 Open Access
Open Access
ARTICLE
CircR-ZC3HC1 mediates MiR-384-5p/SIRT1 axis to promote neuronal autophagy and relieves ischemic stroke
1 Department of Neurology, The First Affiliated Hospital of Ningbo University, Ningbo, 315010, China
2 School of Medicine, Ningbo University, Ningbo, 315211, China
* Corresponding Authors: QINGHUA CAO. Email: ; ZHEZHE SUN. Email:
BIOCELL 2024, 48(3), 491-499. https://doi.org/10.32604/biocell.2023.047640
Received 13 November 2023; Accepted 11 December 2023; Issue published 15 March 2024
Abstract
Objective: Circular RNAs (circRNAs) have been shown to involve in pathological processes of ischemic stroke (IS), including autophagy. This study was designed to explore the effect of circR-ZC3HC1 on neuronal autophagy in IS and the related mechanisms. Methods: Expression of circR-ZC3HC1 in blood samples of IS patients and healthy controls was detected. Hippocampal neurons were treated with oxygen and glucose deprivation (OGD) to establish IS in vitro model. The expression of LC3 and p62 and the number of autophagosomes were examined to evaluate the autophagy level of OGD induced neurons using western blotting and transmission electron microscope. Cell apoptosis rate and the expression of cleaved caspase-3, Bax, and Bcl-2 were assessed by flow cytometry and western blotting. The binding relationships among circR-ZC3HC1, miR-384-5p, and SIRT1 were predicted and verified. Results: Low expression of circR-ZC3HC1 was found in blood samples of IS patients and OGD-treated neurons. Overexpressed circR-ZC3HC1 or inhibited miR-384-5p expression promoted autophagy and inhibited apoptosis of OGD-treated neurons, which could be reversed by further 3-MA treatment. Mechanistically, circR-ZC3HC1 targeted miR-384-5p to mediate SIRT1 expression. miR-384-5p overexpression or SIRT1 knockdown in the presence of circR-ZC3HC1 overexpression in OGD-treated neurons lead to reduced autophagy and enhanced apoptosis. Conclusion: Collectively, circR-ZC3HC1 promoted neuronal autophagy to attenuate IS via miR-384-5p/SIRT1 axis.Keywords
Ischemic stroke (IS), caused by blockages in a blood vessel that reduces blood supply to the brain, is a leading reason for death and disability in humans [1]. At present, the acute treatments for IS including intravenous thrombolysis and mechanical thrombolysis can only satisfy a limited number of patients, so it is urgent to develop new neuroprotective methods [2]. Autophagy participates in IS pathogenesis as a double-edged sword [3]. Appropriate autophagy can have a protective effect on ischemic nerve tissue by removing damaged organelles and excess cell components, while excessive autophagy may lead to cell death [4]. Besides, there are many pathological changes involved in the ischemic process alone or together to trigger neuronal death, including apoptosis, ferroptosis, necroptosis, pyroptosis, and phagoptosis [5]. However, the molecular mechanisms underlying neuronal autophagy and apoptosis after IS are far from clear.
Circular RNAs (circRNAs) are a newly discovered class of endogenous non-coding RNAs characterized by backsplicing caused by covalently closed continuous rings [6]. In mammals, circRNAs are expressed at high levels in the brain and have emerged as pivotal regulators in numerous nervous system diseases, including IS [7,8]. For instance, circTLK1 exacerbated neuronal injury and neurological deficits after IS via miR-335-3p/TIPARP [9]. Overexpression of circUCK2 improved cell survival rate and ameliorated IS-induced neuronal injury via miR-125b-5p/GDF11 signaling [10]. So far, there are no relevant studies on circR-ZC3HC1 in IS. In this paper, we discovered that the expression of circR-ZC3HC1 in the blood of patients with IS was significantly reduced through the analysis of GSE133768 dataset. Hence, we speculated that circR-ZC3HC1 expression might link to the development of IS.
In addition to circRNAs, microRNAs (miRNAs) are widely implicated in IS, serving as possible targets for potential neuroprotective treatments [11]. Recently, Zhu et al. pointed out that isoflurane post-conditioning alleviated ischemic neuronal injury may associate with miR-384-5p/ATG5-mediated autophagy [12]. A prior study illustrated that miR-384 was upregulated in chronic cerebral ischemia-induced HT22 cells and hippocampal tissues, and knockdown of miR-384 inhibited chronic IS-induced neuron apoptosis [13]. In our study, it was predicted that circR-ZC3HC1 could target miR-384-5p. Therefore, we hypothesized that circR-ZC3HC1 may play a role by regulating miR-384-5p in IS. The silent information regulator sirtuin 1 (SIRT1), belonging to the sirtuin family, is closely associated with IS process [14]. A prior paper has exhibited that upregulating SIRT1 inhibited nerve cell apoptosis and improved nerve cell damage, playing a protective role in IS [15]. Moreover, miR-384-5p repressed SIRT1 expression through specific binding [16]. Through bioinformatics analysis, we noted that SIRT1 may be a target of miR-384-5p. Based on the above discoveries, we hypothesized that circR-ZC3HC1 might upregulate the expression of SIRT1 by regulating miR-384-5p, thereby promoting autophagy and inhibiting IS. If this scientific hypothesis is confirmed, circR-ZC3HC1 may become an important target for the treatment of IS, providing an important solid theoretical foundation for the cure of IS in the future.
On the basis of standard procedure, blood samples were collected from 16 patients with IS (IS group) and 10 healthy subjects (control group) from the First Affiliated Hospital of Ningbo University (from May 2019 to March 2021). Notably, fresh peripheral blood was collected within 24 h after IS. IS patients were diagnosed with IS by computed tomography and/or magnetic resonance imaging (ICD-9-CM codes 433, 434, and 436). All included IS patients had complete clinical data and were free from malignancy. IS patients with transient ischemic attack or hemorrhagic stroke caused by tumor, blood disease, traumatic brain injury, or cerebrovascular malformation were excluded. In the control group, the participants were free from IS, obvious ischemic changes, stroke symptoms, malignant tumors, severe hepatic and renal insufficiency, or immune diseases. The protocol of our study was permitted by the Ethics Committee of the First Affiliated Hospital of Ningbo University (No. 2022KYYS119), and informed consent forms were signed by all participants. Clinical trials followed the Declaration of Helsinki.
Mouse hippocampal neuron HT22 cells were purchased from MINGZHOUBIO (Ningbo, China) and cultivated in Dulbecco’s modified Eagle medium (DMEM; Item No. 12491015, Thermo Fisher, Waltham, MA, USA) added with 10% fetal bovine serum (FBS) and 10% penicillin-streptomycin at 37°C with 5% CO2. shRNAs targeting SIRT1 (sh-SIRT1#1-3), circR-ZC3HC1 overexpression plasmid (p-ZC3HC1, pcDNA3.1 vector), miR-384-5p inhibitor, miR-384-5p mimic, and their controls were all derived from Vector Builder (Guangzhou, China). The transient transfection method was used for overexpression and knockdown, and the transfection dose was 2 μg. Follow-up experiments were performed after 48-h transfection.
To inhibit autophagy in mouse neurons, HT22 cells were treated with autophagy inhibitor 3-methyladenine (3-MA, 3 mM, HY-19312, Medchemexpress) for 72 h [17]. Afterward, the autophagy level of neurons was then detected and subsequent experiments were performed.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
From blood samples or cells, the RNA samples were extracted and then diluted to a suitable concentration, followed by RT. Afterward, cDNA was synthesized from miRNA or mRNA utilizing Tthe TaqMan™ MicroRNA RT kit (4366597,) or RevertAid RT kit (K1691) (all from Thermo Fisher). The gene expression was examined using TB Green® Premix Ex Taq™ II (RR820Q, Takara, Tokyo, Japan) on LightCycler 480 (Roche, Indianapolis, IN, USA) fluorescence quantitative PCR, including pre-denaturation at 95°C for 5 min, denaturation at 95°C for 10 s, annealing at 60°C for 10 s, and extension at 72°C for 20 s, with a total of 40 cycles. The relative expression of target genes was measured by means of 2−ΔΔCt method [18], with β-actin or U6 as the internal reference. The primer sequences are exhibited in Table 1.
Oxygen and glucose deprivation (OGD)
HT22 cells were inoculated into glucose-free DMEM and then maintained in anoxic environment containing 94% N2, 1% O2, and 5% CO2 for 2 h at 37°C. Next, the medium was changed to a normal DMEM, and the cells were cultured under normal conditions (at 37°C with 5% CO2) to simulate reperfusion for 24 h. Under the standard conditions of 90% humidity, 5% CO2, and 37°C, the cells cultured with DMEM with 10% FBS were used for control group [19].
Flow cytometry for cell apoptosis
Cell apoptosis was assessed using the Annexin V-fluorescein isothiocyanate (FITC)/propidium iodide (PI) apoptosis kit (C1062S, Beyotime, Shanghai, China). The cells were rinsed in precooled PBS and then resuspended in 100 μL of binding buffer. Afterward, the cells were mixed with 5 μL of Annexin V-FITC and PI at ambient temperature for 15 min. At last, FITC and PI fluorescence were detected by a flow cytometer, followed by analysis of cell apoptotic rate.
The total proteins were extracted using RIPA lysis buffer (89901, Thermo Fisher), after which the obtained proteins were separated by 12% SDS-PAGE and transferred to PVDF membranes (IPVH00010, Merck, Darmstadt, Germany). Next, the membranes were blocked with 5% bovine serum albumin at ambient temperature for 2 h, followed by overnight incubation (4°C) with primary antibodies including anti-LC3A/B (1:1000, ab62721, Abcam, Cambridge, UK), anti-p62 (1:10000, ab109012, Abcam), anti-cleaved caspase-3 (1:500, GTX86952, Genetex, Irvine, CA, USA), anti-Bax (1:1000, ab32503, Abcam), anti-Bcl-2 (1:2000, ab182858, Abcam), anti-SIRT1 (1:1000, ab233398, Abcam), and anti-β-actin (1:1000, ab5694, Abcam). Afterward, the membranes were incubated with the secondary antibody at ambient temperature for 2 h. The expression level of proteins was examined through development after Tris-buffered saline-tween buffer washing.
Transmission electron microscopy (TEM)
The cells to be tested were seeded onto 6-well plates at a density of 1 × 105 cells/well overnight. After the medium was discarded, the cells were washed with PBS thrice. Following fixation in 2.5% glutaraldehyde, the cells were treated with 1% osmium tetroxide, followed by dehydration, embedding, sectioning, and TEM staining. Finally, TEM images were captured with an HT7700 transmission electron microscope to observe and record the number of autophagosomes.
Dual luciferase reporter gene assay
Dual-Luciferase® Reporter Assay System (E1910, Promega, Madison, WI, USA) was employed for the dual luciferase reporter gene assay. In short, HEK293T cells were co-transfected with miR-384-5p mimic and the 3′-UTR luciferase reporter vector, containing wild-type (WT circR-ZC3HC1 or WT SIRT1) or mutant-type (MUT circR-ZC3HC1 or MUT SIRT1) of circR-ZC3HC1 or SIRT1. Forty-eight h after transfection, the cells were lysed with 20 μL lysis buffer and luciferase activity was assessed.
All statistical analyses were conducted using GraphPad prism 8.02 software. Data from at least three independent experiments were depicted in the form of mean ± standard deviation. Differences between two groups were comprised using the paired t-test or unpaired t-test, and differences among multiple groups using the one-way analysis of variance test, with Tukey’s multiple comparisons test used for post hoc multiple comparisons. Pearson correlation analysis is utilized to detect the correlation between two variables. Shapiro-Wilk test or Kolmogorov-Smirnov test was employed for the normality test. For samples that did not conform to normal distribution, Mann-Whitney U test was used for two independent samples, and Kruskal-Wallis was used for statistical analysis for multiple independent samples. Statistical significance was declared when p < 0.05.
Low expression of circR-ZC3HC1 in IS
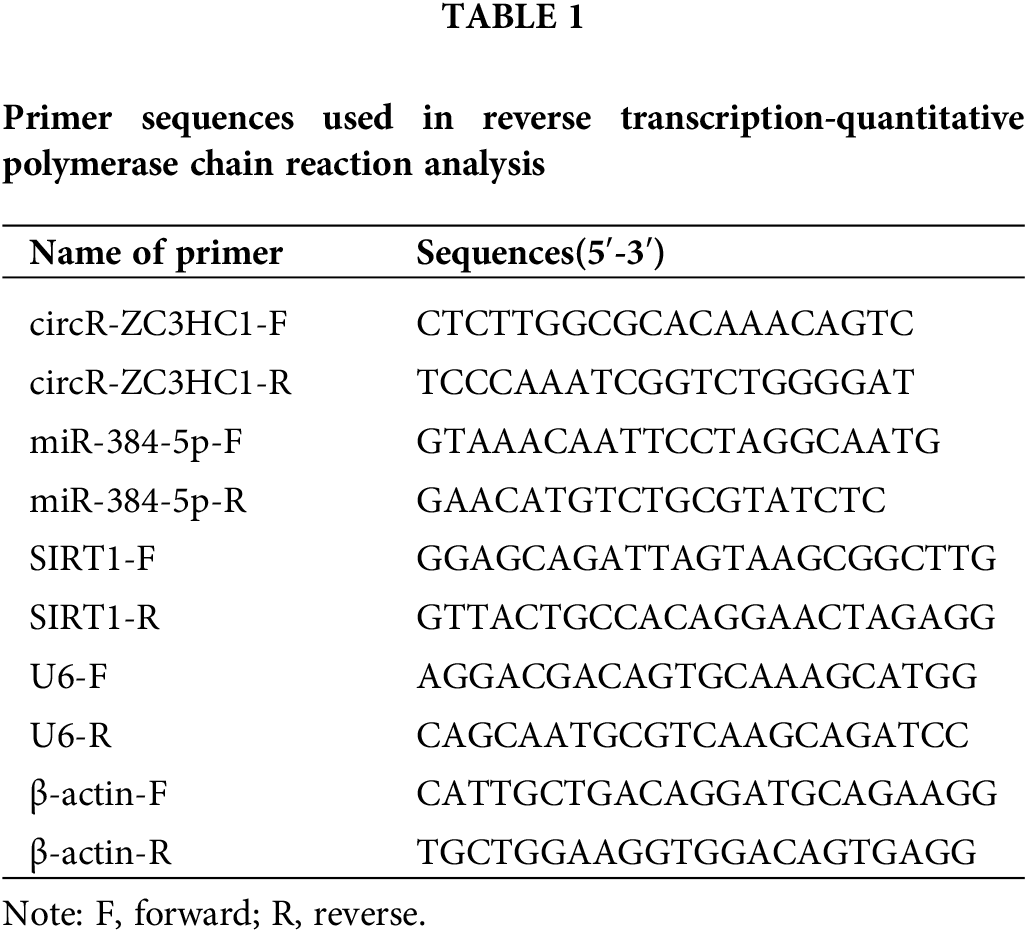
Reportedly, circRNAs are enriched in brain tissue and play important roles in the regulation of neuronal function [20,21]. First, we downloaded the GSE133768 dataset from the GEO database, and subsequently used the Limma program package in R language to perform differential expression analysis on the chip data. The screening condition for differential circRNAs was |log2 (Fold Change)| > 1, and the corrected p < 0.05. The results showed that circR-ZC3HC1 was lowly expressed in IS patients (Figs. 1A and 1B). In addition, common variant in ZC3HC1 gene contributed to an increase risk for IS [22]. However, the role of circR-ZC3HC1 in IS remains unclear. Thus, it is reasonable to speculate the possible relation between circR-ZC3HC1 expression and IS.
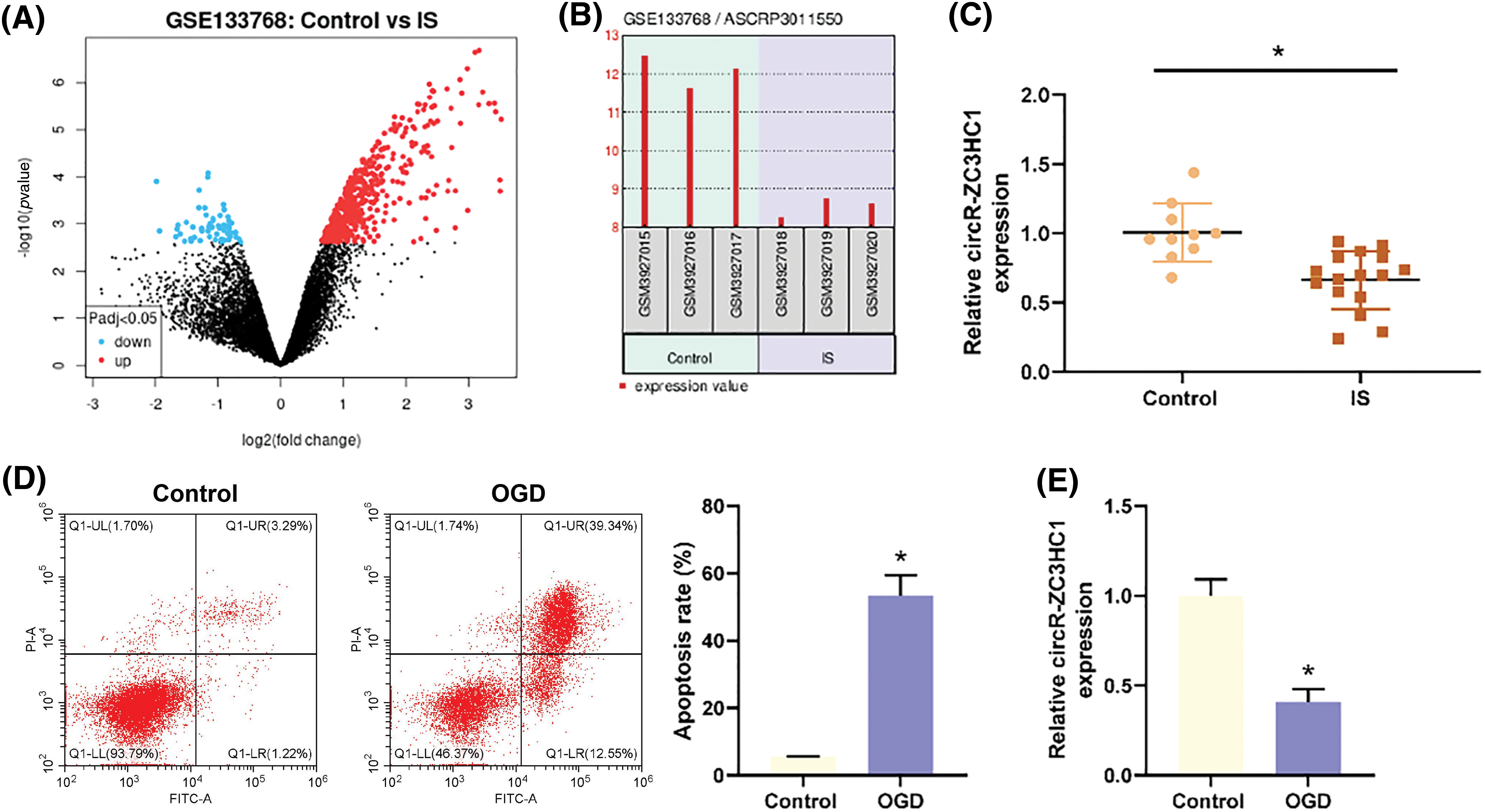
Figure 1: circR-ZC3HC1 was decreased in IS. Notes: (A and B) the expression of circR-ZC3HC1 in GSE133768 dataset was analyzed by GEO database (ASCRP3011550 [circRNA ID: hsa_circ_0082319; Gene symbol: circR-ZC3HC1]). (C) The expression of circR-ZC3HC1 in blood samples of 16 patients with IS and 10 healthy participants was detected by RT-qPCR. After hippocampal neuron HT22 cells underwent OGD treatment, (D) cell apoptosis was examined by flow cytometry. (E) circR-ZC3HC1 expression in HT22 cells was determined via RT-qPCR. Each cell experiment was repeated thrice independently), *p < 0.05.
Next, we designed RT-qPCR to assess the expression of circR-ZC3HC1 in clinical blood samples, which manifested that the IS patients had lower expression of circR-ZC3HC1 than the healthy subjects (Fig. 1C). Thereby, hippocampal neuron HT22 cells underwent OGD treatment, followed by flow cytometry for cell apoptosis and RT-qPCR for circR-ZC3HC1 expression. Compared with the control group, enhanced cell apoptosis (Fig. 1D) and decreased expression of circR-ZC3HC1 (Fig. 1E) were noted in the OGD group.
Overexpression of circR-ZC3HC1 promoted autophagy and inhibited apoptosis of OGD-treated neurons
It was reported that autophagy could inhibit neuronal cell apoptosis caused by traumatic brain injury [23]. To verify the effect of circR-ZC3HC1 on neurons, hippocampal neuron HT22 cells treated with OGD were transfected with p-ZC3HC1. Results of RT-qPCR revealed an increase of ZC3HC1 expression in the p-ZC3HC1 group as compared to the p-NC group (Fig. 2A). Subsequently, the HT22 cells were treated with 3-MA, after which the expression of autophagy marker LC3 and autophagy flux marker p62 and the number of autophagosomes were monitored using western blotting and TEM, respectively. In contrast to the p-NC group, the p-ZC3HC1 group had elevated LC3II expression and autophagosome number and reduced p62 expression, while 3-MA treatment reversed these trends (Figs. 2B and 2C). Flow cytometry results elicited that the apoptotic rate of OGD-treated HT22 cells was repressed in the p-ZC3HC1 group (vs. the p-NC group), which could be reversed by 3-MA treatment (Fig. 2D). Additionally, western blotting for detections of the expression of pro-apoptotic factors (cleaved caspase-3 and Bax) and anti-apoptotic factor (Bcl-2) displayed that p-ZC3HC1 transfection reduced the expression of cleaved caspase-3 and Bax, but elevated Bcl-2 expression, while further 3-MA treatment reversed these trends (Fig. 2E). Taken together, overexpression of circR-ZC3HC1 accelerated autophagy and restrained apoptosis of OGD-treated neurons, which were reversed by 3-MA treatment. From the above results, it was concluded that circR-ZC3HC1 inhibited OGD-treated neuron apoptosis by promoting autophagy.
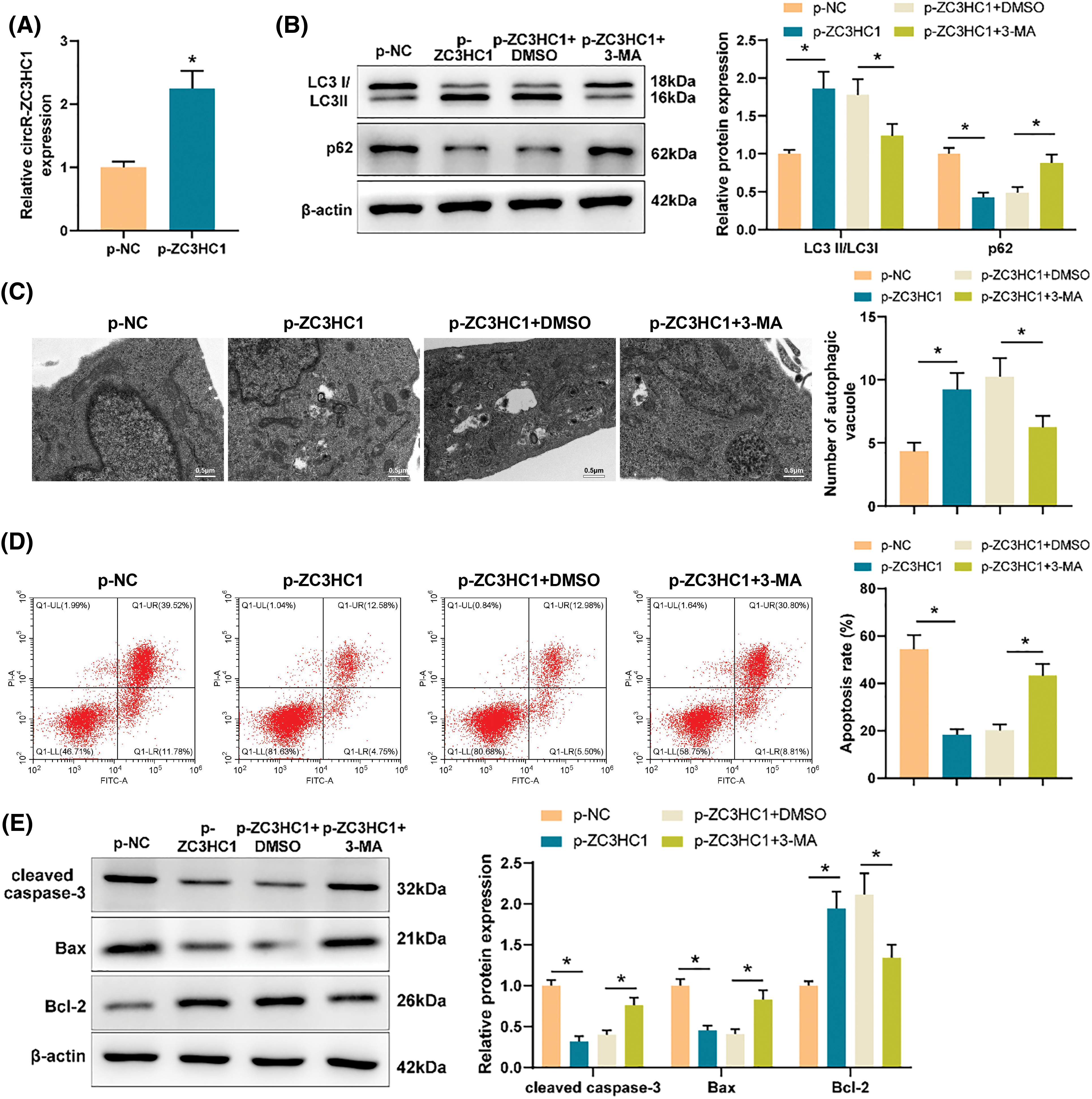
Figure 2: Overexpressed circR-ZC3HC1 inhibited apoptosis of OGD-treated neurons by promoting autophagy. Notes: After hippocampal neuron HT22 cells treated with OGD were transfected with p-ZC3HC1, (A) circR-ZC3HC1 expression was measured using RT-qPCR. Then, the HT22 cells were treated with 3-MA, (B) the expression of autophagy marker LC3 and autophagy flux marker p62 was assessed by western blotting. (C) The number of autophagosomes was observed through TEM. (D) Cell apoptosis was tested by flow cytometry. (E) The expression of pro-apoptotic factors (cleaved caspase-3 and Bax) and anti-apoptotic factor (Bcl-2) was examined by western blotting. Each assay was repeated three times, *p < 0.05.
circR-ZC3HC1 targeted miR-384-5p to upregulate SIRT1 expression
Through bioinformatics analysis, we found that circR-ZC3HC1 could target miR-384-5p (Fig. 3A) and also noticed the binding sites of miR-384-5p and SIRT1 (Fig. 3B). Therefore, we conjectured that circR-ZC3HC1 might assume a role in regulating autophagy of neurons by targeting miR-384-5p to mediate SIRT1 expression.
Figure 3: circR-ZC3HC1 upregulated SIRT1 expression by targeting miR-384-5p. Notes: (A) the binding relationship between circR-ZC3HC1 and miR-384-5p was predicted through bioinformatics analysis. (B) The binding relationship between miR-384-5p and SIRT1 was predicted through bioinformatics analysis. (C and D) Dual luciferase reporter gene assay was used to verify the binding relationship between circR-ZC3HC1 and miR-384-5p or between miR-384-5p and SIRT1. (E and F) The expression of miR-384-5p and SIRT1 was examined by RT-qPCR and western blotting. (G) The transfection efficiency of miR-384-5p mimic was assessed by RT-qPCR. (H and I) The expression of SIRT1 was detected by RT-qPCR and western blotting. Each cell experiment was repeated thrice independently, *p < 0.05.
The data of dual luciferase reporter gene assay unveiled that miR-384-5p mimic significantly reduced the luciferase activities of WT circR-ZC3HC1 and WT SIRT1 (Figs. 3C and 3D), which suggested that circR-ZC3HC1 could specifically bind to miR-384-5p, and miR-384-5p could bind to SIRT1. RT-qPCR and western blotting results showed that upregulated miR-384-5p and downregulated SIRT1 were discovered in the OGD group compared with the control group (Figs. 3E and 3F). Thereby, we transfected miR-384-5p mimic into cells overexpressed with circR-ZC3HC1, and then measured the transfection efficiency by RT-qPCR (Fig. 3G). Western blotting and RT-qPCR results exhibited that SIRT1 was significantly increased after circR-ZC3HC1 overexpression, but decreased after further overexpression of miR-384-5p (Figs. 3H and 3I). These findings demonstrated that circR-ZC3HC1 elevated SIRT1 expression by targeting miR-384-5p.
Knockdown of miR-384-5p facilitated autophagy and repressed apoptosis of OGD-treated neurons
To explore the effect of miR-384-5p on neurons, OGD-treated neuron HT22 cells were transfected with miR-384-5p inhibitor. RT-qPCR revealed that miR-384-5p knockdown reduced the expression of miR-384-5p but enhanced the expression of SIRT1 (Fig. 4A). As reflected in western blotting results, miR-384-5p knockdown increased the expression of SIRT1 and LC3II but declined the expression of p62 (Fig. 4B). Through TEM, increased autophagosomes were observed following miR-384-5p knockdown (Fig. 4C). As expected, the apoptosis level of OGD-treated HT22 cells was inhibited after miR-384-5p knockdown (Fig. 4D), accompanied by decreased cleaved caspase-3 and Bax expression, and increased Bcl-2 expression (Fig. 4E). In short, deficiency of miR-384-5p could promote autophagy and suppress apoptosis of OGD-treated neurons.
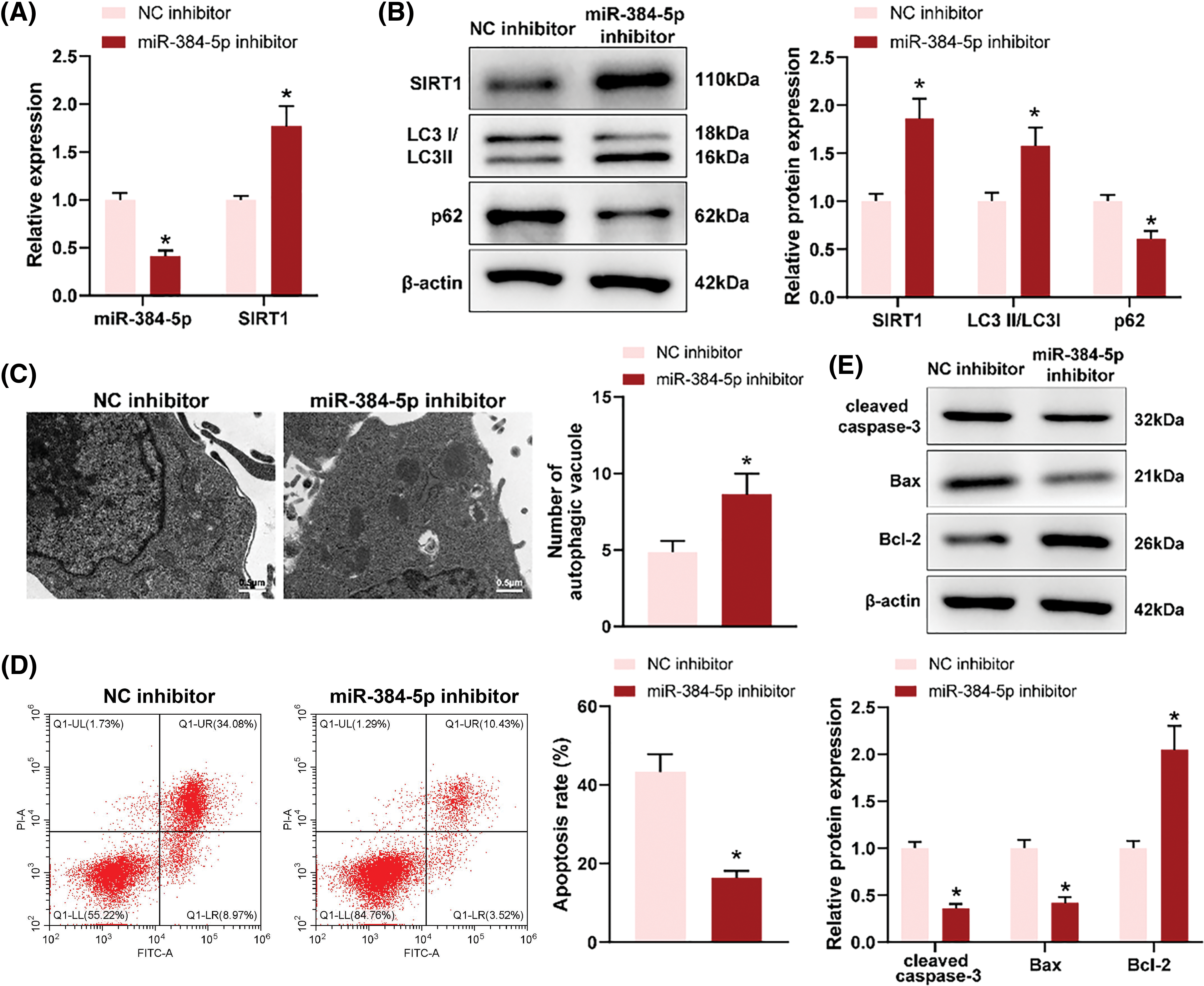
Figure 4: Loss of miR-384-5p promoted autophagy and suppressed apoptosis of OGD-treated neurons. Notes: After OGD-treated neuron HT22 cells were transfected with miR-384-5p inhibitor, (A) RT-qPCR was employed for the assessment of the expression of miR-384-5p and SIRT1. (B) The protein expression of SIRT1, LC3, and p62 was evaluated using western blotting. (C) The number of autophagosomes was observed through TEM. (D) Cell apoptosis was tested by flow cytometry. (E) The expression of pro-apoptotic factors (cleaved caspase-3 and Bax) and anti-apoptotic factor (Bcl-2) was examined by western blotting. Each cell experiment was duplicated thrice, *p < 0.05.
circR-ZC3HC1 promoted autophagy and suppressed apoptosis of OGD-treated neurons by upregulating SIRT1
To verify that circR-ZC3HC1 mediated autophagy and apoptosis of neurons through up-regulation of SIRT1, we combined SIRT1 inhibition in OGD-treated neuron HT22 cells overexpressing circR-ZC3HC1. The transfection efficiency of SIRT1 was detected by RT-qPCR and western blotting, and sh-SIRT1#3, which had the best knockdown efficiency, was selected for subsequent experiments (Figs. 5A and 5B). Discoveries from western blotting, TEM, and TEM showed that, after further SIRT1 inhibition, the autophagy level was weakened, shown by decreased LC3II and increased p62 (Fig. 5C) and reduced autophagosomes (Fig. 5D); the apoptosis level was enhanced (Fig. 5E), accompanied by elevated cleaved caspase-3 and Bax, and reduced Bcl-2 (Fig. 5F). Overall, circR-ZC3HC1 promoted autophagy and restrained apoptosis of OGD-treated neurons by upregulating SIRT1.
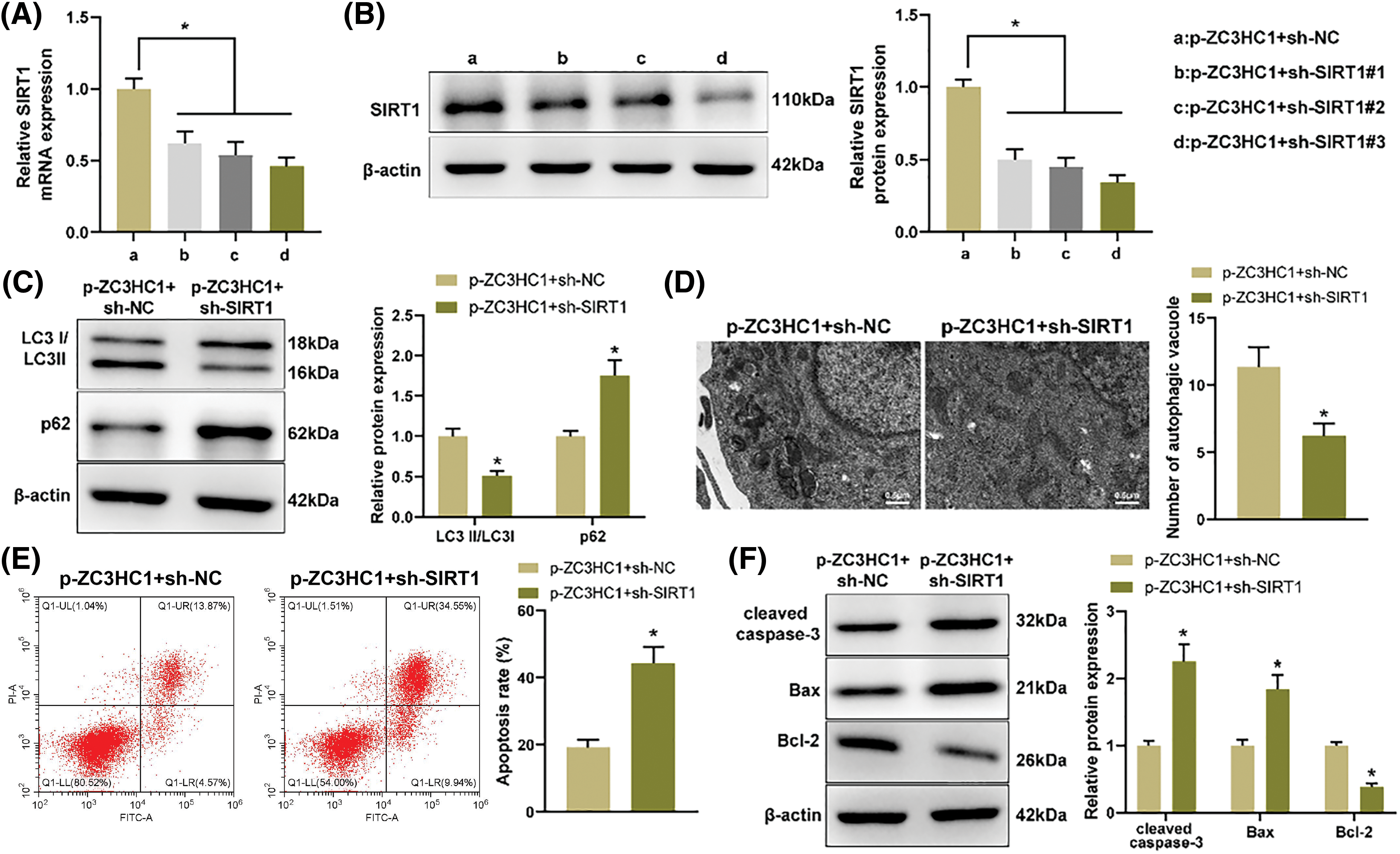
Figure 5: circR-ZC3HC1 enhanced autophagy and inhibited apoptosis of OGD-treated neurons by increasing SIRT1. Notes: After OGD-treated neuron HT22 cells overexpressing circR-ZC3HC1 were transfected with sh-SIRT1, (A and B) RT-qPCR and western blotting were adopted for detection of SIRT1 expression. (C) Western blotting was designed to evaluate the protein expression of LC3 and p62. (D) The number of autophagosomes was observed through TEM. (E) Cell apoptosis was tested by flow cytometry. (F) The expression of pro-apoptotic factors (cleaved caspase-3 and Bax) and anti-apoptotic factor (Bcl-2) was evaluated by western blotting. Experimentation was repeated thrice independently, *p < 0.05.
IS is a fatal disease with high disability and mortality rates, resulting in a heavy burden on society and family [24]. During IS, autophagy is activated in various cell types of the brain, including microvascular cells, neurons, and glial cells [25]. Nowadays, the modulation of autophagy may be a promising target in the treatment of IS [26]. Previously, researchers have shown that as a new type of non-coding RNA, circRNAs are involved in IS-induced autophagy and various pathologic processes after IS [27]. In this research, we discovered downregulated circR-ZC3HC1 in IS patients via bioinformatics analysis, which was subsequently confirmed by the functional experiments. Further assays verified that overexpressed circR-ZC3HC1 promoted autophagy and inhibited apoptosis of OGD-treated neurons by targeting miR-384-5p to upregulate SIRT1 expression.
With the continuous development of technology, bioinformatics tools have obvious advantages in predicting the structure, product, function and evolution of unknown genes and proteins, which have been adopted by more and more researchers [28,29]. Through this method, we predicted that circR-ZC3HC1 was closely associated with IS, and then discovered low expression of circR-ZC3HC1 in blood samples of IS patients and OGD-treated hippocampal neuron HT22 cells. Moreover, overexpression of circR-ZC3HC1 facilitated autophagy and repressed apoptosis of OGD-treated neuron HT22 cells, which could be abrogated by 3-MA treatment. A previous research displayed that autophagy could restrain neuronal cell apoptosis caused by spinal cord injury [30]. Here, we concluded that overexpressed circR-ZC3HC1 inhibited OGD-treated neuron apoptosis by promoting autophagy. Although there are no relevant studies on circR-ZC3HC1 in IS, there are many studies on the effects of other circRNAs on the development of IS by regulating autophagy and apoptosis. For example, circ-0025984 protected astrocytes against ischemia-induced apoptosis and autophagy by modulating the miR-143-3p/TET1 pathway, and finally repressed IS-induced cerebral injury [31]. Circ-SHOC2 suppressed neuronal apoptosis by regulating autophagy and ameliorated ischemic brain injury via the miR-7670-3p/SIRT1 axis [15]. From the above data, it could be concluded that circRNAs may regulate IS-induced autophagy and apoptosis by targeting some miRNAs.
Next, we deeply probed the downstream targets of circR-ZC3HC1. miR-384-5p was found to be targeted by circR-ZC3HC1 via bioinformatics analysis. RT-qPCR results showed high expression of miR-384-5p in OGD-treated neurons, and further assays unraveled that knockdown of miR-384-5p could promote autophagy and inhibit apoptosis of OGD-treated neuron HT22 cells. In hippocampal neurons, miR-384-5p downregulation exerted neuroprotection by restraining apoptosis and reactive oxygen species activity in neurons [32]. Another study stated that isoflurane post-treatment relieved ischemic neuronal injury through miR-384-5p-mediated autophagy [12]. Also, we noticed that miR-384-5p could bind to SIRT1 through bioinformatics method. A lot of studies implied that SIRT1 has neuroprotective effect on cerebral ischemic injury and is considered as a valuable candidate gene for IS [33,34]. A prior research described that the activation of SIRT1/AMPK pathway attenuated the IS-induced neuronal damage [35]. Tian et al. proposed that lncRNA Snhg8 relieved microglial inflammation and blood-brain barrier damage in IS by regulating miR-425-5p mediated SIRT1/NF-κB signaling [36]. In our work, we verified that circR-ZC3HC1 acted as a sponge for miR-384-5p and promotes SIRT1 expression to promote autophagy and repress apoptosis of OGD-treated neuron HT22 cells, thereby mitigating IS.
Our results demonstrated for the first time that circR-ZC3HC1 protects neurons from ischemic-induced autophagy and apoptosis by targeting miR-384-5p/SIRT1 axis and might inhibit IS process. Of course, this article has some limitations to consider. Firstly is the small sample size for clinical experiments due to limitations of time and region. Second, we only conducted relevant studies at the cellular level, and validation in the in vivo experiments is required if more budget is accessible. In the future, future research needs to address these deficiencies through animal and clinical trials. In any case, our findings contribute to a better understanding of the complex mechanism behind neuronal autophagy and apoptosis in IS and provide a rapidly expanding field for the development of new effective therapeutic targets for IS.
Acknowledgement: None.
Funding Statement: Supported by Ningbo Health Technology Project, Nos. 2020Y12 and 2022Y12.
Author Contributions: The authors confirm contribution to the paper as follows: study conception and design: SM; data collection: SM, XXM, SGL, WLZ, YT, SH and WW; analysis and interpretation of results: SM, XXM, CQH and SZZ; draft manuscript preparation: SM, XXM, SGL and WLZ. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.
Ethics Approval: The protocol of our study was permitted by the Ethics Committee of the First Affiliated Hospital of Ningbo University (No. 2022KYYS119), and informed consent forms were signed by all participants. Clinical trials followed the Declaration of Helsinki.
Conflicts of Interest: The authors declare there is no conflict of interest regarding this study.
References
1. Iadecola C, Buckwalter MS, Anrather J. Immune responses to stroke: mechanisms, modulation, and therapeutic potential. J Clin Invest. 2020;130(6):2777–88. doi:10.1172/JCI135530. [Google Scholar] [PubMed] [CrossRef]
2. Haupt M, Gerner ST, Bahr M, Doeppner TR. Neuroprotective strategies for ischemic stroke-future perspectives. Int J Mol Sci. 2023;24(5):4334. doi:10.3390/ijms24054334. [Google Scholar] [PubMed] [CrossRef]
3. Shi Q, Cheng Q, Chen C. The role of autophagy in the pathogenesis of ischemic stroke. Curr Neuropharmacol. 2021;19(5):629–40. doi:10.2174/1570159X18666200729101913. [Google Scholar] [PubMed] [CrossRef]
4. Ajoolabady A, Wang S, Kroemer G, Penninger JM, Uversky VN, Pratico D, et al. Targeting autophagy in ischemic stroke: from molecular mechanisms to clinical therapeutics. Pharmacol Ther. 2021;225:107848. doi:10.1016/j.pharmthera.2021.107848. [Google Scholar] [PubMed] [CrossRef]
5. Tuo QZ, Zhang ST, Lei P. Mechanisms of neuronal cell death in ischemic stroke and their therapeutic implications. Med Res Rev. 2022;42(1):259–305. doi:10.1002/med.21817. [Google Scholar] [PubMed] [CrossRef]
6. Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi:10.15252/embj.2018100836. [Google Scholar] [PubMed] [CrossRef]
7. Sekar S, Liang WS. Circular RNA expression and function in the brain. Noncoding RNA Res. 2019;4(1):23–9. doi:10.1016/j.ncrna.2019.01.001. [Google Scholar] [PubMed] [CrossRef]
8. Liu Z, Zhou Y, Xia J. CircRNAs: key molecules in the prevention and treatment of ischemic stroke. Biomed Pharmacother. 2022;156(25):113845. doi:10.1016/j.biopha.2022.113845. [Google Scholar] [PubMed] [CrossRef]
9. Wu F, Han B, Wu S, Yang L, Leng S, Li M, et al. Circular RNA TLK1 aggravates neuronal injury and neurological deficits after ischemic stroke via miR-335-3p/TIPARP. J Neurosci. 2019;39(37):7369–93. doi:10.1523/JNEUROSCI.0299-19.2019. [Google Scholar] [PubMed] [CrossRef]
10. Chen W, Wang H, Feng J, Chen L. Overexpression of circRNA circUCK2 attenuates cell apoptosis in cerebral ischemia-reperfusion injury via miR-125b-5p/GDF11 signaling. Mol Ther Nucleic Acids. 2020;22:673–83. doi:10.1016/j.omtn.2020.09.032. [Google Scholar] [PubMed] [CrossRef]
11. Gugliandolo A, Silvestro S, Sindona C, Bramanti P, Mazzon E. MiRNA: involvement of the MAPK pathway in ischemic stroke. A promising therapeutic target. Medicina. 2021;57(10):1053. doi:10.3390/medicina57101053. [Google Scholar] [PubMed] [CrossRef]
12. Zhu X, Yang M, Yang L. Isoflurane postconditioning alleviates ischemic neuronal injury via MiR-384-5p regulated autophagy. Neuroscience. 2023;517:26–36. doi:10.1016/j.neuroscience.2023.01.018. [Google Scholar] [PubMed] [CrossRef]
13. Liu J, An P, Xue Y, Che D, Liu X, Zheng J, et al. Mechanism of Snhg8/miR-384/Hoxa13/FAM3A axis regulating neuronal apoptosis in ischemic mice model. Cell Death Dis. 2019;10(6):441. doi:10.1038/s41419-019-1631-0. [Google Scholar] [PubMed] [CrossRef]
14. Zhang JF, Zhang YL, Wu YC. The role of Sirt1 in ischemic stroke: pathogenesis and therapeutic strategies. Front Neurosci. 2018;12:833. doi:10.3389/fnins.2018.00833. [Google Scholar] [PubMed] [CrossRef]
15. Chen W, Wang H, Zhu Z, Feng J, Chen L. Exosome-shuttled circSHOC2 from IPASs regulates neuronal autophagy and ameliorates ischemic brain injury via the miR-7670-3p/SIRT1 axis. Mol Ther Nucleic Acids. 2020;22:657–72. doi:10.1016/j.omtn.2020.09.027. [Google Scholar] [PubMed] [CrossRef]
16. Tao H, Liu Y, Hou Y. miRNA‐384‐5p regulates the progression of Parkinson’s disease by targeting SIRT1 in mice and SH‐SY5Y cell. Int J Mol Med. 2020;45(2):441–50. doi:10.3892/ijmm.2019.4426. [Google Scholar] [PubMed] [CrossRef]
17. Cheng L, Chen Y, Guo D, Zhong Y, Li W, Lin Y, et al. mTOR-dependent TFEB activation and TFEB overexpression enhance autophagy-lysosome pathway and ameliorate Alzheimer’s disease-like pathology in diabetic encephalopathy. Cell Commun Signal. 2023;21(1):91. doi:10.1186/s12964-023-01097-1. [Google Scholar] [PubMed] [CrossRef]
18. Li P, Xing J, Zhang J, Jiang J, Liu X, Zhao D, et al. Inhibition of long noncoding RNA HIF1A-AS2 confers protection against atherosclerosis via ATF2 downregulation. J Adv Res. 2020;26(4):123–35. doi:10.1016/j.jare.2020.07.015. [Google Scholar] [PubMed] [CrossRef]
19. Xu ZQ, Zhang JJ, Kong N, Zhang GY, Ke P, Han T, et al. Autophagy is involved in neuroprotective effect of Alpha7 nicotinic acetylcholine receptor on ischemic stroke. Front Pharmacol. 2021;12:676589. doi:10.3389/fphar.2021.676589. [Google Scholar] [PubMed] [CrossRef]
20. Hanan M, Soreq H, Kadener S. CircRNAs in the brain. RNA Biol. 2017;14(8):1028–34. doi:10.1080/15476286.2016.1255398. [Google Scholar] [PubMed] [CrossRef]
21. Akhter R. Circular RNA and Alzheimer’s disease. Adv Exp Med Biol. 2018;1087:239–43. doi:10.1007/978-981-13-1426-1. [Google Scholar] [CrossRef]
22. Jafaripour S, Sasanejad P, Dadgarmoghaddam M, Sadr-Nabavi A. ADAMTS7 and ZC3HC1 share genetic predisposition to coronary artery disease and large artery ischemic stroke. Crit Rev Eukaryot Gene Expr. 2019;29(4):351–61. doi:10.1615/CritRevEukaryotGeneExpr.2019028209. [Google Scholar] [PubMed] [CrossRef]
23. Chen X, Pan Z, Fang Z, Lin W, Wu S, Yang F, et al. Omega-3 polyunsaturated fatty acid attenuates traumatic brain injury-induced neuronal apoptosis by inducing autophagy through the upregulation of SIRT1-mediated deacetylation of Beclin-1. J Neuroinflammation. 2018;15(1):310. doi:10.1186/s12974-018-1345-8. [Google Scholar] [PubMed] [CrossRef]
24. Su XQ, Chen YH, Yuan XX. The autophagy in ischemic stroke: a regulatory role of non-coding-RNAs. Cell Signal. 2023;104:110586. doi:10.1016/j.cellsig.2022.110586. [Google Scholar] [PubMed] [CrossRef]
25. Zhang Y, Cao Y, Liu C. Autophagy and ischemic stroke. Adv Exp Med Biol. 2020;1207:111–34. doi:10.1007/978-981-15-4272-5. [Google Scholar] [CrossRef]
26. Lu X, Zhang J, Ding Y, Wu J, Chen G. Novel therapeutic strategies for ischemic stroke: recent insights into autophagy. Oxid Med Cell Longev. 2022;2022(2):3450207. doi:10.1155/2022/3450207. [Google Scholar] [PubMed] [CrossRef]
27. Li X, Li L, Si X, Zhang Z, Ni Z, Zhou Y, et al. The regulatory roles of circular RNAs via autophagy in ischemic stroke. Front Neurol. 2022;13:963508. doi:10.3389/fneur.2022.963508. [Google Scholar] [PubMed] [CrossRef]
28. Rincon-Riveros A, Morales D, Rodriguez JA, Villegas VE, Lopez-Kleine L. Bioinformatic tools for the analysis and prediction of ncRNA interactions. Int J Mol Sci. 2021;22(21):11397. doi:10.3390/ijms222111397. [Google Scholar] [PubMed] [CrossRef]
29. Ma L, Li H, Lan J, Hao X, Liu H, Wang X, et al. Comprehensive analyses of bioinformatics applications in the fight against COVID-19 pandemic. Comput Biol Chem. 2021;95:107599. doi:10.1016/j.compbiolchem.2021.107599. [Google Scholar] [PubMed] [CrossRef]
30. Ren XD, Wan CX, Niu YL. Overexpression of lncRNA TCTN2 protects neurons from apoptosis by enhancing cell autophagy in spinal cord injury. FEBS Open Bio. 2019;9(7):1223–31. doi:10.1002/2211-5463.12651. [Google Scholar] [PubMed] [CrossRef]
31. Zhou D, Huang Z, Zhu X, Hong T, Zhao Y. Circular RNA 0025984 ameliorates ischemic stroke injury and protects astrocytes through miR-143-3p/TET1/ORP150 pathway. Mol Neurobiol. 2021;58(11):5937–53. doi:10.1007/s12035-021-02486-8. [Google Scholar] [PubMed] [CrossRef]
32. Yang Q, Long F. MiRNA-384-5p targets GABRB1 to regulate ketamine-induced neurotoxicity in neurons. Turk Neurosurg. 2022. doi:10.5137/1019-5149.JTN.36367-21.2. [Google Scholar] [PubMed] [CrossRef]
33. Tang H, Wen J, Qin T, Chen Y, Huang J, Yang Q, et al. New insights into Sirt1: potential therapeutic targets for the treatment of cerebral ischemic stroke. Front Cell Neurosci. 2023;17:1228761. doi:10.3389/fncel.2023.1228761. [Google Scholar] [PubMed] [CrossRef]
34. Jiao F, Gong Z. The beneficial roles of SIRT1 in neuroinflammation-related diseases. Oxid Med Cell Longev. 2020;2020(3):6782872. doi:10.1155/2020/6782872. [Google Scholar] [PubMed] [CrossRef]
35. Liang H, Chang X, Xia R, Wu W, Guo H, Yang M. Magnoflorine attenuates cerebral ischemia-induced neuronal injury via autophagy/Sirt1/AMPK signaling pathway. Evid Based Complement Alternat Med. 2022;2022:2131561. doi:10.1155/2022/2131561. [Google Scholar] [PubMed] [CrossRef]
36. Tian J, Liu Y, Wang Z, Zhang S, Yang Y, Zhu Y, et al. LncRNA Snhg8 attenuates microglial inflammation response and blood-brain barrier damage in ischemic stroke through regulating miR-425-5p mediated SIRT1/NF-κB signaling. J Biochem Mol Toxicol. 2021;35(5):e22724. doi:10.1002/jbt.22724. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools