 Open Access
Open Access
ARTICLE
Densely Convolutional BU-NET Framework for Breast Multi-Organ Cancer Nuclei Segmentation through Histopathological Slides and Classification Using Optimized Features
1 Artificial Intelligence & Data Analytics Lab, College of Computer & Information Sciences (CCIS), Prince Sultan University, Riyadh, 11586, Saudi Arabia
2 Centre of Real Time Computer Systems, Kaunas University of Technology, Kaunas, LT-51386, Lithuania
3 Department of Mathematical Sciences, College of Science, Princess Nourah bint Abdulrahman University, Riyadh, 11671, Saudi Arabia
* Corresponding Author: Robertas Damasevicius. Email:
(This article belongs to the Special Issue: Emerging Artificial Intelligence Technologies and Applications)
Computer Modeling in Engineering & Sciences 2024, 141(3), 2375-2397. https://doi.org/10.32604/cmes.2024.056937
Received 02 August 2024; Accepted 10 October 2024; Issue published 31 October 2024
Abstract
This study aims to develop a computational pathology approach that can properly detect and distinguish histology nuclei. This is crucial for histopathological image analysis, as it involves segmenting cell nuclei. However, challenges exist, such as determining the boundary region of normal and deformed nuclei and identifying small, irregular nuclei structures. Deep learning approaches are currently dominant in digital pathology for nucleus recognition and classification, but their complex features limit their practical use in clinical settings. The existing studies have limited accuracy, significant processing costs, and a lack of resilience and generalizability across diverse datasets. We proposed the densely convolutional Breast U-shaped Network (BU-NET) framework to overcome the mentioned issues. The study employs BU-NET’s spatial and channel attention methods to enhance segmentation processes. The inclusion of residual blocks and skip connections in the BU-NEt architecture enhances the process of extracting features and reconstructing the output. This enhances the robustness of training and convergence processes by reducing the occurrence of vanishing gradients. The primary objective of BU-NEt is to enhance the model’s capacity to acquire and analyze more intricate features, all the while preserving an efficient working representation. The BU-NET experiments demonstrate that the framework achieved 88.7% average accuracy, 88.8% F1 score for Multi-Organ Nuclei Segmentation Challenge (MoNuSeg), and 91.2% average accuracy, 91.8% average F1 for the triple-negative breast cancer (TNBC) dataset. The framework also achieved 93.92 Area under the ROC Curve (AUC) for TNBC. The results demonstrated that the technology surpasses existing techniques in terms of accuracy and effectiveness in segmentation. Furthermore, it showcases the ability to withstand and recover from different tissue types and diseases, indicating possible uses in medical treatments. The research evaluated the efficacy of the proposed method on diverse histopathological imaging datasets, including cancer cells from many organs. The densely connected U-NEt technology offers a promising approach for automating and precisely segmenting cancer cells on histopathology slides, hence assisting pathologists in improving cancer diagnosis and treatment outcomes.Keywords
Breast cancer, which causes abnormal cell development in breast tissue, is a worldwide health concern that affects women [1]. The risk factors include age, family history, reproductive traits, hormone replacement therapy, and lifestyle choices, in addition to mutations like breast cancer gene 1 (BRCA1) and breast cancer gene 2 (BRCA2). Mammography, clinical breast examination, and breast self-examination are screening techniques that are essential for early detection [2]. The most accurate method for identifying various subtypes of breast cancer is the histopathological examination of breast tissue. Chemotherapy is the most prevalent treatment for triple-negative breast cancer (TNBC), which has a poorer prognosis and fewer available therapeutic alternatives [3]. Histopathology is a crucial field in medicine, involving the evaluation of tissue samples for diagnosing illnesses and understanding disease progression. It is essential for diagnosing a wide range of diseases, including malignant tumors and infectious disorders, by separating distinct cellular components, particularly nuclei [4]. Nuclei store critical information on cell health and functioning, and abnormalities suggest pathological disorders. The use of computer technologies has transformed the field of pathology, increasing precision in diagnosis, simplifying procedures, and enabling personalized treatment regimens [5]. Research and development efforts have focused on creating automated techniques for nucleus segmentation, aiming to reduce manual labor and provide accurate and consistent findings. However, tissue structures are complex and staining patterns may vary, making nucleus segmentation challenging due to differences in staining patterns [6]. Traditional methods require significant human effort and are sensitive to changes in data evaluation. Therefore, we recommend avoiding these methods and developing automated systems that can effectively handle various image features and atypical conditions [7].
Tissue samples are more intricate due to the influence of several cell types, tissue compositions, and pathological conditions on the morphological characteristics and staining qualities of nuclei. Deep learning networks such as convolutional neural networks (CNNs) have demonstrated high efficiency in the processing of histopathology images, notably in tasks such as nucleus segmentation [8]. They possess distinctive attributes of both scalability and accuracy. The proliferation of multi-modal imaging techniques has led to an expansion in the range of computer approaches accessible for nucleus segmentation. Fluorescence microscopy and multi-stain histology are among the methods employed in this study. This advancement has facilitated the attainment of enhanced accuracy in localization and segmentation. In addition to the segmentation of nuclei, the analysis of histological pictures includes a range of tasks. These occupations encompass the classification of tissues, the identification of neoplasms, and the assessment of prognostic indicators. The integration of nucleus segmentation with subsequent analytical processes facilitates the acquisition of therapeutically valuable information that aids in illness diagnosis, prognosis assessment, and therapy selection [9].
Artificial intelligence and digital pathology are used to improve therapies and diagnose diseases. Understanding these technologies is crucial for their impact on scientific inquiry. Deep neural network topologies and annotated datasets are used in computer-aided detection systems like feature improvements [10], feature tracking [11], CT scans [12], TNBC liposomes [13], and Liver [14]. However, there is a shortage of pathologists in economically disadvantaged regions and countries, highlighting the need for more efficient cancer screening methods [15]. The study [16] evaluated the long-term therapeutic efficacy of anti-human epidermal growth factor receptor 2 (HER2)-targeted medications in combination with craniocerebral irradiation for patients with HER2-positive breast cancer.
The development of therapeutic and diagnostic techniques for breast cancer relies on accurate segmentation of cell nuclei in disease images. Machine learning and deep learning techniques are used to determine the borders of nuclei, while image processing methods like edge detection and segmentation algorithms are used. Data from the region of interest is collected to gather information about the nucleus’s size, shape, texture, color, and other important qualities. This approach leads to the development of pathomic parameters. Conventional methods often rely on human feature engineering, which can be time-consuming and not capture subtle patterns in data [17].
Breast cancer is one of the most epidemic malignancies affecting women worldwide. Timely diagnosis and identification are critical for improving survival rates. Histopathological investigation, which involves microscopic examination of tissue samples, is essential to ensure the diagnosis and evaluation of the severity of breast cancer. Histopathology aims to analyze tissue samples to detect abnormalities in structures like cell nuclei. Cellular dimensions, morphology, and structural irregularities often indicate the presence of cancer or other pathological conditions. By contrast, manually examining histological images is time-consuming, susceptible to human fallibility, and may provide contradictory findings from many observers. Thus, there is an increasing interest in developing automated systems capable of segmenting cells and detecting anomalies, which are essential for accurate diagnosis.
Contributions
Precise segmentation of cell nuclei in breast cancer images is crucial for the advancement of diagnostic and treatment algorithms. This proposed technique effectively reduces the transfer of non-essential features by controlling the transmission of information via skip connections from the contraction (encoder) to the expansion (decoder) side. The following are the contributions of this study:
• We propose a novel densely convolutional Breast Enhanced U-shaped Network (BU-NET) framework for classifying breast cancer multi-organ nuclei using histopathology slides. The BU-NET design follows the contraction path from the left side of the U shape, which contains convolution and Max pooling two-dimensional blocks for the extraction of spatial features. The expansion path from the right side functions as a decoder, merging the features and upsampling the blocks, while the center of the U shape compresses the bottleneck feature.
• We resized the images as per the model requirements, normalized the image pixels to increase the model training in the preprocessing and applied several image augmentation approaches like cropping, rotating, zooming, etc., to address the issue of overfitting and enhance the model’s performance.
• An evaluation of the framework’s effectiveness is conducted using numerous performance indicators, including recall, accuracy, precision, and F1 score. The achieved findings showed a substantial improvement in comparison to the existing methodologies.
Researchers use a range of conventional image processing, basic machine learning, and deep learning methods to automatically identify histological lesions and isolated nuclei in images of breast cancer. Clustering techniques such as thresholding, shape processes, and K-means segmentation kernels were included in an early approach. These approaches often encounter blurring, cell overlap, and uneven patterns, thereby resulting in imprecise outcomes.
2.1 Conventional Machine Learning
Researchers have used several machine learning methods such as Support Vector Machines (SVM) and Random Forests. Despite the enhanced efficiency, its generalisability to different datasets is limited due to its resource-intensive nature. Hu et al. [18] suggested an automated quantitative image analysis technique could enhance BC images. The top-bottom hat transformation’s goal was to optimize image quality for nuclei segmentation. By combining wavelet decomposition with multi-scale region-growing, it becomes possible to generate regions of interest sets, allowing for precise location estimation. They were able to classify cell nuclei by extracting 177 textural features from color spaces and four shape-based characteristics. Support vector machines generated the most optimal set of features. The study [19] introduced a novel segmentation method for the accurate localisation of nuclei in breast histopathology images stained with haematoxylin and eosin. This paper takes a loopy back propagation approach in a Markov random field to identify nucleus borders from the event map, and then it employs tensor voting to estimate nucleus event maps. Using this method, they were able to analyse whole slides and picture frames from breast cancer histopathology.
Accurate determination of cell counts on histological images is essential for cancer diagnosis, staging, and prognosis. Heuristic classification based on morphological features increases computer accuracy and reduces human diagnostic errors. The present study proposes a watershed segmentation technique that uses a two-stage embryo segmentation model to distinguish non-carcinoma and carcinoma in a dataset. After destaining, captured images of histology sections serve as the technique’s foundation [20]. Researchers in [21] discussed the early results of classifying cell nuclei using the Hausdorff distance. Using the K-Nearest Neighbors (KNN) Hausdorff distance, the authors achieved a classification accuracy rate of 75% for individual cell nuclei. This work [22] presented an automated breast cancer classification approach using support vector machine (SVM) classification based on the included features. We used two publicly available datasets: BreakHiZ and UCSB. It was also recommended that a classification comparison module using histopathology images be included to find the best classification for breast cancer detection. Artificial Neural Network (ANN), Random Forest (RF), SVM, and KNN are all part of this module.
ConDANet is a neural network that uses contourlet-based techniques to improve single-cell analysis in histological images. The main goal was to accurately identify and delineate the boundaries of individual nuclei. The network employs a unique attention mechanism and a sampling strategy to preserve information and extract the edge properties of nucleus areas. In paper [23], control signals are made using wavelet pooling, contourlet transform, and wavelet multi-scale time-frequency localization to keep the nuclei’s complex texture in histopathology pictures. The technique had been evaluated on three publicly available histopathological datasets, achieving dice scores of 88.9%, 81.71%, and 75.12%, surpassing state-of-the-art methods. This novel technique aims to enhance the precision of single-cell analysis in histopathology images. Computer-aided diagnosis (CAD) algorithmic method was developed to assist pathologists in assessing the density of cancer cells on breast histopathology slides [24]. An advanced neural network technique [25] was implemented to classify malignancies and detect and differentiate between cell nuclei using lung tissue scans. This study suggests that during model training, parts of the image that were not important were given less weight because the tumor type classifier segments and keeps the nuclei in the input image. By prioritizing the borders of the nuclear region to emphasize shape properties, this approach effectively mitigates the overall computational burden of training. In essence, it has a favorable influence on the accuracy of data classification. The convolution block attention modules (CBAM)-Residual deep learning architecture was used to extend the capabilities of the utility network (U-NEt). The main goal was to enhance the durability, accuracy, and flexibility of the generalized segmentation algorithm when applied to various tissue types and staining techniques. The design utilizes CBAM and a ResConv to analyze the deep and surface aspects of the images, respectively. The attention mechanism in the CBAM module helps it divide raw input patterns into useful parts by focusing on things like the shape, texture, and intensity of the cell nucleus. The suggested design requires a minimal number of trainable parameters, resulting in reduced computational and temporal costs. Three publicly available datasets, namely Triple-Negative Breast Cancer (TNBC), Data Science Bowl (DSB) 2018, and GlaS, underwent a comprehensive review and evaluation method. The results indicate that the model being evaluated surpasses existing cutting-edge techniques for accurately identifying cellular borders, particularly in terms of detailed segmentation [26].
The work [27] used pixel-level analysis to segment cell nuclei from histopathology images using the star convex polygon approach. The validation Intersection over Union (IoU) score of 87.56% demonstrates that the research shows this technique produces a segmentation conclusion that was more accurate. During the testing phase, this technique demonstrates a high degree of true positive rate and pixel-level shape correspondence detection. They offer a novel modified UNet architecture that makes use of spatial-channel attention and embeds ResNet blocks in the encoder layers. Tissue variability was solved because the UNet baseline efficiently maintains both large-scale and small-scale properties. Authors show that the proposed model may be applied to 20 distinct cancer sites, which was more than the number reported in the literature so far. When compared to craniotomy, this technique for draining intracerebral hemorrhage is effective and may lessen the risk of iatrogenic injuries [28].
Nuclei are segmented using the Densely Convolutional Spatial Attention Network (DCSA-Net) model, which was based on the utility network (U-NEt). Multi-Organ Nuclei Segmentation Challenge (MoNuSeg), an external multi-tissue dataset, was used to assess the model. Much data was needed in order to create deep learning systems that can segment nuclei effectively. Two hospitals provided hematoxylin and eosin-stained imaging data sets, which were used to train the model with a variety of nuclear appearances. The work proposes an improved method that outperforms existing algorithms for nuclei segmentation from two distinct image datasets. Additionally, the technique provides enhanced segmentation of cell nuclei. In order to ascertain the tumor-to-stroma ratio of invasive breast cancer, the researchers want to offer a thorough data collection of breast and prostate cancer tissue from a variety of individuals, utilizing a region-based segmentation technique. They intend to look at a segmentation technique based on an attention network with convolutional long-term and short-term memory. Although the method is a major advance over current ones, further study is required to increase the accuracy of nucleus segmentation. By adding certain improvements to DCSA-Net, the suggested model can perform better. In order to increase treatment planning and efficiency, the study looks at nuclei segmentation, employing nuclei of various sizes retrieved from multiscale images for training [29]. The optimal approach for achieving nucleus segmentation tasks involves the utilization of a two-stage deep learning network. The network employed in the initial step is utilized for coarse segmentation, whereas in the subsequent stage, it was employed for precise segmentation. In the context of nucleus segmentation, their methodology demonstrates superior performance compared to conventional network topologies [30].
The authors present two deep learning frameworks, USegTransformer-P and USegTransformer-S, designed for medical image segmentation. These frameworks help accelerate diagnosis processes and are valuable for medical practitioners. They also show the effectiveness of combining transformer-based encoding with FCN-based encoding and two distinct methods for combining transformer-based and FCN-based segmentation models [31]. The authors [32] used the R2U-NEt for the first time to partition nuclei using a publicly accessible dataset from the 2018 Data Science Bowl Grand Challenge. The model achieved a segmentation accuracy of around 92.15% during testing, indicating its resilience in effectively addressing the nuclei segmentation challenge. Deep learning algorithms have gained popularity for cell nuclei segmentation, surpassing traditional methods. CNN was used to autonomously acquire complex visual properties for tasks like segmentation, detection, and classification [33,34]. Dathar [35] employed semantic segmentation by transferring and optimizing learned representations from classification networks like GoogleNet, VGGNet, and AlexNet. End-to-end deep neural networks were employed for the first time to do semantic image segmentation. Medical image segmentation algorithms have progressed from manual to semi-automated to fully automated [36].
2.3 Existing Shortcomings and Proposed Solution
Recent approaches for separating cell nuclei include clustering methods such as K-means, thresholding, and shape functions. These methodologies often provide imprecise results as a result of geographical discrepancies, overlapping cells, and inconsistent patterns. The researchers used SVM and KNN classifiers to distinguish between the tissues. A significant contribution to poor performance is the presence of hand-crafted features, which limits the ability to generalize across various data sets. Recent years have seen a rise in the use of CNN and other deep learning-based approaches for the analysis of histopathology images. For segmentation and classification tasks, they used models like Residual Networks (ResNet), U-NEt, and VGG16. The variability of histopathological images may be attributed to differences in staining techniques, tissue preparation processes, and imaging medium. Existing models have poor generalisability due to their lack of resilience to these changes, especially those based on classical or hierarchical machine learning methodology. Many deep learning models lack transparency, providing high accuracy but limited comprehension of the underlying features that drive their predictions. Given that confidence in automated testing assessment techniques relies on interpretation, the absence of transparency in this regard may hinder the evaluation process.
The proposed method improves cell segmentation by using a deep learning architecture that integrates the BUNet model, attention techniques, and skipped connections. In order to address problems such as overlapping cells and uncertain boundaries, the model may use the attention mode to highlight relevant regions of the image. To address the breast tissue image decomposition model, we conduct training on an enhanced dataset that considers both cancer features and tissue architecture. The generalisability of the model to different clinical settings is ensured. A novel strategy is proposed to tackle the problem of group heterogeneity by integrating adaptive data augmentation techniques with a loss function. This method is especially advantageous for complex classification problems and diverse classes. The proposed methodology eliminates the limitations of previous methods while simultaneously improving classification accuracy and the ability to distinguish between histopathological images.
The methodology presented in this study encompasses many stages, including dataset preparation, normalization, augmentation, the highly convolutional BU-NET framework, and key evaluation measures. The subsections provide a detailed description of the sequential stages of the proposed method. The flow diagram is presented in Fig. 1.

Figure 1: Flow diagram for the proposed framework. This research presents a technique that incorporates a number of steps, such as the preparation of the dataset, the normalization of the dataset, the augmentation of the dataset, the highly convolutional BU-NET framework, and fundamental evaluation metrics
Fluorescence and brightfield microscopy are often used as microscopy methods for examining individual molecules and cellular architecture. Nevertheless, these approaches provide images that exhibit diverse contrast, color statistics, and intensity levels. In order to address these disparities, it is necessary to convert these images into ones that possess uniform intensity and color statistics. The normalization of stains is an essential procedure in the preparation of tissue engineering, and the quality of histopathological images is affected by several aspects. Computational pathology approaches use CNN to employ various strategies for the detection, classification, and segmentation of images, depending on their texture and color [37]. Here are a few preparation steps used in this work:
• To get the desired size, the image, and the associated labeled mask must first be scaled. This is significant since constant input measurements are often needed for deep learning models. Different-sized images and masks may be used, but the model’s fit can be assessed by scaling them to the identical target size (for example,
• Following resizing, the image will shrink back to its original size. In the majority of image collections, pixel values vary from 0 to 255. Normalization converts pixel data into the range [0, 1] by tightly limiting all input values to a small and consistent range, therefore facilitating quicker learning of the model. Perform a normalization by dividing each pixel value by 255. An essential stage in the training process is to fine-tune the model by modifying the input level.
• Following resizing and normalization, the ultimate step involves transforming the image and labeled mask into Numpy arrays. This modification is particularly noteworthy since several deep learning frameworks, like TensorFlow and PyTorch, need inputs in the form of Numpy arrays to facilitate fast data processing and block processing. Numpy arrays provide efficient computations and offer the necessary adaptability for training the models.
CNN algorithms regularly demonstrate efficacy in tumor segmentation during the pre-processing phase; however, this efficacy is contingent upon the implementation of stain normalization. Vahadane et al. [38] introduced a technique for color normalization that preserves the original image’s structure while adjusting the color to meet the requirements of the target domain. This approach is crucial for doing more in-depth image analysis and provides exceptional results in the stain normalization procedure. A stain normalization methodology was used for nuclei segmentation, providing two methods to prevent irregularities in the staining process and enhance the precision of quantitative analysis. A dense BU-NET framework is shown in Algorithm 1.
Data augmentation refers to the act of incorporating more data samples into an already existing dataset. Deep learning techniques benefit from data augmentation by generating novel and distinct instances for training datasets, hence enhancing their efficiency and performance. In order for a learning model to operate efficiently and accurately, it must have access to a large and sufficient dataset. Random augmentation packets were used in this work to supplement the data. We first rotated the image at its centre, cropped it, and then, by tracking a line that extended horizontally or vertically, we reversed the mirrored image. The two main filters used are sharpness and blur. Blurring obscures intricate features and boundaries, whereas sharpening enhances them.
3.3 Densely Convolutional BU-NET Framework
Thomas Brox, Olaf Ronneberger, and Philipp Fischer created the U-NEt architecture in 2015 with the intention of segmenting biological images using CNN [39]. The design comprises a diminishing encoder route and an expanding decoder path. The U-shaped building is called such because of its arrangement in the form of the letter U. The expanding pathway is used to merge localization data with the segmentation map, while the contracting pathway gathers background information and creates a feature representation of the input image [40]. The contracting path, often referred to as the encoder, is an essential component of the U-NEt design. It is responsible for extracting hierarchical characteristics from input images while simultaneously decreasing spatial resolution. A densely convolutional BU-NET framework is shown in Fig. 2, and dense blocks for breast cancer are in Fig. 3.

Figure 2: Densely convolutional BU-NET framework

Figure 3: Dense blocks for the breast cancer

The system employs convolutional layers to extract information by using adaptable filters, progressively representing input in more intricate structures. Each convolutional layer is followed by the addition of a rectified linear unit (ReLU) activation function. This is done to introduce non-linearity and facilitate the identification of intricate connections. The Contracting Path additionally uses max-pooling layers to decrease the spatial dimensions of feature maps while retaining crucial attributes. This procedure partitions each feature map into non-overlapping segments, with the exception of the maximum value inside each segment. The approach uses convolutional layers and max-pooling processes to reduce the input size, acquiring local and global information. It simplifies segmentation by encoding abstract representations, decreasing spatial resolution, and improving feature channels.
The decoder, also known as the enlarged route, is a crucial component in BU-NEt design, responsible for acquiring geographical data and producing high-resolution segmentation maps. It uses convolutional processes, interpolation algorithms, and upsampling layers to extract the spatial dimensions of feature maps and align them with the initial supplied picture. The decoder uses skip connections to construct direct connections between layers representing contraction and expansion stages, enabling the network to obtain high-resolution feature maps from earlier system phases. The network’s accuracy in segmentation and localization operations can be attributed to its properties.
In Eq. (1), Conv indicates convolutional, ifm indicates input feature map, bt indicates bias term in the architecture, and ReLU indicates the rectified linear unit and convolutional filter (CF).
Eq. (2) concatenate the contraction and expansion paths of feature inputs.
Eq. (3) presents the last layer where ifm shows the input feature map of the layer and others indicate the sum of all feature maps.
In Eq. (4), cross-entropy loss is used, where
Upsampling is performed on each individual construction element that makes up the Expansive Path. It is then followed by the application of two
In order to overcome the limitations of U-NEt, we introduce BU-NEt, an extension of the U-Net architecture that incorporates a bottom-up feature extraction approach. The primary objective of BU-NEt is to enhance the model’s capacity to acquire and analyze more intricate features, all the while preserving an efficient working representation. The bottom-up method of BU-NEt is designed to enhance the administration of intricate infrastructure, the process of service acquisition, and the execution of services inside the network.
Within the U-NeT architecture, the encoder first decreases the spatial parameters during the feature learning process, while the decoder subsequently enhances these factors to reconstruct the image. Yet, this conventional hierarchical method becomes less effective as the network becomes more complex. BU-NEt addresses this issue by using a meticulously designed feature extraction technique that ensures the collection and propagation of extremely detailed information across the network.
An evaluation of the effectiveness of the proposed approach was conducted using Google Colab and two datasets obtained from open source. This section evaluates the comparison analysis and other performance indicators.
The Triple Negative Breast Cancer (TNBC) cell histopathological dataset is a collection of digitized histopathology images of breast cancer tissue samples, aiming to aid in the development of algorithms for identifying and categorizing different subtypes of breast cancer [41]. The dataset includes 81 digital images from individuals diagnosed with triple-negative breast cancer, which lacks the human epidermal growth factor receptor 2, progesterone receptor, and estrogen receptor. The collection includes fifty images and four thousand annotated cells, including invasive carcinomatous cells, fibroblasts, endothelial cells, adipocytes, macrophages, and inflammatory cells. Images and their predicted masks using TNBC data are presented in Fig. 4.

Figure 4: Images and its predicted masks using TNBC dataset
The second dataset analyzed in this paper is the Multi-Organ Nuclei Segmentation Challenge (MoNuSeg). The collection consists of a diverse range of tissue slides stained with hematoxylin-eosin (HE), totaling around 22,000 images of cell nuclei. Boundary annotations are supplied for training reasons. Additionally, there are 14 photographs available for testing, each including 7000 nuclear boundary annotations. The histology photographs in this collection have a resolution of

Figure 5: Images and its predicted masks using MoNuSeg dataset
The main objective of this work is to develop a deep-learning model that can accurately segment histopathology images. MoNuSeg and TNBC datasets were perfectly designed for this paradigm. One of the many stages of the process involves preparing data, selecting a model architecture, fine-tuning hyperparameters, training, and assessing performance. The suggested approach focuses on addressing each aspect individually to ensure accurate and reliable segmentation results. To ensure that the datasets’ input sizes and image resolutions remain constant, the initial steps involve making precise adjustments. On a positive note, the pictures obtained via MoNuSeg were resized to
The ability of a model to generalize is enhanced by increasing the variety of training data through the use of data augmentation procedures. The model is made more resistant to changes in input data by simulating many points of view. Improving the neural network’s performance relies heavily on well-chosen hyperparameters. Hyperparameters such as learning rate, optimizer selection, and decay rate are fine-tuned by empirical study. Here, we employ the Adam optimizer with a 0.0001 learning rate. The goal of training is to reduce segmentation loss as much as possible by repeatedly tweaking the model parameters with the help of the training dataset. To make sure the model learned optimally and converged, it was trained for 150 epochs. Faster training and more efficient computation were the results of the hardware accelerator, which was the Google Cloud Platform. The model’s accuracy, precision, recall, and overall performance are evaluated by looking at how well it performs on the datasets.
Accuracy is the number of properly identified pixels as a percentage of the total number of pixels in an image. This includes both true positive cases (TPC) and false negative cases (TNC).
Precision, which is also called positive predictive value, is a number that shows how many true positive cases (TPC) there are out of all the positive estimates the model makes. This is a number that shows how many properly labeled true positive cases there were compared to the total number of true positive cases in the ground truth.
The specificity of a model measures its ability to accurately identify true negative cases. This implies that a portion of accurately detected negative data will be inaccurately interpreted as positive; this occurrence is referred to as a false positive case (FPC).
To calculate the recall, divide the total of all positive samples by the number of samples that were correctly recognized. A recall metric quantifies the accuracy of a model in correctly detecting positive samples. Identification of additional high-quality samples gets increasingly straightforward as recall improves.
The harmonic mean of precision and recall is used to figure out the F1 score [43].
4.4 Comparative Analysis Using MoNuSeg Dataset
The comparison of several U-NET-based techniques employing the MoNuSeg dataset is shown in Table 1. The U-NET single approach achieved an average accuracy of 74.983%, precision of 75.014%, recall of 74.757%, and an F1 score of 74.885%. The VGG-16 U-NET model achieved an average accuracy of 77.353%, precision of 77.067%, recall of 76.453%, and a F1 score of 76.758%. The ResNet-50 U-NET model achieved an average accuracy of 81.532%, precision of 82.578%, recall of 80.389%, and a F1 score of 81.469%. The Inception-V3 U-NET model achieved an average accuracy of 79.645%, precision of 80.842%, recall of 81.934%, and a F1 score of 81.384%. The Mask R CNN achieved an average accuracy of 72.431%, precision of 72.368%, recall of 73.797%, and a F1 score of 73.076%.

The Random Forest model achieved an average accuracy of 66.984%, precision of 66.854%, recall of 65.283%, and a F1 score of 67.454%. The EfficientNet-B0 U-NET achieved an average accuracy of 84.263%, precision of 87.460%, recall of 86.858%, and a F1 score of 87.153%. The proposed method attained an average accuracy of 88.783%, a precision of 89.753%, a recall of 88.034%, and an F1 score of 88.885%. Comparative analysis using several performance measures with MoNuSeg data is shown in Fig. 6.

Figure 6: Comparative analysis using several performance measures with TNBC data
4.5 Comparative Analysis Using TNBC Dataset
The comparative analysis of several U-NET-based methods using the TNBC dataset is represented in Table 2. The U-NET single method attained 76.432% average accuracy, 77.235% precision, 76.346% recall, and 76.787% F1 score. VGG-16 U-NET attained 78.675% average accuracy, 77.565% precision, 79.564% recall, and a 78.551% F1 score. ResNet-50 U-NET attained 85.235% average accuracy, 86.346% precision, 86.898% recall, and an 86.621% F1 score. Inception-V3 U-NET attained 87.568% average accuracy, 85.768% precision, 81.934% recall, and an 86.773% F1 score. Mask R CNN attained 72.457% average accuracy, 74.346% precision, 75.235% recall, and 74.787% F1 score. Random Forest attained 73.764% average accuracy, 73.564% precision, 74.176% recall, and 73.868% F1 score. EfficientNet-B0 U-NET attained 85.343% average accuracy, 84.897% precision, 86.824% recall, and an 85.849% F1 score.
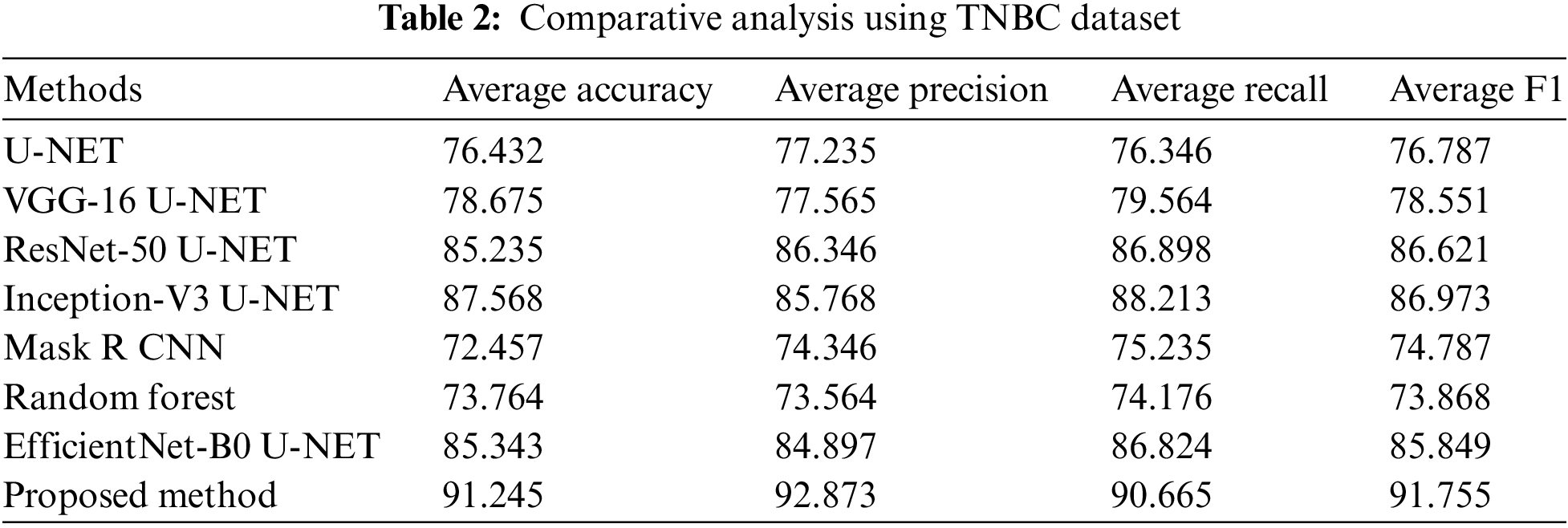
The proposed method attained 91.245% average accuracy, 92.873% precision, 90.665% recall, and 91.755% F1 score. Comparative analysis using several performance measures with TNBC data is shown in Fig. 7.
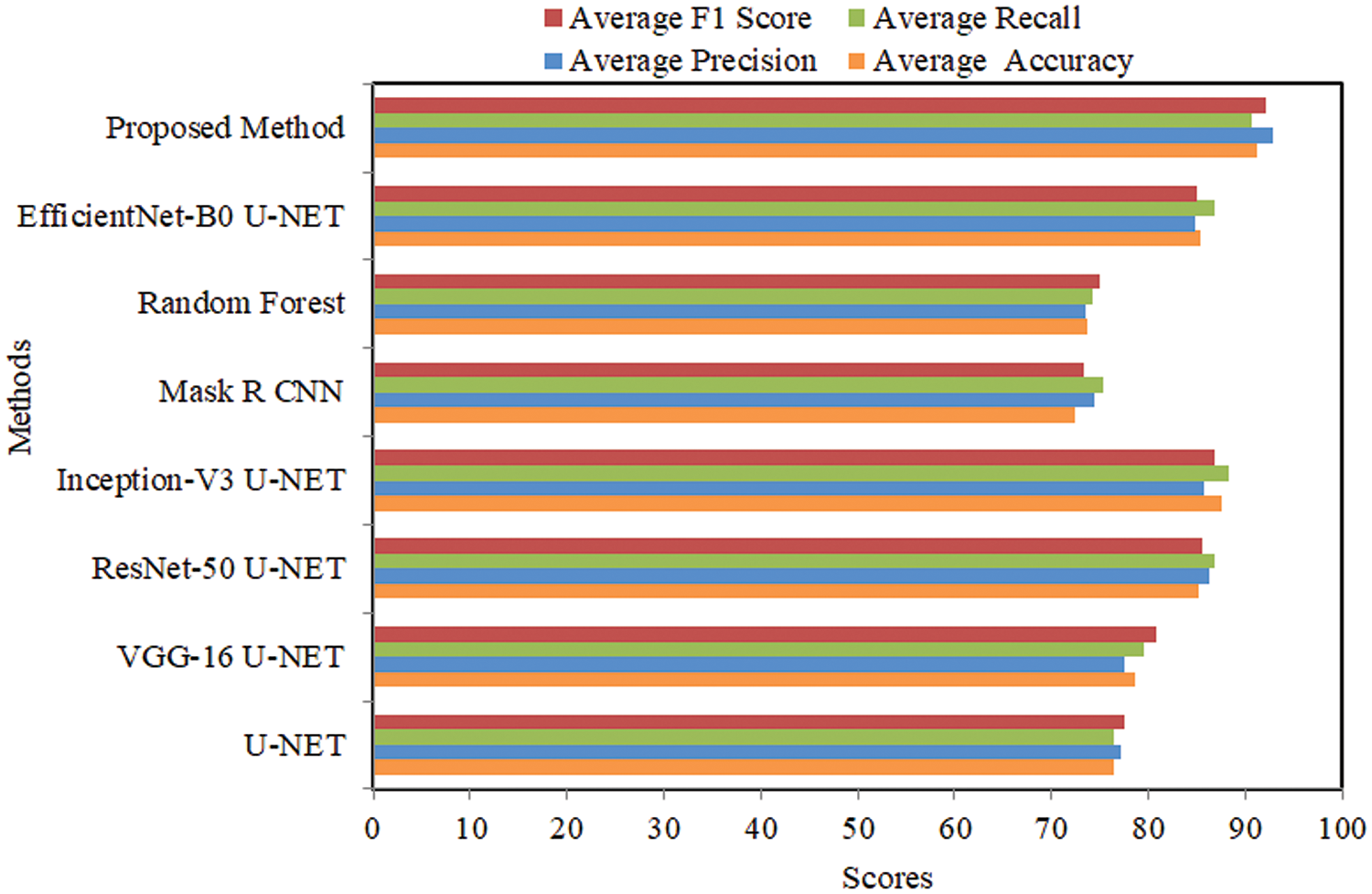
Figure 7: Comparative analysis using several performance measures wit MoNuSeg data
Validation accuracy curves are crucial in assessing the effectiveness of a neural network model during training epochs on a validation dataset. These curves show how the model’s accuracy evolves over time, detecting whether it is overfitting or underfitting. Optimization of parameters during an epoch, or full loop around the training dataset, is a common approach to enhancing performance. The validation accuracy curve is illustrated in Fig. 8. The validation dataset, a subset of the dataset, is used to evaluate the model’s performance during training. The test aims to determine if the model can acquire new information independently and identify issues like inadequate or excessive fitting. The validation accuracy curve, represented by each point on the curve, is determined by applying the model to the validation dataset after completing a certain number of training epochs. The accuracy may rise in the early stages of training as the model generalizes from its training set. If it stays the same or drops as training progresses, it may indicate that the training program is too suitable for the individual.
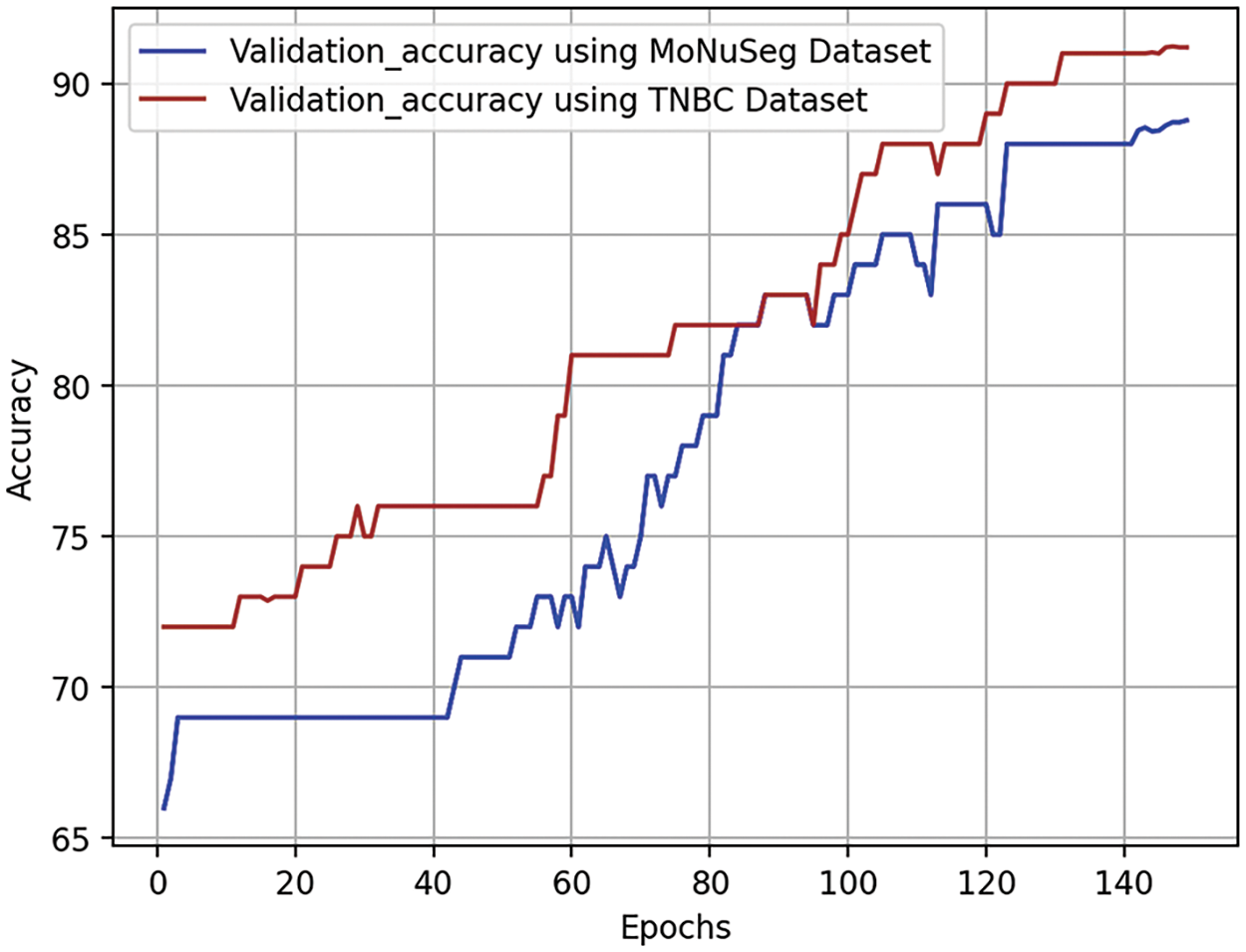
Figure 8: Visual representation of validation curves for densely convolutional BU-NET framework
The continuous growth of validation accuracy without reaching a ceiling indicates that the model is learning well from the training set and performing well on new data.
One way to evaluate a binary classification model’s efficacy across different threshold values is via a receiver operating characteristic curve, often known as an ROC curve. To find out if an event is positive or negative, it analyzes the true positive rate and false positive rate at different levels. ROC curves for densely convolutional BU-NET framework are shown in Fig. 9. A ROC curve is obtained by comparing TPR to FPR over different thresholds. The Area under the ROC Curve (AUC) is a metric used to measure the overall effectiveness of a binary classification model. A 0.5 AUC indicates model accuracy is comparable to random speculation, while a 1.0 AUC indicates flawless classification.

Figure 9: Visual representation of ROC curves for densely convolutional BU-NET framework
One very efficient approach for evaluating the efficacy of a model is to apply masks to actual data. In the context of image classification tasks, the ground truth refers to the tangible feature generated by reliable sources, whereas the used feature is the numerical outcome. To generate the overlay predicted mask, we take both ground truth and predicted masks and align them with the original image. By precisely overlaying two masks over the same image, a projection is produced as presented in Fig. 10. To efficiently distinguish between the ground truth and prediction masks, a third color, such as white, may be used by assigning them separate hues (e.g., blue for prediction and green for ground truth). The operational effectiveness of the locations and their shortcomings may be clearly seen using this prediction model. We will promptly identify any absent or extensively scattered areas. With the aim of helping the program identify areas for improvement, this method provides a rapid and straightforward means of evaluating strengths and weaknesses.
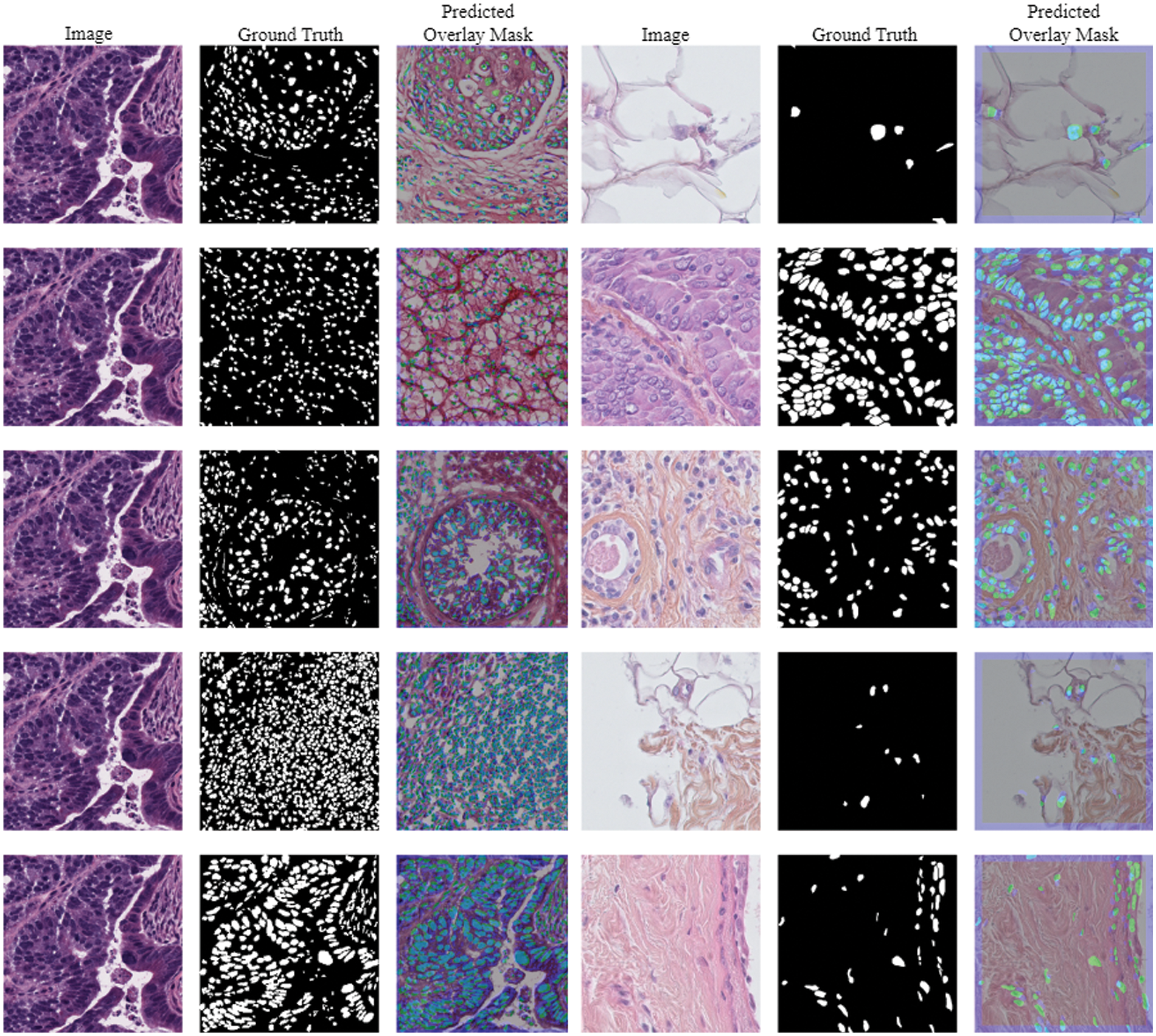
Figure 10: Visual difference between overlay predicted mask with ground truth data
The proposed BU-NEt database eliminates the need for including deep and costly computational layers often seen in traditional deep learning models, due to its highly structured feature extraction approach. In this systematic approach, the need for useless computations is reduced.
U-NEt requires more processing resources than BU-NEt because of its complex encoding and decoding structure and many parameters. However, BU-NEt maintains comparable or superior performance with fewer training parameters and more efficient processing. The results of our studies indicate that BU-NEt maintains or improves segmentation accuracy while the proposed model takes 179 to 230 s per step, depending upon the resources. The time savings and decreased computing cost make BU-NEt more suitable for applications in resource-constrained environments. In addition, BU-NEt reduces the amount of data that has to be processed at each network level. Implementing large-scale applications may result in significant cost savings because of the reduced memory and processing power required.
4.9 Comparison with Existing Particular Studies
This work conducted a comparative analysis of the proposed model with existing studies, particularly that focused on the TNBC and MoNuSeg datasets. Using the same two datasets as our approaches, Imtiaz et al. [23] used a Wavelet-based pooling technique to segment the cancer border as illustrated in Table 3. They obtained an accuracy of 79.9% for the MoNuSeg dataset and 72.0% for the TNBC dataset. The artificial network developed by Jaisakthi et al. [25] based on NuCLS obtained a relatively low accuracy rate of 77%. Using identical TNBC data, Shah et al. [26] developed a CBAM-Residual u-shaped network and attained a precision of 82.2%. Furthermore, another research [44] used the EfficietNet-based U-NEt approach with identical TNBC data and obtained a very low F1 score of 56.1%. The previous research used intricate U-NEt and artificial networks, which resulted in higher computational expenses and time needed and yielded poor performance for segmentation tasks. The proposed method demonstrated exceptional accuracy and higher performance metrics for both datasets when compared to other studies.
The current study utilized a densely convolutional BU-NET architecture to effectively segregate breast cancer. This was achieved by the implementation of various preprocessing approaches and advanced augmentation methods. The study utilizes two extremely valuable datasets, namely TNBC and MoNuSeg, which pertain to breast cancer. The results of the study are assessed using essential performance metrics such as accuracy, precision, recall, F1 score, validation accuracy, and ROC curves. Furthermore, a comparison analysis is performed on the present results of the BU-NET research in relation to the most recent state-of-the-art studies. The results of this study indicate that the proposed method attained an accuracy of 91.24% when applied to the TNBC dataset and 88.78% when applied to the MoNuSeg dataset. This study’s approach demonstrated an ROC-AUC of 90.18 for the MuNuSeg dataset and 93.92 for the TNBC dataset, signifying superior performance. In the future, there will be an increase in the collection and integration of datasets in order to improve performance. We will then employ advanced feature engineering and segmentation approaches.
Acknowledgement: This research was supported by Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. The authors would also like to acknowledge the support of Prince Sultan University, Riyadh, Saudi Arabia.
Funding Statement: This research was funded by Princess Nourah bint Abdulrahman University and Researchers supporting Project number (PNURSP2024R346), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Muhammad Mujahid, Amjad Rehman; data collection: Tanzila Saba, Faten S Alamri; analysis and interpretation of results: Tanzila Saba, Muhammad Mujahid, Robertas Damasevicius; draft manuscript preparation: Amjad Rehman, Muhammad Mujahid; supervision: Robertas Damasevicius, Amjad Rehman; project administration: Faten S Alamri. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets that were used in the experimentation are cited in References [41,42]. The datasets are also available at the following links: TNBC: https://www.kaggle.com/datasets/mahmudulhasantasin/tnbc-nuclei-segmentation-original-dataset. Monuseg: https://www.kaggle.com/datasets/tuanledinh/monuseg2018, accessed on 09 October 2024.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Yin L, Duan JJ, Bian XW, Sc Yu. Triple-negative breast cancer molecular subtyping and treatment progress. Breast Cancer Res. 2020;22(1):1–13. doi:10.1186/s13058-020-01296-5 [Google Scholar] [PubMed] [CrossRef]
2. Ogundokun RO, Misra S, Douglas M, Damaševičius R, Maskeliūnas R. Medical internet-of-things based breast cancer diagnosis using hyperparameter-optimized neural networks. Future Internet. 2022;14(5):153. doi:10.3390/fi14050153. [Google Scholar] [CrossRef]
3. Shokooh K, Mahsa F, Jeong S. Bio-inspired and smart nanoparticles for triple negative breast cancer microenvironment. Pharmaceutics. 2021;13(2):1–24. doi:10.3390/pharmaceutics13020287 [Google Scholar] [PubMed] [CrossRef]
4. Obayya M, Maashi MS, Nemri N, Mohsen H, Motwakel A, Osman AE, et al. Hyperparameter optimizer with deep learning-based decision-support systems for histopathological breast cancer diagnosis. Cancers. 2023;15(3):885. doi:10.3390/cancers15030885 [Google Scholar] [PubMed] [CrossRef]
5. Ghosh S, Guha P. A rare case report of acanthomatous ameloblastoma based on aspiration cytology with histopathological confirmation. CytoJournal. 2023;20. doi:10.25259/Cytojournal. [Google Scholar] [CrossRef]
6. Huang PW, Ouyang H, Hsu BY, Chang YR, Lin YC, Chen YA, et al. Deep-learning based breast cancer detection for cross-staining histopathology images. Heliyon. 2023;9(2):e13171. doi:10.1016/j.heliyon.2023.e13171 [Google Scholar] [PubMed] [CrossRef]
7. Chan RC, To CKC, Cheng KCT, Yoshikazu T, Yan LLA, Tse GM. Artificial intelligence in breast cancer histopathology. Histopathology. 2023 Jan;82(1):198–210. doi:10.1111/his.14820 [Google Scholar] [PubMed] [CrossRef]
8. Kaur A, Kaushal C, Sandhu JK, Damaševičius R, Thakur N. Histopathological image diagnosis for breast cancer diagnosis based on deep mutual learning. Diagnostics. 2024;14(1):1–16. doi:10.3390/diagnostics14010095 [Google Scholar] [PubMed] [CrossRef]
9. Ding K, Zhou M, Wang H, Gevaert O, Metaxas D, Zhang S. A large-scale synthetic pathological dataset for deep learning-enabled segmentation of breast cancer. Sci Data. 2023 Apr;10(1):231. doi:10.1038/s41597-023-02125-y [Google Scholar] [PubMed] [CrossRef]
10. Liu Y, Tian J, Hu R, Yang B, Liu S, Yin L, et al. Improved feature point pair purification algorithm based on SIFT during endoscope image stitching. Front Neurorobot. 2022;16:840594. doi:10.3389/fnbot.2022.840594 [Google Scholar] [PubMed] [CrossRef]
11. Lu S, Liu S, Hou P, Yang B, Liu M, Yin L, et al. Soft tissue feature tracking based on deep matching network. Comput Model Eng Sci. 2023;136(1):363–79. doi:10.32604/cmes.2023.025217. [Google Scholar] [CrossRef]
12. Yang C, Sheng D, Yang B, Zheng W, Liu C. A dual-domain diffusion model for sparse-view ct reconstruction. IEEE Signal Process Lett. 2024;31:1279–128. doi:10.1109/LSP.2024.3392690. [Google Scholar] [CrossRef]
13. Lan J, Chen L, Li Z, Liu L, Zeng R, He Y, et al. Multifunctional biomimetic liposomes with improved tumor-targeting for TNBC treatment by combination of chemotherapy, anti-angiogenesis and immunotherapy. Adv Healthc Mater. 2024;11:2400046. doi:10.1002/adhm.202400046 [Google Scholar] [PubMed] [CrossRef]
14. Sun T, Lv J, Zhao X, Li W, Zhang Z, Nie L. In vivo liver function reserve assessments in alcoholic liver disease by scalable photoacoustic imaging. Photoacoustics. 2023;34(8):100569. doi:10.1016/j.pacs.2023.100569 [Google Scholar] [PubMed] [CrossRef]
15. Shafi S, Parwani AV. Artificial intelligence in diagnostic pathology. Diagn Pathol. 2023;18(1):109. doi:10.1186/s13000-023-01375-z [Google Scholar] [PubMed] [CrossRef]
16. Tang L, Zhang W, Chen L. Brain Radiotherapy combined with targeted therapy for HER2-positive breast cancer patients with brain metastases. Breast Cancer: Targets Ther. 2024;16:379–92. doi:10.2147/BCTT.S460856 [Google Scholar] [PubMed] [CrossRef]
17. Shihabuddin AR, Sabeena BK. Multi CNN based automatic detection of mitotic nuclei in breast histopathological images. Comput Biol Med. 2023 May;158:106815. doi:10.1016/j.compbiomed.2023.106815 [Google Scholar] [PubMed] [CrossRef]
18. Hu H, Qiao S, Hao Y, Bai Y, Cheng R, Zhang W, et al. Breast cancer histopathological images recognition based on two-stage nuclei segmentation strategy. PLoS One. 2022;17(4):e0266973. doi:10.1371/journal.pone.0266973 [Google Scholar] [PubMed] [CrossRef]
19. Paramanandam M, O’Byrne M, Ghosh B, Mammen JJ, Manipadam MT, Thamburaj R, et al. Automated segmentation of nuclei in breast cancer histopathology images. PLoS One. 2016;11(9):e0162053. doi:10.1371/journal.pone.0162053 [Google Scholar] [PubMed] [CrossRef]
20. Wang P, Hu X, Li Y, Liu Q, Zhu X. Automatic cell nuclei segmentation and classification of breast cancer histopathology images. Signal Process. 2016;122(5):1–13. doi:10.1016/j.sigpro.2015.11.011. [Google Scholar] [CrossRef]
21. Skobel M, Kowal M, Korbicz J. Breast cancer computer-aided diagnosis system using k-NN algorithm based on hausdorff distance. In: Current Trends in Biomedical Engineering and Bioimages Analysis, University of Zielona Góra, Poland: Springer; 2020. p. 179–88. doi:10.1007/978-3-030-29885-2_16. [Google Scholar] [CrossRef]
22. Aswathy M, Jagannath M. An SVM approach towards breast cancer classification from H&E-stained histopathology images based on integrated features. Med Biolog Eng Comput. 2021;59(9):1773–83. doi:10.1007/s11517-021-02403-0 [Google Scholar] [PubMed] [CrossRef]
23. Imtiaz T, Fattah SA, Saquib M. Contourlet driven attention network for automatic nuclei segmentation in histopathology images. IEEE Access. 2023;11:129321–30. doi:10.1109/ACCESS.2023.3321799. [Google Scholar] [CrossRef]
24. Altini N, Puro E, Taccogna MG, Marino F, De Summa S, Saponaro C, et al. Tumor cellularity assessment of breast histopathological slides via instance segmentation and pathomic features explainability. Bioengineering. 2023 Mar;10(4):1–18. [Google Scholar]
25. Jaisakthi SM, Desingu K, Mirunalini P, Pavya S, Priyadharshini N. A deep learning approach for nucleus segmentation and tumor classification from lung histopathological images. Netw Model Anal Health Inform Bioinform. 2023;12(1):4551. doi:10.1007/s13721-023-00417-2. [Google Scholar] [CrossRef]
26. Shah HA, Kang JM. An optimized multi-organ cancer cells segmentation for histopathological images based on CBAM-residual U-net. IEEE Access. 2023;11:111608–21. doi:10.1109/ACCESS.2023.3295914. [Google Scholar] [CrossRef]
27. Nelson AD, Krishna S. An effective approach for the nuclei segmentation from breast histopathological images using star-convex polygon. Procedia Comput Sci. 2023;218(3):1778–90. doi:10.1016/j.procs.2023.01.156. [Google Scholar] [CrossRef]
28. Zhang C, Ge H, Zhang S, Liu D, Jiang Z, Lan C, et al. Hematoma evacuation via image-guided para-corticospinal tract approach in patients with spontaneous intracerebral hemorrhage. Neurol Ther. 2021;10(2):1001–13. doi:10.1007/s40120-021-00279-8 [Google Scholar] [PubMed] [CrossRef]
29. Rashadul S, Bhattacharjee YB, Hwang H, Rahman HC, Kim WS, Ryu DM, et al. Densely convolutional spatial attention network for nuclei segmentation of histological images for computational pathology. Front Oncol. 2023;13:1009681. doi:10.3389/fonc.2023.1009681 [Google Scholar] [PubMed] [CrossRef]
30. Jian P, Kamata SI. A two-stage refinement network for nuclei segmentation in histopathology images. In: 2022 4th International Conference on Image, Video and Signal Processing, 2022; New York, NY, USA: ACM; p. 8–13. [Google Scholar]
31. Dhamija T, Gupta A, Gupta S, Anjum, Katarya R, Singh G. Semantic segmentation in medical images through transfused convolution and transformer networks. Appl Intell. 2023;53(1):1132–48. doi:10.1007/s10489-022-03642-w [Google Scholar] [PubMed] [CrossRef]
32. Alom MZ, Yakopcic C, Taha TM, Asari VK. Nuclei segmentation with recurrent residual convolutional neural networks based U-net (R2U-net). In: NAECON 2018-IEEE National Aerospace and Electronics Conference, 2018; Dayton, OH, USA: IEEE; p. 228–33. doi:10.1109/NAECON.2018.8556686. [Google Scholar] [CrossRef]
33. Kowal M, Skobel M, Gramacki A, Korbicz J. Breast cancer nuclei segmentation and classification based on a deep learning approach. Int J Appl Math Comput Sci. 2021;31(1):85–106. doi:10.34768/amcs-2021-0007. [Google Scholar] [CrossRef]
34. Krishnakumar B, Kousalya K. Optimal trained deep learning model for breast cancer segmentation and classification. Inf Technol Control. 2023;52(4):915–34. [Google Scholar]
35. Dathar H. A modified convolutional neural networks model for medical image segmentation. TEST Eng Manage. 2020;83:16798–808. [Google Scholar]
36. Shang H, Zhao S, Du H, Zhang J, Xing W, Shen H. A new solution model for cardiac medical image segmentation. J Thorac Dis. 2020 Dec;12(12):7298–312 [Google Scholar] [PubMed]
37. Robin M, John J, Ravikumar A. Breast tumor segmentation using U-NET. In: 2021 5th International Conference on Computing Methodologies and Communication (ICCMC), 2021, Erode, India: IEEE; p. 1164–7. doi:10.1109/ICCMC51019.2021.9418447. [Google Scholar] [CrossRef]
38. Vahadane A, Peng T, Albarqouni S, Baust M, Steiger K, Schlitter AM, et al. Structure-preserved color normalization for histological images. In: 2015 IEEE 12th International Symposium on Biomedical Imaging (ISBI), 2015; Brooklyn, NY, USA: IEEE; p. 1012–5. doi:10.1109/ISBI.2015.7164042. [Google Scholar] [CrossRef]
39. Soulami KB, Kaabouch N, Saidi MN, Tamtaoui A. Breast cancer: one-stage automated detection, segmentation, and classification of digital mammograms using UNet model based-semantic segmentation. Biomed Signal Process Control. 2021;66:102481. [Google Scholar]
40. Baccouche A, Garcia-Zapirain B, Castillo Olea C, Elmaghraby AS. Connected-UNets: a deep learning architecture for breast mass segmentation. npj Breast Cancer. 2021;7(1):151 [Google Scholar] [PubMed]
41. Naylor P, Laé M, Reyal F, Walter T. Segmentation of nuclei in histopathology images by deep regression of the distance map. IEEE Trans Med Imaging. 2018;38(2):448–59. doi:10.1109/TMI.2018.2865709 [Google Scholar] [PubMed] [CrossRef]
42. Kumar N, Verma R, Anand D, Zhou Y, Onder OF, Tsougenis E, et al. A multi-organ nucleus segmentation challenge. IEEE Trans Med Imaging. 2019;39(5):1380–91. doi:10.1109/TMI.2019.2947628 [Google Scholar] [PubMed] [CrossRef]
43. Alam T, Shia WC, Hsu FR, Hassan T. Improving breast cancer detection and diagnosis through semantic segmentation using the Unet3+ deep learning framework. Biomedicines. 2023;11(6):1536. doi:10.3390/biomedicines11061536 [Google Scholar] [PubMed] [CrossRef]
44. Dinh TL, Kwon SG, Lee SH, Kwon KR. Breast tumor cell nuclei segmentation in histopathology images using EfficientUnet++ and multi-organ transfer learning. J Korea Multimed Soc. 2021;24(8):1000–11. [Google Scholar]
45. Mahmood T, Owais M, Noh KJ, Yoon HS, Koo JH, Haider A, et al. Accurate segmentation of nuclear regions with multi-organ histopathology images using artificial intelligence for cancer diagnosis in personalized medicine. J Pers Med. 2021;11(6):515. doi:10.3390/jpm11060515 [Google Scholar] [PubMed] [CrossRef]
46. Kanadath A, Angel Arul Jothi J, Urolagin S. Multilevel multiobjective particle swarm optimization guided superpixel algorithm for histopathology image detection and segmentation. J Imaging. 2023;9(4):78. doi:10.3390/jimaging9040078 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF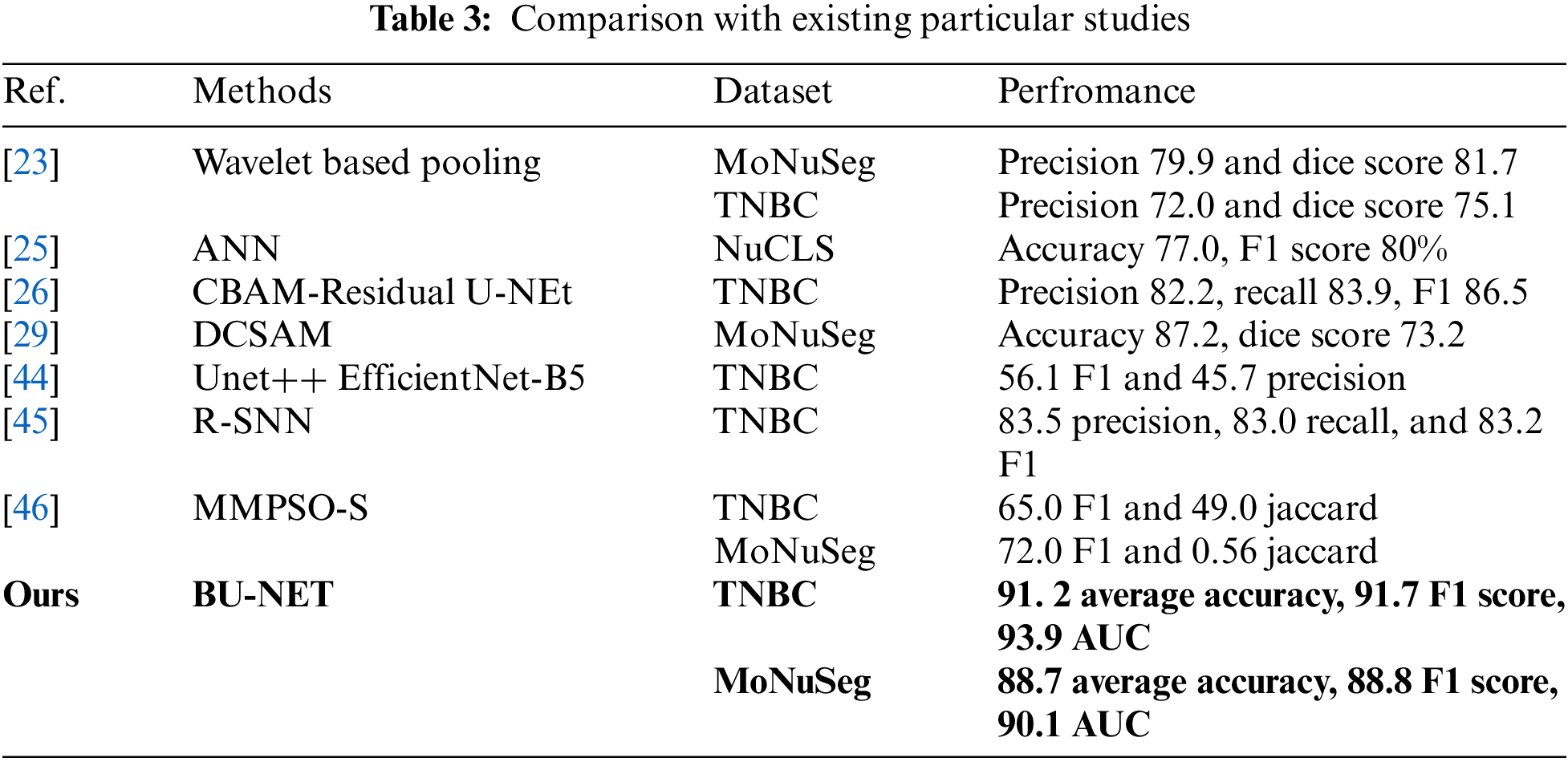
 Downloads
Downloads
 Citation Tools
Citation Tools