 Open Access
Open Access
ARTICLE
Mutations in epigenetic regulator KMT2C detected by liquid biopsy are associated with worse survival in prostate cancer patients
Department of Urology, Institute of Urology, West China Hospital, Sichuan University, Chengdu, 610041, China
* Corresponding Authors: PENGFEI SHEN. Email: ; HAO ZENG. Email:
Oncology Research 2023, 31(4), 605-614. https://doi.org/10.32604/or.2023.028321
Received 12 December 2022; Accepted 17 April 2023; Issue published 25 June 2023
Abstract
Background: KMT2 (lysine methyltransferase) family enzymes are epigenetic regulators that activate gene transcription. KMT2C is mainly involved in enhancer-associated H3K4me1, and is also one of the top mutated genes in cancer (6.6% in pan-cancer). Currently, the clinical significance of KMT2C mutations in prostate cancer is understudied. Methods: We included 221 prostate cancer patients diagnosed between 2014 and 2021 in West China Hospital of Sichuan University with cell-free DNA-based liquid biopsy test results in this study. We investigated the association between KMT2C mutations, other mutations, and pathways. Furthermore, we evaluated the prognostic value of KMT2C mutations, measured by overall survival (OS) and castration resistance-free survival (CRFS). Also, we explored the prognostic value of KMT2C mutations in different patient subgroups. Lastly, we investigated the predictive value of KMT2C mutations in individuals receiving conventional combined anti-androgen blockade (CAB) and abiraterone (ABI) as measured by PSA progression-free survival (PSA-PFS). Results: The KMT2C mutation rate in this cohort is 7.24% (16/221). KMT2C-mutated patients showed worse survival than KMT2C-wild type (WT) patients regarding both CRFS and OS (CRFS: mutated: 9.9 vs. WT: 22.0 months, p = 0.015; OS: mutated: 71.9 vs. WT 137.4 months, p = 0.012). KMT2C mutations were also an independent risk factor in OS [hazard ratio: 3.815 (1.461, 9.96), p = 0.006] in multivariate analyses. Additionally, we explored the association of KMT2C mutations with other genes. This showed that KMT2C mutations were associated with Serine/Threonine-Protein Kinase 11 (STK11, p = 0.004) and Catenin Beta 1 (CTNNB1, p = 0.008) mutations. In the CAB treatment, KMT2C-mutated patients had a significantly shorter PSA-PFS compared to KMT2C-WT patients. (PSA-PFS: mutated: 9.9 vs. WT: 17.6 months, p = 0.014). Moreover, KMT2C mutations could effectively predict shorter PSA-PFS in 10 out of 23 subgroups and exhibited a strong trend in the remaining subgroups. Conclusions: KMT2C-mutated patients showed worse survival compared to KMT2C-WT patients in terms of both CRFS and OS, and KMT2C mutations were associated with STK11 and CTNNB1 mutations. Furthermore, KMT2C mutations indicated rapid progression during CAB therapy and could serve as a potential biomarker to predict therapeutic response in prostate cancer.Keywords
Supplementary Material
Supplementary Material FileProstate cancer is the most frequent non-skin malignancy in men [1]. Next-generation sequencing techniques facilitate the possibility of tailoring clinical treatment based on various molecular characteristics of patients, which is especially important in the case of prostate cancer since it is a highly heterogeneous disease. Therefore, deciphering the mutations associated with treatment responses or patient outcomes is essential in targeted therapy.
KMT2 (lysine methyltransferase) family enzymes regulate gene transcription via post-translational histone methylation. More specifically, KMT2 enzymes methylate histone 3 lysine 4 (H3K4) on promoters (di- and tri-methylation, me2/me3) or enhancers (mono-methylation, me1), leading to increased genome accessibility and thereby activating gene transcription [2]. The transcription activation function of the KMT2 family is essential in normal cell physiology and mammalian development [3]. However, cancer cells are known to hijack normal development-related machinery for their own malignant evolution and proliferation [4]. Frequent dysregulation of KMT2 family function in cancer is precisely an example of this malignant transformation [5]. The KMT2 family in Homo sapiens (originally named mixed-lineage leukemia (MLL) family) are a highly conserved group of proteins, including MLL1/KMT2A, MLL2/KMT2B, MLL3/KMT2C, MLL4/KMT2D, KMT2F (SET1A), and KMT2G (SET1B) [5]. KMT2C/KMT2D are mainly involved in enhancer-associated H3K4me1 [5], and they are also the most frequently mutated genes in cancer [6], especially KMT2C, with a mutation rate in pan-cancer being 6.6% [7].
Missense mutations are the most common KMT2C mutation type. They contribute to tumorigenesis through dysregulating genome enhancer activity, which could lead to a previously fine-tuned network regulated by oncoproteins and tumor suppressors being disrupted [5]. KMT2C mutations have been reported to correlate with patient prognosis in many cancer types. However, it is currently unclear whether KMT2C works as a tumor suppressor or as an oncogene. In most cancers, e.g., medulloblastoma [8], lung cancer [9], head and neck cancer [10], gastric cancer [11], squamous cell carcinoma [12], KMT2C mutations or low expression of KMT2C correlate with worse survival outcomes, whereas in tumors like breast cancer and pancreatic ductal adenocarcinoma, conflicting prognostic evidence exists [13–17].
Besides prognosis significance, mechanistically, KMT2C may promote the transcription of DNA damage response (DDR) pathway genes and PD-L1 [18,19]. Thus its loss of function may confer sensitivity to poly (ADP-ribose) polymerase inhibitors (PARPis) and immune checkpoint inhibitors (ICIs). There are also clinical observations about the predictive value of KMT2C mutations for PARPis and ICIs [9,18,20,21].
Meanwhile, although KMT2C dysregulation plays an oncogenic role and is also a top mutated gene in prostate cancer (7%) [22,23], the clinical significance of KMT2C mutations in prostate cancer is currently understudied. Generally, there have been few attempts to explore the clinical value of KMT2C mutations in prostate cancer cohorts. One recent study investigated the biological consequences of KMT2C alterations in prostate cancer cells. Here, they used data from the International Cancer Genome Consortium (ICGC) and found that KMT2C truncating mutations were associated with reduced disease-free survival in prostate cancer [24]. However, probably due to the heterogeneity of data sources and lack of complete follow-up information, the mutation rate of ICGC (1.5%) does not reflect the actual distribution of KMT2C mutations in prostate cancer [22]. Thus, this finding needs further validation.
Our institute has a large database of prostate cancer patients with liquid biopsy genomic test results. We keep records of their baseline information, disease progression, treatment outcomes, and genomic data. Thus, inspired by the research mentioned above, we analyzed the association between KMT2C mutation status and patients’ survival using data from our institute to further address the significance of KMT2C mutations in prostate cancer. Our results clearly suggest an adverse predictive and prognostic value of KMT2C mutations. Furthermore, we also discovered two gene mutations associated with KMT2C mutations. Lastly, KMT2C mutations were significantly associated with poor survival in patients with specific signaling pathway aberrations, suggesting further research should focus on the KMT2C-mediated epigenetic regulation of these pathways.
Prostate cancer patients diagnosed between 2014 and 2021 in West China Hospital of Sichuan University with liquid biopsy test results were included in this study. Other inclusion criteria are available from detailed pathological reports in our institute, having acceptable patient compliance and being willing to complete the follow-up regularly. Exclusion criteria are concurrent second or multiple tumors from other origins, pathological subtype as neuroendocrine prostate cancer, and being unable to give informed consent. A total of 221 patients fulfilled our requirements and were included in this study. All included patients’ demographic and clinicopathological data, including date of birth, baseline prostate-specific antigen (PSA), de novo metastatic status, pathological reports, etc., were collected from the electronic medical record system. Venous blood was taken from each patient at the time of disease diagnosis. All patients provided written informed consent to participate in this study, which was conducted following the Declaration of Helsinki. The protocols in this study had the approval of the West China Hospital institutional review board in December 2021 (version number: 2021-1703).
Blood samples were collected in cell-free DNA tubes (BEAVER, 43803, Jiangsu, China). The plasma was harvested by centrifuging at 1600 g for 10 min, aliquoted in 1.5 mL microtubes, followed by another centrifugation for 10 min of 16000 g, and then stored at −80°C. We used QIAamp Circulating Nucleic Acid Kit (Qiagen, Hilden, Germany) to extract the cell-free DNA (cfDNA) from plasma. The buffy coat was used for genomic DNA (gDNA) extraction. QIAamp DNA Mini Kit (Qiagen) was used to extract DNA from the white blood cells.
Kapa Hyper Prep Kit (Kapa Biosystems, Wilmington, MA, USA) was applied to construct the sequencing library using 30–60 ng cfDNA, combined with Unique Molecular Identifier (UMI) to lower the false-positive rate of variant calls and increase variant detection sensitivity. Final libraries were sequenced on Illumina Nextseq500 (PE 75) (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions.
For the liquid biopsy, cfDNA was estimated by a targeted sequencing strategy capturing all exons of 150 PCa-related genes (Suppl. Table 1). NGS was performed in 3Dmed Clinical Laboratory, a College of American Pathologists (CAP) and Clinical Laboratory Improvement Amendments (CLIA) certified laboratory of 3D Medicines Inc. All mutations were interpreted as pathogenic or likely pathogenic mutations according to American College of Medical Genetics and Genomics criteria. The supplementary table contains specific information about which signaling pathway the gene belongs to (Suppl. Table 1). Additionally, the details of the pathogenic (and probably pathogenic) variants detected in this study are provided in the Suppl. Table 2.
The paired-end reads were mapped by BWA-MEM algorithm [25]. A filtering model for the accuracy of mutation calling was applied as described before [26]. Single-nucleotide variations (SNVs), small insertion/deletions (indels) were called using MuTect [26], the union of Varscan 2 [27] and Pindel [28]. To increase the result accuracy, the mutations were further reviewed by Integrative Genomics Viewer (IGV) [29]. All mutations were manually reviewed using IGV to eliminate false-positive results further. We calculated mutant ctDNA and wild-type (WT) cfDNA fragments’ probability density distributions by Gaussian kernel smoothing using StatsModels [30], a Python library (v0.13.5 under python v3.7).
Patients were required to visit our department every three weeks for routine history-taking, physical examinations, and PSA tests. Computerized tomography (CT), Magnetic resonance imaging (MRI), and single-photon emission computed tomography (SPECT) were performed when necessary.
This study used PSA progression-free survival (PSA-PFS), castration resistance-free survival (CRFS), and overall survival (OS) as primary endpoints. According to the PCWG3 criteria [31], PSA-progression was defined as two consecutive rises in the PSA level of 25% or more above the nadir (and by ≥2 ng/ml) after the treatment initiation. We used the chi-square and rank-sum tests to compare baseline characteristic differences between groups. Kaplan-Meier curve and log-rank test were used for survival curves. We used the Cox regression model for univariate and multivariate survival analyses. The impact of all variables on the outcome of the patients was determined through multivariate analyses using the “enter” method. All tests were two-sided. All statistical analyses were performed by R version 4.0.5 using the R package tableone v0.13.2, readxl v1.4.1, survival v3.4.0, plyr v1.8.6, forestplot v3.1.0, survminer v0.4.9, ggplot2 v3.3.5, gridExtra v2.3, patchwork v1.1.2. A p < 0.05 was considered significant.
Study design and patient characteristics
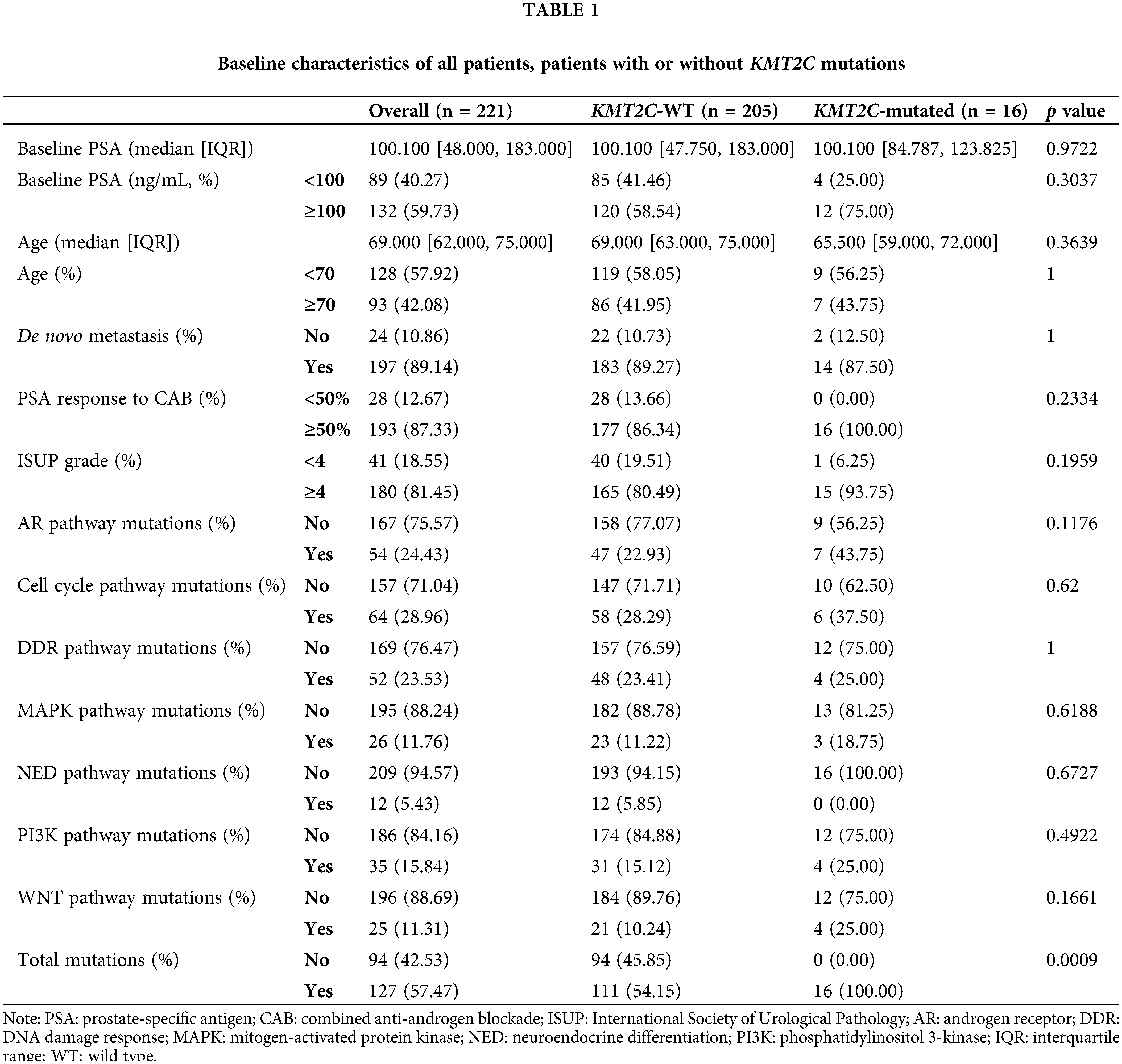
We retrospectively enrolled 221 prostate cancer patients with liquid biopsy genomic test results in this study. Most patients belonged to high-risk or advanced groups: baseline PSA >100 ng/mL: 61.40% (132/221); de novo metastasis: 89.14% (197/221); International Society of Urological Pathology (ISUP) grade ≥4: 81.45% (180/221). All baseline characteristics were balanced between KMT2C-WT and KMT2C-mutated groups, including baseline PSA, age, metastasis at different sites, and ISUP grade (Table 1).
In the hormone-sensitive prostate cancer (HSPC) stage, 185 patients were treated with conventional combined anti-androgen blockade (CAB). At the end of the follow-up, 164 patients had progressed to castration-resistant prostate cancer (CRPC), 137 of whom had received ABI treatment. Baseline characteristics were balanced and comparable between KMT2C-WT and KMT2C-mutated groups in patients who were still in the HSPC stage as well as those who had advanced to the CRPC stage (Suppl. Table 3).
The KMT2C mutation rate in this cohort is 7.24% (16/221), consistent with a previous report with an extensive database of 1,013 prostate cancer exome sequencing data [22]. Also, mutations in other pathways, including androgen receptor (AR) signaling pathway, cell cycle pathway, DDR pathway, mitogen-activated protein kinase (MAPK) pathway, neuroendocrine differentiation (NED) related pathway, phosphatidylinositol 3-kinase (PI3K) pathway, WNT pathway, did not differ between these two groups (Table 1).
However, a correlation between KMT2C mutations and Serine/Threonine-Protein Kinase 11 (STK11, p = 0.004) mutations and Catenin Beta 1 (CTNNB1, p = 0.008) mutations was found (Suppl. Table 4). Interestingly, when we split the entire cohort into two groups based on the patient’s current stage, we found that KMT2C mutations were associated with STK11 (p = 0.043) mutation in the patients who had not progressed. Moreover, KMT2C mutations were correlated with CTNNB1 (p = 0.012) and peckle-type poxvirus and zinc-finger protein (SPOP, p = 0.007) mutations in those that had advanced to CRPC (Suppl. Table 4).
KMT2C-mutated patients showed worse survival than KMT2C-WT patients in terms of both CRFS and OS (CRFS: mutated: 9.9 vs. WT: 22.0 months, p = 0.015; OS: mutated: 71.9 vs. WT: 137.4 months, p = 0.012, Fig. 1).
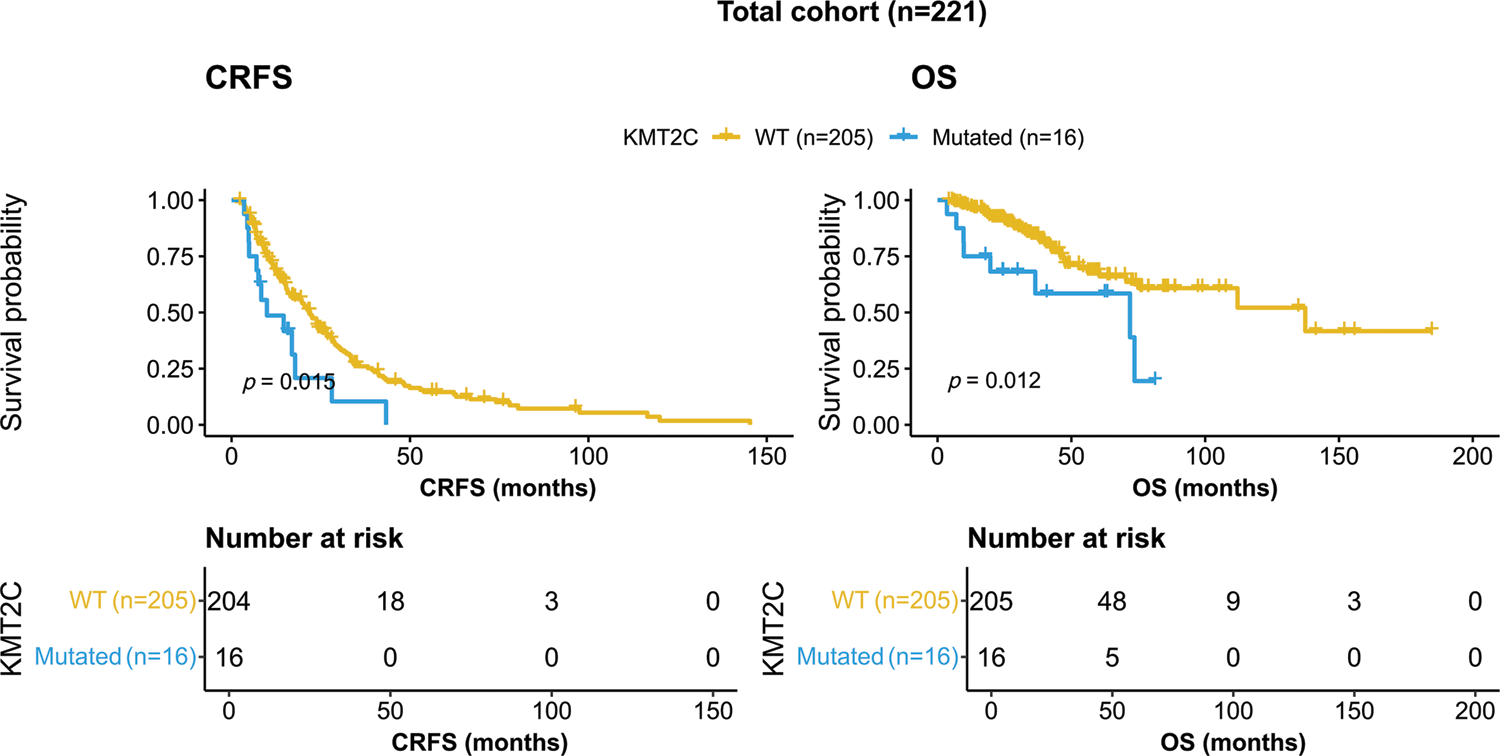
Figure 1: Kaplan-Meier curves of patients with or without KMT2C mutations. CRFS: castration-resistance free survival; OS: overall survival; WT: wild type.
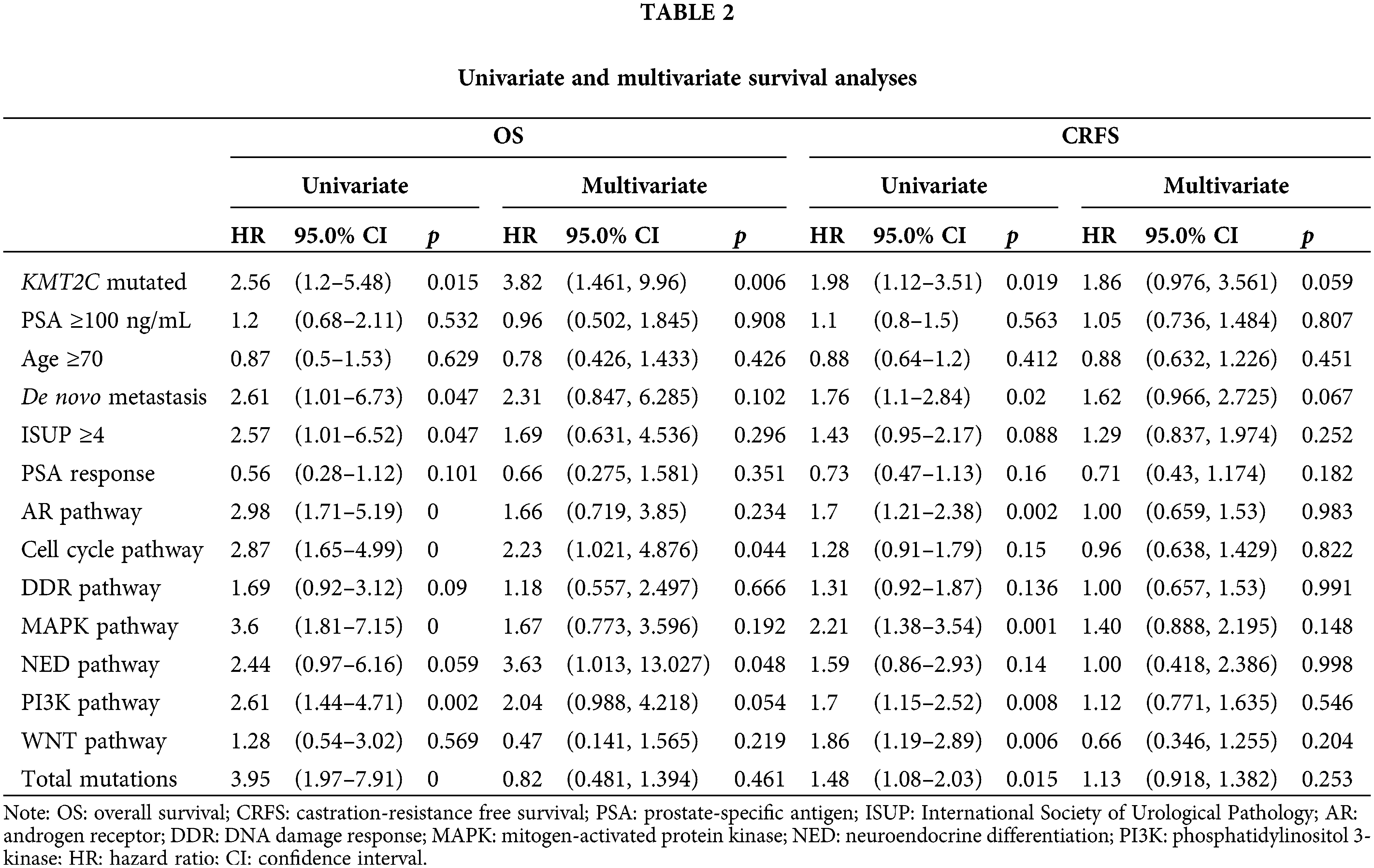
Furthermore, we did univariate and multivariate Cox regression analyses to explore the prognostic significance of KMT2C mutations, mutations in other pathways, and other clinicopathological indexes. The results showed that KMT2C mutations were associated with shorter CRFS and OS in univariate analyses, and were also an independent risk factor in OS [hazard ratio (HR): 3.815 (95% confidence interval (95% CI): 1.461, 9.96), p = 0.006], along with mutations in cell cycle pathway [HR: 2.231 (95% CI: 1.021, 4.876), p = 0.044] and neuroendocrine differentiation [HR: 3.633 (95% CI: 1.013, 13.027), p = 0.048] (Table 2). Although multiple factors were associated with shorter CRFS in univariate analyses, such as KMT2C mutations (p = 0.019), de novo metastasis (p = 0.020), AR pathway mutations (p = 0.002), MAPK pathway mutations (p = 0.001), PI3K pathway mutations (p = 0.008), WNT pathway mutations (p = 0.006), and total mutations (p = 0.015), none of them remained the prognostic significance in multivariate analyses for CRFS (only KMT2C mutations and de novo metastasis displayed borderline significance; p = 0.059 and p = 0.067).
Next, we explored the prognostic value of KMT2C mutations in different patient subgroups. For OS, KMT2C mutations were associated with survival in patient subgroups with baseline PSA <100 ng/mL (HR: 6.37, p = 0.018), age <70 (HR: 3.26, p = 0.016), de novo metastasis (HR: 2.80, p = 0.013), ISUP grade ≥4 (HR: 2.67, p = 0.012), PSA response to CAB ≥50% (HR: 2.86, p = 0.008), no DDR pathway mutation (HR: 2.65, p = 0.031), without MAPK pathway mutation (HR: 3.01, p = 0.009), without NED pathway mutation (HR: 2.78, p = 0.009), with PI3K pathway mutation (HR: 6.64, p = 0.009), with WNT pathway mutation (HR: 2.44, p = 0.043) (Fig. 2).
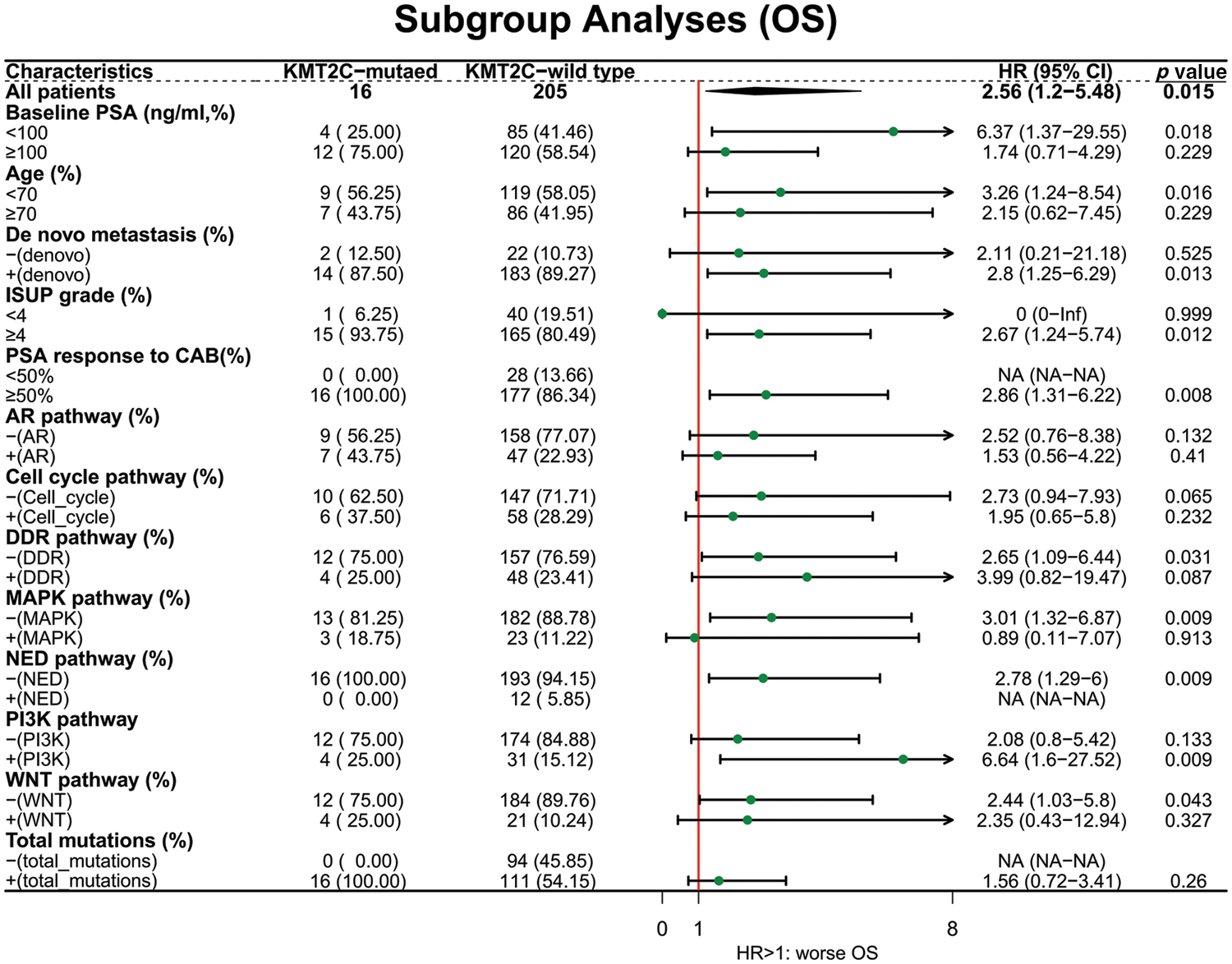
Figure 2: The prognostic value of KMT2C mutations in different clinicopathological subgroups. OS: overall survival; PSA: prostate-specific antigen; CAB: combined anti-androgen blockade; ISUP: International Society of Urological Pathology; AR: androgen receptor; DDR: DNA damage response; MAPK: mitogen-activated protein kinase; NED: neuroendocrine differentiation; PI3K: phosphatidylinositol 3-kinase; HR: hazard ratio; CI: confidence interval.
KMT2C mutations and therapeutic response
We further evaluated the prognostic significance of KMT2C mutations in predicting the treatment efficacy at different disease stages. KMT2C-mutated patients had a significantly shorter PSA-PFS compared to KMT2C-WT patients when treated with CAB therapy during the HSPC phase (CAB PSA-PFS: mutated: 9.9 vs. WT: 17.6 months, p = 0.014, Fig. 3). KMT2C mutations were a significant prognosticator of PSA-PFS in the univariate analysis and had a borderline p value (p = 0.09) as well as the highest hazard ratio (HR = 1.84) in the multivariate analysis (Table 3). The predictive value of KMT2C mutations was also validated in patients of different subgroups. It could predict shorter PSA-PFS in 10 out of 23 subgroups while exhibiting a strong trend in the remaining subgroups (Fig. 4). In the ABI treatment, the CRPC patients with KMT2C mutations experienced a numerically shorter PSA-PFS (ABI PSA-PFS: mutated: 8.1 vs. WT 12.5 months, p = 0.32, Fig. 3). However, KMT2C mutations were not an independent factor in Cox regression analyses (Table 3). In subgroup analyses, the predictive significance of KMT2C mutations was only evident in the subgroup with age <70 (HR: 3.54, p = 0.022, Suppl. Fig. 1).
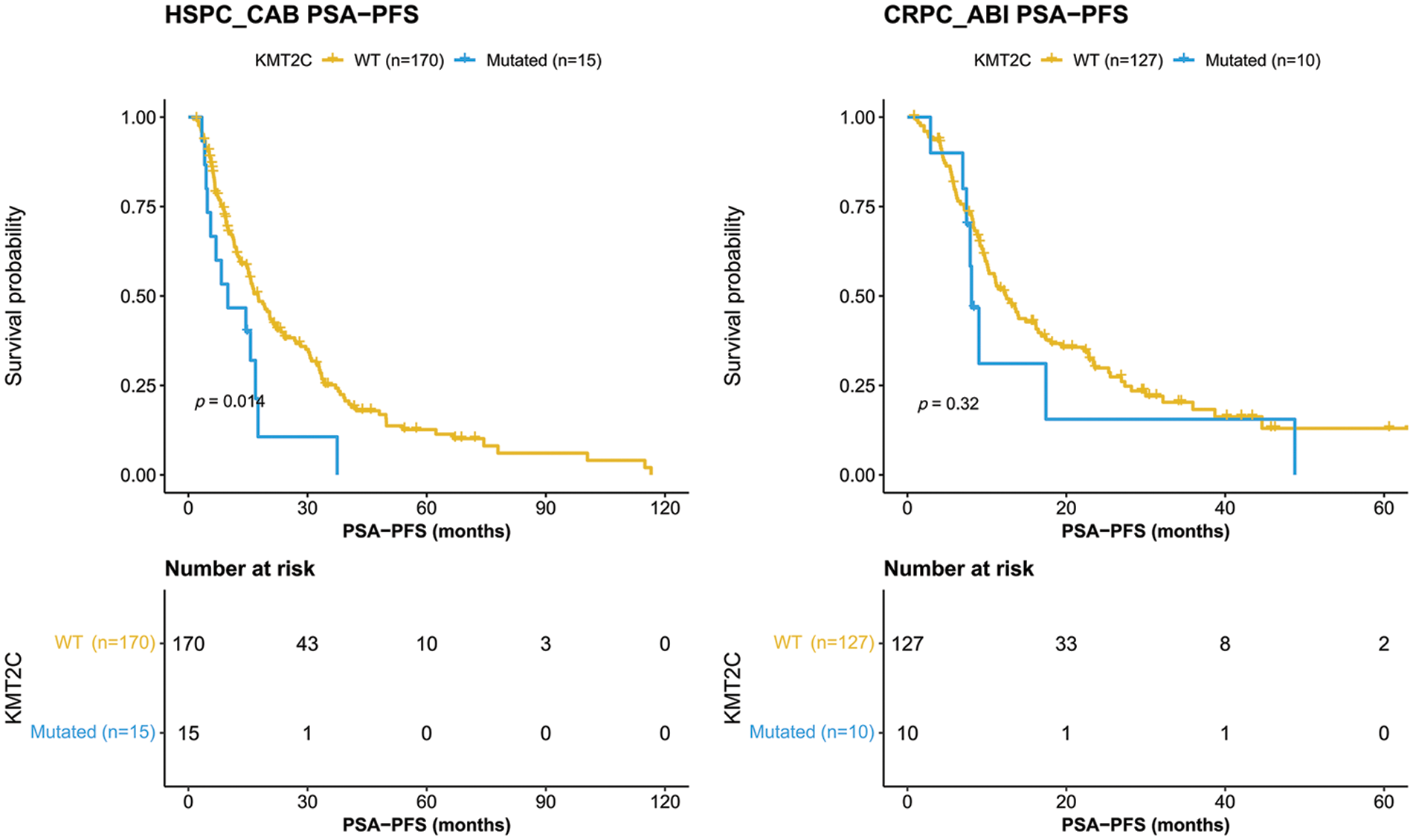
Figure 3: Kaplan-Meier curves of patients with or without KMT2C mutations who were treated combined anti-androgen blockade or abiraterone treatment. HSPC: hormone-sensitive prostate cancer; CRPC: castration-resistant prostate cancer; CAB: combined antiandrogen blockade; ABI: abiraterone; PSA: prostate-specific antigen; PSA-PFS: PSA progression-free survival.
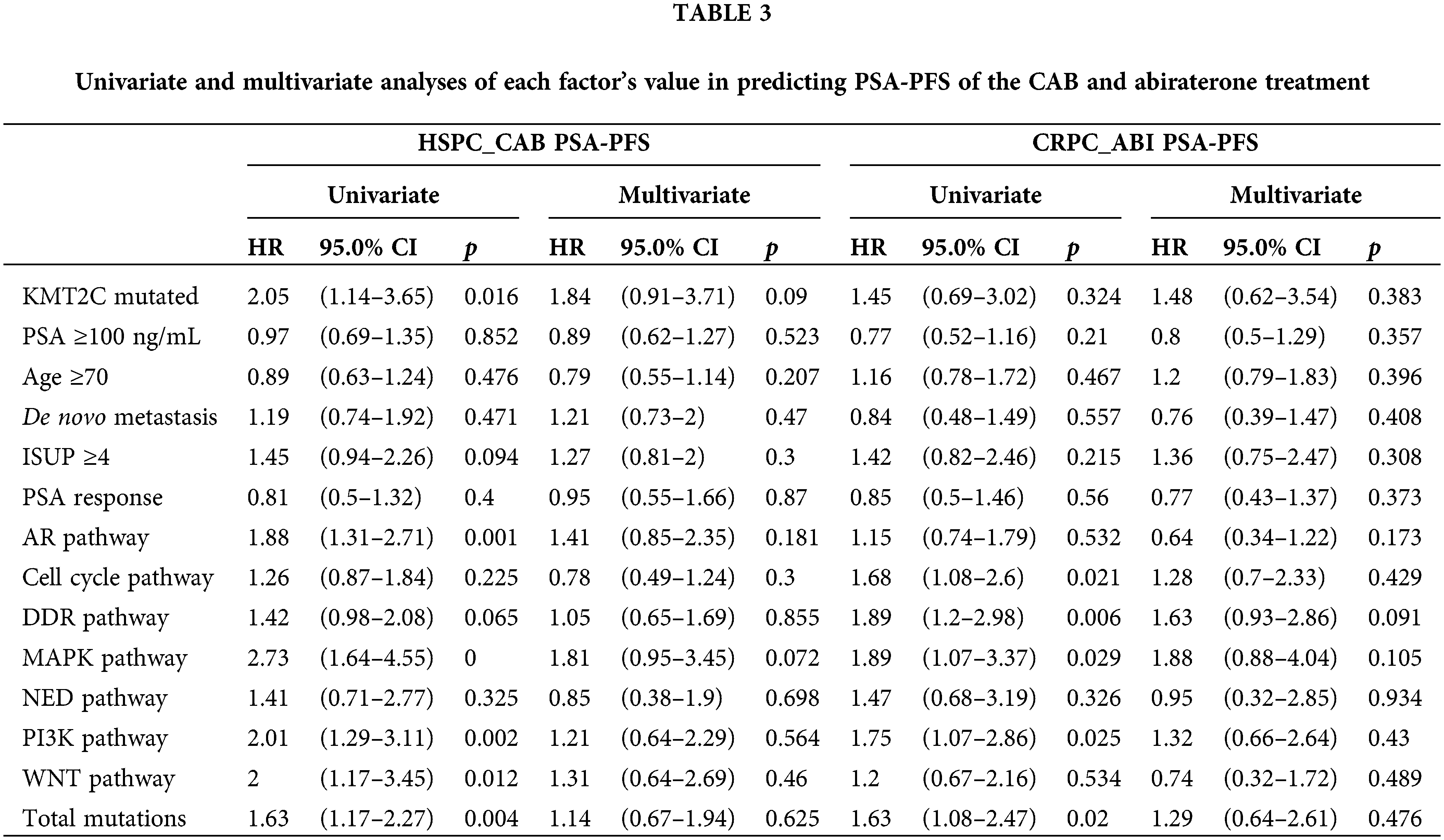
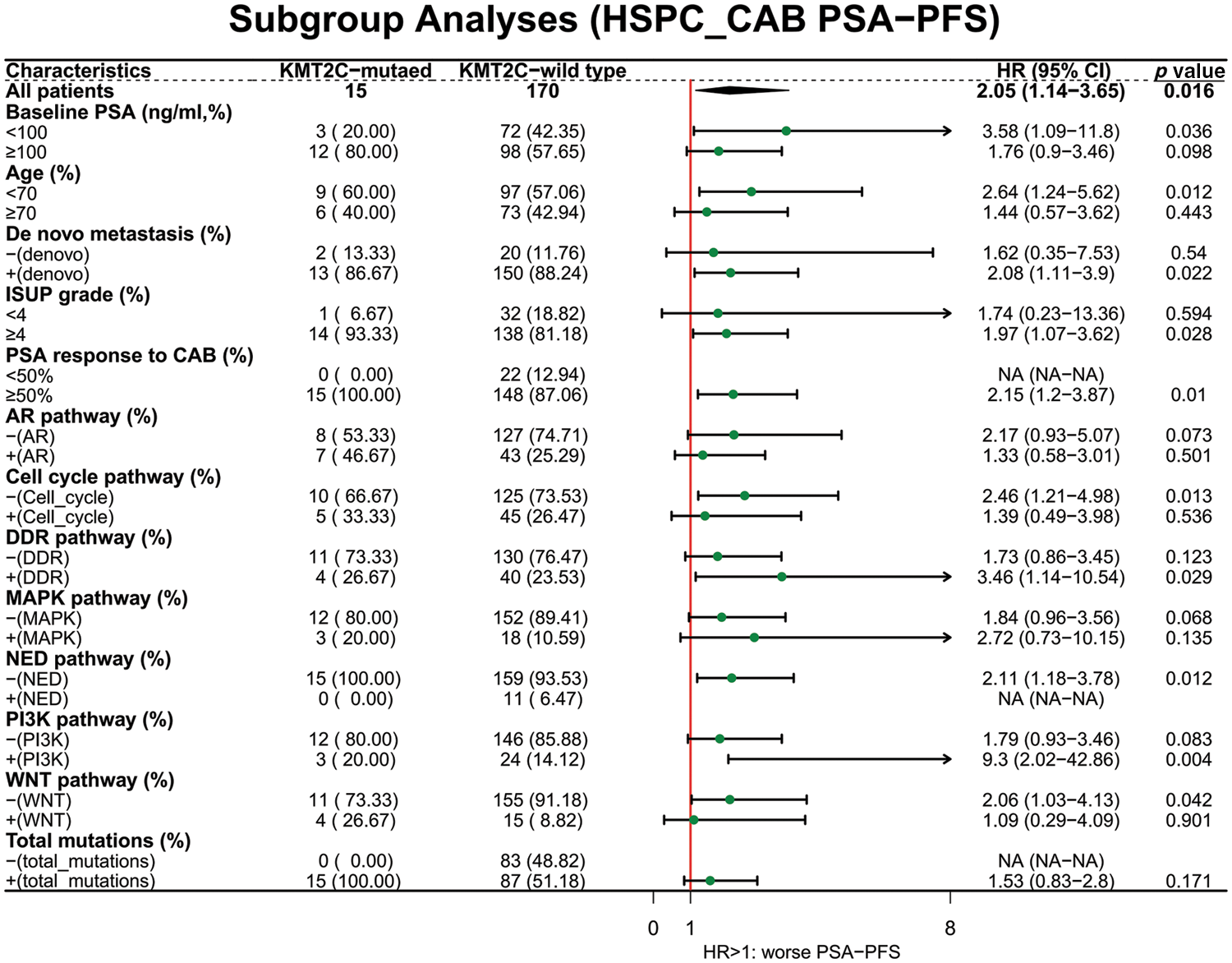
Figure 4: The prognostic value of KMT2C mutations in predicting the therapeutic efficacy of combined anti-androgen blockade in different clinicopathological subgroups. HSPC: hormone-sensitive prostate cancer; CAB: combined antiandrogen blockade; PSA: prostate-specific antigen; PSA-PFS: PSA progression-free survival; ISUP: International Society of Urological Pathology; AR: androgen receptor; DDR: DNA damage response; MAPK: mitogen-activated protein kinase; NED: neuroendocrine differentiation; PI3K: phosphatidylinositol 3-kinase; HR: hazard ratio; CI: confidence interval.
This study investigates the prognostic significance of KMT2C mutations in a prostate cancer cohort. Our results suggested that KMT2C mutations were associated with STK11 and CTNNB1 mutations. KMT2C-mutated patients showed worse survival than KMT2C-WT patients in terms of both CRFS and OS, and KMT2C mutations were an independent risk factor in OS. Besides, KMT2C mutations were significantly associated with survival in certain patient subgroups, such as patients with PI3K and WNT pathway mutation, etc. Furthermore, KMT2C mutations were also associated with the treatment efficacy of prostate cancer patients.
KMT2C loss may exert a dual impact depending on the tumor context. Previous studies have shown that KMT2C mutations are linked to clinical outcomes and can be used for patient stratification for many tumors. In most cases, KMT2C mutations predict worse survival but can also indicate a better prognosis in certain cancer types [8–17]. Currently, there is no conclusion on the impact of KMT2C mutations in prostate cancer patients. Two in vivo studies showed conflicting results: one claimed KMT2C has a carcinogenic role and reported an overexpression in prostate cancer than in normal tissues (did not show survival data) [23]; another one claimed the truncated KMT2C a driver of tumor development and used public data to display a negative prognostic indication of KMT2C-loss [24]. Based on a relatively large population with balanced baseline characteristics, our results demonstrated that KMT2C mutations predicted poorer survival in prostate cancer patients and were an independent risk factor for OS. However, the discrepancies regarding the prognostic indication of KMT2C mutations in different cancer types and different experiment contexts require further exploration.
Our data also showed that KMT2C mutations were correlated with an unfavorable therapeutic response to androgen deprivation therapy and novel anti-androgen therapy. Interestingly, the association of KMT2C mutations with rapid progression and drug resistance in AR-directed treatment was only found in the HSPC stage, which prompts the use of liquid biopsy for early detection of KMT2C mutations in prostate cancer in order to provide in-time treatment outcome prediction. KMT2C is a known epigenetic regulator of gene expression. KMT2C regulates the DDR by its direct recruitment to DNA damage sites and KMT2C mutations are shown to disrupt homologous recombination (HR)–mediated DNA double-strand break repair, thus conferring PARPi sensitivity [18]. KMT2C also promotes the transcription of PD-L1 by binding to its enhancer and promoter in prostate cancer cells and regulating immune evasion through PD-L1 downregulation in mice [19]. Therefore, its loss-of-function possibly also sensitizes tumors to ICIs. Thus, we postulate that KMT2C-mutated cancer cells are likely to accumulate more DNA damage, possibly leading to high mutation loads and further promoting rapid progression and drug resistance. Therefore, with our knowledge of the complex impact of KMT2C mutations on therapeutic outcome, it is crucial to carefully consider treatment choices for this group of patients, thus stressing the significance of early detecting KMT2C mutations in prostate cancer patients.
We also noticed two gene mutations associated with KMT2C mutations: STK11 and CTNNB1. STK11 encodes a liver serine/threonine B1 kinase that is essential in many cellular processes and is involved in malignant metabolic transformations [32]. STK11 is also considered a tumor suppressor in prostate cancer and is targeted by the diabetes drug metformin which has been shown to have anti-cancer activity [33,34]. CTNNB1 encodes the pivotal protein β-catenin in the canonical WNT signaling, which is also frequently mutated in CRPC [35]. Further investigation showed that the association between KMT2C and CTNNB1 existed only in patients who advanced to CRPC but not those who remained hormone-sensitive. This result indicates the necessity of looking into the synergistic effect and possible pharmaceutical combinations of KMT2C, STK11, and CTNNB1 mutations. Also, the prognostic value of KMT2C mutations was more prominent in patients with PI3K and WNT pathway mutation. This finding reinforces the need for future studies to focus on KMT2C-mediated epigenetic regulation on PI3K and WNT pathways, both critical pathways in progressing to CRPC [32,35].
We used cfDNA-based liquid biopsy to detect the KMT2C mutations in this study. Several advantages of this method, including sensitivity, and the non-invasive and economical nature, have made it more prevalent than traditional tumor biopsies. Nowadays, in the context of personalized medicine, the need to weaponize liquid biopsies in favor of dynamic disease management is increasing. This study provides another illustration of using liquid biopsy results in clinical practice: KMT2C mutations could serve as a biomarker to predict unfavorable therapeutic efficacy and clinical outcomes; therefore, more aggressive treatment and closer follow-up will be needed for this group of patients.
Acknowledgement: The authors sincerely thank Alexander Thouin, a Ph.D. candidate at Netherlands Cancer Institute, for his careful revision of this manuscript.
Funding Statement: This work was supported by the Natural Science Foundation of China (NSFC 81902577) and the Research Foundation for the Postdoctoral Program of Sichuan University (2021SCU12014).
Author Contributions: Sha Zhu and Nanwei Xu designed this study, analyzed the data and drafted the manuscript. Jiayu Liang, Jinge Zhao, Xingming Zhang, Junru Chen, Guangxi Sun participated in study design. Fengnian Zhao, Zilin Wang, Yuchao Ni, Jindong Dai helped the data collection. Pengfei Shen and Hao Zeng supervised this project and provided funding. All authors read and approved the final manuscript.
Availability of Data and Materials: The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics Approval: The protocols in this study had the approval of the West China Hospital institutional review board in December 2021 (version number: 2021-1703).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Siegel, R. L., Miller, K. D., Fuchs, H. E., Jemal, A. (2022). Cancer statistics. CA: A Cancer Journal for Clinicians, 72(1), 7–33. [Google Scholar] [PubMed]
2. Calo, E., Wysocka, J. (2013). Modification of enhancer chromatin: What, how, and why? Molecular Cell, 49(5), 825–837. https://doi.org/10.1016/j.molcel.2013.01.038 [Google Scholar] [PubMed] [CrossRef]
3. Crump, N. T., Milne, T. A. (2019). Why are so many MLL lysine methyltransferases required for normal mammalian development? Cellular and Molecular Life Sciences, 76(15), 2885–2898. https://doi.org/10.1007/s00018-019-03143-z [Google Scholar] [PubMed] [CrossRef]
4. Hanahan, D. (2022). Hallmarks of cancer: New dimensions. Cancer Discovery, 12(1), 31–46. https://doi.org/10.1158/2159-8290.CD-21-1059 [Google Scholar] [PubMed] [CrossRef]
5. Rao, R. C., Dou, Y. (2015). Hijacked in cancer: The KMT2 (MLL) family of methyltransferases. Nature Reviews Cancer, 15(6), 334–346. https://doi.org/10.1038/nrc3929 [Google Scholar] [PubMed] [CrossRef]
6. Fagan, R. J., Dingwall, A. K. (2019). COMPASS ascending: Emerging clues regarding the roles of MLL3/KMT2C and MLL2/KMT2D proteins in cancer. Cancer Letters, 458, 56–65. https://doi.org/10.1016/j.canlet.2019.05.024 [Google Scholar] [PubMed] [CrossRef]
7. Kandoth, C., McLellan, M. D., Vandin, F., Ye, K., Niu, B. et al. (2013). Mutational landscape and significance across 12 major cancer types. Nature, 502(7471), 333–339. https://doi.org/10.1038/nature12634 [Google Scholar] [PubMed] [CrossRef]
8. Wong, G. C., Li, K. K., Wang, W. W., Liu, A. P., Huang, Q. J. et al. (2020). Clinical and mutational profiles of adult medulloblastoma groups. Acta Neuropathologica Communications, 8(1), 191. https://doi.org/10.1186/s40478-020-01066-6 [Google Scholar] [PubMed] [CrossRef]
9. Mastoraki, S., Balgkouranidou, I., Tsaroucha, E., Klinakis, A., Georgoulias, V. et al. (2021). KMT2C promoter methylation in plasma-circulating tumor DNA is a prognostic biomarker in non-small cell lung cancer. Molecular Oncology, 15(9), 2412–2422. https://doi.org/10.1002/1878-0261.12848 [Google Scholar] [PubMed] [CrossRef]
10. Machnicki, M. M., Rzepakowska, A., Janowska, J. I., Pepek, M., Krop, A. et al. (2022). Analysis of mutational profile of hypopharyngeal and laryngeal head and neck squamous cell carcinomas identifies KMT2C as a potential tumor suppressor. Frontiers in Oncology, 12, 768954. https://doi.org/10.3389/fonc.2022.768954 [Google Scholar] [PubMed] [CrossRef]
11. Li, B., Liu, H. Y., Guo, S. H., Sun, P., Gong, F. M. et al. (2014). Association of MLL3 expression with prognosis in gastric cancer. Genetics and Molecular Research, 13(3), 7513–7518. https://doi.org/10.4238/2014.September.12.18 [Google Scholar] [PubMed] [CrossRef]
12. Pickering, C. R., Zhou, J. H., Lee, J. J., Drummond, J. A., Peng, S. A. et al. (2014). Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clinical Cancer Research, 20(24), 6582–6592. https://doi.org/10.1158/1078-0432.CCR-14-1768 [Google Scholar] [PubMed] [CrossRef]
13. Bortolini Silveira, A., Bidard, F. C., Tanguy, M. L., Girard, E., Trédan, O. et al. (2021). Multimodal liquid biopsy for early monitoring and outcome prediction of chemotherapy in metastatic breast cancer. NPJ Breast Cancer, 7(1), 115. https://doi.org/10.1038/s41523-021-00319-4 [Google Scholar] [PubMed] [CrossRef]
14. Liu, X., Qiu, R., Xu, M., Meng, M., Zhao, S. et al. (2021). KMT2C is a potential biomarker of prognosis and chemotherapy sensitivity in breast cancer. Breast Cancer Research and Treatment, 189(2), 347–361. https://doi.org/10.1007/s10549-021-06325-1 [Google Scholar] [PubMed] [CrossRef]
15. Chen, X., Zhang, G., Chen, B., Wang, Y., Guo, L. et al. (2019). Association between histone lysine methyltransferase KMT2C mutation and clinicopathological factors in breast cancer. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie, 116(1), 108997. https://doi.org/10.1016/j.biopha.2019.108997 [Google Scholar] [PubMed] [CrossRef]
16. Sato, K., Akimoto, K. (2017). Expression levels of KMT2C and SLC20A1 identified by information-theoretical analysis are powerful prognostic biomarkers in estrogen receptor-positive breast cancer. Clinical Breast Cancer, 17(3), e135–e142. https://doi.org/10.1016/j.clbc.2016.11.005 [Google Scholar] [PubMed] [CrossRef]
17. Dawkins, J. B., Wang, J., Maniati, E., Heward, J. A., Koniali, L. et al. (2016). Reduced expression of histone methyltransferases KMT2C and KMT2D correlates with improved outcome in pancreatic ductal adenocarcinoma. Cancer Research, 76(16), 4861–4871. https://doi.org/10.1158/0008-5472.CAN-16-0481 [Google Scholar] [PubMed] [CrossRef]
18. Chang, A., Liu, L., Ashby, J. M., Wu, D., Chen, Y. et al. (2021). Recruitment of KMT2C/MLL3 to DNA damage sites mediates DNA damage responses and regulates PARP inhibitor sensitivity in cancer. Cancer Research, 81(12), 3358–3373. https://doi.org/10.1158/0008-5472.CAN-21-0688 [Google Scholar] [PubMed] [CrossRef]
19. Xiong, W., Deng, H., Huang, C., Zen, C., Jian, C. et al. (2019). MLL3 enhances the transcription of PD-L1 and regulates anti-tumor immunity. Biochimica et Biophysica Acta. Molecular Basis of Disease, 1865(2), 454–463. https://doi.org/10.1016/j.bbadis.2018.10.027 [Google Scholar] [PubMed] [CrossRef]
20. Shi, Y., Lei, Y., Liu, L., Zhang, S., Wang, W. et al. (2021). Integration of comprehensive genomic profiling, tumor mutational burden, and PD-L1 expression to identify novel biomarkers of immunotherapy in non-small cell lung cancer. Cancer Medicine, 10(7), 2216–2231. https://doi.org/10.1002/cam4.3649 [Google Scholar] [PubMed] [CrossRef]
21. Bai, X., Wu, D. H., Ma, S. C., Wang, J., Tang, X. R. et al. (2020). Development and validation of a genomic mutation signature to predict response to PD-1 inhibitors in non-squamous NSCLC: A multicohort study. Journal for Immunotherapy of Cancer, 8(1), e000381. https://doi.org/10.1136/jitc-2019-000381 [Google Scholar] [PubMed] [CrossRef]
22. Armenia, J., Wankowicz, S. A. M., Liu, D., Gao, J., Kundra, R. et al. (2018). The long tail of oncogenic drivers in prostate cancer. Nature Genetics, 50(5), 645–651. https://doi.org/10.1038/s41588-018-0078-z [Google Scholar] [PubMed] [CrossRef]
23. Lian, J., Xu, C., Chen, X., Huang, S., Wu, D. (2022). Histone methyltransferase KMT2C plays an oncogenic role in prostate cancer. Journal of Cancer Research and Clinical Oncology, 148(7), 1627–1640. https://doi.org/10.1007/s00432-022-03968-5 [Google Scholar] [PubMed] [CrossRef]
24. Limberger, T., Schlederer, M., Trachtová, K., Garces de Los Fayos Alonso, I., Yang, J. et al. (2022). KMT2C methyltransferase domain regulated INK4A expression suppresses prostate cancer metastasis. Molecular Cancer, 21(1), 89. https://doi.org/10.1186/s12943-022-01542-8 [Google Scholar] [PubMed] [CrossRef]
25. Li, H., Durbin, R. (2010). Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics, 26(5), 589–595. https://doi.org/10.1093/bioinformatics/btp698 [Google Scholar] [PubMed] [CrossRef]
26. Yang, N., Li, Y., Liu, Z., Qin, H., Du, D. et al. (2018). The characteristics of ctDNA reveal the high complexity in matching the corresponding tumor tissues. BMC Cancer, 18(1), 319. https://doi.org/10.1186/s12885-018-4199-7 [Google Scholar] [PubMed] [CrossRef]
27. Koboldt, D. C., Zhang, Q., Larson, D. E., Shen, D., McLellan, M. D. et al. (2012). VarScan 2: Somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Research, 22(3), 568–576. https://doi.org/10.1101/gr.129684.111 [Google Scholar] [PubMed] [CrossRef]
28. Ye, K., Schulz, M. H., Long, Q., Apweiler, R., Ning, Z. (2009). Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics, 25(21), 2865–2871. https://doi.org/10.1093/bioinformatics/btp394 [Google Scholar] [PubMed] [CrossRef]
29. Chabon, J. J., Simmons, A. D., Lovejoy, A. F., Esfahani, M. S., Newman, A. M. et al. (2016). Circulating tumour DNA profiling reveals heterogeneity of EGFR inhibitor resistance mechanisms in lung cancer patients. Nature Communications, 7(1), 11815. https://doi.org/10.1038/ncomms11815 [Google Scholar] [PubMed] [CrossRef]
30. Seabold, S., Perktold, J. (2010). Statsmodels: Econometric and statistical modeling with python. 9th Python in Science Conference, vol. 57, pp. 92–96. Austin, Texas, USA. [Google Scholar]
31. Scher, H. I., Morris, M. J., Stadler, W. M., Higano, C., Basch, E. et al. (2016). Trial design and objectives for castration-resistant prostate cancer: Updated recommendations from the prostate cancer clinical trials working group 3. Journal of Clinical Oncology, 34(12), 1402–1418. https://doi.org/10.1200/JCO.2015.64.2702 [Google Scholar] [PubMed] [CrossRef]
32. Bitting, R. L., Armstrong, A. J. (2013). Targeting the PI3K/Akt/mTOR pathway in castration-resistant prostate cancer. Endocrine-Related Cancer, 20(3), R83–R99. https://doi.org/10.1530/ERC-12-0394 [Google Scholar] [PubMed] [CrossRef]
33. Shaw, R. J., Lamia, K. A., Vasquez, D., Koo, S. H., Bardeesy, N. et al. (2005). The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science, 310(5754), 1642–1646. https://doi.org/10.1126/science.1120781 [Google Scholar] [PubMed] [CrossRef]
34. Ahn, H. K., Lee, Y. H., Koo, K. C. (2020). Current status and application of metformin for prostate cancer: A comprehensive review. International Journal of Molecular Sciences, 21(22), 8540. https://doi.org/10.3390/ijms21228540 [Google Scholar] [PubMed] [CrossRef]
35. Murillo-Garzón, V., Kypta, R. (2017). WNT signalling in prostate cancer. Nature Reviews Urology, 14(11), 683–696. https://doi.org/10.1038/nrurol.2017.144 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
SUPPLEMENTARY FIGURE 1. The prognostic value of KMT2C mutations in predicting the therapeutic efficacy of abiraterone in different clinicopathological subgroups.
SUPPLEMENTARY TABLE 1. Gene sequencing panel and list of genes included in each pathway.
SUPPLEMENTARY TABLE 2. Pathogenic (and likely pathogenic) variants detected in this study.
SUPPLEMENTARY TABLE 3. Baseline characteristics of patients who were still in HSPC stage and who had progressed to CRPC stage.
SUPPLEMENTARY TABLE 4. The association between KMT2C mutations and other gene mutations.
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools