 Open Access
Open Access
REVIEW
Advances in Grapevine Breeding: Integrating Traditional Selection, Genomic Tools, and Gene Editing Technologies
1 Facultad de Ciencias Agrícolas y Forestales, Universidad Autónoma de Chihuahua, Km 2.5 Carretera a Rosales, Campus Delicias, Delicias, C.P. 33000, Chihuahua, Mexico
2 Facultad de Posgrado, Universidad Técnica de Manabí, Portoviejo, 130105, Ecuador
3 Instituto Politécnico Nacional, CIIDIR Unidad Sinaloa, Blvd. Juan de Dios Bátiz-Paredes 250, San Joachín, Guasave, C.P. 81101, Sinaloa, Mexico
4 Centro de Investigación en Alimentación y Desarrollo (CIAD), Unidad Delicias, Av. Cuarta Sur 3828, Fracc. Vencedores del Desierto, Delicias, C.P. 33089, Chihuahua, Mexico
* Corresponding Author: Sandra Pérez-Álvarez. Email:
(This article belongs to the Special Issue: Adaptation Mechanisms of Grapevines to Growing Environments and Agricultural Strategies)
Phyton-International Journal of Experimental Botany 2025, 94(12), 3749-3803. https://doi.org/10.32604/phyton.2025.072135
Received 20 August 2025; Accepted 07 November 2025; Issue published 29 December 2025
Abstract
Grape (Vitis vinifera L.) cultivation has progressed from early domestication and clonal propagation to modern, data-driven breeding that is reshaping viticulture and wine quality. Yet climatic and biotic constraints still impose heavy losses—downy mildew can reduce yields by ≈75% in humid regions and gray mold by 20–50%—sustaining the need for resistant cultivars. Producer selection, interspecific crossing, and formal improvement programs have generated ~10,000 varieties, although only a few dozen dominate global acreage. Conventional breeding has delivered fungus-resistant “PIWI” cultivars that retain ≥85% of the V. vinifera genome; in Austria, national PIWI varieties are gaining acceptance for combined resistance to downy and powdery mildew and strong enological quality, while in Brazil, using ‘BRS Isis’ as a male parent produced a high proportion of seedless progeny. Over the past two decades, mapping studies have identified >30 resistance loci to Plasmopara viticola (Rpv) and 15 to Erysiphe necator (Ren/Run), enabling MAS and locus pyramiding; widely deployed loci include Rpv1, Rpv3 haplotypes, Rpv10, Rpv12, Run1, Ren1, Ren3, and Ren9. Gene editing further expands options: CRISPR knockout of VvMLO3 confers powdery-mildew resistance, whereas VvPR4b knockout increases susceptibility to P. viticola, highlighting both opportunity and gene-specific risk. To date, no consolidated program- or country-level percentages exist for MAS/CRISPR adoption in grape. Instead, proxy indicators—MAS screening throughput, the number of programs employing MAS, and CRISPR’s laboratory/pilot status with no commercial releases—suggest broad operational MAS and early-stage CRISPR implementation; for example, Germany reported >23 disease-resistant grapevine varieties developed with MAS and the loci above by 2022. Finally, this review analyzes the future of grapevine breeding, with a particular emphasis on the adoption of novel approaches to multi-omics, AI in breeding models, and sustainability for improving breeding schemes. An interdisciplinary effort will be required to find future solutions, as viticulture has entered a precision breeding revolution to address the challenges posed by the industry and the fight for long-term sustainability of grape production.Keywords
The grapevine (Vitis vinifera L.) originated in the Caucasus region, from where it spread to the Mediterranean, the rest of Europe, and later to other parts of the world. Its domestication began at least around 6000 years ago, and its uses include the production of wine, vinegar, and juice, as well as consumption as fresh fruit or dried as raisins. Grapevine is the fruit that occupies the largest cultivated area on the planet, with more than 7 million hectares [1].
It is estimated that there are around 10,000 grape varieties worldwide [2], resulting from traditional selection by winemakers and scientific breeding efforts that began in the 19th century. This has led to a remarkable diversity in the physiological and agronomic characteristics of grapevines, as well as in the organoleptic properties of their fruits. However, large-scale commercial production has resulted in only a few dozen varieties—such as Cabernet Sauvignon, Chardonnay, and Merlot—occupying the largest cultivated areas. Consequently, the actual genetic base of global viticulture has eroded [3], reducing the adaptability of varieties to new challenges.
Among the main contemporary threats to viticulture are bacterial diseases such as Pierce’s disease, caused by Xylella fastidiosa [4], and fungal diseases such as gray mold (Botrytis cinerea), downy mildew (Plasmopara viticola), and powdery mildew (Erysiphe necator) [1]. Climate change events are altering plant growth patterns and their yield [5] as well as wine quality [6]. In addition, the intensification of extreme conditions caused by climate change exacerbates the effects of water deficit susceptibility in grapevine varieties [7] and increases the damage caused by pathogenic microorganisms.
Genetic improvement offers a medium- and long-term solution to overcome these challenges, and encompasses a wide range of techniques and approaches. Hybridization among V. vinifera varieties has mainly aimed to increase yield and obtain wines with specific properties, while interspecific hybridization is of great interest to breeders because germplasm from other Vitis species can provide genes for resistance to biotic and abiotic factors [8]. Clonal selection of outstanding genotypes has focused on obtaining individuals distinguished by their enological qualities and/or their tolerance to different types of stress [9,10]. The use of rootstocks, although not considered by some authors as a genetic improvement technique per se, enhances fruit production and wine quality and contributes to pest and disease management [11].
The conservation and recovery of wild Vitis varieties and related taxa are contributing both to the development of genotypes tolerant to biotic and abiotic stress factors and to their use as parents in breeding programs, owing to the valuable genes they contribute to the progeny [8,12].
Recent advances in molecular biology have been increasingly applied to support genetic improvement. Genomic studies [13], research on genetic variability using molecular markers [3], genome editing with CRISPR/cas9 [14], and the identification of genes for resistance to fungal diseases [1] are some examples of the contribution of molecular biology to the development of promising genotypes.
This article reviews the progress achieved in the genetic improvement of Vitis vinifera L. through conventional breeding methods, traditional propagation, and in vitro culture techniques, as well as advances in the use of marker-assisted selection, genome modification, and gene editing. Particular emphasis is placed on the application of multi-omics technologies in breeding, and the recent introduction of artificial intelligence for this purpose. In this way, an integrated overview is provided of the achievements in grapevine improvement and its future prospects.
2 Traditional Breeding Methods
The beginning of genetic improvement in V. vinifera can be traced back to the late 19th century. The introduction of varieties from North America to Europe brought with it a dangerous insect (Daktulosphaira vitifoliae), commonly known as phylloxera, capable of destroying entire vineyards. Its appearance in 1863 in France and Great Britain was followed by a rapid spread across Europe, Australia, and American countries. The immediate solution was the use of North American Vitis species, resistant to phylloxera, as rootstocks, primarily V. berlandieri Planch, V. riparia Michx., and V. rupestris Scheele [15].
Grafting is not considered, in itself, a genetic improvement technique, since there is no genetic exchange between the rootstock and the scion, nor is a hybrid progeny obtained. However, several authors include this practice in discussions of genetic improvement for the following reasons:
- ➣It has been shown that in grafted plants, the genetic characteristics of the rootstock are expressed, such as enhanced absorption of nutrients like nitrogen, phosphorus, potassium, calcium, and magnesium, among others [16,17], which improves performance and leads to higher yields.
- ➣Grafting modifies the expression of genes associated with stress resistance [18,19], resulting in better plant behavior against biotic and abiotic factors.
- ➣The vascular connection between rootstock and scion enables communication through chemical and hormonal signals, which in turn modifies the scion’s transcriptome [20]. Consequently, although no genetic changes occur in the scion, the expression of rootstock-derived genes can be detected in it.
Rootstocks commonly used belong to species of the Vitis genus, primarily distinguished by their tolerance to biotic and abiotic factors. In Brazil, the rootstocks IAC 313 and IAC 572, developed through hybridization at the Instituto Agronômico de Campinas between 1950 and 1970, are regarded as the cornerstone of tropical viticulture in that country [21]. Their introduction enabled grapevine cultivation under Brazil’s warm and humid climate, to which other European rootstocks were poorly adapted.
Species such as V. labrusca, V. aestivalis, V. rupestris, V. smalliana, V. caribaea, V. shuttleworthii, and V. berlandieri have been, and continue to be, used as rootstocks to confer on the scion desirable traits such as resistance to viruses, nematodes, soil acidity, drought, and salinity, among other biotic and abiotic stresses [22]. In China, the rootstocks LDP-191 and LDP-294 (V. piasezkii Maxim. var. pagnucii) have provided cold tolerance to the table grape cultivars ‘Fujiminori’ and ‘Red Globe’ [23]. In Florida, grafting onto Vitis vulpina L. has enhanced resistance to nematodes, whereas Vitis champinii Planch. is known as a rootstock resistant to Pierce’s disease, drought, and nematodes [24].
Other Vitis species, such as V. rotundifolia, V. coignetiae, and V. thunbergii, although recognized for their strong resistance to fungal pathogens, nematodes, and temperature extremes [25], cannot be employed as rootstocks for V. vinifera due to graft incompatibility. Nevertheless, the success of grafting V. vinifera cultivars onto other compatible rootstocks led, by the 1980s, to the widespread replacement of own-rooted vineyards with grafted vines [26]. An interesting breeding strategy involves interspecific hybridization between Euvitis and Muscadinia, aimed at developing rootstocks with enhanced adaptive traits [27], although the progeny from such crosses generally exhibits low viability because of chromosomal and vascular incompatibilities.
In order to obtain rootstocks that confer desired characteristics to the scion, hybridization programs between Vitis species have been developed, and outstanding hybrids have been selected, for both direct use as rootstocks and as parents [22,23,26,27,28,29].
In traditional breeding through hybridization, the main objectives, either separately or together, have been those examined below.
Disease Resistance
In general, commercial varieties of Vitis vinifera are, to a greater or lesser extent, susceptible to diseases such as downy mildew (Plasmopara viticola) and powdery mildew (Erysiphe necator). The strategy for improvement has been the hybridization of European commercial varieties with North American Vitis species. Based on the studies conducted so far, the use of V. rotundifolia and V. piasezkii in crossing confers complete resistance to the progeny [30].
The varieties obtained through this method are called “PIWI”, a term derived from the German “Pilzwiderstandsfähig” (fungus-resistant). The acceptance of these new varieties is growing, as it has been demonstrated that they retain at least 85% of the original V. vinifera genome. Among these varieties are the German “Regent” and “Solaris”, which combine proven resistance with the production of wines of adequate quality [31]. A detailed list of PIWI varieties is publicly available and can be consulted at the International Piwi organization (https://piwi-international.org).
In Brazil [32] resistance to P. viticola in three PIWI varieties obtained in Germany was significantly higher than that of the susceptible control. Ten PIWI varieties evaluated in Italy showed an average 93% reduction in P. viticola infection compared to the susceptible control variety “Pinot Grigio” [33]. In Austria, three national PIWI varieties are gaining great acceptance for their resistance to both diseases and their high enological quality [34]. In La Rioja, Spain, the introduction of five white PIWI varieties and four red ones has also yielded good results [35].
Climate Adaptation and Tolerance to Abiotic Stress
Among the challenges already faced by this crop are spring frosts, which occur beyond the winter season, and excessive heat at the beginning of autumn. Frosts can damage the shoots, preventing the plant from forming grape clusters, whereas excessively high temperatures lead to over-ripening, producing musts with excessive sugar and consequently high alcohol content. The solution lies in the search for varieties with late budding and ripening, to prevent these phenological phases from coinciding with frosts and off-season heat [36].
Fortunately, there is considerable diversity in traits related to climate change adaptation, such as polyphenol content in fruits [37] and drought tolerance [38]. It has also been observed that there is a positive correlation between fruit size at maturity and malic acid content, and a negative correlation with tartaric acid content in a set of 33 genotypes [39]. The study of these traits and their expression under adverse conditions can contribute to the development of breeding programs to address climate change.
Traits such as cold tolerance are also of interest for genetic improvement. Selection in the progeny of crosses between V. vinifera and other species has led to the development of varieties suited to the harsh winter conditions of northern states in the U.S. [40]. Since 2011, North Dakota State University has conducted a hybridization program with this goal, and recently, genetic-molecular tools have been incorporated into the research [41].
An interesting result was obtained [42] when investigating daytime and nighttime transpiration of two varieties (“Syrah” and “Grenache”) and 186 descendants from their cross. A negative correlation was observed between nighttime transpiration (resulting from poor stomatal closure) and plant growth, both under normal water supply and under water deficit conditions. However, more basic studies are needed to better understand the mechanisms associated with tolerance. For instance, it is known that the response of grapevine varieties to drought involves both abscisic acid-dependent and -independent mechanisms, but the role of each and their interaction remain unclear, as does, the genetic regulation of root system performance, xylem, and other factors contributing to tolerance [43]. Nonetheless, some recent studies have reported success in breeding for drought stress [44], salinity [45] and heat tolerance [46,47].
Agronomic Characteristics and Agricultural Yield
Today, yield is not a priority for breeding; the most pressing issue has been solving other problems previously mentioned, such as disease resistance and adaptation to abiotic stress. Of course, obtaining varieties with these characteristics implies maintaining adequate yield and commercial quality, so evaluating these parameters is essential for achieving crop sustainability [48].
Grafting, used as a practice to improve resistance or tolerance to biotic and abiotic factors, indirectly influences agricultural yield, as it provides the scion with benefits such as more efficient nutrient absorption [16,17] and enhanced defense mechanisms against stress [18,19]. Additionally, grafting has been observed to directly impact yield: scions of Cabernet Sauvignon and Chardonnay grafted onto 15 different rootstocks increased their yield by approximately 50%, due to higher fruit weight [49]. The authors suggest that this effect likely results from rootstocks enhancing water absorption by the plants.
A review of research conducted in countries with a long tradition of grapevine genetic improvement reveals that the main breeding efforts have historically focused on other primary objectives [50]. While yield remains an essential trait that must be maintained in newly developed varieties, the focus has expanded to encompass crop sustainability through the management of multiple factors.
Fruit Quality and Chemical Composition
Among the parameters considered to evaluate quality are sugars, which determine the alcohol content in the wine; polyphenols, which provide body and color to the must; organic acids, which influence acidity; cations, which affect pH; and other compounds related to aroma and flavor. Although the exact ways in which temperature and other factors affect the composition of the fruit are not yet fully understood [51], genetic improvement considers this one of its main objectives.
Not all crosses aimed at improving tolerance to adverse climatic conditions result in fruits capable of producing wines of adequate quality. For example, the progeny of V. vinifera × V. riparia, while producing descendants able to withstand the low temperatures of the northern states of the U.S., fails to meet the quality standards required by viticulturists [40,52]. Crosses of V. vinifera × V. amurensis to obtain cold-tolerant progeny in China have also produced some varieties [53], though their enological quality remains low.
It seems that hybridization of V. vinifera with other species, while tending to produce individuals with some environmental plasticity, often reduces wine quality. In contrast, crosses among V. vinifera varieties known for their enological quality can yield new varieties whose properties match or even surpass those of their parents. The hybridization of the varieties “Graciano” and “Tempranillo” generated several offspring producing grapes with high polyphenol content, adequate acidity, intense color, and excellent aroma [54].
As in any hybridization strategy, combining parents in grapevine faces issues related to pollen fertility and offspring viability. These variables were studied in six V. vinifera varieties and their progeny during 2020 and 2021 [55]. “Ecolly” was the variety with the highest pollen fertility, suggesting its use as a male parent. Conversely, the variety “Marselan” exhibited a self-fertilization index between 0.88 and 0.93, indicating that its use as a female parent may be challenging.
Seedless Grapes
The absence of seeds is a highly valued trait in table grapes and raisins [56], and breeding programs have been developed with this goal in mind.
In Brazil, 200 descendants from 38 combinations were evaluated over six seasons between 2018 and 2021 [57]. The crosses involved 30 cultivars of V. vinifera and interspecific hybrids. The fruits of the progenies were classified as seedless, soft-seeded, and seeded fruits. Around 20% of the hybrids produced seedless fruits; the use of the Brazilian variety “BRS Isis” as a male parent resulted in a high proportion of seedless descendants.
In Turkey, one of the world’s leading producers of table grapes, hybridization programs have also been developed to continue improving fruit quality. For this purpose, seedless V. vinifera varieties, either Turkish or American, have been used as male parents, while both V. vinifera and V. labrusca varieties have served as female parents [58]. The progeny has been characterized, and several outstanding cultivars have been recommended for evaluation in different regions of the country.
It is known that inheritance of this trait is determined by three independent recessive genes controlled by the dominant gene SdI [59]. This knowledge has enabled the use of more precise biotechnological tools for the development of seedless grape varieties, such as embryo rescue and marker-assisted selection [56].
3 Clonal Propagation and Selection
Traditional Cloning
Grapevine cloning dates back thousands of years [60], and according to Walter [61] a clone is defined as the offspring of a crop, in this case grapevine, selected for its varietal purity, phenotypic characteristics and health status.
Since the 1960s, the focus of clonal selection in grapevines has been on developing high-yield, virus-free clones that have higher sugar content in the fruit. However, the quality potential of these certified clones can differ based on the grape variety. While clonal selection is generally embraced for its benefits, it does come with two main downsides: first, the limited qualitative impact of clonal selection in certain varieties like Cabernet Franc and Sémillon, as well as many secondary varieties such as Carmenère, Malbec, Petit Verdot or Cot; and second, the significant loss of genetic diversity within grapevine populations when clonal selection is the only method used to enhance plant material [62,63]. Fig. 1 shows the clonal selection process for grapevine cultivation.
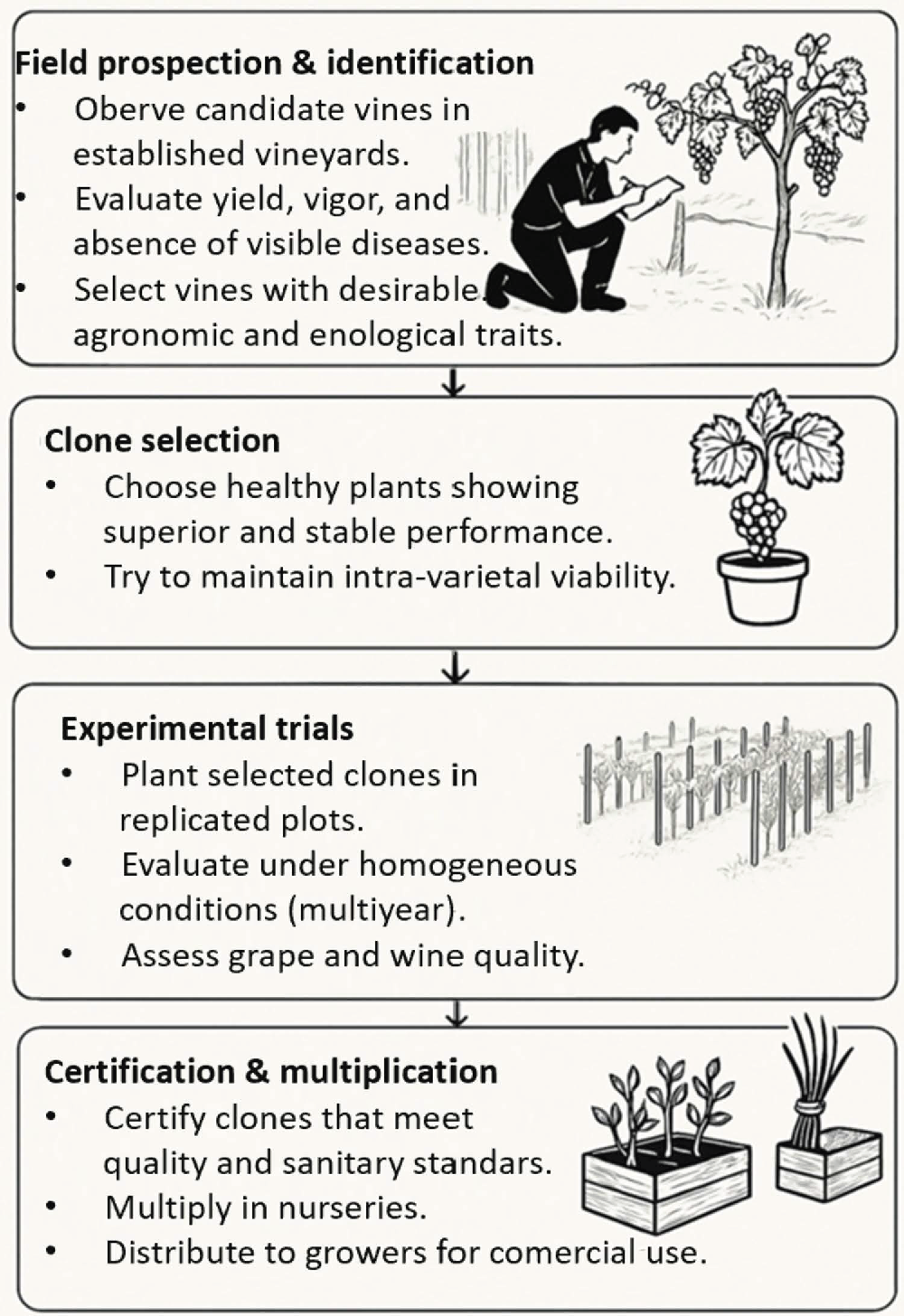
Figure 1: Traditional clonal selection workflow for grapevine cultivation.
According to Fig. 1 the clonal selection process for grapevine cultivation begins with the identification of promising clones, which involves detailed assessments of agronomic traits (such as yield, vigor, and growth habit) and enological characteristics (including sugar content, acidity, and flavor profile) to determine their potential for high-quality grape and wine production; secondly, the selection of the clone process involves two main stages that are genetic and sanitary selection, which together consider genetic diversity while maintaining varietal purity prevent the propagation of virus-infected or disease-prone plants and mitigates the negative effects of mutations in future vineyards [64,65]; thirdly, only the healthiest and most promising clones are chosen to ensure long-term sustainability. These clones are tested under uniform conditions to determine their capacity for producing high-quality grapes and to support their certification and availability to growers [66]. Finally, the certification of a grapevine clone involves a thorough evaluation to ensure it meets strict standards for genetic identity, agronomic performance, and phytosanitary status (e.g., free from viruses and other diseases). Once certified, the clone is approved for commercial propagation and can be distributed to nurseries and growers for large-scale cultivation, ensuring consistency in vine quality and performance across vineyards [67].
A major disadvantage of cloning is that all offspring will have the same characteristics as the mother plant, so any biotic or abiotic factor that affects that vineyard will cause all plants to suffer the same consequences. Therefore, it is important to have a large number of selected clones to obtain a good response to the pressure of natural selection (new pests, droughts, excessive rainfall, salinity, climate changes, etc.), improve the quality of the wines and not affect the genetic variability of the cultivars. The best technique to achieve this is to use old vineyards for preselection, taking into account first of all the number of vineyards and not the number of plants per vineyard, thus being able to have greater variability in the selected phenotypes [62]. Nevertheless, once clones propagate the line, they accumulate somatic mutations. Some of these mutations have become a new resource for genetic improvement [68,69]. Fig. 2 presents the benefits and risks of grape propagation by clones, aspects that are analyzed in this section theoretically or through case studies.

Figure 2: Benefits and risks of clonal propagation in grapevine.
By the 20th century, these somatic changes became the foundation for clonal selection in viticulture. Through clonal propagation for over nine centuries, a number of variants of Pinot Noir have emerged via spontaneous bud mutation mechanisms, such as Pinot Meunier, Pinot Grigio, Pinot Blanc, and Pinot Gris [70]. The first reference genome for grapevines was built using PN40024, a highly homozygous line derived from PN [71].
The Institute for Grapevine Breeding at Geilweilerhof (JKI) in Germany has demonstrated the significant impact of genetic disease resistance on sustainable grape production through its breeding programs. A prime example is the ‘Regent’ cultivar, released in 1994, which possesses genetic resistance to downy mildew and powdery mildew. Despite moderate resistance levels now due to evolving pathogens, ‘Regent’ vines in 2021 trials still showed a 50% reduction in fungicide applications compared to susceptible cultivars, even under intense downy mildew pressure. This highlights the long-term potential of resistance breeding for environmentally friendly and economically viable viticulture. The JKI’s work underscores the importance of investing in grapevine breeding for durable disease resistance to reduce environmental impact, ensure economic viability for growers, promote biodiversity, and safeguard consumer health [72].
Complementing resistance breeding, traditional clonal selection has been pivotal for shaping regional wine styles and ensuring consistency. In Champagne (region in northeastern France), clonal breeding is the basis of grape growing in order to achieve wine style and varietal expression. Over decades of the clonal selection of Pinot Meunier, Pinot Noir, and Chardonnay vineyards, uniformity of yields has been attained, along with performance consistency, thus assisting in sustaining the world fame of the appellation. At the same time, clonal preservation of less-used cultivars—Pinot Blanc, Pinot Gris, Arbane, Petit Meslier—is now proving strategic: recent research indicates these minor varieties may ripen more appropriately in warmer vintages, and their reintroduction into blends could help maintain freshness and acidity under climate change [73]. Hence, clonal selection ensures varietal identity while simultaneously widening the adaptive range of the grower.
Extending the evidence base, the long-term trials in the Carpathian Basin highlighted the quantitative gains obtainable through clonal selection in multiple cultivars. For Welsch Riesling, the Vojvodina clone SK. 54 and Hungarian B. 20 outperformed unselected populations over 13–14 years, the traits being improved at a level of ~10% and yields were 30% more. Previously undisclosed cases obtained similar results for Pinot Gris: for yield-related traits, clones B. 10 (Hungary) and Gm. 2-54 (Germany) ranged from being 22 to 62% superior. These differences in yield-related traits further extended to large differences between clone effects seen in Chardonnay-French 75 being 83% greater in cluster yield and 57% greater in cluster size compared with Italian VCR. 4, whereas for White Riesling, clone Gm. 2-54 yielded 55% more than local unselected vines. Thus, these results prove that clonal selection may increase productivity and agronomic performance while maintaining varietal typicity [74].
Thus, after the clonal lines have been fixed, sanitation can improve final performance by sanitizing the viral constraints. Removing Grapevine leafroll virus (GLRaV-1, GLRaV-3) in clones already in use has been shown to promote vine vigor and better physiological functioning so as to improve grape and wine qualities, whereas in some cases, a virus-free clone of the same genotype offers better results despite likely not showing an increase in yield [75]. In summary, combining resistance breeding together with clonal selections and sanitation could create a coherent pathway toward a sustainable pool of viticulture, high quality, and lower-input.
In addition to winemaking, table grape cultivation has become a key source of agricultural income for communities in various parts of the world. Despite a general decline in global vineyard areas, grape production has risen, largely due to expanded table grape plantations and the improved productivity of newer cultivars [76]. Countries such as Morocco, Egypt, India, and China have seen notable growth in table grape production areas. Recently, the popularity of seedless grapes has driven market trends, with nearly all new grape varieties being seedless [77]. In response to evolving consumer preferences, numerous nations have launched breeding programs aimed at producing new grape varieties. These programs focus on developing seedless grapes with larger berries, unique flavors, and resistance or tolerance to various stress factors [78].
The clonal propagation efficiency was evaluated in a newly developed microvine variety for the production of seedless table grapes, considering two factors: (a) position of the cutting on the shoot and (b) length of the misting period (between 3 and 7 weeks). Some of the rooted cuttings were subsequently transferred to the hydroponic system under controlled conditions and monitored for establishment, initial growth, flowering, and fruit development. After 3 weeks, 83.7% of the cuttings rooted, regardless of which part of the shoot they came from or the duration of misting; 16.7% of the cuttings did not survive. After transplanting to the hydroponic system, 100% of the rooted cuttings were successfully established, and the fruit yield and quality did not vary among the cutting sources. The whole production cycle has a duration of 208 days, with 63 days in seedling development and 145 days from transplanting until the first harvest. Flowering occurred at about 33.9 days, while veraison started after around 116 days. Under these conditions, fruits complied with the sugar content requirements for the Australian market [79].
Developed by the Commonwealth Scientific and Industrial Research Organisation-like agency, the table grape breeding program in Australia focuses on cultivars customized to consumer needs. Seedlessness, large-size berries, good texture, and flavor are major attributes under consideration, along with resistance to the more common diseases. Varieties assessing early and late ripening are also included to increase the harvest window. It has been important, as well, to develop grapes that can live longer on the shelf, thus being stored and exported to faraway markets. As an example, being the first CSIRO variety offered to market, Marroo Seedless was born of a cross between Ruby Seedless and Carolina Blackrose, and it is a very productive large berries: seedless, crisp, black, sweet-flavored, downy mildew tolerant, but powdery mildew susceptible. In the US, it became an important variety with over 1.3 million boxes produced in the 1990s. Similarly, in selecting high-yielding Sultana clones that represented more than half of Australia’s total grape crop in the 1970s for wine and raisin production, the CSIRO played a pivotal role [76].
Anamaria et al. [80] conducted a study in which four varieties of table grapes—Moldova, Bican Roz, Victoria, and Centennial Seedless—were tested. In the first year, 10 clonal selections were made for each variety; in the years that followed, five elite clones per variety were selected. Clones derived from this selection exhibited normal development and showed a greater grape weight than those of non-selected clones. The clonal selections of Moldova and Bican Roz were well suited for transport and storage; in contrast, Victoria and Centennial Seedless were much better suited for fresh consumption. Additional uses of Centennial Seedless included raisining.
The clonal selection controls viral diseases and allows the targeted selection of genotypes with regard to agronomic, viticultural, or enological traits. On the other hand, while clonal selection may provide uniformity and stability, obtaining asexual clones also limits recombination, thus generating cases of genetic erosion. Genetic erosion in clonally propagated grapevines arises when reliance on a narrow set of elite genotypes limits recombination and gradually narrows the adaptive pool. Although vegetative propagation confers relative genetic stability, somatic mutations still occur [81]. On the positive side, such mutations can yield superior attributes or distinctive characters—occasionally giving rise to sports or new cultivars—but they may also disrupt key quality traits [82]. Crucially, asexual propagation fixes these mutations, so vineyards accumulate intra-varietal diversity over time, generating multiple clonal lines with distinct phenotypes within the same named variety [83]. This duality—stability with drift—means that, without deliberate conservation (massal selection, broader clonal portfolios, and genomic monitoring), clonal agriculture can erode genetic breadth even as it preserves typicity, ultimately reducing resilience to new pathogens and climate stresses. Studies have shown that excessive reliance on clonal reproduction may reduce the genetic resources available for biotic and abiotic stress adaptation. For example, Vondras et al. [84] also showed that long-term propagation of Zinfandel clones had led to thousands of somatic mutations accumulating, representing both the possibilities for divergence within the variety and the narrowing of genetic options through time. It was concluded in the study that intergenic repetition is a key factor in the genomic diversification of clones, as well as the accumulation of presumably deleterious mutations.
Case studies highlight the menace of erosion that arises with decades of vegetative propagation of grape cultivars. Strioto et al. [85] documented for the popular table grape, ‘Italia’ how extensive clonal propagation gave somatic variation that destabilized key agronomic traits like berry size and fertility. This shows how a long-term reliance on a few selected clones possibly can undermine the same uniformity it attempts to protect while simultaneously diminishing genetic receptivity to new viticultural challenges.
According to Roby et al. [86], the preservation of grapevine genetic resources requires more than just institutional clonal selection. Authors highlight the importance of combining clonal-type practices with those of massal selection in order to conserve intra-varietal diversity. Further, Pelsy [87] studied another paradox of clonality: it maintains desirable traits but limits wider levels of genetic exchange, potentially reducing adaptive ability. This tension implies that viticulture must walk a fine line between safeguarding clonal stability on the one hand and fostering diversity in the long run on the other. Taken together, the research stresses that the future of viticulture can be saved only by engaging in a deliberate act of balancing the positives of clonal propagation with strategies against genetic erosion induced by climate change and new disease pressures.
in vitro Cloning
Grapes, like all crops, are under various conditions of biotic and abiotic stress that affect their yield and fruit quality, and also demand has increased not only for fresh and dried fruit, but also for wines (increase in wine-producing industries) [88]. The issues mentioned above are generally addressed by generating and providing healthy propagation material, along with implementing genetic enhancement and introducing genotypes—such as exclusive varieties—that offer resistance or tolerance to pests, diseases, and varying soil and climate conditions [89,90,91].
In vitro culture, grounded in the theory of totipotency—the concept that complete plants can be regenerated from somatic cells [91]—enables the mass propagation of plants without requiring sexual reproduction (cloning). This method also yields plant material with superior phytosanitary quality compared to conventional propagation techniques. Among various biotechnological tools, in vitro culture stands out as a key technique for the propagation, conservation, and genetic improvement of plant species [92].
Genetic improvement programs for perennial species like grapevines are inherently slow and complex due to their extended life cycle [93]. As a result, more than 80% of grapevines have historically been propagated through vegetative means, a practice sustained for centuries to preserve desirable traits [94].
Some authors claim that depending on the table and wine grape cultivars, standardized culture media and their optimal conditions for in vitro propagation should be set up [95]. The major factors that can influence their propagation response include the cultivar type [96], the pruning method [97], genotype and the culture medium used [98], and the inappropriate or correct use of plant growth regulators [99].
As part of the breeding program at the Kazakh Scientific Research Institute of Fruit Growing and Viticulture, an in vitro collection was established to safely preserve grapevine hybrids, while also evaluating their resistance to Plasmopara viticola through the presence of the Rpv3 and Rpv12 loci. For in vitro initiation, plant material (18 selected hybrids of grapevines) consisted of either shoots collected directly from field-grown vines or budwood cuttings forced to sprout indoors. Disinfection of field-harvested shoots was most effective with a treatment of 0.1% mercuric chloride (HgCl2) for 5–7 min, achieving a moderate viability rate, with 17–21% of explants establishing successfully across all tested genotypes. The optimal growth medium was Murashige and Skoog (MS) [100] basal medium supplemented with 1 mg L−1 BAP, 0.1 mg L−1 IBA, and 0.1 mg L−1 gibberellic acid (GA) at pH 5.7, resulting in 4 to 4.4 shoots per explant. When using shoots from budwood cuttings sprouted in the laboratory, establishment was less effective. Although contamination rates decreased with longer HgCl2 exposure (from 65% at 5 min to 6% at 10 min), viability also declined, with high rates of necrosis (10% at 5 min and 87% at 10 min) and poor regeneration. Ultimately, 16 grapevine accessions were identified as possessing P. viticola resistance associated with the Rpv3 and Rpv12 loci [101].
Another in vitro way to improve grape cultivation and also to conserve resources is somatic embryogenesis (SE). In this process, somatic cells are stimulated to generate cells with embryogenic potential, which produce structures that give rise to a complete plant [102,103].
In short, SE using floral-origin explants is a fantastic way to regenerate healthy grapevine plants that are free from a host of viruses. While SE is a bit more complex and takes more time than the usual virus-cleaning methods [104], it has shown to be incredibly effective for grapevines. For chimeric grapevine cultivars, viruses can be removed through traditional techniques like meristem tip culture paired with thermotherapy, or by using cryotherapy, often requiring a mix of these methods [105,106].
Alongside traditional clonal selection in the vineyard, in vitro approaches have been instrumental in generating enhanced grapevine varieties, whether through sanitation, somaclonal variation, embryo rescue, or induced mutagenesis. Early studies illustrate the potential of these methods: the first establishment of somatic embryogenesis from cultured anthers was achieved in the 1970s [107,108], with germinable embryos later recorded in the interspecific hybrid Gloryvine. By 1977, vines derived from somatic embryos of cv. Seyval were already being planted commercially in Maryland [109], and in 1985 a U.S. patent formalized a grape SE protocol [110].
Sanitation via in vitro regeneration has also been reported. For example, embryos and plantlets obtained from ovaries of V. vinifera cv. Roobernet infected with Grapevine fanleaf virus (GFLV) tested negative for all leafroll-associated viruses, indicating successful elimination. However, when the same technique was applied to anthers of infected V. rupestris cv. Rupestris du Lot, regenerated plants remained infected [111], highlighting both the potential and the limitations of in vitro sanitation.
In parallel, embryo rescue has become a cornerstone of modern grapevine breeding. Over the past five years, nearly 60% of newly released seedless cultivars have been obtained through this technique, which recovers otherwise abortive embryos. Landmark achievements include the first stenospermocarpic, seedless hybrid between V. vinifera and V. rotundifolia [112]. More recently, programs combining stenospermocarpic female parents with seeded, cold-tolerant male parents produced cold-resistant seedless grapes, with 91 progenies showing seedlessness and 18 carrying cold tolerance [113].
Finally, induced mutagenesis has further expanded the scope of in vitro breeding strategies. Defined as the generation of new cultivars through novel genetic variation induced by physical or chemical agents [114], this technique has been tested in several grapevine cultivars. Munir et al. [115] reported that gamma irradiation enhanced plant height more effectively than chemical mutagens in V. vinifera cvs. Desi, Sundar Khani, and Chinese grape, with sodium azide being effective only at low concentrations (0.1%), while higher doses caused explant browning. Similarly, Kuksova et al. [116] demonstrated in V. vinifera cv. Podarok Magaracha that somaclonal variation and in vitro mutagenesis produced notable genetic diversity: 2.5% of regenerants were spontaneous tetraploids, gamma irradiation (5–100 Gy) increased tetraploid frequency to about 7% and induced some aneuploids, whereas colchicine failed to induce tetraploids. Subsequent field evaluations confirmed additional phenotypic variability among regenerants, underlining the capacity of in vitro mutagenesis to generate agronomically useful diversity.
Finally, a comparative summary of the main features of grape traditional breeding, traditional cloning and in vitro cloning is provided in Table 1.
Table 1: Comparative summary of grapevine improvement strategies: traditional breeding, traditional cloning, and in vitro cloning.
| Grapevine Breeding Method | Definition | Positive Aspects | Negative Aspects |
|---|---|---|---|
| Traditional breeding | Hybridizes existing grape varieties to combine desirable traits. | Can introduce new desired traits like seedlessness or disease resistance. | Slow due to the long-life cycle of grapes; difficult to create new seedless varieties using traditional methods, as embryos are often lost; grape varieties often lack resistance to diseases and environmental stresses; relies on existing genetic variability within the V. vinifera genus; high genetic variability; low propagation efficiency. |
| Traditional cloning | This is the offspring of a crop, in this case grapevine, selected for its varietal purity, phenotypic characteristics and health status. | Maintains the desirable characteristics of a specific cultivar; can be used to select for improved yield, resistance, or quality; very low genetic variability; moderate to high propagation efficiency. | The limited qualitative impact of clonal selection in certain varieties; the significant loss of genetic diversity within grapevine populations. |
| In vitro cloning | Complete plants can be regenerated from somatic cells. | The mass propagation of plants without requiring sexual reproduction; plant material with superior phytosanitary quality; rapid multiplication of desired genotypes; very low genetic variability; high propagation efficiency. | Can induce somaclonal variations or mutations, potentially altering the genetic makeup of the regenerated plants; the induction of somatic embryogenesis can be low and highly dependent on genotype. |
These approaches, taken together, illustrate the continuum of strategies found in grapevine breeding: from producing new cultivars to improving already existing cultivars and finally to applying biotechnological tools that more quickly cone propagation and adaptation.
Plant breeding has evolved parallelly with human populations, since the beginning of civilization when the human began to culture its own food, they started to select plants with desired characters, based on observations of individual plant phenotypes in order to produce offspring with desired traits [117]. Targeting plant breeding began implementing morphological markers, not only to visually distinguish and select desired characters, such as fruit, seed, flower and stem related traits, but also perceptible important agronomic traits, such as flavor and nutritional and sugar content. Additionally, main concerns in commercial and intensive grapevine production have focused on selecting characters related to improve yields and enhance biotic and abiotic stress resistance [118]. Morphological markers have consistently proven to be a valuable and readily available tool in plant cultivation, facilitating plant breeding initiatives without necessitating specialized expertise, methodologies, or equipment. Despite their utility, these markers present certain drawbacks, including the requirement for considerable, sustained effort and the potential for ambiguity. A primary concern with conventional plant breeding methods is their lack of specificity, as phenotypic traits can be influenced more by environmental factors than by genotype.
Advances in biotechnology and molecular tools have transformed plant breeding to molecular plant breeding, turning traits selection by genotype rather than phenotype-based tools, giving place to Marker-Assisted Selection (MAS). In MAS morphological/phenotypical traits are selected based on the genotype or molecular markers. A molecular marker is a small fragment of DNA sequence that acts as sign or flag closely related with changes in specific genes. These changes in DNA are majorly controlling gene expression of a particular gene that results in improved agronomic traits [119]. Once the molecular markers have been proven, this information is valuable to be used not only for targeting plant breeding but also to plant transformation by incorporating desirable agronomic traits either from other species or from wild-related species by introgression. Many of the agronomic traits are polygenic, which means that more than one gene/marker should be analyzed to increase the accuracy of the association. The advent of new technologies for massive DNA sequencing released the opportunity to analyze multiple molecular markers in parallel. This advance in DNA sequencing technology has revolutionized MAS by reducing time and efforts, enhancing targeting plant breeding efficiency [120].
With the advent of Next generation sequencing (NGS), a bunch of DNA molecular markers have been developed and are available to be successfully implemented in MAS for many crops and for genetic conservation of wild populations [121]. Application of MAS began around the 1980s but it was accelerated a decade later due to the emergence of the Polymerase Chain Reaction (PCR). Before NGS, most of research on MAS lied to a single gene amplified by PCR; variations in amplified DNA sequence were not only located in specific chromosome locations but also related to changes in agronomic traits, this technique was known as quantitative trait loci (QTLs) [117]. The number of publications with the terms QTL and MAS increased tremendously at least during the next two decades [120]. Despite the vast number of available molecular markers for plant breeding, choosing the right molecular marker is fundamental to have better results, the more polymorphic, the more informative to identify useful variations in the nucleotide sequences of DNA, enabling to identify alleles, either in the nucleotide composition or in the longitude of the sequence. Polymorphisms in DNA sequences are revealed by electrophoresis and molecular techniques such as restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), random amplified DNA polymorphism (RAPD), simple sequence repeats (SSRs), single-nucleotide polymorphism (SNP), among others. NGS enhance tremendously the genotyping technologies making possible to analyze hundreds of thousands of SNP markers by genotyping technologies such as genotyping by sequencing (GBS), restriction site associated DNA sequencing (RADSeq) diversity arrays technology (DArT) and others [122], allowing differentiation among individuals or populations more efficiently.
Since the huge pallet of molecular markers available, choosing the appropriate marker technique must respond to the research question. However, sometimes the choice of the molecular techniques is based on the previous knowledge of the species, such as genome size, genetic diversity, frequency of repetitive DNA in the genome, among others. Furthermore, considerations regarding technical know-how, infrastructure, funding research, are also crucial to choose the right markers to be working with [123]. MAS might be performed directly or indirectly by identifying polymorphisms in DNA sequences that are closely linked to underlying genes. When the association is robust, the polymorphisms serve as practical flags that signal the gene or surrounding gene associated with agronomical traits independently if the gene is known or not [124]. Advances on NGS technologies have revolutionized the way of genotyping and massive SNP marker analysis. Plant genome sequencing is now available for many crop species, which represents unprecedented advances for MAS and generation of elite varieties for many crops. This advance in knowledge and technical capability might enhance the way to identify specific and nonspecific genes, and molecular markers associated with desirable agronomical traits. MAS is a powerful tool that enables plant breeders to make more accurate selection as early as first stages of plant development, enhancing tremendously the efficiency to develop elite varieties, especially in long-life and woody perennial plants such as grapevine. Whole genome sequencing has enhanced the understanding of structural and functional genomics, stating valuable knowledge on the genetic architecture of plants. This knowledge makes it possible to develop complex statistical models for genomic selection that can be very informative for predicting the performance of a given molecular marker for plant breeding [118]. One of the most known techniques for association markers is the Genome-wide association study (GWAS), which is an approach that involves scanning markers across the genomes of many plants/cultivars and integrates statistical models to help plant breeders to find genetic variations such as SNP markers associated with desired agronomical traits. Though GWAS and other association marker studies such as QTLs enable identifying associated SNP markers or QTL statistically linked to traits, these regions are not always causative. It has been reported that SNP markers identified by GWAS techniques as highly associated to traits, often are not causal one but they are in linkage disequilibrium (LD) with main causative genes [125,126]. LD is referred to the non-random association of gene polymorphisms at different genetic loci in a population, meaning certain alleles appear together often and this is due to the physical proximity of these alleles on a chromosome rather than real association between the genotype with the phenotype.
The advances in high-throughput genotyping technologies and statistical models have tremendously enhanced the accuracy of GWAS in plant breeding research. However, the more complexity of data, the more resources and bioinformatics expertise resources are needed for analysis and biological interpretation. Models on GWAS have increased the accuracy to identify SNP markers, QTL or other DNA variations that are strongly associated with desirable agronomic traits by analyzing and comparing genomes of more diverse species or breeding populations [127,128]. Recent evidence suggests that a huge number of desirable traits for plant breeding, including disease or pest resistance have been identified through GWAS, including complex traits enriched near genes or polygenic trait-associated genetic variants [129]. Hence, trait-associated gene discovery can be empowered by elucidating the aggregated effect of a set of variants not only within but also around a gene, where transcriptional factors play fundamental roles through gene regulation. For polygenic traits as seedlessness in grapevine varieties, conducting gene-based association mapping should be integrated by analyzing GWAS information, Linkage Disequilibrium (LD) and molecular QTL data, complemented by populations genetics and comparative genomics from a reference sample [127,129,130].
Grapevines (Vitis vinifera) is an important fruit crop; it has been a source of food and wine since their domestication. Due to its economic importance, grapevines have been highly studied, becoming a model organism for studying perennial fruit crops. The whole genome sequence of V. vinifera was one the pioneers in fruit crops, the first draft genome sequence was released in 2007 by “The French–Italian Public Consortium for Grapevine Genome Characterization”, PN40024 [71]; since then, it has been used as genome reference for thousands of GWAS, trait-gene related and transcriptomic studies. A new, more complete reference genome, developed with advanced long-read sequencing, offers unprecedented detail and accuracy, overcoming limitations of previous techniques in repetitive and complex regions [131]. This resource is expected to significantly impact various fields, including research into biotic and abiotic stress in grapevine cultivars [132]. Gene annotation in grapevines has been in continuous evolution for both structural and functional genomics, however, a high complexity of the genome makes it difficult to fully understand the grapevine genome [133]. Now, technologies of massive sequencing and bioinformatics are fastly evolving [134]. V. vinifera has shown a very high genetic diversity with a high number of commercial cultivars, some of them have already genome assembled [135,136,137]. The genomes of most commercial grapevine varieties are available on a web portal for grape research (https://www.grapegenomics.com/index.php). However, it is still a challenge to understand complex traits, such as disease and pest-resistance, berry metabolites, morphology, seeds related traits and other oenological characteristics. Genomic regions associated with these traits have been widely identified through GWAS, LD studies and QTL analyses. However, most of the traits of interest for grapevine breeding are polygenic and show a complex quantitative trait-association [138,139] making MAS for grapevines a difficult and laborious process.
The integration of MAS with advances in agriculture of precision have increased the development of new cultivars adapted to climate change, higher yield and other desirable traits. Grapevine cultivars serve various purposes, either by consumption in fresh or processed in different products such as wine, raisins, juice and other applications. The development of products obtained from grapevine has become increasingly diversified, nowadays there are a high number of commercial cultivars that are product-specific, growing in different regions. However industrial production demands for new cultivars of even more productive and high-value products, promoting the development of new strategies for grapevine breeding [139]. Genetic mapping for QTL detection has been one of the widely used and preferred methods for MAS in grapevines. The high-density genotyping and genetic mapping have tremendously increased with the advent of massive sequencing, giving place to Association Mapping (AM) studies. However, as stated before, strategies for dissecting and selecting complex and polygenic traits should be integrative, including tools such as LD, GWAS, QTL and AM [140]. AM studies consider a big and diverse collection of germplasm, which might include commercial cultivars, inbreeding lines and/or wild populations. This set of diverse genotypes comprise an association mapping for discriminating new QTLs for targeting breeding, increasing the number of alleles available at each locus for a selected trait [138]. However, AM studies must be taken cautiously since the mix of geographically distinct genotypes with different pedigree might result in false-positive marker trait association by confounding effects of population structure [141]. Fortunately, many statistical methods such as estimation of false discovery rate (FDR), and Bonferroni correction have been developed to reduce and correct the false-positive associated traits in AM studies [142].
AM provides higher resolution compared to inaccurate QTL methods, especially in germplasm with high genetic diversity and low LD such as grapevines [140]. Thus, AM has been considered as the most widely accepted path to disentangle the right association between genotype and phenotype diversity for a high number of crops [142]. In grapevines, AM studies have been a useful tool to explain the genetic basis for complex traits of agronomic interest, such as yield, disease and pest resistance [143], cold tolerance [144], berry metabolites, berry morphology, phenology, vegetative traits, seed related traits [129], cluster related traits and abiotic stress to enhance resilience facing global warming [142]. Despite the limitations, the process of developing new grapevine cultivars through AM studies have shown very effective results by reducing the time of dissecting meaningful genetic markers effectively linked to the phenotype. Integrative approaches have shown better and more accurate results in plant breeding research. Technical advances in knowledge, technologies and the generation of more specific models to reduce the error rate enables new approaches to resolve previous QTL limitations in grapevine research.
Recent strategies have focused on identifying genetic markers associated with advantageous agricultural characteristics. Genomic Selection (GS) and Marker-Assisted Selection (MAS) are both standard methodologies utilized in plant breeding for the identification of genetic markers associated with agronomic traits. While both serve this purpose, they differ significantly in their approaches, primarily concerning the number of markers utilized per trait. MAS, an older and more established technique, employs a considerably smaller number of markers. This makes it effective for simple traits controlled by a limited number of QTL markers. In contrast, GS is a more recent and powerful tool. It considers a large set of markers distributed across the entire genome, offering a statistically more robust approach for identifying loci associated with desired characteristics. Consequently, while MAS is efficient for simple traits, GS demonstrates a superior success rate for complex traits that are controlled by multiple QTLs.
The publication of the V. vinifera pangenome represents a significant advancement for viticultural research and breeding [145]. This comprehensive genetic resource encapsulates most of the genetic diversity of Vitis spp. furnishing a powerful instrument for comprehending characteristics such as disease resistance, fruit quality, and climate adaptability. In contrast to preceding single reference genomes, the pangenome offers a more exhaustive genomic perspective, thereby enabling breeders to identify and leverage a broader spectrum of advantageous alleles. This augmented genetic understanding is projected to accelerate the development of novel grapevine cultivars that exhibit greater resilience to environmental exigencies, necessitate reduced chemical interventions, and produce superior quality fruit, ultimately benefiting the global wine and grape industries.
In conclusion, grapevine breeding research has experienced remarkable advancements in recent years, primarily driven by the synergy of NGS and bioinformatics, alongside the comprehensive integration of various omics approaches. These include genomics, transcriptomics, proteomics, metabolomics, and epigenomics, all of which contribute to a holistic understanding of grapevine biology. The availability of high-quality and integrative genomic information is pivotal. It not only enhances the resolution and precision of MAS but also provides novel frameworks for unraveling the intricate mechanisms of phenotype-genotype interactions. Understanding how specific genetic variations translate into observable traits is crucial for targeted breeding efforts.
Moreover, it is essential to analyze the results from MAS, GS, and AM in conjunction with data generated through integrative genomics. This combined approach allows for a more profound comprehension of the functional metabolic pathways that underpin complex traits in grapevines. By correlating genetic markers and genomic regions with expression patterns, protein profiles, metabolic fluxes, and epigenetic modifications, researchers can identify key regulatory elements and biological processes influencing desired characteristics such as disease resistance, fruit quality, and climate adaptability. This comprehensive integration of diverse data types is critical for developing more resilient and productive grapevine varieties, ultimately contributing to the sustainability and advancement of viticulture.
Despite the advances in plant breeding technologies, food security is still insufficient due to the increasing worldwide population. Advances in biotechnology and MAS have boosted crop yield by not only identifying and selecting but also by incorporating new traits into plants. The emergence of recombinant DNA techniques in the 1970s marked the birth of genetically modified (GM) crops, which claimed the ability of improved pest/disease resistance, yield and nutritional value [146]. GM crops are plants whose DNA has been genetically engineered and utilized in agriculture. The Plant DNA of these plants is manipulated by insertions of specific segments of foreign DNA into the host plant genome, mediated either by Agrobacterium tumefaciens or by direct gene transfer through recombinant technology. The genes inserted into the genome of the host plant can be naturally present in the same or related species (cisgenesis) or different species (transgenesis). Cisgenesis involves no foreign DNA, thus might not be considered as a strict plant transformation since genes are or were already present in the same or closely related species, conserving genetic diversity naturally present [147]. Transgenesis, on the other hand, is more controversial since it involves modification on the DNA by the introducing foreign genes from non-related species, which often implies ecological and biosafety concerns. Despite advances in knowledge, GM technology is still questionable, main concerns are over potential environmental and health impacts that avoids the complete acceptance and difficult its regulation [148].
Notwithstanding the concerns about using GM crops, the adoption of GM technology is quickly increasing. The pioneer GM crops were mostly herbicide and/or pesticide-resistant in the early 1990s, stating the beginning of the “Gene revolution” era. First commercially available GM crops were introduced to the USA in 1994. By 2014, the USA had become the largest producer of GM crops, first cultivated transgenic crops in the USA were represented by maize (Zea mays L.), soybean (Glycine max), and cotton (Gossypium hirsutum) plants, representing more than 90% of its production. GM crops were also worldwide accepted, representing an increase from only 1.7 million ha cultivated in 1996 to 190.4 million ha by 2019 [148]. For grapevine, first efforts in GM were mainly focused on incorporating resistance genes for susceptibility to fungus attack. Resistant genes were isolated from Trichoderma spp. and Arabidopsis thaliana to be incorporated by using double gene constructs engineering grapevine, which conferred resistance to powdery mildew fungus in Vitis spp. [149]. Despite the controversy, using transgenic crops has tremendously boosted world agricultural profits, a global meta-analysis of the impact of GM crop adoption has estimated that transgenic technology has increased farmer profits by 68%. Furthermore, GM crops have reduced chemical pesticide use by 37% and increased crop yields by 22%. This information expands the knowledge of GM technologies by giving valuable information about ecological and economic benefits that GM crops have provided since they emerged [150]. Advances in biotechnology and MAS have provided a huge amount of tools for GM crops development, more effort have been applied to develop GM crops tolerant to herbicides such as glyphosate, however pest/disease resistance, abiotic stress tolerance and nutritional enhancement are also important traits to be working with [150,151].
Initially, genetic modification strategies were based on a single trait [152]. Advances in technology of recombinant DNA and whole genome sequencing make it now possible to easily introduce multiple traits within the same GM plant by gene stacking [153]. Gene stacking is a promissory tool in grapevine aiming to increase breeding efficiency for resistance to downy mildew and powdery mildew since it takes advantage of already known resistance genes or QTLs which have previously isolated and validated. Using gene stacking for grapevine breeding allowed it to have resistance genes on many varieties of Vitis spp. worldwide cultured [149]. Stacked plants increased tremendously in their planted area from 2000 until 2020 about 2 to 90% in corn and from 30 to 95% in cotton [150]. More commercialized GM crops are maize, cotton, potato, soybean, brinjal, rice, poplar, tomato, sugarcane, beans, canola, carnation, sugar beet, wheat, safflower, papaya, squash, plum and sweet pepper. Aiming to dismiss public concerns and uncertainties surrounding transgenic technology have led to a search for alternative approaches, such as cisgenesis, intragenesis and genome editing. With these techniques, in spite of a genetic modification being involved during its development, the end products are free of any foreign gene (transgene). Therefore, crop plants developed by those approaches are not genetically different from plants developed by way of plant breeding, thus, they will be more easily accepted by the society compared to the transgenic crops and would be approved by worldwide legislation in a shorter time [154].
These new technologies for genetically modified plants represent promising advances for genetic plant breeding and the development of new improved commercial cultivars; however, these techniques present biological limitations in certain woody and fruit plants, such as grapevines. These limitations include the incidence of somaclonal variation and recalcitrance to regeneration, which hinder the full progression of these approaches [155]. Moreover, the application of cisgenesis in grape breeding is currently limited by the lack of effective and robust promoters for Vitis species. Promoters are essential for controlling gene expression, and without efficient ones, the potential of cisgenesis to introduce desirable traits like disease resistance or improved fruit quality remains unfulfilled. Identifying and characterizing native Vitis promoters with strong and reliable activity is therefore crucial for advancing cisgenic approaches in grape biotechnology [149].
Regeneration is the process by which plants reestablish not only their cells but also their organs and tissues after an injury. A reduced number of crop species have the capacity of natural regeneration, however, lots of them can regenerate plantlets in vitro if explants are cultivated on a nutrient-enriched medium supplemented with auxin and cytokinin [156]. Tissue culture is a very useful technique for clonal propagation of many crop species, plant regeneration might start from various types of source organs, such as stem, roots, leaves, and sexual organs. Thus, regeneration is fully exploited to transmit the desired genetic modification to new cultivars, starting from different sources of explants [155]. Understanding the complex molecular processes that control plant regeneration in grapevines is extremely important. This knowledge is crucial for understanding cell and developmental biology, and has significant implications for viticulture and agricultural biotechnology. The investigation of genetic and biochemical pathways in grapevine regeneration provides valuable insights into plant developmental plasticity, the regulation of meristematic activity, and cellular reprogramming signals. This knowledge has the potential to facilitate improved propagation methods, the development of disease-resistant varieties, and the enhancement of desirable traits in Vitis spp. [156].
Genomic design is a cutting-edge approach in grapevine breeding involving high-throughput sequencing, genomic selection, and a predictive model for speeding up the design of superior cultivars. It measures genome-wide markers along with phenotypic traits to identify favorable combinations of alleles for complex traits such as resistance to disease, fruit quality, and climate resilience [157,158]. Genomic design in grapevines, which has a long juvenile phase and a high degree of heterozygosity, brings transformation possibilities to overcome traditional limitations imposed by breeding efficiency and to enhance genetic gain [159].
The Vitis genus (2n = 38) accommodates vast genetic diversity comprising wild subspecies, hybrids, and cultivated varieties [160,161]. This diversity has remained due to its mode of asexual propagation through cloning [162]. Also, commercially up to 1200 grapevine cultivars have found their origin through crossing of domesticated varieties with wild Vitis species [163]. Hence, the grapevine germplasm comprises some 15,000 named cultivars, many of which are either synonyms and are genetically the same but called by different names, or homonyms having the same name but are genetically different [164].
Whole-genome resequencing (WGR) as a whole-genome resequencing (WGR) application became popular in grapevine research after in 2007 the first draft genome (8X) of V. vinifera, derived from a highly homozygous (PN40024) and heterozygous (ENTAV115) Pinot Noir accession, came out [131,165]. This resource was later developed with more refined versions (12X.v0 and 12X.v2) that allowed serious studies on genetic variation within and between species of grapevines [166,167]. WGR also allows genotyping of extremely high resolution at a clonal level that cannot be achieved through traditional markers such as SSRs and SNPs [168,169]. Using second-generation sequencing technologies like Illumina and Roche 454, many works have been applied in grapevine studies and stand as the best choice for DNA variation identification, with respective powers of generating millions of sequences simultaneously. Carrier et al. [170] for instance, with the 454 sequencing, studied three Pinot Noir clones, identifying transposable elements as the most noteworthy source of somatic mutations. Carbonell-Bejerano et al. [171] were instead interested in a structural rearrangement causing white berries in Tempranillo using Illumina sequencing, while Gambino et al. [172] re-sequenced three highly divergent Nebbiolo clones and identified thousands of clone-specific SNVs.
Next Generation Sequencing (NGS) allowed for the use of genomic methods to genetically explore complex phenotypes through different approaches, such as GWAS or GS. Disappointingly, methods like GWAS and GS that leverage markers across the genome to predict phenotypes are not easy to apply in highly heterozygous species like grapevine [173], however there are a few examples of GS application for grapevine breeding. In this case Fodor et al. [173] assessed the ability of GWAS-GS approaches to predict structured traits using four grapevine training populations comprising 1000 individuals each. Results demonstrated that the combined GWAS-GS model gave the highest prediction accuracy, reaching as much as 0.9. On the basis of these results, the authors suggest utilizing this integrated prediction model with the core collection as the training population to further grapevine breeding or breeding programs of other economically important crops bearing similar traits.
Evidence from another study indicates that in order to enhance the understanding of the genomic variation of agronomic traits in table grape populations for future MAS and GS needs, a molecular marker set associated with variation in genes was used to detect several Quantitative Trait Loci (QTLs), whereas the QTL method is imprecise and must yield to a more powerful lookup of the genetic architectures of the studied population done using an alternative genomic analysis known as Bayesian Lasso (BLasso, Bayesian Least absolute shrinkage and selection operator), which is a statistical method used in GS for predicting complex and polygenic traits, such as grapevine breeding, reducing the many minor effects of markers and thus inferring more efficient utility for the acceleration of selection for agronomic trait studies in table grapes as compared with QTL analyses [174]. In an additional study focused on genomic prediction and QTL detection for drought-related traits in grapevine, penalized regression methods applied on data collected from biparental progeny of 14 traits under two irrigation environments showed high predictive accuracy within populations (with a maximum of 0.68), leading to the detection of new QTL and candidate genes and thus establishing the promise of genomic prediction in grapevine breeding [175].
A related study assessed the genomic prediction accuracy for 15 key traits in grapevine, covering phenology, yield, vigor, and berry composition, at both cross and individual levels, while uncovering that whereas prediction accuracy for cross means averaged 0.6 (up to 0.7), and for individual values averaged 0.26. Results emphasize the importance of genetic distance, heritability, and cross effects on predictive power, which provide a great insight toward optimizing training sets and implementing genomic selection in breeding programs [176]. Another point discussed by Flutre et al. [177], is that a genetic diversity panel with 279 V. vinifera cultivars assessed between numerous years for 127 traits over 5 vineyard blocks was genotyped for about 63,000 SNPs from combined microarray and sequencing data. They detected 489 robust and novel QTLs-many of these previously undetected in bipolar studies-with average prediction accuracies above 0.42 for half of the traits studied and provided valuable insights regarding the genomic architecture of complex agro-economic and quality traits.
The advent of third-generation sequencing (TGS) technology allows the generation of long reads, from tens to hundreds of kilobases, for the overspilling analysis of structural variation (SV). Hence, they are equipped with more theoretical possibilities for in-depth SV analyses. In 2016, Chin et al. [135] created a new reference for the heterozygous cultivar Cabernet Sauvignon, stitching together SMRT sequencing with the FALCON assembler into a much more contiguous assembly (contig N50 = 2.17 Mb) compared to the previous PN40024 genome (contig N50 = 102.7 kbp) and releasing the first haplotype-phased sequence of the diploid V. vinifera genome. Recently, Vondras et al. [84] exercised whole-genome PacBio sequencing on 15 Zinfandel clones and found evidence that clonal propagation incites the accumulation of deleterious mutations, mostly in non-coding regions such as introns and intergenic regions. Besides long-read sequencing are more useful in complex genomes with highly repetitive gene cluster regions, allowing higher resolution among predicted alleles [178], TGS platforms have two major drawbacks for large-scale genotyping: they cost more than short-read sequencing and are prone to even more errors, ranging from 10% to 20% in PacBio and Nanopore technologies, compared to up to 0.1% in Illumina, limiting their usefulness for the detection of small variants, especially in repetitive regions of the genome [136].
Recent advancements in DNA sequencing technologies, genomics, transcriptomics and bioinformatics have substantially enhanced our comprehension of the genetic foundations of various crucial traits in grapevines, including stress tolerance, fruit quality, and overall yield. These technological progressions present an unparalleled opportunity to investigate the molecular mechanisms governing these intricate characteristics. The strategic integration of these modern technologies with traditional physiological and biochemical resources, alongside established conventional breeding methodologies, holds considerable promise for the future trajectory of grapevine research and development. This synergistic approach facilitates a more comprehensive and efficient pathway towards the development of improved grapevine varieties. A particularly compelling avenue within this integrated framework is the utilization of cutting-edge CRISPR/Cas9 gene-editing technology. This revolutionary tool offers the capability for precise and targeted modification of pivotal genes directly associated with grapevine stress resilience. By accurately editing these specific genes, researchers can potentially augment the natural defenses of Vitis spp. against a broad spectrum of environmental stressors, such as drought, extreme temperatures, and various pathogens. This precision gene-editing approach creates opportunities for developing grapevines that are not only more robust and adaptable to changing climatic conditions but also maintain or enhance desirable fruit quality and yield, ultimately contributing to more sustainable viticulture practices globally.
Genome editing technologies have been developed as powerful tools to improve traits in grapevines, with more precision and efficiency than traditional breeding methods. Among others, CRISPR/Cas9 has been used to edit mutated genes for resistance against diseases, fruit quality, and tolerance against stresses [179]. Due to the complexity in the genome and a long generation time of the grapevine Vitis vinifera L., these given techniques represent an attractive alternative to speeding up genetic improvement to meet challenges of climate change and new pathogens [14,180].
While many valuable crop varieties have evolved via precision breeding, the acceptance of genetically modified plants has remained highly limited. Plants modified through genome editing, especially those without foreign DNA insertion, gained broader acceptance as they are often not considered transgenic. Countries such as Argentina, Brazil, U.S. and Australia, allow such edited crops to enter the market without going through the long and strict safety evaluation typically applied to GMOs, given that they carry no foreign genetic material [181].
Some genome editing tools involve such enzymes as transcription activator-like effector nucleases (TALENs) [182], zinc finger nucleases (ZFNs) [183] and the Cas9 nuclease being associated with clustered regularly interspaced short palindromic repeats (CRISPR) [184] to catalyze the production of site-specific double-strand breaks (DSBs) within DNA molecules. Among the best genome editing tools, CRISPR-Cas9 is regarded to have the highest efficacy due to its higher specific nature and minimal off-target effects [185]. This requires a guide RNA (gRNA) carrying the spacer sequence complementary to the desired DNA sequence. The gRNA and Cas9 enzyme complexes look for complementary double-stranded DNA sequences in the genome [186]. The nuclease subsequently recognizes the adjacent protospacer motif (PAM) and produces a DSB within the gene sequence of interest. Hence, genome editing via the CRISPR-Cas9 system requires a PAM sequence downstream of the target gene and the appropriate guide RNAs with an appropriate design based upon gene sequences coding for traits of importance [187].
CRISPR-Cas9 has emerged as a powerful tool among new plant breeding techniques for enhancing resistance to both biotic and abiotic stressors [188,189]. Key factors include target recognition, gRNA design, and the frequency of homologous recombination repair events, as well as the action of anti-CRISPR proteins that can inactivate Cas9 [185]. Although CRISPR-Cas9 offers significant advantages over ZFNs and TALENs, a major limitation of this technology is the potential for off-target mutations [190]. CRISPR-Cas9 mediated genome editing in grapevine was first reported by Ren et al. [191], since then, the use of this technology for plant breeding has increased.
In this context Villette et al. [14] optimized zCas9i for the edition of the grapevine genome. In the course of this study, the protocol was optimized with gene editing for ‘Chardonnay’, as they used fluorescence microscopy to facilitate better selection of transformants. By trying different Cas9 constructs and promoters [Human codon optimized Cas9 construction (GB0575), Zea mays L. codon optimized Cas9 (zCas9i) and RPS5a (At3g11940) promoter], they determined that zCas9i carrying 13 introns affords high levels of editing efficiency, even producing full biallelic mutations. These results seemed tightly correlated to the expression levels of Cas9. Overall, this study sets the stage for a reliable protocol for obtaining homozygous knockout lines in grapevine that will fulfill both gene function analysis and further breeding efforts.
The greater majority of the studies that deal with genome editing of grapevine using CRISPR-Cas9 technology, were carried out to increase the resistance to Botrytis cinerea [192] and powdery mildew [193,194], to increase tartaric acid production [191], and to manipulate the carotenoid biosynthesis pathway while also generating the albino phenotype [195,196].
Throughout the growing period, grapevines undergo attack by the phytopathogenic fungus B. cinerea, which is a long-standing issue [197,198]. Infection by the pathogen begins near flowering; also, actual flower abortion and poor fruit growth are often the result. In the early stages of development, the infection may go undetected, but further on, during berry ripening, the infection is activated much more frequently [197]. In doing so, surface moisture and high sugar content from ripening berries created excellent conditions for the development of the fungus. Whereas B. cinerea can penetrate intact berry tissue directly with its hyphae, shallow wounds enormously boost and accelerate the infection process [199]. In an effort to confer resistance against this fungus to grapevine, Wang et al. [192] employed the genome editing system CRISPR/Cas9 with four gRNAs targeting the VvWRKY52 transcription factor in Thompson Seedless grape embryos and found that deletion of VvWRKY52 enhanced resistance to B. cinerea, supporting this gene’s role in susceptibility and the potential of CRISPR/Cas9 as a tool to increase fungal resistance in grapevine without adverse effects.
Another important pathogen in grapes is Plasmopara viticola that causes downy mildew. This fungus inhibits various young and susceptible tissues of grapevine, such as leaves, shoots, flower clusters, petioles, and developing berries [200]. The initial symptoms of the infection consist of translucent yellowish spots affecting the leaf surface, which slowly develop into white fungal growth on the leaf’s underside. In more advanced infection, however, spots expand and coalesce to form irregular patches. As time goes on, inhibition occurs in photosynthesis, leading to a decreased accumulation of sugars in berries and dormant buds, delayed ripening of fruits, reduction in fruit set, impairment of grape quality, and ultimately, substantial yield losses [201]. Li et al. [193] investigated the role of VvPR4b in grapevine resistance to Plasmopara viticola (Rpv genes). Their study involved knocking out the VvPR4b gene in the ‘Thompson Seedless’ cultivar using CRISPR/Cas9 and comparing these lines to wild-type grapevines. The results demonstrated that VvPR4b-deficient lines were more susceptible to P. viticola, exhibiting increased pathogen abundance and reduced accumulation of reactive oxygen species around stomata. These findings unequivocally establish VvPR4b as a crucial component of grapevine defense mechanisms against downy mildew.
Wan et al. [194] employed CRISPR/Cas9 technology to modify the VvMLO3 and VvMLO4 genes in the ‘Thompson Seedless’ grapevine cultivar. These genes, identified as Ren genes, confer resistance to Erysiphe necator, the causative agent of powdery mildew. The objective of this research was to introduce powdery mildew resistance into grapevines. The edited lines exhibited various mutations, including homozygous, biallelic, and heterozygous alterations. Notably, plants with VvMLO3 mutations demonstrated enhanced resistance to powdery mildew. These plants also displayed augmented host cell death, elevated hydrogen peroxide accumulation, and reinforced cell wall fortification in four instances. Erysiphe necator presents a significant global threat to V. vinifera, impacting both table and wine grapes [202,203]. This pathogen leads to substantial reductions in grape yields and negatively affects the quality of table grapes, wine grapes, must, and the final wine product [204,205,206]. Outbreaks typically originate from fungal mycelia that survive within infected buds or from cleistothecia that overwinter on fallen leaves and the exfoliating bark of grapevines [207,208].
In addition to its application to obtain resistance in grape plants to important pathogens, this editing technology was employed to study the tartaric acid content of this cultivar. Therefore, the IdnDH gene, responsible for the actual accumulation of tartaric acid in grape cells, was deliberately subjected to targeted genome editing (TGE) in suspended cells of the Chardonnay cultivar. Using A. tumefaciens, two sgRNA expression cassettes with homologous recombination sites were simultaneously introduced into a pCACRISPR/Cas9 binary vector and used for plant transformation. PCR analysis confirmed T-DNA integration, with an average transformation efficiency of 37.78%. Indeed, indel mutations at the target site were observed using CEL I endonuclease assays and sequencing, with a mutation frequency of 100% achieved in both transgenic cell masses and regenerated plants expressing sgRNA1/Cas9. Furthermore, tartaric acid levels and the expression of Cas9 and sgRNA were assessed to confirm gene editing. No off-target mutations were observed, demonstrating the precision and reliability of CRISPR/Cas9 for grapevine genome editing beyond pathogen-related traits [191].
Two other contributions to grape genome editing using the CRISPR/Cas9 system are related to carotene biosynthesis in the Neo Muscat, Chardonnay and 41B varieties. In the first research Nakajima et al. [195], cotransformed embryonic calli from the Neo Muscat variety with Cas9 and synthetic sgRNA targeting the VvPDS (phytoene desaturase) gene. The regenerated plants had albino leaves, and the sequence of the target region of the VvPDS gene showed mutations. Interestingly, a higher percentage of mutated cells was found in older lower leaves compared to younger upper leaves, suggesting an accumulation of edited cells in mature tissues, as aging could involve repeated inductions of DSBs or, alternatively, less effective DSB repair in older grapevine cells. The second experiment intended to analyze three variables that may improve targeted genome editing (TGE) efficiency in grapevine [196]. The first variable assessed was the GC content of single-guide RNAs (sgRNAs) designed to target the VvPDS gene encoding phytoene desaturase, a crucial enzyme in the carotenoid biosynthesis pathway. The disruption of this gene causes a loss of pigment and the expression of an albino, dwarf phenotype, which visually distinguishes the edited plants from the non-mutants. The second factor addressed was the choice of grapevine cultivar used during transformation, while the third involved assaying for the expression levels of SpCas9 in the transgenic CMs. To investigate these variables, four sgRNAs of varying GC content were designed to target exon regions of the VvPDS gene, and transformations were carried out on suspension cells from ‘Chardonnay’ and ‘41B’. The T7 endonuclease I and PCR/restriction enzyme assays revealed a positive correlation between higher GC content in sgRNAs and editing efficiency; GC content at 65% was noted to provide the most consistent results across both genotypes. The other finding of note was that editing efficiency was observed to be better in ‘41B’ compared to ‘Chardonnay’. Further, while differences in SpCas9 expression levels among the constructs could be detected, sgRNA GC content exerted a far greater influence on genome editing efficiency than that of Cas9 expression.
The process of protoplast culture paired with CRISPR-Cas editing technologies presents an excellent means-for highly efficient genome modification in plants. Protoplasts are plant cells from which the cell wall has been removed, allowing direct delivery of reagents into the cell for transient expression and also reduction of transgene integration. Tile approach allows to quickly measure the editing efficiency and analyze gene function while regenerating edited, non-transgenic plants. In all of these, a great approach for crop improvement and functional genomics.
A highly efficient protocol for the regeneration onto whole plants from protoplasts of a variety of grapes was presented by Tricoli and Debernardi [209]. Calcium alginate beads enclose the protoplasts while they are co-cultured along with feeder cultures, allowing the protoplasts to divide and give callus colonies that regenerate into embryos and eventually plants. This particular protocol has been shown to be successful for various wine and table grape varieties (V. vinifera), grape rootstocks as well as in the grapevine wild relative, V. arizonica. Further, by transfecting protoplasts with CRISPR-plasmid RNP complexes, albino plants with edits in the VvPHYTOENE DESATURASE gene were generated in three varieties as well as in V. arizonica. In another report authors present the first successful demonstration of DNA-free genome editing in grapevine by direct delivery of CRISPR-Cas9 RNP complexes to isolated protoplasts and regeneration of the complete plant. Using a transgenic Thompson Seedless line expressing GFP as the model system for genome editing, the authors obtained targeted knockout of the GFP gene and assessed editing efficiencies by loss of fluorescence and sequencing of insertion mutations. The regenerated plants grew normally and exhibited normal morphology in vitro, without any signs of somaclonal variation or chimerism. The approach avoids the integration of transgenic DNA and thus gets around most regulatory concerns over genetically modified organisms [210].
In a previous study Scintilla et al. [211] mention successful cases of targeted mutagenesis of several genes in grapevine (V. vinifera cv. Sugraone and Crimson seedless), mostly targeting genes responsible for disease resistance, fruit quality, and metabolic pathways. The transformation methodologies discussed are Agrobacterium-mediated delivery and protoplast transfection, with guidelines on gRNA design, Cas9 expression, and off-targets thrown into the mix. The fully regenerated, non-chimera plants containing the edits targeted to the expected susceptibility genes of downy mildew (VviDMR6) and powdery mildew (VviMlo6) were introduced as either single or double mutants. The potential of CRISPR/Cas9 in speeding up functional genomics and breeding in grapevine, on the other hand, is put forth by the study, especially if linked with improved tissue culture and regeneration systems, despite some technical constraints such as low transformation efficiency and regeneration bottlenecks in plants.
Table 2 summarizes everything discussed above.
Table 2: Targeted gene editing in grapevine: Current applications of CRISPR/Cas9 for trait improvement.
| Effector | Target | Plant Material | Results | Reference |
|---|---|---|---|---|
| zCas9i | Various genes | V. vinifera (‘Chardonnay’) | Homozygous knockout lines in grapevine. | [14] |
| CRISPR/Cas9 | IdnDH | Chardonnay cell suspensions | Edit genes to increase tartaric acid content. | [191] |
| CRISPR/Cas9 (4 gRNAs) | VvWRKY52 | Thompson Seedless embryos | Improve resistance to B. cinerea. | [192] |
| CRISPR/Cas9 | VvPR4b | Thompson Seedless | Assess role in defense against P. viticola (downy mildew) | [193] |
| CRISPR/Cas9 | VvMLO3 and VvMLO4 | Thompson Seedless | Increase resistance to powdery mildew (Erysiphe necator) | [194] |
| CRISPR/Cas9 | VvPDS | Neo Muscat | Induce albino phenotype to study carotenoid biosynthesis. | [195] |
| CRISPR/Cas9 | VvPDS | Chardonnay and 41B suspension cells | Albino, dwarf phenotype. | [196] |
| CRISPR-Cas9 RNP | VvPDS | Various grape varieties and V. arizonica | Albino plants by DNA-free genome editing via protoplasts. | [209] |
| CRISPR-Cas9 RNP | GFP | Thompson Seedless protoplasts expressing GFP | Demonstrate DNA-free editing without transgene integration and protoplast regeneration. | [210] |
| CRISPR/Cas9 | VviDMR6 and VviMlo6 | Sugraone and Crimson Seedless | Non-chimera plants resistant to downy mildew and powdery mildew. | [211] |
CRISPR/Cas9 technology has made substantial advances in functional genomics and breeding strategies for grapevine research. Studies have been successful in editing a number of genes related to pathogen susceptibility that include VvWRKY52, VvPR4b, VvMLO3, VvMLO4, VviDMR6, and VviMlo6 to enhance resistance against Botrytis cinerea, Plasmopara viticola, and Erysiphe necator. Moreover, this technology has been used to edit traits concerning tartaric acid accumulation and carotenoid biosynthesis. Lately, CRISPR-based editing methods combined with protoplast culture techniques have given rise to non-transgenic genome-edited plants. Together, these studies highlight that CRISPR/Cas9 can be an accurate and highly versatile tool to improve grapevine traits bearing agronomic importance.
Notwithstanding advances in CRISPR-Cas9 genome editing in Vitis spp. there are complications for applying most of the recent CRISPR technologies in grapevine, such as efficient transformation due to recalcitrance, lack of efficient system for testing Cas effectors, limited available explants, delivery of CRISPR-Cas reagents and plant regeneration [212]. Despite the limitations, these authors believe that advances in CRISPR-Cas9 technology might boost grapevine genome engineering due to the predicted innovation in CRISPR-Cas9 technology in the decade ahead. The economic importance of grapevine cultivation is also boosting breeders and growers to adopt new grapevine breeding innovations despite their current limitations. Furthermore, there is a need for analyzing public politics to inform the advantages such as reducing the pesticides by using disease-resistant cultivars [77] and disadvantages such as public acceptance, regulatory restrictions of using genetic modified/edited grapevine varieties of grapevine breeding in the near future and long-term viability and resilience of viticulture [213].
The development of new fruit varieties is fundamentally based on improvements in abiotic factors, which have intensified with climate change, and biotic factors, especially due to the adaptation of pathogens to new climatic conditions and their resistance to the chemicals continually applied to them [214]. In the case of grapevines, the genetic diversity of modern cultivars is significantly limited due to decades of domestication and selective breeding, so as a result, they are highly susceptible to various stress factors, including gray rot, downy mildew, and cold temperatures. This vulnerability presents serious challenges for viticulture and the wine industry as a whole [145].
To meet the upsurge in demands brought about by a growing global population, crop improvement is increasingly taking the road of innovative technologies—multi-omics and artificial intelligence—for sustainable breeding strategies. The topics to be discussed in this theme are summarized in Fig. 3.
Figure 3: Novel breeding methods in grape.
The improvement in omics technologies revolutionized the characterization and correct identification of biomolecules and genes, and thus, very specialized databases and tools have been generated for various crops. These technologies have provided infinite possibilities for more advanced and novel options in plant breeding [215]. Omics technologies give insights into and interpretations of plant metabolism at a molecular level with more precision. New breeding strategies stem mainly from studies conducted through genomics, proteomics, metabolomics, and transcriptomics [216,217,218]. Most importantly, with a better understanding of molecular mechanisms, the investigation of grapevine responses to environmental stresses has come to the forefront, especially through high-throughput sequencing methods and integrated multi-omics platforms [219].
Genomics
Genomics, being a segment of omics sciences, looks at a plant’s genome, thus providing important knowledge to plant breeders in the understanding of domestication processes and genetic improvement. Furthermore, genomics provides practically oriented scientific tools in marker-assisted selection for the development of new cultivars and the introduction of traits never thought of in association with plants before, such as the production of biopharmaceuticals and industrial compounds [220,221].
Whole-genome resequencing is a strong and widely adopted method for assessing genetic variation in different crop populations, identifying necessary traits linked to loci, performing genome comparisons, and assessing evolutionary relationships [222,223]. Since the grapevine genome’s completion in 2007 [131,165], several genes related to growth and development, metabolic pathways, and biotic and abiotic stress responses have been identified. Whole-genome sequencing has helped reveal genomic characteristics responsible for phenotypic variation among grapevine cultivars and to study their level of genetic diversity [224].
In order to fight against diseases, improvement programs for many crops focus on developing resistant varieties, grapevines being no exception. Particularly in viticulture, the need is to develop cultivars that are resistant to Pierce’s disease, downy mildew, and powdery mildew. Genomics has played a crucial role in understanding disease resistance in grapes. This has been achieved through identifying resistance loci in wild grape species, phasing resistance haplotypes, conducting association analyses, and performing gene expression experiments [225]. For example, resistance to powdery and downy mildew in Muscadinia rotundifolia was mapped to chromosome 12. This resistance was attributed to two homologous TIR-NB-LRR genes, MrRUN1 and MrRPV1, which are the first cloned grapevine resistance genes. When transferred into V. vinifera, these genes confer strong resistance [226]. Additionally, thirty pathogen genomes from 19 species that cause grapevine diseases have been sequenced and assembled. This extensive genomic database provides insights into potential virulence factors and offers initial clues about the evolution of these pathogens [225].
Run1.2 was the first R locus analyzed in a diploid grape genome and is linked to powdery mildew resistance introgressed from Muscadinia rotundifolia Trayshed [227]. Comparative analyses of NLR (nucleotide-binding site leucine-rich repeat) gene content found that there were more TIR-NBS-LRR genes in Muscadinia relative to the susceptible V. vinifera cv. Cabernet Sauvignon. Then, by expression profiling of NLR genes at the Run1.2b and Run2.2 loci, several genes were shown to be constitutively expressed; a TIR-NBS gene, two TIR-NBS-LRR genes, and a CC-NBS-LRR gene from Run1.2b and a TIR-NBS-LRR gene from Run2.2 were thus proposed as candidate genes for resistance [225]. In examining Pierce’s disease resistance (PDR) in V. arizonica, four extracellular receptor genes were pinpointed as prospective R genes in a region on chromosome 14 [4], corroborating the view held by earlier workers that PdR1 may be the only locus conferring Pierce’s disease resistance in V. arizonica and its hybrids [228,229]. In the same way for downy mildew, two resistance loci were found, called Rpv12 and Rpv33, respectively [230,231] and candidate resistance genes were identified in both loci by considering resistant haplotypes compared with alternate haplotypes linked to susceptibility and the gene expression analyses. At Rpv12, the candidate gene encodes a CC-NBS-LRR protein. In contrast, at Rpv33, candidate genes encompass three genes of the NLR family [232].
Genomic analysis was conducted by Guo et al. [145] for 72 Vitis accessions comprising 25 wild ones and 47 cultivated varieties, with a fully resolved haplotype genome of V. vinifera. It was revealed from the study that European cultivars possess few genes for disease resistance of the NLR-type that are highly susceptible to the devastating powdery mildew fungal disease in grapes. NLR genes, which are important in plant immune responses to pathogens, display a high degree of polymorphism among different accessions of the same species. Out of this, structurally, 79 variations and genes of the proteins are highly associated with the resistance against downy mildew. This information from the study could play a vital role in the fast genetic improvement of grapevine and provide a deeper insight into the evolution and biology of grapevine. According to Shi et al. [131], earlier grapevine reference genomes had quite a few fragmented regions, as well as being telomere- and centromere-deficient, so a study on the inheritance of important agronomic traits would be impeded by it. To allow an unobstructed view of its genome, the scientists developed a gap-free telomere-to-telomere (T2T) genome assembly measuring 503.9 Mb with 9018 genes additional to the old version 12X.v0. This improved assembly was able to annotate 67% of the repetitive sequences, identify 19 centromeres and 36 telomeres, and transfer gene annotations from previous versions. Moreover, the study found 377 gene clusters associated with complex traits such as aroma and disease resistance. Another research on genome annotation for grapevine improvement is that of Ritter et al. [233], who produced highly complete assemblies of two V. vinifera cultivars, Dakapo and Rubired—both descendants of Teinturier du Cher but likely derived from distinct clonal lineages. The Dakapo genome (508.5 Mbp) was assembled using Nanopore and Illumina sequencing, resulting in 36,940 annotated genes, while the Rubired genome was generated with PacBio HiFi reads and phased into haplotypes of 474.7–476.0 Mbp, with 56,681 annotated genes. Together, these genomic resources offer new opportunities for investigating Teinturier varieties and for supporting breeding programs aimed at enhancing anthocyanin-rich berry flesh pigmentation and other agronomic traits.
Until 2007, a major obstacle in studying sex determination in Vitis was that the V. vinifera reference genome simply consisted of an incompletely assembled F (female) haplotype [131]. On the other hand, according to Picq et al. [234] the gene or genes responsible for the shift to hermaphroditism in V. vinifera were located in a genomic segment of approximately 150 kb on chromosome 2. This shift in flower morphology was a key event in domestication: the wild subspecies sylvestris is dioecious, while the cultivated V. vinifera is mainly hermaphroditic. In Vitis, sex is determined by three alleles located at a single locus-hermaphrodite (H), female (F), and male (M), with a dominance hierarchy of M > H > F. The consensus is that the hermaphroditic alleles arise from male alleles in wild vines by a recombination event. As reported by the authors, the evidence thus provided for multiple domestications and introgression from other Asian Vitis species into the genetic background of cultivated grapevines. Based on the findings of Massonnet et al. [235] for the sex determination twenty haplotypes from hermaphrodite, female, and male grapevines were analyzed, and researchers identified sex-linked genomic regions, including a putative mutation in VviINP1 for male sterility, and a likely involvement of the transcription factor VviYABBY3 in female sterility. According to the authors, dioecy probably disappeared during domestication due to a rare recombination event between male and female haplotypes. This work presents a major step forward in the comprehension of genetic mechanisms underlying sex determination in Vitis and valuable tools for the rapid identification of sex types in grape breeding programs.
Structural variations and their evolutionary dynamics were addressed in relation to clonal propagation in cultivated grapevines and their wild, outcrossing ancestors by examining the genome assemblies of Chardonnay and Cabernet Sauvignon. The study shows that purifying selection acts against structural variations; except that in clonal lineages, these variants accumulate as recessive heterozygotes. The study also unraveled anomalously divergent genomic regions between the wild and cultivated grapevines, thereby implying their role in domestication. It is intriguing that among these are the sex determination locus and the berry color locus—the very regions targeted by massive, complicated, and independent evolutionary processes, which resulted in parallel phenotypic evolution [236].
Finally, it is important to address some genomic advances in resistance to some abiotic stresses. The FAR1-RELATED SEQUENCE (FRS) genes consist of semitransposase-derived transcription factors that are key to plant development via activities like regulating light signaling, hormone responses, and stress adaptation. Whereas grapes enjoy a high ecological and commercial standing, there have been fewer intensive studies on FRS genes in this species. In the research conducted by Yao et al. [237] 43 genes of VvFRS distributed on 13 grape chromosomes were identified and they were assigned names VvFRS1 to VvFRS43, depending on the position on chromosomes and corresponding Locus ID. A study was further carried out on the physiological and biochemical characteristics of the proteins. On average, the VvFRS proteins had 602 amino acids (ranging from 209, in VvFRS23, to 985 in VvFRS1), with molecular weights between 24.05 and 113.74 kDa. Most had a slightly acidic character and were considered generally stable. The aliphatic index ranged between 63.26 (VvFRS1) and 85.56 (VvFRS3) and the family was mostly hydrophilic, with VvFRS24 showing maximum hydrophilicity (−0.923). Importantly, about half were predicted to localize in the nucleus. In subsequent research Wang et al. [238] studied the β-1,3-glucanase gene in grapevine. These genes are greatly involved in plant development and stress responses. Still, until now, however, the identification and expression profiling of the β-1,3-glucanase gene family in grapevine had never been reported. In this study, 42 VviBG genes were identified in V. vinifera, all having GH-17 domains with variable C-terminal regions. Phylogenetically, the VviBG genes were divided into three principal clades: α, β, and γ, each further subdivided into six groups (A to F), with members within every subgroup sharing conserved motifs, domain characteristics, and intron/exon arrangements. Expression analyses show that most VviBG genes respond to stress treatments, such as wounding, UV, downy mildew, cold, salinity, and drought stresses, with a strong response from eight subgroup-A γ clade genes. These results were confirmed by RT-qPCR analysis, which demonstrated that eight genes possess an upregulated status when exposed to abiotic stress, and PEG6000 as well as NaCl treatments induced higher levels of expression than cold stress [238].
Stress-associated proteins (SAPs) essentially mediate plant responses to biotic and abiotic stresses. 15 genes for the A20/AN1-type zinc-finger protein were located through the mining of assorted grapevine genomic and proteomic databases. According to their structural characteristics and phylogenetic correlations, the SAP genes fell into three main clusters that suggested a rather conserved evolutionary history. Group I contained two Arabidopsis SAPs (AtSAPs) and three grapevine SAPs (VvSAPs). Group II contained seven AtSAPs, six VvSAPs, and four rice SAPs (OsSAPs), whereas Group III consisted of five AtSAPs, fourteen OsSAPs, and six VvSAPs [239].
Fig. 4 summarizes the cited genomic research.
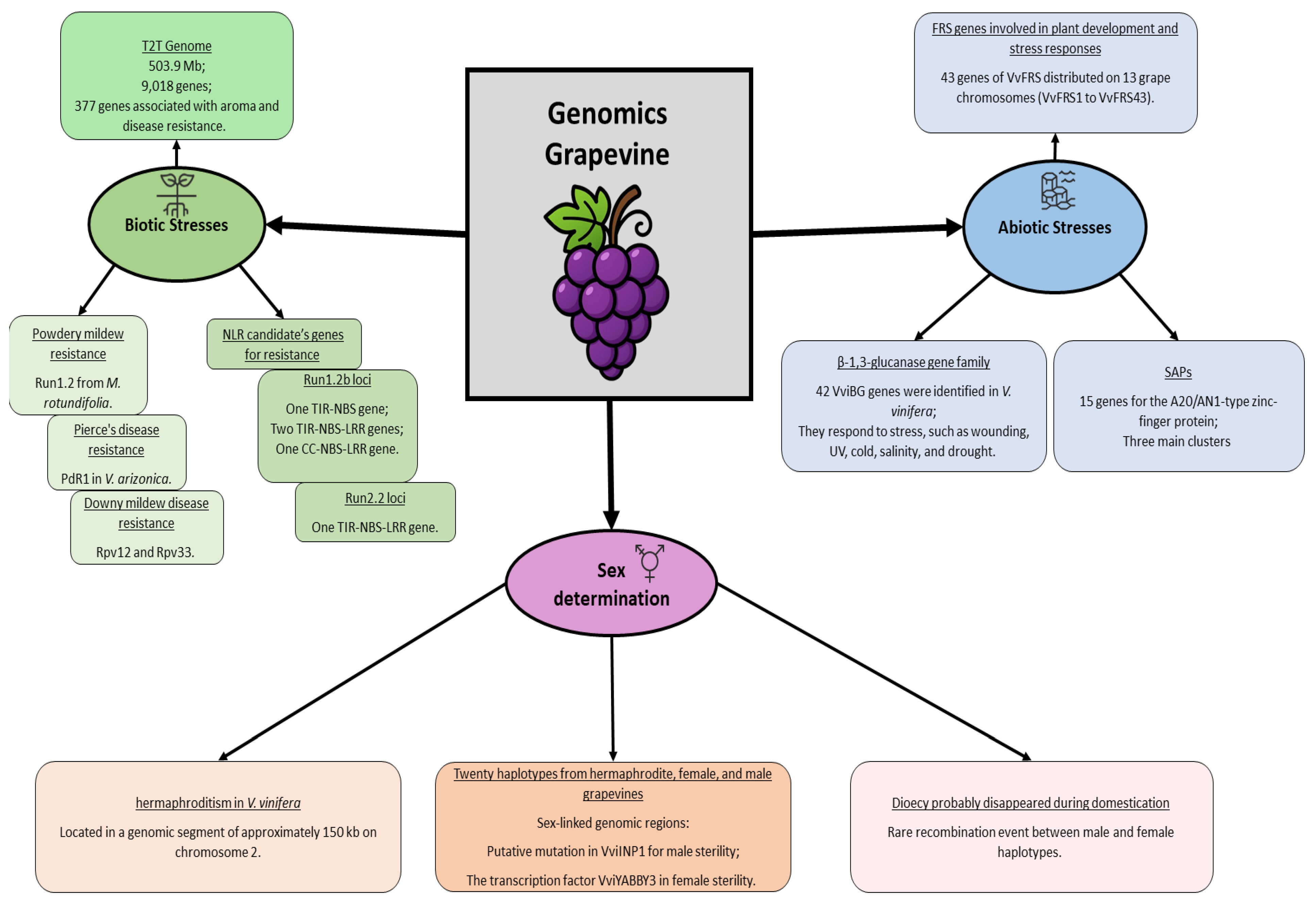
Figure 4: Summary of some genomic studies in grapes related to stress and sex determination.
Genomics-related research in the grapevine system has given us deeper insights into important traits linked to disease resistance, stress tolerance, and domestication. Usage of high-resolution genome assemblies, gene expression profiling, and comparative analyses has led to the identification of significant resistance loci, candidate genes, and structural variations governing the genetic architecture of the cultivated and wild Vitis species. These findings, in improvement programs, accelerate selection for commercial production of grapevine cultivars that are more resistant because they are well adjusted to modern agricultural and environmental constraints.
Proteomics
Proteomics comprises the quantitative and qualitative study of the proteome, of all the proteins in a cell. This type of analysis is challenging in eukaryotic organisms because of the numerous types of post-translational modifications that can exist through different modes at different sites [240,241]. Being a hallmark of omics technology, proteomics sheds light on gene function and expression variability. More significantly, the proteins form the core of biological processes; hence, their abundance within an organism is also regulated by molecular mechanisms acting at translational levels apart from mRNA expression levels [242].
To date, at least six major proteomic investigations have been conducted, directly related to the genetic improvement of grapevine, focusing upon stress response (drought, cold, heat), fruit quality (anthocyanins), and seed development (seedless grapes). These studies, being on the lighter side, have presented the required data and candidate proteins to breeding programs for stress tolerance, yield, and fruit quality improvement.
One of the investigations that addresses the issue of abiotic stress using proteomics is the one developed by Prinsi et al. [243] studied the proteomic and metabolomic responses triggered in two rootstocks, M4 and 101.14, which are differentially susceptible to water stress. It was found that water stress altered protein profiles in both genotypes, 17% of proteins in 101.14 and 22% in M4 being affected. Metabolomics studies indicated that M4 responded to water deficit by inducing a higher oxidative stress response, thereby protecting cellular structural features and maintaining cell turgor pressure. In contrast, the 101.14 roots suffered prominent damage associated with the stress. Proteins associated with energy metabolism, hormonal routes, protein turnover, secondary metabolism, and stress responses were differentially expressed herein. In compatible solute accumulation behavior, M4 evidently showed its stronger side by accumulation of higher levels of proline and several sugar alcohols. Hydrolysis-induced water stress significantly increased the concentration of polyols such as mannitol, inositol, galactinol, and erythritol in the roots. The tolerant M4 genotype yields much higher amounts compared to the less tolerant one, underscoring the importance of these compounds in the plant’s stress adaptations. In addition, the results exhibited that, in M4, the water-stress tolerance mechanism strongly relies on carbon metabolism regulation, osmotic adjustment, specific stress-responsive pathways, and mitochondrial functionality, especially in root tissues, thereby providing some insights into the mechanism of drought tolerance in Vitis.
Given that few proteins expressed in grapevine roots have been characterized for cold stress, Chen et al. [244] carried out a proteomic study using an iTRAQ approach to analyze root protein expression in a cold-resistant hybrid (V. riparia × V. labrusca, T1) and cold-sensitive cultivar (Cabernet Sauvignon, T3) at −4°C; with the resistant variety also being analyzed at −15°C (T2). The results revealed greater activity in the cold-resistant genotype compared to the sensitive genotype, though this decreased with decreasing temperature. Twenty-five proteins were differentially co-expressed between T2 versus T1 and T1 versus T3 concerning their function in stress responses, signal transduction, metabolism, energy production, and protein translation. These distinct translation patterns in grapevine roots affected by spatiotemporal cold treatments provide important clues with respect to the molecular basis for cold tolerance in varying grapevine genotypes.
In a subsequent study Zhang et al. [245] searched for different regulatory factors involved in the formation of seedless grapes, thus much favored by consumers. A proteomics approach was used to screen for proteins and antigens that accumulated differentially during grape development. Antigenic signals greatly varied between seeded and seedless types and 2587 differentially accumulated proteins were, therefore, identified. After validation by immunoblotting, 71 antigens were immunoprecipitated and identified by MS, which revealed changes in metabolic pathways—especially carbon metabolism and glycolysis. To further support these results, VvDUF642, a previously uncharacterized DUF642 domain-containing protein in grapevine but associated with seed formation, was ectopically expressed in Solanum lycopersicum ‘MicroTom’ (Tomato), resulting in a significant decrease in seed production, thus revealing both VvDUF642 and VvPAE (pectin acetylesterase) as promising targets for breeding programs to obtain seedless grapes and maybe other seedless fruit crops.
A notable proteomic investigation addressing grape berry quality is that of Murcia et al. [246], who demonstrated that exogenous ABA and GA3 applications in Malbec grapes modulate ripening by influencing H2O2 and sugar levels, thereby altering anthocyanin dynamics. Their analysis revealed that both hormones enhanced antioxidant defense proteins while downregulating phenylpropanoid biosynthesis, which resulted in reduced levels of specific anthocyanins (petunidin-3-G and peonidin-3-G) at the nearly ripe stage. At the same time, the accumulation of non-anthocyanin compounds such as E-viniferin and quercetin increased, suggesting a compensatory role in oxidative stress mitigation. Overall, the study highlighted a common molecular mechanism by which ABA and GA3 regulate berry pigmentation and composition during ripening. In conclusion, proteomic studies in grapevines have significantly contributed to the understanding of the molecular mechanisms underlying important agronomic traits, such as stress tolerance, fruit development, and seedlessness. By identifying key proteins and pathways involved in these processes, proteomics offers valuable information that complements genomic data and supports more precise and efficient breeding strategies. As this field of research continues to advance, it has great potential to accelerate grapevine improvement and the development of superior cultivars adapted to changing environmental conditions and market demands.
Metabolomics
The metabolites synthesized by grapevines largely determine fruit quality and are strongly influenced by environmental conditions. Investigations of the plant metabolome provide valuable insights into underlying metabolic processes and are essential for enhancing stress tolerance while improving crop quality and productivity [247]. As part of these processes, secondary metabolites play a central role in plant defense mechanisms. Given that V. vinifera is highly susceptible to many diseases, other Asian and American Vitis species, which show varying degrees of resistance, serve as valuable genetic resources for improvement. In this regard, metabolites can act as biomarkers of susceptibility or resistance. Supporting this, Maia et al. [248] profiled the foliar metabolome of 11 Vitis genotypes differing in resistance to powdery mildew, gray mold, and downy mildew, and identified 190 metabolites. They reported that compounds such as caffeic acid, catechin/epicatechin, hexadecanoic acid derivatives, dodecanoic acid, and leucocyanidin were more abundant in susceptible V. vinifera cultivars, whereas dihydroquercetin, quercetin 3-O-glucoside (isoquercitrin), and other flavonol 3-O-glucosides predominated in resistant or partially resistant genotypes.
In another investigation analyzing metabolites related to plant defense, Yu et al. [249] applied metabolomic profiling to compare berry responses in two cultivars with contrasting tolerance to powdery mildew: ‘Guipu’ No. 6 (GP6, Vitis sp.), which showed greater leaf tolerance, and the susceptible V. vinifera cultivar ‘Marselan’. After infection, both cultivars accumulated flavonoids, phenolic acids, stilbenes, and terpenoids, suggesting that resistance mechanisms are linked to phenylpropanoid–flavonoid metabolism. Notably, GP6 displayed higher stilbene accumulation, while Marselan showed greater flavonoid accumulation, highlighting distinct metabolic strategies in their defense against powdery mildew.
In this context, Ciubotaru et al. [250] investigated the alterations in primary and secondary metabolism triggered by E. necator inoculation in mono-locus resistant genotypes, pyramided resistant genotypes, and a susceptible cultivar. Their analysis revealed several metabolites that accumulated specifically in resistant genotypes but were absent in the susceptible one. Among the ten compounds that differentiated resistant from susceptible plants, pallidol, oleic acid + cis-vaccenic acid, and astringin had already been proposed as potential biomarkers of resistance to P. viticola [251]. Overall, the study suggests a clear connection between genotype and/or resistance loci and the metabolic response of grapevine cultivars to pathogen attack. Interestingly, although mono-locus and pyramided genotypes sharing the same loci displayed similar metabolomic profiles, the presence of multiple resistance loci did not directly translate into higher production of the identified biomarker metabolites.
Further insights were obtained from a metabolomic study quantifying primary metabolite sugars, designed to evaluate how early leaf removal (ELR) influences grapevine physiology and fruit quality through four distinct defoliation treatments: pre-flowering mechanical defoliation (PB-Mech), pre-flowering manual defoliation of six basal leaves (PB-Man), post-flowering manual defoliation of six basal leaves (AB-Man), and post-flowering mechanical defoliation (AB-Mech). Correlations were also studied between anthocyanins and their glycosides. Intermediate anthocyanins, such as syringetin, laricitrin, and myricetin, as well as acylated malvidin derivatives and p-coumarylated malvidin derivatives, accounted for the final products of the anthocyanin biosynthetic pathway that renders color to red grape berries. Therefore, these results show that ELR favors the accumulation of specialized color compounds and links them to sugar metabolism. The initial problems in interpreting fruit metabolic profiles and vine response variables under different treatments of leaf removal were solved through Multivariate Analysis of Variance (MANOVA) and sparse Partial Least Squares-Discriminant Analyses (sPLS-DA analyses). According to MANOVA, leaf removal prior to bloom appeared to be a stable means to affect phenolic compounds as opposed to sugars across years, suggesting sugar accumulation is a process more affected by seasonal fluctuations in growing degree days (GDD) and average daily temperature. sPLS-DA, on the other hand, identified prebloom leaf removal as a good method to improve both technological maturity and phenolic development of Merlot (V. vinifera) from a cool climate. The practice is thus advised as a reliable way of enhancing fruit quality in cool-climate viticulture [252].
In 2024, Gomez et al. [253] performed a meta-analysis combining metabolic profiling with genetic data to assess the effects of genetic distance on juice composition in several grapevine genotypes. A non-targeted metabolomics strategy was employed, allowing for the identification of 147 metabolites present in grape juices, including 30 volatiles, 21 phenolics, and 96 features using ultrahigh-performance liquid chromatography–mass spectrometry (UHPLC-MS). Late-ripening cultivars contained higher amounts of beneficial compounds, such as trans-resveratrol, catechin, and luteolin. These cultivars also had a more diverse array of volatiles, with ‘BRS Magna’ presenting 29 unique ones. Notably, the most dissimilar UHPLC-MS metabolic profile was that of ‘BRS Carmem’, a mid-season cultivar. Through targeted hybrid selection and controlled crosses, traits besting soluble solids, total phenolics, and anthocyanins were improved. Coalesced data analysis highlighted that juice metabolite profiles strongly correspond to the genetic distances between cultivars, whereas both ‘BRS Violeta’ and ‘BRS Magna’ stood out as particularly rich in health-beneficial phenolics and unique volatiles, illustrating a perfect example of how breeding has succeeded in ameliorating nutritional and sensory qualities.
Metabolomic studies in grapevine are powerful tools applied to breeding improvement and hence, provide detailed insights into biochemical diversity underlying traits related to fruit quality, stress tolerance, and disease resistance. By identifying key metabolites and correlating them with genetic backgrounds, these studies contribute toward targeted breeding to promote favorable traits. In essence, metabolomics serves as a complementary tool to genomics and proteomics in the screening and development of better grape cultivars suited to changing agricultural and market needs.
Integration of multiple Omics tools
The combined use of multi-omics tools, including genomics, transcriptomics, proteomics, and metabolomics, has become a powerful approach toward accelerating grapevine breeding by separating complex traits. Some studies combining two or more of these tools have yielded favorable results.
In response to climate-driven overripening in grapes, especially in hot, arid regions, a multi-omics study (metabolomics, transcriptomics, proteomics) was conducted on V. vinifera cv. Cabernet Sauvignon berries over two seasons to understand the molecular mechanisms behind this phenomenon. Results showed that delayed harvest significantly altered gene and protein expression: stress-responsive pathways were upregulated while metabolic functions were downregulated, suggesting a shift in berry function away from active metabolite production during overripening. Key genes and proteins related to sugars, aromatics acids, amino acids, and resveratrol metabolism were identified (Table 3) [254].
Table 3: Some of the key genes and proteins identified by Shi et al. [254] in overripe grape berries.
| Metabolic function | Genes | Proteins |
|---|---|---|
| Sugar Metabolism Glycolysis pathway | 1 VviENO (g235450) and 2 VviPK (g122800 and g382840) were up-regulated. | |
| In sucrose inversion pathway | 5 β-fructofuranosidase genes (g390650, g589680, g350940, g589650, and g130750) were up-regulated. | Protein (g551810.m01) |
| Aromatics Metabolism During the overripening stage: | VviLOX (g096070) was up-regulated. | VviHPL |
| The terpene synthesis pathway: | VviDXS was down-regulated; VviHMGCR (g308990) was up-regulated. | |
| Amino Acid Metabolism | 2 genes of glutamate dehydrogenase (g487110 and g267500) were up-regulated. | The glutamate decarboxylase protein (g559380.m02). |
| The proline dehydrogenase genes (g401310 and g137760) were up-regulated. | The glutamine synthetase protein (g190380.m01) and prolyl 4-hydroxylase protein (g001030.m04). |
In another research, the authors used an integrated metabolomic and transcriptomic approach for the identification of 55 aroma volatiles in ‘Queen Nina’ grapes, with esters emerging as the dominant compounds at maturity. Five key volatiles (benzenacetaldehyde, 2,5-dimethyl-4-hydroxy-3(2H)-furanone, ethyl octanoate, ethyl crotonate, and ethyl (E, Z)-2,4-decadienoate) were pinpointed as metabolic indicators of fruit ripening, contributing to the cultivar’s distinctive sensory profile, including notes of strawberry, acidity, and a subtle hint of rum. To decipher the regulatory basis of ester biosynthesis, transcriptome analysis revealed 29 candidate genes involved in ester metabolism, mainly within the α-linolenic and linoleic acid pathways. Notably, gene families such as LOX, HPL, ADH, and AAT were associated with ester synthesis, while the CXE gene family was identified for the first time in grapes as participating in ester degradation [255].
Building on the insights from flavor-related multi-omics studies, another integrative analysis combining proteomics and metabolomics in Cabernet Sauvignon revealed how elevated temperatures significantly disrupt berry physiology. High-temperature (HT) exposure altered the abundance of 592 proteins out of 2279 identified, particularly those involved in stress response, secondary metabolism, protein regulation, and cell wall remodeling. The study showed that heat stress impairs carbohydrate and energy metabolism in a development-dependent manner and underscored a poor correlation between transcript and protein levels, emphasizing the added value of proteomics for understanding complex responses to abiotic stress [256].
According to another report Orduña et al. [257] revealed that MYB14, MYB15, and MYB13 transcription factors play a central role in regulating resveratrol (primary stilbene synthesized in grape tissues with antioxidant and antiviral properties) biosynthesis in grapevine by directly binding to numerous genes in the stilbenoid pathway. Using DAP-Seq and gene co-expression networks, the researchers identified high-confidence targets, including 30 of the 47 stilbene synthase (STS) genes, as well as genes involved in upstream pathways (PAL, C4H, 4CL, and shikimate), confirming their broad regulatory influence on primary and secondary metabolism.
In a further multi-omics study integrating transcriptomics and metabolomics, researchers identified 14 key genes (including CXE, TPS, LOX, and AAT families) involved in the synthesis and regulation of aroma-related VOCs in ten grape cultivars. Metabolomic profiling revealed 613 volatile compounds with distinct accumulation patterns among varieties, such as linalool, β-phellandrene, and rose-related VOCs like trans-rose oxide [258].
In general, this multi-omics investigation provides relevant views on the molecular control of flavor formation in grape berries and paves the way for directed breeding programs strengthening aromatic traits. The presence of genes and metabolites of fruitiness and floral notes provides valuable targets for breeding purposes to improve grape aroma quality. The identification of candidate protein markers also sets the basis for understanding temperature-dependent changes in metabolism and for supporting the development of grapevines resilient to climate change. Altogether, a fine-tuning of harvesting timing with an aim to retain grape quality under warming conditions could be derived from these results; and also, it shows how powerful an integrated approach can be for establishing a framework for mapping transcriptional regulation, especially in non-model plants.
6.2 Artificial Intelligence in Breeding Models
For many years, genetics and conventional plant breeding have been at the forefront of agricultural advancement, primarily through processes like hybridization and selection. Despite their success, these traditional approaches are inherently time-consuming, costly, and dependent on long-term field evaluations. The advent of genomic technologies revolutionized breeding by providing unprecedented access to detailed genetic information. However, the sheer volume and complexity of this data introduced a new challenge: how to efficiently interpret and utilize it in breeding programs. In this context, Artificial Intelligence (AI)—which emulates human cognitive functions such as learning, reasoning, perception, and prediction—has emerged as a transformative tool. AI, particularly through machine learning (ML), enables rapid genomic and phenomic selection by identifying key traits associated with disease resistance. Moreover, AI facilitates the integration of multi-omics datasets to enhance predictive models for resilient plant phenotypes [259,260]. Beyond breeding, AI algorithms can also integrate data from satellites, drones, and soil sensors to anticipate disease outbreaks, accurately forecast yields, and deliver prescriptive recommendations for vineyard management, thus advancing viticulture from a merely “precision” approach to a truly “predictive” paradigm [261]. Some of the technologies that use AI in grapes include:
Machine Learning (ML) and Deep Learning (DL)
Machine learning is that branch of AI concerned with allowing a system to learn from data and to make decisions or predictions-based decisions from these data, without explicit programming to perform those activities, and Deep learning systems use multilayer neural networks to model complex patterns in data. Convolutional Neural Networks (CNNs) and Recurrent Neural Networks (RNNs) are two principal deep learning architectures and are so named because one is for image data and the other for sequence data [262,263].
It is increasingly AI technology, which involves Machine Learning (ML) and Deep Learning (DL), that is starting to improve plant breeding by altering the way things work [264]. For example, in the research of Gan et al. [265], a wide-ranging approach was undertaken that integrates DL, ML, transcriptomics, quantitative genetics, and plant phenomics to improve Genomic Selection (GS) for pest resistance in grapevines. The study proposed a Deep Convolutional Neural Network (DCNN) capable of scoring pest damage on grape leaves with an accuracy of 95.3% using the VGG16 model (this is a famous DCNN for image classification, commanding attention for its comparatively simple and uniform architecture with excellent performance in visual recognition tasks). Further, correlations of 0.94 were achieved in regression using the DCNN-PDS (Pest Damage Score or Pest Damage Severity) system. Pest damage was treated as both a binary and a continuous trait in QTL mapping. Finally, whole-genome resequencing data from 231 grapevine accessions were employed in a GWAS, identifying a total of 69 QTLs and 139 candidate genes in pest resistance pathways, such as those of jasmonic acid, salicylic acid, and ethylene signaling. When coupled with transcriptomic data, a cluster of key pest-resistance genes were identified involving ACA12 and CRK3 as hubs for herbivore defense responses. With ML-based GS, the models exhibit very high prediction accuracies (95.7%) and correlation (0.90) for binary and continuous resistance traits, respectively.
Herzog et al. [266] described the development of AI-powered sensor platforms for vineyard canopy health and disease resilience analysis. Such developments, coupled with ML applied with Self-Organizing Maps (SOMs) through a tool named “SOMmelier” were used to extract and visualize very complex genomic information of hundreds of grapevine accessions. Using this technique, aspects of genetic diversity, domestication history, and putative traits such as disease resistance can be analyzed in an easier and more interpretable way. It shows how ML can be used to tackle enormous-size genomic data and gives some ideas about where this branch of AI can be taken into grapevine transcriptomics and ampelography [267]. Likewise, a huge volume of data created using technologies like molecular biology, phenotyping, and genomics can now get combined into a data-integrated infrastructure geared towards crop improvement using AI [260].
Izquierdo-Bueno et al. [268] conducted independent research performing a systematic literature survey regarding AI applications along the entire wine production chain, from vineyard cultivation to bottling, emphasizing on the use of AI in traceability and food safety. The review also showed that AI technologies optimize the grape cultivation, fermentation, bottling processes, quality control, and the importance of AI in microbiological risk management, most especially in the detection and control of mycotoxins. Another key topic addressed by the authors is how AI enables the detection of plant diseases with various tools, such as the integration of Transfer Learning with Vision Transformer (TLMViT), which works with images, and PlantVillage, which analyses information of more than 54,000 images, thus allowing rates of identification above 98% [269,270,271]. In related research Kunduracioglu and Pacal [272] applied DL to classify grapevine leaves and diseases, where fine-tuning pre-trained convolutional neural network (CNN) and vision transformer models—particularly Swinv2-Base—achieved 100% accuracy in identifying four disease classes (Black Rot, Leaf Blight, Healthy, and Esca) and five classes for leave recognition (Ak, Alaidris, Buzgulu, Dimnit, and Nazli) across two datasets, underscoring the potential of DL for early disease detection and precise cultivar identification in viticulture. According to authors in DL, CNNs are widely used in image recognition, employing convolutional layers to extract features, pooling layers to reduce dimensionality, and fully connected layers for classification or prediction. In contrast, vision transformers (ViTs) apply a transformer-based architecture originally developed for natural language processing, using self-attention mechanisms to capture long-range dependencies in images—an approach that has proven highly effective for visual recognition and object detection tasks.
According to other reports, the Deep Convolutional Neural Networks (DCNN) based on VGG16 architecture (vision model being the highest technology available today)-further extended by more CNN layers and data augmentation-has succeeded in grape leaf disease detection with accuracy of 99.18% on training data and 99.04% on test data, thereby showing a strong potential of being used as a quick and reliable decision support mechanism for disease management in viticulture. In addition, the DCNN-based classifier model turns out to be an effective decision support tool for farmers in fast and accurate identification of grapevine diseases for timely and appropriate intervention. This study has proved the reliability of the model and emphasized a promising application of the model in managing diseases in agriculture [273]. CNN architectures were also used for the detection of plant stress with grapevine RGB images taken under real field conditions with natural variability in parameters such as light, shadows, insects, etc. Amongst five models tried, EfficientNetB3 (deep learning model that offers high accuracy and computational efficiency for image classification tasks, making it ideal for detecting grapevine stress or diseases in real-world field conditions) yielded the best results with 97.2 percent classification accuracy, 0.996 ROC (Receiver Operating Characteristic curve) AUC (Area Under the Curve), and 0.958 average precision, thus showing promise as a potential tool for helping winegrowers in the detection of stress beyond the traditional visual inspection method [274].
Related to low temperature stress Konecny et al. [275] developed a ML approach based on SOMs that analyze gene expression patterns under diverse temperature regimes. It was in this study that the fruitful application of SOM was shown in the analysis of complex transcriptomic datasets, and in providing insights into the molecular mechanism of cold stress responses in grapevines. Notably, the analysis revealed a link between temperature-regulated preservation and the biosynthesis of vitamin B1 (thiamine). Additionally, the identification of potential epigenetic events suggested a regulatory role in modulating gene expression in response to low-temperature exposure, highlighting the relevance of integrative ML strategies in grapevine stress physiology.
Other DL investigation was conducted by Mashharawi et al. [276] for the identification of grape type using 4565 images with 2393 allocated for training, 1026 for validation, and 1146 for testing—corresponding to a 70%/30% split between training and validation datasets. Remarkably, the trained model achieved 100% accuracy. This study represents the first attempt to classify grape species based on color variation (including black, crimson, yellow, dark blue, green, and pink).
Together, these investigations provide major statements on the applications that AI, particularly ML and DL, can have in transforming grapevine breeding, stress physiology, disease control, and cultivar identification. The enhancements brought in the field of genomic selection for pest resistance with DCNN-based scoring systems stand to truly change the paradigm from predictive viticulture to precision viticulture aided by AI applications, where disease diagnosis and grape varieties classification can be achieved with an accuracy of almost 100%, employing CNNs and Vision Transformers. AI-based tools such as SOM-based transcriptome analysis, DCNNs based on the VGG16 architecture, and EfficientNetB3-based models provide reliable decision support systems with higher accuracy than traditional methodologies in field conditions. Furthermore, with the help of integrated AI approaches, transcriptomic, genomic, phenomic, and environmental data have been integrated to figure out stress-responsive genes, candidate QTLs, and epigenetic mechanisms in response to heat and cold stresses. These novel developments are immediately useful for vineyard management, early disease detection, and rapid diagnosis, while a long-term strategy for producing climate-resilient and high-quality grape cultivars is underway. To conclude, an AI and DL application enhances accuracy and speed in viticulture and makes the very foundation of the data-driven future for sustainable grape production.
Drone Technology and Sensors
Sensors are devices designed to detect and respond to environmental stimuli such as light, heat, motion, moisture, pressure, or chemical composition. These inputs are typically converted into electrical signals that can be recorded, analyzed, or interpreted by humans or machines [277]. Drones, or unmanned aerial vehicles (UAVs), are remotely operated aircraft equipped with advanced sensors and high-resolution cameras, enabling the efficient acquisition of spatial and environmental data for agricultural applications [278].
With the sensor technologies and platforms for drones evolving, it is now possible to monitor grapevines and breeding techniques. Sharma et al. [279] discussed how low-cost Vis-NIR cameras are effective in evaluating physiological traits in grapevines, while Sawyer et al. [280] validated hyperspectral imaging as a pre-symptomatic virus differentiation tool in precision viticulture. Engler et al. [281] and Moreno and Andújar [282] considered 2.5D and 3D phenotyping tools as complementary to alleviate background noise through depth maps and enhance the morphological characterization of canopy architecture and yield parameters. In field applications on vineyards, Šupčík et al. [283] estimated yield from canopy measurements using UAV-based RGB cameras, and López et al. [284] treated the hyperspectral data’s curse of dimensionality via spatial attention modules and inception blocks to improve biophysical and yield trait retrieval for genetic selection.
Field-based yield prediction studies also benefited from UAV photogrammetry. Torres-Sánchez et al. [285] accurately estimated yields with 3-point clouds and color indices for two grape cultivars under contrasting defoliation treatments in Navarra, Spain. In a similar fashion, Ballesteros et al. [286] and Miranda-Fuentes et al. [287] mention that obtaining canopy data at key stages increases yield accuracy-and yet the 3D processing continues to be the core heavy computational task. In another study, Bendel et al. [288] used Vis-NIR sensors to diagnose endophytic diseases like grapevine yellows, showing the application of imaging tools for early disease detection and health monitoring.
On the disease management side, UAVs were put to use by Albetis et al. [289] to photogrammetrically detect Flavescence dorée and Grapevine Trunk Diseases in French vineyards, thereby cutting down manual scouting and permitting targeted pesticide application. In contrast, MacDonald et al. [290] developed spectral bands to detect grapevine leaf curl virus in real time. Such methods are applied not only to the management of diseases but also to the identification of resistant genotypes for breeding. Patil and Thorat [291] created a sensor- and ML-based system that worked on an IoT platform to monitor leaf temperature and humidity, which were early indicators of concern, plus it sent real-time alerts to the farmer, such that it could be cited as a practical instance of predictive viticulture. Kiani and Seyyedabbasi [277] further explored the wider potential of IoT in agriculture by defining it as a network of interconnected devices that trade data in order to arrive at autonomous decisions.
In breeding applications, Kicherer et al. [292] created an autonomous robot capable of imaging 250 vines per hour and analyzing fruit size and color through IMAGEdata to generate high-throughput phenotypic data needed for selecting genotypes with desirable traits. This earlier classic work by Lorenz et al. [293] highlighted the difficulty of phenotyping large, genetically diverse grapevine populations wherein high-resolution imaging remains a key to distinguishing subtle genotypic differences within breeding blocks.
In more general terms, UAVs have been solicited for quick gathering of high-resolution data on crops for assessing their health, nutrient status, water requirement, and soil variability. Studies by Barasa et al. [294] and Zarco-Tejada et al. [295] show how UAVs optimize inputs, minimize chemical use, and increase productivity, while review papers by Petkovics et al. [296] and Deng et al. [297] underline the operational and agronomic advantages of UAVs in precision agriculture.
The integration of drones and advanced sensing technologies-from hyperspectral and Vis-NIR imaging to 3D mapping and IoT platforms-provide a dynamic, non-invasive toolset for real-time monitoring, stress detection, and phenotyping of the grapevine. One of the newest developments in grapevine breeding is the simultaneous, precise, and high-throughput selection of genotypes capable of enhanced yield, stress tolerance, and fruit quality. Other than the non-destructive, real-time assessment of genotype phenotypic structure and traits such as canopy development, berry growth, and disease symptoms-extending across large vineyard areas-drones and sensors have brought cheap and fast high-throughput phenotyping into the vineyard. This, in turn, reduces human manipulation, thereby lowering cost and time investment, i.e., feeding back in data-driven decision-making initiatives within breeding programs, thus increasing both the efficiency and pace of genetic enhancement. The marriage of remote sensing and AI-driven analytics thus becomes the bedrock of another era-a precision breeding and sustainable viticulture.
Finally, Fig. 5 illustrates the mechanisms and stages of adoption of AI and drone technologies in the genetic improvement of grapevines.
Figure 5: Mechanism and adoption of AI and drone technology in grape improvement.
The figure depicts the implementation of artificial intelligence (AI) and drone technologies in grapevine breeding, revealing not only the mechanisms behind it but also the stages of adoption. Drones outfitted with high-performance sensors perform real-time, high-throughput phenotyping by monitoring and reporting the most significant data factors related to tree row—structure, disease, and fruit. The gathered data goes to AI-enabled models—comprising machine learning and deep learning algorithms—which are used for the aforementioned tasks as genotype selection, stress detection, yield forecasting, and early disease diagnosis. The combination of these technologies, therefore, not only creates a perfect ground for breeding but also speeds up the decision-making process and lowers the need for conventional fieldwork. The adoption of these technologies is thus taking the wine-producing environment from traditional to a data-powered, eco-friendly, and more efficient one in terms of grapevine genetic improvement.
While this review has highlighted the promising applications of novel grape breeding methods, it is important to acknowledge that these strategies are not without limitations (Table 4).
Table 4: Main limitations and challenges in the integration of omics, artificial intelligence, and sensor technologies for grapevine breeding applications.
| Technology | Limitation | Challenge | References |
|---|---|---|---|
| Multi-omics | Individual omics offer partial insights; only integrated multi-omics can elucidate complete phenotype regulation. | Integration of diverse omics datasets remains computationally and methodologically complex. | [298,299] |
| AI | High-dimensional datasets require advanced and specialized computational tools for analysis. | Lack of user-friendly, domain-specific platforms for non-expert breeders. | [300,301,302] |
| AI | DL and ML models often act as “black boxes”, lacking transparency. | Interpreting AI predictions in biologically meaningful ways is difficult. | [303] |
| AI | Annotation of agricultural images requires expert input, increasing cost and limiting scalability. | Scarcity of large, high-quality labeled datasets in the agricultural domain. | [304] |
| AI/multi-omics | Building biological interaction networks requires complex multi-layered data interpretation. | Understanding cross-talk between genomic, transcriptomic, and phenotypic layers remains limited. | [305] |
| Genomic/AI | Genomic and sensor data raise concerns about privacy and equitable access. | Ethical and regulatory frameworks are still evolving in agricultural genomics. | [306] |
| AI | High initial costs of sensor and imaging platforms. | Adoption in low-resource settings is limited due to lack of infrastructure and training. | [307] |
Despite countless scientific breakthroughs for omics integration, AI, and sensor-based technologies aimed at grapevine improvement, equally critical challenges remain. They include heterogeneity across omics data platforms, severe unavailability of high-quality annotated datasets, and the inferences that are made from the complex AI models. Inability to ensure computational scalability, to calibrate sensors properly, and having environmental variability also affect prediction reproducibility and generalizability. Furthermore, considerations regarding ethics related to data ownership and the privileges of well-guarded and developing breeding programs need to be considered. Overcoming these limitations will require synergic efforts and agreements toward standardization of protocols in data acquisition and sharing, alongside with building highly robust and transparent analytic pipelines. Once all are in place, the multi-omics and AI-oriented sensing technological framework will promote breeders increasingly at developing climate-resilient, high-quality grapevine cultivars in a fair and sustainable manner.
7 Future Trends and Opportunities
In the genetic improvement of Vitis vinifera L., climate change, the emergence and spread of new diseases, and the demands of constantly evolving markets constitute growing challenges [1,5,6]. In this context, several trends and opportunities can be envisaged that should guide research and development in the coming years.
The introduction of resistant genes from wild species and local varieties to create climate-resilient cultivars is one of the most important first steps. The genetic resources represented by these wild species and local varieties are significant alleles for tolerance to the extreme conditions of drought, frost, and heat. Consequently, these genetic resources are gradually increasing the genetic base in breeding programs [60,159].
Complementarily, the integration of traditional breeding methods with advanced biotechnological tools, such as MAS [117,118,119,120,143], gene editing through CRISPR/Cas9 [14,191,192,193,194,195,196], and genomic design [157,158,159], will accelerate the development of varieties with greater tolerance to biotic and abiotic factors, without sacrificing wine or fresh consumption quality. The use of wild species and local varieties, which have so far been underutilized, will be key to broadening the genetic base and improving grapevine resilience [3,8,12].
In this context, the coupling of MAS with genomic prediction is a very significant step in the right direction that allows polygenic traits like yield, phenology, and abiotic stress tolerance to be targeted with greater efficiency. The potential of genomic prediction to hasten the identification of superior genotypes has been supported by recent studies in grapevine populations that have demonstrated high levels of accuracy across multiple traits [173].
Clonal diversity is yet another important factor, which not only provides a source of genetic variation but also of epigenetic variation. The application of different clones along with genomic design strategies is a leap towards not only resilience but also enological quality. The recent literature calls for the merging of clonal diversity with multi-omics platforms in order to discover the functionally relevant variants [60].
In this regard, multi-omics technologies (genomics, transcriptomics, proteomics, metabolomics, and epigenomics) will become pillars for understanding the genetic architecture of complex traits, identifying key metabolic pathways, and, on that basis, designing more precise selection strategies [142,145]. The increasing availability of pangenomes and high-quality reference sequences [135,142,145], together with high-resolution association analyses [127,138,140], will facilitate the future identification of genes and alleles of interest.
In parallel, gene editing, particularly in DNA-free modalities [209,210], opens opportunities for the development of improved varieties that could gain greater public and regulatory acceptance [181]. This will make it possible to precisely target genes associated with disease resistance, fruit quality, and adaptation to climate stress [191], thereby reducing the development and introduction times of new varieties compared to conventional programs.
In addition, in vitro propagation and somatic embryogenesis will remain essential tools for large-scale multiplication and conservation of elite genotypes, as well as for the regeneration of edited or genetically transformed plants [92,102]. Their optimization will contribute to the production of pathogen-free plant material and the preservation of at-risk varieties [105,106].
In the future, the merging of artificial intelligence and machine learning with genetic prediction models will lead to new possibilities for improving the understanding of genotype × environment interactions and selecting the best parents. For instance, the latest studies can reach 97% accuracy with these tools in the case of complicated traits like seedlessness [308].
Besides, the growing demand for seedless grapes [56,57,58,59], with differentiated sensory profiles and longer postharvest life, will open new market niches and stimulate the development of specific programs that integrate quality, productivity, and sustainability [81,82,83]. Future viticulture will need to strike a balance between technological innovation, the preservation of genetic diversity, and adaptation to the expectations of consumers and producers [86,140]. Finally, AI and predictive modeling, leveraging field and molecular data, will make it possible to anticipate genotype performance in different production and climatic scenarios, guiding selection and management decisions.
It is crucial to examine the challenges inherent in combining conventional breeding, modern molecular tools, and AI/drone technologies in grape improvement. Traditional breeding still faces long generation times, limited recombination for complex traits, and high cost of field trials. Molecular and genomic methods (such as MAS, genomic selection) accelerate progress, but they suffer from issues of prediction accuracy across environments, marker–trait decay, and limited ability to capture epistasis or genotype × environment interactions. For example, the integration of large-scale omics datasets into predictive models is hampered by data heterogeneity, missing values, and computational burdens [309].
When adding AI and drone technologies, new bottlenecks emerge. While UAV-based sensing and AI models enable high-throughput phenotyping and early disease detection, their adoption is constrained by high startup costs, battery and flight constraints, need for calibration across diverse sensor platforms, and limited generalization of AI models to new vineyards or climates [310]. In viticulture specifically, remote sensing has shown promise for disease detection and variety classification [311], but the translation of these methods into breeding decisions is still nascent. Overcoming these hurdles will require standardized data pipelines, robust model transferability, and integration across scales—from leaf to vineyard—so that drones and AI tools truly support varietal improvement in the field rather than simply monitoring it.
In summary, the integration of multi-omics approaches, clonal diversity, pangenomic resources, and artificial intelligence is redefining the landscape of grapevine breeding. These combined advances lay the foundation for precision genome editing and the development of more sustainable cultivars, capable of addressing current sanitary, environmental, and market challenges through enhanced data-driven selection and predictive modeling [79,312,313].
Grapevine genetic improvement is currently at a point of convergence between traditional methods of hybridization, cloning, and grafting and a new era of innovation based on genomic precision, gene editing, and integrated multi-omic approaches. Molecular and biotechnological tools, along with the exploitation of available genetic diversity, offer new possibilities for the development of varieties that respond to current and future challenges, without losing sight of the quality and organoleptic characteristics demanded by the market.
Advances in analysis and selection technologies, as well as the integration of approaches that combine field and laboratory data, will accelerate the development of varieties adapted to changing climatic conditions and new health pressures. The efficient propagation of elite genotypes and the conservation of genetic resources will be essential to ensure the sustainability and resilience of the wine sector.
Therefore, it will be essential to advance concrete strategies to strengthen breeding programs, such as the integration of marker-assisted selection with genomic prediction for polygenic traits, the development of climate-resilient cultivars that take advantage of the diversity of wild species and local varieties, and the combination of clonal diversity with genomic design supported by multi-omics platforms. From now on and in the future, the trend is the use of artificial intelligence and advanced predictive models, which will improve the interpretation of genotype-environment interactions and accelerate the selection of superior genotypes. The incorporation of multi-omics approaches and the use of pangenomes will provide a more solid foundation for the identification of key genes and the implementation of precision gene editing. Therefore, the inclusion of sustainability criteria in breeding objectives will be essential to ensure not only productivity and quality, but also the resilience and sustainability of viticulture in the context of climate change.
Despite these achievements, critical gaps in our understanding of the genotype × environment × management interaction persist. The poor standardization of multi-omics data, limited validation of AI predictions under field conditions, and insufficient exploration of the vineyard-associated microbiome are priority areas for research. Furthermore, the transition to selection programs assisted by genomic prediction models requires strengthening bioinformatics infrastructure and multidisciplinary training in computational biology, quantitative genetics, and applied viticulture.
In short, it can be said that the viticulture of the future must balance productivity, sustainability, and innovation, integrating solutions that address both production efficiency and biodiversity preservation. The ability to anticipate and adapt to environmental conditions, supported by cutting-edge scientific and technological tools, will be key to maintaining competitiveness and meeting the expectations of consumers and producers. Furthermore, the integration of multi-omics platforms with machine learning models and the construction of dynamic pangenomes will allow for a more precise deciphering of the genetic basis of traits of interest in grapevines. These approaches will not only transform the efficiency of breeding programs but will also contribute to designing strategies for adaptation to climate change and the genetic diversification of plantations.
In conclusion, the future of grapevine breeding will depend on the ability to combine the empirical tradition of viticulture with the digital and biotechnology revolution. The challenge and opportunity lie in translating genomic information into intelligent agronomic decisions that ensure the sustainability, resilience, and quality of grapevine cultivation in the coming decades.
Acknowledgement:
Funding Statement: The authors received no specific funding for this study.
Author Contributions: The authors confirm contribution to the paper as follows: Conceptualization, Sandra Pérez-Álvarez; writing—original draft preparation, Sandra Pérez-Álvarez, Eduardo Fidel Héctor-Ardisana and Luisa Patricia Uranga-Valencia; writing—review and editing, Sandra Pérez-Álvarez, Eduardo Fidel Héctor-Ardisana, Eduardo Sandoval Castro and Erick H. Ochoa-Chaparro. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: No data was used for the research described in the article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
References
1. Li Z , Wu R , Guo F , Wang Y , Nick P , Wang X . Advances in the molecular mechanism of grapevine resistance to fungal diseases. Mol Hortic. 2025; 5: 1. doi:10.1186/s43897-024-00119-x. [Google Scholar] [CrossRef]
2. Vignani R , Scali M . Grapevine origin and diversity. In: Martin-Lopes P , editor. Grapevine: from origin to the vineyard. advances in botanical research, 110. Cambridge, MA, USA: Academic Press; 2024. p. 1– 25. [Google Scholar]
3. Ikten A , Sari D , Sabir A , Mydan H , Mutlu N . Estimating genetic diversity among selected wild grapevine genotypes from Southern Turkey by simple sequence repeat (SSR) and inter-Primer Binding Site(iPBS) markers. Genet Resour Crop Evol. 2025; 72: 2361– 77. doi:10.1007/s10722-024-02102-3. [Google Scholar] [CrossRef]
4. Morales-Cruz A , Aguirre-Liguori J , Massonet M , Minio A , Zaccheo M , Cochetel N , et al. Multigenic resistance to Xylella fastidiosa in wild grapes (Vitis sps.) and its implications within a changing climate. Commun Biol. 2023; 6: 580. doi:10.1038/s42003-023-04938-4. [Google Scholar] [CrossRef]
5. Lippa MN , Tarolli P , Straffelini E . Climate change impacts and the reshaping of Canadian viticulture. iScience. 2025; 26–28( 3): 111941. doi:10.1016/j.isci.2025.111941. [Google Scholar] [CrossRef]
6. Gerbi V , De Paolis C . The effects of climate change on wine composition and winemaking processes. Ital J Food Sci. 2025; 37( 1): 246– 60. doi:10.15586/ijfs.v37i1.2775. [Google Scholar] [CrossRef]
7. Alonso-Forn D , Buesa I , Flor L , Sabater A , Medrano H , Escalona JM . Implications of root morphology and anatomy for water deficit tolerance and recovery of grapevine rootstocks. Front Plant Sci. 2025; 16: 1541523. doi:10.3389/fpls.2025.1541523. [Google Scholar] [CrossRef]
8. Atak A . Vitis species for stress tolerance/resistance. Genet Resour Crop Evol. 2025; 72: 2425– 44. doi:10.1007/s10722-024-02106-z. [Google Scholar] [CrossRef]
9. Mendes Lemos A , Machado N , Egea-Cortines M , Barros AI . Assessment of quality parameters and phytochemical content of thirty “Tempranillo” grape clones for varietal improvement in two distinct sub-regions of Douro. Sci Hortic. 2020; 262: 109096. doi:10.1016/j.scienta.2019.109096. [Google Scholar] [CrossRef]
10. Tortosa I , Escalona JM , Toro G , Douthe C , Medrano H . Clonal behavior in response to soil water availability in Tempranillo grapevine cv: From plant growth to water use efficiency. Agronomy. 2020; 10( 6): 862. doi:10.3390/agronomy10060862. [Google Scholar] [CrossRef]
11. Chen Y , Fei Y , Howell K , Chen D , Clingeleffer P , Zhang P . Rootstocks for grapevines now and into the future: Selection of rootstocks based on drought tolerance, soil nutrient availability, and soil pH. Aust J Grape Wine Res. 2024; 2024: 6704238. doi:10.1155/2024/6704238. [Google Scholar] [CrossRef]
12. Kui L , Tang M , Duan S , Wang S , Dong X . Identification of selective sweeps in the domesticated table and wine grape (Vitis vinifera L.). Front Plant Sci. 2020; 11: 572. doi:10.3389/fpls.2020.00572. [Google Scholar] [CrossRef]
13. Liu Z , Wang N , Su Y , Long Q , Peng Y , Shangguan L , et al . Grapevine pangenome facilitates trait genetics and genomic breeding. Nat Genet. 2024; 56: 2804– 14. doi:10.1038/s41588-024-01967-5. [Google Scholar] [CrossRef]
14. Villette J , Lecourieux F , Bastiancig E , Héloir MC , Poinssot B . New improvements in grapevine genome editing: high efficiency biallelic homozygous knock-out from regenerated plantlets by using an optimized zCas9i. Plant Methods. 2024; 20( 1): 45. doi:10.1186/s13007-024-01173-8. [Google Scholar] [CrossRef]
15. Arnold C , Schnitzler A . Ecology and genetics of natural populations of North American Vitis Species used as rootstocks in European grapevine breeding programs. Front Plant Sci. 2020; 11: 866. doi:10.3389/fpls.2020.00866. [Google Scholar] [CrossRef]
16. Gautier A , Cookson SJ , Lagalle L , Ollat N , Marguerit E . Influence of the three main genetic backgrounds of grapevine rootstocks on petiolar nutrient concentrations of the scion, with a focus on phosphorus. OENO One. 2020; 54( 1): 1– 13. doi:10.20870/oeno-one.2020.54.1.2458. [Google Scholar] [CrossRef]
17. Provost C , Campbell A , Dumont F . Rootstocks impact yield, fruit composition, nutrient deficiencies, and winter survival of hybrid cultivars in eastern Canada. Horticulturae. 2021; 7( 8): 237. doi:10.3390/horticulturae7080237. [Google Scholar] [CrossRef]
18. Cochetel N , Ghan R , Toups HS , Degu A , Tillett RL , Schlauch KA , et al. Drought tolerance of the grapevine, Vitis champinii cv. Ramsey, is associated with higher photosynthesis and greater transcriptomic responsiveness of abscisic acid biosynthesis and signaling. BMC Plant Biol. 2020; 20( 1): 55. doi:10.1186/s12870-019-2012-7. [Google Scholar] [CrossRef]
19. Prinsi B , Simeoni F , Galbiati M , Meggio F , Tonelli C , Scienza A , et al. Grapevine rootstocks differently affect physiological and molecular responses of the scion under water deficit condition. Agronomy. 2021; 11( 2): 289. doi:10.3390/agronomy11020289. [Google Scholar] [CrossRef]
20. Chitarra W , Perrone I , Avanzato CG , Minio A , Boccacci P , Santini D , et al. Grapevine grafting: scion transcript profiling and defense-related metabolites induced by rootstocks. Front Plant Sci. 2017; 8: 654. doi:10.3389/fpls.2017.00654. [Google Scholar] [CrossRef]
21. Camargo UA . Grape breeding for the subtropical and tropical regions of Brazil. Acta Hortic. 2000; 528( 68): 473– 7. doi:10.17660/ActaHortic.2000.528.68. [Google Scholar] [CrossRef]
22. Burger P , Bouquet A , Striem MJ . Grape Breeding. In: Breeding plantation tree crops: tropical species. Berlin/Heidelberg, Germany: Springer; 2009. p. 161– 89. doi:10.1093/aob/mcp206. [Google Scholar] [CrossRef]
23. Zhang JL , Cao ZY , Ma JF . Screening of cold-resistant seedlings of a Chinese wild grape (Vitis piasezkii Maxim var. pagnucii) native to loess plateau of eastern Gansu province, China, as rootstocks. Sci Hortic. 2009; 122( 1): 125– 8. doi:10.1016/j.scienta.2009.04.002. [Google Scholar] [CrossRef]
24. Halbrooks MC , Mortensen JA . Origin and significance of Florida hybrid bunch grapes and rootstocks. Hort Sci. 1989; 24( 4): 546– 50. doi:10.21273/HORTSCI.24.4.546. [Google Scholar] [CrossRef]
25. Conner PJ . A century of muscadine grape (Vitis rotundifolia Michx) breeding at the University of Georgia [ dissertation]. Athens, GA, USA: Center for Agribusiness and Economic Development, University of Georgia; 2010. p. 1– 7. [Google Scholar]
26. Alleweldt G , Possingham JV . Progress in grapevine breeding. Theoret Appl Genet. 1988; 75: 669– 73. doi:10.1007/BF00265585. [Google Scholar] [CrossRef]
27. Olien WC . The Muscadine grape: Botany, viticulture, history, and current industry. Hort Sci. 1990; 25( 7): 732– 9. doi:10.21273/HORTSCI.25.7.732. [Google Scholar] [CrossRef]
28. Alleweldt G , Spiegel-Roy P , Reisch B . Grapes (Vitis). Acta Hortic. 1991; 290: 291– 330. doi:10.17660/actahortic.1991.290.7. [Google Scholar] [CrossRef]
29. Riaz S , Tenscher AC , Graziani R , Krivanek AF , Ramming DW , Walker MA . Using marker-assisted selection to breed pierce’s disease-resistant grapes. Am J Enol Vitic. 2009; 60( 2): 199– 207. doi:10.5344/ajev.2009.60.2.199. [Google Scholar] [CrossRef]
30. Merdinoglu D , Schneider C , Prado E , Wiedemann-Merdinoglu S , Mestre P . Breeding for durable resistance to downy and powdery mildew in grapevine. OENO One. 2018; 52( 3): 203– 9. doi:10.20870/oeno-one.2018.52.3.2116. [Google Scholar] [CrossRef]
31. Villano C , Aversano R . Towards grapevine (Vitis vinifera L.) mildews resistance: molecular defence mechanisms and new breeding technologies. Italus Hortus. 2020; 27: 1– 17. doi:10.26353/j.itahort/2020.3.0117. [Google Scholar] [CrossRef]
32. Zanghelini JA , Bogo A , Dal Vesco LL , Gomes BR , Mecabô CV , Herpich CH , et al. Response of PIWI grapevine cultivars to downy mildew in highland region of southern Brazil. Eur J Plant Pathol. 2019; 154: 1051– 8. doi:10.1007/s10658-019-01725-y. [Google Scholar] [CrossRef]
33. De Bem BP , Bogo A , Brighenti AF , Bruz DA , Allebrand R , Stefanini M , et al. Temporal dynamics of grapevine downy mildew in Piwi varieties in San Michele all’Adige Region, Trentino-Italy. Summa Phytopathol. 2020; 46( 3): 212– 20. doi:10.1590/0100-5405/230013. [Google Scholar] [CrossRef]
34. Regner F , Endler A , Ferschel E , Hack R , Rockenbauer A , Eisenheld C . The change to ‘Piwi’ cultivars in the Austrian viticulture. Acta Hortic. 2025; 1418: 205– 16. doi:10.17660/ActaHortic.2025.1418.26. [Google Scholar] [CrossRef]
35. Blanco-González SI , Hernández MM , Menéndez CM . ‘Piwi’ cultivars for La Rioja: innovation for improved sustainability. Acta Hortic. 2025; 1418: 217– 22. doi:10.17660/ActaHortic.2025.1418.27. [Google Scholar] [CrossRef]
36. Weinmann E , Boos M , Ehret P , Flubacher L , Schneider C , Veith L . Breeding of new disease-tolerant grape varieties—viticulture in times of climatic change. Bio Web Conf. 2019; 15: 01035. doi:10.1051/bioconf/20191501035. [Google Scholar] [CrossRef]
37. Koyama K , Kamigakiuchi H , Iwashita K , Mochioka R , Goto-Yamamoto N . Polyphenolic diversity and characterization in the red–purple berries of East Asian wild Vitis species. Phytochem. 2017; 134: 78– 86. doi:10.1016/j.phytochem.2016.10.003. [Google Scholar] [CrossRef]
38. Wolkovich EM , García de Cortázar-Atauri I , Morales-Castilla I , Nicholas KA , Lacombe T . From pinot to Xinomavro in the world’s future wine-growing regions. Natu Clim Change. 2018; 8: 29– 37. doi:10.1038/s41558-017-0016-6. [Google Scholar] [CrossRef]
39. Bigard A , Berhe DT , Maoddi E , Sire Y , Boursiquot J-M , Ojeda H , et al. Vitis vinifera L. fruit diversity to breed varieties anticipating climate changes. Front Plant Sci. 2018; 9: 455. doi:10.3389/fpls.2018.00455. [Google Scholar] [CrossRef]
40. Atucha A , Hedtcke J , Workmaster BA . Evaluation of cold-climate interspecific hybrid wine grape cultivars for the upper midwest. J Am Pomol Soc. 2018; 2( 2): 80– 93. [Google Scholar]
41. Svyantek A , Stenger J , Auwarter C , Shikanai A , Köse B , Wang Z , et al. This is how we chill from ‘23’ til: breeding cold hardy grapevines for unprecedented and unpredictable climate challenges. Acta Hortic. 2024; 1385: 127– 38. doi:10.17660/ActaHortic.2024.1385.17. [Google Scholar] [CrossRef]
42. Coupel-Ledru A , Lebon E , Christophe A , Gallo A , Gago P , Pantin F , et al. Reduced nighttime transpiration is a relevant breeding target for high water-use efficiency in grapevine. Proc Natl Acad Sci U S A. 2016; 113( 32): 8963– 8. doi:10.1073/pnas.1600826113. [Google Scholar] [CrossRef]
43. Gambetta GA , Herrera JC , Dayer S , Feng Q , Hochberg U , Castellarin SD . The physiology of drought stress in grapevine: towards an integrative definition of drought tolerance. J Exp Bot. 2020; 71( 16): 4658– 76. doi:10.1093/jxb/eraa245. [Google Scholar] [CrossRef]
44. Jiao S , Zeng F , Huang Y , Zhang L , Mao J , Chen B . Physiological, biochemical and molecular responses associated with drought tolerance in grafted grapevine. BMC Plant Biol. 2023; 23: 110. doi:10.1186/s12870-023-04109-x. [Google Scholar] [CrossRef]
45. Buesa I , Pérez-Pérez JG , Visconti F , Strah R , Intrigliolo DS , Bonet L , et al. Physiological and transcriptional responses to saline irrigation of young ‘Tempranillo’ vines grafted onto different rootstocks. Front Plant Sci. 2022; 13: 1011533. doi:10.3389/fpls.2022.1011533. [Google Scholar] [CrossRef]
46. Venios X , Korkas E , Nisiotou A , Banilas G . Grapevine responses to heat stress and global warming. Plants. 2020; 9( 12): 1754. doi:10.3390/plants9121754. [Google Scholar] [CrossRef]
47. Magon G , de Rosa V , Martina M , Falchi R , Acquadro A , Barcaccia G , et al. Boosting grapevine breeding for climate-smart viticulture: from genetic resources to predictive genomics. Front Plant Sci. 2023; 14: 1293186. doi:10.3389/fpls.2023.1293186. [Google Scholar] [CrossRef]
48. Santiago-Brown I , Metcalfe A , Jerram C , Collins C . Sustainability assessment in wine-grape growing in the new world: Economic, environmental, and social indicators for agricultural businesses. Sustainability. 2015; 7( 7): 8178– 204. doi:10.3390/su7078178. [Google Scholar] [CrossRef]
49. Migicovsky Z , Cousins P , Jordan LM , Myles S , Striegler RK , Verdegaal P , et al. Grapevine rootstocks affect growth-related scion phenotypes. Plant Direct. 2021; 5: e00324. doi:10.1002/pld3.324. [Google Scholar] [CrossRef]
50. Bokzurt A , Yağci A . A general overview of grape breeding in consideration of literature from past to present. In: Kunter B , Keskin N , editors. Chapters on viticulture. Ankara, Turkey: IKSAD Publishing House; 2022. p. 3– 24. [Google Scholar]
51. Bigard A , Romieu C , Sire Y , Torregrosa L . Vitis vinifera L. Diversity for cations and acidity is suitable for breeding fruits coping with climate warming. Front Plant Sci. 2020; 11: 01175. doi:10.3389/fpls.2020.01175. [Google Scholar] [CrossRef]
52. Hatterman HM , Auwarter CP , Stenger JE . Evaluation of cold-hardy grape cultivars for North Dakota and the North Dakota State University germplasm enhancement project. Acta Hortic. 2016; 115: 13– 22. doi:10.17660/ActaHortic.2016.1115.3. [Google Scholar] [CrossRef]
53. Wang ZL , Xue TT , Gao FF , Zhang L , Han X , Wang Y , et al. Intraspecific recurrent selection in V. vinifera: an effective method for breeding of high quality, disease-, cold-, and drought-resistant grapes. Euphytica. 2021; 217: 111. doi:10.1007/s10681-021-02851-7. [Google Scholar] [CrossRef]
54. Manso-Martínez C , Sáenz-Navajas MP , Hernández MM , Menéndez CM . Sensory profiling and quality assessment of wines derived from Graciano X Tempranillo selections. LWT. 2020; 127: 109394. doi:10.1016/j.lwt.2020.109394. [Google Scholar] [CrossRef]
55. Wang ZL , Yao F , Hui M , Wu D , Wang Y , Han X , et al. Fertility analysis of intraspecific hybrids in Vitis vinifera and screening of superior hybrid combinations. Front Plant Sci. 2022; 13: 940540. doi:10.3389/fpls.2022.940540. [Google Scholar] [CrossRef]
56. Akkurt M , Tahmaz H , Veziroğlu S . Recent developments in seedless grapevine breeding. S Afr J Enol Vitic. 2019; 40( 2): 260– 5. doi:10.21548/40-2-3342. [Google Scholar] [CrossRef]
57. Leão PCS , Carvalho JN . Assessment of table grape progenies and correlation between seedlessness and other agronomic traits. Rev Bras Frutic. 2023; 45: e-225. doi:10.1590/0100-29452023225. [Google Scholar] [CrossRef]
58. Atak A , Akgün M , Sen A , Göksel Z , Uzun A . Evaluation of quality characteristics and cultivar candidate potentials of hybrid genotypes obtained from different Vitis species. Hortic. 2025; 11( 4): 354. doi:10.3390/horticulturae11040354. [Google Scholar] [CrossRef]
59. Doligez A , Bouquet A , Danglot Y , Lahogue F , Riaz S , Meredith CP , et al. Genetic mapping of grapevine (Vitis vinifera L.) applied to the detection of QTLs for seedlessness and berry weight. Theor Appl Genet. 2002; 105( 5): 780– 95. doi:10.1007/s00122-002-0951-z. [Google Scholar] [CrossRef]
60. Callipo P , Schmidt M , Strack T , Robinson H , Vasudevan A , Vossfels KP . Harnessing clonal diversity in grapevine: from genomic insights to modern breeding applications. Theor Appl Genet. 2025; 138: 196. doi:10.1007/s00122-025-04986-w. [Google Scholar] [CrossRef]
61. Walter B . Virus et viroses de la vigne: diagnostic et méthodes de lute (Virus and virus-deseases of the grapevine: diagnosis and control methods). Virologie. 1998; 2: 435– 44. [Google Scholar]
62. Lacombe T , Boursiquot JM , Audeguin L . The prospection, conservation and evaluation of vine clones in France. Bull OIV. 2004; 77( 885): 799– 810. [Google Scholar]
63. Forget D , Dufour MC , Lusseau T . Bilan et perspectives pour la s’election clonale des principaux cèpages du Bordelais. Prog Agric Vitic. 2002; 119: 199– 206. [Google Scholar]
64. van Leeuwen C , Destrac-Irvine A , Dubernet M , Duchêne E , Gowdy M , Marguerit E , et al. An update on the impact of climate change in viticulture and potential adaptations. Agronomy. 2019; 9( 9): 514. doi:10.3390/agronomy9090514. [Google Scholar] [CrossRef]
65. Rühl E , Konrad H , Lindner B , Bleser E . Quality criteria and targets for clonal selection in grapevine. Acta Hortic. 2004; 652: 29– 33. doi:10.17660/ActaHortic.2004.652.1. [Google Scholar] [CrossRef]
66. Loureiro M , Moreno-Sanz P , Suárez B . Clonal preselection of grapevine cultivars of the appellation “Cangas Quality Wine” (Asturias, Spain). Hort Sci. 2011; 38( 2): 71– 80. doi:10.17221/87/2010-HORTSCI. [Google Scholar] [CrossRef]
67. Šikuten I , Preiner D , Marković Z , Andabaka Ž , Stupić D , Štambuk P , et al. Individual clonal selection of Croatian native cultivars. In: Proceedings of the 22nd Int’l Geisenheim Conference on Grapevine Propagation; 2018 Jun 28–30; Geisenheim, Njemačka. [Google Scholar]
68. Yu L , Boström C , Franzenburg S , Bayer T , Dagan T , Reusch TBH . Somatic genetic drift and multilevel selection in a clonal seagrass. Nat Ecol Evol. 2020; 4( 7): 952– 62. doi:10.1038/s41559-020-1196-4. [Google Scholar] [CrossRef]
69. López EH , Palumbi SR . Somatic mutations and genome stability maintenance in clonal coral colonies. Mol Biol Evol. 2020; 37( 3): 828– 38. doi:10.1093/molbev/msz270. [Google Scholar] [CrossRef]
70. Vezzulli S , Leonardelli L , Malossini U , Stefanini M , Velasco R , Moser C . Pinot blanc and Pinot gris arose as independent somatic mutations of Pinot noir. J Exp Bot. 2012; 63( 18): 6359– 69. doi:10.1093/jxb/ers290. [Google Scholar] [CrossRef]
71. The French–Italian Public Consortium for Grapevine Genome Characterization . The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007; 449( 7161): 463– 7. doi:10.1038/nature06148. [Google Scholar] [CrossRef]
72. Trapp O , Avia K , Borrelli C , Eibach R , Merdinoglu D , Töpfer R . More sustainability in Europe’s vineyards—using resistant grapevine varieties to reduce the input of pesticides. Plants People Planet. 2025; 7( 6): 1621– 8. doi:10.1002/ppp3.70038. [Google Scholar] [CrossRef]
73. Didier A , Teles-Campos I . The four less-used grapes of Champagne: their history, use and their reflection to the terroir of the region [ dissertation]. Umeå, Sweden: UMEÅ Universitet; 2024. 35 p. [Google Scholar]
74. Hajdu E , Korać N , Cindrić P , Ivaniševic D , Medić M . The importance of clonal selection of grapevine and the role of selected clones in production of healthy propagating stocks. Int J Hortic Sci. 2011; 17( 3): 15– 24. [Google Scholar]
75. Mannini F . Clonal selection in grapevine: interactions between genetic and sanitary strategies to improve propagation material. Acta Hortic. 2000; 528: 703– 12. doi:10.17660/ActaHortic.2000.528.106. [Google Scholar] [CrossRef]
76. Dry IB , Davies C , Dunlevy JD , Smith HM , Thomas MR , Walker AR , et al. Development of new wine-, dried- and table grape scions and rootstocks for Australian viticulture: past, present and future. Aust J Grape Wine Res. 2022; 28( 2): 177– 95. doi:10.1111/ajgw.12552. [Google Scholar] [CrossRef]
77. Ibáñez J , Carreño J , Yuste J , Martínez-Zapater JM . Grapevine breeding and clonal selection programmes in Spain. In: Reynolds A , editor. Woodhead publishing series in food science, technology and nutrition, grapevine breeding programs for the wine industry. Cambridge, UK: Woodhead Publishing; 2015. p. 183– 209. doi:10.1016/B978-1-78242-075-0.00009-0. [Google Scholar] [CrossRef]
78. Puglisi D , Las Casas G , Ferlito F , Nicolosi E , Di Guardo M , Scollo F , et al. Parents’ selection affects embryo rescue, seed regeneration and the heredity of seedless trait in table grape breeding programs. Agriculture. 2022; 12( 8): 1096. doi:10.3390/agriculture12081096. [Google Scholar] [CrossRef]
79. Malai ES , O’Sullivan CA , Grant TJ , Sreekantan L , Mellor VA , Schmidt S , et al. Propagation, establishment, and early fruit production of table grape microvines in an LED-Lit hydroponics system: a demonstration case study. Plant-Environ Interact. 2024; 5( 6): e70018. doi:10.1002/pei3.70018. [Google Scholar] [CrossRef]
80. Anamaria N , Botu M , Aurora R , Stroe CT , Ionica D . Behavior of clonal elites for table grapes, under the influence of climatic conditions of the year 2020, in the ecosystem of the Murfatlar wine center. J Hortic Biotehnol. 2020; 24( 4): 43– 9. [Google Scholar]
81. Meneghett S , Calo A , Bavaresco L . A strategy to investigate the intravarietal genetic variability in Vitis vinifera L. for clones and biotypes identification and to correlate molecular profiles with morphological traits or geographic origins. Mol Biotechnol. 2012; 52( 1): 68– 81. doi:10.1007/s12033-011-9475-6. [Google Scholar] [CrossRef]
82. Anhalt UCM , Martinez SC , Ruhl E , Forneck A . Dynamic grapevine clones—an AFLP–marker study of the Vitis vinifera cultivar Riesling comprising 86 clones. Tree Genet. Genomes. 2011; 7( 4): 739– 46. doi:10.1007/s11295-011-0370-x. [Google Scholar] [CrossRef]
83. Franks T , Botta R , Thomas MR , Franks J . Chimerism in grapevines: implications for cultivar identity, ancestry and genetic improvement. Theor Appl Genet. 2002; 104: 192– 9. doi:10.1007/s001220100683. [Google Scholar] [CrossRef]
84. Vondras AM , Minio A , Blanco-Ulate B , Figueroa-Balderas R , Penn MA , Zhou Y , et al. The genomic diversification of grapevine clones. BMC Genom. 2019; 20( 972): 972. doi:10.1186/s12864-019-6211-2. [Google Scholar] [CrossRef]
85. Strioto DK , Carrasco AP , Gibim TE , Carniatto GM , Mangolin CA , Belizário LC , et al. Clonal propagation of cv. Italy grapes and the generation of genetic divergence among vineyards. Sci Hortic. 2019; 244: 263– 9. doi:10.1016/j.scienta.2018.09.049. [Google Scholar] [CrossRef]
86. Roby J-P , van Leeuwen C , Gonçalves E , Graça A , Martins A . The preservation of genetic resources of the vine requires cohabitation between institutional clonal selection, mass selection and private clonal selection. BIO Web Conf. 2014; 3: 01018. doi:10.1051/bioconf/20140301018. [Google Scholar] [CrossRef]
87. Pelsy F . Molecular and cellular mechanisms of diversity within grapevine varieties. Heredity. 2010; 104: 331– 40. doi:10.1038/hdy.2009.161. [Google Scholar] [CrossRef]
88. Fayek MA , Jomaa AH , Shalaby AB , Al-Dhaher AM . Meristem tip culture for in vitro eradication of grapevine leaf roll associated virus-1 (GLRaV-1) and grapevine fan leaf virus (GFLV) from infected flame seedless grapevine plantlets. Iniciación A La Investig. 2009;( 4): 1– 11. [Google Scholar]
89. Kulzhanov SN , Kazybayeva SZ , Tazhibaev TS , Azhitaeva LA , Yessenaliyeva A . Effect of climatic conditions on the productive and biochemical characteristics of grape varieties grown on sierozem soil. Eurasian J Soil Sci. 2022; 11( 2): 174– 83. doi:10.18393/ejss.1057156. [Google Scholar] [CrossRef]
90. Pozharskiy AS , Aubakirova KP , Gritsenko DA , Tlevlesov NI , Karimov NZ , Galiakparov NN , et al. Genotyping and morphometric analysis of Kazakhstani grapevine cultivars versus Asian and European cultivars. Genet Mol Res. 2020; 19( 1): gmr18482. doi:10.4238/gmr18482. [Google Scholar] [CrossRef]
91. Haberlandt G . Culturversuche mit isolierten Pflanzenzellen. In: Laimer M , Rücker W , editors. Plant Tissue Culture. Vienna, Austria: Springer; 2003. doi:10.1007/978-3-7091-6040-4_1. [Google Scholar] [CrossRef]
92. Valdés-Infante J , Rodríguez N , González L , Velázquez J , Rodríguez D , Sourd D , et al. La biotecnología como herramienta para la propagación, conservación y el mejoramiento genético del guayabo. Rev Colomb Biotecnol. 2014; XIV( 2): 7– 19. [Google Scholar]
93. Bouquet A . Interet des techniques de culture in vitro pour l’amelioration genetique de la vigne. Bull L’OIV. 1989; 697–698: 179– 92. [Google Scholar]
94. Meredith CP . Grapevine genetics: Probing the Past and Facing the Future. Agric Conspec Sci. 2001; 66( 1): 21– 5. [Google Scholar]
95. Melyan G , Sahakyan A , Harutyunyan A . Micropropagation of grapevine (Vitis vinifera L.) seedless cultivar “Parvana” through lateral bud development. Vitis-J Grapevine Res. 2015; 54: 253– 5. [Google Scholar]
96. Hewstone NO , Valenzuela JB , Muñoz CS . Efecto de la variedad en el desarrollo de embriones in vitro de vides estenospermocárpicas. Agric Téc. 2006; 66( 2): 124– 32. [Google Scholar]
97. Ponce M , Ocvirk M , Agüero C . Efecto de la intensidad de la poda en el desarrollo in vitro de embriones de vides estenospermocárpicas. Revista FCA. 2009; XLI( 1): 45– 53. [Google Scholar]
98. Valdéz JG . Immature embryo rescue of grapevine (Vitis vinifera L) after an extended period of seed trace culture. Vitis-J Grapevine Res. 2005; 44( 1): 17– 23. [Google Scholar]
99. Nookaraju A , Barreto SM , Agrawal DC . Rapid in vitro propagation of grapevine cv. Crimson Seedless—influence of basal media and plant growth regulators. J Appl Hortic. 2008; 10( 1): 44– 9. doi:10.37855/jah.2008.v10i01.09. [Google Scholar] [CrossRef]
100. Murashige T , Skoog F . A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant. 1962; 15( 3): 473– 97. doi:10.1111/j.1399-3054.1962.tb08052.x. [Google Scholar] [CrossRef]
101. Romadanova NV , Aralbayeva MM , Zemtsova AS , Alexandrova AM , Kazybayeva SZ , Mikhailenko NV , et al. In vitro collection for the safe storage of grapevine hybrids and identification of the presence of Plasmopara viticola resistance genes. Plants. 2024; 13( 8): 1089. doi:10.3390/plants13081089. [Google Scholar] [CrossRef]
102. Wang F-X , Shang G-D , Wu L-Y , Xu Z-G , Zhao X-Y , Wang J-W . Chromatin accessibility dynamics and a hierarchical transcriptional regulatory network structure for plant somatic embryogenesis. Dev Cell. 2020; 54( 2): 742– 57. doi:10.1016/j.devcel.2020.07.003. [Google Scholar] [CrossRef]
103. Ikeuchi M , Ogawa Y , Iwase A , Sugimoto K . Plant regeneration: cellular origins and molecular mechanisms. Development. 2016; 143( 9): 1442– 51. doi:10.1242/dev.134668. [Google Scholar] [CrossRef]
104. Martinelli L , Gribaudo I . Strategies for effective somatic embryogenesis in grapevine: An appraisal. In: Roubelakis-Angelakis KA , editor. Grapevine molecular physiology & biotechnology. Dordrecht, The Netherlands: Springer; 2009. p. 461– 93. doi:10.1007/978-90-481-2305-6_17. [Google Scholar] [CrossRef]
105. Bi W-L , Hao X-Y , Cui Z-H , Pathirana R , Volk GM , Wang Q-C . Shoot tip cryotherapy for efficient eradication of grapevine leafroll-associated virus-3 from diseased grapevine in vitro plants. Ann Appl Biol. 2018; 173( 3): 261– 70. doi:10.1111/aab.12459. [Google Scholar] [CrossRef]
106. Bettoni JC , Dalla-Costa M , Gardin JPP , Kretzschmar AA , Pathirana R . Cryotherapy: a new technique to obtain grapevine plants free of viruses. Rev Bras Frutic. 2016; 38( 2): e-833. doi:10.1590/0100-29452016833. [Google Scholar] [CrossRef]
107. Gresshoff PM , Doy CH . Derivation of a haploid cell line from Vitis vinifera and the importance of the stage of meiotic development of anthers for haploid culture of this and other genera. Z Pflanzenphysiol. 1974; 73( 2): 132– 41. doi:10.1016/S0044-328X(74)80084-X. [Google Scholar] [CrossRef]
108. Hirabayashi T , Kozaki I , Akihama T . In vivo differentiation of shoots from anther callus in Vitis grape research. HortScience. 1976; 11( 5): 511– 2. [Google Scholar]
109. Krul ER , Mowbray GH . Grapes. In: Sharp WR , Evans D , Ammirato PV , Yamada Y , editors. Handbook of plant cell cultures. Vol. II. Crop Species. New York, NY, USA: Macmillan; 1984. p. 396– 434. [Google Scholar]
110. Krul WR , Narragansett RI , inventors; U.S. Patent and Trademark Office, assignee. In vitro propagation of vine. United States patent US 4,532,733A. 1985 Aug 6. [Google Scholar]
111. Goussard PG , Wiid J , Kasdorf GGF . The effectiveness of in vitro somatic embryogenesis in eliminating fanleaf virus and leafroll associated viruses from grapevines. S Afr J Enol Vitic. 1991; 12( 2): 77– 81. doi:10.21548/12-2-2214. [Google Scholar] [CrossRef]
112. David WR , Richard LE , Ronald ET . A stenospermocarpic, seedless Vitis vinifera × Vitis rotundifolia hybrid developed by embryo rescue. HortScience. 2000; 35( 4): 732– 4. doi:10.21273/HORTSCI.35.4.732. [Google Scholar] [CrossRef]
113. Chu Y , Li M , Li R , Zhang K , Qiu P , Yuan X , et al. Embryo rescue breeding of new cold-resistant, seedless grapes. Horticulturae. 2023; 9( 9): 992. doi:10.3390/horticulturae9090992. [Google Scholar] [CrossRef]
114. Sangle SM , Lad JS . Review on mutation breeding for improvement of food legumes—past and recent. Int J Res Anal. 2020; 7( 1): 476– 81. [Google Scholar]
115. Munir N , Safdar I , Naz S . Effect of induced mutation for varietal improvement in some local grapevine cultivars. J Anim Plant Sci. 2015; 25( 1): 234– 42. [Google Scholar]
116. Kuksova VB , Piven NM , Gleba YY . Somaclonal variation and in vitro induced mutagenesis in grapevine. Plant Cell Tissue Organ Cult. 1997; 49: 17– 27. doi:10.1023/A:1005830305206. [Google Scholar] [CrossRef]
117. Baidyussen A , Khassanova G , Utebayev M , Jatayev S , Kushanova R , Khalbayeva S , et al. Assessment of molecular markers and marker-assisted selection for drought tolerance in barley (Hordeum vulgare L.). J Integr Agric. 2024; 23( 1): 20– 38. doi:10.1016/j.jia.2023.06.012. [Google Scholar] [CrossRef]
118. Mangal V , Verma LK , Singh SK , Saxena K , Roy A , Karn A , et al. Triumphs of genomic-assisted breeding in crop improvement. Heliyon. 2024; 10( 15): e35513. doi:10.1016/j.heliyon.2024.e35513. [Google Scholar] [CrossRef]
119. Nadeem MA , Nawaz MA , Shahid MQ , Doğan Y , Comertpay G , Yıldız M , et al. DNA molecular markers in plant breeding: current status and recent advancements in genomic selection and genome editing. Biotechnol Biotechnol Equip. 2017; 32( 2): 261– 85. doi:10.1080/13102818.2017.1400401. [Google Scholar] [CrossRef]
120. Xu Y , Crouch JH . Marker-assisted selection in plant breeding: from publications to practice. Crop Sci. 2008; 48( 2): 391– 407. doi:10.2135/cropsci2007.04.0191. [Google Scholar] [CrossRef]
121. Amiteye S . Basic concepts and methodologies of DNA marker systems in plant molecular breeding. Heliyon. 2021; 7( 10): e08093. doi:10.1016/j.heliyon.2021.e08093. [Google Scholar] [CrossRef]
122. Hasan N , Choudhary S , Naaz N , Sharma N , Laskar RA . Recent advances in molecular marker-assisted selection and amplifications in plant breeding programmes. J Genet Eng Biotechnol. 2021; 19( 1): 128. doi:10.1186/s43141-021-00231-1. [Google Scholar] [CrossRef]
123. Chenuil A . Choosing the right molecular genetic markers for studying biodiversity: from molecular evolution to practical aspects. Genetica. 2006; 127( 1–3): 101– 20. doi:10.1007/s10709-005-2485-1. [Google Scholar] [CrossRef]
124. Redd BV , Reddy CVCM , Sekhar AC , Reddy PCO , Rajasekhar P , Srinivasulu K . Role of molecular based markers methods and their applications in crop improvement. Plant Cell Biotechnol Mol Biol. 2021; 22( 23–24): 38– 54. [Google Scholar]
125. Sesia M , Bates S , Candès E , Marchini J , Sabatti C . False discovery rate control in genome-wide association studies with population structure. Proc Natl Acad Sci U S A. 2021; 118( 40): e2105841118. doi:10.1073/pnas.2105841118. [Google Scholar] [CrossRef]
126. Kaňovská I , Biová J , Škrabišová M . New perspectives of post-GWAS analyses: From markers to causal genes for more precise crop breeding. Curr Opin Plant Biol. 2024; 82( 102658): 1369– 5266. doi:10.1016/j.pbi.2024.102658. [Google Scholar] [CrossRef]
127. Qi T , Song L , Guo Y , Chen C , Yang J . From genetic associations to genes: methods, applications, and challenges. Trends Genet. 2024; 40( 8): 642– 67. doi:10.1016/j.tig.2024.04.008. [Google Scholar] [CrossRef]
128. Liu N , Guan M , Ma B , Chu H , Tian G , Zhang Y , et al. Unraveling genetic mysteries: a comprehensive review of GWAS and DNA insights in animal and plant pathosystems. Int J Biol Macromol. 2025; 285: 138216. doi:10.1016/j.ijbiomac.2024.138216. [Google Scholar] [CrossRef]
129. Wang X , Liu Z , Zhang F , Xiao H , Cao S , Xue H , et al. Integrative genomics reveals the polygenic basis of seedlessness in grapevine. Curr Biol. 2024; 34( 16): 3763– 77. doi:10.1016/j.cub.2024.07.022. [Google Scholar] [CrossRef]
130. Tello J , Ibáñez J . Review: status and prospects of association mapping in grapevine. Plant Sci. 2023; 327: 111539. doi:10.1016/j.plantsci.2022.111539. [Google Scholar] [CrossRef]
131. Shi X , Cao S , Wang X , Huang S , Wang Y , Liu Z , et al. The complete reference genome for grapevine (Vitis vinifera L.) genetics and breeding. Hortic Res. 2023; 410( 5): uhad061. doi:10.1093/hr/uhad061. [Google Scholar] [CrossRef]
132. Konecny T , Asatryan A , Binder H . Responding to stress: Diversity and resilience of grapevine in a changing climate under the perspective of omics research. Int J Mol Sci. 2025; 26: 7877. doi:10.3390/ijms26167877. [Google Scholar] [CrossRef]
133. Grimplet J , Cramer GR . The grapevine genome annotation. In: Cantu D , Walker MA , editors. The grape genome. Cham, Switzerland: Springer International Publishing; 2019. p. 89– 101. doi:10.1007/978-3-030-18601-2_6. [Google Scholar] [CrossRef]
134. Katara A , Chand S , Chaudhary H , Chaudhary V , Chandra H , Dubey RC . Evolution and applications of Next Generation Sequencing and its intricate relations with chromatographic and spectrometric techniques in modern day sciences. J Chromatogr Open. 2024; 5: 100121. doi:10.1016/j.jcoa.2024.100121. [Google Scholar] [CrossRef]
135. Chin CS , Peluso P , Sedlazeck FJ , Nattestad M , Concepcion GT , Clum A , et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat Methods. 2016; 13( 12): 1050– 4. doi:10.1038/nmeth.4035. [Google Scholar] [CrossRef]
136. Roach MJ , Johnson DL , Bohlmann J , van Vuuren HJJ , Jones SJM , Pretorius IS , et al. Population sequencing reveals clonal diversity and ancestral inbreeding in the grapevine cultivar Chardonnay. PLoS Genet. 2018; 14( 11): e1007807. doi:10.1371/journal.pgen.1007807. [Google Scholar] [CrossRef]
137. Minio A , Massonnet M , Figueroa-Balderas R , Castro A , Cantu D . Diploid genome assembly of the wine grape carménère. G3. 2019; 9( 5): 1331– 7. doi:10.1534/g3.119.400030. [Google Scholar] [CrossRef]
138. Vezzulli S , Doligez A , Bellin D . Molecular mapping of grapevine genes. In: Cantu D , Walker MA , editors. The Grape genome. Cham, Switzerland: Springer International Publishing; 2019. p. 103– 36. doi:10.1007/978-3-030-18601-2_7. [Google Scholar] [CrossRef]
139. Moura MF , de Oliveira GL , Sousa-Rodrigues C , Paioli-Pires EJ . The role of plant breeding in grapevine production. In: Martins-Lopes P , editor. Advances in botanical research. Cambridge, MA, USA: Academic Press; 2024. p. 255– 94. doi:10.1016/bs.abr.2024.01.001. [Google Scholar] [CrossRef]
140. Zadokar A , Sharma P , Sharma R . Comprehensive insights on association mapping in perennial fruit crops breeding—its implications, current status and future perspectives. Plant Sci. 2025; 350: 112281. doi:10.1016/j.plantsci.2024.112281. [Google Scholar] [CrossRef]
141. Myles S , Boyko AR , Owens CL , Brown PJ , Grassi F , Aradhya MK , et al. Genetic structure and domestication history of the grape. Proc Natl Acad Sci U S A. 2011; 108( 9): 3530– 5. doi:10.1073/pnas.1009363108.}. [Google Scholar] [CrossRef]
142. Tibbs-Cortes L , Zhang Z , Yu J . Status and prospects of genome-wide association studies in plants. TPG. 2021; 14( 1): e20077. doi:10.1002/tpg2.20077. [Google Scholar] [CrossRef]
143. Zhang Y , Feng L , Fan X , Jiang J , Zheng X , Sun H , et al. Genome-wide assessment of population structure, linkage disequilibrium and resistant QTLs in Chinese wild grapevine. Sci Hort. 2017; 215: 59– 64. doi:10.1016/j.scienta.2016.12.014. [Google Scholar] [CrossRef]
144. Ren C , Fan P , Li S , Liang Z . Advances in understanding cold tolerance in grapevine. Plant Physiol. 2023; 192( 3): 1733– 46. doi:10.1093/plphys/kiad092. [Google Scholar] [CrossRef]
145. Guo L , Wang X , Ayhan DH , Rhaman MS , Yan M , Jiang J , et al. Super pangenome of Vitis empowers identification of downy mildew resistance genes for grapevine improvement. Nat Genet. 2025; 57( 3): 741– 53. doi:10.1038/s41588-025-02111-7. [Google Scholar] [CrossRef]
146. Hamdan MF , Tan BC . Genetic modification techniques in plant breeding: a comparative review of CRISPR/Cas and GM technologies. Hortic Plant J. 2025; 11( 5): 1807– 29. doi:10.1016/j.hpj.2024.02.012. [Google Scholar] [CrossRef]
147. Vasudevan SN , Pooja SK , Raju TJ , Damini CS . Cisgenics and intragenics: boon or bane for crop improvement. Front Plant Sci. 2023; 14: 1275145. doi:10.3389/fpls.2023.1275145. [Google Scholar] [CrossRef]
148. Hamdan MF , Mohd-Noor SN , Abd-Aziz N , Pua T-L , Tan BC . Green revolution to gene revolution: technological advances in agriculture to feed the world. Plants. 2022; 11( 10): 1297. doi:10.3390/plants11101297. [Google Scholar] [CrossRef]
149. Pirrello C , Magon G , Palumbo F , Farinati S , Lucchin M , Barcaccia G , et al. Past, present, and future of genetic strategies to control tolerance to the main fungal and oomycete pathogens of grapevine. J Exp Bot. 2023; 74( 5): 1309– 30. doi:10.1093/jxb/erac487. [Google Scholar] [CrossRef]
150. Klümper W , Qaim M . A Meta-analysis of the impacts of genetically modified crops. PLoS ONE. 2014; 9( 11): e111629. doi:10.1371/journal.pone.0111629. [Google Scholar] [CrossRef]
151. Zhang C , Wohlhueter R , Zhang H . Genetically modified foods: a critical review of their promise and problems. Food Sci Hum Well. 2016; 5( 3): 116– 23. doi:10.1016/j.fshw.2016.04.002. [Google Scholar] [CrossRef]
152. Que Q , Chilton MD , de Fontes CM , He C , Nuccio M , Zhu T , et al. Trait stacking in transgenic crops: challenges and opportunities. GM Crops. 2010; 1( 4): 220– 9. doi:10.4161/gmcr.1.4.13439. [Google Scholar] [CrossRef]
153. Chen W , Ow DW . Precise, flexible and affordable gene stacking for crop improvement. Bioengineered. 2017; 8( 5): 451– 6. doi:10.1080/21655979.2016.1276679. [Google Scholar] [CrossRef]
154. Kumar K , Gambhir G , Dass A , Tripathi AK , Singh A , Jha AK , et al. Genetically modified crops: current status and future prospects. Planta. 2020; 251( 4): 1– 27. doi:10.1007/s00425-020-03372-8. [Google Scholar] [CrossRef]
155. Nuzzo F , Gambino G , Perrone I . Unlocking grapevine in vitro regeneration: issues and perspectives for genetic improvement and functional genomic studies. PPB. 2022; 193: 99– 109. doi:10.1016/j.plaphy.2022.10.027. [Google Scholar] [CrossRef]
156. Ikeuchi M , Favero DS , Sakamoto Y , Iwase A , Coleman D , Rymen B , et al. Molecular mechanisms of plant regeneration. Annu Rev Plant Biol. 2019; 70: 377– 406. doi:10.1146/annurev-arplant-050718-100434. [Google Scholar] [CrossRef]
157. Varshney RK , Bohra A , Yu J , Graner A , Zhang Q , Sorrells ME . Designing future crops: genomics-assisted breeding comes of age. Trends Plant Sci. 2021; 26( 6): 631– 49. doi:10.1016/j.tplants.2021.03.010. [Google Scholar] [CrossRef]
158. Voss-Fels KP , Cooper M , Hayes BJ . Accelerating crop genetic gains with genomic selection. Theor Appl Genet. 2019; 132: 669– 86. doi:10.1007/s00122-018-3270-8. [Google Scholar] [CrossRef]
159. Brault C , Segura V , Roques M , Lamblin P , Bouckenooghe V , Pouzalgues N , et al. Enhancing grapevine breeding efficiency through genomic prediction and selection index. G3. 2024; 14( 4): jkae038. doi:10.1093/g3journal/jkae038. [Google Scholar] [CrossRef]
160. Rusjan D . Genetic and phenotypic diversity and relations between grapevine varieties: slovenian germplasm. In: Poljuha D , Sladonja B , editors. The mediterranean genetic code-grapevine and olive. London, UK: IntechOpen; 2013. doi:10.5772/51773. [Google Scholar] [CrossRef]
161. Villano C , Cigliano AR , Esposito S , D’Amelia V , Iovene M , Carputo D , et al. DNA-Based Technologies for grapevine biodiversity exploitation: State of the art and future perspectives. Agronomy. 2022; 12( 2): 491. doi:10.3390/agronomy12020491. [Google Scholar] [CrossRef]
162. Emanuelli F , Lorenzi S , Grzeskowiak L , Catalano V , Stefanini M , Troggio M , et al. Genetic diversity and population structure assessed by SSR and SNP markers in a large germplasm collection of grape. BMC Plant Biol. 2013; 13: 39. doi:10.1186/1471-2229-13-39. [Google Scholar] [CrossRef]
163. Lacombe T , Audeguin L , Boselli M , Bucchetti B , Cabello F , Chatelet P , et al. Grapevine European catalogue: towards a comprehensive list. Vitis. 2011; 50( 2): 65– 8. doi:10.5073/vitis.2011.50.65-68. [Google Scholar] [CrossRef]
164. Jackson RS . Grape species and varieties. In: Ronald SJ , editor. Food science and technology, wine science. 4th ed. Cambridge, MA, USA: Academic Press; 2014. p. 21– 67. doi:10.1016/B978-0-12-381468-5.00002-6. [Google Scholar] [CrossRef]
165. Velasco R , Zharkikh A , Troggio M , Cartwright DA , Cestaro A , Pruss D , et al. A high-quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007; 2( 12): e1326. doi:10.1371/journal.pone.0001326. [Google Scholar] [CrossRef]
166. Adam-Blondon A-F , Martinez-Zapater J-M , Kole C . Genetics, genomics, and breeding of grapes. Boca Raton, FL, USA: CRC Press; 2016. 404 p. [Google Scholar]
167. Canaguier A , Grimplet J , Di Gaspero G , Scalabrin S , Duchêne E , Choisne N , et al. A New version of the grapevine reference genome assembly (12X. v2) and of its annotation (VCost. V3). Genom Data. 2017; 14: 56– 62. doi:10.1016/j.gdata.2017.09.002. [Google Scholar] [CrossRef]
168. Mercati F , De Lorenzis G , Brancadoro L , Lupini A , Abenavoli MR , Barbagallo MG , et al. High-throughput 18K SNP array to assess genetic variability of the main grapevine cultivars from Sicily. Tree Genet Genomes. 2016; 12: 1– 15. doi:10.1007/s11295-016-1021-z. [Google Scholar] [CrossRef]
169. Pelsy F , Bevilacqua L , Blanc S , Merdinoglu D . A Molecular marker set combining a retrotransposon insertion and SSR polymorphisms is useful for assessing diversity in Vitis. OENO One. 2021; 55( 2): 403– 14. doi:10.20870/oeno-one.2021.55.2.4473. [Google Scholar] [CrossRef]
170. Carrier G , Le Cunff L , Dereeper A , Legrand D , Sabot F , Bouchez O , et al. Transposable elements are a major cause of somatic polymorphism in Vitis vinifera L. PLoS One. 2012; 7: e32973. doi:10.1371/journal.pone.0032973. [Google Scholar] [CrossRef]
171. Carbonell-Bejerano P , Royo C , Mauri N , Ibáñez J , Zapater JMM . Somatic variation and cultivar innovation in grapevine. In: Advances in grape and wine biotechnology. London, UK: IntechOpen; 2019. p. 1– 22. [Google Scholar]
172. Gambino G , Dal Molin A , Boccacci P , Minio A , Chitarra W , Avanzato CG , et al. Whole-Genome Sequencing and SNV genotyping of ‘Nebbiolo’ (Vitis vinifera L.) clones. Sci Rep. 2017; 7: 17294. doi:10.1038/s41598-017-17405-y. [Google Scholar] [CrossRef]
173. Fodor A , Segura V , Denis M , Neuenschwander S , Fournier-Level A , Chatelet P , et al. Genome-wide prediction methods in highly diverse and heterozygous species: Proof-of-concept through simulation in grapevine. PLoS ONE. 2014; 9( 11): e110436. doi:10.1371/journal.pone.0110436. [Google Scholar] [CrossRef]
174. Viana AP , Resende MDV , de Riaz S , Walker MA . Genome selection in fruit breeding: application to table grapes. Sci Agric. 2016; 73( 2): 142– 9. doi:10.1590/0103-9016-2014-0323. [Google Scholar] [CrossRef]
175. Brault C , Doligez A , Cunff L , Coupel-Ledru A , Simonneau T , Chiquet J , et al. Harnessing multivariate, penalized regression methods for genomic prediction and QTL detection of drought-related traits in grapevine. G3. 2021; 11( 9): jkab248. doi:10.1093/g3journal/jkab248. [Google Scholar] [CrossRef]
176. Brault C , Segura V , This P , Le Cunff L , Flutre T , François P , et al. Across-population genomic prediction in grapevine opens up promising prospects for breeding. Hortic Res. 2022; 9: uhac041. doi:10.1093/hr/uhac041. [Google Scholar] [CrossRef]
177. Flutre T , Le Cunff L , Fodor A , Launay A , Romieu C , Berger G , et al. A genome-wide association and prediction study in grapevine deciphers the genetic architecture of multiple traits and identifies genes under many new QTLs. G3. 2022; 12( 7): jkac103. doi:10.1093/g3journal/jkac103. [Google Scholar] [CrossRef]
178. Wilkerson D , Zou C , Sun Q , Nigar Q , Paineau M , Cantu D , et al. Comparative genomics of Rpv3, a multiallelic downy mildew resistance locus in grapevine (Vitis sp.). OENO One. 2025; 59( 1): 8244. doi:10.20870/oeno-one.2025.59.1.8244. [Google Scholar] [CrossRef]
179. Osakabe Y , Liang Z , Ren C , Nishitani C , Osakabe K , Wada M , et al. CRISPR–Cas9-mediated genome editing in apple and grapevine. Nat Protoc. 2018; 13: 2844– 63. doi:10.1038/s41596-018-0067-9. [Google Scholar] [CrossRef]
180. Malnoy M , Viola R , Jung M-H , Koo O-J , Kim S , Kim J-S , et al. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci. 2016; 7: 1904. doi:10.3389/fpls.2016.01904. [Google Scholar] [CrossRef]
181. Butiuc-Keul A , Coste A . Biotechnologies and strategies for grapevine improvement. Horticulturae. 2023; 9( 1): 62. doi:10.3390/horticulturae9010062. [Google Scholar] [CrossRef]
182. Li L , Piatek MJ , Atef A , Piatek A , Wibowo A , Fang X , et al. Rapid and highly efficient construction of TALE-based transcriptional regulators and nucleases for genome modification. Plant Molec Biol. 2012; 78( 4–5): 407– 16. doi:10.1007/s11103-012-9875-4. [Google Scholar] [CrossRef]
183. Carroll D . Genome engineering with zinc-finger nucleases. Genetics. 2011; 188( 4): 773– 82. doi:10.1534/genetics.111.131433. [Google Scholar] [CrossRef]
184. Jinek M , Chylinski K , Fonfara I , Hauer M , Doudna JA , Charpentier E . Programmable dual-RNA–guided DNA endonuclease in adaptive bacterial immunity. Science. 2012; 337( 6096): 816– 21. doi:10.1126/science.1225829. [Google Scholar] [CrossRef]
185. Kaul T , Sony SK , Verma R , Motelb KFA , Prakash AT , Eswaran M , et al. Revisiting CRISPR/Casmediated crop improvement: Special focus on nutrition. J Biosci. 2020; 45: 1– 37. doi:10.1007/s12038-020-00094-7. [Google Scholar] [CrossRef]
186. van der Oost J , Westra ER , Jackson RN , Wiedenheft B . Unravelling the structural and mechanistic basis of CRISPR-Cas systems. Nat Rev Microbiol. 2014; 12: 479– 92. doi:10.1038/nrmicro3279. [Google Scholar] [CrossRef]
187. Xie K , Zhang J , Yang Y . Genome-wide prediction of highly specific guide RNA spacers for CRISPR-Cas9-mediated genome editing in model plants and major crops. Mol Plant. 2014; 7( 5): 923– 6. doi:10.1093/mp/ssu009. [Google Scholar] [CrossRef]
188. Ren C , Lin Y , Liang Z . CRISPR/Cas genome editing in grapevine: recent advances, challenges and future prospects. Fruit Res. 2022; 2( 7): 1– 9. doi:10.48130/FruRes-2022-0007. [Google Scholar] [CrossRef]
189. Moffa L , Mannino G , Bevilacqua I , Gambino G , Perrone I , Pagliarani C , et al. CRISPR/Cas9-driven double modification of grapevine MLO6-7 imparts powdery mildew resistance, while editing of NPR3 augments powdery and downy mildew tolerance. Plant J. 2025; 122( 2): e17204. doi:10.1111/tpj.17204. [Google Scholar] [CrossRef]
190. Cong L , Ran FA , Cox D , Lin S , Barretto R , Habib N , et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013; 339( 6121): 819– 23. doi:10.1126/science.123114. [Google Scholar] [CrossRef]
191. Ren C , Liu X , Zhang Z , Wang Y , Duan W , Li S , et al. CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci Rep. 2016; 6: 32289. doi:10.1038/srep32289. [Google Scholar] [CrossRef]
192. Wang X , Tu M , Wang D , Liu J , Li Y , Li Z , et al. CRISPR/Cas9-mediated efficient targeted mutagenesis in grape in the first generation. Plant Biotechnol J. 2018; 16( 4): 844– 55. doi:10.1111/pbi.12832. [Google Scholar] [CrossRef]
193. Li MY , Jiao YT , Wang YT , Zhang N , Wang BB , Liu RQ , et al. CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic Res. 2020; 7: 149. doi:10.1038/s41438-020-00371-4. [Google Scholar] [CrossRef]
194. Wan DY , Guo Y , Cheng Y , Hu Y , Xiao SY , Wang YJ , et al. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic Res. 2020; 7: 116. doi:10.1038/s41438-020-0339-8. [Google Scholar] [CrossRef]
195. Nakajima I , Ban Y , Azuma A , Onoue N , Moriguchi T , Yamamoto T , et al. CRISPR/Cas9-mediated targeted mutagenesis in grape. PLoS ONE. 2017; 12( 5): e0177966. doi:10.1371/journal.pone.0177966. [Google Scholar] [CrossRef]
196. Ren F , Ren C , Zhang Z , Duan W , Lecourieux D , Li S , et al. Efficiency Optimization of CRISPR/Cas9-Mediated Targeted Mutagenesis in Grape. Front Plant Sci. 2019; 10: 612. doi:10.3389/fpls.2019.00612. [Google Scholar] [CrossRef]
197. Van Leeuwen C , Seguin G . The concept of terroir in viticulture. J Wine Res. 2006; 17( 1): 1– 10. doi:10.1080/09571260600633135 [Google Scholar] [CrossRef]
198. González-Fernández E , Piña-Rey A , Fernández-González M , Aira MJ , Rodríguez-Rajo FJ . Identification and evaluation of the main risk periods of Botrytis cinerea infection on grapevine based on phenology, weather conditions and airborne conidia. J Agric Sci. 2020; 158( 1–2): 88– 98. doi:10.1017/S0021859620000362. [Google Scholar] [CrossRef]
199. Coertze S , Holz G , Sadie A . Germination and establishment of infection on grape berries by single airborne conidia of Botrytis cinerea. Plant Dis. 2001; 85( 6): 668– 77. doi:10.1094/PDIS.2001.85.6.668. [Google Scholar] [CrossRef]
200. Clippinger JI , Dobry EP , Laffan I , Zorbas N , Hed B , Campbell MA . Traditional and emerging approaches for disease management of Plasmopara viticola, causal agent of downy mildew of grape. Agriculture. 2024; 14( 3): 406. doi:10.3390/agriculture14030406. [Google Scholar] [CrossRef]
201. Oerke E-C , Juraschek L , Steiner U . Hyperspectral mapping of the response of grapevine cultivars to Plasmopara viticola infection at the tissue scale. J Exp Bot. 2023; 74( 1): 377– 95. doi:10.1093/jxb/erac390. [Google Scholar] [CrossRef]
202. Bulit J , Lafon R . Powdery mildew of the vine. In: The powdery mildews. London, UK: Academic Press; 1978. p. 525– 48. [Google Scholar]
203. Jarvis WR , Gubler WD , Grove GG . Epidemiology of powdery mildews in agricultural patho-systems. In: The powdery mildews: a comprehensive treatise. St. Paul, MN, USA: The American Phytopathological Society; 2002. p. 169– 99. doi:10.5555/20023170355. [Google Scholar] [CrossRef]
204. Latorre BA , Pszczólkowski P , Torres R , Broome JC . Efectividad de los ácidos grasos e inhibidores de esteroles contra el oídio de la vid y acción sobre la vinificación en Chile. Fitopatología. 1996; 31( 1): 52– 8. [Google Scholar]
205. Campbell P , Bendek C , Latorre BA , Torres R . Pronóstico y efecto del ambiente en el desarrollo del oídio de la vid. Aconex. 2003;( 81): 5– 10. [Google Scholar]
206. Calonnec A , Cartolaro P , Poupot C , Dubourdieu D , Darriet P . Effects of Uncinula necator on the yield and quality of grapes (Vitis vinifera) wine. Plant Pathol. 2004; 53( 4): 434– 45. doi:10.1111/j.0032-0862.2004.01016.x. [Google Scholar] [CrossRef]
207. Grove GG . Perennation of Uncinula necator in vineyards of eastern Washington. Plant Dis. 2004; 88( 3): 242– 7. doi:10.1094/PDIS.2004.88.3.242. [Google Scholar] [CrossRef]
208. Rumbolz J , Gubler WD . Susceptibility of grapevine buds to infection by powdery mildew Erysiphe necator. Plant Pathol. 2005; 54( 4): 535– 48. doi:10.1111/j.1365-3059.2005.01212.x. [Google Scholar] [CrossRef]
209. Tricoli DM , Debernardi JM . An efficient protoplast-based genome editing protocol for Vitis species. Hortic Res. 2024; 11( 1): uhad266. doi:10.1093/hr/uhad266. [Google Scholar] [CrossRef]
210. Najafi S , Bertini E , D’Incà E , Fasoli M , Zenoni S . DNA-Free genome editing in grapevine using CRISPR/Cas9 ribonucleoprotein complexes followed by protoplast regeneration. Hortic Res. 2023; 10( 1): uhac240. doi:10.1093/hr/uhac240. [Google Scholar] [CrossRef]
211. Scintilla S , Salvagnin U , Giacomelli L , Zeilmaker T , Malnoy MA , Rouppe van der Voort J , et al. Regeneration of non-chimeric plants from DNA-free edited grapevine protoplasts. Front Plant Sci. 2022; 13: 1078931. doi:10.3389/fpls.2022.1078931. [Google Scholar] [CrossRef]
212. Ren C , Mohamed MSM , Aini N , Kuang Y , Liang Z . CRISPR/Cas in Grapevine Genome Editing: The Best Is Yet to Come. Horticulturae. 2024; 10( 9): 965. doi:10.3390/horticulturae10090965. [Google Scholar] [CrossRef]
213. Schmidt M , Strack T , Andrews H , Hickey LT , Crisp PA , Voss-Fels KP . A new climate for genomic and epigenomic innovation in grapevine. Mol Horticult. 2025; 5( 1): 44. doi:10.1186/s43897-025-00171-1. [Google Scholar] [CrossRef]
214. Campa M , Miranda S , Licciardello C , Lashbrooke JG , Dalla Costa L , Guan Q , et al. Application of new breeding techniques in fruit trees. Plant Physiol. 2024; 194( 3): 1304– 22. doi:10.1093/plphys/kiad374. [Google Scholar] [CrossRef]
215. Zhang W , Ramautar R . CE-MS for metabolomics: developments and applications in the period 2018–2020. Electrophoresis. 2021; 42( 4): 381– 401. doi:10.1002/elps.202000203. [Google Scholar] [CrossRef]
216. Chen J , Hu X , Shi T , Yin H , Sol D , Hao YF , et al. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol J. 2020; 18( 8): 1722– 35. doi:10.1111/pbi.13335. [Google Scholar] [CrossRef]
217. Maruvada P , Lampe JW , Wishart DS , Barupal D , Chester DN , Dodd D , et al. Perspective: dietary biomarkers of intake and exposure—exploration with omics approaches. Adv Nutr. 2020; 11( 2): 200– 15. doi:10.1093/advances/nmz075. [Google Scholar] [CrossRef]
218. Mookkan M , Nelson-Vasilchik K , Hague J , Zhang ZJ , Kausch AP . Selectable marker independent transformation of recalcitrant maize inbred B73 and sorghum P898012 mediated by morphogenic regulators BABY BOOM and WUSCHEL2. Plant Cell Rep. 2017; 36( 9): 1477– 91. doi:10.1007/s00299-017-2169-1. [Google Scholar] [CrossRef]
219. Zeng X , Yuan H , Dong X , Peng M , Jing XY , Xu Q , et al. Genome-wide dissection of co-selected UV-B responsive pathways in the UV-B adaptation of qingke. Mol Plant. 2020; 13( 1): 112– 27. doi:10.1016/j.molp.2019.10.009. [Google Scholar] [CrossRef]
220. Lin T , Zhu G , Zhang J , Xu X , Yu Q , Zheng Z , et al. Genomic analyses provide insights into the history of tomato breeding. Nat Genet. 2014; 46( 11): 1220– 6. doi:10.1038/ng.3117. [Google Scholar] [CrossRef]
221. Ahmad P , Ashraf M , Younis M , Xiangyang H , Kumar A , Akram NA , et al. Role of transgenic plants in agriculture and biopharming. Biotechnol Adv. 2012; 30( 3): 524– 40. doi:10.1016/j.biotechadv.2011.09.006. [Google Scholar] [CrossRef]
222. Zhou Z , Jiang Y , Wang Z , Gou Z , Lyu J , Li W , et al. Resequencing 302 wild and cultivated accessions identifies genes related to domestication and improvement in soybean. Nat Biotechnol. 2015; 33( 4): 408– 14. doi:10.1038/nbt.3096. [Google Scholar] [CrossRef]
223. Alaimo S , Marceca GP , Giugno R , Ferro A , Pulvirenti A . Current knowledge and computational techniques for grapevine meta-omics analysis. Front Plant Sci. 2018; 8: 2241. doi:10.3389/fpls.2017.02241. [Google Scholar] [CrossRef]
224. Tabidze V , Pipia I , Gogniashvili M , Kunelauri N , Ujmajuridze L , Pirtskhalava M , et al. Whole genome comparative analysis of four Georgian grape cultivars. Mol Genet Genomics. 2017; 292( 6): 1377– 89. doi:10.1007/s00438-017-1353-x. [Google Scholar] [CrossRef]
225. Paineau M , Zaccheo M , Massonnet M , Cantu D . Advances in grape and pathogen genomics toward durable grapevine disease resistance. J Exp Bot. 2025; 76( 11): 3059– 70. doi:10.1093/jxb/erae450. [Google Scholar] [CrossRef]
226. Foria S , Copetti D , Eisenmann B , Magris G , Vidotto M , Scalabrin S , et al. Gene duplication and transposition of mobile elements drive evolution of the Rpv3 resistance locus in grapevine. Plant J. 2020; 101( 3): 529– 42. doi:10.1111/tpj.14551. [Google Scholar] [CrossRef]
227. Cochetel N , Minio A , Massonnet M , Vondras AM , Figueroa-Balderas R , Cantu D . Diploid chromosome-scale assembly of the Muscadinia rotundifolia genome supports chromosome fusion and disease resistance gene expansion during Vitis and Muscadinia divergence. G3. 2021; 11( 4): jkab033. doi:10.1093/g3journal/jkab033. [Google Scholar] [CrossRef]
228. Riaz S , Huerta-Acosta K , Tenscher AC , Walker MA . Genetic characterization of Vitis germplasm collected from the southwestern US and Mexico to expedite Pierce’s disease-resistance breeding. Theor Appl Genet. 2018; 131( 7): 1589– 602. doi:10.1007/s00122-018-3100-z. [Google Scholar] [CrossRef]
229. Huerta-Acosta KG , Riaz S , Tenscher A , Walker MA . Genetic characterization of Pierce’s disease resistance in a Vitis arizonica/monticola wild grapevine. Am J Enol Vitic. 2022; 74: 0740003. doi:10.5344/ajev.2022.22021. [Google Scholar] [CrossRef]
230. Frommer B , Müllner S , Holtgräwe D , Viehöver P , Huettel B , Töpfer R , et al. Phased grapevine genome sequence of an Rpv12 carrier for biotechnological exploration of resistance to Plasmopara viticola. Front Plant Sci. 2023; 14: 1180982. doi:10.3389/fpls.2023.1180982. [Google Scholar] [CrossRef]
231. Zou C , Sapkota S , Figueroa-Balderas R , Glaubitz J , Cantu D , Kingham BF , et al. A multitiered haplotype strategy to enhance phased assembly and fine mapping of a disease resistance locus. Plant Physiol. 2023; 193( 4): 2321– 36. doi:10.1093/plphys/kiad494. [Google Scholar] [CrossRef]
232. Sargolzaei M , Maddalena G , Bitsadze N , Maghradze D , Bianco PA , Failla O , et al. Rpv29, Rpv30 and Rpv31: three novel genomic loci associated with resistance to Plasmopara viticola in Vitis vinifera. Front Plant Sci. 2020; 11: 562432. doi:10.3389/fpls.2020.562432. [Google Scholar] [CrossRef]
233. Ritter EJ , Cochetel N , Minio A , Cousins P , Cantu D , Niederhuth C . The assembly and annotation of two teinturier grapevine varieties, Dakapo and Rubired. Gigabyte. 2025; 2025: gigabyte149. doi:10.46471/gigabyte.149. [Google Scholar] [CrossRef]
234. Picq S , Santoni S , Lacombe T , Latreille M , Weber A , Ardisson M , et al. A small XY chromosomal region explains sex determination in wild dioecious V. vinifera and the reversal to hermaphroditism in domesticated grapevines. BMC Plant Biol. 2014; 14: 229. doi:10.1186/s12870-014-0229-z. [Google Scholar] [CrossRef]
235. Massonnet M , Cochetel N , Minio A , Vondras AM , Lin J , Muyle A , et al. The genetic basis of sex determination in grapes. Nat Commun. 2020; 11( 1): 2902. doi:10.1038/s41467-020-16700-z. [Google Scholar] [CrossRef]
236. Zhou Y , Minio A , Massonnet M , Solares E , Lv Y , Beridze T , et al. The population genetics of structural variants in grapevine domestication. Nat Plants. 2019; 5( 9): 965– 79. doi:10.1038/s41477-019-0507-8. [Google Scholar] [CrossRef]
237. Yao H , Zheng Y , Sun Y , Liu Y . Genome-wide identification and expression analysis of the FAR1-RELATED SEQUENCE (FRS) gene family in grape (Vitis vinifera L.). Int J Mol Sci. 2025; 26( 10): 4675. doi:10.3390/ijms26104675. [Google Scholar] [CrossRef]
238. Wang L , Li R , Li K , Qu Z , Zhou R , Lu G , et al. Genome-wide identification of the grapevine β-1,3-glucanase gene (VviBG) family and expression analysis under different stresses. BMC Plant Biol. 2024; 24( 1): 911. doi:10.1186/s12870-024-05597-1. [Google Scholar] [CrossRef]
239. Sun X , Xia X , Guan X . Genome-wide identification and characterisation of stress-associated protein gene family to biotic and abiotic stresses of grapevine. Pathogens. 2022; 11( 12): 1426. doi:10.3390/pathogens11121426. [Google Scholar] [CrossRef]
240. Domon B , Aebersold R . Mass spectrometry and protein analysis. Science. 2006; 312( 5771): 212– 7. doi:10.1126/ciencia.1124619. [Google Scholar] [CrossRef]
241. Krishna RG , Wold F . Post-translational modifications of proteins. In: Methods in protein sequence analysis. Boston, MA, USA: Springer; 1993. p. 167– 72. doi:10.1007/978-1-4899-1603-7_21. [Google Scholar] [CrossRef]
242. Canales RD , Luo Y , Willey JC , Austermiller B , Barbacioru CC , Boysen C , et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006; 24( 9): 1115– 22. doi:10.1038/nbt1236. [Google Scholar] [CrossRef]
243. Prinsi B , Negri AS , Failla O , Scienza A , Espen L . Root proteomic and metabolic analyses reveal specific responses to drought stress in differently tolerant grapevine rootstocks. BMC Plant Biol. 2018; 18( 1): 126. doi:10.1186/s12870-018-1343-0. [Google Scholar] [CrossRef]
244. Chen S , Su H , Xing H , Mao J , Sun P , Li M . Comparative proteomics reveals the difference in root cold resistance between Vitis.riparia × V. labrusca and Cabernet Sauvignon in response to freezing temperature. Plants. 2022; 11( 7): 971. doi:10.3390/plants11070971. [Google Scholar] [CrossRef]
245. Zhang Y , Wang Y , Liu R , Fei Z , Fan X , Jiang J , et al. Antibody array-based proteome approach reveals proteins involved in grape seed development. Plant Physiol. 2024; 195( 1): 462– 78. doi:10.1093/plphys/kiad682. [Google Scholar] [CrossRef]
246. Murcia G , Alonso R , Berli F , Arias L , Bianchimano L , Pontin M , et al. Quantitative proteomics analysis of ABA- and GA3-treated Malbec berries reveals insights into H2O2 scavenging and anthocyanin dynamics. Plants. 2024; 13( 17): 2366. doi:10.3390/plants13172366. [Google Scholar] [CrossRef]
247. Li T , Wang YH , Liu JX , Feng K , Xu ZS , Xiong AS . Advances in genomic, transcriptomic, proteomic, and metabolomic approaches to study biotic stress in fruit crops. Crit Rev Biotechnol. 2019; 39( 5): 680– 92. doi:10.1080/07388551.2019.1608153. [Google Scholar] [CrossRef]
248. Maia M , Ferreira AEN , Nascimento R , Monteiro F , Traquete F , Marques AP , et al. Integrating metabolomics and targeted gene expression to uncover potential biomarkers of fungal/oomycetes-associated disease susceptibility in grapevine. Sci Rep. 2020; 10( 1): 15688. doi:10.1038/s41598-020-72781-2. [Google Scholar] [CrossRef]
249. Yu H , Li H , Wei R , Cheng G , Zhou Y , Liu J , et al. Widely targeted metabolomics profiling reveals the effect of powdery mildew on wine grape varieties with different levels of tolerance to the disease. Foods. 2022; 11( 16): 2461. doi:10.3390/foods11162461. [Google Scholar] [CrossRef]
250. Ciubotaru RM , Garcia-Aloy M , Masuero D , Franceschi P , Zulini L , Stefanini M , et al. Semi-targeted profiling of the lipidome changes induced by Erysiphe necator in disease-resistant and Vitis vinifera L. varieties. Int J Mol Sci. 2023; 24( 4): 4072. doi:10.3390/ijms24044072. [Google Scholar] [CrossRef]
251. Ciubotaru RM , Franceschi P , Zulini L , Stefanini M , Škrab D , Rossarolla MD , et al. Mono-locus and pyramided resistant grapevine cultivars reveal early putative biomarkers upon artificial inoculation with Plasmopara viticola. Front Plant Sci. 2021; 12: 693887. doi:10.3389/fpls.2021.693887. [Google Scholar] [CrossRef]
252. Medina-Meza IG , Vander-Weide J , Torres-Palacios C , Sabbatini P . Quantitative metabolomics unveils the impact of agricultural practices in the grape metabolome. ACS Agric Sci Technol. 2021; 1: 253– 61. doi:10.1021/acsagscitech.1c00043. [Google Scholar] [CrossRef]
253. Gomez HAG , Niederauer GF , Minatel IO , Antunes ERM , Carneiro MJ , Sawaya ACHF , et al. Metabolite profiling reveals the influence of grapevine genetic distance on the chemical signature of juices. J Sci Food Agric. 2024; 104( 4): 2383– 97. doi:10.1002/jsfa.13124. [Google Scholar] [CrossRef]
254. Shi N , Cheng J , Gao XT , Lu H-C , Tian M-B , Li M-Y , et al. Deciphering metabolic regulation in overripe grapes through multi-omics analysis of non-targeted metabolome, proteome, and transcriptome. BMC Plant Biol. 2025; 25( 1): 1183. doi:10.1186/s12870-025-07235-w. [Google Scholar] [CrossRef]
255. Wang J , Zhang C , Li M , Fan X , Zhang Y , Jiang J , et al. Multi-omics analysis unravels the dynamics of flavor profile in ‘Queen Nina’ berries and the regulatory mechanism underlying volatile esters. Food Chem-Mol Sci. 2025; 10: 100248. doi:10.1016/j.fochms.2025.100248. [Google Scholar] [CrossRef]
256. Lecourieux D , Kappel C , Claverol S , Pieri P , Feil R , Lunn JE , et al. Proteomic and metabolomic profiling underlines the stage- and time-dependent effects of high temperature on grape berry metabolism. Integr Plant Biol. 2020; 62( 8): 1132– 58. doi:10.1111/jipb.12894. [Google Scholar] [CrossRef]
257. Orduña L , Li M , Navarro-Payá D , Zhang C , Santiago A , Romero P , et al. Direct regulation of shikimate, early phenylpropanoid, and stilbenoid pathways by Subgroup 2 R2R3-MYBs in grapevine. Plant J. 2022; 110( 2): 529– 47. doi:10.1111/tpj.15686. [Google Scholar] [CrossRef]
258. Xun Z , Guo X , Ma X , Huang L , Wang M , Dong Z , et al. Combined multi-omics analysis revealed the aroma characteristics and the formation mechanism in different grape varieties. Appl Food Res. 2025; 5( 1): 100974. doi:10.1016/j.afres.2025.100974. [Google Scholar] [CrossRef]
259. Prasanna GSS , Joshi JL , Ganesan S , Shoba D , Muralidharan A . Advances in genetics and plant breeding with artificial intelligence, data science and machin learning. Int J Agric Food Sci. 2025; 7( 9): 399– 403. doi:10.33545/2664844X.2025.v7.i9e.772. [Google Scholar] [CrossRef]
260. Farooq MA , Gao S , Hassan MA , Huang Z , Rasheed A , Hearne S , et al. Artificial intelligence in plant breeding. Trends Genet. 2024; 40( 10): 891– 908. doi:10.1016/j.tig.2024.07.001. [Google Scholar] [CrossRef]
261. Alex B . From terroir to technology: The evolution and future of precision viticulture. Int J Sci Res Arch. 2025; 16( 03): 690– 4. doi:10.30574/ijsra.2025.16.3.2631. [Google Scholar] [CrossRef]
262. Xu D , Liu B , Wang J , Zhang Z . Bibliometric analysis of artificial intelligence for biotechnology and applied microbiology: Exploring research hotspots and frontiers. Front Bioeng Biotechnol. 2022; 10: 998298. doi:10.3389/fbioe.2022.998298. [Google Scholar] [CrossRef]
263. Cardon P . Searching for the Right Metaphors to Understand and Interrogate the AI Age. Bus Commun Res Pract. 2023; 6( 2): 65– 9. doi:10.22682/bcrp.2023.6.2.65. [Google Scholar] [CrossRef]
264. Eftekhari M , Ma C , Orlov YL . Editorial: Applications of artificial intelligence, machine learning, and deep learning in plant breeding. Front Plant Sci. 2024; 15: 1420938. doi:10.3389/fpls.2024.1420938. [Google Scholar] [CrossRef]
265. Gan Y , Liu Z , Zhang F , Xu Q , Wang X , Xue H , et al. Deep learning empowers genomic selection of pest-resistant grapevine. Hortic Res. 2025; 12( 8): uhaf128. doi:10.1093/hr/uhaf128. [Google Scholar] [CrossRef]
266. Herzog K , Kicherer A , Malagol N , Hausmann L , Trapp O , Töpfer R . How sensor technologies combined with artificial intelligence increase the efficiency in grapevine breeding (research): current developments and future perspectives. IVES Conference Series, Open GPB. Villenave, France: International Viticulture and Enology Society; 2024. doi:10.58233/Qivjoi0k. [Google Scholar] [CrossRef]
267. Magaryan K , Nikoghosyan M , Baloyan A , Gasoyan H , Hovhannisyan E , Galstyan L , et al. Machine learned-based visualization of the diversity of grapevine genomes worldwide and in Armenia using SOMmelier. BIO Web Conf. 2023; 68: 01009. doi:10.1051/bioconf/20236801009. [Google Scholar] [CrossRef]
268. Izquierdo-Bueno I , Moraga J , Cantoral JM , Carbú M , Garrido C , González-Rodríguez VE . Smart Viniculture: Applying artificial intelligence for improved winemaking and risk management. Appl Sci. 2024; 14( 22): 10277. doi:10.3390/app142210277. [Google Scholar] [CrossRef]
269. Prajapati S , Qureshi S , Rao Y , Nadkarni S , Retharekar M , Avhad A . Plant disease identification using Deep Learning. In: Proceedings of the 2023 4th International Conference for Emerging Technology, INCET 2023; 2023 May 26–28; Belgaum, India. p. 1– 5. [Google Scholar]
270. Elbasi E , Mostafa N , Alarnaout Z , Zreikat AI , Cina E , Varghese G , et al. Artificial Intelligence Technology in the Agricultural Sector: a Systematic Literature Review. IEEE Access. 2023; 11: 171– 202. doi:10.1109/ACCESS.2022.3232485. [Google Scholar] [CrossRef]
271. Chohan M , Khan A , Chohan R , Katper S , Mahar M . Plant disease detection using Deep Learning. Int J Recent Technol Eng. 2020; 9( 1): 909– 14. doi:10.35940/ijrte.A2139.059120. [Google Scholar] [CrossRef]
272. Kunduracioglu I , Pacal I . Advancements in deep learning for accurate classification of grape leaves and diagnosis of grape diseases. J Plant Dis Prot. 2024; 131: 1061– 80. doi:10.1007/s41348-024-00896-z. [Google Scholar] [CrossRef]
273. Prasad KV , Vaidya H , Rajashekhar C , Swamy KK , Sali R , Sooppy KN . Multiclass classification of diseased grape leaf identification using deep convolutional neural network (DCNN) classifier. Sci Rep. 2024; 14( 1): 9002. doi:10.1038/s41598-024-59562-x. [Google Scholar] [CrossRef]
274. Cándido-Mireles M , Hernández-Gama R , Salas J . Detecting vineyard plants stress in situ using deep learning. Comput Electron Agric. 2023; 210: 107837. doi:10.1016/j.compag.2023.107837. [Google Scholar] [CrossRef]
275. Konecny T , Nikoghosyan M , Binder H . Machine learning extracts marks of thiamine’s role in cold acclimation in the transcriptome of Vitis vinifera. Front Plant Sci. 2023; 14: 1303542. doi:10.3389/fpls.2023.1303542. [Google Scholar] [CrossRef]
276. Mashharawi L , Qasim H , Alshawwa IA , Elkahlout M . Grape type classification using deep learning. Int J Acad Eng Res. 2019; 3( 12): 41– 5. [Google Scholar]
277. Kiani F , Seyyedabbasi A . Wireless sensor network and Internet of Things in precision agriculture. Int J Adv Comput Sci Appl. 2018; 9: 1– 5. doi:10.14569/IJACSA.2018.090614. [Google Scholar] [CrossRef]
278. Saikumar N , Emmanuel N , Krishna KSP , Chinnabbai C , Krishna KU . Artificial intelligence for classification and detection of major insect pests of Brinjal. Ind J Entomol. 2023; 85( 3): 563– 6. doi:10.55446/IJE.2023.1388. [Google Scholar] [CrossRef]
279. Sharma P , Thilakarathna I , Fennell A . Hyperspectral imaging and artificial intelligence enhance remote phenotyping of grapevine rootstock influence on whole vine photosynthesis. Front Plant Sci. 2024; 15: 1409821. doi:10.3389/fpls.2024.1409821. [Google Scholar] [CrossRef]
280. Sawyer E , Laroche-Pinel E , Flasco M , Cooper ML , Corrales B , Fuchs M , et al. Phenotyping grapevine red blotch virus and grapevine leafroll-associated viruses before and after symptom expression through machine-learning analysis of hyperspectral images. Front Plant Sci. 2023; 14: 1117869. doi:10.3389/fpls.2023.1117869. [Google Scholar] [CrossRef]
281. Engler H , Gauweiler P , Huber F , Krause J , Fischer B , Hoffmann B , et al. Phenoquad: a new multi sensor platform for field phenotyping and screening of yield relevant characteristics within grapevine breeding research. VITIS J Grapevine Res. 2023; 62: 41– 8. doi:10.5073/VITIS.2023.62.SPECIAL-ISSUE.41-48. [Google Scholar] [CrossRef]
282. Moreno H , Andújar D . Proximal sensing for geometric characterization of vines: a review of the latest advances. Comput Electron Agric. 2023; 210: 107901. doi:10.1016/j.compag.2023.107901. [Google Scholar] [CrossRef]
283. Šupčík A , Milics G , Matečný I . Predicting grape yield with vine canopy morphology analysis from 3D point clouds generated by UAV imagery. Drones. 2024; 8( 6): 216. doi:10.3390/drones8060216. [Google Scholar] [CrossRef]
284. López A , Ogayar CJ , Feito FR , Sousa JJ . Classification of grapevine varieties using UAV Hyperspectral Imaging. Remote Sens. 2024; 16( 12): 2103. doi:10.3390/rs16122103. [Google Scholar] [CrossRef]
285. Torres-Sánchez J , Mesas-Carrascosa FJ , Santesteban L-G , Jiménez-Brenes FM , Oneka O , Villa-Llop A , et al. Grape cluster detection using UAV photogrammetric point clouds as a low-cost tool for yield forecasting in vineyards. Sensors. 2021; 21( 9): 3083. doi:10.3390/s21093083. [Google Scholar] [CrossRef]
286. Ballesteros R , Intrigliolo DS , Ortega JF , Ramírez-Cuesta JM , Buesa I , Moreno MA . Vineyard yield estimation by combining remote sensing, computer vision and artificial neural network techniques. Precis Agric. 2020; 21: 1242– 62. doi:10.1007/s11119-020-09717-3. [Google Scholar] [CrossRef]
287. Miranda-Fuentes A , Llorens J , Gamarra-Diezma JL , Gil-Ribes JA , Gil E . Towards an optimized method of olive tree crown volume measurement. Sensors. 2015; 15( 2): 3671– 87. doi:10.3390/s150203671. [Google Scholar] [CrossRef]
288. Bendel N , Backhaus A , Kicherer A , Köckerling J , Maixner M , Jarausch B , et al. Detection of two different grapevine yellows in Vitis vinifera using hyperspectral imaging. Remote Sens. 2020; 12( 24): 4151. doi:10.3390/rs12244151. [Google Scholar] [CrossRef]
289. Albetis J , Jacquin A , Goulard M , Poilvé H , Rousseau J , Clenet H , et al. On the potentiality of UAV multispectral imagery to detect flavescence dorée and grapevine trunk diseases. Remote Sens. 2018; 11( 1): 23. doi:10.3390/rs11010023. [Google Scholar] [CrossRef]
290. MacDonald SL , Staid M , Staid M , Cooper ML . Remote Hyperspectral Imaging of grapevine leafroll-associated virus 3 in Cabernet Sauvignon vineyards. Comput Electron Agric. 2016; 130: 109– 17. doi:10.1016/j.compag.2016.10.003. [Google Scholar] [CrossRef]
291. Patil SS , Thorat SA . Early detection of grapes diseases using machine learning and IoT. In: Proceedings of the 2nd Second International Conference on Cognitive Computing and Information Processing (CCIP); 2016 Aug 12–13; Mysuru, India. p. 1– 5. doi:10.1109/CCIP.2016.7802887. [Google Scholar] [CrossRef]
292. Kicherer A , Herzog K , Pflanz M , Wieland M , Rüger P , Kecke S , et al. An Automated Field Phenotyping Pipeline for Application in Grapevine Research. Sensors. 2015; 15( 3): 4823– 36. doi:10.3390/s150304823. [Google Scholar] [CrossRef]
293. Lorenz DH , Eichhorn KW , Bleiholder H , Klose R , Meier U , Weber E . Growth stages of the grapevine: Phenological growth stages of the grapevine (Vitis vinifera L. ssp. Vinifera)-Codes and descriptions according to the extended BBCH scale. Aust J Grape Wine Res. 1995; 1: 100– 3. doi:10.1111/j.1755-0238.1995.tb00085.x. [Google Scholar] [CrossRef]
294. Barasa PM , Botai CM , Botai JO , Mabhaudhi T . A review of climate-smart agriculture research and applications in Africa. Agronomy. 2021; 11( 6): 1255. doi:10.3390/agronomy11061255. [Google Scholar] [CrossRef]
295. Zarco-Tejada PJ , Diaz-Varela R , Angileri V , Loudjani P . Tree height quantification using very high resolution imagery acquired from an unmanned aerial vehicle (UAV) and automatic 3D photo-reconstruction methods. Eur J Agron. 2014; 55: 89– 99. doi:10.1016/j.eja.2014.01.004. [Google Scholar] [CrossRef]
296. Petkovics I , Petkovic D , Petkovics A . IoT Devices vs. Drones for Data Collection in Agriculture. In: Katalinic B , editor. DAAAM international scientific book. Vienna, Austria: DAAAM International; 2017. p. 63– 80. doi:10.2507/daaam.scibook.2017.06. [Google Scholar] [CrossRef]
297. Deng L , Mao Z , Li X , Hu Z , Duan F , Yan Y . UAV-based multispectral remote sensing for precision agriculture: a comparison between different cameras. ISPRS J Photogramm Remote Sens. 2018; 146: 124– 36. doi:10.1016/j.isprsjprs.2018.09.008. [Google Scholar] [CrossRef]
298. Jamil IN , Remali J , Azizan KA , Nor Muhammad NA , Arita M , Goh H-H , et al. Systematic multi-omics integration (MOI) approach in plant systems biology. Front Plant Sci. 2020; 11: 944. doi:10.3389/fpls.2020.00944. [Google Scholar] [CrossRef]
299. Li J , Wang Y , Suh JH . Multi-omics approach in tea polyphenol research regarding tea plant growth, development and tea processing: Current technologies and perspectives. Food Sci Hum Wellness. 2022; 11( 3): 524– 36. doi:10.1016/j.fshw.2021.12.010. [Google Scholar] [CrossRef]
300. Jung J , Maeda M , Chang A , Bhandari M , Ashapure A , Landivar-Bowles J . The potential of remote sensing and artificial intelligence as tools to improve the resilience of agriculture production systems. Curr Opin Biotechnol. 2021; 70: 15– 22. doi:10.1016/j.copbio.2020.09.003. [Google Scholar] [CrossRef]
301. Isewon I , Apata O , Oluwamuyiwa F , Aromolaran O , Oyelade J . Machine learning algorithms: their applications in plant omics and agronomic traits’ improvement. F1000Research. 2022; 11: 1256. doi:10.12688/f1000research.125425.1. [Google Scholar] [CrossRef]
302. Yan J , Wang X . Unsupervised and semi-supervised learning: the next frontier in machine learning for plant systems biology. Plant J. 2022; 111( 6): 1527– 38. doi:10.1111/tpj.15905. [Google Scholar] [CrossRef]
303. Graziani M , Dutkiewicz L , Calvaresi D , Amorim JP , Yordanova K , Vered M , et al. A global taxonomy of interpretable AI: Unifying the terminology for the technical and social sciences. Artif Intell Rev. 2023; 56( 4): 3473– 504. doi:10.1007/s10462-022-10256-8. [Google Scholar] [CrossRef]
304. Jiang Y , Li C . Convolutional neural networks for image-based high-throughput plant phenotyping: a review. Plant Phenomics. 2020; 2020: 4152816. doi:10.34133/2020/4152816. [Google Scholar] [CrossRef]
305. Baduge SK , Thilakarathna S , Perera JS , Arashpour M , Sharafi P , Teodosio B , et al. Artificial intelligence and smart vision for building and construction 4.0: machine and deep learning methods and applications. Autom Constr. 2022; 141: 104440. doi:10.1016/j.autcon.2022.104440. [Google Scholar] [CrossRef]
306. Thriveni K , Teotia J , Hazra S , Bharti T , Kumar M , Lallawmkimi MC , et al. A review on integrating bioinformatics tools in modern plant breeding. Arch Curr Res Int. 2024; 24( 9): 293– 308. doi:10.9734/acri/2024/v24i9894. [Google Scholar] [CrossRef]
307. Portela F , Sousa JJ , Araújo-Paredes C , Peres E , Morais R , Pádua L . A systematic review on the advancements in remote sensing and proximity tools for grapevine disease detection. Sensors. 2024; 24( 24): 8172. doi:10.3390/s24248172. [Google Scholar] [CrossRef]
308. Feng W , Gao P , Wang X . AI breeder: Genomic predictions for crop breeding. New Crops. 2024; 1( 3): 100010. doi:10.1016/j.ncrops.2023.12.005. [Google Scholar] [CrossRef]
309. Gatou P , Tsiara X , Spitalas A , Sioutas S , Vonitsanos G . Artificial intelligence techniques in grapevine research: a comparative study with an extensive review of datasets, diseases, and techniques evaluation. Sensors. 2024; 24( 19): 6211. doi:10.3390/s2419621. [Google Scholar] [CrossRef]
310. Agrawal J , Arafat MY . Transforming farming: a review of AI-powered UAV technologies in precision agriculture. Drones 2024; 8( 11): 664. doi:10.3390/drones8110664. [Google Scholar] [CrossRef]
311. Molnár S , Tamás L . Close proximity aerial image for precision viticulture. A review. J Plant Dis Prot. 2025; 132: 57. doi:10.1007/s41348-024-01050-5. [Google Scholar] [CrossRef]
312. Migicovsky Z . Genomic resources for crop wild relatives are critical for perennial fruit breeding and conservation. Am J Bot. 2025; 112( 7): e70068. doi:10.1002/ajb2.70068. [Google Scholar] [CrossRef]
313. Laucou V , Launay A , Bacilieri R , Lacombe T , Adam-Blondon AF , Bérard A , et al. Extended diversity analysis of cultivated grapevine Vitis vinifera with 10K genome-wide SNPs. PLoS One. 2018; 13( 2): e0192540. doi:10.1371/journal.pone.0192540. [Google Scholar] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools