 Open Access
Open Access
REVIEW
Molecular Mechanism Underlying Plant Response to Cold Stress
College of Biology and the Environment, Nanjing Forestry University, Nanjing, China
* Corresponding Author: Liming Yang. Email:
Phyton-International Journal of Experimental Botany 2023, 92(9), 2665-2683. https://doi.org/10.32604/phyton.2023.024929
Received 14 June 2022; Accepted 13 September 2022; Issue published 28 July 2023
Abstract
Low temperature stress is one of the most important factors limiting plant growth and geographical distribution. In order to adapt to low temperature, plants have evolved strategies to acquire cold tolerance, known as, cold acclimation. Current molecular and genomic studies have reported that annual herbaceous and perennial woody plants share similar cold acclimation mechanisms. However, woody perennials also require extra resilience to survive cold winters. Thus, trees have acquired complex dynamic processes to control the development of dormancy and cold resistance, ensuring successful tolerance during the coldest winter season. In this review, we systemically described how woody plants perceive and transduce cold stress signals through a series of physiological changes such as calcium signaling, membrane lipid, and antioxidant changes altering downstream gene expression and epigenetic modification, ultimately bud dormancy. We extended the discussion and reviewed the processes endogenous phytohormones play in regulating the cold stress. We believe that this review will aid in the comprehension of underlying mechanisms in plant acclimation to cold stress.Keywords
Cold stress is an important abiotic stress that limits the geographic distribution of plants and agricultural yields by affecting plant growth and survival [1]. Therefore, cold stress may be defined by an average maximum temperature of <0°C, over most of the Earth’s land area (about 64%), and by at least −10°C in 48% of the land [2]. However, in mid-to-high latitudes cold stress damage may increase due to global warming, affecting partially or fully open buds during the flowering period of cover trees [3], and premature bud break may occur more frequently [4]. In economic crops such as Oryza sativa and Hylocereus undulatus, low temperature will damage the fruit plasma membrane, leading to the loss of solute and affecting the seed germination rate [5,6]. Interestingly, most temperate plants can acquire cold tolerance by brief exposure to low but not freezing temperatures through a process known as cold acclimation [7]. Some plants such as Arabidopsis and barley (Hordeum vulgare) cannot tolerate non-freezing temperatures without cold acclimation [8]. Likewise, sandalwood (Santalum album) and Himalayan birch (Betula utilis), as the dominant vegetation in cold environments (eastern Nepal), are also sensitive to cold stress during the growing season [9,10]. Tropical and subtropical plants are sensitive to low temperature stress and lack the ability to adapt to cold, for instance: rice (O. sativa), corn (Zea mays), soybeans (Glycine max), etc. Nonetheless, when the low temperature period extends more than 4 days, these seeds will germinate poorly, grow slowly, and sometimes even die [11].
Relatively speaking, woody plants have the strongest cold tolerance among all other plants, and their lowest limit of cold tolerance depends on the stage of acclimation, the rate and extent of temperature drop, and the heritability of tissues to adapt to extracellular freezing and concomitant dehydration stress [12]. These factors aid woody plants in cold adaptation, and species population perpetuation in a specific niche for a prolonged duration. Therefore, trees have evolved the abilities to pause growth and development through dormancy upon sensing changes in day length to withstand the harsh winter [13].
Understanding how plants respond to cold stress will provide valuable information and genetic resources for improving plant cold tolerance. In this review, we summarize the recent research progress in the regulation mechanism of cold stress tolerance, especially the achievements and problems related to forest trees.
Unraveling the mechanisms by which plants sense cold signals is critical to understanding how plants avoid damage caused by low temperatures. Researchers have long focused on the fundamental question of how plants sense cold signals. These studies have all concurred to that cold stress is not sensed by only one protein, but by various proteins at different sensory levels [14].
2.1 Membrane Modification in Response to Cold Signals
The plasma membrane of an organism is the first line of defense against cold stress, acting like a safety net, protecting and maintaining the shape of cells, recognizing and exchanging signaling molecules and other substances required for various physiological and developmental processes [15]. These processes are facilitated by open spaces in the plasma membrane or by different transporters, pumps and gated channels [16]. For example, during cold acclimation, plants express tonoplast monosaccharide transporters (TMT) to increase sugar levels and maintain cellular homeostasis [17]. Another example is that low temperature induces the expression of annexin in poplar leaves. Annexin can mediate the transport of cytosolic free Ca2+ concentration in response to the reactive oxygen species (ROS) generated by cold stress [18].
The major components of the plasma membrane include lipids, proteins and carbohydrates. Changes in the physicochemical properties of the plasma membrane are part of the mechanism by which plants respond to cold stress (Fig. 1) [19]. Pharmacological analysis showed that at 25°C, the expression of cold responsive (COR) gene was induced by membrane stiffening, whereas at 4°C, it was inhibited by membrane fluidization. Plasma membrane fluidity is related to the proportion of desaturated fatty acids. For example, in H. undulatus, cold stress leads to severe oxidation of unsaturated fatty acids at the plasma membrane, leading to cellular damage and apoptosis [20]. Fatty acid desaturase 2 (FAD2) encodes an oleate desaturase enzyme necessary for membrane fluidity. Mutation of FAD2 disrupts some physiological responses to cold stress, such as leaf number and hypocotyl length [21]. Another study has found that the content of phosphatidylglycerol, dipalmitoyl plus the l-palmitoyl-2-(trans-3-hexadecenoyl) in cold-tolerant plants and sensitive plants is significantly different. The content of phosphatidylglycerol, dipalmitoyl plus the l-palmitoyl-2-(trans-3-hexadecenoyl) in cold-tolerant plants is about 3%–19%, in sensitive plants constitutes~26%–65% [22]. In addition, the lipid composition of the plasma membrane and chloroplast envelope changes in cold-acclimated plants, by increasing the unsaturated fatty acid content of the cold-adapted membranes, making them more fluid, consequently increasing cold resistance [23]. Therefore, changes in membrane fluidity can be used to represent the cold signal [24]. In Norway spruce, although there are differences between different varieties, short-term temperature changes has been shown to induce more significant changes in cold resistance than the overall climatic conditions [25]. The primary input of cold signaling may be tiny mechanical forces induced by membrane stiffening. Cold signaling is integrated by the microtubule receptor system and amplified by COLD1-dependent signaling pathways [26]. Some members of the MAP65 family have been shown to reduce the stiffness of microtubules, directly affecting the force transmission of membrane rigidification, possibly by increasing the abundance of unsaturated fatty acids and adaptive reflow of the membrane by microtubules related [27].
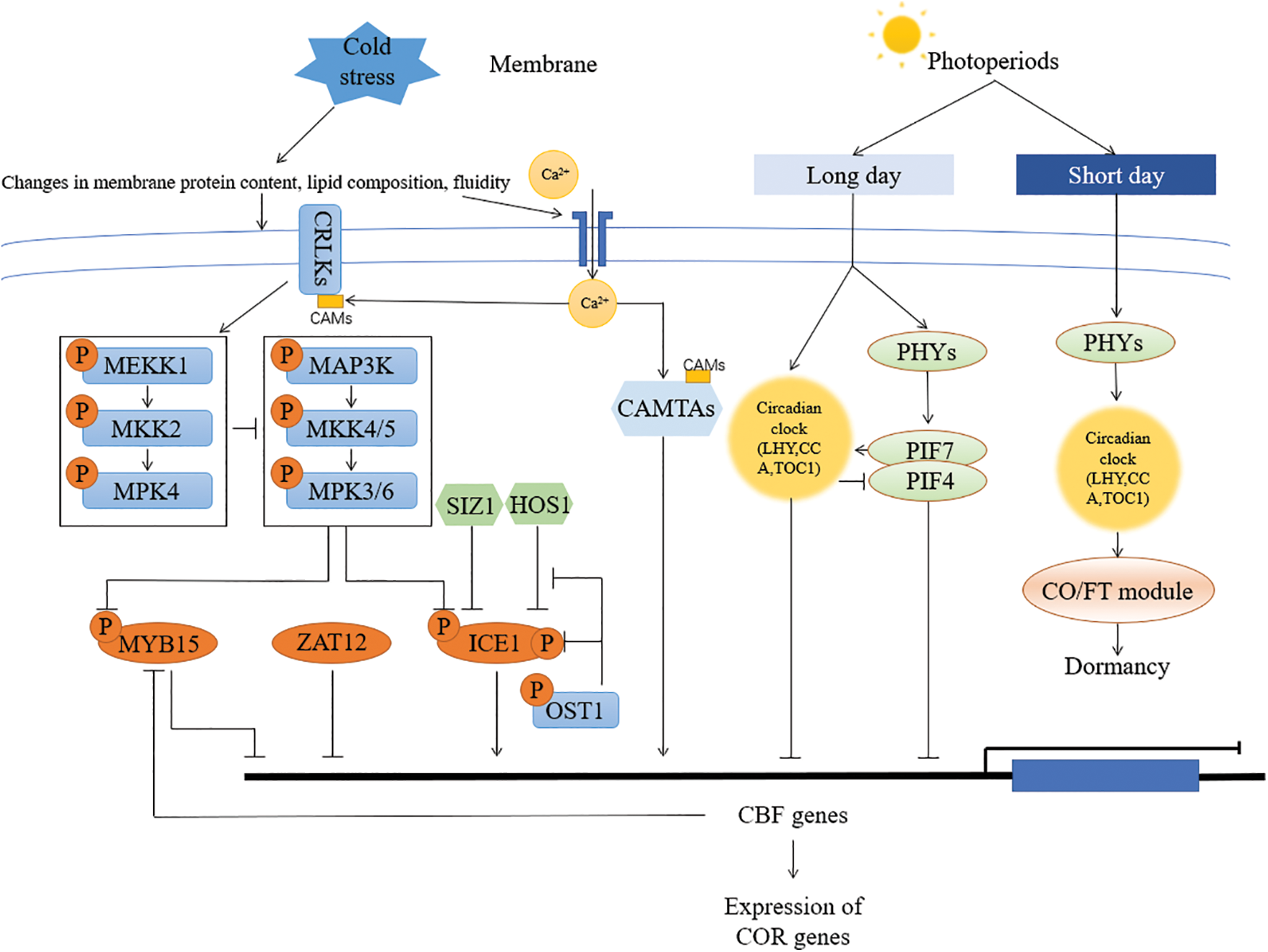
Figure 1: Schematic representation of the molecular response mechanism to cold stress in Arabidopsis. Cold stress-induced changes in membrane modifications and calcium ion concentrations lead to downstream signaling events. Calcium ion signals are sensed by calcium sensors such as CAM and CRLK, which are present in the cytoplasm, plasma membrane, chloroplast membrane, mitochondria, vacuoles, and nucleus, among others. These signals further activate other components of the cold stress response mechanism, such as the MAPK cascade, which ultimately regulates the expression levels of ICE1, CBF, and other cold-regulated genes. CCA is involved in the control of phytochrome interacting factors (PIFs) that mediate the cold response, integrating photoperiod and temperature cues [58,73]. PIF7 was shown to suppress CBF expression under long photoperiods and to promote growth under short photoperiods [13]
Paclitaxel, a compound that stabilizes microtubules, was found to inhibit the development of frost resistance. This suggests that effective activation of frost resistance requires certain microtubule dynamics [1]. Although microtubules do not transduce cold signals, they play a role in induce cold hardening [28]. Deeper research has also shown that when the membrane rigidify rapidly, the reduction in microtubule flexural rigidity (contributed by MAP65-2) prevents microtubule rupture under cold stress, while microtubule bundling is expected to improve force transmission to calcium channels as an actual sensory structure [27].
2.2 Calcium2+ Signaling in Response to Cold Stress
Calcium (Ca2+) is a universal second messenger involved in many aspects of plant development and environmental responses. Abiotic and biotic stresses, such as cold, heat, salt, and pathogen infection, act on Ca2+ permeation channels, resulting in transient increases in cytosolic free Ca2+ concentration ([Ca2+]cyt). These Ca2+ signals convert external signals into a variety of intracellular biochemical reactions [29].
Since trees are very large plants, rapid signal transduction in tree systems is critical. Fromm et al. [30] found that fast signals coordinate the physiological activities of trees and that calcium signals are the first step in action potential-dependent fast signals. Trauma-triggered electrical signals can travel long distances and alter photosynthesis in distant leaves. In trees grown under calcium-deficient conditions, the excitability of phloem cells was completely blocked after leaf injury [31]. Calcium-deficient trees also had no apparent gas exchange response after injury. These findings evidenced that calcium channels are involved in the induction of electrical signals at different distances in trees.
After cold stress alters the fluidity of the plasma membrane, it is sensed by Ca2+ channels and other plasma membrane proteins, and induces actin filaments depolymerization, triggering an increase in [Ca2+] cyt and activating the cold stress response [32,33]. It was found that the G-protein regulator Chilling Tolerance Divergence 1 (COLD1) binds to rice G protein α subunit 1 (RGA1), mediates cold-induced Ca2+ influx, and provides cold induction in rice [34]. In spruce mesophyll cells, plasma membrane-associated calcium accumulates strongly in early autumn and then declines after the first frost as cold resistance develops [35]. Ca2+ transmits low temperature signals by activating the signaling sensor calmodulin (CAM), which in turn mediates a range of downstream pathways, including development, defense against pathogens, and responses to abiotic stresses such as temperature, drought, salinity, and light [36]. Calcium-related signal transduction usually involves calcium-dependent protein kinases (CDPKs). The expression level of SiCDPK24 in Foxtail millet was significantly increased under cold stress [37]. Populus euphratica overexpressing CDPKs showed enhanced drought and freezing resistance [38]; however, Z. mays overexpressing ZmCPK1 showed reduced cold tolerance, indicating that CDPKs play different roles in cold signal transduction.
Calmodulin-binding transcription activators (CAMTA) proteins belong to a class of transcription factors conserved in eukaryotes that respond to calcium signals by binding to CAM. CAMTA protein senses the increase of Ca2+ level and interacts with various calmodulins as a response to cold stress [39]. There are 6 CAMTA genes in Arabidopsis thaliana, among which AtCAMTA3 can act as a positive regulator of AtCBF2 transcription factor by binding to the cis element (CM2 motif) of AtCBF2. In the Atcamta1 Atcamta3 double mutant, the expression of the CBF gene family was reduced by approximately 50% [40].
It has been reported that Ca2+/CaM-regulated receptor-like kinases 1 (CRLK1), which encodes a PM-associated serine/threonine kinase regulated by Ca2+ signaling, plays a key role in plant responses to cold stress. Under freezing temperature, crlk1 knockout mutant plants showed stronger sensitivity than WT, and the expression of some cold-responsive genes was significantly reduced. These results suggest that CRLK1 positively regulates the cold response in Arabidopsis. Further studies showed that Ca2+/CaM is required for the activation of AtCRLK1 kinase (Fig. 1). In the presence of Ca2+, AtCRLK1 kinase activity increased with increasing CAM concentration, whereas the CAM antagonist CPZ inhibited CAM-stimulated AtCRLK1 kinase activity. Furthermore, the CaM-binding domain (positions 369–390) at the C-terminus of AtCRLK1 is required for CaM-stimulated kinase activity, and only specific CAM isoforms are responsible for AtCRLK1 activation. These observations suggest the existence of a cold-response pathway for Ca2+ signaling regulated by AtCRLK1 [41]. Ca2+ signaling is involved in multiple signaling pathways and plays a key role in plant responses to cold stress.
In trees, calcium has been implicated in lignification and dormancy in addition to its involvement in signal transduction, and a reduction in calcium content has led to a reduction in lignin, with concomitant changes in the hardness and elasticity of the secondary wall [42]. At the onset of tree dormancy, calcium is recovered from the leaves to the stem, and calcium levels in the shoot apical meristem and dormant cambium become very low, and when dormancy is broken, activation of the cambium and flushing of the bud induces a marked rise in meristem calcium content, which is associated with the onset of cell division, suggesting that calcium plays a significant role in the activation of this process [43].
2.3 Optical Signaling Regulates CBF and Dormancy
The red-to-far-red light ratio (F/FR) is another factor that interacts with components of the circadian oscillator to regulate AtCBF 1 to 3 expression and cold resistance. Arabidopsis mutant studies have shown that phytochrome B (AtPHYB), D (AtPHYD), and AtPHYB interactors, phytochrome interacting factor 4 (AtPIF4) and 7 (AtPIF7), are involved in this process. Under long-day (LD) conditions, phytochrome activation triggers interaction of AtPIF7 with the circadian oscillator Timing of Cab Expression 1 (AtTOC1), followed by binding to the G-box sequence (CAGTG) in the CBF promoter, thereby downregulating CBF Expression. Under short-day (SD) conditions in autumn, this inhibition is alleviated, and PIF7 cooperates with PIF4 to regulate CBF and cold resistance [44]. In-silico gene expression analyses have shown light induction of the CBF pathway to be related to the phytochrome AtPHYA, as a link between AtPHYA and the AtICE1 signaling pathway [45]. However, long-term exposure to SD was reported not to increase the cold tolerance of PHYA-overexpressing poplars [46].
Phytochrome gene expression has also been associated with dormancy; transgenic poplars overexpressing the Avena sativa oat PHYA gene were shown to be insensitive to SDs even within 6 h, whereas wild-type plants stopped growth and germination under a 14-h photoperiod. In Poplar, PttPHYA gene knockout has ceased growth and terminal buds were formed earlier than WT plants at the critical day length of 15.5 h, indicating that SDs-induced seasonal dormancy and cold tolerance of poplars were mediated by the PHYA light signaling pathway. Furthermore, quantitative trait locus analysis revealed that PHYB in poplar is a candidate gene involved in photoperiod sensing and is associated with shoot formation time [13].
3 Gene Expression Changes during Cold Stress
3.1 The CBF Pathway Mediates Response of the Cold Stress
The main regulators of transcriptional cold response networks in higher plants are transcription factors of the CBF/DREB1 family, which were originally reported in A. thaliana. CBFs, also known as dehydration responsive element binding (DREB) proteins are highly conserved ABA-independent transcription factors family encoding transcription factors closely related to AP2/ERF DNA-binding protein family members that are rapidly induced upon exposure to 4°C [47,48]. CBF proteins are plant-specific transcription factors. A. thaliana encodes three cold-induced CBF genes, AtCBF1, AtCBF2, and AtCBF3. Transgenic Arabidopsis constitutively expressing AtCBF1, AtCBF2, or AtCBF3 results in increased cold tolerance of plants without exposure to low temperatures [49]. The AtCBF4 gene is not induced by cold, but can also enhance the cold tolerance of plants after overexpression [50]. The homologous gene of DREB1/CBF has been isolated from Triticum aestivum, Brassica napus, O. sativa, H. vulgare, Z. mays and other plants. Overexpression of Arabidopsis DREB1/CBF gene in transgenic B. napus or Nicotiana tabacum can induce the homologous gene of A. thaliana CBF/DREB1 homologous gene and improve the cold and drought tolerance of transgenic plants [51]. In Ipomoea batatas, IbCBF3 can significantly activate downstream IbCORs and IbSINA5 under cold stress, IbSINA5 can also induce multiple IbCORs [52].
The CBF genes are controlled by upstream transcription factors which includes: Inducer of CBF expression (AtICE), calmodulin binding transcription activator 3 (AtCAMTA3), ZAT12, Ethylene Insensitive 3 (AtEIN3), etc. [10]. AtICE1 and AtICE2 bind to the promoter regions of AtCBF1-3 to actively regulate their expression [53]. AtICE1 is respectively subjected to SUMOylation and polyubiquitination mediated by the SUMO E3 ligase AtSIZ1 and the ubiquitin E3 ligase AtHOS1, followed by proteasomal degradation [10]. SIZ1-mediated SUMOylation of AtICE1 controls AtCBF expression by stabilizing AtICE1 [54]. In Arabidopsis, the CBF family is upstream regulated by AtICE1, and under low temperature conditions, AtICE1 induces AtCBF3 by inhibiting transcription of AtMYB15, a repressor of AtCBF [47,48], AtMYB15 (R2R3-MYB family member) also negatively regulates AtCBF gene expression in Arabidopsis [55]. AtMYB15 can express in the absence of cold stress and inhibits AtCBF expression by binding to the AtMYB recognition site of the AtCBF promoter [41]. In addition, AtZAT12, a C2H2 zinc finger transcription factor, also acts as a negative regulator of CBF genes. In Arabidopsis, overexpressing AtZAT12 reduced the expression level of AtCBF under cold stress [56]. As discussed above, several regulatory pathways contribute to fine regulation of CBF-dependent signaling during cold stress. However, the ICE-CBF pathway has been challenged in recent years. Studies have shown that neither Atice1-1 mutant nor AtICE1 overexpression could regulate AtDREB1A/AtICE3 expression. This finding caused an uproar among cold stress researchers, but their results also suggested that hypermethylation in the AtDREB1A promoter region was due to another transgene, not AtICE1. Therefore, although the ICE-CBF pathway has been well studied and established in cold stress signaling, there is still great value in revalidating the pathways and findings associated with this signaling cascade [34,57].
Homologous CBF is also found in woody angiosperms, including Populus, Betula, Citrus reticulata, Prunus persica and Malus pumila. Cold-induced ICE1 and CBF homologues were also identified in several conifers, including Picea glauca, P. sitchensis, and Cryptomeria japonica. The P. persica CBF gene PpCBF1 was overexpressed in M. pumila trees that are normally insensitive to photoperiod, resulting in increased sensitivity to short photoperiods and accelerated growth arrest and leaf senescence. Apparently, the CBF pathway is also integrated into the photoperiodic signaling pathway [58].
The regulation of CBFs in woody plants is complex, showing gene-, tissue- and age-related specificities, as well as temporal differences in induction time, which are not observed in herbaceous plants [59]. Benedict et al. [60] reported different expression patterns of poplar CBF genes in annual and perennial tissues. Xiao et al. [61,62] reported a similar phenomenon in grapes, grape CBFs1-3 were only expressed in young tissues in cold response, while grape CBF4 was expressed in both young and old tissues in cold response. Under LD, birch trees produced CBFs within 15 min of exposure to low temperature [63]. However, CBF induction was delayed and up-regulated for a longer period of time in the SD. Welling et al. [63] further demonstrated that exposure of dormant birch to freezing temperatures (−10°C) after tree thawing only induces CBFs expression and COR gene expression. PpCBFs1-4 were induced by circadian rhythms, especially in leaves, but less so in bark. Differences in expression timing and patterns among peach CBFs may reflect their underlying regulatory complexity [64].
3.2 Mitogen-Activated Protein Kinase Regulate the Cold Stress Response via the ICE1 Pathway
Mitogen-activated protein kinases (MAPKs) play an integral role in integrating multiple intracellular signals transmitted by various stress-induced secondary messengers, thereby activating or inactivating proteins through post-translational phosphorylation of target proteins. In general, the MAPK cascade consists of three kinases acting in succession, with inactive MAPKKKs (MEKKs) activated by stimuli or signaling messengers. The MAPKKKs are activated by phosphorylating MAPKKs at conserved serine/threonines. Activated MAPKKs (MKKs) further activate MAPKs (MPKs) through phosphorylating. Recently, studies have found that MAPK signaling regulates the cold stress response in Arabidopsis through the ICE1 pathway [65].
Teige et al. [66] found that Arabidopsis AtMKK2 is activated by cold stress and controls COR gene expression and plant tolerance to cold stress. AtMKK2 is located downstream of AtMEKK1 and upstream of AtMPK4 and AtMPK6, which are activated by various stresses such as low temperature. Atmkk2 mutant plants were more sensitive to cold stress than WT, while plants overexpressing AtMKK2 exhibited stronger cold tolerance. Furthermore, plants overexpressing AtMKK2 could induce the transcriptional expression of AtCBF even at room temperature. Further studies found that low temperature activates Ca2+ signaling-mediated AtCRLK1, which in turn phosphorylates and activates AtMEKK1, an upstream component in the AtMKK2. Activation of AtMKK2 by AtMEKK1 induces the phosphorylation of AtMPK4 and AtMPK6 [67,68].
AtMPK3, AtMPK4, and AtMPK6 were induced within 30 min of cold exposure, suggesting their involvement in cold stress response [65]. In Arabidopsis, AtMPK6 phosphorylates AtMYB15 and reduces its binding to the CBF promoter, thereby releasing its inhibitory effect on AtCBF expression. However, two reports have provided strong genetic and biochemical evidence that AtMPK3/6 are negative regulators of CBF signaling. In Arabidopsis, cold-activated AtMPK3/6 interacts with and phosphorylates AtICE1, reducing its stability and binding activity to the AtCBF3 promoter, resulting in reduced cold tolerance [52,69]. And the AtMEKK1-AtMKK1/2-AtMPK4 cascade promotes the stability of AtICE1 by antagonizing AtMPK3/AtMPK6, thereby regulating the cold-responsive transcription factors AtCAMTA and AtMYB, consequently, enhancing cold tolerance [70].
Calcium signaling is also involved in the MAPK cascade pathway in response to cold stress. Two calcium/calmodulin-regulated receptor-like kinases, AtCRLK1 and AtCRLK2, induce AtMPK4 to inhibit the activation of AtMPK3/6, thereby positively regulating freezing tolerance [69].
In addition to phosphorylation, ubiquitination mediates protein degradation pathways in cold signaling. So far, the most suitable E3 ubiquitin ligase in the CBF signaling pathway is high expression of osmotically responsive gene1 (AtHOS1). AtHOS1 interacts with AtICE1 in the nucleus and promotes its degradation under cold stress. Transgenic plants overexpressing AtHOS1 have shown higher freezing sensitivity. Interestingly, open stomata 1 (AtOST1) mediates phosphorylation of AtICE1 interferes with its interaction with AtHOS1. Thereby, increasing the stability of AtICE1 through antagonizing the AtHOS1 E3 ubiquitin ligase (Fig. 1) [71]. In contrast, AtMPK3/6-mediated degradation of AtICE1 does not affect the AtHOS-AtICE interaction and may act through a different path [52].
Additionally, SUMO E3 ligase-mediated SUMOylation often protects proteins from degradation. In Arabidopsis, AtSIZ1 (SAP and Miz) encodes the SUMO E3 ligase, which is required for cold tolerance. AtSIZ1 can phosphorylate and stabilize AtICE1, thereby promoting the expression of AtCBF. The siz1 null mutant exhibits cold-induced CBF expression and impaired freezing tolerance [72].
Like AtICE1 in Arabidopsis, OsICE1 in rice is also degraded by OsHOS1. Unlike Arabidopsis, OsMPK3-mediated phosphorylation inhibits the degradation of OsICE1. It also disrupts the interaction of OsICE1 with the E3 ubiquitin ligase OsHOS1. This in turn enhances the stability of the OsICE1 protein and its ability to bind to trehalose-6-phosphate phosphatase1 (OsTPP1), which encodes a key enzyme that catalyzes trehalose biosynthesis, thereby enhancing cold tolerance. These findings suggest that MAPK3 plays different roles in cold tolerance in different species, and that the Ca2+ and MAPK cascades are important cold signal transducers [14].
As perennials, besides the synchronization of phenology and growth process with climatic rhythms, flexible responses to environmental stimuli are also crucial. Trees in temperate regions need to cope with climate change to survive the winter, while in boreal regions, trees are more resistant to cold, so in addition to cold acclimation, trees develop the ability to bud dormancy [13,74]. Bud dormancy and cold acclimation are independent processes, but both are induced by a combination of short photoperiod and low temperature [75]. In trees, the cessation of growth and the development of cold resistance usually coincide with bud formation and lead to bud dormancy. The role of photoperiod-induced budding has been reported in a variety of trees, including temperate deciduous trees, and boreal evergreen coniferous trees [58]. The development of bud formation and dormancy is regulated by hormones such as ABA, GA, and controlled by an integrated pathway network of photoperiod response, ABA response, and circadian clock [76]. Endodormancy is the initial stage of bud dormancy. At this stage, bud development is inhibited by meristem endogenous factors. Endodormancy occurs in late summer to early fall and coincides with the initial development of cold tolerance; the cold tolerance of fully dormant buds is further deepened and peaks when dormancy is about to be broken or after it has been broken [77].
From the beginning to the end of dormancy, photoreceptors such as phytochromes and the biological clock system sense and regulate changes in the light environment within the Arabidopsis. The core of the plant biological clock system comprise two MYB genes, late elongated hypocotyl l (AtLHY) circadian clock associated 1 (AtCCA1) and a pseudo-response regulator 1/timing of cab2 expression 1 (AtPRR1/AtTOC1) [70]. The master switch of cold-responsive genes, AtCBFs, is also regulated by the circadian clock system, and the mutual regulation of CBF and circadian clock in Arabidopsis contributes to the improvement of cold resistance. AtCCA1 and AtLHY can bind to the evening element (EE) and related DNA regulatory motifs present within the AtCBF locus and induce high-level expression of the AtCBFs. AtPIF4 and AtPIF7 inhibit the expression of AtCBFs. And the oscillation in AtPIF4 transcript levels is disrupted by constitutive overexpression of AtCCA1 [78].
In many tree species, growth arrest means that the activity of the apical bud meristem stops as the day length decreases, starting in late summer and early fall, and eventually stops growing and enters a dormant state [79]. In most trees, the increase in tree cold tolerance parallels the cessation of growth caused by the short-day (SD) photoperiod. Prolonged exposure to SD environments significantly improves cold tolerance, whereas subsequent exposure to low and freezing temperatures rapidly increases cold tolerance, which is necessary to increase the maximum cold tolerance [80]. However, a reduction in photoperiod did not cause growth to stop in all tree species. For example, apples and pears only respond to low temperature and are not sensitive to photoperiod [81]. The occurrence of dormancy in forest trees and the molecular mechanism of cold tolerance improvement during dormancy need to be further studied. The dormancy of trees in nature is accompanied by hardening of the overwintering tissue, and the de-hardening potential of early-maturing varieties (Gooseberry and Prunus) is higher than that of late-maturing varieties (Fagus and Quercus). Under the same phenological stage and dormancy period, the early-maturing cultivars exhibit stronger cold tolerance than the late-maturing cultivars, and have a greater hardening potential at subzero freezing temperatures. The potential for hardening is higher in active tissue and lower in mature tissue [82]. The degree of hardening is not proportional to the cold tolerance, because the cold resistance of the mature tissue itself is higher than that of the active tissue, indicating that hardening is an acquired ability during dormancy, and the tree’s overwintering tissue can obtain a certain degree of cold resistance through this change [52].
The establishment of dormancy involves the induction of a large number of genes, so the identification of transcription factors that play a role in the regulation of gene expression by dormancy will be an important step in understanding this process. The similarity of dormancy-inducing transcription factors to signature proteins in other systems provides clues to the function of transcription factors in cambium dormancy. For example, a poplar gene highly similar to CBF1 was induced 6.7-fold in the dormant cambium. The CBF gene family regulates gene expression under osmotic stress and low temperature stress [83].
5 Epigenetic Regulation of Cold Acclimation
The extraordinary phenotypic plasticity of forest trees allows them to grow and survive long lifespans in diverse environments. Epigenetic phenomena, or persistent changes in gene function during meiosis or mitosis, while the sequence of the genome remains unchanged, can contribute to the plasticity of trees. DNA methylation, post-translational changes in histones, and the actions of many small RNAs can shape the epigenetic phenomena at the molecular level. Thus, the regulation of epigenetic modifications provides an organism with additional flexibility, enabling plants to tune their gene expression, stress responses, and ultimately their phenotype throughout whole lifetime and even across generations. Epigenetic marks can respond to both external and internal signals, and their role in the development of stress responses has been recognized [58].
Post-translational histone modifications and DNA methylation are associated with gene expression levels. Histone acetylation/deacetylation catalyzed by histone acetyltransferases (HATs) and histone deacetylases (HDAs) catalyzed histone acetylation/deacetylation play a role in cold reactions in plants [40]. Arabidopsis histone deacetylase6 (AtHDA6) is up-regulated under cold stress and positively regulates cold tolerance [84]. Cold-induced COR expression correlates with histone modification levels. In Arabidopsis, H3K27me3 in two COR genes, AtCOR15A and AtGOLS3, was gradually decreased after cold treatment [84].
To date, epigenetic responses in trees have been poorly studied and remain a largely under-studied area. There is evidence that, in conifers, temperature and photoperiod control growth arrest and bud dormancy can reproduce through epigenetic mechanisms [58]. Johnsen et al. [85] observed that growth arrest and late bud dormancy in Norway. spruce seedlings from northern clones grown in southern nurseries, consistent with the more southern breed phenotype. These observations inspired a series of experiments revealing that temperature and photoperiod conditions at the time of flowering in females and at the time of seed development affect the cold tolerance of offspring [85]. Recent studies have shown that this epigenetic memory in N. spruce has unique gene expression patterns and microRNA profiles, as well as potentially other epigenetic marks [86,87]. These results suggest epigenetic control of plant cold acclimation and budding phenology, and that epigenesis itself may be an adaptive trait in trees. The study of tree epigenetic phenomena may provide a strong basis for further research on adaptive traits, biotechnology and ecosystem protection against climate change.
6 Antioxidants during the Cold Stress Response
Previous research has evidenced that ROS and nitric oxide (NO) are involved in regulating plant responses to cold stress, and ROS, including superoxide (O2˙-), hydroxyl radicals (OH-), and hydrogen peroxide (H2O2), produced in plant responses to various stresses. ROS has a dual role in plant cells: on one hand, they induce gene expression and protein synthesis, protecting cells from stress; on the other hand, they induce oxidative stress [6,88].
Under cold stress, plants accumulate H2O2, and excess H2O2 can have deleterious effects on plant cells. The H2O2 scavenging system is activated by the conversion of GSH (reduced glutathione) to GSSG (oxidized glutathione) in plants. And the promoter regions of some COR genes contain antioxidant response elements or GSSG binding sites, suggesting that redox signaling chains may regulate gene expression in response to cold stress [89]. Therefore, it will be important to identify COR genes with downstream functions in redox signaling. The Arabidopsis frostbit1 (Atfro1) mutant, which constitutively accumulates high levels of ROS, exhibits impaired COR gene expression and hypersensitivity to low temperature and freezing. FRO1 encodes the Fe-S subunit of respiratory electron transport chain complex I (NADH dehydrogenase) in mitochondria, the disruption of which results in high levels of ROS production [90]. Furthermore, as mentioned before, calcium signaling is involved in the regulation of cold stress. There is evidence that there is a close link between Ca2+ and ROS. For example, low ROS concentrations promote the influx of Ca2+ into the cytoplasm [91]. Thus, Ca2+ regulates the production of ROS in plants under various stimuli, such as drought and high relative humidity stress [92]. Calcium signaling may be involved in the response to cold stress in concert with ROS.
7 Phytohormones Involved in the Cold Stress Response
Abscisic acid (ABA) is one of the most important hormones in cold stress. ABA increases when stress affects plants. Furthermore, the aba-dependent pathway and the ICE-CBF-COR pathway have overlapping connections in cold tolerance [93]. Numerous studies have shown that ABA can induce an increase in the transcription level of CBF genes, possibly through binding to CRT/DRE elements. It has the potential to promote the activation of CBF. Research in Arabidopsis has shown that, AtHAP5A regulates cold tolerance by binding to the CCAAT motif of AtXTH21, and plants overexpressing AtHAP5A and AtXTH21 are more cold-tolerant than wild-type plants, but less sensitive to ABA [94]. In addition, ABA also plays an important role during cold acclimation and can induce the expression of COR genes [2]. Some genes cooperate with ABA, GA and ethylene to up-regulate COR genes [95]. The ABA signaling pathway also has a positive regulator, the BpUVR8 gene, which regulates the expression of a subset of ABA-responsive genes in ABA-treated Arabidopsis and birch [73].
Li et al. [52] showed in the transcriptome study of pear that the expression of the ABA receptor PYL and the positive regulator SnRK2s were up-regulated at the dormant entry; while the inhibitor of ABA signaling, PP2Cs, remained low during dormancy and decreased with decreasing ABA content and increase until dormancy cessation. In addition, exogenous ABA treatment promoted the expression of SnRK2s and suppressed the expression of PP2Cs.
Gibberellins (GAs) are thought to be involved in plant dormancy. GA1, a biologically active gibberellin, controls Salix subapical meristem responses to SD and LD, leading to growth arrest and growth initiation respectively. SD sensed by PHYA has been shown to block certain steps of GA1 biosynthesis, resulting in the growth arrest of poplar. In addition, it was found that the transcription level of PttRGA1 gene in poplar was highly up-regulated in the dormant cambium, and the PttRGA1 gene was highly similar to AtRGA1, a repressor of gibberellin response. This shows that not only GA levels are altered during dormancy, but also that GA signaling is also regulated in overwintering tissues. Transcript levels of the ABA biosynthesis gene, 9-cis-epoxycarotenoid dioxygenase, did not differ between the dormant cambium and the active cambium [77].
GA is also thought to be involved in the CBF pathway. Under cold stress, CBFs are partially suppressed in the context of GA-related gene mutation. DELLA proteins are repressors of the GA hormone signaling pathway, mainly by regulating the activity of transcription factors in plants. GAs induce DELLA to inhibit protein degradation, thereby controlling many key developmental processes and responses to stresses such as cold. Recently, it has been suggested that AtCBFs promote DELLA stabilization by activating the GA catabolism gene AtGA2ox7 (GA2 OXIDASE7) [96].
Other researches have shown that chemical inhibition of gibberellin biosynthesis combined with lower nighttime temperatures was unable to induce dormancy in hybrid poplar under LD photoperiods [97], suggesting that GAs may play a role in SD photoperiod sensing-mediated sleep initiation pathway functions downstream. The role of GAs (especially GA3 and GA4) in promoting shoot dormancy release is also evident and has been demonstrated in several woody species. In poplar, exogenous GA4 was found to replace the effects of cold stress by upregulating several low temperature-responsive genes, and inducing bud burst. Whereas CBFs overexpression led to upregulation of DELLA and decreased apple growth [98]. So far, it has not been clear whether these GA signaling genes respond directly to environmental signals or whether their expression simply reflects developmental events during dormancy.
Studies have shown that jasmonicacid (JA) positively regulates the ICE-CBF pathway to enhance the cold tolerance of Arabidopsis [99]. Exogenous application of MeJA significantly improved the cold tolerance of plants with or without cold acclimation. Conversely, blocking JA biosynthesis and signaling leads to hypersensitivity to freezing stress. Under cold stress, the biosynthesis of endogenous JA is activated, which is consistent with the regulatory role of JA on cold tolerance. Further mechanistic analysis revealed that several repressors of AtJAZ physically interact with the AtICE1 transcription factor to inhibit its transcriptional activity. Additionally, the overexpression of AtJAZ1 or AtJAZ4 in Arabidopsis inhibited the ICE-CBF signaling pathway and response to cold, and JA was concluded to actively regulate the cold response as an upstream signal of the ICE-CBF pathway. In addition to the ICE-CBF cascade, JA also regulates the expression of some ICE-CBF pathway-independent genes. For example, under cold stress, some genes encoding secondary metabolite biosynthesis enzymes (such as polyamines, glutathione, and anthocyanins) are regulated by JA signaling [100].
The AtJAZ1/2 homologous gene MdJAZ1/2 in apple has a similar function, which can negatively regulate cold tolerance by inhibiting the binding of B-BOX protein MdBBX37 to MdCBF1/4 promoter and inhibiting the interaction between BBX37 and ICE1, evidencing that the exogenous application of promoting inhibition of MdJAZ1/2 [101]. JA has also been implicated in the cold storage (e.g., 5°C or 7°C) of several tropical or subtropical fruits, for example, the application of exogenous MeJA has been shown to induce low temperature tolerance (7°C) in banana (Musa acuminata) fruit [102]. Further analysis showed that two homologous genes of AtMYC2 in bananas (MaMYC2a and MaMYC2b) were rapidly induced after MeJA treatment during cold storage. A yeast two-hybrid analysis indicated that MaMYC2 interacted with MaICE1. Consistent with this, the expression of ICE-CBF cold-responsive pathway genes MaCBF1, MaCBF2, MaCOR1, MaRD2, MaRD5 and MaKIN2 were significantly increased in MeJA-treated banana fruits stored at low temperature (7°C) [103].
8 Conclusions and Future Perspective
The process of woody plants cold acclimation is complex, since perennial woody plants need to adapt to cyclical climate changes, their mechanisms for adapting to cold are more diverse. In addition to being directly stimulated by temperature, they are also affected by hormones, biological clocks, light and other various factor control, so the internal molecular mechanism is equally complex. The cold acclimation of natural forest trees starts with the shortening of light, which is sensed by phytochromes, transmitted through Ca2+ signal, ABA, GA and other signaling pathways, and activates the CBF-COR pathway with CBF as the core. As an important transcription factor upstream of AtCBFs, AtICE1 not only directly induces AtCBFs, but also negatively regulates the upstream negative regulator AtMYB15 of AtCBFs. The protein level and activity of AtICE1 are post-translationally regulated by AtHOS1-mediated ubiquitination and AtSIZ1-mediated ubiquitination.
SD photoperiod not only activates cold acclimation, but also activates bud dormancy in forest trees. However, the current molecular research basis for cold acclimation and dormancy mainly relies on annual herbs (such as Arabidopsis) or deciduous trees (such as poplar), and there are still few studies on evergreen trees [104]. In addition, studies have shown that the cold-induced genes of different organs are very different, 86% of the cold-induced genes are not shared between roots and leaves [105]. Within relatively small distances within the same plant, the mechanisms responsible for cold resistance are sometimes different, and in some cases, even adjacent cells are different. For instance, the bark tissue in woody plants is cold-resistant, protecting the xylem tissue from freezing due to deep supercooling, and the cold tolerance of the root system is much lower than that of the aerial parts of the plant [59]. This means that in different plant tissues, there may be different low temperature regulatory networks. Besides, forest trees have significant phenotypic plasticity, and epigenetic phenomena are also involved in adaptation to cold. Research in these areas may become important directions for further research on adaptive traits, biotechnology and ecosystem protection to adapt to changing climatic conditions. Under the circumstance of global climate change, it is extremely important to study the ability of plants to adapt to cold in time.
Acknowledgement: This researchers would like to extend their sincere appreciation to the reviewers for the effort invest towards this review manuscript.
Funding Statement: This research was funded by the National Natural Science Foundation of China (No. 31971682), and the Research Startup Fund for High-Level and High-Educated Talents of Nanjing Forestry University.
Author Contributions: The authors confirm their contribution to the paper as follows: study conception and design: Yiwei Cao and Liming Yang; data collection: Yuanlin Guan; draft manuscript preparation: Yiwei Cao; draft manuscript revision: Delight Hwarari, Yasmina Radani and Liming Yang; funding acquisition: Liming Yang. All authors reviewed the results and approved the final version of the manuscript.
Conflicts of Interest: The authors declare that they have no conflicts of interest to report regarding the present study.
References
1. Abdrakhamanova, A., Wang, Q. Y., Khokhlova, L., Nick, P. (2003). Is microtubule disassembly a trigger for cold acclimation? Plant and Cell Physiology, 44(7), 676–686. https://doi.org/10.1093/pcp/pcg097 [Google Scholar] [PubMed] [CrossRef]
2. Rihan, H. Z., Al-Issawi, M., Fuller, M. P. (2017). Advances in physiological and molecular aspects of plant cold tolerance. Journal of Plant Interactions, 12(1), 143–157. https://doi.org/10.1080/17429145.2017.1308568 [Google Scholar] [CrossRef]
3. Albertos, P., Wagner, K., Poppenberger, B. (2019). Cold stress signaling in female reproductive tissues. Plant, Cell & Environment, 42(3), 846–853. [Google Scholar]
4. Augspurger, C. K. (2013). Reconstructing patterns of temperature, phenology, and frost damage over 124 years: Spring damage risk is increasing. Ecology, 94(1), 41–50. https://doi.org/10.1890/12-0200.1 [Google Scholar] [PubMed] [CrossRef]
5. Anwar, M. P., Khalid, M. A. I., Islam, A. K. M., Yeasmin, S., Ahmed, S. et al. (2021). Potentiality of different seed priming agents to mitigate cold stress of winter rice seedling. Phyton-International Journal of Experimental Botany, 90(5), 1491–1506. https://doi.org/10.32604/phyton.2021.015822 [Google Scholar] [CrossRef]
6. Qi, J., Song, C. P., Wang, B., Zhou, J., Kangasjärvi, J. et al. (2018). Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. Journal of Integrative Plant Biology, 60(9), 805–826. https://doi.org/10.1111/jipb.12654 [Google Scholar] [PubMed] [CrossRef]
7. Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V. et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. The Plant Cell, 19(4), 1403–1414. https://doi.org/10.1105/tpc.106.048397 [Google Scholar] [PubMed] [CrossRef]
8. Zhao, C., Lang, Z., Zhu, J. K. (2015). Cold responsive gene transcription becomes more complex. Trends in Plant Science, 20(8), 466–468. https://doi.org/10.1016/j.tplants.2015.06.001 [Google Scholar] [PubMed] [CrossRef]
9. Zhang, X., Teixeira da Silva, J. A.,Niu, M., Li, M., He, C. et al. (2017). Physiological and transcriptomic analyses reveal a response mechanism to cold stress in Santalum album L. leaves. Scientific Reports, 7(1), 1–18. https://doi.org/10.1038/srep42165 [Google Scholar] [PubMed] [CrossRef]
10. Norén, L., Kindgren, P., Stachula, P., Rühl, M., Eriksson, M. E. et al. (2016). Circadian and plastid signaling pathways are integrated to ensure correct expression of the CBF and COR genes during photoperiodic growth. Plant Physiology, 171(2), 1392–1406. [Google Scholar]
11. Najeeb, S., Mahender, A., Anandan, A., Hussain, W., Li, Z. et al. (2021). Genetics and breeding of low-temperature stress tolerance in rice. In: Ali, J., Wani, S. H. (Eds.Rice improvement, pp. 221–280. Germany: Springer. [Google Scholar]
12. George, M. F., Becwar, M. R., Burke, M. J. (1982). Freezing avoidance by deep undercooling of tissue water in winter-hardy plants. Cryobiology, 19(6), 628–639. https://doi.org/10.1016/0011-2240(82)90192-4 [Google Scholar] [PubMed] [CrossRef]
13. Arakawa, K., Kasuga, J., Takata, N. (2018). Mechanism of overwintering in trees. In: Iwaya-Inoue, M., Sakurai, M., Uemura, M. (Eds.Survival strategies in extreme cold and desiccation, pp. 129–147. Singapore USA: Springer. [Google Scholar]
14. Ding, Y., Shi, Y., Yang, S. (2019). Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytologist, 222(4), 1690–1704. https://doi.org/10.1111/nph.15696 [Google Scholar] [PubMed] [CrossRef]
15. Chrispeels, M. J., Crawford, N. M., Schroeder, J. I. (1999). Proteins for transport of water and mineral nutrients across the membranes of plant cells. The Plant Cell, 11(4), 661–675. https://doi.org/10.1105/tpc.11.4.661 [Google Scholar] [PubMed] [CrossRef]
16. Seidel, T., Siek, M., Marg, B., Dietz, K. J. (2013). Energization of vacuolar transport in plant cells and its significance under stress. International Review of Cell and Molecular Biology, 304(32), 57–131. https://doi.org/10.1016/B978-0-12-407696-9.00002-6 [Google Scholar] [PubMed] [CrossRef]
17. Neuhaus, H. E. (2007). Transport of primary metabolites across the plant vacuolar membrane. FEBS Letters, 581(12), 2223–2226. https://doi.org/10.1016/j.febslet.2007.02.003 [Google Scholar] [PubMed] [CrossRef]
18. Morgan, R. O., Martin-Almedina, S., Garcia, M., Jhoncon-Kooyip, J., Fernandez, M. P. (2006). Deciphering function and mechanism of calcium-binding proteins from their evolutionary imprints. Biochimica et Biophysica Acta (BBA)-Molecular Cell Research, 1763(11), 1238–1249. https://doi.org/10.1016/j.bbamcr.2006.09.028 [Google Scholar] [PubMed] [CrossRef]
19. Uemura, M., Steponkus, P. L. (1999). Cold acclimation in plants: Relationship between the lipid composition and the cryostability of the plasma membrane. Journal of Plant Research, 112(2), 245–254. https://doi.org/10.1007/PL00013882 [Google Scholar] [CrossRef]
20. Sheng, K., Wei, S., Mei, J., Xie, J. (2021). Chilling injury, physicochemical properties, and antioxidant enzyme activities of red pitahaya (Hylocereus polyrhizus) fruits under cold storage stress. Phyton-International Journal of Experimental Botany, 90(1), 291–305. https://doi.org/10.32604/phyton.2020.012985 [Google Scholar] [CrossRef]
21. Martinière, A., Shvedunova, M., Thomson, A. J., Evans, N. H., Penfield, S. et al. (2011). Homeostasis of plasma membrane viscosity in fluctuating temperatures. New Phytologist, 192(2), 328–337. https://doi.org/10.1111/j.1469-8137.2011.03821.x [Google Scholar] [PubMed] [CrossRef]
22. Murata, N. (1983). Molecular species composition of phosphatidylglycerols from chilling-sensitive and chilling-resistant plants. Plant and Cell Physiology, 24(1), 81–86. https://doi.org/10.1093/oxfordjournals.pcp.a076516 [Google Scholar] [CrossRef]
23. Uemura, M., Joseph, R. A., Steponkus, P. L. (1995). Cold acclimation of Arabidopsis thaliana (effect on plasma membrane lipid composition and freeze-induced lesions). Plant Physiology, 109(1), 15–30. https://doi.org/10.1104/pp.109.1.15 [Google Scholar] [PubMed] [CrossRef]
24. Los, D. A., Murata, N. (2004). Membrane fluidity and its roles in the perception of environmental signals. Biochimica et Biophysica Acta (BBA)-Biomembranes, 1666(1–2), 142–157. https://doi.org/10.1016/j.bbamem.2004.08.002 [Google Scholar] [PubMed] [CrossRef]
25. Wellburn, A. R. (1997). Environmental factors and genotypic changes in lipids contribute to the winter hardiness of Norway spruce (Picea abies). The New Phytologist, 135(1), 115–121. https://doi.org/10.1046/j.1469-8137.1997.00617.x [Google Scholar] [PubMed] [CrossRef]
26. Wang, L., Sadeghnezhad, E., Nick, P. (2020). Upstream of gene expression: What is the role of microtubules in cold signalling? Journal of Experimental Botany, 71(1), 36–48. https://doi.org/10.1093/jxb/erz419 [Google Scholar] [PubMed] [CrossRef]
27. Portran, D., Zoccoler, M., Gaillard, J., Stoppin-Mellet, V., Neumann, E. et al. (2013). MAP65/Ase1 promote microtubule flexibility. Molecular Biology of the Cell, 24(12), 1964–1973. https://doi.org/10.1091/mbc.e13-03-0141 [Google Scholar] [CrossRef]
28. Wang, L., Sadeghnezhad, E., Riemann, M., Nick, P. (2019). Microtubule dynamics modulate sensing during cold acclimation in grapevine suspension cells. Plant Science, 280(1), 18–30. https://doi.org/10.1016/j.plantsci.2018.11.008 [Google Scholar] [PubMed] [CrossRef]
29. Liu, Q., Ding, Y., Shi, Y., Ma, L., Wang, Y. et al. (2021). The calcium transporter ANNEXIN1 mediates cold-induced calcium signaling and freezing tolerance in plants. The EMBO Journal, 40(2), e104559. https://doi.org/10.15252/embj.2020104559 [Google Scholar] [PubMed] [CrossRef]
30. Fromm, J., Meyer, A. J., Weisenseel, M. H. (1997). Growth, membrane potential and endogenous ion currents of willow (Salix viminalis) roots are all affected by abscisic acid and spermine. Physiologia Plantarum, 99(4), 529–537. https://doi.org/10.1111/j.1399-3054.1997.tb05353.x [Google Scholar] [CrossRef]
31. Lautner, S., Grams, T. E. E., Matyssek, R., Fromm, J. (2005). Characteristics of electrical signals in poplar and responses in photosynthesis. Plant Physiology, 138(4), 2200–2209. https://doi.org/10.1104/pp.105.064196 [Google Scholar] [PubMed] [CrossRef]
32. Lv, F., Wang, S., Tian, R., Wang, P., Liu, K. (2021). Villin family members associated with multiple stress responses in cotton. Phyton-International Journal of Experimental Botany, 90(6), 1645–1660. https://doi.org/10.32604/phyton.2021.016947 [Google Scholar] [CrossRef]
33. Zhu, J. K. (2016). Abiotic stress signaling and responses in plants. Cell, 167(2), 313–324. https://doi.org/10.1016/j.cell.2016.08.029 [Google Scholar] [PubMed] [CrossRef]
34. Ma, Y., Dai, X., Xu, Y., Luo, W., Zheng, X. et al. (2015). COLD1 confers chilling tolerance in rice. Cell, 160(6), 1209–1221. https://doi.org/10.1016/j.cell.2015.01.046 [Google Scholar] [PubMed] [CrossRef]
35. DeHayes, D. H., Schaberg, P. G., Hawley, G. J., Borer, C. H., Cumming, J. R. et al. (1997). Physiological implications of seasonal variation in membrane-associated calcium in red spruce mesophyll cells. Tree Physiology, 17(11), 687–695. https://doi.org/10.1093/treephys/17.11.687 [Google Scholar] [PubMed] [CrossRef]
36. Conde, A., Chaves, M. M., Gerós, H. (2011). Membrane transport, sensing and signaling in plant adaptation to environmental stress. Plant and Cell Physiology, 52(9), 1583–1602. https://doi.org/10.1093/pcp/pcr107 [Google Scholar] [PubMed] [CrossRef]
37. Saleem, S., Mushtaq, N. U., Shah, W. H., Rasool, A., Hakeem, K. R. et al. (2021). Morpho-physiological, biochemical and molecular adaptation of millets to abiotic stresses: A review. Phyton-International Journal of Experimental Botany, 90(5), 1363–1385. https://doi.org/10.32604/phyton.2021.014826 [Google Scholar] [CrossRef]
38. Chen, J., Xue, B., Xia, X., Yin, W. (2013). A novel calcium-dependent protein kinase gene from Populus euphratica, confers both drought and cold stress tolerance. Biochemical and Biophysical Research Communications, 441(3), 630–636. https://doi.org/10.1016/j.bbrc.2013.10.103 [Google Scholar] [PubMed] [CrossRef]
39. Bouché, N., Scharlat, A., Snedden, W., Bouchez, D., Fromm, H. (2002). A novel family of calmodulin-binding transcription activators in multicellular organisms. Journal of Biological Chemistry, 277(24), 21851–21861. https://doi.org/10.1074/jbc.M200268200 [Google Scholar] [PubMed] [CrossRef]
40. Doherty, C. J., Van Buskirk, H. A., Myers, S. J., Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. The Plant Cell, 21(3), 972–984. https://doi.org/10.1105/tpc.108.063958 [Google Scholar] [PubMed] [CrossRef]
41. Yang, T., Chaudhuri, S., Yang, L., Du, L., Poovaiah, B. W. (2010). A calcium/calmodulin-regulated member of the receptor-like kinase family confers cold tolerance in plants. Journal of Biological Chemistry, 285(10), 7119–7126. https://doi.org/10.1074/jbc.M109.035659 [Google Scholar] [PubMed] [CrossRef]
42. Wimmer, R., Lucas, B. N. (1997). Comparing mechanical properties of secondary wall and cell corner middle lamella in spruce wood. Iawa Journal, 18(1), 77–88. https://doi.org/10.1163/22941932-90001463 [Google Scholar] [CrossRef]
43. Lautner, S., Fromm, J. (2010). Calcium-dependent physiological processes in trees. Plant Biology, 12(2), 268–274. https://doi.org/10.1111/j.1438-8677.2009.00281.x [Google Scholar] [PubMed] [CrossRef]
44. Deng, G., Ying, S., Jing, S., Zhou, J., Lu, S. et al. (2021). AFP2 coordinates the activity of PIF7 for thermomorphogenesis in Arabidopsis seedlings. Phyton-International Journal of Experimental Botany, 90(4), 1089–1101. https://doi.org/10.32604/phyton.2021.016217 [Google Scholar] [CrossRef]
45. Benedict, C., Geisler, M., Trygg, J., Huner, N., Hurry, V. (2006). Consensus by democracy. Using meta-analyses of microarray and genomic data to model the cold acclimation signaling pathway in Arabidopsis. Plant Physiology, 141(4), 1219–1232. https://doi.org/10.1104/pp.106.083527 [Google Scholar] [PubMed] [CrossRef]
46. Welling, A., Palva, E. T. (2006). Molecular control of cold acclimation in trees. Physiologia Plantarum, 127(2), 167–181. https://doi.org/10.1111/j.1399-3054.2006.00672.x [Google Scholar] [CrossRef]
47. Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: Insights gained from studying the CBF cold response pathway. Plant Physiology, 154(2), 571–577. https://doi.org/10.1104/pp.110.161794 [Google Scholar] [PubMed] [CrossRef]
48. Hwarari, D., Guan, Y., Ahmad, B., Movahedi, A., Min, T. (2022). ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. International Journal of Molecular Sciences, 23(3), 1549. https://doi.org/10.3390/ijms23031549 [Google Scholar] [PubMed] [CrossRef]
49. Jaglo-Ottosen, K. R., Gilmour, S. J., Zarka, D. G., Schabenberger, O., Thomashow, M. F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science, 280(5360), 104–106. https://doi.org/10.1126/science.280.5360.104 [Google Scholar] [PubMed] [CrossRef]
50. Haake, V., Cook, D., Riechmann, J., Pineda, O., Thomashow, M. F. et al. (2002). Transcription factor CBF4 is a regulator of drought adaptation in Arabidopsis. Plant Physiology, 130(2), 639–648. https://doi.org/10.1104/pp.006478 [Google Scholar] [PubMed] [CrossRef]
51. Yamaguchi-Shinozaki, K., Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annual Review of Plant Biology, 57(1), 781–803. https://doi.org/10.1146/annurev.arplant.57.032905.105444 [Google Scholar] [PubMed] [CrossRef]
52. Li, H., Ding, Y., Shi, Y., Zhang, X., Zhang, S. et al. (2017). MPK3- and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Developmental Cell, 43(5), 630–642. https://doi.org/10.1016/j.devcel.2017.09.025 [Google Scholar] [PubMed] [CrossRef]
53. Lee, B. H., Henderson, D. A., Zhu, J. K. (2005). The Arabidopsis cold-responsive transcriptome and its regulation by ICE1. The Plant Cell, 17(11), 3155–3175. https://doi.org/10.1105/tpc.105.035568 [Google Scholar] [PubMed] [CrossRef]
54. Miura, K., Furumoto, T. (2013). Cold signaling and cold response in plants. International Journal of Molecular Sciences, 14(3), 5312–5337. https://doi.org/10.3390/ijms14035312 [Google Scholar] [PubMed] [CrossRef]
55. Agarwal, P. K., Agarwal, P., Reddy, M. K., Sopory, S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reports, 25(12), 1263–1274. https://doi.org/10.1007/s00299-006-0204-8 [Google Scholar] [PubMed] [CrossRef]
56. Vogel, J. T., Zarka, D. G., van Buskirk, H. A., Fowler, S. G., Thomashow, M. F. (2005). Roles of the CBF2 and ZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. The Plant Journal, 41(2), 195–211. https://doi.org/10.1111/j.1365-313X.2004.02288.x [Google Scholar] [PubMed] [CrossRef]
57. Kidokoro, S., Kim, J. S., Ishikawa, T., Suzuki, T., Shinozaki, K. et al. (2020). DREB1A/CBF3 is repressed by transgene-induced DNA methylation in the Arabidopsis ICE1-1 mutant. The Plant Cell, 32(4), 1035–1048. https://doi.org/10.1105/tpc.19.00532 [Google Scholar] [PubMed] [CrossRef]
58. Chang, C. Y. Y., Bräutigam, K., Hüner, N. P., Ensminger, I. (2021). Champions of winter survival: Cold acclimation and molecular regulation of cold hardiness in evergreen conifers. New Phytologist, 229(2), 675–691. https://doi.org/10.1111/nph.16904 [Google Scholar] [PubMed] [CrossRef]
59. Wisniewski, M., Nassuth, A., Teulieres, C., Marque, C., Rowland, J. et al. (2014). Genomics of cold hardiness in woody plants. Critical Reviews in Plant Sciences, 33(2–3), 92–124. https://doi.org/10.1080/07352689.2014.870408 [Google Scholar] [CrossRef]
60. Benedict, C., Skinner, J. S., Meng, R., Chang, Y., Bhalerao, R. et al. (2006). The CBF1-dependent low temperature signalling pathway, regulon and increase in freeze tolerance are conserved in Populus spp. Plant, Cell & Environment, 29(7), 1259–1272. https://doi.org/10.1111/j.1365-3040.2006.01505.x [Google Scholar] [PubMed] [CrossRef]
61. Xiao, H., Siddiqua, M., Braybrook, S., Nassuth, A. (2006). Three grape CBF/DREB1 genes respond to low temperature, drought and abscisic acid. Plant, Cell & Environment, 29(7), 1410–1421. https://doi.org/10.1111/j.1365-3040.2006.01524.x [Google Scholar] [PubMed] [CrossRef]
62. Xiao, H., Tattersall, E. A., Siddiqua, M. K., Cramer, G. R., Nassuth, A. (2008). CBF4 is a unique member of the CBF transcription factor family of Vitis vinifera and Vitis riparia. Plant, Cell & Environment, 31(1), 1–10. [Google Scholar]
63. Welling, A., Palva, E. T. (2008). Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiology, 147(3), 1199–1211. https://doi.org/10.1104/pp.108.117812 [Google Scholar] [PubMed] [CrossRef]
64. Artlip, T. S., Wisniewski, M. E., Bassett, C. L., Norelli, J. L. (2013). CBF gene expression in peach leaf and bark tissues is gated by a circadian clock. Tree Physiology, 33(8), 866–877. https://doi.org/10.1093/treephys/tpt056 [Google Scholar] [PubMed] [CrossRef]
65. Aslam, M., Fakher, B., Ashraf, M. A., Cheng, Y., Wang, B. et al. (2022). Plant low-temperature stress: Signaling and response. Agronomy, 12(3), 702. https://doi.org/10.3390/agronomy12030702 [Google Scholar] [CrossRef]
66. Teige, M., Scheikl, E., Eulgem, T., Dóczi, R., Ichimura, K. et al. (2004). The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Molecular Cell, 15(1), 141–152. https://doi.org/10.1016/j.molcel.2004.06.023 [Google Scholar] [PubMed] [CrossRef]
67. Furuya, T., Matsuoka, D., Nanmori, T. (2013). Phosphorylation of Arabidopsis thaliana MEKK1 via Ca2+ signaling as a part of the cold stress response. Journal of Plant Research, 126(6), 833–840. https://doi.org/10.1007/s10265-013-0576-0 [Google Scholar] [PubMed] [CrossRef]
68. Furuya, T., Matsuoka, D., Nanmori, T. (2014). Membrane rigidification functions upstream of the MEKK1-MKK2-MPK4 cascade during cold acclimation in Arabidopsis thaliana. FEBS Letters, 588(11), 2025–2030. https://doi.org/10.1016/j.febslet.2014.04.032 [Google Scholar] [PubMed] [CrossRef]
69. Zhao, C., Wang, P., Si, T., Hsu, C. C., Wang, L. et al. (2017). MAP kinase cascades regulate the cold response by modulating ICE1 protein stability. Developmental Cell, 43(5), 618–629. https://doi.org/10.1016/j.devcel.2017.09.024 [Google Scholar] [PubMed] [CrossRef]
70. Kim, Y., Park, S., Gilmour, S. J., Thomashow, M. F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. The Plant Journal, 75(3), 364–376. https://doi.org/10.1111/tpj.12205 [Google Scholar] [PubMed] [CrossRef]
71. Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proceedings of the National Academy of Sciences of the United States of America, 103(21), 8281–8286. https://doi.org/10.1073/pnas.0602874103 [Google Scholar] [PubMed] [CrossRef]
72. Miura, K., Ohta, M., Nakazawa, M., Ono, M., Hasegawa, P. M. (2011). ICE1 Ser403 is necessary for protein stabilization and regulation of cold signaling and tolerance. The Plant Journal, 67(2), 269–279. https://doi.org/10.1111/j.1365-313X.2011.04589.x [Google Scholar] [PubMed] [CrossRef]
73. Zhao, C., Mao, K., You, C. X., Zhao, X. Y., Wang, S. H. et al. (2016). Molecular cloning and functional analysis of a UV-B photoreceptor gene, MdUVR8 (UV Resistance Locus 8from apple. Plant Science, 247(5), 115–126. https://doi.org/10.1016/j.plantsci.2016.03.006 [Google Scholar] [PubMed] [CrossRef]
74. Sakai, A., Larcher, W. (2012). Frost survival of plants: Responses and adaptation to freezing stress, vol. 62. Germany: Springer Science & Business Media. [Google Scholar]
75. Arora, R., Wisniewski, M. E., Scorza, R. (1992). Cold acclimation in genetically related (sibling) deciduous and evergreen peach (Prunus persica [L.] BatschI. Seasonal changes in cold hardiness and polypeptides of bark and xylem tissues. Plant Physiology, 99(4), 1562–1568. https://doi.org/10.1104/pp.99.4.1562 [Google Scholar] [PubMed] [CrossRef]
76. Cooke, J. E., Eriksson, M. E., Junttila, O. (2012). The dynamic nature of bud dormancy in trees: Environmental control and molecular mechanisms. Plant, Cell & Environment, 35(10), 1707–1728. [Google Scholar]
77. Welling, A., Moritz, T., Palva, E. T., Junttila, O. (2002). Independent activation of cold acclimation by low temperature and short photoperiod in hybrid aspen. Plant Physiology, 129(4), 1633–1641. https://doi.org/10.1104/pp.003814 [Google Scholar] [PubMed] [CrossRef]
78. Lee, C. M., Thomashow, M. F. (2012). Photoperiodic regulation of the C-repeat binding factor (CBF) cold acclimation pathway and freezing tolerance in Arabidopsis thaliana. Proceedings of the National Academy of Sciences of the United States of America, 109(37), 15054–15059. https://doi.org/10.1073/pnas.1211295109 [Google Scholar] [PubMed] [CrossRef]
79. Rinne, P. L., Kaikuranta, P. M., van Der Schoot, C. (2001). The shoot apical meristem restores its symplasmic organization during chilling-induced release from dormancy. The Plant Journal, 26(3), 249–264. https://doi.org/10.1046/j.1365-313X.2001.01022.x [Google Scholar] [PubMed] [CrossRef]
80. Junttila, O., Kaurin, Å. (1990). Environmental control of cold acclimation in Salix pentandra. Scandinavian Journal of Forest Research, 5(1–4), 195–204. https://doi.org/10.1080/02827589009382605 [Google Scholar] [CrossRef]
81. Heide, O. M., Prestrud, A. K. (2005). Low temperature, but not photoperiod, controls growth cessation and dormancy induction and release in apple and pear. Tree Physiology, 25(1), 109–114. https://doi.org/10.1093/treephys/25.1.109 [Google Scholar] [PubMed] [CrossRef]
82. Hänninen, H. (2016). Boreal and temperate trees in a changing climate biometeorology. Dordrecht, Netherlands: Springer. [Google Scholar]
83. Schrader, J., Moyle, R., Bhalerao, R., Hertzberg, M., Lundeberg, J. et al. (2004). Cambial meristem dormancy in trees involves extensive remodelling of the transcriptome. The Plant Journal, 40(2), 173–187. https://doi.org/10.1111/j.1365-313X.2004.02199.x [Google Scholar] [PubMed] [CrossRef]
84. Kwon, C. S., Lee, D., Choi, G., Chung, W. I. (2009). Histone occupancy-dependent and-independent removal of H3K27 trimethylation at cold-responsive genes in Arabidopsis. The Plant Journal, 60(1), 112–121. https://doi.org/10.1111/j.1365-313X.2009.03938.x [Google Scholar] [PubMed] [CrossRef]
85. Johnsen, Ø., Kvaalen, H., Yakovlev, I. A., Dæhlen, O. G., Fossdal, C. G. et al. (2009). An epigenetic memory from time of embryo development affects climatic adaptation in Norway spruce. In: Plant cold hardiness: From the laboratory to the field, pp. 99–107. Wallingford: CABI. [Google Scholar]
86. Yakovlev, I. A., Fossdal, C. G. (2017). In silico analysis of small RNAs suggest roles for novel and conserved miRNAs in the formation of epigenetic memory in somatic embryos of Norway spruce. Frontiers in Physiology, 8, 674. https://doi.org/10.3389/fphys.2017.00674 [Google Scholar] [PubMed] [CrossRef]
87. Yakovlev, I. A., Lee, Y., Rotter, B., Olsen, J. E., Skrøppa, T. et al. (2014). Temperature-dependent differential transcriptomes during formation of an epigenetic memory in Norway spruce embryogenesis. Tree Genetics & Genomes, 10(2), 355–366. https://doi.org/10.1007/s11295-013-0691-z [Google Scholar] [CrossRef]
88. Heidarvand, L., Maali-Amiri, R. (2013). Physio-biochemical and proteome analysis of chickpea in early phases of cold stress. Journal of Plant Physiology, 170(5), 459–469. https://doi.org/10.1016/j.jplph.2012.11.021 [Google Scholar] [PubMed] [CrossRef]
89. Kocsy, G., Tóth, B., Berzy, T., Szalai, G., Jednákovits, A. et al. (2001). Glutathione reductase activity and chilling tolerance are induced by a hydroxylamine derivative BRX-156 in maize and soybean. Plant Science, 160(5), 943–950. https://doi.org/10.1016/S0168-9452(01)00333-8 [Google Scholar] [PubMed] [CrossRef]
90. Chinnusamy, V., Zhu, J., Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends in Plant Science, 12(10), 444–451. https://doi.org/10.1016/j.tplants.2007.07.002 [Google Scholar] [PubMed] [CrossRef]
91. Rihan, H. Z., Al-Issawi, M., Fuller, M. P. (2017). Upregulation of CBF/DREB1 and cold tolerance in artificial seeds of cauliflower (Brassica oleracea var. botrytis). Scientia Horticulturae, 225(3), 299–309. https://doi.org/10.1016/j.scienta.2017.07.017 [Google Scholar] [CrossRef]
92. Wang, W. H., He, E. M., Guo, Y., Tong, Q. X., Zheng, H. L. (2016). Chloroplast calcium and ROS signaling networks potentially facilitate the primed state for stomatal closure under multiple stresses. Environmental and Experimental Botany, 122, 85–93. https://doi.org/10.1016/j.envexpbot.2015.09.008 [Google Scholar] [CrossRef]
93. Wang, D. Z., Jin, Y. N., Ding, X. H., Wang, W. J., Zhai, S. S. et al. (2017). Gene regulation and signal transduction in the ICE-CBF–COR signaling pathway during cold stress in plants. Biochemistry, 82(10), 1103–1117. https://doi.org/10.1134/S0006297917100030 [Google Scholar] [PubMed] [CrossRef]
94. Shi, H., Ye, T., Zhong, B., Liu, X., Jin, R. et al. (2014). AtHAP5A modulates freezing stress resistance in Arabidopsis through binding to CCAAT motif of AtXTH21. New Phytologist, 203(2), 554–567. https://doi.org/10.1111/nph.12812 [Google Scholar] [PubMed] [CrossRef]
95. Wang, M., Zhang, X., Liu, J. H. (2015). Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf.) in response to cold stress. BMC Genomics, 16(1), 1–20. https://doi.org/10.1186/s12864-015-1629-7 [Google Scholar] [PubMed] [CrossRef]
96. Lantzouni, O., Alkofer, A., Falter-Braun, P., Schwechheimer, C. (2020). GROWTH-REGULATING FACTORS interact with DELLAs and regulate growth in cold stress. The Plant Cell, 32(4), 1018–1034. https://doi.org/10.1105/tpc.19.00784 [Google Scholar] [PubMed] [CrossRef]
97. MØLMANN, J. A., Asante, D. K., Jensen, J. B., Krane, M. N., Ernstsen, A. et al. (2005). Low night temperature and inhibition of gibberellin biosynthesis override phytochrome action and induce bud set and cold acclimation, but not dormancy in PHYA overexpressors and wild-type of hybrid aspen. Plant, Cell & Environment, 28(12), 1579–1588. https://doi.org/10.1111/j.1365-3040.2005.01395.x [Google Scholar] [CrossRef]
98. Liu, J., Sherif, S. M. (2019). Hormonal orchestration of bud dormancy cycle in deciduous woody perennials. Frontiers in Plant Science, 10, 6484. https://doi.org/10.3389/fpls.2019.01136 [Google Scholar] [PubMed] [CrossRef]
99. Wang, J., Song, L., Gong, X., Xu, J., Li, M. (2020). Functions of jasmonic acid in plant regulation and response to abiotic stress. International Journal of Molecular Sciences, 21(4),1446. [Google Scholar] [PubMed]
100. Hu, Y., Jiang, Y., Han, X., Wang, H., Pan, J. et al. (2017). Jasmonate regulates leaf senescence and tolerance to cold stress: Crosstalk with other phytohormones. Journal of Experimental Botany, 68(6), 1361–1369. https://doi.org/10.1093/jxb/erx004 [Google Scholar] [PubMed] [CrossRef]
101. An, J. P., Wang, X. F., Zhang, X. W., You, C. X., Hao, Y. J. (2021). Apple B-box protein BBX37 regulates jasmonic acid mediated cold tolerance through the JAZ-BBX37-ICE1-CBF pathway and undergoes MIEL1-mediated ubiquitination and degradation. New Phytologist, 229(5), 2707–2729. https://doi.org/10.1111/nph.17050 [Google Scholar] [PubMed] [CrossRef]
102. Zhao, M. L., Wang, J. N., Shan, W., Fan, J. G., Kuang, J. F. et al. (2013). Induction of jasmonatesignalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant, Cell & Environment, 36(1), 30–51. https://doi.org/10.1111/j.1365-3040.2012.02551.x [Google Scholar] [PubMed] [CrossRef]
103. Ba, L. J., Kuang, J. F., Chen, J. Y., Lu, W. J. (2016). MaJAZ1 attenuates the MaLBD5-mediated transcriptional activation of jasmonate biosynthesis gene MaAOC2 in regulating cold tolerance of banana fruit. Journal of Agricultural and Food Chemistry, 64(4), 738–745. https://doi.org/10.1021/acs.jafc.5b05005 [Google Scholar] [PubMed] [CrossRef]
104. Neale, D. B., Kremer, A. (2011). Forest tree genomics: Growing resources and applications. Nature Reviews Genetics, 12(2), 111–122. https://doi.org/10.1038/nrg2931 [Google Scholar] [PubMed] [CrossRef]
105. Kreps, J. A., Wu, Y., Chang, H. S., Zhu, T., Wang, X. et al. (2002). Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiology, 130(4), 2129–2141. https://doi.org/10.1104/pp.008532 [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2023 The Author(s). Published by Tech Science Press.
Copyright © 2023 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools