 Open Access
Open Access
ARTICLE
Identification of Secondary Metabolites of Lycium ruthenicum Murray by UPLC-QTOF/MS and Network Pharmacology of Its Anti-Inflammatory Properties
Anhui Provincial Engineering Laboratory for Efficient Utilization of Featured Resource Plants, College of Life Sciences, Huaibei Normal University, Huaibei, 235000, China
* Corresponding Author: Chen Chen. Email:
# These authors contributed equally to this work
Phyton-International Journal of Experimental Botany 2025, 94(3), 793-807. https://doi.org/10.32604/phyton.2025.063549
Received 17 January 2025; Accepted 20 February 2025; Issue published 31 March 2025
Abstract
Lycium ruthenicum Murray, a plant widely cultivated in northwestern China, is integral to traditional Chinese medicine, with applications in treating menstrual disorders, cardiovascular diseases, and menopausal symptoms. Despite its recognized medicinal value and use as a functional food, comprehensive knowledge of its metabolites and their pharmacological effects remains limited. This study presents an innovative approach using ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry (UPLC–QTOF/MS) to conduct a detailed analysis of both wild and cultivated L. ruthenicum samples. A total of 62 peaks were detected in the total ion current profile, with 59 metabolites identified based on accurate mass and MS/MS fragmentation patterns. Multivariate analyses revealed distinct chemical profiles that effectively differentiate between wild and cultivated samples, identifying six key chemical markers crucial for the classification of L. ruthenicum varieties. Furthermore, a comprehensive interaction network was constructed, highlighting the top 20 significant pathways, which elucidates the components–targets–pathways–disease relationships. These findings not only provide a robust methodology for quality assessment and geographical discrimination of L. ruthenicum but also lay a theoretical foundation for its future exploration in traditional Chinese medicine, thereby enhancing its potential as both a medicinal and functional food source.Keywords
Lycium ruthenicum Murray is a functional and medicinal plant widely cultivated in the saline-alkali deserts of northwestern China, renowned for its resilience to salt and drought. This hardy species plays a pivotal role in mitigating soil desertification and salinization, making it valuable for ecological restoration [1]. In addition to its environmental significance, L. ruthenicum has been used in traditional medicine for centuries, as documented in classic Tibetan medical books such as The Four Medical Tantras and Jing Zhu Ben Cao. It has been employed to treat conditions like abnormal menstruation, cardiovascular disease, and menopausal symptoms, providing a rich basis for its exploration in modern pharmacological research [2–4].
Phytochemical studies have identified bioactive compounds in L. ruthenicum, including flavonoids, phenolic acids, anthocyanins, and alkaloids [5–7]. While much of the research has focused on anthocyanins, a key contributor to its health benefits, comprehensive profiling of its secondary metabolites is still lacking [8–10]. This knowledge gap hinders the full potential of L. ruthenicum as a functional food and therapeutic product. To ensure food safety and maximize its applications, further research into the plant’s secondary metabolites is essential. Despite the increasing popularity of L. ruthenicum, the differences in the metabolite compositions of wild and cultivated fruits remain unexplored. The need for reliable, accurate methodologies to analyze these metabolites for quality control has never been more urgent. Among them, ultra–performance liquid chromatography coupled with quadrupole time–of–flight mass spectrometry (UPLC–QTOF/MS) effectively combines the advantages of UPLC and high–resolution MS and is a powerful tool to identify compounds in foods and herbs.
Modern pharmacological studies have confirmed that L. ruthenicum possesses a wide range of beneficial properties, including anti-fatigue, radiation resistance, antioxidant, immune-enhancing, memory disorder relief, gouty arthritis alleviation, and anti-inflammatory effects [11–14]. Among these, its anti-inflammatory activity has garnered significant interest. Research has shown that L. ruthenicum exhibits intestinal anti-inflammatory effects in mice [15–17]. For example, Wang et al. demonstrated that the plant could downregulate the expression of IL-1 and IL-2, alleviating inflammation caused by oxidative damage [18]. Yan et al. reported that L. ruthenicum significantly improved acute gouty arthritis in rats through its anti-inflammatory effects [19]. Wen et al. found that the plant reduced serum inflammatory markers TNF-α and IL-6, thereby protecting the neurological function of rats after acute spinal cord injury [20]. L. ruthenicum has the characteristics of complex chemical components and excellent anti-inflammatory effects, resulting in problems such as an ambiguous foundation, unclear mechanism of action, and disconnection between research basis and clinical application
In recent years, network pharmacology has emerged as a frontier in traditional Chinese medicine research. This approach constructs a multilevel network of components, targets, and pathways, providing a more holistic view of herbal medicines’ mechanisms of action. Unlike traditional pharmacological studies, which often focus on isolated compounds or single biological pathways, network pharmacology enables the identification of multiple, interconnected targets and their associated pathways. While the application of network pharmacology to L. ruthenicum is still limited, its potential to provide deeper insights into the plant’s multi-target, multi-pathway therapeutic effects is significant.
This study aims to bridge the gap between traditional uses and modern pharmacological understanding of L. ruthenicum by combining UPLC-QTOF/MS with network pharmacology. We employed UPLC-QTOF/MS to analyze the chemical profiles of wild and cultivated L. ruthenicum and performed multivariate analyses, including principal component analysis (PCA) and orthogonal partial least squares discriminant analysis (OPLS-DA), to identify marker compounds and guide the discrimination of different varieties. Additionally, we explored inflammation-related components, targets, and pathways to elucidate the plant’s mechanism of action against inflammation. This integrated approach offers a comprehensive framework for understanding L. ruthenicum’s therapeutic potential and lays the foundation for its future application in both traditional and modern medical contexts.
Methanol and formic acid (FA) were purchased from Sigma, and MS–grade acetonitrile was purchased from Merck. Fresh L. ruthenicum fruits were collected in Qinghai and Xinjiang Provinces, China.
The fruits were lyophilized and subsequently ground into a fine powder. For extraction, 200 mg of the powder was subjected to ultrasound-assisted extraction with 10 mL of 75% methanol for 30 min. After extraction, the mixture was centrifuged at 5000× g for 10 min, and the supernatant was filtered through a 0.22 μm filter prior to UPLC-QTOF/MS analysis. Three replicates were prepared for each sample to assess consistency.
UPLC-QTOF/MS analysis was performed using a Waters system coupled with a QTOF mass spectrometer (Waters Corporation, Milford, MA, USA). The flow rate was set at 0.3 mL/min. The chromatographic separation was achieved using a UHPLC column (250 mm × 2.1 mm, 1.8 μm). The mobile phase consisted of phase A (water with 0.1% formic acid, FA) and phase B (acetonitrile with 0.1% FA), with the following gradient elution: 3%–15% B for 0–20 min, 15%–30% B for 20–40 min, 30%–95% B for 40–45 min, and 95% B for 45–55 min. Peak identification was performed using a PDA detector set to 280 nm, with UV–vis spectra acquired from 190 to 600 nm.
The mass spectrometry parameters were as follows: Electrospray ionization (ESI) in negative ion mode, source temperature of 100°C, cone gas flow of 50 L/h, desolvation temperature of 400°C, capillary voltage of 2.50 kV, mass range of 50–1500 m/z, and a scan rate of 0.25 s. High-resolution mass data were collected at both high and low collision energies, and both fragments and precursor ions were analyzed simultaneously.
The raw data obtained from the samples were processed using Progenesis QI software (Nonlinear Dynamics, UK) for peak detection and automatic alignment. Compound identification was initially performed by querying the METLIN database, followed by further confirmation through the comparison of MS/MS fragmentation patterns and accurate masses. Where possible, identifications were cross-referenced with known standards and existing literature. To further validate the results, reference compounds were employed, and the findings were compared with previously reported data on similar compounds. Multivariate statistical analysis was conducted using SIMCA-P 14.1 (Umetrics, Sweden), which included Principal Component Analysis (PCA) and Orthogonal Partial Least Squares Discriminant Analysis (OPLS-DA).
The secondary metabolites identified through UPLC-QTOF/MS analysis were imported into the Swiss Target Prediction database (http://SwissTargetPrediction) to predict the targets of each compound. The resulting targets were then merged for further analysis. To identify target proteins associated with inflammation, the keyword “inflammation” was searched in the DRUGBANK and DisGeNET databases, and gene names were retrieved from the UniProt database. The candidate component targets and disease-related targets were imported into the STRING platform to construct a protein–protein interaction (PPI) network. This PPI network was then analyzed using Cytoscape and the NetworkAnalyzer tool for network topology. The degree values of the nodes were calculated to rank the genes, with genes having degree values above the average threshold considered key targets. In total, 64 key targets were identified and further analyzed for their potential roles within the network. These key targets were imported into the DAVID database for Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses to obtain biological information and explore potential pharmacological mechanisms of action.
Under the UPLC conditions described above, 12 samples from wild and cultivated plants were analyzed. Peaks consistently observed across all chromatograms were assigned, demonstrating the reproducibility and reliability of the UPLC separation. Fig. 1a shows the UPLC chromatographic profile at 280 nm, highlighting the separation of metabolites based on retention time. In contrast, Fig. 1b presents the total ion chromatogram (TIC) acquired in negative ion mode, offering an overview of the detected metabolites and their relative intensities in mass spectrometry.
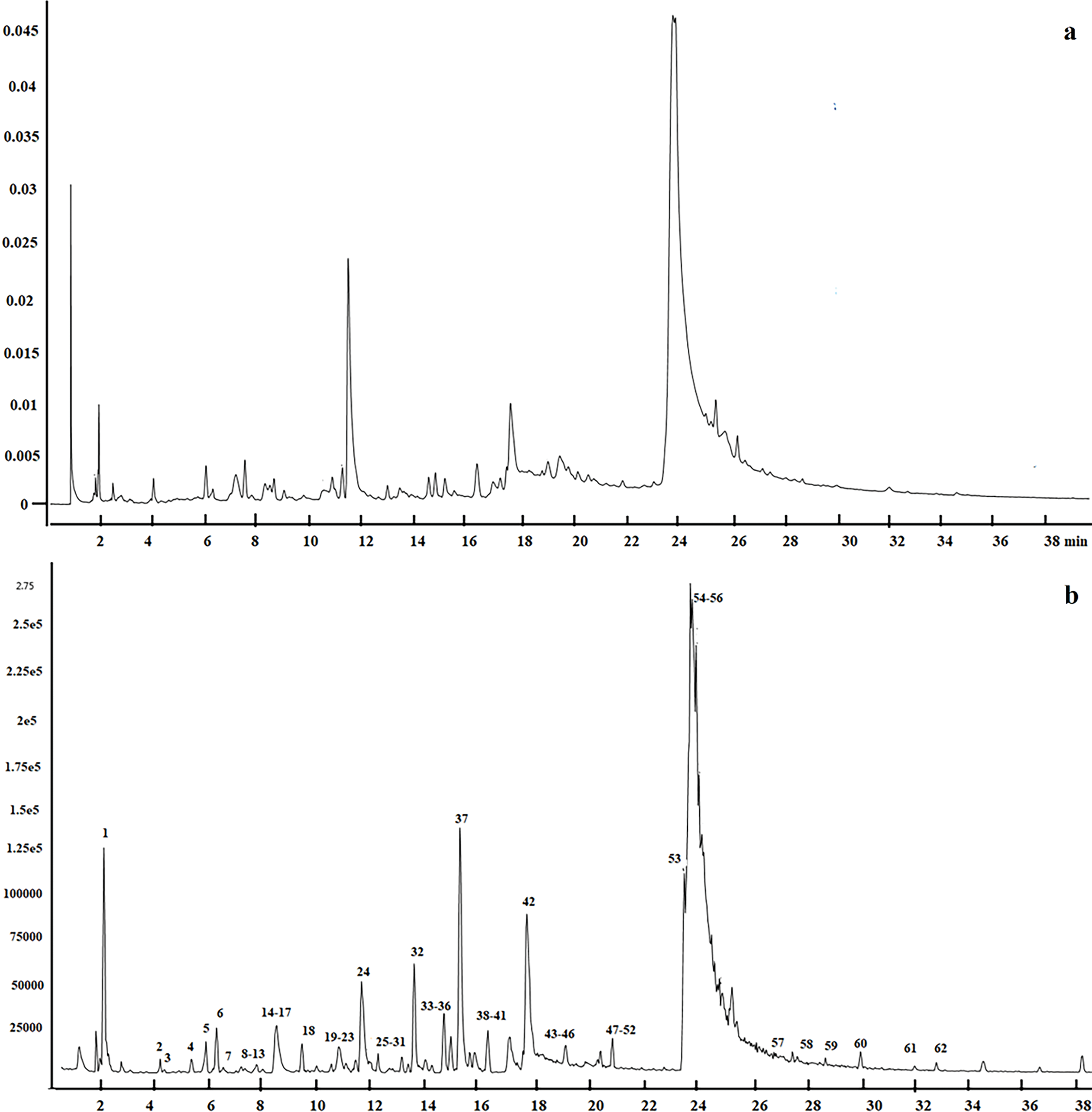
Figure 1: UPLC-QTOF/MS analysis of L. ruthenicum. (a) Chromatographic profile at 280 nm showing the separation of metabolites. (b) Total ion chromatogram (TIC) in negative ion mode, displaying the overall metabolite ion distribution obtained by mass spectrometry
3.2 Identification of the Metabolites
The validation of the UPLC-QTOF/MS analytical method enabled the identification and authentication of compounds in L. ruthenicum. A total ion chromatogram in negative ESI mode under optimal conditions is shown in Fig. 1b, with compound formulas, accurate masses, fragment ions, and retention times of the major peaks listed in Table 1. To ensure the reliability of metabolite identification, the mass accuracy (expressed as ppm error) and identification confidence levels were included. A total of 62 peaks were detected, primarily consisting of alkaloids, anthocyanins, phenolic acids and derivatives, and flavonoids.
Peaks 1, 6, 9, 11, 15, 17–19, 24, 26, 27, and 32 were identified as phenolic acids and derivatives. MS data showed ions at m/z 191 ([quinic acid–H]−) and 179 ([caffeic acid–H]−), suggesting the presence of quinic acid and caffeoyl moieties, respectively. Peak 1 was identified as quinic acid, peaks 9 and 24 as chlorogenic acid isomers, and peaks 15, 16, 17, and 25 as dicaffeoylquinic acid isomers [22]. Peak 4 was identified as protocatechuic acid glucoside, with a protonated molecular ion at m/z 315.0716 and fragment ions at m/z 153 (protocatechuic acid) and m/z 162 (glucoside) [23].
A total of 11 alkaloids (10, 12, 23, 30, 37, 39, 42, 43, 44, 47, and 48) were detected. Tryptophan (peak 10) showed a protonated molecular ion at m/z 203.2083, with fragment ions at m/z 159, 142, and 116 ([M–H–CO2]−, [M–H–NH3]−, [M–H–CO2–NH3–C2H2]−) [21]. Peak 45, with a parent ion at m/z 245.0928, was identified as N–acetyl–tryptophan [24]. Peak 23 matched the spectrum of N,N–bis(dihydrocaffeoyl) spermine from L. ruthenicum [4]. Peaks 28 and 31, with a parent ion at m/z 794.3343, were identified as N–caffeoyl, N–dihydrocaffeoyl spermidine dihexose isomers [25]. Peak 32 was identified as lycibarbarspermidine H, based on the published spectrum from L. barbarum [26]. Peaks 40, 41, and 42 were identified as caffeoyl (dihydrocaffeoyl) spermidine isomers ([M–H]− at m/z 470), while peak 35, with a parent ion at m/z 632, was identified as dihydrocaffeoyl-caffeoyl spermidine hexose, matching the fragmentation pattern of peak 42. Peak 37, with a parent ion at m/z 472.2452, was identified as N,N–bis-dihydrocaffeoyl spermidine [4]. Peak 48, with a parent ion at m/z 468.2131, was identified as N,N–dicaffeoylspermidine [4].
Fourteen flavonoid compounds were identified (peaks 27, 34, 36, 39, 47, 48, 49, 51–54, 58, 60, 61). Peak 36, with a parent ion at m/z 771.1979 ([M–H]−), produced fragment ions at m/z 609 and 301, which indicated it was quercetin-rutinoside-hexose [27]. Peaks 39, 48, 58, and 61 were identified as isorhamnetin-glucoside-gentiobioside, kaempferol-rutinoside-glucoside, isorhamnetin 3-rutinoside, and kaempferol-caffeylsophoroside, respectively, based on coelution with standards and MS confirmation. Peak 47, with a parent ion at m/z 625.1405, was identified as myricetin 3-rutinoside [28]. Peak 49, with a parent ion at m/z 579.1717 and a fragment ion at m/z 271, was identified as naringin. Peak 51, with a parent ion at m/z 597.1815, was identified as phloretin-diglucoside [29]. Peak 52, with a parent ion at m/z 493.0983 and a typical fragment at m/z 331, was identified as laricitrin 3-glucoside. Peak 60, with a parent ion at m/z 433.1131, was identified as helichrysin A, matching a previous study on L. ruthenicum [3].
Anthocyanins, which contribute to the plant’s vibrant coloration, were also abundant in L. ruthenicum. Eight anthocyanins (peaks 29, 43, 44, 50, 55–57, and 59) were identified by matching their mass spectra with those reported in previous studies of L. ruthenicum [7]. These included several 3,5–diglycoside derivatives of petunidin, acylated with phenolic acids, as well as trans–cis isomeric forms.
3.3 Multivariate Analyses of L. ruthenicum
To compare the metabolomic profiles among wild and cultivated L. ruthenicum fruits, multivariate analyses, including PCA and OPLS–DA, were performed on the MS data. Ion features (retention time, m/z) for L. ruthenicum were extracted from the raw MS data. The PCA score plots are shown in Fig. 2, with clear separation of three distinct groups in the PC1/PC2 score plot. These groups corresponded to the wild Qinghai, wild Xinjiang, and cultivated Qinghai samples. This separation indicates that the chemical profile of L. ruthenicum is a significant factor influencing fruit quality.
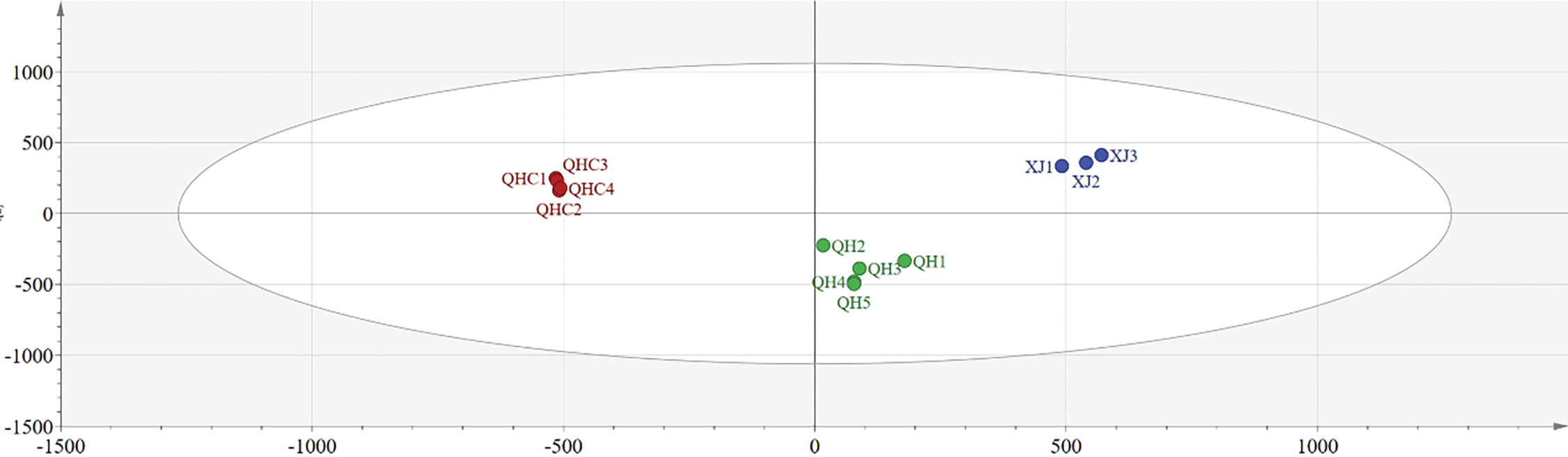
Figure 2: PCA score plots of L. ruthenicum, illustrating the differentiation of wild and cultivated samples based on their chemical profiles
OPLS–DA was employed to compare the chemical profiles of L. ruthenicum from different collection and cultivation sites. The R2Y value of the model was 0.983, and the Q2 was 0.957, indicating that the model was well established and had strong predictive ability [8,30]. The OPLS–DA score chart (Fig. 3) clearly classified the samples into three distinct groups, consistent with the PCA results, based on their chemical profiles.
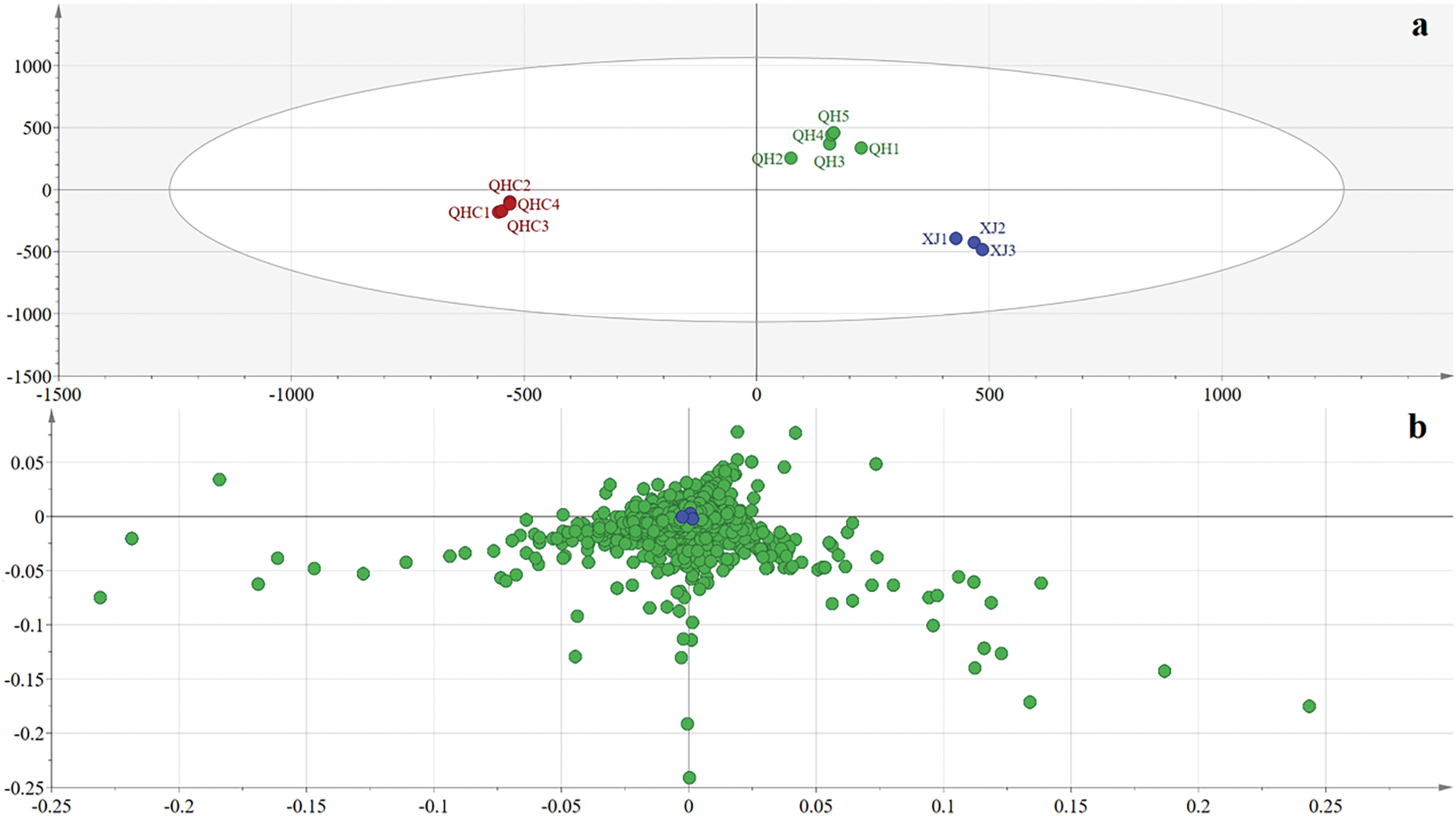
Figure 3: OPLS–DA analysis of L. ruthenicum. (a) Score plot of L. ruthenicum samples, demonstrating the separation between different collection and cultivation areas. (b) Loading plot of L. ruthenicum, highlighting the variables contributing to the differentiation
Potential sources of variation that could influence the results of the multivariate analysis include biological factors (such as differences in fruit maturity and environmental conditions) and inconsistencies in sample preparation. To minimize these variations, all samples were processed following the same protocol and analyzed under identical instrumental conditions. These measures effectively reduced the impact of experimental noise and enhanced the robustness of the model.
The identification of compounds with high VIP values provides valuable insights into the metabolites that drive the differentiation of L. ruthenicum samples. Compounds 30, 35, 39, 41, 53, and 55, based on their accurate mass data and retention times, serve as key markers for distinguishing between the different samples. These findings suggest that these specific compounds can be utilized not only to authenticate L. ruthenicum but also to potentially monitor quality and assess variations in plant samples from different origins or cultivation practices. The use of these compounds in further studies could help refine the authentication process and provide additional insights into the metabolic profile of L. ruthenicum.
The interplay between ecological factors and cultivation practices plays a pivotal role in shaping the chemical composition and overall quality of L. ruthenicum. The distinct environmental conditions found in regions such as Qinghai and Xinjiang provide a natural basis for variations in metabolite profiles. Qinghai’s plateau environment, with its prolonged sunshine durations and saline soils, offers an ideal setting for L. ruthenicum to flourish, which may contribute to the superior quality of the plants produced there [31,32]. Furthermore, the differences in longitude, latitude, altitude, and precipitation between Qinghai and Xinjiang further influence the chemical composition of L. ruthenicum, underscoring the complexity of plant responses to their environmental context. As noted, cultivation practices such as fertilizer use, irrigation, and farming techniques also play a critical role in shaping the metabolite profiles of the plant. These anthropogenic factors can lead to distinctions between wild and cultivated L. ruthenicum, a trend that is clearly reflected in the PCA and OPLS-DA models, where samples from these two groups were distinctly segregated [33].
These observations suggest that a holistic approach, integrating both ecological and cultivation factors, is essential for understanding and optimizing the quality of L. ruthenicum. Adjusting cultivation practices to replicate the ideal environmental conditions may enhance the medicinal properties and chemical composition of cultivated plants, offering a promising avenue for improving consistency and quality in production.
3.4 Network Pharmacology of the Anti-Inflammatory Properties
A total of 44 anti-inflammatory metabolites were identified from the 59 metabolites obtained, and 247 potential targets were selected based on a database search. Using intersecting targets, the STRING database was employed to construct a protein–protein interaction (PPI) network, as shown in Fig. 4. This network comprises 247 nodes and 691 edges, with an average degree of 5.6. The node size is proportional to its degree, and the intensity of the node color correlates with its degree, where a redder hue indicates a higher value. The edge thickness and color represent the combined scores between interacting nodes (Fig. 4). Topological analysis identified several key anti-inflammatory targets, including SRC (degree = 41), STAT3 (degree = 40), MAPK1 (degree = 37), and HSP90AA1 (degree = 37), all of which exhibit significant roles within the network, highlighting their importance in the anti-inflammatory effects of L. ruthenicum.
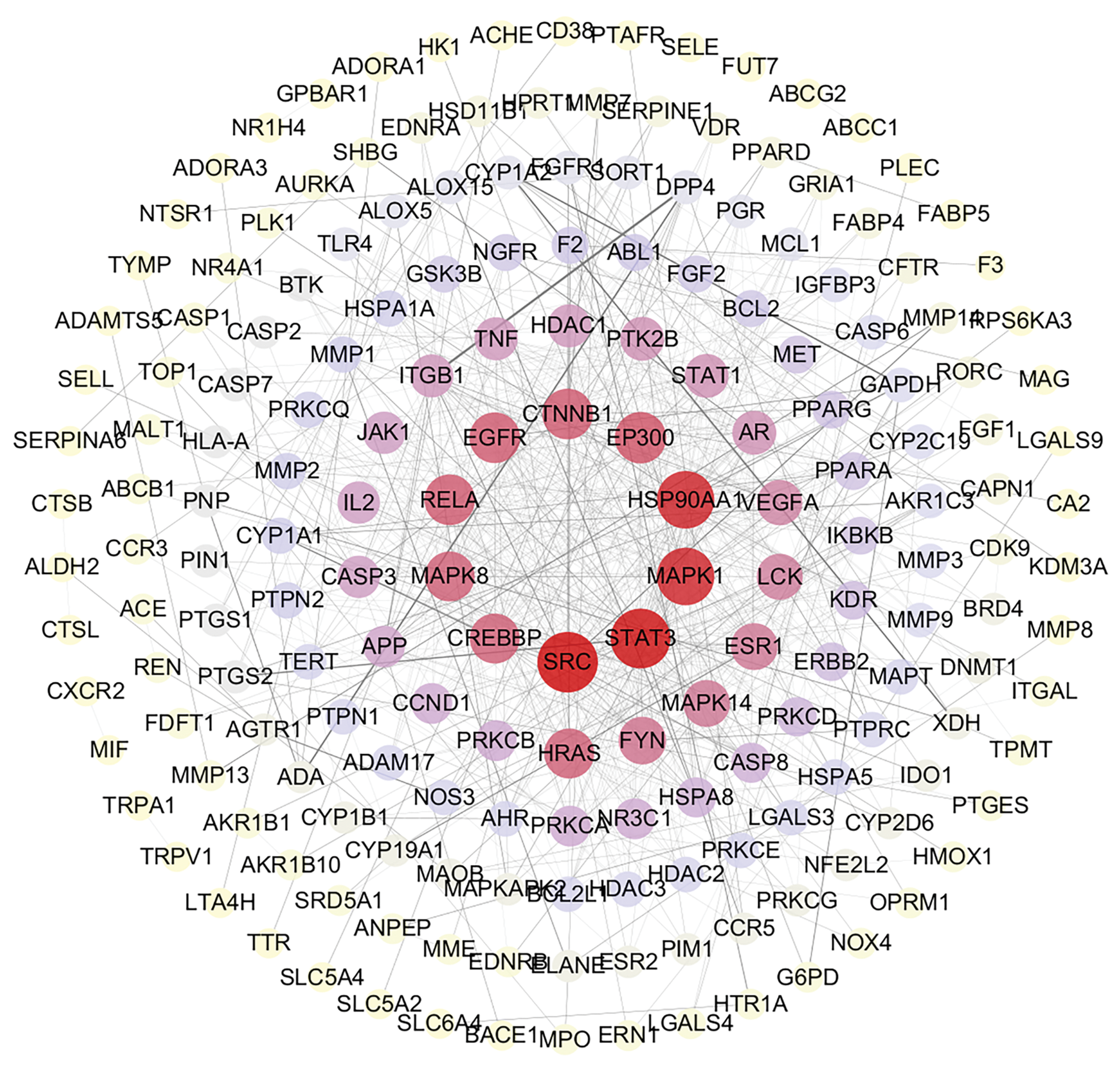
Figure 4: PPI network of the intersection targets of L. ruthenicum, showing protein–protein interactions and key proteins involved in the anti-inflammatory mechanism
GO enrichment analysis revealed a total of 2497 biological processes, 163 cellular components, and 226 molecular functions. The top ten terms from each category are illustrated in Fig. 5. The most prominent biological process was the response to molecules of bacterial origin, followed by the response to lipopolysaccharide and regulation of the inflammatory response. In the cellular component category, the most significant terms included membrane raft, membrane microdomain, and membrane region. In molecular function, endopeptidase activity, DNA-binding transcription factor binding, amide binding, and protein serine/threonine kinase activity ranked highest.
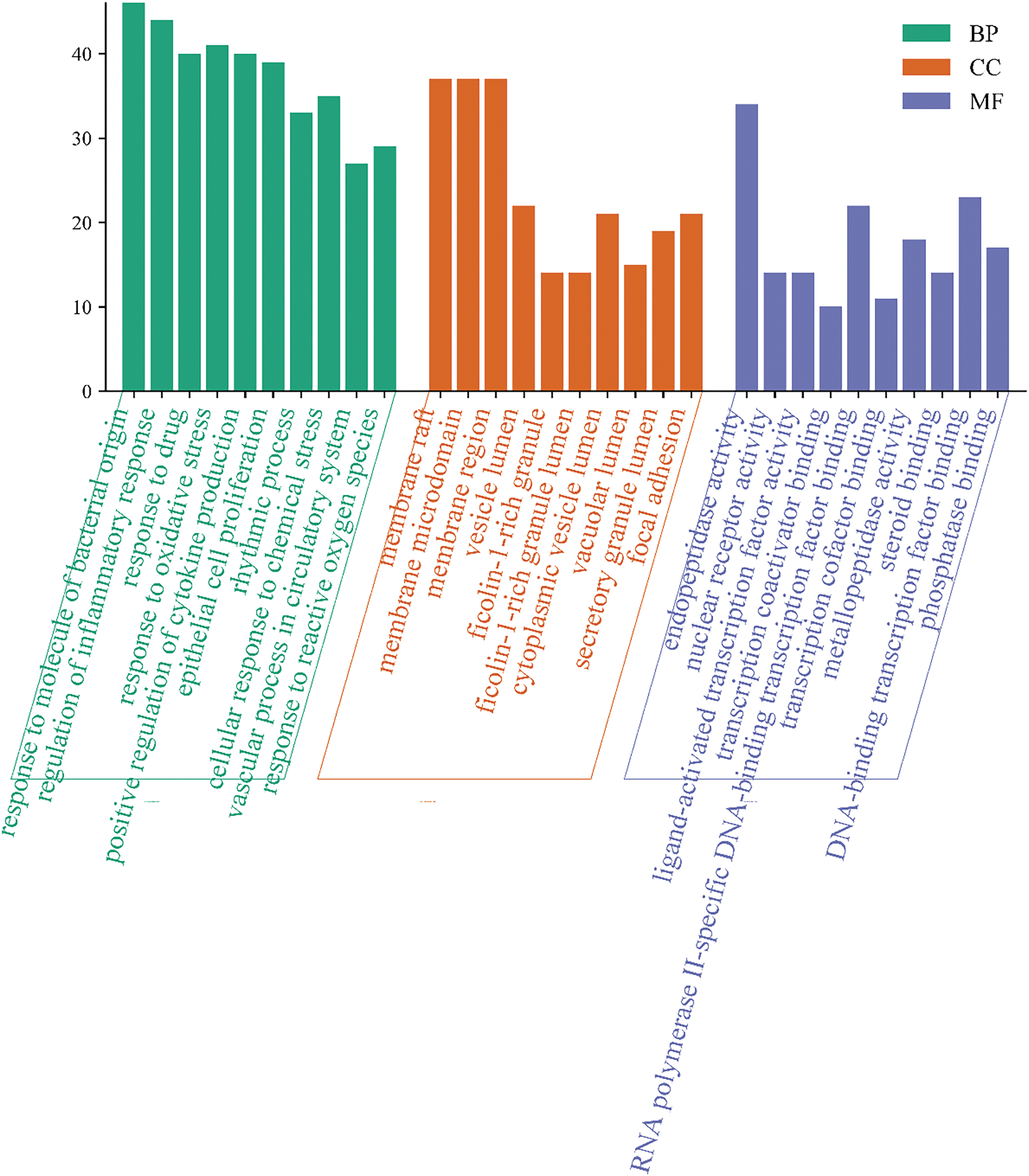
Figure 5: GO functional analysis histogram, depicting the top biological processes, cellular components, and molecular functions enriched in the intersection targets of L. ruthenicum
KEGG enrichment analysis identified 156 signaling pathways, with the top twenty pathways listed in Fig. 6. Among the inflammation-related pathways, the top ten pathways exhibiting the highest degrees of enrichment included the lipid and atherosclerosis pathway, neuroactive ligand-receptor interaction pathway, calcium signaling pathway, microRNAs in cancer pathway, chemical carcinogenesis-receptor activation pathway, MAPK signaling pathway, human cytomegalovirus infection pathway, proteoglycans in cancer pathway, and PI3K-Akt signaling pathway.
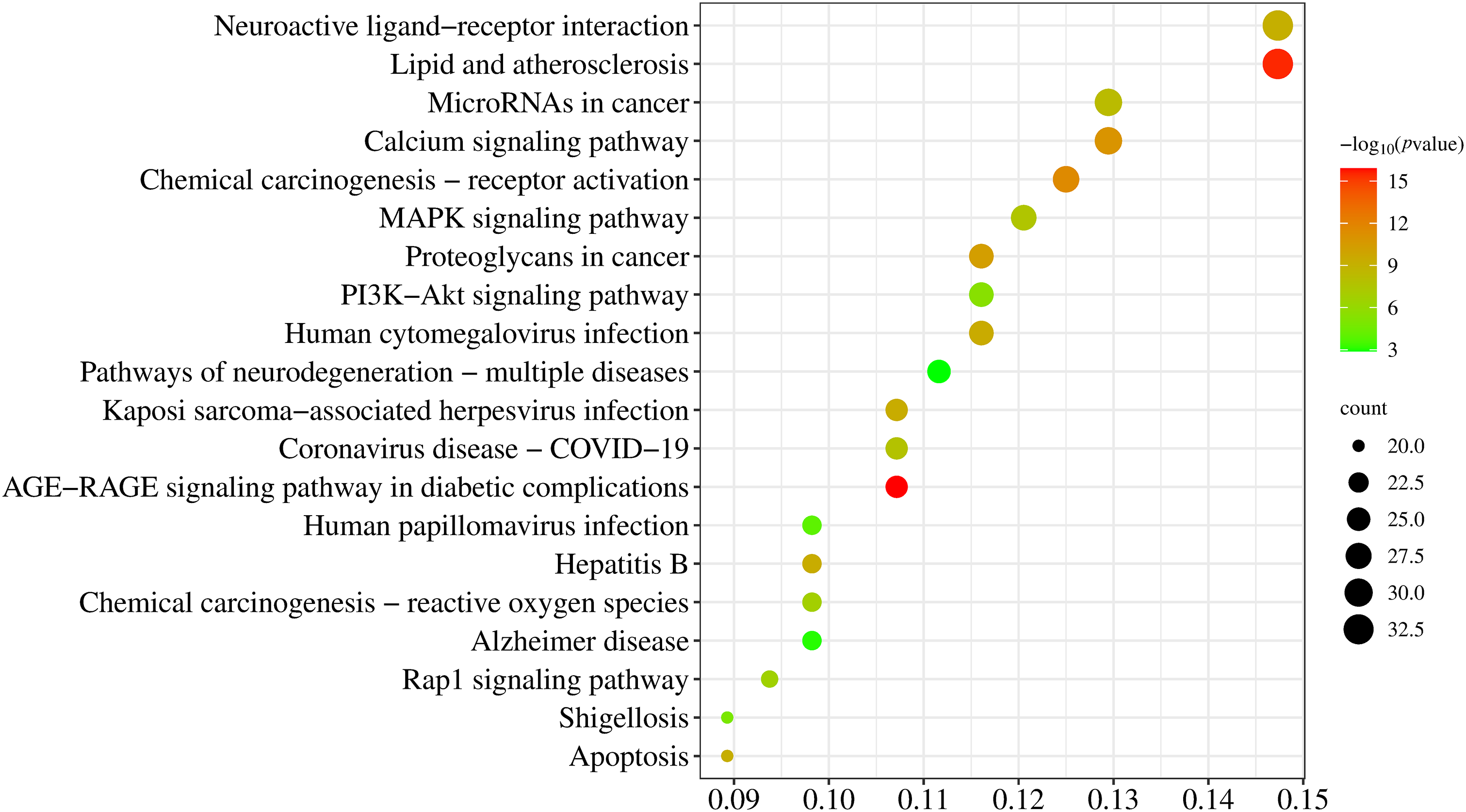
Figure 6: Dot plot of the KEGG pathway enrichment analysis, visualizing the top 20 pathways significantly enriched by the metabolites and their potential roles in the pharmacological effects of L. ruthenicum
As illustrated in Fig. 7, Cytoscape software was used to map the relationships between metabolites, targets, pathways, and diseases. In the map, red represents the targets of L. ruthenicum metabolites, blue denotes the metabolites themselves, green highlights the top twenty pathways, and yellow represents the associated diseases. The anti-inflammatory effects of L. ruthenicum are likely mediated through each active metabolite, its related targets, and the corresponding pathways. This study comprehensively captured the multimetabolite, multitarget, and multipathway mechanisms underlying the anti-inflammatory action of L. ruthenicum.
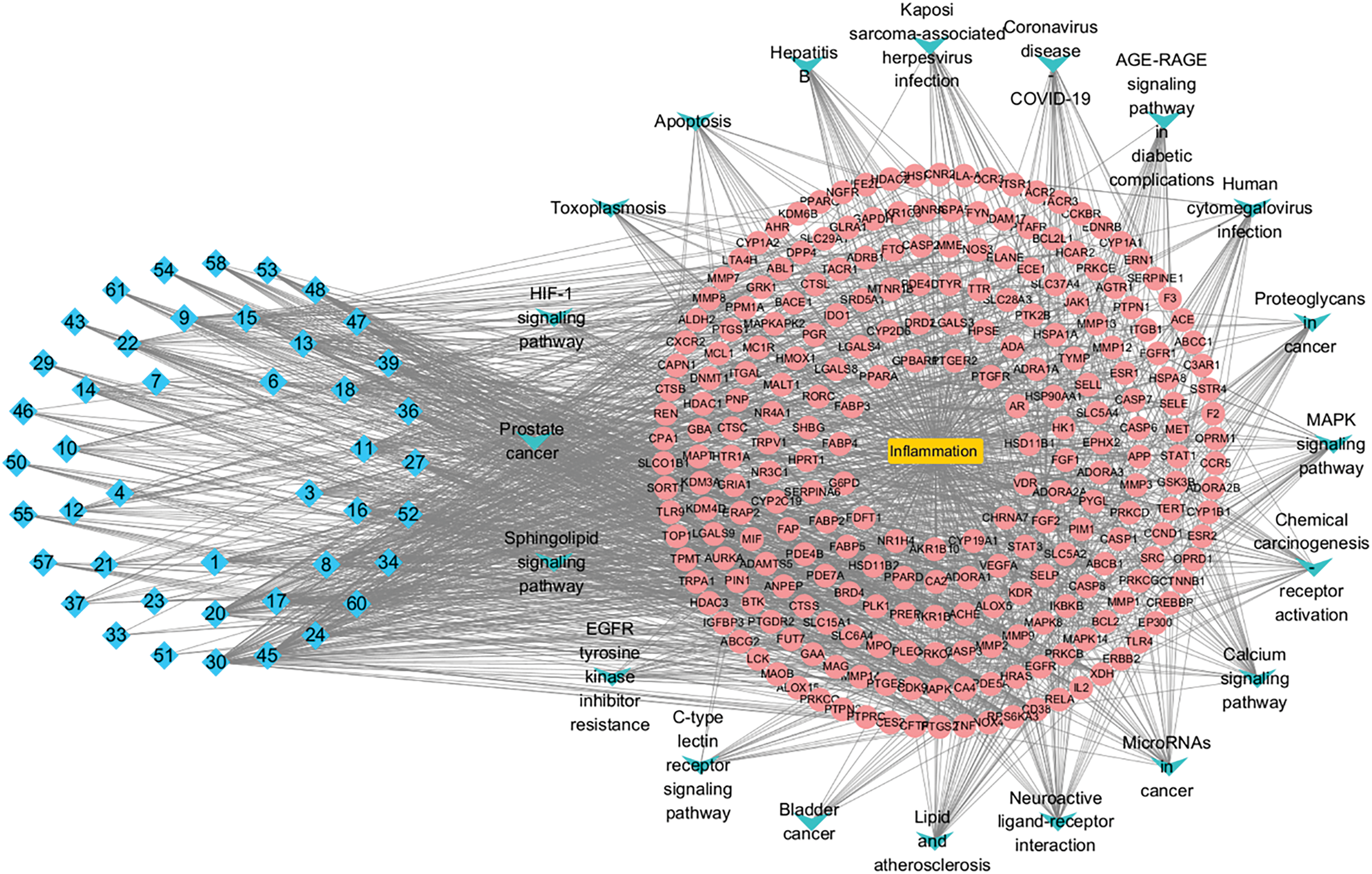
Figure 7: Ingredients-targets-pathways-disease network diagram, illustrating the relationships between the active ingredients of L. ruthenicum, their target proteins, relevant pathways, and associated diseases
As one of the most prominent compounds in L. ruthenicum, anthocyanins are widely recognized for their anti-inflammatory properties. These metabolites modulate the MAPK and NF-κB signaling pathways, which are crucial in regulating pro-inflammatory cytokines such as TNF-α and IL-1β. Anthocyanins inhibit the activation of MAPKs, reducing the production of inflammatory cytokines and alleviating inflammation-related diseases. Additionally, phenolic acids, including caffeic acid and chlorogenic acid, were identified as significant contributors to the anti-inflammatory effects of L. ruthenicum. These compounds inhibit the expression of inflammatory mediators by suppressing the NF-κB signaling pathway, reducing the secretion of pro-inflammatory cytokines. Phenolic acids also possess antioxidant properties, further enhancing their anti-inflammatory activity by neutralizing reactive oxygen species that contribute to inflammation.
The inflammatory response is a complex process involving multiple genes and signaling pathways. By integrating the PPI network with the drug-active ingredient-target-disease network, we identified key genes—SRC, STAT3, MAPK1, HSP90AA1, EP300, CTNNB1, EGFR, and RELA—that exhibit high centrality in the network (degree ≥ 4 times the average value). These genes are hypothesized to be core targets mediating the anti-inflammatory effects of L. ruthenicum.
KEGG pathway enrichment analysis revealed that these core targets were primarily enriched in 20 signaling pathways, which can be categorized into three functional groups. The first group includes inflammation-related pathways such as neuroactive ligand-receptor interaction, calcium signaling, MAPK signaling, PI3K-Akt signaling, and Rap1 signaling pathways. The second group encompasses pathways related to infectious diseases, including human cytomegalovirus infection, Kaposi sarcoma-associated herpesvirus infection, COVID-19, and hepatitis B. The third group comprises cancer-associated pathways such as microRNAs in cancer, chemical carcinogenesis-receptor activation, proteoglycans in cancer, and chemical carcinogenesis-reactive oxygen species.
TNF-α and IL-1β are key regulators in various chronic inflammatory diseases. The activation of MAPKs triggers transcriptional cascades that promote the production of pro-inflammatory cytokines like TNF-α and IL-1β, which, in turn, activate a range of enzymes associated with the inflammatory response. Inhibiting MAPK kinases or blocking the overproduction of pro-inflammatory cytokines could mitigate chronic inflammatory diseases, as inflammation is closely linked to the development of numerous conditions. Based on these findings, we speculate that L. ruthenicum exerts its anti-inflammatory effects through the regulation of the MAPK signaling pathway.
The role of inflammation in cancer development has garnered significant attention in recent years. While inflammation generally serves as a protective response to injury or infection, prolonged or uncontrolled inflammation can increase the likelihood of genomic mutations and facilitate the overexpression of inflammatory mediators, creating an environment conducive to cancer development. Our analysis of the filtered signaling pathways indicates that cancer-related pathways are prominently involved in the pharmacological effects of L. ruthenicum. Although these pathways are not directly linked to inflammation regulation, they highlight the potential of L. ruthenicum in cancer therapy through its anti-inflammatory actions.
On one hand, these findings support the connection between inflammation and various diseases, suggesting that L. ruthenicum may be beneficial in the clinical treatment of cancer via its anti-inflammatory effects. On the other hand, our results also underscore the limitations of network pharmacology. While network pharmacology has become a powerful tool for studying the complex mechanisms of traditional Chinese medicine (TCM), it still has inherent limitations. Mathematical models and network simulations often fail to fully capture the intricacies of biological systems. Therefore, the reliability of data derived from network analyses requires further validation through pharmacological and pharmacodynamic experiments.
In this study, we demonstrated that UPLC-QTOF/MS coupled with multivariate analysis is an effective and innovative approach for evaluating the chemical composition and quality of L. ruthenicum from different sources. The comprehensive characterization revealed that L. ruthenicum is particularly rich in anthocyanins and phenolic acids, bioactive compounds with potential therapeutic effects. To the best of our knowledge, this is one of the first studies to apply network pharmacology to elucidate the underlying mechanisms responsible for the anti-inflammatory effects of L. ruthenicum, providing new insights into its molecular targets and signaling pathways. This integrated approach not only enhances our understanding of the multifaceted biological activities of L. ruthenicum, but also lays the groundwork for exploring its therapeutic potential.
The results suggest that L. ruthenicum holds promise for use in nutraceuticals, pharmaceuticals, and cosmetics. Furthermore, the use of multivariate analysis to study samples from various geographical regions represents a novel method that could be employed to optimize and standardize the production of bioactive metabolites, ensuring consistent quality and efficacy. Future studies should validate these findings through in vivo models and clinical trials to confirm the therapeutic potential and safety of L. ruthenicum for human health.
Acknowledgement: We thank Yun Shao and Huilan Yue (Chinese Academy of Sciences Key Laboratory of Tibetan Medicine Research, Northwest Institute of Plateau Biology, Xining, China) for sampling assistance.
Funding Statement: This work was supported by Qinghai Science and Technology Achievement Transformation Project (2021-SF-149), awarded to C. Chen; Innovation and Entrepreneurship Training Program for College Students (S202410373040), awarded to C. Chen; Anhui Provincial College Student Innovation and Entrepreneurship Project (20221502032), awarded to C. Chen.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Chen Chen; methodology, Chen Chen; software, Chunli Li; validation, Qianhong Li and Luyao Li; investigation, Fengqin Liu; resources, Chen Chen; data curation, Tengfei Li; writing—original draft preparation, Chunli Li; writing—review and editing, Chen Chen; visualization, Chen Chen; supervision, Fengqin Liu; funding acquisition, Chen Chen. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data available on request from the authors. The data that support the findings of this study are available from the corresponding author, Chen Chen, upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Chen C, Wen H, Zhao X, Tao Y, Shao Y, Mei L. Fast determination of phenolic acids in Lycium ruthenicum Murr juice by solid phase extraction and HPLC. China J Chin Mater Med. 2011;36(7):896–8. [Google Scholar]
2. Jiao XL, Chi XF, Dong Q, Xiao YC, Hu FZ. Analysis of the nutritional components of Lycium ruthenicum. Biotic Resour. 2011;33(3):60–2. (In Chinese). doi:10.3969/j.issn.1006-8376.2011.03.016. [Google Scholar] [CrossRef]
3. Qi JJ, Yan YM, Wang CX, Cheng YX. Compounds from Lycium ruthenicum. Nat Prod Res Dev. 2018;30:345–53. [Google Scholar]
4. Wu T, Lv H, Wang F, Wang Y. Characterization of polyphenols from Lycium ruthenicum fruit by UPLC-Q-TOF/MS(E) and their antioxidant activity in Caco-2 cells. J Agric Food Chem. 2016;64(11):2280–8. doi:10.1021/acs.jafc.6b00035. [Google Scholar] [PubMed] [CrossRef]
5. Zheng J, Ding C, Wang L, Li G, Shi J, Li H, et al. Anthocyanins composition and antioxidant activity of wild Lycium ruthenicum Murr. from Qinghai-Tibet Plateau. Food Chem. 2011;126(3):859–65. doi:10.1016/j.foodchem.2010.11.052. [Google Scholar] [CrossRef]
6. Wang Z, Yan Y, Nisar T, Zou L, Yang X, Niu P, et al. Comparison and multivariate statistical analysis of anthocyanin composition in Lycium ruthenicum Murray from different regions to trace geographical origins: the case of China. Food Chem. 2018;246(4):233–41. doi:10.1016/j.foodchem.2017.11.030. [Google Scholar] [PubMed] [CrossRef]
7. Jin H, Liu Y, Yang F, Wang J, Fu D, Zhang X, et al. Characterization of anthocyanins in wild Lycium ruthenicum Murray by HPLC-DAD/QTOF-MS/MS. Anal Methods. 2015;7(12):4947–56. doi:10.1039/c5ay00612k. [Google Scholar] [CrossRef]
8. Wu X, Li Y, Wang Q, Li W, Feng Y. Effects of berberine and pomegranate seed oil on plasma phospholipid metabolites associated with risks of type 2 diabetes mellitus by U-HPLC/Q-TOF-MS. J Chromatogr B Analyt Technol Biomed Life Sci. 2015;1007:110–20. doi:10.1016/j.jchromb.2015.11.008. [Google Scholar] [PubMed] [CrossRef]
9. Yossa Nzeuwa IB, Xia Y, Qiao Z, Feng F, Bian J, Liu W, et al. Comparison of the origin and phenolic contents of Lycium ruthenicum Murr. by high-performance liquid chromatography fingerprinting combined with quadrupole time-of-flight mass spectrometry and chemometrics. J Sep Sci. 2017;40(6):1234–43. doi:10.1002/jssc.201601147. [Google Scholar] [PubMed] [CrossRef]
10. Luo Z, Yu G, Chen X, Liu Y, Zhou Y, Wang G, et al. Integrated phytochemical analysis based on UHPLC-LTQ-Orbitrap and network pharmacology approaches to explore the potential mechanism of Lycium ruthenicum Murr. for ameliorating Alzheimer’s disease. Food Funct. 2020;11(2):1362–72. doi:10.1039/c9fo02840d. [Google Scholar] [PubMed] [CrossRef]
11. Chen C, Zhao XH, Wen HX, Tao YD, Shao Y, Mei LJ. Determination of antioxidant compositions in Lycium ruthenicum Murr and its oxygen radical absorbance capacity assay. Chin J Hosp Pharm. 2011;31:1305–6. [Google Scholar]
12. Chen S, Zhou H, Zhang G, Meng J, Deng K, Zhou W, et al. Anthocyanins from Lycium ruthenicum Murr. ameliorated d-galactose-induced memory impairment, oxidative stress, and neuroinflammation in adult rats. J Agric Food Chem. 2019;67(11):3140–9. doi:10.1021/acs.jafc.8b06402. [Google Scholar] [PubMed] [CrossRef]
13. Ni W, Gao T, Wang H, Du Y, Li J, Li C, et al. Anti-fatigue activity of polysaccharides from the fruits of four Tibetan Plateau indigenous medicinal plants. J Ethnopharmacol. 2013;150(2):529–35. doi:10.1016/j.jep.2013.08.055. [Google Scholar] [PubMed] [CrossRef]
14. Zhang G, Chen S, Zhou W, Meng J, Deng K, Zhou H, et al. Anthocyanin composition of fruit extracts from Lycium ruthenicum and their protective effect for gouty arthritis. Ind Crops Prod. 2019;129(4):414–23. doi:10.1016/j.indcrop.2018.12.026. [Google Scholar] [CrossRef]
15. Tian B, Zhao J, Zhang M, Chen Z, Ma Q, Liu H, et al. Lycium ruthenicum anthocyanins attenuate high-fat diet-induced colonic barrier dysfunction and inflammation in mice by modulating the gut microbiota. Mol Nutr Food Res. 2021;65(8):e2000745. doi:10.1002/mnfr.202000745. [Google Scholar] [PubMed] [CrossRef]
16. Peng Y, Yan Y, Wan P, Chen D, Ding Y, Ran L, et al. Gut microbiota modulation and anti-inflammatory properties of anthocyanins from the fruits of Lycium ruthenicum Murray in dextran sodium sulfate-induced colitis in mice. Free Radic Biol Med. 2019;136:96–108. doi:10.1016/j.freeradbiomed.2019.04.005. [Google Scholar] [PubMed] [CrossRef]
17. Zong S, Yang L, Park HJ, Li J. Dietary intake of Lycium ruthenicum Murray ethanol extract inhibits colonic inflammation in dextran sulfate sodium-induced murine experimental colitis. Food Funct. 2020;11(4):2924–37. doi:10.1039/d0fo00172d. [Google Scholar] [PubMed] [CrossRef]
18. Wang G, Luo WJ, Ye J, Yan HL. Anti-skin aging effects of proanthocyanidins in Lycium ruthenicum from Chaidamu by antioxidant and inflammatory factors in mice. Chin High-Alti Medi Bio. 2018;39:175–8. [Google Scholar]
19. Yan SP, Wang S, Chen T, Shen C. Effect of anthocyanin extract of Lycium ruthenicum Murr. on rats with acute gouty arthritis. J Food Sci Technol. 2022;40:105–11. [Google Scholar]
20. Wen CL, Liu CH, Sun WW, Wang Y. Effects of Lycium ruthenicum Murr. anthocyanin combined with zinc gluconate on histopathology and inflammatory factors of spinal cord in rat model with acute spinal cord injury. J Gansu Univer Chin Medi. 2019;36:1–6. [Google Scholar]
21. Wang LS, Zhang MD, Tao X, Zhou YF, Liu XM, Pan RL, et al. LC-MS/MS-based quantification of tryptophan metabolites and neurotransmitters in the serum and brain of mice. J Chromatogr B Analyt Technol Biomed Life Sci. 2019;1112:24–32. doi:10.1016/j.jchromb.2019.02.021. [Google Scholar] [PubMed] [CrossRef]
22. Zhou W, Shan J, Wang S, Ju W, Meng M, Cai B, et al. Simultaneous determination of caffeic acid derivatives by UPLC-MS/MS in rat plasma and its application in pharmacokinetic study after oral administration of Flos lonicerae-Fructus Forsythiae herb combination. J Chromatogr B. 2014;949(3):7–15. doi:10.1016/j.jchromb.2013.12.035. [Google Scholar] [PubMed] [CrossRef]
23. Kiselova-Kaneva Y, Galunska B, Nikolova M, Dincheva I, Badjakov I. High resolution LC-MS/MS characterization of polyphenolic composition and evaluation of antioxidant activity of Sambucus ebulus fruit tea traditionally used in Bulgaria as a functional food. Food Chem. 2022;367(1):130759. doi:10.1016/j.foodchem.2021.130759. [Google Scholar] [PubMed] [CrossRef]
24. Agrawal V, Baghel R, Singh AK, Pathak DP, Sandal N. Development and validation of LC-MS method for the estimation of N-acetyl-tryptophan and its impurities under stress conditions. Chromatographia. 2019;82(10):1467–77. doi:10.1007/s10337-019-03776-z. [Google Scholar] [CrossRef]
25. Ahad H, Jin H, Liu Y, Wang J, Sun G, Liang X, et al. Chemical profiling of spermidines in goji berry by strong cation exchange solid-phase extraction (SCX-SPE) combined with ultrahigh-performance liquid chromatography-quadrupole time-of-flight mass spectrometry (UPLC-Q-TOF/MS/MS). J Chromatogr B Analyt Technol Biomed Life Sci. 2020;1137:121923. doi:10.1016/j.jchromb.2019.121923. [Google Scholar] [PubMed] [CrossRef]
26. Zhou ZQ, Fan HX, He RR, Xiao J, Tsoi B, Lan KH, et al. Lycibarbarspermidines A-O, new dicaffeoylspermidine derivatives from wolfberry, with activities against Alzheimer’s disease and oxidation. J Agric Food Chem. 2016;64(11):2223–37. doi:10.1021/acs.jafc.5b05274. [Google Scholar] [PubMed] [CrossRef]
27. Agregán R, Munekata PES, Feng X, Astray G, Gullón B, Lorenzo JM. Recent advances in the extraction of polyphenols from eggplant and their application in foods. LWT. 2021;146(7):111381. doi:10.1016/j.lwt.2021.111381. [Google Scholar] [CrossRef]
28. Sójka M, Guyot S, Kołodziejczyk K, Król B, Baron A. Composition and properties of purified phenolics preparations obtained from an extract of industrial blackcurrant (Ribes nigrum L.) pomace. J Hortic Sci Biotechnol. 2009;84(6):100–6. doi:10.1080/14620316.2009.11512604. [Google Scholar] [CrossRef]
29. Seididamyeh M, Phan ADT, Sivakumar D, Netzel ME, Mereddy R, Sultanbawa Y. Valorisation of three underutilised native Australian plants: Phenolic and organic acid profiles and in vitro antimicrobial activity. Foods. 2023;12(3):623. doi:10.3390/foods12030623. [Google Scholar] [PubMed] [CrossRef]
30. Qi Y, Pi Z, Liu S, Song F, Lin N, Liu Z. A metabonomic study of adjuvant-induced arthritis in rats using ultra-performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry. Mol Biosyst. 2014;10(10):2617–25. doi:10.1039/c4mb00131a. [Google Scholar] [PubMed] [CrossRef]
31. He C, Han T, Tan L, Li X. Effects of dark septate endophytes on the performance and soil microbia of Lycium ruthenicum under drought stress. Front Plant Sci. 2022;13:898378. doi:10.3389/fpls.2022.898378. [Google Scholar] [PubMed] [CrossRef]
32. Li Y, Zhang T, Zhang Z, He K. The physiological and biochemical photosynthetic properties of Lycium ruthenicum Murr in response to salinity and drought. Sci Hortic. 2019;256:108530. doi:10.1016/j.scienta.2019.05.057. [Google Scholar] [CrossRef]
33. Liu Z, Dong B, Liu C, Zong Y, Shao Y, Liu B, et al. Variation of anthocyanin content in fruits of wild and cultivated Lycium ruthenicum. Ind Crops Prod. 2020;146(4):112208. doi:10.1016/j.indcrop.2020.112208. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF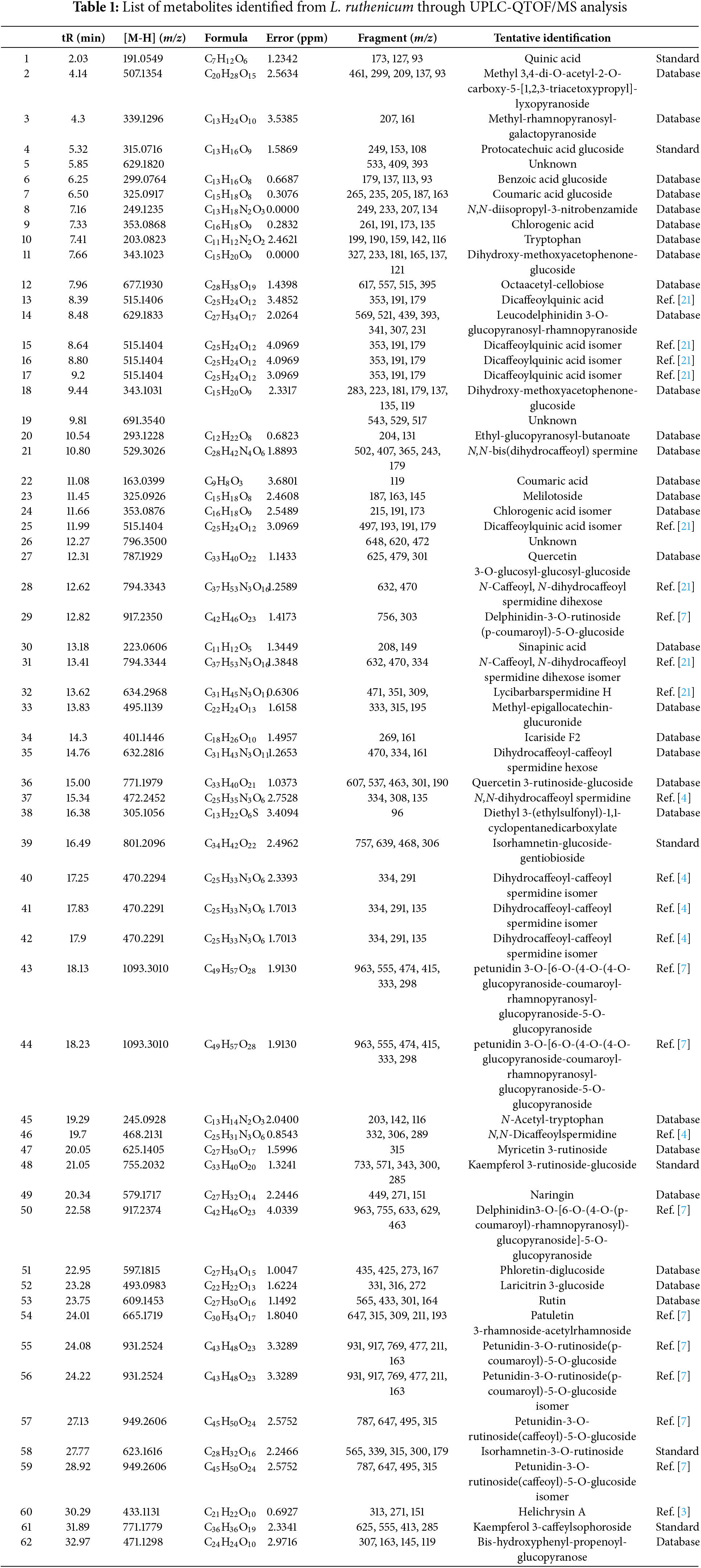
 Downloads
Downloads
 Citation Tools
Citation Tools