 Open Access
Open Access
ARTICLE
A Novel Meta-Heuristic Optimization Algorithm in White Blood Cells Classification
Mathematics Department, Faculty of Science, Al-Azhar University, Cairo, 11884, Egypt
* Corresponding Author: Humam K. Yaseen. Email:
Computers, Materials & Continua 2023, 75(1), 1527-1545. https://doi.org/10.32604/cmc.2023.036322
Received 26 September 2022; Accepted 14 December 2022; Issue published 06 February 2023
Abstract
Some human diseases are recognized through of each type of White Blood Cell (WBC) count, so detecting and classifying each type is important for human healthcare. The main aim of this paper is to propose a computer-aided WBCs utility analysis tool designed, developed, and evaluated to classify WBCs into five types namely neutrophils, eosinophils, lymphocytes, monocytes, and basophils. Using a computer-artificial model reduces resource and time consumption. Various pre-trained deep learning models have been used to extract features, including AlexNet, Visual Geometry Group (VGG), Residual Network (ResNet), which belong to different taxonomy types of deep learning architectures. Also, Binary Border Collie Optimization (BBCO) is introduced as an updated version of Border Collie Optimization (BCO) for feature reduction based on maximizing classification accuracy. The proposed computer aid diagnosis tool merges transfer deep learning ResNet101, BBCO feature reduction, and Support Vector Machine (SVM) classifier to form a hybrid model ResNet101-BBCO-SVM an accurate and fast model for classifying WBCs. As a result, the ResNet101-BBCO-SVM scores the best accuracy at 99.21%, compared to recent studies in the benchmark. The model showed that the addition of the BBCO algorithm increased the detection accuracy, and at the same time, decreased the classification time consumption. The effectiveness of the ResNet101-BBCO-SVM model has been demonstrated and beaten in reasonable ratios in recent literary studies and end-to-end transfer learning of pre-trained models.Keywords
A peripheral blood smear consists of Red Blood Cells (RBCs), White Blood Cells (WBCs), and platelets [1]. White blood cells (WBCs) primarily consist of granulocytes, monocytes, and lymphocytes, crucial human blood components and essential elements in the immune system. According to their morphological properties, when stained with Wright’s stain, neutrophils, eosinophils, and basophils are the three subtypes of granulocytes, a kind of leukocyte with granules in its cytoplasm [2,3]. Notably, changes in the ratio number of white blood cells have been associated with blood diseases. For example Leukopenia is a condition in which the number of WBCs is lower than the reference value [4]. Detecting the type of WBC is a crucial diagnostic step because various blood disorders are distinguished by morphological variation and modifications in the ratio of different WBC types [5]. Granulocytes with the three types of neutrophil, eosinophil, and basophil count for about 60% of the total number of white blood cells, lymphocytes about 30%, while monocytes with a minor percentage about 10% [2]. So, according to the brief introduction, the main problem discussed in this paper is the detection and classification of WBC types fast and accurately. The automatic detection of the WBC type is essential for creating an automated WBC analysis approach.
Dyeing contaminants, cytoplasm with low picture contrast, and changing appearances under different staining circumstances contribute to WBC types detection difficulties [6]. Hence, the WBC detection problem can be considered a computer vision recognition problem. Numerous literature studies concentrate on classifying WBC images according to cell types using artificial intelligence supervised learning (machine learning and deep learning) algorithms. Deep learning (DL) approaches have high image representation capabilities. They have drawn significant attention to computer vision and medical image analysis in recent years [6]. The main drawback facing the deep learning algorithms is the time consumption for the classification. Traditional classifiers achieve high classification accuracy on large-scale image datasets in terms of design speed requirement, but cannot extract the features from the images efficiently like deep learning. Due to the contradictions of representation complexity for deep learning algorithms and the high accuracy rate of machine learning algorithms, there is a feature reduction problem. Medical images have many features that require computational analysis, leading to model complexity regarding resources and time [7]. Hence, the feature reduction problem can be overcome using one of the robust swarm-based optimization algorithms. In turn, extracting features of WBC images by deep learning with high image representation capabilities leads to multiple features that can be selected using the feature selection method. Also, combining deep learning as a feature extractor with a regular classification algorithm introduces a stable learning model regarding classification accuracy, speed, and resource complexity in WBC detection.
So, the main motivation for the paper is how the detection of the type of WBC may apply to human healthcare and the speed of this detection.
The primary research objective focuses on designing a hybrid classification learning pipeline based on pre-trained deep learning model(s) as feature extraction, swarm-based feature subset selection, and support vector machine as a machine learning classifier to recognize WBC quickly and accurately.
Various contributions have been proposed in this study, including:
• Suggesting a model to classify the five types of white blood cells.
• Proposing a novel Binary Border Collie Optimization (BBCO) algorithm derived from Border Collie Optimization to act as a feature selection.
• Building self-controlled and transferred learning to create plausible and unbiased representations of features, reducing the risk of overfitting when learning from limited, labeled data.
• Reducing the used number of features by the classification model, the total resources and the time consumed.
• Benchmarking the state-of-the-art methods vs. other models and classifiers using the same dataset(s).
• Supporting physician process according to the outcomes of WBC classifications and presenting informed conclusions.
Section 2 of this paper will discuss the most recent studies, Section 3 overviews the methods and techniques and introduces the proposed model, while Section 4 contains the experiment, discussion, and a brief overview of the dataset used. Section 5 concludes the paper.
Different efforts have been made to design tools, methods, and techniques for WBC analysis and detection. Significant research contributions have been introduced and developed on shallow learning or deep learning, which are discussed and illustrated in the subsequent paragraphs.
Gupta et al. [8] introduced the Optimized Binary Bat Algorithm (OBBA) as a feature selection to reduce the features extracted from the Leukocyte Images for Segmentation and Classification (LISC) white blood cell dataset. The authors employed four machine learning algorithms for the classification: logistic regression, decision tree, K-Nearest Neighbor (KNN), and random forest.
Tavakoli et al. [9] proposed a model for classifying white blood cells from three different datasets. The model has three steps. In the first step, the segmentation was used to find the nucleus and cytoplasm. In step two, there was the creation and extraction of four color and three shape features, while step three is the classification step using the support vector machine algorithm.
An end-to-end convolutional deep architecture called WBCsNet was presented in [10]. This architecture was built from scratch, and the overall accuracy was 96.1% for the five types of white blood cells.
In [11], a dataset of white blood cells from normal peripheral blood was created and called Raabin-WBC. It is a multifunctional dataset that may be used as a benchmark for various machine-learning tasks, like classification, detection, segmentation, and localization. In addition, the author tries to classify the five types of white blood cells using several pre-trained deep learning models.
Sharma et al. [12] proposed a Leukemia classification model in their paper. They used the histogram of oriented gradients descriptor to extract the features and four different scenarios to segment the nucleus. Finally, Convolutional Neural Network (CNN) was used as a classifier.
A novel deep-learning method to classify WBC was proposed in [13]. This model integrates the advantages of ResNet and Densely Connected Convolutional Network. The model includes a Spatial Attention Module (SAM) to highlight the fundamental features and a Channel Attention Module (CAM) to detect features in the feature maps. Finally, four datasets were used for the classification performance.
The authors in [14] proposed a strategy for segmenting blood leucocytes from static microscope images. In the proposal, the authors used SVM to spot the position of the Region Of Interest (ROI); after which filtering was done using histogram analysis to avoid objects with no ROI. Finally, recognize blood cells using the texture feature.
Almost all studies achieved a high accuracy rate using deep learning-based models. Even though these models have high performance, they consume hardware resources and time complexity. Some of them used optimization algorithms to reduce complexity but with low accuracy. Hence, this propose a model that reduces processing time and complexity, feature sparse matrix representation in memory, and at the same time takes accuracy in mind which is crucial issue in the WBC classification.
Many contributions, algorithms, methods, and techniques have been introduced to detect and recognize WBC. The most recent ones will be used and applied for this study, exploring the pros and cons of each and their importance for the proposed model to overcome the mined challenges.
Today, ML is considered one of the fastest growing technical areas. ML sits at the intersection of computer science and statistics and is the core of artificial intelligence and data science. ML is a broad phrase that describes computational algorithms to enhance functionality or produce accurate predictions regarding performance metrics [15]. Experience in this context refers to the learner’s prior knowledge, frequently represented through electronically gathered data available for analysis [16]. Major ML algorithms are categorized into supervised and unsupervised learning algorithms. Unsupervised algorithms are based on the variation and pattern of data, whereas supervised algorithms consider the labels and classifications of the dataset [17–19].
Data scientists often remind us that there is no “one size fits all” approach to resolving an issue. Hence, ML uses a range of algorithms to handle data problems. The problem you are attempting to solve, the number of variables involved, the most appropriate model to utilize, and other factors, influence the method used [20]. There are numerous ML algorithms; the support vector machine is a crucial algorithm interested in WBC detection and recognition.
3.1.1 Support Vector Machine (SVM)
SVM is an ML algorithm that has become very popular recently for various classification problems [21]. Due to its relative simplicity and flexibility in handling multiple classification issues, SVM provides balanced predicted performance, even in studies with limited sample sets [22]. The SVM classifier is based on the concept of the most appropriate hyper-planes employed to distinguish among classes [15,23]. Due to the goals of SVM, the decision boundary might have to be very close to one specific class to correctly label all data points in the training set [23]. SVM tries to minimize the number of misclassified examples due to the high penalty added by the c-parameter, meaning the results in a decision boundary would be a smaller margin. Also, SVM depends on kernel functions, such as the Radial Basis Function (RBF), a similarity measure that becomes linearly separable. The RBF depends on gamma, leading to overfitting models with very large gamma. Hence, it is important to normalize the feature matrix so that features are compatible and on the same scale [24].
Deep learning, a subset of machine learning, has recently risen to prominence due to massive data, parallel and distributed computing, and advanced algorithms. Since the early 2000s, academic interest in deep learning has grown significantly, overcoming past limits [6].
Convolutional Neural Network (CNN) comes from deep learning. It consists mainly of convolutional, pooling, filters, fully connected, and soft-max layers [23]. CNN specializes in dealing with images due to its ability to extract images’ features. Many CNN algorithms exist, such as AlexNet, VGG, ResNet, etcetera.
Machine learning and deep learning algorithms have generally been intended to operate independently. These algorithms have been trained to address particular problems. Once the feature-space distribution is modified, the models must be recreated from scratch [25]. Transfer learning is a strategy that transfers the acquired knowledge of one domain (the source domain) to another domain (the target domain) when data is (often) sparse or rapid training is required [26]. The most significant benefits of transfer learning are: a) to overcome the notion of solitary learning, b) to utilize the knowledge gained from one task to solve a related problem, c) to utilize a small dataset dependent on an extensive dataset used for training, d) to reach the model’s optimal parameters quickly, and e) to enhance sample efficiency and decrease the computing demands of new tasks.
As noted in the abovementioned statements, image analysis using a computer-based tool is considered a supervised machine learning application. So, extracting the features from the image is essential for fitting image classification [27]. Shallow classifiers depend on the quality of the feature extraction method and the derived features map in terms of the high dimensionality of the images, which leads to complex models that consume resources and time. Convolutional layers in deep learning solve this problem by reducing the dimension of the image and extracting the desired features [23].
In recent decades, the rising complexity and difficulty of real-world issues have necessitated the development of more trustworthy optimization approaches, particularly meta-heuristic optimization algorithms. These methods are primarily stochastic and estimate the best solutions to various optimization problems [28,29].
The two search strategies are exploration and exploitation, which are used by meta-heuristic optimization algorithms [30]. Exploration is the capacity to comb through all of the search space. This skill is related to escaping entrapped local optima and avoiding local optima. On the other hand, exploitation means looking for suitable options close by and improving them locally [31]. All population-based algorithms use these properties differently [28].
Evolutionary algorithms, swarm intelligence algorithms, physics-based methods, and human-based methods are some of the most common meta-heuristic classifications [32]. Evolutionary algorithms use operators inspired by biological characteristics like crossover and mutation to imitate natural evolution habits. The Genetic Algorithm (GA), motivated by Darwinian evolutionary theories, is a recurrent evolutionary algorithm [28]. Swarm intelligence algorithms are a type of meta-heuristic that mimics the behavior of animals in groups, such as those in which they are moving or hunting [33]. The fundamental feature of this group is sharing all animals’ organism information via the optimization course [28].
Dutta et al. introduce Border Collie Optimization (BCO), a new swarm algorithm that imitates the herding behavior of Border Collie dogs [34,35]. The Border collie dogs’ herding approach, capacity to judge a situation and ability to make adaptable decisions inspire the development of a BCO algorithm. BCO primarily employs three herding methods: Gathering, Stalking, and Eyeing, representing different approaches to herding than other breeds [34]. Fig. 1 shows the herding methods. The minimum initialization of a population is three dogs and sheep. Velocity, acceleration, and time determine the sheep and dogs’ distance traveled and direction [34]. The effectiveness of BCO is assessed using 35 test functions. The capabilities for exploration, exploitation, and local minima avoidance are compared to those of seven state-of-the-art algorithms, and the results prove the BCO algorithm’s superiority [34]. Many researchers mentioned border collie optimization, like Vijayakumar et al. [36] and Chandol et al. [37].
Figure 1: Border collie dogs’ herding approach
The research focuses on developing a computer-aided tool for computer vision and recognition of human WBCs by employing different Artificial Intelligent (AI) algorithms to help physicians and specialists detect, recognize, and identify diseases with a high precision rate, accuracy, and low computation complexity. Fig. 2 represents a standard pipeline for building software-based tuned machine learning WBCs images utility tool. It includes collect, prepare and preprocess of the data, feature engineering including feature extraction and reduction, model training controlled by tuning, model analysis and validation, and model deployment. This study focuses on utilizing the feature engineering process and model training to enhance model analysis and validation using a mixture of deep learning feature mining, swarm-based feature reduction, and classification. As illustrated in Fig. 3, the proposed model contains four main phases which are: data pre-processing, feature extraction, feature subset selection, and classification. The following sub-section explains these phases in detail.
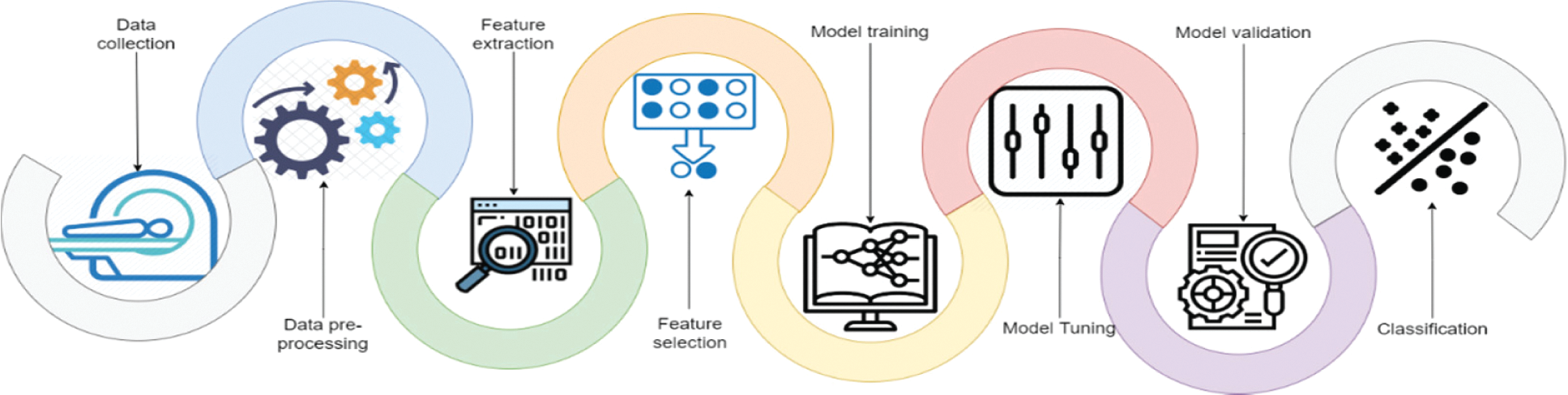
Figure 2: WBC images pipelines classification
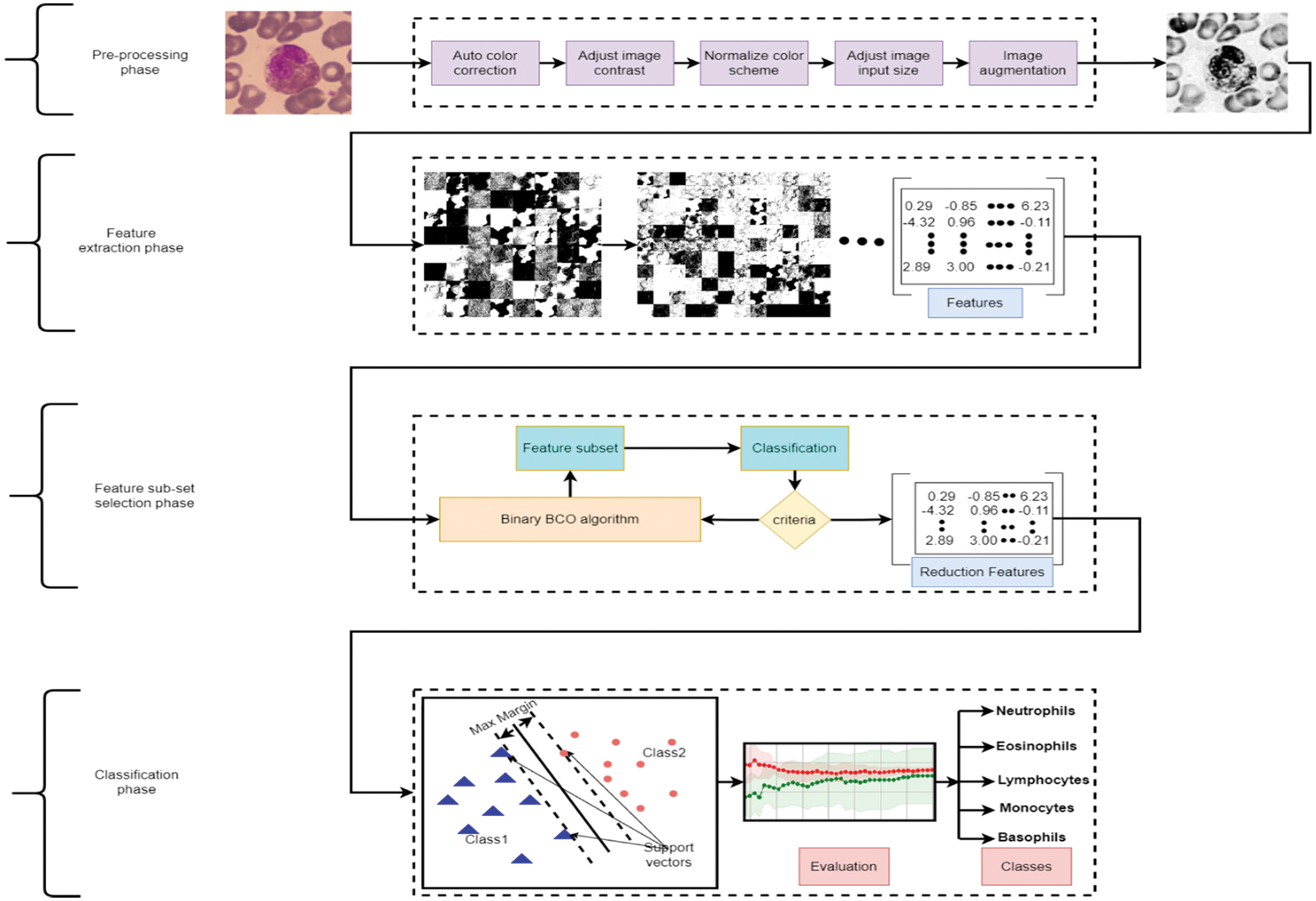
Figure 3: Proposed model
The medical data is generally captured without processing or transforming, and keeps an immutable record related to the original data stream. Such data can be captured from multiple sources, including streaming or on-demand from different services. Generally, a data collection service to captures data and then processes and stores it, offering high throughput, reliability, and low latency.
Most medical images come from multiple sources and suffer from varying noises like blurring, unclear vision, image rarity, different image sizes, etcetera. Hence, the pre-processing stage is essential that must be considered for any system or model dealing with medical images.
So, WBCs images can be pre-processed for analysis by applying five procedures. A color correction to enhance the color, adjusting the color contrast of the images, normalizing the color scheme, making augmentations from different angles, and finally, modifying the image size to match what the pre-trained models need. Fig. 4 represents the order sequence illustrating a sample input and the corresponding output.
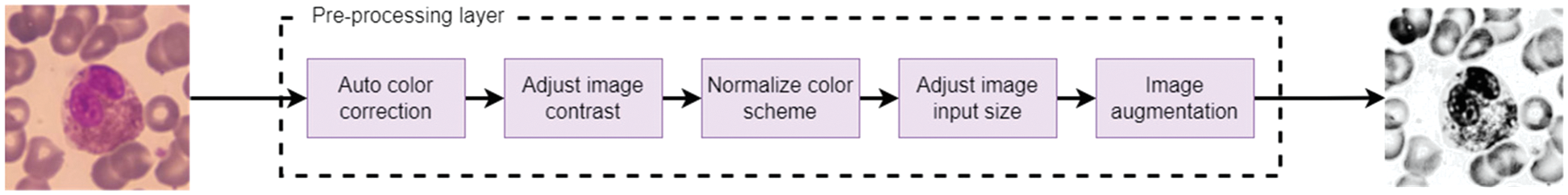
Figure 4: Pre-processing sequences for handling WBC image
4.3 Feature Extraction Using Deep Learning
Deep learning algorithms have an essential property represented as the power to extract the features from images without computation consumption. Common, well-known deep learning models can be re-used as feature extraction, including AlexNet, VGG16, VGG19, ResNet50, and ResNet101. Such models are primarily used to extract essential features from images for resolving the main problem of image classification. Hence, deep learning algorithms were used to extract the best features and detect the edges to extract WBC images. Fig. 5 represents the subsequent transformation steps to derive the features from instance input image(s) and produce them as a numeric matrix representing most of the image features to be used later in the classification problem.
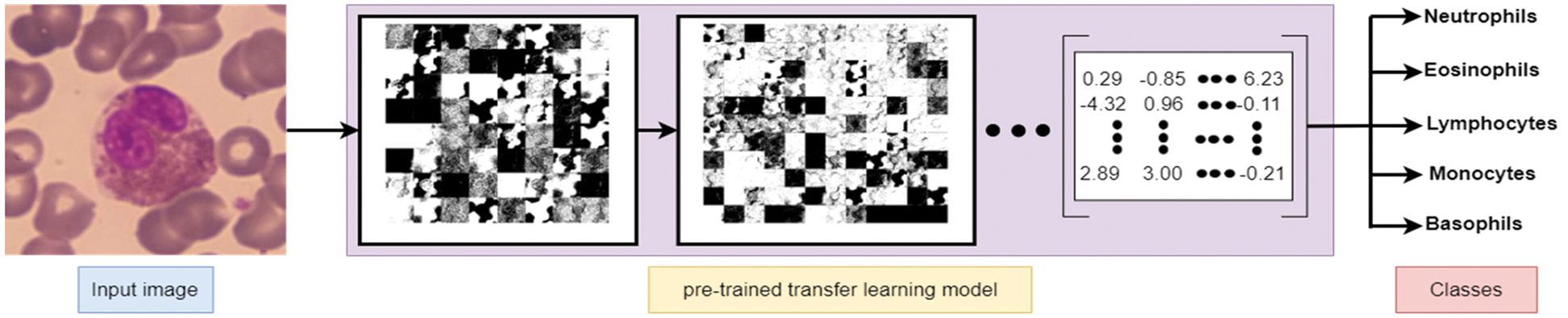
Figure 5: Feature extraction using pre-trained deep learning net
The crucial process of choosing a subset of relevant characteristics from the whole feature space to improve a model is called feature selection [38]. Data features are not always unrelated; they can share some information, so there is usually dummy data in the data pattern. A crucial aspect that machine learning suffers from is feature selection. The primary goal of the feature selection problem is to decrease the dimension of the feature set while retaining performance accuracy [39]. The primary purpose of dimensionality reduction is to improve data classification or to make data visualization easier by selecting the most pertinent features [17,40].
There is no best way to perform tasks for machine learning models. Many strong recommendations say that learning-based methods for choosing features are better than other methods.
As mentioned, one of the classification algorithm problems is the high dimensionality of the image data, especially in colorful images. The optimization algorithms can handle such a problem by selecting the most critical features [41]. Optimization algorithms estimate the optimal decision for diverse optimization problems in a stochastic manner [28].
4.5 Binary Border Collie Optimization (BBCO)
The BBCO is a learning-based feature selection method that extends the standard BCO algorithm. It is a novel algorithm introduced as feature selection, and it determines the subset feature set by maximizing the accuracy of the classifier in each dataset. The BBCO has non-linear constraints, large search spaces, and is non-convex. Hence, BBCO works on a precise balance between the two search strategies for excellent performance. BBCO is efficient, flexible, and has fast convergence behavior since all instances are learned from the right, left, and lead dog solutions, which could easily fall into the local optimum. The BBCO strategy ensured that the leader dog did not get stuck in the local optimum. Right and left dogs alter inertia weight, which makes it easier to search locally and globally, leading to high exploration and employing the position-based learning strategy to improve the way the features are chosen. Since BBCO is an extension of BCO, which includes gathering, stalking, and eyeing approaches, all these approaches are defined using mathematical simulation, and Table 1 utilizes the reference parameters of the simulation. For gathering, procedures can be defined mathematically for sheep as:
The following mathematical equations define stalking procedures as
and finally, the eyeing procedures can be defined using:
The simulation of a minimum initialization of a population containing three dogs can be defined as:
Fig. 6 demonstrates the pseudo-code of the novel BBCO selection method, which is designed to extract relevant features for classification, especially for WBC analysis and recognition.
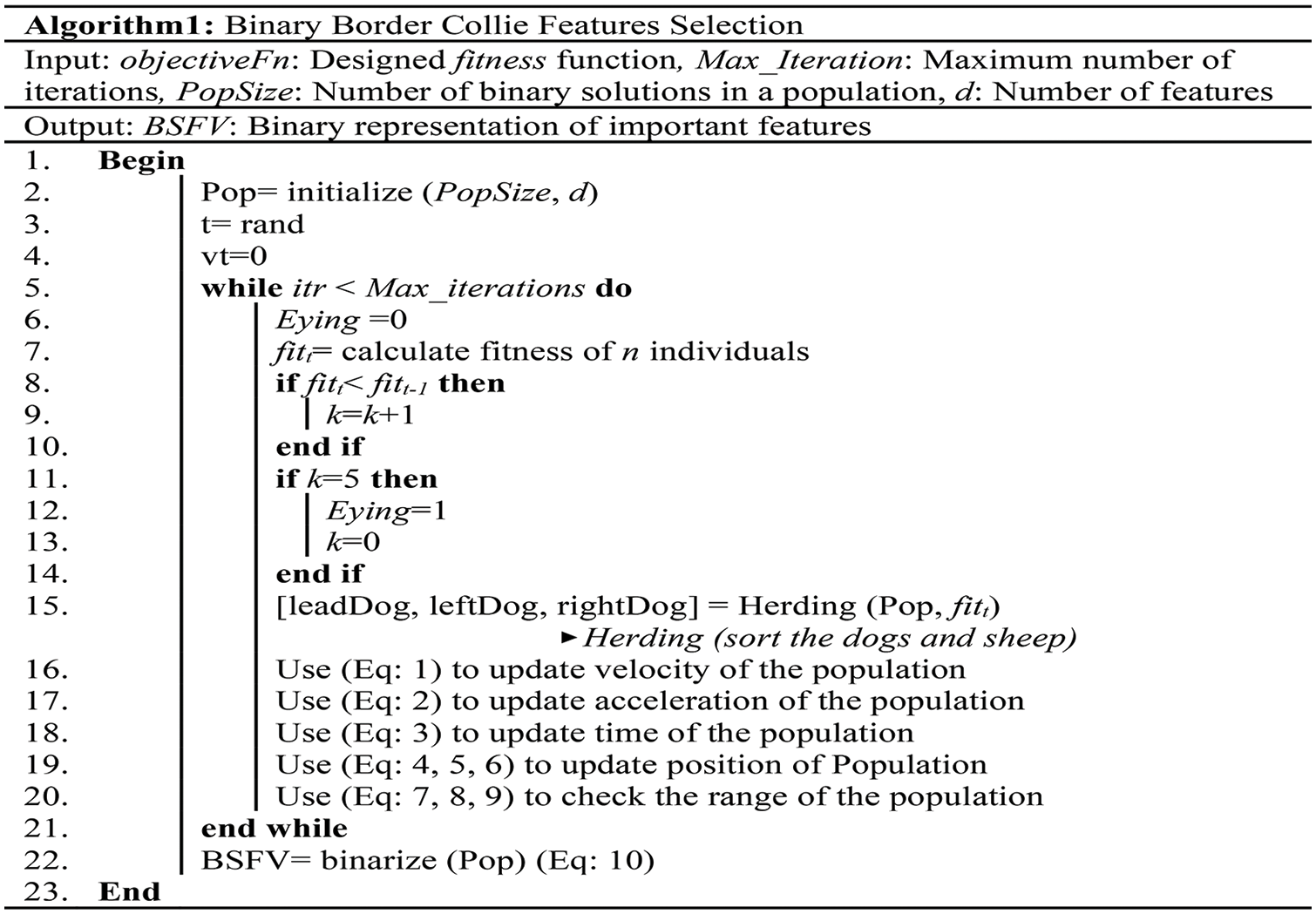
Figure 6: Pseudo-code of the BBCO feature selection method
During the different stages of BBCO, one hundred population solutions are handled over fifteen iterations to derive a new population and enhance the quality of the solution using exploration and exploitation strategies. Finally, the best solution is used to reduce the feature vector during the ensemble learning selection method, which is binarized using:
where
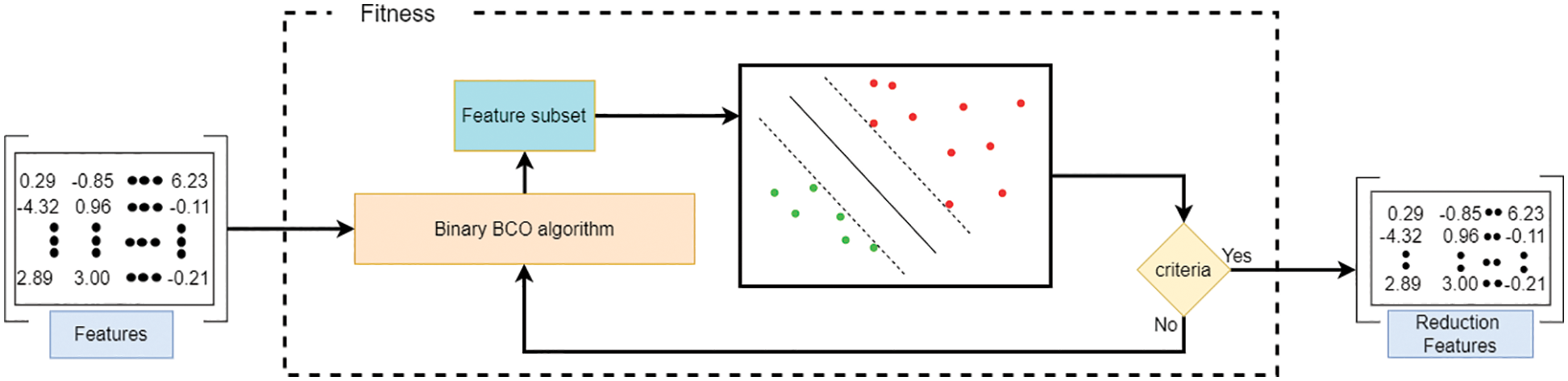
Figure 7: Feature selection
4.6 Classification Learning Algorithms
The classification phase in this paper is twofold:
In the first one, the system depends on the five deep learning pre-trained algorithms in combination with the transfer learning technique: AlexNet, VGG16, VGG19, ResNet50, and ResNet101. These algorithms are updated to classify five types of WBC. Fig. 5 illustrates this type of classification.
In the second one, the Support Vector Machine (SVM) is used, which is a shallow machine learning method for classification. We use two input feature matrixes for this algorithm: the feature matrix extracted directly from the pre-trained models and the reduced feature matrix from the BBCO algorithm. Fig. 8 illustrates the shallow classification type.
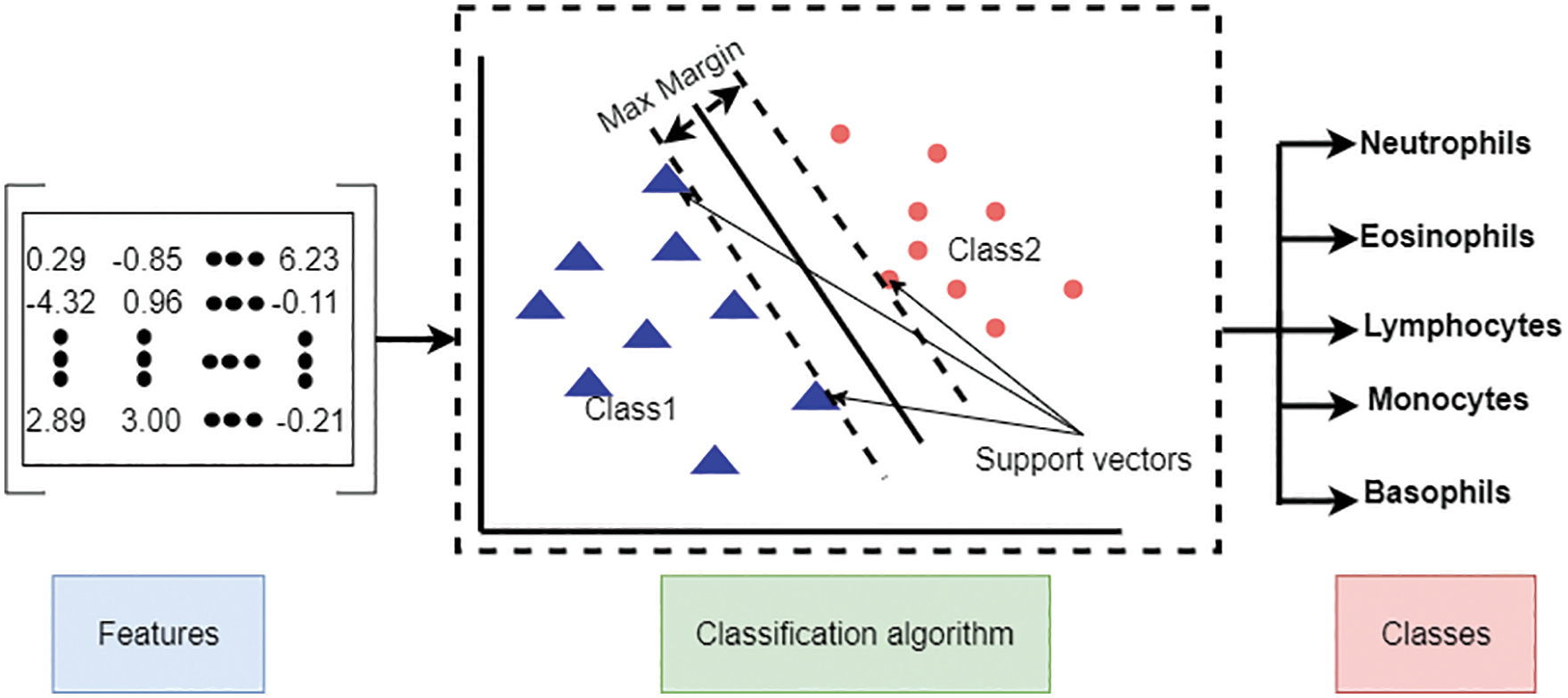
Figure 8: Shallow machine learning classification
The proposed model is thoroughly examined to demonstrate the solutions’ efficacy and investigate the impact of combining transfer learning, self-controlled learning, and optimization. We will detail the dataset used for the proposed model, the experimental environment, parameters, and performance metrics results in the following subparagraphs.
Computer vision, image processing, neural networks, deep learning, and machine learning are used by the help of MatLab R2020a and deep learning toolboxes to implement the suggested model. The model runs on an HP Zbook workstation with Windows 10 64-bit, an i7-6820HQ processor, 32 GB of Random Access Memory (RAM), and an eight-gigabyte Double Data Rate (DDR5) Graphic Processing Unit (GPU).
The Raabin-WBC dataset [11] is used to evaluate the model. This dataset contains 16633 images available online, and two experts labelled these images. Various smears were captured using two separate cameras and two different microscopes to give the necessary diversity. The evaluation supported the holdout approach using an 80% train set and a 20% test set. Table 2 shows the number of images per class, while Fig. 9 shows the distribution of WBC type in percent of the total images in the dataset.
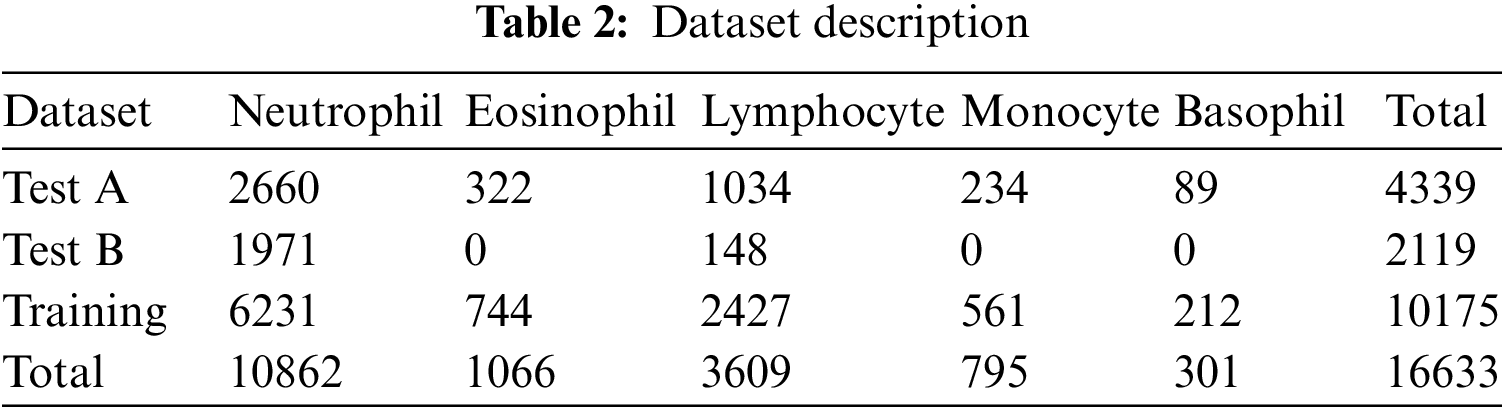
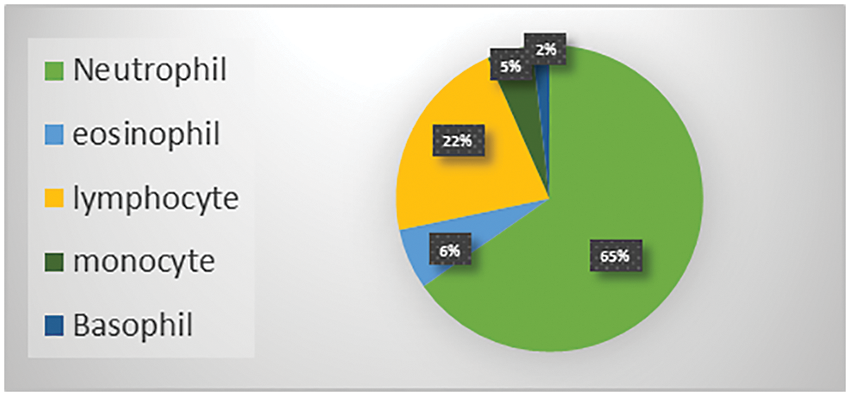
Figure 9: WBC types percent in the Raabin-WBC dataset
The experimental design includes two distinct scenarios using the benchmark dataset. The first scenario depends on utilizing pre-trained deep learning model(s) for end-to-end software WBC analysis and recognition. Five state-of-the-art pre-trained algorithms are used separately to extract features and classify WBC. Such software is considered an image classification problem, and deep learning is the most appropriate tool. Fig. 10 represents the workflow of the designed end-to-end classification model which also illustrates how the features resulted from the last layer of the model before the classification, in which a set of pre-trained models has been tuned using stochastic gradient descent. The second scenario is the experimental evaluation workflow of the proposed research methods. Fig. 11 represents the subsequent stages of the feature matrix-based classification supported by feature reduction. Table 3 represents the evaluation results of the first scenario using a deep learning pipeline.
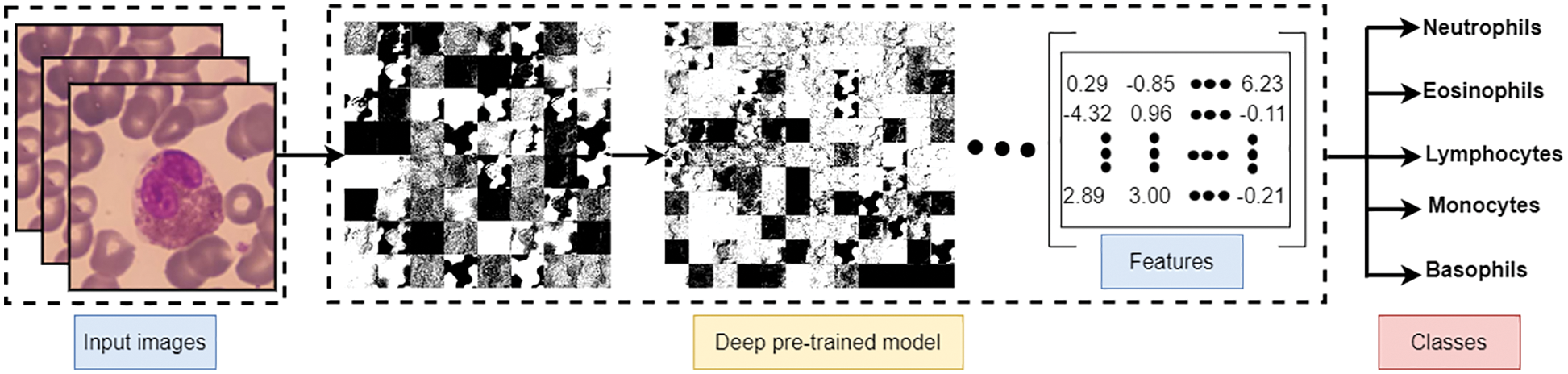
Figure 10: End-to-End evaluation classification process
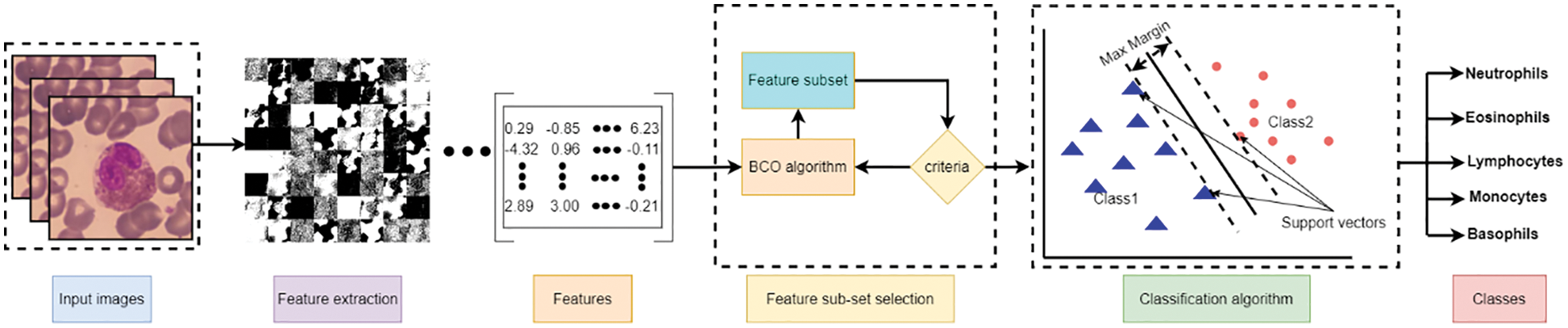
Figure 11: Feature-based evaluation classification process
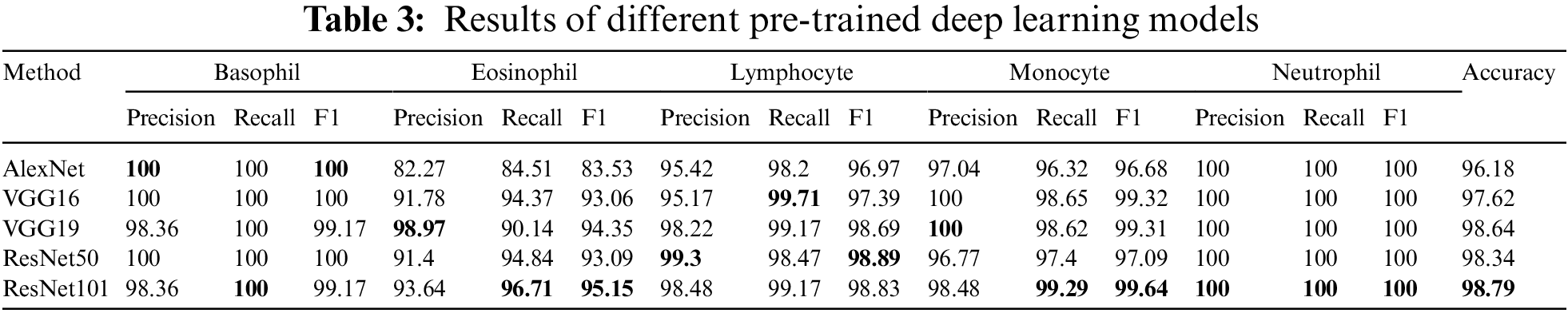
Overall, the results are amazing, and all methods have an acceptable outcome on the test set. The pre-trained ResNet101 has a superior recall of Basophil, Eosinophil, Monocyte, and Neutrophil than other models except for Lymphocyte, where VGG16 is better a little bite. Also, ResNet101 beats others in terms of accuracy by 0.15% of the pre-trained VGG19 model. For the F1-score, the pertained ResNet101 achieves higher rates for Eosinophil, Monocyte, and Neutrophil, with the highest precision for Neutrophil. Otherwise, VGG19 achieves higher precision in Eosinophil, Monocyte, as well as Neutrophil classes. ResNet50 scores better rates in precision and F1-score for the Lymphocyte. AlexNet, Vgg16, and ResNet50 reach 100% in precision, recall, and F1 score for the Basophil class. Finally, all model takes 100% for all metrics in Neutrophil as illustrated in Table 3.
Table 4 represents the evaluation results of using the features based on the tuned SVM classifier. The table includes the number of features used in the training model. The rate of feature reduction ranges from 48.58% to 50.73% of original features. The experiments include, using features matrix extracted via different pre-trained models and the reduced features produced after performing the BBCO feature selection method. The table represents the inter-comparison between the performance of the SVM classifier before and after feature reduction. Overall, SVM models have better outcomes using the reduced feature-matrix than the raw extracted feature-matrix. Generally, ResNet101-SVM beats all alternative models in terms of accuracy and reaches better precision, recall, and F1-score for Basophil, Monocyte, and Neutrophil classes. Also, it scores the best precision and F1-score for the Lymphocyte class. VGG16 is better than others in classifying Eosinophil in terms of precision and F1-score using raw feature-matrix and reduced feature-matrix in terms of recall for the Eosinophil and Lymphocyte. In addition, all models with the SVM classifier reach 100% precision for Basophil and 100% for all metrics in the Neutrophil class. Generally, it is safe to say that the feature-matrix-based algorithms can extract more meaningful features from cell images than the deep neural networks, especially after applying feature reduction using BBCO. For the training time of creating and tuning the model, deep learning has substantial time consumption, followed by the hybrid of features-based classification. The lowest time consumption is the features-based classifier after performing feature reduction by BBCO. The time-saving rate between using original features verse reduced features equals 50% on average. Fig. 12 compares different accuracy rates using BBCO for feature reduction. The proposed BBCO enhances accuracy rates overall and beats especially the recently introduced model in 2022 using ResNext50. Fig. 13 represents a comparative image illustrating the number of features per classifier. The figure confirms that using the most relevant features helps enhance the training procedure and score higher accuracy rates.
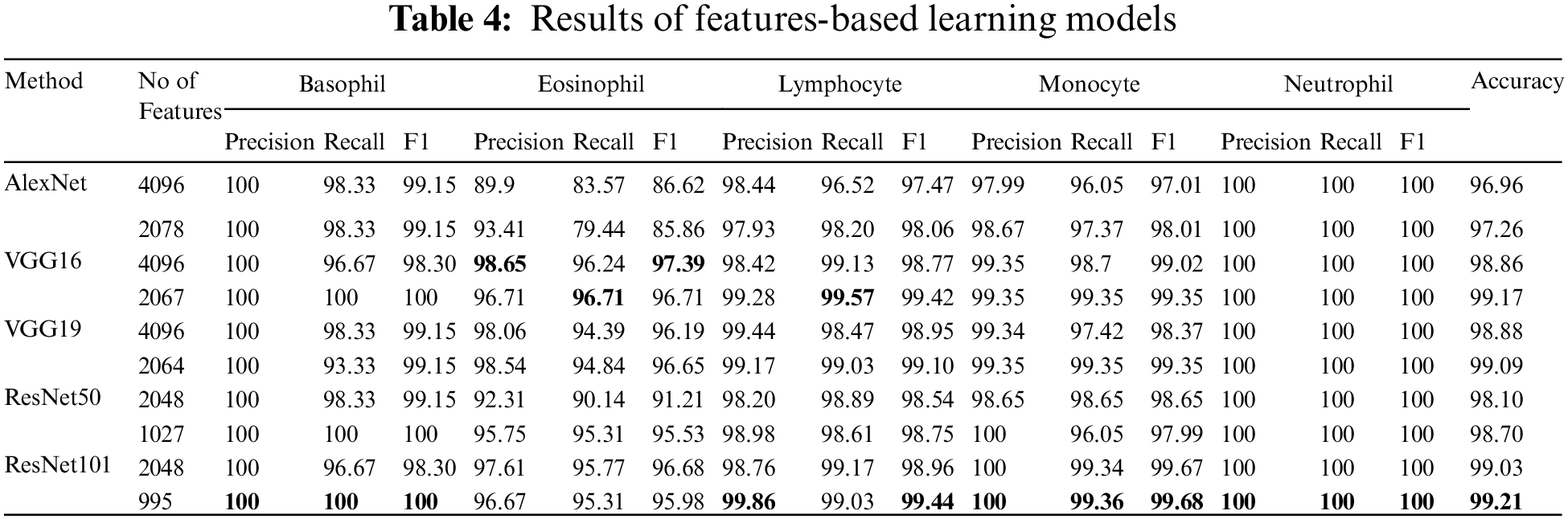
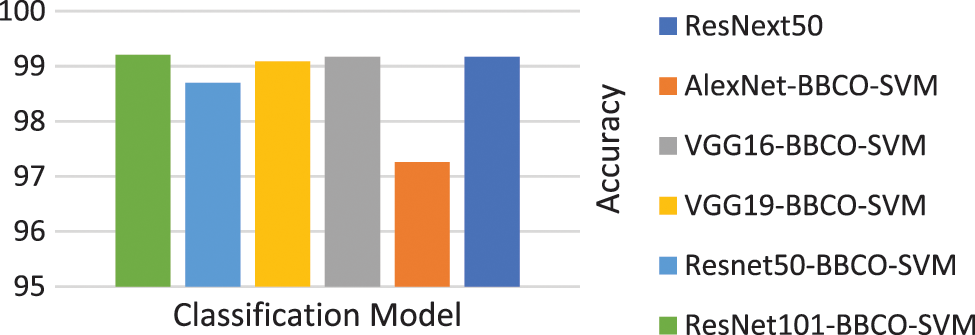
Figure 12: Comparison of accuracy rates for classification model
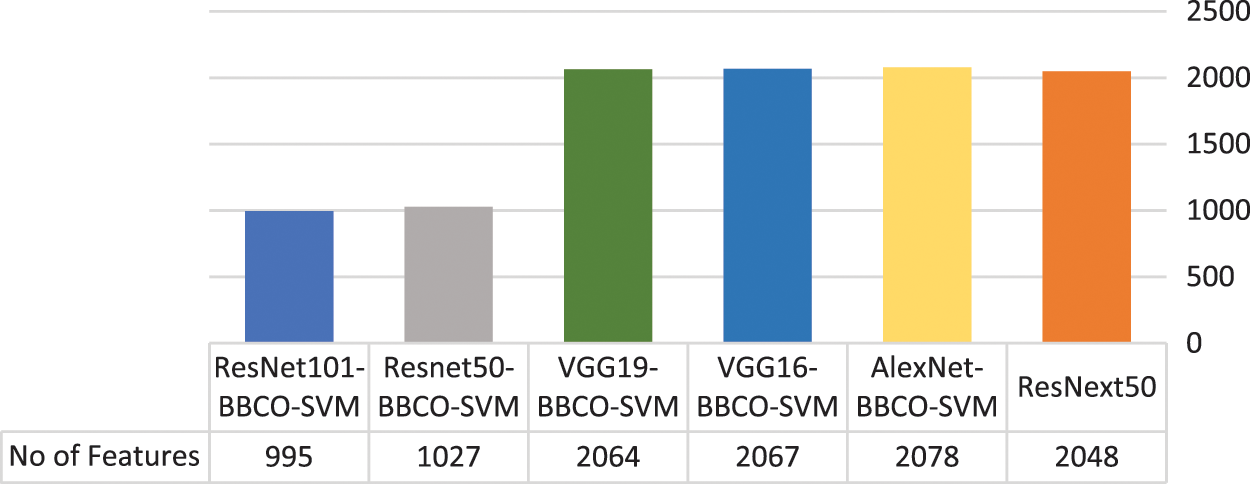
Figure 13: Number of used features per classification model
In this section, we aim to compare our proposed model (ResNet101-BBCO-SVM) with recent literature studies that deal with the same dataset (Raabin-WBC) on the detection and analysis of WBCs, as shown in Table 5. Our proposed model uses ResNet101 to extract features, the novel BBCO for selecting, and the SVM for classification. The proposed model achieved an accuracy of 99.21%, which is better than other recent works for analyzing five types of WBCs (like Basophil, Eosinophil, Lymphocyte, Monocyte, and Neutrophil) as the confusion matrix shown in Fig. 14.
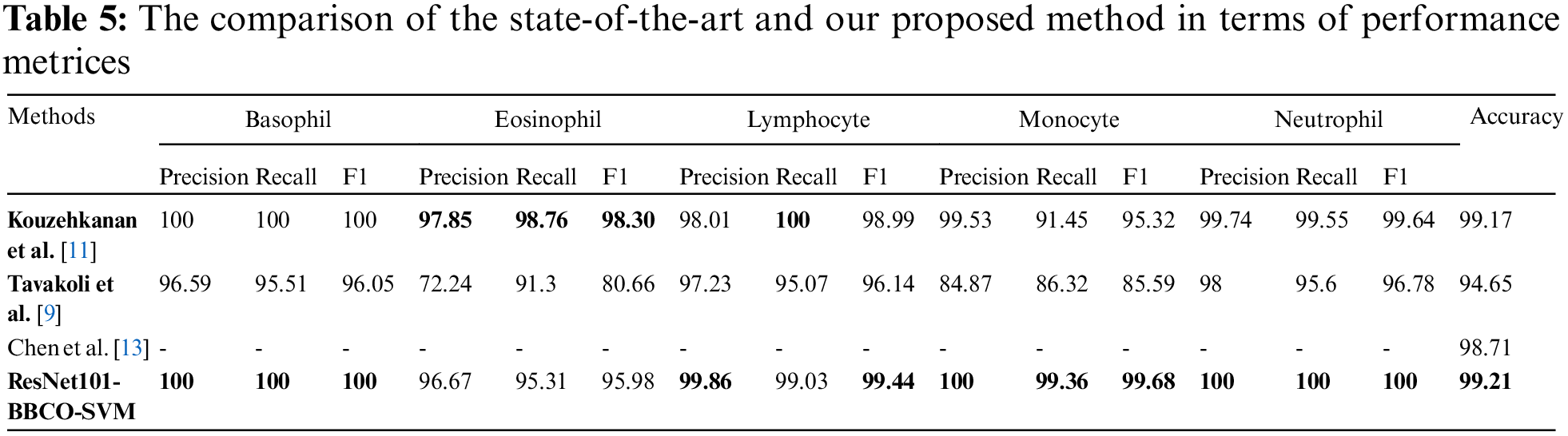
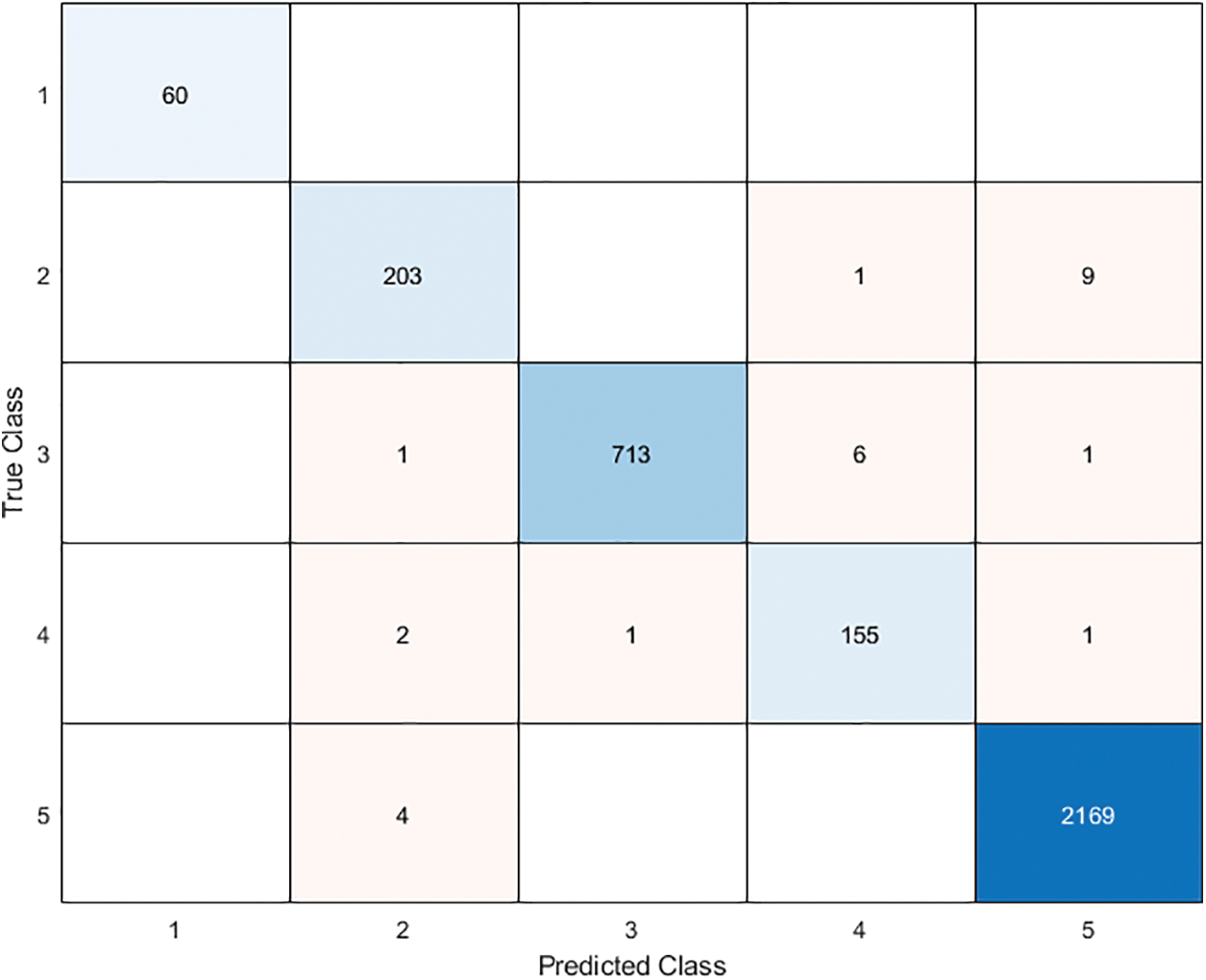
Figure 14: The proposed model ResNet101-BBCO-SVM confusion matrix
Furthermore, the proposed model beats others in recognition in terms of precision, recall, and F1 score for Basophil, Monocyte, and Neutrophil classes. Moreover, it exceeds the others in precision and F1 score in the Lymphocyte class. While in the Eosinophil class analysis, Kouzehkanan et al. achieved the best values compared to other models in terms of all metrics and recall for the Lymphocyte class. So, as the above comparison and Table 5, it is clear that our model ResNet101-BBCO-SVM is better than the other recent work on the same dataset for either the overall accuracy or the precision, recall, and F1 score metrics for each class.
The proposed model using the BBCO reduces the time used for the classification by about half, as illustrated in Table 6. This reduction decreases the complexity of the model and thus decreases the resource used by the model. Fig. 15 shows the time reduction after using the BBCO.

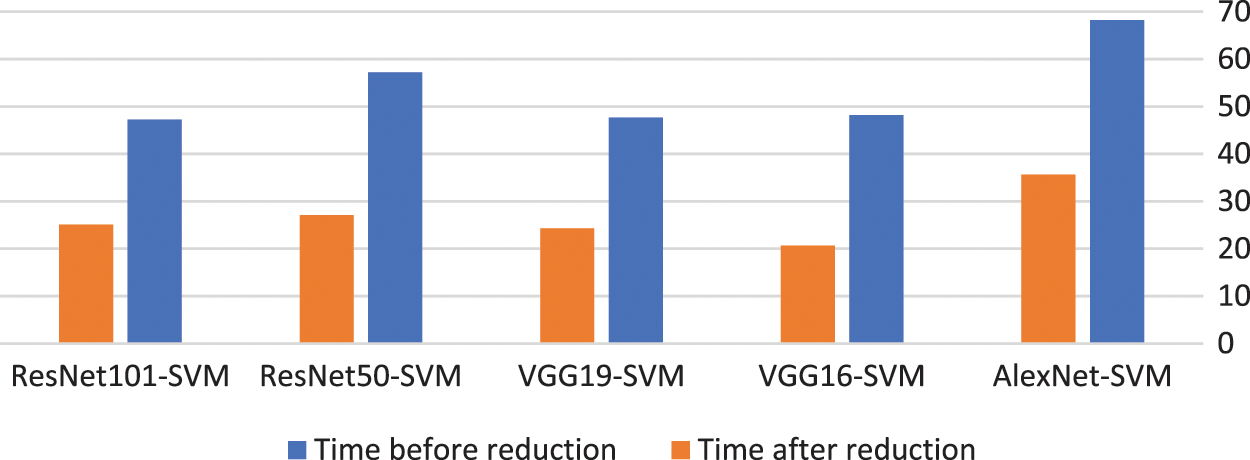
Figure 15: Time before and after using BBCO
While deep learning models are better in feature extraction, the comparison between Tables 3 and 4 shows advance for SVM in classification for each WBC class or for the entire classification. For example, in Table 4, we notice that the precision of the Basophile class for all the models reaches 100%.
One of the limitations of the proposed model is depending on one dataset. In future work, we will evaluate the proposed on different datasets.
The WBC is considered a vital issue in the blood smear domain as an indication of human health status. Since analysis of WBC depends on image processing and classification, it requires feature extraction and accuracy enhancement in minimum response time. Hence, this research focus on optimizing the extracted features, classification accuracy, and precision for indicating the types of WBC. This paper introduces a hybrid meta-heuristic features extraction with a shallow classifier based on a novel method called Border Collie Optimization (BCO) to fit the intention. The proposed model excludes irrelevant features using binary BCO optimization to increase classification accuracy. Also, the proposed model optimizes precision, and time using an SVM classifier. Hence, the proposed overall reaches superiority rates in accuracy, precision, and execution time.
The proposed model classifies various types of WBC that are started with pre-processing the images to enhance them and make them more clearly and suitable for modelling. Features extraction and reduction is performed in combination with the transfer learning technique, deep learning algotithms, and binary BCO. The pre-trained deep learning models were used to extract the features, and binary BCO to remove irrelevant features. The classification can be performed using the tuned pre-trained models including AlexNet, VGG16, VGG19, ResNet50, and ResNet101 or by using the SVM classifier for the proposed hybrid model. The BCO was binarized to select the most relevant features to reduce the feature dimension and increase classifier performance. Thus, it reduces the high resource and time consumption for training the models. The selected features were used by the classifier to measure the optimization efficiency.
This paper focuses on the features extracted from the images and mining the knowledge from them with the best classification accuracy and minimum time and resource consumption. The last one is done by the novel BBCO algorithm suggested in this paper.
The Raabin-WBC dataset was used to evaluate the proposed model in terms of feature reduction rate, classification metrics, training, and testing execution time. The benchmark included a comparison with recent literature, fine-tuned pre-trained deep learning models, and also a comparison between SVM classification before and after the feature reduction. The model ResNet101-BBCO-SVM showed an accuracy rate of 99.21%.
In future work, we will perform BBCO on different datasets belonging to different diseases. Also, different classifiers will apply to evaluate the model.
Acknowledgement: The authors thank TopEdit (www.topeditsci.com) for its linguistic assistance during the preparation of this manuscript.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. R. B. Hegde, K. Prasad, H. Hebbar and B. M. K. Singh, “Comparison of traditional image processing and deep learning approaches for classification of white blood cells in peripheral blood smear images,” Biocybernetics and Biomedical Engineering, vol. 39, no. 2, pp. 382–392, 2019. [Google Scholar]
2. J. Yao, X. Huang, M. Wei, W. Han, X. Xu et al., “High-efficiency classification of white blood cells based on object detection,” Journal of Healthcare Engineering, vol. 2021, no. Special Issue, pp. 1–11, 2021. [Google Scholar]
3. H. Kutlu, E. Avci and F. Özyurt, “White blood cells detection and classification based on regional convolutional neural networks,” Medical Hypotheses, vol. 135, pp. 109472–109489, 2020. [Google Scholar]
4. C. L. Kumar, A. V. Juliet, B. Ramakrishna, S. Chakraborty, M. A. Mohammed et al., “Computational microfluidic channel for separation of escherichia coli from blood-cells,” CMC Comput. Mater. Contin., vol. 67, no. 2, pp. 1369–1384, 2021. [Google Scholar]
5. Q. Wang, J. Wang, M. Zhou, Q. Li, Y. Wen et al., “A 3D attention networks for classification of white blood cells from microscopy hyperspectral images,” Optics & Laser Technology, vol. 139, pp. 106931–106939, 2021. [Google Scholar]
6. Y. Lu, X. Qin, H. Fan, T. Lai and Z. Li, “WBC-Net: A white blood cell segmentation network based on UNet++ and ResNet,” Applied Soft Computing, vol. 101, pp. 107006–107017, 2021. [Google Scholar]
7. A. A. Abdulsahib, M. A. Mahmoud, H. Aris, S. S. Gunasekaran and M. A. Mohammed, “An automated image segmentation and useful feature extraction algorithm for retinal blood vessels in fundus images,” Electronics, vol. 11, no. 9, pp. 1295, 2022. [Google Scholar]
8. D. Gupta, J. Arora, U. Agrawal, A. Khanna and V. H. C. de Albuquerque, “Optimized binary bat algorithm for classification of white blood cells,” Measurement, vol. 143, pp. 180–190, 2019. [Google Scholar]
9. S. Tavakoli, A. Ghaffari, Z. M. Kouzehkanan and R. Hosseini, “New segmentation and feature extraction algorithm for classification of white blood cells in peripheral smear images,” Scientific Reports, vol. 11, no. 1, pp. 1–13, 2021. [Google Scholar]
10. A. I. Shahin, Y. Guo, K. M. Amin and A. A. Sharawi, “White blood cells identification system based on convolutional deep neural learning networks,” Computer Methods and Programs in Biomedicine, vol. 168, pp. 69–80, 2019. [Google Scholar]
11. Z. M. Kouzehkanan, S. Saghari, S. Tavakoli, P. Rostami, M. Abaszadeh et al., “A large dataset of white blood cells containing cell locations and types, along with segmented nuclei and cytoplasm,” Scientific Reports, vol. 12, no. 1, pp. 1–14, 2022. [Google Scholar]
12. A. Sharma, D. Prashar, A. A. Khan, F. A. Khan and S. Poochaya, “Automatic leukaemia segmentation approach for blood cancer classification using microscopic images,” CMC-Computers Materials & Continua, vol. 73, no. 2, pp. 3629–3648, 2022. [Google Scholar]
13. H. Chen, J. Liu, C. Hua, J. Feng, B. Pang et al., “Accurate classification of white blood cells by coupling pre-trained ResNet and DenseNet with SCAM mechanism,” BMC Bioinformatics, vol. 23, no. 1, pp. 1–20, 2022. [Google Scholar]
14. E. Abdulhay, M. A. Mohammed, D. A. Ibrahim, N. Arunkumar and V. Venkatraman, “Computer aided solution for automatic segmenting and measurements of blood leucocytes using static microscope images,” Journal of Medical Systems, vol. 42, no. 4, pp. 1–12, 2018. [Google Scholar]
15. A. A. Alhabshy, B. I. Hameed and K. A. Eldahshan, “An ameliorated multiattack network anomaly detection in distributed big data system-based enhanced stacking multiple binary classifiers,” IEEE Access, vol. 10, pp. 52724–52743, 2022. [Google Scholar]
16. M. Mohri, A. Rostamizadeh and A. Talwalkar, Foundations of Machine Learning, 2nd ed. (Adaptive computation and machine learning series). Cambridge, Massachusetts: MIT press, 2018, pp. 1–412. [Google Scholar]
17. B. Ghojogh, M. N. Samad, S. A. Mashhadi, T. Kapoor, W. Ali et al., “Feature selection and feature extraction in pattern analysis: A literature review,” arXiv preprint arXiv:1905.02845, 2019. [Google Scholar]
18. A. Latif, A. Rasheed, U. Sajid, J. Ahmed, N. Ali et al., “Content-based image retrieval and feature extraction: A comprehensive review,” Mathematical Problems in Engineering, vol. 2019, no. 8, pp. 1–21, 2019. [Google Scholar]
19. A. A. Abdulsahib, M. A. Mahmoud, M. A. Mohammed, H. H. Rasheed, S. A. Mostafa et al., “Comprehensive review of retinal blood vessel segmentation and classification techniques: Intelligent solutions for green computing in medical images, current challenges, open issues, and knowledge gaps in fundus medical images,” Network Modeling Analysis in Health Informatics and Bioinformatics, vol. 10, no. 1, pp. 1–32, 2021. [Google Scholar]
20. B. Mahesh, “Machine learning algorithms-a review,” International Journal of Science and Research (IJSR). [Internet], vol. 9, no. 1, pp. 381–386, 2020. [Google Scholar]
21. C. Cortes and V. Vapnik, “Support-vector networks,” Machine Learning, vol. 20, no. 3, pp. 273–297, 1995. [Google Scholar]
22. D. A. Pisner and D. M. Schnyer, “Support vector machine,” in Machine Learning, Elsevier, Academic Press, pp. 101–121, 2020. [Google Scholar]
23. M. T. Abou-Kreisha, H. K. Yaseen, K. A. Fathy, E. A. Ebeid and K. A. ElDahshan, “Multisource smart computer-aided system for mining COVID-19 infection data,” Healthcare, 2022, vol. 10, no. 1, pp. 109–129, Multidisciplinary Digital Publishing Institute. [Google Scholar]
24. I. S. Al-Mejibli, J. K. Alwan and H. Abd Dhafar, “The effect of gamma value on support vector machine performance different kernels,” International Journal of Electrical and Computer Engineering, vol. 10, no. 5, pp. 5497–5506, 2020. [Google Scholar]
25. D. Sarkar, R. Bali and T. Ghosh, Hands-on Transfer Learning with Python: Implement Advanced Deep Learning and Neural Network Models Using TensorFlow and Keras, Packt Publishing Ltd., 2018. [Google Scholar]
26. B. Neyshabur, H. Sedghi and C. Zhang, “What is being transferred in transfer learning?” Advances in Neural Information Processing Systems, in Vancouver, Canada, vol. 33, pp. 512–523, 2020. [Google Scholar]
27. M. K. Abd Ghani, M. A. Mohammed, N. Arunkumar, S. A. Mostafa, D. A. Ibrahim et al., “Decision-level fusion scheme for nasopharyngeal carcinoma identification using machine learning techniques,” Neural Computing and Applications, vol. 32, no. 3, pp. 625–638, 2020. [Google Scholar]
28. L. Abualigah, A. Diabat, S. Mirjalili, M. Abd Elaziz and A. H. Gandomi, “The arithmetic optimization algorithm,” Computer Methods in Applied Mechanics and Engineering, vol. 376, pp. 113609–113647, 2021. [Google Scholar]
29. A. S. Desuky, M. A. Cifci, S. Kausar, S. Hussain and L. M. El Bakrawy, “Mud ring algorithm: A new meta-heuristic optimization algorithm for solving mathematical and engineering,” IEEE Access, vol. 10, pp. 50448–50466, 2022. [Google Scholar]
30. L. Abualigah, A. Diabat and Z. W. Geem, “A comprehensive survey of the harmony search algorithm in clustering applications,” Applied Sciences, vol. 10, no. 11, pp. 3827–3853, 2020. [Google Scholar]
31. L. Abualigah, “Group search optimizer: A nature-inspired meta-heuristic optimization algorithm with its results, variants, and applications,” Neural Computing and Applications, vol. 33, no. 7, pp. 2949–2972, 2021. [Google Scholar]
32. S. H. Atawneh, W. A. Khan, N. N. Hamadneh and A. M. Alhomoud, “Application of metaheuristic algorithms for optimizing longitudinal square porous fins,” CMC Computers Materials & Continua, vol. 67, no. 1, pp. 73–87, 2021. [Google Scholar]
33. A. H. Gandomi and A. H. Alavi, “Krill herd: A new bio-inspired optimization algorithm,” Communications in Nonlinear Science and Numerical Simulation, vol. 17, no. 12, pp. 4831–4845, 2012. [Google Scholar]
34. T. Dutta, S. Bhattacharyya, S. Dey and J. Platos, “Border collie optimization,” IEEE Access, vol. 8, pp. 109177–109197, 2020. [Google Scholar]
35. R. K. Godi and S. Janakiraman, “Border collie optimization algorithm-based node clustering technique in vehicular ad hoc networks,” Transactions on Emerging Telecommunications Technologies, vol. 33, no. 5, pp. 1–24, 2022. [Google Scholar]
36. A. Vijayakumar and D. Uma, “A GBDT-BCO technique based cost reduction and energy management between electric vehicle and electricity distribution system,” Energy Sources, Part A: Recovery, Utilization, and Environmental Effects, vol. 43, no. 15, pp. 1838–1852, 2021. [Google Scholar]
37. M. K. Chandol and M. K. Rao, “Border collie cat optimization for intrusion detection system in healthcare IoT network using deep recurrent neural network,” The Computer Journal, vol. 64, no. 1, pp. 1–154, 2021. [Google Scholar]
38. S. Mahajan and A. K. Pandit, “Hybrid method to supervise feature selection using signal processing and complex algebra techniques,” Multimedia Tools and Applications, vol. 80, pp. 1–22, 2021. [Google Scholar]
39. S. Mahajan, L. Abualigah, A. K. Pandit and M. Altalhi, “Hybrid aquila optimizer with arithmetic optimization algorithm for global optimization tasks,” Soft Computing, vol. 26, no. 10, pp. 4863–4881, 2022. [Google Scholar]
40. S. Liu, D. Maljovec, B. Wang, P. -T. Bremer and V. Pascucci, “Visualizing high-dimensional data: Advances in the past decade,” IEEE Transactions on Visualization and Computer Graphics, vol. 23, no. 3, pp. 1249–1268, 2016. [Google Scholar]
41. S. Mahajan and A. K. Pandit, “Image segmentation and optimization techniques: A short overview,” Medicon Eng Themes, vol. 2, no. 2, pp. 47–49, 2022. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools