 Open Access
Open Access
ARTICLE
Stigma-Specific Comparative Proteomic Analysis Reveals the Distyly Response to Self-Incompatibility in Plumbago auriculata Lam
1 School of Fine Arts and Calligraphy, Sichuan Normal University, Chengdu, 610000, China
2 Sichuan Certification and Accreditation Association, Chengdu, 610000, China
3 College of Landscape Architecture, Sichuan Agricultural University, Chengdu, 610000, China
4 School of Design, Chongqing Industry Polytechnic College, Chongqing, 401120, China
* Corresponding Author: Shouli Yi. Email:
Phyton-International Journal of Experimental Botany 2024, 93(4), 681-697. https://doi.org/10.32604/phyton.2024.049166
Received 29 December 2023; Accepted 08 March 2024; Issue published 29 April 2024
Abstract
In plants, heteromorphic self-incompatibility (HetSI) is a strategy for avoiding self-pollination and promoting outcrossing, and during this process, numerous protein-protein interaction events occur between the pistil and pollen. Previous studies in Primula and Fagopyrum that focused on HetSI systems have provided interesting insights; however, the molecular mechanism underlying HetSI remains largely unknown. In this study, we profiled the proteome of Plumbago auriculata stigmas before and after self-incompatible (SI) and self-compatible (SC) pollination. Comparative analyses were conducted by 4D-DIA (Four-dimensional data independent acquisition), a promising technology that increases the sensitivity and reduces the spectral complexity of proteomic analysis by adding a fourth dimension, ion mobility. The results revealed 33387 peptides and 5311 proteins in all samples. The pathways in which the differentially expressed proteins (DEPs) identified in the P × P (Pin style self-pollinated with pin pollen) vs. PS (Pin style) and T × T (Thrum style self-pollinated with thrum pollen) vs. TS (Thrum style) comparisons were significantly enriched were biosynthesis of secondary metabolites and pentose and glucuronate interconversions. In the P × T (Pin style cross-pollinated with thrum pollen) vs. PS and T × P (Thrum style cross-pollinated with pin pollen) vs. TS comparison, the top three pathways were biosynthesis of secondary metabolites, pentose and glucuronate interconversions, and phenylpropanoid biosynthesis. The phenylpropanoid biosynthesis, cutin, suberine and wax biosynthesis, and flavonoid biosynthesis pathways were enriched in the P × T vs. P × P comparison, and starch and sucrose metabolism, glycerophospholipid metabolism, and alpha-linolenic acid metabolism were abundant in the T × T vs. T × P comparison. The enriched pathways between PS and TS were the biosynthesis of secondary metabolites, phenylpropanoid biosynthesis, and pentose and glucuronate interconversion. Self-incompatibility protein S1 (SI S1), Mitogen-activated protein kinase 3/4 (MPK3/4), Mitogen-activated protein kinase kinase 2/3 (M2K2/3), Exocyst complex component EXO70A1 (E70A1) and Thioredoxin H1/2 (TRXH1/2) were found to be HetSI-related candidates, and O-fucosyltransferase 23 (OFT23), 3-ketoacyl-CoA synthase 6 (KCS6), Receptor-like protein kinase FERONIA (FERON), Fimbrin-5 (FIMB5), Pollen-specific leucine-rich repeat extensin-like protein 4 (PLRX4), Transcription initiation factor IIB-2 (TF2B2) and Pectinesterase 1 (AL11A), etc., were identified as other regulatory transducers. These findings combined with our morphological and reactive oxygen species (ROS) intensity analyses indicate that P. auriculata has typical dry-stigmas and that the HetSI mechanism might differ between the pin and thrum. SI S1 might be the key factor in HetSI, and ROS are overexpressed during SC pollination to rapidly activate the mitogen-activated protein kinase (MAPK)-mediated phosphorylation of E70A1 to maintain stigma receptivity in plants with HetSI.Keywords
Supplementary Material
Supplementary Material FileSelf-incompatibility (SI) is a mating system characterized by a high level of heterozygosity resulting from the avoidance of inbreeding and overcoming outcrossing [1]. SI can be classified as homomorphic (HomSI) (plants with a single type of floral morphology) or heteromorphic (HetSI) (plants with two or three types of floral morphology) [2]. Based on differences in genetic determination, HomSI can be divided into sporophytic self-incompatibility (SSI, the phenotype is determined by the S genotype of the sporophyte) and gametophytic self-incompatibility (GSI, the phenotype is determined by the haploid genes of the pollen itself) [3]. A study on the SSI of Brassicaceae demonstrated that the SI system is generally controlled by SRK (S-locus receptor kinase) in the papilla cell of the stigma and by SCR/SP11 (S-locus cysteine-rich protein/S-locus protein 11) in the pollen coat [4]. Activated SRK interacts with ARC1 through the phosphorylation of ARC1 (Arm repeat containing 1), and phosphorylated ARC1 then promotes the ubiquitination and degradation of EXO70A1 (Exocyst complex component EXO70A1)/GLO1 (Glyoxalase 1) to inhibit self-pollination [5]. THL1/2 (Thioredoxin h-like proteins) and MLPK (M-locus protein kinase) are also involved downstream of the SI signaling response. MLPK localizes to the papilla cell membrane and interacts with SRK/ARC1 to transduce the SI signal [6]. THLs can inhibit the autophosphorylation of SRK [7]. Moreover, a recent study demonstrated SRK binds to Receptor-like protein kinase FERONIA (FERON)-NADPH oxidases (RBOHs) to further increase reactive oxygen species (ROS) accumulation in pollen and disrupt F-actin (actin filaments) to lead to SI pollen rejection in Brassica [8]. There are two distinct GSI systems, one is based on the SLF (S-locus F-box protein)/S-RNase system. Research on Pyrus pyrifolia has shown that a high level of S-RNase protein selectively inhibits the growth of pollen tubes with SI [9]. Another one is based on PrsS1 (Self-incompatibility protein S1, SI S1)-PrpS1 (Pollen S) system in Papaver with a requirement of GPI-APs (Glycosyl-phosphatidy linositol-anchored proteins). After cognate recognition, ROS was reported to increase to induce programmed cell death (PCD) in SI pollen and prevent self-fertilization [8]. Research on the molecular mechanisms of HetSI has demonstrated that HetSI in both plant types (pin and thrum) shares the same HetSI controller-S locus and exhibits similar pollen-stigma interactions [10]. Studies of HetSI in Averrhoa carambola have demonstrated that proteins specific to the stamen and style exist in both style morphs [11]. However, in Turnera, specific proteins were identified only in short styles [12]. In Fagopyrum esculentum, specific protein groups found in self-pollinated short pistils are believed to play a role in pollen adhesion to the stigma surface or the inhibition of pollen tube elongation in the HetSI response [13]. In Primula, P450 CYP734A50 in thrum morphs can alter female compatibility by repressing brassinosteroid (BR) levels [14]. Moreover, researchers have hypothesized that different flower types might have varied HetSI mechanisms [15]. While HetSI has attracted long-standing attention, molecular-level data remain scarce, and the mechanism of HetSI is still unclear. This research aims to propose new insights at the molecular level of HetSI and provide a reference to help address SI-related reproductive isolation challenges in production and new cultivar breeding in crops and ornamental plants.
Plumbaginaceae contains approximately 650–1000 species and is an ideal system for investigating heterostyly in the presence of various distylous species, including the genera Armeria, Acantholimon, Ceratostigma, Dyerophytum, Goniolimon, Limonium, Limoniastrum, and Plumbago [16]. Plumbago auriculata Lam. is a critical HetSI plant with two floral morphs: pin (flowers with long styles and short stamens) and thrum (flowers with short styles and long stamens). Our previous studies on the morphology and metabolomes of P. auriculata revealed the following: (1) regarding interpollination, pollen germinated within 2 h, the pollen tube reached the ovule within 4 h, and pollen germination and tube growth were faster in thrum styles than in pin flowers; (2) regarding intrapollination, pollen adhesion failed, and SI occurred on the stigma surface. (3) The metabolic results demonstrated that a large amount of energy-related nutrients were present in thrum styles, supporting faster pollen tube growth, which is consistent with our morphological data, furthermore, nutrient deficiency might not cause HetSI [17,18].
In contrast to conventional proteomics methods based on the retention time (RT), the mass-to-charge ratio (m/z), and ion intensity for identifying and quantifying peptides, 4D-DIA (Four-dimensional data independent acquisition) is a promising technology that adds a fourth dimension, ion mobility, to increase the sensitivity and reduce the spectral complexity in proteomics analysis [19]. Here, we performed 4D-DIA comparative stigmatic proteomic analysis, scanning electron microscopy (SEM), and ROS intensity analyses to detect specific proteins involved in the HetSI of P. auriculata to evaluate the abovementioned studies and further elucidate the interaction between pollen and the stigma in HetSI.
Seeds of Plumbago auriculata Lam. were imported from PanAmerica Seeds company (U. S.) and plants were cultivated in the greenhouse of Sichuan Normal University, Sichuan, China, with an average temperature of 30°C during the day and 25°C at night, alongside a 14-h photoperiod. Pin and thrum flowers (50 plants each) were cultivated separately to avoid pollinating insects. The flowers were randomly emasculated and bagged before dehiscence for self- and cross-pollination. Fresh stigmas were collected 2 h after artificial pollination at 10:00 AM, and styles (50 mg) were collected for each sample. The following samples were used in the analyses (Fig. 1): PS, pin style; TS, thrum style; P × P, pin stigma pollinated with pin pollen; P × T, pin stigma pollinated with thrum pollen; T × T, thrum stigma pollinated with thrum pollen; and T × P, thrum stigma pollinated with pin pollen. The samples for protein extraction were maintained in liquid nitrogen, and fresh styles were prepared for SEM and ROS intensity analyses [17,20,21].
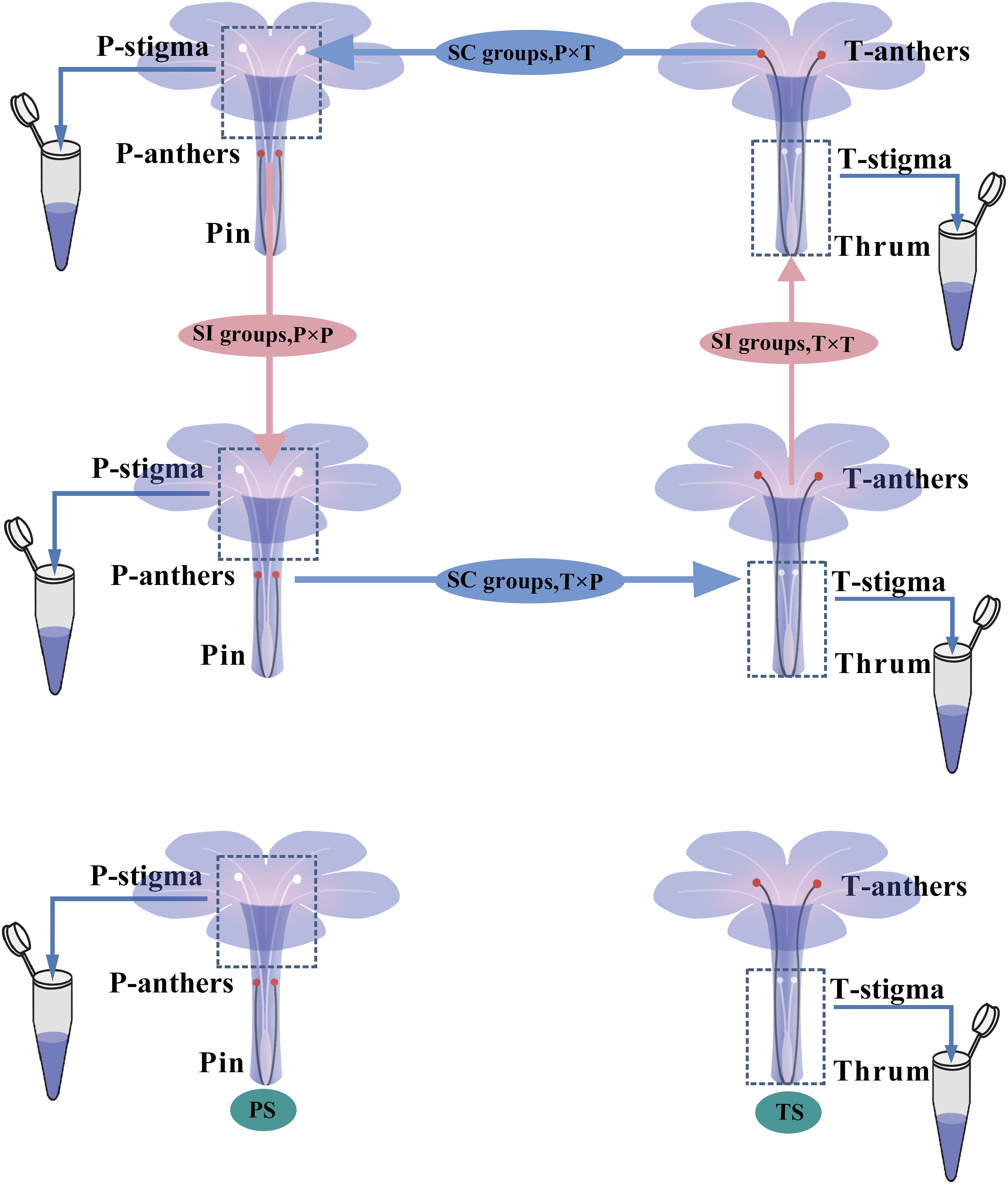
Figure 1: Pin and thrum of P. auriculata and pollination method for 4D-DIA, SEM and ROS intensity analyses. SI groups: P × P, pin stigma pollinated with pin pollen; and T × T, thrum stigma pollinated with thrum pollen. Self-compatibility (SC) groups: P × T, pin stigma pollinated with thrum pollen; and T × P, thrum stigma pollinated with pin pollen. Unpollinated groups: PS, unpollinated pin style; TS, unpollinated thrum style. Styles within the dashed line were used for 4D-DIA, SEM and ROS intensity analyses
2.2 Protein Extraction and Digestion
The samples were ground into powder, and extracts were obtained via lysis buffer containing 1.5% sodium dodecyl sulfate (SDS) and 100 mM Tris(hydroxymethyl)methyl aminomethane-HCl (Tris-Cl). The mixture was subsequently centrifuged for 10 min at 7100 × g at 4°C, and the supernatant was mixed with chilled 8 M urea and 100 mM Tris-Cl. This step was followed by reduction and alkylation with 5 mM dithiothreitol (DTT) for 1 h at 55°C. After the samples were cooled on ice to room temperature, treatment with 10 mM iodoacetamide (IAA) was conducted for 30 min in darkness at room temperature. Afterward, the samples were diluted by adding 100 mM Tris-Cl to a urea concentration of less than 2 M. The enzymolysis diluent was added at a protein: enzyme ratio of 50:1 (m/m), and the mixture was subsequently incubated for digestion at 37°C for 12 h. The enzymatic reaction was stopped by the addition of trifluoroacetic acid (TFA). After centrifugation, the digested samples were desalted on a Sep-Pak C18 column (Waters, U.S.), dried, and stored at −20°C.
All analyses were performed with a TimsTOF Pro2 (Bruker, USA) mass spectrometer equipped with an UltiMate 3000 RSLCnano (Thermo Fisher Scientific, USA). The samples were loaded on a trap column (75 μm × 20 mm, particle size of 2 μm, pore size of 100 Å; Thermo Fisher Scientific, USA), resolved and separated on an analytical column (75 μm × 250 mm, particle size of 1.6 μm, pore size of 100 Å, IonOpticks, Australia) over a 60-min organic gradient by mixing solvent A (0.1% formic acid in H2O) and solvent B (0.1% formic acid in acetonitrile). The flow rate was 300 nL/min. Peptides were ionized by CaptiveSpray nanoelectrospray ionization. MS analysis was conducted with the diaPASEF method and TIMS scans.
DIA-NN (v.1.8) was utilized for diaPASEF analysis, configured to perform an ORF prediction on our Plumbago auriculata de novo transcriptome assembly (30763 entries). The software, in its library-free mode, alongside MBR quantification, facilitated the normalization of protein levels. The search parameters included carbamidomethyl (C) as a fixed modification and oxidaton (M) along with protein N-terminal acetylation as variable modifications. Analyses were conducted adhering to a false discovery rate of 1%. The MaxLFQ algorithm was implemented to normalize protein intensities. The R programming language was utilized for subsequent bioinformatics analysis. Significantly differentially expressed proteins (DEPs) identified based on a p-value < 0.05 and fold changes > 1.5 or < 0.67 (1/1.5).
To study the functions of the obtained proteins, we determined the functions of the proteins by Gene Ontology (GO) and Cluster of Orthologous Groups of Proteins (COG) enrichment analyses. Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis was used to investigate the biological pathways involved.
2.5 Scanning Electron Microscopy (SEM)
An FEI Quanta 250 microscope (FEI, USA) was used to observe the pollen and stigma polymorphisms. Fresh samples of pin and thrum styles and pollen were collected. For observation, the pressure was set to 150 Pa in a low vacuum to prevent stigma deformation. A higher vacuum was set to 3.13 × 10−3 Pa with a conductive coating for pollen, and the tension was set to 12.5 kV for all the samples. Each experiment was replicated three times.
For analysis of the stigmatic ROS levels, DCFH-DA (2,7-Dichlorodihydrofluorescein diacetate) staining was conducted. The samples were soaked in PBS buffer, stained with 50 μM DCFH-DA for 2 h, and washed at least 5 times before observation. A Leica Stellaris 5 system (Leica, Germany) was used for imaging. The prepared samples were observed based on GFP. Analysis with Image Plus 6.0 and SPSS 27.0 was carried out to quantify the ROS concentration in the stigma. Each sample was replicated three times.
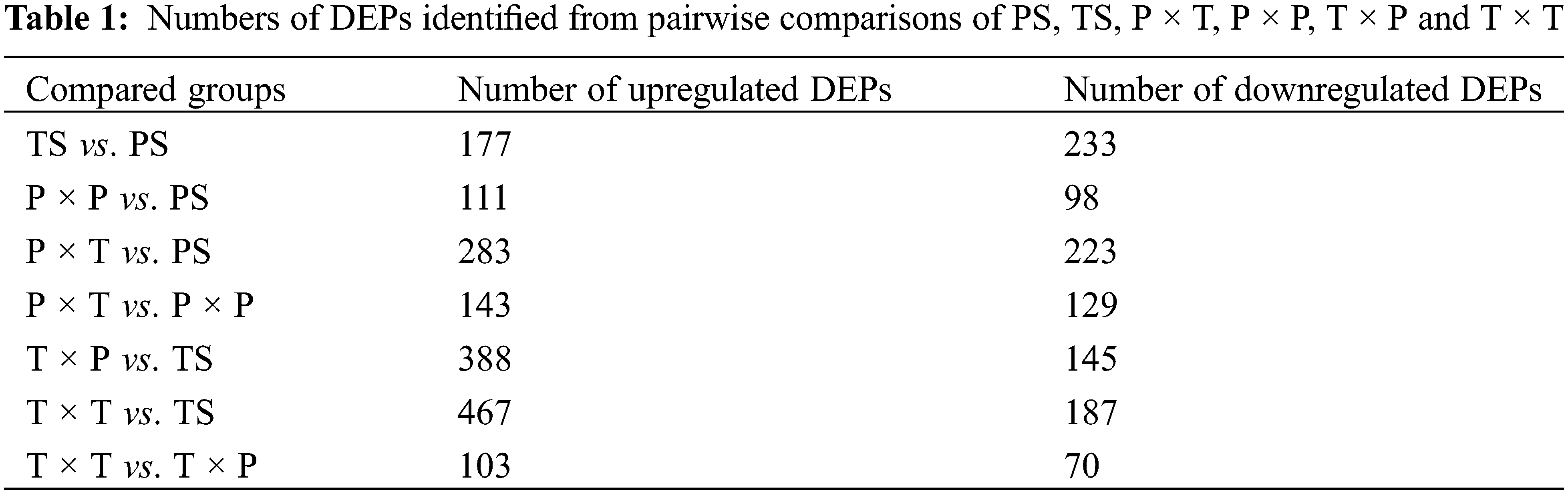
The 4D-DIA proteomic analysis assessed 33387 peptides and 5311 proteins (Fig. 2a). Cutoffs of p-value < 0.05 and fold changes > 1.5 for upregulation or < 0.67 (1/1.5) for downregulation were used for the identification of differentially expressed proteins (DEPs) (Fig. 2b). Within the styles (PS vs. TS), 177 upregulated and 233 downregulated DEPs were identified; the P × P vs. PS comparison identified 111 upregulated and 98 downregulated DEPs, the P × T vs. PS comparison revealed 283 upregulated and 223 downregulated DEPs, the T × P vs. TS comparison identified 388 upregulated and 145 downregulated DEPs, and the T × T vs. TS comparison identified up to 467 upregulated DEPs and 187 downregulated DEPs. The P × T vs. P × P revealed 143 upregulated DEPs, which was greater than the number of downregulated DEPs, and in the T × T vs. T × P comparison, the number of upregulated DEPs was greater than the number of downregulated DEPs (Table 1).
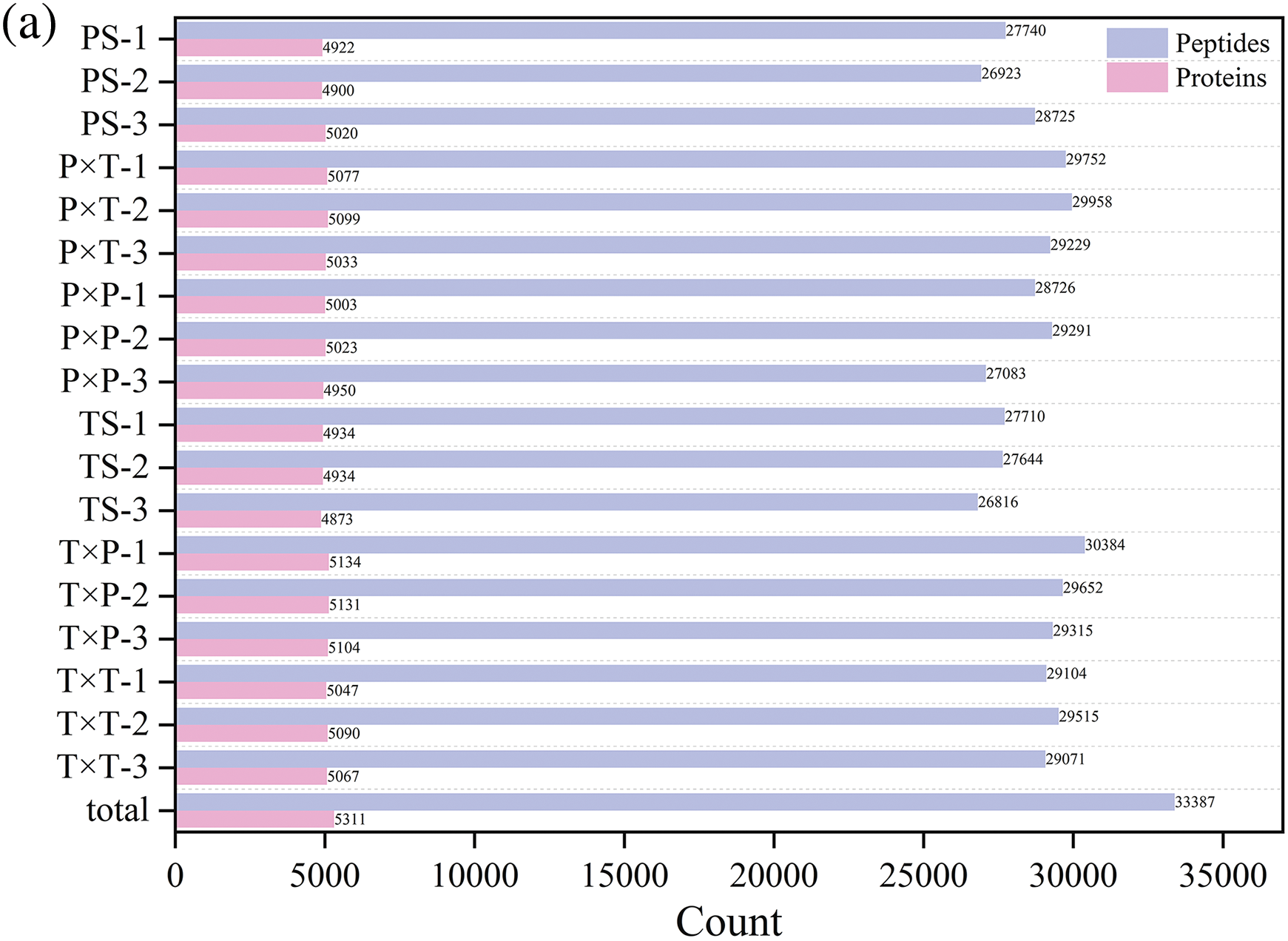
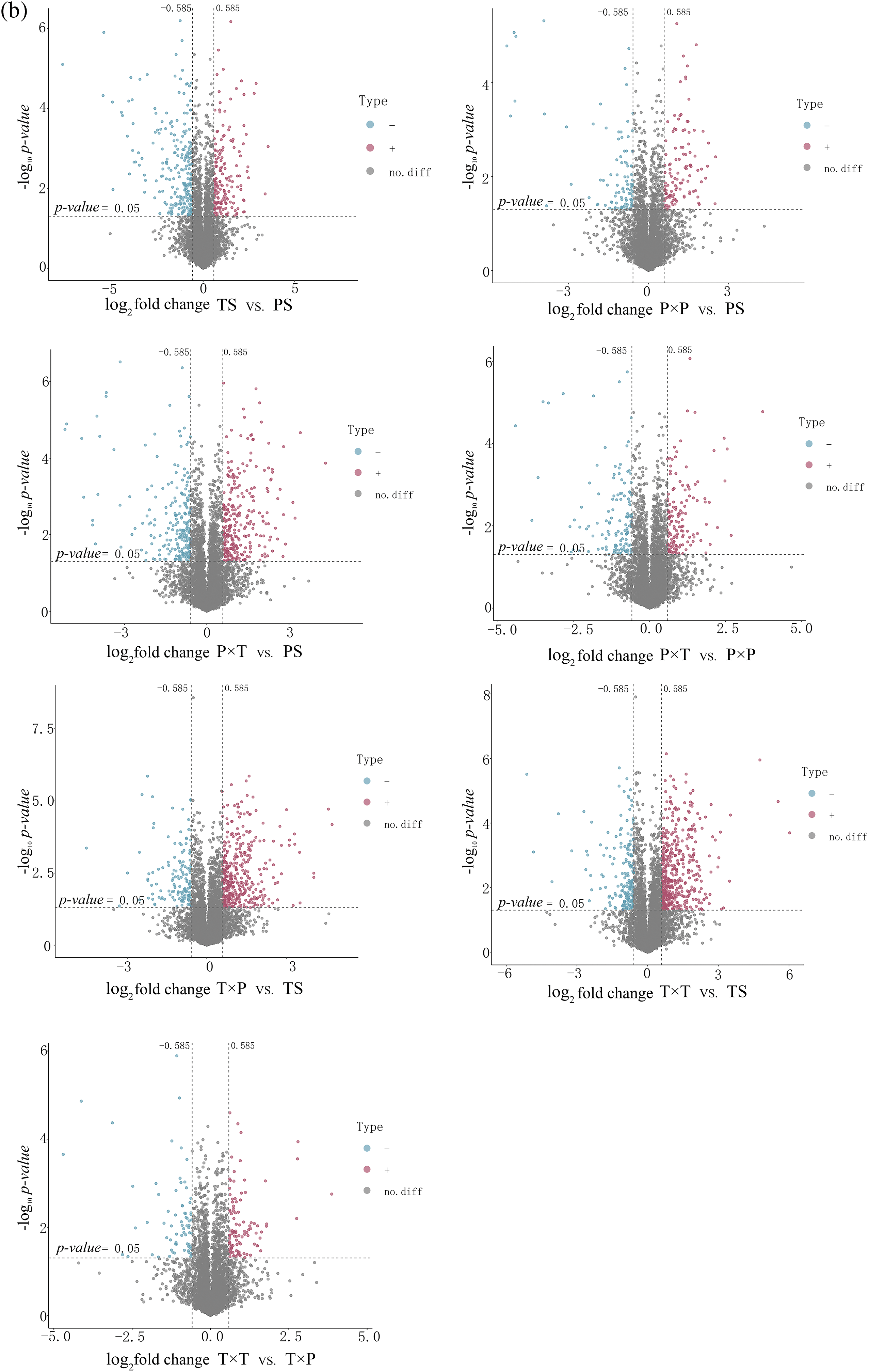
Figure 2: (a) Number of peptides and proteins in unpollinated and pollinated styles. (b) Volcano plots of DEPs in unpollinated and pollinated styles
All DEPs were annotated by GO, COG, and KEGG analyses, and the number of DEPs associated with each term is shown in Table 2. The DEPs were annotated with biological process (BP), cellular component (CC), and molecular function (MF) terms in the GO analysis. According to the BP data, the fertilization-related DEPs in the unpollinated vs. pollinated styles (T × T vs. TS, P × P vs. PS, T × P vs. TS, and P × T vs. PS) were involved in the “response to stimulus”, “reproduction”, “reproductive processes”, “growth” and “interspecies interactions between organism”. The DEPs identified from both comparisons of thrum flower (T × P vs. TS and T × T vs. TS) were annotated with the terms “NADP metabolic process”, “phenylpropanoid metabolic process” and “hormone catabolic process”. Interestingly, among the comparisons of pin flowers (P × T vs. PS and P × P vs. PS), the term “biological adhesion” was found only for the P × T vs. PS comparison. We can infer that pollen-stigma adhesion might fail during SI pollination. The DEPs identified from the comparisons of the SI and SC pollinations (T × T vs. T × P and P × T vs. P × P) were enriched in “response to stimulus”, “developmental process”, “multicellular organismal process”, “reproduction”, “reproductive process”, “growth” and “immune system process”. In the MF category, the top three terms enriched in the proteins identified from the P × P vs. PS comparison were “transferase activity”, “oxidoreductase activity” and “enzyme regulator activity”. The subterms enriched in the DEPs identified in the P × T vs. PS comparisons were “hydrolase activity”, “transmembrane transporter activity” and “enzyme regulator activity”. We can infer that pollen-stigma interactions are related to enzymatic interactions and transmembrane transport. In the thrum comparative groups (T × T vs. TS, T × P vs. TS), the DEPs were enriched in “hydrolase activity”, “oxidoreductase activity” and “enzyme regulator activity” in T × T vs. TS. “Oxidoreductase activity, “enzyme regulator activity”, “amide binding” in T × P vs. TS. Interestingly,“transferase activity” were abundant in the P × T vs. P × P comparison, and “hydrolase activity” were enriched in the T × T vs. T × P comparison. Moreover, DEPs related to “signal recognition particles” and “signal recognition particle receptor complexes” in CC were found only in the comparisons of thrum samples and not those of pin samples. The DEPs identified from the PS vs. TS comparison were enriched in the following BP terms: “response to stimulus”, “developmental process” and “multicellular organismal process”. “Response to abiotic stimulus”, “response to external stimulus” and “catabolic process” terms were frequently identified among BP sub-layer. In the CC category, most of the DEPs were enriched in the “cell periphery”, “external encapsulating structure” and “extracellular region” terms, and the terms “transferase activity”, “oxidoreductase activity” and “enzyme regulator activity” terms were enriched in the MF category (Supplementary Material Tables S1–S7).
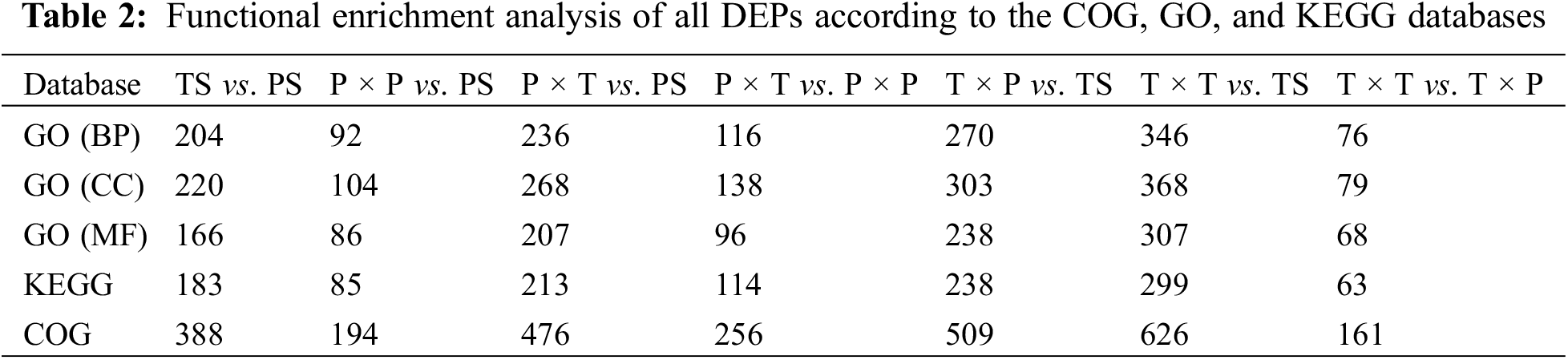
According to the KEGG pathway analysis, the three pathways with the greatest enrichment of DEPs identified from the SI pollination vs. style comparisons (T × T vs. TS and P × P vs. PS) were “biosynthesis of secondary metabolites”, “pentose and glucuronate interconversions” and “two-component systems”. For the SC pollination vs. style comparisons (T × P vs. TS, P × T vs. PS), the top three pathways were “biosynthesis of secondary metabolites”, “pentose and glucuronate interconversions” and “phenylpropanoid biosynthesis”. The “phenylpropanoid biosynthesis”, “cutin, suberine, and wax biosynthesis” and “flavonoid biosynthesis pathways” were enriched by the DEPs identified from the P × T vs. P × P comparison, and “starch and sucrose metabolism”, “glycerophospholipid metabolism” and “alpha-linolenic acid metabolism” were enriched by the DEPs identified from the T × T vs. T × P comparison. The pathways enriched by the DEPs identified from the comparison of PS with TS included “biosynthesis of secondary metabolites”, “phenylpropanoid biosynthesis” and “pentose and glucuronate interconversion” (Supplementary Material Table S8).
According to the COG database, the DEPs identified from the PS vs. TS comparison were enriched in the terms “carbohydrate transport and metabolism”, “posttranslational modification, protein turnover, chaperones” and “secondary metabolites biosynthesis, transport, and catabolism”. A comparison of the SI pollination styles with unpollinated styles (T × T vs. TS and P × P vs. PS) revealed the enrichment of the terms “signal transduction mechanisms” and “energy production and conversion”. DEPs were more abundant in the thrum comparative group than in the pin in these terms. From the comparisons of SC pollination with unpollinated styles (T × P vs. TS and P × T vs. PS) and SC pollination with SI pollination (P × T vs. P × P and T × P vs. T × T) “signal transduction mechanisms”, “energy production and conversion”, “amino acid transport and metabolism” and “lipid transport and metabolism” were enriched for pollen tube growth (Supplementary Material Table S9).
According to the GO database, 21 candidates in pollination term (GO:0009856) were found to be overexpressed. Receptor-like protein kinase FERONIA (FERON), 3-ketoacyl-CoA synthase 6 (KCS6), O-fucosyltransferase 23 (OFT23), Fimbrin-5 (FIMB5), Cysteine synthase (CYSK) and Protein SABRE (SAB) are involved in pollen stigma recognition, pollen hydration, pollen tube germination and penetration. Pollen-specific leucine-rich repeat extensin-like protein 4 (PLRX4), Transcription initiation factor IIB-2 (TF2B2), Pyruvate dehydrogenase E1 component subunit beta-3 (ODPB3), Probable protein kinase At2g41970 (Y2197) and Pectinesterase 1 (AL11A) are involved in pollen tube growth in SI. Basic blue proteins (BABL) accumulate in the stigma to affect pollen tube penetration. Ent-kaurenoic acid oxidase 2 (KAO2), Purple acid phosphatase 15 (PPA15), and Probable dolichyl-diphosphooligosaccharide--protein (OST3B) benefit fertilization through plant hormone regulation (Table 3, Supplementary Material Table S10).
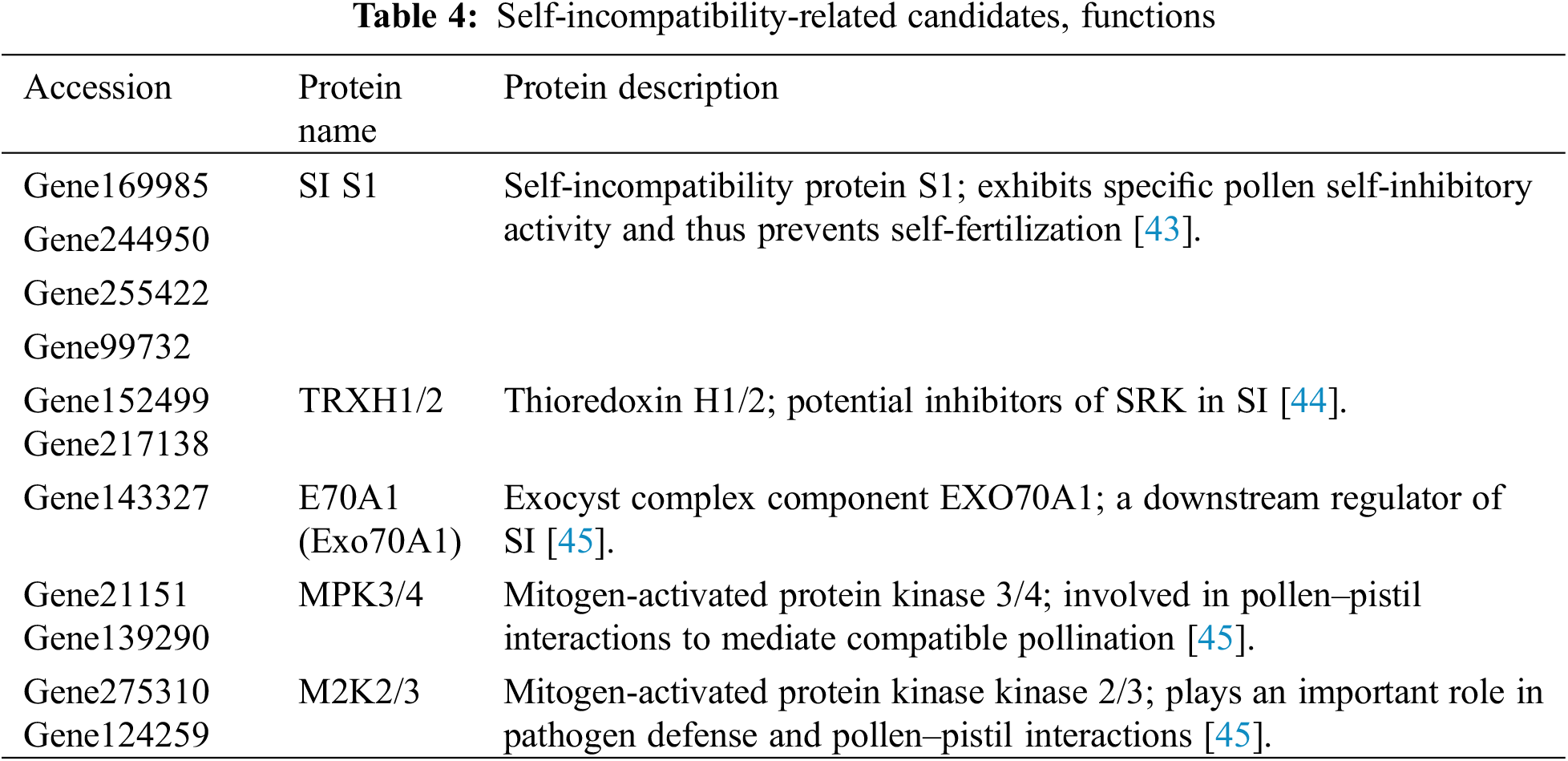
Self-incompatibility protein S1 (SI S1), Mitogen-activated protein kinase 3/4 (MPK3/4), Mitogen-activated protein kinase kinase 2/3 (M2K2/3), Thioredoxin H1/2 (TRXH1/2) and Exocyst complex component EXO70A1 (E70A1) were found to be involved in the HetSI of P. auriculata. SI S1 was expressed at a high level in all the samples and was involved in preventing the self-fertilization of plants with SI. TRXH1/2, E70A1, MPK3/4 and M2K2/3 were reported as the downstream factors participating in pollen-pistil interactions in SI (Table 4, Supplementary Material Table S10).
To further investigate the mechanism of HetSI, we observed the differences in stigma and pollen morphology between the pin and thrum morphotypes by SEM. The SEM results showed that the pin and thrum styles were clawed with clustered papilla cells, and these papilla cells were densely clustered at the apex. Compared with those on the pin style, the clustered papillae on the thrum stigma were more intense. No surface secretions were found on the pistil, and we can infer that P. auriculata has a typical dry-type stigma. The pollen exine sculptures differed between the pin and thrum morphotypes, and the wart-shaped thrum pollen surface was comparatively smooth (Fig. 3).
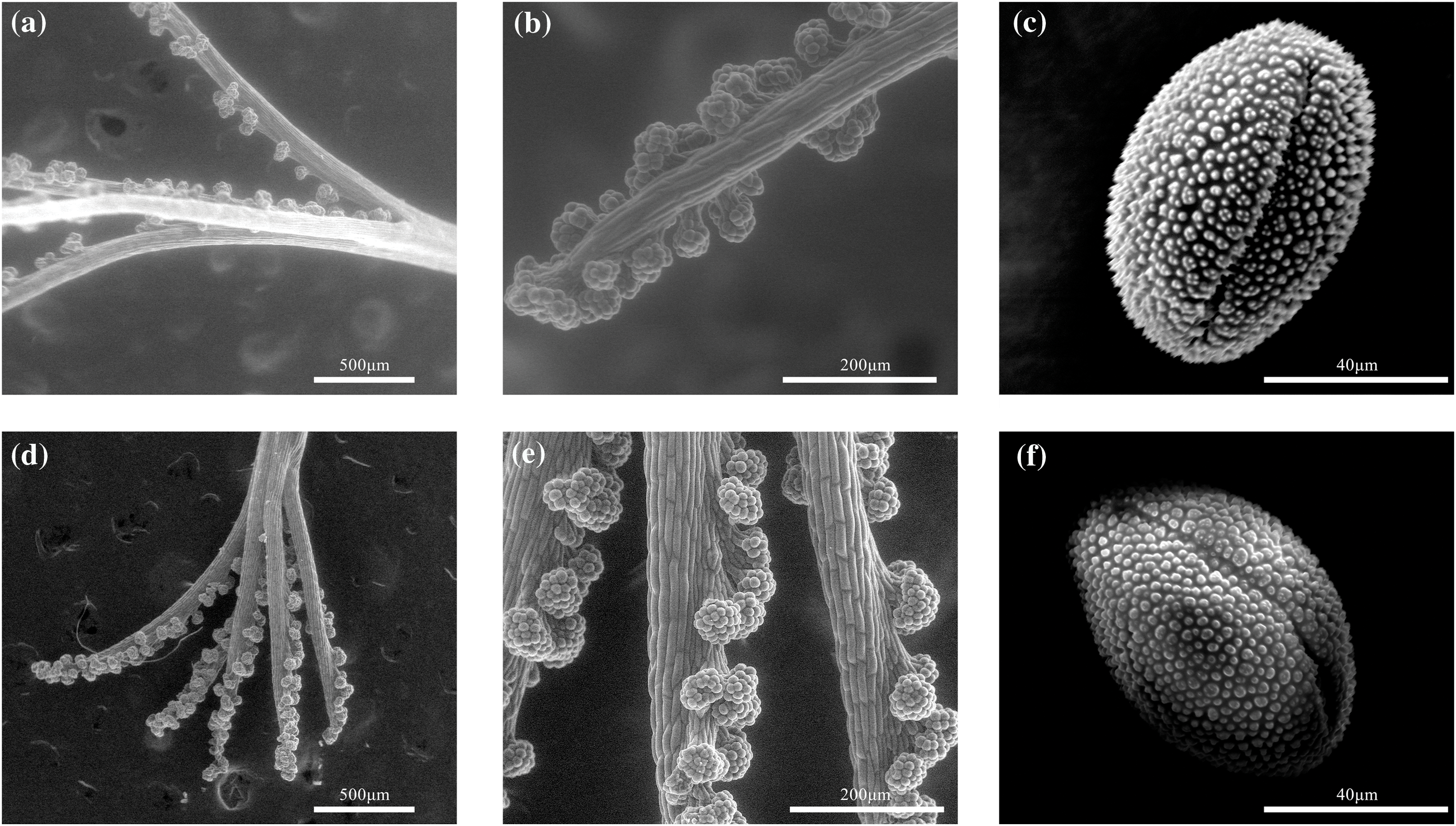
Figure 3: Pollen and unpollinated stigma (UPS) SEM images: (a and b) images of pin stigma; (c) images of pin pollen; (d and e) images of thrum stigma; (f) images of thrum pollen
We conducted a ROS intensity analysis after proteomic profiling to further determine whether ROS are involved in the HetSI of P. auriculata. We stained unpollinated stigma (UPS), SC-pollinated stigma (P × T and T × P), and SI-pollinated (P × P and T × T) stigma. The results showed that the ROS levels in thrum UPS were greater than those in pin UPS. After SI pollination, the ROS level markedly decreased in both pin and thrum flowers, whereas after SC pollination, the ROS level increased sharply in both pin and thrum flowers. According to our proteomic results, the ROS level was consistent with the expression of FERON after SC and SI pollination (Fig. 4); however, the opposite trend was observed in Zhang’s SI-related study of Brassica rapa [46]. HetSI might have a different mechanism than HomSI.
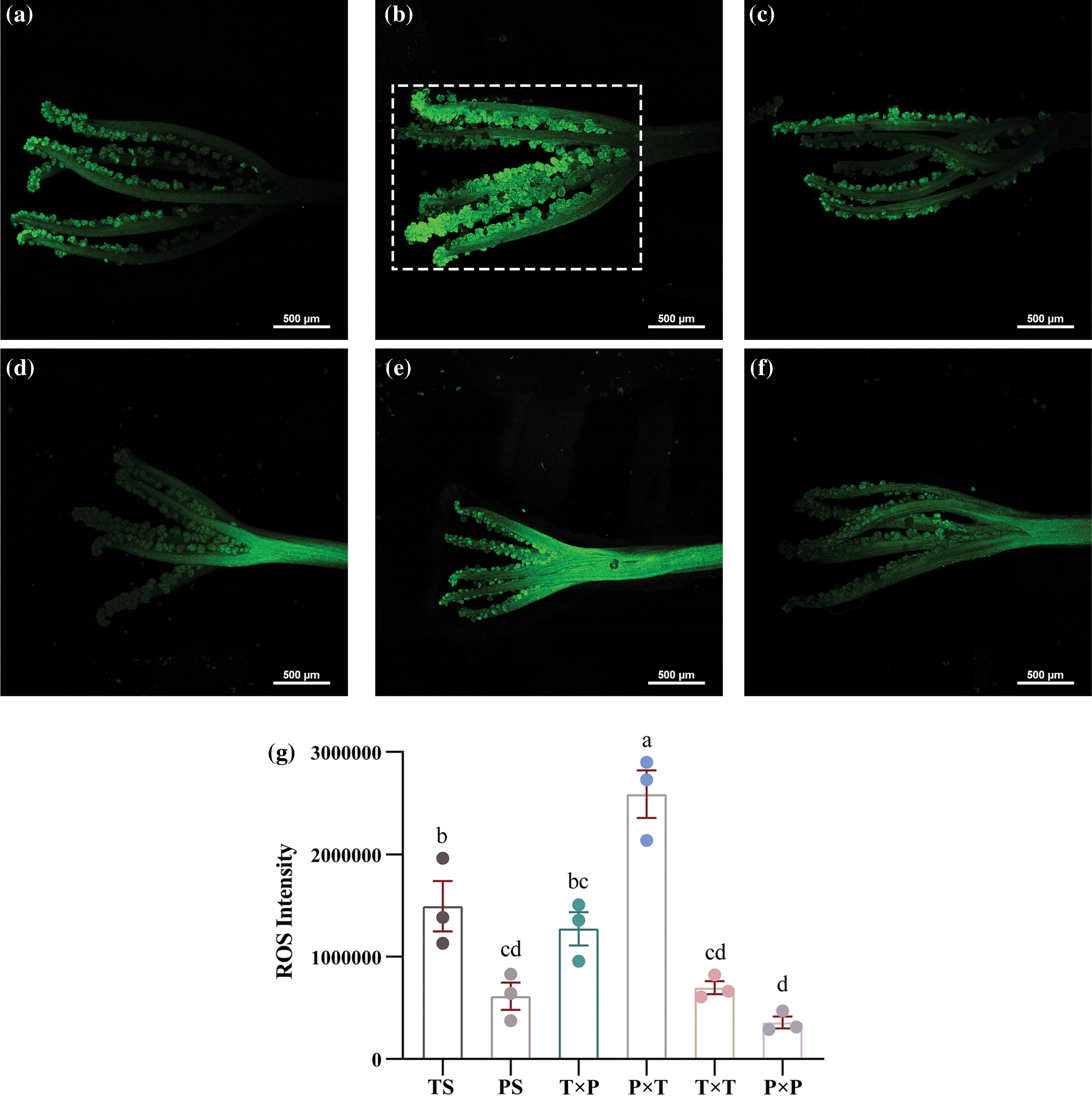
Figure 4: The stigmatic ROS levels decrease 2 h after SI pollination and increase 2 h after SC pollination (measured within the dashed line for all samples): (a) stigmatic ROS levels of unpollinated pin flowers; (b) stigmatic ROS levels of SC-pollinated pin flowers; (c) stigmatic ROS levels of SI-pollinated pin flowers; (d) stigmatic ROS levels of unpollinated thrum flowers; (e) stigmatic ROS levels of SC-pollinated thrum flowers; (f) stigmatic ROS levels of SI-pollinated thrum flowers; (g) ROS intensity (stained with DCFH-DA). The average ROS signals were determined with Image Plus Pro 6.0 and SPSS 27.0. The different lowercase letters on the histogram indicate significant differences (p < 0.05) between different treatments
4.1 Increased Reproduction-Related Protein Expression in Thrum Styles for Faster SC Pollen Germination
According to the metabolic and morphological results, SC pollen germination on the thrum stigma might be faster because more energy-related compounds accumulate in the thrum styles [17]. In the present study, both the pollen and stigma of the pin and thrum flowers showed distinct features. According to the proteomic analysis, DEPs identified from the PS vs. TS comparisons, such as TIP31 (aquaporin TIP3-1), FERON, OLEO (Oleosin), and BABL, which were annotated to the reproduction term GO (GO:0000003), showed markedly higher expression in TS. TIPs are reportedly regulators of water flow across cellular membranes between pollen and stigma, and TIP31 promotes faster cross-pollen hydration and germination [47]. OLEO, which is localized on the pollen coat, might also mediate hydration and water transfer. GRP17, an OLEO domain protein, is involved in the rapid initiation of pollen hydration in Arabidopsis [48]. Based on the abovementioned information, we can infer that higher expression of TIP31 and OLEO might be the factors causing faster SC pollen hydration and germination on the thrum style. Interestingly, BABL (Basic blue protein) was found to be an extracellular matrix protein in the stigma. In the BABL-overexpression mutant, the wild-type pollen tube is arrested on the stigma surface, and the pollen tube fails to penetrate the papilla cell [28]. According to our results, BABL was enriched in the thrum style, and we can infer that in thrum flowers, a lower position leads to more self-pollen falling on the stigma. BABL was found to be expressed at a higher level to avoid selfing. In addition, the ROS regulator FERON was expressed at higher levels in TS due to greater ROS intensity (Fig. 4).
Moreover, the KEGG enrichment showed that the DEPs identified from the P × T vs. P × P comparison were enriched in the “phenylpropanoid biosynthesis”, “cutin, suberine, and wax biosynthesis” and “flavonoid biosynthesis pathways” pathways; however, the DEPs identified from the T × P vs. T × T comparison were enriched in completely different pathways, namely, “starch and sucrose metabolism”, “glycerophospholipid metabolism” and “alpha-linolenic acid metabolism”. HetSI might play distinct roles in pin and thrum.
4.2 Self-Incompatibility Protein S1 Might Be the Key Factor of HetSI
SI S1 might participate in pollen–stigma recognition, which is usually controlled by the S-locus with allelic variants and is found in Citrus reticulata and Papaver rhoeas [43,49]. These plants are typical GSI plants that have been reported to have wet stigmas, and pollen rejection occurs in the style. According to our morphological results, P. auriculata has typical dry stigmas in both pin and thrum flowers and pollen rejection occurs on stigmas [17], similar to SSI in HomSI. SI S1 was found for the first time in plants with HetSI. SRK, which is reportedly the key factor in SSI, was not found in our proteomic analysis [4]. We can infer that SRK might be phosphorylated and degraded during the pollen–stigma recognition process or that the SSI-like system may have diverse mechanisms in the HetSI of P. auriculata and that SRK might be replaced by SI S1, which plays a similar role in pollen–stigma recognition.
MPK3/4, M2K2/3, E70A1 (EXO70A1) and TRXH1/2 are also known as factors in the SI signaling cascade. Moreover, MPK3/4 and M2K2/3 were found to be self-incompatible candidates. MPK3/4 belongs to the mitogen-activated protein kinase (MAPK) family and constitutes a functionally redundant pair needed for mediating stigma receptivity to accept compatible pollen in Arabidopsis. The loss of these two proteins could cause pollen germination reduction and a decrease in the pollen tube growth rate. M2K2/3 was also found in styles as an upstream activator of MPK3/4, and the loss or suppression of M2K2/3 was associated with a decrease in pollen attachment. Moreover, E70A1, which is a factor that regulates pollen hydration and germination, was found in our study. MPK3/4 phosphorylates E70A1 at the single consensus phosphorylation motif to localize E70A1 at plasma membranes and regulate exocytosis, thus maintaining stigma receptivity [45]. In addition, TRXH1/2 belongs to the thioredoxin H family and was previously found to be a potential inhibitors of SRK in SI [44].
4.3 ROS Are Highly Abundant during SC Pollination
Dry stigmas often contain associated peroxidases [50], which are very important for stigma function. Strong evidence of the role of ROS in SI has emerged, but the relationship between the differential patterns of ROS accumulation in the stigma and SI has not been determined [51]. Citrus reticulata and Citrus × limon display similar SI systems but have different ROS patterns. In contrast, distinct SI systems in Brassica oleracea. and Citrus × limon share the same ROS pattern [50]. In our study, after SC pollination, ROS accumulated in the pollinated styles of both pin and thrum flowers. In the SI system, the ROS concentration decreased sharply (Fig. 4). FERON is considered a stigmatic ROS regulator in Brassica, and FER-Rboh signaling determines pollen acceptance or rejection on the stigma surface through the regulation of the ROS levels [46]. Interestingly, compared with that in response to SI pollination, the FERON protein was upregulated in response to SC pollination. We can infer that ROS are involved in pollen–pistil interactions in the HetSI of P. auriculata. Moreover, ROS are reportedly strong activators of MAPKs. After successful pollination, FERON was overexpressed leading to a high level of ROS and the rapidly activated MAPK phosphorylates E70A1 to enhance SC pollen–stigma receptivity [45].
4.4 Other Regulatory Transducers of the HetSI
OFT23, KCS6, FIMB5, PLRX4, TF2B2, and AL11A were found to accumulate in pollen and are important for pollen tube growth in HetSI. The pollen coat regulates water transfer through the stigma to the pollen, and KCS6 is involved in the accumulation of pollen coat lipids, which are important for pollen hydration. The triple mutant (kcs7/15/21) leads to a reduction in pollen coat lipids and defective pollen hydration in Arabidopsis [26]. KCS6 was identified as a downregulated protein in the P × T vs. P × P comparisons. We can infer that the increment of KCS6 may increase pollen coat lipids and subsequently enhance pollen hydration during SC pollination in pin flowers. The O-fucosyltransferase (OFT) family of proteins may facilitate pollen tube penetration through the stigma–pistil interface, and the oft1 mutant exhibited a reduced ability to penetrate the stigma surface in Arabidopsis. OFT23 was identified in our research and may regulate pollen tube penetration [42]. Research in Arabidopsis has shown that FIMB5 is involved in the regulation of pollen germination and tube growth by regulating the organization of actin structures within the subapical region of the pollen tube. FIMB5 loss-of-function mutants fail to organize apical actin structures in pollen tubes, resulting in failure of pollen germination and tube growth [23]. According to the T × T vs. TS and P × P vs. PS comparisons, FIMB5 was downregulated in both the pin and thrum of P. auriculata. We can infer that FIMB5 is involved in the SI pollen–pistil interaction. PLRX4 is a pollen-specific protein that reportedly interacts with rapid alkalinization factors (RALFs), and proteome analysis revealed that this protein sustains pollen tube growth in Arabidopsis [52]. Moreover, PLRX4 (PEX4) was found to be involved in the SI of Dendrobium officinale [53] and is similar to PEX1 in Zea mays. Several studies have shown that ZmPEX1 is involved in pollen tube growth and may be an important signaling molecule in pollen stigma recognition [54]. TF2B2, AL11A, and BABL were found to be factors that affect pollen tube reception, growth, and guidance [22,28,32].
HetSI is a very complex physiological process that promotes interpollination and prevents intrapollination. Major advancements have been made toward obtaining a comprehensive understanding of the molecular foundation of HetSI, but the mechanism of HetSI has not been elucidated. To explore the mechanism underlying HetSI, we conducted a 4D-DIA analysis of P. auriculata dry stigmas before and after pollination. A total of 33387 peptides and 5311 proteins were detected, and among them, SI S1, MPK3/4, M2K2/3, E70A1, and TRXH1/2 were identified as HetSI-related candidates in our study. In addition, other regulatory transducers were identified. From the results, we can infer that SI S1 might be the key factor in HetSI and that ROS are overexpressed to enhance SC pollen–stigma receptivity during SC pollination.
Acknowledgement: This work was supported by Science & Technology Department of Sichuan Province (2021YJ0497).
Funding Statement: Scientific Research Foundation of Science & Technology Department of Sichuan Province (2021YJ0497).
Author Contributions: The authors confirm the following contributions to the paper: study conception and design: Di Hu, Shouli Yi and Suping Gao; data collection: Di Hu and Tingdan Xu; analysis and interpretation of the results: Di Hu, Wenji Li, and Ting Lei; draft manuscript preparation: Di Lin, Songlin Jiang and Suping Gao. All the authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All the data generated or analyzed during this study are included in this published article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2024.049166.
References
1. Kitasiba H, Nishio T. Self-incompatibility. In: Biotechnology of crucifers. New York, NY: Springer; 2013. p. 187–208. [Google Scholar]
2. Jain S, Maurya P, Jain S, Kumar V, Kiran B, Subhasmita S, et al. Incompatibility systems in fruit crops: applications and achievements. Int J Environ Clim Change. 2023;13(9):2653–63. [Google Scholar]
3. Wang F, Li Y, Li G, Chen S. Genetic components of self-incompatibility in Brassica vegetables. Hortic. 2023;9(2):265. [Google Scholar]
4. Banović B, Miljuš-Đukić J, Konstantinović M, Maksimović V. A search of Brassica SI-involved orthologs in buckwheat leads to novel buckwheat sequence identification: MLPK possibly involved in SI response. Arch Biol Sci. 2010;62(2):315–21. [Google Scholar]
5. Kenney P, Sankaranarayanan S, Balogh M, Indriolo E. Expression of Brassica napus GLO1 is sufficient to breakdown artificial self-incompatibility in Arabidopsis thaliana. Plant Reprod. 2020;33:159–71. [Google Scholar] [PubMed]
6. Hou S, Zhao T, Yang Z, Yang D, Li Q, Liang L. Molecular cloning and yeast two-hybrid provide new evidence for unique sporophytic self-incompatibility system of Corylus. Plant Biol. 2022;24(1):104–16. [Google Scholar] [PubMed]
7. Azibi T, Hadj-Arab H, Lodé M, Ferreira de Carvalho J, Trotoux G, Negre S, et al. Impact of whole genome triplication on the evolutionary history and the functional dynamics of regulatory genes involved in Brassica self-incompatibility signalling pathway. Plant Reprod. 2020;33:43–58. [Google Scholar] [PubMed]
8. Goring DR, Bosch M, Franklin-Tong VE. Contrasting self-recognition rejection systems for self-incompatibility in Brassica and Papaver. Curr Biol. 2023;33(11):R530–42. [Google Scholar] [PubMed]
9. Hiratsuka S, Zhang SL, Nakagawa E, Kawai Y. Selective inhibition of the growth of incompatible pollen tubes by S-protein in the Japanese pear. Sex Plant Reprod. 2001;13:209–15. [Google Scholar]
10. Matsui K, Yasui Y. Buckwheat heteromorphic self-incompatibility: genetics, genomics and application to breeding. Breed Sci. 2020;70(1):32–8. [Google Scholar] [PubMed]
11. Wong KC, Watanabe M, Hinata K. Protein profiles in pin and thrum floral organs of distylous Averrhoa carambola L. Sex Plant Reprod. 1994;7:107–15. [Google Scholar]
12. Tamari F, Shore JS. Allelic variation for a short-specific polygalacturonase in Turnera subulata: is it associated with the degree of self-compatibility? Int J Plant Sci. 2006;167(1):125–33. [Google Scholar]
13. Wang Y, Scarth R, Campbell C. Sh and Sc—Two complementary dominant genes that control self-compatibility in buckwheat. Crop Sci. 2005;45(4):1229–34. [Google Scholar]
14. Huu CN, Plaschil S, Himmelbach A, Kappel C, Lenhard M. Female self-incompatibility type in heterostylous Primula is determined by the brassinosteroid-inactivating cytochrome P450 CYP734A50. Curr Biol. 2022;32:671–6. [Google Scholar] [PubMed]
15. Miljuš-Đukić J, Ninković S, Radović S, Maksimović V, Brkljačić J, Nešković M. Detection of proteins possibly involved in self-incompatibility response in distylous buckwheat. Biol Plant. 2004;48:293–6. [Google Scholar]
16. Caperta AD, Rois AS, Teixeira G, Garcia-Caparros P, Flowers TJ. Secretory structures in plants: lessons from the Plumbaginaceae on their origin, evolution and roles in stress tolerance. Plant, Cell Environ. 2020;43(12):2912–31. [Google Scholar] [PubMed]
17. Hu D, Li W, Gao S, Lei T, Hu J, Shen P, et al. Untargeted metabolomic profiling reveals that different responses to self and cross pollination in each flower morph of the heteromorphic plant Plumbago auriculata. Plant Physiol Biochem. 2019;144:413–26. [Google Scholar] [PubMed]
18. Hu D, Gao S, Li W, Lei T, Liu Y, Li Q, et al. Dissecting the distyly response to pollination using metabolite profiling in heteromorphic incompatibility system interactions of Plumbago auriculata Lam. Acta Physiol Plant. 2020;42:1–6. [Google Scholar]
19. Chen M, Zhu P, Wan Q, Ruan X, Wu P, Hao Y, et al. High-coverage four-dimensional data-independent acquisition proteomics and phosphoproteomics enabled by deep learning-driven multidimensional predictions. Anal Chem. 2023;95(19):7495–7502. [Google Scholar] [PubMed]
20. Jiang X, Gao Y, Zhou H, Chen J, Wu J, Zhang S. Apoplastic calmodulin promotes self-incompatibility pollen tube growth by enhancing calcium influx and reactive oxygen species concentration in Pyrus pyrifolia. Plant Cell Rep. 2014;33:255–63. [Google Scholar] [PubMed]
21. Wang Y, Li X, Wu Y, Cai Z, Li B, Xie Z. Observation of flowering process of grape (Vitis vinifera L.)—insight into the starting of pollination of flower hiding in Calyptras (Cap). J Exp Bot. 2023;92(6):1859–71. [Google Scholar]
22. Tang C, Zhu X, Qiao X, Gao H, Li Q, Wang P, et al. Characterization of the pectin methyl-esterase gene family and its function in controlling pollen tube growth in pear (Pyrus bretschneideri). Genom. 2020;112(3):2467–77. [Google Scholar]
23. Wu Y, Yan J, Zhang R, Qu X, Ren S, Chen N, et al. Arabidopsis FIMBRIN5, an actin bundling factor, is required for pollen germination and pollen tube growth. Plant Cell. 2010;22(11):3745–63. [Google Scholar] [PubMed]
24. Baumberger N, Doesseger B, Guyot R, Diet A, Parsons RL, Clark MA, et al. Whole-genome comparison of leucine-rich repeat extensins in Arabidopsis and rice. A conserved family of cell wall proteins form a vegetative and a reproductive clade. Plant Physiol. 2003;131(3):1313–26. [Google Scholar] [PubMed]
25. Pocsfalvi G, Turiák L, Ambrosone A, Del Gaudio P, Puska G, Fiume I, et al. Protein biocargo of citrus fruit-derived vesicles reveals heterogeneous transport and extracellular vesicle populations. J Plant Physiol. 2018;229:111–21. [Google Scholar] [PubMed]
26. Zhang Z, Zhan H, Lu J, Xiong S, Yang N, Yuan H, et al. Tapetal 3-Ketoacyl-Coenzyme A synthases are involved in pollen coat lipid accumulation for pollen–stigma interaction in Arabidopsis. Front Plant Sci. 2021;12:770311. [Google Scholar] [PubMed]
27. Birke H, Heeg C, Wirtz M, Hell R. Successful fertilization requires the presence of at least one major O-acetylserine(thiol)lyase for cysteine synthesis in pollen of Arabidopsis. Plant Physiol. 2013;163:959–72. [Google Scholar] [PubMed]
28. Dong J, Kim ST, Lord EM. Plantacyanin plays a role in reproduction in Arabidopsis. Plant Physiol. 2005;138(2):778–89. [Google Scholar] [PubMed]
29. Gong P, He C. Uncovering divergence of rice exon junction complex core heterodimer gene duplication reveals their essential role in growth, development, and reproduction. Plant Physiol. 2014;165(3):1047–61. [Google Scholar] [PubMed]
30. Escobar-Restrepo JM, Huck N, Kessler S, Gagliardini V, Gheyselinck J, Yang WC, et al. The FERONIA receptor-like kinase mediates male–female interactions during pollen tube reception. Science. 2007;317(5838):656–60. [Google Scholar] [PubMed]
31. Pan J, Li D, Shu Z, Jiang X, Ma W, Wang W, et al. CsPDC-E1α, a novel pyruvate dehydrogenase complex E1α subunit gene from Camellia sinensis, is induced during cadmium inhibiting pollen tube growth. Can J Plant Sci. 2017;98(1):62–70. [Google Scholar]
32. Zhou JJ, Liang Y, Niu QK, Chen LQ, Zhang XQ, Ye D, et al. The Arabidopsis general transcription factor TFIIB1 (AtTFIIB1) is required for pollen tube growth and endosperm development. J Exp Bot. 2013;64(8):2205–18. [Google Scholar] [PubMed]
33. Christensen CA, Gorsich SW, Brown RH, Jones LG, Brown J, Shaw JM, et al. Mitochondrial GFA2 is required for synergid cell death in Arabidopsis. Plant Cell. 2002;14(9):2215–32. [Google Scholar] [PubMed]
34. Masoomi-Aladizgeh F, Najeeb U, Hamzelou S, Pascovici D, Amirkhani A, Tan DK, et al. Pollen development in cotton (Gossypium hirsutum) is highly sensitive to heat exposure during the tetrad stage. Plant, Cell Environ. 2021;44(7):2150–66. [Google Scholar] [PubMed]
35. Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ. The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci. 2001;98(4):2065–70. [Google Scholar] [PubMed]
36. Röhricht H, Przybyla-Toscano J, Forner J, Boussardon C, Keech O, Rouhier N, et al. Mitochondrial ferredoxin-like is essential for forming complex I-containing supercomplexes in Arabidopsis. Plant Physiol. 2023;191(4):2170–84. [Google Scholar]
37. Zhang W, Gruszewski HA, Chevone BI, Nessler CL. An Arabidopsis purple acid phosphatase with phytase activity increases foliar ascorbate. Plant Physiol. 2008;146(2):431–40. [Google Scholar] [PubMed]
38. Nazemof N, Couroux P, Xing T, Robert LS. Proteomic analysis of the mature Brassica stigma reveals proteins with diverse roles in vegetative and reproductive development. Plant Sci. 2016;250:51–8. [Google Scholar] [PubMed]
39. Chen W, Salari H, Taylor MC, Jost R, Berkowitz O, Barrow R, et al. NMT1 and NMT3 N-methyltransferase activity is critical to lipid homeostasis, morphogenesis, and reproduction. Plant Physiol. 2018;177(4):1605–28. [Google Scholar] [PubMed]
40. Procissi A, Guyon A, Pierson ES, Giritch A, Knuiman B, Grandjean O, et al. KINKY POLLEN encodes a SABRE-like protein required for tip growth in Arabidopsis and conserved among eukaryotes. Plant J. 2003;36(6):894–904. [Google Scholar] [PubMed]
41. Liao HZ, Zhu MM, Cui HH, Du XY, Tang Y, Chen LQ, et al. MARIS plays important roles in Arabidopsis pollen tube and root hair growth. J. Integr. Plant Biol. 2016;58(11):927–40. [Google Scholar] [PubMed]
42. Smith DK, Jones DM, Lau JB, Cruz ER, Brown E, Harper JF, et al. A putative protein O-fucosyltransferase facilitates pollen tube penetration through the stigma-style interface. Plant Physiol. 2018;176(4):2804–18. [Google Scholar] [PubMed]
43. Miao H, Ye Z, Hu G, Qin Y. Comparative transcript profiling of gene expression between self-incompatible and self-compatible mandarins by suppression subtractive hybridization and cDNA microarray. Molec Breed. 2015;35(1):47. [Google Scholar]
44. Baumann U, Juttner J. Plant thioredoxins: the multiplicity conundrum. Cell Mol Life Sci. 2002;59:1042–57. [Google Scholar] [PubMed]
45. Jamshed M, Sankaranarayanan S, Abhinandan K, Samuel MA. Stigma receptivity is controlled by functionally redundant MAPK pathway components in Arabidopsis. Molec Plant. 2020;13(11):1582–93. [Google Scholar]
46. Zhang L, Huang J, Su S, Wei X, Yang L, Zhao H, et al. FERONIA receptor kinase-regulated reactive oxygen species mediate self-incompatibility in Brassica rapa. Curr Biol. 2021;31(14):3004–16. [Google Scholar] [PubMed]
47. Soto G, Alleva K, Mazzella MA, Amodeo G, Muschietti JP. AtTIP1; 3 and AtTIP5; 1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 2008;582(29):4077–82. [Google Scholar] [PubMed]
48. Mayfield JA, Preuss D. Rapid initiation of Arabidopsis pollination requires the oleosin-domain protein GRP17. Nat Cell Biol. 2000;2(2):128–30. [Google Scholar] [PubMed]
49. Wheeler MJ, Armstrong SA, Franklin-Tong VE, Franklin FCH. Genomic organization of the Papaver rhoeas self-incompatibility S1 locus. J Exp Bot. 2003;54(380):131–9. [Google Scholar] [PubMed]
50. Zafra A, Rejón JD, Hiscock SJ, Alché JDD. Patterns of ROS accumulation in the stigmas of angiosperms and visions into their multi-functionality in plant reproduction. Front Plant Sci. 2016;7:1112. [Google Scholar] [PubMed]
51. McInnis SM, Desikan R, Hancock JT, Hiscock SJ. Production of reactive oxygen species and reactive nitrogen species by angiosperm stigmas and pollen: potential signalling crosstalk? New Phytol. 2006;172(2):221–8. [Google Scholar] [PubMed]
52. Mecchia MA, Santos-Fernandez G, Duss NN, Somoza SC, Boisson-Dernier A, Gagliardini V, et al. RALF4/19 peptides interact with LRX proteins to control pollen tube growth in Arabidopsis. Science. 2017;358(6370):1600–3. [Google Scholar] [PubMed]
53. Chen Y, Hu B, Zhang F, Luo X, Xie J. Cytological observation and transcriptome comparative analysis of self-pollination and cross-pollination in Dendrobium officinale. Genes. 2021;12(3):432. [Google Scholar] [PubMed]
54. Rubinstein AL, Broadwater AH, Lowrey KB, Bedinger PA. Pex1, a pollen-specific gene with an extensin-like domain. Proc Natl Acad Sci. 1995;92(8):3086–90. [Google Scholar] [PubMed]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools