 Open Access
Open Access
ARTICLE
Circ_0053943 complexed with IGF2BP3 drives uveal melanoma progression via regulating N6-methyladenosine modification of Epidermal growth factor receptor
1 Department of Ophthalmology, The First Affiliated Hospital with Nanjing Medical University, Nanjing, 210029, China
2 The First Clinical Medical College, Nanjing Medical University, Nanjing, 211166, China
3 Department of Ophthalmology, Nanjing First Hospital, Nanjing Medical University, Nanjing, 210006, China
* Corresponding Authors: HU LIU. Email: ; XUEJUAN CHEN. Email:
# These authors contributed equally to this paper
Oncology Research 2024, 32(5), 983-998. https://doi.org/10.32604/or.2024.045972
Received 13 September 2023; Accepted 22 December 2023; Issue published 23 April 2024
Abstract
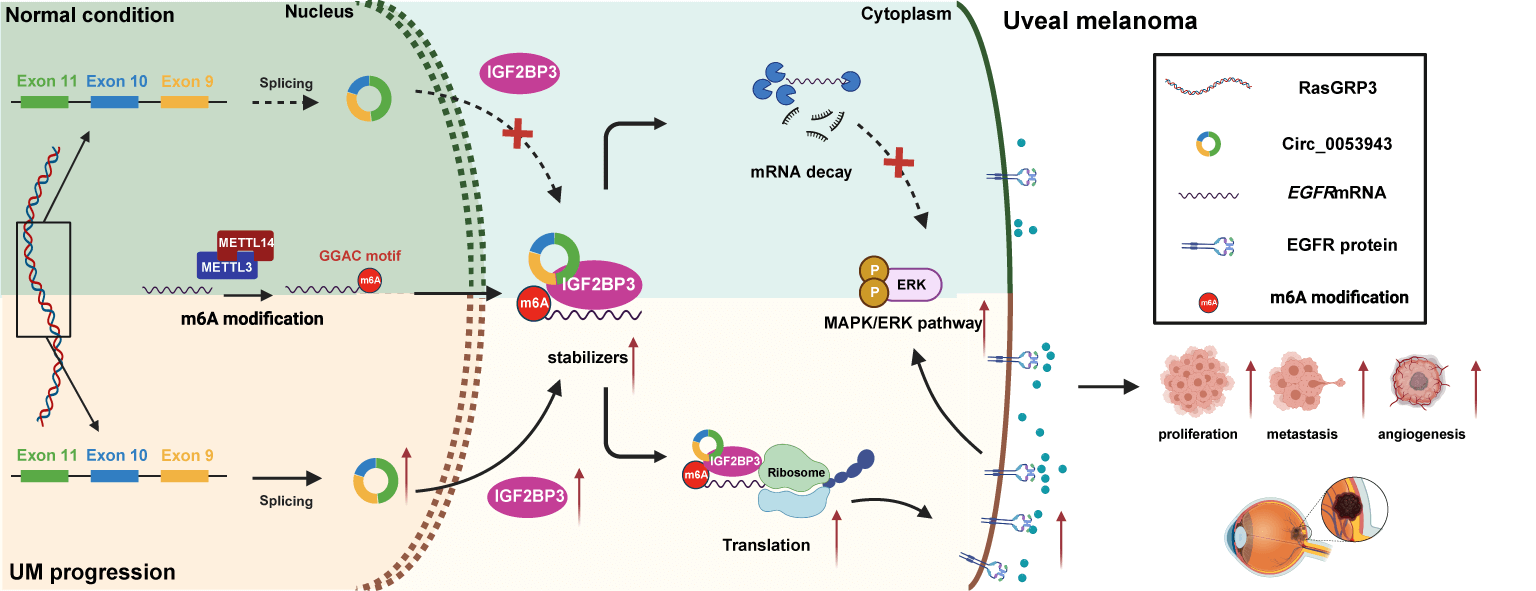
Numerous studies have characterized the critical role of circular RNAs (circRNAs) as regulatory factors in the progression of multiple cancers. However, the biological functions of circRNAs and their underlying molecular mechanisms in the progression of uveal melanoma (UM) remain enigmatic. In this study, we identified a novel circRNA, circ_0053943, through re-analysis of UM microarray data and quantitative RT-PCR. Circ_0053943 was found to be upregulated in UM and to promote the proliferation and metastatic ability of UM cells in both in vitro and in vivo settings. Mechanistically, circ_0053943 was observed to bind to the KH1 and KH2 domains of insulin-like growth factor 2 mRNA-binding protein 3 (IGF2BP3), thereby enhancing the function of IGF2BP3 by stabilizing its target mRNA. RNA sequencing assays identified epidermal growth factor receptor (EGFR) as a target gene of circ_0053943 and IGF2BP3 at the transcriptional level. Rescue assays demonstrated that circ_0053943 exerts its biological function by stabilizing EGFR mRNA and regulating the downstream mitogen-activated protein kinase/extracellular signal-regulated kinase (MAPK/ERK) signaling pathway. Collectively, circ_0053943 may promote UM progression by stabilizing EGFR mRNA and activating the MAPK/ERK signaling pathway through the formation of a circ_0053943/IGF2BP3/EGFR RNA-protein ternary complex, thus providing a potential biomarker and therapeutic target for UM.Graphic Abstract
Keywords
Supplementary Material
Supplementary Material FileUveal melanoma (UM) stands out as the most prevalent primary intraocular malignant tumor in adults, originating from melanocytes of the uveal tract, which includes the iris, ciliary body, and choroid [1]. Owing to its ocular location, UM presents unique challenges for diagnosis, often complicating direct visualization. Consequently, clinicians rely on a combination of clinical examinations and advanced imaging modalities to confirm the presence of the tumor and characterize its properties, including ultrasound (US) echography and the standardized A-scan technique [2,3]. Despite aggressive management of the primary tumor, approximately 50% of patients eventually develop distant metastasis, particularly in the liver, leading to a grim prognosis within 5 months [4,5]. Given this bleak long-term clinical outlook and the limitations of current treatments, there exists an unmet medical need to investigate the mechanisms of UM tumorigenesis and assess novel effective therapies.
Several studies have identified an abundance of genetic aberrancies in UM, with mutations in BAP1, SF3B1, and EIF1AX playing essential roles in progression and metastasis, while activating mutations in Gα11/Q are closely related to UM oncogenesis in most cases [6,7]. Nevertheless, the precise molecular mechanisms underlying UM remain primarily unclear. Circular RNAs (circRNAs), a recently discovered subclass of noncoding RNAs characterized by higher resistance to RNase R due to their closed continuous loop structures, exhibit increased stability, making them more suitable for diagnostic biomarkers and therapeutic targets compared to other RNA types [8]. Indeed, mounting evidence suggests that circRNAs are involved in the metastasis and proliferation of various cancers by acting as microRNA (miRNA) sponges, interacting with RNA-binding proteins (RBPs), splicing pre-mRNA transcripts, and even encoding small peptides or proteins [9–11]. However, the number of published studies investigating circRNAs in UM is limited compared to other cancers [12,13]. Recently, microarray analysis identified several differentially expressed circRNAs between UM and normal uveal tissues [12]. Subsequently, another study verified and demonstrated that the upregulated circ_0119872, derived from the host gene RAS guanyl-releasing protein 3 (RasGRP3), could promote UM tumorigenesis via sponging miRNA and serve as an important prognostic biomarker [13]. Therefore, to gain a better understanding of UM development and progression, we aim to explore novel insights into the role and mechanism of circRNAs in UM. Considering that upregulated circRNAs can be potential therapeutic targets, we have selected circ_0053943, also derived from the host gene RasGRP3 and listed among the top 10 overregulated circRNAs [12], for further in-depth investigation.
N6-methyladenosine (m6A) represents the most abundant epitranscriptomic modification found in messenger RNAs (mRNAs), contributing to the tumorigenesis of multiple cancers [14–16]. This modification is governed by a methyltransferase complex (“writers”), demethylases (“erasers”), and RNA-binding proteins (“readers”). Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs, including IGF2BP1/2/3) are newly identified distinct m6A readers, facilitating the stability and translation of m6A-modified transcripts [17,18]. Previous studies have demonstrated that non-coding RNAs (ncRNAs) might participate in IGF2BPs-mediated functions and modulate the expression of target transcripts [19–21]. However, the involvement of IGF2BPs in UM tumorigenesis remains to be elucidated concerning how underlying ncRNAs mediate these functional biological processes.
In this study, we identified a novel circRNA (hsa_circ_0053943) serving as an RNA m6A reader-cooperator, assisting the m6A reader (IGF2BP3) in guarding m6A-modified epidermal growth factor receptor (EGFR) against decay and promoting tumorigenesis and metastasis in UM. Our findings suggest that circ_0053943 may cooperate with IGF2BP3 in the post-transcriptional regulation of EGFR, highlighting the functional importance of the circ_0053943/IGF2BP3/EGFR mRNA-protein ternary complex in the invasion and metastasis of UM.
A total of five UM tissues and five human normal melanocyte tissues were acquired from The First Affiliated Hospital of Nanjing Medical University. Two experienced histopathologists independently assessed the histological features of all specimens. The clinicopathological characteristics can be found in Suppl. Table 1. The study involving patients received approval from the Human Ethics Committee of The First Affiliated Hospital of Nanjing Medical University under the number 2022-SRFA-334, adhering to the Declaration of Helsinki. Written informed consent was obtained from each patient. Subsequently, all specimens were promptly frozen in liquid nitrogen and stored at −80°C until utilized.
Following a prior investigation [13], six ocular melanoma cell lines (MUM2B [RRID: CVCL_3447], C918 [RRID: CVCL_8471], Mel 270 [RRID: CVCL_C302], Omm2.5 [RRID: CVCL_C307], OCM-1 [RRID: CVCL_6934], and OCM-1A [RRID: CVCL_6935]) and the human retinal pigment epithelial cell line (ARPE-19 [RRID: CVCL_0145]) were selected and procured from the Cell Bank of Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Human primary umbilical vein endothelial cells (HUVECs [RRID: CVCL_2959]) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). Before the commencement of the study, all cell lines underwent confirmation using a short tandem repeat method, ensuring they were free of mycoplasma contamination. According to the manufacturers’ guidelines, all cell lines were cultured in recommended media and incubated at 37°C in a humidified incubator with 5% CO2.
RNA extraction and quantitative real-time polymerase chain reaction (qRT–PCR)
The total RNA of cell lines was extracted using TRIzol reagent (Invitrogen, Waltham, MA, USA) and subsequently reverse transcribed into complementary DNA (cDNA) using the PrimeScript RT Reagent Kit (Takara, Osaka, Japan), following the manufacturer’s instructions. The subsequent qRT-PCR, using the SYBR Green Kit (Takara), was conducted on the StepOnePlus Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The −ΔΔCT method was employed to assess the transcript levels of mRNA, with either glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6 serving as an internal control. The primer sequences used can be found in Suppl. Table 2.
RiboBio (Guangzhou, China) synthesized recombinant lentivirus containing short hairpin RNA targeting circ_0053943 (sh-circ_0053943#1/2/3) and IGF2BP3 (sh-IGF2BP3#1/2), along with the full-length targeting sequence of circ_0053943 and IGF2BP3, including the corresponding negative control. Obio (Shanghai, China) synthesized the IGF2BP3 full-length (FL) and truncation-mutation plasmids with a C-terminus 3× Flag tag. Small interfering RNA (siRNA) oligonucleotides targeting methyltransferase-like 3 (METTL3) and methyltransferase-like 14 (METTL14) were also synthesized by RiboBio. Transfection of short hairpin (shRNAs) and plasmid vectors was carried out using Lipofectamine 3000 (Invitrogen). The transfection efficiency was subsequently confirmed through qRT-PCR. Detailed sequences of shRNAs and siRNAs (for silencing) can be found in Suppl. Table 3.
Cell proliferation assay and flow cytometry assay
Cell proliferation/growth was assessed using a Cell Counting Kit-8 (CCK-8; Dojindo Laboratories, Dojindo, Japan) and the 5-ethynyl-2-deoxyuridine (EDU) assay with the Cell-Light EdU Apollo 488 In Vitro Kit (RiboBio), following the manufacturer’s protocols.
For the cell cycle assay, treated cells were fixed in 75% alcohol overnight at −20°C. After three washes, fixed cells were stained with propidium iodide (PI) buffer using a Cell Cycle Analysis Kit (Beyotime, Shanghai, China). Cells were incubated with H2O2 (1 mM) for 4 h to stimulate apoptosis for cell apoptosis assays. Subsequently, cells were stained with Annexin VAPC/7-AAD Apoptosis Detection Kit (KeyGEN, Jiangsu, China) following the manufacturer’s protocol. Cell cycle distribution percentage and apoptotic rates were determined by flow cytometry (CytoFLEX; Beckman Coulter, Brea, CA, USA) and analyzed with FlowJo v7.6.1 software.
Cell migration and invasion assays
Transwell assay was performed to evaluate cell migration and invasion abilities using Transwell chambers (CORNING Life Sciences, Corning, MA, USA). For the migration assay, 2 × 104 transfected UM cells were suspended in a 200 μL serum-free medium in the upper chamber, while the lower chamber contained 800 μL medium with 10% fetal bovine serum. After 24 or 48 h (based on different cell lines), the upper chamber was washed, fixed, stained with 0.25% crystal violet, and counted under a light microscope. Cell numbers were counted in three random fields of view. For the invasion assay, the same steps were followed, with Matrigel (BD Biosciences, Franklin Lakes, NJ, USA) pre-coated onto the upper layer for 60 min at 25°C before the experiment.
For the wound healing assay, a 200-µL pipette tip was used to create a vertical scratch wound in the middle slide after cells were transfected and seeded at least 90% confluence into 6-well plates. Pictures of the wound were taken at the same position under a microscope at 0 and 24 h. Migration ability was analyzed by quantitatively evaluating the gap distance using ImageJ software.
Fluorescence in situ hybridization (FISH) and immunofluorescence
RNA FISH was conducted to assess the subcellular localization of circ_0053943 in MUM2B, C918, and OCM-1A cells using a FISH Kit (RiboBio) following the manufacturer’s instructions. Circ_0053943-specific Cy3-labeled probes were designed and synthesized by RiboBio. Briefly, cells were fixed with 4% paraformaldehyde, permeabilized with 0.5% Triton X-100 in phosphor-buffered saline (PBS), and blocked with a prehybridization buffer. Subsequently, the cells were incubated in a hybridization buffer containing a FISH probe overnight. After rinsing with sodium citrate buffer, cells were incubated with Hoechst 33342 (ThermoFisher Scientific, MA, USA) for nuclear staining. For dual RNA-FISH and immunofluorescence, cells were blocked with an immunostaining blocking solution (Beyotime) after incubation with the hybridization buffer containing the FISH probe. Subsequently, cells were incubated with primary antibodies overnight and labeled with fluorescence-conjugated secondary antibodies for 1 h in the dark. Hoechst 33342 (ThermoFisher Scientific) was added for nuclear visualization. Images were acquired using the Nikon A1Si Laser Scanning Confocal Microscope (Nikon Instruments, Tokyo, Japan).
Tissue or cellular protein was lysed in RIPA lysis buffer (Beyotime) following the instructions, and the protein concentration was detected using the BCA kit (Vazyme, Nanjing, China). Western blotting experiments and signal quantification were performed according to the manufacturer’s protocols. Antibody information is provided in Suppl. Table 4.
The biotin-labeled pulldown probe targeting circ_0053943 and the negative control were designed and synthesized by RiboBio as detailed in Suppl. Table 5. The RNA-pulldown assay was conducted with the Pierce Magnetic RNA-Protein Pull-Down Kit according to the protocol (ThermoFisher Scientific). Lysates from UM cells were incubated with transcribed biotin-labeled circ_0053943 and pulled down with streptavidin beads. Eluted proteins from the RNA pulldown assay were subjected to mass spectrometric analysis or western blot.
The luciferase reporter assay was employed to evaluate the direct binding specificity between EGFR 3′UTR and IGF2BP3. For the luciferase reporter assays, a wild (EGFR–3’UTR) or mutant (EGFR–3′UTR-Mut #1 or #2) fragment was inserted into a pmirGLO vector (Promega, Madison, WI, USA) containing the firefly luciferase gene (hLuc+) and renilla luciferase gene (hRluc). UM cells cultured in 24-well plates at 60%–80% confluency were transfected with the pmirGLO wildtype or mutant reporter vector along with circ_0053943 plasmid or vectors using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s protocol. The relative values of hLuc+ and hRluc were detected by Centro LB960 XS3 (Berthold, Bad Wildbad, German), and each measurement was repeated three times.
RNA-seq libraries were constructed in circ_0053943-silenced MUM2B cells. Initially, RNA integrity was assessed using the Agilent 2100 Bioanalyzer (Genesky, Shanghai, China) after quality inspection with starting RNA at 2 μg. Subsequently, Oligo-dT magnetic beads were employed for mRNA purification and fragmentation to 100–300 bp. Following this, first-and second-strand cDNA was synthesized, followed by end repair and adenylate 3′ ends. Finally, after ligating adapters, fragment size selection, and PCR amplification, a HiSeq system (Illumina, San Diego, CA, USA) in Pair End mode was used for high-throughput RNA sequencing. Differentially expressed genes (DEGs) between two groups were identified using the Deseq2 package in R v3.6.3. A cutoff criterion of the absolute value of fold change (|log2FC|) > 1 and p-value < 0.05 was selected for DEG identification.
A RIP Kit (Millipore, Burlington, MA, USA) was acquired for RIP assays. Initially, cells were lysed with RIP lysis buffer, along with protease and RNase inhibitors. Subsequently, magnetic beads were mixed with 10 μg anti-IGF2BP3, anti-Flag, or anti-IgG antibodies. Cell lysates were then mixed with magnetic beads and antibodies at 4°C overnight. Finally, the immunoprecipitated RNA was extracted for qRT-PCR after RNA purification.
Relevant RNA level was measured by qRT-PCR after the extracted RNA of UM cells was incubated with or without RnaseR (3 U/μg) for 10 min at 37°C. For actinomycin D treatment, the total RNA of UM cells was harvested every 6 h after treatment (5 μg/mL). The variation trends of circ_0053943 and RasGRP3 were detected by qRT-PCR.
For the orthotopic and subcutaneous xenografted model, 1 × 106 transfected cells were resuspended in PBS and subcutaneously injected into the armpits of BALB/c nude mice (four weeks old, male) obtained from the Animal Center of Nanjing Medical University (Nanjing, China) for the xenograft model. A total of 48 mice were randomly divided into eight research groups, each consisting of six mice. The tumor volume was calculated every week, and all mice were sacrificed after 4 weeks. Tumors were resected and fixed in formalin for subsequent immunohistochemistry detection.
For the tumor metastasis assay in vivo, 1 × 106 relevant cells resuspended in 50 μL PBS were injected into the mice spleen via the left epigastric incision. A total of 40 mice were grouped into eight groups, with five mice per group. The livers of the mice were resected and fixed after 6 weeks. All animal experiments were performed according to the institutional guidelines of the Committee on the Ethics of Animal Experiments of Nanjing Medical University under the number IACUC-2108017.
Every experiment was repeated at least three times. SPSS v29.0 software (Chicago, IL, USA) and GraphPad Prism software (La Jolla, CA, USA) were used for the statistical analyses. Before determining appropriate statistical tests or procedures, we conducted Shapiro-Wilk tests to detect the normality distribution of variables. Variation analysis of two groups was performed with Student’s t-test, while analysis of variance was used to analyze the difference between more than two groups.
Verification of circ_0053943 in UM
The present study focused on upregulated circRNAs based on the published circRNA microarray data between UM and normal uveal tissues [12]. Then, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis was performed to detect these upregulated circRNAs in UM and normal tissues for further validation. Circ_0053943 was identifeid as the most significantly differentially expressed circ RNAs upregulated in UM tissues (Fig. 1A), similar to the previously confirmed circ_0119872 (Suppl. Fig. S1A) [12,13]. Subsequently, we assessed the level of circ_0053943 in UM cell lines; as shown in Fig. 1B, its level in ARPE-19 cells was significantly lower than that in UM cell lines.
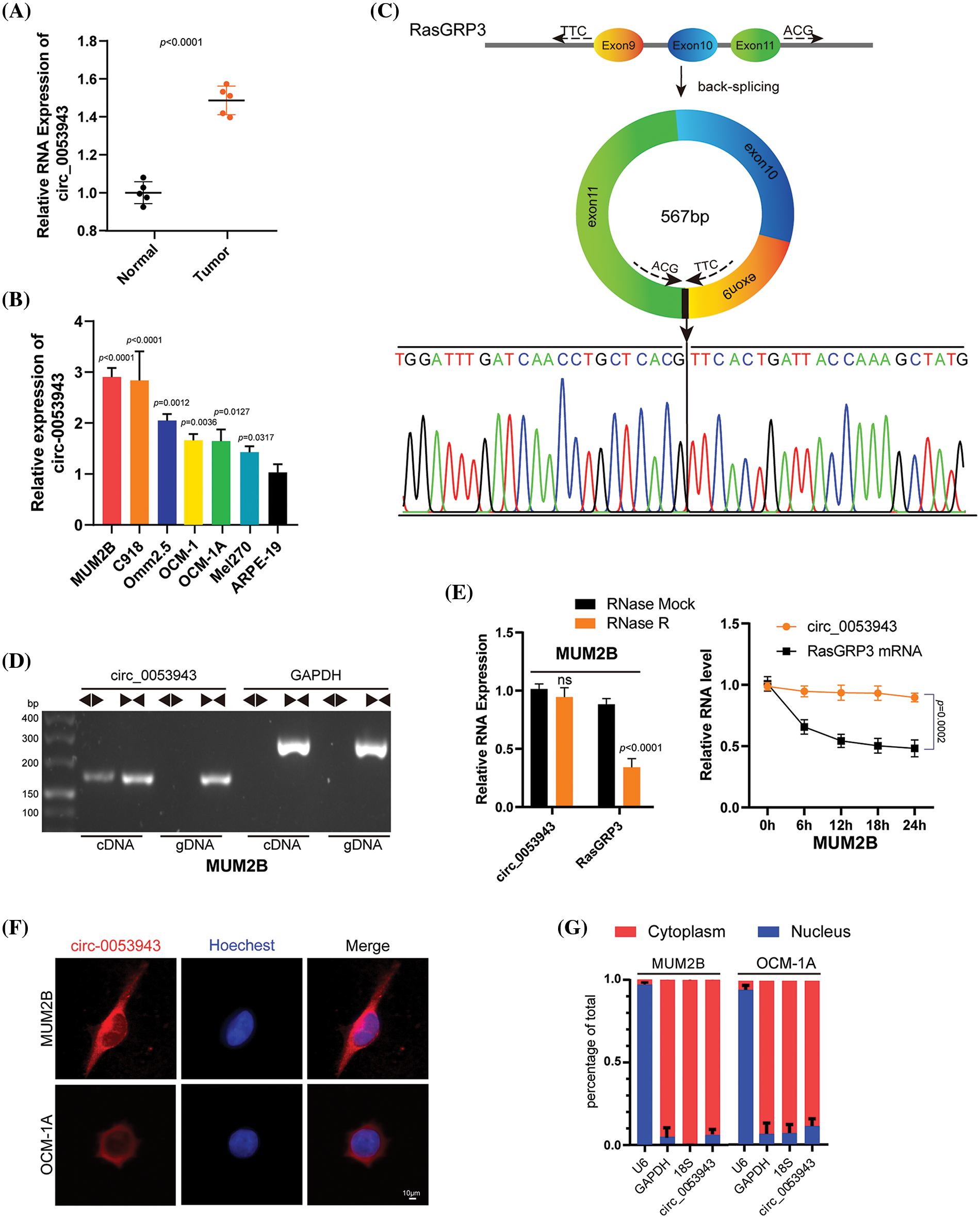
Figure 1: Characterization of circ_0053943 in uveal melanoma (UM). (A) Expression of circ_0053943 in 5 UM and 5 human normal melanocyte tissues in our cohort. (B) Expression of circ_0053943 in human retinal pigment epithelial cell line (ARPE-19) and 6 UM cell lines. (C) The schematic illustration showed the back splicing of circ_0053943, and sanger sequence validated the splicing site. (D) PCR and agarose gel electrophoresis confirmed the circular formation of circ_0053943, using divergent and convergent primers in gDNA and cDNA of MUM2B cells. GAPDH was used as a negative control. (E) Circ_0053943 and linear RasGRP3 expression levels were detected after RNase R in MUM2B. (F, G) RNA fluorescence in situ hybridization (FISH) (F) and subcellular fractionation assays. Scale bars, 50 μm. (G) Indicated that circ_0053943 was predominately localized in the cytoplasm of UM cells. nsp > 0.05.
Circ_0053943 originates from exons 9, 10, and 11 of RasGRP3, containing 567 nucleotides, according to the circBase annotation (Fig. 1C) [21]. To confirm its circular nature, convergent and divergent primers were designed to amplify linear and back-splicing products based on cDNA and genomic DNA (gDNA). Agarose gel electrophoresis demonstrated that convergent primers successfully amplified products of the expected size for circ_0053943 and GAPDH from both cDNA and gDNA. However, divergent primers for circ_0053943 only yielded a PCR product from cDNA, not gDNA (Fig. 1D and Suppl. Fig. S1B). Additionally, an RNase R treatment assay confirmed the higher resistance of circ_0053943 to RNase R and actinomycin D treatment compared to linear RasGRP3 mRNA (Fig. 1E and Suppl. Fig. S1C). RNA-FISH and RNA nucleus/cytoplasm separation revealed that circ_0053943 was predominantly localized in the cytoplasm (Figs. 1F, 1G and Suppl. Figs. S1D–S1E). Three shRNAs targeting circ_0053943 were transfected into MUM2B, Omm2.5, and C918 cells, while Mel270, OCM-1, and OCM-1A cells were developed with an overexpression lentivirus. Notably, the level of RasGRP3 mRNA remained unchanged in cells with reduced or increased circ_0053943 levels (Suppl. Figs. S1F, S1G).
Circ_0053943 promotes the proliferation and metastasis of UM cells
To elucidate the biological role of circ_0053943 in UM, shRNA#1 and #2, along with a control shRNA, were chosen for cell phenotype assays due to their greater knockdown efficiency. Cell proliferation was assessed using the CCK8 and EDU staining assays. Downregulation of circ_0053943 significantly suppressed UM cell proliferation, while upregulation of circ_0053943 promoted cell proliferation (Figs. 2A, 2B and Suppl. Figs. S2A, S2B). Transwell assays and scratch wound healing assays revealed that circ_0053943 knockdown significantly decreased tumor cell migration and invasion ability. In contrast, circ_0053943 upregulation enhanced these abilities in UM cells (Figs. 2C, 2D and Suppl. Figs. S2C, S2D). Flow cytometric assays of cell cycle distribution indicated that downregulation of circ_0053943 increased the percentage of cells in the G0/G1 phase and decreased the population in the S phase of the cell cycle (Fig. 3A). Conversely, upregulation of circ_0053943 promoted the progression of G1-to-S phase transition remarkably in UM cells (Fig. 3B and Suppl. Fig. S2E). Apoptosis assay demonstrated that cells transfected with shRNAs exhibited higher apoptotic rates than the control, while transfection with the overexpressed lentivirus reduced apoptotic cells (Figs. 3C, 3D and Suppl. Fig. S2F). Western blot showed that reducing circ_0053943 led to decreased protein levels of Cyclin D1, Cyclin-dependent kinase 4 (CDK4), and B-cell lymphoma 2 (Bcl-2) while increasing the level of Bcl2-associated X (Bax). These changes were consistent with the results obtained from flow cytometric assays. Conversely, the upregulation of circ_0053943 had opposite effects (Figs. 3E, 3F and Suppl. Fig. S2G).
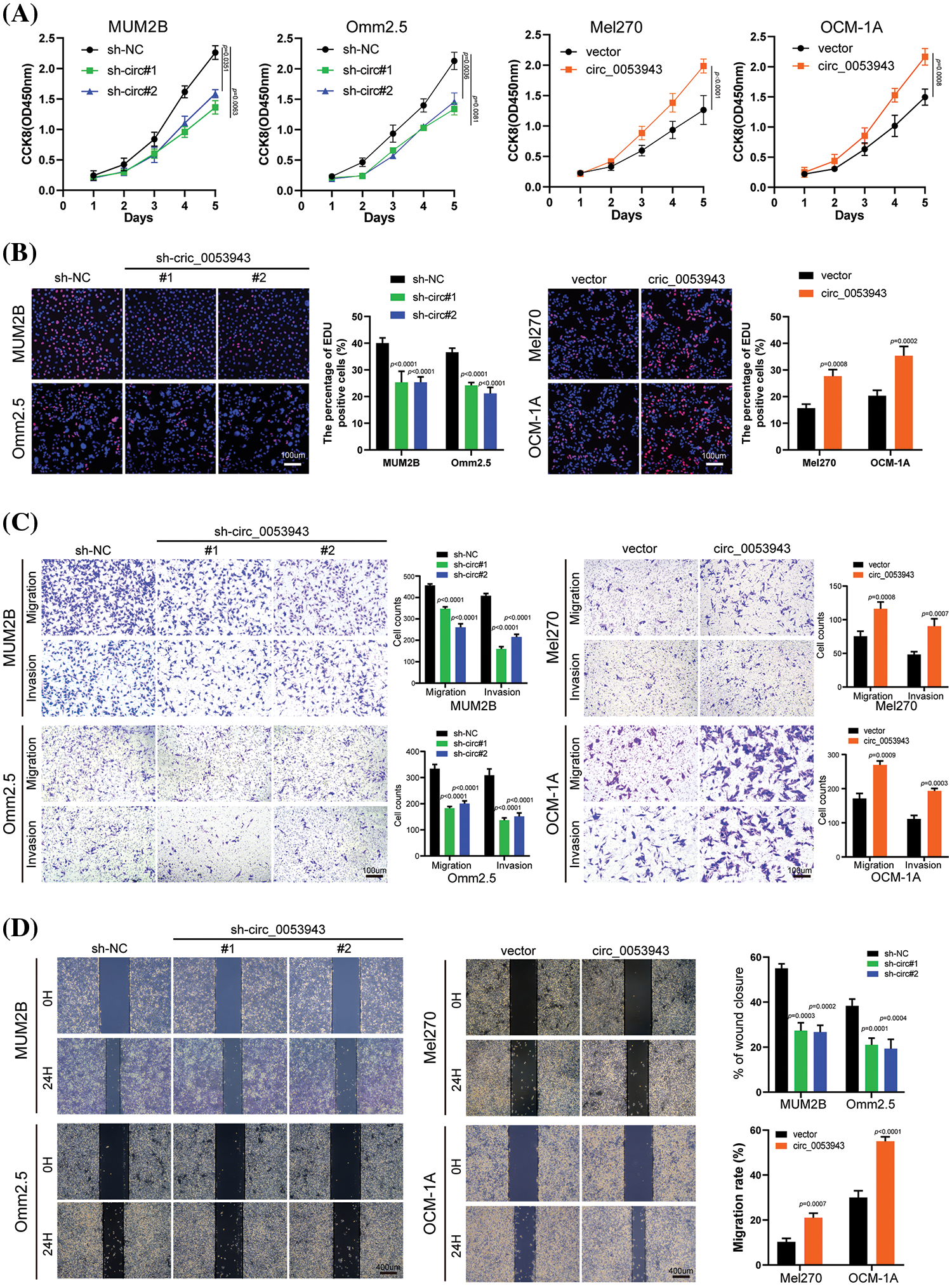
Figure 2: Circ_0053943 promotes the proliferation, migration, and invasion of UM cells in vitro. (A) Cell Counting Kit-8 (CCK8) assays were applied to determine the growth curves of circ_0053943 knockdown or overexpression cells. (B) 5-ethynyl-2-deoxyuridine (EDU) assays were performed to assess the cell proliferation ability. Scale bars, 100 μm. (C) Transwell migration and invasion assays were applied to evaluate the migration and invasion abilities of UM cells. Scale bars, 100 μm. (D) Cell migration ability was assessed by wound healing assay. Scale bars, 400 μm.
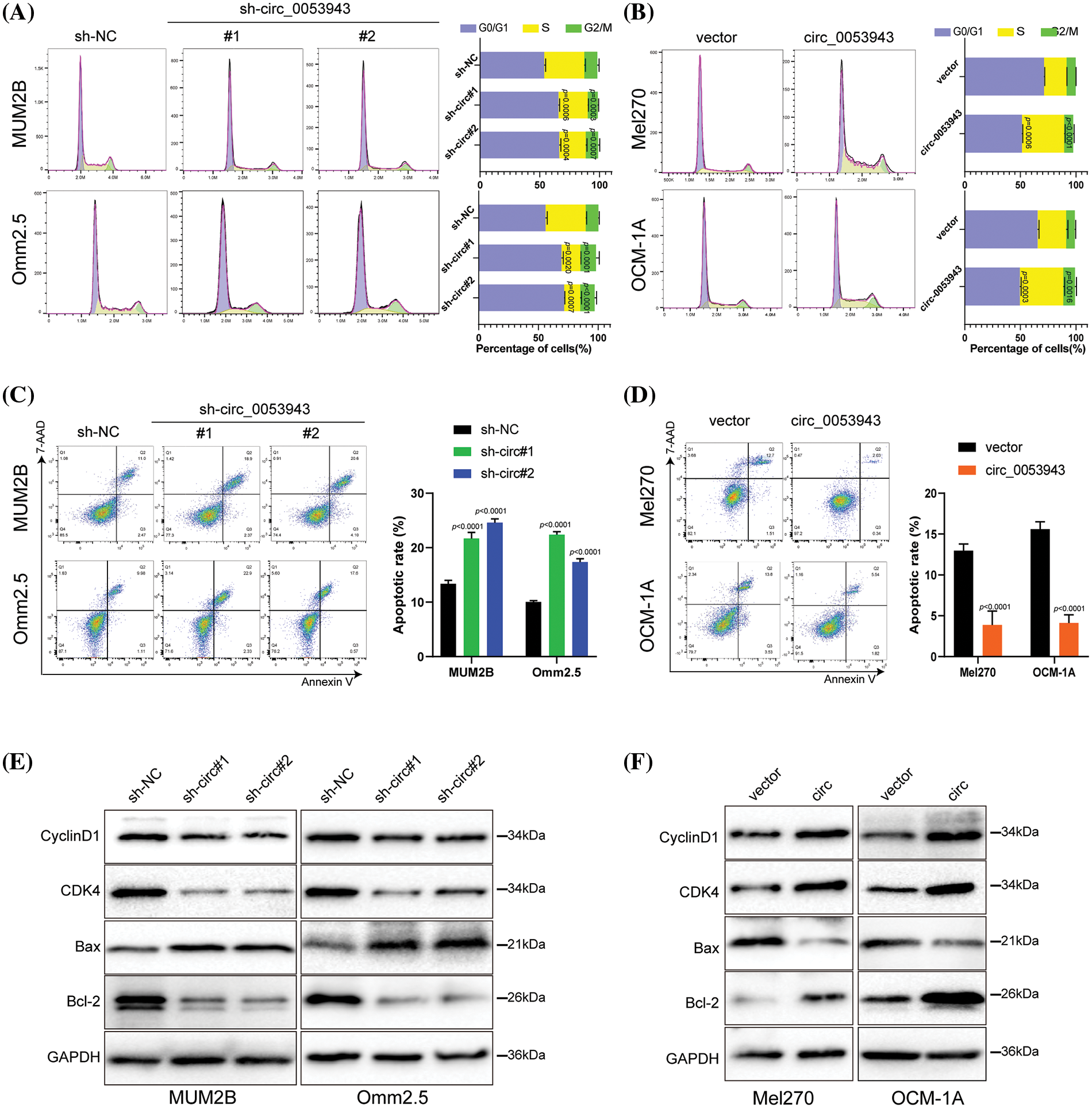
Figure 3: Circ_0053943 regulates the cell cycle and apoptosis of UM cells. (A, B) Cell cycle distributions were detected by flow cytometry in circ_0053943 knockdown or overexpression cells. (C, D) The apoptotic rates were performed and analyzed after cells were treated with 0.5 mM H2O2 for 4 h. All data are presented as the means ± SD of three independent experiments. (E, F) The expression of cell cycle and apoptosis makers (Cyclin D1, CDK4, Bcl-2, and Bax) were detected by western blot in relatively treated cells.
Circ_0053943 and IGF2BP3 cooperate to play oncogenic roles
The study explored the functional role of circ_0053943 and its interaction with IGF2BP3 in UM progression. It was observed that circ_0053943 may not act as a miRNA sponge in UM, as it was not significantly enriched by the Argonaute 2 (AGO2) antibody (Suppl. Fig. S3A). To identify potential protein interactors of circ_0053943, an RNA pulldown assay coupled with mass spectrometry MS was conducted, revealing 167 differential proteins between the sense and antisense circ_0053943 transcript pulldown groups in MUM2B cells (Suppl. Table 6). Among these proteins, 50 were identified as potential partners with RBP characteristics (Suppl. Table 7). Given the predominant cytoplasmic localization of circ_0053943, a biotin-labeled RNA pulldown assay using cytoplasmic protein extracts from MUM2B cells was performed. The sense-specific band at 55~70 kDa, obtained from sodium dodecyl sulfate-polyacrylamide gel electrophoresis, was excised for further analysis (Fig. 4A and Suppl. Fig. S3B). Among the proteins identified, only IGF2BP3 was detected in both the input group and circ_0053943 pulldown products (Fig. 4B and Suppl. Fig. S3C). Furthermore, the study investigated the clinical relevance of IGF2BP3 levels in UM, revealing that patients with higher IGF2BP3 levels exhibited shorter overall survival and disease-free survival rates (Suppl. Fig. S3D). To confirm the interaction between circ_0053943 and IGF2BP3, a RIP assay was performed using an anti-IGF2BP3 antibody. The results showed that the anti-IGF2BP3 antibody specifically enriched circ_0053943 compared to the anti-IgG antibody (Suppl. Fig. S3E). Additionally, dual RNA-FISH and immunofluorescence assay demonstrated the co-localization of circ_0053943 and IGF2BP3 in MUM2B and OCM-1A cells, providing further evidence of their interaction (Fig. 4C).
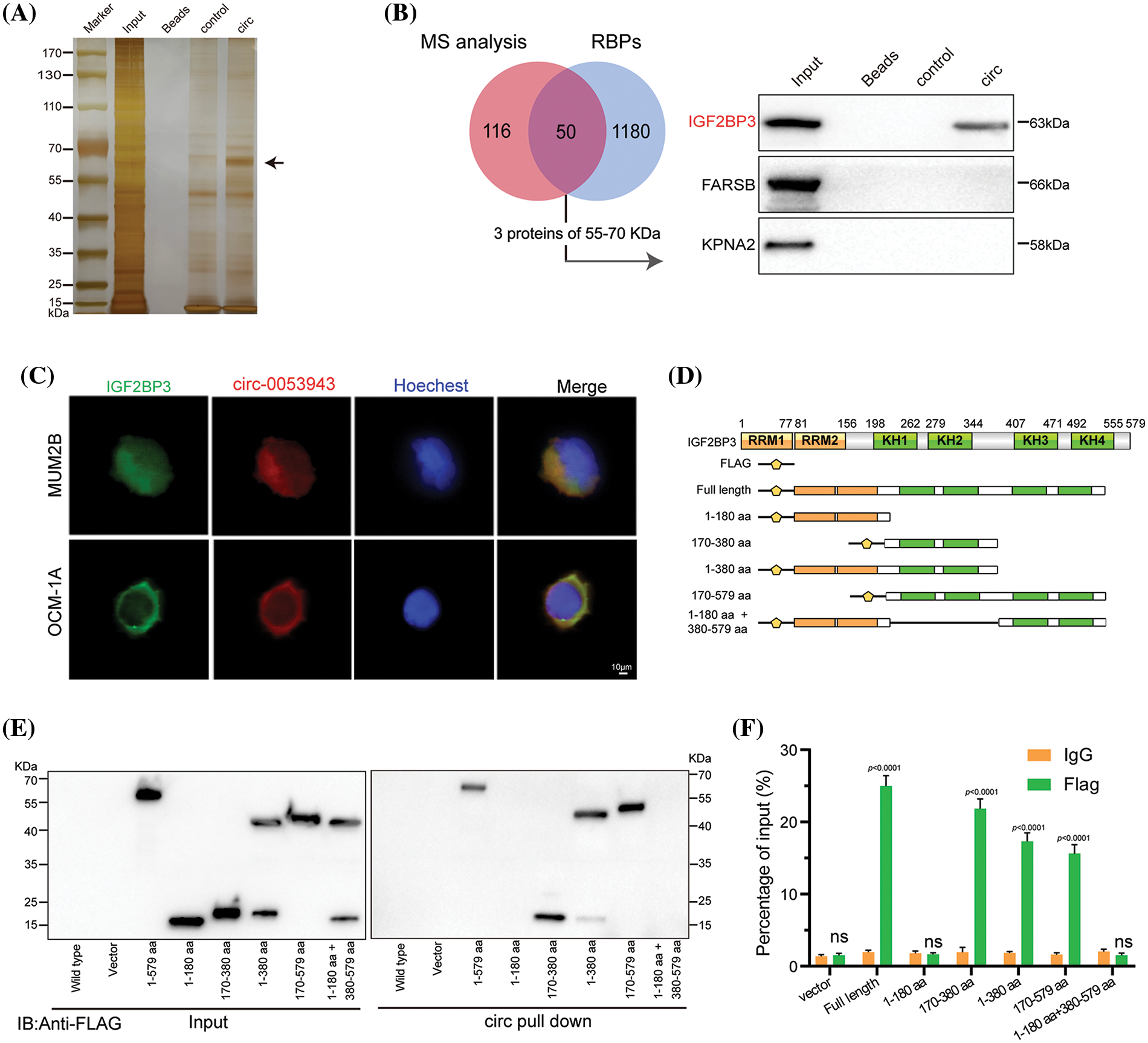
Figure 4: Circ_0053943 binds to the KH1 and KH2 domains of IGF2BP3. (A, B) RNA pulldown assay followed by silver staining and western blot indicated that circ_0053943 specifically interacted with IGF2BP3. (C) Dual RNA FISH and immunofluorescence assays showed that circ_0053943 and IGF2BP3 colocalized in the cytoplasm of MUM2B and OCM-1A. Scale bars, 50 μm. (D) Functional domain and truncated mutation annotation of IGF2BP3. (E, F) RIP assay and RNA pulldown assay confirmed that circ_0053943 characteristically interacted with the KH1 and KH2 domains of IGF2BP3. nsp > 0.05.
Next, six truncated IGF2BP3 plasmids targeting its functional domains were designed. Protein domain mapping and RIP assay revealed that the KH1 and KH2 domains were essential for the interaction between IGF2BP3 and circ_0053943 (Figs. 4D–4F). Moreover, modulation of circ_0053943 level did not affect the mRNA and protein levels of IGF2BP3 (Suppl. Figs. S3F–S3G), and knockdown of IGF2BP3 did not significantly alter the level of circ_0053943 (Suppl. Figs. S3H–S3I). Importantly, IGF2BP3 depletion abolished the circ_0053943-induced cell proliferation, migration, and invasion. Conversely, upregulation of IGF2BP3 could not affect cell proliferation and metastatic ability when circ_0053943 was knocked down (Suppl. Figs. S4A–S4F). These findings highlight the potential collaboration between circ_0053943 and IGF2BP3 in driving UM oncogenesis.
EGFR identified as a downstream target of circ_0053943 and IGF2BP3
To elucidate the mechanism by which circ_0053943 promotes UM progression through IGF2BP3, the study focused on the role of IGF2BP3 in stabilizing target mRNA transcripts, as previously reported in various cancers [22–24]. RNA-sequence analysis was performed to profile global gene expression changes upon circ_0053943 knockdown in MUM2B cells. The analysis revealed a total of 4,293 dysregulated genes, including 2,308 upregulated and 1,985 downregulated genes (Fig. 5A). Pathway enrichment analysis based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database showed that the mitogen-activated protein kinase (MAPK) pathway was the most enriched (Fig. 5B). The key components of the MAPK pathway, including P38, extracellular signal-regulated kinase (ERK), c-Jun NH2-terminal kinase (JNK), and their phosphorylation forms, were evaluated using western blot. Interestingly, while the levels of P38, ERK1/2, and JNK did not show significant changes upon circ_0053943 silencing, the phosphorylated forms of ERK significantly reduced (Fig. 5C).
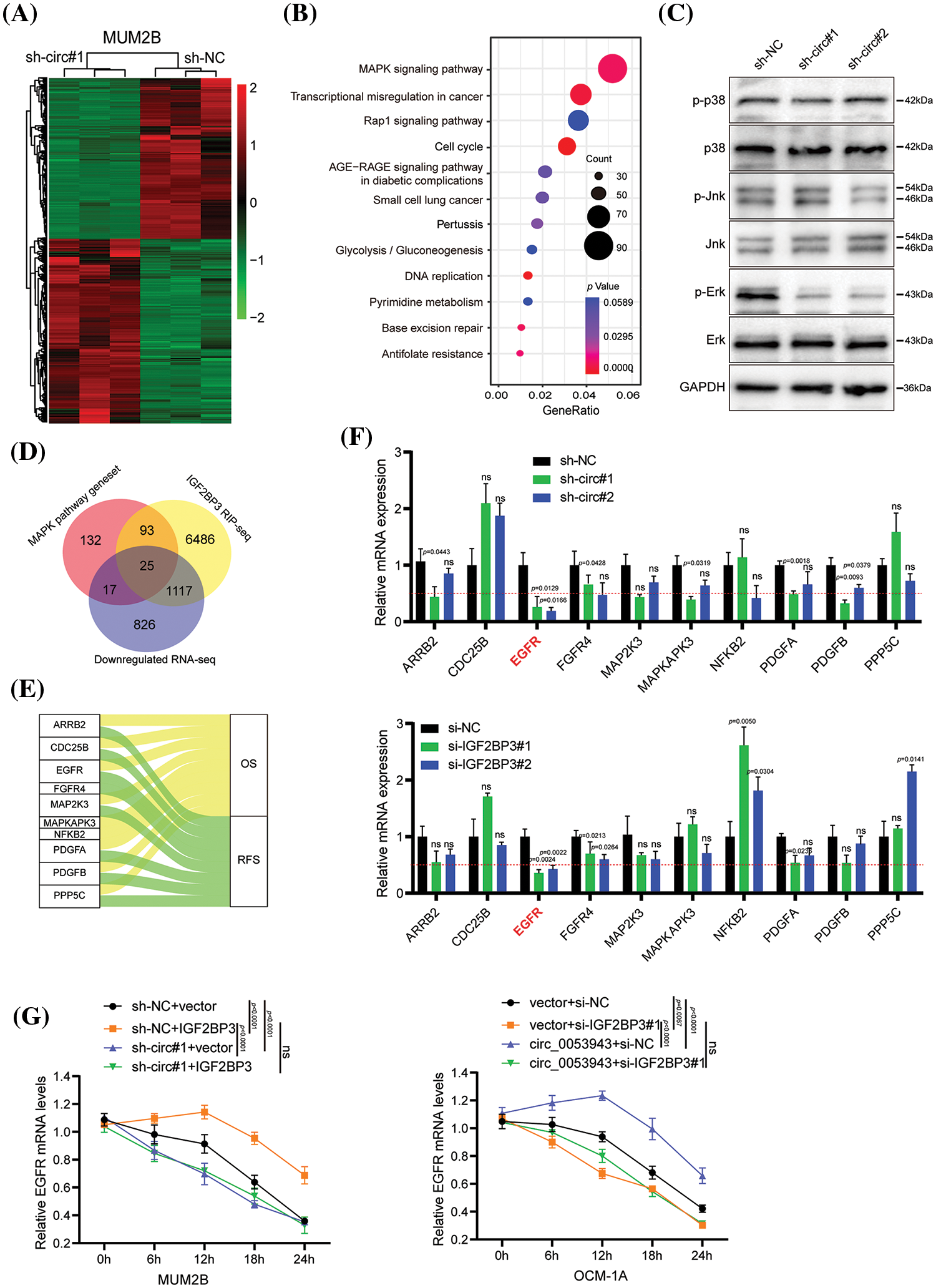
Figure 5: EGFR is downregulated by circ_0053943 and correlated with UM progression. (A) Heatmap showed the RNA-seq result in circ_0053943 knockdown and control MUM2B cells. (B) The top 12 enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) terms for DEGs. (C) Western blot indicated total and phosphorylated proteins of MAPK signaling pathway. (D) Venn diagram showing the 25 overlapping genes by downregulated genes of silencing circ_0053943 RNA-sequence data and MAPK pathway gene set together with IGF2BP3 binding genes of IGF2BP3 RIP-sequence data (GSE90642 and GSE90639). (E) Sankey diagram representing 10 overlapping genes associated with UM progression. (F) The transcript levels of these 10 genes in the circ_0053943 knockdown (upper) and IGF2BP3 knockdown (down) MUM2B cells using qRT-PCR. (G) The half-life of EGFR mRNA after treatment with 5 μM actinomycin D for the indicated times in the silencing circ_0053943 (left) and IGF2BP3 (right) cells with ectopically expressed IGF2BP3 and circ_0053943. All data are presented as the means ± SD of three independent experiments. nsp > 0.05.
A circ_0053943 integrated analysis of the RNA-sequence data, two published IGF2BP3 RIP-sequence datasets [17], and critical components in the MAPK signaling pathway identified 25 potential target genes (Fig. 5D). Survival analysis of these genes by GEPIA 2 [24], revealed 10 genes that showed prognostic impacts in UM (Fig. 5E and Suppl. Figs. S5A–S5B). Validation in circ_0053943 and IGF2BP3 knockdown cells showed that ARRB2, EGFR, FGFR4, PDGFA, and PDGFB exhibited decreased transcript levels upon circ_0053943 depletion, with EGFR demonstrating a significant decrease of more than 50% in both circ_0053943 and IGF2BP3 depletion cells (Fig. 5F).
EGFR was selected as the IGF2BP3-bound target altered by circ_0053943 for further investigation. Western blot analysis confirmed that knockdown of either circ_0053943 or IGF2BP3 reduced the protein levels of EGFR (Suppl. Fig. S6A). Additionally, qRT-PCR and western blot demonstrated that knocking down circ_0053943 abrogated the long-lasting effect of IGF2BP3 upregulation on the EGFR transcript. Conversely, reducing IGF2BP3 abolished the half-life, and the mRNA level increased by circ_0053943 overexpression (Fig. 5G and Suppl. Figs. S6B, S6C). These findings suggested a functional interdependency between circ_0053943 and IGF2BP3 in stabilizing the EGFR transcript, highlighting EGFR as a downstream target in the circ_0053943/IGF2BP3 axis contributing to UM progression.
Circ_0053943 cooperates with IGF2BP3 to stabilize EGFR mRNA in an m6A-dependent manner
To investigate whether circ_0053943 and IGF2BP3 regulate the stability of EGFR mRNA in an m6A-dependent manner, the study identified potential m6A-modified regions of EGFR using m6A RIP-sequence data. The canonical “GGAC” m6A motif was found in these m6A peaks, confirming that EGFR mRNA is indeed modified by m6A methylation (Fig. 6A).
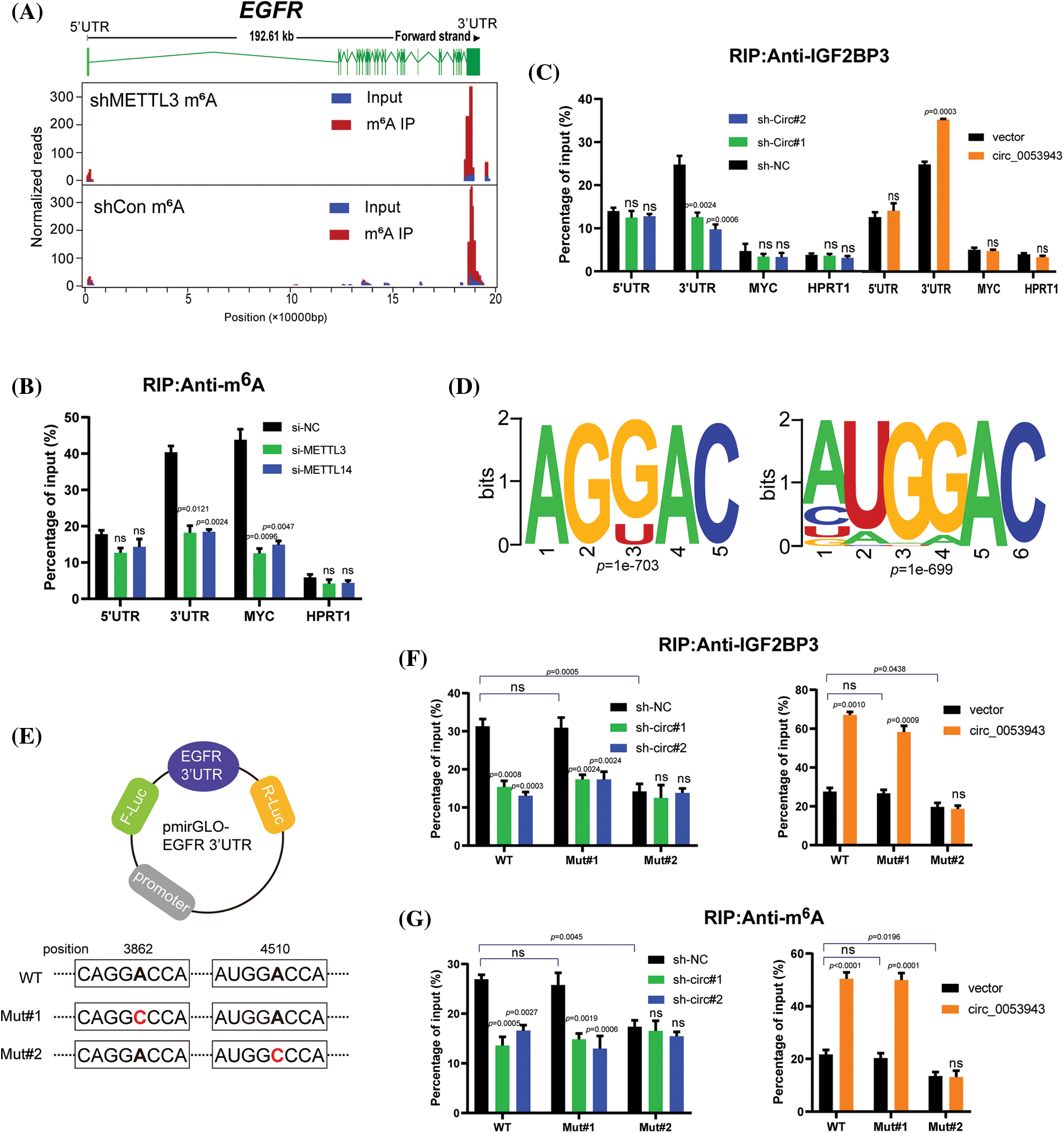
Figure 6: Circ_0053943 cooperates with IGF2BP3 to regulate EGFR in an m6A-dependent manner. (A) Distribution of m6A peaks across the EGFR transcript based on m6A RIP-sequence data. (B) RIP qRT-PCR showing the enrichment of m6A modification in the EGFR 5′ UTR/3′ UTR regions in the METTL3 and METTL14 knockdown MUM2B cells. MYC CRD was a positive control. HPRT1 was a negative control. (C) RIP qRT-PCR detecting the enrichment of IGF2BP3 in the EGFR 3′UTR, 5′UTR, MYC CRD, and HPRT1 in circ_0053943 knockdown and overexpressed cells. (D) Two top consensus sequences of IGF2BP3-binding sites and the m6A motif detected by SRAMPA and RMBase V2.0 motif analysis. (E) Schematic representation of wild-type (WT) and mutated (MUT) EGFR 3′UTR of the pmirGLO vector. (F, G) RIP qRT-PCR detection of the enrichment of IGF2BP3 and m6A in the EGFR 3′UTR WT and MUT luciferase reporters in the circ_0053943 knockdown and overexpression cells. All data are presented as the means ± SD of three independent experiments. nsp > 0.05.
M6A-specific RIP was then performed to determine the m6A methylation level of EGFR in METTL3 and METTL14 knockdown cells (Suppl. Fig. S7A). A reduction in the coding region instability determinant (CRD) of MYC (positive control), but not in HPRT1 (negative control), confirmed that EGFR was modified by m6A (Fig. 6B). Since IGF2BP3 preferentially binds to the “GGAC” m6A core motif of its targets [17], the study examined whether circ_0053943 affects the binding of IGF2BP3 to the m6A-modified EGFR. IGF2BP3 RIP qRT-PCR assays showed that knockdown of circ_0053943 significantly reduced the binding of IGF2BP3 to the m6A site in the 3′UTR region of EGFR, while overexpression of circ_0053943 facilitated this binding. This effect was not observed in the EGFR 5′UTR or MYC CRD (Fig. 6C). Moreover, an RNA pulldown assay with in vitro-transcribed circ_0053943 confirmed that the binding ability of circ_0053943 to the 3′UTR of EGFR was largely abolished following IGF2BP3 knockdown and increased after IGF2BP3 overexpression (Suppl. Fig. S7B). These findings confirm that circ_0053943 may bind to the 3′UTR of EGFR in an IGF2BP3-dependent manner.
In the EGFR 3′UTR region, two “GGAC” m6A motifs were predicted by m6A modification site predictors SRAMPA and RMBase v2.0 (Fig. 6D and Suppl. Table 8). Luciferase reporters containing the wild-type EGFR 3′UTR (WT-3′UTR), mutant #1, and #2 3′UTR (Mut#1-and Mut#2-3′UTR; changing GGAC to GGCC) were constructed to elucidate the potential roles of circ_0053943 in m6A modification of EGFR (Fig. 6E). Both IGF2BP3 and m6A RIP qRT-PCR assays showed higher enrichment with the WT and Mut#1-3′UTR reporters compared to Mut#2-3′UTR. Overexpression of circ_0053943 or IGF2BP3 remarkably increased the binding of IGF2BP3 and m6A in the WT and Mut#1-3′UTR but not in the Mut#2-3′UTR report. Conversely, knocking down circ_0053943 and IGF2BP3 independently reduced this binding except in the Mut#2-3′UTR report (Figs. 6F, 6G). Furthermore, luciferase reporter assays demonstrated that the relative luciferase activities were decreased by knockdown of circ_0053943 or IGF2BP3 in the WT and Mut#1-3′UTR but not in Mut#2-3′UTR reporter. Similarly, the relative luciferase activity of MUT#2-3′UTR reporter was also not altered by upregulated circ_0053943 or IGF2BP3 (Figs. 6H, 6I). Thus, methylation of the m6A-modified site c.4510A in EGFR 3′UTR contributed to EGFR expression.
Circ_0053943 promotes UM proliferation, metastasis, and angiogenesis through the upregulation of EGFR in vitro
EGFR-specific shRNA (sh-EGFR) and the negative control (sh-Ctrl), as well as an EGFR overexpression plasmid, were transfected into UM cells (Suppl. Fig. S8A). Cell proliferation assays revealed that EGFR silencing significantly inhibited the proliferation ability of circ_0053943 upregulated cells. Conversely, EGFR overexpression promoted UM cell proliferation in circ_0053943 knockdown cells (Figs. 7A, 7B, and Suppl. Figs. S8B, S8C). Transwell assays and wound healing assays indicated that metastatic abilities of circ_0053943 knockdown cells were significantly increased in sh-EGFR cells compared to sh-Ctrl cells (Figs. 7C, 7D and Suppl. Figs. S8D, S8E).
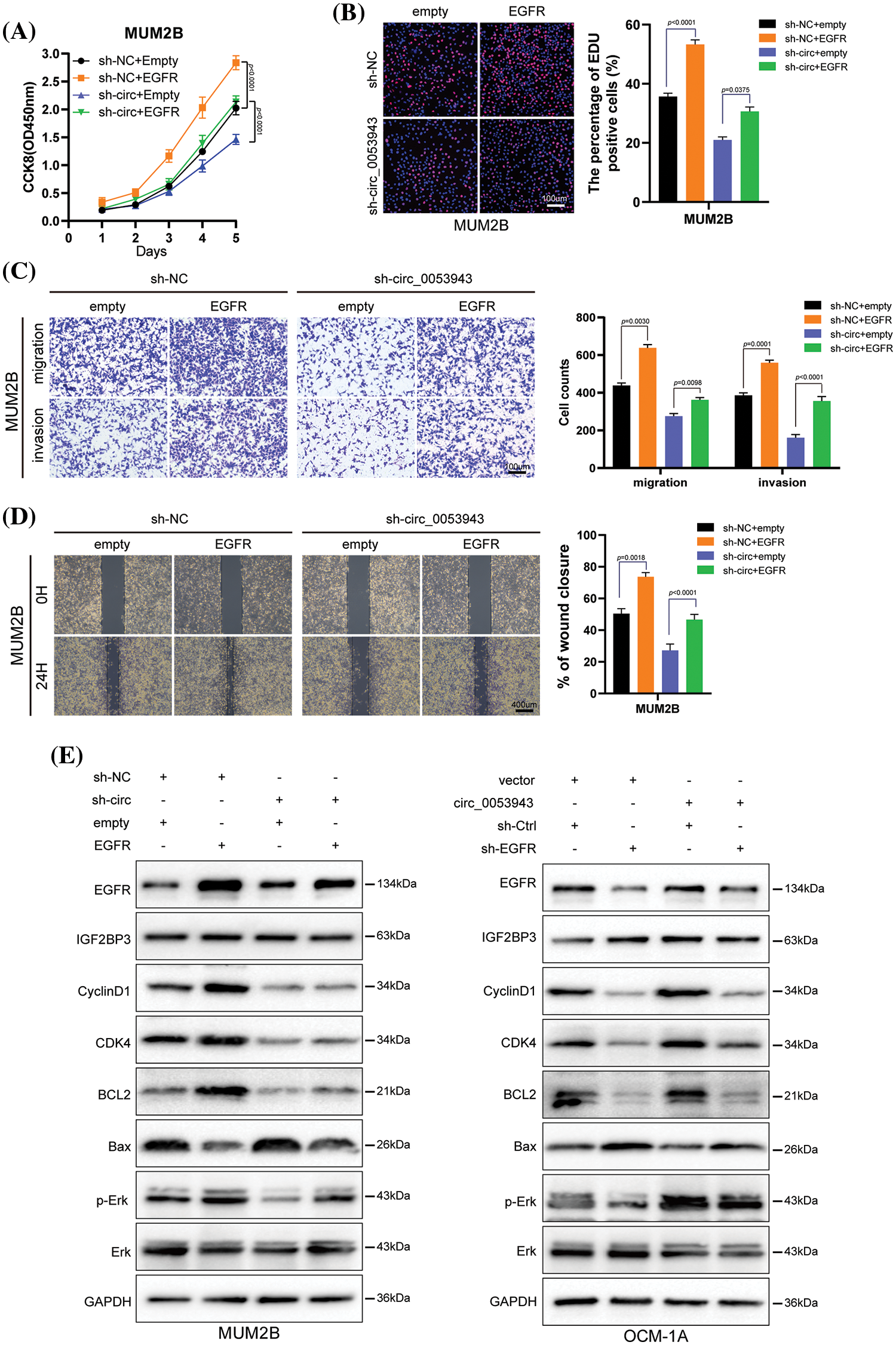
Figure 7: Circ_0053943 regulates UM proliferation, metastasis, cell cycle, and apoptosis via upregulating EGFR expression in vitro. (A, B) CCK8 and EdU were used to detect UM cell proliferation ability in circ_0053943 knockdown cells with ectopically expressed EGFR. Scale bars, 100 μm. (C, D) Transwell and wound healing assays detected UM cell metastasis capacity in circ_0053943 knockdown cells with ectopically expressed EGFR. Scale bars, 100 μm (C), 400 μm (D). (E) The expression of cell cycle and apoptosis makers (Cyclin D1, CDK4, Bcl-2, and Bax) along with EGFR, IGF2BP3, Erk, and p-Erk were detected by western blot in relatively treated cells. All data are presented as the means ± SD of three independent experiments.
Considering the role of EGFR in tumor angiogenesis, the potential implication of angiogenesis was analyzed using the HUVEC tube formation assay. Conditioned medium (CM) from sh-circ_0053943 tumor cells dramatically reduced the formation of capillary-like structures by HUVECs. Conversely, EGFR overexpression reversed the decreased capillary-like structure formation caused by circ_0053943 depletion. Meanwhile, CM from circ_0053943 UM cells showed promoting effects on capillary-like structure formation, which was inhibited by silencing EGFR (Suppl. Figs. S8F, S8G).
Western blotting results showed that the knockdown of circ_0053943 impaired the G1 to S transition by downregulating cycle-related proteins (Cyclin D1, CDK4), and the overexpression of EGFR could rescue it. Circ_0053943 upregulation promoted the G1 to S transition, while silencing EGFR impaired this transition. Alterations in apoptosis-related proteins (Bcl-2, Bax) demonstrated that the anti-apoptotic effect of circ_0053943 was weakened upon EGFR knockdown (Figs. 7E, 7F).
Circ_0053943 promotes UM proliferation, metastasis and angiogenesis through upregulating EGFR in vivo
To investigate the effects of circ_0053943 on UM proliferation and metastasis in vivo, subcutaneous xenograft nude mice models were established. MUM2B and OCM-1A cells transfected with sh-circ_0053943/sh-EGFR/overexpression plasmids, along with the respective control group and co-transfection group, were separately injected into nude mice.
Results showed that tumor growth was repressed by circ_0053943 knockdown, with smaller tumor volume and weight compared to the control groups. Overexpression of EGFR reversed the growth inhibition caused by circ_0053943 depletion (Figs. 8A, 8B). In circ_0053943 upregulated cells, the opposite effects were observed due to the repression by EGFR depletion (Figs. 8C, 8D).
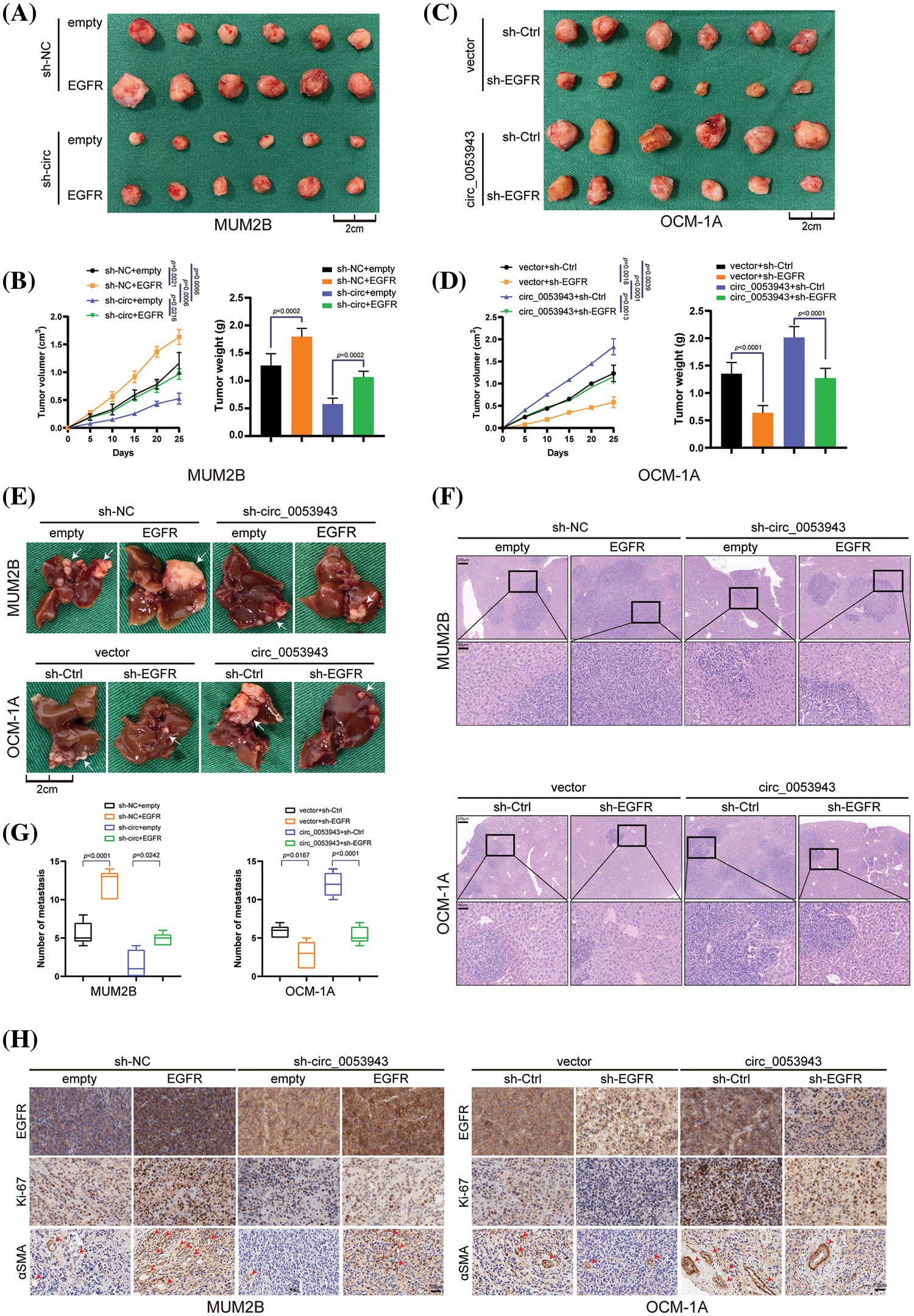
Figure 8: Circ_0053943 promotes UM proliferation and metastasis via upregulating EGFR expression in vivo. (A, B) Representative photographs of subcutaneous xenograft tumors were obtained from the different groups of nude mice transfected MUM2B cells with knockdown circ_0053943 and ectopically expressed EGFR. Tumors were observed by tumor size and average weight. (C, D) Representative photographs of subcutaneous xenograft tumors were obtained from the different groups of nude mice transfected OCM-1A cells with knockdown EGFR and ectopically expressed circ_0053943. Tumors were observed by tumor size and average weight. (E–G) Representative photographs (E) and HE staining (F) of liver metastases were obtained from nude mice transfected with relatively treated cells, and the number of metastases (G) was measured. Scale bars, 200 μm (up), 50 μm (down). (H) Protein levels of EGFR, Ki-67 and α-SMA in the tumor samples were determined by IHC. All data are presented as the means ± SD of three independent experiments. Scale bars, 100 μm.
Anatomical results confirmed that metastatic nodules were reduced in the circ_0053943 knockdown group compared with the control group. Conversely, metastatic lesions at the liver surface were more abundant in the EGFR overexpression group. Further analysis showed that the reduction of metastatic nodules caused by circ_0053943 depletions was increased by EGFR overexpression. In contrast, the opposite effect was observed in nude mice injected with circ_0053943/sh-EGFR cells (Figs. 8E–8G).
Immunohistochemistry results demonstrated that levels of EGFR, Ki-67 (indicating tumor proliferation), and α-SMA (indicating angiogenesis) were decreased in the circ_0053943 depletion group and increased in the overexpression group (Fig. 8H).
Collectively, these results confirmed that circ_0053943 upregulation enhanced the proliferation, metastasis, and angiogenesis capacity of UM by upregulating EGFR in vivo.
The present study has demonstrated the crucial oncogenic role of circ_0053943, a highly expressed circRNA in UM cells. Our phenotype experiment reveals that circ_0053943 promotes proliferation and metastasis in UM, including proliferation, migration, invasion, and angiogenesis of UM cells in vitro and tumorigenesis and hepatic metastasis in vivo. Notably, our study unveils a novel regulatory mechanism of circ_0053943 in promoting UM progression through its interaction with a protein. Specifically, increased circ_0053943 cooperates with IGF2BP3, forming a circ_0053943/IGF2BP3/EGFR mRNA-protein ternary complex. Consequently, it enhances the stability of EGFR mRNA, leading to the upregulation of EGFR and activation of MAPK/ERK signaling pathways in UM.
While cutaneous melanoma and UM share some similarities, it is essential to note that UM exhibits minimal overlapping genetic signatures with cutaneous melanoma [7,25,26]. In UM, genetic aberrancies are believed to drive tumor carcinogenesis through the abnormal activation of the Gα11/Q pathway [27,28]. Specifically, genes including GNAQ, GNA11, PLCB4, and CYSTLR2, and three secondary driver genes (BAP1, SF3B1, EIF1AX) have been reported to be frequently mutated in a substantial fraction of UM tissues [27,29–31]. However, somatic mutations above were observed only in limited UM cases, and GNAQ and GNA11 are relatively weak oncoproteins without co-mutated secondary driver genes [32]. Previous studies have emphasized the importance of aberrant ncRNA expressions in oncogenesis, providing a unique opportunity to optimize treatment paradigms and establish new therapeutic options for UM [33–36]. Therefore, further studies on the pathological functions of ncRNAs are required to obtain a complete picture of the undeveloped regulatory mechanisms under these somatic mutations.
CircRNAs represent a novel class of endogenous ncRNAs characterized by a covalently closed loop structure. Accumulating evidence suggests that dysregulated circRNA is associated with clinical and pathological features in various tumors, confirming its potential biological significance [37,38]. To date, only two studies have investigated the dysregulation of circRNAs in UM. Yang et al. identified an abnormal expression profile of circRNAs in UM tissues but did not explore the underlying mechanisms [12]. Building on these identified circRNAs, Liu et al. demonstrated that upregulated cytoplasmic circ_0119872 could promote UM tumorigenesis by acting as a miRNA sponge, modulating the activity of miRNAs on target genes [14]. Interestingly, circ_0053943, investigated in this study, was also predominantly located in the cytoplasm of UM cells, suggesting a potential link to post-transcriptional modifications. Moreover, it was unexpectedly found to be unable to bind with AGO2, ruling out the possibility of functioning as a miRNA sponge. To date, other mechanisms of circRNAs in tumor progression, particularly in UM, have not been thoroughly explored. This study identified that circ_0053943 could interact with the KH1 and KH2 domains of IGF2BP3, enhancing its biological function.
Insight into the functions of IGF2BPs (IGF2BP1/2/3) as a newly discovered family of the m6A “reader,” regulating mRNA stability and translation, has grown [39,14]. Despite several findings demonstrating the critical role of m6A modification in UM tumorigenesis, little is known about the effects of IGF2BPs in this disease [40,41]. Wan et al. reported widespread upregulation of IGF2BP3 in various ocular cancers, including UM [42]. Survival analysis using The Cancer Genome Atlas further indicates that IGF2BP3 can independently predict prognosis for patients with UM. Our findings suggest a latent mechanism by which IGF2BP3 facilitates tumor development, and circ_0053943 enhances this function in UM. Mechanistically, under the control of circ_0053943, IGF2BP3 directly binds to the m6A site, forming a circ_0053943/IGF2BP3/EGFR mRNA-protein ternary complex that enhances mRNA stability or translation of EGFR in an m6A-dependent manner. However, the molecular mechanism underlying IGF2BP3 upregulation in UM remains enigmatic.
EGFR, a 170-kDa transmembrane tyrosine kinase receptor, is detected in various malignant cells and has been reported to play a vital role in tumor development and progression [43]. Additionally, EGFR has been reported to regulate various downstream signaling pathways, including phosphatidylinositol-3-kinases/protein kinase B (PI3K/Akt) and MAPK/ERK signaling pathways [44–47]. Accumulating evidence shows that EGFR upregulation is significantly associated with tumor development and indicates poor prognosis in UM [48,49]. However, most studies have primarily focused on verifying how the upregulation of EGFR affects cancer cell proliferation and migration. This, however, is only part of the story, as the cause of EGFR upregulation in UM has not been thoroughly investigated. Therefore, our study demonstrated that upregulated circ_0053943 could enhance EGFR expression through the circ_0053943/IGF2BP3/EGFR complex, ultimately activating the MAPK/ERK pathway. These findings were consistent with a previous study [50] and offered insights into possible upstream regulators.
Our study provides evidence that circ_0053943 and IGF2BP3 cooperatively regulate the m6A modification of EGFR in UM, shedding new light on post-transcriptional EGFR expression and emphasizing the essential role of circRNAs in RNA metabolism. However, the mechanisms that upregulate circ_0053943 and mediate its export circ_0053943 from the nucleus to the cytoplasm require further investigation. Additionally, we did not determine how circ_0053943 mediated the interaction between IGF2BP3 and EGFR by specifically binding to the KH1 and KH2 domains of IGF2BP3. RNA-seq and subsequent qRT-PCR verification demonstrated that the function of circ_0053943 on IGF2BP3-modified target mRNA is selective. The specificity of ncRNA function remains elusive, but it aligns with a recent study based on clear cell renal cell carcinoma [51]. Furthermore, it is important to note that the published IGF2BP3 RIP-sequence data mentioned in this study are not from UM cell lines [17]. Therefore, the identification of the IGF2BP3 binding molecular spectrum based on UM cells needs further exploration. Another strength of our study is that we substantiated our results using an in vivo metastasis animal model. While the animal model may not fully replicate the complexity of UM in humans, our findings from this model partly support the clinical data and in vitro function of circ_0053943 in promoting UM development. To date, the use of MAPK/ERK kinase (MEK) inhibitors in the management of metastatic uveal melanoma remains controversial, considering inevitable side effects [51,52].
In summary, we identified a novel circRNA, hsa_circ_0053943, overexpressed in UM and associated with a poor prognosis. Upregulating circ_0053943 exerted effect in stabilizing EGFR mRNA by forming a circ_0053943/IGF2BP3/EGFR RNA-protein ternary complex, which finally promotes the proliferation and aggressiveness of UM cells. Overall, our finding may provide a new target, circ_0053943, for the diagnosis and treatment of UM.
Acknowledgement: We are grateful to all of the patients who participated in this study.
Funding Statement: This work is supported by the National Natural Science Foundation of China (Nos. 82273159 and 82171838) and the Jiangsu Province’s Science and Technology Project (No. BE2020722). The sponsor or funding organization had no role in the design or conduct of this research.
Author Contributions: CXJ and LH conceived of the study and carried out its design. ZAD and WY performed the experiments. WZJ, QS, QG, HZ and SSY collected clinical samples. CXJ, ZAD, WY and WZJ conducted the statistical analysis. ZAD wrote the paper and CXJ revised the paper. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data supporting the conclusions of this study have been included in this article. Please contact the authors for requests for raw data.
Ethics Approval: The study protocol for patients was approved by the Human Ethics Committee of The First Affiliated Hospital of Nanjing Medical University under the number 2022-SRFA-334. It was conducted in accordance with the Declaration of Helsinki. All patients provided written informed consent. The protocol for the animal experiments was approved by the Institutional Animal Care and Use Committee of Nanjing Medical University under the number IACUC-2108017.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/or.2023.045972.
References
1. Chattopadhyay, C., Kim, D. W., Gombos, D. S., Oba, J., Qin, Y. et al. (2016). Uveal melanoma: From diagnosis to treatment and the science in between. Cancer, 122(15), 2299–2312. [Google Scholar] [PubMed]
2. Binkley, E. M., King, B. A., Hyer, D. E., Javed, A., Milhem, M. M. et al. (2023). Postoperative echography for optimization of radiation dosimetry in patients with uveal melanoma treated with plaque brachytherapy. Ophthalmol Retina, 7(7), 620–627. [Google Scholar] [PubMed]
3. Rosa, N., de Bernardo, M., di Stasi, M., Cione, F., Capaldo, I. (2022). A-scan ultrasonographic evaluation of patients with idiopathic intracranial hypertension: Comparison of optic nerves. Journal of Clinical Medicine, 11(20), 6153. [Google Scholar] [PubMed]
4. Spagnolo, F., Caltabiano, G., Queirolo, P. (2012). Uveal melanoma. Cancer Treatment Reviews, 38(5), 549–553. [Google Scholar] [PubMed]
5. Jager, M. J., Shields, C. L., Cebulla, C. M., Abdel-Rahman, M. H., Grossniklaus, H. E. et al. (2020). Uveal melanoma. Nature Reviews Disease Primers, 6(1), 24. [Google Scholar] [PubMed]
6. Smit, K. N., Jager, M. J., de Klein, A., Kili, E. (2020). Uveal melanoma: Towards a molecular understanding. Progress in Retinal and Eye Research, 75, 100800. [Google Scholar] [PubMed]
7. Smit, K. N., van Poppelen, N. M., Vaarwater, J., Verdijk, R., van Marion, R., et al. (2018). Combined mutation and copy-number variation detection by targeted next-generation sequencing in uveal melanoma. Modern Pathology, 31(5), 763–771. [Google Scholar] [PubMed]
8. Kristensen, L. A. O., Jakobsen, T., Hager, H., Kjems, J. (2021). The emerging roles of circRNAs in cancer and oncology. Nature Reviews. Clinical Oncology, 19(3), 188–206. [Google Scholar] [PubMed]
9. Goodall, G. J., Wickramasinghe, V. A. O. (2021). RNA in cancer. Nature Reviews Cancer, 21(1), 22–36. [Google Scholar] [PubMed]
10. Zeng, Z., Xia, L., Fan, S., Zheng, J., Qin, J. et al. (2021). Circular RNA CircMAP3K5 acts as a MicroRNA-22-3p sponge to promote resolution of intimal hyperplasia via TET2-mediated smooth muscle cell differentiation. Circulation, 143(4), 354–371. [Google Scholar] [PubMed]
11. Schneider, T., Bindereif, A. (2017). Circular RNAs: Coding or noncoding? Cell Research, 27(6), 724–725. [Google Scholar] [PubMed]
12. Yang, X., Li, Y., Liu, Y., Xu, X., Wang, Y. et al. (2018). Novel circular RNA expression profile of uveal melanoma revealed by microarray. Chinese Journal of Cancer Research, 30(6), 656–668. [Google Scholar] [PubMed]
13. Liu, S., Chen, L., Chen, H., Xu, K., Peng, X. et al. (2021). Circ_0119872 promotes uveal melanoma development by regulating the miR-622/G3BP1 axis and downstream signalling pathways. Journal of Experimental & Clinical Cancer Research, 40(1), 66. [Google Scholar]
14. He, L., Li, H., Wu, A., Peng, Y., Shu, G. et al. (2019). Functions of N6-methyladenosine and its role in cancer. Molecular Cancer, 18(1), 176. [Google Scholar] [PubMed]
15. Wang, X., Lu, Z., Gomez, A., Hon, G. C., Yue, Y. et al. (2014). N6-methyladenosine-dependent regulation of messenger RNA stability. Nature, 505(7481), 117–120. [Google Scholar] [PubMed]
16. Lu, H., Xie, Y., Tran, L., Lan, J., Yang, Y. et al. (2020). Chemotherapy-induced S100A10 recruits KDM6A to facilitate OCT4-mediated breast cancer stemness. The Journal of Clinical Investigation, 130(9), 4607–4623. [Google Scholar] [PubMed]
17. Huang, H., Weng, H., Sun, W., Qin, X., Shi, H. et al. (2018). Recognition of RNA N6-methyladenosine by IGF2BP proteins enhances mRNA stability and translation. Nature Cell Biology, 20(3), 285–295. [Google Scholar] [PubMed]
18. Paramasivam, A., George, R., Priyadharsini, J. V. (2021). Genomic and transcriptomic alterations in m6A regulatory genes are associated with tumorigenesis and poor prognosis in head and neck squamous cell carcinoma. American Journal of Cancer Research, 11(7), 3688–3697. [Google Scholar] [PubMed]
19. Xie, F., Huang, C., Liu, F., Zhang, H., Xiao, X. et al. (2021). CircPTPRA blocks the recognition of RNA N6-methyladenosine through interacting with IGF2BP1 to suppress bladder cancer progression. Molecular Cancer, 20(1), 68. [Google Scholar] [PubMed]
20. Yu, Y. Z., Lv, D. J., Wang, C., Song, X. L., Xie, T. et al. (2022). Hsa_circ_0003258 promotes prostate cancer metastasis by complexing with IGF2BP3 and sponging miR-653-5p. Molecular Cancer, 21(1), 12. [Google Scholar] [PubMed]
21. Glažar, P., Papavasileiou, P., Rajewsky, N. (2014). circBase: A database for circular RNAs. RNA, 20(11), 1666–1670. [Google Scholar] [PubMed]
22. Li, Z., Peng, Y., Li, J., Chen, Z., Chen, F. et al. (2020). N6-methyladenosine regulates glycolysis of cancer cells through PDK4. Nature Communications, 11(1), 2578. [Google Scholar] [PubMed]
23. Chen, R. X., Chen, X., Xia, L. P., Zhang, J. X., Pan, Z. Z. et al. (2019). N6-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nature Communications, 10(1), 4695. [Google Scholar] [PubMed]
24. Tang, Z., Kang, B., Li, C., Chen, T., Zhang, Z. (2019). GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Research, 47(W1), W556–W560. [Google Scholar] [PubMed]
25. Rimoldi, D., Salvi, S., Liénard, D., Lejeune, F. J., Speiser, D. et al. (2003). Lack of BRAF mutations in uveal melanoma. Cancer Research, 63(18), 5712–5715. [Google Scholar] [PubMed]
26. Van Poppelen, N. M., Vaarwater, J., Mudhar, H. S., Sisley, K., Rennie, I. G. et al. (2018). Genetic background of iris melanomas and iris melanocytic tumors of uncertain malignant potential. Ophthalmology, 125(6), 904–912. [Google Scholar] [PubMed]
27. Yavuzyigitoglu, S., Koopmans, A. E., Verdijk, R. M., Vaarwater, J., Eussen, B. et al. (2016). Uveal melanomas with SF3B1 mutations: A distinct subclass associated with late-onset metastases. Ophthalmology, 123(5), 1118–1128. [Google Scholar] [PubMed]
28. Vader, M., Madigan, M., Versluis, M., Suleiman, H., Gezgin, G. et al. (2017). GNAQ and GNA11 mutations and downstream YAP activation in choroidal nevi. British Journal of Cancer, 117(6), 884–887. [Google Scholar] [PubMed]
29. Chen, X., Wu, Q., Depeille, P., Chen, P., Thornton, S. et al. (2017). RasGRP3 mediates MAPK pathway activation in GNAQ mutant uveal melanoma. Cancer Cell, 31(5), 685–696.E6. [Google Scholar] [PubMed]
30. Van Essen, T. H., van Pelt, S. I., Versluis, M., Bronkhorst, I. H., van Duinen, S. G. et al. (2014). Prognostic parameters in uveal melanoma and their association with BAP1 expression. British Journal of Ophthalmology, 98(12), 1738–1743. [Google Scholar] [PubMed]
31. Martin, M., Maßhöfer, L., Temming, P., Rahmann, S., Metz, C. et al. (2013). Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nature Genetics, 45(8), 933–936. [Google Scholar] [PubMed]
32. Shain, A. A. O., Bagger, M. M., Yu, R., Chang, D. A. O., Liu, S. et al. (2019). The genetic evolution of metastatic uveal melanoma. Nature Genetics, 51(7), 1123–1130. [Google Scholar] [PubMed]
33. Chai, P., Jia, R., Li, Y., Zhou, C., Gu, X. et al. (2021). Regulation of epigenetic homeostasis in uveal melanoma and retinoblastoma. Progress in Retinal and Eye Research, 38, 101030. [Google Scholar]
34. Zhang, L., He, X., Li, F., Pan, H., Huang, X. et al. (2018). The miR-181 family promotes cell cycle by targeting CTDSPL, a phosphatase-like tumor suppressor in uveal melanoma. Journal of Experimental & Clinical Cancer Research, 37(1), 15. [Google Scholar]
35. Li, P., He, J., Yang, Z., Ge, S., Zhang, H. et al. (2020). ZNNT1 long noncoding RNA induces autophagy to inhibit tumorigenesis of uveal melanoma by regulating key autophagy gene expression. Autophagy, 16(7), 1186–1199. [Google Scholar] [PubMed]
36. Xing, Y., Wen, X., Ding, X., Fan, J., Chai, P. et al. (2017). CANT1 lncRNA triggers efficient therapeutic efficacy by correcting aberrant lncing cascade in malignant uveal melanoma. Molecular Therapy, 25(5), 1209–1221. [Google Scholar] [PubMed]
37. Peng, C., Tan, Y., Yang, P., Jin, K., Zhang, C. et al. (2021). Circ-GALNT16 restrains colorectal cancer progression by enhancing the SUMOylation of hnRNPK. Journal of Experimental & Clinical Cancer Research, 40(1), 272. [Google Scholar]
38. Li, B., Zhu, L., Lu, C., Wang, C., Wang, H. et al. (2021). circNDUFB2 inhibits non-small cell lung cancer progression via destabilizing IGF2BPs and activating anti-tumor immunity. Nature Communications, 12(1), 295. [Google Scholar] [PubMed]
39. Lin, S., Choe, J., Du, P., Triboulet, R., Gregory, R. I. (2016). The m6A methyltransferase METTL3 promotes translation in human cancer cells. Molecular Cell, 62(3), 335–345. [Google Scholar] [PubMed]
40. Tang, J., Wan, Q., Lu, J. (2020). The prognostic values of m6A RNA methylation regulators in uveal melanoma. BMC Cancer, 20(1), 674. [Google Scholar] [PubMed]
41. Jia, R., Chai, P., Wang, S., Sun, B., Xu, Y. et al. (2019). m6A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Molecular Cancer, 18(1), 161. [Google Scholar] [PubMed]
42. Wan, Q., Tang, J. (2019). Exploration of potential key pathways and genes in multiple ocular cancers through bioinformatics analysis. Graefe’s Archive for Clinical and Experimental Ophthalmology, 257(10), 2329–2341. [Google Scholar] [PubMed]
43. Fang, R., Chen, X., Zhang, S., Shi, H., Ye, Y. et al. (2021). EGFR/SRC/ERK-stabilized YTHDF2 promotes cholesterol dysregulation and invasive growth of glioblastoma. Nature Communications, 12(1), 177. [Google Scholar] [PubMed]
44. Hino, N., Rossetti, L., Marin-Llaurado, A., Aoki, K., Trepat, X. et al. (2020). ERK-mediated mechanochemical waves direct collective cell polarization. Developmental Cell, 53(6), 646–660.E8. [Google Scholar] [PubMed]
45. Ma, G., Liang, Y., Chen, Y., Wang, L., Li, D. et al. (2020). Glutamine deprivation induces PD-L1 expression via activation of EGFR/ERK/c-jun signaling in renal cancer. Molecular Cancer Research, 18(2), 324–339. [Google Scholar] [PubMed]
46. Pei, T. M., Meng, F. Z., Xiao, P., Han, J. H., Song, R. P. et al. (2020). MUC13 promotes intrahepatic cholangiocarcinoma progression via EGFR/PI3K/AKT pathways. Journal of Hepatology, 72(4), 761–773. [Google Scholar]
47. Zhangyuan, G., Wang, F., Zhang, H., Jiang, R., Tao, X. et al. (2020). VersicanV1 promotes proliferation and metastasis of hepatocellular carcinoma through the activation of EGFR-PI3K-AKT pathway. Oncogene, 39(6), 1213–1230. [Google Scholar] [PubMed]
48. Topcu-Yilmaz, P., Kiratli, H., Saglam, A., Soylemezoglu, F., Hascelik, G. (2010). Correlation of clinicopathological parameters with HGF, c-Met, EGFR, and IGF-1R expression in uveal melanoma. Melanoma Research, 20(2), 126–132. [Google Scholar] [PubMed]
49. Amaro, A., Mirisola, V., Angelini, G., Musso, A., Tosetti, F. et al. (2013). Evidence of epidermal growth factor receptor expression in uveal melanoma: Inhibition of epidermal growth factor-mediated signalling by gefitinib and cetuximab triggered antibody-dependent cellular cytotoxicity. European Journal of Cancer, 49(15), 3353–3365. [Google Scholar] [PubMed]
50. Hurks, H. M., Metzelaar-Blok, J. A., Barthen, E. R. et al. (2000). Expression of epidermal growth factor receptor: Risk factor in uveal melanoma. Investigative Ophthalmology & Visual Science, 41(8), 2023–2027. [Google Scholar]
51. Gu, Y., Niu, S., Wang, Y., Duan, L., Pan, Y. et al. (2021). DMDRMR-mediated regulation of m6A-modified CDK4 by m6A reader IGF2BP3 drives ccRCC progression. Cancer Research, 81(4), 923–934. [Google Scholar] [PubMed]
52. Chen, X., Wu, Q., Tan, L., Porter, D., Jager, M. J. et al. (2014). Combined PKC and MEK inhibition in uveal melanoma with GNAQ and GNA11 mutations. Oncogene, 33(39), 4724–4734. [Google Scholar] [PubMed]
Supplementary Materials
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools