 Open Access
Open Access
REVIEW
Understanding the Molecular Mechanisms of Nitrogen Assimilation in C3 Plants under Abiotic Stress: A Mini Review
1 State Key Laboratory of Subtropical Silviculture & College of Environment and Resources, College of Carbon Neutrality, Zhejiang A&F University, Hangzhou, 311300, China
2 Guangxi Colleges and Universities Key Laboratory for Cultivation and Utilization of Subtropical Forest Plantation, College of Forestry, Guangxi University, Nanning, 530004, China
* Corresponding Authors: Izhar Ali. Email: ,
(This article belongs to the Special Issue: Metabolic Mechanisms of Plant Responses to Stress)
Phyton-International Journal of Experimental Botany 2025, 94(4), 1029-1045. https://doi.org/10.32604/phyton.2025.064608
Received 19 February 2025; Accepted 03 April 2025; Issue published 30 April 2025
Abstract
Nitrogen (N) assimilation is crucial for the growth and development of C3 plants, as it converts inorganic N into organic forms, important for protein synthesis, nucleic acids and other vital biomolecules. However, abiotic stressors such as drought, salinity, extreme temperatures and others significantly impact N uptake and utilization, thereby hindering plant growth and development. Recent advances in molecular biology have illuminated the complex networks that govern N assimilation under these stressful conditions, emphasizing the role of transcription factors, regulatory genes, and stress-responsive pathways. This review provides an integrated perspective on the latest research in nitrogen metabolism under abiotic stress, focusing on the intricate regulatory mechanisms involving gene expression, signaling pathways, and enzymes that modulate N uptake and assimilation. Specifically, it highlights the recent findings on how hormones, reactive oxygen species production, N metabolism and calcium signaling are regulated under stress conditions. In addition, recent advancements in genomics and transcriptomics have further clarified the dynamic regulation of genes linked to N absorption and other metabolic processes. Understanding these mechanisms is important for developing strategies to enhance the N use efficiency and stress tolerance in C3 crops, thereby promoting sustainable agriculture and food security. Future research should focus on exploring the genetic and molecular bases of N metabolism in relation to abiotic stress, with the ultimate goal of enhancing crop performance in challenging environments.Keywords
Nitrogen assimilation is one of the most vital processes required for plant growth and development. It, therefore, involves the process of assimilation of inorganic nitrogen, mainly nitrate (NO3−) and ammonium (NH4+) ions, into organic nitrogen forms required for the synthesis of proteins, nucleic acids and other biomolecules [1,2]. This process is important for plant growth and yield, notably for C3 plants such as wheat, rice and barley. Furthermore, C3 plant possess a specific photosynthetic pathway, in which Calvin cycle is used to assimilate CO2 into three C molecules of 3-phosphoglyceric acid [3]. This process defines the C3 photosynthetic pathway, which is directly linked to nitrogen metabolism. The Calvin cycle not only drives carbon fixation but also influences the overall nitrogen economy of the plant by integrating carbon and nitrogen metabolic pathways. The integration is essential for coordinating the synthesis of amino acids, proteins, and other nitrogen-containing compounds ultimately impacting plant growth, development and yield [4].
A number of abiotic stress conditions, such as drought, excessive salt, and severe temperatures, have a major impact on C3 plants ability to assimilate nitrogen. Water availability is reduced under drought stress, which lowers stomatal conductance and CO2 absorption. This upsets the carbon-to-nitrogen ratio and hinders nitrogen metabolism [5]. Nitrate reductase (NR) and glutamine synthetase (GS), two essential enzymes involved in nitrogen absorption that are crucial in the conversion of nitrate and ammonium to amino acids, are impacted by high salinity due to ionic disruption and osmotic stress [6]. By reducing the effectiveness of metabolic pathways, extreme temperatures might modify protein stability and enzyme activity, which further complicates the absorption of nitrogen [7].
Recent advancements in molecular biology and genomics have brought new insights into the regulation of nitrogen utilization under stress conditions. Research has demonstrated the existence of stress-responsive pathways and transcription factors that influence nitrogen metabolism in response to environmental changes [8,9]. Consequently, these findings are vital for the establishment of measures to boost the resilience of plants under different stress conditions. Moreover, knowledge of the interaction between nitrogen assimilation and stress response mechanisms will help in the breeding of crops that can efficiently use nitrogen, which is crucial in sustainable crop production and food security [10].
This review aims to summarize the latest research on the molecular mechanisms underlying nitrogen assimilation in C3 plants under abiotic stress. By examining recent research findings, we will delve into the complex connections between nitrogen metabolism, stress responses, and plant adaptation strategies. Furthermore, we will highlight areas where our current knowledge is lacking and suggest potential research avenues to advance our understanding of these processes.
2 Molecular Regulation of Nitrogen Assimilation in C3 Plants
Nitrogen assimilation is a fundamental physiological process in C3 plants that plays a pivotal role in the uptake of inorganic nitrogen, principally as nitrate NO3− and ammonium NH4+ ions, into the shoot and subsequently into other organs for the synthesis of nitrogen-containing compounds such as amino acids, proteins, nucleic acids, and chlorophyll [11]. Recent studies have provided new insights into the regulatory networks involved in nitrogen assimilation under both optimal and stress conditions. Advancements in genomics and transcriptomics have identified key nitrogen transporter genes (e.g., NRT1.1, NRT2.1, AMT1.1) and their regulation under abiotic stresses such as drought and salinity. In particular, the role of nitrogen transporter genes in the redistribution of nitrate from roots to shoots has been linked to improved nitrogen use efficiency (NUE) in challenging environments. Additionally, transcription factors like NLP7 and TCP20 have been found to modulate nitrate signaling and integration with other metabolic pathways.
Recent research has also shed light on the cross-talk between nitrogen metabolism and other signaling pathways, such as reactive oxygen species (ROS) and phytohormones like abscisic acid (ABA) and cytokinin, which influence nitrogen uptake and assimilation during stress. For instance, studies show that calcium signaling, through CNGC channels, plays a critical role in regulating nitrate transport and stress responses, thus contributing to the optimization of nitrogen metabolism under stress conditions. Understanding these molecular mechanisms enhances our comprehension of how C3 plants cope with environmental stressors and efficiently manage nitrogen, which is critical for plant health and productivity. These findings pave the way for developing strategies to improve nitrogen use efficiency and stress resilience in C3 crops, ultimately promoting sustainable agriculture.
2.1 Nitrate Uptake and Transport
Nitrate uptake in C3 plants initiates through specialized transport proteins known as nitrate transporters (NRTs), particularly the NRT1 and NRT2 families. These transporters are embedded on the plasma membranes of root cells, facilitate the transport of nitrate ions from the soil into the plant. NRT1.1 is of particular importance as it comprises a transport and a signaling function; it transports nitrates and also regulates its transport depending on the nitrogen status of the plant [3,10,11]. When soil nitrate levels are elevated, the expression of NRT1.1 increases, enhancing the plant’s nitrate absorption capability. NRTs are not merely passive transporters; they also modulate signaling pathways leading to transcriptional regulation of genes relevant to nitrogen assimilation. Upon nitrate uptake, NRTs trigger downstream signaling cascades involving Nitrate Responsive Transcription Factors (NRTFs), whose expression is regulated by multiple genes involved in nitrogen metabolism, including Nitrate Reductase (NR) and Glutamine Synthetase (GS) [12]. Nitrate active transport is energy-dependent, relying on the proton motive force generated by the plasma membrane H+-ATPase to maintain a favorable gradient for nitrate influx. The interplay of C3 plants’ intricate transport mechanisms allows them to effectively utilize available nitrate resources, essential for growth and productivity (Fig. 1).
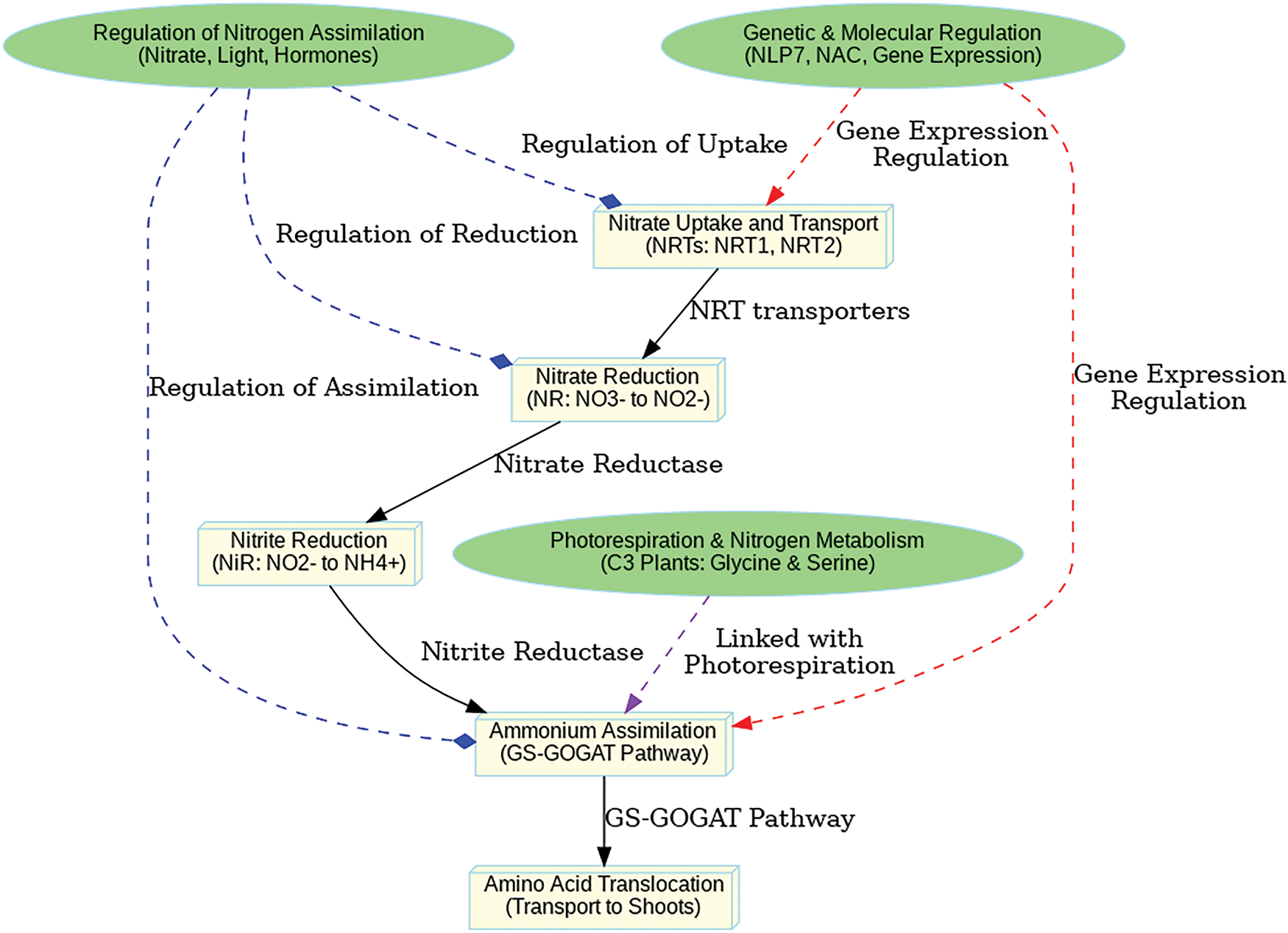
Figure 1: Nitrate is taken up by nitrate transporters (NRT1, NRT2) and reduced to nitrite by nitrate reductase (NR). Nitrite is further reduced to ammonium by nitrite reductase (NiR) in chloroplasts or plastids. Ammonium is assimilated into organic compounds through the GS-GOGAT pathway, producing glutamine and glutamate, which are then transported from roots to shoots for protein synthesis. Nitrogen assimilation is regulated by nitrate availability, light, hormones, and genetic factors like NLP7. Nitrogen metabolism is also linked to photorespiration, recycling ammonium through the production of glycine and serine
The reduction of nitrate to nitrite is a critical step after nitrate uptake, catalyzed by Nitrate Reductase (NR) in the cytoplasm, with NADH as the electron donor. The enzyme Nitrate Reductase controls nitrogen metabolism as a primary regulatory step while responding to external factors comprising environmental nitrate amounts together with light exposure levels and plant physiological state. Research demonstrates how protein modifications like phosphorylation and ubiquitination control NR activity so plants can quickly regulate their nitrogen assimilation through environmental changes [13,14]. After formation nitrite carries into plastids or chloroplasts for NiR to turn it into ammonium (NH4+). Nickel oxide players control this conversion process since nitrite acts as an intermediate that has toxic potential but needs efficient reduction for cellular protection. Nitrogen assimilation, including the conversion of nitrite to ammonium, utilizes electrons from ferredoxin, linking it to photosynthetic electron transport. Recent research highlights the chloroplast redox state as a key regulator of nitrogen assimilation, as redox conditions influence the efficiency of electron utilization in this process. Under high light conditions, nitrogen assimilation serves as an essential electron sink, contributing to redox balance and protecting photosynthetic efficiency [15]. Research findings show that abscisic acid (ABA), together with cytokinins controls NR and NiR activities alongside direct nitrate regulation of these processes. The hormones ABA along with cytokinins act as controllers of NR expression levels throughout drought stress because ABA reduces NR activity to save energy but cytokinins increase NR functionality for maintaining nitrogen processes [16]. The precise regulation of nitrate reduction by photosynthetic processes with hormonal control demonstrates how complicated nitrogen assimilation operates in C3 plants yet offers new aspects for maximizing crop nitrogen efficiency.
Excess nitrite can disrupt cellular homeostasis by inhibiting enzymatic processes and generating reactive nitrogen species, making its rapid reduction a critical step in nitrogen assimilation. Once inside the chloroplasts, Nitrite Reductase (NiR) catalyzes the conversion of nitrite to ammonium (NH4+), a process that requires electrons from ferredoxin, a key component of the photosynthetic electron transport chain. The ammonium produced is subsequently assimilated into organic molecules, primarily through the glutamine synthetase (GS) and glutamate synthase (GOGAT) cycle, forming essential amino acids and other nitrogenous compounds necessary for plant growth and development [17,18]. The proper regulation of nitrite levels by C3 plants serves to preserve nitrogen homeostasis because high levels damage metabolic functions and disrupt redox balance while producing oxidative stress, which deteriorates plant health and reduces productivity. The chloroplast’s redox status represents a vital regulatory mechanism that controls NiR activity by matching nitrite reduction to energy requirements and the metabolic progress of the plant system. ABSICIC ACID (ABA) and cytokinin hormonal signaling pathways control nitrite reduction activities that occur under various stress scenarios, such as nitrogen deficiency and drought conditions. Under unfavorable circumstances NiR activity decreases through ABA-mediated mechanisms, which preserves energy consumption, but cytokinins increase NiR activity when plants need to assimilate nitrogen throughout their active growth phase. Nitrate reduction shows essential interdependence with light reactions of photosynthesis, therefore demanding coordinated carbon and nitrogen metabolic control. NiR depends on ferredoxin made from light-dependent photosynthesis reactions thus any changes in light resources directly affect nitrogen assimilation rates [19,20]. The intricate regulatory network adjusts nitrogen metabolism through the plants metabolic needs and energy status to optimize nitrogen use efficiency for sustainable growth across diverse environmental contexts.
Ammonium assimilation primarily involves the Glutamine Synthetase–Glutamate Synthase (GS–GOGAT) pathway, where ammonium is incorporated into organic molecules. After the reduction of nitrite to ammonium, it is rapidly assimilated into amino acids such as glutamine and glutamate, which are essential for protein synthesis and other metabolic functions [21]. The GS enzyme catalyzes the ATP-dependent reaction of ammonium and glutamate to form glutamine, a key nitrogen donor in various metabolic pathways. Glutamate Synthase (GOGAT) converts glutamine and 2-oxoglutarate into two glutamate molecules, which are critical for further glutamine assimilation and amino acid production [22]. The enzymatic pathway functions significantly during nitrogen recycling procedures within senescing leaves and root nodules together with responses to nitrogen availability changes. Studies have identified multiple GS/GOGAT isoforms that show specific regulatory patterns with respect to abiotic stress situations including drought as well as salinity. The two enzyme forms GS1 (cytosolic) and GS2 (chloroplastic) operate independently because GS2 functions most efficiently in photosynthetic areas, yet GS1 conducts nitrogen remobilization processes throughout stress periods. Increased GS1 levels lead to better nitrogen use efficiency and stress adaptation in C3 crop plants. GS activity is regulated through the TOR (Target of Rapamycin) signaling pathway, which integrates nitrogen availability data with growth and metabolic modulation mechanisms. Under diverse environmental conditions the C/N ratio functionally controls GS and GOGAT expression to enable efficient ammonium assimilation. Furthermore, ammonium transporters (AMTs) play a pivotal role in the uptake and intracellular distribution of ammonium, influencing GS–GOGAT activity. Studies on AMT1;1 and AMT1;2 have shown their significance in ammonium homeostasis, particularly under high-nitrogen or stress conditions. The enhanced activity of both GS and GOGAT in response to environmental variations highlights the significance of ammonium assimilation in maintaining plant health and productivity. Understanding these regulatory mechanisms provides new avenues for improving nitrogen use efficiency and developing stress-resilient C3 crops, thereby promoting sustainable agriculture.
2.5 Translocation of Amino Acids
After nitrogen assimilation, the transport of amino acids from root to shoot is essential in C3 plants. Glutamine and asparagine, synthesized in the roots, support protein synthesis and serve as nitrogen sources for various metabolic processes [7]. Specialized amino acid transporters (AATs) located in the membranes of root and shoot cells facilitate amino acid movement within C3 plants. Several amino acid transporter families, including Lysine–Histidine Transporters (LHTs) and Amino Acid Permeases (AAPs), as well as Proline Transporters (ProTs) have been identified to perform essential functions in nitrogen remobilization during conditions of limited nitrogen availability. The proton gradient produced by H+-ATPase powers PATs for membrane transport and the root growth and vascular differentiation and nitrogen allocation processes are influenced by auxins and cytokinins and abscisic acid (ABA). The expression levels of AAP2 and AAP6 genes increase under drought stress and salinity in addition to nitrogen limitation, enabling better nitrogen transfer to plant leaves and shoots. Under stress conditions GABA (γ-aminobutyric acid) transporters contribute to signaling pathways that maintain metabolic balance between nitrogen transport systems. [22,23]. The sophisticated regulatory system optimizes nitrogen distribution primarily during times of high nutrient requirement while providing possibilities to boost nitrogen efficiency and farming productivity for sustainable agricultural practices.
2.6 Genetic and Molecular Regulation
The regulatory processes of C3 plant nitrogen assimilation function through diverse genetic mechanisms and molecular networks with many components yet to be fully understood. During nitrogen uptake and assimilation the NAC transcription factors together with NIN-like protein 7 (NLP7) act as primary controllers of gene expression [24]. NLP7 functions as a nitrate detection system to control nitrate transporter and assimilatory enzyme gene transcription, which facilitates synchronization between nitrogen assimilation and environmental nitrogen availability fluctuations. NLP6 and NLP9 as well as NLP7 form part of the NIN-like protein family which functions to precisely control the expression of nitrogen-responsive genes. Research indicates that that post-translational modifications, including phosphorylation and ubiquitination, further influence NLP7 activity, affecting nitrogen signaling pathways [25,26].
NAC transcription factors regulate gene expression against environmental stressors to help plants adapt nitrogen assimilation procedures while experiencing diverse environmental conditions. Additionally, MYB and bZIP transcription factors have been identified as key regulators of ammonium assimilation, influencing the expression of glutamine synthetase (GS) and glutamate synthase (GOGAT) [27]. DNA methylation alongside histone acetylation represents two epigenetic processes that regulate genes reacting to nitrogen levels in C3 plant systems. These regulatory networks ensure efficient nitrogen uptake and assimilation, optimizing plant growth and stress resilience under diverse environmental conditions. Knowledge expansion about these regulatory systems presents the possibility of developing improved nitrogen-use approaches to maximize crop productivity in sustainable farming systems.
2.7 Regulation of Nitrogen Assimilation
The assimilation of nitrogen in C3 plants depends on regulatory actions from internal plant elements together with external factors while nitrate functions both as sustenance supply and important signaling molecule to influence gene expression for nitrogen uptake and metabolic activities. The studies recently demonstrated that NRT1.1 and NRT2.1 perform a regulatory function to monitor nitrate availability, which results in optimal nitrogen usage across diverse environmental settings. Plant ammonium assimilation requires two proteins called Nitrite Reductase (NiR) and Glutamine Synthetase (GS) that work based on precise nitrogen levels together with metabolic requirements of the plant. Light functions as a key variable that intensively affects the processes of nitrogen metabolism. Research indicates the circadian clock controls nitrogen-related gene expression, which results in optimized nitrogen assimilation rates during daylight hours when photosynthetic energy reaches its peak [28]. The synchronized relationship between carbon and nitrogen cycles leads to efficient plant growth performance that becomes more noticeable in changing light and nutrient situations. The three phytohormones cytokinins auxins and abscisic acid (ABA) control the process of nitrogen assimilation through their ability to merge developmental signaling with nutrient levels. Under drought stress ABA restricts nitrogen processing but stimulates root development and nutrient relationship through gene activation of nitrate transmitters while cytokinins regulate root growth to optimize energy intake. Recent findings present strigolactones and ethylene as chemical compounds which refine nitrogen signaling process when plants encounter stressful environments. This hormonal regulation helps plants adapt to environmental changes and ensures optimal growth and development [29,30].
2.8 Photorespiration and Nitrogen Metabolism in C3 Plants
Nitrogen assimilation in C3 plants is closely linked to photorespiration, a process in which ribulose bisphosphate carboxylase/oxygenase (Rubisco) reacts with oxygen instead of carbon dioxide, particularly under conditions of high light and temperature. This leads to the production of glycine and serine, amino acids essential for nitrogen metabolism. Ammonia produced during photorespiration can be recycled through various transamination reactions, contributing to the plants nitrogen pool [31]. Maintaining nitrogen balance, especially during periods of high photorespiratory activity, requires a close interaction between nitrogen metabolism and photorespiration. Nitrogenous compounds formed during these processes serve as sources for the synthesis of critical biomolecules like proteins and nucleotides [32]. Efficient nitrogen management in C3 plants ensures sufficient nitrogen availability for growth and development. Photorespiration also influences nitrogen assimilation efficiency because its byproducts can regulate nitrogen-assimilating enzymes. For example, increased levels of glycine may stimulate glutamine synthetase activity, promoting the incorporation of ammonium into organic compounds. The complexity of this metabolism mark its interdependence, as photosynthetic and nitrogen assimilation pathways must be coordinately regulated for optimal growth and maximum productivity.
3 Signaling Pathways Influencing Nitrogen Assimilation
The signaling pathways regulate nitrogen intake and assimilation efficiency through complex and intertwined mechanisms involving hormonal regulation, gene transcription, calcium signaling, reactive oxygen species (ROS) generation, protein kinase signaling, nutrient sensing, and autophagy mechanisms (Fig. 2). Among the aforementioned hormones, abscisic acid (ABA) emerges as a critical hormones affecting nitrogen metabolism processes. During drought stress, ABA levels rise and suppresses the genes involved in nitrogen assimilation enzymes such as NADPH-dependent nitrate reductase (NR) and glutamine synthetase (GS), which are essential for converting nitrate and ammonium into amino acids. The inhibition of these enzymes under stress conditions adversely affects the plant’s nitrogen status, compromising growth and productivity [20,33]. This ABA-mediated regulation highlights the complex balance that plants must maintain between stress responses and nutrient assimilation [34]. Besides ABA, cytokinins play a crucial role in regulating nitrogen metabolism in plants, particularly in the modulation of nitrate reductase (NR) activity, which is essential for nitrogen assimilation under favorable non-stress conditions [35]. Research indicates that cytokinins are involved in the regulation of gene expression related to NR, influencing the overall nitrogen utilization efficiency in plants. Moreover, the interaction between nitrous oxide (NO) and cytokinin signaling is significant, as NO acts as a potential second messenger in these pathways, facilitating the rapid response of plants to nitrogen availability [35]. These findings highlight the ability of C3 plants to adapt their nitrogen metabolism dynamically according to environmental conditions, allowing them to optimize growth and development when resources are plentiful [36]. The integration of cytokinin-mediated regulation and nitrogen uptake reflects the intricate balance plants maintain between nutrient demands and available resources.
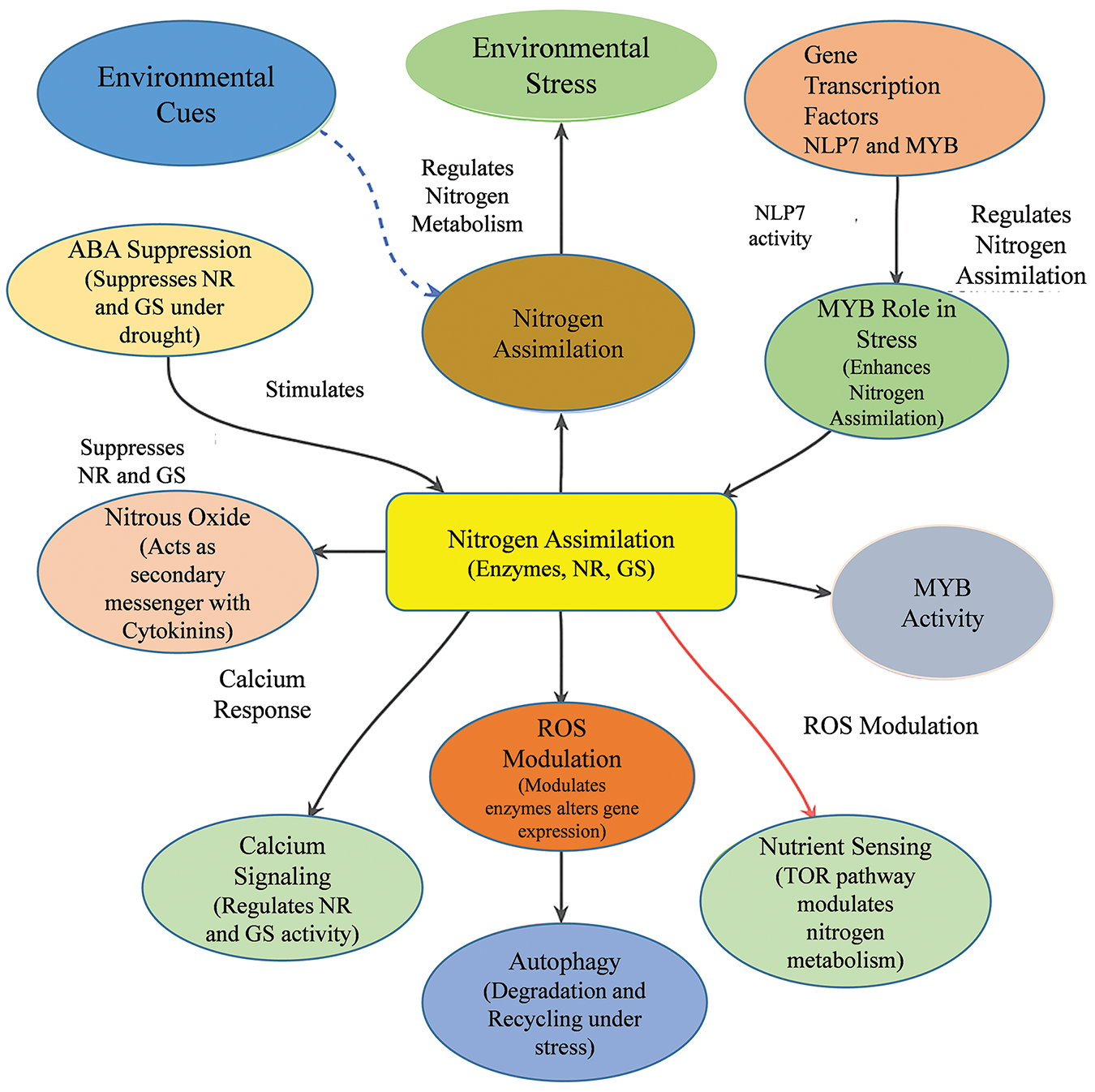
Figure 2: The nitrogen assimilation process in plants, showing key signaling pathways and regulatory mechanisms. The chart includes hormonal regulation (Abscisic Acid, Cytokinins), gene transcription (NLP7, MYB), calcium signaling, reactive oxygen species (ROS) involvement, MAPK pathways, nutrient sensing (TOR signaling pathway), and autophagy mechanisms. The interplay between environmental cues (drought, nitrogen availability) and nitrogen assimilation enzymes (nitrate reductase, glutamine synthetase) is shown, demonstrating how these factors collectively contribute to nitrogen metabolism and plant adaptation
In C3 plants, nitrogen assimilation is regulated primarily through transcriptional mechanisms, with key regulators such as NLP7 and MYB transcription factors playing essential roles [37]. NLP7 plays an important role in regulating the plant’s reaction to nitrogen status and stress conditions and is involved in the regulation of expression of several genes involved in nitrogen metabolism. Furthermore, MYB transcription factors play a role in improving nitrogen assimilation through genes that respond to stress allowing C3 plants to optimize nitrogen uptake and usage under stress conditions [38]. Calcium signaling acts as a vital secondary messenger in C3 plants, mediating the plants ability to respond to various abiotic stressors, including drought and salinity. It also regulates the activities of nitrogen-related enzymes such as nitrate reductase (NR) and glutamine synthetase (GS) [33,37]. By interpreting environmental cues, calcium signaling allows for rapid adjustments in metabolic pathways, ensuring that nitrogen assimilation processes are optimized under fluctuating conditions. Moreover, reactive oxygen species (ROS) play a dual role, acting as both signaling molecules and mediators of stress responses [39]. Under abiotic stress, ROS influence nitrogen metabolism by modulating the activity of critical enzymes and altering gene expression associated with nitrogen assimilation [40]. This regulatory aspect enables plants to control their metabolic activities in a way that would enable them to cope with stress and maintain efficient nitrogen utilization.
The Mitogen-activated protein kinases (MAPKs), which control essential nitrogen assimilation enzymes and transcription factors are vital in transmitting signaling pathways related to abiotic stress in C3 plants [41]. By integrating various environmental signals, MAPK signaling pathways enable C3 plants to fine-tune their nitrogen metabolism, ensuring that uptake and assimilation processes are aligned with the plants current physiological state. Furthermore, nutrient sensing mechanisms, especially the Target of Rapamycin (TOR) signaling pathway, is important in modulating nitrogen metabolism in C3 plants under abiotic stress conditions [42]. These pathways enable plants to monitor their nutrient status and adjust metabolic processes as necessary. In the case of nutrient deficiency or abundance, the TOR pathway regulates nitrogen assimilation and other processes, allowing C3 plants to ration resources and sustain themselves during periods of stress. Similarly, autophagy pathways play a significant role in managing nitrogen metabolism in C3 plants, particularly under abiotic stress. By facilitating the degradation and recycling of cellular components, autophagy helps in the turnover of proteins, thus ensuring that nitrogen resources are utilized effectively during nutrient scarcity [43]. This regulatory mechanism is essential for maintaining nitrogen homeostasis in C3 plants, allowing them to adapt to and thrive in various stress conditions.
4 Impact of Abiotic Stress on Nitrogen Assimilation and Adaptive Responses
Abiotic stresses such as drought, salinity, and extreme temperature significantly impact nitrogen assimilation processes in C3 plants, affecting their growth and productivity. These stresses interfere with various stages of nitrogen metabolism, from uptake to assimilation, leading to reduced nitrogen use efficiency and overall plant health (Table 1).
Drought is one of the major abiotic stresses that affect nitrogen assimilation by adversely affecting the root function and reducing soil water, hinders the uptake of nitrate and ammonium [44,45]. Thus, drought stress reduces the expression and activity of NR and GS enzymes [46]. The decrease in NR activity impairs the conversion of nitrate to nitrite and eventually ammonium. Likewise, drought stress decreases the GS activity, which in turn decreases the synthesis of glutamine. This reduction in glutamine synthesis disrupts key metabolic pathways, including protein synthesis and nitrogen storage, ultimately hindering plant growth. These reductions also impact overall protein synthesis as well as plant growth. At the molecular level, there is regulation of genes closely related to nitrogen metabolism in plants under drought stress. For example, the transcription factor MYB42, which is involved in nitrogen and carbon metabolism, is upregulated under drought conditions, while the expression of NR and GS genes is downregulated, further exacerbating nitrogen assimilation issues [47]. Furthermore, drought stress can also trigger osmotic stress conditions, which involve signaling pathways such as the abscisic acid (ABA) pathway [48], which modulate nitrogen assimilation processes [44]. ABA has been shown to influence the expression of genes involved in both nitrogen uptake and assimilation, thus linking water stress directly to nutrient stress in plants.
Salinity stress, caused by high concentrations of soluble salts, leads to osmotic stress and ion toxicity, affecting water uptake and nitrate assimilation. High concentrations of salts in the soil decrease the soils water potential, impairing nitrate uptake and limiting the activity of NR [41]. Salinity stress can have direct impacts on nitrogen assimilation enzymes such as GS via changes in the stability and functionality of these enzymes. Furthermore, it can affect the mechanisms regulating the expression of salt tolerance and nitrogen-metabolizing genes. For example, genes such as SOS1 (Salt Overly Sensitive 1) and NHX1 (Na+/H+ Exchanger 1) are involved in ion homeostasis and may indirectly influence nitrogen assimilation under saline conditions [49,50]. Recent studies have highlighted that the interaction between SOS1 and NHX1 plays a crucial role in modulating the expression of nitrogen-related genes, such as those involved in nitrate transport and assimilation, in response to salinity [49,50]. Additionally, the salt-induced activation of the MAPK (Mitogen-Activated Protein Kinase) signaling pathway is also thought to regulate key genes involved in nitrogen metabolism, underscoring the complex signaling network in plants under salinity stress.
Nitrogen uptake in plants is also affected by the extreme temperatures that induce changes in the plants physiological and biochemical status. High temperatures can cause coagulation of proteins and enzymes involved in nitrogen metabolism, such as NR and GS [50]. This denaturation affects enzyme activity and disrupted nitrogen assimilation pathways. Elevated temperatures can also alter the expression of heat shock proteins (HSPs) and stress-induced genes, involved in nitrogen metabolisms [51]. Conversely, low temperatures can slow down metabolic activities, affecting assimilation of nitrogen and its efficiency [52]. Cold stress affects root growth and function, which in turn affects the plants ability to uptake nitrogen. Secondly, low temperature affects regulating genes to cold acclimation and nitrogen metabolism, which changes the overall nitrogen contents in plant tissues [52,53]. In summary, the abiotic stresses affect nitrogen assimilation through a complex interplay of impaired nutrient uptake, a decrease in nitrogen metabolizing enzymes and changes in gene regulation. Understanding these effects is crucial in finding ways to enhance plant resilience to withstand different abiotic stresses and nutrient conditions in the context of modern agriculture.
C3 plants have developed various mechanisms to improve nitrogen use efficiency, under abiotic stress conditions which demonstrates the plants ability in managing scarcity of resources and other adverse factors [44]. Some of them include changes in root systems, metabolism rate, nutrient cycling process, and utilization of genetic variation to enhance stress tolerance [54]. In conditions like drought or nutrient limitation, C3 plants are able to adapt their root architecture to enhance nitrogen uptake [55]. This includes improvement in number and extent of root branching that enhances the plants ability to acquire nitrogen from deeper soil layers and explore the rhizosphere. For instance, plants with deeper roots can access the moisture and nutrients not accessible by those with shallow root structures [55,56]. Also, changes in root morphology, for instance, more lateral roots and root hairs make nutrients uptake easier under stressful situations [57]. Under stress conditions, plants may increase the production of amino acids like proline and glutamine. Proline helps protect the plant by maintaining osmotic balance, stabilizing proteins and cell structures, while glutamine serves as a central nitrogen carrier within the plant, essential for maintaining nitrogen balance [55]. These metabolic changes enable plants to maintain cellular functions and adapt to environmental changes.
Furthermore, when examining the genetic basis of C3 plants there is evidence of varietal differences in abiotic stress tolerance [58]. Some of the crop varieties show an enhanced ability to fix nitrogen and better stress tolerance compared to others [58,59]. This shows the potential of utilizing genetic variation to increase NUE and stress resilience [58]. Breeding programs focused on new varieties with enhanced root architecture, nitrogen use efficiency, and stress tolerance can contribute significantly to improving crop yield under adverse climatic conditions [58,59]. Technological advances in molecular biology, including marker assisted selection and genetic engineering, present new opportunities for improving NUE and coping with biotic and abiotic stress.
5 Recent Advances in Understanding Nitrogen Assimilation
Recent advances in nitrogen assimilation in C3 plants under abiotic stress have benefited greatly from genomic and transcriptomic approaches [72]. These advancements have given a better perspective on how nitrogen metabolism occurs and how plants respond to stress factors in their environment [73]. One major development is the application of RNA sequencing (RNA-seq), which has greatly enhanced the capacity to study gene expression at a much higher resolution [74]. In nitrogen metabolism, RNA-seq has revealed large-scale regulatory connections and dynamic gene expression changes under different stress conditions [75]. For instance, in the current studies, it has been revealed that drought stress affects the transcription of genes that are related to nitrogen acquisition and assimilation, like nitrate reductase (NR) and glutamine synthetase (GS) [63]. These studies have shown that drought stress not only inhibits these important enzymes but also affects related transcriptional factors and signaling pathways, such as those involving abscisic acid (ABA), which mediates stress signaling and regulation of nitrogen metabolism [64,76]. The current transcriptome studies have also revealed that abiotic stresses such as salinity and temperature extremes affect the genes regulation involved in nitrogen metabolism. High salinity conditions increase the expression of stress-responsive genes and increase the accumulation of osmoprotectants such as proline [77,78]. This response helps mitigate the impact of salinity stress on nitrogen assimilation by regulating the activity of the enzyme and nitrogen homeostasis.
Likewise, experiments on temperature stress have demonstrated that high temperature affects the differential gene expression associated with nitrogen assimilation, which affects the stability as well as the functionality of the enzymes [79]. Apart from gene expression studies, the identification of new regulatory sequences and metabolic networks has also added to the knowledge of nitrogen assimilation. Some of the emerging studies have been directed towards the identification of microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) in stress management of nitrogen metabolism. For instance, miR169 has been identified to target genes that are involved in nitrogen uptake and its assimilation into the plant, and, therefore, acts as a layer of post-transcriptional regulation of plant responses to changes in the environment [78]. The exploration of metabolic pathways has also revealed new aspects of how plants coordinate nitrogen assimilation with other biochemical processes. For instance, recent studies have revealed important metabolic centers, including the carbon and nitrogen balance and the interplay between nitrogen assimilation and photosynthesis. These observations indicate that nitrogen metabolism is a rather complex process, where plants regulate nitrogen assimilation and other metabolic processes to optimize growth under stress conditions [49,69]. Thus, these recent findings in the genomic and transcriptomic studies are important for formulating a holistic view of the C3 plants in nitrogen metabolism under abiotic stress.
6 Implications for Future Research
Subsequent research should be directed towards the elucidation of the genetic and molecular mechanisms of nitrogen assimilation in C3 plants under the context of enhancing abiotic stress tolerance. Genetic engineering has shown significant promise in enhancing nitrogen use efficiency (NUE) in crops, as demonstrated by several case studies. The overexpression of specific genes and the use of CRISPR/Cas9 technology have been pivotal in improving nitrogen uptake and assimilation, leading to increased crop yields under nitrogen-limited conditions. In rice (Oryza sativa), overexpression of OsNRT2.3b, a high-affinity nitrate transporter gene, enhanced nitrogen uptake efficiency and improved grain yield under limited nitrogen conditions [80]. Similarly, CRISPR/Cas9-mediated knockout of OsHHO3, a transcriptional repressor, resulted in increased ammonium uptake and improved NUE, growth, and yield in rice by upregulating ammonium transporter genes [81]. In wheat (Triticum aestivum), transgenic lines overexpressing TaNRT2.1 exhibited increased nitrate uptake, leading to higher biomass production and improved nitrogen utilization efficiency [82]. While genetic engineering offers promising solutions for improving NUE, integrating genetic interventions with agronomic practices and breeding innovations is essential to fully realize the potential of these technologies. Additionally, environmental and regulatory aspects must be carefully managed to ensure the sustainable deployment of genetically modified crops in agricultural systems [83]. Further, the possibility of using synthetic biology methods to create novel regulatory networks for optimizing nitrogen metabolism could be a promising idea. This could entail designing plants to produce stress-inducible promoters that enhance the expression of nitrogen-assimilation genes only in conditions of stress to increase nitrogen uptake, thereby conserving energy under normal conditions. Marker assisted selection (MAS) and genomic selection represent two important molecular breeding techniques that require focus. Through marker technology, researchers can develop high-yielding crop varieties which show tolerance to conditions that are less than perfect since markers reflect genes that affect NUE and stress tolerance. Quantitative trait locus (QTL) mapping serves to find genomic locations holding NUE relationships under stressed environments, which will direct future breeding strategies.
Researchers must focus their future investigations on studying how epigenetic mechanisms regulate nitrogen metabolism in stress-related environments. Among all epigenetic mechanisms involved in stress-related gene expression, the major ones are DNA methylation and histone modifications. Knowledge of these modifications would enable breeding scientists to produce new approaches that enhance plant nitrogen use efficiency under stress through epigenetic editing and epigenetic marks selection in breeding programs. The discussion about nitrogen metabolism needs to be integrated within concepts of plant physiology and ecology. The knowledge of the specific environmental changes and microbial communities affecting plant nitrogen processes is vital for developing holistic approaches to managing nitrogen in agricultural systems. The role of soil microbiomes in nitrogen cycling, particularly plant-microbe symbioses such as rhizobia in legumes and endophytic bacteria in non-legumes, warrants further exploration. Harnessing beneficial microbial consortia could provide natural solutions for enhancing nitrogen uptake and stress resilience. This might involve the study of plant-microbe interactions, where beneficial soil microbes that increase nitrogen availability could be harnessed to increase crop resistance and NUE. Given the challenges posed by climate change, it is also essential to develop predictive models that integrate genetic, molecular, and environmental data to forecast how crops will perform under future climate scenarios. Such models could guide the development of crops that are not only resilient to current stressors but also adaptable to future environmental changes. Advanced computational techniques, such as machine learning and systems biology approaches, could play a critical role in deciphering the complex regulatory networks of nitrogen metabolism under stress conditions. These models could also help identify novel gene targets for improving NUE. Ultimately, this knowledge will be pivotal in creating sustainable farming practices and implementing precision agriculture techniques that optimize nitrogen management, reduce environmental impacts, and enhance crop productivity. Exploring nitrogen assimilation within the context of regenerative agriculture and climate-smart cropping systems could further improve sustainability and resilience. By focusing on these advanced genetic and molecular approaches, future research can contribute significantly to global food security and the sustainability of agricultural systems in the face of mounting environmental challenges.
A deep understanding of the molecular mechanisms governing nitrogen assimilation in C3 plants under abiotic stress is essential for improving agricultural resilience and sustainability. Recent advances in genomic and transcriptomic technologies have uncovered the complex regulatory networks that govern nitrogen metabolism in these plants when subjected to environmental stressors such as drought, salinity, and temperature extremes. Recent molecular advancements reveal the adaptive capacities of C3 plants to optimize nitrogen assimilation and uptake processes under challenging growth environments. Additionally, these findings provide prospects for genetically modifying crops with enhanced nitrogen utilization efficiency while improving their stress tolerance capabilities. By targeting key nitrogen assimilation pathways and utilizing cutting-edge genetic tools like CRISPR/Cas9, it is possible to engineer crops that are both high-yielding and resilient to abiotic stresses. The obtained knowledge helps establish more effective nitrogen management methods to lower fertilizer impact while making agriculture sustainable. Understanding these molecular mechanisms stands as a fundamental requirement for developing crop varieties that can thrive in harsh environments to maintain food security while advancing sustainable farming practices worldwide.
Acknowledgement: Not applicable.
Funding Statement: Not applicable.
Author Contributions: Study conception and design: Izhar Ali and Saif Ullah; writing original draft preparation: Saif Ullah; review and editing: Izhar Ali and Saif Ullah. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: No data was used for the research described in the article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Zayed O, Hewedy OA, Abdelmoteleb A, Ali M, Youssef MS, Roumia AF, et al. Nitrogen journey in plants: from uptake to metabolism, stress response, and microbe interaction. Biomolecules. 2023;13(10):1443. doi:10.3390/biom13101443. [Google Scholar] [PubMed] [CrossRef]
2. The SV, Snyder R, Tegeder M. Targeting nitrogen metabolism and transport processes to improve plant nitrogen use efficiency. Front Plant Sci. 2021;11:628366. doi:10.3389/fpls.2020.628366. [Google Scholar] [PubMed] [CrossRef]
3. Opoku E, Sahu PP, Findurová H, Holub P, Urban O, Klem K. Differential physiological and production responses of C3 and C4 crops to climate factor interactions. Front Plant Sci. 2024;15:1345462. doi:10.3389/fpls.2024.1345462. [Google Scholar] [PubMed] [CrossRef]
4. Zhang G, He C. Understanding the interdependence of nitrogen and carbon metabolism in plants. Front Plant Sci. 2021;12:650–65. [Google Scholar]
5. Sallam A, Alqudah AM, Dawood MFA, Stephen Baenziger P, Börner A. Drought stress tolerance in wheat and barley: advances in physiology, breeding and genetics research. Int J Mol Sci. 2019;20(13):3137. doi:10.3390/ijms20133137. [Google Scholar] [PubMed] [CrossRef]
6. Ashraf M, Shahzad SM, Imtiaz M, Rizwan MS. Salinity effects on nitrogen metabolism in plants-focusing on the activities of nitrogen metabolizing enzymes: a review. J Plant Nutr. 2018;41(8):1065–81. doi:10.1080/01904167.2018.1431670. [Google Scholar] [CrossRef]
7. Tejada-Jimenez M, Llamas A, Galván A, Fernández E. Role of nitrate reductase in NO production in photosynthetic eukaryotes. Plants. 2019;8(3):56. doi:10.3390/plants8030056. [Google Scholar] [PubMed] [CrossRef]
8. Liao HS, Chung YH, Hsieh MH. Glutamate: a multifunctional amino acid in plants. Plant Sci. 2022;318:111238. doi:10.1016/j.plantsci.2022.111238. [Google Scholar] [PubMed] [CrossRef]
9. Govindasamy P, Muthusamy SK, Bagavathiannan M, Mowrer J, Jagannadham PTK, Maity A, et al. Nitrogen use efficiency–a key to enhance crop productivity under a changing climate. Front Plant Sci. 2023;14:1121073. doi:10.3389/fpls.2023.1121073. [Google Scholar] [PubMed] [CrossRef]
10. Akhtar K, Ain NU, Prasad PVV, Naz M, Aslam MM, Djalovic I, et al. Physiological, molecular, and environmental insights into plant nitrogen uptake, and metabolism under abiotic stresses. Plant Genome. 2024;17(2):e20461. doi:10.1002/tpg2.20461. [Google Scholar] [PubMed] [CrossRef]
11. Ueda Y, Konishi M, Yanagisawa S. Molecular basis of the nitrogen response in plants. Soil Sci Plant Nutr. 2017;63(4):329–41. doi:10.1080/00380768.2017.1360128. [Google Scholar] [CrossRef]
12. Maeda Y, Konishi M, Kiba T, Sakuraba Y, Sawaki N, Kurai T, et al. A NIGT1-centred transcriptional cascade regulates nitrate signalling and incorporates phosphorus starvation signals in Arabidopsis. Nat Commun. 2018;9(1):1376. doi:10.1038/s41467-018-03832-6. [Google Scholar] [PubMed] [CrossRef]
13. Yoneyama T, Suzuki A. Exploration of nitrate-to-glutamate assimilation in non-photosynthetic roots of higher plants by studies of 15N-tracing, enzymes involved, reductant supply, and nitrate signaling: a review and synthesis. Plant Physiol Biochem. 2019;136:245–54. doi:10.1016/j.plaphy.2018.12.011. [Google Scholar] [PubMed] [CrossRef]
14. Yanagisawa S. Transcription factors involved in controlling the expression of nitrate reductase genes in higher plants. Plant Sci. 2014;229:167–71. doi:10.1016/j.plantsci.2014.09.006. [Google Scholar] [PubMed] [CrossRef]
15. Gilad G, Sapir O, Hipsch M, Waiger D, Ben-Ari J, Zeev BB, et al. Nitrogen assimilation plays a role in balancing the chloroplastic glutathione redox potential under high light conditions. Plant Cell Environ. 2025;48(5):3559–72. doi:10.1111/pce.15368. [Google Scholar] [PubMed] [CrossRef]
16. Zlobin I, Efimova M, Permykova N, Sokolova I, Kuznetsov V, Deineko E. The modification of abscisic acid and cytokinin signaling with genome editing to increase plant drought tolerance. In: Plant physiology annual volume 2023. London, UK: IntechOpen; 2023. doi:10.5772/intechopen.113980. [Google Scholar] [CrossRef]
17. Sanz-Luque E, Chamizo-Ampudia A, Llamas A, Galvan A, Fernandez E. Understanding nitrate assimilation and its regulation in microalgae. Front Plant Sci. 2015;6(28):899. doi:10.3389/fpls.2015.00899. [Google Scholar] [PubMed] [CrossRef]
18. Suárez MF, Avila C, Gallardo F, Cantón FR, García-Gutiérrez A, Claros MG, et al. Molecular and enzymatic analysis of ammonium assimilation in woody plants. J Exp Bot. 2002;53(370):891–904. doi:10.1093/jexbot/53.370.891. [Google Scholar] [PubMed] [CrossRef]
19. Wahid I, Kumari S, Ahmad R, Hussain SJ, Alamri S, Siddiqui MH, et al. Silver nanoparticle regulates salt tolerance in wheat through changes in ABA concentration, ion homeostasis, and defense systems. Biomolecules. 2020;10(11):1506. doi:10.3390/biom10111506. [Google Scholar] [PubMed] [CrossRef]
20. Sahay S, Robledo-Arratia L, Glowacka K, Gupta M. Root NRT, NiR, AMT, GS, GOGAT and GDH expression levels reveal NO and ABA mediated drought tolerance in Brassica juncea L. Sci Rep. 2021;11(1):7992. doi:10.1038/s41598-021-86401-0. [Google Scholar] [PubMed] [CrossRef]
21. Tegeder M, Rentsch D. Uptake and partitioning of amino acids and peptides. Mol Plant. 2010;3(6):997–1011. doi:10.1093/mp/ssq047. [Google Scholar] [PubMed] [CrossRef]
22. Yao X, Nie J, Bai R, Sui X. Amino acid transporters in plants: identification and function. Plants. 2020;9(8):972. doi:10.3390/plants9080972. [Google Scholar] [PubMed] [CrossRef]
23. Yu LH, Wu J, Tang H, Yuan Y, Wang SM, Wang YP, et al. Overexpression of Arabidopsis NLP7 improves plant growth under both nitrogen-limiting and-sufficient conditions by enhancing nitrogen and carbon assimilation. Sci Rep. 2016;6(1):27795. doi:10.1038/srep27795. [Google Scholar] [PubMed] [CrossRef]
24. Bian C, Demirer GS, Oz MT, Cai Y, Witham SS, Mason GA, et al. Conservation and divergence of regulatory architecture in nitrate-responsive plant gene circuits. bioRxiv. 2023;23:7. doi:10.1101/2023.07.17.549299. [Google Scholar] [CrossRef]
25. Krämer K, Brock J, Heyer AG. Interaction of nitrate assimilation and photorespiration at elevated CO2. Front Plant Sci. 2022;13:897924. doi:10.3389/fpls.2022.897924. [Google Scholar] [PubMed] [CrossRef]
26. Choi SJ, Lee Z, Jeong E, Kim S, Seo JS, Um T, et al. Signaling pathways underlying nitrogen transport and metabolism in plants. BMB Rep. 2023;56(2):56–64. doi:10.5483/BMBRep.2022-0178. [Google Scholar] [PubMed] [CrossRef]
27. Kojima S, Minagawa H, Yoshida C, Inoue E, Takahashi H, Ishiyama K. Coregulation of glutamine synthetase1;2 (GLN1;2) and NADH-dependent glutamate synthase (GLT1) gene expression in Arabidopsis roots in response to ammonium supply. Front Plant Sci. 2023;14:1127006. doi:10.3389/fpls.2023.1127006. [Google Scholar] [PubMed] [CrossRef]
28. Porco S, Yu S, Liang T, Snoeck C, Hermans C, Kay SA. The clock-associated LUX ARRHYTHMO regulates high-affinity nitrate transport in Arabidopsis roots. Plant J. 2024;120(5):1786–97. doi:10.1111/tpj.17080. [Google Scholar] [PubMed] [CrossRef]
29. Ehleringer JR, Cerling TE. C3 and C4 photosynthesis. Encycl Glob Environ Change. 2002;2(4):186–90. [Google Scholar]
30. Nunes-Nesi A, Fernie AR, Stitt M. Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol Plant. 2010;3(6):973–96. doi:10.1093/mp/ssq049. [Google Scholar] [PubMed] [CrossRef]
31. Jose E, Soni KKB, Alex S, Shalini PPP, Beena R, Stephen R. Molecular frameworks of nitrogen response in plants: a review. Int J Environ Clim Change. 2023;13(12):380–90. doi:10.9734/ijecc/2023/v13i123694. [Google Scholar] [CrossRef]
32. Ye JY, Tian WH, Jin CW. Nitrogen in plants: from nutrition to the modulation of abiotic stress adaptation. Stress Biol. 2022;2(1):4. doi:10.1007/s44154-021-00030-1. [Google Scholar] [PubMed] [CrossRef]
33. Tun NN, Holk A, Scherer GFE. Cytokinin, nitrate reductase, and nitric oxide (NONew insights into regulatory processes in nitrogen metabolism. In: Plant nutrition: food security and sustainability of agro-ecosystems through basic and applied research. Dordrecht, The Netherland: Springer; 2001. p. 128–9. [Google Scholar]
34. Kamada-Nobusada T, Makita N, Kojima M, Sakakibara H. Nitrogen-dependent regulation of de novo cytokinin biosynthesis in rice: the role of glutamine metabolism as an additional signal. Plant Cell Physiol. 2013;54(11):1881–93. doi:10.1093/pcp/pct127. [Google Scholar] [PubMed] [CrossRef]
35. Maurya J, Bandyopadhyay T, Prasad M. Transcriptional regulators of nitrate metabolism: key players in improving nitrogen use in crops. J Biotechnol. 2020;324(4):121–33. doi:10.1016/j.jbiotec.2020.10.001. [Google Scholar] [PubMed] [CrossRef]
36. Yang Y, Gao S, Su Y, Lin Z, Guo J, Li M, et al. Transcripts and low nitrogen tolerance: regulatory and metabolic pathways in sugarcane under low nitrogen stress. Environ Exp Bot. 2019;163(10):97–111. doi:10.1016/j.envexpbot.2019.04.010. [Google Scholar] [CrossRef]
37. Sámano ML, Nanjareddy K, Arthikala MK. NIN-like proteins (NLPs) as crucial nitrate sensors: an overview of their roles in nitrogen signaling, symbiosis, abiotic stress, and beyond. Physiol Mol Biol Plants. 2024;30(7):1209–23. doi:10.1007/s12298-024-01485-y. [Google Scholar] [PubMed] [CrossRef]
38. Jagodzik P, Tajdel-Zielinska M, Ciesla A, Marczak M, Ludwikow A. Mitogen-activated protein kinase cascades in plant hormone signaling. Front Plant Sci. 2018;9:1387. doi:10.3389/fpls.2018.01387. [Google Scholar] [PubMed] [CrossRef]
39. Li D, Ding Y, Cheng L, Zhang X, Cheng S, Ye Y, et al. Target of rapamycin (TOR) regulates the response to low nitrogen stress via autophagy and hormone pathways in Malus hupehensis. Hortic Res. 2022;9:uhac143. doi:10.1093/hr/uhac143. [Google Scholar] [PubMed] [CrossRef]
40. Su T, Li X, Yang M, Shao Q, Zhao Y, Ma C, et al. Autophagy: an intracellular degradation pathway regulating plant survival and stress response. Front Plant Sci. 2020;11:164. doi:10.3389/fpls.2020.00164. [Google Scholar] [PubMed] [CrossRef]
41. Iqbal N, Sadras VO, Denison RF, Zhou Y, Denton MD. Clade-dependent effects of drought on nitrogen fixation and its components–number, size, and activity of nodules in legumes. Field Crops Res. 2022;284:108586. doi:10.1016/j.fcr.2022.108586. [Google Scholar] [CrossRef]
42. Naikwade PV. Plant responses to drought stress: morphological, physiological, molecular approaches, and drought resistance. In: Plant metabolites under environmental Stress. Palm Bay, FL, USA: Apple Academic Press; 2023. p. 149–83. [Google Scholar]
43. Xu Y, Fu X. Reprogramming of plant central metabolism in response to abiotic stresses: a metabolomics view. Int J Mol Sci. 2022;23(10):5716. doi:10.3390/ijms23105716. [Google Scholar] [PubMed] [CrossRef]
44. Sajjad N, Bhat EA, Shah D, Manzoor I, Noor W, Shah S, et al. Nitrogen uptake, assimilation, and mobilization in plants under abiotic stress. In: Transporters and plant osmotic stress. Cambridge, MA, USA: Academic Press; 2021. p. 215–33. doi:10.1016/B978-0-12-817958-1.00015-3. [Google Scholar] [CrossRef]
45. Anjana Umar S, Iqbal M. Factors responsible for nitrate accumulation: a review. In: Sustainable agriculture. Dordrecht, The Netherland: Springer; 2009. p. 533–49. [Google Scholar]
46. Xia H, Xu T, Zhang J, Shen K, Li Z, Liu J. Drought-induced responses of nitrogen metabolism in Ipomoea batatas. Plants. 2020;9(10):1341. doi:10.3390/plants9101341. [Google Scholar] [PubMed] [CrossRef]
47. Wang X, Wei H, Wang K, Tang X, Li S, Zhang N, et al. MYB transcription factors: acting as molecular switches to regulate different signaling pathways to modulate plant responses to drought stress. Ind Crops Prod. 2025;226:120676. doi:10.1016/j.indcrop.2025.120676. [Google Scholar] [CrossRef]
48. Haq SU, Khan A, Ali M, Khattak AM, Gai WX, Zhang HX, et al. Heat shock proteins: dynamic biomolecules to counter plant biotic and abiotic stresses. Int J Mol Sci. 2019;20(21):5321. doi:10.3390/ijms20215321. [Google Scholar] [PubMed] [CrossRef]
49. Xu G, Fan X, Miller AJ. Plant nitrogen assimilation and use efficiency. Annu Rev Plant Biol. 2012;63(1):153–82. doi:10.1146/annurev-arplant-042811-105532. [Google Scholar] [PubMed] [CrossRef]
50. Theocharis A, Clément C, Barka EA. Physiological and molecular changes in plants grown at low temperatures. Planta. 2012;235(6):1091–105. doi:10.1007/s00425-012-1641-y. [Google Scholar] [PubMed] [CrossRef]
51. Kiba T, Krapp A. Plant nitrogen acquisition under low availability: regulation of uptake and root architecture. Plant Cell Physiol. 2016;57(4):707–14. doi:10.1093/pcp/pcw052. [Google Scholar] [PubMed] [CrossRef]
52. Soualiou S, Duan F, Li X, Zhou W. Nitrogen supply alleviates cold stress by increasing photosynthesis and nitrogen assimilation in maize seedlings. J Exp Bot. 2023;74(10):3142–62. doi:10.1093/jxb/erad073. [Google Scholar] [PubMed] [CrossRef]
53. Guy C. Cold acclimation and freezing stress tolerance: role of protein metabolism. Annu Rev Plant Physiol Plant Mol Biol. 1990;41(1):187–223. doi:10.1146/annurev.arplant.41.1.187. [Google Scholar] [CrossRef]
54. Bhatla SC, Lal MA. Plant physiology, development and metabolism. 2nd ed. Singapore: Springer; 2023. [Google Scholar]
55. Sandhu N, Sethi M, Kumar A, Dang D, Singh J, Chhuneja P. Biochemical and genetic approaches improving nitrogen use efficiency in cereal crops: a review. Front Plant Sci. 2021;12:657629. doi:10.3389/fpls.2021.657629. [Google Scholar] [PubMed] [CrossRef]
56. Lebedev VG, Popova AA, Shestibratov KA. Genetic engineering and genome editing for improving nitrogen use efficiency in plants. Cells. 2021;10(12):3303. doi:10.3390/cells10123303. [Google Scholar] [PubMed] [CrossRef]
57. Zhou R, Yu X, Li X, Mendanha Dos Santos T, Rosenqvist E, Ottosen CO. Combined high light and heat stress induced complex response in tomato with better leaf cooling after heat priming. Plant Physiol Biochem. 2020;151:1–9. doi:10.1016/j.plaphy.2020.03.011. [Google Scholar] [PubMed] [CrossRef]
58. Lobell DB, Hammer GL, Chenu K, Zheng B, McLean G, Chapman SC. The shifting influence of drought and heat stress for crops in northeast Australia. Glob Chang Biol. 2015;21(11):4115–27. doi:10.1111/gcb.13022. [Google Scholar] [PubMed] [CrossRef]
59. Saud S, Fahad S, Chen Y, Ihsan MZ, Hammad HM, Nasim W, et al. Effects of nitrogen supply on water stress and recovery mechanisms in Kentucky bluegrass plants. Front Plant Sci. 2017;8:983. doi:10.3389/fpls.2017.00983. [Google Scholar] [PubMed] [CrossRef]
60. Bista DR, Heckathorn SA, Jayawardena DM, Mishra S, Boldt JK. Effects of drought on nutrient uptake and the levels of nutrient-uptake proteins in roots of drought-sensitive and-tolerant grasses. Plants. 2018;7(2):28. doi:10.3390/plants7020028. [Google Scholar] [PubMed] [CrossRef]
61. He J, Hu W, Li Y, Zhu H, Zou J, Wang Y, et al. Prolonged drought affects the interaction of carbon and nitrogen metabolism in root and shoot of cotton. Environ Exp Bot. 2022;197:104839. doi:10.1016/j.envexpbot.2022.104839. [Google Scholar] [CrossRef]
62. Staszel K, Lasota J, Błońska E. Effect of drought on root exudates from Quercus petraea and enzymatic activity of soil. Sci Rep. 2022;12(1):7635. doi:10.1038/s41598-022-11754-z. [Google Scholar] [PubMed] [CrossRef]
63. Redden R. New approaches for crop genetic adaptation to the abiotic stresses predicted with climate change. Agronomy. 2013;3(2):419–32. doi:10.3390/agronomy3020419. [Google Scholar] [CrossRef]
64. Gaju O, Allard V, Martre P, Snape JW, Heumez E, LeGouis J, et al. Identification of traits to improve the nitrogen-use efficiency of wheat genotypes. Field Crops Res. 2011;123(2):139–52. doi:10.1016/j.fcr.2011.05.010. [Google Scholar] [CrossRef]
65. Wang H, Zhang M, Guo R, Shi D, Liu B, Lin X, et al. Effects of salt stress on ion balance and nitrogen metabolism of old and young leaves in rice (Oryza sativa L.). BMC Plant Biol. 2012;12(1):194. doi:10.1186/1471-2229-12-194. [Google Scholar] [PubMed] [CrossRef]
66. Wang L, Zheng J, Zhou G, Li J, Qian C, Lin G, et al. Moderate nitrogen application improved salt tolerance by enhancing photosynthesis, antioxidants, and osmotic adjustment in rapeseed (Brassica napus L.). Front Plant Sci. 2023;14:1196319. doi:10.3389/fpls.2023.1196319. [Google Scholar] [PubMed] [CrossRef]
67. Zhang X, He P, Guo R, Huang K, Huang X. Effects of salt stress on root morphology, carbon and nitrogen metabolism, and yield of Tartary buckwheat. Sci Rep. 2023;13(1):12483. doi:10.1038/s41598-023-39634-0. [Google Scholar] [PubMed] [CrossRef]
68. Farooq M, Bramley H, Palta JA, Siddique KHM. Heat stress in wheat during reproductive and grain-filling phases. Crit Rev Plant Sci. 2011;30(6):491–507. doi:10.1080/07352689.2011.615687. [Google Scholar] [CrossRef]
69. Tian J, Pang Y, Zhao Z. Drought, salinity, and low nitrogen differentially affect the growth and nitrogen metabolism of Sophora japonica (L.) in a semi-hydroponic phenotyping platform. Front Plant Sci. 2021;12:715456. doi:10.3389/fpls.2021.715456. [Google Scholar] [PubMed] [CrossRef]
70. Goel P, Singh AK. Abiotic stresses downregulate key genes involved in nitrogen uptake and assimilation in Brassica juncea L. PLoS One. 2015;10(11):e0143645. doi:10.1371/journal.pone.0143645. [Google Scholar] [PubMed] [CrossRef]
71. Seleiman MF, Al-Suhaibani N, Ali N, Akmal M, Alotaibi M, Refay Y, et al. Drought stress impacts on plants and different approaches to alleviate its adverse effects. Plants. 2021;10(2):259. doi:10.3390/plants10020259. [Google Scholar] [PubMed] [CrossRef]
72. Cerda A, Alvarez JM. Insights into molecular links and transcription networks integrating drought stress and nitrogen signaling. New Phytol. 2024;241(2):560–6. doi:10.1111/nph.19403. [Google Scholar] [PubMed] [CrossRef]
73. Goel P, Sharma NK, Bhuria M, Sharma V, Chauhan R, Pathania S, et al. Transcriptome and co-expression network analyses identify key genes regulating nitrogen use efficiency in Brassica juncea L. Sci Rep. 2018;8(1):7451. doi:10.1038/s41598-018-25826-6. [Google Scholar] [PubMed] [CrossRef]
74. Mehta D, Vyas S. Comparative bio-accumulation of osmoprotectants in saline stress tolerating plants: a review. Plant Stress. 2023;9(5):100177. doi:10.1016/j.stress.2023.100177. [Google Scholar] [CrossRef]
75. Dikilitas M, Simsek E, Roychoudhury A. Role of proline and glycine betaine in overcoming abiotic stresses. In: Protective chemical agents in the amelioration of plant abiotic stress: biochemical and molecular perspectives. Hoboken, NJ, USA: John Wiley & Sons, Inc.; 2020. p. 1–23. [Google Scholar]
76. Lopez-Delacalle M, Camejo DM, García-Martí M, Nortes PA, Nieves-Cordones M, Martínez V, et al. Using tomato recombinant lines to improve plant tolerance to stress combination through a more efficient nitrogen metabolism. Front Plant Sci. 2020;10:1702. doi:10.3389/fpls.2019.01702. [Google Scholar] [PubMed] [CrossRef]
77. Singh P, Kumar K, Jha AK, Yadava P, Pal M, Rakshit S, et al. Global gene expression profiling under nitrogen stress identifies key genes involved in nitrogen stress adaptation in maize (Zea mays L.). Sci Rep. 2022;12(1):4211. doi:10.1038/s41598-022-07709-z. [Google Scholar] [PubMed] [CrossRef]
78. Singh A, Jain D, Pandey J, Yadav M, Bansal KC, Singh IK. Deciphering the role of miRNA in reprogramming plant responses to drought stress. Crit Rev Biotechnol. 2023;43(4):613–27. doi:10.1080/07388551.2022.2047880. [Google Scholar] [PubMed] [CrossRef]
79. Yuan Y, Liu L, Gao Y, Yang Q, Dong K, Liu T, et al. Comparative analysis of drought-responsive physiological and transcriptome in broomcorn millet (Panicum miliaceum L.) genotypes with contrasting drought tolerance. Ind Crops Prod. 2022;177:114498. doi:10.1016/j.indcrop.2021.114498. [Google Scholar] [CrossRef]
80. Reddy MB, Sravani P, Kumar S, Rajawat MVS, Jaiswal DK, Dhar S, et al. Nitrogen use efficiency reimagined: advancements in agronomic, ecophysiological, and molecular strategies. J Plant Nutr. 2025;360(3):1–27. doi:10.1080/01904167.2024.2447840. [Google Scholar] [CrossRef]
81. Liu K, Sakuraba Y, Ohtsuki N, Yang M, Ueda Y, Yanagisawa S. CRISPR/Cas9-mediated elimination of OsHHO3, a transcriptional repressor of three AMMONIUM TRANSPORTER1 genes, improves nitrogen use efficiency in rice. Plant Biotechnol J. 2023;21(11):2169–72. doi:10.1111/pbi.14167. [Google Scholar] [PubMed] [CrossRef]
82. Kumar V, Rithesh L, Raghuvanshi N, Kumar A, Parmar K. Advancing nitrogen use efficiency in cereal crops: a comprehensive exploration of genetic manipulation, nitrogen dynamics, and plant nitrogen assimilation. S Afr N J Bot. 2024;169(633):486–98. doi:10.1016/j.sajb.2024.04.040. [Google Scholar] [CrossRef]
83. Sharma N, Jaiswal DK, Kumari S, Dash GK, Panda S, Anandan A, et al. Genome-wide urea response in rice genotypes contrasting for nitrogen use efficiency. Int J Mol Sci. 2023;24(7):6080. doi:10.3390/ijms24076080. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools