 Open Access
Open Access
ARTICLE
Cloning of and analysis of cadmium resistant in Potentilla sericea
1 College of Landscape Architecture, Northeast Forestry University, Harbin, 150040, China
2 Research Center for Economic Plant Pests, Heilongjiang Forest Protection Institute, Harbin, 150040, China
* Corresponding Author: JIANHUI WU. Email:
(This article belongs to the Special Issue: Physiology and Molecular Biology of Plant Stress Tolerance)
BIOCELL 2023, 47(7), 1571-1582. https://doi.org/10.32604/biocell.2023.029106
Received 02 February 2023; Accepted 20 March 2023; Issue published 21 June 2023
Abstract
Background:Potentilla sericea is a heavy metal hyperaccumulator landscaping plant. MYB transcription factors play an important role in regulating plant stress response to adversity. However, there are few studies on MYB transcription factors in stress tolerance in Potentilla sericea. In this study, the gene was successfully cloned from Potentilla sericea. Methods: Bioinformatic analysis and real-time quantitative PCR (qPCR) methods were used to evaluate this gene. The transgenic A. thaliana were obtained by flower dipping and the gene function was identified by determining physiological indicators under cadmium stress. Results: The open reading frame of is 942 bp, which encodes 313 amino acids (aa) and belongs to the R2R3 MYB transcription factor. The plant overexpression vector PBI121-PsMYB62-GFP was constructed and successfully transferred into A. thaliana. The relative expression level of PsMYB62was significantly increased by CdCl2, NaCl, ABA, and mannitol treatments. The germination rate of transgenic seeds was higher than those of wild type (WT) and empty vector (EV) under different concentrations of cadmium treatment. Upon treatment with 100 μmol·L-1 of CdCl2·2.5H2O, the activities of superoxide dismutase (SOD), peroxidase (POD), and catalase (CAT) in the transgenic plants were significantly higher than those in the WT and EV. The contents of H2O2, O2·- and malondialdehyde (MDA) in transgenic lines were increased, but lower than those in WT and EV. The expression levels of AtGSH, AtPCS, and AtNAS4 that were related to the regulation of cadmium were increased, but the expression levels of transgenic lines were higher than those of WT and EV. Conclusion: The above results showed that could be induced by cadmium and could improve the cadmium resistance of plants.Keywords
Soil heavy metal pollution is becoming increasingly serious, posing a serious threat to food safety, food security, and human health (Zhou et al., 2017). Excessive levels of the heavy metal cadmium (Cd) in the soil due to human activities such as mining, agricultural production, and heavy industry (Stephanie and Li, 2021). Excess cadmium affects plant growth by inducing excessive production of reactive oxygen species (ROS) and mediating molecular and cellular damage. This in turn leads to morphological changes and effects on physiological metabolic processes (Ahmad et al., 2019; Kohli et al., 2019; Mansoor et al., 2022).
Potentilla sericea is a perennial herb that belongs to the Potentilla genus in the Rosaceae family. It has the characteristics of cold resistance, drought resistance, barren resistance, extensive management resistance, and strong stress resistance, and has a long green period and flowering landscaping. This species is also a potential heavy metal hyper-enriched plant. The research on Potentilla sericea is mainly focused on the domestication of introduced species and its medicinal uses (Zhang, 2014; Yao et al., 2019; Wu et al., 2022). Although there are studies on the effects of its ultrastructure and physiology under abiotic stresses (Wu et al., 2016, 2017; Qi et al., 2018; Zhang, 2020), there is still a lack of studies on the plant at the molecular level under cadmium stress.
Transcription factors are protein molecules with specific structures that initiate gene expression and play an important role in the regulation of plant responses to stresses (Yamasaki et al., 2008). As one of the largest families of transcription factors in higher plants, MYB is a type of transcription factor family with highly conserved DNA binding domains. Studies have shown that the MYB transcription factor family is widely involved in plant growth and development, secondary metabolism, hormone regulation, and biotic and abiotic stress processes (Albert et al., 2015; Qiu et al., 2020; Wang et al., 2021a; Wei and Lan, 2022). As of now, there have been studies on the response of MYB transcription factors to heavy metal cadmium stress in plants. For example, AtMYB49 positively regulates the expression of bHLH38 and bHLH101, which are required for cadmium uptake in A. thaliana (Zhang et al., 2019). AtMYB59 promotes the uptake of cadmium into the cells by calcium transport proteins and enhances the sensitivity of plants to cadmium (Suo et al., 2003). The Juglans regia transcription factor JrMYB2 directly binds to the MYB core element of the JrVHAG1 (G-subunit of vacuolar H+-ATPase) promoter to enhance resistance to cadmium stress in A. thaliana (Xu et al., 2018). In another report, Wang et al. (2021c) found that the BvMYB44 gene of Beta vulgaris was significantly up-regulated in leaves and roots to varying degrees after cadmium stress treatment. Further, it is hypothesized that there is a certain response relationship between BvMYB44 and cadmium stress.
At present, studies have shown that the MYB family of genes can regulate plant response to Cd stress. Our previous studies isolated and identified an MYB transcription factor—PsMYB2 from Potentilla sericea that regulates the plant response to Cd stress. According to the MYB family analysis of Fan (2021), this study cloned an MYB transcription factor gene from the transcriptome data of Potentilla sericea and named it PsMYB62, which belongs to the same subfamily as MYB2. We obtained transgenic overexpression strains of A. thaliana by Agrobacterium transformation and further investigated its response mechanism in response to Cd stress, aiming to provide genetic resources and a theoretical basis for breeding Cd-tolerant plants.
The seedlings of Potentilla sericea with consistent growth trends were taken from the nursery of the College of Landscape Architecture, Northeast Forestry University for hydroponic culture. The seedlings were transplanted in 1/2 Hoagland nutrient solution with pH = 5.8 for 2 weeks, and the nutrient solution was changed every 2 days for subsequent use. The seeds of wild-type A. thaliana (Columbia-0 type) were preserved at the College of Landscape Architecture, Northeast Forestry University.
Extraction of RNA from the roots of Potentilla sericea
The total RNA was extracted from the roots of Potentilla sericea using the E.Z.N.A. Total RNA Kit I (OMEGA, Lilburn, USA) provided by Harbin Yuze Technology. The cDNA was obtained by reverse transcription using ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, Ōsaka, Japan).
The PsMYB62 gene conventional PCR primer PsMYB62-F/R (Table 1) was designed using Primer Premier 5.0 according to the principle of primer design. PsMYB62 was amplified by KOD-Plus (TOYOBO, Ōsaka, Japan) polymerase with cDNA as the template and PsMYB62-F/R as primers according to the manufacturer’s protocol. The products were examined using agarose gel electrophoresis (140 V, 400 mA, 12 min), and the gels containing the correct band were recovered using the Gel Extraction Kit D2500 (OMEGA, Lilburn, USA). The gel recovery product was transformed into E. coli DH-5α (Weidi Biotech, Shanghai, China) and sequenced by Harbin RuiboXingke Biotechnology Co., Ltd., China.
Bioinformatics analysis of the PsMYB62 gene
The conserved domain of the PsMYB62 protein was predicted by InterPro (InterPro (Ebi.ac.UK)), and eight homologous protein sequences were searched by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). DNAMAN 8.0.8 was used for protein multiple sequence alignment. The selected protein sequences were aligned by Clustal W, and the phylogenetic tree was constructed by Neighbor-Joining in MEGA7.0. Protein physicochemical properties were analyzed using the Prot-Param (https://web.expasy.org/resources/protparam) and hydrophobicity was analyzed using the ProtScale (https://web.expasy.org/resources/protscale). The DeepTMHMM (https://dtu.biolib.com/DeepTMHMM) was used to predict the transmembrane structural domains. The Protein secondary and tertiary structures were predicted using online tools SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=npsa_sopma.html) and SWISS-MODEL (http://swissmodel.expasy.org/interactive).
Analysis of the expression pattern of the PsMYB62 gene
The healthy Potentilla sericea seedlings with uniform growth were selected and replaced with nutrient solution containing CdCl2·2.5H2O at 0, 100 μmol·L−1, containing NaCl at 0, 200, and 0, 400 mmol·L−1 mannitol, and treated with 0, 100 μmol·L-1Abscisic Acid (ABA). At 0, 3, 6, 12, 24, and 48 h, 0.1 g sample of root, stem, and leaf of each treatment were taken and the sampling was repeated three times. The qPCR analysis followed the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (Bustin et al., 2009). RNA was extracted from each sample and reverse transcribed into cDNA according to the instruction manual, and the product cDNA was diluted 10-fold as a template for qPCR. The internal reference gene was referenced to the β-actin gene (β-actin, ACTB) commonly used in strawberries (Zhang et al., 2016). The reaction was carried out using the Ultra SYBR Mixture (Low ROX) (CWBIO, Beijing, China) on a LightCycler® 96 (Roche, Munich, Germany). The sequences of primers PsMYB62-qPCR-F/R and β-actin-F/R used are shown in Table 1. The 2−ΔΔCt method was used for data analysis and expression levels of PsMYB62. SPSS 22.0 was used for significance analysis.
Construction of plant expression vector and identification of transgenic lines
Seeds of wild-type A. thaliana were sown into plug trays and cultured until flowering and bolting about 10 cm.
Plasmids of Blunt-PsMYB62 and PBI121-GFP vectors with enzymatic sites were digested with BamH I and Sal I restriction enzymes. The two target fragments recovered from the gel were ligated with T4-DNA ligase overnight at 16°C, and then the ligation product was transformed into E. coli DH-5α. After shaking and amplification at 37°C, the recombinant plasmid was extracted. The PBI121-PsMYB62-GFP overexpression vector was constructed and transformed into Agrobacterium tumefaciens EHA105 (Weidi Biotech, Shanghai, China) for expanded culture, and subsequently transformed into A. thaliana by the flower dipping method.
A. thaliana continued to be cultured until the pods matured, and the seeds were collected and screened for resistance using 1/2 MS solid medium containing 50 mg·L−1 Kana. Seedlings that could grow normally on the medium were transferred into soil for further culture. The DNA of the leaf was extracted using the HP Plant DNA Kit D2485 (OMEGA, Lilburn, USA) as a template, and PsMYB62-GFP-F/R was used as a primer (Table 1) to identify trans-PsMYB62 positive lines by PCR. The expression level of the PsMYB62 gene was verified by qPCR, and stable transgenic overexpression lines were screened out. The screening process was repeated to obtain lines with a stable inheritance of resistance for 3 generations.
Analysis of stress resistance of trans-PsMYB62 A. thaliana under Cd stress at the germination stage
The wild-type, empty, and transgenic A. thaliana seeds were disinfected and evenly seeded on 1/2 MS plates containing 0, 50, 100, 150 μmol·L−1 CdCl2·2.5H2O. Three replicates were done for each concentration. The dishes with four equal parts were first vernalized in a refrigerator at 4°C for 3 d, and then placed in a culture chamber for 7 d. The germination rate of A. thaliana seeds under Cd stress was observed and measured using the following formula: the rate of emergence (%) = the number of seeds germinated/total number of seeds × 100% (Lv et al., 2021).
Determination of physiological indicators of trans-PsMYB62 A. thaliana under Cd stress
One wild-type line (WT), one empty line (EV), and two transgenic lines of A. thaliana with good growth and similar growth patterns were selected for the experiment. For the control group, daily watering with 50 ml was done while the other group was treated with 50 ml of 100 μmol·L−1 CdCl2·2.5H2O each day. After 14 days, the phenotype of A. thaliana was observed and the leaves were sampled as three biological replicates. The activity of superoxide dismutase (SOD), peroxidase (POD), catalase (CAT), and the content of malondialdehyde (MDA) was determined by using commercial kits (Grace Bio-Tek, Suzhou, China). The determination of the chlorophyll content was as reported by Wang (2006). The relative conductivity was determined following the method of Wang et al. (2021b). The contents of H2O2 and O2·− were determined using the H2O2-2-Y and SA-2-G kits (Comin Bio-Tek, Suzhou, China).
Expression of related genes under Cd stress
Four lines were treated with 50 ml of 100 μmol·L−1 CdCl2·2.5H2O each day. After 7 days, the leaves were sampled as three biological replicates. Referring to the manufacturer’s protocol, the total RNA of the sample was extracted and reverse transcribed. The expression of AtGSH1, AtPCS1, AtNAS4, and AtHMA2 under CdCl2 treatment was analyzed by qPCR using A. thaliana Actin11 as the internal reference gene. Primer sequences are given in Table 1.
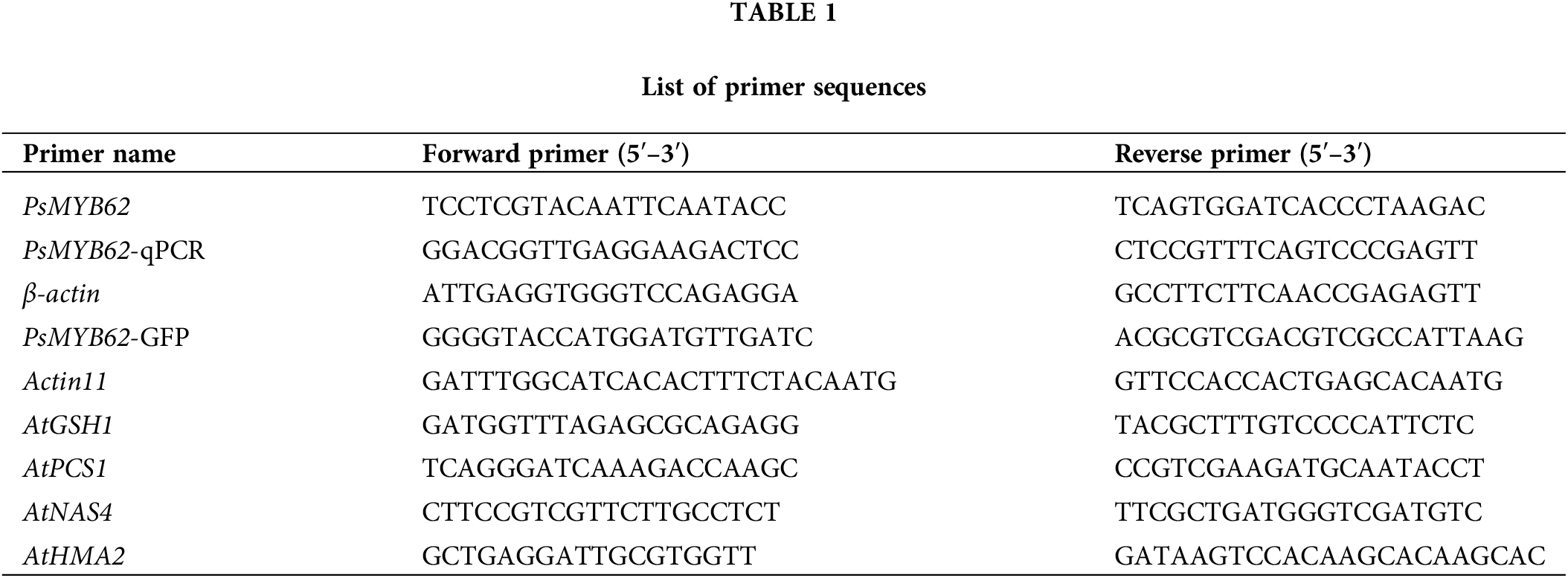
PCR amplification was performed using the cDNA of Potentilla sericea roots as the template and PsMYB62-F/R as a primer. The bands between 750 and 1000 bp were recovered (Fig. 1). The sequencing results were compared with BLAST in NCBI, and the comparison was correct. Therefore, we successfully cloned the PsMYB62 gene with an ORF length of 942 bp, encoding 313 amino acids (aa). The GenBank accession number was OQ158992.
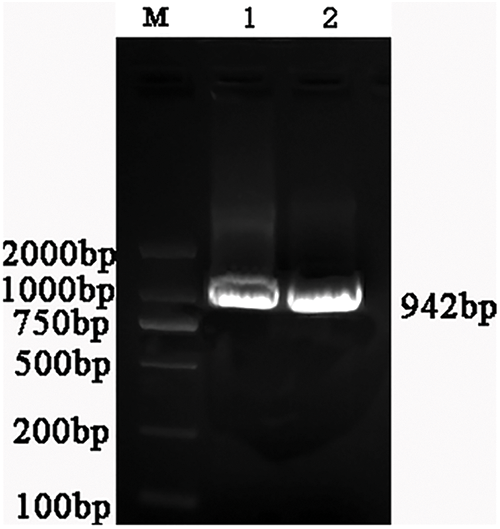
Figure 1: PCR results of the cloning of PsMYB62. Note: M: DL2000 Marker (Takara, Ōsaka, Japan); 1, 2: Bands with different Tm.
Bioinformatics analysis of the PsMYB62 gene
The conserved domain analysis of the protein showed that PsMYB62 had a SANT motif at 32–79 amino acids and 84–129 amino acids respectively, and each SANT is an MYB domain. The homologous proteins were seen in Rosa chinencis, Potentilla anserina, Prunus mume, Prunus dulcis, Prunus persica, Prunus avium, Pyrus betulifolia, and Malus domestica. Homology analysis using DNAMAN 8.0.8 showed that the PsMYB62 gene had the highest homology with Rosa chinensis. The phylogenetic tree was constructed using MEGA7.0 (Fig. 2). PsMYB62 clustered with Rosa chinensis and Potentilla anserina, with the closest evolutionary distance and the highest homology.
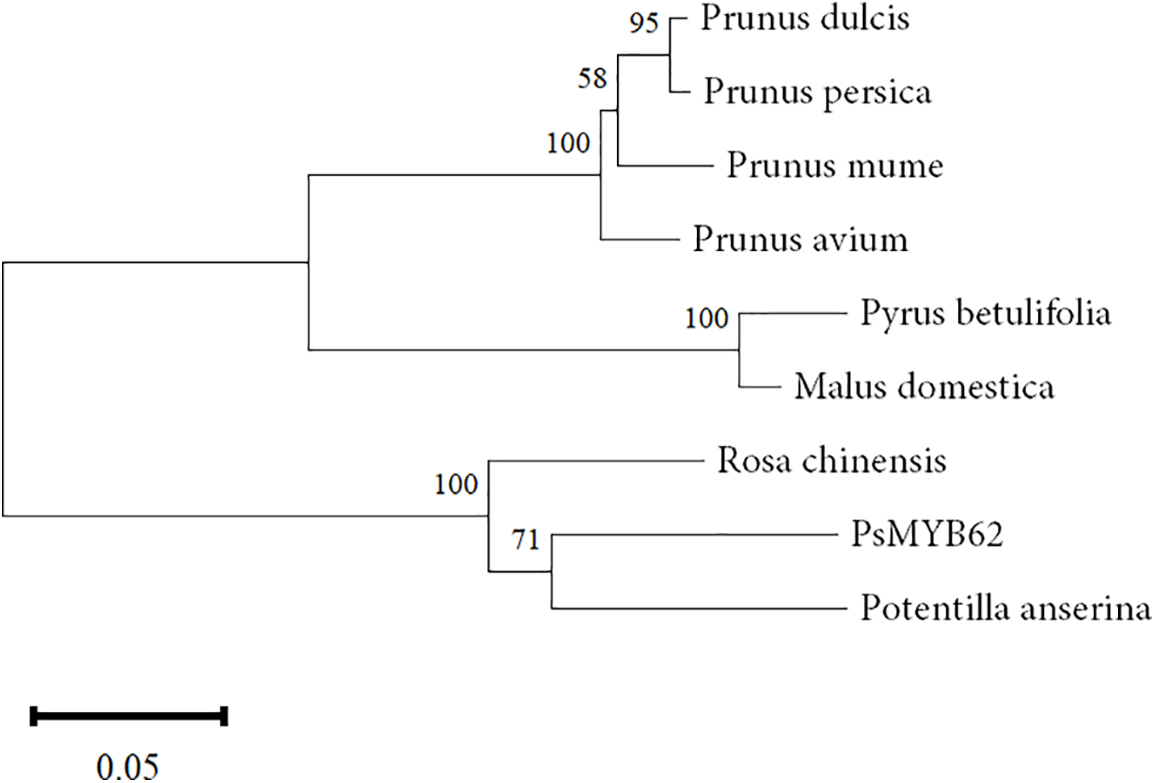
Figure 2: Phylogenetic tree analysis of PsMYB62.
The analysis of the physical and chemical properties of 313 aa encoded by the PsMYB62 gene showed that the protein has a relative molecular mass of 76.91 kDa, a theoretical isoelectric point of 5.08, and an instability coefficient of 50.83. Hence, it was speculated to be an unstable protein. The hydrophilic prediction results showed that the PsMYB62 protein contained 272 hydrophilic regions and 41 hydrophobic regions. It has the strongest hydrophobicity at 183aa and the strongest hydrophilicity at 297aa with an average coefficient of hydrophilicity of −1.073, which is presumed to be a hydrophilic protein (Fig. 3).
Figure 3: Hydrophilicity analysis of PsMYB62.
The secondary structure of the PsMYB62 protein was predicted by (self-optimized prediction method with alignment) SOPMA2.0. In the secondary structure of PsMYB62 protein, the alpha-helix accounted for 34.94%, the beta-turn accounted for 2.88%, the extended strand accounted for 7.05%, and the random coil accounted for 55.13%, belonging to R2R3 MYB transcription factor (Fig. 4A). The three-dimensional homology modeling of PsMYB62 was consistent with the secondary structure (Fig. 4B).
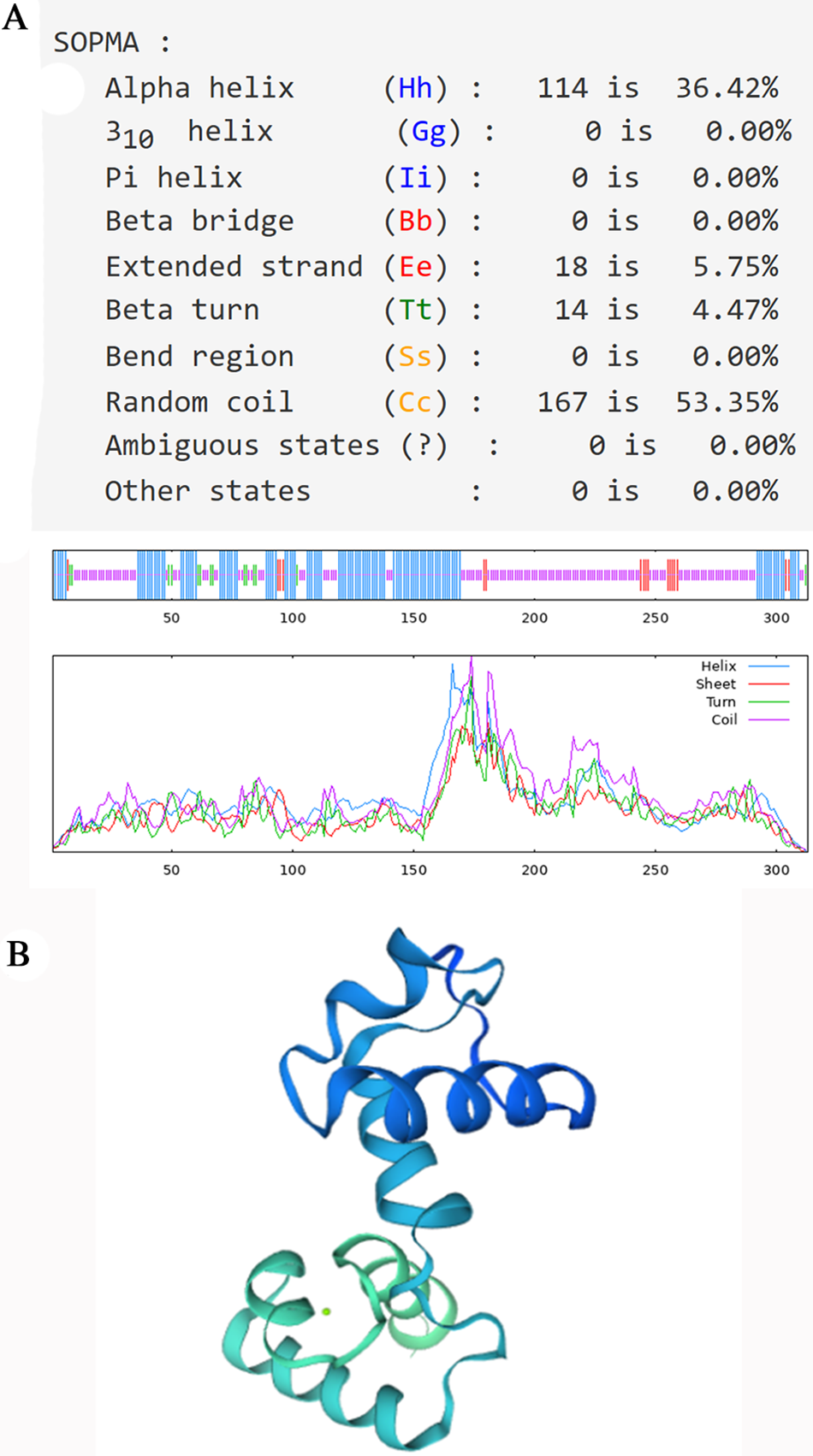
Figure 4: PsMYB62 protein structure prediction. (A) Prediction of the secondary structure of PsMYB62 protein; (B) Prediction of the tertiary structure of PsMYB62 protein.
Analysis of the expression pattern of the PsMYB62 gene
The results of the qPCR analysis showed that the expression of the PsMYB62 gene in different parts of Potentilla sericea was up-regulated to different degrees under different treatments (Figs. 5A–5D).
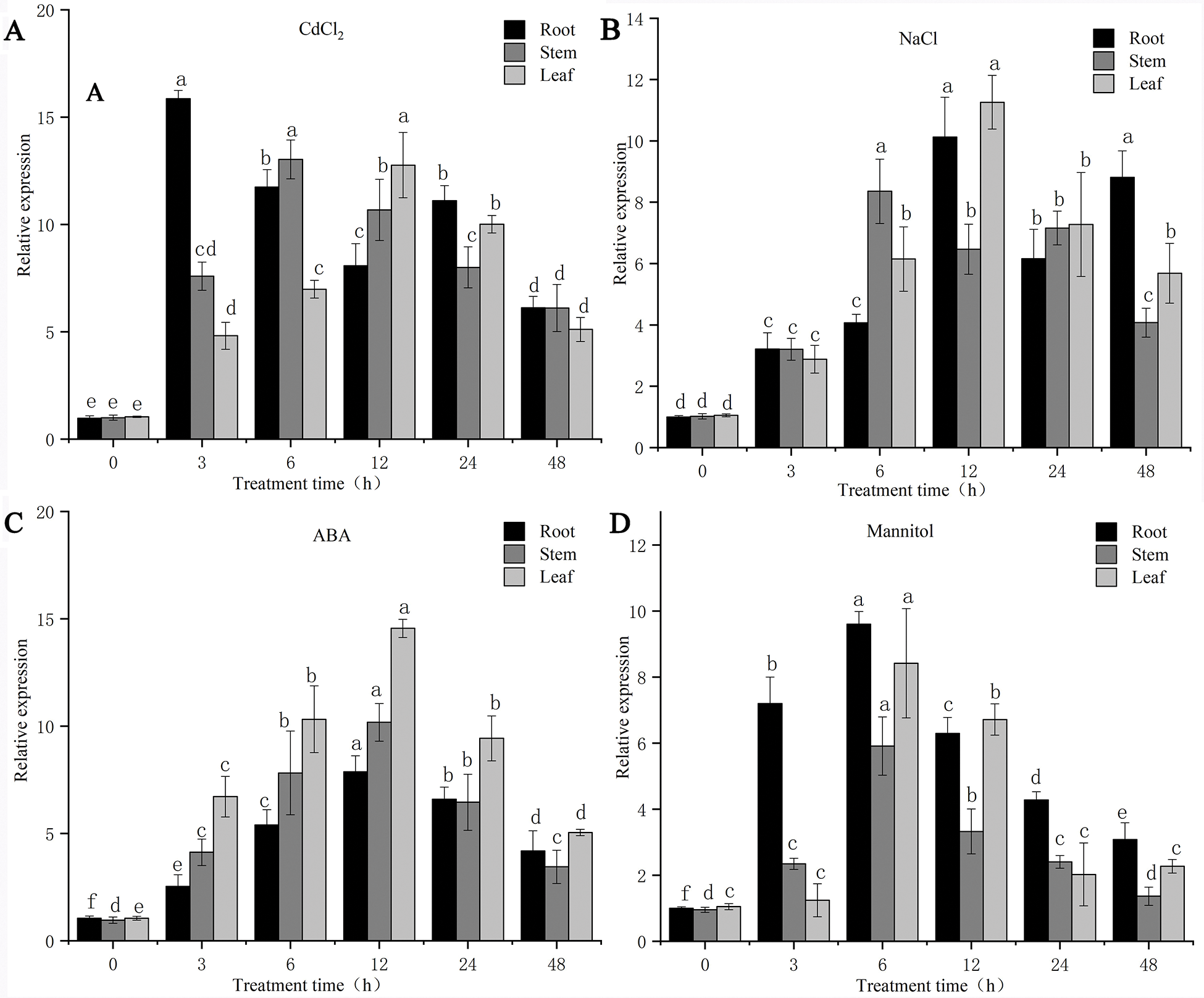
Figure 5: Expression patterns of PsMYB62 under different abiotic stress treatments.
Compared with the NaCl treatment and the mannitol treatment, the expression of the PsMYB62 gene increased to a higher degree under the CdCl2 treatment and ABA treatment. Under Cd treatment, the expression of the PsMYB62 gene in roots reached the maximum at 3 h, which was 15.85 times that of the control. After 12 h of ABA and NaCl treatment, the expression of the gene in leaves reached the highest value of 14.55 and 11.26, respectively. The maximum expression of this gene under mannitol treatment was 9.60 and was detected in the roots at 6 h. Under the four different treatments, the relative expression of the PsMYB62 gene showed an increasing trend which then decreased with the extension of treatment time. Further, the relative expression across different parts of Potentilla sericea dropped to a lower level at 48 h. The root was most sensitive to Cd treatment. The expression of the PsMYB62 gene reached the highest value at 3 h, then decreased in a fluctuating manner. The highest expression was at 6 h under mannitol treatment while the highest level was reached at 12 h under NaCl and ABA treatments. The expression level of PsMYB62 in the stem was up-regulated to the highest level at 12 h under Cd treatment, NaCl treatment, and ABA treatment, while it reached the maximum at 6 h under mannitol treatment. PsMYB62 was most sensitive to mannitol treatment in leaves, and its expression reached the maximum at 6 h while it reached the highest level at 12 h under the other three treatments. Although the relative expression of the PsMYB62 gene had the same trend under different treatments, the expression level was higher under Cd treatment and ABA treatment. This indicated that PsMYB62 played an important role in response to Cd and ABA stress. The gene expression reached the highest level at 6 h in the root and was most sensitive to Cd, probably because the root had stronger resistance to Cd than the stem and leaf.
Construction of the plant expression vector and identification of transgenic lines
The status of the constructed recombinant plasmid PBI121-PsMYB62-GFP was verified again using BamH I and Sal I restriction endonucleases. The recombinant plasmid was cut into two bands, and the lower band was close to 1000 bp, which was consistent with the length of the target band. Therefore, the PBI121-PsMYB62-GFP recombinant plasmid was successfully constructed.
Seedlings that were able to grow normally on 1/2 MS solid medium containing 50 mg·L−1 Kana were transferred to soil for culture. Six strains of OE-1~OE-6 were selected for DNA extraction for PCR identification and qPCR detection. The results showed that the OE-1~OE-6 lines were all PsMYB62 transgenic positive lines. Compared with the wild type (WT), the relative expression levels of the gene in each line were 11.63, 27.86, 21.76, 5.98, 23.81, and 17.59. The trans-PsMYB62 lines OE-2 and OE-5 were selected for subsequent experiments.
Analysis of stress resistance of trans-PsMYB62 A. thaliana under Cd stress at the germination stage
The WT, EV, and transgenic strains seeds could germinate and grow normally without significant differences on the medium without CdCl2. With the increase of CdCl2 concentration, the germination rate of A. thaliana seeds was inhibited, and the higher the concentration of CdCl2, the more obvious the inhibitory effect. However, the WT and EV were more significantly inhibited than the transgenic plants (Fig. 6).
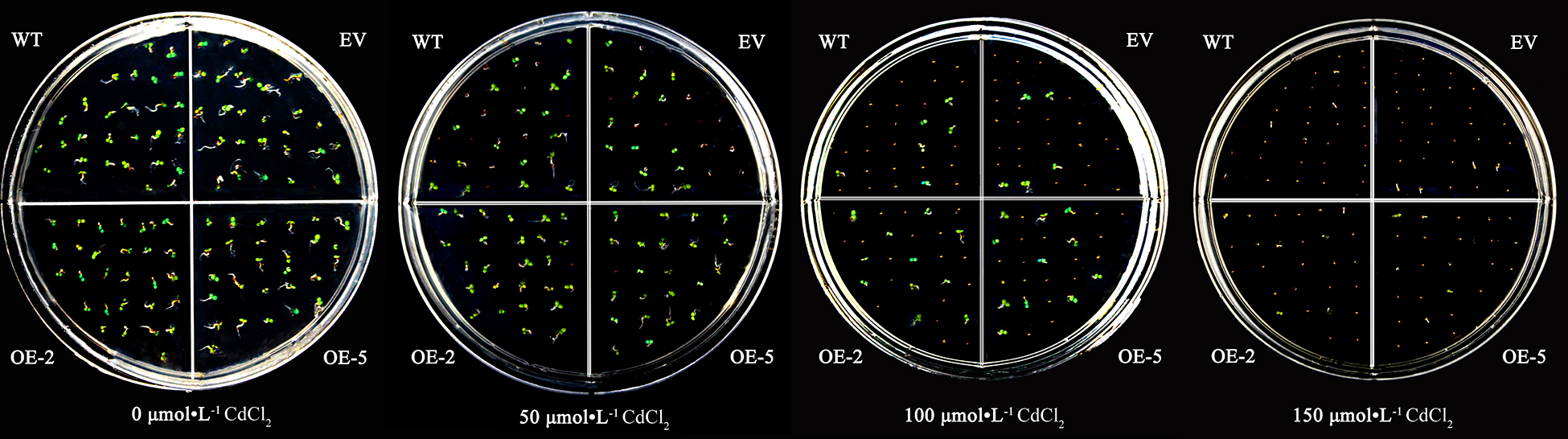
Figure 6: The germination of A. thaliana seeds transformed with the PsMYB62 gene under cadmium stress.
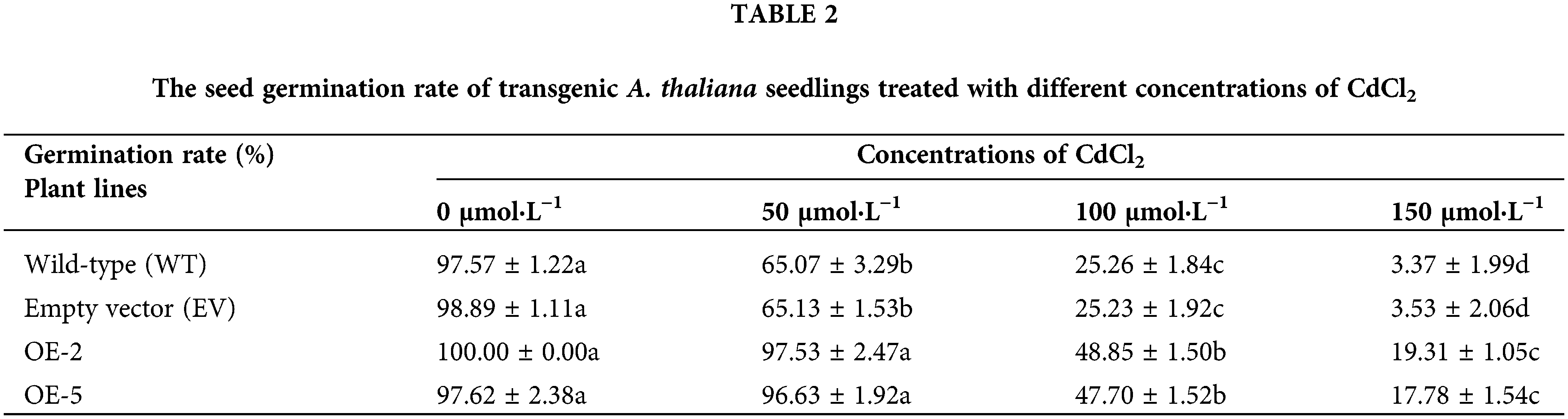
Before treatment, the seed germination rate was more than 97% for the WT, EV, and transgenic lines, with no significant difference. When treated with 50 μmol·L−1 CdCl2, the germination rate of transgenic lines remained above 96%, while the germination rate of WT and EV seeds decreased to 65%. After 100 μmol·L−1 CdCl2 treatment, nearly half of the seeds of the transgenic strains were able to germinate, while the germination rate of WT and EV was only 25%. When the concentration of CdCl2 treatment increased to 150 μmol·L−1, the germination rate of WT and EV was only 3%, with almost no germination (Table 2; Fig. 4). It could be seen that in a certain concentration of cadmium treatment, overexpression of PsMYB62 gene could enhance the cadmium resistance of A. thaliana. Subsequently, we treated A. thaliana seedlings with 100 μmol·L−1 CdCl2 to further verify the mitigation effect of the PsMYB62 gene on cadmium stress.
Analysis of physiological indicators of trans-PsMYB62 A. thaliana under Cd stress
Phenotypic changes with regards to the shooting were observed in the treated A. thaliana every 7 d. Before treatment, A. thaliana grew normally, the leaves were green, and the rosette leaves unfolded naturally. There was no significant difference in the phenotype among the four lines. At day 7, all four strains were able to maintain normal growth. After 14 d of treatment, the leaves of WT and EV plants yellowed, development stagnated, and most plants withered. Though the growth and development of transgenic lines were inhibited compared with those before treatment, and the old leaves began to yellow, the growth of WT and EV plants was more seriously inhibited. After 21 d, the leaves of transgenic lines withered and were damaged significantly, half of the plants withered and died, and could not maintain normal growth. All WT and EV plants withered and died (Fig. 7).
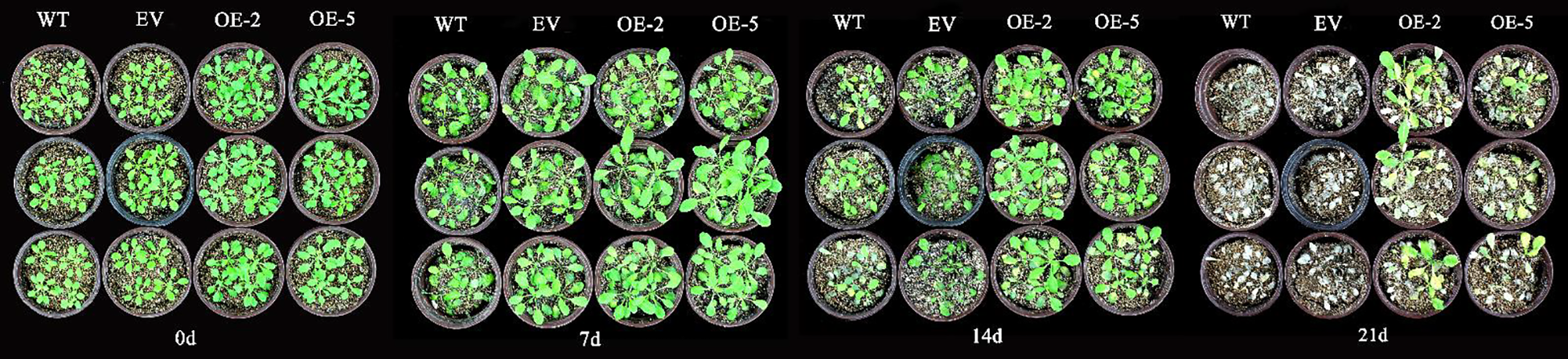
Figure 7: Growth states of transgenic A. thaliana under Cd stress.
Antioxidant enzyme activity and chlorophyll content of trans-PsMYB62 A. thaliana under Cd stress
There was no significant difference in the activity levels of SOD, POD, CAT, and chlorophyll content between the WT, EV and transgenic strains before Cd treatment (Fig. 8). After Cd treatment, the activity levels of SOD, POD, and CAT in WT, EV, and the transgenic lines increased significantly. There was no significant difference in the activity of three enzymes between EV and WT, while the activity of three enzymes in transgenic lines was significantly higher than that in WT and EV. The greatest increase in the level of POD activity was observed in the transgenic strains OE-2 and OE-5, where the POD activity was 2.64 and 3.37 times higher than that before treatment, respectively. The activities of SOD, POD, and CAT in the WT were 2.14, 1.32 and 1.41 times higher than those before treatment, respectively. However the three enzyme activities in OE-2 were lower in the two transgenic lines, which were 3.07, 2.64, and 1.88 times higher than those before treatment, respectively (Figs. 6A–6C). The chlorophyll content of the four lines decreased after Cd treatment, but the chlorophyll content of the transgenic lines after treatment was still significantly higher than that of WT and EV (Fig. 6D). It can be seen that trans-PsMYB62 plants under Cd stress have stronger antioxidant capacity than wild-type plants. The PsMYB62 gene could assist plants to alleviate the damage caused by heavy metal ions to the intracellular environment, which enhances the Cd tolerance of transgenic A. thaliana.
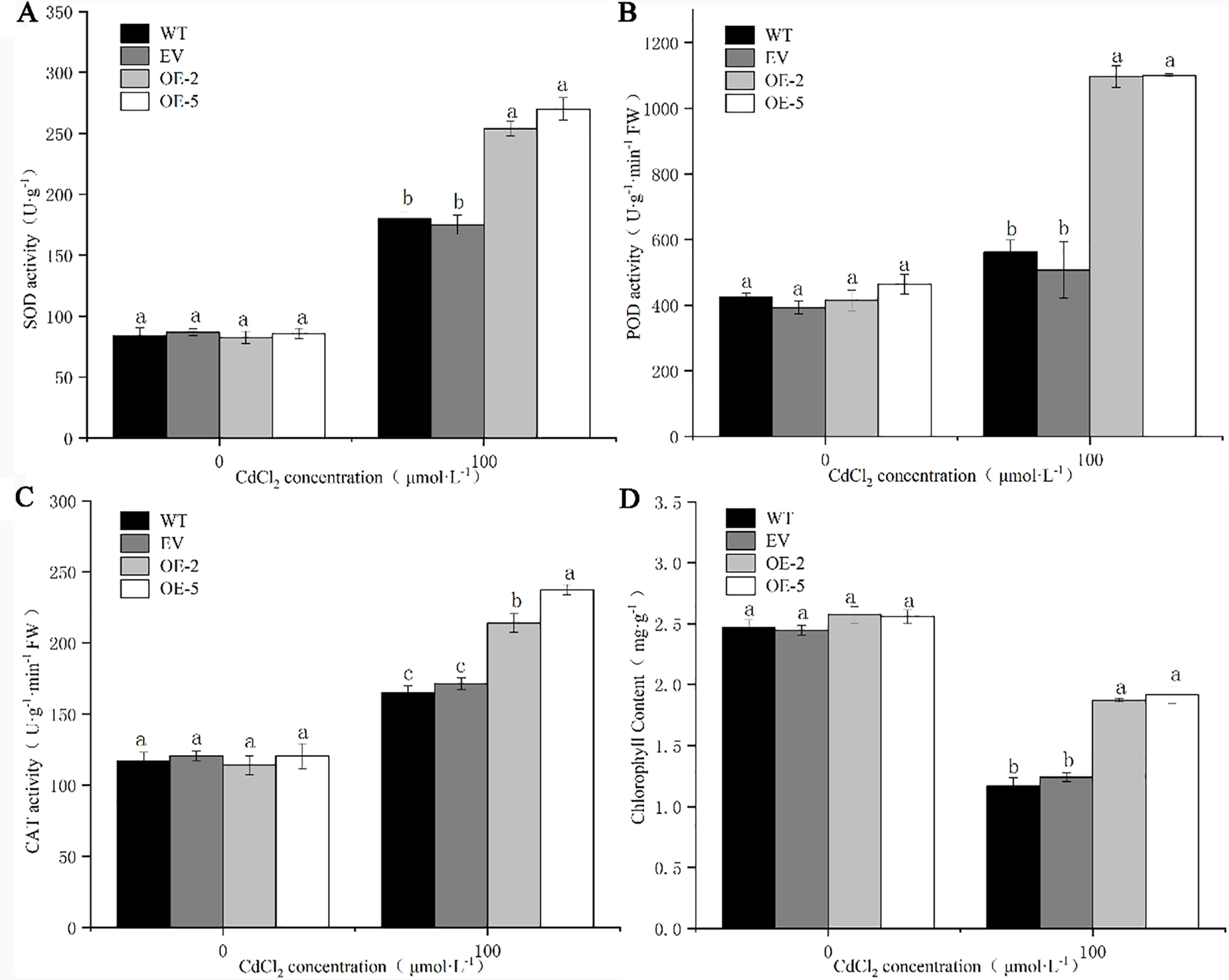
Figure 8: Antioxidative enzyme activity and chlorophyll content of trans-PsMYB62 A. thaliana under Cd stress. Note: (A–D): Superoxide dismutase (SOD) activity, peroxidase (POD) activity, catalase (CAT) activity, and the chlorophyll content of wild-type (WT), empty vector (EV), and transgenic lines before and after treatment with 100 μmol·L−1 CdCl2.
Oxidative stress analysis of trans-PsMYB62 A. thaliana under Cd stress
As an important basis for reflecting the degree of peroxidation and damage of plant cells, the four indicators used: O2·−, H2O2, MDA content, and relative conductivity showed a consistent trend after the four lines were subjected to Cd stress. After Cd treatment, the indicator levels of the four lines were significantly increased, and the levels in EV were not significantly different from WT. However, the indicator levels in transgenic lines were significantly lower than WT and EV. The relative conductivity of transgenic lines OE-2 and OE-5 were 2.67 and 2.81 times higher than that before treatment, respectively. The content of O2·− was least elevated, and the content of O2·− in the transgenic strains OE-2 and OE-5 was 1.33 and 1.35 times higher, respectively than before treatment. The O2·−, H2O2, MDA content, and relative conductivity of WT were 1.77, 3.16, 3.01, and 6.80 times higher, respectively than those before treatment. The large increase in each index in OE-2 in the two transgenic strains was 1.33, 1.91, 1.88, and 2.67 times lower than that of WT before treatment (Fig. 9). It was proved that WT and EV plants suffered more serious damage than trans-PsMYB62 plants under Cd stress.
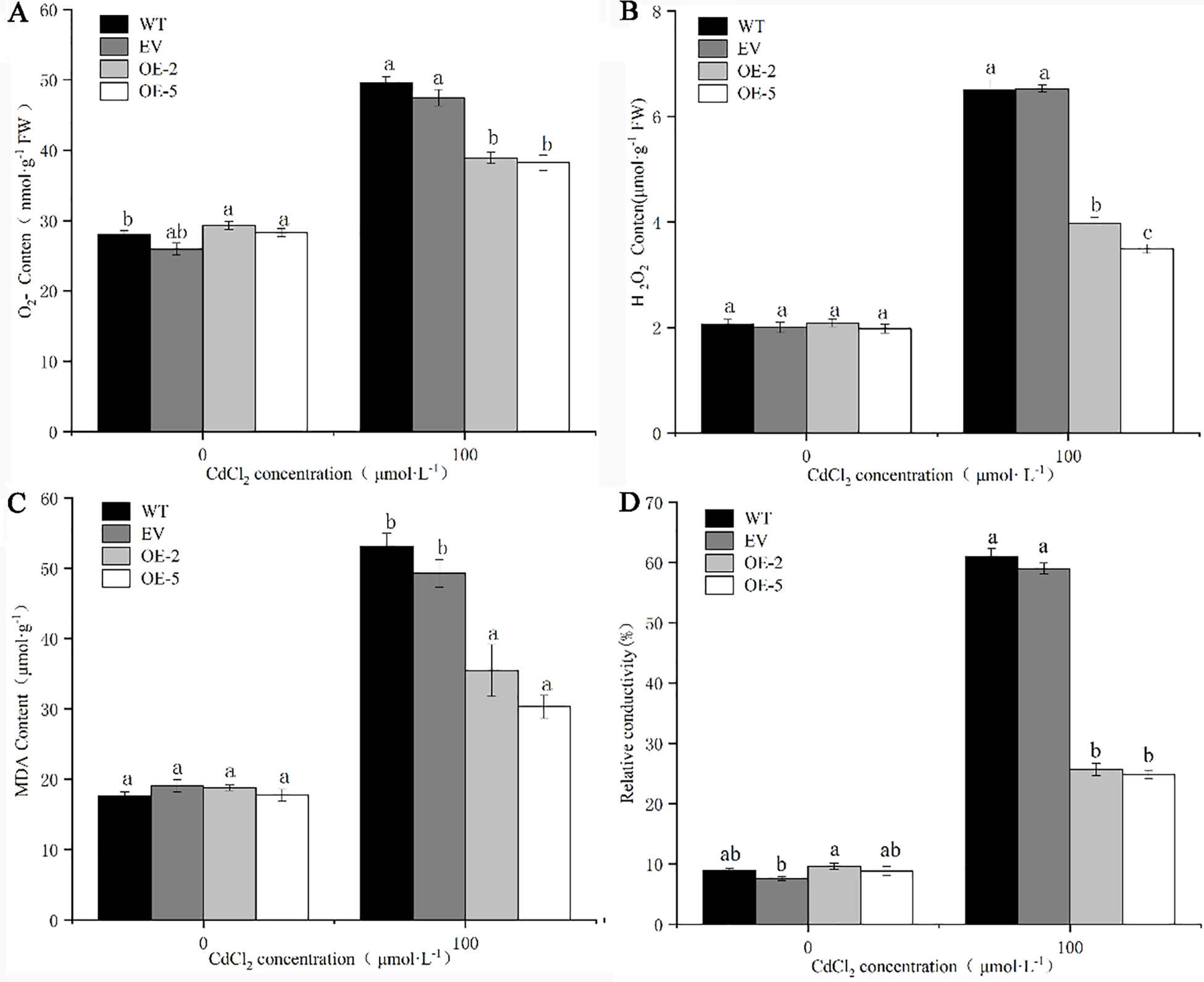
Figure 9: Oxidative stress conditions in trans-PsMYB62 A. thaliana under Cd stress. Note: (A–D): O2·− content, H2O2 content, and the malondialdehyde (MDA) content, and the relative conductivity of wild-type (WT) and transgenic lines before and after treatment with 100 μmol·L−1 CdCl2.
Expression of heavy metal resistance-related genes in trans-PsMYB62 A. thaliana under Cd stress
Plants enhance their tolerance to heavy metals by chelating metals or vacuolar compartmentation, in which AtGSH1, AtPCS1, AtNAS4, and AtHMA2 are involved. In order to further verify the tolerance of PsMYB62 to cadmium, the expression levels of several heavy metal resistance-related genes were determined in transgenic plants, WT plants, and EV plants after Cd stress. Before treatment, there was no significant difference in the expression levels of AtGSH1, AtPCS1, AtNAS4, and AtHMA2 in the WT, EV, and transgenic lines. After Cd treatment, the expression levels of AtGSH1, AtPCS1, and AtNAS4 were significantly up-regulated while that of AtHMA2 was down-regulated and was significantly down-regulated in transgenic lines (Fig. 10). It is hypothesized that PsMYB62 reduces Cd2+ flow from the vacuole to the cytoplasm by enhancing Cd2+ chelation and inhibits Cd transfer from the root to the bud.
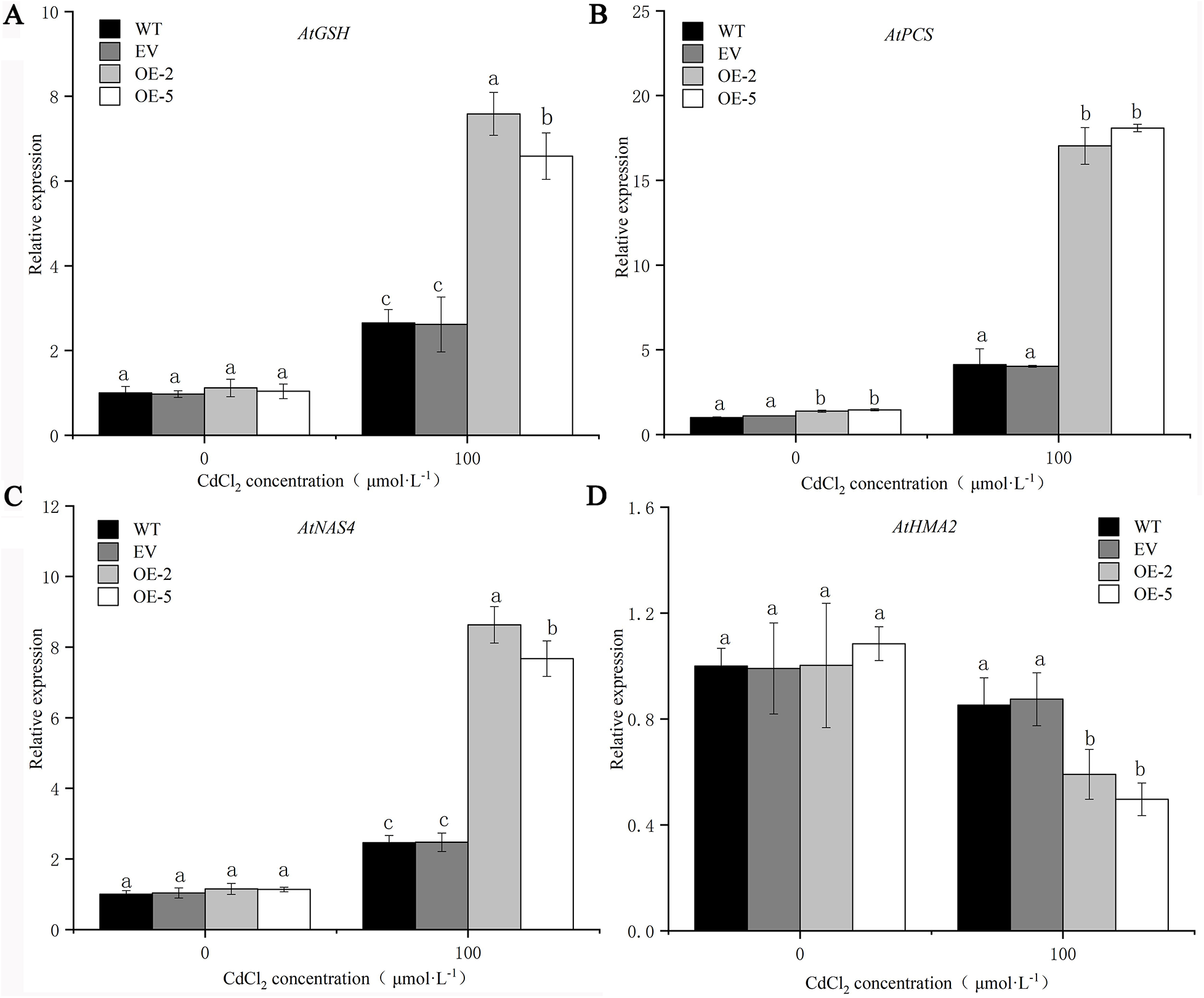
Figure 10: Resistant to heavy metals-related gene expression of trans-PsMYB62 A. thaliana under Cd stress. Note: (A–D) Expression of AtGSH1, AtPCS1, AtNAS4, and AtHMA2 in A. thaliana before and after 100 μmol·L−1 CdCl2 treatment.
Cadmium is a well-known toxic metal for plants, which inhibits root growth and uptake of essential micronutrients, affects photosynthesis, and induces oxidative stress and DNA damage (Sinha and Mukherjee, 2008; Andresen and Kupper, 2013; Wang et al., 2014; Haider et al., 2021). Studies have shown that MYB transcription factors are widely involved in the response of plants to Cd stress (Ding et al., 2018; Meng et al., 2022b; Zhang, 2022), but there are few reports on MYB transcription factors in Potentilla sericea. The AtMYB2 gene in A. thaliana subgroup 20 is functionally related to the response to heavy metals, and we found that the PsMYB62 gene in the same subfamily as it, thus indicating that they are functionally similar (Jia et al., 2020). This study successfully cloned the PsMYB62 from Potentilla sericea and verified its function. In this study, we treated the seeds of the A. thaliana transformed with the PsMYB62 gene with CdCl2·2.5H2O for stress and counted the germination rate of the seeds. The results showed that the seedlings of the A. thaliana trans-PsMYB62 gene responded to cadmium, and the results confirmed the above hypothesis.
In this study, PsMYB62 was up-regulated by Cd, salt, drought, and ABA stresses, and showed different expression trends in roots, stems, and leaves. The expression levels of this gene under CdCl2 and ABA treatments were relatively higher than those under NaCl and mannitol treatments. Among these, PsMYB62 was the fastest induced and reached the highest relative expression level in roots under Cd stress. A study showed that the expression of MaMYB genes in Morus alba were up-regulated under abiotic stress and preferentially expressed in roots or stems (Liu et al., 2022). This is consistent with our experimental results, but the mechanism of ABA signal regulating plant cadmium stress response needs further research and verification.
By studying the changes in germination rate, growth status, and physiological indicators of trans-PsMYB62 A. thaliana under Cd treatment, the results showed that the growth damage of trans-PsMYB62 lines was lower with increased SOD, POD, and CAT activity to remove hazardous substances than that of WT and EV. The trans-PsMYB62 gene showed its mitigation ability in A. thaliana under cadmium stress. Excessively high concentrations of Cd2+ destroy the intracellular redox balance and produce excessive reactive oxygen species (ROS). This leads to cell membrane lipid peroxidation, electrolyte leakage, and protein damage, thus affecting plant growth and development (Zhang et al., 2007; Howladar, 2014; Zouari et al., 2016). ROS includes O2·− and H2O2, their content with the MDA content and the relative conductivity reflect the degree of membrane lipid peroxidation of plant cells (Zhao et al., 2016; Chen et al., 2022). In this study, under Cd stress, the O2·−, H2O2, and MDA contents and the relative conductivity in WT, EV, and transgenic lines all increased, indicating that Cd stress caused membrane lipid peroxidation and increased intracellular ROS accumulation in plants. However, the O2·−, H2O2, and MDA content and relative conductivity of transgenic lines were significantly lower than those of WT and EV, indicating that transgenic lines can better maintain the membrane lipid structure and reduce the degree of plant damage under Cd stress. This is consistent with the results of Feng et al. (2020). When the intracellular oxygen metabolism is imbalanced, SOD in the antioxidant enzyme system can catalyze the dismutation of O2·− to produce O2 and H2O2, which continues to be degraded by POD and CAT to O2 and H2O, thereby inhibiting the increase of ROS content (Siddiqui et al., 2012). Additionally, Liu et al. (2023) showed that ScCAM up-regulated the expression of antioxidant enzymes to improve the activity of antioxidant enzymes in plants to resist external stress. In this study, the activities of SOD, POD, and CAT in WT, EV, and transgenic lines increased under Cd stress, and the enzyme activities of transgenic lines were higher than that of WT and EV. It was speculated that PsMYB62 had a certain activation effect on the plant antioxidant enzyme system. Plants can reflect the strength of their photosynthesis through chlorophyll content. It was found that the chlorophyll structure of plants was destroyed and the chlorophyll content was significantly reduced under Cd stress (Yu et al., 2010; Peng et al., 2015). This result was also documented in this study, which may have contributed to the loss of greenish-yellowing of the leaves. In order to further investigate the mechanism of PsMYB62 in plant resistance to Cd stress, this study examined the expression levels of heavy metal-related resistance genes in WT, EV, and transgenic A. thaliana after Cd stress. It is generally believed that cadmium exists in plants in the free form (Cd2+) and chelated forms (such as GSH-Cd, PC-Cd, and NA-Cd) (Verbruggen et al., 2009). Plant chelation synthase (PCS) uses GSH as a substrate, which is encoded by PCS genes (PCS1 and PCS2) and induced by Cd stress. GSH and PCS play an important role in plant cadmium tolerance (Seth et al., 2012). A. thaliana mutants lacking nicotianamine synthase 4 (NAS4) function have significantly lower nicotianamine (NA) levels and exhibit sensitivity to cadmium stress (Schuler et al., 2012; Emmanuel et al., 2013). In this study, the expression levels of AtGSH1, AtPCS1, and AtNAS4 in trans-PsMYB62 lines under Cd stress were significantly up-regulated compared with WT and EV. It is hypothesized that free Cd2+ in trans-PsMYB62 lines was chelated and entered the vacuole in the form of GSH-Cd, PC-Cd, and NA-Cd chelates, so that lesser Cd levels remained in the cell, indicating that trans-PsMYB62 lines showed stronger cadmium tolerance. P1B-type ATPases HMA2 and HMA4 are transporters located on the plasma membrane, expressed in vascular tissues, and promote the transport of Cd from roots to shoots (Wong and Cobbett, 2009). In this study, the expression level of AtHMA2 was down-regulated in transgenic lines. It is suggested that PsMYB62 may inhibit the long-distance transport of Cd from roots to buds, making Cd more concentrated in the roots. This is consistent with the results of Meng et al. (2022a). Therefore, PsMYB62 can be involved in the regulation of Cd resistance genes to enhance the tolerance of plants to Cd stress. However, the specific Cd accumulation in roots and leaves of plants and other mechanisms of the gene expression under Cd stress need to be further studied and verified.
In this study, an R2R3 transcription factor of the MYB transcription factor family, PsMYB62, was successfully cloned from Potentilla sericea, which could be induced to be expressed by Cd stress. Overexpression of the PsMYB62 gene could significantly increase the activities of antioxidant enzymes in transgenic A. thaliana to enhance the ability of plant cells to scavenge ROS and alleviate the effects of Cd stress on plant growth and development. Subsequently, we found that PsMYB62 up-regulated the expression of AtGSH1, AtPCS1, and AtNAS4, and down-regulated the expression of AtHMA2. This finally improved the cadmium tolerance of transgenic A. thaliana.
Acknowledgement: We thank the Biotechnology Company (Ruiboxingke Biotechnology Co., Ltd., Harbin, China) for the technical support, and we are sincerely thankful for the help from Pengfei Gao and Weifang Fan.
Funding Statement: The research was funded by the Natural Science Foundation of Heilongjiang Province (LH2020C045).
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: Zhenghong Feng, Jianhui Wu; data collection: Yu Gao; analysis and interpretation of results: Bing Gao, Zhenghong Feng; draft manuscript preparation: Zhenghong Feng. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
Ahmad P, Tripathi DK, Deshmukh R, Pratap Singh V, Corpas FJ (2019). Revisiting the role of ROS and RNS in plants under changing environment. Environmental and Experimental Botany 161: 1–3. https://doi.org/10.1016/j.envexpbot.2019.02.017 [Google Scholar] [CrossRef]
Albert N, Griffiths A, Cousins G, Verry I, Williams W (2015). Anthocyanin leaf markings are regulated by a family of R2R3-MYB genes in the genus Trifolium. New Phytologist 205: 882–893. https://doi.org/10.1111/nph.13100 [Google Scholar] [PubMed] [CrossRef]
Andresen E, Kupper H (2013). Cadmium toxicity in plants. Metal Ions in Life Sciences 11: 395–413. https://doi.org/10.1007/978-94-007-5179-8 [Google Scholar] [CrossRef]
Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J et al. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry 55: 611–622. https://doi.org/10.1373/clinchem.2008.112797 [Google Scholar] [PubMed] [CrossRef]
Chen SY, Cui LL, Wang XH (2022). A plant cell wall-associated kinase encoding gene is dramatically down regulated during nematode infection of potato. Plant Signaling & Behavior 17: 2004026. https://doi.org/10.1080/15592324.2021.2004026 [Google Scholar] [PubMed] [CrossRef]
Ding J, Zhang XN, Piao CL, Cui ML, Gao ZR (2018). Analysis of R2R3-MYB genes in soybean roots in response to cadmium stress. Journal of Ecology 37: 2030–2039. [Google Scholar]
Emmanuel K, Angélique B, Duc C, Astier J, Gravot A, Richaud P, Lamotte O, Boucherez J, Gaymard F, Wendehenne D (2013). Arabidopsis thaliana nicotianamine synthase 4 is required for proper response to iron deficiency and to cadmium exposure. Plant Science 209: 1–11. https://doi.org/10.1016/j.plantsci.2013.04.006 [Google Scholar] [PubMed] [CrossRef]
Fan WF (2021). Transcriptome Analysis of Potentilla Sericea and Preliminary Study on PsMYB2 Function under Cadmium Stress (Master Thesis). Northeast Forestry University, China. [Google Scholar]
Feng YW, Lei W, Gu R, Zhao P, Ni SJ, Lei NF (2020). Differential effects of ammonium and nitrate on growth performance of glechoma longituba under heterogeneous Cd stress. Phyton-International Journal of Experimental Botany 89: 667–679. https://doi.org/10.32604/phyton.2020.010160 [Google Scholar] [CrossRef]
Haider FU, Cai LQ, Coulter JA, Cheema SA, Wu J, Zhang RZ, Ma WJ, Farooq M (2021). Cadmium toxicity in plants: impacts and remediation strategies. Ecotoxicology and Environmental Safety 211: 111887. https://doi.org/10.1016/j.ecoenv.2020.111887 [Google Scholar] [PubMed] [CrossRef]
Howladar SM (2014). A novel Moringa oleifera leaf extract can mitigate the stress effects of salinity and cadmium in bean (Phaseolus vulgaris L.) plants. Ecotoxicology and Environmental Safety 100: 69–75. https://doi.org/10.1016/j.ecoenv.2013.11.022 [Google Scholar] [PubMed] [CrossRef]
Jia TQ, Zhang KD, Li F, Huang YF, Fan M, Huang T (2020). The AtMYB2 inhibits the formation of axillary meristem in Arabidopsis by repressing RAX1 gene under environmental stresses. Plant Cell Reports 39: 1755–1765. https://doi.org/10.1007/s00299-020-02602-3 [Google Scholar] [PubMed] [CrossRef]
Kohli SK, Khanna K, Bhardwaj R, Abd AE, Ahmad P, Corpas FJ (2019). Assessment of subcellular ROS and NO metabolism in higher plants: Multifunctional signaling molecules. Antioxidants 8: 641. https://doi.org/10.3390/antiox8120641 [Google Scholar] [PubMed] [CrossRef]
Liu L, Chao N, Yidilisi K, Kang X, Cao X (2022). Comprehensive analysis of the MYB transcription factor gene family in Morus alba. BMC Plant Biology 22: 281. https://doi.org/10.1186/s12870-022-03626-5 [Google Scholar] [PubMed] [CrossRef]
Liu JX, Feng JF, Zhang C, Ren YJ, Su WH et al. (2023). Overexpression of a sugarcane ScCaM gene negatively regulates salinity and drought stress responses in transgenic Arabidopsis thaliana. BIOCELL 47: 215–225. https://doi.org/10.32604/biocell.2022.022477 [Google Scholar] [CrossRef]
Lv Z, Zhu F, Jin D, Wu Y, Wang S (2021). Seed germination and seedling growth of Dendrocalumus brandisii in vitro, and the inhibitory mechanism of colchicine. Frontiers in Plant Science 12: 784581. https://doi.org/10.3389/fpls.2021.784581 [Google Scholar] [PubMed] [CrossRef]
Mansoor S, Ali WO, Lone JK, Manhas S, Kour N, Alam P, Ahmad A, Ahmad P (2022). Reactive oxygen species in plants: From source to sink. Antioxidants 11: 225. https://doi.org/10.3390/antiox11020225 [Google Scholar] [PubMed] [CrossRef]
Meng YT, Zhang XL, Wu Q, Shen RF, Zhu XF (2022a). Transcription factor ANAC004 enhances Cd tolerance in Arabidopsis thaliana by regulating cell wall fixation, translocation and vacuolar detoxification of Cd, ABA accumulation and antioxidant capacity. Journal of Hazardous Materials 436: 129121. https://doi.org/10.1016/j.jhazmat.2022.129121 [Google Scholar] [PubMed] [CrossRef]
Meng SQ, Zhong XA, Wang M, Xing W, Liu DL (2022b). Characterization analysis of sugar beet MYB transcription factors under cadmium stress. Molecular Plant Breeding 20: 4917–4930. [Google Scholar]
Peng L, Tian XP, Yang J, Qv CJ, Zhao XH (2015). Alleviation of cadmium stress on root tip of rape seedlings by selenium. Acta Scientiae Circumstantiae 35: 2597–2604. [Google Scholar]
Qi JY, Niu J, Zhang J, Di XL, Tang S, Hua CQ, Jiang CR, Kong X, Wu JH (2018). Effects of heavy metal lead on photosynthetic characteristics and fluorescence parameters of Potentilla sericea. Acta Agrestia Sinica 26: 447–452. [Google Scholar]
Qiu WY, Wang SY, Li XF, Xu H, Zhang H, Zhu Y, Wang LC (2020). Functions of plant MYB transcription factors in response to abiotic stress and plant hormones. Acta Agriculturae Zhejiangensis 32: 1317–1328. [Google Scholar]
Schuler M, Rellan-Alvarez R, Fink-Straube C, Abadia J, Bauer P (2012). Nicotianamine functions in the Phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. The Plant Cell 24: 2380–2400. https://doi.org/10.1105/tpc.112.099077 [Google Scholar] [PubMed] [CrossRef]
Seth CS, Remans T, Keunen E, Jozefczak M, Gielen H, Opdenakker K, Weyens N, Vangronsveld J, Cuypers A (2012). Phytoextraction of toxic metals: A central role for glutathione. Plant Cell and Environment 35: 334–346. https://doi.org/10.1111/j.1365-3040.2011.02338.x [Google Scholar] [PubMed] [CrossRef]
Siddiqui MH, Al-Whaibi MH, Sakran AM, Basalah MO, Ali HM (2012). Effect of calcium and potassium on antioxidant system of Vicia faba L. under cadmium stress. International Journal of Molecular Sciences 13: 6604–6619. https://doi.org/10.3390/ijms13066604 [Google Scholar] [PubMed] [CrossRef]
Sinha S, Mukherjee SK (2008). Cadmium-induced siderophore production by a high Cd-resistant bacterial strain relieved Cd toxicity in plants through root colonization. Current Microbiology 56: 55–60. https://doi.org/10.1007/s00284-007-9038-z [Google Scholar] [PubMed] [CrossRef]
Stephanie WA, Li J (2021). Evaluating the environmental and economic impact of mining for post-mined land restoration and land-use: A review. Journal of Environmental Management 1: 279. https://doi.org/10.1016/j.jenvman.2020.111623 [Google Scholar] [PubMed] [CrossRef]
Suo JF, Liang X, Pu L, Zhang YS, Xue YB (2003). Identification of GhMYB109 encoding a R2R3 MYB transcription factor that expressed specifically in fiber initials and elongating fibers of cotton (Gossypium hirsutum L.). Biochimica et Biophysica Acta (BBA)—Gene Structure and Expression 1630: 25–34. https://doi.org/10.1016/j.bbaexp.2003.08.009 [Google Scholar] [PubMed] [CrossRef]
Verbruggen N, Hermans C, Schat H (2009). Molecular mechanisms of metal hyperaccumulation in plants. New Phytologist 181: 759–776. https://doi.org/10.1111/j.1469-8137.2008.02748.x [Google Scholar] [PubMed] [CrossRef]
Wang XK (2006). Principles and techniques of plant physiological and biochemical experiments. Beijing: Higher Education Press. [Google Scholar]
Wang XP, Niu YL, Zheng Y (2021a). Multiple functions of MYB transcription factors in abiotic stress responses. International Journal of Molecular Sciences 22: 6125. https://doi.org/10.3390/ijms22116125 [Google Scholar] [PubMed] [CrossRef]
Wang RY, Wen WW, Zhao EH, Zhou P, An Y (2021b). Cloning and salt-tolerance analysis of MsWRKY11 in alfalfa. Acta Prataculturae Sinica 30: 157–169. [Google Scholar]
Wang QL, Zhang LT, Zou JH, Liu DH, Yue JY (2014). Effects of cadmium on root growth, cell division and micronuclei formation in root tip cells of Allium cepa var. agrogarum L. Phyton-International Journal of Experimental Botany 83: 291–298. https://doi.org/10.32604/phyton.2014.83.291 [Google Scholar] [CrossRef]
Wang M, Zhou WT, Zhou X, Li SQ, Wang XQ, Wang LH, Li WS, Li JJ, Gao Z, Liu DL (2021c). Transcription factor BvMYB44 gene in energy beet: expression characteristics in response to cadmium stress and lts bioinformatics analysis. Chinese Agricultural Science Bulletin 37: 25–33. [Google Scholar]
Wei XX, Lan HY (2022). Advances in the regulation of plant MYB transcription factors in secondary metabolism and stress response. Biotechnology Bulletin 38: 12–23. [Google Scholar]
Wong CKE, Cobbett CS (2009). HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana. New Phytologist 181: 71–78. https://doi.org/10.1111/j.1469-8137.2008.02638.x [Google Scholar] [PubMed] [CrossRef]
Wu JH, Lan F, Zhang J, Niu J, Wang L (2017). Effect of Pb stress on ultrastructure and physiological characteristics of Potentilla sericea. Pratacultural Science 34: 1383–1389. [Google Scholar]
Wu JH, Liu JX, Zhang J, Hao HJ, Zhao QY (2016). The response of Potentilla sericea ultrastructure and physiological characteristics to cadmium stress. Acta Agrestia Sinica 24: 1278–1282. [Google Scholar]
Wu J, Zhang ZQ, Yu HH, Huang FB, Chen ZL, Chu LL, Li B, Wang W (2022). Research progress on chemical constituents and pharmacological activities of Potentilla. China Journal of Chinese Materia Medica 47: 1509–1538. [Google Scholar] [PubMed]
Xu Z, Ge Y, Zhang W, Zhao Y, Yang G (2018). The walnut JrVHAG1 gene is involved in cadmium stress response through ABA-signal pathway and MYB transcription regulation. BMC Plant Biology 18: 19. https://doi.org/10.1186/s12870-018-1231-7 [Google Scholar] [PubMed] [CrossRef]
Yamasaki K, Kigawa T, Inoue M, Watanabe S, Tateno M, Seki M, Shinozaki K, Yokoyama S (2008). Structures and evolutionary origins of plant-specific transcription factor DNA-binding domains. Plant Physiology and Biochemistry 46: 394–401. https://doi.org/10.1016/j.plaphy.2007.12.015 [Google Scholar] [PubMed] [CrossRef]
Yao GX, Yang SH, Ji Y, Ren GL, Luo ZW (2019). The edible value and cultivation technology of Potentilla fragarioides. Special Economic Animals and Plants 22: 37+40. [Google Scholar]
Yu FM, Tang YT, Chou RL, Zhou XY, Ying RR, Hu PJ, Zhang T (2010). Antioxidative responses to cadmium stress in the hyperaccumulator Arabis paniculata Franch. Acta Scientiae Circumstantiae 30: 409–414. [Google Scholar]
Zhang YF (2014). Study on the domestication of Potentilla anserina. Journal of Qinghai Normal University (Natural) 30: 46–51. [Google Scholar]
Zhang J (2020). Effects of Drought Stress on Root Structure and Physiology of Two Kinds of Potentilla (Master Thesis). Northeast Forestry University, China. [Google Scholar]
Zhang W (2022). Cadmium Resistance and Regulatory Pathway of BpTT2-overexpressed Broussonetia Papyrifera (Ph.D. Thesis). Central South University of Forestry and Technology, China. [Google Scholar]
Zhang P, Wang R, Ju Q, Li WQ, Tran LP, Xu J (2019). The R2R3-MYB transcription factor MYB49 regulates cadmium accumulation. Plant Physiology 180: 529–542. https://doi.org/10.1104/pp.18.01380 [Google Scholar] [PubMed] [CrossRef]
Zhang FQ, Wang YS, Lou ZP, Dong JD (2007). Effect of heavy metal stress on antioxidative enzymes and lipid peroxidation in leaves and roots of two mangrove plant seedlings (Kandelia candel and Bruguiera gymnorrhiza). Chemosphere 67: 44–50. https://doi.org/10.1016/j.chemosphere.2006.10.007 [Google Scholar] [PubMed] [CrossRef]
Zhang Q, Xing Y, Cao QQ, Qin L (2016). Application of optimized fluorescence quantitative PCR technology for the research of strawberry. Journal of Beijing University of Agriculture 31: 21–25. [Google Scholar]
Zhao X, Yang XW, Pei SQ, He G, Wang XY, Tang Q, Jia CL, Lu Y, Hu RB, Zhou GK (2016). The miscanthus NAC transcription factor MlNAC9 enhances abiotic stress tolerance in transgenic Arabidopsis. Gene 586: 158–169. https://doi.org/10.1016/j.gene.2016.04.028 [Google Scholar] [PubMed] [CrossRef]
Zhou L, Zheng XQ, Ding YZ, Huang HK, Zheng SA, Shi RG, Li XG, Feng RW, Wang RG (2017). Prevention and control of cadmium and arsenic contamination of agricultural land and crop safety planting technology discussion. Journal of Agricultural Environmental Science 36: 613–619. [Google Scholar]
Zouari M, Ben AC, Elloumi N, Bellassoued K, Delmail D, Labrousse P, Ben AF, Ben RB (2016). Impact of proline application on cadmium accumulation, mineral nutrition and enzymatic antioxidant defense system of Olea europaea L. cv Chemlali exposed to cadmium stress. Ecotoxicology and Environmental Safety 128: 195–205. https://doi.org/10.1016/j.ecoenv.2016.02.024 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools