 Open Access
Open Access
ARTICLE
Silencing of peroxiredoxin 2 suppresses proliferation and Wnt/β-catenin pathway, and induces senescence in hepatocellular carcinoma
1 Department of Interventional Radiology, Sichuan Cancer Hospital and Institute, Sichuan Cancer Center, School of Medicine, University of Electronic Science and Technology of China, Radiation Oncology Key Laboratory of Sichuan Province, Chengdu, 610041, China
2 Department of Interventional Radiology, The First Affiliated Hospital of Sun Yat-sen University, Guangzhou, 510080, China
3 Department of Interventional Radiology, Affiliated Hospital of Guizhou Medical University, Guiyang, 550004, China
4 School of Basic Medical Science, Guizhou Medical University, Guiyang, 550002, China
* Corresponding Authors: TIANZHI AN. Email: ; ZHI HUANG. Email:
# These authors contributed equally to this work
(This article belongs to the Special Issue: Role of Reactive Oxygen Species and DNA Damage in Tumor Immunological Responses)
Oncology Research 2024, 32(1), 213-226. https://doi.org/10.32604/or.2023.030768
Received 21 April 2023; Accepted 03 August 2023; Issue published 15 November 2023
Abstract
Hepatocellular carcinoma (HCC), a common malignancy worldwide, still lacks effective clinical treatment. The study aimed to investigate the oncogenes that affect the progression of HCC and their possible mechanisms. In our study, we initially confirmed a higher level of PRDX2 in the bile of HCC patients compared to those with choledocholithiasis by 2-DE, LC-MS, and ELISA. Subsequently, we demonstrated the high expression of peroxiredoxin 2 (PRDX2) in HCC based on the TCGA database and clinical sample analysis. Furthermore, PRDX2 overexpression enhanced the viability of HCC cells. And PRDX2 silencing induced senescence of HCC cells. In vivo, knockdown of PRDX2 significantly reduced the weight of xenograft tumors. PRDX2 also was found to activate the Wnt/β-catenin pathway by inducing β-catenin nuclear translocation. Consequently, we proved that silencing PRDX2 could inhibit proliferation and Wnt/β-catenin pathway while promoting senescence in HCC cells.Keywords
Hepatocellular carcinoma (HCC) is currently the sixth most common malignant cancer worldwide [1,2]. The majority of HCC cases (>80%) occur in sub-Saharan Africa and Eastern Asia [3]. China, in particular, is considered a high-risk regions for HCC, primarily due to factors such as chronic hepatitis B virus (HBV) infection or aflatoxin exposure [4,5]. Despite various treatment techniques available for HCC patients in clinical practice, long-term survival outcomes remain unsatisfactory [6]. The challenges in HCC treatment largely stem from our limited understanding of the molecular-level tumorigenic mechanism of HCC. In addition, early diagnosis of HCC remains challenging due to the high incidence of asymptomatic disease, resulting in a reported 5-year survival rate of approximately 10% [7]. Therefore, further exploration of the pathogenesis and mechanism underlying HCC is of great significance to reduce HCC-related mortality.
Recent studies have demonstrated the utility of proteomic analysis for quantifying proteins in bile and exploring potential biomarkers [8,9]. Through screening with two-dimensional electrophoresis (2-DE), we identified peroxiredoxin 2 (PRDX2) as a potential therapeutic marker for HCC. PRDX2, a key member of the PRDX family, functions as a vital intracellular antioxidant [10]. It actively participates in the cellular antioxidant defense system, effectively mitigating cellular damage caused by oxidative stress and promoting tumor cell proliferation and survival [11]. Furthermore, PRDX2 plays a crucial role in signal transduction, cell differentiation, apoptosis, metastasis, radiotherapy resistance, and other processes [12,13]. High expression of PRDX2 has also been reported in various cancer cells and tissues, contributing to cancer progression [14,15]. Increasing evidence suggests that PRDX2 has a regulatory role in tumorigenesis [14,16,17]. Several studies have also proved the involvement of PRDX2 in HCC progression [18–22], making it a valuable candidate for assessing therapeutic efficacy and cancer prognosis. However, the precise effects of PRDX2 on HCC cell senescence and the underlying mechanisms remain incompletely understood.
Accumulating evidence suggests that dysregulation of Wnt/β-catenin pathway plays a crucial role in the tumorigenesis and development of certain solid tumors and hematological malignancies [23–25]. Activation of Wnt/β-catenin pathway occurs through the accumulation of β-catenin protein in the nucleus [26,27]. It has been reported that approximately 40%–70% of HCC patients exhibit nuclear accumulation of β-catenin protein [28,29]. The activation of Wnt/β-catenin pathway results in extensive transcription of downstream target genes [30]. Moreover, research has also manifested that PRDX2 could promote the progression of esophageal squamous cell carcinoma through the Wnt/β-catenin pathway [31]. However, whether PRDX2 regulates Wnt/β-catenin pathway in HCC cells is unclear.
In this study, we conducted further investigations to explore the effects of PRDX2 on the proliferation and senescence of HCC cells both in vitro and in vivo. Additionally, we sought to verify the effects of PRDX2 on Wnt/β-catenin pathway. Consequently, our findings support the proposition that that PRDX2 may serve as a potential biomarker for HCC.
In this study, bile fluid samples were collected from 26 patients at the Affiliated Hospital of Guizhou Medical University, China, between January 2011 and August 2013. All patients were followed up for a period of 3 to 6 months, with a minimum follow-up duration of one year. Additionally, serum samples from 35 patients constituted an independent “validation” cohort. HCC was diagnosed based on the criteria established by the American Association for the Study of Liver Diseases (AASLD) [32]. None of the HCC patients received chemotherapy. Choledocholithiasis was diagnosed through magnetic resonance imaging, endoscopic ultrasound, endoscopic retrograde cholangiography (ERCP), and other diagnosis methods. This study was approved by the local ethics committee of Guizhou Medical University. The written informed consent was obtained from all participants.
Bile fluid samples were obtained from patients undergoing ERCP for choledocholithiasis (n = 14) or percutaneous transhepatic cholangial drainage (PTCD) for HCC (n = 12). Up to 20 ml of bile fluid samples were collected in prechilled, ice-cold tubes and immediately centrifuged at 16000 g for 15 min at 4°C to remove cell debris, nucleic acid and mucins. The resulting supernatant was then stored in 1 ml aliquots at −80°C until further analysis. Serum samples were also collected and stored accordingly. For immunohistochemistry analysis, a total of 9 pairs of HCC and para-carcinoma tissues were obtained from the Department of Surgery, Affiliated Hospital of Guiyang Medical College, China.
Pretreatment of bile prior to proteomic analysis
To prepare the bile samples for proteomic analysis, 1 ml of preliminary separated bile samples were chilled on the ice and mixed with 250 μl of cleanascite™ (Biotech Support Group, NJ, USA). After 1 h of rotation, samples were centrifuged to remove lipid micelles. Subsequently, the resulting supernatant was transferred to a Millipore YM-3 Centurion Filter Unit (Bedford, MA, USA), where it underwent centrifugation and filtration [33]. Finally, 200 μl of MilliQ water was added, and the centrifugation process was repeated three times, following previously described protocols [34].
Two-dimensional electrophoresis (2-DE)
Based on previous studies [35,36], a total of eight patients (choledocholithiasis, n = 4; HCC, n = 4) were included in this study. Bile samples were mixed with pre-cooled acetone at a four-fold volume of precipitation. After incubating the mixture at 20°C for 2 h, the precipitated proteins were collected by centrifugation. Acetone was discarded, and the protein pellet was subjected to ice drying. Subsequently, protein pellets were re-solubilized using a lysis buffer containing 4% CHAPS, 0.001% bromophenol blue, 2 M thiourea, 65 mM dithiothreitol, 7 M urea, 0.5% pH 3–10 and IPG buffer. Protein concentrations were determined using the Bio-Rad DC system. Each 1 mg of purified sample was diluted to 350 μl with the buffer. Protein samples were then loaded onto Immobilized pH gradient (IPG) strips (17 cm; Bio-Rad, USA) for isoelectric focusing (IEF) with a commercial device (Protean IEF Cell, Bio-Rad, USA). The focusing step was covered with mineral oil (Bio-Rad, USA). Isoelectric focusing was performed following the manufacturer’s instructions. After isoelectric focusing, IPG strips were equilibrated by immersing them in an equilibration buffer for 15 min. SDS-PAGE separation was then performed with a vertical electrophoresis system (Bio-Rad, Protein II, USA) at a current of 5 mA/gel for 30 min, followed by 25 mA/gel for 5 h.
In accordance with the study [37], the gels were fixed with a solution containing 40% methanol and 10% acetic acid for 1 h. Following fixation, the gels were stained with 0.025% Komas Blue R-350 (Imperial Chemical Industries, London, UK) and subsequently heated to 80°C–90°C. For gel destaining, a 10% acetic acid solution was employed. Subsequently, the gels were scanned using a commercial imaging program (Bio-Rad Labroratories, GS-800, USA). The acquired images were analyzed with PDQuest 2D software (Bio-Rad, USA). Protein spots exhibiting a fold-change in intensity of at least 2-fold were selected, subjected to digestion, and analyzed using MALDI-TOF-MS.
Identification of protein by MALDI-TOF/TOF-MS analysis
Protein identification was conducted following previously reported protocols [38]. Protein spots on the 2D gels were manually selected, excised, and subjected to digestion. To decontaminated the gel spots, a solution of 25 mM ammonium bicarbonate and 50% acetonitrile (ACN) was employed at 37°C, followed by ACN dehydration. Subsequently, the gel blocks were digested and treated with 25 mm ammonium bicarbonate for 8 h at 37°C. After elution, the peptides were solubilized and analyzed using MALDI-TOF/TOF-MS. The MALDI-TOF and MALDI-TOF/TOF spectra were acquired using an Ultraflex III TOF/TOF mass spectrometer (Bruker Daltonics, Bremen, Germany). Peptide mass fingerprint (PMF) data were compared with Swiss-Prot or NCBInr databases using MASCOT [26,27], and the confidence levels of the identified proteins were determined. A MASCOT scores greater than 62 indicates statistical significance (p < 0.05).
The tissues were ground, and cells from each group were collected. Total RNA was extracted using the RNeasy Mini Kit (QIAGEN, Hilden, Germany). Subsequently, cDNA synthesis was performed using a reverse transcription kit (Promega). RT-qPCR was conducted using the SYBR-Green PCR kit (TransGen Biotech, Inc., Beijing, China). The data was analyzed using the 2−ΔΔCt method to determine relative gene expression levels.
A total of 20 μg of proteins from each patient’s bile (choledocholithiasis, n = 14; HCC, n = 12) were separated by SDS-gel electrophoresis and transferred to PVDF membranes (Millipore, USA). The membranes were then incubated overnight at 4°C with primary antibodies, including anti-PRDX2 (1:1000, Abcam, ab109367), anti-CyclinD1 (1:1000, Abcam, ab16663), anti-cMYC (1:1000, Abcam, ab32072), anti-c-Jun (1:1000, Abcam, ab40766), anti-fra1 (1:1000, Abcam, ab252421), anti-β-catenin (1:1000, Abcam, ab32572), anti-Lamin B1 (1:1000, Abcam, ab16048), anti-GAPDH (1:2000, Abcam, ab76523) and anti-β-actin (1:1000, Abcam, ab8227). Following primary antibody incubation, the membranes were washed and incubated with peroxidase-conjugated secondary antibody (1:2000, Abcam, Cambridge, UK). Subsequently, substrate development was performed using ECL chemiluminescence (Millipore, Bedford, Mass, USA).
Immunohistochemistry (IHC) assay
HCC and para-tumor tissues were fixed, dehydrated, cleared, immersed in wax, embedded, and sectioned. Paraffin sections were then subjected to gradient ethanol dehydration and antigen retrieval using sodium citrate. After blocking, the sections were incubated overnight at 4°C with primary antibodies against PRDX2 (Abcam, ab59539, 1:50). Then, HRP-labeled secondary antibodies (1:200, Abcam) were applied and incubated at 37°C for 30 min. The sections were subsequently stained with diamino-benzidine (DAB, Maxim Biotech, Fuzhou, China) and hematoxylin (Solarbio, China; cat. no. H8070), followed by differentiation, hydration, and making them transparent. Finally, the slides were sealed, and the results were observed under a microscope (Nikon, Tokyo, Japan).
Enzyme-linked immunosorbent assay (ELISA) assay
ELISA was performed to determine the levels of PRDX2 using a kit supplied by CUSABIO (Wuhan, Hubei, China) according to the manufacturers’ protocols.
The SNU-387, PLC.5, SK-Hep-1, Huh7, and HepG2 (Purchased from ATCC) cells were cultured in DMEM (Gibco, cat. no. 1859228) supplemented with 10% fetal bovine serum (FBS, Gibco), 100 units/ml penicillin G, and 100 mg/ml streptomycin. The cells were incubated at 37°C in a cell culture incubator with 5% CO2. Prior to the experiment, the cells were collected for further analysis.
The day before transfection, cells were plated in 6-well plate with normal growth medium without antibiotics at a density of 2 × 105 cells/well. On the day of transfection, when the cell density reaches 80%, the medium was replaced with Opti-MEM (Invitrogen). The DNA or Lipofectamine 2000 was mixed separately with Opti-MEM and incubated for 5 min. Subsequently, the diluted DNA and diluted Lipofectamine 2000 (Invitrogen) were gently mixed together and left at room temperature for 20 min before adding to cells.
HuH-7 cells were transfected with indicated plasmids. Following transfection, cells were subjected to puromycin selection at a concentration of 1 μg/mL for 4 days before conducting the CCK-8 assay. For the CCK-8 assay, 100 μl of transfected HuH-7 cells (2 × 103 cells) were transferred to each well of a 96-well plate and incubated for 24 h. Afterward, 10 μl of CCK-8 solution (Beyotime, China) was added to each well, and the plate was incubated at 37°C for 1.5 h. The absorbance values were then measured using a microplate reader.
HuH-7 cells were transfected with indicated plasmids. After transfection, puromycin selection was applied at a concentration of 1 μg/mL for 4 days before performing colony forming assay. For the assay, 500 transfected cells were plated into 6-well plate containing medium with 10% FBS and 1% Penicillin-Streptomycin. Cells were then cultured for approximately 2 weeks, fixed with cold methanol, and stained with 0.1% crystal violet.
Coverslips were plated in 6-well plates, and HepG2 cells were added onto them. The following day, cells were fixed with 4% paraformaldehyde, then permeabilized by treating them with PBS containing 0.1% Triton X-100 for 15 min. The slides were washed three times with PBS, then blocked with 2% BSA for 30–45 min at room temperature, The slides were incubated overnight with the following primary antibodies: anti-PRDX2 rabbit monoclonal antibody (1:250, ab109367, Abcam) and anti-β-catenin mouse monoclonal antibody (1:200, ab19381, Abcam) at 4 degrees. The next day, the slides were washed three times with PBST for 5 min each time. Then, the slides were incubated with fluorescent secondary antibody: Goat anti-rabbit-DyLight® 488 (1:1000, ab96899, Abcam) and Goat anti-mouse-DyLight® 647 (1:1000, ab150115, Abcam) for one hour at room temperature in the dark. Afterwards, the slides were washed three times with PBST and observed using a fluorescent microscope.
Top-flash reporter plasmid and PRDX2 expression plasmid were co-transfected into HEK293 cells, following the transfection procedure. After 2 days of transfection, a dual luciferase assay kit (Promega) was used to measure the luciferase activity of the cells.
A total of 16 BALB/c nude mice (8 weeks old) were purchased from Jiangsu Jicui Yaokang Biotechnology Co., Ltd. (Nanjing, China). All mice were individually housed in an SPF animal laboratory. Following a period of 1 to 2 weeks of adaptive feeding, HepG2-shGFP (shGFP) and HepG2-PRDX2-shRNA#2 (shRNA#2) cells (2 × 106) were separately suspended in Matrigel. Subsequently, these cells were subcutaneously implanted into the flanks of 8-week-old BALB/c nude mice. Tumor volume was monitored on days 5, 8, 11, 14, 17, and 20. This study was approved by the Research Ethics Committee of the Guizhou Medical University.
HepG2 cells were treated with shGFP or sh#2 for 48 h and collected. After centrifugation, the β-Gal activity was analyzed using the β-Gal assay kit (Solarbio, Beijing, China) following the instructions provided with the Kit.
After washing twice with pre-cooled PBS, the cells were lysed with pre-cooled RIPA buffer. The lysates were then centrifuged at 14000 g for 15 min at 4°C, and the resulting supernatant was transferred to a new centrifuge tube. The supernatant was incubated overnight at 4°C with 1 μg of primary antibody/20 μl of protein A/G beads (Abcam). The next day, samples were centrifuged at 14000 g for 15 min at 4°C. The supernatant was discarded, and the beads were washed with loading buffer. Subsequently, the beads were boiled at 100°C for 5 min, and Western blotting was performed.
The data of this study were obtained from three replicate experiments, and statistical analysis was performed using Graph Pad Prism. The parametric tests used in this study included Mann-Whitney U-test and Student’s t test. Measurement data were presented as Mean ± SD. Unpaired t test and Bonferroni post hoc test were used to compare two groups of data with normal distribution and homogeneity of variance. Pearson correlation analysis was used to analyze the relationship between variables. Multiple regression analyses were performed using SPSS version 16.0 (SPSS, Chicago, IL, USA). Immunohistochemical markers were evaluated using Fisher’s exact test. Statistical significance was defined as p < 0.05.
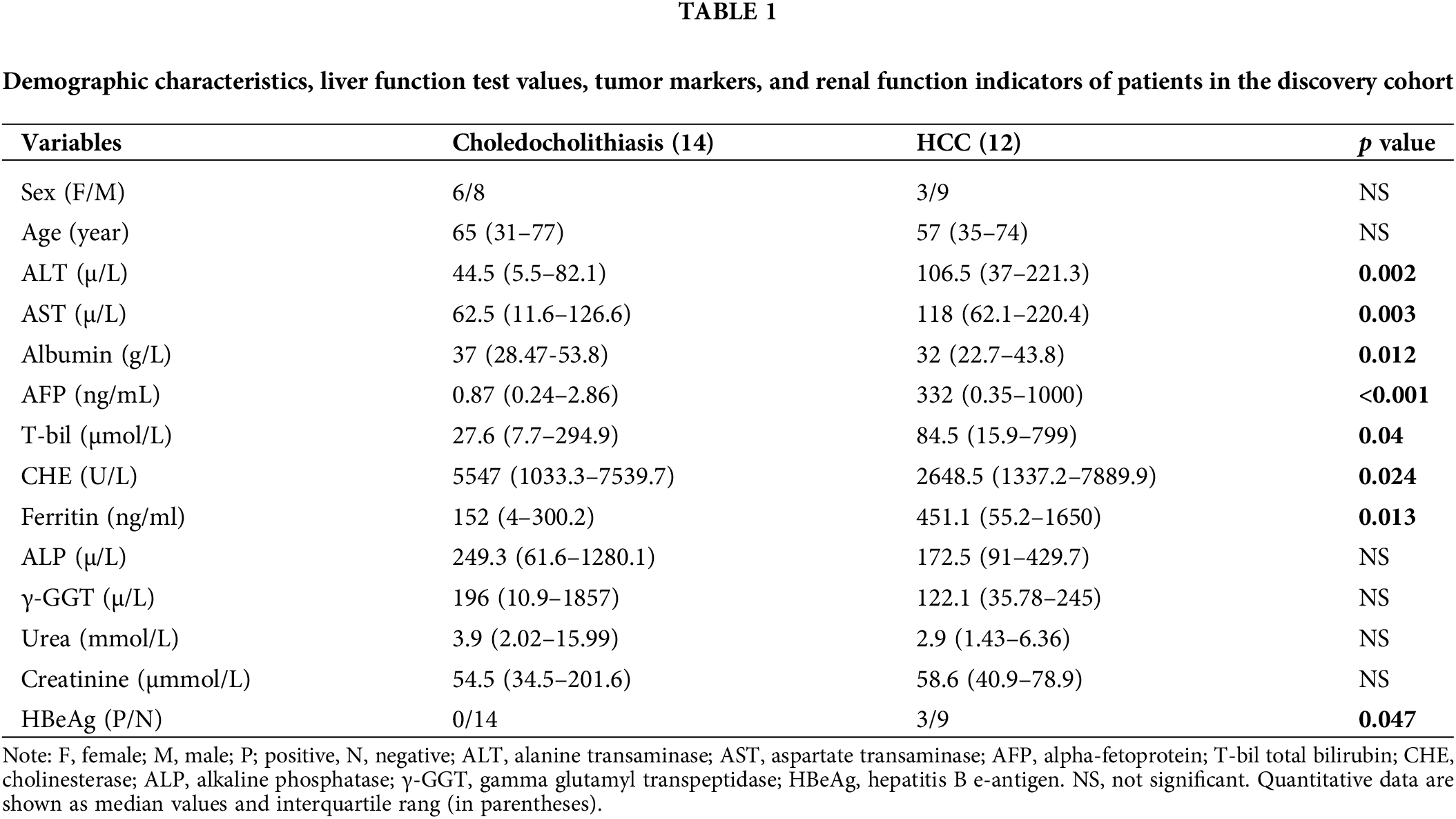
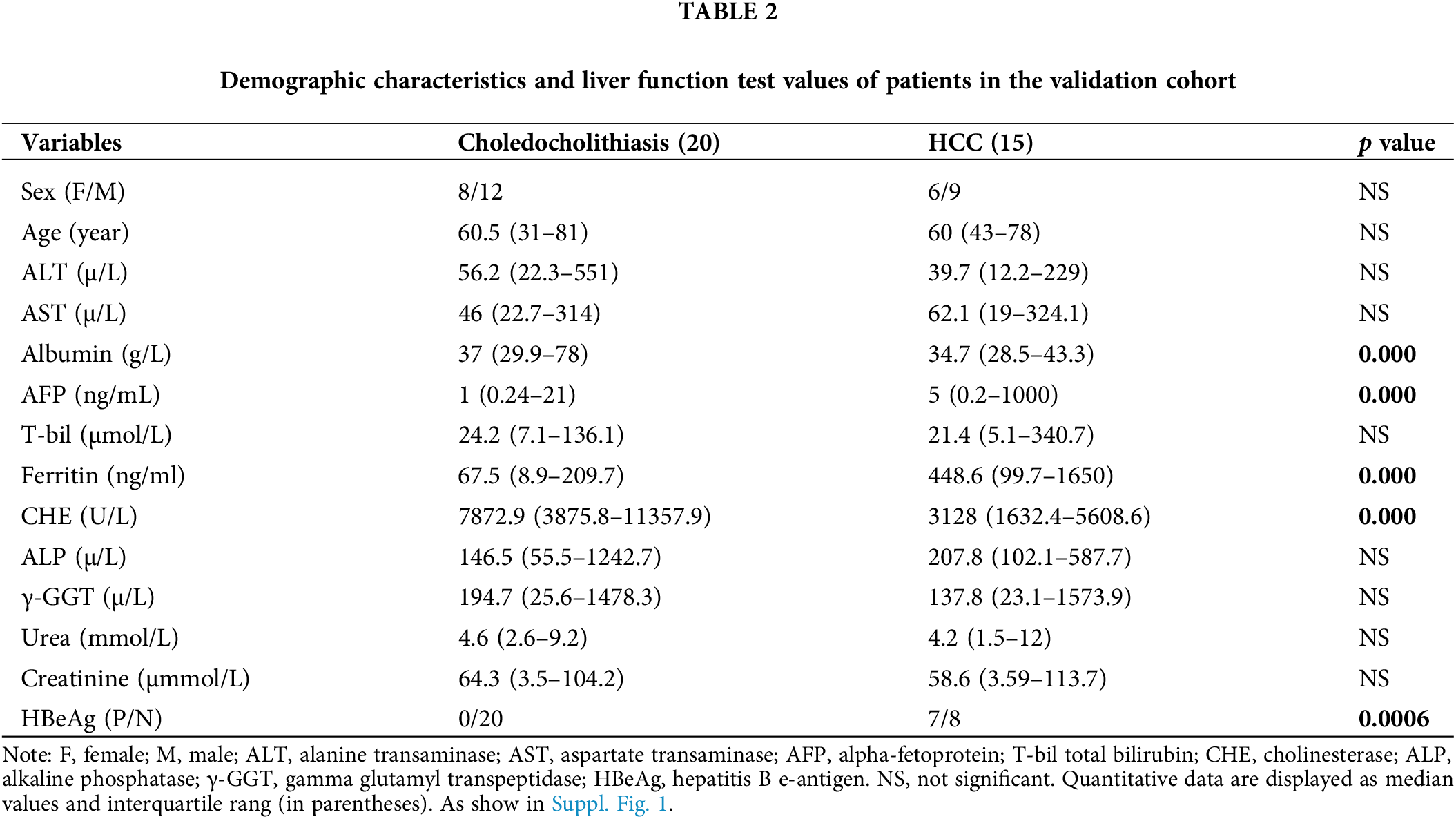
Basic information about the patients
We initially collected bile fluid samples from 26 patients. The demographic data of these patients revealed that 14 samples were obtained from choledocholithiasis patients (6 females, mean age 65 with a range of 31 to 77) and 12 samples were obtained from HCC patients (3 females, mean age 57 with a range of 35 to 74). The levels of ALT, AST, AFP, T-bil, Ferritin, and HBeAg were significantly elevated, while the levels of Albumin and CHE were markedly reduced in HCC patients compared to choledocholithiasis patients. There were no significant differences in sex, age, ALP, γ-GGT, urea, and creatinine between the two groups of patients (Table 1). Furthermore, we collected serum samples from additional 35 patients. The data from these 35 patients indicated that 20 samples were obtained from choledocholithiasis patients (8 females, mean age 60.5 with the range of 31–81), and 15 samples were obtained from HCC patients (6 females, mean age 60 with the range of 43–78). The levels of AFP, Ferritin, and HBeAg were notably increased, while the levels of Albumin and CHE were significantly decreased in HCC patients compared to choledocholithiasis patients. There were no significant changes in sex, age, ALT, AST, T-bil, ALP, γ-GGT, urea, and creatinine between the two groups of patients (Table 2).
PRDX2 is a potential diagnostic biomarker of HCC
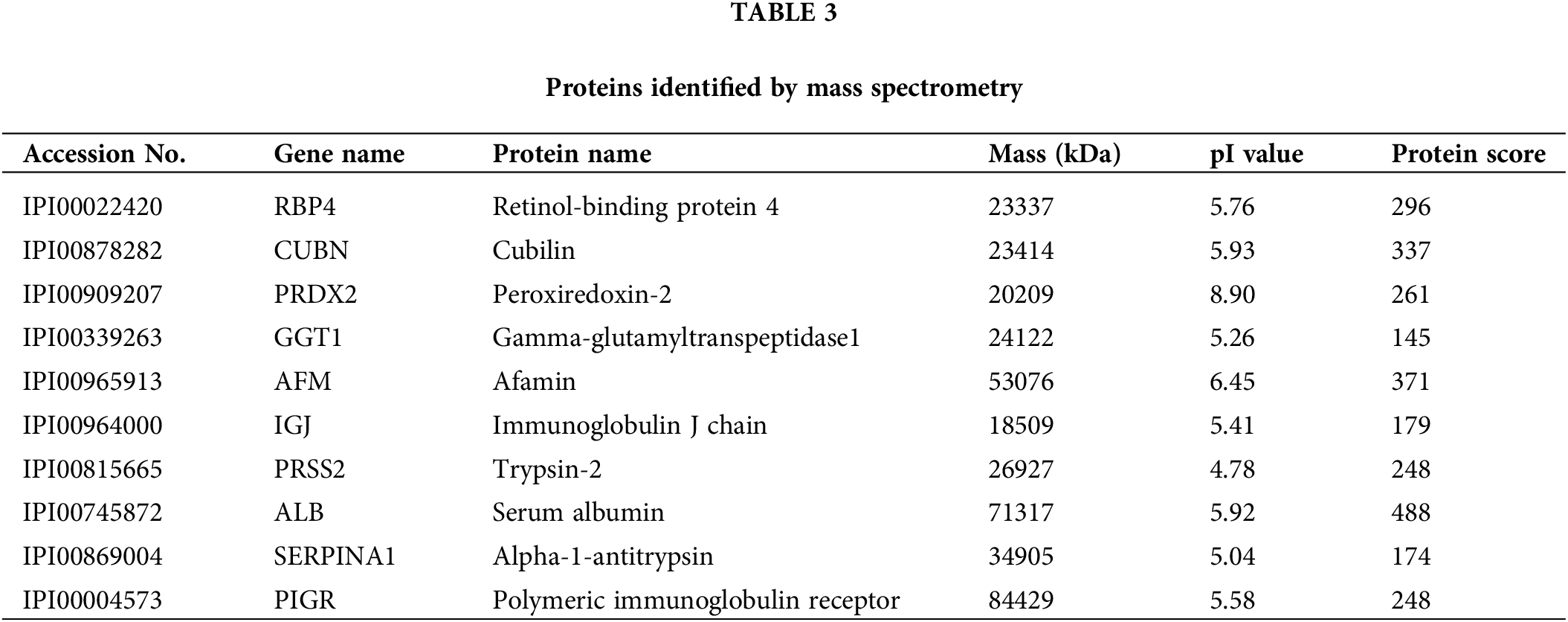
To investigate the potential diagnostic and therapeutic markers for HCC, we analyzed the related proteins extracted from bile samples of four HCC and four paired choledocholithiasis patients in the “discovery” cohort. The proteins were separated using 2-DE and stained with Coomassie brilliant blue. By employing MALDI-TOF-MS and peptide mass matching, we identified the differentially expressed protein spots in four sample pairs (Fig. 1A). And there was a total of 10 identified differentially expressed proteins. Among them, three proteins (Peroxiredoxin-2, Afamin, and Alpha-1-antitrypsin) were up-regulated in HCC, while seven were down-regulated (Table 3). The molecular mass/pI value of PRDX2 was determined to be 20.209 kDa/8.9, corresponding to spot 5 on the 2-DE gel. The MS/MS spectrum of PRDX2 was shown in Suppl. Fig. 1. Further experiments illustrated a significantly higher PRDX2 level in both bile and serum samples from the full set of the “validation” cohort. In bile, the expression level of PRDX2 in HCC patients was markedly higher than that in choledocholithiasis patients (median level 9,395 vs. 4,858 ng/ml, p = 0.000) (Fig. 1B). Similarly, the level of PRDX2 in serum of HCC patients was notably higher than that in choledocholithiasis patients (median level 5,780 vs. 3,743 ng/ml, p = 0.000) (Fig. 1C). Taken together, these data indicated that PRDX2 held potential as a diagnostic biomarker of HCC.
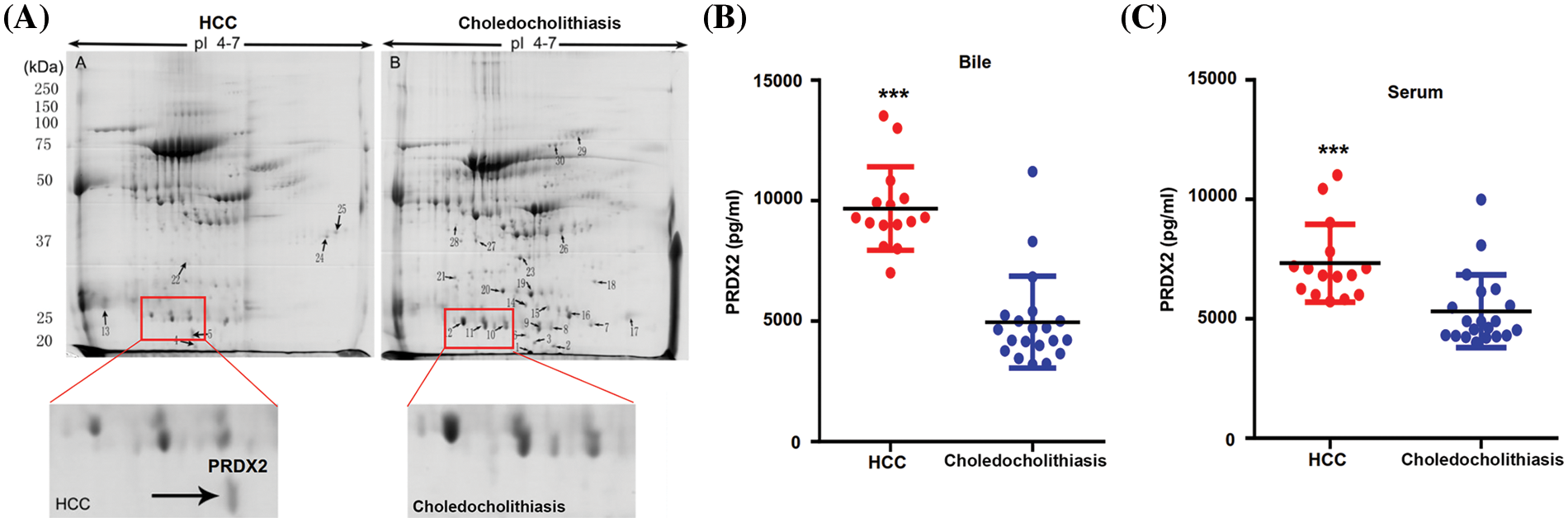
Figure 1: PRDX2 was a potential diagnostic biomarker of HCC. (A) Differential proteomic analysis of bile from HCC (left panel) and choledocholithiasis (right panel) using 2-DE gels. Total protein lysates were separated by isoelectric focusing (IEF) using IPG strips (17 cm, pH4-7) in the first dimension followed by 8% SDS-PAGE in the second dimension. Coomassie-stained gels were analyzed using the PD-Quest 2D analysis software. Totally, 30 differentially expressed spots were identified by MS/MS analysis (upper); 2-DE gel images of PRDX2 in bile from HCC and choledocholithiasis (lower). PRDX2 was upregulated more than 2-fold in bile from HCC when compared with the choledocholithiasis. (B and C) PRDX2 expression was measured by ELISA in the bile (B) and serum (C) of patients with HCC and choledocholithiasis. Horizontal lines indicate the medians. ***p < 0.001.
PRDX2 is a clinic relevant oncogene in HCC
To investigate the potential oncogenic role of PRDX2 in HCC, we analyzed TCGA data and examined the expression change of PRDX2 in the tissues of HCC patients, including tumor tissues and adjacent non-tumor tissues. Results revealed that the expression level of PRDX2 in HCC tumor tissues was significantly higher than that in para-tumor tissues (Fig. 2A). Furthermore, we conducted IHC analysis on HCC tissues and para-tumor tissues obtained from the same HCC patients (n = 9). As shown in Fig. 2B, IHC results demonstrated that the expression of PRDX2 was significantly increased in HCC tumor tissues compared to that in para-tumor tissues. Western blot results also confirmed the increased protein level of PRDX2 in HCC tumor tissues (Fig. 2C). Furthermore, the high expression of PRDX2 was also found to be associated with poor survival of HCC patients (Fig. 2D). To sum up, these results indicated that PRDX2 was a clinically relevant oncogene in HCC.
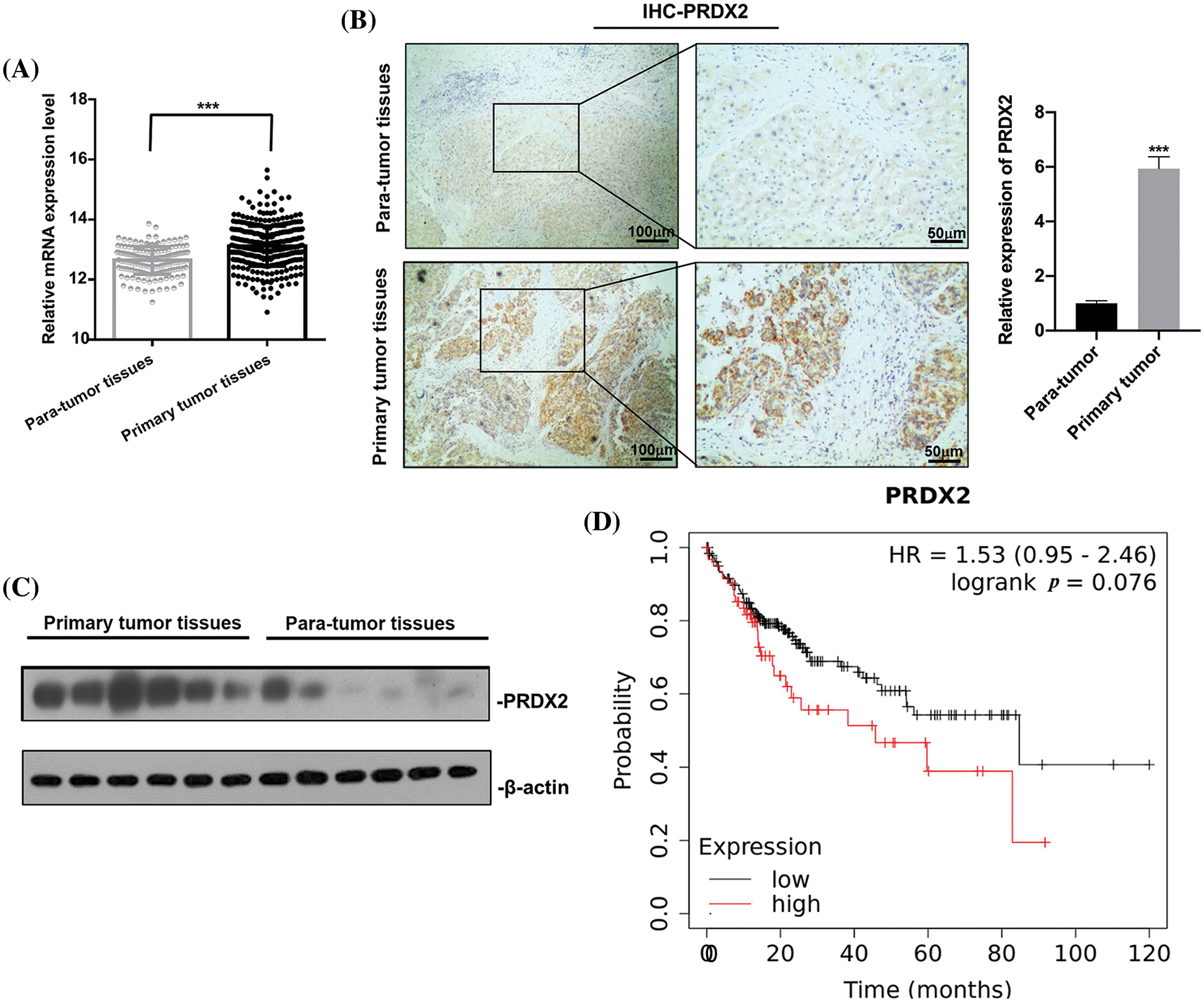
Figure 2: PRDX2 is a clinic relevant oncogene in HCC. (A) PRDX2 mRNA expression in liver cancer tissue and para-tumoral tissue among the HCC patients from TCGA database. (B) IHC results of PRDX2 expression in HCC tissues (primary tumor tissues) and adjacent non-tumor/normal liver tissues (para-tumor tissues), scale bars: 50, 100 μm; magnification: 100× or 400×. (C) Western blotting analysis of PRDX2 expression in primary tumor tissues and para-tumor tissues; 20 μg protein of crude bile from each specimen was loaded onto a homemade 12% SDS-PAGE for separation. Proteins were electroblotted onto a nitrocellulose membrane. Western blot was performed using rabbit monoclonal anti-PRDX2. Lanes 1–6, protein of HCC tissues; lanes 7–12, protein of para-tumor tissues. (D) K-M survival analysis of PRDX2 in HCC patients using TCGA database. ***p < 0.001.
PRDX2 promotes the proliferation of HUH-7 cells
To investigate the oncogene function of PRDX2 in vitro, we first analyzed PRDX2 expression in different HCC cell lines. We found that the level of PRDX2 was relatively higher in HepG2 cells than that in other cells including SNU-387, HUH-7, PLC-PRF-5, and SK-HEP1 cells, and the expression level of PRDX2 was lowest in HUH-7 cells (Figs. 3A and 3B). Subsequently, we transfected HUH-7 cells with a PRDX2 overexpression plasmid or a control vector, followed by puromycin selection. Western blotting data confirmed that compared to the vector group, there was a significant increase in PRDX2 expression in the PRDX2-overexpression group (Fig. 3C). To further assess the impact of PRDX2 on the proliferation of HUH-7 cells, we performed CCK-8 and colony forming assays. As shown in Figs. 3D and 3E, overexpression of PRDX2 promoted the growth and colony forming ability of HUH-7 cells.
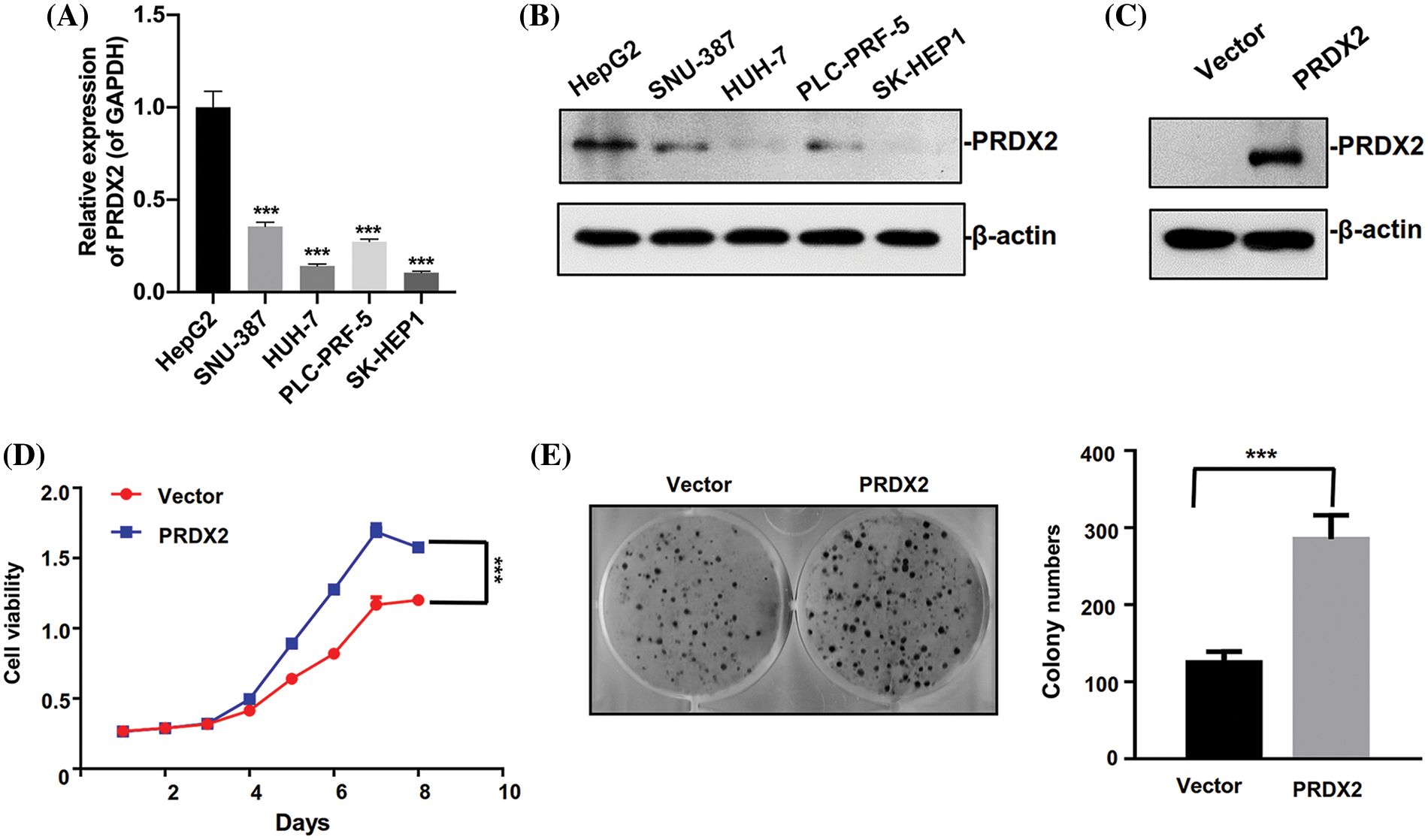
Figure 3: Ectopic expression of PRDX2 promotes HUH7 cells aggressive behavior. (A) RT-qPCR analysis of PRDX2 expression in HepG2, SNU-387, HUH-7, PLC-PRF-5, and SK-HEP1 cells. (B) PRDX2 expression was analyzed using western blot in HepG2, SNU-387, HUH-7, PLC-PRF-5, and SK-HEP1 cell lines. (C) HUH-7 cells were transfected with Vector and PRDX2 for 48 h, and then the cells were treated with puromycin (1 μg/ml) for 48 h, PRDX2 expression was identified with western blot. (D) HUH-7 cells were treated with PRDX2, followed by puromycin (1 μg/ml), and CCK-8 was adopted to assess the effect of PRDX2 overexpression on the viability of HUH-7 cells. OD450 was determined as indicated time points. (E) Effect of PRDX2 overexpression on colony forming ability of HUH-7 cells. After processing with PRDX2 and puromycin, colony forming assay was conducted. ***p < 0.001.
Silence of PRDX2 prevents the proliferation of HepG2 cells
Based on the above results, the expression of PRDX2 was found to be highest in HepG2 cells relative to other HCC cell lines. Therefore, the HepG2 cell line was chosen to investigate the impact of PRDX2 knockdown on the relevant function of HCC cells. HepG2 cells were transfected with PRDX2-shRNA-#1 (sh#1) and PRDX2-shRNA-#2 (sh#2). Western blot results indicated that knockdown of PRDX2 efficiently reduced its expression in HepG2 cells, especially with the transfection of sh#2 (Fig. 4A). Subsequently, our results demonstrated that knockdown of PRDX2 led to a decrease in the viability and colony forming ability of HepG2 cells, especially with sh#2 (Figs. 4B and 4C). Taken together, our results showed that PRDX2 contributed to the proliferation of HCC cells.
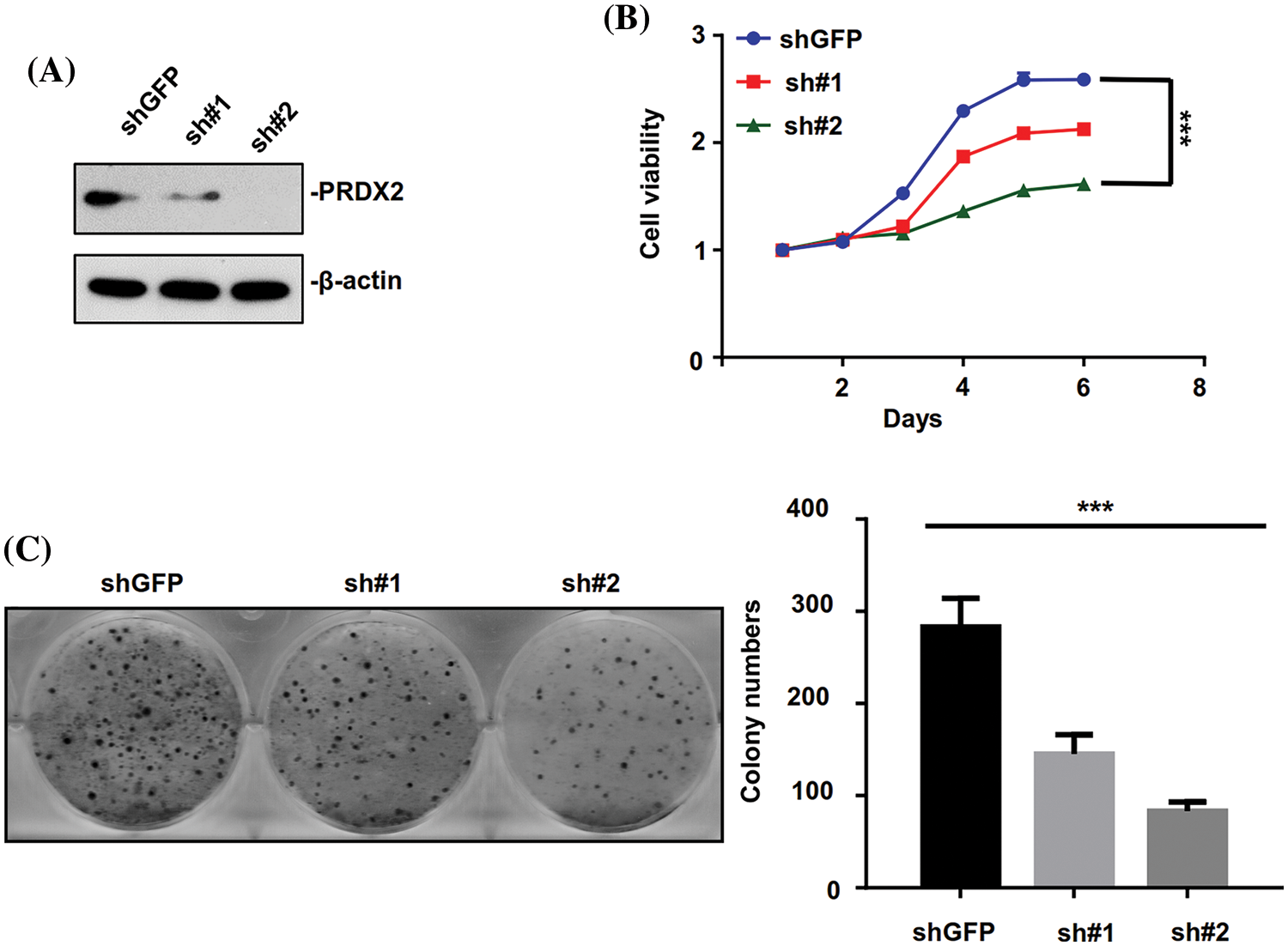
Figure 4: PRDX2 silencing inhibits the aggressive behavior of HepG2 cells. (A) HepG2 cells were transfected with shGFP, pRDX2-shRNA-#1 (sh#1), and PRDX2-shRNA-#2 (sh#2) for 48 h, and then the cells were treated with puromycin (1 μg/ml), PRDX2 expression was monitored with western blot. (B) After processing with sh#1, sh#2, or puromycin, cell viability was confirmed with CCK-8. (C) The proliferation ability was examined using colony forming assay in HepG2 cells after treatment with shGFP, sh#1, sh#2, or puromycin. ***p < 0.001.
Silence of PRDX2 induces senescence and weakens activation of the Wnt/β-catenin pathway in HCC cells
We further investigated the relevant molecular mechanism of PRDX2 and its impact on the senescence of HCC cells. Through the detection of cycle-related genes (cyclin-B1, cyclin-D1, and cyclin-E1), RT-qPCR data first revealed that the level of cylinD1 was significantly reduced in the PRDX2 silencing group compared to that of the shGFP group (Fig. 5A). And Western blot results also signified that silence of PRDX2 markedly downregulated PRDX2 and cylinD1 in HepG2 cells (Fig. 5B). Since cylinD1 is a downstream transcriptional target gene of the Wnt/β-catenin pathway, we speculated that PRDX2 may be involved in the regulation of this pathway. RT-qPCR and Western blot results further confirmed that knockdown of PRDX2 resulted in a significant downregulation of Wnt/β-catenin pathway-related proteins (c-Myc, c-jun, and fra-1) in HepG2 cells (Figs. 5C and 5D). Furthermore, we found that there was a marked increase in cellular senescence (β-Gal activity) of HepG2 cells after PRDX2 silencing (Fig. 5E). Taken together, these data indicated that PRDX2 silencing could induce the senescence and inhibit the Wnt/β-catenin pathway in HCC cells.
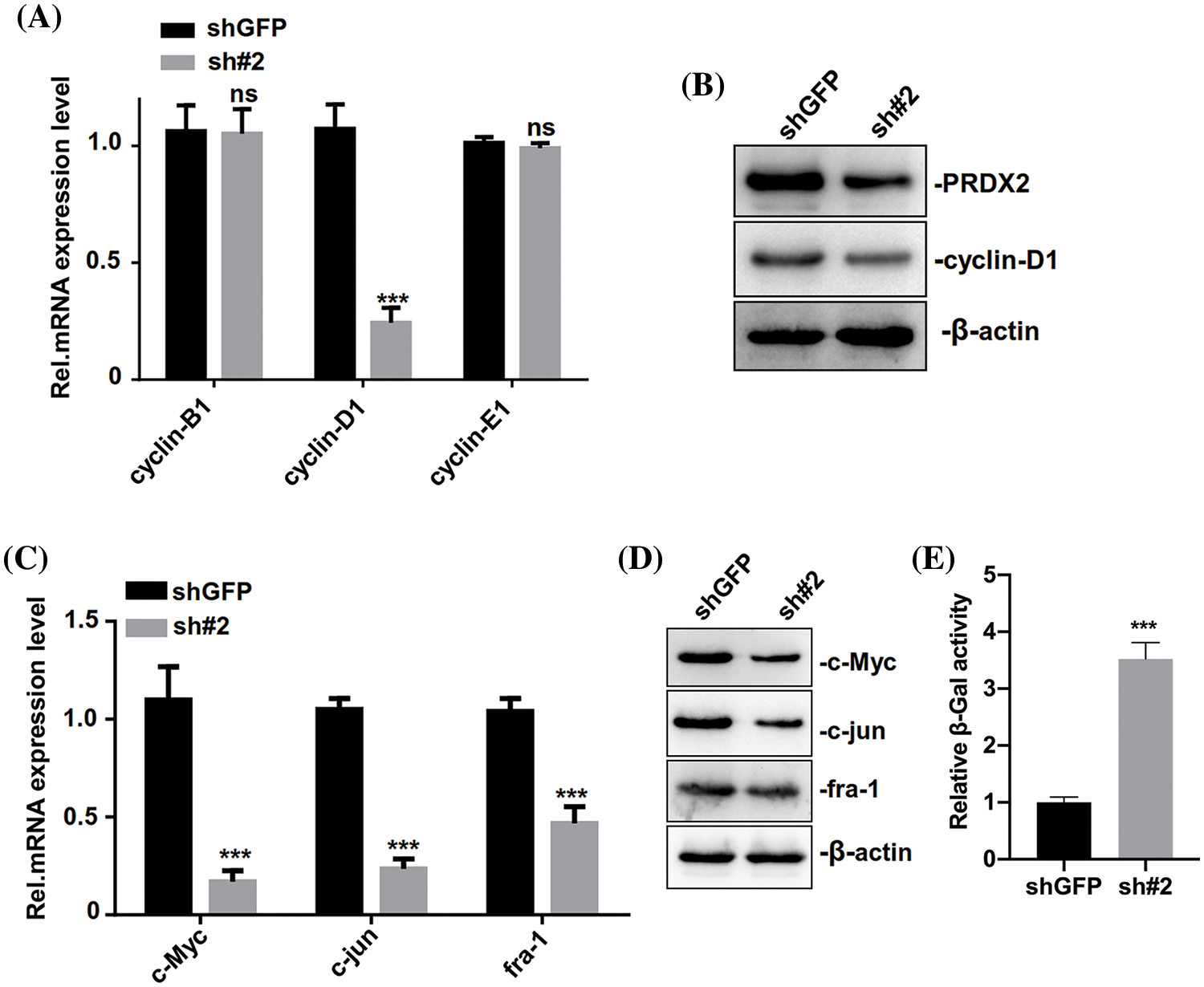
Figure 5: PRDX2 silencing regulates the senescence of HCC cells. (A) HepG2 cells were addressed with shGFP or sh#2 and puromycin, then the levels of cell cycle-related proteins (cyclin-B1, cyclin-D1, and cyclin-E1) were detected by western blot. (B) PRDX2 and cyclin-D1 expressions were determined using western blot in the treated HepG2 cells. The levels of c-Myc, c-jun, and fra-1 were also analyzed with RT-qPCR (C) and western blot (D) in the processed HepG2 cells. (E) The senescence (β-Gal activity) of HepG2 cells was assessed using a β-Gal assay kit. ***p < 0.001.
PRDX2 silencing inhibits HCC tumors growth in vivo
To further investigate the in vivo function of PRDX2, we inoculated stable HepG2-shGFP (shGFP) and shRNA#2 cell lines into nude mice subcutaneously. Tumor volume was measured every three days and when it reached approximately 80 mm3, the mice were euthanized by cervical dislocation 20 days after subcutaneous injection (Fig. 6A). As displayed in Fig. 6B, PRDX2 silencing significantly inhibited tumor growth in mice. Consistently, the tumor size and weight in the shRNA#2 group were significantly smaller than that in the shGFP group (Figs. 6C and 6D). Importantly, the levels of cylinD1, c-Myc, c-jun, fra-1, and PRDX2 were notably down-regulated in the sh#2 group compared to the shGFP group (Fig. 6E). These data strongly argued that PRDX2 silencing inhibited HCC development in a mouse model of HCC.
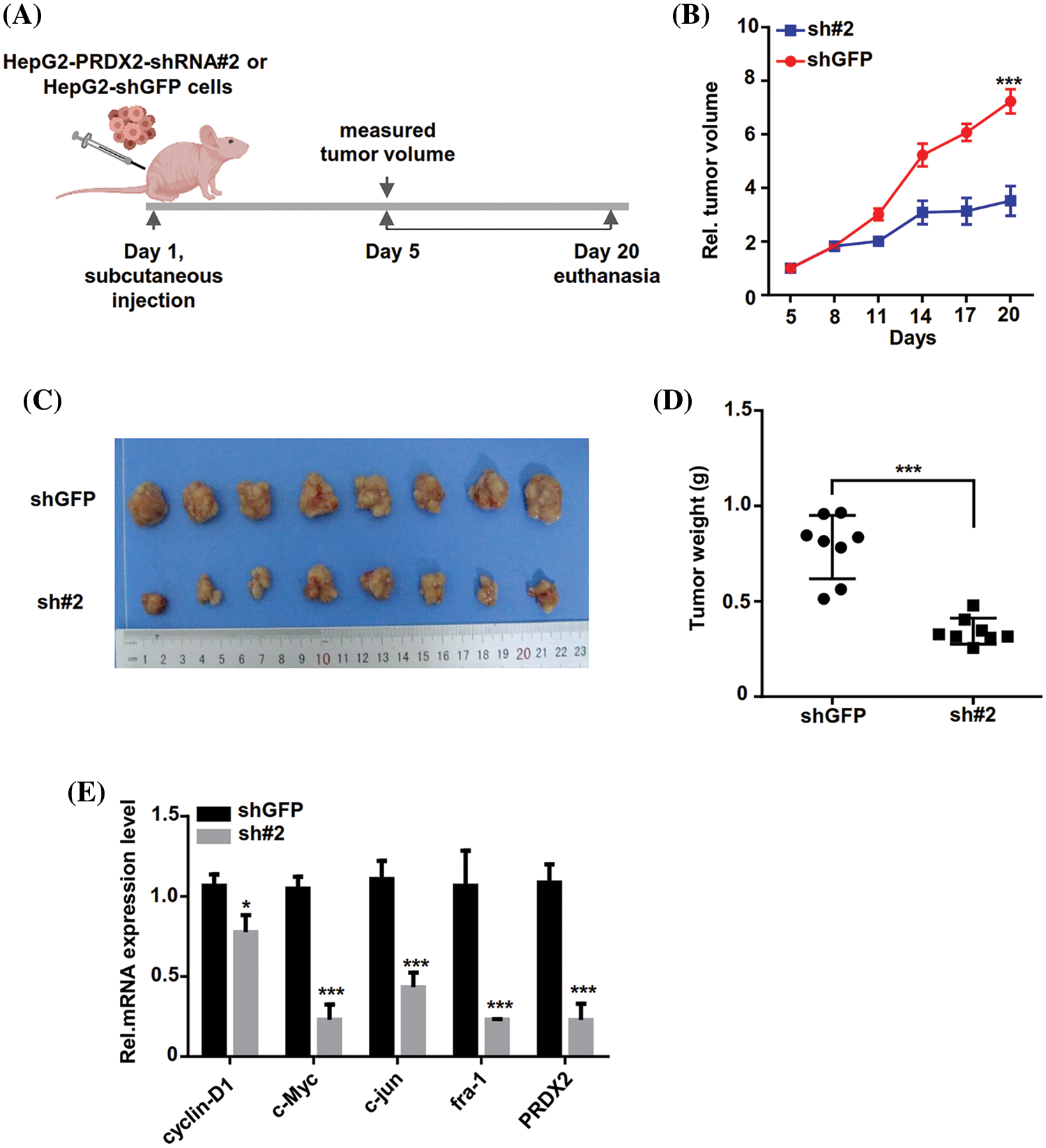
Figure 6: PRDX2 silencing inhibits HCC tumors growth in vivo. (A) Schematic illustration of the experimental design of the xenografted tumor in nude mice. (B and C) Effect of sh-PRDX2 on HCC xenografted tumors. Stably HepG2-shGFP (shGFP) and HepG2-sh#2 cell lines were subcutaneously inoculated into nude mice. 5 days week later, when the tumor volume was about 80 mm3, the tumor size was measured every three days and the tumor growth curve was drawn. The tumor volume and tumors were displayed. Tumor volume (mm3) = d2/2 × D (D is the longest, d is the shortest diameter). (D) Statistics of the transplanted tumors weight. (E) The levels of cyclin-D1, c-Myc, c-jun, fra-1, and PRDX2 were monitored using RT-qPCR in the xenografts. *p < 0.05, ***p < 0.001.
PRDX2 promotes β-catenin nuclear translocation
Then we further investigated the underlying molecular mechanism of PRDX2 in regulating the Wnt/β-catenin pathway. We first performed Top-flash assay to assess the effect of PRDX2 on the transcriptional activity of β-catenin. A Top-flash assay results showed a noticeable promotion of β-catenin’s transcriptional activity by PRDX2 (Fig. 7A). Furthermore, Co-IP data manifested the interaction between PRDX2 and β-catenin in HepG2 cells (Fig. 7B). Immunofluorescence results demonstrated the co-localized of PRDX2 and β-catenin in the cytoplasm of HepG2 cells (Fig. 7C). Additionally, the data disclosed that overexpression of PRDX2 markedly increased the expression of β-catenin in the nucleus of HepG2 cells, suggesting that PRDX2 markedly facilitated the translocation of β-catenin into the nucleus (Fig. 7D). Taken together, these data suggested that PRDX2 promoted the activation of the Wnt/β-catenin pathway by inducing the nuclear translocation of β-catenin in HCC cells.
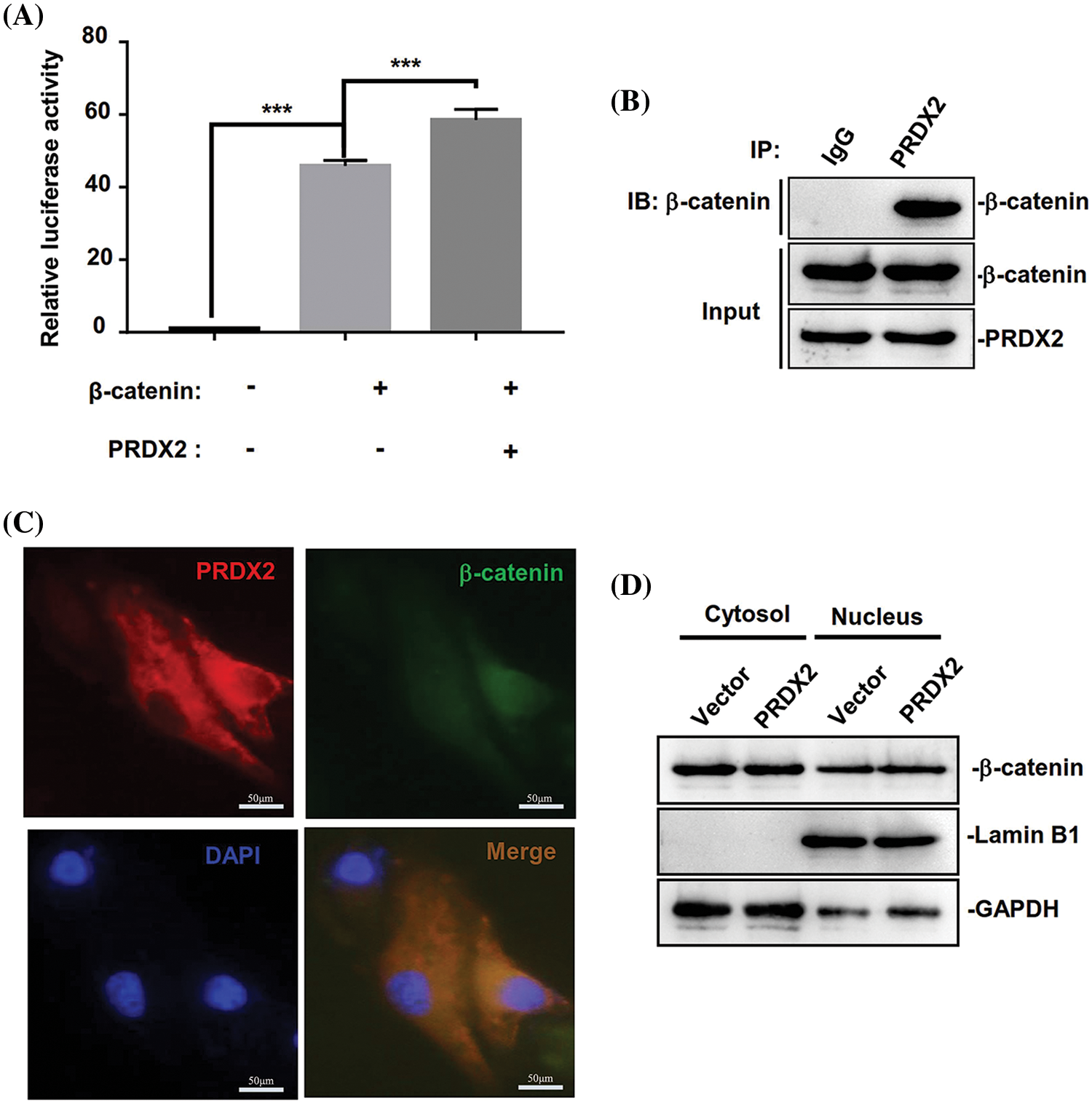
Figure 7: PRDX2 regulates Wnt/β-catenin pathway by promoting β-catenin nuclear translocation. (A) The impact of PRDX2 on β-catenin transcriptional activity. (B) The interaction between PRDX2 and β-catenin was detected by Co-IP experiment. (C) The immunofluorescence results of PRDX2 and β-catenin co-localized in the cytoplasm of HepG2 cells. Red: PRDX2, green: β-catenin, blue: nucleus; scale bars: 50 μm; Magnification: 400×. (D) β-catenin expression in the nucleus and cytoplasm was assessed using western blot after PRDX2 overexpression. ***p < 0.001.
The main reason for the lack of sufficient therapy options for HCC patients is our limited understanding of the molecular mechanism underlying HCC pathogenesis [2]. In our study, we first performed 2-DE and LC-MS on a subset of 26 patients (HCC: n = 12, choledocholithiasis (choledocholithiasis): n = 14) to identify differentially expressed proteins between HCC and choledocholithiasis patients. We aimed to discover a potential HCC diagnostic biomarker in bile. Our results revealed a significantly higher expression of PRDX2 in the bile of HCC patients compared to choledocholithiasis patients. Similarly, in serum samples, PRDX2 level was notably elevated in HCC causes compared to choledocholithiasis. Furthermore, immunostaining analysis demonstrated distinct differences in staining intensity and the number of positively stained cells between HCC tissues and non-tumor tissues. Therefore, we hypothesized that PRDX2 may function as an oncogene in HCC.
PRDX2 has been identified as a vital cellular defense system against oxidative stress due to its catalytic activity and protein sequence [39]. Accumulating evidence suggests that PRDX2 expression is elevated in a variety of human malignancies [15,16]. Several studies have confirmed the oncogenic role of PRDX2 in different types of cancer. For instance, PRDX2 silencing has been shown to inhibit proliferation and invasion in non-small cell lung cancer cells [14,40], promote growth, survival, and resistance to cisplatin in gastric cancer cells [11,15] and impair proliferation, cell cycle, and autophagy in colorectal cancer cells [11,16]. Additionally, PRDX2 has been implicated in HCC, where its silencing increases ROS production induced by H2O2, thereby contributing to carcinogenesis [41]. ROS also plays a critical role in the initiation and progression of HCC [42]. Consistent with previous research, our study proved the upregulation of PRDX2 in HCC tissues and its ability to enhance proliferation, supporting its oncogenic function in HCC. Furthermore, considering the involvement of cellular senescence in suppressing HCC cell proliferation, it is plausible that PRDX2 may be implicated in senescence [43]. Our findings align with this hypothesis, as we observed that knockdown of PRDX2 induced senescence in HepG2 cells.
Studies also revealed that PRDX2 possesses the ability to inhibit ROS production in HCC cells by activating Wnt/β-catenin pathway [44]. The Wnt/β-catenin pathway is a classical pathway that regulates the stability and nuclear localization of β-catenin [23]. This pathway plays a key role in cell proliferation, differentiation, and tissue homeostasis maintenance [45]. Aberrations in this pathway can promote cancer stem cell renewal, cell proliferation, and differentiation, thereby significantly contributing to tumorigenesis and treatment response [46]. β-catenin serves as a positive regulator of the Wnt pathway. Upong entering the nucleus, it binds to target genes and initiates the transcription process, regulating the expression of specific genes [47]. The ultimate outcome of Wnt pathway is the binding of β-catenin to T cell transcription factor (Tcf) and subsequent activation of related transcription processes. Studies have proved that introducing exogenous Wnt signaling or mutant genes into normal cells can lead to the formation of β-catenin-Tcf complex, greatly increasing the transcriptional activity of cells. The binding of β-catenin to Tcf may drive the transformation of normal cells into cancerous cells. The accumulation of free β-catenin protein in the cytoplasm and its translocation into the nucleus are key factors in the carcinogenic process associated with aberrant Wnt signaling pathway [48]. Studies also have identified c-Myc as one of the vital oncogenes regulated by aberrant Wnt signaling in the nucleus [49]. Several studies have also revealed the involvement of Wnt/β-catenin pathway activation in the development of HCC [28,50]. In our study, we further revealed that PRDX2 interacted with β-catenin. This interaction stabilized β-catenin and enables its entry into the nucleus of HCC cells, thereby activating Wnt/β-catenin pathway.
In summary, the findings of this study supported the role of PRDX2 as an oncogene in HCC and suggested its involvement in β-catenin nuclear translocation. These results contributed to the theoretical understanding of HCC pathogenesis and provided a foundation for the development of potential therapeutic targets. However, it is important to acknowledge the limitations of this study. The precise mechanism by which PRDX2 affects HCC progression through β-catenin nuclear translocation remains unclear and requires further investigation, which could be addressed through future rescue experiments.
Acknowledgement: None.
Funding Statement: This study was supported by National Nature Science Foundation of China (Nos. 81960118, 81860115, 81760116 and 82060116), Guizhou Science and Technology Project: Qiankehe Foundation (No. (2020)1Y300), Natural Science Foundation of Sichuan (No. 2022NSFSC0837), and Science and Technology Project of Chengdu (No. 2022-YF05-01811-SN), Science and Technology Project of Guizhou Province (No. YQK(2023)032), Guizhou Medical University Doctoral Start-Up Fund (No. gyfybsky-2021-27), Guizhou Medical University Doctoral Start-Up Fund (No. gyfybsky-2021-26), Guizhou Science and Technology Department (No. (2019)1259), Guizhou Science and Technology Department Guizhou Science and Technology Platform Talents (No. (2017)5718), Science and Technology Fund of Guizhou Provincial Health Commission (No. gzwki2021-382), and The Affiliated Hospital of Guizhou Medical University Excellent Reserve Talent in 2023 (No. gyfyxkrc-2023-06).
Author Contributions: Xuegang Yang and Xianhong Xiang designed and performed experimental work; Shi Zhou conducted literature research; Kaili Wu and Guohui Xu participated in the experimental work. Tianzhi Huang and Zhi Huang performed manuscript editing. All authors read and approved the final manuscript.
Availability of Data and Materials: The original contributions presented in the study are included in the article/Supplementary Materials. Further inquiries can be directed to the corresponding author.
Ethics Approval: Studies involving human participants were reviewed and approved by the Research Ethics Committees of the Guizhou Medical University. The animal study was reviewed and approved by the Animal Care and Use Committee of the Guizhou Medical University.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Khan, A., Zhang, X. (2022). Function of the long noncoding RNAs in hepatocellular carcinoma: Classification, molecular mechanisms, and significant therapeutic potentials. Bioengineering, 9, 406. [Google Scholar] [PubMed]
2. Andolfi, C., Tiribelli, C., Pascut, D. (2022). Recent hints on the dual role of discs large MAGUK scaffold protein 5 in cancers and in hepatocellular carcinoma. Frontiers in Bioscience-Landmark, 27, 164. [Google Scholar]
3. Kedar Mukthinuthalapati, V. V. P., Sewram, V., Ndlovu, N., Kimani, S., Abdelaziz, A. O. et al. (2021). Hepatocellular carcinoma in Sub-Saharan Africa. JCO Global Oncology, 7, 756–766. [Google Scholar] [PubMed]
4. Sung, H., Ferlay, J., Siegel, R. L., Laversanne, M., Soerjomataram, I. et al. (2020). Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer Journal for Clinicians, 71, 209–249. [Google Scholar]
5. Sun, W., Shen, J., Liu, J., Han, K., Liang, L. et al. (2022). Gene signature and prognostic value of ubiquitin-specific proteases members in hepatocellular carcinoma and explored the immunological role of USP36. Frontiers in Bioscience-Landmark, 27, 190. [Google Scholar]
6. Ganesan, P., Kulik, L. M. (2023). Hepatocellular carcinoma: New developments. Clinics in Liver Disease, 27, 85–102. [Google Scholar] [PubMed]
7. Sarveazad, A., Agah, S., Babahajian, A., Amini, N., Bahardoust, M. (2019). Predictors of 5 year survival rate in hepatocellular carcinoma patients. Journal of Research in Medical Sciences, 24, 86. [Google Scholar] [PubMed]
8. Kan, M., Chiba, T., Konno, R., Kouchi, Y., Mishima, T. et al. (2022). Bile proteome analysis by high-precision mass spectrometry to examine novel biomarkers of primary sclerosing cholangitis. Journal of Hepatobiliary Pancreat Science, 30, 914–923. [Google Scholar] [PubMed]
9. Son, K. H., Ahn, C. B., Kim, H. J., Kim, J. S. (2020). Quantitative proteomic analysis of bile in extrahepatic cholangiocarcinoma patients. Journal of Cancer, 11, 4073–4080. [Google Scholar] [PubMed]
10. Chen, X., Zhao, Y., Luo, W., Chen, S., Lin, F. et al. (2020). Celastrol induces ROS-mediated apoptosis via directly targeting peroxiredoxin-2 in gastric cancer cells. Theranostics, 10, 10290–10308. [Google Scholar] [PubMed]
11. Wang, S., Chen, Z., Zhu, S., Lu, H., Peng, D. et al. (2020). PRDX2 protects against oxidative stress induced by H. pylori and promotes resistance to cisplatin in gastric cancer. Redox Biology, 28, 101319. [Google Scholar] [PubMed]
12. Peng, L., Xiong, Y., Wang, R., Xiang, L., Zhou, H. et al. (2021). The critical role of peroxiredoxin-2 in colon cancer stem cells. Aging, 13, 11170–11187. [Google Scholar] [PubMed]
13. Gu, C., Luo, J., Lu, X., Tang, Y., Ma, Y. et al. (2019). REV7 confers radioresistance of esophagus squamous cell carcinoma by recruiting PRDX2. Cancer Science, 110, 962–972. [Google Scholar] [PubMed]
14. Chen, Y., Yang, S., Zhou, H., Su, D. (2020). PRDX2 promotes the proliferation and metastasis of non-small cell lung cancer in vitro and in vivo. BioMed Research International, 2020, 8359860. [Google Scholar] [PubMed]
15. Zhang, S., He, J., Tang, M., Sun, H. (2020). Prdx2 upregulation promotes the growth and survival of gastric cancer cells. Pathology Oncology Research, 26, 1869–1877. [Google Scholar] [PubMed]
16. Wang, W., Wei, J., Zhang, H., Zheng, X., Zhou, H. et al. (2021). PRDX2 promotes the proliferation of colorectal cancer cells by increasing the ubiquitinated degradation of p53. Cell Death Disease, 12, 605. [Google Scholar] [PubMed]
17. Shi, J., Zhou, L., Huang, H. S., Peng, L., Xie, N. et al. (2022). Repurposing oxiconazole against colorectal cancer via PRDX2-mediated autophagy arrest. International Journal of Biological Sciences, 18, 3747–3761. [Google Scholar] [PubMed]
18. Park, Y. H., Kim, S. U., Kwon, T. H., Kim, J. M., Song, I. S. et al. (2016). Peroxiredoxin II promotes hepatic tumorigenesis through cooperation with Ras/Forkhead box M1 signaling pathway. Oncogene, 35, 3503–3513. [Google Scholar] [PubMed]
19. Xu, M., Xu, J., Zhu, D., Su, R., Zhuang, B. et al. (2021). Expression and prognostic roles of PRDXs gene family in hepatocellular carcinoma. Journal of Translational Medicine, 19, 126. [Google Scholar] [PubMed]
20. Kwon, T., Bak, Y., Park, Y. H., Jang, G. B., Nam, J. S. et al. (2016). Peroxiredoxin II is essential for maintaining stemness by redox regulation in liver cancer cells. Stem Cells, 34, 1188–1197. [Google Scholar] [PubMed]
21. Zhao, S., Su, G., Yang, W., Yue, P., Bai, B. et al. (2017). Identification and comparison of differentiationrelated proteins in hepatocellular carcinoma tissues by protemics. Technology in Cancer Research & Treatment, 16, 1092–1101. [Google Scholar]
22. Hou, J. K., Huang, Y., He, W., Yan, Z. W., Fan, L. et al. (2014). Adenanthin targets peroxiredoxin I/II to kill hepatocellular carcinoma cells. Cell Death Disease, 5, e1400. [Google Scholar] [PubMed]
23. Liu, J., Xiao, Q., Xiao, J., Niu, C., Li, Y. et al. (2022). Wnt/β-catenin signalling: Function, biological mechanisms, and therapeutic opportunities. Signal Transduction and Targeted Therapy, 7, 3. [Google Scholar] [PubMed]
24. Yu, F., Yu, C., Li, F., Zuo, Y., Wang, Y. et al. (2021). Wnt/β-catenin signaling in cancers and targeted therapies. Signal Transduction and Targeted Therapy, 6, 307. [Google Scholar] [PubMed]
25. Zhou, Y., Xu, J., Luo, H., Meng, X., Chen, M. et al. (2022). Wnt signaling pathway in cancer immunotherapy. Cancer Letter, 525, 84–96. [Google Scholar]
26. Helaehil, J. V., Helaehil, L. V., Alves, L. F., Huang, B., Santamaria, J. M. et al. (2023). Electrical stimulation therapy and HA/TCP composite scaffolds modulate the wnt pathways in bone regeneration of critical-sized defects. Bioengineering, 10, 2023. [Google Scholar]
27. Afify, S. M., Hassan, G., Osman, A., Calle, A. S., Nawara, H. M. et al. (2019). Metastasis of cancer stem cells developed in the microenvironment of hepatocellular carcinoma. Bioengineering, 6, 73. [Google Scholar] [PubMed]
28. Xu, C., Xu, Z., Zhang, Y., Evert, M., Calvisi, D. F. et al. (2022). β-catenin signaling in hepatocellular carcinoma. Journal of Clinical Investigation, 132, 1536. [Google Scholar]
29. Leung, R. W. H., Lee, T. K. W. (2022). Wnt/β-catenin signaling as a driver of stemness and metabolic reprogramming in hepatocellular carcinoma. Cancers, 14, 5468. [Google Scholar] [PubMed]
30. Najafi, S. M. A. A. (2020). The canonical Wnt signaling (Wnt/β-Catenin PathwayA potential target for cancer prevention and therapy. Iranian Biomedical Journal, 24, 269–280. [Google Scholar]
31. Feng, A. L., Han, X., Meng, X., Chen, Z., Li, Q. et al. (2020). PRDX2 plays an oncogenic role in esophageal squamous cell carcinoma via Wnt/β-catenin and AKT pathways. Clinical and Translational Oncology, 22, 1838–1848. [Google Scholar] [PubMed]
32. Huang, D. Q., Lee, D. H., Le, M. H., Le, A., Yeo, Y. H. et al. (2023). Liver complications in untreated treatment-ineligible versus treated treatment-eligible patients with hepatitis B. Digestive Diseases, 41, 115–123. [Google Scholar] [PubMed]
33. Barbhuiya, M. A., Sahasrabuddhe, N. A., Pinto, S. M., Muthusamy, B., Singh, T. D. et al. (2011). Comprehensive proteomic analysis of human bile. Proteomics, 11, 4443–4453. [Google Scholar] [PubMed]
34. Li, Z. J., Abudumijiti, A., Xu, D. Q., Youlidouzi, M., Silafu, A. et al. (2020). Quantitative proteomics analysis of Fructus Psoraleae-induced hepatotoxicity in rats. Chinese Journal of Natural Medicines, 18, 123–137. [Google Scholar] [PubMed]
35. Chen, Y., Feng, Y., Chen, S. (2022). HSP90B1 overexpression is associated with poor prognosis in tongue squamous cell carcinoma. Journal of Stomatology, Oral and Maxillofacial Surgery, 123, e833–e838. [Google Scholar] [PubMed]
36. Zhang, X., Xing, M., Ma, Y., Zhang, Z., Qiu, C. et al. (2023). Oridonin induces apoptosis in esophageal squamous cell carcinoma by inhibiting cytoskeletal protein LASP1 and PDLIM1. Molecules, 28, 805. [Google Scholar] [PubMed]
37. Wen, J., Zou, J., Huang, X., Wen, H., Suo, X. et al. (2019). Identification of candidate antigens by 2-DE Immunoblotting for diagnosis of Toxoplasma gondii infection in chickens and rabbits. Experimental Parasitology, 204, 107723. [Google Scholar] [PubMed]
38. Huarachi, S. F., Petroselli, G., Erra-Balsells, R., Audisio, M. C. (2022). Antibacterial activity against enterovirulent Escherichia coli strains from Bacillus amyloliquefaciens B31 and Bacillus subtilis subsp. subtilis C4: MALDI-TOF MS profiling and MALDI TOF/TOF MS structural analysis on lipopeptides mixtures. Journal of Mass Spectrometry, 57, e4896. [Google Scholar] [PubMed]
39. Stancill, J. S., Corbett, J. A. (2021). The role of thioredoxin/peroxiredoxin in the β-cell defense against oxidative damage. Frontiers in Endocrinology, 12, 718235. [Google Scholar] [PubMed]
40. Jing, X., Du, L., Niu, A., Wang, Y., Wang, Y. et al. (2020). Silencing of PRDX2 inhibits the proliferation and invasion of non-small cell lung cancer cells. Biomed Research International, 2020, 1276328. [Google Scholar] [PubMed]
41. Zhou, S., Han, Q., Wang, R., Li, X., Wang, Q. et al. (2016). PRDX2 protects hepatocellular carcinoma SMMC-7721 cells from oxidative stress. Oncology Letters, 12, 2217–2221. [Google Scholar] [PubMed]
42. Hashim, Z., Ilyas, A., Zarina, S. (2020). Therapeutic effect of hydrogen peroxide via altered expression of glutathione S-transferase and peroxiredoxin-2 in hepatocellular carcinoma. Hepatobiliary Pancreat Diseases International, 19, 258–265. [Google Scholar] [PubMed]
43. Ma, Y., Liang, Y., Liang, N., Zhang, Y., Xiao, F. (2021). Identification and functional analysis of senescence-associated secretory phenotype of premature senescent hepatocytes induced by hexavalent chromium. Ecotoxicology and Environmental Safety, 211, 111908. [Google Scholar] [PubMed]
44. Bai, B., Lin, Y., Hu, J., Wang, H., Li, L. et al. (2019). Peroxiredoxin2 downregulation enhances hepatocellular carcinoma proliferation and migration, and is associated with unfavorable prognosis in patients. Oncology Reports, 41, 1539–1548. [Google Scholar] [PubMed]
45. Zhang, Y., Wang, X. (2020). Targeting the Wnt/β-catenin signaling pathway in cancer. Journal Hematology Oncology, 13, 165. [Google Scholar]
46. Huang, P., Yan, R., Zhang, X., Wang, L., Ke, X. et al. (2019). Activating Wnt/β-catenin signaling pathway for disease therapy: Challenges and opportunities. Pharmacology Therapeutics, 196, 79–90. [Google Scholar] [PubMed]
47. Wang, Z., Li, Z., Ji, H. (2021). Direct targeting of β-catenin in the Wnt signaling pathway: Current progress and perspectives. Medicinal Research Reviews, 41, 2109–2129. [Google Scholar] [PubMed]
48. Hayat, R., Manzoor, M., Hussain, A. (2022). Wnt signaling pathway: A comprehensive review. Cell Biology International, 46, 863–877. [Google Scholar] [PubMed]
49. Kazi, A., Xiang, S., Yang, H., Delitto, D., Trevino, J. et al. (2018). GSK3 suppression upregulates β-catenin and c-Myc to abrogate KRas-dependent tumors. Nature Communications, 9, 5154. [Google Scholar] [PubMed]
50. He, S., Tang, S. (2020). WNT/β-catenin signaling in the development of liver cancers. Biomedicine & Pharmacotherapy, 132, 110851. [Google Scholar]
Supplementary Materials
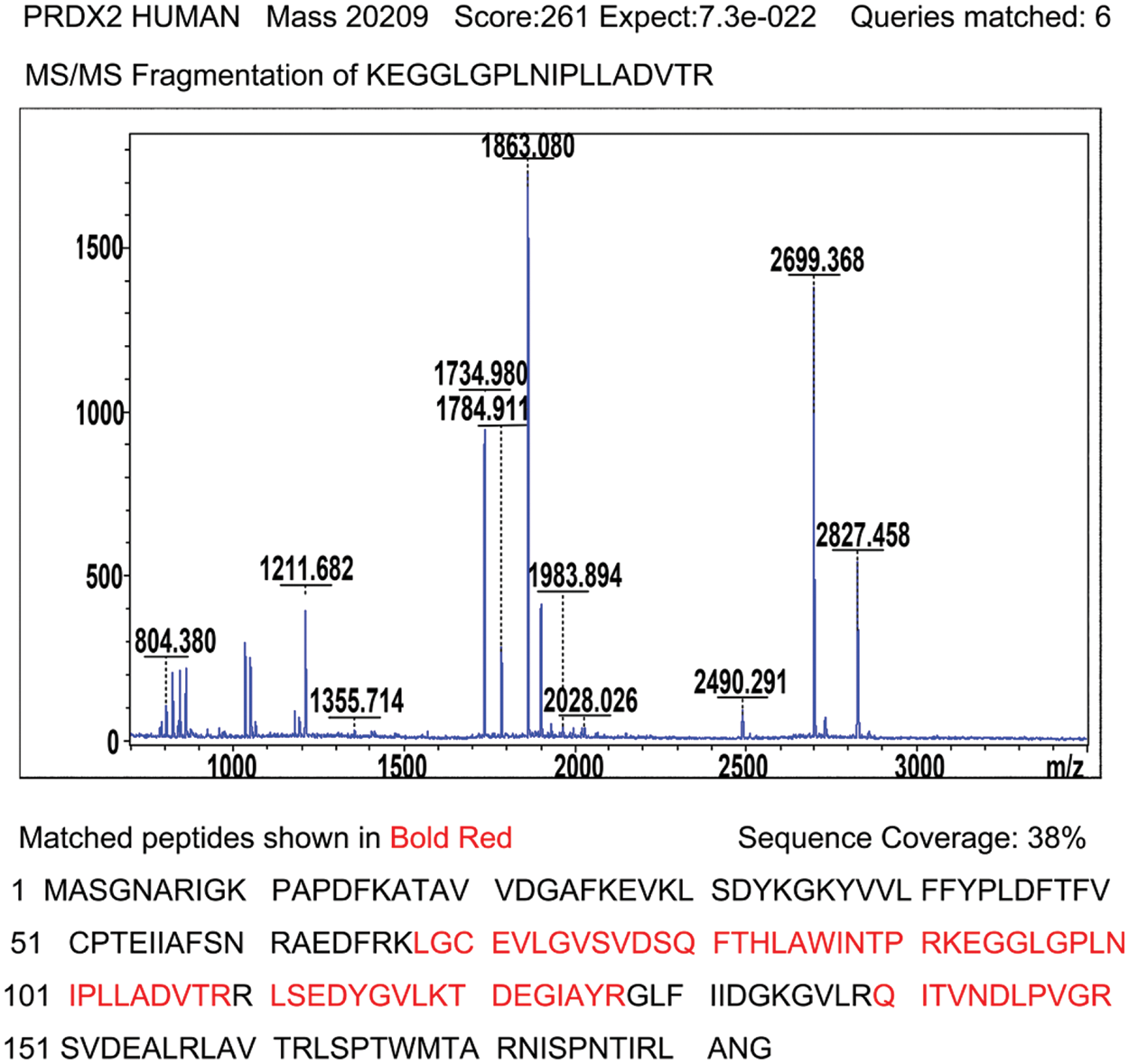
Supplementary Figure 1: Identification of PRDX2 by MALDI-TOF tandem mass spectrometry. After enzyme digestion, protein identification with high score (261) shows that spot mentioned above corresponds to peroxiredoxin-2; acquired spectra were search against a Mascot Search engine based on the Swiss-Prot protein database; MS/MS spectrum of peptide 92KEGGLGPLNIPLLADVTR109 with corresponding peak values was identified as PRDX2; Schematic representation of the matched peptides in a reference sequence shows 38% of protein coverage.
Cite This Article
 Copyright © 2024 The Author(s). Published by Tech Science Press.
Copyright © 2024 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools