 Open Access
Open Access
ARTICLE
Molecular Cloning, Subcellular Localization and Expression Analyses of PdbHLH57 Transcription Factor in Colored-Leaf Poplar
1 Jiangsu Key Laboratory for the Research and Utilization of Plant Resources, Institute of Botany, Jiangsu Province and Chinese Academy of Sciences (Nanjing Botanical Garden Mem. Sun Yat-Sen), Nanjing, 210014, China
2 College of Life Sciences, Nanjing Agricultural University, Nanjing, 210095, China
3 College of Horticulture, Nanjing Agricultural University, Nanjing, 210095, China
* Corresponding Authors: Weibing Zhuang. Email: ; Zhong Wang. Email:
Phyton-International Journal of Experimental Botany 2025, 94(4), 1211-1223. https://doi.org/10.32604/phyton.2025.063647
Received 20 January 2025; Accepted 13 March 2025; Issue published 30 April 2025
Abstract
bHLH transcription factors, widely exist in various plants, and are vital for the growth and development of these plants. Among them, many have been implicated in anthocyanin biosynthesis across various plants. In the present study, a PdbHLH57 gene, belonging to the bHLH IIIf group, was characterized, which was isolated and cloned from the colored-leaf poplar ‘Zhongshancaiyun’ (ZSCY). The cDNA sequence of PdbHLH57 was 1887 base pairs, and the protein encoded by PdbHLH57 had 628 amino acids, the isoelectric point and molecular weight of which were 6.26 and 69.75 kDa, respectively. Through bioinformatics analysis, PdbHLH57 has been classified into the IIIf bHLH subgroup, with many members of this subgroup known to participate in anthocyanin biosynthesis. The subcellular localization analysis conducted in the leaf protoplasts of ‘ZSCY’ revealed that the PdbHLH57 protein is specifically localized in the nucleus. The transcription activation analysis was also conducted, and the results showed that the PdbHLH57 protein had self-transcription activation. To better explore the functions of the PdbHLH57 protein, two parts of this protein (PdbHLH57-1, PdbHLH57-2) were split to detect their transcriptional activation activity. The results indicated that PdbHLH57-1 (1-433aa) had self-transcription activation, and PdbHLH57-2 (433-628aa) had no transcription activation. The expression of PdbHLH57 peaked in June during different developmental stages in ‘ZSCY’, and it was most highly expressed in the phloem among various tissues. These findings offer a basis for understanding the role of PdbHLH57 in colored-leaf poplar.Keywords
Supplementary Material
Supplementary Material FileColored-leaf plants have emerged as significant components of modern landscapes due to their tripartite value in socio-economic development, environmental sustainability, and aesthetic enhancement. Colored-leaf poplars, as part of this group, offer numerous advantages such as superior quality, strong adaptability, and high disease resistance, making them widely utilized for courtyard decoration and roadside greenery [1–3]. Recently, several varieties of colored-leaf poplars have been developed, including ‘Quanhong Poplar’ (QHP), ‘Jinhong Poplar’ (JHP), ‘Zhongshancaiyun’ (ZSCY), ‘Caihong Poplar’ (CHP), and ‘Zhonghong Polar’ (ZHP) [4–6]. Among these colored-leaf poplars, ‘ZSCY’ has been widely used due to its rich and unique colors and longer ornamental period. However, the pathways of pigment formation in ‘ZSCY’ are not yet understood. Consequently, it is crucial to explore the molecular mechanisms of pigment formation in ‘ZSCY’.
Transcription factors (TFs) serve as master regulators of plant growth, development, and environmental adaptation, orchestrating processes ranging from signaling to secondary metabolism. Plant bHLH transcription factors are a large family of transcription factors characterized by a basic helix-loop-helix (bHLH) domain. This domain enables them to bind to specific DNA sequences and regulate the expression of target genes. Besides the bHLH domain, the overall consensus amino acid sequence between bHLH proteins is poorly conserved. This is partly because of the presence of additional domains necessary to modulate their activity and/or DNA-binding specificity, such as the leucine-zipper motif, the MYB-interacting region (MIR), or the aspartate kinase, chorismate mutase, and TyrA-like (ATC-like) domain [7,8]. Many bHLH transcription factors have been identified in various species, which play important roles in various aspects of plant growth and development, such as cell differentiation, development, and responses to environmental stresses. In Arabidopsis, more than 162 members of bHLH proteins have been identified, which were further clustered into 12 distinctive subfamilies [8]. Among them, IIIf subfamily bHLH members might be involved in flavonoid biosynthesis, such as AtbHLH0012/MYC1 and AtbHLH042/TT8 [9]. More and more bHLH TF associated with flavonoid synthesis have been identified [9,10]. Lc in maize was the first plant bHLH TF associated with flavonoid synthesis [11]. In the Asiatic hybrid lily (Lilium spp.), LhbHLH2 has been shown to play a role in the anthocyanin biosynthesis of lily buds when they suffer from light [12]. MdbHLH3, isolated from apple (Malus domestica Borkh.), promoted fruit coloration and the accumulation of anthocyanin when they were exposed to low temperatures [13]. CmbHLH2, clustered in the IIIf bHLH subgroup, could combine with the promoter of CmDFR to enhance the accumulation of anthocyanin when co-expressed with CmMYB6 in chrysanthemum (Chrysanthemum morifolium Ramat.) [14]. FcbHLH42, a gene isolated from fig (Ficus carica L.), was transiently expressed in tobacco, leading to a noticeable accumulation of anthocyanins [15]. AcB2, one of the IIIf bHLH subfamily members, could cooperate and physically interact with AcMYB1 to regulate the expression levels of structural genes associated with anthocyanin biosynthesis, including AcANS and AcF3H1, which increased the accumulation of anthocyanins in onion (Allium cepa L.) [16]. However, fewer bHLH TFs related to the accumulation of anthocyanins have been discovered in poplar. PdTT8 could significantly enhance the expression level of anthocyanin biosynthesis genes, which were isolated from Populus deltoids [17]. In poplar (Populus trichocarpa), bHLH131 could form the MBW complex with another two TF (PtrMYB57 and PtrTTG1), which could bind to the promoter of flavonoid genes, such as ANS1, DFR1, CHS4, and 4CL5, thereby inhibiting the biosynthesis of anthocyanin [18]. In the future, more bHLH TFs related to the biosynthesis of anthocyanin need to be identified.
In our previous study, the anthocyanin contents between green-leaf poplar and colored-leaf poplar were determined, and the genome-wide identification and characterization of PdbHLH transcription factors in colored-leaf poplar were also conducted. Combined with the transcriptome analysis between green-leaf poplar and colored-leaf poplar, PdbHLH57 from subgroup III(f) might be involved in anthocyanin biosynthesis [19]. However, the functional and structural characterization of this gene remains unclear. In this experiment, PdbHLH57 was isolated from the leaves of ‘ZSCY’, and structural and functional analysis was conducted to obtain the basic information of PdbHLH57. The transcriptional activation assay and subcellular localization analysis were also conducted. Additionally, the expression pattern of PdbHLH57 was evaluated in poplar leaves at 2-month intervals from 15 April to 15 October 2023. The spatial expression pattern of PdbHLH57 in ‘ZSCY’ was also explored, including immature leaves, mature leaves, petioles, phloem, xylem, and roots. In addition, anthocyanin contents of ‘ZSCY’ leaves at different developmental stages were measured. The results acquired in the present study could establish some foundations to better analyze functions of PdbHLH57 in poplar.
The deciduous colored-leaf poplar cultivar ‘ZSCY’ used in the present study was cultivated at Nanjing Botanical Garden Mem. Sun Yat-Sen. The leaf color of ‘ZSCY’ is dark red (RHS 47B) in spring, while the leaf color of the upper leaves was RHS 67B; the middle and lower leaves were RHS 189C and RHS N189B in summer. Field management practices such as irrigation and fertilization were conducted according to common practices. Leaves of ‘ZSCY’ were collected at 2-month intervals from 15 April to 15 October 2023. Specific tissues (young leaves, mature leaves, petioles, phloem, xylem, and roots) were harvested on 15 June 2024. The experiments were conducted with three biological replicates, each consisting of three technical replicates. Collected samples were immediately frozen in liquid nitrogen and stored at −80°C for subsequent analyses.
2.2 Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from ‘ZSCY’ leaves using a commercial RNA extraction kit. RNA quality was assessed by 1% agarose gel electrophoresis, with quantification performed using a NanoDrop 2000 spectrophotometer. First-strand cDNA was synthesized using the PrimeScript™ RT Reagent Kit (RR047A, Dalian, China) following manufacturer’s protocols.
2.3 Identification and Cloning of PdbHLH57
The cDNA template from ‘ZSCY’ leaves was used to obtain the sequence of the PdbHLH57 gene with specific primers (Table S1). The reference sequences of PdbHLH57 were acquired from the database of Phytozome, and SnapGene software was used to design primers. Polymerase chain reaction was used to amplify the ORF sequence of PdbHLH57 in ‘ZSCY’ leaves. The products obtained were subsequently purified and subcloned into a vector with different kits (FastPure Gel DNA Extraction Mini Kit and pTOPO001 Blunt Simple Vector). Successfully converted monoclonal antibodies were used to be sequenced by GeneralBiol (Anhui, China).
2.4 Gene Structure and Bioinformatics Analysis of PdbHLH57
The online tool ProtParam was used to calculate the theoretical isoelectric point and molecular weight. The subcellular localization of PdbHLH57 was predicted with the online tool CELLO. The sequence alignments between PdbHLH57 protein and other bHLH proteins were conducted with the software DNAMAN, and these sequences can be downloaded from the database of Arabidopsis Information Resource (TAIR) and National Center for Biotechnology Information (NCBI). With neighbor-joining (NJ) method in MEGA11 software, phylogenetic trees were built using 1000 bootstrap replicates. The prediction of transmembrane domain in PdbHLH57 protein was carried out by the online tool TMHMM. The online tool STRING was used to predict the interaction network of candidate genes with option value >0.400. The online tools PSIPRED and Phyre2 were used to predict secondary structure and tertiary structure of PdbHLH57.
2.5 Subcellular Localization of PdbHLH57
The coding sequences of PdbHLH57 were cloned into pAN580 vector with specific primers (Table S1) to obtain PdbHLH57-GFP construct, which was driven by the double 35S promoter, and included a GFP tag to detect its subcellular localization. Protoplast preparation and transformation were carried out by a previously established method with some modification [20]. The protoplasts used for introducing the PdbHLH57-GFP construct were generated from tender leaves of ‘ZSCY’. Before examination, the protoplasts used for introducing the PdbHLH57-GFP construct were incubated for 16 h at 28°C. The confocal laser scanning microscope (LSM980, Carl Zeiss, Oberkochen, Germany) was used to determine the subcellular localization of PdbHLH57, and the emission wavelength and excitation wavelength used to record their GFP signals were 509 and 488 nm, respectively.
2.6 Transcriptional Activation Assay of PdbHLH57
According to the manual instructions, Matchmaker® Gold Yeast Two-Hybrid System was used to determine the transcriptional activity of PdbHLH57 (Clontech, Mountain View, CA, USA). The different parts of PdbHLH57 sequences (1–628 aa, PdbHLH57; 1–425 aa, PdbHLH57-1;426–628 aa, PdbHLH57- 2) were independently fused in the pGBKT7 vector with a GAL4 DNA-binding region, and these fused pGBKT7 vectors were introduced into the yeast strain Y2H. In addition, the co-transformation between pGADT7-T and pGBKT7-Lam served as negative control, while the co-transformation between pGADT7-T and pGBKT7-53 served as positive control. The transformants were incubated for 3–5 days at 30°C, and the activity of transcriptional activation was evaluated on the selective solid medium plate SD/−Trp, supplemented with 200 ng/mL AbA. The presence of blue colonies on the selective solid medium plate indicated the candidate proteins had a self-transactivation activity in yeast cells.
2.7 Anthocyanin Content Measurement
The contents of ‘ZSCY’ leaves anthocyanin were determined with a previous method with some modification [6]. The fresh leaves (about 1.0 g) were placed into 10 mL ethanol mixture with 1% HCl (v/v), which was maintained 30 min at 60°C. The supernatant was collected after centrifuging the mixture for 5 min at 13,000× g, and the absorbance of the supernatant was measured using a spectrophotometer at 650, 620, and 530 nm. The contents of anthocyanin were calculated using the formula [(A530 − A620 − 0.1 × (A650 − A620)) × 2500/0.462] per gram of fresh weight, and the unit of anthocyanin content was μg/g fresh weight.
2.8 Tissue Specificity and Spatial Specificity of Expression
To better understand the function of PdbHLH57, the expression level of PdbHLH57 at different growth stages and in various tissues of ‘ZSCY’ including young leaves, mature leaves, petioles, phloem, xylem and roots were further evaluated. The procedure and data analysis of qTR-PCR were conducted by the previous method [21]. Primers used in the study were listed in Table S1, and Actin2, EF1 and UBQ genes were used as reference genes [22].
3.1 Cloning of PdbHLH57 in ‘ZSCY’ Leaves and Sequence Analysis
The sequences of PdbHLH57 were obtained in the leaves of ‘ZSCY’ with the specific primers (Table S1). An expected band was amplified by RT-PCR using the cDNA of ‘ZSCY’ leaves, which was 1887 bp in length encoded 628 amino acids (Fig. S1, Table S3). According to the bioinformatics analysis, the PdbHLH57 protein had an Mw of 69.75 kDa, and the pI was 6.26. In addition, the PdbHLH57 protein also contained a typical bHLH domain (Fig. 1a).

Figure 1: Protein sequence alignment and phylogenetic analysis between PdbHLH57 in ‘ZSCY’ and other bHLH proteins from other plants. The other bHLH proteins are as follows: Arabidopsis thaliana AtTT8 (Q9FT81), AtGL3 (NP_680372), AtEGL3 (Q9CAD0), AtMYC1 (BAA11933), AtbHLH061 (AAM10950), AtbHLH116 (AAL84972), AtDYT1 (O81900), AtbHLH021 (NP_179283), AtbHLH027 (AAS79544), AtbHLH035 (NP_974948), AtJAM2 (Q9LNJ5), AtbHLH017 (AAM19778), AtMYC2 (Q39204), and AtbHLH028 (AAL55721); Malus domestica MdbHLH33 (ABB84474) and MdbHLH3 (ADL36597); Vitis vinifera VvMYCA1 (ABM92332) and VvMYC1 (ACC68685); Medicago truncatula MtTT8 (KM892777); Oryza rufipogon OsRc (ABB17166); Petunia hybrida PhAN1 (AAG25928) and PhJAF13 (AAC39455); Lilium hybrid LhbHLH2 (BAE20058); Antirrhinum majus AmDEL (AAA32663.1); Ipomoea nil InDEL(BAE94393.1); Dahlia pinnata DvIVS (BAJ33516.1); Ipomoea tricolor ItbHLH2 (BAD18984.1); Allium cepa AcB2 (AUG71567); and Zea mays ZmR (P13027)
A neighbor-joining phylogenetic tree between PdbHLH57 and other bHLH proteins from other plants was constructed using MEGA 11 software. The results showed that there was a close relationship between PdbHLH57 and AtGL3, AtEGL3, which belonged to the subgroup bHLH IIIf family involved with anthocyanin biosynthesis (Fig. 1b).
The secondary structure of the PdbHLH57 protein was further predicted using PSIPRED software (Fig. 2a). The results indicated that PdbHLH57 protein included 52.55% random coil, 37.42% alpha-helix, and 10.03% extended strand, with the highest number of random coil secondary structures. To better understand the functions of the PdbHLH57 protein, the tertiary structure was predicted with Phyre2 software (Fig. 2b). Through homology comparison analysis, the crystal structure of transcription factor egl3 was used as a template. 256 residues could be modeled at >90% confidence using multiple templates, indicating that the simulated tertiary structure of PdbHLH57 protein can be reliably predicted.
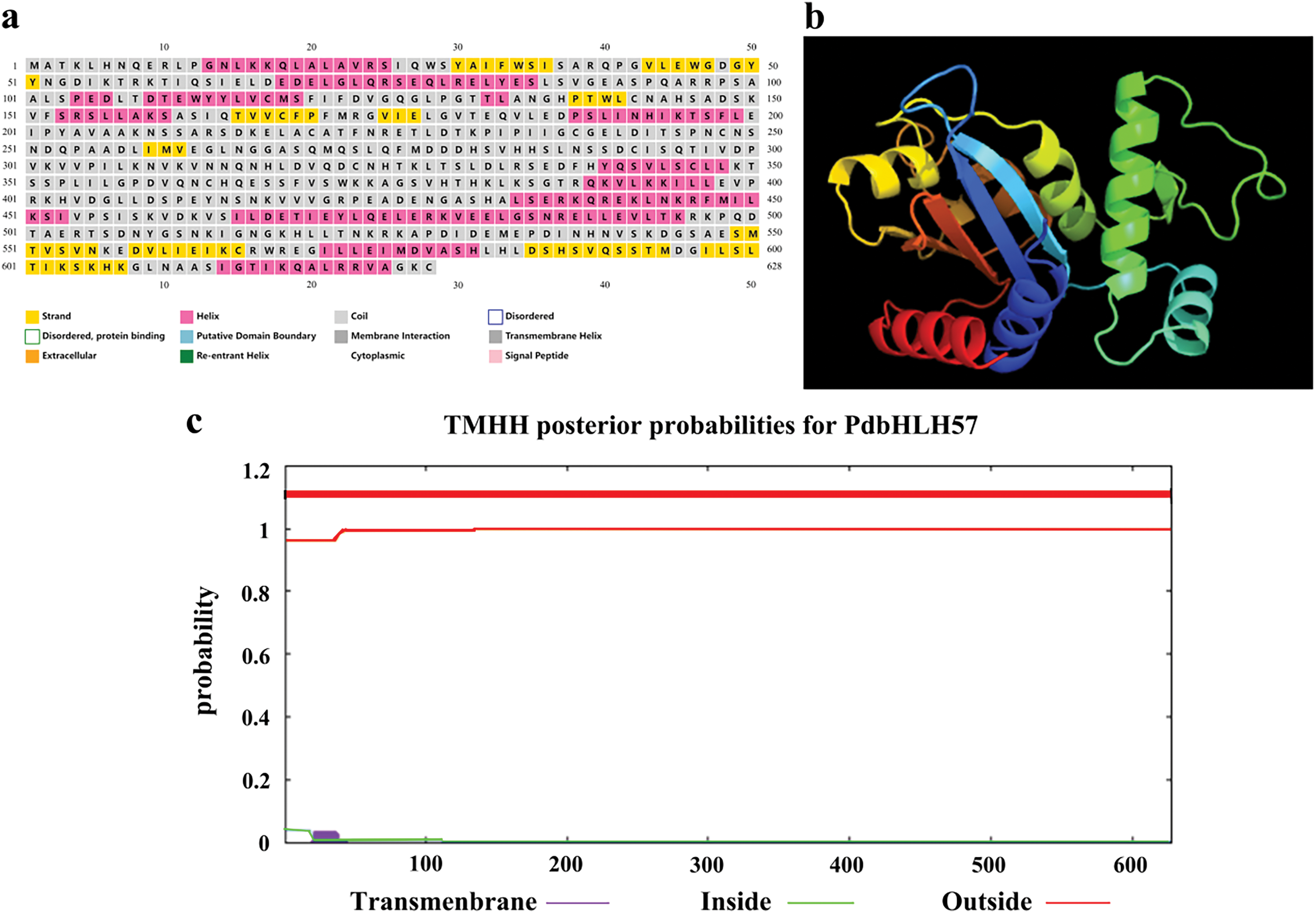
Figure 2: The prediction of the structure for PdbHLH57. (a) The prediction of secondary structure. (b) The prediction of tertiary structure. (c) Prediction of transmembrane structure in protein PdbHLH57
Furthermore, the transmembrane helices in protein PdbHLH57 were also predicted by TMHMM (Fig. 2c). The results showed that the first 60 AAs Exp number, AAs in TMHs Exp number, the predicted TMHs number is 0, 0.71, and 0.63, respectively, indicating that PdbHLH57 does not contain a transmembrane domain.
The prediction of the interaction network associated with PdbHLH57 was conducted with STRING. In the present study, PtrGL3 from Populus trichocarpa, homologous to PdbHLH57, was used to be a key node to predict the interaction network between PdbHLH57 and the corresponding proteins (Fig. S2). PdbHLH57 was predicted to form an interaction network with multiple proteins, including MYB-LIKE, WD-REPEAT, and bHLH92 (Table S2). Functional annotations of the PdbHLH57 were inferred from the known biological functions of proteins, providing some references for the prediction of potential functions of PdbHLH57 in color-leaf poplar.
3.2 Subcellular Localization Analysis and Transcriptional Activation Detection for PdbHLH57
To figure out the subcellular localization of the PdbHLH57 protein, the CELLO online tools were first used to predict. When PdbHLH57 protein is localized in the nucleus, the predicted score was 3.717, much higher than that for other locations (<0.8), indicating that PdbHLH57 is localized in the nucleus. To further verify the subcellular localization of PdbHLH57 protein through experiment, the fused vector containing green fluorescent protein and coding sequence of PdbHLH57 was constructed and transiently expressed in ‘ZSCY’ protoplasts to confirm its subcellular localization. The PAN580-GFP was used as a control. There was a ubiquitous distribution for the fluorescence signal of 35S:GFP protein, whereas there was a specific distribution localized to the cell nucleus for the fluorescence signal of PAN580-PdbHLH57-GFP (Fig. 3), which further indicated that PdbHLH57 was a nuclear protein.
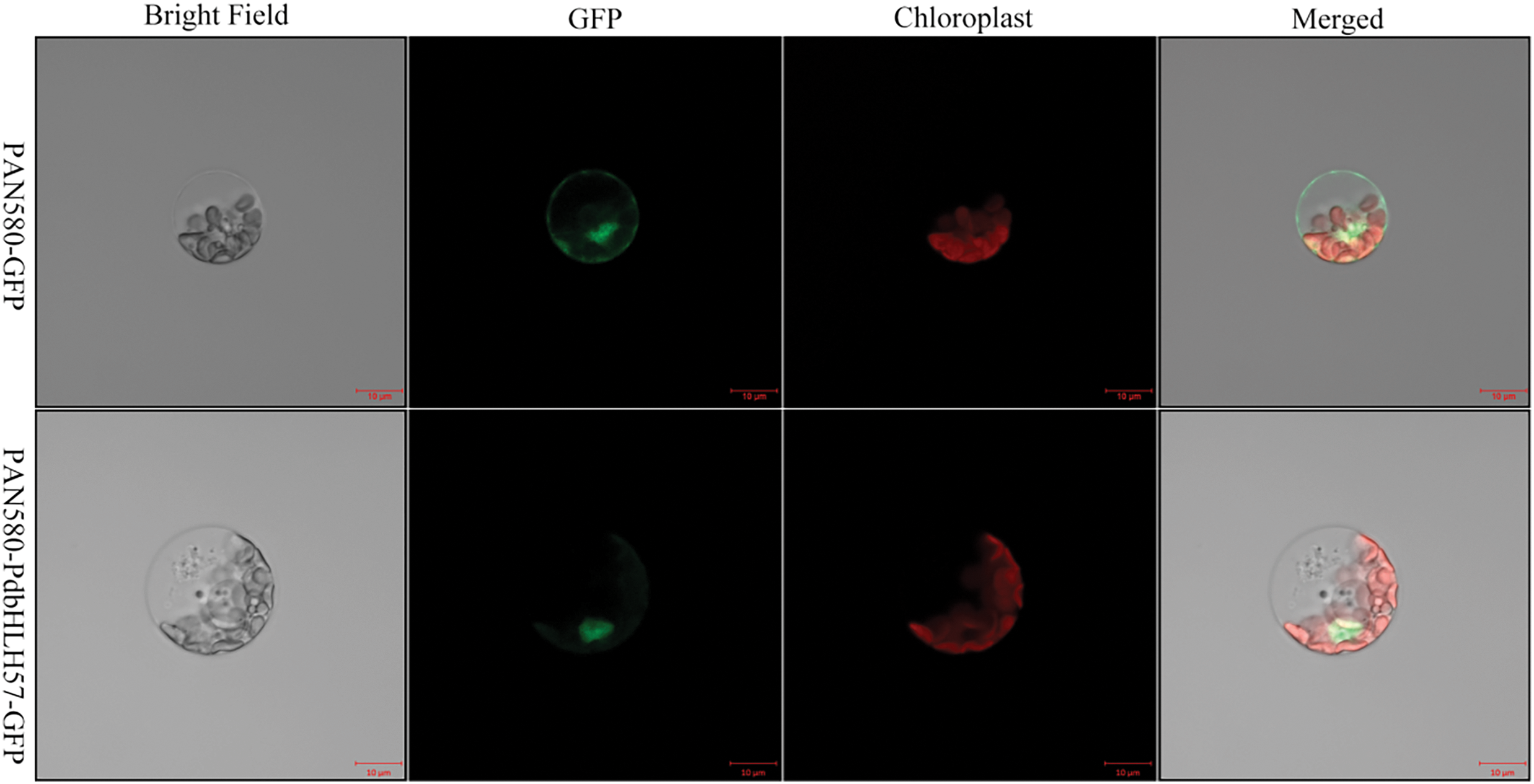
Figure 3: Subcellular localization of PdbHLH57 protein. The vector PAN580-PdbHLH57-GFP or PAN580-GFP was incubated in the protoplasts of the ‘ZSCY’ leaf. The red and green fluorescence were excited and visualized with 488 and 561 nm lasers, respectively
To better explore the functions of PdbHLH57, the transactivation activity of PdbHLH57 was detected in Y2H yeast cells with Y2H assays (Fig. 4). Compared with the negative and positive control, the transformed yeast cells containing pGBKT7-PdbHLH57 can grow well under the selection medium, indicating that PdbHLH57 exhibited self-transactivation activity (Fig. 4b). Two truncated variants were divided to further explore which part of PdbHLH57 had transcriptional transactivation (Fig. 4a). Compared with the negative and positive control, the transformed yeast cells containing pGBKT7-PdbHLH57-1 can grow well under selection medium, and the yeast cells containing pGBKT7-PdbHLH57-2 cannot grow well under selection medium, indicating that PdbHLH57-1 had a self-transactivation activity, and PdbHLH57-2 can be used to construct the yeast two-hybrid cDNA library to further screen the proteins interacted with PdbHLH57 (Fig. 4c).

Figure 4: Self-activation test of PdbHLH57 protein in yeast. PdbHLH57-1(1-432aa). PdbHLH57-2 (433-628aa). pGADT7-largeT/pGBKT7-p53 (Positive control); pGADT7-largeT/pGBKT7-laminC (Negative control); SD-ALWH: -ade, -his, -leu, -trp; SD-LW: -leu, -trp
3.3 Expression Analysis of PdbHLH57 among Different Development Stages in the Leaves of ‘ZSCY’
To better explore the roles of PdbHLH57 in the leaves of ‘ZSCY’, the expression levels of PdbHLH57 among different development stages were evaluated by qRT-PCR analysis (Fig. 5a). The expression level of PdbHLH57 increased significantly from April to June, and decreased significantly continuously from June to October, indicating that PdbHLH57 may play an important role at different developmental stages. As PdbHLH57 might regulate anthocyanin biosynthesis (Fig. 1), the total anthocyanin contents of leaves from ‘ZSCY’ leaves at four different development stages were also conducted. The total anthocyanin content decreased significantly from April to June, increased significantly from June to August, and decreased significantly from August to October (Fig. 5b). The above results indicated that PdbHLH57 might partially participate in the anthocyanin biosynthesis.
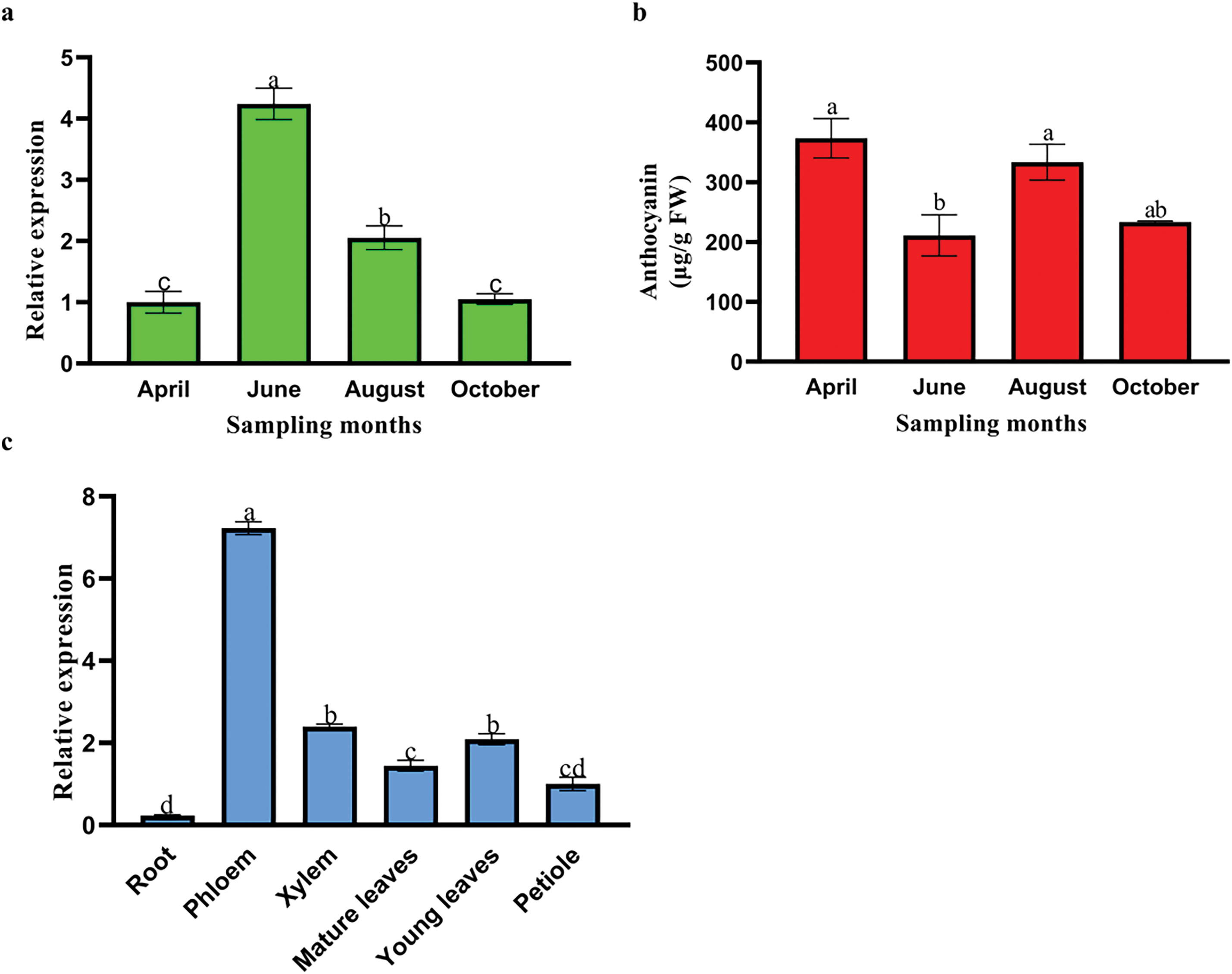
Figure 5: The expression levels of PdbHLH57 among different development stages (a). The total anthocyanin contents of leaves from ‘ZSCY’ leaves at four different development stages (b). Spatial expression pattern of PdbHLH57 in ‘ZSCY’ (c). The expression levels of PdbHLH57 in different organs: young leaves, mature leaves, petioles, phloem, xylem, and roots. Different letters show significant differences with Duncan tests (p < 0.05), and error bars represent standard deviations
3.4 Spatial Expression Pattern of PdbHLH57 in ‘ZSCY’
The function of genes is usually reflected through their expression patterns. To characterize the spatial expression pattern of PdbHLH57 in ‘ZSCY’, the expression level of PdbHLH57 was evaluated in different tissues, including xylem, mature leaves, young leaves, petioles, phloem, and roots (Fig. 5c). The expression of PdbHLH57 can be detected among all the tissues of ‘ZSCY’ with different transcription levels. For example, there was a highest expression level for PdbHLH57 in phloem, and a lowest expression level in roots, which has a 31.12 times difference in the expression level of PdbHLH57 between phloem and roots. While no significant difference was observed among young leaves, mature leaves, and xylem for the expression level of PdbHLH57 in ‘ZSCY’. The above results showed that PdbHLH57 may exert various roles in different tissues of ‘ZSCY’.
The bHLH family genes are involved in plant growth and development, which include the biosynthesis of anthocyanin. Although some researchers have reported the influences of bHLH genes on anthocyanin biosynthesis, our understanding of this topic in poplar remains limited. In present results, PdbHLH57, closely related to AtGL3, and AtEGL3, belongs to the bHLH IIIf subfamily (Fig. 1). In previous studies, most anthocyanin biosynthesis-related bHLH transcription factors belong to the bHLH IIIf subfamily. PhJAF13, AtEGL3, and AtGL3 have also been reported to participate in the biosynthesis of anthocyanin [23–25]. MdbHLH3, isolated from apple (Malus domestica Borkh.), promoted the accumulation of anthocyanin and the coloration of fruit under low temperatures [13]. MdbHLH33 could reduce the inhibitory effect of MdMYB16 on the biosynthesis of anthocyanin by interacting with MdMYB16 [26]. In grapevine (Vitis vinifera), VvMYC1 physically interacted with MYBPA1, MYBA1/A2, MYB5b, and MYB5a, could regulate gene expression, and enhance their proanthocyanidin and/or anthocyanin accumulation [27]. Combined with our results, PdbHLH57 might play an important role in anthocyanin synthesis.
To explore the functions of transcription factors, it is essential to verify whether a transcription factor has self-activation activity or not. Some bHLH transcription factors have been reported to show self-activation activity. For example, in tomato, there was a self-activation activity for SlbHLH96, and no self-activation activity for C-terminal segments of SlbHLH96 (200–441 bp), which include the conserved bHLH domain [28]. However, other transcription factors do not have self-activation activity. CmbHLH2 from Chrysanthemums (Chrysanthemum morifolium Ramat.) and AcB2 from onion (Allium cepa L.) showed no self-activation activity [14,16]. In our results, there was a self-activation activity for PdbHLH57, and no self-activation activity for C-terminal segments of PdbHLH57 (PdbHLH57-2, 426–628) with conserved bHLH domain, which can be used for its functional identification.
Various spatial expression patterns occurred for bHLH genes in different plant species. In Solanum lycopersicum, SlbHLH041 was expressed at a higher level in leaves than in other organs [29]. The expression of GhPAS1 in Gossypium spp. occurred in various tissues, with a preference for leaves [30]. In eggplant (Solanum melongena L.), bHLH genes exhibited diverse spatial expression patterns. SmbHLH1 was expressed highly in the flowers, petioles, leaves, and peels of purple-peel eggplant, but lower in green and white tissues. Conversely, SmbHLH117 was more highly expressed in green tissues like stems and leaves, but less in flowers and peels [31]. In Prunus mume, several bHLH Ib (2) subfamily genes had higher expression levels in roots, while PmbHLH58 and PmbHLH33 were expressed much higher in stems and leaves, respectively, indicating that the gene function in the same family gradually diverged during the evolutionary process [32]. In the present study, PdbHLH57 was expressed significantly higher in the phloem than in other tissues, suggesting that PdbHLH57 may regulate the growth and development of ‘ZSCY’ special in the phloem tissue.
Different bHLH genes have various expression patterns at different developmental stages in plants. For example, the contents of anthocyanin in Malus hupehensis increased from the flower developmental stage S1 to S2 and decreased from S2 to S3. The expression levels of some genes associated with anthocyanin synthesis (PAL, CHI, CHS, FLS) had a similar pattern from S1 to S3, while the expression level of MYB always increased from S1 to S3, which is inconsistent with the changes of anthocyanin contents [33]. In Chinese bayberry, the anthocyanin contents increased continuously at different fruit development (78, 82, and 86 days after full bloom). The expression level of MrbHLH1 increased from 78 days after full bloom to 82 days after full bloom, and decreased from 82 days after full bloom to 86 days after full bloom, while the expression level of MrMYB1 increased continuously at different fruit development [34]. In blueberries, anthocyanin contents continuously increased during fruit development stages, while the expression level of VcbHLH1-2 declined from the green to the red stage and increased from red to blue stages, which is inconsistent with the changes in anthocyanin contents [35]. LibHLH members of the same subfamily (XII subfamily) have different expression patterns in Lagerstroemia indica, such as LibHLH45, LibHLH50, and LibHLH62, indicating that LibHLH members have diverse expression patterns, and might play a role in the synthesis of anthocyanin [36]. In the present results, the expression pattern of PdbHLH57 in ‘ZSCY’ leaves was inconsistent with the accumulation of anthocyanin (Fig. 5), indicating that PdbHLH57 might partially regulate anthocyanin biosynthesis.
In the present study, a PdbHLH57 gene, belonging to the IIIf group of bHLH, was characterized, which was isolated and cloned from the colored-leaf poplar ‘ZSCY’. PdbHLH57 protein was localized to the nucleus in the leaf protoplasts of ‘ZSCY’ according to subcellular localization analysis. PdbHLH57-2 (433-628aa) lacked transcription activation, which can be utilized to construct a yeast two-hybrid cDNA library for further screening of proteins that interact with PdbHLH57. The expression levels of different developmental stages and spatial patterns of PdbHLH57 in ‘ZSCY’ were also evaluated, which indicated its potential characteristics. The findings of the present work provided references for further functional identification of PdbHLH57 in colored-leaf poplar.
Acknowledgement: Not applicable.
Funding Statement: This research was supported by the Natural Science Foundation of Jiangsu Province, China (BK20242007), the Natural Science Foundation of China (32271916), and the Jiangsu Agricultural Science and Technology Innovation Fund [CX(24)3048].
Author Contributions: The authors confirm their contributions to the paper as follows: study conception and design: Weibing Zhuang and Zhong Wang; data collection: Yuhang Li, Xiaochun Shu and Li Sun; analysis and interpretation of results: Yuhang Li, Tao Wang and Tengyue Yan; draft manuscript preparation: Yuhang Li; writing—review and editing: Zhihong Gao, Bingjun Yu, Weibing Zhuang and Zhong Wang. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: This article included all the data and materials.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2025.063647.
References
1. Zhang F, Wan XQ, Zheng YX, Sun LX, Chen QB, Guo YL, et al. Physiological and related anthocyanin biosynthesis genes responses induced by cadmium stress in a new colored-leaf plant “Quanhong Poplar”. Agrofor Syst. 2014;88(2):343–55. doi:10.1007/s10457-014-9687-4. [Google Scholar] [CrossRef]
2. Chen MJ, Li H, Zhang W, Huang L, Zhu J. Transcriptomic analysis of the differences in leaf color formation during stage transitions in Populus × euramericana ‘Zhonghuahongye’. Agronomy. 2022;12(10):2396. doi:10.3390/agronomy12102396. [Google Scholar] [CrossRef]
3. Chen MJ, Chang CF, Li H, Huang L, Zhou ZS, Zhu J, et al. Metabolome analysis reveals flavonoid changes during the leaf color transition in Populus × euramericana ‘Zhonghuahongye’. Front Plant Sci. 2023;14:1162893. doi:10.3389/fpls.2023.1162893. [Google Scholar] [PubMed] [CrossRef]
4. Zhu YL, Wang N, Cheng XJ, Zhou CS. A new poplar red foliar variety ‘Quanhong’. Sci Silvae Sin. 2012;48(2):188. [Google Scholar]
5. Zhu YL, Cheng XJ. A new poplar red foliar variety ‘Zhonghong’. Sci Silvae Sin. 2008;44(10):173–4. [Google Scholar]
6. Zhuang WB, Shu XC, Lu XY, Wang T, Zhang FJ, Wang N, et al. Ornamental poplar ‘Zhongshancaiyun’. HortScience. 2021;56(10):1291–2. doi:10.21273/HORTSCI16016-21. [Google Scholar] [CrossRef]
7. Toledo-Ortiz G, Huq E, Quail PH. The Arabidopsis basic/helix-loop-helix transcription factor family. Plant Cell. 2003;15(8):1749–70. doi:10.1105/tpc.013839. [Google Scholar] [PubMed] [CrossRef]
8. Heim MA. The basic helix-loop-helix transcription factor family in plants: a genome-wide study of protein structure and functional diversity. Mol Biol Evol. 2003;20(5):735–47. doi:10.1093/molbev/msg088. [Google Scholar] [PubMed] [CrossRef]
9. Li M, Sun L, Gu H, Cheng D, Guo X, Chen R, et al. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in spine grapes (Vitis davidii). Sci Rep. 2021;11(1):6863. doi:10.1038/s41598-021-85754-w. [Google Scholar] [PubMed] [CrossRef]
10. Baudry A, Heim MA, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L. TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J. 2004;39(3):366–80. doi:10.1111/j.1365-313X.2004.02138.x. [Google Scholar] [PubMed] [CrossRef]
11. Ludwig SR, Habera LF, Dellaporta SL, Wessler S. Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc Natl Acad Sci. 1989;86(18):7092–96. doi:10.1073/pnas.86.18.7092. [Google Scholar] [PubMed] [CrossRef]
12. Nakatsuka A, Yamagishi M, Nakano M, Tasaki K, Kobayashi N. Light-induced expression of basic helix-loop-helix genes involved in anthocyanin biosynthesis in flowers and leaves of Asiatic hybrid lily. Sci Hortic. 2009;121(1):84–91. doi:10.1016/j.scienta.2009.01.008. [Google Scholar] [CrossRef]
13. Xie XB, Li S, Zhang RF, Zhao J, Chen YC, Zhao Q, et al. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Env. 2012;35(11):1884–97. doi:10.1111/j.1365-3040.2012.02523.x. [Google Scholar] [PubMed] [CrossRef]
14. Xiang LL, Liu XF, Li X, Yin XR, Grierson D, Li F, et al. A novel bHLH transcription factor involved in regulating anthocyanin biosynthesis in Chrysanthemums (Chrysanthemum morifolium Ramat.). PLoS One. 2015;10(11):e0143892. doi:10.1371/journal.pone.0143892. [Google Scholar] [PubMed] [CrossRef]
15. Song M, Wang H, Wang Z, Huang H, Chen S, Ma H. Genome-wide characterization and analysis of bHLH transcription factors related to anthocyanin biosynthesis in Fig (Ficus carica L.). Front Plant Sci. 2021;12:730692. doi:10.3389/fpls.2021.730692. [Google Scholar] [PubMed] [CrossRef]
16. Li X, Cao L, Jiao B, Yang H, Ma C, Liang Y. The bHLH transcription factor AcB2 regulates anthocyanin biosynthesis in onion (Allium cepa L.). Hortic Res. 2022;9:uhac128. doi:10.1093/hr/uhac128. [Google Scholar] [PubMed] [CrossRef]
17. Wang H, Wang X, Yu C, Wang C, Jin Y, Zhang H. MYB transcription factor PdMYB118 directly interacts with bHLH transcription factor PdTT8 to regulate wound-induced anthocyanin biosynthesis in poplar. BMC Plant Biol. 2020;20(1):173. doi:10.1186/s12870-020-02389-1. [Google Scholar] [PubMed] [CrossRef]
18. Wan S, Li C, Ma X, Luo K. PtrMYB57 contributes to the negative regulation of anthocyanin and proanthocyanidin biosynthesis in poplar. Plant Cell Rep. 2017;36(8):1263–76. doi:10.1007/s00299-017-2151-y. [Google Scholar] [PubMed] [CrossRef]
19. Wang XJ, Peng XQ, Shu XC, Li YH, Wang Z, Zhuang WB. Genome-wide identification and characterization of PdbHLH transcription factors related to anthocyanin biosynthesis in colored-leaf poplar (Populus deltoids). BMC Genomics. 2022;23(1):244. doi:10.1186/s12864-022-08460-5. [Google Scholar] [PubMed] [CrossRef]
20. Yang P, Sun Y, Sun X, Li Y, Wang L. Optimization of preparation and transformation of protoplasts from Populus simonii × P. nigra leaves and subcellular localization of the major latex protein 328 (MLP328). Plant Methods. 2024;20(1):3. doi:10.1186/s13007-023-01128-5. [Google Scholar] [PubMed] [CrossRef]
21. Zhuang WB, Wang HX, Liu TY, Wang T, Zhang FJ, Shu XC, et al. Integrated physiological and genomic analysis reveals struct ural variations and expression patterns of candidate genes for colored- and green-leaf poplar. Sci Rep. 2019;9(1):11150. doi:10.1038/s41598-019-47681-9. [Google Scholar] [PubMed] [CrossRef]
22. Wang S, Yao W, Wei H, Jiang T, Zhou B. Expression patterns of ERF genes underlying abiotic stresses in di-haploid Populus simonii × P. nigra. Sci World J. 2014;2014:1–10. [Google Scholar]
23. Tominaga-Wada R, Masakane A, Wada T. Effect of phosphate deficiency-induced anthocyanin accumulation on the expression of Solanum lycopersicum GLABRA3 (SlGL3) in tomato. Plant Signal Behav. 2018;13(6):e1477907. doi:10.1080/15592324.2018.1477907. [Google Scholar] [PubMed] [CrossRef]
24. Chiu LW, Li L. Characterization of the regulatory network of BoMYB2 in controlling anthocyanin biosynthesis in purple cauliflower. Planta. 2012;236(4):1153–64. doi:10.1007/s00425-012-1665-3. [Google Scholar] [PubMed] [CrossRef]
25. D’Amelia V, Aversano R, Batelli G, Caruso I, Castellano MM, Castro-Sanz AB, et al. High AN1 variability and interaction with basic helix-loop-helix co-factors related to anthocyanin biosynthesis in potato leaves. Plant J. 2014;80(3):527–40. doi:10.1111/tpj.12653. [Google Scholar] [PubMed] [CrossRef]
26. Xu H, Wang N, Liu J, Qu C, Wang Y, Jiang S, et al. The molecular mechanism underlying anthocyanin metabolism in apple using the MdMYB16 and MdbHLH33 genes. Plant Mol Biol. 2017;94(1–2):149–65. doi:10.1007/s11103-017-0601-0. [Google Scholar] [PubMed] [CrossRef]
27. Hichri I, Heppel SC, Pillet J, Leon C, Czemmel S, Delrot S, et al. The basic helix-loop-helix transcription factor MYC1 is involved in the regulation of the flavonoid biosynthesis pathway in grapevine. Mol Plant. 2010;3(3):509–23. doi:10.1093/mp/ssp118. [Google Scholar] [PubMed] [CrossRef]
28. Liang Y, Ma F, Li B, Guo C, Hu T, Zhang M, et al. A bHLH transcription factor, SlbHLH96, promotes drought tolerance in tomato. Hortic Res. 2022;9:uhac198. doi:10.1093/hr/uhac198. [Google Scholar] [PubMed] [CrossRef]
29. Sun H, Fan HJ, Ling HQ. Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics. 2015;16(1):9. doi:10.1186/s12864-014-1209-2. [Google Scholar] [PubMed] [CrossRef]
30. Wu H, Ren Z, Zheng L, Guo M, Yang J, Hou L, et al. The bHLH transcription factor GhPAS1 mediates BR signaling to regulate plant development and architecture in cotton. Crop J. 2021;9(5):1049–59. doi:10.1016/j.cj.2020.10.014. [Google Scholar] [CrossRef]
31. Tian S, Li L, Wei M, Yang F. Genome-wide analysis of basic helix-loop-helix superfamily members related to anthocyanin biosynthesis in eggplant (Solanum melongena L.). PeerJ. 2019;7:e7768. doi:10.7717/peerj.7768. [Google Scholar] [PubMed] [CrossRef]
32. Ding A, Ding A, Li P, Wang J, Cheng T, Bao F, et al. Genome-wide identification and low-temperature expression analysis of bHLH genes in Prunus mume. Front Genet. 2021;12:762135. doi:10.3389/fgene.2021.762135. [Google Scholar] [PubMed] [CrossRef]
33. Zhang Y, Liu F, Wang B, Wu H, Wu J, Liu J, et al. Identification, characterization and expression analysis of anthocyanin biosynthesis-related bHLH genes in Blueberry (Vaccinium corymbosum L.). Int J Mol Sci. 2021;22(24):13274. doi:10.3390/ijms222413274. [Google Scholar] [PubMed] [CrossRef]
34. Liu XF, Yin XR, Allan AC, Lin WK, Shi YN, Huang YJ, et al. The role of MrbHLH1 and MrMYB1 in regulating anthocyanin biosynthetic genes in tobacco and Chinese bayberry (Myrica rubra) during anthocyanin biosynthesis. Plant Cell Tissue Organ Cult. 2013;115(3):285–98. doi:10.1007/s11240-013-0361-8. [Google Scholar] [CrossRef]
35. Han M, Yang C, Zhou J, Zhu J, Meng J, Shen T, et al. Analysis of flavonoids and anthocyanin biosynthesis-related genes expression reveals the mechanism of petal color fading of Malus hupehensis (Rosaceae). Braz J Bot. 2020;43(1):81–9. doi:10.1007/s40415-020-00590-y. [Google Scholar] [CrossRef]
36. Yu M, Bai M, Chen M, Zhang G, zhao Y, Ma Q, et al. Identification of bHLH transcription factors and screening of anthocyanin-related genes in Lagerstroemia indica. Genetica. 2024;152(4–6):179–97. doi:10.1007/s10709-024-00215-2. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools