 Open Access
Open Access
REVIEW
Advances in CRISPR-based gene editing technology and its application in nucleic acid detection
1 Translational Medicine Institute, the First People’s Hospital of Chenzhou, Hengyang Medical School, University of South China, Chenzhou, 423000, China
2 The First Clinical College of Xiangnan University, The First Affiliated Hospital of Xiangnan University, Chenzhou, 423000, China
3 National Engineering Research Center of Personalized Diagnostic and Therapeutic Technology, Hunan University of Chinese Medicine, Changsha, 410208, China
4 State Key Laboratory of Digital Medical Engineering, School of Biological Science and Medical Engineering, Southeast University, Nanjing, 210096, China
* Corresponding Author: ZHENG HU. Email:
# These authors contributed equally to this work
BIOCELL 2025, 49(1), 21-43. https://doi.org/10.32604/biocell.2024.056698
Received 28 July 2024; Accepted 27 September 2024; Issue published 24 January 2025
Abstract
Nucleic acid analysis is a key technique that enables accurate detection of various microorganisms. Conventional nucleic acid testing typically requires access to specialized laboratories, equipment, and trained personnel, which hinders the widespread use of on-site testing for DNA and RNA targets. However, integrating gene editing technology with traditional nucleic acid detection methods, especially isothermal amplification technology, can help overcome the limitations associated with on-site testing. This combination can accomplish precise and swift detection of nucleic acid sequences, offering a robust tool for on-site detection. The Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated proteins (CRISPR/Cas) technology, which comprises the CRISPR system and Cas effector proteins, is a powerful tool that is advancing the field of nucleic acid detection. Specifically, Cas12, Cas13, and Cas14 proteins have emerged as straightforward, effective, precise, sensitive, and cost-effective methods for in vitro nucleic acid detection because of their “collateral cleavage” characteristics. When combined with the “collateral cleavage” ability of Cas protein and isothermal amplification, CRISPR/Cas systems have great potential to advance nucleic acid detection. This article summarizes the research progress of different CRISPR/Cas systems and their applications in nucleic acid detection and future perspectives.Keywords
Abbreviation List
| CRISPR | Clustered Regularly Interspaced Short Palindromic Repeats |
| Cas | CRISPR-associated proteins |
| OMEGA | Obligate mobile element–guided activity |
| FLSHclust | Fast locality-sensitive hashing-based clustering |
| Agos | Argonautes |
| MARPS | Mesophilic Argonaute Report-based single millimeter Polystyrene Sphere |
| PAM | Protospacer adjacent motif |
| crRNA | CRISPR RNA |
| dsDNA | Double-stranded DNA |
| SSB | Single strand DNA-binding protein |
| ssRNA | Single-stranded RNA |
| ZFNs | Zinc-finger nucleases |
| TALENs | Transcription activator-like effector nucleases |
| HNH | Histidine asparagine histidine |
| sgRNA | Single guide RNA |
| HEPN | Higher eukaryotes and prokaryotes nucleotide-binding domain |
| ssRNA | Single-stranded RNA |
| ORF | Open reading frame |
| ssDNA | Single-stranded DNA |
| IAT | Isothermal amplification technology |
| PCR | Polymerase Chain Reaction |
| LAMP | Loop-mediated isothermal amplification |
| SDA | Strand displacement amplification |
| RPA | Recombinase polymerase amplification |
| CPA | Crossing priming amplification |
| RCA | Rolling circle amplification |
| HDA | Helicase-dependent amplification |
| NGS | Next generation sequencing |
| RNA-Seq | RNA sequencing |
| ChIP-Seq | Chromatin immunoprecipitation-sequencing |
| Hi-C | Chromosome conformation capture coupling with high-throughput sequencing |
| SMRT | Single-molecule, real-time sequencing |
| FISH | Fluorescence in situ hybridization |
| gRNA | Guide RNA |
| tracrRNA | Trans-activating CRISPR RNA |
| Nease | Endonuclease |
| NASBACC | Nucleic Acid Sequence-Based Amplification-CRISPR Cleavage |
| ZIKV | Zika virus |
| HIV-1 | Human Immunodeficiency Virus-1 |
| NASBA | Nucleic Acid Sequence-Based Amplification |
| LTR | Long terminal repeats |
| CAS-EXPAR | CRISPR/Cas9-isothermal exponential amplification reaction |
| FLASH | Finding low abundance sequences by hybridization |
| Cas9 RNPs | Cas9 ribonucleoprotein complexes |
| ROI | Region of interest |
| gDNA | Genomic DNA |
| Cas9nAR | Cas9 nickase-based amplification reaction |
| nCATS | Nanopore Cas9-targeted sequencing |
| EGFR | Epidermal growth factor receptor gene |
| SMART | Small molecule-activated allosteric aptamer regulating |
| dCas9 | Dead Cas9 |
| PC | Paired dCas9 |
| MTB | Mycobacteriu m tuberculosis |
| RCH | Rolling circle amplification-CRISPR-split-horseradish peroxidase |
| sHRP | Split-HRP |
| TMB | Tetramethyl benzidine |
| gFET | Graphene-based field-effect transistor |
| REs | Restriction enzymes |
| DETECTR | DNA Endonuclease-Targeted CRISPR Trans Reporter |
| HOLMES | One-hour low-cost multipurpose highly efficient system |
| RT-RPA | Reverse Transcription and Recombinase Polymerase Isothermal Amplification |
| AIOD-CRISPR | All-in-one dual CRISPR-Cas12a |
| SHERLOCK | Specific high sensitivity enzymatic reporter unlocking platform |
| RT-LAMP | Reverse transcription loop-mediated isothermal amplification |
| Cas12a-based sPAMC | Suboptimal PAM of Cas12a-based test |
| HCMV | Human cytomegalovirus |
| RT-CORDS | RT-PCR combined with CRISPR on-site rapid detection system |
| CRISPR-SPADE | CRISPR Single Pot Assay for Detecting Emerging VOCs |
| POC | Point-of-care |
| SPEEDi-CRISPR | Solid-Phase Extraction and Enhanced Detection Assay using CRISPR-Cas12a |
| LFA | Lateral flow assay |
| POCT | Point-of-care testing |
| RAA | Recombinase-aided amplification |
| Cas12c-DETECTOR | Cas12c-based nucleic acid detection platform |
| SNPs | Single nucleotide polymorphisms |
| PFS | Protospacer-flanking site |
| DENV | Dengue virus |
| HUDSON | Heating unextracted diagnostic samples to obliterate nucleases |
| EBV | Epstein-Barr virus |
| STV | Streptavidin |
| ASFV | African swine fever virus |
| PAC-MAN | Prophylactic antiviral CRISPR in human cells |
| ECL | Electrochemiluminescence |
| 2D-pMOFs | 2D porphyrin metal-organic framework nanosheets |
| MC-LR | Microcystin-LR |
| SMN1 | Survival motor neuron 1 gene |
| SMA | Spinal Muscular Atrophy |
CRISPR/Cas is a vital component in the bacterial acquired immune system of prokaryotes and comprises Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) and CRISPR-associated proteins (Cas) [1]. The discovery of these repeating sequences along with spacer sequences in Escherichia coli by Ishino et al. in 1987 was a groundbreaking moment in the study of CRISPR/Cas [2]. Subsequent findings of this sequence in other organisms, such as Haloferax in 1995 [3], Cyanobacteria in 1996 [4], and even archaea in 2000, solidified the importance of CRISPR sequences in prokaryotic genomes [5]. Jansen et al. termed these repeated nucleotide sequences with spacers as “CRISPR sequences” in 2002, marking a significant milestone in the understanding of the CRISPR/Cas system [6]. Then in 2007, Barrangou et al. revealed the immune mechanism mediated by the CRISPR/Cas system [7]. This system functions by recognizing and cleaving exogenous viruses and plasmid DNA that enter bacteria. The Cas proteins within the CRISPR system are responsible for this recognition and cleavage process, ultimately leading to the formation of an acquired immune system [8]. Upon reinvasion by the pathogen, the bacteria’s CRISPR/Cas system is activated to cut off and degrade the exogenous DNA [9]. Recently, it was discovered through molecular pedigree analysis that the TnpB protein in the obligate mobile element–guided activity (OMEGA) family may be the ancestor of CRISPR/Cas12 endonuclease and eukaryotic protein Fanzor [10]. In 2021, the OMEGA family was reported as a new class of RNA-guided nucleases [10]. Fanzor is found in eukaryotes but, it does not have the property of “collateral activity”. Fanzor is an OMEGA-like and a CRISPR-like gene editing system, which demonstrates that RNA-directed endonucleases exist not only in prokaryotes but also in eukaryotes [11]. In 2023, researchers introduced the fast locality-sensitive hashing-based clustering (FLSHclust) algorithm; 188 novel CRISPR/Cas-related systems were discovered and new biochemical functions linked to adaptive immunity were unveiled [12](Fig. 1).
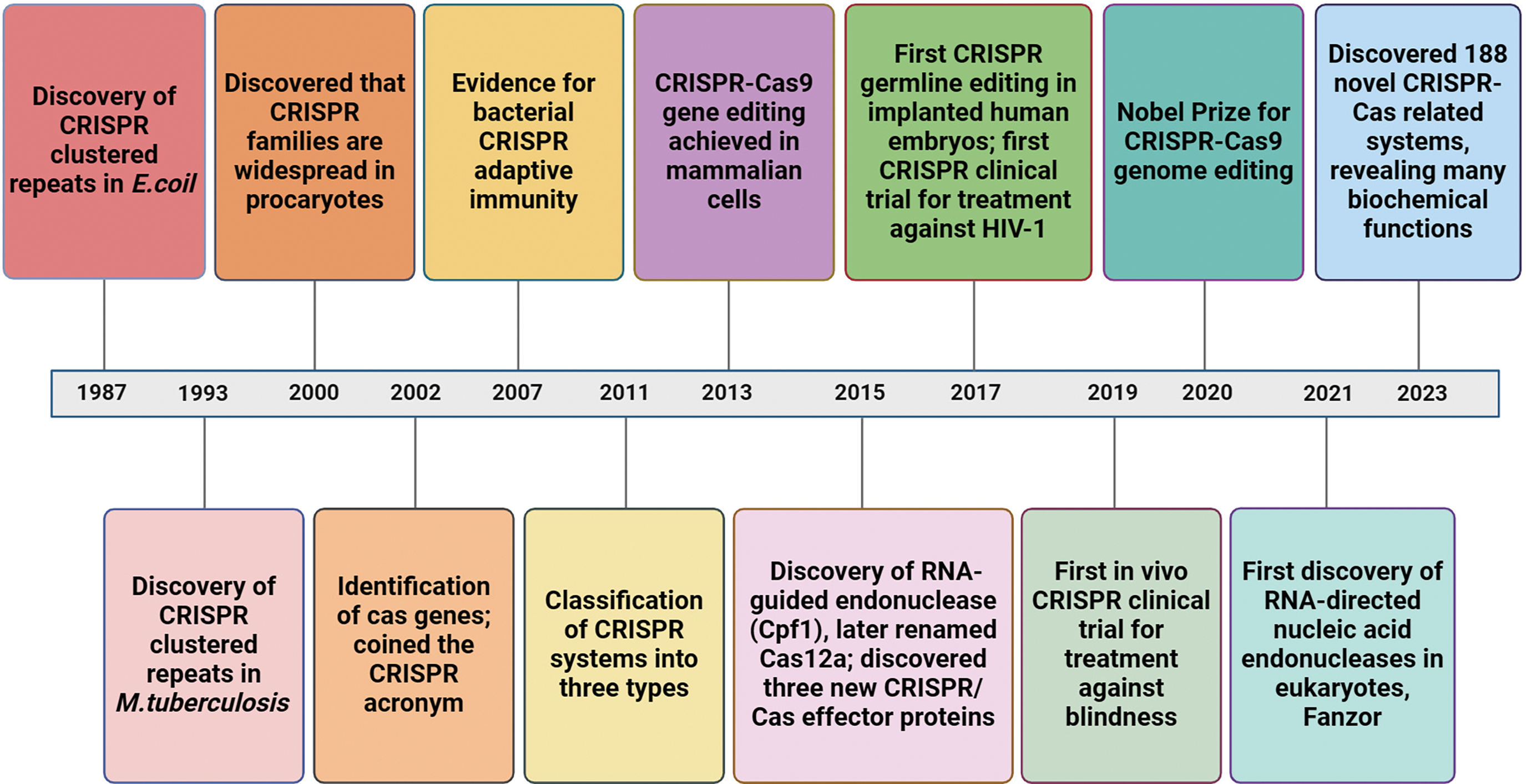
Figure 1: Timeline diagram of key milestones in the clustered regularly interspaced palindromic repeats (CRISPR)/Cas system.
The development of gene editing technology progressed through three stages. The initial stage featured zinc finger nucleases (ZFNs) [13], followed by transcription activator-like effector nucleases (TALENs) in the subsequent stage [14]. The third generation of gene editing technology, which is the most widely used, is represented by the CRISPR/Cas system. Both ZFNs and TALENs rely on FokI nuclease and engage with their target sequences through protein-DNA interactions, whereas the CRISPR/Cas9 system identifies its target sites based on RNA-DNA interactions [15]. ZFN is smaller and easier to edit, but it is difficult to construct functional ZFNs, and the off-target activity is high [16]. While it is not possible to completely eliminate off-target effects, the CRISPR/Cas9 system offers a more streamlined process and benefits from the presence of a protospacer adjacent motif (PAM) sequence, which enhances efficient targeting and reduces off-target activity. Within nucleic acid amplification processes, CRISPR/Cas systems provide advantages compared to recombinases and nucleases (e.g., nicking and restriction endonucleases) to improve nucleic acid identification and molecular testing. By altering the CRISPR RNA (crRNA), the CRISPR/Cas9 and CRISPR/Cas12 systems can cut either both strands or a single strand of double-stranded DNA (dsDNA) with different sequences. However, when recombinase is used, single-strand DNA-binding protein (SSB) must be involved, while incision and restriction enzymes are only applicable to dsDNA sequences with or near particular recognition sites [17]. Argonautes (Agos) are next-generation nucleic acid endonucleases that facilitate simultaneous and multiple detection through one enzyme. This enzyme is not restricted to nucleic acid detection, but extends to the detection of diverse non-nucleic acid targets by the Mesophilic Argonaute Report-based single millimeter Polystyrene Sphere (MARPS) platform. In contrast to the constraints of the CRISPR/Cas system in this context, this platform signifies a significant progression in utilizing gene editing systems for non-nucleic acid target identification [18].
CRISPR/Cas systems are divided into two main classes, each containing various types and subtypes. Effector protein complexes make up Class I systems, where each protein has a specific function in the CRISPR process. Class II CRISPR/Cas systems are distinguished by a single effector protein that performs multiple roles in the CRISPR process [19]. CRISPR/Cas effector proteins are now known to exist in various forms, with differences in structure, size, target and composition. The CRISPR system, specifically the Class II type II (CRISPR/Cas9) system, is currently under intense research and is a versatile tool for gene editing and numerous other applications [20,21].
The CRISPR/Cas system is a crucial technology within the realm of life sciences. It serves a vital function in fundamental research and exhibits significant promise in clinical diagnostics [22–25], food-borne pathogen detection [26–29], agricultural applications [30–33], and other fields [34–36] (Fig. 2). As the technology continues to advance and improve, we can expect CRISPR/Cas systems to play an even greater role in these fields. This paper reviews the advancements in research and utilization of the CRISPR/Cas system for nucleic acid detection, as well as the prospects of CRISPR/Cas systems.
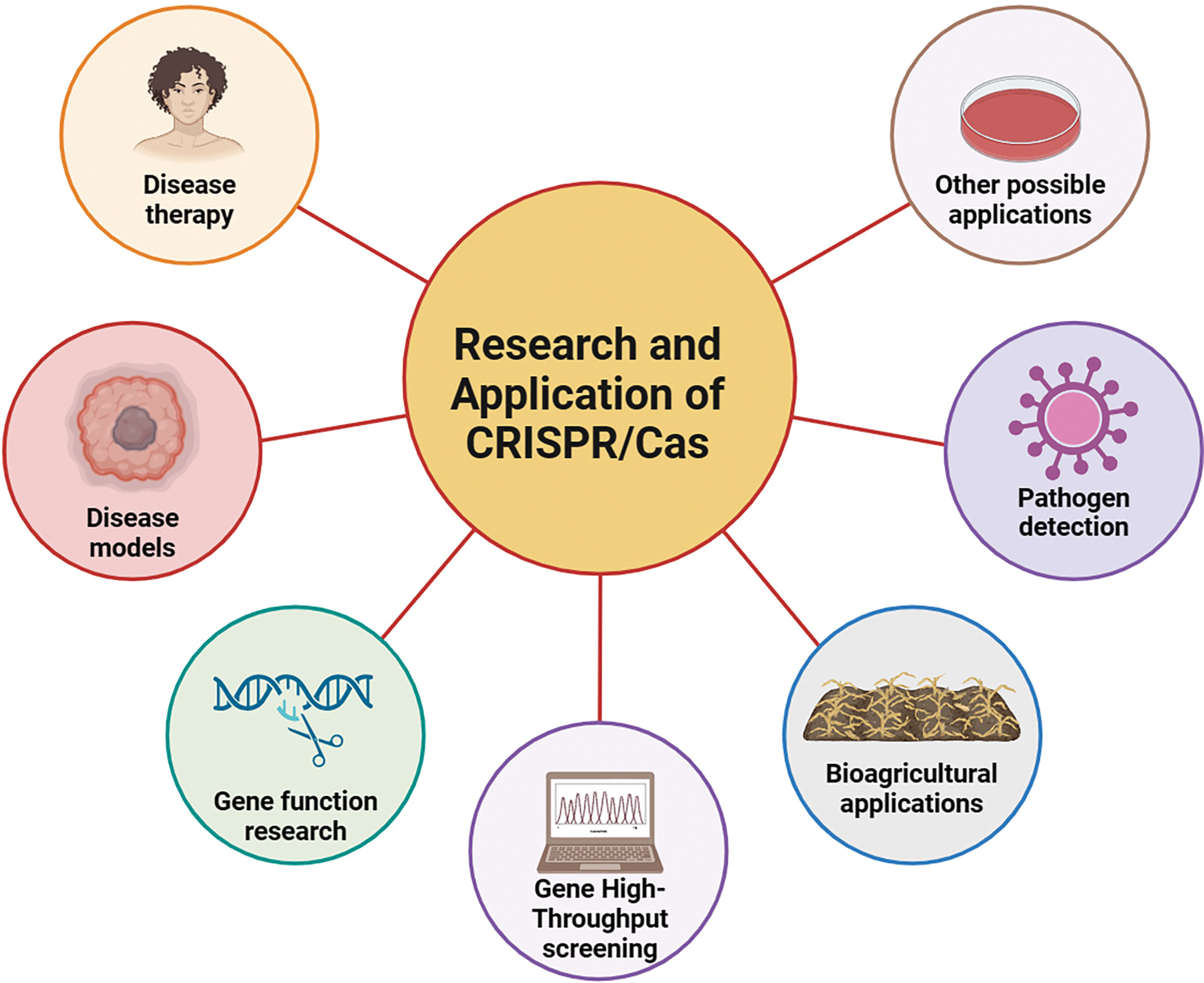
Figure 2: Research and application of CRISPR/Cas.
Classification of CRISPR/Cas Systems
The CRISPR/Cas system is classified into two categories depending on the Cas proteins: a multi-subunit protein Cas complex in Class I and a solitary Cas effector protein in Class II [19]. Class I systems feature numerous Cas enzymes working together to target DNA. Because of the complexity involved in engineering and delivering multiple Cas enz42ymes to cells, Class I systems are not commonly employed for genome editing [37]. In contrast, Class II Cas proteins only need a single multifunctional Cas enzyme and are therefore more adaptable [38,39].
Class II systems include the type II system of Cas9, the Cas12 type V system, and the Cas14 system, as well as the Cas13 type VI system. The Cas9 nuclease contains two functional domains for the cleavage of DNA, known as histidine asparagine histidine (HNH) and RuvC. Specifically, the HNH domain cleaves the target DNA in the region where the single guide RNA (sgRNA) interacts with the specific DNA, while the RuvC domain cleaves the non-target strand [40]. Cas12a employs only the RuvC domain to cleave both DNA strands in cis and non-target DNA in trans [41,42]. This distinct mechanism sets Cas12 apart from Cas9 and provides a unique approach to precise DNA cleavage. The primary type VI system within Class II is Cas13, which lacks RuvC or HNH nuclease domains. Rather, Cas13 is equipped with two Higher eukaryotes and prokaryotes nucleotide-binding domain (HEPN) domains specifically for targeting RNA, rather than DNA, offering a distinct pathway for epigenetic editing [43]. The promiscuous dual HEPN domains of Cas13 have collateral RNase activity and can non-specifically cleave target transcripts and nearby transcripts after single-stranded RNA (ssRNA) and crRNA pairing. The Cas13 system is divided into four subtypes, differentiated by adaptation genes (Cas1/Cas2), additional open reading frame (ORF) regions and associated accessory proteins: Cas13a (formerly referred to as C2c2) [38,44], Cas13b [45], Cas13c [46], and Cas13d [47]. The CRISPR/Cas14 system is differs from Cas9 in the guides required [48]. The Cas14 protein does not rely on a PAM site to target single-stranded DNA (ssDNA). This unique characteristic makes Cas14 an effective tool for detecting ssDNA-based pathogens. The CRISPR/Cas system is widely used in the field of nucleic acid detection, including the detection of SARS-CoV-2. Different Cas proteins in the CRISPR/Cas system have unique mechanisms for detecting nucleic acids. Cas9 recognizes targets via designing specific sgRNAs, while Cas12, Cas13 and Cas14 rely on collateral cleavage activity [49]. Only cas14 currently has no research showing that it can be used to detect SARS-CoV-2. Other Cas proteins have been combined with different technologies, such as isothermal amplification technology (IAT), to improve the sensitivity and specificity of detecting SARS-CoV-2 [50–53]. Combined with IAT is versatile and suitable for many places, providing on-site detection, rapid diagnosis, and preliminary screening. As shown in Table 1, a variety of nucleic acid detection techniques utilizing the CRISPR/Cas system have been developed in many fields. The various types of CRISPR/Cas mechanisms for nucleic acid detection are described below.
Conventional Nucleic Acid Testing Technology
CRISPR usually needs to be combined with conventional detection techniques for nucleic acid detection to ensure precision and sensitivity in pathogen detection. An increasing number of diagnostic platforms have been created using the CRISPR/Cas system in conjunction with detection methods such as the polymerase chain reaction (PCR) and IAT (Table 2).
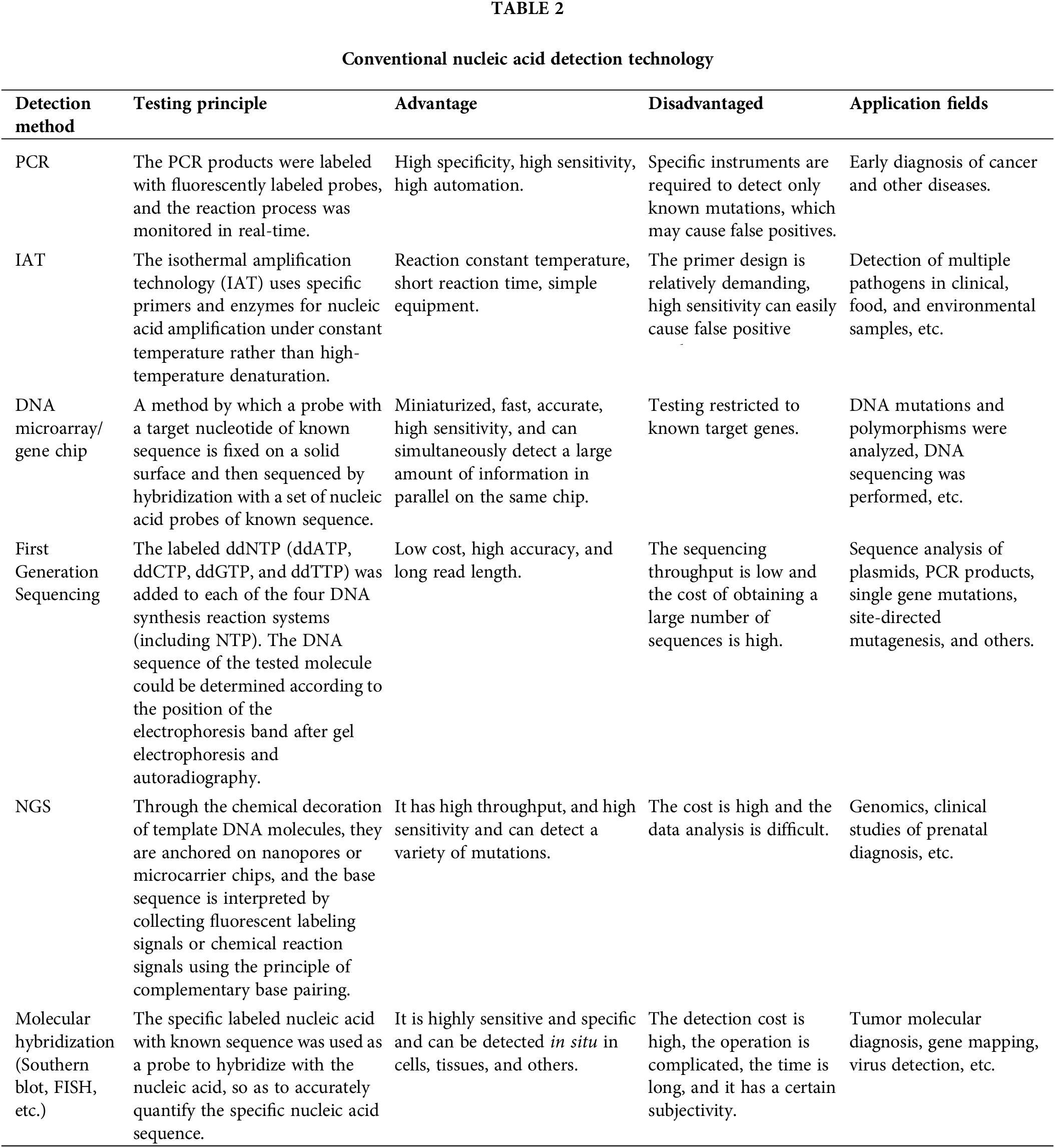
PCR involves the amplification of target DNA fragments by multiple cycles of denaturation at high temperatures, annealing at low temperatures, and primer extension with the assistance of DNA polymerase. It includes conventional real-time PCR (RT-PCR) [82], multiple PCR technology [83,84], digital PCR [85,86], nested PCR [87], real-time fluorescence quantitative PCR [88], and asymmetric PCR technology [89] technologies. These PCR techniques have extensive applications in nucleic acid detection and other areas [90–92]. IAT is a novel isothermal technology for nucleic acid amplification. It encompasses loop-mediated isothermal amplification (LAMP) [93], strand displacement amplification (SDA) [94], recombinase polymerase amplification (RPA) [95], crossing priming amplification (CPA) [96], rolling circle amplification (RCA) [97], helicase-dependent amplification (HDA) [98]. The amplified outputs can be analyzed through real-time fluorescence quantification, electrophoresis, and naked-eye interpretation of a transverse flow dipstick. Isothermal amplification technology allows for the direct amplification of DNA and RNA, demonstrating high efficiency in amplification and offering a broad scope of applicability. Nucleic acid sequencing technology is divided into first-generation sequencing [99], next-generation sequencing (NGS) [100] and third-generation sequencing [101]. The first-generation sequencing method is Sanger sequencing, which uses synthetic termination. Many mainstream sequencing technologies, such as RNA sequencing (RNA-Seq) [102], chromatin immunoprecipitation-sequencing (ChIP-Seq) [103], and chromosome conformation capture coupling with high-throughput sequencing (Hi-C) [104] are based on NGS which is a powerful and versatile tool in the fields of clinical and basic biomedical research, particularly for the diagnosis and surveillance of RNA viruses [105–108]. Compared with the first two generations of sequencing technology, third-generation sequencing technology can sequence single molecules and has an extremely long sequencing length. The third-generation sequencing technology is mainly represented by Oxford Nanopore technology and Pacific Biosciences’ single-molecule, real-time sequencing (SMRT) technology [109,110]. DNA microarray or gene chip technology sequences DNA by hybridization with a set of nucleic acid probes of known sequence [111]. Hybridization technology is used to identify proteins or nucleic acid sequences, such as traditional Southern blotting [112] and Northern blotting [113]. Traditional hybridization technology is limited by its ability to analyze only a single gene at a time. In contrast, microarray technology allows high-throughput analysis and offers increased specificity and sensitivity [114]. This advancement has made microarray technology a crucial tool in both molecular biology and clinical research [115]. Fluorescence in situ hybridization (FISH) is an effective method for detecting and pinpointing specific DNA sequences within cells or tissues. This method involves the use of fluorescently labeled probes that bind target nucleic acids, allowing for accurate identification and visualization [116]. The advantage of FISH lies in the in situ fluorescent signal that can be directly observed and measured under a fluorescence microscope. FISH is the most commonly used detection method for specific chromosomal abnormalities [117]. In the clinic, FISH is most commonly used in tumor diagnosis, gene location analysis, and antenatal examinations [118–120].
Application of CRISPR/Cas Gene Editing Technology in Nucleic Acid Detection
Application of the CRISPR/Cas9 system in nucleic acid detection
The CRISPR/Cas9 system is composed of two main components: the Cas9 protein and the guide RNA (gRNA) complex, which includes gRNA consisting of trans-activating CRISPR RNA (tracrRNA) and crRNA. This system has been successfully implemented in a wide range of model bacteria, such as E. coli, Mycobacterium spp., Lactobacillus spp., and Streptomyces spp. [121]. When pathogen DNA invades bacteria, the Cas9 system first synthesizes a gRNA that incorporates an RNA sequence that matches the target site. When DNA binds to the gRNA complex, it is degraded by the Cas9 protein. Fig. 3 illustrates how Cas9 binds to the target nucleic acid and activates the corresponding signal molecules. Nucleic acid detection using the Cas9 system depends on this characteristic. A unique gRNA is designed according to the target DNA and can thus detect different genes [122].
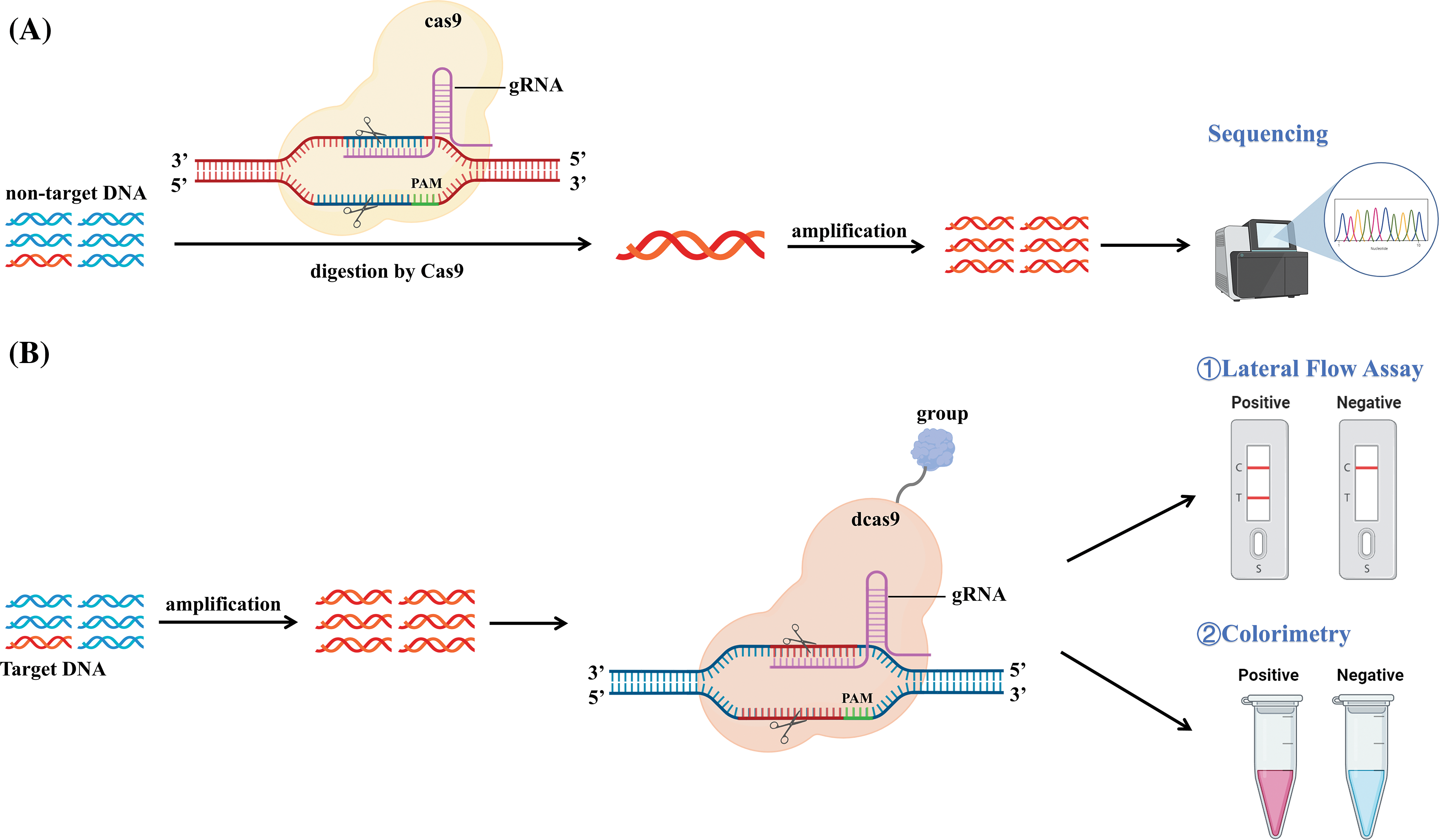
Figure 3: Schematic representation of the CRISPR/Cas9 mechanism. (A) The non-target DNA was digested by Cas9 and cleaved by PAM under the guidance of gRNA. The remaining uncleaved target DNA was specifically amplified by PCR, isothermal amplification, or other methods of amplification, and finally detected by sequencing. (B) The target DNA was amplified using PCR, isothermal amplification, or other methods of amplification. dCas9 with biotin/digoxin and other groups precisely recognizes and binds to target DNA. Capture the complex, and these trapped complexes can be detected by lateral flow detection, visual observation of color changes, and other detection methods.
Many detection platforms for CRISPR/Cas9 have been developed and utilize its cleavage and endonuclease (NEase) activity. Nucleic Acid Sequence-Based Amplification-CRISPR Cleavage (NASBACC) successfully differentiated between Zika virus (ZIKV) strains in America and Africa using the PAM region in the target sequence for Cas9. Through Nucleic Acid Sequence-Based Amplification (NASBA) amplification, the sensor detected the amplicon of the American ZIKV, which includes the PAM region and was cleaved by Cas9, while the African ZIKV, which lacks the PAM region, was not cleaved [54].
Attempts to treat Human Immunodeficiency Virus-1 (HIV-1) with CRISPR/Cas9-based therapies were first made in 2013. CRISPR/Cas9 could inhibit the expression of HIV-1 genes by targeting the HIV-1 long terminal repeats (LTR) [123]. The ability of CRISPR to eliminate integrated viral genes within the infected host chromosome implies that Cas9 could present a promising approach for addressing AIDS [124]. In addition, a colorimetric technique for identifying viruses using the CRISPR/Cas9 system was reported in 2020 [125]. This approach involves directly detecting RNA from the viral lysate using the CRISPR/dCas9 system combined with a biotin PAM presenting oligonucleotide (PAMmer). Under the guidance of biotin-PAMmer, gRNA can recognize RNA in viral lysates, and streptavidin-horseradish peroxidase (HRP) and biotin-PAMmer react to induce color changes. SARS-CoV-2, drug-resistant influenza A virus (pH1N1/H275Y), and influenza A virus (pH1N1) were successfully identified by observation with the naked eye [125].
The CRISPR/Cas9-isothermal exponential amplification reaction (CAS-EXPAR) method is based on isothermal amplification utilizing CRISPR/Cas9 technology. PAM presents oligonucleotides to activate Cas9-specific cleavage activity for product cleavage, and then EXPAR amplifies and cleaves the product. The amplification product is combined with fluorescent dye to produce fluorescence. The CAS-EXPAR detection technology is a highly adaptable method for identifying various types of nucleic acids, including DNA, RNA, and methylated DNA, and it has been successfully used to detect DNA methylation and total RNA from L. monocytogenes [55]. Integration of CRISPR/Cas9 technology with methylation sequencing techniques offers a powerful tool for studying DNA methylation patterns and their functional significance. CRISPR/Cas systems can be engineered to target specific genomic loci and introduce DNA methylation changes. This can be achieved by fusing catalytically inactive Cas proteins (dCas) with DNA methyltransferase domains or recruiting endogenous DNA methyltransferases to targeted loci using gRNAs. By precisely manipulating DNA methylation levels at specific genomic regions, researchers can investigate the functional consequences of methylation changes on gene expression, chromatin structure, and cellular phenotype [126].
In 2019, a nucleic acid detection platform called “finding low abundance sequences by hybridization” (FLASH) was established. This platform combines Cas9 cleavage, PCR amplification, and high-throughput NGS [24]. The gRNA was specifically designed to direct the Cas9 nuclease to a targeted gene sequence for cleavage. The spliced sequences were amplified by PCR and then sequenced by high-throughput sequencing. FLASH-NGS could effectively detect antimicrobial resistance genes in various samples in a highly multiplexed manner (Fig. 4C).
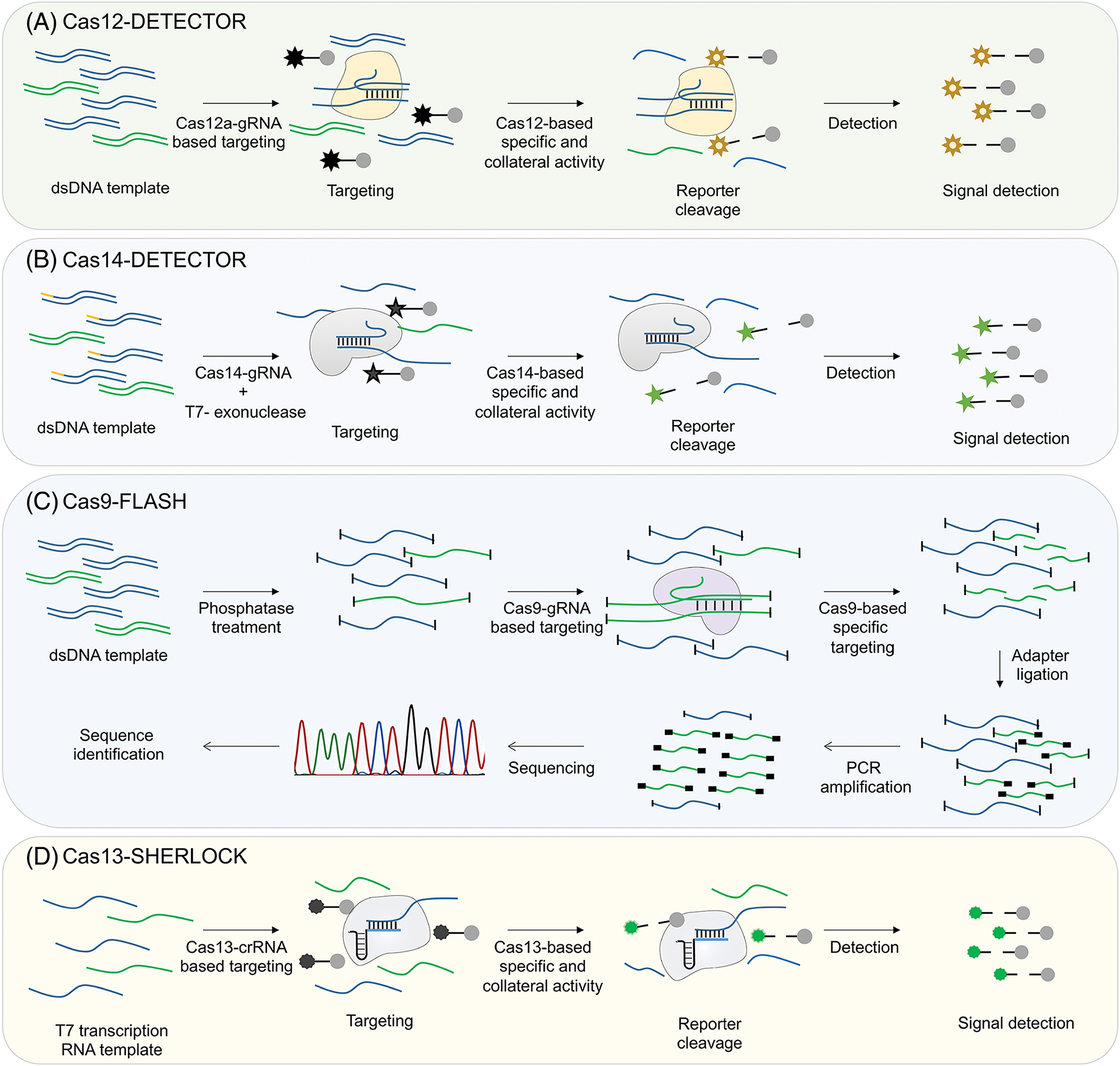
Figure 4: Schematic illustrating the process of CRISPR/Cas detection approaches. (A) Detection of nucleic acids using Cas12. DNA templates amplified by RPA are utilized directly as targets for the Cas12-gRNA complex that is specific to the target. When the target is recognized and cleaved, the collateral activity of Cas12 leads to the degradation of the fluorescent reporter, allowing for the detection of the target’s presence. (B) Detection of nucleic acids using Cas14. Cas14 is able to efficiently recognize and bind single-stranded DNA substrates. Specific single-stranded DNA is amplified through RPA with primers that are adjusted to modify one end of the resulting double-stranded DNA products (yellow end). The DNA strand with the modified end is then protected from degradation by T7 exonuclease, which selectively degrades the unmodified DNA strand. This allows the modified strand to remain intact for interaction with the Cas14-gRNA complex. Once the target is identified and cleaved, the collateral activity of Cas14 will lead to the breakdown of the fluorescent reporter molecule, allowing for the detection of the presence of the target. (C) Detection of nucleic acids using Cas9. A group of double-stranded DNA fragments treated with phosphatase undergoes specific cleavage of the target sequence mediated by Cas9. Unlike the DNA that remains uncut, the ends of the cleaved DNA are subsequently connected to adapters. PCR primers specific to the adapters are utilized for amplifying the cleaved product, which is then sequenced to determine the target sequence. (D) Detection of nucleic acids using Cas13. After the RNA substrate is reverse transcribed, the DNA templates that are amplified by RPA undergo transcription in vitro using T7 RNA polymerase, resulting in the production of RNA templates. These RNA templates then interact with complexes of Cas13 and gRNA. When the target is identified and cleaved, Cas13 collateral activity leads to the breakdown of the fluorescent reporter, allowing for the subsequent detection of the target’s presence. Reproduced with permission from Aman et al., published by American Chemical Society, 2020 [122].
Another important approach of CRISPR/Cas9 for methylation sequencing is targeted methylation sequencing, especially using nanopore technologies. Unlike most other sequencing methods that require amplification of the target gene, which can lead to amplification bias, or require nucleic acid conversion as in bisulfite sequencing approaches, nanopore sequencing technology can directly identify various base methylations bioinformatically. However, these non-amplifying methods require large amounts of DNA to ensure sufficient genome coverage [127–130]. Many studies have showcased the effectiveness of combining CRISPR/Cas9 with long-read sequencing for nanopore analysis. The application of the CRISPR/Cas9 platform has demonstrated promising results in achieving the enhanced sequence coverage necessary for the precise identification of genetic variations. This breakthrough has facilitated the discovery of distinct methylation patterns between afflicted and unaffected individuals, leading to the detection of novel hypomethylated regions within the genome. For instance, Gilpatrick and colleagues developed an innovative approach: nanopore Cas9-targeted sequencing (nCATS) that eliminates the need for amplification by using the CRISPR-Cas9 system to target and cleave a specific region of the genome, and the enriched fragments are subjected to long-read nanopore sequencing [127] (Fig. 5).
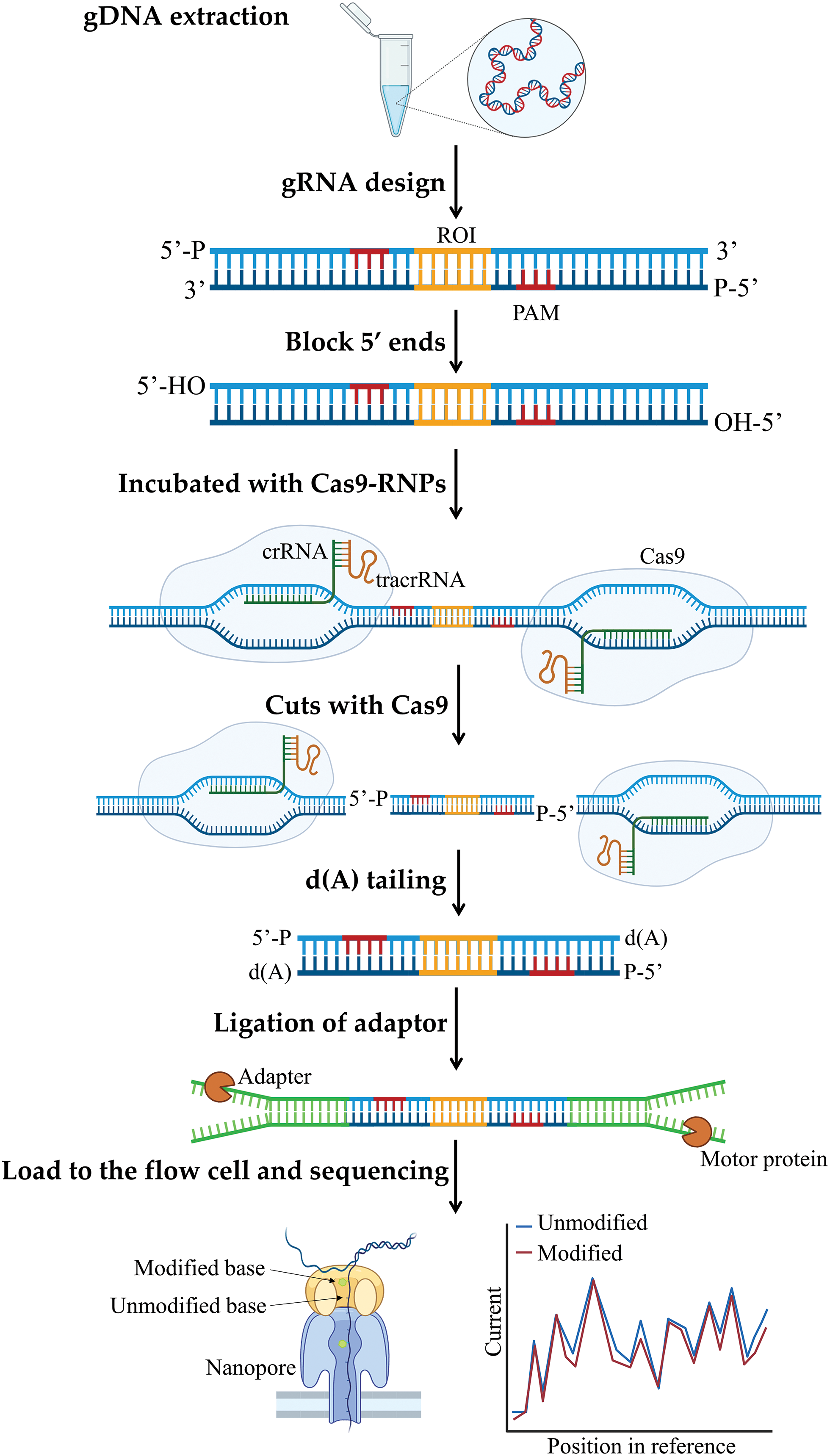
Figure 5: Cas9 enrichment workflow for targeted nanopore sequencing. Cas9 ribonucleoprotein complexes (Cas9 RNPs) were used to cut the region of interest (ROI) from genomic DNA (gDNA) using a designed gRNA. Sequencing adapters were attached to the excised region surrounding the ROI. Nanopore sequencing can detect alterations in the DNA.
A novel approach for amplifying DNA that utilizes the Cas9 nickase-based amplification reaction (Cas9nAR) technique, allows for the simultaneous amplification of Salmonella typhimurium and Escherichia coli, two prevalent foodborne pathogens. This innovative method is capable of dual-target detection without the need for specialized equipment. The entire process can be completed in just three hours showcasing exceptional specificity and sensitivity. The detection limits for genomic DNAs and bacteria are both set at 100 copies and 100 CFU/mL, respectively. The results are visually displayed on a lateral flow strip [26,131]. CRISPR/Cas9 technology has found another use in detecting gene mutations. An assay with high sensitivity is designed to identify deletions in the epidermal growth factor receptor gene (EGFR) [132]. This approach aims to increase the presence of rare mutant alleles by removing wild-type fragments through the CRISPR complex. Subsequently, the mutant fragments are selectively amplified via PCR to achieve the detection sensitivity required for NGS. The results indicate that CRISPR/Cas9 in conjunction with PCR amplification can effectively enrich a variety of EGFR exon19 deletion mutations [132]. This may have implications for early cancer screening [133,134]. It is also possible to induce mutagenesis to mitigate resistance to cancer therapy [135,136]. Moreover, the combination of CRISPR/Cas9 and aptamers holds significant potential for applications in biotechnology and medicine. Aptamer mediated delivery of the CRISPR/Cas9 system is a delivery method that may reduce off-target effects. This method involves the modulation of CRISPR sgRNA through aptamers. For example, an investigation by Kundert et al. analyzed the possibility of regulating sgRNA using aptamers to produce ligand-responsive sgRNAs that activate and deactivate CRISPR/Cas9 function with theophylline [137]. Lin et al. also devised a comparable approach with small molecule-activated allosteric aptamer regulating (SMART) sgRNAs [138]. Aptamers in combination with CRISPR/Cas9 offer a promising solution to the lack of cell specificity for in vivo tumor delivery because of their impressive sensitivity and specificity, and minimal immunogenicity [139–141].
In addition to Cas9, dead Cas9 (dCas9) without nuclease activity is also used for nucleic acid detection. The innovative paired dCas9 (PC) detection platform was established with dCas9 [56]. Following amplification, the targeted DNA binds with dCas9 to form an active recombinant luciferase, which generates a fluorescent signal for detection. The PC detection system can be used to detect Mycobacterium tuberculosis DNA. Another rolling circle amplification-CRISPR-split-HRP (RCH) system based on dCas9 detection uses a pair of dCas9 to fuse the two domains of split-HRP (sHRP) after the target miRNAs are amplified by RCA technology [57]. Finally, the miRNAs are detected by adding the chromogenic substrate tetramethyl benzidine (TMB). The RCH system is mainly used to detect miRNAs. Detection methods accomplished through dCas9-mediated techniques cover a wide range of disciplines, including electrochemistry, fluid dynamics, and microfluidics. The CRISPR-Chip, a handheld device utilizing a graphene-based field-effect transistor (gFET) sensor combined with dCas9, enables quick and specific on-chip electrical tests of unamplified target genes within genomes without the need for amplification [58].
Previous restriction enzymes (REs) only recognized specific DNA sequences, while wild-type CRISPR/Cas9 nucleases require the PAM sequence. Consequently, current in vitro DNA cleavage and molecular cloning techniques remain incapable of precisely cleaving the directly neighboring target DNA base. A CRISPR-SpCas9 mutant, SpRY, which has no PAM sequence requirement, can cut DNA in almost any sequence on the genome [142]. In 2022, Walton et al. performed SpRYDNA digestion (SpRYgests) on multiple PAMs using over 130 gRNAs and found that SpRY can cut DNA of almost any sequence without PAM in vitro, including the hard-to-cut sites of wild-type SpCas9, which promises to simplify, accelerate, and improve the precision of molecular applications [142].
Application of the CRISPR/Cas12 system in nucleic acid detection
Cas12 is an essential RNA-guided endonuclease in gene editing second only to Cas9 in importance [143]. TracrRNA is crucial for the activity of Cas12b, while Cas12a functions independently of tracrRNA. However, more research is needed to fully understand the role of tracrRNA in Cas12c activity [38]. As illustrated in Fig. 6, Cas12 specifically recognizes target DNA under the guidance of gRNA, leading to specific cis-cleavage activity as well as non-specific trans-cleavage activity. This non-specific collateral cleavage activity is used in the development of nucleic acid detection technology.
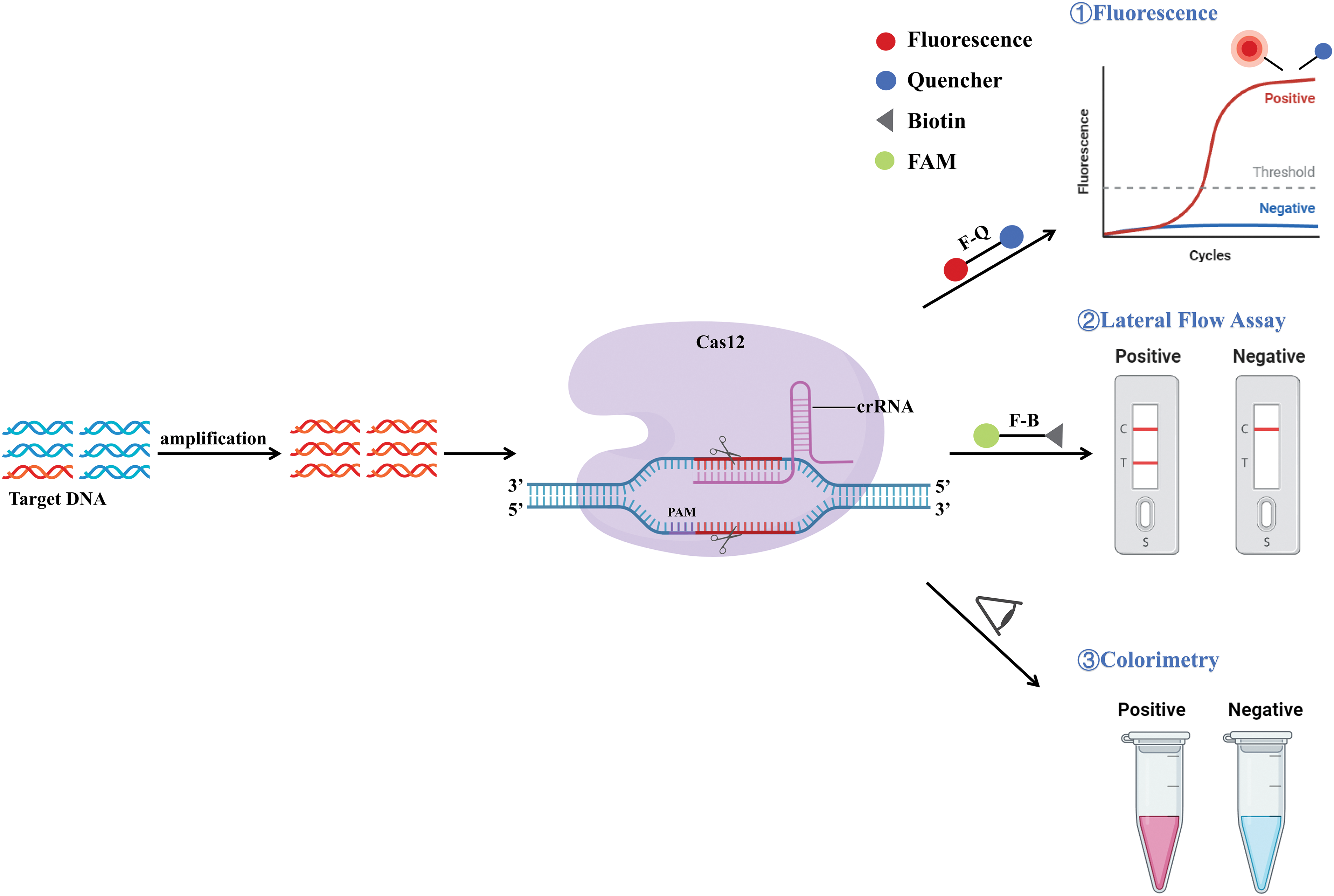
Figure 6: Schematic representation of the mechanism of CRISPR/Cas12. The DNA target was amplified using PCR, isothermal amplification, or other methods of amplification. The guide sequence of crRNA is designed to target a distinct area within the target DNA. With the mediation of crRNA and PAM, Cas12 can specifically recognize the target DNA sequence, and activate specific cis-specificity and non-specific collateral cleavage. This results in the cleavage of reporter groups and these cleaved reporter groups can be detected by detection methods such as fluorescence signals, lateral flow detection, and visual observation of color changes.
The development of DNA Endonuclease-Targeted CRISPR Trans Reporter (DETECTR) technology and one-hour low-cost multipurpose highly efficient system (HOLMES) technology is rooted in the ssDNA trans-cleavage activity of Cas12. HOLMES was achieved through the utilization of the cis and trans-cutting functions of Cas12a, along with self-quenched-fluorescent ssDNA molecules as probes [59]. Chen et al. proposed the new detection method DETECTR by integrating the Cas12a trans-cleavage activity with IAT [60]. The process involves amplifying the target DNA through RPA or PCR, adding Cas12a and gRNA, and then introducing a fluorescence-quenched ssDNA probe. The Cas12a-gRNA complex targets and cleaves the DNA, triggering collateral cleavage activity which in turn cleaves the ssDNA probe, resulting in a fluorescent signal (Fig. 4A). DETECTR, based on CRISPR Cas12, offers a faster and more visually intuitive detection of SARS-CoV-2 using lateral flow chromatography [51]. DETECTR can also identify human papillomavirus (HPV) 16 and HPV18 within various subtypes of HPV [60].
The detection platform OR-DETECTR for identification of SARS-CoV-2 incorporates Reverse Transcription and Recombinase Polymerase Isothermal Amplification (RT-RPA) along with DETECTR technology [61]. In addition, a lateral flow assay was developed by Sun et al. based on OR-DETECTR for the detection of SARS-CoV-2. Operating through a lateral flow strip, the OR-DETECTR test offers a visual method of testing. The assay involves using activated Cas12a to cut the Carboxyfluorescein-biotin (FAM-biotin) reporter, generating a signal at the test line for initial detection. The uncut reporters are then captured at the control line. This design allowed for visual visualization of the trial results. The platform, which can be enclosed in a tube, is accurate, rapid, user-friendly, and does not require bulky equipment, making it suitable for at-home COVID-19 detection. During the COVID-19 outbreak in 2019, there was a critical need for accurate, rapid, and efficient testing methods. In response, scientists developed the all-in-one dual CRISPR/Cas12a (AIOD-CRISPR) method, which combines RPA technology with the Cas12a mechanism in a single-step testing platform [62]. Amplification and cutting are performed simultaneously in one system, which avoids contamination and reduces reaction time. AIOD-CRISPR produces visible fluorescence via the transverse cleavage ability of Cas12a, enabling the detection of SARS-CoV-2.
Another one-pot detection system is the specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) platform, which is used in the SHERLOCK Testing in One Pot Covid, version 2 (STOPCovid.v2) assay. This one-pot assay platform integrates Cas12b with reverse transcription loop-mediated isothermal amplification (RT-LAMP) [63]. The procedure of STOPCovid.v2 is not complicated. First, viral nucleic acid is extracted, DNA extraction solution is added, and then RT-LAMP reagent and Cas12b reagent are added. When the reaction is over, the lateral flow test strip can be used for detection. The STOPCovid.v2 method does not need to extract viral RNA, nor does it need complex laboratory equipment. Operating this equipment is easy and quick, making it ideal for detecting SARS-CoV-2 in the field [72].
In order to make nucleic acid detection faster and more convenient, a suboptimal PAM of the Cas12a-based test (Cas12a-based sPAMC) was proposed in 2022. This method enables one-step CRISPR detection by IAT and Cas12a cleavage simultaneously in a single tube. By utilizing crRNAs that target substrates with suboptimal PAMs instead of standard PAMs, the reaction time is expedited by two to three times. The sPAMC approach can rapidly detect the human cytomegalovirus (HCMV) DNA virus within 10 min or the RNA virus SARS-CoV-2 in only 15 min using unextracted samples. Importantly, the sPAMC technique achieves detection limits comparable to traditional quantitative PCR (qPCR) in both cases [64].
The growing number of SARS-CoV-2 variants exhibiting spike mutations has sparked concerns due to increased transmission rates. Thus, a novel detection system for monitoring SARS-CoV-2 variants, known as Cas12a-based RT-PCR combined with CRISPR on-site rapid detection system (RT-CORDS), was created. Upon binding of Cas12a/crRNA to the mutant target, the enzymatic cleavage activity of Cas12a is triggered, enabling non-specific cleavage of the single-stranded DNA reporter (FAM-ssDNA-BHQ or Biotin-ssDNA-Digoxin). Ultimately, the presence of the mutant target is identified through fluorescence detection or using a paper strip [144]. Cas12a in combination with the LAMP assay can also detect respiratory viral point mutations, including the mutant SARS-CoV-2 spike N501Y [145]. In 2022, a detection system called CRISPR Single Pot Assay for Detecting Emerging A thermostable Cas12b from Brevibacillus leverages one-pot discrimination of SARS-CoV-2 variants of concern (VOCs) (CRISPR-SPADE) was developed, which combines RT-LAMP and CRISPR in one reaction. The critical aspect of this approach lies in using a heat-resistant Cas12b effector endonuclease derived from Brevibacillus sp. BrCas12b demonstrates robust performance in an IAT buffer and thus integrating BrCas12b into an RT-LAMP based all-in-one reaction system is an effective method to differentiate SARS-CoV-2 VOCs in specimens from patients. This method enables continuous monitoring of the reaction in real time [65]. Compared with the detection of only one viral pathogen, the RPA/CRISPR-Cas12a system, when used in conjunction with lateral flow chromatography, enables the simultaneous identification of SARS-CoV-2, influenza A, and influenza B. This approach offers increased efficiency and effectiveness, serving as a robust tool for on-the-spot testing [66].
The use of CRISPR and similar technologies in cancer research has provided insight into previously challenging issues related to cancer genetics, the noncoding genome, and tumor diversity [146]. Ethical approval was granted in China for a 2016 clinical trial involving gene editing to address lung cancer, suggesting a potential breakthrough in developing an effective genome editing approach for combating cancer [147,148]. In 2021, the EasyCatch method for detecting mutations in cancer samples was developed. RPA technology was used to amplify the mutant sequences of cancer samples. The Cas12a-crNA complex first cleaved the target DNA after incorporating a fluorescence-quenched ssDNA probe and then cleaved the ssDNA probe to produce fluorescence [67].
Cas12a binds crRNA and the target DNA to form complexes and, upon Cas12a/crRNA binding to African swine fever virus (ASFV) DNA, this complex becomes activated and degrades the fluorescent ssDNA reporter in the system. When coupled with a point-of-care (POC) system, achieved a detection limit of 2 pM within 1 h without nucleic acid amplification [68]. Another detection technique that does not require amplification is Solid-Phase Extraction and Enhanced Detection Assay using CRISPR-Cas12a (SPEEDi-CRISPR). This method is based on using cas12a-coated magnetic beads to enrich target DNA, which can ultimately be read visually through smartphones or lateral flow analysis. SPEEDi-CRISPR can detect HPV18 and distinguish it from HPV16 and parvovirus B19. Additionally, it meets the requirements for point-of-care testing (Fig. 7) [69].
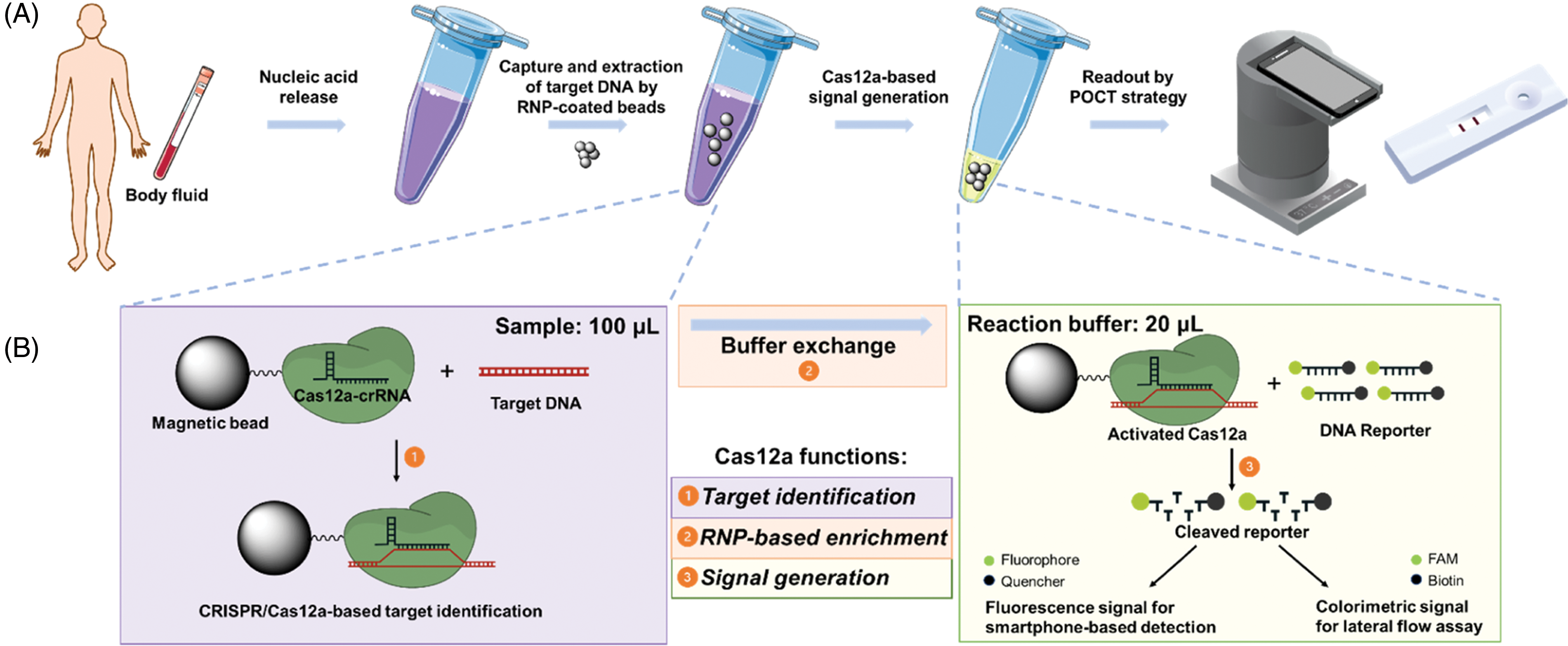
Figure 7: SPEEDi-CRISPR assay. (A) Process of SPEEDi-CRISPR for point-of-care diagnosis. Samples are lysed to release nucleic acids by heat; then, beads covered in Cas12a-crRNA RNP are introduced to a substantial sample volume for the selective capturing and enrichment of target DNA. Following enrichment, the liquid component is removed, and the reaction buffer with reporters is added to induce signal production. The outcome is quantifiable using a laboratory instrument and can also be connected to a portable tool or lateral flow assay (LFA) for a visual readout in point of care testing (POCT). (B) The SPEEDi-CRISPR assay operates on the principle that the N-terminal His-tag of Cas12a interacts with cobalt ions located on magnetic beads. This interaction leads to the formation of a layer of Cas12a-crRNA RNP on the surface of the beads. These customized beads capture the target DNA through the sequence recognition process of Cas12a-crRNA RNP. Post-extraction, the activated Cas12a enzyme will cleave the reporters in the reaction buffer, thus producing a signal. Reproduced with permission from Tian et al., published by American Chemical Society, 2024 [69].
CRISPR/Cas12a’s trans-cleavage function was incorporated into an electrochemical biosensor (E-CRISPR), along with recombinase-aided amplification (RAA). By combining RAA and CRISPR, an electrochemical biosensor platform was developed for the identification of Listeria monocytogenes. This system shows a remarkable sensitivity with a detection threshold of 0.68 aM and exceptional specificity [70].
The CRISPR/Cas12c system requires the PAM sequence to bind the DNA in a target-specific fashion without cleavage. Using this feature, the Cas12c-based nucleic acid detection platform (Cas12c-DETECTOR) was developed. This platform was coupled to an optimized sgRNA for precise identification of single nucleotide polymorphisms (SNPs). By integrating pre-amplification techniques and either visual fluorescence detection methods or lateral flow strips, it can be employed for the detection of human and plant pathogens [71].
Application of the CRISPR/Cas13 system in nucleic acid detection
CRISPR proteins Cas12 and Cas13 have shown promising results in the detection and treatment of ssRNA viruses [65]. In particular, Cas13 can create a complex with numerous crRNAs, enabling targeted cleavage at various points along an RNA transcript with exceptional accuracy [149]. Cas13-crRNA complexes can be used to target specific sites within an ssRNA transcript, resulting in the cleavage of the RNA and subsequent inhibition of post-transcriptional gene expression. Cas13-mediated gene inhibition can counteract immune evasion mechanisms and enhance anti-cancer defenses [150].
Fig. 8 illustrates the activity of Cas13 in cleaving collateral RNA, with the Cas13-gRNA complex binding to the target RNA. This target RNA includes a specific protospacer-flanking site (PFS) located at the 3′ end [151]. By utilizing the collateral cleavage function of the CRISPR/Cas13 system, the nucleic acid detection method SHERLOCK was created. SHERLOCK offers quick and precise diagnostic tests relying on CRISPR/Cas13 for the detection of emerging COVID-19 [152]. SHERLOCK is similar to the DETECTR mechanism, but the target recognized by Cas13a is RNA, so when the target is DNA, the DNA needs to be transcribed into RNA for detection [73]. The initial step involves IAT of the target DNA using RPA technology, followed by transcription of the target DNA into target RNA by T7 transcript in vitro. When the Cas13a-crRNA complex is added, the non-specific trans-cleavage activity of Cas13a is triggered at the same time as its binding to the target RNA. The added fluorescent reporter RNA molecular probe is cleaved to produce a fluorescent signal (Fig. 4D). The SHERLOCK nucleic acid detection technology has proven to be effective in detecting ZIKV and dengue virus (DENV). Since the SHERLOCK platform can only detect a single nucleic acid at a time, the second-generation SHERLOCK (SHERLOCK v2) platform was created that can simultaneously detect four different target RNAs. In addition, a CRISPR-related enzyme (Csm6) was added to improve the sensitivity, and the test strip of the SHERLOCKv2 platform was further developed to detect the results intuitively [72].
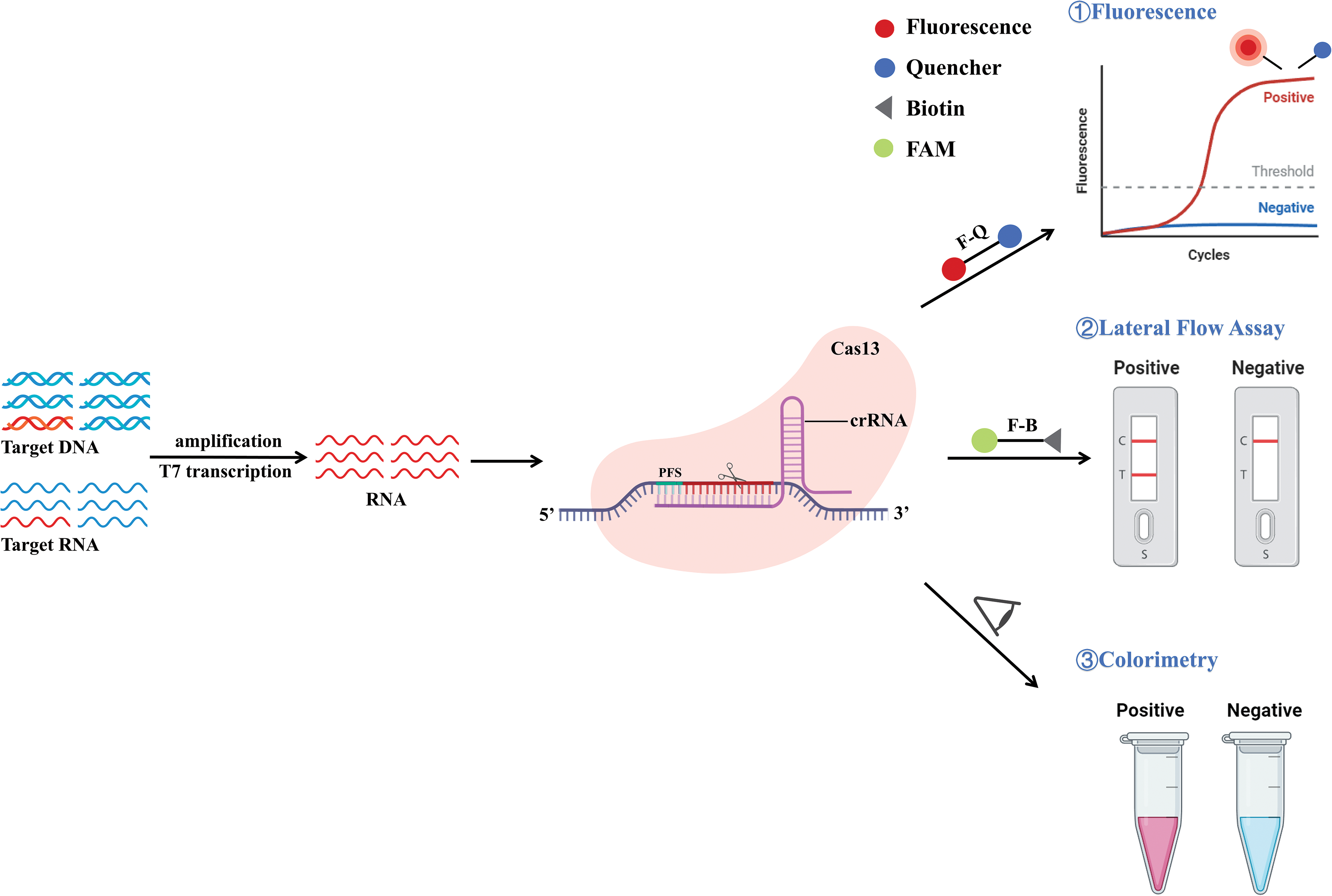
Figure 8: Schematic representation of the mechanism of CRISPR/Cas13. The target DNA/RNA is specifically amplified by PCR, isothermal amplification, or other amplification methods and then converted into RNA by T7 transcription. Cas13 recognizes RNA and binds to it as a complex. The target sequence consists of a protospacer-flanking site (PFS) at the 3′ end. After binding to PFS, crRNA is recognized by Cas13 and complements with PFS to cleave RNA, and activate collateral cleavage, leading to cleavage of RNA reporter groups. These cleaved reporter groups can be detected by detection methods such as fluorescence signals, lateral flow detection, and visual observation of color changes.
To enhance the SHERLOCK platform’s detection capability, a technique called heating unextracted diagnostic samples to obliterate nucleases (HUDSON) was designed. This method allows for the direct release of viral nucleic acids from clinical samples for detection, rather than relying on conventional nucleic acid extraction methods [74]. Combining the SHERLOCK platform with HUDSON technology could directly detect DENV and ZIKV in blood, plasma, serum, saliva, and urine samples. Besides the above applications of Cas13, a SHERLOCK nucleic acid test was also constructed in 2019 to detect human Epstein-Barr virus (EBV) DNA [153]. The principle is that the activation of the Cas13 enzyme upon binding to the targeted viral RNA leads to the cleavage of the RNA and triggers collateral RNase activity [154]. This innovative platform provides a promising solution for efficient and accurate diagnosis of SARS-CoV-2, particularly in settings where immediate testing is required.
The SHERLOCK method is not only capable of detecting viruses but can also be used for malaria diagnosis [155,156]. By combining RPA technology, CRISPR-RNA base pairing, and Cas13a activity, SHERLOCK can detect all known Plasmodium species that cause malaria disease. Furthermore, SHERLOCK can be used to detect and drug-resistance genotyping in mosquitoes [156]. Lee et al. showcased the effectiveness of the SHERLOCK technique in detecting and differentiating different Plasmodium parasites [157]. They employed portable fluorescence detection or lateral flow strip detection methods, which are more intuitive and suitable for immediate detection.
SHERLOCK technology can also be used for the swift identification of soybean crop traits by amplifying and detecting nucleic acid targets from crude soybean extracts. The SHERLOCK technique is easier to operate than other detection methods, processes samples more quickly, and does not require complex instruments. These principles can also be extended to other agricultural detection methods [33].
The Cas13a protein has been enhanced for nucleic acid identification through the collateral lytic function of Leptotrichia wadei Cas13a (LwCas13a). A method called CRISPR/Cas13a-LFD was developed for quickly and accurately detecting ASFV by integrating RAA with a lateral flow strip [75]. CRISPR/Cas13a-LFD displayed precise detection abilities and can be used for on-site clinical ASFV identification with no expensive equipment. An alternative CRISPR/Cas13a trial exists that eliminates the need for amplification, enabling direct detection of SARS-CoV-2 in nasal swab RNA. This technology allows results to be interpreted using a smartphone microscope [76]. This method increases Cas13a activation by binding multiple crRNAs and analyzes fluorescence changes over time, offering a rapid, precise, compact, and economical solution for POC SARS-CoV-2 screening.
A platform developed by combining the collateral cleavage properties of Cas12a and Cas13a with HRP as an enzymatic reporter can generate signals detected by colorimetric, fluorescent, or chemiluminescent methods. This method, which does not require amplification, can detect both nucleic acid and non-nucleic acid targets and is a powerful dual-amplification sensing platform (Fig. 9). This technology has shown promise in quickly detecting SARS-CoV-2 viral nucleic acid markers [77].
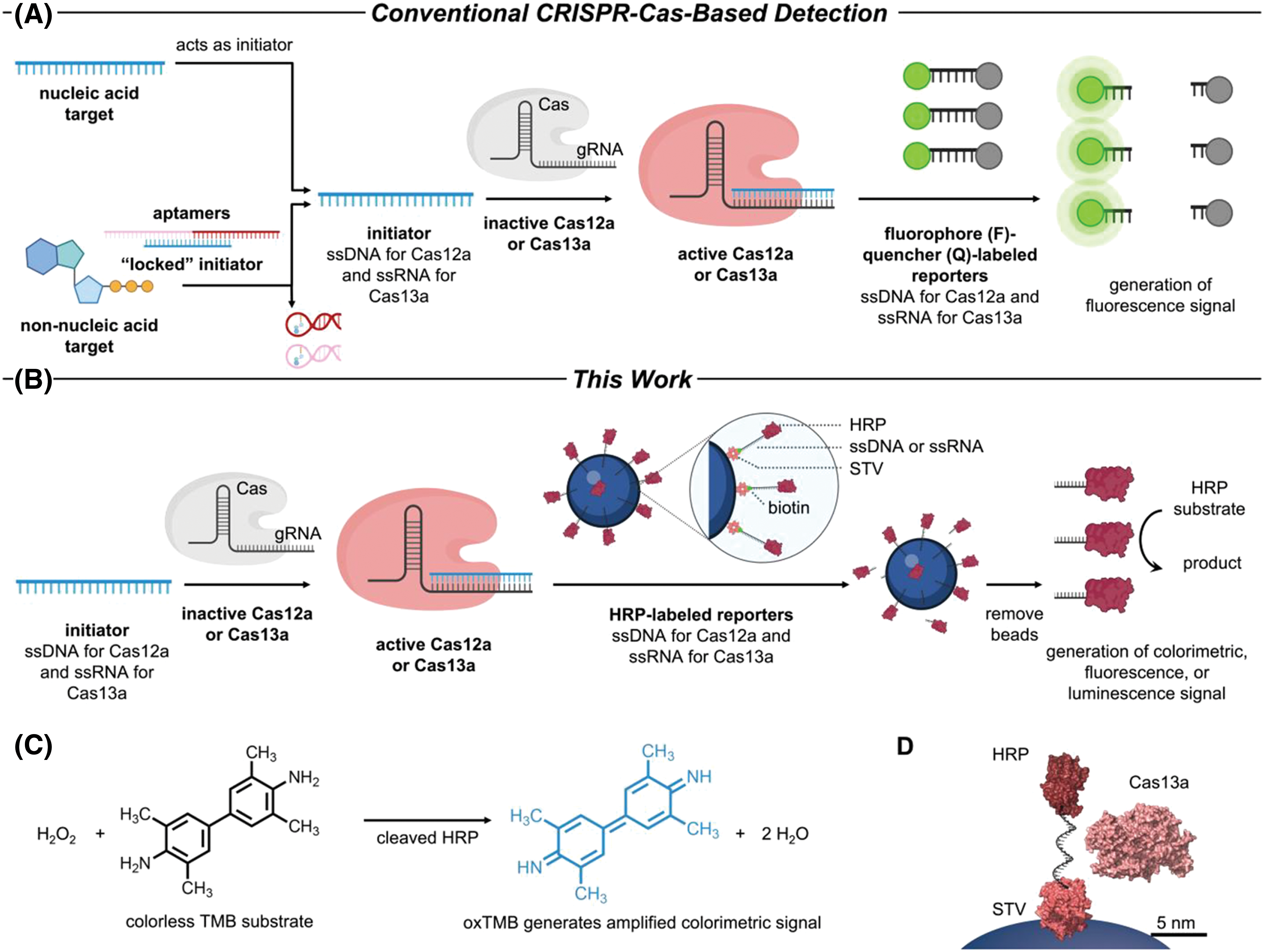
Figure 9: (A) Using conventional CRISPR-Cas12a/Cas13a sensing method that incorporates reporters tagged with fluorophore/quencher. (B) Amplifying CRISPR-Cas12a/Cas13a-based detection with reporters labeled by enzymes. (C) The chemical reaction involving TMB oxidation is mediated by HRP. (D) Illustration (drawn accurately) depicting the proportional dimensions of RNA modified by HRP, streptavidin (STV), and Cas13a bound to its gRNA. Reproduced with permission from Samanta et al., published by American Chemical Society, 2022 [77].
Class II CRISPR/Cas systems demonstrate a wide range of structures and functions, offering flexibility in genome editing and nucleic acid detection [72]. The efficient Cas13d effectors display strong cleavage of targets and collateral RNase activity. The system, derived from Ruminococcus flavefaciens XPD3002, belongs to Class II type VI-D CRISPR/Cas13d [47,158]. The prophylactic antiviral CRISPR in human cells (PAC-MAN) technology has been reported to be useful against SARS-CoV-2, influenza A virus (IAV), and possibly all coronavirus strains [159]. There is a report of a CRISPR live-cell FISH tool based on Cas13d [160]. In this method, they used catalytically deactivated Cas13d and dCas13d defused with crRNA labeled with fluorescence [161].
In 2021, researchers created a technique to analyze SARS-CoV-2 and its altered forms using a method that uses RNA aptamers to signal CRISPR/Cas13 amplification [78]. Specificity of sequences was achieved through the ligation process and recognition by Cas13a/crRNA, enabling the detection of mutations in viral RNA. RNA aptamers that light up are unique RNA molecules capable of binding specifically to dyes and stabilizing their structure to create a complex with a fluorescent signal. The unlabeled RNA aptamer that lights up enables the sensitive generation of amplification signals through the ribonuclease activity of CRISPR/Cas13a triggered by the target [162,163].
The PCR-CRISPR platform is a Cas13a-based platform for Hepatitis B virus DNA (HBV) that can detect one copy of each HBV DNA and drug-resistant mutations [79], making it a valuable tool for early detection of HBV infection, monitoring drug resistance, and providing guidance for treatment.
Cas13a has shown potential in detecting miRNA biomarkers for early cancer diagnosis. A portable ECL chip (PECL)-CRISPR platform for detecting miRNAs was constructed by the introduction of Cas13a electrochemiluminescence (ECL) [80]. The target miRNAs were directly identified by Cas13a, and then the reaction was placed on an electrochemical luminescence to detect miRNAs in tumor cells on an electrochemical luminescence chip [164]. This technology offers a promising approach in the field of clinical diagnostics.
Application of the CRISPR/Cas14 system in nucleic acid detection
The Cas14 protein requires tracrRNA and crRNA to identify and target ssDNA without the need for a PAM site and activates Cas14 protein-specific cis-cleavage activity as well as non-specific collateral cleavage activity (Fig. 10). While Cas14 primarily targets ssDNA, it can also target dsDNA through a T-rich PAM sequence [165,166]. Based on the characteristics of Cas14, Cas14a-DETECTR technology was developed for the identification of human high-fidelity DNA SNP sites and viral DNA (Fig. 4B) [48]. Integrating the CRISPR/Cas14a system with 2D porphyrin metal-organic framework nanosheets (2D-pMOFs) has led to the creation of a novel and adaptable fluorescence sensor known as the Cas14-pMOFs fluorescence sensor, which allows for highly selective and sensitive detection of microcystin-LR (MC-LR) levels [81].
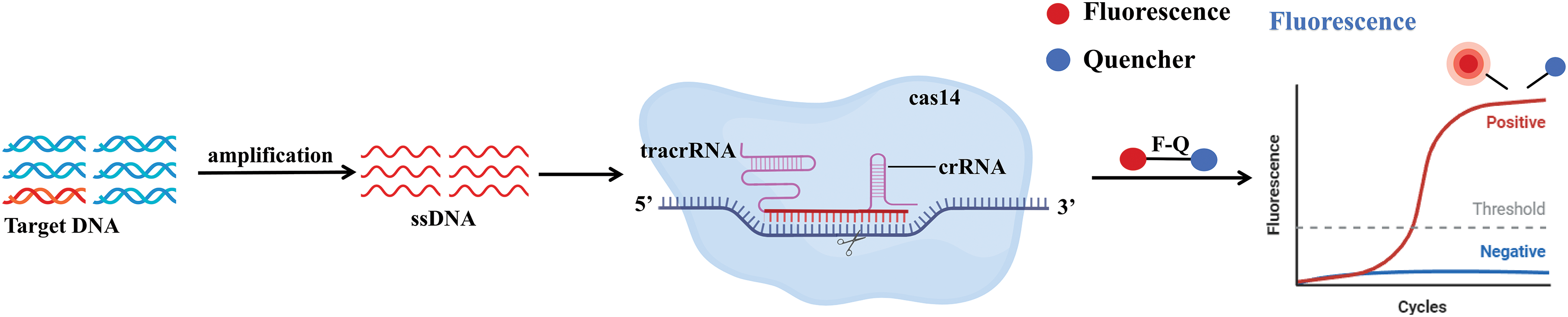
Figure 10: Schematic representation of the mechanism of CRISPR/Cas14. The target DNA is specifically amplified to single-stranded DNA by PCR, isothermal amplification, or other amplification methods. With the help of tracrRNA and crRNA, in the absence of the PAM region present, cleaves the single-stranded DNA, activating Cas14-specific cis-cleavage activity and nonspecific collateral cleavage activity, leading to cleavage of reporter groups. These cleaved reporter groups can be detected by fluorescence signals.
By integrating Cas14a1 with asymmetric PCR, Hu et al. created a technique to identify homozygous deletion of exon 7 of the survival motor neuron 1 gene (SMN1) in patients with spinal muscular atrophy (SMA). In this testing method, specific crRNA-guided Cas14a1 endonuclease amplifies target DNA. Once activated, Cas14a1 breaks down the ssDNA-FQ probe via non-specific trans-cleavage activity, leading to the production of a fluorescence signal [167]. The engineered effector CasMINI, based on the naturally found Cas12f (Cas14) system, was created using CRISPR/Cas technology. Experimental findings revealed that while the original Cas12f and its corresponding sgRNA were inactive in mammalian cells, the modified Cas12f protein and sgRNAs demonstrated effective control over gene expression and editing, offering the potential for genome engineering tasks [42]. Compared to Cas9, Cas14 has the advantage of targeting a wider range, but whether this might cause higher off-target effects is yet to be addressed.
In 2013, the mammalian gene editing community made a groundbreaking discovery with the invention of the CRISPR/Cas9 system. Thereafter, CRISPR technology has transformed the landscape of gene editing because of its remarkable efficiency, simplicity, and affordability. Through extensive research on the CRISPR system, various CRISPR-based DNA and RNA editing tools and gene expression regulation tools have been developed. The CRISPR/Cas system is valuable not just for gene editing, but also for nucleic acid detection. This article outlines the potential of using CRISPR technology to detect a wide range of nucleic acids. The clinical applications of CRISPR/Cas systems targeting nucleic acids are extensive and diverse, including the use of RNA knockdown to treat genetic diseases, and the treatment of diseases associated with mRNA misplacing.
The fusion of Cas technology with PCR amplification and NGS has found widespread use in the field of nucleic acid detection. Examples include FLASH and PC based on Cas9, HOLMES based on Cas12, and PCR-CRISPR based on Cas13. However, these require time and energy, as well as special instruments and laboratories, and operators need professional training. Moreover, such technology failed to fulfill the criteria for rapid on-site detection and operability in disease categorization and pathogenic microorganism screening. Recently, progress in nucleic acid IAT, especially the combination of nucleic acid IAT with Cas gene editing technology and nucleic acid visualization technology such as colloidal gold or fluorescence, has opened up the applications of CRISPR in nucleic acid detection. For example, DETECTR based on Cas12 uses RPA to amplify target genes and fluorescence visualization to display detection results. The integration of IAT which is characterized by rapid processing times and constant temperature, with the high specificity of the CRISPR/Cas system addresses several limitations inherent in traditional molecular diagnostic techniques. This combination significantly enhances the specificity of detection results and makes it more suitable for on-site detection, thereby meeting the requirements for POC testing. Additionally, it offers advantages such as lower costs, ease of operation, and simplified instrumentation. However, it is important to note that this combination may lead to reduced sensitivity and carries a risk of off-target effects. Further research is necessary to strike a balance between the sensitivity of IAT and the specificity needed for nucleic acid testing, while also preventing sample contamination during field operations. Many studies have made good attempts, such as improving the reaction process to realize the one-tube operation of gene editing and IAT visualization as well as optimizing Cas enzymes and crRNA.
Future research should focus on developing methods that require fewer operational steps and utilize smaller instruments by integrating IAT with one-pot detection methods. The one-pot method offers several advantages, including enhanced efficiency and streamlined workflows. For example, combining the strengths of CRISPR technology with IAT, as demonstrated by systems like AIOD-CRISPR and STOPCovid.v2, which employ a one-pot approach. In these systems, amplification and cleavage occur simultaneously within a single platform, followed by detection through fluorescence or lateral flow readouts. However, it is essential to address the nucleic acid extraction step in the one-pot method. Simplifying this step is crucial, as direct detection of samples can significantly improve the sensitivity and specificity of the assays while also reducing costs and pollution. The advancement of technology has led to the development of various nucleic acid detection methods that use the CRISPR/Cas system. These methods offer numerous advantages and hold significant value in applications such as POC detection of pathogenic microorganisms, early cancer screening, agriculture, parasitic infections, antibiotic resistance studies, and genotyping. However, the existing detection methods are limited by PAM sites, off-target effects, and time consumption. Currently, various strategies are being explored to reduce the off-target effects associated with the CRISPR/Cas system. It is crucial to optimize gRNA, use aptamers to mediate the delivery of the CRISPR/Cas system and develop new CRISPR/Cas mutants to reduce the off-target effects of CRISPR/Cas. Additionally, CRISPR/Cas mutants exhibit advantages in reduced off-target activity as well as cleavage efficiency, significantly improving nucleic acid detection methods based on CRISPR/Cas technology. Many Cas proteins depend on distinct PAMs for target recognition, which may pose challenges to their broader implementation. For example, the newly developed variant of SpCas9, SpRY, can cut DNA of almost any sequence without PAM in vitro. If gRNA can be synthesized quickly and easily, it will be easier to implement SpRYgest, which will be useful in the field of molecular cloning. Optimizing or employing efficient Cas proteins and enhancing the sequence stability of CRISPR/Cas can lead to improved targeting efficiency. False positives pose a significant challenge, particularly due to off-target effects associated with the CRISPR/Cas system. Combining IAT technology can achieve highly sensitive detection, but is affected by aerosol contamination, which can also lead to false positives. While RT-qPCR is recognized as the most dependable technique, the inconsistent quality of commercial kits can result in diminished sensitivity and specificity, ultimately causing false negative findings. Looking ahead, the development of a one-tube method, combined with lateral chromatography test strips or portable fluorescence detection, will reduce the occurrence of false positives. In addition to this, pre-amplification may improve the sensitivity but may also prolong the detection time because of the need for additional procedures. Future efforts should focus on refining these systems into more straightforward and versatile detection approaches, as well as advancing CRISPR/Cas systems capable of simultaneous multiple detections for large sample sets. Furthermore, new detection systems, such as Agos, can perform multiple assays can be performed simultaneously with one enzyme.
Despite the limitations of the novel nucleic acid detection systems, with the development of CRISPR technology and research into Cas proteins, we believe the problems faced by CRISPR/Cas technology will be overcome. It is expected that CRISPR/Cas technology will have a wide range of applications in nucleic acid detection in the future.
Acknowledgement: This work was supported by the image owner, who granted rights for reprints and quotations. We also extend our gratitude to Biorender and Medpeer for their assistance.
Funding Statement: This work was supported by the Natural Science Foundation of Hunan Province (Grant Nos. 2021JJ30050 and 2023JJ50368); Science and Technology Program of Hunan Province (Grant No. 2021SK50313); the Research Project of Hunan Provincial Health Commission (Grant Nos. 202203102912 and 202203103105, W20243264); the Science and Technology Program of Chenzhou (Grant No. ZDYF2020011); the Key Project of the First People’s Hospital of Chenzhou (Grant No. CZYY202203); the Innovative Team Project of the First People’s Hospital of Chenzhou (Grant No. CX202103).
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Zheng Hu; draft manuscript preparation: Liujie Chen, Lili Duan, and Zheng Hu; review and editing: Duanfang Liao, Nongyue He, Kai Li, and Zheng Hu; visualization: Liujie Chen, Lili Duan, Jia Li, and Jun Chen; supervision: Zheng Hu. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data sharing not applicable to this article as no datasets were generated or analyzed during the current study.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Sorek R, Kunin V, Hugenholtz P. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6(3):181–6. doi:10.1038/nrmicro1793. [Google Scholar] [PubMed] [CrossRef]
2. Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–33. doi:10.1128/jb.169.12.5429-5433.1987. [Google Scholar] [PubMed] [CrossRef]
3. Mojica FJ, Ferrer C, Juez G, Rodríguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17(1):85–93. doi:10.1111/mmi.1995.17.issue-1. [Google Scholar] [CrossRef]
4. Masepohl B, Görlitz K, Böhme H. Long tandemly repeated repetitive (LTRR) sequences in the filamentous cyanobacterium Anabaena sp. PCC 7120. Biochim Biophys Acta. 1996;1307(1):26–30. doi:10.1016/0167-4781(96)00040-1. [Google Scholar] [PubMed] [CrossRef]
5. Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36(1):244–6. doi:10.1046/j.1365-2958.2000.01838.x. [Google Scholar] [PubMed] [CrossRef]
6. Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–75. doi:10.1046/j.1365-2958.2002.02839.x. [Google Scholar] [PubMed] [CrossRef]
7. Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315(5819):1709–12. doi:10.1126/science.1138140. [Google Scholar] [PubMed] [CrossRef]
8. Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–8. doi:10.1038/nature10886. [Google Scholar] [PubMed] [CrossRef]
9. Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–70. doi:10.1126/science.1179555. [Google Scholar] [PubMed] [CrossRef]
10. Altae-Tran H, Kannan S, Demircioglu FE, Oshiro R, Nety SP, Mckay LJ, et al. The widespread IS200/IS605 transposon family encodes diverse programmable RNA-guided endonucleases. Science. 2021;374(6563):57–65. doi:10.1126/science.abj6856. [Google Scholar] [PubMed] [CrossRef]
11. Saito M, Xu P, Faure G, Maguire S, Kannan S, Altae-Tran H, et al. Fanzor is a eukaryotic programmable RNA-guided endonuclease. Nature. 2023;620(7974):660–8. doi:10.1038/s41586-023-06356-2. [Google Scholar] [PubMed] [CrossRef]
12. Altae-Tran H, Kannan S, Suberski AJ, Mears KS, Demircioglu FE, Moeller L, et al. Uncovering the functional diversity of rare CRISPR-Cas systems with deep terascale clustering. Science. 2023;382(6673):eadi1910. doi:10.1126/science.adi1910. [Google Scholar] [PubMed] [CrossRef]
13. Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11(9):636–46. doi:10.1038/nrg2842. [Google Scholar] [PubMed] [CrossRef]
14. Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8. doi:10.1038/nbt.1755. [Google Scholar] [PubMed] [CrossRef]
15. Nemudryi AA, Valetdinova KR, Medvedev SP, Zakian SM. TALEN and CRISPR/Cas genome editing systems: tools of discovery. Acta Naturae. 2014;6(3):19–40. doi:10.32607/20758251-2014-6-3-19-40. [Google Scholar] [CrossRef]
16. Katayama S, Watanabe M, Kato Y, Nomura W, Yamamoto T. Engineering of zinc finger nucleases through structural modeling improves genome editing efficiency in cells. Adv Sci. 2024;11(23):e2310255. doi:10.1002/advs.v11.23. [Google Scholar] [CrossRef]
17. Zhang K, Deng R, Teng X, Li Y, Sun Y, Ren X, et al. Direct visualization of single-nucleotide variation in mtDNA using a CRISPR/Cas9-mediated proximity ligation assay. J Am Chem Soc. 2018;140(36):11293–301. doi:10.1021/jacs.8b05309. [Google Scholar] [PubMed] [CrossRef]
18. Lu Y, Wen J, Wang C, Wang M, Jiang F, Miao L, et al. Mesophilic argonaute-based single polystyrene sphere aptamer fluorescence platform for the multiplexed and ultrasensitive detection of non-nucleic acid targets. Small. 2023;20(20):e2308424. [Google Scholar] [PubMed]
19. Makarova KS, Wolf YI, Iranzo J, Shmakov SA, Alkhnbashi OS, Brouns SJJ, et al. Evolutionary classification of CRISPR-Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(2):67–83. doi:10.1038/s41579-019-0299-x. [Google Scholar] [PubMed] [CrossRef]
20. Wang M, Zhang R, Li J. CRISPR/cas systems redefine nucleic acid detection: principles and methods. Biosens Bioelectron. 2020;165:112430. doi:10.1016/j.bios.2020.112430. [Google Scholar] [PubMed] [CrossRef]
21. Tiwari PK, Ko TH, Dubey R, Chouhan M, Tsai LW, Singh HN, et al. CRISPR/Cas9 as a therapeutic tool for triple negative breast cancer: from bench to clinics. Front Mol Biosci. 2023;10:1214489. doi:10.3389/fmolb.2023.1214489. [Google Scholar] [PubMed] [CrossRef]
22. Ramachandran A, Huyke DA, Sharma E, Sahoo MK, Huang C, Banaei N, et al. Electric field-driven microfluidics for rapid CRISPR-based diagnostics and its application to detection of SARS-CoV-2. Proc Natl Acad Sci USA. 2020;117(47):29518–25. doi:10.1073/pnas.2010254117. [Google Scholar] [PubMed] [CrossRef]
23. Trung NT, Son LHP, Hien TX, Quyen DT, Bang MH, Song LH. CRISPR-Cas12a combination to alleviate the false-positive in loop-mediated isothermal amplification-based diagnosis of Neisseria meningitidis. BMC Infect Dis. 2022;22(1):429. doi:10.1186/s12879-022-07363-w. [Google Scholar] [PubMed] [CrossRef]
24. Quan J, Langelier C, Kuchta A, Batson J, Teyssier N, Lyden A, et al. FLASH: a next-generation CRISPR diagnostic for multiplexed detection of antimicrobial resistance sequences. Nucleic Acids Res. 2019;47(14):e83. doi:10.1093/nar/gkz418. [Google Scholar] [PubMed] [CrossRef]
25. Li T, Li S, Kang Y, Zhou J, Yi M. Harnessing the evolving CRISPR/Cas9 for precision oncology. J Transl Med. 2024;22(1):749. doi:10.1186/s12967-024-05570-4. [Google Scholar] [PubMed] [CrossRef]
26. Wang L, Shen X, Wang T, Chen P, Qi N, Yin BC, et al. A lateral flow strip combined with Cas9 nickase-triggered amplification reaction for dual food-borne pathogen detection. Biosens Bioelectron. 2020;165(6313):112364. doi:10.1016/j.bios.2020.112364. [Google Scholar] [PubMed] [CrossRef]
27. Liu H, Wang J, Zeng H, Liu X, Jiang W, Wang Y, et al. RPA-Cas12a-FS: a frontline nucleic acid rapid detection system for food safety based on CRISPR-Cas12a combined with recombinase polymerase amplification. Food Chem. 2021;334:127608. doi:10.1016/j.foodchem.2020.127608. [Google Scholar] [PubMed] [CrossRef]
28. Chen R, Zhao J, Han M, Dong Y, Jiang F, Chen Y. DNA extraction- and amplification-free nucleic acid biosensor for the detection of foodborne pathogens based on CRISPR/Cas12a and argonaute protein-mediated cascade signal amplification. J Agric Food Chem. 2023;71(46):18037–45. doi:10.1021/acs.jafc.3c06530. [Google Scholar] [PubMed] [CrossRef]
29. Wu J, Mukama O, Wu W, Li Z, Habimana JD, Zhang Y, et al. A CRISPR/Cas12a based universal lateral flow biosensor for the sensitive and specific detection of african swine-fever viruses in whole blood. Biosensors. 2020;10(12):203. doi:10.3390/bios10120203. [Google Scholar] [PubMed] [CrossRef]
30. Yao K, Peng D, Jiang C, Zhao W, Li G, Huang W, et al. Rapid and visual detection of Heterodera schachtii using recombinase polymerase amplification combined with Cas12a-mediated technology. Int J Mol Sci. 2021;22(22):12577. doi:10.3390/ijms222212577. [Google Scholar] [PubMed] [CrossRef]
31. Hu JJ, Liu D, Cai MZ, Zhou Y, Yin WX, Luo CX. One-pot assay for rapid detection of benzimidazole resistance in Venturia carpophila by combining RPA and CRISPR/Cas12a. J Agric Food Chem. 2023;71(3):1381–90. doi:10.1021/acs.jafc.2c06549. [Google Scholar] [PubMed] [CrossRef]
32. Guo H, Zhang Y, Kong F, Wang C, Chen S, Wang J, et al. A Cas12a-based platform combined with gold nanoparticles for sensitive and visual detection of Alternaria solani. Ecotoxicol Environ Saf. 2023;263:115220. doi:10.1016/j.ecoenv.2023.115220. [Google Scholar] [PubMed] [CrossRef]
33. Abudayyeh OO, Gootenberg JS, Kellner MJ, Zhang F. Nucleic acid detection of plant genes using CRISPR-Cas13. CRISPR J. 2019;2(3):165–71. doi:10.1089/crispr.2019.0011. [Google Scholar] [PubMed] [CrossRef]
34. Yin D, Yin L, Wang J, Shen X, Pan X, Hou H, et al. Visual detection of duck tembusu virus with CRISPR/Cas13: a sensitive and specific point-of-care detection. Front Cell Infect Microbiol. 2022;12:848365. doi:10.3389/fcimb.2022.848365. [Google Scholar] [PubMed] [CrossRef]
35. Chen Z, Yang X, Xia H, Wu C, Yang J, Dai T. A frontline, rapid, nucleic acid-based Fusarium circinatum detection system using CRISPR/Cas12a combined with recombinase polymerase amplification. Plant Dis. 2023;107(6):1902–10. doi:10.1094/PDIS-05-22-1234-RE. [Google Scholar] [PubMed] [CrossRef]
36. Pérez AA, Tobin A, Stechly JV, Ferrante JA, Hunter ME. A minimally invasive, field-applicable CRISPR/Cas biosensor to aid in the detection of Pseudogymnoascus destructans, the causative fungal agent of white-nose syndrome in bats. Mol Ecol Resour. 2024;24(2):e13902. doi:10.1111/men.v24.2. [Google Scholar] [CrossRef]
37. Koonin EV, Makarova KS. Origins and evolution of CRISPR-Cas systems. Philos Trans Royal Soc B: Biol Sci. 2019;374(1772):20180087. doi:10.1098/rstb.2018.0087. [Google Scholar] [PubMed] [CrossRef]
38. Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60(3):385–97. doi:10.1016/j.molcel.2015.10.008. [Google Scholar] [PubMed] [CrossRef]
39. Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, Van Der Oost J. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science. 2016;353(6299):aad5147. doi:10.1126/science.aad5147. [Google Scholar] [PubMed] [CrossRef]
40. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. doi:10.1126/science.1225829. [Google Scholar] [PubMed] [CrossRef]
41. Yan WX, Hunnewell P, Alfonse LE, Carte JM, Keston-Smith E, Sothiselvam S, et al. Functionally diverse type V CRISPR-Cas systems. Science. 2019;363(6422):88–91. doi:10.1126/science.aav7271. [Google Scholar] [PubMed] [CrossRef]
42. Xu X, Chemparathy A, Zeng L, Kempton HR, Shang S, Nakamura M, et al. Engineered miniature CRISPR-Cas system for mammalian genome regulation and editing. Mol Cell. 2021;81(20):4333–45.e4334. doi:10.1016/j.molcel.2021.08.008. [Google Scholar] [PubMed] [CrossRef]
43. Li C, Brant E, Budak H, Zhang B. CRISPR/Cas: a Nobel Prize award-winning precise genome editing technology for gene therapy and crop improvement. J Zhejiang Univ-Sci B. 2021;22(4):253–84 (In Chinese). doi:10.1631/jzus.b2100009. [Google Scholar] [PubMed] [CrossRef]
44. East-Seletsky A, O’connell MR, Knight SC, Burstein D, Cate JH, Tjian R, et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature. 2016;538(7624):270–3. doi:10.1038/nature19802. [Google Scholar] [PubMed] [CrossRef]
45. Smargon AA, Cox DBT, Pyzocha NK, Zheng K, Slaymaker IM, Gootenberg JS, et al. Cas13b is a type VI-B CRISPR-Associated RNA-Guided RNase differentially regulated by accessory proteins Csx27 and Csx28. Mol Cell. 2017;65(4):618–30.e617. doi:10.1016/j.molcel.2016.12.023. [Google Scholar] [PubMed] [CrossRef]
46. Shmakov S, Smargon A, Scott D, Cox D, Pyzocha N, Yan W, et al. Diversity and evolution of class 2 CRISPR-Cas systems. Nat Rev Microbiol. 2017;15(3):169–82. doi:10.1038/nrmicro.2016.184. [Google Scholar] [PubMed] [CrossRef]
47. Yan WX, Chong S, Zhang H, Makarova KS, Koonin EV, Cheng DR, et al. Cas13d Is a compact RNA-targeting type VI CRISPR effector positively modulated by a WYL-domain-containing accessory protein. Mol Cell. 2018;70(2):327–39.e325. doi:10.1016/j.molcel.2018.02.028. [Google Scholar] [PubMed] [CrossRef]
48. Harrington LB, Burstein D, Chen JS, Paez-Espino D, Ma E, Witte IP, et al. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. Science. 2018;362(6416):839–42. doi:10.1126/science.aav4294. [Google Scholar] [PubMed] [CrossRef]
49. Li Y, Li S, Wang J, Liu G. CRISPR/Cas systems towards next-generation biosensing. Trends Biotechnol. 2019;37(7):730–43. doi:10.1016/j.tibtech.2018.12.005. [Google Scholar] [PubMed] [CrossRef]
50. Aouida M, Saifaldeen M, Al-Ansari DE, Taleb S, Hssain AA, Ramotar D. A CRISPR-based approach using dead Cas9-sgRNA to detect SARS-CoV-2. Front Mol Biosci. 2023;10:1201347. doi:10.3389/fmolb.2023.1201347. [Google Scholar] [PubMed] [CrossRef]
51. Broughton JP, Deng X, Yu G, Fasching CL, Servellita V, Singh J, et al. CRISPR-Cas12-based detection of SARS-CoV-2. Nat Biotechnol. 2020;38(7):870–4. doi:10.1038/s41587-020-0513-4. [Google Scholar] [PubMed] [CrossRef]
52. Patchsung M, Jantarug K, Pattama A, Aphicho K, Suraritdechachai S, Meesawat P, et al. Clinical validation of a Cas13-based assay for the detection of SARS-CoV-2 RNA. Nat Biomed Eng. 2020;4(12):1140–9. doi:10.1038/s41551-020-00603-x. [Google Scholar] [PubMed] [CrossRef]
53. Chandrasekaran SS, Agrawal S, Fanton A, Jangid AR, Charrez B, Escajeda AM, et al. Rapid detection of SARS-CoV-2 RNA in saliva via Cas13. Nat Biomed Eng. 2022;6(8):944–56. doi:10.1038/s41551-022-00917-y. [Google Scholar] [PubMed] [CrossRef]
54. Pardee K, Green AA, Takahashi MK, Braff D, Lambert G, Lee JW, et al. Rapid, low-cost detection of zika virus using programmable biomolecular components. Cell. 2016;165(5):1255–66. doi:10.1016/j.cell.2016.04.059. [Google Scholar] [PubMed] [CrossRef]
55. Huang M, Zhou X, Wang H, Xing D. Clustered regularly interspaced short palindromic Repeats/Cas9 triggered isothermal amplification for site-specific nucleic acid detection. Anal Chem. 2018;90(3):2193–200. doi:10.1021/acs.analchem.7b04542. [Google Scholar] [PubMed] [CrossRef]
56. Zhang Y, Qian L, Wei W, Wang Y, Wang B, Lin P, et al. Paired design of dCas9 as a systematic platform for the detection of featured nucleic acid sequences in pathogenic strains. ACS Synth Biol. 2017;6(2):211–6. doi:10.1021/acssynbio.6b00215. [Google Scholar] [PubMed] [CrossRef]
57. Qiu XY, Zhu LY, Zhu CS, Ma JX, Hou T, Wu XM, et al. Highly effective and low-cost microRNA detection with CRISPR-Cas9. ACS Synth Biol. 2018;7(3):807–13. doi:10.1021/acssynbio.7b00446. [Google Scholar] [PubMed] [CrossRef]
58. Hajian R, Balderston S, Tran T, Deboer T, Etienne J, Sandhu M, et al. Detection of unamplified target genes via CRISPR-Cas9 immobilized on a graphene field-effect transistor. Nat Biomed Eng. 2019;3(6):427–37. doi:10.1038/s41551-019-0371-x. [Google Scholar] [PubMed] [CrossRef]
59. Li SY, Cheng QX, Wang JM, Li XY, Zhang ZL, Gao S, et al. CRISPR-Cas12a-assisted nucleic acid detection. Cell Discov. 2018;4(1):4–20. doi:10.1038/s41421-018-0028-z. [Google Scholar] [PubMed] [CrossRef]
60. Chen JS, Ma E, Harrington LB, Da Costa M, Tian X, Palefsky JM, et al. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity. Science. 2018;360(6387):436–9. doi:10.1126/science.aar6245. [Google Scholar] [PubMed] [CrossRef]
61. Sun Y, Yu L, Liu C, Ye S, Chen W, Li D, et al. One-tube SARS-CoV-2 detection platform based on RT-RPA and CRISPR/Cas12a. J Transl Med. 2021;19(1):74. doi:10.1186/s12967-021-02741-5. [Google Scholar] [PubMed] [CrossRef]
62. Ding X, Yin K, Li Z, Lalla RV, Ballesteros E, Sfeir MM, et al. Ultrasensitive and visual detection of SARS-CoV-2 using all-in-one dual CRISPR-Cas12a assay. Nat Commun. 2020;11(1):4711. doi:10.1038/s41467-020-18575-6. [Google Scholar] [PubMed] [CrossRef]
63. Joung J, Ladha A, Saito M, Kim NG, Woolley AE, Segel M, et al. Detection of SARS-CoV-2 with SHERLOCK one-pot testing. N Engl J Med. 2020;383(15):1492–4. doi:10.1056/NEJMc2026172. [Google Scholar] [PubMed] [CrossRef]
64. Lu S, Tong X, Han Y, Zhang K, Zhang Y, Chen Q, et al. Fast and sensitive detection of SARS-CoV-2 RNA using suboptimal protospacer adjacent motifs for Cas12a. Nat Biomed Eng. 2022;6(3):286–97. doi:10.1038/s41551-022-00861-x. [Google Scholar] [PubMed] [CrossRef]
65. Nguyen LT, Macaluso NC, Pizzano BLM, Cash MN, Spacek J, Karasek J, et al. A thermostable Cas12b from Brevibacillus leverages one-pot discrimination of SARS-CoV-2 variants of concern. eBioMedicine. 2022;77(15):103926. doi:10.1016/j.ebiom.2022.103926. [Google Scholar] [PubMed] [CrossRef]
66. Wang Y, Wu L, Yu X, Wang G, Pan T, Huang Z, et al. Development of a rapid, sensitive detection method for SARS-CoV-2 and influenza virus based on recombinase polymerase amplification combined with CRISPR-Cas12a assay. J Med Virol. 2023;95(11):e29215. doi:10.1002/jmv.29215. [Google Scholar] [PubMed] [CrossRef]
67. Liu Y, Chen Y, Dang L, Liu Y, Huang S, Wu S, et al. EasyCatch, a convenient, sensitive and specific CRISPR detection system for cancer gene mutations. Mol Cancer. 2021;20(1):157. doi:10.1186/s12943-021-01456-x. [Google Scholar] [PubMed] [CrossRef]
68. He Q, Yu D, Bao M, Korensky G, Chen J, Shin M, et al. High-throughput and all-solution phase African Swine Fever Virus (ASFV) detection using CRISPR-Cas12a and fluorescence based point-of-care system. Biosens Bioelectron. 2020;154:112068. doi:10.1016/j.bios.2020.112068. [Google Scholar] [PubMed] [CrossRef]
69. Tian Z, Yan H, Zeng Y. Solid-phase extraction and enhanced amplification-free detection of pathogens integrated by multifunctional CRISPR-Cas12a. ACS Appl Mater Interfaces. 2024;16(12):14445–56. doi:10.1021/acsami.3c17039. [Google Scholar] [PubMed] [CrossRef]
70. Li F, Ye Q, Chen M, Zhou B, Zhang J, Pang R, et al. An ultrasensitive CRISPR/Cas12a based electrochemical biosensor for Listeria monocytogenes detection. Biosens Bioelectron. 2021;179:113073. doi:10.1016/j.bios.2021.113073. [Google Scholar] [PubMed] [CrossRef]
71. Wang Z.Zhong C. Cas12c-DETECTOR: a specific and sensitive Cas12c-based DNA detection platform. Int J Biol Macromol. 2021;193(Pt A):441–9. [Google Scholar]
72. Gootenberg JS, Abudayyeh OO, Kellner MJ, Joung J, Collins JJ, Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018;360(6387):439–44. doi:10.1126/science.aaq0179. [Google Scholar] [PubMed] [CrossRef]
73. Gootenberg JS, Abudayyeh OO, Lee JW, Essletzbichler P, Dy AJ, Joung J, et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017;356(6336):438–42. doi:10.1126/science.aam9321. [Google Scholar] [PubMed] [CrossRef]
74. Myhrvold C, Freije CA, Gootenberg JS, Abudayyeh OO, Metsky HC, Durbin AF, et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science. 2018;360(6387):444–8. doi:10.1126/science.aas8836. [Google Scholar] [PubMed] [CrossRef]
75. Wei N, Zheng B, Niu J, Chen T, Ye J, Si Y, et al. Rapid detection of genotype II african swine fever virus using CRISPR Cas13a-based lateral flow strip. Viruses. 2022;14(2):179. doi:10.3390/v14020179. [Google Scholar] [PubMed] [CrossRef]
76. Fozouni P, Son S, Díaz De León Derby M, Knott GJ, Gray CN, D’ambrosio MV, et al. Amplification-free detection of SARS-CoV-2 with CRISPR-Cas13a and mobile phone microscopy. Cell. 2021;184(2):323–33.e329. doi:10.1016/j.cell.2020.12.001. [Google Scholar] [PubMed] [CrossRef]
77. Samanta D, Ebrahimi SB, Ramani N, Mirkin CA. Enhancing CRISPR-Cas-mediated detection of nucleic acid and non-nucleic acid targets using enzyme-labeled reporters. J Am Chem Soc. 2022;144(36):16310–5. doi:10.1021/jacs.2c07625. [Google Scholar] [PubMed] [CrossRef]
78. Wang Y, Zhang Y, Chen J, Wang M, Zhang T, Luo W, et al. Detection of SARS-CoV-2 and its mutated variants via CRISPR-Cas13-based transcription amplification. Anal Chem. 2021;93(7):3393–402. doi:10.1021/acs.analchem.0c04303. [Google Scholar] [PubMed] [CrossRef]
79. Wang S, Li H, Kou Z, Ren F, Jin Y, Yang L, et al. Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system. Clin Microbiol Infect. 2021;27(3):443–50. doi:10.1016/j.cmi.2020.04.018. [Google Scholar] [PubMed] [CrossRef]
80. Zhou T, Huang R, Huang M, Shen J, Shan Y, Xing D. CRISPR/Cas13a powered portable electrochemiluminescence chip for ultrasensitive and specific MiRNA detection. Adv Sci. 2020;7(13):1903661. doi:10.1002/advs.v7.13. [Google Scholar] [CrossRef]
81. Wu P, Ye X, Wang D, Gong F, Wei X, Xiang S, et al. A novel CRISPR/Cas14a system integrated with 2D porphyrin metal-organic framework for microcystin-LR determination through a homogeneous competitive reaction. J Hazard Mater. 2022; 424:127690. doi:10.1016/j.jhazmat.2021.127690. [Google Scholar] [PubMed] [CrossRef]
82. Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1(3):1559–82. doi:10.1038/nprot.2006.236. [Google Scholar] [PubMed] [CrossRef]
83. Waggoner JJ, Stittleburg V, Pond R, Saklawi Y, Sahoo MK, Babiker A, et al. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg Infect Dis. 2020;26(7):1633–5. doi:10.3201/eid2607.201285. [Google Scholar] [PubMed] [CrossRef]
84. Kudo E, Israelow B, Vogels CBF, Lu P, Wyllie AL, Tokuyama M, et al. Detection of SARS-CoV-2 RNA by multiplex RT-qPCR. PLoS Biol. 2020;18(10):e3000867. doi:10.1371/journal.pbio.3000867. [Google Scholar] [PubMed] [CrossRef]
85. Vogelstein B, Kinzler KW. Digital PCR. Proc Natl Acad Sci U S A. 1999;96(16):9236–41. doi:10.1073/pnas.96.16.9236. [Google Scholar] [PubMed] [CrossRef]
86. Pohl G, Shih Ie M. Principle and applications of digital PCR. Expert Rev Mol Diagn. 2004;4(1):41–7. doi:10.1586/14737159.4.1.41. [Google Scholar] [PubMed] [CrossRef]
87. Li Q, Xia Y, Liao D, Nie H, Zhang M, Wang T, et al. Dual-target one-step nested PCR for sensitive detection of SARS-CoV-2 nucleic acids. Prep Biochem Biotechnol. 2022;52(4):471–7. doi:10.1080/10826068.2021.1964084. [Google Scholar] [PubMed] [CrossRef]
88. Kubista M, Andrade JM, Bengtsson M, Forootan A, Jonák J, Lind K, et al. The real-time polymerase chain reaction. Mol Asp Med. 2006;27(2–3):95–125. doi:10.1016/j.mam.2005.12.007. [Google Scholar] [PubMed] [CrossRef]
89. Reddy Banda S, Klapproth H, Smit N, Bednar S, Brandstetter T, Rühe J. An advanced and efficient asymmetric PCR method for microarray applications. Front Bioeng Biotechnol. 2022;10:1045154. doi:10.3389/fbioe.2022.1045154. [Google Scholar] [PubMed] [CrossRef]
90. Kang BH, Lee Y, Yu ES, Na H, Kang M, Huh HJ, et al. Ultrafast and real-time nanoplasmonic on-chip polymerase chain reaction for rapid and quantitative molecular diagnostics. ACS Nano. 2021;15(6):10194–202. doi:10.1021/acsnano.1c02154. [Google Scholar] [PubMed] [CrossRef]
91. Mocellin S, Rossi CR, Pilati P, Nitti D, Marincola FM. Quantitative real-time PCR: a powerful ally in cancer research. Trends Mol Med. 2003;9(5):189–95. doi:10.1016/S1471-4914(03)00047-9. [Google Scholar] [PubMed] [CrossRef]
92. Liu Y, Wu H, Zhou Q, Song Q, Rui J, Zou B, et al. Digital quantification of gene methylation in stool DNA by emulsion-PCR coupled with hydrogel immobilized bead-array. Biosens Bioelectron. 2017;92(9):596–601. doi:10.1016/j.bios.2016.10.054. [Google Scholar] [PubMed] [CrossRef]
93. Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, et al. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000;28(12):E63. doi:10.1093/nar/28.12.e63. [Google Scholar] [PubMed] [CrossRef]
94. Walker GT, Fraiser MS, Schram JL, Little MC, Nadeau JG, Malinowski DP. Strand displacement amplification–an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20(7):1691–6. doi:10.1093/nar/20.7.1691. [Google Scholar] [PubMed] [CrossRef]
95. Daher RK, Stewart G, Boissinot M, Bergeron MG. Recombinase polymerase amplification for diagnostic applications. Clin Chem. 2016;62(7):947–58. doi:10.1373/clinchem.2015.245829. [Google Scholar] [PubMed] [CrossRef]
96. Fang R, Li X, Hu L, You Q, Li J, Wu J, et al. Cross-priming amplification for rapid detection of Mycobacterium tuberculosis in sputum specimens. J Clin Microbiol. 2009;47(3):845–7. doi:10.1128/JCM.01528-08. [Google Scholar] [PubMed] [CrossRef]
97. Ali MM, Li F, Zhang Z, Zhang K, Kang DK, Ankrum JA, et al. Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine. Chem Soc Rev. 2014;43(10):3324–41. doi:10.1039/c3cs60439j. [Google Scholar] [PubMed] [CrossRef]
98. Barreda-García S, Miranda-Castro R, De-Los-Santos-Álvarez N, Miranda-Ordieres AJ, Lobo-Castañón MJ. Helicase-dependent isothermal amplification: a novel tool in the development of molecular-based analytical systems for rapid pathogen detection. Anal Bioanal Chem. 2018;410(3):679–93. doi:10.1007/s00216-017-0620-3. [Google Scholar] [PubMed] [CrossRef]
99. Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463–7. doi:10.1073/pnas.74.12.5463. [Google Scholar] [PubMed] [CrossRef]
100. Metzker ML. Sequencing technologies-the next generation. Nat Rev Genet. 2010;11(1):31–46. doi:10.1038/nrg2626. [Google Scholar] [PubMed] [CrossRef]
101. Van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C. The third revolution in sequencing technology. Trends Genet. 2018;34(9):666–81. doi:10.1016/j.tig.2018.05.008. [Google Scholar] [PubMed] [CrossRef]
102. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi:10.1038/nrg2484. [Google Scholar] [PubMed] [CrossRef]
103. Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316(5830):1497–502. doi:10.1126/science.1141319. [Google Scholar] [PubMed] [CrossRef]
104. Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93. doi:10.1126/science.1181369. [Google Scholar] [PubMed] [CrossRef]
105. Uhlen M, Quake SR. Sequential sequencing by synthesis and the next-generation sequencing revolution. Trends Biotechnol. 2023;41(12):1565–72. doi:10.1016/j.tibtech.2023.06.007. [Google Scholar] [PubMed] [CrossRef]
106. Ibañez-Lligoña M, Colomer-Castell S, González-Sánchez A, Gregori J, Campos C, Garcia-Cehic D, et al. Bioinformatic tools for NGS-based metagenomics to improve the clinical diagnosis of emerging, re-emerging and new viruses. Viruses. 2023;15(2):587. doi:10.3390/v15020587. [Google Scholar] [PubMed] [CrossRef]
107. Gu W, Miller S, Chiu CY. Clinical metagenomic next-generation sequencing for pathogen detection. Annu Rev Pathol. 2019;14(1):319–38. doi:10.1146/pathmechdis.2019.14.issue-1. [Google Scholar] [CrossRef]
108. Ghareeb AA, Sazan Moffaq A. Genetic analysis of SARS-CoV-2 spike gene using next generation sequencing from COVID-19 patients in Erbil/Iraq. Cell Mol Biol. 2024;70(6):7–13. doi:10.14715/cmb/2024.70.6.2. [Google Scholar] [PubMed] [CrossRef]
109. Ying YL, Hu ZL, Zhang S, Qing Y, Fragasso A, Maglia G, et al. Nanopore-based technologies beyond DNA sequencing. Nat Nanotechnol. 2022;17(11):1136–46. doi:10.1038/s41565-022-01193-2. [Google Scholar] [PubMed] [CrossRef]
110. Jain M, Olsen HE, Paten B, Akeson M. The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community. Genome Biol. 2016;17(1):239. doi:10.1186/s13059-016-1103-0. [Google Scholar] [PubMed] [CrossRef]
111. Miller MB, Tang YW. Basic concepts of microarrays and potential applications in clinical microbiology. Clin Microbiol Rev. 2009;22(4):611–33. doi:10.1128/CMR.00019-09. [Google Scholar] [PubMed] [CrossRef]
112. Southern E. Southern blotting. Nat Protoc. 2006;1(2):518–25. doi:10.1038/nprot.2006.73. [Google Scholar] [PubMed] [CrossRef]
113. Schwarzkopf M, Pierce NA. Multiplexed miRNA northern blots via hybridization chain reaction. Nucleic Acids Res. 2016;44(15):e129. [Google Scholar] [PubMed]
114. Chen Q, Li J, Deng Z, Xiong W, Wang Q, Hu YQ. Comprehensive detection and identification of seven animal coronaviruses and human respiratory coronavirus 229E with a microarray hybridization assay. Intervirology. 2010;53(2):95–104. doi:10.1159/000264199. [Google Scholar] [PubMed] [CrossRef]
115. Oneda B, Rauch A. Microarrays in prenatal diagnosis. Best Pract Res Clin Obstet Gynaecol. 2017;42:53–63. doi:10.1016/j.bpobgyn.2017.01.003. [Google Scholar] [PubMed] [CrossRef]
116. Chen AY, Chen A. Fluorescence in situ hybridization. J Invest Dermatol. 2013;133(5):e8. [Google Scholar] [PubMed]
117. Gelali E, Girelli G, Matsumoto M, Wernersson E, Custodio J, Mota A, et al. iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture. Nat Commun. 2019;10(1):1636. doi:10.1038/s41467-019-09616-w. [Google Scholar] [PubMed] [CrossRef]
118. Döhner H, Stilgenbauer S, Benner A, Leupolt E, Kröber A, Bullinger L, et al. Genomic aberrations and survival in chronic lymphocytic leukemia. N Engl J Med. 2000;343(26):1910–6. doi:10.1056/NEJM200012283432602. [Google Scholar] [PubMed] [CrossRef]
119. Stumm M, Klopocki E, Gasiorek-Wiens A, Knoll U, Wirjadi D, Sarioglu N, et al. Molecular cytogenetic characterisation of an interstitial deletion 12p detected by prenatal diagnosis. Prenat Diagn. 2007;27(5):475–8. doi:10.1002/pd.1703. [Google Scholar] [PubMed] [CrossRef]
120. Liehr T, Othman MA, Rittscher K, Alhourani E. The current state of molecular cytogenetics in cancer diagnosis. Expert Rev Mol Diagn. 2015;15(4):517–26. doi:10.1586/14737159.2015.1013032. [Google Scholar] [PubMed] [CrossRef]
121. Sorek R, Lawrence CM, Wiedenheft B. CRISPR-mediated adaptive immune systems in bacteria and archaea. Annu Rev Biochem. 2013;82:237–66. doi:10.1146/biochem.2013.82.issue-1. [Google Scholar] [CrossRef]
122. Aman R, Mahas A, Mahfouz M. Nucleic acid detection using CRISPR/Cas biosensing technologies. ACS Synth Biol. 2020;9(6):1226–33. doi:10.1021/acssynbio.9b00507. [Google Scholar] [PubMed] [CrossRef]
123. Ebina H, Misawa N, Kanemura Y, Koyanagi Y. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510. doi:10.1038/srep02510. [Google Scholar] [PubMed] [CrossRef]
124. Xiao Q, Guo D, Chen S. Application of CRISPR/Cas9-based gene editing in HIV-1/AIDS therapy. Front Cell Infect Microbiol. 2019;9:69. doi:10.3389/fcimb.2019.00069. [Google Scholar] [PubMed] [CrossRef]
125. Moon J, Kwon HJ, Yong D, Lee IC, Kim H, Kang H, et al. Colorimetric detection of SARS-CoV-2 and drug-resistant pH1N1 using CRISPR/dCas9. ACS Sens. 2020;5(12):4017–26. doi:10.1021/acssensors.0c01929. [Google Scholar] [PubMed] [CrossRef]
126. Katayama S, Shiraishi K, Gorai N, Andou M. A CRISPR/Cas9-based method for targeted DNA methylation enables cancer initiation in B lymphocytes. Adv Genet. 2021;2(1):e10040. doi:10.1002/ggn2.v2.1. [Google Scholar] [CrossRef]
127. Gilpatrick T, Lee I, Graham JE, Raimondeau E, Bowen R, Heron A, et al. Targeted nanopore sequencing with Cas9-guided adapter ligation. Nat Biotechnol. 2020;38(4):433–8. doi:10.1038/s41587-020-0407-5. [Google Scholar] [PubMed] [CrossRef]
128. Mcdonald TL, Zhou W, Castro CP, Mumm C, Switzenberg JA, Mills RE, et al. Cas9 targeted enrichment of mobile elements using nanopore sequencing. Nat Commun. 2021;12(1):3586. doi:10.1038/s41467-021-23918-y. [Google Scholar] [PubMed] [CrossRef]
129. Lopatriello G, Maestri S, Alfano M, Papa R, Di Vittori V, De Antoni L, et al. CRISPR/Cas9-mediated enrichment coupled to nanopore sequencing provides a valuable tool for the precise reconstruction of large genomic target regions. Int J Mol Sci. 2023;24(2):1076. doi:10.3390/ijms24021076. [Google Scholar] [PubMed] [CrossRef]
130. Leitner K, Motheramgari K, Borth N, Marx N. Nanopore Cas9-targeted sequencing enables accurate and simultaneous identification of transgene integration sites, their structure and epigenetic status in recombinant Chinese hamster ovary cells. Biotechnol Bioeng. 2023;120(9):2403–18. doi:10.1002/bit.v120.9. [Google Scholar] [CrossRef]
131. Wang T, Liu Y, Sun HH, Yin BC, Ye BC. An RNA-Guided Cas9 nickase-based method for universal isothermal DNA amplification. Angewandte Chemie. 2019;58(16):5382–6. doi:10.1002/anie.v58.16. [Google Scholar] [CrossRef]
132. Wang L, Jiang J, Li X, Li K, He R, Li J, et al. Improved EGFR mutation detection sensitivity after enrichment by Cas9/sgRNA digestion and PCR amplification. Acta Biochim Biophys Sin. 2020;52(12):1316–24. doi:10.1093/abbs/gmaa123. [Google Scholar] [PubMed] [CrossRef]
133. Zhao WS, Yan WP, Chen DB, Dai L, Yang YB, Kang XZ, et al. Genome-scale CRISPR activation screening identifies a role of ELAVL2-CDKN1A axis in paclitaxel resistance in esophageal squamous cell carcinoma. Am J Cancer Res. 2019;9(6):1183–200. [Google Scholar] [PubMed]
134. He C, Han S, Chang Y, Wu M, Zhao Y, Chen C, et al. CRISPR screen in cancer: status quo and future perspectives. Am J Cancer Res. 2021;11(4):1031–50. [Google Scholar] [PubMed]
135. Zhang Z, Wang H, Yan Q, Cui J, Chen Y, Ruan S, et al. Genome-wide CRISPR/Cas9 screening for drug resistance in tumors. Front Pharmacol. 2023;14:1284610. doi:10.3389/fphar.2023.1284610. [Google Scholar] [PubMed] [CrossRef]
136. Mintz RL, Lao YH, Chi CW, He S, Li M, Quek CH, et al. CRISPR/Cas9-mediated mutagenesis to validate the synergy between PARP1 inhibition and chemotherapy in BRCA1-mutated breast cancer cells. Bioeng Transl Med. 2020;5(1):e10152. doi:10.1002/btm2.10152. [Google Scholar] [PubMed] [CrossRef]
137. Kundert K, Lucas JE, Watters KE, Fellmann C, Ng AH, Heineike BM, et al. Controlling CRISPR-Cas9 with ligand-activated and ligand-deactivated sgRNAs. Nat Commun. 2019;10(1):2127. doi:10.1038/s41467-019-09985-2. [Google Scholar] [PubMed] [CrossRef]
138. Lin B, An Y, Meng L, Zhang H, Song J, Zhu Z, et al. Control of CRISPR-Cas9 with small molecule-activated allosteric aptamer regulating sgRNAs. Chem Commun. 2019;55(81):12223–6. doi:10.1039/C9CC05531B. [Google Scholar] [PubMed] [CrossRef]
139. Liang C, Li F, Wang L, Zhang ZK, Wang C, He B, et al. Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials. 2017;147(Suppl 1):68–85. doi:10.1016/j.biomaterials.2017.09.015. [Google Scholar] [PubMed] [CrossRef]
140. Xing S, Lu Z, Huang Q, Li H, Wang Y, Lai Y, et al. An ultrasensitive hybridization chain reaction-amplified CRISPR-Cas12a aptasensor for extracellular vesicle surface protein quantification. Theranostics. 2020;10(22):10262–73. doi:10.7150/thno.49047. [Google Scholar] [PubMed] [CrossRef]
141. Li H, Xing S, Xu J, He Y, Lai Y, Wang Y, et al. Aptamer-based CRISPR/Cas12a assay for the ultrasensitive detection of extracellular vesicle proteins. Talanta. 2021;221:121670. doi:10.1016/j.talanta.2020.121670. [Google Scholar] [PubMed] [CrossRef]
142. Walton RT, Christie KA, Whittaker MN, Kleinstiver BP. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368(6488):290–6. doi:10.1126/science.aba8853. [Google Scholar] [PubMed] [CrossRef]
143. Pickar-Oliver A, Gersbach CA. The next generation of CRISPR-Cas technologies and applications. Nat Rev Mol Cell Biol. 2019;20(8):490–507. doi:10.1038/s41580-019-0131-5. [Google Scholar] [PubMed] [CrossRef]
144. He C, Lin C, Mo G, Xi B, Li AA, Huang D, et al. Rapid and accurate detection of SARS-CoV-2 mutations using a Cas12a-based sensing platform. Biosens Bioelectron. 2022;198:113857. doi:10.1016/j.bios.2021.113857. [Google Scholar] [PubMed] [CrossRef]
145. Zhang T, Zhao W, Zhao W, Si Y, Chen N, Chen X, et al. Universally stable and precise CRISPR-LAMP detection platform for precise multiple respiratory tract virus diagnosis including mutant SARS-CoV-2 spike N501Y. Anal Chem. 2021;93(48):16184–93. doi:10.1021/acs.analchem.1c04065. [Google Scholar] [PubMed] [CrossRef]
146. Katti A, Diaz BJ, Caragine CM, Sanjana NE, Dow LE. CRISPR in cancer biology and therapy. Nat Rev Cancer. 2022;22(5):259–79. doi:10.1038/s41568-022-00441-w. [Google Scholar] [PubMed] [CrossRef]
147. Baylis F, Mcleod M. First-in-human Phase 1 CRISPR gene editing cancer trials: are we ready? Curr Gene Ther. 2017;17(4):309–19. [Google Scholar] [PubMed]
148. Cyranoski D. Chinese scientists to pioneer first human CRISPR trial. Nature. 2016;535(7613):476–7. doi:10.1038/nature.2016.20302. [Google Scholar] [PubMed] [CrossRef]
149. Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353(6299):aaf5573. doi:10.1126/science.aaf5573. [Google Scholar] [PubMed] [CrossRef]
150. Alves E, Taifour S, Dolcetti R, Chee J, Nowak AK, Gaudieri S, et al. Reprogramming the anti-tumor immune response via CRISPR genetic and epigenetic editing. Mol Ther Methods Clin Dev. 2021;21:592–606. doi:10.1016/j.omtm.2021.04.009. [Google Scholar] [PubMed] [CrossRef]
151. Zhang J, You Y. CRISPR-Cas13a system: a novel approach to precision oncology. Cancer Biol Med. 2020;17(1):6–8. doi:10.20892/j.issn.2095-3941.2019.0325. [Google Scholar] [PubMed] [CrossRef]
152. Udugama B, Kadhiresan P, Kozlowski HN, Malekjahani A, Osborne M, Li VYC, et al. Diagnosing COVID-19: the disease and tools for detection. ACS Nano. 2020;14(4):3822–35. doi:10.1021/acsnano.0c02624. [Google Scholar] [PubMed] [CrossRef]
153. Wu Y, Liu SX, Wang F, Zeng MS. Room temperature detection of plasma epstein-barr virus DNA with CRISPR-Cas13. Clin Chem. 2019;65(4):591–2. doi:10.1373/clinchem.2018.299347. [Google Scholar] [PubMed] [CrossRef]
154. Kellner MJ, Koob JG, Gootenberg JS, Abudayyeh OO, Zhang F. SHERLOCK: nucleic acid detection with CRISPR nucleases. Nat Protoc. 2019;14(10):2986–3012. doi:10.1038/s41596-019-0210-2. [Google Scholar] [PubMed] [CrossRef]
155. Khan WA, Barney RE, Tsongalis GJ. CRISPR-cas13 enzymology rapidly detects SARS-CoV-2 fragments in a clinical setting. J Clin Virol. 2021;145:105019. doi:10.1016/j.jcv.2021.105019. [Google Scholar] [PubMed] [CrossRef]
156. Cunningham CH, Hennelly CM, Lin JT, Ubalee R, Boyce RM, Mulogo EM, et al. A novel CRISPR-based malaria diagnostic capable of Plasmodium detection, species differentiation, and drug-resistance genotyping. eBioMedicine. 2021;68:103415. doi:10.1016/j.ebiom.2021.103415. [Google Scholar] [PubMed] [CrossRef]
157. Lee RA, Puig H, Nguyen PQ, Angenent-Mari NM, Donghia NM, Mcgee JP, et al. Ultrasensitive CRISPR-based diagnostic for field-applicable detection of Plasmodium species in symptomatic and asymptomatic malaria. Proc Natl Acad Sci U S A. 2020;117(41):25722–31. doi:10.1073/pnas.2010196117. [Google Scholar] [PubMed] [CrossRef]
158. Konermann S, Lotfy P, Brideau NJ, Oki J, Shokhirev MN, Hsu PD. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell. 2018;173(3):665–76.e614. doi:10.1016/j.cell.2018.02.033. [Google Scholar] [PubMed] [CrossRef]
159. Abbott TR, Dhamdhere G, Liu Y, Lin X, Goudy L, Zeng L, et al. Development of CRISPR as an antiviral strategy to combat SARS-CoV-2 and influenza. Cell. 2020;181(4):865–76.e812. doi:10.1016/j.cell.2020.04.020. [Google Scholar] [PubMed] [CrossRef]
160. Wang H, Nakamura M, Abbott TR, Zhao D, Luo K, Yu C, et al. CRISPR-mediated live imaging of genome editing and transcription. Science. 2019;365(6459):1301–5. doi:10.1126/science.aax7852. [Google Scholar] [PubMed] [CrossRef]
161. Wang F, Wang L, Zou X, Duan S, Li Z, Deng Z, et al. Advances in CRISPR-Cas systems for RNA targeting, tracking and editing. Biotechnol Adv. 2019;37(5):708–29. doi:10.1016/j.biotechadv.2019.03.016. [Google Scholar] [PubMed] [CrossRef]
162. Paige JS, Nguyen-Duc T, Song W, Jaffrey SR. Fluorescence imaging of cellular metabolites with RNA. Science. 2012;335(6073):1194. doi:10.1126/science.1218298. [Google Scholar] [PubMed] [CrossRef]
163. Ying ZM, Wu Z, Tu B, Tan W, Jiang JH. Genetically encoded fluorescent RNA sensor for ratiometric imaging of microRNA in living tumor cells. J Am Chem Soc. 2017;139(29):9779–82. doi:10.1021/jacs.7b04527. [Google Scholar] [PubMed] [CrossRef]
164. Bruch R, Baaske J, Chatelle C, Meirich M, Madlener S, Weber W, et al. CRISPR/Cas13a-powered electrochemical microfluidic biosensor for nucleic acid amplification-free miRNA diagnostics. Adv Mater. 2019;31(51):e1905311. doi:10.1002/adma.201905311. [Google Scholar] [PubMed] [CrossRef]
165. Wu Z, Zhang Y, Yu H, Pan D, Wang Y, Wang Y, et al. Programmed genome editing by a miniature CRISPR-Cas12f nuclease. Nat Chem Biol. 2021;17(11):1132–8. doi:10.1038/s41589-021-00868-6. [Google Scholar] [PubMed] [CrossRef]
166. Karvelis T, Bigelyte G, Young JK, Hou Z, Zedaveinyte R, Budre K, et al. PAM recognition by miniature CRISPR-Cas12f nucleases triggers programmable double-stranded DNA target cleavage. Nucleic Acids Res. 2020;48(9):5016–23. doi:10.1093/nar/gkaa208. [Google Scholar] [PubMed] [CrossRef]
167. Hu Z, Chen M, Zhang C, Li Z, Feng M, Wu L, et al. Cas14a1-mediated nucleic acid diagnostics for spinal muscular atrophy. Biosensors. 2022;12(5):268. doi:10.3390/bios12050268. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF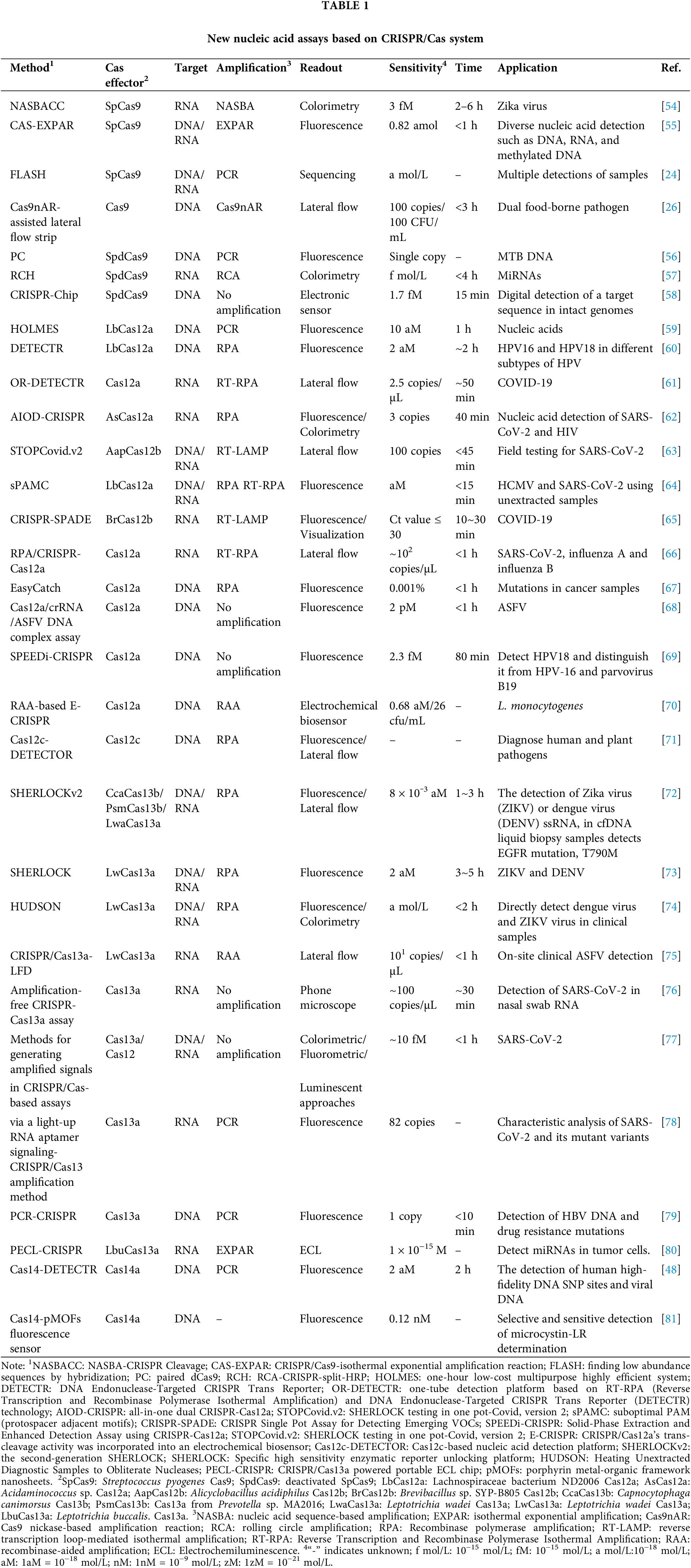
 Downloads
Downloads
 Citation Tools
Citation Tools