 Open Access
Open Access
ARTICLE
Combining Transcriptomics and Metabolomics to Uncover the Effects of High-Energy Lithium-Ion Beam Irradiation on Capsicum annuum L.
1 Horticultural Sub-Academy, Heilongjiang Academy of Agricultural Sciences, Harbin, 150069, China
2 Institute of Crop Science, Chinese Academy of Agricultural Sciences National Center of Space Mutagenesis for Crop Improvement, Beijing, 10008, China
3 Heihe Agricultural and Rural Bureau, Heihe, 164300, China
* Corresponding Authors: Luxiang Liu. Email: ,
(This article belongs to the Special Issue: Multi-omics Approach to Understand Plant Stress Tolerance)
Phyton-International Journal of Experimental Botany 2023, 92(11), 2947-2964. https://doi.org/10.32604/phyton.2023.042919
Received 16 June 2023; Accepted 07 August 2023; Issue published 24 October 2023
Abstract
Hot pepper (Capsicum annuum L.) is consumed as one of the oldest domesticated crops all over the world. Although mutation breeding using radiation has been performed in hot peppers, little is known about the comparative analysis of mutagenic effects at the molecular level by ion beam irradiation. To comprehend the response mechanism of hot pepper to the ion beam, we used a mutant with favorable economic characteristics induced by lithium-ion beam irradiation to investigate the biological effects. The results indicated that the lithium-ion beam had a positive effect on important agronomic traits, particularly yield unit, but had a negligible effect on the photosynthetic rate of hot pepper, with a specific influence on chlorophyll b rather than chlorophyll a. By RNA-Seq analysis, 671 up-regulated and 376 down-regulated genes were identified as differentially expressed genes (DEGs) between irradiated and unirradiated hot pepper. Based on GO and KEGG network analysis, the auxin metabolic process was the common pathway in these two networks. A total of 118 potential reactive oxygen species (ROS) scavenging genes and 262 signal transduction genes were identified, suggesting a balance between antioxidant enzymes and enhanced ROS transduction. The amounts of 15 metabolite, involved in GABA pathways, secondary metabolism, carbohydrate metabolism, shikimate pathways, TCA cycles, nitrogen metabolism, glycerol metabolism and acetate pathways, were significantly changed in the ion beam irradiated sample. These results highlighted that the enriched pathways could play important roles in response to ion beam irradiation in hot pepper plants. In summary, these data provide valuable information for future research on ion beam irradiation and genomic studies in hot pepper.Keywords
Supplementary Material
Supplementary Material FileDue to the complex environmental conditions in space, such as cosmic radiation, microgravity, weak magnetism, high vacuum, and ultralow temperature, spaceflight mutagenesis has been employed to induce stress response and alter DNA or gene expression in plants, leading to heritable changes. Due to the large investment, high technical requirements, and limited test opportunities in outer space, research involving ground-based simulation of aerospace environmental factors is of great significance. Currently, heavy ion beams, which are powerful and efficient mutagenic tools with high linear energy transfer radiation and the ability to cause more DNA double-stranded breaks, have shown promising results in mutation breeding studies compared to conventional radiation methods [1]. The ion beam technique can induce broad-spectrum mutations at high frequencies, resulting in pronounced biological effects while minimizing damage to target species and ensuring stable inheritance of mutations, making it an ideal method for breeding new cultivars [2,3]. So far, this method has already been widely applied to conduct mutagenesis studies of various plants, including maize, rice, wheat, and Arabidopsis [4–6]. However, few studies have used ion beam irradiation technology to breed new hot pepper cultivars.
Mutations are an important resource in phenotypic screening, gene isolation, and function mining, in forward and reverse genetics [7,8]. Heavy-ion beam irradiation has higher linear energy transfer and can generate mutations with high mutation efficiency and a broad mutation spectrum [9]. Therefore, this random mutagenesis technique could produce various types of mutants [10]. Compared to other ion beams, lithium-ion beams could produce a greater number of single base substitutions (SBSs) than short insertions and deletions (InDels) [11]. Moreover, radiation damage to DNA occurs in injury caused by thermal ionization, as well as indirect injury caused by the cytotoxic reaction to energy deposition [12–14]. In the last century, the characterization of mutations induced by radiation focused on phenotype, chromosome aberration, genetic polymorphism, or specific gene sequencing analysis [7,15]. Although mutations induced by ion beam radiation have been applied to improve the yield and quality of crops [16], the biological effect mechanism remains unclear. Transcriptomics and metabolomics are powerful tools for providing a far more precise measurement of levels of transcripts and metabolites than other molecular methods [17]. Many kinds of researches have used these novel techniques to understand the complexity of plant response to ion beams, including in Oryza sativa L. [18], Vitis vinifera L. [19], and Zea mays [20], but no related findings have been reported in hot pepper.
Hot pepper (Capsicum annuum L.), a member of the Solanaceae family, is consumed as one of the oldest domesticated crops in several parts of the world [21]. As an important vegetable crop, it is not only used as a condiment with a long history but also possesses nutritional and health-care effects on the human body [22]. Its fruits are rich in vitamins C and E, provitamin A, and carotenoids-compounds with antioxidant properties [23]. Materska et al. isolated phenolics, flavonoids, and capsaicinoids and studied their antioxidant capacity [24]. The whole-genome sequencing and assembly of this species has been completed and served as a platform for improving the nutritional and medicinal values of Capsicum species [25]. Additionally, based on spatial mutagenesis, we identified differentially expressed genes and conducted comparative proteome analysis in hot pepper after space flight in our previously published papers [26,27]. However, the gene expression and metabolomic changes in mutants induced by ion beams are still unknown.
Nowadays, no studies have been reported to investigate the response of hot pepper in mutagenesis by combining physiology, transcriptomics, and metabolomics analyses. Therefore, we compared transcriptomics and metabolomics under lithium-ion beam-induced mutation to uncover how this species responds to lithium-ion beams (LIB). Our findings will help us better understand the molecular variations induced by lithium-ion beams in hot pepper and provide novel approaches for mutation breeding research.
2.1 Lithium-Ion Beam Irradiation and Mutant Screening
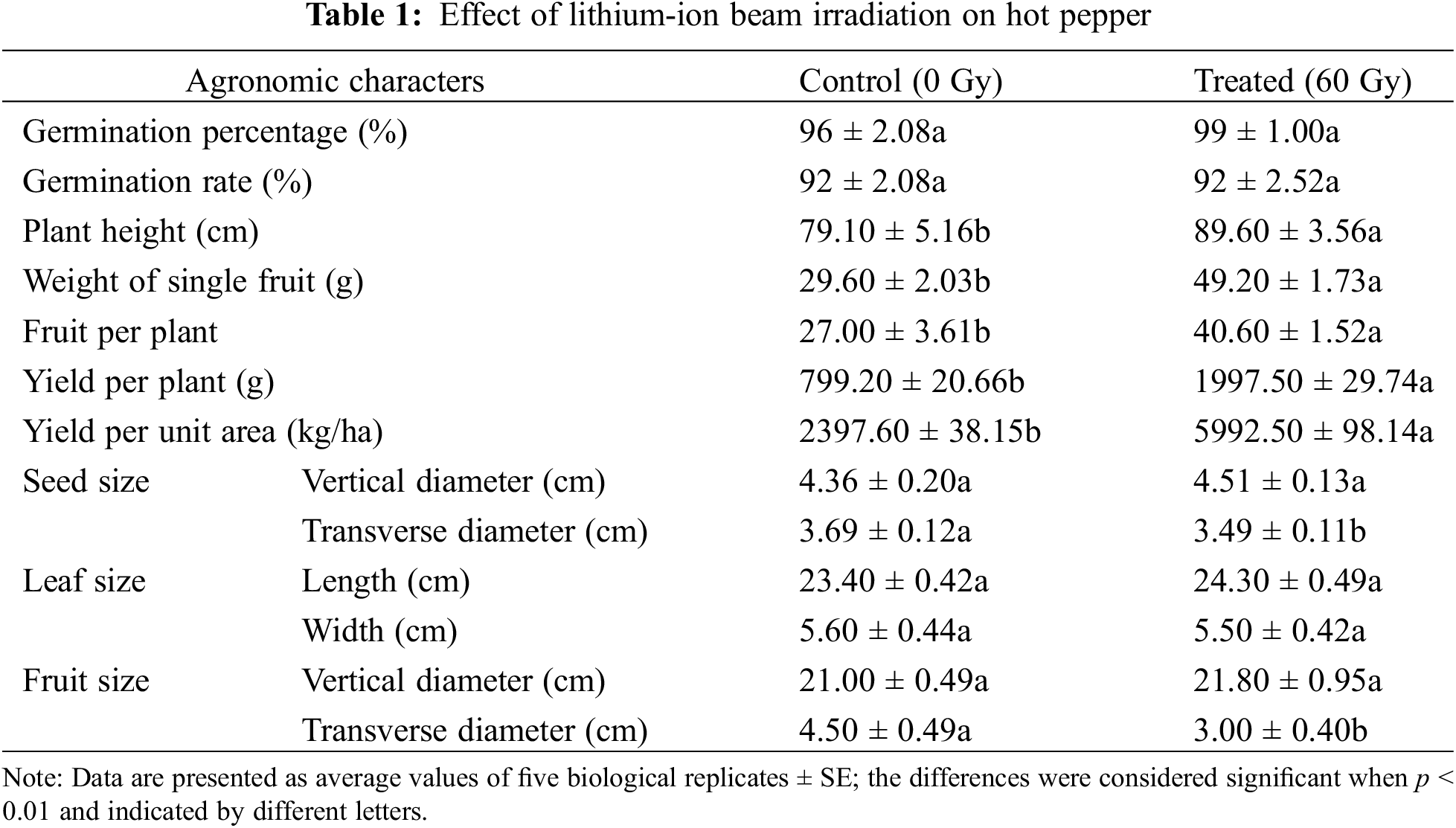
The seeds of hot pepper (Accession: Longshu No. 5) were collected from the Heilongjiang Forest Botanical Garden (E 126°16′, N 45°45′), China [28]. Approximately 300 seeds were exposed to lithium-ions at 60 Gy (energy, 42.3 MeV/nucleon), generated by the HI-3 tandem accelerator at the China Institute of Atomic Energy, with three replications. The 50 irradiated (60 Gy) and 50 unirradiated seeds were placed in 90 mm Petri dishes for germination and grown in an individual pot at 28°C with five replicates per treatment. We compared the effects of LIB on germination percentage (Germination Percentage = (Number of germinated seeds/Total number of tested seeds) × 100%) and germination rate (Germination Rate = (Number of germinated seeds/Total number of tested seeds)/Number of days) of irradiated and unirradiated seeds to determine the surviving fraction. After 2–3 days, the germination percentage and rate of the hot pepper were recorded. Given its importance as a vegetable crop, its detailed agronomic characteristics were assessed after growth for 30 days, including seed and leaf sizes, plant height, the weight of single fruits, yield per plant, and per unit area.
2.2 Photosynthetic Parameter, Chlorophyll Fluorescence, and Content Analysis
Net photosynthetic rate (Pn), stomatal conductance (Gs), transpiration rate (Tr), and intercellular CO2 concentration (Ci) of leaves were determined using a portable open flow gas exchange photosynthesis system, LI-6400 XT (Li-Cor, Inc., Lincoln, NE, USA). Additionally, chlorophyll fluorescence was measured to calculate the maximal efficiency of PSII photochemistry (Fv/Fm), actual photochemistry efficiency (ФPSII), electron transport rate (ETR), and photochemical quenching (qP) [29]. The content of chlorophyll a, chlorophyll b, and chlorophyll a+b were measured following the methods described in a previous paper [30]. Measurements were repeated six times for each blade from the same position of five plants in each treatment group, and the averages were recorded.
2.3 RNA Sample Preparation, cDNA Library Preparation, and Illumina Hiseq Sequencing
The hot pepper leaves in the control group (without ion beam) and treated samples (with ion beam irradiation) were collected 30 days after sowing at Heilongjiang Forest Botanical Garden, China. The leaf tissues were washed with distilled water, followed by surface drying with filter paper. Harvested tissues were frozen in liquid nitrogen and stored at −80°C until further analysis. Three completely independent harvesting experiments were repeated as biological replicates.
Total RNA was extracted from the leaves using Trizol in accordance with the manufacturer’s protocol (TIANGEN, Beijing, China). The purity and integrity of the obtained RNA were assessed using agarose gel electrophoresis and a Nanophotometer spectrophotometer (IMPLEN, CA, USA). A total of 1.5 μg RNA per sample was used for cDNA synthesis with the PrimeScript™ RT Reagent Kit and gDNA Eraser (TaKaRa, Tokyo, Japan). A cDNA library was used for sequencing analysis via an Illumina HiSeq 4,000 sequencing platform (Illumina Inc., San Diego, CA, USA) and 150 bp paired-end reads were generated.
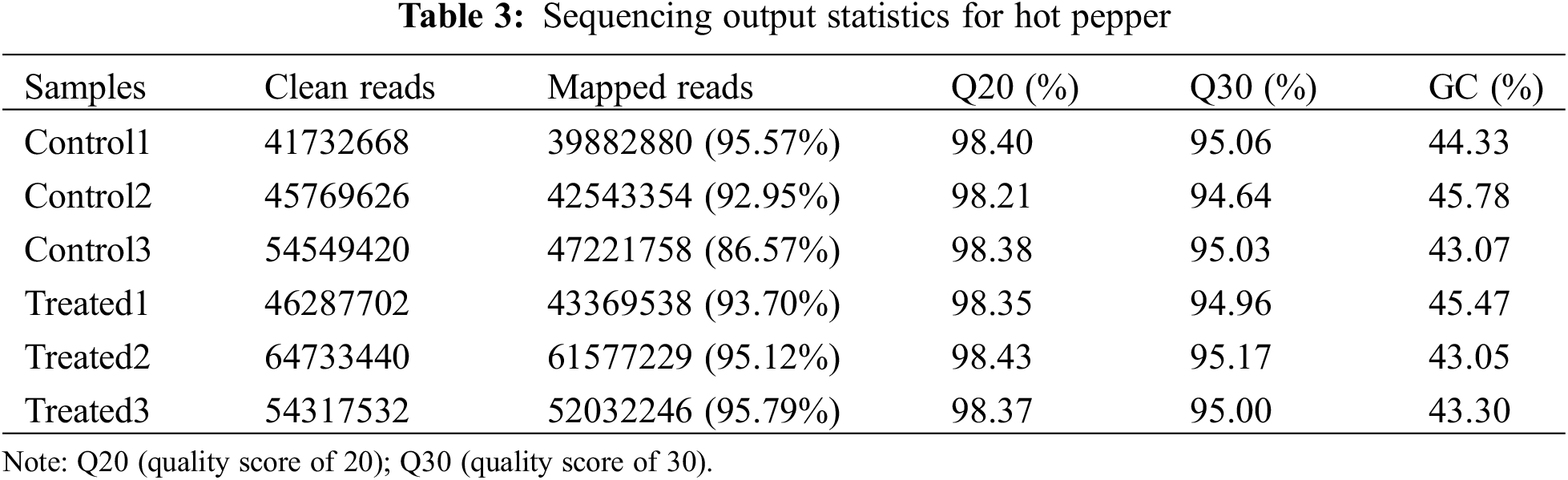
Raw data in fastq format was first processed through in-house Perl scripts. In this step, clean data were obtained by removing reads containing adapters, poly-N, and reads of low quality. All downstream analyses were based on high-quality clean data. Next, Q20 (quality score of 20), Q30 (quality score of 30), GC-content, and sequence duplication levels of clean reads were calculated in each sample. The clean reads were aligned to the Capsicum annuum cultivar: UCD-10X-F1 genome (PRJNA814299) as the reference genome using HISAT2 [31], and the aligned reads were assembled using StringTie [32]. All genes were then subjected to a blast search (E-value < 10−5) and annotated against various databases, including the Kyoto Encyclopedia of Genes and Genomes Ortholog (KEGG) and Gene Ontology (GO) databases.
2.5 Identification and Annotation of Differentially Expressed Genes
Differential expression analysis between control and treated samples was performed using the edgeR (1.10.1) [33]. The adjusted p-values (q-value) were calculated using the Benjamini and Hochberg method to control the false discovery rate. For differentially expressed genes (DEGs), the number of unigenes per sample was normalized and counted with TMM (Trimmed Mean of M values) using edgeR package. DEGs were considered if they had a q-value < 0.05, as identified by DESeq, and exhibited a 2-fold or greater change in TMM-normalized fragments per kilobase of gene per million mapped reads (FPKM) using the R package limma (Linear Models for Microarray and RNA-Seq Data) with upper-quantile normalization [34]. Regarding the functional annotation of DEGs, GO enrichment analysis was implemented by the GOseq R packages based on the Wallenius non-central hypergeometric distribution [35]. Finally, KOBAS software was used to test the statistical enrichment of DEGs in KEGG pathways [36].
2.6 Metabolite Extraction and Gas Chromatography-Mass Spectrometry (GC-MS) Analysis
A total of 60 mg of leaves were collected and placed into 1.5 mL microtubes. The samples were freeze-dried, and the metabolites were dissolved in 0.5 mL 80% methanol. Six parallel experiments were performed and analyzed. For GC-MS detection, metabolites were extracted and samples derivatized following Erban et al. [37]. One microliter aliquot of the derivatized solution was injected in splitless mode into the Agilent 7890A-5975C GC-MS system (Agilent, USA). The separation was carried out on a non-polar DB-5 capillary column (30 m × 250 μm I.D., J&W Scientific, Folsom, CA), with high purity helium as the carrier gas at a constant flow rate of 1.0 mL/min.
2.7 Metabolomics Data Processing
The acquired MS data from GC-MS were analyzed by ChromaTOF software (v 4.34, LECO, St Joseph, MI). Briefly, after alignment with the Statistic Compare component, a CSV file was obtained with three-dimensional datasets, including sample information, retention time-m/z, and peak intensities. The detectable peaks of samples in GC-MS were 550 in total, and the internal standard was used for data quality control (reproducibility). After removing internal standards and any known pseudo-positive peaks, such as peaks caused by noise, column bleed, and the BSTFA derivatization procedure, the detectable metabolite was combined with 250 in total. The dataset was normalized using the sum intensity of the peaks in each sample.
The data sets resulting from GC-MS were separately imported into the SIMCA-P+ 14.0 software package (Umetrics, Umeå, Sweden). (Orthogonal) partial least-squares-discriminant analysis (O)PLS-DA was carried out to visualize the metabolic alterations among experimental groups after mean centering and unit variance scaling. Variable importance in the projection (VIP) ranked the overall contribution of each variable to the (O)PLS-DA model, and those variables with VIP > 1.0 were considered relevant for group discrimination [38].
All the differentially expressed compounds in the treated group were selected by comparing the compounds in the treated group with the control using the multivariate statistical method and the Wilcoxon-Mann-Whitney test. Metabolites with both multivariate and univariate statistical significance (VIP > 1.0 and p < 0.05) were annotated with the aid of available reference standards in our lab and the NIST 11 standard mass spectral databases and the Feinh databases linked to ChromaTOF software. A similarity of more than 70% could be considered a reference standard.
2.8 Data Analysis and Statistics
One-way ANOVA was used to test the differences between the control and each treatment. The differences between mean values were considered statistically significant when p < 0.05. All statistical analyses were carried out using SPSS software (Version 15.0, SPSS Inc., USA).
3.1 Effects of Lithium-Ion Beams (LIB) Radiation on Plant Growth
In comparison with the control samples, several traits in the irradiated plants showed significant differences after ion beam irradiation. Although the germination percentage and rate of seeds were similar between the irradiated group and the control (Table 1), plant height, weight of single fruit, fruit per plant, yield per plant, and per unit area in the irradiated samples were significantly higher than those of the control (p < 0.05). Importantly, yield per unit area, which is one of the most important indicators at agricultural production levels, was 5992.5 kg/ha in the irradiated group, while that of the control was only 2397.6 kg/ha. In addition, the transverse diameters of seeds and fruits from irradiated hot pepper were slightly decreased (p < 0.05). However, the vertical diameters of seeds and fruits, and the leaf length and width were similar to irradiated and un-irradiated hot pepper (p > 0.05).
3.2 Photosynthetic Rate and Chlorophyll Fluorescence
We determined the photosynthetic parameters in hot pepper in control and treated samples (Table 2). In contrast with control plants, the net photosynthetic rate (Pn) was significantly reduced, but intercellular CO2 concentration (Ci) significantly increased in the leaves of irradiated plants (p < 0.05). However, stomatal conductance (Gs) and transpiration rate (Tr) were not markedly changed after irradiation. The maximal efficiency of the photosystem (Fv/Fm), actual photochemistry efficiency (ФPSII), electron transport rate (ETR), and photochemical quenching (qP) were determined on leaves (Table 2). The values of chlorophyll fluorescence indexes (Fv/Fm, ФPSII, ETR, qP) were not significantly different between the control and irradiated plants, suggesting that the capacity of photosynthesis was not changed after LIB irradiation.
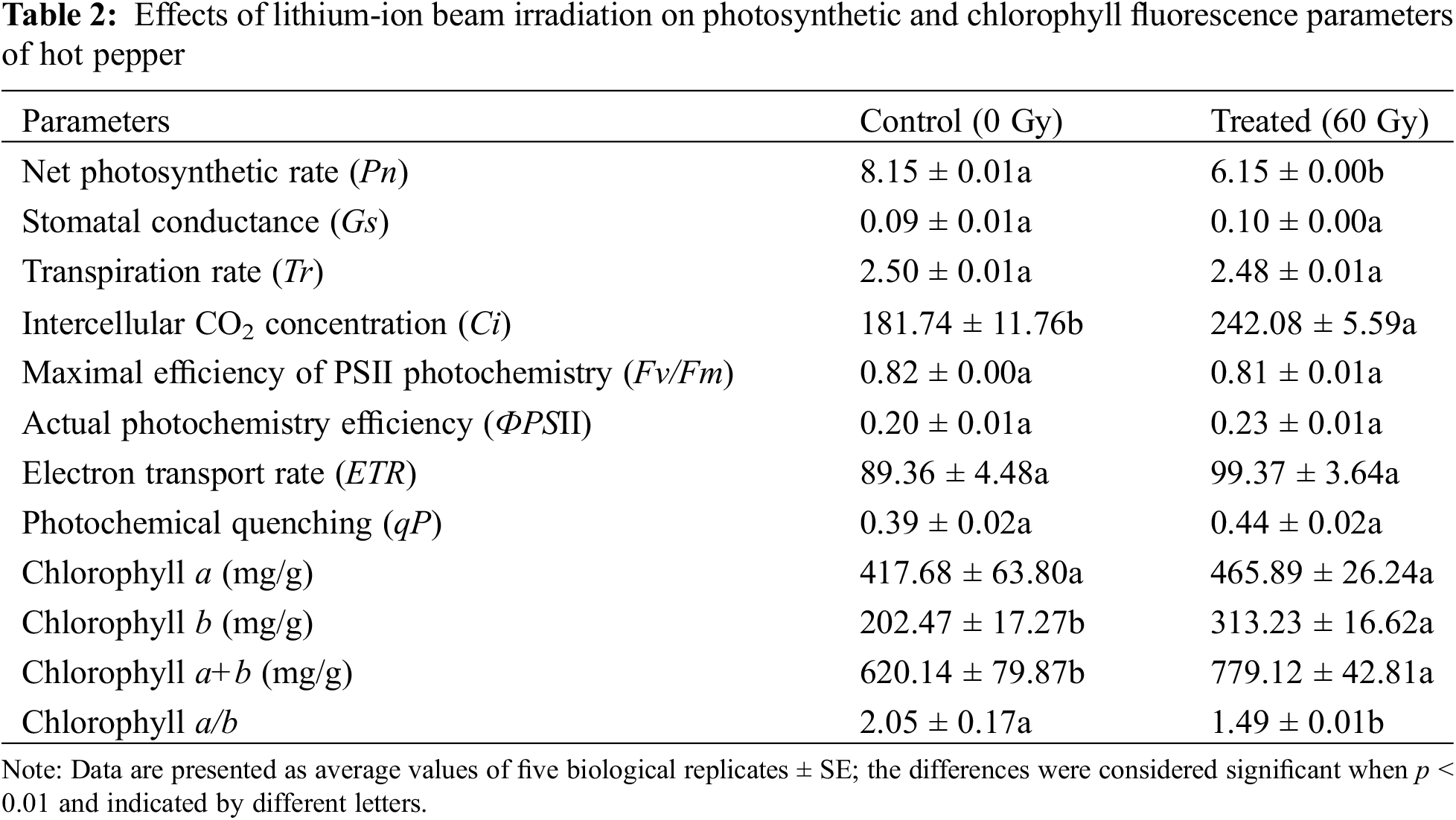
Chlorophyll is a well-known pigment reflecting the efficiency of the absorption, transfer, and conversion of light energy in plant photosynthesis [39]. The contents of chlorophyll b and a+b significantly increased in the irradiated group, whereas the chlorophyll a/b content decreased significantly in the irradiated group compared to that of the control group (p < 0.05).
3.3 De novo Assembly and Quality Assessment of Transcriptome
To determine the molecular characteristics and mechanisms of lithium-ion beam irradiation, we conducted transcriptome sequence analysis in irradiated and control groups. In summary, a total of 307,390,388 paired-end reads were obtained with a sequencing error rate of less than 0.01% (Table 3). The Q20 and Q30 values were over 98.21% and 94.64%, respectively. The GC content among the reads varied from 43.05% to 45.78%. After removing adapters, reads containing poly-N and low-quality reads, a total of 286,627,005 mapped reads were obtained from the raw reads, with the mapped percentage from 86.57% to 95.79% (Table 3).
3.4 Differentially Expressed Genes (DEGs) in Hot Pepper
To better understand the genes associated with LIB irradiation in hot pepper, we identified DEGs between the control and samples from irradiation using the DESeq R package. In total, 1,047 genes were identified as DEGs, including 671 up-regulated and 376 down-regulated genes (Supplementary Fig. S1 and Table S1).
These DEGs were then successfully assigned GO annotations and classified into three major metabolic pathways (Supplementary Fig. S2). The up-regulated DEGs were mainly involved in metabolic process (273 DEGs), cell (173), and catalytic process (219) in biological processes, cellular components, and molecular function, respectively. For the down-regulated DEGs, metabolic process (133), cell (83), and catalytic process (104) were also the largest subcategories in biological processes, cellular component, and molecular function, respectively. Comparing the DEGs in up- and down-regulated groups, antioxidant activity (4/0), electron carrier activity (6/0), enzyme regulator activity (7/0), guanyl-nucleotide exchange factor activity (1/0), molecular transducer activity (9/0), and receptor activity (6/0) were only enriched by up-regulated genes.
In addition, we analyzed the networks of pathways enriched by the DEGs. In Fig. 1A, the up-regulated genes mainly participated in five pathways, including regulation of the cellular ketone metabolic process, glutamine family amino acid metabolic process, alcohol biosynthetic process, auxin metabolic process, and nucleocytoplasmic transport. Among them, short-chain alcohol dehydrogenase (ABA2) and abscisic-aldehyde oxidase (AAO3) genes were related to three and two different pathways, respectively. Additionally, to better understand the biological function of DEGs involved in LIB radiation in hot pepper, KEGG pathway analysis was performed with five classes, including environmental information processing, genetic information processing, cellular processes, organismal systems, and metabolism (Supplementary Fig. S3). In this network analysis, a total of six pathways were detected based on KEGG, including the regulation of cellular proliferation, auxin metabolic processes, glutamine family amino acid metabolic processes, unsaturated fatty acid metabolic processes, lipid biosynthetic processes, and cellular response to jasmonic acid stimulus (Fig. 1B).
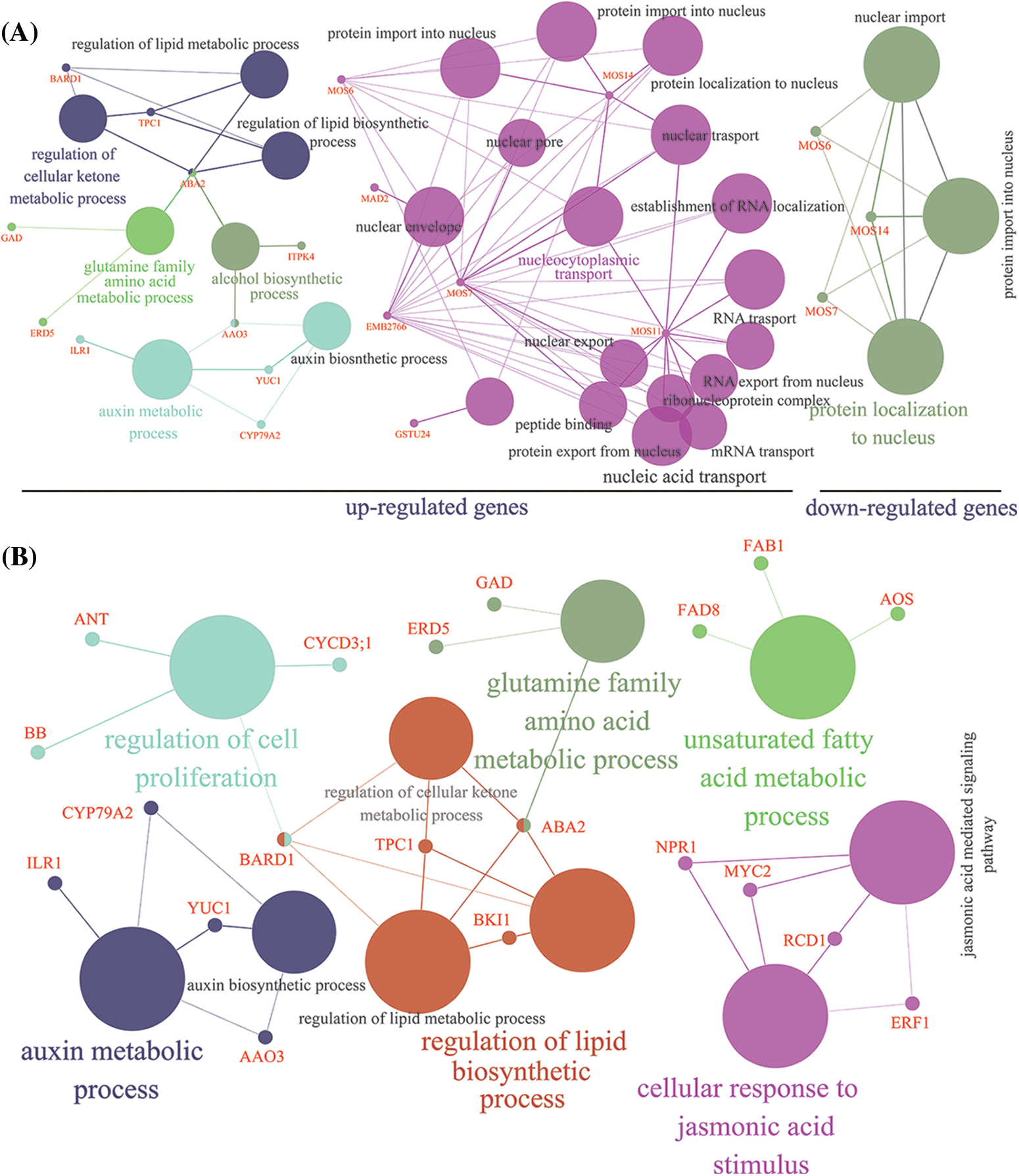
Figure 1: Functional annotation in GO network (A) and KEGG network (B) of differentially expressed genes based on transcriptome analysis. The size of the circle represents the total number of genes in GO or KEGG pathways. The genes detected in this study (Supplementary Table S1) were colored red
Antioxidant processes play important roles in response to heavy ion beam irradiation. We listed the expression of antioxidant genes in the ROS-scavenging network in Table 4. A total of 118 ROS-scavenging genes were annotated into six subgroups, including catalase (CAT), NADPH oxidase, Ascorbate peroxidase (APX), Superoxide dismutase (SOD), Glutathione peroxidase (GPX) and Thioredoxin (Trx) (Tables 4 and Supplementary Table S2). GPX1 was up-regulated after irradiation by 1.35-fold, whereas NADPH oxidase was the largest down-regulated group, including NADPH oxidase RBOHB (−1.58 fold), ferric-chelate reductase (FRO1), ferric-chelate reductase (FRO5) (−2 fold) and ferric-chelate reductase (FRO7) (−1.34 fold). Interestingly, all APXs were slightly up-regulated by the ion beam treatment, whereas all CATs were down-regulated. In SOD families, Cu/ZnSOD was the largest up-regulated group by 0.88-fold in irradiated samples compared with the control group. Additionally, three types of thioredoxin were somewhat changed after radiation.
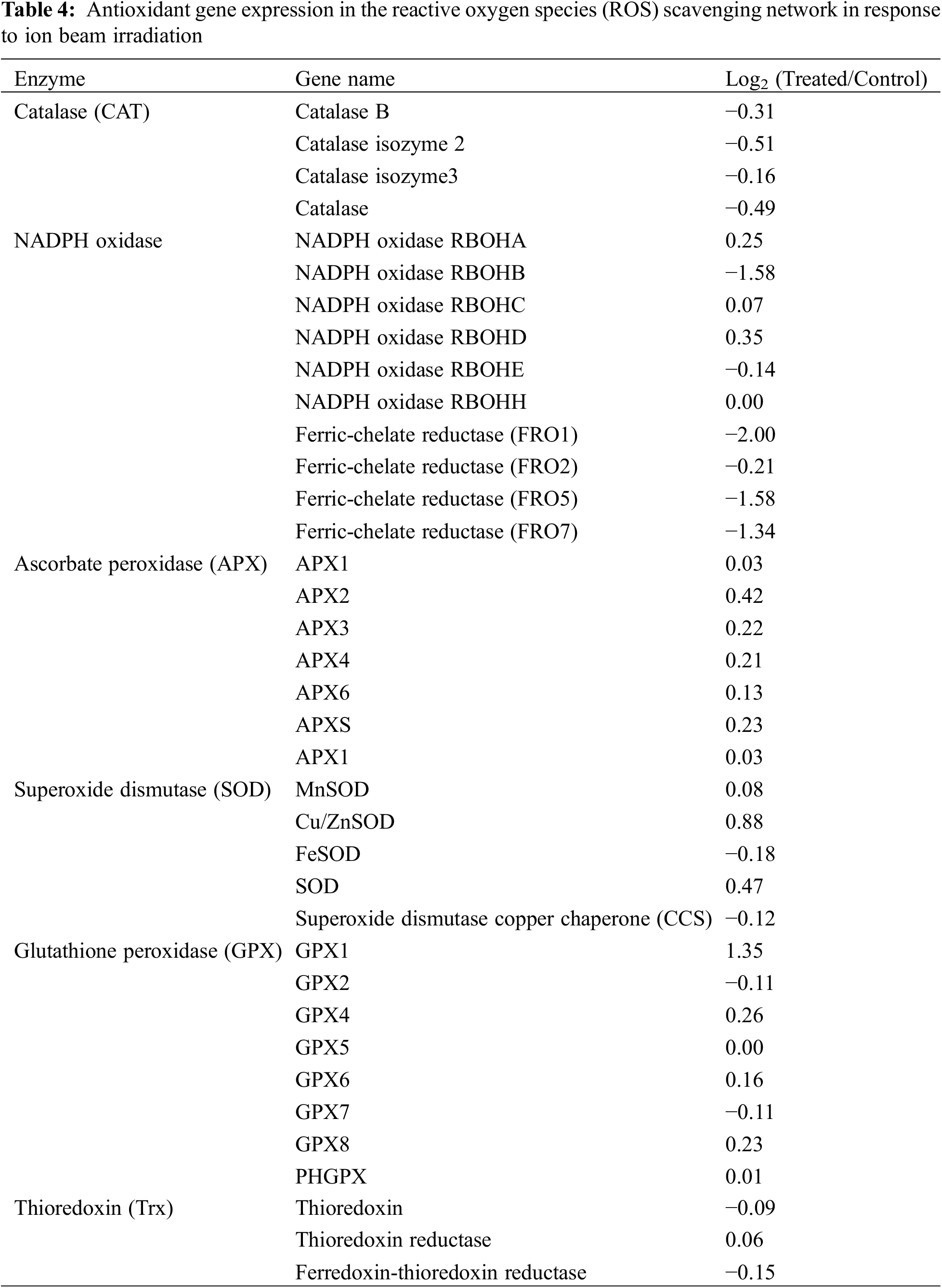
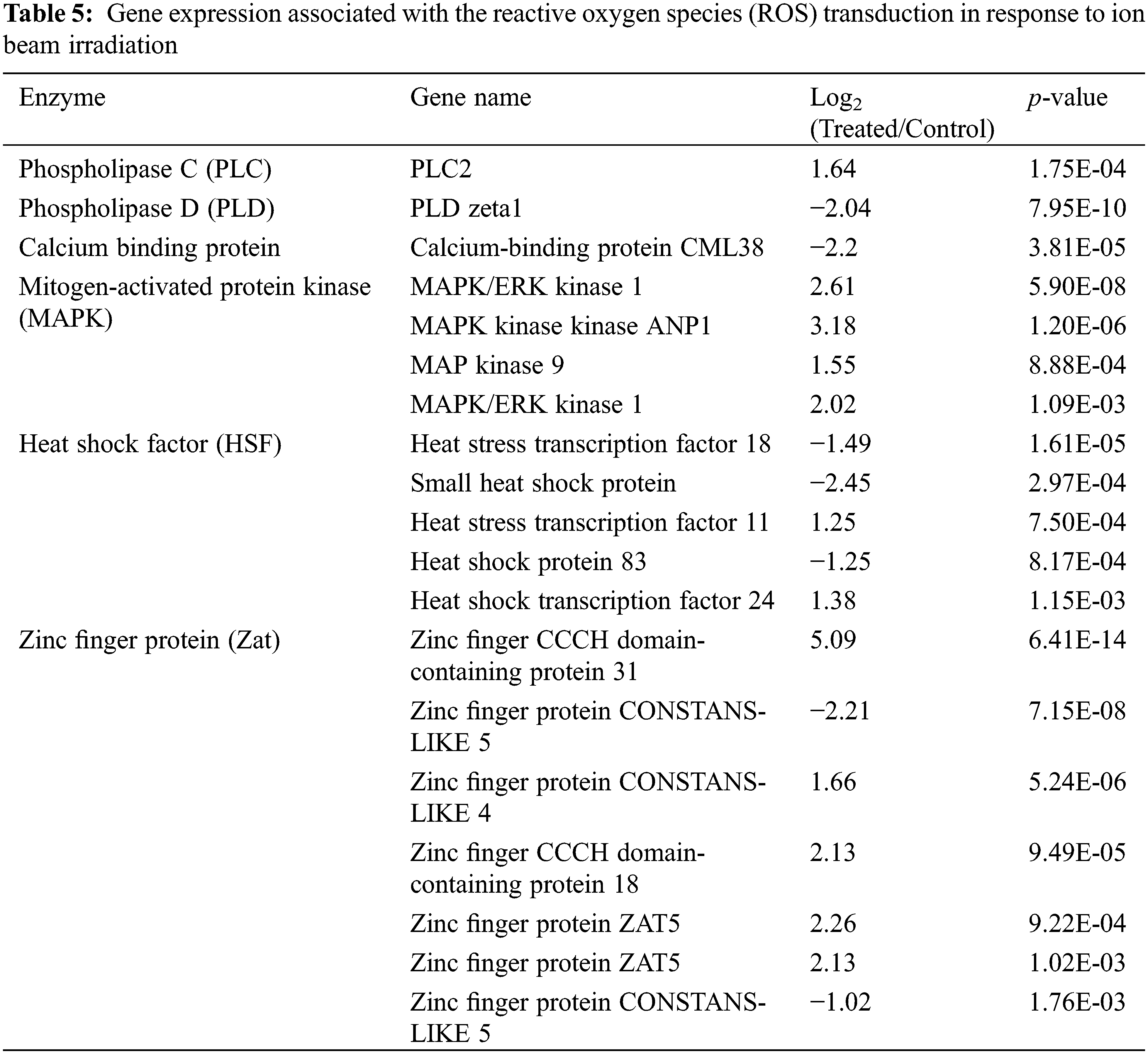
Except for detecting gene expression in the ROS scavenging network, ROS signaling transduction-associated genes are listed in Tables 5 and Supplementary Table S3. A total of six types of genes (262 genes) were significantly up-regulated or down-regulated after LIB irradiation, including phospholipase C (PLC), phospholipase D (PLD), calcium-binding protein, mitogen-activated protein kinase (MAPK), heat shock factor (HSF), and zinc finger protein (Zat). Among these genes, all MAPKs were highly up-regulated by treatment, with changes of more than 2-fold. Three out of five HSF genes were strikingly down-regulated, and five out of seven Zat genes were up-regulated after irradiation. Importantly, Zinc finger CCCH domain-containing protein 31 and MAPK kinase ANP1 were highly up-regulated with 5.09 and 3.18 expression fold changes (Log2), respectively. These results suggested the up-regulated expression level of ROS transduction-associated genes responding to ion beam irradiation.
3.5 Metabolites Extraction and Metabolomic Profiling Analysis
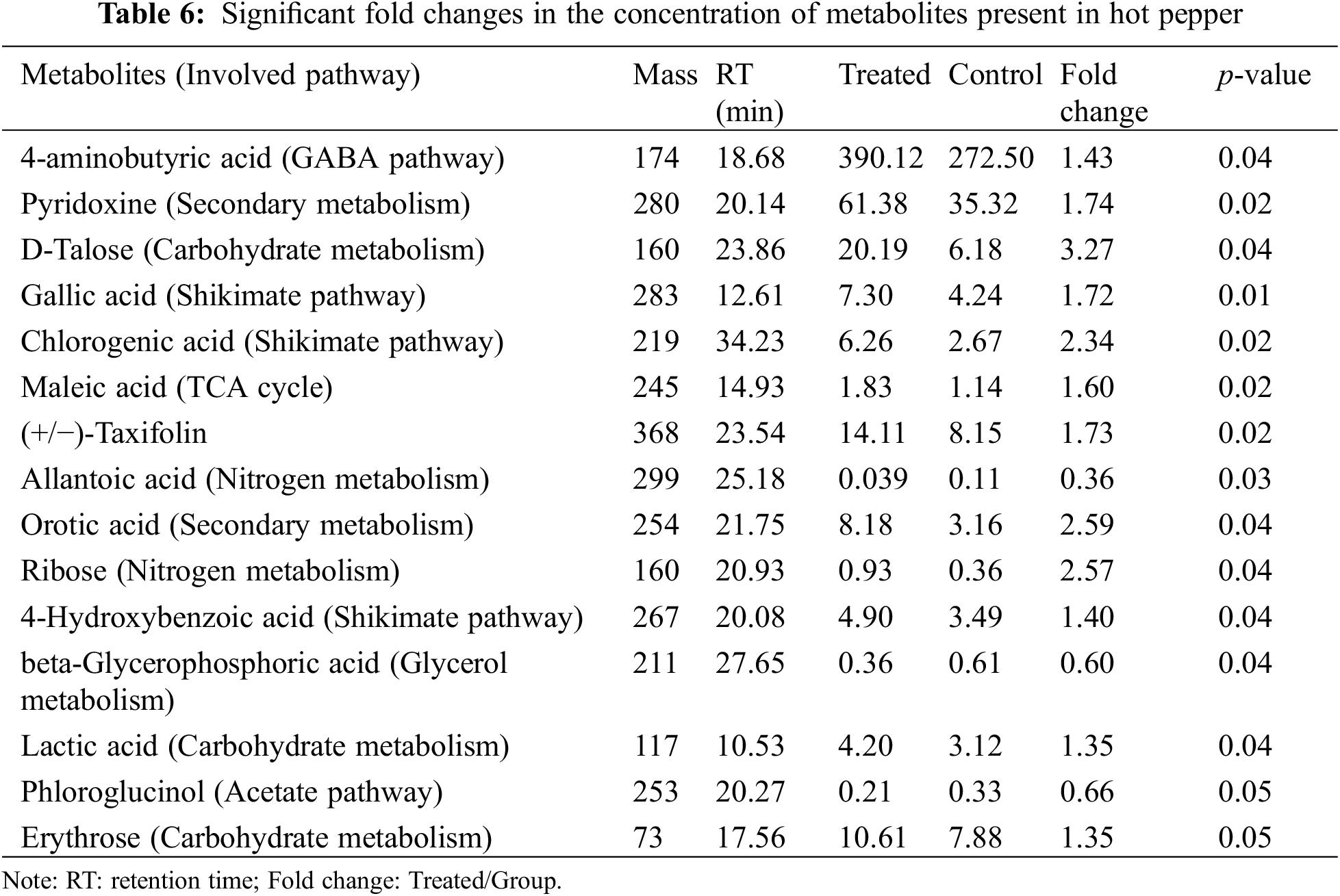
GC-MS was applied to compare the amount of intracellular metabolites in hot pepper with or without LIB irradiation. A loading plot was constructed based on the OPLS-DA model. Using this model, all samples were classified into two groups (Fig. 2). These values illustrated that a significant variation in the statistical data was attributable to the variation of metabolites between the two groups. A total of 15 metabolites were identified as significant changes in these two groups (Table 6). After being exposed to the ion beam, almost all metabolites were greatly increased, except for allantoic acid, beta-glycerophosphoric acid, and phloroglucinol, suggesting that ion beams might enhance some metabolism pathways. All the significantly changed metabolites were involved in GABA pathways, secondary metabolism, carbohydrate metabolism, shikimate pathways, TCA cycles, nitrogen metabolism, glycerol metabolism, and acetate pathways, effectively reflecting the intracellular metabolic characteristics of hot pepper.
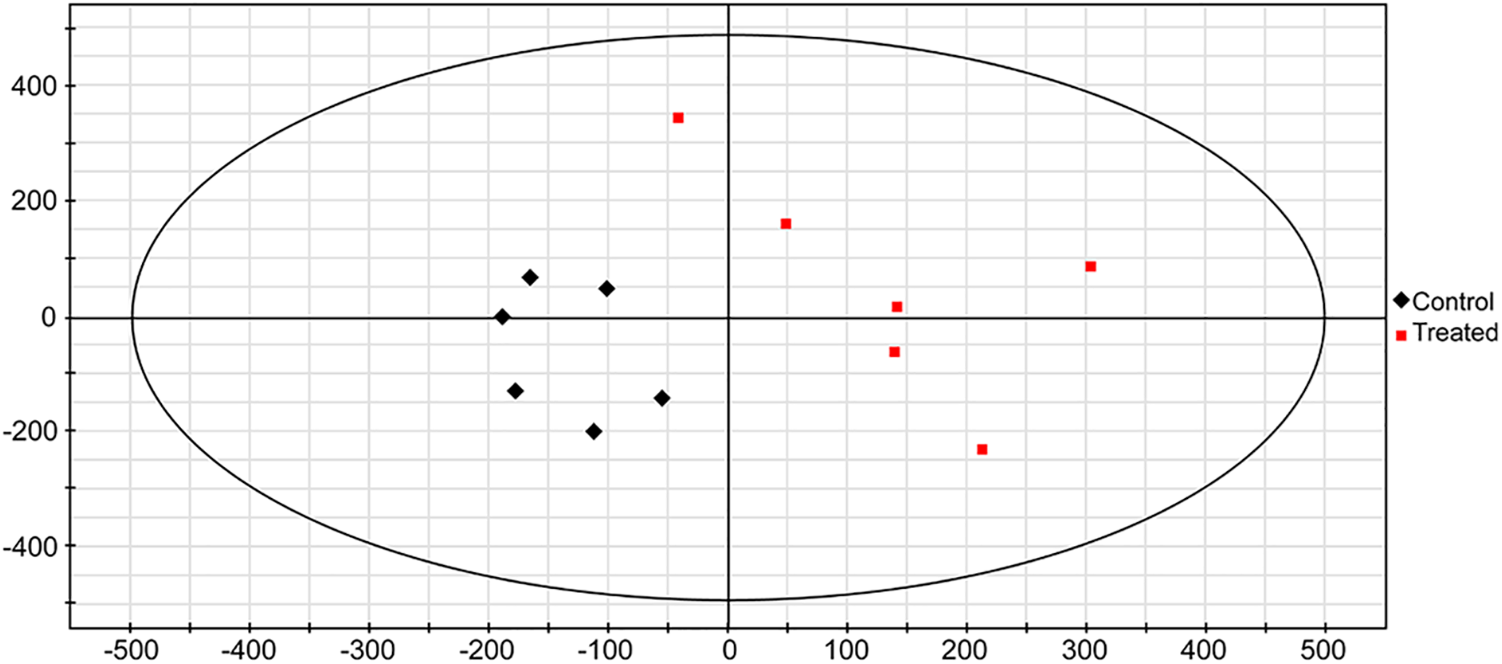
Figure 2: Score scatter plot of (O)PLS-DA model for the group treated by irradiation vs. control. The scatter color represents different experimental samples
4.1 Physiological Changes in Response to Lithium-Ion Beams
Since the 1960s, researchers have applied low-energy ion beams in plant mutagenesis breeding [40]. Compared with X-ray, electron beam, and laser mutagenesis, ion beams have the advantages of minor risk of physical damage, high mutation frequency, and a wide range of mutations [41]. Ion beams could cause great variations in cell ultrastructure and chromosome structure [42]. A previous study suggested that lithium-ion beam has a higher biological impact on energy deposition, momentum transferring, and mass deposition compared to traditional radiation [43]. Ion beam irradiation strongly affected plant growth and development. A significant change in phenotypes has been observed in the model plant Arabidopsis thaliana using carbon-ion beam irradiation [7]. For many agricultural crops like pepper, agronomic traits, such as yield and disease resistance, are of interest to the researchers. In our study, the results showed that the main economic characteristics (plant height, fruit weight, yield) were greatly increased after LIB irradiation (Table 1). Compared to traditional breeding, ion beam treatment could obtain desirable characteristics in a short time [44].
Chlorophyll contents have important functions in photosynthetic efficiency and are often used to evaluate plant response to stresses [45]. In our results, the chlorophyll b and a+b contents in ion-beam-treated samples increased by 1.55-fold and 1.26-fold, respectively. Consistent with the present study, Ling et al. [46] found that a 40 Gy ion beam had a stimulating effect on the chlorophyll content of rice. Kim et al. [18] compared three different types of ionizing radiation and suggested that the chlorophyll contents were slightly lower in rice irradiated by gamma rays and cosmic rays, whereas in the ion beam-irradiated rice, it was significantly increased. However, other studies reported that ion beam radiation inhibited chlorophyll biosynthesis in plants due to the breakdown of the porphyrin ring of the chlorophyll molecule [47,48]. Still, it is difficult to compare the plant physiological and morphological responses to irradiation as the parameters of each experiment varied greatly. For example, the type and dose of irradiation, plant species, and developmental stage could all differ among studies.
4.2 Transcriptome Changes in Response to Lithium-Ion Beam
Compared to the control samples, 671 up-regulated and 376 down-regulated genes were identified as DEGs, suggesting that these genes were involved in response to ion beam irradiation. After several database annotations, some functional proteins were obtained. Five genes were identified as phosphatidylinositol phosphokinase, which is activated by osmotic stress or when quiescent cells are stimulated with growth factors [49,50]. Uroporphyrinogen decarboxylase (UROD, EC 4.1.1.37) is a key regulatory enzyme for the biosynthesis of protoheme and chlorophyll in plants, catalyzing the formation of coproporphyrinogen from uroporphyrinogen [51]. One gene (gene 29706) was annotated as UROD (Supplementary Table S1), and its expression was much higher in the treated groups than in the control. Eighteen genes encode protein related to thionin, which is widely distributed in plant cells, could inhibit or kill pathogenic microorganisms, and has been applied in plant disease resistance breeding in recent years [52]. Six genes encode zinc finger protein, which are important for regulating gene expression. Previous reports suggested that bZIP genes in wheat were strongly induced by multiple abiotic stresses through the abscisic acid (ABA) signaling pathway [53]. All these findings suggested that the induced variety might harbor more resistance and tolerance than the control.
The phytohormone auxin is a key metabolite in regulating plant growth and development stages [54]. Indole-3-acetic acid (IAA) has been recognized as the most abundant auxin in plants [55]. In this study, we detected a gene (gene32888, Supplementary Table S1) annotated to IAA-amino acid hydrolase ILR1-L like, which helps regulate free IAA concentrations in Arabidopsis by cleaving their conjugates [56,57]. Indole-3-acetaldehyde (IAId) oxidase-like (gene31897, Supplementary Table S1) was up-regulated in irradiated samples. This enzyme is involved in the synthesis of IAId, which is a precursor of IAA [58].
Jasmonates are important lipid regulators in plants that mediate responses to both mechanical trauma and pathogenesis and play pivotal roles in reproduction and metabolic regulation [59]. Plants employ ubiquitin-proteasome system-dependent proteolysis of MYC transcription factor (MYC2) as a mechanism to control plant responses to various environmental stresses in the jasmonic acid pathway [60], while the down-regulated MYC2 (gene4128, Table 4) was detected in the treated group. Ethylene response factor1 (ERF1) functions as a transcription factor that integrates signals from ethylene and jasmonic acid pathway [61]. The up-regulated ERF1 (gene15698, Table 4) in irradiated samples suggested the involvement of ERF1 in ethylene response to abiotic stress.
Reactive oxygen species (ROS) have been recognized as key regulators in the complex signaling network in plants in response to abiotic stress [62]. Kim et al. found 33 and 42 genes associated with ROS scavenging and signal transduction pathways, respectively, were induced or repressed in Arabidopsis [63]. After different doses of ion beam treatments in rice, 118 and 813 rice genes involved in ROS scavenging and signal transduction pathways showed induction or repression after ionization treatment [18]. Similarly, we also identified the differentially expressed genes in ROS scavenging pathways and signaling transduction (Tables 4 and 5). Most of the genes associated with ROS transduction were up-regulated under irradiated samples, suggesting that the exposure of the plants to ion beams directly or indirectly influenced ROS-related processes. In addition, Matsumoto et al. [64] suggested that the generation of ROS was imbalanced at the molecular level after carbon-ion induction. The carbon ion beam could modulate the physiological response and cause high expression of cold signaling genes to mitigate cold stress in Arabidopsis [65]. The activities of antioxidant enzymes induced by ROS played important roles in growth promotion in Arabidopsis after ion beam irradiation [66]. Zhang et al. indicated that ion implantation activated antioxidant enzymes and inhibited the ROS system, thus enhancing cell viability and division activity [67].
4.3 Metabolic Changes in Response to Lithium-Ion Beam
Furthermore, we identified 15 metabolites with significant differences in amount between these two groups. Allantoin and allantoic acid are regarded as important transport compounds of fixed nitrogen from soybean nodules to other parts of the plant [68]. Therefore, allantoic acid plays a vital role in nitrogen metabolism. Allantoic acid increased significantly after irradiation, suggesting that ion beam radiation could change nitrogen metabolism in hot pepper. Beta-glycerophosphoric acid is the initial substrate of glycerophospholipids, which are the main constituent in the cellular membrane. Phloroglucinol is naturally found in certain plants and has various useful biological properties, such as anti-inflammatory, anticancer, anti-microbial, antioxidant, and neuro-regenerative activities [69].
Gamma-aminobutyric acid (GABA) metabolism has been found in plants under many abiotic stress conditions, and its metabolism is regulated by the GABA shunt pathway [70]. Earlier studies have demonstrated that GABA metabolism increased under various abiotic stresses [71]. A similar finding was detected in our observation that aminobutyric acid was markedly increased after irradiation (Table 6). A study demonstrated that an enhanced GABA shunt pathway might provide intermediates for the TCA cycle, thus maintaining metabolic homeostasis during abiotic stress [72].
The shikimic acid pathway has been identified for the biosynthesis of most aromatic amino acids in plants, and it mainly links carbohydrate metabolism to aromatic compound biosynthesis [73]. Chlorogenic acid is a phenylpropanoid compound produced by the shikimate pathway during aerobic respiration [74]. Here, chlorogenic acid was increased (2.3-fold) in the ion beam irradiated sample (Table 6). Recently, transcriptomic analysis revealed changes in chlorogenic acid after frost in Mulberry [74]. Additionally, metabolomic studies showed a significant increase in chlorogenic acid as a response of sugarcane to biotic stress [75].
In summary, physiological, transcriptome, and metabolic analyses of hot pepper under lithium-ion beam irradiation demonstrated that 60 Gy of ion beam exposure could significantly improve all-important agronomic traits (particularly yield unit) through the regulation of multiple physiological processes and metabolic pathways, including the improvement of chlorophyll content, the balance of antioxidant enzymes and the enhancement of ROS transduction adjustment, as well as the accumulation of amino acids, carbohydrates, and organic acids. The enhanced carbohydrate metabolism, shikimic acid pathways, GABA shunts, and TCA cycles could play important roles in ion beam-irradiated plants.
Acknowledgement: We thank Mu Peng (Hubei Minzu University) for his assistance in revising the paper and the submission process.
Funding Statement: This project was supported by the China Atomic Energy Authority (Crop Varietal Improvement and Insect Pests Control by Nuclear Radiation), Key Technological Innovation Key Project of Agricultural Science and Technology Breakthrough in Heilongjiang Province-Breakthrough Variety Breeding and Industrial Application of Vegetables.
Author Contributions: Luxiang Liu contributed to conceiving and designing the experiments. Chunmei Xu, Yongdun Xie, Hongchun Xiong, and Xinchun Han performed the experiments and data analysis. Libo Xie and Xue Wang drew the figures and wrote sections of the manuscript. Mu Guo revised the manuscript.
Availability of Data and Materials: All data included in this study are available upon request by contact with the corresponding author.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declared that they have no conflicts of interest in this work.
Supplementary Materials: The supplementary material is available online at https://doi.org/10.32604/phyton.2023.042919.
References
1. Song, X., Zhang, Y., Zhu, X., Wang, Y., Chu, J. et al. (2017). Mutation breeding of high avermectin B1a-producing strain by the combination of high energy carbon heavy ion irradiation and sodium nitrite mutagenesis based on high throughput screening. Biotechnology and Bioprocess Engineering, 22, 539–548. [Google Scholar]
2. Luo, S., Zhou, L., Li, W., Du, Y., Yu, L. et al. (2016). Mutagenic effects of carbon ion beam irradiations on dry Lotus japonicus seeds. Nuclear Instruments & Methods in Physics Research, 383, 123–128. [Google Scholar]
3. Du, Y., Luo, S., Yu, L., Cui, T., Chen, X. et al. (2018). Strategies for identification of mutations induced by carbon-ion beam irradiation in Arabidopsis thaliana by whole genome re-sequencing. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis, 807, 21–30. [Google Scholar] [PubMed]
4. Suprasanna, P., Mirajkar, S., Bhagwat, S. (2015). Induced mutations and crop improvement. In: Plant biology and Biotechnology, pp. 593–617. New York City: Springer. [Google Scholar]
5. Li, C., Tang, J., Hu, Z., Wang, J., Yu, T. et al. (2020). A novel maize dwarf mutant generated by Ty1-copia LTR-retrotransposon insertion in Brachytic2 after spaceflight. Plant Cell Reports, 39, 393–408. [Google Scholar] [PubMed]
6. Zeng, D., Cui, J., Yin, Y., Dai, C., Zhao, H. et al. (2022). Combining proteomics and metabolomics to analyze the effects of spaceflight on rice progeny. Frontiers in Plant Science, 13, 900143. [Google Scholar] [PubMed]
7. Du, Y., Luo, S., Li, X., Yang, J., Cui, T. et al. (2017). Identification of substitutions and small insertion-deletions induced by carbon-ion beam irradiation in Arabidopsis thaliana. Frontiers in Plant Science, 8, 1851. [Google Scholar] [PubMed]
8. Magori, S., Tanaka, A., Kawaguchi, M., Meksem, K., Kahl, G. (2010). Physically induced mutation: Ion beam mutagenesis. In: The handbook of plant mutation screening. KGaA, Weinheim: WILEY-VCH Verlag GmbH & Co. [Google Scholar]
9. Permata, T. B. M., Sato, H., Gu, W., Kakoti, S., Uchihara, Y. et al. (2021). High linear energy transfer carbon-ion irradiation upregulates PD-L1 expression more significantly than X-rays in human osteosarcoma U2OS cells. Journal of Radiation Research, 62, 773–781. [Google Scholar] [PubMed]
10. Shikazono, N., Tanaka, A., Kitayama, S., Watanabe, H., Tano, S. (2002). LET dependence of lethality in Arabidopsis thaliana irradiated by heavy ions. Radiation & Environmental Biophysics, 41, 159–162. [Google Scholar]
11. Xiong, H., Guo, H., Xie, Y., Gu, J., Zhao, L. et al. (2020). Comparative transcriptome analysis of two common wheat varieties induced by 7Li-ion beam irradiation reveals mutation hotspot regions and associated pathways. Radiation Physics and Chemistry, 170, 108650. https://doi.org/10.1016/j.radphyschem.2019.108650 [Google Scholar] [CrossRef]
12. Tokuyama, Y., Furusawa, Y., Ide, H., Yasui, A., Terato, H. (2015). Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation. Journal of Radiation Research, 56, 446–455. [Google Scholar] [PubMed]
13. Alizadeh, E., Sanz, A. G., García, G., Sanche, L. (2013). Radiation damage to DNA: The indirect effect of low energy electrons. Journal of Physical Chemistry Letters, 4, 820–825. [Google Scholar] [PubMed]
14. Becker, D., Sevilla, M. D. (1993). 3-The chemical consequences of radiation damage to DNA. Advances in Radiation Biology, 17, 121–180. [Google Scholar]
15. Du, Y., Li, W., Yu, L., Chen, G., Liu, Q. et al. (2014). Mutagenic effects of carbon-ion irradiation on dry Arabidopsis thaliana seeds. Mutation Research/Genetic Toxicology & Environmental Mutagenesis, 759, 28–36. [Google Scholar]
16. Zhang, L., Chen, Q., Su, M., Yan, B., Zhang, X. et al. (2016). High-molecular-weight glutenin subunit-deficient mutants induced by ion beam and the effects of Glu-1 loci deletion on wheat quality properties. Journal of the Science of Food and Agriculture, 96(4), 1289–1296. https://doi.org/10.1002/jsfa.7221 [Google Scholar] [PubMed] [CrossRef]
17. Wang, Z., Gerstein, M., Snyder, M. (2009). RNA-seq: A revolutionary tool for transcriptomics. Nature Reviews Genetics, 10, 57–63. [Google Scholar] [PubMed]
18. Kim, S. H., Song, M., Lee, K. J., Hwang, S. G., Jang, C. S. et al. (2012). Genome-wide transcriptome profiling of ROS scavenging and signal transduction pathways in rice (Oryza sativa L.) in response to different types of ionizing radiation. Molecular Biology Reports, 39, 11231. [Google Scholar]
19. Pontin, M. A., Piccoli, P. N., Francisco, R., Bottini, R., Martinezzapater, J. M. et al. (2010). Transcriptome changes in grapevine (Vitis vinifera L.) cv. Malbec leaves induced by ultraviolet-B radiation. BMC Plant Biology, 10, 224. [Google Scholar] [PubMed]
20. Casati, P., Walbot, V. (2004). Rapid transcriptome responses of maize (Zea mays) to UV-B in irradiated and shielded tissues. Genome Biology, 5, R16. [Google Scholar] [PubMed]
21. Austin, D. F. (2012). Domestication of plants in the old world. The origin and spread of domesticated plants in South-West Asia, Europe, and the Mediterranean Basin. In: Zohary, D., Hopf, M., Weiss, E. (Eds.Economic botany, 4th Editionvol. 66, pp. 420–421. New York: Oxford University Press. [Google Scholar]
22. Motukuri, S. K., Jaswanthi, N. (2020). Hot pepper (Capsicum annuum L.An alternative food to reduce micronutrient deficiencies in human. In: Capsicum. London: IntechOpen. [Google Scholar]
23. Hervert-Hernandez, D., Sayago-Ayerdi, S. G., Goni, I. (2010). Bioactive compounds of four hot pepper varieties (Capsicum annuum L.antioxidant capacity, and intestinal bioaccessibility. Journal of Agricultural and Food Chemistry, 58, 3399–3406. [Google Scholar] [PubMed]
24. Materska, M., Perucka, I. (2005). Antioxidant activity of the main phenolic compounds isolated from hot pepper fruit (Capsicum annuum L.). Journal of Agricultural & Food Chemistry, 53, 1750–1756. [Google Scholar]
25. Kim, S., Park, M., Yeom, S. I., Kim, Y. M., Lee, J. M. et al. (2014). Genome sequence of the hot pepper provides insights into the evolution of pungency in Capsicum species. Nature Genetics, 46, 270–278. [Google Scholar] [PubMed]
26. Xie, L., Wang, X., Peng, M., Meng, F., Zhou, Y. et al. (2014). Isolation and detection of differential genes in hot pepper (Capsicum annuum L.) after space flight using AFLP markers. Biochemical Systematics & Ecology, 57, 27–32. [Google Scholar]
27. Xie, L. B., Wang, X., Peng, M., Zhou, Y., Chen, L. X. et al. (2017). Comparative proteome analysis in hot pepper (Capsicum annuum L.) after space flight. Phyton-International Journal of Experimental Botany, 86, 236–245. https://doi.org/10.32604/phyton.2017.86.236 [Google Scholar] [PubMed] [CrossRef]
28. Guo, Y. (1997). Longshu No. 5, a big claw pepper species. Northern Horticulture, 6, 67. [Google Scholar]
29. Larbi, A., Abadía, A., Abadía, J., Morales, F. (2006). Down co-regulation of light absorption, photochemistry, and carboxylation in Fe-deficient plants growing in different environments. Photosynthesis Research, 89, 113–126. [Google Scholar] [PubMed]
30. Inskeep, W. P., Bloom, P. R. (1985). Extinction coefficients of chlorophyll a and b in N,N-dimethylformamide and 80% acetone. Plant Physiology, 77, 483–485. [Google Scholar] [PubMed]
31. Kim, D., Langmead, B., Salzberg, S. L. (2015). HISAT: A fast spliced aligner with low memory requirements. Nature Methods, 12, 357–360. [Google Scholar] [PubMed]
32. Trapnell, C., Roberts, A., Loyal, G., Geo, P., Daehwan, K. et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols, 7, 562–578. [Google Scholar] [PubMed]
33. Robinson, M. D., McCarthy, D. J., Smyth, G. K. (2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics, 26, 139–140. [Google Scholar] [PubMed]
34. Ritchie, M. E., Phipson, B., Wu, D., Hu, Y., Law, C. W. et al. (2015). Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research, 43(7), e47. https://doi.org/10.1093/nar/gkv007 [Google Scholar] [PubMed] [CrossRef]
35. Benjamini, Y., Hochberg, Y. (1995). Controlling the false discovery rate—A practical and powerful approach to multiple testing. Journal of the Royal Statistical Society, 57, 289–300. [Google Scholar]
36. Mao, X. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics, 21(21), 3787–3793. [Google Scholar] [PubMed]
37. Erban, A., Schauer, N., Fernie, A. R., Kopka, J. (2007). Nonsupervised construction and application of mass spectral and retention time index libraries from time-of-flight gas chromatography-mass spectrometry metabolite profiles. In: Metabolomics, pp. 19–38. New York: Springer. [Google Scholar]
38. Westerhuis, J. A., Velzen, E. J. J. V., Hoefsloot, H. C. J., Smilde, A. K. (2010). Multivariate paired data analysis: Multilevel PLSDA versus OPLSDA. Metabolomics, 6(1), 119–128. [Google Scholar] [PubMed]
39. Huang, Y., Huang, Q., Li, J., Yin, Y., Jiao, Z. (2021). Photosynthetic physiology and molecular response mechanisms of Indica-Japonica intersubspecific tetraploid rice seedlings to ion beams. Journal of Plant Growth Regulation, 40(2), 722–735. [Google Scholar]
40. Sax, K. (1963). The stimulation of plant growth by ionizing radiation. Radiation Botany, 3, 179–186. [Google Scholar]
41. Wu, L., Yu, Z. (2001). Radiobiological effects of a low-energy ion beam on wheat. Radiation and Environmental biophysics, 40, 53–57. [Google Scholar] [PubMed]
42. Dwiranti, A., Takata, H., Uchiyama, S., Fukui, K. (2016). The effect of magnesium ions on chromosome structure as observed by scanning electron microscopy (SEM) and scanning transmission electron microscope (STEM) tomography. Chromosome Science, 19, 19–23. [Google Scholar]
43. Salah, N., Lochab, S., Kanjilal, D., Sahare, P., Aleynikov, V. (2007). Effect of high-energy 7Li2+ ions on the TL behavior of LiF: Mg, Cu, P detectors. Radiation Measurements, 42, 1294–1300. [Google Scholar]
44. Liu, L., Guo, H., Zhao, L., Wang, J., Gu, J. et al. (2009). Achievements and perspectives of crop space breeding in China. Induced Plant Mutations in the Genomics Era, 7, 213–215. [Google Scholar]
45. Cui, T., Luo, S., Du, Y., Yu, L., Yang, J. et al. (2019). Research of photosynthesis and genomewide resequencing on a yellow-leaf Lotus japonicus mutant induced by carbon ion beam irradiation. Grassland Science, 65(1), 41–48. https://doi.org/10.1111/grs.12216 [Google Scholar] [CrossRef]
46. Ling, A. P. K., Ung, Y. C., Hussein, S., Harun, A. R., Tanaka, A. et al. (2013). Morphological and biochemical responses of Oryza sativa L. (cultivar MR219) to ion beam irradiation. Journal of Zhejiang University SCIENCE B, 14, 1132–1143. [Google Scholar] [PubMed]
47. Jia, C. F., Li, A. L. (2008). Effect of gamma radiation on mutant induction of Fagopyrum dibotrys Hara. Photosynthetica, 46(3), 363. https://doi.org/10.1007/s11099-008-0066-0 [Google Scholar] [CrossRef]
48. Hao, J., Ruan, S., Lin, S., Xue, M., Zhang, T. et al. (2015). A comparative study on chlorophyll fluorescence and chlorophyll content characteristics effects of Ti+ and Fe+ ion implantation in Chinese fir seedling. Journal of Radiation Research and Radiation Processing, 33, 7. [Google Scholar]
49. Roche, S., Koegl, M., Courtneidge, S. A. (1994). The phosphatidylinositol 3-kinase alpha is required for DNA synthesis induced by some, but not all, growth factors. Proceedings of the National Academy of Sciences of the United States of America, 91, 9185–9189. [Google Scholar] [PubMed]
50. Dove, S. K., Cooke, F. T., Douglas, M. R., Sayers, L. G., Parker, P. J. et al. (1997). Osmotic stress activates phosphatidylinositol-3, 5-bisphosphate synthesis. Nature, 390, 187–192. [Google Scholar] [PubMed]
51. Chen, T. C., Miller, G. W. (1974). Purification and characterization of uroporphyrinogen decarboxylase from tobacco leaves. Plant and Cell Physiology, 15, 993–1005. [Google Scholar]
52. Plattner, S., Gruber, C., Stadlmann, J., Widmann, S., Gruber, C. W. et al. (2015). Isolation and characterization of a thionin proprotein processing enzyme from Barley. Journal of Biological Chemistry, 290(29), 18056–18067. [Google Scholar] [PubMed]
53. Zhang, L., Zhang, L., Xia, C., Zhao, G., Liu, J. et al. (2015). A novel wheat bZIP transcription factor, TabZIP60, confers multiple abiotic stress tolerances in transgenic Arabidopsis. Physiologia Plantarum, 153, 538–554. [Google Scholar] [PubMed]
54. Mashiguchi, K., Tanaka, K., Sakai, T., Sugawara, S., Kawaide, H. et al. (2011). The main auxin biosynthesis pathway in Arabidopsis. Proceedings of the National Academy of Sciences of the United States of America, 108, 18512–18517. [Google Scholar] [PubMed]
55. Zhao, Y. (2010). Auxin biosynthesis and its role in plant development. Annual Review of Plant Biology, 61, 49–64. [Google Scholar] [PubMed]
56. Cooke, T. J., Poli, D., Sztein, A. E., Cohen, J. D. (2002). Evolutionary patterns in auxin action. In: Auxin molecular biology, pp. 319–338. New York: Springer. [Google Scholar]
57. LeClere, S., Tellez, R., Rampey, R. A., Matsuda, S. P., Bartel, B. (2002). Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. Journal of Biological Chemistry, 277, 20446–20452. [Google Scholar] [PubMed]
58. Percival, F. W., Purves, W. K., Vickery, L. E. (1973). Indole-3-ethanol oxidase: Kinetics, inhibition, and regulation by auxins. Plant Physiology, 51, 739–743. [Google Scholar] [PubMed]
59. Robin, L., Edward, E. F. (2002). The jasmonate pathway. Science, 296, 1649–1650. [Google Scholar]
60. Zhai, Q., Yan, L., Tan, D., Chen, R., Sun, J. et al. (2013). Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genetics, 9, e1003422. [Google Scholar] [PubMed]
61. Shinshi, H. (2008). Ethylene-regulated transcription and crosstalk with jasmonic acid. Plant Science, 175, 18–23. [Google Scholar]
62. You, J., Chan, Z. (2015). ROS regulation during abiotic stress responses in crop plants. Frontiers in Plant Science, 6, 1092. https://doi.org/10.3389/fpls.2015.01092 [Google Scholar] [PubMed] [CrossRef]
63. Kim, D. S., Kim, J. B., Goh, E. J., Kim, W. J., Kim, S. H. et al. (2011). Antioxidant response of Arabidopsis plants to gamma irradiation: Genome-wide expression profiling of the ROS scavenging and signal transduction pathways. Journal of Plant Physiology, 168, 1960–1971. [Google Scholar] [PubMed]
64. Matsumoto, K. I., Nyui, M., Ueno, M., Ogawa, Y., Nakanishi, I. (2019). A quantitative analysis of carbon-ion beam-induced reactive oxygen species and redox reactions. Journal of Clinical Biochemistry and Nutrition, 65, 18–34. [Google Scholar]
65. Wang, L., Ma, R., Yin, Y., Jiao, Z. (2018). Role of carbon ion beams irradiation in mitigating cold stress in Arabidopsis thaliana. Ecotoxicology and Environmental Safety, 162, 341–347. [Google Scholar] [PubMed]
66. Wang, L., Ma, R., Yin, Y., Jiao, Z. (2018). Antioxidant response of Arabidopsis thaliana seedlings to oxidative stress induced by carbon ion beams irradiation. Journal of Environmental Radioactivity, 195, 1–8. [Google Scholar] [PubMed]
67. Zhang, L., Qi, W., Xu, H., Wang, L., Jiao, Z. (2016). Effects of low-energy N+-beam implantation on root growth in Arabidopsis seedlings. Ecotoxicology and Environmental Safety, 124, 111–119. [Google Scholar] [PubMed]
68. Triplett, E. W., Blevins, D. G., Randall, D. D. (1980). Allantoic acid synthesis in soybean root nodule cytosol via xanthine dehydrogenase. Plant Physiology, 65, 1203–1206. [Google Scholar] [PubMed]
69. Abdel-Ghany, S. E., Day, I., Heuberger, A. L., Broeckling, C. D., Reddy, A. S. N. (2016). Production of phloroglucinol, a platform chemical, in Arabidopsis using a bacterial gene. Scientific Reports, 6(1), 38483. https://doi.org/10.1038/srep38483 [Google Scholar] [PubMed] [CrossRef]
70. Jalil, S. U., Khan, M. I. R., Ansari, M. I. (2019). Role of GABA transaminase in the regulation of development and senescence in Arabidopsis thaliana. Current Plant Biology, 19, 100119. [Google Scholar]
71. Shi, S. Q., Shi, Z., Jiang, Z. P., Qi, L. W., Sun, X. M. et al. (2010). Effects of exogenous GABA on gene expression of Caragana intermedia roots under NaCl stress: Regulatory roles for H2O2 and ethylene production. Plant, Cell & Environment, 33, 149–162. [Google Scholar]
72. Li, Z., Yu, J., Peng, Y., Huang, B. (2016). Metabolic pathways regulated by γ-aminobutyric acid (GABA) contributing to heat tolerance in creeping bentgrass (Agrostis stolonifera). Scientific Reports, 6, 30338. [Google Scholar] [PubMed]
73. Noda, S., Shirai, T., Mori, Y., Oyama, S., Kondo, A. (2017). Engineering a synthetic pathway for maleate in Escherichia coli. Nature Communications, 8, 1153. [Google Scholar] [PubMed]
74. Zhao, L., Wang, D., Liu, J., Yu, X., Wang, R. et al. (2019). Transcriptomic analysis of key genes involved in chlorogenic acid biosynthetic pathway and characterization of MaHCT from Morus alba L. Protein Expression and Purification, 156, 25–35. [Google Scholar] [PubMed]
75. Naranjo Pinta, M., Montoliu, I., Aura, A. M., Seppänen-Laakso, T., Barron, D. et al. (2018). In vitro gut metabolism of [U-13C]-quinic acid, the other hydrolysis product of chlorogenic acid. Molecular Nutrition & Food Research, 62, 1800396. [Google Scholar]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools