 Open Access
Open Access
ARTICLE
Resveratrol Preserves Mitochondrial DNA Integrity and Long-Term Memory without Decreasing Amyloid-β Levels in Alzheimer’s Disease Mouse Models
1 Department of Genetics, Cytology and Bioengineering, Voronezh State University, Voronezh, 396018, Russia
2 A.N. Belozersky Institute of Physico-Chemical Biology, Lomonosov Moscow State University, Moscow, 119899, Russia
* Corresponding Author: EGOR Y. PLOTNIKOV. Email:
(This article belongs to the Special Issue: Autophagy and Aging: Mechanisms and Implications)
BIOCELL 2025, 49(5), 873-892. https://doi.org/10.32604/biocell.2025.063557
Received 17 January 2025; Accepted 18 April 2025; Issue published 27 May 2025
Abstract
Background: Mitochondrial dysfunction plays a critical role in the pathogenesis of Alzheimer’s disease (AD). Resveratrol is a promising compound for the treatment of various neurodegenerative diseases, including AD. Aims: To investigate mitochondrial damage and the effects of resveratrol on inflammation, cognitive function, and mitochondrial quality control in APP/PS1 mice. Methods: Comparative analysis of mitochondrial DNA (mtDNA) damage was conducted between 10-month-old APP/PS1 mice and age-matched C57BL/6 mice. Assessments included measurement of amyloid-β levels, inflammatory markers, swimming distance in the Morris water maze, and gut microbiome composition. Resveratrol’s effects on cytokine expression, mtDNA levels in plasma, and activation of Nuclear factor erythroid 2-related factor 2/Antioxidant response element (Nrf2/ARE) and phosphoinositide 3-kinase/protein kinase B (also known as Akt)/mechanistic target of rapamycin complex 1 (PI3K/Akt/mTORC1) signaling pathways were also evaluated. Results: APP/PS1 mice exhibited significantly increased mtDNA damage in the prefrontal cortex, midbrain, and cerebellum, alongside higher amyloid-β levels and inflammatory markers. Resveratrol treatment led to reduced expression of pro-inflammatory cytokines, a decrease in Proteobacteria levels, and lower cell-free mtDNA in plasma. Partial improvement in long-term spatial memory was observed in APP/PS1 mice following resveratrol treatment, likely due to its anti-inflammatory properties. Activation of the Nrf2/ARE signaling pathway and markers of PI3K/Akt/mTORC1 axis activation were noted, with the latter regulating long-term potentiation. Conclusion: Resveratrol demonstrates potential in mitigating inflammation and improving mitochondrial quality control in APP/PS1 mice, but it does not reduce amyloid-β levels, highlighting the complexity of AD pathology and the need for further research.Keywords
Cognitive dysfunction refers to specific changes in a human’s mental abilities, including memory impairment, intellectual disability, attention deficits, language disorders, and other impairments. The most severe cases of irreversible cognitive decline fall into the category of dementia [1]. Among various dementia disorders, Alzheimer’s disease (AD) manifests the most severe clinical presentation [2]. The dominant hypothesis attributes its development to progressive amyloid-β peptide aggregation in brain structures [3]. AD is accompanied by mitochondrial dysfunction, which can arise from the direct effects of amyloid-β on mitochondrial proteins and membranes. It is known that there is a decrease in the activity of the Krebs cycle, mitochondrial respiratory chain, and ATP production in the brains of patients with AD [4]. Neuropathological examinations reveal significant mitochondrial genome abnormalities in Alzheimer’s disease. Quantitative analysis shows that cortical mitochondrial DNA (mtDNA) in AD patients below 75 years contains 15 times more deletions than neurologically intact individuals of comparable age [5]. Deletions in mtDNA result in a deficiency of cytochrome c oxidase in the brains of patients with AD [6]. Patients have 63% more mutations than healthy individuals, and notably, these mutations tend to accumulate in the regulatory regions of mtDNA [7]. The number of oxidized bases, especially 8-oxoguanine, was also approximately ten times higher in patients with AD compared to control subjects. Similarly, mtDNA had ten times more oxidative damage than nuclear DNA [8].
Proteolytic processing of APP by β-secretase and γ-secretase generates amyloid-β peptides, with presenilin proteins (PSEN1/2) forming the active site of γ-secretase [9]. AD predominantly (>90%) occurs as sporadic late-onset disease, though rare familial cases result from pathogenic variants in APP or presenilin genes [10]. Although laboratory animals, particularly mice, do not naturally develop AD, transgenic models have been created in which human mutant APP and PSEN1/2 genes are expressed [11]. In the APP/PS1 mouse model, progressive amyloid-β aggregation triggers neuroinflammatory cascades and ultimately leads to cognitive dysfunction [12]. Research indicates that these genetically modified mice develop mitochondrial abnormalities comparable to those found in individuals with AD [13]. It has also been shown that a large number of mitochondrial DNA deletions accumulate in the brains of mice in an AD model expressing an inducible mitochondrial-targeted endonuclease [14]. However, the accumulation of mutations, particularly mtDNA deletions, is not always preceded by the accumulation of oxidative damage [15]. Currently, there is no data on changes in the amount of mtDNA damage in animal models of AD.
Many signaling pathways that regulate mitochondrial metabolism and the clearance of amyloid-β are considered promising targets for pharmacological agents. In particular, nuclear factor erythroid 2-related factor 2 (Nrf2) plays a key role in the adaptive response to oxidative stress and can inhibit ferroptosis, which is a relatively new avenue in Alzheimer’s disease therapy [16]. Nrf2 has a close interplay with other signaling pathways. Specifically, Nrf2 is capable of regulating the expression of the gene that encodes a key component of the mechanistic target of rapamycin complex 1 (mTORC1). mTORC1-dependent pathways for regulating autophagy are also considered interesting targets for Alzheimer’s disease therapy [17]. Furthermore, these pathways are involved in maintaining synaptic plasticity and regulating long-term potentiation, thereby directly influencing cognitive functions [18]. Some polyphenolic compounds, including resveratrol, are considered promising for potentially slowing down the pathogenesis of AD. Resveratrol possesses anti-inflammatory properties and can activate signaling pathways that potentially protect brain mitochondria from damage caused by amyloid-β accumulation [19,20]. However, it should be noted that many polyphenols, including resveratrol, have limited bioavailability as they are largely metabolized in the intestine. The oral absorption of resveratrol in humans is approximately 75% through trans-epithelial diffusion. However, intensive metabolism in the gut and liver results in the bioavailability of resveratrol being significantly less than 1% when taken orally [21]. Resveratrol’s pleiotropic actions necessitate investigation within the gut-brain axis framework, recognizing this intricate signaling system coordinates neural, hormonal, and immunological communication between the intestine and brain. This axis is modulated through interconnected mechanisms involving neuronal signaling, endocrine pathways, immune responses, and metabolic regulation. The effects on the gut microbial composition may have remote consequences, including on inflammatory processes in the brain, and consequently, on cognitive functions [22]. The goal of this study was to investigate the ability of resveratrol to impact cognitive functions in APP/PS1 mice, amyloidogenesis, markers of inflammation, including in the gut microbiome, the level of mtDNA damage in the brain, as well as the expression level of genes that may influence cognitive functions and protect mitochondria from damage.
The experiment involved 6-month-old male and female mice from two groups: wild-type C57BL/6 (n = 7) and transgenic APP/PS1 (n = 13). Their initial weights varied from 27 to 32 g. The C57BL/6 strain was procured from Stolbovaya breeding laboratory (Moscow region, Russia), whereas the APP/PS1 mice were supplied by Pushchino Nursery for Laboratory Animals (Moscow region, Russia). The animals were kept under standardized conditions featuring a 12-h photocycle, constant 25°C ambient temperature, with unlimited access to water and commercial rodent diet (Ssniff-Spezialdiäten GmbH). APP/PS1 mice starting from the age of 6 months were divided into two groups. The first group (n = 7) continued to receive water. The second group of APP/PS1 mice (n = 6) received resveratrol (Sigma-Aldrich, 501-36-0, St. Louis, MA, USA) at a concentration of 20 mg/kg/day with drinking water. This concentration was previously tested in an experiment on middle-aged non-diseased mice [23]. The amount of water consumed with dissolved resveratrol was monitored daily. The treatment lasted for 4 months. Long-term spatial memory function was quantitatively analyzed using the Morris water maze paradigm following the experimental treatment phase. Subsequently, the mice were euthanized for molecular and genetic analyses. Brain tissue was extracted and dissected, with the cerebral cortex isolated for DNA/RNA extraction and protein profiling. Hippocampus, ventral midbrain, thalamus, cerebellum, and mice feces were used for the DNA extraction only.
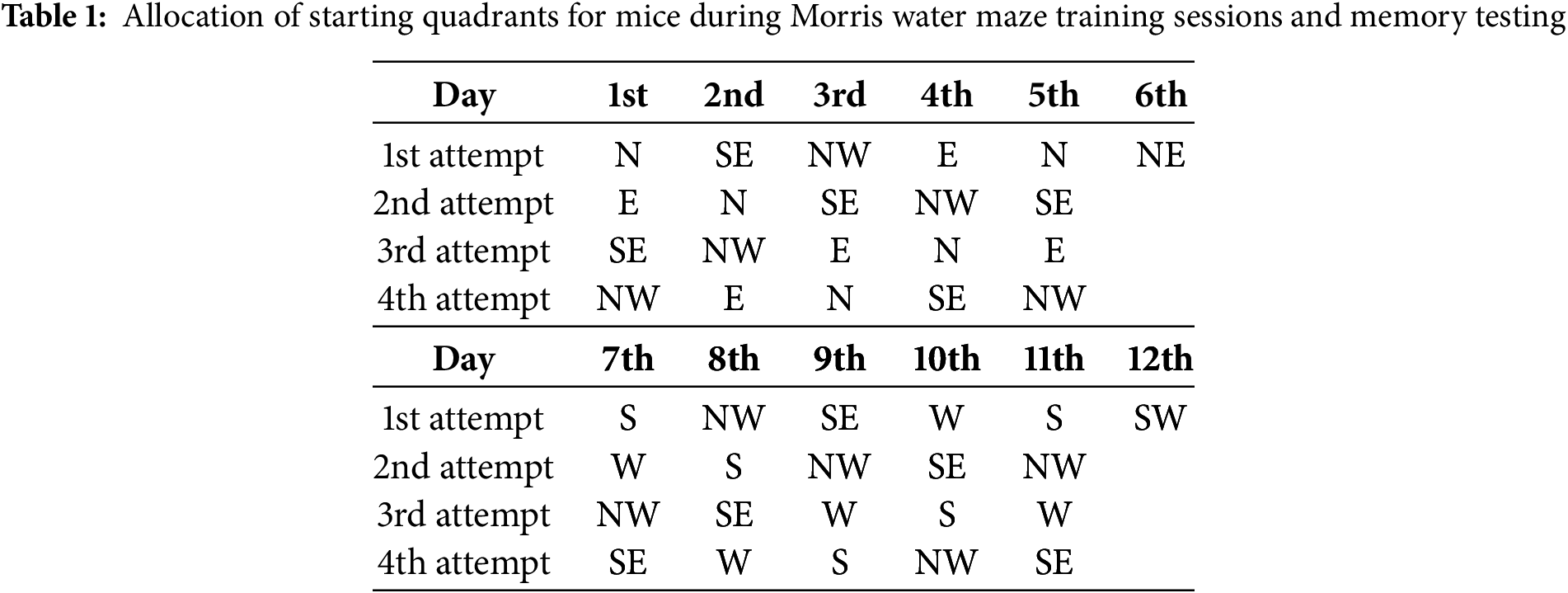
The Morris water maze (MWM) test was employed to evaluate the cognitive functions of mice, following the protocol described by Vorhees and Williams (2006) [24]. The apparatus consisted of a circular pool made of rubber, measuring 147 cm in diameter and 33 cm in height. The pool was filled to approximately half its height with water maintained at room temperature and rendered opaque by the addition of food-grade titanium dioxide dye to obscure the platform from view. The pool was conceptually divided into four quadrants: North (N), South (S), East (E), and West (W). Visual cues, positioned consistently throughout the experiment, were utilized as spatial references, and the experimenter maintained a fixed position to minimize external variables. The assessment of spatial long-term memory was conducted over a 12-day protocol, divided into four distinct phases. During the initial phase (days 1–5), mice underwent training to establish reference memory, with the platform consistently placed in the Southwest (SW) quadrant. Each mouse performed four trials per day, with each trial lasting 60 s. On day 6, a probe trial was conducted, during which the platform remained in the SW quadrant, but each mouse was given a single 60-s trial starting from the Northeast (NE) quadrant (see Table 1). Subsequently, a reversal training phase was implemented over the next five days (days 7–11), wherein the platform was relocated to the opposite Northeast (NE) quadrant. This phase aimed to evaluate the animals’ ability to extinguish the previously learned spatial memory and acquire a new navigational strategy. On day 12, a reversal probe trial was conducted, with the starting position set to the SW quadrant (Table 1). Spatial long-term memory was assessed based on two primary parameters: the latency to locate the platform and the total distance swam by the mice during the search.
Genomic DNA was purified from murine brain regions and fecal samples using the Proba-GS extraction kit (DNA Technology, P-003/1, Moscow, Russia) according to the manufacturer’s protocol. Cortical RNA was isolated in parallel using the ExtractRNA system (Evrogen, BC032, Moscow, Russia). Nucleic acid integrity was verified by electrophoretic separation in 2% agarose/TAE buffer.
2.4 Estimation of the Bacterial Composition of the Gut Microbiome
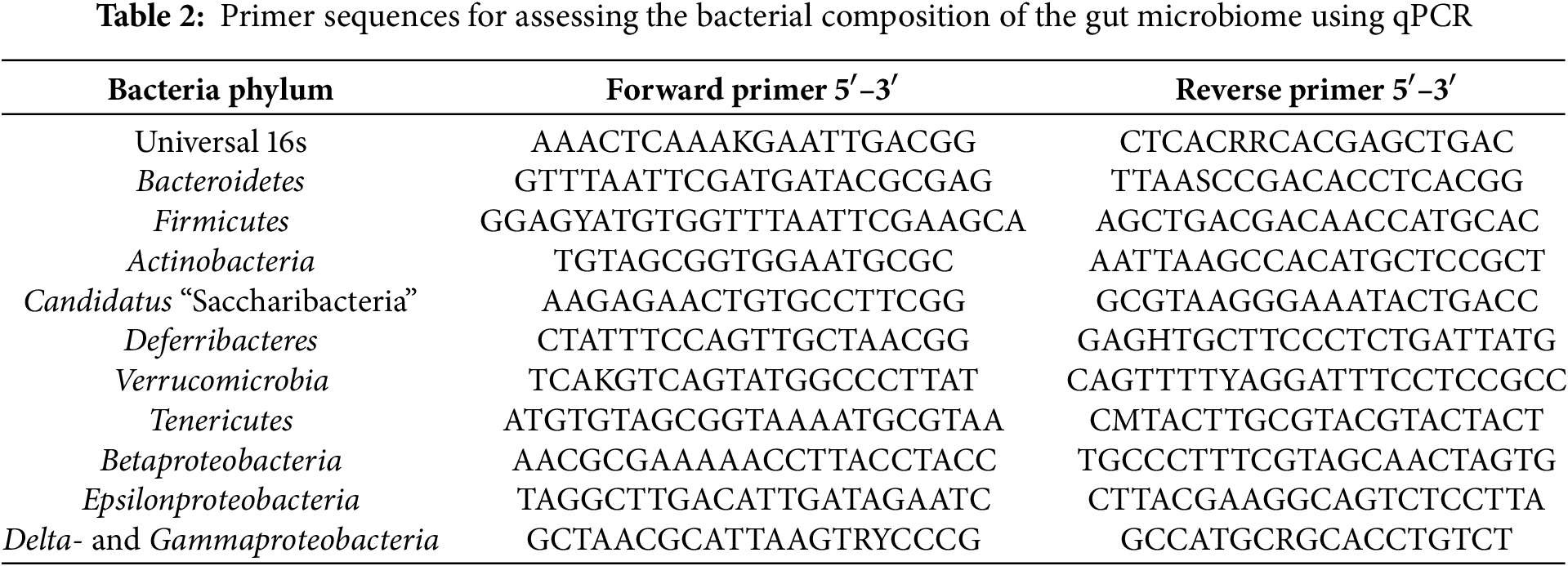
A quantitative PCR (qPCR) technique was employed to evaluate the bacterial composition of the gut microbiome [25]. The qPCR was conducted on a CFX96TM Real-Time System thermocycler (Bio-Rad, C1000, Hercules, CA, USA) using a reaction mixture comprising 1× qPCRmix-HS SYBR (Evrogen, PK147S, Moscow, Russia), a 20 pM concentration of forward and reverse primer combination (Evrogen, SP001, Moscow, Russia), and 10 ng of DNA template. Primer sequences are presented in Table 2. The proportion of bacterial phylum content was calculated using the formula:
2.5 Measurement of the Number of mtDNA Damages
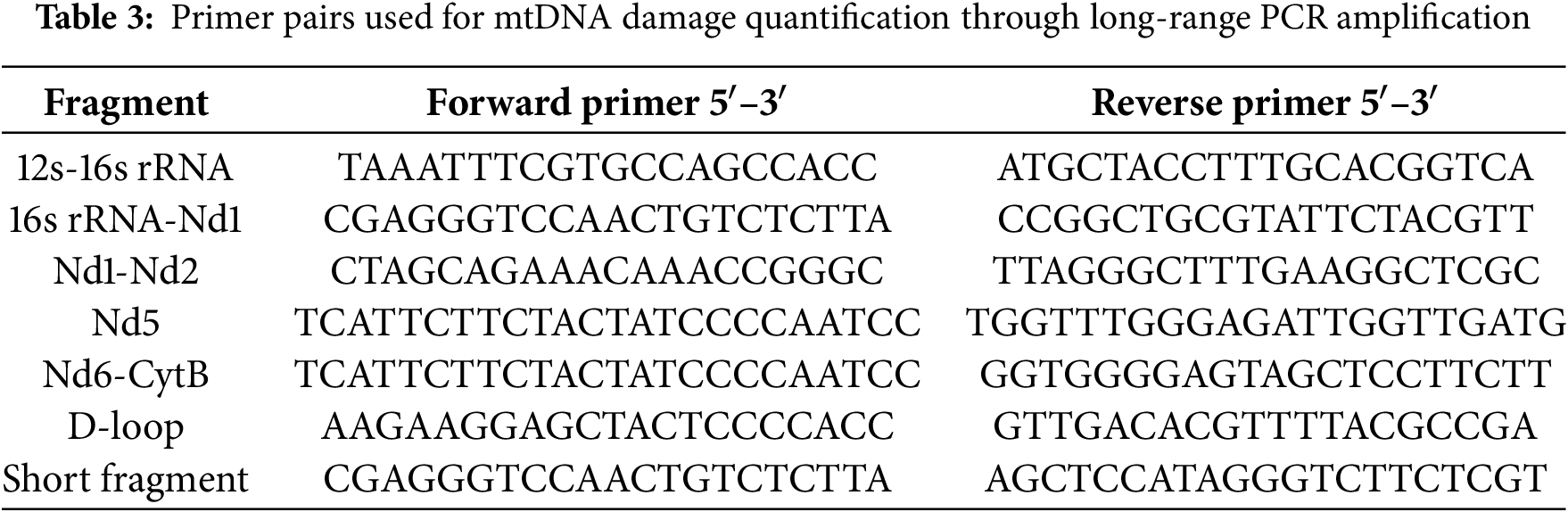
The frequency of mtDNA lesions was determined by measuring amplification efficiency of long mitochondrial fragments using Encyclo polymerase (Evrogen, PK002S, Moscow, Russia) on a Bio-Rad CFX96™ Real-Time PCR System (Bio-Rad, C1000, Hercules, CA, USA), with comparison to short reference amplicons. This approach relies on the principle that DNA lesions—including single-strand breaks, base modifications, or adducts—inhibit DNA polymerase progression, thereby reducing PCR product yield. Consequently, the amplification efficiency of target DNA segments shows an inverse correlation with the level of DNA damage. Specific primer panels were designed for mice, allowing amplification of approximately 2 kb fragments (Table 3). Previous studies have demonstrated that this PCR product size is optimal for assessing the heterogeneity of damage distribution throughout the mtDNA structure. Additionally, increasing the fragment length reduces the efficiency and linearity of PCR, making result interpretation more challenging [26]. Short (~100 bp) fragments were simultaneously amplified and used for normalizing the level of damage to the mtDNA copy number. The primers were carefully selected to minimize non-specific amplification from nuclear pseudogenes, which are abundant in mammalian nuclear DNA [27].
The PCR protocol consisted of an initial denaturation (95°C, 3 min), followed by 35 cycles of: 95°C for 10 s (denaturation), 59°C for 30 s (annealing), and 72°C for 4.5 min (elongation). Relative mtDNA damage was determined by comparing ΔCq values between long and short fragments (reference), with lesion frequency calculated per 10 kb using the formula:
2.6 Gene Expression Estimation
cDNA synthesis was performed using the RIVERTA-L reverse transcription kit (AmpliSens, 2008/03994, Moscow, Russia) on an Eppendorf Mastercycler personal thermocycler (Eppendorf, PF-217186, Enfield, MA, USA). Subsequent quantitative PCR analysis was carried out on a Bio-Rad CFX96™ Real-Time System with qPCRmix-HS SYBR master mix (Evrogen, PK147S, Moscow, Russia). Transcript levels were normalized to Gapdh reference gene expression and quantified according to the comparative Cq (2(−ΔΔCq)) method. Data visualization included heatmaps generated using Bio-Rad CFX Manager software (v2.1). Primer sequences are provided in Table 4.

2.7 Assessment of mtDNA Copy Number in the Plasma
Measurement and quantification of cell-free mtDNA were performed using the method described by Lindqvist et al. (2016) [28]. Blood was collected prior to the mice’s sacrifice. The blood was centrifuged at 1700× g for 5 min to obtain plasma. Isolated DNA was then amplified using a pair of primers specific for amplifying a short fragment of mtDNA (Table 3).
Brain hemispheres were homogenized in PBS (pH 7.4) (Amresco, Am-E404-100, Solon, OH, USA). The homogenization solution also contained 1 mM of the protease inhibitor phenylmethane sulfonyl fluoride (PMSF) (Amresco, G2008-1ML, Solon, OH, USA). The concentration of proteins in the samples was determined using the bicinchoninic acid assay (Sigma, 12352106, St. Louis, MO, USA). The brain homogenate samples were applied to Tris-glycine polyacrylamide gels with a gradient composition of 5%–20%. Each lane of the gels contained a total of 10 µg of protein. Following electrophoretic separation, proteins were transferred to PVDF membranes (Amersham Pharmacia Biotech, 41105339, Amersham, Buckinghamshire, UK) using standard wet transfer conditions. Membranes were subsequently blocked with 5% non-fat dry milk (SERVA, HS: 04021019, Heidelberg, Germany) in PBST with 0.05% Tween-20 (Panreac, 556608.0922, Barcelona, Spain) for 1 h at room temperature, followed by overnight incubation with primary antibodies at 4°C (Table 5).

Following primary antibody incubation, membranes were probed for 1 h at 36°C with species-specific HRP-conjugated secondary antibodies (anti-rabbit IgG (P-GAR Iss) or anti-mouse IgG (P-GAM Iss), 1:5000 dilution; IMTEK, Russia). Protein bands were detected using Advansta Western Bright™ ECL substrate (K-12045-C20, San Jose, CA, USA) and imaged on a Bio-Rad V3 Western Blot Imager system (Bio-Rad, Hercules, CA, USA). Quantitative analysis was performed using Bio-Rad’s Image Lab software for densitometric measurements. B-actin was used as an internal control. Graphs show the signal intensity of bands.
Data analysis was performed using Statistica 12 (StatSoft, Tulsa, OK, USA), with results expressed as mean ± SEM. A minimum of six biological replicates were used for physiological experiments, mtDNA damage assessments, and gut microbiome analyses, while Western blotting experiments included at least three technical replicates. Intergroup comparisons were analyzed using the Kruskal-Wallis non-parametric test, with a statistical significance threshold set at p < 0.05.
3.1 The Effect of Resveratrol on the Cognitive Functions of APP/PS1 Mice
Spatial memory retention was evaluated by quantifying platform search time and path length during probe trials conducted on days 6 and 12, following a 5-day acquisition training period in the Morris water maze. On the 6th day of testing, APP/PS1 mice spent 78% more time searching for the platform compared to wild-type mice. On the 12th day, transgenic mice spent 75% more time searching, but the differences were not statistically significant (Fig. 1). On the 6th test day, the APP/PS1 mice swam a distance 7 times greater than the C57BL/6 mice of the same age (p < 0.001). It is worth noting that the differences in the distance covered while searching for the platform between the C57BL/6 mice and the APP/PS1 mice that received resveratrol for 4 months were not statistically significant (p = 0.051), which may indicate partial improvement in cognitive functions compared to the control transgenic mice. On the 12th test day, the APP/PS1 mice swam a distance 4 times greater than the control C57BL/6 mice (p < 0.05) (Fig. 2A). Representative trajectories of mice for each experimental group on the 6th and 12th testing days are presented in Fig. 2B.
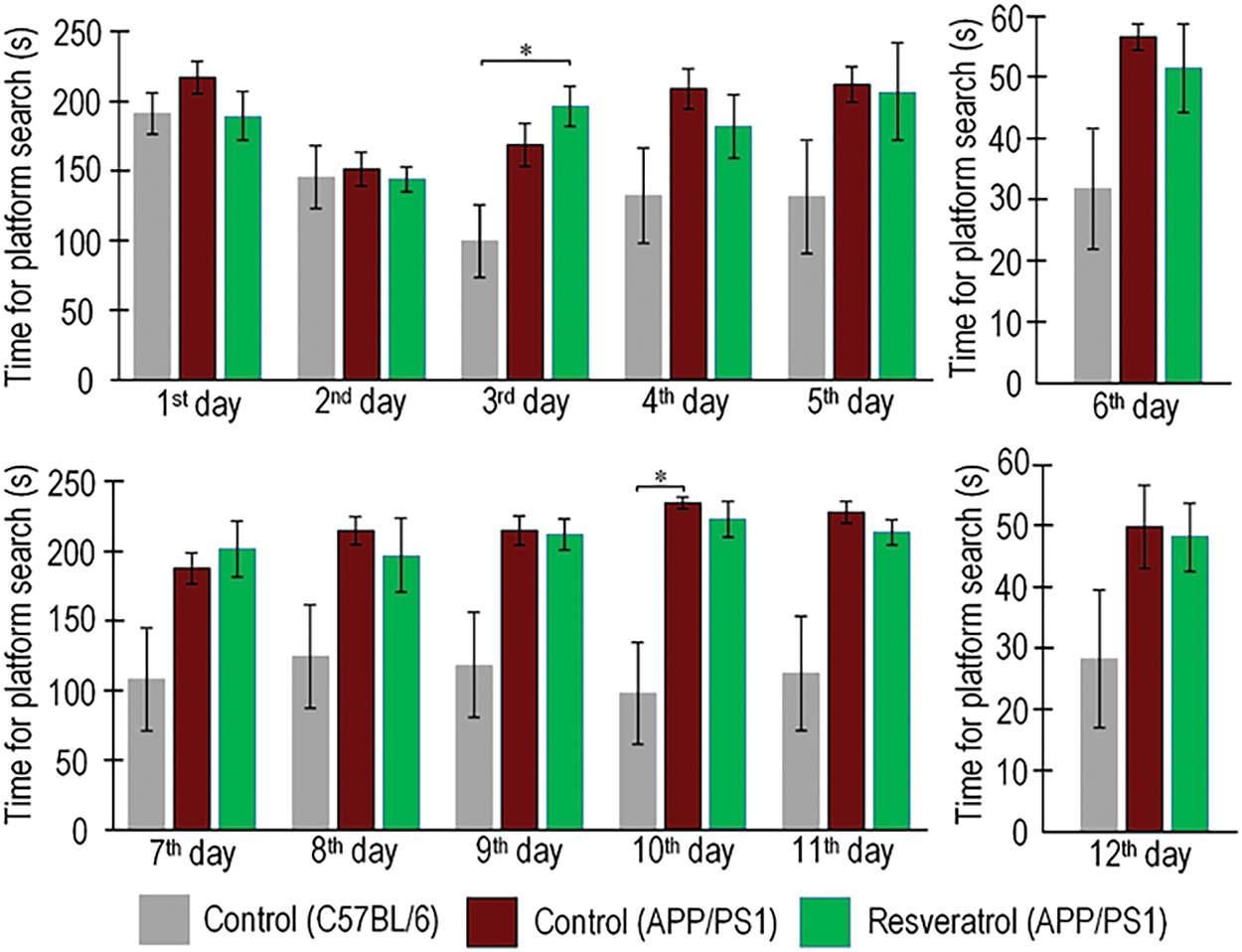
Figure 1: Values of long-term spatial memory in wild-type C57BL/6 mice, transgenic APP/PS1 mice, and transgenic mice administered resveratrol. Time spent by mice searching for the platform during training days from the 1st to 5th day and from 7th to 11th day, as well as during test days 6th and 12th. Control C57BL/6 (n = 7), control (APP/PS1) (n = 7), resveratrol (APP/PS1) (n = 6). Significance of differences between groups: *p < 0.05 (Kruskal-Wallis test)
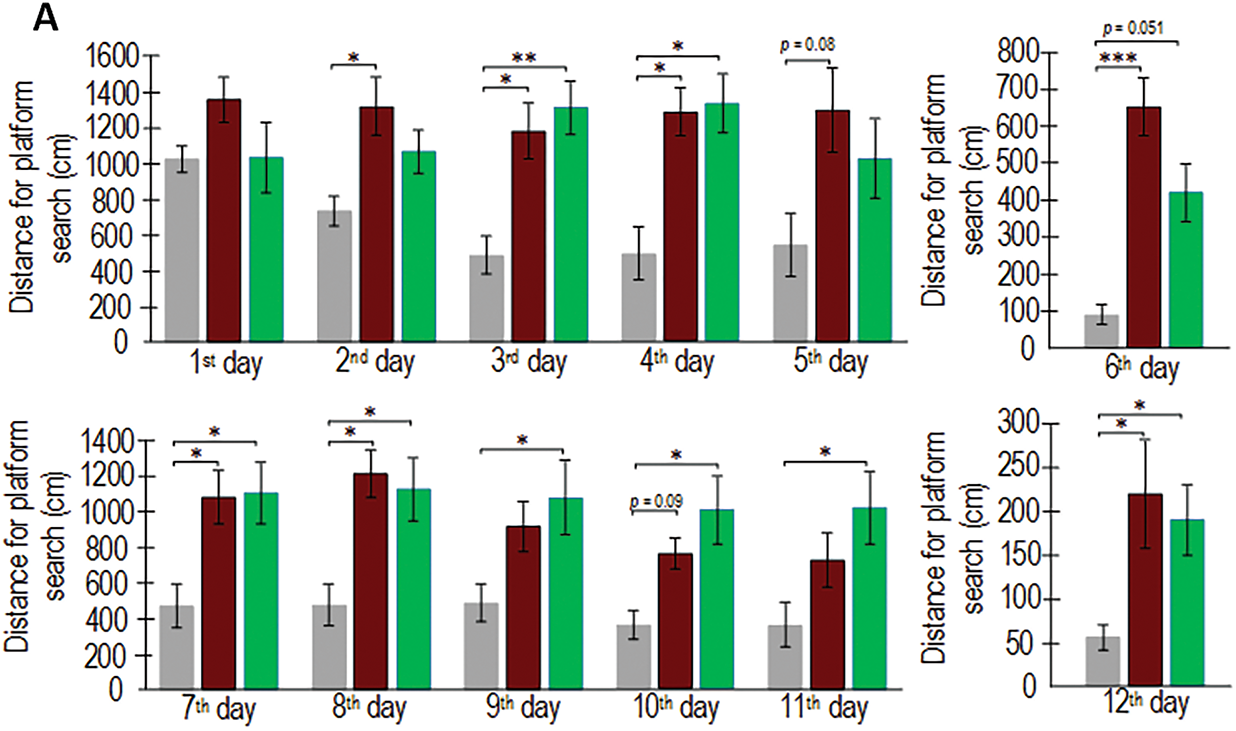
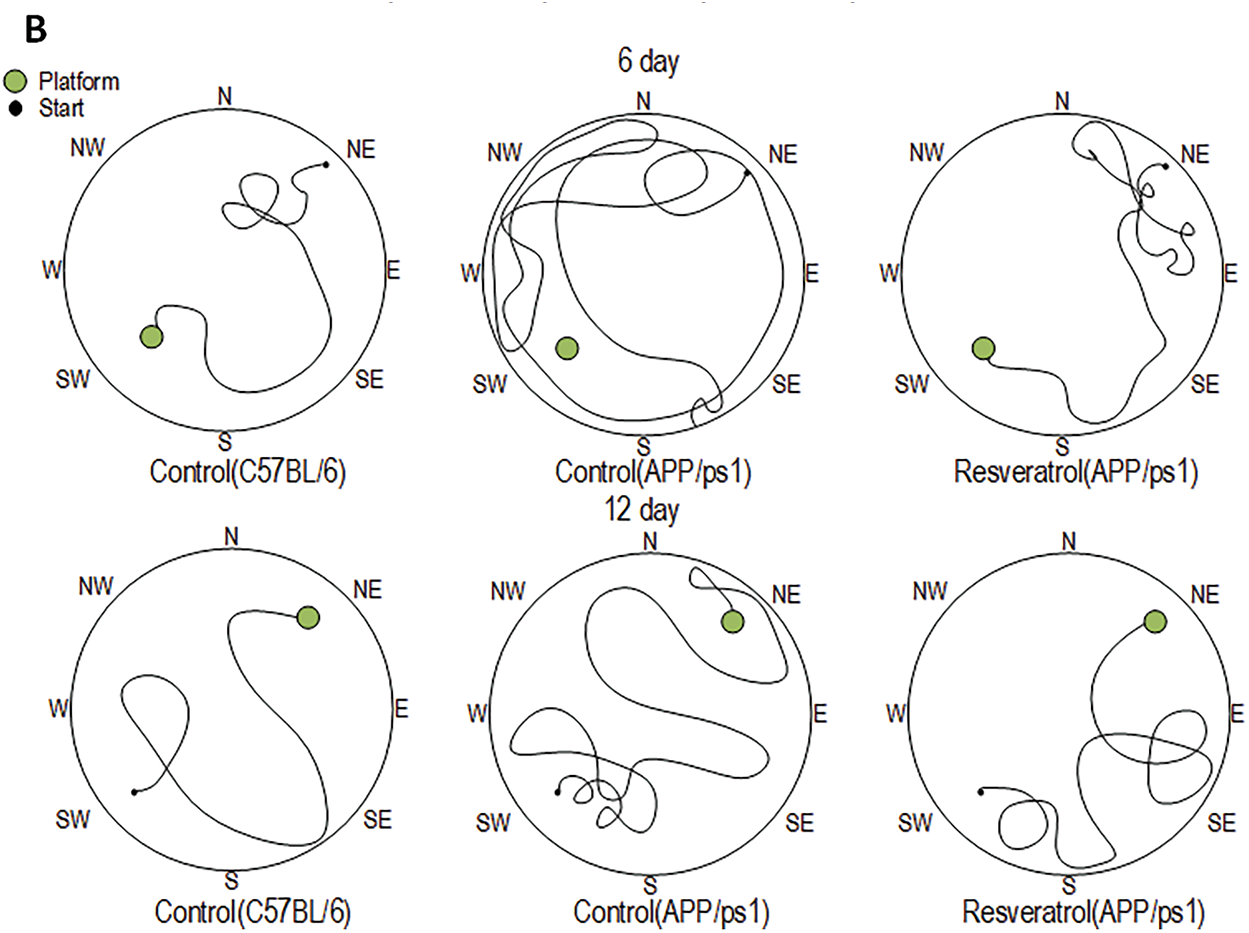
Figure 2: Values of long-term spatial memory in wild-type C57BL/6 mice, transgenic APP/PS1 mice, and transgenic mice administered resveratrol. The distance covered by mice in search of the platform during training days from the 1st to 5th day and from 7th to 11th day, as well as during test days 6th and 12th. Control C57BL/6 (n = 7), control (APP/PS1) (n = 7), resveratrol (APP/PS1) (n = 6) (A). Representative behavioral trajectory during test attempts (B). Significance of differences between groups: *p < 0.05, ** p < 0.01; ***p < 0.001 (Kruskal-Wallis test)
The analysis of the training days yields inconclusive findings. The analysis of the time spent searching for the platform revealed that on the 10th training day, the APP/PS1 mice spent twice as much time as the control C57BL/6 mice of the same age (p < 0.05). However, no statistically significant differences were observed between the resveratrol-treated APP/PS1 mice and the wild-type mice (Fig. 1), partially confirming the hypothesis that resveratrol mitigates cognitive deficits in transgenic mice. On one hand, the distance covered in search of the platform by the resveratrol-treated APP/PS1 mice did not differ from the control C57BL/6 mice on the 2nd and 5th training days, whereas differences were observed between the control C57BL/6 and APP/PS1 mice (Fig. 2). On the other hand, the opposite results were observed on the 9th, 10th, and 11th days, where the resveratrol-treated APP/PS1 mice swam a greater distance in search of the platform compared to the control mice of the same mutant genotype.
3.2 The Amount of mtDNA Damage in Different Brain Regions
MtDNA damage, serving as a biomarker of oxidative stress, demonstrated significant regional variations in APP/PS1 mice compared to C57BL/6 controls. The cerebellum exhibited the most pronounced damage increase (2.3-fold, p < 0.001), followed by the ventral midbrain (+57%, p < 0.05) and prefrontal cortex (+24%, p < 0.05). Resveratrol treatment specifically reduced cortical mtDNA damage by 50% in APP/PS1 mice (p < 0.05), while showing no significant protective effects in other examined brain regions. These results highlight both the region-specific vulnerability to oxidative stress in AD-model mice and the selective neuroprotective capacity of resveratrol (Fig. 3).
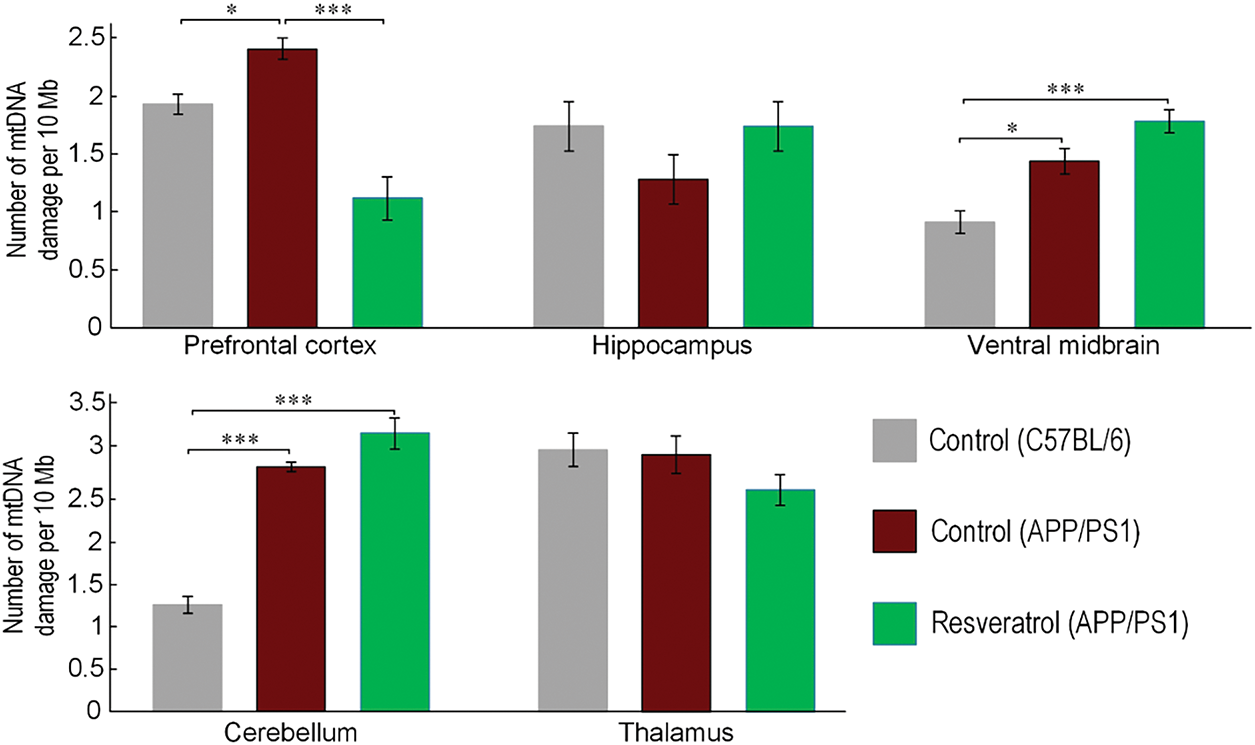
Figure 3: Comparison of the amount of mtDNA damage in different brain regions between C57BL/6 and APP/PS1 mice, as well as mice receiving resveratrol. Control C57BL/6 (n = 7), control (APP/PS1) (n = 7), resveratrol (APP/PS1) (n = 6). Significance of differences between groups: *p < 0.05; ***p < 0.001 (Kruskal-Wallis test)
3.3 The Effect of Resveratrol on Amyloidogenesis
We determined that both experimental groups of 10-month-old APP/PS1 mice exhibited a 44%–50% higher level of amyloid precursor protein (APP) compared to age-matched C57BL/6 mice. Resveratrol did not influence the level of APP in the cortex of transgenic mice (Fig. 4A). In 10-month-old APP/PS1 mice, the level of amyloid-β was 6.7 times higher than in C57BL/6 mice of the same age. In transgenic mice that received resveratrol, the amount of amyloid-β increased by 5.8 times (p < 0.001). Therefore, resveratrol reduced the levels of amyloid-β by 15%, although the differences were not statistically significant between the experimental groups of transgenic mice (Fig. 4B).
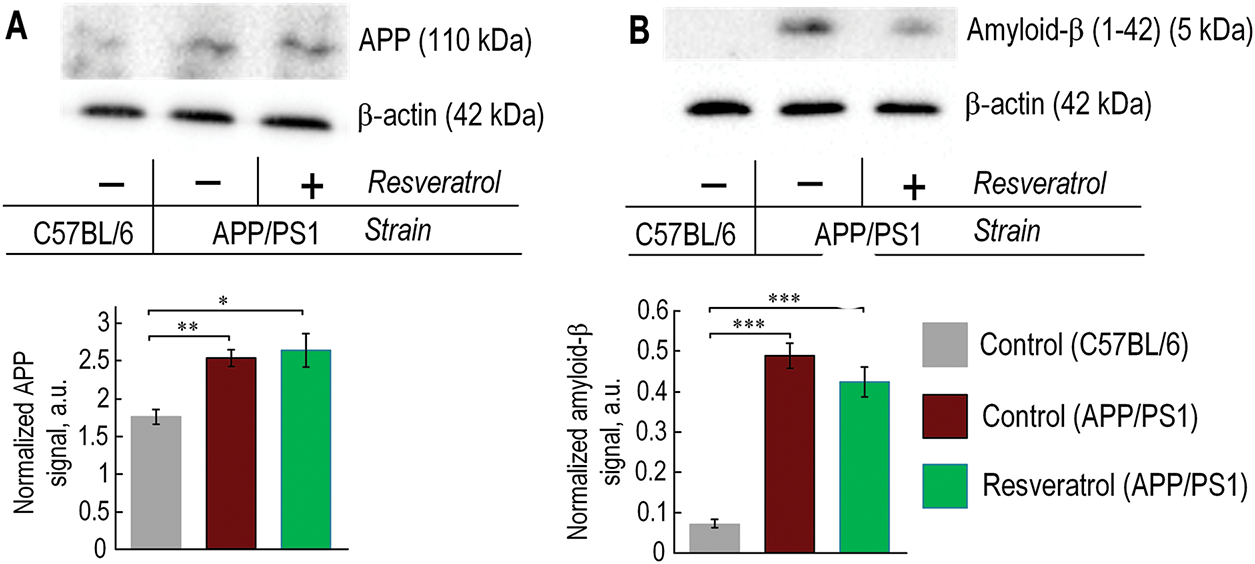
Figure 4: In 10-month-old APP/PS1 mice, there was an increased level of amyloid-β and its precursor APP, regardless of resveratrol treatment. (A) The representative western blot results for APP and relative units are presented, normalized to b-actin signal. (B) The representative western blot results for amyloid-β and relative units are presented, normalized to β-actin signal. The number of repetitions was at least 3. Significance of differences between groups: *p < 0.05; **p < 0.01; ***p < 0.001 (Kruskal-Wallis test)
3.4 The Effect of Mutant Genotype and Resveratrol on the Levels of Inflammatory Markers
We observed a significant increase in the expression of Il6 and Tnf genes—classical markers of inflammation, in the prefrontal cortex of APP/PS1 mice. Within the same gene cluster, Ptgs2, which encodes prostaglandin-endoperoxide synthase, a key marker of inflammation, was also localized. Its level in the cortex of APP/PS1 mice was three times higher compared to C57BL/6 mice. Additionally, the expression level of Gfap, a marker of astrocyte status, was significantly increased in the brains of APP/PS1 mice. Mice receiving resveratrol showed lower expression levels of Gfap, Il6, Tnf, and Ptgs2 compared to control mice of the same genotype, with reductions of 2.1, 3.4, 6.7, and 6-fold, respectively. However, the expression level of Il1b did not correlate with the expression levels of the other pro-inflammatory markers and was localized in a different gene cluster (Fig. 5A).
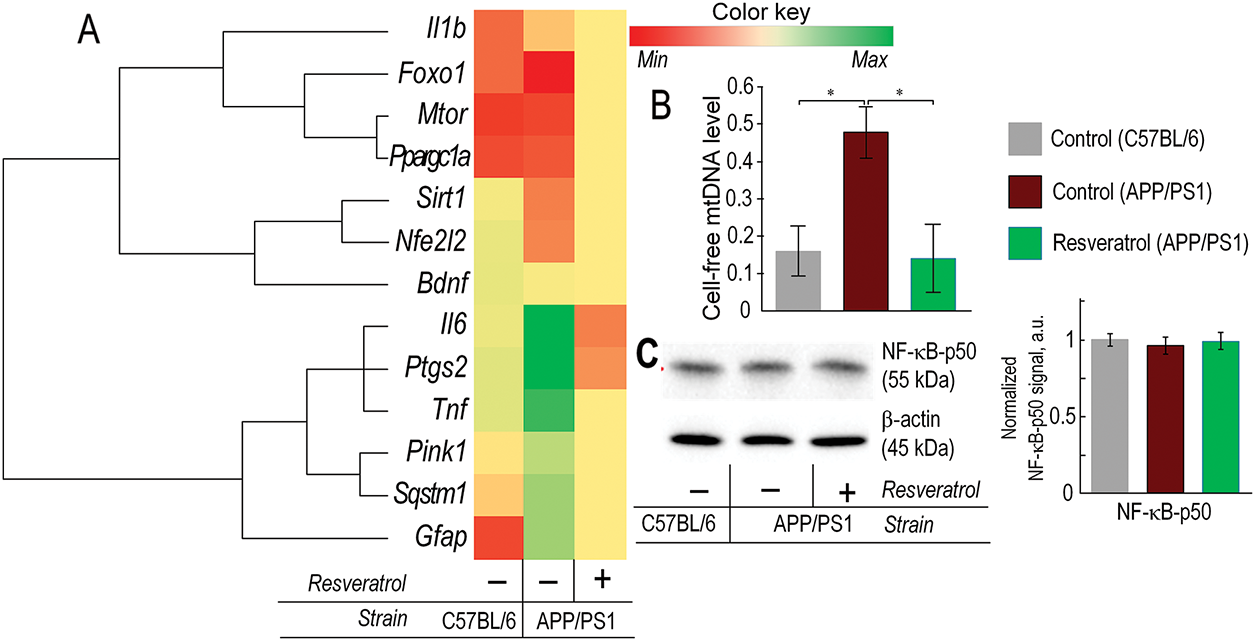
Figure 5: In 10-month-old APP/PS1 mice, there was an increase in pro-inflammatory markers, while resveratrol contributed to a reduction in inflammation. (A) Heatmap of gene expression in the prefrontal cortex. The expression of genes associated with inflammation (Il1b, Il6, Ptgs2, Tnf, Gfap), genes involved in maintaining mitochondrial quality control (Foxo1, Ppargc1a, Sirt1, Nfe2l2, Pink1, Sqstm1), and signaling through the Bdnf-mTORC1 pathway was evaluated. (B) Level of extracellular mtDNA in plasma. (C) The representative Western blot results for NF-κB-p50, with relative units presented normalized to the β-actin signal. The number of repetitions was at least 3. Significance of differences between groups: *p < 0.05 (Kruskal-Wallis test)
We also measured the level of extracellular mtDNA in the plasma as a pro-inflammatory marker. In 10-month-old APP/PS1 transgenic mice, the amount of extracellular mtDNA was three times higher compared to C57BL/6 mice of the same age (p < 0.05). However, in mice with the mutant genotype receiving resveratrol, the level of cell-free circulating mtDNA was comparable to that of control C57BL/6 mice (Fig. 5B). The Level of NF-κB protein subunit was unchanged in the brain of transgenic mice (Fig. 5C).
3.5 The Effect of Genotype and Resveratrol on the Expression Levels of Nrf2/ARE and PI3K/AKT/mTORC1 Signaling Genes
The expression level of the Nfe2l2 gene was significantly reduced in control APP/PS1 mice compared to wild-type mice of the same age. The expression level of the Nfe2l2 gene was comparable to that of C57BL/6 mice in APP/PS1 mice receiving resveratrol (Fig. 5A). Western blot analysis confirmed the gene expression data. The protein level of NRF2 in control APP/PS1 mice was 18% lower compared to wild-type mice (p < 0.05). The protein level of NRF2 in mice receiving resveratrol did not increase compared to control APP/PS1 mice, but there were no statistically significant differences compared to C57BL/6 mice (Fig. 6A). The expression of mitophagy-related genes Pink1 and Sqstm1 was approximately twofold higher in the frontal cortex of control APP/PS1 mice compared to wild-type mice. However, in transgenic mice receiving resveratrol, the expression level of these genes was not increased (Fig. 5A).
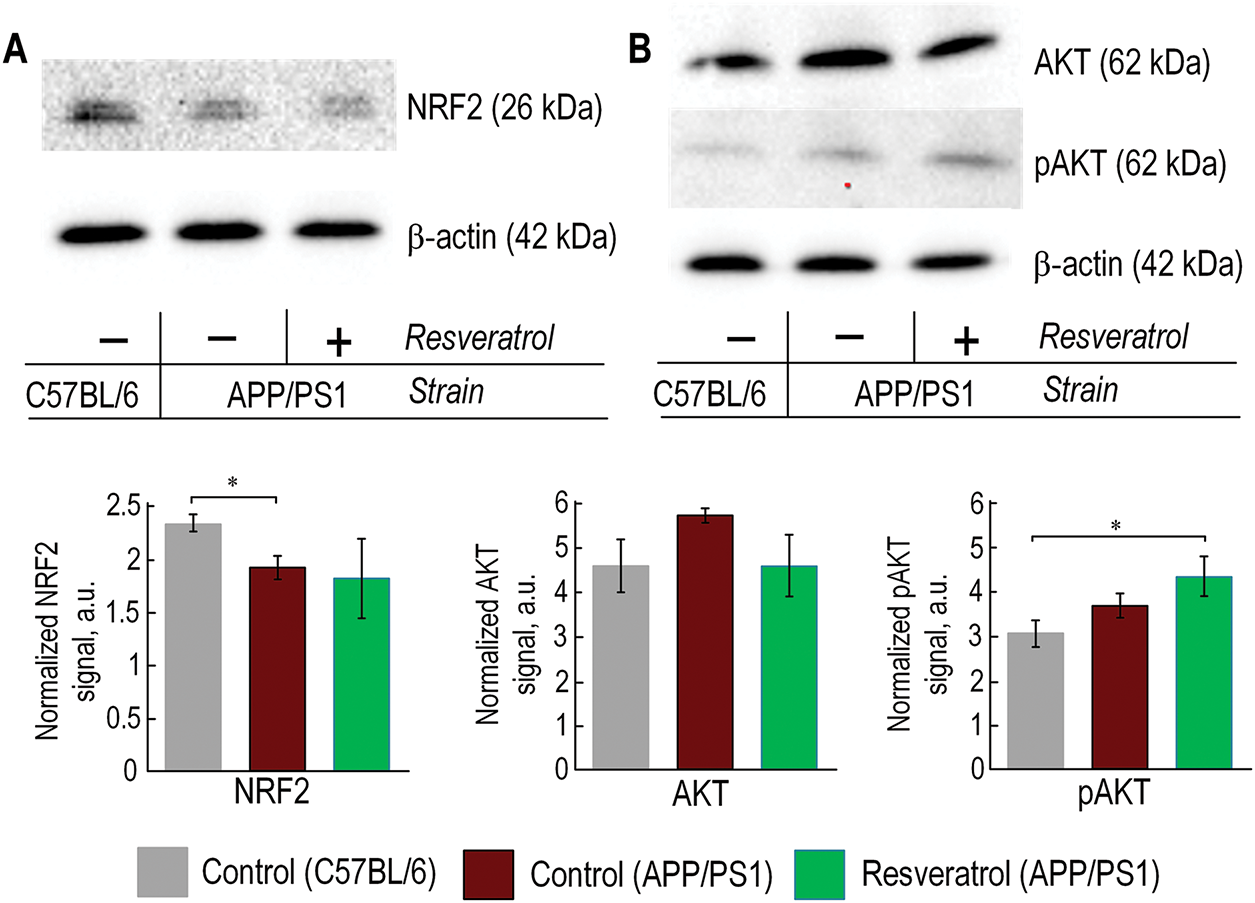
Figure 6: In APP/PS1 mice, a decrease in NRF2 protein was observed, while resveratrol facilitated an increase in AKT phosphorylation. The representative Western blot results for NRF2 (A), AKT, and pAKT (B) are presented, with relative units normalized to the β-actin signal. The number of repetitions was at least 3. Significance of differences between groups: *p < 0.05 (Kruskal-Wallis test)
Ppargc1a, Sirt1, Foxo1, Mtor, and Bdnf genes were also clustered with the Nfe2l2 gene. Its expression was increased in the group of transgenic mice receiving resveratrol, except for Bdnf (Fig. 5A). Western blot analysis of AKT and phosphorylated AKT (pAKT) protein levels was performed to assess PI3K/AKT/mTORC1 pathway activation. While total AKT expression showed no significant differences between experimental groups, a non-significant increasing trend was observed in APP/PS1 control mice compared to wild-type (C57BL/6) animals. Notably, resveratrol-treated APP/PS1 mice exhibited significantly elevated pAKT levels relative to C57BL/6 controls (p < 0.05), indicating enhanced pathway activation following treatment. These results demonstrate that resveratrol specifically modulates AKT phosphorylation status without affecting total AKT protein abundance (Fig. 6B).
3.6 Bacterial Composition of the Gut Microbiome
The phyla Bacteroidetes and Firmicutes accounted for 93% to 95% of the total bacterial population in the mouse gut microbiome across all experimental groups. While resveratrol contributed to a shift in the Bacteroidetes/Firmicutes ratio towards an increase in Firmicutes levels, the differences were not statistically significant (Fig. 7A). Among the minor phyla, it is worth noting a significant increase in Proteobacteria levels in the microbiome of APP/PS1 mice compared to C57BL/6 mice. However, in APP/PS1 mice receiving resveratrol, the level of Proteobacteria was decreased (Fig. 7A,B).
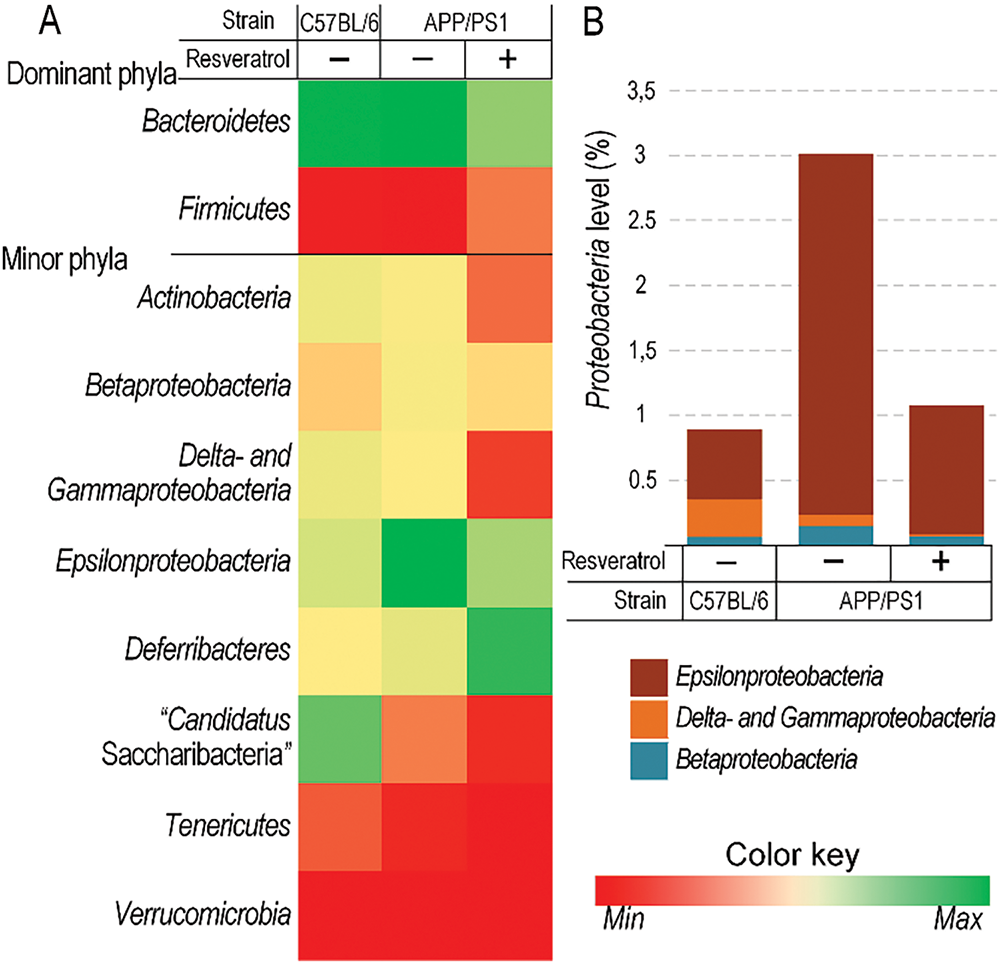
Figure 7: The level of Proteobacteria was higher in APP/PS1 mice compared to C57BL/6 mice, while resveratrol contributed to a reduction in their abundance. Changes in the level of dominant and minor phyla of bacteria (A) and Proteobacteria levels (B) in C57BL/6 and APP/PS1 mice receiving either water or resveratrol. Control C57BL/6 (n = 7), control (APP/PS1) (n = 7), resveratrol (APP/PS1) (n = 6)
Neuronal inflammation is one of the characteristic features of AD pathogenesis. Chronic neuroinflammation observed in the brains of patients with AD can be attributed to activated microglial cells and the release of numerous cytokines [29,30]. Over the past decade, a substantial amount of experimental and clinical data has been obtained, indicating the involvement of inflammation in the pathogenesis of AD. Pro-inflammatory cytokine analysis revealed significantly elevated levels of IL-1, IL-6, TNF-α, and IL-18 in AD model systems compared to controls [31–34]. We found that the expression of Il-6 and Tnf genes was significantly increased in the prefrontal cortex of APP/PS1 mice (Fig. 5A). In the same gene cluster, we identified Ptgs2 (Fig. 5A), encoding cyclooxygenase-2, a crucial marker of inflammation [35]. Furthermore, the expression level of Gfap, a key biomarker for astrocytic activity and central nervous system (CNS) integrity [36], was significantly increased in the brains of APP/PS1 mice (Fig. 5A). The expression of these pro-inflammatory factors is regulated by the NF-κB signaling pathway. However, we did not observe an increase in the level of p50, a subunit of NF-κB (Fig. 5C). Although previous studies have shown an increase in the phosphorylated form of p65 (another subunit of NF-κB) in the brains of APP/PS1 mice [37,38]. It is likely that assessing the level of p65 is a more indicative marker of inflammatory pathway activation through the NF-κB signaling.
Previously, it has been shown that resveratrol can partially prevent the development of age-related cognitive dysfunction by reducing microglial activation [39]. In our study, resveratrol decreased the expression levels of the inflammatory markers, confirming its anti-inflammatory role (Fig. 5C). Moreover, when transgenic mice were given resveratrol, the amount of the cell-free mtDNA in blood plasma was similar to that of wild-type control mice (Fig. 5B). It is known that cell-free circulating mtDNA can act as a mediator of inflammatory processes. MtDNA functions as a damage-associated molecular pattern (DAMP) that can activate innate immune DNA sensors. When released during cellular or tissue damage, mtDNA serves as a potent signaling molecule that triggers inflammatory responses [40]. Levels of certain inflammatory markers, such as TNFα and Il6, are correlated with the level of extracellular mtDNA [40]. Multiple clinical studies have established a significant correlation between elevated circulating mtDNA concentrations and adverse clinical outcomes in acute respiratory distress syndrome (ARDS) patients, including increased mortality rates and prolonged ventilator dependence, sepsis [41], and COVID-19 [42]. Therefore, we observed an elevated level of cell-free mtDNA in the plasma of APP/PS1 mice, while resveratrol resulted in a significant reduction of its level (Fig. 5B), further confirming its anti-inflammatory properties. The significant increase in Proteobacteria levels in the gut microbiome of APP/PS1 mice (Fig. 7) further confirms the fact that inflammatory processes are more intense in transgenic mice compared to C57BL/6 mice. Increased levels of Proteobacteria are usually associated with dysbiosis, which is often linked to an elevation of pro-inflammatory markers due to disruption of the gut barrier function [43]. However, the level of Proteobacteria in resveratrol-treated APP/PS1 mice was equivalent to that of C57BL/6 mice (Fig. 7). Previous studies have demonstrated that resveratrol can reduce Proteobacteria levels in mice with induced obesity [44]. This suggests that the reduction in Proteobacteria levels could be considered as a result of the anti-inflammatory action of resveratrol.
It is known that resveratrol can activate the Nrf2/ARE signaling pathway [45]. We found that the transcript and protein Nrf2 level were decreased in 10-month-old APP/PS1 mice compared to wild-type mice of the same age (Figs. 5A and 6A). Previous studies have also demonstrated that the Nrf2/ARE pathway is attenuated in the brains of APP/PS1 transgenic mice during amyloid-β plaque deposition [46]. Downregulation of Nrf2/ARE signaling has also been observed in the brains of patients with AD [47]. This decrease may be associated with inflammatory processes. It is known that certain inflammatory cytokines, such as TNF-α, have a bidirectional effect on Nrf2/ARE signaling. Excessive production of TNF-α reduces the expression of antioxidant genes by inhibiting Nrf2/ARE signaling, while at lower concentrations, this pro-inflammatory cytokine is associated with Nrf2 activation [48].
The Nrf2/ARE signaling pathway is closely associated with maintaining the integrity of mtDNA. Nrf2 regulates the expression of many antioxidant proteins targeted to mitochondria [49], as well as genes involved in mitochondrial turnover [50], mtDNA repair [51], and enzymes for detoxification of drugs and xenobiotics [52], which are often targeted to mitochondria and mtDNA [53,54]. Indeed, we found that the ventral midbrain, cerebellum, and prefrontal cortex of APP/PS1 mice had significantly more mtDNA damage compared to C57BL/6 mice (Fig. 3). Similar findings were obtained in post-mortem brain analyses of patients with AD. It has been shown that the number of oxidized bases, particularly 8-oxoguanine, was approximately ten times higher in patients with AD compared to control patients. Specifically, mtDNA had ten times more oxidative damage than nuclear DNA [55]. In the cortex of patients up to 75 years old, the amount of mtDNA deletions was fifteen times higher than in healthy individuals of the same age. Interestingly, in patients over 75 years old, the level of deletions was slightly lower than in the control group [56]. Subsequently, it was revealed that deletions in mtDNA caused a deficiency of cytochrome c oxidase in the hippocampus of patients with AD [57]. It is worth noting that no increase in mtDNA damage was observed in the hippocampus (Fig. 3). This suggests that alterations in mtDNA integrity are not the primary cause of cognitive function impairments in the AD model.
In transgenic mice treated with resveratrol, the level of mtDNA damage in the prefrontal cortex was reduced (Fig. 3). It is known that resveratrol induces conformational changes in SIRT1, which affect its activity and specificity towards acetylated substrates [58]. SIRT1, in turn, deacetylates Nrf2, leading to its activation [59]. Several studies have demonstrated that the regulation of Nrf2 and SIRT1 is bidirectional. Under conditions of depressive-like behavior induced by systemic inflammation with lipopolysaccharides, melatonin reduced the level of inflammation and oxidative stress in mitochondria. The addition of siRNA targeting both Nrf2 and SIRT1 neutralized the positive effects of melatonin [60]. Earlier studies have shown that Nrf2 positively influences the deacetylase activity of SIRT1 towards proteins such as fibronectin (FN) and transforming growth factor-β1 (TGF-β1) [61]. It has not been previously explored whether Nrf2 can increase the expression of the Sirt1 gene. Computational analysis using FIMO (p value threshold < 1 × 10−5) identified a high-probability antioxidant response element (ARE) at position −9407/−9393 bp relative to the Sirt1 transcription start site, with a statistically significant match (p < 0.00001) to the canonical ARE consensus sequence. The fact that the expression of Nfe2l2 and Sirt1 genes (Fig. 5A) was found in the same cluster suggests the existence of a feedback loop in which Sirt1 expression depends on Nrf2/ARE signaling.
Resveratrol is also associated with other signaling pathways that may be involved in protecting mitochondria and mtDNA. SIRT1-mediated deacetylation of PGC-1α serves as a critical regulatory mechanism governing metabolic homeostasis and mitochondrial biogenesis, facilitating the restoration of mitochondrial function under stress conditions [62]. Resveratrol-induced activation of SIRT1 can initiate antioxidant pathways through the SIRT1/FOXO1 axis [63]. Notably, all these genes (Nfe2l2, Sirt1, Pparagc1a, Foxo1) are clustered together, and their expression is downregulated in APP/PS1 mice, which is partially reversed by resveratrol in the brain (Fig. 5A). The Mtor gene is also located in this cluster, and its product is a core component of the mTORC1 complex. The mTORC1 complex is responsible for ribosome assembly and protein synthesis, which are necessary for the formation of new neuronal connections that underlie long-term memory formation [64], which may explain the partial improvement in memory observed in transgenic mice treated with resveratrol (Figs. 1 and 2). Another indicator supporting this hypothesis is the increased level of phosphorylated AKT (Fig. 6B). The PI3K/AKT signaling pathway stimulates mTORC1 activity by inhibiting tuberous sclerosis complex 2 (TSC2), a negative regulator of mTORC1 [65]. The total level of AKT remained unchanged, but the level of phosphorylated AKT was increased (Fig. 6B), confirming the idea that resveratrol activates the PI3K/AKT/mTORC1 axis. Previous data on the role of resveratrol in activating the PI3K/AKT/mTORC1 axis have been somewhat contradictory. In some studies, it has been shown that resveratrol inhibits the phosphorylation of AKT, but these results were mostly obtained in tumor tissues [66–68]. On the other hand, resveratrol can activate the PI3K/AKT/mTORC1 axis in models of cardiac ischemic diseases [69] and Parkinson’s disease [70]. Additionally, there is cross-talk between the PI3K/AKT and Nrf2/ARE signaling pathways through the inhibition of GSK3β. Previous studies have shown downregulation of these signaling pathways in the AD pathogenesis [71].
However, the activation of the PI3K/AKT/mTORC1 pathway may have negative consequences in the context of potential AD therapy. The mTORC1 complex inhibits the process of autophagy by suppressing autophagic membrane formation [72]. Nevertheless, autophagy is crucial for amyloid-β clearance. In recent years, it has been suggested that inhibition of mTORC1 could serve as a therapeutic approach for AD by promoting autophagy and removing amyloid-β [73]. Indeed, we observed partial improvement in cognitive function of APP/PS1 mice treated with resveratrol (Figs. 1 and 2). At the same time, resveratrol only contributed to a 15% reduction in the accumulation of amyloid-β in the brains of mice (Fig. 4B). Additionally, APP/PS1 mice treated with resveratrol showed reduced expression of Sqstm1 and Pink1 compared to untreated mice of the same genotype (Fig. 5A). The mentioned proteins are key participants in mitophagy, which is a specific type of autophagy. Mitophagy is also crucial for the removal of dysfunctional mitochondria, which are affected in AD [74].
The principal limitation of our study resides in the insufficient examination of the resveratrol-associated mechanisms pertaining to amyloid-β. Our investigation predominantly concentrated on the mTORC1-dependent regulatory pathway of autophagy, while alternative pathways involved in the clearance of amyloid-β were not explored within the scope of this experiment.
Therefore, due to the potential mTORC1-dependent inhibition of autophagy, resveratrol only slightly contributed to the reduction of amyloid-β levels. However, the observed resveratrol-induced amelioration of cognitive deficits in the AD model may be attributed to the activation of the PI3K/AKT/mTORC1 and Nrf2/ARE signaling pathways, which directly participate in the formation of long-term memory and the mitigation of mitochondrial dysfunction (Fig. 8). Thus, we believe that the use of resveratrol may modulate cognitive properties not only in the context of Alzheimer-type dementias but potentially also in other memory disorders.
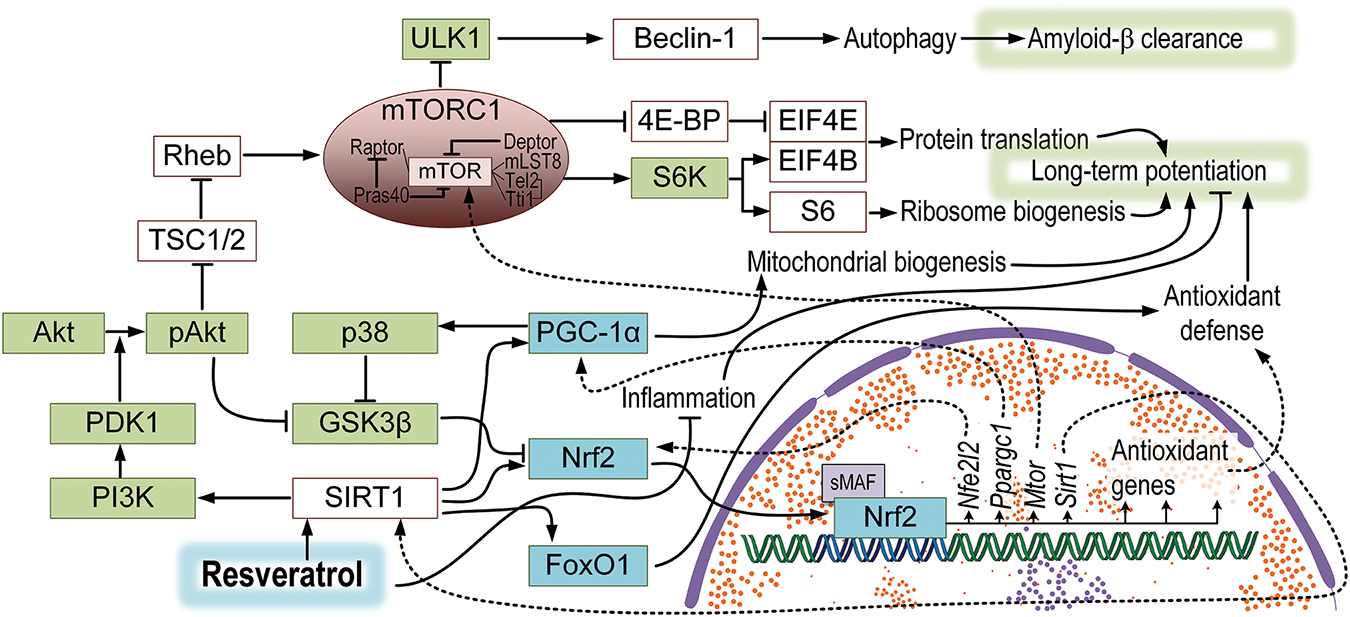
Figure 8: A hypothetical model of the effect of resveratrol on signaling pathways in the brain of APP/PS1 mice. Resveratrol may lead to the activation of SIRT1, which is associated with a range of other signaling pathways. The PI3K/Akt/mTORC1 pathway is linked to the improvement of long-term potentiation but also the suppression of autophagy, resulting in a slowdown of amyloid-β clearance. The deacetylation of PGC-1α may induce an increase in mitochondrial biogenesis, while FoxO1 may enhance antioxidant defense. SIRT-1 dependent activation of Nrf2 is also associated with increased antioxidant protection and forms feedback loops with PGC-1α and SIRT1, which could be connected to mTORC1 activation
Acknowledgement: None.
Funding Statement: This work was supported by the Russian science foundation (grant #22-74-00115 to A.P.G.).
Author Contributions: Study conception and design: Artem P. Gureev, Egor Y. Plotnikov; data collection: Irina S. Sadovnikova, Ekaterina V. Chernyshova, Ekaterina P. Krutskikh, Irina B. Pevzner, Ljubava D. Zorova, Veronika V. Nesterova, Polina I. Babenkova; analysis and interpretation of results: Artem P. Gureev, Irina S. Sadovnikova, Irina B. Pevzner; draft manuscript preparation: Artem P. Gureev, Egor Y. Plotnikov. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Data available on request from the authors.
Ethics Approval: The study was conducted in accordance with the ARRIVE guidelines, and approved by the ethical commission of the Voronezh State University (Section of Animal Care and Use, 94 protocol 42-03 of October 8, 2020).
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Zenimoto T, Takahashi M. Effect of a standardized extract of asparagus officinalis stem (ETAS®50) on cognitive function, psychological symptoms, and behavior in patients with dementia: a randomized crossover trial. Evid Based Complement Alternat Med. 2023;1:9960094. doi:10.1155/2023/9960094. [Google Scholar] [PubMed] [CrossRef]
2. Quan XY, Ma XT, Li GD, Fu XQ, Li JT, Zeng LL. Exploring exosomes to provide evidence for the treatment and prediction of Alzheimer’s disease. BIOCELL. 2023;47(10):2163–76. doi:10.32604/biocell.2023.031226. [Google Scholar] [CrossRef]
3. Pereira MV, Marques AC, Oliveira D, Martins R, Moreira FTC, Sales MGF, et al. Paper-based platform with an in situ molecularly imprinted polymer for β-amyloid. ACS Omega. 2020;5(21):12057–66. doi:10.1021/acsomega.0c00062. [Google Scholar] [PubMed] [CrossRef]
4. Ashleigh T, Swerdlow RH, Beal MF. The role of mitochondrial dysfunction in Alzheimer’s disease pathogenesis. Alzheimers Dement. 2023;19(1):333–42. doi:10.1002/alz.12683. [Google Scholar] [PubMed] [CrossRef]
5. Onyango IG, Bennett JP, Stokin GB. Mitochondrially-targeted therapeutic strategies for Alzheimer’s disease. Curr Alzheimer Res. 2021;18(10):753–71. doi:10.2174/1567205018666211208125855. [Google Scholar] [PubMed] [CrossRef]
6. Wang W, Zhao F, Ma X, Perry G, Zhu X. Mitochondria dysfunction in the pathogenesis of Alzheimer’s disease: recent advances. Mol Neurodegener. 2020;15(1):30. doi:10.1186/s13024-020-00376-6. [Google Scholar] [PubMed] [CrossRef]
7. Coskun PE, Beal MF, Wallace DC. Alzheimer’s brains harbor somatic mtDNA control-region mutations that suppress mitochondrial transcription and replication. Proc Natl Acad Sci U S A. 2004;101(29):10726–31. doi:10.1073/pnas.0403649101. [Google Scholar] [PubMed] [CrossRef]
8. Wong KY, Roy J, Fung ML, Heng BC, Zhang C, Lim LW. Relationships between mitochondrial dysfunction and neurotransmission failure in Alzheimer’s disease. Aging Dis. 2020;1(5):1291–316. doi:10.14336/AD.2019.1125. [Google Scholar] [PubMed] [CrossRef]
9. Pahrudin AA, Shukri SNS, Mohd MN, Ahmad AB, Wan WZ, Ahmad DH, et al. Alpha- and gamma-tocopherol modulates the amyloidogenic pathway of amyloid precursor protein in an in vitro model of Alzheimer’s disease: a transcriptional study. Front Cell Neurosci. 2022;16:846459. doi:10.3389/fncel.2022.846459. [Google Scholar] [PubMed] [CrossRef]
10. Piaceri I, Nacmias B, Sorbi S. Genetics of familial and sporadic Alzheimer’s disease. Front Biosci-Elite. 2013;5(1):167–77. doi:10.2741/e605. [Google Scholar] [PubMed] [CrossRef]
11. LLabre JE, Gil C, Amatya N, Lagalwar S, Possidente B, Vashishth D. Degradation of bone quality in a transgenic mouse model of Alzheimer’s disease. J Bone Miner Res. 2022;37(12):2548–65. doi:10.1002/jbmr.4723. [Google Scholar] [PubMed] [CrossRef]
12. Wirths O, Zampar S. Neuron loss in Alzheimer’s disease: translation in transgenic mouse models. Int J Mol Sci. 2020;21(21):8144. doi:10.3390/ijms21218144. [Google Scholar] [PubMed] [CrossRef]
13. Consoli DC, Brady LJ, Bowman AB, Calipari ES, Harrison FE. Ascorbate deficiency decreases dopamine release in gulo-/- and APP/PSEN1 mice. J Neurochem. 2021;157(3):656–65. doi:10.1111/jnc.15151. [Google Scholar] [PubMed] [CrossRef]
14. Pinto M, Pickrell AM, Fukui H, Moraes CT. Mitochondrial DNA damage in a mouse model of Alzheimer’s disease decreases amyloid beta plaque formation. Neurobiol Aging. 2013;34(10):2399–407. doi:10.1016/j.neurobiolaging.2013. [Google Scholar] [CrossRef]
15. Gureev AP, Nesterova VV, Sadovnikova IS. Long-range PCR as a tool for evaluating mitochondrial DNA damage: principles, benefits, and limitations of the technique. DNA Repair. 2025;146(4):103812. doi:10.1016/j.dnarep.2025.103812. [Google Scholar] [PubMed] [CrossRef]
16. Wang Y, Lv MN, Zhao WJ. Research on ferroptosis as a therapeutic target for the treatment of neurodegenerative diseases. Ageing Res Rev. 2023;91:102035. doi:10.1016/j.arr.2023.102035. [Google Scholar] [PubMed] [CrossRef]
17. Sose PM, Doshi GM, Kale PP. An update on autophagy as a target in the treatment of Alzheimer’s disease. Curr Drug Targets. 2023;24(7):547–67. doi:10.2174/1389450124666230417104325. [Google Scholar] [PubMed] [CrossRef]
18. Khamsing D, Lebrun S, Fanget I, Larochette N, Tourain C, de Sars V, et al. A role for BDNF- and NMDAR-induced lysosomal recruitment of mTORC1 in the regulation of neuronal mTORC1 activity. Mol Brain. 2021;14(1):112. doi:10.1186/s13041-021-00820-8. [Google Scholar] [PubMed] [CrossRef]
19. Al-Edresi S, Alsalahat I, Freeman S, Aojula H, Penny J. Resveratrol-mediated cleavage of amyloid β1-42 peptide: potential relevance to Alzheimer’s disease. Neurobiol Aging. 2020;94:24–33. doi:10.1016/j.neurobiolaging.2020.04.012. [Google Scholar] [PubMed] [CrossRef]
20. Pyo IS, Yun S, Yoon YE, Choi JW, Lee SJ. Mechanisms of aging and the preventive effects of resveratrol on age-related diseases. Molecules. 2020;25(20):4649. doi:10.3390/molecules25204649. [Google Scholar] [PubMed] [CrossRef]
21. Walle T. Bioavailability of resveratrol. Ann New York Acad Sci. 2011;1215(1):9–15. doi:10.1111/j.1749-6632.2010.05842.x. [Google Scholar] [PubMed] [CrossRef]
22. Chung JY, Jeong JH, Song J. Resveratrol modulates the gut-brain axis: focus on glucagon-like peptide-1, 5-ht, and gut microbiota. Front Aging Neurosci. 2020;12:588044. doi:10.3389/fnagi.2020.588044. [Google Scholar] [PubMed] [CrossRef]
23. Sadovnikova IS, Gureev AP, Ignatyeva DA, Gryaznova MV, Chernyshova EV, Krutskikh EP, et al. Nrf2/ARE activators improve memory in aged mice via maintaining of mitochondrial quality control of brain and the modulation of gut microbiome. Pharmaceuticals. 2021;14(7):607. doi:10.3390/ph14070607. [Google Scholar] [PubMed] [CrossRef]
24. Vorhees CV, Williams MT. Morris water maze: procedures for assessing spatial and related forms of learning and memory. Nat Protoc. 2006;1(2):848–58. doi:10.1038/nprot.2006.116. [Google Scholar] [PubMed] [CrossRef]
25. Yang AL, Kashyap PC. A clinical primer of the role of gut microbiome in health and disease. Trop Gastroenterol. 2015;36(1):1–13. doi:10.7869/tg.238. [Google Scholar] [PubMed] [CrossRef]
26. Gureev AP, Shaforostova EA, Starkov AA, Popov VN. Simplified qPCR method for detecting excessive mtDNA damage induced by exogenous factors. Toxicology. 2017;382:67–74. doi:10.1016/j.tox.2017.03.010. [Google Scholar] [PubMed] [CrossRef]
27. Malik AN, Shahni R, Rodriguez-de-Ledesma A, Laftah A, Cunningham P. Mitochondrial DNA as a non-invasive biomarker: accurate quantification using real time quantitative PCR without co-amplification of pseudogenes and dilution bias. Biochem Biophys Res Commun. 2011;412(1):1–7. doi:10.1016/j.bbrc.2011.06.067. [Google Scholar] [PubMed] [CrossRef]
28. Lindqvist D, Fernström J, Grudet C, Ljunggren L, Träskman-Bendz L, Ohlsson L, et al. Increased plasma levels of circulating cell-free mitochondrial DNA in suicide attempters: associations with HPA-axis hyperactivity. Transl Psychiatry. 2016;6(12):e971. doi:10.1038/tp.2016.236. [Google Scholar] [PubMed] [CrossRef]
29. Kinney JW, Bemiller SM, Murtishaw AS, Leisgang AM, Salazar AM, Lamb BT. Inflammation as a central mechanism in Alzheimer’s disease. Alzheimer’s Dement Transl Res Clin Interv. 2018;4(1):575–90. doi:10.1016/j.trci.2018.06.014. [Google Scholar] [PubMed] [CrossRef]
30. Zhao JF, Jiang YR, Guo TL, Jiao YQ, Wang X. Exploring the vital role of microglial membrane receptors in Alzheimer’s disease pathogenesis: a comprehensive review. BIOCELL. 2024;48(7):1011–22. doi:10.32604/biocell.2024.050120. [Google Scholar] [CrossRef]
31. Wang WY, Tan MS, Yu JT, Tan L. Role of pro-inflammatory cytokines released from microglia in Alzheimer’s disease. Ann Transl Med. 2015;3(10):136. doi:10.3978/j.issn.2305-5839.2015.03.49. [Google Scholar] [PubMed] [CrossRef]
32. Sutinen EM, Pirttilä T, Anderson G, Salminen A, Ojala JO. Pro-inflammatory interleukin-18 increases Alzheimer’s disease-associated amyloid-β production in human neuron-like cells. J Neuroinflamm. 2012;9(1):199. doi:10.1186/1742-2094-9-199. [Google Scholar] [PubMed] [CrossRef]
33. De Gaetano A, Solodka K, Zanini G, Selleri V, Mattioli AV, Nasi M, et al. Molecular mechanisms of mtDNA-mediated inflammation. Cells. 2021;10(11):2898. doi:10.3390/cells10112898. [Google Scholar] [PubMed] [CrossRef]
34. Rani V, Verma R, Kumar K, Chawla R. Role of pro-inflammatory cytokines in Alzheimer’s disease and neuroprotective effects of pegylated self-assembled nanoscaffolds. Curr Res Pharmacol Drug Discov. 2022;4:100149. doi:10.1016/j.crphar.2022.100149. [Google Scholar] [PubMed] [CrossRef]
35. Li R, Xie J, Xu W, Zhang L, Lin H, Huang W. LPS-induced PTGS2 manipulates the inflammatory response through trophoblast invasion in preeclampsia via NF-κB pathway. Reprod Biol. 2022;22(4):100696. doi:10.1016/j.repbio.2022.100696. [Google Scholar] [PubMed] [CrossRef]
36. Abdelhak A, Foschi M, Abu-Rumeileh S, Yue JK, D’Anna L, Huss A, et al. Blood GFAP as an emerging biomarker in brain and spinal cord disorders. Nat Rev Neurol. 2022;18(3):158–72. doi:10.1038/s41582-021-00616-3. [Google Scholar] [PubMed] [CrossRef]
37. He Y, Ruganzu JB, Zheng Q, Wu X, Jin H, Peng X, et al. Silencing of LRP1 exacerbates inflammatory response via TLR4/NF-κB/MAPKs signaling pathways in APP/PS1 transgenic mice. Mol Neurobiol. 2020;57(9):3727–43. doi:10.1007/s12035-020-01982-7. [Google Scholar] [PubMed] [CrossRef]
38. Chen S, Liu H, Wang S, Jiang H, Gao L, Wang L, et al. The neuroprotection of verbascoside in Alzheimer’s disease mediated through mitigation of neuroinflammation via blocking NF-κB-p65 signaling. Nutrients. 2022;14(7):1417. doi:10.3390/nu14071417. [Google Scholar] [PubMed] [CrossRef]
39. Kodali M, Parihar VK, Hattiangady B, Mishra V, Shuai B, Shetty AK. Resveratrol prevents age-related memory and mood dysfunction with increased hippocampal neurogenesis and microvasculature, and reduced glial activation. Sci Rep. 2015;5(1):8075. doi:10.1038/srep08075. [Google Scholar] [PubMed] [CrossRef]
40. Nidadavolu LS, Feger D, Chen D, Wu Y, Grodstein F, Gross AL, et al. Associations between circulating cell-free mitochondrial DNA, inflammatory markers, and cognitive and physical outcomes in community dwelling older adults. Immun Ageing. 2023;20(1):24. doi:10.1186/s12979-023-00342-y. [Google Scholar] [PubMed] [CrossRef]
41. Faust HE, Reilly JP, Anderson BJ, Ittner CAG, Forker CM, Zhang P, et al. Plasma mitochondrial DNA levels are associated with ARDS in trauma and sepsis patients. Chest. 2020;157(1):67–76. doi:10.1016/j.chest.2019.09.028. [Google Scholar] [PubMed] [CrossRef]
42. Scozzi D, Cano M, Ma L, Zhou D, Zhu JH, O’Halloran JA, et al. Circulating mitochondrial DNA is an early indicator of severe illness and mortality from COVID-19. JCI Insight. 2021;6(4):e143299. doi:10.1172/jci.insight.143299. [Google Scholar] [PubMed] [CrossRef]
43. Shin NR, Whon TW, Bae JW. Proteobacteria: microbial signature of dysbiosis in gut microbiota. Trends Biotechnol. 2015;33(9):496–503. doi:10.1016/j.tibtech.2015.06.011. [Google Scholar] [CrossRef]
44. Sung MM, Kim TT, Denou E, Soltys CM, Hamza SM, Byrne NJ, et al. Improved glucose homeostasis in obese mice treated with resveratrol is associated with alterations in the gut microbiome. Diabetes. 2017;66(2):418–25. doi:10.2337/db16-0680. [Google Scholar] [PubMed] [CrossRef]
45. Farkhondeh T, Folgado SL, Pourbagher-Shahri AM, Ashrafizadeh M, Samarghandian S. The therapeutic effect of resveratrol: focusing on the Nrf2 signaling pathway. Biomed Pharmacother. 2020;127(17):110234. doi:10.1016/j.biopha.2020.110234. [Google Scholar] [PubMed] [CrossRef]
46. Kanninen K, Malm TM, Jyrkkänen HK, Goldsteins G, Keksa-Goldsteine V, Tanila H, et al. Nuclear factor erythroid 2-related factor 2 protects against beta amyloid. Mol Cell Neurosci. 2008;39(3):302–13. doi:10.1016/j.mcn.2008.07.010. [Google Scholar] [PubMed] [CrossRef]
47. Ramsey CP, Glass CA, Montgomery MB, Lindl KA, Ritson GP, Chia LA, et al. Expression of Nrf2 in neurodegenerative diseases. J Neuropathol Exp Neurol. 2007;66(1):75–85. doi:10.1097/nen.0b013e31802d6da9. [Google Scholar] [PubMed] [CrossRef]
48. Shanmugam G, Narasimhan M, Sakthivel R, Kumar R, Davidson C, Palaniappan S, et al. A biphasic effect of TNF-α in regulation of the Keap1/Nrf2 pathway in cardiomyocytes. Redox Biol. 2016;9:77–89. doi:10.1016/j.redox.2016.06.004. [Google Scholar] [PubMed] [CrossRef]
49. Deng Y, Zhu J, Mi C, Xu B, Jiao C, Li Y, et al. Melatonin antagonizes Mn-induced oxidative injury through the activation of keap1-Nrf2-ARE signaling pathway in the striatum of mice. Neurotox Res. 2015;27(2):156–71. doi:10.1007/s12640-014-9489-5. [Google Scholar] [PubMed] [CrossRef]
50. Esteras N, Abramov AY. Nrf2 as a regulator of mitochondrial function: energy metabolism and beyond. Free Radic Biol Med. 2022;189(2):136–53. doi:10.1016/j.freeradbiomed.2022.07.013. [Google Scholar] [PubMed] [CrossRef]
51. Amirinejad R, Shirvani-Farsani Z, Naghavi B, Sahraian MA, Mohammad SB, Behmanesh M. Vitamin D changes expression of DNA repair genes in the patients with multiple sclerosis. Gene. 2021;781(10):145488. doi:10.1016/j.gene.2021.145488. [Google Scholar] [PubMed] [CrossRef]
52. Shin S, Wakabayashi N, Misra V, Biswal S, Lee GH, Agoston ES, et al. NRF2 modulates aryl hydrocarbon receptor signaling: influence on adipogenesis. Mol Cell Biol. 2007;27(20):7188–97. doi:10.1128/MCB.00915-07. [Google Scholar] [PubMed] [CrossRef]
53. Chen T, Tan J, Wan Z, Zou Y, Afewerky HK, Zhang Z, et al. Effects of commonly used pesticides in China on the mitochondria and ubiquitin-proteasome system in Parkinson’s disease. Int J Mol Sci. 2017;18(12):2507. doi:10.3390/ijms18122507. [Google Scholar] [PubMed] [CrossRef]
54. Samoylova NA, Gureev AP, Popov VN. Methylene blue induces antioxidant defense and reparation of mitochondrial DNA in a Nrf2-dependent manner during cisplatin-induced renal toxicity. Int J Mol Sci. 2023;24(7):6118. doi:10.3390/ijms24076118. [Google Scholar] [PubMed] [CrossRef]
55. Wang J, Xiong S, Xie C, Markesbery WR, Lovell MA. Increased oxidative damage in nuclear and mitochondrial DNA in Alzheimer’s disease. J Neurochem. 2005;93(4):953–62. doi:10.1111/j.1471-4159.2005.03053.x. [Google Scholar] [PubMed] [CrossRef]
56. Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, McKee AC, Beal MF, et al. Marked changes in mitochondrial DNA deletion levels in Alzheimer brains. Genomics. 1994;23(2):471–6. doi:10.1006/geno.1994.1525. [Google Scholar] [PubMed] [CrossRef]
57. Krishnan KJ, Ratnaike TE, De Gruyter HL, Jaros E, Turnbull DM. Mitochondrial DNA deletions cause the biochemical defect observed in Alzheimer’s disease. Neurobiol Aging. 2012;33(9):2210–4. doi:10.1016/j.neurobiolaging.2011.08.009. [Google Scholar] [PubMed] [CrossRef]
58. Borra MT, Smith BC, Denu JM. Mechanism of human SIRT1 activation by resveratrol. J Biol Chem. 2005;280(17):17187–95. doi:10.1074/jbc.M501250200. [Google Scholar] [PubMed] [CrossRef]
59. Ding YW, Zhao GJ, Li XL, Hong GL, Li MF, Qiu QM, et al. SIRT1 exerts protective effects against paraquat-induced injury in mouse type II alveolar epithelial cells by deacetylating NRF2 in vitro. Int J Mol Med. 2016;37(4):1049–58. doi:10.3892/ijmm.2016.2503. [Google Scholar] [CrossRef]
60. Arioz BI, Tastan B, Tarakcioglu E, Tufekci KU, Olcum M, Ersoy N, et al. Melatonin attenuates LPS-induced acute depressive-like behaviors and microglial NLRP3 inflammasome activation through the SIRT1/Nrf2 pathway. Front Immunol. 2019;10:1511. doi:10.3389/fimmu.2019.01511. [Google Scholar] [PubMed] [CrossRef]
61. Huang K, Gao X, Wei W. The crosstalk between Sirt1 and Keap1/Nrf2/ARE anti-oxidative pathway forms a positive feedback loop to inhibit FN and TGF-β1 expressions in rat glomerular mesangial cells. Exp Cell Res. 2017;361(1):63–72. doi:10.1016/j.yexcr.2017.09.042. [Google Scholar] [PubMed] [CrossRef]
62. Zhou Y, Wang S, Li Y, Yu S, Zhao Y. SIRT1/PGC-1α signaling promotes mitochondrial functional recovery and reduces apoptosis after intracerebral hemorrhage in rats. Front Mol Neurosci. 2018;10:443. doi:10.3389/fnmol.2017.00443. [Google Scholar] [PubMed] [CrossRef]
63. Ren CZ, Wu ZT, Wang W, Tan X, Yang YH, Wang YK, et al. SIRT1 exerts anti-hypertensive effect via FOXO1 activation in the rostral ventrolateral medulla. Free Radic Biol Med. 2022;188:1–13. doi:10.1016/j.freeradbiomed.2022.06.003. [Google Scholar] [PubMed] [CrossRef]
64. Hoeffer CA, Klann E. mTOR signaling: at the crossroads of plasticity, memory and disease. Trends Neurosci. 2010;33(2):67–75. doi:10.1016/j.tins.2009.11.003. [Google Scholar] [PubMed] [CrossRef]
65. Dan HC, Ebbs A, Pasparakis M, Van Dyke T, Basseres DS, Baldwin AS. Akt-dependent activation of mTORC1 complex involves phosphorylation of mTOR (mammalian target of rapamycin) by IκB kinase α (IKKα). J Biol Chem. 2014;289(36):25227–40. doi:10.1074/jbc.M114.554881. [Google Scholar] [PubMed] [CrossRef]
66. Jiang H, Shang X, Wu H, Gautam SC, Al-Holou S, Li C, et al. Resveratrol downregulates PI3K/Akt/mTOR signaling pathways in human U251 glioma cells. J Exp Ther Oncol. 2009;8(1):25–33. [Google Scholar] [PubMed]
67. Innets B, Thongsom S, Petsri K, Racha S, Yokoya M, Moriue S, et al. Akt/mTOR targeting activity of resveratrol derivatives in non-small lung cancer. Molecules. 2022;27(23):8268. doi:10.3390/molecules27238268. [Google Scholar] [PubMed] [CrossRef]
68. Bian P, Hu W, Liu C, Li L. Resveratrol potentiates the anti-tumor effects of rapamycin in papillary thyroid cancer: PI3K/AKT/mTOR pathway involved. Arch Biochem Biophys. 2020;689(3):108461. doi:10.1016/j.abb.2020.108461. [Google Scholar] [PubMed] [CrossRef]
69. Yu D, Xiong J, Gao Y, Li J, Zhu D, Shen X, et al. Resveratrol activates PI3K/AKT to reduce myocardial cell apoptosis and mitochondrial oxidative damage caused by myocardial ischemia/reperfusion injury. Acta Histochem. 2021;123(5):151739. doi:10.1016/j.acthis.2021.151739. [Google Scholar] [PubMed] [CrossRef]
70. Huang N, Zhang Y, Chen M, Jin H, Nie J, Luo Y, et al. Resveratrol delays 6-hydroxydopamine-induced apoptosis by activating the PI3K/Akt signaling pathway. Exp Gerontol. 2019;124:110653. doi:10.1016/j.exger.2019.110653. [Google Scholar] [PubMed] [CrossRef]
71. Sotolongo K, Ghiso J, Rostagno A. Nrf2 activation through the PI3K/GSK-3 axis protects neuronal cells from Aβ-mediated oxidative and metabolic damage. Alzheimers Res Ther. 2020;12(1):13. doi:10.1186/s13195-019-0578-9. [Google Scholar] [PubMed] [CrossRef]
72. Rabanal-Ruiz Y, Otten EG, Korolchuk VI. mTORC1 as the main gateway to autophagy. Essays Biochem. 2017;61(6):565–84. doi:10.1042/EBC20170027. [Google Scholar] [PubMed] [CrossRef]
73. Subramanian A, Tamilanban T, Alsayari A, Ramachawolran G, Wong LS, Sekar M, et al. Trilateral association of autophagy, mTOR and Alzheimer’s disease: potential pathway in the development for Alzheimer’s disease therapy. Front Pharmacol. 2022;13:1094351. doi:10.3389/fphar.2022.1094351. [Google Scholar] [PubMed] [CrossRef]
74. Mary A, Eysert F, Checler F, Chami M. Mitophagy in Alzheimer’s disease: molecular defects and therapeutic approaches. Mol Psychiatry. 2023;28(1):202–16. doi:10.1038/s41380-022-01631-6. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools