 Open Access
Open Access
REVIEW
The bacterial small RNAs: The new biomarkers of oral microbiota-associated cancers and diseases
1 Department of Endodontics, Shanghai Ninth People’s Hospital, College of Stomatology, Shanghai Jiao Tong University School of Medicine, Shanghai, China
2 National Clinical Research Center for Oral Diseases, Shanghai, China
3 Shanghai Key Laboratory of Stomatology & Shanghai Research Institute of Stomatology, Shanghai, China
4 Department of Nursing, Shanghai Ninth People’s Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai, China
* Corresponding Author: ZHENGWEI HUANG. Email:
# These authors have contributed equally to this work
BIOCELL 2023, 47(10), 2187-2193. https://doi.org/10.32604/biocell.2023.042357
Received 27 May 2023; Accepted 27 July 2023; Issue published 08 November 2023
Abstract
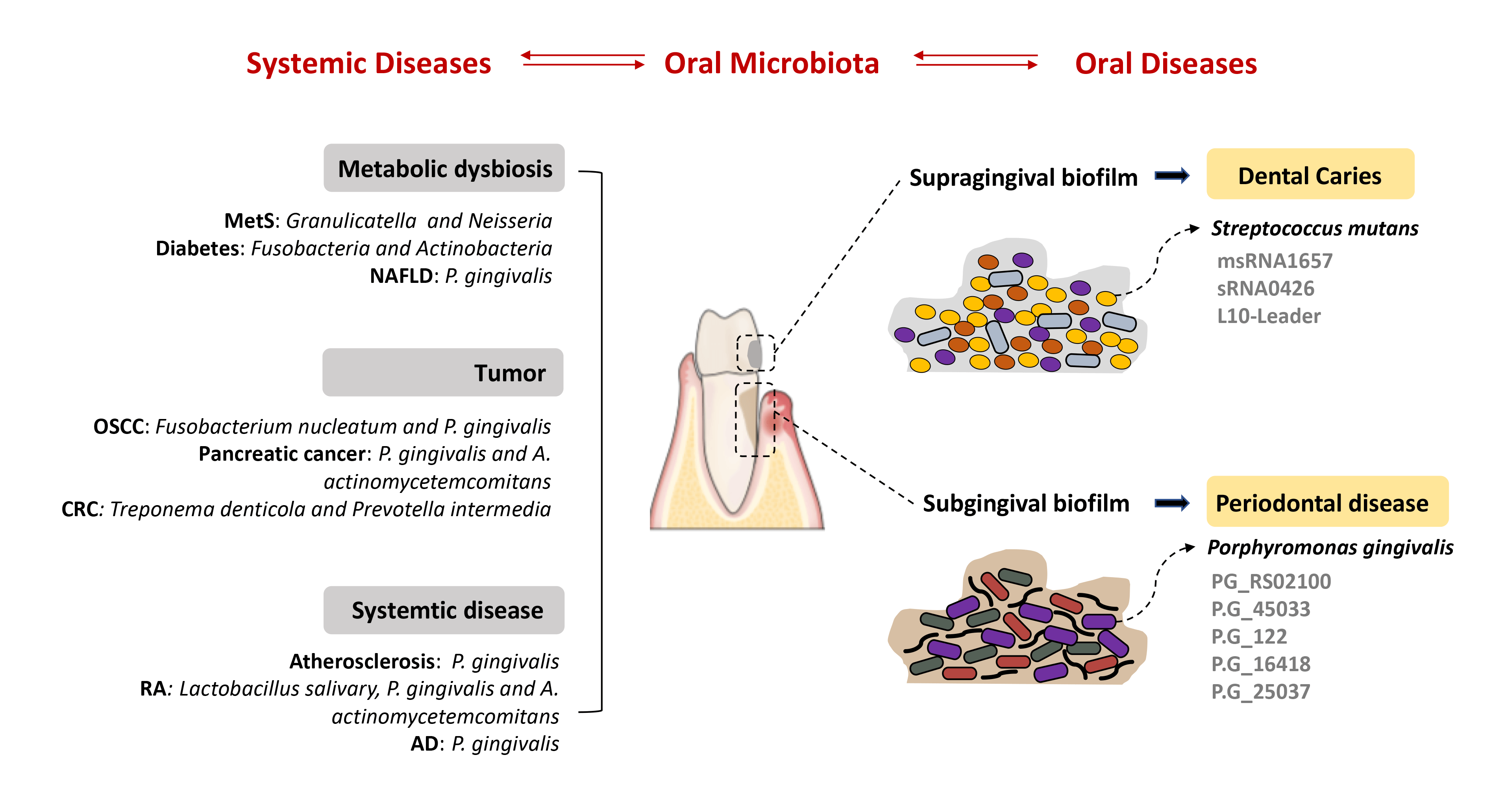
The oral microbiota is a vital part of the human microbiota that functions in various physiological processes and is highly relevant to cancers and other diseases. With the alterations of host immune competence, the homeostatic balance existing between the oral microbiota and host may be disturbed and result in the development of diseases. Numerous observations have suggested that small RNAs are key regulators of bacterial pathogenesis and bacteria-host interactions. Further, bacterial small RNAs are considered to be promising biomarkers for the development of novel, and efficacious therapies for oral dysbiosis. Mechanistic insights into how oral pathogens communicate with other bacteria or host cells in oral cancers via small RNAs are hot topics of research. Current studies also have begun to elucidate the key role of oral microbiota in the development of systemic diseases. This article discusses existing findings and nascent mechanisms governing the small RNA-based interactions between oral microbiota and associated diseases. The knowledge of such interactions is key in planning approaches to reverse dysbiosis to achieve health.Graphic Abstract
Keywords
List of Abbreviations
| OSCC | Oral squamous cell carcinoma |
| sRNA | Small RNA |
| P. gingivalis | Porphyromonas gingivalis |
| F. nucleatum | Fusobacterium nucleatum |
| OMV | Outer membrane vesicle |
| DSC2 | Desmocollin-2 |
| CRC | Colorectal carcinoma |
| A. actinomycetemcomitans | Aggregatibacter actinomycetemcomitans |
| EPS | Extracellular polymeric substances |
| S. mutans | Streptococcus mutans |
| msRNAs | microRNA-size small RNAs |
| GCF | Gingival crevicular fluid |
| MetS | Metabolic syndrome |
| MAFLD | Metabolic-associated fatty liver disease |
| RA | Rheumatoid arthritis |
The oral microbiota refers to the collection of various microorganisms that colonize the human oral cavity (Zhang et al., 2015). This microbiota is receiving increasing attention from scholars and doctors given its relevant accessibility and complex interactions with cancers and many other diseases (Krishnan et al., 2017). The complex structure and dynamic aspects of oral microbiota are pathogenic factors of oral diseases such as periodontitis and dental caries. They are also considered highly relevant to oral squamous cell carcinoma (OSCC) (Lamont et al., 2018). Substantial evidence has implicated the mechanistic role of oral microbiota in inflammation and immune responses. As the unique microenvironment of the oral cavity, the majority of oral microbiota tend to form a biofilm against the mechanical action of mastication and fluid shear stress (Flemming et al., 2016; Yin et al., 2019). Within the biofilm, oral microbiota uses various means to communicate with one another and their eukaryotic hosts (Bassler and Losick, 2006). Bacterial small RNAs (sRNAs) are a significant number of RNA molecules between 40 and 500 nucleotides in length. They have been found to serve as post-transcriptional regulators that affect almost every field of bacterial physiology, including quorum sensing, environmental stimulus-response, and biofilm formation (Bossi and Figueroa-Bossi, 2016; Carroll et al., 2016; Lei et al., 2019; Merritt et al., 2014). Many sRNAs are found to be formed through interactions with host cells and, in turn, affect their viability in the host (Papenfort and Vogel, 2010; Storz et al., 2011). Therefore, bacterial sRNAs are considered novel biomarkers for oral microbiota-associated cancer therapy. The question of whether sRNAs function as the functional regulator or signal transduction in bacteria or host cells is an active area of discovery (Irfan et al., 2020; Zhou et al., 2019). Recent studies have explored the interaction of oral pathogens with host cells in cancers and oral diseases via sRNAs. This article discusses existing findings and nascent mechanisms governing the sRNAs-based interactions between oral microbiota and associated diseases.
The Role of Oral Microbiota in Oral Health
The oral cavity has distinct microenvironments where the oral microbiota located in the supragingival and subgingival communities vary dramatically in composition and spatial organization (Baker et al., 2017; Lamont et al., 2018). Oral microbiota in a healthy physiological state is considered a stable component of the ecosystem, which can compete with and resist exogenous pathogens and contribute to the development of normal tissues and the immune system (Graves et al., 2019; Mira et al., 2017; Willis and Gabaldon, 2020). However, with the alterations in host and environmental factors, the microbiome can shift to a state of dysbiosis and finally lead to oral diseases (Costalonga and Herzberg, 2014). In particular, it is reported that poor oral hygiene could lead to a remarkable increase in the relative abundance of potential cariogenic Leptotrichia and a decrease in Streptococcus (Belstrom et al., 2018). Further, smoking appears to be a major environmental factor associated with oral disease progression (Yu et al., 2017). Studies in recent years have shown that smoking may affect the balance of the microbiome by changing biofilm organization, local microenvironment, immune homeostasis, or direct contact with the microorganisms contained in it, which may be involved in the occurrence of diverse oral diseases (Huang and Shi, 2019). Thus, the oral microbiome is home to various microorganisms that have established mutually beneficial relationships with their hosts under healthy conditions. Additionally, the approaches that trigger a shift from the dysbiosis of oral microbiota and host to eubiosis are active areas of discovery.
Small RNA-Based Interaction of Oral Pathogens and Cancers
OSCC is the sixth most prevalent cancer worldwide and the most common head and neck malignant tumor with a poor prognosis and survival rate (Lafuente Ibanez de Mendoza et al., 2020). Several studies have shown periodontitis to be a major risk factor for OSCC, and periodontal pathogens have similarly been associated with the lesions (Lamont et al., 2022; Shin et al., 2019). For example, the pattern is one of a dysbiotic microbial community enriched in potentially carcinogenic organisms such as Porphyromonas gingivalis (P. gingivalis) and with an underrepresentation of homeostatic commensals, contributing to the development of OSCC. A theoretical mechanistic framework for a bacterial contribution to carcinogenesis includes modulation of the balance of host cell proliferation and death, disruption of immune surveillance, and alteration of the metabolism of host-produced compounds, nutritional substrates, or drugs (Fitzsimonds et al., 2020).
According to a report, P. gingivalis was shown to promote the development of OSCC and could modulate the expression of microRNAs (miRs) in host cells. P. gingivalis could increase OSCC cell proliferation by regulating cyclin D1 expression via the miR-21/programmed cell death 4 (PDCD4)/AP-1 negative feedback signaling pathway (Chang et al., 2019). In another study, it was observed P.gingivalis worked as an enhancer of OSCC cell invasiveness by the dysregulation of the expression of the zinc-finger E-box-binding homeobox 1 (ZEB1)/CK13/miR-200 circuit (Sztukowska et al., 2016). While Fusobacterium nucleatum (F. nucleatum) is prevalent in healthy microbiota, several studies have found that F. nucleatum is significantly enriched in patients with head and neck cancer (McIlvanna et al., 2021). Further, Yost et al. (2018) profiled the RNA expression in the oral microbiome in OSCC and reported that F. nucleatum showed the highest upregulation of putative virulence factors for tumor sites (Yost et al., 2018). Similar to P. gingivalis, it was found that F. nucleatum could upregulate the expression of miR-21 to promote the proliferation of cancer cells.
Recent evidence suggests that outer membrane vesicles (OMVs) released by P. gingivalis that contain differently packaged sRNAs may function in the development of OSCC. The OMV production can be found in all growth phases of bacterial culture, and is considered to play a significant role in the interaction between the bacteria and the host (Choi et al., 2017b). For instance, it was reported that sRNA23392 was abundant in P. gingivalis OMVs and it promoted the invasion and migration of OSCC cells by targeting desmocollin-2 (DSC2). This study revealed a novel P. gingivalis OMVs/sRNA23392/DSC2 pathway-dependent tumor migration and invasion, whereby P. gingivalis promotes OSCC by delivering sRNAs into host cells (Liu et al., 2021).
Other oral microbiota-associated cancers
Oral microbiota is located at the starting point of the digestive tract and plays an important role in regulating nutrient absorption, substance metabolism, and immune response (Gao et al., 2018). Diseases or changes in the human environment often accompany changes in oral microbiota. Further, oral microbiota is also related to a greater risk of some gastrointestinal tumors. Landmark publications in 2012 from two independent groups reported that F. nucleatum infection was prevalent in human colorectal carcinoma (CRC) (Castellarin et al., 2012; Kostic et al., 2013). Additionally, F. nucleatum DNA was higher in colorectal polyp tissue than in healthy tissue from controls. P. gingivalis and A. actinomycetemcomitans were confirmed to be associated with an increasing risk of pancreatic cancer (Fan et al., 2018). Further, Treponema denticola and Prevotella intermedia, two common oral pathogens, were also reported to have distinctive enrichment in CRC in a case-control study (Yang et al., 2019).
Indeed, microscopic imaging and molecular biology analyses have demonstrated that bacteria can deliver their sRNAs to and interact with eukaryotic cells via OMVs (Dauros-Singorenko et al., 2018). Recently, using the method of small RNA Metagenomics by Sequencing (sMETASeq), a research team showed that bacterial sRNAs were expressed more widely and consistently than previously expected (Mjelle et al., 2020). In the human body, bacterial sRNAs have been confirmed in plasma and serum, which has been reported in other tissues in more and more studies (Wang et al., 2012). Up to now, secreted RNAs were confirmed in many bacteria, including P. gingivalis, Streptococcus sp., Vibrio cholerae, Pseudomonas aeruginosa and Helicobacter pylori (Ghosal, 2018; Koeppen et al., 2016; Zhang et al., 2020). The description of sRNA-based host-pathogen interactions has been carried out mainly for oral tumors (Pita et al., 2020). The roles of such interactions in the pathogenesis of some oral microbiota-associated systemic cancers need to be explored in the future. The sRNA-based interaction of oral pathogens and cancers are depicted in Fig. 1.
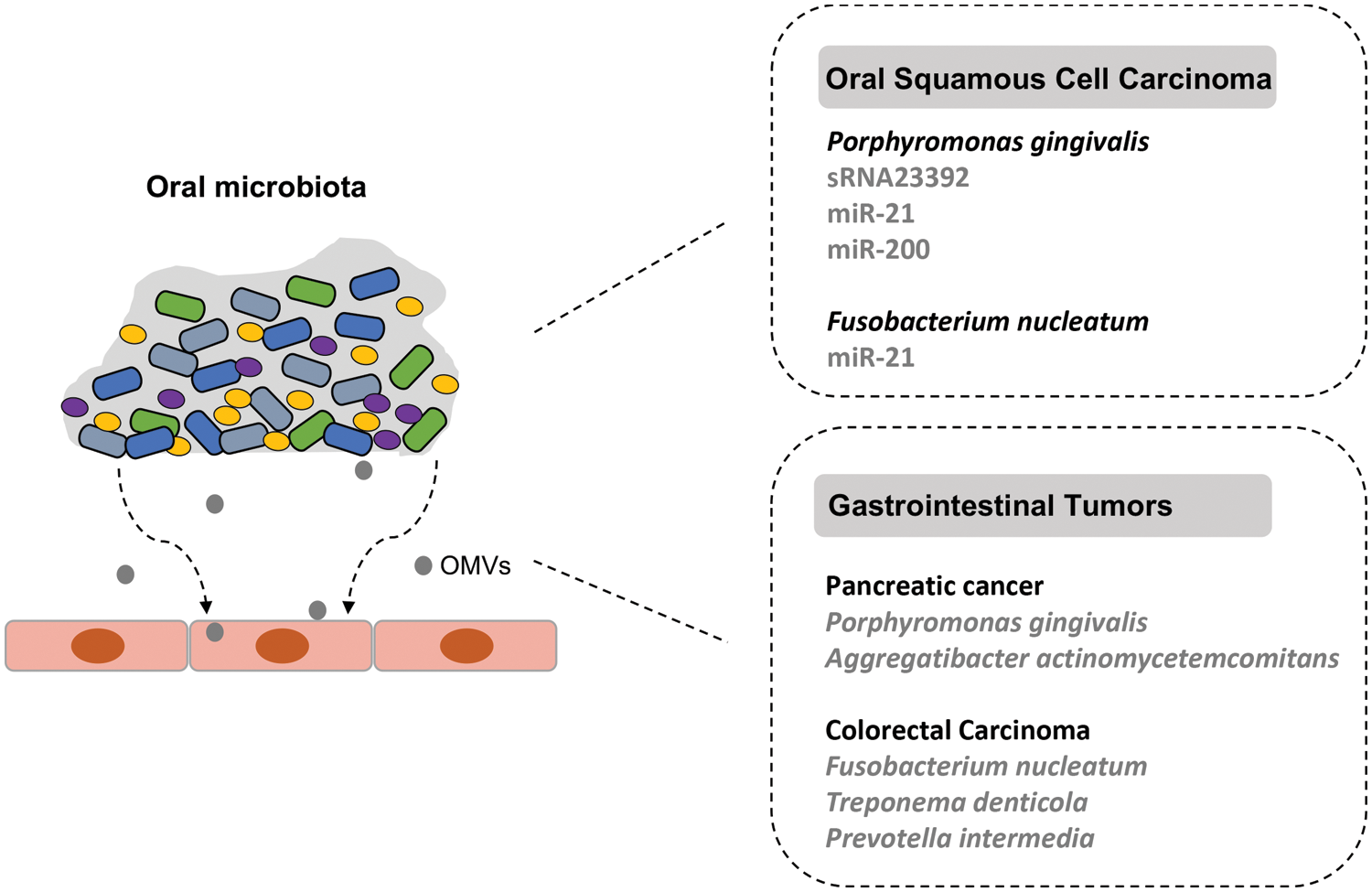
Figure 1: The small RNA (sRNA)-based interaction of oral microbiota with associated cancers. The dysbiotic oral microbial community is related to the development of oral squamous cell carcinoma (OSCC) and gastrointestinal tumors. The periodontal pathogen P. gingivalis can release outer membrane vesicles (OMVs) that contain different packaged sRNAs and function in the development of OSCC.
Small RNA-Based Interaction of Oral Pathogens and Diseases
Dental caries is a multifactorial and dynamic disease with oral microorganisms as the initiating factor, which causes the staged demineralization and remineralization of dental hard tissues (Bowen, 2015; Pitts et al., 2017). Early colonizers are commensal bacteria, such as Streptococcus sanguinis and Streptococcus mitis, which adhere more firmly to the teeth surfaces and grow faster than cariogenic bacteria, maintain microbial homeostasis and stability via antagonism against pathogens through various mechanisms (Bowen et al., 2018; Maske et al., 2017). Excessive dietary carbohydrate intake can promote the production of bacterial extracellular polymers substance (EPS) and acid metabolites, leading to the accumulation of acid-producing and acidophilic microorganisms (Tanner et al., 2018). The formation of an EPS-rich biofilm accelerates acid production and protects the pathogenic microorganisms from rapid buffering by saliva (Bowen et al., 2018; Marsh and Zaura, 2017).
As one of the recognized principal pathogens of caries, Streptococcus mutans (S. mutans) plays a vital pathogenic role in the production of extracellular polysaccharides and acid metabolites and a strong acid tolerance (Klein et al., 2015). A great number of genes encode relative virulence factors and enable the competition mechanisms of S. mutans, which are linked with lipoteichoic acids (dltABCD, SMU_775c), exopolysaccharides (gtfBCD, gbpB, dexA, vicRKX) and extracellular DNA (lytST, lrgAB, ccpA) (Florez Salamanca and Klein, 2018; Lei et al., 2018; Senadheera et al., 2005; Smith and Spatafora, 2012). However, acquiring virulence genes is only the first step to disease development. Numerous studies have reported that sRNAs regulated the pathogenicity of cariogenic S. mutans. For instance, A novel group of microRNA-size small RNAs (msRNAs) was found in S. mutans where these msRNAs could down-regulate exopolysaccharides by directly binding to the 5′ untranslated region (UTR) region of the vicR gene (Lei et al., 2020; Mao et al., 2016). In another analysis, S. mutans were exposed to 1% or 5% sucrose, and bacterial RNA libraries were established to assess the expression levels of virulence-related sRNAs and target genes (gtfB, gtfC, and spaP). Several differentially expressed sRNAs were obtained, suggesting that sucrose can induce a series of sRNAs and might be involved in regulating biofilm formation (Liu et al., 2019, 2017). Several other sRNAs taking part in the intricate regulatory networks of biofilm formation were identified by using deep sequencing. These include sRNA0426, which takes part in metabolic pathways, especially carbon metabolism (Yin et al., 2020), and L10-Leader, which is related to growth and stress response (Xia et al., 2012). In general, sRNAs of cariogenic S. mutans mainly function in the fine regulation of cellular processes that help it adapt to environmental changes and stress responses.
In periodontal disease, the polymicrobial community induces a chaotic and destructive host reaction through a complex mechanism, causing the progression of periodontal inflammatory disease (Ebersole et al., 2017; Proctor et al., 2020). The dysbiosis of oral microbiota is clearly related to the development of periodontal inflammation (Curtis et al., 2020). It is certain that bacteria could cause gingivitis as their removal results in the reversal of inflammatory conditions (Huang et al., 2014). The development of periodontitis is accompanied by dramatic changes in the composition of subgingival communities, with different Gram-negative species, rather than those that accumulate during gingivitis, gaining an ecological advantage over health-related groups (Abusleme et al., 2013). P. gingivalis, Tannerella forsythia, and Treponema denticola (red complex) were recognized as such main pathogenic microorganisms (Holt and Ebersole, 2005). Changes in the host microenvironment may drive this dysregulation of the microbiome, and one possible driver of such changes is the host inflammatory response. In particular, inflammation exacerbates environmental dysfunction and tissue destruction, increases the flow of gingival crevicular fluid (GCF), carries degraded tissue collagen, blood, and other nutrients into the gingival crevicular canal. It also facilitates the growth and aggregation of pathogenic bacteria, especially those proteolytic and hydrolytic bacteria with iron acquisition ability. In contrast, health-related (symbiotic) species are less likely to survive in the new environment and are competed out (Diaz et al., 2016). This imbalance leads to further ecological imbalances that exacerbate inflammation and ultimately lead to periodontitis in susceptible populations (Hajishengallis, 2015; Lamont et al., 2018).
sRNAs play a critical role in post-transcriptional regulation, especially in rapid-changing environments such as periodontitis progression. For instance, it is reported that 20 Rfam sRNA families are overexpressed during periodontitis progression. These differentially expressed sRNAs are involved in various gene ontology activities, including carbohydrate metabolism, amino acid metabolism, ethanolamine catabolism, control of plasmid copy number, intron splicing, response to stress, and signal recognition particle-dependent cotranslational protein targeting to membrane (Duran-Pinedo et al., 2015). These findings indicate that sRNAs are crucial regulatory elements of the dysbiotic process resulting in periodontitis. The newly identified PG_RS02100, a small conserved transcript, has been confirmed to regulate the survival of P. gingivalis, particularly against oxidative stress (Phillips et al., 2018). Moreover, deep sequencing and bioinformatic analysis identified a group of msRNAs was also identified in the periodontal pathogens Aggregatibacter asctinomycetemcomitans (A.A_11134, A.A_20050, A.A_30457, and A.A_25585), P. gingivalis (P.G_45033, P.G_122, P.G_16418, and P.G_25037) and Treponema denticola (T.D_2161, T.D_16563, and T.D_15612). Additionally, msRNAs were detected to be secreted and delivered into host cells via bacterial OMVs, which suppressed the expression of certain cytokines in host cells (Choi et al., 2017a). It has been proposed that sRNAs may serve as a new bacterial signaling molecule that mediates the interaction between bacteria and humans. The sRNA-based interaction of oral pathogens and oral diseases are depicted in Fig. 2.
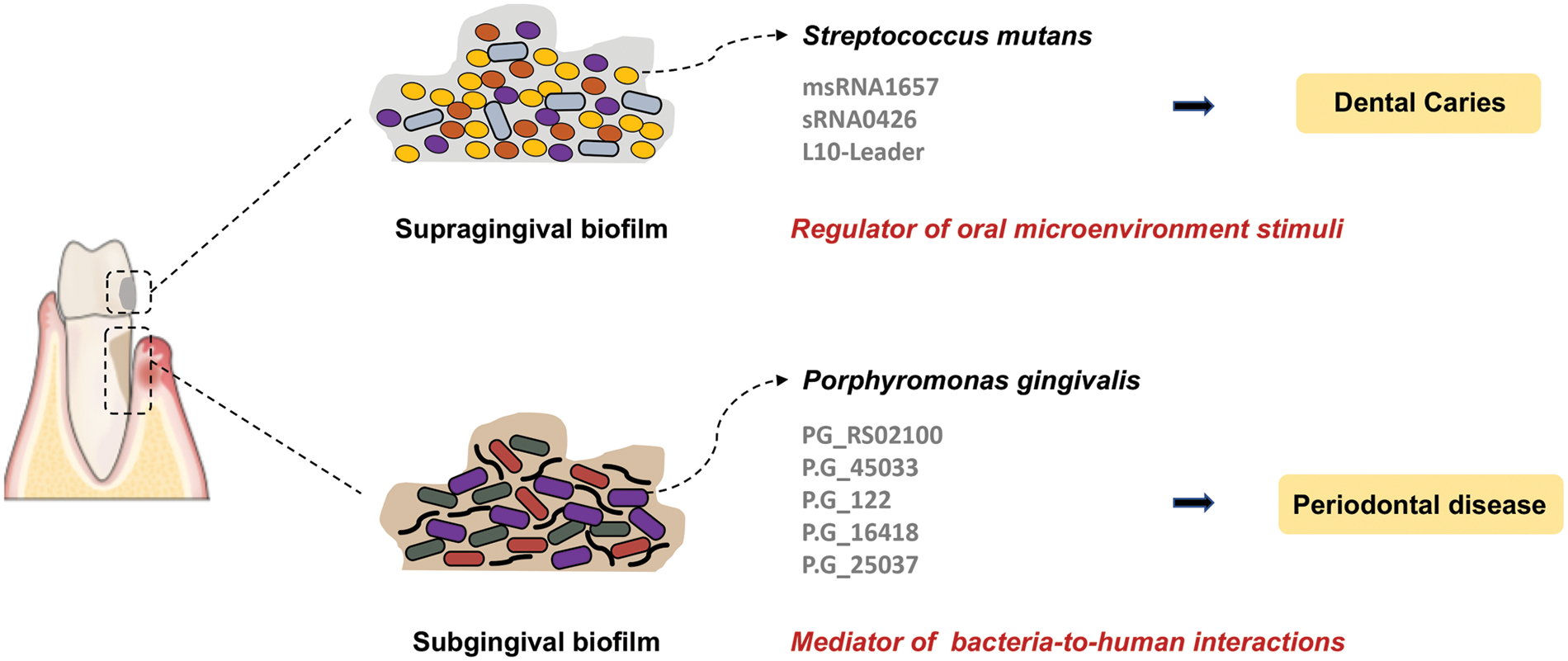
Figure 2: The small RNA (sRNA)-based interaction of oral microbiota with dental caries and periodontal disease. The dysbiosis of supragingival and subgingival biofilms leads to the development of dental caries and periodontal disease, respectively. Further, the small RNAs are key regulators of bacterial pathogenesis and bacteria-host interactions.
Many studies have shown the immense changes the oral microbiome has undergone in most oral diseases, such as periodontitis and dental caries, which are also common in various systemic diseases. An analysis of the oral microbiome and metabolic syndrome (MetS) identified Granulicatella and Neisseria as distinctive genera in subjects with MetS and Peptococcus distinctive in the healthy controls (Si et al., 2017). High serum thyroid-stimulating hormone (TSH) levels are associated with many metabolic disorders. For example, a recent study reported that the salivary microbiome showed significantly higher diversity in individuals with higher TSH levels (Dong et al., 2021). Further, Fusobacteria and Actinobacteria were detected as significantly more abundant in subjects with diabetes, while Proteobacteria were less abundant (Matsha et al., 2020). It was also reported that the diversity of the oral resident microbes in metabolic-associated fatty liver disease (MAFLD) patients was significantly higher than in healthy individuals (Zhao et al., 2020). Furthermore, dysbiosis of oral microbiota was detected in rheumatoid arthritis (RA) patients and partially resolved after RA treatment (Zhang et al., 2015). There was evidence that periodontitis-related pathogens such as P. gingivalis and A. actinomycetemcomitans may promote autoantibody production and autoimmune exacerbation in RA (Maeda and Takeda, 2019). Indeed, substantial evidence has implicated the mechanistic role of the oral microbiome, including transmigration, cytokine release, molecular mimicry, toxin release, and translocation of oral bacteria to the gut in various systemic diseases (Read et al., 2021; Tonelli et al., 2023). Unlike oral diseases and cancers, the knowledge of whether sRNAs are involved in pathogenesis is not clear. It is worth studying whether the bacterial sRNAs can be considered promising biomarkers for developing novel and efficacious therapies for oral dysbiosis.
As an important part of the human microbiota, oral microorganisms are involved in various physiological processes, including the development of oral and systemic diseases. The pathological state affects the composition of oral microbiota, and then the dysbiosis of oral microbiota exacerbates the development of the disease. The mechanisms may involve inflammation and immune responses, where sRNAs may play an important regulatory role during this process. Numerous observations have suggested that sRNAs are key regulators of bacterial pathogenesis and bacteria-host interactions. Further research is required to explore the molecular mechanisms involved to illuminate the key roles of oral microbiota.
Acknowledgement: None.
Funding Statement: This work was supported by the National Natural Science Foundation of China (Nos. 82071104 and 81900988), the Clinical Research Program of 9th People’s Hospital, Shanghai Jiao Tong University School of Medicine (No. JYLJ201908).
Author Contributions: The authors confirm contribution to the paper as follows: conceptualization, MYM, and TD; resources, YJL; writing-original draft preparation, TD; writing-review and editing, MYM; visualization, KYY, QQJ, and PFZ; supervision, ZWH; project administration, ZWH; funding acquisition, ZWH, and MYM. All authors have read and agreed to the published version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI (2013). The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME Journal 7: 1016–1025. [Google Scholar] [PubMed]
Baker JL, Bor B, Agnello M, Shi W, He X (2017). Ecology of the oral microbiome: Beyond bacteria. Trends in Microbiology 25: 362–374. [Google Scholar] [PubMed]
Bassler BL, Losick R (2006). Bacterially speaking. Cell 125: 237–246. [Google Scholar] [PubMed]
Belstrom D, Sembler-Moller ML, Grande MA, Kirkby N, Cotton SL, Paster BJ, Twetman S, Holmstrup P (2018). Impact of oral hygiene discontinuation on supragingival and salivary microbiomes. JDR Clinical & Translational Research 3: 57–64. [Google Scholar]
Bossi L, Figueroa-Bossi N (2016). Competing endogenous RNAs: A target-centric view of small RNA regulation in bacteria. Nature Reviews Microbiology 14: 775–784. [Google Scholar] [PubMed]
Bowen WH (2015). Dental caries—not just holes in teeth! A perspective. Molecular Oral Microbiology 31: 228–233. [Google Scholar] [PubMed]
Bowen WH, Burne RA, Wu H, Koo H (2018). Oral biofilms: Pathogens, matrix, and polymicrobial interactions in microenvironments. Trends in Microbiology 26: 229–242. [Google Scholar] [PubMed]
Carroll RK, Weiss A, Broach WH, Wiemels RE, Mogen AB, Rice KC, Shaw LN (2016). Genome-wide annotation, identification, and global transcriptomic analysis of regulatory or small RNA gene expression in Staphylococcus aureus. mBio 7: e01990-01915. [Google Scholar]
Castellarin M, Warren RL, Freeman JD, Dreolini L, Krzywinski M et al. (2012). Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Research 22: 299–306. [Google Scholar] [PubMed]
Chang C, Wang H, Liu J, Pan C, Zhang D, Li X, Pan Y (2019). Porphyromonas gingivalis infection promoted the proliferation of oral squamous cell carcinoma cells through the miR-21/PDCD4/AP-1 negative signaling pathway. ACS Infectious Diseases 5: 1336–1347. [Google Scholar] [PubMed]
Choi JW, Kim SC, Hong SH, Lee HJ (2017a). Secretable small RNAs via outer membrane vesicles in periodontal pathogens. Journal of Dental Research 96: 458–466. [Google Scholar] [PubMed]
Choi JW, Um JH, Cho JH, Lee HJ (2017b). Tiny RNAs and their voyage via extracellular vesicles: Secretion of bacterial small RNA and eukaryotic microRNA. Experimental Biology and Medicine 242: 1475–1481. [Google Scholar] [PubMed]
Costalonga M, Herzberg MC (2014). The oral microbiome and the immunobiology of periodontal disease and caries. Immunology Letters 162: 22–38. [Google Scholar] [PubMed]
Curtis MA, Diaz PI, Van Dyke TE (2020). The role of the microbiota in periodontal disease. Periodontology 83: 14–25. [Google Scholar]
Dauros-Singorenko P, Blenkiron C, Phillips A, Swift S (2018). The functional RNA cargo of bacterial membrane vesicles. FEMS Microbiology Letters 365: fny023. https://doi.org/10.1093/femsle/fny023 [Google Scholar] [CrossRef]
Diaz PI, Hoare A, Hong BY (2016). Subgingival microbiome shifts and community dynamics in periodontal diseases. Journal of the California Dental Association 44: 421–435. [Google Scholar] [PubMed]
Dong T, Zhao F, Yuan K, Zhu X, Wang N, Xia F, Lu Y, Huang Z (2021). Association between serum thyroid-stimulating hormone levels and salivary microbiome shifts. Frontiers in Cellular and Infection Microbiology 11: 603291. [Google Scholar] [PubMed]
Duran-Pinedo AE, Yost S, Frias-Lopez J (2015). Small RNA transcriptome of the oral microbiome during periodontitis progression. Applied and Environmental Microbiology 81: 6688–6699. [Google Scholar] [PubMed]
Ebersole JL, Dawson D, Emecen-Huja P, Nagarajan R, Howard K et al. (2017). The periodontal war: Microbes and immunity. Periodontology 75: 52–115. [Google Scholar]
Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ et al. (2018). Human oral microbiome and prospective risk for pancreatic cancer: A population-based nested case-control study. Gut 67: 120–127. [Google Scholar] [PubMed]
Fitzsimonds ZR, Rodriguez-Hernandez CJ, Bagaitkar J, Lamont RJ (2020). From beyond the pale to the pale riders: The emerging association of bacteria with oral cancer. Journal of Dental Research 99: 604–612. [Google Scholar] [PubMed]
Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S (2016). Biofilms: An emergent form of bacterial life. Nature Reviews Microbiology 14: 563–575. [Google Scholar] [PubMed]
Florez Salamanca EJ, Klein MI (2018). Extracellular matrix influence in Streptococcus mutans gene expression in a cariogenic biofilm. Molecular Oral Microbiology 33: 181–193. [Google Scholar] [PubMed]
Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F (2018). Oral microbiomes: More and more importance in oral cavity and whole body. Protein & Cell 9: 488–500. [Google Scholar]
Ghosal A (2018). Secreted bacterial RNA: An unexplored avenue. FEMS Microbiology Letters 365: fny036. https://doi.org/10.1093/femsle/fny036 [Google Scholar] [CrossRef]
Graves DT, Correa JD, Silva TA (2019). The oral microbiota is modified by systemic diseases. Journal of Dental Research 98: 148–156. [Google Scholar] [PubMed]
Hajishengallis G (2015). Periodontitis: From microbial immune subversion to systemic inflammation. Nature Reviews Immunology 15: 30–44. [Google Scholar] [PubMed]
Holt SC, Ebersole JL (2005). Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: The “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontology 2000 38: 72–122. [Google Scholar] [PubMed]
Huang S, Li R, Zeng X, He T, Zhao H et al. (2014). Predictive modeling of gingivitis severity and susceptibility via oral microbiota. The ISME Journal 8: 1768–1780. [Google Scholar] [PubMed]
Huang C, Shi G (2019). Smoking and microbiome in oral, airway, gut and some systemic diseases. Journal of Translational Medicine 17: 225. [Google Scholar] [PubMed]
Irfan M, Delgado RZR, Frias-Lopez J (2020). The oral microbiome and cancer. Frontiers in Immunology 11: 591088. [Google Scholar] [PubMed]
Klein MI, Hwang G, Santos PH, Campanella OH, Koo H (2015). Streptococcus mutans-derived extracellular matrix in cariogenic oral biofilms. Frontiers in Cellular and Infection Microbiology 5: 10. [Google Scholar] [PubMed]
Koeppen K, Hampton TH, Jarek M, Scharfe M, Gerber SA et al. (2016). A novel mechanism of host-pathogen interaction through sRNA in bacterial outer membrane vesicles. PLoS Pathogens 12: e1005672. [Google Scholar] [PubMed]
Kostic AD, Chun E, Robertson L, Glickman JN, Gallini CA et al. (2013). Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host & Microbe 14: 207–215. [Google Scholar]
Krishnan K, Chen T, Paster BJ (2017). A practical guide to the oral microbiome and its relation to health and disease. Oral Diseases 23: 276–286. [Google Scholar] [PubMed]
Lafuente Ibanez de Mendoza I, Maritxalar Mendia X, Garcia de la Fuente AM, Quindos Andres G, Aguirre Urizar JM (2020). Role of Porphyromonas gingivalis in oral squamous cell carcinoma development: A systematic review. Journal of Periodontal Research 55: 13–22. [Google Scholar] [PubMed]
Lamont RJ, Fitzsimonds ZR, Wang H, Gao S (2022). Role of Porphyromonas gingivalis in oral and orodigestive squamous cell carcinoma. Periodontology 89: 154–165. [Google Scholar]
Lamont RJ, Koo H, Hajishengallis G (2018). The oral microbiota: Dynamic communities and host interactions. Nature Reviews Immunology 16: 745–759. [Google Scholar]
Lei L, Stipp RN, Chen T, Wu SZ, Hu T, Duncan MJ (2018). Activity of Streptococcus mutans VicR is modulated by antisense RNA. Journal of Dental Research 97: 1477–1484. [Google Scholar] [PubMed]
Lei L, Yang Y, Yang Y, Wu S, Ma X, Mao M, Hu T (2019). Mechanisms by which small RNAs affect bacterial activity. Journal of Dental Research 98: 1315–1323. [Google Scholar] [PubMed]
Lei L, Zhang B, Mao M, Chen H, Wu S, Deng Y, Yang Y, Zhou H, Hu T (2020). Carbohydrate metabolism regulated by antisense vicR RNA in cariogenicity. Journal of Dental Research 99: 204–213. [Google Scholar] [PubMed]
Liu D, Liu S, Liu J, Miao L, Zhang S, Pan Y (2021). sRNA23392 packaged by Porphyromonas gingivalis outer membrane vesicles promotes oral squamous cell carcinomas migration and invasion by targeting desmocollin-2. Molecular Oral Microbiology 36: 182–191. [Google Scholar] [PubMed]
Liu S, Zhou Y, Tao Y, Zhuang P, Pang L, Zhi Q, Lin H (2019). Effect of different glucose concentrations on small RNA levels and adherence of Streptococcus mutans. Current Microbiology 76: 1238–1246. [Google Scholar] [PubMed]
Liu SS, Zhu WH, Zhi QH, Liu J, Wang Y, Lin HC (2017). Analysis of sucrose-induced small RNAs in Streptococcus mutans in the presence of different sucrose concentrations. Applied Microbiology and Biotechnology 101: 5739–5748. [Google Scholar] [PubMed]
Maeda Y, Takeda K (2019). Host-microbiota interactions in rheumatoid arthritis. Experimental & Molecular Medicine 51: 1–6. [Google Scholar]
Mao MY, Yang YM, Li KZ, Lei L, Li M et al. (2016). The rnc gene promotes exopolysaccharide synthesis and represses the vicRKX gene expressions via MicroRNA-Size small RNAs in Streptococcus mutans. Frontiers in Microbiology 7: 687. [Google Scholar] [PubMed]
Marsh PD, Zaura E (2017). Dental biofilm: Ecological interactions in health and disease. Journal of Clinical Periodontology 44: S12–S22. [Google Scholar] [PubMed]
Maske TT, van de Sande FH, Arthur RA, Huysmans M, Cenci MS (2017). In vitro biofilm models to study dental caries: A systematic review. Biofouling 33: 661–675. [Google Scholar] [PubMed]
Matsha TE, Prince Y, Davids S, Chikte U, Erasmus RT, Kengne AP, Davison GM (2020). Oral microbiome signatures in diabetes mellitus and periodontal disease. Journal of Dental Research 99: 658–665. [Google Scholar] [PubMed]
McIlvanna E, Linden GJ, Craig SG, Lundy FT, James JA (2021). Fusobacterium nucleatum and oral cancer: A critical review. BMC Cancer 21: 1212. [Google Scholar] [PubMed]
Merritt J, Chen Z, Liu N, Kreth J (2014). Posttranscriptional regulation of oral bacterial adaptive responses. Current Oral Health Reports 1: 50–58. [Google Scholar] [PubMed]
Mira A, Simon-Soro A, Curtis MA (2017). Role of microbial communities in the pathogenesis of periodontal diseases and caries. Journal of Clinical Periodontology 44: S23–S38. [Google Scholar] [PubMed]
Mjelle R, Aass KR, Sjursen W, Hofsli E, Saetrom P (2020). sMETASeq: Combined profiling of microbiota and host small RNAs. iScience 23: 101131. [Google Scholar] [PubMed]
Papenfort K, Vogel J (2010). Regulatory RNA in bacterial pathogens. Cell Host & Microbe 8: 116–127. [Google Scholar]
Phillips PL, Reyes L, Sampson EM, Murrell EA, Whitlock JA, Progulske-Fox A (2018). Deletion of a conserved transcript PG_RS02100 expressed during logarithmic growth in Porphyromonas gingivalis results in hyperpigmentation and increased tolerance to oxidative stress. PLoS One 13: e0207295. [Google Scholar] [PubMed]
Pita T, Feliciano JR, Leitao JH (2020). Extracellular RNAs in bacterial infections: From emerging key players on host-pathogen interactions to exploitable biomarkers and therapeutic targets. International Journal of Molecular Sciences 21: 9634. [Google Scholar] [PubMed]
Pitts NB, Zero DT, Marsh PD, Ekstrand K, Weintraub JA, Ramos-Gomez F, Tagami J, Twetman S, Tsakos G, Ismail A (2017). Dental caries. Nature Reviews Disease Primers 3: 17030. [Google Scholar] [PubMed]
Proctor DM, Shelef KM, Gonzalez A, Davis CL, Dethlefsen L et al. (2020). Microbial biogeography and ecology of the mouth and implications for periodontal diseases. Periodontology 82: 26–41. [Google Scholar]
Read E, Curtis MA, Neves JF (2021). The role of oral bacteria in inflammatory bowel disease. Nature Reviews Gastroenterology & Hepatology 18: 731–742. [Google Scholar]
Senadheera MD, Guggenheim B, Spatafora GA, An YCCLG, Choi J, Hung DCI, Treglown JS, Goodman SD, Ellen RP, Cvitkovitch DG (2005). A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. Journal of Bacteriology 187: 4064–4076. [Google Scholar] [PubMed]
Shin YJ, Choung HW, Lee JH, Rhyu IC, Kim HD (2019). Association of periodontitis with oral cancer: A case-control study. Journal of Dental Research 98: 526–533. [Google Scholar] [PubMed]
Si J, Lee C, Ko G (2017). Oral microbiota: Microbial biomarkers of metabolic syndrome independent of host genetic factors. Frontiers in Cellular and Infection Microbiology 7: 516. [Google Scholar] [PubMed]
Smith EG, Spatafora GA (2012). Gene regulation in S. mutans: Complex control in a complex environment. Journal of Dental Research 91: 133–141. [Google Scholar] [PubMed]
Storz G, Vogel J, Wassarman KM (2011). Regulation by small RNAs in bacteria: Expanding frontiers. Molecular Cell 43: 880–891. [Google Scholar] [PubMed]
Sztukowska MN, Ojo A, Ahmed S, Carenbauer AL, Wang Q, Shumway B, Jenkinson HF, Wang H, Darling DS, Lamont RJ (2016). Porphyromonas gingivalis initiates a mesenchymal-like transition through ZEB1 in gingival epithelial cells. Cellular Microbiology 18: 844–858. [Google Scholar] [PubMed]
Tanner ACR, Kressirer CA, Rothmiller S, Johansson I, Chalmers NI (2018). The caries microbiome: Implications for reversing dysbiosis. Advances in Dental Research 29: 78–85. [Google Scholar] [PubMed]
Tonelli A, Lumngwena EN, Ntusi NAB (2023). The oral microbiome in the pathophysiology of cardiovascular disease. Nature Reviews Cardiology 20: 386–403. [Google Scholar] [PubMed]
Wang K, Li H, Yuan Y, Etheridge A, Zhou Y, Huang D, Wilmes P, Galas D (2012). The complex exogenous RNA spectra in human plasma: An interface with human gut biota? PLoS One 7: e51009. [Google Scholar] [PubMed]
Willis JR, Gabaldon T (2020). The human oral microbiome in health and disease: From sequences to ecosystems. Microorganisms 8: 308. [Google Scholar] [PubMed]
Xia L, Xia W, Li S, Li W, Liu J et al. (2012). Identification and expression of small non-coding RNA, L10-Leader, in different growth phases of Streptococcus mutans. Nucleic Acid Therapeutics 22: 177–186. [Google Scholar] [PubMed]
Yang Y, Cai Q, Shu XO, Steinwandel MD, Blot WJ, Zheng W, Long J (2019). Prospective study of oral microbiome and colorectal cancer risk in low-income and African American populations. International Journal of Cancer 144: 2381–2389. [Google Scholar] [PubMed]
Yin W, Wang Y, Liu L, He J (2019). Biofilms: The microbial “protective clothing” in extreme environments. International Journal of Molecular Sciences 20: 3423. [Google Scholar] [PubMed]
Yin L, Zhu W, Chen D, Zhou Y, Lin H (2020). Small noncoding RNA sRNA0426 is involved in regulating biofilm formation in Streptococcus mutans. Microbiologyopen 9: e1096. [Google Scholar] [PubMed]
Yost S, Stashenko P, Choi Y, Kukuruzinska M, Genco CA, Salama A, Weinberg EO, Kramer CD, Frias-Lopez J (2018). Increased virulence of the oral microbiome in oral squamous cell carcinoma revealed by metatranscriptome analyses. International Journal of Oral Science 10: 32. [Google Scholar] [PubMed]
Yu G, Phillips S, Gail MH, Goedert JJ, Humphrys MS, Ravel J, Ren Y, Caporaso NE (2017). The effect of cigarette smoking on the oral and nasal microbiota. Microbiome 5: 3. [Google Scholar] [PubMed]
Zhang X, Zhang D, Jia H, Feng Q, Wang D et al. (2015). The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nature Medicine 21: 895–905. [Google Scholar] [PubMed]
Zhang H, Zhang Y, Song Z, Li R, Ruan H, Liu Q, Huang X (2020). sncRNAs packaged by Helicobacter pylori outer membrane vesicles attenuate IL-8 secretion in human cells. International Journal of Medical Microbiology 310: 151356. [Google Scholar] [PubMed]
Zhao F, Dong T, Yuan KY, Wang NJ, Xia FZ, Liu D, Wang ZM, Ma R, Lu YL, Huang ZW (2020). Shifts in the bacterial community of supragingival plaque associated with metabolic-associated fatty liver disease. Frontiers in Cellular and Infection Microbiology 10: 581888. [Google Scholar] [PubMed]
Zhou LY, Qin Z, Zhu YH, He ZY, Xu T (2019). Current RNA-based therapeutics in clinical trials. Current Gene Therapy 19: 172–196. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools