 Open Access
Open Access
ARTICLE
Multi-Stage Vision Transformer and Knowledge Graph Fusion for Enhanced Plant Disease Classification
1 Computer Science, Faculty of Computer Science & Information Technology, University of Karbala, Karbala, 56001, Iraq
2 Law College, Karbala University, Karbala, 56001, Iraq
* Corresponding Author: Wafaa H. Alwan. Email:
Computer Systems Science and Engineering 2025, 49, 419-434. https://doi.org/10.32604/csse.2025.064195
Received 08 February 2025; Accepted 03 April 2025; Issue published 30 April 2025
Abstract
Plant diseases pose a significant challenge to global agricultural productivity, necessitating efficient and precise diagnostic systems for early intervention and mitigation. In this study, we propose a novel hybrid framework that integrates EfficientNet-B8, Vision Transformer (ViT), and Knowledge Graph Fusion (KGF) to enhance plant disease classification across 38 distinct disease categories. The proposed framework leverages deep learning and semantic enrichment to improve classification accuracy and interpretability. EfficientNet-B8, a convolutional neural network (CNN) with optimized depth and width scaling, captures fine-grained spatial details in high-resolution plant images, aiding in the detection of subtle disease symptoms. In parallel, ViT, a transformer-based architecture, effectively models long-range dependencies and global structural patterns within the images, ensuring robust disease pattern recognition. Furthermore, KGF incorporates domain-specific metadata, such as crop type, environmental conditions, and disease relationships, to provide contextual intelligence and improve classification accuracy. The proposed model was rigorously evaluated on a large-scale dataset containing diverse plant disease images, achieving outstanding performance with a 99.7% training accuracy and 99.3% testing accuracy. The precision and F1-score were consistently high across all disease classes, demonstrating the framework’s ability to minimize false positives and false negatives. Compared to conventional deep learning approaches, this hybrid method offers a more comprehensive and interpretable solution by integrating self-attention mechanisms and domain knowledge. Beyond its superior classification performance, this model opens avenues for optimizing metadata dependency and reducing computational complexity, making it more feasible for real-world deployment in resource-constrained agricultural settings. The proposed framework represents an advancement in precision agriculture, providing scalable, intelligent disease diagnosis that enhances crop protection and food security.Keywords
Agriculture has been the backbone of human subsistence, developing through innovations that have propelled the production of enough food to meet the demands of a constantly growing population [1–5]. Yet, food security is still precariously affected by a combination of many factors, including climate change, a decrease in the population of pollinators, and plant diseases. Among those, plant diseases pose not only a serious threat to the world’s food security but also have serious economic impacts, particularly on smallholder farmers who depend almost entirely on crop yields [6–10]. Crop losses from plant diseases, often accentuated by insects along with other environmental stresses, have spurred a number of solutions to help reduce destruction [11–13]. The development of integrated pest management systems, which emphasize sustainable and targeted methods over conventional blanket chemical applications, has been important in this [14–18]. More recently, the rapid proliferation of mobile technologies has facilitated access to such tools on a previously unimaginable scale through the rapid adoption of smartphones globally [19,20].
Accurate classification of plant diseases is crucial for timely interventions that guarantee agricultural productivity and food security.
However, present approaches suffer serious limitations, mainly when dealing with datasets that involve inter-class similarities and intra-class variations (see Related Work). An example of such challenges is the Plant Diseases Dataset, which encompasses about 87,000 RGB images of healthy and diseased crop leaves for 38 classes. Variability in disease expression, visual overlaps between diseases, and environmental inconsistencies cause difficulties in determining the exact presence of diseases using even state-of-the-art models of machine learning. Most of the current techniques rely on convolutional neural network (CNN)s, which, although very popular, have very limited capabilities for capturing global relationships in images and are not really capable of incorporating domain-specific knowledge. Besides, most state-of-the-art solutions consider only image-based features and do not include contextual information that might lead to a significant enhancement in the classification performance. This calls for a gap in terms of a robust, interpretable framework that integrates advanced feature extraction with domain knowledge in handling such complexities.
This study addresses this gap by proposing a hybrid framework for classification, which fuses the EfficientNet-B8, Vision Transformer (ViT), and knowledge graph fusion (KGF), as shown in Fig. 1. The convolutional structure of EfficientNet-B8 extracts fine-grained local features from the leaf images, while the global perspective is ensured by the capture of high semantic features and long-range dependencies, enabled by a self-attention mechanism in the ViT. Complementary to these deep features, the domain-specific contextual insights are imbued into the model through the knowledge graph extracted from disease metadata on crop types, environmental factors, and inter-disease relationships. It enables the integration of a framework that merges both local and global visual patterns with metadata-driven correlations for a full-fledged, robust classification of plant diseases.
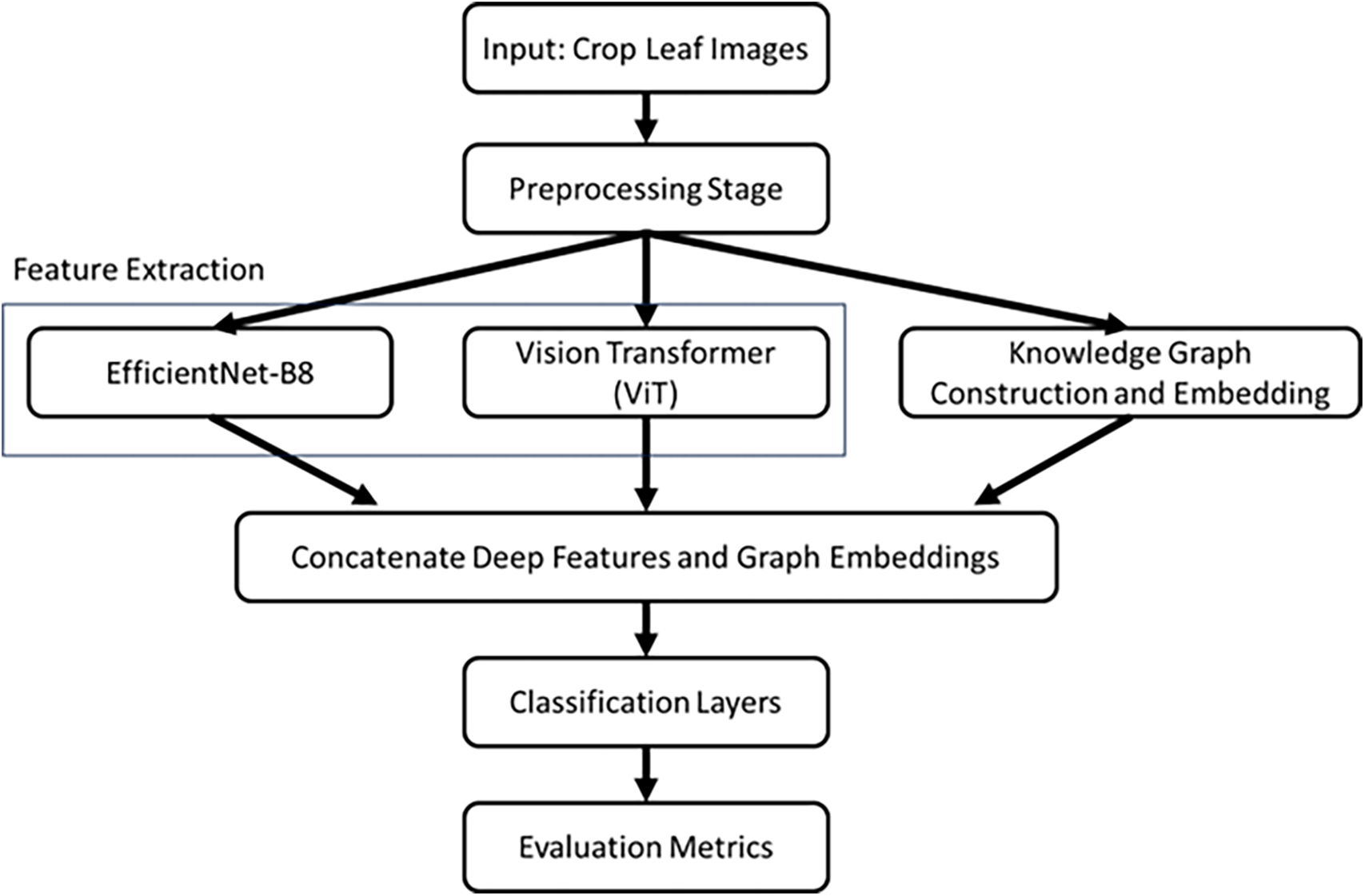
Figure 1: Flowchart of the proposed hybrid classification framework
The key novel contributions of this study are as follows. First, we introduce a hybrid deep learning framework that synergistically integrates convolution-based EfficientNet-B8, self-attention-driven ViT, and domain-specific KGF, providing a comprehensive feature representation that surpasses conventional CNN-based models. Unlike existing approaches that rely solely on image-based features, our framework effectively incorporates contextual knowledge, allowing for improved interpretability and enhanced classification performance. Second, the proposed method addresses the critical challenge of inter-class similarities and intra-class variations in plant disease classification by leveraging the complementary strengths of EfficientNet-B8 and ViT, where the former captures intricate local details and the latter extracts high-level semantic relationships through long-range dependencies.
Sanath Rao et al. [21] have illustrated the use of transfer learning techniques for the endorsement of leaf disease recognition in grape and mango plants. Besides considering Visual Geometry Group 16 (VGG-16) and ResNet-50 models, certain aspects of the two models were contrasted as well. From the findings of this study, it was noted that CNN models that had been fine-tuned were capable of achieving high accuracy in identifying the diseases in the images of mango and grape leaves. The computations of the VGG-16 model portrayed an accuracy of 96.13% and 96.25% for grape and mango leaves, respectively, while 97.34% and 97.17% were calculated for the ResNet-50 (Residual Network) model on grape and mango leaves, respectively. According to Goyal et al. [22], a deeper CNN architecture was suggested for the classification and detection of leaf and spike wheat diseases. The authors state that their proposed architecture compares unfavorably with VGG-16 and ResNet-50 on any of these criteria and thus is superior when it comes to accuracy, precision, and recall. As for the wheat-diseased dataset, the researchers fine-tuned their CNN model using transfer learning methods, which ultimately improved its accuracy in diagnosing the disease. Out of all the various diseases, the authors classified them into seven; powdery tan spot and mildew being some of them. For detecting and classifying wheat diseases, a classification accuracy of 96.5% was attained by the proposed model. It was proven through this work that these models based on deep CNN could identify the intricate disorders in wheat. A deep learning approach to classifying cassava leaf disease was suggested by Ayu et al. [23]. A CNN was trained on a set of 4238 images, which consisted of both healthy and infected cassava leaves utilizing transfer learning. The accuracy achieved by the model to classify the cassava leaf infections was 97.37 percent. This focus excelled other models in terms of accuracy; however, the authors do. The study concluded that deep learning had the capability to accurately and reliably detect infections in plants. Overall, the research reviewed a deep learning-based approach for diagnosing cassava leaf diseases and improving disease management techniques in agriculture. As Zhuang (2021) [24] used the ViT model, images of cassava leaves containing 4 diseased classes along with 1 healthy class were collected by the authors. Untouched images were subsequently processed before introducing them to a model—Vision Transformer. An accuracy rate of 96.23% was achieved using these methods; however, the method still surpassed most achievements. Nishad et al. [25] in another instance also applied k-means segmentation followed by deep learning networks in identifying and classifying potato leaf diseases. K-means was used for the segmentation of images into regions with diseased and healthy tissues. Then the classification of the diseases was done with deep models including ResNet-50 and VGG-19, where VGG-19 combined with k-means segmentation performed best, achieving an accuracy result of 97.34%. The proposed system would make it easy and cheap for users to diagnose potato diseases in the field efficiently as well as cost-effectively. More datasets can be considered for the further testing of the system, and in this way augmenting the accuracy of the system, the article also proposes. Pan et al. [26] came up with a diagnosis system for northern corn leaf blight that is based on deep learning. The model is trained and tested using images of the leaves of corn. The authors have fine-tuned a pre-trained Inception V3 model to be a deep feature extraction model. Model performance was quantified based on metrics of accuracy, precision, and F1 score with an accuracy of 96.43%. This study concluded that the proposed model was able to detect leaf diseases and therefore is suitable for use in precision agriculture crop disease diagnosis. Umamageswari et al. [27] designed six models to express 18 classes related to the diseases of banana plants based on the images of respective plant portions. They tested several models, including ResNet-50, MobileNetV1, and Inception V2, and the former two have a greater success rate than the latter. The proposed single shot detector (SSD) MobileNet V1 was more than 90% accurate in detecting diseases and pests in banana plants.
Ishengoma et al. [28] tackled the challenge of maize plant disease detection by proposing a hybrid CNN model. Their system, which fused InceptionV3 and VGG16 architectures in a parallel structure, outperformed existing CNN models (VGG16, XceptionNet, ResNet50) by achieving a 16%–44% reduction in training time and an accuracy of 96.98%. Despite its high accuracy, the hybrid model increased system cost and computational complexity.
Sharma et al. [29] developed a deeper lightweight multiclass CNN architecture (DLMC-Net) for detecting diseases in citrus, cucumber, grape, and tomato leaves. By integrating collective blocks and a passage layer, the model mitigated the vanishing gradient problem and reduced parameter size through low-rank matrix approximation. The model attained accuracies of 93.56% for citrus, 92.34% for cucumber, 99.50% for grapes, and 95.56% for tomatoes, highlighting its effectiveness in multi-class classification.
Kaya and Gürsoy (2023) [30] proposed a multi-headed DenseNet-based architecture that fuses segmented and RGB (red, green and blue) images for plant disease detection. Their evaluation of the PlantVillage dataset, consisting of 54,183 images across 38 classes, yielded an average accuracy, recall, precision, and F1-score of 98.17%, 98.17%, 98.16%, and 98.12%, respectively. While their model demonstrated high resilience, its complexity could hinder real-time applications.
Dai et al. (2023) [31] introduced the ITF-WPI (Image and text based cross-modal feature fusion model for wolfberry pest recognition) cross-modal feature fusion model, leveraging a transformer-based structure with Pyramid Squeezed Attention (PSA) to improve multi-scale feature extraction. Their model achieved 97.98% accuracy, 93.19% F1-score, and reduced computational complexity by 30% compared to CNN-LSTM-based models, making it a competitive alternative in deep learning-based plant disease detection.
Gogoi et al. (2023) [32] suggested a 3-stage CNN architecture with transfer learning. After assessment, the acquired findings emphasized an outstanding accuracy of 94% with tenfold cross-validation, surpassing prior approaches in order to stress the efficacy of the suggested method for disease detection in rice.
Chakrabarty et al. (2024) [33] addressed the challenge of rice leaf disease identification by combining lightweight CNNs with optimized Bidirectional Encoder Representations from Transformers (BEiT). Their model surpassed previous deep learning and transformer-based approaches, including ViT, Xception, InceptionV3, DenseNet169, VGG16, and ResNet50, by achieving an F1-score of 0.97. The incorporation of local interpretable segmentation techniques further enhanced model explainability, making it a promising tool for real-time agricultural disease detection.
Shwetha et al. (2024) [34] introduce a MobileNetV3-based CNN for detecting leaf spot disease in jasmine plants. The model integrates depthwise convolution layers and max pooling for feature extraction, while conditional generative adversarial network (GAN)-based data augmentation and Particle Swarm Optimization enhance robustness. It achieves 97% training accuracy and 94%–96% test accuracy under real-world conditions, demonstrating resilience to varied lighting and camera angles. Compared to existing CNN classifiers, LeafSpotNet offers improved generalization and computational efficiency, making it a promising tool for real-time plant disease detection.
Adopting depthwise separable convolution with inverted residual blocks for the detection of illnesses in leaves in plants, Thanjaivadivel et al. (2025) [35] presented an enhanced convolutional neural network. The model is developed with regard to the morphological characteristics of color, intensity, and size for data categorization. Reaching an accuracy of 99.87% using a dataset of 39 classes of plants.
The dataset used in this work is available on Kaggle [36]. It consists of about 87k images of healthy and diseased leaves, categorized into 38 distinct classes, as shown in Fig. 2. The dataset has been structured in a way that will allow efficient training and validation processes. The dataset is divided into an 80%:20% ratio of a training set and a validation set, respectively. Care has been taken to maintain the directory structure such that all images are organized class-wise. Later, a separate directory containing 33 test images was created for prediction purposes, allowing independent evaluation of the model. The training set consists of a wide variety of crop-leaf images with different disease conditions. For example, the training set contains 2016 images of apple leaves infected with apple scab, 1987 images infected with black rot, 1760 images infected with cedar apple rust, and 2008 images of healthy apple leaves. Similarly, the dataset includes healthy and diseased images of other crops such as blueberries (1816 healthy images), cherries (1683 images with powdery mildew and 1826 healthy images), and corn (e.g., 1642 images with Cercospora leaf spot and 1907 images with common rust). Other notable entries include 1888 images of grape leaves affected by black rot, 2010 images of orange leaves infected with Huanglongbing, also known as citrus greening, and 1920 images of tomato leaves affected by early blight. This ensures a wide coverage of the conditions of the plants regarding their diseases and healthy state, hence being a strong dataset for classification purposes. For testing purposes, the dataset contains a similar number of images distributed across the same categories. For instance, the testing set consists of 504 images of apple leaves infected by scab of apples, 497 infected with black rot, 440 with cedar apple rust, and 502 healthy apple leaf images. Similarly, it consists of 454 images of healthy blueberry leaves, 421 images of cherry leaves infected with powdery mildew, and 456 healthy cherry leaf images. Other examples include grape leaves with black rot, where there are 472 images; orange leaves suffering from Huanglongbing at 503; and 480 images of tomato leaves suffering from early blight.

Figure 2: Class distribution in the datasets
3.2 Proposed Classification Approach
The proposed classification methodology in this work is a multistage framework to improve the accuracy and interpretability of plant disease detection. The first stage of the classification methodology differentiates between healthy and diseased leaves. This binary classification step lays the foundation for further steps by reducing the overall task complexity. Let the input image be denoted as
Subsequent stages further categorize the diseased leaves into specific disease types after binary classification. This kind of hierarchical classification technique will ensure subtle inter-class variations are captured by the model. Further processing of the extracted features is performed through fully connected layers, mapping the feature space onto the disease categories. For a given class c, the probability
where wc and bc are the weight vector and bias term for class c, and C is the total number of classes. This probabilistic formulation facilitates robust multi-class classification.
A key innovation in this framework is the integration of deep feature representations from EfficientNet-B8 [40,41] and ViT with domain knowledge via KGF as shown in Fig. 3. EfficientNet-B8 captures fine-grained, local visual patterns through convolutional operations, providing a rich feature vector
where
which fuses local features, global patterns, and domain knowledge.
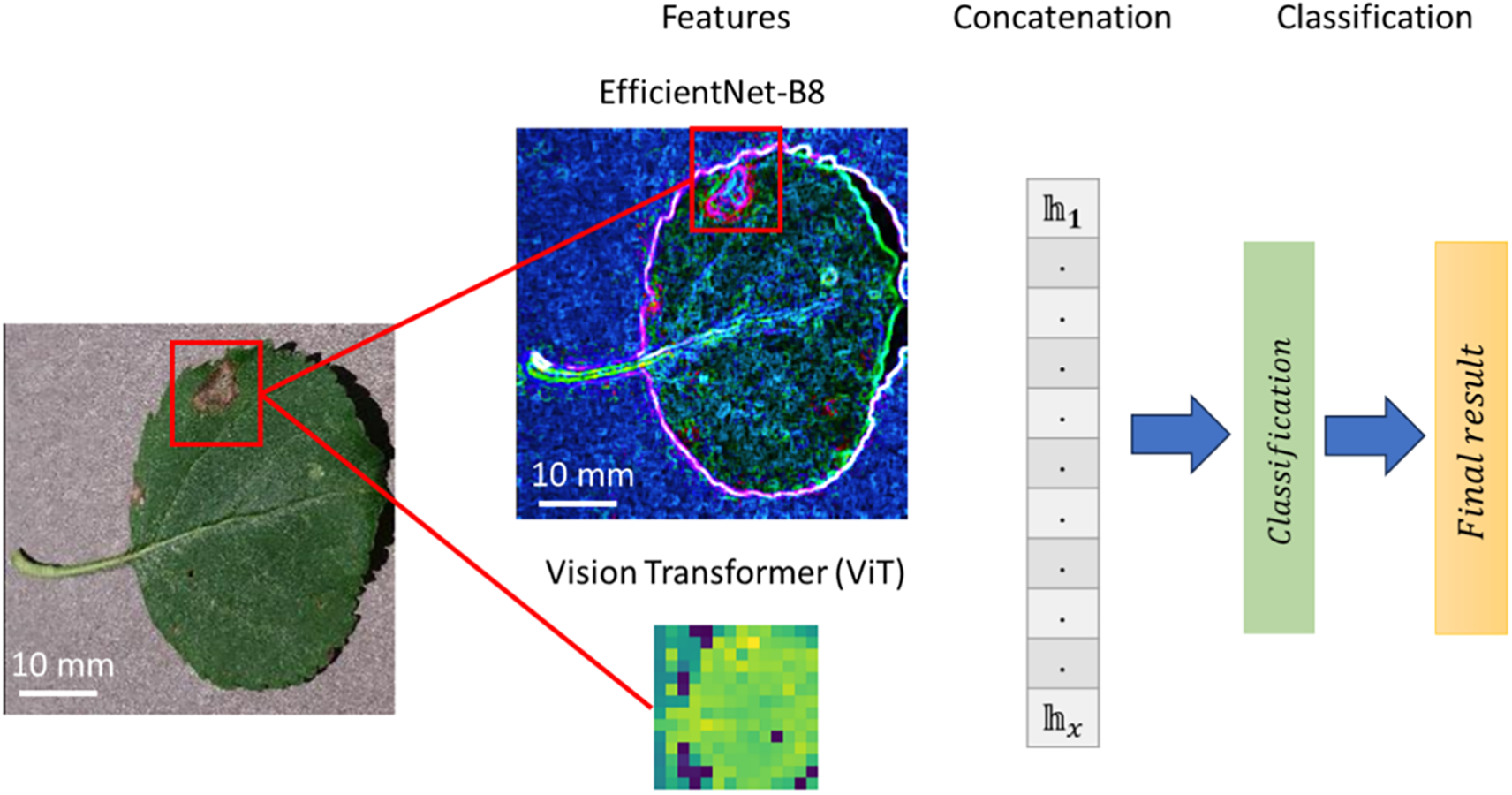
Figure 3: Illustration of the hybrid classification framework for plant disease detection
This concatenated representation hx is passed through fully connected dense layers to map it to class probabilities. For a given class c, the probability P(c∣x) is computed using the softmax function:
Cross-resolution augmentation is performed before training to handle variability in the quality of images and for robustness. This technique preprocesses images in different resolutions to improve the generalization capability of the model. The network was trained on an 80/20 train-validation split and the categorical cross-entropy loss:
where
The experiments and results presented in this study were conducted using Python in Google Colab, a cloud-based computational environment that provides access to high-performance GPUs and TPUs. The platform utilized an NVIDIA Tesla T4 or A100 GPU with a memory allocation of up to 32 GB RAM and seamless integration with Google Drive for data storage. The deep learning models were implemented using Python 3.9, with TensorFlow 2.9 and PyTorch 1.12 as the primary deep learning frameworks, while Keras was employed for high-level model construction.
The performance of the model has been evaluated using accuracy, precision, recall, and the F1-score [21,35,42]. Additionally, a confusion matrix is generated to analyze classification errors. The explainability of Grad-CAM is done to verify that the decision of the model agrees with clinically relevant features in the images.
Accuracy is the ratio of correctly classified samples to the total number of samples. It is defined as:
where:
• TP stands for True Positives (accurately anticipated positive instances).
• TN stands for True Negatives (refers to accurately anticipated negative instances).
• FP stands for False Positives (refers to mistakenly projected positive instances).
• FN stands for False Negatives (refers to improperly anticipated negative situations).
Precision refers to the fraction of accurately predicted positive samples among all anticipated positive samples. It is defined as:
Recall, also known as sensitivity, is the fraction of properly anticipated positive samples among all positive samples. It is defined as:
The F1-score:
The confusion matrix contains a thorough breakdown of predictions, including the number of TN, FN, TP, and FP classifications for each class.
Fig. 4 represents the training vs. validation accuracy and loss for the proposed model through 30 epochs. It can be analyzed that the accuracy of training increased linearly, reaching almost 100% around the 25th epoch and then remaining at 100% in the last epochs with minor fluctuations. Similarly, the validation accuracy increased gradually, reaching 99.3% at the end of the 30th epoch, which reflects that the model has generalized well on unseen data. The loss curve displays a persistent training loss decline from a high at the first epochs to stabilization in later epochs around approximately 0.1, whereas the validation loss comes down consistently and stabilizes around 0.15, although there is very slight overfitting. The close alignment of the training and validation curves underlines the robustness of the model and shows the efficiency of the proposed framework to handle such a complex dataset.
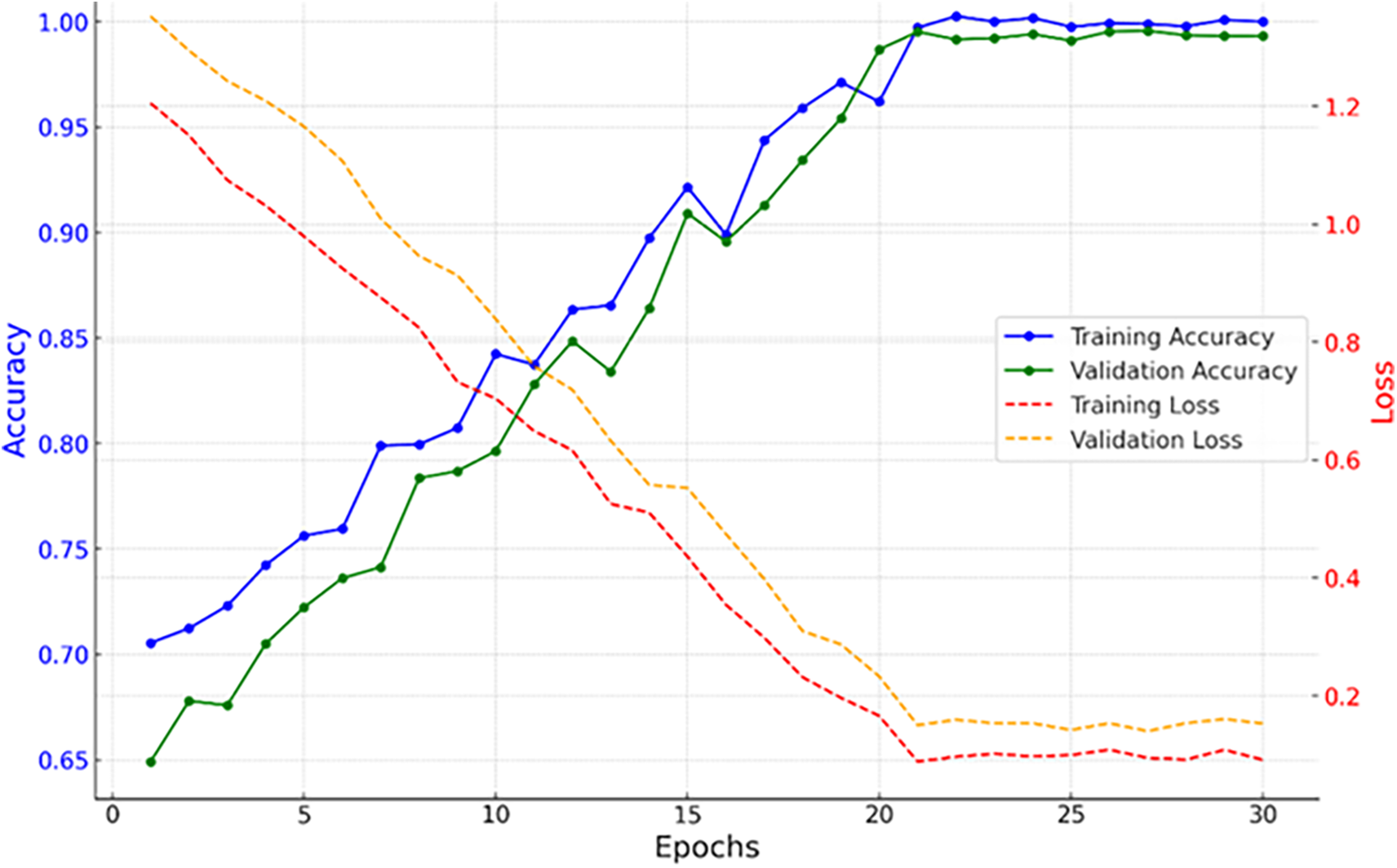
Figure 4: Training and validation performance over 30 epochs, showing accuracy and loss trends with dual y-axes
Fig. 5 illustrates the different performance metrics obtained on average for both the training and testing phases using the proposed model. In this case, the model showed outstanding results on the training with 99.7% accuracy, 99.26% precision, 99.58% recall, and 99.61% F1-score. It assures high values of these measures that support the model learned nicely from the training dataset for correct classification for all disease categories with consistency. It also fares pretty well during the testing phase with an accuracy of 99.3%, a precision of 98.9%, a recall of 98.92%, and an F1-score of 98.67%. These results indicate that the model generalizes to unseen data quite well, along with high predictive accuracy and balanced precision and recall. The slight differences in the metrics for training and testing suggest expected generalization behavior, as the model performance is almost exactly the same for both datasets. The balance here ensures the model does not overfit and thus remains reliable when applied in practical scenarios.

Figure 5: Average performance metrics of the proposed model during training and testing, represented in terms of accuracy, recall, and F1-score
Fig. 6 presents the class-wise precision and recall of the proposed plant disease classification framework in detail. The high precision and recall values from all 38 classes show the effectiveness of the model in terms of accurate and complete identification of diseases. For most of the classes, the values of precision and recall are more than 98%, indicating the efficiency of the model in reducing both false positives and false negatives. Notable examples include “Corn (maize)/Cercospora leaf spot” and “Blueberry/healthy”, both of which reach or approximate perfect scores, indicating the model is highly proficient in those respective categories. The close alignment between precision and recall values for each class further underlines the balance achieved by the model in making sure both specificity and sensitivity are attained for disease detection. Although there are minor variations within some classes, such as “Tomato/Bacterial spot” and “Apple/Black rot”, the metrics remain very well above 94%, thus justifying the model’s reliability even for challenging categories with possible inter-class similarities.

Figure 6: Class-wise precision and recall values for the proposed model, showcasing its performance across diverse plant disease categories
Table 1 shows a comparison of the proposed model performance against the state-of-the-art techniques. On the other hand, the proposed technique has used an integrated hybrid network architecture comprising EfficientNet-B8, Vision Transformer, and Knowledge Graph Fusion and performed well compared to the state-of-the-art architectures for different types of crops and diseases. The paper proposed by Nishad et al. 2022 concentrates on potato leaf diseases with VGG and ResNet models, along with k-means segmentation. While the model proposed by Nishad et al. achieved a very good accuracy of 97.34%, it had a very limited scope, and no domain-specific knowledge was integrated into it. Pan et al. (2022) proposed a fine-tuned Inception V3 model for northern corn leaf blight detection and achieved an accuracy of 96.43%. In contrast, the proposed work not only achieves a higher accuracy of 99.3% in testing but also generalizes to 38 disease classes, which proves scalability and robustness. Similarly, Umamageswari et al. (2022) utilized MobileNetV1 and SSD MobileNetV1 for banana plant disease detection, achieving 90% accuracy with SSD MobileNetV1. The proposed framework significantly outperforms this work by leveraging ViT’s ability to capture global image features and incorporating knowledge graphs for context-aware classification. Gogoi et al. (2023) introduced a 3-stage CNN with transfer learning for rice disease detection, achieving 94% accuracy through tenfold cross-validation. However, these CNNs are limited to small datasets and lack domain-specific knowledge integration, narrowing their applicability. The proposed model overcomes such limitations by considering domain-specific metadata through knowledge graph fusion and gains not only higher accuracy but also much better explainability. Thanjaivadivel et al. (2025) proposed an enhanced CNN architecture that achieved a higher accuracy of 99.87% across 39 classes with the help of advanced convolutional techniques. While true, this does not employ knowledge-based methods that contextualize the relationship between diseases or metadata. Competitive in performance at 99.3% test accuracy, the model proposes a new fusion of visual and insight-driven, knowledge-based approaches that are arguably more robust and more interpretable. Summarily, the work outperforms or at least matches the accuracies of the earlier approaches with the added advantages consequent on its hybrid design. It brings high performance, scalability, and contextual understanding together with EfficientNet-B8, ViT, and knowledge graph fusion in a way that is different from previous methods.

The proposed hybrid framework had a very promising result for plant disease classification. It has attained 99.7% for training accuracy and 99.3% for testing accuracy. Also, the precision, recall, and F1-score of all classes are relatively well-balanced, which states that the model is strong enough and scalable for various classes of plant diseases. It integrates local feature extraction with EfficientNet-B8, global pattern capture with ViT, and contextual insights with Knowledge Graph Fusion, thereby allowing the framework to outperform the existing approaches. This architecture achieves state-of-the-art accuracy with added interpretability by integrating domain knowledge, which is often missing in traditional deep learning models.
Contrasting the proposed framework with related work, the framework does not share its weaknesses. Various models, such as that proposed by Nishad et al. (2022) and that by Pan et al. (2022), did report high values of accuracy in the case of a few crops or diseases but did have limited scopes and did not make use of contextual knowledge. Similarly, works like Gogoi et al. (2023) and Umamageswari et al. (2022) could hardly be generalized when working on a particular dataset or crop. The advanced CNN architecture was proposed by Thanjaivadivel et al. (2025) with very high performance; however, they failed to provide any knowledge-based approach to enhance the contextual understanding of diseases. It overcame such lacunas by effectively combining advanced feature extraction with metadata-driven knowledge graph embeddings and thus has shown high accuracy and scalability for plant diseases into 38 classes. The comprehensive design thus lets the model handle complex patterns in diseases and similarities between classes with much efficiency.
Despite these strengths, the proposed model has certain limitations that need to be addressed in future work. In fact, knowledge graph integrations enhance the understanding of contexts; however, such a graph construction itself relies on rich metadata, which might not be accessible in all crop types or regions. Moreover, the computational cost of training and inference using both EfficientNet-B8 and ViT models can be very high, which may challenge their deployment on low-resource devices in rural farming areas. Further studies must be directed to optimizing the framework for computational efficiency without compromising its performance. Besides, the knowledge graph can further be extended for more comprehensive results by including different metadata like time-varying patterns of diseases, environmental factors enhancing the classification result, and broad applicability of models to a wide scenario.
The proposed multi-component approach advances the field by leveraging different strengths of deep learning architectures and domain knowledge, providing insights beyond traditional image-based classification. EfficientNet-B8, with its self-attention mechanisms, excels at capturing small yet critical details in leaf images, such as lesion shapes, color variations, and texture anomalies, while ViT complements this by identifying global spatial patterns, enabling the model to detect disease progression stages and structural damage in plants. KGF introduces domain-specific knowledge into the model, incorporating relationships between crop types, disease symptoms, and environmental factors, which enhances the interpretability of results and aligns with plant pathology principles. Unlike black-box deep learning models, KGF provides contextual explanations by linking predicted diseases to known agricultural phenomena, such as disease co-occurrence in certain climates or susceptibility of specific plant species. The model can distinguish visually similar diseases that previously required expert intervention, improving early detection and precision. By combining image-based learning with structured knowledge, our approach aligns predictions with plant physiology, such as how stress-induced symptoms differ from fungal or bacterial infections. This hybrid framework not only improves classification accuracy across 38 plant disease classes but also enhances interpretability, making it a valuable tool for researchers and agricultural experts.
For inference, our model demonstrates efficient real-time performance, with an average testing time of 1.1 s per image, making it feasible for deployment in real-world agricultural applications. The integration of KGF enhances interpretability without significantly increasing inference time, as the additional metadata is preprocessed and utilized efficiently. To address scalability concerns, we plan to explore lighter versions of the model by leveraging EfficientNet variants with lower computational footprints (e.g., EfficientNet-B0 to B5) and experimenting with pruned Vision Transformer architectures. Additionally, knowledge distillation techniques could be employed to optimize the model further for edge devices.
To mitigate this limitation, our framework is designed to be flexible and adaptable to varying levels of metadata availability. When metadata is missing or incomplete, the model still functions effectively using only image-based features from EfficientNet-B8 and ViT, as KGF serves as an enhancement rather than a strict requirement. Additionally, the knowledge graph can be dynamically updated and expanded over time as more agricultural data becomes available.
While our model achieves high accuracy across a diverse dataset, its real-world applicability may be influenced by variations in environmental conditions, imaging devices, and regional disease patterns. To enhance generalization, future work should validate performance on external datasets and employ domain adaptation techniques such as transfer learning and fine-tuning on region-specific data. Additionally, synthetic data augmentation and semi-supervised learning could mitigate metadata limitations, improving robustness in data-scarce environments. Our framework focuses on single-image diagnosis and does not incorporate temporal dynamics, which are crucial for tracking disease progression over time. Future work could integrate recurrent models, such as LSTMs or TCNs, to analyze sequential image data and improve long-term disease monitoring. Additionally, leveraging remote sensing or time-series vegetation indices could enhance predictive capabilities for better disease management.
The integration of temporal data, and multi-modal inputs, such as satellite imagery combined with close-up images of leaves, and environmental sensors, is another promising direction in the search for a truly robust model with respect to ever-changing conditions. Testing the scalability of the framework within diverse geographies and crop types would further demonstrate its adaptability and scalability. Overcoming these limitations and further exploring these future directions might finally allow even more robust, interpretable, and resource-efficient models for plant disease diagnosis, which could enable precision agriculture with food security.
This study presents a novel hybrid framework integrating EfficientNet-B8, Vision Transformer (ViT), and Knowledge Graph Fusion (KGF) for plant disease classification across 38 distinct disease classes. The proposed model effectively leverages deep learning and domain-specific knowledge, achieving high accuracy, precision, recall, and F1-score, demonstrating its robustness and scalability in real-world agricultural applications. EfficientNet-B8 excels at detecting fine-grained image details, while ViT captures global structural features, and KGF enhances interpretability by integrating contextual knowledge of plant species, disease relationships, and environmental factors.
The experimental results indicate that our framework significantly outperforms existing approaches by efficiently handling inter-class similarities and intra-class variations, which are major challenges in plant disease classification. Unlike traditional deep learning models, which often operate as black-box classifiers, our approach incorporates agricultural knowledge graphs, making the system more interpretable and valuable for precision agriculture applications. The ability to contextualize disease predictions ensures that the model’s decisions align with agronomic and plant pathology principles, further increasing its reliability.
Despite the strong performance, certain limitations remain. The model’s dependency on metadata availability and computational cost may limit its real-time deployment in resource-constrained environments. Additionally, while knowledge graph integration improves model explainability, constructing and maintaining an extensive and domain-specific knowledge base requires continuous updates and expert input.
For future research, optimization techniques such as model pruning and quantization can be explored to reduce computational overhead, making the framework more efficient for deployment on edge devices. Further work could also focus on integrating multi-modal data sources, such as hyperspectral imaging, climate data, and soil conditions, to enhance disease diagnosis beyond visual symptoms. Extensive field testing in diverse geographic regions with different crop varieties would further validate the model’s generalizability and adaptability to real-world agricultural settings.
This study lays the foundation for intelligent and scalable plant disease diagnostic systems, contributing to sustainable agriculture and global food security. By integrating advanced deep learning with domain-specific knowledge, the proposed framework not only enhances classification performance but also provides a more interpretable and practical solution for early disease detection in crops.
Acknowledgement: Not applicable.
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Wafaa H. Alwan, Sabah M. Alturfi; data collection: Sabah M. Alturfi; analysis and interpretation of results: Wafaa H. Alwan; draft manuscript preparation: Wafaa H. Alwan, Sabah M. Alturfi. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Pawlak K, Kołodziejczak M. The role of agriculture in ensuring food security in developing countries: considerations in the context of the problem of sustainable food production. Sustainability. 2020;12(13):5488. doi:10.3390/su12135488. [Google Scholar] [CrossRef]
2. Wijerathna-Yapa A, Pathirana R. Sustainable agro-food systems for addressing climate change and food security. Agriculture. 2022;12(10):1554. doi:10.3390/agriculture12101554. [Google Scholar] [CrossRef]
3. Alt KW, Al-Ahmad A, Woelber JP. Nutrition and health in human evolution-past to present. Nutrients. 2022;14(17):3594. doi:10.3390/nu14173594. [Google Scholar] [PubMed] [CrossRef]
4. Assan N. Socio-cultural, economic, and environmental implications for innovation in sustainable food in Africa. Front Sustain Food Syst. 2023;7:1192422. doi:10.3389/fsufs.2023.1192422. [Google Scholar] [CrossRef]
5. Sarfraz S, Ali F, Hameed A, Ahmad Z, Riaz K. Sustainable agriculture through technological innovations. In: Sustainable agriculture in the era of the OMICs revolution. Berlin/Heidelberg, Germany: Springer; 2023. p. 223–39. [Google Scholar]
6. Chakraborty S, Newton AC. Climate change, plant diseases and food security: an overview. Plant Pathol. 2011;60(1):2–14. doi:10.1111/j.1365-3059.2010.02411.x. [Google Scholar] [CrossRef]
7. Marshman J, Blay-Palmer A, Landman K. Anthropocene crisis: climate change, pollinators, and food security. Environments. 2019;6(2):22. doi:10.3390/environments6020022. [Google Scholar] [CrossRef]
8. Singh BK, Delgado-Baquerizo M, Egidi E, Guirado E, Leach JE, Liu H, et al. Climate change impacts on plant pathogens, food security and paths forward. Nat Rev Microbiol. 2023;21(10):640–56. doi:10.1038/s41579-023-00900-7. [Google Scholar] [PubMed] [CrossRef]
9. Angelotti F, Hamada E, Bettiol W. A comprehensive review of climate change and plant diseases in Brazil. Plants. 2024;13(17):2447. doi:10.3390/plants13172447. [Google Scholar] [PubMed] [CrossRef]
10. Elad Y, Pertot I. Climate change impacts on plant pathogens and plant diseases. J Crop Improv. 2014;28(1):99–139. doi:10.1080/15427528.2014.865412. [Google Scholar] [CrossRef]
11. Oliveira CM, Auad AM, Mendes SM, Frizzas MR. Crop losses and the economic impact of insect pests on Brazilian agriculture. Crop Prot. 2014;56(4):50–4. doi:10.1016/j.cropro.2013.10.022. [Google Scholar] [CrossRef]
12. Lahlali R, Taoussi M, Laasli SE, Gachara G, Ezzouggari R, Belabess Z, et al. Effects of climate change on plant pathogens and host-pathogen interactions. Crop Environ. 2024;3(3):159–70. doi:10.1016/j.crope.2024.05.003. [Google Scholar] [CrossRef]
13. Gai Y, Wang H. Plant disease: a growing threat to global food security. Agronomy. 2024;14(8):1615. doi:10.3390/agronomy14081615. [Google Scholar] [CrossRef]
14. Finger R, Sok J, Ahovi E, Akter S, Bremmer J, Dachbrodt-Saaydeh S, et al. Towards sustainable crop protection in agriculture: a framework for research and policy. Agric Syst. 2024;219(11):104037. doi:10.1016/j.agsy.2024.104037. [Google Scholar] [CrossRef]
15. Helepciuc FE, Todor A. Greener European agriculture? evaluating EU member states’ transition efforts to integrated pest management through their national action plans. Agronomy. 2022;12(10):2438. doi:10.3390/agronomy12102438. [Google Scholar] [CrossRef]
16. Karlsson Green K, Stenberg JA, Lankinen Å. Making sense of integrated pest management (IPM) in the light of evolution. Evol Appl. 2020;13(8):1791–805. doi:10.1111/eva.13067. [Google Scholar] [PubMed] [CrossRef]
17. Zhou W, Arcot Y, Medina RF, Bernal J, Cisneros-Zevallos L, Akbulut MES. Integrated pest management: an update on the sustainability approach to crop protection. ACS Omega. 2024;9(40):41130–47. doi:10.1021/acsomega.4c06628. [Google Scholar] [PubMed] [CrossRef]
18. Deguine JP, Aubertot JN, Flor RJ, Lescourret F, Wyckhuys KAG, Ratnadass A. Integrated pest management: good intentions, hard realities. A review. Agron Sustain Dev. 2021;41(3):38. doi:10.1007/s13593-021-00689-w. [Google Scholar] [CrossRef]
19. Kariyanna B, Sowjanya M. Unravelling the use of artificial intelligence in management of insect pests. Smart Agric Technol. 2024;8(6):100517. doi:10.1016/j.atech.2024.100517. [Google Scholar] [CrossRef]
20. Christakakis P, Papadopoulou G, Mikos G, Kalogiannidis N, Ioannidis D, Tzovaras D, et al. Smartphone-based citizen science tool for plant disease and insect pest detection using artificial intelligence. Technologies. 2024;12(7):101. doi:10.3390/technologies12070101. [Google Scholar] [CrossRef]
21. Sanath Rao U, Swathi R, Sanjana V, Arpitha L, Chandrasekhar K, Naik PK. Deep learning precision farming: grapes and mango leaf disease detection by transfer learning. Glob Transitions Proc. 2021;2(2):535–44. doi:10.1016/j.gltp.2021.08.002. [Google Scholar] [CrossRef]
22. Goyal L, Sharma CM, Singh A, Singh PK. Leaf and spike wheat disease detection & classification using an improved deep convolutional architecture. Inform Med Unlocked. 2021;25(2):100642. doi:10.1016/j.imu.2021.100642. [Google Scholar] [CrossRef]
23. Ayu HR, Surtono A, Apriyanto DK. Deep learning for detection cassava leaf disease. J Phys Conf Ser. 2021;1751(1):012072. doi:10.1088/1742-6596/1751/1/012072. [Google Scholar] [CrossRef]
24. Zhuang L. Deep-learning-based diagnosis of cassava leaf diseases using vision transformer. In: 2021 4th Artificial Intelligence and Cloud Computing Conference; 2021 Dec 17–19; Kyoto, Japan. p. 74–9. doi:10.1145/3508259.3508270. [Google Scholar] [CrossRef]
25. Nishad MAR, Mitu MA, Jahan N. Predicting and classifying potato leaf disease using K-means segmentation techniques and deep learning networks. Procedia Comput Sci. 2022;212(1):220–9. doi:10.1016/j.procs.2022.11.006. [Google Scholar] [CrossRef]
26. Pan SQ, Qiao JF, Wang R, Yu HL, Wang C, Taylor K, et al. Intelligent diagnosis of northern corn leaf blight with deep learning model. J Integr Agric. 2022;21(4):1094–105. doi:10.1016/S2095-3119(21)63707-3. [Google Scholar] [CrossRef]
27. Umamageswari A, Deepa S, Raja K. An enhanced approach for leaf disease identification and classification using deep learning techniques. Meas Sens. 2022;24(S1):100568. doi:10.1016/j.measen.2022.100568. [Google Scholar] [CrossRef]
28. Ishengoma FS, Rai IA, Ngoga SR. Hybrid convolution neural network model for a quicker detection of infested maize plants with fall armyworms using UAV-based images. Ecol Inform. 2022;67(12):101502. doi:10.1016/j.ecoinf.2021.101502. [Google Scholar] [CrossRef]
29. Sharma V, Tripathi AK, Mittal H. DLMC-Net: deeper lightweight multi-class classification model for plant leaf disease detection. Ecol Inform. 2023;75(7):102025. doi:10.1016/j.ecoinf.2023.102025. [Google Scholar] [CrossRef]
30. Kaya Y, Gürsoy E. A novel multi-head CNN design to identify plant diseases using the fusion of RGB images. Ecol Inform. 2023;75:101998. doi:10.1016/j.ecoinf.2023.101998. [Google Scholar] [CrossRef]
31. Dai G, Fan J, Dewi C. ITF-WPI: image and text based cross-modal feature fusion model for wolfberry pest recognition. Comput Electron Agric. 2023;212(2):108129. doi:10.1016/j.compag.2023.108129. [Google Scholar] [CrossRef]
32. Gogoi M, Kumar V, Begum S, Sharma N, Kant S. Classification and detection of rice diseases using a 3-stage CNN architecture with transfer learning approach. Agriculture. 2023;13(8):1505. doi:10.3390/agriculture13081505. [Google Scholar] [CrossRef]
33. Chakrabarty A, Ahmed ST, Islam MFU, Aziz SM, Maidin SS. An interpretable fusion model integrating lightweight CNN and transformer architectures for rice leaf disease identification. Ecol Inform. 2024;82(20):102718. doi:10.1016/j.ecoinf.2024.102718. [Google Scholar] [CrossRef]
34. Shwetha V, Bhagwat A, Laxmi V. LeafSpotNet: a deep learning framework for detecting leaf spot disease in jasmine plants. Artif Intell Agric. 2024;12:1–18. doi:10.1016/j.aiia.2024.02.002. [Google Scholar] [CrossRef]
35. Thanjaivadivel M, Gobinath C, Vellingiri J, Kaliraj S, Femilda Josephin JS. EnConv: enhanced CNN for leaf disease classification. J Plant Dis Prot. 2024;132(1):32. doi:10.1007/s41348-024-01033-6. [Google Scholar] [CrossRef]
36. Kaggle. New plant diseases dataset. [cited 2024 Feb 19]. Available from: https://www.kaggle.com/datasets/vipoooool/new-plant-diseases-dataset. [Google Scholar]
37. Dosovitskiy A, Beyer L, Kolesnikov A, Weissenborn D, Zhai X, Unterthiner T, et al. An image is worth 16 × 16 words: transformers for image recognition at scale. arXiv:2010.11929. 2020. [Google Scholar]
38. Huo Y, Jin K, Cai J, Xiong H, Pang J. Vision transformer (ViT)-based applications in image classification. In: 2023 IEEE 9th International Conference on Big Data Security on Cloud (BigDataSecurityIEEE International Conference on High Performance and Smart Computing (HPSC) and IEEE International Conference on Intelligent Data and Security (IDS); 2023 May 6–8; New York, NY, USA. p. 135–40. doi:10.1109/BigDataSecurity-HPSC-IDS58521.2023.00033. [Google Scholar] [CrossRef]
39. Borhani Y, Khoramdel J, Najafi E. A deep learning based approach for automated plant disease classification using vision transformer. Sci Rep. 2022;12(1):11554. doi:10.1038/s41598-022-15163-0. [Google Scholar] [PubMed] [CrossRef]
40. Shen A, Chen R, Zhu Y, Hu R. Segmentation of multi-organ functional tissue units using UNet-EfficientNet-B8. In: Proceedings of the 2023 4th International Symposium on Artificial Intelligence for Medicine Science; 2023 Oct 20–22; Chengdu, China. p. 232–5. doi:10.1145/3644116.3644158. [Google Scholar] [CrossRef]
41. Xie C, Tan M, Gong B, Wang J, Yuille AL, Le QV. Adversarial examples improve image recognition. In: Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR); 2020 Jun 13–19; Seattle, WA, USA. p. 816–25. doi:10.1109/cvpr42600.2020.00090. [Google Scholar] [CrossRef]
42. Naser MZ, Alavi AH. Error metrics and performance fitness indicators for artificial intelligence and machine learning in engineering and sciences. Archit Struct Constr. 2023;3(4):499–517. doi:10.1007/s44150-021-00015-8. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools