 Open Access
Open Access
ARTICLE
Effect of Algae on Melon (Cucumis melo subsp. agrestis var. conomon) Growth and Development under Drought-Stress Conditions
1 Department of Horticulture, Faculty of Agriculture, Siirt University, Siirt, 56100, Türkiye
2 Department of Agricultural Biotechnology Faculty of Agriculture, Siirt University, Siirt, 56100, Türkiye
* Corresponding Author: Behcet İNAL. Email:
Phyton-International Journal of Experimental Botany 2023, 92(11), 3177-3193. https://doi.org/10.32604/phyton.2023.030979
Received 07 May 2023; Accepted 18 September 2023; Issue published 24 October 2023
Abstract
Due to climate change, it is necessary to develop plant varieties that are resilient to climate conditions and resistant to abiotic and biotic stresses. The use of microalgae, which are microorganisms that contain carbohydrates, proteins, lipids, and vitamins, against drought tolerance is a new approach. The aim of the current study was to determine the drought-related mechanisms in the conomon melon genotype and develop drought-tolerant melon cultivars. Morphological, physiological, pomological, and molecular analyses were carried out on the algae-treated genotypes. It has been determined that commercial algae application provides the best results in leaf temperature, leaf relative water contents (LRWC), plant height, fruit length, fruit diameter, and yield, while Cag Cag (a special river in the region) stream algae application gives better results in main stem diameter. It was determined that the number of nodes in the control (without algae) plots was higher than in the other two treatments. Yield and LRWC and plant height values of genotype 7 were the best values among other genotypes. The leaf temperature measurement was lower on genotype 9 than on the other genotypes. While the highest fruit length value was measured in genotype 1, genotype 8 was superior in the main stem diameter, fruit diameter, and the number of nodes among the remaining plant materials. In terms of yield, it was determined that the interaction between genotype 2 and the commercial algae resulted in the best outcomes. In addition, the results of the genetic evaluation revealed that the materials used were not genetically distant from each other and more detailed genetic evaluations are needed. The molecular kinship analysis revealed that the genotypes used in the study were divided into three distinct groups, with individuals within each group exhibiting a high degree of relatedness to one another. As a result of the study, it was found that the application of microalgae had significant effects on improving the drought tolerance of Cucumis melo subsp. agrestis var. conomon genotypes.Keywords
Nomenclature
| AFLP | Amplified Fragment Length Polymorphism |
| DNA | Deoxyribonucleic Acid |
| ICP-MS | Inductively Coupled Plasma Mass Spectrometry |
| IPCC | Intergovernmental Panel on Climate Change |
| ITS | Internal Transcribed Spacer |
| LRWC | Leaf Relative Water Contents |
| LSD | The Least Significant Difference |
| Ng | Nanogram |
| NJ | Neigbour-Joining |
| PCR | Polymerase Chain Reaction |
| RAPD | Random Amplified Polymorphic DNA |
| SNP | Single Nucleotide Polymorphism |
| SSR | Simple Sequence Repeat |
| μL | Microliter |
| °C | Degrees centigrade |
| % | Percentage |
Drought is undoubtedly the most destructive among the abiotic stresses, as it is primarily caused by insufficient moisture in the growing medium. When drought is combined with high temperature, transpiration, evapotranspiration, and evaporation, water loss accelerates [1]. Under such conditions, plants that cannot find sufficient water become vulnerable to all other stresses. As a result, the plant is unable to fulfill normal functions such as seed germination nitrogen assimilation, and even protein synthesis. In limited water conditions, the main purpose of the plant will not be to increase or produce high yield or yield components. The plant will primarily focus on sustaining minimal life processes. With the disruption of cell elongation and slowed development, reductions in the number of tillers, root width, length, fresh and dry weights, turgor pressure, and leaf area will be observed [2–4].
The most practical way to supply enough food for the increasing world population is to increase the yield and quality of agricultural products. However, due to abiotic stress conditions such as salinity and drought, it has become necessary to implement new policies aimed at developing resistant varieties that can thrive in arid conditions. In the final declaration of the Intergovernmental Panel on Climate Change (IPCC) in 2013, it was stated that there would be an increase in the average temperature by 1.8 to 4.0 degrees Celsius by 2100. According to the 2020 determinations, approximately 55 million people are suffering from drought-related economic difficulties [5]. On the other hand, it is projected that around 700 million people will be compelled to relocate their settlements by 2030 [6]. Additionally, water resources are anticipated to be inadequate by 2050 due to the expected global population of 9 billion [4].
Conomon, which is eaten both cooked and raw, is called sugar-free cucumber and is a melon variety. It is called pickled melon in Japanese and English [7]. Conomon, being capable of cultivation without the need for water, particularly in arid regions, has garnered close attention from both scientific studies and rural producers, as mentioned by Swamy [7]. In the Southeast Anatolia region of Turkey, conomon is grown without the need for irrigation, even under scorching temperatures exceeding 50 degrees Celsius in direct sunlight. Solving the evolutionary development and cultivation process may guide the understanding of the genetic diversity of this valuable vegetable and ultimately the development of new varieties [7].
As the moisture in the soil decreases, both plant productivity [8] and the quality of the product are negatively affected under stress conditions [9,10]. High temperature, salinity, and drought are major abiotic stresses that both indirectly impact and directly threaten agricultural activities. Despite the insufficient water resources, the increasing need for food is an enigma that is difficult to solve. Finding solutions to existing problems by using all technological possibilities is easy but expensive. Developing a variety that is resistant or tolerant to stress is considered one of the most effective and sustainable solutions. In situations where there is a shortage of water (moisture) required by the plant in its growing environment, the plant responds by reducing chlorophyll content, diminishing flower production, shedding leaves, and ultimately experiencing a decline in seed yield and overall productivity [9].
Algae are considered environmentally friendly and offer several advantages over chemical fertilizers due to their multifunctional benefits [11,12]. In terms of sustainable agriculture, the use of algae-based products instead of chemical fertilizers may help prevent the harm of chemical fertilizers [13,14] to the environment. The excessive use of chemical fertilizers, combined with climate change and pesticide usage, amplifies the adverse effects of abiotic stresses on plant productivity and leads to ecosystem degradation [15]. Whereas algae extracts help to clear existing ROS by increasing the synthesis of antioxidant enzymes [16].
DNA-based molecular markers are widely used in taxonomy, physiology, embryology, genetic engineering, etc. They are versatile tools used in all fields of life sciences [17]. In recent years, studies on plant diversity have increasingly utilized molecular techniques to gain insights into the genetic composition of various species. These techniques include random amplified polymorphic DNA (RAPD), amplified fragment length polymorphism (AFLP), microsatellites, single nucleotide polymorphisms (SNPs), and nuclear-organelle genes [18]. In a previous study, Simple Sequence Repeats (SSR) and internal transcribed spacer regions (ITS1 and ITS2) of the ribosomal RNA genes were used as nuclear markers for the phylogenetic analysis of Cucumis sp. classification [19,20]. However, subsequent molecular phylogenetic studies, based on DNA sequences from chloroplast genes (rbcL, matK, rpl20-rps12, trnL, and trnL-F), nuclear ITS, and plastid trnS-trnG regions, indicated that species from closely related genera such as Cucumella, Dicaelospermum, Mukia, Myrmecosicyos, and Oreosyce should be incorporated into the Cucumis genus [21]. Furthermore, an extensive phylogenetic study was conducted on Cucumis, examining the diversity of both nuclear and chloroplastic DNA [22,23]. Therefore, the objective of this study was to identify the most drought-tolerant wild melon genotypes by assessing their development, yield, and molecular parameters under drought-stress conditions. To achieve this, ten different genotypes of Cucumis melo subsp. agrestis var. conomon (C. melo var. conomon) were subjected to drought stress and the effects of applying various algae varieties were measured.
2.1 Plant Material and Growth Conditions
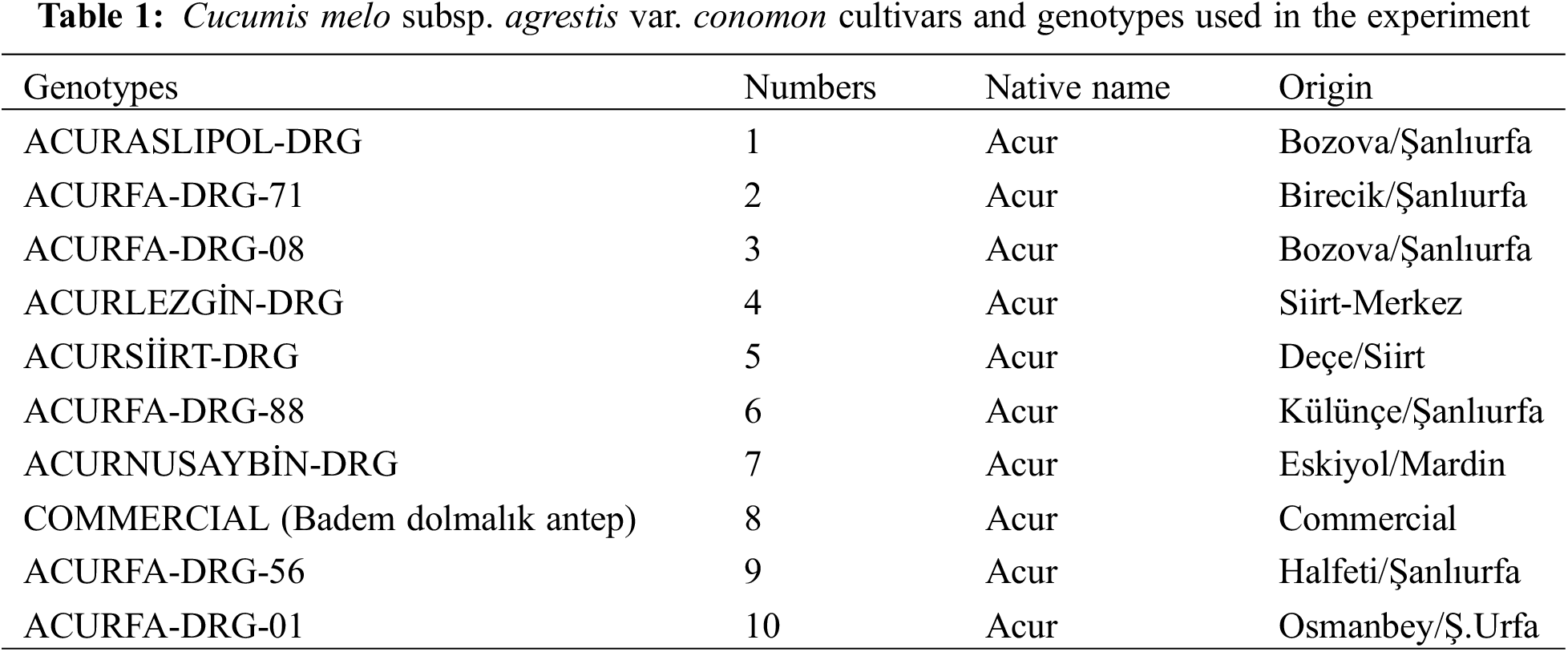
The study was carried out at Siirt University, Faculty of Agriculture, Department of Horticulture, between 2017 and 2018. In the study, nine different C. melo var. conomon genotypes and one commercial C. melo var. conomon variety was used. All ten genotypes were selected specifically from the ones that can be grown under arid conditions. The origin of the genotypes and the places where they were collected are given in Table 1. The experiment was established in nested-factorial experimental design with 3 replications, including algae, control, local genotypes, and commercial cultivars.
2.2 Experimental Design Algae Treatments
Blue-green algae (Anabaena vaginicola L.) obtained from the Cag Cag stream in Nusaybin, along with commercially available algae extract (Mc Crop), were used as the algae extract in the study. The experiment was conducted in the field under drought conditions, and it followed a nested-factorial experimental design with three replications. Seeds were sown using traditional methods by direct sowing in the third week of April. The plants were cultivated with a row spacing of 2 m, and there was a spacing of 60 cm between plants within each row. The experiment was planned with 15 plants per replication per genotype (GNP). Before the throat-filling stage, algae were applied in such a way that 10 grams of algae came to the root zone of the plant.
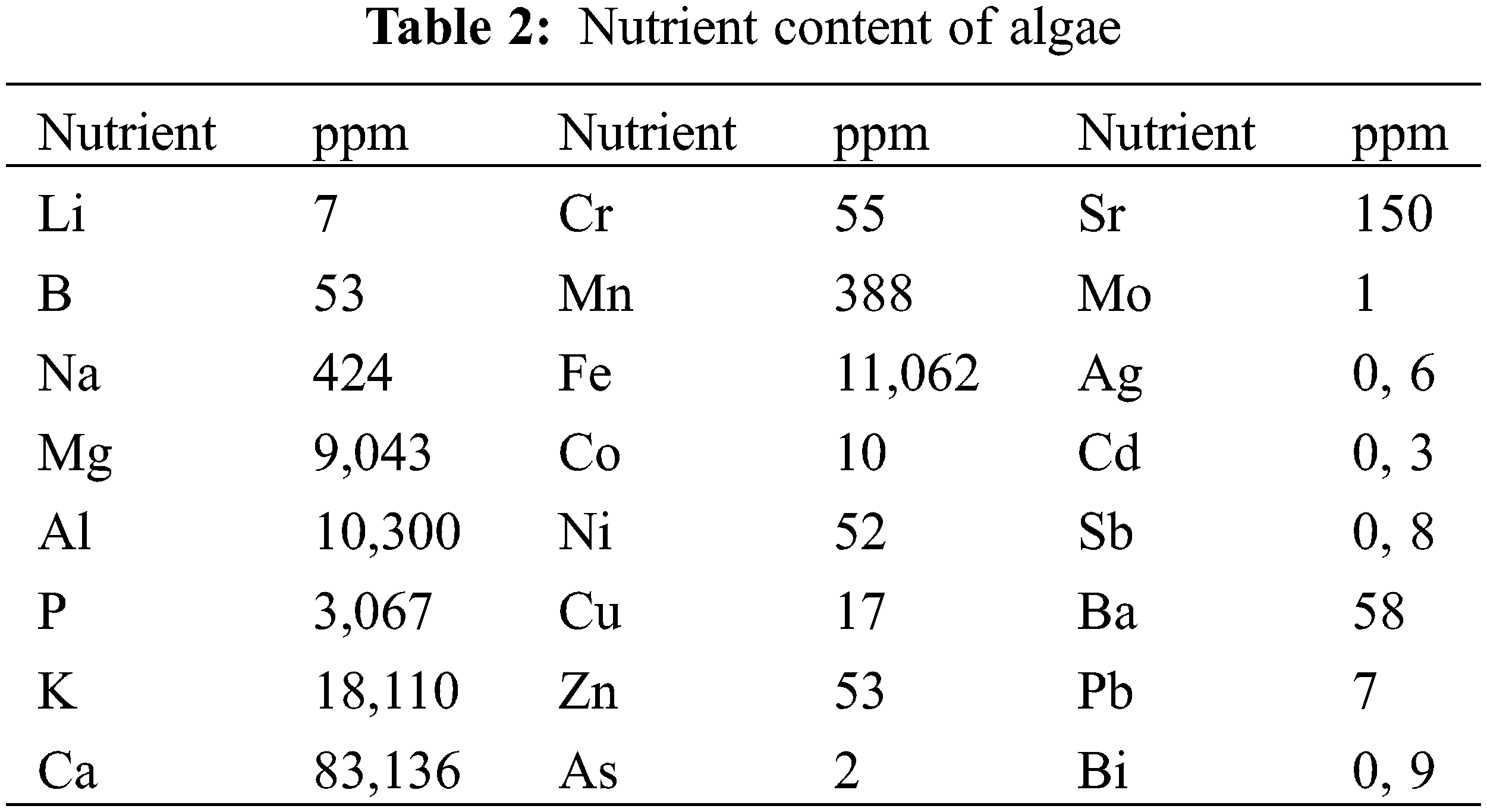
2.3.1 Algae Nutrient Content Analysis
The nutrient analysis of the blue-green algae obtained from the Nusaybin Cag Cag stream, which was used in the experiment, was conducted using an ICP-MS (Inductively Coupled Plasma Mass Spectrometry, Thermo) apparatus. The standard torch used in the analysis included a 1.5-mm-diameter injector. Analysis was achieved at Siirt University Science and Technology Application and Research Center.
2.4 Morphological, Physiological and Pomological Evaluations
Leaf temperature measurements were taken on all four sides of the plant during July. The measurements were conducted at 11:00 am. Three plants of each genotype were selected for the measurements, and an infrared thermometer was used to obtain the leaf temperature readings.
2.4.2 LRWC (Leaf Relative Water Content)
LRWC was carried out on the 20th day of each month (a total of 3 months). Three biological replicates were used for each genotype. Firstly, fresh weight was taken, and leaves were kept in distilled water for four hours. Then, turgor weights were determined (Fig. 1). To determine the dry weight of the leaf samples, the leaves were weighed after drying at 85°C for 24 h. Fresh and dry leaf weights were determined as % with the following formula LRWC = (Fresh Weight-Dry Weight)/(Turgor Weight-Dry Weight) × 100 according to Ackley [24] and Smart et al. [25].

Figure 1: Determination of leaf relative water content (LRWC)
Plant height was measured in three plants in each plot, and the main stem lengths were measured three times at 1-month intervals after planting [26] (Fig. 2).

Figure 2: Plant height measurements
The number of nodes that emerged on the main stem, beginning from the cotyledon leaves, was counted and documented as the number of nodes per plant in the plants where plant height measurements were obtained [26].
The main stem diameters were measured with the help of a caliper just above the root collar in plants where height was measured [26].
Fruit size was determined manually using a ruler, and three fruits from each plot were included in the measurement for each genotype [26] (Fig. 3).

Figure 3: Fruit size measurements
Fruit diameter measurements were conducted using a ruler, with three fruits per plot assessed for each genotype [26]. Fruit diameter was measured from the widest point of the fruit’s width (Fig. 3).
Fruits that reached the optimum stage of harvest maturity were collected. Due to the rapid growth of melons, daily harvests were performed. The total yield was calculated by dividing the total amount of harvested fruits by the plot area [26].
Genomic DNA isolation from the plants was done following Karaca et al. [27] using the CTAB method. The quality and quantity of the isolated DNAs were measured with the Thermo nanodrop device.
2.5.2 PCR (Polymerase Change Reaction) Process
To determine the kinship relationships among melon genotypes, the trnL-F gene region was amplified by PCR. Following Primer sequences were used trnL-c 5’ CGAAATCGGTAGACGCTACG 3’, trnF-f 5’ ATTTGAACTGGTGACACGAG 3’ [28]. The total reaction volume for PCR was prepared as 50 µl. For this purpose, 6 μL of 25 mM MgCl2, 1 μL of 10 mM dNTP, 2 μL of each 50 pmol primer, 0.7 μL of Taq DNA Polymerase, 5 μL of Taq buffer, 2 μL of BSA, and 3 µL (120 ng) template DNA were used and the total volume of PCR was adjusted to 50 µL by taking ddH2O. In addition, negative controls were used to check for contamination. The PCR cycling parameters were as follows: an initial denaturation step at 95°C for 15 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 51°C for 1 min, and extension at 72°C for 2 min. Finally, the reaction was incubated at 72°C for 10 min.
2.5.3 Agarose Gel Electrophoresis
1.5% agarose gel was prepared for gel electrophoresis to visualize the PCR bands. To prepare the gel, 0.6 grams of agarose gel was measured and dissolved in 40 ml of 1X TBE buffer by boiling it in a microwave oven. The mixture was cooled to 50°C and 3 µL of ethidium bromide dye (10 mg/mL in H2O) was added. Samples and DNA ladder (100 bp, Thermo Fisher UK, Cat # SM0241) were run at 90 volts for 30 min. Then, the gel was observed under UV light with the help of a computer program, and its photograph was taken and recorded.
2.5.4 Sequence Analysis and Kinship
PCR products were sent to a commercial company (MEDSANTEK) to obtain the DNA sequences of trnL-F amplified regions. BioEdit sequencer software was used to process and trim the sequences. Consensus sequences were created by processing DNA sequences in this software. Then, these sequences were copied to text format and converted to Fasta format, ready for sequence alignment. ClustalW software [29] was used to align the sequences. The MEGAX [30] software was used to construct the Neigbour-Joining (NJ) tree with a 500 bootstrap value for learning the kinship relationships of the cultivars whose sequences were aligned.
The analysis of the data was done using the SAS 9.1 statistical software [31] according to the nested-factorial experimental design [32]. In this experiment, the algae treatment was nested within the levels of time, while genotype and algae were arranged in a factorial design. Thus, this design had both nested and factorial factors. The linear model for this design was
where
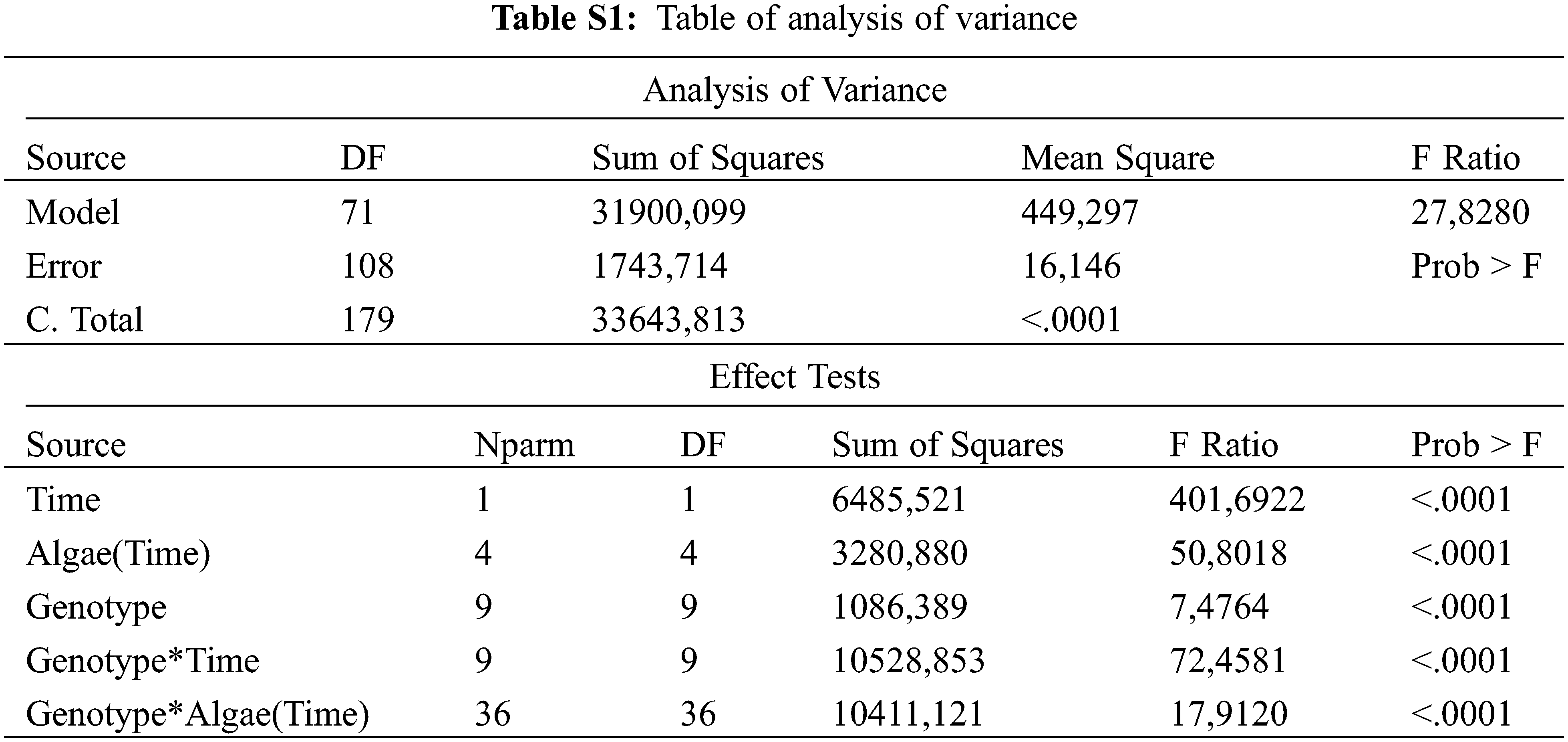
The least significant difference (LSD) multiple comparison test was used for comparing the treatment means. Table of analysis of variance was given as supplementary in Table S1.
The LRWC value is naturally high in the seedling stages in plants under arid growth conditions. In addition to the LRWC (Leaf Relative Water Content) values of tolerant or resistant plants, the LRWC values of other plants generally exhibit a tendency to decrease over time. It was determined that the LRWC value in the first month was higher than the LRWC value in the second month. LRWC values of commercial algae were found to be high. Such a result is expected since commercial algae are quickly dissolved in the soil which affects plant growth. The interaction between the GNP10 genotype and commercial algae treatment resulted in the highest level of genotype interaction. It is normal to have high LRWC values in the early measurements, and the second LRWC measurements provide separation of tolerant genotypes. The second LRWC value of GNP5 was higher than other genotypes. The initial and subsequent LRWC measurements of tolerant genotypes are not inferior to the previously identified genotypes that exhibited relatively higher LRWC values. Multiple interactions (genotype × algae × time) are explained in more detail and integration (Fig. 4). When the results were examined, it was determined that the result obtained from multiple interactions was closer to the expected and more transferrable to practice. The same graph also reveals that an excessive number of groups were formed, indicating a high level of variability or complexity in the data. It was determined that the highest LRWC value was obtained from GNP10 × commercial algae × plant group (93.07%) in the first measurement interactions, and the lowest LRWC value (31.0%) was obtained in GNP4 × Cag Cag stream algae × first-month LRWC measurement value. It was concluded that the absorption of Cag Cag stream algae in the soil takes a long time (Fig. 4). The analysis results are presented in Table 2.
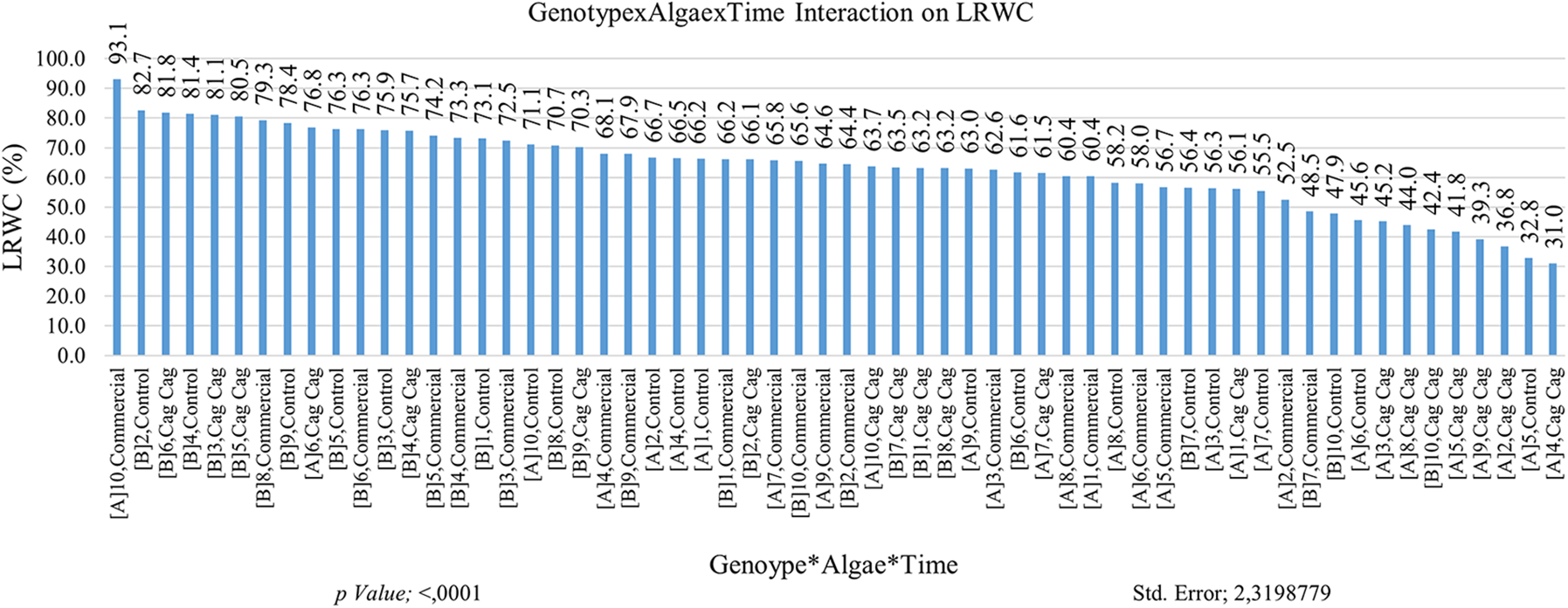
Figure 4: Effect of genotype × algae × time interaction on leaf relative water content (LRWC). Each histogram is the mean of n = 3. Histograms were compared using the least significant difference (LSD) test. Histograms that do not share the same lowercase letter are considered statistically significant at p < 0.001. (A and B) time points for the LRWC measurements, 1–10: Cucumis melo subsp. agrestis var. conomon genotypes are given in Table 1
The highest yield (4.87 kg) was obtained from GNP7 among the genotypes in the study. The yield value of GNP6 (4.6 kg) was in the same statistical group as the yield value of GNP7. GNP3 with a yield value of 1.17 kg per decare and GNP8 with a yield value of 1.88 kg were the genotypes with the lowest yield value. The highest yield per decare was obtained in GNP7 with 1352.8 kg. The lowest yield was obtained in GNP3 as 325 kg per decare. The yield value of the commercial variety, GNP8, was determined as 894.4 kg per decare. When examining the effects of algae on yield values, it was observed that the highest yield (3.91 kg) was obtained from the plots where commercial algae was applied. On the other hand, the lowest yield (2.65 kg) was obtained from the control plots without any algae application. The algae applications were statistically categorized into different groups based on their effects on yield. The variety × algae interaction effect on yield values highlights the influence of algae applications and makes their effects more apparent. The highest yield value (5.87 kg) was obtained from GNP2 and the commercial algae group. Yield values of plants in commercial algae interaction of genotypes 6 and 7 (5.50 and 5.37 kg, respectively) were found to be close to the yield values of GNP2. The lowest yield value (0.93 kg) was recorded in the plants of GNP3 in the control plots (Fig. 5).
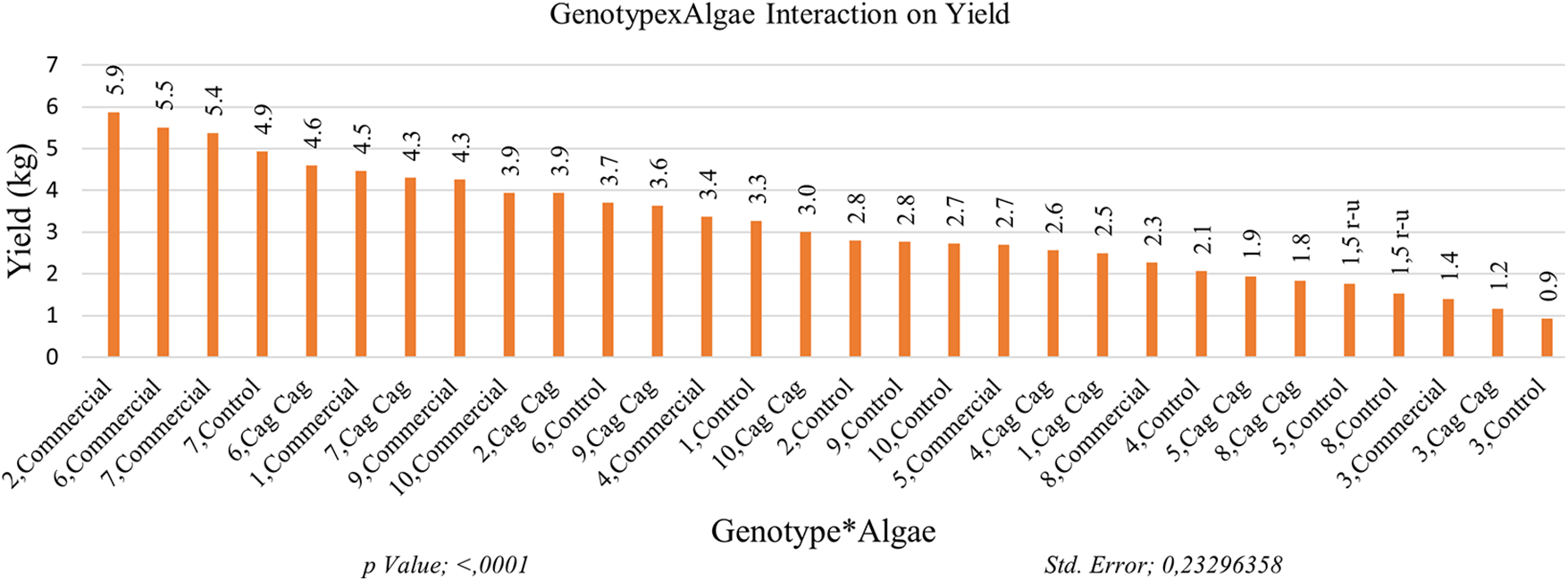
Figure 5: Impact of algae on yield of Cucumis melo subsp. agrestis var. conomon. Each histogram is the mean of n = 3. Histograms were compared using the LSD test. Histograms that do not share the same lowercase letter are statistically significant at p < 0.001
One of the determining physiological characteristics of drought tolerance is leaf temperature. Leaf temperature increases in drought-affected plants. The highest leaf temperature value measured in cultivars/genotypes was in GNP8 (33.38°C), on the other hand, the lowest leaf temperature value was recorded in GNP9 as 29.16°C. The highest leaf temperature value (33.58°C) was measured in the control group and the lowest leaf temperature value (28.37°C) was measured in the commercial algae group. It is plausible to assume that algae applications improve plant root development and improve water retention capacity in the soil [13,33]. According to the genotype × application interaction, the highest leaf temperature value (36.7°C) was recorded in the GNP7 × control group, and the lowest leaf temperature value (25°C) was recorded in the plants in the GNP1 × commercial algae interaction group (Fig. 6).
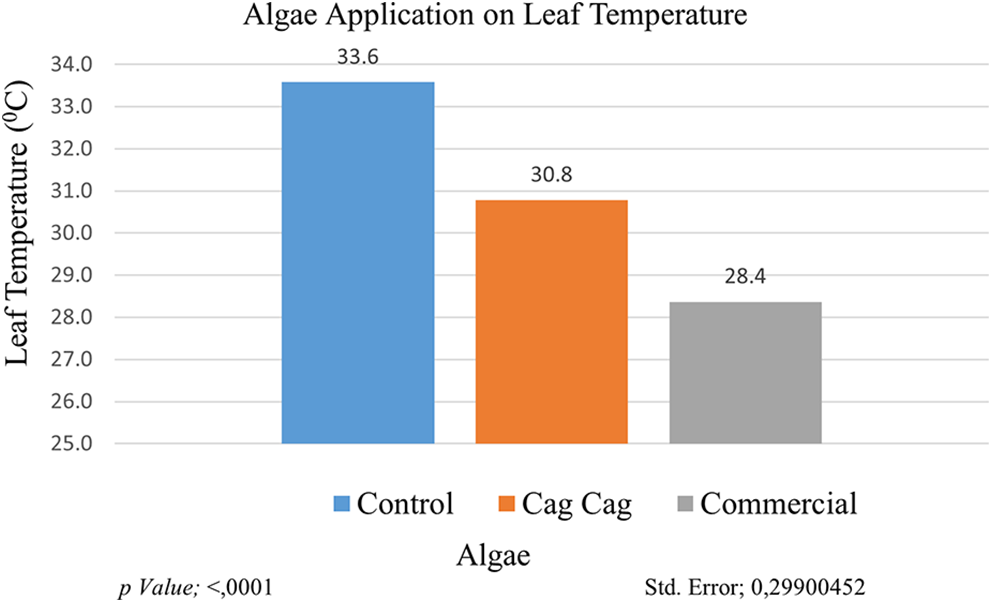
Figure 6: Effect of algae application on leaf temperature. Each histogram is the mean of n = 3. Histograms were compared using the LSD test. Histograms that do not share the same lowercase letter are statistically significant at p < 0.001
When examining the interactions of commercial algae with genotypes, it was observed that commercial algae had a positive effect on the morphological properties of C. melo var. conomon. Cag Cag algae was the second group, and the interaction group formed with all genotypes by the control group, except for the number of nodes, which was in the last place. Commercial algae had the highest values for fruit length (15.2 cm), fruit width (7.97 cm), and plant height (157.67) ensuring the formation of the interaction group that gave the highest values. The highest number of nodes (20.5 pcs.) was recorded in the plants in the interaction group formed by the control group with GNP8 (Table 3).
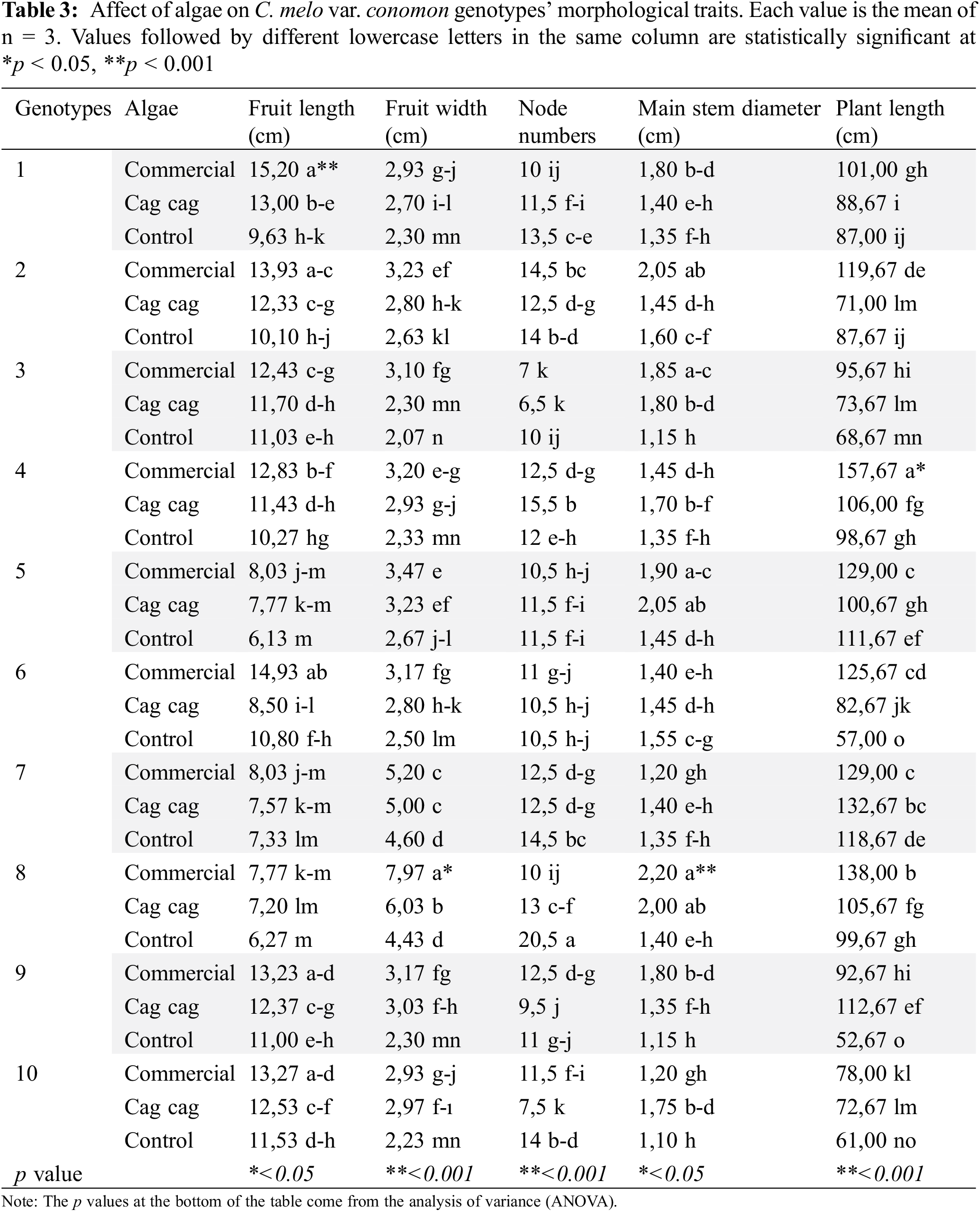
Algae accelerate the growth and development of melons, tomato, cucumbers, strawberries, peppers, carrots, and onions [34–42]. In our study, commercial algae increased all morphological traits of plants except the number of nodes. Researchers stated that algae increases the defense mechanism of plants against stress and their development [15,38,40,43–47].
In our study, the main stem diameter, plant height, and LRWC were increased with algae applications. The application of algae to melon plants grown under abiotic stressed conditions resulted in increased shoot length, fresh and dry weight, leaf area, and leaf number compared to the control group [46]. In the study that utilized algae extracts, it was reported that the application of these extracts resulted in early flowering and fruit formation in tomato plants. Additionally, the fruit weight of the tomatoes was also observed to be affected positively by the algae extracts [35]. It was determined that tomato fruit size (width and length) was higher in algae-treated plots [35,42]. In a previous report, Algae extracts resulted in higher shoot/root lengths in rice compared to the control [44]. According to Patel et al. [48], the shoot lengths, root lengths, and biomass amounts of three durum wheat cultivars increased with algae application. Similarly, in the research conducted by Al-Ghamdi et al. [49] on asparagus, algae application led to an increase in shoot lengths, root lengths, and biomass amounts. In another study, algae increased the fresh and dry weights and shoot lengths of wheat under salt-stressed conditions [50]. Dookie et al. [51] applied the water algae to the tomato and observed an increased number of flower buds, the number of flowers, and the number of fruits. It was also determined that the algae applied to the onion increased the onion head weight, head dry weight, and head and root collar diameters [39]. Additionally, Elsharkawy et al. [52] determined that plant height, number of leaves, and leaf dry weight of artichoke plants increased with seaweed application. Hassan et al. [47] reported that there was a significant improvement in the fruits of cucumber with the application of algae. In a separate study, it was observed that larger and a greater number of pea and melon fruits were obtained in the plots where seaweed extracts were applied. The application of seaweed extracts positively influenced the size and yield of both pea and melon fruits [41,53].
According to previous reports, which are consistent with our findings, the yield and yield components of pepper and cucumber increased, as did the yield of tomato, with algae application [14,47]. Abbas et al. [39] applied seaweed to the onion and Elsharkawy et al. [52] applied seaweed to artichoke, both studies reported increased yields in seaweed applications.
According to Gitau et al. [35], it was determined that the leaf temperature in tomato plants treated with algae was within the normal range. The leaf temperature was found to vary based on leaf thickness. Similarly, lower leaf temperature values were determined in the commercial and Cag Cag stream algae-treated plots in our study, compared to the plants in the non-algae-treated plots.
Algae also had a positive effect on post-harvest quality. Products obtained from algae-treated plants had a long shelf life and high chemical properties [42,47]. Additionally, Elsharkawy et al. [52] determined that some chemical components of artichoke also increased with algae application. The uptake of a larger amount of nutrients in the algae-applied section of the tomato plant demonstrates that algae play a role in enhancing the development of plant roots [38]. Plants with enhanced root morphology can accumulate non-structural carbohydrates, which leads to increased energy storage, improved metabolism, and enhanced water adaptations. This enables them to develop resistance against abiotic stresses and utilize available nutrients from the soil more efficiently, resulting in higher overall performance. It also affects the defense mechanism against all kinds of pathogens [40,54]. It has been reported that the uptake rate of algae extracts by plants is higher than chemical fertilizers [55]. Algae also provide efficient use of soil water uptake due to its effect on root development [38]. It was determined that C. melo conomon plants grown in arid conditions exhibited increased resistance, which can be attributed to the application of algae. Because algae-based agricultural solutions have great potential against abiotic stresses and improve overall agricultural sustainability, the algae mechanism is more resistant to stresses [56,57]. According to Lakshmi et al. [58], the use of algae-based biofertilizers and bio-stimulants offers several benefits, including increased plant growth, productivity, and tolerance to various abiotic stresses. Because algae (Chlorella TLD6B) grow rapidly and increase adaptability under abiotic stress (extreme desert environment) conditions [23]. According to previous reports, algae affect the internal responses of plants, and there are strong relationships between proline and algae, which promotes the biochemical and enzymatic responses of plants with positive effects against abiotic stress conditions [59].
Trees formed by using NJ methods of the trnL-f region gave significant results in revealing the genetic relationship of C. melo var. conomon genetic resources collected from different regions. When the tree results were evaluated, 10 C. melo var. conomon cultivars and 3 different kinship groups were formed. The first group consisted of Nevzet Tic, Nevzet Yerli, Acurfa71 DRG, the second group, ACURASLIPOL-DRG, Acurfa56 DRG, Lezgin Siirt in the last 3rd group, Acurfa01 DRG, Acurfa08 DRG, Acurfa88 DRG, and Acur Dese Village genotypes. When the results were examined, it was seen that the commercial and domestic varieties were not very different from each other in terms of kinship. It is estimated that the two different varieties (local and commercial) are genetically very similar. Because the genetic similarity was very high, the marker gene used did not separate them into commercially and locally distinct clusters (Fig. 7). Several studies conducted on the non-coding regions of chloroplasts have indicated increased variation and frequent mutations specifically within the Ophioglossaceae family [60]. A molecular-based investigation utilizing optimized DNA isolation and amplification of the trnL-F and trnl-R intergenic spacer was carried out in cucumber Mercy F1 hybrids and Mercy-Mutant populations. The resulting DNA bands were observed to be pure, with minimal contaminants, making them suitable for subsequent procedures [61]. Among species of seed plants, the tRNALeu intron and trnL-F spacer contain multiple variable and informative sites. Consequently, these sequences have gained recognition as effective markers for phylogenetic analysis, both among species and within closely related species [62].
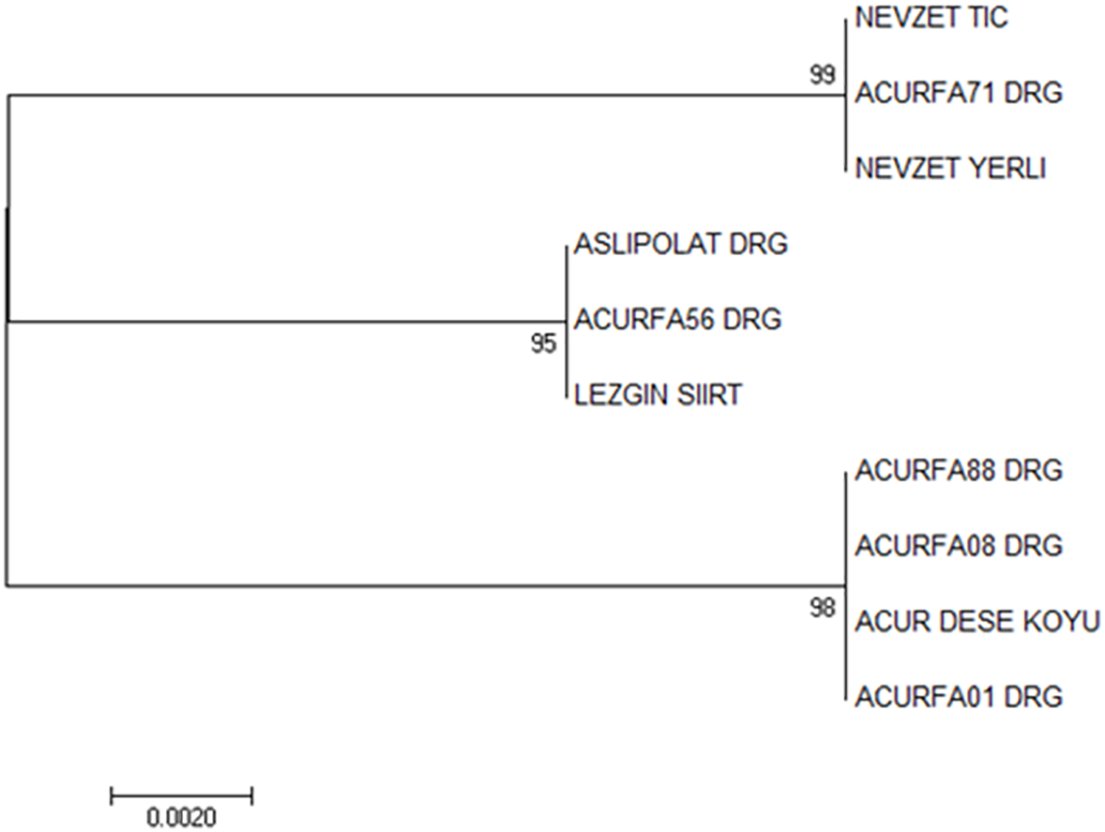
Figure 7: The tree of the trnL-f region resulting from the neighbor-joining analysis (The numbers in the nodes of the phylogenetic tree show the bootstrap values)
It was determined that the algal content might be used to nourish plants. Lower leaf temperature is induced by algae. It was found that algal applications with interaction groups based on genotypes had a positive effect on LRWC, plant height, and main stem diameter values. The application of algae had the best results in terms of fruit length, fruit diameter, and yield parameters. However, it has been found that compared to other applications, algal applications have a smaller effect on the number of nodes. In terms of kinship, the commercial and domestic varieties of C. melo var. conomon were not very dissimilar from each other.
Acknowledgement: We would like to express our gratitude to the Siirt University Science and Technology Application and Research Center for their invaluable assistance in analyzing the ICP-MS samples.
Funding Statement: This study was supported by the Scientific Research Coordinatorship of Siirt University with the Project Number 2018-SİÜFEB-022.
Author Contributions: Behcet İNAL played a pivotal role in conceptualizing the study’s design, conducting molecular and ICP-MS sample experiments, instrumental analysis, and contributing significantly to the manuscript’s writing and review. M. Zeki KARİPÇİN was responsible for executing field experimental patterns, conducting physiological and agronomic experiments, managing material harvesting, and performing statistical analysis. All authors actively participated in revising and granting final approval for this article.
Availability of Data and Materials: The data used in the study are available upon request from the corresponding author.
Ethics Approval: We want to emphasize that we adhered to standard procedures and took appropriate precautions to ensure personnel safety and minimize environmental impacts, which included measures to reduce waste generation and prevent water contamination.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Kulak, M., Ozkan, A., Bindak, R. (2019). A bibliometric analysis of the essential oil-bearing plants exposed to the water stress: How long way we have come and how much further? Scientia Horticulturae, 246, 418–436. [Google Scholar]
2. Bernardo, L., Carletti, P., Badeck, F. W., Rizza, F., Morcia, C. et al. (2019). Metabolomic responses triggered by arbuscular mycorrhiza enhance tolerance to water stress in wheat cultivars. Plant Physiology and Biochemistry, 137, 203–212. [Google Scholar] [PubMed]
3. Das, D., Prakash, P., Rout, P. K., Bhaladhare, S. (2021). Synthesis and characterization of superabsorbent cellulose-based hydrogel for agriculture application. Starch-Stärke, 73(1–2), 1900284. [Google Scholar]
4. Zia, R., Nawaz, M. S., Siddique, M. J., Hakim, S., Imran, A. (2021). Plant survival under drought stress: Implications, adaptive responses, and integrated rhizosphere management strategy for stress mitigation. Microbiological Research, 242, 126626. [Google Scholar] [PubMed]
5. WHO (2020). https://www.who.int/ 1 March 2023 [Google Scholar]
6. Yadav, B., Jogawat, A., Rahman, M. S., Narayan, O. P. (2021). Secondary metabolites in the drought stress tolerance of crop plants: A review. Gene Reports, 23, 101040. [Google Scholar]
7. Swamy, K. R. M. (2017). Origin, distribution and systematics of culinary cucumber (Cucumis melo subsp. agrestis var. conomon). Journal of Horticultural Sciences, 12(1), 1–22. [Google Scholar]
8. Andic, M. (2020). Investigation of the effects of hydrogen sulfide (H2S) in providing drought tolerance in tomato (Solanum lycopersicum L.) plant at biochemical and gene expression level (Doctoral Dissertation). Necmettin Erbakan University, Turkey. [Google Scholar]
9. Yang, X., Lu, M., Wang, Y., Wang, Y., Liu, Z. et al. (2021). Response mechanism of plants to drought stress. Horticulturae, 7(3), 50. [Google Scholar]
10. Kiran, S. (2019). Effects of vermicompost on some morphological, physiological and biochemical parameters of lettuce (Lactuca sativa var. crispa) under drought stress. Notulae Botanicae Horti Agrobotanici Cluj-Napoca, 47(2), 352–358. [Google Scholar]
11. Ghosh, D., Ghorai, P., Debnath, S., Indrama, T., Kondi, V. et al. (2022). Algal biofertilizer towards green sustainable agriculture. In: New and future developments in microbial biotechnology and bioengineering, pp. 27–45. San Diego: Elsevier. [Google Scholar]
12. Shahi Khalaf Ansar, B., Kavusi, E., Dehghanian, Z., Pandey, J., Asgari Lajayer, B. et al. (2022). Removal of organic and inorganic contaminants from the air, soil, and water by algae. Environmental Science and Pollution Research, 29, 1–29. [Google Scholar]
13. Emadodin, I., Reinsch, T., Rotter, A., Orlando-Bonaca, M., Taube, F. et al. (2020). A perspective on the potential of using marine organic fertilizers for the sustainable management of coastal ecosystem services. Environmental Sustainability, 3, 105–115. [Google Scholar]
14. Ashour, M., Hassan, S. M., Elshobary, M. E., Ammar, G. A., Gaber, A. et al. (2021). Impact of commercial seaweed liquid extract (TAM®) biostimulant and its bioactive molecules on growth and antioxidant activities of hot pepper (Capsicum annuum). Plants, 10(6), 1045. [Google Scholar] [PubMed]
15. Kumari, R., Bhatnagar, S., Mehla, N., Vashistha, A. (2022). Potential of organic amendments (AM fungi, PGPR, vermicompost and seaweeds) in combating salt stress—A review. Plant Stress, 6, 100111. [Google Scholar]
16. El Boukhari, M. E. M., Barakate, M., Bouhia, Y., Lyamlouli, K. (2020). Trends in seaweed extract based biostimulants: Manufacturing process and beneficial effect on soil-plant systems. Plants, 9(3), 359. [Google Scholar] [PubMed]
17. Schlötterer, C. (2004). The evolution of molecular markers—just a matter of fashion? Nature Reviews Genetics, 5(1), 63–69. [Google Scholar]
18. Zhang, J., Yang, J., Lv, Y., Zhang, X., Xia, C. et al. (2023). Genetic diversity analysis and variety identification using SSR and SNP markers in melon. BMC Plant Biology, 23(1), 1–10. [Google Scholar]
19. Matsumoto, Y., Miyagi, M., Watanabe, N., Kuboyama, T. (2012). Temperature-dependent enhancement of pollen tube growth observed in interspecific crosses between wild Cucumis spp. and melon (C. melo L.). Scientia Horticulturae, 138, 144–150. [Google Scholar]
20. Moing, A., Allwood, J. W., Aharoni, A., Baker, J., Beale, M. H. et al. (2020). Comparative metabolomics and molecular phylogenetics of melon (Cucumis melo, Cucurbitaceae) biodiversity. Metabolites, 10(3), 121. [Google Scholar] [PubMed]
21. Renner, S. S., Schaefer, H., Kocyan, A. (2007). Phylogenetics of Cucumis (CucurbitaceaeCucumber (C. sativus) belongs in an Asian/Australian clade far from melon (C. melo). BMC Ecology and Evolution, 7, 58. [Google Scholar]
22. Sebastian, P., Schaefer, H., Telford, I. R. H., Renner, S. S. (2010). Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proceedings of the National Academy of Sciences of the United States of America, 107, 14269–14273. [Google Scholar] [PubMed]
23. Zhou, C., Wang, P., Zeng, Q., Zeng, R., Hu, W. et al. (2023). Comparative chloroplast genome analysis of seven extant Citrullus species insight into genetic variation, phylogenetic relationships, and selective pressure. Scientific Reports, 13(1), 6779. [Google Scholar] [PubMed]
24. Ackley, W. B. (1954). Seasonal and diurnal changes in the water contents and water deficits of bartlett pear leaves. Plant Physiology, 29(5), 445–448. [Google Scholar] [PubMed]
25. Smart, R. E., Bingham, G. E. (1974). Rapid estimates of relative water content. Plant Physiology, 53(2), 258–260. [Google Scholar] [PubMed]
26. Karipçin, M. Z. (2009). Determination of drought tolerance in native and wild watermelon genotypes (Doctoral Thesis). Çukurova university graduate school of natural and applied sciences, Adana. [Google Scholar]
27. Karaca, M., İnce, A. G., Elmasulu, S. Y., Onus, A. N., Turgut, K. (2005). Coisolation of genomic and organelle DNAs from 15 genera and 31 species of plants. Analytical Biochemistry, 343(2), 353–355. [Google Scholar] [PubMed]
28. Taberlet, P., Gielly, L., Pautou, G., Bouvet, J. (1991). Universal primers for amplification of three noncoding regions of chloroplast DNA. Plant Molecular Biology, 17, 1105–1109. [Google Scholar] [PubMed]
29. Thompson, J. D., Higgins, D. G., Gibson, T. J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673–4680. [Google Scholar] [PubMed]
30. Kumar, S., Stecher, G., Li, M., Knyaz, C., Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549. [Google Scholar] [PubMed]
31. SAS Institute (2011). SAS/IML 9.3 user’s guide. Cary, North Carolina: SAS Institute. [Google Scholar]
32. Montgomery, D. C. (2017). Design and analysis of experiments. Ninth edition. Hoboken, NJ: John Wiley & Sons, Inc. [Google Scholar]
33. Zhang, X., Koehler, H. (2022). Soil algae for combating soil degradation—greenhouse experiment with different soil amendments. Soil Research, 61(1), 70–82. [Google Scholar]
34. Negreiros, A. M., JúNior, R. S., Francisco, F., Silva, R. B., Costa, J. A. et al. (2019). Lithothamnion calcareum nanoparticles increase growth of melon plants. Notulae Botanicae Horti Agrobotanici Cluj-Napoca, 47(2), 426–431. [Google Scholar]
35. Gitau, M. M., Farkas, A., Ördög, V., Maróti, G. (2022). Evaluation of the biostimulant effects of two chlorophyta microalgae on tomato (Solanum lycopersicum). Journal of Cleaner Production, 364, 132689. [Google Scholar]
36. Bajpai, S., Shukla, P. S., Asiedu, S., Pruski, K., Prithiviraj, B. (2019). A biostimulant preparation of brown seaweed Ascophyllum nodosum suppresses powdery mildew of strawberry. The Plant Pathology Journal, 35, 406–416. [Google Scholar] [PubMed]
37. De Saeger, J., van Praet, S., Vereecke, D., Park, J., Jacques, S. et al. (2020). Toward the molecular understanding of the action mechanism of Ascophyllum nodosum extracts on plants. Journal of Applied Psychology, 32(1), 573–597. [Google Scholar]
38. Ali, O., Ramsubhag, A., Jayaraj, J. (2021). Biostimulant properties of seaweed extracts in plants: Implications towards sustainable crop production. Plants, 10, 1–27. [Google Scholar]
39. Abbas, M., Anwar, J., Zafar-ul-Hye, M., Iqbal Khan, R., Saleem, M. et al. (2020). Effect of seaweed extract on productivity and quality attributes of four onion cultivars. Horticulturae, 6(2), 28. [Google Scholar]
40. Keya Tudu, C., Dey, A., Pandey, D. K., Panwar, J. S., Nandy, S. (2022). Role of plant derived extracts as biostimulants in sustainable agriculture: A detailed study on research advances, bottlenecks and future prospects. In: New and future developments in microbial biotechnology and bioengineering. pp. 159–179. Amsterdam, The Netherlands: Elsevier. [Google Scholar]
41. Yang, S., Gil, Y., Oh, H., Koo, Y. (2020). Melon growth enhancement by fucoidan and fucoidan decomposing bacteria. Korean Journal of Environmental Agriculture, 39(1), 20–25. [Google Scholar]
42. Ali, O., Ramsubhag, A., BennJr, D., Ramnarine, S., Jayaraman, J. (2022). Transcriptomic changes induced by applications of a commercial extract of Ascophyllum nodosum on tomato plants. Scientific Reports, 12(1), 1–13. [Google Scholar]
43. Craigie, J. S., Cornish, M. L., Deveau, L. E. (2019). Commercialization of Irish moss aquaculture: The Canadian experience. Botanica Marina, 62(5), 411–432. [Google Scholar]
44. Liu, H., Chen, X., Song, L., Li, K., Zhang, X. et al. (2019). Polysaccharides from Grateloupia filicina enhance tolerance of rice seeds (Oryza sativa L.) under salt stress. International Journal of Biological Macromolecules, 124, 1197–1204. [Google Scholar] [PubMed]
45. Rouphael, Y., Colla, G. (2020). Biostimulants in agriculture. Frontiers in Plant Science, 11, 40. [Google Scholar] [PubMed]
46. Kusvuran, S., Tuna, U. T. (2021). Microalgae (Chlorella vulgaris)-mediated seconder metabolites and antioxidative defense system improve plant growth and salt tolerance in melon. International Journal of Agricultural and Natural Sciences, 14(1), 33–46. [Google Scholar]
47. Hassan, S. M., Ashour, M., Sakai, N., Zhang, L., Hassanien, H. A. et al. (2021). Impact of seaweed liquid extract biostimulant on growth, yield, and chemical composition of cucumber (Cucumis sativus). Agriculture, 11(4), 320. [Google Scholar]
48. Patel, K., Agarwal, P., Agarwal, P. K. (2018). Kappaphycus alvarezii sap mitigates abiotic-induced stress in Triticum durum by modulating metabolic coordination and improves growth and yield. Journal of Applied Phycology, 30(4), 2659–2673. [Google Scholar]
49. Al-Ghamdi, A. A., Elansary, H. O. (2018). Synergetic effects of 5-aminolevulinic acid and Ascophyllum nodosum seaweed extracts on asparagus phenolics and stress related genes under saline irrigation. Plant Physiology and Biochemistry, 129, 273–284. [Google Scholar] [PubMed]
50. Zou, P., Yang, X., Yuan, Y., Jing, C., Cao, J. et al. (2021). Purification and characterization of a fucoidan from the brown algae Macrocystis pyrifera and the activity of enhancing salt-stress tolerance of wheat seedlings. International Journal of Biological Macromolecules, 180, 547–558. [Google Scholar] [PubMed]
51. Dookie, M., Ali, O., Ramsubhag, A., Jayaraman, J. (2021). Flowering gene regulation in tomato plants treated with brown seaweed extracts. Scientia Horticulturae, 276, 109715. [Google Scholar]
52. Elsharkawy, G. A., Ibrahim, H. A. H., Salah, A. H., Akrami, M., Ali, H. M. et al. (2021). Early and total yield enhancement of the globe artichoke using an ecofriendly seaweed extract-based biostimulant and PK fertilizer. Agronomy, 11(9), 1819. [Google Scholar]
53. Duarte, I. J., Hernández, S. H. Á., Ibañez, A. L., Canto, A. R. (2018). Macroalgae as soil conditioners or growth promoters of Pisum sativum (L). Annual Research & Review in Biology, 27(6), 1–8. [Google Scholar]
54. Dalal, A., Bourstein, R., Haish, N., Shenhar, I., Wallach, R. et al. (2019). Dynamic physiological phenotyping of drought-stressed pepper plants treated with “productivity-enhancing” and “survivability-enhancing” biostimulants. Frontiers in Plant Science, 10, 905. [Google Scholar] [PubMed]
55. Al-Naqeeb, S. N. Y., Merza, T. K. (2020). Effect of spraying with nano seaweed extract and quality of irrigation water on growth of volkamer lemon rootstock saplings. Plant Archives, 20(2), 3952–3954. [Google Scholar]
56. Abideen, Z., Waqif, H., Munir, N., El-Keblawy, A., Hasnain, M. et al. (2022). Algal-mediated nanoparticles, phycochar, and biofertilizers for mitigating abiotic stresses in plants: A review. Agronomy, 12(8), 1788. [Google Scholar]
57. Ferrari, M., Muto, A., Bruno, L., Cozza, R. (2023). DNA Methylation in algae and its impact on abiotic stress responses. Plants, 12(2), 241. [Google Scholar] [PubMed]
58. Lakshmi, P. K., Meenakshi, S. (2022). Micro and macroalgae: A potential biostimulant for abiotic stress management and crop production. In: New and future developments in microbial biotechnology and bioengineering. pp. 63–82. Australia: Elsevier. [Google Scholar]
59. da Cunha, J. G., Cavalcante, Í. H. L., da Silva, M. A., e Amariz, R. A., do Carmo, R. N. et al. (2022). Proline and algal extract to alleviate the abiotic stress in mango ‘Tommy Atkins’ in the Tropical Semiarid. Erwerbs-Obstbau, 64(1), 115–126. [Google Scholar]
60. Small, L. R., Lickey, E. B., Shaw, J., Hauk, W. D. (2005). Amplification of noncoding chloroplast DNA for phylogenetic studies in lycophytes and monilophytes with a comparative example of relative phylogenetic utility from Ophioglossaceae. Molecular Phylogenetics and Evolution, 36, 509–522. [Google Scholar] [PubMed]
61. Fathurrahman, F. (2023). Growth and genetic characteristics of cucumber (Cucumis sativus L.) cultivar mercy f1 hybrid and mutant populations. SABRAO Journal of Breeding and Genetics, 55(2), 485–494. https://doi.org/10.54910/sabrao2023.55.2.20 [Google Scholar] [CrossRef]
62. Almutairi, Z. M. (2022). Genetic diversity and phylogeny of pearl millets [Pennisetum glaucum (L.) R. Br.] based on chloroplast trnL-F region. Genetic Resources and Crop Evolution, 69(8), 2849–2859. [Google Scholar]
Supplementary Materials
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools