 Open Access
Open Access
ARTICLE
Genome-Wide Identification, Evolution and Expression Analyses of GA2ox Gene Family in Brassica napus L.
1 Chongqing Academy of Agricultural Sciences, Chongqing, 401329, China
2 Chongqing Municipal Agricultural School, Chongqing, 401329, China
3 Fengdu Agricultural Technology Service Center, Chongqing, 408201, China
* Corresponding Authors: Taocui Huang. Email: ; Yan Zhou. Email:
# These authors contributed equally to this paper as first authors
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(3), 815-835. https://doi.org/10.32604/phyton.2023.023928
Received 18 May 2022; Accepted 09 September 2022; Issue published 29 November 2022
Abstract
Gibberellin 2-oxidases (GA2ox) are important enzymes that maintain the balance of bioactive GAs in plants. GA2ox genes have been identified and characterized in many plants, but these genes were not investigated in Brassica napus. Here, we identified 31 GA2ox genes in B. napus and 15 of these BnaGA2ox genes were distributed in the A and C subgenomes. Subcellular localization predictions suggested that all BnaGA2ox proteins were localized in the cytoplasm, and gene structure analysis showed that the BnaGA2ox genes contained 2–4 exons. Phylogenetic analysis indicated that BnGA2ox family proteins in monocotyledons and dicotyledons can be divided into four groups, including two C19-GA2ox and two C20-GA2ox clades. Group 4 is a C20-GA2ox Class discovered recently. Most BnaGA2ox genes had a syntenic relationship with AtGA2ox genes. BnaGA2ox genes in the C subgenome had experienced stronger selection pressure than genes in the A subgenome. BnaGA2ox genes were highly expressed in specific tissues such as those involved in growth and development, and most of them were mainly involved in abiotic responses, regulation of phytohormones and growth and development. Our study provided a valuable evolutionary analysis of GA2ox genes in monocotyledons and dicotyledons, as well as an insight into the biological functions of GA2ox family genes in B. napus.Keywords
Gibberellins (GAs) are diterpenoid compounds with biological activity in plants. They are factors that help to regulate plant growth and development, including seed germination, fruit development, flowering time, leaf morphology and internode elongation [1,2]. The biosynthetic pathways and the main regulatory mechanisms of GAs are understood in Arabidopsis thaliana [3–5]. Most of the genes involved in GAs biosynthesis and regulation of GAs have been cloned in plants such as A. thaliana [6], rice (Oryza sativa) [7] and pea (Pisum sativum) [8]. Deletion or mutation of the genes associated with the biosynthesis of GAs can result in dwarf or semi-dwarf plant phenotypes [8–11]. GAs have been manipulated in crop plants to increase elongation of stems in sugarcane (Saccharum officinarum) and retard seed growth in cotton (Gossypium hirsutum) [11].
GA2ox, GA3ox and GA20ox are key oxidases in GA biosynthesis. They belong to the 20G-Fe (II) oxygenase superfamily, and their deletion or mutation can cause dwarf or semi-dwarf plant phenotypes [8–12]. According to the number of carbon (C) atoms, GAs are classified into different groups, including C19-GAs (such as GA9, GA20, GA1, and GA4), and C20-GAs (such as GA12, GA15, GA24, GA53, GA44, and GA19). GA2-oxidases can act on bioactive GA1 and GA4 to affect C-2 hydroxylation to transform them into inactive GA8 and GA34, and the GA2ox proteins are also classified into C19-GA2ox or C20-GA2ox according to its roles for C20-GA or C19-GA substrates. However, the synthesis of active GA1 and GA4 depends on GA12 and GA53 [2,11,13]. GA2ox is a key factor maintaining the balance between bioactive GAs and intermediates in plants [14]. It was first identified in bean (Phaseolus coccineus) [15] and then identified in other plants such as rice [16,17], A. thaliana [11,18], maize (Zea mays) [19], grape (Vitis vinifera) [20] and tea tree (Camellia lipoensis) [21]. Overexpression of GA2ox genes without tissue or species-specific regulation results in growth and reproductive abnormalities such as dwarfism and sterility. For example, overexpression of bean GA2ox1 in rye (Secale cereale) and ryegrass (Lolium perenne) resulted in a dwarf phenotypes owing to the reduction of bioactive GA1 and GA4 in the transgenic plants [15]. Similar dwarf phenotypes have been observed in tobacco (Nicotiana tabacum) by overexpressing tea GA2ox1 or GA2ox3. These transgenic plants showed delayed flowering, reduced growth, and smaller, rounder and darker green leaves [21]. In addition to the regulation of plant height, GA2ox genes also affect responses to abiotic stresses and to photomorphogenesis. Overexpression of OsGA2ox5 improved resistance to high salt in rice [22]. Induction of GA2ox expression reduced bioactive GA4 and promoted photomorphogenesis in A. thaliana [23]. Some AtGA2ox proteins also function in catalyzing the binding of certain phytohormones to amino acids, such as indoleacetic acid and salicylic acid [24,25].
B. napus (rapeseed) is an important oilseed crop worldwide and is formed from two diploid species Brassica rapa and B. oleracea [26,27]. Ectopic expression of B. napus GA2ox6 in A. thaliana resulted in late flowering and increased chlorophyll content in leaf [28]. This suggested that the GA pathway regulation via the GA2ox genes may have potential for B. napus improvement. However, there are little studies focused on the biosynthetic activity of GA2ox genes in B. napus compared to rice and wheat, and the GA2ox family genes in B. napus still remain unknown. Here, we identified BnaGA2ox genes using a reciprocal Basic Local Alignment Search Tool Protein (BLASTP) with AtGA2ox genes as query sequences. We analyzed their protein characters and gene structure, as well as the cis-elements in promoter regions. We also examined the evolution among GA2ox members from seven species and investigated the spatiotemporal expression patterns of BnaGA2ox genes. The results provide insights into the evolution of GA2ox proteins in plants and help to elucidate the roles of GA2ox in B. napus.
2.1 Data Acquisition and GA2ox Genes Identification
The sequence files, including genomic, protein, coding sequences of A. thaliana were obtained from the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/), and that information from B. oleracea (JZS) and B. rapa (Chiifu) was obtained from the Brassicaceae Database (http://brassicadb.cn). The B. napus pan-genome Information Resource (BnPIR, http://cbi.hzau.edu.cn/bnapus/) was used to retrieve the sequences files of B. napus (ZS11), and the sequence files of tomato (Solanum lycopersicum) were downloaded from the Solanaceae Database (https://solgenomics.net). Sequence files of O. sativa (Japonica) and Z. mays (Zm-B73) were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/t).
GA2ox genes were identified following two steps: First, using nine GA2ox protein sequences from A. thaliana as query sequences, the candidate GA2ox genes were identified in a local reciprocal BLASTP [29] with the screening parameters e-value < 1e-5 and qcover ≥50% [30]. Then, all candidate GA2ox proteins were studied through the online PfamScan (http://www.ebi.ac.uk/Tools/pfa/pfamscan/) to validate their functional domain of 2OG-FeII_Oxy (PF03171.20).
The online website ExPASy (http://web.expasy.org/protparam/) was used to calculate and predict the characteristics of BnaGA2ox proteins, including molecular weight (MW, kDa), number of amino acids (AA) and isoelectric point (pI). The SignalP 5.0 server was (http://www.cbs.dtu.dk/services/SignalP/) used to predict each BnaGA2ox protein signal peptide with default parameters, and their subcellular localizations were analyzed through the Plant-mPLoc server (http://www.csbio.sjtu.edu.cn/bioinf/plant-multi/) with default parameters.
2.3 Phylogenetic and Evolution Analysis
The multiple GA2ox protein sequences were aligned by MUSCLE software [31] with default arguments. The Molecular Evolutionary Genetics Analysis (MEGA) 7 program [32] was used to construct the phylogenetic trees using the Neighbor-Joining (NJ) method, with the patterns setting to number of different + G+I substitution model + 1,000 bootstrap replications. The phylogenetic tree was displayed and modified using FigTree v1.4.3 (http://tree.bio.ed.ac.uk/software/figtree/). To compare the selective pressure on GA2ox genes, the coding sequence alignments between B. napus and A. thaliana were imported into KaKs_Calculator 2.0 [33] to evaluate the non-synonymous mutation rate (Ka) and synonymous mutation rate (Ks) with GY-HKY model.
2.4 Gene Structure and Protein Motif Identification Analysis
The exon/intron structures of BnaGA2ox were displayed using the Gene Structure Display Server (GSDS2.0, http://gsds.gao-lab.org/). The conserved motifs of BnaGA2ox proteins were identified using the online Multiple Em for Motif Elicitation (MEME, https://meme-suite.org/meme/tools/meme) through the following arguments: (1) optimum width of motif ranging from 6 to 100; (2) maximum number of motif sets to 10; and (3) e-value > 1e-10. The conserved motifs were displayed using TBtools [34].
2.5 Chromosomal Location, Syntenic Relationships and Cis-Acting Elements
The chromosomal locations of BnaGA2ox were obtained based on the annotation file in B. napus of ZS11 from BnPIR. The syntenic relationships of GA2ox between B. napus and A. thaliana were acquired from the Brassicaceae database (http://brassicadb.org/brad/), and displayed using TBtools. To analyze the cis-acting elements in BnaGA2ox promoter regions, the 1500-bp genomic DNA sequences upstream of BnaGA2ox genes were obtained using TBtools and submitted to the online PlantCARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/).
2.6 Expression Patterns of BnaGA2ox Genes and Quantitative Reverse-Transcription PCR
To compare and analyze the expression levels of BnaGA2ox genes in multiple tissues, we downloaded their expression files in cultivar ZS11 from B. napus transcriptome information resource (BnTIR, http://yanglab.hzau.edu.cn/BnTIR) [35]. There were 91 different tissues used in this study including roots, stems, leaves and sepals, petals, pollen, siliques and seeds at different stages (a detailed schematic diagram of tissues is shown in Supplementary Fig. S1). Meanwhile, we also downloaded the expression patterns of BnaGA2ox genes under phytohormone and abiotic treatments at the seedling leaves (SLs) and seedling roots (SRs) from BnTIR, including seven phytohormones and six abiotic treatments. All gene expression levels of BnaGA2ox genes were normalized by Log2(TPM+1).
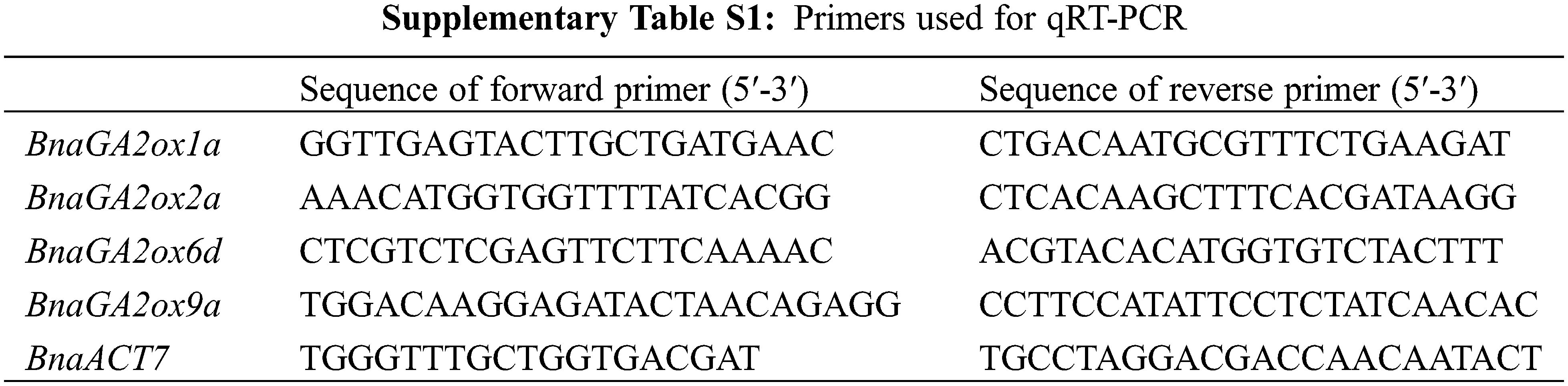
To verify the RNA-seq results, four genes (BnaGA2ox1a, BnaGA2ox2a, BnaGA2ox6d, BnaGA2ox9a) were selected for qRT-PCR analysis. Total RNA was extracted from tissues using an RNA Extraction Kit (Tiangen, Beijing, China), including roots, stems, leaves and buds at the bolting stages, and seeds at different stages. Their cDNA was synthesized according to the manufacturer’s instructions using M-MLV transcriptase (TaKaRa Biotechnology, Dalian, China). A Bio-Rad CFX96 Real Time System (USA) was used to perform the Reverse transcription quantitative PCR (RT-qPCR), and gene-specific primers were designed using Geneious 10 (Biomatters, Auckland, New Zealand) (Supplementary Table S1). The qRT-PCR reactions were performed as described in the MIQE guidelines [36], with three technical replicates for each sample. Relative expression levels were calculated using the 2−∆∆Ct method, with BnACT7 as internal controls.
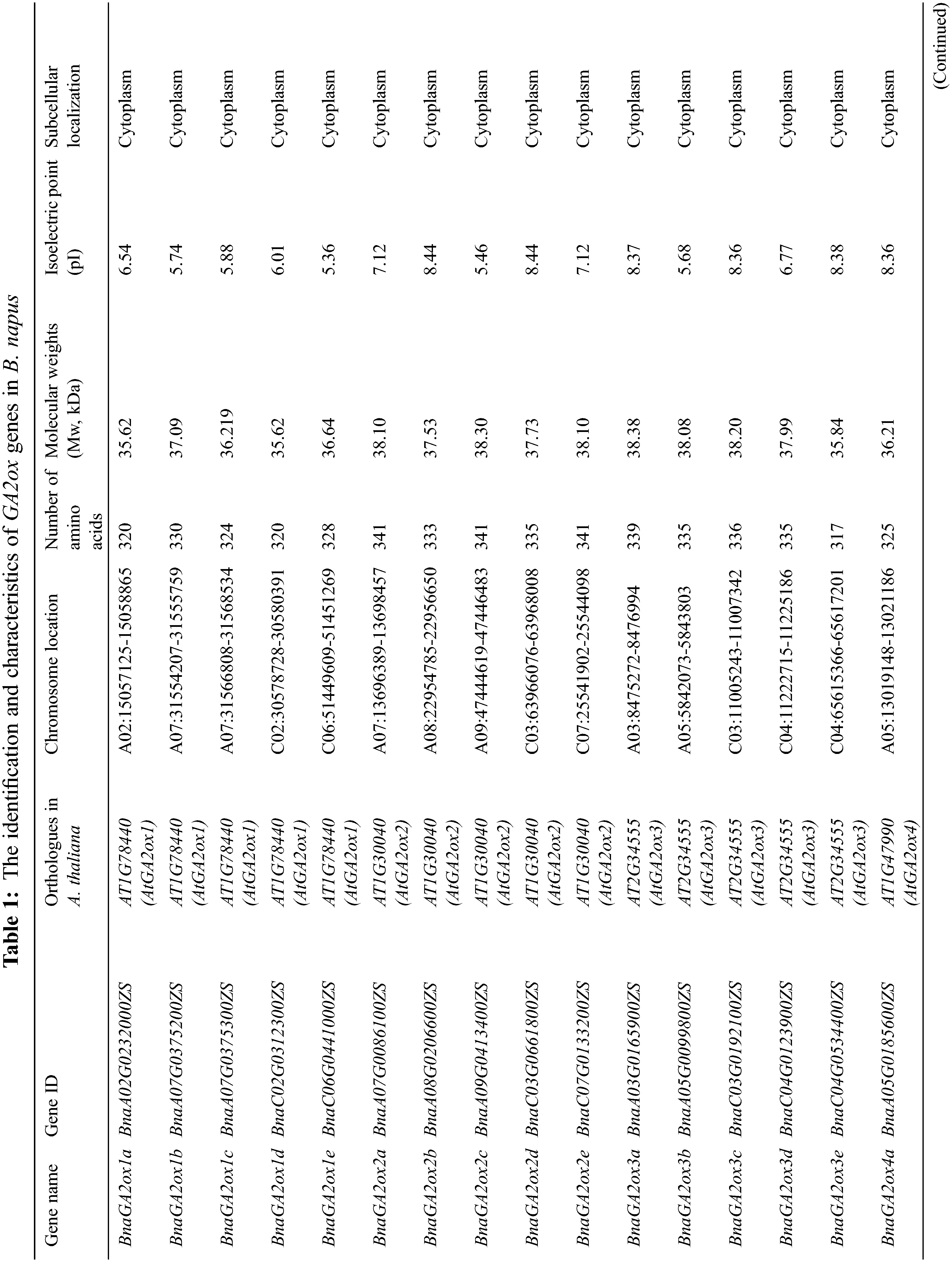
3.1 Identification and Characterization of BnGA2ox Genes
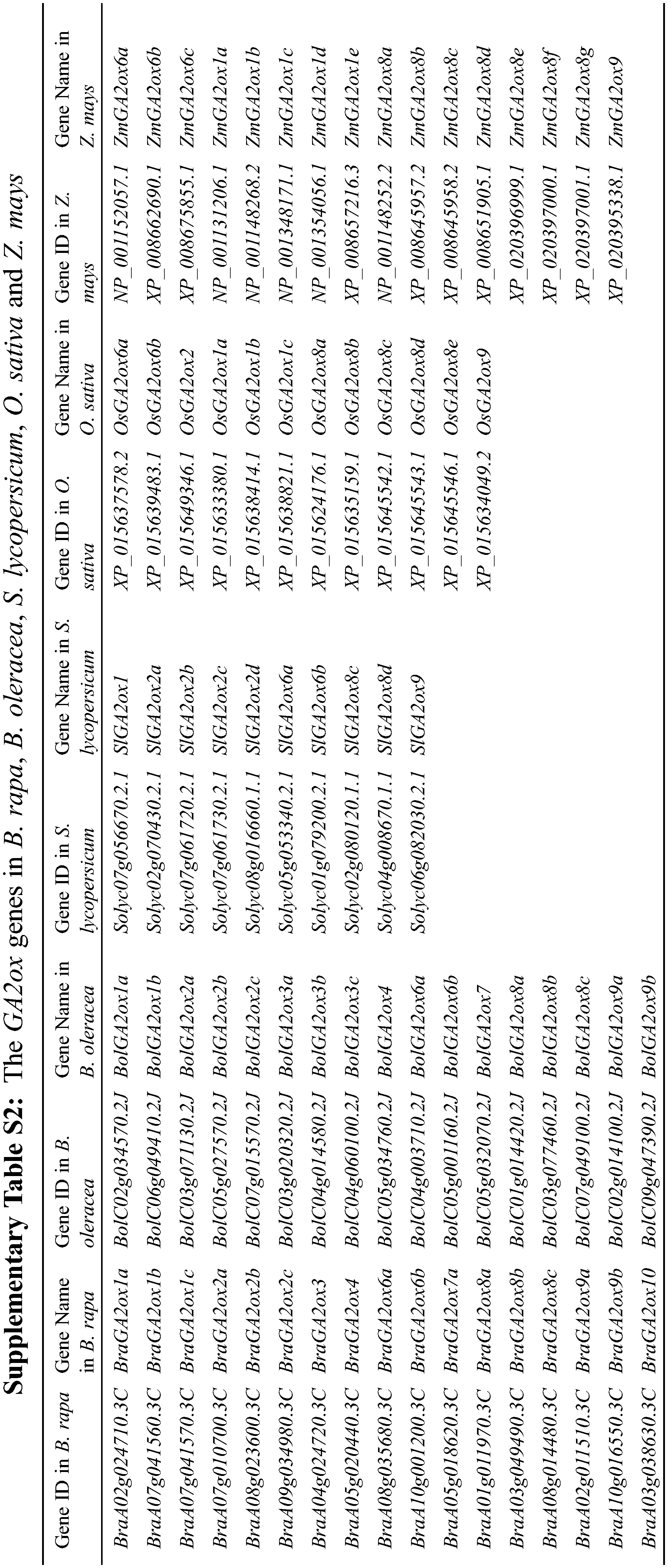
We found 10 GA2ox genes in A. thaliana by searching the GA2ox name from TAIR. However, AtGA2ox5 (AT3G17203) is a pseudogene. Using the remaining nine AtGA2ox protein sequences as query, a total of 31 BnaGA2ox genes were identified using a combination between the reciprocal BLASTP and 2OG-FeII_Oxy domain screening (Table 1). We named the putative BnaGA2ox genes according to their homologous AtA2ox genes. For most of the AtGA2ox genes, two to five homologous BnaGA2ox genes were identified; however, no homolog was identified for AtGA2ox10 in B. napus genome. Among all BnaGA2ox proteins, the number of amino acids ranged from 213 (BnaGA2ox7b) to 379 aa (BnaGA2ox9c), with an average of 327.6 aa. The Mw (molecular weights) were found to range from 24.63 (BnaGA2ox7b) to 42.47 kDa (BnaGA2ox9c), with an average of 36.99 kDa, and the pI (isoelectric point) ranged from 4.80 (BnaGA2ox9c) to 8.98 (BnaGA2ox7b), with an average of 6.84 (Table 1). All BnaGA2ox proteins were suggested to be localized in the cytoplasm, while no signal peptide was observed in any BnaGA2ox protein (Table 1). In addition, using the same methods, we also identified 12 GA2ox genes in O. sativa, 16 GA2ox genes in Z. mays, 11 GA2ox genes in S. lycopersicum, 17 GA2ox genes in B. oleracea and 17 GA2ox genes in B. rapa (Supplementary Table S2), respectively.
3.2 Phylogenetic Analysis of GA2ox Protein Members
To study the phylogenetic relationships of GA2ox proteins in B. napus, as well as in monocotyledons and dicotyledons, we constructed a NJ tree using GA2ox proteins from B. rapa, B. oleracea, B. napus, A. thaliana, O. sativa, Z. mays and S. lycopersicum (Fig. 1, Supplementary Table S2). The GA2ox proteins in these seven species were divided into four groups based on phylogenetic topology and bootstrap values (Fig. 1). They were named Group 1 (GA2ox1/2/3 subfamily), Group 2 (GA2ox4/6 subfamily), Group 3 (GA2ox7/8 subfamily) and Group 4 (GA2ox9/10 subfamily). Group 1 and Group 2 belong to the C20-GA2ox Class, and Group 3 and Group 4 belong to the C19-GA2ox Class. Among the four groups, the largest clade was Group 1, containing 47 GA2ox proteins, including 33 GA2ox proteins of Brassicaceae, and the smallest clade was Group 4, containing 13 GA2ox proteins, including 10 GA2ox proteins of Brassicaceae, and each clade presented at least one GA2ox member per plant. Further analysis revealed that the GA2ox members from B. oleracea, B. rapa, B. napus, O. sativa and Z. mays in Group 1 and Group 3 were larger than those in Group 2 and Group 4. This indicated an expansion of GA2ox members in the Brassica species of Group 1 and Group 3, as well as in the monocotyledons. In each group, the AtGA2oxs always first clustered with its orthologues from the Brassica to form a Brassicaceae cluster, and then clustered with the GA2oxs in S. lycopersicum to form a dicotyledon cluster that finally joined with GA2ox members in O. sativa and Z. mays to form a specific GA2ox clade (Fig. 1). These results suggest that the homology of GA2ox proteins in the species was consistent with previous perceived species relationships. We also hypothesized that there should be two ancient copies of GA2ox in the common ancestral species of monocotyledons and dicotyledons based on the GA2ox phylogenetic relationships. One as the ancestral copy for C20-GA2ox Class and the other as the ancestral copy for C19-GA2ox Class.
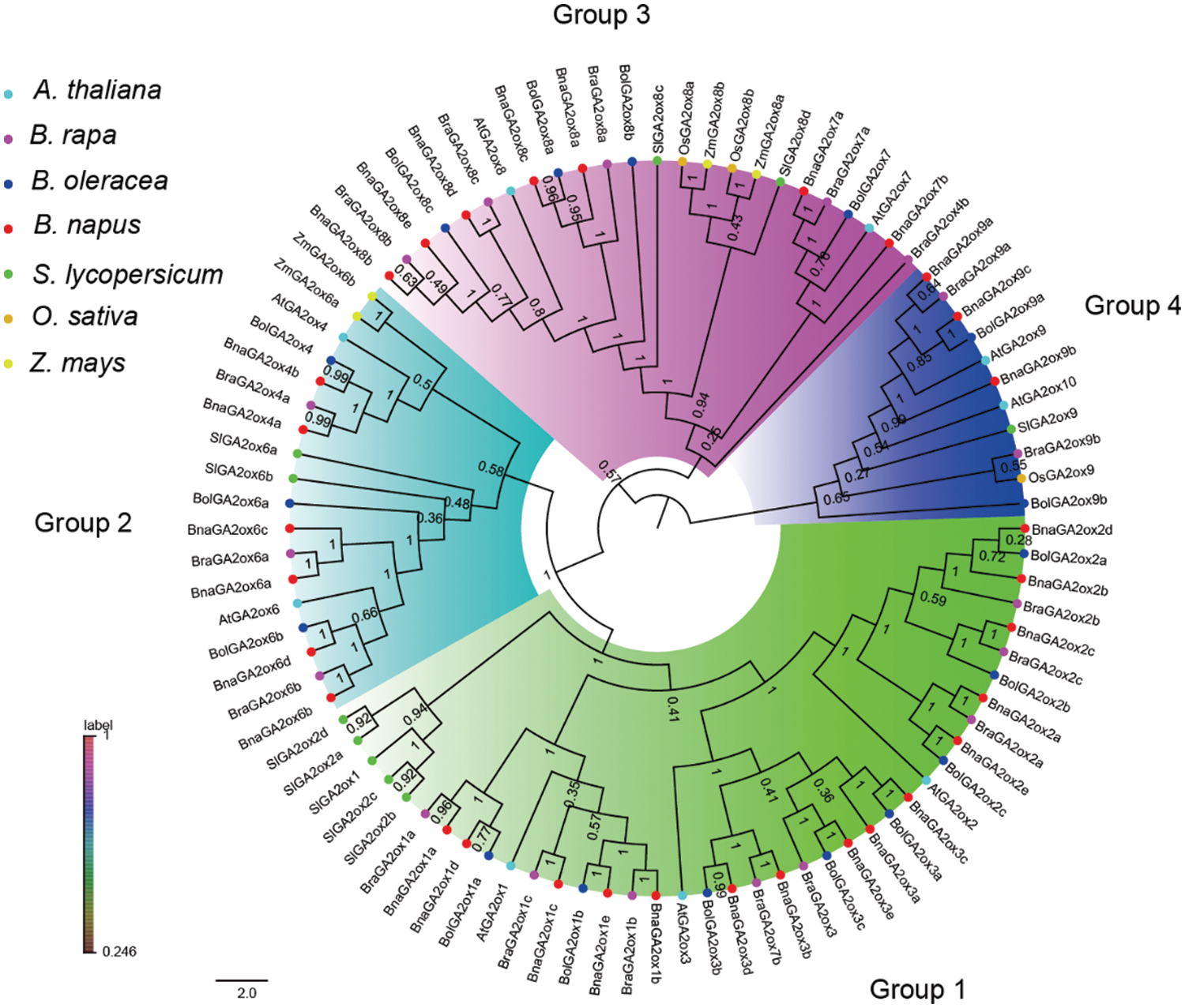
Figure 1: Phylogenetic relationship among GA2ox proteins in plants
Note: The NJ tree was generated with bootstrap analysis (1,000 replicates) using aligned GA2ox protein sequences from seven species and the tree was displayed with FigTree v1.4.0. At: A. thaliana; Bna: B. napus; Bra: B. rapa; Bol: B. oleracea; Sl: S. lycopersicum; Os: O. sativa; Zm; Z. mays.
3.3 Gene Structure and Conserved Motif Analysis of BnaGA2ox Family Members
To analyze and compare the intron/exon structures of BnaGA2ox family genes, their CDSs were aligned with the corresponding genomic sequences. The intron/exon structures among BnaGA2ox genes showed a wide difference, particularly the members belonging to different groups, while the BnaGA2ox genes of the same clade also showed certain exon segments and distributions (Fig. 2). All of the BnaGA2ox genes contained less exons and introns and none had more than four exons or introns. The single exon fragment in BnaGA2ox genes was less than one kilobyte. However, BnaGA2ox8d contained an intron of about eight kilobytes, BnaGA2ox8c had an intron of about five kilobytes, and BnaGA2ox8a contained an intron of about three kilobytes. The remaining BnaGA2ox8a genes had a single intron fragment smaller than two kilobytes.
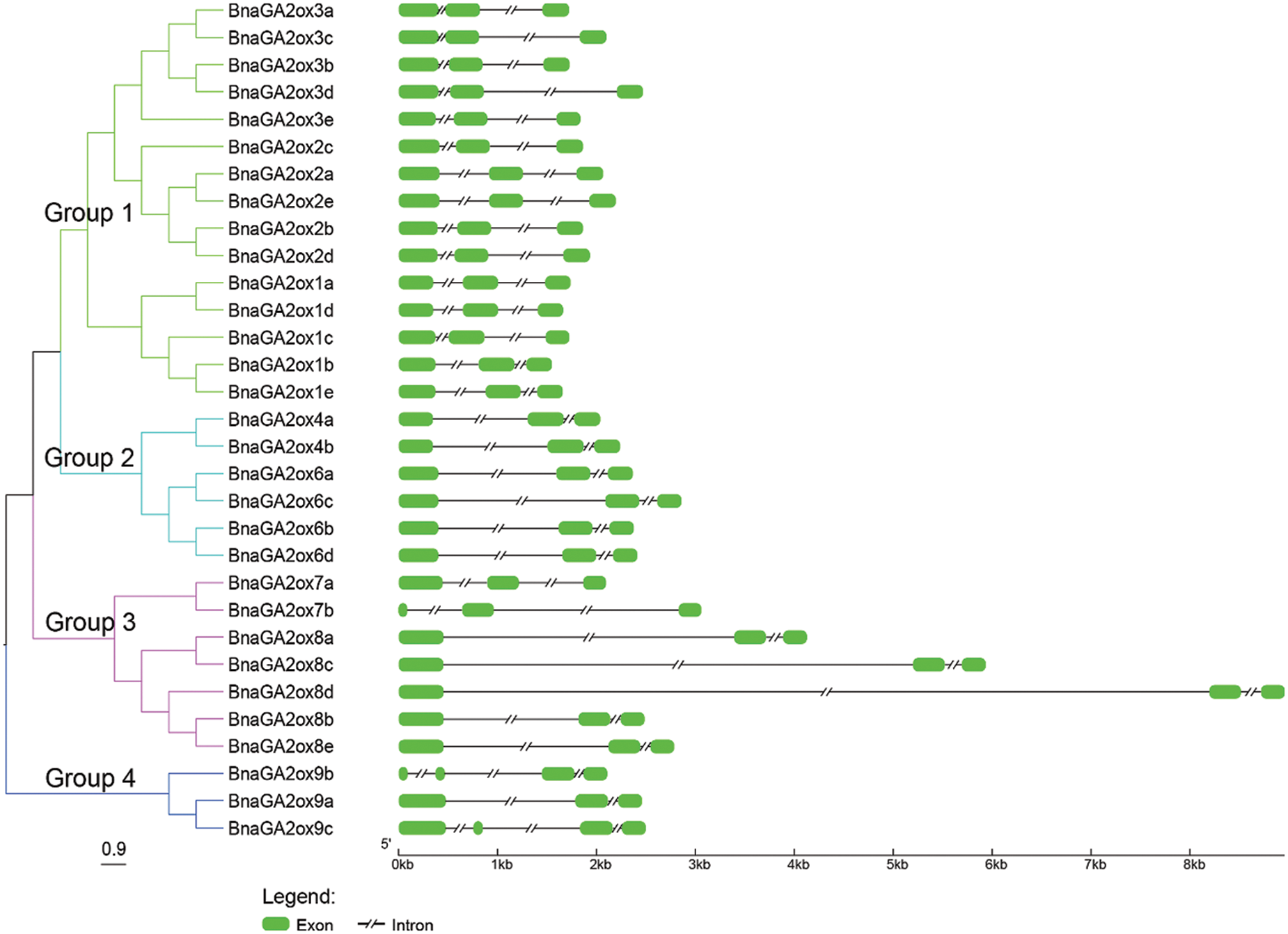
Figure 2: Phylogenic relationship and gene structure of BnaGA2ox genes
Note: The NJ tree was generated with bootstrap analysis (1,000 replicates) using aligned BnaGA2ox protein sequences, and the tree was displayed with FigTree v1.4.0.
To study protein structural diversity of BnaGA2ox members, the conserved motifs were analyzed by online MEME/MAST tools. We predicted ten putative protein motifs in BnaGA2ox proteins. Motif 1 and Motif 3 were detected in all 31 BnaGA2ox proteins (Fig. 3), and these motifs belonged to the 2OG-FeII_Oxy structural domain. Motif 2 was also present in all BnaGA2ox proteins, with the exception of BnaGA2ox9a protein, this indicates that the GA2ox family proteins are highly conserved in B. napus. BnaGA2ox proteins from different groups also contained some unique motifs. For example, Motif 10 was only detected in some BnaGA2ox proteins belonging to Group 2 and Group 3, and Motif 5 and Motif 6 were only present in the BnaGA2ox proteins belonging to Group 1 and Group 2. Motif 7 and Motif 8 were only detected in the BnaGA2ox proteins from Group 3 and Group 4. BnaGA2ox members from the same clade or subgroup showed a similar structural distribution, which was opposite to the gene structure distribution.

Figure 3: Phylogenic relationship and motif prediction of BnaGA2ox proteins
Note: The NJ tree was generated with bootstrap analysis (1,000 replicates) using aligned BnaGA2ox protein sequences and the tree is displayed with FigTree v1.4.0.
3.4 Chromosomal Location and Syntenic Analysis of BnGA2ox Genes
To study chromosomal location of BnaGA2ox genes, we searched for their information in the published BnPIR database and visualized them on chromosomes using TBtools. A total of 30 BnaGA2ox genes were found on 14 chromosomes of B. napus, and one gene (BnaGA2ox9c) was on undefined Bnascaffold0025. Among them, 15 BnaGA2ox genes were located on the A and C subgenomes. No BnaGA2ox were found on chromosomes A04, A06, C08 and C09, and only one BnaGA2ox gene was found on each chromosome of A01, A09, C01 and C06 (Fig. 4). Two BnaGA2ox genes were identified on each of the A02, A03, A08, C02 and C07 chromosomes, and three BnaGA2ox genes were found on each of the A05, A07, C03, C04 and C05 chromosomes.
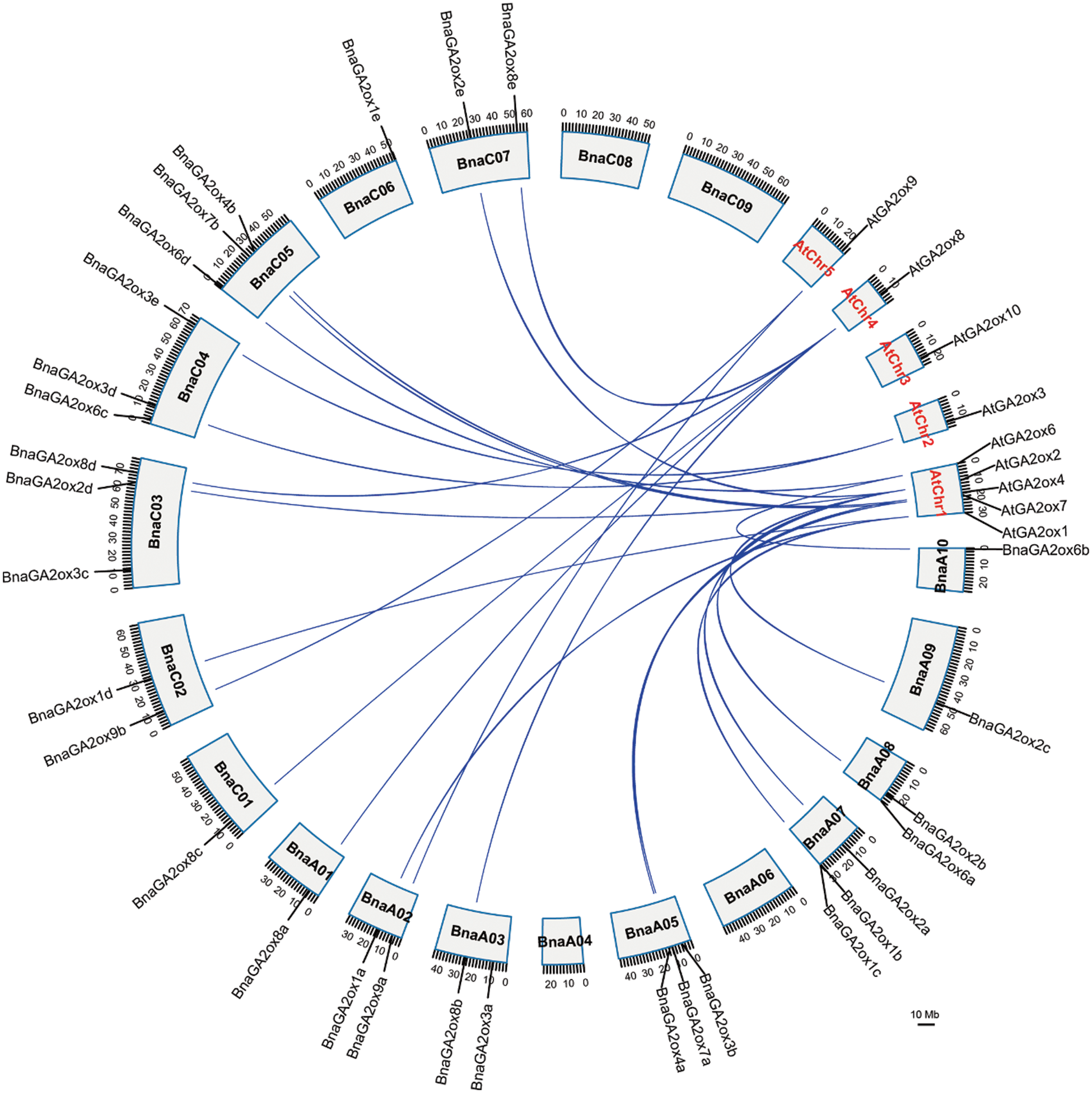
Figure 4: Chromosome location and syntenic relationship of GA2ox genes of A. thaliana and B. napus
Through syntenic analysis of GA2ox genes between B. napus and A. thaliana, eight AtGA2ox genes showed a syntenic relationship with 25 BnaGA2ox genes (Fig. 4). Most of the syntenic AtGA2ox genes were located on chromosome A05, while the distribution of the syntenic BnGA2ox genes was relatively scattered. Among them, AtGA2ox2 and AtGA2ox8 corresponded to five syntenic BnGA2ox genes, and AtGA2ox1 had four syntenic BnGA2ox genes, while the remaining AtGA2ox2 showed two syntenic BnGA2ox genes. We observed a tandem duplicated event between BnaGA2ox1b and BnaGA2ox1c in the B. napus genome.
3.5 Selection Pressure Analysis
To study the selection pressure on BnaGA2ox genes, the Ka/Ks ratios were calculated between BnaGA2ox genes and its orthologous AtGA2ox genes, with Ka/Ks < 1 indicating purifying selection, Ka/Ks = 1 indicating neutral selection and Ka/Ks > 1 indicating positive selection [37]. The Ka/Ks ratios of BnaGA2ox genes ranged from 0.1066 (BnaGA2ox6b) to 0.3191 (BnaGA2ox9c) (Table 2). Comparing the Ka/Ks ratios between the A and C subgenomes, we found that the average Ka/Ks ratio of BnaGA2ox genes of the A subgenome was less than that of the C subgenome. This suggested that BnaGA2ox genes from the C subgenome underwent greater selection pressure during the evolution of B. napus. Moreover, we found that the average Ka/Ks ratio of BnaGA2ox genes of Group 4 was the highest, while the average Ka/Ks ratio of BnaGA2ox genes of Group 2 was the lowest among the four groups (Table 2). Among the BnaGA2ox genes belonging to Group 1, the Ka/Ks ratio of BnaGA2ox1c was lower than its homologous copies, and that of BnaGA2ox2a and BnaGA2ox3a were higher than their homologous copies. However, the Ka/Ks ratios of BnaGA2ox showed little difference among homologous copies in the other three groups, indicating that different evolutionary pressures may have been imposed to the BnaGA2ox homologous copies in Group 1 after the genome-wide triploidization of B. napus.
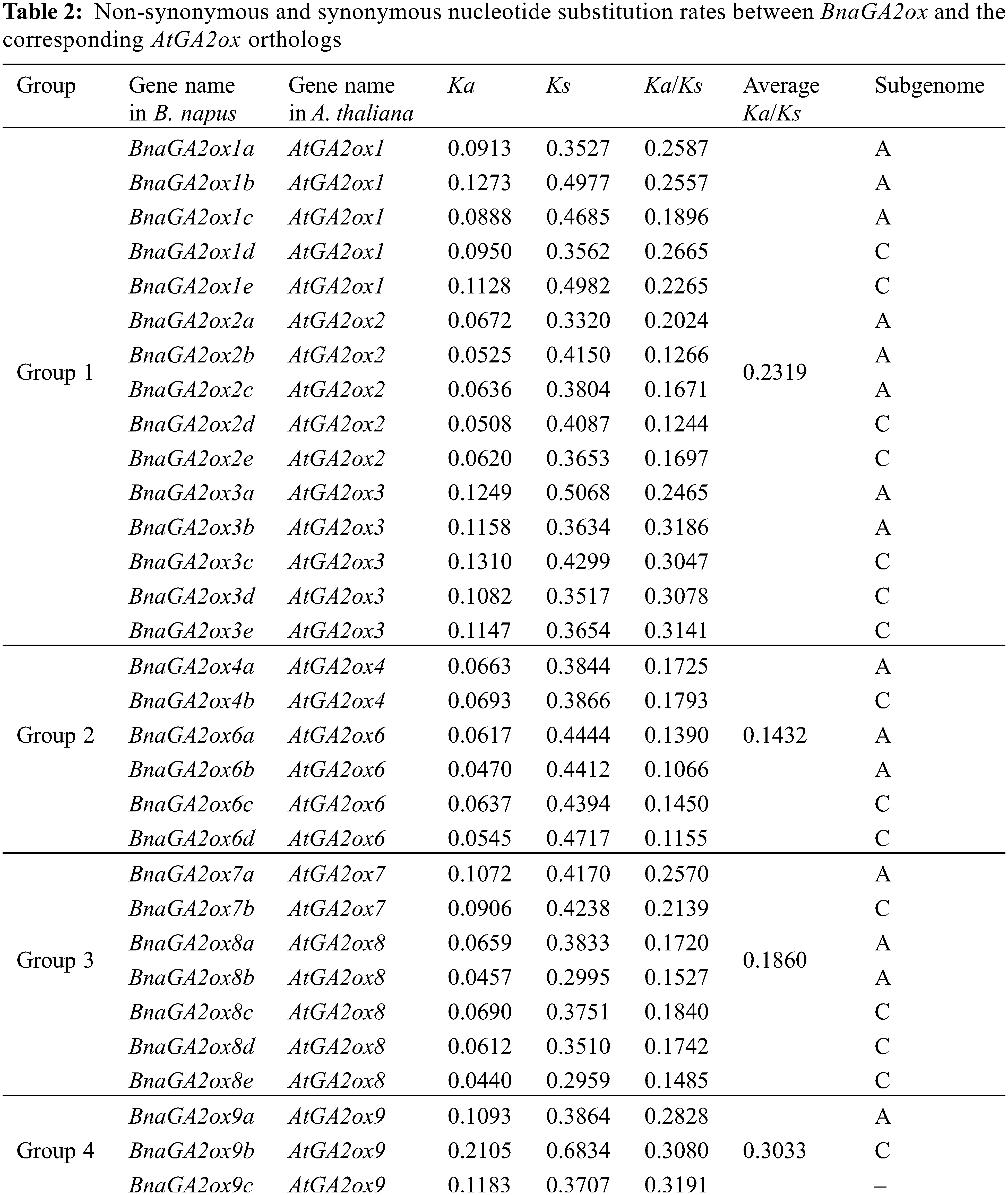
3.6 Putative Cis-Acting Elements in the Promoter Regions of BnaGA2ox Genes
To study and explore the potential transcriptional regulation of BnaGA2ox genes, the cis-acting elements were identified using the PlantCARE database. All BnaGA2ox genes contained the basic conserved functional motifs in their promoter regions, such as TATA-box and CAAT-box, as well as numerous important elements. To study the potential function of BnaGA2ox genes, we screened the cis-acting elements involved in light-responsive, biological or abiotic elements and phytohormones (Fig. 5). The light response element was the most abundant. Most BnaGA2ox genes contained it in their promoter region, but it was mainly concentrated in the BnaGA2ox promoter region in Group 1. Two phytohormone cis-acting elements, the auxin (AUXRR-core/TGA-element) elements, were discovered in the promoter regions of BnaGA2ox1a, BnaGA2ox1c, BnaGA2ox1d, BnaGA2ox8e, BnaGA2ox9a, BnaGA2ox9b and BnaGA2ox9c. Gibberellin response elements, such as P-box, GARE-motif and TATC-box were found in the promoter regions of BnaGA2ox4a, BnaGA2ox4b, BnaGA2ox6d, BnaGA2ox7a, BnaGA2ox8b, BnaGA2ox8e, BnaGA2ox9a, BnaGA2ox9b and BnaGA2ox9c. Moreover, defense and abiotic stress, anaerobic induction and low temperature were detected in the promoter of BnaGA2ox genes in Groups 1 and 4. Drought-inducibility and MeJA and abscisic acid responsive elements were also examined in the promoter region of most BnaGA2ox genes. We also detected biological clock-related elements (circadian) in the promoter regions of BnaGA2ox2b, BnaGA2ox2c and BnaGA2ox3c.
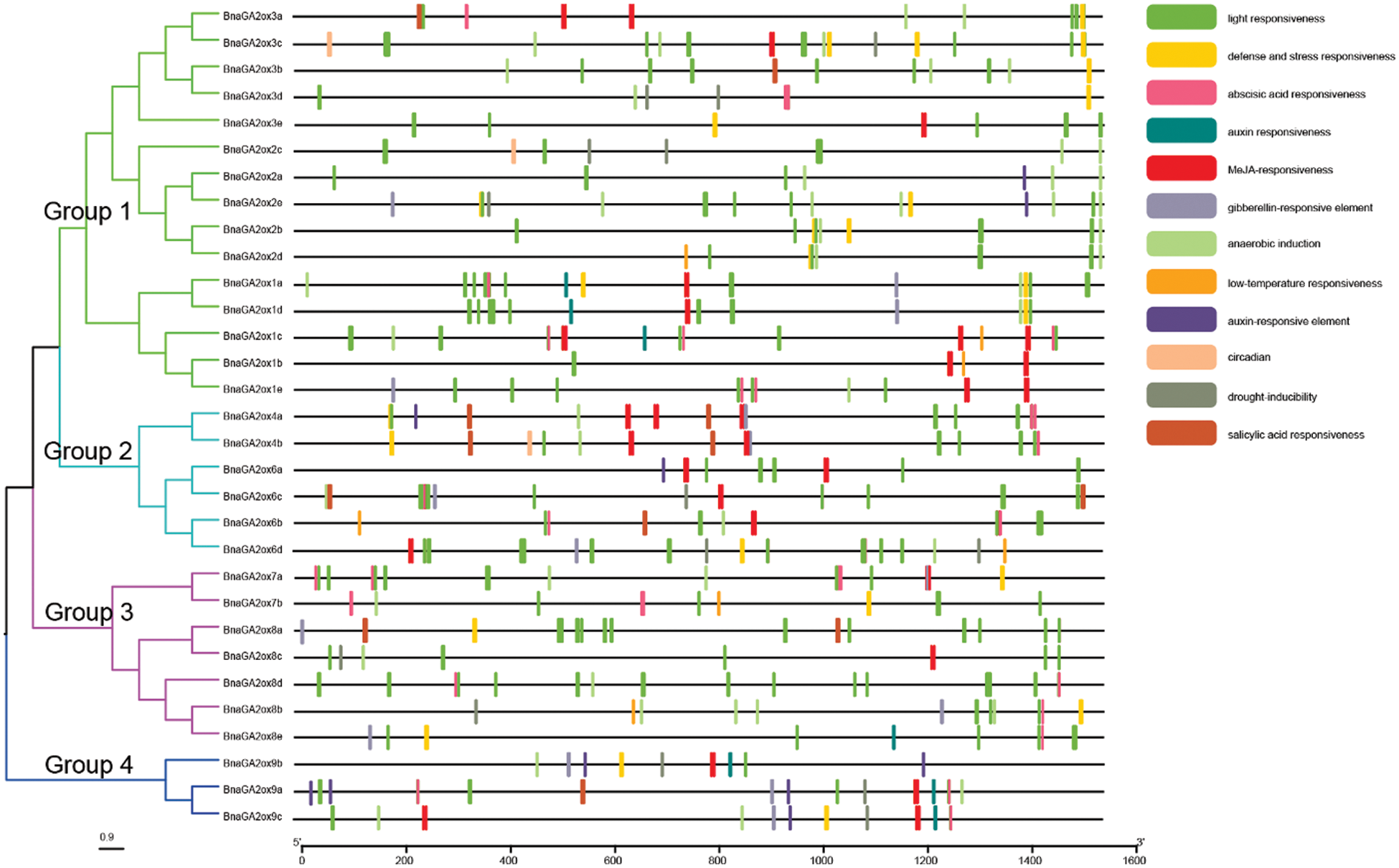
Figure 5: Genomic distribution of cis-acting elements in promoter regions of BnaGA2ox genes
Note: Different cis-acting elements are indicated by different colors. The cis-acting elements in promoter regions were predicted using PlantCARE.
3.7 Gene Expression Pattern Analysis of BnaGA2ox Family Genes
To analyze and compare the gene expression levels of BnaGA2ox family genes, we downloaded their expression patterns in 90 tissues in different developmental stages from BnTIR. In B. napus, BnaGA2ox genes showed low or no expression in the majority of tissues but some displayed tissue-specific expression (Fig. 6). Among them, BnaGA2ox2b, BnaGA2ox2d, BnaGA2ox8a and BnaGA2ox8e were highly expressed in roots, while the three copies of BnaGA2ox8, BnaGA2ox8b, BnaGA2ox8c and BnaGA2ox8d were highly expressed in stalks. BnaGA2ox6d was highly expressed in most detected leaves and BnaGA2ox2c, BnaGA2ox6b and BnaGA2ox6d were highly expressed in flowers. During silique development, BnaGA2ox6d highly expressed in the silique wall at all detected stages. BnaGA2ox9a and BnaGA2ox9c were consistently highly expressed in seeds at all detected stages. To verify the RNA-seq results, we evaluated the expression of four BnaGA2ox by qRT-PCR analysis, and the qRT-PCR results were in accordance with their RNA-seq results (Supplementary Fig. S2).
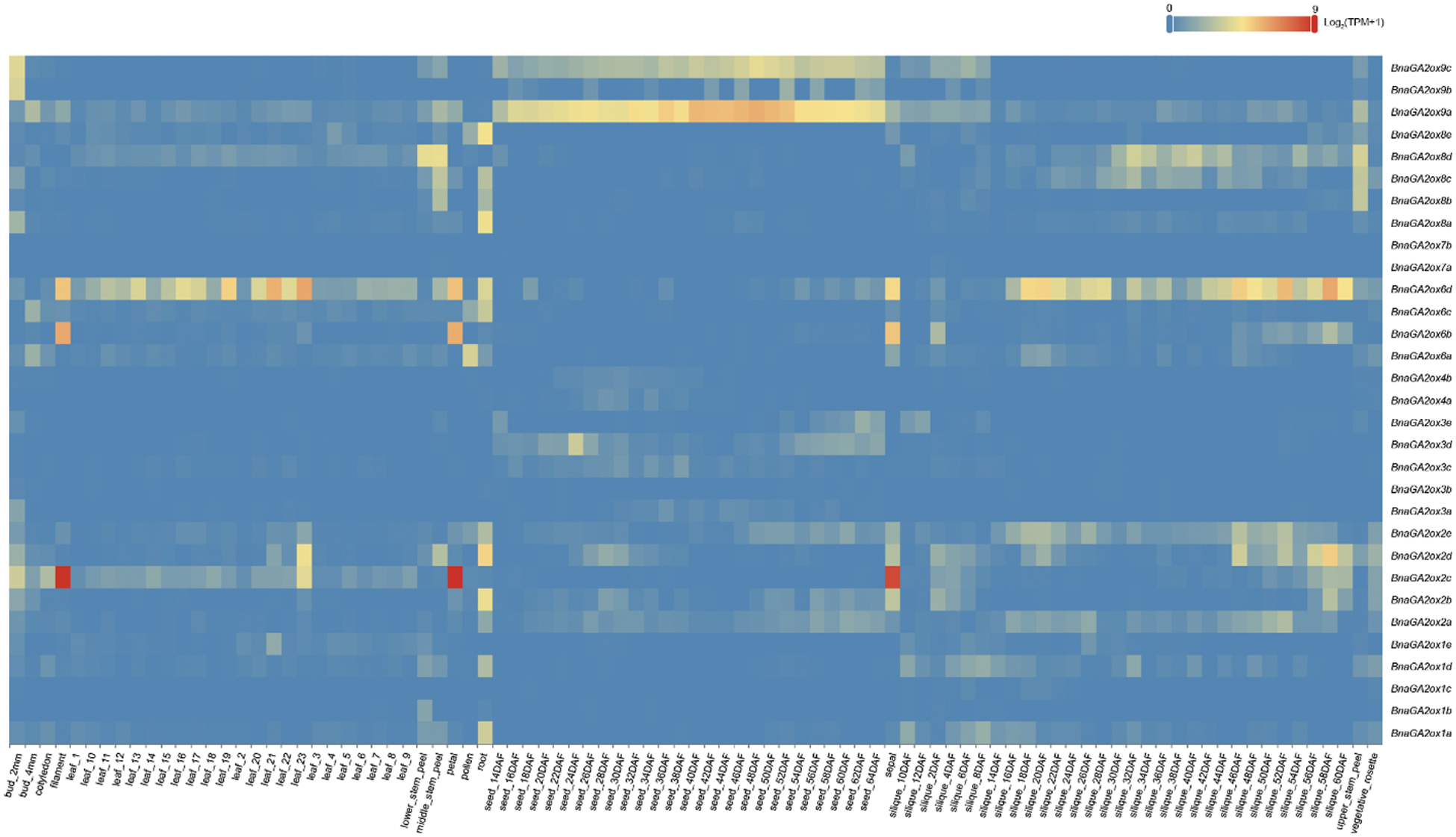
Figure 6: Expression patterns of BnaGA2ox family genes
Note: The color bar inthe figure represents the log2 expression values, and the expression values were normalized using TPM+1.
3.8 Gene Expression Patterns of BnaGA2ox Family Genes under Phytohormone and Abiotic Treatment
To investigate the expression patterns of BnaGA2ox genes under phytohormone and abiotic treatments, their expressions at SLs and SRs were examined at various time points (Fig. 7). In SLs under phytohormone treatments, most of BnaGA2ox genes were not obviously induced by phytohormones, while BnaGA2ox6d were upregulated under aminocyclopropane carboxylic acid (ACC) treatment, and three BnaGA2ox2 members, BnaGA2ox2b, BnaGA2ox2c and BnaGA2ox2d, were evidently induced by jasmonic acid (JA) (Fig. 7A). In SRs, most of the BnaGA2ox genes were unregulated or downregulated under phytohormone treatments (Fig. 7B). For example, BnaGA2ox6d were obviously unregulated under indoleacetic acid (IAA) and ACC treatments, while those were distinctly downregulated by gibberellin (GA), trans-Zeatin (TZ) and abscisic acid (ABA) treatments. Meanwhile, BnaGA2ox2d were also evidently induced by JA in root, and suppressed by GA and ACC, and BnaGA2ox8e were obviously induced by ACC and JA.
Figure 7: Expression patterns of BnaGA2ox family genes under phytohormone and abiotic treatments. (A) Expression patterns of BnaGA2ox family genes under phytohormone treatment in SLs. (B) Expression patterns of BnaGA2ox family genes under phytohormone treatment in SRs. (C) Expression patterns of BnaGA2ox family genes under abiotic treatment in SLs. (D) Expression patterns of BnaGA2ox family genes under abiotic treatment in SRs
Note: The color bar in the figure represents the log2 expression values, and the expression values were normalized using TPM+1.
Under abiotic treatments, BnaGA2ox6d were found to be obviously upregulated by freezing, cold and osmotic treatments in SLs, but that was obviously upregulated by salt, drought, freezing and osmotic treatment in SRs (Figs. 7C, 7D). In SLs, we also discovered that BnaGA2ox1d, BnaGA2ox2a, BnaGA2ox2e, BnaGA2ox2b and BnaGA2ox6c were upregulated under cold treatment, and BnaGA2ox1e and BnaGA2ox2d were upregulated under drought treatment. In SRs, BnaGA2ox2b, BnaGA2ox2d, BnaGA2ox2e and BnaGA2ox8e were also found to be distinctly upregulated under drought treatment, and BnaGA2ox8a was upregulated by freezing induction.
GAs play pivotal roles in plant development and growth. Decreasing and increasing levels of active GAs can both result in dwarf or semi-dwarf phenotypes. As key enzymes regulating GAs activity in plants, GA2ox family genes have been identified in multiple species including A. thaliana [11,18], O. sativa [16,17] and Z. mays [19]. We identified 31 BnaGA2ox genes in B. napus, as well as 18 BraGA2ox and 17 BolGA2ox genes, respectively, in its two parental species, B. oleracea and B. rapa. Consistent with previous studies [20], GA2ox genes had fewer exons in B. napus and showed similar protein motif distributions among homologous copies in each subfamily. This was consistent with previous studies on GA oxidase [38]. Most cis-acting elements were also detected in the promoter regions of multiple BnaGA2ox genes, including some specific cis-acting elements, such as defense and abiotic stress, low temperature, and drought-inducibility. This suggests that BnaGA2ox genes not only play important functions in plant development and growth, but also have potential functions in response to biotic or abiotic stresses, as well as some specific signals.
Compared to the genomes of O. sativa and Z. mays, Brassica species experienced an extra genome-wide triploidization event in their genomes during their evolution [27,28]. However, there was no obvious difference among B. rapa, B. oleracea, O. sativa and Z. mays when comparing the gene number of GA2ox. Homology analysis found that three genes, GA2ox1, GA2ox6 and GA2ox8, from O. sativa and Z. mays had multiple homologous copies through comparison with their homologous genes AtGA2ox (Supplementary Table S1). Previous studies revealed that polyploidization was the main force promoting the appearance of OsGA2ox copies during evolution [39]. Although numerous copy genes have been lost by diploidization after experiencing the tetraploid stage in the evolution of maize, approximately 30% of the genes retain multiple copies in the maize genome [40,41]. Certain GA2ox genes have two copies in the maize genome [19]. GA2ox genes can be divided into three groups in monocotyledon and dicotyledon plants [19,42], including two C19-GA2ox Classes and one C20-GA2ox Class. In this study, we added one new C20-GA2ox Class, which was recently identified in A. thaliana and includes AtGA2ox9 (At5g58660) and AtGA2ox10 (At3g47190) [43], as well as their homologs identified in B. rapa, B. oleracea, B. napus, O. sativa and Z. mays, S. lycopersicum. Phylogeny evidence indicates that GA2ox genes can be classified into four taxa, two C20-GA2ox Classes and two C19-GA2ox Classes. Among them, GA2ox genes are in an expansion state in Group 1 of C20-GA2ox Class and Group 3 of C19-GA2ox Class.
The expression patterns of GA2ox genes have been detected in several plants. In A. thaliana, the expressions of seven AtGA2ox genes suggest strong tissue-specific expression of GA2ox genes [44]. The tissue-specific expressions of AtGA2ox2 and AtGA2ox4 verified their importance in maintaining the levels of bioactive GAs in shoot apical meristem [45,46]. Consistent with their results, similar findings were found in O. sativa [47]. In S. lycopersicum, SlGA2ox2 was expressed in flowers, roots, stems, leaves and immature fruits [48,49]. In Z. mays, the GA2ox genes are highly expressed in the shoot apical meristem and primary roots [19]. In this study, we found that some BnaGA2ox genes also had strong tissue-specific expression. For example, BnaGA2ox2c, BnaGA2ox6b and BnaGA2ox6d had high expression levels in sepals, petals and filaments of floral organs, and BnaGA2ox9a and BnaGA2ox9c were highly expressed in seeds at all detected stages, especially at 42–52 days after flowering. These data suggest that the balance of bioactive GAs and their intermediates, which are maintained through GA2ox enzymes, may play a crucial role in the precise regulation of plant growth. However, most of BnaGA2ox genes had no or low expression levels in almost all studied tissues. Consistent with the cis-elements analysis, we found that several BnaGA2ox genes were obviously induced under phytohormone or abiotic treatments, such as BnaGA2ox6b which was markedly induced by ACC, IAA, GA, freezing, cold, salt and drought. However, there was a wide difference in induction of phytohormone or abiotic treatments at SRs and SLs, and most of BnaGA2ox genes are more easily induced or suppressed at SRs under phytohormone or abiotic treatments. Moreover, most of BnaGA2ox genes were not induced by GA, and some of them were suppressed under GA treatment in SRs, suggesting that plants need to reduce the GA synthesis to maintain a dynamic balance of bioactive GAs in shoot apical meristems when spraying exogenous GAs [46]. The response to other phytohormones on BnaGA2ox genes indicates mutual regulation between GAs and other phytohormones. Surprisingly, we found that the light response of relative cis-elements occurs in most GA2ox promoter regions, but the expression results found that only BnaGA2ox6b was highly expressed in the petiolate leaves and silique pericarps of B. napus. Although we can not currently verify whether the expression of BnaGA2ox is induced by lighting, while previous studies have suggested that silique pericarps have replaced leaves as the main photosynthetic organs during the middle and late development of siliques in B. napus [50,51], indicating that BnaGA2ox6b may play a key role in B. napus photosynthesis.
Acknowledgement: We thank Yonghai Fan from Southwest University (China) for technical assistances and critical reading of the manuscript.
Authorship: The authors confirm contribution to the paper as follows: YZ and TH conceived and designed the experiments; YL, HH, YS, SH, PZ and XD performed the sampling and experiments; YL, TL, CX, XT and YZ contributed to data analysis and interpretation; YL and HH wrote the paper. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Materials: Supplementary materials can be found at the end on Figs. S1 and S2 and Tables S1 and S2.
Funding Statement: This work was supported by the Chongqing Academy of Agricultural Sciences Youth Innovation Team Project (NKY-2018QC01), Chongqing Finance Special Project (NKY-2022AC002), the Natural Science Foundation Project of Yongchuan (2021yc-jckx20013), the Technology Innovation and Application Development (Surface) Project of Yongchuan (2021yc-cxfz30007), and the National Oilseed Rape Industrial Technology System Sanxia Comprehensive Experiment Station Project (CARS-13).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Hedden, P., Thomas, S. G. (2002). Gibberellin biosynthesis and its regulation. Biochemical Journal, 444(1), 11–25. [Google Scholar]
2. Giacomelli, L., Rota-Stabelli, O., Masuero, D., Acheampong, A. K., Moretto, M. et al. (2013). Gibberellin metabolism in Vitis vinifera L. during bloom and fruit-set: Functional characterization and evolution of grapevine gibberellin oxidases. Journal of Experimental Botany, 64(14), 4403–4419. [Google Scholar]
3. Yamaguchi, S., Smith, M. W., Brown, R. G. S., Kamiya, Y. J., Sun, T. P. (1998). Phytochrome regulation and differential expression of gibberellin 3β-hydroxylase genes in germinating Arabidopsis seeds. The Plant Cell, 10(12), 2115–2126. [Google Scholar]
4. Achard, P., Gong, F., Cheminant, S., Alioua, M., Hedden, P. et al. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. The Plant Cell, 20(8), 2117–2129. [Google Scholar]
5. Peters, R. J. (2012). Gibberellin phytohormone metabolism. In: Isoprenoid synthesis in plants and microorganisms, pp. 233–249. New York, USA: Springer. [Google Scholar]
6. Peng, J. R., Carol, P., Richards, D. E., King, K. E., Cowling, R. J. et al. (1997). The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes & Development, 11(23), 3194–3205. [Google Scholar]
7. Ross, J. J., Reid, J. B., Swain, S. M., Hasan, O., Poole, A. T. et al. (1995). Genetic regulation of gibberellin deactivation in Pisum. The Plant Journal, 7(3), 513–523. [Google Scholar]
8. El-Sharkawy, I., Kayal, W., Prasath, D., Fernandez, H., Bouzayen, M. et al. (2012). Identification and genetic characterization of a gibberellin 2-oxidase gene that controls tree stature and reproductive growth in plum. Journal of Experimental Botany, 63(3), 1225–1239. [Google Scholar]
9. Lee, D. H., Lee, I. C., Kim, K. J., Kim, D. S., Na, H. J. et al. (2014). Expression of gibberellin 2-oxidase 4 from Arabidopsis under the control of a senescence-associated promoter results in a dominant semi-dwarf plant with normal flowering. Journal of Plant Biology, 57(2), 106–116. [Google Scholar]
10. Fukazawa, J., Mori, M., Watanabe, S., Miyamoto, C., Ito, T. et al. (2017). DELLA-GAF1 complex is a main component in gibberellin feedback regulation of GA20 oxidase 2. Plant Physiology, 175(3), 1395–1406. [Google Scholar]
11. Hedden, P., Phillips, A. L. (2000). Manipulation of hornmone biosynthetic genes in transgenic plants. Current Opinion Biotechnology, 11(2), 130–137. [Google Scholar]
12. Zhou, P., Ren, B., Zhang, X. M., Wang, Y., Wei, C. H. et al. (2010). Stable ex-pression of rice dwarf virus Pns10 suppresses the post-transcriptional gene silencing in transgenic Nicotiana benthamiana plants. Acta Virologica, 54(2), 99–104. [Google Scholar]
13. Shi, J. B., Wang, J., Wang, N., Zhou, H., Xu, Q. H. et al. (2018). Overexpression of StGA2ox1 gene increases the tolerance to abiotic stress in transgenic potato (Solanum tuberosum L.) plants. Applied Biochemistry Biotechnology, 187(4), 1204–1219. [Google Scholar]
14. Wuddineh, W. A., Mazarei, M., Zhang, J. Y., Poovaiah, C. R., Mann, D. G. J. et al. (2015). Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnology Journal, 13(5), 636–647. [Google Scholar]
15. Dijkstra, C., Adams, E., Bhattacharya, A., Page, A. F., Anthony, P. et al. (2008). Over-expression of a gibberellin 2-oxidase gene from Phaseolus coccineus L. enhances gibberellin inactivation and induces dwarfism in Solanum species. Plant Cell Reports, 27(3), 463–470. [Google Scholar]
16. Sakai, M., Sakamoto, T., Saito, T., Matsuoka, M., Tanaka, H. et al. (2003). Expression of novel rice gibberellin 2-oxidase gene is under homeostatic regulation by biologically active gibberellins. Journal of Plant, 116(2), 161–164. [Google Scholar]
17. Lo, S. F., Yang, S. Y., Chen, K. T., Hsing, Y. I., Zeevaart, J. A. et al. (2008). A novel class of gibberellin 2-oxidases control semi dwarfism, tillering, and root development in rice. The Plant Cell, 20(10), 2603–2618. [Google Scholar]
18. Schomburg, F. M., Bizzell, C. M., Lee, D. J., Zeevaart, J. A., Amasino, R. M. (2003). Overexpression of a novel class of gibberellin 2-oxidases decreases gibberellin levels and creates dwarf plants. The Plant Cell, 15(1), 151–163. [Google Scholar]
19. Li, Y. D., Shan, X. H., Jiang, Z. L., Zhao, L., Jin, F. X. (2021). Genome-wide identification and expression analysis of the GA2ox gene family in maize (Zea mays L.) under various abiotic stress conditions. Plant Physiology and Biochemistry, 166, 621–633. [Google Scholar]
20. He, H. H., Liang, G. P., Lu, S. X., Wang, P. P., Liu, T. et al. (2019). Genome-wide identification and expression analysis of GA2ox, GA3ox, and GA20ox are related to gibberellin oxidase genes in grape (Vitis vinifera L.). Genes, 10(9), 680. [Google Scholar]
21. Xiao, Z., Fu, R. P., Li, J. Y., Fan, Z. Q., Yin, H. F. (2016). Overexpression of the Gibberellin 2-Oxidase gene from Camellia lipoensis induces dwarfism and smaller flowers in Nicotiana tabacum. Plant Molecular Biology Reporter, 34(1), 182–191. [Google Scholar]
22. Shan, C., Mei, Z., Duan, J., Chen, H., Feng, H. et al. (2014). OsGA2ox5, a gibberellin metabolism enzyme, is involved in plant growth, the root gravity response and salt stress. PLoS One, 9(1), e87110. [Google Scholar]
23. Zhao, X. Y., Yu, X. H., Foo, E., Gregory, M., Symons, S. M. et al. (2007). A study of gibberellin homeostasis and cryptochrome-mediated blue light inhibition of hypocotyl elongation. Plant Physiology, 145(1), 106–118. [Google Scholar]
24. Staswick, P. E., Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. The Plant Cell, 16(8), 2117–2127. [Google Scholar]
25. Staswick, P. E., Serban, B., Rowe, M., Tiryaki, I., Maldonado, M. T. et al. (2005). Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. The Plant Cell, 17(2), 616–627. [Google Scholar]
26. Nagaharu, U., Nagaharu, N. (1935). Genome analysis in Brassica with special reference to the experimental formation of B. napus and peculiar mode of fertilization. Journal of Japanese Botany, 7, 389–452. [Google Scholar]
27. Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H. et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science, 345(6199), 950–953. [Google Scholar]
28. Yan, J. D., Liao, X. Y., He, R. Q., Zhong, M., Feng, P. P. et al. (2017). Ectopic expression of GA 2-oxidase 6 from rapeseed (Brassica napus L.) causes dwarfism, late flowering and enhanced chlorophyll accumulation in Arabidopsis thaliana. Plant Physiology and Biochemistry, 111, 10–19. [Google Scholar]
29. Altschul, S. F., Gish, W., Miller, W., Myers, E. W., Lipman, D. J. (1990). Basic local alignment search tool. Molecular Biology, 215(3), 403–410. [Google Scholar]
30. Fan, Y. H., Yu, M. N., Liu, M., Zhang, R., Sun, W. et al. (2017). Genome-wide identification, evolutionary and expression analyses of the galactinol synthase gene family in rapeseed and tobacco. International Journal of Molecular Sciences, 18(12), 2768. [Google Scholar]
31. Edgar, R. C. (2004). MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792–1797. [Google Scholar]
32. Sudhir, K., Glen, S., Koichiro, T. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Molecular Biology and Evolution, 33(7), 1870–1874. [Google Scholar]
33. Wang, D., Zhang, Y., Zhang, Z., Zhu, J., Yu, J. (2010). KaKs_Calculator 2.0: A toolkit incorporating gamma-series methods and sliding window strategies. Genomics, Proteomics, Bioinformatics, 8(1), 77–80. [Google Scholar]
34. Chen, C. J., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H. et al. (2020). TBtools: An integrativetoolkit developed for interactive analyses of big biological data. Molecular Plant, 13(8), 1194–1202. [Google Scholar]
35. Liu, D., Yu, L., Wei, L., Yu, P., Wang, J. et al. (2021). BnTIR: An online transcriptome platform for exploring RNA-seq libraries for oil crop Brassica napus. Plant Biotechnology Journal, 19(10), 1895–1897. [Google Scholar]
36. Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J. et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55, 611–622. [Google Scholar]
37. Nekrutenko, A., Makova, K. D., Li, W. H. (2002). The Ka/Ks ratio test for assessing the protein-coding potential of genomic regions: An empirical and simulation study. Genome Research, 12(1), 198–202. [Google Scholar]
38. Huang, Y., Wang, X., Ge, S., Rao, G. Y. (2015). Divergence and adaptive evolution of the gibberellin oxidase genes in plants. BMC Evolution Biology, 15(1), 1–15. DOI 10.1186/s12862-015-0490-2. [Google Scholar] [CrossRef]
39. Han, F., Zhu, B. (2011). Evolutionary analysis of three gibberellin oxidase genes in rice, Arabidopsis, and soybean. Gene, 473(1), 23–35. DOI 10.1016/j.gene.2010.10.010. [Google Scholar] [CrossRef]
40. Schnable, P. S., Ware, D., Fulton, R. S., Stein, J. C., Wei, F. et al. (2009). The B73 maize genome: Complexity, diversity, and dynamics. Science, 326(5956), 1112–1115. DOI 10.1126/science.1178534. [Google Scholar] [CrossRef]
41. Schnable, J. C., Springer, N. M., Freeling, M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. PNAS, 108(10), 4069–4074. DOI 10.1073/pnas.1101368108. [Google Scholar] [CrossRef]
42. Cheng, J., Ma, J., Zheng, X., Lv, H., Zhang, M. et al. (2021). Functional analysis of the Gibberellin 2-oxidase gene family in Peach. Frontiers in Plant Science, 12, 619158. DOI 10.3389/fpls.2021.619158. [Google Scholar] [CrossRef]
43. Lange, T., Kramer, C., Pimenta Lange, M. J. (2020). The class III gibberellin 2-oxidases AtGA2ox9 and AtGA2ox10 contribute to cold stress tolerance and fertility. Plant Physiology, 184(1), 478–486. DOI 10.1104/pp.20.00594. [Google Scholar] [CrossRef]
44. Li, C., Zheng, L., Wang, X., Hu, Z., Zheng, Y. et al. (2019). Comprehensive expression analysis of Arabidopsis GA2-oxidase genes and their functional insights. Plant Science, 285, 1–13. DOI 10.1016/j.plantsci.2019.04.023. [Google Scholar] [CrossRef]
45. Hay, A., Kaur, H., Phillips, A., Hedden, P., Hake, S. et al. (2002). The gibberellin pathway mediates KNOTTED1-type homeobox function in plants with different body plans. Current Biology, 12(18), 1557–1565. DOI 10.1016/S0960-9822(02)01125-9. [Google Scholar] [CrossRef]
46. Tanaka-Ueguchi, M., Itoh, H., Oyama, N., Koshioka, M., Matsuoka, M. (1998). Over expression of a tobacco homeobox gene, NTH15, decreases the expression of a gibberellin biosynthetic gene encoding GA 20-oxidase. The Plant Journal, 15(3), 391–400. DOI 10.1046/j.1365-313X.1998.00217.x. [Google Scholar] [CrossRef]
47. Sakamoto, T., Kobayashi, M., Itoh, H., Tagiri, A., Kayano, T. et al. (2001). Expression of a gibberellin 2-oxidase gene around the shoot apex is related to phase transition in rice. Plant Physiology, 125(3), 1508–1516. DOI 10.1104/pp.125.3.1508. [Google Scholar] [CrossRef]
48. Xiao, J. H., Zhang, J. H., Zhang, Y. Y., Wang, T. T., Chen, R. G. et al. (2007). Isolation and expression of GA 2-oxidase2 in tomato. DNA Sequence, 18(6), 474–479. [Google Scholar]
49. Ding, Q., Wang, F., Xue, J., Yang, X., Fan, J. et al. (2020). Identification and expression analysis of hormone biosynthetic and metabolism genes in the 2OGD family for identifying genes that may be involved in tomato fruit ripening. International Journal of Molecular Science, 21(15), 5344. [Google Scholar]
50. Wang, C., Hai, J., Tian, J., Yang, J., Zhao, X. (2014). Influence of silique and leaf photosynthesis on yield and quality of seed of oilseed rape (Brassica napus L.) after flowering. Xibei Zhiwu Xuebao, 34(8), 1620–1626 (in Chinese). [Google Scholar]
51. Wang, C., Hai, J., Yang, J., Tian, J., Chen, W. J. et al. (2016). Influence of leaf and silique photosynthesis on seeds yield and seeds oil quality of oilseed rape (Brassica napus L.). European Journal of Agronomy, 74, 112–118. [Google Scholar]
Appendix
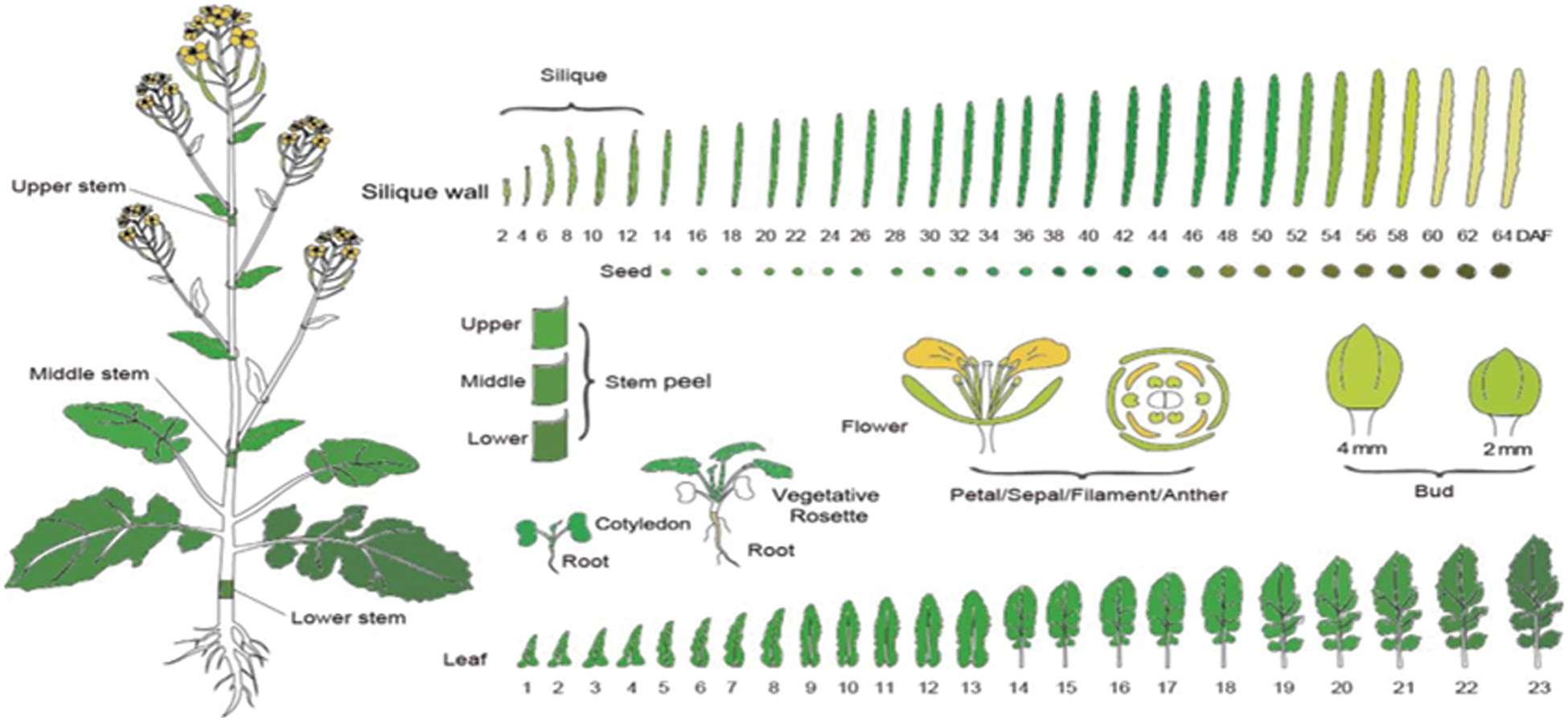
Supplementary Figure S1: The diagram of 90 tissues in B. napus. The Figure downloaded from BnTIR database
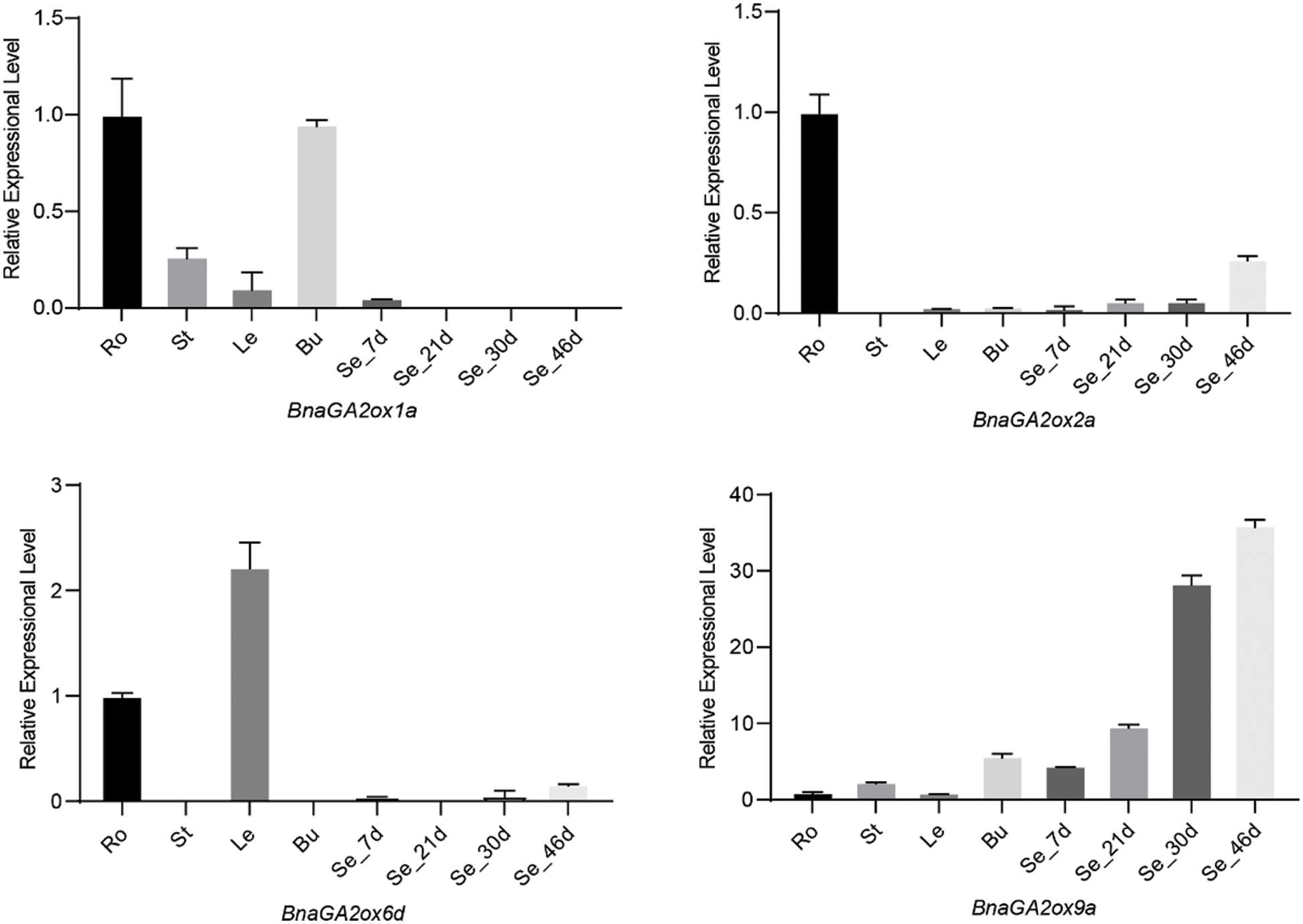
Supplementary Figure S2: The qRT-PCR results for four selected genes
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools