 Open Access
Open Access
ARTICLE
The Improvement of Soybean Salt Tolerance by Overexpressed GmPAO1
College of Agronomy, Jilin Agricultural University, Changchun, 130118, China
* Corresponding Author: Piwu Wang. Email:
(This article belongs to the Special Issue: Abiotic and Biotic Stress Tolerance in Crop)
Phyton-International Journal of Experimental Botany 2023, 92(4), 1109-1124. https://doi.org/10.32604/phyton.2023.025503
Received 17 July 2022; Accepted 17 October 2022; Issue published 06 January 2023
Abstract
Polyamines play an important regulatory role during plant growth and development and adversity stress, and polyamine oxidase (PAO) is involved in polyamine catabolism. In this study, an up-regulated polyamine oxidase gene GmPAO1 was obtained by transcriptome sequencing analysis and screening at soybean seedling stages. Also, its expression pattern and function were analyzed. The identification results of transgenic GmPAO1 soybean positive lines showed that the relative expression level of GmPAO1 in the overexpressed lines was increased under salt stress. With increasing stress concentration, the seed germination rate decreased. However, the seed germination rate of the overexpressed lines was significantly higher than that of the control lines, and the phenotypic character of the root systems was also better than that of the control lines. The measurement of superoxide dismutase (SOD) and peroxidase (POD) activities and malondialdehyde and hydrogen peroxide contents revealed that the overexpressed soybean lines significantly increased the SOD and POD activities, significantly reducing the malondialdehyde content. Although the hydrogen peroxide content in the transformed plants gradually increased, the hydrogen peroxide content in the overexpression lines was still lower than that in the gene editing lines. Based on this, it was preliminarily judged that GmPAO1 can improve soybean salt tolerance.Keywords
1.1 The Effects of Salt Stress on Soybean Growth and Development during Germination and Seedling Stages
Soil salinization could affect plant normal growth and development, which was one of the important factors leading to yield decline. Also, it was one of the important abiotic factors affecting the ecological environment. Currently, about more than 100 countries and regions worldwide were suffering from the harm caused by land salinization. According to statistics, the total area of saline land in China was about 6 million hectares, accounting for 4.88% of the available land area [1]. Soybeans were a moderately salt-tolerant crop. Studies have shown that when the salt content in the soil exceeds 5 ds/m, soybean growth and development can be significantly affected, resulting in a decrease in soybean yield and quality [2]. A high concentration of salt caused different degrees of damage to the whole life cycle of soybean seeds such as germination, vegetative growth and reproductive growth stages [3]. Also, there were differences in soybean salt tolerance in different growth and development stages. The germination stage, one of the most critical links in soybean production, was the primary one in the soybean growth cycle and also one of the growth stages in which soybean was more sensitive to salt stress [4]. Kan et al. [5] used soybean seeds as materials under four different environments to study the seed salt tolerance during germination. The experimental results showed that the germination rate and index were greatly affected by salt stress, while the imbibition rate was less affected by salt stress. Under salt stress, the relative salty damage imbibition index of soybeans was significantly negatively correlated with their germination rate and index. The ionic toxicity caused by salt stress reduces the activity of the peroxidase (POD) system, resulting in a massive accumulation of reactive oxygen species (ROS) and the inhibition of budding growth. Presently, great progress has been made in the research on the salt tolerance of soybean seedling stages at home and abroad. Also, it was confirmed that a series of physiological changes could occur in the soybean body under salt stress at the seedling stage. Hosseini et al. [6] obtained the following findings in studies: under 160 mmol/l NaCl stress, 97% of soybean seeds could germinate normally, but under the same salt concentration stress, the growth rate of soybean seedlings was only 14% of that of the control; when the NaCl concentration increased to 330 mmol, the growth rate of the seedlings was almost zero 3 days after germination, and the soybean seeds were more salt-tolerant at the germination stage than at the seedling stage.
1.2 The Role of Polyamine Oxidase (PAO) in Plant Growth and Development and Adversity Stress
Polyamines were a class of low molecular mass aliphatic nitrogen-containing bases with biological activities, which played an important role in the plant growth and adaptation to environmental changes [7]. The most common polyamines in the plant body were putrescine (Put), spermidine (Spd) and spermine (Spm); polyamine content in the plant body was regulated by biosynthesis and transportation; polyamine oxidative degradation was catalyzed by diamine oxidase (DAO) and PAO [8]. During plant growth, DAO and PAO regulated different kinds of polyamines. Under the same adversity, different organs of plants also had different regulatory mechanisms for polyamines. PAO was a monomeric soluble protein. As a coenzyme, flavin adenine dinucleotide (FAD) molecule was non-covalently bound to PAO protein and could catalyze the oxidation of polyamines and acetylpolyamines. The main physiological function of PAO was to catalyze the polyamine oxidative degradation in cells, regulate polyamine levels in the plant body and maintain the balance of polyamine content. Based on the difference in PAO involved in the catabolism process of polyamine, it could be divided into two categories. In the first category, PAO catalyzed the involvement of Spd and Spm in terminal catabolism of polyamine, and in the second one, PAO catalyzed the reverse conversion of Spm into Spd and Spm [9]. Amar et al. [10] found in studies that in NaCl-treated tomato leaves, polyamine catabolism was closely related to proline accumulation, which was one of the most common metabolic responses of tomato leaves to water or salt stress and related to the decline of Put and Spd and the partial rise of diaminopimelic acid (DAP), and after CuAO inhibition treatment, proline accumulation decreased. In transgenic tobacco with the overexpressed S-Adenosylmethionine Decarboxylase (SAMDC), with DAO activity increasing, the plants had relatively high salt and drought tolerance and also could resist fungal wilt disease [11]. In studies, Scalet et al. [12] obtained the findings as follows: External damage could increase the content of Put and cadmium(Cad) in chickpea seedlings, and PAO and POD activity also increased; after the increased PAO activity, H2O2 and a large number of amino aldehydes would be produced, which could resist microbial invasion; meanwhile, the increased PAO activity could also catalyze POD, which could participate in the lignification process with the ability to regulate damage repair.
1.3 The Research Progress in PAO-Related Genes
So far, the genes encoding PAO have been identified in many plants, and PAO genes have also been successfully cloned in many kinds of plants, with their functional verification performed. Studies have shown that PAO is encoded by one or more PAO genes, which have different localization and catalytic activities, and their gene functions are also different [13]. Wang et al. [14] found in studies that under salt stress, the germination speed and rate of the transgenic CsPAO4 tobacco were higher, compared with the control, but the vegetative growth and root system were inhibited, and the H2O2 accumulation increased, resulting in the PCD phenomenon. Chen et al. [15] found 11 PAO homologous genes (OsPAO1-OsPAO11) in the rice genome, which could be divided into 4 subfamilies. In the experiment, they obtained the following findings: The expression level of OsPAO5 was significantly up-regulated in germinating seeds; H2O2 produced by PAO was involved in the germination process of rice seeds; the expression of OsPAO5 genes was the main source of PAO activity in the germination process of rice seeds. Liu et al. [16] cloned OsPAO3 genes at the germination stage of rice seeds by the map-based cloning method. Studies showed as follows: The expression of OsPAO3 genes was up-regulated during germination under salt stress; overexpressed OsPAO3 genes increased PAO activity and polyamine content in the seed coleoptile; the increased polyamine content might enhance the activity of ROS scavenging enzymes in the seed coleoptile, which could scavenge the excessively accumulated H2O2, reduce Na+ content, maintain ionic homeostasis, weaken Na+ damage, and improve rice salt tolerance rice during germination.
So far, the identification of PAO gene family members has been studied in such plants as Arabidopsis thaliana, rice, barley, corn, tobacco, sweet orange and upland cotton. However, there have been few reports on PAO genes in soybeans. The root system was an important organ for soybean growth and development, and its development state could directly affect the growth of aboveground parts. In this study, GmPAO1 genes were screened from the transcriptome sequencing data obtained from the salt stress test on the developed material M18 of soybean root systems. Also, Molecular detection and salt tolerance identification were performed by transgenic GmPAO1 positive plants. The expression of GmPAO1 genes in different tissues was analyzed by salt stress induction to verify their salt tolerance in soybeans, thus verifying the functions of GmPAO1 genes and laying a foundation for cultivating new salt-tolerant soybean varieties.
2.1 Construction and Genetic Transformation of the Target Gene Expression Vector
Construction and genetic transformation of the target gene expression vector were performed by double digestion of pCAMBIA3301 plasmid with restriction enzymes Bgl II and BstE II, and the GUS gene was cut to linearize the vector. According to the upstream primer 5′-3′ GmPAO1-S (agaacacgggggactcttga CCATGGTAATGGCAGAGGCAGTAC) and the downstream primer GmPAO1-AS (cgatcggggaaattcgagct GGTCACCTCATAGTGAGTTTCTTAGCACTACTTG), the target gene was amplified by PCR. The GUS gene with a fragment length of 2024 bp was replaced with the target gene GmPAO1 with a fragment length of 582 bp to construct an overexpression vector pCAMBIA3301-GmPAO1 with the Bar gene as the selection marker. The CRISPR/Cas9 vector was synthesized by Baige Biotechnology. The target gene overexpression vector and CRISPR/Cas9 expression vector were transferred into soybean variety Jinong 74 by Agrobacterium-mediated method. The harvested grains were added to the T2 generation for subsequent experiments.
2.2 Bioinformatics Analysis of GmPAO1
According to the full-length sequence of the GmPAO1 gene, its open reading frame of GmPAO1 gene was found by using the online website, and the open reading frame sequence was translated into amino acid sequence by online software Expasy. According to the amino acid sequence, the online software SOPMA, SWISS-MODEL and ProtScale were used to predict and analyze the protein secondary structure, protein tertiary structure, and protein hydrophilicity and hydrophobicity encoded by GmPAO1 gene. At the same time, the homologous sequences of 18 plants were downloaded from the NCBI database, and the phylogenetic tree was constructed by using the software MEGA6.0.
2.3 Polyamine Oxidase Activity Assay
1 g fresh soybean leaves were cut into slices, and the extract (0.1 mol/L phosphate buffer, 0.15 mol/L sodium chloride, pH 6.5) was added for grinding, filtration and centrifugation. Add 2 ml phosphate buffer (PBS, 0.1 mol/L, pH 6.5), 0.2 mL guaiacol solution (25 mmol/mL), 0.1 mL POD solution (1 mg/mL), and cultivate them at 4°C Incubate for 2 min. Then 0.1 mL Spd with a concentration of 30 mmol/L was added respectively, and the obtained mixture was water-bathed at 30°C for 2 min, and the change in optical density at 475 nm was measured. The change in optical density value per minute was 0.01 as 1 unit of enzyme activity.
2.4 Determination of Polyamine Content
Fresh soybean leaves were ground into powder by adding liquid nitrogen, then 1.5% pectinase, 1% cellulase and 5 mL 5% perchloric acid solution were added overnight for extraction, and the extract was centrifuged at 6000 r/min for 30 min at low temperature; Then, 500 μL of the centrifuged supernatant was added to 15 μL of benzoyl chloride for acylation reaction, and 1 mL of NaOH was added to adjust the pH and fully vortexed for 20 s. After 30 min of water bath at 37°C, 2 mL of ether and 2 mL of saturated NaCl were added. Centrifugation at low temperature at 6000 r/min for 10 min and extraction of 1 mL diethyl ether phase with a syringe. The ether phase was blown dry with nitrogen in a nitrogen blower, and then 200 μL of chromatographic grade methanol was added to dissolve it, and a syringe was used to pass the 0.22 um organic phase filter membrane for UPLC detectio.
UPLC detection parameters: chromatographic column parameters were Agilent XDB-C18 reversed-phase column (4.6 mm× 250 mm); mobile phase was methanol: water (70:30 V/V); flow rate was 1 mL/min; injection volume was 10 μL; detection temperature was 30°C; detection Wavelength was 230 nm; retention time was 20 min.
2.5 PCR Detection of T2 Generation Transgenic GmPAO Soybean Positive Plants
The plant materials M18 and Jinong 74 were provided by the Biotechnology Center of Jilin Agricultural University. Plant expression vectors and transgenic positive plants were obtained from the prophase experiments. To verify the differences in the expression patterns of GmPAO1 among different strains, the RNAs of OEA1~OEA4 and KO1~KO3 were extracted respectively, with three repetitions performed for each line, and the cDNA was obtained by reverse transcription and subjected to the fluorescent quantitative PCR to identify the lines with high expression of target genes for subsequent experiments, according to the upstream primer 5′-3′ qGmPAO1-S(ATCTACCTTTACAGCGTTTGGCG) and the downstream primer qGmPAO1-AS (CTTCCCCGATGCTCTATTTCC). The genomes of the above screened positive transformed lines were extracted for routine PCR detection, the DNA of overexpressed positive lines taken as the template, and the upstream primer 5′-3′ Bar-S(TCAAATCTCGGTGACGGGC) and the downstream primer Bar-AS(ATGAGCCCAGAACGACGCC) were used to amplify selective marker bar genes, with the annealing temperature (58°C), 30 s and 35 cycles. According to the specific primers 5′-3′ CRISPR/Cas9-GmPAO1-S: TCCCAGTCACGACGTTGTAA, 5′-3′ CRISPR/Cas9-GmPAO1-AS: GCCATTTGTCTGCAGAATTG, PCR was performed on the genome of the transformed plant with the gene editing vector, and the product was sent for sequencing. The results determine whether there is a base mutation in the target.
2.6 The Salt Stress Treatment for Germination of Transgenic Positive Soybean Seeds
The control materials Jinong 74 (WT), the overexpressed lines OEA1 and OEA2, and the gene-edited lines KO1 and KO2 were selected. 20 grains of each line were soaked in 75% alcohol and 5% NaCl for 2 min, then washed twice with distilled water and placed in a 9 cm Petrie dish. Subsequently, two layers of sterile filter paper were placed above and below, respectively, and the experiment was repeated 3 times. Next, four different concentrations of NaCl salt solution of 0, 50, 100, and 200 mmol/L were prepared, followed by the addition of 20 mL stress solution for stress treatment, respectively, and the control group was treated with the same amount of distilled water. After that, the Petrie dish was in the artificial climate incubator, and the germination experiment was carried out at a constant temperature of 25°C in the dark. After 6 days, the germination experiment was completed to determine the seed germination rate. The data obtained from the experiment were made statistics by Excel, and the significantly differential analysis was performed by GraphPad software. The Dunnet control method was used for single factor tests to compare and analyze the significance of the difference between the control material Jinong 74 and transgenic lines (*P < 0.05, **P < 0.01).
2.7 The Determination of Root System Phenotype of Transgenic GmPAO1 Soybean Plants under Salt Stress
DJ-GXG02 root system imaging analysis software was used to analyze the root system of soybeans from these aspects: total root length, mean connection projection area, mean connection surface area, root volume, mean root diameter, total root tip number, total furcation number and total crossing number. The obtained experimental data were analyzed by SPSS and GraphPad software.
2.8 The Determination of Enzyme Activities and Physiological Indexes of Transgenic GmPAO1 Soybean Plants under Salt Stress
The control materials Jinong 74, the overexpressed lines OEA1 and OEA2, and the gene-edited lines KO1 and KO2 were selected. And they were separately planted in the flowerpots in which vermiculite was mixed with soil (1:1 ratio of vermiculite to soil), with 15 pots for each line and three plants for each pot. They were all planted in the tissue culture room to ensure sufficient water and light. After the first complete trifoliate compound leaf of soybeans was unfolded, the experimental materials were subjected to stress treatment with two different concentrations of NaCl salt solution of 100 and 200 mmol/L in turn. 10 pots of soybeans with the same concentration were placed in a plastic pot of 30 L. And they were irrigated with 1 L NaCl solution of the corresponding concentration for salt stress treatment. The control group was irrigated with the same amount of distilled water for treatment. NaCl stress was carried out every 3 days and the experiment ended after day 9. Then, the relevant indexes of soybean leaves were measured, including the relative water content, chlorophyll content, malondialdehyde (MDA) content, POD activity (POD), superoxide dismutase (SOD) activity and hydrogen peroxide (H2O2) content. The experimental data were analyzed by Spss statistics. Also, the physiological and biochemical indexes at Soybean Seedling stages under the stress of different concentrations of NaCl solution were analyzed by one-way ANOVA.
3.1 Bioinformatics Analysis of GmPAO1
According to the prediction results of the protein hydrophobicity and hydrophobicity of the online software ProtScale, the highest score in the 85th place was 2.567, the most hydrophobic; At the 165th position, the lowest score was −3.356, and the hydrophilicity was the strongest. On the whole, the GmPAO1 amino acid sequence showed far more hydrophilic amino acids than the hydrophobic amino acid sequence. The protein encoded by the GmPAO1 gene contained more hydrophilic regions and had a certain hydrophilic ability (Fig. 1A).
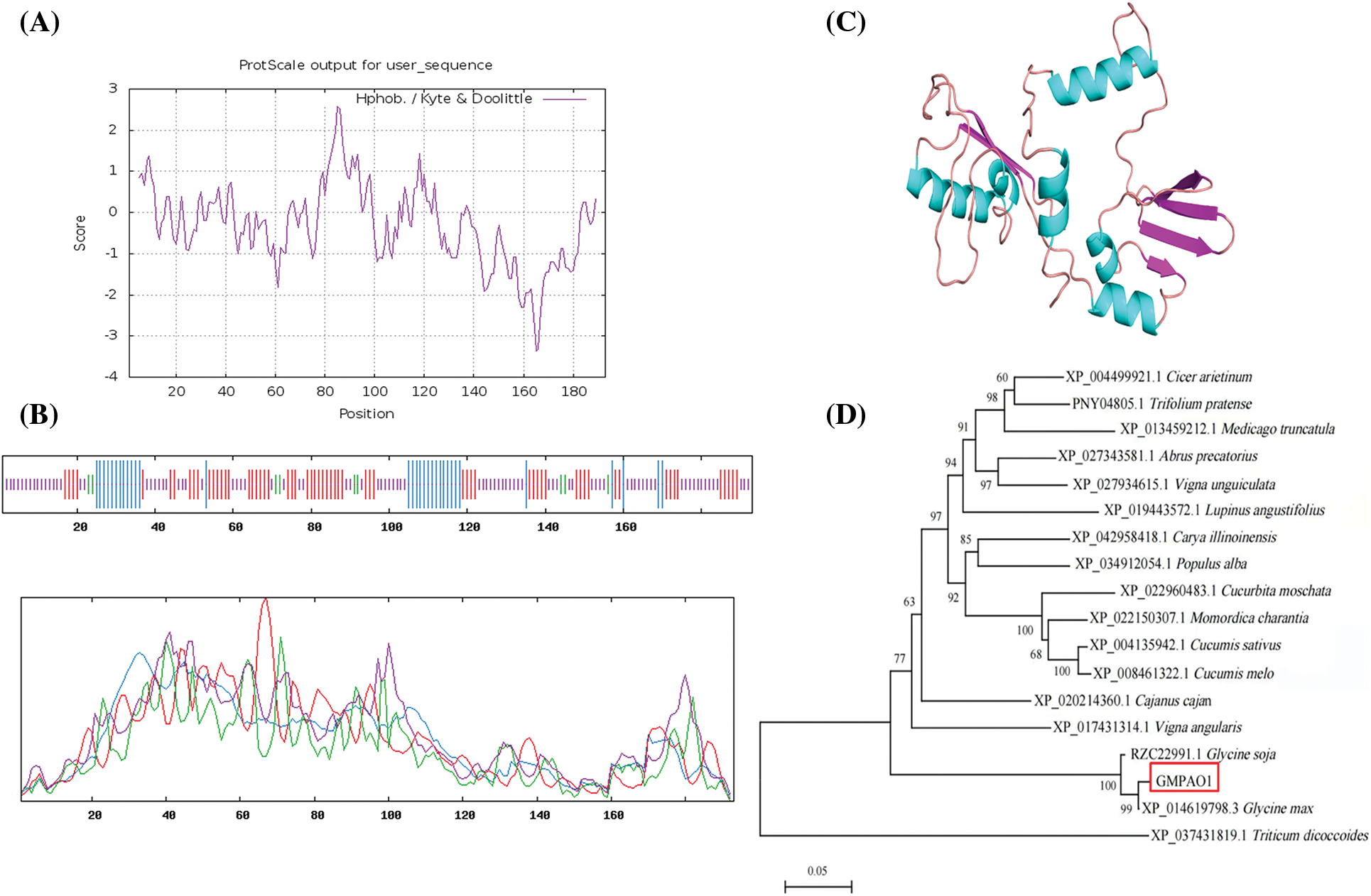
Figure 1: GnPAO1 bioinformatics analysis. (A) Hydrophilic and hydrophobic prediction of GmPAO1. (B) Secondary structure analysis of proteins. (C) 3D modeling of the tertiary structure of proteins. (D) The phylogenetic tree of homologous sequences of GmPAO1
The secondary structure of the protein was predicted by using the SOPMA online software (Fig. 1B). The secondary structure of the protein encoded by the entire GmPAO1 gene was mainly divided into four parts: α helix, β sheet, extended chain and random coil, of which random coil was 46.11%. Followed by the extension chain accounted for 32.12%, α helix accounted for 17.1%, and β angle accounted for 4.66%. Secondly, by using SWISS-MODEL software, 3D homology modeling was performed on the amino acid sequence of GmPAO1 gene, and the protein tertiary structure prediction results (Fig. 1C) were shown.
Homologous sequences were searched by Blast in the NCBI website, and finally 17 plant homologous sequences were downloaded. The phylogenetic tree analysis of GmPAO1 homologous proteins in different species was performed using MEGA 6.0 software (Fig. 1D). GmPAO1 was the most closely related to soybean and wild-type soybean.
3.2 Gene Expression Pattern Analysis of GmPAO1 in Wild-Type Soybean
In order to explore the response of GmPAO1 gene to abiotic stress, the changes of GmPAO1 gene expression in soybean leaves under abiotic stress of 10% 100 mmol/L NaCl solution were detected, and fluorescence quantitative PCR was performed with the expression of GmPAO1 gene under unstressed conditions as the control detection (Fig. 2A). Under the stress of 100 mmol/L NaCl solution, the relative expression of GmPAO1 in soybean leaves increased first and then decreased. The relative expression reached the highest at 8 h of stress, and then began to decline, indicating that GmPAO1 could respond to salt stress and up-regulated expression.
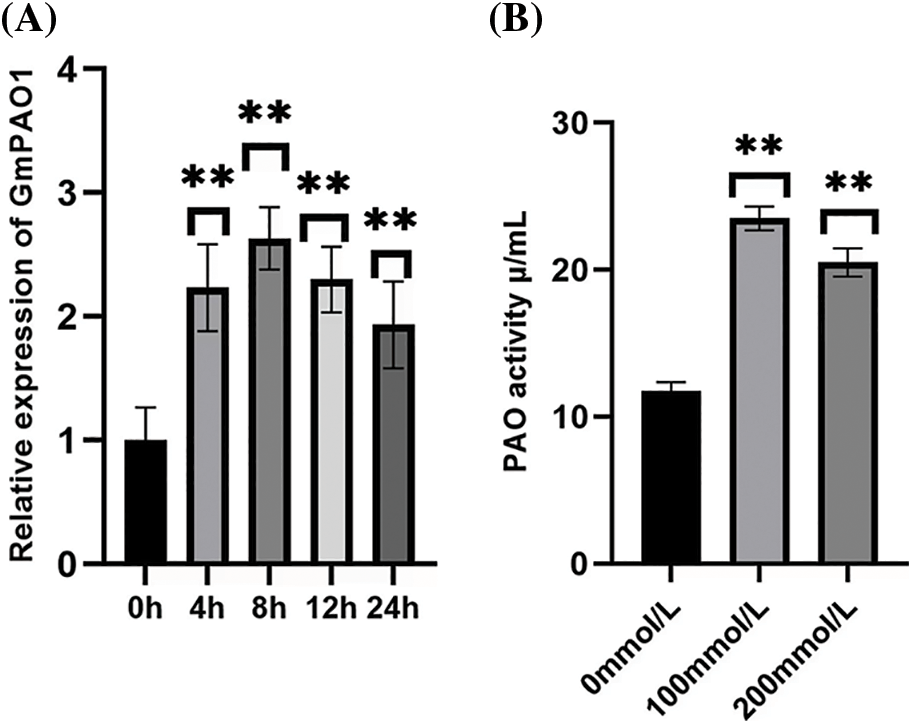
Figure 2: Gene expression pattern analysis and enzyme activity detection of GmPAO1 in wild-type soybean. (A) Gene expression patterns of GmPAO1 in wild-type soybean. (B) Polyamine oxidase activity in plants under different concentrations of salt solution stress (*P < 0.05, **P < 0.01)
3.3 Enzyme Activity Detection of Polyamine Oxidase
Wild-type plants were treated with 0, 100, and 200 mmol/L NaCl salt solutions for 8 h, and the activity of polyamine oxidase was measured in soybean plants (Fig. 2B). The results showed that the activity of polyamine oxidase was the highest under the stress of 100 mmol/L salt solution, and the activity decreased slightly after the stress of 200 mmol/L salt solution. This indicated that polyamine oxidase had a certain activity under salt stress. With the increase of stress concentration, the activity of polyamine oxidase showed a trend of increasing first and then decreasing.
3.4 The Expression Pattern Analysis of GmPAO1 Genes in Overexpressed Positive Soybean Transformed Plants
To screen out the transgenic lines with relatively high expression of the target gene GmPAO1, the fluorescent quantitative PCR detection of the transgenic plants was performed (Fig. 3). The results showed that the relative expression levels of OEA1 and OEA2 were the highest among the four overexpressed lines OEA1~OEA4 and the relative expression levels of the target genes KO1 and KO2 were the lowest among the gene-edited lines KO1~KO3.
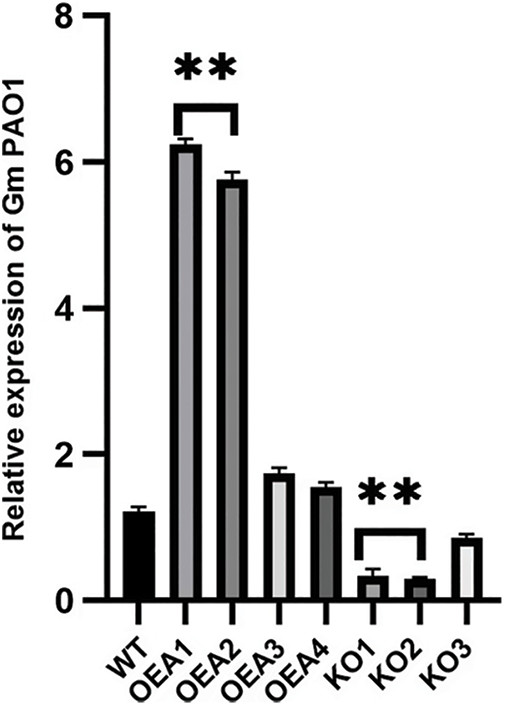
Figure 3: Relative expression level of target gene in transformed plants (*P < 0.05, **P < 0.01)
3.5 The Molecular Detection of T2 Generation Positively Transformed Plants at the Genome Level
The genomic DNA was extracted from the leaves of T2 generation overexpressed, gene-edited and recipient plants, and PCR was used to detect the selective marker gene bar of overexpressed plants, with a fragment length of 500 bp (Fig. 4A). The comparison results of the detection targets of gene-edited plants were shown in Fig. 4B: One base in target 1 of the KO1 line was missing; one base in target 2 of the KO2 line was replaced; one base in both targets 1 and 2 of the KO3 line was replaced.
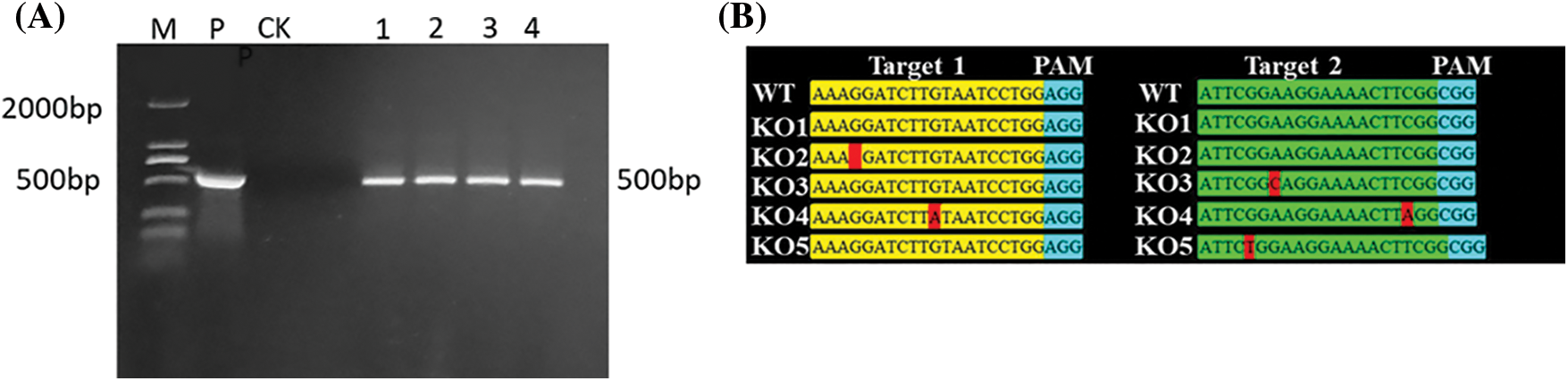
Figure 4: Molecular detection of T2 generation positive transformed plants. (A) PCR detection of overexpression positive plants (B) Target detection results of T2 generation gene editing plants
3.6 The Determination of the Germination Rate of Transgenic GmPAO1 Soybean Seeds under NaCl Stress
There was no significant difference in the soybean germination rate among various lines under no stress or 50 mmol/L salt stress. When the NaCl concentration reached 100 and 200 mmol/L, the germination rates of the overexpressed lines OEA1 and OEA2 were both significantly higher than those of the control group, and the germination rates of the gene-edited lines KO1 and KO2 were both significantly lower than those of the control group (Figs. 5A, 5C). This indicated that overexpressed GmPAO1 genes could significantly improve the germination rate of soybean seeds under salt stress.
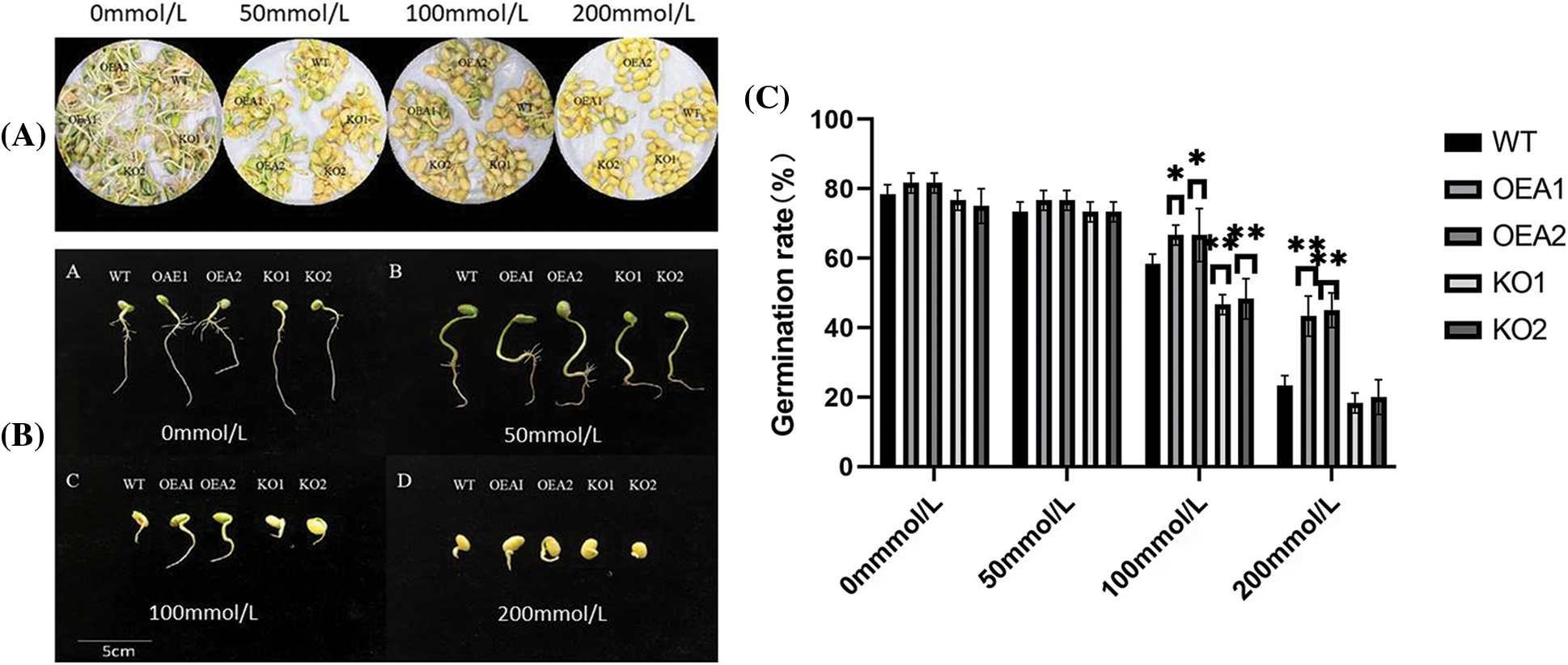
Figure 5: Seed germination (A) and (B) germination phenotypes of seeds under different concentrations of salt stress. (C) Seed germination rate under different concentrations of salt stress (*P < 0.05, **P < 0.01)
3.7 The Phenotypic Identification of Transgenic GmPAO1 Soybean Seeds at the Germination Stage
After 6 days of germination, there was no significant difference in the root length between WT, OEA1, OEA2, KO1 and KO2 seeds under no stress or the stress of 50 mmol/L NaCl solution (Fig. 5B). However, the number of lateral roots in the overexpressed transgenic lines OEA1 and OEA2 was larger than that in the control lines; the number of lateral roots in gene-edited lines was significantly smaller than that in the control lines; the growth of lateral roots in the gene-edited lines KO1 and KO2 was inhibited. Under the stress of 100 or 200 mmol/L NaCl solution, the bud length of the overexpressed transgenic lines OEA1 and OEA2 was significantly higher than that of the control lines. The bud length of the gene-edited lines KO1 and KO2 was significantly lower than that of the control. With the aggravation of NaCl solution stress, the lateral root growth and bud length of seeds in each line were significantly inhibited.
3.8 Phenotypic Differences in Root Systems of Soybean Positively Transformed Plants under Different Concentrations of Salt Stress
Under no stress, the total root length, mean connection projection area, root volume, mean root diameter, total root tip number, total furcation number and total crossing number of the overexpressed transgenic lines were all extremely significantly higher, compared with the control lines; there was no significant difference in the mean connection surface area; all phenotypic indexes of the root systems in the gene-edited lines were extremely significantly lower than those of the control lines. After the stress of NaCl solution, the phenotypic indexes of the root system in the overexpressed transgenic lines were all extremely significantly higher than those of the control lines (Fig. 6).
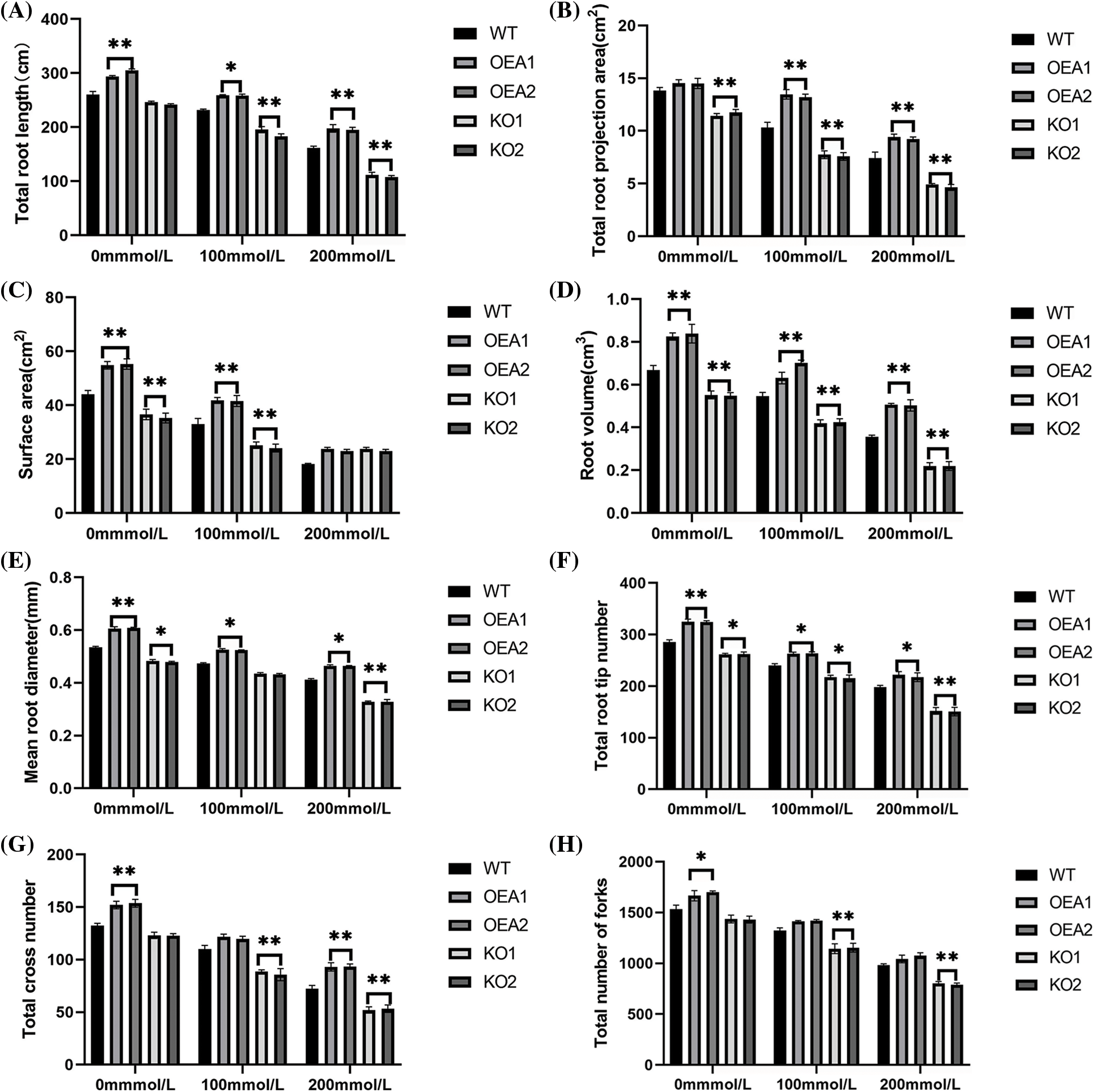
Figure 6: Root phenotype (A) total root length (B) total root projection area (C) surface area (D) root volume (E) mean root diameter (F) total root tip number (G) total cross number (H) total number of forks (*P < 0.05,**P < 0.01)
3.9 The Changes in Relevant Physiological and Biochemical Indexes and Enzyme Activities of Positively Transformed Soybean Plants in Response to Salt Stress of Different Concentrations
Under no stress, the leaves of soybean plants in each strain were thick green and plump. With the aggravation of salt stress, the leaves dried up and turned yellow. The degree of damage to the leaves of the overexpressed lines was significantly smaller than that of the control lines. By measuring the relative water content of leaves, it was found that there was no significant difference in the relative water content of leaves of each strain under no stress, which was all about 86%–89%. After NaCl stress, the relative water content of leaves in each strain all decreased significantly. Under the stress of 100 or 200 mmol/L NaCl, the relative water content of the overexpressed lines OEA1 and OEA2 was extremely significantly higher than that of the control lines, and the relative water content of leaves in the gene-edited lines KO1 and KO2 was extremely significantly lower than that in the control lines (Figs. 7A, 7B).
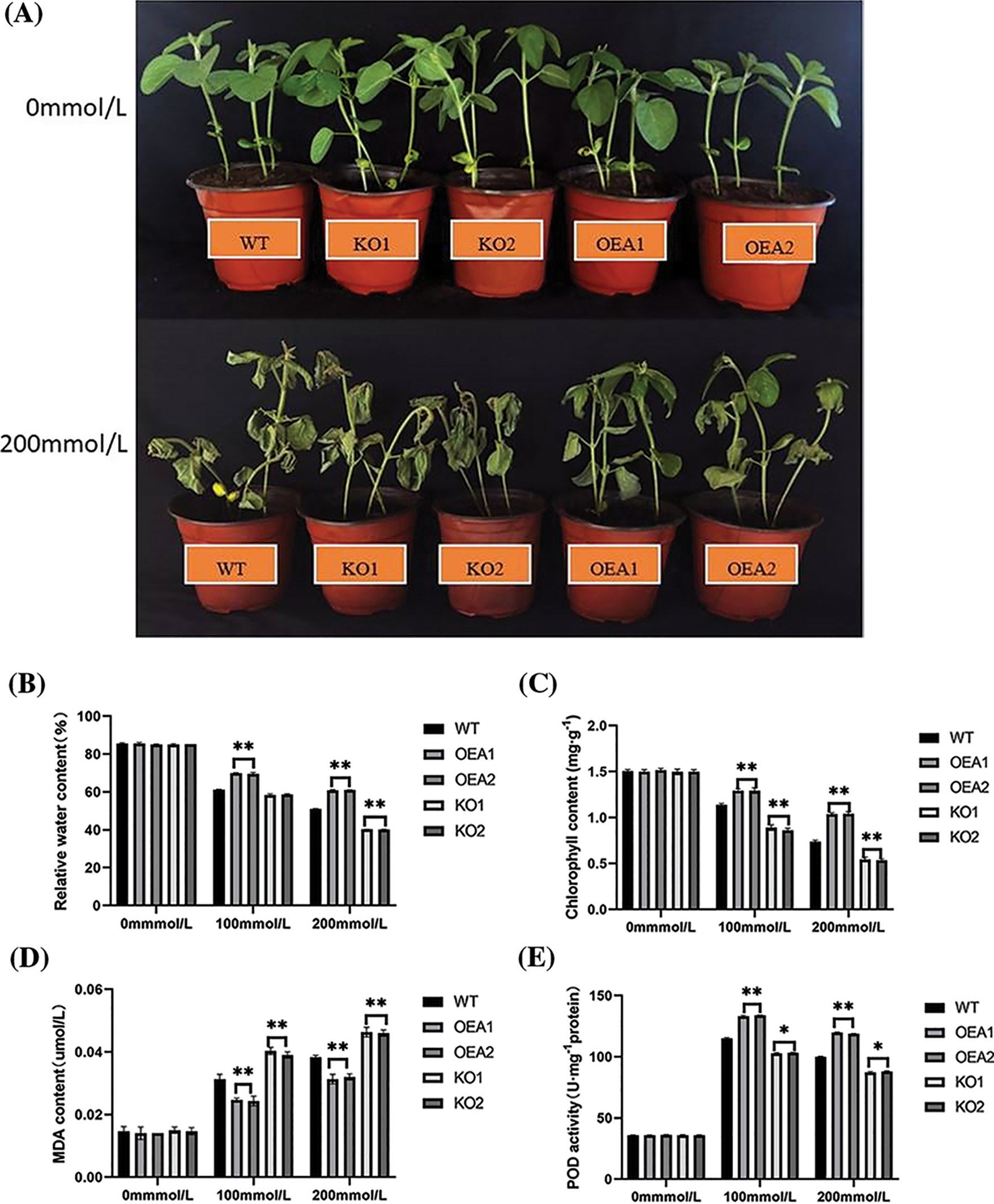
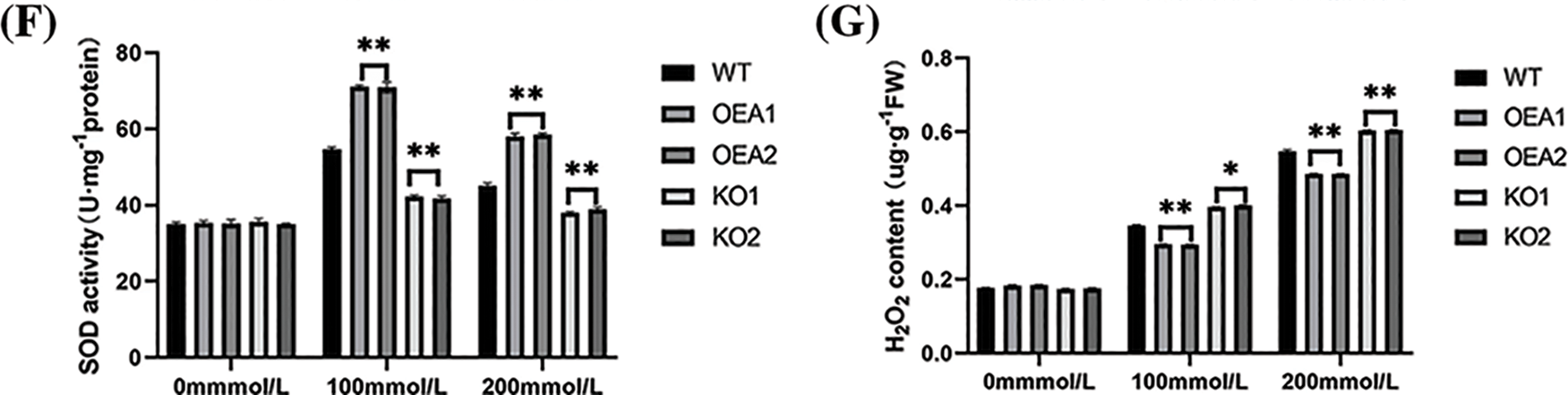
Figure 7: Plant phenotype and related physiological and biochemical indicators and enzyme activities under salt stress. (A) Plant phenotype under different concentrations of salt stress (B) relative water content under 0 mmol/L NaCl, 100 mmol/L NaCl, and 200 mmol/L NaCl stress (C) chlorophyll content under 0 mmol/L NaCl, 100 mmol/L NaCl, and 200 mmol/L NaCl stress (D) MDA content under 0 mmol/L NaCl, 100 mmol/L NaCl, and 200 mmol/L NaCl stress (E) POD activity under 0 mmol/L NaCl, 100 mmol/L NaCl, and 200 mmol/L NaCl stress (F) SOD activity under 0 mmol/L NaCl, 100 mmol/L NaCl, and 200 mmol/L NaCl stress (G) H2O2 content under 0 mmol/L NaCl, 100 mmol/L NaCl, and 200 mmol/L NaCl stress (*P < 0.05, **P < 0.01)
Chlorophyll was the primary pigment for soybean photosynthesis. The degree of soybean salt stress could be judged by measuring the change in chlorophyll content. The experimental results showed no significant difference in the chlorophyll content of soybean plants in each strain under no stress, which was all about 1.6 mg/g. Under the stress of 100 mmol/L NaCl solution, the chlorophyll content of soybean plants in each strain all decreased, and that of the overexpressed lines OEA1 and OEA2 increased by 13.68% and 13.68%, respectively, compared with the controls. The chlorophyll content of the gene-edited lines KO1 and KO2 decreased by 11.97% and 12.82%, respectively, compared with the controls. When the concentration of NaCl solution reached 200 mmol/L, the chlorophyll content of soybean plants in each strain was the lowest (Fig. 7C).
Under no stress, there was no significant difference in malondialdehyde (MDA) content among all strains, which were all at a low level. Under the stress of 100 mmol/L NaCl solution, the MDA content of overexpressed lines was extremely significantly lower than that of the controls. The MDA content of gene-edited lines was extremely significantly higher than that of the controls. Under the stress of 200 mmol/L NaCl concentration, the MDA content of all strains reached the highest level. The MDA content of the overexpressed lines OEA1 and OEA2 decreased by 18.42% and 15.79%, respectively, compared with the control, and the MDA content of the gene-edited lines KO1 and KO2 increased by 21.05% and 21.05%, respectively. With NaCl concentration increasing, the MDA content of all lines showed an upward trend, but the increase of MDA in the overexpressed lines was less than that in the control group (Fig. 7D).
POD and SOD were important protective enzymes for plant cells to resist ROS damage, thereby reducing the damage to membrane structure caused by adversity stress and improving the self-protection and regulation ability of soybeans under adversity. Studies showed that POD activity was positively correlated with plant salt tolerance. There was no significant difference in SOD and POD activities among strains under no stress. With NaCl concentration increasing, SOD and POD activities in all strains showed a trend of increasing first and then decreasing. Under the stress of 100 or 200 mmol/L NaCl solution, the SOD and POD activities of the overexpressed lines were extremely significantly higher than those of the control lines, and the SOD and POD activities of the gene-edited lines KO1 and KO2 were extremely significantly lower than those of control lines (Figs. 7E, 7F).
Under no stress, the hydrogen peroxide content of overexpressed lines was slightly higher than that of control plants, but the difference was insignificant. Under the stress of 100 or 200 mmol NaCl solution, the hydrogen peroxide content of overexpressed lines was extremely significantly lower than that of control lines, and the hydrogen peroxide content of gene-edited lines was extremely significantly higher than that of control lines (Fig. 7G).
3.10 Changes of Polyamines Content in Wild-Type and Transgenic Lines
The polyamines in plants mainly include Put, spermidine Spd, and spermine Spm. We measured the polyamine content in wild-type and transgenic plants. Under normal water growth conditions, the content of Spd was slightly higher than that of the other two polyamines. The polyamine content of transgenic overexpression lines OEA1~OEA2 was slightly higher than that of wild-type plants. The polyamine content of the gene editing lines KO1~KO2 was slightly lower than that of the wild type (Fig. 8A); Under the stress of 100 mmol/L NaCl solution, the content of Spd was slightly lower than that of Put and Spm, and the polyamine content in the overexpression lines OEA1~OEA2 was significantly higher than that in the wild type plants (Fig. 8B); Under the stress of 200 mmol/L NaCl solution, the content of Spd remained slightly lower than that of the other two polyamines, and the polyamine content in the overexpression lines OEA1~OEA2 was also significantly higher than that in the wild-type plants (Fig. 8C). Overall, with the increase of stress concentration, the content of three polyamines showed an upward trend, but when the concentration reached 200 mmol/L, the accumulation of polyamines in plants was limited to a certain extent.
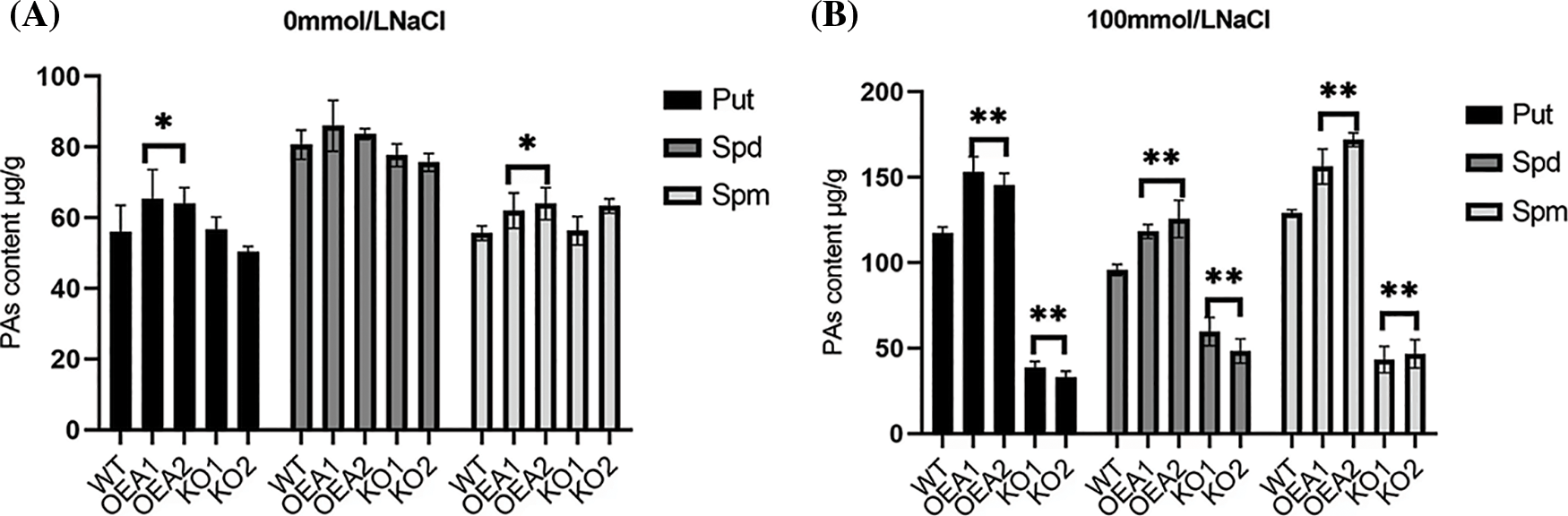
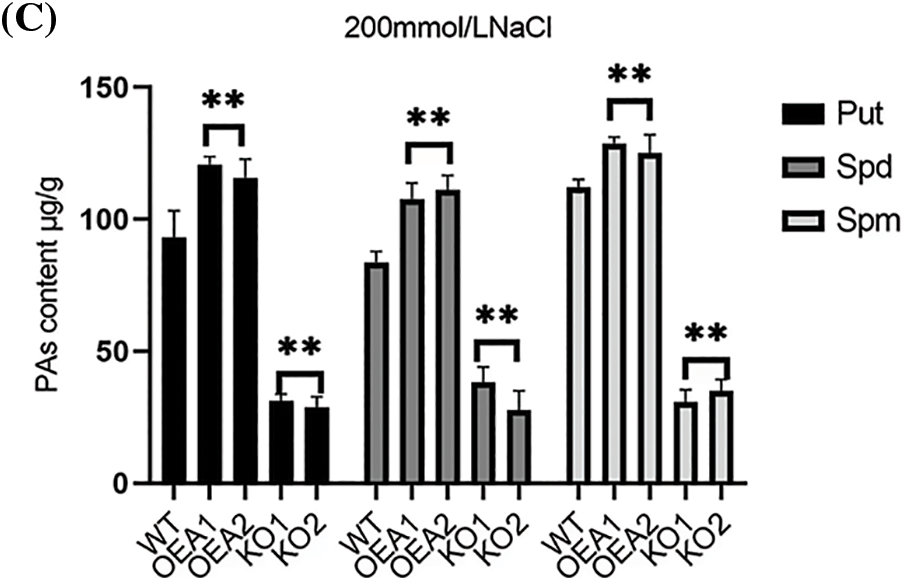
Figure 8: Polyamines content of wild type and transgenic lines. (A) Polyamines content in wild-type WT and transgenic overexpression lines OEA1~OEA2 and gene editing lines KO1~KO2 under the stress of 0 mmol/L NaCl solution. (B) Polyamines content in wild-type WT and transgenic overexpression lines OEA1~OEA2 and gene editing lines KO1~KO2 under the stress of 100 mmol/L NaCl solution. (C) Polyamines content in wild-type WT and transgenic overexpression lines OEA1~OEA2 and gene editing lines KO1~KO2 under the stress of 200 mmol/L NaCl solution
Soil salinization seriously restricts the yield and quality of crops in China, and a high salt environment could destroy the ionic homeostasis and osmotic balance inside and outside plant cells, thus inhibiting plant growth and development [17]. The salt-sensitive period of soybean was mainly in the seed germination and seedling stages. The indexes such as the seed germination rate, germination vigor, germination index and seed vigor index could all reflect the germination speed. Therefore, the salt tolerance analysis of different transgenic soybeans at the germination stage was performed. The experimental results showed that overexpressed GmPAO1 genes improved the salt tolerance of soybean seeds. The research results in this experiment were similar to those obtained by Liu et al. [18]. Under normal conditions, the germination rate of overexpressed CsPAO3 lines was higher than that of the wild type on days 2–4 after sowing, and it remained high under NaCl stress. It was concluded that overexpressed CsPAO3 can induce seeds to germinate earlier and improve the germination rate under salt stress.
The salt tolerance at the germination stage could not represent that at the seedling stage and other growth cycles. The salt tolerance at the soybean seedling stage continued to be analyzed. The growth and development of soybean root systems could directly affect the growth of aboveground parts; the root system was the organ that could feel the adverse environment earliest; soybean plants with developed and strong root systems and many lateral roots had strong salt tolerance and could resist the damage from the adverse environment [19]. Therefore, the relevant phenotypic indexes related to soybean root systems were measured. The results showed that the relevant phenotypic indexes such as the developed root system, total root length and total root tip number of the plants with overexpressed GmPAO1 genes were all significantly higher, compared with the control plants. It was speculated that overexpressed GmPAO1 genes promote the growth and development of soybean root systems.
A high-salt environment could cause the disorder of ROS metabolism in the soybean body, the intensified membrane lipid oxidation, and the increased cytoplasmic membrane permeability, and thus, the relative water content, chlorophyll content, MDA content, POD and SOD activities, and hydrogen peroxide content of soybean leaves at the seedling stage could all reflect the stress resistance level to a certain extent [20]. After salt stress, the chlorophyll content of soybean leaves was significantly reduced, and photosynthesis can be hindered, thereby inhibiting soybean growth and development and reducing the yield [21]. In this experiment, it was found that chlorophyll and leaf relative water contents of overexpressed GmPAO1 gene lines were both higher than that of the controls after salt stress, indicating that overexpressed transgenic lines had a better growth status and strong photosynthetic capacity. SOD was a key enzyme in the antioxidant enzyme system. Xie et al. [22] found that the SOD activity of transgenic GmGT-2B soybeans was significantly higher than that of the controls, and that under salt stress, the cell membrane of transgenic soybeans was slightly damaged by oxidation, which improved the self-protection and regulation ability of soybeans under salt stress. POD could reduce the formation of hydroxyl radical, which played a critical role in protecting the membrane system from damage. In this experiment, it was found that the SOD and POD activities of overexpressed transgenic lines were significantly higher than those of the control lines after salt stress, and that their H2O2 content was significantly lower than that of the controls, indicating that overexpressed GmPAO1 lines could improve the salt tolerance of soybeans, which helped alleviate salt stress-induced growth inhibition. Due to the dual role of H2O2 in plant stress responses, it was speculated that the H2O2 content produced by the metabolism of GmPAO1 genes was not enough to damage them, and instead, but can act as a signal molecule to promote the growth and development of soybean roots.
Polyamines had protective effects on plants at high salt concentrations, improving K/Na balance by limiting Na influx to roots and preventing K loss from shoots [23]. It was reported that polyamines effectively prevented NaCl-induced K efflux by blocking nonselective cation channels and activating plasma membrane H-ATPase, thereby restoring the membrane potential. Polyamines could also scavenge free radicals and effectively prevent damage caused by ROS under salt stress [24]. In our study, the polyamine content in the leaves of transgenic plants and wild-type plants was measured, and it was found that the content of spermidine (Spd) was relatively low under the stress of 100 mmol/L NaCl solution. This was consistent with previous studies that AtPAO1 and AtPAO4 could catalyze the reverse conversion of spermidine to spermine and the conversion of spermidine to Put in Arabidopsis thaliana [25]. In addition, after the overexpression of GmPAO1, the theoretical polyamines were not degraded and decreased, but the contents of the three polyamines increased to varying degrees under the condition of increasing salt stress. Therefore, we speculated that overexpression of GmPAO1 could degrade higher-order polyamines and convert them to lower-order polyamines, rather than directly oxidizing lower-order polyamines in plants to participate in stress resistance.
Under salt stress, overexpression of GmPAO1 gene can increase the seed germination rate, increase polyamine content in transgenic plants, promote root development, and reduce the damage of ROS to plant cells, thereby improving the salt tolerance of soybean. This study lays a theoretical foundation for further understanding the biological function and molecular mechanism of GmPAO1 when soybean is under abiotic stress, and also provides a reference for breeding new salt-tolerant soybean varieties.
Authorship: The authors confirm their contribution to the paper as follows: draft manuscript preparation: Yeyao Du; literature review: Ye Zhang, Hanzhu Zhang; data collection: Sujie Fan; genetic transformationsults: Yang Song, Zhuo Zhang, Kun Wang, Boran Yuan; overall conception: Piwu Wang. All authors reviewed the results and approved the final version of the manuscript.
Acknowledgement: We would like to thank Jilin Agricultural University, Plant Biotechnology Center for facilitating the working environment to conduct the experiment.
Funding Statement: This work was supported by Jilin Province Science and Technology Development Plan Project, Grant No. 20190103120JH. Jilin Province Science and Technology Development Plan-Outstanding Young Talents Fund Project, Grant No. 20190103120J. The fourth batch of Jilin Province Youth Science and Technology Talent Support Project, Grant No. QT202020 and National Natural Science Foundation of China Projects, Grant No. 31801381.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Zhang, H., Yang, C., Guo, H., Hou, G. (2022). Characteristics of land use conversion in soda saline-alkali soil region of central Northeast China. Global Geology, 25(2), 116–125. [Google Scholar]
2. Shelke, D. B., Pandey, M., Nikalje, G. C., Zaware, B. N., Suprasanna, P. et al. (2017). Salt responsive physiological, photosynthetic and biochemical attributes at early seedling stage for screening soybean genotypes. Plant Physiology and Biochemistry, 118(4), 519–528. DOI 10.1016/j.plaphy.2017.07.013. [Google Scholar] [CrossRef]
3. Phang, T. H., Shao, G., Lam, H. M. (2008). Salt tolerance in soybean. Journal of Integrative Plant Biology, 50(10), 1196–1212. DOI 10.1111/j.1744-7909.2008.00760.x. [Google Scholar] [CrossRef]
4. Dinler, B. S., Gunduzer, E., Tekinay, T. (2016). Pre-treatment of fulvic acid plays a stimulant role in protection of soybean (Glycine max L.) leaves against heat and salt stress. Acta Biologica Cracoviensia Series Botanica, 58(1), 29–41. DOI 10.1515/abcsb-2016-0002. [Google Scholar] [CrossRef]
5. Kan, G., Zhang, W., Yang, W., Ma, D., Zhang, D. et al. (2015). Association mapping of soybean seed germination under salt stress. Molecular Genetics and Genomics, 290(6), 2147–2162. DOI 10.1007/s00438-015-1066-y. [Google Scholar] [CrossRef]
6. Hosseini, M. K., Powell, A. A., Bingham, I. J. (2002). Comparison of the seed germination and early seedling growth of soybean in saline conditions. Seed Science Research, 12(3), 165–172. [Google Scholar]
7. Shi, H., Chan, Z. (2014). Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. Journal of Integrative Plant Biology, 56(2), 114–121. DOI 10.1111/jipb.12128. [Google Scholar] [CrossRef]
8. Masson, P. H., Takahashi, T., Angelini, R. (2017). Editorial: Molecular mechanisms underlying polyamine functions in plants. Frontiers in Plant Science, 8, 14. DOI 10.3389/fpls.2017.00014. [Google Scholar] [CrossRef]
9. Yu, Z., Jia, D., Liu, T. (2019). Polyamine oxidases play various roles in plant development and abiotic stress tolerance. Plants, 8(6), 184. DOI 10.3390/plants8060184. [Google Scholar] [CrossRef]
10. Amar, T., Nourredine, Y., Fatima-Zohra, F. L. (2016). Morphometric variability and biochemical analysis of growth seedlings under salt stress in tomato (Lycopersicon esculentum Mill.) cultivars. Plant Breeding, 7(4), 1–9. [Google Scholar]
11. Mellidou, I., Moschou, P. N., Ioannidis, N. E., Pankou, C., Gėmes, K. et al. (2016). Silencing s-adenosyl-l-methionine decarboxylase (SAMDC) in Nicotiana tabacum points at a polyamine-dependent trade-off between growth and tolerance responses. Frontiers in Plant Science, 7(132), 379. DOI 10.3389/fpls.2016.00379. [Google Scholar] [CrossRef]
12. Scalet, M., Federico, R., Angelini, R. (1991). Time courses of diamine oxidase and peroxidase activities, and polyamine changes after mechanical injury of chick-pea seedlings. Journal of Plant Physiology, 137(5), 571–575. [Google Scholar]
13. Sagor, G., Inoue, M., Kusano, T., Berberich, T. (2021). Expression profile of seven polyamine oxidase genes in rice (Oryza sativa) in response to abiotic stresses, phytohormones and polyamines. Physiology and Molecular Biology of Plants, 27(6), 1353–1359. DOI 10.1007/s12298-021-01006-1. [Google Scholar] [CrossRef]
14. Wang, W., Liu, J. H. (2016). CsPAO4 of Citrus sinensis functions in polyamine terminal catabolism and inhibits plant growth under salt stress. Scientific Reports, 6(1), 31384. DOI 10.1038/srep31384. [Google Scholar] [CrossRef]
15. Chen, B. X., Li, W. Y., Gao, Y. T., Chen, Z. J., Zhang, W. N. et al. (2016). Involvement of polyamine oxidase-produced hydrogen peroxide during coleorhiza-limited germination of rice seeds. Frontiers in Plant Science, 7, 1219. DOI 10.3389/fpls.2016.01219. [Google Scholar] [CrossRef]
16. Liu, G., Jiang, W., Tian, L., Fu, Y., Tan, L. et al. (2022). Polyamine oxidase 3 is involved in salt tolerance at the germination stage in rice. Journal of Genetics and Genomics, 49(5), 458–468. DOI 10.1016/j.jgg.2022.01.007. [Google Scholar] [CrossRef]
17. Zhao, W., Zhou, Q., Tian, Z., Cui, Y., Liang, Y. et al. (2020). Apply biochar to ameliorate soda saline-alkali land, improve soil function and increase corn nutrient availability in the Songnen plain. The Science of the Total Environment, 722, 137428. DOI 10.1016/j.scitotenv.2020.137428. [Google Scholar] [CrossRef]
18. Liu, T., Kim, D. W., Niitsu, M., Berberich, T., Kusano, T. (2014). Oryza sativa polyamine oxidase 1 back-converts tetraamines, spermine and thermospermine, to spermidine. Plant Cell Reports, 33(1), 143–151. DOI 10.1007/s00299-013-1518-y. [Google Scholar] [CrossRef]
19. Xiong, R., Liu, S., Considine, M. J., Siddique, K., Lam, H. M. et al. (2021). Root system architecture, physiological and transcriptional traits of soybean (Glycine max L.) in response to water deficit: A review. Physiologia Plantarum, 172(2), 405–418. DOI 10.1111/ppl.13201. [Google Scholar] [CrossRef]
20. Zhang, M., He, S., Zhan, Y., Qin, B., Jin, X. et al. (2019). Exogenous melatonin reduces the inhibitory effect of osmotic stress on photosynthesis in soybean. PLoS One, 14(12), e0226542. DOI 10.1371/journal.pone.0226542. [Google Scholar] [CrossRef]
21. Li, M., Xu, J., Wang, X., Fu, H., Zhao, M. et al. (2018). Photosynthetic characteristics and metabolic analyses of two soybean genotypes revealed adaptive strategies to low-nitrogen stress. Journal of Plant Physiology, 229, 132–141. DOI 10.1016/j.jplph.2018.07.009. [Google Scholar] [CrossRef]
22. Xie, Z. M., Zou, H. F., Lei, G., Wei, W., Zhou, Q. Y. et al. (2009). Soybean Trihelix transcription factors GmGT-2A and GmGT-2B improve plant tolerance to abiotic stresses in transgenic Arabidopsis. PLoS One, 4(9), e6898. DOI 10.1371/journal.pone.0006898. [Google Scholar] [CrossRef]
23. Yamaguchi, K., Takahashi, Y., Berberich, T., Imai, A., Miyazaki, A. et al. (2006). The polyamine spermine protects against high salt stress in Arabidopsis thaliana. FEBS Letters, 580(30), 6783–6788. DOI 10.1016/j.febslet.2006.10.078. [Google Scholar] [CrossRef]
24. Recalde, L., Vázquez, A., Groppa, M. D., Benavides, M. P. (2018). Reactive oxygen species and nitric oxide are involved in polyamine-induced growth inhibition in wheat plants. Protoplasma, 255(5), 1295–1307. DOI 10.1007/s00709-018-1227-z. [Google Scholar] [CrossRef]
25. Hummel, I., Gouesbet, G., El Amrani, A., Aïnouche, A., Couée, I. (2004). Characterization of the two arginine decarboxylase (polyamine biosynthesis) paralogues of the endemic subantarctic cruciferous species Pringlea antiscorbutica and analysis of their differential expression during development and response to environmental stress. Gene, 342(2), 199–209. DOI 10.1016/j.gene.2004.08.024. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools