 Open Access
Open Access
VIEWPOINT
Microfluidic methods used in exosome isolation
1 Department of Mechanical Engineering, Aydın Adnan Menderes University, Aydın, 09010, Turkiye
2 Department of Biochemistry, School of Medicine, Aydın Adnan Menderes University, Aydın, 09010, Turkiye
* Corresponding Author: ADEM OZCELIK. Email:
BIOCELL 2023, 47(5), 959-964. https://doi.org/10.32604/biocell.2023.028371
Received 14 December 2022; Accepted 31 January 2023; Issue published 10 April 2023
Abstract
Exosomes are important biomarkers for clinical diagnosis. It is critical to isolate secreted exosomes from bodily fluids such as blood, saliva, breast milk, and urine for liquid biopsy applications. The field of microfluidics provides numerous benefits for biosample processing, diagnostics, and prognostics. Several microfluidics-based methods have been employed for the isolation and purification of exosomes in the last ten years. These microfluidic methods can be grouped into two categories based on passive and active isolation mechanisms. In the first group, inertial and hydrodynamic forces are employed to separate exosomes based on their size differences. In the second group, external forcefields are integrated into microfluidic platforms to actively isolate exosomes from other bioparticles. In this paper, the application of microfluidic methods in exosome isolation is discussed, and future perspectives on this field are highlighted.Keywords
Cells maintain tissue homeostasis through the extracellular vesicles that they secrete to communicate with different cells and to ensure the transport of molecules to different locations (Panfoli et al., 2022; You and Qian, 2022). Cells use the extracellular vesicles derived from endosomes and plasma membranes for such types of communications. Among these extracellular vesicles are exosomes that are secreted from cells, which are the biological structures having the smallest sizes (30–100 nm). Exosomes can be isolated from all bodily fluids (such as urine, serum, synovial fluid, cerebrospinal fluid, and saliva).
The contents of exosomes reflect the composition of the donor cell from which they originate, and their formation occurs via cellular stimuli (Valadi et al., 2007). Exosomes are used to diagnose or treat many diseases, from cardiovascular diseases to diabetes, cancer to infectious diseases, and neurodegenerative diseases to immune deficiencies (Wang et al., 2021a; Lai et al., 2022). The transport of miRNAs by exosomes has drawn attention in the pathogenesis of cardiovascular diseases. For instance, it has been shown that miRNAs carried by exosomes released by cardiomyocytes isolated from diabetic rats have antiangiogenic roles. It was also suggested that exosomes carrying these miRNAs increase the risks of cardiovascular diseases in diabetes by blocking the repair of damaged blood vessels (Wang et al., 2014).
Owing to the widespread use of exosomes in diagnosis, researchers are searching for new ways to isolate them and make them functional by inducing modifications and also use them as drug carriers (Abas et al., 2022; Uslu et al., 2022; Cenik et al., 2022). The isolation of exosomes is generally done using ultracentrifugation, ultrafiltration, isolation kits, chromatographic methods, and immunoaffinity capture (Li et al., 2022). The isolation of exosomes using these techniques presents advantages or disadvantages depending on the sample and required amount (Zheng et al., 2022). Although ultracentrifugation is the gold standard method to isolate exosomes, there are disadvantages owing to the high cost of the required instrumentation, and the long extraction time involved (Momen-Heravi et al., 2012). Size exclusion chromatography (SEC) is also widely used to isolate high-quality exosomes and minimize damage to exosomes. However, SEC is preferred for industrial exosomal production because the large sizes of the columns used cause few restrictive problems (Konoshenko et al., 2018). While ultrafiltration is widely used in the isolation of exosomes from bodily fluids given that it is ideal and fast for large volumes of samples, however, the extraction efficiency of the exosomal isolation is lowered because of filter clogging (Shirejini and Inci, 2022). Finally, in the immunoaffinity capture technique, exosomes are captured through specific antibodies, aptamers, and recombinant proteins that are immobilized on the matrix and subsequently collected with the help of magnetic beads (Wang et al., 2019). Due to the high sensitivity of this approach, it is widely used in diagnostic studies, but it is however not suitable when the sample volume is large (Wang et al., 2019). Isolation kits entail the use of polymers with significant hydrophilic properties that allow the passage of exosomes to the hydrophobic microenvironment. However, the problem of purity is often encountered in this approach. In order to overcome such disadvantages of traditional exosomal isolation methods, new techniques have been developed using microfluidic platforms.
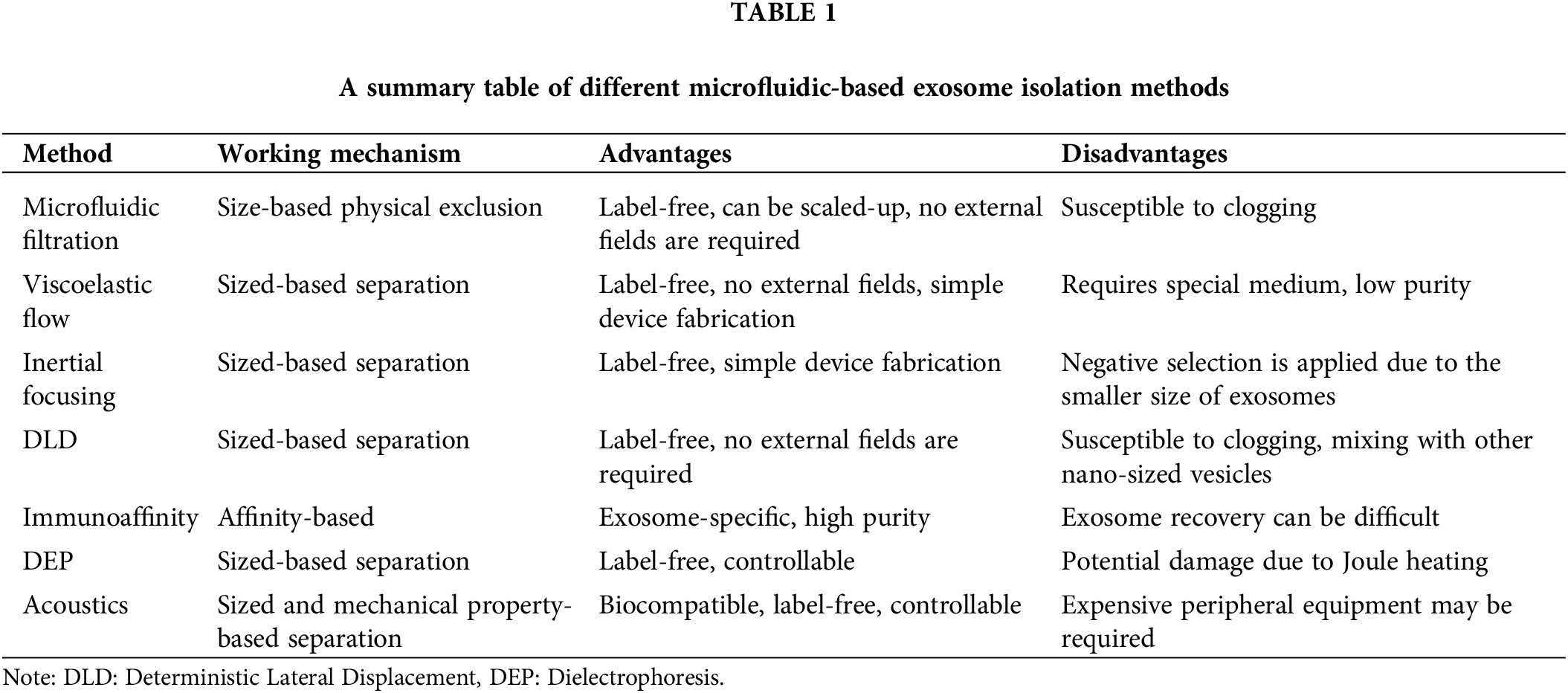
Microfluidics is important for biological and medical studies because it allows for precise control and manipulation of small fluid volumes (Akkoyun and Özçelik, 2022; Ozcelik, 2022). Additionally, microfluidic devices can be used to create microenvironments that mimic those found in the body, which can be useful to study disease mechanisms and the effectiveness of potential therapies (Ozcelik et al., 2022a). Microfluidic devices can also be used to isolate exosomes based on their size and surface markers. A few examples of microfluidic-based exosome isolation methods are depicted in Fig. 1. Exosome isolation using microfluidics-based platforms is highly efficient and potentially allows for high-throughput isolation of exosomes with minimal sample loss, while also providing a high degree of purity. This can be especially useful for basic research and drug discovery, as well as for clinical applications such as diagnostics and therapeutics. A general summary of the characteristics of microfluidic exosome isolation methods is given in Table 1. In this paper, different microfluidic methods for exosome isolation are discussed to highlight their strengths and weaknesses.
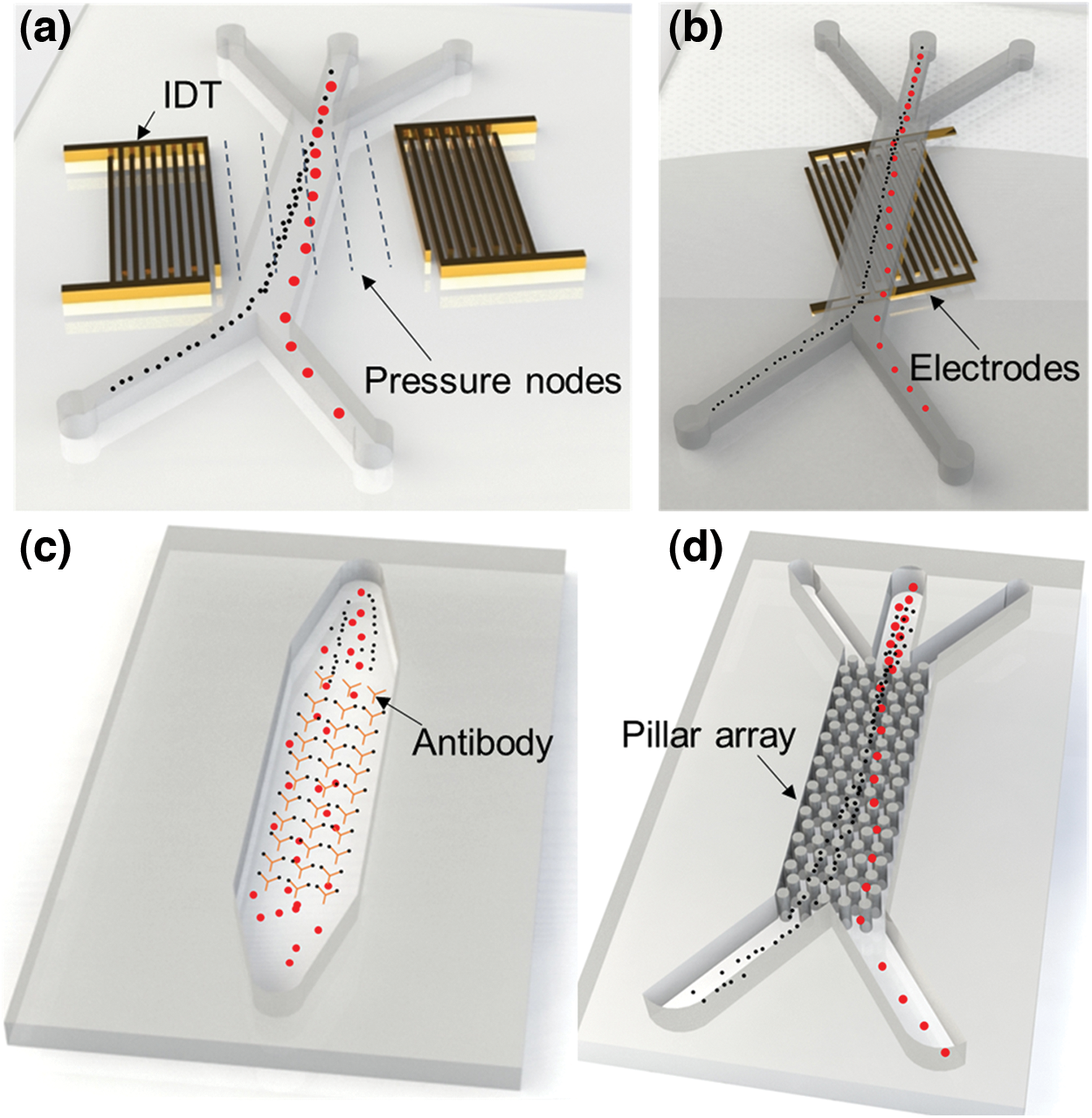
Figure 1: Schematic depiction of examples of microfluidic exosome isolation methods. (a) Acoustic-based, (b) DEP-based, (c) Immunoaffinity-based, and (d) DLD-based exosome isolation methods.
Passive Microfluidic Exosome Isolation
Passive microfluidic exosome isolation methods do not require an external actuation mechanism and hence are simpler and more straightforward compared to active methods. The main passive microfluidic isolation methods include filtration, inertial focusing, immunoaffinity, lateral displacement, and viscoelastic flow (Liga et al., 2015; Tayebi et al., 2020; Shirejini and Inci, 2022; Theel and Schwaminger, 2022). Except for the immunoaffinity-based isolation methods, the other passive methods are label-free and based on the physical properties of the exosomes.
Filtration, in general, is one of the most widely researched methods for the isolation of target bioparticles. Microfluidic exosome filtration has been performed by two common approaches; membrane integrated microfluidics (Liang et al., 2017) and nanowire trapping (Le and Fan, 2021). Generally, exosomes show a relatively wide range of size distribution varying from 30 to 150 nm (Ding et al., 2021; Wu et al., 2017). Such a size variation limits the use of filtration-based methods in terms of the isolation efficiency and the recovery of exosomes. The fixed geometry of the microfluidic filters restricts their target separation window. A wider target-size filtering approach can increase the contamination of the isolated exosomes with other nanoparticles falling within the same size distribution of the exosomes (Shirejini and Inci, 2022). Nevertheless, microfluidic filtering is a simple approach to isolate exosomes from various bodily fluids that do not require extensive user training or special equipment. The most important shortcoming of the microfluidic filtering methods is device clogging that reduces the isolation efficiency and can render the device unusable (Choe et al., 2021). A recent discovery has addressed this issue by using mechanical vibrations to apply negative pressure oscillations and prevent membrane clogging (Chen et al., 2021). An exosome recovery rate of about 90% was obtained by the authors which is higher compared to most of the conventional microfluidic filtering methods where the yield efficiency was between 40% to 70% (Wang et al., 2013; Le and Fan, 2021; Wang et al., 2021b). This is a good example of using a combined strategy for achieving better functionality and exosome separation efficiency compared to the traditional filtering approaches.
Inertial focusing takes advantage of inertial lift forces to push particles to specific locations based on their physical properties within a microfluidic channel. This is a continuous process that can enable high throughput isolation and avoids clogging (Lin et al., 2021; Teoh et al., 2022). For instance, a spiral device geometry has been commonly employed for the manipulation and isolation of submicron microvesicles and microparticles by taking advantage of inertial microfluidics (Tay et al., 2017). However, due to the smaller sizes of exosomes, inertial focusing is not an effective approach to manipulating exosomes. Thus, rather than direct manipulation, a negative selection approach is employed in which larger cells and particles can be separated from the sample flow leaving the nanosized bioparticles including exosomes behind. To improve the efficiency of the inertial focusing method for isolating exosomes, a combined strategy of immuno-binding and inertial focusing has been developed. Here, surface-functionalized microbeads are incubated with a solution containing exosomes, and the exosome-bound microbeads are isolated by inertial focusing (Dudani et al., 2015; Shirejini and Inci, 2022). This approach is more efficient due to the much larger sizes of the microbeads that can experience stronger inertial effects.
Immunoaffinity-based exosome-capturing methods have been applied in microfluidics through direct immobilization of exosome-specific antibodies on the surfaces of the microfluidic channels or microbeads (Shirejini and Inci, 2022). Magnetic particles are commonly used to capture exosomes and achieve antibody-specific exosome isolation (Li et al., 2021). The magnetic approach is also extensively used commercially due to its effectiveness and separation efficiency. Despite these advantages of the magnetic immunoassay binding method, the process is time-consuming and requires multiple steps.
Deterministic lateral displacement (DLD) is a sized-based continuous-flow passive isolation method. Pillar arrays of different sizes and shapes are employed in this method to laterally separate target bioparticles (McGrath et al., 2014). Nanoscale DLD has been also enabled by fabricating pillar arrays with nanometer-size geometries that allowed the separation of exosomes and nanoparticles that were as small as 20 nm (Wunsch et al., 2016; Smith et al., 2018). Nanoscale DLD is a promising approach to continuous-flow exosome separation. However, several disadvantages need attention to improve DLD-based exosome isolation. For example, DLD can have a low yield of exosomes, especially when working with low-concentration samples. In addition, DLD can be non-specific and can result in the isolation of other types of vesicles and particles. These include liposomes, microvesicles, and protein aggregates, which can lead to contamination of the isolated exosome samples. Further study of this approach is necessary to demonstrate its application for clinically relevant exosome isolation from various biofluids.
The viscoelastic flow-based exosome separation method relies on the elastic lift forces that increase with the volume of the particles (Yuan et al., 2018). In a recent example, this method was applied in exosome and bioparticle separation by modifying the sample medium with diluted poly(oxyethylene) achieving more than 90% exosome purity and a recovery rate of 80% (Liu et al., 2017). Repetitions of the microchannel geometry were later shown to improve the device performance by using periodically reversed Dean secondary flows to yield a purity of more than 92% (Zhou et al., 2019). Similar to the majority of the other passive microfluidic exosome separation methods, the viscoelastic flow-based separation method requires validation with various bodily fluids to prove the potential of this method for medical diagnostic applications.
There are several ways to improve viscoelastic flow-based exosome isolation methods. For example, the flow conditions, such as the flow rate, pressure, and channel geometry can be optimized to improve the efficiency and specificity of exosome isolation. In addition, combining viscoelastic flow-based isolation with other methods, such as size exclusion, dielectrophoresis, or magnetic bead-based isolation, can enhance the purity and yield of exosomes. Machine learning and artificial intelligence can also be applied to optimize the isolation process, by analyzing the flow characteristics, particle size, and surface markers.
Active Microfluidic Exosome Isolation
Active exosome isolation methods generally employ external force fields to manipulate particles and separate exosomes through negative or positive selection. The typical methods in this category are acoustic and dielectrophoretic isolation methods (Lin et al., 2021; Su et al., 2019). Both of these methods enable continuous-flow exosome isolation and are dynamically controllable. Even though active methods are more complex and demanding, there is a considerable research effort to actively improved exosome isolation strategies (Wang et al., 2021b).
The acoustic exosome isolation approach usually employs surface acoustic wave-based devices that generate mechanical waves on a piezoelectric surface (Wu et al., 2019). In the acoustic methods, depending on the interplay of acoustic radiation and streaming forces, bioparticles have been separated from each other based on their mechanical and physical differences (Ozcelik and Huang, 2020; Zhang et al., 2020; Ozcelik et al., 2022b). An important milestone in acoustic exosome isolation was the demonstration of exosome isolation from whole blood using an integrated microfluidic chip (Wu et al., 2017). In this approach, a cell removal module and an exosome separation module were functionally fused to isolate blood-derived exosomes in a continuous process with an efficiency of 98.4%. A similar approach was used for salivary exosomes to detect cancer (Wang et al., 2020). Even though acoustic exosome isolation offers dexterity and relatively higher efficiencies compared to most passive isolation methods, the complexity of the system demands a high level of user expertise. In addition, the equipment required for acoustic methods can be expensive and may not be accessible to all laboratories. To reduce the complexity of the isolation process, one can use commercially available kits and devices that are designed specifically for acoustic exosome isolation. These kits can simplify the isolation process and make the approach more accessible to researchers.
Dielectrophoresis (DEP) is another commonly explored particle isolation method that depends on medium and sample conductivity (Lee et al., 2020). In this method, externally applied alternating or direct currents can be applied to induce polarization for particle separation. DEP is employed to enrich and isolate exosomes from small sample volumes (Ibsen et al., 2017). In a study, DEP was utilized in two separate sections to identify two subpopulations of exosomes of varying sizes. Exosomes were captured by introducing a 2000 V electric potential differential for 20 s. Once the exosomes were captured, they were recovered using an electroosmotic flow (Ayala-Mar et al., 2019). While similar examples of DEP applications also exist in literature with good separation efficiencies, an important drawback of DEP-based isolation methods is the undesired Joule heating that can be detrimental to the biospecimen. In addition, device fabrication and operation are generally more intricate and costly in contrast to passive exosome isolation methods.
Conclusion and Future Perspectives
Both active and passive microfluidics platforms present viable alternatives to traditional benchtop exosome isolation methods that rely on expensive facilities, are time-consuming, and are potentially harmful to the exosomes due to extremely high g forces generated during ultra-centrifugation. Numerous microfluidic approaches have been implemented in exosome isolation studies with different advantages and disadvantages. In general, passive microfluidic exosome isolation methods are simpler in terms of the required peripheral equipment and user expertise. On the other hand, passive methods do not allow the dynamic control that active isolation methods offer. Both acoustic and DEP-based isolation methods have been demonstrated to yield high exosome isolation efficiencies. However, these methods are far from being the industrial standard for nanoparticle or exosome isolation because of their inherited limitations and current challenges.
An important limitation of microfluidic platforms is the lack of standardization. The majority of the reported microfluidic exosome isolation examples are proof of concept demonstrations that rely on niche architectures and distinct working mechanisms without universal standards. This prevents the improvement of microfluidic platforms for desired standards in terms of user experience and performance. This is also a problem in the global benchmarking of microfluidic isolation technologies. Commercialization is the only viable option for the widespread adoption of these microfluidic tools in life sciences and medical applications, but the lack of standardization limits the number of commercial products. Finally, the efficiencies of the microfluidic exosome isolation methods have been generally demonstrated in simple applications including standard samples. These platforms should be tested with a variety of bodily fluids and sample concentrations for simulating real-life clinical applications. This validation is also critical for faster commercialization of the documented microfluidic platforms.
Acknowledgement: We thank Dr. Sinan GUCLUER for the valuable discussions.
Funding Statement: This research was funded by The Scientific and Technological Research Council of Turkiye (TUBITAK), Grant Number 221S982.
Author Contributions: Study conception and: A.O. and O.C.; draft manuscript writing and editing: A.O. and O.C.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Abas BI, Demirbolat GM, Cevik O (2022). Wharton jelly-derived mesenchymal stem cell exosomes induce apoptosis and suppress EMT signaling in cervical cancer cells as an effective drug carrier system of paclitaxel. PLoS One 17: e0274607. https://doi.org/10.1371/journal.pone.0274607 [Google Scholar] [PubMed] [CrossRef]
Akkoyun F, Özçelik A (2022). A battery-powered fluid manipulation system actuated by mechanical vibrations. Actuators 11: 116. https://doi.org/10.3390/act11050116 [Google Scholar] [CrossRef]
Ayala-Mar S, Perez-Gonzalez VH, Mata-Gómez MA, Gallo-Villanueva RC, González-Valdez J (2019). Electrokinetically driven exosome separation and concentration using dielectrophoretic-enhanced PDMS-based microfluidics. Analytical Chemistry 91: 14975–14982. https://doi.org/10.1021/acs.analchem.9b03448 [Google Scholar] [PubMed] [CrossRef]
Cenik M, Abas BI, Kocabiyik B, Demirbolat GM, Cevik O (2022). Development of a new drug delivery system from HELA-derived exosomes and the effect of docetaxel-loaded exosomes on mitochondrial apoptosis. Journal of Pharmaceutical Innovation 17: 931–939. https://doi.org/10.1007/s12247-021-09566-1 [Google Scholar] [CrossRef]
Chen Y, Zhu Q, Cheng L, Wang Y, Li M et al. (2021). Exosome detection via the ultrafast-isolation system: EXODUS. Nature Methods 18: 212–218. https://doi.org/10.1038/s41592-020-01034-x [Google Scholar] [PubMed] [CrossRef]
Choe S, Kim B, Kim M (2021). Progress of microfluidic continuous separation techniques for Micro-/Nanoscale bioparticles. Biosensors 11: 464. https://doi.org/10.3390/bios11110464 [Google Scholar] [PubMed] [CrossRef]
Ding L, Yang X, Gao Z, Effah CY, Zhang X, Wu Y, Qu L (2021). A holistic review of the state-of-the-art microfluidics for exosome separation: An overview of the current status, existing obstacles, and future outlook. Small 17: 2007174. https://doi.org/10.1002/smll.202007174 [Google Scholar] [PubMed] [CrossRef]
Dudani JS, Gossett DR, Tse HTK, Lamm RJ, Kulkarni RP, Carlo DD (2015). Rapid inertial solution exchange for enrichment and flow cytometric detection of microvesicles. Biomicrofluidics 9: 014112. https://doi.org/10.1063/1.4907807 [Google Scholar] [PubMed] [CrossRef]
Ibsen SD, Wright J, Lewis JM, Kim S, Ko SY et al. (2017). Rapid isolation and detection of exosomes and associated biomarkers from plasma. ACS Nano 11: 6641–6651. https://doi.org/10.1021/acsnano.7b00549 [Google Scholar] [PubMed] [CrossRef]
Konoshenko MY, Lekchnov EA, Vlassov AV, Laktionov PP (2018). Isolation of extracellular vesicles: General methodologies and latest trends. BioMed Research International 2018: 1–27. https://doi.org/10.1155/2018/8545347 [Google Scholar] [PubMed] [CrossRef]
Lai JJ, Chau ZL, Chen S, Hill JJ, Korpany KV et al. (2022). Exosome processing and characterization approaches for research and technology development. Advanced Science 9: 2103222. https://doi.org/10.1002/advs.202103222 [Google Scholar] [PubMed] [CrossRef]
Le MCN, Fan HZ (2021). Exosome isolation using nanostructures and microfluidic devices. Biomedical Materials 16: 022005. https://doi.org/10.1088/1748-605X/abde70 [Google Scholar] [PubMed] [CrossRef]
Lee S, Roh SM, Lee E, Park Y, Lee BC, Kwon Y, Kim HJ, Kim J (2020). Applications of converged various forces for detection of biomolecules and novelty of dielectrophoretic force in the applications. Sensors 20: 3242. https://doi.org/10.3390/s20113242 [Google Scholar] [PubMed] [CrossRef]
Li X, Su L, Zhang X, Chen Q, Wang Y et al. (2022). Recent advances on the function and purification of milk exosomes: A review. Frontiers in Nutrition 9: 474. https://doi.org/10.3389/fnut.2022.871346 [Google Scholar] [PubMed] [CrossRef]
Li G, Zhu N, Zhou J, Kang K, Zhou X, Ying B, Yi Q, Wu Y (2021). A magnetic surface-enhanced Raman scattering platform for performing successive breast cancer exosome isolation and analysis. Journal of Materials Chemistry B 9: 2709–2716. https://doi.org/10.1039/D0TB02894K [Google Scholar] [PubMed] [CrossRef]
Liang LG, Kong MQ, Zhou S, Sheng YF, Wang P et al. (2017). An integrated double-filtration microfluidic device for isolation, enrichment and quantification of urinary extracellular vesicles for detection of bladder cancer. Scientific Reports 7: 46224. https://doi.org/10.1038/srep46224 [Google Scholar] [PubMed] [CrossRef]
Liga A, Vliegenthart ADB, Oosthuyzen W, Dear JW, Kersaudy-Kerhoas M (2015). Exosome isolation: A microfluidic road-map. Lab on a Chip 15: 2388–2394. https://doi.org/10.1039/C5LC00240K [Google Scholar] [PubMed] [CrossRef]
Lin B, Lei Y, Wang J, Zhu L, Wu Y, Zhang H, Wu L, Zhang P, Yang C (2021). Microfluidic-based exosome analysis for liquid biopsy. Small Methods 5: 2001131. https://doi.org/10.1002/smtd.202001131 [Google Scholar] [PubMed] [CrossRef]
Liu C, Guo J, Tian F, Yang N, Yan F, Ding Y, Wei J, Hu G, Nie G, Sun J (2017). Field-free isolation of exosomes from extracellular vesicles by microfluidic viscoelastic flows. ACS Nano 11: 6968–6976. https://doi.org/10.1021/acsnano.7b02277 [Google Scholar] [PubMed] [CrossRef]
McGrath J, Jimenez M, Bridle H (2014). Deterministic lateral displacement for particle separation: A review. Lab on a Chip 14: 4139–4158. https://doi.org/10.1039/C4LC00939H [Google Scholar] [PubMed] [CrossRef]
Momen-Heravi F, Balaj L, Alian S, Tigges J, Toxavidis V, Ericsson M, Distel RJ, Ivanov AR, Skog J, Kuo WP (2012). Alternative methods for characterization of extracellular vesicles. Frontiers in Physiology 3: 1–8. https://doi.org/10.3389/fphys.2012.00354 [Google Scholar] [PubMed] [CrossRef]
Ozcelik A (2022). 3D printed device for separation of cells and particles by tilted bulk acoustic wave actuation. Actuators 11: 249. https://doi.org/10.3390/act11090249 [Google Scholar] [CrossRef]
Ozcelik A, Abas BI, Erdogan O, Cevik E, Cevik O (2022a). On-chip organoid formation to study CXCR4/CXCL-12 chemokine microenvironment responses for renal cancer drug testing. Biosensors 12: 1177. https://doi.org/10.3390/bios12121177 [Google Scholar] [PubMed] [CrossRef]
Ozcelik A, Huang TJ (2020). Acoustic tweezers for single-cell manipulation. In: Handbook of Single Cell Technologies, pp. 1–27. Singapore: Springer. [Google Scholar]
Ozcelik A, Rich J, Huang TJ (2022b). Fundamentals and applications of acoustics in microfluidics. In: Multidisciplinary Microfluidic and Nanofluidic Lab-on-a-Chip, pp. 297–321. Amsterdam: Elsevier. [Google Scholar]
Panfoli I, Bruschi M, Candiano G (2022). Exosomes: Key tools for cancer liquid biopsy. BIOCELL 46: 2167–2176. https://doi.org/10.32604/biocell.2022.020154 [Google Scholar] [CrossRef]
Shirejini SZ, Inci F (2022). The Yin and Yang of exosome isolation methods: Conventional practice, microfluidics, and commercial kits. Biotechnology Advances 54: 107814. https://doi.org/10.1016/j.biotechadv.2021.107814 [Google Scholar] [PubMed] [CrossRef]
Smith JT, Wunsch BH, Dogra N, Ahsen ME, Lee K et al. (2018). Integrated nanoscale deterministic lateral displacement arrays for separation of extracellular vesicles from clinically-relevant volumes of biological samples. Lab on a Chip 18: 3913–3925. https://doi.org/10.1039/C8LC01017J [Google Scholar] [PubMed] [CrossRef]
Su W, Li H, Chen W, Qin J (2019). Microfluidic strategies for label-free exosomes isolation and analysis. TrAC—Trends in Analytical Chemistry 118: 686–698. https://doi.org/10.1016/j.trac.2019.06.037 [Google Scholar] [CrossRef]
Tay HM, Kharel S, Dalan R, Chen ZJ, Tan KK, Boehm BO, Loo SCJ, Hou HW (2017). Rapid purification of sub-micrometer particles for enhanced drug release and microvesicles isolation. NPG Asia Materials 9: e434. https://doi.org/10.1038/am.2017.175 [Google Scholar] [CrossRef]
Tayebi M, Zhou Y, Tripathi P, Chandramohanadas R, Ai Y (2020). Exosome purification and analysis using a facile microfluidic hydrodynamic trapping device. Analytical Chemistry 92: 10733–10742. https://doi.org/10.1021/acs.analchem.0c02006 [Google Scholar] [PubMed] [CrossRef]
Teoh BY, Lim YM, Chong WY, Subramaniam M, Tan ZZ, Misran M, Suk VRE, Lo KW, Lee PF (2022). Isolation of exosome from the culture medium of Nasopharyngeal cancer (NPC) C666-1 cells using inertial based Microfluidic channel. Biomedical Microdevices 24: 12. https://doi.org/10.1007/s10544-022-00609-z [Google Scholar] [PubMed] [CrossRef]
Theel EK, Schwaminger SP (2022). Microfluidic approaches for affinity-based exosome separation. International Journal of Molecular Sciences 23: 9004. https://doi.org/10.3390/ijms23169004 [Google Scholar] [PubMed] [CrossRef]
Uslu D, Abas BI, Demirbolat GM, Cevik O (2022). Effect of platelet exosomes loaded with doxorubicin as a targeted therapy on triple-negative breast cancer cells. Molecular Diversity 201: 785. https://doi.org/10.1007/s11030-022-10591-6 [Google Scholar] [PubMed] [CrossRef]
Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nature Cell Biology 9: 654–659. https://doi.org/10.1038/ncb1596 [Google Scholar] [PubMed] [CrossRef]
Wang T, Chen C, Larcher LM, Barrero RA, Veedu RN (2019). Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development. Biotechnology Advances 37: 28–50. https://doi.org/10.1016/j.biotechadv.2018.11.001 [Google Scholar] [PubMed] [CrossRef]
Wang X, Huang W, Liu G, Cai W, Millard RW, Wang Y, Chang J, Peng T, Fan GC (2014). Cardiomyocytes mediate anti-angiogenesis in type 2 diabetic rats through the exosomal transfer of miR-320 into endothelial cells. Journal of Molecular and Cellular Cardiology 74: 139–150. https://doi.org/10.1016/j.yjmcc.2014.05.001 [Google Scholar] [PubMed] [CrossRef]
Wang C, Li Z, Liu Y, Yuan L (2021a). Exosomes in atherosclerosis: Performers, bystanders, biomarkers, and therapeutic targets. Theranostics 11: 3996–4010. https://doi.org/10.7150/thno.56035 [Google Scholar] [PubMed] [CrossRef]
Wang Z, Li F, Rufo J, Chen C, Yang S et al. (2020). Acoustofluidic salivary exosome isolation: A liquid biopsy compatible approach for human papillomavirus-associated oropharyngeal cancer detection. Journal of Molecular Diagnostics 22: 50–59. https://doi.org/10.1016/j.jmoldx.2019.08.004 [Google Scholar] [PubMed] [CrossRef]
Wang J, Ma P, Kim DH, Liu BF, Demirci U (2021b). Towards microfluidic-based exosome isolation and detection for tumor therapy. Nano Today 37: 101066. https://doi.org/10.1016/j.nantod.2020.101066 [Google Scholar] [PubMed] [CrossRef]
Wang Z, Wu H, Fine D, Schmulen J, Hu Y, Godin B, Zhang JXJ, Liu X (2013). Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab on a Chip 13: 2879–2882 https://doi.org/10.1039/c3lc41343h [Google Scholar] [PubMed] [CrossRef]
Wu M, Ouyang Y, Wang Z, Zhang R, Huang PH et al. (2017). Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proceedings of the National Academy of Sciences of the United States of America 114: 10584–10589. https://doi.org/10.1073/pnas.1709210114 [Google Scholar] [PubMed] [CrossRef]
Wu M, Ozcelik A, Rufo J, Wang Z, Fang R, Jun Huang T (2019). Acoustofluidic separation of cells and particles. Microsystems Nanoengineering 5: 32. https://doi.org/10.1038/s41378-019-0064-3 [Google Scholar] [PubMed] [CrossRef]
Wunsch BH, Smith JT, Gifford SM, Wang C, Brink M, Bruce RL, Austin RH, Stolovitzky G, Astier Y (2016). Nanoscale lateral displacement arrays for the separation of exosomes and colloids down to 20 nm. Nature Nanotechnology 11: 936–940. https://doi.org/10.1038/nnano.2016.134 [Google Scholar] [PubMed] [CrossRef]
You B, Qian H (2022). MSCs-exosomes in regeneration medicine: Current evidence and future perspectives. BIOCELL 46: 1459–1463. https://doi.org/10.32604/biocell.2022.018378 [Google Scholar] [CrossRef]
Yuan D, Zhao Q, Yan S, Tang S-Y, Alici G, Zhang J, Li W (2018). Recent progress of particle migration in viscoelastic fluids. Lab on a Chip 18: 551–567. https://doi.org/10.1039/C7LC01076A [Google Scholar] [PubMed] [CrossRef]
Zhang P, Bachman H, Ozcelik A, Huang TJ (2020). Acoustic microfluidics. Annual Review of Analytical Chemistry 13: 17–43. https://doi.org/10.1146/annurev-anchem-090919-102205 [Google Scholar] [PubMed] [CrossRef]
Zheng C, Xie L, Qin H, Liu X, Chen X, Lv F, Wang L, Zhu X, Xu J (2022). The role of extracellular vesicles in systemic lupus erythematosus. Frontiers in Cell and Developmental Biology 10: 835566. https://doi.org/10.3389/fcell.2022.835566 [Google Scholar] [PubMed] [CrossRef]
Zhou Y, Ma Z, Tayebi M, Ai Y (2019). Submicron particle focusing and exosome sorting by wavy microchannel structures within viscoelastic fluids. Analytical Chemistry 91: 4577–4584. https://doi.org/10.1021/acs.analchem.8b05749 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools