 Open Access
Open Access
ARTICLE
Genomic Characterization of BvMLO Genes in Sugar Beet Focusing on BvMLO2 BvMLO7 Responses to Cercospora beticola and Abiotic Stress
1 College of Modern Agriculture and Ecological Environment, Heilongjiang University, Harbin, 150080, China
2 Sugar Beet Engineering Research Center of Heilongjiang Province, Heilongjiang University, Harbin, 150080, China
* Corresponding Author: Guangzhou Ding. Email:
# These authors contributed equally to this work
Phyton-International Journal of Experimental Botany 2025, 94(3), 875-895. https://doi.org/10.32604/phyton.2025.061550
Received 27 November 2024; Accepted 26 February 2025; Issue published 31 March 2025
Abstract
Mildew resistance locus O (MLO) proteins are extensively found in various plant species and are essential for multiple biological functions. The characterization and analysis of MLO genes have been conducted across numerous species. However, the functions and features of MLO genes inside sugar beet remain poorly understood. In the present research, we conducted a comprehensive analysis of the structural features of MLO genes, physicochemical characteristics of proteins, evolutionary connections, and expression profiles in sugar beet. A total of 13 BvMLO genes containing MLO structural domains were detected and renamed based on their locations on chromosomes within the sugar beet genome. According to the classification of AtMLO genes, the evolutionary analysis revealed that these 13 BvMLO genes were classified into three subgroups and unevenly located across four chromosomes. Synteny and collinearity analysis confirmed that gene clusters occurred during the evolution of the BvMLO gene family. Examination of cis-regulatory elements revealed specific stress-induced and hormone-associated components within the regulatory regions of BvMLOs. We also found that the expression levels of BvMLO2 and BvMLO7 cloned from sugar beet plants inoculated by Erysiphe betae (Vanha) were significantly regulated by Cercospora beticola Sacc (C. beticola), which indicated that they might both participate in some disease resistance processes. Moreover, quantitative real-time PCR (qRT-PCR) results confirmed that BvMLO2 and BvMLO7 were involved in plant resistance to various biotic and abiotic stress factors. Overall, this research provides a fundamental basis for upcoming studies on the functions and control mechanisms of BvMLO genes within sugar beet. These research findings help advance the progress of disease-resistant breeding in sugar beet and enhance the effectiveness of its resistance breeding.Keywords
Supplementary Material
Supplementary Material FileA distinct type of plant-specific CaM-binding protein gene, MLO genes [1–3], was first identified in barley [4]. After discovering the first MLO gene, MLO-encoding genes have been detected in various plant species. For example, seven MLO genes have been identified in wheat [5], while twelve are found in rice [6]. These genes code for MLO proteins that possess a C-terminal CaM-binding domain (CaMBD), as well as a conserved structure that includes seven transmembrane (TM) domains [2,3]. The interaction between the CaM-binding domain (CaMBD) and MLO proteins mediates a calcium-dependent (Ca2+) response to pathogenic attacks [3,7,8], establishing MLO genes as early responders in Ca2+-dependent plant resistance mechanisms. The first discovery of an HvMLO gene mutation resulting in loss of function occurred in barley, marking the earliest known case of MLO-related resistance in plants [9]. The enduring resistance effect, broad protection, and recessive inheritance traits are key characteristics of MLO loss-of-function mutations, conferring resistance to plants. Similarly, various crops, such as wheat, pea, and tomato, have been found to carry loss-of-function mutations, indicating that MLO-mediated resistance is more widely distributed than previously thought.
MLO proteins are essential for regulating a variety of physiological processes in plants. As a key factor in pollen tube function within the embryo sac, AtMLO7 plays a role in pollen tube recognition; however, its mutation leads to reduced fertility [10,11]. Moreover, AtMLO4 and AtMLO11 contribute to root development, as their complete loss of function results in irregular root growth and pronounced bending [11–13]. Increasing evidence suggests that specific MLO genes play roles in various biological stress responses, including participation in light-signaling pathways and heat tolerance [14,15].
Globally, Beta vulgaris L., commonly known as sugar beet, is a key sugar-producing crop. It belongs to the Amaranthaceae family, which was previously classified under Chenopodiaceae. In the global context of sugar production, it holds the second position, trailing only behind Saccharum officinarum L. (sugarcane) [16]. Although advancements in cultivation techniques and breeding have significantly improved sugar beet yield and production over the past few decades, the crop remains highly susceptible to various abiotic and biotic stressors that limit its growth and overall productivity. Cercospora leaf spot (CLS) is considered the most severe disease affecting sugar beet foliage [17]. This disease is caused by the airborne fungal pathogen Cercospora beticola Sacc (C. beticola) [18], which thrives in warm and humid cultivation environments, leading to substantial damage. C. beticola not only affects sugar beet but also infects a variety of crops [19,20], causing significant yield and quality losses. The infection process of C. beticola follows two main pathways. The first type is localized direct infection, where asexual fungal spores adhere to the surface of the leaf, enter through the stomata, infiltrate the spaces between cells, and cause damage to the nearby tissue cells [21,22]. The second pathway is systemic invasion, where C. beticola breaches the epidermal cells and rapidly proliferates within the tissue, particularly targeting meristematic regions [23]. Several strategies have been implemented to manage CLS, including fungicide application, crop rotation, and breeding for resistant varieties [24–26]. However, achieving effective disease control remains a major challenge, highlighting the need for continued research and the development of integrated disease management strategies.
For years, plant scientists and breeding specialists have collaborated to improve CLS resistance in sugar beet. Wild sea beet (B. vulgaris subsp. maritima) has long been an important contributor of genes for CLS resistance. The introduction of CLS resistance in breeding programs was facilitated by the precise mapping of resistance QTLs, which also enhanced the efficiency of marker-assisted selection (MAS). Research findings indicate that CLS resistance is controlled by multiple QTLs, with at least four major loci identified. Improving mapping precision enhances the resolution needed to separate linked resistance loci, thereby facilitating more efficient breeding strategies [25]. In recent years, in addition to employing precise QTL mapping to facilitate MAS for CLS resistance-related traits, researchers have also begun to explore specific genes associated with CLS resistance. Key genes linked to CLS resistance include members of the SP and SE families, as well as QTL-associated loci, which serve as potential molecular targets for resistance enhancement through breeding [27].
MLO genes have been identified in various species, with their functions extensively characterized, particularly in A. thaliana [12,13,28]. Studying the MLO gene family in sugar beet has been particularly challenging. Our laboratory previously conducted an analysis of sugar beet powdery mildew, providing foundational insights into its genetic basis. Building upon these findings, we conducted a comprehensive genomic investigation of the MLO gene family in sugar beet to provide a genetic framework for future functional studies. These findings hold significant promise for advancing the improvement of sugar beet cultivars.
The genome sequence of sugar beet (Beta vulgaris L.) was accessed via the Ensembl Plants database (https://plants.ensembl.org) as of 03 December 2023. MLO sequences from Arabidopsis thaliana were retrieved from the Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org) on 10 December 2023. The Hidden Markov Model (HMM) was used to identify representative MLO protein sequences in sugar beet. Using the domain profiles of MLO (PF03094) retrieved from Pfam (http://pfam.xfam.org/) on 20 December 2023, the sequences were analyzed using HMMER software [29]. To validate the reliability of the predicted gene candidates, the Conserved Domain Database (CDD) [30] was employed for analysis. Furthermore, BvMLO proteins were screened via SMART (https://smart.embl.de/, retrieved 28 December 2023).
2.2 Analysis of Physicochemical Traits and Subcellular Positioning of BvMLOs
The ProtParam online tool (https://www.expasy.org/) facilitated the examination of the predicted characteristics of BvMLO proteins to evaluate their physicochemical properties. The tool was accessed on 30 December 2023. The analysis included molecular weight, theoretical isoelectric point (pI), and the grand average of hydropathicity (GRAVY) index [31]. Additionally, the subcellular localization of BvMLO proteins was predicted using the Cell-PLoc 2.0 online tool [32]. Table S1 summarizes the analysis results of BvMLO genes.
2.3 Phylogenetic Tree Construction and Sequence Alignment
The full-length sequences of MLO proteins from nine plant species, including sugar beet, Cucumis melo, Citrullus lanatus, Gossypium raimondii, Gossypium arboreum, Gossypium hirsutum, rice, tomato and A.thaliana, were subjected to alignment using the MUSCLE algorithm with Clustal Omega’s default settings (https://www.ebi.ac.uk/Tools/msa/clustalo/, retrieved 10 January 2024) [33]. The resulting evolutionary tree was displayed through the web-based platform iTOL (Interactive Tree of Life) (https://itol.embl.de/, retrieved 12 January 2024). All identified and examined BvMLO proteins were grouped into subfamilies based on the phylogenetic framework of A. thaliana.
2.4 Investigation of BvMLO Gene Architecture, Domains, and Conserved Motifs
The exon-intron distribution patterns of BvMLO genes were determined from GFF3 annotation files and visualized using TBtools software [34]. The identification of conserved domains was performed through the NCBI CD-Search batch tool (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi, retrieved 20 January 2024) [30], applying default parameters. Motif analysis of all BvMLO proteins was performed using MEME Suite, with the maximum number of motifs set to 10 while maintaining default settings for other parameters. Results were visualized using TBtools software [34].
2.5 Genomic Distribution and Comparative Collinearity Analysis
The chromosomal locations of BvMLO genes were obtained through the annotation data in the GFF3 format from the Ensembl Plants database (https://plants.ensembl.org/, retrieved 20 February 2023) and mapped using TBtools software. Gene duplication events were identified using the Advanced Circos module in TBtools with default parameters. Furthermore, a comparative homology analysis of MLO genes across sugar beet and four other crops—Cucumis melo, tomato, Citrullus lanatus, and A. thaliana, was conducted with the assistance of TBtools software.
2.6 Analysis of Cis-Acting Elements in BvMLO Genes
To investigate cis-regulatory element composition, the 2-kb upstream region of the ATG start codon of each sugar beet MLO gene was analyzed via the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed on 18 March 2023) [35].
2.7 Prediction and Analysis of Three Dimensional (3D) Structures of BvMLO Proteins
The amino acid sequences of all MLO family proteins were submitted to SWISS-MODEL (https://www.swissmodel.expasy.org/, accessed on 25 March 2024) for three-dimensional structure prediction. SWISS-MODEL employs the ProMod3 modeling engine within the OpenStructure computational framework [36] to construct structural models.
2.8 Observation of the Sugar Beet Leaf Surface
Samples were collected at three time points (8, 12, and 18 h) daily for 15 days post-inoculation. The outer leaf layers of sugar beet were peeled off to prepare temporary slides, and a light microscope was used to observe powdery mildew infection on the leaf surface.
2.9 Analysis of the BvMLO Gene Family’s Role in Response to Pathogen Resistance
2.9.1 Plant Materials and Treatments
All experimental materials for this study were supplied by the research lab at Heilongjiang University. KWS9147 (highly susceptible to CLS) is a sugar beet variety produced by KWS SAAT SE & Co. KGaA (KWS) Company in Germany. It has good yield performance but poor disease resistance, especially to C. beticola.
2.9.2 Sugar Beet Seedling Cultivation
The plant kernels were sown in small containers filled with peat soil at a depth of 1–1.5 cm and placed in a controlled cultivation environment with a 16-h light and 8-h dark cycle at 26°C and 60% air moisture.
2.9.3 Preparation of Spore Suspension
The suspension of laboratory-preserved strains was serially diluted 10−1, 10−2 and 10−3, and 100 μL of each dilution was transferred onto a PDA medium and incubated at 28°C in a temperature-controlled room. To facilitate the purification and cultivation of C. beticola, 50 mL of sterile water was added to a petri dish containing fungal cultures, and the spores were gently scraped off using a sterile scalpel. After filtration with sterilized filter paper, it was transferred to a sterilized triangular flask, shaken, and mixed well to prepare the mother liquor. The fungi solution was diluted according to the 10-fold dilution method, and the number of conidia was counted with a blood cell counting plate under the microscope, and the 5 plates were repeated. Then add an appropriate amount of sterile water to dilute the conidial concentration to 1 × 106 spores/mL for later use. Spore concentration (spore/mL) = average number of spores per cell × 4 × 106 × dilution factor.
2.9.4 Sampling and Processing of Powdery Mildew Experiments
All materials utilized in this research were exclusively from the KWS9147 variety. In the three-true-leaf stage, a spore suspension (1 × 106 spores/mL) was sprayed onto both surfaces of sugar beet leaves using a spray inoculation method. Following inoculation, the leaves were incubated under controlled light conditions at 25°C and 90% relative humidity.
2.9.5 Sampling and Processing of C. beticola Experiments
The experimental materials used in this phase were from the KWS9147 variety. The treatment procedure for the materials was as follows: leaves at the three-true-leaf stage, exhibiting uniform growth and no mechanical damage, were selected. These leaves were disinfected with 75% ethanol and then rinsed thoroughly with sterile water. After drying, the leaves were sprayed with a C. beticola spore suspension at a concentration of 1 × 106 conidia/mL for inoculation. Following inoculation, the petioles were wrapped with sterilized, cotton-soaked material and placed on a solidified water-agar medium for incubation. The progress of the disease was monitored on the excised leaves, which were cultured on water-agar plates for up to 30 days post-inoculation.
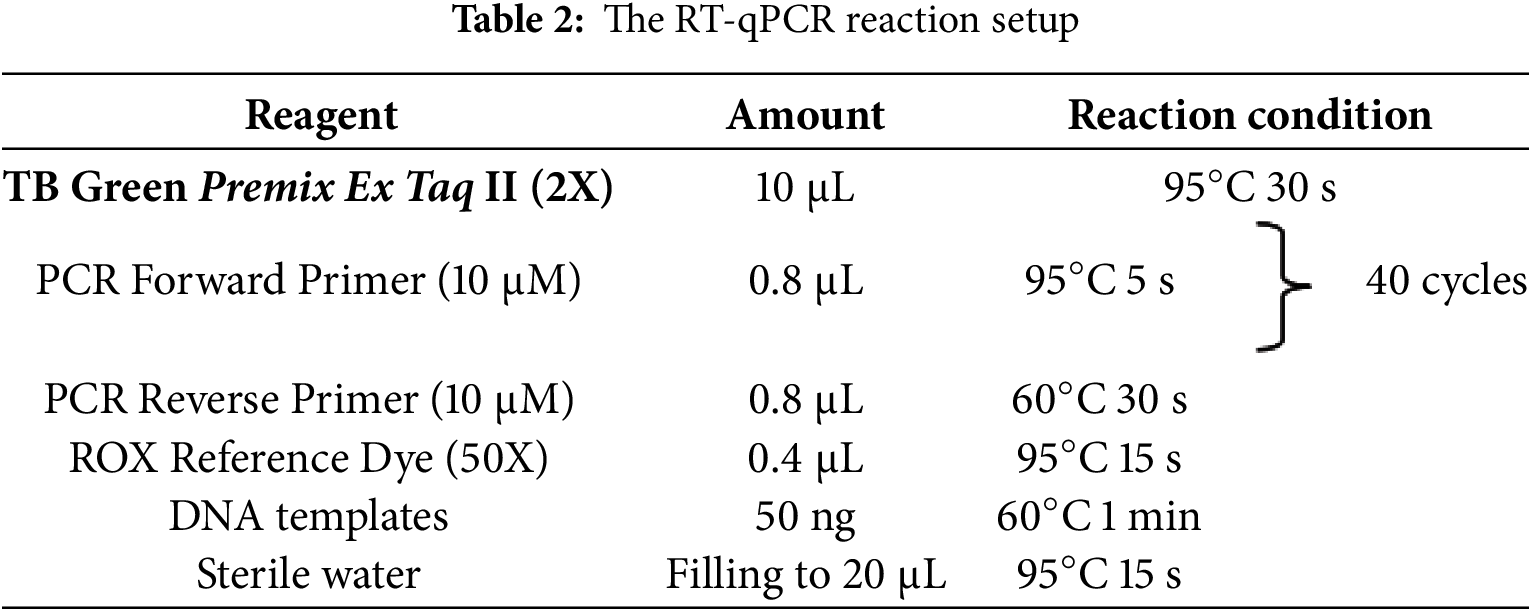
2.9.6 RNA Extraction, Reverse Transcription to cDNA, and Quantitative Real-Time PCR (qRT-PCR)
Leaf tissues from different time points after treatment were collected and stored. Leaf samples (1 g each) were collected from three plants at the same developmental stage and under identical treatment conditions. Every collected specimen was placed into an individual centrifuge tube and stored at −80°C for RNA extraction, cDNA synthesis, and subsequent qRT-PCR analysis. Total RNA was extracted from sugar beet leaves using the RNAprep Pure Plant Kit (TIANGEN, China), following the manufacturer’s instructions. The FastKing One-Step Reverse Transcription Kit (TIANGEN, China) was utilized for cDNA synthesis. The cDNA samples were stored at −20°C before PCR. qRT-PCR was performed using the Bio-Rad CFX96TM Real-Time System (Bio-Rad, USA) and SuperReal PreMix Plus (SYBR Green) (TIANGEN, China). The BvGAPDH gene from sugar beet served as the internal control gene [37]. Primer sequences are listed in Table 1, and the reaction conditions are presented in Table 2. The PCR amplification program included an initial step at 95°C for 30 s, followed by 40 cycles of 95°C for 5 s, and 60°C for 60 s for annealing/extension. Each sample had three biological replicates, and each replicate was performed in triplicate. The relative abundance of MLO gene transcripts was determined through comparative analysis, employing the 2−ΔΔCt approach [38]. Statistical evaluations were performed with SPSS (V26.0) and Microsoft Excel 2019.
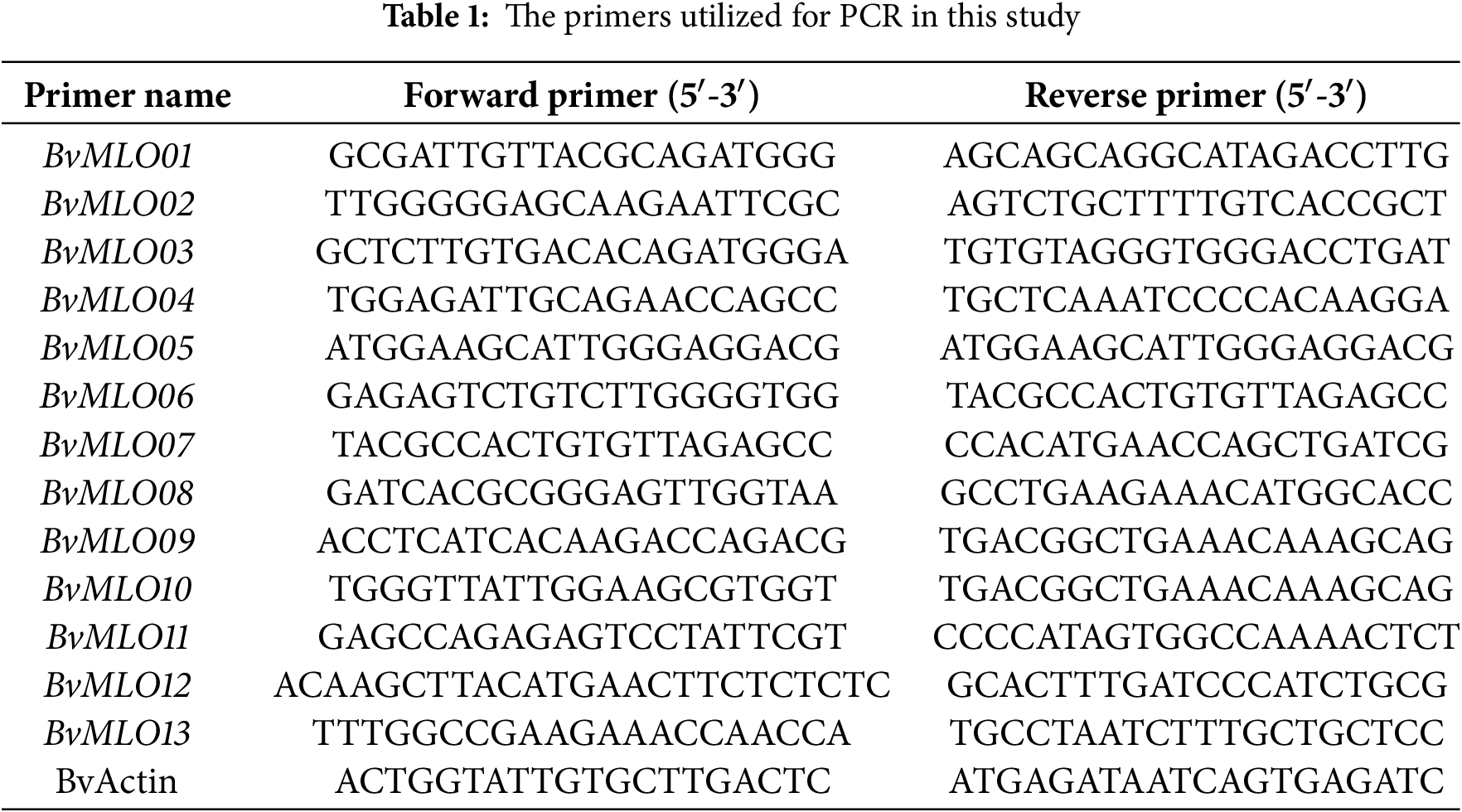
2.9.7 Analysis of BvMLO2 and BvMLO7 Expression in Response to Abiotic Stress
To explore the dynamic changes in BvMLO2 and BvMLO7 gene expression at multiple time points under various abiotic stress conditions (including methyl jasmonate (MeJA), salicylic acid (SA), NaCl to simulate salt stress, and mannitol to represent drought stress), KWS9147 seedlings at the three-leaf stage were chosen as experimental materials. The cultivated plants were subjected to treatments with 100 μM MeJA, 100 μM SA, 100 mM NaCl, and 100 mM mannitol, while an equal volume of distilled water was applied to the control group. Samples were collected at seven different time intervals, with 0 h serving as the reference. For each time interval, three independent biological repeats were used, and every sample underwent three assessments. By mapping the 7-time intervals on the horizontal coordinate and their corresponding expression measurements on the vertical coordinate, the dynamic expression patterns were illustrated. qRT-PCR was used to verify gene activity under these treatments, ensuring the accuracy and reliability of the results.
3.1 Comprehensive Identification and Evolutionary Assessment of BvMLO Genes across the Genome
By conducting a homology analysis, we identified 13 MLO genes within the genome sequence of sugar beet (Table 1). BvMLO1 to BvMLO13 were assigned as names based on their respective chromosomal positions. To investigate the evolutionary associations of these genes, phylogenetic analyses were conducted, and phylogenetic trees were generated for sugar beet, Cucumis melo, Citrullus lanatus, Gossypium raimondii, Gossypium arboreum, Gossypium hirsutum, rice, tomato, and the model species A. thaliana, utilizing their protein sequences. Using the Neighbor-Joining technique, the protein sequences were categorized into six phylogenetic subgroups: I, II, III, IV, V, and VI (Fig. 1). Notably, no sugar beet genes were found in subfamilies III, V, or VI (Fig. 1). Among the BvMLO subfamilies, group I contained the highest number of genes, comprising six members, followed by subfamily II with five and subfamily IV with two (Fig. 1). According to predictions from Cell-PLoc 2.0, most BvMLO proteins were primarily located in the inner membrane (Table S1). Additionally, multiple sequence alignment of amino acid sequences revealed that the highly conserved structural regions of BvMLOs, including seven transmembrane (TM) helices, were positioned at both the N-terminal and C-terminal regions, where they interacted with the cell membrane (Fig. S1). In the characteristic structural domain representation of sugar beet MLOs, the letter height in the sequence logo indicates the degree of conservation, with taller letters signifying more conserved residues (Fig. 2).
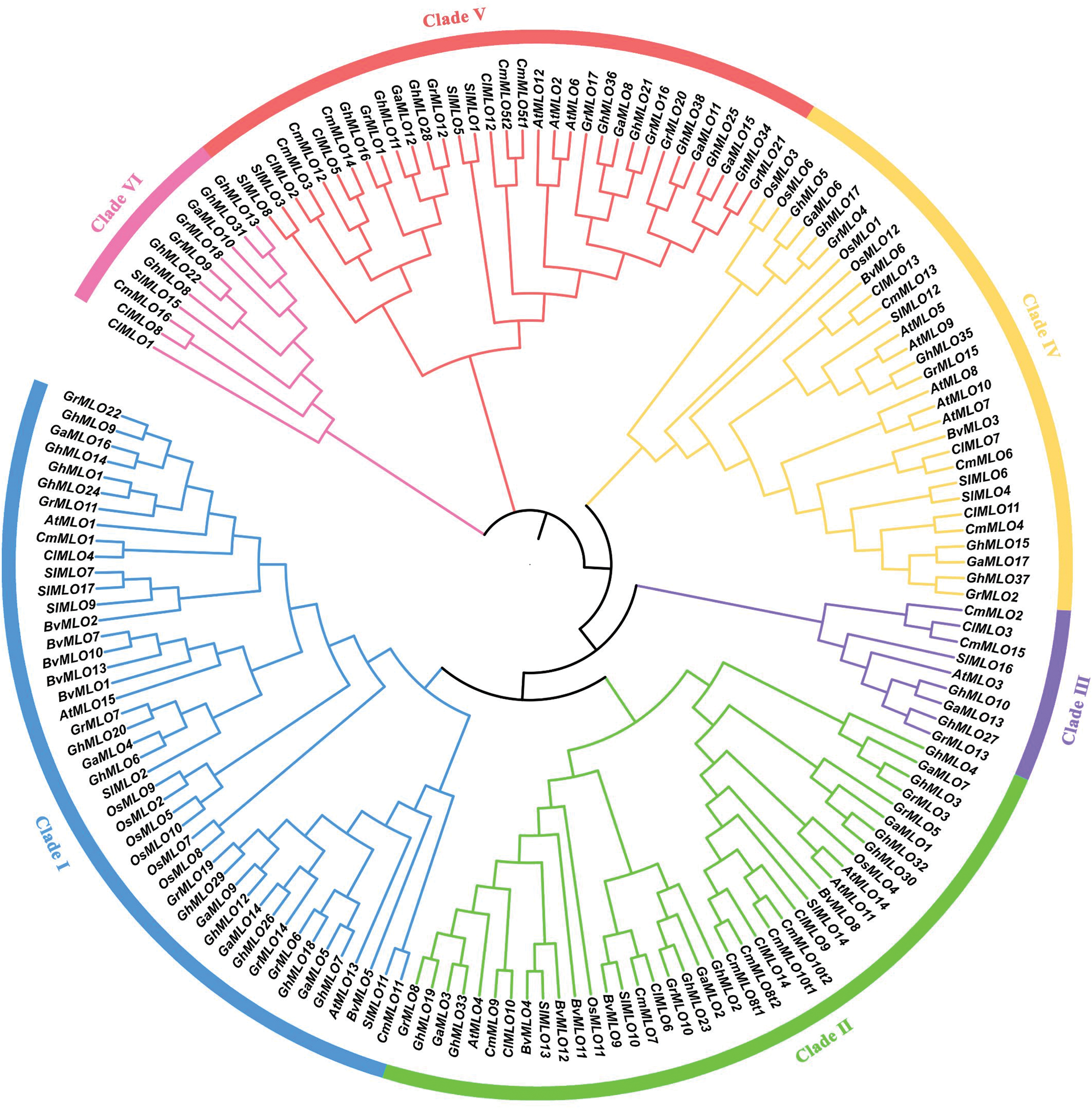
Figure 1: The evolutionary relationships of the MLO gene family in Cucumis melo, Citrullus lanatus, Gossypium raimondii, Gossypium arboreum, Gossypium hirsutum, rice, tomato, A. thaliana, and sugar beet. The six evolutionary subgroups were identified and represented by distinct colors
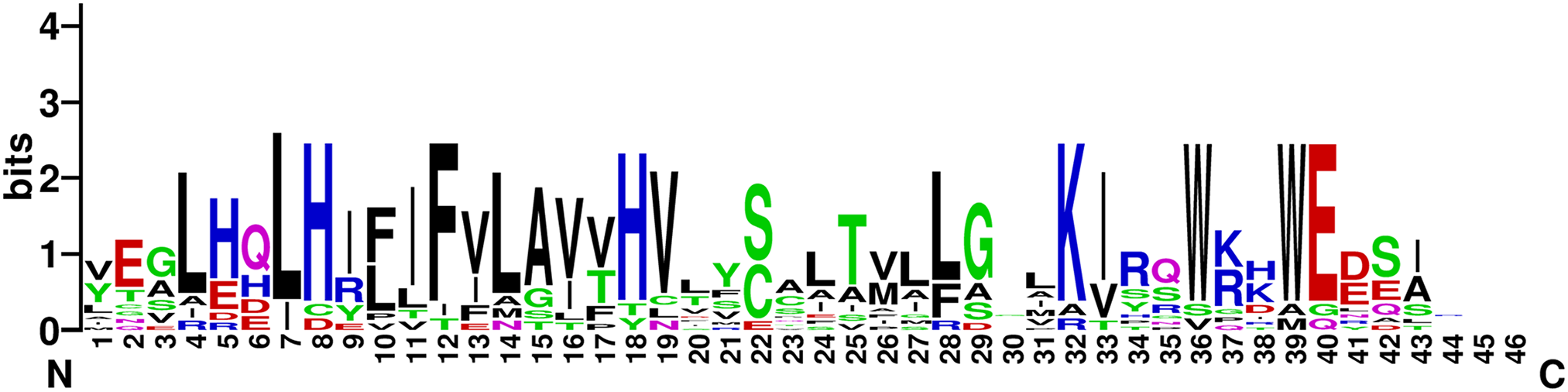
Figure 2: Sequence logo representation of conserved motifs in MLO proteins of sugar beet. The total stack height reflects sequence conservation at each position, while the bit score denotes the relative occurrence of specific amino acid residues
3.2 The Potential Functional Diversity of the MLO Gene Family
Gene structure diversity plays a pivotal role in the evolution of multigene families. In this study, we analyzed the structure of BvMLO genes (Fig. 3) and found that these genes consist of approximately 3 to 20 exons and 2 to 19 introns. A total of 13 sugar beet MLO proteins were identified, encompassing 10 conserved motifs, labeled Motif1 to Motif10. Our results revealed that while most BvMLOs exhibited similar structural features, a few displayed differentiations.
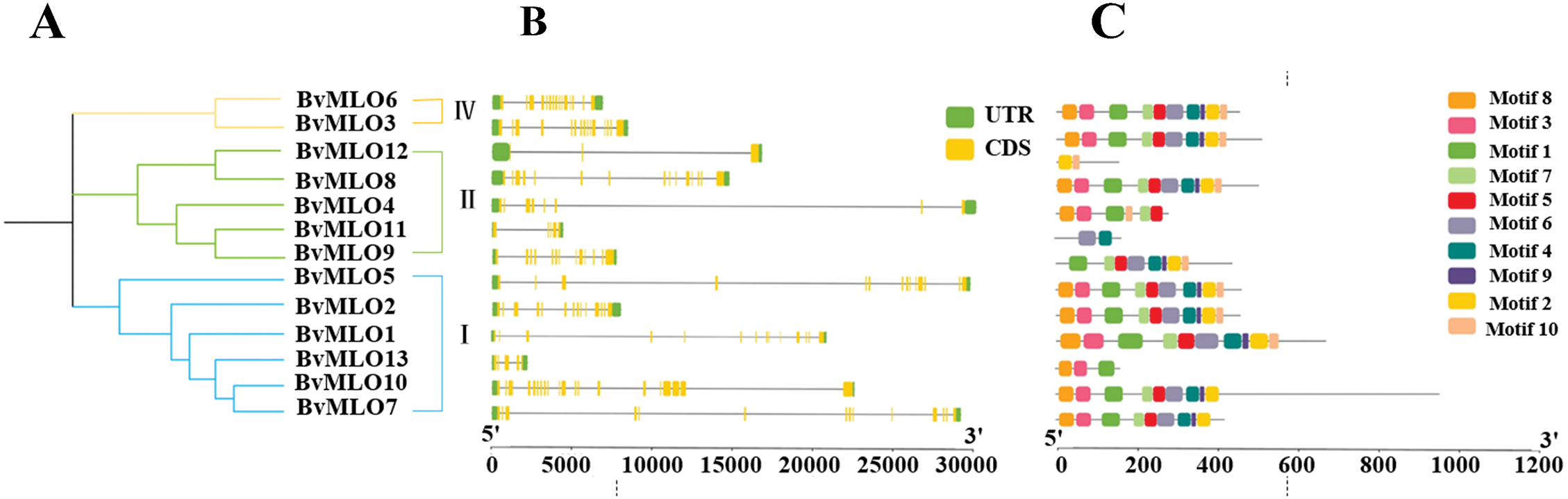
Figure 3: Structural characteristics and preserved sequence patterns of the BvMLO gene family: (A) A phylogenetic tree without a defined root was generated. (B) The exon-intron arrangement of BvMLO genes. (C) The distribution of conserved sequence motifs in BvMLO genes
To better understand the variation and conservation of MLO protein structures in sugar beet, conserved motifs were analyzed within the protein sequences of the MLO gene family using the MEME server. This analysis identified 10 conserved motifs, which were visualized using TBtools software (Fig. 3C). Notably, BvMLO1, BvMLO2, BvMLO5, and BvMLO7 possessed an identical count of exons and introns. Furthermore, it was noted that a highly conserved domain, Motif 8, was present in the C-terminal regions of all 13 BvMLO proteins, except for BvMLO9, BvMLO11, and BvMLO12.
Interestingly, the quantity of conserved motifs differs among subfamilies. For example, the first phylogenetic subfamily possessed the greatest count of conserved motifs, totaling around 10. This observation implies that proteins with a close evolutionary relationship generally exhibit similar conserved motif patterns. To summarize, our findings suggest that although the presence and organization of conserved structural domains in sugar beet MLO genes are generally consistent within the same phylogenetic clade, they differ significantly among various clades. Moreover, the conserved structure of exons and introns, along with consistent domain patterns, supports the phylogenetic grouping of MLO genes in beet.
3.3 BvMLO Gene Family: Chromosomal Positioning and Collinearity
The chromosomal mapping revealed that nine MLO genes in sugar beet were distributed across four different chromosomes. As illustrated in the figure, Chr6 contains the highest number of MLO genes, with 6, whereas Chr1, Chr3, and Chr5 each harbor only 1 gene, representing the lowest count. The chromosomal positions of four MLO genes (BvMLO3, BvMLO7, BvMLO8, BvMLO10) remained undetermined, as they were mapped to unanchored scaffolds (Fig. 4), this nonuniform distribution of MLOs was also observed in other plants including Lagenaria siceraria [38]. In investigating the evolutionary aspects of the MLO gene family in sugar beet, a natural gene cluster was identified on chromosome 6, consisting of three MLO genes: BvMLO4, BvMLO11, and BvMLO12 (Fig. 5A). Studies suggest that the formation of gene clusters in plants may result from various evolutionary mechanisms, including gene duplication, gene relocation, neofunctionalization, or independent gene evolution to acquire specific metabolic functions [39]. To validate this hypothesis, we systematically analyzed the gene cluster using the Advance Circos module in TBtools with default parameters [34]. The results revealed that genes closely located on chromosomes often exhibit co-expression patterns [40], and these gene clusters may function as genetic units for developmental regulation and defense responses, contributing to environmental adaptation and heritability. To gain deeper insights into the evolutionary relationships of MLO genes in sugar beet, we generated four syntenic maps comparing sugar beet with four representative species (A. thaliana, Cucumis melo, tomato, and Citrullus lanatus) (Fig. 5B). Notably, our analysis revealed that sugar beet and tomato exhibit the highest number of duplicated gene pairs, whereas A. thaliana possesses the fewest.
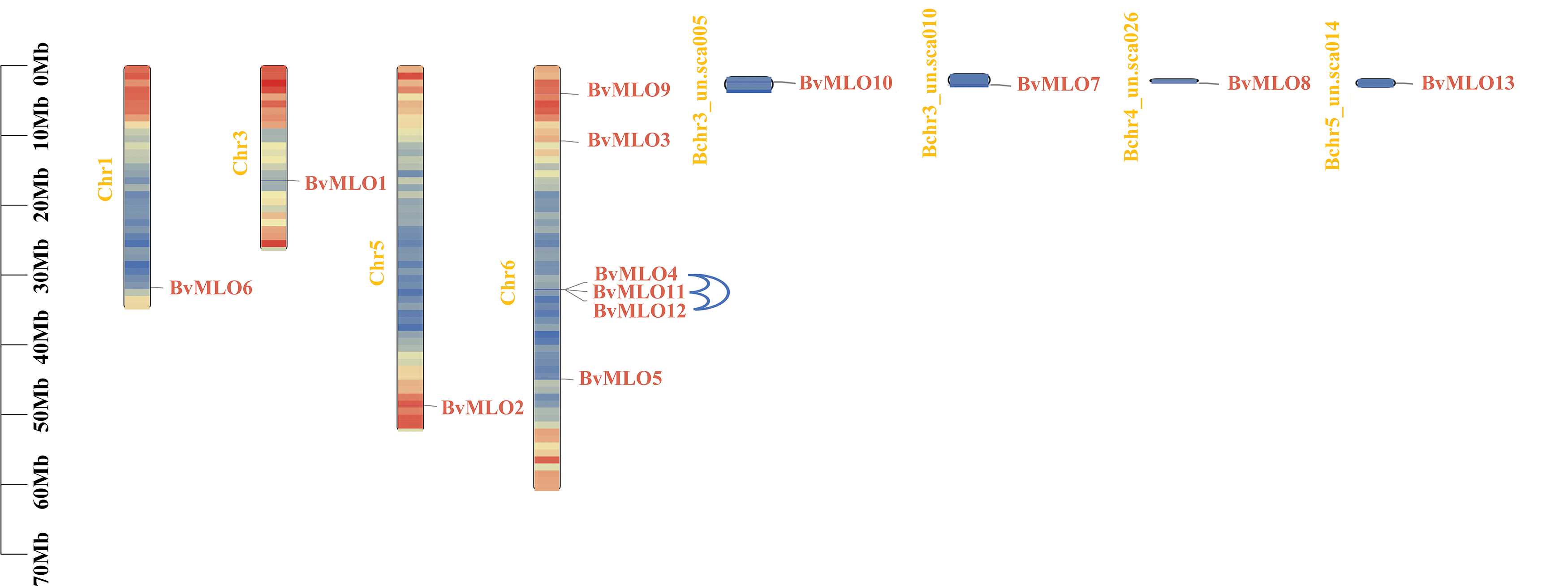
Figure 4: Distribution of MLO genes in sugar beet across four chromosomes. The positions of the genes and chromosome lengths are represented by the left-side scale. The chromosome-scale is measured in megabases (Mb). Blue lines link gene clusters
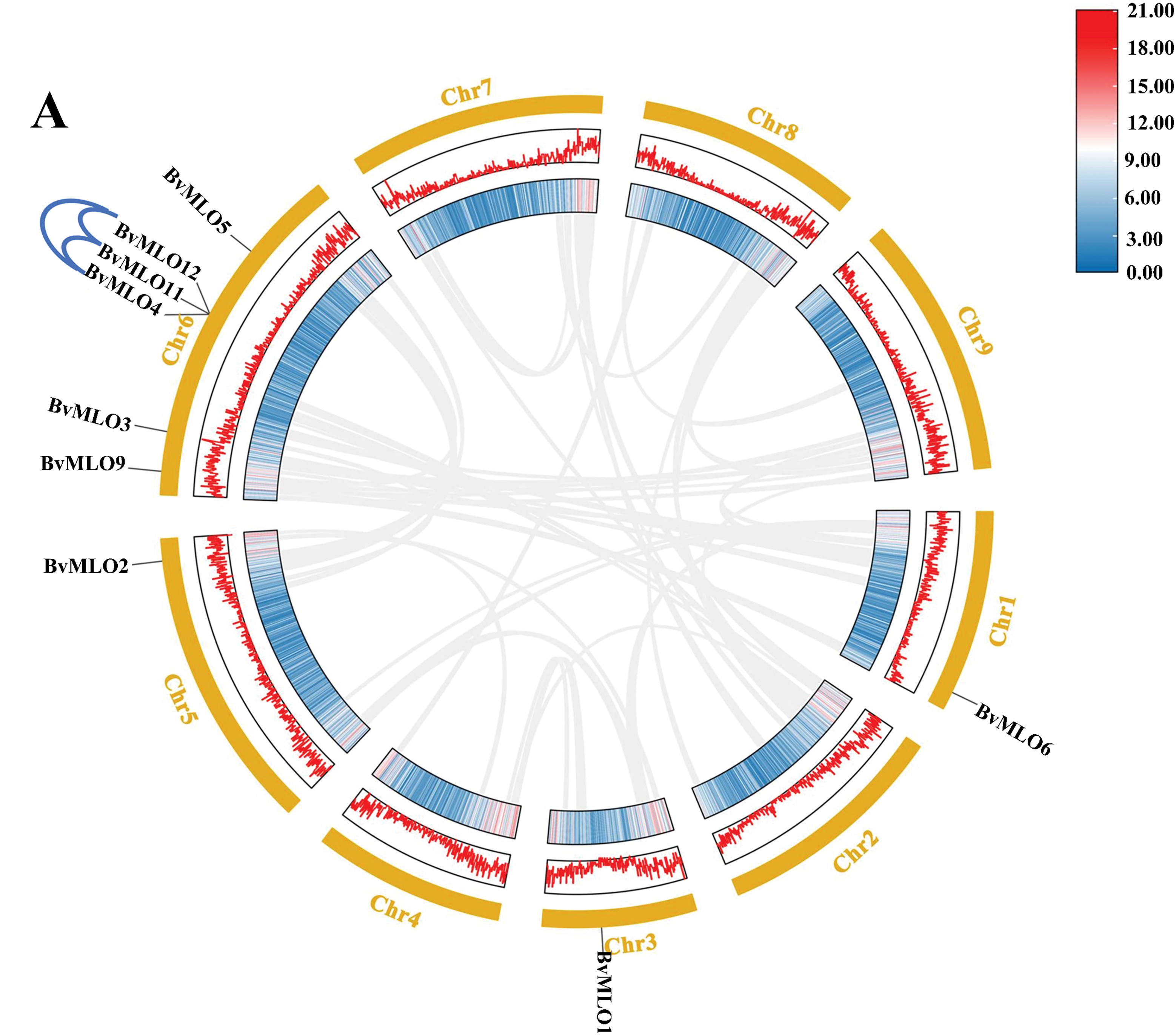
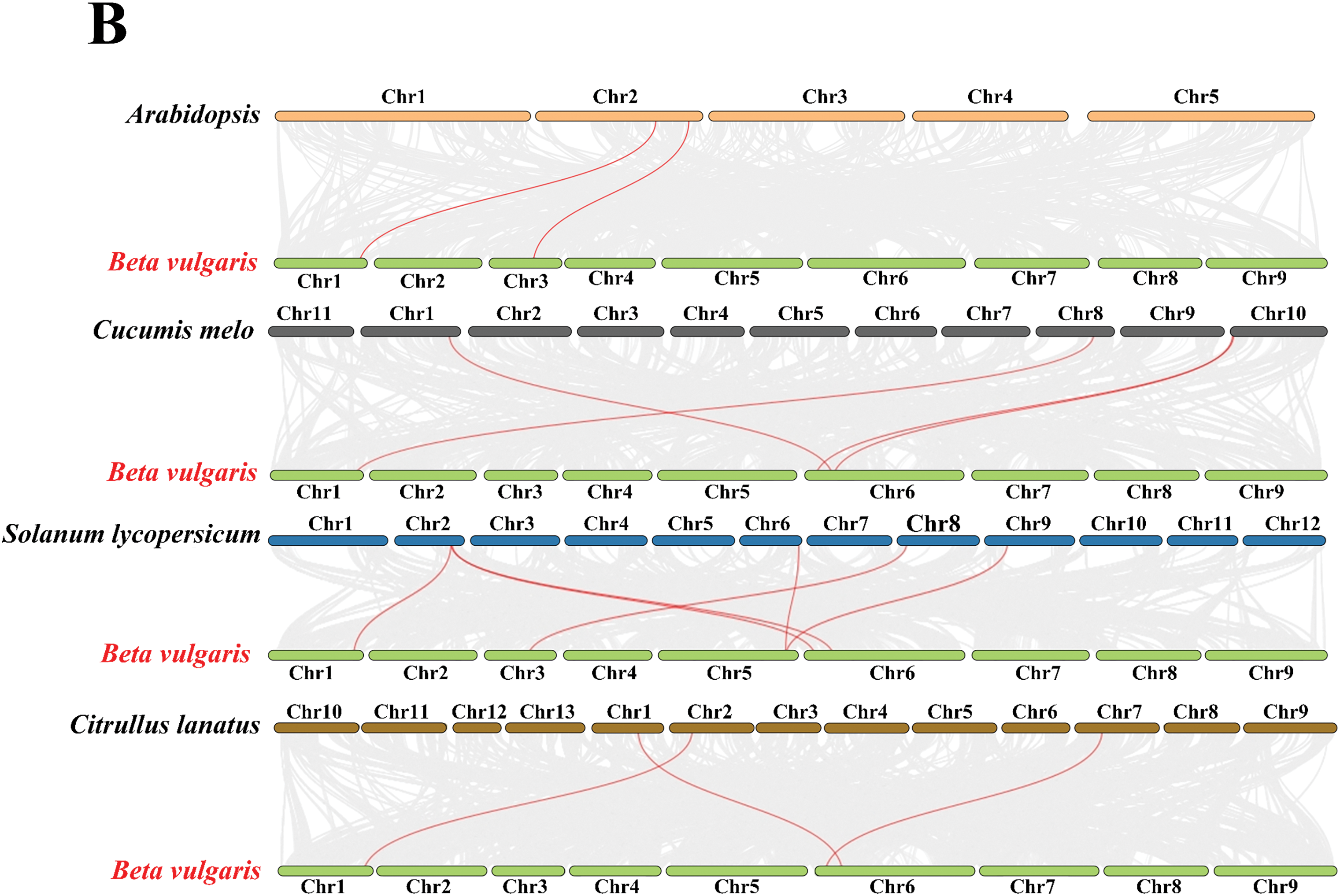
Figure 5: Analysis of the collinearity patterns of the MLO gene family in sugar beet (A) Yellow rectangles represent chromosomes 1–9, with gene density depicted using lines, heatmaps, and histograms along each chromosome. Gray lines indicate syntenic blocks within the sugar beet genome, while blue lines highlight gene clusters. (B) Synteny analysis between sugar beet MLO genes and those in other selected species (A. thaliana, Cucumis melo, tomato, Citrullus lanatus). Gray lines denote collinear genome blocks, whereas red lines specifically indicate syntenic MLO gene pairs across species
3.4 BvMLO Gene Family: Analysis of the Cis-Regulatory Elements
Cis-regulatory elements are fundamental in governing gene networks, particularly in response to diverse stimuli. By interacting with promoter regions, they influence tissue-specific regulation and modulate stress-induced gene expression patterns [41]. Based on the findings from PlantCARE, we detected several cis-regulatory elements related to phytohormone responsiveness within the sugar beet MLO gene family, and they were present in most members. Specifically, GA-responsive elements were identified in 8 BvMLOs, SA-responsive elements in six, and MeJA-responsive elements in eleven (Fig. 6).
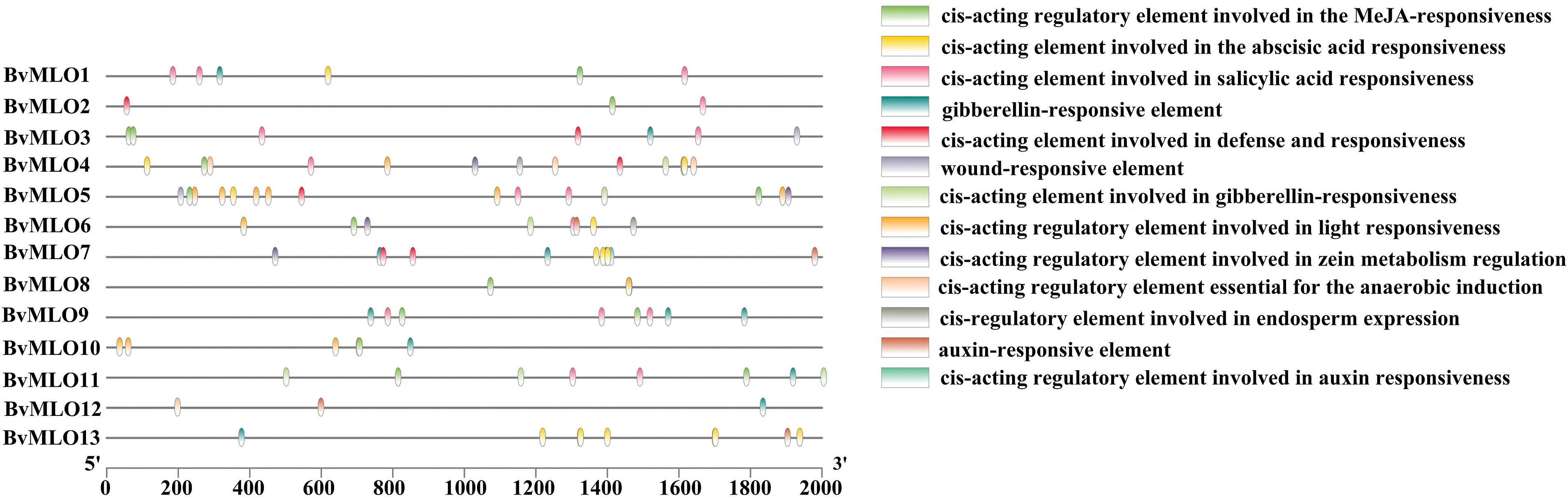
Figure 6: Analysis of cis-regulatory elements in the promoters of sugar beet MLO genes
Furthermore, all BvMLOs contained a minimum of two distinct stress-associated cis-regulatory elements, with the exception of BvMLO1, BvMLO9, and BvMLO13. This suggests that BvMLOs may play a role in responding to various environmental stressors (Fig. 6). Additionally, cis-elements linked to plant physiological and metabolic processes were identified. Four categories of cis-regulatory elements linked to environmental stress factors were identified, including those involved in anaerobic induction, elements responsive to defense and stress, regulatory sequences associated with light responsiveness, and wound-responsive elements. Notably, BvMLO4, BvMLO5, BvMLO6, and BvMLO7 contained regulatory elements involved in zein metabolism regulation. The findings underline the key function of cis-regulatory sequences in genes, especially in processes like hormone signaling, stress adaptation, and light-mediated regulation.
3.5 Prediction and Analysis of Three-Dimensional (3D) Structures of BvMLOs
The functional analysis of proteins largely depends on their three-dimensional (3D) structures, which are essential for deciphering their interactions with other molecules or proteins and revealing their possible functions in regulating key biological processes related to plant development and growth. The three-dimensional structures of BvMLO proteins in sugar beet were modeled through SWISS-MODEL, and validation results showed that all predicted structures achieved a quality factor exceeding 70%, suggesting that these models closely resemble their native conformations. SWISS-MODEL was employed to visually model the protein 3D structures (Fig. 7), and upon visualization, it was observed that these structures exhibited similar configurations of alpha helices and extended strands across all proteins (Fig. 7). A simple evaluation of modeling quality is based on the Global Model Quality Estimation (GMQE) value, which ranges from 0 to 1; the closer the value is to 1, the better the modeling quality. In summary, the predicted 3D structures of the BvMLO family proteins were validated to be of high quality, with consistent structural features and satisfactory GMQE values.
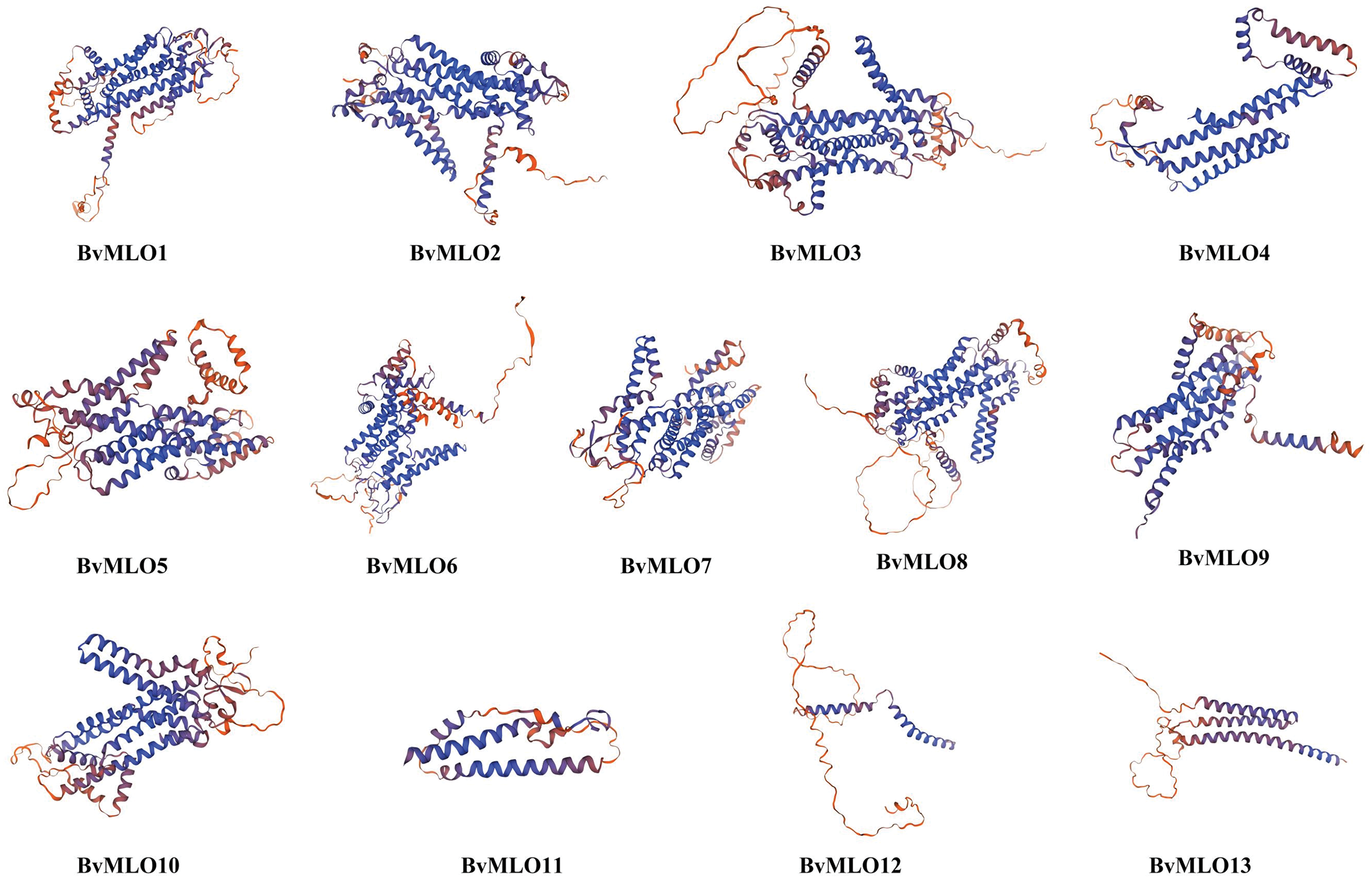
Figure 7: 3D structures of putative BvMLOs in sugar beet
3.6 Genetic Resistance Mechanisms in Sugar Beet against Powdery Mildew
The MLO gene initially discovered in barley, is a well-established recessive monogenic trait that confers long-lasting and broad-spectrum resistance against powdery mildew induced by Blumeria hordei. Among these, broad-spectrum resistance comprises multiple forms of plant pathogen resistance, such as targeted resistance achieved through the aggregation of QTLs in mature plants, as well as general or non-host resistance, which provides protection against multiple diseases [42–44]. Building on these established concepts, our laboratory has conducted studies on sugar beet powdery mildew. This type of resistance, which closely resembles that identified in barley, serves as a basis for elucidating the dynamic relationship between host plants and pathogens. To further investigate this, we observed that the sugar beet leaves were infected with powdery mildew after inoculation, white velvety spots approximately 1 cm in size form on the leaves, followed by the appearance of a white hyphal layer, which eventually forms a powdery layer comprising the pathogen’s mycelium and conidia (Fig. 8A). It illustrates the typical process of sugar beet powdery mildew infection, from the initial visible symptoms to the formation of the powdery spore layer, while also hinting at the potential for studying resistance mechanisms similar to those in barley. To build on the observation of the typical infection process, we further examined the sugar beet powdery mildew pathogen at a microscopic level. We observed the pathogen under an electron microscope, where at 200× magnification, The conidia of sugar beet powdery mildew are typically oval, round, or elongated in shape. They vary in size, generally ranging from 10 to 30 μm. The surface is smooth and has a certain degree of transparency. (Fig. 8B). It can be concluded that this demonstrates the similarity of the resistance mechanisms in sugar beet to those found in barley, and provides potential for further resistance studies
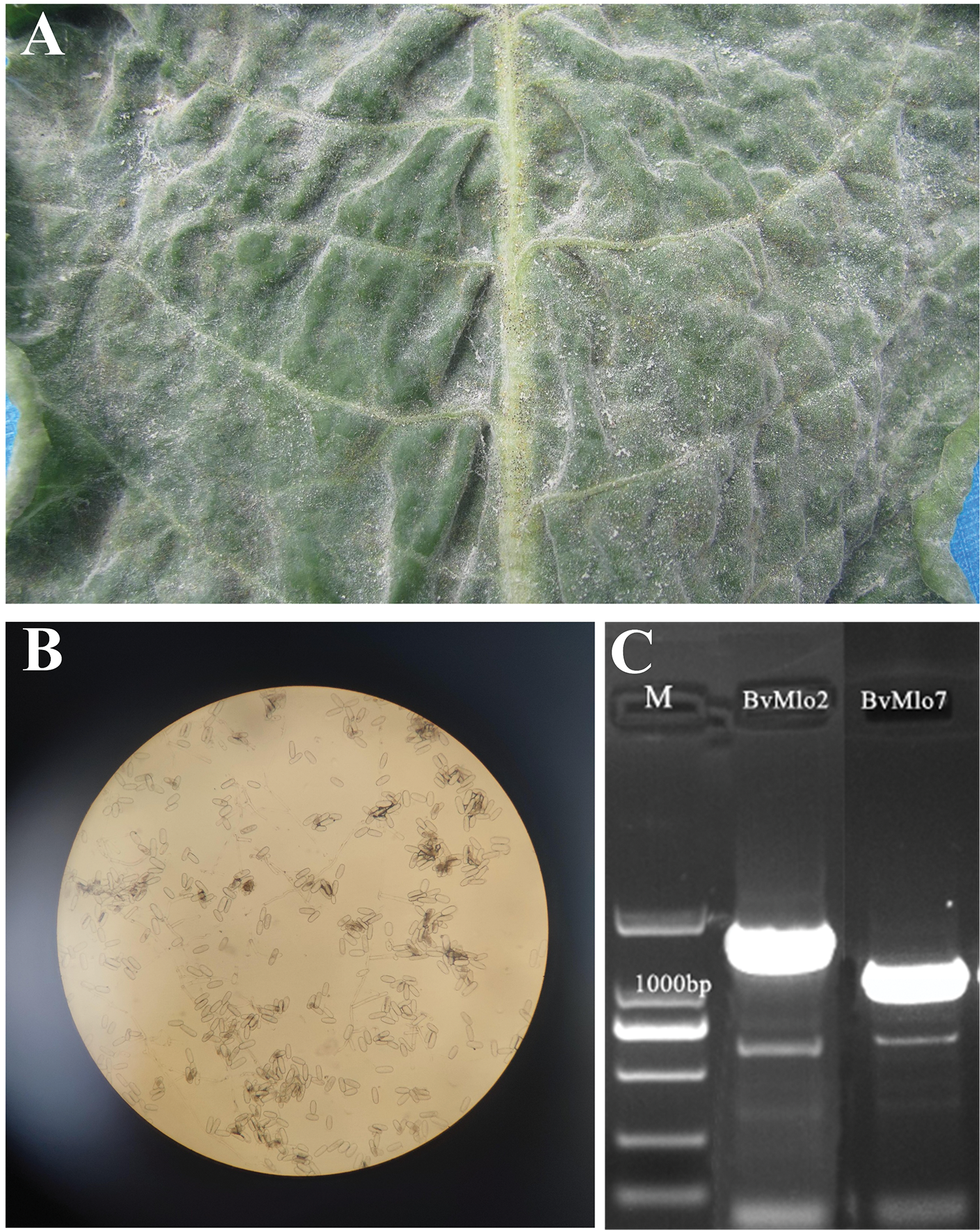
Figure 8: Cloning of sugar beet BvMLO2 and BvMLO7 genes and morphology of pathogen spores of sugar beet powdery mildew. (A) Inoculation performance of sugar beet susceptible materials. (B) Beet powdery mildew and pathogen spores 200× under optical microscope. (C) Cloning of BvMLO2 and BvMLO7 genes from sugar beet
Considering the resemblance between sugar beet and barley in their reactions to powdery mildew infection, we proposed that the MLO gene family within sugar beet could likewise contribute to resistance. To explore this possible genetic resistance, we examined the expression patterns for BvMLO genes in sugar beet after infection by Blumeria hordei. We noted a substantial increase in BvMLO2 and BvMLO7 gene expression within the susceptible variety KWS9147. By utilizing specific primers, we successfully amplified the target genes, generating two PCR fragments of roughly 1300 and 800 bp. The purified PCR fragments were inserted into the pCE-Zero vector, and the resulting recombinant constructs were introduced into competent DH5α cells through transformation. After culturing on Luria-Bertani (LB) solid medium, we selected well-grown single colonies for further shaking culture in LB liquid medium and subsequent bacterial liquid PCR reactions. Electrophoresis results (Fig. 8C) showed that the two target bands were slightly larger than expected, measuring approximately 1800 bp and 900 bp, thus confirming the presence of positive recombinant plasmids, suggesting that these genes were involved in sugar beet’s resistance mechanisms against powdery mildew.
3.7 Gene Expression Changes of BvMLO2 and BvMLO7 in Sugar Beet in Response to C. beticola Infection
CLS (Cercospora leaf spot) is the most important leaf disease of sugar beet in the world. It occurs in sugar beet planting countries all over the world. The main cause of C. beticola is the infection of Mycosphaerella [45,46]. It is crucial to explore the genetic factors involved in the plant’s response to this pathogen. In this study, as shown in the images, the structure and color of a healthy sugar beet leaf are normal and clear (Fig. 9A). On the 5th day after inoculation, spots began to appear on the leaves (Fig. 9B). By the 10th day, patchy lesions had developed (Fig. 9C), and by the 15th day, extensive lesions and necrosis were evident (Fig. 9D). This indicates that the disease develops rapidly and is destructive, it is essential to examine the characteristics of the pathogen itself. Therefore, we also investigated the colony morphology of C. beticola, the causative agent of CLS, the colonies of C. beticola are round, grayish-white, with a diameter ranging from 0.5 to 6 mm. The dark-colored, blackish substrate is visible against a grayish background (Fig. 9E,F).
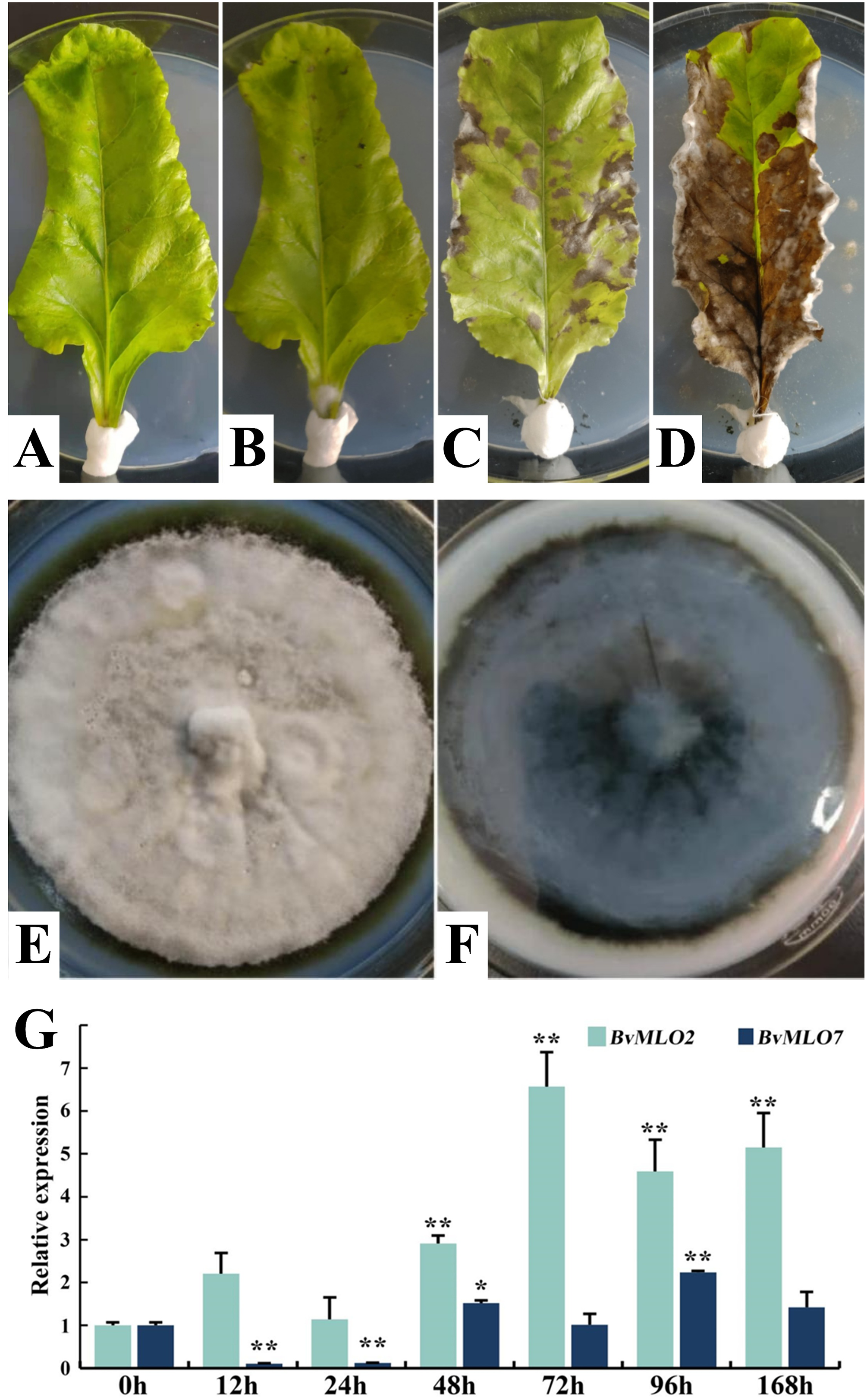
Figure 9: Sugar beet leaf response and gene expression following C. beticola inoculation. (A) Healthy, non-inoculated leaves. (B) Leaves observed 5 days post-C. beticola inoculation. (C) Leaves were examined 10 days after inoculation. (D) Leaves assessed 15 days following inoculation. (E, F) Morphological characteristics of C. beticola colonies. (G) Relative transcript levels of BvMLO2 and BvMLO7 in response to C. beticola infection. Gene expression levels were normalized to BvGAPDH and presented relative to the 0-h time point. The 2−ΔΔCt method was applied to quantify the expression levels of BvMLO2 and BvMLO7 across different time intervals. Statistical significance was determined using one-way ANOVA with GraphPad Prism, where asterisks indicate significant differences (*p < 0.05; **p < 0.01)
Given the swift progression and severe impact of CLS, along with the necessity of comprehending both the pathogen’s traits and the plant’s genetic reaction, additional studies are crucial to investigate how sugar beet contributes to resistance against CLS. We examined how gene expression altered due to C. beticola infection. Specifically, we analyzed the upregulated expression of sugar beet genes upon C. beticola infection and found that BvMLO2 and BvMLO7 exhibited a marked increase in expression levels in response to C. beticola. When subjected to biotic stress caused by C. beticola, the expression level of BvMLO2 was significantly upregulated, peaking at 72 h post-infection before declining. Conversely, BvMLO7 expression was almost undetectable until 24 h post-infection, after which it steadily increased (Fig. 9G). The results (Fig. 9G) show that the expression levels of BvMLO2 and BvMLO7 are not consistent, and there are also certain differences in their relative expression levels.
Overall, following Brevipila betae inoculation, both BvMLO2 and BvMLO7 exhibited increased expression at various time intervals in comparison to the untreated control. Additionally, the downregulation of BvMLO7 expression at 12 and 24 h post-infection raises the question of whether it is due to the plant’s own defense mechanisms, which warrants further investigation.
3.8 Expression of BvMLO2 and BvMLO7 under Abiotic Stress in Sugar Beet
Considering the distinct expression patterns of BvMLO2 and BvMLO7 following C. beticola infection, further research is necessary to determine whether these genes also contribute to sugar beet’s adaptation to abiotic stress conditions. Given that environmental stressors like water deficiency or high salinity can also influence plant defense responses, investigating BvMLO2 and BvMLO7 expression under these conditions will offer a more comprehensive understanding of their roles in overall stress resilience.
We investigated how BvMLO2 and BvMLO7 responded to MeJA, SA, NaCl, and mannitol-induced stress conditions (Fig. 10A, Table S2). Under MeJA and SA treatment, BvMLO2 showed a continuous decline in expression, whereas BvMLO7 reacted rapidly to both stresses, exhibiting a significant increase as early as 4 h post-treatment. Notably, its expression reached the highest level at 12 h under MeJA treatment and peaked at 8 h following SA exposure (Fig. 10B,C). It indicated that BvMLO7 had a strong response to MeJA and SA, predicting that BvMLO7 might regulate sugar beet disease resistance through these two hormones. Under NaCl and Mannitol stresses, BvMLO2 and BvMLO7 showed irregular changes, which were different from the above two treatments. Under the NaCl stress, BvMLO2 and BvMLO7 were rapidly induced and highly significantly upregulated, reaching a peak at 4 h after treatment, followed by a trend of downregulation and then upregulation again. Under the Mannitol stress, the change trends of both BvMLO2 and BvMLO7 expression levels were similar to those of NaCl stress, except for the fact that BvMLO7 expression peaked at 120 h after treatment. These results suggested that NaCl and Mannitol have a selective effect on the expression of BvMLO2 and BvMLO7 (Fig. 10D,E). Overall, BvMLO2 and BvMLO7 displayed unique transcriptional responses to different environmental challenges, both biotic and abiotic, highlighting their potential roles in plant resistance mechanisms. The differences in their gene expression across various adverse environments suggest that these genes may contribute to stress adaptation through distinct functional and regulatory pathways.
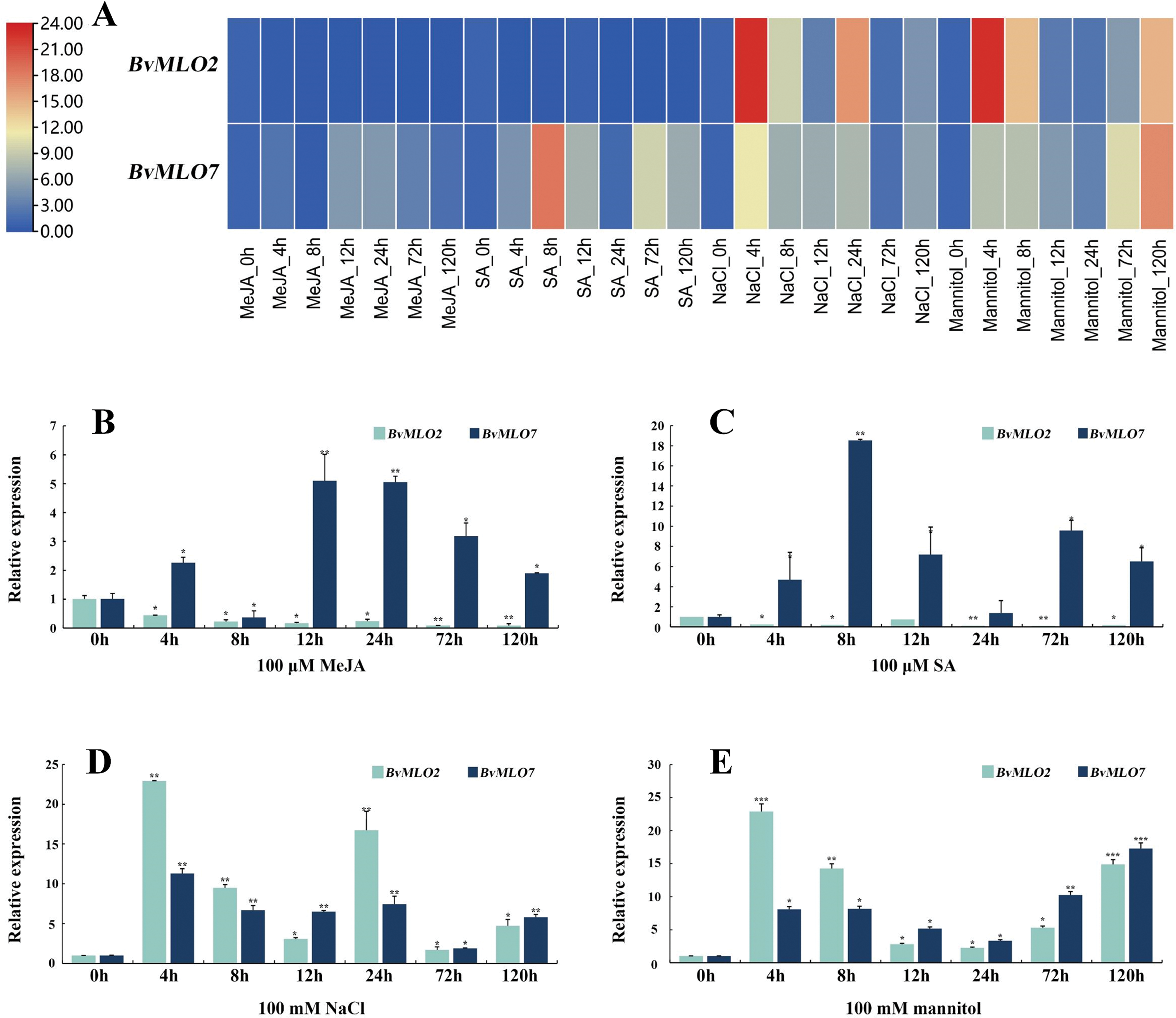
Figure 10: Modulation of BvMLO2 and BvMLO7 transcription in leaf tissues under four distinct abiotic stress conditions. (A) Expression dynamics of BvMLO genes in response to MeJA, SA, NaCl, and mannitol treatments. (B) Relative transcript levels of BvMLO2 and BvMLO7 in leaf tissue following exposure to 100 μM MeJA. (C) Expression variations of BvMLO2 and BvMLO7 in leaf samples under 100 μM SA treatment. (D) Changes in BvMLO2 and BvMLO7 expression in leaves subjected to 100 mM NaCl. (E) BvMLO2 and BvMLO7 expression levels in leaves treated with 100 mM mannitol. The expression of both genes was normalized against BvGAPDH and represented relative to the 0 h time point. The 2−ΔΔCt method was applied to determine expression fluctuations across different time intervals. Statistical significance was assessed using one-way ANOVA in GraphPad Prism, with asterisks denoting significant differences (*p < 0.05; **p < 0.01; ***p < 0.001)
Numerous studies indicate that MLO genes conferring broad-spectrum resistance are plant-specific disease resistance factors that have been utilized across various crop species, including tomato [47], A. thaliana [28], Citrullus lanatus [48], Cucumis melo [49], tobacco [50], Rosa [51], eggplant [52], wheat [53], and so on. The genome sequence of sugar beet was released in 2014 [54]. Although the MLO gene family has been well studied in several crops, there is no report relevant information about the MLO gene family in sugar beet. In marked contrast to the findings of previous studies in A. thaliana [29], Cucumis sativus [55], the BvMLOs are not evenly distributed across chromosomes. Studies have indicated that the uneven distribution of genes along chromosomes can reveal insights into their evolutionary history [56]. The physicochemical analysis of MLO proteins revealed that most were alkaline and inherently unstable, resembling the characteristics of tomato MLO proteins [47]. This analysis offers a theoretical foundation for further functional studies of BvMLO proteins.
The MLO proteins were classified into seven subfamilies in most studies, but the results of different studies were not entirely consistent [52,54,55]. To further elucidate the classification and structural characteristics of BvMLO proteins, sequence logos were employed to visualize positional variability and conservation across the family. The height of each stack overall in the sequence logo reflects the degree of conservation at a given position, with taller stacks indicating higher conservation. This analysis highlights conserved regions essential for BvMLO functionality while also identifying highly variable sites, which may signify evolutionary divergence or potential adaptive significance. Phylogenetic analysis classified BvMLO proteins into three subgroups, revealing collinear relationships with Arabidopsis MLO proteins, consistent with the evolutionary proximity between sugar beet and Arabidopsis, both belonging to dicotyledons. The functional properties of proteins are closely linked to their three-dimensional (3D) structures, and the conserved structural features of BvMLO proteins further support their phylogenetic classification and domain conservation. Motif analysis identified 10 conserved motifs, with a highly conserved domain (Motif 8) located in the C-terminal region. Within subgroup I, all BvMLO proteins, except for BvMLO7, BvMLO10, and BvMLO13, retained all 10 motifs. Similarly, BvMLO3 and BvMLO6, clustered within subgroup IV, exhibited identical motif structures. Notably, BvMLO proteins within the same phylogenetic clade shared similar motif compositions, suggesting a conserved evolutionary pattern within the gene family. Sequence alignment further revealed that BvMLO proteins exhibit relatively low sequence similarity, a hallmark of evolutionary divergence (Table S3, Fig. S1), Specifically, five members—BvMLO4, BvMLO7, BvMLO10, BvMLO11, and BvMLO13—lacked the conserved Motif 10, underscoring the structural complexity and evolutionary diversity of the BvMLO gene family.
The pathogen infection process involves initial contact and invasion of the host plant, followed by expansion and reproduction within the host, ultimately leading to disease development and visible symptoms. Simultaneously, the host plant triggers a sequence of responses to infection, including defense mechanisms, resistance strategies, and adaptive modifications, which influence symptom manifestation. However, studies on the infection process of C. beticola remain limited. Throughout this process, plant hormones, as crucial endogenous signaling molecules, integrate environmental stress signals to precisely regulate plant growth, development, and disease resistance under stress conditions [57]. According to the analysis of Fig. 6, most genes contain response elements for MeJA and SA, suggesting that plants may activate defense mechanisms through the MeJA [58] and SA [59] signaling pathways. Additionally, response elements related to external environmental stress were identified, indicating that the plant’s defense response to pathogen infection may intersect with its response to external environmental stress. This further reveals that disease resistance and stress tolerance in plants may share partial regulatory pathways. Therefore, analyzing the infection process in sugar beet suggests that the series of responses triggered by pathogen invasion are closely linked to the regulation of plant hormone response elements.
Research indicates that MLO family members play a role in regulating plant resistance to powdery mildew and influencing cell death pathways by mediating Ca2+ signaling responses [2,3,60]. Previous inoculation experiments conducted in our laboratory on the MLO gene of sugar beet powdery mildew fungus revealed significant induced changes in its expression. However, the response mechanisms of the BvMLO gene family to C. beticola remain unexplored. In this study, visible lesions on sugar beet leaves appeared within 15 days post-inoculation. Further analysis revealed significant variations in the expression levels of BvMLO2 and BvMLO7 across different time points and under various abiotic stress conditions. As critical regulators of plant growth and development, MeJA and SA exhibited pronounced response characteristics during the mid-to-late stages of treatment, indicating that BvMLO2 and BvMLO7 could be instrumental in systemic acquired resistance of sugar beet to C. beticola by mediating the MeJA and SA signaling pathways. This observation aligns with the distribution of MeJA- and SA-responsive elements in the promoter regions (Fig. 6), reinforcing the crucial role of these two genes in regulating hormone signaling. Additionally, NaCl stress significantly disrupts the dynamic balance of Ca2+ in plants [61,62]. Studies have found that elevated Ca2+ concentrations in epidermal cells can suppress plant disease resistance. During the early stages of pathogen infection, defense responses are activated, triggering Ca2+ influx, but MLO gene expression is suppressed. In later stages, as MLO protein levels recover, cytosolic Ca2+ concentrations rise sharply, and calmodulin (CaM) levels increase significantly. This leads to enhanced MLO protein activity under CaM regulation, ultimately resulting in suppressed resistance and susceptibility [3]. Therefore, the significant expression of BvMLO2 and BvMLO7 under NaCl stress may be closely related to the regulation of Ca2+ signaling pathways. Furthermore, mannitol, a key osmotic regulator and antioxidant in plants is crucial for defense mechanisms and stress responses [63]. This study revealed that BvMLO2 and BvMLO7 exhibited a marked increase in expression under mannitol stress, suggesting their possible role in plant defense and stress regulatory networks. Given the strong association between their transcription profiles and environmental challenges, it is reasonable to deduce that BvMLO2 and BvMLO7 play crucial roles in plant adaptations to external stresses and pathogen attacks. Increasing evidence suggests that MLO gene expression is regulated by hormones and non-biological stresses. For instance, studies on cotton GhMLOs have shown that MLO proteins can be suppressed or induced by ABA, ethylene (ETH), JA, and SA [64]. Four GmMLOs in soybeans respond to various non-biological stresses and phytohormone treatments [65]. The silencing of CaMLO2 in pepper via virus-induced gene suppression, along with its overexpression in Arabidopsis, revealed that CaMLO2 plays a role in drought stress regulation and functions as a negative regulator of ABA signaling [66]. Under non-biological stress conditions, the expression levels of these two genes are regulated to varying degrees, highlighting the conserved and diverse roles of the MLO family in plant hormone and stress responses.
In this research, we identified the 13 MLO genes in sugar beet. We divided 13 BvMLOs into three subgroups based on the phylogenetic analysis. Further, the evolutionary history and genetic variation of sugar beet MLO genes were systematically examined, including the evolutionary relationships, chromosomal positioning, synteny patterns, structural features of the genes, and their expression pattern. The expression levels of BvMLO2 and BvMLO7 in response to several biotic stress and abiotic stresses were analyzed and results suggested that both BvMLO2 and BvMLO7 might be involved in the stress resistance process of sugar beet. This study will contribute to further analysis of the mechanism of defense-related genes and the breeding of highly tolerant sugar beet varieties.
Acknowledgement: Not applicable.
Funding Statement: This study was supported by the collaborative innovation system project for the economic crop industry technology in Heilongjiang Province (2018711).
Author Contributions: Conceptualization, Guangzhou Ding; methodology, Guangzhou Ding; software, Ran Li, Liuhong Chen, and Yu Liu; validation, Ran Li, Liuhong Chen, and Yu Liu; formal analysis, Ran Li, Liuhong Chen, and Yu Liu; investigation, Chunlei Zhao and Yanli Li; resources, Guangzhou Ding; data curation, Ran Li, Liuhong Chen, and Yu Liu; writing—original draft preparation, Ran Li; writing—review and editing, Ran Li and Guangzhou Ding; visualization, Ran Li, Liuhong Chen, and Yu Liu; supervision, Guangzhou Ding; project administration, Guangzhou Ding; funding acquisition, Guangzhou Ding. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All relevant data is provided in the article or Supplementary Materials. For further questions, contact the corresponding author.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2025.061550. Figure S1: Multiple sequence comparison of 13 BvMLO proteins; Table S1: Features of sugar beet MLO transcription factors; Table S2: The relative expression levels of BvMLO2 and BvMLO7 in leaves under biotic stress and abiotic stresses by qRT-PCR; Table S3: The similarities among 13 BvMLO proteins of sugar beet.
References
1. Wolter M, Hollricher K, Salamini F, Schulze-Lefert P. The mlo resistance alleles to powdery mildew infection in barley trigger a developmentally controlled defence mimic phenotype. Mol Gen Genet MGG. 1993;239(1–2):122–8. doi:10.1007/bf00281610. [Google Scholar] [PubMed] [CrossRef]
2. Kim MC, Lee SH, Kim JK, Chun HJ, Choi MS, Chung WS, et al. Mlo, a modulator of plant defense and cell death, is a novel calmodulin-binding protein: isolation and characterization of a rice Mlo homologue. J Biol Chem. 2002;277(22):19304–14. doi:10.1074/jbc.M108478200. [Google Scholar] [PubMed] [CrossRef]
3. Kim MC, Panstruga R, Elliott C, Müller J, Devoto A, Yoon HW, et al. Calmodulin interacts with MLO protein to regulate defence against mildew in barley. Nature. 2002;416(6879):447–51. doi:10.1038/416447a. [Google Scholar] [PubMed] [CrossRef]
4. Büschges R, Hollricher K, Panstruga R, Simons G, Wolter M, Frijters A, et al. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88(5):695–705. doi:10.1016/s0092-8674(00)81912-1. [Google Scholar] [PubMed] [CrossRef]
5. Konishi S, Sasakuma T, Sasanuma T. Identification of novel Mlo family members in wheat and their genetic characterization. Genes Genet Syst. 2010;85(3):167–75. doi:10.1266/ggs.85.167. [Google Scholar] [PubMed] [CrossRef]
6. Liu Q, Zhu H. Molecular evolution of the MLO gene family in Oryza sativa and their functional divergence. Gene. 2008;409(1–2):1–10. doi:10.1016/j.gene.2007.10.031. [Google Scholar] [PubMed] [CrossRef]
7. Iqbal Z, Shariq Iqbal M, Singh SP, Buaboocha T. Ca2+/calmodulin complex triggers CAMTA transcriptional machinery under stress in plants: signaling cascade and molecular regulation. Front Plant Sci. 2020;11:598327. doi:10.3389/fpls.2020.598327. [Google Scholar] [PubMed] [CrossRef]
8. von Bongartz K, Sabelleck B, Baquero Forero A, Kuhn H, Leissing F, Panstruga R. Comprehensive comparative assessment of the Arabidopsis thaliana MLO2-CALMODULIN2 interaction by various in vitro and in vivo protein-protein interaction assays. Biochem J. 2023;480(20):1615–38. doi:10.1042/BCJ20230255. [Google Scholar] [PubMed] [CrossRef]
9. Jørgensen IH. Discovery, characterization and exploitation of Mlo powdery mildew resistance in barley. Euphytica. 1992;63(1–2):141–52. doi:10.1007/BF00023919. [Google Scholar] [CrossRef]
10. Kessler SA, Shimosato-Asano H, Keinath NF, Wuest SE, Ingram G, Panstruga R, et al. Conserved molecular components for pollen tube reception and fungal invasion. Science. 2010;330(6006):968–71. doi:10.1126/science.1195211. [Google Scholar] [PubMed] [CrossRef]
11. Davis TC, Jones DS, Dino AJ, Cejda NI, Yuan J, Willoughby AC, et al. Arabidopsis thaliana MLO genes are expressed in discrete domains during reproductive development. Plant Reprod. 2017;30(4):185–95. doi:10.1007/s00497-017-0313-2. [Google Scholar] [CrossRef]
12. Jacott CN, Ridout CJ, Murray JD. Unmasking mildew resistance locus O. Trends Plant Sci. 2021;26(10):1006–13. doi:10.1016/j.tplants.2021.05.009. [Google Scholar] [PubMed] [CrossRef]
13. Zhu L, Zhang XQ, Ye D, Chen LQ. The mildew resistance locus O 4 interacts with CaM/CML and is involved in root gravity response. Int J Mol Sci. 2021;22(11):5962. doi:10.3390/ijms22115962. [Google Scholar] [PubMed] [CrossRef]
14. Nguyen VN, Vo KT, Park H, Jeon JS, Jung KH. A systematic view of the MLO family in rice suggests their novel roles in morphological development, diurnal responses, the light-signaling pathway, and various stress responses. Front Plant Sci. 2016;7:1413. doi:10.3389/fpls.2016.01413. [Google Scholar] [PubMed] [CrossRef]
15. Yang S, Shi Y, Zou L, Huang J, Shen L, Wang Y, et al. Pepper CaMLO6 negatively regulates Ralstonia solanacearum resistance and positively regulates high temperature and high humidity responses. Plant Cell Physiol. 2020;61(7):1223–38. doi:10.1093/pcp/pcaa052. [Google Scholar] [PubMed] [CrossRef]
16. Porcel R, Bustamante A, Ros R, Serrano R, Mulet Salort JM. BvCOLD1: a novel aquaporin from sugar beet (Beta vulgaris L.) involved in boron homeostasis and abiotic stress. Plant Cell Environ. 2018;41(12):2844–57. doi:10.1111/pce.13416. [Google Scholar] [PubMed] [CrossRef]
17. Heitefuss R. Cercospora leaf spot of sugar beet and related species. J Phytopathol. 2011;159(4):325–5. doi:10.1111/j.1439-0434.2010.01762.x. [Google Scholar] [CrossRef]
18. Refat AAH, Ghaffar MSA. The economic impact of sugar beet cultivation in new lands (study of Al-Salam Canal area status). Aust J Basic Appl Sci. 2010;4:1641–9. [Google Scholar]
19. Vale PAS, de Resende MLV, dos Santos Botelho DM, de Andrade CCL, Alves E, Ogoshi C, et al. Epitypification of Cercospora coffeicola and its involvement with two different symptoms on coffee leaves in Brazil. Eur J Plant Pathol. 2021;159(2):399–408. doi:10.1007/s10658-020-02170-y. [Google Scholar] [CrossRef]
20. Terensan S, Fernando HNS, Silva JN, Perera SCN, Kottearachchi NS, Weerasena OJ, et al. Morphological and molecular analysis of fungal species associated with blast and brown spot diseases of Oryza sativa. Plant Dis. 2022;106(6):1617–25. doi:10.1094/PDIS-04-21-0864-RE. [Google Scholar] [CrossRef]
21. Vaghefi N, Nelson SC, Kikkert JR, Pethybridge SJ. Genetic structure of Cercospora beticola populations on Beta vulgaris in New York and Hawaii. Sci Rep. 2017;7(1):1726. doi:10.1038/s41598-017-01929-4. [Google Scholar] [PubMed] [CrossRef]
22. Nsibo DL, Barnes I, Omondi DO, Dida MM, Berger DK. Population genetic structure and migration patterns of the maize pathogenic fungus, Cercospora zeina in East and Southern Africa. Fungal Genet Biol. 2021;149(578):103527. doi:10.1016/j.fgb.2021.103527. [Google Scholar] [PubMed] [CrossRef]
23. Arens N, Backhaus A, Döll S, Fischer S, Seiffert U, Mock HP. Non-invasive presymptomatic detection of Cercospora beticola infection and identification of early metabolic responses in sugar beet. Front Plant Sci. 2016;7:1377. doi:10.3389/fpls.2016.01377. [Google Scholar] [PubMed] [CrossRef]
24. El-Fawy MM, El-Sharkawy RMI, Abo-Elyousr KAM. Evaluation of certain Penicillium frequentans isolates against Cercospora leaf spot disease of sugar beet. Egypt J Biol Pest Control. 2018;28(1):1–7. doi:10.1186/s41938-018-0053-0. [Google Scholar] [CrossRef]
25. Rangel LI, Spanner RE, Ebert MK, Pethybridge SJ, Stukenbrock EH, de Jonge R, et al. Cercospora beticola: the intoxicating lifestyle of the leaf spot pathogen of sugar beet. Mol Plant Pathol. 2020;21(8):1020–41. doi:10.1111/mpp.12962. [Google Scholar] [CrossRef]
26. Xie Y, Ouyang Y, Han S, Se J, Tang S, Yang Y, et al. Crop rotation stage has a greater effect than fertilisation on soil microbiome assembly and enzymatic stoichiometry. Sci Total Environ. 2022;815(4):152956. doi:10.1016/j.scitotenv.2022.152956. [Google Scholar] [PubMed] [CrossRef]
27. Misra V, Mall AK, Pandey H, Srivastava S, Sharma A. Advancements and prospects of CRISPR/Cas9 technologies for abiotic and biotic stresses in sugar beet. Front Genet. 2023;14:1235855. doi:10.3389/fgene.2023.1235855. [Google Scholar] [PubMed] [CrossRef]
28. Chen Z, Hartmann HA, Wu MJ, Friedman EJ, Chen JG, Pulley M, et al. Expression analysis of the AtMLO gene family encoding plant-specific seven-transmembrane domain proteins. Plant Mol Biol. 2006;60(4):583–97. doi:10.1007/s11103-005-5082-x. [Google Scholar] [PubMed] [CrossRef]
29. Johnson LS, Eddy SR, Portugaly E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010;11(1):1–8. doi:10.1186/1471-2105-11-431. [Google Scholar] [PubMed] [CrossRef]
30. Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR. CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res. 2020;48(D1):D265–8. doi:10.1093/nar/gkz991. [Google Scholar] [PubMed] [CrossRef]
31. Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, Appel RD, et al. Protein identification and analysis tools in the ExPASy server. Methods Mol Biol. 1999;112:531–52. doi:10.1385/1-59259-584-7:. [Google Scholar] [CrossRef]
32. Chou KC, Shen HB. Cell-PLoc: a package of Web servers for predicting subcellular localization of proteins in various organisms. Nat Protoc. 2008;3(2):153–62. doi:10.1038/nprot.2007.494. [Google Scholar] [PubMed] [CrossRef]
33. Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–45. doi:10.1002/pro.3290. [Google Scholar] [PubMed] [CrossRef]
34. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. doi:10.1016/j.molp.2020.06.009. [Google Scholar] [PubMed] [CrossRef]
35. Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30(1):325–7. doi:10.1093/nar/30.1.325. [Google Scholar] [PubMed] [CrossRef]
36. Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46(W1):W296–303. doi:10.1093/nar/gky427. [Google Scholar] [PubMed] [CrossRef]
37. Li J, Liu X, Xu L, Li W, Yao Q, Yin X, et al. Low nitrogen stress-induced transcriptome changes revealed the molecular response and tolerance characteristics in maintaining the C/N balance of sugar beet (Beta vulgaris L.). Front Plant Sci. 2023;14:1164151. doi:10.3389/fpls.2023.1164151. [Google Scholar] [PubMed] [CrossRef]
38. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25(4):402–8. doi:10.1006/meth.2001.1262. [Google Scholar] [PubMed] [CrossRef]
39. Nützmann HW, Huang A, Osbourn A. Plant metabolic clusters—from genetics to genomics. New Phytol. 2016;211(3):771–89. doi:10.1111/nph.13981. [Google Scholar] [PubMed] [CrossRef]
40. Elizondo LI, Jafar-Nejad P, Clewing JM, Boerkoel CF. Gene clusters, molecular evolution and disease: a speculation. Curr Genomics. 2009;10(1):64–75. doi:10.2174/138920209787581271. [Google Scholar] [PubMed] [CrossRef]
41. Abdullah M, Cao Y, Cheng X, Meng D, Chen Y, Shakoor A, et al. The sucrose synthase gene family in Chinese pear (Pyrus bretschneideri Rehd.structure, expression, and evolution. Molecules. 2018;23(5):1144. doi:10.3390/molecules23051144. [Google Scholar] [PubMed] [CrossRef]
42. Moolhuijzen P, Ge C, Palmiero E, Ellwood SR. A unique resistance mechanism is associated with RBgh2 barley powdery mildew adult plant resistance. Theor Appl Genet. 2023;136(6):145. doi:10.1007/s00122-023-04392-0. [Google Scholar] [PubMed] [CrossRef]
43. Wu Y, Sexton W, Yang B, Xiao S. Genetic approaches to dissect plant nonhost resistance mechanisms. Mol Plant Pathol. 2023;24(3):272–83. doi:10.1111/mpp.13290. [Google Scholar] [PubMed] [CrossRef]
44. Dreiseitl A. Mlo-mediated broad-spectrum and durable resistance against powdery mildews and its current and future applications. Plants. 2024;13(1):138. doi:10.3390/plants13010138. [Google Scholar] [PubMed] [CrossRef]
45. Imbusch F, Liebe S, Erven T, Varrelmann M. Dynamics of cercospora leaf spot disease determined by aerial spore dispersal in artificially inoculated sugar beet fields. Plant Pathol. 2021;70(4):853–61. doi:10.1111/PPA.13337. [Google Scholar] [CrossRef]
46. Bublitz DM, Hanson LE, McGrath JM. Weather conditions conducive for the early-season production and dispersal of Cercospora beticola spores in the Great Lakes region of North America. Plant Dis. 2021;105(10):3063–71. doi:10.1094/PDIS-09-20-2004-RE. [Google Scholar] [PubMed] [CrossRef]
47. Chen Y, Wang Y, Zhang H. Genome-wide analysis of the mildew resistance locus o (MLO) gene family in tomato (Solanum lycopersicum L.). Plant Omics. 2014;7(2):87–93. doi:10.1007/s10658-015-0833-2. [Google Scholar] [CrossRef]
48. Iovieno P, Andolfo G, Schiavulli A, Catalano D, Ricciardi L, Frusciante L, et al. Structure, evolution and functional inference on the Mildew Locus O (MLO) gene family in three cultivated Cucurbitaceae spp. BMC Genom. 2015;16(1):1–13. doi:10.1186/s12864-015-2325-3. [Google Scholar] [PubMed] [CrossRef]
49. Howlader J, Kim HT, Park JI, Ahmed NU, Robin AH, Jung K, et al. Expression profiling of MLO family genes under podosphaera xanthii infection and exogenous application of phytohormones in Cucumis melo L. J Life Sci. 2016;26(4):419–30. doi:10.5352/jls.2016.26.4.419. [Google Scholar] [CrossRef]
50. Appiano M, Pavan S, Catalano D, Zheng Z, Bracuto V, Lotti C, et al. Identification of candidate MLO powdery mildew susceptibility genes in cultivated Solanaceae and functional characterization of tobacco NtMLO1. Transgenic Res. 2015;24(5):847–58. doi:10.1007/s11248-015-9878-4. [Google Scholar] [CrossRef]
51. Qiu X, Wang Q, Zhang H, Jian H, Zhou N, Ji C, et al. Antisense RhMLO1 gene transformation enhances resistance to the powdery mildew pathogen in Rosa multiflora. Plant Mol Biol Report. 2015;33(6):1659–65. doi:10.1007/s11105-015-0862-1. [Google Scholar] [CrossRef]
52. Bracuto V, Appiano M, Ricciardi L, Göl D, Visser RG, Bai Y, et al. Functional characterization of the powdery mildew susceptibility gene SmMLO1 in eggplant (Solanum melongena L.). Transgenic Res. 2017;26(3):323–30. doi:10.1007/s11248-016-0007-9. [Google Scholar] [PubMed] [CrossRef]
53. Acevedo-Garcia J, Spencer D, Thieron H, Reinstädler A, Hammond-Kosack K, Phillips AL, et al. mlo-based powdery mildew resistance in hexaploid bread wheat generated by a non-transgenic TILLING approach. Plant Biotechnol J. 2017;15(3):367–78. doi:10.1111/pbi.12631. [Google Scholar] [CrossRef]
54. Dohm JC, Minoche AE, Holtgräwe D, Capella-Gutiérrez S, Zakrzewski F, Tafer H, et al. The genome of the recently domesticated crop plant sugar beet (Beta vulgaris). Nature. 2014;505(7484):546–9. doi:10.1038/nature12817. [Google Scholar] [PubMed] [CrossRef]
55. Zhou SJ, Jing Z, Shi JL. Genome-wide identification, characterization, and expression analysis of the MLO gene family in Cucumis sativus. Genet Mol Res. 2013;12(4):6565–78. doi:10.4238/2013.december.11.8. [Google Scholar] [CrossRef]
56. Mushtaq N, Munir F, Gul A, Amir R, Paracha RZ. Genome-wide analysis, identification, evolution and genomic organization of dehydration responsive element-binding (DREB) gene family in Solanum tuberosum. PeerJ. 2021;9(2):e11647. doi:10.7717/peerj.11647. [Google Scholar] [PubMed] [CrossRef]
57. Ryu H, Cho YG. Plant hormones in salt stress tolerance. J Plant Biol. 2015;58(3):147–55. doi:10.1007/s12374-015-0103-z. [Google Scholar] [CrossRef]
58. Fonseca S, Chico JM, Solano R. The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol. 2009;12(5):539–47. doi:10.1016/j.pbi.2009.07.013. [Google Scholar] [PubMed] [CrossRef]
59. Ding P, Ding Y. Stories of salicylic acid: a plant defense hormone. Trends Plant Sci. 2020;25(6):549–65. doi:10.1016/j.tplants.2020.01.004. [Google Scholar] [PubMed] [CrossRef]
60. Piffanelli P, Zhou F, Casais C, Orme J, Jarosch B, Schaffrath U, et al. The barley MLO modulator of defense and cell death is responsive to biotic and abiotic stress stimuli. Plant Physiol. 2002;129(3):1076–85. doi:10.1104/pp.010954. [Google Scholar] [PubMed] [CrossRef]
61. Niu X, Bressan RA, Hasegawa PM, Pardo JM. Ion homeostasis in NaCl stress environments. Plant Physiol. 1995;109(3):735–42. doi:10.1104/pp.109.3.735. [Google Scholar] [PubMed] [CrossRef]
62. Ramoliya PJ, Patel HM, Pandey AN. Effect of salinization of soil on growth and macro- and micro-nutrient accumulation in seedlings of Salvadora persica (Savadoraceae). For Ecol Manag. 2004;202(1–3):181–93. doi:10.1016/j.foreco.2004.07.020. [Google Scholar] [CrossRef]
63. Patel TK, Williamson JD. Mannitol in plants, fungi, and plant-fungal interactions. Trends Plant Sci. 2016;21(6):486–97. doi:10.1016/j.tplants.2016.01.006. [Google Scholar] [PubMed] [CrossRef]
64. Wang X, Ma Q, Dou L, Liu Z, Peng R, Yu S. Genome-wide characterization and comparative analysis of the MLO gene family in cotton. Plant Physiol Biochem. 2016;103:106–19. doi:10.1016/j.plaphy.2016.02.031. [Google Scholar] [PubMed] [CrossRef]
65. Liu PL, Huang Y, Shi PH, Yu M, Xie JB, Xie L. Duplication and diversification of lectin receptor-like kinases (LecRLK) genes in soybean. Sci Rep. 2018;8(1):5861. doi:10.1038/s41598-018-24266-6. [Google Scholar] [PubMed] [CrossRef]
66. Lim CW, Lee SC. Functional roles of the pepper MLO protein gene, CaMLO2, in abscisic acid signaling and drought sensitivity. Plant Mol Biol. 2014;85(1–2):1–10. doi:10.1007/s11103-013-0155-8. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools