 Open Access
Open Access
REVIEW
The tumor suppressor role and ceRNA network of miR-1294 in cancer
1 Department of Clinical Medicine, School of Medicine, Zhejiang University City College, Hangzhou, 310000, China
2 Sir Run Run Shaw Hospital, College of Medicine, Zhejiang University, Hangzhou, 310016, China
* Corresponding Authors: Feng Zhu, ; Shiwei Duan,
# These authors contributed equally to this work
Oncology Research 2023, 31(1), 1-12. https://doi.org/10.32604/or.2022.027359
Received 22 October 2022; Accepted 31 January 2023; Issue published 01 March 2023
Abstract
miRNAs are endogenous small RNAs that are important regulators of gene expression. miR-1294 was found to be significantly down-regulated in 15 cancers and regulated by 21 upstream regulators. miR-1294 affects the proliferation, migration, invasion, and apoptosis of cancer cells. The target genes of miR-1294 are involved in the PI3K/AKT/mTOR, RAS, and JAK/STAT signaling pathways. Six target genes of miR-1294 are the targets of a variety of drugs. Low expression of miR-1294 is associated with resistance to cisplatin and TMZ and a poorer prognosis in patients with ESCC, GC, EOC, PDAC, or NSCLC. Therefore, this work outlines the molecular mechanisms and provides a basis for the clinical significance of the tumor suppressor miR-1294 in cancer.Keywords
Abbreviations
| ATO | Arsenic trioxide |
| AKT1 | AKT serine/threonine kinase 1 |
| BC | Breast cancer |
| BLCA | Bladder urothelial carcinoma |
| BRCA | Breast invasive carcinoma |
| CC | Cervical cancer |
| ccRCC | Clear cell renal cell carcinoma |
| ceRNA | Competitive endogenous RNA |
| CHOL | Cholangiocarcinoma |
| CircRNA | Circular RNA |
| c-Myc | MYC proto-oncogene, bHLH transcription factor |
| DFS | Disease-free survival |
| EC | Esophageal cancer |
| EGFR | Epidermal factor receptor |
| ENO1 | Enolase 1 |
| ESCA | Esophageal carcinoma |
| ESCC | Esophageal squamous cell carcinoma |
| FGFR1 | Fibroblast growth factor 1 |
| FOXK1 | Forkhead box K1 |
| GBM | Glioblastoma multiforme |
| GC | Gastric cancer |
| GM | Glioma |
| GRAMD1A | GRAM domain containing 1A |
| HCC | Hepatocellular carcinoma |
| HMGA1 | High mobility group AT-hook 1 |
| HNSC | Head and neck squamous cell carcinoma |
| HOXA6 | Homeobox A6 |
| ICMT | Isoprenylcysteine carboxyl methyltransferase |
| IGF1R | Insulin-like growth factor 1 receptor |
| IRGQ | Immunity related GTPase Q |
| JAK | Janus kinase |
| KICH | Kidney chromophobe |
| KIRC | Kidney renal clear cell carcinoma |
| KIRP | Kidney renal papillary cell carcinoma |
| LASP1 | LIM and SH3 protein 1 |
| LIHC | Liver hepatocellular carcinoma |
| LncRNA | Long non-coding RNA |
| LSCC | Laryngeal squamous cell carcinoma |
| LUAD | Lung adenocarcinoma |
| LUSC | Lung squamous cell carcinoma |
| mRNA | Messenger RNA |
| miRNA | MicroRNA |
| MPM | Malignant pleural mesothelioma |
| mTOR | Mechanistic target of rapamycin kinase |
| NSCLC | Non-small cell lung cancer |
| OC | Ovarian cancer |
| OS | Osteosarcoma |
| OSCC | Oral squamous cell carcinoma |
| PCOS | Polycystic ovary syndrome |
| PDAC | Pancreatic ductal adenocarcinoma |
| PIM1 | Pim-1 proto-oncogene, serine/threonine kinase |
| PI3K | Phosphatidylinositol 3-kinase, putative |
| PKM2 | Pyruvate kinase M1/2 |
| PRAD | Prostate adenocarcinoma |
| STAD | Stomach adenocarcinoma |
| STAT | Signal transducer and activator of transcription |
| TEAD1 | TEA domain transcription factor 1 |
| THCA | Thyroid carcinoma |
| TPX2 | TPX2 microtubule nucleation factor |
| UCEC | Uterine corpus endometrial carcinoma |
| YWHAZ | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta |
As endogenous small RNAs, microRNAs (miRNAs) bind to the 3′ UTRs of their target messenger RNAs (mRNAs) to inhibit their expression, thereby affecting the development, differentiation, and progression of diseases [1,2]. Competitive endogenous RNAs (ceRNAs) such as long non-coding RNAs (lncRNAs) and circular RNAs (circRNAs) can compete with miRNAs [3], and thus regulate the expression of miRNAs and their targeted inhibition of protein-coding genes [4].
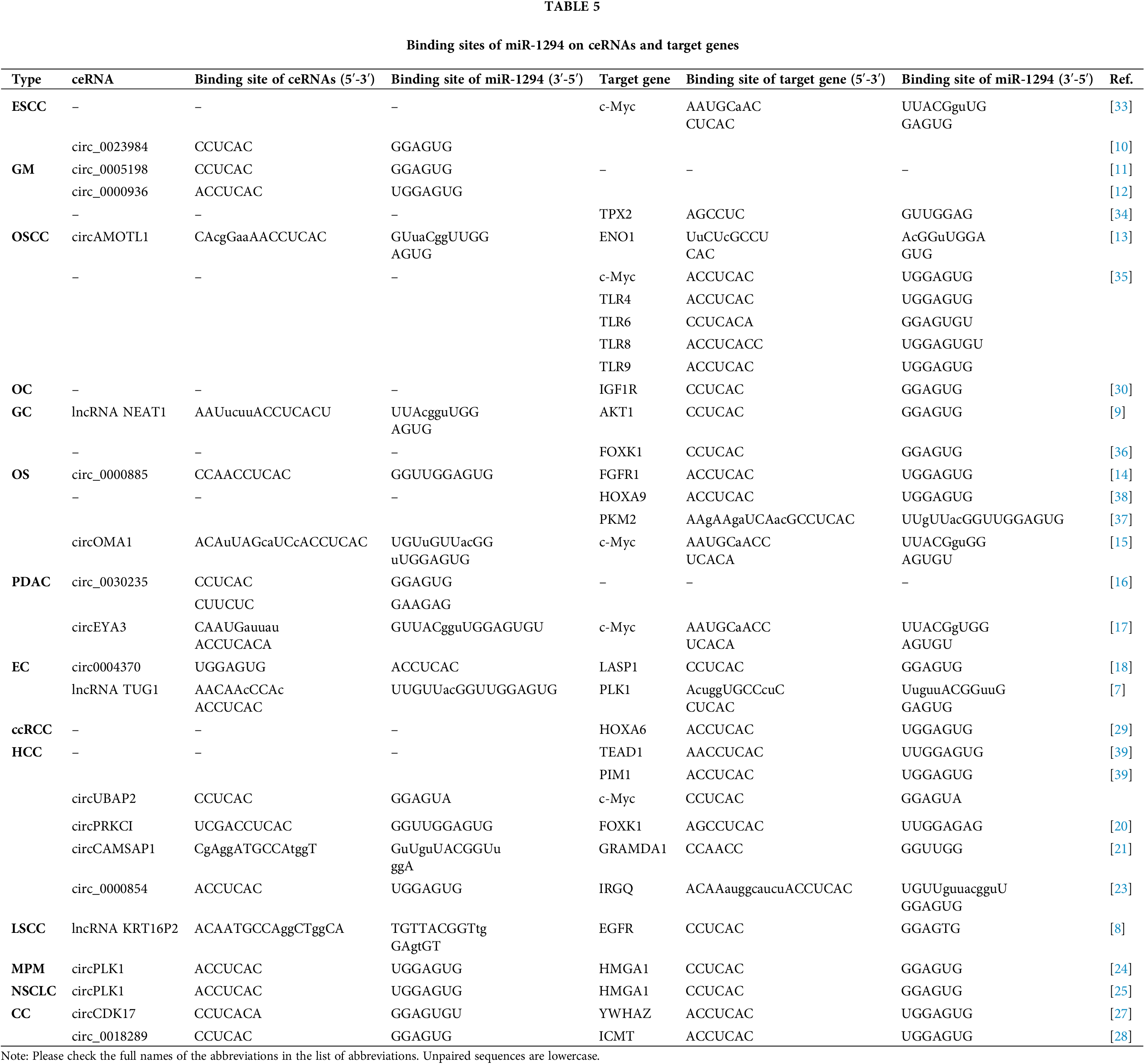
There are at least 19 target genes of miR-1294. The regulation of miR-1294 by ceRNA in various cancers can affect the expression of downstream target genes and various cellular behaviors of cancer cells. The downstream genes of miR-1294 are involved in the regulation of the phosphatidylinositol 3-kinase (PI3K)/AKT/mechanistic target of rapamycin kinase (mTOR), RAS, and Janus kinase (JAK)/signal transducer and activator of transcription (STAT) signaling pathways. Six target genes of miR-1294 are the targets of a variety of known drugs. This work provides a comprehensive summary of miR-1294, which provides potential directions for future research.
Dysregulated miR-1294 in cancer
Previous studies have shown that miR-1294 is downregulated in 15 cancers, suggesting that elevated expression of miR-1294 may have anticancer potential. CeRNAs can compete with miRNAs to regulate the expression of protein-coding genes at the post-transcriptional level [5,6]. The ceRNAs of miR-1294 are highly expressed in 11 tumors, and by inhibiting the expression of miR-1294, they promote the occurrence and development of cancer (Tables 1 and 2). These ceRNAs are 3 lncRNAs and 16 circRNAs, including lncRNA TUG1 [7] in esophageal cancer (EC), KRT16P2 [8] in laryngeal squamous cell carcinoma (LSCC), and NEAT1 [9] in gastric cancer (GC); circRNAs include circ_0023984 in esophageal squamous cancer (ESCC) [10], circ_0005198 [11] and circ_0000936 [12] in glioma (GRAMD1A), and circAMOTL1 in oral squamous cell carcinoma (OSCC) [13], circ_0000885 [14] and circOMA1 [15] in osteosarcoma (OS), circ_0030235 [16] and circEYA3 [17] in pancreatic ductal adenocarcinoma (PDAC), circ_0004370 [18] in EC, circ_0000854 [19], circPRKCI [20], circCAMSAP1 [21], circUBAP2 [22], and circ_0000854 [23] in hepatocellular carcinoma (HCC), circPLK1 in malignant pleural mesothelioma (MPM) [24], circPLK1 [25] and circSHKBP1 [26] in non-small cell lung cancer (NSCLC), and circCDK17 [27], circ_0018289 [28] in cervical cancer (CC). Furthermore, miR-1294 was downregulated in ovarian cancer (OC) and clear cell renal cell carcinoma (ccRCC), thereby relaxing its repressive effects on insulin-like growth factor 1 receptor (IGF1R) and homeobox A6 (HOXA6).
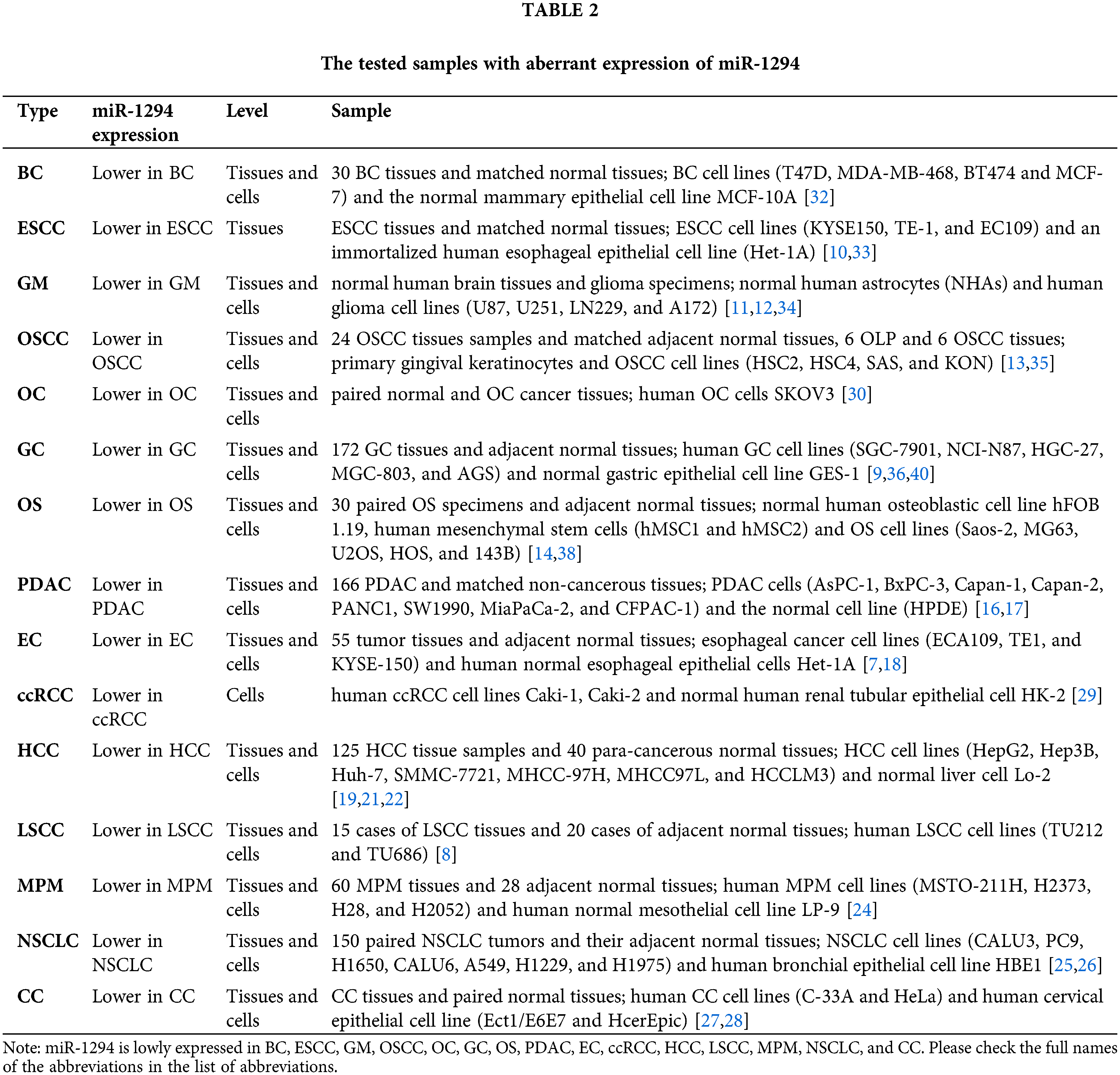
Notably, ceRNAs of miR-1294 have not been found in Breast cancer (BC), ccRCC, OC, and low expression of miR-1294 can relax the repression of HOXA6 [29], IGF1R [30], thereby promoting cancer risk. In addition, low expression of circLDLR in ovarian fluid significantly upregulated the expression of miR-1294, which was associated with the risk of polycystic ovary syndrome (PCOS) [31].
Pan-cancer analysis of miR-1294
We downloaded the TCGA (pan-cancer) dataset from the UCSC Xena database (https://xenabrowser.net/). After removing cancer species without control samples, we performed a log2(x+1) transformation of the extracted miR-1294 expression data (RPM) in the samples, and we finally obtained miR-1294 expression data for 15 cancer types. In addition, we calculated the median expression of all miRNAs in each of the 15 cancers and calculated the quantile ranking of miR-1294 among all non-zero-expressed miRNAs. As shown in Fig. 1a, miR-1294 was highly expressed in 9 tumors including lung adenocarcinoma (LUAD), thyroid carcinoma (THCA), head and neck squamous cell carcinoma (HNSC), kidney chromophobe (KICH), stomach adenocarcinoma (STAD), uterine corpus endometrial carcinoma (UCEC), kidney renal clear cell carcinoma (KIRC), cholangiocarcinoma (CHOL), and esophageal carcinoma (ESCA) (0.5–0.75 quantile, Q3). miR-1294 was moderately expressed in 6 tumors (bladder urothelial carcinoma (BLCA), breast invasive carcinoma (BRCA), kidney renal papillary cell carcinoma (KIRP), liver hepatocellular carcinoma (LIHC), lung squamous cell carcinoma (LUSC), and prostate adenocarcinoma (PRAD)) (0.25–0.5 quantile, Q2). Finally, we calculated the difference in miR-1294 expression between normal and tumor samples of 15 cancers (unpaired Wilcoxon test).
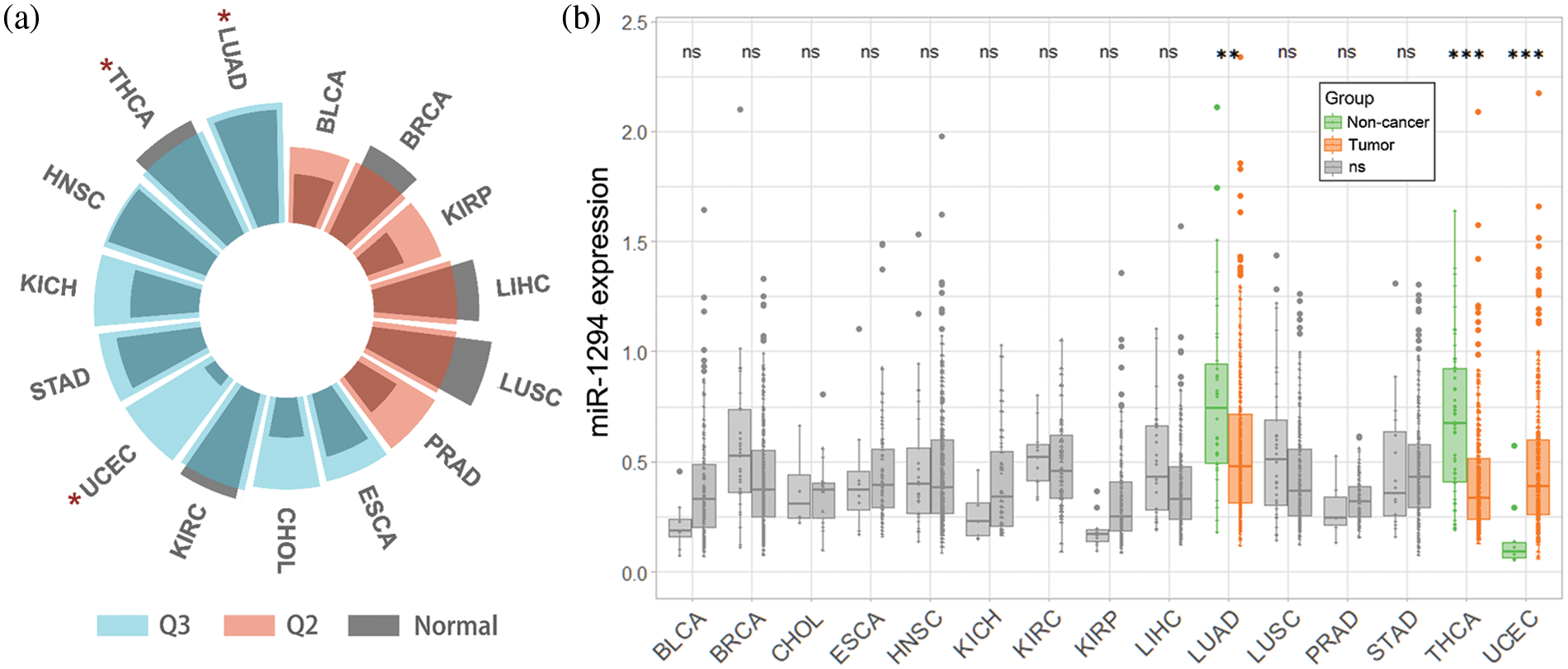
Figure 1: Pan-cancer analysis of miR-1294 using TCGA database. Please check the full names of the abbreviations in the list of abbreviations. a: * means there is a significant difference (p < 0.05) in the expression of miR-1294 between tumor and non-tumor samples. b: miR-1294 expression was Log2(RPM+1) transformed. *** means p < 0.001; ** means p < 0.01; * means p < 0.05; ns means no significant difference.
Pan-cancer analysis showed that miR-1294 was downregulated in TCGA-LUAD and TCGA-THCA (Figs. 1a and 1b), which further validated the anticancer effect of miR-1294. Notably, miR-1294 was upregulated in TCGA-UCEC. Due to the small number of noncancerous samples involved (n = 10), the cancer-promoting effect of miR-1294 in TCGA-UCEC needs to be treated with caution.
Studies have shown that the expression of miR-1294 is significantly down-regulated in 7 cancers including BC, ESCC, EC, GC, ccRCC, HCC, and NSCLC. However, there was no significant association of miR-1294 expression with cancer risk among the corresponding TCGA cancer types (BRCA, ESCA, STAD, KIRC, LIHC, and LUSC) (Table 3).
These inconsistencies may be due to the following reasons. First, miR-1294-related studies are mostly based on cell lines with controlled heterogeneity. However, the heterogeneity of the cancerous and paracancerous tissues in TCGA was high. The proportion of cancer cells also varied among TCGA cancer tissue samples. Second, the expression level of miR-1294 was lower in various cancer types of TCGA (Q2-Q3). The expression of miR-1294 in TCGA was detected by RNA-seq technology. However, the existing miR-1294-related research generally uses qRT-PCR technology to amplify the target gene, and this method can detect very low expression of miR-1294. In addition, cell line-based studies involve more target RNA content and are more suitable for studying miR-1294, which is less expressed. Third, there may be highly expressed tissue-specific regulatory factors or ceRNAs, which significantly inhibit the expression level of miR-1294. And this affects the differential analysis of miR-1294 expression between cancerous and paracancerous tissues in TCGA. Taken together, the differences in the association results between miR-1294 expression and cancer risk may be related to different cancer tissue samples, gene expression detection methods, differences in sample numbers, and the presence of tissue-specific regulators such as ceRNAs. The anticancer effect of miR-1294 in more samples needs to be further verified in the future.
Molecular mechanisms of miR-1294 affecting cancer cell behaviors
The low expression of miR-1294 in cancer cells can relieve its inhibitory effect on downstream protein-coding genes, and then regulate the proliferation, apoptosis, invasion, and migration of cancer cells, and finally lead to the occurrence and development of cancer (Fig. 2).
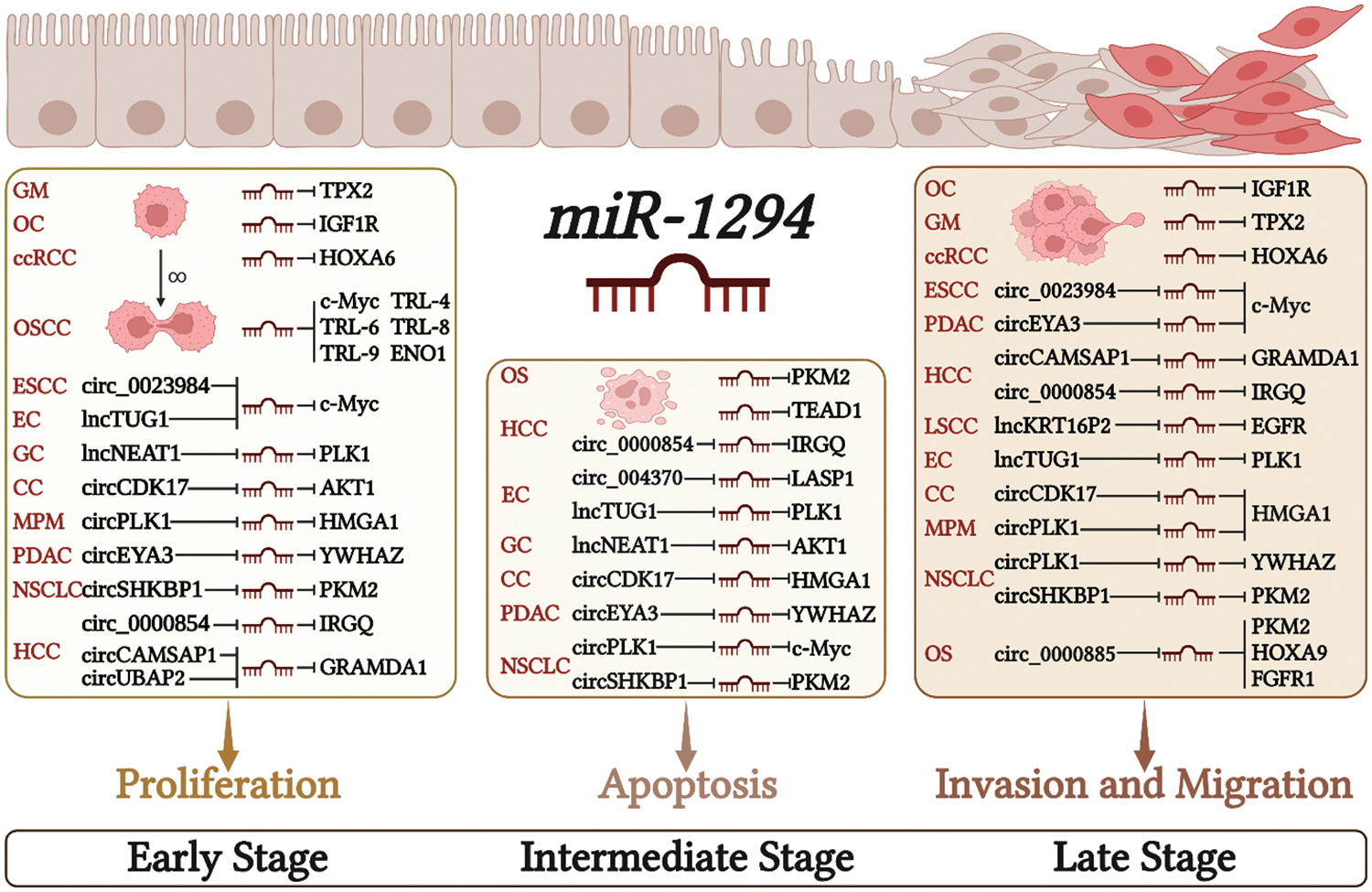
Figure 2: Molecular mechanisms by which miR-1294 affects cancer cell behaviors. Downregulation of miR-1294 promotes cell proliferation, invasion, and migration, and inhibits cancer cell apoptosis by regulating the expression of various target genes. Please check the full names of the abbreviations in the list of abbreviation.
Cell proliferation is an essential component of cell growth and differentiation [41]. Low expression of miR-1294 can up-regulate the expression of downstream protein-coding genes microtubule nucleation factor (TPX2) [34], IGF1R [30], MYC proto-oncogene, bHLH transcription factor (c-Myc) [33,35] and TRL4, TRL6, TRL8, TRL9 [35], enolase 1 (ENO1) [13], thereby promoting the proliferation of various tumor cells. In ESCC, PDAC, EC, GC, MPM, CC, NSCLC, and HCC, The highly expressed ceRNAs increase the expression of downstream protein-coding genes by inhibiting miR-1294, thereby promoting the proliferation of cancer cells. These ceRNA/miRNA/PCG signaling axes include circ_0023984/miR-1294/c-Myc in ESCC [10], circEYA3/miR-1294/c-Myc in PDAC [17], lncTUG1/miR-1294/PLK1 in EC [7], lncNEAT1/miR-1294/AKT serine/threonine kinase 1 (AKT1) in GC [9], circPLK1/miR-1294/high mobility group AT-hook 1 (HMGA1) in MPM [24], circCDK17/miR-1294/tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein zeta (YWHAZ) in CC [27], circSHKBP1/miR-1294/pyruvate kinase M2 (PKM2) in NSCLC [26], circCAMSAP1| CircUBAP2/miR-1294/GRAM domain containing 1A (GRAMD1) [21,22] and circ_0000854/miR-1294/immunity related GTPase Q (IRGQ) [23] in HCC.
Apoptosis is a form of programmed cell death that removes damaged cells in an orderly and efficient manner. Dysregulation of apoptosis machinery is a hallmark of cancer [42]. Low expression of miR-1294 inhibited cancer cell apoptosis (Fig. 2). In OS and HCC, under-expressed miR-1294 inhibits cancer cell apoptosis by upregulating pyruvate kinase M2 (PKM2) in OS [37] and TEA domain transcription factor 1 (TEAD1) and pim-1 proto-oncogene in HCC [39]. These ceRNA/miRNA/PCG signaling axes that inhibit cancer cell apoptosis include lncTUG1/miR-1294/PLK1 [7] and circ_0004370/miR-1294/LIM and SH3 protein 1 (LASP1) [18] in EC, lncNEAT1/miR-1294/AKT1 in GC [9], circPLK1/miR-1294/HMGA1 [25] and circSHKBP1/miR-1294/PKM2 [26] in NSCLC, circCDK17/miR-1294/YWHAZ in CC [27], and CircEYA3/miR-1294/c-Myc in PDAC [17].
Metastasis of cancer cells is a major cause of cancer death, and its initial steps are cancer cell migration and invasion into surrounding tissues and vasculature [43]. miR-1294 is closely associated with cell migration and invasion in cancer (Fig. 2). The low expression of miR-1294 can up-regulate the downstream target genes c-Myc [33], TPX2 [34], IGF1R [30], and HOXA6 [29] to promote the invasion and migration of ESCC, GM, OC, ccRCC tumor cells. These ceRNA/miRNA/PCG signaling axes that can promote tumor cell invasion and migration include circEYA3/miR-1294/c-Myc in PDAC [17], circ_0023984/miR-1294/c-Myc in ESCC [10], circCAMSAP1/miR-1294/GRAMD1 [21] and circ_0000854/miR-1294/IRGQ [23] in HCC, lncKRT16P2/miR- 1294/epidermal factor receptor (EGFR) in LSCC [8], lncTUG1/miR-1294/PLK1 in EC [7], circCDK17/miR-1294/YWHAZ in CC [27], circPLK1/miR-1294/HMGA1 in NSCLC [25] and MPM [24], and circSHKBP1/miR-1294/PKM2 in NSCLC [26].
miR-1294-related signaling pathways
miR-1294 inhibits the expression of at least 18 target genes (Fig. 3). Among them, five target genes (c-Myc, IGF1R, AKT, fibroblast growth factor 1 (FGFR1), and pim-1 proto-oncogene, serine/threonine kinase (PIM1)) are involved in the regulation of the PI3K/AKT/mTOR, RAS, JAK/STAT signaling pathways (Fig. 4), thereby affecting the proliferation, apoptosis, invasion, and progression of cancer cells.
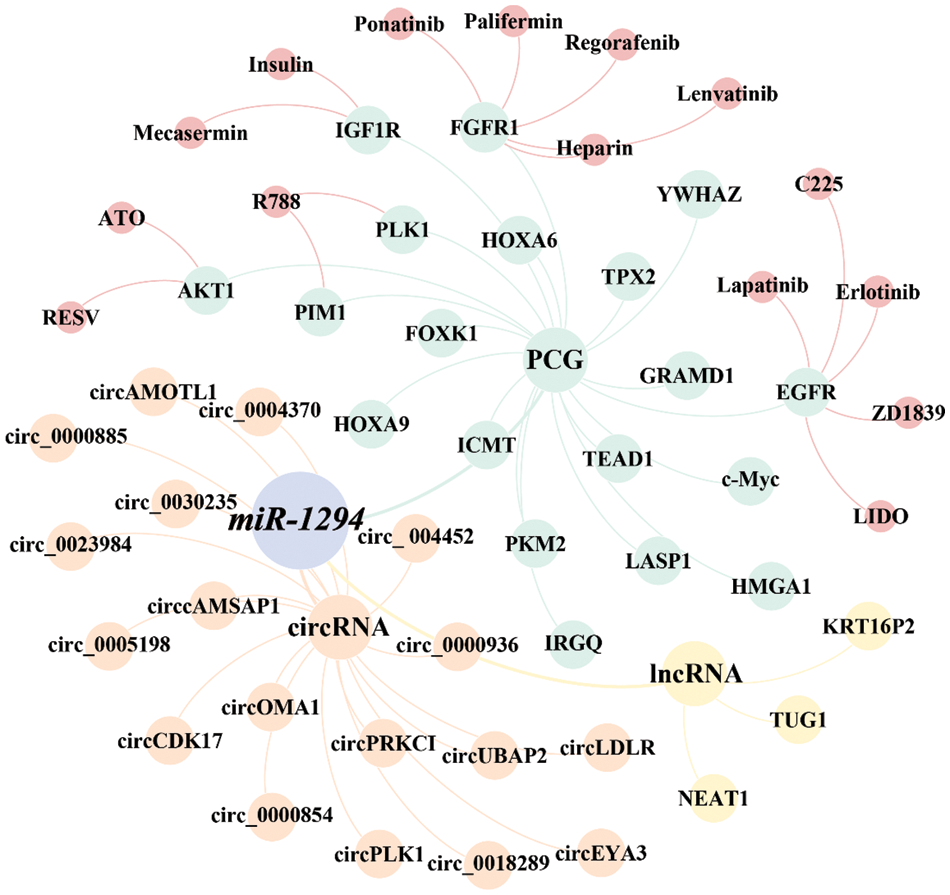
Figure 3: The ceRNA network and the druggable PCGs of miR-1294. The ceRNA network of miR-1294 includes 3 lncRNAs, 18 circRNAs, and 18 downstream PCGs, of which 6 PCGs have targeted drugs.
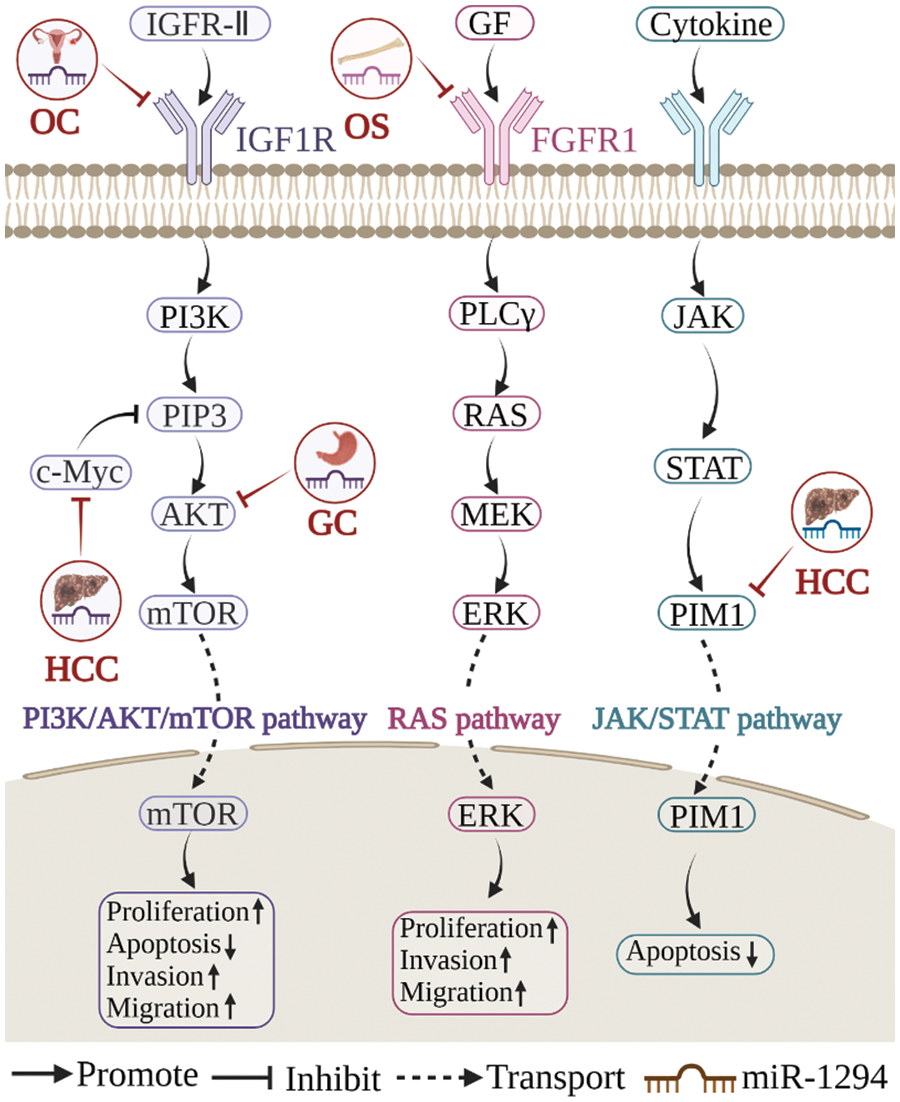
Figure 4: Three signaling pathways related to miR-1294. miR-1294 participates in three signaling pathways of PIK3/AKT/mTOR, RAS, and JAK/STAT to regulate cell biological processes.
miR-1294 and PI3K/AKT/mTOR signaling pathway
The PI3K/AKT/mTOR signaling pathway is a master regulator of cancer [44], which is frequently activated in various cancers and is considered a promising therapeutic target [45]. In cisplatin-resistant tissues and cell lines (SKOV3/DDP) of OC, low expression of miR-1294 can increase the expression level of IGF1R, thereby mediating the activation of the PI3K/AKT/mTOR signaling pathway and promoting the proliferation, migration, and invasion of OC cells [30]. In HCC, CircUBAP2 acts as a sponge for miR-1294, upregulates c-Myc expression, and inhibits PI3P, thereby inhibiting the PI3K/AKT/mTOR signaling pathway and promoting tumorigenesis [22]. In GC, LncRNA NEAT1 increased the expression level of AKT1 by sponging miR-1294, mediated the activation of the PI3K/AKT/mTOR signaling pathway, promoted the proliferation and migration of GC cells, and inhibited apoptosis [9].
miR-1294 and RAS signaling pathway
The RAS signaling pathway can control cell growth, survival, and differentiation by integrating extracellular signals. Aberrant activation of the RAS pathway is a highly prevalent major oncogenic event [46]. Circ_0000885, which is highly expressed in OS, can restore the expression level of FGFR1 by targeting miR-1294, thereby mediating the activation of the RAS signaling pathway and promoting the progression of OS [14].
miR-1294 and JAK/STAT signaling pathway
The JAK/STAT signaling pathway is a mechanism by which extracellular factors regulate gene expression and is involved in many key biological processes such as cell proliferation, differentiation, apoptosis, and immune regulation [47]. Arsenic trioxide (ATO) is the most toxic compound in traditional Chinese medicine and has been shown to effectively inhibit cancer cell processes. In HCC, ATO induced the upregulation of miR-1294, decreased the expression level of PIM1, and inhibited the JAK/STAT signaling pathway, thereby promoting the apoptosis of HCC cells [39].
The clinical significance of miR-1294
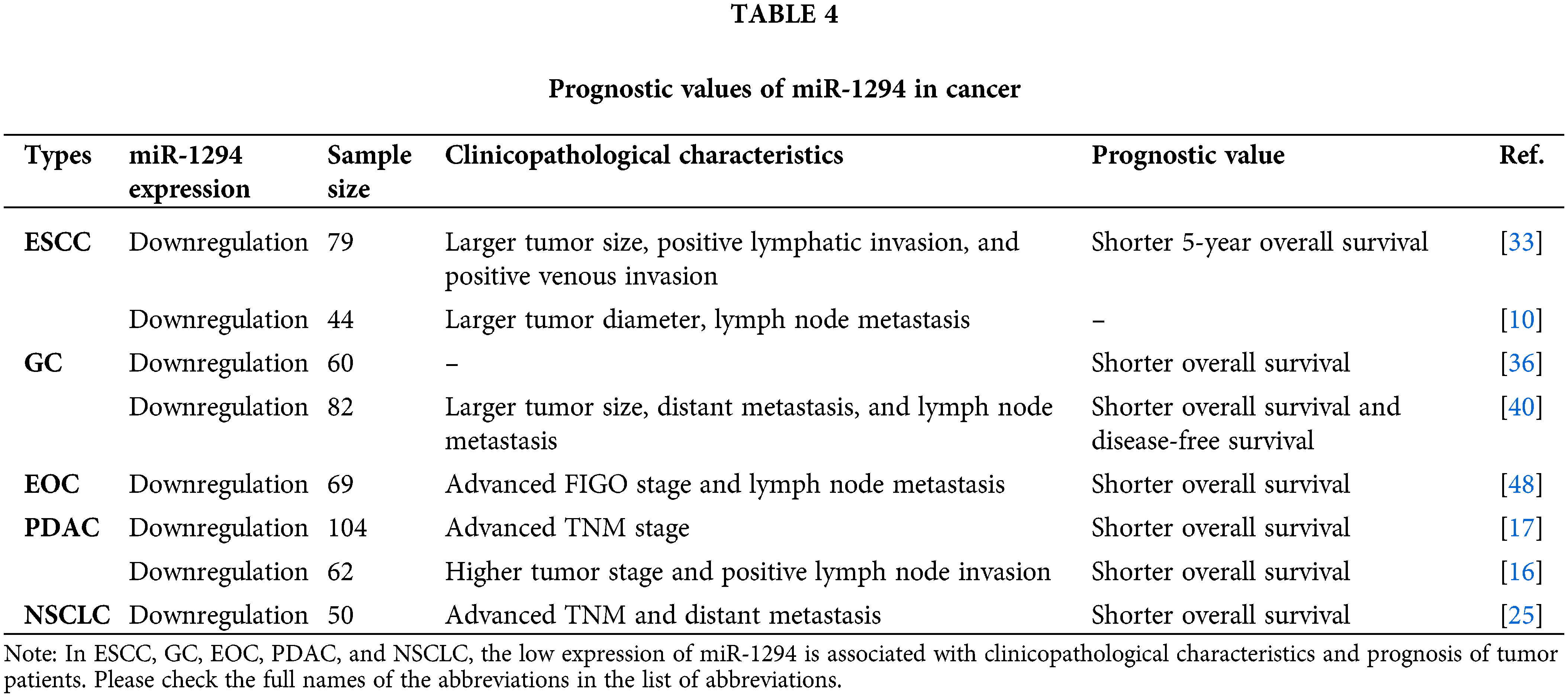
As shown in Table 4, the abnormal expression of miR-1294 in cancer is not only correlated with tumor prognostic indicators but also closely related to the clinicopathological phenotype of cancer patients. Cancer therapeutic drugs can target cancer by targeting the downstream genes of miR-1294. In addition, studies have also shown that low expression of miR-1294 is also associated with resistance to cisplatin and TMZ.
The diagnostic and prognostic value of miR-1294
As shown in Table 4, miR-1294 was down-regulated in most cancers, and its abnormal expression correlated with prognostic indicators of tumors. In ESCC, the overall survival rate of the miR-1294-low-expression group was significantly lower than that of the miR-1294-high-expression group [33]. In GC, patients with low miR-1294 expression had significantly shorter overall survival [36,40] and disease-free survival (DFS) [40] than patients with high expression of miR-1294 [36]. In EOC, the overall survival rate of the miR-1294-low-expression group was lower compared with the miR-1294-high-expression group [48]. The expressions of CircEYA3 and Circ_0030235 were significantly up-regulated in PDAC tissues compared with adjacent normal tissues. Survival analysis showed that the overall survival rate of PDAC patients with high expression of CircEYA3 and Circ_0030235 group was lower [17,16], thus indicating that in PDAC, the group with low expression of miR-1294 had lower overall survival rate. In NSCLC, the overall survival rate was lower in the miR-1294-low-expression group compared with the miR-1294-high-expression group [25].
The relationship between miR-1294 and clinicopathological characteristics
As shown in Table 4, the expression level of miR-1294 was closely related to the clinicopathological phenotype of cancer patients. In ESCC, low expression of miR-1294 was associated with larger tumors, positive lymphatic infiltration, lymph node metastasis, and positive venous infiltration [33,10]. In GC, low expression of miR-1294 was associated with larger tumors, lymph node metastasis, and distant metastasis [40]. In EOC, low expression of miR-1294 was associated with advanced FIGO stage and lymph node metastasis [48]. In PDAC, low expression of miR-1294 was associated with advanced TNM stage [17], higher tumor stage, and positive lymph node invasion [16]. In NSCLC, low expression of miR-1294 was associated with advanced TNM staging and distant metastasis in NSCLC patients [25].
As shown in Fig. 3, we found that currently listed drugs can target 6 downstream genes of miR-1294 via the CADDIE website (https://exbio.wzw.tum.de/caddie/drug-lookup) [49]. These drugs are Palifermin, Heparin, Regorafenib, Ponatinib, and Lenvatinib targeting FGFR1, R788 (Fostamatinib) targeting PLK1 and PIM1, C225 (Cetuximab), LIDO (Lidocaine), (ZD1839) Gefitinib targeting EGFR, Erlotinib, and Lapatinib; Insulin and Mecasermin targeting IGF1R, and ATO and RESV (Resveratrol) targeting AKT1. In the future, it is necessary to confirm whether miR-1294 interacts with these drugs (Table 5).
miR-1294 was closely associated with cisplatin and TMZ resistance in cancer cells (Fig. 5). miR-1294 can affect the drug resistance of tumor cells by regulating targets, activating signaling pathways, or changing the normal behavior of molecules in two tumor cells.
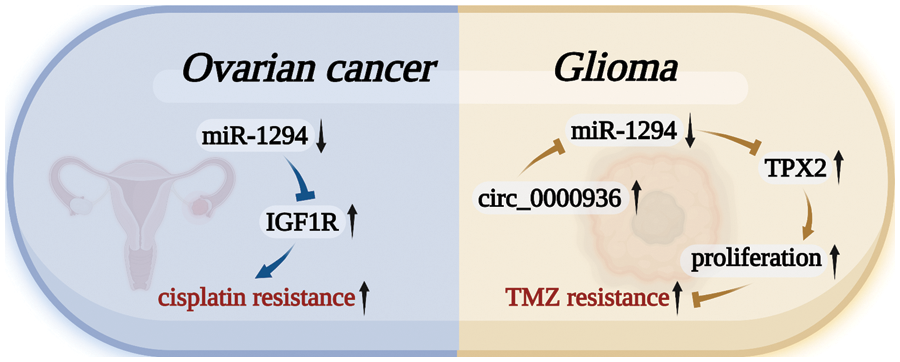
Figure 5: miR-1294 affects cellular drug resistance by inhibiting target genes. In OC tumor cells, the low expression of miR-1294 increased the resistance of OC cancer cells to cisplatin by upregulating the expression of the target gene IGF1R. In GM, the highly expressed Circ_0000936 can down-regulate the expression level of miR-1294 to up-regulate the expression of TPX2, promote the proliferation of GM cancer cells, and increase the resistance of GM cancer cells to TMZ.
Cisplatin is a well-known chemotherapy drug that has been used to treat a variety of human cancers [50]. The development of cisplatin chemoresistance can lead to the failure of cisplatin therapy [51]. In OC, miR-1294 was significantly decreased in tissues of cisplatin-resistant patients compared with cisplatin-sensitive patients. In vitro, miR-1294 also showed low expression in cisplatin-resistant cell lines (SKOV3/DDP) compared with OC SKOV3 cells. Low expression of miR-1294 can restore the expression level of the target gene IGF1R and activate the PI3K/AKT/mTOR signaling pathway, thereby upregulating the cisplatin resistance of OC cells [30].
Temozolomide is used as an oral alkylating agent in the treatment of glioblastoma multiforme (GBM) and astrocytoma [52]. miR-1294 expression was lower in high-grade gliomas than in low-grade gliomas. Low-expressed miR-1294 upregulates the expression of TPX2, which promotes the proliferation, migration, and invasion of GM cells, and reduces the chemosensitivity of GM cells to temozolomide [34]. Meanwhile, the expression of Circ_0000936 in temozolomide-resistant GM tissues was higher than that in temozolomide-sensitive GM tissues. The highly expressed Circ_0000936 can down-regulate the expression level of miR-1294, thereby increasing the resistance of GM cells to TMZ [12].
Available evidence indicates that miR-1294 expression is downregulated in 15 tumors, including BC, ESCC, OC, ccRCC, GM, OSCC, GC, OS, PDAC, EC, HCC, LSCC, MPM, NSCLC, and CC. miR-1294 has 21 upstream ceRNAs (including 18 circRNAs and 3 lncRNAs) and 19 downstream target genes (Fig. 6). Low expression of miR-1294 can promote the proliferation, apoptosis, invasion, and migration of cancer cells, and can participate in the activation of PI3K/AKT/mTOR, RAS, JAK/STAT signaling pathways, and promote the development of cancer. Down-regulation of miR-1294 was associated with poorer prognosis in ESCC, GC, EOC, PDAC, and NSCLC. In addition, low expression of miR-1294 was also associated with resistance to cisplatin and TMZ.
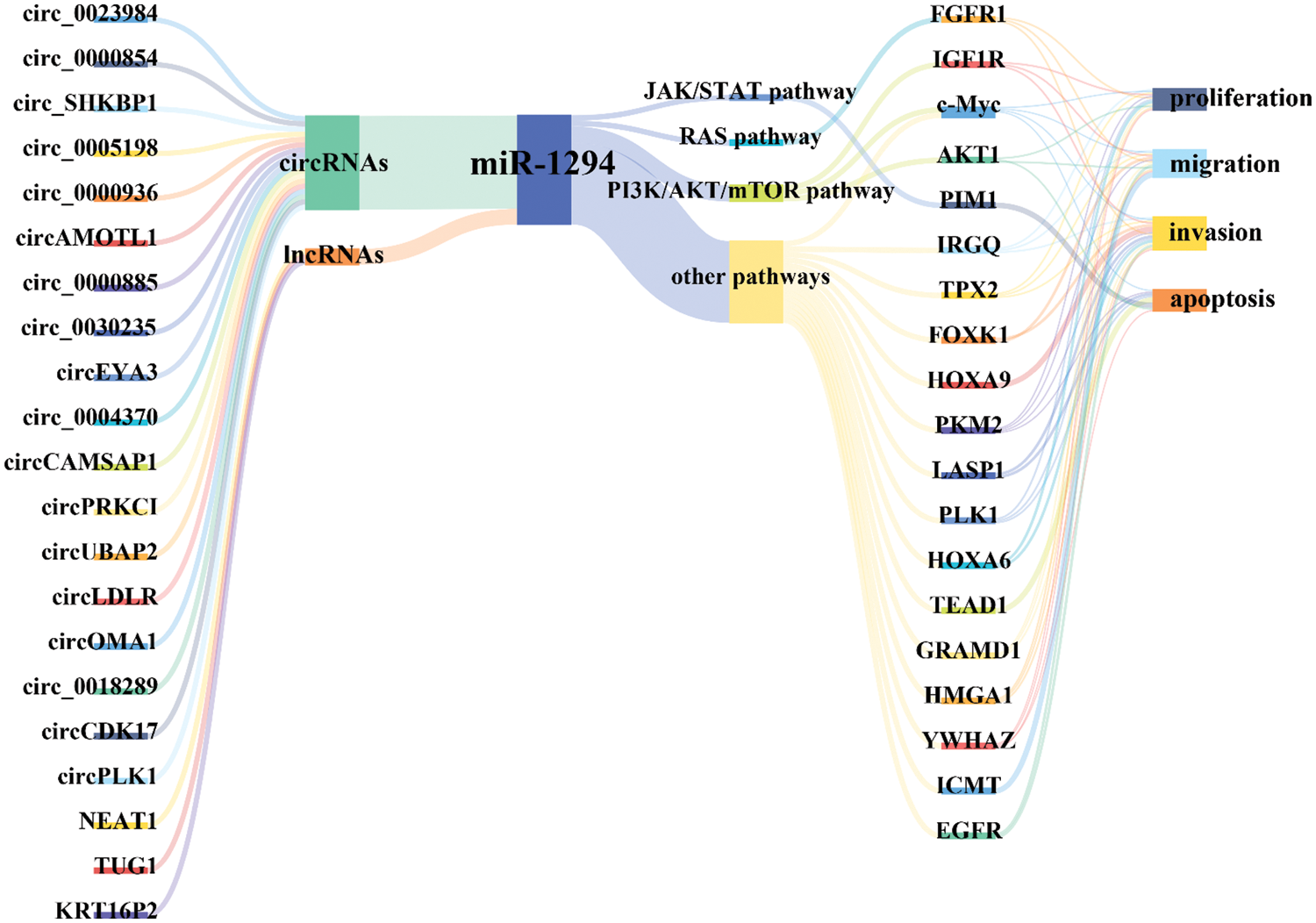
Figure 6: Molecular mechanism of miR-1294-centered ceRNA network. Under the regulation of multiple ceRNAs, inhibition of miR-1294 can relax the down-regulation of its target genes, thereby regulating the migration, proliferation, invasion, and apoptosis of cancer cells. Please check the full names of the abbreviations in the list of abbreviations.
Notably, the analysis of TCGA also found that miR-1294 was down-regulated in TCGA-LUAD and TCGA-THCA, while its expression was up-regulated in TCGA-UCEC. Furthermore, miR-1294 expression was upregulated in the noncancerous disease PCOS. The functional differences of miR-1294 may be related to mechanisms such as differences in samples, miRNA detection methods, differences in sample numbers, and the presence of tissue-specific regulators such as ceRNAs.
Low expression of miR-1294 in ovarian cancer and glioma is associated with TMZ and cisplatin resistance. Porous lyotropic liquid crystal nanoparticles are promising delivery vehicles for cancer therapy [19]. The use of targeted nanomedicine to deliver miR-1294 may have great potential for cancer therapy.
However, there are still many deficiencies in the current research on miR-1294. First, the number of current research samples is small, and relevant results need to be verified in larger samples and other populations. Secondly, some studies on the biological functions of miR-1294 are limited to in vitro cell experiments, and it is necessary to strengthen the verification of in vivo animal experiments in the future. Finally, the molecular mechanism of miR-1294 in disease is still not fully understood, and more in-depth research is needed in the future to provide a theoretical basis for miR-1294-targeted therapeutic regimens.
As a tumor suppressor, the low expression of miR-1294 has an important molecular regulatory mechanism in cancer cell behavior and carcinogenesis. In addition, the overview of miR-1294 in cancer diagnosis, prognosis, and treatment is expected to provide potential clues and directions for miR-1294-related clinical research.
Acknowledgement: The authors would like to thank the PubMed, TCGA, KEGG, and CADDIE databases for useful information, and BioRender (https://biorender.com/) for rendering Figs. 2, 4 and 5.
Funding Statement: This study was supported by Qiantang Scholarship in Zhejiang University City College, Hangzhou Agricultural and Social Development Research Project (2020ZDSJ0637).
Author Contributions: YM, JS, LF, and FZ collected and analyzed the literature, drafted the figures, and wrote the paper; SD and FZ conceived and gave the final approval of the submitted version. All authors have read and agreed to the published version of the manuscript.
Availability of Data and Materials: All data generated or analyzed during this study are included in the article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Michlewski, G., Cáceres, J. F. (2019). Post-transcriptional control of miRNA biogenesis. RNA, 25(1), 1–16. https://doi.org/10.1261/rna.068692.118 [Google Scholar] [PubMed] [CrossRef]
2. Syeda, Z. A., Langden, S. S. S., Munkhzul, C., Lee, M., Song, S. J. (2020). Regulatory mechanism of MicroRNA expression in cancer. International Journal of Molecular Sciences, 21(5), 1723. https://doi.org/10.3390/ijms21051723 [Google Scholar] [PubMed] [CrossRef]
3. Zhong, C., Xie, Z., Zeng, L. H., Yuan, C., Duan, S. (2022). MIR4435-2HG is a potential pan-cancer biomarker for diagnosis and prognosis. Frontiers in Immunology, 13, 855078. https://doi.org/10.3389/fimmu.2022.855078 [Google Scholar] [PubMed] [CrossRef]
4. Mao, M., Zhang, J., Xiang, Y., Gong, M., Deng, Y. et al. (2022). Role of exosomal competitive endogenous RNA (ceRNA) in diagnosis and treatment of malignant tumors. Bioengineered, 13(5), 12156–12168. https://doi.org/10.1080/21655979.2022.2073130 [Google Scholar] [PubMed] [CrossRef]
5. Li, H., Li, Y., Tian, D., Zhang, J., Duan, S. (2021). miR-940 is a new biomarker with tumor diagnostic and prognostic value. Molecular Therapy Nucleic Acids, 25(7, Suppl), 53–66. https://doi.org/10.1016/j.omtn.2021.05.003 [Google Scholar] [PubMed] [CrossRef]
6. Shen, J., Wu, Y., Ruan, W., Zhu, F., Duan, S. (2022). miR-1908 dysregulation in human cancers. Frontiers in Oncology, 12, 857743. https://doi.org/10.3389/fonc.2022.857743 [Google Scholar] [PubMed] [CrossRef]
7. Zong, M., Feng, W., Wan, L., Yu, X., Yu, W. (2020). LncRNA TUG1 promotes esophageal cancer development through regulating PLK1 expression by sponging miR-1294. Biotechnology Letters, 42(12), 2537–2549. https://doi.org/10.1007/s10529-020-02984-0 [Google Scholar] [PubMed] [CrossRef]
8. Yang, T., Li, S., Liu, J., Yin, D., Yang, X. et al. (2020). Long non-coding RNA KRT16P2/miR-1294/EGFR axis regulates laryngeal squamous cell carcinoma cell aggressiveness. American Journal of Translational Research, 12(6), 2939–2955 [Google Scholar] [PubMed]
9. Wu, D., Li, H., Wang, J., Li, H., Xiao, Q. et al. (2020). LncRNA NEAT1 promotes gastric cancer progression via miR-1294/AKT1 axis. Open Medicine, 15(1), 1028–1038. https://doi.org/10.1515/med-2020-0218 [Google Scholar] [PubMed] [CrossRef]
10. Liang, W., Wang, C., Wang, J., Zhang, M. (2022). Hsa_circ_0023984 regulates cell proliferation, migration, and invasion in esophageal squamous cancer via regulating miR-1294/PI3K/Akt/c-Myc pathway. Applied Biochemistry and Biotechnology, 194(9), 1–16. https://doi.org/10.1007/s12010-022-03935-3 [Google Scholar] [PubMed] [CrossRef]
11. Wang, J., Li, J., Wang, H., Lv, L., Sun, J. (2019). Overexpression of circ_0005198 sponges miR-1294 to regulate cell proliferation, apoptosis, migration, and invasion in glioma. Journal of Cellular Biochemistry, 120(9), 15538–15545. https://doi.org/10.1002/jcb.28820 [Google Scholar] [PubMed] [CrossRef]
12. Hua, L., Huang, L., Zhang, X., Feng, H. (2020). Downregulation of hsa_circ_0000936 sensitizes resistant glioma cells to temozolomide by sponging miR-1294. Journal of Biosciences, 45(1), 101. https://doi.org/10.1007/s12038-020-00072-z [Google Scholar] [CrossRef]
13. Liu, J., Yang, Q., Sun, H., Wang, X., Saiyin, H. et al. (2020). The circ-AMOTL1/ENO1 axis implicated in the tumorigenesis of OLP-associated oral squamous cell carcinoma. Cancer Management and Research, 12, 7219–7230. https://doi.org/10.2147/CMAR.S251348 [Google Scholar] [PubMed] [CrossRef]
14. Chen, Y., Zhang, S., Bai, C., Guan, Z., Chen, W. (2020). Circ_0000885 enhances osteosarcoma progression by increasing FGFR1 expression via sponging miR-1294. Cancer Management and Research, 12, 6441–6452. https://doi.org/10.2147/CMAR.S244382 [Google Scholar] [PubMed] [CrossRef]
15. Shi, Y., Tian, Y., Sun, X., Qiu, Y., Zhao, Y. (2022). Silencing circOMA1 inhibits osteosarcoma progression by sponging miR-1294 to regulate c-Myc expression. Frontiers in Oncology, 12, 889583. https://doi.org/10.3389/fonc.2022.889583 [Google Scholar] [PubMed] [CrossRef]
16. Xu, Y., Yao, Y., Gao, P., Cui, Y. (2019). Upregulated circular RNA circ_0030235 predicts unfavorable prognosis in pancreatic ductal adenocarcinoma and facilitates cell progression by sponging miR-1253 and miR-1294. Biochemical and Biophysical Research Communications, 509(1), 138–142. https://doi.org/10.1016/j.bbrc.2018.12.088 [Google Scholar] [PubMed] [CrossRef]
17. Rong, Z., Shi, S., Tan, Z., Xu, J., Meng, Q. et al. (2021). Circular RNA CircEYA3 induces energy production to promote pancreatic ductal adenocarcinoma progression through the miR-1294/c-Myc axis. Molecular Cancer, 20(1), 106. https://doi.org/10.1186/s12943-021-01400-z [Google Scholar] [PubMed] [CrossRef]
18. Zhang, Z., Lin, W., Gao, L., Chen, K., Yang, C. et al. (2019). Hsa_circ_0004370 promotes esophageal cancer progression through miR-1294/LASP1 pathway. Bioscience Reports, 39(5), BSR20182377. https://doi.org/10.1042/BSR20182377 [Google Scholar] [PubMed] [CrossRef]
19. Pramanik, A., Xu, Z., Shamsuddin, S. H., Khaled, Y. S., Ingram, N. et al. (2022). Affimer tagged cubosomes: Targeting of carcinoembryonic antigen expressing colorectal cancer cells using in vitro and in vivo models. ACS Applied Materials & Interfaces, 14(9), 11078–11091. https://doi.org/10.1021/acsami.1c21655 [Google Scholar] [PubMed] [CrossRef]
20. Chen, W., Li, Y., Zhong, J., Wen, G. (2021). circ-PRKCI targets miR-1294 and miR-186-5p by downregulating FOXK1 expression to suppress glycolysis in hepatocellular carcinoma. Molecular Medicine Reports, 23(6), 464. https://doi.org/10.3892/mmr.2021.12103 [Google Scholar] [PubMed] [CrossRef]
21. Luo, Z., Lu, L., Tang, Q., Wei, W., Chen, P. et al. (2021). CircCAMSAP1 promotes hepatocellular carcinoma progression through miR-1294/GRAMD1A pathway. Journal of Cellular and Molecular Medicine, 25(8), 3793–3802. https://doi.org/10.1111/jcmm.16254 [Google Scholar] [PubMed] [CrossRef]
22. Yu, M. C., Ding, G. Y., Ma, P., Chen, Y. D., Zhu, X. D. et al. (2021). CircRNA UBAP2 serves as a sponge of miR-1294 to increase tumorigenesis in hepatocellular carcinoma through regulating c-Myc expression. Carcinogenesis, 42(10), 1293–1303. https://doi.org/10.1093/carcin/bgab068 [Google Scholar] [PubMed] [CrossRef]
23. Lin, G., Li, J., Chen, K., Wang, A., Guo, C. (2022). Circ_0000854 regulates the progression of hepatocellular carcinoma through miR-1294/IRGQ axis. Clinical Immunology, 238, 109007. https://doi.org/10.1016/j.clim.2022.109007 [Google Scholar] [PubMed] [CrossRef]
24. Zhang, Q., Wang, Z., Cai, H., Guo, D., Xu, W. et al. (2022). CircPLK1 acts as a carcinogenic driver to promote the development of malignant pleural mesothelioma by governing the miR-1294/HMGA1 pathway. Biochemical Genetics, 60(5), 1527–1546. https://doi.org/10.1007/s10528-022-10186-8 [Google Scholar] [PubMed] [CrossRef]
25. Li, C., Wang, G., Ma, X., Tao, T., Li, Q. et al. (2022). Upregulation of exosomal circPLK1 promotes the development of non-small cell lung cancer through the miR-1294/high mobility group protein A1 axis. Bioengineered, 13(2), 4185–4200. https://doi.org/10.1080/21655979.2022.2026727 [Google Scholar] [PubMed] [CrossRef]
26. Chen, W., Tang, D., Lin, J., Huang, X., Lin, S. et al. (2022). Exosomal circSHKBP1 participates in non-small cell lung cancer progression through PKM2-mediated glycolysis. Molecular Therapy Oncolytics, 24, 470–485. https://doi.org/10.1016/j.omto.2022.01.012 [Google Scholar] [PubMed] [CrossRef]
27. Chen, R., Liang, F., Yan, J., Wang, Y. (2022). CircCDK17 knockdown inhibits tumor progression and cell glycolysis by downregulaing YWHAZ expression through sponging miR-1294 in cervical cancer. Journal of Ovarian Research, 15(1), 24. https://doi.org/10.1186/s13048-022-00952-y [Google Scholar] [PubMed] [CrossRef]
28. Li, Y., Gao, X., Yang, C., Yan, H., Li, C. (2022). CircRNA hsa_circ_0018289 exerts an oncogenic role in cervical cancer progression through miR-1294/ICMT axis. Journal of Clinical Laboratory Analysis, 36(5), e24348. https://doi.org/10.1002/jcla.24348 [Google Scholar] [PubMed] [CrossRef]
29. Pan, W., Pang, L. J., Cai, H. L., Wu, Y., Zhang, W. et al. (2019). MiR-1294 acts as a tumor suppressor in clear cell renal cell carcinoma through targeting HOXA6. European Review for Medical and Pharmacological Sciences, 23(9), 3719–3725. https://doi.org/10.26355/eurrev_201905_17797 [Google Scholar] [PubMed] [CrossRef]
30. Zhang, Y., Huang, S., Guo, Y., Li, L. (2018). MiR-1294 confers cisplatin resistance in ovarian cancer cells by targeting IGF1R. Biomedicine & Pharmacotherapy, 106(1), 1357–1363. https://doi.org/10.1016/j.biopha.2018.07.059 [Google Scholar] [PubMed] [CrossRef]
31. Huang, X., Wu, B., Chen, M., Hong, L., Kong., P. et al. (2020). Depletion of exosomal circLDLR in follicle fluid derepresses miR-1294 function and inhibits estradiol production via CYP19A1 in polycystic ovary syndrome. Aging, 12(15), 15414–15435. https://doi.org/10.18632/aging.103602 [Google Scholar] [PubMed] [CrossRef]
32. Chen, K., Xiao, X., Xu, Z. (2022). MiR-1294 inhibits the progression of breast cancer via regulating ERK signaling. Bulletin du Cancer, 109(10), 999–1006. https://doi.org/10.1016/j.bulcan.2022.02.017 [Google Scholar] [PubMed] [CrossRef]
33. Liu, K., Li, L., Rusidanmu, A., Wang, Y., Lv, X. (2015). Down-regulation of miR-1294 is related to dismal prognosis of patients with esophageal squamous cell carcinoma through elevating C-MYC expression. Cellular Physiology and Biochemistry, 36(1), 100–110. https://doi.org/10.1159/000374056 [Google Scholar] [PubMed] [CrossRef]
34. Chen, H., Liu, L., Li, X., Shi, Y., Liu, N. (2018). MicroRNA-1294 inhibits the proliferation and enhances the chemosensitivity of glioma to temozolomide via the direct targeting of TPX2. American Journal of Cancer Research, 8(2), 291–301 [Google Scholar] [PubMed]
35. Wang, Z., Yan, J., Zou, T., Gao, H. (2018). MicroRNA-1294 inhibited oral squamous cell carcinoma growth by targeting c-Myc. Oncology Letters, 16(2), 2243–2250. https://doi.org/10.3892/ol.2018.8967 [Google Scholar] [PubMed] [CrossRef]
36. Wang, Y., Liu, G., Sun, S., Qin, J. (2020). miR-1294 alleviates epithelial-mesenchymal transition by repressing FOXK1 in gastric cancer. Genes & Genomics, 42(2), 217–224. https://doi.org/10.1007/s13258-019-00899-3 [Google Scholar] [PubMed] [CrossRef]
37. Yuan, Q., Yu, H., Chen, J., Song, X., Sun, L. (2020). Antitumor effect of miR-1294/Pyruvate kinase M2 signaling cascade in osteosarcoma cells. OncoTargets and Therapy, 13, 1637–1647. https://doi.org/10.2147/OTT [Google Scholar] [CrossRef]
38. Zhang, Z. F., Li, G. R., Cao, C. N., Xu, Q., Wang, G. D. et al. (2018). MicroRNA-1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. European Review for Medical and Pharmacological Sciences, 22(24), 8582–8588. https://doi.org/10.26355/eurrev_201812_16621 [Google Scholar] [PubMed] [CrossRef]
39. Cai, X., Yu, L., Chen, Z., Ye, F., Ren, Z. et al. (2020). Arsenic trioxide-induced upregulation of miR-1294 suppresses tumor growth in hepatocellular carcinoma by targeting TEAD1 and PIM1. Cancer Biomarkers, 28(2), 221–230. https://doi.org/10.3233/CBM-190490 [Google Scholar] [PubMed] [CrossRef]
40. Shi, Y. X., Ye, B. L., Hu, B. R., Ruan, X. J. (2018). Expression of miR-1294 is downregulated and predicts a poor prognosis in gastric cancer. European Review for Medical and Pharmacological Sciences, 22(17), 5525–5530. https://doi.org/10.26355/eurrev_201809_15813 [Google Scholar] [PubMed] [CrossRef]
41. Díaz-Coránguez, M., Liu, X., Antonetti, D. A. (2019). Tight junctions in cell proliferation. International Journal of Molecular Sciences, 20(23), 5972. https://doi.org/10.3390/ijms20235972 [Google Scholar] [PubMed] [CrossRef]
42. Pistritto, G., Trisciuoglio, D., Ceci, C., Garufi, A., D’Orazi, G. (2016). Apoptosis as anticancer mechanism: Function and dysfunction of its modulators and targeted therapeutic strategies. Aging, 8(4), 603–619. https://doi.org/10.18632/aging.100934 [Google Scholar] [PubMed] [CrossRef]
43. Duff, D., Long, A. (2017). Roles for RACK1 in cancer cell migration and invasion. Cellular Signalling, 35, 250–255. https://doi.org/10.1016/j.cellsig.2017.03.005 [Google Scholar] [PubMed] [CrossRef]
44. Xia, P., Xu, X. Y. (2015). PI3K/Akt/mTOR signaling pathway in cancer stem cells: From basic research to clinical application. American Journal of Cancer Research, 5(5), 1602–1609 [Google Scholar] [PubMed]
45. Aoki, M., Fujishita, T. (2017). Oncogenic roles of the PI3K/AKT/mTOR axis. Current Topics in Microbiology and Immunology, 407, 153–189. https://doi.org/10.1007/978-3-319-61804-3 [Google Scholar] [CrossRef]
46. Masliah-Planchon, J., Garinet, S., Pasmant, E. (2016). RAS-MAPK pathway epigenetic activation in cancer: miRNAs in action. Oncotarget, 7(25), 38892–38907. https://doi.org/10.18632/oncotarget.6476 [Google Scholar] [PubMed] [CrossRef]
47. Xin, P., Xu, X., Deng, C., Liu, S., Wang, Y. et al. (2020). The role of JAK/STAT signaling pathway and its inhibitors in diseases. International Immunopharmacology, 80, 106210. https://doi.org/10.1016/j.intimp.2020.106210 [Google Scholar] [PubMed] [CrossRef]
48. Guo, T. Y., Xu, H. Y., Chen, W. J., Wu, M. X., Dai, X. (2018). Downregulation of miR-1294 associates with prognosis and tumor progression in epithelial ovarian cancer. European Review for Medical and Pharmacological Sciences, 22(22), 7646–7652. https://doi.org/10.26355/eurrev_201811_16381 [Google Scholar] [PubMed] [CrossRef]
49. Hartung, M., Anastasi, E., Mamdouh, Z. M., Nogales, C., Schmidt, H. et al. (2022). Cancer driver drug interaction explorer. Nucleic Acids Research, 50(W1), W138–W144. https://doi.org/10.1093/nar/gkac384 [Google Scholar] [PubMed] [CrossRef]
50. Dasari, S., Tchounwou, P. B. (2014). Cisplatin in cancer therapy: Molecular mechanisms of action. European Journal of Pharmacology, 740, 364–378. https://doi.org/10.1016/j.ejphar.2014.07.025 [Google Scholar] [PubMed] [CrossRef]
51. Galluzzi, L., Senovilla, L., Vitale, I., Michels, J., Martins, I. et al. (2012). Molecular mechanisms of cisplatin resistance. Oncogene, 31(15), 1869–1883. https://doi.org/10.1038/onc.2011.384 [Google Scholar] [PubMed] [CrossRef]
52. Lee, S. Y. (2016). Temozolomide resistance in glioblastoma multiforme. Genes & Diseases, 3(3), 198–210. https://doi.org/10.1016/j.gendis.2016.04.007 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF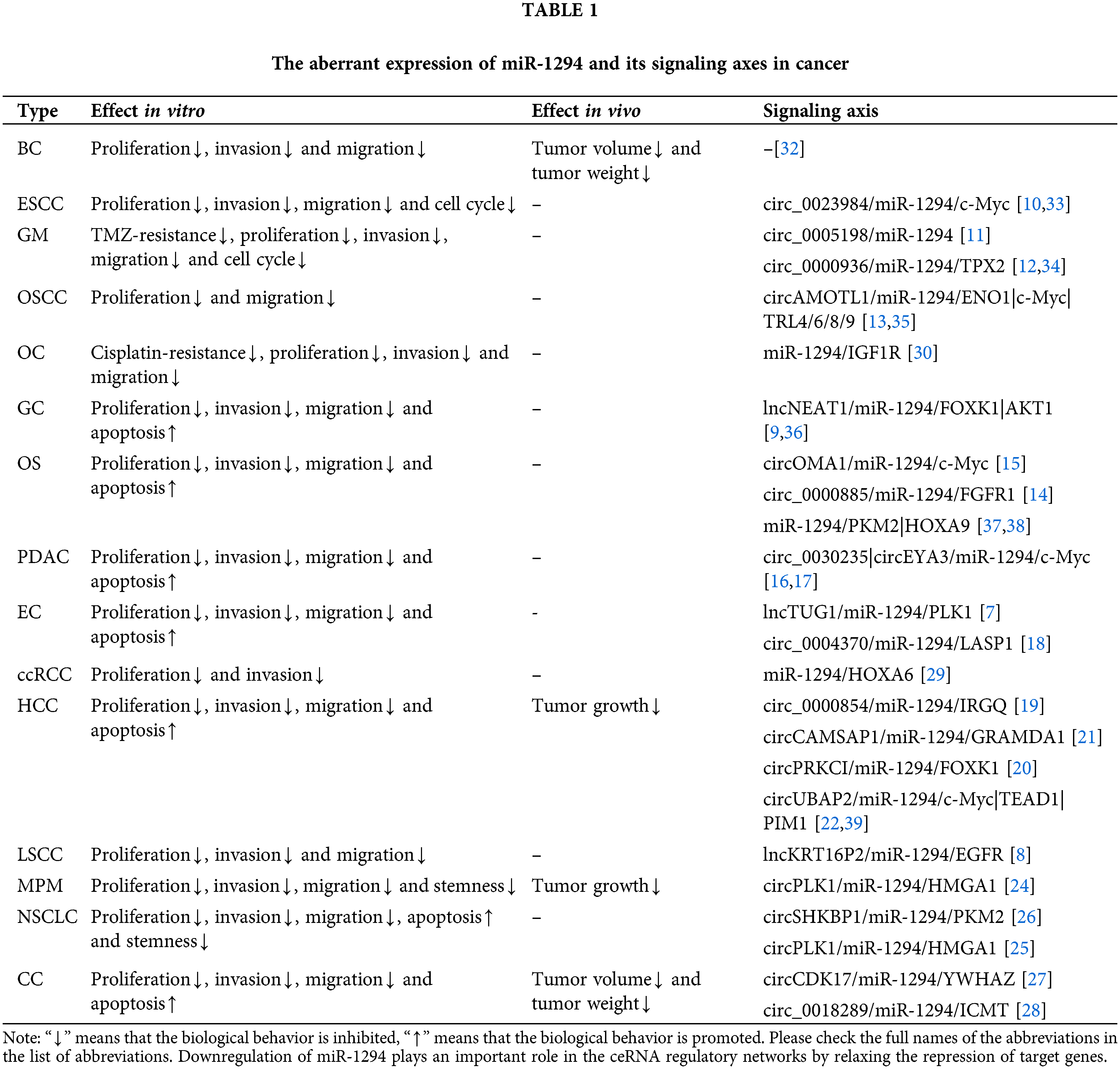
 Downloads
Downloads
 Citation Tools
Citation Tools