 Open Access
Open Access
ARTICLE
Intron Retention Fine-Tunes the Resistance of the Rice Mutant pls4 to Rice Sheath Blight (Rhizotonia solani AG I.1a)
Key Laboratory of Crop Physiology, Ecology and Genetic Breeding, Ministry of Education, Jiangxi Agricultural University, Nanchang, 330045, China
* Corresponding Authors: Dahu Zhou. Email: ; Haihui Fu. Email:
(This article belongs to the Special Issue: Plant Omics in Challenging Environment)
Phyton-International Journal of Experimental Botany 2023, 92(7), 2035-2049. https://doi.org/10.32604/phyton.2023.028296
Received 10 December 2022; Accepted 29 March 2023; Issue published 29 May 2023
Abstract
OsPLS4 encodes a β-ketoacyl carrier protein reductase (KAR). The role of OsPLS4 in rice sheath blight (Rhizoctonia solani) remains unclear. Our preliminary studies showed that premature leaf senescence mutants (pls4) were highly susceptive to sheath blight in the early stage of rice development. To explore the role of this gene in the development of rice sheath blight, the transcriptome profiles of the rice pls4 mutant and wild type were compared by RNA-seq. The results revealed 2,569 differentially expressed genes (DEGs). The down-regulated genes were significantly enriched in the defense response-related biological processes. These down-regulated genes included the chitinase genes and WRKY genes, which were significantly changed in pls4 mutants. Furthermore, 467 genes induced significant alternative splicing (AS) events. Among them, intron retention (IR) affected gene expression levels and functions of the vitamin B6 (VB6) metabolism pathway related to sheath blight. This result suggests that IR plays an important role in the sheath blight resistance of mutant pls4. Together, these results indicate that pls4 could be involved in the biological process of sheath blight via DEGs and the fine-tuning of IR. The present study provides a molecular basis for further investigation of the resistance of rice to sheath blight.Keywords
Rhizoctonia solani, one of the three major diseases (including rice blast, bacterial blight and sheath blight) affecting rice, has a significant influence on rice yield [1]. Two innate immune systems, i.e., pattern-triggered immunity (PTI) and effector-triggered immunity (ETI), have evolved in plants to resist exogenous pathogen infection [2–4]. PTI originates from the exposure of pathogen-associated molecular patterns (PAMPs) to pattern recognition receptors (PRRs) on plant cell surfaces. PAMPs contain important components, including bacterial lipopolysaccharides and fungal chitin cell walls, which are evolutionarily conserved [5]. However, the innate ETI response is more intense [6]. R proteins in plant cells recognize effector molecules secreted by pathogens or respond to specific non-toxic Avr proteins [7]. Therefore, changes in the gene expression patterns of the PTI and ETI pathways likely play essential roles in the sheath blight resistance of rice.
PTI is the main early defence response of the indica rice variety and involves activation of pathogenesis-related (PR) gene expression. For example, over-expression of the chitinase gene in indica rice enhances its resistance to sheath blight [8]. The co-expression of chitinase and thaumatin-like proteins in rice confers enhanced resistance to bacterial blight, as compared with lines expressing a single gene [9]. Moreover, transgenic plants with CHI11, TLP and Xa21 show enhanced resistance to sheath blight and bacterial blight [10]. Other reports indicate that the co-expression of the chitinase and β-1,3-glucanase genes enhances the plant’s resistance to sheath blight but also inversely affects rice seed germination [11]. In another study, the rice oxalate oxidase gene driven by a green tissue-specific promoter was found to improve the tolerance of transgenic rice to sheath blight pathogens [12]. Similarly, some reports indicate that green tissue-specific co-expression of the chitinase and oxalate oxidase 4 genes in rice enhances sheath blight resistance [1]. In other work, stable expression of polygalacturonase RNAi constructs in rice was able to efficiently silence AG1IA_04727 and suppress sheath blight [13]. It has been reported that ETI can trigger reactive oxygen species (ROS) burst, hypersensitive response (HR), cyclin-dependent kinase protein (CDKP) and mitogen-activated protein kinase (MAPK) signaling pathways, and related transcription factors initiate gene expression, regulate jasmonic acid (JA), endogenous salicylic acid (SA) and other disease-resistant hormone genes, thereby promoting resistance to disease infection [4].
It should be noted that the over-expression of genes other than the chitinase gene can also affect the resistance of rice to sheath blight. The over-expression of Dm-AMP1 isolated from Dahlia merckii was found to enhance indica rice resistance to sheath blight and rice blast [14]. A new gene, Bsr2, which encodes an unidentified cytochrome P450 protein belonging to the CYP78A family, has also been found to be related to disease resistance, plant growth rate and seed size of rice [15].
Plant hormones play a vital role in plant growth and development, defence response and secondary metabolism. Ethylene is a hormone with low content in plants. The synthesis of ethylene is significantly enhanced under pressure and over-expression of the ethylene biosynthesis gene OsACS2 in rice confers broad-spectrum resistance to sheath blight and rice blast [16,17]. Transcription factors, such as the WRKY family, can also regulate multiple resistance responses, achieving resistance to rice blast through gene expression and hormonal changes; the expression changes in these transcription factors result from the upstream ETI immune response. Over-expression of WRKY13 in rice plants was found to enhance the resistance to sheath blight [18]. Over-expressions of OsWRKY4 and OsWRKY80 in rice plants can also enhance the expression of disease course-related genes and improve the resistance of rice to sheath blight by activating the JA and ethylene signalling pathways [19]. Plant SA and PR proteins were found to induce plant defence response manifested as a hypersensitive response (HR) and SA accumulation [20]. OsWRKY30 was also found to promote endogenous JA accumulation and PR gene expression, enhancing the disease resistance of rice [21]. Subsequently, a previous study showed that SA treatment inhibits the development of R. solani [22].
At present, RNA-Seq technology has been used to study the changes in gene expression of plants after pathogen infection, providing a theoretical base for studying the genetic mechanism of plant immune defences [23]. For example, comparative analyses of leaf transcriptomes were conducted with a medium-resistant cultivar and a susceptible cultivar at 12, 24, 36, 48, and 72 h after infection with R. solan; 4,802 DEGs were identified, with respiration, JA and phenylpropane metabolism playing important roles in disease resistance [24]. In another study, 32 and 26 modules, respectively, were found in TeQing and Lemont rice by Weighted correlation network analysis (WGCNA) [25]. In a recent study, the molecular responses of the resistant cultivar Yanhui-888 and the susceptible cultivar Jiang-30 to sheath blight were similar, but there were differences in timing and the expression levels of ethylene-sensitive protein 2, the transcription factor WRKY33, and trans-cinnamate 4-monooxygenase [24]. In another study, 177 miRNAs were identified by miRNA-sequencing and bioinformatics; the putative pathogenic miRNA Rhi-milR-16, which negatively regulated target gene expression, and 23 candidate rice miRNAs involved in plant immunity to R. solani, were identified [26]. In another study, 45 differentially expressed proteins (DEPs) were analysed and identified by mass spectrometry; these proteins were associated with stress, resistance and pathogenesis, cell wall metabolism, photosynthesis and cytoskeleton development [27]. The relative proteome and metabolome profiles of untransformed wild-type and AtNPR1 transgenic rice lines before and after R. solani infection were investigated with the results revealing an increased abundance of various metabolites and a new protein, 14-3-3gf14f, which was up-regulated in transgenic rice plant leaves after infection with sheath blight [28]. Through genome-wide association analysis combined with transcriptome data and resistance QTL locus analysis, 653 genes were found to be significantly associated with rice sheath blight resistance; overexpression of OsRSR1 and OsRLCK5 was found to significantly improve the resistance of rice to sheath blight. On the contrary, interference with OsRSR1 and OsRLCK5 was found to significantly reduce the resistance of rice to sheath blight [29].
Previous studies have shown that OsPLS4 (LOC_Os04g30760) encodes a β-ketoacyl carrier protein reductase (KAR) [30]. Gene annotation showed that this gene mainly affected the synthesis of palmitic acid and linolenic acid in vivo, thereby affecting the formation of intima [31]. The knockout mutant showed high temperature sensitivity under heat treatment, but low sensitivity to salt and drought treatment [32]. To date, OsPLS4 has not been reported to be involved in the stress-related process of pathogens. In the current study, The pls4 mutants were constructed the resistance phenotypes of the rice mutants to sheath blight were examined, and their transcriptome changes were compared with the wild-type rice using high-throughput sequencing technology. The expression patterns of DEGs in resistance-related genes and WRKY family factors were analysed and the genome-wide alternative splicing (AS) was analysed in the mutants. This study provides a theoretical scientific basis for interpreting important biological processes involved in plant immune mechanisms under biotic stress, and also offers new gene resources for cultivating novel varieties resistant to sheath blight.
The premature leaf senescence (pls4) mutant utilized in this study was an Ethylmethane sulfonate (EMS) homozygous mutant from ZH11 rice (O. sativa japonica). Its DNA sequence has a single nucleotide substitution (G-A) within the ninth exon of the fourth ORF, LOC_Os04g30760 [32]. All plants were grown at the Nanchang Experimental Base of Jiangxi Agricultural University. At the tillering stage, an appropriate number of leaves were removed and placed in tin foil, then quickly placed in liquid nitrogen. The leaves were transferred to a −80°C refrigerator for future analysis.
The Rhizoctonia solani AG-1-IA strain GD118 was provided by South China Agricultural University. According to the method of Park [33], 5 mm mycelium discs were embedded in the inner side of the top 3rd leaf sheaths of the rice tillers (after 30 days of incubation at 28°C). The inoculation sites were wrapped with aluminium foil immediately after inoculation. Three tillers per plant of each variety were inoculated, for a total of three plants. The temperature was controlled at about 28°C and the humidity was about 90%. After three days, the aluminium foil was removed, and the condition of plants were investigated after 1, 8 days when most plants showed symptoms of infection.
2.2 OsPLS4 Expression Analysis
Transcripts per million (TPM) values of OsPLS4 24, 36, 48 and 72 h after sheath blight infection in resistant (TeQing, T) and susceptible (Lemont, L) varieties of rice are stored under accession number SRP11364 [34].
2.3 RNA Extraction, RNA-Seq Library Construction and Sequencing
RNA was isolated according to the Trizol Reagent kit protocol and purification was performed (Invitrogen). The RNA-seq library was prepared with a TruSeq RNA Sample Preparation Kit v2. Sequencing was completed by Guangzhou GENE NENOVO using an Illumina platform.
Firstly, low-quality reads were removed, leaving only clean reads. The alignment software Tophat2 (2.1.1) was used to align the clean reads to the reference genome of rice and unique alignments were extracted for subsequent analysis. Using the default parameters, Gene Ontology (GO) enrichment analysis of the DEGs was conducted with the online analysis software PlantRegMap. Correlation analysis between biological replicates for each sample was carried out using ggplot2(R function). Then, clustering and principal component analysis of Fragments Per Kilobase per Million (FPKM) values and reads per gene for two samples (three biological replicates per sample) were performed using fviz_dend and DESeq2, respectively. DESeq2 software was used for differential analysis of gene expression between groups, and the False discovery rate (FDR) and log2FC values were used to screen differential genes. The screening conditions were as: FDR < 0.05 and |log2FC| > 2. Finally, a Venn diagram was constructed using the online tool Venny 2.1.0 (http://bioinfogp.cnb.csic.es/tools/venny/).
Analysis of alternative splicing (AS) events was performed using rMATS software (http://rnaseqmats.sourceforge.net/index.html). Taking each comparison group for differential AS analysis as a unit, the types and quantities of AS events that occurred were counted and the expression levels of each type of AS event were then calculated. Splicing events were analyzed by differential analysis (p < 0.01). Specific criteria were used to identify IR events. The deletion of more than two IR events in a gene was conducted. We only used “junction count only” for subsequent analysis, which resulted in only counted reads from the AS regions, i.e., the slashed regions were in the AS event classification graph. The threshold for significant IR events was p-value < 0.05. Compared with the wild type, one type of mutant had increased introns while other mutants had decreased. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis of AS and IR genes was conducted using the online analysis software KOBAS [35].
3.1 Transcriptomic Profile Across Samples
To test the resistance level of the mutant pls4 to sheath blight, the roots of both the mutant and wild-type rice were inoculated with rice husks carrying R. solani mycelia. The mutant pls4 exhibited obvious susceptibility at the seedling stage compared with the wild type (Fig. 1a, Table 1). Further, OsPLS4 was significantly up-regulated at 24, 36, 48 and 72 h after infection with sheath blight in the resistant (TeQing, T) and susceptible (Lemont, L) varieties. The largest increase was observed 24 h after infection (Fig. 1b). This indicates that OsPLS4 plays an important role in rice sheath blight resistance. Thus, we were interested in the function of the pls4 gene in the process of sheath blight resistance.
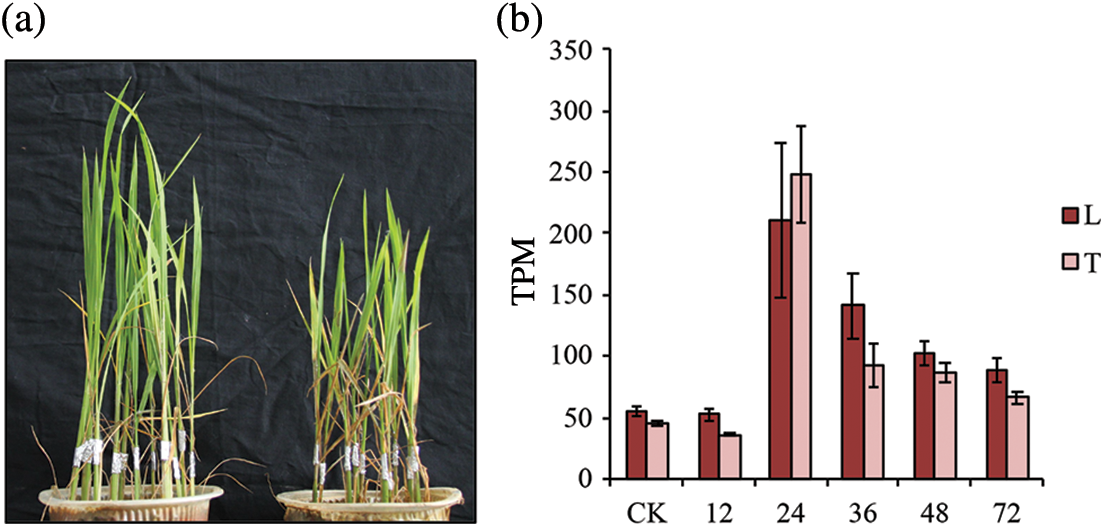
Figure 1: Phenotypic and expression patterns in the mutant pls4. (a) Phenotypic pls4 mutant and wild type after inoculation with R. solani. The foil wrap is where the sheath blight infection started. Left is the wild type (Wt) and right is the pls4 mutant (Mt). (b) The expression pattern of pls4 after inoculation with sheath blight in the resistant (TeQing, T) and susceptible (Lemont, L) varieties. CK is 0 h after infection with sheath blight and 12, 24, 36, 48 and 72 h later. TPM is an abbreviation for transcripts per million and is an algorithm for measuring gene expression levels
3.2 Down-Regulated Genes Involved in Defence Response-Related Biological Processes
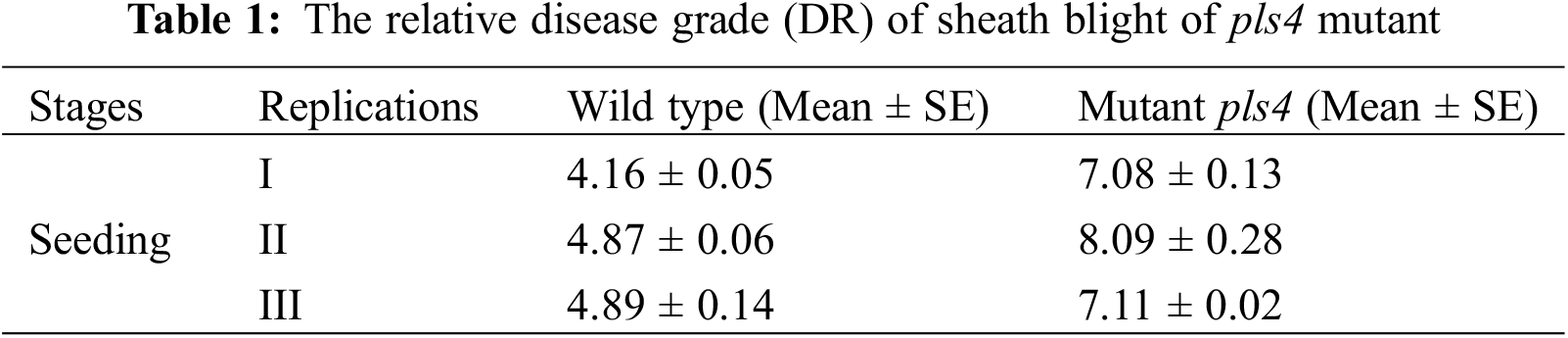
To explore the novel resistance function of pls4, transcriptome sequencing was performed with the mutant and wild-type rice. Six samples from the two sets of data were mapped to the Japonica rice genome. The mapping rate of the reads across six samples was above 90% (Supplemental Table 1). The reads of three biological replicates exhibited good correlations (Supplemental Fig. 1; R ≤ 1, p < 0.05). Moreover, the three biological replicates of each sample were clustered together, and the difference of principal component 2 was set as 99% (Fig. 2). Together, these results suggest that the transcriptome data was of good reliability and repeatability, and was suitable to be used for further analyses.
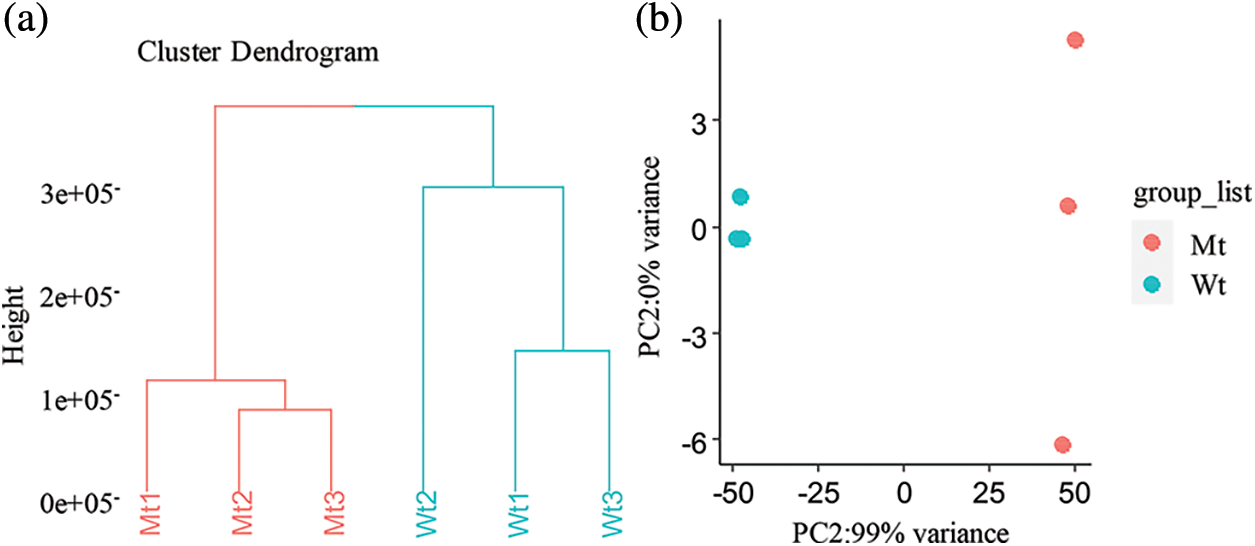
Figure 2: Transcriptomic profile of the rice. (a) Cluster map of DEGs across samples. Mt is the pls4 mutant while Wt is the wild type. (b) PCA analysis of samples
First, the DEGs between the pls4 mutant and wild-type plants were analysed. There were 1,173 up-regulated genes and 1396 down-regulated genes (FDR < 0.001, |log2FC| > 2, p > 0.05, Fig. 3a). Second, the underlying roles of the up- and down-regulated genes in rice sheath blight were investigated, and GO enrichment analysis was conducted for both the up- and down-regulated genes. Interestingly, the down-regulated genes were significantly enriched in fatty acid biosynthesis, secondary metabolism, oxidation-reduction and defence response processes, among others (Fig. 3), and the up-regulated genes were not enriched in the above processes.
Figure 3: Analysis of DEGs between mutant and wild-type rice. (a) Volcano map of DEGs. FDR < 0.05 and |log2FoldChange| > 2. (b) GO analysis of down-regulated genes
3.3 Chitin Was Markedly Down-Regulated in Mutant pls4
There were 2,569 DEGs between the mutant and wild type, and some of the genes functioned in the two innate immune pathways, PTI and ETI. There were significant changes in the expression levels of genes associated with sheath blight resistance, such as chitinase-related genes, pathogen-related genes, and the WRKY gene family (p ≤ 0.05). As a whole, the expression level of chitinase-related genes was down-regulated (Log10(FC) < 0), except for chi 8 and chi11, which were up-regulated (Fig. 4). These results indicate that OsPLS4 affects the expression of several direct effector genes in PTI innate immunity.
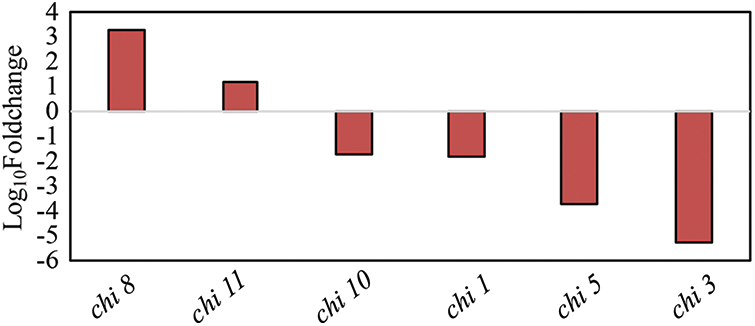
Figure 4: The fold change in the FPKM of the chi gene between the wild type and mutant. FDR < 0.05. FPKM is a measure of relative gene expression after normalization
3.4 Expression Pattern of the WRKY Transcription Factor (TF) Family
Transcription factors can regulate the expression of several stress-related genes during plant stress signal transduction, such as the WRKY TF family. Therefore, the transcription profile of the WRKY gene family was compared between the pls4 mutant and wild type. Very interestingly, the expression level of the WRKY gene family in mutants displayed an overall up-regulated trend in comparison to that in the wild type. According to the heat map of WRKY gene family expression (Q value ≤ 0.05), 20 WRKY genes were significantly up-regulated in the mutant. Some of these WRKY transcription factors were related to rice sheath blight resistance, such as WRKY13, WRKY71 and WRKY30, with the WRKY104 expression level showing the largest change. A small number of WRKY genes, e.g., WRKY125, WRKY33, WRKY74, WRKY76, and WRKY55, were significantly down-regulated, with WRKY125 exhibiting a large variation range (Fig. 5). These results suggest that the expression pattern of WRKY family TFs was changed in the mutant pls4.
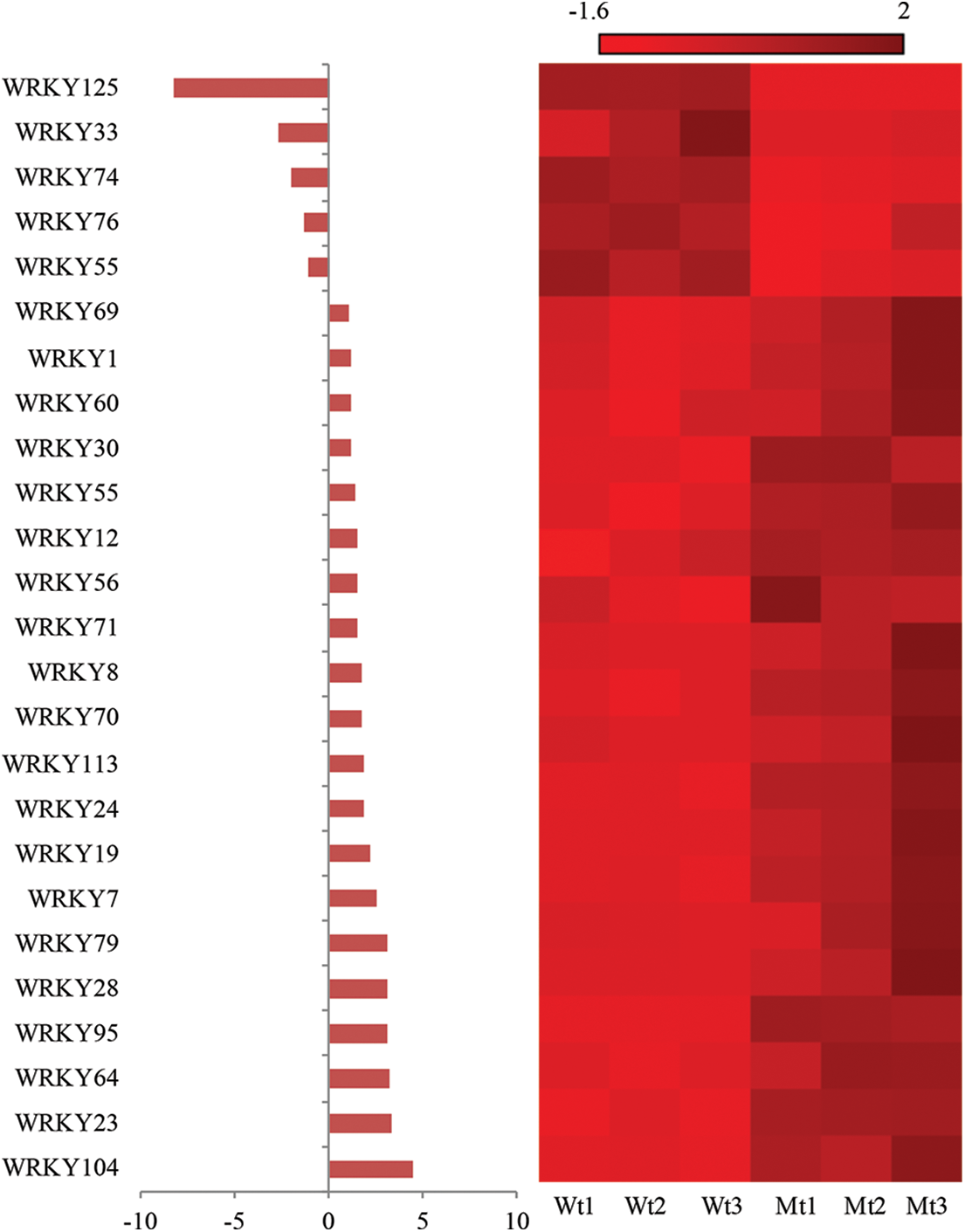
Figure 5: Pattern expression of WRKY family transcription factors. FDR < 0.05 and |log2FoldChange| > 1
3.5 AS Genes Were Significantly Changed in the Mutant pls4
AS is a post-transcriptional regulation mode involved in plant development and resistance. The AS analysis was performed in mutant varieties. The results showed that there were five AS patterns in the mutant, which changed significantly compared with the wild type. AS events occurred in 337 genes of the mutant shown by the current analysis. Among the AS events, Skip Exon (SE) was the main mode (Fig. 6a), accounting for 61.59%, and IR was the second mode, accounting for 16.82%; the other modes accounted for no more than 10%. Then, the potential biological functions of these AS genes were investigated, and KEGG enrichment analysis of these AS genes was performed. The gene enrichment pathways in the different splicing modes were also distinct (p < 0.05). Genes with SE were only enriched in ubiquinone and other terpenoid-quinone biosynthesis biological processes. Genes with mutually exclusive exons (MXE) were enriched in glycosaminoglycan degradation and cutin and suberine and wax biosynthesis. Genes with alternative 3’ splice sites (A3SS) were enriched in aminoacyl-tRNA biosynthesis and inositol phosphate metabolism. Genes with alternative 5’ splice sites (A5SS) were enriched in valine, leucine and isoleucine degradation, fatty acid degradation, proteasome, and fatty acid metabolism. IR genes were enriched in carbon metabolism, VB6 metabolism, amino sugar and nucleotide sugar metabolism, butanoate metabolism, and propanoate metabolism (Fig. 6b). These findings illustrate that different splicing modes perform different biological functions in the mutant pls4. Although the genes with IR were not the most common, the pathways with IR genes exhibited the greatest enrichment, suggesting that IR might have significantly changed in mutants and affected gene expression levels (Fig. 6c). Consequently, the relationships between these IR-producing genes and the corresponding gene expression levels were calculated.
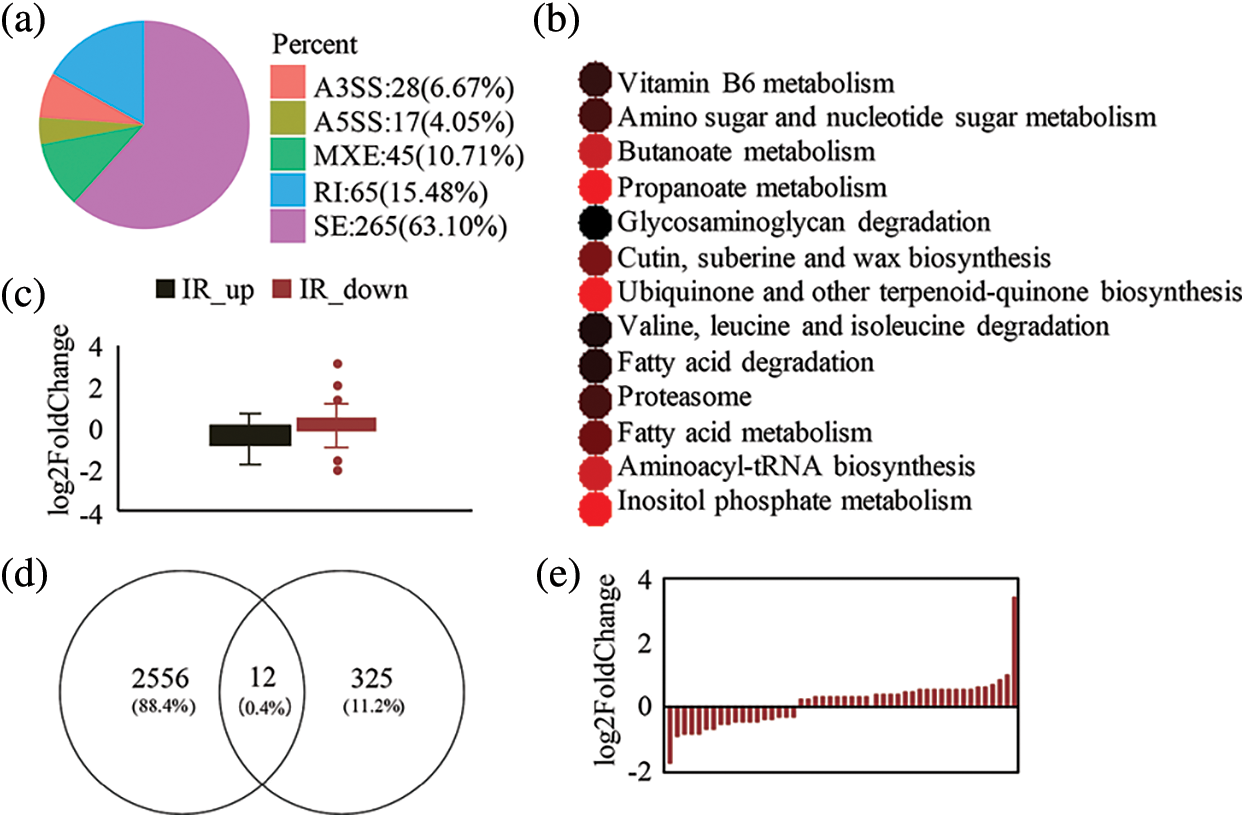
Figure 6: AS analyses between Mt and Wt. (a) Typical AS type statistics. p-value < 0.01. (b) Enrichment analysis of different types of AS pathways. p-value < 0.05. (c) IR gene expression boxplot. (d) Venn analysis of DEGs and AS genes. (e) Pattern of expression of intron splicing (IS) factor
It was well known that AS can affect gene expression, and thus, a Venn diagram was constructed (Fig. 6d). Among the 2,556 DEGs, only 12 genes overlapped with 325 AS genes, which indicates AS eventshad little effect on gene expression levels. Similarly, in the mutant pls4, the enriched KEGG pathways for DEGs differed from those for AS genes. Two genes had IR AS events, two genes had MXE AS events, and eight genes had SE AS events. Then, GO annotation of these genes was performed. The results demonstrated that four of these genes were hypothetical proteins and eight genes were involved in 4Fe-4S ferredoxin, cyclin, oxidoreductase, acyl carrier, peptidase C1A, cellular response to nitrate, transporter-like, and RNA recognition. This suggests that the DEGs and the genes in which AS events presented were involved in multiple biological processes in the mutant pls4.
3.6 IR Affected the Gene Expression Level by Fine-Tuning Regulation and Was Mainly Involved in the VB6 Metabolism Pathway
As an important form of AS, IR is ubiquitous in the post-transcriptional regulation of cellular genes. Most IR results in premature termination codons (PTC) for transcripts and thus, affects gene expression levels [36]. To explore the relationship between changes in IR and the corresponding genes, IR was divided into increased IR and reduced IR in mutants, as compared to the wild type. There was a greater number of reduced IR in mutants than increased IR. Moreover, the gene expression levels with increased IR were globally down-regulated in the mutant pls4, and, in contrast, were up-regulated with reduced IR (Fig. 6d). These findings suggest that IR affects gene expression levels.
At the same time, the DEG enrichment pathways were found to be significantly different from the differential AS gene enrichment pathways. Again, AS had little effect on DEGs. So, what is the function of differential AS genes? To answer this, we investigated the potential roles of several genes in IR-enriched pathways. Very interestingly, Os03g0115500 that a gene involved in the VB6 metabolism pathway and encoding the pyridoxine 5’-phosphate oxidase-related protein, significantly inhibited IR and was slightly up-regulated in the mutant pls4. This suggests that the VB6 content might be affected in the mutant. Os01g0796400 that encodes 3-hydroxy acyl-CoA dehydrogenase was significantly up-regulated. It reduced the IR of mutant pls4 and played a role in β-oxidation and butyrate metabolic pathways OsUXE2 (Os04g0618200), which encodes UDP-xylose epimerase, was significantly up-regulated and reduced IR in the mutant pls4; it was enriched in the amino sugar and nucleotide sugar metabolism pathway. VB6 metabolism and genes related to the metabolic pathways were found to be affected by IR AS events. It is well known that VB6 is an important antioxidant and plays a crucial role in plant stress response and co-expression. Furthermore, the pathogen-derived VB6 gene is crucial in the development of R. solani [37].
Intron splicing (IS) plays an important role in IR events, and the expression pattern of IS factors in the mutant pls4 was detected. Interestingly, IS factors were significantly up-regulated overall in the mutant pls4 (p-value < 0.05; Fig. 6e). This indicates that IR events might be associated with the overall upregulation of this factor in the mutant pls4.
This study investigated transcriptome-level gene changes in the mutant pls4, which is susceptible to sheath blight at the seedling stage. Down-regulated genes were significantly enriched in known biological processes related to defence responses, with most genes enriched in biological processes related to plant immunity. In addition, 467 genes had AS events, and IR, one of the AS events, affected the corresponding gene expression levels in the mutant pls4. Gene-enriched pathways were associated with sheath blight resistance.
One of the most important findings in this research is that down-regulated genes in pls4 mutants were significantly enriched in defence biological processes such as the oxidation-reduction process and defence response. This suggests that there is significant down-regulation of genes in these pathways related to rice sheath blight resistance, which might be the main reason for the early susceptibility of these mutants to sheath blight. Cytochrome P450 (Os06g0599200), which acts upon the defence response, exhibited 380-fold down-regulation in the mutant pls4. A previous study demonstrated that overexpression of this gene enhances resistance to rice sheath blight [15]. Thus, the significant down-regulation of this gene might be an important reason for the susceptibility of the mutant pls4 to sheath blight. According to reports, cytochrome P450 is involved in the biosynthesis of the plant diterpenoid hormone gibberellin [38]. Interestingly, the encoding gene of ent-kaurene synthase was significantly down-regulated in the mutant pls4 and was involved in the biological process of defence response. Similarly, BAK1 (Os06g0274500) showed distinct down-regulation in mutant pls4 and was also enriched in the defence response. The brassinosteroid mutant bri1-D is more susceptible to sheath blight [39]. Pathogenesis-related (PR) genes are direct response genes that respond to the invasion of external pathogens in plants [40,41]. Here, chitinase-related genes in the plant PTI immune pathway were mainly down-regulated, which is in contrast to previous research results where chitinase proteins were found to play a role in plant disease resistance. Chitinase5 (Os04g0494100) involved in defense response was also significantly down-regulated in this study. Previous studies have illustrated that PR-5 gene overexpression in rice enhances the plant’s resistance to sheath blight [42]. JIOsPR10 (Os03g0300400), which encodes the jasmonate inducible pathogenesis-related class 10 protein, was also significantly down-regulated in the mutant pls4 and involved in the defence response. Although no studies have reported that this gene may be involved in sheath blight resistance, it is related to the occurrence of rice blast [18]. Peroxiredoxin (Os02g0192700), which encodes an antioxidant protein, was also down-regulated in pls4, but was up-regulated during sheath blight infection [43]. This indicates that the down-regulation of defence response-related genes in the mutant pls4 confers the main susceptibility to sheath blight.
Combined with the pls4 phenotype, the differential changes in the above genes seem to be well explained. In our previous study, OsPLS4 was found to be involved in the fatty acid synthesis and the regulation of cuticular wax synthesis in rice. Plants need wax as the first protective layer to resist various biotic and abiotic stresses. A base mutation in OsPLS4 disrupts the wax synthesis pathway, alters cuticle synthesis and leaf senescence in pls4 mutants, and confers sensitivity to chilling stress. Therefore, defence-related genes are significantly down-regulated in pls4 mutants, and many factors contribute to the susceptibility of the mutants to sheath blight. The above analysis indicates that OsPLS4 has important potential value in modifying rice agrologic traits.
This study found significant upregulation of the WRKY transcription factor family in the mutant rice variety. Thousands of genes involved in innate immunity, including ETI and PTI, exist in plants and interact with each other to jointly regulate multiple anti-stress biological processes, including the WRKY transcription factor family. Previous studies have reported that three of these WRKY families are associated with fungal pathogens, e.g., Magnaporthe oryzae and Rhizoctonia solani [21,24]. For example, overexpression of WRKY30 in transgenic rice enhanced the plant’s resistance to sheath blight by activating JA-related gene biosynthesis and increasing endogenous JA accumulation [21]. Moreover, WRKY80 was found to grant sheath blight resistance by activating WRKY4 and the JA/ET pathway [19]. Note that these WRKY family genes were found to be significantly up-regulated in the non-infection period of R. solani in the mutant pls4; however, early-stage resistance to rice sheath blight in pls4 was not conferred. It is speculated that the main reason for this might be that the up-regulation of these genes was not large enough compared with the overexpression of WRKY TFs in transgenic rice plants.
The other important finding in this study was that IR events affected the expression levels of the corresponding genes in the mutant pls4, and genes with IR were involved in VB6 metabolism. AS is a process by which pre-mRNA can be cleaved to produce different proteins. It has been shown to be a key post-transcriptional regulatory mechanism during plant development and stress. A recent study reported that the tomato Russet Burbank is sensitive to R. solani, perhaps due to a reduction in VB6 content [44], and the VB6 salvage pathway seemed to be involved in the oxidative stress response to R. solani [45]. The above studies suggest that VB6 metabolism is an important emergency response to R. solani infection in plants. Further, R. solani infection in potato significantly activates both fungal and de novo VB6 genes in both the host and the pathogen [46]. Interestingly, previous studies have shown that there are three VB6 genes with IR events in R. solani AG3, and different intron numbers exist in different VB6 de novo biosynthetic genes. [47,48]. This suggests that IR events are widespread in the VB6 metabolic pathway in hosts and pathogens. Os05g0157300 homologous gene affects 4Fe-4S ferredoxin. Silencing of this gene can lead to necrotic lesions with typical cell death symptoms and ROS production in leaves, which enhances tomato resistance to Pseudomonas syringae PV. [32]. This also implies that some genes might be involved in mutant resistance through AS and differential expression. Overall, the expression levels of genes with reduced IR tended to be up-regulated in the mutant pls4 and vice versa; however, the expression levels of individual IR genes were not significantly altered. Moreover, only 12 DEGs had AS events, of which, only two genes had IR. This indicates that the IR event is a fine-tuning regulation method, which is different from gene transcription inheritance and produces low protein yield. Therefore, IR may play a fine-tuning role in the sensitivity of R. solani. In summary, pls4 mutations and AS events were found to be significantly enriched in the VB6 metabolic pathway in pls4 mutants. It is hypothesized that a base mutation of OsPLS4 may cause the occurrence of IR events in VB6 metabolic pathway genes, which indirectly leads to a reduction in VB6 in the pls4 mutant. This may also fine-tune the susceptibility of pls4 mutants to R. solani.
The breeding and application of disease-resistant rice varieties are the most economical and effective means to prevent and control sheath blight. However, there are no reports of rice resources with high resistance or immunity to sheath blight, and the main disease-resistance genes are unknown. This study identified susceptible gene resources in pls4 mutants from high-throughput data. This approach not only allowed for the analysis of gene expression changes, like traditional transcriptomics, but also revealed that IR in AS can fine-tune the susceptibility of pls4 mutants to R. solani at the post-transcriptional regulatory level. The findings of this study not only provide genetic resources for rice molecular breeding but also offer a theoretical foundation for the study of plant AS.
Acknowledgement: We are very grateful to Professor Ting Ni of Fudan University for his suggestions on the IR data analysis and we also thank other laboratory members for their discussions and suggestions.
Funding Statement: This work was supported by a grant from the Double Thousand Plan of Jiangxi Province (No. jxsq2019101057) and the National Natural Science Foundation of China (No. 3196150349) to Haihui Fu and Xu Jie, respectively.
Author Contributions: Haihui Fu and Dahu Zhou conceived the research ideas and wrote the paper, Dahu Zhou provided experimental materials, and Jiamin Hu, Shaochun Liu, Xu Jie, Junru Fu and Haohua He participated in data analysis and paper writing.
Availability of Data and Materials: The raw data from this study have been submitted to NCBI’s GEO database and have been deposited in the NCBI under Accession Number GSE203661.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Karmakar, S., Molla, K. A., Chanda, P. K., Sarkar, S. N., Datta, S. K. et al. (2016). Green tissue-specific co-expression of chitinase and oxalate oxidase 4 genes in rice for enhanced resistance against sheath blight. Planta, 243(1), 115–130. https://doi.org/10.1007/s00425-015-2398-x [Google Scholar] [PubMed] [CrossRef]
2. Schreiber, K. J., Baudin, M., Hassan, J. A., Lewis, J. D. (2016). Die another day: Molecular mechanisms of effector-triggered immunity elicited by type III secreted effector proteins. Seminars in Cell Developmental Biology, 56, 124–133. https://doi.org/10.1016/j.semcdb.2016.05.001 [Google Scholar] [PubMed] [CrossRef]
3. Li, S., Peng, X., Wang, Y., Hua, K., Xing, F. et al. (2019). The effector AGLIP1 in Rhizoctonia solani AG1 IA triggers cell death in plants and promotes disease development through inhibiting PAMP-triggered immunity in Arabidopsis thaliana. Frontiers in Microbiology, 10, 2228. https://doi.org/10.3389/fmicb.2019.02228 [Google Scholar] [PubMed] [CrossRef]
4. Liu, W., Liu, J., Ning, Y., Ding, B., Wang, X. et al. (2013). Recent progress in understanding PAMP-and effector-triggered immunity against the rice blast fungus Magnaporthe oryzae. Molecular, Plant, 6(3), 605–620. https://doi.org/10.1093/mp/sst015 [Google Scholar] [PubMed] [CrossRef]
5. Liu, W., Liu, J., Triplett, L., Leach, J. E., Wang, G. L. (2014). Novel insights into rice innate immunity against bacterial and fungal pathogens. Annual Review of Phytopathology, 52, 213–241. https://doi.org/10.1146/annurev-phyto-102313-045926 [Google Scholar] [PubMed] [CrossRef]
6. Cui, H., Tsuda, K., Parker, J. E. (2015). Effector-triggered immunity: From pathogen perception to robust defence. Annual Review of Plant Biology, 66, 487–511. https://doi.org/10.1146/annurev-arplant-050213-040012 [Google Scholar] [PubMed] [CrossRef]
7. Lin, W., Anuratha, C. S., Datta, K., Potrykus, I., Muthukrishnan, S. et al. (1995). Genetic engineering of rice for resistance to sheath blight. Bioresouce Technology, 13(7), 686–691. https://doi.org/10.1038/nbt0795-686 [Google Scholar] [CrossRef]
8. Datta, K., Tu, J., Oliva, N., Ona, I., Velazhahan, I. et al. (2001). Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Science, 160(3), 405–414. https://doi.org/10.1016/s0168-9452(00)00413-1 [Google Scholar] [PubMed] [CrossRef]
9. Richa, K., Tiwari, I. M., Devanna, B. N., Botella, J. R., Sharma, V. et al. (2017). Novel chitinase gene LOC_Os11g47510 from Indica rice tetep provides enhanced resistance against sheath blight pathogen Rhizoctonia solani in rice. Frontiers in Plant Science, 8, 596. https://doi.org/10.3389/fpls.2017.00596 [Google Scholar] [PubMed] [CrossRef]
10. Maruthasalam, S., Kalpana, K., Kumar, K. K., Loganathan, M., Poovannan, K. et al. (2007). Pyramiding transgenic resistance in elite indica rice cultivars against the sheath blight and bacterial blight. Plant Cell Reports, 26(6), 791–804. https://doi.org/10.1007/s00299-006-0292-5 [Google Scholar] [PubMed] [CrossRef]
11. Mao, B., Liu, X., Hu, D., Li, D. (2014). Co-expression of RCH10 and AGLU1 confers rice resistance to fungal sheath blight Rhizoctonia solani and blast Magnorpathe oryzae and reveals impact on seed germination. World Journal of Microbiology and Biotechnology, 30(4), 1229–1238. https://doi.org/10.1007/s11274-013-1546-3 [Google Scholar] [PubMed] [CrossRef]
12. Molla, K. A., Karmakar, S., Chanda, P. K., Ghosh, S., Sarkar, S. N. et al. (2013). Rice oxalate oxidase gene driven by green tissue-specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Molecular Plant Pathology, 14(9), 910–922. https://doi.org/10.1111/mpp.12055 [Google Scholar] [PubMed] [CrossRef]
13. Rao, T. B., Chopperla, R., Methre, R., Punniakotti, E., Venkatesh, V. et al. (2019). Pectin induced transcriptome of a Rhizoctonia solani strain causing sheath blight disease in rice reveals insights on key genes and RNAi machinery for development of pathogen derived resistance. Plant Molecular Biology, 100(1–2), 59–71. https://doi.org/10.1007/s11103-019-00843-9 [Google Scholar] [PubMed] [CrossRef]
14. Jha, S., Tank, H. G., Prasad, B. D., Chattoo, B. B. (2009). Expression of Dm-AMP1 in rice confers resistance to Magnaporthe oryzae and Rhizoctonia solani. Transgenic Research, 18(1), 59–69. https://doi.org/10.1007/s11248-008-9196-1 [Google Scholar] [PubMed] [CrossRef]
15. Maeda, S., Dubouzet, J. G., Kondou, Y., Jikumaru, Y., Seo, S. et al. (2019). The rice CYP78A gene BSR2 confers resistance to Rhizoctonia solani and affects seed size and growth in Arabidopsis and rice. Scientific Reports, 9(1), 587. https://doi.org/10.1038/s41598-018-37365-1 [Google Scholar] [PubMed] [CrossRef]
16. Kessler, A., Baldwin, I. T. (2002). Plant responses to insect herbivory the emerging molecular analysis. Annual Review of Plant Biology, 53(1), 299–328. https://doi.org/10.1146/annurev.arplant.53.100301.135207 [Google Scholar] [PubMed] [CrossRef]
17. Helliwell, E. E., Wang, Q., Yang, Y. (2013). Transgenic rice with inducible ethylene production exhibits broad-spectrum disease resistance to the fungal pathogens Magnaporthe oryzae and Rhizoctonia solani. Plant Biotechnology Journal, 11(1), 33–42. https://doi.org/10.1111/pbi.12004 [Google Scholar] [PubMed] [CrossRef]
18. John Lilly, J., Subramanian, B. (2019). Gene network mediated by WRKY13 to regulate resistance against sheath infecting fungi in rice (Oryza sativaL.). Plant Science, 280, 269–282. https://doi.org/10.1016/j.plantsci.2018.12.017 [Google Scholar] [PubMed] [CrossRef]
19. Peng, X., Wang, H., Jang, J. C., Xiao, T., He, H. et al. (2016). OsWRKY80-OsWRKY4 module as a positive regulatory circuit in rice resistance against Rhizoctonia solani. Rice, 9(1), 63. https://doi.org/10.1186/s12284-016-0137-y [Google Scholar] [PubMed] [CrossRef]
20. Zribi, I., Ghorbel, M., Brini, F. (2021). Pathogenesis related proteins (PRs) from cellular mechanisms to plant defense. Current Protein and Peptide Science, 22(5), 1–17. https://doi.org/10.2174/1389203721999201231212736 [Google Scholar] [PubMed] [CrossRef]
21. Peng, X., Hu, Y., Tang, X., Zhou, P., Deng, X. et al. (2012). Constitutive expression of rice WRKY30 gene increases the endogenous jasmonic acid accumulation, PR gene expression and resistance to fungal pathogens in rice. Planta, 236(5), 1485–1498. https://doi.org/10.1007/s00425-012-1698-7 [Google Scholar] [PubMed] [CrossRef]
22. Kouzai, Y., Kimura, M., Watanabe, M., Kusunoki, K., Osaka, D. et al. (2018). Salicylic acid-dependent immunity contributes to resistance against Rhizoctonia solani, a necrotrophic fungal agent of sheath blight, in rice and Brachypodium distachyon. New Phytologist, 217(2), 771–783. https://doi.org/10.1111/nph.14849 [Google Scholar] [PubMed] [CrossRef]
23. Wang, C., Tariq, R., Ji, Z., Wei, Z., Zheng, K. et al. (2019). Transcriptome analysis of a rice cultivar reveals the differentially expressed genes in response to wild and mutant strains of Xanthomonas oryzae pv. oryzae. Scientific Reports, 9(1), 3757. https://doi.org/10.1038/s41598-019-39928-2 [Google Scholar] [PubMed] [CrossRef]
24. Shi, W., Zhao, S. L., Liu, K., Sun, Y. B., Ni, Z. B. et al. (2020). Comparison of leaf transcriptome in response to Rhizoctonia solani infection between resistant and susceptible rice cultivars. BMC Genomics, 21(1), 245. https://doi.org/10.1186/s12864-020-6645-6 [Google Scholar] [PubMed] [CrossRef]
25. Zhang, J., Zhao, W., Fu, R., Fu, C., Wang, L. et al. (2018). Comparison of gene co-networks reveals the molecular mechanisms of the rice (Oryza sativaL.) response to Rhizoctonia solani AG1 IA infection. Functional Integrative Genomics, 18(5), 545–557. https://doi.org/10.1007/s10142-018-0607-y [Google Scholar] [PubMed] [CrossRef]
26. Lin, R., He, L., He, J., Qin, P., Wang, Y. et al. (2016). Comprehensive analysis of microRNA-Seq and target mRNAs of rice sheath blight pathogen provides new insights into pathogenic regulatory mechanisms. DNA Research, 23(5), 415–425. https://doi.org/10.1093/dnares/dsw024 [Google Scholar] [PubMed] [CrossRef]
27. Prathi, N. B., Palit, P., Madhu, P., R., M., Laha, G. S. et al. (2018). Proteomic and transcriptomic approaches to identify resistance and susceptibility related proteins in contrasting rice genotypes infected with fungal pathogen Rhizoctonia solani. Plant Physiology and Biochemistry, 130, 258–266. https://doi.org/10.1016/j.plaphy.2018.07.012 [Google Scholar] [PubMed] [CrossRef]
28. Karmakar, S., Datta, K., Molla, K. A., Gayen, D., Das, K. et al. (2019). Proteo-metabolomic investigation of transgenic rice unravels metabolic alterations and accumulation of novel proteins potentially involved in defence against Rhizoctonia solani. Scientific Reports, 9(1), 10461. https://doi.org/10.1038/s41598-019-46885-3 [Google Scholar] [PubMed] [CrossRef]
29. Wang, A., Shu, X., Jing, X., Jiao, C., Zheng, A. (2021). Identification of rice (Oryza sativaL.) genes involved in sheath blight resistance via a genome-wide association study. Plant Biotechnology Journal, 19(8), 1553–1566. https://doi.org/10.1111/pbi.13569 [Google Scholar] [PubMed] [CrossRef]
30. Chen, F., Dong, G., Wang, F., Shi, Y., Zhu, J. et al. (2021). A β-ketoacyl carrier protein reductase confers heat tolerance via the regulation of fatty acid biosynthesis and stress signaling in rice. The New Phytologist, 232(2), 655–672. https://doi.org/10.1111/nph.17619 [Google Scholar] [PubMed] [CrossRef]
31. Lim, C. J., Kim, W. B., Lee, B. S., Lee, H. Y., Kwon, T. H. et al. (2010). Silencing of SlFTR-c, the catalytic subunit of ferredoxin: Thioredoxin reductase, induces pathogenesis-related genes and pathogen resistance in tomato plants. Biochemical and Biophysical Research Communications, 399(4), 750–754. https://doi.org/10.1016/j.bbrc.2010.08.016 [Google Scholar] [PubMed] [CrossRef]
32. Zhou, D., Li, T., Yang, Y., Qu, Z., Ouyang, L. et al. (2020). OsPLS4 Is involved in cuticular wax biosynthesis and affects leaf senescence in rice. Frontiers in Plant Science, 11, 782. https://doi.org/10.3389/fpls.2020.00782 [Google Scholar] [PubMed] [CrossRef]
33. Park, D. S., Sayler, R. J., Hong, Y. G., Nam, M. H., Yang, Y. (2008). A method for inoculation and evaluation of rice sheath blight disease. Plant Disease, 92(1), 25–29. https://doi.org/10.1094/PDIS-92-1-0025 [Google Scholar] [PubMed] [CrossRef]
34. Cheng, Y., Li, S., Deng, Q., Wang, S., Zhu, J. et al. (2017). Transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Frontiers in Plant Science, 8, 1422. https://doi.org/10.3389/fpls.2017.01422 [Google Scholar] [PubMed] [CrossRef]
35. Bu, D., Luo, H., Huo, P., Wang, Z., Zhang, S. et al. (2021). KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Research, 49(W1), W317–W325. https://doi.org/10.1093/nar/gkab447 [Google Scholar] [PubMed] [CrossRef]
36. Gui, X., Zhang, P., Wang, D., Ding, Z., Wu, X. et al. (2022). Phytophthora effector PSR1 hijacks the host pre-mRNA splicing machinery to modulate small RNA biogenesis and plant immunity. Plant Cell, 34(9), 3443–3459. https://doi.org/10.1093/plcell/koac176 [Google Scholar] [PubMed] [CrossRef]
37. Jwa, N. S., Kumar Agrawal, G., Rakwal, R., Park, C. H., Prasad Agrawal, V. (2001). Molecular cloning and characterization of a novel Jasmonate inducible pathogenesis-related class 10 protein gene, JIOsPR10, from rice (Oryza sativaL.) seedling leaves. Biochemical and Biophysical Research Communications, 286(5), 973–983. https://doi.org/10.1006/bbrc.2001.5507 [Google Scholar] [PubMed] [CrossRef]
38. Mafu, S., Jia, M., Zi, J., Morrone, D., Wu, Y. et al. (2016). Probing the promiscuity of ent-kaurene oxidases via combinatorial biosynthesis. Proceedings of the National Academy of Sciences of the United States of America, 113(9), 2526–2531. https://doi.org/10.1073/pnas.1512096113 [Google Scholar] [PubMed] [CrossRef]
39. Yuan, P., Zhang, C., Wang, Z. Y., Zhu, X. F., Xuan, Y. H. (2018). RAVL1 activates brassinosteroids and ethylene signaling to modulate response to sheath blight disease in rice. Phytopathology, 108(9), 1104–1113. https://doi.org/10.1094/PHYTO-03-18-0085-R [Google Scholar] [PubMed] [CrossRef]
40. Sridevi, G., Parameswari, C., Sabapathi, N., Raghupathy, V., Veluthambi, K. (2008). Combined expression of chitinase and β-1,3-glucanase genes in indica rice (Oryza sativaL.) enhances resistance against Rhizoctonia solani. Plant Science, 175(3), 283–290. https://doi.org/10.1016/j.plantsci.2008.04.011 [Google Scholar] [CrossRef]
41. Datta, K., Tu, J., Oliva, N., Ona, I. I., Velazhahan, R. et al. (2001). Enhanced resistance to sheath blight by constitutive expression of infection-related rice chitinase in transgenic elite indica rice cultivars. Plant Science, 160(3), 405–414. https://doi.org/10.1016/s0168-9452(00)00413-1 [Google Scholar] [PubMed] [CrossRef]
42. Datta, K., Velazhahan, R., Oliva, N., Ona, I., Mew, T. et al. (1999). Over-expression of the cloned rice thaumatin-like protein (PR-5) gene in transgenic rice plants enhances environmental friendly resistance to Rhizoctonia solani causing sheath blight disease. Theoretical and Applied Genetics, 98(6–7), 1138–1145. https://doi.org/10.1007/s001220051178 [Google Scholar] [CrossRef]
43. Sruthilaxmi, C. B., Babu, S. (2020). Proteome responses to individual pathogens and abiotic conditions in rice seedlings. Phytopathology, 110(7), 1326–1341. https://doi.org/10.1094/PHYTO-11-19-0425-R [Google Scholar] [PubMed] [CrossRef]
44. Samsatly, J., Bayen, S., Jabaji, S. H. (2020). Vitamin B6 is under a tight balance during disease development by Rhizoctonia solani on different cultivars of potato and on Arabidopsis thaliana mutants. Frontiers in Plant Science, 11, 875. https://doi.org/10.3389/fpls.2020.00875 [Google Scholar] [PubMed] [CrossRef]
45. Samsatly, J., Chamoun, R., Gluck-Thaler, E., Jabaji, S. (2016). Genes of the de novo and salvage biosynthesis pathways of vitamin B6 are regulated under oxidative stress in the plant pathogen Rhizoctonia solani. Frontiers in Microbiology, 6, 1429. https://doi.org/10.3389/fmicb.2015.01429 [Google Scholar] [PubMed] [CrossRef]
46. Samsatly, J., Copley, T. R., Jabaji, S. H. (2018). Antioxidant genes of plants and fungal pathogens are distinctly regulated during disease development in different Rhizoctonia solani pathosystems. PLoS One, 13(2), e0192682. https://doi.org/10.1371/journal.pone.0192682 [Google Scholar] [PubMed] [CrossRef]
47. Ehrenshaft, M., Daub, M. E. (2001). Isolation of PDX2, a second novel gene in the pyridoxine biosynthesis pathway of eukaryotes, archaebacteria, and a subset of eubacteria. Journal of Bacteriology, 183(11), 3383–3390. https://doi.org/10.1128/JB.183.11.3383-3390.2001 [Google Scholar] [PubMed] [CrossRef]
48. Benabdellah, K., Azcón-Aguilar, C., Valderas, A., Speziga, D., Fitzpatrick, T. B. et al. (2009). GintPDX1 encodes a protein involved in vitamin B6 biosynthesis that is up-regulated by oxidative stress in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytologist, 184(3), 682–693. https://doi.org/10.1111/j.1469-8137.2009.02978.x [Google Scholar] [PubMed] [CrossRef]
Supplementary Materials
Supplementary Figure 1: The correlation of reads across three biological replicatives
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools