 Open Access
Open Access
ARTICLE
Raloxifene-loaded and aptamer-bonded exosomes induce autophagic and apoptotic death in HeLa cells by enhancing the lysosomotropic effect
1 Department of Medical Biochemistry, School of Medicine, Aydin Adnan Menderes University, Aydin, 09010, Turkey
2 Department of Pharmaceutical Technology, Faculty of Pharmacy, Acibadem Mehmet Ali Aydinlar University, Istanbul, 34758, Turkey
* Corresponding Author: OZGE CEVIK. Email:
BIOCELL 2023, 47(5), 1051-1063. https://doi.org/10.32604/biocell.2023.028129
Received 01 December 2022; Accepted 28 February 2023; Issue published 10 April 2023
Abstract
Background: Raloxifene, a selective estrogen receptor modulator, is also known to be a lysosomotropic agent. The bioavailability of raloxifene is around 2% due to extensive hepatic transport. Exosomes are nanosized vesicles that are naturally released from cells. Method: In this study, exosomes released from HeLa cervical cancer cells were loaded with raloxifene to increase its bioavailability, and an aptamer was attached to the exosome membrane for targeting only HeLa cells. Characterization of exosomes isolated from HeLa cells was performed by transmission electron microscopy, zeta sizer, and western blotting. In addition, the cytotoxic, apoptotic, autophagic, and lysosomotropic effects of the prepared Exo-Apt-Ral formulation on HeLa cervical cancer cells were investigated. Results: According to zeta analysis, the sizes of the empty exosome and Exo-Apt-Ral formulation were measured as 66 ± 12 and 120 ± 21 nm, respectively. There was a rise in the lysosomal permeability of HeLa cells after the Exo-Apt-Ral application. In addition, both apoptotic and autophagic death mechanisms were triggered in HeLa cells after the Exo-Apt-Ral application. Conclusion: This study showed that raloxifene functionalized by loading into aptamer-bound exosomes can be a new targeted drug carrier system for cervical cancer.Keywords
Conventional cancer therapy approaches such as radiotherapy, chemotherapy, photodynamic therapy, and photothermal therapy can induce severe side effects in tissues other than the target tissue. As a result, research on targeted treatment methods to minimize these side effects has greatly accelerated. One of the primary clinical approaches used for targeted cancer therapy is aptamers. Aptamers are DNA and RNA oligonucleotides that selectively bind to their target ligands with high affinity. Although their molecular weights are about one-tenth smaller than monoclonal antibodies, they interact closely with target molecules due to their complex tertiary 3D structure. Therefore, aptamers are ideal agents for protein and cell targeting (Barbas et al., 2010; Sun et al., 2014; Yang et al., 2022).
Autophagy is one of the most studied cellular processes in which intracellular components are directed to lysosomes or other vacuoles for degradation. The substrates of autophagy are proteins and organelles that have completed their normal physiological process or are damaged. The degradation products of these substrates are recycled to help the cell survive under different stress circumstances such as nutrient shortage, growth factor deprivation, hypoxia, and infections (Levine, 2007).
Lysosomes are membrane-enclosed cytoplasmic vesicles found in all eukaryotic cells. They play critical roles in cellular pathways such as autophagy, endocytosis, exocytosis, and phagocytosis. The main feature that distinguishes lysosomes from other intercellular vesicles is their high intramembrane acidity. Lysosomal membrane proton pumps provide an acidic medium for the three-dimensional structures of biological macromolecules, including proteins, to relax and become available for degradation and, at the same time to establish an optimal pH for the activity of lysosomal acid hydrolases. The deterioration of biological macromolecules within the lysosome is facilitated by more than 50 hydrolases, including proteases, nucleases, lipases, glycosidases, phosphatases, and sulfatases (Zhao et al., 2020). The most crucial molecular change in lysosome-based cell death mechanisms is the deformation of the lysosomal membrane structure.
Lysosomal membrane permeability can be increased by several agents that were named “lysosomotropic” by Duve in 1974 (de Duve et al., 1974). Once lysosomotropic agents cross the lysosomal membrane, they become protonated and accumulate in the lysosome cytoplasm. When the concentration of the lysosomotropic agent reaches a critical level, the agent acquires membranolytic characteristics, allowing the lysosomal contents to be released into the cell cytoplasm. The leakage of lysosomal acid hydrolases triggers cell death mechanisms in the cytoplasm (Ndolo et al., 2012). The lysosomotropic activity of raloxifene, an estrogen receptor modulator for the cure of osteoporosis in the postmenopausal period, has been reported. However, taken orally in tablet form, raloxifene is catabolized due to extensive hepatic transmission, and its absolute bioavailability is reduced to 2% (Selyunin et al., 2021). Therefore, it is essential for lysosomotropic drugs, including raloxifene, to be targeted to cells and formulations with delivery systems in cancer therapy. In this study, raloxifene was formulated utilizing exosomes, which are part of cells and function as natural carriers. The autophagic effects were investigated by directing it to cervical cancer cells via aptamers.
HeLa human cervical cancer cells and L929 mouse fibroblast cells provided from the American Type Culture Collection (CCL-2) were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM) medium with 10% fetal bovine serum, 1% penicillin/streptomycin containing L-glutamine and HEPES, in 25 cm2 flasks, at 5% CO2 and 37°C ambient conditions. When the cells filled the flasks 80%–90%, they were passaged into new 25 and 75 cm2 flasks, and the stocks were multiplied for use in subsequent experiments (Erdogan et al., 2019).
Exosome isolation from HeLa cervical cancer cells
The HeLa human cervical cancer cells were seeded into 75 cm2 flasks. They were cultured at 5% CO2 and in DMEM medium containing 10% fetal bovine serum and 1% penicillin-streptomycin. When the cells reached 90% occupancy, they were washed twice with Dulbecco’s phosphate buffered saline (DPBS). To induce exosome release, low glucose DMEM medium containing only 1% penicillin-streptomycin was added to the cells. This medium was collected every two days. To remove cell debris from the medium, the collected medium was first centrifuged at 4.000 rpm for 30 min and then at 30.000 rpm for 30–60 min. The supernatant was passed through a 0.22 µm diameter filter. Then, it was filled into ultracentrifuge tubes and the caps of the tubes were melted with a capping machine and closed. It was centrifuged at 90.000 rpm for 7 h in an ultracentrifuge. The supernatant was poured and the collapsed pellet was dissolved in 200 µL of phosphate buffer (pH 7.4). Total protein was measured by spectrophotometer in the UV region (260 nm) (Cenik et al., 2022).
Loading of raloxifene into exosomes by electroporation technique
An electroporator was used to load raloxifene into exosomes. For this, exosome and raloxifene at different concentrations (1–1000 µM) were added to 400 µL of PBS. This prepared mixture was loaded into a 400 µL electroporation cuvette. The protocol specific to HeLa cells was loaded into the electroporator (voltage = 400 V, capacitance = 125 µF, resistance = ∞Ω). The electroporation cuvette was placed in its chamber in the device and loading was performed after the given impulses. The samples taken from the cuvette were transferred to microfuge tubes (Isolab GmbH, Eschau, Germany). The samples were precipitated with ultracentrifuge at 90.000 rpm for 3 h to remove the drug that was not loaded on the exosome in the medium. The supernatant was poured and the pellet was dissolved in PBS. It was stored at −20°C until use (Cenik et al., 2022).
Calculation of loading doses of raloxifene into exosomes
Loading doses of raloxifene on exosomes were calculated spectrophotometrically as earlier reported by Nagaraju et al. (2014). After measuring the optical density at 288 nm of 5, 10, 15, 20, and 25 µM doses of raloxifene, the standard graph was drawn. The entrapment efficiency of raloxifene was calculated from the standard graph by taking the optical density of the exosome-drug medium before and after loading (Demirbolat et al., 2022b).
Assays for aptamers binding to exosomes
For drug targeting, the Apt13 and Apt20 aptamers, which specifically bind to the L1 capsid protein found on the membrane of HeLa cervical cancer cells, were selected from the literature (Graham and Zarbl, 2012). The sequences of the aptamers are 5′GGGACAGACGGAAGATGAGAATTGTGGGCTTAGTATAGTGAGGTGCGGT-3′ for Apt13 and 5′GGGGAGGGAGACACA GTCATGGAGCAGTTATTAGGGTGTACCGGGTGTAGT-3′ for Apt20.
In aptamer binding to exosomes released from HeLa cells, aptamers were dissolved in tris- ethylenediaminetetraacetate (EDTA) buffer and a modified binding buffer (pH 7.4) was prepared in accordance with earlier protocols (Embark, 2016; Siddiqui and Yuan, 2021). For the binding experiment, optimization studies were performed using 1–500 µg protein-containing exosomes and 1–1000 nM aptamers. Eppendorf tubes were incubated at 37°C for 0–240 min. The amount of single-stranded DNA (ssDNA) in the samples was measured in the nanodrop and the amount of aptamer bound was calculated.
Binding assays of Exo-Apt-Ral to HeLa cells
The binding affinities of aptamer-bound exosomes (Exo-Apt-Ral) to HeLa and L929 cells were demonstrated by fluorescence microscopy. For this, carboxyfluorescein-labeled aptamers were used. 1 × 106 HeLa and L929 cells were seeded in 6-well plates. After 24 h of incubation for the cells to adhere to the plate base, FAM-labeled aptamer-bound exosomes (100 µg) were added to the medium. After 12 h of incubation, the cells were imaged under a fluorescent microscope (Graham and Zarbl, 2012).
Uptake of exosomes by HeLa cells
HeLa cells were seeded in 12-well plates at 1 × 105 cells per well. The cells were incubated in a carbon dioxide oven for 24 h to adhere to the bottom of the plate. The cells were incubated for 24 h with the half maximal inhibitory concentration (IC50) dose of Exo-Apt-Ral formulation. After incubation, PKH67 exosome dye was added to the medium and after 60 min of incubation, images of cells were taken under a fluorescent microscope.
The release profile of raloxifene was carried out by dialysis bag diffusion procedure. After the dialysis bag (MW 12 kDa) was absorbed in the phosphate buffered saline (PBS), pH 7.4 overnight, a specific amount of Exo-Apt-Ral was put into the dialysis bag. The dialysis bags were closed from both sides, and were placed in bakers including 100 mL of PBS of pH 7.4, and incubated in a water bath set at 37°C ± 0.1°C. The system was repeatedly stirred at 100 rpm. At prearranged time periods, 1 mL of the media was withdrawn and refilled with fresh release medium at the same temperature. Samples were measured with a UV spectrophotometer, and the concentration of each sample was assayed. All the release studies were carried through in triplicate and the mean values with standard deviation were calculated (Demirbolat et al., 2022a).
HeLa cell viability measurement
HeLa cells were added in 96-well plates at a density of 1 × 104. After the cells adhered to the bottom of the plate, 1,10,100 and 1000 µM doses of raloxifene and Exo-Apt-Ral formulations were added to the wells. After 24 h of incubation, 10 µL of 3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyl tetrazolium bromide (MTT) dye was added to each well. After 4 h of incubation, the medium was discharged and 100 µL of DMSO was added to all wells. The absorbance was read at 570 nm (Erdoğan et al., 2021).
HeLa cells were seeded in 12-well plates at a density of 1 × 105. After 24 h of incubation, a line was drawn in the middle of each well with a pipette tip. The IC50 doses of raloxifene and Exo-Apt-Ral were added to each well. Images of cells were taken at 0, 24, 48, and 72 h. The migration amount was measured with the ImageJ program (Paşa et al., 2021).
HeLa cells were seeded in 12-well plates at 1 × 105 cells per well. The cells were incubated in a carbon dioxide oven for 24 h to adhere to the bottom of the plate. The cells were incubated for 24 h with the IC50 dose of the raloxifene and the Exo-Apt-Ral formulation. After incubation, lysotracker DND-26 dye (50 nM) was supplied to the medium and after 30 min of incubation, images of cells were taken under a fluorescent microscope and changes in lysosomal functions were observed (Ye et al., 2018).
HeLa cells were seeded in 6-well plates with the 1 × 106 cell density. They were incubated for 24 h for cells to adhere. The IC50 doses of raloxifene and Exo-Apt-Ral were added to each well. After 24 h of incubation, cells were removed with 200 µL of trypsin and transferred to microfuge tubes. After centrifugation at 1200 rpm for 5 min, the supernatant was discarded. The cells were dissolved in 100 µL of medium and 100 µL of annexin V reagent was added. After 20 min of incubation in the dark, apoptosis was measured with the Muse Cell Analyzer (Türk et al., 2020).
This experiment was performed to investigate the safety of raloxifene and Exo-Apt-Ral formulations for intravenous administration. Freshly collected venous blood was taken into tubes containing EDTA. Tubes were centrifuged at 1000 × g for 15 min. Precipitated erythrocytes were washed 3 times with pH 7.4 phosphate buffer. To prepare a 10% erythrocyte solution, 100 µL of red blood cells were taken and 900 µL of phosphate buffer (pH 7.4) was added. 50 µL of this prepared erythrocyte solution was taken and 450 µL of raloxifene and Exo-Apt-Ral were added at concentrations of (1, 10, 100, and 1000 µM). NaCl (0.9%) was used as the negative control and distilled water was used as a positive control. Microfuge tubes were incubated at 37°C for 30 min and then centrifuged at 1000 × g for 15 min. The absorbance of the color formed by taking 100 µL from the supernatant was measured at 540 nm (Demirbolat et al., 2022b).
Apoptotic and autophagic protein expression measurement by western blotting
HeLa cells were seeded in 6-well plates at 1 × 106 cells per well. The cells were incubated in a carbon dioxide oven for 24 h to adhere to the bottom of the plate. Cells were incubated with IC50 doses of raloxifene and Exo-Apt-Ral formulations for 24 h. After 24 h of incubation, the cells were removed and the cells were lysed by adding 350 µL of 2X sample loading buffer. Using solutions of pH 6.8, 0.5 M Tris, pH 8.8 1.5 M Tris, 30% acrylamide-bisacrylamide, 10% ammonium persulfate, 10% sodium dodecyl sulfate (SDS) and TEMED, 4% stacking, and 12% separation gel were poured. The samples containing 5–10 µg of protein were loaded onto a gel and run with a running buffer (5 mM Tris, 38.4 mM glycine, 1% SDS) at 100 V constant current for 1–2 h. After electrophoresis, immunoblotting was performed with a semi-dry system to transfer the protein bands in the gel to the polyvinylidene fluoride (PVDF) membrane. The gel was carefully placed on the membrane, and a nitrocellulose layer wetted with a transfer buffer was placed on it again. In a semi-dry transfer device (Biorad Transblot Turbo), blotting was performed at 25 V and 1 Amp constant current for 30 min. After transferring the proteins to the PVDF membrane, the membrane was washed three times with TBST (20 mM Tris, 154 mM NaCl, 0.1% Tween 20) and blocked with 2.5% BSA for 2 h. After washing three times with TBST, the PVDF membrane was incubated with the following primary antibodies BCL-2-associated X protein (Bax, Santa Cruz SC7480), BCL-2 (Santa Cruz SC7382), LC3AB (Cell Signaling 12741), Beclin-1 (Cell Signaling 3495), and B-actin (Santa Cruz SC47778) at 4°C overnight on an orbital shaker. After incubation, the membrane was washed three times with TBST for 10 min. Subsequently, the membrane was incubated with horse radish peroxidase-conjugated secondary antibody for 2 h at room temperature, the membrane was incubated with chemiluminescence reagent (ECL, Santa Cruz) for 1 min in the dark. The images of the bands were taken with the imaging system (Syngene GBOX). Densitometric analysis of the bands was done with the ImageJ program (Paşa et al., 2021).
Apoptotic and autophagic gene expressions measurements by quantitative polymerase chain reaction
The total RNA content was isolated from 5 × 106 of HeLa cells as previously described (Paşa et al., 2021). 1 μg total RNA was reverse transcribed as the template for cDNA synthesis using a high-capacity cDNA Reverse Transcription Kit (Applied Biosytems Waltham, USA). Quantitative real-time PCR (qRT-PCR) was carried out with Bcl-2, Bax, LC3B, Beclin-1, and GAPDH primers. The primer sequences are given here: Bcl-2: 5′-GACAGAAGATCATGCCGTCC-3′ (forward), 5′-GGTACCAATGGCACTTCAAG-3′ (reverse); Bax: 5′-GCCCTTTTGCTTCAGGGTTT-3′ (forward), 5′-TCCAATGTCCAGCCCATGAT-3′ (reverse); LC3B: 5′-ACTTTGTTGTTTGGCAGAAGC-3′ (forward), 5′-TTTGTCCCGAGCCTTCATT-3′ (reverse); Beclin1: 5′-CGGAAACCATTCATATCTGG AG-3′ (forward), 5′-TCCCAGAAAAACCGCAAC-3′ (reverse); GAPDH: 5′-TGCACCACC AACTGCTTAGC-3′ (forward), 5′-GGCATGGACTGTGGTCATGAG-3′ (reverse). 100 nanograms of cDNA were amplified by Sybr Green PCR Master Mix (Applied Biosystem) on the ABI StepOne Plus detection system. The program parameter for amplification was 95°C for 10 min, then 40 cycles of 95°C for 15 s, and 60°C for 1 min. The qPCR data were resolved using StepOne Software v2.3 (Applied Biosystems, Foster City, CA), and the genes of interest were normalized to the corresponding GAPDH results. Data plotted as fold change vs. control (Erdogan et al., 2019).
Data from three independent experiments are presented as mean ± SD. Variations between groups were assessed by the One Way ANOVA test. Statistical analysis was carried out with GraphPad Prism version 7.0 software. Statistical importance was considered as follows: *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Characterization of HeLa exosomes
The expression of CD9, CD63 and CD81 proteins expressed in the membrane of the HeLa cervical cancer cells exosomes isolated by the ultracentrifugation method was demonstrated by western blotting. The expression of CD9, CD63, and CD81 proteins in cell lysates was less pronounced than the expression of CD9, CD63, and CD81 proteins in the exosome, according to the amount of equally loaded protein (Fig. 1A). Dimensional analysis of exosomes isolated from HeLa cervical cancer cells was performed with transmission electron microscopy (TEM) and zeta sizer. Dimensions of the Exo-Apt-Ral formulation were analyzed with a zeta-sizer. According to the zeta results, the size of the pure exosome was measured as 66 ± 12 nm, while the size of the Exo-Apt-Ral was 120 ± 21 nm (Fig. 1B). When the TEM image was examined, spherical structures of approximately 52.34 and 116.1 nm for empty exosome and raloxifene loaded exosome were observed, respectively (Figs. 1C and 1D).
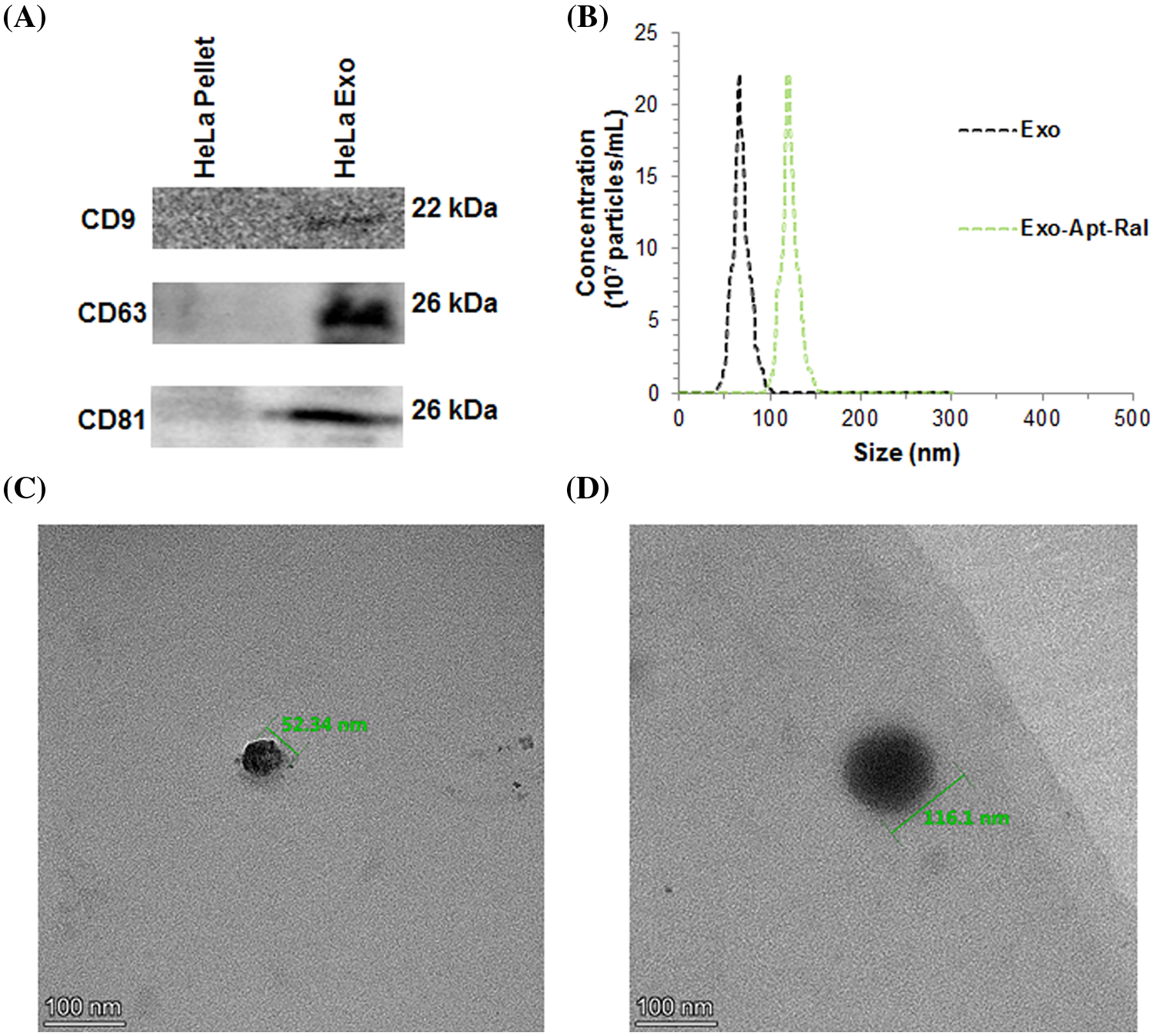
Figure 1: Characterizations of exosomes. (A) Representation of Western blot bands of specific biomarkers expression from exosomes. (B) Size distribution of exosomes and raloxifene-loaded exosomes measuring with zeta sizer. (C) Transmission electron microscopy (TEM) image of an empty exosome (D) TEM image of a raloxifene-loaded exosome (Scale bar = 100 nm).
Entrapment efficiency and raloxifene drug release
The entrapment efficiency of raloxifene was analyzed by the spectrophotometric method. The linear regression equation of raloxifene was y = 43.03x − 1.6011, r2 = 0.998 (Suppl. Fig. S1). The entrapment efficiency of raloxifene was calculated to be 72% ± 2.15%.
The release profile of raloxifene in the Exo-Apt-Ral formulation in PBS at different pH is shown in Fig. 2D. The prepared formulation released more raloxifene at pH = 5.0. In PBS at pH = 5.0, pH = 7.4, and pH = 8.2, raloxifene was found to be maximally released at approximately 40 h.
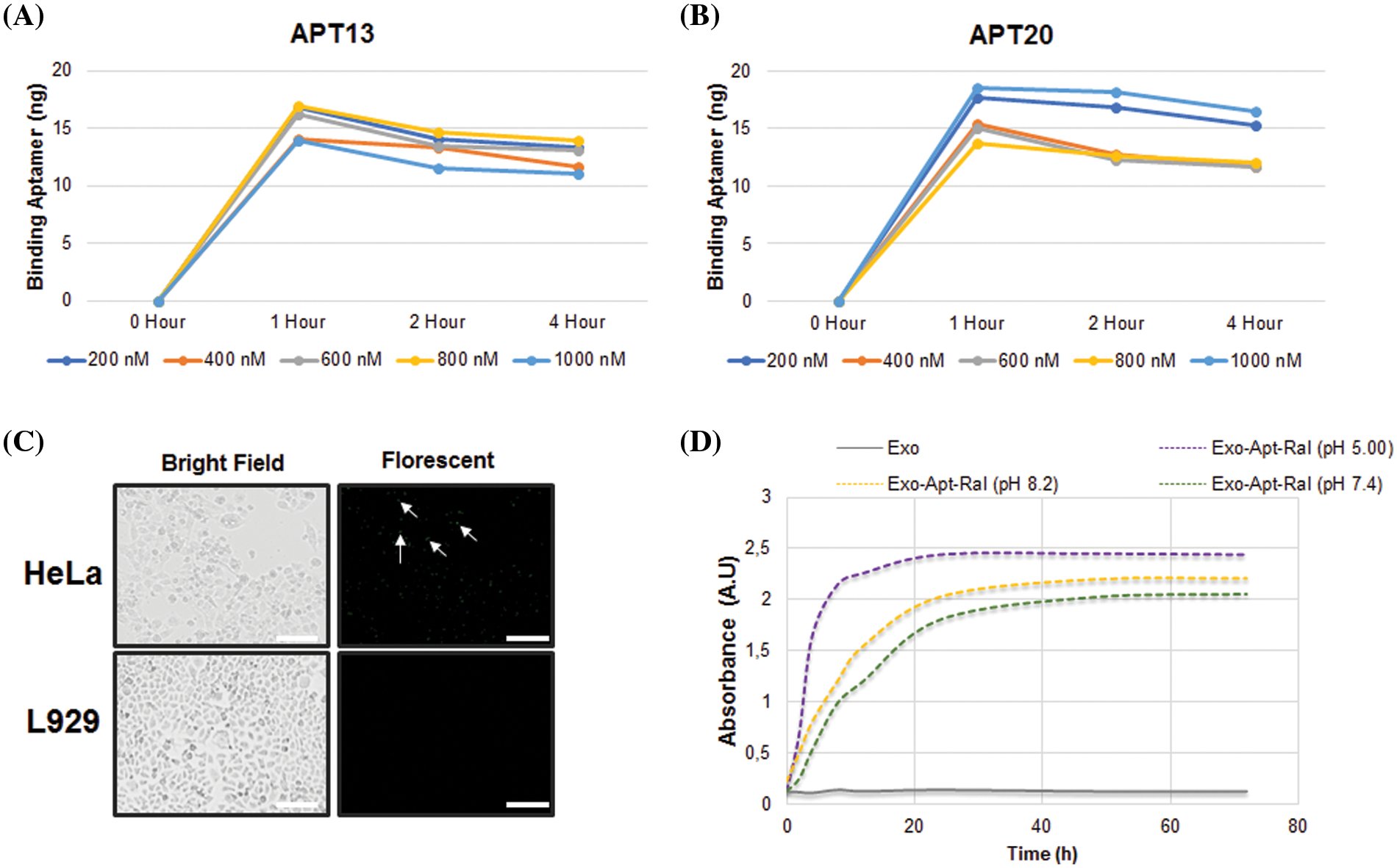
Figure 2: Aptamer binding and drug release studies. (A) Time-dependent binding assay of Apt13 aptamers to exosomes. (B) Time-dependent binding assay of Apt20 aptamers to exosomes. (C) Demonstration of Exo-Apt-Ral binding to HeLa and L929 cells with fluorescently-labeled aptamers (Scale bar = 200 µm). (D) Cumulative raloxifene release in PBS buffer at different pH (5.0, 7.4, and 8.2) (Each experiment was done in triplicate).
The binding levels of Apt13 and Apt20 aptamers to the exosomes, which specifically bind to the L1 capsid protein in the membrane of HeLa cervical cancer cells, were measured spectrophotometrically. While Apt13 and Apt20 aptamers seemed to bind maximally for up to 1 h depending on the concentration, the Apt20 aptamer retained its binding over time. For this reason, Apt20 aptamer was preferentially selected in formulation studies of raloxifene because it is more stable than Apt13 (Figs. 2A and 2B).
To demonstrate that Exo-Apt-Ral specifically binds to HeLa cells, a raloxifene-loaded exosome coupled with fluorescently labeled Apt20 aptamer was used. The Exo-Apt-Ral formulation entering the cells after incubation with Exo-Apt-Ral HeLa cells has been shown (Fig. 2C) with a fluorescent microscope. L929 cells were used as a negative control. Fluorescently labeled formulations in HeLa cells are indicated by the tip of the arrows.
Effects of Exo-Apt-Ral on HeLa and L-929 cell viability
The exosome uptake behavior of HeLa cells was determined by PKH67 exosome staining (Fig. 3A). Observations of an increase in green fluorescence and lumen formation supported the uptake of external exosomes into cells.
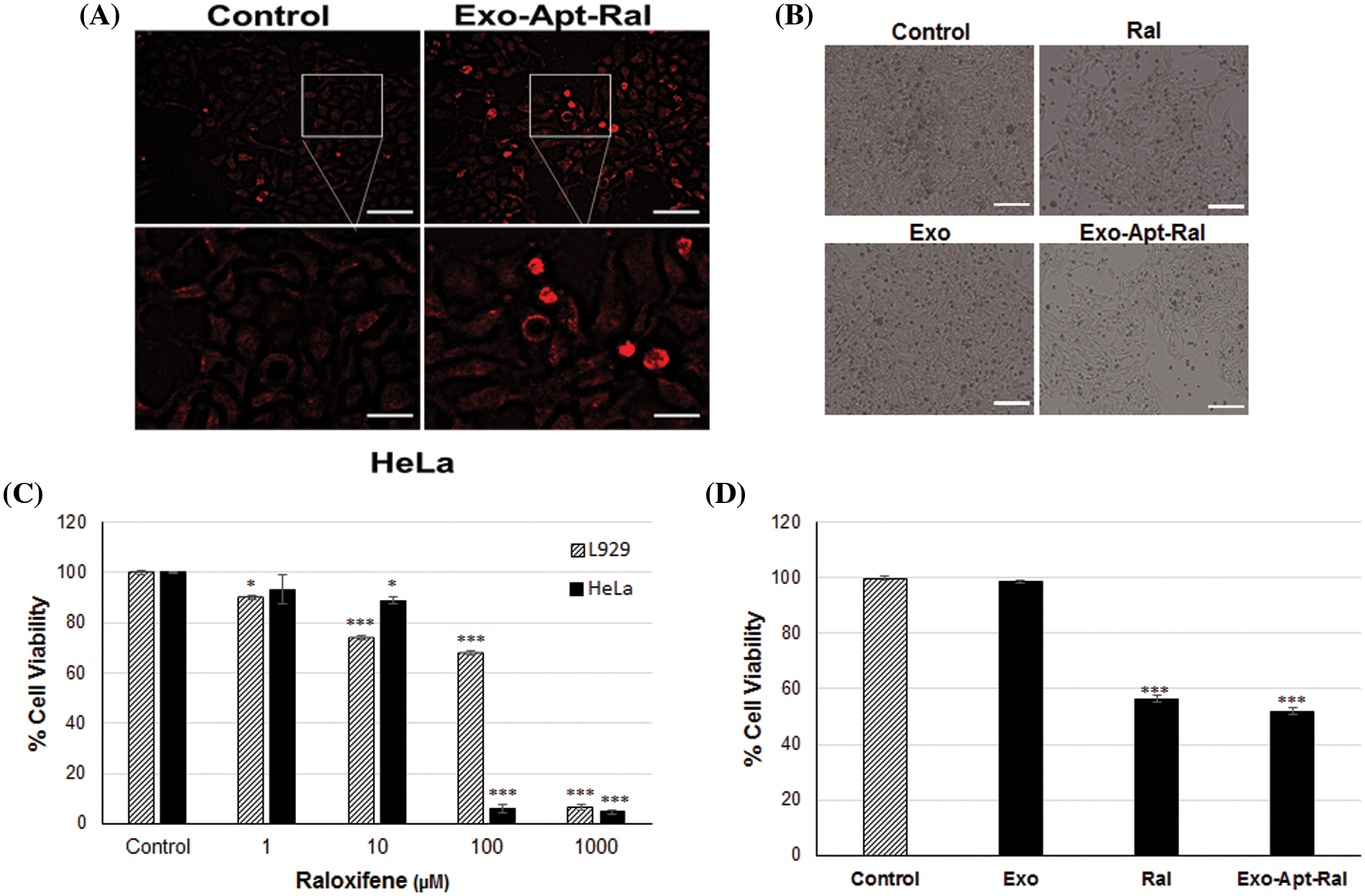
Figure 3: The uptake experiments and cytotoxic effects of exosomes, raloxifene, and Exo-Apt-Ral. (A) The illustration of the uptake of exosomes by HeLa Cells with PKH67 exosome staining (Scale bar = 100 µm for the top two images, scale bar = 50 µm for the bottom two images). (B) Morphological images of HeLa cells treated with raloxifene and Exo-Apt-Ral formulations (Scale bar = 200 µm). (C) Cell viability graphs of HeLa and L929 cells of raloxifene at 24 h of incubation. (D) Cell viability graphs of exosome, raloxifene, and Exo-Apt-Ral administered groups at 24 h of incubation (Each group was studied in triplicate for cytotoxicity experiments, *, p < 0.05; ***, p < 0.001 compared to the control group).
The changes in cell morphology after the treatment of raloxifene and Exo-Apt-Ral formulations to HeLa cells were examined with an inverted microscope. It was determined that the cells in the Exo-Apt-Ral treated groups moved away from each other and their cell size decreased compared to the cells in the control group (Fig. 3B). The 24-h cytotoxic effects of 1, 10, 100, and 1000 µM doses of raloxifene on HeLa and L929 cells were investigated by the MTT method. The IC50 values of cells treated with raloxifene were determined as 22.19 ± 1.37 µM and 107.40 ± 8.81 µM for HeLa and L929, respectively (Fig. 3C). It was determined that when Exo-Apt-Ral was incubated with 22.19 µM of raloxifene, cell viability was significantly reduced compared to cells treated with empty exosomes (p < 0.001) (Fig. 3D).
Effects of Exo-Apt-Ral on HeLa cell migration
The formed wounds at 0 h and the healing progression at 24, 48, and 72 h in HeLa cells are shown in Fig. 4A. In cells treated with raloxifene, the migration rate decreased to 55.84% after 72 h (p < 0.001). In cells treated with Exo-Apt-Ral, the migration rate decreased to 46.14% after 72 h (p < 0.001) (Fig. 4B).
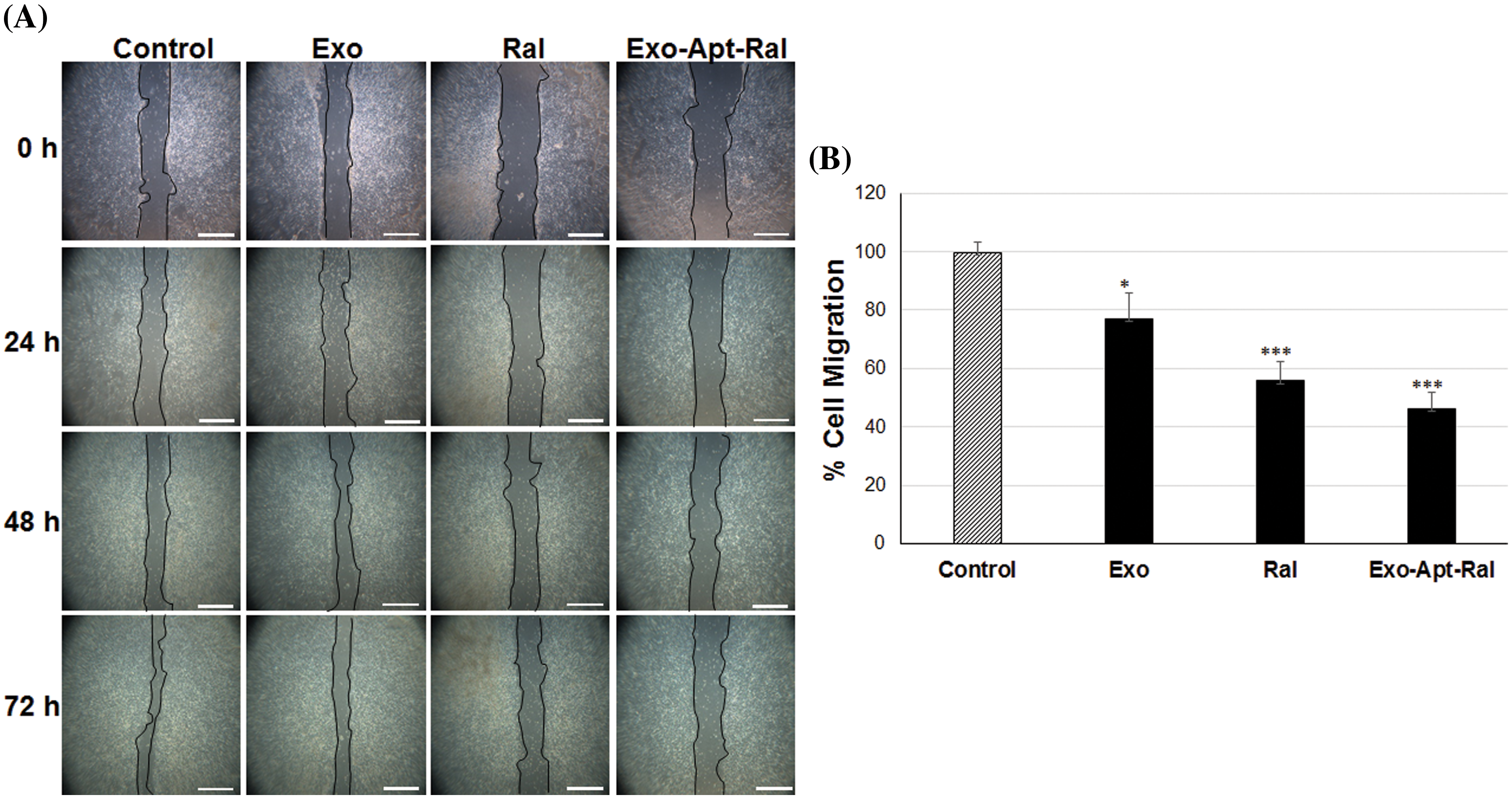
Figure 4: The effects of Exo-Apt-Ral on cell migration in HeLa cells. (A) Cell migration was measured with the scratch wound healing assay of wounded HeLa cells at 0, 24, 48, and 72 h (Scale bar = 100 μm). (B) Cell migration graphs of HeLa cells treated with exosome, raloxifene, and Exo-Apt-Ral for the three biological replicates within each group (*p < 0.05; ***, p < 0.001 compared to the control group).
Effects of Exo-Apt-Ral on erythrocytes
An in vitro hemolysis experiment was carried out to investigate whether the prepared formulation is safe for intravenous administration. As a result of the hemolysis experiment (Suppl. Fig. S2A), raloxifene and Exo-Apt-Ral (200 μg/mL) significantly reduced the hemolysis rates of red blood cells compared to the positive control (p < 0.001) (Suppl. Fig. S2B).
Effects of the Exo-Apt-Ral formulation on mitochondrial apoptotic proteins in HeLa cells
The apoptotic cell rates in HeLa cells were determined with annexin-V binding. The percentages of apoptotic cell rates were increased while the percentages of live cell rates were decreased after treatment with Exo-Apt-Ral formulations (Figs. 5A and 5B).
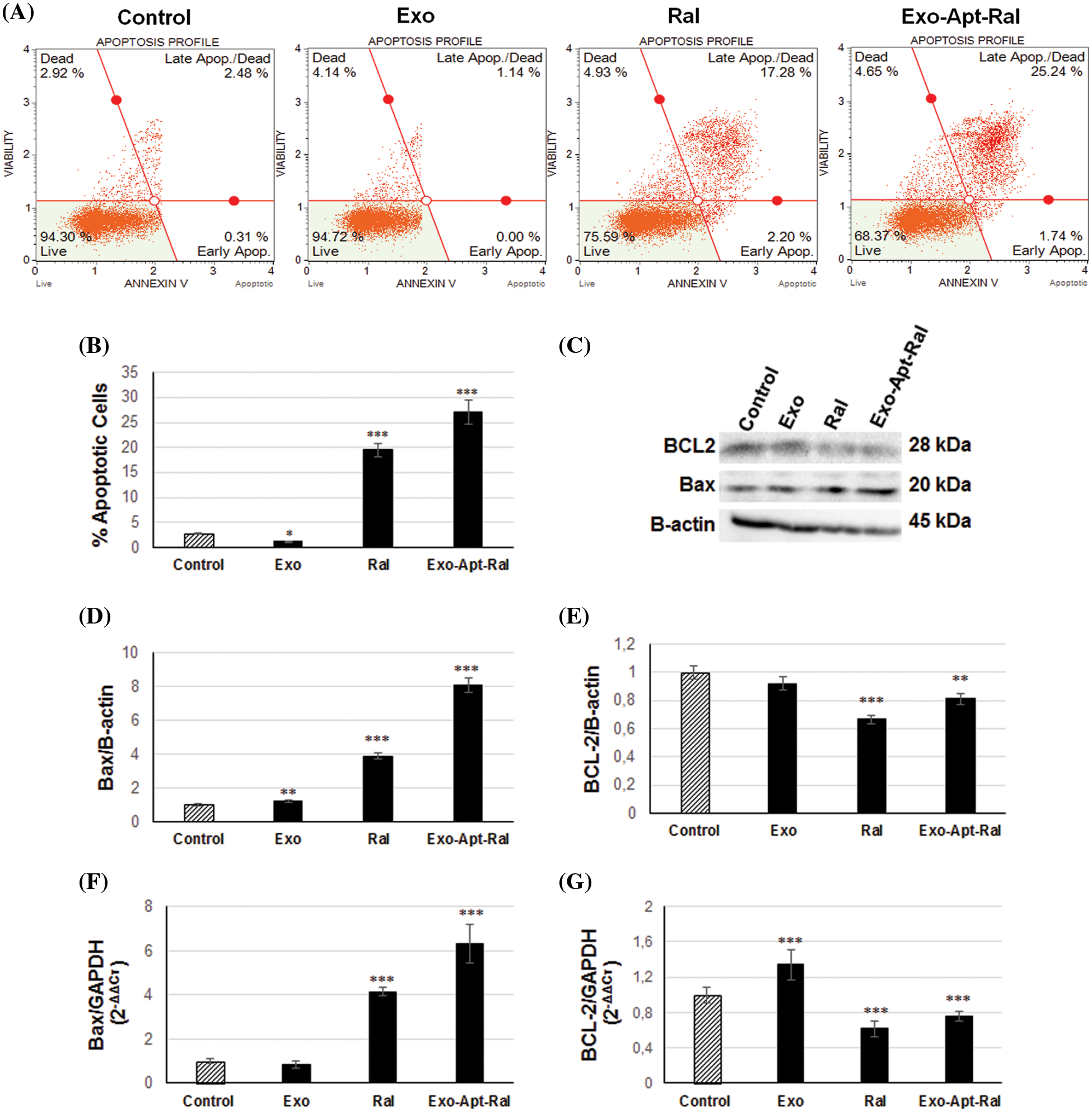
Figure 5: Effect of Exo-Apt-Ral on HeLa cell apoptosis. (A) The apoptosis profile of HeLa cells was measured by Annexin V after treatment with exosome, raloxifene, and Exo-Apt-Ral. (B) The percentage of apoptotic HeLa cells treated with exosome, raloxifene, and Exo-Apt-Ral. (C) Western blotting showing bands of Bax, BCL-2, and B-actin in HeLa cells treated with exosome, raloxifene, and Exo-Apt-Ral. (D) Graphs of Bax protein expression after exosome, raloxifene, and Exo-Apt-Ral treatments. (E) BCL-2 protein expression after exosome, raloxifene, and Exo-Apt-Ral treatments. (F) Bax gene expression of HeLa cells after exosome, raloxifene, and Exo-Apt-Ral treatments. (G) BCL-2 gene expression of HeLa cells of exosome, raloxifene, and Exo-Apt-Ral treatments (each group was studied in triplicate for apoptosis experiments, *, p < 0.05; **, p < 0.01; ***, p < 0.001 compared to the control group).
To determine the apoptotic effect of the Exo-Apt-Ral formulation on HeLa cells, the expression of anti-apoptotic BCL-2 and pro-apoptotic Bax proteins were examined by western blotting (Fig. 5C). The expression level of the pro-apoptotic protein Bax was dramatically increased (Fig. 5D), whereas the expression of the anti-apoptotic protein Bcl-2 was reduced (Fig. 5E) after treating HeLa cells with raloxifene and Exo-Apt-Ral. Further qPCR assessment revealed that the expression of the anti-apoptotic BCL-2 gene in HeLa cells decreased while that of the pro-apoptotic Bax increased compared to the control group after raloxifene and Exo-Apt-Ral treatments (Figs. 5F and 5G).
Effects of the Exo-Apt-Ral formulation on lysosomal and autophagic proteins in HeLa cells
The tendency of raloxifene and Exo-Apt-Ral formulation to accumulate in the lysosomes of HeLa cervical cancer cells was examined by lysotracker DND-26 staining. When the images taken at 40× magnification were examined under a fluorescent microscope, it was seen that the cells treated with Exo-Apt-Ral appeared to have more fluorescence than the cells treated with raloxifene (Fig. 6A). Also, the intensity of the lysotracker green color of HeLa cells treated with exosomes, raloxifene, and Exo-Apt-Ral formulations was more compared with the control group cells. The lysotracker green color intensity increased in cells treated with only exosomes compared to the control group (p < 0.01). Likewise, it was determined that the intensity of lysotracker increased in cells treated with raloxifene and Exo-Apt-Ral (p < 0.001) (Fig. 6B).
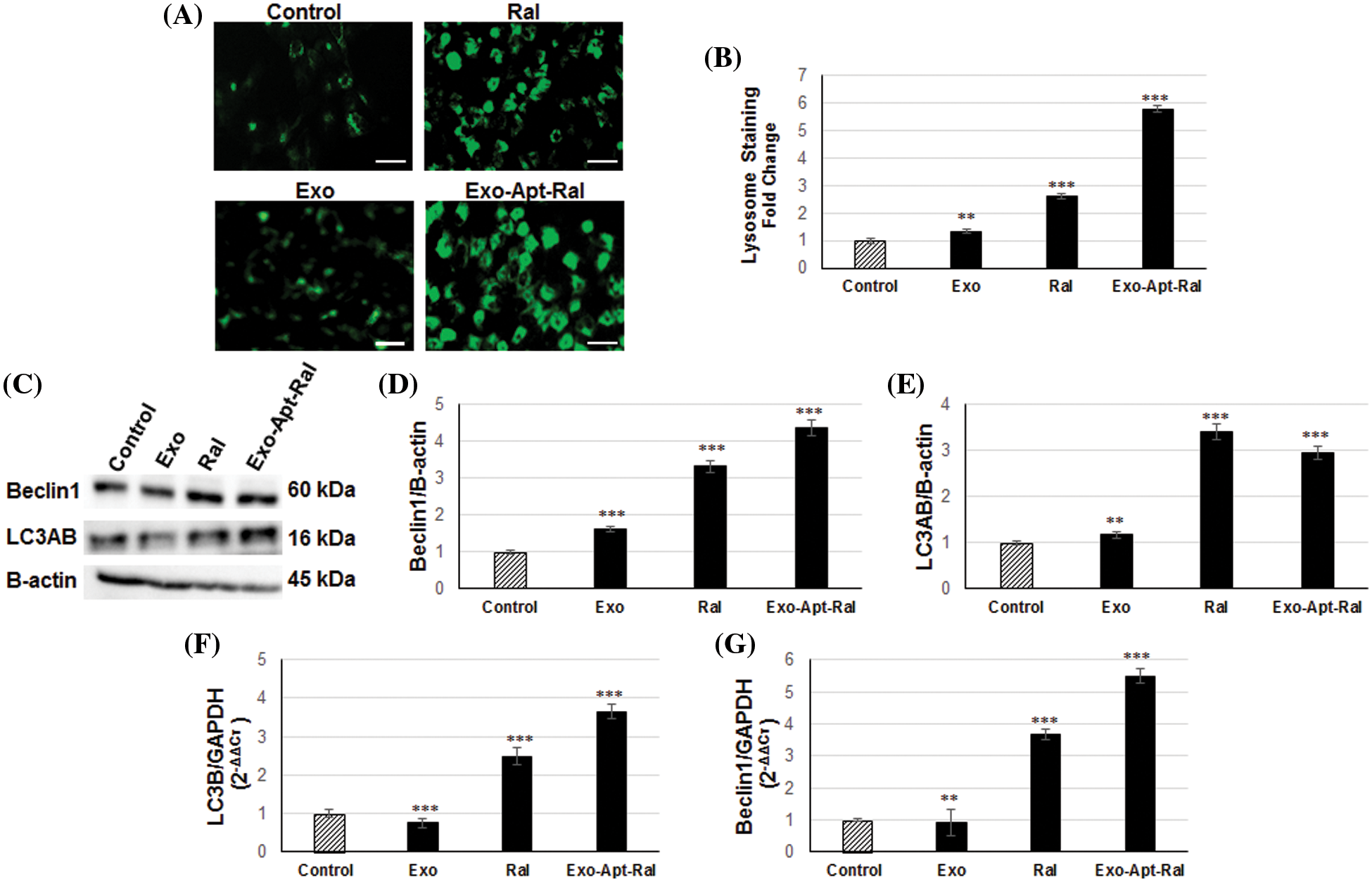
Figure 6: Effect of Exo-Apt-Ral on the HeLa cell lysosome and autophagy pathway (A) DND-26 lysotracker staining images of HeLa cells treated with exosome, raloxifene, and Exo-Apt-Ral formulations (Scale bar = 50 µm). (B) Graphical illustration of the lysotracker density in HeLa cells treated with exosome, raloxifene, and Exo-Apt-Ral. (C) Western blotting bands of Beclin1, LC3AB, and B-actin in HeLa cells treated with exosome, raloxifene, and Exo-Apt-Ral. (D) Graphs of beclin1 protein expression in HeLa cells after exosome, raloxifene, and Exo-Apt-Ral treatments. (E) Graphs of LC3AB protein expression after exosome, raloxifene, and Exo-Apt-Ral treatments. (F) Graphs of LC3B gene expression in HeLa cells after exosome, raloxifene, and Exo-Apt-Ral treatments. (G) Graphs of Beclin1 gene expression in HeLa cells after exosome, raloxifene, and Exo-Apt-Ral treatments (Each group was studied in triplicates for lysosome and autophagy experiments, **, p < 0.01; ***, p < 0.001 compared to the control group).
LC3AB and Beclin-1 protein expression was examined by western blotting to determine the autophagic effects of the Exo-Apt-Ral formulation on HeLa cells (Fig. 6C). The expression level of Beclin-1 and LC3AB increased in HeLa cells treated with raloxifene and Exo-Apt-Ral compared to the control group (Figs. 6D and 6E).
In addition, there was an increase in the expressions of autophagic genes Beclin-1 and LC3B in HeLa cells treated with raloxifene and Exo-Apt-Ral when compared to the control group (Figs. 6F and 6G).
Cervical cancer is reported as the second most common cancer that threatens the health of women. The most critical risk factor causing cervical cancer is the human papilloma virus (HPV). Various treatment methods, including drugs and vaccines, have been developed to treat cervical cancer, which is an avoidable and curable disease. The most widespread treatment procedure is chemotherapy with anti-metabolites (e.g., methotrexate) and DNA interacting agents (doxorubicin, cisplatin) along with some cytotoxic drugs. However, because these drugs travel in the bloodstream and spread to all body parts, cancer patients routinely experience various side effects. These include nausea, hair loss, damage to other non-target tissues, toxicity, neurotoxicity, multi-drug resistance, anemia, and neutropenia. These side effects restrict and/or put at risk the effectiveness of therapeutic procedures.
For this reason, there is an immediate necessity for the development of new anticancer drugs with better effectiveness and fewer side effects. The controlled release of existing drugs in target tissue with more effective formulations is also essential (Ordikhani et al., 2016; Hassanpour and Dehghani, 2017; Wang et al., 2021).
Exosomes secreted by all cell types in the body have attracted great scientific interest as drug delivery systems. Some of the most important benefits of exosomes include a long half-life, improved delivery, increased drug circulation time, controlled and continuous release of the drug, the versatility of route of administration, the increased intracellular concentration of the drug, and increased bioavailability of insufficient soluble drugs. After such exosomes are isolated, their characterization is performed by techniques such as western blotting, flow cytometry, zeta-potential analysis, atomic force microscopy, scanning electron microscopy, and transmission electron microscopy (TEM) (Zhou et al., 2020). Although exosomes have variable sizes, those with a certain size range (30–150 nm) are known to contain exosome biomarkers (Doyle and Wang, 2019). In a study on exosomes released from cancer cells, Zhang et al. (2019) isolated the exosomes released from CaSki cervical cancer cells by the ultracentrifugation method and showed that the CD9, CD63, and HSP70 protein expression was specific to these exosomes by Western blotting. Furthermore, they measured the size of exosomes released from CaSki cells to be in the range of 50–180 nm by TEM analysis. In our study, after the exosomes released from HeLa cervical cancer cells were isolated by the ultracentrifugation method, we detected the expression of CD9, CD63, and CD81 proteins in the exosome membrane by Western blotting. Both TEM and zeta potential analysis results have shown us that exosomes can be purified in the desired range. In the zeta potential analysis of our study, while the dimensions of the initially purified exosomes were 66 ± 12 nm, it was determined that the size increased slightly (120 ± 21 nm) when loaded with raloxifene, but the exosome integrity did not change. Similar results were also documented in the TEM analysis. It has been shown in studies that the size of exosomes previously isolated from Hela cells increased slightly with docetaxel loading (Cenik et al., 2022). At the same time, it has been reported that when paclitaxel was loaded in exosomes isolated from mesenchymal stem cells, their sizes increase by 36 nm with drug loading compared to the previous size and their integrity was not impaired (Abas et al., 2022).
Aptamers are RNA and DNA oligonucleotides that selectively bind to their target ligands with high affinity. Aptamers have been developed that specifically bind to various proteins found in the membranes of cancer cells. For example, a study identified two RNA aptamers that specially bind to prostate-specific membrane antigen (PSMA) after six rounds of Systematic evolution of ligands by exponential enrichment (SELEX). They showed that these aptamers, xPSM-9 and xPSM-10, added fluorescent probes to the end of the aptamers, bound to PSMA-positive LNCaP cells, and did not bind to PSMA-negative PC-3 cells (Lupold et al., 2002). There are two studies in the literature targeting cervical cancer cells. In the first study, Graham and Zarbl selected 11 different design aptamers that selectively bind to HPV-transformed cervical cancer cells (SiHa) with the SELEX method. Among these aptamers, 13, 14, 20, and 28 coded aptamers were shown to bind to SiHa cells with high affinity by fluorescence microscopy (Graham and Zarbl, 2012). In the second aptamer study on cervical cancer, it was shown that the 51 base-long C-9S DNA aptamer specifically binds to SiHa and HeLa cervical cancer cells using flow cytometry and confocal fluorescence microscopy analytical techniques (Wang et al., 2019). Since the 13 and 20 coded aptamers used in this study are specific to Hela cells, we bound these aptamers to the surface of exosomes and transported raloxifene to HeLa cells by exosomes. HeLa-specific aptamers have previously been shown to only bind to these cells documented in the study by Graham and Zarbl. Our study showed that when raloxifene is loaded with aptamer-bound exosomes by electroporation, it is a target-specific carrier system that binds to HeLa cells rather than L929 cells. We also detected this effect as raloxifene specifically changes the acidity by accumulating in lysosomes.
The acidic inner environment of lysosomes makes them sensitive to the accumulation of weak bases capable of crossing the lysosomal membrane. Once they reach this compartment, these weak bases are protonated, and their back-diffusion into the cytosol as charged molecules is heavily inhibited. Such substances are called lysosomotropic agents, and through this mechanism, they can accumulate in lysosomes at concentrations one hundred times than their cytosolic concentrations. The lysosomotropic abilities of many well-known clinically used drugs including chloroquine, antipsychotics (chlorpromazine, aripiprazole, and thioridazine), antidepressants (desipramine, clomipramine, and imipramine) have been documented (Nadanaciva et al., 2011). The lysosomotropic activity of raloxifene, which is used as an estrogen receptor modulator for the treatment of osteoporosis in the postmenopausal period has also been reported (Selyunin et al., 2021). Oral administration of raloxifene in the pill form is catabolized due to extensive hepatic transit, and its absolute bioavailability is reduced to 2%. Therefore, various formulations based on liposomal, chitosan and microemulsion have been developed to increase the bioavailability of raloxifene (Golmohammadzadeh et al., 2017). A research team prepared a lipid-based microemulsified formulation of raloxifene and reported that the lipid-based microemulsion of raloxifene had a more significant outstanding in vitro intestinal transit than commercial formulations on the market (Thakkar et al., 2011). Saini et al. (2015) developed a chitosan-based nanoparticulate formulation of raloxifene and calculated the raloxifene loading efficiency of this nanoparticle formulation to be 23.89%. In vivo studies emphasized that nasal administration significantly increased the amount of raloxifene in plasma compared to oral administration. For example, in one study, raloxifene was loaded with 79% efficiency into the micelles they prepared using Pluronic F68 and Gelucire 44/14. In their in vitro cytotoxicity experiments, the IC50 value of the micelle formulation of raloxifene in MCF-7 human breast cancer cells was calculated as 22.5 µg/mL. They also found that the in vivo bioavailability of the micelle formulation of raloxifene increased 1.5 times compared to pure raloxifene (Kanade et al., 2018). Exosome-based formulation of raloxifene to increase its bioavailability and target-specific orientation was demonstrated for the first time in this study. In our study, the exosome-based formulation of raloxifene with 72% ± 2.15% entrapment efficiency had a similar loading capacity as in other lipid-based studies. Lipid-based formulations of raloxifene have been administered orally in in vivo studies (Murthy et al., 2020). Since exosomes are lipids of biological origin that are naturally released from cells, the exosome-derived formulation of raloxifene has the potential to be administered orally in the future. The prepared Exo-Apt-Ral disrupted the lysosomal integrity of HeLa cervical cancer cells and allowed the lysosomal contents to pass into the cell cytoplasm to trigger cell death mechanisms. Autophagy plays a duplicate role in tumor growth and suppression and also facilitates cancer cell development and proliferation. Therefore, chemotherapy combined with autophagy is a more effective strategy to target cancer cells. Unnatural autophagy restricts the degradation of harmed components or proteins in oxidatively stressed cells, leading to cancer development (Levine, 2007). It has been reported that there is a close relationship between exosome release and the autophagy pathway, and both mechanisms are triggered in conditions such as nutrient deprivation and oxidative stress. In addition, it has been emphasized that exosomes trigger autophagy (Xing et al., 2021). In this study, it was shown that the autophagy mechanism was triggered by increasing LC3AB and Beclin1 protein expression in HeLa cervical cancer cells after exosome treatment. Decreased autophagy and increased cell proliferation have been reported with BECN1 deletion in cancer cell lines and animal models (Yun and Lee, 2018). Kim et al. (2015) reported that the expression of LC3B, Beclin-1, ATG5, and ATG12 autophagic proteins was increased in MCF-7 human breast cancer cells treated with 10 µM raloxifene and autophagic death of MCF-7 cells was triggered. This study determined that the autophagic death mechanism was triggered by an increase in the expression of LC3AB and Beclin-1 autophagic proteins in HeLa cervical cancer cells treated with Exo-Apt-Ral.
Accumulation of chemotherapeutics in undesirable tissues is one of the most significant disadvantages of cancer treatment. Focusing on the development of cervical cancer-specific therapeutic release strategies, this study demonstrated that drugs could be transported to target cells with binding aptamers to drug-loaded functionalized exosomes. The data obtained from this study will contribute to the development of specific delivery systems with high biocompatibility in targeted and personalized therapies. This would contribute to the redefinition of biotechnological drug development strategies in the future, and constitute preliminary data for clinical applications.
Funding Statement: This study has been supported by a Grant (221S945) from the Scientific and Technological Research Council of Turkey (TUBITAK) and an Aydin Adnan Menderes University Research Grant (ADU-TPF-20041).
Author Contributions: The authors confirm their contributions to the paper as follows: study conception and design: OE, OC; data collection: OE, GMD; analysis and interpretation of results: OE, GMD; draft manuscript preparation: OE, OC. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abas BI, Demirbolat GM, Cevik O (2022). Wharton jelly-derived mesenchymal stem cell exosomes induce apoptosis and suppress EMT signaling in cervical cancer cells as an effective drug carrier system of paclitaxel. PLoS One 17: e0274607. https://doi.org/10.1371/journal.pone.0274607 [Google Scholar] [PubMed] [CrossRef]
Barbas AS, Mi J, Clary BM, White RR (2010). Aptamer applications for targeted cancer therapy. Future Oncology 6: 1117–1126. https://doi.org/10.2217/fon.10.67 [Google Scholar] [PubMed] [CrossRef]
Cenik M, Abas BI, Kocabiyik B, Demirbolat GM, Cevik O (2022). Development of a new drug delivery system from HELA-derived exosomes and the effect of docetaxel-loaded exosomes on mitochondrial apoptosis. Journal of Pharmaceutical Innovation 17: 931–939. https://doi.org/10.1007/s12247-021-09566-1 [Google Scholar] [CrossRef]
de Duve C, de Barsy T, Poole B, Tulkens P (1974). Lysosomotropic agents. Biochemical Pharmacology 23: 2495–2531. https://doi.org/10.1016/0006-2952(74)90174-9 [Google Scholar] [PubMed] [CrossRef]
Demirbolat GM, Aktas E, Coskun GP, Erdogan O, Cevik O (2022a). New approach to formulate methotrexate-loaded niosomes: In vitro characterization and cellular effectiveness. Journal of Pharmaceutical Innovation 17: 622–637. https://doi.org/10.1007/s12247-021-09539-4 [Google Scholar] [CrossRef]
Demirbolat GM, Erdoğan Ö, Coşkun GP, Çevik Ö (2022b). PEG4000 modified liposomes enhance the solubility of quercetin and improve the liposome functionality: In vitro characterization and the cellular efficacy. Turkish Journal of Chemistry 46: 1011–1023. https://doi.org/10.55730/1300-0527.3411 [Google Scholar] [CrossRef]
Doyle LM, Wang MZ (2019). Overview of extracellular vesicles, their origin, composition, purpose, and methods for exosome isolation and analysis. Cells 8: 727. https://doi.org/10.3390/cells8070727 [Google Scholar] [PubMed] [CrossRef]
Embark H (2016). In vitro selection and binding studies of a DNA aptamer targeting phosphatidylserine (PS) using non-fluorescence measurements. Assiut Veterinary Medical Journal 62: 113–118. https://doi.org/10.21608/avmj.2016.170014 [Google Scholar] [CrossRef]
Erdogan O, Abbak M, Demirbolat GM, Birtekocak F, Aksel M, Pasa S, Cevik O (2019). Green synthesis of silver nanoparticles via Cynara scolymus leaf extracts: The characterization, anticancer potential with photodynamic therapy in MCF7 cells. PLoS One 14: e0216496. https://doi.org/10.1371/journal.pone.0216496 [Google Scholar] [PubMed] [CrossRef]
Erdoğan Ö., Paşa S, Demirbolat GM, Çevik Ö. (2021). Green biosynthesis, characterization, and cytotoxic effect of magnetic iron nanoparticles using Brassica Oleracea var capitata sub var rubra (red cabbage) aqueous peel extract. Turkish Journal of Chemistry 45: 1086–1096. https://doi.org/10.3906/kim-2102-2 [Google Scholar] [PubMed] [CrossRef]
Golmohammadzadeh S, Farhadian N, Biriaee A, Dehghani F, Khameneh B (2017). Preparation, characterization and in vitro evaluation of microemulsion of raloxifene hydrochloride. Drug Development and Industrial Pharmacy 43: 1619–1625. https://doi.org/10.1080/03639045.2017.1328430 [Google Scholar] [PubMed] [CrossRef]
Graham JC, Zarbl H (2012). Use of cell-SELEX to generate DNA aptamers as molecular probes of HPV-associated cervical cancer cells. PLoS One 7: e36103. https://doi.org/10.1371/journal.pone.0036103 [Google Scholar] [PubMed] [CrossRef]
Hassanpour SH, Dehghani M (2017). Review of cancer from perspective of molecular. Journal of Cancer Research and Practice 4: 127–129. https://doi.org/10.1016/j.jcrpr.2017.07.001 [Google Scholar] [CrossRef]
Kanade R, Boche M, Pokharkar V (2018). Self-assembling raloxifene loaded mixed micelles: Formulation optimization, in vitro cytotoxicity and in vivo pharmacokinetics. AAPS PharmSciTech 19: 1105–1115. https://doi.org/10.1208/s12249-017-0919-6 [Google Scholar] [PubMed] [CrossRef]
Kim DE, Kim Y, Cho DH, Jeong SY, Kim SB, Suh N, Kim CS (2015). Raloxifene induces autophagy-dependent cell death in breast cancer cells via the activation of AMP-activated protein kinase. Molecules and Cells 38: 138–144. https://doi.org/10.14348/molcells.2015.2193 [Google Scholar] [PubMed] [CrossRef]
Levine B (2007). Autophagy and cancer. Nature 446: 745–747. https://doi.org/10.1038/446745a [Google Scholar] [PubMed] [CrossRef]
Lupold SE, Hicke BJ, Lin Y, Coffey DS (2002). Identification and characterization of nuclease-stabilized RNA molecules that bind human prostate cancer cells via the prostate-specific membrane antigen. Cancer Research 62: 4029–4033. https://doi.org/10.1158/0008-5472.CAN-12-2152 [Google Scholar] [CrossRef]
Murthy A, Rao Ravi P, Kathuria H, Malekar S (2020). Oral bioavailability enhancement of raloxifene with nanostructured lipid carriers. Nanomaterials 10: 1085. https://doi.org/10.3390/nano10061085 [Google Scholar] [PubMed] [CrossRef]
Nadanaciva S, Lu S, Gebhard DF, Jessen BA, Pennie WD, Will Y (2011). A high content screening assay for identifying lysosomotropic compounds. Toxicology in Vitro 25: 715–723. https://doi.org/10.1016/j.tiv.2010.12.010 [Google Scholar] [PubMed] [CrossRef]
Nagaraju P, VenuGopal K, Murali Krishna N, Bhargavi V, Srinivasulu N (2014). UV-spectrophotometric method development and validation for determination of raloxifene in pharmaceutical dosage form. Research and Reviews: Journal of Pharmaceutical Analysis 3: 27–31. [Google Scholar]
Ndolo RA, Luan Y, Duan S, Forrest ML, Krise JP (2012). Lysosomotropic properties of weakly basic anticancer agents promote cancer cell selectivity in vitro. PLoS One 7: e49366. https://doi.org/10.1371/journal.pone.0049366 [Google Scholar] [PubMed] [CrossRef]
Ordikhani F, Erdem Arslan M, Marcelo R, Sahin I, Grigsby P, Schwarz JK, Azab AK (2016). Drug delivery approaches for the treatment of cervical cancer. Pharmaceutics 8: 23. https://doi.org/10.3390/pharmaceutics8030023 [Google Scholar] [PubMed] [CrossRef]
Paşa S, Erdogan O, Cevik O (2021). Design, synthesis and investigation of procaine based new Pd complexes as DNA methyltransferase inhibitor on gastric cancer cells. Inorganic Chemistry Communications 132: 108846. https://doi.org/10.1016/j.inoche.2021.108846 [Google Scholar] [CrossRef]
Saini D, Fazil M, Ali MM, Baboota S, Ameeduzzafar Ali J (2015). Formulation, development and optimization of raloxifene-loaded chitosan nanoparticles for treatment of osteoporosis. Drug Delivery 22: 823–836. https://doi.org/10.3109/10717544.2014.900153 [Google Scholar] [PubMed] [CrossRef]
Selyunin AS, Nieves-Merced K, Li D, McHardy SF, Mukhopadhyay S (2021). Tamoxifen derivatives alter retromer-dependent endosomal tubulation and sorting to block retrograde trafficking of Shiga toxins. Toxins 13: 424. https://doi.org/10.3390/toxins13060424 [Google Scholar] [PubMed] [CrossRef]
Siddiqui S, Yuan J (2021). Binding characteristics study of DNA based aptamers for E. coli O157: H7. Molecules 26: 204. https://doi.org/10.3390/molecules26010204 [Google Scholar] [PubMed] [CrossRef]
Sun H, Zhu X, Lu PY, Rosato RR, Tan W, Zu Y (2014). Oligonucleotide aptamers: New tools for targeted cancer therapy. Molecular Therapy-Nucleic Acids 3: e182. https://doi.org/10.1038/mtna.2014.32 [Google Scholar] [PubMed] [CrossRef]
Thakkar H, Nangesh J, Parmar M, Patel D (2011). Formulation and characterization of lipid-based drug delivery system of raloxifene-microemulsion and self-microemulsifying drug delivery system. Journal of Pharmacy and Bioallied Sciences 3: 442. https://doi.org/10.4103/0975-7406.84463 [Google Scholar] [PubMed] [CrossRef]
Türk S, Tok F, Erdoğan Ö, Çevik Ö, Tok TT, Koçyiğit-Kaymakçıoğlu B, Karakuş S (2020). Synthesis, anticancer evaluation and in silico ADMET studies on urea/thiourea derivatives from gabapentin. Phosphorus, Sulfur, and Silicon and the Related Elements 196: 382–388. https://doi.org/10.1080/10426507.2020.1845678 [Google Scholar] [CrossRef]
Wang J, Gao T, Luo Y, Wang Z, Zhang Y, Zhang Y, Zhang Y, Pei R (2019). In vitro selection of a DNA aptamer by cell-SELEX as a molecular probe for cervical cancer recognition and imaging. Journal of Molecular Evolution 87: 72–82. https://doi.org/10.1007/s00239-019-9886-8 [Google Scholar] [PubMed] [CrossRef]
Wang X, Liu S, Guan Y, Ding J, Ma C, Xie Z (2021). Vaginal drug delivery approaches for localized management of cervical cancer. Advanced Drug Delivery Reviews 174: 114–126. https://doi.org/10.1016/j.addr.2021.04.009 [Google Scholar] [PubMed] [CrossRef]
Xing H, Tan J, Miao Y, Lv Y, Zhang Q (2021). Crosstalk between exosomes and autophagy: A review of molecular mechanisms and therapies. Journal of Cellular and Molecular Medicine 25: 2297–2308. https://doi.org/10.1111/jcmm.16276 [Google Scholar] [PubMed] [CrossRef]
Yang C, Jiang Y, Hao SH, Yan XY, Naranmandura H (2022). Aptamers: An emerging navigation tool of therapeutic agents for targeted cancer therapy. Journal of Materials Chemistry B 10: 20–33. https://doi.org/10.1039/D1TB02098F [Google Scholar] [PubMed] [CrossRef]
Ye Z, Xiong C, Pan J, Su D, Zeng L (2018). Highly photostable, lysosome-targeted BODIPYs with green to near-infrared emission for lysosome imaging in living cells. Dyes and Pigments 155: 30–35. https://doi.org/10.1016/j.dyepig.2018.03.015 [Google Scholar] [CrossRef]
Yun CW, Lee SH (2018). The roles of autophagy in cancer. International Journal of Molecular Sciences 19: 3466. https://doi.org/10.3390/ijms19113466 [Google Scholar] [PubMed] [CrossRef]
Zhang L, Li H, Yuan M, Li M, Zhang S (2019). Cervical cancer cells-secreted exosomal microRNA-221-3p promotes invasion, migration and angiogenesis of microvascular endothelial cells in cervical cancer by down-regulating MAPK10 expression. Cancer Management and Research 11: 10307. https://doi.org/10.2147/CMAR [Google Scholar] [CrossRef]
Zhao Q, Gao SM, Wang MC (2020). Molecular mechanisms of lysosome and nucleus communication. Trends in Biochemical Sciences 45: 978–991. https://doi.org/10.1016/j.tibs.2020.06.004 [Google Scholar] [PubMed] [CrossRef]
Zhou M, Weber SR, Zhao Y, Chen H, Sundstrom JM (2020). Methods for exosome isolation and characterization. Exosomes: 23–38. https://doi.org/10.1016/B978-0-12-816053-4.00002-X [Google Scholar] [CrossRef]
Supplementary Materials
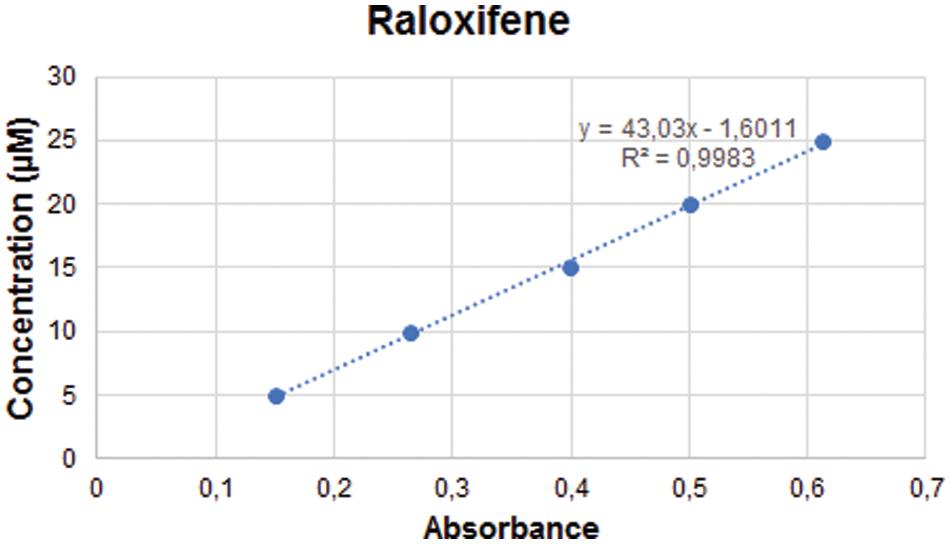
Figure S1: The standard curve of Raloxifene.
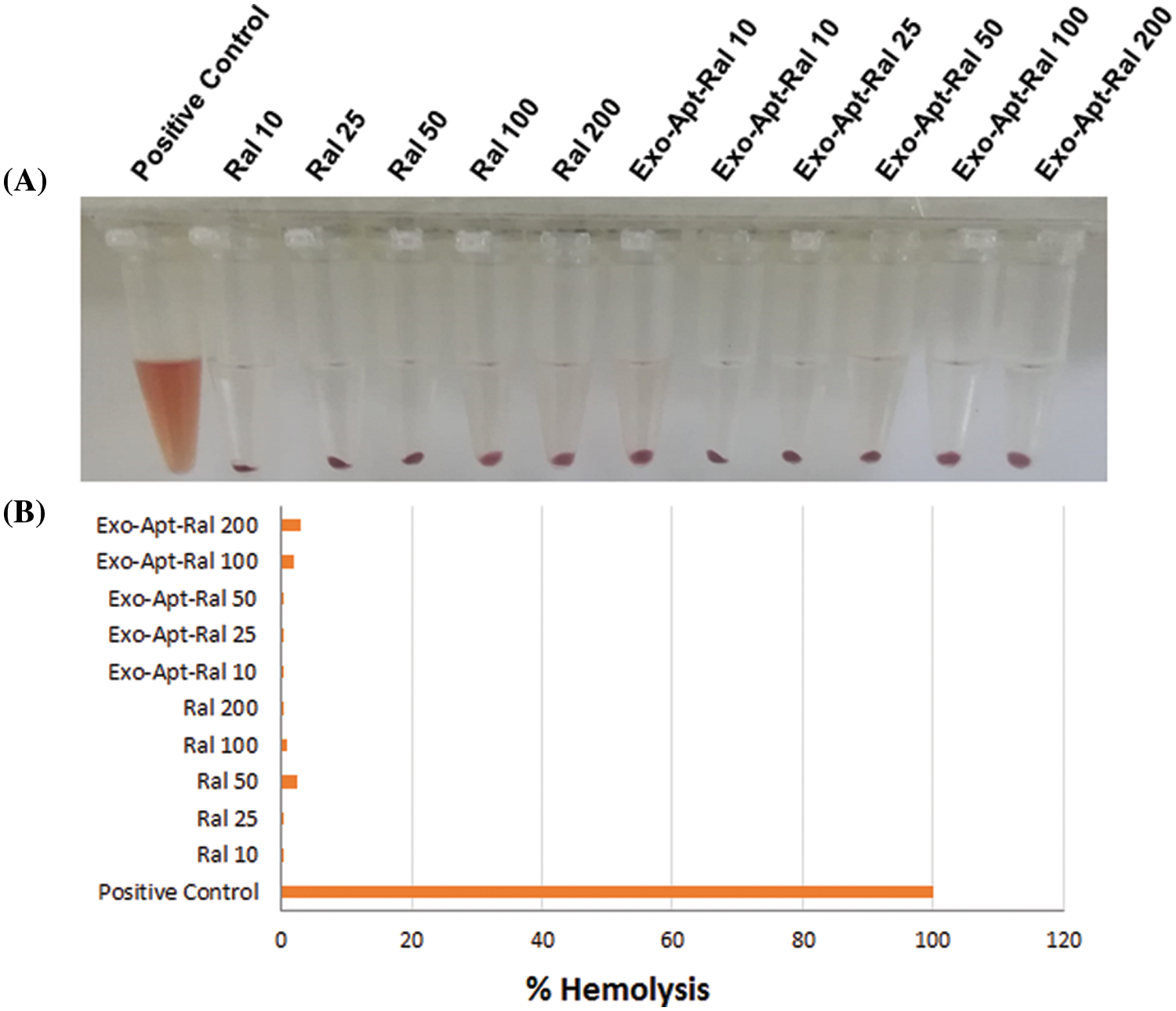
Figure S2: (A) The images of RBC cells treated with the raloxifene and Exo-Apt-Ral concentrations. (B) The graph (% hemolysis) of cells treated with the raloxifene and E xo-Apt-Ral concentrations.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools