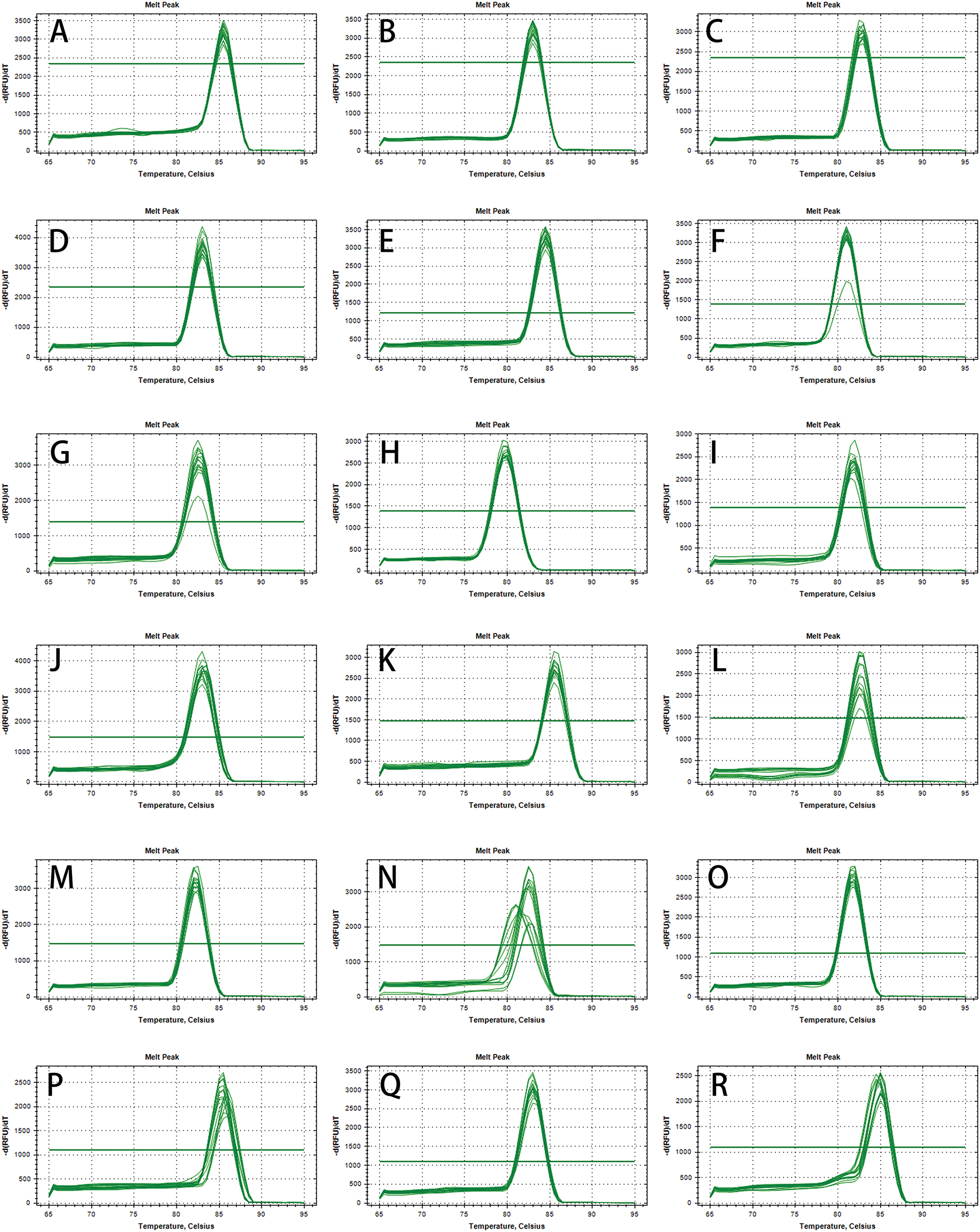
Figure 2: The melting curve analysis for each of the potential reference genes in quantitative PCR (qPCR). (A) Cluster-23533.35198. (B) Cluster-23533.40208. (C) Cluster-23533.42968. (D) Cluster-23533.45179. (E) Cluster-23533.37961. (F) Cluster-23533.21203. (G) Cluster-23533.34641. (H) Cluster-23533.48337. (I) Cluster-23533.37693 (J) Cluster-23533.42950. (K) Cluster-23533.45159. (L) Cluster-23533.39931. (M) Cluster-23533.39576. (N) Cluster-23533.40653. (O) Cluster-23533.38592. (P) Cluster-23533.33992. (Q) Cluster-23533.45050. (R) Cluster-23533.37654
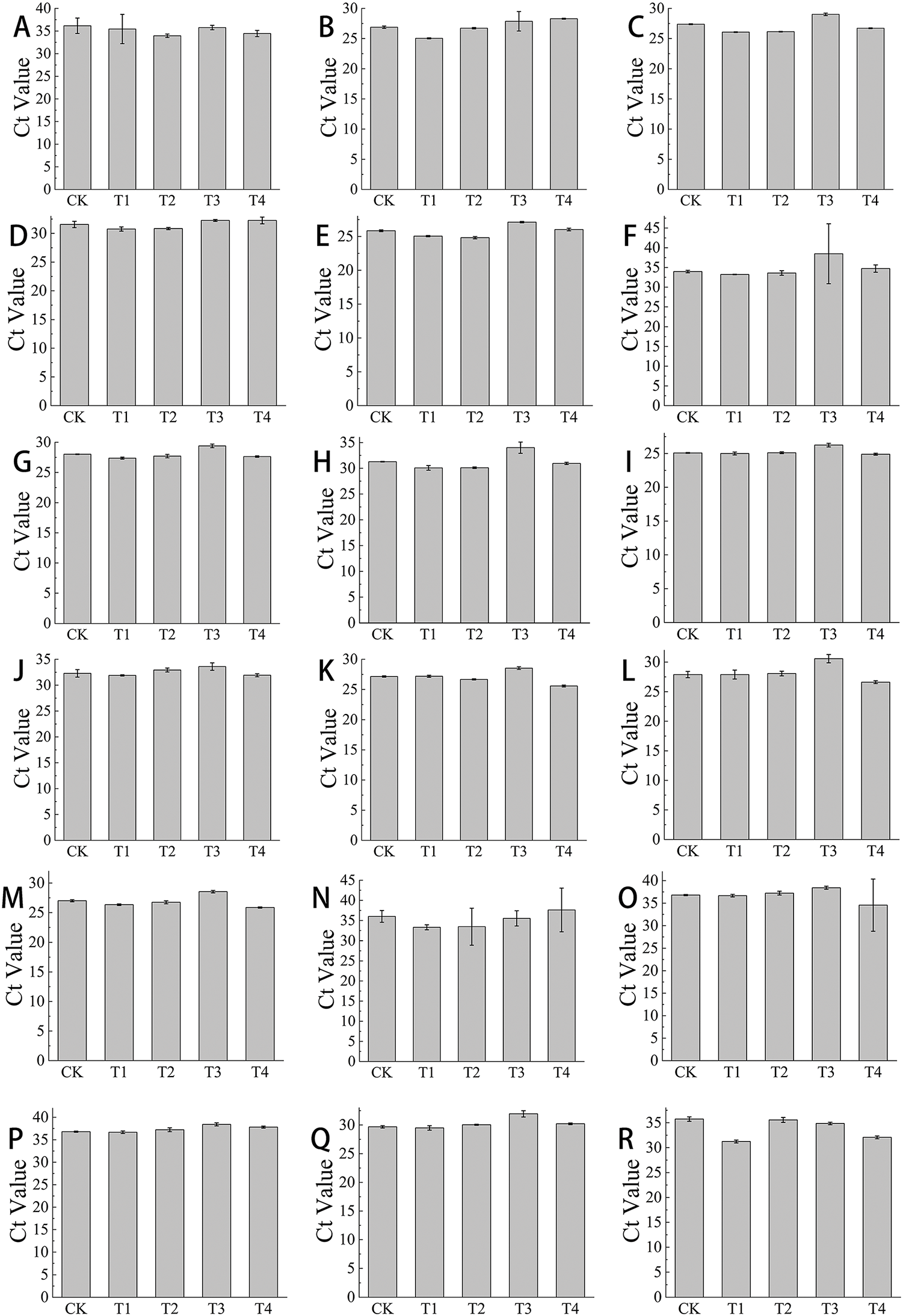
Figure 3: The Ct value of all candidate genes in qPCR (A) Cluster-23533.35198. (B) Cluster-23533.40208. (C) Cluster-23533.42968. (D) Cluster-23533.45179. (E) Cluster-23533.37961. (F) Cluster-23533.21203. (G) Cluster-23533.34641. (H) Cluster-23533.48337. (I) Cluster-23533.37693 (J) Cluster-23533.42950. (K) Cluster-23533.45159. (L) Cluster-23533.39931. (M) Cluster-23533.39576. (N) Cluster-23533.40653. (O) Cluster-23533.38592. (P) Cluster-23533.33992. (Q) Cluster-23533.45050. (R) Cluster-23533.37654
3.4 Assessment of Candidate Reference Gene Expression Stability
The stability of 18 potential reference genes was evaluated across various conditions drought stress levels was assessed using four methods based on the result of qPCR.
According to the Genorm analysis, the average expression stability (represented by numerical M value) of the 18 candidate reference genes ranged from 0.351 (Cluster-23533.34641) to 2.647 (Cluster-23533.37654).Cluster-23533.34641 and Cluster-23533.37693, Cluster-23533.39576 were identified as the most stable, as they exhibited the lowest M value among the tested genes. Conversely, Cluster-23533.40653, cluster-23533.21203 and Cluster-23533.37654, these three genes with the highest M values were identified as the most unstable (Table 3). In the result of NormFinder, the stability (represented by numerical M value) of 18 candidate genes ranged from 0.175 (Cluster-23533.34641) to 8.772 (Cluster-23533.37654), the Cluster-23533.34641 (0.175), Cluster-23533.42968 (0.178), and Cluster-23533.37961 (0.225) were the three top stable genes, The three most unstable genes identified were Cluster-23533.21203 (3.055), Cluster-23533.40653 (3.045) and Cluster-23533.37654 (8.772) (Table 3). Nearly the same as the result of Genorm and NormFinder, the result of Delta Ct analysis showed that the Cluster-23533.34641 (1.70), Cluster-23533.37961 (1.76) and Cluster-23533.42968 (1.77) were the top three stable genes, and Cluster-23533.40653 (3.61), Cluster-23533.21203 (3.68) and Cluster-23533.37654 (8.88) were the top three unstable genes (Table 3). Slightly different from the result of Genorm and NormFinder, the result of BestKeeper (represented by numerical SD and CV value) showed that Cluster-23533.37693, Cluster-23533.34641 and Cluster-23533.45179 (Table 3).
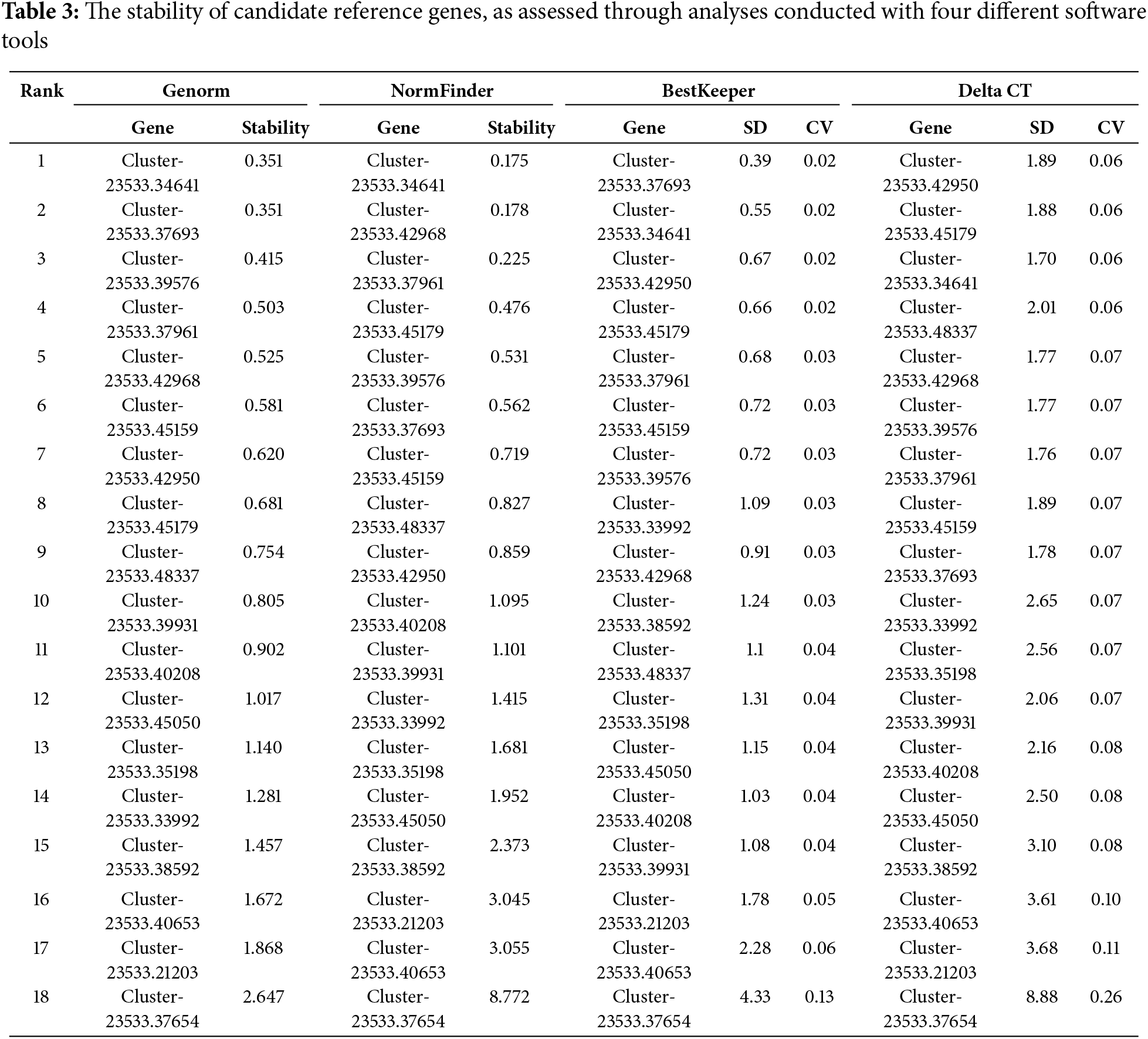
Veen diagram of the most stable reference genes based on four analyzing methods showed except the Cluster-23533.40208, Cluster-23533.48337 and Cluster-23533.39931, the rest of the candidate genes are common to the results of the four analysis methods. According to their stability, Cluster-23533.34641, Cluster-23533.37693 and Cluster-23533.39576 were the top three stable genes (Fig. 4). Based on a comprehensive comparison of several analysis methods, we recommend Cluster-23533.34641 as the priority reference gene for M. transitoria under drought stress.
Figure 4: The Veen diagram of candidate genes
With climate change, resistant plant breeding is receiving increasing attention from researchers, in particular, the functional research on resistance-related genes is the most critical for breeding. Developing and assessing the most appropriate reference gene is the key step and fundamental for gene function research.
Booming sequencing technology affords convenience for gene function research, and reference gene development and assessment also hitched a ride on technological development. Based on the Transcriptome sequencing, four new reference genes were developed and validated in drought-stressed soybeans [31]. Fifteen candidate reference genes were found through mining the publicly available transcriptional sequencing of Solanum lycopersicum [32].
The Cluster-23533.34641 was annotated as the putative vacuolar protein sorting-associated protein 24 (VPS) (Table 1), VPS including numerous family members, such as VPS34, VPS35, VPS38 and other members. These members are related to intracellular transport [33], sorting of vacuolar proteins [34], calcium-dependent lipid binding [35], perivacuolar compartment [36], and even related to the salt resistance of plants [37]. However, there is no report about the VPS 24 in M. transitoria, and its function needs to be verified.
The Cluster-23533.37693 was annotated as the protein deletion of SUV3 suppressor 1 in NR, and related to proteasome assembly, and mRNA export from the nucleus in bioprocess. The Cluster-23533.39576 was annotated as serine/threonine-protein kinase, most serine/threonine-protein kinases act as a regulator of plants when tolerance to stress, for example, SNF1-type serine/threonine protein kinase could enhance stress tolerance in Arabidopsis in drought, salt, and freezing stresses [38], the same result also found in rice [39]. In addition, serine threonine-protein kinase is related to the plant disease resistance gene [40,41]. The Cluster-23533.37961 was annotated as the E3 ubiquitin-protein ligase RHC1A, according to previous research it acted as a negative regulatory factor of LpbZIP40, it may be related to the high-temperature resistance mechanism of ryegrass [42]. The Cluster-23533.42968 was annotated as WRKY transcription factor 20, in some plants it is considered a conserved drought-up-regulated transcription factor [43]. Its over-expression could promote drought tolerance in Arabidopsis thaliana [44]. However, Cluster-23533 (Probable WRKY transcription factor 20) were stably expressed in M. transitoria.
According to this result, we suspected that the function of the above-mentioned candidate genes is not the same as that in other species. However, due to the lack of a reference genome, we cannot determine the functions of these genes in M. transitoria, so further research into gene function is needed.
In a word, the Cluster-23533.34641 was validated as the preferred reference gene for M. transitoria and for other related species under drought stress, and the rest reference genes could be combined and used as double internal references.
Acknowledgement: None.
Funding Statement: This research was supported by the Natural Science Foundation of Qinghai Province (2022-ZJ-902).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Defang Zhang; data collection: Ting Li, Jun Xv; analysis and interpretation of results: Defang Zhang, Ting Li; draft manuscript preparation: Defang Zhang, Ting Li. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are openly available in the National Center for Biotechnology Information (NCBI) SRA database (Accession number PRJNA1172450).
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Abbreviation
| Sample | Raw Reads |
| qRT-PCR | quantitative Reverse Transcription PCR |
| UBQ | Ubiquitin extension |
| EF1 | Elongation Factor 1 |
| 18S rRNA | 18S ribosomal RNA |
| GAPDH | Glyceraldehyde-3-phosphate dehydrogenase |
| ACTB | Actin isoform B |
| ACT | Actin |
| UBQ-1 | Ubiquitin-1 |
| PP2A | phosphatase 2A |
| RPL2 | Ribosomal protein L2 |
| TUB | Tubulin |
| MDH | Malate dehydrogenase |
| THFS | formate-tetrahydrofolate ligase |
| RPL4 | Ribosomal protein L4 |
| TMp1 | type 1 membrane protein |
| RPL18 | Ribosomal protein L18 |
| HIS3 | Histone 3 |
| TUA3 | Tubulin α |
| MdH1 | Histone 1 |
Appendix A
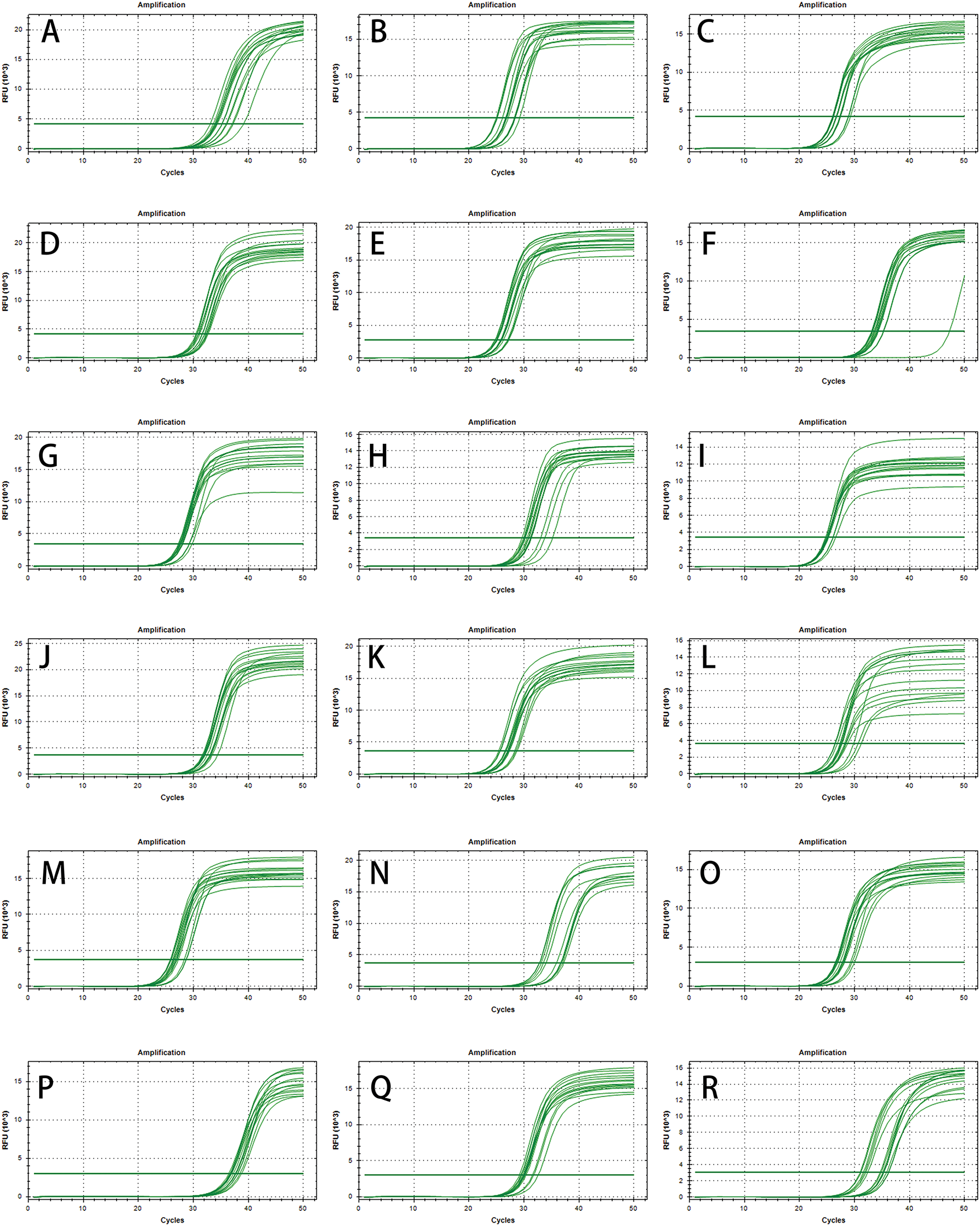
Figure A1: The amplification curve of all candidate reference gene in qPCR. (A) Cluster-23533.35198. (B) Cluster-23533.40208. (C) Cluster-23533.42968. (D) Cluster-23533.45179. (E) Cluster-23533.37961. (F) Cluster-23533.21203. (G) Cluster-23533.34641. (H) Cluster-23533.48337. (I) Cluster-23533.37693 (J) Cluster-23533.42950. (K) Cluster-23533.45159. (L) Cluster-23533.39931. (M) Cluster-23533.39576. (N) Cluster-23533.40653. (O) Cluster-23533.38592. (P) Cluster-23533.33992. (Q) Cluster-23533.45050. (R) Cluster-23533.37654. This picture is automatically generated by the machine when doing experiments, can not be modified for people, please forgive me
References
1. Yu DJ.Flora reipublicae popularis sinicae. Vol. 36. Beijing, China: Science Press; 1982. [Google Scholar]
2. Li XY, Xu HX, Chen JW.Rapid identification of red-flesh loquat cultivars using EST-SSR markers based on manual cultivar identification diagram strategy. Genet Mol Res. 2014;13(2):3384–94. doi:10.4238/2014.April.29.17. [Google Scholar] [PubMed] [CrossRef]
3. Guo X, Zhang D, Bai L.Development of EST-SSR markers and population genetic structure and genetic diversity of the Malus transitoria (Batalin) CK Schneider in Qinghai-Tibetan Plateau. Genet Resour Crop Evol. 2023;70(3):919–33. doi:10.1007/s10722-022-01477-5. [Google Scholar] [CrossRef]
4. Shi WJ, Zhang DF, Guo XD. Analysis and prediction of suitable area of Malus transitoria (Batalin) C, Schneider under the background of climate change. J Qinghai Univ. 2023;41(6):24–31. [Google Scholar]
5. Cong PH. Screening of reference genes for real-time fluorescence quantitative PCR in apple (Malus × domestica). J Fruit Sci. 2012;29(6):6–11. [Google Scholar]
6. Yan JW, Yuan FR, Long GY, Qin L, Deng ZN. Selection of reference genes for quantitative real-time RT-PCR analysis in citrus. Mol Biol Rep. 2012;39:1831–8. [Google Scholar] [PubMed]
7. Wei TL, Wang H, Pei MS, Liu HN, Yu YH, Jiang JF, et al. Identification of optimal and novel reference genes for quantitative real-time polymerase chain reaction analysis in grapevine. Grape Wine Res. 2021;27(3):325–33. doi:10.1111/ajgw.12483. [Google Scholar] [CrossRef]
8. Ren F, Zhang ZP, Fan XD, Hu GJ, Li C, Dong YF. Screening and validation of reference genes for real-time fluorescence quantitative PCR in grapevine. Molecul Plant Breed. 2019;17(23):7801–10. [Google Scholar]
9. Zhu LF, Yang CQ, You YH, Liang W, Wang NN, Ma FW, et al. Validation of reference genes for qRT-PCR analysis in peel and flesh of six apple cultivars (Malus domestica) at diverse stages of fruit development. Sci Hortic. 2019;244:165–71. [Google Scholar]
10. Fan LM, Wang C, Liu GS, Yuan YB. Screening and validation of reference genes for real-time fluorescence quantitative PCR during coloring period in apple (Malus domestica). Plant Physiology Communis. 2014;50(12):1903–11. [Google Scholar]
11. Kumar G, Singh AK. Reference gene validation for qRT-PCR based gene expression studies in different developmental stages and under biotic stress in apple. Sci Hortic. 2015;197:597–606. doi:10.1016/j.scienta.2015.10.025. [Google Scholar] [CrossRef]
12. Perini P, Pasquali G, Margis PM, Oliviera PRD, Revers LF. Reference genes for transcriptional analysis of flowering and fruit ripening stages in apple (Malus×domestica Borkh.). Mol Breed. 2014;34:829–42. doi:10.1007/s11032-014-0078-3. [Google Scholar] [CrossRef]
13. Lin SK, Xu SC, Huang LY, Qiu FX, Zheng YH, Liu QH, et al. Selection and validation of reference genes for normalization of RT-qPCR analysis in developing or abiotic-stressed tissues of loquat (eriobotrya japonica). Phyton-Int J Experim Bot. 2023;92(4):1185–201. doi:10.32604/phyton.2023.026752. [Google Scholar] [CrossRef]
14. Sheng ZP, Luan YT, Xu C, Tao J, Zhao DQ. Selection of stable reference genes for quantitative real-time PCR on herbaceous peony (paeonia lactiflora pall.) in response to drought stress. Phyton-Int J Experim Bot. 2023;92(3):801–14. doi:10.32604/phyton.2023.024953. [Google Scholar] [CrossRef]
15. Zhou YJ, Xia H, Liu XL, Lin ZY, Guo YQ, Deng HH, et al. Identification of suitable reference genes for qRT-PCR normalization in kiwifruit. Horticulturae. 2022;8(2):170. doi:10.3390/horticulturae8020170. [Google Scholar] [CrossRef]
16. Nguyen DQ, Nguyen NL, Nguyen VT, Nguyen THG, Nguyen TH, Nguyen TKL, et al. Reliable reference genes for accurate gene expression profiling across different tissues and genotypes of rice seedlings (Oryza sativa L.) under salt stress. Russ J Plant Physiol. 2023;70(5):104. doi:10.1134/S102144372360068X. [Google Scholar] [CrossRef]
17. Ma L, Wu JY, Qi WL, Coulter JA, Fang Y, Li XC, et al. Screening and verification of reference genes for analysis of gene expression in winter rapeseed (Brassica rapa L.) under abiotic stress. PLoS One. 2020;15(9):e0236577. doi:10.1371/journal.pone.0236577. [Google Scholar] [PubMed] [CrossRef]
18. Sun HY, Li CP, Li SY, Ma JX, Li S, Li X, et al. Identification and validation of stable reference genes for RT-qPCR analyses of Kobresia littledalei seedlings. BMC Plant Biol. 2024;24:389. doi:10.1186/s12870-024-04924-w. [Google Scholar] [PubMed] [CrossRef]
19. Tang Q, Zhou GC, Liu SJ, Li W, Wang YL, Xu GY, et al. Selection and validation of reference genes for qRT-PCR analysis of gene expression in Tropaeolum majus it (Nasturtium). Horticulturae. 2023;9(11):1176. doi:10.3390/horticulturae9111176. [Google Scholar] [CrossRef]
20. Bowen JH, Schaffer RJ, Johnston J, Ireland HS, Croehurst R, Luo ZW, et al. Use of mRNA-seq data to select Malus × domestica (apple) genes for use as quantitative PCR reference genes. Acta Hortic. 2014;1110:179–84. doi:10.17660/ActaHortic.2016.1110.26. [Google Scholar] [CrossRef]
21. Bowen J, Ireland HS, Crowhurst R, Luo ZW, Watson AE, Foster TS, et al. Selection of low-variance expressed Malus × domestica (apple) genes for use as quantitative PCR reference genes (housekeepers). Tree Gene Gen. 2014;10:751–59. doi:10.1007/s11295-014-0720-6. [Google Scholar] [CrossRef]
22. Storch TT, Pegoraro C, Finatto T, Quecini V, Rombaldi CV, Girardi CL. Identification of a novel reference gene for apple transcriptional profiling under postharvest conditions. PLoS One. 2015;10(3):e0120599. doi:10.1371/journal.pone.0120599. [Google Scholar] [PubMed] [CrossRef]
23. Gutierrez L, Mauriat M, Guénin S, Pelloux J, Lefebvre JF, Louvet R, et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnol J. 2008;6(6):609–18. doi:10.1111/j.1467-7652.2008.00346.x. [Google Scholar] [PubMed] [CrossRef]
24. Vandesompele J, Preter KD, Pattyn F, Poppe B, Roy NV, Paepe AD, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3(7):1–12. doi:10.1186/gb-2002-3-7-research0034. [Google Scholar] [PubMed] [CrossRef]
25. Huggett J, Dheda K, Bustin S, Zumla A. Real-time RT-PCR normalization; strategies and considerations. Genes Immun. 2005;6(4):279–84. doi:10.1038/sj.gene.6364190. [Google Scholar] [PubMed] [CrossRef]
26. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nat Biotechnol. 2013;29(7):644–52. doi:10.1038/nbt.1883. [Google Scholar] [PubMed] [CrossRef]
27. Liu Y, Shi W, Zhang D.Development and evaluation of suitable reference genes for qRT-PCR normalization of hybrids derived from Lycium barbarum and Lycium ruthenicum.Mol Biol Rep.2024;51(1):922.doi:10.1007/s11033-024-09848-0. [Google Scholar] [PubMed] [CrossRef]
28. Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 2004;64(15):5245–50. doi:10.1158/0008-5472.CAN-04-0496. [Google Scholar] [PubMed] [CrossRef]
29. Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett. 2004;26(6):509–15. doi:10.1023/B:BILE.0000019559.84305.47. [Google Scholar] [PubMed] [CrossRef]
30. Wang Q, Ishikawa T, Michiue T, Zhu BL, Guan DW, Maeda H. Stability of endogenous reference genes in postmortem human brains for normalization of quantitative real-time PCR data: comprehensive evaluation using geNorm, NormFinder, and BestKeeper. Int J Legal Med. 2012;126(6):943–52. doi:10.1007/s00414-012-0774-7. [Google Scholar] [PubMed] [CrossRef]
31. Marcolino-Gomes J, Rodrigues FA, Fuganti-Pagliarini R, Nakayama TJ, Reis RR, Bouças Farias JR, et al. Transcriptome-wide identification of reference genes for expression analysis of soybean responses to drought stress along the day. PLoS One. 2015;10(9):e0139051. doi:10.1371/journal.pone.0139051. [Google Scholar] [PubMed] [CrossRef]
32. Duan YK, Han R, Su Y, Wang AY, Li S, Sun H, et al. Transcriptional search to identify and assess reference genes for expression analysis under stress and hormone treatment conditions. J Integr Agric. 2022;21(11):3216–29. doi:10.1016/j.jia.2022.07.051. [Google Scholar] [CrossRef]
33. Marshall RS, Vierstra RD. Autophagy: the master of bulk and selective recycling. Annu Rev Plant Biol. 2018;69(1):173–208. doi:10.1146/annurev-arplant-042817-040606. [Google Scholar] [PubMed] [CrossRef]
34. Liu F, Hu WM, Li FQ, Marshall RS, Zarza X, Munnik T, et al. AUTOPHAGY-RELATED 14 and its associated phosphatidylinositol 3-kinase complex promote autophagy in Arabidopsis. Plant Cell. 2020;32(12):3939–60. doi:10.1105/tpc.20.00285. [Google Scholar] [PubMed] [CrossRef]
35. Welters P, Takegawa K, Emr SD, Chrispeels MJ. AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc Natl Acad Sci U S A. 1994;91(24):11398–402. doi:10.1073/pnas.91.24.11398. [Google Scholar] [PubMed] [CrossRef]
36. Nodzynski T, Feraru MI, Hirsch S, De Rycke R, Niculaes C, Boerjan W, et al.Retromer subunits VPS35A and VPS29 mediate prevacuolar compartment (PVC) function in Arabidopsis.Mol Plant.2013;6(6):1849–62. doi:10.1093/mp/sst044. [Google Scholar] [PubMed] [CrossRef]
37. Xia ZL, Wei YY, Sun KL, Wu JY, Wang YX, Wu K. The maize AAA-type protein SKD1 confers enhanced salt and drought stress tolerance in transgenic tobacco by interacting with Lyst-interacting protein. PLoS One. 2013;8:e69787. doi:10.1371/journal.pone.0069787. [Google Scholar] [PubMed] [CrossRef]
38. Mao XG, Zhang HY, Jing R.TaSnRK2.4, an SNF1-type serine/threonine protein kinase of wheat (Triticum aestivum L.confers enhanced multistress tolerance in Arabidopsis.J Exp Bot.2010;61(3):683–96.doi:10.1093/jxb/erp331. [Google Scholar] [PubMed] [CrossRef]
39. Diédhiou CJ, Popova OV, Dietz KJ, Golldack D. The SNF1-type serine-threonine protein kinase SAPK4 regulates stress-responsive gene expression in rice. BMC Plant Biol. 2008;8(1):49. doi:10.1186/1471-2229-8-49. [Google Scholar] [PubMed] [CrossRef]
40. Song WY, Wang GL, Chen LL, Kim HS, Pi LY, Holsten T, et al.A receptor kinase-like protein encoded by the rice disease resistance gene, Xa21. Science. 1995;270(5243):1804–6. doi:10.1126/science.270.5243.1804. [Google Scholar] [PubMed] [CrossRef]
41. Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, et al. Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science. 1993;262(5138):1432–6. doi:10.1126/science.7902614. [Google Scholar] [PubMed] [CrossRef]
42. Fang ZF.Molecular regulatory mechanism of LpbZIP40 enhanced heat tolerance in perennial ryegrass [Ph.D. thesis]. Wuhan, China: Huazhong Agricultural University; 2023. [Google Scholar]
43. Luo X, Bai X, Sun X, Zhu D, Liu B, Ji W, et al. Expression of wild soybean WRKY20 in Arabidopsis enhances drought tolerance and regulates ABA signalling. J Exp Bot. 2013;64(8):2155–69. doi:10.1093/jxb/ert073. [Google Scholar] [PubMed] [CrossRef]
44. Feng Y, Liang CL, Li BB, Wan T, Liu T, Cai YL. Differential expression profiles and pathways of genes in drought resistant tree species Prunus mahaleb roots and leaves in response to drought stress. Sci Hortic. 2017;226:75–84. doi:10.1016/j.scienta.2017.07.057. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools