 Open Access
Open Access
ARTICLE
Identification of a Novel OsCYP2 Allele that Was Involved in Rice Response to Low Temperature Stress
1
College of Agriculture, Northeast Agricultural University, Harbin, 150030, China
2
Hulin Agricultural Technology Extension Center, Hulin, 158400, China
3
Liaoning Academy of Agricultural Sciences, Shenyang, 110161, China
4
Zhejiang University, Hangzhou, 310000, China
* Corresponding Author: Zhongchen Zhang. Email:
# The author died prior to the submission of this paper. Other authors express their great gratitude and remembrance to him
(This article belongs to the Special Issue: Identification of Genetic/Epigenetic Components Responding to Biotic and Abiotic Stresses in Crops)
Phyton-International Journal of Experimental Botany 2023, 92(6), 1743-1763. https://doi.org/10.32604/phyton.2023.026516
Received 09 September 2022; Accepted 12 December 2022; Issue published 11 April 2023
Abstract
Cyclophilin (CYP) plays an important role in plant response to stress, and OsCYP2, one gene of cyclophlilin family, is involved in auxin signal transduction and stress signaling in rice. However, the mechanism that OsCYP2 is involved in rice response to low temperature is still unclear. We identified a new OsCYP2 allelic mutant, lrl3, with fewer lateral roots, and the differences in shoot height, primary root length and adventitious root length increased with the growth process compared to the wild-type plant. Auxin signaling pathway was also affected and became insensitive to gravity. The transgenic rice plants with over-expression of OsCYP2 were more tolerant to low temperature than the wild-type plants, suggesting that OsCYP2 was involved in the low temperature response in rice. In addition, OsCYP2 negatively regulated the expression of OsTPS38, a terpene synthase gene, and was dependent on the OsCDPK7-mediated pathway in response to low temperature stress. OsTPS38- overexpressed transgenic line ox-2 was more sensitive to low temperature. Therefore, OsCYP2 may negatively regulate OsTPS38 through an OsCDPK7-dependent pathway to mediate the response to low temperature in rice. These results provide a new basis for auxin signaling genes to regulate rice response to low temperature stress.Keywords
Supplementary Material
Supplementary Material FilePlants resist low temperature through a series of complex mechanisms [1]. Ca2+ is an important signaling molecule that can participate in the sensing mechanism of low temperature in plants [2–4]. The calcium dependent protein kinase (CDPK) system is an important Ca2+ transduction pathway in cells and can sense changes in external signals [5–12]. Previous studies have confirmed that CDPK is widely distributed. When plants are under low temperature stress, Ca2+ binds to the regulatory region of CDPK, activates the activity of CDPK and transmits signals to the downstream [13–18]. CDPK can positively regulate the expression of stress-related genes in the signal transduction pathway under low temperature stress in rice [19]. Other studies have shown that the expression of OsCDPK genes has a circadian rhythm, which is not obviously induced in the early stage under low temperature treatment, but is activated after 18–24 h under low temperature treatment, and gradually plays its function with the increase of stress time [18]. CDPK7 is a relatively conserved gene in the CDPK family and can be induced to express under low temperature stress [20]. Moreover, the sense OsCDPK13/OsCDPK7 transgenic lines in rice had higher recovery rates after cold treatment than the control [21]. According to the previous studies, CDPK mediated the mechanism of plant response to low temperature by sensing Ca2+ signal.
Cyclophilin (CYP), a member of the immunophilic family, has the PPIase activity and can limit the folding and processing of rate-limiting proteins [22]. Cyclophilin is the target protein of cyclosporin A, which can bind to Ca2+ in the cytoplasm and plays an important role in Ca2+ signal transduction pathway [23]. Clones of cyclinoid fragments from beans, maize, Arabidopsis thaliana and rice indicate that cyclinoids are involved in the regulation of cell division and transcriptional regulation, and mediate signal transduction pathways, as well as play a key role in plant response to stresses [24–28]. Functional loss of OsCYP20-2 protein in rice can make the mutant sensitive to low temperature stress, and OsCYP20-2 protein in rice chloroplasts can promote the formation of homo-dimer of OsFSD2 to eliminate the effect on ROS under low temperature stress [29]. CyPs1 plays a regulatory role in the infection and development of the pathogen, and 29 Cyclophilin genes found in A. thaliana are involved in the processes of photosynthesis, plant stress resistance and mRNA splicing [30]. Moreover, CYP19-4s (AtCYP19-4 and OsCYP19-4) may affect the polarity of auxin transport and PIN localization [31]. AtCYP18-3 is involved in the process of seedling decolorization [32]. The dgt (LeCyp1 gene mutant) mutant in tomato showed few lateral roots [33,34]. The lrt2 (lateral rootless 2, namely OsCYP2 allele) mutant in rice showed similar phenotypes of auxin signal mutants, and OsCYP2 interacts with OsSGT1 to participate in auxin signal transduction [35–37]. OsCYP2 is believed to be a key regulator of ROS levels by regulating the activity of antioxidant enzymes at translational level, and may be involved in circadian rhythm regulation and signaling pathways of stress such as salt, heat, cold or ABA [38]. However, the mechanism of OsCYP2 in response to low temperature in rice is still unclear.
Terpene synthase (TPS) is an important enzyme in terpene biosynthesis and has circadian rhythm [39–41], which is derived from mevalonic acid and can participate in plant defense response to stress [41,42]. 16 TPS genes have been identified in rice, of which OsTPS37 (LOC_Os08g04500), OsTPS38 (LOC_Os07g11790) and OsTPS40 (LOC_Os08g07100) are mainly responsible for the synthesis of rice sesquiterpenes and participate in the defense system of rice [43]. At present, the molecular mechanism of OsTPS38 involved in low temperature response in rice is not clear.
In this study, we identified a novel OsCYP2 allelic mutant, lrl3, with few lateral roots. The auxin signaling pathway in lrl3 was affected and OsCYP2 regulated the low temperature response of OsTPS38. Moreover, OsCYP2 negatively regulated OsTPS38 expression and mediated the low temperature response mechanism in rice through the OsCDPK7-dependent pathway. These results will provide a new basis for auxin signaling genes to regulate rice response to low temperature stress.
2.1 lrl3 Mutant Showed Few Lateral Roots and Insensitive Gravitropism
We screened the T-DNA insertion mutant pools of zh11 in japonica rice, and identified a lateral rootless mutant lrl3 (lateral rootless 3) compared with the wild-type plant zh11, and further functional studies were conducted using homozygous mutants and BC1F2 generations.
In order to study the function of lrl3 mutant in details, we performed phenotypic analysis on the wild-type plant zh11 and the mutant lrl3 cultured in normal rice (Oryza sativa) medium for 7 days, and found that compared with the wild-type plant zh11, the mutant lrl3 had short shoot and long primary roots, but a few adventitious roots and few lateral roots (Fig. 1A). Further stereoscopic observation revealed that no significant differences were showed between lrl3 and zh11 in root hairs, root tips and root caps, but lateral roots were significantly absent in lrl3 (Fig. 1B). After statistical analysis, it was found that significant or extremely significant differences between lrl3 and zh11 were detected in shoot height, primary root length, number of lateral roots and number of adventitious roots (p < 0.05 or p < 0.01) (Fig. 1D). These differences were further increased when lrl3 and zh11 were cultured for 14 days (Fig. 1C).

Figure 1: Identification and analysis of the phenotype of the wild-type plant and the mutant. (A) Phenotype of the wild-type zh11 and the mutant lrl3 at 7-day-old seeding stage. Bar is 5 cm. (B) Root hair observation at 7-day-old seeding stage. Bar is 1 mm. The first line is the rhizome junction; the second lined type zh11 is the mature region of primary root; the third line is the primary root tip; the fourth line is primary root cap. (C) Phenotype of the wild-type zh11 and the mutant lrl3 at 14-day-old seeding stage. Bar is 5 cm. (D) The phenotype parameters of the wild-type zh11 and the mutant lrl3 cultured in normal nutrient solution for 7 (top line) and 14 (bottom line) days, * and ** indicate significant differences at 5% and 1% levels, respectively
These results suggest that the mutation of lrl3 controls shoot elongation, primary shoot, and adventitious root elongation, as well as the number of adventitious roots and lateral roots, and that the differences are more significant along with the growth process.
To investigate the root gravitropism, we horizontally placed the primary root of lrl3 T2 lines and zh11 two days after sowing, and found that lrl3 was slow in response to gravity and barely bent compared with zh11 (see Supplementary Fig. 1). This suggests that the mutation site of lrl3 regulates the response of rice root to gravity.
2.2 The Auxin Signaling Pathway of lrl3 Was Disrupted
To understand the physiological mechanism of root development, we treated lrl3 and zh11 with auxin polar transport inhibitor N-1-naphthylphthalamic (NPA) and Synthetic hormones 1-naphthlcetic acid (NAA). The results presented that lrl3 and zh11 showed similar responses to NPA, while lrl3 showed significant resistance to NAA compared with zh11 under the treatment of 0.1 μM NAA (Fig. 2). These results suggest that the mutation locus of lrl3 is involved in auxin signaling during lateral root formation in rice, rather than auxin polar transport.

Figure 2: Phenotype of the wild-type plant and the mutant under NPA and NAA treatments. (A) and (C) Phenotype of the wild type zh11 and the mutant lrl3 under 1 μM NPA and 0.1 μM NAA treatment at 7-day-old seeding stage. Bar is 2 cm. (B) and (D) Phenotype of the wild-type zh11 and the mutant lrl3 under 1 μM NPA and 0.1 μM NAA treatment at 7-day-old seeding stage. The top, middle and bottom line are root hair phenotype of rhizome junction, mature zone and root tip of primary root, respectively. Bar is 1 mm
2.3 LRL3 Gene Mapping and Genetic Complementation Verification
Genetic analysis of BC1F2 populations of lrl3 and zh11 showed that the ratio of wild-type individuals to mutant individuals was in accordance with the expected value of 3:1, suggesting that lrl3 lateral rootless mutation is controlled by a single recessive gene (Table 1).
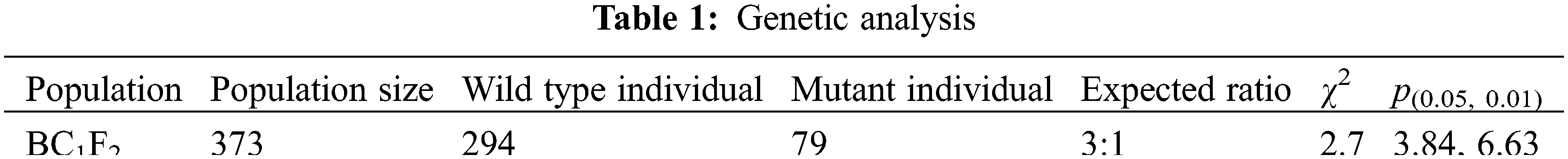
We employed 30 mutant individuals in F2 genetic population to detect polymorphisms by SIS2 (M2) and RM12368 markers in close linkage with OsCYP2 gene regulating lateral root formation. The lrl3 band was detected in all the 30 mutant individuals at SIS2 (M2) and in all the 29 mutant individuals at RM12368. The results indicated that the LRL3 gene, the candidate gene OsCYP2 for lrl3 mutation, was preliminarily located near the molecular markers SIS2 (M2) and RM12368 on the chromosome 2 (Fig. 3A). As reported, the gene OsCYP2 encoding rice cyclophilin, participates in the protein folding, which is thought to be associated with auxin signaling [35], so the OsCYP2 gene is regarded as the candidate gene for lrl3 mutation. The coding region of the LRL3/OsCYP2 gene is 519 bp between 1116066 bp and 1116971 bp on chromosome 2. Agarose gel electrophoresis and sequencing analysis on the mutant lrl3 indicated that 59 basepairs of nucleotides were missing at LRL3/OsCYP2 (Figs. 3B–3C). Further bioinformatics analysis showed that the 59-bp deletion at LRL3/OsCYP2 led to the deletion of glutamate from 15th to 35th and frame-coding mutations, so that the LRL3/OsCYP2 gene in lrl3 encodes only 94 amino acids (ΔOsCYP2 in Fig. 3C) because of frame-shift mutation. To confirm that lrl3 is an OsCYP2 allelic mutant, we crossed lrl3 (deletion of 59 bp) with lrt2 (deletion of 50 bp), and obtained positive double-mutant F1 plants. Phenotypic analysis of the wild-type plant zh11, lrl3, double mutant F1 between lrl3 and lrt2, lrt2 and the wild-type plant Nipponbare showed that the double mutant F1, lrl3 and lrt2 all exhibited lateral rootless phenotype. This result of genetic complementarity suggests that lrl3 lateral rootless phenotype is caused by OsCYP2 deletion mutation (Figs. 3D–3E). In addition, the homology analysis showed that the four amino acids of OsCYP2 were only present in the 12 plants analyzed, but not in human sapiens and yeast, suggesting that these four conserved amino acids are unique to plants during the evolution of the Cyclophilin family (see Supplementary Fig. 2).

Figure 3: Gene mapping and genetic complementation analysis of LRL3/OsCYP2. (A) Map-based cloning of LRL3 gene that is the candidate gene OsCYP2 for lrl3 mutation. (B) Agarose gel electrophoresis detection of OsCYP2 allelic mutant. MW, DNA Marker DL2000; A the wild type plant zh11; B the mutant lrl3. (C) Schematic diagram of the OsCYP2 allele structure. (D) The poly-acrylamide gel electrophoresis detection for genetic complementation analysis. M, DL2000 Marker; 1, zh11; 2, lrl3; 3, F1; 4, lrt2; 5, Nipponbare. (E) Phenotype of 7-year-old seedlings of zh11, lrl3, F1 of lrl3 and lrt2, lrt2, and Nipponbare for genetic complementation analysis. a, zh11; b, lrl3; c, F1 of lrl3 and lrt2; d, lrt2; e, Nipponbare. Bar is 2 cm
2.4 OsCYP2 is Involved in Low Temperature Response
There was no significant difference in leaf morphology and color between zh11 and lrl3 before low temperature treatment. However, compared with the wild-type plant zh11, lrl3 was more severe curled of leaf, yellowing of leaf tip and needle-like leaf shape after low temperature treatment (Figs. 4A–4B). Furthermore, because of lrl3 low seed-setting, we investigated the response of the OsCYP2 gene to low temperature using OsCYP2-overexpressed lines SSBM-OE and the wild-type plants SSBM. To clarify the different response of the OsCYP2-overexpressed lines SSBM-OE and the wild-type plants SSBM to low temperature stress, we carried out low temperature treatment of the OsCYP2-overexpressed lines SSBM-OE and the wild-type plants SSBM for 27 days, all the wild-type plants SSBM died while some of the OsCYP2-overexpressed lines SSBM-OE were still alive (Fig. 5). These results indicated that OsCYP2 positively regulated resistance to low temperature.

Figure 4: Phenotype of the wild-type plant and the mutant under low temperature treatment. (A) and (B) Phenotype of the wild-type plant zh11 and mutant lrl3 before and after low temperature treatment, bar is 5 cm. WT, the wild-type plant zh11; MT, mutant lrl3; CK, before low temperature treatment; TM, after low temperature treatment

Figure 5: Phenotype of the OsCYP2-overexpressed lines SSBM-OE, the wild-type plants SSBM before and after under low temperature. The plants in the pot were the OsCYP2-overexpressed lines SSBM-OE (left), the wild-type plants SSBM (right). (A) Before low temperature; (B) After low temperature, bar is 2 cm. CK, before low temperature treatment; TM, after low temperature treatment
2.5 OsCYP2 Negatively Regulates OsTPS38 Response to Low Temperature through OsCDPK7-Dependent Pathway
In order to detect downstream genes regulated by OsCYP2, we performed transcriptome analysis on zh11 and lrl3 before and after low temperature treatment, and found that a TPS gene OsTPS38 (Os08g0139700) showed a significant difference in expression between zh11 and lrl3 after low temperature treatment, which was contrary to the expression pattern of OsCYP2 gene, confirmed by qRT-PCR (Figs. 6A–6B). This suggests that OsTPS38 is negatively regulated by OsCYP2 to mediate the mechanism of low temperature response in rice. Meanwhile, OsCYP2 regulates the low temperature response mechanism in rice through OsCDPK7-dependent pathway (Fig. 6C).

Figure 6: Expression of OsCYP2, OsTPS38 and OsCDPK7 in the wild-type and mutant under low temperature treatment by transcriptome analysis and qRT-PCR. WTCK_2, zh11 before low temperature; WTCOLD_2, zh11 after low temperature; MTCK_2, lrl3 before low temperature; MTCOLD_2, lrl3 after low temperature; RPKM, reads per kilobase per million mapped reads
2.6 OsTPS38 is Involved in Low Temperature Response
To further clarify the molecular mechanism of OsTPS38 in response to low temperature, the overexpression lines ox-2 of OsTPS38 and the wild-type plant zh11 were used to undergo low temperature treatment for 3 days at three-leaf stage, and it was found that the degree of leaf curling in ox-2 was higher than that in zh11 (Figs. 7A–7B). These results suggest that OsTPS38 may negatively regulate the low temperature response in rice.

Figure 7: Response of OsTPS38 to low temperature. (A) and (B) Phenotypes of transgenic lines and zh11 before and after low temperature treatment. WT, the wild-type plant zh11; OX-2, overexpression lines ox-2 of OsTPS38; CK, before low temperature treatment; TM, after low temperature treatment. Bar is 2 cm
3.1 The Mechanism of Auxin Pathway Regulating Low Temperature Response
Auxin is the first plant hormone that has been recognized and studied by humans. It almost participates in the whole process of plant growth and development, and plays an important role in the hormonal regulatory network at all stages of growth and development [44–46]. Interestingly, the auxin-signaling mutants axr1, tir1, gps2-1, gps1, gps2 and gps3, which show a decreased gravity response, responded to low temperature treatment [47–49]. In our study, lrl3 mutant was also affected by gravity response and involved in response to low temperature, suggesting that OsCYP2 may be involved in the low temperature response mediated by the auxin pathway. Low temperature stress may affect the function of the Pin-formed proteins (PINs) by inhibiting the transport or localization of intracellular proteins, and eventually cause slow root growth and gravitation anomaly [50–53]. However, we found significant differences in the expression of OsPIN10a in OsCYP2 mutant compared with the wild type (see Supplementary Fig. 3). These results may suggest that OsCYP2 also responds to low temperature through the auxin pathway. There are few studies on the mechanism of auxin pathway regulating low temperature response, but WES1, which encodes IAA amino acid synthase, is up-regulated in low temperature stress, and activates the expression of stress-related genes Pathogenesis-related protein 1 (PR-1) and C-repeat binding factors (CBFs) by inactivating IAA, this indicates that the expression of the key genes CBFs in low temperature is directly regulated by auxin [54]. It has been reported that low temperature can affect the differential expression of some auxin response genes, such as IAA20 [55,56], while we also found that the expression of IAA20 gene was obviously affected by OsCYP2 under exogenous IAA treatment (see Supplementary Fig. 4). Otherwise, another study has confirmed that OsCYP2 can interact with OsIAA11 [37]. In conclusion, we speculate that OsCYP2 may be involved in the regulation of auxin response genes so as to mediate the cold response in rice (Fig. 8). However, the molecular mechanism of OsCYP2 regulating rice response to low temperature through auxin pathway needs to be further improved.

Figure 8: Cold sensing and responsive pathway. Solid arrows indicate positive regulation; T-shaped lines indicate negative regulation; broken arrows indicate predicted activation. The genes in the oval frame are from rice; the genes in the hexagonal frame are from A. thaliana
3.2 The Low Temperature Response Pathway Depending on the ICE-CBF-COR Transcriptional Cascade to Sense Ca2+ Signals
Plants can regulate the corresponding physiological and biochemical responses to adapt to low temperature by activating signal transduction pathways, and the response to low temperature can be divided into ABA-dependent pathway, Ca2+ sensing pathway and ROS sensing pathway [57]. The low temperature environment causes the hardening of the plasma membrane in rice, which leads to the Ca2+ channel opening and the extracellular Ca2+ entering the cytoplasm, resulting in the difference of intracellular and extracellular Ca2+ concentration. However, protein kinases such as CDPKs can sense the change of endogenous Ca2+ level and carry out phosphorylation reaction and activate the transcription factors of each downstream family, ultimately affecting the expression of COR genes. In this experiment, OsCYP2 is involved in the pathway in response to low temperature by CDPK7-mediated Ca2+ signaling, which is also known as the ABA-independent ICE-CBF-COR transcriptional cascade pathway [58,59]. Under low temperature stress, the AP2 conserved domain of CBFs can bind CCGAC (CRT, C-repeat), the core elements of CORs initiation region, to activate CORs at the transcriptional level, while CBF genes are usually regulated by ICE1. The transcription factor OsDREB1F may be activated by OsICE1, which then activates the expression of downstream low-temperature responsive COR genes harboring DRE/CRT domain [60]. In addition, OsICE1 is regulated by phosphorylation, sumoylation, and ubiquitination mediated by E3 ubiquitin ligase, and simultaneously binds to cis-elements on the promoter of CBF3/DREB1 to participate in the low temperature response [61]. However, some studies have suggested that CBF3 inhibition in ice1-1 is gene silencing caused by T-DNA-triggered methylation [62]. OsICE2 over-expression is involved in low temperature response of CBF1/DREB1 [63]. CBF2 is a negative regulator of CBF1 and CBF3, but not depending on the expression of CBF1 and CBF3 [64–66]. OsMYB3R-2 and OsMYB2 involved in cold resistance up-regulates the expression of OsDREB2A, while MYBS3 negatively down-regulates the expression of OsDREB1 and OsDREB2A [67–69]. Taken together, we propose that rice response to low temperature may occur through the following pathways (Fig. 8).
4.1 Plant Materials and Growth Conditions
The rice seeds of zh11 (the wild-type cultivar in japonica background), lrl3, Oscyp2-1, SSBM (the wild-type cultivar in japonica background), SSBM-OE, Kasalath (the wild-type cultivar in indica background) and Oscyp2-2 in cold treatment were provided by Professor Xiaorong Mo of Zhejiang University, SSBM-OE transgenic lines were obtained by introducing the OsCYP2 CDS sequence into the vector 35S-pCAMBIA1300 (see Supplementary Fig. 5) and then the transformation of the above construct into SSBM wild-type calli by EHA105, the seeds of lrt2 and Nipponbare in allelic verification were provided by Professor Jianru Zuo of Chinese Academy of Sciences. The F1 of lrl3 crossed by lrt2 was obtained in the Growth Chamber of Northeast Agricultural University.
In the hydroponic experiment, the pH was adjusted to 5.5 with 1 N NaOH in normal rice (O. sativa) medium [70]. Rice seeds were washed with distilled water, treated to break dormancy by 0.6% HNO3 for 16 h at room temperature, and transferred to germinate in the incubator at 37°C. The germinated seeds were planted in normal rice (O. sativa) medium (3 L) on a nylon mesh floating, in rice culture chamber (day/night: 30°C/22°C, 12/12 h; Rh80%, 450 μmol photons m−2 s−1).
In order to study the effect of auxin on root growth and development, the seeds of zh11 and lrl3 were cultured under 0.1μM NAA (Sigma Aldrich, http://www.sigmaaldrich.com/) liquid nutritious medium. IAA and NPA were used in short-term auxin treatment.
The seeds of the wild-type zh11, lrl3 mutant, OsCYP2-overexpressed lines SSBM-OE and the wild-type plants SSBM were germinated and cultured in hydroponic solution. Then the one-leaf seedlings were transplanted into 6 cm × 6 cm × 6 cm culture pots in the incubator (HPG-280, Harbin Donglian Electronic Technology Development Co., Ltd., Harbin, China). The culture conditions were as follows: the diurnal temperature was 25°C/22°C, 12/12 h, and the relative humidity was 80%. Sampling for 0 h was conducted before chilling treatment, and zh11 and lrl3 were subsequently treated at 17°C at three-leaf stage. The culture conditions were as follows: the day and night temperature was maintained at 17°C/17°C, 12/12 h, and the relative humidity was 80%.
Seedling roots of zh11 and lrl3 were sampled on 7 and 14 days after sowing, and 5 samples were taken from each treatment for statistical analysis. Shoot height, primary root length and adventitious root length of the plants were measured with a scale. The number of adventitious roots was counted visually. Transmission Scanner STD1600 Scanner (Epson, Nagano Prefecture, Japan) was used to scan the primary roots of rice. WinRhizo (Regent Instruments Inc., Quebec, Canada) image analysis system was employed to calculate the number and length of lateral roots of the root system.
4.4 Analysis of Root Gravitation Response
The seeds of zh11 and lrl3 were germinated and cultured on a nylon mesh in 5 L rice culture solution for two days, then the seeds were transferred to a plastic plate (11.7 cm × 11.7 cm) with the nutritious solution soaked in blue phosphorus-free paper, and the main roots were placed in parallel horizontally.
4.5 LRL3 Gene Mapping and Genetic Complementation Verification
F1 individuals were obtained by crossing lrl3 mutant (japonicarice zh11) as female parent and indica rice Kasalath as male parent. F1 individuals self-cross to obtain F2 genetic population. In 30 F2 mutant plants, LRL3 gene responsible for lrl3 mutation was mapped to molecular markers SIS2 and RM12368. OsCYP2 controlling lateral root formation was detected in this region and named the candidate gene LRL3/OsCYP2. Supplementary Tables 1–2 provide details of all the mapping markers.
To test the function of LRL3/OsCYP2 for lateral root formation of lrl3 mutant, we crossed lrl3 by lrt2 mutant with 50 bp deletion in OsCYP2 to obtain double mutant F1, and to verify the allelism of lrl3 and lrt2 genetically.
ClustalX 1.81 and Genedoc3.2 software were used to analyze the protein sequences of OsCYP2 with the homologous proteins of the biological cyclophilin-ABH-like domain, and the homology of their conserved domains was compared. Then the phylogenetic tree was constructed by Neighbor-joining method with MEGA3.1 software. The evolutionary standard bootstrap was 1000, and the results were output with MEGA3.1 software.
4.7 Semi-Quantitative and Quantitative RT-PCR
Total RNA was extracted using Trizol D0410 reagent according to the manufacturer’s instructions (Invitrogen, http://www.invitrogen.com/). The first cDNA strand was synthesized after 5 μg of total RNA was treated with Invitrogen II reverse transcriptase. Semi-quantitative RT-PCR and quantitative RT-PCR are described above [70]. Details of primers are shown in Supplementary Table 3. RT-PCR was performed at least three times with independent biological replicates, qRT-PCR was performed at least three times with independent technical replicates.
Rice seedlings were treated at low temperature and the leaves of zh11 and lrl3 seedlings under the normal and chilling treatment were sampled and quickly frozen in liquid nitrogen and stored at −80°C for later analysis. Novogene Bioinformatics Technology Co., Ltd., China was entrusted to complete RNA extraction, quality control, database construction and Illumina HiSeQTM sequencing.
4.9 Construction of OsTPS38-Overexpressed Vector
According to the known sequence of OsTPS38 gene (http://rice.uga.edu/cgi-bin/ORF_infopage.cgi?orf=LOC_Os08g04500.1) and the cloning site of plant expression vector pBWA(V)HU-45001OE-Gus, specific primers (45001OE-F: cagtCACCTGCaaaacaacatggcaacctctgttccgagtgtacta; 45001OE-R: cagtCACCTGCaaaatacattaaacagagaggatgtagatggagtg) were designed. Then the OsTPS38 gene was amplified and introduced into the vector pBWA(V)HU-45001OE-Gus (see Supplementary Fig. 6). The construction and genetic transformation of OsTPS38-overexpressed vector were entrusted to Wuhan Biorun Biological Technology Co., Ltd. (China) to obtain OsTPS38-overexpressed positive lines in zh11 background.
Acknowledgement: We thank Prof. Xiaorong Mo of Zhejiang University and Prof. Jianru Zuo of Chinese Academy of Sciences for the materials provided for this study.
Funding Statement: This study was financially supported by the Natural Science Foundation of Heilongjiang Province (LH2020C006) and the Heilongjiang Postdoctoral Scientific Research Developmental Fund (LBH-Q17031).
Author Contributions: Hongxiu Gao, Tianqi Liu, Lin Zhu and Zhenxing Zhu performed phenotypic observation and measurement. Hongxiu Gao, Tianqi Liu, Lin Zhu, Haitao Lv and Zhongchen Zhang performed gene mapping and data analysis. Lin Zhu and Zhongchen Zhang performed genetic complementation verification. Tianqi Liu and Lin Zhu performed RNA data analysis. Tianqi Liu, Xueyu Leng, Wei Xie and Zhongchen Zhang wrote the manuscript. Hongxiu Gao and Lin Zhu contributed to the article equally. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: NCBI-SRA database under the BioProject no. PRJNA732107 and accession nos. SRR14629497, SRR14629496, SRR14629495, and SRR14629494 for the RNA-seq data.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Guy, C. L. (1990). Cold acclimation and freezing stress tolerance: Role of protein metabolism. Annual Review of Plant Physiology and Plant Molecular Biology, 41, 187–223. https://doi.org/10.1146/annurev.pp.41.060190.001155 [Google Scholar] [CrossRef]
2. Perochon, A., Aldon, D., Galaud, J. P., Ranty, B. (2011). Calmodulin and calmodulin-like proteins in plant calcium signaling. Biochimie, 93(12), 2048–2053. https://doi.org/10.1016/j.biochi.2011.07.012 [Google Scholar] [PubMed] [CrossRef]
3. Knight, M. R., Knight, H. (2012). Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytologist, 195(4), 737–751. https://doi.org/10.1111/j.1469-8137.2012.04239.x [Google Scholar] [PubMed] [CrossRef]
4. Finka, A., Cuendet, A. F., Maathuis, F. J., Saidi, Y., Goloubinoff, P. (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. The Plant Cell, 24(8), 3333–3348. https://doi.org/10.1105/tpc.112.095844 [Google Scholar] [PubMed] [CrossRef]
5. Ludwig, A. A., Romeis, T., Jones, J. D. G. (2004). CDPK-mediated signalling pathways: Specificity and cross-talk. Journal of Experimental Botany, 55(395), 181–188. https://doi.org/10.1093/jxb/erh008 [Google Scholar] [PubMed] [CrossRef]
6. Romeis, T., Ludwig, A. A., Martin, R., Jones, J. D. G. (2001). Calcium-dependent protein kinases play an essential role in a plant defence response. The EMBO Journal, 20, 5556–5567. https://doi.org/10.1093/emboj/20.20.5556 [Google Scholar] [PubMed] [CrossRef]
7. Ludwig, A. A., Saitoh, H., Felix, G., Freymark, G., Miersch, O. et al. (2005). Ethylene-mediated cross-talk between calcium-dependent protein kinase and MAPK signaling controls stress responses in plants. Proceedings of the National Academy of Sciences of the United States of America, 102(30), 10736–10741. https://doi.org/10.1073/pnas.0502954102 [Google Scholar] [PubMed] [CrossRef]
8. Mori, I. C., Murata, Y., Yang, Y. Z., Munemasa, S., Wang, Y. F. et al. (2006). CDPKs CPK6 and CPK3 function in ABA regulation of guard cell S-type anion- and Ca2+-permeable channels and stomatal closure. PLoS Biology, 4(10), 1749–1762. https://doi.org/10.1371/journal.pbio.0040327 [Google Scholar] [PubMed] [CrossRef]
9. Zhu, S. Y., Yu, X. C., Wang, X. J., Zhao, R., Li, Y. et al. (2007). Two calcium-dependent protein kinases, CPK4 and CPK11, regulate abscisic acid signal transduction in Arabidopsis. The Plant Cell, 19(10), 3019–3036. https://doi.org/10.1105/tpc.107.050666 [Google Scholar] [PubMed] [CrossRef]
10. Ishida, S., Yuasa, T., Nakata, M., Takahashi, Y. (2009). A tobacco calcium-dependent protein kinase, CDPK1, regulates the transcription factor repression of shoot growth in response to gibberellins. The Plant Cell, 20(12), 3273–3288. https://doi.org/10.1105/tpc.107.057489 [Google Scholar] [PubMed] [CrossRef]
11. Kobayashi, M., Ohura, I., Kawakita, K., Yokota, N., Fujiwara, M. et al. (2007). Calcium-dependent protein kinases regulate the production of reactive oxygen species by potato NADPH oxidase. The Plant Cell, 19(3), 1065–1080. https://doi.org/10.1105/tpc.106.048884 [Google Scholar] [PubMed] [CrossRef]
12. Kudla, J., Batistic, O., Hashimoto, K. (2010). Calcium signals: The lead currency of plant information processing. The Plant Cell, 22(3), 541–563. https://doi.org/10.1105/tpc.109.072686 [Google Scholar] [PubMed] [CrossRef]
13. Lu, S. X., Hrabak, E. M. (2002). An Arabidopsis calcium-dependent protein kinase is associated with the endoplasmic reticulum. Plant Physiology, 128(3), 1008–1021. https://doi.org/10.1104/pp.010770 [Google Scholar] [PubMed] [CrossRef]
14. Myers, C., Romanowsky, S. M., Barron, Y. D., Garg, S., Azuse, C. L. et al. (2009). Calcium-dependent protein kinases regulate polarized tip growth in pollen tubes. The Plant Journal, 59(4), 528–539. https://doi.org/10.1111/j.1365-313X.2009.03894.x [Google Scholar] [PubMed] [CrossRef]
15. Zhang, H. F., Liu, W. Z., Zhang, Y. P., Deng, M., Niu, F. F. et al. (2014). Identification, expression and interaction analyses of calcium-dependent protein kinase (CPK) genes in canola (Brassica napus L.). BMC Genomics, 15(211), 1471–2164. https://doi.org/10.1186/1471-2164-15-211 [Google Scholar] [PubMed] [CrossRef]
16. Liu, H. L., Che, Z. J., Zeng, X. R., Zhou, X. Q., Sitoe, H. M. et al. (2016). Genome-wide analysis of calcium-dependent protein kinases and their expression patterns in response to herbivore and wounding stresses in soybean. Functional Integrative Genomics, 16(5), 481–493. https://doi.org/10.1007/s10142-016-0498-8 [Google Scholar] [PubMed] [CrossRef]
17. Li, A. L., Zhu, Y. F., Tan, X. M., Wang, X., Wei, B. et al. (2008). Evolutionary and functional study of the CDPK gene family in wheat (Triticum aestivum L.). Plant Molecular Biology, 66(4), 429–443. https://doi.org/10.1007/s11103-007-9281-5 [Google Scholar] [PubMed] [CrossRef]
18. Xu, X. W., Liu, M., Lu, L., He, M., Qu, W. Q. et al. (2015). Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Molecular Genetics and Genomics, 290(4), 1403–1414. https://doi.org/10.1007/s00438-015-1002-1 [Google Scholar] [PubMed] [CrossRef]
19. Saijo, Y., Hata, S., Kyozuka, J., Shimamoto, K., Lzui, K. (2000). Over-expression of a single Ca2+-dependent protein kinase confers both cold and salt/drought tolerance on rice plants. The Plant Journal, 23(3), 319–327. https://doi.org/10.1046/j.1365-313x.2000.00787.x [Google Scholar] [PubMed] [CrossRef]
20. Geng, S. F., Zhao, Y. L., Tang, L. C., Zhang, Z. R., Sun, M. H. et al. (2011). Molecular evolution of two duplicated CDPK genes CPK7 and CPK12 in grass species: A case study in wheat (Triticum aestivum L.). Gene, 475(2), 94–103. https://doi.org/10.1016/j.gene.2010.12.015 [Google Scholar] [PubMed] [CrossRef]
21. Abbasi, F., Onodera, H., Toki, S., Tanaka, H., Komatsu, S. (2004). OsCDPK13, a calcium-dependent protein kinase gene from rice, is induced by cold and gibberellin in rice leaf sheath. Plant Molecular Biology, 55(4), 541–552. https://doi.org/10.1007/s11103-004-1178-y [Google Scholar] [PubMed] [CrossRef]
22. Brandts, J. F., Halvorson, H. R., Brennan, M. (1975). Consideration of the possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry, 14(22), 4953–4963. https://doi.org/10.1021/bi00693a026 [Google Scholar] [PubMed] [CrossRef]
23. Takahashi, N., Hayano, T., Suzuki, M. (1989). Peptidyl-prolyl cis-trans isomerase is the cyclosporin A-binding protein cyclophilin. Nature, 337(6206), 473–475. https://doi.org/10.1038/337473a0 [Google Scholar] [PubMed] [CrossRef]
24. Brazin, K. N., Mallis, R. J., Fulton, D. B., Andreotti, A. H. (2002). Regulation of the tyrosine kinase Itk by the peptidyl-prolyl isomerase cyclophilin A. Proceedings of the National Academy of Sciences of the United States of America, 99(4), 1899–1904. https://doi.org/10.1073/pnas.042529199 [Google Scholar] [PubMed] [CrossRef]
25. Rycyzyn, M. A., Clevenger, C. V. (2002). The intranuclear prolactin/cyclophilin B complex as a transcriptional inducer. Proceedings of the National Academy of Sciences of the United States of America, 99(10), 6790–6795. https://doi.org/10.1073/pnas.092160699 [Google Scholar] [PubMed] [CrossRef]
26. Godoy, A. V., Lazzaro, A. S., Casalongué, C. A., Segundo, B. S. (2000). Expression of a Solanum tuberosum cyclophilin gene is regulated by fungal infection and abiotic stress conditions. Plant Science, 152(2), 123–134. https://doi.org/10.1016/S0168-9452(99)00211-3 [Google Scholar] [CrossRef]
27. Chen, A. P., Wang, G. L., Qu, Z. L., Lu, C. X., Liu, N. et al. (2007). Ectopic expression of ThCYP1, a stress-responsive cyclophilin gene from Thellungiella halophila, confers salt tolerance in fission yeast and tobacco cells. Plant Cell Reports, 26(2), 237–245. https://doi.org/10.1007/s00299-006-0238-y [Google Scholar] [PubMed] [CrossRef]
28. Marivet, J., Margis-Pinheiro, M., Frendo, P., Burkard, G. (1994). Bean cyclophilin gene expression during plant development and stress conditions. Plant Molecular Biology, 26(4), 1181–1189. https://doi.org/10.1007/BF00040698 [Google Scholar] [PubMed] [CrossRef]
29. Ge, Q., Zhang, Y. Y., Xu, Y. Y., Bai, M. Y., Luo, W. et al. (2020). Cyclophilin OsCYP20-2 with a novel variant integrates defense and cell elongation for chilling response in rice. New Phytologist, 225(6), 2453–2467. https://doi.org/10.1111/nph.16324 [Google Scholar] [PubMed] [CrossRef]
30. Romano, P. G. N., Horton, P., Gray, J. E. (2004). The Arabidopsis cyclophilin gene family. Plant Physiology, 134(4), 1268–1282. https://doi.org/10.1104/pp.103.022160 [Google Scholar] [PubMed] [CrossRef]
31. Li, B. B., Xu, W. Z., Xu, Y. Y., Zhang, Y. Y., Wang, T. et al. (2010). Integrative study on proteomics, molecular physiology, and genetics reveals an accumulation of cyclophilin-like protein, TaCYP20-2, leading to an increase of Rht protein and dwarf in a novel GA-insensitive mutant (gaid) in wheat. Journal of Proteome Research, 9(8), 4242–4253. https://doi.org/10.1021/pr100560v [Google Scholar] [PubMed] [CrossRef]
32. Trupkin, S. A., Mora-García, S., Casal, J. J. (2012). The cyclophilin ROC1 links phytochrome and cryptochrome to brassinosteroid sensitivity. Plant Journal, 71(5), 1–12. https://doi.org/10.1111/j.1365-313X.2012.05013 [Google Scholar] [CrossRef]
33. Rice, M. S., Lomax, T. L. (2000). The auxin-resistant diageotropica mutant of tomato responds to gravity via an auxin-mediated pathway. Planta, 210(6), 906–913. https://doi.org/10.1007/s004250050696 [Google Scholar] [PubMed] [CrossRef]
34. Oh, K. C., Ivanchenko, M. G., White, T. J., Lomax, T. L. (2006). The diageotropica, gene of tomato encodes a cyclophilin: A novel player in auxin signaling. Planta, 224(1), 133–144. http://hdl.handle.net/1957/31803 [Google Scholar] [PubMed]
35. Kang, B., Zhang, Z. C., Wang, L. L., Zheng, L. B., Mao, W. H. et al. (2013). OsCYP2, a chaperone involved in degradation of auxin-responsive proteins, plays crucial roles in rice lateral root initiation. The Plant Journal, 74(1), 86–97. https://doi.org/10.1111/tpj.12106 [Google Scholar] [PubMed] [CrossRef]
36. Zheng, H. K., Li, S. J., Ren, B., Zhang, J., Ichii, M. et al. (2013). LATERAL ROOTLESS2, a cyclophilin protein, regulates lateral root initiation and auxin signaling pathway in rice. Molecular Plant, 6(5), 1719–1721. https://doi.org/10.1093/mp/sst052 [Google Scholar] [PubMed] [CrossRef]
37. Jing, H., Yang, X., Zhang, J., Liu, X., Zheng, H. et al. (2015). Peptidyl-prolyl isomerization targets rice Aux/IAAs for proteasomal degradation during auxin signalling. Nature Communications, 6(1), 7395–7404. https://doi.org/10.1038/ncomms8395 [Google Scholar] [PubMed] [CrossRef]
38. Ruan, S. L., Ma, H. S., Wang, S. H., Fu, Y. P., Xin, Y. et al. (2011). Proteomic identification of OsCYP2, a rice cyclophilin that confers salt tolerance in rice (Oryza sativa L.) seedlings when overexpressed. BMC Plant Biology, 11(1), 1–15. https://doi.org/10.1186/1471-2229-11-34 [Google Scholar] [PubMed] [CrossRef]
39. Cole, M. D., Bridge, P., Dellar, J., Cole, M. D., Bridge, P. D. et al. (1991). Antifungal activity of neo-clerodane diterpenoids from Scutellaria. Phytochemistry, 30(4), 1125–1127. https://doi.org/10.1016/S0031-9422(00)95186-0 [Google Scholar] [CrossRef]
40. Penuelasz, J., Llusia, J. (2002). Linking photorespiration, monoterpenes and thermotolerance in Quercus. New Phytoogist, 155(2), 227–237. https://doi.org/10.1046/j.1469-8137.2002.00457.x [Google Scholar] [CrossRef]
41. Dudareva, N., Martin, D., Kish, C. M., Kolosova, N., Gorenstein, N. et al. (2003). (E)-β-Ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. The Plant Cell, 15(5), 1227–1241. https://doi.org/10.1105/tpc.011015 [Google Scholar] [PubMed] [CrossRef]
42. Tholl, D., Chen, F., Petri, J., Gershenzon, J., Eran, P. (2005). Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. The Plant Journal, 42(5), 757–771. https://doi.org/10.1111/j.1365-313X.2005.02417.x [Google Scholar] [PubMed] [CrossRef]
43. Yuan, J. S., Köllner, T. G., Wiggins, G., Grant, G., Feng, C. (2008). Molecular and genomic basis of volatile-mediated indirect defense against insects in rice. The Plant Journal, 55(3), 491–503. https://doi.org/10.1111/j.1365-313X.2008.03524.x [Google Scholar] [PubMed] [CrossRef]
44. Swarup, R., Parry, G., Graham, N., Allen, T., Bennett, M. (2002). Auxin cross-talk: Integration of signalling pathways to control plant development. Plant Molecular Biology, 49(3–4), 411–426. https://doi.org/10.1023/A:1015250929138 [Google Scholar] [CrossRef]
45. Chandler, J. W. (2009). Auxin as compère in plant hormone crosstalk. Planta, 231(1), 1–12. https://doi.org/10.1007/s00425-009-1036-x [Google Scholar] [PubMed] [CrossRef]
46. Depuydt, S., Hardtke, C. S. (2011). Hormone signalling crosstalk in plant growth regulation. Current Biology, 21(9), R365–R373. https://doi.org/10.1016/j.cub.2011.03.013 [Google Scholar] [PubMed] [CrossRef]
47. Shibasaki, K., Uemura, M., Tsurumi, S., Rahman, A. (2009). Auxin response in Arabidopsis under cold stress: Underlying molecular mechanisms. The Plant Cell, 21(12), 3823–3838. https://doi.org/10.1105/tpc.109.069906 [Google Scholar] [PubMed] [CrossRef]
48. Wyatt, S. E., Rashotte, A. M., Shipp, M. J., Robertson, D., Muday, G. K. (2002). Mutations in the gravity persistence signal loci in Arabidopsis disrupt the perception and/or signal transduction of gravitropic stimuli. Plant Physiology, 130(3), 1426–1435. https://doi.org/10.1111/j.1365-313X.2012.05013.x [Google Scholar] [PubMed] [CrossRef]
49. Nadella, V., Shipp, M. J., Muday, G. K., Wyatt, S. E. (2006). Evidence for altered polar and lateral auxin transport in the gravity persistent signal (gps) mutants of Arabidopsis. Plant Cell Environment, 29(4), 682–690. https://doi.org/10.1111/j.1365-3040.2005.01451.x [Google Scholar] [PubMed] [CrossRef]
50. Friml, J., Wiśniewska, J., Benková, E., Mendgen, K., Palme, K. (2002). Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature, 415(6873), 806–809. https://doi.org/10.1038/415806a [Google Scholar] [PubMed] [CrossRef]
51. Paciorek, T., Zažímalová, E., Ruthardt, N., Petrásek, J., Stierhof, Y. et al. (2005). Auxin inhibits endocytosis and promotes its own efflux from cells. Nature, 435(30), 1251–1256. https://doi.org/10.1038/nature03633 [Google Scholar] [PubMed] [CrossRef]
52. Harrison, B. R., Masson, P. H. (2008). ARL2, ARG1 and PIN3 define a gravity signal transduction pathway in root statocytes. The Plant Journal, 53(2), 380–392. https://doi.org/10.1111/j.1365-313X.2007.03351.x [Google Scholar] [PubMed] [CrossRef]
53. Sukumar, P., Edwards, K. S., Rahman, A., Delong, A., Muday, G. et al. (2009). PINOID kinase regulates root gravitropism through modulation of PIN2-dependent basipetal auxin transport in Arabidopsis. Plant Physiology, 150(2), 722–735. https://doi.org/10.1104/pp.108.131607 [Google Scholar] [PubMed] [CrossRef]
54. Park, J. E., Park, J. Y., Kim, Y. S., Staswick, P. E., Jeon, J. et al. (2007). GH3-mediated auxin homeostasis links growth regulation with stress adaptation response in Arabidopsis. Journal of Biological Chemistry, 282(13), 10036–10046. https://doi.org/10.1074/jbc.M610524200 [Google Scholar] [PubMed] [CrossRef]
55. Du, H., Liu, H. B., Xiong, L. Z. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Frontiers in Plant Science, 4, 397. https://doi.org/10.3389/fpls.2013.00397 [Google Scholar] [PubMed] [CrossRef]
56. Yang, Y. T., Yu, Q., Yang, Y. Y., Su, Y. C., Ahmad, W. et al. (2018). Identification of cold-related miRNAs in sugarcane by small RNA sequencing and functional analysis of a cold inducible ScmiR393 to cold stress. Environmental and Experimental Botany, 155, 464–476. https://doi.org/10.1016/j.envexpbot.2018.07.030 [Google Scholar] [CrossRef]
57. Zhang, Q., Chen, Q. H., Wang, S. L., Hong, Y. H., Wang, Z. L. (2014). Rice and cold stress: Methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice, 7(24), 1–12. https://doi.org/10.1186/s12284-014-0024-3 [Google Scholar] [PubMed] [CrossRef]
58. Komatsu, S., Yang, G. X., Khan, M., Onodera, H., Toki, S. (2007). Over-expression of calcium-dependent protein kinase 13 and calreticulin interacting protein 1 confers cold tolerance on rice plants. Molecular Genetics and Genomics, 277(6), 713–723. https://doi.org/10.1007/s00438-007-0220-6 [Google Scholar] [PubMed] [CrossRef]
59. Huang, G. T., Ma, S. L., Bai, L. P., Zhang, L., Ma, H. et al. (2012). Signal transduction during cold, salt, and drought stresses in plants. Molecular Biology Reports, 39(2), 969–987. https://doi.org/10.1007/s11033-011-0823-1 [Google Scholar] [PubMed] [CrossRef]
60. Chinnusamy, V., Zhu, J. H., Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends in Plant Science, 12(10), 1360–1385. https://doi.org/10.1016/j.tplants.2007.07.002 [Google Scholar] [PubMed] [CrossRef]
61. Miura, K., Furumoto, T. (2013). Cold signaling and cold response in plants. International Journal of Molecular Sciences, 14(3), 5312–5337. https://doi.org/10.3390/ijms14035312 [Google Scholar] [PubMed] [CrossRef]
62. Kidokoro, S., Kim, J. S., Ishikawa, T., Suzuki, T., Yamaguchi-Shinozaki, K. (2020). DREB1A/CBF3 is repressed by transgene-induced DNA methylation in the Arabidopsis ice1-1 mutant. The Plant Cell, 32(4), 1035–1048. https://doi.org/10.1105/tpc.19.00532 [Google Scholar] [PubMed] [CrossRef]
63. Fursova, O. V., Pogorelko, G. V., Tarasov, V. A. (2009). Identification of ICE2, a gene involved in cold acclimation which determines freezing tolerance in Arabidopsis thaliana. Gene, 429(1–2), 98–103. https://doi.org/10.1016/j.gene.2008.10.016 [Google Scholar] [PubMed] [CrossRef]
64. Ruelland, E., Vaultier, M. N., Zachowski, A., Hurry, A. V. (2009). Cold signalling and cold acclimation in plants. Advances in Botanical Research, 49, 36–149. https://doi.org/10.1016/S0065-2296(08)00602-2 [Google Scholar] [CrossRef]
65. Novillo, F., Alonso, J. M., Ecker, J. R., Salinas, J. (2004). CBF2/DREB1C is a negative regulator of CBF1/DREB1B and CBF3/DRE1A expression and plays a central role in stress tolerance in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 101(11), 3985–3990. https://doi.org/10.1073/pnas.0303029101 [Google Scholar] [PubMed] [CrossRef]
66. Novillo, F., Medina, J., Salinas, J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proceedings of the National Academy of Sciences of the United States of America, 104(52), 21002–21007. https://doi.org/10.1073/pnas.0705639105 [Google Scholar] [PubMed] [CrossRef]
67. Dai, X. Y., Xu, Y. Y., Ma, Q. B., Xu, W. Y., Wang, T. et al. (2007). Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought, and salt stress in transgenic Arabidopsis. Plant Physiology, 143, 1739–1751. https://doi.org/10.1104/pp.106.094532 [Google Scholar] [PubMed] [CrossRef]
68. Su, C. F., Wang, Y. C., Hsieh, T. H., Lu, C. A., Tseng, T. H. (2010). A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiology, 153(1), 145–158. https://doi.org/10.1104/pp.110.153015 [Google Scholar] [PubMed] [CrossRef]
69. Yang, A., Dai, X., Zhang, W. H. (2012). A R2R3-type MYB gene, OsMYB2, is involved in dehydration tolerance in rice. Environmental And Experimental Botany, 63, 2541–2556. https://doi.org/10.1093/jxb/err431 [Google Scholar] [PubMed] [CrossRef]
70. Zhu, Z. X., Liu, Y., Liu, S. J., Mao, C. Z., Wu, Y. R. (2012). A gain-of function mutation in OsIAA11 affects lateral root development in rice. Molecular Plant, 5(1), 154–161. https://doi.org/10.1093/mp/ssr074 [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials

Supplementary Figure 1: Gravitropic response of the wild-type plant zh11 and the lrl3 T2 generation. (A) and (B) are 12 h and 24 h primary root horizontal phenotypes of two-day-old seedlings, respectively. On the left are the wild-type seedlings zh11, and on the right are lrl3 T2 generation lines (arrows indicate lrl3 mutant, and others are wild-type individuals)

Supplementary Figure 2: Protein alignment between rice and other organisms with cyclophilin_ABH_like domain. AtCYP19-2, A. thaliana (gi: 98960923); BnCYP, Brassica napus (gi: 1345921); DlCYP, Digitalis lanata (gi: 1563719); hCYPA, Human sapiens (gi: 13543666); KCCYP1, Kandelia candel (gi: 37722431); LeCYP1, Lycopersicon esculentum (gi: 170439); OsCYP1, Oryza sativa (gi: 600764); OsCYP2, Oryza sativa (gi: 600768); Cpr1, S. cerevisiae (gi: 6320359); SSCCYP1, Solanum commersonii (gi: 1928938); StCyP, Solanum tuberosum (gi: 62529356); ThCYP1, Thellungiella halophila (gi: 38708271); TaCYP1, Triticum aestivum (gi: 13925734); CYP/ROT1, Zea mays (gi: 118104)

Supplementary Figure 3: Semi-quantitative RT-PCR of OsPIN10a in Kasalath and Oscyp2-2. WT and MT are seedling roots of Kasalath and Oscyp2-2 under normal condition (control) and 10 μM IAA (IAA (3h)) for 3 h

Supplementary Figure 4: Quantitative RT-PCR expression in roots of Kasalath and Oscyp2-2. WT-CK and MT-CK are seedling roots of Kasalath and Oscyp2-2 under normal condition, WT-IAA and MT-IAA are seedling roots of Kasalath and Oscyp2-2 under 10 μM IAA for 3 h

Supplementary Figure 5: The vector of 35S-pCAMBIA1300

Supplementary Figure 6: The vector of pBWA(V)HU-45001OE-Gus
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools