 Open Access
Open Access
REVIEW
From Model Organism to Pharmaceutical Powerhouse: Innovative Applications of Yeast in Modern Drug Research
1 Department of Science and Industry, Chongqing Medical and Pharmaceutical College, Chongqing, 401331, China
2 School of Public Health, Chongqing Medical University, Chongqing, 400016, China
3 NHC Key Laboratory of Diagnosis and Treatment on Brain Functional Diseases, The First Affiliated Hospital of Chongqing Medical University, Chongqing, 401122, China
* Corresponding Authors: Jingxin Mao. Email: ; Xuemei Li. Email:
BIOCELL 2025, 49(5), 813-832. https://doi.org/10.32604/biocell.2025.062124
Received 10 December 2024; Accepted 24 February 2025; Issue published 27 May 2025
Abstract
Yeast-based models have become a powerful platform in pharmaceutical research, offering significant potential for producing complex drugs, vaccines, and therapeutic agents. While many current drugs were discovered before fully understanding their molecular mechanisms, yeast systems now provide valuable insights for drug discovery and personalized medicine. Recent advancements in genetic engineering, metabolic engineering, and synthetic biology have improved the efficiency and scalability of yeast-based production systems, enabling more sustainable and cost-effective manufacturing processes. This paper reviews the latest developments in yeast-based technologies, focusing on their use as model organisms to study disease mechanisms, identify drug targets, and develop novel therapies. We highlight key platforms such as the yeast two-hybrid system, surface display technologies, and optimized expression systems. Additionally, we explore the future integration of yeast engineering with artificial intelligence (AI), machine learning (ML), and advanced genome editing technologies like CRISPR/Cas9, which are expected to accelerate drug discovery and enable personalized therapies. Furthermore, yeast-based systems are increasingly employed in large-scale drug production, vaccine development, and therapeutic protein expression, offering promising applications in clinical and industrial settings. This paper discusses the practical implications of these systems and their potential to revolutionize drug development, paving the way for safer, more effective therapies.Keywords
Yeasts are eukaryotic, single-celled microorganisms classified as members of the fungus kingdom. The first yeast originated hundreds of millions of years ago, and over 1500 species have been identified to date, with new species continuously being discovered through ongoing taxonomic research and molecular techniques [1,2]. Yeasts are estimated to constitute about 1% of all described fungal species. As a eukaryotic model organism, yeasts were the first eukaryotic organism to have its genome sequenced, ushering in a new era for humankind’s in-depth understanding of the functions of eukaryotic cells and the molecular mechanisms of their life activities [3,4].
Compared to mammalian cells, yeast (e.g., Saccharomyces cerevisiae, Hansenula polymorpha, Yarrowia lipolytica, Pichia pastoris) offers several unique advantages in biotechnology. Saccharomyces cerevisiae (baker’s yeast), a genetic model organism model organism in genetics, is widely used due to its well-characterized genetic background, ease of culture, and comprehensive gene manipulation tools. Its ability to undergo both asexual (budding) and sexual reproduction makes it particularly versatile for genetic studies. It has been used extensively in genetic research, protein production, and fermentation processes and is a cornerstone in industrial biotechnology [5,6]. Hansenula polymorpha, a methylotrophic yeast, is especially valued for its ability to utilize methanol as a sole carbon source, making it ideal for the high-level expression of recombinant proteins. It is commonly used to produce biopharmaceuticals and enzymes [7]. Yarrowia lipolytica, an oleaginous yeast, is renowned for its ability to accumulate lipids, making it useful in biofuel production and the synthesis of fatty acids. It is also employed in the production of organic acids and industrial enzymes [8]. Pichia pastoris, another methylotrophic yeast, has gained widespread use in biotechnology due to its strong, tightly regulated alcohol oxidase (AOX1) promoter, which facilitates the high-level expression of recombinant proteins. It is especially useful for producing proteins with complex post-translational modifications, such as glycosylation, and is commonly used in both research and industrial applications [9].
These yeasts offer the advantage of being genetically tractable and capable of efficient heterologous gene expression. Their protein glycosylation pathways are highly conserved with those in mammalian cells, making them suitable for producing human therapeutic proteins with the desired post-translational modifications [10,11]. Additionally, the yeast system benefits from mature tools and extensive databases. E.g., the S. cerevisiae knockout collection, maintained by the Saccharomyces Genome Database (SGD) (https://www.yeastgenome.org/, accessed on 12 December 2024), provides a comprehensive resource for genome-wide knockout studies and protein interaction analyses [12]. This collection contains strains where each gene in the S. cerevisiae genome has been systematically deleted, enabling large-scale functional genomics studies (Table 1) [13].
Further, S. cerevisiae and other yeast species, such as Pichia pastoris, Yarrowia lipolytica, and Hansenula polymorpha, have fully sequenced genomes, offering an invaluable resource for exploring gene regulatory regions and identifying potential targets for genetic manipulation [14]. The S. cerevisiae genome was the first eukaryotic genome to be sequenced, and subsequent efforts have led to the creation of various databases, including the SGD and the Yeast Gene Order Browser, which catalog gene annotations, regulatory regions, and gene interactions [15,16]. These resources enable researchers to conduct in-depth analyses of yeast genetics, regulatory networks, and metabolic pathways, facilitating advancements in synthetic biology and metabolic engineering [16].
However, when using yeast in new pharmaceutical research, there are also some drawbacks: firstly, there is limited complexity: yeast cells have lower complexity than human cells, which may restrict the study and understanding of specific drug effects [14,17]. Secondly, there is a lack of tissue structure: yeast, a single-celled organism, lacks cellular tissue structure compared to complex multicellular tissues, which may affect the testing and assessment of certain drugs’ effects [18]. Thirdly, there are limited metabolic pathways: yeast cells have metabolic pathways that differ from human cells, meaning they may not fully mimic conditions within the human body when assessing drug metabolism and drug interactions, as presented in Table 1 [5].
Nevertheless, advances in yeast technology have paved the way for a variety of new genome-wide screening approaches for new drug discovery. In the past, studies using yeasts enabled breakthroughs in understanding basic cellular and molecular processes [22,28]. In recent years, yeasts are experiencing a ‘rebirth’ in fundamental and applied pharmaceutical research [16]. Various experimental strategies employing yeast aim to elucidate disease-related molecular events and uncover novel drugs [17,18]. This paper summarizes the impact of yeast as an experimental tool for new pharmaceutical research and evaluates biomedical research utilizing the yeast two-hybrid system. The recently applied and promising approach of yeast surface display (YSD) technology in new drug discovery is also discussed.
2 Yeast Genome Analysis and New Targets for Pharmaceutical Research
In 1996, Goffeau et al. [29] summarized the genome of baker’s yeast Saccharomyces cerevisiae laboratory strain S288c, consisting of 12 million base pairs and over 6000 genes. This marked significant progress in understanding eukaryotic organisms and established yeast as a model organism with a well-defined genetic background in biomedical research. The availability of more than 6000 mutant strains of Saccharomyces cerevisiae, created through various mutation methods, has greatly facilitated research across different fields.
Through the analysis of open reading frames (ORFs) in the Saccharomyces cerevisiae genome, researchers later found that the functions of 60% of the ORFs were unknown [30]. When these ORFs were compared with the functional genes of humans and other mammals (e.g., rats, mice, cows, and sheep) reported in GenBank, it was discovered that 31% of them had high homology with mammalian functional genes. Notable examples include the yeast genes RAS1 and RAS2 (which are homologous to mammalian RAS genes), MSH2 (yeast homolog: MSH2, mammalian homolog: MSH2), MLH1 (yeast homolog: MLH1, mammalian homolog: MLH1), IRA2 (yeast homolog: IRA2, mammalian homolog: IRAK2), SGS1 (yeast homolog: SGS1, mammalian homolog: WRN), and TEL1 (yeast homolog: TEL1, mammalian homolog: ATM) [31,32]. This indicates a significant level of similarity between these yeast ORFs and their mammalian counterparts. The homology percentage was calculated at the DNA level and is estimated to be between 60% and 85%, depending on the gene. By constructing deletion mutants of these homologous genes in yeast, it becomes possible to screen compounds for activity, leveraging the conserved functions between yeast and mammalian systems [33,34]. Lum et al. [35] established a yeast mutant library containing 3503 heterozygous allelic deletions. They used the principle that different mutants have varying sensitivities to drugs, thus affecting their growth, to analyze 78 antimicrobial and antiviral drugs, including 5-fluorocytosine. They screened some of these drugs for their targets. They found that the enzyme lanosterol synthase in the sterol biosynthesis pathway was the target of anti-cardiovascular drugs. At the same time, Ribosomal ribonucleic acid (rRNA) processing was identified as a potential target of the cell growth inhibitor 5-fluorouracil [35–37]. Establishing a method for screening drug targets using heterozygous alleles in yeast has provided new insights for drug research and accelerated the study of drug activity, side effects, and chemical toxicity. Besides, the study of yeast genes such as GAL4, LEU, TRP, HIS, ADE, and CDC25 has provided additional screening markers for establishing a yeast two-hybrid system, yeast surface display system, and yeast expression system [35,38].
Additionally, recent advances in genome editing technologies, such as CRISPR-Cas9, could facilitate the use of yeast as a model organism for advancing drug discovery. CRISPR-Cas9 requires whole genome sequences to design and guide RNA (gRNAs) in targeting relevant genes. This system is very effective in Saccharomyces cerevisiae and other yeasts, such as Pichia pastoris, Yarrowia lipolytica, and Hansenula polymorpha, primarily due to their efficient homology-directed DNA repair mechanisms. Interestingly, other unconventional yeasts have also reported high success rates, suggesting that many genomes can be modified using the CRISPR-Cas9 system (Fig. 1) [39,40]. For example, in the heat-tolerant methylotrophic yeast Ogataea polymorpha, CRISPR-Cas9-assisted multiplexed genome editing successfully introduced all the genes required for resveratrol biosynthesis, as well as genes for the biosynthesis of human serum albumin and cadaverine [39]. These new biotechnological applications provide a promising avenue for developing subsequent drugs [41].
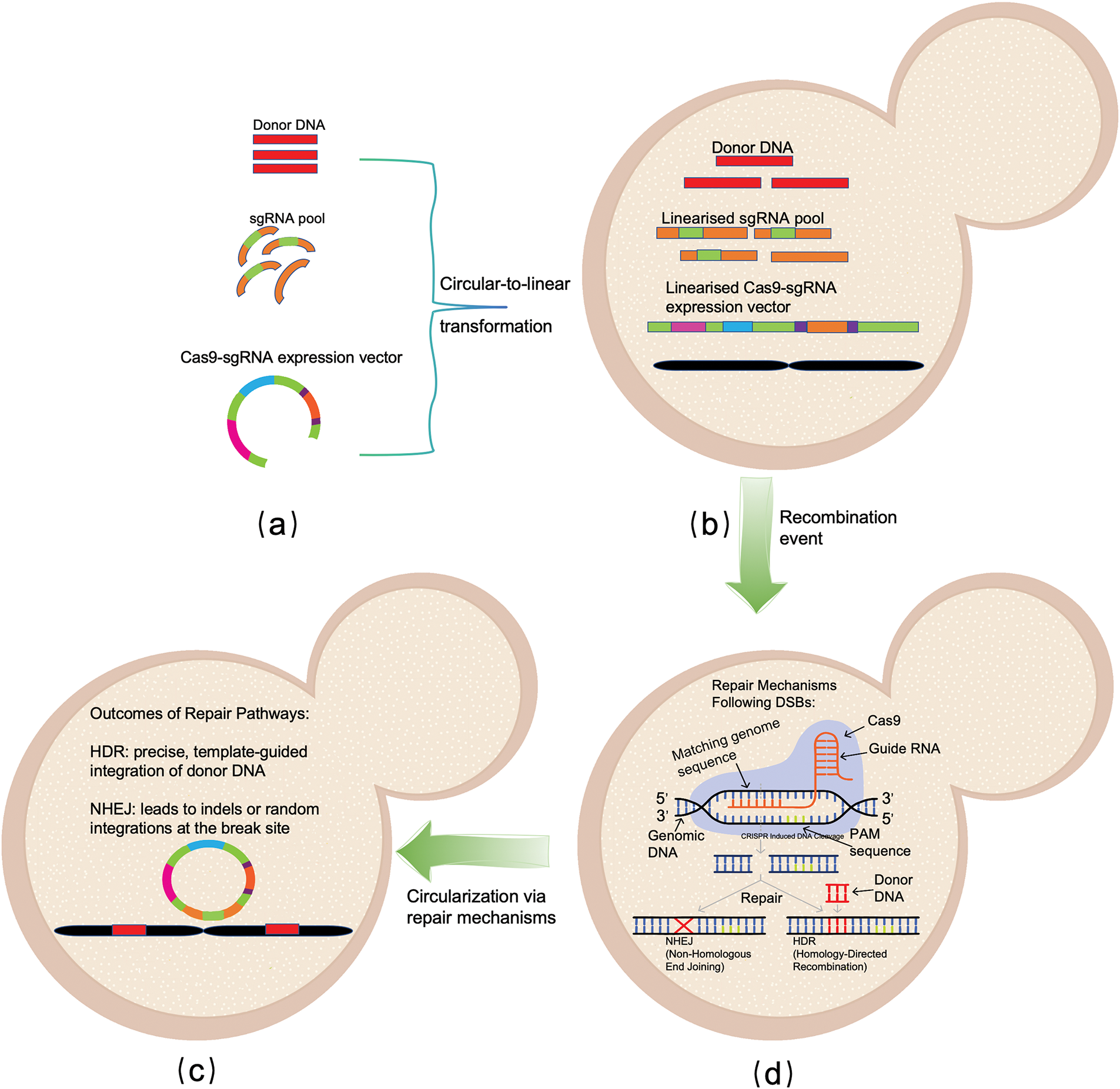
Figure 1: Schematic Overview of Cas9-Mediated DNA Editing and Repair Pathways in Yeast. a. Input: the initial inputs include donor DNA templates for homology-directed repair (HDR), a pool of single guide Ribonucleic acid (sgRNAs) targeting specific loci, and a Cas9-sgRNA expression vector. These constructs are typically circular before the transformation process. b. Transformation: circular DNA constructs are linearized and introduced into yeast cells through transformation. Linearized donor DNA, sgRNA pools, and Cas9-sgRNA expression vectors are prepared for subsequent recombination events. c. DSB Induction and Repair: the Cas9-sgRNA complex induces double-strand breaks (DSBs) at target genomic sites. Cellular repair mechanisms are activated with two main pathways: (1) Homology-Directed Repair (HDR), which uses donor DNA as a template for precise integration, and (2) Non-Homologous End Joining (NHEJ), which is error-prone and may result in insertions or deletions. d. Repair Outcomes: depend on the pathway utilized. HDR facilitates precise integration of donor DNA, while NHEJ leads to random or imprecise modifications. The circularization of DNA may occur as part of the repair mechanisms, ensuring vector stability
3 Yeast Two-Hybrid (Y2H) System in Biomedical Research
Brückner et al. [42] established a yeast two-hybrid system (Y2H) to detect protein-protein interactions by using the Gal4 transcriptional activator of the yeast Saccharomyces cerevisiae. The Gal4 protein activates the transcription of a gene involved in galactose utilization, forming the basis of selection [43]. In a Y2H system, the premise is that the activation of the downstream reporter gene(s) occurs through the binding of a transcription factor to an upstream activating sequence (UAS) [44]. The transcription factor is split into two fragments: the DNA-binding domain (BD) and the activating domain (AD). The BD binds to the UAS, while the AD activates transcription [45]. A known functional protein is typically used as the ‘Bait,’ and proteins that may interact with the ‘Bait’ are used as the ‘Prey.’ The interaction between the ‘Bait’ and the ‘Prey’ is verified by the expression and detection of reporter genes [46].
To date, yeast two-hybrid technology has been expanded to study a broader range of intermolecular interactions [47]. Variants include the reverse two-hybrid system for studying interactions between proteins and small-molecule inhibitors, the single-hybrid system for studying interactions between DNA and proteins, the three-hybrid system for studying interactions between RNA and proteins, and another three-hybrid system for studying interactions between small molecules and proteins in a ligand-dependent manner [48]. The differences among these systems lie in selecting reporter genes and the ‘Prey.’ Additionally, they differ in selecting the reporter gene and the ‘Bait’ or ‘Prey’ expression vector. Yeast two-hybrid systems have been widely used in biomedical research [49]. Their widespread application is expected due to their relative affordability, lack of need for specialized large equipment, and feasibility in any molecular biology laboratory with reasonable throughput (Fig. 2) [50].
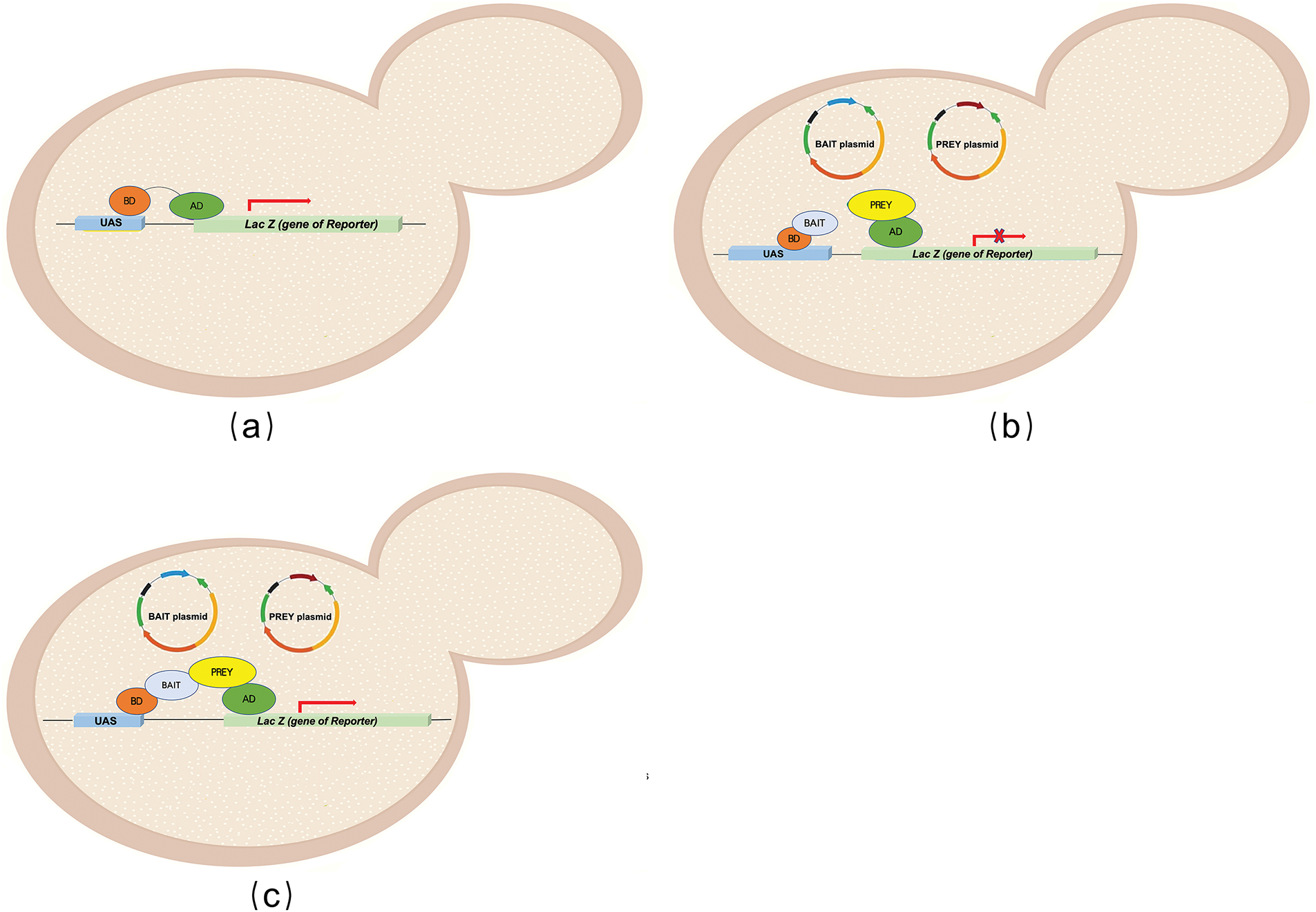
Figure 2: Overview of yeast two-hybrid system (Y2H), a genetic technique that detects protein-protein interactions: in a Y2H system, the expression of a reporter gene requires activation through the binding of a transcription factor to an upstream activating sequence (UAS). The UAS consists of two independent domains: the DNA-binding domain (BD) and the activation domain (AD). The BD binds to the UAS, while the AD activates transcription (a). The protein fused to the BD is called the Bait, and the one fused to the AD is referred to as the Prey. Without Bait-Prey interaction, the AD domain cannot initiate gene expression (b). However, when the Bait and Prey interact, the BD binds to the DNA, localizing the AD upstream of the reporter gene, resulting in the expression of the reporter gene (c)
One of the primary applications of the yeast two-hybrid (Y2H) system in drug discovery is the identification of protein-protein interactions that are critical for disease progression [51]. By mapping these interactions, researchers can pinpoint potential drug targets. The yeast two-hybrid system continues to be a cornerstone in the field of drug discovery. Its ability to uncover protein-protein interactions and facilitate the screening of potential inhibitors makes it an essential tool for identifying and validating new drug targets [52]. As technology advances, its role in drug discovery is likely to become even more prominent, contributing to the development of more effective and targeted therapies [53]. In the following section, we will discuss the latest advancements and progress in yeast two-hybrid systems within biomedical research.
3.1 Study of Protein Interactions
Protein-protein interactions (PPIs) occur throughout a cell and are essential for understanding cellular functions. Studying these interactions in model organisms enhances our understanding of biological processes, aids in deciphering disease mechanisms, and helps in identifying potential drug targets and screening new therapeutics [54]. By constructing DB-Bait and AD-Prey yeast expression vectors, interactions between proteins can be detected using reporter genes. In this context, we will refer to the protein used in the DB vector as ‘Bait’ and the protein used in the AD vector as ‘Prey’ in the subsequent sections. Additionally, indirect interactions between different proteins can be investigated through host endogenous protein-mediated methods [55]. For example, early studies demonstrated that the HIV-encoded Rev protein interacts with the yeast nucleoprotein Rip1 [56]. Further research using the yeast two-hybrid system revealed that this interaction is mediated by Crm1p in yeast [54].
3.2 Discovery of New Proteins and New Functions of Proteins
Using a protein with a known function as ‘Bait’, new proteins can be discovered from cDNA libraries as ‘Prey’, and new functions of known proteins can also be identified. For example, Fañanás-Pueyo et al. [57] transformed all predicted open reading frames (ORFs) in yeast into MATa (mating type a) and MATα (mating type alpha) strains as ‘Bait’ or ‘Prey’ by ligating them with DB or AD, respectively. In S.cerevisiae, MATa and MATα represent the two mating types, where MATa is the a-type and MATα is the alpha-type, each having distinct roles in the mating process. These strains are used to facilitate the identification of protein interactions in the yeast two-hybrid system. They then transformed these constructs into MATa and MATα yeasts to form ‘Bait pools’ and ‘Prey pools’. By selecting 96 positive clones from each pool and systematically comparing the results, they identified 183 pairs of interacting proteins, more than half of which were previously unreported [58]. These new protein interactions may be related to different transport modes of substances in yeast [59]. Besides, other targets for screening aptamers using yeast two-hybrid systems include the transcription factor signal transducer and activator of transcription 3 (Stat3) and the tyrosine kinase avian erythroblastic leukemia viral oncogene homolog 2 (ErbB2). Constitutive activation of Stat3 has been observed in various tumors, while ErbB2 is overexpressed in numerous cancers, including breast, ovarian, bladder, and lung cancers [60]. Therefore, investigating the oncogenic functions of these two proteins through a yeast two-hybrid system may open new therapeutic avenues for cancer research [61].
3.3 Antigen-Antibody Interaction Studies
Existing techniques for detecting antigen-antibody interactions are based on in vitro immunoreactivity. However, the interactions of drugs, especially peptides and proteins, with normal components in the body are highly complex and challenging to determine using conventional immunological studies [62]. Therefore, evaluating drug immunotoxicity and/or immunogenicity during safety assessments is difficult with standard methods. Mehta et al. [63] linked various tumor suppressor p53 and T antigen mutants (produced via PCR) to DB or AD to form fusion proteins, using LacZ as the reporter gene to establish a new reliable detection method. They detected interactions between 34 types of mutant p53 and the T antigen, and these mutant p53 proteins were also found in tumor patients. The gradual improvement of this method is expected to bring significant advancements to the preclinical safety evaluation of drugs.
Zhang et al. [62] developed an antigen-antibody co-display (AACD) system for detecting interactions between G-protein-coupled receptors (GPCRs) and single-chain variable fragments (scFvs) using a split-ubiquitin-based yeast two-hybrid (YTH) system. This system was engineered by fusing a transmembrane peptide to anchor scFv antibodies to cell membranes, allowing co-display of the GPCRs on cell membranes. They further optimized the topology and key elements of the scFv fusion proteins, creating an AACD system that can rapidly determine the association between GPCRs and their candidate antibodies. This innovation shortens the research cycle for off-target detection and epitope recognition [64].
The transfer process of pathogenic bacteria or viruses after invasion is related to specific proteins on their surfaces. Some proteins on the surface of host cells may become the ‘targets’ for these pathogens. Compounds that bind to these specific proteins on bacteria or viruses could potentially prevent them from invading normal tissues or cells, achieving therapeutic and preventive purposes [65]. Using various screening methods to identify compounds that bind to specific proteins on the surface of these pathogens makes it possible to prevent their invasion of normal tissues or cells. This targeted binding can serve both therapeutic and preventive functions [66].
A gene fragment encoding a specific protein from a pathogen can be used as ‘Bait’ by linking it to a DNA-binding domain (DB), while a cDNA library or random sequences serve as ‘Prey’ by attaching to an activation domain (AD). These constructs are transformed into yeast, enabling the identification of ‘Prey’ proteins or peptides that bind to the bait. This approach was first demonstrated by Zhu et al. [67] using a yeast two-hybrid system to screen for hormone-receptor interactions, identifying a proinsulin mutant with high binding affinity to the Insulin-like Growth Factor 1 (IGF-1) receptor, a potential candidate for drug screening in diabetes treatment. Similarly, researchers have also optimized a high-throughput assay for HBx-DDB1 interactions in S. cerevisiae, suggesting its potential for discovering therapeutic agents for chronic hepatitis B [68].
4 Yeast Surface Display System and New Drug Screening
Yeast surface display (YSD) is a “whole-cell” platform used for the heterologous expression of proteins immobilized on the yeast’s cell surface. From 10,000 to 1,000,000 copies of the fusion protein can be expressed on the surface of each yeast cell [69]. Substances that interact with these gene products can be identified by displaying target gene products with unknown functions or products related to specific pathological processes on the yeast cell wall [70]. This approach helps to study the functions of unknown genes and can lead to the discovery of new drug targets or the screening of new lead compounds for disease treatment. YSD is a promising technology that is not yet optimized for biotechnological applications [71].
Yeast surface display technology is based on the study by Teparić et al. [72] on a glycoprotein in the yeast cell wall. They showed that a- and α-agglutinin are two types of mannose proteins in the yeast cell wall. The a-agglutinin consists of two parts: Aga1 and Aga2. Aga1 is covalently linked to β-glucan in the inner layer of the yeast cell wall, while Aga2 binds to Aga1 through two disulfide bonds and is displayed on the cell surface. The most commonly used yeast surface display system originally developed by Wang, a single-chain variable fragment (scFv) was genetically engineered to be fused to the C-terminus of Aga2, with a GAL4 promoter inserted upstream of Aga2 [73]. Under galactose induction, the scFv was expressed and displayed on the surface of the yeast. This setup allowed the corresponding ligands to be easily detected from a library of random mutants using flow cytometry (Fig. 3) [73]. Orr et al. [74] further developed this method by ligating the Vbeta8 domain of a soluble T-cell receptor and an antibody mutant targeting the c-myc epitope to the C-terminus of scFv, displaying it on the surface of yeast cells. They then used flow cytometry with a fluorescent marker to detect scFv mutants with higher affinity for the T-cell receptor [74]. These studies demonstrated that this method can be used to analyze antigenic epitopes, particularly for binding sites of eukaryotic secretory or cell surface proteins.
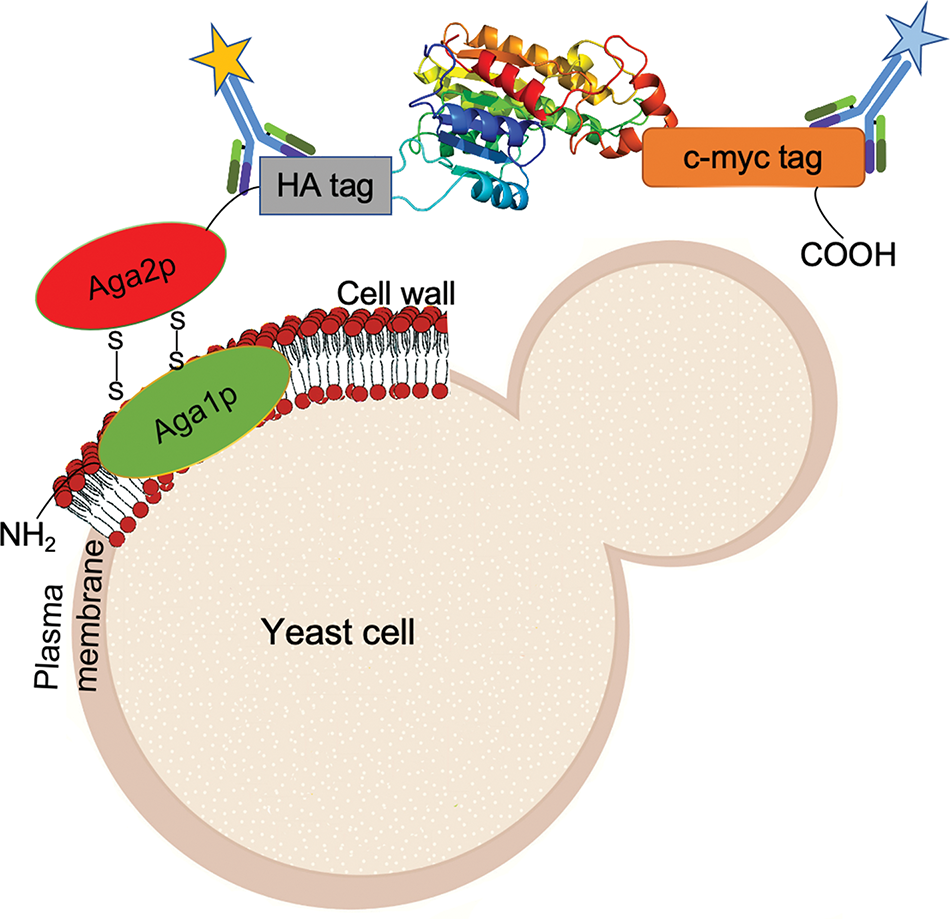
Figure 3: Overview of the Yeast Surface Display (YSD) system, a biotechnological technique used for the cell surface expression of heterologous proteins, which are fused to the C-terminus of the mating agglutinin protein Aga2. YSD construct includes two epitope tags: a hemagglutinin (HA) tag between Aga2p as well as a c-myc tag. Aga1p is anchored to the inner layer of the yeast cell wall, while Aga2p binds to Aga1p through two disulfide bonds. Induction of protein expression results in surface display of the fusion protein through disulfide bond formation of Aga2p to Aga1p
Other applications for YSD include library screening, whole-proteome studies, bioremediation, vaccine and antibiotic development, biosensor production, ethanol production, biocatalysis, and new drug discovery [75]. In a recent study, Wang et al. [73] developed a new method for high-throughput rapid extracellular antigen profiling using yeast surface display technology to discover autoantibodies against medically relevant autoimmune targets. By displaying 2688 barcoded human extracellular proteins on the yeast surface and comparing them to patient serum immunoglobulins, they identified relevant antigens from sorted yeast using next-generation sequencing. This approach led to the identification of several autoantibodies present in autoimmune polyglandular syndrome type 1 and systemic lupus erythematosus conditions in autoimmune patients [76,77]. In addition, Lopez-Morales et al. [78] used YSD to study the evolution of the SARS-CoV-2 receptor-binding domain (RBD) and the propensity of mutations to evade immunological recognition. By displaying a library of receptor-binding domains (RBDs) on the surface of yeast cells, researchers can analyze the antibody-binding properties of various viral variants through deep mutation scanning and next-generation sequencing. In the case of SARS-CoV-2, the RBD is part of the spike protein, which binds to the angiotensin-converting enzyme 2 (ACE2) receptor on human cells. By mapping mutations in the RBD, researchers have identified several mutations that enhance ACE2 binding, providing insights into the potential for viral immune escape. This approach has been particularly valuable for studying SARS-CoV-2 variants. By optimizing the YSD workflow, researchers have mapped binding affinities between the SARS-CoV-2 RBD and different classes of antibodies, offering predictive insights into how mutations may impact antibody recognition and suggesting potential future variants of concern. The study of viral mutations and their capacity to evade immune recognition is critical, and yeast models offer a robust platform for investigating these mutations at a high throughput. Additionally, the ability to screen for novel antibodies or vaccine candidates in yeast systems may help mitigate the risks associated with emerging viral variants.
Interestingly, yeast two-hybrid and Yeast surface display techniques focus on protein-protein interactions (including ligand-receptor interactions), and the combination of these two techniques will facilitate the development of vaccines as well as new pharmaceutical research [27]. E.g., to obtain a soluble antibody with high stability and correct folding after intracellular expression, Tristán-Manzano et al. [27] attached the scFv fragment of the antibody to the activation domain (AD) of the transcription factor VP16 and the antigen to the DNA-binding domain (DB) of LexA. They established a yeast surface display method to demonstrate the interaction between the antigen and the antibody using His3 and LacZ as reporter genes. When an antigen-antibody interaction occurs, the combination of AD and DB initiates the expression of the reporter genes, which can be detected using a nutrient-deficient medium or the blue-white spotting method. This method allows for the screening of antigen-specific antibody fragments from the scFv mutant display library, providing a valuable platform for vaccine development.
5 Optimizing the Yeast Expression System for New Drug Development
The use of genetic engineering to address the issue of ‘drug sourcing’ is a focal point and a hot topic in the field of biomedicine. Various peptides or proteins have been successfully expressed in prokaryotic cells, eukaryotic cells, animal mammary glands, plants, and other systems [79]. Yeast, as a model organism with extensive genetic background research, has emerged as a prominent system for exogenous gene expression. Its rapid growth, ease of genetic manipulation, and ability to complete post-translational processing of eukaryotic proteins or peptides make yeast a major platform for exogenous gene expression [80]. Brooks et al. [81] were the first to express an exogenous gene in Saccharomyces cerevisiae. They ligated the Liver-expressed antimicrobial peptide D (LeIF-D) gene upstream of the ethanol dehydrogenase I gene, resulting in the expression of biologically active LeIF-D in yeast at a rate of 1 × 106 molecules/cell. Saccharomyces cerevisiae has been utilized for expressing heterologous proteins for a significant period, owing to its extensive history in the fermentation industry. The completion of the yeast’s full sequence analysis further solidified its position as a popular expression system. In a study by Bhattacharya et al. [82], a S.cerevisiae mutant deficient in four enzymes related to ergosterol metabolism was genetically engineered. This mutant was then modified to incorporate an enzyme derived from Hypericum perforatum (St. John’s Wort), a plant known for its use in hydrocortisone synthesis, enabling the yeast to produce hydrocortisone. When cultured in a simple carbon medium, the mutant exhibited high expression levels. Subsequently, the cultured mutant produced a steroid hormone suitable for treating patients with arthritis and adrenocortical insufficiency. This innovative work holds promise for producing small molecule compounds through biotransformation technology.
However, researchers have identified some limitations in using Saccharomyces cerevisiae as a model organism for expressing exogenous proteins: the exogenous gene vector commonly used in the transgenic system of Saccharomyces cerevisiae is a 2 μm add-on plasmid. This vector is genetically unstable and prone to loss during cell proliferation, leading to instability in engineered strains and ultimately affecting the production of recombinant proteins [83]. Additionally, Saccharomyces cerevisiae contains an α1-3 glycosidic bond at the end of its core polysaccharide. The presence of this glycosidic bond increases the antigenicity of recombinant proteins, posing a significant challenge in developing these proteins or peptides into drugs [84].
Recently, the yeast Pichia pastoris has become more widely used in the production of exogenous proteins. This yeast features a robust promoter, the alcohol oxidase 1 (AOX1) promoter, which is activated in the absence of a repressive carbon source such as glucose and utilizes methanol as the sole carbon source. This promoter ensures strict regulation and high efficiency in the expression of exogenous genes [85]. Additionally, the expression system often employs integration plasmids as vectors for exogenous genes, allowing the transfected genes to integrate into the yeast genome and replicate synchronously with genomic DNA. This integration reduces the likelihood of gene loss, resulting in stable engineered strains [86].
Moreover, since this expression system commonly utilizes integration plasmids as exogenous gene vectors, the transferred genes can integrate into the yeast genome and replicate in parallel with genomic DNA, reducing the likelihood of loss and resulting in relatively stable engineered strains capable of achieving high expression levels [87]. For example, the novel vector pIB4α developed by Gurkan et al. [88] enables the expression of C6.5, a functional single-chain antibody fragment targeting the surface glycoprotein HER2 (human epidermal growth factor receptor 2) found on ovarian and breast cancer cells, at up to 70 mg/L. This level of expression holds promise for commercial production applications.
Castro et al. [89] observed that although the N-terminus of human interferon β1 secreted and expressed in Pichia pastoris was identical to that of the natural product, the secreted protein was partially N-glycosylated, potentially affecting its biological activity. To address this issue, Hamilton et al. [90] developed a strategy to delete endogenous yeast genes associated with the glycosylation pathway. They then introduced five enzymes or proteins, including mannosidases (I and II), N-acetylglucosaminyltransferases (I and II), and UDP-N-acetylglucosaminyltransferase, into the yeast genome. This led to the formation of a human-like glycosylation pathway, specifically the N-acetylglucosamine (GlcNAc)2-mannose (Man)3-N-acetylglucosamine (GlcNAc)2 (GlcNAc2Man3GlcNAc2) pathway, within the yeast cells. This modification facilitated the expression of human N-glycosylated glycoproteins, improving the quality of the expressed proteins. This establishment of the expression system serves as a foundation for the large-scale production of human glycoproteins for medicinal purposes and offers a valuable tool for studying the conformational relationships of glycoproteins.
Furthermore, since the plasmid vector with a yeast gene AOX promoter was utilized in the expression system of Pichia pastoris, requiring methanol for inducing the expression of exogenous genes, it posed challenges for industrial production due to methanol’s flammability and volatility [91]. To address this issue, Arjmand (2024) [92] proposed replacing the AOX promoter with the Formaldehyde Dehydrogenase 1 (FLD1) promoter. This promoter can induce the expression of exogenous genes when methanolamine is the sole nitrogen source. Additionally, the FLD1 promoter offers advantages such as high tunability, making it a promising alternative for industrial applications.
In addition to protein and peptide expression and production, transgenic yeast holds potential for various applications such as vaccine development, drug screening model construction, and pathogenesis studies. For instance, pathogenic bacteria and fungi frequently form biofilms upon invading host tissues, which provide protection and render drug treatments ineffective [93]. Biofilm formation is a well-documented mechanism in pathogens like Pseudomonas aeruginosa and Candida albicans, contributing to their resistance to both the host immune response and antimicrobial therapies. Further research into biofilm formation mechanisms could aid in developing new antimicrobial drugs [94]. Bouyx et al. [95] investigated biofilm formation and genesis using Saccharomyces cerevisiae as a model and found that yeast adhesion is related to its surface glycoprotein. Interestingly, when knocking out this glycoprotein-encoding gene (FLO11) along with its regulatory gene (FLO8), the adhesion between mutant yeast and other yeasts was eliminated. Since homologues of the FLO11 gene also exist in some pathogenic fungi like Candida albicans, Saccharomyces cerevisiae could serve as a model for studying pathogenic bacteria invasion and pathogenesis, as well as for the rapid screening of antimicrobial drugs [96].
The rapid advancements in engineering yeast for pharmaceutical research have revolutionized the field of drug discovery and development. The comprehensive analysis of the yeast genome has provided invaluable insights into new targets for pharmaceutical research, leveraging the remarkable genetic similarities between yeast and higher eukaryotes [97]. The identification and functional analysis of yeast genes homologous to human disease genes have opened new avenues for understanding disease mechanisms and identifying potential drug targets [98].
The Y2H system has emerged as a powerful tool for studying protein-protein interactions, offering a high-throughput method to map complex interaction networks. This system has significantly contributed to identifying drug targets, understanding viral pathogenesis, and exploring the molecular underpinnings of various diseases, including neurodegenerative disorders and cancer [99]. The integration of Y2H with next-generation sequencing and CRISPR-Cas9 technology further enhances its potential, providing a more detailed and dynamic view of protein interactions and genetic interactions [70].
The YSD system has become an essential platform for new drug screening and development. By displaying peptides and proteins on the yeast cell surface, this system allows for the rapid screening of large libraries for binding affinity and specificity, facilitating the identification of therapeutic candidates. The YSD system’s ability to mimic mammalian post-translational modifications and present complex antigens has made it an invaluable tool in vaccine development and antibody engineering [100].
Optimizing the yeast expression system has been crucial for producing high yields of recombinant proteins, essential for developing new drugs and therapeutic agents. Advances in metabolic engineering, synthetic biology, and genome editing have led to more efficient and scalable production processes [23]. The flexibility and robustness of yeast as a production platform make it ideal for manufacturing a wide range of pharmaceuticals, from simple peptides to complex biologics [20].
In conclusion, engineering yeast has become a cornerstone of modern pharmaceutical research, offering versatile and powerful platforms for drug discovery, screening, and production. The continued development and refinement of yeast-based technologies promise to enhance the efficiency and sustainability of pharmaceutical manufacturing processes, ultimately leading to safer and more effective therapies for treating human diseases. As research progresses, the integration of these yeast-based systems with emerging technologies will likely yield even more significant breakthroughs, paving the way for innovative therapeutic solutions and transforming the landscape of modern medicine.
The future of engineering yeast for pharmaceutical research is poised to bring transformative changes to drug discovery and development. As we continue to harness the power of yeast, several promising avenues and emerging technologies are expected to shape the next generation of pharmaceutical research.
7.1 Integration with Artificial Intelligence and Machine Learning
The integration of Artificial Intelligence (AI) and Machine Learning (ML) with yeast-based systems is rapidly transforming the landscape of pharmaceutical research. By harnessing the power of AI and ML, researchers can analyze vast and complex datasets generated from yeast genome studies, protein-protein interaction networks, and high-throughput screening assays [101]. These technologies enable the identification of novel drug targets, prediction of therapeutic efficacy, and optimization of experimental workflows, ultimately accelerating the drug discovery process.
AI and ML algorithms are particularly adept at uncovering hidden patterns and correlations in large-scale biological data. For example, ML models can analyze genetic sequences and protein interactions to predict the functional outcomes of specific genetic modifications in yeast [102]. This ability to predict the effects of genetic alterations in yeast models enables a more targeted approach to drug development and pathway engineering. Furthermore, AI-driven approaches are being used to optimize yeast strains for the efficient production of therapeutic proteins, improving yields and reducing costs associated with biopharmaceutical manufacturing [103].
One of the key areas where AI and ML are making an impact is in the design of synthetic biology circuits within yeast. These circuits often involve complex gene interactions, and predicting their behavior is a challenging task. AI and ML models, particularly deep learning techniques, can simulate these gene regulatory networks, providing insights into how synthetic constructs might behave in vivo. This allows for the design of more robust and predictable yeast-based biosystems for drug production [102].
In high-throughput drug screening, AI-powered tools can enhance the analysis of large datasets by identifying potential drug candidates with higher accuracy and efficiency. These tools can analyze patterns in compound-library screening results, identify promising drug-like molecules, and predict their interactions with yeast-expressed target proteins [101]. Additionally, AI algorithms can assist in the identification of off-target effects, toxicity risks, and the potential for drug resistance-critical factors in the drug development process.
The integration of AI and ML also enables adaptive learning in yeast engineering processes. As data from experimental trials are collected, ML models can continuously learn from these experiments, improving their predictive capabilities over time. This iterative approach can lead to more refined yeast strains and optimized fermentation processes, enhancing the scalability and productivity of therapeutic molecule production [103].
Moreover, the combination of AI with next-generation sequencing and genome editing technologies such as CRISPR-Cas9 has the potential to revolutionize the precision of yeast-based genetic engineering. AI tools can predict the most effective CRISPR guide RNAs, thereby reducing off-target effects and improving the efficiency of gene editing. This synergy between AI and advanced genome-editing techniques is likely to play a crucial role in the development of next-generation yeast platforms for pharmaceutical applications [104].
In summary, AI and ML have the potential to significantly enhance the capabilities of yeast-based systems in drug discovery and production. Their ability to analyze complex biological data, predict genetic outcomes, and optimize experimental designs will not only streamline the drug development process but also enable more efficient and sustainable manufacturing of therapeutics. As these technologies continue to evolve, they will play an increasingly vital role in shaping the future of pharmaceutical research and biotechnology.
7.2 Advanced Genome Editing Techniques
The ongoing advancements in genome editing technologies, particularly CRISPR-Cas systems, will further enhance the precision and efficiency of genetic modifications in yeast. These tools will enable more complex and targeted manipulation of yeast genomes, facilitating the study of intricate gene functions and interactions [39]. Additionally, the development of novel CRISPR variants and delivery methods will expand the applicability of genome editing in diverse yeast strains [87].
7.3 Synthetic Biology and Metabolic Engineering
The field of synthetic biology will continue to play a crucial role in engineering yeast for pharmaceutical applications. The design and construction of synthetic gene circuits and metabolic pathways will enable the production of novel and complex biomolecules. Innovations in metabolic engineering will optimize yeast metabolism for higher yields and improved scalability of biopharmaceuticals [105]. The development of orthogonal systems and synthetic promoters will provide greater control over gene expression and metabolic flux.
7.4 Expanding the Yeast Toolkit
The exploration and utilization of non-conventional yeast species will diversify the toolkit available for pharmaceutical research. Yeast species such as Pichia pastoris, Yarrowia lipolytica, and Kluyveromyces lactis offer unique metabolic capabilities that make them particularly valuable for various biotechnological applications [106]. Pichia pastoris is known for its high capacity for methanol utilization, making it a robust expression system for recombinant protein production, particularly for complex proteins requiring post-translational modifications such as glycosylation. Yarrowia lipolytica, an oleaginous yeast, excels in lipid metabolism, allowing it to efficiently produce lipids and biofuels, as well as valuable pharmaceuticals like biopolymers and enzymes [107]. Additionally, Kluyveromyces lactis is a strong candidate for dairy-related applications, capable of efficiently fermenting lactose and producing recombinant proteins with high yield [108]. By leveraging these species’ specific metabolic advantages, researchers can develop tailored solutions for a wide range of pharmaceutical applications, including protein production, vaccine development, and bioremediation [109].
In conclusion, the potential of engineering yeast for pharmaceutical research remains vast and promising. As technological advancements continue to evolve, yeast-based systems are expected to become more refined, offering enhanced capabilities for the production of safer, more effective, and scalable therapeutic agents. By leveraging these innovations, researchers can accelerate the development of novel pharmaceutical treatments, optimizing drug discovery processes and improving the overall efficiency of therapeutic development [110]. These advancements hold the key to overcoming current challenges and driving progress in modern medicine, ultimately facilitating the creation of more targeted and personalized therapies [111].
Acknowledgement: We would like to extend our heartfelt appreciation to Mr. Chenying He and Mr. Zhenjun Xi for their invaluable contributions to the data collection and interpretation. Their meticulous efforts ensured the accuracy and reliability of our research findings.
Funding Statement: This research was funded by 2024 Scientific Research Project of Chongqing Medical and Pharmaceutical College (No. ygzrc2024101), Chongqing Education Commission Natural Science Foundation (No. KJQN202402821), Chongqing Shapingba District Science and Technology Bureau Project (No. 2024071), and 2024 Chongqing Medical and Pharmaceutical College Innovation Research Group Project (No. ygz2024401) and Chongqing Science and Health Joint Medical Research Project (No. 2024SQKWLHMS051), respectively.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Xiaobing Li, Jingxin Mao, Xuemei Li; draft manuscript preparation: Xiaobing Li, Yongsheng Liu, Limin Wei, Li Rao, Jingxin Mao, Xuemei Li; review and editing: Xiaobing Li, Jingxin Mao, Xuemei Li; visualization: Xiaobing Li, Yongsheng Liu, Limin Wei, Li Rao, Jingxin Mao, Xuemei Li; supervision: Jingxin Mao, Xuemei Li. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data that support the findings of this study are available from the corresponding authors, Jingxin Mao and Xuemei Li, upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Wood V, Lock A, Harris MA, Rutherford K, Bahler J, Oliver SG. Hidden in plain sight: what remains to be discovered in the eukaryotic proteome? Open Biol. 2019;9(2):180241. doi:10.1098/rsob.180241. [Google Scholar] [PubMed] [CrossRef]
2. Lahue C, Madden AA, Dunn RR, Smukowski Heil C. History and domestication of Saccharomyces cerevisiae in bread baking. Front Genet. 2020;11:584718. doi:10.3389/fgene.2020.584718. [Google Scholar] [PubMed] [CrossRef]
3. Gaikani HK, Stolar M, Kriti D, Nislow C, Giaever G. From beer to breadboards: yeast as a force for biological innovation. Genome Biol. 2024;25(1):10. doi:10.1186/s13059-023-03156-9. [Google Scholar] [PubMed] [CrossRef]
4. Vanderwaeren L, Dok R, Voordeckers K, Nuyts S, Verstrepen KJ. Saccharomyces cerevisiae as a model system for eukaryotic cell biology, from cell cycle control to DNA damage response. Int J Mol Sci. 2022;23(19):11665. doi:10.3390/ijms231911665. [Google Scholar] [PubMed] [CrossRef]
5. Madhavan A, Arun KB, Sindhu R, Krishnamoorthy J, Reshmy R, Sirohi R, et al. Customized yeast cell factories for biopharmaceuticals: from cell engineering to process scale up. Microb Cell Fact. 2021;20(1):124. doi:10.1186/s12934-021-01617-z. [Google Scholar] [PubMed] [CrossRef]
6. Shi S, Chen Y, Nielsen J. Metabolic engineering of yeast. Annu Rev Biophys. 2025. doi:10.1146/annurev-biophys-070924-103134. [Google Scholar] [PubMed] [CrossRef]
7. Manfrao-Netto JHC, Gomes AMV, Parachin NS. Advances in using hansenula polymorpha as chassis for recombinant protein production. Front Bioeng Biotechnol. 2019;7:94. doi:10.3389/fbioe.2019.00094. [Google Scholar] [PubMed] [CrossRef]
8. Chai L, Che J, Liu X, Wang Z, Qi Q, Hou J. Secretory and metabolic engineering of squalene in Yarrowia lipolytica. Bioresour Technol. 2025;421(12):132171. doi:10.1016/j.biortech.2025.132171. [Google Scholar] [PubMed] [CrossRef]
9. Ingram Z, Patkar A, Oh D, Zhang KK, Chung C, Lin-Cereghino J, et al. Overcoming obstacles in protein expression in the yeast Pichia pastoris: interviews of leaders in the pichia field. Pac J Health. 2021;4(1):2. doi:10.56031/2576-215x.1010. [Google Scholar] [PubMed] [CrossRef]
10. Tullio V. Yeast genomics and its applications in biotechnological processes: what is our present and near future? J Fungi. 2022;8(7):752. doi:10.3390/jof8070752. [Google Scholar] [PubMed] [CrossRef]
11. Popova LG, Khramov DE, Nedelyaeva OI, Volkov VS. Yeast heterologous expression systems for the study of plant membrane proteins. Int J Mol Sci. 2023;24(13):10768. doi:10.3390/ijms241310768. [Google Scholar] [PubMed] [CrossRef]
12. Cherry JM. The Saccharomyces genome database: a tool for discovery. Cold Spring Harb Protoc. 2015;2015(12):pdb.top083840. doi:10.1101/pdb.top083840. [Google Scholar] [PubMed] [CrossRef]
13. Messner CB, Demichev V, Muenzner J, Aulakh SK, Barthel N, Rohl A, et al. The proteomic landscape of genome-wide genetic perturbations. Cell. 2023;186(9):2018–34. doi:10.1016/j.cell.2023.03.026. [Google Scholar] [PubMed] [CrossRef]
14. Bandbe CD, Patil KS, Pathan EK. Tuning fungal promoters for the expression of eukaryotic proteins. World J Microbiol Biotechnol. 2024;40(12):400. doi:10.1007/s11274-024-04198-2. [Google Scholar] [PubMed] [CrossRef]
15. Lang OW, Nash RS, Hellerstedt ST, Engel SR, Project SGD. An introduction to the Saccharomyces genome database (SGD). Methods Mol Biol. 2018;1757(20):21–30. doi:10.1007/978-1-4939-7737-6_2. [Google Scholar] [PubMed] [CrossRef]
16. Douglass AP, Byrne KP, Wolfe KH. The methylotroph gene order browser (MGOB) reveals conserved synteny and ancestral centromere locations in the yeast family Pichiaceae. FEMS Yeast Res. 2019;19(6):foz058. doi:10.1093/femsyr/foz058. [Google Scholar] [PubMed] [CrossRef]
17. Gwon Y, So KK, Chun J, Kim DH. Metabolic engineering of Saccharomyces cerevisiae for the biosynthesis of a fungal pigment from the phytopathogenic fungus Cladosporium phlei. J Biol Eng. 2024;18(1):33. doi:10.1186/s13036-024-00429-0. [Google Scholar] [PubMed] [CrossRef]
18. Schuh BRF, Bernardi A, Daley VL, Fernandes SR, de Freitas JA. Can the supplementation of autolyzed yeast (Saccharomyces cerevisiae) affect the diet digestibility, feeding behavior, levels of blood metabolites, and performance of Dorper x Santa Ines lambs finished in feedlot? Trop Anim Health Prod. 2025;57(1):24. doi:10.1007/s11250-024-04268-5. [Google Scholar] [PubMed] [CrossRef]
19. Stepchenkova EI, Zadorsky SP, Shumega AR, Aksenova AY. Practical approaches for the yeast Saccharomyces cerevisiae genome modification. Int J Mol Sci. 2023;24(15):11960. [Google Scholar]
20. Martinic Cezar T, Mardetko N, Trontel A, Paic A, Slavica A, Teparic R, et al. Engineering Saccharomyces cerevisiae for the production of natural osmolyte glucosyl glycerol from sucrose and glycerol through Ccw12-based surface display of sucrose phosphorylase. J Biol Eng. 2024;18(1):69. doi:10.1186/s13036-024-00468-7. [Google Scholar] [PubMed] [CrossRef]
21. Praveen M, Brogi S. Microbial fermentation in food and beverage industries: innovations, challenges, and opportunities. Foods. 2025;14(1):114. doi:10.3390/foods14010114. [Google Scholar] [PubMed] [CrossRef]
22. Heslop R, Gao M, Brito Lira A, Sternlieb T, Loock M, Sanghi SR, et al. Genome-wide libraries for protozoan pathogen drug target screening using yeast surface display. ACS Infect Dis. 2023;9(5):1078–91. doi:10.1021/acsinfecdis.2c00568. [Google Scholar] [PubMed] [CrossRef]
23. Maneira C, Chamas A, Lackner G. Engineering Saccharomyces cerevisiae for medical applications. Microb Cell Fact. 2025;24(1):12. doi:10.1186/s12934-024-02625-5. [Google Scholar] [PubMed] [CrossRef]
24. Waneskog M, Hoch-Schneider EE, Garg S, Kronborg Cantalapiedra C, Schafer E, Krogh Jensen M, et al. Accurate phenotype-to-genotype mapping of high-diversity yeast libraries by heat-shock-electroporation (HEEL). mBio. 2024;16(2):e0319724. doi:10.1128/mbio.03197-24. [Google Scholar] [PubMed] [CrossRef]
25. Torres-Garcia S, Huang Y, Dewornu FS, Tong P, Yeboah R, Allshire R, et al. Genome-wide profiling of histone modifications in fission yeast using CUT&Tag. Methods Mol Biol. 2025;2862(19):309–20. doi:10.1007/978-1-0716-4168-2_22. [Google Scholar] [PubMed] [CrossRef]
26. Qi Q, Li F, Yu R, Engqvist MKM, Siewers V, Fuchs J, et al. Different routes of protein folding contribute to improved protein production in Saccharomyces cerevisiae. mBio. 2020;11(6):e02743–20. doi:10.1128/mBio.02743-20. [Google Scholar] [PubMed] [CrossRef]
27. Tristan-Manzano M, Justicia-Lirio P, Maldonado-Perez N, Cortijo-Gutierrez M, Benabdellah K, Martin F. Externally-controlled systems for immunotherapy: from bench to bedside. Front Immunol. 2020;11:2044. doi:10.3389/fimmu.2020.02044. [Google Scholar] [PubMed] [CrossRef]
28. Grosjean N, Le Jean M, Ory J, Blaudez D. Yeast deletomics to uncover gadolinium toxicity targets and resistance mechanisms. Microorganisms. 2023;11(8):2113. doi:10.3390/microorganisms11082113. [Google Scholar] [PubMed] [CrossRef]
29. Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, et al. Life with 6000 genes. Science. 1996;274(5287):546, 63–7. doi:10.1126/science.274.5287.546. [Google Scholar] [PubMed] [CrossRef]
30. Stojkovic L, Gligorovski V, Geramimanesh M, Labagnara M, Rahi SJ. Automated plasmid design for marker-free genome editing in budding yeast. G3: Genes Genom Genet. 2024;2016(6):jkae297. doi:10.1093/g3journal/jkae297. [Google Scholar] [PubMed] [CrossRef]
31. Alcover-Sanchez B, Garcia-Martin G, Paleo-Garcia V, Quintas A, Dopazo A, Gruart A, et al. R-Ras1 and R-Ras2 regulate mature oligodendrocyte subpopulations. Glia. 2025;73(4):701–19. doi:10.1002/glia.24643. [Google Scholar] [PubMed] [CrossRef]
32. Muthye V, Lavrov DV. Multiple losses of MSH1, gain of mtMutS, and other changes in the MutS family of DNA repair proteins in animals. Genome Biol Evol. 2021;13(9):evab191. doi:10.1093/gbe/evab191. [Google Scholar] [PubMed] [CrossRef]
33. Fromont-Racine M, Khanna V, Jacquier A, Badis G. YLR419W is the homolog of the mammalian translation initiation factor DHX29. MicroPubl Biol. 2024. doi:10.17912/micropub.biology.001112. [Google Scholar] [PubMed] [CrossRef]
34. Gardner EC, Tramont C, Bachanova P, Wang C, Do H, Boutz DR, et al. Engineering a human P2X2 receptor with altered ligand selectivity in yeast. J Biol Chem. 2024;300(5):107248. doi:10.1016/j.jbc.2024.107248. [Google Scholar] [PubMed] [CrossRef]
35. Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, et al. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116(1):121–37. doi:10.1016/S0092-8674(03)01035-3. [Google Scholar] [PubMed] [CrossRef]
36. Pawlowski PH, Szczesny P, Rempola B, Poznanska A, Poznanski J. Combined in silico and 19F NMR analysis of 5-fluorouracil metabolism in yeast at low ATP conditions. Biosci Rep. 2019;39(12):BSR20192847. doi:10.1042/BSR20192847. [Google Scholar] [PubMed] [CrossRef]
37. Szachnowski U, Sallou O, Boudet M, Bretaudeau A, Wery M, Morillon A, et al. The 5-fluorouracil RNA expression viewer (5-FU(R)) facilitates interpreting the effects of drug treatment and RRP6 deletion on the transcriptional landscape in yeast. Yeast. 2024;41(10):629–40. doi:10.1002/yea.3982. [Google Scholar] [PubMed] [CrossRef]
38. Zhang J. Use of a yeast three-hybrid system to clone bridging proteins. Methods Enzymol. 2000;328:103–10. doi:10.1016/s0076-6879(00)28393-6. [Google Scholar] [PubMed] [CrossRef]
39. Wang L, Deng A, Zhang Y, Liu S, Liang Y, Bai H, et al. Efficient CRISPR-Cas9 mediated multiplex genome editing in yeasts. Biotechnol Biofuels. 2018;11(1):277. doi:10.1186/s13068-018-1271-0. [Google Scholar] [PubMed] [CrossRef]
40. Tavakoli K, Pour-Aboughadareh A, Kianersi F, Poczai P, Etminan A, Shooshtari L. Applications of CRISPR-Cas9 as an advanced genome editing system in life sciences. BioTech. 2021;10(3):14. doi:10.3390/biotech10030014. [Google Scholar] [PubMed] [CrossRef]
41. Meng J, Qiu Y, Shi S. CRISPR/Cas9 systems for the development of Saccharomyces cerevisiae cell factories. Front Bioeng Biotechnol. 2020;8:594347. doi:10.3389/fbioe.2020.594347. [Google Scholar] [PubMed] [CrossRef]
42. Bruckner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10(6):2763–88. doi:10.3390/ijms10062763. [Google Scholar] [PubMed] [CrossRef]
43. Rajeshkannan, Mahilkar A, Saini S. GAL regulon in the yeast S. cerevisiae is highly evolvable via acquisition in the coding regions of the regulatory elements of the network. Front Mol Biosci. 2022;9:801011. doi:10.3389/fmolb.2022.801011. [Google Scholar] [PubMed] [CrossRef]
44. Masuda R, Phyu Thant KP, Kawahara K, Oki H, Kadonosono T, Kobayashi Y, et al. A yeast two-hybrid system to obtain triple-helical ligands from combinatorial random peptide libraries. J Biol Chem. 2024;300(11):107794. doi:10.1016/j.jbc.2024.107794. [Google Scholar] [PubMed] [CrossRef]
45. Cluet D, Amri I, Vergier B, Leault J, Audibert A, Grosjean C, et al. A quantitative tri-fluorescent yeast two-hybrid system: from flow cytometry to in cellula affinities. Mol Cell Proteomics. 2020;19(4):701–15. doi:10.1074/mcp.TIR119.001692. [Google Scholar] [PubMed] [CrossRef]
46. Huang S, Zhang H, Chen W, Wang J, Wu Z, He M, et al. Screening of tnfaip1-interacting proteins in zebrafish embryonic cDNA libraries using a yeast two-hybrid system. Curr Issues Mol Biol. 2023;45(10):8215–26. doi:10.3390/cimb45100518. [Google Scholar] [PubMed] [CrossRef]
47. Watanabe S, Kanno Y, Seo M. Screening of ABA transporters by a yeast two-hybrid system-based screening using the receptor complex as a sensor. Methods Mol Biol. 2022;2462:85–99. doi:10.1007/978-1-0716-2156-1. [Google Scholar] [CrossRef]
48. Cho SK, Hannapel DJ. The yeast three-hybrid system for screening RNA-binding proteins in plants. Methods Mol Biol. 2018;1794:207–24. doi:10.1007/978-1-4939-7871-7. [Google Scholar] [CrossRef]
49. Hackney JF, Broatch JE, Dallal RA, Brotherson C, Livingston S, Sabir Z, et al. Rexinoids induce differential gene expression in human glioblastoma cells and protein-protein interactions in a yeast two-hybrid system. ACS Chem Neurosci. 2024;15(15):2897–915. doi:10.1021/acschemneuro.4c00286. [Google Scholar] [PubMed] [CrossRef]
50. Zhu L, Li Y, Qiu L, Chen X, Guo B, Li H, et al. Screening of genes encoding proteins that interact with Nrf2: probing a cDNA library from Mytilus coruscus using a yeast two-hybrid system. Fish Shellfish Immunol. 2023;142(1):109112. doi:10.1016/j.fsi.2023.109112. [Google Scholar] [PubMed] [CrossRef]
51. Conde JN. Yeast two-hybrid system for mapping novel dengue protein interactions. Methods Mol Biol. 2022;2409:119–32. doi:10.1007/978-1-0716-1879-0. [Google Scholar] [CrossRef]
52. Jia Y, Kowalski P, Lopez I. Using yeast two-hybrid system and molecular dynamics simulation to detect venom protein-protein interactions. Curr Res Toxicol. 2021;2(Suppl. 13):93–8. doi:10.1016/j.crtox.2021.02.006. [Google Scholar] [PubMed] [CrossRef]
53. Murphy RO, Beckmann JF. Using baker’s yeast to determine functions of novel wolbachia (and other prokaryotic) effectors. Methods Mol Biol. 2024;2739:321–36. doi:10.1007/978-1-0716-3553-7. [Google Scholar] [CrossRef]
54. Verheggen C, Bertrand E. CRM1 plays a nuclear role in transporting snoRNPs to nucleoli in higher eukaryotes. Nucleus. 2012;3(2):132–7. doi:10.4161/nucl.19266. [Google Scholar] [PubMed] [CrossRef]
55. Lin JS, Lai EM. Protein-protein interactions: yeast two hybrid. Methods Mol Biol. 2024;2715:235–46. doi:10.1007/978-1-0716-3445-5. [Google Scholar] [CrossRef]
56. Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82(3):495–506. doi:10.1016/0092-8674(95)90438-7. [Google Scholar] [PubMed] [CrossRef]
57. Fananas-Pueyo I, Anhel AM, Goni-Moreno A, Onate-Sanchez L, Carrera-Castano G. Workflow to select functional promoter DNA baits and screen arrayed gene libraries in yeast. Curr Protoc. 2024;4(11):e70059. doi:10.1002/cpz1.70059. [Google Scholar] [PubMed] [CrossRef]
58. Badura J, Bernardi B, Muno-Bender J, Matti K, Zimmer K, Wendland J. Isolation and characterization of haploid heterothallic beer yeasts. Appl Microbiol Biotechnol. 2025;109(1):17. doi:10.1007/s00253-024-13397-8. [Google Scholar] [PubMed] [CrossRef]
59. Bigey F, Menatong Tene X, Wessner M, Pradal M, Aury JM, Cruaud C, et al. Differential adaptation of the yeast Candida anglica to fermented food. Food Microbiol. 2024;123:104584. doi:10.1016/j.fm.2024.104584. [Google Scholar] [PubMed] [CrossRef]
60. Cheng H, Chen L, Huang C. Advances of signal transducer and activator of transcription 3 inhibitors in acute myeloid leukemia (Review). Oncol Lett. 2025;29(3):134. doi:10.3892/ol.2025.14881. [Google Scholar] [PubMed] [CrossRef]
61. Xia S, Huo X, Zheng C, Chen J. Yeast two-hybrid assay for investigating antiviral innate immunity. Methods Mol Biol. 2025;2854:213–20. doi:10.1007/978-1-0716-4108-8_21. [Google Scholar] [PubMed] [CrossRef]
62. Zhang Y, Wu BJ, Yu X, Luo P, Ye H, Yu Y, et al. Development of an antigen-antibody co-display system for detecting interaction of G-protein-coupled receptors and single-chain variable fragments. Int J Mol Sci. 2021;22(9):4711. doi:10.3390/ijms22094711. [Google Scholar] [PubMed] [CrossRef]
63. Mehta N, Maddineni S, Kelly RL, Lee RB, Hunter SA, Silberstein JL, et al. An engineered antibody binds a distinct epitope and is a potent inhibitor of murine and human VISTA. Sci Rep. 2020;10(1):15171. doi:10.1038/s41598-020-71519-4. [Google Scholar] [PubMed] [CrossRef]
64. Su J, Zhu B, Inoue A, Oyama H, Morita I, Dong J, et al. The patrol yeast: a new biosensor armed with antibody-receptor chimera detecting a range of toxic substances associated with food poisoning. Biosens Bioelectron. 2023;219:114793. doi:10.1016/j.bios.2022.114793. [Google Scholar] [PubMed] [CrossRef]
65. Chaturvedi S, Sarethy IP. GC-MS-based metabolite fingerprinting reveals the presence of novel anticancer compounds in the microcolonial fungus Aureobasidium sp. TD-062 from the under-explored Thar Desert. Nat Prod Res. 2024;1(4):1–7. doi:10.1080/14786419.2024.2418450. [Google Scholar] [PubMed] [CrossRef]
66. Cabello-Donayre M, Cabello-Donayre I, Guerra D, Orrego LM, Morales JC, Cautain B, et al. A yeast-based high-throughput screen identifies inhibitors of trypanosomatid HRG heme transporters with potent leishmanicidal and trypanocidal activity. Int J Antimicrob Agents. 2024;63(3):107092. doi:10.1016/j.ijantimicag.2024.107092. [Google Scholar] [PubMed] [CrossRef]
67. Zhu J, Kahn CR. Analysis of a peptide hormone-receptor interaction in the yeast two-hybrid system. Proc Natl Acad Sci U S A. 1997;94(24):13063–8. doi:10.1073/pnas.94.24.13063. [Google Scholar] [PubMed] [CrossRef]
68. Glazer B. Building a high-throughput yeast two-hybrid assay to screen for novel chronic hepatitis B drugs [dissertation/master’s thesis]. Appleton, WI, USA: Lawrence University; 2023. [Google Scholar]
69. Raeeszadeh-Sarmazdeh M, Boder ET. Yeast surface display: new opportunities for a time-tested protein engineering system. Methods Mol Biol. 2022;2491:3–25. doi:10.1007/978-1-0716-2285-8. [Google Scholar] [CrossRef]
70. Teymennet-Ramirez KV, Martinez-Morales F, Trejo-Hernandez MR. Yeast surface display system: strategies for improvement and biotechnological applications. Front Bioeng Biotechnol. 2021;9:794742. doi:10.3389/fbioe.2021.794742. [Google Scholar] [PubMed] [CrossRef]
71. Xu C, Meng X, Chai P, Liu H, Duan Z, Tsai YH, et al. Directed evolution of multicyclic peptides using yeast display for sensitive and selective fluorescent analysis of CD28 on the cell surface. Anal Chem. 2025;97(7):4031–40. doi:10.1021/acs.analchem.4c05681. [Google Scholar] [PubMed] [CrossRef]
72. Teparic R, Lozancic M, Mrsa V. Evolutionary overview of molecular interactions and enzymatic activities in the yeast cell walls. Int J Mol Sci. 2020;21(23):8996. doi:10.3390/ijms21238996. [Google Scholar] [PubMed] [CrossRef]
73. Wang EY, Dai Y, Rosen CE, Schmitt MM, Dong MX, Ferre EMN, et al. High-throughput identification of autoantibodies that target the human exoproteome. Cell Rep Methods. 2022;2(2):100172. doi:10.1016/j.crmeth.2022.100172. [Google Scholar] [PubMed] [CrossRef]
74. Orr BA, Carr LM, Wittrup KD, Roy EJ, Kranz DM. Rapid method for measuring ScFv thermal stability by yeast surface display. Biotechnol Prog. 2003;19(2):631–8. doi:10.1021/bp0200797. [Google Scholar] [PubMed] [CrossRef]
75. Li Y, Wang X, Zhou NY, Ding J. Yeast surface display technology: mechanisms, applications, and perspectives. Biotechnol Adv. 2024;76:108422. doi:10.1016/j.biotechadv.2024.108422. [Google Scholar] [PubMed] [CrossRef]
76. Clarke T, Du P, Kumar S, Okitsu SL, Schuette M, An Q, et al. Autoantibody repertoire characterization provides insight into the pathogenesis of monogenic and polygenic autoimmune diseases. Front Immunol. 2023;14:1106537. doi:10.3389/fimmu.2023.1106537. [Google Scholar] [PubMed] [CrossRef]
77. Savvateeva EN, Yukina MY, Nuralieva NF, Filippova MA, Gryadunov DA, Troshina EA. Multiplex autoantibody detection in patients with autoimmune polyglandular syndromes. Int J Mol Sci. 2021;22(11):5502. doi:10.3390/ijms22115502. [Google Scholar] [PubMed] [CrossRef]
78. Lopez-Morales J, Vanella R, Utzinger T, Schittny V, Hirsiger J, Osthoff M, et al. Multiplexed on-yeast serological assay for immune escape screening of SARS-CoV-2 variants. iScience. 2023;26(5):106648. doi:10.1016/j.isci.2023.106648. [Google Scholar] [PubMed] [CrossRef]
79. Miserez A, Yu J, Mohammadi P. Protein-based biological materials: molecular design and artificial production. Chem Rev. 2023;123(5):2049–111. doi:10.1021/acs.chemrev.2c00621. [Google Scholar] [PubMed] [CrossRef]
80. Buda De Cesare G, Sauer FM, Kolecka A, Stavrou AA, Verrips TC, Boekhout T, et al. The development of single-domain VHH nanobodies that target the Candida albicans cell surface. Microbiol Spectr. 2024;12(11):e0426923. doi:10.1128/spectrum.04269-23. [Google Scholar] [PubMed] [CrossRef]
81. Brooks N, McHugh PJ, Lee M, Hartley JA. Alteration in the choice of DNA repair pathway with increasing sequence selective DNA alkylation in the minor groove. Chem Biol. 2000;7(9):659–68. doi:10.1016/s1074-5521(00)00010-7. [Google Scholar] [PubMed] [CrossRef]
82. Bhattacharya S, Esquivel BD, White TC. Overexpression or deletion of ergosterol biosynthesis genes alters doubling time, response to stress agents, and drug susceptibility in Saccharomyces cerevisiae. mBio. 2018;9(4):e01291–18. doi:10.1128/mBio.01291-18. [Google Scholar] [PubMed] [CrossRef]
83. Li X, Cordat E, Schmitt MJ, Becker B. Boosting endoplasmic reticulum folding capacity reduces unfolded protein response activation and intracellular accumulation of human kidney anion exchanger 1 in Saccharomyces cerevisiae. Yeast. 2021;38(9):521–34. doi:10.1002/yea.3652. [Google Scholar] [PubMed] [CrossRef]
84. Sarder HAM, Li X, Funaya C, Cordat E, Schmitt MJ, Becker B.Saccharomyces cerevisiae: first steps to a suitable model system to study the function and intracellular transport of human kidney anion exchanger 1. mSphere. 2020;5(1):e00802–19. doi:10.1128/mSphere.00802-19. [Google Scholar] [PubMed] [CrossRef]
85. Yang J, Wang R, Ren N, Zhao DG, Huang X, Zhao Y. Exogenous application of Eucommia ulmoides β-1, 4-glucanase promotes propagation by increasing the expression of wound healing genes. Sci Rep. 2024;14(1):30398. doi:10.1038/s41598-024-81695-2. [Google Scholar] [PubMed] [CrossRef]
86. Lin X, Ding W, Zheng S, Wu L, Chen X, Xie C, et al. Novel transcriptional regulation of the GAP promoter in Pichia pastoris towards high expression of heterologous proteins. Microb Cell Fact. 2024;23(1):206. doi:10.1186/s12934-024-02435-9. [Google Scholar] [PubMed] [CrossRef]
87. Wang X, Li Y, Jin Z, Liu X, Gao X, Guo S, et al. A novel CRISPR/Cas9 system with high genomic editing efficiency and recyclable auxotrophic selective marker for multiple-step metabolic rewriting in Pichia pastoris. Synth Syst Biotechnol. 2023;8(3):445–51. doi:10.1016/j.synbio.2023.06.003. [Google Scholar] [PubMed] [CrossRef]
88. Gurkan C, Ellar DJ. Expression of the bacillus thuringiensis Cyt2Aa1 toxin in Pichia pastoris using a synthetic gene construct. Biotechnol Appl Biochem. 2003;38(Pt 1):25–33. doi:10.1042/BA20030017. [Google Scholar] [PubMed] [CrossRef]
89. Castro LS, Lobo GS, Pereira P, Freire MG, Neves MC, Pedro AQ. Interferon-based biopharmaceuticals: overview on the production, purification, and formulation. Vaccines. 2021;9(4):328. doi:10.3390/vaccines9040328. [Google Scholar] [PubMed] [CrossRef]
90. Hamilton SR, Gerngross TU. Glycosylation engineering in yeast: the advent of fully humanized yeast. Curr Opin Biotechnol. 2007;18(5):387–92. doi:10.1016/j.copbio.2007.09.001. [Google Scholar] [PubMed] [CrossRef]
91. Singh A, Narang A. P (AOX1) expression in mixed-substrate continuous cultures of Komagataella phaffii (Pichia pastoris) is completely determined by methanol consumption regardless of the secondary carbon source. Front Bioeng Biotechnol. 2023;11:1123703. doi:10.3389/fbioe.2023.1123703. [Google Scholar] [PubMed] [CrossRef]
92. Arjmand S. Promoters in Pichia pastoris: a toolbox for fine-tuned gene expression. Methods Mol Biol. 2024;2844(4):159–78. doi:10.1007/978-1-0716-4063-0_11. [Google Scholar] [PubMed] [CrossRef]
93. Guo J, Liu H, Xu Y, Li L, Xin C. Ectopic expression of the yeast Mn2+ transporter SMF2 enhances tolerance and resistance to cadmium and arsenic in transgenic Arabidopsis. Int J Phytoremediation. 2024;26(13):2103–12. doi:10.1080/15226514.2024.2373974. [Google Scholar] [PubMed] [CrossRef]
94. Jenjitwanich A, Marx H, Sauer M. Characterization of the metabolism of the yeast Yarrowia lipolytica growing as a biofilm. FEMS Microbes. 2024;5:xtae026. doi:10.1093/femsmc/xtae026. [Google Scholar] [PubMed] [CrossRef]
95. Bouyx C, Schiavone M, Teste MA, Dague E, Sieczkowski N, Julien A, et al. The dual role of amyloid-beta-sheet sequences in the cell surface properties of FLO11-encoded flocculins in Saccharomyces cerevisiae. Elife. 2021;10:e68592. doi:10.7554/eLife.68592. [Google Scholar] [PubMed] [CrossRef]
96. Kraft L, Ribeiro VST, Petroski LP, Herai RH, Peronni KC, Figueiredo DLA, et al. Saprochaete clavata invasive infection: characterization, antifungal susceptibility, and biofilm evaluation of a rare yeast isolated in Brazil. Rev Inst Med Trop Sao Paulo. 2023;65(7):e12. doi:10.1590/S1678-9946202365012. [Google Scholar] [PubMed] [CrossRef]
97. de Carvalho BT, Subotic A, Vandecruys P, Deleu S, Vermeire S, Thevelein JM. Enhancing probiotic impact: engineering Saccharomyces boulardii for optimal acetic acid production and gastric passage tolerance. Appl Environ Microbiol. 2024;90(6):e0032524. doi:10.1128/aem.00325-24. [Google Scholar] [PubMed] [CrossRef]
98. Han B, Yue F, Zhang X, Xu K, Zhang Z, Sun Z, et al. Genetically engineering of Saccharomyces cerevisiae for enhanced oral delivery vaccine vehicle. Fish Shellfish Immunol. 2024;146(2):109425. doi:10.1016/j.fsi.2024.109425. [Google Scholar] [PubMed] [CrossRef]
99. Jia D, Chen S. Yeast paves the way for cancer immunotherapy. Cell Chem Biol. 2025;32(1):9–11. doi:10.1016/j.chembiol.2024.12.011. [Google Scholar] [PubMed] [CrossRef]
100. Grubbe WS, Zhang B, Kauffman A, Bylehn F, Padol K, Jung HG, et al. Structural studies of the IFNlambda4 receptor complex using cryoEM enabled by protein engineering. Nat Commun. 2025;16(1):818. doi:10.1038/s41467-025-56119-y. [Google Scholar] [PubMed] [CrossRef]
101. Li XY, Zhou MH, Zeng DW, Zhu YF, Zhang FL, Liao S, et al. Membrane transport engineering for efficient yeast biomanufacturing. Bioresour Technol. 2024;418(6):131890. doi:10.1016/j.biortech.2024.131890. [Google Scholar] [PubMed] [CrossRef]
102. Soleimani M, Esmaili K, Rahdar A, Aminizadeh M, Cheraqpour K, Tabatabaei SA, et al. From the diagnosis of infectious keratitis to discriminating fungal subtypes; a deep learning-based study. Sci Rep. 2023;13(1):22200. doi:10.1038/s41598-023-49635-8. [Google Scholar] [PubMed] [CrossRef]
103. Xu L, Bai X, Joong Oh E. Strategic approaches for designing yeast strains as protein secretion and display platforms. Crit Rev Biotechnol. 2024;30:1–18. doi:10.1080/07388551.2024.2385996. [Google Scholar] [PubMed] [CrossRef]
104. Qiu Y, Wei GW. Artificial intelligence-aided protein engineering: from topological data analysis to deep protein language models. Brief Bioinform. 2023;24(5):bbad289. doi:10.1093/bib/bbad289. [Google Scholar] [PubMed] [CrossRef]
105. Goold HD, Kroukamp H, Erpf PE, Zhao Y, Kelso P, Calame J, et al. Construction and iterative redesign of synXVI a 903 kb synthetic Saccharomyces cerevisiae chromosome. Nat Commun. 2025;16(1):841. doi:10.1038/s41467-024-55318-3. [Google Scholar] [PubMed] [CrossRef]
106. O.’Riordan NM, Juric V, O’Neill SK, Roche AP, Young PW. A yeast modular cloning (MoClo) toolkit expansion for optimization of heterologous protein secretion and surface display in Saccharomyces cerevisiae. ACS Synth Biol. 2024;13(4):1246–58. doi:10.1021/acssynbio.3c00743. [Google Scholar] [PubMed] [CrossRef]
107. Jach ME, Malm A. Yarrowia lipolytica as an alternative and valuable source of nutritional and bioactive compounds for humans. Molecules. 2022;27(7):2300. doi:10.3390/molecules27072300. [Google Scholar] [PubMed] [CrossRef]
108. Bilal M, Ji L, Xu Y, Xu S, Lin Y, Iqbal HMN, et al. Bioprospecting Kluyveromyces marxianus as a robust host for industrial biotechnology. Front Bioeng Biotechnol. 2022;10:851768. doi:10.3389/fbioe.2022.851768. [Google Scholar] [PubMed] [CrossRef]
109. Petrucco CA, Crocker AW, D.’Alessandro A, Medina EM, Gorman O, McNeill J, et al. Tools for live-cell imaging of cytoskeletal and nuclear behavior in the unconventional yeast, Aureobasidium pullulans. Mol Biol Cell. 2024;35(4):br10. doi:10.1091/mbc.E23-10-0388. [Google Scholar] [PubMed] [CrossRef]
110. Singh AK, Goar H, Vashist N, Sinha P, Bachhawat AK. Efficient assembly of a synthetic attenuated SARS-CoV-2 genome in Saccharomyces cerevisiae using multi-copy yeast vectors. J Genet. 2024;103:9. [Google Scholar]
111. Slavny P, Hegde M, Doerner A, Parthiban K, McCafferty J, Zielonka S, et al. Advancements in mammalian display technology for therapeutic antibody development and beyond: current landscape, challenges, and future prospects. Front Immunol. 2024;15:1469329. doi:10.3389/fimmu.2024.1469329. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF
 Downloads
Downloads
 Citation Tools
Citation Tools