 Open Access
Open Access
ARTICLE
STC2+ Malignant Cell State Associated with EMT, Tumor Microenvironment Remodeling, and Poor Prognosis Revealed by Single-Cell and Spatial Transcriptomics in Colorectal Cancer
1 School of Laboratory Medicine, Chongqing Medical University, Key Laboratory of Clinical Laboratory Diagnostics, Ministry of Education, Chongqing, 400016, China
2 Department of Clinical Laboratory, Women and Children’s Hospital of Chongqing Medical University, Chongqing, 401147, China
3 Department of Clinical Laboratory, Chongqing Health Center for Women and Children, Chongqing, 401147, China
* Corresponding Authors: Wuxian Li. Email: ; Min Tang. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Machine Learning for Disease Subtyping, from Molecular to Clinical Features)
Oncology Research 2026, 34(1), 24 https://doi.org/10.32604/or.2025.070143
Received 09 July 2025; Accepted 24 September 2025; Issue published 30 December 2025
Abstract
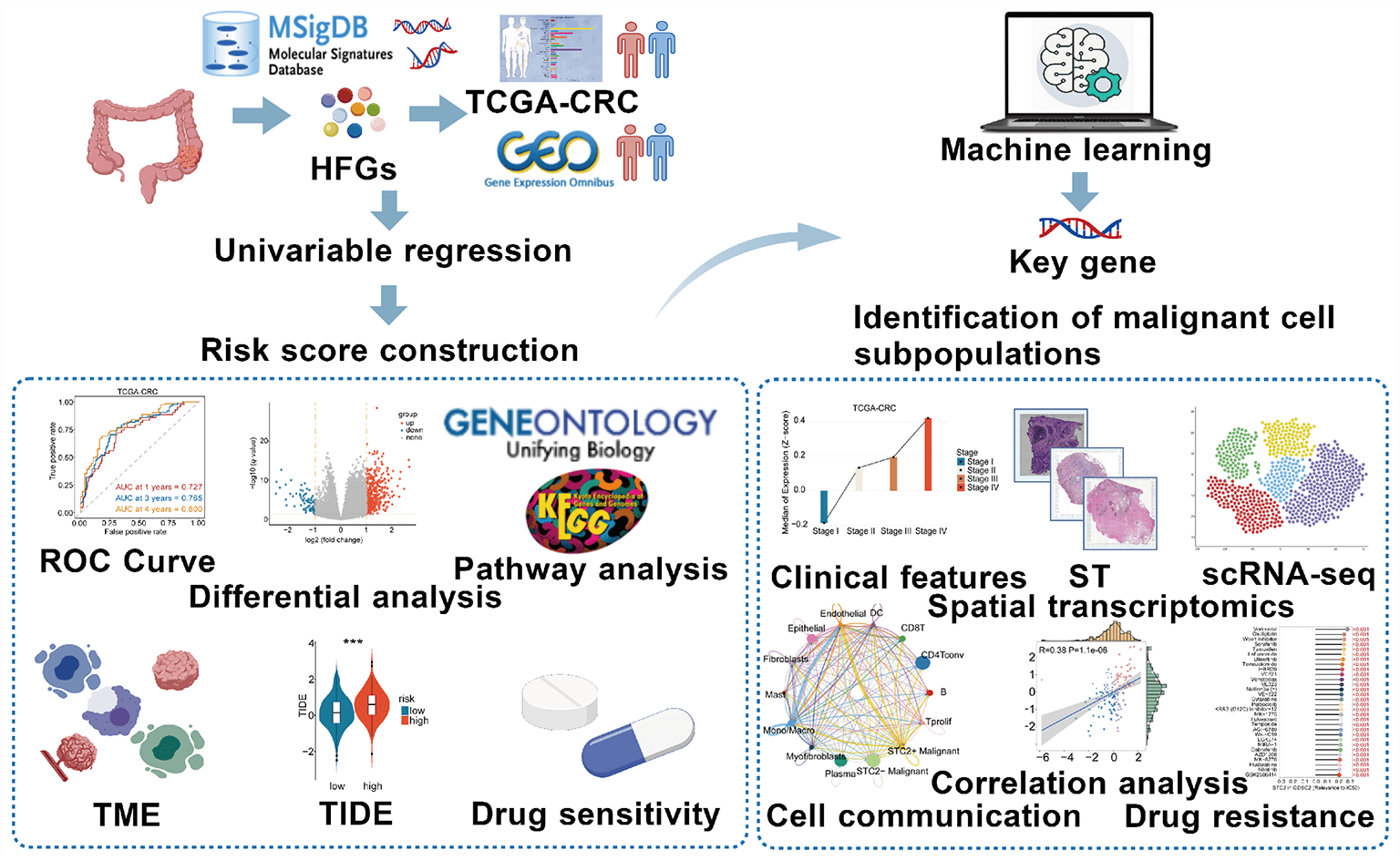
Objectives: The mechanism by which specific tumor subsets in colorectal cancer (CRC) use alternative metabolic pathways, particularly those modulated by hypoxia and fructose, to alter the tumor microenvironment (TME) remains unclear. This study aimed to identify these malignant subpopulations and characterize their intercellular signaling networks and spatial organization through an integrative multi-omics approach. Methods: Leveraging bulk datasets, single-cell RNA sequencing, and integrative spatial transcriptomics, we developed a prognostic model based on hypoxia-and fructose metabolism-related genes (HFGs) to delineate tumor cell subpopulations and their intercellular signaling networks. Results: We identified a specific subset of stanniocalcin-2 positive (STC2+) malignant cells spatially enriched within tumor regions and strongly associated with poor prognosis. This subset served as a key signaling hub in the TME, exhibiting increased epithelial–mesenchymal transition activity. STC2+ cells engage in two spatially organized ligand–receptor interactions: the growth differentiation factor 15 (GDF15)—transforming growth factor beta receptor 2 (TGFBR2) pathway targeting endothelial cells and the migration inhibitory factor (MIF)—(cluster of differentiation 74 [CD74]+C-X-C motif chemokine receptor 4 [CXCR4]) pathway targeting macrophages. Conclusion: This study identified a malignant cell state in CRC that is metabolically defined and spatially limited, including liver metastases, and is characterized by elevated STC2 expression and active immune-stromal interactions. Given the interplay between metabolic reprogramming and TME remodeling, STC2+ malignant cells are a functionally significant subpopulation and a potential therapeutic target.Graphic Abstract
Keywords
Supplementary Material
Supplementary Material FileCite This Article
 Copyright © 2026 The Author(s). Published by Tech Science Press.
Copyright © 2026 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools