 Open Access
Open Access
ARTICLE
Identifying Cancer Disease Using Softmax-Feed Forward Recurrent Neural Classification
Department of Computer Science and Engineering, Sathyabama Institute of Science and Technology, Chennai, India
* Corresponding Author: P. Saranya. Email:
Intelligent Automation & Soft Computing 2023, 36(1), 1137-1149. https://doi.org/10.32604/iasc.2023.031470
Received 18 April 2022; Accepted 12 July 2022; Issue published 29 September 2022
Abstract
In today’s growing modern world environment, as human food activities are changing, it is affecting human health, thus leading to diseases like cancer. Cancer is a complex disease with many subtypes that affect human health without premature treatment and cause death. So the analysis of early diagnosis and prognosis of cancer studies can improve clinical management by analyzing various features of observation, which has become necessary to classify the type in cancer research. The research needs importance to organize the risk of the cancer patients based on data analysis to predict the result of premature treatment. This paper introduces a Maximal Region-Based Candidate Feature Selection (MRCFS) for early risk diagnosing using Soft-Max Feed Forward Neural Classification (SMF2NC) to solve the above problem. The predictive model is based on a different relational feature learning model, which is possessed to candidate selection to reduce the dimensionality. The redundant features are processed marginal weight rates for observing similar features’ variants and the absolute value. Softmax neural hidden layers are trained using the Sigmoid Activation Function (SAF) to create the logical condition for feed-forward layers. Further, the maximal features are introduced to invite a deep neural network constructed on the Feed Forward Recurrent Neural Network (FFRNN). The classifier produces higher classification accuracy than the previous methods and observes the cancer detection, which is recommended for early diagnosis.Keywords
Identifying the early stage of cancer is essential for diagnosing and treating cancer to reduce disease growth from the risk to the patient health. Due to the different gene expressions of cancer, the type may be varied. So the risk identification is varied due to features of the cancer category. The cancer classification using Personal Health Record (PHR) data can be used as a valuable feature source by observing more data from the patient. However, using the health care profile, the physical features are directly observed from patients, and accurate cancer classification remains challenging with the original high-dimensional features and small data model size. So we need feature reduction to reduce the dimensionality from non-related parts.
Feature selection and classification are essential in knowledge learning analyses to make decisions in real-world datasets. It is critical in identifying the disease and predicting results to categorize the dataset for Early Risk Prediction (ERP). Mainly the implementations are carried out to supportive for biomedical datasets. Feature selection (FS) and classification are well supported for biomedical data processing in the high imperative high dimensional dataset. Due to various research and implementation is not well sufficient to predict classification accuracy. We need a dedicative approach using the neural network based on the classification model to solve the classification problem. The feature mining result takes Systematic Decisions (SD) or a human thinkable approach based on the data statistics or input resources observed from the cancer patients. Due to the accurate word entitles, the data processes are challenging to process because the raw data contain high dimensionality datasets.
Fig. 1 shows the essential operation of dataset classification using the feature selection approach. Data mining in medical fields is most useful for the knowledge process for medical diagnosis to classify the results. Based on NLP (Natural language processing), the preprocessing is initialized to do datasets, or PHR information is ready to organize for optimization results. The feature selection holds the information or sets rules for selecting a type definition of data. Classification revives the type of definition to produce the optimized result.
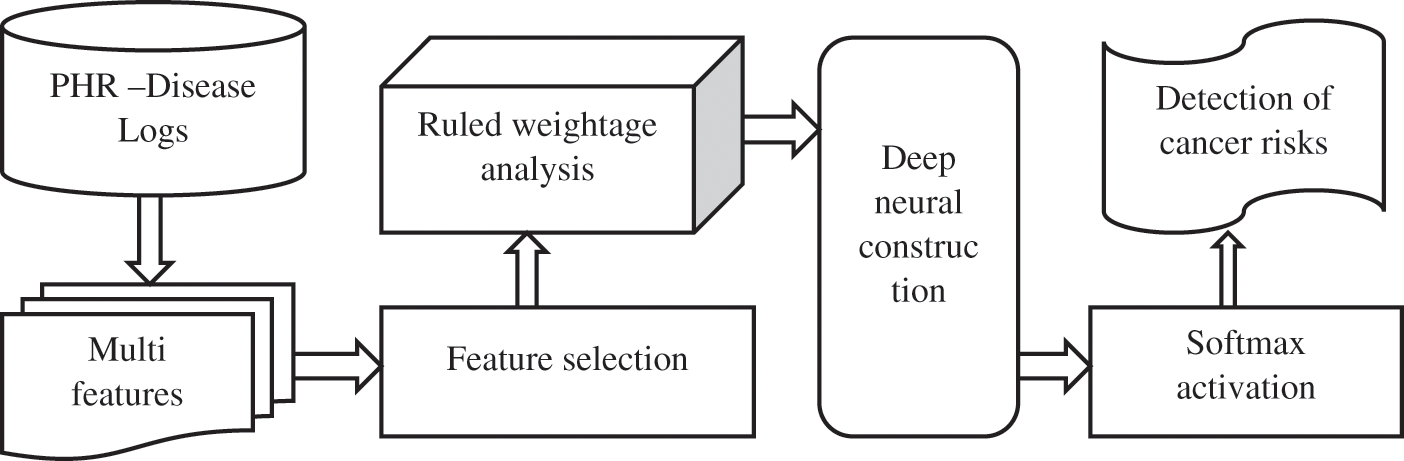
Figure 1: Process of disease prediction based on feature selection and classification
The Candidate Feature Selection Model (CFSM) frequently selects the attribute to specify the filter case. Finding the proper subset of prescient features is critical in its own particular right. For instance, the doctor may settle on a choice in light of the chosen elements, whether a risky medical procedure is essential for treatment or not. Candidate selection in directed learning has been very much considered to select the data based on disease relation. The principal objective is to discover a feature subset that produces to support higher classification exactness.
The candidate features are labelled into classes that carry the type definition using Subset Feature Classification (SFC), type data identifiers, and a wrapping class container. Finally, the feature selection has the relational disease weight information rules to define the category. The classification categorizes the class labels for the information type under the neural network. Generally, non-redundant features dropping the quantity of redundant/repetitive elements can drastically diminish the learning calculations’ running time and yields a broader classifier for accuracy. This guides in enhancing comprehension of the concealed thought of a genuine classification issue. The Artificial Neural Network (ANN) contains three processing parts, starting from input processing called the input layer, which includes multi-process to take input based on the feature selection. The centralized layer is called the hidden layer, and this may be the single or multi-perspective model set with the number of neurons trained by deep neural through a soft-max activation function. The final layer produces the output with neurons projected classification form of class. Artificial intelligence frameworks have discovered far-reaching use in disease classification, expanding feature classification.
Cancer is a serious worldwide health problem usually associated with genetic abnormalities that cause severe disease. In [1], these cancer cells form particular molecules called biomarkers, which indicate the presence of anomalies and release them into the cardiovascular system. In [2] author discusses. However, there is still a lack of tools or models to automate the analysis of human cells to determine the presence of cancer. In [3], the Author defined C-reactive protein (CRP), the serum marker of inflammation, as associated with a decrease in Heart Rate Variability (HRV), a standard medical tool for assessing autonomic function.
In [4], the Author discussed Acute Lymphoblastic Leukemia (ALL), a type of cancer in which the bone marrow produces more lymphocytes. On the other hand, Multiple Myeloma (MM) is another type of cancer in which cancer cells accumulate in the bone marrow rather than being released into the bloodstream. Therefore, they are congested and interfere with producing healthy blood cells. Traditionally, this process is done manually by skilled professionals for a considerable period. In [5], the Author describes the classifier selection and decision tree by choosing the right features to categorize cancer risk. Also, this uses the clustering ensemble approach in the minor factor considering non-important features to classify the results. In [6], the parts are classified based on the nearest neighbour classification to produce the category to categorize the data. This provides an attribute weighting factor to choose the features compared with K-Nearest Neighbor (KNN) and decision tree algorithms.
In [7], the Support Vector Machine (SVM) intends to define cancer recognition based on bagged ensembles. Still, the support values don’t produce effective feature selection because non-dimensionality leads to none related feature selection to reduce the classification accuracy. In [8], the Author described lung cancer prediction based on hidden knowledge-based techniques like naïve Bayes, Artificial neural Networks to discover the secret principle of the feature. It makes it a difficult time to predict the data. In [9], the Author describes the type-2 fuzzy logic system Fuzzy C-Means (FCM) based on the wavelet feature selection depending on the hybrid learning process. In [10], the Author describes the image processing principle-based lung cancer detection using the feature segmentation approach; the segmented value is a feature consideration to classify the weights in a feed-forward backpropagation network. The features increase the deviation principle for redundant dataset decisions, leading to inaccuracy in classification.
The automated decision and classification were based on the oral lesion using a deep learning model to detect cancer [11]. The variety makes decision forums under different Weightage to predict the result, but complex patterns were derived for tackling this difficult task.
The Self-training subspace clustering algorithm was used to find the features, and the accurate identification of data considers the low-rank features for finding the match case parts. The non-relevant features lead to inaccurate classification [12].
Multi-modality medical imaging techniques are increasingly used in clinical practice and research. Related multimedia image analysis and group learning programs are overgrowing, adding unique value to clinical applications. They were inspired by the recent success of using in-deep learning techniques in medical image processing [13]. Then select the principle of redundant features, but this doesn’t ensemble the classifier to predict relation data produce inaccuracy.
Reference [14], describes the image processing based on a multi-model deep learning process with the ensemble learning process. It selects the features based on a cross-fusion feature selection model trained with a Convolutional Neural Network. (CNN) [15].
Reference [16] introduces a 3D-classification system using neural networks and feature selection to classify heart disease based on imaging. By reducing the number of features, the number of diagnostic tests minimizes the patient’s need for a physician, but the collective feature needs the data for classification. In [17], Intensity-based statistical feature selection methods are used to predict the results used in this study of heart disease databases, showing that accuracy is superior to older classification techniques.
Health research minimizes the importance of various data processing technologies based on taxonomic features [18]. In Machine-to-Machine (M2M) correspondence from cell configurations, systems that participate in creating distances far from vehicle configurations and sensor configurations [19]. The reviews and methods compare the inaccuracy of the feature selection model under the redundant forms discussed and issues of health data mining technology [20]. In this study, Non-cellular Small lung cancer (NSCLC) is high-risk cancer usually scanned for diagnosis, diagnosis and treatment by PET-CT. However, PET-CT scans should produce at least 640 images per patient in the existing hospital setting. In [21], An optical biosensor design with high-resolution detection based on vibration holes is proposed for a Two-Domain Photonic Crystal (2DPhC) with a free spectral range near FSR = 630 nm. This sensor analyses the internal components of blood.
3 Maximal Region-Based Feature Selection and Feed-Forward Neural Classification
The contribution is to implement an early cancer prediction through a deep learning approach for indenting cancer disease from PHR data. These intent methods reduce the problematic consideration to resolve the feature selection and classification mitigations. Because the high dimensional dataset in medical diagnoses using an artificial neural network makes an easy prediction and classification system. This proposed implementation intends a new method of Identifying Cancer Disease Based on Maximal Region-Based Candidate Feature Selection Using Soft-max Feed Forward Neural Classification (SMF2NC) for Early Risk Diagnosing.
To improve the feature selection, make an efficient approach in preprocessing to point the data to reduce dimensionality, category as the classes’ label. The Multi-Attribute Case Prediction Model (MACPM) is used for categorization using the artificial neural network in profound moral to improve the classification accuracy. To deploy a Deep Neural Classier (DNC) for classification using the relational cluster based on the optimal feature selection model. The optimal feature defines the Radial Basis Function (RBF) to identify the marginal closest weight. The proposed scheme is easy to implement by redundant features that carry the performance of the present approach and is significantly superior to the competing research work for classification accuracy.
Fig. 2 shows initially, and the PHR dataset contains the multiple records collected from hospitals for cancer data analytics. It is observed in preprocessing to reduce dimensionality using Stable Auction Mapping (SAT) to prepare a noiseless null record dataset for feature selection. This selects the relevance identified portion to predict the features on varying weights to make a single cluster. At first, a couple of relational features are obtained by finding the comparability of attributes between every single member value in the dataset. Selection connectives are used in light of Maximal Stable External Region (MSER) to identify the relation to featured clustering. An additional feature selection rule is used to reduce the high-dimensional dataset.
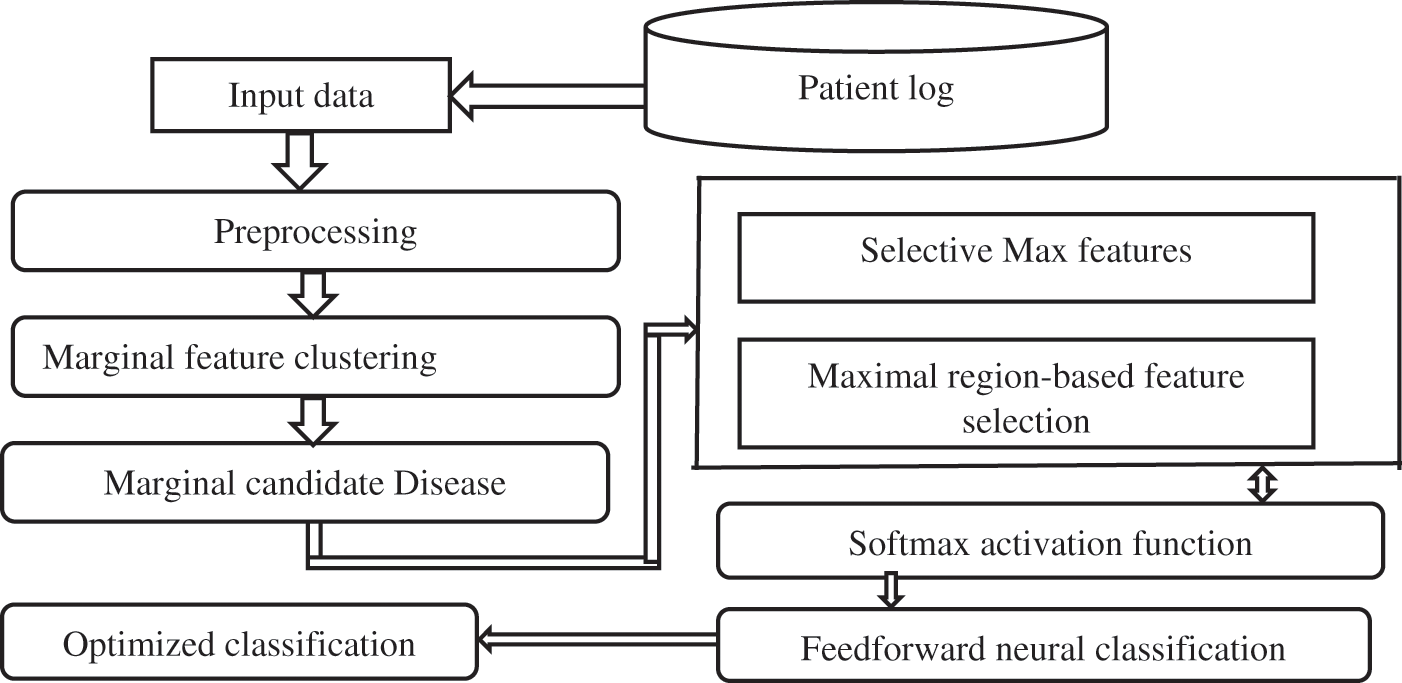
Figure 2: Architectural process SMF2NC
Further, the selected features are connected to an optimized Deep Artificial Neural Network classifier based on a feed-forward neural network. The means for each component are predicted, and a mean grid trained with multiple iterated neural in hidden layers, utilizing the average of the classification structures, is built. The features are segmented into the classification network based on the average mean value.
3.1 Marginal Feature Clustering
Initially, to process the relational feature based on marginal values, which consider the multi attributes have the same similarity. Select the identical unique features, as similar on another feature nature weights are obtained by correlated weight margins. Then create a fuzzy membership function-based rule and estimate the total mean rate between lower and upper bound limits to check the condition belongs to the feature limits and defined selected features. It creates a decision tree traversal by shifting max weights closest to the individual class.
The attribute feature selection is proposed for the interior focuses.

The above algorithm has been constructed to analyse the result’s significance and dependency. Furthermore, lower and upper approximations in fuzzy preference investigation can be interpreted as the positive and negative decision class
3.2 Marginal Candidate Disease Influence Rate
In this stage, the marginal influence rate is estimated using Ri absolute features weight. This method uses the event similarity to find the presence of features occurrence or event support measures measured according to candidate support value. The most affected influence rate is calculated by the mean rate observed from most infection rates in a cardiac feature. Also, the mean rate is referred to as features present in each event class. According to the value of Event Support, a specific event has been selected and produced as a result.
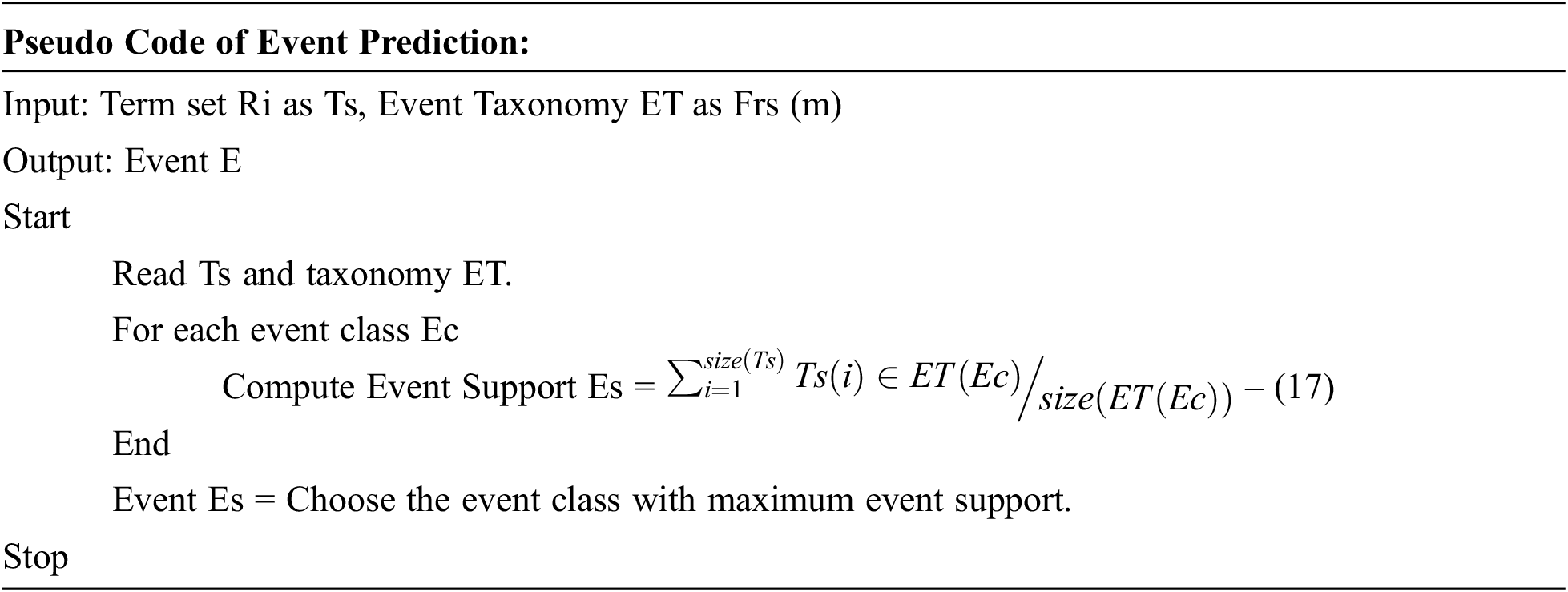
The process of occurrence event prediction is presented above the pseudocode, and the method computes the event support for various event classes. Finally, a single event has been selected as a possible event to produce a result.
3.3 Maximal Region-Based Feature Selection
This stage considers the maximal region similarity measurement by covering the boundary limit of scaling values estimated from the Event class. This creates rough groups to set the fuzzy rules for the non-deterministic as lower features. All over the relational features are maximum weight, which is closest to cardiac principle features weights are clustered. The multi attributes have the same similarity, identical to nature considered unique features similar group weightage features, identical to another feature nature. The relational features are disease-relevant margins as essential features like ESR and CRP gone with immunity weights.

Every segment vector is then utilized as an input vector, while the normal of every attribute at maximal features. Calculate the minimum
3.4 Soft-Max Feed Forward Neural Network
At this stage, the classifier finalizes the result for medical recommendation by the risk of patients and its categorized class based on the sigmoid activation function. The slight weight control initialized all the feature weights fed into the hidden layer through the softmax activation function. The activation function trains the neurons to tune the resultant clusters into mean depth values to create a logical rule for the neurons.
The feed-forward network is optimized with an adaptive forager search algorithm rule obtained as a training rule into a recurrent neural network, tuning neurons to the closest weight prediction. During this phase, the neural network can adjust the connection weights to match its output with the actual production in an iterative process until a desirable result is reached. Based on the sinusoidal weight, the importance of the Maximum feature is attained to the neural decision gradually increased; if the redundant weight is found, they spread the value into the nearest class without backpropagation to avoid link broken during training—this reduced search optimization of relevance feature fed forward into the next layer.
The activation function trains remains the f(x) =
We begin to function W approximately b. We track all the information using each corrugation function to find and repeat the corresponding prediction result and find and calculate the square error loss.
A fully integrated feed-forward of the neuron is illustrated in the construction of the network
The frequent neurons of Weightage are optimized in constant τi based on the current unit ia s valid training weightage based on the Intraclass logistic activation function. The input X (i) and Y (i) bet activated based on the weightage w (i).

The input features are trained with the classification structures. They are then labelled with neural weights, represent the classes, and the input factors aid in separating the area learning about relational features from the hidden neurons. The activation function tunes each neuron to optimize the classification accuracy by class by reference.
The Recurrent neural algorithm consists of artificial neurons activating functions that involve a hidden layer due to binary classification problems. For each neuron, the perceptron uses the activation function. Therefore, repetitive nerve cells spread relative weights along the cluster group nature to find the optimal class based on the nerve structure. The activation function reduces the number of layers in the two layers by changing the weight assigned to the perceptron that determines each neuron’s weighted input. Cognitive classes are bias weights for classifying cancer disease classes based on feature substation weights.
The proposed Maximal Region-Based Candidate Feature Selection Using Soft max-Feed Forward Neural Classification-based cancer prediction algorithms have been implemented and tested for their efficiency Python deep learning framework. The resultant parameters are tested with the confusion matrix to produce the result performance. The implementation provides higher efficiency than the previous test case results in the method Fuzzy C-Means (FCM), Convolution Neural Network (CNN). Tab. 1. shows the details of the bio-medical dataset that are processed to test the performance of the proposed systems.
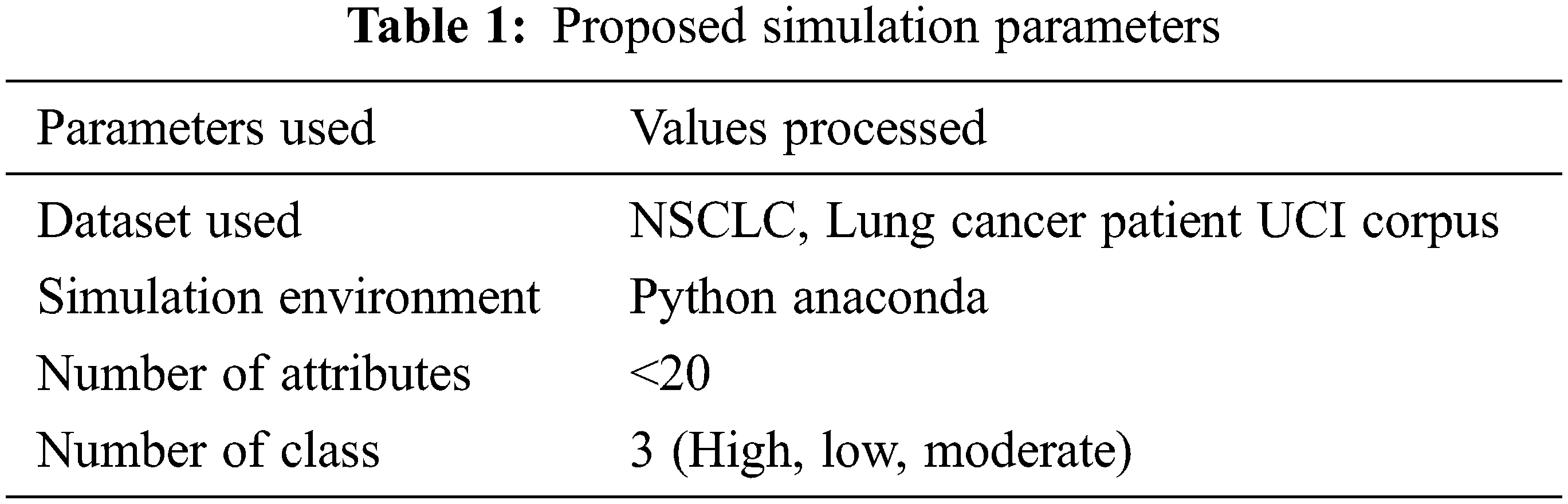
The details used for evaluating performance produced by different methods are presented in Tab. 1. Accordingly, the procedures are measured for their execution in various parameters. The results obtained have been presented in detail in this section.
The confusion matrix calculate based on True Positive (TP), False Negative (FP), True Negative (TN), and False Negative (FN).
The accuracy in classification from the big data toward disease prediction is presented in Tab. 2, where the proposed SMF2NC approach has produced higher clustering accuracy than other methods.
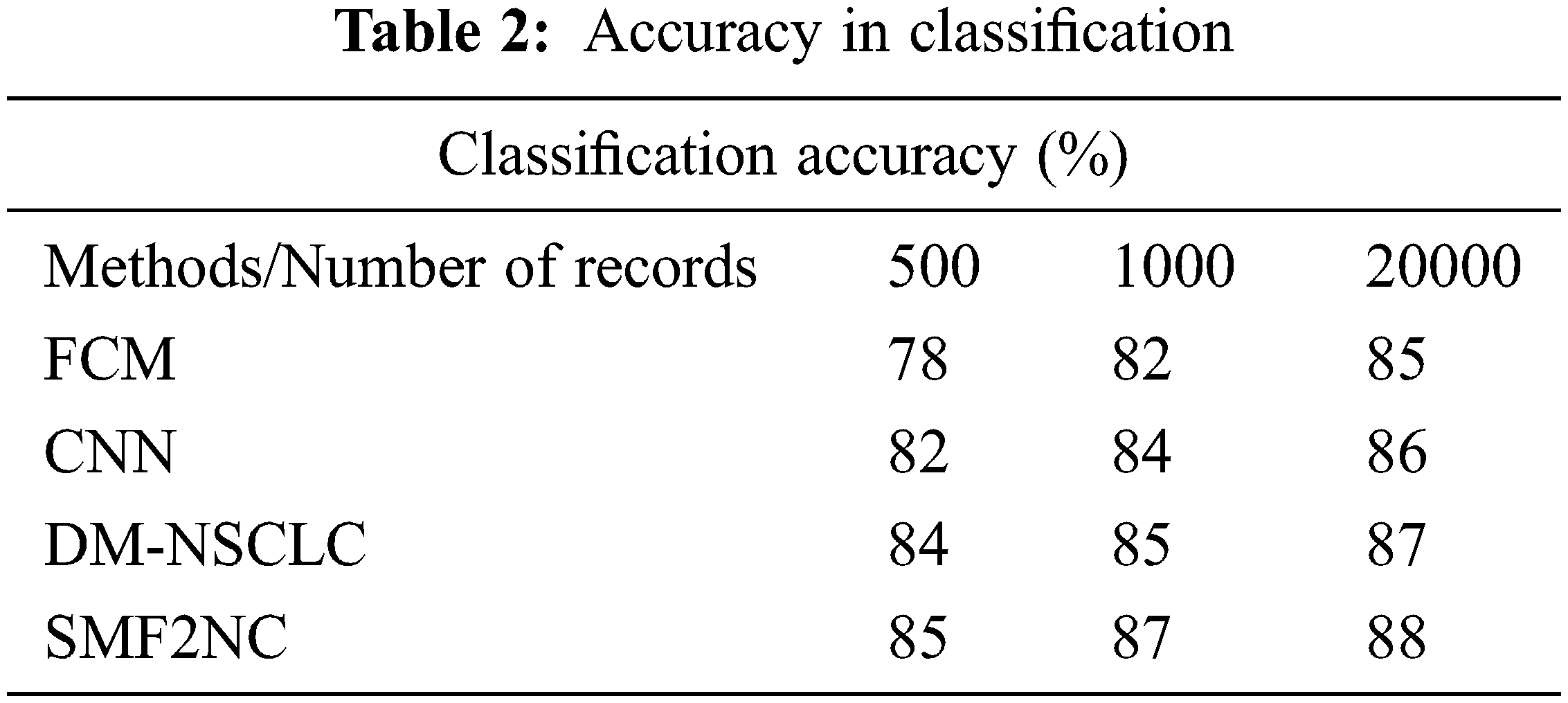
The classification accuracy performance is produced by mean rate precision and recall rate compared with different methods, as presented in Fig. 3. The proposed SMF2NC approach has made higher clustering accuracy under various diseases considered.
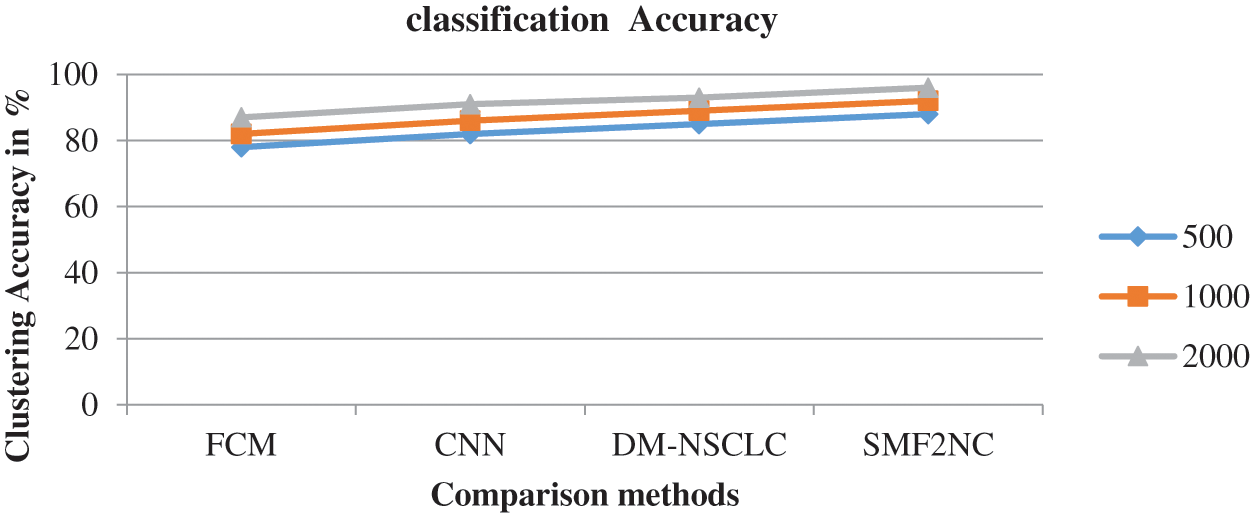
Figure 3: Accuracy in classification
The performance in disease prediction and its accuracy have been measured by considering the different disease classes. The results obtained are presented in Tab. 3. The proposed SMF2NC approach has produced higher disease prediction accuracy than other methods.
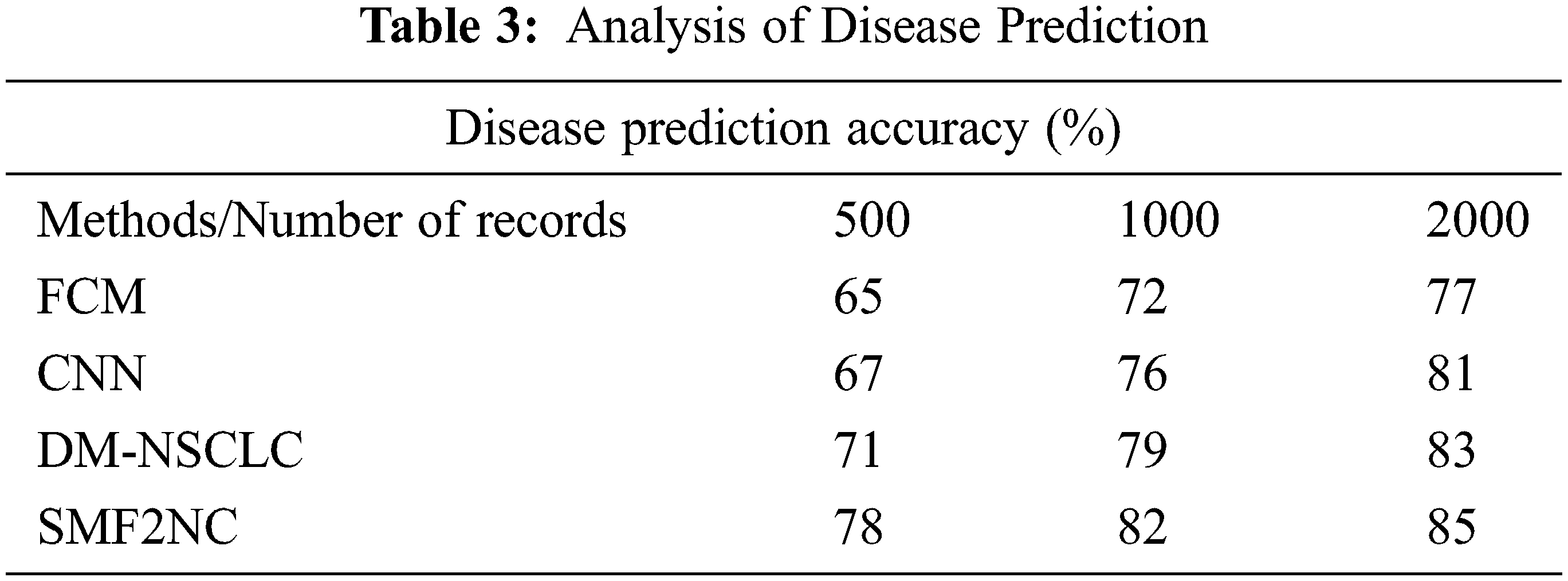
The accuracy of disease prediction produced by different methods is measured and presented in Fig. 4. The proposed SMF2NC approach has made higher disease predictions than other approaches.
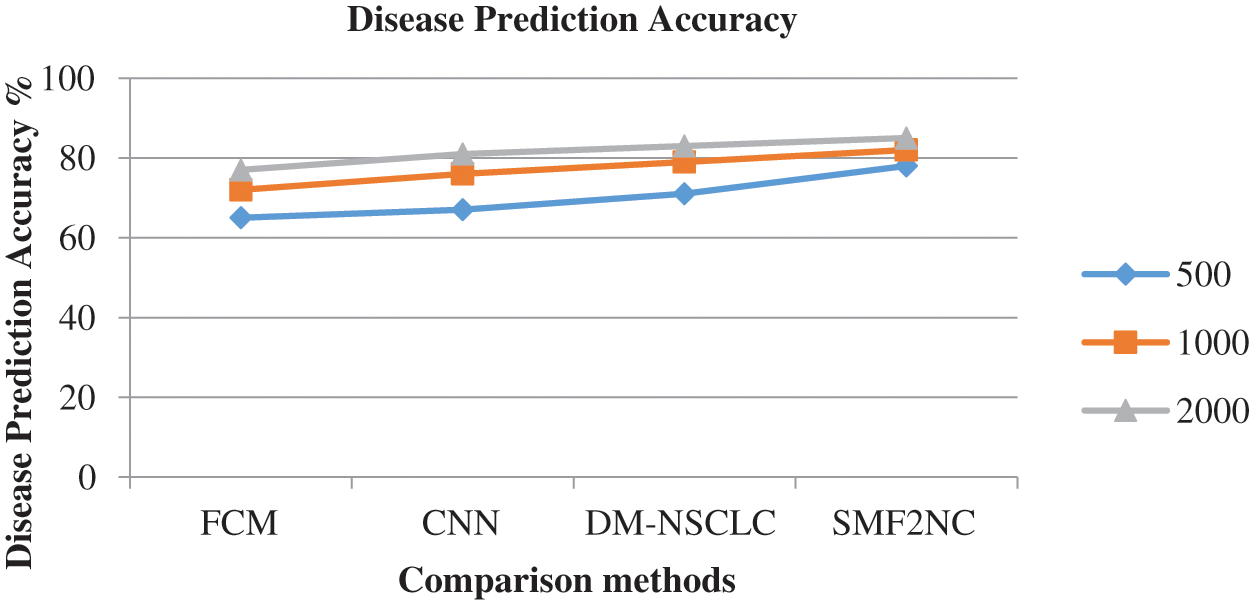
Figure 4: Analysis of disease prediction accuracy
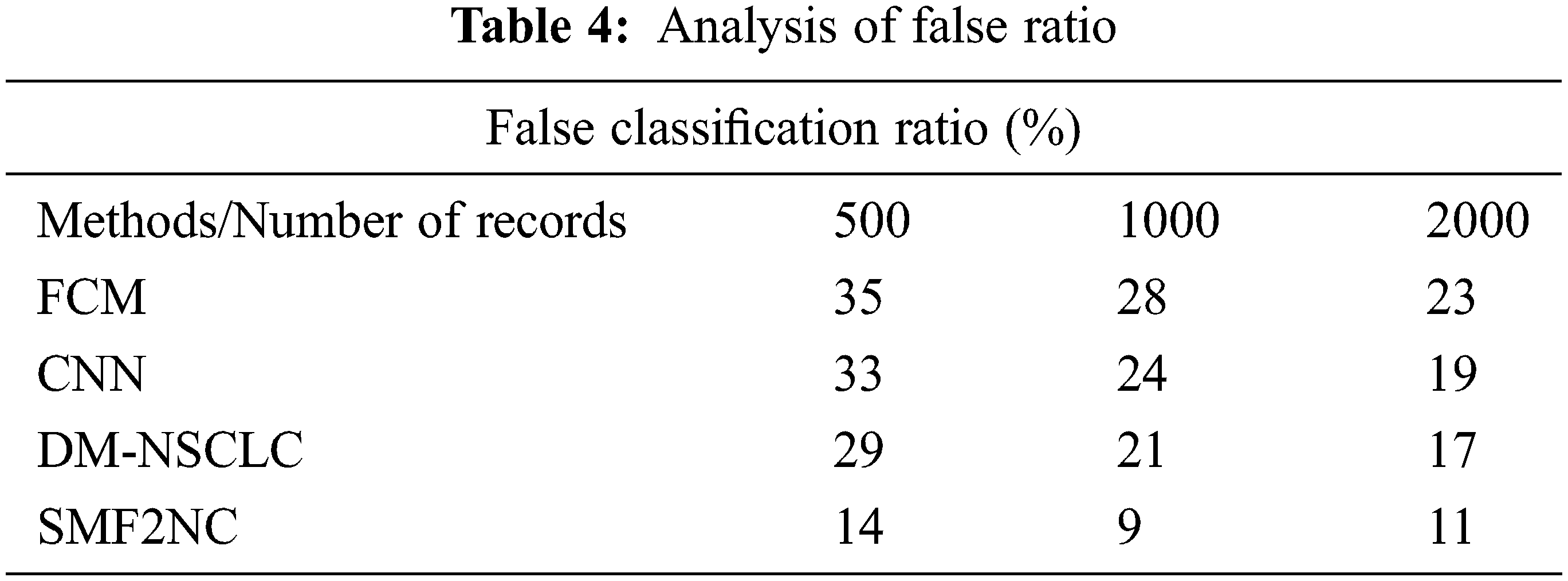
The ratio of false classification introduced by different methods is measured and presented in Tab. 4, where the proposed SMF2NC approach has produced fewer incorrect ratios than other methods.
The accuracy of disease prediction produced by different methods is measured and presented in Fig. 5. The proposed SMF2NC approach has made higher disease predictions than other approaches in each class.
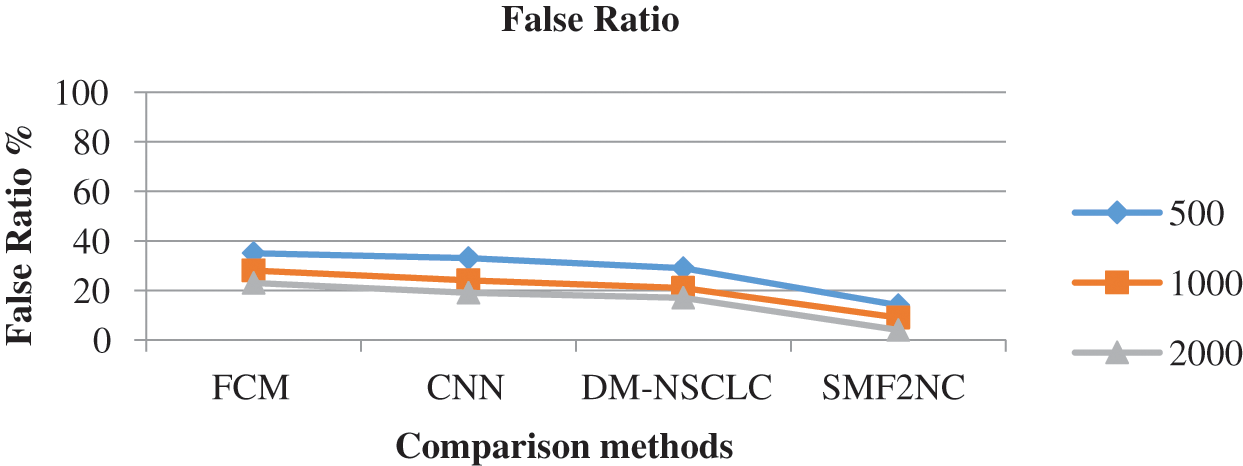
Figure 5: Analysis of false classification ratio
To conclude, cancer prediction reduces the dimensionality of the data analysis nature. It also provides its users with predictive results that give them the status that produces a premature influence rate for early diagnosis treatment. The Identifying Cancer Disease Based on Maximal Region-Based Candidate Feature Selection Using Soft max-Feed Forward Neural Classification for Early Risk Diagnosing algorithm provides higher performance based on the input provided by the feature-based classification approach. Suppose the system increases the disease prediction accuracy up to 94% well than other dissimilar methods. In that case, the percentage of people who have a portion who die from cancer will eventually decrease, even if their current state of mind is provided with their awareness.
Funding Statement: The authors received no specific funding for this study.
Conflicts of Interest: The authors declare they have no conflicts of interest to report regarding the present study.
References
1. R. Mosayebi, A. Ahmadzadeh, W. Wicke, V. Jamali, R. Schober et al., “Early cancer detection in blood vessels using mobile nanosensors,” IEEE Transactions on NanoBioscience, vol. 18, no. 2, pp. 103–116, 2019. [Google Scholar]
2. L. C. Soto-Ayala and J. A. Cantoral-Ceballos, “Automatic blood-cell classification via convolutional neural networks and transfer learning,” IEEE Latin America Transactions, vol. 19, no. 12, pp. 2028–2036, 2021. [Google Scholar]
3. L. Wang, B. Shi, P. Li, G. Zhang, M. Liu et al., “Short-term heart rate variability and blood biomarkers of gastric cancer prognosis,” IEEE Access. vol. 8, pp. 15159–15165, 2020. [Google Scholar]
4. D. Kumar, J. Nikita, A. Khurana S. Mittal, S. C. Suresh et al., “Automatic detection of white blood cancer from bone marrow microscopic images using convolutional neural networks,” IEEE Access. vol. 8, pp. 142521–142531, 2020. [Google Scholar]
5. C. Wu, Chengyue Wu, D. A. Hormuth, T. A. Oliver, F. Pineda et al., “Patient-specific characterization of breast cancer hemodynamics using image-guided computational fluid dynamics,” IEEE Transactions on Medical Imaging, vol. 39, no. 9, pp. 2760–2771, 2020. [Google Scholar]
6. F. Wu, M. H. Shen, J. Yang, H. Wang, R. Mikhaylov et al., “An enhanced tilted-angle acoustofluidic chip for cancer cell manipulation,” IEEE Electron Device Letters, vol. 42, no. 4, pp. 577–580, 2021. [Google Scholar]
7. J. Sun, Y. Yang, Y. Wang, L. Wang, X. Song et al., “Survival risk prediction of esophageal cancer based on self-organizing maps clustering and support vector machine ensembles,” IEEE Access, vol. 8, pp. 131449–131460, 2020. [Google Scholar]
8. A. Javadi, F. Keighobadi, V. Nekoukar and M. Ebrahimi, “Finite-set model predictive control of melanoma cancer treatment using signaling pathway inhibitor of cancer stem cell,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 18, no. 4, pp. 1504–1511, 2021. [Google Scholar]
9. G. Li and H. Li, “Linear model selection and regularization for serum prostate-specific antigen prediction of patients with prostate cancer using R,” IEEE Access, vol. 9, no. 1, pp. 97591–97602, 2021. [Google Scholar]
10. R. A. Welikala, R. A. Welikala, P. Remagnino, J. H. Lim, C. S. Chan et al., “Automated detection and classification of oral lesions using deep learning for early detection of oral cancer,” IEEE Access, vol. 8, pp. 132677–132693, 2020. [Google Scholar]
11. C. -Q. Xia, K. Han, Y. Qi, Y. Zhang and D. -J. Yu, “A self-training subspace clustering algorithm under low-rank representation for cancer classification on gene expression data,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 15, no. 4, pp. 1315–1324, 2018. [Google Scholar]
12. K. Mandal, R. Sarmah and D. K. Bhattacharyya, “Biomarker identification for cancer disease using biclustering approach: An empirical study,” IEEE/ACM Transactions on Computational Biology and Bioinformatics, vol. 16, no. 2, pp. 490–509, 2019. [Google Scholar]
13. Z. Guo, X. Li, H. Huang, N. Guo and Q. Li, “Deep learning-based image segmentation on multimodal medical imaging,” IEEE Trans. Radiat. Plasma Med. Sci, vol. 3, no. 2, pp. 162–169, 2019. [Google Scholar]
14. W. Sun, G. C. Zhang, X. R. Zhang, X. Zhang and N. N. Ge, “Fine-grained vehicle type classification using a lightweight convolutional neural network with feature optimization and joint learning strategy,” Multimedia Tools and Applications, vol. 80, no. 20, pp. 30803–30816, 2021. [Google Scholar]
15. W. Sun, X. Chen, X. R. Zhang, G. Z. Dai, P. S. Chang et al., “A multi-feature learning model with enhanced local attention for vehicle re-identification,” Computers, Materials & Continua, vol. 69, no. 3, pp. 3549–3561, 2021. [Google Scholar]
16. S. N. Tukimin, S. B. Karman, M. Y. Ahmad and W. S. Wan Kamarul Zaman, “Polarized light-based cancer cell detection techniques: A review,” IEEE Sensors Journal, vol. 19, no. 20, pp. 9010–9025, 2019. [Google Scholar]
17. G. Zhang, Z. Mei, Y. Zhang, X. Ma, B. Lo et al., “A noninvasive blood glucose monitoring system based on smartphone PPG signal processing and machine learning,” IEEE Transactions on Industrial Informatics, vol. 16, no. 11, pp. 7209–7218, 2020. [Google Scholar]
18. M. Sharma, J. S. Bhatt and M. V. Joshi, “Early detection of lung cancer from CT images: Nodule segmentation and classification using deep learning,” in Proc. ICMV, Vienna, Austria, pp. 1–10, 2018. [Google Scholar]
19. S. Kiranyaz, O. Avci, O. Abdeljaber, T. Ince, M. Gabbouj et al., “1D convolutional neural networks and applications: A survey,” arXiv preprint arXiv, pp. 1905.03554, 2019. [Google Scholar]
20. J. Wu, Y. Tan, Z. Chen and M. Zhao, “Decision based on big data research for non-small cell lung cancer in medical artificial system in developing country,” Computer Methods and Programs in Biomedicine, vol. 159, pp. 87–101, 2018. [Google Scholar]
21. M. H. Sani and S. Khosroabadi, “A novel design and analysis of high-sensitivity biosensor based on nano-cavity for detection of blood component, diabetes, cancer and glucose concentration,” IEEE Sensors Journal, vol. 20, no. 13, pp. 7161–7168, 2020. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools