 Open Access
Open Access
ARTICLE
Hsa_circ_0036740 in familial adenomatous polyposis: Immune regulation and neutrophil effects in CRC based on high-throughput assay
Department of Gastrointestinal Surgery, Meizhou People’s Hospital, Meizhou, 514031, China
* Corresponding Author: EN LI. Email:
(This article belongs to the Special Issue: Bioinformatics Study of Diseases)
BIOCELL 2023, 47(11), 2409-2422. https://doi.org/10.32604/biocell.2023.031186
Received 19 May 2023; Accepted 21 June 2023; Issue published 27 November 2023
Abstract
Familial adenomatous polyposis (FAP) is an autosomal dominant disease with a high probability of becoming cancerous. Many RNAs potentially associated with FAP have not been identified. In this study, a circRNA (circular RNA) expression profile of FAP was established using a circRNA microarray, and differentially expressed circRNAs were verified by RT-qPCR. The effects of hsa_circ_0036740 on the malignant behavior of tumor cells (proliferation, apoptosis, and epithelial mesenchymal transition) and the levels of C3A complement protein expression were evaluated. Moreover, neutrophils were isolated and co-cultured with colorectal cancer cells (CRCs), followed by measurements of MPO-DNA, citrullinated histone H3, interleukin (IL)-1β, IL-6, and IL-8 levels. Nuclear translocation of arginine deiminase 4 (PAD4) was observed using immunofluorescence assays. Based on the high-throughput assay, 238 downregulated circRNAs, and 38 upregulated circRNAs were identified. A Kyoto Encyclopedia of Genes and Genomes enrichment analysis suggested that immune regulation might be involved in FAP. A total of 10 DECs (differentially expressed circular RNAs) were identified by RT-qPCR, and among them, hsa_circ_0036740 showed the highest fold-change in upregulation. Results of gain-of- and loss-of-function studies revealed that hsa_circ_0036740 enhanced the malignant behavior of tumor cells, such as metastasis, proliferation, and apoptosis, with an increasing level of C3A complement. Moreover, hsa_circ_0036740 also significantly increased neutrophil extracellular trap formation and inflammation in neutrophils, as shown by an increased expression of PAD4. In conclusion, this study revealed the expression profiles of circRNAs in FAP and confirmed the possible involvement of hsa_circ_0036740 in the immune regulation mediated by neutrophils. Finally, hsa_circ_0036740 was suggested as a new therapeutic target for CRC.Graphic Abstract
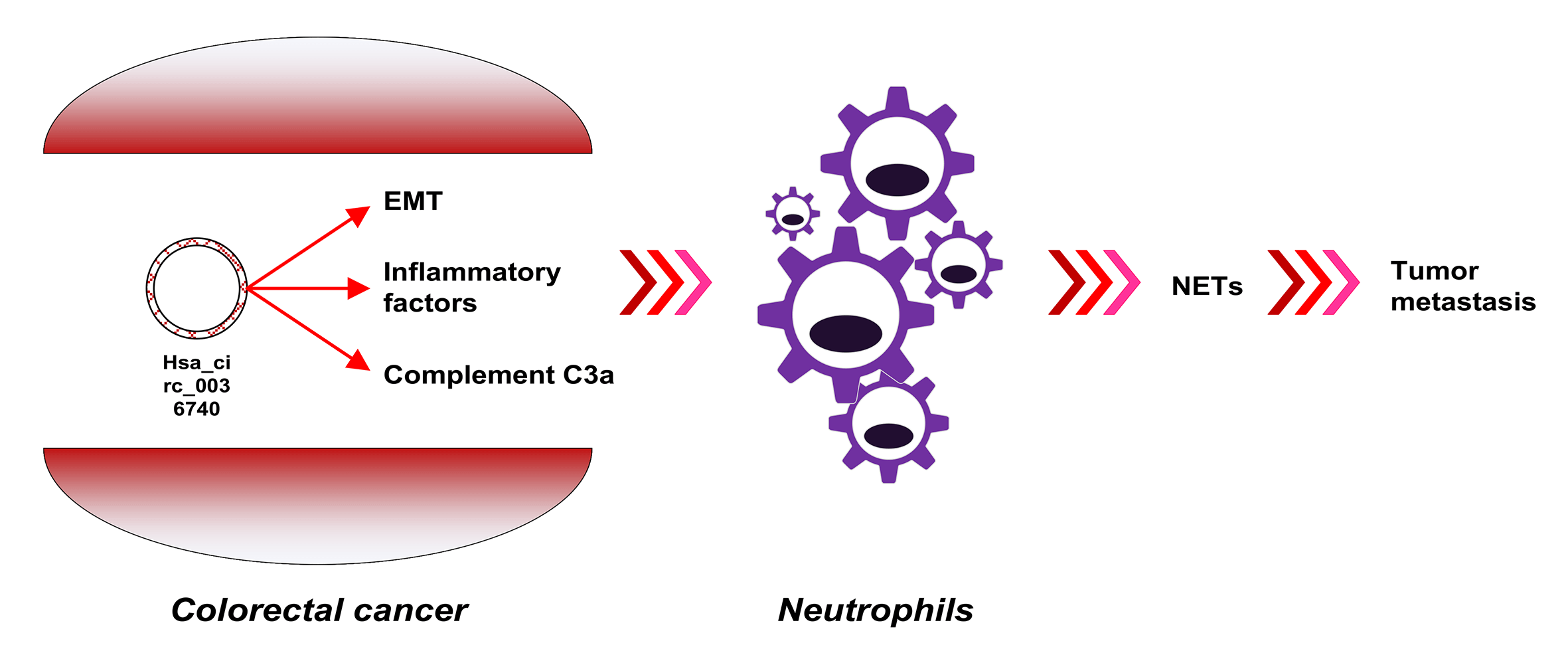
Keywords
Supplementary Material
Supplementary Material FileFamilial adenomatous polyposis (FAP) is an autosomal dominant disorder that results from a germline mutation in the adenomatous polyposis coli (APC) gene on chromosome 5 (Bhurwal et al., 2020). Its typical clinical presentation is the appearance of hundreds of adenomatous polyps in the colorectal mucosa starting from adolescence (Dinarvand et al., 2019). Without effective medical intervention, the probability of FAP becoming cancerous is almost 100% (Kemp Bohan et al., 2021). FAP accounts for 0.2% to 1% of all colorectal cancers (CRCs), and CRC is the number one cause of death among FAP patients (Mirinezhad et al., 2019). In recent years, the number of CRC patients with FAP carcinogenesis has increased each year (Karstensen et al., 2019). Therefore, it is important to investigate the molecular mechanism of FAP carcinogenesis and improve the prognosis of FAP patients.
Circular RNAs (circRNAs) comprise a group of non-coding RNAs with a circular structure (Jakobi and Dieterich, 2019). CircRNAs are more resistant to degradation by exonucleases and have a more stable structure than linear RNAs (Huang et al., 2020). CircRNAs have been reported to function via multiple mechanisms, such as regulating gene splicing or transcription, acting as microRNA (miRNA) sponges, interacting with proteins, or regulating epigenetic mechanisms (Kristensen et al., 2019; Li et al., 2020). Furthermore, circRNAs can also regulate the expression of upstream genes or other genes (Zheng et al., 2020). Current studies have confirmed that circRNAs are specifically expressed in multiple malignancies, including CRC, and are involved in cancer progression (Zhang et al., 2021a). However, the differentially expressed circRNAs associated with FAP have not been reported.
Neutrophils are white blood cells that make up the largest proportion of peripheral blood (Wu et al., 2020). When local tissues are attacked by pathogens, neutrophils cross the vessel wall into the inflamed tissue, where they accumulate due to the action of chemokines and other factors (Silvestre-Roig et al., 2020). Neutrophils can phagocytose and digest the invading pathogens and function as the first line of immune defense (Ravindran et al., 2019). A neutrophil extracellular trap (NET) is an extracellular fibrous meshwork comprising of a DNA backbone and various active proteins embedded in neutrophils (Castanheira and Kubes, 2019). NETs are mainly composed of circulating free DNA (cfDNA), myeloperoxidase (MPO), citrullinated histone H3 (CitH3), neutrophil elastase (NE), histones, and other substances (Tan et al., 2021). Protein arginine deiminase 4 (PAD4), as a key immune factor, can participate in the NET-mediated bacterial killing pathway (Rohrbach et al., 2012). Thus, MPO-DNA, CitH3, NE, and PAD4 can all be used to assess the numbers of NETs. In addition to their bactericidal role, NETs are also associated with a variety of diseases, such as immune disorders, blood clots, and tumors (Masucci et al., 2020). In recent years, several studies have proven that NETs are highly expressed in multiple tumors and that tumors and their microenvironments can promote the production of NETs (Papayannopoulos, 2018; Lee et al., 2019). Moreover, NETs are also involved in CRC processes, such as cell proliferation and metastasis, which provides a new idea for the clinical management of CRC (Demkow, 2021; Khan et al., 2021). However, the information on regulatory mechanisms of circRNAs and NETs in CRC are far from adequate.
Complement C3a is a small peptide molecule in the complement system that can be formed by non-enzymatic cleavage of C3 protein in the plasma. When complement C3a binds to its receptor (C3aR), it can stimulate and activate inflammatory and immune cells, thus mediating and regulating inflammatory responses and immune responses. A recent study discloses a significant correlation between the levels of C3a and lymph node metastasis in breast cancer (Hameed et al., 2022). Complement also promotes chronic inflammation, maintaining the immune-suppressive state of the microenvironment (Senent et al., 2021). We also noted that the complement system can regulate the immune microenvironment by intervening in neutrophil behavior, thereby playing a role in regulating tumor growth (Nabizadeh et al., 2016).
Therefore, in this study, we screened for circRNAs that were differentially expressed in FAP tissues by microarray to provide a theoretical basis for the therapy of FAP patients. Our data revealed that hsa_circ_0036740 expression was significantly upregulated in FAP tissues. We also investigated the function of hsa_circ_0036740 in CRC cells, including its role in NET formation. Our results may provide new potential targets for preventing FAP carcinogenesis.
Samples of FAP (n = 20) and matched peritumoral tissue (Pt) tissue (n = 20) were collected from patients with FAP via endoscopic resection at Meizhou People’s Hospital between March 2021 and November 2021. The above tissues were pathologically confirmed and stored in an ultra-low temperature (−80°C) freezer for later testing. All study subjects provided their signed informed consent. The study protocol was approved by the Ethics Committee of Meizhou People’s Hospital.
Total RNA was extracted from FAP and Pt tissues by using a Trizol kit (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. Each RNA extract was examined for purity and integrity, and linear RNA was removed by incubation with nucleic acid exonuclease (Epicenter, Biotechnologies, Madison, WI, USA). The Super RNA Labeling method (Arraystar, Rockville, MD, USA) was used for cDNA amplification and fluorescence labeling. The labeled probes were aligned and hybridized with Human circRNA Array (Arraystar, Rockville, MD, USA). The Cy3 fluorescence intensity of each specimen was scanned using an Agilent Microarray Scanner (G2505C), and the images were entered into Agilent Feature Extraction Software for grid alignment and data analysis. The R package was used to identify differentially expressed circRNAs (DECs). Volcano plots and a heatmap were drawn based on the DECs screened by the microarray. Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses were conducted via DAVID (https://david.ncifcrf.gov/) on the parental genes of the obtained circRNAs (Li et al., 2021; Shen et al., 2019; Klopfenstein et al., 2018).
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) analysis
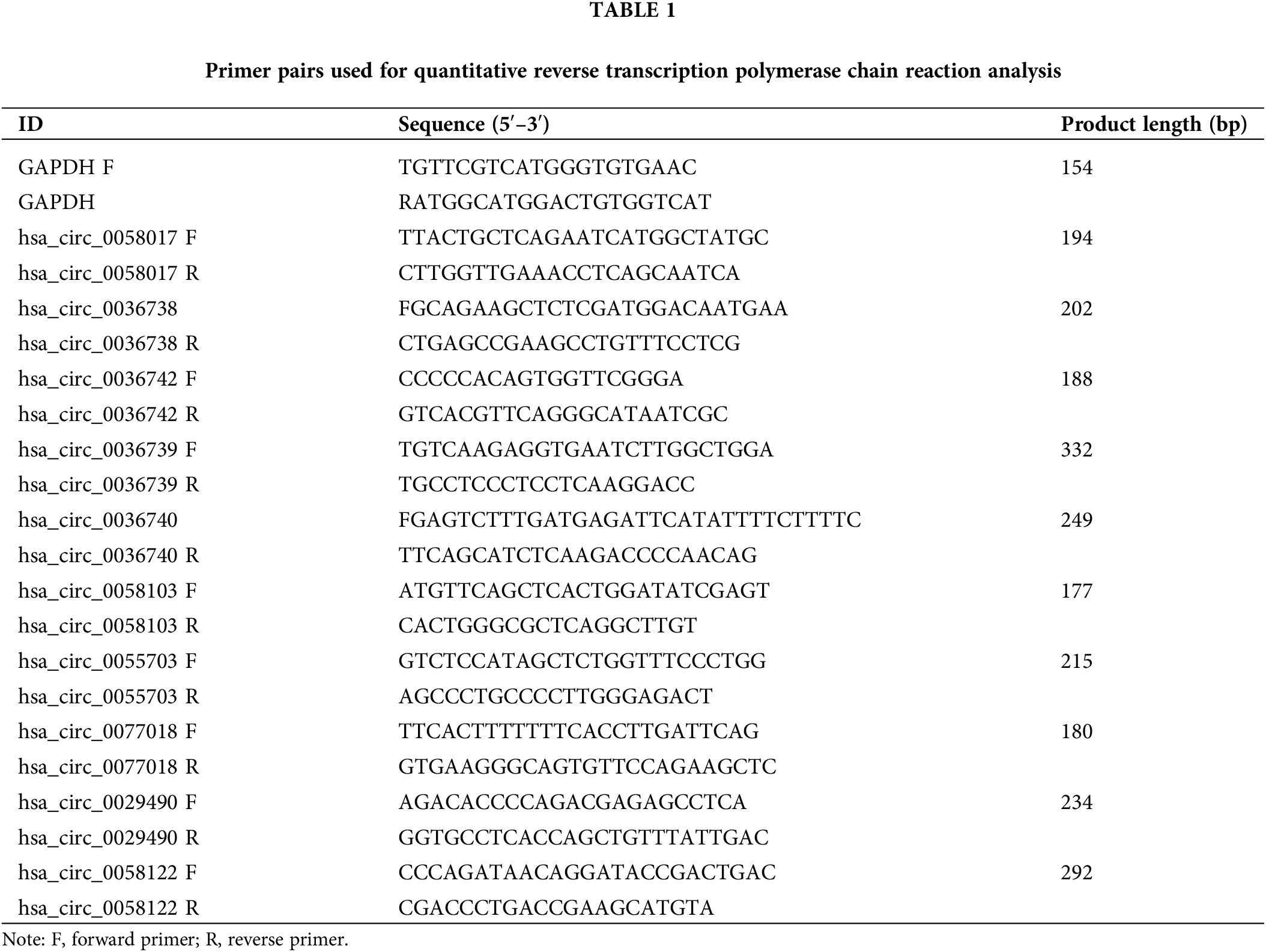
Total RNA was collected from each tissue after grinding, and reverse transcription was performed using a Prime ScriptRT reagent Kit (Takara, Shiga, Japan). RT-qPCR was performed with a SYBR® Premix Ex Taq™ kit (Takara Biotechnology, Shiga, Japan) to confirm the expression of circRNAs screened by microarray. Gene primers were designed and synthesized by Genepharma (China), and the sequences of the primers are shown in Table 1. The relative expression of each differentially circRNA was quantitated using the 2−ΔΔCT method.
Fluorescence in situ hybridization (FISH)
Each group of CRC cells was fixed and treated sequentially with DEPC, 30% H2O2+pure methanol, 0.25% HCl, proteinase K (20 min), and 0.1 mol/L glycine. After washing, the cells were fixed again and processed with pre-hybridization at 65°C for 1 h; after which 500 ng/mL digoxigenin-labeled probes were added, and the mixture was incubated at 65°C for 48 h. After blocking for 30 min, biotin and anti-digoxigenin antibody (Roche) was added to the cells, and the mixture was incubated at 37°C for 1 h, followed by incubation with FITC-labeled antibody at 37°C for 30 min. After DAPI staining of the nuclei, the samples were blocked and observed with a fluorescence microscope. The probes for FISH were designed and synthesized by Wuhan BioService Biological Technology Co., Ltd. (China).
HCT-15 and SW480 cells (two cell lines with APC mutation) were selected for this study. The cell lines were purchased from the National Infrastructure of Cell Line Recourse of China. The HCT-15 cells were cultured in DMEM (Gibco, Waltham, MA, USA), and the SW480 cells were incubated in the L-15 medium at 37°C with 5% CO2. Both media contained 10% fetal bovine serum (Gibco, Waltham, MA, USA).
Neutrophils were cultured in a serum-free medium. For co-culture, neutrophils were seeded in the lower chamber of a Transwell chamber, and tumor cells were cultured at a 5:1 ratio in the upper chamber. The co-culture time was 5 h, after which relevant tests were performed on the tumor cells.
Negative control (NC) and hsa_circ_0036740 siRNAs were designed and synthesized by Genepharm (Shanghai, China). Empty vector (EV) and hsa_circ_0036740 overexpression plasmids were provided by Integrated Biotech Solutions (Shanghai, China). HCT-15 and SW480 cells were transfected with the above plasmids or siRNAs using Lipofectamine 3000 (Invitrogen, Waltham, MA, USA) as described in the manufacturer’s instructions. Actinomycin D (2 mg/mL; Merck, Darmstadt, Germany) was added during the processing of HCT-15 cells to block transcription.
For migration, the digested cells in each group were suspended in an FBS-free medium at a concentration of 2 × 106/mL. Next, an aliquot of suspended cells was inoculated into each upper chamber of a Transwell plate (Corning, New York, NY, USA), while 600 μL of medium containing 10% FBS was added to each lower chamber. The 24-well plates were then incubated at 37°C in a 5% CO2 atmosphere for 24 h. After washing, the non-migrated cells were gently wiped off, and the migrated cells were fixed with paraformaldehyde for 30 min and then stained with 0.1% crystal violet (Sigma-Aldrich, St. Louis, MO, USA) for 20 min. After washing, the cells were observed under a microscope, and five fields of view were randomly selected for photography and cell counting. For invasion, Matrigel (Corning) was diluted with culture medium at a ratio of 1:8, Then, 50 μL of the prepared Matrigel was evenly added to each upper chamber of a Transwell plate at 37°C for 1 h. The other steps were consistent with the migration experiment.
Cell counting kit (CCK)-8 assay
HCT-15 and SW480 cells were inoculated into 96-well plates at a density of 4,000 cells per well and then treated according to the experimental protocol. Following treatment, CCK-8 solution (Beyotime, Shanghai, China) was added to each well, and the cells were incubated for 0, 24, 48, and 72 h. After 3 h, the optical density (OD) ’f each well at 450 nm was measured using a microplate reader.
5‑Ethynyl‑2'‑deoxyuridine (EdU) staining
An EdU kit was used to assess cell proliferation. Cells from each group were inoculated into 96-well plates (1 × 103 cells/well), and incubated for 72 h. Next, EdU solution (50 μmol/L) was added to each well, and the cells were incubated for an additional 2 h. The culture medium was then discarded, and the cells were fixed with 100 μL of 4% paraformaldehyde for 15 min and then permeabilized with 100 μL of 0.5% Triton NX-100 for 30 min. Next, the cells were incubated with 100 μL of Hoechst 33342 (Sigma-Aldrich) for 20 min in the dark. After washing, different fields of view were randomly selected for photography using a fluorescence microscope.
Samples of peripheral anticoagulated blood were collected from FAP patients and mixed with an equal volume of dextran T2500 (30 g/mL) for 30 min. The plasma layer was harvested and separated by the addition of a lymphocyte separation solution. The leukocyte membrane layer was collected and hemolyzed with ammonium chloride hemolysate. After centrifugation, the neutrophils were isolated, and their activity and purity were estimated to be >95%. The Neutrophils were then suspended at 2 × 106/mL and incubated in a CO2 incubator.
Cells were inoculated onto sterile coverslips in 6-well plates and examined for evidence of crawling. After processing, the cells were fixed with 4% paraformaldehyde and permeabilized with 0.1% Triton X-100. Subsequently, the cells were covered for 30 min, exposed to anti-PAD4 (1: 20; Abcam, Cambridge, MA, USA) overnight at 4°C, and fluorescently labeled with a secondary antibody (1: 20; Abcam, Cambridge, MA, USA) for 2 h. After washing, the nuclei were stained with DAPI for 2 min. A fluorescence microscope was used to observe the results.
Flow cytometry to determine apoptosis
Cell apoptosis was examined with a FITC/PI kit (Cat. No. G1511, Servicebio, Wuhan, China). Briefly, CRC cells were processed and diluted to single cell suspensions of 2 × 105 cells/mL. After centrifugation, the CRC cells were resuspended in 100 μL of binding buffer and stained with 5 μL of Annexin V-FITC and 5 μL of PI for 15 min in the dark. The apoptotic cells were detected by flow cytometry (Beckman Coulter, Inc., Brea, CA, USA). The sum of the ratio of the first quadrant and the fourth quadrant is defined as the apoptosis ratio.
A 3 μL aliquot of CD16-PE (BD Bioscience, San Jose, CA, USA) was added to cultured neutrophils (1 × 105/tube) and thoroughly mixed for 30 min. After washing, 3 mL of ammonium chloride hemolysate was added to the mixture, which was then protected from light for 15 min. After centrifugation, CD16 expression on the surface of neutrophils was assessed by flow cytometry (BD Biosciences).
The total protein was extracted from processed HCT-15 and SW480 cells using cold RIPA buffer. After quantification and denaturation, a 50 μg sample of protein from each extract was separated by 10% SDS-PAGE and the protein bands were transferred onto PVDF membranes. After blocking, the membranes were exposed to anti-CitH3 (Abcam), anti-E-cadherin (Abcam), anti-N-cadherin (Abcam), anti-NE (Abcam), anti-PAD4 (Abcam), or anti-β-actin (Abcam). (1:1000) at 4°C overnight and then subsequently exposed to HRP-conjugated secondary antibodies (1:2000) for 2 h. The immunostained protein bands were visualized using ECL reagents (Invitrogen, Waltham, MA, USA).
Enzyme-linked immunosorbent assay (ELISA)
Treated HCT-15 and SW480 cells were cultured, and the medium supernatant was collected. Next, the supernatant levels of IL-1β, IL-6, and IL-8 were quantified using an ELISA Kit (R&D Systems, Minneapolis, MN, USA) as described in the manufacturer’s protocol. CitH3 levels were analyzed using a commercial kit (Cayman Chemical, cat. no. 501620). Likewise, samples of whole blood from CRC and non-tumor patients were collected, and the serum was separated by centrifugation. The levels of C3A in patient serum were examined using a C3A ELISA kit (Quidel®, San Diego, CA, USA).
Results are presented as a mean value ± SD of data obtained from three replicate samples. Statistical data were analyzed using IBM SPSS Statistics for Windows, Version 23.0 software (IBM Corp., Armonk, NY, USA). Comparisons between two groups were performed using the t-test, and comparisons among multiple groups were performed using one-way ANOVA. While, for data that did not follow a normal distribution, non-parametric tests were used for difference analysis. A p-value < 0.05 indicated statistical significance.
Screening of differentially expressed circRNAs by microarray analysis
Three randomly selected FAP (Familial adenomatous polyposis) patient samples and their paired Pt (peritumoral) tissue samples were used for circRNA microarray analysis. In total, 1970 circRNAs were downregulated and 783 were upregulated (Suppl. Table S1). After screening for differentially expressed factors (|Fold change| > 4, p-value < 0.05), our data showed that 1970 circRNAs were downregulated and 783 circRNAs were upregulated (Suppl. Tables S2 and S3). A heatmap and volcano plot showed all the circRNAs with differential expression between the FAP and Pt tissues (Figs. 1A and 1B). Subsequently, we normalized the raw data to facilitate our data analysis (Fig. 1C). The top 10 circRNAs with the highest fold-changes in upregulation and downregulation are displayed in a heatmap (Fig. 1D).
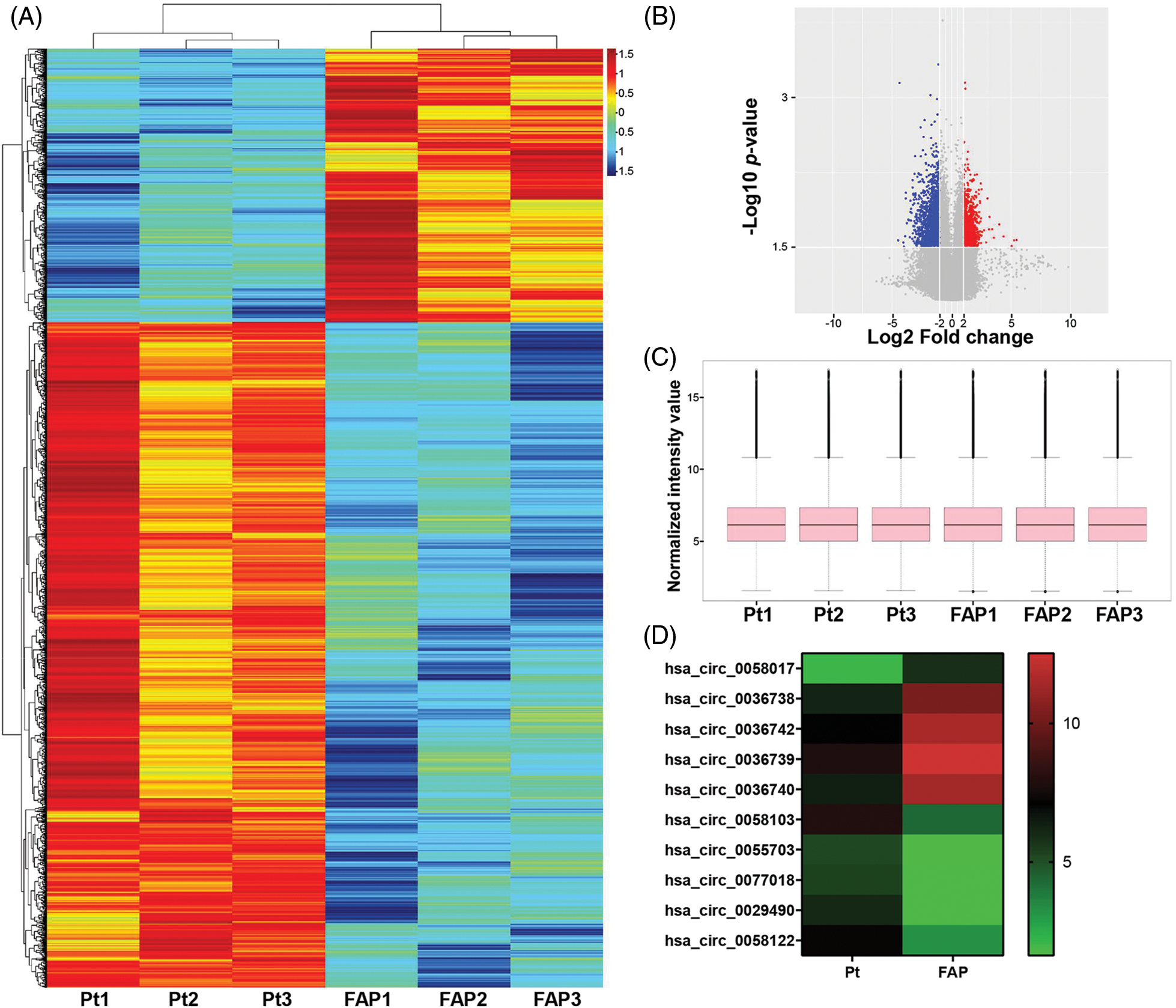
Figure 1: Screening of differentially expressed circRNAs (DECs) by microarray analysis. DECs in Familial adenomatous polyposis (FAP) (n = 3) and Pt tissues (n = 3) were analyzed using circRNA microarray. (A and B) A heatmap and volcano plot showed the DEGs between FAP and Pt tissues (log2FC > 1.5 or log2FC < −1.5; p < 0.05). (C) After normalizing the raw data, a Box plot was used to analyze the normalized data and observe the distribution characteristics of the individual sample data. (D) The top 10 circRNAs with the highest fold-changes in upregulation or downregulation are displayed in the heatmap.
Functional analysis of differential circRNA parent gene mRNAs
To further confirm the related functions of the DECs between FAP and Pt tissues, we performed a KEGG enrichment analysis of the parental genes for differentially expressed circRNAs. The KEGG results showed that the pathways with high enrichment of circRNA parental genes included immune system, immune diseases, cancers, cell motility, cell growth and death, and signal transduction (Fig. 2). Based on the functional analysis, we hypothesized that these differentially expressed circRNAs might be strongly associated with cell functions, such as proliferation, apoptosis, and metastasis.
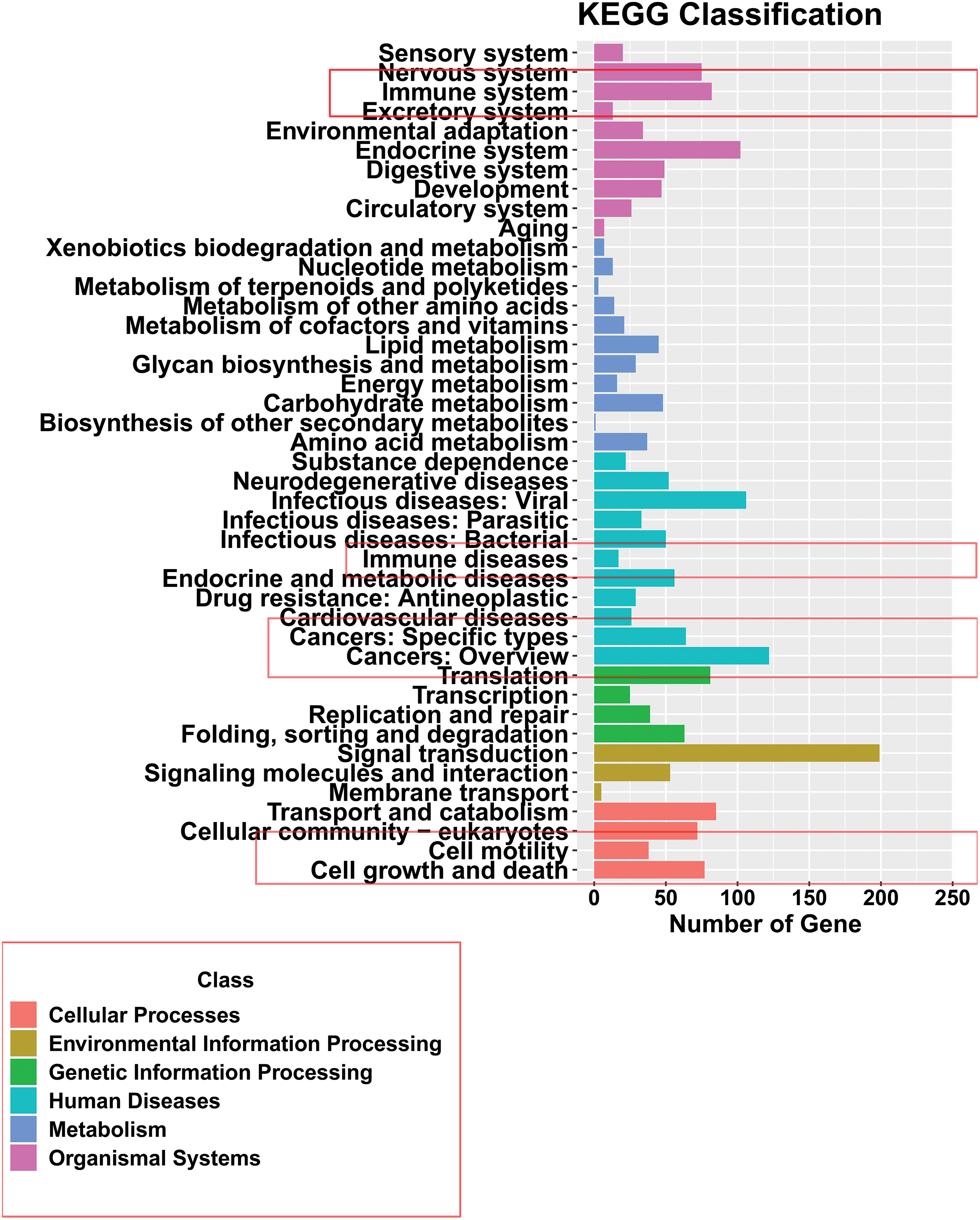
Figure 2: Functional analysis of differentially expressed circRNA parent gene mRNAs. Kyoto Encyclopedia of Genes and Genomes analysis of the parent gene mRNAs of differentially expressed DECs in familial adenomatous polyposis and Pt tissues.
Validation of hsa_circ_0036740 in colorectal cancer
Subsequently, we used RT-qPCR to validate the 10 screened circRNAs in the tissue samples from 20 FAP patients. The baseline of patients included in this study was shown in Table 2. The results showed that the change trends of those 10 circRNAs were consistent with the microarray results. Specifically, relative to the Pt group, the levels of hsa_circ_0058017, hsa_circ_0036738, hsa_circ_0036742, hsa_circ_0036739, and hsa_circ_0036740 were markedly higher in the FAP tissues (Fig. 3A), and the levels of hsa_circ_0058103, hsa_circ_0055703, hsa_circ_0077018, hsa_circ_0029490, and hsa_circ_0058122 were markedly lower in the FAP tissues (Fig. 3B). Based on those validation data and a literature analysis, we selected the circRNA (hsa_circ_0036740) with the greatest expression difference as the target of our study. Meanwhile, RT-qPCR data showed that ANPEP (the host gene of hsa_circ_0036740) expression was significantly and time-dependently reduced by actinomycin D in HCT-15 cells, whereas hsa_circ_0036740 exhibited greater stability (Fig. 3C). In addition, FISH results revealed that hsa_circ_0036740 was expressed and localized in the cytoplasm of HCT-15 and SW480 cells (Fig. 3D).
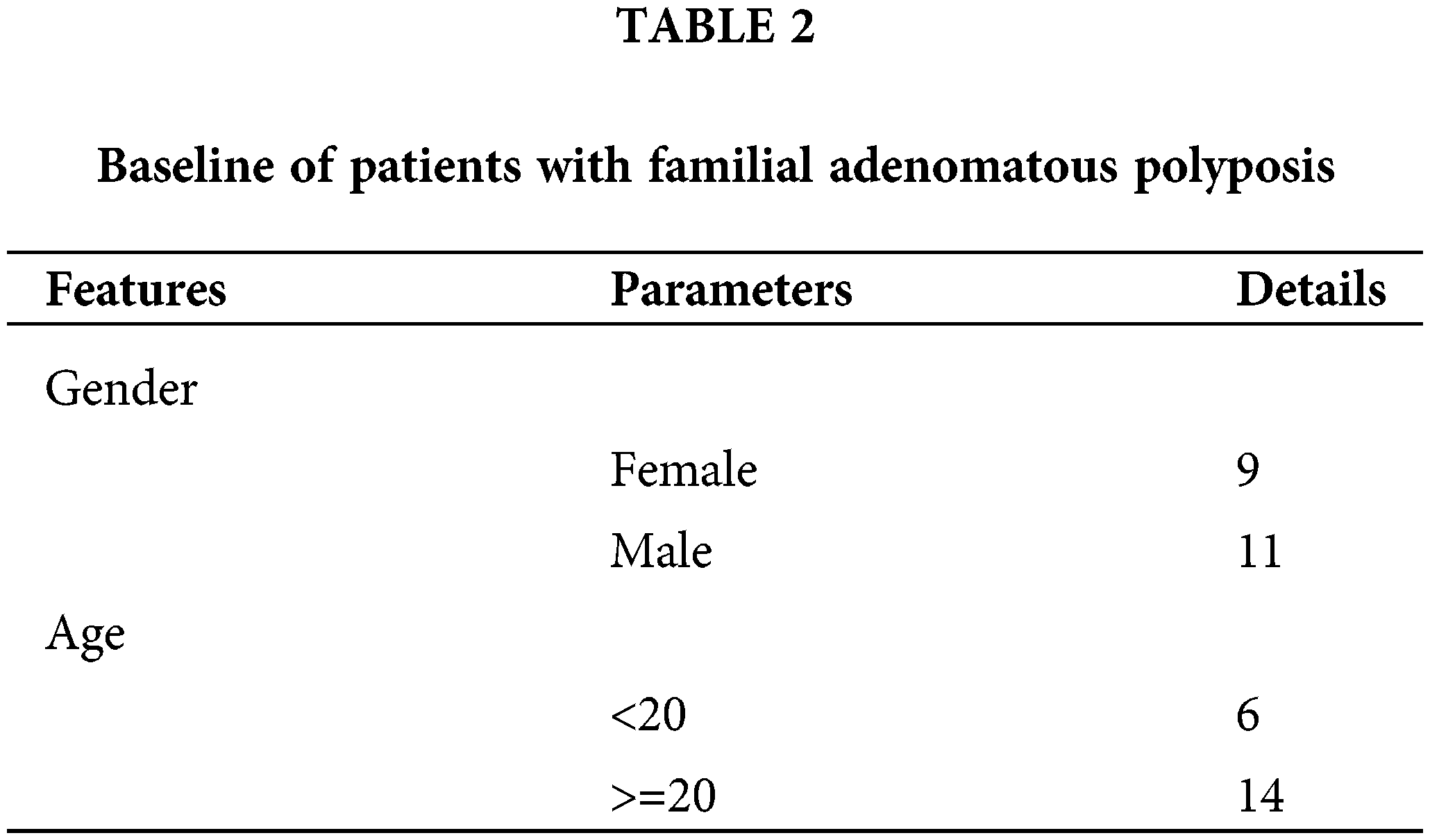
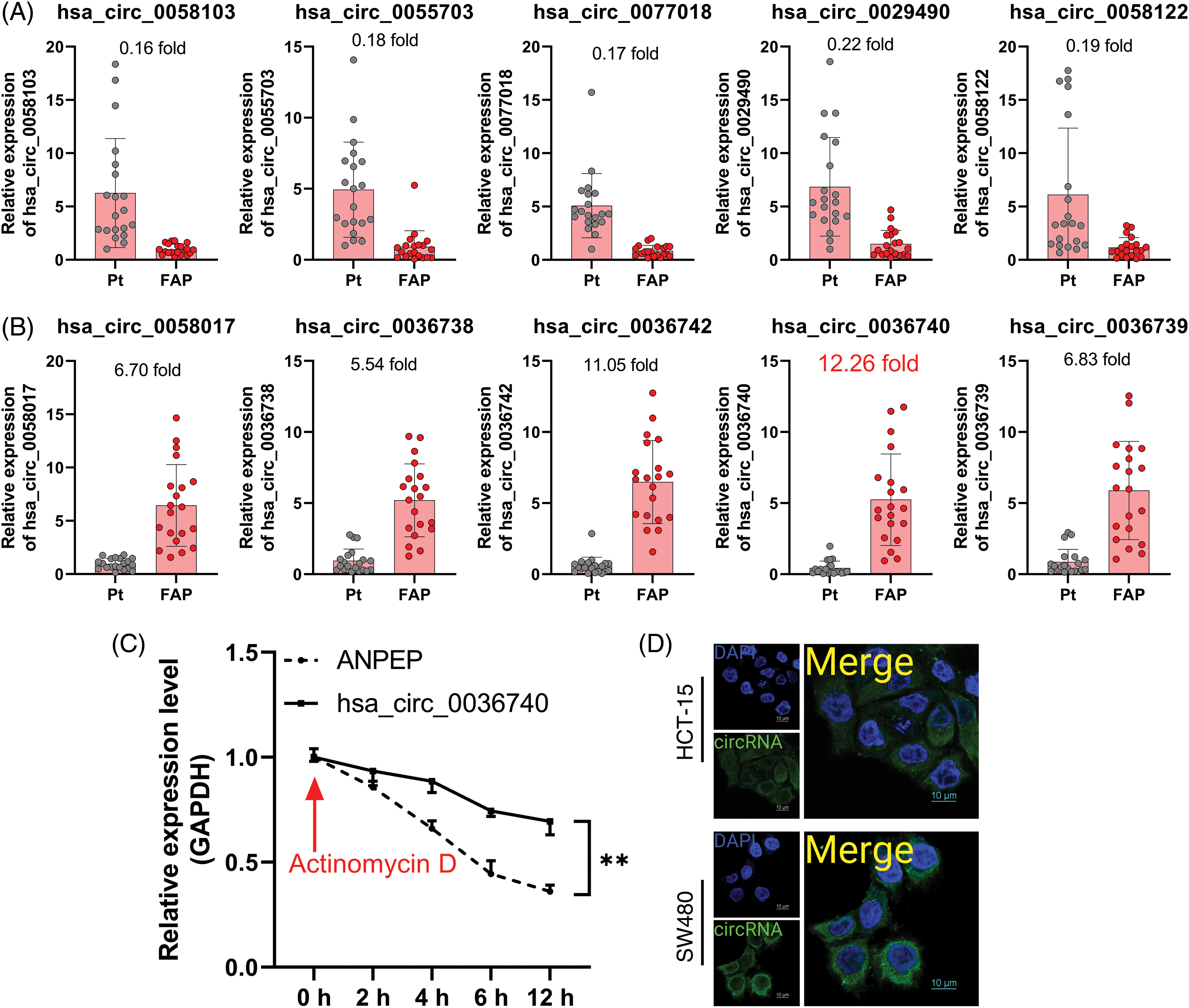
Figure 3: The screening and identification of hsa_circ_0036740. (A) RT-qPCR was used to confirm changes in the expression of 5 up-regulated circRNAs (hsa_circ_0058017, hsa_circ_0036738, hsa_circ_0036742, hsa_circ_0036739, and hsa_circ_0036740) in familial adenomatous polyposis (FAP; n = 20) and Pt (n = 20 tissues). (B) Quantitative reverse transcription polymerase chain reaction (RT-qPCR) was also used to determine changes in the expression of 5 down-regulated circRNAs (hsa_circ_0058103, hsa_circ_0055703, hsa_circ_0077018, hsa_circ_0029490, and hsa_circ_0058122) in FAP (n = 20) and Pt (n = 20) tissues. (C) The stability of hsa_circ_0036740 and ANPEP mRNA in CRC cells was verified via RT-qPCR after examining the effects of actinomycin D in HCT-15 cells. (D) Fluorescence in-situ hybridization assays for hsa_circ_0036740 in HCT-15 and SW480 cells. **p < 0.01. Magnification, x400.
Complement C3A was highly expressed and positively correlated with hsa_circ_0036740 in colorectal cancer tissues
Additionally, we also analyzed the concentrations of C3A in the serum of CRC patients. ELISA data showed that C3A levels were notably elevated in the serum of CRC patients vs. those in non-tumor patients (Fig. 4A). Based on the database (https://www.proteinatlas.org/) information, we also found that C3A expression in tumor tissues was higher than in non-tumor tissue as determined by IHC results (Fig. 4B). A correlation analysis revealed a positive correlation between hsa_circ_0036740 expression and C3A levels in CRC (Fig. 4C). Therefore, we hypothesized that hsa_circ_0036740 might be associated with the levels of C3A.
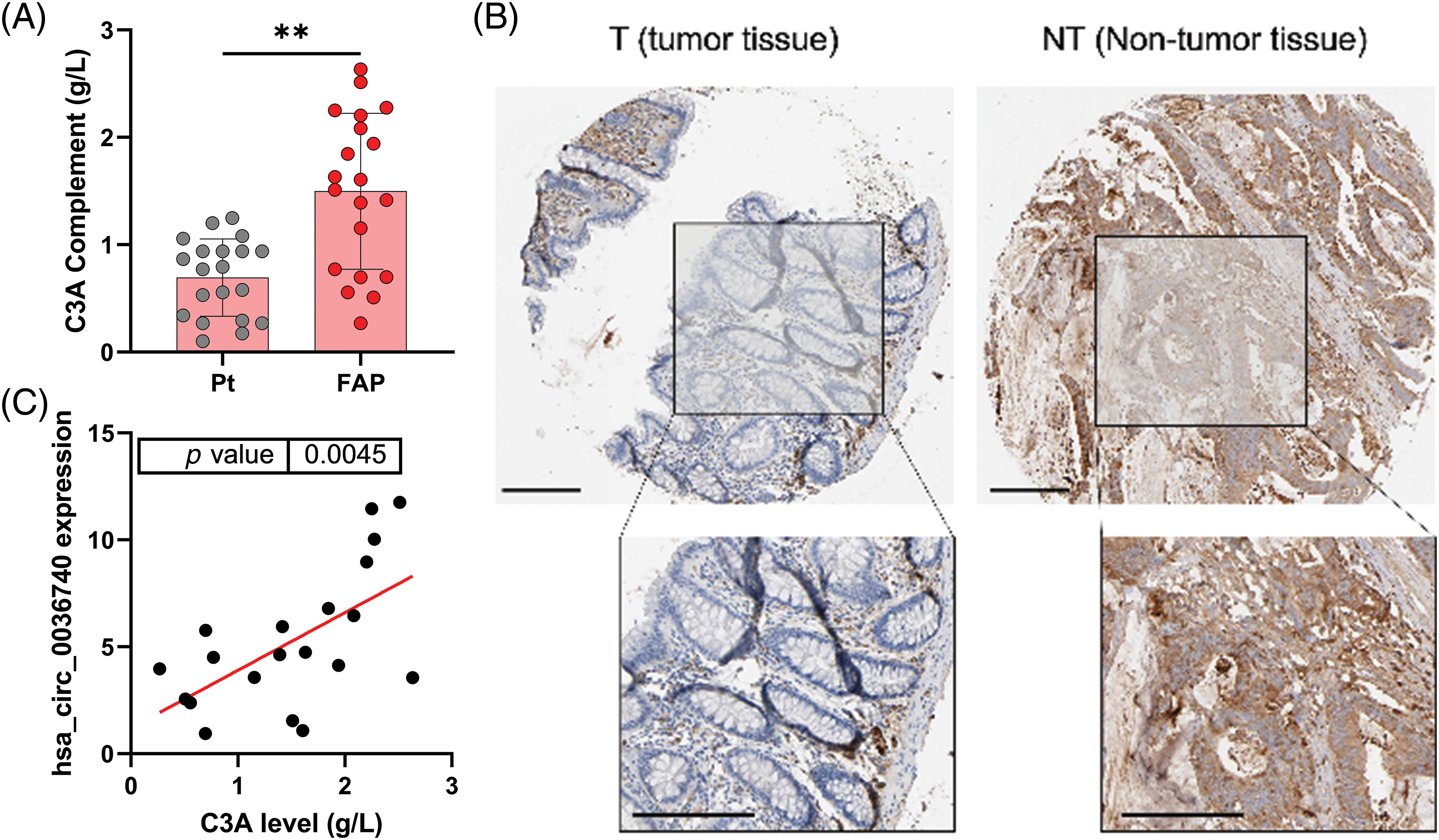
Figure 4: Complement C3A was highly expressed and related to hsa_circ_0036740 in CRC tissues. (A) The concentrations of C3A in the serum of patients with colorectal cancer were detected by enzyme-linked immunosorbent assay. (B) IHC data for C3A as obtained from the database (https://www.proteinatlas.org/), (C) The correlation between hsa_circ_0036740 expression and C3A levels. **p < 0.01. Scale bar: 200 μm.
Hsa_circ_0036740 overexpression accelerated colorectal cancer cell migration, invasion, and epithelial-mesenchymal transition (EMT)
Functionally, we first validated the effects of hsa_circ_0036740 on the migration and invasion of CRC cells by forcing the overexpression or silencing of hsa_circ_0036740 in CRC cells (HCT-15 and SW480). RT-qPCR data indicated that hsa_circ_0036740 overexpression could cause a significant increase in hsa_circ_0036740 levels, and hsa_circ_0036740 silencing could lead to a significant decrease in hsa_circ_0036740 levels in HCT-15 and SW480 cells (Figs. 5A and 5B). Meanwhile, Transwell results indicated that hsa_circ_0036740 overexpression strongly induced the migration and invasion of HCT-15 and SW480 cells, and hsa_circ_0036740 silencing dramatically suppressed the migration and invasion of those two cell lines (Figs. 5C–5E). Additionally, we further explored the role of hsa_circ_0036740 in the EMT process of CRC cells. Western blotting was performed to validate the expression levels of EMT-related markers. Those results showed that hsa_circ0036740 overexpression notably downregulated E-Cadherin levels and upregulated N-cadherin levels in HCT-15 and SW480 cells, and hsa_circ_0036740 silencing strongly upregulated E-cadherin levels and downregulated N-Cadherin levels in those two cell lines (Fig. 5F). Overall, these data indicated that hsa_circ_0036740 had a significant facilitative effect on the metastatic process of CRC cells.
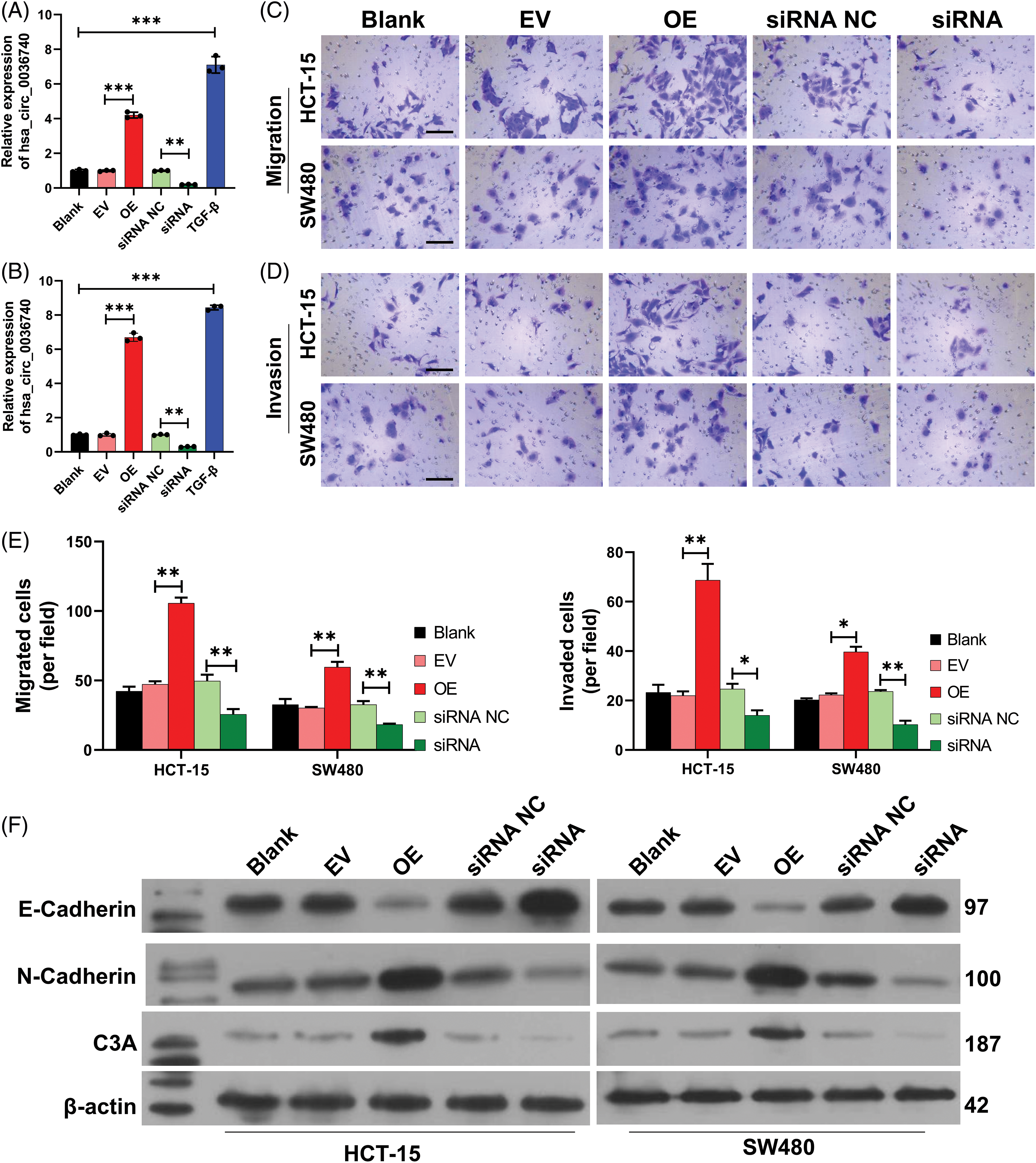
Figure 5: Effects of hsa_circ_0036740 overexpression or silencing on colorectal cancer (CRC) cell migration, invasion, and epithelial-mesenchymal transition. CRC cells (HCT-15 and SW480) were treated with hsa_circ_0036740 overexpression plasmids or hsa_circ_0036740 siRNA. The levels of hsa_circ_0036740 in the treated HCT-15 (A) and SW480 cells (B) were identified via quantitative reverse transcription polymerase chain reaction. Transwell assays were conducted to monitor the migration (C) and invasion (D) of the treated CRC cells. Magnification, x200. (E) The migrated and invaded cells were quantified. (F) Changes in E-cadherin and N-cadherin expression were assessed by western blotting. *p < 0.05; **p < 0.01; ***p < 0.001. Scale bar: 100 μm.
Hsa_circ_0036740 markedly induced colorectal cancer cell proliferation and prevented apoptosis
Similarly, we also evaluated the effect of hsa_circ_0036740 on the apoptosis of CRC cells. Flow cytometry results showed that relative to the control group (EV or siRNA NC), cell apoptosis was markedly attenuated in the hsa_circ_0036740 overexpression (OE) group and enhanced in the hsa_circ_0036740 silencing (siRNA) group (Fig. 6A). Next, we further verified the effect of hsa_circ_0036740 on CRC cell proliferation by performing Edu staining and CCK-8 assays. Edu staining results showed that after transfection of HCT-15 and SW480 cells, the proliferation ability of cells in the hsa_circ_0036740 overexpression group (OE) was markedly enhanced when compared to cells in the control (EV) group. Furthermore, the proliferation ability of cells in the hsa_circ_0036740 silenced group (siRNA) was notably attenuated when compared to cells in the control (siRNA NC) group (Fig. 6B). CCK-8 results further verified that hsa_circ_0036740 overexpression promoted the proliferation and hsa_circ_0036740 silencing inhibited the proliferation of HCT-15 and SW480 cells (Fig. 6C). Thus, we revealed that hsa_circ_0036740 induces the proliferation and inhibits the apoptosis of CRC cells.
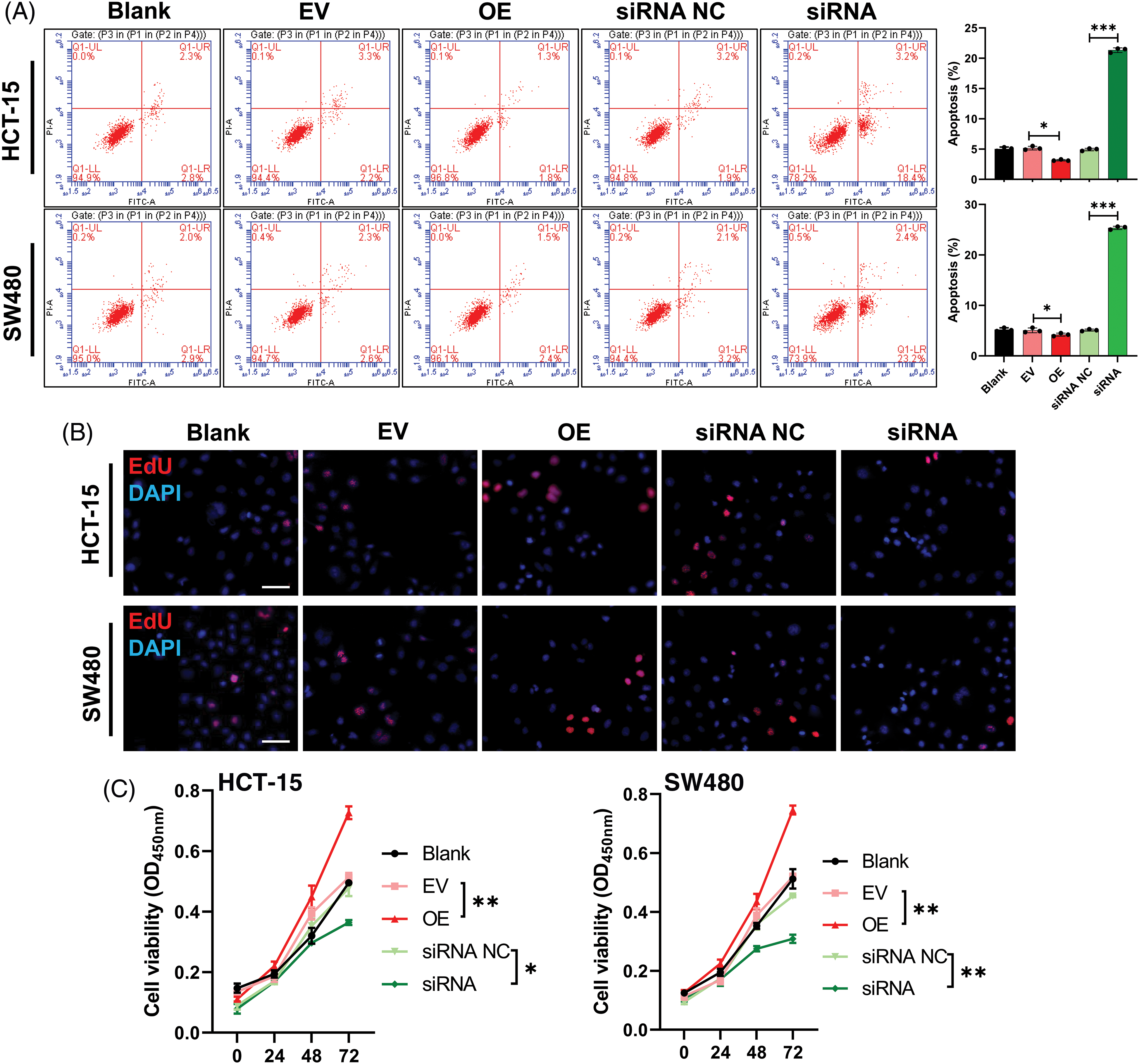
Figure 6: Hsa_circ_0036740 significantly inhibited apoptosis and induced the proliferation of colorectal cancer (CRC) cells. (A) The apoptosis of HCT-15 and SW489 cells after hsa_circ_0036740 overexpression or treatment was monitored by flow cytometry. (B) Edu staining of HCT-15 and SW480 cells after hsa_circ_0036740 overexpression or silencing. Magnification, x100. (C) Cell Counting Kit-8 assays were performed to verify the viability of hsa_circ_0036740 overexpressing or silenced CRC cells. *p < 0.05; **p < 0.01; ***p < 0.001. Scale bar: 50 μm.
Hsa_circ_0036740 markedly induced neutrophil extracellular trap formation and inflammation in colorectal cancer cells
More importantly, we further investigated the effects of hsa_circ_0036740 on NETs in CRC cells. Western blot results showed that the levels of NET marker protein (H3Cit) were significantly increased in FAP tissues when compared with those in Pt tissues (Fig. 7A). Next, we isolated neutrophils and used flow cytometry to analyze neutrophil CD16 expression. Those data showed that the isolated neutrophils were positive for CD16, suggesting their successful separation (Fig. 7B). Furthermore, we co-cultured the neutrophils with hsa_circ_0036740 overexpressing or silenced HCT-15 and SW480 cells. Our data showed that hsa_circ_0036740 overexpression increased MPO-DNA levels, and hsa_circ_0036740 silencing notably decreased MPO-DNA levels in neutrophils (Fig. 7C). Meanwhile, ELISA data further showed that hsa_circ_0036740 overexpression could also dramatically increase the levels of CitH3, IL-1β, IL-6, and IL-8, while hsa_cirsc_0036740 silencing significantly decreased the levels of CitH3, IL-1β, IL-6, and IL-8 in culture medium (Figs. 7D–7G). Western blot studies revealed that relative to the EV or siRNA NC group, the levels of NE and PAD4 expression were upregulated in the OE group and downregulated in the siRNA group (Fig. 7H). Likewise, IF staining results indicated that hsa_circ_0036740 overexpression markedly increased PAD4 expression, and hsa_circ_0036740 silencing decreased PAD4 expression in neutrophils (Fig. 7I). These findings indicated that hsa_circ_0036740 could induce NET formation and an inflammatory response.
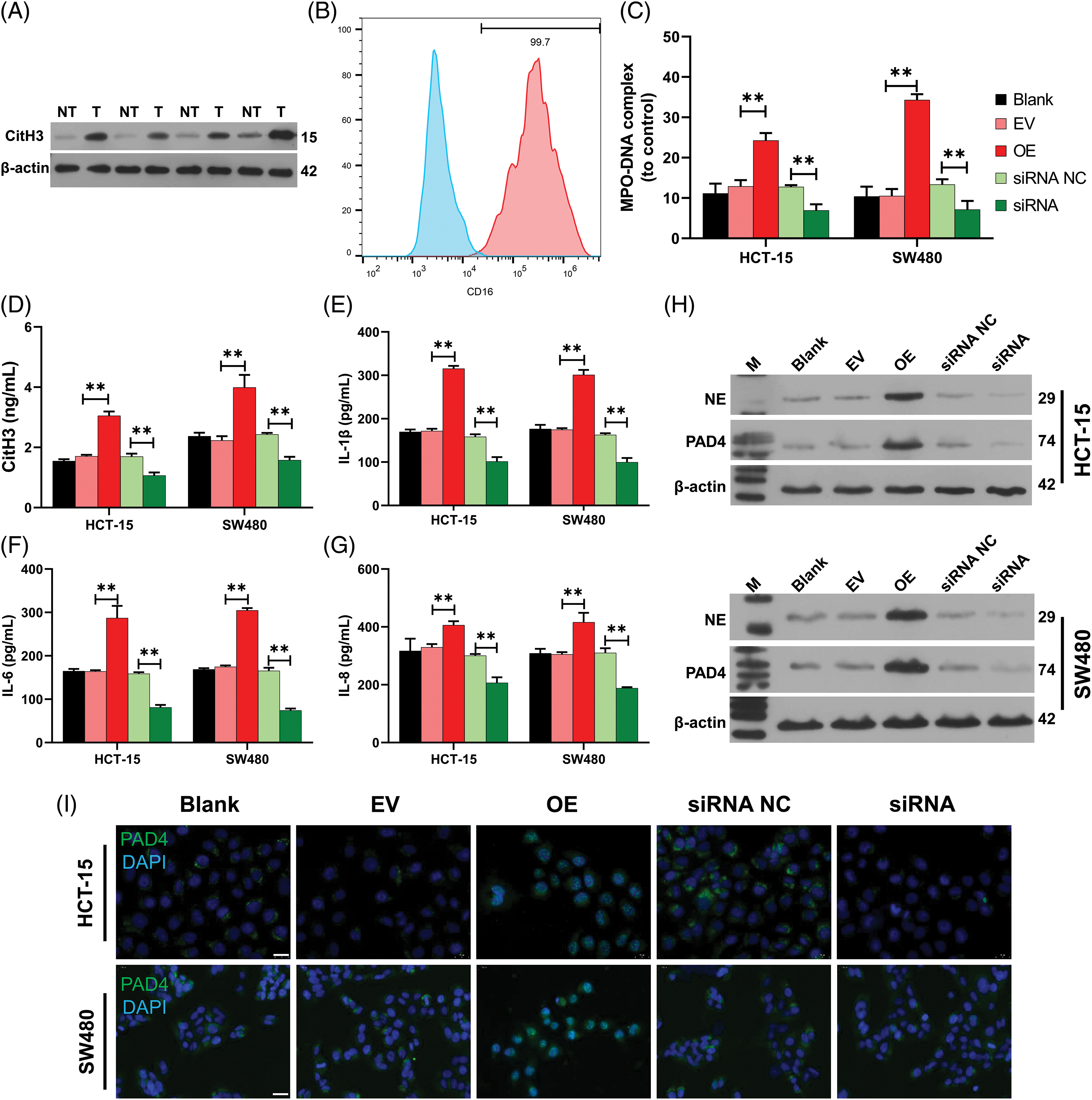
Figure 7: Hsa_circ_0036740 markedly induced neutrophil extracellular trap (NETs) formation and inflammation. (A) The levels of citrullinated histone H3 (CitH3) in familial adenomatous polyposis (FAP) (T) and Pt tissues were confirmed by western blotting. (B) Neutrophils were isolated and neutrophil CD16 was analyzed by flow cytometry. (C) Hsa_circ_0036740 was overexpressed or silenced in HCT-15 and SW480 cells by transfection, and the CRC cells were co-cultured with neutrophils. The levels of myeloperoxidase (MPO)-DNA in neutrophils were detected using a commercial kit. (D–G) After co-culture, enzyme-linked immunosorbent assay kits were used to examine the levels of CitH3, interleukin (IL)-1β, IL-6, and IL-8 in the culture medium. (H) Western blotting analysis of neutrophil elastase (NE) and arginine deiminase 4 (PAD4) protein expression in neutrophils. (I) IF staining of PAD4 in hsa_circ_0036740 overexpressing or silenced neutrophils. **p < 0.01. Scale bar: 20 μm.
FAP is a familial aggregation of polyps, that, if left untreated, has a nearly 100% lifetime risk of developing into cancer (Ml et al., 2017). Age of onset, the number and size of adenomas, and their histological types are all relevant factors affecting FAP carcinogenesis (Cyrany et al., 2018). The main causes of death among FAP patients are CRC, duodenal cancer, and fibroma durum (Martin et al., 2021). The mean age for developing CRC is 39 years, and 7% of FAP patients are diagnosed with CRC before the age of 21 years (Zhan et al., 2020). Currently, sulforaphane combined with erlotinib and endoscopic resection combined with low-dose aspirin have been identified as potentially effective treatment options for FAP (Ulusan et al., 2021). However, the molecular mechanism of FAP remains unclear.
The structural characteristics of circRNA make it less susceptible to degradation by nucleases in cells than linear RNA, and the expression pattern denotes tissue, cellular, and developmental stage specificity (Zhu et al., 2021). The half-life of circRNAs is longer than that of linear RNAs (Kristensen et al., 2019). Therefore, circRNAs are ideal for use as biomarkers for multiple diseases, including cancer (Chen and Shan, 2021). Several studies have screened DECs in CRC patient tissues by gene microarray (Liu et al., 2021; Wang et al., 2021; Zhang et al., 2017). However, the circRNA expression profile associated with FAP has not been reported. In our study, we collected FAP patient tissues and their corresponding paraneoplastic tissues and performed high-throughput detection of circRNAs by use of circRNA microarray. We then screened out circRNAs that were up- or down-regulated in FAP tissues. Based on the GO and KEGG enrichment analysis, we also discovered that the parental genes with differential circRNAs are closely associated with cell proliferation and metastasis processes. Additionally, RT-qPCR studies were performed to validate 10 differentially expressed circRNAs. Based on the expression differences of those 10 circRNAs, we finally selected hsa_circ_0036740 as a focus of our research.
Tumor metastasis is the primary cause of failure of anticancer therapy and patient death (Bakir et al., 2020). The tumor microenvironment is inseparable from tumorigenesis and metastasis (Mowers et al., 2018). Immune cells play a crucial role in tumor progression by secreting various cytokines (Lei et al., 2020). Neutrophils, a type of immune cell, account for approximately 60% of circulating leukocytes (Shaul and Fridlender, 2019). Neutrophils have been reported to have chemotactic, phagocytic, and bactericidal effects (Wu et al., 2020). As a key component of intrinsic immunity, neutrophils are usually involved in the defense and protection process against infectious diseases (Russell et al., 2019). NETs are complex substances released from neutrophils and consist of a DNA backbone and multiple embedded proteins (Rada, 2019). The main pathogenic substances of NETs are CitH3, MPO, and other protein components, which have pro-inflammatory and cytotoxic effects (Kopytek et al., 2019). Various biomolecules produced by organisms or pathogens can induce the activation of neutrophils and the production of NETs (Ravindran et al., 2019). NET levels also have been discovered to correlate with the stage of tumors (Masucci et al., 2020; Lee et al., 2019). NET levels are significantly elevated in patients with cancers, such as lung, gastric, and esophageal cancers, and are higher in patients with advanced cancers than early-stage cancers (Chu et al., 2021; Shao et al., 2021). Therefore, biomarkers of NET can play an important role in predicting tumor progression and prognosis. Currently, drugs targeting NETs and their components have shown good efficacy in animal experiments, but the exact effects of NETs on an organism have not been fully elucidated. Therefore, further studies on the related regulatory mechanisms of NETs are still needed to provide new ideas for the future development of therapeutic regimens and targeted drugs for clinically relevant diseases.
The NET formation is triggered by receptor-ligand binding events and is regulated by a series of intracellular signaling pathways (Rosazza et al., 2021). The receptors on neutrophils are essential for mediating NETosis (Chen et al., 2021). The main receptors that trigger NETosis are pattern recognition receptors (PRRs), complement receptors (CRs), and chemokine receptors (CXCRs) (Huang et al., 2022). CRs play a vital role in the pathophysiology of NET-mediated autoimmune diseases. Among them, C3a is mainly produced in the liver and can bind to receptors on neutrophils (Hsu et al., 2020). Research has shown that NETs released from neutrophils are associated with the upregulation of complement C3a protein in cancer (Guglietta et al., 2016; Masucci et al., 2020; Nolan and Malanchi, 2020). Our study also confirmed that C3A is highly expressed in the serum of CRC patients, and C3A is positively correlated with hsa_circ_0036740. More importantly, we proved that the levels of NET marker protein (H3Cit) were obviously elevated in FAP tissues when compared with their levels in Pt tissues, indicating that NETs are relevant to FAP. Our study also showed that hsa_circ_0036740 overexpression could increase the levels of NET markers (CitH3, MPO-DNA, NE, and PAD4), as well as inflammation-associated cytokines. Thus, we speculated that hsa_circ_0036740 could accelerate NET formation in CRC, which is related to C3A.
Recent studies have also confirmed that NETs are critical for CRC progression and metastasis (Khan et al., 2021). For instance, prevention of NET formation can improve PD-1 blockade immunotherapy in CRC (Zhang et al., 2021b). NETs also contribute to the procoagulant activity in patients with CRC (Zhang et al., 2019) and can induce epithelial-mesenchymal transition (EMT) in CRC cells (Stehr et al., 2022). EMT refers to the process by which epithelial cells with basal polarity transform into mesenchymal cells (Mittal, 2018). In addition to the change in cell phenotype that occurs during EMT, the surface markers of the cells are also altered. For example, E-cadherin expression was found to be decreased, and N-cadherin expression was increased (Zhang and Weinberg, 2018). E-cadherin, as an adhesion linker, can maintain epithelial tissue stability (Wong et al., 2018), and upregulation of N-cadherin was found to be positively correlated with cell proliferation and migration (Cao et al., 2019). Further, we found that hsa_circ_0036740 overexpression could downregulate E-cadherin expression and upregulate N-cadherin expression, suggesting that hsa_circ_0036740 also has a significant induction effect on the EMT process in CRC cells. In addition, our data showed that overexpression of hsa_circ_0036740 could enhance metastasis and proliferation and prevent the apoptosis of CRC cells. Meanwhile, hsa_circ_0036740 silencing could prevent CRC progression, including NET formation.
In summary, our study identified a large number of DECs that are associated with FAP. After screening and validation, we found that hsa_circ_0036740 overexpression has a significant accelerating effect on CRC processes, including induction of NET formation. Thus, the current results confirmed that hsa_circ_0036740 plays a crucial role in FAP carcinogenesis and suggest hsa_circ_0036740 as a diagnostic and therapeutic marker for FAP carcinogenesis. Future relevant studies should explore the involvement of other immune cells. Finally, we should recognize that targeting circRNAs is currently an undertaking with potential risks. There remains a great deal of research and clinical work to be done in the future.
Acknowledgement: Not applicable.
Funding Statement: This work was supported by The Scientific Research Cultivation Project of Meizhou People’s Hospital (No. PY-C2019001).
Author Contributions: All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by LZW, YH, LYP, WWS, and ZHJ. Interpretation of results and the first draft of the manuscript was written by LZW and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Availability of Data and Materials: All data generated during this study are included in this published article (and its supplementary information files).
Ethics Approval: All the study participants signed an informed consent for the inclusion in the study. The study was approved by the ethics committee of Meizhou People’s Hospital (No. 2020-CY-01).
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
Supplementary Materials: Supplementary material is available online at https://doi.org/10.32604/biocell.2023.031186.
References
Bakir B, Chiarella AM, Pitarresi JR, Rustgi AK (2020). EMT, MET, plasticity, and tumor metastasis. Trends in Cell Biology 30: 764–776. [Google Scholar] [PubMed]
Bhurwal A, Prochilo G, Patel AV (2020). Familial adenomatous polyposis. American Journal of the Medical Sciences 359: 392–393. [Google Scholar] [PubMed]
Cao ZQ, Wang Z, Leng P (2019). Aberrant N-cadherin expression in cancer. Biomedicine & Pharmacotherapy 118: 109320. [Google Scholar]
Castanheira FVS, Kubes P (2019). Neutrophils and NETs in modulating acute and chronic inflammation. Blood 133: 2178–2185. [Google Scholar] [PubMed]
Chen T, Li Y, Sun R, Hu H, Liu Y, Herrmann M, Zhao Y, Muñoz LE (2021). Receptor-mediated NETosis on neutrophils. Frontiers in Immunology 12: 775267. [Google Scholar] [PubMed]
Chen L, Shan G (2021). CircRNA in cancer: Fundamental mechanism and clinical potential. Cancer Letters 505: 49–57. [Google Scholar] [PubMed]
Chu ZQ, Zhang KC, Chen L (2021). Neutrophil extracellular traps in gastrointestinal cancer. World Journal of Gastroenterology 27: 5474–5487. [Google Scholar] [PubMed]
Cyrany J, Bureš J, Rejchrt S, Kopáčová M (2018). Familial adenomatous polyposis: Complex patient management. Vnitřní Lékařství 64: 635–641. [Google Scholar] [PubMed]
Demkow U (2021). Neutrophil extracellular traps (NETs) in cancer invasion, evasion and metastasis. Cancers 13: 4495. [Google Scholar] [PubMed]
Dinarvand P, Davaro EP, Doan JV, Ising ME, Evans NR, Phillips NJ, Lai J, Guzman MA (2019). Familial adenomatous polyposis syndrome: An update and review of extraintestinal manifestations. Archives of Pathology & Laboratory Medicine 143: 1382–1398. [Google Scholar]
Guglietta S, Chiavelli A, Zagato E, Krieg C, Gandini S, Ravenda PS, Bazolli B, Lu B, Penna G, Rescigno M (2016). Coagulation induced by C3aR-dependent NETosis drives protumorigenic neutrophils during small intestinal tumorigenesis. Nature Communications 7: 11037. [Google Scholar] [PubMed]
Hameed BH, Abdulsatar Al-Rayahi I, Muhsin SS (2022). The preoperative serum levels of the anaphylatoxins C3a and C5a and Their association with clinico-pathological factors in breast cancer patients. Archives of Razi Institute 77: 1873–1879. [Google Scholar] [PubMed]
Hsu BE, Roy J, Mouhanna J, Rayes RF, Ramsay L, Tabaries S, Annis MG, Watson IR, Spicer JD, Costantino S (2020). C3a elicits unique migratory responses in immature low-density neutrophils. Oncogene 39: 2612–2623. [Google Scholar] [PubMed]
Huang J, Hong W, Wan M, Zheng L (2022). Molecular mechanisms and therapeutic target of NETosis in diseases. MedComm 3: e162. [Google Scholar] [PubMed]
Huang A, Zheng H, Wu Z, Chen M, Huang Y (2020). Circular RNA-protein interactions: Functions, mechanisms, and identification. Theranostics 10: 3503–3517. [Google Scholar] [PubMed]
Jakobi T, Dieterich C (2019). Computational approaches for circular RNA analysis. Wiley Interdisciplinary Reviews RNA 10: e1528. [Google Scholar] [PubMed]
Karstensen JG, Burisch J, Pommergaard HC, Aalling L, Højen H, Jespersen N, Schmidt PN, Bülow S (2019). Colorectal cancer in individuals with familial adenomatous polyposis, based on analysis of the danish polyposis registry. Clinical Gastroenterology and Hepatology 17: 2294–2300.e1. [Google Scholar] [PubMed]
Kemp Bohan PM, Mankaney G, Vreeland TJ, Chick RC, Hale DF et al. (2021). Chemoprevention in familial adenomatous polyposis: Past, present and future. Familial Cancer 20: 23–33. [Google Scholar] [PubMed]
Khan U, Chowdhury S, Billah MM, Islam KMD, Thorlacius H, Rahman M (2021). Neutrophil extracellular traps in colorectal cancer progression and metastasis. International Journal of Molecular Sciences 22: 7260. [Google Scholar] [PubMed]
Klopfenstein D, Zhang L, Pedersen BS, Ramírez F, Vesztrocy AW, Naldi A, Mungall CJ, Yunes JM, Botvinnik O, Weigel M (2018). GOATOOLS: A python library for gene ontology analyses. Scientific Reports 8: 10872. [Google Scholar] [PubMed]
Kopytek M, Kolasa-Trela R, Ząbczyk M, Undas A, Natorska J (2019). NETosis is associated with the severity of aortic stenosis: Links with inflammation. International Journal of Cardiology 286: 121–126. [Google Scholar] [PubMed]
Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J (2019). The biogenesis, biology and characterization of circular RNAs. Nature Reviews Genetics 20: 675–691. [Google Scholar] [PubMed]
Lee W, Ko SY, Mohamed MS, Kenny HA, Lengyel E, Naora H (2019). Neutrophils facilitate ovarian cancer premetastatic niche formation in the omentum. Journal of Experimental Medicine 216: 176–194. [Google Scholar] [PubMed]
Lei X, Lei Y, Li JK, Du WX, Li RG, Yang J, Li J, Li F, Tan HB (2020). Immune cells within the tumor microenvironment: Biological functions and roles in cancer immunotherapy. Cancer Letters 470: 126–133. [Google Scholar] [PubMed]
Li J, Sun D, Pu W, Wang J, Peng Y (2020). Circular RNAs in cancer: Biogenesis, function, and clinical significance. Trends Cancer 6: 319–336. [Google Scholar] [PubMed]
Li CB, Wang HF, Feng ZK, Fu YB, Zhang J, Qin JY (2021). Identification of immune-related genes for Hepatocellular Carcinoma: A study based on TCGA data. Journal of Men’s Health 17: 101–113. [Google Scholar]
Liu X, Liu Y, Liu Z, Lin C, Meng F et al. (2021). CircMYH9 drives colorectal cancer growth by regulating serine metabolism and redox homeostasis in a p53-dependent manner. Molecular Cancer 20: 114. [Google Scholar] [PubMed]
Martin I, Roos VH, Anele C, Walton SJ, Cuthill V et al. (2021). Gastric adenomas and their management in familial adenomatous polyposis. Endoscopy 53: 795–801. [Google Scholar] [PubMed]
Masucci MT, Minopoli M, Del Vecchio S, Carriero MV (2020). The emerging role of neutrophil extracellular traps (NETs) in tumor progression and metastasis. Frontiers in Immunology 11: 1749. [Google Scholar] [PubMed]
Mirinezhad SK, Moaddab SY, Bonyadi MJ, Shirmohammadi M, Eftekharsadat AT, Somi MH (2019). Survival of familial adenomatous polyposis coexistence colorectal cancer in Iran. Journal of Cancer Research and Therapeutics 15: 87–91. [Google Scholar] [PubMed]
Mittal V (2018). Epithelial mesenchymal transition in tumor metastasis. Annual Review of Pathology: Mechanisms of Disease 13: 395–412. [Google Scholar]
Ml MLDE, Tonelli F, Quaresmini D, Lovero D, Della-Morte D, Silvestris F, Guadagni F, Palmirotta R (2017). Desmoid tumors in familial adenomatous polyposis. Anticancer Research 37: 3357–3366. [Google Scholar]
Mowers EE, Sharifi MN, Macleod KF (2018). Functions of autophagy in the tumor microenvironment and cancer metastasis. The FEBS Journal 285: 1751–1766. [Google Scholar] [PubMed]
Nabizadeh JA, Manthey HD, Steyn FJ, Chen W, Widiapradja A, Md Akhir FN, Boyle GM, Taylor SM, Woodruff TM, Rolfe BE (2016). The complement C3a contributes to melanoma tumorigenesis by inhibiting neutrophil and CD4+ T cell responses. The Journal of Immunology 196: 4783–4792. [Google Scholar] [PubMed]
Nolan E, Malanchi I (2020). Neutrophil ‘safety net’ causes cancer cells to metastasize and proliferate. Nature 583: 32–33. [Google Scholar] [PubMed]
Papayannopoulos V (2018). Neutrophil extracellular traps in immunity and disease. Nature Reviews Immunology 18: 134–147. [Google Scholar] [PubMed]
Rada B (2019). Neutrophil extracellular traps. Methods in Molecular Biology 1982: 517–528. [Google Scholar] [PubMed]
Ravindran M, Khan MA, Palaniyar N (2019). Neutrophil extracellular trap formation: Physiology, pathology, and pharmacology. Biomolecules 9: 365. [Google Scholar] [PubMed]
Rohrbach AS, Slade DJ, Thompson PR, Mowen KA (2012). Activation of PAD4 in NET formation. Frontiers in Immunology 3: 360. [Google Scholar] [PubMed]
Rosazza T, Warner J, Sollberger G (2021). NET formation-mechanisms and how they relate to other cell death pathways. The FEBS Journal 288: 3334–3350. [Google Scholar] [PubMed]
Russell CD, Parajuli A, Gale HJ, Bulteel NS, Schuetz P, de Jager CPC, Loonen AJM, Merekoulias GI, Baillie JK (2019). The utility of peripheral blood leucocyte ratios as biomarkers in infectious diseases: A systematic review and meta-analysis. Journal of Infection 78: 339–348. [Google Scholar] [PubMed]
Senent Y, Ajona D, González-Martín A, Pio R, Tavira B (2021). The complement system in ovarian cancer: An underexplored old path. Cancers 13: 3806. [Google Scholar] [PubMed]
Shao BZ, Yao Y, Li JP, Chai NL, Linghu EQ (2021). The role of neutrophil extracellular traps in cancer. Frontiers in Oncology 11: 714357. [Google Scholar] [PubMed]
Shaul ME, Fridlender ZG (2019). Tumour-associated neutrophils in patients with cancer. Nature Reviews Clinical Oncology 16: 601–620. [Google Scholar] [PubMed]
Shen S, Kong J, Qiu Y, Yang X, Wang W, Yan L (2019). Identification of core genes and outcomes in hepatocellular carcinoma by bioinformatics analysis. Journal of Cellular Biochemistry 120: 10069–10081. [Google Scholar] [PubMed]
Silvestre-Roig C, Braster Q, Ortega-Gomez A, Soehnlein O (2020). Neutrophils as regulators of cardiovascular inflammation. Nature Reviews Cardiology 17: 327–340. [Google Scholar] [PubMed]
Stehr AM, Wang G, Demmler R, Stemmler MP, Krug J et al. (2022). Neutrophil extracellular traps drive epithelial-mesenchymal transition of human colon cancer. Journal of Pathology 256: 455–467. [Google Scholar] [PubMed]
Tan C, Aziz M, Wang P (2021). The vitals of NETs. Journal of Leukocyte Biology 110: 797–808. [Google Scholar] [PubMed]
Ulusan AM, Rajendran P, Dashwood WM, Yavuz OF, Kapoor S et al. (2021). Optimization of erlotinib plus sulindac dosing regimens for intestinal cancer prevention in an Apc-mutant model of familial adenomatous polyposis (FAP). Cancer Prevention Research 14: 325–336. [Google Scholar] [PubMed]
Wang J, Wang T, Hu S, Li J, Ni C, Ye M (2021). Hsa_circ_0043278 inhibits tumorigenesis and is downregulated in colorectal cancer. Cancer Management and Research 13: 965–975. [Google Scholar] [PubMed]
Wong SHM, Fang CM, Chuah LH, Leong CO, Ngai SC (2018). E-cadherin: Its dysregulation in carcinogenesis and clinical implications. Critical Reviews in Oncology/Hematology 121: 11–22. [Google Scholar] [PubMed]
Wu L, Saxena S, Singh RK (2020). Neutrophils in the tumor microenvironment. Advances in Experimental Medicine and Biology 1224: 1–20. [Google Scholar] [PubMed]
Zhan Q, Wang L, Xu X, Sun Y, Li L et al. (2020). An APC mutation in a large Chinese kindred with familial adenomatous polyposis was identified using both next generation sequencing and simple STR marker haplotypes. Frontiers in Genetics 11: 191. [Google Scholar] [PubMed]
Zhang S, Sun J, Gu M, Wang G, Wang X (2021a). Circular RNA: A promising new star for the diagnosis and treatment of colorectal cancer. Cancer Medicine 10: 8725–8740. https://doi.org/10.1002/cam4.4398 [Google Scholar] [PubMed] [CrossRef]
Zhang H, Wang Y, Onuma A, He J, Wang H et al. (2021b). Neutrophils extracellular traps inhibition improves PD-1 blockade immunotherapy in colorectal cancer. Cancers 13: 5333. [Google Scholar] [PubMed]
Zhang Y, Wang C, Yu M, Zhao X, Du J et al. (2019). Neutrophil extracellular traps induced by activated platelets contribute to procoagulant activity in patients with colorectal cancer. Thrombosis Research 180: 87–97. [Google Scholar] [PubMed]
Zhang Y, Weinberg RA (2018). Epithelial-to-mesenchymal transition in cancer: Complexity and opportunities. Frontiers of Medicine 12: 361–373. [Google Scholar] [PubMed]
Zhang P, Zuo Z, Shang W, Wu A, Bi R, Wu J, Li S, Sun X, Jiang L (2017). Identification of differentially expressed circular RNAs in human colorectal cancer. Tumor Biology 39: 1010428317694546. [Google Scholar] [PubMed]
Zheng SL, Li L, Zhang HP (2020). Progress on translation ability of circular RNA. Yi Chuan = Hereditas 42: 423–434. [Google Scholar] [PubMed]
Zhu G, Chang X, Kang Y, Zhao X, Tang X, Ma C, Fu S (2021). CircRNA: A novel potential strategy to treat thyroid cancer (Review). International Journal of Molecular Medicine 48: 5–201. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools