 Open Access
Open Access
ARTICLE
Regulation of Dendritic Cell Function by RFX5 through Interaction with HDAC2 and Its Mechanism in Pediatric Asthma
1 Department of Pediatrics, Ji’an Hospital, Shanghai East Hospital, Ji’an, 343000, China
2 Department of Pediatrics, Shanghai East Hospital, Shanghai, 200120, China
* Corresponding Author: Fang Liu. Email:
(This article belongs to the Special Issue: Subcellular Organelles and Cellular Molecules: Localization, Detection, Prediction, and Diseases)
BIOCELL 2025, 49(4), 701-720. https://doi.org/10.32604/biocell.2025.061289
Received 21 November 2024; Accepted 08 April 2025; Issue published 30 April 2025
Abstract
Background: Dendritic cells (DCs) play a pivotal role in antigen presentation and regulating adaptive immune responses in asthma pathophysiology. However, the underlying molecular mechanisms remain incompletely understood. Methods: Bioinformatics analysis of the GSE27011 dataset identified differentially expressed genes associated with pediatric asthma. An ovalbumin (OVA)-induced asthma mouse model and an Rfx5 knockdown model were established. RFX5 expression was assessed in DCs from patients with asthma and asthmatic mouse lung tissues using qRT-PCR, Western blotting, and immunohistochemistry. The regulatory effects of regulatory factor X5 (RFX5) on histone deacetylase 2 (HDAC2), class II major histocompatibility complex transactivator (CIITA), and major histocompatibility complex class II molecules (MHC II) expression, as well as its influence on lung tissue integrity, airway resistance, cytokine profiles, and immune cell infiltration, were analyzed. Co-immunoprecipitation and chromatin immunoprecipitation assays were performed to explore the interaction between RFX5 and HDAC2. Results: During asthma progression, RFX5 expression was upregulated, while HDAC2 levels were reduced in DCs. Rfx5 knockdown significantly alleviated lung pathology and inflammation, decreased granulocyte and lymphocyte counts, and lowered levels of pro-inflammatory cytokines interleukin 6 (IL-6), tumor necrosis factor-alpha (TNF-α), and interleukin-1β (IL-1β). In contrast, the expression of anti-inflammatory cytokines such as interleukin 4 (IL-4), interleukin 10 (IL-10), interleukin 18 (IL-18), and prostaglandin E2 (PGE2) was elevated, along with an increase in CIITA and MHC II gene transcription. Further analysis revealed a direct association between HDAC2 and the RFX5 promoter region. Conclusion: During asthma pathogenesis, allergens may upregulate RFX5 expression in DCs, enhancing its interaction with HDAC2, thereby alleviating the HDAC2-mediated effect. This process promotes the transcription of MHC II-associated genes and facilitates antigen presentation, ultimately driving asthma initiation and progression. This study elucidates the role of the RFX5/HDAC2 signaling pathway in the regulation of antigen presentation in pediatric asthma.Keywords
Asthma, a chronic inflammatory airway disorder, primarily manifests in childhood, a critical developmental stage marked by particularly high incidence and prevalence rates. Emerging evidence indicates that childhood asthma may significantly impair airway development, potentially leading to irreversible pulmonary function deficits, with early-onset asthma resulting in more pronounced long-term consequences compared to late-onset forms [1]. From a pathophysiological standpoint, asthma is classified into neutrophilic and eosinophilic subtypes, both predominantly driven by type 2 helper T (Th2) lymphocyte-mediated inflammatory pathways. The Th2 cell-driven inflammatory cascade triggered by inhaled allergens typically begins during early childhood, a critical immunological window characterized by heightened susceptibility to allergic sensitization [2,3]. This persistent inflammation induces airway wall edema, excessive mucus secretion, and bronchial smooth muscle hyperresponsiveness, which together contribute to progressive airflow limitation and the hallmark symptoms of asthma [4]. Eosinophilic asthma, often coexisting with allergic asthma, is a treatment-responsive phenotype, whereas neutrophilic asthma displays greater therapeutic resistance. Mechanistic studies have shown that eosinophilic asthma is characterized by the recruitment of stromal macrophages and myeloid dendritic cells (mDCs), while neutrophilic asthma is primarily mediated by plasmacytoid dendritic cells (pDCs), exudative macrophages, and GL7+ activated B cells [5]. Despite these differences, both subtypes share common immunological features, particularly the activation and recruitment of macrophages and DCs.
Research has demonstrated that mDCs are central to asthma pathogenesis by initiating a Th2-polarized immune response upon allergen exposure, thereby triggering eosinophilic airway inflammation [6]. In neutrophilic asthma, pDCs play a key role in antigen processing and presentation, promoting T cell activation and inflammatory responses [7]. Asthma management typically includes long-term control and quick-relief medications, with inhaled corticosteroids (ICS) being the primary and most commonly used treatment. ICS work by binding to glucocorticoid receptors (GR) with co-activators, recruiting histone deacetylase 2 (HDAC2) to the inflammatory gene transcription complex and reversing histone acetylation, thereby suppressing activated inflammatory genes [8]. However, prolonged ICS use in pediatric populations is associated with adverse effects, including decreased bone mineral density, impaired growth velocity [9,10], and increased risks of ocular complications such as cataracts and glaucoma [11]. Additionally, a subset of patients exhibits suboptimal responses to ICS, resulting in inadequate disease control and potentially contributing to progressive airway remodeling and persistent airflow limitation. These pathological changes can heighten susceptibility to comorbid conditions like obstructive sleep apnea, recurrent pneumonia, and gastroesophageal reflux disease [12]. Emerging evidence suggests that reduced HDAC2 expression plays a pivotal role in corticosteroid resistance in severe asthma. Lower HDAC2 levels directly impair glucocorticoid receptor signaling, exacerbating uncontrolled airway inflammation in treatment-resistant patients [13–15]. Studies have shown significantly reduced HDAC2 expression in asthma model mice compared to controls [16,17]. Therefore, HDAC2 presents a key target for asthma therapy.
Extensive research efforts have been dedicated to developing novel therapeutic strategies for pediatric asthma, with a particular focus on elucidating the cellular and molecular mechanisms driving disease pathogenesis and progression. Comprehensive genomic analyses have identified distinct patterns of differentially expressed genes (DEGs) in children with severe and mild asthma compared to healthy controls. Functional annotation and pathway analysis have revealed the prominent involvement of X-box binding protein-related pathways, particularly highlighting significant enrichment in antigen processing and presentation processes mediated by CD4 and regulatory factor X (RFX) family genes [18]. Within the RFX family, RFX5 collaborates with HDAC2 to suppress collagen promoter activity, while HDAC1 enhances RFX1’s inhibitory effect on the same promoter. The transcription of collagen genes plays a vital role in developmental damage and repair processes [19]. Bioinformatic analysis of the GEO database (GSE27011 microarray dataset) demonstrates significant upregulation of RFX5 expression in asthmatic samples. However, the potential interaction between RFX family proteins and HDAC2 in pediatric asthma pathogenesis, as well as their underlying molecular mechanisms, remains inadequately understood. Additionally, the specific cooperative relationship between RFX5 and HDAC2 in childhood asthma development requires further exploration. This study aims to elucidate the molecular mechanisms by which DC-mediated regulation of helper T cell differentiation is controlled by RFX transcription factors and HDAC2-dependent epigenetic modulation, with the ultimate objective of identifying potential therapeutic targets for the development of innovative treatment strategies in pediatric asthma management.
2.1 Identification of Potential Disease-Associated Targets
To identify novel targets associated with the disease of interest, microarray data were accessed and downloaded from the NCBI GEO database (GSE27011, http://www.ncbi.nlm.nih.gov/geo/, accessed on 10 November 2024). The dataset was analyzed using Bioconductor packages (http://www.bioconductor.org/, accessed on 10 November 2024) for differential expression and clustering analyses. Gene expression profiles obtained from the GSE27011 dataset were systematically evaluated using microarray technology to identify DEGs exhibiting statistically significant alterations across comparative experimental conditions [20]. Differential expression analysis was performed using the limma package in R (R 3.6.1 software), with genes meeting the criteria of fold change ≥ 2 and adjusted p-value < 0.05 being classified as differentially expressed. The results were visualized through volcano plots to highlight upregulated and downregulated genes with statistical significance. To explore gene expression patterns across samples in GSE27011, clustering analysis was conducted. Hierarchical clustering and principal component analysis (PCA) were performed using the stats and pcaMethods packages in R, enabling the identification of unique gene expression clusters linked to various experimental conditions or disease states.
2.2 Isolation and Induction of PBMCs and DCs
Between March and May 2023, twenty participants from our institution were prospectively enrolled, including ten pediatric patients diagnosed with acute-phase asthma and ten age-matched healthy controls undergoing routine health assessments. The study population demonstrated a balanced gender distribution, with an age range of 5 to 13 years in both groups. The asthma cohort had an average age of 8.11 ± 2.33 years, while the healthy control cohort had an average age of 8.67 ± 1.36 years, with no significant difference in baseline characteristics. Inclusion criteria were: (1) diagnosis of pediatric asthma based on clinical guidelines; (2) age between 4 and 14 years; and (3) informed consent obtained from the patient’s guardian. Exclusion criteria included: (1) presence of other respiratory diseases or developmental abnormalities; (2) severe systemic or debilitating conditions; and (3) a recent history of respiratory tract infection or corticosteroid use within the last four weeks. Human peripheral blood was collected with informed consent and approval from the Ethics Committees of Shanghai East Hospital (Approval number: 2024YS-139).
Venous blood samples were aseptically collected from both healthy volunteers and patients with asthma using heparin-anticoagulated vacuum tubes. Following collection, the samples were processed within 4 h. Whole blood was aliquoted into 15 mL sterile centrifuge tubes and gently mixed with an equal volume of phosphate-buffered saline (PBS) using pipetting to maintain cellular integrity. Peripheral blood mononuclear cells (PBMCs) were isolated using a lymphocyte separation medium (Solarbio, Beijing, China, P8610) and transferred into new 15 mL sterile centrifuge tubes. After centrifugation at 2000 rpm for 20 min (Luxiang, TDZ4B-WS, Shanghai, China), the PBMC layer was carefully collected and washed twice with sterile PBS by centrifugation at 1500 rpm for 10 min. The cell pellet was treated with red blood cell lysis buffer (Solarbio, Beijing, China, R1010), followed by centrifugation, discarding the supernatant, and resuspending the cells in sterile PBS. PBMCs were counted under a microscope (OLYMPUS, Tokyo, Japan, IX71), aliquoted into sterile EP tubes, centrifuged, and the supernatant removed. The cell pellet was resuspended and cultured in dishes with granulocyte-macrophage colony-stimulating factor (GM-CSF, MCE, HY-P7361, USA), interleukin-4 (IL-4, MCE, HY-P70445, USA), and tumor necrosis factor-alpha (TNF-α, MCE, HY-P1860, USA) to induce differentiation into DCs for subsequent experiments. PBMCs induced to differentiate into DCs from healthy controls (A) and asthma patients (B) were processed as follows: cells were collected and centrifuged at 1500 rpm for 5 min to remove the supernatant, followed by resuspension in PBS. Cells were washed twice with PBS at 1500 rpm for 5 min each, then resuspended in PBS and incubated with specific antibodies at 37°C for 30 min. After centrifugation at 1500 rpm for 5 min, the cells were washed twice with PBS and resuspended in PBS for flow cytometry analysis using a BD FACSVerse system (USA). The markers used for DC phenotyping were FITC-CD11c (Abcam Ab22540, USA) [21]. For lymphocyte phenotyping, the markers used were CD45+CD3+ [22].
The day before transfection, approximately 300,000 cells per well were seeded into a six-well plate to reach 70%–80% confluence the following day. Each well of the six-well plate was replaced with 2 mL of fresh culture medium. To transfect the cells in each well of the six-well plate, 125 μL of basic culture medium without antibiotics and serum was added. Then, 100 pmol of siRNA (mus-si NC: UUCUCCGAACGUGUCACGUTT; mus-Rfx5-si1: CGGACAACGACAAGCUGUACCTT GGUACAGCUUGUCGUUGUCCGTT; mus-Rfx5-si2: GGUCAUUAUGUAAGUCCAAGATT UCUUGGACUUACAUAAUGACCTT; mus-Rfx5-si3: GGAUACUACAGUAAGUGAAGCTT GCUUCACUUACUGUAGUAUCCTT) was added. The mixture was then gently mixed by pipetting. Next, 4 μL of Lipo8000TM transfection reagent (Beyotime, Shanghai, China, C0533) was added, and the mixture was gently mixed. A 125 μL mixture of Lipo8000TM transfection reagent and siRNA was uniformly added to each well of the six-well plate and gently mixed. After approximately 2 days of continuous culture, the downregulation of target genes by siRNA was assessed via RT-PCR.
2.3 Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, 15596018, Waltham, MA, USA) according to the manufacturer’s protocol. Complementary DNA was synthesized with the TRUEscript RT MasterMix (Aidlab, PC5801, Beijing, China). qRT-PCR was conducted using SYBR Green qPCR Mix (Aidlab, PC3302, Beijing, China) on an ABI 7500 Real-Time PCR System (Thermo Fisher Scientific, ABI7500, Waltham, MA, USA). The following primers were sourced from Genecefe Biotechnology Co., Ltd.: RFX5(Human)-RT-F (5′-GTGGCATAAGGAGGAAGAC-3′), RFX5(Human)-RT-R (5′-CAACGATGGAACTGAAGGA-3′) for RFX5 amplification; HDAC2(Human)-RT-F (5′-AACCGACAACAGACTGATAT-3′), HDAC2(Human)-RT-R (5′-AAGGATGGCAAGCACAAT-3′) for HDAC2 amplification; GAPDH(Human)-RT-F (5′-GGAGCGAGATCCCTCCAAAAT-3′), GAPDH(Human)-RT-R (5′-GGCTGTTGTCATACTTCTCATGG-3′) for GAPDH amplification. Additionally, the following primers were used: Rfx5(Mouse)-RT-F (GCAGCAGCATCTCATCTC), Rfx5(Mouse)-RT-R (ACGACATTCTTGGACTTACA) for Rfx5; HDAC2(Mouse)-RT-F (CTGAGGATGAAGGTGAAGG), HDAC2(Mouse)-RT-R (GTTGCTGAGTTGTTCTGAC) for HDAC2; Ciita(Mouse)-RT-F (CCTACATCTCTACCACCTCT), Ciita(Mouse)-RT-R (GTCTCTTCATCCAGTTCCAT) for Ciita; Mhcii(Mouse)-RT-F (GCCTACGGTGACTGTGTA), Mhcii(Mouse)-RT-R (TCTCCTCCTTGCCATTCC) for Mhcii; Gapdh(Mouse)-RT-F (GGTGAAGGTCGGTGTGAACG), Gapdh(Mouse)-RT-R (CTCGCTCCTGGAAGATGGTG) for Gapdh amplification. mRNA expression levels were normalized to GAPDH or Gapdh, with relative expression calculated using the 2−ΔΔCT method.
For interleukin 6 (IL-6, mlbio, mlC30294-1, Shanghai, China) detection, samples were prepared by adding diluent to blank wells, with standard samples added to other wells. The reaction plate was mixed and incubated at 37°C for 50 min. After washing three times with 1 × wash solution per well, a biotinylated antibody diluent was added to blank wells, and a 1 × biotinylated antibody working solution was added to others. Incubation occurred at 37°C for 40 min. Following another wash, 100 μL of SABC complex working solution was added to each well and incubated at 37°C for 30 min. After washing, TMB chromogenic substrate was added and incubated at 37°C in the dark for 25 min. The reaction was terminated with a stop solution, and absorbance was measured at 450 nm within 30 min using a microplate reader (Thermo Fisher Scientific, Multiskan FC, Waltham, MA, USA). Standard curves were generated using known concentrations. Similar procedures were followed for TNF-α (MlBio, mlC30498, Shanghai, China), IL-1β (MlBio, mlC30270, Shanghai, China), IL-4 (MlBio, mlC30292, Shanghai, China), IL-10 (MlBio, mlC30246, Shanghai, China), and IL-18 (MlBio, mlC30261, Shanghai, China), with appropriate adjustments for standard curve concentrations and specific biotinylated antibodies for each cytokine.
2.5 Paraffin Sectioning and Hematoxylin-Eosin Staining
Tissue dehydration was achieved through a graded ethanol series (SinoPharm, 10009218, Beijing, China): 75% ethanol for 4 h, 85% for 2 h, 90% for 1.5 h, 95% for 1 h, and absolute ethanol I and II for 0.5 h each. Tissues were then cleared using xylene (SinoPharm, 10023418, Beijing, China) (1:1 with absolute ethanol), followed by two additional treatments with xylene I and II for 10 min each. Paraffin infiltration (SinoPharm, 69019361, Beijing, China) was performed in three steps at 60°C for 1 h each. After embedding, sections were cut at 5 μm thickness, floated in a 40°C water bath, and transferred to glass slides. Following dewaxing, sections were stained with Mayer’s hematoxylin (Sigma, H9627) for 5 min, rinsed, counterstained with 1% eosin (SinoPharm, 71014544, Beijing, China) for 5 min, dehydrated, cleared, and mounted with neutral resin (SinoPharm, 10004160, Beijing, China) for microscopic examination.
2.6 Immunohistochemical Staining
For antigen retrieval, dewaxed paraffin sections (SinoPharm, 69019361, Beijing, China) were treated with xylene I, II, and III, followed by dehydration in graded alcohols (95%, 90%, 80%, 70%) and distilled water. Antigen retrieval was performed by microwaving in citrate buffer (Beyotime, P0086, Shanghai, China) for 15 min. 1% Triton X-100 is added to the slice to destroy the cell membrane and allow the membrane to penetrate so that the antibody can enter the cell. Endogenous peroxidase activity was blocked using 3% hydrogen peroxide for 15 min, followed by a 30-min incubation with normal goat serum (Beyotime, C0265, Shanghai, China) at room temperature. Primary antibodies, including HDAC2 (1/250, Thermo Fisher Scientific, 51-51000, Waltham, MA, USA) and RFX5 (1/1000, Immunoway, YT7052, Shenzhen, China), were applied and incubated overnight at 4°C. After washing with PBS, sections were incubated with a Rabbit kit (ZSGB-Bio, PV-6001, Beijing, China) or Mouse kit (ZSGB-Bio, PV-6002, Beijing, China) for 30 min at 37°C. DAB substrate (ZSGB-Bio, ZLI-9018, Beijing, China) was used for visualization, and sections were counterstained with Mayer’s hematoxylin (Sigma, H9627, St. Louis, MO, USA) for 2 min before mounting. Immunohistochemical analysis was performed using average optical density.
For protein extraction, samples were homogenized with RIPA Lysis Buffer (Beyotime, Shanghai, China, P0013B) supplemented with protease inhibitor PMSF (RUIBIO, Wuxi, China, BP2655) and phosphatase inhibitors (Beyotime, Shanghai, China, S1873) at 4°C for 30 min. After centrifugation, supernatants were collected, and protein concentrations were determined using a bicinchoninic acid assay (Biosharp, BL521A, Hefei, China). Proteins (20 μg) were separated by SDS-PAGE (Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis) and transferred onto nitrocellulose membranes (Millipore, Darmstadt, Germany, IPVH00010). The membrane was rinsed with TBST for 3 times for 5 min each time, and then slowly shook with 5% skim milk powder at 37°C for 2 h to block the non-specific binding site. Primary antibodies for HDAC2 (Thermo Fisher Scientific, Waltham, MA, USA, 51-5100, 1/250), RFX5 (Immunoway, Plano, TX, USA, YT7052, 1/1000), CIITA (Class II Transactivators, abnova, Taiwan, PAB19313, 1/1000), MHC II (Major Histocompatibility Complex Class II, Abcam, Waltham, MA, USA, ab139365, 1/1000), and GAPDH (Proteintech, Wuhan, China, 60004-1-Ig, 1/5000) were applied according to the manufacturer’s instructions. Secondary antibodies, including horseradish peroxidase-conjugated goat anti-rabbit IgG (Beyotime, Shanghai, China, A0208, 1/1000), goat anti-mouse IgG (Beyotime, Shanghai, China, A0216, 1/1000), and goat anti-rat IgG (Beyotime, Shanghai, China, A0192, 1/1000), were used. Protein bands were visualized with a chemiluminescence imager (CLINX, Shanghai, China, ChemiScope 5300 Pro) and enhanced chemiluminescence (7sea Biotech, Shanghai, China, E03-500). Quantitative analysis was conducted using ImageJ software 2.0 (National Institutes of Health, Bethesda, MA, USA).
2.8 Animal Modeling and Treatment
Male C57 mice (7–8 weeks, Jiangsu Huachuang Xinnuo, Taizhou, China) were housed under a 12/12-h light-dark cycle with unrestricted access to food and water at room temperature (22°C–24°C). A total of 32 mice were allocated into four groups: Control, ovalbumin (OVA) model, OVA model + si-NC (OVA + si-NC), and OVA model + RFX5 knockdown (OVA + si-RFX5). On Day 0, mice were intraperitoneally injected with 10 μg OVA (Solarbio, A8041, Beijing, China) and 1 mg Al(OH)3 (Sigma, 239186, St. Louis, MO, USA). Mice in the OVA + si-NC and OVA + si-RFX5 groups received tail vein injections of RFX5 siRNA or NC control siRNA (10 μg/mouse, thrice weekly), mixed with in vivo-jetPEI (Polyplus Transfection, 101000030, Illkirch, France). From Day 21, mice were nebulized with 1% OVA for 30 min per day over one week [23,24]. The OVA + si-RFX5 + SAHA group was injected four times with 300 mg/kg SAHA (dissolved in 10% DMSO in 0.1 mL saline). All animal procedures were approved by the Ethics Committee of Experimental Animal Management at Tongji University (Approval number: TJBB08224101).
2.9 Airway Resistance Measurement
Airway resistance was measured using a whole-body plethysmography system (TOW-INT Tech, Shanghai, China, WBP-8M) across the four groups: Control, OVA model, OVA model + si-NC (OVA + si-NC), and OVA model + RFX5 knockdown (OVA + si-RFX5). The respiratory rate was set at 140–150 breaths/min, with a tidal volume of 0.13–0.15 mL. Mice were exposed to saline (baseline) and acetyl-β-methylcholine (Beijing Vokai Biotechnology Co., Ltd., Beijing, China, O135659-5g) at concentrations of 3.125, 6.25, 12.5, 25, and 50 mg/mL for 30 s each. Penh values were recorded for 3 min, with 4-min recovery intervals, and expressed as percentages of baseline (Penh/NS%) to assess airway reactivity.
For antibody immobilization, 20 μL of AminoLink Plus Coupling Resin (Thermo Fisher Scientific, 26149, Waltham, MA, USA) was washed with 1× coupling buffer, then incubated with 10 μg of antibody in 200 μL buffer containing 3 μL sodium cyanoborohydride for 90–120 min. After incubation, the resin was washed, quenched, and re-washed. For IP, the resin was washed with IP wash buffer and incubated with protein lysate for 2 h at room temperature. The resin was then washed with lysis/wash buffer, and bound proteins were eluted with elution buffer. The eluates were collected for further analysis.
2.11 Chromatin Isolation and Immunoprecipitation (ChIP)
Cells (2.5 × 107) were cross-linked with 1% formaldehyde (Beyotime, P0099, Shanghai, China), quenched with glycine (Bioforxx, 1275GR500), and washed with ice-cold PBS containing 1 mM PMSF (RUIBIO, BP2655, Guangzhou, China). The cell pellets were lysed using SDS lysis buffer (P2078-11), and chromatin was sonicated into fragments ranging from 200 to 1000 bp (Shunma Tech, SM-150A, Nanjing, China). The lysate was diluted with ChIP dilution buffer (Beyotime, P2078, Shanghai, China), pre-cleared using the Universal UNlQ-10 Column DNA Purification Kit (Sangon Biotech, B511281, Shanghai, China), and incubated overnight with 10 µg of HDAC2 antibody (Thermo Fisher Scientific, 51-5100, Waltham, MA, USA). Complexes were precipitated, washed sequentially with low salt, high salt, LiCl, and TE buffers, and then eluted with an elution buffer. Cross-links were reversed at 65°C for 4 h, and DNA was purified for subsequent PCR analysis. The detection primers for ChIP were ChIP1-F (GGGTACTGGTTAGTTCATATTG) and ChIP1-R (AAGAGGACAAGCCGACTT).
All experiments were performed at least in triplicate. Data analysis was conducted using GraphPad Prism 8.0 (GraphPad Software, Boston, MA, USA), with results presented as mean ± SD differences between groups were analyzed using ANOVA followed by Tukey’s post hoc test. Statistical significance was set at p < 0.05.
3.1 Transcriptome Sequencing Analysis of the Association between RFX Family Genes and Childhood Asthma
This study explored the relationship between RFX family gene expression patterns and childhood asthma by analyzing transcriptomic data from 53 pediatric subjects across three cohorts: 17 severe asthmatics (SA group), 19 mild asthmatics (MA group), and 17 healthy controls (HC group). Utilizing advanced bioinformatics tools and robust statistical analyses, differential gene expression profiles were systematically examined across the groups. The analysis revealed statistically significant (p < 0.05) alterations in transcriptional patterns in SA patients compared to both HC and MA groups, with these findings being consistently validated through multiple analytical approaches and statistical methods. Volcano plot (Fig. 1A) and clustering analysis (Fig. 1B) identified 81 genes with significant differences. Notably, CD4 signaling pathway molecules and RFX showed pronounced differential expression, particularly in antigen presentation pathways. These results highlight the potential role of RFX family genes, especially in CD4 signaling, in childhood asthma pathogenesis. Further investigation of these molecular targets could offer new insights into therapeutic strategies for managing pediatric asthma.
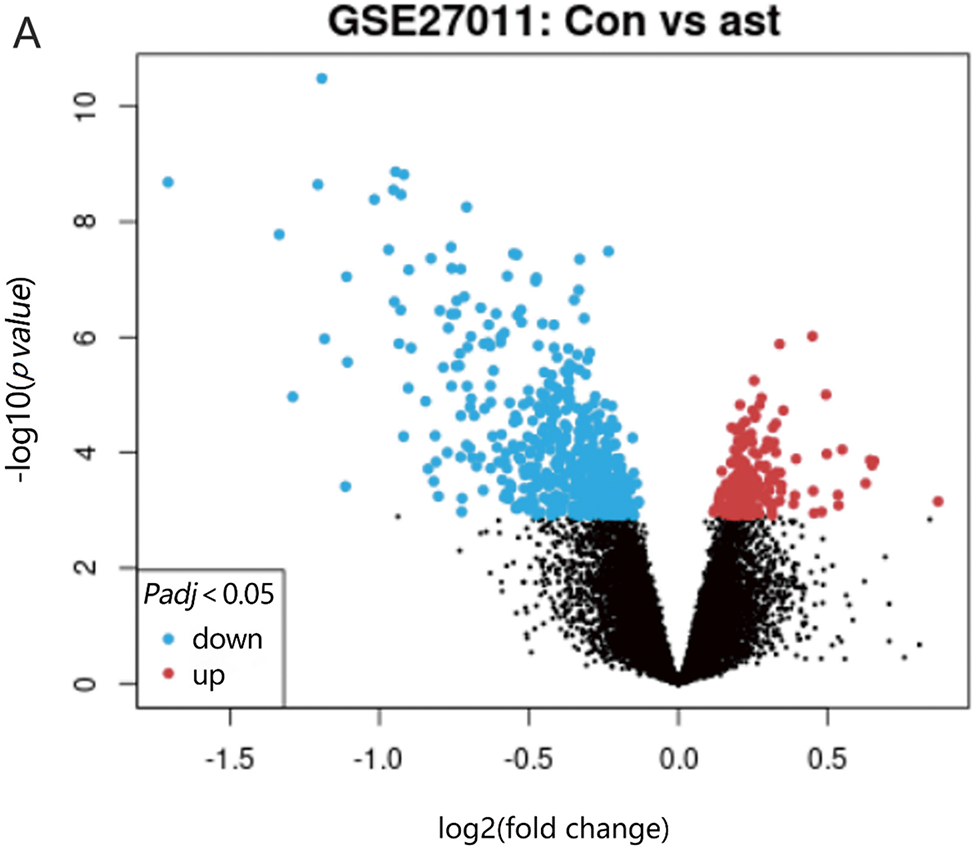
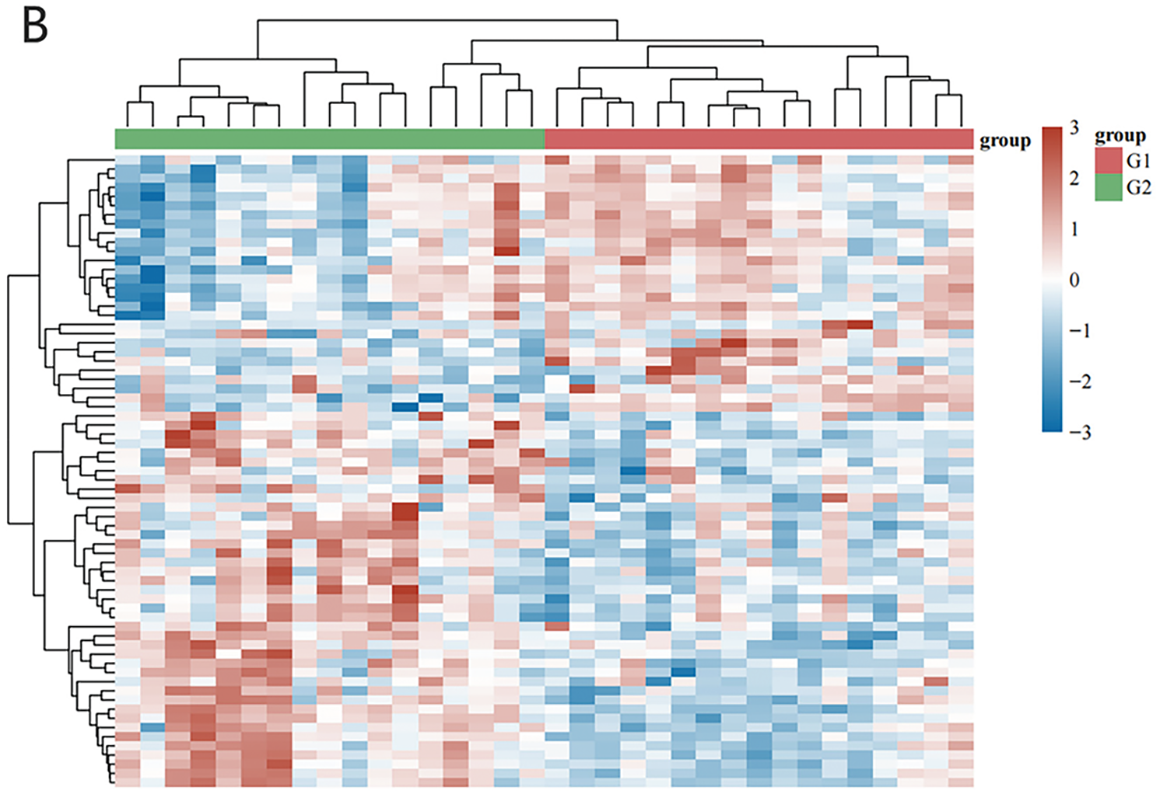
Figure 1: Bioinformatics analysis was conducted to search for targeted gene RFX5. (A) Volcano plot analysis of differentially expressed genes in GSE27011 volcano plot depicting the differential gene expression analysis from the GSE27011 microarray dataset. The x-axis represents the log2 fold change, while the y-axis displays the negative log10 adjusted p-value (FDR). Each gene is depicted as a point, colored red for upregulated and blue for downregulated, with a horizontal dashed line marking the statistical significance threshold (adjusted p-value < 0.05). (B) Cluster analysis of the GSE27011 dataset reveals gene expression patterns across samples. The heatmap depicts hierarchical clustering of samples according to gene expression profile similarities. Genes are represented by rows, while columns correspond to individual samples. Colors represent expression levels compared to the mean, with red indicating high expression and blue indicating low expression
3.2 PBMCs-Induced DCs Expression of RFX5 Gene in Asthmatic Patients Correlates Clearly with Asthma and Asthma Severity
The downregulation of histone deacetylase 2 (HDAC2) in asthmatic patients has been mechanistically linked to enhanced differentiation of CD4+ T lymphocytes into a Th17 phenotype, a key pathway in asthma pathogenesis [8]. Additionally, bioinformatics analysis (Fig. 1) emphasizes the potential involvement of RFX5 in the CD4+ T cell signaling pathway. Therefore, this study investigated the expression of RFX5 and HDAC2 genes in DCs induced from PBMCs of asthma patients compared to healthy controls. Results demonstrated a significant increase in RFX5 expression and a marked decrease in HDAC2 expression in asthma patients, confirmed by qRT-PCR analysis (Fig. 2B). These molecular changes suggest dysregulation in key epigenetic regulatory mechanisms, particularly those governing transcriptional control and chromatin remodeling processes essential for immune homeostasis. The identified molecular signatures correlated strongly with clinical asthma severity indices, indicating their potential as reliable biomarkers for disease progression and promising targets for precision therapeutic interventions. Flow cytometry analysis (Fig. 2A) further validated the successful induction of DCs from PBMCs in both healthy and asthma groups, showing high positivity rates for DC surface markers, affirming the experimental rigor. The observed upregulation of pro-inflammatory cytokines (IL-6, TNF-α, and IL-1β) coupled with the downregulation of anti-inflammatory cytokines (IL-4, IL-10, and IL-18) (Fig. 2C) underscores the inflammatory milieu characteristic of asthma pathogenesis. The dysregulated expression of RFX5 and HDAC2 appears to play a pivotal role in disrupting immune homeostasis and sustaining chronic inflammatory processes in asthma. These results highlight the need for detailed mechanistic studies to clarify their pathophysiological contributions and assess their therapeutic potential through targeted interventions.
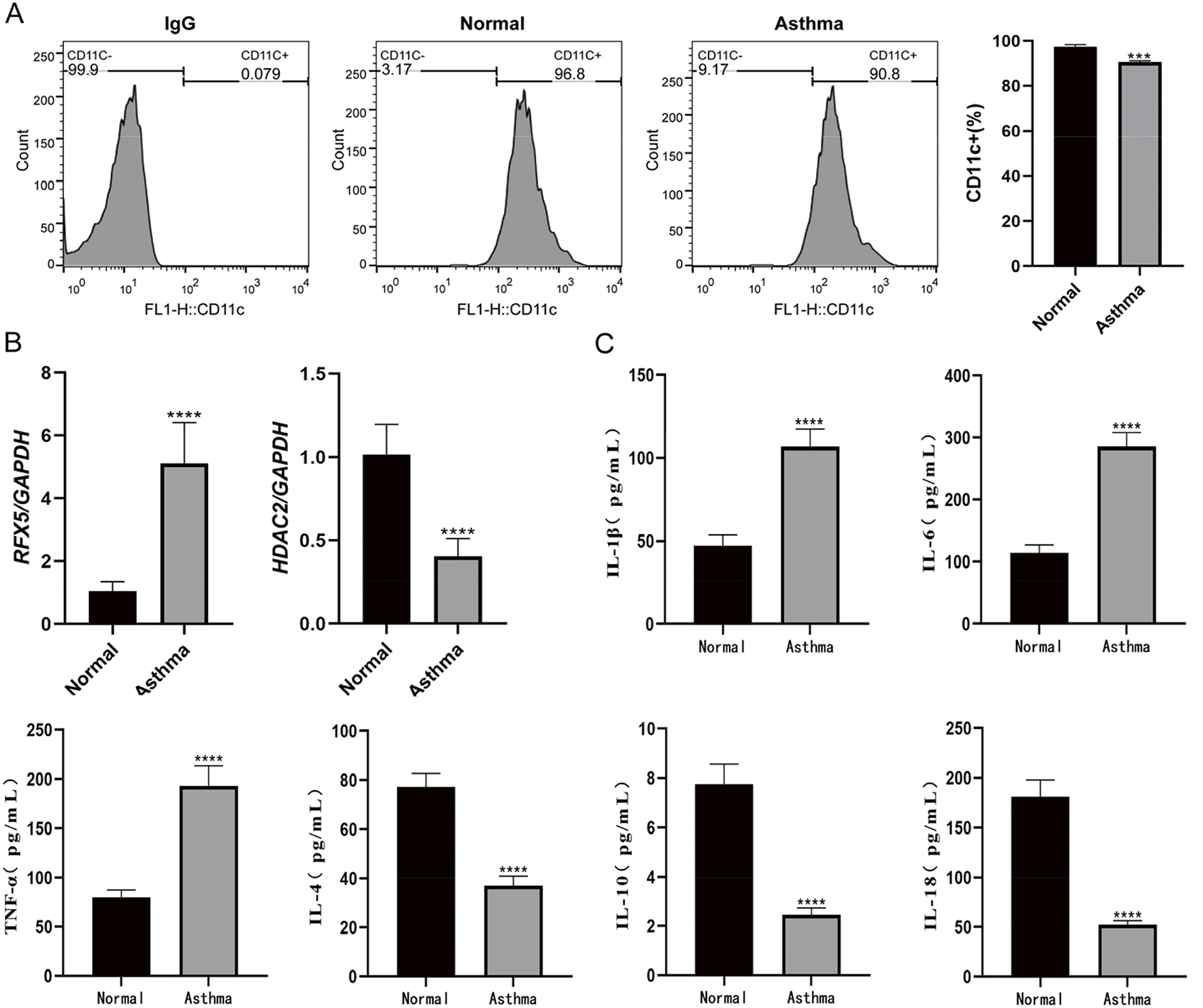
Figure 2: Peripheral blood mononuclear cells-induced DCs expression analysis. (A) Flow cytometry analysis of peripheral blood mononuclear cells-induced dendritic cells from healthy controls and asthma patients. The positivity rate for surface molecular markers on DCs exceeded 90% in both groups, indicating successful DC induction. (B) qRT-PCR analysis of RFX5 and HDAC2 gene expression in peripheral blood mononuclear cells from asthma patients compared to normal controls. RFX5 expression was significantly upregulated, while HDAC2 expression was significantly downregulated in the asthma group. (C) ELISA kit used to detect the secretion of inflammatory factors in the normal group and the asthma group (*vs. Normal Group: ***p < 0.001, ****p < 0.0001)
3.3 RFX5 Promotes Inflammation and Airway Remodeling while HDAC2 Inhibits Inflammation
The dysregulated expression of RFX5 and HDAC2 has been mechanistically implicated in the modulation of inflammatory pathways and the progression of airway remodeling in asthma pathogenesis. Our results consistently demonstrate the upregulation of RFX5 and downregulation of HDAC2 across multiple assays—histological (Fig. 3A), immunohistochemical (Fig. 3B), qRT-PCR (Fig. 3C), and Western blotting analyses (Fig. 3D)—in an OVA-induced asthma mouse model. These findings establish RFX5 as a key transcriptional regulator mediating both inflammatory responses and airway remodeling mechanisms while identifying HDAC2 as a compensatory epigenetic modulator that may counterbalance these pathological changes through its regulatory functions. Therapeutically, modulating these molecular targets could provide promising intervention strategies for mitigating asthma-related pulmonary pathology and controlling chronic inflammation. Our results highlight the complex molecular network underlying asthma pathogenesis, particularly emphasizing the dysregulation of transcriptional regulators (RFX5) and epigenetic modulators (HDAC2). Comprehensive mechanistic studies are essential to fully elucidate the roles of these molecular regulators in both the initiation and progression of asthma, paving the way for targeted therapeutic strategies that could not only alleviate disease severity but also improve long-term clinical outcomes in pediatric asthma patients.
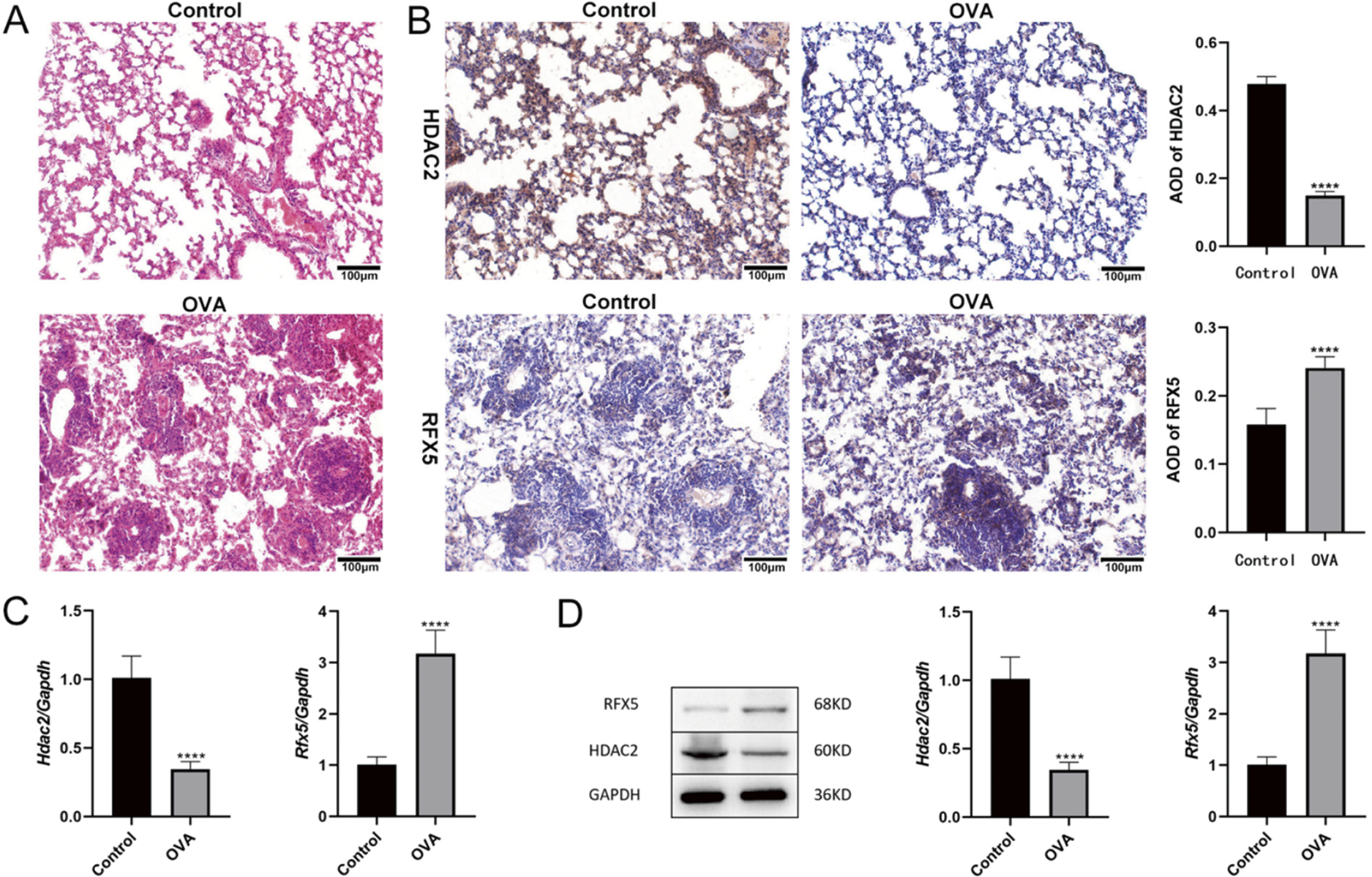
Figure 3: Expression of RFX5 and HDAC2 in an asthma mouse model. (A) Histological examination of lung tissues from OVA-induced asthma model mice. Hematoxylin and eosin (HE) staining (200× magnification; scale bar, 100 µm) shows disrupted alveolar structure, widened lung interstitium, bronchial wall damage, alveolar and interstitial congestion, and increased inflammatory factors compared to the Control group. (B) Immunohistochemical analysis of lung tissues (200× magnification; scale bar, 100 µm). Compared to the Control group, OVA-exposed mice exhibit significantly elevated expression of RFX5 and decreased expression of HDAC2 in lung tissues. (C) qRT-PCR analysis of lung tissues. Quantitative qRT-PCR results demonstrate significantly increased expression of Rfx5 and decreased expression of HDAC2 in lung tissues of the OVA-induced asthma model mice compared to controls. (D) Western blot analysis of lung tissues. Protein levels of RFX5 are significantly elevated, while HDAC2 levels are markedly decreased in lung tissues from OVA-exposed mice compared to controls (*vs. Normal Group: ****p < 0.0001)
3.4 The Knockdown of RFX5 Reduces Asthma Symptoms
In investigating the role of RFX5 in asthma using a murine model, qRT-PCR results (Fig. 4A) revealed a significant upregulation of Rfx5 and a marked downregulation of HDAC2 in the OVA-induced asthma group. These results suggest that RFX5 plays a pivotal role in asthma pathogenesis by modulating HDAC2 expression, a key regulator of inflammation and immune responses. Airway resistance measurements (Fig. 4B) demonstrated significantly heightened airway hyperresponsiveness (AHR) in the OVA group compared to the Control group, a hallmark of asthma. Therapeutic administration of siRFX5 led to a significant attenuation of AHR, indicating that targeted suppression of RFX5 could serve as an effective strategy for mitigating asthma-related pathological manifestations. Flow cytometry analysis (Fig. 4C) revealed a decrease in lymphocyte counts in the OVA model group, indicative of an inflammatory response. The reversal of these changes upon siRFX5 transfection suggests that RFX5 modulates immune cell distribution and potentially inflammatory responses in asthma. ELISA results (Fig. 4D) further corroborated RFX5’s pro-inflammatory role, showing increased levels of pro-inflammatory cytokines (IL-6, TNF-α, IL-1β) and decreased levels of anti-inflammatory cytokines (IL-4, IL-10, IL-18) in the OVA group. Transfection with siRFX5 normalized these cytokine levels, indicating a shift toward a less inflammatory state. Histological analysis (Fig. 4E) revealed significant pathological alterations in the lung tissue of the OVA group, including disrupted alveolar structures and increased inflammatory infiltration. These changes were alleviated by siRFX5 treatment, suggesting a protective effect of RFX5 knockdown on lung tissue integrity.
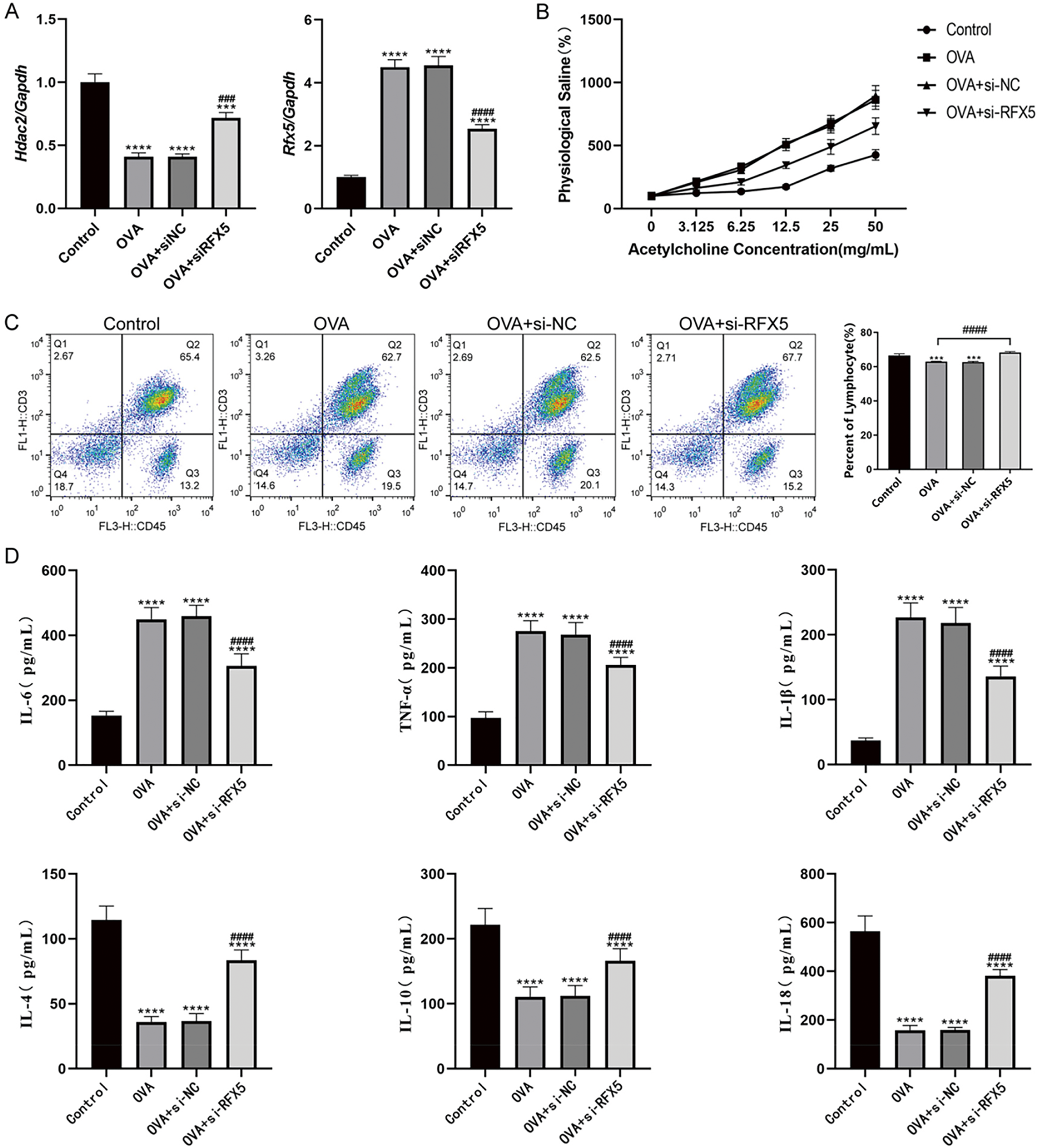
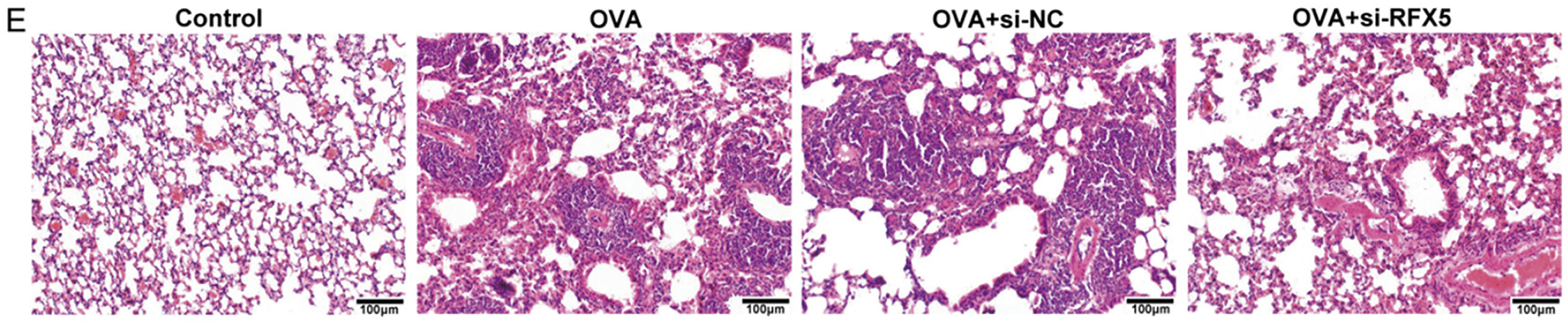
Figure 4: Effects of RFX5 on asthma in animal models. (A) qRT-PCR results demonstrate significant upregulation of Rfx5 and downregulation of HDAC2 in the OVA group compared to the Control group. Transfection with siRFX5 significantly decreased Rfx5 expression and increased HDAC2 expression. (B) Airway resistance measurements show significantly increased airway reactivity in the OVA group compared to the Control group. This heightened reactivity was significantly reduced following siRFX5 administration. (C) Flow cytometry analysis reveals a decrease in lymphocyte count in the OVA group compared to the Control group. Transfection with siRFX5 reversed these changes. (D) ELISA results indicate significantly elevated levels of IL-6, TNF-α, and IL-1β and reduced levels of IL-4, IL-10, and IL-18 in the OVA group compared to the Control group. Transfection with siRFX5 significantly decreased IL-6, TNF-α, and IL-1β levels and increased IL-4, IL-10, and IL-18 levels. (E) HE staining results (200× magnification; scale bar, 100 µm) show disrupted alveolar structure, widened alveolar septa, damaged bronchial walls, and increased infiltration of inflammatory cells in the OVA group compared to the Control group. Administration of siRFX5 mitigated these pathological changes (*vs. Normal Group: ***p < 0.001, ****p < 0.0001); (#vs. OVA Group: ###p < 0.001, ####p < 0.0001)
3.5 Effects of RFX5 on Inflammatory Cytokine Secretion by DC Cells and Its Molecular Mechanism
This study rigorously investigated the regulatory function of RFX5 in DC-mediated inflammatory cytokine production through a series of in vitro assays, while dissecting the associated molecular signaling pathways and transcriptional regulatory mechanisms. Flow cytometry results (Fig. 5A) demonstrated a marked reduction in CD80, CD86, and MHCII levels in the OVA-induced asthma model compared to wild-type controls. However, siRFX5 transfection reversed these changes, highlighting the critical role of RFX5 in modulating DC activation. qRT-PCR analysis (Fig. 5B) revealed a significant decrease in Ciita and Mhcii expression levels in the OVA group, suggesting a potential impairment in antigen presentation and adaptive immune response. Transfection with siRFX5 led to a significant increase in the expression of Ciita and Mhcii, indicating that RFX5 knockdown enhances the antigen presentation capabilities of DCs. Western blotting analysis (Fig. 5C) supported the qRT-PCR findings, showing a reduction in CIITA and MHCII protein levels in the OVA group, which were significantly upregulated following siRFX5 transfection. These results demonstrate that RFX5 functions as a key transcriptional regulator of immune modulators, controlling the expression of critical molecules essential for immune homeostasis and inflammatory response regulation.
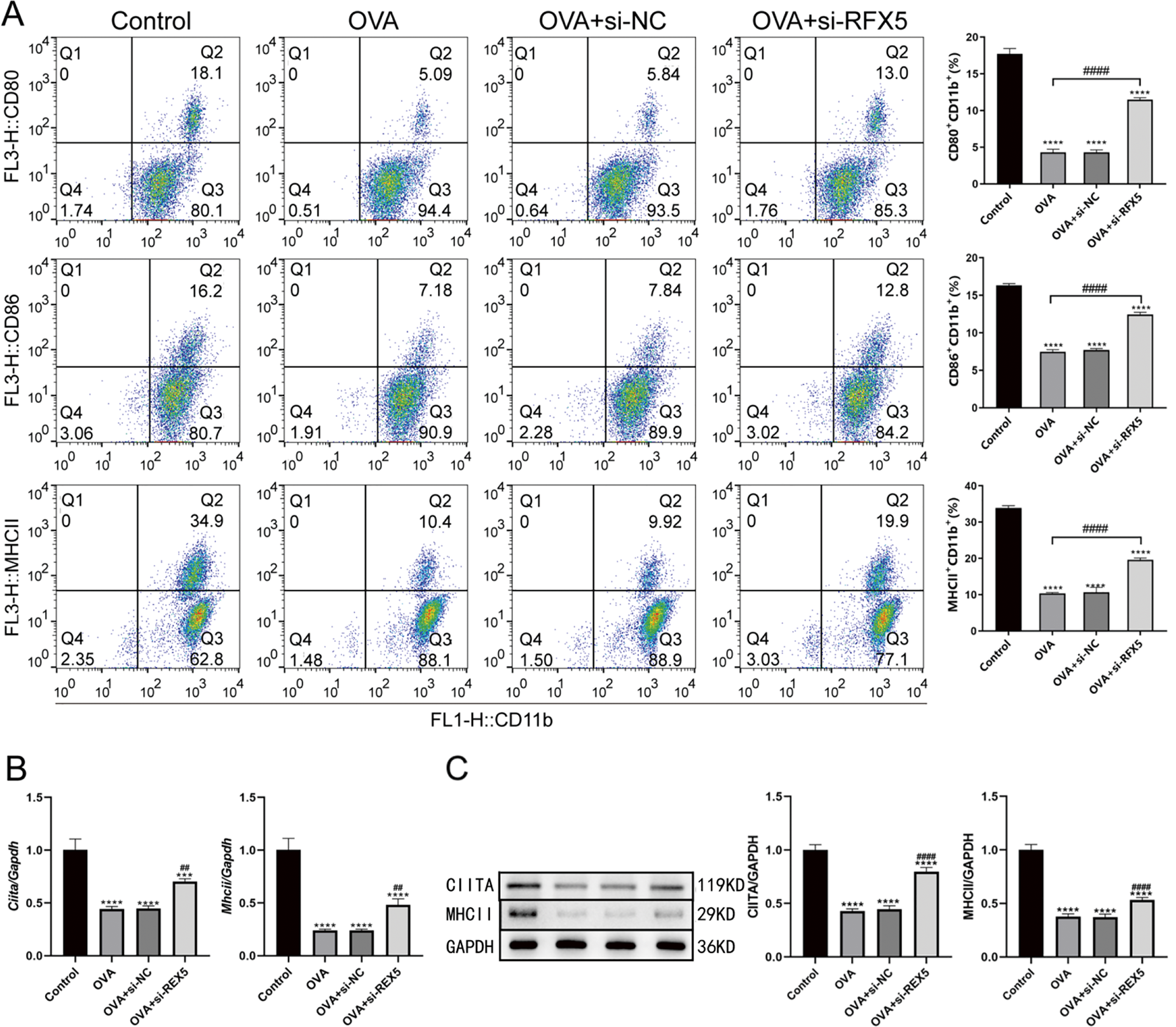
Figure 5: Effect of RFX5 gene knockout on the composition of alveolar lavage fluid in OVA model asthma. (A) Flow cytometry analysis shows that in the OVA model group, the levels of CD80, CD86, and MHCII are decreased compared to wild-type mice. This decrease is reversed upon si-RFX5 transfection. (B) qRT-PCR results indicate that CIITA and MHCII expression is significantly reduced in the OVA group compared to the Control group. Transfection with siRFX5 results in a significant increase in CIITA and MHCII expression. (C) Western blot analysis demonstrates that CIITA and MHCII expression are significantly decreased in the OVA group compared to the Control group. This decrease is reversed upon siRFX5 transfection, showing increased CIITA and MHCII expression (*vs. Control Group: ***p < 0.001, ****p < 0.0001); (#vs. OVA Group: ##p < 0.01, ####p < 0.0001)
3.6 Relationship between RFX5 and HDAC2 in Wild-Type Mouse DC Cells
The study further delved into the molecular interactions between RFX5 and HDAC2 in DCs isolated from wild-type murine models, emphasizing their protein-protein interactions and regulatory network dynamics in immune modulation. Co-IP experiments (Fig. 6A) revealed a physical interaction between HDAC2 and RFX5, suggesting a potential regulatory mechanism at the protein-protein interaction level. ChIP assays (Fig. 6B) provided additional insights, showing that HDAC2 binds to the promoter region of the Rfx5 gene, as evidenced by the successful amplification of a 234 bp fragment. This suggests that HDAC2 may directly regulate Rfx5 expression through promoter binding. qRT-PCR analysis (Fig. 6C) confirmed the effectiveness of siRFX5 transfection, revealing a significant reduction in Rfx5 expression. Additionally, qRT-PCR analysis (Fig. 6D) demonstrated that the OVA-induced asthma model group exhibited increased Rfx5 expression and decreased Hdac2 expression relative to the Control group. siRFX5 transfection reversed these effects, suggesting a negative regulatory relationship between Rfx5 and HDAC2. Western blotting analysis (Fig. 6E) corroborated these results, showing a significant reduction in HDAC2 protein levels in the OVA group compared to the Control group, which was significantly reversed following siRFX5 transfection, leading to a marked upregulation of HDAC2 expression at both the transcriptional and protein levels.
Figure 6: Relationship between RFX5 and HDAC2 in wild-type mouse DC cells. (A) Co-immunoprecipitation using HDAC2 antibody demonstrates the interaction between HDAC2 and RFX5 proteins in DC cells. (B) Chromatin immunoprecipitation (ChIP) using HDAC2 antibody, followed by PCR amplification of the Rfx5 promoter region, shows successful amplification of a 234 bp fragment, indicating binding of HDAC2 to the Rfx5 promoter. (C) qRT-PCR results show that Rfx5 expression is significantly reduced after siRFX5 transfection compared to the Control group. (D) qRT-PCR results indicate that in the OVA group, Rfx5 expression is significantly increased while HDAC2 expression is significantly decreased compared to the Control group. Transfection with siRFX5 leads to a significant decrease in Rfx5 expression and an increase in HDAC2 expression. (E) Western blot results show that HDAC2 expression is significantly decreased in the OVA group compared to the Control group. HDAC2 expression is significantly increased after siRFX5 transfection. (*vs. Control Group: **p < 0.01, ****p < 0.0001); (#vs. OVA Group: ###p < 0.001, ####p < 0.0001; Lane M in the left picture of B represents DL500 DNA Marker; Lane 1 represents negative control and lane 2 represents IgG immunoprecipitated DNA; Lane 3 represents IgG immunoprecipitated DNA; Lane 4 represents PCR amplification of Input products; Lane M on the right of figure (B) represents DL500 DNA Marker; Lane 1 represents negative control and lane 2 represents IgG immunoprecipitated DNA; Lane 3 represents PCR amplification of input products, and lane 4 represents PCR amplification of CHIP products)
3.7 The Knockdown of RFX5 Reduces Asthma Symptoms
qRT-PCR analysis (Fig. 7A) revealed a significant increase in HDAC2 expression in the OVA + siRFX5 group compared to the OVA + siNC group, indicating that RFX5 knockdown leads to HDAC2 upregulation. In contrast, treatment with SAHA, an HDAC inhibitor, significantly reduced HDAC2 expression, suggesting tight regulation of HDAC2 activity, which can be modulated pharmacologically. Western blotting analysis (Fig. 7B) confirmed these observations, showing elevated HDAC2 protein levels in the OVA + siRFX5 group, which were notably decreased upon SAHA treatment. These results highlight the critical role of HDAC2 in the regulatory network affected by RFX5 knockdown. ELISA results (Fig. 7C) further explored the functional consequences of this molecular interaction. The OVA + siRFX5 group displayed significantly reduced levels of pro-inflammatory cytokines IL-6, PGE2, TNF-α, and IL-1β, along with elevated levels of anti-inflammatory cytokines IL-4, IL-10, and IL-18, compared to the OVA + siNC group. This shift in the cytokine profile suggests that RFX5 knockdown promotes an anti-inflammatory state. SAHA treatment reversed these effects, increasing IL-6, PGE2, TNF-α, and IL-1β levels while reducing IL-4, IL-10, and IL-18 levels, demonstrating the pivotal role of HDAC2 in regulating these inflammatory processes. These results suggest that RFX5 and HDAC2 collaborate to control the expression of inflammatory cytokines in DCs. RFX5 knockdown appears to upregulate HDAC2, resulting in a reduction of pro-inflammatory cytokines and an increase in anti-inflammatory cytokines. This newly identified regulatory axis presents a promising therapeutic target for modulating inflammatory responses in asthma and other related inflammatory disorders. The efficacy of SAHA in regulating this molecular interaction further emphasizes the translational potential of targeted pharmacological interventions within this pathway.
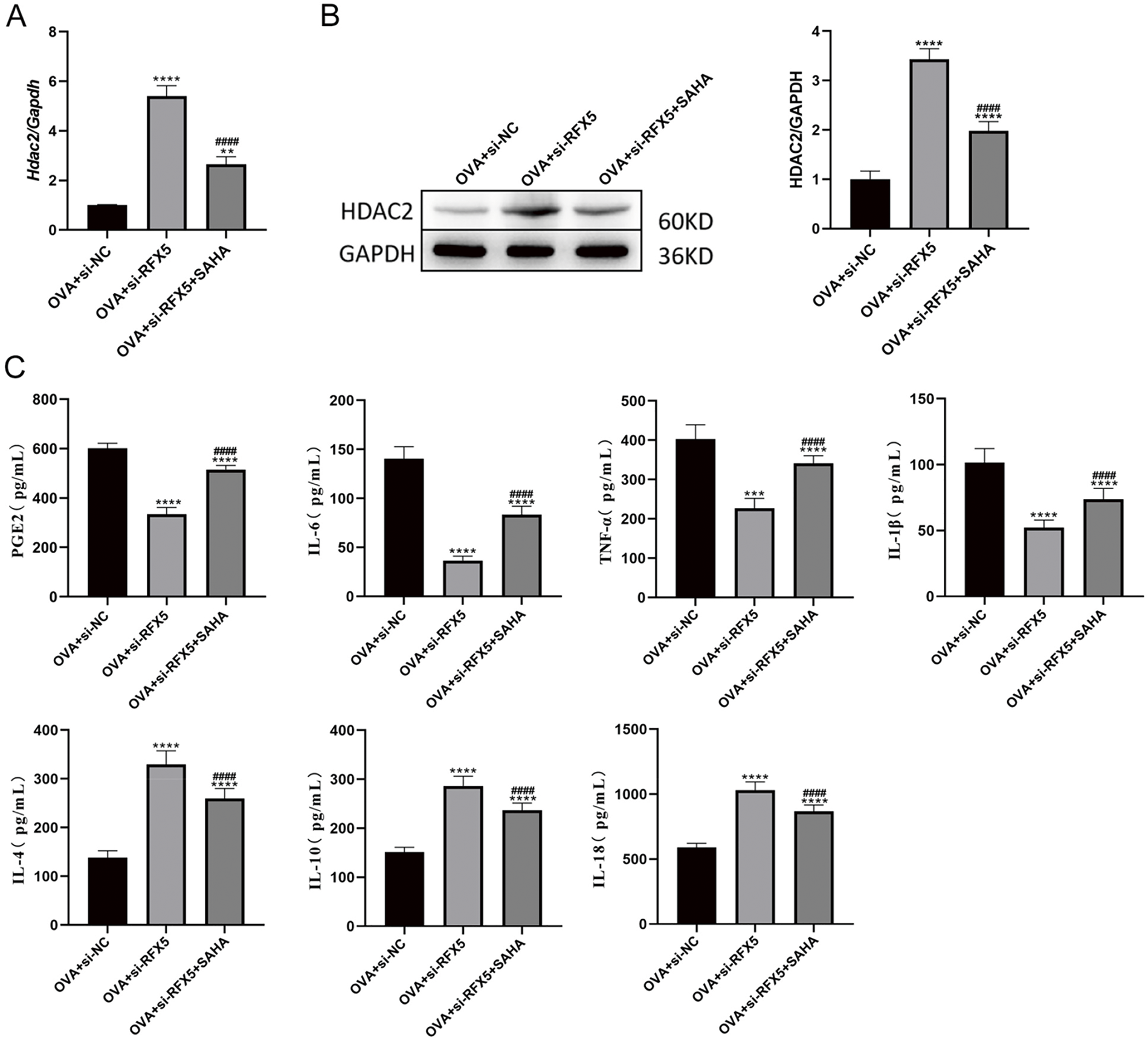
Figure 7: Knockdown of RFX5 regulates inflammatory cytokine expression through Interaction with HDAC2. (A) qRT-PCR results indicate that HDAC2 expression is significantly increased in the OVA + siRFX5 group compared to the OVA + siNC group. Treatment with SAHA significantly reduces HDAC2 expression. (B) Western blot results show that HDAC2 protein levels are significantly elevated in the OVA + siRFX5 group compared to the OVA + siNC group. SAHA treatment results in a significant decrease in HDAC2 protein levels. (C) ELISA results demonstrate that, compared to the OVA + siNC group, the OVA + siRFX5 group exhibits significantly lower levels of IL-6, TNF-α, and IL-1β, and significantly higher levels of IL-4, IL-10, IL-18, and PGE2. SAHA treatment leads to a significant increase in IL-6, TNF-α, and IL-1β levels, and a significant decrease in IL-4, IL-10, IL-18, and PGE2 levels (*vs. Normal Group: **p < 0.01, ***p < 0.001, ****p < 0.0001) (#vs. OVA + siRFX5 Group: ####p < 0.0001)
Asthma, a chronic respiratory disorder characterized by recurrent exacerbations and airway inflammation, poses a significant pediatric health challenge, impeding normal childhood development and imposing substantial socioeconomic burdens on affected families and healthcare systems [25]. Pediatric allergic asthma, predominantly mediated by DC-activated Th2 lymphocytes, leads to the secretion of key cytokines (IL-4, IL-5, and IL-13) that drive eosinophilic airway inflammation and bronchial hyperresponsiveness [26,27]. Since 2016, emerging therapeutic strategies have been integrated into revised treatment guidelines, reflecting international consensus and regional adaptations [28], marking significant advancements over the 2008 recommendations. Nevertheless, ICS remains the cornerstone of current clinical management approaches [29].
The RFX family, distinguished by highly conserved DNA binding domains, consists of seven members in mammals (RFX1-7), two in yeast, and one in C. elegans [30,31]. In humans, RFX1, RFX2, RFX3, RFX5, and RFX7 exhibit widespread expression across various tissues, whereas RFX4 is highly specific to the brain, spinal cord, and testis, and RFX6 is predominantly expressed in the gastrointestinal tract [32]. Structurally, RFX1, RFX2, and RFX3 share significant homology, containing DNA binding domains, activation domains, domains B and C, and dimerization domains, whereas RFX5 and RFX7 possess only DNA binding domains. RFX1 has been implicated in regulating transcription in various cancers, including brain tumors and hepatocellular carcinoma, where it functions as a tumor suppressor by downregulating the oncogene c-myc. It also modulates the activity of enhancer I of the hepatitis B virus [33]. RFX2 knockdown in Xenopus laevis and zebrafish embryos results in neural tube closure defects and impaired left-right axis patterning [34]. Elevated RFX4 expression correlates with tumor progression and poor clinical outcomes in glioblastoma patients [35]. RFX5 promotes liver cancer progression by activating tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta and inhibiting apoptosis [36]. These studies underscore the involvement of RFX family members in various human diseases, although their role in immune antigen presentation in asthma remains underexplored. Our research team focuses on two key areas in pediatric asthma: identifying the specific RFX family member(s) involved in antigen presentation and evaluating the potential of RFX proteins as novel diagnostic biomarkers or therapeutic targets for asthma management.
In asthma, immune cell involvement in inflammatory responses forms a complex network. DCs, as specialized antigen-presenting cells, play a pivotal role as initiators, participants, and regulators in the asthma immune response [37]. In addition to their essential role in initiating T cell proliferation and differentiation, DCs significantly contribute to asthma pathogenesis by producing pro-inflammatory mediators crucial for the onset and maintenance of chronic airway inflammation [38]. Research has shown that among the RFX family members, only RFX1, 2, 3, 5, and 7 are expressed in human lungs, with RFX1 exhibiting the highest expression. Gene knockout of Rfx5 in mice results in severe immunodeficiency, primarily due to a deficiency of MHC class II molecules in the thymus, which profoundly impacts the expression of these molecules in DCs [39]. The precise regulation of MHC class II expression is vital for maintaining a balance between effective defense against foreign antigens and preventing immune-mediated damage. While CIITA is the primary regulator of MHC class II expression, emerging evidence suggests that epigenetic factors also play a significant role. For example, HDAC2 directly binds to the CIITA promoter, altering histone acetylation and suppressing CIITA promoter activation, thereby downregulating CIITA expression [40]. Furthermore, HDAC2 modulates inflammation by directly interacting with the IL-6 promoter region in DCs, leading to the suppression of IL-6 expression [41]. Our experimental findings support these observations, showing significant upregulation of RFX5 expression in DCs differentiated from PBMCs of asthmatic patients and in lung tissue from OVA-induced murine asthma models. Notably, targeted suppression of RFX5 in OVA-challenged mice resulted in substantial improvements in asthma parameters, including reduced airway hyperresponsiveness, decreased granulocyte infiltration, and ameliorated inflammation. Further investigation into the molecular mechanisms of RFX5 in antigen presentation revealed that RFX5 inhibition led to a marked increase in HDAC2 expression while simultaneously promoting CIITA and MHC II expression. Additionally, ChIP assays confirmed a direct interaction between RFX5 and HDAC2. These results are consistent with those of Kong et al. [42], who demonstrated that RFX5 interacts with HDAC2 to downregulate collagen promoter activity. Based on these findings, it is hypothesized that RFX5 modulates DC function through its interaction with HDAC2. Given (1) the pivotal role of DCs in asthma, (2) the limited research on the RFX family in asthma and its impact on DC function, and (3) the direct interaction between RFX5 and HDAC2, which regulates CIITA and IL-6 expression, an OVA asthma mouse model appears to be an ideal model to investigate this further. Preliminary studies show significant upregulation of RFX1 and RFX5 in DCs from asthma models compared to controls, with minimal or no expression of other RFX family members. Given the significant alterations in RFX5 expression, further investigation into its role and mechanism in asthma DCs is warranted. Rfx5 knockout mice have been successfully generated, and an OVA-induced asthma model has been established. Comparative analysis revealed substantial attenuation of pulmonary inflammation in Rfx5 knockout asthma models compared to wild-type mice. Upon isolating DCs from the asthma model mice, a comparative analysis of inflammatory marker expression demonstrated a notable reduction in IL-6 and IL-13 in the Rfx5 knockout DCs relative to controls. These observations support the hypothesis that allergens may upregulate RFX5 expression in DCs, with elevated RFX5 interacting with HDAC2 to inhibit its binding to CIITA and the IL-6 promoter, maintaining acetylation levels at the promoter regions. This, in turn, facilitates the expression of MHC II-related genes and IL-6, promoting Th0 differentiation into Th2 cells and driving the onset and progression of asthma. As an important transcription factor, RFX5 regulates various biological processes, including immune responses and gene expression. Knocking out RFX5 in the asthma model may broadly affect gene expression, complicating the interpretation of the results. Moreover, compensatory mechanisms in gene knockout models may obscure the true role of RFX5 in asthma pathogenesis.
In conclusion, this study examines RFX5 gene expression in clinical samples from pediatric asthma patients. Using a combination of cellular assays, RNA sequencing, bioinformatics, and ChIP assays, this study aims to clarify how RFX5 influences asthma’s molecular mechanisms through its interaction with HDAC2. This approach will provide critical insights into the diagnosis and treatment of childhood asthma.
Acknowledgement: None.
Funding Statement: This project was supported by the 2021 Jiangxi Provincial Department of Science and Technology Applied Research Cultivation Plan Project (No. 20212BAG70005, Yahui Wu) and 2021 Science and Technology Special Project and Social Development Project in Ji’an City, Jiangxi Province (No. 20211-025242, Yahui Wu).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Yahui Wu, Fang Liu; Administrative support: Jingwen Qin, Jian Guo; Provision of study materials or patients: Yahui Wu, Fang Liu; Data collection: Jitao Fan, Jun Mei; Analysis and interpretation of results: Tiansheng Dai, Xiaoli Li; Draft manuscript preparation: Yahui Wu, Fang Liu; Revise articles and polish the language: Tiansheng Dai. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data used to support the findings of this study are included within the article.
Ethics Approval: All methods were performed in accordance with the relevant guidelines and regulations. Human peripheral blood was collected following informed consent and with approval from the Ethics Committees of Shanghai East Hospital (Approval number: 2024YS-139). All animal studies were approved by the Ethics Committee of Experimental Animal Management at Tongji University (Approval number: TJBB08224101).
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Dharmage SC, Perret JL, Custovic A. Epidemiology of asthma in children and adults. Front Pediatr. 2019;7:246. doi:10.3389/fped.2019.00246. [Google Scholar] [PubMed] [CrossRef]
2. Habib N, Pasha MA, Tang DD. Current understanding of asthma pathogenesis and biomarkers. Cells. 2022;11(17):2764. doi:10.3390/cells11172764. [Google Scholar] [PubMed] [CrossRef]
3. Moldaver DM, Larché M, Rudulier CD. An update on lymphocyte subtypes in asthma and airway disease. Chest. 2017;151(5):1122–30. doi:10.1016/j.chest.2016.10.038. [Google Scholar] [PubMed] [CrossRef]
4. Mims JW. Asthma: definitions and pathophysiology. Int Forum Allergy Rhinol. 2015;5(S1):S2–6. doi:10.1002/alr.21609. [Google Scholar] [PubMed] [CrossRef]
5. Ozkan M, Eskiocak YC, Wingender G. Macrophage and dendritic cell subset composition can distinguish endotypes in adjuvant-induced asthma mouse models. PLoS One. 2021;16(6):e0250533. doi:10.1371/journal.pone.0250533. [Google Scholar] [PubMed] [CrossRef]
6. Lajiness JD, Cook-Mills JM. Catching our breath: updates on the role of dendritic cell subsets in asthma. Adv Biol. 2023;7(6):2200296. doi:10.1002/adbi.202200296. [Google Scholar] [PubMed] [CrossRef]
7. Chairakaki AD, Saridaki MI, Pyrillou K, Mouratis MA, Koltsida O, Walton RP, et al. Plasmacytoid dendritic cells drive acute asthma exacerbations. J Allergy Clin Immunol. 2018;142(2):542–56. doi:10.1016/j.jaci.2017.08.032. [Google Scholar] [PubMed] [CrossRef]
8. Ouyang LH, Su GM, Quan JY, Xiong ZL, Lai TW. Emerging roles and therapeutic implications of HDAC2 and IL-17A in steroid-resistant asthma. Chin Med J Pulm Crit Care Med. 2023;1(2):108–12. doi:10.1016/j.pccm.2023.04.003. [Google Scholar] [PubMed] [CrossRef]
9. Axelsson I, Naumburg E, Prietsch SO, Zhang L. Inhaled corticosteroids in children with persistent asthma: effects of different drugs and delivery devices on growth. Cochrane Database Syst Rev. 2019;6(6):CD010126. doi:10.1002/14651858.CD010126.pub2. [Google Scholar] [PubMed] [CrossRef]
10. Pruteanu AI, Chauhan BF, Zhang L, Prietsch SO, Ducharme FM, Ducharme FM. Inhaled corticosteroids in children with persistent asthma: dose-response effects on growth. Evid Based Child Health. 2014;9(4):931–1046. doi:10.1002/ebch.1989. [Google Scholar] [PubMed] [CrossRef]
11. Osman S, Charlotte SU. Inhaled corticosteroid exposure and risk of cataract in patients with asthma and copd: a systematic review and meta-analysis. J Ophthalmol. 2023;1(1):8209978. doi:10.1155/2023/8209978. [Google Scholar] [PubMed] [CrossRef]
12. Devonshire AL, Kumar R. Pediatric asthma: principles and treatment. Allergy Asthma Proc. 2019;40(6):389–92. doi:10.2500/aap.2019.40.4254. [Google Scholar] [PubMed] [CrossRef]
13. Lai T, Wu M, Zhang C, Che L, Xu F, Wang Y, et al. HDAC2 attenuates airway inflammation by suppressing IL-17A production in HDM-challenged mice. Am J Physiol Lung Cell Mol Physiol. 2019;316(1):L269–79. doi:10.1152/ajplung.00143.2018. [Google Scholar] [PubMed] [CrossRef]
14. Liang Z, Michael R, Eleni P, Michael T, Daiana S. Expression of glucocorticoid receptor and HDACs in airway smooth muscle cells is associated with response to steroids in COPD. Respir Res. 2024;25(1):227. doi:10.1186/s12931-024-02769-3. [Google Scholar] [PubMed] [CrossRef]
15. Mishra R, Chaturvedi R, Hashim Z, Nath A, Khan A, Gupta M, et al. Role of P-gp and HDAC2 and their reciprocal relationship in uncontrolled asthma. Curr Pharm Biotechnol. 2021;22(3):408–13. doi:10.2174/1389201021666200529104042. [Google Scholar] [PubMed] [CrossRef]
16. Qian L, Lijuan H, Chen B, Luxia K, Jiannan H, Chao L, et al. Inhibition of spleen tyrosine kinase restores glucocorticoid sensitivity to improve steroid-resistant asthma. Front Pharmacol. 2022;13:885053. doi:10.3389/fphar.2022.885053. [Google Scholar] [PubMed] [CrossRef]
17. Hao M, Lin J, Shu J, Zhang X, Luo Q, Pan L, et al. Clarithromycin might attenuate the airway inflammation of smoke-exposed asthmatic mice via affecting HDAC2. J Thorac Dis. 2015;7(7):1189–97. doi:10.3978/j.issn.2072-1439.2015.05.18. [Google Scholar] [PubMed] [CrossRef]
18. Wu Y, Zhang JF, Xu T, Xu L, Qiao J, Liu F, et al. Identification of therapeutic targets for childhood severe asthmatics with DNA microarray. Allergol Immunopathol Madr. 2016;44(1):76–82. doi:10.1016/j.aller.2015.03.002. [Google Scholar] [PubMed] [CrossRef]
19. Xu Y, Sengupta PK, Seto E, Smith BD. Regulatory factor for X-box family proteins differentially interact with histone deacetylases to repress collagen alpha2(I) gene (COL1A2) expression. J Biol Chem. 2006;281(14):9260–70. doi:10.1074/jbc.M511724200. [Google Scholar] [PubMed] [CrossRef]
20. Pietras CO, James A, Konradsen JR, Nordlund B, Söderhäll C, Pulkkinen V, et al. Transcriptome analysis reveals upregulation of bitter taste receptors in severe asthmatics. Eur Respir J. 2013;42(1):65–78. doi:10.1183/09031936.00077712. [Google Scholar] [PubMed] [CrossRef]
21. Wu L, Luo ZH, Liu YT, Jia L, Jiang YY, Du J, et al. Aspirin inhibits RANKL-induced osteoclast differentiation in dendritic cells by suppressing NF-κB and NFATc1 activation. Stem Cell Res Ther. 2019;10(1):375. doi:10.1186/s13287-019-1500-x. [Google Scholar] [PubMed] [CrossRef]
22. Parikh A, Shin JH, Faquin W, Lin DT, Tirosh I, Sunwoo JB, et al. Malignant cell-specific CXCL14 promotes tumor lymphocyte infiltration in oral cavity squamous cell carcinoma. J Immuno Ther Cancer. 2020;8(2):e001048. doi:10.1136/jitc-2020-001048. [Google Scholar] [PubMed] [CrossRef]
23. Tang LL, Zhang XN, Xu XQ, Liu L, Sun XH, Wang BH, et al. BMAL1 regulates MUC1 overexpression in ovalbumin-induced asthma. Mol Immunol. 2023;156(Pt 2):77–84. doi:10.1016/j.molimm.2023.03.004. [Google Scholar] [PubMed] [CrossRef]
24. Thakur VR, Khuman V, Beladiya JV, Chaudagar KK, Mehta AA. An experimental model of asthma in rats using ovalbumin and lipopolysaccharide allergens. Heliyon. 2019;5(11):e02864. doi:10.1016/j.heliyon.2019.e02864. [Google Scholar] [PubMed] [CrossRef]
25. Simoneau T, Cloutier MM. Controversies in pediatric asthma. Pediatr Ann. 2019;48(3):e128–34. doi:10.3928/19382359-20190226-01. [Google Scholar] [PubMed] [CrossRef]
26. George S, Seblewongel A, Jeanne A, Wei KL, Kirsten N, Dylan B et al. IL-4 and IL-13, not eosinophils, drive type 2 airway inflammation, remodeling and lung function decline. Cytokine. 2023;162:15609. doi:10.1016/j.cyto.2022.156091. [Google Scholar] [PubMed] [CrossRef]
27. Wang CM, Chang CB, Wu SF. Differential DNA methylation in allergen-specific immunotherapy of asthma. Cell Mol Immunol. 2020;17(9):1017–8. doi:10.1038/s41423-020-0476-x. [Google Scholar] [PubMed] [CrossRef]
28. Chen B, Feng S, Yin XW. Effect of obesity on treatment outcome of asthma predictive index-positive infants and young children with wheezing. Chin J Contemp Pediatr. 2016;18(10):991–4. doi:10.7499/j.issn.1008-8830.2016.10.015. [Google Scholar] [PubMed] [CrossRef]
29. Zhu H, Liu H, Sui Z, Yu J, Zheng Q, Li L. Quantitative comparison of different inhaled corticosteroids in the treatment of asthma in children. Pediatr Res. 2023;93:31–8. doi:10.1038/s41390-022-02095-8. [Google Scholar] [PubMed] [CrossRef]
30. Katjana S, Luis C, Konstantin R, Erika KS, Norman R, Häckes D, et al. Multi-omics analysis identifies RFX7 targets involved in tumor suppression and neuronal processes. Cell Death Discov. 2023;9(1):80. doi:10.1038/s41420-023-01378-1. [Google Scholar] [PubMed] [CrossRef]
31. Marissa LL, Milja K, Mailo JA, Shailly JG. Phenotype expansion and neurological manifestations of neurobehavioural disease caused by a variant in RFX7. Eur J Med Genet. 2023;66(1):104657. doi:10.1016/j.ejmg.2022.104657. [Google Scholar] [PubMed] [CrossRef]
32. Sugiaman TD, Vitezic M, Jouhilahti EM, Mathelier A, Lauter G, Misra S, et al. Characterization of the human RFX transcription factor family by regulatory and target gene analysis. BMC Genom. 2018;19(1):181. doi:10.1186/s12864-018-4564-6. [Google Scholar] [PubMed] [CrossRef]
33. Reinhold W, Emens L, Itkes A, Blake M, Ichinose I, Zajac-Kaye M. The myc intron-binding polypeptide associates with RFX1 in vivo and binds to the major histocompatibility complex class II promoter region, to the hepatitis B virus enhancer, and to regulatory regions of several distinct viral genes. Mol Cell Biol. 1995;15(6):3041–8. doi:10.1128/MCB.15.6.3041. [Google Scholar] [PubMed] [CrossRef]
34. Shawlot W, Vazquez-Chantada M, Wallingford JB, Finnell RH. Rfx2 is required for spermatogenesis in the mouse. Genesis. 2015;53(9):604–11. doi:10.1002/dvg.22880. [Google Scholar] [PubMed] [CrossRef]
35. Jeong HY, Kim HJ, Kim CE, Lee S, Choi MC, Kim SH. High expression of RFX4 is associated with tumor progression and poor prognosis in patients with glioblastoma. Int J Neurosci. 2021;131(1):7–14. doi:10.1080/00207454.2020.1732969. [Google Scholar] [PubMed] [CrossRef]
36. Chen BD, Zhao YJ, Wang XY, Liao WJ, Chen P, Deng KJ, et al. Regulatory factor X5 promotes hepatocellular carcinoma progression by transactivating tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein theta and suppressing apoptosis. Chin Med J Engl. 2019;132(13):1572–81. doi:10.1097/CM9.0000000000000296. [Google Scholar] [PubMed] [CrossRef]
37. Froidure A, Shen C, Pilette C. Dendritic cells revisited in human allergic rhinitis and asthma. Allergy. 2016;71(2):137–48. doi:10.1111/all.12770. [Google Scholar] [PubMed] [CrossRef]
38. Brugha R, Mushtaq N, McCarthy NE, Stagg AJ, Grigg J. Respiratory tract dendritic cells in paediatric asthma. Clin Exp Allergy. 2015;45(3):624–31. doi:10.1111/cea.12457. [Google Scholar] [PubMed] [CrossRef]
39. Clausen BE, Waldburger JM, Schwenk F, Barras E, Mach B, Rajewsky K, et al. Residual MHC class II expression on mature dendritic cells and activated B cells in RFX5-deficient mice. Immunity. 1998;8(2):143–55. doi:10.1016/S1074-7613(00)80467-7. [Google Scholar] [PubMed] [CrossRef]
40. Nash G, Paidimuddala B, Zhang L. Structural aspects of the MHC expression control system. Biophys Chem. 2022;284(1):106781. doi:10.1016/j.bpc.2022.106781. [Google Scholar] [PubMed] [CrossRef]
41. Zhang Q, Zhao K, Shen Q, Han YM, Gu Y, Li X, et al. Tet2 is required to resolve inflammation by recruiting Hdac2 to specifically repress IL-6. Nature. 2015;525(7569):389–93. doi:10.1038/nature15252. [Google Scholar] [PubMed] [CrossRef]
42. Kong X, Fang M, Li P, Fang F, Xu Y. HDAC2 deacetylates class II transactivator and suppresses its activity in macrophages and smooth muscle cells. J Mol Cell Cardiol. 2009;46(3):292–9. doi:10.1016/j.yjmcc.2008.10.023. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools