 Open Access
Open Access
ARTICLE
The characterization of transmembrane protein 59-like (TMEM59L) reveals its role in the regulating the level of the GDI protein family
1 School of Environmental Engineering, Central Plains Specialty Food Engineering & Technology Research Center, Kaifeng Key Laboratory of Food Composition and Quality Assessment, Yellow River Conservancy Technical Institute, Kaifeng, 475004, China
2 Laboratory of Molecular Neural Biology, School of Life Sciences, Shanghai University, Shanghai, 200444, China
* Corresponding Authors: HAIFENG WANG. Email: ; TIEQIAO WEN. Email:
BIOCELL 2022, 46(12), 2615-2624. https://doi.org/10.32604/biocell.2022.021247
Received 04 January 2022; Accepted 05 May 2022; Issue published 10 August 2022
Abstract
The characterization and functions of transmembrane protein 59-like (TMEM59L), a type I transmembrane protein, are not clearly understood until now. Some TMEM59L and fluorescent fusion proteins constructs were transfected in cell lines and liposomes, and their localization was observed. The effects of protein constructs were studied by fluorescence microscopy and western blotting. This study reports a novel function of human TMEM59L (hTMEM59L) related to the expression and location of some proteins. In addition, we report two novel splice variants of human TMEM59L (hTMEM59L). The localization of mutants of this protein, lacking a middle region, and a C-terminal deletion, markedly differed from that of full-length hTMEM59L. Intracellular movement assessment in living cells showed the localization of TMEM59L to vesicular structures in the Golgi bodies, and the cell membrane was observed in living cells. Overexpression of TMEM59L markedly increased the level of amyloid precursor protein because of TMEM59L-mediated inhibition of cell membrane transport but did not affect the expression of beta-secretase 2. TMEM59L overexpression also significantly increased the levels of Rab GDP dissociation inhibitor alpha and Rab GDP dissociation inhibitor beta proteins. These results suggest that TMEM59L is involved in the packaging of acidic vesicles and thereby functions in the trafficking and processing of intracellular proteins.Keywords
Transmembrane protein 59-like (TMEM59L, also known as brain-specific membrane-anchored protein (BSMAP), is a membrane protein. This protein is preferentially expressed in the brain, suggesting its important role in the central nervous system. TMEM59L modulates O-glycosylation and complex N-glycosylation steps during Golgi maturation and shedding of amyloid precursor protein (APP) (Ullrich et al., 2010). APP is a type I transmembrane glycoprotein that plays a pivotal role in the pathogenesis of Alzheimer’s disease (AD). Beta-secretase 2 (BACE2) cleaves APP between residues 690 and 691 to generate beta-cleaved soluble APP (Hussain et al., 2000). However, amyloid β peptide (Aβ) is not generated in transit when transport of APP is blocked by tau protein (Goldsbury et al., 2010), and its production is affected by the trafficking and subcellular localization of APP. Aβ is mainly generated in endosomes (Huse et al., 2002; Steinhilb et al., 2010). The adaptor protein sorting nexin 17 is associated with the trafficking and processing of APP in early endosomes (Lee et al., 2008). Presenilin 1 and APP are co-transported in the same vesicular compartment, resulting in cleavage of APP and thus generation of Aβ (Kim et al., 2001). Endogenous low-density lipoprotein receptor-related protein is required for delivery of APP to lipid rafts, and Aβ is primarily generated in the endocytic, rather than the secretory, pathway (Yoon et al., 2007).
Intracellular transport of proteins involves continuous movement of the cytoskeleton, which requires a large amount of energy. APP is found in Rab5-positive early endosomes but not Rab7-positive late endosomes (Jin et al., 2004). Anterograde transport of APP requires Rab3A GTPase activity for assembly of the transport vesicle (Szodorai et al., 2009). Rab GDP dissociation inhibitor alpha (GDI1) and Rab GDP dissociation inhibitor beta (GDI2) function in the vesicular trafficking of proteins between cellular organelles by regulating GDP-GTP exchange (Bächner et al., 1995).
To date, Aβ is assumed to be generated in vesicles or the endosomal-lysosomal pathway. Based on the existing knowledge, we hope to further study the protein function of TMEM59L with a special focus on the interaction between proteins. However, it is unclear whether this process involves GDIs-regulated Aβ peptide production, although it is assumed that Aβ formation may occur in the secretory or endosomal-lysosomal pathways (Koo and Squazzo, 1994). Maturation of protein is a complex problem special associate the location and function modified of protein, this study reports that human TMEM59L (hTMEM59L) localized to acidic vesicles and moved bi-directionally between the Golgi apparatus and plasma membrane. In addition, Overexpression of hTMEM59L increased protein expression of GDI1 and GDI2 but did not change the BACE2 protein level. These results suggest that TMEM59L affects the maturation and shedding of target proteins by mediating their transport. Based on the existing knowledge, we hope to further study the function of the proteins that may involve mediating proteins maturation.
cDNA cloning and construction of plasmid harboring cTMEM59L gene
Full-length hTMEM59L cDNA was amplified from the human brain cDNA library (BioDev) and was cloned into pEGFP-N2 between the EcoRI and SmaI sites using the primers 5’-GTCGAATTCACCATGGCTGCGGTGGCGCT-3’ and 5’-GCACCCGGGCAGCTTGGTCAGGTCCAGC-3’ to generate a plasmid that expressed hTMEM59L-enhanced green fluorescent protein (EGFP). The hTMEM59L fragment was amplified from a U251 cDNA library and sequenced. To generate the TMEM59L mutant with a C-terminal deletion (cTMEM59L) and then cloned in pEGFP-N2 expression vector to generate a plasmid expressing a cTMEM59L-EGFP fusion protein was preserved in the laboratory. Full-length mouse TMEM59L (mTMEM59L) was PCR-amplified from a mouse brain cDNA library and cloned in pEGFP-N2 between the EcoRI and BamHI restriction sites using primers 5’-GTCGAATTCACCATGGCTGCGGTAGCATTGC-3’ and 5’-GCAGGATCCGCAGCGTGGTCAAATCAAGC-3’. Human APP was PCR amplified using the primers 5’-GACAAGCTTATGCTGCCCGGTTTGG-3’ and 5’-GATCCCGGGGTTCTGCATCTGCTCAAAGAACT-3’ and cloned into pEGFP-N2 at the HindIII and SmaI restriction sites. Human GDI1 was PCR amplified using the primers 5’-GATAAGCTTATGGACGAGGAATACGAT-3’ and 5’-GTAGGATCCGCTGCTCAGCTTCTCCAAAG-3’ and cloned into pEGFP-N2 at the HindIII and BamHI sites. Human GDI2 was PCR amplified using the primers 5’-GATCTCGAGATGAATGAGGAGTACGAC-3’ and 5’-GTAGGATCCGGTCTTCCCCATAGATGTCA-3’ and cloned into pEGFP-N2 between the Xho I and BamHI sites. Human BACE2 was PCR amplified using the primers 5’-GATCTCGAGATGGGCGCACTGGC-3’ and 5’-GTAGGATCCGTTTCCAGCGATGTCTGACC-3’ and cloned into pEGFP-N2 at the Xho I and BamHI sites. In addition, hTMEM59L and mTMEM59L were also cloned into pcDNA3.1myc/his(A). hTMEM59L was additionally cloned into pDsRed-Express-N1. PCR products digested with two enzymes were purified using the AxyprepTM PCR Cleanup kit (AxyGEN, USA). RNA was isolated using the RNAisoTM Plus kit according to the manufacturer’s specifications. The RNA concentration was determined by measuring the absorbance ratio at 260/280 nm using a spectrophotometer (Eppendorf, Hamburg, Germany). cDNA was synthesized by reverse-transcribing 5 µg of total RNA using ReverTra Ace, and oligo (dT) were used for first-stand synthesis following the manufacturer’s instructions. Then DNase I was added to the system and incubated at 37°C for 30 min and stored at −80°C until used.
U251 and U87 human glioma cells, human embryonic kidney (HEK) 293T cells, 4T1 mouse mammary tumor cells, and U87 human glioma cells were purchased from the Chinese Academy of Sciences (Shanghai, China). U251, HEK293T, and U87 cells were cultured in high-glucose Dulbecco’s modified Eagle’s medium (DMEM, Gibco) with 10% fetal calf serum (FCS, Gibco). 4T1 cells were cultured in Roswell Park Memorial Institute-1640 (Gibco) medium with 10% FCS. The C17.2 mouse neural stem cell (NSC) line was cultured in high-glucose DMEM containing 10% FCS and 5% horse serum (Gibco). All cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C. U251, HEK293T, 4T1, U87, and C17.2 cells were transiently transfected with respective plasmid constructs using Lipofectamine 2000 reagent (Invitrogen) when they reached 60%–70% confluency. Cells were exposed to transfection complexes for 4 h in a serum-free medium, and then a normal culture medium was added, in accordance with the manufacturer’s instructions.
Analyses of subcellular localization of the proteins
Twenty-four hours after transfection of the vectors containing EGFP-tagged TMEM59L, cells were washed three times with phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde for 30 min, and then washed again with PBS. U251 cells were also transfected with vTMEM59L and cTMEM59L. Nuclei were stained with DAPI for 5 min at room temperature. Golgi-Tracker Red, Lyso-Tracker Red, and Mito-Tracker Green (Beyotime Biotechnology Company, China) were used for staining. U251 and C17.2 cells were transiently transfected with hTMEM59L-EGFP for 24 h and then stained with Golgi-Tracker Red or Lyso-Tracker Red to label the Golgi apparatus and acidic organelles, respectively. U251 cells were transiently transfected with hTMEM59L-DsRed for 24 h and then stained with Mito-Tracker Green to stain the mitochondria.
To analyze the dynamics of TMEM59L in live cells, fluorescence was observed at 48 h after transfection of plasmids using a Nikon Eclipse TE2000-U fluorescence microscope. Images were acquired using a SPOT CCD digital camera.
HEK293T cells were co-transfected with EGFP-APP, EGFP-GDI1, EGFP-GDI2, EGFP-BACE2, together with pcDNA3.1myc/his(A), pcDNA3.1myc/his(A)-hTMEM59L, or pcDNA3.1myc/his(A)-mTMEM59L. Cells were harvested by scraping, washed with cold PBST (PBS containing 0.5% Tween 20), and incubated in lysis buffer according to the manufacturer’s instructions (KeyGEN BioTECH, Nanjing, China). Samples were mixed with protein loading buffer, boiled for 5 min, electrophoresed in 12% sodium dodecyl sulfate-polyacrylamide gels, and transblotted to cellulose nitrate membranes (Whatman, Maidstone, Kent, UK) for 1 h at 25 V. Membranes were blocked with PBST containing 1% bovine serum albumin at room temperature and then incubated with an anti-GFP, anti-myc, or anti-actin antibody (1:2000; Santa Cruz) for 2 h at room temperature. Thereafter, membranes were rinsed, incubated with a secondary antibody conjugated with a far-infrared dye (1:12000; LI-COR, Lincoln, NE, USA) for 1 h, and scanned using an Odyssey scanner (LI-COR).
Localization of TMEM59L to vesicular structures
U251, HEK293T, C17.2, 4T1, and U87 cells were transfected with a plasmid harboring hTMEM59L-EGFP. The pEGFP-N2 plasmid was used as a negative control. hTMEM59L localized to vesicular structures in the cytoplasm and perinuclear region but was not detected in the nucleus (Fig. 1).
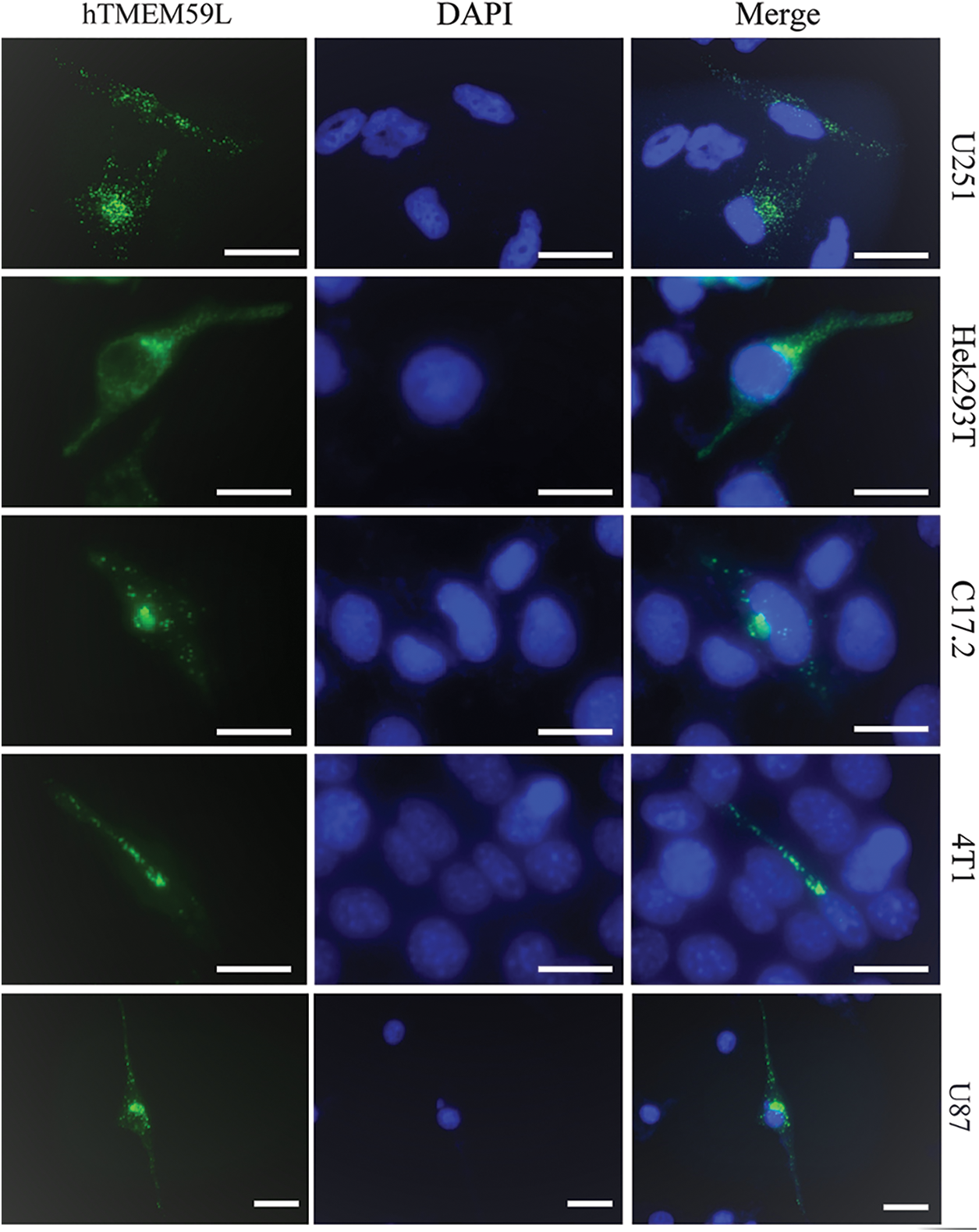
Figure 1: Intracellular localization of the hTMEM59L in different cells. U251, Hek293T, C17.2, 4T1, and U87 cells were transiently transfected with pEGFP-N2-hTMEM59L. After 24 h, the cells were observed under the fluorescence microscope, and photographs were taken. Scale bars = 20 μm. The data show the localization of hTMEM59L to vesicular structures in the cytoplasm and the perinuclear region.
Deletion of the middle or C-terminal region of hTMEM59L affects its localization
Twenty colony clones of full-length hTMEM59L were rescreened by PCR, one of which was a splice variant comprising exons 1, 2, 3, and 8 of hTMEM59L (named vTMEM59L). vTMEM59L contains 534 nucleotides and encodes a protein of 178 amino acids, meaning that a middle region of the hTMEM59L gene was deleted (Supplement Fig. 1. To determine which regions of hTMEM59L are functionally important, we deleted the C-terminal region to generate cTMEM59L. This mutant is 808 nucleotides long and encodes a protein of 269 amino acids (Supplement Fig. 2). Fluorescence was detected in U251 cells transfected with vTMEM59L, and cTMEM59L, indicating the expression of proteins; however, the fluorescence intensity was markedly lower than in cells transfected with hTMEM59L. More importantly, the number of vesicular structures was markedly lower in cells transfected with vTMEM59L and cTMEM59L than in cells transfected with hTMEM59L, illustrating that the middle and C-terminal regions of hTMEM59L are crucial for its expression and localization (Fig. 2).
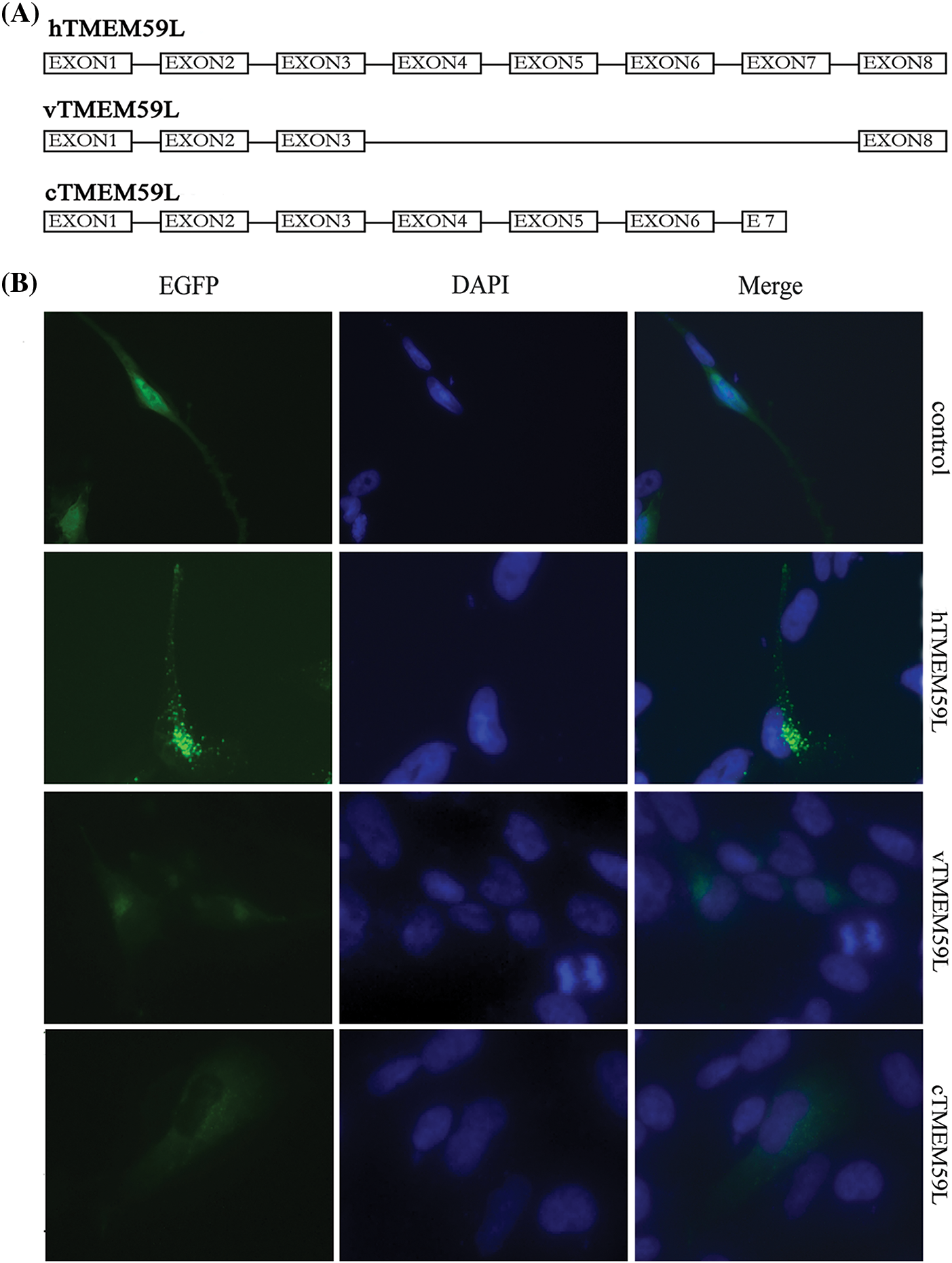
Figure 2: Localization of vTMEM59L and cTMEM59L in U251 cells. (A) Sequence of three different hTMEM59L forms: hTMEM59L, full-length hTMEM59L; vTMEM59L, a novel alternative splicing isoform of hTMEM59L; cTMEM59L, C-terminal deletion mutant of hTMEM59L. (B) U251 cells were transiently transfected with pEGFP-N2-hTMEM59L, pEGFP-N2-vTMEM59L, and pEGFP-N2-cTMEM59L. After 24 h, the cells were observed under a fluorescence microscope, compared to hTMEM59L, vTMEM59L and cTMEM59L, mainly localized in the perinuclear region, and the vesicular structures significantly decreased.
The role of hTMEM59L in intracellular vesicle movement
The subcellular localization of hTMEM59L in U251 and C17.2 cells was further assessed by staining cells with Golgi-Tracker Red and Lyso-Tracker Red dyes and observed that hTMEM59L mainly localized in lysosomes and acidic vesicular structures. Dynamic punctuated fluorescent signals were observed in the neurites and cytoplasm of living U251 cells transiently transfected with the plasmid harboring hTMEM59L-EGFP (Fig. 3).
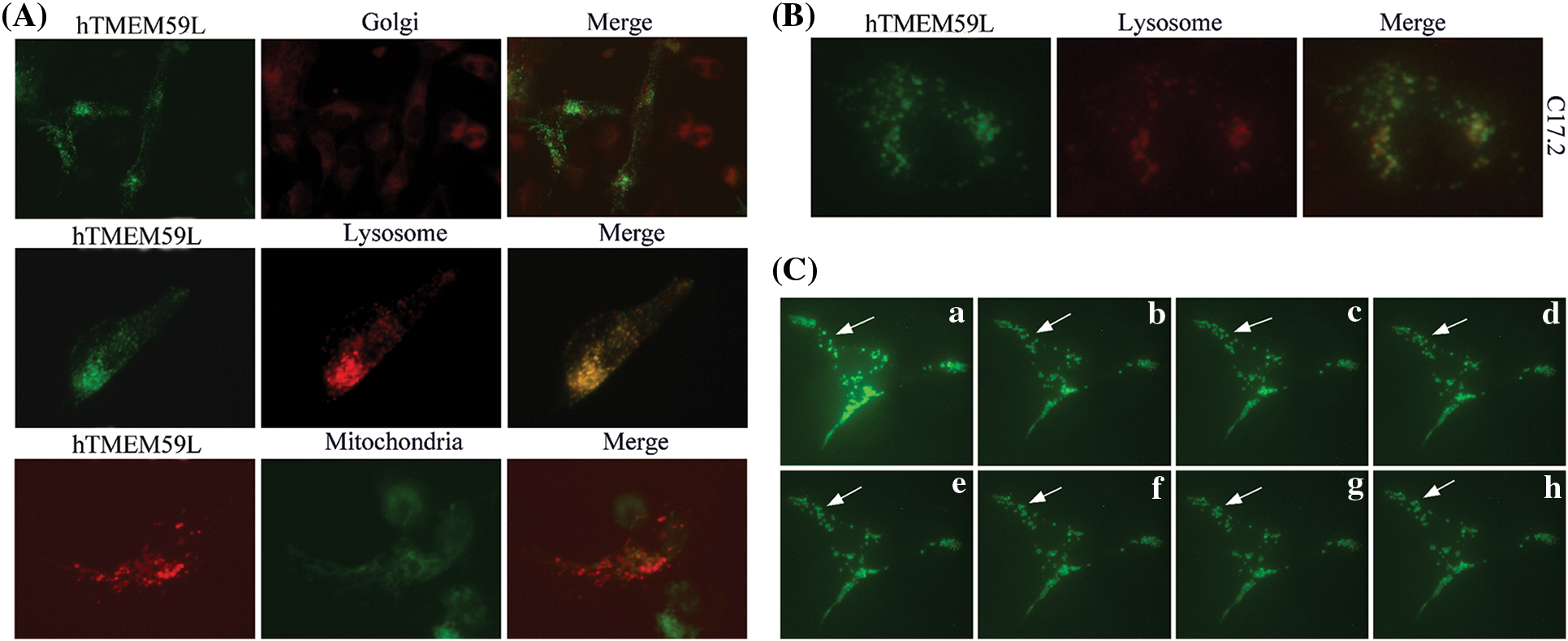
Figure 3: Subcellular localization of the hTMEM59L in U251 cells. (A) hTMEM59L-EGFP and hTMEM59L-DsRed fusion protein were transiently expressed in U251 cells. Golgi-Tracker Red, Lyso-Tracker Red, and Mito-Tracker Green staining are shown in parallel. All merged pictures are shown in the rightmost panels. (B) The hTMEM59L-EGFP fusion protein was transiently expressed in C17.2 cells. Lyso-Tracker Red staining is shown in parallel. Similar to (A), partial co-localization of hTMEM59L in the lysosome and acidic vesicular structures was observed. (C) (a–h) Images of living C17.2 cells expressing hTMEM59L-EGFP following a specific interval. The arrow points to punctuated fluorescent signals in the neurite and the cytoplasm, whereas the fluorescent signal also appeared dense and clustered in perinuclear regions.
TMEM59L-mediated increase in GDI1 and GDI2 protein levels, but no effect was seen on BACE2
In control transfected HEK293T cells, EGFP-APP fluorescence was observed in the cytoplasm and the Golgi bodies. In contrast, in cells transfected with TMEM59L, EGFP-APP fluorescence in the Golgi bodies significantly decreased. Further co-localization analysis showed significant localization of APP and TMEM59L to Golgi bodies (Fig. 4). HEK293T cells were transiently co-transfected with GDI1 and TMEM59L, and a significantly enhanced fluorescence intensity was observed and the cells appeared more spherical. Co-localization studies showed that GDI1 distribution was not significantly affected by TMEM59L (Fig. 5). However, western blotting demonstrated that overexpression of TMEM59L markedly increased the level of APP protein. Overexpression of TMEM59L protein further increased GDI1 and GDI2 levels but did not affect BACE2 levels (Figs. 6, 7, Supplement Fig. 3).
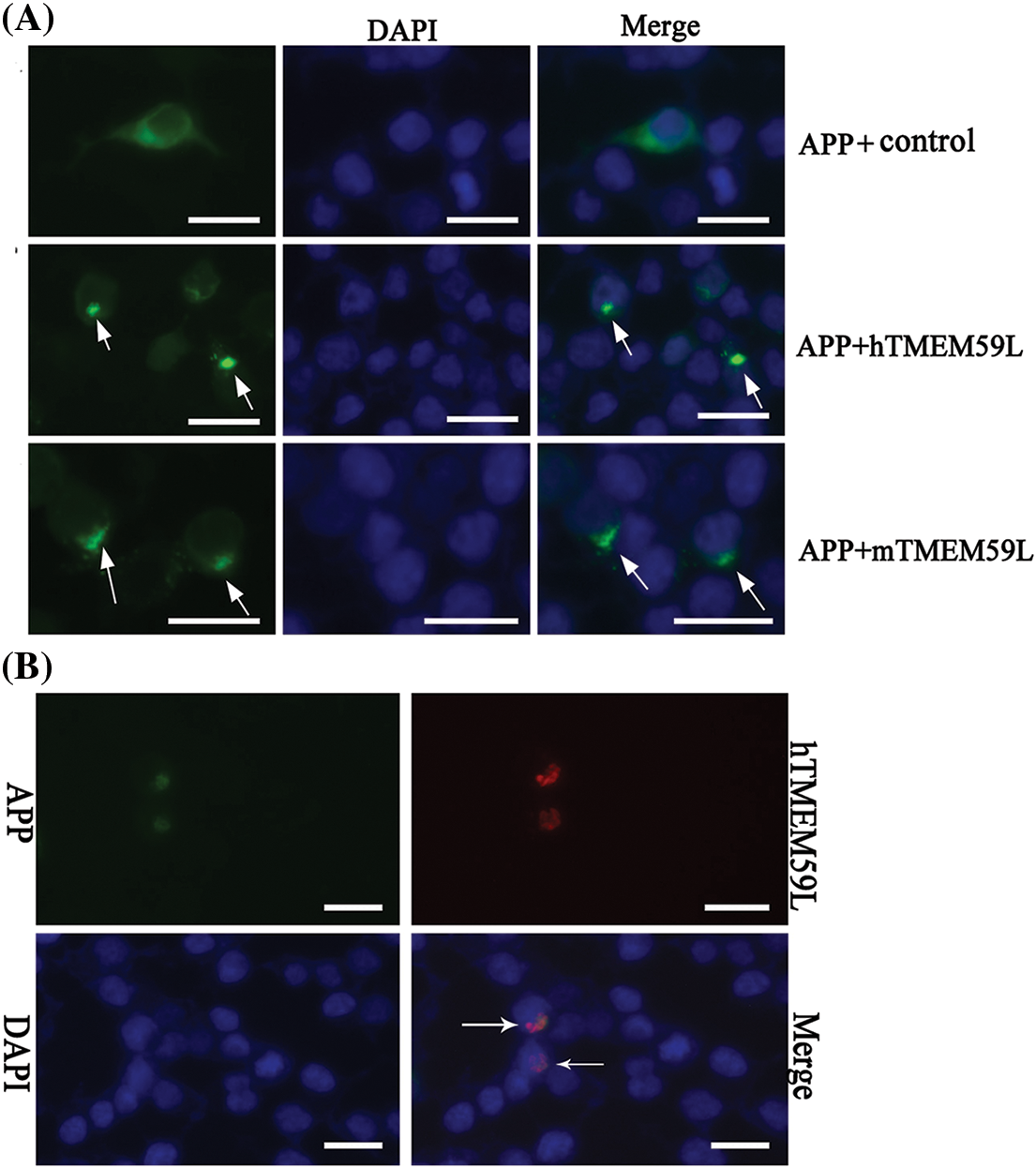
Figure 4: TMEM59L inhibited the transport and increased the production of APP in HEK293T cells. (A) HEK293T cells were transiently co-transfected with EGFP-APP and either the control plasmid or hTMEM59L/mTMEM59L. The arrows indicate the APP, with significant retention in Golgi bodies. (B) Co-localization of APP and human TMEM59L following the expression of APP-EGFP and human TMEM59L-DsRed. The arrows indicate the co-localization of APP and TMEM59L. Scale bars = 20 μm. APP, amyloid precursor protein.
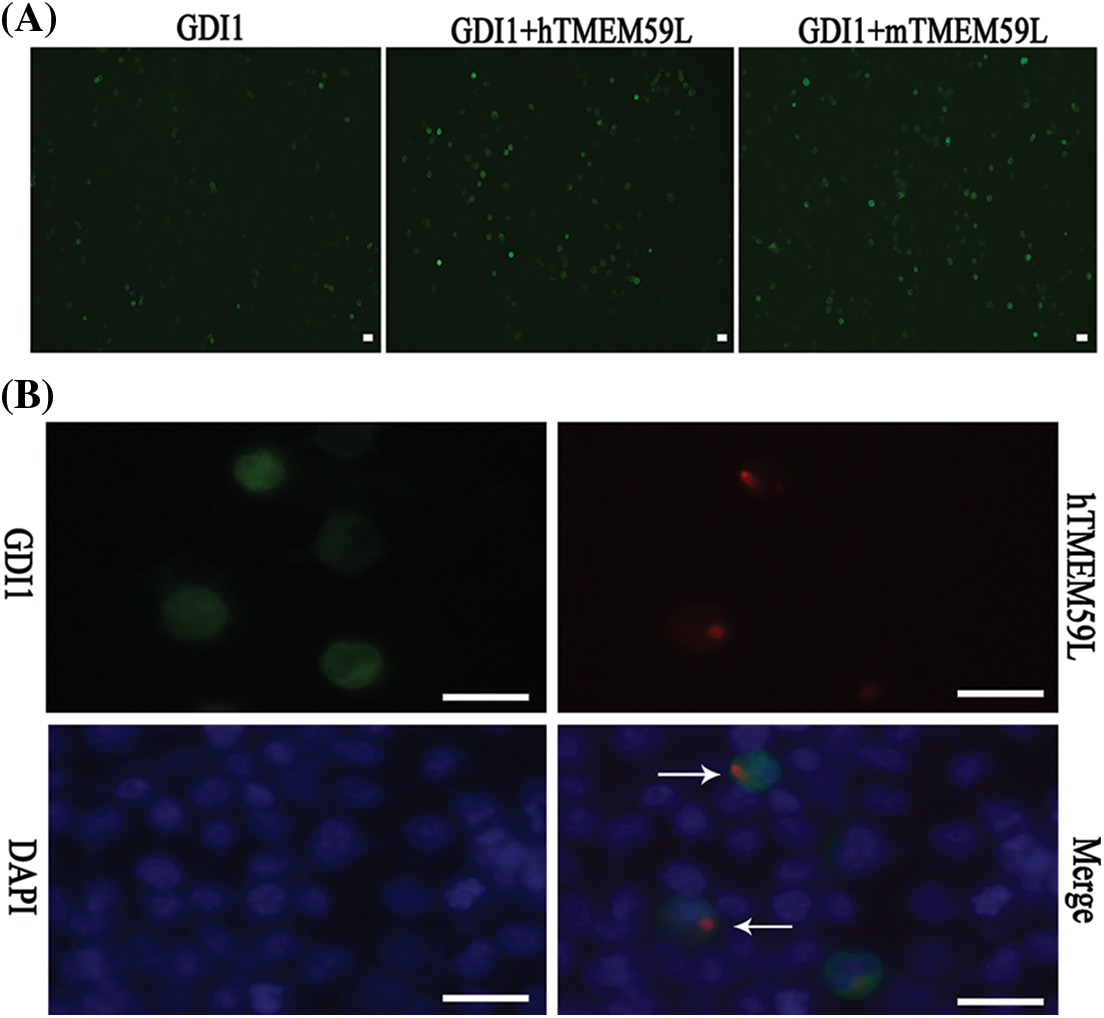
Figure 5: TMEM59L increased the expression of GDI1. (A) EGFP-GDI1 and either control plasmid or hTMEM59L/mTMEM59L were transiently co-transfected in HEK293T cells, and after TMEM59L overexpression, cells tend to be more spherical. (B) GDI1-EGFP and human TMEM59L-DsRed were transiently co-transfected in HEK293T cells that showed TMEM59L-induced GDI1 accumulation in the cytoplasm. However, the distribution of GDI1 did not change significantly due to TMEM59L. Scale bars = 20 μm. GDI1, Rab GDP dissociation inhibitor alpha.
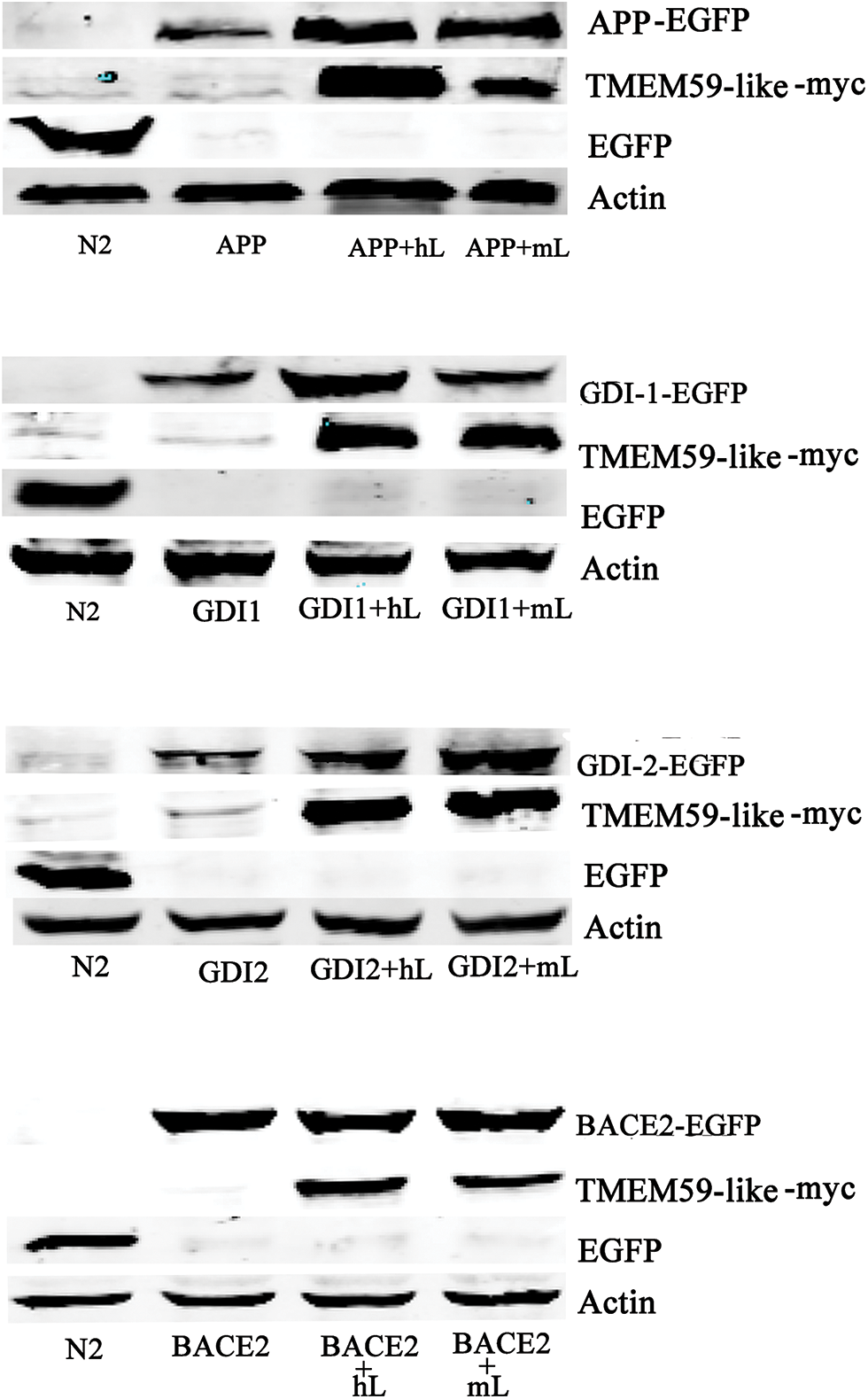
Figure 6: TMEM59L increased GDI1 and GDI2 protein expressions. Target proteins and either control plasmid or hTMEM59L/mTMEM59L were transiently co-transfected in HEK293T cells; enhanced APP, GDI1, and GDI2 expression and no change in BACE2 expression due to TMEM59L overexpression. GDI1, Rab GDP dissociation inhibitor alpha; GDI2, Rab GDP dissociation inhibitor beta.
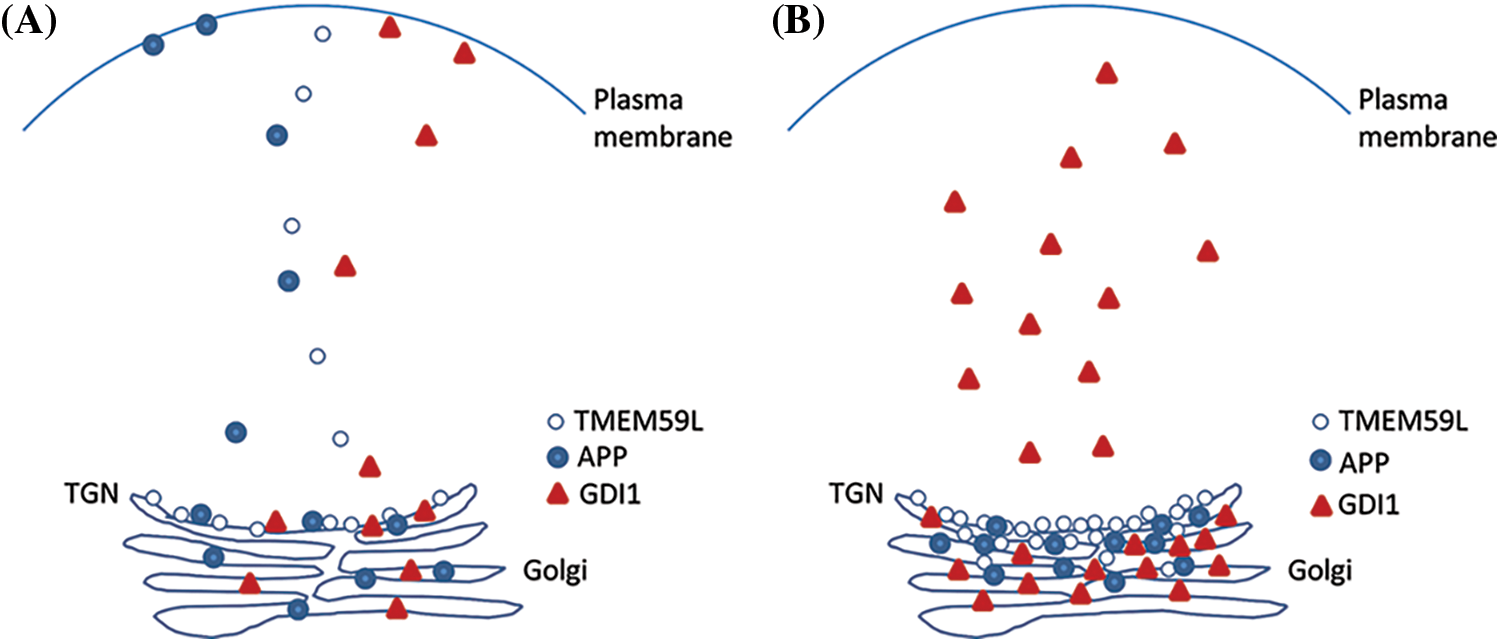
Figure 7: TMEM59L plays an important function in the expression of GDI1 protein. Under normal conditions that TMEM59L, APP and GDI1 involved assemble into vesicular structures and distribution to cytoplasm. Furthermore, APP and GDI1 also can localization to the cell membrane, overexpression of TMEM59L promote APP rention to Golgi may be duo to GDI1 mianly accumulate to Golgi and cytoplasm.
Only a few studies have investigated the functions of TMEM59L. We previously found that TMEM59L has a distant homolog called TMEM59 (transmembrane protein 59, also known as DCF1) and that upregulation of TMEM59 maintains NSCs in an undifferentiated state, whereas these cells tend to differentiate into neurons and astrocytes when TMEM59 is silenced using small interfering RNA (Wang et al., 2008). TMEM59 is a membrane-bound glycoprotein that localizes to the Golgi apparatus. Overexpression of TMEM59 inhibits glycosylation of APP in the Golgi apparatus and blocks cleavage of APP by α- and β-secretases, thereby inhibiting the generation of Aβ peptides, which form amyloid plaques in AD; however, the precise function of TMEM59 is still not known (Mao et al., 1998; Ullrich et al., 2010). In a recent study, TMEM59 was found to contribute to AD based on the identification of TMEM59 regulators during the differentiation of mouse NSCs from a compendium of expression profiles (Zhang et al., 2011). All these findings provide clues regarding the function of TMEM59L. Characterization of transmembrane proteins indicates that those with an N-terminal signal sequence localize to internal cell membranes, secretory vesicles, and the plasma membrane (Kveine et al., 2002). TMEM59L localizes to the Golgi apparatus and inhibits the maturation and shedding of APP (Ullrich et al., 2010). Our results confirmed the effect of TMEM59L on APP. However, it is unclear whether TMEM59L participates in the formation of specialized vesicles during the processing of APP. In mammalian cells, the major acidic organelles are the trans-Golgi network (TGN), lysosomes, and recycling endosomes. The current study demonstrated a punctate distribution of hTMEM59L in the perinuclear region due to its accumulation in the Golgi apparatus and rapidly moved between the Golgi apparatus and plasma membrane. However, we cannot rule out the possibility that TMEM59L also localizes to other vesicular structures. Newly synthesized plasma membrane proteins are sorted in the TGN and endosomal compartments (Perret et al., 2005). The TGN is a well-defined acidic compartment in the secretory pathway. Aβ is suggested to form in the Golgi/TGN rather than in post-TGN vesicles (Xu et al., 1997). The current study demonstrated that vTMEM59L and cTMEM59L, which lacked a middle and C-terminal regions, respectively, did not localize correctly and consequently markedly decreased the number of vesicular structures. These results indicate that the middle and C-terminal regions of TMEM59L are required for its correct localization and expression. GDIs play an important role in membrane trafficking and movement of functional proteins to vesicular pools following detachment from target membranes (Garrett et al., 1994). GDI1 and GDI2 are related to brain development and protein trafficking. GDI1 is involved in the regulation of Rab GTPases, and mutation of this protein is associated with X-linked intellectual disability (Strobl-Wildemann et al., 2011). We evaluated the relationship between TMEM59L and Rab GDIs. Overexpression of TMEM59L increased the levels of GDI1 and GDI2 proteins, indicating that TMEM59L affects these two GDIs and thereby modulates cytoskeletal transport of protein. By contrast, overexpression of TMEM59L did not markedly affect BACE2 expression. Therefore, the function of TMEM59L must be investigated further.
In conclusion, our results provide insight into the function of TMEM59L. We confirmed that TMEM59L is closely associated with a variety of membranous structures such as Golgi apparatus, lysosome, endosomes, and transport vesicles in several cell lines. Further, the overexpression of TMEM59L increases the expression of GDI1 and GDI2 proteins but decreases that of BACE2. Taken together, the results of this study suggest that TMEM59L mediates the transport and processing of proteins via modulating GDIs.
Acknowledgement: The C17.2 mouse neural stem cell (NSC) line was provided by Dr. Evan Y. Snyder.
Availability of Data and Materials:All data generated or analyzed during this study are included in this published article.
Author Contribution:The authors confirm contribution to the paper as follows: study conception and design: Tieqiao Wen; data collection: Haifeng Wang; analysis and interpretation of results: Haifeng Wang, Juan Guo; draft manuscript preparation: Haifeng Wang. All authors reviewed the results and approved the final version of the manuscript.
Ethics Approval: Not applicable.
Funding Statement:This work was supported by the Science and Technology Development Plan of Kaifeng in 2020 (2003048) and the National Natural Science Foundation of China (Grant No. 31070954).
Conflicts of Interest:The authors declare that they have no conflicts of interest to report regarding the present study.
References
Bächner D, Sedlacek Z, Korn B, Hameister H, Poustka A (1995). Expression patterns of two human genes coding for different rab GDP-dissociation inhibitors (GDIsextremely conserved proteins involved in cellular transport. Human Molecular Genetics 4: 701–708. DOI 10.1093/hmg/4.4.701. [Google Scholar] [CrossRef]
Garrett MD, Zahner JE, Cheney CM, Novick PJ (1994). GDI1 encodes a GDP dissociation inhibitor that plays an essential role in the yeast secretory pathway. EMBO Journal 13: 1718–1728. DOI 10.1002/j.1460-2075.1994.tb06436.x. [Google Scholar] [CrossRef]
Goldsbury C, Mocanu MM, Thies E, Kaether C, Haass C et al. (2010). Inhibition of APP trafficking by tau protein does not increase the generation of amyloid-beta peptides. Traffic 7: 873–888. DOI 10.1111/j.1600-0854.2006.00434.x. [Google Scholar] [CrossRef]
Huse JT, Liu K, Pijak DS, Carlin D, Lee VM et al. (2002). Beta-secretase processing in the trans-Golgi network preferentially generates truncated amyloid species that accumulate in Alzheimer’s disease brain. Journal of Biological Chemistry 277: 16278–16284. DOI 10.1074/jbc.M111141200. [Google Scholar] [CrossRef]
Hussain I, Powell DJ, Howlett DR, Chapman GA, Gilmour L et al. (2000). ASP1 (BACE2) cleaves the amyloid precursor protein at the beta-secretase site. Molecular & Cellular Neuroscience 16: 609–619. DOI 10.1006/mcne.2000.0884. [Google Scholar] [CrossRef]
Lee J, Retamal C, Cuitiño L, Caruano-Yzermans A, Shin JE et al. (2008). Adaptor protein sorting nexin 17 regulates amyloid precursor protein trafficking and processing in the early endosomes. Journal of Biological Chemistry 283: 11501–11508. DOI 10.1074/jbc.M800642200. [Google Scholar] [CrossRef]
Jin LW, Shie FS, Maezawa I, Vincent I, Bird T (2004). Intracellular accumulation of amyloidogenic fragments of amyloid-beta precursor protein in neurons with Niemann-Pick type C defects is associated with endosomal abnormalities. American Journal of Pathology 164: 975–985. DOI 10.1016/S0002-9440(10)63185-9. [Google Scholar] [CrossRef]
Kim SH, Leem JY, Lah JJ, Slunt HH, Levey AI et al. (2001). Multiple effects of aspartate mutant presenilin 1 on the processing and trafficking of amyloid precursor protein. Journal of Biological Chemistry 276: 43343–43350. DOI 10.1074/jbc.M108245200. [Google Scholar] [CrossRef]
Koo EH, Squazzo SL (1994). Evidence that production and release of amyloid beta-protein involves the endocytic pathway. Journal of Biological Chemistry 269: 17386–17389. DOI 10.1016/S0021-9258(17)32449-3. [Google Scholar] [CrossRef]
Kveine M, Tenstad E, Døsen G, Funderud S, Rian E (2002). Characterization of the novel human transmembrane protein 9 (TMEM9) that localizes to lysosomes and late endosomes. Biochemical & Biophysical Research Communications 297: 912–917. DOI 10.1016/S0006-291X(02)02228-3. [Google Scholar] [CrossRef]
Mao M, Fu G, Wu JS, Zhang QH, Zhou J et al. (1998). Identification of genes expressed in human CD34+ hematopoietic stem/progenitor cells by expressed sequence tags and efficient full-length cDNA cloning. PNAS 95: 8175–8180. DOI 10.1073/pnas.95.14.8175. [Google Scholar] [CrossRef]
Perret E, Lakkaraju A, Deborde S, Schreiner R, Rodriguez-Boulan E (2005). Evolving endosomes: How many varieties and why? Current Opinion in Cell Biology 17: 423–434. DOI 10.1016/j.ceb.2005.06.008. [Google Scholar] [CrossRef]
Steinhilb ML, Turner RS, Gaut JR (2010). ELISA analysis of β-secretase cleavage of the Swedish amyloid precursor protein in the secretory and endocytic pathways. Journal of Neurochemistry 80: 1019–1028. DOI 10.1046/j.0022-3042.2002.00764.x. [Google Scholar] [CrossRef]
Strobl-Wildemann G, Kalscheuer VM, Hu H, Wrogemann K, Ropers HH et al. (2011). Novel GDI1 mutation in a large family with nonsyndromic X-linked intellectual disability. American Journal of Medical Genetics Part A 155: 3067–3070. DOI 10.1002/ajmg.a.34291. [Google Scholar] [CrossRef]
Szodorai A, Kuan YH, Hunzelmann S, Engel U, Sakane A et al. (2009). APP anterograde transport requires Rab3A GTPase activity for assembly of the transport vesicle. Journal of Neuroscience 29: 14534–14544. DOI 10.1523/JNEUROSCI.1546-09.2009. [Google Scholar] [CrossRef]
Ullrich S, Münch A, Neumann S, Kremmer E, Tatzelt J et al. (2010). The novel membrane protein TMEM59 modulates complex glycosylation, cell surface expression, and secretion of the amyloid precursor protein. Journal of Biological Chemistry 285: 20664–20674. DOI 10.1074/jbc.M109.055608. [Google Scholar] [CrossRef]
Wang L, Wang J, Wu Y, Wu J, Pang S et al. (2008). A novel function of dcf1 during the differentiation of neural stem cells in vitro. Cellular & Molecular Neurobiology 28: 887–894. DOI 10.1007/s10571-008-9266-1. [Google Scholar] [CrossRef]
Xu H, Sweeney D, Wang R, Thinakaran G, Sisodia SS et al. (1997). Generation of alzheimer β-amyloid protein in the trans-golgi network in the apparent absence of vesicle formation. PNAS 94: 3748–3752. DOI 10.1073/pnas.94.8.3748. [Google Scholar] [CrossRef]
Yoon IS, Chen E, Busse T, Repetto E, Lakshmana MK et al. (2007). Low-density lipoprotein receptor-related protein promotes amyloid precursor protein trafficking to lipid rafts in the endocytic pathway. FASEB 21: 2742–2752. DOI 10.1096/fj.07-8114com. [Google Scholar] [CrossRef]
Zhang L, Ju X, Cheng Y, Guo X, Wen T (2011). IdentifyingTmem59related gene regulatory network of mouse neural stem cell from a compendium of expression profiles. BMC Systems Biology 5: 152. DOI 10.1186/1752-0509-5-152. [Google Scholar] [CrossRef]

SUPPLEMENT FIGURE 1: Alignment of hTMEM59L and vTMEM59L amino acid sequences.

SUPPLEMENT FIGURE 2: Alignment of hTMEM59L and cTMEM59L amino acid sequences.
SUPPLEMENT FIGURE 3: Crude gel images of the western blot assay.
Cite This Article
 Copyright © 2022 The Author(s). Published by Tech Science Press.
Copyright © 2022 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools