 Open Access
Open Access
ARTICLE
Exploring the attenuation mechanisms of Dalbergia odorifera leaves extract on cerebral ischemia-reperfusion based on weighted gene co-expression network analysis
1 Department of Pharmacy, The First Affiliated Hospital of Nanchang University, Nanchang, 330006, China
2 Department of Pharmacy, The Second Affiliated Hospital of Nanchang University, Nanchang, 330006, China
3 Key Laboratory of Modern Preparation of TCM, Ministry of Education, Jiangxi University of Traditional Chinese Medicine, Nanchang, 330006, China
4 Jiangxi Provincial Institute of Translational Medicine, The First Affiliated Hospital of Nanchang University, Nanchang, 330006, China
* Corresponding Author: YANNI LV. Email:
# Co-first authors: Jinfang Hu, Jiangen Ao, and Longsheng Fu contributed equally
(This article belongs to the Special Issue: Bioinformatics Study of Diseases)
BIOCELL 2023, 47(7), 1611-1622. https://doi.org/10.32604/biocell.2023.028684
Received 01 January 2023; Accepted 10 March 2023; Issue published 21 June 2023
Abstract
Background: The attenuation function of Dalbergia odorifera leaves on cerebral ischemia-reperfusion (I/R) is little known. The candidate targets for the Chinese herb were extracted from brain tissues through the high-affinity chromatography. The molecular mechanism of D. odorifera leaves on cerebral I/R was investigated. Methods: Serial affinity chromatography based on D. odorifera leaves extract (DLE) affinity matrices were applied to find specific binding proteins in the brain tissues implemented on C57BL/6 mice by intraluminal middle cerebral artery occlusion for 1 h and reperfusion for 24 h. Specific binding proteins were subjected to mass-spectrometry to search for the differentially expressed proteins between control and DLE-affinity matrices. The hub genes were screened based on weighted gene co-expression network analysis (WGCNA). Then, predictive biology and potential experimental verification were performed for the candidate genes. The protective role of DLE in blood-brain barrier damage in cerebral I/R mice was evaluated by the leakage of Evans blue, western blotting, immunohistochemistry, and immunofluorescent staining. Results: 952 differentially expressed proteins were classified into seven modules based on WGCNA under soft threshold 6. Based on WGCNA, AKT1, PIK3CA, NOS3, SMAD3, SMAD1, IL6, MAPK1, TGFBR2, TGFBR1, MAPK3, IGF1R, LRG1, mTOR, ROCK1, TGFB1, IL1B, SMAD2, and SMAD5 18 candidate hub proteins were involved in turquoise module. TGF-β, MAPK, focal adhesion, and adherens junction signaling pathway were associated with candidate hub proteins. Gene ontology analysis demonstrated that candidate hub proteins were related to the TGF-β receptor signaling pathway, common-partner SMAD protein phosphorylation, etc. DLE could significantly reduce the leakage of Evans blue in mice with cerebral I/R, while attenuating the expression of occludin, claudin-5, and zonula occludens-1. Western blotting demonstrated that regulation of TGF-β/SMAD signaling pathway played an essential role in the protective effect of DLE. Conclusion: Thus, a number of candidate hub proteins were identified based on DLE affinity chromatography through WGCNA. DLE could attenuate the dysfunction of blood-brain barrier in the TGF-β/SMAD signaling pathway induced by cerebral I/R.Keywords
Supplementary Material
Supplementary Material FileStroke is the first reason of death worldwide, with the increase in its incidence and the rate of mortality (Li et al., 2019); it is also the first cause of death for Chinese people. Plants are naturally available source of food supplements or drugs for stroke therapy around the globe; for instance, leaves of Ginkgo biloba L. (Li et al., 2021) or Acanthopanax senticosus (Wang et al., 2020) are attractive materials for treating ischemic stroke. Dalbergia odorifera is mainly distributed in the Hainan and Guangdong provinces of China. D. odorifera is used for preparing drugs for treating cerebral infarction, thrombosis, and coronary heart diseases (Wang et al., 2020). D. odorifera grows very slowly, for approximately 15 years until maturity; however, it has numerous fallen leaves. A large number of components have been extracted from D. odorifera leaves (DO leaves) (Ma et al., 2020), including a high content of flavone, isoflavone compounds, etc. One previous research group confirmed that 70% ethanol extract of D. odorifera leaves could exert a hypolipidemic effect on the hyperlipidemic rat model (Mei et al., 2021). These studies were related to flavone compounds as the main component for treating cardiac and cerebral vascular diseases. However, little is known about the medicinal value, and the attenuation function of D. odorifera leaves on cerebral I/R.
Earlier, the high-affinity proteins were extracted through a method combined with serial affinity chromatography and shotgun proteomics analysis for traditional Chinese medicine (Lv and Fu, 2018). The integrated method was also applied to mine the target information for D. odorifera leaves. Weighted gene co-expression network analysis (WGCNA) is a method to describe correlation patterns between different genetic samples. WGCNA facilitates a complete investigation of the relationship between genes and their phenotypes (Sonoda et al., 2019) and aids in screening for key genes for the identification of candidate biomarkers or therapeutic targets (Shuai et al., 2021). Here, a number of differentially expressed proteins were obtained from the affinity resin of D. odorifera leaves extract (DLE) for cerebral I/R. The WGCNA-based approach was used to identify candidate protein modules associated with differentially expressed proteins to study the role and mechanisms of DLE on cerebral I/R.
DO leaves were collected from the Jianfengling area, Sanya City, Hainan Province, China. A voucher specimen (No. 20200123) was stored in the Key Laboratory of Modern Preparation of Traditional Chinese Medicine, Ministry of Education, Jiangxi University of Traditional Chinese Medicine, China.
Preparation of 70% ethanol extract of Dalbergia odorifera leaves
The D. odorifera (500 g) extract (DLE) was prepared using air-dried leaves and 70% ethanol under a refluxing extraction (2 h, 5 L × twice) and filtered. The combined extracts were then rotary evaporated at 45°C and lyophilized (Shao et al., 2016). The ethanol extract was dissolved with 30% methanol and stored at 4°C until use. Rutin was accurately weighed to 5.41 mg and dissolved with 30% methanol to obtain the reference solution (0.541 mg/mL). The general flavonoid content of DLE was determined using the colorimetric method. Rutin was used as the standard. The absorbance of samples was measured at 500 nm.
HPLC analysis for general flavones of 70% ethanol extract of Dalbergia odorifera leaves
The air-dried leaves of D. odorifera (100 g) were used to prepare 70% ethanol under refluxing extraction (2 h, 1 L × twice) and filtered. The combined extracts were then rotary evaporated at 45°C and lyophilized. The extract ethanol was dissolved with methanol and stored at 4°C until use. Separation was performed on a 250 × 4.6 mm Phenomenex C18 (5 μm, California, USA) at 30°C, and the UV absorbance was monitored at 260 nm on Agilent 1100. The flow rate was 1.0 mL/min. The solvent system used the improved method published earlier (Li et al., 2016): composed of acetonitrile (A) and 0.1% phosphoric acid (B) using an optimized gradient program: 0–10 min 35% A; 10–15 min 35%–48% A; 15–30 min 48%–49% A. The injection volume was 10 μL. The DLE in 70% ethanol was accurately weighed and dissolved in a 2 mL volumetric flask with methanol. The final solution was filtered through a 0.22 µm microfilter to obtain a reference solution.
Animals and ischemia-reperfusion
C57BL/6J male mice (8~10 weeks old; weighing 18~22 g) were purchased from the College of Veterinary Medicine YanZhou University of Comparative Medicine (certificate No. SCXK (SU) 2022-0009) and placed in the laboratory of the First Affiliated Hospital of Nanchang University. All efforts were made to minimize animal suffering and the number of animals used. C57BL/6 mice were anesthetized with 1.2% isoflurane during the ischemia period. The environment temperature was maintained at 37°C during the ischemia and recovery period. A blunt tip 6–0 nylon monofilament was inserted into the neck vessel and advanced 8–10 mm forwards. After 1 h of occlusion, the monofilament was withdrawn, then 24 h reperfusion was immediately followed. Mice in the sham-operated group underwent the same surgical incision, but the nylon monofilament was not advanced into the internal carotid artery. Animals whose blood flow dropped below 30% of pre-ischemia levels were used for further study.
The administration dose was determined according to earlier studies (Mei et al., 2021), while the dose given to rats was converted corresponding to that of mice. The DLE treatment group was administered intragastrically with 0.6, 1.2, or 2.4 g/kg 1 h of the onset of cerebral ischemia and every 24 h during the experimental period, whereas the sham-operated group was administered intragastrically with a solution of equivalent volume NaCl. Mice were randomly categorized into six groups (8 mice for each group): (1) sham-operated mice (sham); (2) mice exposed to 1 h of ischemia and 24 h of reperfusion stroke (I/R); (3) mice exposed to 1 h of ischemia and 24 h of reperfusion stroke and treatment with a low dose of DLE (0.6 g/kg) (I/R+0.6 g/kg DLE); (4) mice exposed to 1 h of ischemia 24 h of reperfusion stroke, and treated with a middle dose of DLE (1.2 g/kg) (I/R+1.2 g/kg DLE); (5) mice exposed to 1 h of ischemia and 24 h of reperfusion stroke and treatment with a high dose of DLE (2.4 g/kg) (I/R+2.4 g/kgDLE); (6) 1 h of ischemia stroke, 24 h of reperfusion stroke and treatment with 10 mg/kg nimodipine (I/R+10 mg/kg NM) (Lv and Fu, 2018). At the end of the cerebral I/R period, the mice were examined for leakage of Evans blue, immunostaining, immunohistochemistry, and western blot analysis.
Preparation of mice brain lysate and Dalbergia odorifera leaves extract affinity resin
The epoxy-activated sepharose™6B (EAS-6B) was from GE Healthcare, Delaware, USA. The affinity resin was prepared using the earlier published formula (Lv and Fu, 2018). The leaf extract was dissolved in DMSO, then added into EAS-6B, which was fully expanded before 300 mmoL, soaked at 25°C for 24 h, lightly filtered with double distilled water for 15 min, drained, and preserved at 4°C under dry conditions. Brain tissue homogenates were centrifuged at 9,500 rpm for 10 min and 50,000 rpm for 30 min and diluted in lysate buffer to a total protein concentration of about 7 mg/mL. The homogenate was shaken gently at 4°C for 4 h with an affinity medium of 60 mL. This step was repeated five times. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; Bio-Rad, California, USA) was performed with the supernatant solution, and silver staining was performed using Quantity One software 4.6.2. The proteins were identified in the mass spectrometry database from the SEQUEST output file.
Screened out differential expressed proteins
Limma and Impute package in R software (4.0.4) were installed, and each group of data in section ‘Materials and chemicals’ to matrix were processed, and the missing matrix filled. The log2 value of the data set was taken and corrected to the same level. The up-regulated and down-regulated proteins compared between control-affinity proteins or DLE-affinity proteins were screened out, and with differentially expressed proteins, the truncated values were set to log2F ≥ 1 and p-value < 0.05. The volcano map and heat map were used to display the differential expressed proteins. In R software, define log2FC as the x-axis and the p-value as the y-axis to draw the volcano map.
Construction of weighted gene co-expression network
The screened differential expressed proteins were processed in R Studio 4.0.4 software using the WGCNA package to construct co-expression networks. First, the scale-free topology model Fitsigned R^2 and mean connectivity were calculated to determine the soft threshold (power) for the protein co-expression network. According to the compatible soft threshold, the degree of fitting was considered between the chosen genes and putative proteins. Then, the protein expression similarity matrix was converted into an adjacency matrix; then, the adjacency matrix was converted into a topological overlap matrix (TOM). The TOM assay was applied to convert the protein expression similarity matrix to the adjacency matrix. Hierarchical clustering based on TOM was constructed using a dynamic tree-cutting algorithm, Connectivity Cut was set as 1,000, and the preset number of modules was set as height 0.8 and minsize 3. After determining the protein modules, the eigenvector values of each module were calculated for clustering analysis, and the modules that were closer were merged into new modules. Genes with similar expression patterns were considered specific branch modules marked with a unique color.
Gene ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) singling pathway analysis
GO enrichment and KEGG signaling pathway analysis were performed on gene modules using the annotation and database DAVID (https://david.ncifcrf.gov/). Cut-off criteria were set as count ≥10, and p < 0.05 was chosen as the critical value, while false discovery rate (FDR) correction was applied for multiple test corrections. The significance level was set to 0.05, and classification information was set to functional classification and human annotation. The biological processes (BP), cellular components (CC), and molecular functions (MF) module were applied to analyze the biological function of proteins, and −Log2FC was defined to make the biological function map for the y-axis. The protein function was enriched by Fisher’s rectified p-value ≤ 0.01, and the signaling pathways with similar functions were classified according to the enrichment score.
Evaluation of blood-brain barrier permeability
0.1 mL/10 g of 2% Evans blue (Sigma Aldrich Co., Saint Louis, USA) was administered through a caudal vein 2 h before euthanasia. To quantitatively measure the leakage of Evans blue, the ipsilateral hemisphere was homogenized and centrifuged at 12,000 rpm for 20 min. The absorbance of the supernatant was measured by spectrophotometer at 620 nm, and the concentration of Evans blue was determined.
Histological assessment of tight junction proteins
The I/R brain tissue was soaked with 4% paraformaldehyde for 24 h and then embedded with paraffin after dehydration. Anti-occludin, claudin-5, and zonula occludens-1 (ZO-1; rabbit polyclonal IgG, 1:50, Zyemd, Wuhan, China) antibodies were incubated with brain tissues. The occludin, claudin-5, and ZO-1 positive boundaries were quantitatively measured in a 20 μm enlarged view. The data were expressed as a percentage of the number of positive boundaries in cerebral I/R mice.
Immunofluorescent staining of tight junction proteins
Brain tissues in each group were cut into 5 mm pieces, washed with phosphate-buffered saline, immobilized with cold ethanol at 4°C for 10 min, and fixed with 10% normal goat serum, 3% bovine serum albumin, and 0.1% Triton X-100 in phosphate-buffered saline for 1 h. Then, the tissue pieces were incubated with the occludin (ab216327, Abcam, Cambridge, UK), claudin-5 (ab131259, Abcam, Cambridge, UK), and ZO-1 (ab96587, Abcam, Cambridge, UK) primary antibodies (IF for 1:100) for 48 h at 4°C. Brain tissues were incubated overnight with Alexa Fluor® 488 conjugated donkey anti-goat IgG (H+L) antibody (ab150129, Abcam, Cambridge, UK; 1:2000) at 4°C, while the nuclei of all cells were washed and incubated with 4′6-diamidino-2-phenylindole (DAPI, ab104139, Abcam, Cambridge, UK; 1:1000). Briefly, confocal images were used to observe the fluorescence images of each group (ChemiScope 6100BZ, Shanghai, China).
The protein concentration was determined by the BCA method. The membranes separated by SDS-PAGE electrophoresis were transferred onto polyvinylidene difluoride membranes. The membranes were incubated for blocking with 5% nonfat dry milk at room temperature for 2 h, then incubated with anti-leucine-rich α-2 glycoprotein 1 (LRG1; ab178698, Abcam, Cambridge, UK), anti-transforming growth factor beta 1 (TGF-β1; ab235578, Abcam, Cambridge, UK), rho-associated protein kinase1 (ROCK1; ab134181, Abcam, Cambridge, UK), phosphorylated-mechanistic target of rapamycin (p-mTOR; ab109268, Abcam, Cambridge, UK), mTOR (ab13490, Abcam, Cambridge, UK), anti-phosphorylated-SMAD2 (p-SMAD2; ab280888, Abcam, Cambridge, UK), anti-SMAD2 (ab40855, Abcam, Cambridge, UK), anti-p-SMAD3 (ab52903, Abcam, Cambridge, UK), anti-SMAD3 (ab40854, Abcam, Cambridge, UK), anti-occludin (ab216327, Abcam, Cambridge, UK), anti-claudin-5 (ab131259, Abcam, Cambridge, UK), anti-ZO-1 (ab276131, Abcam, Cambridge, UK), and β-actin (ab8226, Abcam, Cambridge, UK) primary antibodies (1:1000), and then incubated with secondary antibody (HRP-conjugated secondary antibody, 1:2000, ab6728; Abcam, Cambridge, UK) for 50 min. Enhanced chemiluminescence was used for image capture through a bioimage analysis system (Bio-Rad, California, USA).
All values are expressed as mean ± SD. The data were analyzed based on a two-tailed student’s t-test (two groups) or a two-way ANOVA followed by Bonferroni’s test (three or more groups) through Graph Prism 5.2. The significance level was set at p < 0.05 and p < 0.01.
High-performance liquid chromatography (HPLC) detection of general flavones from Dalbergia odorifera leaves
HPLC method was used for the detection of general flavones from D. odorifera leaves. At wavelengths of 211, 254, 270, and 360 nm, the absorption peak of fingerprint of general flavonoids of 70% ethanol extract of D. odorifera leaves was detected (Fig. 1). The mass proportion of general flavonoids was 140.74 ± 2.16 mg/g (Fig. 1).
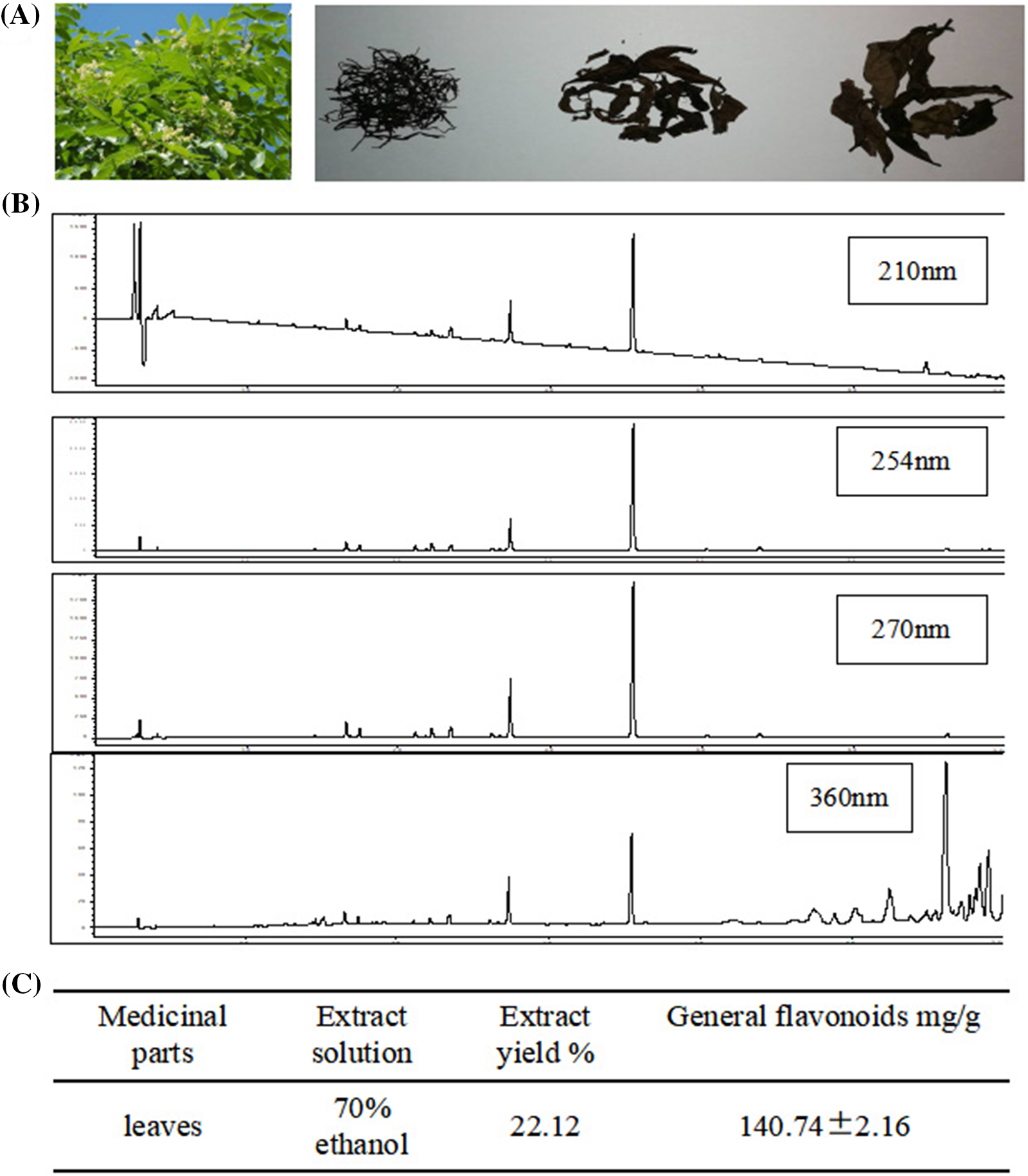
Figure 1: High-performance liquid chromatography (HPLC) for the determination of general flavones of Dalbergia odorifera leaves. (A) The shape of fresh and air-dried D. odorifera leaves; (B) HPLC fingerprint of general flavonoids of 70% ethanol extracted from D. odorifera leaves; (C) The mass proportion of general flavonoids in D. odorifera leaves.
Identification of differential expressed proteins
Eight decreased expression protein bands were singled out from cerebral I/R mice treated with control-affinity resin or DLE (70% ethanol extract of D. odorifera leaves)-affinity resin via series affinity chromatography (Figs. 2A and 2B). The identified proteins from the control affinity resin were compared with those protein bands from DLE-affinity resin. In total, 2,190 proteins were identified (Suppl. Tables S1 and S2). For subsequent analysis, 952 differential expression proteins were screened (log2Fold Change > 0.5, adj.p.Val < 0.05) (Suppl. Table S3 and Suppl. Fig. S1). The comparison revealed 644 up-regulated and 308 down-regulated differentially expressed proteins on proteins identified from the control affinity resin.
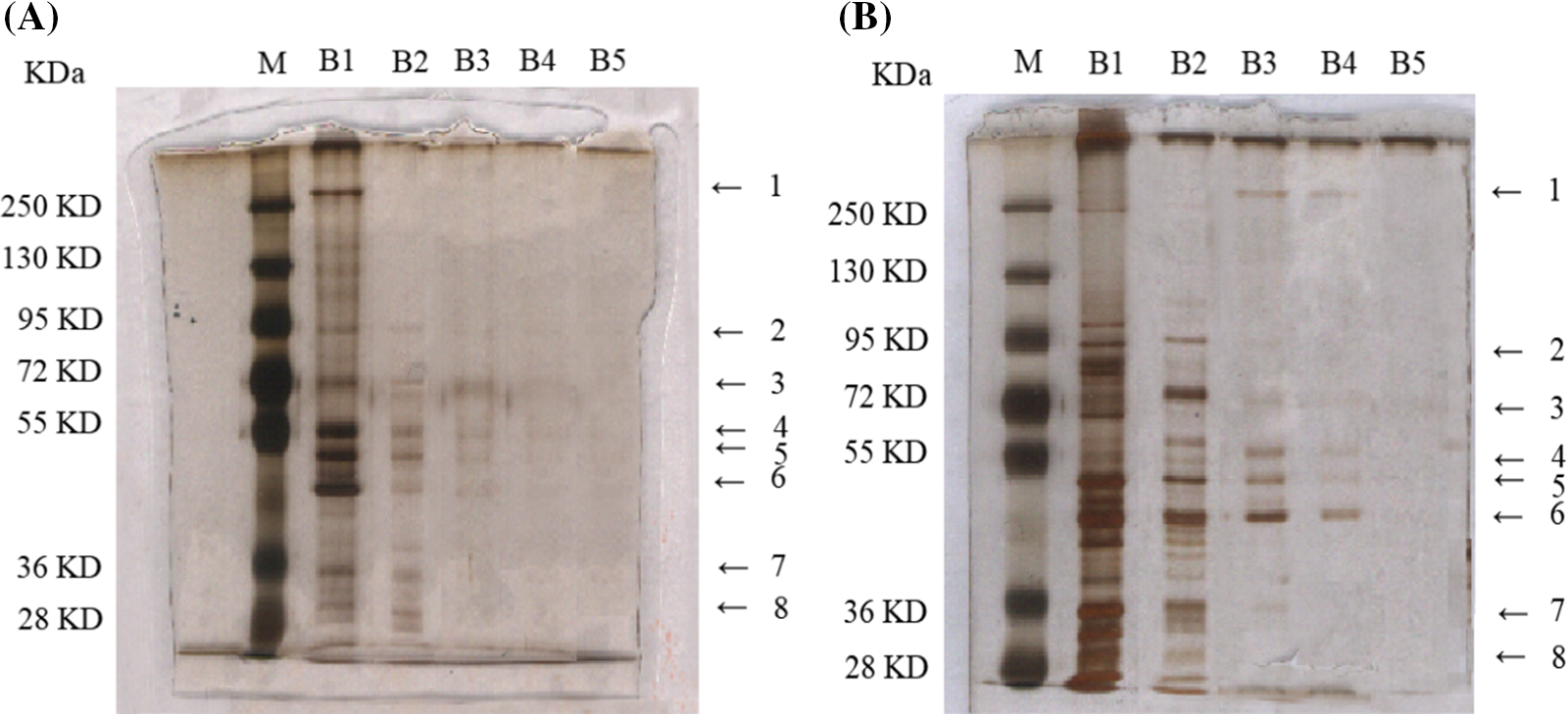
Figure 2: Differential concentration expressed protein bands from mice after 1 h of ischemia and 24 h of reperfusion, based on serial affinity chromatography. (A) Silver staining bands from control-affinity resin via serial affinity chromatography. (B) Silver staining bands from DLE-affinity resin via serial affinity chromatography. The protocol was analyzed with five repeats. Lane M: Marker; Lane B1–B5: After the synthesis of DLE affinity resin, the resin was subjected to incubation with brain tissue homogenates, repeated five times. Lane B numbered 1–5, represents the protein samples from each incubation via silver staining.
Co-expression protein modules extraction based on weighted gene co-expression network analysis
A total of 952 differential expressed proteins were screened by WGCNA. The parameter β was screened between 1 and 20, and the optimal soft threshold value was determined as 6. When the soft threshold power was set as 6, the degree of fitting was scale-free R^2 = 0.95 and trunc.R^2 = 0.95 (Figs. 3A and 3B). According to the soft threshold value, the network approximated the real biological network, and it amounted to the power law distribution. The relationship matrix was translated into an adjacency matrix, and finally, seven expression modules were constructed (Figs. 3D and 3E). Co-expression modules were constructed based on the TOM. The resulting protein tree and corresponding module colors are shown in Fig. 3B, of which the turquoise module had the strongest internal interaction (Fig. 3F).
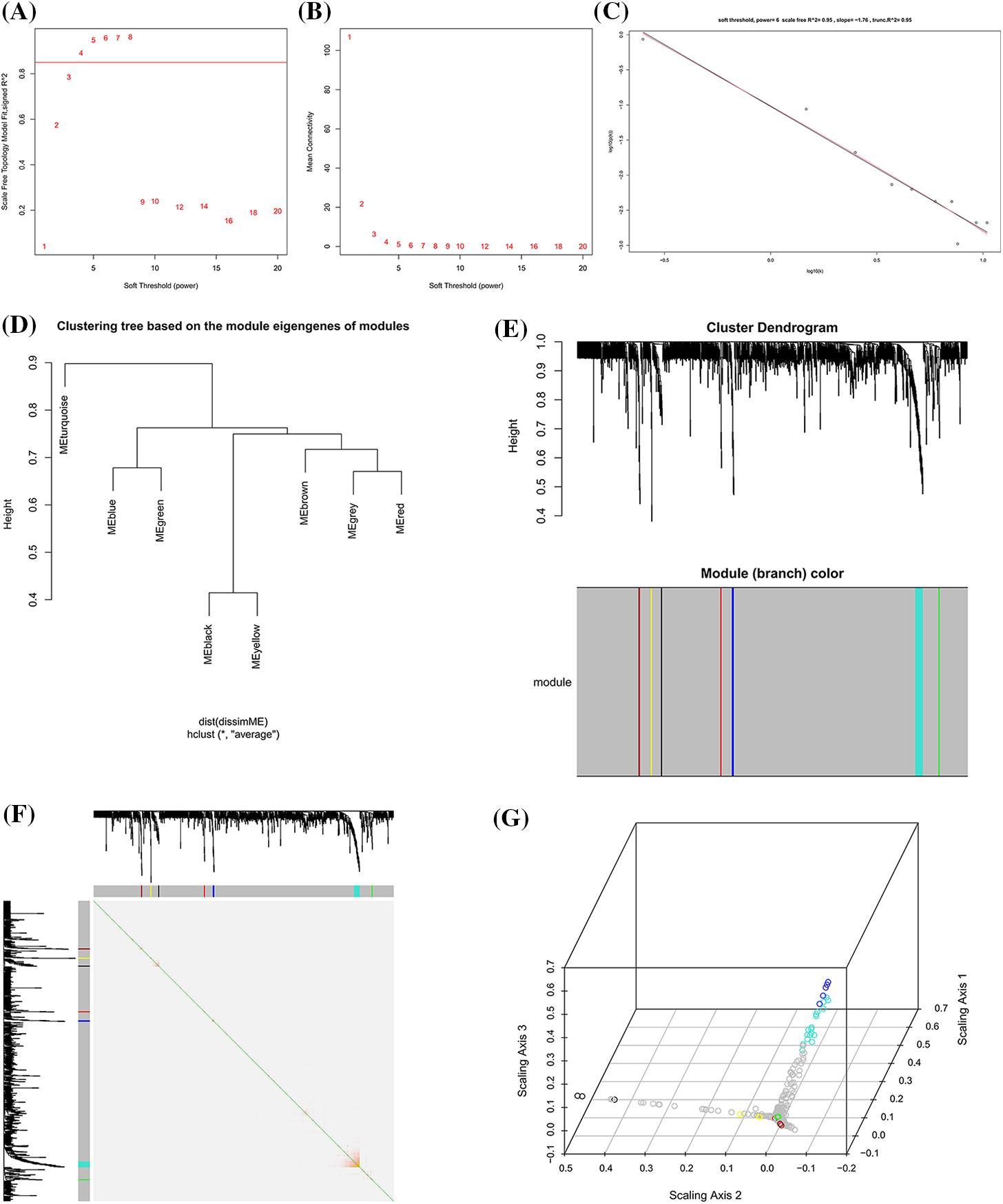
Figure 3: Co-expression protein modules extraction based on weighted gene co-expression network analysis (WGCNA). (A) Soft threshold determination of the WGCNA. (B) Soft threshold power based on the average connectivity. (C) Under the Soft threshold 6, the scale-free network fitted with the ideal curve. (D) Hierarchical clustering dendrograms of identified protein modules with co-expressed proteins. (E) Cluster genes via hierarchical clustering. Different color represents a different module but a similar set of proteins. (F) Correlation matrix between protein modules of adjacency differences adjacency. Each row corresponds to a module feature protein, and each column corresponds to a particular feature. The left and top bars correspond to each row or column. (G) 3D represented each row or column of protein modules.
Prediction through gene ontology biological function and kyoto encyclopedia of genes and genomes signaling pathway analyses
Hub proteins identified via WGCNA were (protein name symbols name represented for protein name) AKT1, PIK3CA, NOS3, SMAD3, SMAD1, ATK1, MAPK1, TGFBR2, TGFBR1, MAPK3, IGF1R, LRG1, mTOR, ROCK1, TGFB1, TGF1, SMAD2, SMAD5 (Fig. 4A). Candidate proteins obtained from hub protein module via WGCNA were subjected to BP, CC, and MF analyses, and −Log10FDR was defined to make the biological function map for the y-axis. BP was related to TGF-β receptor signaling pathway, common-partner SMAD protein phosphorylation, pathway-restricted SMAD protein phosphorylation, etc. CC was associated with positive regulation of pathway-restricted SMAD protein phosphorylation, SMAD protein complex, SMAD2-SMAD3 protein complex, etc. MF was related to the transcription factor complex, inhibitory-SMAD (I-SMAD) binding, SMAD binding, etc. (Fig. 4B). The hub proteins were associated with KEGG signaling pathways as TGF-β signaling pathway, mitogen-activated protein kinases (MAPK) signaling pathway, focal adhesion, adherens junction, etc. (Fig. 4C).
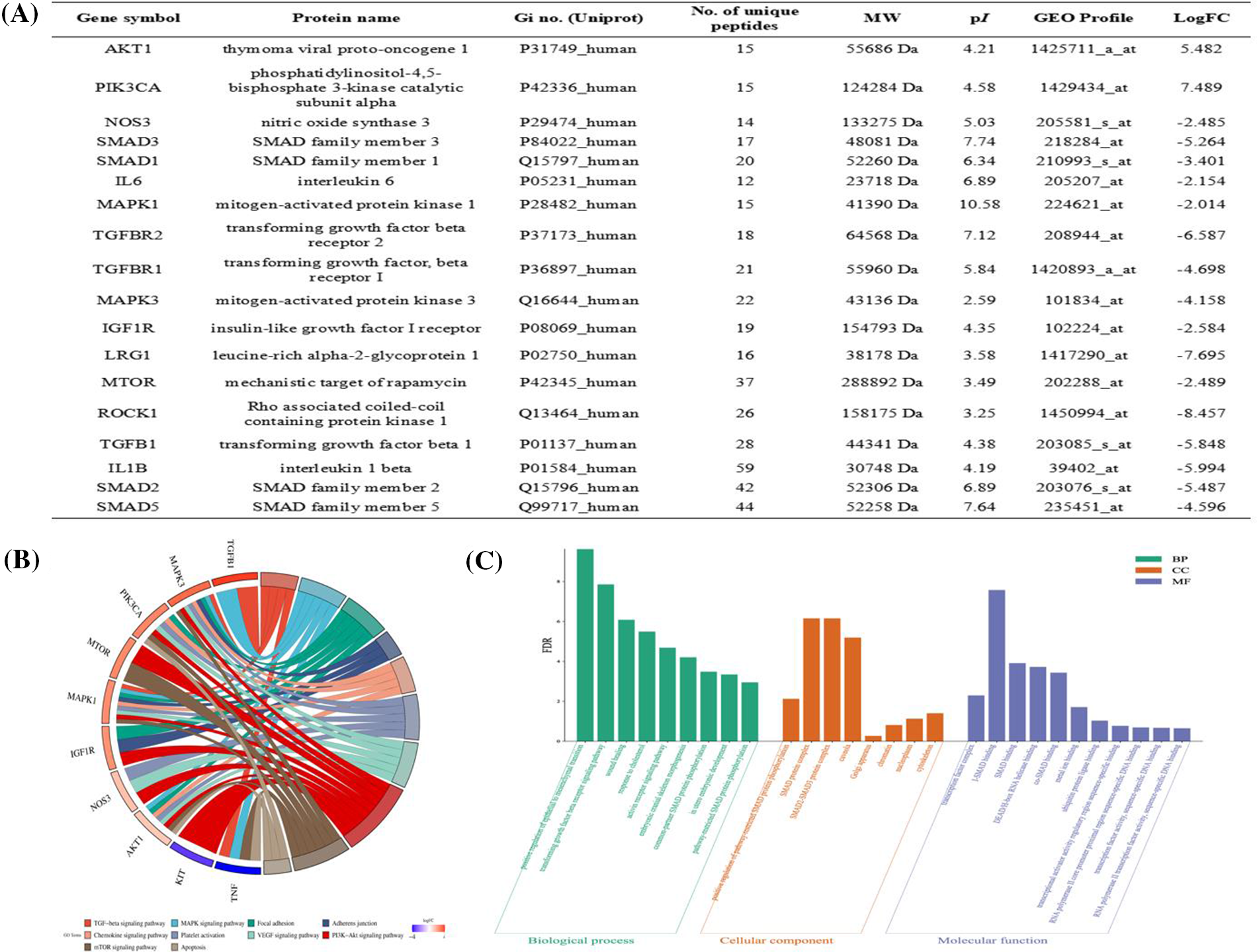
Figure 4: Prediction of hub proteins screened via weighted gene co-expression network analysis (WGCNA) for Gene Ontology (GO) biological function and kyoto encyclopedia of genes and genomes (KEGG) signaling pathway. (A) Hub proteins screened via WGCNA. (B) GO annotation analysis. The top 10 most enriched GO terms respectively represented biological processes (BP), cellular components (CC), and molecular functions (MF). The x-axis represents the enrichment annotation, and the y-axis represents −Log10FDR. (C) The chord figure represents the top 10 enriched KEGG signaling pathways and 10 included proteins. The weight in the chord figure is associated with the −Log10FC relevance protein with signaling pathway enriched through KEGG.
Dalbergia odorifera leaves extract attenuated blood-brain barrier dysfunction
The content of the Evans blue inside cerebral I/R brain tissue was examined to assess the effect of DLE on the blood-brain barrier permeability quantitatively. Quantitative content revealed DLE treatment significantly reduced Evans blue leakage (low, middle, high dose: 2.21 ± 0.24 mg/g, p < 0.01, 1.94 ± 0.21 mg/g, p < 0.01, 1.52 ± 0.24 mg/g, p < 0.01) compared with the cerebral I/R group (2.89 ± 0.26 mg/g, p < 0.01, sham: 0.25 ± 0.29), and 10 mg/kg nimodipine (1.54 ± 0.27 mg/g, p < 0.01) had the same effect (Fig. 5).
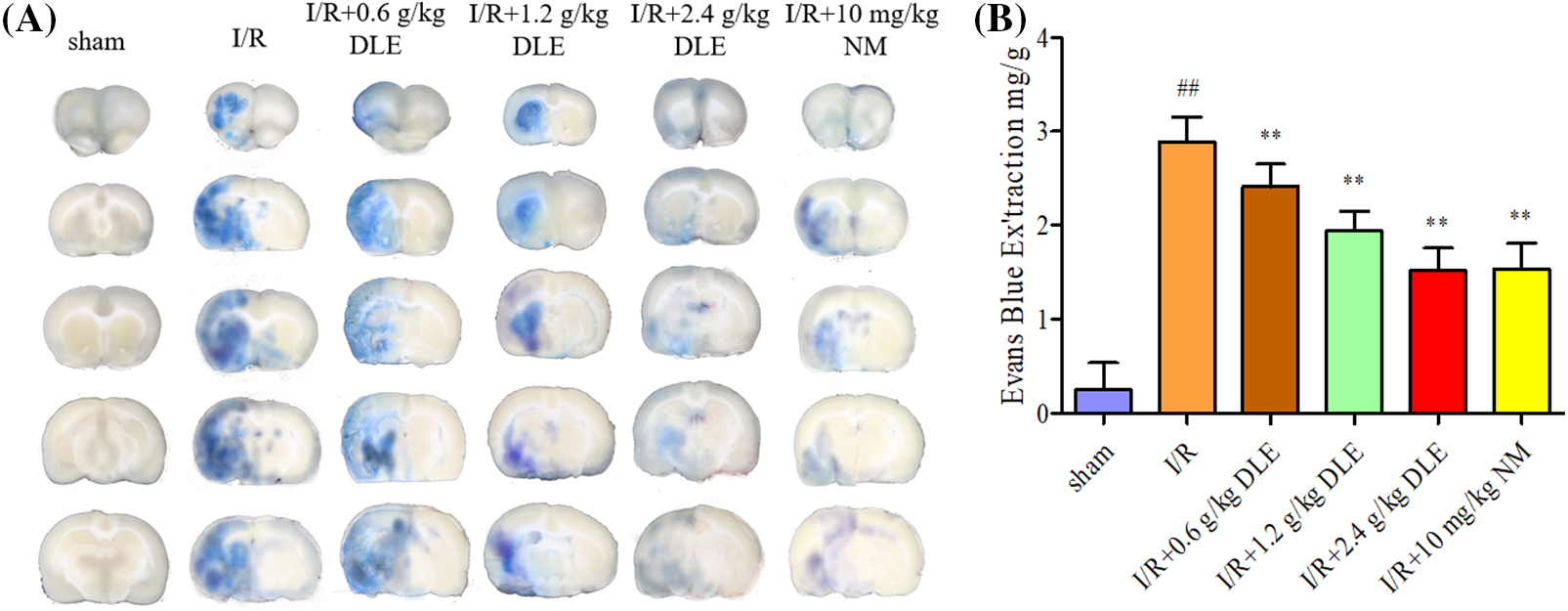
Figure 5: Attenuation mechanism of D. odorifera leaves extract (DLE) on blood-brain barrier permeability in cerebral ischemia-reperfusion (I/R) mice. (A) Representative appearance brain of Evans blue in 1 h of ischemia, 24 h of reperfusion mice. Mice were randomly categorized into six groups: (1) sham-operated mice (sham); (2) mice exposed to 1 h of ischemia, 24 h of reperfusion stroke (I/R); (3) mice exposed to I/R and treatment with 0.6 g/kg DLE (I/R+0.6 g/kg DLE); (4) mice exposed to I/R and treatment with 1.2 g/kg of DLE (I/R+1.2 g/kg DLE); (5) mice exposed to I/R and treatment with 2.4 g/kg of DLE (I/R+2.4 g/kg DLE); (6) mice exposed to I/R and treatment with 10 mg/kg nimodipine (I/R+10 mg/kg NM). (B) Analysis of Evans blue content by quantitative spectrophotometry. The results were presented as eight independent experiments. The data were averages with S.D., n = 8. #p < 0.05, ##p < 0.01 vs. sham-operated mice, *p < 0.05, **p < 0.01 vs. 1 h of ischemia, 24 h of reperfusion stroke.
This linear shape along the tight junction proteins of blood vessels reduced with lower expression in response to I/R (Fig. 6A), with a weaker signal strength of occludin, claudin-5, and ZO-1 (occludin: 10.49 ± 8.77, p < 0.01; claudin-5: 7.49 ± 5.21, p < 0.01; ZO-1: 7.78 ± 10.15, p < 0.01; sham: occludin: 51.49 ± 9.47; claudin-5: 47.59 ± 6.42; ZO-1: 56.88 ± 8.49). The DLE dose-dependent improved the pathological injury of brain tissues in cerebral I/R mice by strengthening immunohistochemical expression intensity of tight junctions (low, middle, high dose DLE: occludin: 14.59 ± 7.44, p < 0.01, 18.48 ± 8.45, p < 0.01, 28.47 ± 5.49, p < 0.01; claudin-5: 14.59 ± 6.34, p < 0.01, 26.47 ± 4.17, p < 0.01, 37.09 ± 3.84, p < 0.01; ZO-1: 12.47 ± 8.55, p < 0.01, 19.09 ± 8.49, p < 0.01, 42.35 ± 9.78, p < 0.01, Figs. 6A and 6B), the integrity of vascular vessel expression was restored to some extent.
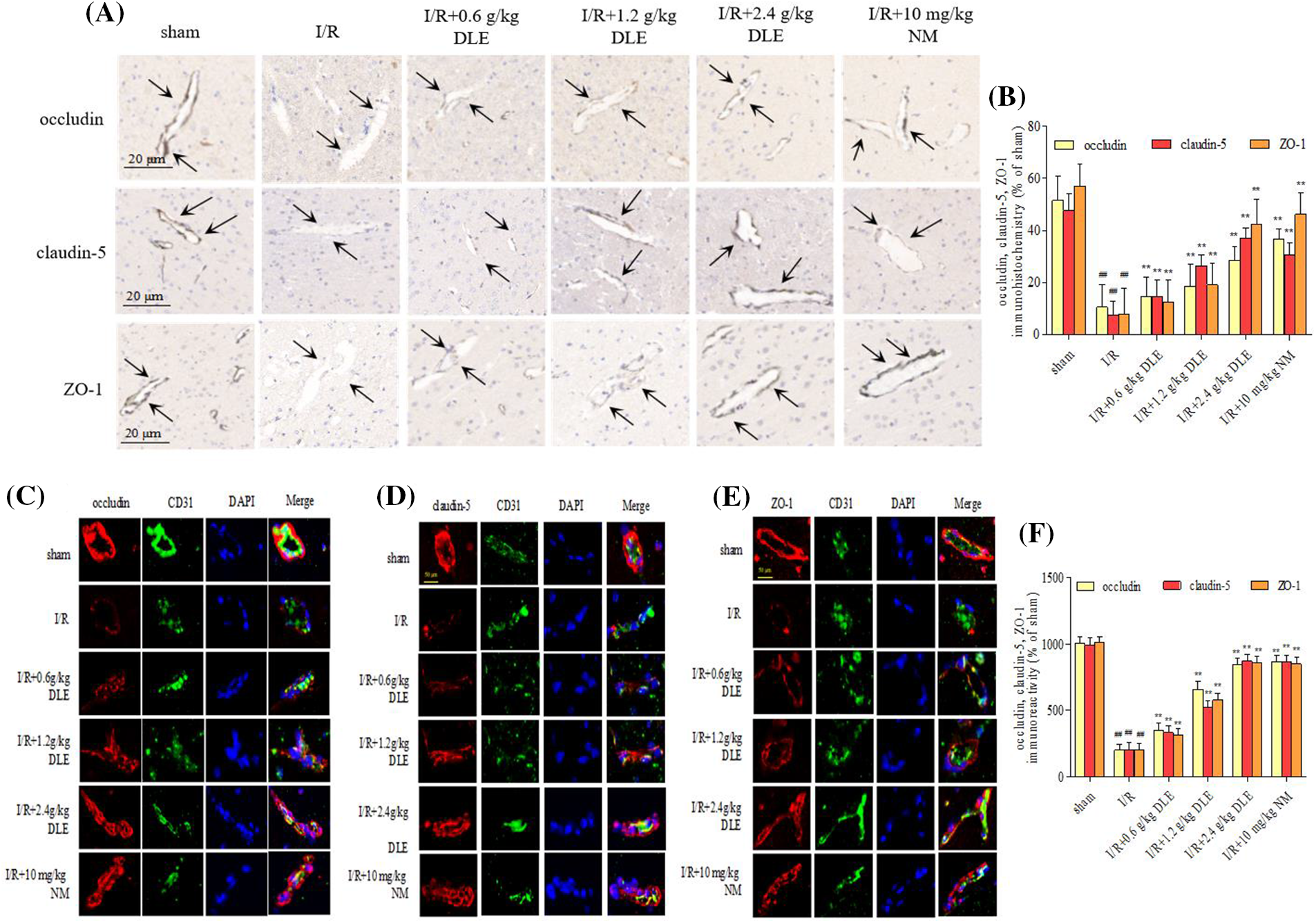
Figure 6: Effect of D. odorifera leaves extract (DLE) on immunohistochemistry staining changes in tight junctions in brain tissues exposed to 1 h of ischemia, 24 h of reperfusion stroke mice. (A) The ischemia-reperfusion (I/R) fields were observed under ChemiScope 6100BZ, bar = 20 μm. Mice were randomly categorized into six groups: (1) sham-operated mice (sham); (2) mice exposed to 1 h of ischemia, 24 h of reperfusion stroke (I/R); (3) mice exposed to I/R and treatment with 0.6 g/kg DLE (I/R+0.6 g/kg DLE); (4) mice exposed to I/R and treatment with 1.2 g/kg of DLE (I/R+1.2 g/kg DLE); (5) mice exposed to I/R and treatment with 2.4 g/kg of DLE (I/R+2.4 g/kg DLE); (6) mice exposed to I/R and treatment with 10 mg/kg nimodipine (I/R+10 mg/kg NM). (B) The tight junction positive boundaries were measured. The data are presented as the percentage of positive boundaries in the cerebral I/R mice. The data were the mean value with SD, n = 8. #p < 0.05, ##p < 0.01 vs. sham-operated mice, *p < 0.05, **p < 0.05 vs. 1 h of ischemia, 24 h of I/R. (C–E) Representative immunofluorescence staining of occludin (red), claudin-5 (red), and ZO-1 (red) in I/R mice. The nucleus was stained with DAPI (blue), and the vascular endothelial cells with CD31 (green). Confocal microscopy images were acquired, bar = 50 μm. The images represent nine individual views from each group. (F) The expression of TGF-β, p-SMAD2, SMAD2, p-SMAD3, and SMAD3, and β-actin in I/R-treated mice were detected via western blotting. The data were the mean value with S.D., n = 8, #p < 0.05, ##p < 0.01 vs. the sham-operated mice, *p < 0.05, **p < 0.01 vs. 1 h of ischemia, 24 h of I/R.
Tight junctions were continuous and well organized in the sham-operated blood vessel edge through immunofluorescent staining (Figs. 6C–6F). After stimulation with I/R, occludin, claudin-5, and ZO-1 revealed structural disturbance with reduced fluorescence compared with the sham-operated group. Moreover, immunocytochemical staining of tight junction proteins along the blood vessel edge was restored to a higher fluorescence expression intensity in response to the treatment of DLE. After treatment with DLE, the protein expression of LRG1, TGF-β1, p-SMAD2/SMAD2, p-SMAD3/SMAD3 were downregulated, while the expressions of ROCK1, p-mTOR/mTOR, occludin, claudin-5, ZO-1 were up-regulated (I/R+2.4 g/kg DLE: LRG1: 1.24 ± 0.02, p < 0.01; TGF-β1: 1.17 ± 0.05, p < 0.01; p-SMAD2/SMAD2: 1.24 ± 0.04, p < 0.01; p-SMAD3/SMAD3: 1.21 ± 0.03, p < 0.01, ROCK1: 1.44 ± 0.04, p < 0.01, p-mTOR/mTOR: 1.46 ± 0.07, p < 0.01, occludin: 1.45 ± 0.09, p < 0.01, claudin-5: 1.47 ± 0.04, p < 0.01, ZO-1: 1.45 ± 0.04, p < 0.01, Fig. 7) compared with those of the I/R group (LRG1: 1.72 ± 0.03, p < 0.01; TGF-β1: 1.66 ± 0.04, p < 0.01; p-SMAD2/SMAD2: 1.79 ± 0.07, p < 0.01; p-SMAD3/SMAD3: 1.74 ± 0.05, p < 0.01, ROCK1: 1.00 ± 0.05, p < 0.01, p-mTOR/mTOR: 1.00 ± 0.05, p < 0.01, occludin: 1.00 ± 0.03, p < 0.01, claudin-5: 1.02 ± 0.04, p < 0.01, ZO-1: 1.02 ± 0.07, p < 0.01, Fig. 7).
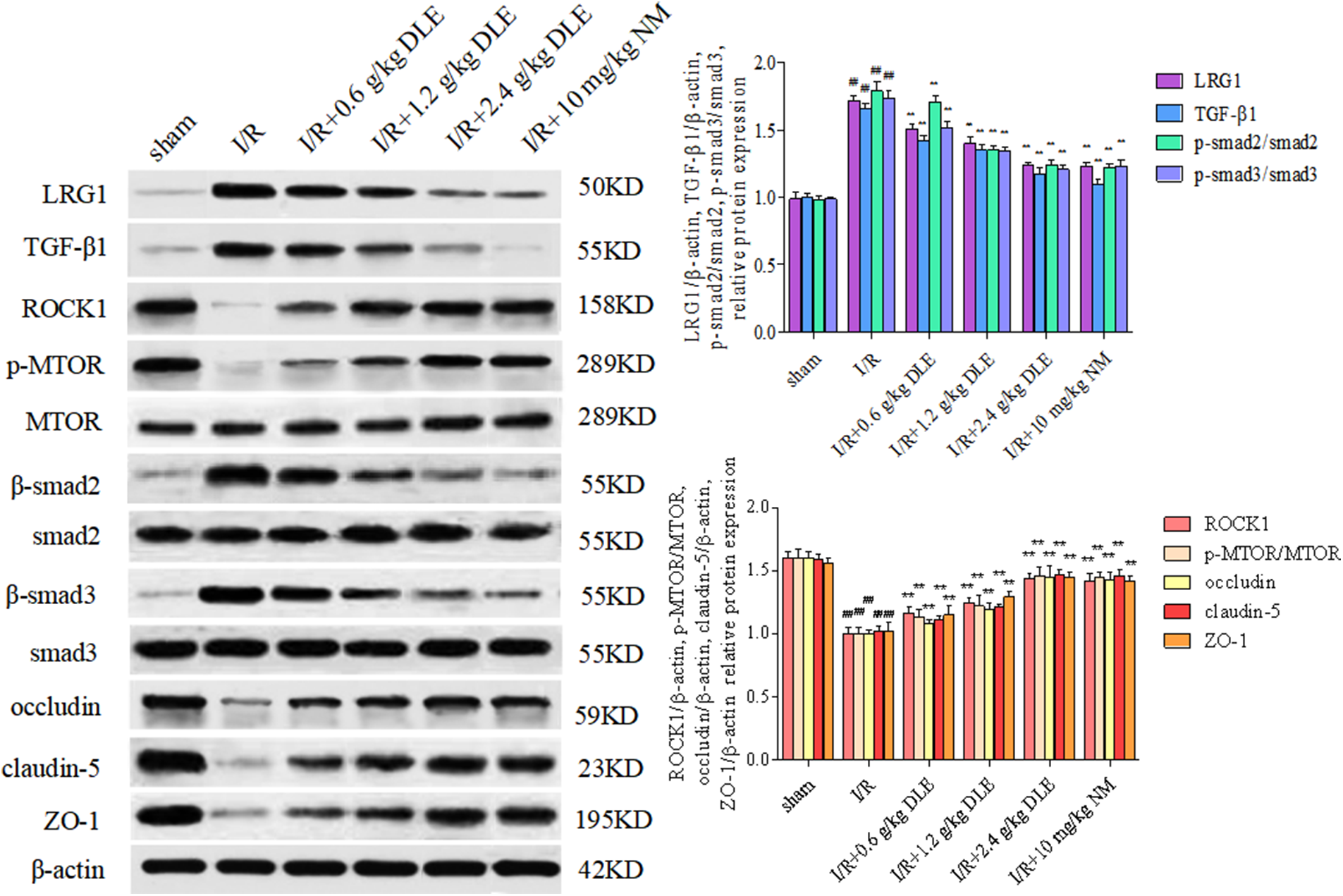
Figure 7: Western blot expression of the treatment of D. odorifera leaves extract (DLE) on tight junctions in brain tissues exposed to 1 h of ischemia, 24 h of reperfusion stroke mice. Mice were randomly categorized into six groups: (1) sham operated mice (sham); (2) mice exposed to 1 h of ischemia, 24 h of reperfusion stroke (I/R); (3) mice exposed to I/R and treatment with 0.6 g/kg DLE (I/R+0.6 g/kg DLE); (4) mice exposed to I/R and treatment with 1.2 g/kg of DLE (I/R+1.2 g/kg DLE); (5) mice exposed to I/R and treatment with 2.4 g/kg of DLE (I/R+2.4 g/kg DLE); (6) mice exposed to I/R and treatment with 10 mg/kg nimodipine (I/R+10 mg/kg NM). The relative expression of LRG1, TGF-β1, p-SMAD2/SMAD, p-SMAD3/SMAD3, ROCK1, p-mTOR/mTOR, occludin, claudin-5, ZO-1, and β-actin was analyzed. Data were averages ± SD, n = 8. ##p < 0.01, #p < 0.05 vs. sham-operated mice, **p < 0.01, *p < 0.05 vs. 1 h of ischemia, 24 h of reperfusion stroke.
Plant parts like rhizome and pith are medicinal plants, whereas leaves, seeds, flowers, and fruits from these plants are all resources that could be developed into food or medicine. Many kinds of leaves from traditional Chinese plants can be used as commercial food or medicine. Previous studies have isolated ~150 components from D. odorifera leaves, containing general flavonoids, exhibiting the highest antioxidant activity in scavenging DPPH·, O2−, ·OH, H2O2, and NO2− (Li and Chen, 2013). General flavonoid components, such as biochanin A and genistein are in abundance and are the main active components in D. odorifera leaves (Li et al., 2016). Biochanin A mitigated dyslipidemia in hyperlipidemic, atherosclerosis, and diabetes in rats (Sarfraz et al., 2020), whereas genistein exerted good hypolipidemic effects on clinical patients and mice (Yang et al., 2021a). Other ingredients, such as tectorigenin and prunetin, were also reported for their pharmaceutical value. Given the amounts of active ingredients contained in D. odorifera leaves and the potential rapid economic benefits, we had good reason to examine the medicinal value of D. odorifera leaves.
The alcohol extraction method yielded a relatively high content of general flavonoids from D. odorifera. leaves. Previously, an integrated approach was successfully used for extracting special binding proteins from Chinese herbal compounds. The integrated method could reveal a more complex protein map for D. odorifera leaves. The identified proteins from the control affinity resin and DLE-affinity resin were compared. The identified differentially expressed proteins were involved in a large network, and a large amount of information was contrary to the accurate experimental verification. WGCNA is suitable for complex transcriptome data, also mapping regulatory networks among gene sets, and identifying key regulatory genes. Using WGCNA. We constructed the gene co-expression network based on the correlation coefficient. Genes/proteins with close relationships through threshold value were delimited and then divided into a closely related module. Different modules were endowed with characteristic values and different colors. Hub modules related to biological problems were screened through the expression mode and function of modules. WGCNA detected 697 unique differentially expressed genes in tonsil allergen sensitization induction, while interleukin (IL)-17 and its regulatory toll-like receptor signaling pathway are more focused on aeroallergen sensitized patients (Hanif et al., 2023). The receiver operating characteristic curves of the four genes through WGCNA demonstrated that these four genes play a critical role and cytoplasmic FMR1 interacting protein 2 (CYFIP2) is strongly associated with the prognosis of patients with rheumatoid arthritis and tumors (Zhao et al., 2022). Cancer-associated fibroblast hub genes have been filtered via WGCNA to construct a prognostic signature for ovarian cancer (Feng et al., 2022). Thus, WGCNA is an advantageous tool to help identify key genes from the vast network.
WGCNA revealed that 952 differential expression proteins were included in the hub module. Therefore, targeted proteins screened by WGCNA might also play a critical role in cerebral I/R. The enriched signaling pathway should be consistent with GO findings. TGF-β is a member of a superfamily, whose most biological jobs include the regulation of cell growth and differentiation; over a third portion of functions were associated with TGF-β1. Human TGF-β1 is localized on chromosome 19q13 and expressed in endothelial cells and connective tissue cells, which affects the regeneration of vascular endothelial cells, epithelial cells, and the hematopoietic system. Here, from the hub protein network, targets of TGFBI, TGFBR2, TGFB1I1, and TGFBRAP1 exhibited high connectivity in the network. Combining SMAD1, SMAD2, SMAD5, SMAD7, and SMAD9 data revealed a close association between GO and KEGG signaling pathway analyses. BP was related to the TGF-β receptor signaling pathway, positive regulation of transcription, SMAD protein signal transduction, etc., while the predicted KEGG signaling pathway was associated to the TGF-β signaling pathway. The activation of the TGF-β/SMAD2/3 signaling pathway increases the release of vascular endothelial growth factor and thus promoted angiogenesis in ischemic stroke brain (Zhang et al., 2022). TGF-β activity plays a key role in mediating neuroplasticity and recovery of neural function through growth arrest and inducing DNA damage after cerebral I/R injury (Zhang et al., 2019). Macrophage inflammatory cascade caused by stroke increased swarming of inflammatory cytokine levels of IL-1β, IL-6, monocyte chemoattractant protein-1 (MCP-1), and TGF-β in the heart might contribute to post-stroke cardiac dysfunction (Yan et al., 2020).
TGF-β is connected with variation in the tight junction and offers a new way to induce exosomal lnc-MMP2-2 to increase the barrier-blood brain permeability to promote brain metastasis due to non-small-cell lung cancer (Wu et al., 2021). Up-regulation of the TGF-β mRNA expression demonstrated barrier-blood brain damage and neurobehavioral changes induced by methyl mercury in the rat cerebellum (Abu-Zeid et al., 2021). Platelet-derived growth factor receptor-α effects might be mediated through TGF-β1, which has a protective effect on the barrier-blood brain (Nguyen et al., 2021). It has also been reported that SMAD2 might be involved in the pathological process of ischemic stroke. Stem cell-derived small extracellular vesicles induce a marked increase in regulatory T cells, while the process depends upon activated TGF-β/SMAD through the transmission of TGF-β, SMAD2, and SMAD4 (Xia et al., 2021). In another study, barrier-blood brain integrity suffered and was destroyed at 12 and 24 h after middle cerebral artery occlusion (MCAO), and the levels of TGF-β1, TGF-βR(II), and p-SMAD2/3 were up-regulated (Yang et al., 2021b) In this study, D. odorifera leaves could attenuate the blood-brain barrier damage, accompanied by TGF-β1 activation and the phosphorylation of SMAD2. Additionally, through western blot assay, we confirmed some potential upstream and downstream targets.
In addition to the TGF-β signaling pathway, we found that focal adhesion and adherens junction were also the predicted signaling pathway. Focal adhesion and adherens junction are the essential ingredients for the blood-brain barrier integrity. Adhesive junctions form “grid” and “lock” shaped tight junctions that maintain the internal environment of brain tissues by preventing the molecules from invading. Previous studies have reported that focal adhesion (Yang et al., 2022) and adherens junction (Cai et al., 2022) have an important function in the blood-brain barrier. Thus, in this study, we conducted the evaluation of blood-brain barrier permeability to verify the focal adhesion and adherens junction pathway and found that the tight junction proteins, occludin, claudin-5, and ZO-1 were downregulated after cerebral I/R, while DLE could restore the downregulation of tight junctions. Also, the pathological staining, immunofluorescence, and western blotting results were in accordance with the Evans blue staining. The WGCNA prediction results agreed well with the experimental observations.
The top ten scoring signaling pathway included oxidative stress, autophagy and apoptosis-associated pathway, MAPK signaling pathway, mTOR signaling pathway, and apoptosis. The hub proteins MAPK3, mTOR, and ROCK1 play an important role in the regulation of cerebral I/R (Xu et al., 2021). Also, the phosphatidylinositol phosphate 3-kinase-Akt signaling pathway (related hub genes: AKT1 and PIK3CA) tends to be a protective pathway, activated after the stimulation of cerebral I/R (Li et al., 2023). In addition, other signaling pathways tightly associated with stroke were the chemokine signaling pathway (related hub gene: NOS3), related to the release of inflammatory factors; the platelet activation signaling pathway (related hub gene: IGF1R) is associated with hyperactivation of clotting function, while the VEGF signaling pathway is relevant to abnormal neovascularization. The literature suggests that these pathways and hub targets closely related to stroke. Due to the limited experimental condition, the more specific experiments could not be verified and need to be confirmed in the future.
LRG1 belongs to the leucine-rich repeat family, containing eight leucine-rich repeats. The LRG1 gene is located on chromosome 19P13.3, with a predicted molecular weight of 38 kD. The vast majority of leucine-rich repeat proteins that have been highly expressed in the nervous system are transmembrane proteins, primarily involved with the formation of the synapse, the growth of neuroprocesses, and the release of neurotransmitters as cell adhesion molecules or interacting proteins (Zhang et al., 2021). There are only a few experimental reports regarding LRG1 and cerebral I/R. LRG1 also exacerbated I/R injury by promoting apoptosis and autophagy via regulating the TGFβ-SMAD1/5 signaling pathway (Jin et al., 2019). Overexpression of LRG1 aggravated cerebral infarction in MCAO rats, enhancing apoptosis and autophagy in the brain tissue (Yamashita et al., 2010). In the MCAO mouse model, LRG1 promoted endothelial cell mitosis and angiogenesis via the activation of the TGF-β pathway (Meng et al., 2016; Cao et al., 2020; Pek et al., 2015). There are only a few reports on the relationship between LRG1 and the tight junction. Studies have shown that LRG1 could regulate the TGF-β signaling pathway (Nakajima et al., 2012); DLE might exert its functions through the LRG1/TGF-β signaling pathway.
This study has certain limitations. Further attempts for experimental verification of the mechanism of other predicted pathways and hub targets might be valuable. Although experiment verification was restricted in the research, we believe the TGF-β signaling pathway is highly correlated with ischemia stroke.
It is evident that an in-depth study of proteins would help to understand the mechanism of the occurrence of I/R and its biological functions. The integrated method used in this study identified the differentially expressed proteins in I/R based on DLE via WGCNA. The exploitation of D. odorifera leaves has great potential economic and environmental value because of its role in the attenuation of blood-brain barrier damage induced by I/R; furthermore, the attenuation mechanism is associated with the regulation of the LRG1/TGF-β signaling pathway.
Acknowledgement: We kindly acknowledge Jiangxi University of Traditional Chinese Medicine for providing the D. odorifera leaves.
Funding Statement: This work was supported by National Natural Science Foundation of China (Nos. 82100417, 81760094, 81760724); The Foundation of Jiangxi Provincial Department of Science and Technology Project (Nos. 20202ACBL206001, 20212BAB206022, 20181BAB205026); Youth Project of Jiangxi Education Department (No. GJJ200217); Open Project of Key Laboratory of Modern of TCM, Ministry of Education Jiangxi University of Traditional Chinese Medicine (TCM-2019010). The funders had no role in the study design, data collection, analysis, or the decision to publish or prepare the manuscript.
Author Contributions: Jinfang Hu and Jiangen Ao wrote the manuscript. Longsheng Fu, Yaoqi Wu, Feng Shao, Tiantian Xu, Mingjin Jiang, and Shaofeng Xiong conducted the experiment and analyzed the experimental data. Yanni Lv wrote the manuscript and made the final complement.
Availability of Data and Materials: All data generated or analyzed during this study are included in this published article.
Ethics Approval: The experimental protocol was established according to the ethical guidelines and was approved by the Ethics Committee of The First Affiliated Hospital of Nanchang University.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at DOI: 10.32604/biocell.2023.028684.
References
Abu-Zeid EH, Khalifa BA, Elewa YHA, Arisha AH, Ismail TA, Hendam BM, Abdel-Hamid SE (2021). Bee venom Apis mellifera lamarckii rescues blood brain barrier damage and neurobehavioral changes induced by methyl mercury via regulating tight junction proteins expression in rat cerebellum. Food and Chemical Toxicology 154: 112309. https://doi.org/10.1016/j.fct.2021.112309 [Google Scholar] [PubMed] [CrossRef]
Cai N, Xu B, Li X, Qin Y, Li M, Chen K, Xu J, Wang H (2022). Roflumilast, a cyclic nucleotide phosphodiesterase 4 inhibitor, protects against cerebrovascular endothelial injury following cerebral ischemia/reperfusion by activating the Notch1/Hes1 pathway. European Journal of Pharmacology 926: 175027. https://doi.org/10.1016/j.ejphar.2022.175027 [Google Scholar] [PubMed] [CrossRef]
Cao JW, Zhong D, Li GZ (2020). LRG1 and TGF after cerebral ischemia in mice-β1. Journal of Brain and Neurological Diseases 28: 595–600. [Google Scholar]
Feng S, Xu Y, Dai Z, Yin H, Zhang K, Shen Y (2022). Integrative analysis from multicenter studies identifies a WGCNA-derived cancer-associated fibroblast signature for ovarian cancer. Frontiers in Immunology 13: 951582. https://doi.org/10.3389/fimmu.2022.951582 [Google Scholar] [PubMed] [CrossRef]
Hanif T, Ivaska LE, Ahmad F, Tan G, Mikola E, Puhakka T, Palomares O, Akdis CA, Toppila-Salmi S, Jartti T (2023). Tonsillar transcriptional profiles in atopic and non-atopic subjects. Allergy 78: 522–536. https://doi.org/10.1111/all.15458 [Google Scholar] [PubMed] [CrossRef]
Jin J, Sun H, Liu D, Wang H, Liu Q, Chen H, Zhong D, Li G (2019). LRG1 promotes apoptosis and autophagy through the TGFβ-smad1/5 signaling pathway to exacerbate ischemia/reperfusion injury. Neuroscience 413: 123–134. https://doi.org/10.1016/j.neuroscience.2019.06.008 [Google Scholar] [PubMed] [CrossRef]
Li JY, Chen DL (2013). Antioxidant activity of the extracts from Dalbergia odorifera T. Chen leaves in vitro. Medicinal Plant 4: 58–61. [Google Scholar]
Li F, Duan J, Zhao M, Huang S, Mu F et al. (2019). A network pharmacology approach to reveal the protective mechanism of Salvia miltiorrhiza-Dalbergia odorifera coupled-herbs on coronary heart disease. Scientific Reports 9: 19343. https://doi.org/10.1038/s41598-019-56050-5 [Google Scholar] [PubMed] [CrossRef]
Li L, Liu JZ, Luo M, Wang W, Huang YY, Efferth T, Wang HM, Fu YJ (2016). Efficient extraction and preparative separation of four main isoflavonoids from Dalbergia odorifera T. Chen leaves by deep eutectic solvents-based negative pressure cavitation extraction followed by macroporous resin column chromatography. Journal of Chromatography B 1033–1034: 40–48. https://doi.org/10.1016/j.jchromb.2016.08.005 [Google Scholar] [PubMed] [CrossRef]
Li Z, Xiao G, Wang H, He S, Zhu Y (2021). A preparation of Ginkgo biloba L. leaves extract inhibits the apoptosis of hippocampal neurons in post-stroke mice via regulating the expression of Bax/Bcl-2 and Caspase-3. Journal of Ethnopharmacology 280: 114481. https://doi.org/10.1016/j.jep.2021.114481 [Google Scholar] [PubMed] [CrossRef]
Li R, Zheng Y, Zhang J, Zhou Y, Fan X (2023). Gomisin N attenuated cerebral ischemia-reperfusion injury through inhibition of autophagy by activating the PI3K/AKT/mTOR pathway. Phytomedicine 110: 154644. https://doi.org/10.1016/j.phymed.2023.154644 [Google Scholar] [PubMed] [CrossRef]
Lv Y, Fu L (2018). The potential mechanism for Hydroxysafflor yellow A attenuating blood-brain barrier dysfunction via tight junction signaling pathways excavated by an integrated serial affinity chromatography and shotgun proteomics analysis approach. Neurochemistry International 112: 38–48. https://doi.org/10.1016/j.neuint.2017.10.012 [Google Scholar] [PubMed] [CrossRef]
Ma RK, Liu H, Shi F, Fu Y, Wei P (2020). The chemical composition and antioxidant activity of essential oils and extracts of Dalbergia odorifera leaves. Holzforschung 74: 755–763. https://doi.org/10.1515/hf-2019-0155 [Google Scholar] [CrossRef]
Mei H, Hu H, Lv Y, Ma G, Tang F, Hong Z, Shao F (2021). The hypolipidemic effect of Dalbergia odorifera T. C. Chen leaf extract on hyperlipidemic rats and its mechanism investigation based on network pharmacology. Evidence-Based Complementary and Alternative Medicine 2021: 3155266. [Google Scholar] [PubMed]
Meng H, Song Y, Zhu J, Liu Q, Lu P, Ye N, Zhang Z, Pang Y, Qi J, Wu H (2016). LRG1 promotes angiogenesis through upregulating the TGF-β1 pathway in ischemic rat brain. Molecular Medicine Reports 14: 5535–5543. https://doi.org/10.3892/mmr.2016.5925 [Google Scholar] [PubMed] [CrossRef]
Nakajima M, Miyajima M, Ogino I, Wada S, Kano T, Kakinuma C, Ogihara T (2012). Brain localization of leucine-rich α2-glycoprotein and its role. Acta Neurochirurgica Supplementum 113: 97–101. https://doi.org/10.1007/978-3-7091-0923-6 [Google Scholar] [CrossRef]
Nguyen QL, Okuno N, Hamashima T, Dang ST, Fujikawa M et al. (2021). Vascular PDGFR-alpha protects against BBB dysfunction after stroke in mice. Angiogenesis 24: 35–46. https://doi.org/10.1007/s10456-020-09742-w [Google Scholar] [PubMed] [CrossRef]
Pek SL, Tavintharan S, Wang X, Lim SC, Woon K, Yeoh LY, Ng X, Liu J, Sum CF (2015). Elevation of a novel angiogenic factor, leucine-rich-α2-glycoprotein (LRG1is associated with arterial stiffness, endothelial dysfunction, and peripheral arterial disease in patients with type 2 diabetes. The Journal of Clinical Endocrinology and Metabolism 100: 1586–1593. https://doi.org/10.1210/jc.2014-3855 [Google Scholar] [PubMed] [CrossRef]
Sarfraz A, Javeed M, Shah MA, Hussain G, Shafiq N et al. (2020). Biochanin A: A novel bioactive multifunctional compound from nature. Science of the Total Environment 722: 137907. https://doi.org/10.1016/j.scitotenv.2020.137907 [Google Scholar] [PubMed] [CrossRef]
Shao F, Gu L, Chen H, Liu R, Huang H, Ren G (2016). Comparation of hypolipidemic and antioxidant effects of aqueous and ethanol extracts of crataegus pinnatifida fruit in high-fat emulsion-induced hyperlipidemia rats. Pharmacognosy Magazine 12: 64–69. https://doi.org/10.4103/0973-1296.176049 [Google Scholar] [PubMed] [CrossRef]
Shuai M, He D, Chen X (2021). Optimizing weighted gene co-expression network analysis with a multi-threaded calculation of the topological overlap matrix. Statistical Applications in Genetics and Molecular Biology 20: 145–153. https://doi.org/10.1515/sagmb-2021-0025 [Google Scholar] [PubMed] [CrossRef]
Sonoda T, Matsuzaki J, Yamamoto Y, Sakurai T, Aoki Y, Takizawa S, Niida S, Ochiya T (2019). Serum microRNA-based risk prediction for stroke. Stroke 50: 1510–1518. https://doi.org/10.1161/STROKEAHA.118.023648 [Google Scholar] [PubMed] [CrossRef]
Wang Y, Liu S, Wang R, Shi L, Liu Z, Liu Z (2020). Study on the therapeutic material basis and effect of Acanthopanax senticosus (Rupr. et Maxim.) Harms leaves in the treatment of ischemic stroke by PK-PD analysis based on online microdialysis-LC-MS/MS method. Food & Function 11: 2005–2016. https://doi.org/10.1039/C9FO02475A [Google Scholar] [PubMed] [CrossRef]
Wu D, Deng S, Li L, Liu T, Zhang T, Li J, Yu Y, Xu Y (2021). TGF-β1-mediated exosomal lnc-MMP2-2 increases blood-brain barrier permeability via the miRNA-1207-5p/EPB41L5 axis to promote non-small cell lung cancer brain metastasis. Cell Death & Disease 12: 721. https://doi.org/10.1038/s41419-021-04004-z [Google Scholar] [PubMed] [CrossRef]
Xia Y, Hu G, Chen Y, Yuan J, Zhang J, Wang S, Li Q, Wang Y, Deng Z (2021). Embryonic stem cell derived small extracellular vesicles modulate regulatory T cells to protect against ischemic stroke. ACS Nano 15: 7370–7385. https://doi.org/10.1021/acsnano.1c00672 [Google Scholar] [PubMed] [CrossRef]
Xu D, Kong T, Zhang S, Cheng B, Chen J, Wang C (2021). Orexin-A protects against cerebral ischemia-reperfusion injury by inhibiting excessive autophagy through OX1R-mediated MAPK/ERK/mTOR pathway. Cellular Signalling 79: 109839. https://doi.org/10.1016/j.cellsig.2020.109839 [Google Scholar] [PubMed] [CrossRef]
Yamashita F, Sasaki M, Takahashi S, Matsuda H, Kudo K, Narumi S, Terayama Y, Asada T (2010). Detection of changes in cerebrospinal fluid space in idiopathic normal pressure hydrocephalus using voxel-based morphometry. Neuroradiology 52: 381–386. https://doi.org/10.1007/s00234-009-0610-z [Google Scholar] [PubMed] [CrossRef]
Yan T, Chen Z, Chopp M, Venkat P, Zacharek A et al. (2020). Inflammatory responses mediate brain-heart interaction after ischemic stroke in adult mice. Journal of Cerebral Blood Flow and Metabolism 40: 1213–1229. https://doi.org/10.1177/0271678X18813317 [Google Scholar] [PubMed] [CrossRef]
Yang J, Hao J, Lin Y, Guo Y, Liao K, Yang M, Cheng H, Yang M, Chen K (2022). Profile and functional prediction of plasma exosome-derived circRNAs from acute ischemic stroke patients. Frontiers in Genetics 13: 810974. https://doi.org/10.3389/fgene.2022.810974 [Google Scholar] [PubMed] [CrossRef]
Yang R, Jia Q, Mehmood S, Ma S, Liu X (2021a). Genistein ameliorates inflammation and insulin resistance through mediation of gut microbiota composition in type 2 diabetic mice. European Journal of Nutrition 60: 2155–2168. https://doi.org/10.1007/s00394-020-02403-0 [Google Scholar] [PubMed] [CrossRef]
Yang X, Liang J, Jia M, Yang T, Deng X et al. (2021b). β-1, 3-galactosyltransferase 2 ameliorates focal ischemic cerebral injury by maintaining blood-brain barrier integrity. Neurochemistry International 144: 104976. https://doi.org/10.1016/j.neuint.2021.104976 [Google Scholar] [PubMed] [CrossRef]
Zhang Y, Mu Y, Ding H, Du B, Zhou M, Li Q, Gong S, Zhang F, Geng D, Wang Y (2022). 1α,25-dihydroxyvitamin D3 promotes angiogenesis after cerebral ischemia injury in rats by upregulating the TGF-β/Smad2/3 signaling pathway. Frontiers in Cardiovascular Medicine 9: 769717. https://doi.org/10.3389/fcvm.2022.769717 [Google Scholar] [PubMed] [CrossRef]
Zhang M, Wang Y, Wang J, Li X, Ma A, Pan X (2021). Serum LRG1 as a novel biomarker for cardioembolic stroke. Clinica Chimica Acta; International Journal of Clinical Chemistry 519: 83–91. https://doi.org/10.1016/j.cca.2021.04.002 [Google Scholar] [PubMed] [CrossRef]
Zhang K, Zhang Q, Deng J, Li J, Li J, Wen L, Ma J, Li C (2019). ALK5 signaling pathway mediates neurogenesis and functional recovery after cerebral ischemia/reperfusion in rats via Gadd45b. Cell Death & Disease 10: 360. https://doi.org/10.1038/s41419-019-1596-z [Google Scholar] [PubMed] [CrossRef]
Zhao Z, He S, Yu X, Lai X, Tang S et al. (2022). Analysis and experimental validation of rheumatoid arthritis innate immunity gene CYFIP2 and pan-cancer. Frontiers in Immunology 13: 954848. https://doi.org/10.3389/fimmu.2022.954848 [Google Scholar] [PubMed] [CrossRef]
Table S1. Identified proteins from cerebral I/R mice treated with control-affinity resin via series affinity chromatography
Table S2. Identified proteins from cerebral I/R mice treated with DLE (70% ethanol extract of D. odorifera leaves)-affinity resin via series affinity chromatography
Table S3. Differential expression proteins for each band screened with the criteria of log2Fold Change > 0.5, adj.p.Val < 0.05 based on the comparison between identified proteins from cerebral I/R mice treated with control-affinity resin and DLE-affinity resin via series affinity chromatography
Figure S1. Volcano map of differential expression proteins based on the comparison between identified proteins from cerebral I/R mice treated with control-affinity resin and DLE-affinity resin via series affinity chromatography.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools