 Open Access
Open Access
ARTICLE
The extracellular secretion of miR-1825 wrapped by exosomes increases CLEC5A expression: A potential oncogenic mechanism in ovarian cancer
1 Department of Gynecology, Clinical Oncology School of Fujian Medical University, Fujian Cancer Hospital, Fuzhou, China
2 Department of Gynecology, Shengli Clinical Medical College of Fujian Medical University, Fuzhou, China
3 Laboratory of Biochemistry and Molecular Biology Research, Department of Clinical Laboratory, Clinical Oncology School of Fujian Medical University, Fujian Cancer Hospital, Fuzhou, China
4 Geneplus-Beijing Institute, Beijing, China
* Corresponding Author: YANG SUN. Email:
BIOCELL 2023, 47(5), 1039-1050. https://doi.org/10.32604/biocell.2023.027494
Received 01 November 2022; Accepted 02 February 2023; Issue published 10 April 2023
Abstract
Background: Ovarian cancer (OC) is a leading cause of gynecological cancer-linked deaths worldwide. Exosomal miR-1825 and its target gene C-type lectin domain family 5 member A (CLEC5A) are associated with tumorigenesis in cancers that was further probed. Methods: Exosomal miR-1825 expression in exosomes and its impact on overall survival (OS) prediction were determined using Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) data. Target genes of miR-1825 were searched in five prediction databases and prognostically significant differentially expressed genes were identified. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were carried out. The ability of CLEC5A to predict OS was evaluated using univariate and multivariate Cox regression analyses and Kaplan-Meier curves. The CLEC5A expression pattern in OC was validated using immunohistochemistry. The CIBERSORT algorithm was used to compare the immune cell landscape, and the results were validated in a GEO cohort. Finally, the predicted half maximal inhibitory concentration (IC50) values for five commonly used chemotherapy agents were also compared. Results: MiR-1825 level was higher in exosomes derived from OC cells and served as a tumor suppressor. The CLEC5A gene was found to be a target of miR-1825, the upregulation of which was correlated with a poor prognosis. M2 macrophage infiltration was significantly enhanced in the CLEC5A high expression group, while T follicular helper cell infiltration was reduced in it. While the predicted IC50 for cisplatin and doxorubicin was higher in the CLEC5A high expression group, that of docetaxel, gemcitabine, and paclitaxel was lower. Conclusion: MiR-1825, a promising OC biomarker, may promote OC progression by increasing CLEC5A expression via exosome-mediated efflux from tumor cells.Keywords
Ovarian cancer (OC) is the second leading cause of gynecological cancer-associated death in the world. For example, in 2020, OC prevalence was expected to be 3.4%, with an annual mortality rate of 4.7% in all female cases (Sung et al., 2021). Comprehensive surgical staging and debulking are advised, followed by systemic chemotherapy according to the National Comprehensive Cancer Network (NCCN) Guidelines on Treatment of OC (http://www.nccn.org) (Armstrong et al., 2021). However, more than 80% of late-stage tumors that were initially responsive to conventional treatment eventually develop resistance (Odunsi, 2017). Therefore, there is an urgent need to identify effective biomarkers for early diagnosis based on a better understanding of the underlying molecular mechanisms of OC development.
Exosomes are extracellular vesicles (EVs) actively secreted by cells. They are approximately 40–160 nm in diameter, and have been implicated in intercellular communication by carrying RNA, lipids, proteins, and other bioactive substances. Their RNA content varies dramatically according to the cell types of origin, reflecting one of the reasons for exosome heterogeneity (Kalluri and LeBleu, 2020).
MicroRNAs (miRNAs), which can inhibit protein synthesis by targeting the complementary sequences located at the 3′-end untranslated regions of messenger RNAs (mRNAs) play an important role in regulating cancer progression (Ali Syeda et al., 2020). Exosomal microRNAs, which are microRNAs encapsulated by exosomes, can promote tumor progression by enhancing mesothelial-to-mesenchymal transition, conferring drug resistance, inducing vascular permeability and angiogenesis, stimulating macrophage M2 polarization, and participating in tumor immune escape by delivering immunosuppressive molecules and factors (Sun et al., 2018; Zhao et al., 2020b). Although the upregulation of miR-1825 has been shown to inhibit the progression of glioblastoma, the roles of miR-1825 and its target genes in OC are yet to be investigated (Lu et al., 2020).
C-type lectin domain family 5 member A (CLEC5A), a member of C-type lectin/C-type lectin-like domain (CTL/CTLD) superfamily is also named C-type lectin superfamily member 5 (CLECSF5) and myeloid DAP12-associating lectin 1 (MDL1) (Bakker et al., 1999). Members of the CTL/CTLD superfamily share a typical protein folding pattern and have diverse functions, such as cell adhesion, cell-cell signaling, glycoprotein turnover, and roles in inflammation and immune response. CLEC5A has been proven to promote brain glioblastoma tumorigenesis and gastric cancer cell proliferation by regulating the phosphoinositide-3-kinase–protein kinase B/Akt (PI3K/Akt) signaling (Fan et al., 2019; Wang et al., 2020). However, the mechanism by which CLEC5A regulates OC awaits further clarification.
The goals of this study were three-fold: (1) to determine the expression pattern and prognostic value of miR-1825 in OC exosomes; (2) to confirm CLEC5A as a target of miR-1825; (3) to assess the performances of the CLEC5A gene in predicting overall survival and relevant characteristics for labeling patients who might benefit from immunotherapy or chemotherapy. In this study, we performed a bioinformatics analysis of the expression patterns of exosomal miRNAs and focused on one target gene CLEC5A. We found that miR-1825, a tumor-suppressor, was highly enriched in OC exosomes, and the downstream oncogenic target gene CLEC5A was highly expressed in OC cells. MiR-1825 impacted the expression of CLEC5A and the latter further led to the change in the immune environment. This ability in transforming the immune environment may be an important mechanism in the development of OC. Further, the suppression of released miRNAs through exosomes from OC cells may be a suitable direction for anti-tumor therapy as decreased expression of miR-1825 in OC cells can promote tumor progression. We also noted that CLEC5A could promote the development of an immune microenvironment in OC and thus has a potential role in the prognosis of OC.
Data retrieval and preprocessing
Affymetrix miRNA microarray data from OC exosomes and cells were available from Gene Expression Omnibus (GEO; series GSE76449) (Kanlikilicer et al., 2016). Exosomal and original samples of chemo-sensitive OC cells and normal ovarian cells (n = 16) were included to rule out the influence of chemotherapy on prediction results. RNA sequencing (RNA-seq) data from normal ovarian tissues were obtained from the Genotype-Tissue Expression (GTEx) portal. RNA-sequencing (RNA-seq) data and clinical data from OC patients were downloaded from GEO (series GSE9891), Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/), International Cancer Genome Consortium (ICGC), and The Cancer Genome Atlas (TCGA), as detailed below (Wang et al., 2016; Zhang et al., 2019).
Bioinformatics analyses of RNA-seq data from TCGA patients were performed using the UCSC bioinformatic pipeline (TOIL RNA-seq). Each dataset was processed identically after removing unavailable data. Clinical data from TCGA were prepared using the TCGA-GDC (Genomic Data Commons) server for subsequent analysis (Goldman et al., 2020).
Finally, all datasets underwent identical processing to achieve an average expression level of samples from the same patient and unavailable data were deleted (NA).
Identification of differentially expressed RNA
In this experiment, RNA-seq data were obtained from TCGA and GTEx. Differentially expressed miRNAs (DEMIs) between exosomal and original samples of OC cells were identified with limma R Bioconductor package (version 3.48.3, p < 0.001 and log2-fold change [logFC] > 4, Student’s t-test) (Ritchie et al., 2015). Differentially expressed mRNAs (DEMs) were screened using the same method (adjusted p < 0.001 and logFC > 3). A boxplot was generated with ggplot2 in R.
Five target gene prediction databases were employed to predict the downstream targets of miR-1825, including TargetScan 7.2, miRDB, TarBasev.8, miRmap, and miRwalk 3.0. Intersections among the five databases were regarded as miR-1825 targets (Agarwal et al., 2015; Chen and Wang, 2020; Karagkouni et al., 2018; Sticht et al., 2018; Vejnar et al., 2013).
Survival data (age, clinical stage, histologic grade, survival status, and length of overall survival) of OC patients were collected from TCGA, GEO, and ICGC and merged with gene expression data. We assessed the prognostic ability of miRNAs or CLEC5A in OC using univariate and multivariate Cox regression hazard analyses combined with Kaplan-Meier survival curves.
Functional enrichment analysis
We performed Gene Ontology Biological Process (GOBP) term and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses with clusterProfiler in R to identify CLEC5A targets (p < 0.05) and explore mechanisms of the role of CLEC5A in OC (Yu et al., 2012). Then, Gene Set Enrichment Analysis (GSEA) was utilized to confirm the selection of target genes by GO or KEGG analyses (p < 0.05 and false discovery rate [FDR] < 0.25) and characterize the activity of KEGG pathways between low and high CLEC5A expression groups in OC, as described in detail elsewhere (Subramanian et al., 2005).
Evaluation of correlation of gene expression with immune characteristics of CLEC5A in OC
We assessed correlations of CLEC5A gene expression with immune cell infiltrates and the expression of immune suppressors to explore the ability of CLEC5A to predict the outcome characteristics in low and high CLEC5A expression groups in OC. TCGA RNA-Seq expression profiles of immune and stromal cells were utilized. Tumor-infiltrating lymphocytes (TILs) and stromal cells were identified with ESTIMATE (Estimation of Stromal and Immune cells in malignant Tumor tissues using Expression data) in R. Correlations of CLEC5A with immune/stromal cell infiltration were assessed, and the results were validated in a GEO cohort of 276 qualified OC cases whose expression and survival data were available for review (GSE9891). This cohort was also used for subsequent immune cell infiltration analysis (Yoshihara et al., 2013).
The infiltration of 22 immune cell subpopulations in OC tissues was estimated using the CIBERSORT algorithm that can accurately quantify TILs in biopsied tumor tissues and the LM22 signature matrix file containing 22 immune cell reference profiles from TCGA and GEO (Chen et al., 2018). Patients were categorized into the low and high CLEC5A expression groups according to the cutoff of CLEC5A gene expression. The immune cell landscape was compared between the two groups. Sixteen tumor immune suppressors have been shown to exert suppressive effects on solid tumors (Cassetta and Pollard, 2018). Among them, interleukin 6 (IL6), tumor necrosis factor (TNF), interferon gamma (IFNG), epidermal growth factor (EGF), major histocompatibility complex class I (MHC-I), CD96 molecule (CD96), leukocyte immunoglobulin like receptor B1 (LIR1), programmed cell death 1 (PDCD1), programmed cell death 1 ligand 2 (PDCD1LG2), CD80 molecule (CD80), and CD86 molecule (CD86), which were frequently reported in OC were selected and their expression levels were compared between the two groups.
Estimation of chemotherapy response
We selected five agents (cisplatin, docetaxel, doxorubicin, gemcitabine, and paclitaxel) from 22 recommended options in the 2020 National Comprehensive Cancer Network (NCCN) clinical practice guidelines in oncology on ovarian cancer (Armstrong et al., 2021). The predicted IC50 was estimated with pRRophetic (version 0.5; https://doi.org/10.1371/journal.pone.0107468) in R and compared between low and high CLEC5A expression OC patient groups from TCGA (Geeleher et al., 2014).
Clinical specimen collection and processing
Patients (n = 9) who had pathologically confirmed OC and had undergone radical excision between May 2019 and May 2021 at the Fujian Medical University Cancer Hospital (or Fujian Cancer Hospital) without prior therapy were selected. Specimens (7 OC and 2 peritumoral tissue specimens) were collected, flash-frozen in liquid nitrogen, and reviewed independently by two senior pathologists. The study protocol was approved by the Ethics Committee of Fujian Cancer Hospital.
Immunohistochemistry (IHC) analysis was performed as previously described (Chen et al., 2021). Briefly, formalin-fixed paraffin-embedded sections were dewaxed, dehydrated, and rehydrated. The slides were incubated overnight at 4°C with an anti-CLEC5A antibody (ab203200, Abcam, China, 1:500). They were then incubated at 37°C with a secondary antibody (ab7090, Abcam, China, 1:500). Immunostaining was performed using a SABC(Strept Avidin-Biotin Complex) kit (SA0025, Solarbio, China). Next, the slides were counterstained with Mayer hematoxylin solution (Solarbio, China) for nuclear staining. Images were obtained using a VANOX microscope (Olympus, Japan) and scored. The scoring was as follows: 0 points (no staining), 1 point (light yellow), 2 points (brownish yellow), and 3 points (brown). For each specimen, fields were randomly selected, and the proportion of CLEC5A-positive (CLEC5A+) cells was calculated and scored as follows (1 point, 0%–25% of CLEC5A+ cells; 2 points, 26%–50% of CLEC5A+ cells; 3 points, 51%–75% of CLEC5A+ cells; 4 points, 76%–100% of CLEC5A+ cells). Specimens with a pathological IHC score greater than the cutoff were identified as high CLEC5A+ OC and low CLEC5A+ OC if the cutoff was otherwise.
Categorical variables were expressed as numbers (percentages) and compared using Spearman’s test. Overall survival data were analyzed using Kaplan-Meier survival curves and compared using the log-rank test. Continuous variables following normal distribution were presented in mean ± standard error of measurement (SEM) and compared using the Student’s t-test. Correlations of CLEC5A gene expression with immune/stromal cell infiltration were assessed using Spearman’s correlation test. All statistical analyses were performed using R 4.1.0, and a two-tailed p-value of < 0.05 was deemed statistically significant.
MiR-1825 expression was high in exosomes from ovarian cancer cells and associated with improved prognosis
We explored the expression patterns in four groups of samples using the GSE76449 dataset. These were: (1) normal ovarian cells; (2) exosomes released from normal ovarian cells; (3) ovarian cancer cells and (4) exosomes released from OC cells. We then identified DEMIs among these four groups. The result indicated that microRNAs exhibited a specific distribution pattern in exosomes and their source cells. Interestingly, the expression levels of miR-1281, miR-1825, miR-6877-3p, and miR-3921 were upregulated in exosomes released from OC cells while they were significantly downregulated in the other three sample groups (Figs. 1A–1E). These observations were also supported by a previous study (Rashed et al., 2017). These miRNAs may be secreted from OC cells after being wrapped in exosomes. Further, the prognostic significance of miR-1281,miR-1825, and miR-6877-3p was evaluated in the high vs. low expression groups using TCGA survival data. MiR-3921 was ruled out as its expression was absent in most OC samples (Fig. 2A). In the Kaplan-Meier survival analysis, miR-1825 upregulation in OC cells was correlated with improved overall survival (p < 0.05) (Figs. 2B–2D). This suggests that it might act as a tumor suppressor in OC.
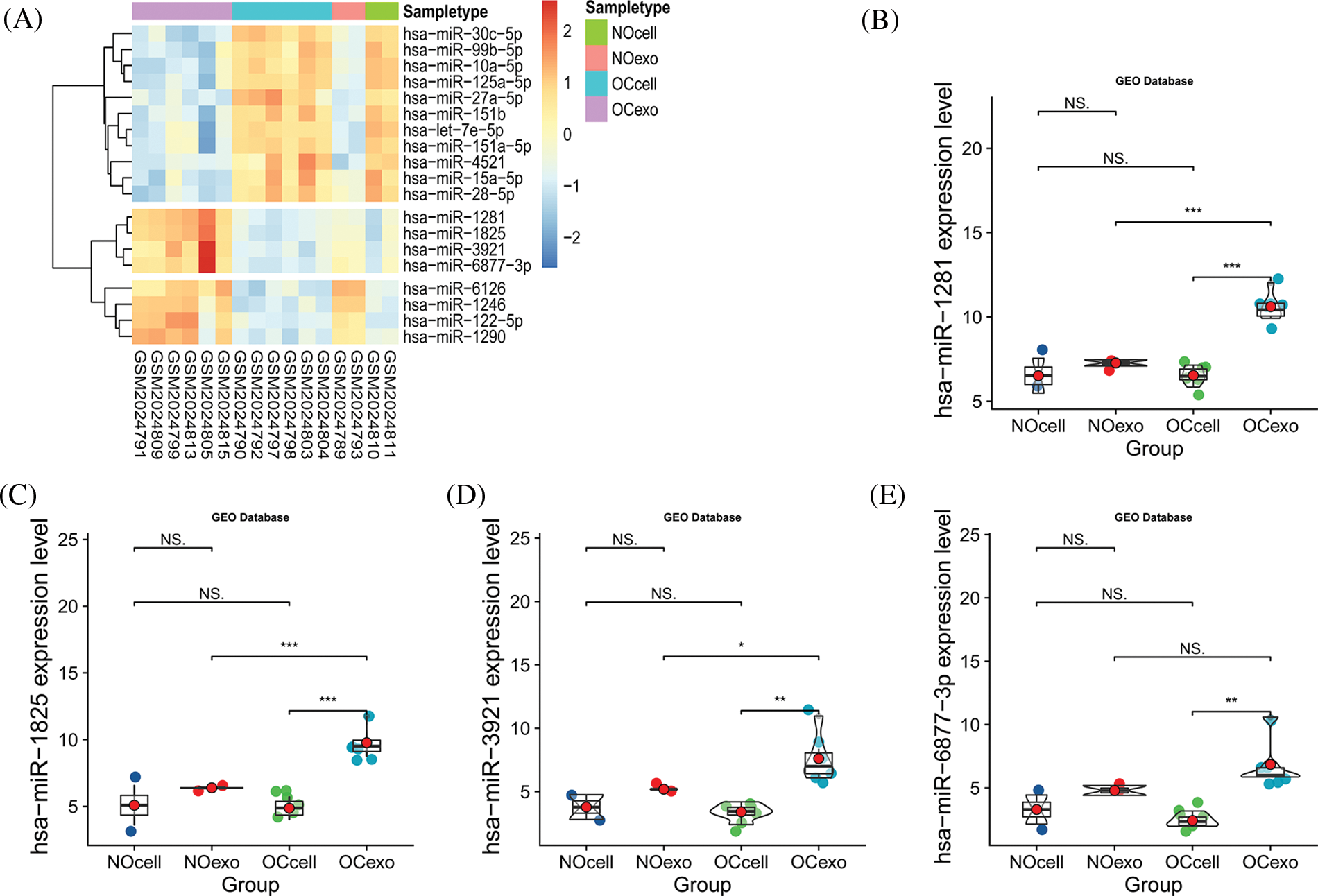
Figure 1: The expression of miR-1825 in ovarian cancer cell-derived exosomes (A) The heatmap shows differences in expressions of differentially expressed microRNAs (DEMIs) in four samples (NOcell, normal ovarian cells; NOexo, exosomes from normal ovarian cells; OCcell, ovarian cancer cells; OCexo, exosomes from ovarian cancer cells); (B) The expression level of miR-1281 in four samples; (C) The expression level of miR-1825 in four samples; (D) The expression level of miR-3921 in four samples; (E) The expression level of miR-6877-3p in four samples (*p < 0.05, **p < 0.01, ***p < 0.001, and NS: Not Statistically Significant).
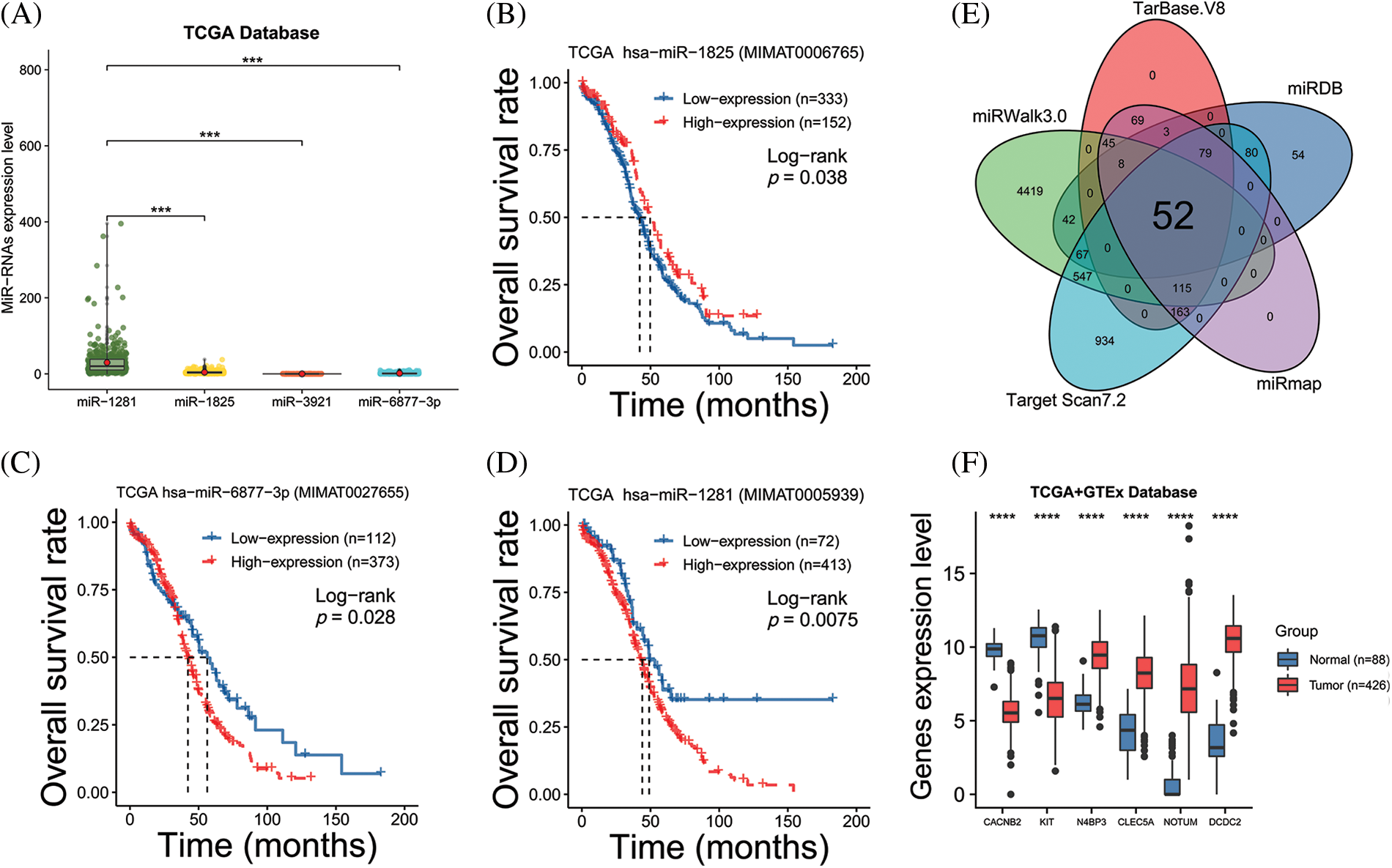
Figure 2: Analyzing the prognostic value of the miRNA and miRNA target genes (A) The expression level of four microRNAs based on The Cancer Genome Atlas (TCGA) database; (B–D) Kaplan-Meier (KM) survival curves show the overall survival of ovarian cancer (OC) patients with high vs. low miR-1825/miR-6877-3p/miR-1281 expression; (E) The Venn diagram reveals the intersection of miR-1825 target genes predicted by five databases; (F) Differential analysis identifies six miR-1825 targets differentially expressed in OC vs. normal ovarian tissues. Genotype-Tissue Expression (GTEx) and The Cancer Genome Atlas (TCGA) gene expression data are processed and analyzed in UCSC Xena (***p < 0.001, ****p < 0.0001).
CLEC5A was a target gene of miR-1825 and had a prognostic value
We performed target gene predictions using TargetScan7.2, miRDB, TarBase v.8, miRmap, and miRwalk3.0 to identify miR-1825 targets. The results displayed 52 genes (Fig. 2E). Differential expression analysis of these 52 target genes using the limma method revealed that the mRNA expression levels of calcium voltage-gated channel auxiliary subunit beta 2 (CACNB2) and KIT proto-oncogene receptor tyrosine kinase (KIT) were downregulated and those of NEDD4 binding protein 3 (N4BP3), CLEC5A, NOTUM palmitoleoyl-protein carboxylesterase (NOTUM), and doublecortin domain containing 2 (DCDC2) were upregulated in the tumor group from the TCGA database in comparison with normal ovarian samples from the GTEx database (|logFC| >3 and adjusted p-value < 0.001) (Fig. 2F). Given the tumor-suppressive effect of miR-1825, the prognostic potential of the four genes with higher expression (N4BP3, CLEC5A, NOTUM, and DCDC2) was assessed. The Kaplan-Meier curves showed that among these four genes, CLEC5A appeared to be a prognostic potential marker for OC, as validated using the validation cohort (GSE9891 and ICGC dataset OV-AU) (Figs. 3A–3C), and was significantly associated with worse overall survival of OC patients (p < 0.05). CLEC5A was a poor prognostic factor in ovarian cancer, cervical cancer, and uterine sarcoma (Fig. 4A). Moreover, CLEC5A could serve as a prognostic biomarker independent of clinicopathological parameters (age, clinical stage, and histologic grade) in OC, as validated using GSE9891 and OV-AU validation cohorts (Fig. 4B). The IHC assays revealed that CLEC5A protein expression was significantly increased in OC vs. normal ovarian tissues (Fig. 4C).
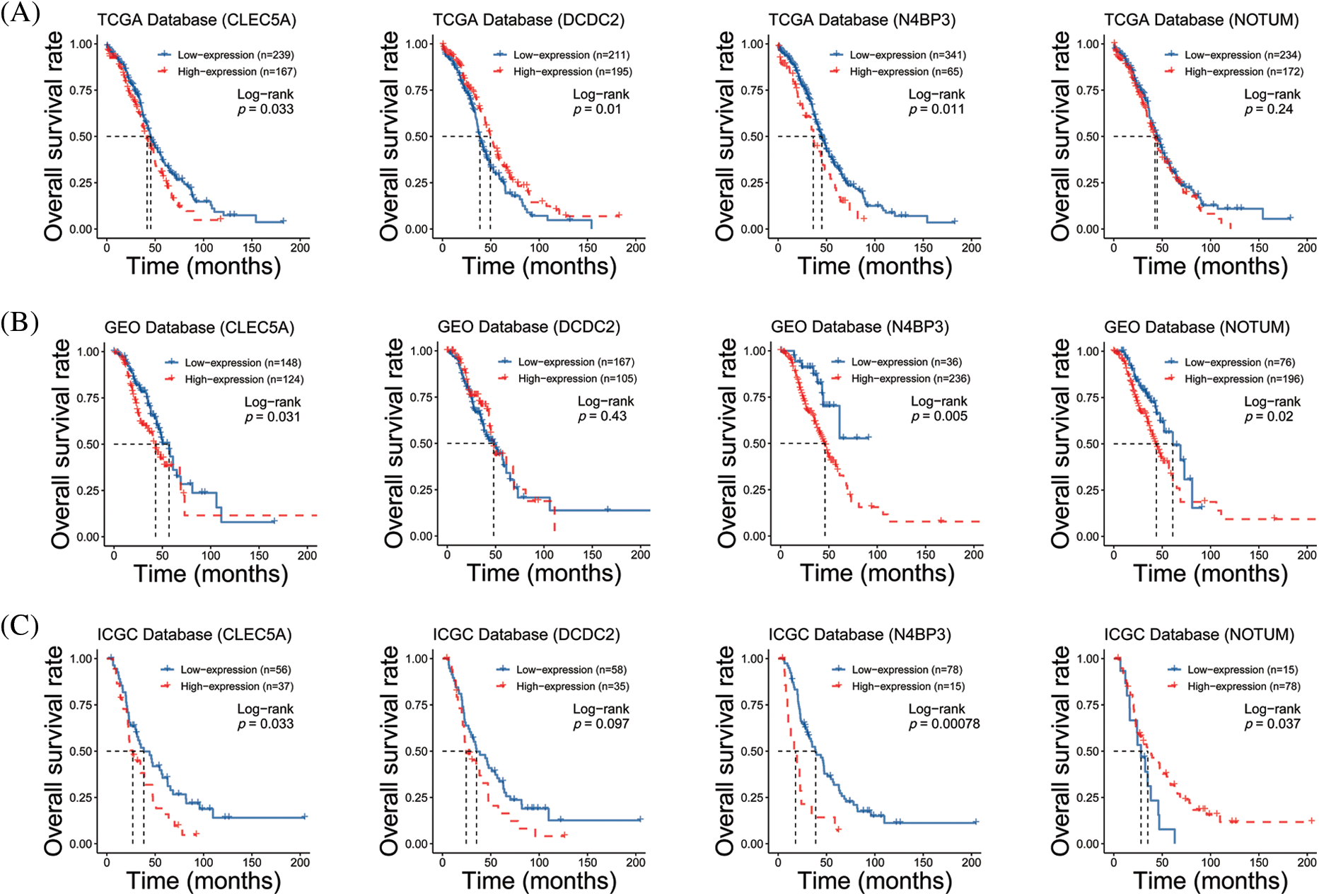
Figure 3: Prognostic analyses of four highly expressed genes (A) Survival curves of CLEC5A, DCDC2, N4BP3, and NOTUM genes based on The Cancer Genome Atlas (TCGA) database; (B) Survival curves of CLEC5A, DCDC2, N4BP3, and NOTUM genes based on the Gene Expression Omnibus (GEO) database; (C) Survival curves of CLEC5A, DCDC2, N4BP3, and NOTUM genes based on The International Cancer Genome Consortium (ICGC) database.
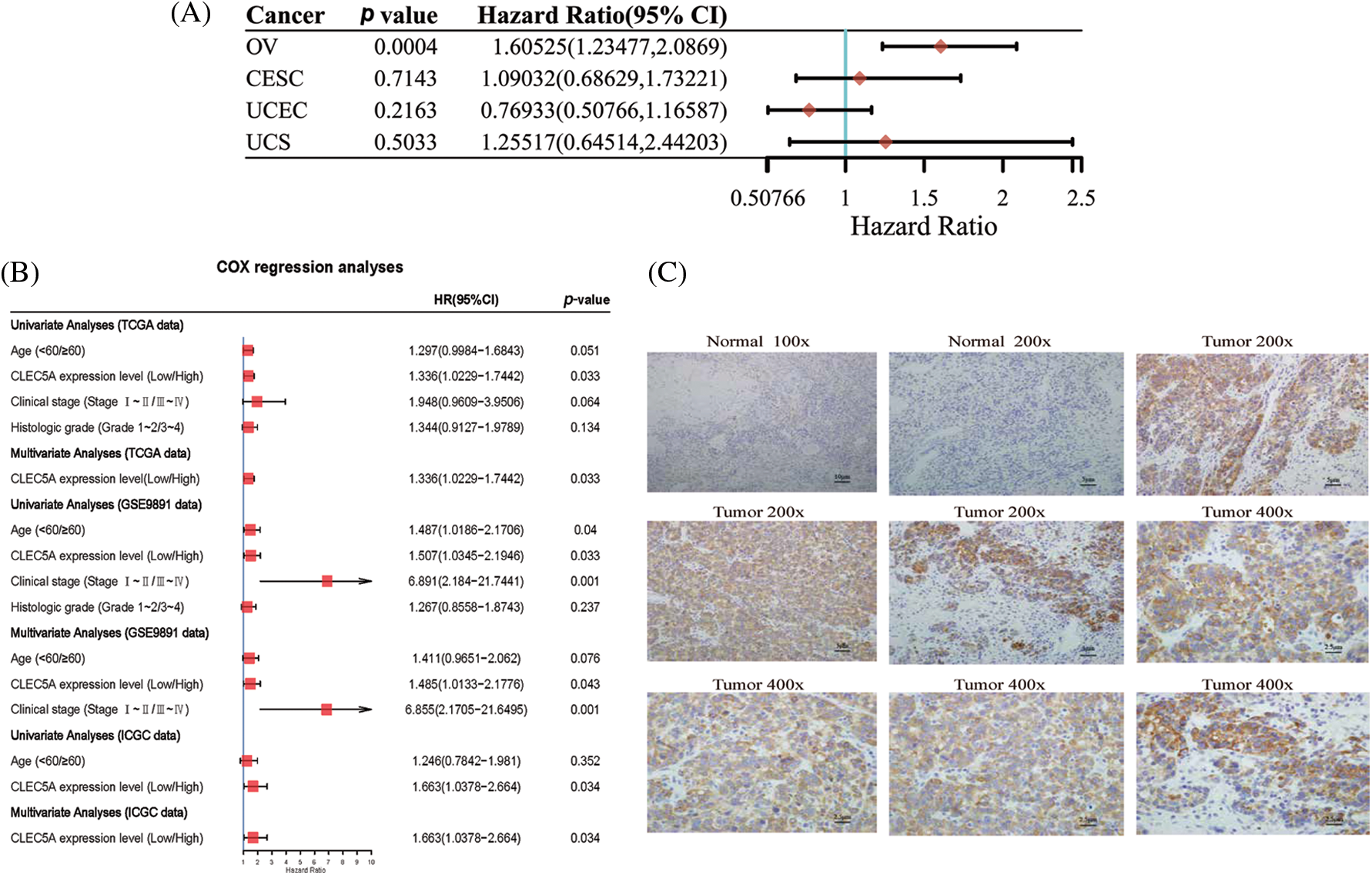
Figure 4: Prognostic value and tissue expression of CLEC5A (A) Prognostic significance of CLEC5A in ovarian cancer, cervical cancer, endometrial cancer, and uterine sarcoma (CI, confidence interval; HR, hazard ratio); (B) Univariate and multivariate Cox regression analyses confirm the independence of CLEC5A in predicting the overall survival of ovarian cancer (OC) patients; (C) Immunohistochemistry (IHC) assays show enhanced expression of CLEC5A protein in ovarian cancer (OC) vs. normal ovarian tissues.
CLEC5A was related to the immune microenvironment
To clarify the functions of CLEC5A expression signatures associated with OC, we assayed biological processes of GO and KEGG pathways enrichment analyses of genes highly correlated to CLEC5A based on the TCGA dataset. As shown in Figs. 5A and 5B, the intersection of GO terms and KEGG pathways showed that a proportion of highly correlated genes was associated with the immune response that plays an important role in tumor inhibition and promotion. These results suggested that immune alterations might contribute to OC occurrence and development. However, the exact mechanism that CLEC5A plays in immunity in OC is not yet clear.
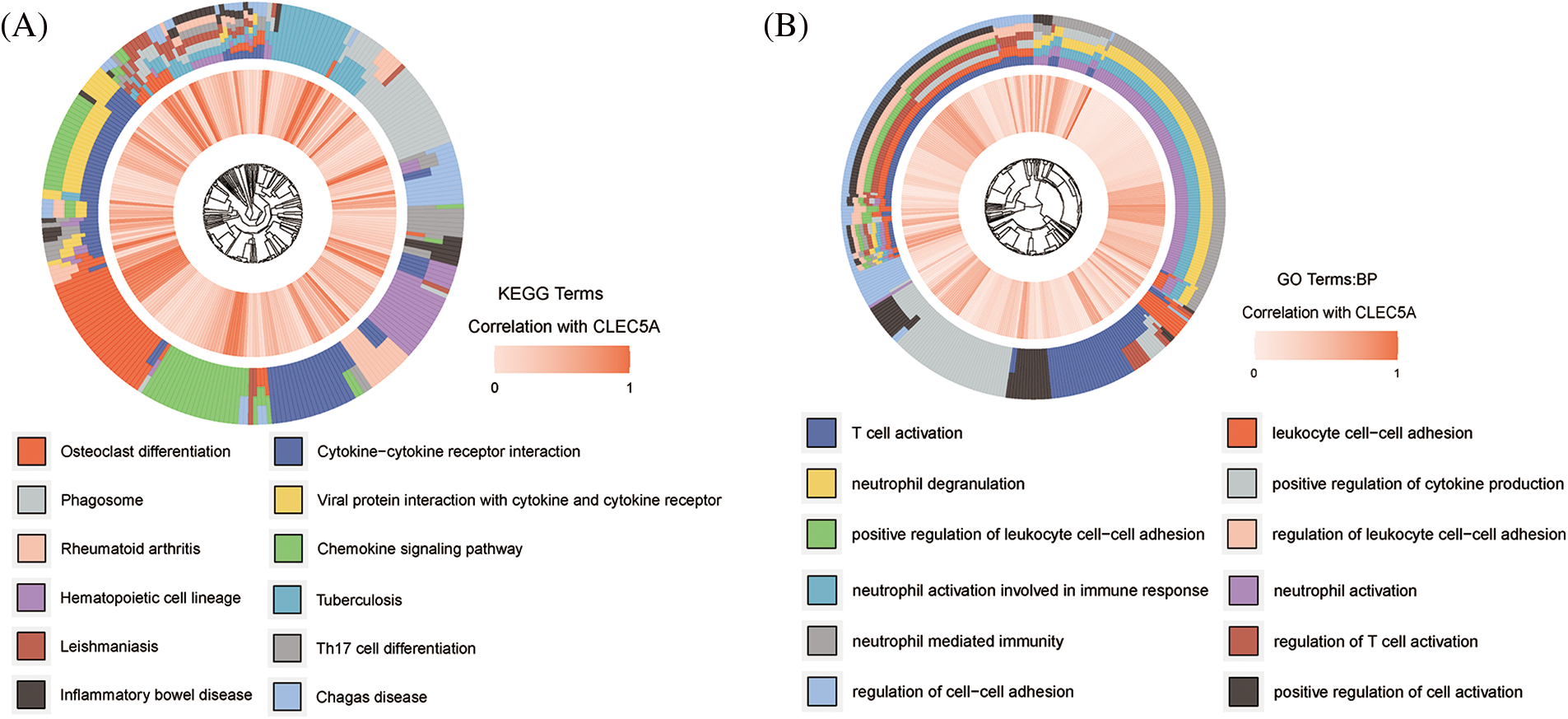
Figure 5: Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis of CLEC5A (A) Signaling pathways that CLEC5A may be involved in; (B) Biological processes that CLEC5A may be involved in (Genes significantly correlated with CLEC5A in ovarian cancer (OC) was explored by Spearman’s correlation analysis; Spearman |R| > 0.4, p < 0.001).
To further verify the results of GO/KEGG enrichment analyses, we conducted a GSEA with the low and high CLEC5A expression datasets. GSEA analyses showed significant differences (p < 0.05) in the enrichment of immune-related pathways and biological processes (Figs. 6A and 6B). Interestingly, CLEC5A also seemed to activate the PI3K/Akt pathway in OC. This activation has been confirmed by experiments in brain glioblastoma by Fan et al. (2019). Given the importance of the PI3K/Akt signaling pathway and the involvement of the immune system on OC, we speculate that the activated PI3K/Akt signaling pathway may contribute in part to the development of the immune environment in OC.
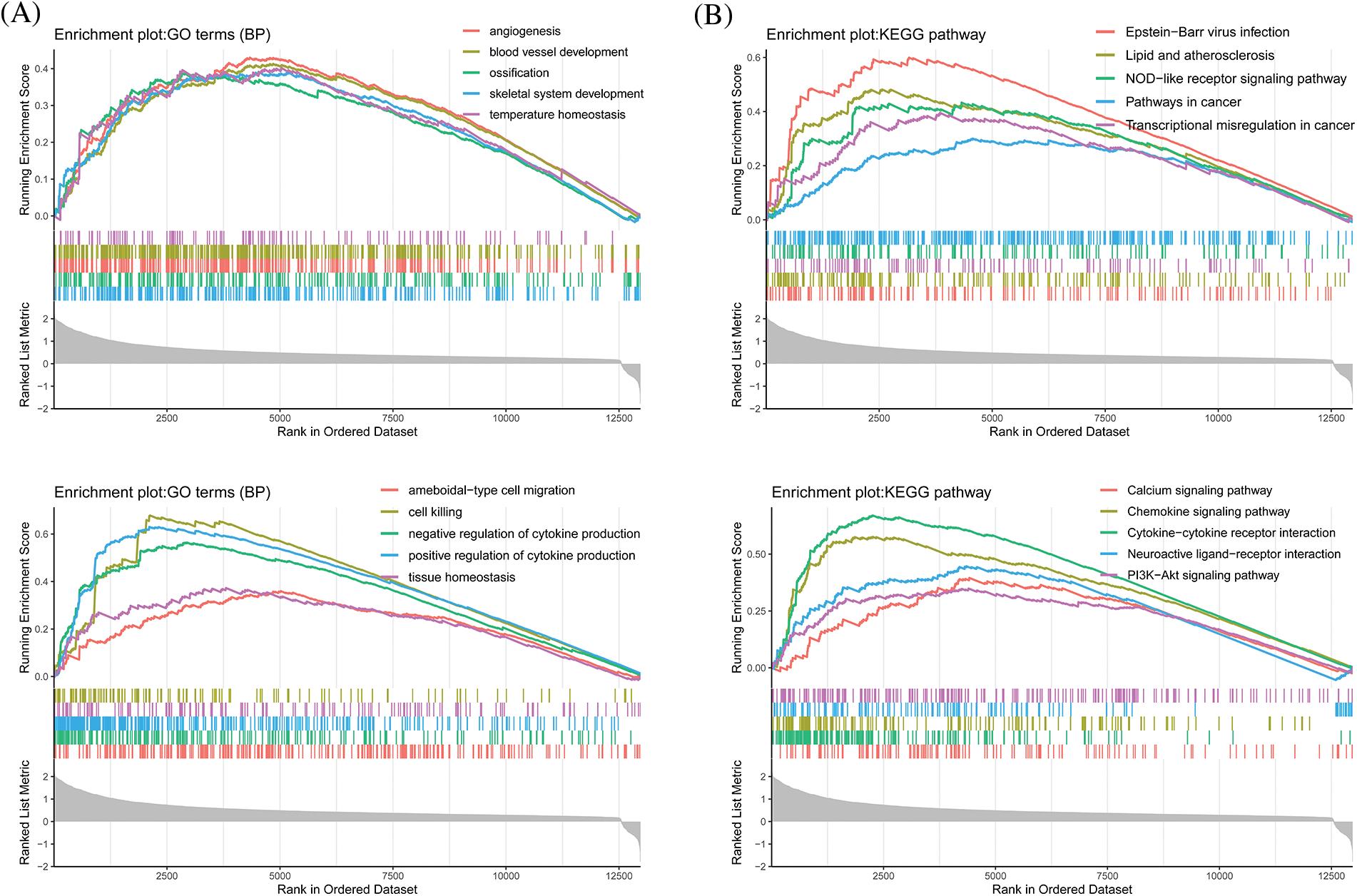
Figure 6: Gene set enrichment analysis for CLEC5A (A) Biological processes that CLEC5A may be involved in; (B) Signaling pathways that CLEC5A may be involved in (All enrichment analyses were performed with clusterProfiler, org.Hs.eg.db, ggstatsplot, enrichplot, and GOplot in R (Walter et al., 2015)).
High CLEC5A expression of was associated with enhanced M2 macrophage infiltration
To further investigate the interaction between CLEC5A and the immune microenvironment in OC, the R package ESTIMATE was used to examine correlations between immune cell and stromal cell infiltration levels and CLEC5A expression. Notably, we observed a positive correlation between CLEC5A expression and immune cell/stromal cell infiltration. A similar result was obtained based on the GSE9891 dataset (Figs. 7A and 7B). These results indicated that CLEC5A might play a role in mediating the immune response and immune cell and stromal cell infiltration in ovarian tumors. To further explore which immune cell subtype correlated with the CLEC5A expression, we utilized CIBERSORT to calculate the infiltration levels of 22 immune cell subtypes. These included B cells (naive B cell, memory B cell, and plasma cell), T cells [CD8 T cell, naive CD4 T cell, memory resting CD4 T cell, memory activated CD4 T cell, follicular helper T cell (Tfh), regulatory T cells (Tregs), and gamma delta T cell], natural killer (NK) cells (resting NK T cell and activated NK cells), and myeloid subsets [monocytes, MO macrophages, M1 macrophages, M2 macrophages, and resting dendritic cells (DC), activated DCs, resting mast cells, activated mast cells, eosinophils, and neutrophils]. After integrating the results from CIBERSORT (p < 0.05) based on the TCGA and GEO databases, CLEC5A was found to be associated with immune cell infiltration in gynecologic tumors. Further, in OC, CLEC5A expression was positively correlated with M2 macrophages and negatively correlated with plasma cells and Tfh cells (Fig. 7C). We found that the proportion of M2 macrophages was higher in the CLEC5A high expression group than that in the CLEC5A low expression group. In contrast, the plasma cells and Tfh cells were lower in the CLEC5A high expression group (Fig. 8).
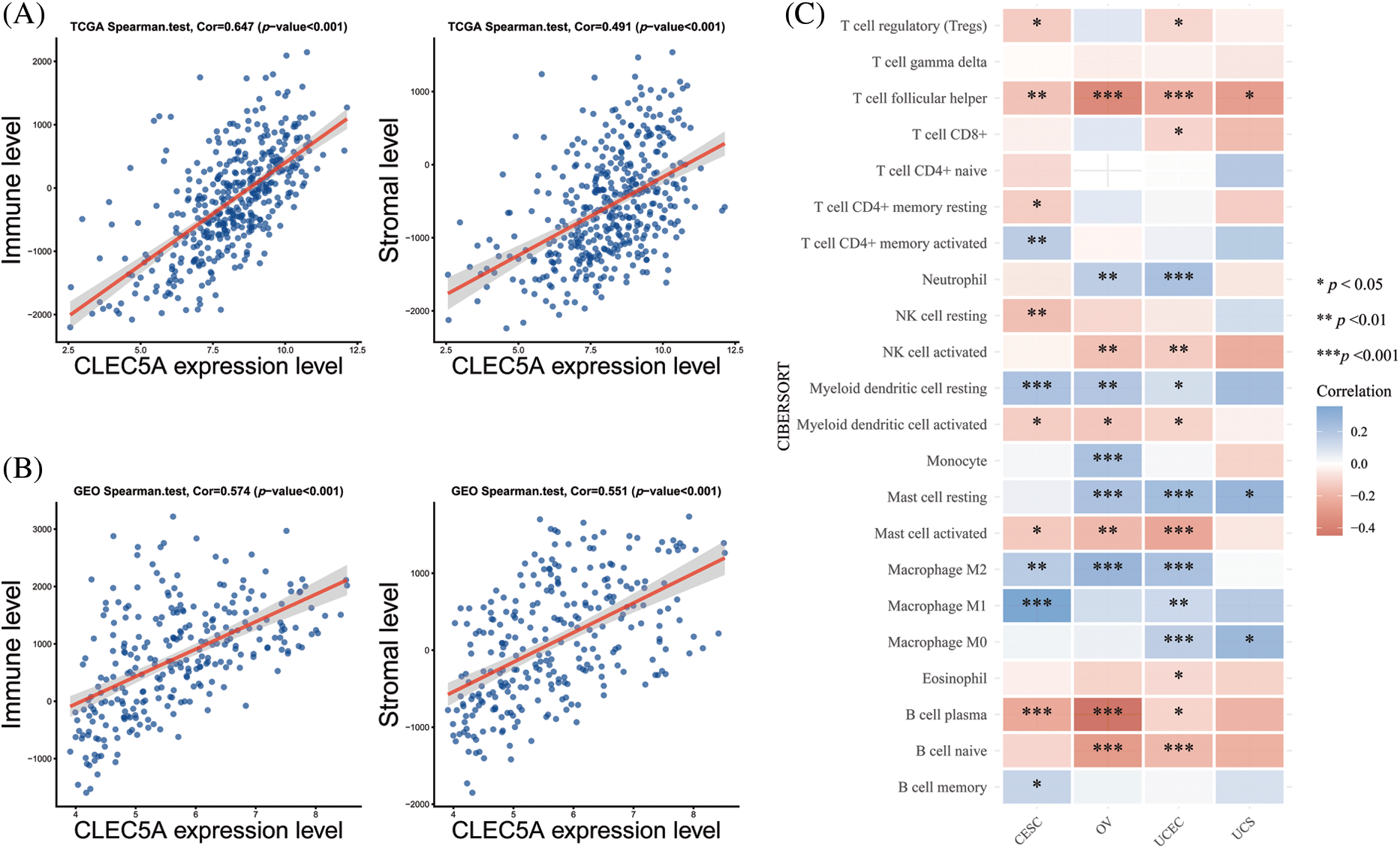
Figure 7: Correlations of CLEC5A with tumor-infiltrating lymphocytes in ovarian cancer (A) Correlations of CLEC5A RNA expression with immune/stromal cell infiltration based on The Cancer Genome Atlas (TCGA) database; (B) Correlations of CLEC5A RNA expression with immune/stromal cell infiltration based on the Gene Expression Omnibus (GEO) database; (C) Correlation of CLEC5A with 22 types of immune cells using the CIBERSORT algorithm in R (*p < 0.05, **p < 0.01, and ***p < 0.001).
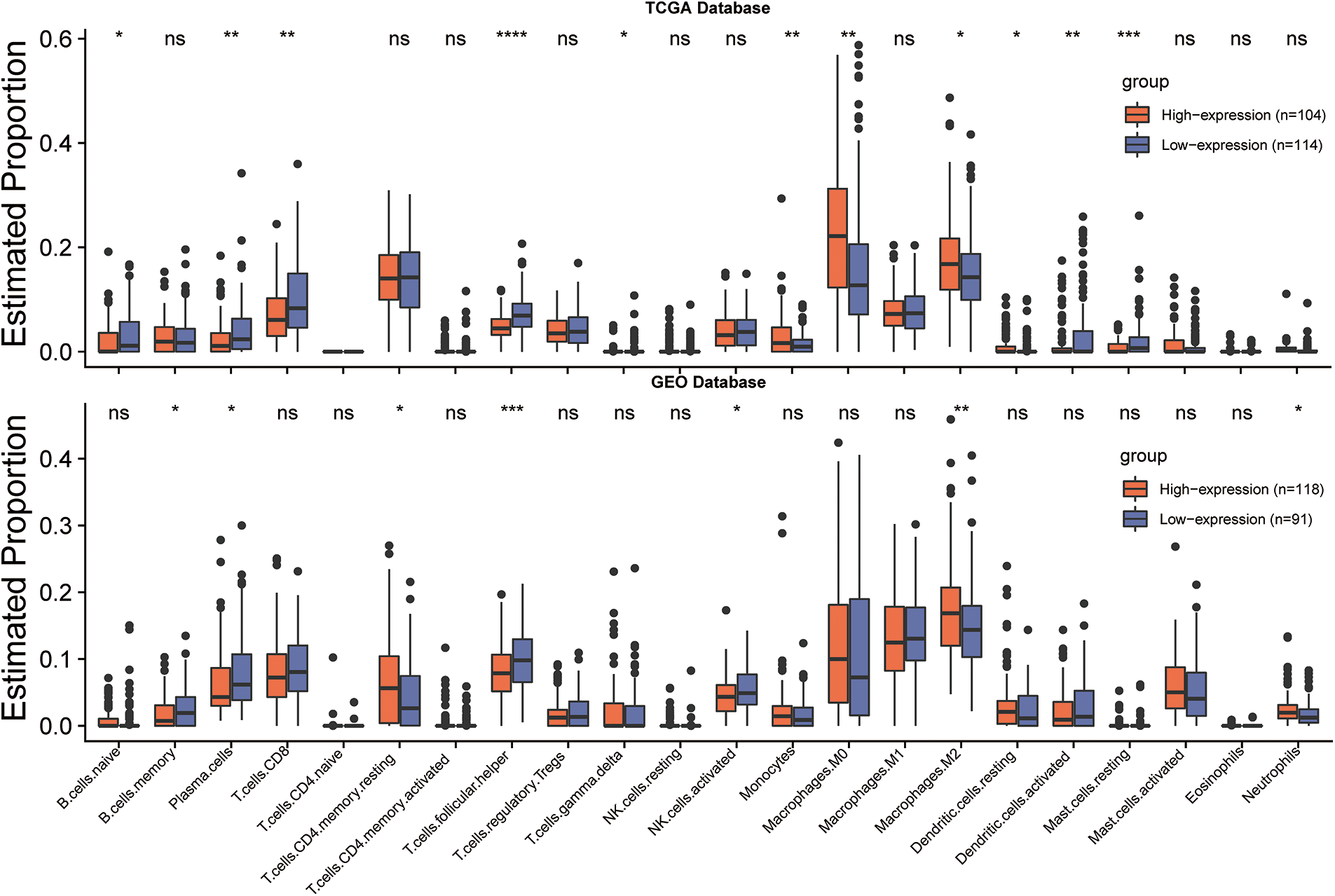
Figure 8: CLEC5A expression levels and tumor-infiltrating lymphocytes in ovarian cancer. Infiltration of 22 immune cell phenotypes in ovarian cancer (OC) using the CIBERSORT algorithm in R (*p < 0.05, **p < 0.01, and ***p < 0.001). Their differences between low and high CLEC5A+ OC are compared with ggplot2 in R.
The accumulation of M2 macrophages has been reported as a pivotal part of promoting tumor progression(Yang and Zhang, 2017). As seen above, the CLEC5A expression level may affect the function of M2 tumor-associated macrophages (TAMs). We thus analyzed the correlations between CLEC5A expression level and 16 factors involved in TAMs- associated with solid tumors. Following our hypothesis, CLEC5A was found to be positively correlated with inhibiting factors based on the TCGA and GEO databases (Figs. 9A and 9B). These results suggest that an increased CLEC5A expression can promote tumors via TAMs.
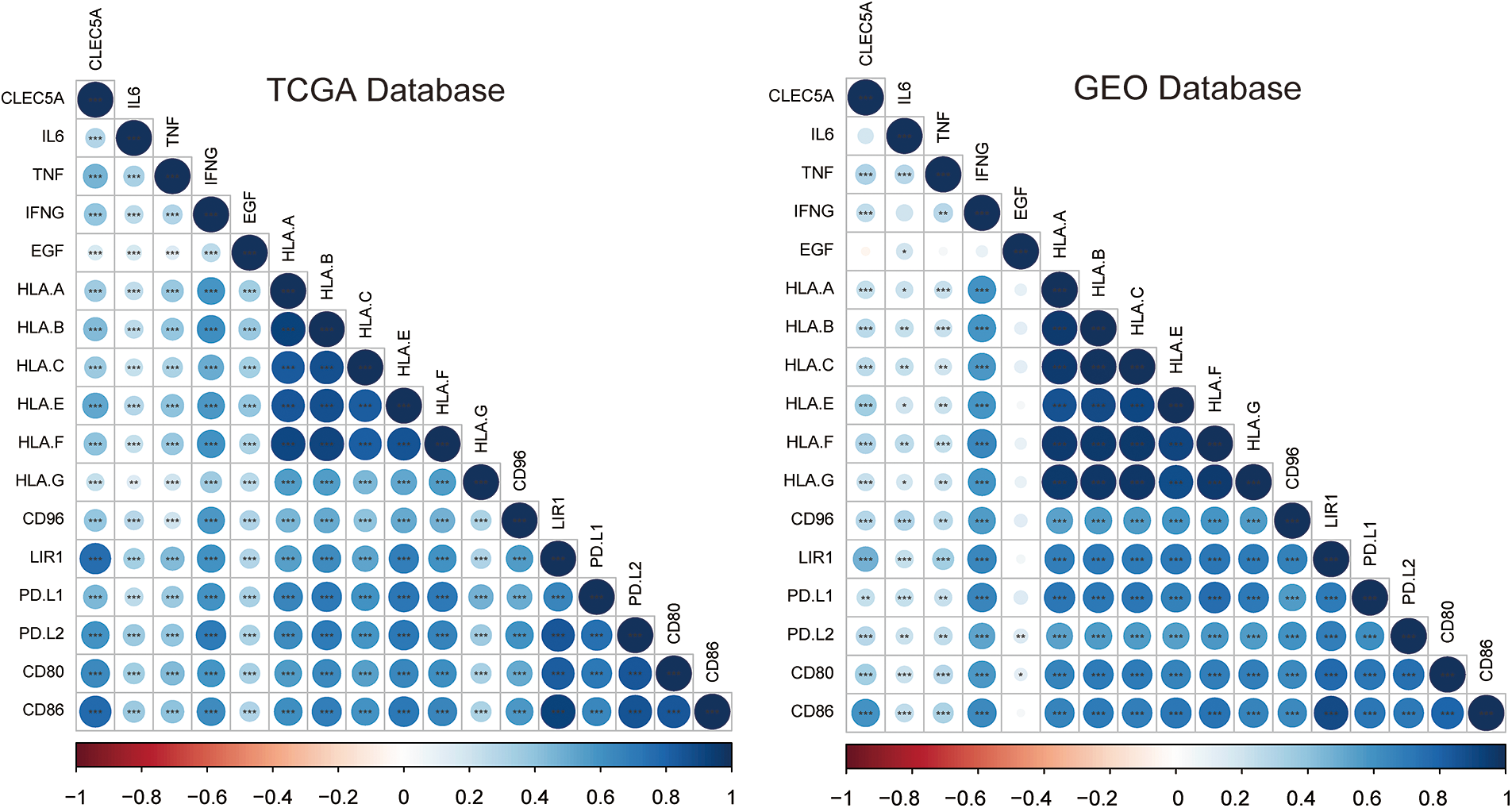
Figure 9: Correlation of CLEC5A with M2 tumor-associated macrophage infiltration. Correlations of CLEC5A with 16 factors associated with M2 tumor-associated macrophage infiltration. The colors represent the magnitude of positive and negative correlations (*p < 0.05, **p < 0.01, and ***p < 0.001).
To summarize, CLEC5A was strongly correlated with the immune environment in OC and its expression level could affect the type of immune cells that infiltrated the tumor. An example is the significant accumulation of M2 macrophages and increased CLEC5A expression. One possible explanation for this accumulation of M2 macrophages might be that the activated PI3K/Akt signaling pathway could cause the macrophage M2 polarization(Vergadi et al., 2017).
New suggestions for clinical drug sensitivity based on CLEC5A expression
We then attempted to offer patients more personalized chemotherapy drug options based on CLEC5A expression. This entailed the evaluation of the responses of the CLEC5A high expression group and low expression group to five chemotherapeutic drugs that were recommended chemotherapeutic regimens. These were cisplatin, docetaxel, doxorubicin, gemcitabine, and paclitaxel. Interestingly, the high CLEC5A expression group showed increased sensitivity to paclitaxel, gemcitabine, and docetaxel with lower half maximal inhibitory concentration (IC50) values. On the contrary, the low CLEC5A expression group was more sensitive to cisplatin and doxorubicin (Fig. 10).
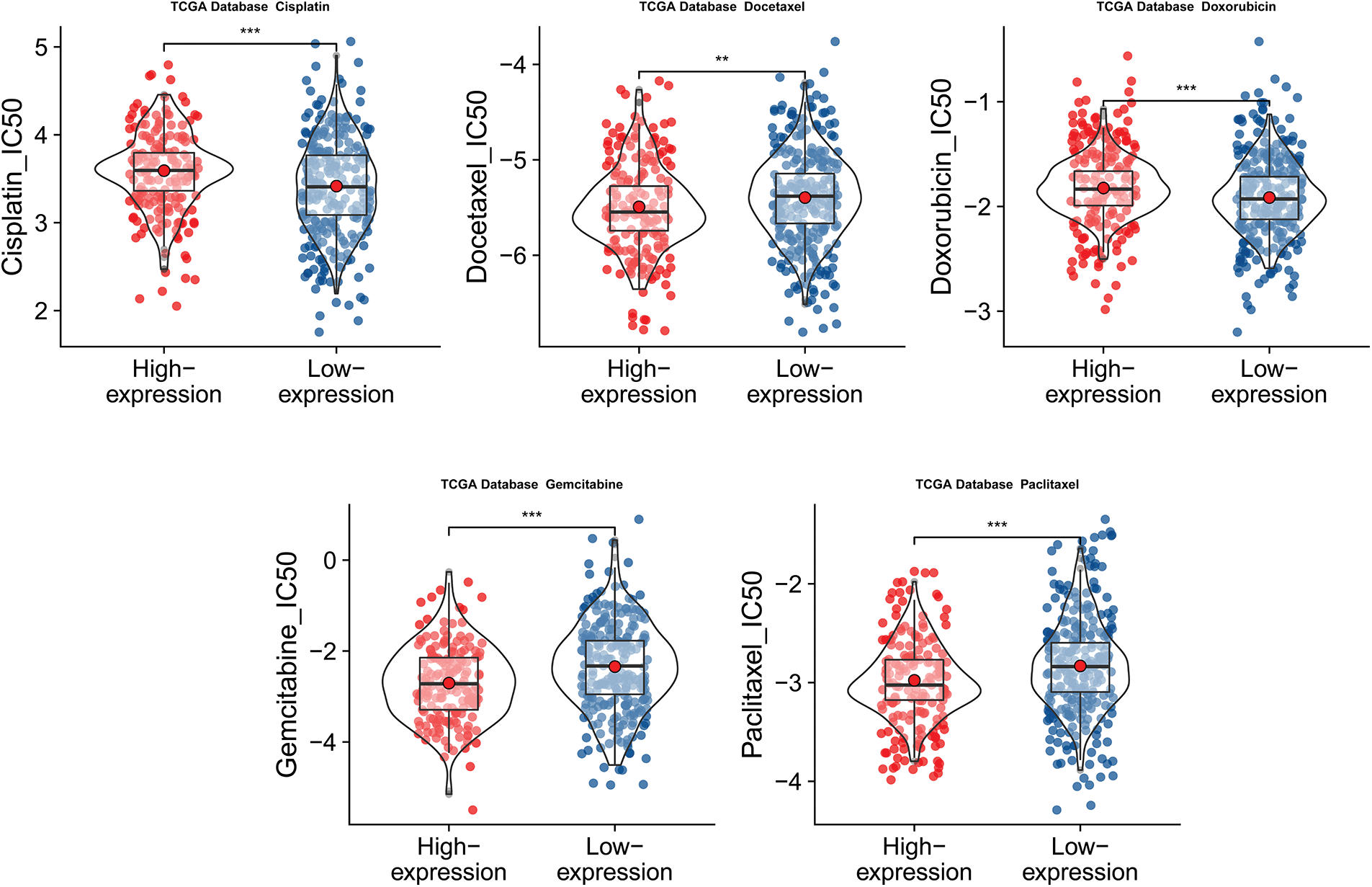
Figure 10: Predicted half maximal inhibitory concentration (IC50) values of five common chemotherapy agents. Predicted half maximal inhibitory concentration (IC50) values of five common chemotherapy agents against low and high CLEC5A expression groups in ovarian cancer (OC) (**p < 0.01 and ***p < 0.001).
In this study, we assayed miRNAs transported by exosomes in the context of OC, explored their influence on the tumor microenvironment, and evaluated their potential prognostic value. We hypothesize that miR-1825 from released OC exosomes leads to elevated expression of its intracellular target gene CLEC5A. In addition, we demonstrated that CLEC5A could not only provides value as a prognostic factor but also presents guidance for immunotherapy and a reference for chemotherapy drug treatment efficacy.
A plethora of studies have implied that exosomal miRNAs can serve as vehicles for intercellular communication. However, the mechanisms of the regulation of the source cells that secrete these exosomes is largely unknown until recently. It has been illustrated that miRNA inhibition promoted oncogenic behavior by stimulating cancer cells to release more exosomes (Kanlikilicer et al., 2016; Ostenfeld et al., 2014; Rashed et al., 2017). In this work, we showed that miR-1825 was highly expressed in exosomes derived from OC cells, but showed low levels in the other three groups (normal ovarian cells, ovarian cancer cells, and exosomes released from normal ovarian cells). MiR-1825 also showed a prognostic value as a potential tumor suppressor. Therefore, we suggest that OC cells secrete the miR-1825 into the extracellular environment via exosomes to maintain their tumorigenic phenotype.
CLEC5A has been found to have a high expression level and the ability to promote cancer in several tumors (Fan et al., 2019; Lu et al., 2019; Wang et al., 2020). However, its expression level and function in OC have not been reported previously. In our cohort, we found that CLEC5A served as a target gene of miR-1825 and was significantly upregulated in OC patients. The upregulation of CLEC5A was further linked to poor OS of OC patients and could also serve as a risk factor. These results indicated that CLEC5A has prognostic value in OC and could be a potential biomarker for OC diagnosis.
Other significant findings of our functional analyses were that CLEC5A played a role in tumor-immune interactions given that the CLEC5A expression was correlated with the immune infiltration level in OC. Our results revealed that there was a significant positive correlation between CLEC5A expression level and the level of infiltration of M2 macrophages and a negative correlation between CLEC5A expression level and the level of infiltration of Tfh cells in OC.
M2 macrophages (also called alternatively activated macrophages) have been involved in the progression of many tumors, including OC (Baek et al., 2020). It has been reported that M2 macrophages are part of tumor-associated macrophages (TAMs) following the view that TAMs are not completely following the M1 and M2 subtypes; they are in general M2-like (Lin et al., 2019; Mehla and Singh, 2019). Increased infiltration of M2 macrophages may be one of the mechanisms by which miR-1825/CLEC5A promotes the development of OC. Recent studies also validate our view that exosomes can deliver tumor suppressor microRNAs to induce M2 macrophage polarization and promote tumor progression (Li et al., 2022). Our study also demonstrated a positive correlation between the high expression level of CLEC5A and the process of TAMs promoting solid tumors.
It has been found that the activation of the PI3K/Akt signaling pathway can promote M2-type polarization in macrophages (Vergadi et al., 2017; Zhao et al., 2020a). Aberrant activation of this signaling network is one of the most frequent events in human cancer and disrupts the control of cell growth, survival, and metabolism from exogenous growth stimuli (Hoxhaj and Manning, 2020). The PI3K/Akt signaling pathway has been shown to exert a pivotal role as an immunomodulator. Blockade of the PI3K pathway might maximize the interferon-gamma (IFN-γ)-mediated anti-tumor effect. On the contrary, activating PI3K/Akt signaling in the neoplastic epithelium can promote nuclear translocation of β-catenin, cellular proliferation, and resistance to apoptosis. Inhibition of PI3K/Akt signaling with immunomodulatory agents may provide a new therapeutic approach for cancer (Bishnupuri et al., 2019; Caforio et al., 2021). In our study, activation of the PI3K/Akt pathway and increased infiltration of M2 macrophages were associated with elevated expression of CLEC5A.
Follicular helper T (Tfh) cells are a subset of CD4+ T cells that specialize in helping B cells to produce antibodies in the face of an antigenic challenge. Their presence in tumor tissue is often associated with a better outcome in several solid tumors (Baumjohann and Brossart, 2021; Song and Craft, 2019). They have also been shown to be positively associated with the long-term survival of humans with breast cancer or non-small cell lung cancer. In our study, the high expression of CLEC5A led to reduced infiltration of Tfh cells. This might be a potential molecular mechanism of protecting tumors by inhibiting ectopic lymphoid structures (ELSs), which are sites of recruitment of CD8+ T cells, NK cells, and macrophages that engage in anti-tumor immunity, or by reducing anti-tumor antibody responses by B cells. However, there is limited data available regarding Tfh cells supporting anti-tumor antibody responses (Crotty, 2019; Gu-Trantien et al., 2013; Ma et al., 2016).
Taken together, our results indicate that efflux of miR-1825 may trigger elevated expression of intracellular target genes and thus induce immune-related behaviors in tumors, especially in the context of OC studied here that result in tumor progression.
Funding Statement: This project was funded by a grant from the National Natural Science Foundation of China (81873045), the Natural Science Foundation of Fujian Province of China (2020J011115), the Medicine Innovation Project of Fujian Province of China (2020CXB007), and the Joint Funds for the Innovation of Science and Technology (2021Y9209).
Author Contributions: The authors confirm their contribution to the paper as follows: YS and QW were responsible for the conception, analysis, and data interpretation. ZC contributed substantially to the conception, design, data interpretation, and revision of the work. All authors wrote different parts of the manuscript, and all of them read and approved the final manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article (and its supplementary information files).
Ethics Approval: This study was approved by the Ethics Committee of the Fujian Cancer Hospital (Reference number: K2022-002-01).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Agarwal V, Bell GW, Nam JW, Bartel DP (2015). Predicting effective microRNA target sites in mammalian mRNAs. eLife 4: e05005. https://doi.org/10.7554/eLife.05005 [Google Scholar] [PubMed] [CrossRef]
Ali Syeda Z, Langden S, Munkhzul C, Lee M, Song SJ (2020). Regulatory mechanism of MicroRNA expression in cancer. International Journal of Molecular Sciences 21: 1723. https://doi.org/10.3390/ijms21051723 [Google Scholar] [PubMed] [CrossRef]
Armstrong DK, Alvarez RD, Bakkum-Gamez JN, Barroilhet L, Behbakht K et al. (2021). Ovarian cancer, version 2.2020, NCCN clinical practice guidelines in oncology. Journal of the National Comprehensive Cancer Network 2: 191–226. https://doi.org/10.6004/jnccn.2021.0007 [Google Scholar] [PubMed] [CrossRef]
Baek SH, Lee HW, Gangadaran P, Oh JM, Zhu L, Rajendran RL, Lee J, Ahn BC (2020). Role of M2-like macrophages in the progression of ovarian cancer. Experimental Cell Research 395: 112211. https://doi.org/10.1016/j.yexcr.2020.112211 [Google Scholar] [PubMed] [CrossRef]
Bakker AB, Baker E, Sutherland GR, Phillips JH, Lanier LL (1999). Myeloid DAP12-associating lectin (MDL)-1 is a cell surface receptor involved in the activation of myeloid cells. Proceedings of the National Academy of Sciences 96: 9792–9796. https://doi.org/10.1073/pnas.96.17.9792 [Google Scholar] [PubMed] [CrossRef]
Baumjohann D, Brossart P (2021). T follicular helper cells: Linking cancer immunotherapy and immune-related adverse events. Journal for Immunotherapy of Cancer 9: e002588. https://doi.org/10.1136/jitc-2021-002588 [Google Scholar] [PubMed] [CrossRef]
Bishnupuri KS, Alvarado DM, Khouri AN, Shabsovich M, Chen B, Dieckgraefe BK, Ciorba MA (2019). IDO1 and kynurenine pathway metabolites activate PI3K-Akt signaling in the neoplastic colon epithelium to promote cancer cell proliferation and inhibit apoptosis. Cancer Research 79: 1138–1150. https://doi.org/10.1158/0008-5472.CAN-18-0668 [Google Scholar] [PubMed] [CrossRef]
Caforio M, de Billy E, De Angelis B, Iacovelli S, Quintarelli C, Paganelli V, Folgiero V (2021). PI3K/Akt pathway: The indestructible role of a vintage target as a support to the most recent immunotherapeutic approaches. Cancers 13: 4040. https://doi.org/10.3390/cancers13164040 [Google Scholar] [PubMed] [CrossRef]
Cassetta L, Pollard JW (2018). Targeting macrophages: Therapeutic approaches in cancer. Nature Reviews Drug Discovery 17: 887–904. https://doi.org/10.1038/nrd.2018.169 [Google Scholar] [PubMed] [CrossRef]
Chen B, Khodadoust MS, Liu CL, Newman AM, Alizadeh AA (2018). Profiling tumor infiltrating immune cells with CIBERSORT. Methods in Molecular Biology 1711: 243–259. https://doi.org/10.1007/978-1-4939-7493-1 [Google Scholar] [CrossRef]
Chen Y, Wang X (2020). miRDB: An online database for prediction of functional microRNA targets. Nucleic Acids Research 48: D127–D131. https://doi.org/10.1093/nar/gkz757 [Google Scholar] [PubMed] [CrossRef]
Chen Y, Xu T, Xie F, Wang L, Liang Z et al. (2021). Evaluating the biological functions of the prognostic genes identified by the Pathology Atlas in bladder cancer. Oncology Reports 45: 191–201. https://doi.org/10.3892/or.2020.7853 [Google Scholar] [PubMed] [CrossRef]
Crotty S (2019). T follicular helper cell biology: A decade of discovery and diseases. Immunity 50: 1132–1148. https://doi.org/10.1016/j.immuni.2019.04.011 [Google Scholar] [PubMed] [CrossRef]
Fan HW, Ni Q, Fan YN, Ma ZX, Li YB (2019). C-type lectin domain family 5, member A (CLEC5A, MDL-1) promotes brain glioblastoma tumorigenesis by regulating PI3K/Akt signalling. Cell Proliferation 52: e12584. https://doi.org/10.1111/cpr.12584 [Google Scholar] [PubMed] [CrossRef]
Geeleher P, Cox N, Huang RS (2014). pRRophetic: An R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One 9: e107468. https://doi.org/10.1371/journal.pone.0107468 [Google Scholar] [PubMed] [CrossRef]
Goldman MJ, Craft B, Hastie M, Repečka K, McDade F et al. (2020). Visualizing and interpreting cancer genomics data via the Xena platform. Nature Biotechnology 38: 675–678. https://doi.org/10.1038/s41587-020-0546-8 [Google Scholar] [PubMed] [CrossRef]
Gu-Trantien C, Loi S, Garaud S, Equeter C, Libin M et al. (2013). CD4+ follicular helper T cell infiltration predicts breast cancer survival. The Journal of Clinical Investigation 123: 2873–2892. https://doi.org/10.1172/JCI67428 [Google Scholar] [PubMed] [CrossRef]
Hoxhaj G, Manning BD (2020). The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nature Reviews Cancer 20: 74–88. https://doi.org/10.1038/s41568-019-0216-7 [Google Scholar] [PubMed] [CrossRef]
Kalluri R, LeBleu VS (2020). The biology, function, and biomedical applications of exosomes. Science 367: eaau6977. https://doi.org/10.1126/science.aau6977 [Google Scholar] [PubMed] [CrossRef]
Kanlikilicer P, Rashed MH, Bayraktar R, Mitra R, Ivan C et al. (2016). Ubiquitous release of exosomal tumor suppressor miR-6126 from ovarian cancer cells. Cancer Research 76: 7194–7207. https://doi.org/10.1158/0008-5472.CAN-16-0714 [Google Scholar] [PubMed] [CrossRef]
Karagkouni D, Paraskevopoulou MD, Chatzopoulos S, Vlachos IS, Tastsoglou S et al. (2018). DIANA-TarBase v8: A decade-long collection of experimentally supported miRNA-gene interactions. Nucleic Acids Research 46: D239–D245. https://doi.org/10.1093/nar/gkx1141 [Google Scholar] [PubMed] [CrossRef]
Li M, Xu H, Qi Y, Pan Z, Li B, Gao Z, Zhao R, Xue H, Li G (2022). Tumor-derived exosomes deliver the tumor suppressor miR-3591-3p to induce M2 macrophage polarization and promote glioma progression. Oncogene 41: 4618–4632. https://doi.org/10.1038/s41388-022-02457-w [Google Scholar] [PubMed] [CrossRef]
Lin Y, Xu J, Lan H (2019). Tumor-associated macrophages in tumor metastasis: Biological roles and clinical therapeutic applications. Journal of Hematology & Oncology 12: 76. https://doi.org/10.1186/s13045-019-0760-3 [Google Scholar] [PubMed] [CrossRef]
Lu J, Chen W, Liu H, Yang H, Liu T (2019). Transcription factor CEBPB inhibits the proliferation of osteosarcoma by regulating downstream target gene CLEC5A. Journal of Clinical Laboratory Analysis 33: e22985. https://doi.org/10.1002/jcla.22985 [Google Scholar] [PubMed] [CrossRef]
Lu F, Li C, Sun Y, Jia T, Li N, Li H (2020). Upregulation of miR-1825 inhibits the progression of glioblastoma by suppressing CDK14 though Wnt/β-catenin signaling pathway. World Journal of Surgical Oncology 18: 147. https://doi.org/10.1186/s12957-020-01927-3 [Google Scholar] [PubMed] [CrossRef]
Ma QY, Huang DY, Zhang HJ, Chen J, Miller W, Chen XF (2016). Function of follicular helper T cell is impaired and correlates with survival time in non-small cell lung cancer. International Immunopharmacology 41: 1–7. https://doi.org/10.1016/j.intimp.2016.10.014 [Google Scholar] [PubMed] [CrossRef]
Mehla K, Singh PK (2019). Metabolic regulation of macrophage polarization in cancer. Trends in Cancer 5: 822–834. https://doi.org/10.1016/j.trecan.2019.10.007 [Google Scholar] [PubMed] [CrossRef]
Odunsi K (2017). Immunotherapy in ovarian cancer. Annals of Oncology 28: viii1–viii7. https://doi.org/10.1093/annonc/mdx444 [Google Scholar] [PubMed] [CrossRef]
Ostenfeld MS, Jeppesen DK, Laurberg JR, Boysen AT, Bramsen JB et al. (2014). Cellular disposal of miR23b by RAB27-dependent exosome release is linked to acquisition of metastatic properties. Cancer Research 74: 5758–5771. https://doi.org/10.1158/0008-5472.CAN-13-3512 [Google Scholar] [PubMed] [CrossRef]
Rashed MH, Kanlikilicer P, Rodriguez-Aguayo C, Pichler M, Bayraktar R et al. (2017). Exosomal miR-940 maintains SRC-mediated oncogenic activity in cancer cells: A possible role for exosomal disposal of tumor suppressor miRNAs. Oncotarget 8: 20145–20164. https://doi.org/10.18632/oncotarget.15525 [Google Scholar] [PubMed] [CrossRef]
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research 43: e47. https://doi.org/10.1093/nar/gkv007 [Google Scholar] [PubMed] [CrossRef]
Song W, Craft J (2019). T follicular helper cell heterogeneity: Time, space, and function. Immunological Reviews 288: 85–96. https://doi.org/10.1111/imr.12740 [Google Scholar] [PubMed] [CrossRef]
Sticht C, de la Torre C, Parveen A, Gretz N (2018). miRWalk: An online resource for prediction of microRNA binding sites. PLoS One 13: e0206239. https://doi.org/10.1371/journal.pone.0206239 [Google Scholar] [PubMed] [CrossRef]
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL et al. (2005). Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America 102: 15545–15550. https://doi.org/10.1073/pnas.0506580102 [Google Scholar] [PubMed] [CrossRef]
Sun Z, Shi K, Yang S, Liu J, Zhou Q, Wang G, Song J, Li Z, Zhang Z, Yuan W (2018). Effect of exosomal miRNA on cancer biology and clinical applications. Molecular Cancer 17: 147. https://doi.org/10.1186/s12943-018-0897-7 [Google Scholar] [PubMed] [CrossRef]
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 Countries. CA: A Cancer Journal for Clinicians 71: 209–249. https://doi.org/10.3322/caac.21660 [Google Scholar] [PubMed] [CrossRef]
Vejnar CE, Blum M, Zdobnov EM (2013). miRmap web: Comprehensive microRNA target prediction online. Nucleic Acids Research 41: W165–168. https://doi.org/10.1093/nar/gkt430 [Google Scholar] [PubMed] [CrossRef]
Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C (2017). Akt signaling pathway in macrophage activation and M1/M2 polarization. Journal of Immunology 198: 1006–1014. https://doi.org/10.4049/jimmunol.1601515 [Google Scholar] [PubMed] [CrossRef]
Walter W, Sánchez-Cabo F, Ricote M (2015). GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 31: 2912–2914. https://doi.org/10.1093/bioinformatics/btv300 [Google Scholar] [PubMed] [CrossRef]
Wang Z, Jensen MA, Zenklusen JC (2016). A practical guide to the cancer genome atlas (TCGA). Methods in Molecular Biology 1418: 111–141. https://doi.org/10.1007/978-1-4939-3578-9 [Google Scholar] [CrossRef]
Wang Q, Shi M, Sun S, Zhou Q, Ding L, Jiang C, Bian T, Jia F, Liu Y, Qin J (2020). CLEC5A promotes the proliferation of gastric cancer cells by activating the PI3K/AKT/mTOR pathway. Biochemical and Biophysical Research Communications 524: 656–662. https://doi.org/10.1016/j.bbrc.2019.10.122 [Google Scholar] [PubMed] [CrossRef]
Yang L, Zhang Y (2017). Tumor-associated macrophages: From basic research to clinical application. Journal of Hematology & Oncology 10: 58. https://doi.org/10.1186/s13045-017-0430-2 [Google Scholar] [PubMed] [CrossRef]
Yoshihara K, Shahmoradgoli M, Martínez E, Vegesna R, Kim H et al. (2013). Inferring tumour purity and stromal and immune cell admixture from expression data. Nature Communications 4: 2612. https://doi.org/10.1038/ncomms3612 [Google Scholar] [PubMed] [CrossRef]
Yu G, Wang LG, Han Y, He QY (2012). ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology 16: 284–287. https://doi.org/10.1089/omi.2011.0118 [Google Scholar] [PubMed] [CrossRef]
Zhang J, Bajari R, Andric D, Gerthoffert F, Lepsa A, Nahal-Bose H, Stein LD, Ferretti V (2019). The international cancer genome consortium data portal. Nature Biotechnology 37: 367–369. https://doi.org/10.1038/s41587-019-0055-9 [Google Scholar] [PubMed] [CrossRef]
Zhao SJ, Kong FQ, Jie J, Li Q, Liu H et al. (2020a). Macrophage MSR1 promotes BMSC osteogenic differentiation and M2-like polarization by activating PI3K/AKT/GSK3β/β-catenin pathway. Theranostics 10: 17–35. https://doi.org/10.7150/thno.36930 [Google Scholar] [PubMed] [CrossRef]
Zhao S, Mi Y, Guan B, Zheng B, Wei P et al. (2020b). Tumor-derived exosomal miR-934 induces macrophage M2 polarization to promote liver metastasis of colorectal cancer. Journal of Hematology & Oncology 13: 156. https://doi.org/10.1186/s13045-020-00991-2 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools