 Open Access
Open Access
ARTICLE
Metabolomic and Transcriptomic Insights into Enhanced Paclitaxel Biosynthesis in Cultivated Taxus cuspidata
School of Science, Liaodong University, Dandong, 118003, China
* Corresponding Author: Dandan Wang. Email:
(This article belongs to the Special Issue: Advances in Molecular Insights of Plant Secondary Metabolites: Biosynthesis, Regulation, and Applications)
Phyton-International Journal of Experimental Botany 2025, 94(4), 1137-1158. https://doi.org/10.32604/phyton.2025.063894
Received 27 January 2025; Accepted 18 March 2025; Issue published 30 April 2025
Abstract
Taxus cuspidata, a rare species of the Taxus genus, and its wild resources are under severe threat. The development of cultivated species has become an important strategy to replace wild species. The objective of this work was to elucidate the differences in secondary metabolite accumulation, particularly in the paclitaxel biosynthesis pathway, between wild and cultivated species. This study employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) and RNA sequencing (RNA-Seq) technologies to conduct integrated metabolomic and transcriptomic analyses of wild and cultivated species of T. cuspidata. The results showed that the content of paclitaxel in cultivated species was significantly higher than in wild species, reaching 1.67 times that of the latter (p < 0.01). Additionally, the content of key paclitaxel precursors, GGPP and 10-deacetylbaccatin III, in cultivated species was 1.94 times (p < 0.05) and 1.71 times (p < 0.01) higher than in wild species, respectively. Transcriptomic analysis identified 2606 differentially expressed genes (DEGs), among which key enzyme genes related to paclitaxel biosynthesis (such as DXS, DXR, GGPS, etc.) were generally upregulated in cultivated species. Multiple key enzyme genes in both the 2-C-methyl-D-erythritol 4-phosphate pathway (MEP) and paclitaxel biosynthesis pathways were significantly upregulated in cultivated species. Conversely, genes and metabolites related to sugar metabolism were found to be higher in content in wild species. These findings reveal the significant advantage of cultivated species in paclitaxel production capacity, providing new insights into the metabolic regulation mechanisms during yew domestication. This has important implications for optimizing paclitaxel biosynthesis and guiding future improvements in T. cuspidata cultivars.Keywords
Supplementary Material
Supplementary Material FileTaxus cuspidata Sieb. et Zucc. is a well-known gymnosperm widely distributed throughout northeastern China, famous for containing paclitaxel, a functional bioactive compound with anticancer properties [1–3]. Additionally, this species is also rich in various active compounds, including terpenes, flavonoids, phenolics, alkaloids, and lipids, which demonstrate remarkable efficacy in enhancing both cardiovascular and immune system functions [4]. As a high-value tree species, although Northeast Yew (T. cuspidata) has been continuously cultivated for centuries, the conservation status of its wild populations remains critical. On the one hand, the growing market demand for paclitaxel and its high commercial value have led to severe destruction of wild resources [5,6]. On the other hand, the poor survival rate of wild seedlings substantially impeded the natural replenishment of the populations. In response to the growing depletion of wild resources, the development of cultivated species has emerged as a key solution to ensure sustainable utilization. However, the growth of cultivated species in artificial environments exhibits a dual nature: on one hand, the stable and controllable environment has optimized certain growth characteristics through artificial selection, such as faster growth rates and morphological features that are more oriented towards ornamental value; on the other hand, the lack of complex and variable survival pressures found in wild environments has resulted in substantial differences between cultivated and wild populations in their morphological characteristics, growth rates, stress tolerance, and secondary metabolite profiles [7,8]. Studies have revealed significant differences between cultivated and wild populations in various aspects, including morphological characteristics [9], growth performance [10], metabolite biosynthesis [11], and genetic diversity [12]. These findings provide valuable insights into their adaptation mechanisms and resource utilization strategies. However, specific differences in the biosynthetic pathway of paclitaxel, a key secondary metabolite, remain largely unexplored.
Paclitaxel, a unique tetracyclic diterpenoid compound extraction from the yew trees, possesses a distinctive 6-8-6-4 tetracyclic skeleton with 11 chiral centers in its core structure and side chain. As one of the most excellent natural antitumor drugs in the pharmaceutical market, its content in yew trees is extremely low (<0.01%) [13]. It is estimated that 2 g of paclitaxel required for one treatment cycle necessitates the felling of 4 century-old wild yew trees, Due to factors such as its complex structure, scarcity of wild resources, and slow growth of yew trees, traditional plant extraction and chemical synthesis methods cannot meet the growing market demand [14]. Currently, significant progress has been made in the research of paclitaxel biosynthesis in Taxus species. The chemical synthesis of paclitaxel was first achieved in 1994 through total synthesis independently by the Holton group [15], but the synthetic routes were complex with extremely low overall yields (<0.1%). Although optimized by teams like Nicolaou’s [16], the efficiency remains low. Present commercial production primarily relies on the semisynthetic route using 10-deacetylbaccatin III (10-DAB) as a precursor. However, this method is limited by the long growth cycle of yew trees and the bottleneck that 10-DAB content only accounts for 0.1% of dry weight. In response to these challenges, researchers have pioneered multiple innovative approaches: First, heterologous biosynthesis of paclitaxel, including production through endophytic fungal fermentation [17], and de novo biosynthesis of key paclitaxel precursors using heterologous chassis organisms such as Escherichia coli [18], Saccharomyces cerevisiae [19], and tobacco plants [20]. Second, progress in yew cell engineering technology, such as discovering that exogenous elicitors like methyl jasmonate (MeJA) promote paclitaxel accumulation in cultured yew cells [21]. Third, detailed elucidation of key enzyme genes in the paclitaxel biosynthetic pathway, including taxadiene synthase (TS) which catalyzes the first step of biosynthesis, hydroxylases (T2αH, T5αH, T13αH, and T10βH) [22], involved in skeleton modification, and transferases (TAT, TBT, DBAT, and DBTNBT) that catalyze late-stage modifications [23]. Fourth, in transcriptional regulation mechanism studies, various transcription factors including MYB, ERF, WRKY, MYC, NAC, and bHLH have been identified for their regulatory roles in paclitaxel biosynthesis [24]. Based on studies of paclitaxel biosynthetic mechanisms, significant breakthroughs have also been achieved in Taxus metabolism research. Tanaka et al. [25] identified and isolated taxane compounds from 1–5 year old Taxus chinensis seedlings using high-resolution multi-stage mass spectrometry (LC-IT-TOF-MS). Yang et al. [26] discovered significant differences in carbohydrate and flavonoid metabolism between wild-type and cultivated T. chinensis. Shao et al. [27] found significant metabolic profile differences between the heartwood and sapwood of T. chinensis. Zhou et al. [28] used metabolomics to reveal that T. cuspidata and Taxus media have more efficient paclitaxel biosynthetic pathways compared to T. chinensis. They identified numerous metabolites associated with paclitaxel biosynthesis and discovered a potential negative correlation between flavonoid metabolism and taxane accumulation. Their study also showed significant variations in the accumulation of bioactive compounds, including paclitaxel, among different Taxus species. Chen et al. [29] compared metabolites in needles of wild-type and cultivated T. chinensis grown in the same habitat. Through widely targeted metabolomics analysis, they identified 71 metabolites with significant differences in concentration, involving both secondary metabolism (flavonoids) and primary metabolism (carbohydrate metabolism and lipid synthesis). Yu et al. [30] conducted an in-depth investigation of the specific accumulation patterns and biosynthetic pathways of taxane compounds in the phloem of Taxus stems. Yu et al. [31] performed comparative metabolomic analyses that revealed differential accumulation patterns of taxoids, flavonoids, and hormones among six Taxus species. Yan et al. [32] demonstrated the dynamic changes in metabolites during different developmental stages (red, yellow, and purple) of Taxus mairei fruits. Zhan et al. [33] used timsTOF flex MALDI-2 analysis to reveal the spatial distribution patterns of bioactive flavonoids, specifically forsythiaside and morin, in the leaves of T. cuspidata. In summary, paclitaxel biosynthesis involves a complex metabolic network requiring coordinated action of multiple key enzymes and transcription factors. Significant metabolic variations exist among different Taxus species and tissues, which may be closely related to the accumulation of secondary metabolites. However, the regulatory mechanisms underlying the differences between wild-type and cultivated species in paclitaxel biosynthesis remain poorly understood. Understanding these differential mechanisms could provide new breakthroughs for enhancing paclitaxel content in cultivated species, offering theoretical basis and genetic resources for directed breeding of high-yield cultivars. This ultimately aims to achieve sustainable development by replacing wild resources with cultivated varieties.
Currently, omics research on wild and cultivated Northeast yew remains relatively scarce. The unique environmental stresses experienced by different ecotypes of Northeastern yew are likely the key factors driving the diversity and abundance changes of their secondary metabolites. However, due to the lack of systematic multi-omics integration analysis, the mechanisms by which environmental stresses influence secondary metabolite accumulation in wild and cultivated species, as well as how these metabolic changes affect their medicinal value and ecological functions, remain unclear. We hypothesize that wild and cultivated species have developed distinct secondary metabolite accumulation patterns, particularly in paclitaxel biosynthesis, through long-term habitat adaptation, likely due to gene regulatory network restructuring. Using samples from Liaoning and Jilin provinces, systematic analysis of differences in paclitaxel biosynthetic networks between wild and cultivated species through transcriptomics and metabolomics approaches, screening and validation of key functional genes this study aims to: (1) identify differential metabolite accumulation patterns, (2) analyze biosynthetic pathways and transcriptional networks, and (3) explore environment-metabolite correlations. Through integrated metabolomics and transcriptomics analysis, we will investigate paclitaxel accumulation mechanisms between wild and cultivated species. This research will enhance our understanding of paclitaxel biosynthesis under different conditions, guide efficient production methods, and support the development of high-yield cultivars for sustainable commercial production.
2.1 Plant Materials and Collection Sites
Due to the scarcity and scattered geographical distribution of wild T. cuspidata resources, this study employed regional mixed sampling analysis. This method not only better reflects regional overall characteristics and reduces the impact of individual differences but also enables obtaining statistically significant and reliable data under limited resource conditions. In May and September 2024, when the paclitaxel content is relatively high [34,35], wild (W) and cultivated (C) samples of T. cuspidata were collected from Northeast China, including two wild samples from Dandong (Liaoning) and Wangqing (Jilin), respectively (Table S1). Three cultivated samples were collected from large T. cuspidata plantations in Liaoning and Jilin, including various genetic variants such as T. cuspidata var. nana, hybrids between T. cuspidata and T. cuspidata var. nana, and golden yew. All selected plants were 10-year-old females with comparable DBH (diameter at breast height) classes and growth characteristics, identified by Professor Zhang Yanwen from Liaodong University. To maintain genetic diversity, samples were collected from individuals at least 100 m apart. Fresh young leaves were flash-frozen in liquid nitrogen and stored at −80°C, and submitted to Shanghai OE Biotech Co., Ltd. (Shanghai, China) for transcriptomics (3 replicates) and metabolomics (6 replicates) analyses.
2.2 Metabolome and Transcriptome Analysis
Leaf tissue (60 mg) from each sample was ground in liquid nitrogen, homogenized with methanol/water (7:3) extraction solution, and centrifuged. The supernatant was diluted, vortexed, and re-centrifuged before LC-MS/MS analysis. Finally, the samples were transferred to LC vials for LC-MS/MS system analysis [35,36]. The LC-MS/MS was performed using a Waters ACQUITY UPLC I-Class system coupled with a Thermo QE high-resolution mass spectrometer at Shanghai OE Biotech Co., Ltd. Mobile phases were 0.1% formic acid in water (A) and 0.35% formic acid in acetonitrile (B), with 0.35 mL/min flow rate. An ACQUITY UPLC HSS T3 column (100 mm × 2.1 mm, 1.8 μm) was maintained at 40°C, with autosampler at 4°C and 3 μL injection volume. Data processing was performed using Progenesis QI (v3.0) software, and metabolite identification was conducted using the HMDB database, Lipidmaps (v2.3), METLIN, and LuMet-Plant3.0 databases. Metabolites were functionally annotated using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. Multivariate statistical analyses were conducted using SIMCA software (version 16.0.2), including principal component analysis (PCA), partial least squares discriminant analysis (PLS-DA), and orthogonal partial least squares discriminant analysis (OPLS-DA). Differentially abundant metabolites were identified using the following criteria: VIP > 1, p < 0.05, and fold change (FC) ≥8.0.
Total RNA was extracted from wild and cultivated species using RNAprep Pure Plant Plus Kit (TIANGEN, China). mRNA was isolated using Oligo (dT) for cDNA library construction, which was quality-checked by Agilent 2100 Bioanalyzer before sequencing on Illumina Novaseq 6000 platform to generate 150 bp paired-end reads. Clean reads obtained through fastp (https://github.com/OpenGene/fastp) (accessed on 17 March 2025).were aligned to the T. chinensis reference genome (GCA_019776745.2) using HISAT2 software (http://ccb.jhu.edu/software/hisat2/index.shtml) (accessed on 17 March 2025). Transcripts were assembled using Stringtie (https://ccb.jhu.edu/software/stringtie/index.shtml) (accessed on 17 March 2025), and FPKM (Fragments Per Kilobase of transcript per Million mapped reads) values were calculated for each gene. HTSeq-count was used for read counting, and RSeQC software was employed for transcriptome data quality evaluation [37]. Gene expression levels were quantified using TPM (Transcripts Per Million), and differential expression analysis was performed using DESeq2 software (version 1.20.0) [38] with thresholds of p < 0.05, q-value < 0.05, and FC ≥ 1.5. GO and KEGG enrichment analyses of DEGs were conducted using EggNOG Mapper and clusterProfiler R package [37].
2.3 Validation of Significant DEGs by qRT-PCR
To verify RNA-Seq data reliability, 10 genes involved in paclitaxel biosynthesis were selected for real-time quantitative PCR (qRT-PCR) analysis. cDNA was synthesized from RNA samples using the TaKaRa PrimeScript™ RT reagent kit with gDNA Eraser. qRT-PCR was performed on the Light Cycler®® 480 system. Primers were designed using Primer3Plus (https://dev.primer3plus.com) (accessed on 17 March 2025) (Table S2). 18S rRNA served as the internal reference gene [39]. The relative expression levels were analyzed using the comparative CT method (2−ΔΔCT method), with results averaged from three biological replicates.
Statistical analyses were performed using SPSS software (version 19.0). Differences in metabolite contents between wild and cultivated species were analyzed using one-way analysis of variance (ANOVA). In the integrated analysis, Pearson correlation analysis was used to explore the relationship between transcriptome and metabolome data. A gene-metabolite association network was constructed using Cytoscape to identify key genes and metabolites associated with paclitaxel biosynthesis. p-values were adjusted for multiple comparisons using the Benjamini-Hochberg procedure to control the false discovery rate. Supervised PLS-DA using metaX distinguished variables between wild and cultivated species.
3.1 Metabolome Analysis of Wild and Cultivated Species of T. cuspidata
LC-MS/MS analysis identified 7030 metabolites in young leaves of wild and cultivated samples, of which 2381 differential metabolites isolated after screening. The metabolite identification was performed using a combination of approaches: (1) comparison with authentic standards for retention time and MS/MS fragmentation patterns; (2) matching against public MS/MS spectral databases including METLIN, MassBank, and HMDB; (3) for compounds without available standards, structural annotation was based on accurate mass measurements, MS/MS fragmentation patterns, and comparison with literature data. The identification confidence levels were assigned according to the Metabolomics Standards Initiative (MSI). Fig. S1 presents the results of PCA and OPLS-DA analyses of metabolites from wild and cultivated T. cuspidata. The cumulative explanation rates for PCA and OPLS-DA are 79.1% and 73.5%, respectively. Both methods show clear separation between wild and cultivated samples, with OPLS-DA providing more distinct differentiation, indicating the growth environment significantly influences metabolite accumulation in T. cuspidata. The OPLS-DA three-dimensional scatter plot further analyzes metabolites in wild and cultivated T. cuspidata from sampling sites including Dandong and Kuandian in Liaoning Province, and Wangqing Nature Reserve in Jilin Province. The cumulative explanation rate of the three principal components is 88.5%, demonstrating the model’s strong explanatory power. Samples from different geographical origins form distinct clusters in the three-dimensional space, highlighting the significant impact of geographical origin on metabolite accumulation. This distribution pattern offers valuable insights for optimizing paclitaxel production and conserving T. cuspidata germplasm resources. Primary metabolites mainly include lipids and lipid-like molecules (38.69%), organic acids and derivatives (13.54%), and nucleosides, nucleotides, and analogues (1.34%), among others. Secondary metabolites, encompass phenylpropanoids and polyketides (13.64%), organoheterocyclic compounds (12.29%), organic oxygen compounds (10.16%), benzenoids (6.97%), organic nitrogen compounds (0.95%), and lignans, neolignans, and related compounds (0.85%) (Fig. 1A). These substances are typically associated with specific biological functions or adaptation to the environment. Lipids and lipid-like molecules have the highest proportion among primary metabolites. Volcano plot analysis of the C/W comparison identified 949 significantly upregulated and 1432 downregulated DAMs (Fig. 1B). The 50 most significant metabolites, screened based on minimum p-values, were visualized in a heatmap. Key intermediates of the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway (D-Glyceraldehyde 3-phosphate, Dimethylallyl diphosphate (DMAPP), Isopentenyl diphosphate (IPP), and Geranylgeranyl Diphosphate (GGPP)) and the paclitaxel biosynthesis pathway (Baccatin III, 10-DAB, and Paclitaxel) were clearly annotated. The heatmap also highlighted 6α-Hydroxypaclitaxel, the primary paclitaxel metabolite catalyzed by cytochrome P450-related genes. All these metabolites showed consistently higher levels in cultivated species compared to wild species, suggesting that cultivation conditions significantly enhanced their accumulation (Fig. 1C).
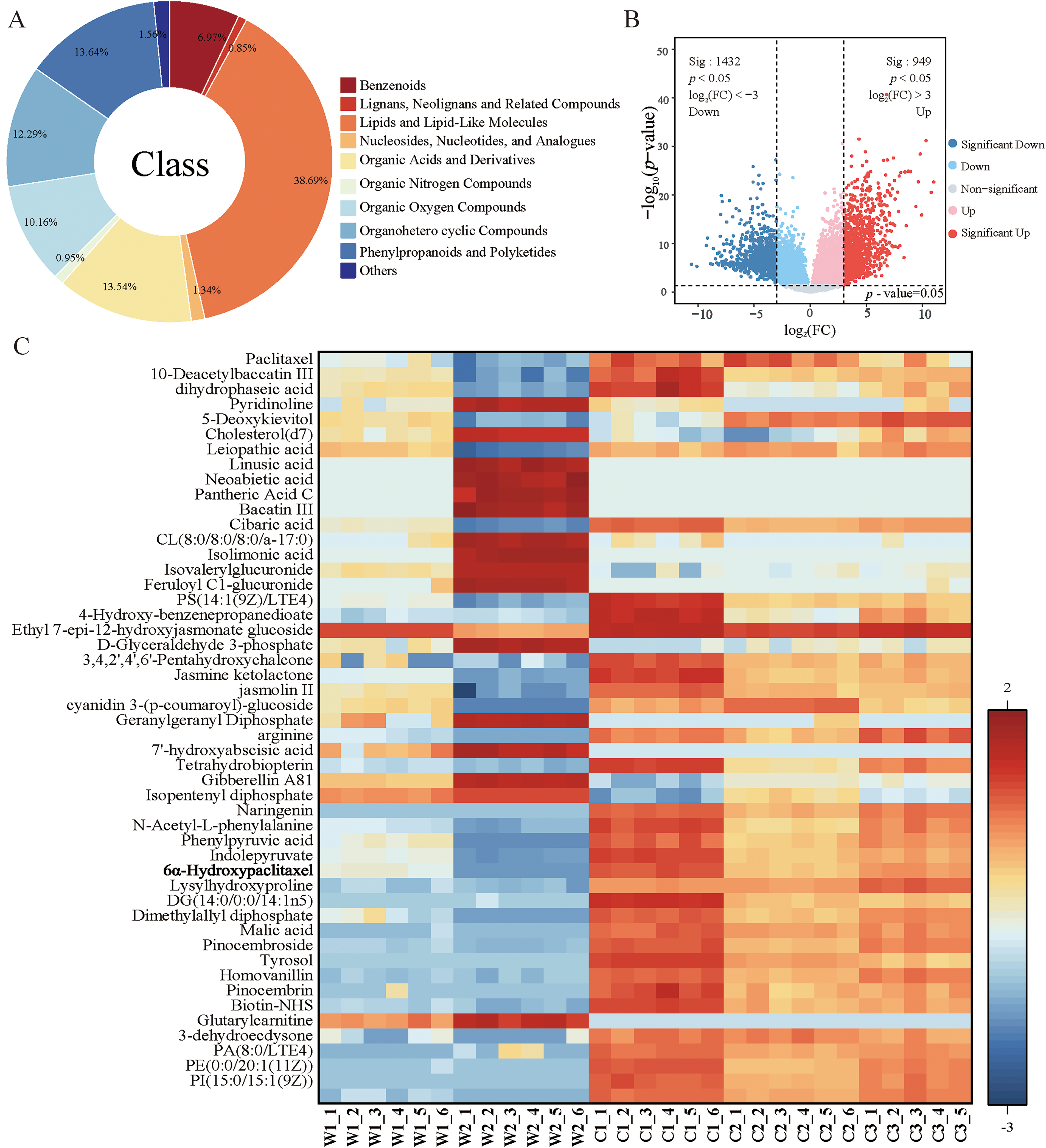
Figure 1: LC-MS/MS analysis of metabolites in wild and cultivated T. cuspidata. (A) Metabolite types and proportions. (B) Volcano plot of differential metabolites, where red dots indicate significantly upregulated metabolites, blue dots indicate significantly downregulated metabolites, and gray dots indicate non-significant metabolites. (C) Heatmap of top 50 metabolites (N = 6, Z-score normalized)
KEGG pathway enrichment analysis revealed that differentially abundant metabolites (DAMs) were significantly enriched in pathways related to primary metabolism, secondary metabolite biosynthesis, and signal transduction (Fig. 2). In the C/W comparison, significant enrichment was observed in isoflavonoid and flavonoid biosynthesis, tryptophan, phenylalanine, cholesterol, glutathione, and tyrosine metabolism, and diterpenoid biosynthesis, etc. (Fig. 2A), which showed an upregulation trend in cultivated species.
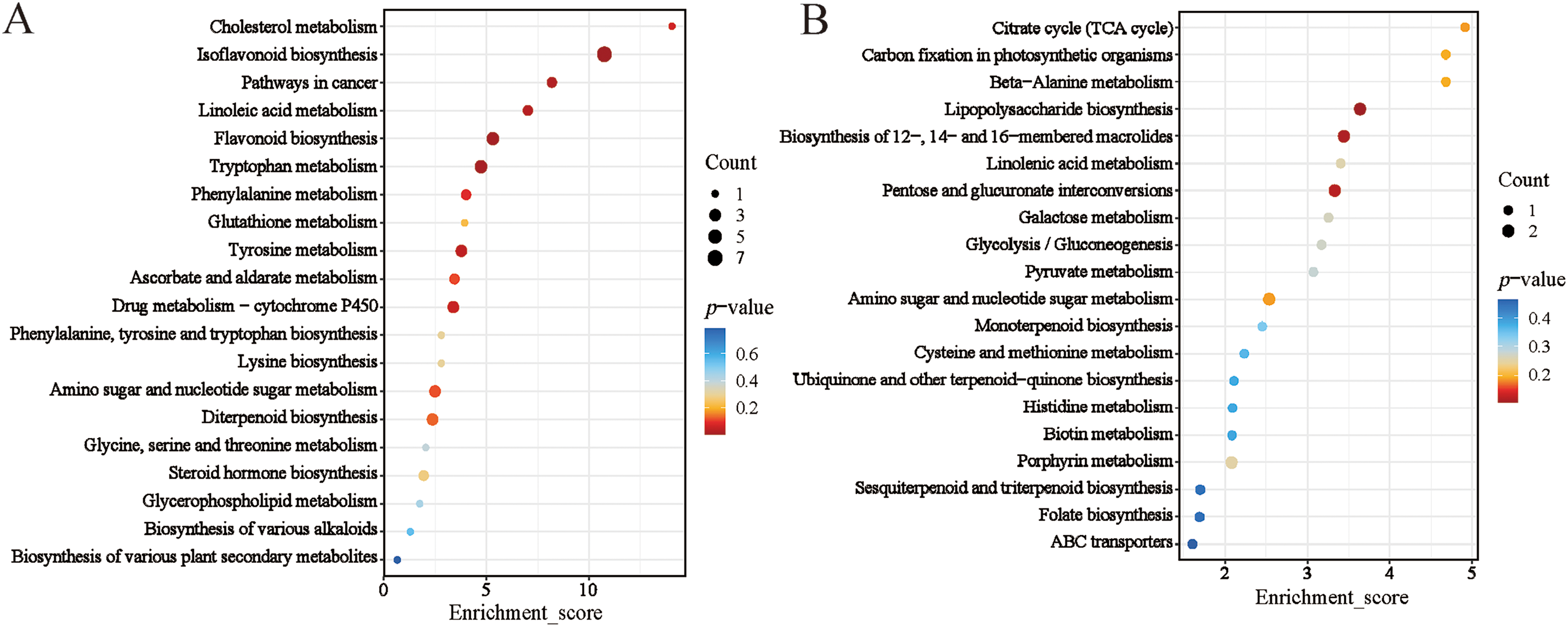
Figure 2: KEGG enrichment analysis of DEGs between wild and cultivated T. cuspidata. (A) Upregulated and (B) Downregulated metabolic pathways in C/W comparison. Bubble size indicates number of differential metabolites; color gradient from blue to red represents decreasing p-value significance. X-axis: Enrichment Score; Y-axis: top pathways
Paclitaxel biosynthesis involves core pathways, such as terpene biosynthesis and phenylalanine metabolism, which directly provide precursors, and auxiliary pathways, like glutathione and tryptophan metabolism, which indirectly support production by regulating the cellular environment and secondary metabolic networks. Terpene biosynthesis is the central metabolic pathway for paclitaxel synthesis, constructing its skeleton from IPP and DMAPP via the MEP pathway and the mevalonate (MVA) pathways. Upregulation of terpene metabolic pathways directly impacts paclitaxel yield. Phenylalanine is one of the key precursors in the biosynthesis of paclitaxel. Phenylalanine, a key precursor, is catalyzed by PAL into cinnamic acid, which converts to p-coumaric acid. This intermediate integrates into phenylpropanoid and terpene pathways, forming critical paclitaxel skeleton compounds. Tyrosine is converted into phenylalanine by transaminase, replenishing the phenylalanine metabolic pool. Tyrosine metabolism products (such as p-hydroxyphenylpyruvic acid) can serve as aromatic precursors in paclitaxel biosynthesis. Through genetic engineering strategies (e.g., overexpression of the PAL gene or optimization of the tyrosine metabolic pathway), the metabolic flux of phenylalanine and tyrosine can be increased, thereby enhancing the production of paclitaxel. During the biosynthesis of paclitaxel, a large amount of reactive oxygen species (ROS) is generated. These ROS are scavenged by glutathione peroxidase (GPX) and glutathione reductase (GR), providing a stable metabolic environment for paclitaxel synthesis. GR utilizes NADPH to reduce oxidized glutathione (GSSG) back to its reduced form (GSH), which not only enhances antioxidant capacity but also supplies the reducing power required for paclitaxel biosynthesis. Additionally, GSH protects key enzymes (such as taxadiene synthase) from oxidative damage, maintains cellular homeostasis, regulates signal transduction, activates gene expression, and promotes paclitaxel accumulation. Studies have shown that exogenous addition of GSH or its precursors (such as cysteine) can significantly increase paclitaxel production. Tryptophan metabolism plays a critical role in regulating signal transduction and stress responses through the synthesis of indole compounds and plant hormones (such as auxin), indirectly promoting the biosynthesis of paclitaxel. As a precursor of indole compounds, tryptophan activates secondary metabolic pathways (such as paclitaxel biosynthesis) to cope with environmental stress. Meanwhile, auxins (such as indole-3-acetic acid, IAA) derived from tryptophan metabolism influence the expression of paclitaxel biosynthesis genes by regulating cell division, differentiation, and gene expression. Conversely, the lipopolysaccharide biosynthesis, pentose and glucuronate interconversions, photosynthetic carbon fixation, TCA cycle, beta-alanine metabolism, linolenic acid metabolism, galactose metabolism, and glycolysis/gluconeogenesis (Fig. 2B), all of which are downregulated in cultivated species. It can be seen that the differences between cultivated and wild species are not only reflected in their paclitaxel biosynthesis capacity but may also involve deficiencies in energy metabolism, photosynthesis, and intracellular and extracellular material transport. This finding provides a metabolic basis for explaining the differences in cold tolerance, growth rate, and pigment characteristics between cultivated and wild species.
3.2 Transcriptome Sequencing Analysis of Wild and Cultivated Species of T. cuspidata
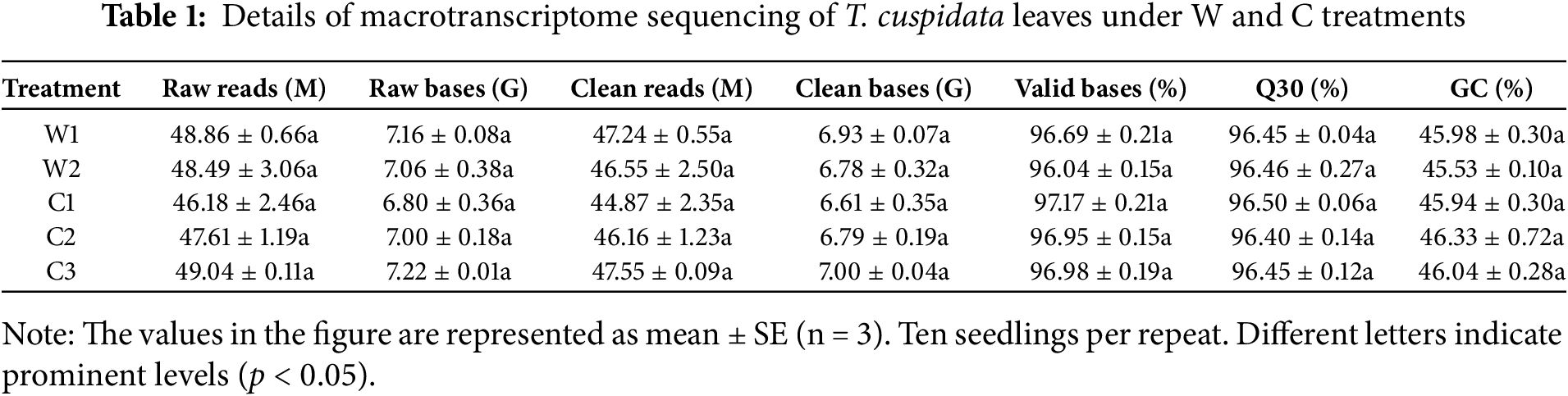
To further investigate gene expression profiles of wild and cultivated species of T. cuspidata, 18 cDNA libraries were constructed for reference transcriptome sequencing, yielding 122.57 Gb of raw reads. After quality control, 121.02 Gb of high-quality clean reads were obtained, with Q30 values ranging from 96.21% to 96.74% and an average GC content of 46.16%, Over 90% of clean reads mapped to the yew reference genome, with biological replicate correlation coefficients exceeding 0.9 (Table 1). The PCA plot clearly showed significant differentiation between cultivated and wild species at the transcriptome level. The PC1 axis (explaining 9.71% of the variance) indicated distinct differences in their primary gene expression patterns, with cultivated species clustering on the right and wild species leaning toward the left. The PC2 axis (explaining 23.3% of the variance) further revealed secondary differences among samples, supporting the significant distinction in gene expression between the two groups. This divergence might have resulted from the long-term artificial selection of cultivated species, bred to adapt to specific agronomic traits, while wild species had to cope with variable and harsh natural environments. Additionally, cultivated species grew in controlled agricultural settings with lower environmental stress, whereas wild species competed for resources in natural habitats, potentially influencing their metabolic pathways and gene expression patterns (Fig. S2A). RNA-seq analysis revealed 2606 differentially expressed genes (DEGs) between wild and cultivated samples, with 1413 genes upregulated and 1193 genes downregulated in the C/W comparison (Fig. S2B).
Gene Ontology (GO) functional enrichment analysis identified 1672 DEGs categorized into biological process (48.2%), molecular function (37.53%), and cellular component (14.27%) in the C/W comparison (Fig. 3). Biological processes were associated with cellular processes, metabolic regulation, response to stimuli, signaling, and immunity. Notably, 12 GO terms were linked to paclitaxel biosynthesis, including paclitaxel biosynthetic process, taxane synthesis, isoprenoid biosynthesis, and terpene synthase activity, with cultivated species showing higher gene expression levels in flavonoid biosynthesis. Conversely, GO analysis directly related to carbohydrate metabolism, including glycolysis, starch biosynthesis, glucose metabolism, and glucose-6-phosphate 1-epimerase activity, showed higher expression in wild species, corroborating metabolomics data (Fig. 3).
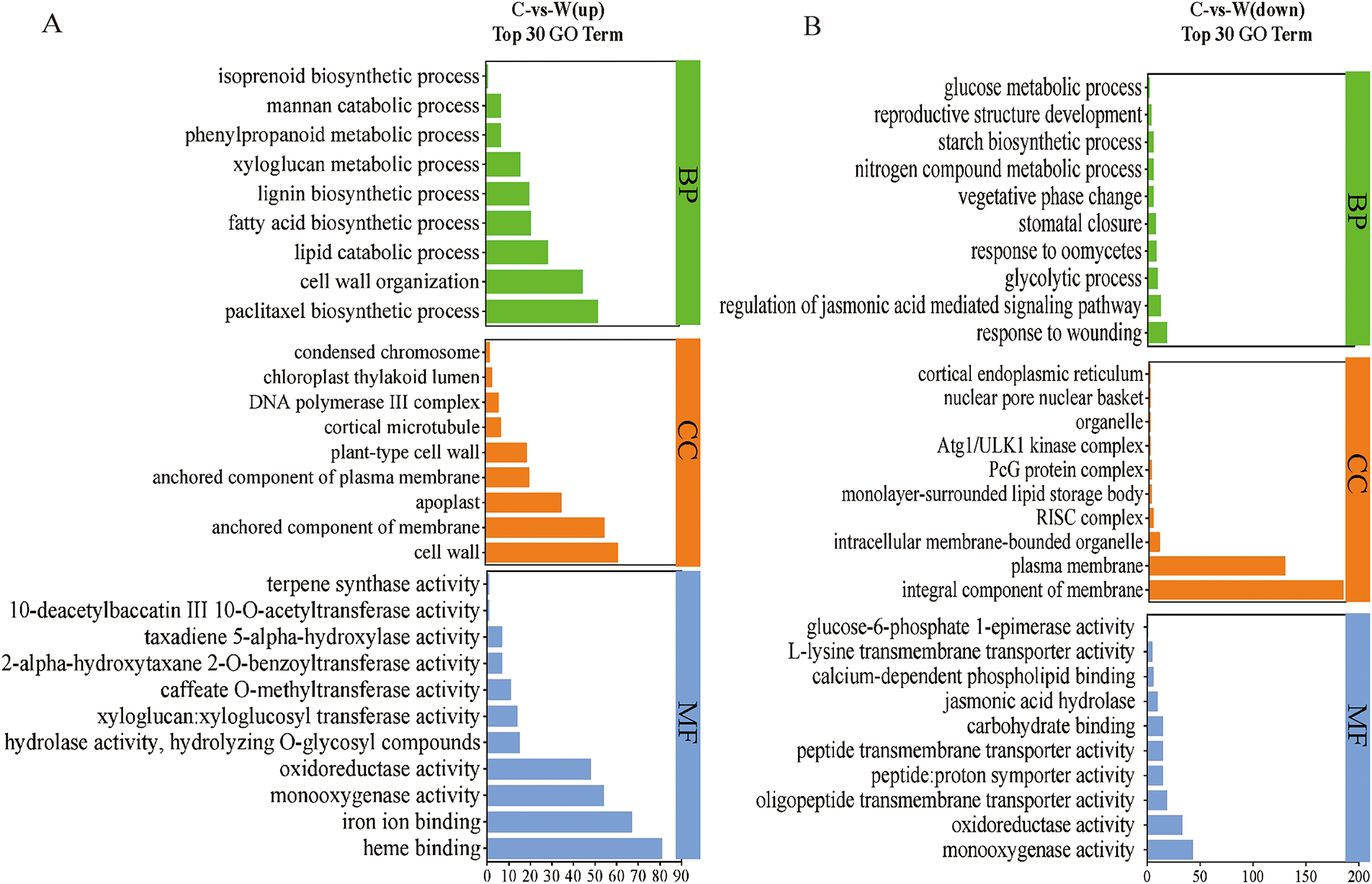
Figure 3: GO enrichment analysis of top 30 DEGs. (A) Upregulated and (B) downregulated pathways
KEGG enrichment analysis results showed that in the C/W comparison, the upregulated DEGs were significantly enriched in five metabolic pathways associated with paclitaxel biosynthesis, including phenylpropanoid biosynthesis, diterpenoid biosynthesis, terpenoid backbone biosynthesis, phenylalanine metabolism, and steroid biosynthesis (Fig. S3). Conversely, the downregulated DEGs were significantly enriched in five major carbohydrate metabolism-related pathways: glycolysis/gluconeogenesis, amino sugar and nucleotide sugar metabolism, starch and sucrose metabolism, pyruvate metabolism, and carbon fixation in photosynthetic organisms. Notably, wild species exhibited significantly higher expression levels of DEGs associated with plant hormone signal transduction and plant-pathogen interaction pathways compared to cultivated species, suggesting potentially stronger adaptability and resistance mechanisms in response to environmental stimuli and pathogen defense in wild species.
3.3 The Impact of the MEP Pathway on Paclitaxel Biosynthesis in Wild and Cultivated Species of T. cuspidata
The MEP pathway, which is fundamental for plant isoprenoid biosynthesis, synthesizes GGPP, an essential C20 precursor that serves as the primary building block for paclitaxel biosynthesis (Fig. 4A). In the MEP pathway, we identified multiple genes encoding key enzymes, including one DXS, two DXRs, two MCTs, two GGPSs, and one GGPPS. Expression analysis revealed that these genes generally exhibited significantly up-regulated expression patterns in cultivated species (Fig. 4B), suggesting that cultivated species might possess higher MEP pathway activity. Metabolomics analysis showed that the key intermediate metabolite of the MEP pathway, GGPP, accumulated substantially in cultivated species, reaching approximately 1.94 times higher than those in wild species (Fig. 4C). Enhanced MEP pathway activity in cultivated species increases isoprenoid precursor (IPP and DMAPP) synthesis, providing more substrates for paclitaxel biosynthesis, likely contributing to their higher paclitaxel yield. Consequently, this elevated MEP pathway activity appears to remodel the metabolic network, channeling more carbon flux toward paclitaxel biosynthesis and enhancing its production.
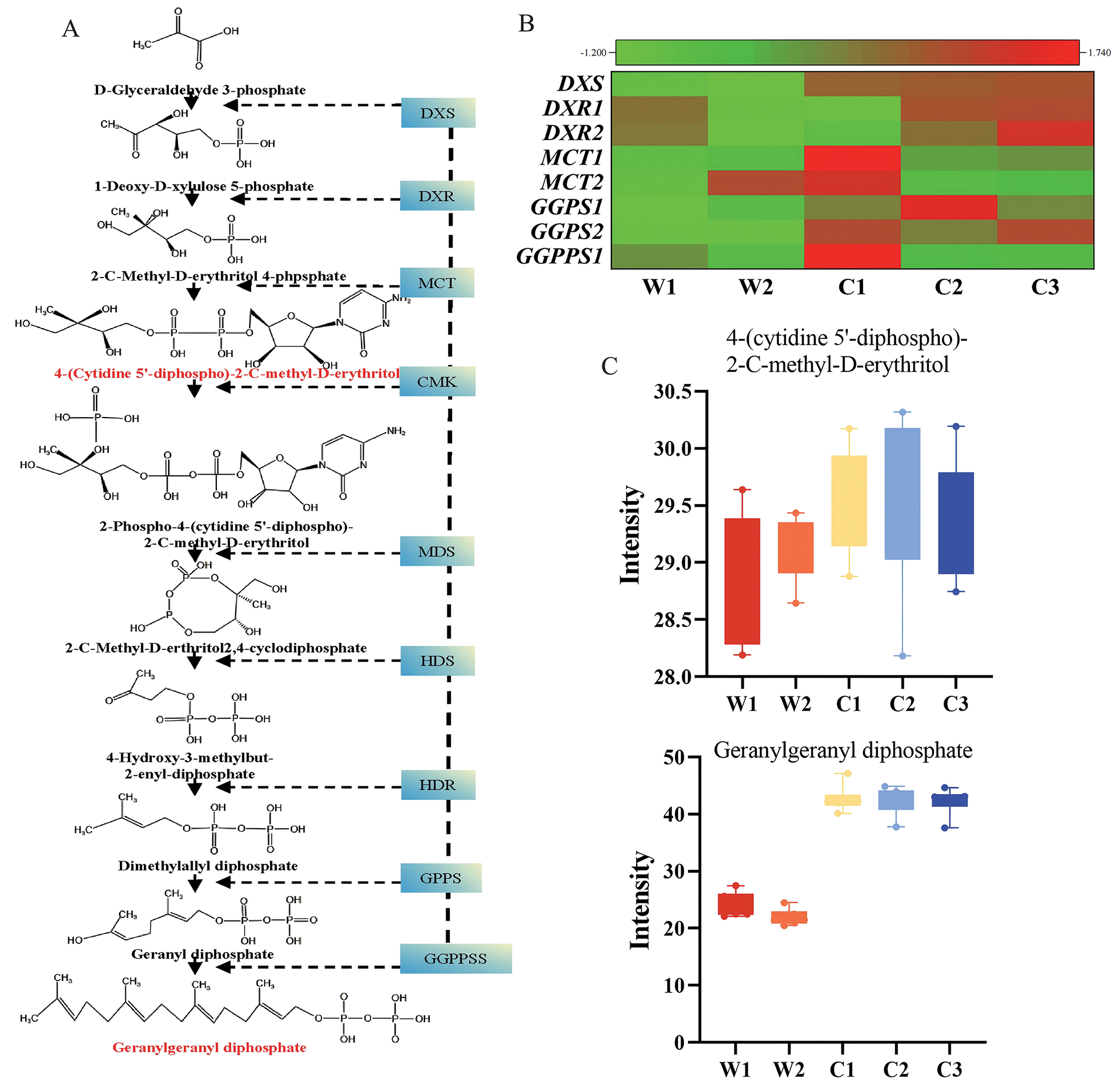
Figure 4: Integrated metabolomic and transcriptomic analysis of MEP pathway. (A) MEP pathway overview, with genes identified by transcriptome (blue background) and metabolites identified by metabolome (red font). (B) Key gene expression in MEP pathway (Z-score normalized). (C) Intermediate metabolite accumulation in MEP pathway
3.4 Enhancement of Paclitaxel Biosynthesis Pathway and Metabolic Remodeling in Cultivated T. cuspidata
The paclitaxel biosynthesis pathway includes three main stages (Fig. 5A,B): (1) formation of the taxane skeleton, where isoprenoid precursors are synthesized via the MEP pathway, three IPP and one DMAPP molecules condense to form GGPP, and taxadiene synthase (TS) catalyzes the cyclization of GGPP to form taxadiene, establishing the tricyclic diterpene skeleton; (2) modification of the taxane skeleton, where taxadiene undergoes a series of hydroxylation, acylation, and ketonization reactions to form baccatin III; and (3) assembly of the C-13 side chain, where BAPT (C-13 phenylpropanoyl-CoA transferase) and DBTNBT (3′-N-debenzoyl-2′-deoxytaxol N-benzoyltransferase) catalyze reactions in the final stage of paclitaxel biosynthesis. Transcriptome and metabolome analyses reveal significant remodeling of paclitaxel biosynthesis under cultivation (Fig. 5C): (1) Enhanced precursor supply, with GGPP content increasing 1.94 times and GGPPS gene expression approximately 3.1 times higher than in wild species. MEP pathway-related gene expression was enhanced, providing sufficient substrates for paclitaxel synthesis. (2) Early steps were enhanced, TS, T5αH, and T13αH expression increased 9.9 times, 8.8 times, and 3.8 times respectively, indicating significantly improved efficiency in early synthesis steps, consistent with findings by Van et al. [40] and Zhang et al. [41] studies. (3) Accelerated intermediate steps, T10βH and TBT expression increased 8.4 times and 5.8 times, promoting 2-α benzoylation and increasing 10-DAB content 1.71 times compare to wild species, aligning with He et al. [42] in cultivated T. yunnanensis, however, this study only investigated the differences in the content of paclitaxel and 10-DAB. (4) DBAT and DBTNBT expressions increased 2.6 times and 6.0 times, with baccatin III and paclitaxel content rising 1.8 times and 1.67 times. DBAT-catalyzed conversion of 10-DAB to baccatin III corroborates findings by Katkovcinova et al. [43] and Chen et al. [44], explaining increased baccatin III in cultivated species. These changes collectively and effectively directed the metabolic flux toward paclitaxel synthesis in cultivated species (Table 2).
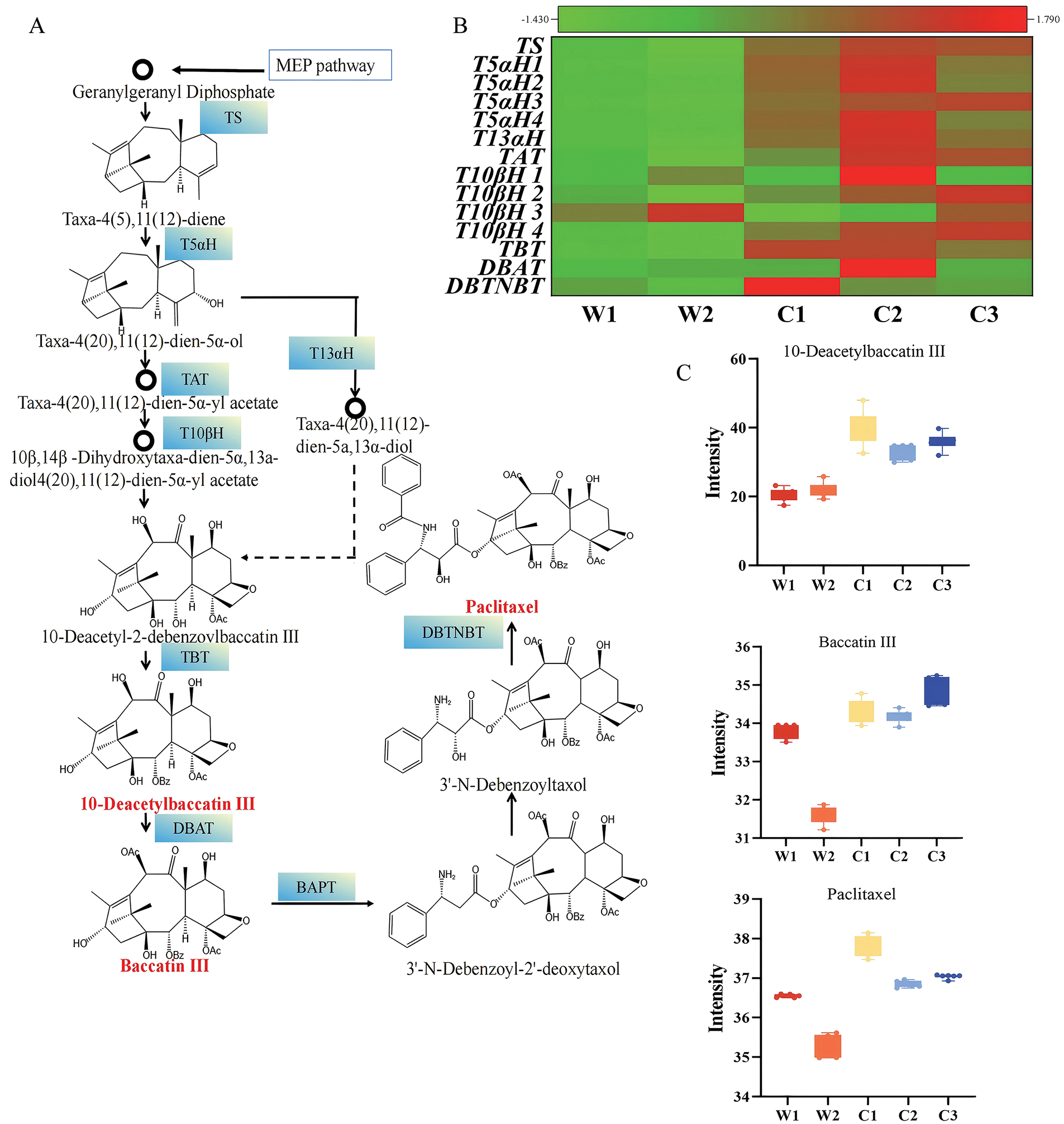
Figure 5: Integrated metabolomic and transcriptomic analysis of paclitaxel biosynthesis pathway. (A) Pathway overview, with genes identified by transcriptome (blue background) and metabolites identified by metabolome (red font). (B) Key gene expression (Z-score normalized). (C) Intermediate metabolite accumulation
By integrating metabolomics and transcriptomics, we identified metabolic pathways of taxusin and taxuyunnanin C closely related to paclitaxel biosynthesis. Our transcriptome identified the encoding genes of T2αH and T7βH that are involved in the metabolism of taxusin metabolites and the encoding gene of CYP450 family [45] that is involved in the biosynthesis of taxuyunnanin C, a classic 14-hydroxylated taxoid (Fig. 6A,B). Notably, taxusin, a vital paclitaxel precursor, shows remarkably higher abundance in cultivated species than in wild ones, indicating a higher capacity for paclitaxel precursor accumulation. Moreover, the key enzyme genes for taxusin synthesis exhibit higher expression levels in cultivated species, further suggesting enhanced paclitaxel synthesis capability. The correlation between key gene expression and metabolite content is evident: T2αOH shows generally higher expression levels in cultivated species, which aligns with the higher Paclitaxel content. Conversely, in wild species, the CYP450 family members (CYP8743 and CYP90A1 genes) associated with the biosynthesis of taxuyunnanin C exhibit a significant trend of up-regulated expression. This differential expression pattern explains why taxuyunnanin C, a paclitaxel derivative, is predominantly found in wild species but occurs at lower levels in cultivated species. This phenomenon suggests that the metabolic flux in cultivated species is more directed toward paclitaxel synthesis (Fig. 6C,D).
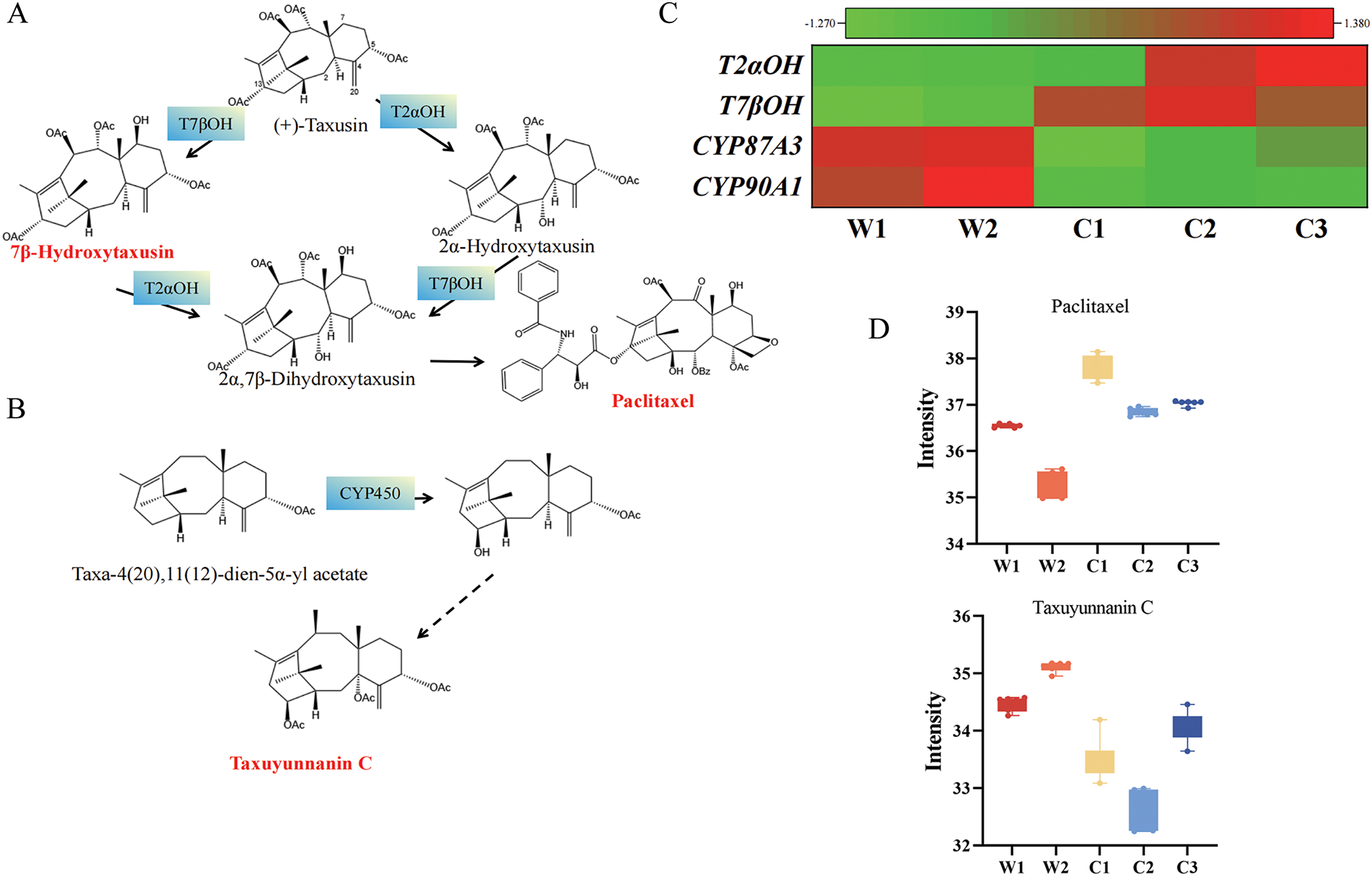
Figure 6: Integrated metabolomic and transcriptomic analysis of paclitaxel biosynthesis branch pathways. Overview of (A) taxusin and (B) taxuyunnanin C metabolism pathways, with genes identified by transcriptome (blue background) and metabolites identified by metabolome (red font). (C) Key gene expression (Z-score normalized). (D) Metabolite accumulation in branch pathways
In this study, we identified 127 differentially expressed transcription factors (TFs) between wild and cultivated T. cuspidata, with 10 TFs associated with paclitaxel biosynthesis. The positively regulating TFs include MYB47 (KI387_038078), MYB3 (KI387_012576), bHLH68 (KI387_021916), WRKY8 (KI387_001399), WRKY12 (KI387_003400), WRKY47 (KI387_036653), and TGA10 (KI387_014643). The negatively regulating TFs comprise WRKY52 (KI387_003376), ERF15 (KI387_010838), ERF12 (KI387_032181), and MYC2 (KI387_029593) (Fig. 7A). These TFs regulate key genes in the paclitaxel biosynthesis pathway, including TS, T5αH, TAT, T10βH, TBT, DBAT, BAPT, and DBTNBT (Fig. 7B).
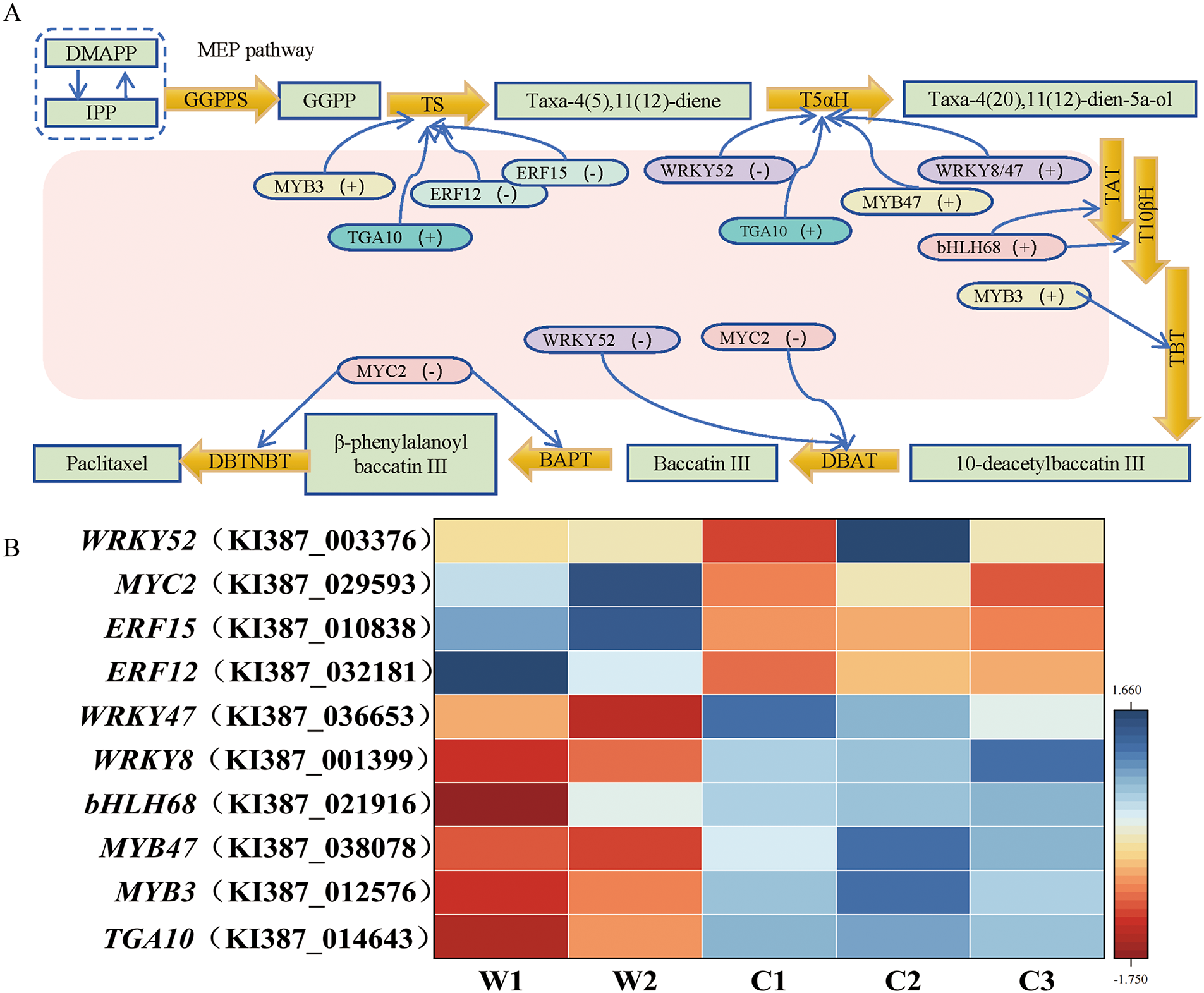
Figure 7: Identification of paclitaxel biosynthesis-related transcription factors (TFs). (A) Predicted transcriptional regulation networks based on specific expression patterns. (B) TF expression analysis (Z-score normalized)
3.5 Expression Validation of Key Genes in the Paclitaxel Pathway
To verify the reliability of DEGs identified by RNA-Seq and further investigate expression differences of key genes in the paclitaxel biosynthesis pathway, we randomly selected 10 related genes for detailed expression analysis (Fig. 8). In cultivated species, most key genes were significantly upregulated compared to wild species (p < 0.05), including DXR (KI387_002164), GGPPS1 (KI387_010989), TS (KI387_028840), T5αH (KI387_028803), TAT (KI387_028565), T10βH (KI387_010542), DBAT (KI387_030977), DBTNBT (KI387_025350), T2αOH (KI387_028803), and T7βH (KI387_028597). In particular, the C2 showed particularly high expression levels (p < 0.05). For instance, T5αH expression in C2 was 3.42 times higher than in C3 and 21.65 times higher than in W1, fully demonstrating C2’s potential advantage in paclitaxel biosynthesis (Fig. 8D). Interestingly, T2αOH displayed a different expression pattern compared to the other genes (Fig. 8I), with significantly higher expression in C3 (p < 0.05), reaching 28.11 times that of W1 and 0.33 times that of C2. This suggests T2αOH’s important role in specific stages of paclitaxel synthesis and offers new directions for investigating regulatory mechanisms, this view is consistent with Walker et al. [46] and Wang et al. [47].
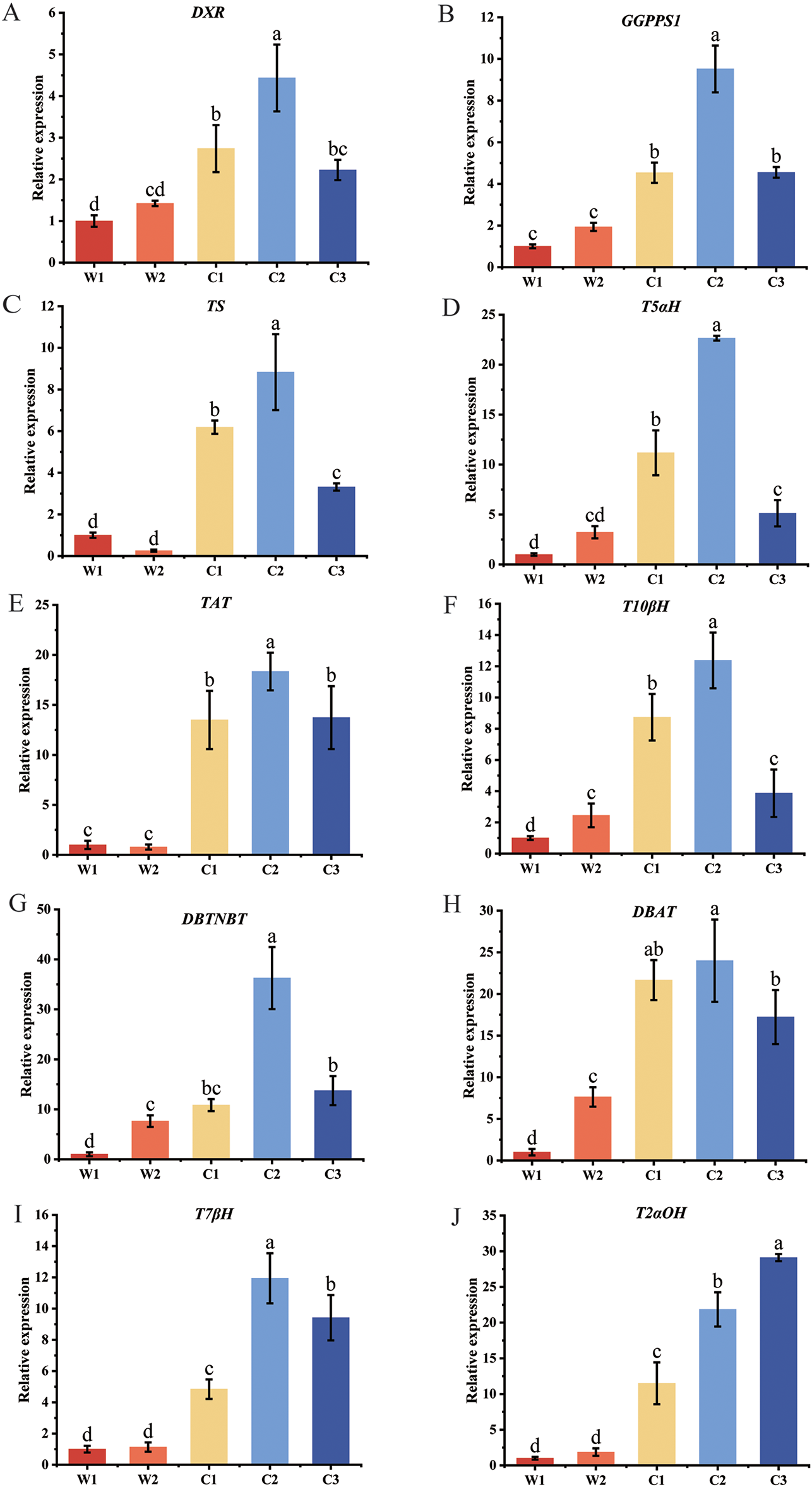
Figure 8: Expression Validation of Ten Key Paclitaxel Pathway Genes. Different lowercase letters denote significant differences between treatments (p < 0.05). (A) DXR, (B) GGPPS1, (C) TS, (D) T5αH, (E) TAT, (F) T10βH, (G) DBTNBT, (H) DBAT, (I) T7βH, (J) T2αH
To date, yew trees remain the sole natural source for paclitaxel extraction, limiting its supply [48]. Paclitaxel biosynthesis involves numerous enzymatic reactions and complex metabolic pathways, with many key enzymes exhibiting low activity and expression restricted to specific plant growth environments. To meet the increasing demand for paclitaxel, developing cultivated species is considered a crucial strategy to replace wild ones. Our research has confirmed significantly higher paclitaxel synthesis capability in cultivated species compared to wild ones. Using an integrated metabolomics and transcriptomics approach, we investigated the species-specific accumulation between cultivated and wild species, providing insights into the molecular mechanisms underlying enhanced paclitaxel production in cultivated yew trees.
Metabolomic analysis revealed significant differences in secondary metabolism between wild and cultivated species, particularly in paclitaxel and 10-DAB accumulation, primarily due to contrasting growth environments. Cultivated species, growing in controlled settings, experiencing less environmental stress, promoting secondary metabolite synthesis, including paclitaxel. Optimized fertilization in cultivation ensures adequate essential elements (such as nitrogen, potassium, calcium, and magnesium) for paclitaxel synthesis [49], resulting in higher 10-DAB content compared to wild species. Conversely, wild species face variable environmental stresses, necessitating balanced energy allocation with more resources devoted to defense and adaptation, which limits paclitaxel synthesis due to their higher carbohydrate metabolism capacity. Cultivated species, developed through selective breeding, show superior growth rates crucial for paclitaxel accumulation. They exhibit elevated levels of growth-related compounds (amino acids, phospholipids, and nucleosides), enhancing cellular processes and tissue development. The upregulation of plant hormones and their biosynthetic precursors (7′-hydroxyabscisic acid [50], jasmolactone [51], and indole-3-pyruvate [52]) facilitates rapid paclitaxel accumulation. In contrast, wild species grow slower due to unstable natural conditions, prioritizing stress defense over paclitaxel production. Metabolomic data reveals 949 upregulated and 1432 downregulated metabolites in cultivated species compared to wild ones. Wild species maintain more diverse metabolic pathways for environmental adaptation, enhancing ecological resilience but potentially limiting specific secondary metabolite synthesis, including paclitaxel, due to metabolism remodeling.
Metabolic remodeling is crucial for efficient paclitaxel production in cultivated species [53]. Compared to wild species, cultivated species show a metabolic pattern favoring paclitaxel synthesis through enhanced key enzyme activities, optimized regulation at metabolic branch points, and adjusted energy allocation. This transition increases paclitaxel yield while significantly altering the content of other metabolites. Taxusin is a precursor to paclitaxel and shares part of its metabolic pathway with other taxane compounds (e.g., taxuyunnanin C). However, at certain branching points, enzyme activity and gene expression levels determine the direction of metabolic flux [54]. Our study revealed that in cultivated species, the key enzymes involved in the conversion of taxusin to paclitaxel (e.g., hydroxylases) generally exhibit higher activity and gene expression levels, which significantly enhance the efficiency of paclitaxel synthesis. The metabolic flux tends to favor the conversion of taxusin to paclitaxel. In contrast, in wild species, taxusin is more likely to be utilized for the synthesis of other taxane compounds (e.g., taxuyunnanin C), thereby limiting the production of paclitaxel to some extent. Simultaneously, moderate suppression of the taxuyunnanin C biosynthesis pathway reduced substrate competition with paclitaxel synthesis. Furthermore, cultivated species exhibited significant changes in carbon flux allocation between primary and secondary metabolism. Primary metabolites such as glucose and sucrose decreased, while intermediates of the diterpenoid pathway, such as 10-DAB, increased. This strategic redistribution of metabolic flux provided sufficient carbon skeletons and energy for increased paclitaxel production in cultivated species, while explaining the reduced levels of certain primary metabolites. These changes demonstrate the intricate regulation and adaptive modifications in plant metabolic networks during domestication, consistent with findings by Kolody et al. [55]. This strategic redistribution of metabolic flux not only provided sufficient carbon skeletons and energy support for increased paclitaxel content in cultivated species but also explained the reduced levels of certain primary metabolites. These metabolic network modifications during domestication demonstrate intricate regulation and adaptation, consistent with findings by Kolody et al. [55].
Paclitaxel biosynthesis primarily relies on isoprenoid units synthesized via the MEP pathway, which generates IPP and DMAPP precursors. The condensation of these compounds initiates paclitaxel biosynthesis [56]. This essential biosynthetic pathway has been systematically elucidated in Taxus species and numerous medicinal plants [57,58]. Geranylgeranyl diphosphate synthase (GGPPS) is a key rate-limiting enzymes in this process. Variations in TcGGPPS, NgGGPPS, and TbGGPPS have led to significant changes in total diterpenoid content in T. Chinensis [59], Ginkgo biloba [60] and Taxus baccata [61], respectively. Furthermore, GGPPS homologous gene transcription levels positively correlate with paclitaxel content in T. baccata [62] and T. wallichiana [63]. This emphasizes the critical role of GGPPS in regulating precursor flux into the paclitaxel biosynthetic pathway, highlighting its potential for genetic manipulation to enhance paclitaxel production. Our metabolomic analysis revealed declining trends in certain primary metabolites in cultivated species, including specific amino acids and carbohydrate metabolism intermediates such as citric acid, isocitric acid, and lipoic acid. Conversely, GGPP, the initial metabolite in paclitaxel biosynthesis, significantly increased to 10.06 times that of wild species. This aligns with the high expression patterns of key enzyme genes involved in paclitaxel precursor synthesis observed in transcriptomic data. High GGPPS1 expression directly leads to GGPP accumulation, providing abundant precursors for paclitaxel biosynthesis. In Taxus species, overexpression of the TS gene significantly increases taxadiene accumulation, a paclitaxel precursor [64]. Taxadiene synthase, the first and rate-limiting enzyme in paclitaxel biosynthesis, catalyzes GGPP cyclization to form taxa-4(5),11(12)-diene. The expression level of the TS gene in cultivated species was significantly higher, reaching 10.21 times that of wild species. This significant difference likely contributes to the higher paclitaxel content in cultivated species. In the metabolic network of T. cuspidata, the allocation of metabolic flux is crucial for synthesizing paclitaxel and other terpene compounds. GGPP, a key substrate for various terpene compounds, directly affects the yield of downstream products. Our study suggests that cultivated species may enhance GGPP metabolic flux towards TS-catalyzed reactions by coordinately upregulating the expression of key enzymes such as TS and GGPPS, thereby improving synthesis efficiency.
Taxol-10-β-acetyltransferase (DBAT) is another crucial enzyme in paclitaxel biosynthesis, catalyzing the final step by transferring an acetyl group to the C-10 position of 10-DAB, generating the immediate precursor baccatin III [65]. DBAT expression level significantly positively correlated with paclitaxel yield, which is consistent with the findings of Walker et al. [66]. As a crucial branching point, DBAT’s efficiency directly affects paclitaxel production. Our study found DBAT expression in cultivated species was 6.28 times higher than in wild species, further explaining the higher paclitaxel content in cultivated species and highlighting DBAT’s core role in regulating paclitaxel biosynthesis. Additionally, multiple hydroxylases also play key roles in paclitaxel biosynthetic pathway. For instance, taxane-5α-hydroxylase catalyzes the first hydroxylation of the taxane skeleton, affecting the efficiency of subsequent steps. Taxol 10β-hydroxylase (T10βH) adds a hydroxyl group to the C-10 position of the paclitaxel precursor, a crucial step in the late-stage synthesis that is essential for the final product structure [67]. Taxane-13α-hydroxylase (T13αH) adds a hydroxyl group to the C-13 position, a necessary modification for forming active paclitaxel [68]. Our results show T5αH and T13αH expression levels in cultivated species were 8.22 and 6.24 times higher than in wild species, respectively (Fig. 7). More importantly, TFs play key roles in regulating paclitaxel biosynthesis. Several members of the MYB, bHLH, WRKY, ERF, MYC, and TGA families, along with their target genes, have been functionally identified across various Taxus species [69–71]. This study identified 10 differentially expressed TFs related to paclitaxel synthesis. In cultivated species, the expression levels of positively regulating factors are elevated, with notable examples being TGA10 and WRKY47, which are 4.91 and 5.50 times higher, respectively, than those in wild species, thereby promoting synthesis. Conversely, negatively regulating TFs like ERF15, ERF12, and MYC2 show markedly decreased expression levels in cultivated species relative to wild species, ERF15 exhibited a 4.13 times higher expression level in wild species compared to cultivated species, suggesting a weaker inhibitory effect on paclitaxel biosynthesis. These findings collectively indicate that cultivated species have a stronger promoting effect on paclitaxel synthesis. In conclusion, this coordinated optimization of gene expression and metabolic flux suggests that cultivated species have redirected more metabolic resources from primary to secondary metabolism, particularly toward the paclitaxel synthesis pathway. This finding reflects the significant evolutionary advantage cultivated species have gained in paclitaxel biosynthesis through natural selection or artificial domestication and provides insights into the plasticity of plant metabolic networks.
In conclusion, cultivated species demonstrate significantly superior paclitaxel production capability compared to wild species. Our comparative analysis reveals comprehensive metabolic remodeling in cultivated species, including enhancement of the MEP pathway, improved taxusin hydroxylation efficiency, inhibited taxuyunnanin C synthesis, and overall optimization of the metabolic network. In cultivated species, the upregulation of key enzyme genes and the redistribution of metabolic flux collectively create an environment conducive to efficient paclitaxel synthesis. This comprehensive metabolic regulation optimizes precursor supply and utilization balance, potentially influencing energy and reducing power distribution within plant cells. These findings deepen our understanding of metabolic regulatory mechanisms during the domestication process of T. cuspidata, providing a theoretical foundation and potential targets for enhancing paclitaxel yield through metabolic engineering.
Acknowledgement: We thank Zhang Chun-Peng, Sui Chang, and Ma Xiao for their assistance in the field.
Funding Statement: This work was supported by grants from the National Science Foundation of China to Yanwen Zhang (32272757, 31972363); and grants from Liaoning Provincial Department of Education Project to Dandan Wang (JYTMS20230698); grants from the Liaoning Provincial Science and Technology. Fund Project to Dandan Wang (2023JH2/101700200).
Author Contributions: Dandan Wang performed the experiments, analyzed the data, and wrote the manuscript. Jiaxin Chen investigated and implemented. Yanwen Zhang conceived and designed the experiments. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data included in this study are available upon request by contacting Wang Dandan.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at 10.32604/phyton.2025.063894.
References
1. Fang W, Wu Y, Zhou J, Chen W, Fang Q. Qualitative and quantitative determination of taxol and related compounds in Taxus cuspidata Sieb et Zucc. Phytochem Anal. 1993;4(3):115–9. doi:10.1002/pca.2800040307. [Google Scholar] [CrossRef]
2. Fengguo D, Banghua W, Xudong W. A preliminary study on the phenology of Taxus cuspidata Sieb. et Zucc. J Northeast For Univ. 1996;7(1):11–6. [Google Scholar]
3. Cheng B, Zheng Y, Sun Q. Genetic diversity and population structure of Taxus cuspidata in the Changbai Mountains assessed by chloroplast DNA sequences and microsatellite markers. Biochem Syst Ecol. 2015;63:157–64. doi:10.1016/j.bse.2015.10.009. [Google Scholar] [CrossRef]
4. Zhang H, Mittal N, Leamy LJ, Barazani O, Song BH. Back into the wild—apply untapped genetic diversity of wild relatives for crop improvement. Evol Appl. 2017;10(1):5–24. doi:10.1111/eva.12434.. [Google Scholar] [PubMed] [CrossRef]
5. Sanchez Munoz R, Bonfill M, Cusidó RM, Palazón J, Moyano E. Advances in the regulation of in vitro paclitaxel production: methylation of a Y-patch promoter region alters BAPT gene expression in Taxus cell cultures. Plant Cell Physiol. 2018;59(11):2255–67. doi:10.1093/pcp/pcy149.. [Google Scholar] [PubMed] [CrossRef]
6. Feng S, Hou K, Zhang H, Chen C, Huang J, Wu Q, et al. Investigation of the role of TmMYB16/123 and their targets (TmMTP1/11) in the tolerance of Taxus media to cadmium. Tree Physiol. 2023;43(6):1009–22. doi:10.1093/treephys/tpad019. [Google Scholar] [PubMed] [CrossRef]
7. Wang H, Vieira FG, Crawford JE, Chu C, Nielsen R. Asian wild rice is a hybrid swarm with extensive gene flow and feralization from domesticated rice. Genome Res. 2017;27(6):1029–38. doi:10.1101/gr.204800.116. [Google Scholar] [PubMed] [CrossRef]
8. Roucou A, Violle C, Fort F, Roumet P, Ecarnot M, Vile D. Shifts in plant functional strategies over the course of wheat domestication. J Appl Ecol. 2018;55(1):25–37. doi:10.1111/1365-2664.13029. [Google Scholar] [CrossRef]
9. Zhang, Y. Taxus chinensis: variation and cultivar selection. Liaoning Science and Technology Press; 2021. [Google Scholar]
10. Wang D, Li X, Zhang Y. Effects of physiology and secondary metabolism between wild and cultivated species of Taxus cuspidata under environmental stress. Acta Agriculturae Boreali-Occidentalis Sinica. 2022;31(8):958–68. doi:10.7606/j.issn.1004-1389.2022.08.003. [Google Scholar] [CrossRef]
11. Liu L, Pu Y, Niu Z, Wu J, Fang Y, Xu J, et al. Transcriptomic insights into root development and overwintering transcriptional memory of Brassica rapa L. grown in the field. Front Plant Sci. 2022;13:900708. doi:10.3389/fpls.2022.900708. [Google Scholar] [PubMed] [CrossRef]
12. Wang D, Li X, Zhang Y. Comparative study of genetic structure and genetic diversity between wild and cultivated populations of Taxus cuspidata, Northeast China. Phyton-Int J Exp Bot. 2024;93(2):355–69. doi:10.32604/phyton.2024.047183. [Google Scholar] [CrossRef]
13. Trynda-Lemiesz L. Paclitaxel-HSA interaction. Binding sites on HSA molecule. Bioorg Med Chem. 2004;12(12):3269–75. doi:10.1016/j.bmc.2004.03.073. [Google Scholar] [PubMed] [CrossRef]
14. Malik S, Cusidó RM, Mirjalili MH, Moyano E, Palazón J, Bonfill M. Production of the anticancer drug taxol in Taxus baccata suspension cultures: a review. Process Biochem. 2011;46(1):23–34. doi:10.1016/j.procbio.2010.09.004. [Google Scholar] [CrossRef]
15. Holton RA, Kim HB, Somoza C, Liang F, Biediger RJ, Boatman PD, et al. First total synthesis of taxol. 2. Completion of the C and D rings. J Am Chem Soc. 1994;116(4):1599–600. doi:10.1021/ja00083a067. [Google Scholar] [CrossRef]
16. Nicolaou K, Yang Z, Liu J, Ueno H, Nantermet P, Guy R, et al. Total synthesis of taxol. Nature. 1994;367(6464):630–4. [Google Scholar] [PubMed]
17. El-Sayed AS, Ali DM, Yassin MA, Zayed RA, Ali GS. Sterol inhibitor Fluconazole enhance the Taxol yield and molecular expression of its encoding genes cluster from Aspergillus flavipes. Process Biochem. 2019;76:55–67. doi:10.1016/j.procbio.2018.10.008. [Google Scholar] [CrossRef]
18. Wu QY, Huang ZY, Wang JY, Yu HL, Xu JH. Construction of an Escherichia coli cell factory to synthesize taxadien-5α-ol, the key precursor of anti-cancer drug paclitaxel. Bioresour Bioprocess. 2022;9(1):82. doi:10.1186/s40643-022-00569-5. [Google Scholar] [PubMed] [CrossRef]
19. Walls LE, Martinez JL. Rios-Solis L: enhancing Saccharomyces cerevisiae taxane biosynthesis and overcoming nutritional stress-induced pseudohyphal growth. Microorganisms. 2022;10(1):163. doi:10.3390/microorganisms10010163. [Google Scholar] [PubMed] [CrossRef]
20. Zhang Y, Wiese L, Fang H, Alseekh S, de Souza LP, Scossa F, et al. Synthetic biology identifies the minimal gene set required for paclitaxel biosynthesis in a plant chassis. Mol Plant. 2023;16(12):1951–61. doi:10.1016/j.molp.2023.10.016. [Google Scholar] [PubMed] [CrossRef]
21. Sabater-Jara AB, Onrubia M, Moyano E, Bonfill M, Palazón J, Pedreño MA, et al. Synergistic effect of cyclodextrins and methyl jasmonate on taxane production in Taxus x media cell cultures. Plant Biotechnol J. 2014;12(8):1075–84. doi:10.1111/pbi.12214. [Google Scholar] [PubMed] [CrossRef]
22. Li H, Horiguchi T, Croteau R, Williams RM. Studies on Taxol biosynthesis: preparation of taxadiene-diol and triol derivatives by deoxygenation of taxusin. Tetrahedron. 2008;64(27):6561–7. doi:10.1016/j.tet.2008.04.008. [Google Scholar] [PubMed] [CrossRef]
23. Kanda Y, Nakamura H, Umemiya S, Puthukanoori RK, Murthy Appala VR, Gaddamanugu GK, et al. Two-phase synthesis of taxol. J Am Chem Soc. 2020;142(23):10526–33. doi:10.1021/jacs.0c03592. [Google Scholar] [PubMed] [CrossRef]
24. Sun F, Dang S, Zheng D, Zhang H, Wang Y, Li Y, et al. Advances in paclitaxel biosynthesis and transcriptional regulation mechanisms. Chin J Biotechnol. 2024;40(5):1380–405. doi:10.13345/j.cjb.230850. [Google Scholar] [PubMed] [CrossRef]
25. Tanaka K, Li F, Morikawa K, Nobukawa T, Kadota S. Analysis of biosynthetic fluctuations of cultured Taxus seedlings using a metabolomic approach. Phytochemistry. 2011;72(14–15):1760–6. doi:10.1016/j.phytochem.2011.06.003. [Google Scholar] [PubMed] [CrossRef]
26. Yang L, Zheng Z-S, Cheng F, Ruan X, Jiang D-A, Pan C-D, et al. Seasonal dynamics of metabolites in needles of Taxus wallichiana var. mairei. Molecules. 2016;21(10):1403. doi:10.3390/molecules21101403. [Google Scholar] [PubMed] [CrossRef]
27. Shao F, Zhang L, Guo J, Liu X, Ma W, Wilson IW, et al. A comparative metabolomics analysis of the components of heartwood and sapwood in Taxus chinensis (Pilger) Rehd. Sci Rep. 2019;9(1):17647. doi:10.1038/s41598-019-53839-2. [Google Scholar] [PubMed] [CrossRef]
28. Zhou T, Luo X, Zhang C, Xu X, Yu C, Jiang Z, et al. Comparative metabolomic analysis reveals the variations in taxoids and flavonoids among three Taxus species. BMC Plant Biol. 2019;19:1–12. [Google Scholar]
29. Chen YM, Wang T, Zhang FJ, Zhuang WB, Shu XC, Wang Z, et al. Comparison of metabolites in the needles of Taxus wallichiana var. mairei and Taxus wallichiana var. mairei cv. ‘Jinxishan’. Acta Bot Boreal-Occid Sin. 2019(5):801-7. [Google Scholar]
30. Yu C, Luo X, Zhang C, Xu X, Huang J, Chen Y, et al. Tissue-specific study across the stem of Taxus media identifies a phloem-specific TmMYB3 involved in the transcriptional regulation of paclitaxel biosynthesis. Plant J. 2020;103(1):95–110. doi:10.1111/tpj.14710. [Google Scholar] [PubMed] [CrossRef]
31. Yu C, Zhan X, Zhang C, Xu X, Huang J, Feng S, et al. Comparative metabolomic analyses revealed the differential accumulation of taxoids, flavonoids and hormones among six Taxaceae trees. Sci Hortic. 2021;285:110196. doi:10.1016/j.scienta.2021.110196. [Google Scholar] [CrossRef]
32. Yan Y, Wen Y, Wang Y, Wu X, Li X, Wang C, et al. Metabolome integrated with transcriptome reveals the mechanism of three different color formations in Taxus mairei arils. Front Plant Sci. 2024;15:1330075. doi:10.1016/j.scienta.2021.110196. [Google Scholar] [CrossRef]
33. Zhan X, Zang Y, Ma R, Lin W, Li XL, Pei Y, et al. Mass spectrometry-imaging analysis of active ingredients in the leaves of Taxus cuspidata. ACS Omega. 2024;9(16):18634–42. doi:10.1021/acsomega.4c01440. [Google Scholar] [PubMed] [CrossRef]
34. Majeed A, Singh A, Sharma RK, Jaitak V, Bhardwaj P. Comprehensive temporal reprogramming ensures dynamicity of transcriptomic profile for adaptive response in Taxus contorta. Mol Genet Genomics. 2020;295:1401–14. doi:10.1007/s00438-020-01709-2. [Google Scholar] [PubMed] [CrossRef]
35. Want EJ, Wilson ID, Gika H, Theodoridis G, Plumb RS, Shockcor J, et al. Global metabolic profiling procedures for urine using UPLC-MS. Nat Protoc. 2010;5(6):1005–18. doi:10.1038/nprot.2010.50. [Google Scholar] [PubMed] [CrossRef]
36. Feng BS, Liu LX, Sun J, Leng P, Wang L, Guo Y, et al. Combined metabolome and transcriptome analyses of quality components and related molecular regulatory mechanisms during the ripening of Huangjin Peach. Sci Hortic. 2024;327:112787. doi:10.1016/j.scienta.2023.112787. Get rights and content. [Google Scholar] [CrossRef]
37. Wu X, Luo D, Zhang Y, Jin L, Crabbe MJC, Qiao Q, et al. Integrative analysis of the metabolome and transcriptome reveals the potential mechanism of fruit flavor formation in wild hawthorn (Crataegus chungtienensis). Plant Divers. 2023;45(5):590–600. doi:10.1016/j.pld.2023.02.001. [Google Scholar] [PubMed] [CrossRef]
38. Liu B, Liu W, Zhao S, Ma L, Zang T, Huang C, et al. Transcriptome sequencing of facial adipose tissue reveals alterations in mRNAs of hemifacial microsomia. Front Pediatr. 2023;11:1099841. doi:10.1016/j.pld.2023.02.001. [Google Scholar] [CrossRef]
39. Amini SA, Shabani L, Afghani L, Jalalpour Z, Sharifi-Tehrani M. Squalestatin-induced production of taxol and baccatin in cell suspension culture of yew (Taxus baccata L.). Turk J Biol. 2014;38(4):528–36. doi:10.3906/biy-1401-47. [Google Scholar] [CrossRef]
40. Van Rozendaal EL, Lelyveld GP, Van Beek TA. Screening of the needles of different yew species and cultivars for paclitaxel and related taxoids. Phytochemistry. 2000;53(3):383–9. doi:10.1016/s0031-9422(99)00094-1. [Google Scholar] [PubMed] [CrossRef]
41. Zhang K, Jiang L, Wang X, Han H, Chen D, Qiu D, et al. Transcriptome-wide analysis of AP2/ERF transcription factors involved in regulating Taxol biosynthesis in Taxus × media. Ind Crops Prod. 2021;171:113972. doi:10.1016/j.indcrop.2021.113972. [Google Scholar] [CrossRef]
42. He CT, Li ZL, Zhou Q, Shen C, Huang YY, Mubeen S, et al. Transcriptome profiling reveals specific patterns of paclitaxel synthesis in a new Taxus yunnanensis cultivar. Plant Physiol Biochem. 2018;122:10–8. doi:10.1016/j.plaphy.2017.10.028. [Google Scholar] [PubMed] [CrossRef]
43. Katkovčinová Z, Lázárová M, Bruňáková K, Košuth J, Čellárova E. Expression of dbat and dbtnbt genes involved in paclitaxel biosynthesis during the growth cycle of Taxus baccata L. callus cultures. Zeitschrift Für Naturforschung C. 2008;63(9–10):721–30. doi:10.1515/znc-2008-9-1018. [Google Scholar] [PubMed] [CrossRef]
44. Chen Y, Zhang M, Zhang WL, Wang YM, Zhang H, Yang L, et al. miR5298b regulated taxol biosynthesis by acting on TcNPR3, resulting in an alleviation of the strong inhibition of the TcNPR3-TcTGA6 complex in Taxus chinensis. Int J Biol Macromol. 2023;248:125909. doi:10.1016/j.ijbiomac.2023.125909. [Google Scholar] [CrossRef]
45. Schneider B. Nuclear magnetic resonance spectroscopy in biosynthetic studies. Prog Nucl Magn Reson Spectrosc. 2007;51(3):155–98. doi:10.1016/j.pnmrs.2007.02.006. [Google Scholar] [CrossRef]
46. Walker K, Long R, Croteau R. The final acylation step in taxol biosynthesis: cloning of the taxoid C13-side-chain N-benzoyltransferase from Taxus. Proc Nat Acad Sci. 2002;99(14):9166–71. doi:10.1073/pnas.082115799. [Google Scholar] [PubMed] [CrossRef]
47. Wang T, Li L, Zhuang W, Zhang F, Shu X, Wang N, et al. Recent research progress in taxol biosynthetic pathway and acylation reactions mediated by Taxus acyltransferases. Molecules. 2021;26(10):2855. doi:10.3390/molecules26102855. [Google Scholar] [PubMed] [CrossRef]
48. Walsh V, Goodman J. The story of taxol: science and politics in the making of an anticancer drug; 2001. [Internet]. [cited 2025 Mar 17]. Available from: http://www.manchester.ac.uk/escholar/uk-ac-man-scw:4b2265. [Google Scholar]
49. Kumar P, Singh B, Thakur V, Thakur A, Thakur N, Pandey D, et al. Hyper-production of taxol from Aspergillus fumigatus, an endophytic fungus isolated from Taxus sp. of the Northern Himalayan region. Biotechnol Rep. 2019;24:e00395. doi:10.1016/j.btre.2019.e00395. [Google Scholar] [PubMed] [CrossRef]
50. Kuang XJ. Studies on the biosynthesis and regulation of diterpenoids, Taxol and tanshinones [dissertation]. Beijing, China: Institute of Medicinal Plant Development; 2020. [Google Scholar]
51. Khan V, Jha A, Seth T, Iqbal N, Umar S. Exploring the role of jasmonic acid in boosting the production of secondary metabolites in medicinal plants: pathway for future research. Ind Crops Prod. 2024;220:119227. doi:10.1016/j.indcrop.2024.119227. [Google Scholar] [CrossRef]
52. Pullaiah T, Karuppusamy S, Swamy MK. Propagation of paclitaxel biosynthesizing plants. In: Paclitaxel. Amsterdam, The Netherlands: Elsevier; 2022. p. 171–202. [Google Scholar]
53. Cusido RM, Onrubia M, Sabater-Jara AB, Moyano E, Bonfill M, Goossens A, et al. A rational approach to improving the biotechnological production of taxanes in plant cell cultures of Taxus spp. Biotechnol Adv. 2014;32(6):1157–67. doi:10.1016/j.biotechadv.2014.03.002. [Google Scholar] [PubMed] [CrossRef]
54. Yu C, Zhang C, Xu X, Huang J, Chen Y, Luo X, et al. Omic analysis of the endangered Taxaceae species Pseudotaxus chienii revealed the differences in taxol biosynthesis pathway between Pseudotaxus and Taxus yunnanensis trees. BMC Plant Biol. 2021;21:1–13. [Google Scholar]
55. Kolody B, Smith S, Zeigler Allen L, McCrow J, Moustafa A, Shi D, et al. Nitrogen and iron availability drive metabolic remodeling and natural selection of diverse phytoplankton during experimental upwelling. Msystems. 2022;7(5). doi:10.1128/msystems.00729-22. [Google Scholar] [PubMed] [CrossRef]
56. He S, Bekhof A-SM, Popova EZ, van Merkerk R, Quax WJ. Improved taxadiene production by optimizing DXS expression and fusing short-chain prenyltransferases. New Biotechnol. 2024;83:66–73. doi:10.1016/j.nbt.2024.06.007. [Google Scholar] [PubMed] [CrossRef]
57. Long RM, Croteau R. Preliminary assessment of the C13-side chain 2′-hydroxylase involved in Taxol biosynthesis. Biochem Biophys Res Commun. 2005;338(1):410–7. doi:10.1016/j.bbrc.2005.08.119. [Google Scholar] [PubMed] [CrossRef]
58. Cusidó RM, Palazón J, Bonfill M, Expósito O, Moyano E, Piñol MT. Source of isopentenyl diphosphate for taxol and baccatin III biosynthesis in cell cultures of Taxus baccata. Biochem Eng J. 2007;33(2):159–67. doi:10.1016/j.bej.2006.10.016. [Google Scholar] [CrossRef]
59. Laskaris G, Van der Heijden R, Verpoorte R. Purification and partial characterisation of geranylgeranyl diphosphate synthase, from Taxus baccata cell cultures: an enzyme that regulates taxane biosynthesis. Plant Sci. 2000;153(2):97–105. doi:10.1016/s0168-9452(99)00263-0. [Google Scholar] [PubMed] [CrossRef]
60. Guo J, Tang W, Tang W, Gao T, Yuan M, Wu Y, et al. Research progress on the types, functions, biosynthesis, and metabolic regulation of ginkgo terpenoids. Plant Physiol Biochem. 2024;212(1):108754. doi:10.1016/j.plaphy.2024.108754. [Google Scholar] [PubMed] [CrossRef]
61. Ding Q, Fang D-M, Li X-H, Gao F. Two new taxane diterpenoids from Taxus baccata. Nat Prod Commun. 2020;15(9):1934578X20953280. doi:10.1177/1934578X20953280. [Google Scholar] [CrossRef]
62. Perez-Matas E, Garcia-Perez P, Miras-Moreno B, Lucini L, Bonfill M, Palazon J, et al. Exploring the interplay between metabolic pathways and taxane production in elicited Taxus baccata cell suspensions. Plants. 2023;12(14):2696. doi:10.3390/plants12142696. [Google Scholar] [PubMed] [CrossRef]
63. Cheng J, Wang X, Liu X, Zhu X, Li Z, Chu H, et al. Chromosome-level genome of Himalayan yew provides insights into the origin and evolution of the paclitaxel biosynthetic pathway. Mol Plant. 2021;14(7):1199–209. doi:10.1016/j.molp.2021.04.015. [Google Scholar] [PubMed] [CrossRef]
64. Soliman S. Tang Y: natural and engineered production of taxadiene with taxadiene synthase. Biotechnol Bioeng. 2015;112(2):229–35. doi:10.1002/bit.25468. [Google Scholar] [PubMed] [CrossRef]
65. Li Y, Yang J, Zhou X, Zhao W, Jian Z. Isolation and identification of a 10-deacetyl baccatin-III-producing endophyte from Taxus wallichiana. Appl Biochem Biotechnol. 2015;175:2224–31. doi:10.1007/s12010-014-1422-0. [Google Scholar] [PubMed] [CrossRef]
66. Walker KD, Klettke K, Akiyama T, Croteau R. Cloning, heterologous expression, and characterization of a phenylalanine aminomutase involved in Taxol biosynthesis. J Biol Chem. 2004;279(52):53947–54. doi:10.1074/jbc.M411215200. [Google Scholar] [PubMed] [CrossRef]
67. Liu JC, De La Peña R, Tocol C, Sattely ES. Reconstitution of early paclitaxel biosynthetic network. Nat Commun. 2024;15(1):1419. doi:10.1038/s41467-024-45574-8. [Google Scholar] [PubMed] [CrossRef]
68. Chakravarthi BV, Singh S, Kamalraj S, Gupta VK, Jayabaskaran C. Evaluation of spore inoculum and confirmation of pathway genetic blueprint of T13αH and DBAT from a Taxol-producing endophytic fungus. Sci Rep. 2020;10(1):21139. doi:10.1038/s41598-020-77605-x. [Google Scholar] [PubMed] [CrossRef]
69. Zhan X, Qiu T, Zhang H, Hou K, Liang X, Chen C, et al. Mass spectrometry imaging and single-cell transcriptional profiling reveal the tissue-specific regulation of bioactive ingredient biosynthesis in Taxus leaves. Plant Commun. 2023;4(5):100630. doi:10.1016/j.xplc.2023.100630. [Google Scholar] [PubMed] [CrossRef]
70. Yu C, Hou K, Zhang H, et al. Integrated mass spectrometry imaging and single-cell transcriptome atlas strategies provide novel insights into taxoid biosynthesis and transport in Taxus mairei stems. Plant J. 2023;115(5):1243–60. doi:10.1111/tpj.16315. [Google Scholar] [PubMed] [CrossRef]
71. Cao X, Xu L, Li L, Wan W, Jiang J. TcMYB29a, an ABA-responsive R2R3-MYB transcriptional factor, upregulates taxol biosynthesis in Taxus chinensis. Front Plant Sci. 2022;13:804593. doi:10.3389/fpls.2022.804593. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools