 Open Access
Open Access
REVIEW
Revolutionizing Metabolic Engineering in Cannabis sativa L.: Harnessing the Power of Hairy Root Culture
1 Department of Applied Biosciences, Kyungpook National University, Daegu, 41566, Republic of Korea
2 Department of Agriculture, Gopalganj Science and Technology University, Gopalganj, 8100, Bangladesh
3 Department of Horticulture, Faculty of Agriculture, Bangladesh Agricultural University, Mymensingh, 2202, Bangladesh
4 Department of Plant Medicals, Andong National University, Andong, 36729, Republic of Korea
5 Institute of Genomics for Crop Abiotic Stress Tolerance, Department of Plant and Soil Science, Texas Tech University, Lubbock, TX 79409, USA
6 Department of Agronomy, Khulna Agricultural University, Khulna, 9100, Bangladesh
7 School of Plant, Environmental, and Soil Sciences, Louisiana State University Agricultural Center, Baton Rouge, LA 70803, USA
8 Institute of Cannabis Biotechnology, Andong National University, Andong, 36729, Republic of Korea
* Corresponding Authors: S. M. Ahsan. Email: ; Hyong Woo Choi. Email:
(This article belongs to the Special Issue: Ethnobotany: Value and Conservation)
Phyton-International Journal of Experimental Botany 2025, 94(12), 3805-3826. https://doi.org/10.32604/phyton.2025.069827
Received 01 July 2025; Accepted 11 November 2025; Issue published 29 December 2025
Abstract
Cannabis sativa is highly valued for its use in fiber production, medicine, and recreational products. Its secondary metabolites (SM) are renowned for their wide range of health benefits and psychoactive properties. While much of the existing research has focused on cannabinoid production in the plant’s aerial parts, particularly the leaves and flowers, the root system remains understudied in terms of its SM profile. One promising in vitro approach for metabolite production involves the use of ‘hairy roots (HRs)’. These roots mimic the phytochemical profile of native roots but grow more efficiently and yield higher quantities of metabolites. HRs are genetically altered root tissues that develop at the site of infection when Agrobacterium rhizogenes is introduced into wounded plant tissues. HRs cultures in Cannabis represent a breakthrough in plant metabolic engineering, offering potential for the controlled biosynthesis of cannabinoids and terpenoids. By utilising genome editing (GE) tools such as CRISPR-based tools, these cultures can produce novel bioactive compounds at an industrial scale. The use of elicitors enhances the production of SM by activating their biosynthetic pathways, further boosting yields. This system provides a sustainable alternative to conventional farming and chemical synthesis, addressing challenges such as pharmaceutical shortages, enhancing climate resilience, and promoting more resource-efficient biomanufacturing. Few studies have explored elicitor-induced HR cultures in Cannabis to enhance terpenoid production. This review highlights research on HRs for SM synthesis and introduces a platform that positions Cannabis as a leader in biomanufacturing and sustainable biotechnology, promoting advancements across the agricultural and pharmaceutical industries globally.Keywords
Industrial hemp (Cannabis sativa L.) is a dioecious, annual herbaceous plant species in the Cannabaceae family. Globally, hemp serves multiple purposes, ranging from food additives and animal feed to textile fibers and traditional medicine applications [1,2]. The therapeutic potential of C. sativa has been explored for its anti-inflammatory, analgesic, anticancer, antioxidant, neuroprotective, and gastrointestinal benefits, as well as its use in epilepsy management [3]. These effects are largely attributed to the plant’s rich and diverse chemical composition, which includes ‘cannabinoids, alkaloids, terpenes, flavonoids, lignans, and steroids’ [4]. To date, over 500 metabolites have been identified in C. sativa, including more than 100 distinct cannabinoids [5]. The main bioactive compounds include ‘Δ9-tetrahydrocannabinol (THC), cannabidiol (CBD), and cannabinolic acid (CBNA)’, with additional contributions from cannabigerolic acid (CBGA), cannabichromenic acid (CBCA), and cannabidiolic acid (CBDA) [6,7]. Remarkably, the highest concentrations of these phytochemicals are typically found in the plant’s aerial parts [1,8,9]. Hemp’s widespread use is due to its versatility, with inflorescence, leaves, stems, and seeds being commonly utilised, while roots are less frequently used [1]. Cannabis is legally permissible for both medical and nonmedical purposes on a global scale. The use of Cannabis and its derivatives have been legalised in the United States, the United Kingdom, Canada, and several European Union (EU) countries [10,11,12]. As comprehension of Cannabis phytochemistry has advanced, different countries have started reassessing their regulatory frameworks. Uruguay was the first nation to legalise Cannabis for both medical and recreational use in 2013. Several countries have emulated this action. Medical Cannabis is now legalised in over 50 countries, including China, Australia, Germany, Israel, Canada and most of the United States. The production of hemp has distinct advantages, and there is an expanding market for hemp-derived goods [10,11,12]. The global Cannabis market is projected to reach US$102 billion by 2026, with cannabinoid demand expected to hit $9.69 billion by 2025 [1,8,9].
While research has primarily focused on glandular trichome metabolites for their therapeutic properties [9,13], other parts of the hemp plant are also valuable sources of specialised metabolites [14]. Monoterpenes and diterpenes are found in inflorescences, sesquiterpenes, triterpenes, alkaloids, and phytosterols in roots, and flavonoids in leaves [9]. Hemp’s root contains a significant amount of pentacyclic triterpenoids [8]. The phytochemical content is responsible for their biological activity [15]. Scientific studies have identified various specialised metabolites in hemp roots, including monoterpenes like carvone and dihydrocarvone; triterpenes like friedelin and epifriedelanol; sterols like “β-sitosterol, campesterol, and sigmasterol; alkaloids like pyrrolidine, piperidine, cannabisativine, and anhydrocannabisativine; and lignanamides like p-coumaroyltyramine” [9,16]. Pre-clinical studies have demonstrated the potential pharmacological properties of C. sativa root extracts, including hepatoprotective, anti-inflammatory, anticancer, antidiabetic, anti-ulcer, antisenescence, anti-diarrheal, analgesic, antipyretic, and antimicrobial effects [1,8]. However, some limitations exist: root growth is slow, harvesting is challenging, and roots may absorb pollutants, including heavy metals [9], which restricts their use [1,8,9].
In vitro cultures have created opportunities to produce phyto-molecules through elicitation strategies, such as hairy root (HRs) cultures [9,17,18,19]. This involves plant transfection with Rhizobium rhizogenes, which induces “hairy root syndrome” in infected hosts [9,19]. When R. rhizogenes transfect a plant, its T-DNA (Transfer DNA) region integrates into the plant genome, leading to the proliferation of rapidly growing root tissues [9,20]. HRs are stable, can grow without added phytohormones in media, develop quickly and can produce higher quantities of metabolites than the roots of the mother plant [8,20,21,22]. Through metabolic engineering, hemp HRs can potentially accumulate novel metabolites that are not found in the parent plants [9,21]. However, the full extent of their chemical diversity has yet to be comprehensively characterised [9,23,24]. This review examines the potential of HRs for the biosynthesis of high-value SMs and biotechnology tools for enhancing SM production in C. sativa. Additionally, it focuses on advancing hemp’s SM production by optimising nutrient media, incorporating precursor feeding, employing elicitation techniques, and utilising molecular strategies. This includes prospects of metabolic engineering through the manipulation of genes (Fig. 1) and different molecular factors.
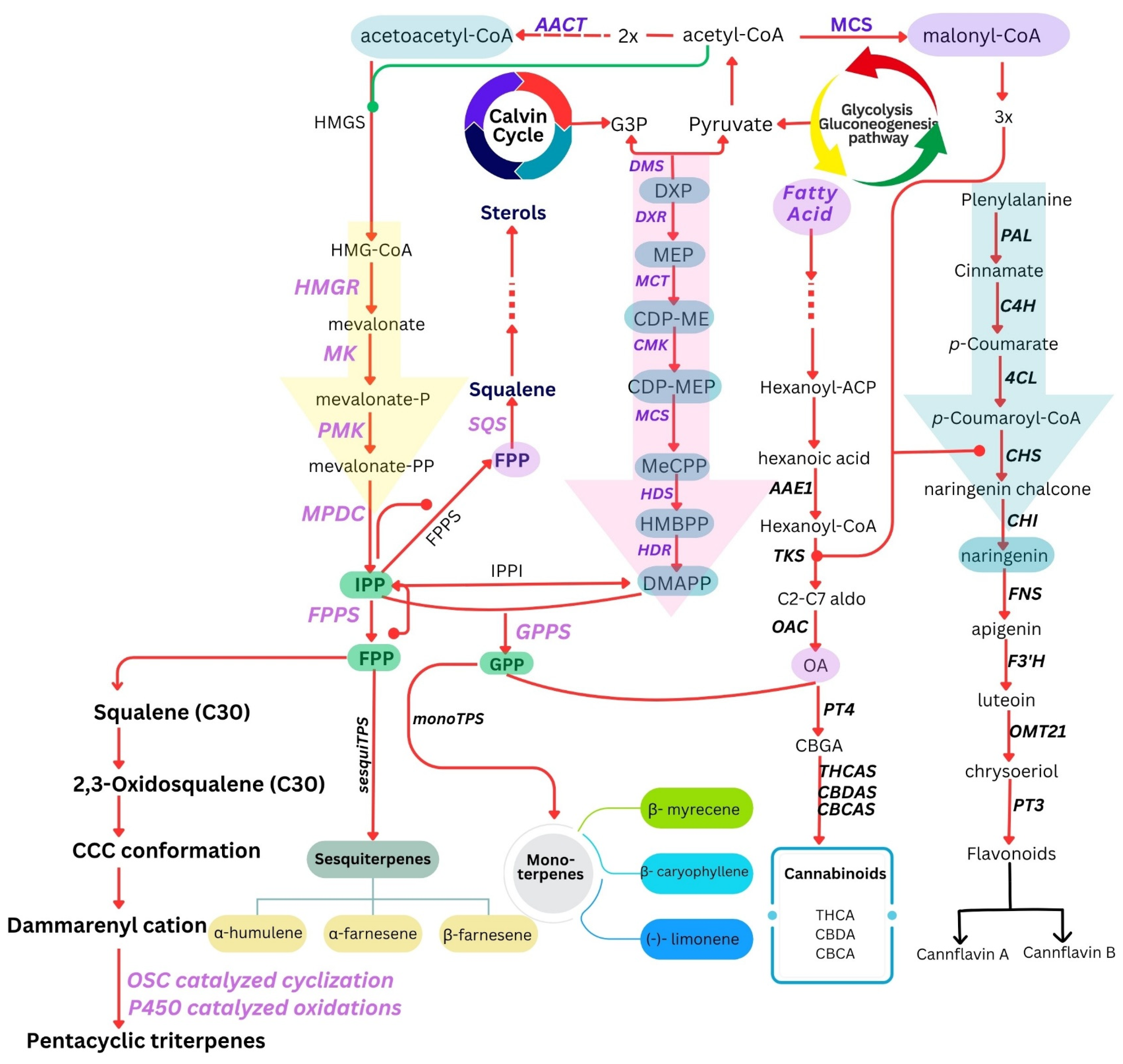
Figure 1: The biosynthesis pathways of specialised metabolites in C. sativa are depicted, focusing on key compounds of medical importance. Cannabinoids, terpenes, and flavonoids are shown as distinct pathways derived from primary metabolism, leading to their respective precursors. The pathways involved in the biosynthesis of these metabolites—specifically the Methylerythritol Phosphate (MEP) pathway, the Mevalonate (MVA) pathway, and the phenylpropanoid pathway. The Calvin cycle and glycolysis/gluconeogenesis pathways are simplified into two circular arrows to represent their involvement in the central metabolism. Abbreviation: “AACT: acetoacetyl-coenzyme A thiolase; MCS: malonyl-CoA synthetase; DMS: 1-deoxy-d-xylulose 5-phosphate synthase; DXR: 1-deoxy-D-xylulose 5-phosphate reductoisomerase; MCT: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; HDS: 4-hydroxy-3-methylbut-2-enyl diphosphate synthase; HDR: (E)-4-hydroxy-3-methylbut-2-enyl diphosphate reductase; HMGS: hydroxymethylglutaryl-CoA synthase; HMGR: 3-hydroxy-3-methyl-glutaryl-coenzyme A reductase; MK: mevalonate kinase; PMK: phosphomevalonate kinase; MPDC: mevalonate pyrophosphate decarboxylase; IPPI: isopentenyl diphosphate isomerase; GPPS: geranyl diphosphate synthase; AAE1: acyl activating enzyme 1; TKS: 3,5,7-Trioxododecanoyl-CoA synthase (polyketide synthase); OAC: olivetolic acid cyclase; FPPS: farnesyl pyrophosphate synthase; PAL: phenylalanine ammonia-lyase; C4H: cinnamate 4-hydroxylase; 4CL: 4-coumaroyl CoA-ligase; CHS: chalcone synthase; CHI: chalcone isomerase; SQS: squalene synthase; PT4: olivetolate geranyl transferase; THCAS: THCA synthase; CBDAS: CBDA synthase; CBCAS: CBCA synthase; sesquiTPS: all sesquiterpene synthases grouped under a single appellation; monoTPS: all monoterpene synthases grouped under a single appellation; FNS: flavone synthase; F3′H: flavonoid 3′-hydroxylase; OMT21: O-methyltransferase 21; PT3: prenyl transferase 3”; OSC: Oxidosqualene cyclase; CCC:-chair–chair–chair.
2 Mechanistic Background of Hairy Root Culture: A Biotechnological Revolution in Plant Secondary Metabolism
In vitro culture methods are crucial in plant propagation, reforestation, conservation, and SMs production [25,26]. Different in vitro culture tactics can be applied to Cannabis to enhance its potential [27]. Compared to the traditional methods, in vitro culture for metabolite production offers several advantages, including stability independent of environmental variables, elimination of pesticide usage, consistent yields and quality, shorter growth cycles, and the capacity to manipulate metabolite pathways through bioengineering approaches [28,29,30].
The engineered production of SMs has been achieved using synthetic biology in in vitro HR culture [29,30]. HRs are root-like structures formed by plants following infection by Agrobacterium rhizogenes [31,32]. These HRs are advantageous due to their robust metabolic synthesis, rapid growth, hormone autotrophy, and genetic stability, making them a popular platform for the biosynthesis of plant SMs [32,33]. The process by which the gram-negative bacterium A. rhizogenes triggers the formation of HRs is akin to how Agrobacterium tumefaciens leads to crown gall disease [34]. The bacterium infects the roots through wounds or natural openings [35]. Four key events initiate this bacterial infection and the development of disease symptoms [18]: (i) chemotactic movement of bacteria toward wounded root cells, followed by attachment, triggered by the release of phenolic compounds like acetosyringone; (ii) T-DNA (which harbors genes involved in phytohormone biosynthesis, includes 18 open reading frames. Among these, the ‘rol’ (rooting locus) genes such as rolA, rolB, rolC, and rolD, are most important for the induction and development of HR formation, processing and T-complex translocation to the plant genome; (iii) T-DNA incorporation and expression; and (iv) development of branched, neoplastic, and ageotropic HRs at infection sites [32,36,37]. The bacterium is attracted to plant cells by “chemotaxis”, a process initiated by phenolic compounds and reducing sugars released from wounded tissue, activating the expression of bacterial virulence genes [38]. The chromosomal virulence genes chvA, chvB, and pscA facilitate bacterial attachment by synthesising and exporting phenolic compounds [18,19,20,32]. The VirA protein detects chemotactic agents, phosphorylating the VirG protein, which binds to the vir regulatory sequence to produce virulence proteins VirA-G, responsible for T-DNA delivery and integration [18,19,20,32] (Fig. 2).
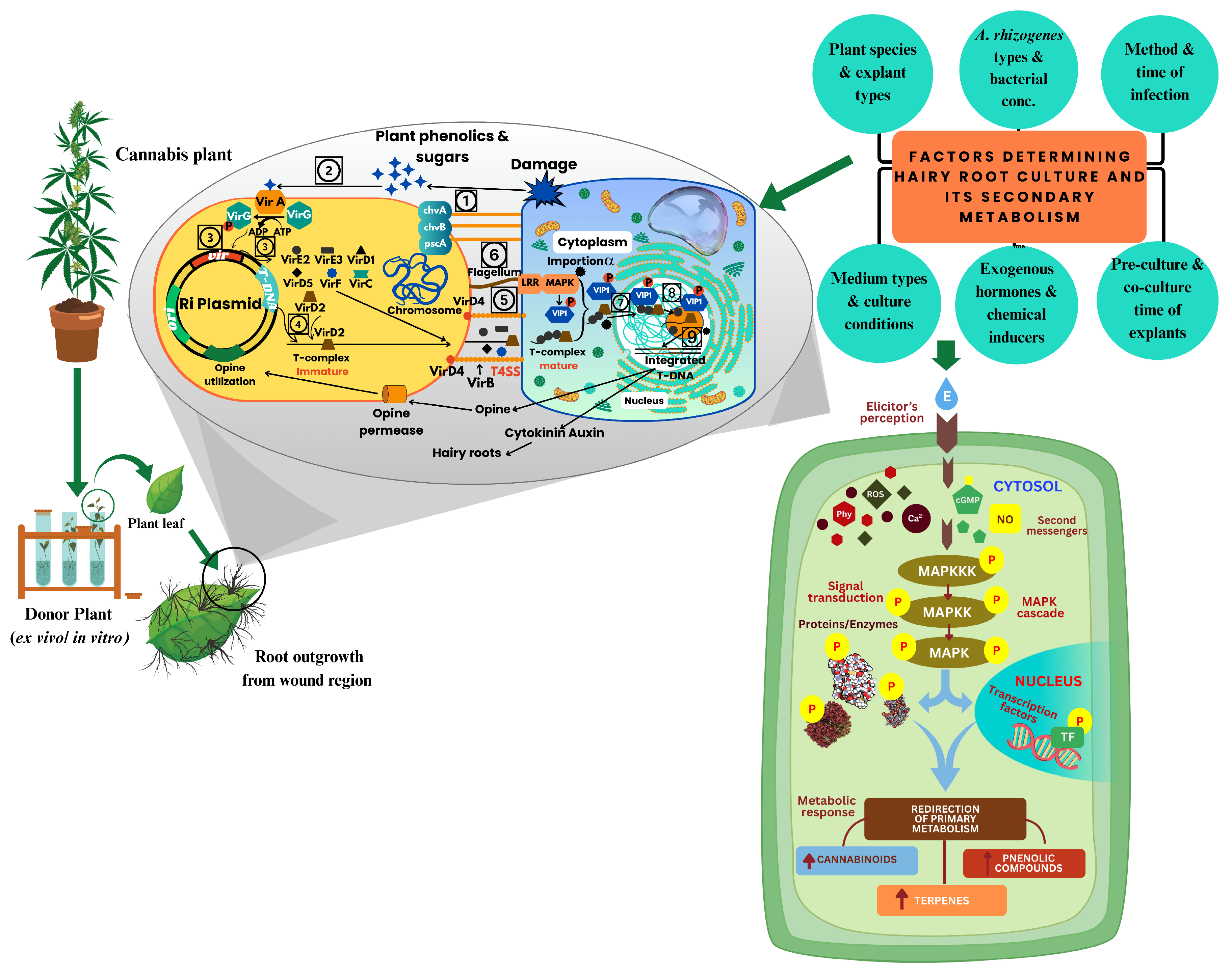
Figure 2: Sterile Cannabis leaf explants are exposed to R. rhizogenes culture for the transfer and integration of T-DNA from the bacterium Ri-plasmid to the plant genome. This process involves ten distinct steps: (1) Agrobacterium attaches to wounded plant cells. (2) The VirA/VirG two-component system detects phenolic signals from the plant and activates the response regulator VirG through phosphorylation. (3) Activated VirG upregulates the expression of other virulence (Vir) genes. (4) The VirD1 and VirD2 proteins process the T-DNA border sequences, producing a single-stranded T-DNA molecule and forming the immature T-DNA complex. (5) This T-DNA, associated with VirD2 and coated by VirE2 proteins, is transported into the plant cell via the Type IV Secretion System (T4SS). (6) Bacterial flagellin triggers a mitogen-activated protein kinase (MAPK) cascade, leading to the phosphorylation of VirE-interacting proteins (VIPs). (7) The T-DNA complex, along with VIPs and importin α, facilitates nuclear import. (8) Once inside the nucleus, VirF and the host ubiquitin-proteasome system remove associated proteins, freeing the T-DNA. (9) The T-DNA is then integrated into the plant genome. (10) Expression of the transferred genes, including those encoding cytokinin and auxin biosynthesis (rol genes), induces uncontrolled cell division and leads to HR formation. Simultaneously, elicitor signals—such as chitin, salicylic acid, and methyl jasmonate—are perceived by membrane-bound receptors and transmitted via secondary messengers including calcium ions (Ca2+), reactive oxygen species (ROS), nitric oxide (NO), cyclic GMP (cGMP), and various phytohormones. These messengers activate protein kinases and MAPK cascades, phosphorylating downstream proteins and transcription factors. This orchestrated signalling network redirects primary metabolism toward the enhanced biosynthesis of secondary metabolites, making HR cultures a powerful platform for phytochemical production.
A. rhizogenes-induced HRs can be generated in vitro and in planta [20,39,40]. The in vitro techniques encompass two ways: (1) Direct inoculation of plants with bacterial cultures and (2) Co-cultivation of plant explants with bacterial suspensions. Effective methodologies, such as vacuum infiltration and sonication, can enhance transformation efficiency [20,41,42,43]. For the biosynthesis of SMs, HRs are initially cultivated in shake flasks and subsequently scaled up in bioreactors [20,43,44,45]. The process involves the translocation of T-DNA and virulence proteins into plant cells, establishing a connection between the bacterial envelope and the host plant cell’s plasma membrane [46,47,48]. Within the plant cell cytoplasm, T-DNA is enveloped by Vir proteins to form a mature T-DNA complex, which subsequently enters the nucleus. Agrobacterium flagellin interacts with leucine-rich repeat (LRR) receptors in plant cells, initiating the MAPK cascade that culminates in the production of phosphorylated VIP proteins [19,32,49,50]. The intracellular transport and nuclear localisation of the T-DNA complex are facilitated by an integrated system involving cytoskeletal elements, the mature T-DNA complex itself, phosphorylated VIPs proteins, and plant importins. Inside the nucleus, Vir proteins, along with the plant’s ubiquitin-proteasome system, strip away bacterial virulence factors and specific host proteins from the T-DNA complex, enabling its efficient integration into the plant genome [19,32,49,50]. However, the integration of T-DNA into the plant genome is not site-specific, and the precise mechanisms governing its incorporation remain incompletely understood [51]. Once integrated, the T-DNA prompts the transformed plant tissue to synthesise cytokinin and auxin, resulting in uncontrolled cell proliferation and the formation of tumours or HRs [52]. Additionally, the transformed tissue produces opine, a nitrogen and carbon source for Agrobacterium [53].
For plants that are challenging to regenerate and have lengthy transformation cycles, Agrobacterium rhizogenes can generate composite plants with transgenic roots and wild-type stems [54]. These composite plants enable the examination of gene functions in root interactions, stress responses, growth, and SMs biosynthesis [55]. HRs are widely employed in plant genetic transformation through several mechanisms: (A) Composite plant generation involves the use of A. rhizogenes, applied via techniques such as stem injection, stem acupuncture, wound coating, and immersion [32,36,56]. (B) In root-suckering species, the cut-dip-budding (CDB) system induces HRs and the subsequent development of transgenic plants under non-sterile conditions. (C) Alternatively, in vitro transformation under sterile conditions can be achieved by either regenerating whole plants from HRs using phytohormones or inducing callus formation from HRs, followed by the generation of transgenic shoots [18,32,36,57]. Composite plants serve as valuable models for investigating the interactions between aerial and root systems and can be cultivated in non-sterile environments, thereby reducing costs [32,36,58,59]. These plants have been developed across over 50 species, including woody, herbaceous, tuberous, and vine types. The induction of HRs is influenced by various factors, such as plant species, explant type, strain type, bacterial components, culture conditions, and hormones [32,60]. For practical applications, it is crucial to select appropriate A. rhizogenes strains and optimise transformation parameters for the target plants [32,61]. SMs are synthesised in plant tissues in response to pathogens or stress. Even in minimal quantities, elicitors can simulate these responses, promoting metabolite accumulation. Elicitors are classified into two categories: abiotic and biotic [62]. Abiotic elicitors originate from non-biological sources, including physical factors (such as temperature, drought, salinity, light, and pH) and chemical stressors (such as metal ions and toxins) [63,64]. In contrast, biotic elicitors originate from biological sources, including microorganisms such as rhizobacteria and fungi, as well as components derived from pathogens or plants following pathogen attacks, such as chitin, chitosan, oligogalacturonides, cellulose, flagellin, and lignin. Additionally, phytohormones and signalling molecules can also act as elicitors by modulating defence responses and secondary metabolism [65,66]. The plant’s response begins with the recognition of elicitors by plasma membrane receptors, initiating signal transduction and leading to the production of intracellular messengers (such as reactive oxygen species, calcium, nitrogen oxide, cGMP, phytohormones) [18,20,23,32,66,67,68,69]. This process activates signalling pathways, including the mitogen-activated protein kinase cascade, which triggers phosphorylation events [18,20,32,69]. Phosphorylated TFs initiate the transcription of defence genes, including those encoding enzymes involved in SMs biosynthesis. Posttranslational phosphorylation may directly activate biosynthetic enzymes [18,20,32,69,70]. Phytohormone signalling pathways interact to regulate plant defence through hormone-responsive transcription factors [68,71,72,73]. Plant hormones and growth regulators enhance immunity against environmental stress, while SMs aid plants in adapting to environmental challenges [20,32,74,75,76]. Throughout their lifespan, plants produce both primary and secondary metabolites [77]. Plant growth regulators, encompassing hormones and signalling molecules, detect stress-induced cellular changes and act as transmitters, facilitating stress-responsive communication between plants and their environment through signalling transductions [78,79,80] in vitro HR culture.
3 Recent Advances of Hairy Root Culture in C. sativa
The experimental settings were optimised to infect hemp, a difficult-to-transform plant, with either Ri or Ti plasmid-bearing agrobacteria strain AR10GUS and create stably transformed tissues [81]. The hypocotyl of intact seedlings was most sensitive, depending on bacterial strain, rol gene types, and plant variety. Nodes and internodes, host tissues with actively proliferating cells, may be attractive targets for Agrobacterium transformation. Since T-DNA impairs auxin response, undamaged seedlings with shoot tips and early leaves are expected to have higher transformation rates. This is the first reported protocol for establishing C. sativa HR cultures [81]. Another research focused on the developmental patterns of HRs in shaking flasks and how various exogenous auxins induced them in callus cultures. A 35-day growth cycle was conducted in darkness at 25°C. HR development grew at regular intervals on solid MS B5 medium supplemented with 4 mg/L naphthalene acetic acid (NAA). The amount of cannabinoids produced by this culture was minimal and persisted below 2.0 μg/g dry weight in this culture [82].
Nevertheless, the conventional in vitro method remains labour-intensive, slow, and challenging to scale up. A new, very effective ex vitro transformation technology has emerged as a solution to these issues, and it has optimised the Cannabis production process [83]. Despite this progress, a major obstacle still stands in the way of a low-cost, quick, simple, efficient and high-throughput HR transformation. Recent groundbreaking research optimised a two-step ex vitro HR transformation process, comparing it to current methods and employing the real mother plant in conjunction with the RUBY system. The real plant achieved a transformation efficiency of 90% using the two-step ex vitro procedure, which was more effective than both the one-step ex vitro and in vitro methods. Reducing the time to produce significant transgenic HR formation by 9–29 days compared to previous approaches, this technology also reveals a quicker and less challenging approach [83].
The primary biotic elicitors that stimulate the production of secondary metabolites include methyl jasmonate (MeJA), jasmonic acid (JA), acetylsalicylic acid, chitosan (CHI), coronatine, pectin, salicylic acid (SA), and yeast extract (YE)’ [80]. These substances enhance terpene accumulation in HRs [1,80]. Given that wild-type C. sativa takes a minimum of three months to mature. Its roots are challenging to separate from the soil. Recent research has explored the application of four elicitors (SA, MeJA, CHI, and YE) to enhance the production of friedelin and epifriedelanol in HR culture as an alternative biosynthetic approach [1]. Friedelin and epifriedelanol are pentacyclic triterpenoids that naturally accumulate in hemp roots [1,8,84].
The highest levels of epifriedelanol (3.79-fold increase) and friedelin (3.25-fold increase) were recorded at the end of the exponential growth phase. After 28 days of cultivation, the maximum substance accumulation occurred when using 3% sucrose in HRs. SA at 100 µM generated the highest epifriedelanol production, and SA at 50 µM produced the maximum friedelin content, which increased production levels by 5.22-fold and 2.88-fold compared to the control after 96 h. Friedelin content and epifriedelanol increased by 2.44 and 3.73 times, respectively, versus control conditions after 24 h of 100 µM MJ treatment [8]. The examination proves that C. sativa plants transformed with rhizogenes wounded roots significantly boost their capability to produce commercial-grade friedelin and epifriedelanol. Multiple aspects influence the development and modification of HRs, including explant type and levels of Rhizobium rhizogenes strain and bacterial density, growth phase duration, infection technique, chemical inducers, and co-cultivation period. According to PCR analysis, the C. sativa HR genome contained proven integration of rolB and rolC genes [8]. A different research study examined triterpenoid production after roots received MeJA, SA, CHI, and YE as elicitors for three and six days. High-Performance Thin-Layer Chromatography (HPTLC) served as a method to screen both triterpenoids before GC-FID measured root tissue levels and verified their identities [1]. The triterpenoid production levels in HRs surpassed natural roots, where salicylic acid at 75 μM promoted 1.95-fold increases in friedelin and 1.4-fold increases in epifriedelanol compared to untreated HRs. The herbal medicine industry could potentially utilise C. sativa HRs instead of natural roots, as these transplanted roots produce higher triterpenoid content while requiring minimal time for growth in a pest-free, heavy metal-tested medium [1].
Bioreactors provide sterile, controlled environments for high-density culture using either liquid nutrient media or liquid/air systems, with key parameters such as agitation, aeration, temperature, and pH precisely regulated to optimise growth and metabolite production [85]. They are broadly categorised into liquid-phase, gas-phase, temporary immersion, and modified types [85]. Liquid-phase bioreactors can be further divided into mechanically and pneumatically agitated types [85]. Metabolite profiling using NMR (Nuclear Magnetic Resonance) and LC/MS of hemp HRs grown in L-type bioreactors, compared with hemp aeroponic roots (HARs), revealed a substantial increase in bioactive compounds. Notably, hemp HRs exhibited a 12-fold increase in cannabisins and a 6-fold increase in acidic triterpenes, along with elevated levels of sterols, phenolic acids, fatty acids, nucleosides, and tyramine derivatives [6,24]. These findings highlight the potential of hemp HRs as a source of specialised metabolites.
Plants initiate diverse immune responses to restrict pathogen infection during Agrobacterium–plant interactions [86]. Phytohormones, vital for plant growth and development, also play a crucial role in regulating immunity [87]. Thus, Agrobacterium virulence is intrinsically influenced by phytohormones and plant-derived metabolites [88]. Despite these insights, the transformation of recalcitrant crops, such as C. sativa, remains challenging due to technical constraints. Enhancing transformation efficiency can be achieved by employing Agrobacterium that overexpresses specific Virulence (Vir) genes [88]. Recent progress in adopting the ternary vector (Tv) system enriched with additional Vir genes has markedly improved both genetic transformation and CRISPR/Cas9-mediated gene editing in hemp [89,90]. The development of this super-infective Tv system represents a significant advancement in overcoming the barriers associated with Agrobacterium-mediated transformation, particularly for recalcitrant crops like Cannabis, and holds promise for future applications in hemp HR cultures [88].
4 Hairy Root Culture Mediated Genetic Transformation: Untapped Potential of Secondary Metabolism in C. sativa
HRs serve as a platform for generating valuable pharmaceuticals, complex SMs, recombinant proteins, and innovative drugs derived from medicinal plants, acting as “green cell factories” [20,91,92]. Recombinant proteins generated by HRs are often secreted extracellularly, allowing purification in a medium lacking protein [91]. HRs play a vital role in areas such as metabolic engineering, bioreactor design, biotransformation research, phytoremediation, and interactions between plants and microbes [93]. These characteristics have unveiled numerous applications, facilitating the industrial-scale production of high-value molecules [20,92,94,95,96].
Combining GE and hairy root transformation provides a practical approach for investigating gene function [97]. Since 1983, interest in gene manipulation has surged following reports of transgenic plant creation [98,99]. DNA modification techniques such as ‘meganucleases, gene/transcription factor overexpression, virus-induced gene silencing (VIGS), RNA interference (RNAi), zinc finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and CRISPR-Cas nucleases’ are prevalent in plant functional genomics [30,36,97,100,101]. Before the advent of CRISPR/Cas, researchers primarily relied on meganucleases, ZFNs, and TALENs [100]. CRISPR/Cas GE was first demonstrated in plant protoplasts and whole plants in 2013, integrating into HR transformation a year later [97]. CRISPR/Cas multiplexing for gene/promoter modifications, chromosomal dissection, CRISPR activation (CRISPRa), CRISPR interference (CRISPRi), CRISPR/Cas13a, and dCas13a for RNA manipulation, activation of cryptic genes, base editing, epigenome engineering, and the development of supervirulent Ag rhizogenes strains can characterize and manipulate cannabinoid biosynthesis in HR culture using various promoters and markers to enhance enzyme efficiency and develop novel Cannabis traits in future [30,36,100,101,102].
Among GE tools, the CRISPR/Cas system has shown advantages in terms of specificity, simplicity, and flexibility compared to earlier GE systems. The emergence of Cas9 mutants like dCas9, which have lost endonuclease activity but retain DNA recognition with guide RNA, offers powerful genetic manipulation tools to knock out, edit, knock in, and downregulate target genes [30,97,102,103]. The CRISPR/Cas system employs nucleases to target specific genome sites, creating double-strand breaks (DSBs) with the help of guide RNAs, which then trigger DNA repair through either the HDR (Homology-Directed Repair) or NHEJ (Non-Homologous End Joining) pathways [104]. Beyond gene deletion or integration, precise control over gene expression is crucial in metabolic engineering [105]. Although siRNA regulation has been applied in plants, dCas9-based CRISPRi is a silencing tool for prokaryotes [30,105,106]. The dCas9 variant, which is catalytically inactive, attaches to genome sites using sgRNA [106]. Upon binding to a genomic location, dCas9 obstructs RNA polymerase from progressing to downstream genes. This interference mechanism can suppress the expression of the target gene by cleaving foreign DNA or RNA, thereby providing precise control of metabolic processes in hemp HR culture [30,106].
Epigenetic modifications are crucial for chromatin remodelling and include mechanisms such as histone posttranslational modifications, DNA methylation, and RNA interference [100,107]. In plants, the biosynthetic gene cluster (BGCs) responsible for secondary metabolite production is typically located in heterochromatin regions, where epigenetic factors regulate their transcription tightly. Small-molecule epigenetic modifiers can alter chromatin architecture, potentially activating these silent gene clusters. This strategy presents a promising avenue for uncovering novel biosynthetic pathways and identifying previously unexpressed bioactive compounds [100,107]. The epigenome comprises sequence-independent biological molecules that influence chromatin structure and gene expression patterns. Epigenomic regulation is achieved through interactions between DNA-binding proteins, biochemical modifications to DNA and histones, and structural changes that affect DNA accessibility. By simply altering the protospacer sequence within guide RNAs (gRNAs), dCas-based effectors have advanced epigenome editing and enhanced the understanding of epigenetic regulation in the secondary metabolism of C. sativa [100,107].
Before the advent of CRISPR-Cas13a, the primary methods for reducing gene expression were RNA interference (RNAi) and CRISPR-Cas9 [108,109]. RNAi suppresses gene expression in mammalian cells, targeting the cytoplasm where the RNA-induced silencing complex resides [109,110,111]. However, RNAi’s effectiveness is limited as it does not interact with nuclear mRNA and necessitates the design of specific siRNA that matches the mRNA sequence, restricting its use [109]. CRISPR-Cas9, on the other hand, can cause DNA off-target effects, and the toxicity of Cas9 may harm essential cellular genes, thus limiting its clinical applications [109]. The CRISPR-Cas13a system, which includes programmable RNA guides, includes two HEPN domains linked to RNases, suggesting its RNA-targeting capability [109,112,113]. Modulating gene expression within cells’ RNA level offers a crucial regulatory mechanism [113]. Editing transcripts provides a novel method for examining gene function, enhancing genetic techniques, and offering insights into RNA-level regulation. CRISPR-Cas13a specifically targets RNA for gene editing [113,114]. Base editing allows for precise genome modifications through single-base conversion without the need for donor templates or double-stranded breaks [113,115].
Chromosomal rearrangements, such as inversions, deletions, and duplications facilitated by Cas9 with paired sgRNAs, are crucial for studying genome variations and regulation, though the mechanisms remain unclear in C. sativa [113,116]. While CRISPR-Cas9 has been at the forefront of GE, the use of CRISPR-Cas12a is on the rise for plant genome engineering [30,103,117]. The growing popularity of CRISPR-Cas12a is due to its versatility and simplified features. Among CRISPR systems, Cas9 and Cas12a, with short guide RNA (42–44 nucleotides of crRNA), are more effective in regulating multiple genes. Although CRISPR-Cas9 remains the most widely used tool for plant GE, CRISPR-Cas12a is being increasingly adopted in metabolic engineering [30,117,118,119,120].
A significant limitation of CRISPR-Cas9 is its restriction to regions with high GC content due to the “G” rich PAM sequence requirements. Despite ongoing efforts to develop nearly “PAM-less” Cas9 variants, their application in plant systems is limited [30,103,117,121]. Cas12a can edit “T” rich PAM regions and create staggered ends that may facilitate site-directed integration [121]. Although Cas12a’s PAM sequence (TTTV) is longer than Cas9′s (NGG), variants with modified PAM specificities have been engineered [30,103,117]. This GE technology could be further expanded by incorporating Cas12a, allowing for more target sites and potentially higher gene integration efficiency in C. sativa HR culture [36,122]. As promoters and introns are AT-rich, Cas12a offers additional engineering flexibility. Although Cas12a exhibits temperature sensitivity, which limits its utility in plant GE, variants with enhanced activities have been developed [30,103,117].
Transcription factors (TFs) regulate gene expression by binding to DNA regulatory sequences, such as enhancers and silencers, thereby either promoting or inhibiting transcription [123,124]. The expression of native or foreign TFs can modify product flow by changing the functions of pathway genes [125]. Various TFs are involved in the transcription of SMs biosynthetic pathways [18,36,97,126,127]. The combination of the dCas9 protein with a transcriptional activator for targeted gene regulation has advanced biotechnology in both medicine and agriculture [128,129]. TFs induced by hormones or elicitors, such as AP2/ERF, WRKY, bHLH, bZIP, MYB, and NAC, facilitate the accumulation of SMs by modulating gene expression through signalling pathways in C. sativa [69,125,130]. CRISPR/dCas9 activation enhances the transcription of endogenous genes by directing TFs to specific promoters [30,103].
The identification of crucial rate-limiting enzymes has enabled metabolic engineering in HRs [131,132]. The synthesis of plant SM involves intermediates and precursors [133]. Genetic manipulation of key genes and TFs can boost the levels of target products. Techniques include gene overexpression and silencing [36,126,127,134,135]. Overexpressing upstream genes increases the supply of precursors, thereby promoting the accumulation of target products, while overexpressing key enzyme genes directly boosts metabolite accumulation [136,137]. To enhance recombinant protein yield in HR cultures, researchers employ several strategies: (1) design efficient expression vectors; (2) incorporate protein stabilizers; (3) develop suitable bioreactors. Various DNA elements, such as constitutive promoters, root-specific promoters, inducible promoters, 5′UTRs, and terminators, regulate gene expression in hemp HR cultures [36,126,127,138,139].
Overall, to fulfil the growing industrial demand for therapeutic plant specialized metabolites (PSMs), advanced approaches such as systems biology, omics technologies, synthetic biology, and GE have become essential tools for unraveling the key molecular components, rate-limiting steps, and regulatory networks governing PSM biosynthesis [105]. Among these, the CRISPR/Cas system has emerged as the most widely adopted GE technology in plants, owing to its simplicity, high precision, adaptability, and capacity for multiplexed gene editing [140]. The application of CRISPR-based strategies in metabolic engineering (ME) holds significant promise for generating Cannabis plants with customized metabolic profiles ranging from nutraceutical to therapeutic compounds through sustainable and eco-friendly approaches, such as HR culture, in the future [30,140].
The plant system offers benefits such as humanised expression, host engineering, growth maintenance, protein design, glycosylation, sialylation, epitope prediction, antibody optimisation, regulatory element prediction, protein stability, reduced risk of pathogenic contamination, and cost-effective production [141,142]. However, expressing recombinant proteins presents challenges like non-human post-translational modifications, protein misfolding, and instability [142,143]. Artificial intelligence (AI) plays a crucial role in biotechnology and plant molecular pharming, enabling increased yield and stability through AI-based strategies to overcome these limitations [30,142,144,145]. Plant-based recombinant protein production can be enhanced using synthetic biology tools and Machine Learning (ML) algorithms for protein folding, stability, and organelle targeting [146]. AI models, including neural networks, support vector machines, linear regression, Gaussian process, and regressor ensembles, predict training data to optimise protein structures, thermostability, catalytic activity, and folding [30,142,144,145,147]. They assist biologists in identifying gene expression patterns by understanding the combinations of factors [148]. Deep Learning (DL) is a supervised learning method that uses multiple layers for high-throughput computing and mapping datasets in plant metabolomics. AI/ML applications include protein engineering, protein interactions, stability, localisation, functional prediction, and catalytic activity [149]. Currently, AI has significantly increased yield and stability in plant molecular pharming [30,142,144,145,147] (Fig. 3). Artificial intelligence (AI) models and optimisation algorithms (OAs) are widely employed in various fields of technology and science, and have recently been applied to enhance different stages of plant in vitro culture [150,151]. The usefulness of AI-OA has been demonstrated in the prediction and optimisation of biomass in plant cell cultures or HR cultures, as well as the optimisation of environmental conditions to achieve maximum productivity from Cannabis HR cultures in the future.
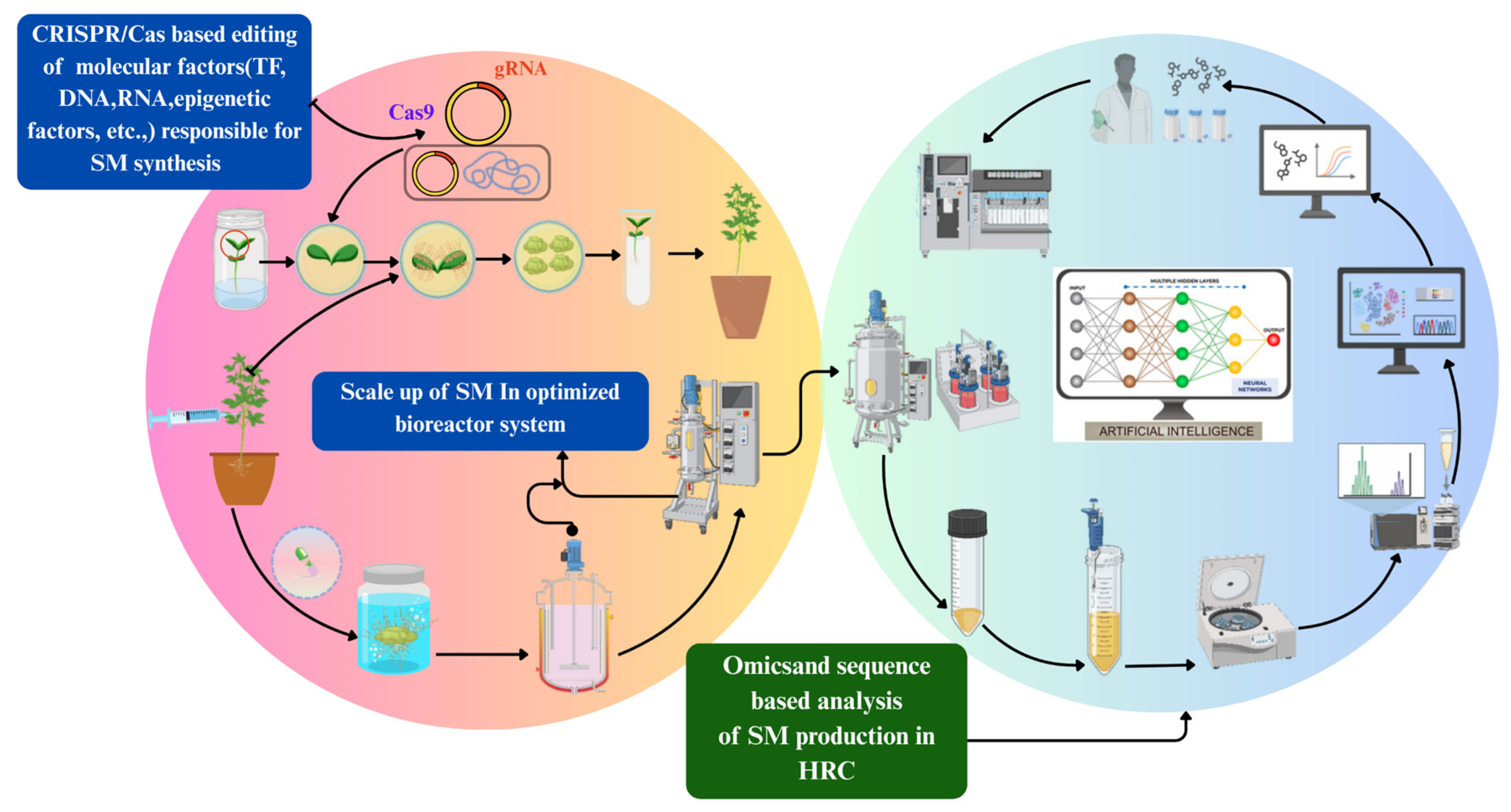
Figure 3: Different CRISPR-based genome and epigenome editing methods for characterising and manipulating the cannabinoid biosynthesis pathway in HR culture. After HR induction, individual lines are established, and high-producing, fast-growing clones are selected. Root cultures exhibiting enhanced production capacity during the shake-flask stage are then transferred to bioreactors with designs optimised for large-scale production. This scaling process can eventually lead to the patenting of the production method. Throughout the shake-flask and bioreactor stages, multiple strategies are applied to enhance secondary metabolite (SM) biosynthesis and optimise process parameters. Additionally, integrating various omics datasets—including genomics, transcriptomics, epigenomics, microRNAomics, proteomics, and metabolomics—facilitates a comprehensive analysis of the cannabinoid biosynthesis pathway in HR cultures. Artificial Intelligence (AI) can be leveraged to model, predict, and optimise HR systems across various stages (development, growth, and production), advancing system biology-based knowledge and enabling more efficient production processes.
5 Bioreactor Technology for HRs Cultivation in C. sativa: Prospects for Small-Scale Cultures to Global Industrial Biomanufacturing
The industrial production of SMs for sectors like health, cosmetics, and food often depends on harvesting non-cultivated endemic species, which threatens genetic resources [152,153,154,155]. Even when these metabolites are present in cultivated species, the extraction rates are frequently too low to be economically feasible [153,154,155,156]. Therefore, developing alternative methods to produce these substances in large quantities is strategically crucial [153,154,155,157,158]. With the rising demand for the health benefits of plant-derived secondary metabolites, large-scale production using bioreactor technology becomes essential [159]. Temporary immersion systems (TIS) are widely used bioreactors that improve plant performance by providing optimal aeration and periodic feeding [153,154,155,160,161]. TISs are effective for both micropropagation and the production of secondary metabolites, especially in HR culture [153,154,155,161].
When transitioning from shake flasks to industrial bioreactors, the environment undergoes changes in both hydrodynamic forces and rheological characteristics [85,161,162]. These alterations impact shear stress, the availability of oxygen, and the composition of gases, which can decrease biomass and secondary metabolite productivity [163]. The complex morphology and uneven growth of HRs pose significant challenges for industrial applications [164]. Their tangled, fibrous structure forms interwoven networks and boundary layers, resulting in non-uniform conditions that hinder the delivery of nutrients and oxygen [85,161]. Oxygen deficiency is a major limitation in HR bioreactors [20,154]. While intense mixing can improve mass transfer, it concurrently raises hydrodynamic shear stress, which adversely affects HRs health and reduces productivity by inducing callus formation [164,165].
Given the specific challenges of HR cultivation, bioreactor design should focus on ensuring adequate mixing with minimal shear stress, improving mass transfer, and lowering hydrodynamic pressure [166]. For successful scale-up, a HR bioreactor must maintain consistent geometry and flow dynamics, supply sufficient nutrients and oxygen, and use scalable parameters that can be reliably applied to larger systems [20,154,167,168,169]. Key issues in bioreactor operation include structure, type of biological entity, CFD simulation, sterilization, parameter configuration, and integration with other bioreactors to control biochemical processes for maximum SM productivity in C. sativa HR culture [20,154,168,169].
The application of HR cultures in C. sativa is not limited to laboratory settings but extends to industrial biomanufacturing [170]. Advanced bioreactors equipped with automated nutrient cycling, environmental management, and real-time metabolite extraction facilitate the continuous production of secondary metabolites [91]. Scaling up to industrial levels is feasible through bioreactor technologies that enhance growth while reducing expenses. Biosensors and AI-driven systems boost production efficiency. Biosensors provide real-time monitoring of metabolite levels [126], while AI systems forecast optimal conditions and extraction timings, elevating terpenoid production to pharmaceutical-grade standards [126]. This precision is crucial for ensuring consistency in large-scale manufacturing, which is vital for adhering to regulatory standards in the pharmaceutical sector [32,126]. The future of HR culture is poised to become the cornerstone of a new bio-industry [171,172]. Plants like Cannabis act as biofactories for producing rare, valuable compounds that are challenging to synthesise chemically [126]. HR cultures provide a sustainable method for generating next-generation pharmaceuticals and bioactive compounds [126,172,173]. This shift from research to global biomanufacturing heralds a new era in industrial biotechnology [172], transforming the production of plant-derived medicines. A significant hurdle in the large-scale production of SMs via HRs cultivation is the lack of comprehensive knowledge about metabolic pathways and their regulation, which diminishes their economic viability in C. sativa [30].
To address this, future strategies should leverage omics technologies, such as genomics, transcriptomics, proteomics, and metabolomics, to gain insights into metabolite synthesis pathways [174,175]. This can be achieved by identifying missing enzymes in these pathways or discovering enzyme variants with enhanced activity, ultimately boosting SM productivity in Cannabis HR culture [176,177]. Initial investments during cultivation are linked to bioreactor design and process parameters [30,142,144]. Disposable bioreactors are favoured for cost reduction, while mathematical models and real-time monitoring can optimise cell cultivation and boost productivity on a commercial scale [178]. Neural network-based ML models with systems engineering could enhance biologics manufacturing across production stages in the Cannabis plant [179]. Omics data is crucial in optimising SMs associated recombinant protein production by providing comprehensive insights into cellular processes. However, further omics-driven studies are essential to fully leverage machine learning tools for predictive modelling and process enhancement [30,142,144].
The research on HRs cultivation in C. sativa has just begun. The effectiveness across other Cannabis cultivars requires validation due to genetic variability. Scaling for commercial purposes may require the use of automation or high-throughput strategies to maintain reproducibility. However, recalcitrant crops such as C. Sativa and prevailing technical challenges continue to serve as significant drawbacks. A promising approach to improving transformation efficiencies involves employing Agrobacterium strains that overexpress specific Vir genes and fine-tuning the protocols to optimize individual cultivars. The potential of HR culture in biotechnology for producing ‘high value, low volume’ SMs is widely acknowledged. Such cultures can be established in various plant species, including those that are endangered or have medicinal values. Through metabolic engineering, HRs clones with high yields of bioactive pharmaceuticals can be developed, which requires a comprehensive understanding of metabolic pathways, regulatory mechanisms, enzymes, genes, and their products. Multipoint metabolic engineering is more effective than single-point approaches in altering metabolic flux. Many SMs’ biosynthetic pathways remain incompletely understood. Current molecular research, utilizing genomics, transcriptomics, proteomics, and next-generation sequencing (NGS) techniques, helps identify genes for metabolic engineering. Functional genomics has identified key genes in cannabinoid biosynthesis, demonstrating that interdisciplinary approaches enable the production of rare cannabinoids, such as cannabigerol (CBG), cannabinol (CBN), cannabichromene (CBC), and tetrahydrocannabivarin (THCV), which have therapeutic potential. Elicitors such as MeJA or salicylic acid SA can boost metabolite production. Introducing multiple genes is challenging due to the unsynchronized expression of these genes. Single-cell omics can explore cellular conditions, revealing interactions within pathways. Systems metabolic engineering combines systems biology, synthetic biology, and traditional methods to identify pathways and mechanisms. HRCs require interdisciplinary strategies for improved production. HRs technology is poised to benefit from advancements in CRISPR/Cas9 technology. Industrial hemp can help with phytoremediation and the hyperaccumulation of organic pollutants. The use of engineered HRs systems in phytoremediation allows the removal of contaminants [180]. They also offer potential for developing stress-resistant Cannabis strains and maintaining production under adverse conditions. While progress has been made in utilizing HRs for valuable products, production remains at the laboratory scale. Bioreactors need optimization for commercial production. Integrating biosensors and artificial intelligence (AI) can improve output by monitoring metabolites.
Acknowledgement:
Funding Statement: This work was supported by a research grant of Gyeongsangbuk-do (No. GBHEMP202504), Republic of Korea.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, S. M. Ahsan and Hyong Woo Choi; Writing—Original draft preparation: S. M. Ahsan and Md. Injamum-Ul-Hoque; Writing—review and editing: S. M. Ahsan, Md. Injamum-Ul-Hoque, Soosan Tavakoli, Shahin Imran, and Mallesham Bulle; Visualization: Md. Mahfuzur Rahman, Nayan Chandra Howlader, and Md. Mezanur Rahman, Validation: Hyong Woo Choi. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Kobtrakul K , Rani D , Binalee A , Udomlarp P , Srichai T , De-Eknamkul W , et al. Elicitation enhances the production of friedelin and epifriedelanol in hairy root cultures of Cannabis sativa L. Front Plant Sci. 2023; 14: 1242584. doi:10.3389/fpls.2023.1242584. [Google Scholar] [CrossRef]
2. Odieka AE , Obuzor GU , Oyedeji OO , Gondwe M , Hosu YS , Oyedeji AO . The medicinal natural products of Cannabis sativa Linn.: a review. Molecules. 2022; 27( 5): 1689. doi:10.3390/molecules27051689. [Google Scholar] [CrossRef]
3. Lazarjani MP , Young O , Kebede L , Seyfoddin A . Processing and extraction methods of medicinal Cannabis: a narrative review. J Cannabis Res. 2021; 3( 1): 32. doi:10.1186/s42238-021-00087-9. [Google Scholar] [CrossRef]
4. Wang Y , Wang D , Li S , Zhai Y , Zhao Y , Zhong F , et al. Methyl jasmonate and salicylic acid enhance the total flavonoid, phenolics, and cannabidiol contents of Cannabis sativa L. adventitious roots. Plant Cell Tissue Organ Cult. 2024; 159( 2): 49. doi:10.1007/s11240-024-02909-z. [Google Scholar] [CrossRef]
5. Kamle M , Mahato DK , Sharma B , Gupta A , Shah AK , Chayan Mahmud MM , et al. Nutraceutical potential, phytochemistry of hemp seed (Cannabis sativa L.) and its application in food and feed: a review. Food Chem Adv. 2024; 4: 100671. doi:10.1016/j.focha.2024.100671. [Google Scholar] [CrossRef]
6. Anokwuru CP , Makolo FL , Sandasi M , Tankeu SY , Elisha IL , Agoni C , et al. Cannabigerol: a bibliometric overview and review of research on an important phytocannabinoid. Phytochem Rev. 2022; 21( 5): 1523– 47. doi:10.1007/s11101-021-09794-w. [Google Scholar] [CrossRef]
7. Lange BM , Zager JJ . Comprehensive inventory of cannabinoids in Cannabis sativa L.: can we connect genotype and chemotype? Phytochem Rev. 2022; 21( 4): 1273– 313. doi:10.1007/s11101-021-09780-2. [Google Scholar] [CrossRef]
8. Mahendran G , Vimolmangkang S . Effect of carbon source and elicitors on biomass production and accumulation of friedelin and epifriedelanol in hairy roots of hemp (Cannabis sativa L.). Plant Cell Tissue Organ Cult PCTOC. 2024; 156( 2): 61. doi:10.1007/s11240-023-02675-4. [Google Scholar] [CrossRef]
9. Kaminsky N , Hubert J , Guerin C , Mazlani M , Kotland A , Pozzobon V , et al. Deciphering the phytochemical potential of hemp hairy roots: a promising source of cannabisins and triterpenes as bioactive compounds. Molecules. 2024; 29( 23): 5792. doi:10.3390/molecules29235792. [Google Scholar] [CrossRef]
10. Shiels D , Prestwich BD , Koo O , Kanchiswamy CN , O’Halloran R , Badmi R . Hemp genome editing-challenges and opportunities. Front Genome Ed. 2022; 4: 823486. doi:10.3389/fgeed.2022.823486. [Google Scholar] [CrossRef]
11. Simiyu DC , Jang JH , Lee OR . Understanding Cannabis sativa L.: current status of propagation, use, legalization, and haploid-inducer-mediated genetic engineering. Plants. 2022; 11( 9): 1236. doi:10.3390/plants11091236. [Google Scholar] [CrossRef]
12. Rohit KG , Ruchi P , Itishree D , Sapana K . Regulatory challenges on cannabis: concern, advantages, and disadvantages. In: Shukla R , Handa M , Singh DP , Dhir A , editors. Cannabis and its Derivatives. Cambridge, MA, USA: Academic Press; 2024. p. 307– 33. doi:10.1016/B978-0-443-15489-8.00011-6. [Google Scholar] [CrossRef]
13. André R , Gomes AP , Pereira-Leite C , Marques-da-Costa A , Monteiro Rodrigues L , Sassano M , et al. The entourage effect in Cannabis Medicinal products: a comprehensive review. Pharmaceuticals. 2024; 17( 11): 1543. doi:10.3390/ph17111543. [Google Scholar] [CrossRef]
14. Jin D , Dai K , Xie Z , Chen J . Secondary metabolites profiled in Cannabis Inflorescences, leaves, stem barks, and roots for medicinal purposes. Sci Rep. 2020; 10( 1): 3309. doi:10.1038/s41598-020-60172-6. [Google Scholar] [CrossRef]
15. Huang S , Li H , Xu J , Zhou H , Seeram NP , Ma H , et al. Chemical constituents of industrial hemp roots and their anti-inflammatory activities. J Cannabis Res. 2023; 5( 1): 1. doi:10.1186/s42238-022-00168-3. [Google Scholar] [CrossRef]
16. Jin D , Henry P , Shan J , Chen J . Identification of chemotypic markers in three chemotype categories of Cannabis Using secondary metabolites profiled in inflorescences, leaves, stem bark, and roots. Front Plant Sci. 2021; 12: 699530. doi:10.3389/fpls.2021.699530. [Google Scholar] [CrossRef]
17. Srivastava S , Srivastava AK . Hairy root culture for mass-production of high-value secondary metabolites. Crit Rev Biotechnol. 2007; 27( 1): 29– 43. doi:10.1080/07388550601173918. [Google Scholar] [CrossRef]
18. Gantait S , Mukherjee E . Hairy root culture technology: applications, constraints and prospect. Appl Microbiol Biotechnol. 2021; 105( 1): 35– 53. doi:10.1007/s00253-020-11017-9. [Google Scholar] [CrossRef]
19. Alcalde MA , Perez-Matas E , Escrich A , Cusido RM , Palazon J , Bonfill M . Biotic elicitors in adventitious and hairy root cultures: a review from 2010 to 2022. Molecules. 2022; 27( 16): 5253. doi:10.3390/molecules27165253. [Google Scholar] [CrossRef]
20. Mirmazloum I , Slavov AK , Marchev AS . The untapped potential of hairy root cultures and their multiple applications. Int J Mol Sci. 2024; 25( 23): 12682. doi:10.3390/ijms252312682. [Google Scholar] [CrossRef]
21. Malarz J , Michalska K , Yudina YV , Stojakowska A . Hairy root cultures as a source of polyphenolic antioxidants: flavonoids, stilbenoids and hydrolyzable tannins. Plants. 2022; 11( 15): 1950. doi:10.3390/plants11151950. [Google Scholar] [CrossRef]
22. Gerszberg A , Wiktorek-Smagur A . Hairy root cultures as a multitask platform for green biotechnology. Plant Cell Tissue Organ Cult PCTOC. 2022; 150( 3): 493– 509. doi:10.1007/s11240-022-02316-2. [Google Scholar] [CrossRef]
23. Pistelli L , Giovannini A , Ruffoni B , Bertoli A , Pistelli L . Hairy root cultures for secondary metabolites production. In: Giardi MT , Rea G , Berra B , editors. Bio-farms for nutraceuticals. Boston, MA, USA: Springer; 2010. p. 167– 84. doi:10.1007/978-1-4419-7347-4_13. [Google Scholar] [CrossRef]
24. Sharma P , Padh H , Shrivastava N . Hairy root cultures: a suitable biological system for studying secondary metabolic pathways in plants. Eng Life Sci. 2013; 13( 1): 62– 75. doi:10.1002/elsc.201200030. [Google Scholar] [CrossRef]
25. Fazili MA , Bashir I , Ahmad M , Yaqoob U , Geelani SN . In vitro strategies for the enhancement of secondary metabolite production in plants: a review. Bull Natl Res Cent. 2022; 46( 1): 35. doi:10.1186/s42269-022-00717-z. [Google Scholar] [CrossRef]
26. Alamgir ANM . Cultivation of herbal drugs, biotechnology, and in vitro production of secondary metabolites, high-value medicinal plants, herbal wealth, and herbal trade. In: Therapeutic use of medicinal plants and their extracts: volume 1: Pharmacognosy. Cham, Switzerland: Springer International Publishing; 2017. p. 379– 452. doi:10.1007/978-3-319-63862-1_9. [Google Scholar] [CrossRef]
27. Monthony AS , Page SR , Hesami M , Jones AMP . The past, present and future of Cannabis sativa tissue culture. Plants. 2021; 10( 1): 185. doi:10.3390/plants10010185. [Google Scholar] [CrossRef]
28. Ndlovu SB , Naidoo D , van Staden J , Gebashe FC . A systematic review of Cannabis sativa L. cultivation techniques: a comprehensive overview of tissue culture innovations and growth optimization. Ind Crops Prod. 2024; 222: 119539. doi:10.1016/j.indcrop.2024.119539. [Google Scholar] [CrossRef]
29. Hesami M , Baiton A , Alizadeh M , Pepe M , Torkamaneh D , Jones AMP . Advances and perspectives in tissue culture and genetic engineering of Cannabis. Int J Mol Sci. 2021; 22( 11): 5671. doi:10.3390/ijms22115671. [Google Scholar] [CrossRef]
30. Hesami M , Pepe M , Baiton A , Jones AMP . Current status and future prospects in cannabinoid production through in vitro culture and synthetic biology. Biotechnol Adv. 2023; 62: 108074. doi:10.1016/j.biotechadv.2022.108074. [Google Scholar] [CrossRef]
31. Hu ZB , Du M . Hairy root and its application in plant genetic engineering. J Integr Plant Biol. 2006; 48( 2): 121– 7. doi:10.1111/j.1744-7909.2006.00121.x. [Google Scholar] [CrossRef]
32. Zhu Y , Zhu X , Wen Y , Wang L , Wang Y , Liao C , et al. Plant hairy roots: induction, applications, limitations and prospects. Ind Crops Prod. 2024; 219: 119104. doi:10.1016/j.indcrop.2024.119104. [Google Scholar] [CrossRef]
33. Halder M , Roychowdhury D , Jha S . A critical review on biotechnological interventions for production and yield enhancement of secondary metabolites in hairy root cultures. In: Srivastava V , Mehrotra S , Mishra S , editors. Hairy roots: an effective tool of plant biotechnology. Singapore: Springer; 2018. p. 21– 44. doi:10.1007/978-981-13-2562-5_2. [Google Scholar] [CrossRef]
34. Georgiev MI , Pavlov AI , Bley T . Hairy root type plant in vitro systems as sources of bioactive substances. Appl Microbiol Biotechnol. 2007; 74( 6): 1175– 85. doi:10.1007/s00253-007-0856-5. [Google Scholar] [CrossRef]
35. Veena V , Taylor CG . Agrobacterium rhizogenes: recent developments and promising applications. Vitro Cell Dev Biol Plant. 2007; 43( 5): 383– 403. doi:10.1007/s11627-007-9096-8. [Google Scholar] [CrossRef]
36. Bagal D , Ali Chowdhary A , Mehrotra S , Mishra S , Rathore S , Srivastava V . Metabolic engineering in hairy roots: an outlook on production of plant secondary metabolites. Plant Physiol Biochem. 2023; 201: 107847. doi:10.1016/j.plaphy.2023.107847. [Google Scholar] [CrossRef]
37. Rogowska A , Szakiel A . Enhancement of phytosterol and triterpenoid production in plant hairy root cultures—simultaneous stimulation or competition? Plants. 2021; 10( 10): 2028. doi:10.3390/plants10102028. [Google Scholar] [CrossRef]
38. Rawat JM , Bhandari A , Raturi M , Rawat B . Agrobacterium rhizogenes mediated hairy root cultures: a promising approach for production of useful metabolites. In: Gupta VK , Pandey A , editors. New and future developments in microbial biotechnology and bioengineering. Amsterdam, The Netherlands: Elsevier; 2019. p. 103– 18. doi:10.1016/b978-0-444-63504-4.00008-6. [Google Scholar] [CrossRef]
39. Vaghari H , Jafarizadeh-Malmiri H , Anarjan N , Berenjian A . Hairy root culture: a biotechnological approach to produce valuable metabolites. In: Meena VS , Mishra PK , Bisht JK , Pattanayak A , editors. Agriculturally important microbes for sustainable agriculture. Volume I: plant-soil-microbe nexus. Singapore: Springer; 2017. p. 131– 60. doi:10.1007/978-981-10-5589-8_7. [Google Scholar] [CrossRef]
40. Ratanasut K , Rod-In W , Sujipuli K . In planta Agrobacterium-mediated transformation of rice. Rice Sci. 2017; 24( 3): 181– 6. doi:10.1016/j.rsci.2016.11.001. [Google Scholar] [CrossRef]
41. Kaur M , Manchanda P , Kalia A , Ahmed FK , Nepovimova E , Kuca K , et al. Agroinfiltration mediated scalable transient gene expression in genome edited crop plants. Int J Mol Sci. 2021; 22( 19): 10882. doi:10.3390/ijms221910882. [Google Scholar] [CrossRef]
42. Trull BN , Sultana MS , Pfotenhauer AC , Stockdale JN , Pantalone V , Zhang B , et al. Robust soybean leaf agroinfiltration. Plant Cell Rep. 2024; 43( 6): 162. doi:10.1007/s00299-024-03245-4. [Google Scholar] [CrossRef]
43. Azizi-Dargahlou S , Pouresmaeil M . Agrobacterium tumefaciens-mediated plant transformation: a review. Mol Biotechnol. 2024; 66( 7): 1563– 80. doi:10.1007/s12033-023-00788-x. [Google Scholar] [CrossRef]
44. Sajc L , Grubisic D , Vunjak-Novakovic G . Bioreactors for plant engineering: an outlook for further research. Biochem Eng J. 2000; 4( 2): 89– 99. doi:10.1016/S1369-703X(99)00035-2. [Google Scholar] [CrossRef]
45. Huang TK , McDonald KA . Bioreactor engineering for recombinant protein production in plant cell suspension cultures. Biochem Eng J. 2009; 45( 3): 168– 84. doi:10.1016/j.bej.2009.02.008. [Google Scholar] [CrossRef]
46. Tzfira T , Citovsky V . From host recognition to T-DNA integration: the function of bacterial and plant genes in the Agrobacterium-plant cell interaction. Mol Plant Pathol. 2000; 1( 4): 201– 12. doi:10.1046/j.1364-3703.2000.00026.x. [Google Scholar] [CrossRef]
47. Azhakanandam K , McCabe MS , Power JB , Lowe KC , Cocking EC , Davey MR . T-DNA transfer, integration, expression and inheritance in rice: effects of plant genotype and Agrobacterium super-virulence. J Plant Physiol. 2000; 157( 4): 429– 39. doi:10.1016/S0176-1617(00)80028-0. [Google Scholar] [CrossRef]
48. Gelvin SB . AGROBACTERIUM and plant genes involved in t-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol. 2000; 51: 223– 56. doi:10.1146/annurev.arplant.51.1.223. [Google Scholar] [CrossRef]
49. Biswas D , Chakraborty A , Mukherjee S , Ghosh B . Hairy root culture: a potent method for improved secondary metabolite production of Solanaceous plants. Front Plant Sci. 2023; 14: 1197555. doi:10.3389/fpls.2023.1197555. [Google Scholar] [CrossRef]
50. Malarz J , Yudina YV , Stojakowska A . Hairy root cultures as a source of phenolic antioxidants: simple phenolics, phenolic acids, phenylethanoids, and hydroxycinnamates. Int J Mol Sci. 2023; 24( 8): 6920. doi:10.3390/ijms24086920. [Google Scholar] [CrossRef]
51. Gelvin SB . Plant DNA repair and Agrobacterium T–DNA integration. Int J Mol Sci. 2021; 22( 16): 8458. doi:10.3390/ijms22168458. [Google Scholar] [CrossRef]
52. Chandra S . Natural plant genetic engineer Agrobacterium rhizogenes: role of T-DNA in plant secondary metabolism. Biotechnol Lett. 2012; 34( 3): 407– 15. doi:10.1007/s10529-011-0785-3. [Google Scholar] [CrossRef]
53. Vladimirov IA , Matveeva TV , Lutova LA . Opine biosynthesis and catabolism genes of Agrobacterium tumefaciens and Agrobacterium rhizogenes. Genetika. 2015; 51( 2): 137– 46. doi:10.1134/S1022795415020167. [Google Scholar] [CrossRef]
54. Hooykaas PJJ . Agrobacterium, a natural metabolic engineer of plants. In: Verpoorte R , Alfermann AW , editors. Metabolic engineering of plant secondary metabolism. Dordrecht, The Netherlands: Springer; 2000. p. 51– 67. doi:10.1007/978-94-015-9423-3_3. [Google Scholar] [CrossRef]
55. Halder M , Sarkar S , Jha S . Elicitation: a biotechnological tool for enhanced production of secondary metabolites in hairy root cultures. Eng Life Sci. 2019; 19( 12): 880– 95. doi:10.1002/elsc.201900058. [Google Scholar] [CrossRef]
56. Singh RS , Chattopadhyay T , Thakur D , Kumar N , Kumar T , Singh PK . Hairy root culture for in vitro production of secondary metabolites: a promising biotechnological approach. In: Kumar N , editor. Biotechnological approaches for medicinal and aromatic plants: conservation, genetic improvement and utilization. Singapore: Springer; 2018. p. 235– 50. doi:10.1007/978-981-13-0535-1_10. [Google Scholar] [CrossRef]
57. Ono NN , Tian L . The multiplicity of hairy root cultures: prolific possibilities. Plant Sci. 2011; 180( 3): 439– 46. doi:10.1016/j.plantsci.2010.11.012. [Google Scholar] [CrossRef]
58. Kim Y , Wyslouzil BE , Weathers PJ . Secondary metabolism of hairy root cultures in bioreactors. Vitro Cell Dev Biol Plant. 2002; 38( 1): 1– 10. doi:10.1079/IVP2001243. [Google Scholar] [CrossRef]
59. Mottaki Z , Rezayian M , Niknam V , Ebrahimzadeh H , Mirmasoumi M . Using hairy roots for production of secondary metabolites in Artemisia. Plant Biotechnol Rep. 2019; 13( 3): 263– 71. doi:10.1007/s11816-019-00534-3. [Google Scholar] [CrossRef]
60. Rency AS , Pandian S , Kasinathan R , Satish L , Swamy MK , Ramesh M . Hairy root cultures as an alternative source for the production of high-value secondary metabolites. In: Akhtar MS , Swamy MK , editors. Natural bio-active compounds. Volume 3: biotechnology, bioengineering, and molecular approaches. Singapore: Springer; 2019. p. 237– 64. doi:10.1007/978-981-13-7438-8_10. [Google Scholar] [CrossRef]
61. Kaur G , Prakash P , Srivastava R , Verma PC . Enhanced secondary metabolite production in hairy root cultures through biotic and abiotic elicitors. In: Ramawat KG , Ekiert HM , Goyal S , editors. Plant cell and tissue differentiation and secondary metabolites: fundamentals and applications. Cham, Switzerland: Springer International Publishing; 2020. p. 1– 36. doi:10.1007/978-3-030-11253-0_38-1. [Google Scholar] [CrossRef]
62. Giri A , Narasu ML . Transgenic hairy roots. recent trends and applications. Biotechnol Adv. 2000; 18( 1): 1– 22. doi:10.1016/s0734-9750(99)00016-6. [Google Scholar] [CrossRef]
63. Wang JW , Wu JY . Effective elicitors and process strategies for enhancement of secondary metabolite production in hairy root cultures. In: Doran PM , editor. Biotechnology of hairy root systems. Berlin/Heidelberg, Germany: Springer; 2013. p. 55– 89. doi:10.1007/10_2013_183. [Google Scholar] [CrossRef]
64. Goel MK , Mehrotra S , Kukreja AK . Elicitor-induced cellular and molecular events are responsible for productivity enhancement in hairy root cultures: an insight study. Appl Biochem Biotechnol. 2011; 165( 5–6): 1342– 55. doi:10.1007/s12010-011-9351-7. [Google Scholar] [CrossRef]
65. Chandra S , Chandra R . Engineering secondary metabolite production in hairy roots. Phytochem Rev. 2011; 10( 3): 371– 95. doi:10.1007/s11101-011-9210-8. [Google Scholar] [CrossRef]
66. Abraham J , Thomas TD . Hairy root culture for the production of useful secondary metabolites. In: Malik S , editor. Biotechnology and production of anti-cancer compounds. Cham, Switzerland: Springer International Publishing; 2017. p. 201– 30. doi:10.1007/978-3-319-53880-8_9. [Google Scholar] [CrossRef]
67. Jeong GA , Park DE . Enhanced secondary metabolite biosynthesis by elicitation in transformed plant root system. Appl Biochem Biotechnol. 2006; 130( 1): 436– 46. doi:10.1385/ABAB:130:1:436. [Google Scholar] [CrossRef]
68. Gharari Z , Bagheri K , Danafar H , Sharafi A . Enhanced flavonoid production in hairy root cultures of Scutellaria bornmuelleri by elicitor induced over-expression of MYB7 and FNSП2 genes. Plant Physiol Biochem. 2020; 148: 35– 44. doi:10.1016/j.plaphy.2020.01.002. [Google Scholar] [CrossRef]
69. Trono D . Elicitation as a tool to improve the accumulation of secondary metabolites in Cannabis sativa. Phytochem Rev. 2025; 24( 4): 3119– 55. doi:10.1007/s11101-024-10019-z. [Google Scholar] [CrossRef]
70. Mishra BN , Ranjan R . Growth of hairy-root cultures in various bioreactors for the production of secondary metabolites. Biotechnol Appl Biochem. 2008; 49( Pt 1): 1– 10. doi:10.1042/BA20070103. [Google Scholar] [CrossRef]
71. Baek S , Ho TT , Lee H , Jung G , Kim YE , Jeong CS , et al. Enhanced biosynthesis of triterpenoids in Centella asiatica hairy root culture by precursor feeding and elicitation. Plant Biotechnol Rep. 2020; 14( 1): 45– 53. doi:10.1007/s11816-019-00573-w. [Google Scholar] [CrossRef]
72. Rachappanavar V , Padiyal A , Sharma JK , Gupta SK . Plant hormone-mediated stress regulation responses in fruit crops—a review. Sci Hortic. 2022; 304: 111302. doi:10.1016/j.scienta.2022.111302. [Google Scholar] [CrossRef]
73. Tripathi D , Singh M , Pandey-Rai S . Crosstalk of nanoparticles and phytohormones regulate plant growth and metabolism under abiotic and biotic stress. Plant Stress. 2022; 6: 100107. doi:10.1016/j.stress.2022.100107. [Google Scholar] [CrossRef]
74. Cheruvathur MK , Thomas TD . Effect of plant growth regulators and elicitors on rhinacanthin accumulation in hairy root cultures of Rhinacanthusnasutus (L.) Kurz. Plant Cell Tissue Organ Cult. 2014; 118( 1): 169– 77. doi:10.1007/s11240-014-0473-9. [Google Scholar] [CrossRef]
75. Kastell A , Schreiner M , Knorr D , Ulrichs C , Mewis I . Influence of nutrient supply and elicitors on glucosinolate production in E. sativa hairy root cultures. Plant Cell Tissue Organ Cult. 2018; 132( 3): 561– 72. doi:10.1007/s11240-017-1355-8. [Google Scholar] [CrossRef]
76. Srivastava S , Srivastava AK . Effect of elicitors and precursors on azadirachtin production in hairy root culture of Azadirachta indica. Appl Biochem Biotechnol. 2014; 172( 4): 2286– 97. doi:10.1007/s12010-013-0664-6. [Google Scholar] [CrossRef]
77. Humbal A , Pathak B . Influence of exogenous elicitors on the production of secondary metabolite in plants: a review (“VSI: secondary metabolites”). Plant Stress. 2023; 8: 100166. doi:10.1016/j.stress.2023.100166. [Google Scholar] [CrossRef]
78. Alvarado AM , Aguirre-Becerra H , Vázquez-Hernández MC , Magaña-Lopez E , Parola-Contreras I , Caicedo-Lopez LH , et al. Influence of elicitors and eustressors on the production of plant secondary metabolites. In: Akhtar MS , Swamy MK , Sinniah UR , editors. Natural bio-active compounds. Volume 1: production and applications. Singapore: Springer; p. 333– 88. doi:10.1007/978-981-13-7154-7_11. [Google Scholar] [CrossRef]
79. Rezazadehfar P , Rezayian M , Niknam V , Mirmasoumi M . Elicitor-enhanced steroidal sapogenin accumulation in hairy root cultures of Trigonella foenum-graecum. Sci Rep. 2024; 14( 1): 19106. doi:10.1038/s41598-024-69625-8. [Google Scholar] [CrossRef]
80. Liu C , Ahmad N , Tao Y , Hussain H , Chang Y , Umar AW , et al. Reprogramming hairy root cultures: a synthetic biology framework for precision metabolite biosynthesis. Plants. 2025; 14( 13): 1928. doi:10.3390/plants14131928. [Google Scholar] [CrossRef]
81. Wahby I , Caba JM , Ligero F . Agrobacteriuminfection of hemp (Cannabis sativa L.): establishment of hairy root cultures. J Plant Interact. 2013; 8( 4): 312– 20. doi:10.1080/17429145.2012.746399. [Google Scholar] [CrossRef]
82. Farag S , Kayser O . Cannabinoids production by hairy root cultures of Cannabis sativa L. Am J Plant Sci. 2015; 6( 11): 1874– 84. doi:10.4236/ajps.2015.611188. [Google Scholar] [CrossRef]
83. Ajdanian L , Torkamaneh D . Mother transformer: a high-throughput, cost-effective in planta hairy root transformation method for Cannabis. BMC Biotechnol. 2025; 25( 1): 60. doi:10.1186/s12896-025-00990-6. [Google Scholar] [CrossRef]
84. Monyela S , Kayoka PN , Ngezimana W , Nemadodzi LE . Evaluating the metabolomic profile and anti-pathogenic properties of Cannabis Species. Metabolites. 2024; 14( 5): 253. doi:10.3390/metabo14050253. [Google Scholar] [CrossRef]
85. Murthy HN , Joseph KS , Paek KY , Park SY . Bioreactor systems for micropropagation of plants: present scenario and future prospects. Front Plant Sci. 2023; 14: 1159588. doi:10.3389/fpls.2023.1159588. [Google Scholar] [CrossRef]
86. Hwang EE , Wang MB , Bravo JE , Banta LM . Unmasking host and microbial strategies in the Agrobacterium-plant defense tango. Front Plant Sci. 2015; 6: 200. doi:10.3389/fpls.2015.00200. [Google Scholar] [CrossRef]
87. Berens ML , Berry HM , Mine A , Argueso CT , Tsuda K . Evolution of hormone signaling networks in plant defense. Annu Rev Phytopathol. 2017; 55: 401– 25. doi:10.1146/annurev-phyto-080516-035544. [Google Scholar] [CrossRef]
88. Jeong JH , Jeon EY , Hwang MK , Song YJ , Kim JY . Development of super-infective ternary vector systems for enhancing the Agrobacterium-mediated plant transformation and genome editing efficiency. Hortic Res. 2024; 11( 9): uhae187. doi:10.1093/hr/uhae187. [Google Scholar] [CrossRef]
89. Kang M , Lee K , Finley T , Chappell H , Veena V , Wang K . An improved Agrobacterium-mediated transformation and genome-editing method for maize inbred B104 using a ternary vector system and immature embryos. Front Plant Sci. 2022; 13: 860971. doi:10.3389/fpls.2022.860971. [Google Scholar] [CrossRef]
90. Anand A , Bass SH , Wu E , Wang N , McBride KE , Annaluru N , et al. An improved ternary vector system for Agrobacterium-mediated rapid maize transformation. Plant Mol Biol. 2018; 97( 1–2): 187– 200. doi:10.1007/s11103-018-0732-y. [Google Scholar] [CrossRef]
91. Atabaki N , Shaharuddin NA , Ahmad SA , Nulit R , Malik S , Vahedi M , et al. Hairy root culture: a reliable bioreactor from transgenic plants. In: Jain A , Malik S , editors. Peptide and protein drug delivery using polysaccharides. New York, NY, USA: Academic Press; 2024. p. 25– 50. doi:10.1016/B978-0-443-18925-8.00013-1. [Google Scholar] [CrossRef]
92. Gupta SK , Shukla P . Gene editing for cell engineering: trends and applications. Crit Rev Biotechnol. 2017; 37( 5): 672– 84. doi:10.1080/07388551.2016.1214557. [Google Scholar] [CrossRef]
93. Aghaali Z , Naghavi MR , Zargar M . Collaboration of hairy root culture and scale-up strategies for enhancing the biosynthesis of medicinal and defensive alkaloids in Papaver sp. Curr Plant Biol. 2024; 40: 100381. doi:10.1016/j.cpb.2024.100381. [Google Scholar] [CrossRef]
94. Banerjee S , Singh S , Ur Rahman L . Biotransformation studies using hairy root cultures—a review. Biotechnol Adv. 2012; 30( 3): 461– 8. doi:10.1016/j.biotechadv.2011.08.010. [Google Scholar] [CrossRef]
95. Zhou ML , Zhu XM , Shao JR , Tang YX , Wu YM . Production and metabolic engineering of bioactive substances in plant hairy root culture. Appl Microbiol Biotechnol. 2011; 90( 4): 1229– 39. doi:10.1007/s00253-011-3228-0. [Google Scholar] [CrossRef]
96. Khalil AM . The genome editing revolution: review. J Genet Eng Biotechnol. 2020; 18( 1): 68. doi:10.1186/s43141-020-00078-y. [Google Scholar] [CrossRef]
97. Kiryushkin AS , Ilina EL , Guseva ED , Pawlowski K , Demchenko KN . Hairy CRISPR: genome editing in plants using hairy root transformation. Plants. 2021; 11( 1): 51. doi:10.3390/plants11010051. [Google Scholar] [CrossRef]
98. Guillon S , Trémouillaux-Guiller J , Pati PK , Rideau M , Gantet P . Hairy root research: recent scenario and exciting prospects. Curr Opin Plant Biol. 2006; 9( 3): 341– 6. doi:10.1016/j.pbi.2006.03.008. [Google Scholar] [CrossRef]
99. Shi M , Liao P , Nile SH , Georgiev MI , Kai G . Biotechnological exploration of transformed root culture for value-added products. Trends Biotechnol. 2021; 39( 2): 137– 49. doi:10.1016/j.tibtech.2020.06.012. [Google Scholar] [CrossRef]
100. Shujat S , Robinson GI , Norouzkhani F , Kovalchuk I . Using advanced biotechnological techniques to improve Cannabis cultivars. Biocatal Agric Biotechnol. 2024; 60: 103250. doi:10.1016/j.bcab.2024.103250. [Google Scholar] [CrossRef]
101. Morey KJ , Peebles CAM . Hairy roots: an untapped potential for production of plant products. Front Plant Sci. 2022; 13: 937095. doi:10.3389/fpls.2022.937095. [Google Scholar] [CrossRef]
102. Bai C , Cao Y , Zhao S , Wu Z , Dai S , Wang H , et al. Generation of CRISPR/Cas9-mediated mutants in Monochasma savatieri using a hairy root system. Ind Crops Prod. 2023; 191: 116008. doi:10.1016/j.indcrop.2022.116008. [Google Scholar] [CrossRef]
103. Ding X , Yu L , Chen L , Li Y , Zhang J , Sheng H , et al. Recent progress and future prospect of CRISPR/cas-derived transcription activation (CRISPRa) system in plants. Cells. 2022; 11( 19): 3045. doi:10.3390/cells11193045. [Google Scholar] [CrossRef]
104. Niazian M , Belzile F , Torkamaneh D . CRISPR/Cas9 in planta hairy root transformation: a powerful platform for functional analysis of root traits in soybean. Plants. 2022; 11( 8): 1044. doi:10.3390/plants11081044. [Google Scholar] [CrossRef]
105. Shelake RM , Jadhav AM , Bhosale PB , Kim JY . Unlocking secrets of nature’s chemists: potential of CRISPR/Cas-based tools in plant metabolic engineering for customized nutraceutical and medicinal profiles. Plant Physiol Biochem. 2023; 203: 108070. doi:10.1016/j.plaphy.2023.108070. [Google Scholar] [CrossRef]
106. Hashemi A . CRISPR-Cas9/CRISPRi tools for cell factory construction in E. coli. World J Microbiol Biotechnol. 2020; 36( 7): 96. doi:10.1007/s11274-020-02872-9. [Google Scholar] [CrossRef]
107. Bind S , Bind S , Sharma AK , Chaturvedi P . Epigenetic modification: a key tool for secondary metabolite production in microorganisms. Front Microbiol. 2022; 13: 784109. doi:10.3389/fmicb.2022.784109. [Google Scholar] [CrossRef]
108. Jiang G , Gao Y , Zhou N , Wang B . CRISPR-powered RNA sensing in vivo. Trends Biotechnol. 2024; 42( 12): 1601– 14. doi:10.1016/j.tibtech.2024.04.002. [Google Scholar] [CrossRef]
109. Zhang Y , Li S , Li R , Qiu X , Fan T , Wang B , et al. Advances in application of CRISPR-Cas13a system. Front Cell Infect Microbiol. 2024; 14: 1291557. doi:10.3389/fcimb.2024.1291557. [Google Scholar] [CrossRef]
110. Zhang X , Li F , Li X , Tian G , Xia Q . Digested crRNA switchable CRISPR-Cas13a system for ultrasensitive detection of RNase H activity. Sens Actuat B Chem. 2025; 442: 138074. doi:10.1016/j.snb.2025.138074. [Google Scholar] [CrossRef]
111. Schindele P , Wolter F , Puchta H . Transforming plant biology and breeding with CRISPR/Cas9, Cas12 and Cas13. FEBS Lett. 2018; 592( 12): 1954– 67. doi:10.1002/1873-3468.13073. [Google Scholar] [CrossRef]
112. Guo C , Ma X , Gao F , Guo Y . Off-target effects in CRISPR/Cas9 gene editing. Front Bioeng Biotechnol. 2023; 11: 1143157. doi:10.3389/fbioe.2023.1143157. [Google Scholar] [CrossRef]
113. Hillary VE , Ceasar SA . CRISPR/Cas system-mediated base editing in crops: recent developments and future prospects. Plant Cell Rep. 2024; 43( 11): 271. doi:10.1007/s00299-024-03346-0. [Google Scholar] [CrossRef]
114. Cheng X , Li Z , Shan R , Li Z , Wang S , Zhao W , et al. Modeling CRISPR-Cas13d on-target and off-target effects using machine learning approaches. Nat Commun. 2023; 14( 1): 752. doi:10.1038/s41467-023-36316-3. [Google Scholar] [CrossRef]
115. Shou J , Li J , Liu Y , Wu Q . Precise and predictable CRISPR chromosomal rearrangements reveal principles of Cas9-mediated nucleotide insertion. Mol Cell. 2018; 71( 4): 498– 509.e4. doi:10.1016/j.molcel.2018.06.021. [Google Scholar] [CrossRef]
116. Zuo E , Huo X , Yao X , Hu X , Sun Y , Yin J , et al. CRISPR/Cas9-mediated targeted chromosome elimination. Genome Biol. 2017; 18( 1): 224. doi:10.1186/s13059-017-1354-4. [Google Scholar] [CrossRef]
117. Paul B , Montoya G . CRISPR-Cas12a: functional overview and applications. Biomed J. 2020; 43( 1): 8– 17. doi:10.1016/j.bj.2019.10.005. [Google Scholar] [CrossRef]
118. Bao J , de Dios Mateos E , Scheller S . Efficient CRISPR/Cas12a-based genome-editing toolbox for metabolic engineering in Methanococcus maripaludis. ACS Synth Biol. 2022; 11( 7): 2496– 503. doi:10.1021/acssynbio.2c00137. [Google Scholar] [CrossRef]
119. Young C , Min W . CRISPRi-dCas12a: a dCas12a-mediated CRISPR interference for repression of multiple genes and metabolic engineering in cyanobacteria. ACS Synth Biol. 2020; 9( 9): 2351– 61. doi:10.1021/acssynbio.0c00091. [Google Scholar] [CrossRef]
120. Yang Z , Edwards H , Xu P . CRISPR-Cas12a/Cpf1-assisted precise, efficient and multiplexed genome-editing in Yarrowia lipolytica. Metab Eng Commun. 2019; 10: e00112. doi:10.1016/j.mec.2019.e00112. [Google Scholar] [CrossRef]
121. Lu L , Shen X , Sun X , Yan Y , Wang J , Yuan Q . CRISPR-based metabolic engineering in non-model microorganisms. Curr Opin Biotechnol. 2022; 75: 102698. doi:10.1016/j.copbio.2022.102698. [Google Scholar] [CrossRef]
122. Bhojiya AA , Joshi H . Crispr gene editing for secondary metabolite production: a review. In: Kumar A , Arora S , Ogita S , Yau Y-Y , Mukherjee K , editors. Gene editing in plants. Singapore: Springer Nature; 2024. p. 437– 75. doi:10.1007/978-981-99-8529-6_17. [Google Scholar] [CrossRef]
123. Vom Endt D , Kijne JW , Memelink J . Transcription factors controlling plant secondary metabolism: what regulates the regulators? Phytochemistry. 2002; 61( 2): 107– 14. doi:10.1016/s0031-9422(02)00185-1. [Google Scholar] [CrossRef]
124. Ahmad Meraj T , Fu J , Ali Raza M , Zhu C , Shen Q , Xu D , et al. Transcriptional factors regulate plant stress responses through mediating secondary metabolism. Genes. 2020; 11( 4): 346. doi:10.3390/genes11040346. [Google Scholar] [CrossRef]
125. Kumar S , Korra T , Thakur R , Arutselvan R , Kashyap AS , Nehela Y , et al. Role of plant secondary metabolites in defence and transcriptional regulation in response to biotic stress. Plant Stress. 2023; 8: 100154. doi:10.1016/j.stress.2023.100154. [Google Scholar] [CrossRef]
126. Khanam MN , Anis M , Javed SB , Mottaghipisheh J , Csupor D . Adventitious root culture—an alternative strategy for secondary metabolite production: a review. Agronomy. 2022; 12( 5): 1178. doi:10.3390/agronomy12051178. [Google Scholar] [CrossRef]
127. Kumar P , Mahato DK , Kamle M , Borah R , Sharma B , Pandhi S , et al. Pharmacological properties, therapeutic potential, and legal status of Cannabis sativa L.: an overview. Phytother Res. 2021; 35( 11): 6010– 29. doi:10.1002/ptr.7213. [Google Scholar] [CrossRef]
128. Pan C , Sretenovic S , Qi Y . CRISPR/dCas-mediated transcriptional and epigenetic regulation in plants. Curr Opin Plant Biol. 2021; 60: 101980. doi:10.1016/j.pbi.2020.101980. [Google Scholar] [CrossRef]
129. Karlson CKS , Mohd-Noor SN , Nolte N , Tan BC . CRISPR/dCas9-based systems: mechanisms and applications in plant sciences. Plants. 2021; 10( 10): 2055. doi:10.3390/plants10102055. [Google Scholar] [CrossRef]
130. Rabeh K , Hnini M , Oubohssaine M . A comprehensive review of transcription factor-mediated regulation of secondary metabolites in plants under environmental stress. Stress Biol. 2025; 5( 1): 15. doi:10.1007/s44154-024-00201-w. [Google Scholar] [CrossRef]
131. Mehrotra S , Rahman LU , Kukreja AK . An extensive case study of hairy-root cultures for enhanced secondary-metabolite production through metabolic-pathway engineering. Biotechnol Appl Biochem. 2010; 56( 4): 161– 72. doi:10.1042/BA20100171. [Google Scholar] [CrossRef]
132. Ludwig-Müller J , Jahn L , Lippert A , Püschel J , Walter A . Improvement of hairy root cultures and plants by changing biosynthetic pathways leading to pharmaceutical metabolites: strategies and applications. Biotechnol Adv. 2014; 32( 6): 1168– 79. doi:10.1016/j.biotechadv.2014.03.007. [Google Scholar] [CrossRef]
133. Zhan X , Chen Z , Chen R , Shen C . Environmental and genetic factors involved in plant protection-associated secondary metabolite biosynthesis pathways. Front Plant Sci. 2022; 13: 877304. doi:10.3389/fpls.2022.877304. [Google Scholar] [CrossRef]
134. Jeena GS , Singh N , Shikha , Shukla RK . An insight into microRNA biogenesis and its regulatory role in plant secondary metabolism. Plant Cell Rep. 2022; 41( 8): 1651– 71. doi:10.1007/s00299-022-02877-8. [Google Scholar] [CrossRef]
135. Upadhyay R , Saini R , Shukla PK , Tiwari KN . Role of secondary metabolites in plant defense mechanisms: a molecular and biotechnological insights. Phytochem Rev. 2025; 24( 1): 953– 83. doi:10.1007/s11101-024-09976-2. [Google Scholar] [CrossRef]
136. Ren H , Yu Y , Xu Y , Zhang X , Tian X , Gao T . GlPS1 overexpression accumulates coumarin secondary metabolites in transgenic Arabidopsis. Plant Cell Tissue Organ Cult. 2023; 152( 3): 539– 53. doi:10.1007/s11240-022-02427-w. [Google Scholar] [CrossRef]
137. Han H , Wang C , Yang X , Wang L , Ye J , Xu F , et al. Role of bZIP transcription factors in the regulation of plant secondary metabolism. Planta. 2023; 258( 1): 13. doi:10.1007/s00425-023-04174-4. [Google Scholar] [CrossRef]
138. Villao-Uzho L , Chávez-Navarrete T , Pacheco-Coello R , Sánchez-Timm E , Santos-Ordóñez E . Plant promoters: their identification, characterization, and role in gene regulation. Genes. 2023; 14( 6): 1226. doi:10.3390/genes14061226. [Google Scholar] [CrossRef]
139. Gharat SA , Tamhane VA , Giri AP , Aharoni A . Navigating the challenges of engineering composite specialized metabolite pathways in plants. Plant J. 2025; 121( 6): e70100. doi:10.1111/tpj.70100. [Google Scholar] [CrossRef]
140. Wei L , Wang Z , She Y , Fu H . CRISPR/cas multiplexed biosensing: advances, challenges, and perspectives. Anal Chem. 2025; 97( 23): 11943– 58. doi:10.1021/acs.analchem.4c04428. [Google Scholar] [CrossRef]
141. Zhao J , Nussinov R , Wu WJ , Ma B . In silico methods in antibody design. Antibodies. 2018; 7( 3): 22. doi:10.3390/antib7030022. [Google Scholar] [CrossRef]
142. Parthiban S , Vijeesh T , Gayathri T , Shanmugaraj B , Sharma A , Sathishkumar R . Artificial intelligence-driven systems engineering for next-generation plant-derived biopharmaceuticals. Front Plant Sci. 2023; 14: 1252166. doi:10.3389/fpls.2023.1252166. [Google Scholar] [CrossRef]
143. Liu H , Timko MP . Improving protein quantity and quality-the next level of plant molecular farming. Int J Mol Sci. 2022; 23( 3): 1326. doi:10.3390/ijms23031326. [Google Scholar] [CrossRef]
144. Silva JCF , Teixeira RM , Silva FF , Brommonschenkel SH , Fontes EPB . Machine learning approaches and their current application in plant molecular biology: a systematic review. Plant Sci. 2019; 284: 37– 47. doi:10.1016/j.plantsci.2019.03.020. [Google Scholar] [CrossRef]
145. Hesami M , Alizadeh M , Jones AMP , Torkamaneh D . Machine learning: its challenges and opportunities in plant system biology. Appl Microbiol Biotechnol. 2022; 106( 9–10): 3507– 30. doi:10.1007/s00253-022-11963-6. [Google Scholar] [CrossRef]
146. Iram A , Dong Y , Ignea C . Synthetic biology advances towards a bio-based society in the era of artificial intelligence. Curr Opin Biotechnol. 2024; 87: 103143. doi:10.1016/j.copbio.2024.103143. [Google Scholar] [CrossRef]
147. Cheng Q , Wang X . Machine learning for AI breeding in plants. Genomics Proteomics Bioinformatics. 2024; 22( 4): qzae051. doi:10.1093/gpbjnl/qzae051. [Google Scholar] [CrossRef]
148. Hesami M , Naderi R , Tohidfar M . Introducing a hybrid artificial intelligence method for high-throughput modeling and optimizing plant tissue culture processes: the establishment of a new embryogenesis medium for Chrysanthemum, as a case study. Appl Microbiol Biotechnol. 2020; 104( 23): 10249– 63. doi:10.1007/s00253-020-10978-1. [Google Scholar] [CrossRef]
149. Kaushik P , Rani M , Khurana N , Pandey P , Payal , Kapoor S . Revolutionizing plant tissue culture: harnessing artificial intelligence for precision propagation and optimization. Nat Prod J. 2025; 15( 3): e040624230642. doi:10.2174/0122103155302871240527094915. [Google Scholar] [CrossRef]
150. Goswami M , Akhtar S , Osama K . Strategies for monitoring and modeling the growth of hairy root cultures: an in silico perspective. In: Srivastava V , Mehrotra S , Mishra S , editors. Hairy roots: an effective Tool OF plant biotechnology. Singapore: Springer; 2018. p. 311– 27. doi:10.1007/978-981-13-2562-5_14. [Google Scholar] [CrossRef]
151. Hesami M , Jones AMP . Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl Microbiol Biotechnol. 2020; 104( 22): 9449– 85. doi:10.1007/s00253-020-10888-2. [Google Scholar] [CrossRef]
152. Wawrosch C , Zotchev SB . Production of bioactive plant secondary metabolites through in vitro technologies-status and outlook. Appl Microbiol Biotechnol. 2021; 105( 18): 6649– 68. doi:10.1007/s00253-021-11539-w. [Google Scholar] [CrossRef]
153. De Carlo A , Tarraf W , Lambardi M , Benelli C . Temporary immersion system for production of biomass and bioactive compounds from medicinal plants. Agronomy. 2021; 11( 12): 2414. doi:10.3390/agronomy11122414. [Google Scholar] [CrossRef]
154. Mirzabe AH , Hajiahmad A , Fadavi A , Rafiee S . Temporary immersion systems (TISs): a comprehensive review. J Biotechnol. 2022; 357: 56– 83. doi:10.1016/j.jbiotec.2022.08.003. [Google Scholar] [CrossRef]
155. Hwang HD , Kwon SH , Murthy HN , Yun SW , Pyo SS , Park SY . Temporary immersion bioreactor system as an efficient method for mass production of in vitro plants in horticulture and medicinal plants. Agronomy. 2022; 12( 2): 346. doi:10.3390/agronomy12020346. [Google Scholar] [CrossRef]
156. Ozyigit II , Dogan I , Hocaoglu-Ozyigit A , Yalcin B , Erdogan A , Yalcin IE , et al. Production of secondary metabolites using tissue culture-based biotechnological applications. Front Plant Sci. 2023; 14: 1132555. doi:10.3389/fpls.2023.1132555. [Google Scholar] [CrossRef]
157. Marchev AS , Yordanova ZP , Georgiev MI . Green (cell) factories for advanced production of plant secondary metabolites. Crit Rev Biotechnol. 2020; 40( 4): 443– 58. doi:10.1080/07388551.2020.1731414. [Google Scholar] [CrossRef]
158. Kusari S , Spiteller M . Are we ready for industrial production of bioactive plant secondary metabolites utilizing endophytes? Nat Prod Rep. 2011; 28( 7): 1203– 7. doi:10.1039/c1np00030f. [Google Scholar] [CrossRef]
159. Krol A , Kokotkiewicz A , Szopa A , Ekiert H , Luczkiewicz M . Bioreactor-grown shoot cultures for the secondary metabolite production. In: Ramawat KG , Ekiert HM , Goyal S , editors. Plant cell and tissue differentiation and secondary metabolites: fundamentals and applications. Cham, Switzerland: Springer International Publishing; 2020. p. 1– 62. doi:10.1007/978-3-030-11253-0_34-1. [Google Scholar] [CrossRef]
160. Georgiev V , Schumann A , Pavlov A , Bley T . Temporary immersion systems in plant biotechnology. Eng Life Sci. 2014; 14( 6): 607– 21. doi:10.1002/elsc.201300166. [Google Scholar] [CrossRef]
161. Valdiani A , Hansen OK , Nielsen UB , Johannsen VK , Shariat M , Georgiev MI , et al. Bioreactor-based advances in plant tissue and cell culture: challenges and prospects. Crit Rev Biotechnol. 2019; 39( 1): 20– 34. doi:10.1080/07388551.2018.1489778. [Google Scholar] [CrossRef]
162. Wilken D , Jiménez González E , Hohe A , Jordan M , Gomez Kosky R , Schmeda Hirschmann G , et al. Comparison of secondary plant metabolite production in cell suspension, callus culture and temporary immersion system. In: Hvoslef-Eide AK , Preil W , editors. Liquid culture systems for in vitro plant propagation. Dordrecht, The Netherlands: Springer; 2005. p. 525– 37. doi:10.1007/1-4020-3200-5_39. [Google Scholar] [CrossRef]
163. Sharma S , Shahzad A . Bioreactors: a rapid approach for secondary metabolite production. In: Shahid M , Shahzad A , Malik A , Sahai A , editors. Recent trends in biotechnology and therapeutic applications of medicinal plants. Dordrecht, The Netherlands: Springer; 2013. p. 25– 49. doi:10.1007/978-94-007-6603-7_2. [Google Scholar] [CrossRef]
164. Mehrotra S , Mishra S , Srivastava V . Bioreactor technology for hairy roots cultivation. In: Pavlov A , Bley T , editors. Bioprocessing of plant in vitro systems. Cham, Switzerland: Springer International Publishing; 2016. p. 1– 25. doi:10.1007/978-3-319-32004-5_10-1. [Google Scholar] [CrossRef]
165. Shiao TL , Doran PM . Root hairiness: effect on fluid flow and oxygen transfer in hairy root cultures. J Biotechnol. 2000; 83( 3): 199– 210. doi:10.1016/s0168-1656(00)00316-3. [Google Scholar] [CrossRef]
166. Murthy HN , Joseph KS , Paek KY , Park SY . Bioreactor configurations for adventitious root culture: recent advances toward the commercial production of specialized metabolites. Crit Rev Biotechnol. 2024; 44( 5): 837– 59. doi:10.1080/07388551.2023.2233690. [Google Scholar] [CrossRef]
167. Palavalli RR , Srivastava S , Srivastava AK . Development of a mathematical model for growth and oxygen transfer in in vitro plant hairy root cultivations. Appl Biochem Biotechnol. 2012; 167( 6): 1831– 44. doi:10.1007/s12010-011-9515-5. [Google Scholar] [CrossRef]
168. Cantarero Rivera FJ , Chen J . Computational fluid dynamics modeling of cell cultures in bioreactors and its potential for cultivated meat production—a mini-review. Future Foods. 2022; 6: 100195. doi:10.1016/j.fufo.2022.100195. [Google Scholar] [CrossRef]
169. Kowalczyk T , Merecz-Sadowska A , Picot L , Brčić Karačonji I , Wieczfinska J , Śliwiński T , et al. Genetic manipulation and bioreactor culture of plants as a tool for industry and its applications. Molecules. 2022; 27( 3): 795. doi:10.3390/molecules27030795. [Google Scholar] [CrossRef]
170. An X , Yang H , Xu J , Jiao J , Yang J , Fu Y . In vitro adventitious roots of hemp in bioreactor culture to efficiently produce non-cannabinoids and cannabidiol phytochemicals. Plant Cell Tissue Organ Cult PCTOC. 2024; 159( 3): 69. doi:10.1007/s11240-024-02922-2. [Google Scholar] [CrossRef]
171. Hwang HG , Ye DY , Jung GY . Biosensor-guided discovery and engineering of metabolic enzymes. Biotechnol Adv. 2023; 69: 108251. doi:10.1016/j.biotechadv.2023.108251. [Google Scholar] [CrossRef]
172. Chakraborty A , Bardhan S , Das S , Chowdhury BR . Chapter 31—Development of biosensors for application in industrial biotechnology. In: Kumar V , Bilal M , Shahi SK , Garg VK , editors. Metagenomics to bioremediation. New York, NY, USA: Academic Press; 2023. p. 737– 53. doi:10.1016/B978-0-323-96113-4.00010-X. [Google Scholar] [CrossRef]
173. Ding N , Yuan Z , Ma Z , Wu Y , Yin L . AI-assisted rational design and activity prediction of biological elements for optimizing transcription-factor-based biosensors. Molecules. 2024; 29( 15): 3512. doi:10.3390/molecules29153512. [Google Scholar] [CrossRef]
174. Wang X , Gu J , Fu J , Wang C , Zhao L , Guo H , et al. Multi-omics analyses of the effect of carbon ion beam irradiation on Cannabis fructus (Cannabis sativa L.) composition. Plant Stress. 2023; 10: 100267. doi:10.1016/j.stress.2023.100267. [Google Scholar] [CrossRef]
175. Aliferis KA , Bernard-Perron D . Cannabinomics: application of metabolomics in Cannabis (Cannabis sativa L.) research and development. Front Plant Sci. 2020; 11: 554. doi:10.3389/fpls.2020.00554. [Google Scholar] [CrossRef]
176. Wiles D , Shanbhag BK , O’Brien M , Doblin MS , Bacic A , Beddoe T . Heterologous production of Cannabis sativa-derived specialised metabolites of medicinal significance—insights into engineering strategies. Phytochemistry. 2022; 203: 113380. doi:10.1016/j.phytochem.2022.113380. [Google Scholar] [CrossRef]
177. Rico S , Garrido J , Sánchez C , Ferreiro-Vera C , Codesido V , Vidal N . A temporary immersion system to improve Cannabis sativa micropropagation. Front Plant Sci. 2022; 13: 895971. doi:10.3389/fpls.2022.895971. [Google Scholar] [CrossRef]
178. Thomas F , Schmidt C , Kayser O . Bioengineering studies and pathway modeling of the heterologous biosynthesis of tetrahydrocannabinolic acid in yeast. Appl Microbiol Biotechnol. 2020; 104( 22): 9551– 63. doi:10.1007/s00253-020-10798-3. [Google Scholar] [CrossRef]
179. Pepe M , Hesami M , Small F , Jones AMP . Comparative analysis of machine learning and evolutionary optimization algorithms for precision micropropagation of Cannabis sativa: prediction and validation of in vitro shoot growth and development based on the optimization of light and carbohydrate sources. Front Plant Sci. 2021; 12: 757869. doi:10.3389/fpls.2021.757869. [Google Scholar] [CrossRef]
180. Moola AK , Balasubramanian P , Satish L , Shamili S , Ramesh M , Kumar TS , et al. Hairy roots as a source for phytoremediation. In: Aravind J , Kamaraj M , Prashanthi Devi M , Rajakumar S , editors. Strategies and tools for pollutant mitigation. Cham, Switzerland: Springer International Publishing; 2021. p. 29– 47. doi:10.1007/978-3-030-63575-6_2. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools