 Open Access
Open Access
ARTICLE
Pathways Related to ROS Production, Clearance, and Signal Transduction during Cold Response in Brassica napus L. with Strong Cold Resistance
1 School of Agriculture and Bioengineering, Longdong University, Qingyang, 745000, China
2 Collaborative Innovation Center for Longdong Dryland Crop Germplasm Improvement and Industrialization, Longdong University, Qingyang, 745000, China
3 Gansu Dryland Research Center of Winter Wheat Germplasm Innovation and Application Engineering, Longdong University, Qingyang, 745000, China
4 Gansu Collaborative Innovation of Academicians and Experts on Dryland Agriculture in the Loess Plateau, Longdong University, Qingyang, 745000, China
5 Agronomy College, Gansu Agriculture University, Lanzhou, 730070, China
6 College of Chemistry and Life Sciences, Chengdu Normal University, Chengdu, 611130, China
* Corresponding Author: Weiliang Qi. Email:
(This article belongs to the Special Issue: Influence of Biotic and Abiotic Stresses Signals on Plants and their Performance at Different Environments)
Phyton-International Journal of Experimental Botany 2025, 94(3), 739-762. https://doi.org/10.32604/phyton.2025.060712
Received 07 November 2024; Accepted 18 February 2025; Issue published 31 March 2025
Abstract
Brassica napus L. (B. napus), recognized as a significant cash and oil crop, faces challenges in popularization and application in northern China due to its limited cold resistance. Clarifying the mechanism of cold stress on gene regulation and signal transduction in B. napus is crucial. To address these issues, we conducted transcriptome sequencing and gene expression analysis, along with gene ontology (GO) and Kyoto encyclopedia of genes and genomes (KEGG) pathway profiling under natural (25°C) and cold (4°C) conditions in cold tolerant 16VHNTS309 and weak cold-resistant Tianyou 2238 B. napus seedlings. Enhanced genomic annotation was achieved through additional sequencing. A total of 6127 and 8531 differentially expressed genes (DEG) were identified in 16VHNTS309 and Tianyou 2238, respectively. The expression patterns of 23 DEGs were validated by quantitative real-time PCR (qRT-PCR), confirming the RNA-Seq results. Under cold stress, 58 pathways in 16VHNTS309 demonstrated significant changes (q-Value < 0.05), compared to 9 pathways in Tianyou 2238 (q-Value < 0.05), highlighting B. napus’ sophisticated regulatory network which aids in managing growth and development challenges. After 48 h of cold stress treatment, genes associated with reactive oxygen species (ROS) clearance, such as those involved in antioxidant VB6, sulfur metabolism, peroxisomes, and phagosomes, were notably up-regulated in 16VHNTS309, indicating its robust ROS clearance capability. Significant gene expressions within Ca2+, MAPK, and transcription factor pathways related to ROS suggest that varieties with strong cold resistance possess a complex signal regulation mechanism. Comprehensive analyses of stomatal cells, physiological parameters of ROS, ABA, and H2S, along with transcriptomic data, revealed that optimal ROS levels interact with ABA and H2S to regulate stomatal closure in B. napus 16VHNTS309 under the influence of antioxidant enzymes.Keywords
Cold stress is one of the primary environmental challenges plants face throughout their life cycle. Cold temperatures negatively impact plant growth and development by causing tissue injury and delaying growth [1], and they significantly restrict the spatial distribution of plants. To adapt, cold-tolerant plants have evolved complex and effective cold-responsive mechanisms, such as changes in leaf tissue structure, accumulation of compatible osmolytes, and both enzymatic and non-enzymatic antioxidative systems [2]. Concurrently, ROS production occurs under cold stress. Prior research has indicated that ROS accumulation can lead to oxidative damage in plant cells, ultimately resulting in cell death. Further investigations into plant signaling molecules have revealed that plants exhibit specific and highly dynamic signaling responses under abiotic stress, with ROS acting as key signaling molecules. Extensive studies have demonstrated the rapid release of ROS under cold stress and their ability to transmit signals over long distances [3,4]. ROS is critical not only for local immune responses but also for cell-to-cell communication. It has been established that ROS interact with other signaling molecules, such as Ca2+ [5], MAPK [6], transcription factors [7] (WRKY, etc.), and ABA [8] among others. These interactions are linked to the differential expression of numerous genes [9]. Cold-responsive genes continue to be identified and annotated through transcriptomic methods across various plant species. For example, significant numbers of genes have been identified as cold-responsive in economically important plants like Arabidopsis [10], Camellia sinensis [11], Poncirus trifoliata [12], Eucalyptus dunnii [13] and Beta vulgaris [14]. Although many cold-regulated genes have been discovered and assigned specific functions across different species, the mechanisms of their cold responses have not been fully elucidated. In addition, genes may be involved in different pathways across species, and various species may have distinct cold-response genes. Therefore, there is a need for further exploration of cold response regulatory pathways and genes across different plants.
Winter rape, an important economic cover crop, offers economic, environmental, and ecological benefits and aids in reducing dust sources in northern China [15]. However, B. napus is primarily distributed in regions such as the Loess Plateau, the middle and lower reaches of the Yangtze River, the Huang-Huai Plain, the Yunnan-Guizhou Plateau, Sichuan Basin, and the coastal areas of south China [16]. The growth and development of B. napus can be hindered by its poor cold resistance [15]. Previous studies have shown that distant hybridization is a highly effective method for promoting gene or chromosomal segment exchanges and enriching germplasm resources, thus breeding new B. napus varieties with biotic or abiotic stress resistance [17]. Zhao et al. [18] and Wang et al. [19] have successfully bred new lines of B.napus with high affinity, good yield, disease, or cold resistance through interspecific hybridization, backcross, and continuous self-cross of B. napus and B. rape. In an effort to overcome the key technical challenges of winter rape overwintering, our research group has developed the cold-resistant B. napus variety 16VHNTS309, which resulted from a cross between B. rapa Longyou7 [20], known for its strong cold resistance, and B. napus. We have studied the cold resistance mechanisms of osmotic adjustment, enzymatic activity, ROS signal generation, and transmission in 16VHNTS309 from physiological, biochemical, and cellular perspectives [4].
To understand the cold resistance mechanism of B. napus, we analyzed the cold-tolerant B. napus 16VHNTS309 and weak cold-resistant B. napus of Tianyou 2238 through transcriptome sequencing, gene expression, GO, and KEGG pathway profiling at normal temperature (25°C) and under cold treatment (4°C) using the Illumina sequencing technique. Based on differential gene expression and GO and KEGG pathway analysis, genes and metabolic pathways related to ROS production, antioxidant systems, and signal transduction were identified. This study aims to enhance our understanding of the molecular-level cold stress response pathways and identify effective strategies to improve cold tolerance in B.napus.
Our group has cultivated a cold-tolerant winter B. rape variety, Longyou7, which can survive in extremely low temperatures (down to −32°C, with an overwinter survival rate of more than 90%) [20]. To develop cold-resistant lines of B.napus, diploid B. rapa Longyou 7 (ArAr, 2n = 2x = 20) was hybridized with amphidiploid B. napus Vision (AnAnCnCn, 2n = 4x = 38) to create the F1 generation (AnArCn, 2n = 3x = 29). Although F1 seeds exhibited heterosis, the rate of self-crossing was very low (0 to 6 seeds per 100 pollinated flowers). After two rounds of self-crossing, F3 plants were crossed with B. napus Vision and backcrossed to B. napus Vision. The BC1F1 generation seeds were self-crossed three times to obtain BC1F4 seeds, which were then subjected to a cold resistance test to select the highly cold-resistant individual plant, 16VHNTS309.
The overwintering rates of strong cold-resistant B. napus 16VHNTS309 and weak cold-resistant B. napus Tianyou 2238 were 85.38% and 18.95%, respectively, provided by Gansu Provincial Key Laboratory of Crop Genetics and Germplasm Improvement, Gansu Agricultural University. The B.napus cultivars 16VHNTS309 and Tianyou 2238 underwent germination in 12 × 8 hole float trays (60 cm × 40 cm × 8 cm), with pots containing a 3:1 ratio of nutritional soil to vermiculite. Cultivation occurred in an illumination incubator at standard conditions (25°C, 16-h photoperiod, 6000 Lx light intensity). When plants reached the four-leaf stage, they were segregated into two groups. The control group remained under standard conditions for 48 h, while the test group was subjected to cold stress in a pre-cooling incubator at 4°C for the same duration. This process was replicated three times to ensure reliability. Post-experiment, samples were preserved at −80°C for subsequent transcriptome and biochemical analysis.
Total RNA extraction was performed utilizing the Trizol total RNA extractor kit (B511311, Sangon, Shanghai, China). RNA samples from 16VHNTS309 and Tianyou2238 of high quality were forwarded to Sangon Biotech (Shanghai) Co., Ltd. (Shanghai, China) for the construction of libraries using HiSeq XTen sequencers (Illumina, San Diego, CA, USA). The quality of the sequenced data was appraised using FastQC (version 0.11.2). HISAT2 (version 2.0) aligned clean reads to the reference genome (https://www.genoscope.cns.fr/brassicanapus/, accessed on 01 November 2024). Statistical evaluations were performed using RSeQC (version 2.6.1), and gene coverage ratios were assessed with BEDTools (version 2.26.0). StringTie (version 1.3.3b) calculated gene expression values. DESeq2 (version 1.12.4) identified DEG, considering genes with a q-Value < 0.001 and |FoldChange| > 2 as significantly expressed. Functional enrichment analysis, which included gene ontology and the Kyoto Encyclopedia of Genes and Genomes, pinpointed DEGs substantially enriched in specific GO terms or metabolic pathways, with a false discovery rate (q-Value) < 0.05 indicating significant alteration. Transcriptome data have been uploaded (SRA data: PRJNA1198720, https://www.ncbi.nlm.nih.gov/sra/PRJNA1198720, accessed on 01 November 2024).
Following cold exposure, the concentrations of VB6 and ABA were quantified using a DDS11A digital conductometer (Leici Instrument Factory, Shanghai, China) and enzyme-linked immunosorbent assay according to the kit specifications provided by Cominbio (Suzhou, China). H2S content was determined following the methodology outlined by Lai et al. [21]. SPSS 19.0 statistical analysis was used to test the different significance of the data, Duncan’s method was used to make multiple comparisons between different treatments, and the difference significance between treatment and control was used by t-test, the significance level was set at p < 0.05.
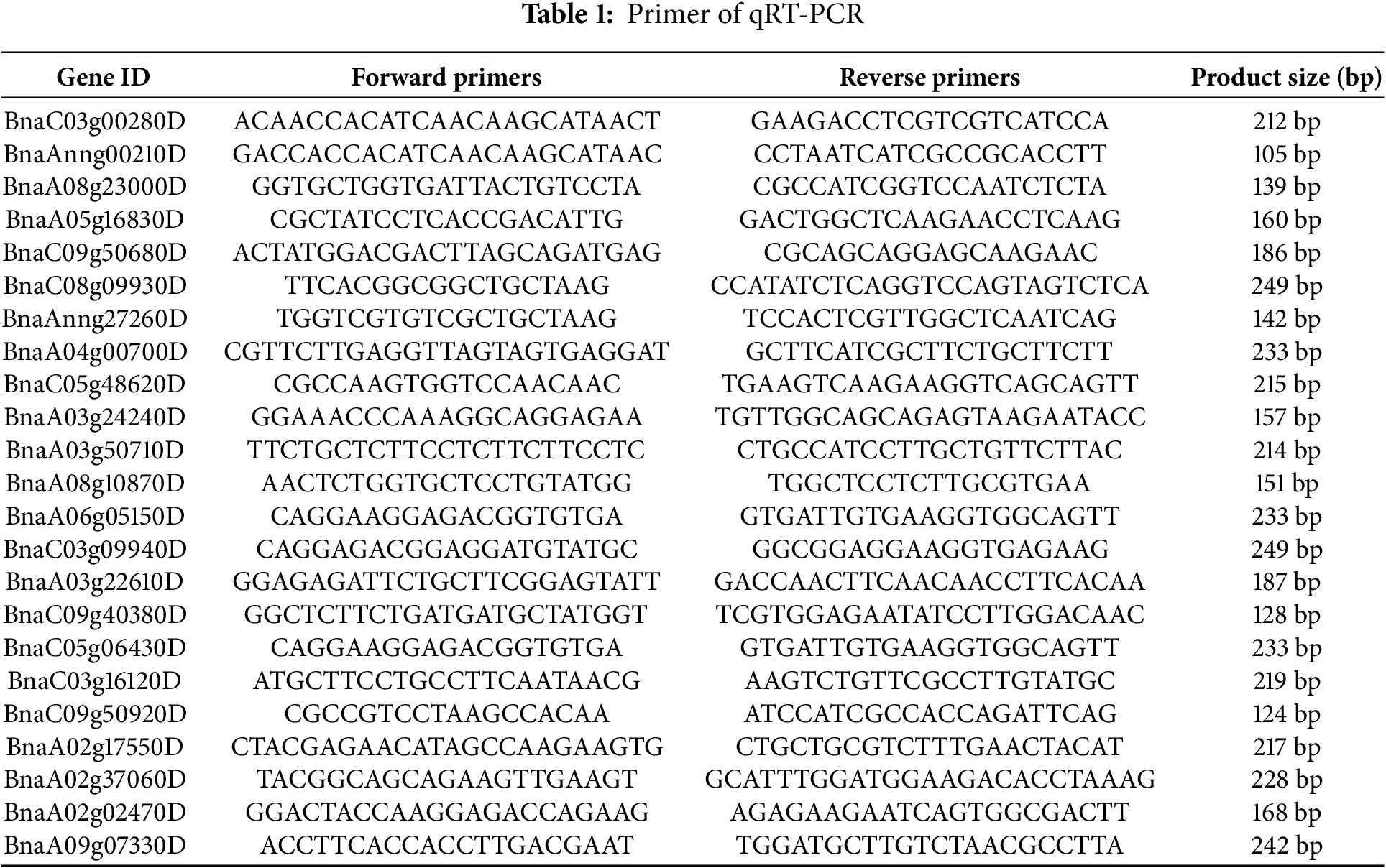
To confirm the accuracy of RNA-Seq data, we selected 23 DEG for qRT-PCR analysis using the Super Real PreMix Plus (SYBR Green) Kit (TIANGEN) on an Applied BioSystems 7500 Real-Time PCR system (Applied BioSystems, Model No. 401511) (Table 1). We quantified gene expression utilizing the 2−ΔΔCt method, with β-actin as the reference gene. Averages and standard deviations (SD) were calculated from three biological replicates.
From the cDNA libraries of 16VHNTS309 and Tianyou2238, we generated high-quality reads, resulting in the creation of 12 libraries. Each library generated approximately 7.55 × 107 raw reads, with a total of 9.06 × 108 raw reads. The Q20 ratios (sequencing error rate <1%) were between 97.13% and 98.49%, and the Q30 ratios (sequencing error rate <0.1%) ranged from 92.75% to 95.96%, reflecting high sequencing quality with a GC content above 47% (Table A1). We utilized trimmomatic software for data preprocessing, which eliminated low-quality reads, yielding 8.77 × 108 clean reads. The mean number of clean reads per library was 7.31 × 107. The Q30 ratio exceeded 92% across all libraries, maintaining a consistent GC content of about 47%. Furthermore, 91% of the clean reads successfully aligned with the B. napus reference genome. The high correlation coefficients (r) nearing 1 and consistent gene expression density curves across samples confirmed the test’s reliability and the suitability of the sample selection.
To investigate gene expression differences in response to cold stress between the cold-tolerant 16VHNTS309 and the weak cold-resistant Tianyou2238, DEGs (q-Value < 0.05 and log2-fold-change > 2) were determined in both strains. The inter-sample correlation heatmap confirmed the high reliability of repeated data (Fig. 1A). Statistical analysis of gene expression density distribution helped in assessing the levels of differential gene expression in 16VHNTS309 and Tianyou2238 over various treatment periods (Fig. 1B). The expression patterns of DEGs in both strains were consistent through various low temperature treatment durations. A Venn diagram displayed the regulation patterns of DEGs, showing up- and down-regulation in both winter rapeseed varieties under cold stress. After 48 h of cold exposure, 6127 DEGs (3427 up-regulated and 2700 down-regulated) in 16VHNTS309 and 8531 DEGs (4738 up-regulated and 3793 down-regulated) in Tianyou2238 were identified (Fig. 2). The higher number of up-regulated DEGs indicates a complex and varied response mechanism in B.napus of both 16VHNTS309 and Tianyou2238.
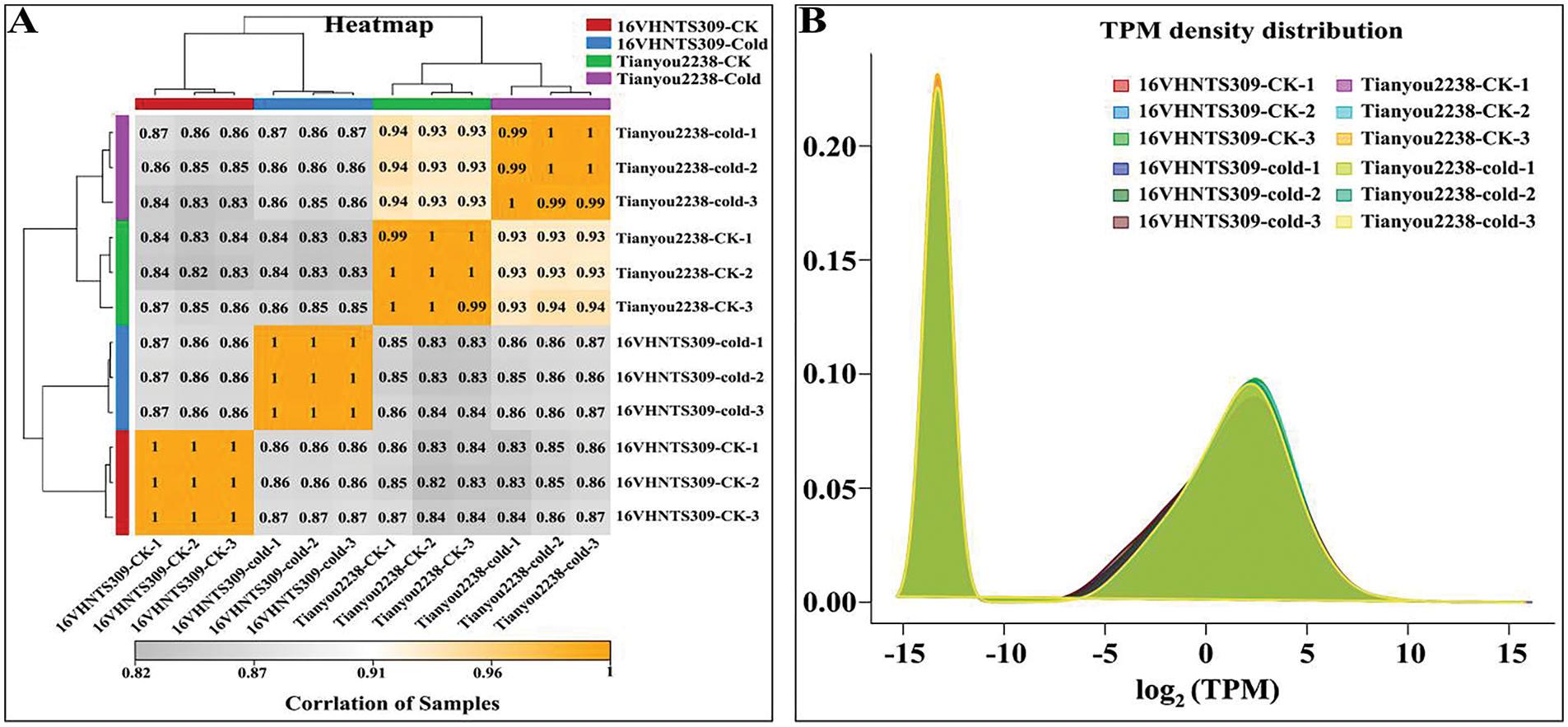
Figure 1: A: Heat-map of correlation analysis between samples. B: Curve graph of gene expression density

Figure 2: List of DEGs and ADEGs in different treatment
Following cold stress treatment, 23 DEGs from B. napus cultivars 16VHNTS309 and Tianyou 2238 underwent qRT-PCR analysis (Fig. 3) to corroborate the transcriptome sequencing findings. The expression patterns of these DEGs in the qRT-PCR confirmed those observed in the RNA-Seq analysis, substantiating the sequencing results’ reliability and demonstrating RNA-Seq’s capability to accurately reflect the transcription levels of B. napus 16VHNTS309 and Tianyou 2238 under cold conditions.
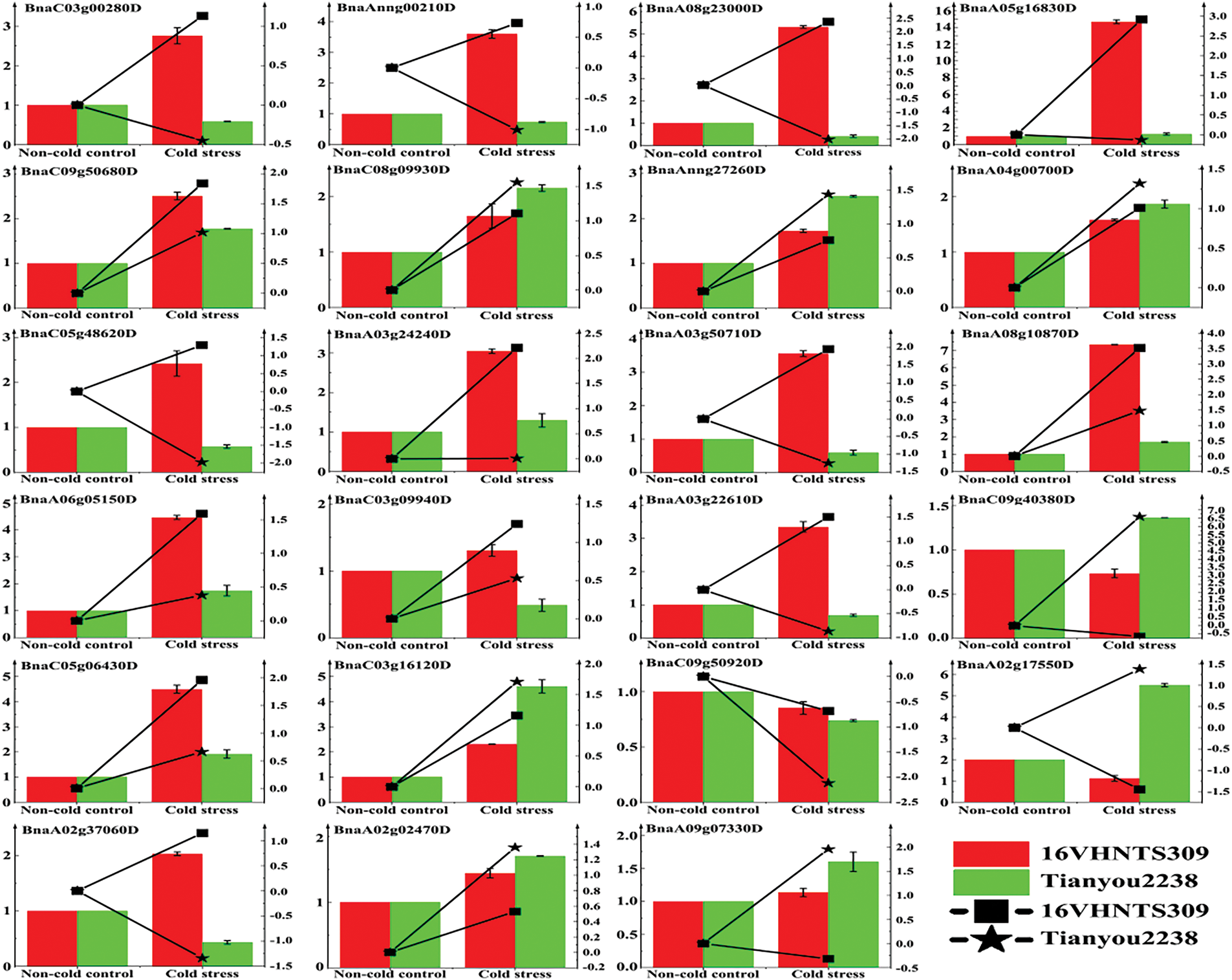
Figure 3: Post 48 h of cold exposure at 4°C, 23 DEGs from B. napus 16VHNTS309 and Tianyou 2238 were subjected to qRT-PCR. Error bars represent the standard errors of the mean values of relative expression levels measured by qRT-PCR (left y-axis). Dashed lines illustrate the changes in transcript levels (log2-fold) based on the FPKM values from RNA-Seq (right y-axis)
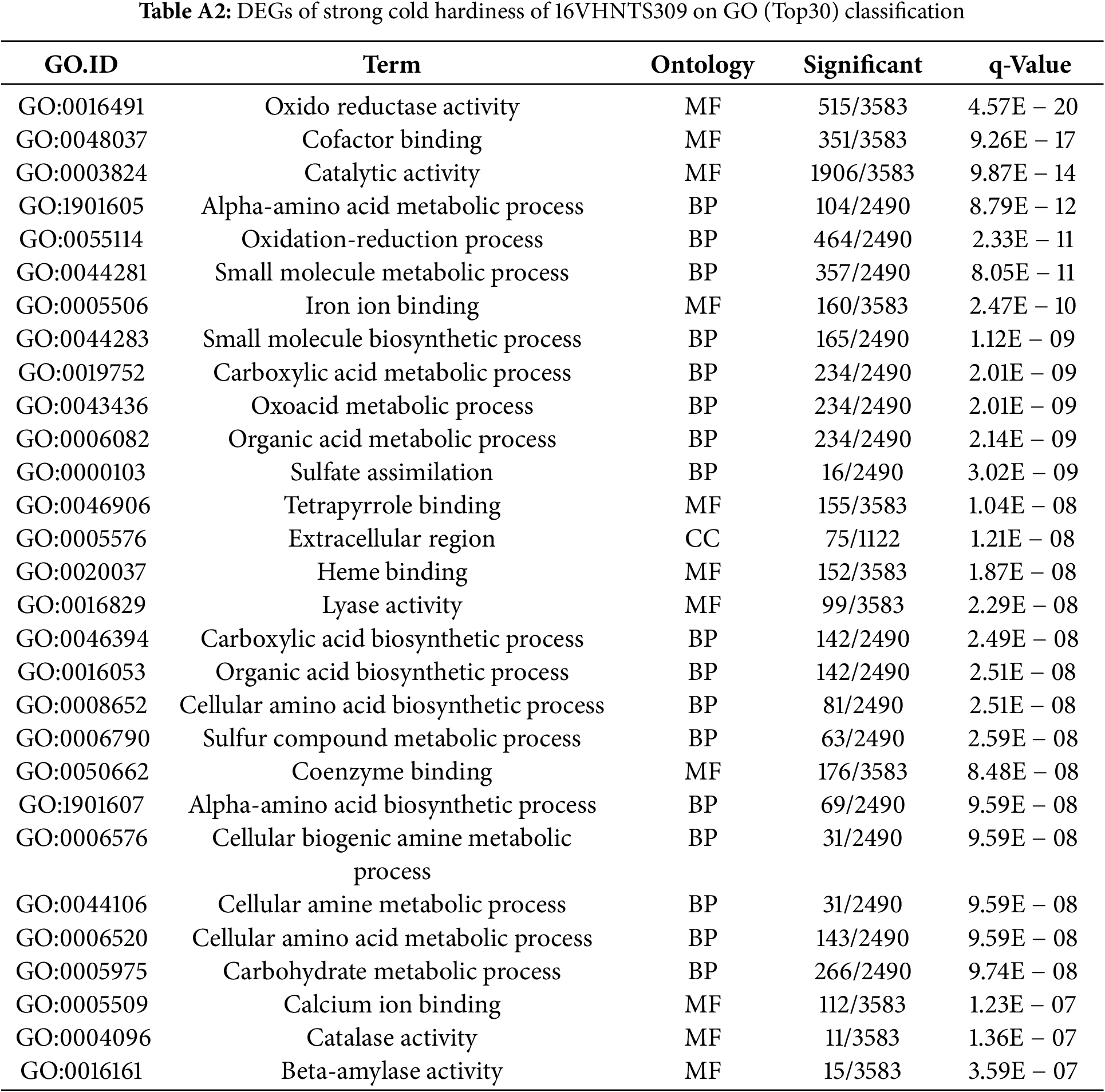
GO analysis was conducted to identify the genes differentially expressed under cold stress in B. napus 16VHNTS309 and Tianyou 2238. This analysis segregated the DEGs into three principal functional categories: molecular function (MF), biological process (BP), and cellular component (CC). DEGs in CC were predominantly found in extracellular regions (GO: 0005576) and others (refer to Table A2). MF DEGs primarily showed enrichment in functions such as oxidoreductase activity (GO: 0016491), catalytic activity (GO: 0003824), and iron ion binding (GO: 0005506). BP DEGs were chiefly involved in alpha-amino acid metabolism (GO: 1901605), oxidation-reduction processes (GO: 0055114), and small molecule metabolism (GO: 0044281).
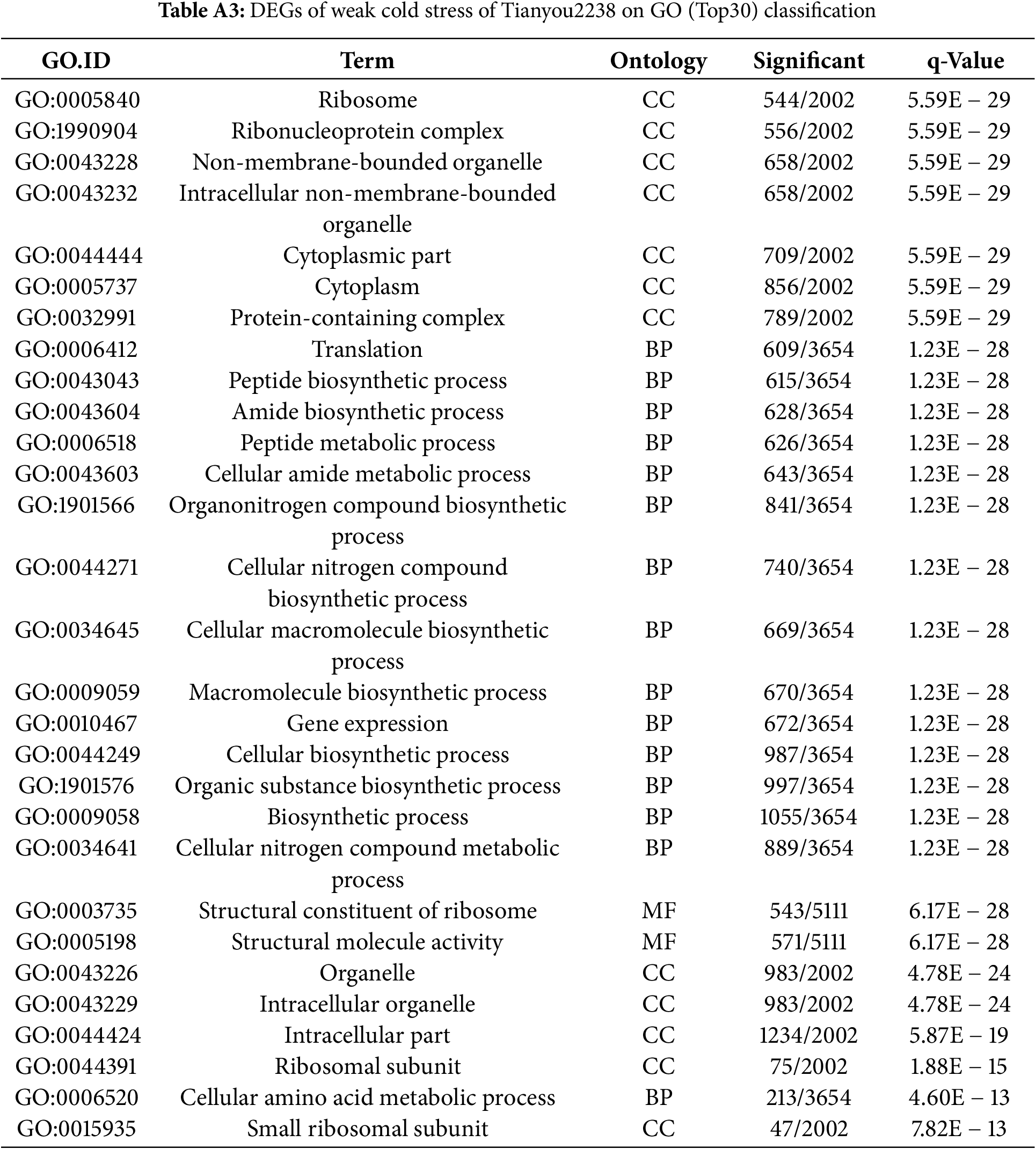
Conversely, in the weak cold-resistant B. napus Tianyou 2238, DEGs in MF categories included structural constituent of ribosome (GO: 0003735) and structural molecule activity (GO: 0005198). CC DEGs were largely concentrated in the ribosome (GO: 0005840) and ribonucleoprotein complex (GO: 1990904), while BP DEGs were mainly involved in translation (GO: 0006412) and peptide biosynthetic processes (GO: 0043043) (Table A3). These findings underscore the distinct roles of genes from 16HNTS309 and 2238 in GO enrichment under cold stress, playing essential roles in ROS production and clearance processes in both BP and MF of cold-resistant B. napus.
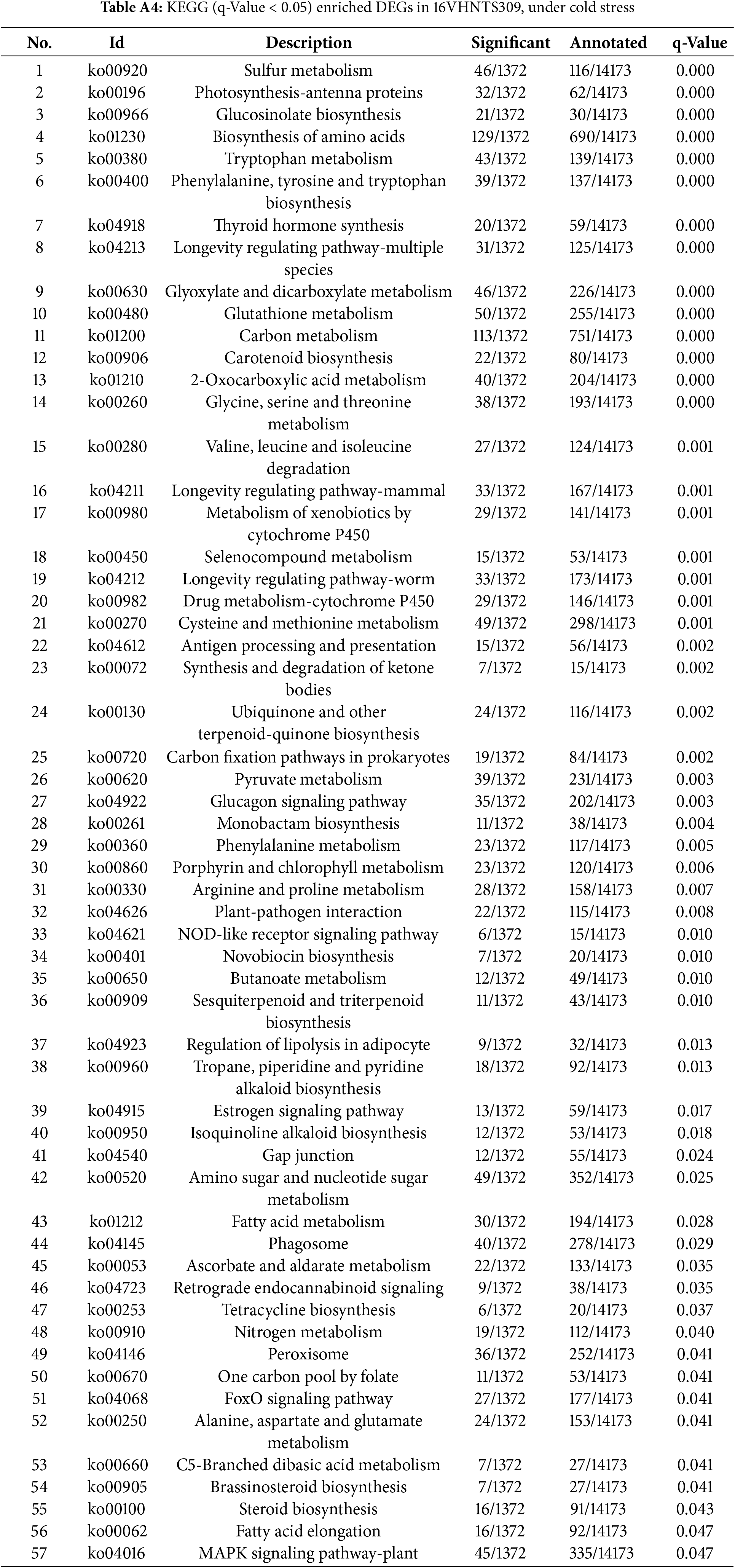
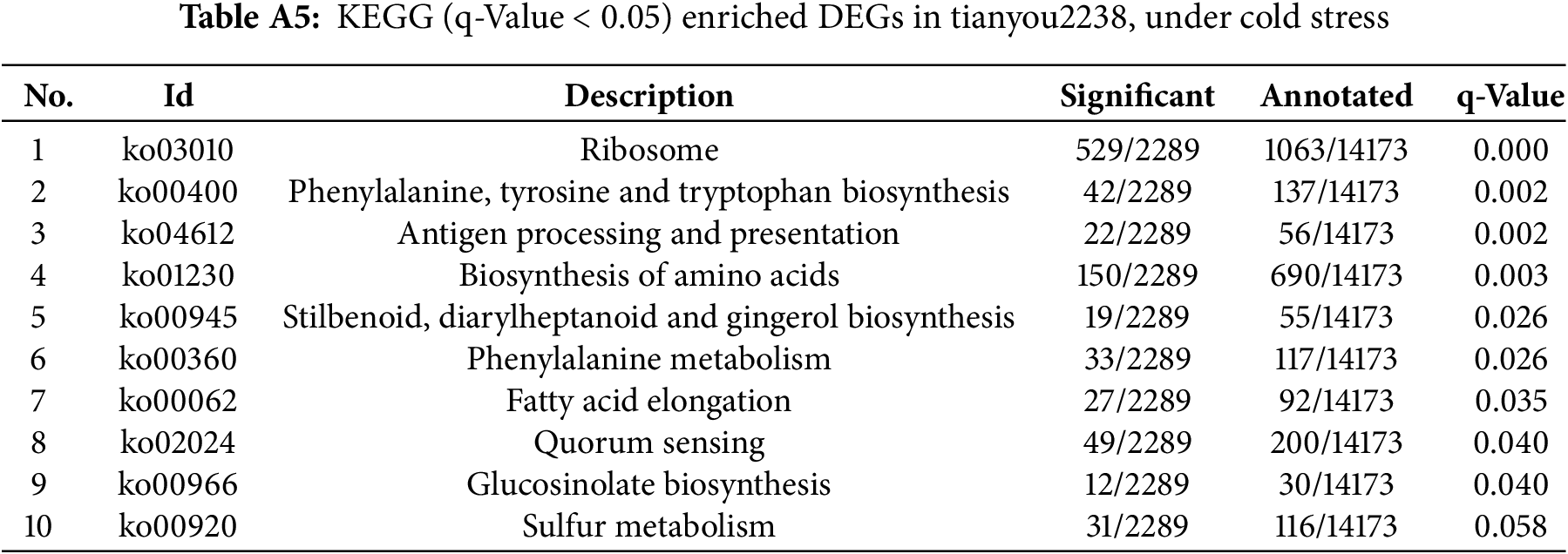
Post-cold stress, there were notable changes in 57 pathways (q-Value <0.05) in the cold-resistant B. napus of 16VHNTS309 (Table A4), as opposed to only 9 pathways in the weak cold-resistant Tianyou 2238 (Table A5). These observations indicate that 16VHNTS309 employs a more intricate mechanism of cold resistance compared to Tianyou 2238. DEGs in 16VHNTS309 were significantly enriched in pathways such as carbon metabolism (Ko, 01200), biosynthesis of amino acids (Ko, 01230), glyoxylate and dicarboxylate metabolism (Ko, 00630), and sulfur metabolism (Ko, 00920) (Fig. 4a). During cold stress, DEGs in Tianyou 2238 were prominently enriched in the ribosome (Ko, 03010), biosynthesis of amino acids (Ko,01230), and sulfur metabolism (Ko, 00920) (Fig. 4b).
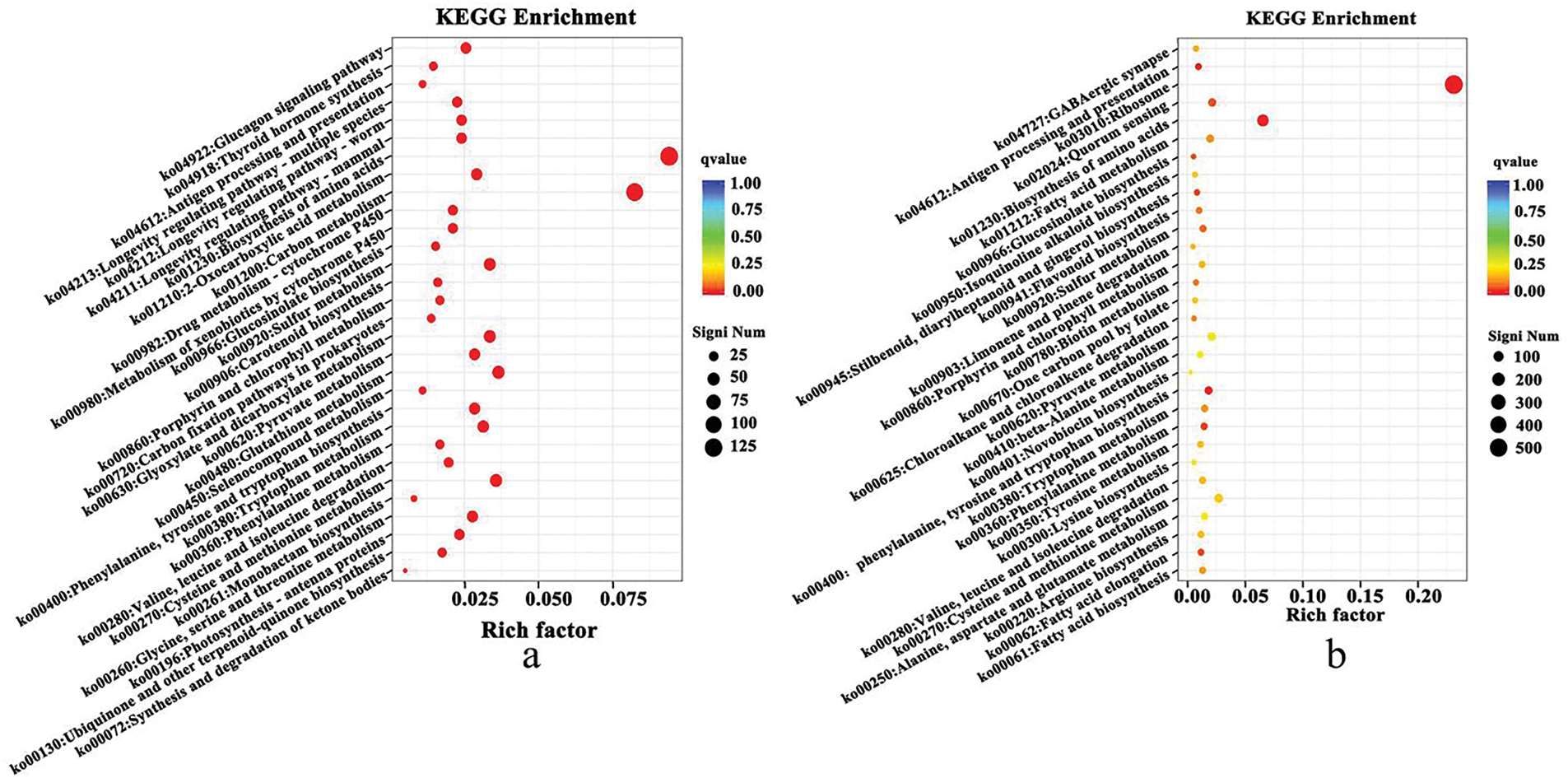
Figure 4: KEGG enriched DEGs in 16VHNTS309 (a) and Tianyou2238 (b) at 48 h of cold stress
3.4 Physiological Index Analysis
There were significant differences in ABA, VB6, and H2S contents among varieties under cold stress. Our study indicated that ABA content in 16VHNTS309 was 377.16 μg/L, significantly higher than in Tianyou2238 (Fig. 5A). The H2S content in the cold tolerant 16VHNTS309 was 11.59 nmol/g, significantly higher than in Tianyou 2238 (Fig. 5B). The VB6 content in 16VHNTS309 was measured at 22.14 μg/g, significantly higher than in Tianyou 2238 (Fig. 5C), suggesting that the cold-tolerant B. napus 16VHNTS309 accumulate more VB6.
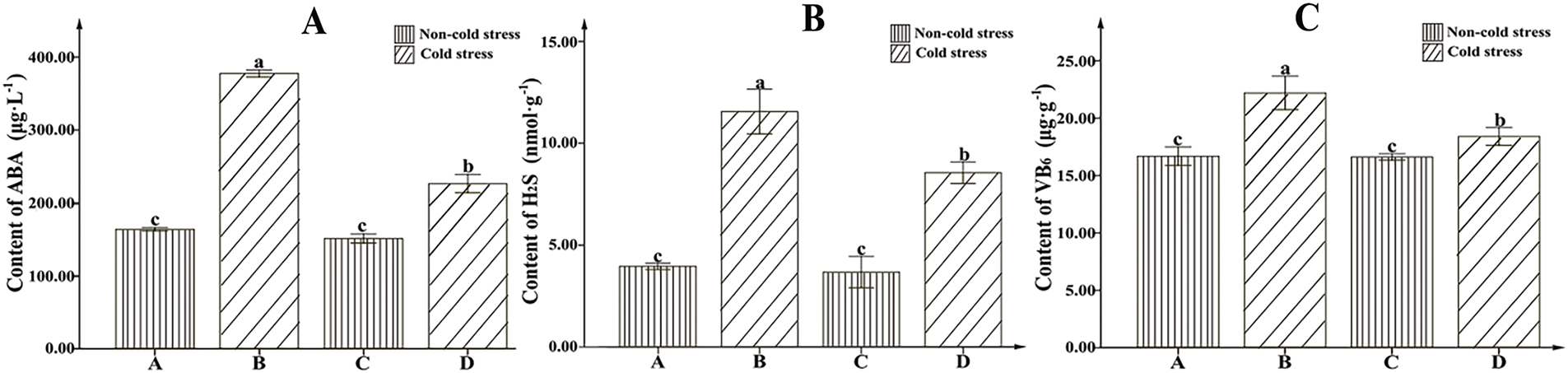
Figure 5: The activity of ABA (A), H2S (B), and VB6 (C) content accumulation in pants of 16VHNTS309 and Tianyou2238 under cold stress at 4°C for 48 h. Uppercase letters denote a significant difference (p < 0.05) in the data between cold-stressed samples and controls
4.1 Synthesis of Vitamin B6 in B. napus and Its Role in ROS Elimination
VB6 plays a pivotal role in plant stress responses. VB6 demonstrates significant antioxidant capabilities, effectively mitigating ROS accumulation in response to diverse environmental stresses [22]. VB6 synthesis in plants predominantly occurs through the DXP-independent pathway, with the critical genes, PDX1 [23] and PDX2 [24], identified in C. nicotianae. These genes are essential for PLP synthase activity; PDX2 catalyzes the production of NH3 from glutamine, while PDX1 facilitates the conversion of glyceraldehyde 3-phosphate, ribose 5-phosphate, and NH3 into PLP [25,26]. Chen et al. [27] isolated a PDX1 homolog, PDX1.3, from yeast under drought and salt stress, with further studies confirming its critical role in biological responses. The expression of PDX1 and PDX2 is up-regulated in Arabidopsis thaliana under abiotic stress [28], leading to increased VB6 content and enhanced antioxidant activity when PDX proteins are overexpressed [29]. Under the cold stress, the VB6 content in 16VHNTS309 was significantly higher than in Tianyou 2238 (Fig. 5C), suggesting that the cold-tolerant B. napus 16VHNTS309 accumulates more VB6, which aids in reducing ROS production. This difference may stem from the distinct VB6 metabolic pathway in response to cold stress between 16VHNTS309 and Tianyou2238. Transcriptome data indicated that PDX1 plays a vital role in regulating PLP synthesis (EC:4.3.3.6), with genes such as BnaC03g00280, BnaAnng00210D, BnaC04g45760D, BnaA10g13320D being up-regulated in 16VHNTS309 following cold stress, but not in Tianyou 2238 (Fig. 6). Consequently, 16VHNTS309 synthesizes more PLP, whereas Tianyou 2238 does not exhibit similar gene expression levels. VB6 is synthesized from PLP via the action of PL phosphatase (EC:3.1.3.74), during which the gene BnaA08g23000D is up-regulated in 16VHNTS309 but down-regulated in Tianyou 2238 by a factor of 2.02. Overall, several genes are up-regulated in the cold-tolerant B. napus 16VHNTS309 under cold stress, unlike in Tianyou 2238, indicating that 16VHNTS309’s cold resistance is linked to the VB6 metabolic pathway, which plays a beneficial role in eliminating excessive ROS in vivo. These results further confirm that PDX1 expression is up-regulated under various stresses, significantly contributing to VB6 synthesis and actively regulating plant stress resistance [30].
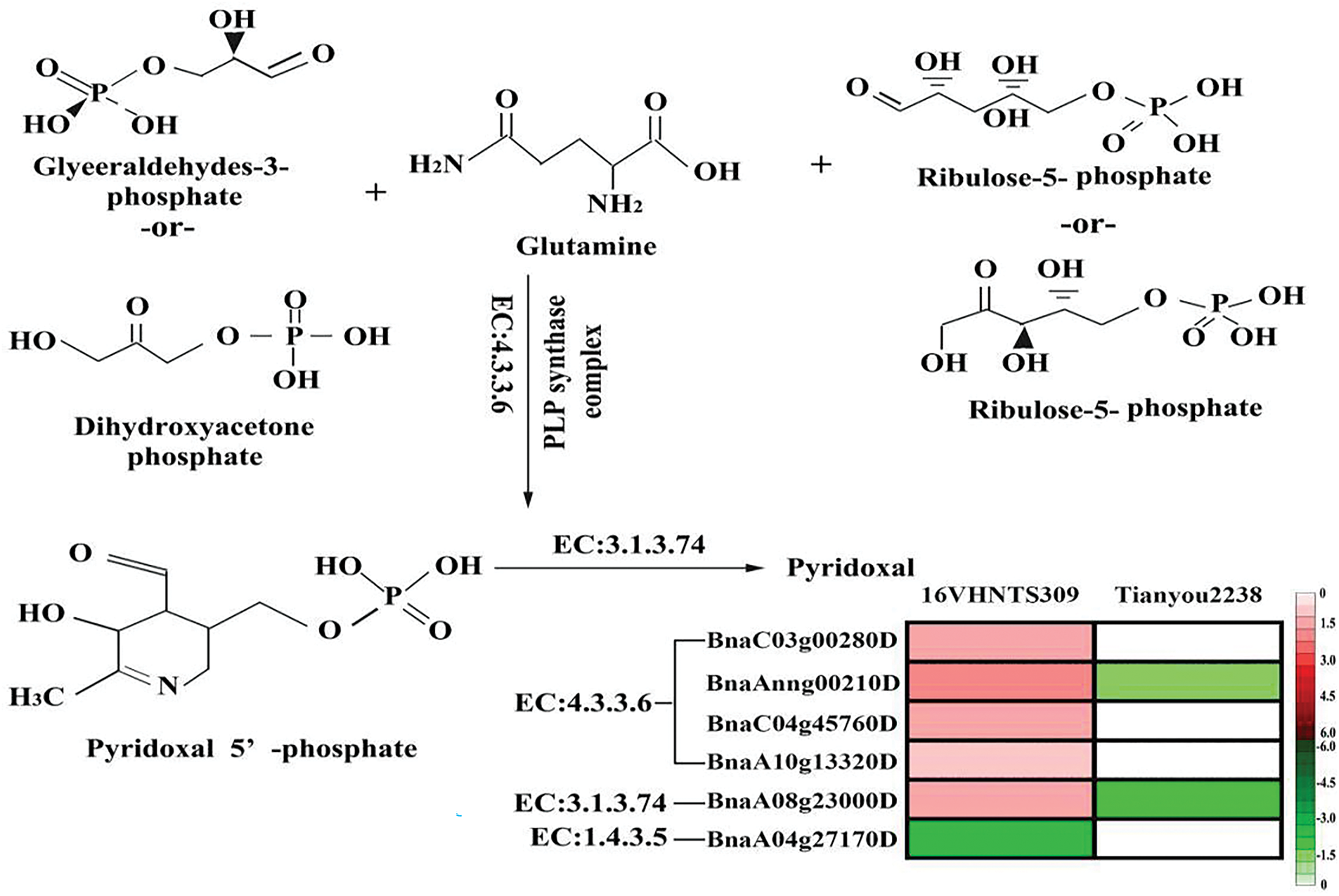
Figure 6: B.napus of DEGs in VB6 pathway, which biosynthetic pathways was quoted from Huang et al. [30]
4.2 Role of Peroxidasome in Maintaining ROS Balance in B.napus
Peroxisomes are crucial for plant development, playing a significant role in controlling H2O2 levels within the ROS scavenging system [31,32]. Immature peroxidase precursors are synthesized in the endoplasmic reticulum. These precursors are then assembled into complete peroxidases by PMP, matrix protein, and PEX. PMP, containing a peroxisomal membrane targeting signal (mPTS), is synthesized at the ribosome and directed into the endoplasmic reticulum by mPTS, mediated by PEX3 and PEX19 [33]. The peroxidase precursors then form complexes with the matrix proteins PTS1-PEX5 and PTS2-PEX7, which are recognized by PEX14 and PEX13 on the membrane, respectively, and enter the peroxisome Matrix [34]. The complexes dissociate, releasing the matrix proteins into the matrix, while empty PEX5 and PEX7 are transferred out of the peroxisome by the translocation complexes (PEX1, PEX10, PEX12) for subsequent transfer [35,36]. Studies have highlighted that the PTS1-PEX5 complex plays a critical role in peroxisome formation [37]. Results indicated that PTS1 genes of BnaC03g09940D, BnaC03g26640D, BnaA03g22610D, BnaC01g36930D, BnaA01g29410D, BnaA05g25050D, BnaC05g39220D were up-regulated in the cold-tolerant B.napus 16VHNTS309 (Fig. 7). In contrast, the expression of PTS1-PEX5 genes was down-regulated or absent in the weakly cold-resistant B.napus Tianyou2238, suggesting that the PEX5-PTS1 complex is vital for peroxisome synthesis under cold stress (Fig. 7).
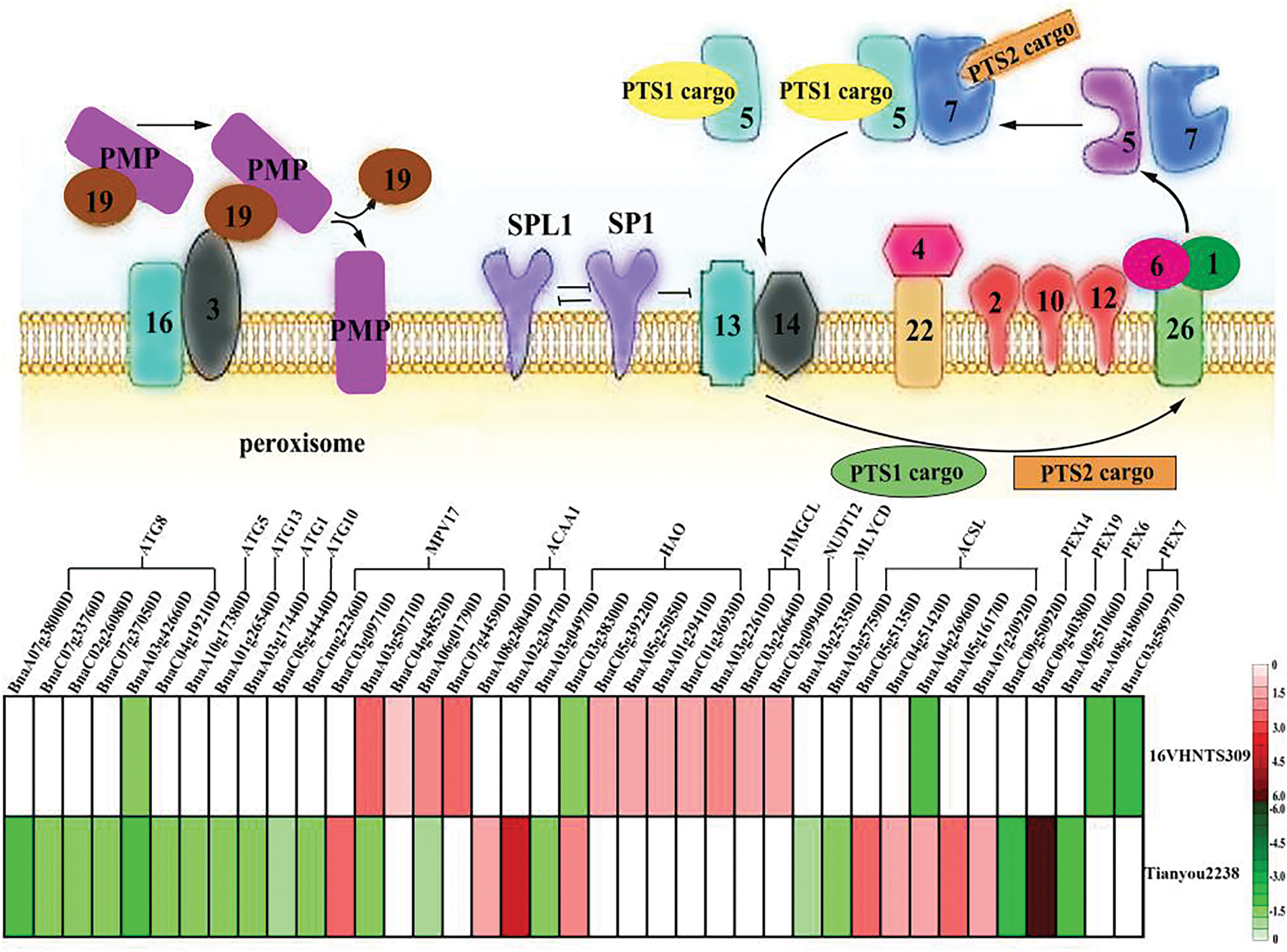
Figure 7: Peroxisomal metabolic pathway quoted from Pan et al. [34]: Analysis of peroxisomal metabolic and autophagosome pathway in B.napus
Mano et al. [38] reported that PTS1-PEX5 could be recognized by Pex14, and regulating the expression of the PEX14 gene enables Arabidopsis thaliana to respond actively to drought stress, ROS metabolism, and metabolic homeostasis. However, the gene BnaC09g50920D regulating PEX14 in the weak cold-resistant B.napus Tianyou2238 was down-regulated by 2.13 following cold stress, indicating a reduced capability of PEX14 to identify and transport PEX5-PTS1. The PEX1-PEX6 complex then interacts with the ubiquitinated PEX5, which is released into the cytoplasm [39]. In Arabidopsis thaliana, mutations in PEX6 or PEX26 have been shown to decrease PEX5 output, further indicating that the stability of the PEX1, PEX6, and PEX26 complexes impacts the stability and function of peroxisomes [40]. Our study showed that the PEX6 gene BnaA09g51060D was down-regulated in the weak cold-resistant B.napus of Tianyou2238, affecting the stability and output capacity of the PEX1, PEX6, and PEX26 complex (Fig. 7).
These findings demonstrate that cellular ROS can regulate PEX gene expression and ultimately influence the increase in peroxisome numbers. Analysis of the peroxidase formation process shows that the control of PEX14 and PEX5 input and output, and related PEX6 genes in the weak cold-resistant B. napus Tianyou2238 were down-regulated, which indicates a reduction in peroxisome production in Tianyou2238 cells, thereby diminishing cold adaptability. It is hypothesized that peroxidase synthesis inhibition is the primary reason for Tianyou2238’s poor cold resistance. The study also confirmed that redundant or damaged peroxisomes could be degraded by autophagosomes following environmental stress [37]. Plant autophagy, regulated by ATG genes, was shown by Ren et al. [41] to be up-regulated in Arabidopsis thaliana following aluminum ion stress; autophagy mutants suffered significant oxidative damage. For example, Arabidopsis ATG2, ATG7, and ATG18 mutants exhibited peroxisome aggregation [42]. Under cold stress, numerous autophagosome genes (ATG1, ATG5, ATG8, ATG10, and ATG13) were down-regulated in weakly cold-resistant B. napus Tianyou2238, indicating an impaired capacity to clear redundant or damaged peroxisomes.
4.3 Effects of Sulfur Metabolism on ROS Scavenging Mechanism in Cold-Resistant B.napus
Previous studies have shown that the absorption, transport, and assimilation of sulfur compounds—including free sulfur (S0), hydrogen sulfide (H2S), glutathione (GSH), and phytochelatins (PCs)—influence plant growth, development, and stress tolerance to varying degrees. Studies reported that aging plants exhibit lower H2S levels but higher malondialdehyde (MDA) levels, suggesting a negative correlation between H2S and MDA levels [43]. Moderate H2S levels enhance the activity of antioxidant enzymes such as superoxide dismutase (SOD), ascorbate peroxidase (APX), peroxidase (POD), and catalase (CAT), thereby enhancing the plants’ antioxidant capacity to remove more ROS. For example, exogenous H2S application has been shown to delay senescence in Chinese rose, Euonymus bungeana, mountain wood, and pomegranate bark [44]. Our results indicate that the H2S content in the cold-tolerant 16VHNTS309 was significantly higher than in Tianyou 2238. Additionally, MDA levels were significantly higher in Tianyou2238 than in 16VHNTS309 [4], further supporting the inverse relationship between H2S and MDA content proposed by Yu et al. [43].
Transcriptome data reveal that the sulfur metabolism pathway plays a crucial role in B.napus under cold stress. Key genes encoding ATP sulfurylases have been cloned in Arabidopsis thaliana [45], and Brassica [46]. ATP sulfurylases (EC 2.7.7.4) catalyze the conversion of SO42− into adenosine 5′-phosphosulfate (APS) [47]. Our findings show that seven genes encoding ATP sulfurylases were upregulated in 16VHNTS309, whereas only Bna05g15510D was upregulated, and BnaA06g36560D and Bna07g17320D were downregulated in Tianyou2238. This suggests that the cold-resistant B.napus 16VHNTS309 exhibits enhanced catalytic activity of ATP sulfurylases, converting SO42− to APS. Subsequently, APS is reduced to SO32− by APS reductase (APR, EC 1.8.99.2) [48]. Our study found that genes encoding APS reductase in 16VHNTS309 (Fig. 8), specifically BnaC07g37060D, BnaA03g59800D, BnaA01g11840D, BnaC04g19270D, BnaA03g45080D, and BnaC01g13420D, were upregulated, indicating a robust capacity to produce SO32−. Conversely, these genes were downregulated in Tianyou2238. Sulfite is further reduced to H2S by sulfite reductase (SIR, EC 1.8.7.1), with genes encoding SIR upregulated in 16VHNTS309 but not significantly expressed in Tianyou 2238. This demonstrates that the sulfur metabolism pathway undergoes significant alterations under cold stress in 16VHNTS309, resulting in increased H2S production. Research also indicates that H2S plays a critical role in the antioxidant enzyme system and in the accumulation of osmotic regulatory substances under stress [49].
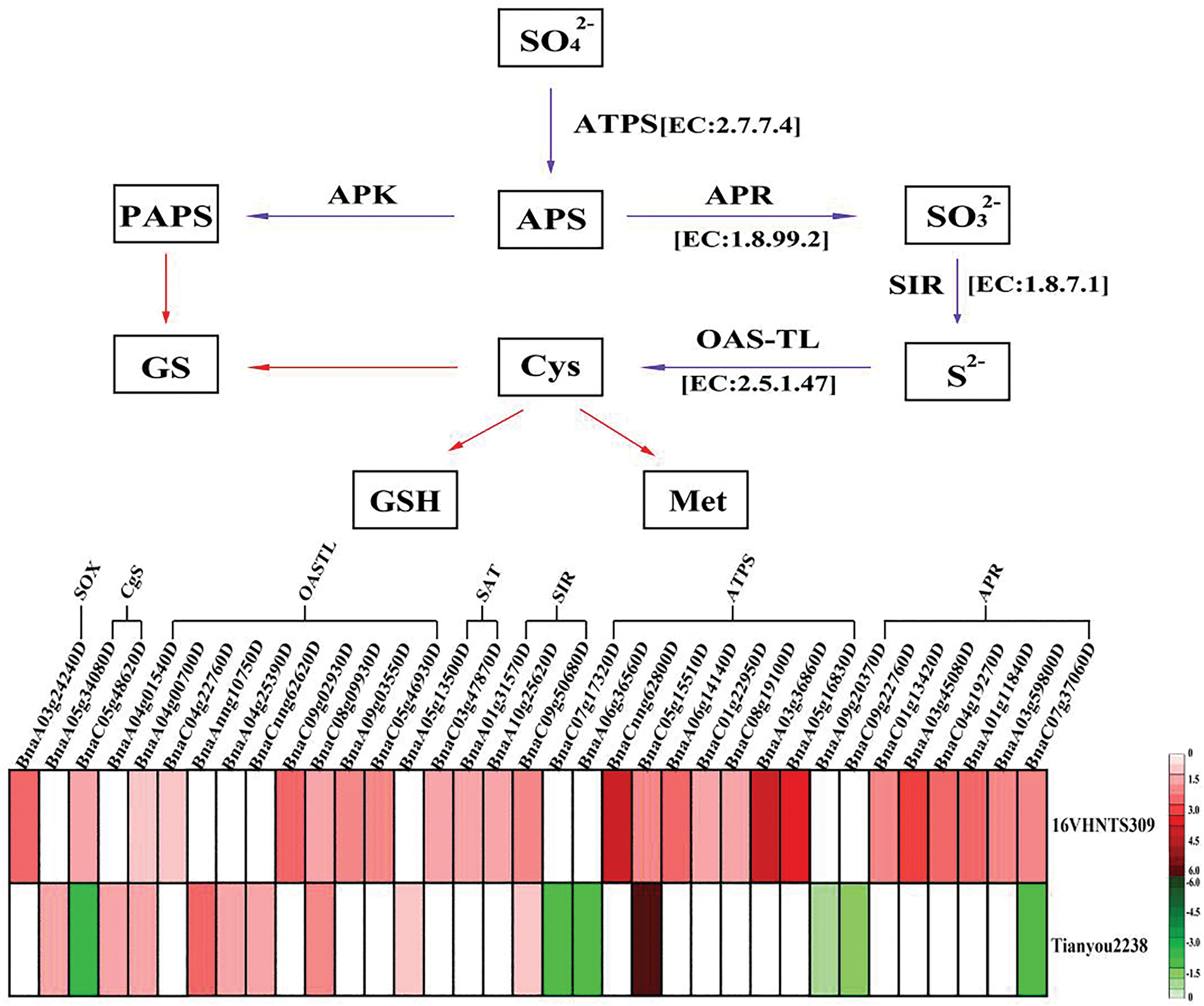
Figure 8: Analysis of sulfur metabolism in B.napus [52]
Cysteine (Cys) synthesis occurs through the action of serine acetyltransferase (SAT, EC: 2.3.1.30) and O-acetylserine (thiol) lyase (OASTL, EC: 2.5.1.47), utilizing H2S and O-acetylserine (OAS). As a central hub in sulfur nutrient metabolism, Cys serves not only as a sulfur donor for vitamins and coenzymes but also as a substrate for the synthesis of the antioxidant GSH [50]. GSH effectively scavenges H2O2 in plants, underscoring Cys’s vital role in oxidative stress mitigation [51]. The present study revealed that numerous cysteine synthetase genes, including those for SAT and OASTL, were upregulated in both 16VHNTS309 and Tianyou 2238, enhancing Cys synthesis in B.napus under cold stress. However, compared to 16VHNTS309, Tianyou 2238 exhibited lower H2S levels and Cys synthesis gene expression after cold exposure. It is speculated that 16VHNTS309, with its strong cold resistance, possesses superior abilities to absorb, transport, and assimilate sulfur elements, subsequently accumulating more Cys for use as sulfur donors for vitamins, coenzyme factors, and GSH.
4.4 The Changes of SOD and CAT Activities in B.napus Are Actively Involved in Regulating ROS Metabolism
An appropriate level of ROS is crucial for the normal growth and development of plants, reliant on a balance between ROS production and elimination. Plants with enhanced cold resistance exhibit higher activities of antioxidant enzymes, which are integral in managing ROS metabolism and mitigating membrane lipid peroxidation. This observation is well-supported in various crops including wheat [53] and rice [54]. The activities of catalase (CAT), peroxidase (POD), and superoxide dismutase (SOD) in the cold-resistant 16VHNTS309 were significantly greater than in Tianyou2238 [4]. Transcriptome analysis revealed that the expression of SOD genes Bna05g06430D, BnaA06g05150D, and BnaA01g14450D was up-regulated in 16VHNTS309, but not in Tianyou2238. Additionally, BnaA08g10870D, BnaC03g65530D, BnaC07g45360D, and BnaA03g53180D showed increased expression levels of 3.52, 3.45, 3.03, and 2.61, respectively, in 16HNTS309. In contrast, BnaC07g45360D and BnaA03g53180D were not significantly expressed in Tianyou2238. The sulfur metabolism pathway further demonstrated that the cold-resistant 16VHNTS309 could synthesize more Cys and promote the synthesis of GSH (Fig. 8). Our findings indicate that minimal ROS accumulated in 16VHNTS309 [4]. The main reason is that the activities of CAT, SOD, and GSH are crucial in reducing ROS production in 16VHNTS309.
4.5 The Strong Cold Resistance of B.napus Was Correlated with the Interaction of ROS and Ca2+ Signals
Ca2+ concentration increases in plants following cold stress. NADPH oxidase is a pivotal enzyme for ROS production, while mutations in RBOHD/F can inhibit the accumulation of both ROS and Ca2+ in cells [55]. This is because Ca2+ binds to the Ca2+ site on NADPH oxidase, altering its conformation and triggering ROS production. Our studies confirm that ROS is mediated by NADPH in B.napus during cold stress [4]. Studies on Arabidopsis thaliana have shown that the expression of calmodulin (CaM), a Ca2+ signaling receptor, increases under cold stress, highlighting the significant role of the CaM protein in plant adaptation to low temperatures [56]. Transcriptome results indicated that several genes encoding CaM and Calcineurin B-like proteins (CBLs) were significantly up-regulated in 16VHNTS309. However, these genes were either down-regulated or not significantly expressed in Tianyou2238, except for BnaA09g03360D and BnaA06g19660D (Fig. 9). This suggests that the Ca2+ signaling pathway plays a crucial role in the strong cold resistance of B.napus 16VHNTS309. It is hypothesized that Ca2+ combines with NADPH, inducing a ROS wave that couples to form a rapid signaling system capable of transmitting signals to neighboring and distal cells.
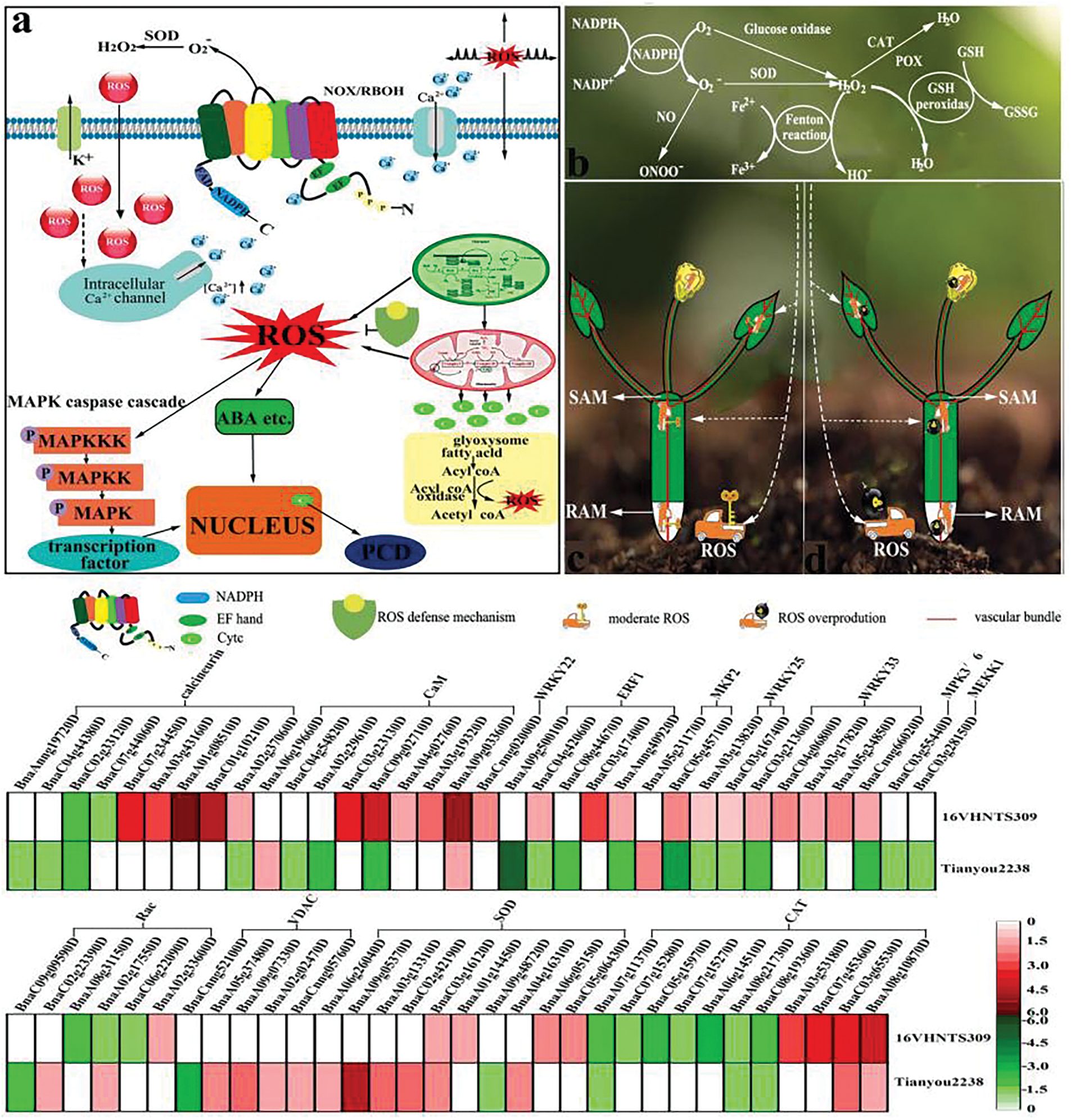
Figure 9: Signal transduction mechanism of ROS in B.napus. These mechanisms include ROS production, ROS clearance, the MAPK cascade, Ca2+ signaling, and VDAC protein, etc. (a), (b): ROS generation and scavenging was quoted from Zhang et al. [66], (c): When the ROS content in the plant is lower than the “threshold”, ROS (like a key) will be conducive to the growth and development of plants. (d): Conversely, an imbalance in ROS homeostasis can lead to oxidative explosions, detrimental to plant growth and development
4.6 The Strong Cold Resistance of B.napus Interacted with ROS, MAPK, and WRKY
Under stress conditions, increased ROS production in plants leads to the activation of Ca2+ signaling channels and the accumulation of phospholipid acid (PA) [57,58]. The accumulated PA and Ca2+ are vital for activating the serine/threonine protein kinase (OXI1), which in turn activates the MAPK cascade (MAPK3/6) [59], thereby influencing plant stress resistance. Thus, the MAPK cascade response plays a crucial role in signal reception, transduction, and nuclear transmission within the signal transduction pathway [59], and it regulates the expression of relevant stress-resistant genes. It has been reported that MKP2, closely associated with oxidative stress signaling in Arabidopsis, actively regulates MAPK3/6 activation and the oxidative stress response in cells [60]. Current results show that the MAPK3/6 gene BnaC03g55440D was down-regulated by 1.01, and the MAPK3/6 related MKP2 and key gene BnaC05g45710D were down-regulated by 2.78 in Tianyou 2238 following cold stress. However, BnaC05g45710D was up-regulated by 1.78 in B.napus 16VHNTS309 (Fig. 9), indicating that MKP2 is actively involved in regulating MAPK3/6 activation, thereby affecting the stress resistance of B.napus.
Increasing evidence suggests that ROS can enhance transcription factor expression, which then regulates the expression of stress-responsive genes to improve plant stress resistance [61]. Among these, the transcription factor WRKY is one of the largest transcription families in plants and is involved in various stress response regulatory pathways [62]. For example, the up-regulation of WRKY33 in Arabidopsis directs phosphorylation targeting MAPK3/6, enhancing plant stress resistance [63]. Our study revealed that several genes (BnaCnng66020, BnaA05g34850D, BnaA03g17820D, BnaC04g06800D, BnaC03g21360D) of WRKY33 were up-regulated in 16VHNTS309 after cold stress, while the same genes BnaCnng66020D, BnaA03g17820D, BnaC03g21360D were down-regulated in Tianyou2238 (Fig. 9). This suggests that WRKY33 plays a significant role in the cold tolerance of 16VHNTS309. It is further suggested that the up-regulated expression of WRKY33 may interact with MAPK3/6 to enhance the cold tolerance of 16VHNTS309.
Studies also indicate that cold tolerance in Arabidopsis thaliana is associated with the up-regulated expression of AtWRKY22, AtWRKY33, and AtWRKY25 [64]. Up-regulation of WRKY22 in wheat and oil palm [65] has been shown to confer significant cold resistance. Our study found that the gene (BnaCnng02000D) of WRKY22 and genes (BnaC03g16740 and BnaA03g13820D) of WRKY25 were up-regulated in 16VHNTS309, whereas the same genes of WRKY22 and WRKY25 were not significantly expressed or were down-regulated, respectively. Based on these findings, it is suggested that the cold tolerance of B.napus 16VHNTS309 is related to the up-regulated expression of WRKY22, WRKY33, and WRKY25 (Fig. 9).
4.7 The Strong Cold Resistance of B. napus Was Correlated with ROS, ABA, and H2S Interactions
ROS also interact with other signaling molecules, forming complex networks that enable plants to effectively respond to chilling injuries, high temperatures, and other stresses. Advances in genetics, molecular biology, biochemistry, and cytology have established that stomatal closure is regulated by ROS and ABA. Studies, including one by Singh et al. [67], have shown that DPI can inhibit ROS production from NADPH oxidase. When treated with DPI for 30 min, ROS fluorescence in stomatal guard cells significantly decreased, and the stomatal closure process was markedly inhibited. This evidence supports the interactions between ROS and ABA in stomatal regulation. The role of ABA in stomatal closure has been extensively studied; PYR/PYL/PCAR are identified as ABA signal receptors in guard cells. Stress treatment inhibits the activity of the PP2C enzyme and changes the conformation of activated PYLs. The interaction between PYLs and PP2C regulates the protein kinase OST1, which is crucial for activating the NADPH enzyme, promoting ROS production, calcium influx, anion outflow, and ultimately stomatal closure (Fig. 10). Our study indicated that ABA content in 16VHNTS309 was significantly higher than in Tianyou2238, suggesting that ABA accumulation aids stomatal closure in B. napus under low-temperature stress, aligning with prior studies. Transcriptome data revealed that genes BnaA10g00540D and Bna04g47050D encoding the PYR/PYL receptors were up-regulated in 16VHNTS309, while 13 receptor genes were down-regulated in Tianyou2238 after cold stress. This resulted in a significant change in the conformation of ABA receptors in 16VHNTS309 cells, activating downstream signals from PP2C and SNRK2, and enhancing ABA accumulation. Additionally, genes BnaC05g41830D, BnaA05g27660D (PP2C), and BnaA06g22800D, BnaC03g50700D, BnaC06g20490D (SNRK2) were up-regulated in 16VHNTS309, but not significantly expressed in Tianyou2238. These physiological, cytological, and transcriptomic findings demonstrate that ABA plays an active role in regulating stomatal closure and cold adaptation in the highly cold-resistant B.napus 16VHNTS309. Under stressful conditions, H2S accumulates and contributes to ABA-induced stomatal closure. Our results show that the sulfur metabolism pathway in 16VHNTS309 is crucial, with significantly higher H2S levels than in Tianyou2238, indicating that the synergy between H2S and ABA is a key mechanism of cold stress response in B. napus.
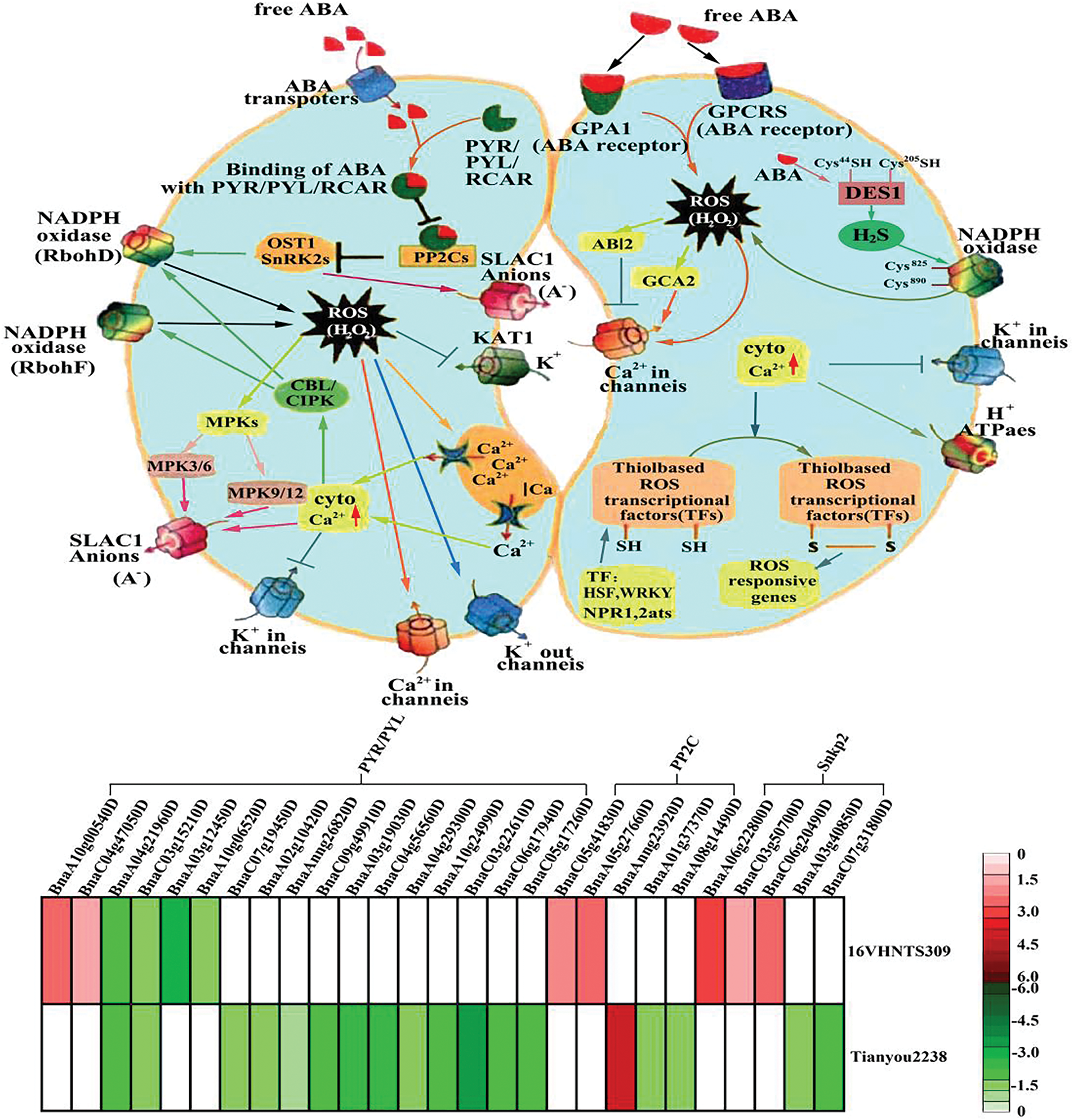
Figure 10: Analysis of the stomatal regulation mechanism of ABA in B.napus (quoted from Singh et al. [67])
4.8 Excessive ROS Can Induce Up-Regulation of VDAC Expression, and Then Trigger PCD
Mitochondrial pore protein (VDAC) has been identified as a key molecule in mitochondrial-mediated PCD. Reduced VDAC expression leads to a 4-fold decrease in ATP synthesis [68], while VDAC knockout directly affects cell metabolism and normal function [69]. VDAC, located in the outer mitochondrial membrane, acts as a “gatekeeper” for metabolite transport between the mitochondria and cytoplasm [70] and is also thought to mediate ROS release into the cytoplasm [69]. Abiotic stresses such as cold, drought, and saline-alkali conditions induce ROS production, with varying VDAC gene expressions reported under these stresses [71]. Studies have also shown that apoptosis is often accompanied by increased ROS production and upregulated VDAC expression, resulting in the VDAC channel switching to a closed state. Our results indicated that cold stress resulted in the relative electrical conductivity of weak cold-resistant B.napus 2238 being significantly higher than that of strong cold-resistant B.napus 16VHNTS309, suggesting a higher frequency of VDAC channel closure in Tianyou2238. qRT-PCR results showed that overexpression of VDAC1 reduced the cold tolerance of Arabidopsis thaliana, and VDAC1 mutant strains performed better than wild-type plants under cold stress, indicating a negative regulatory role for VDAC1 in cold stress response [72]. The current study showed that genes BnaCnng05760D, BnaA02g02470D, BnaA09g07330D, and BnaA05g37480D were up-regulated by 1.35, 1.37, 1.97, and 1.57, respectively. BnaCnng52100D was down-regulated by 2.78, confirming that weak cold-resistant B.napus 2238 accumulated more ROS, leading to enhanced membrane lipid peroxidation and changes in membrane permeability.
Acknowledgement: We thank Zibo Yimore Translation Co., Ltd. provided the English text of a draft of this manuscript.
Funding Statement: This study was financially supported by the National Nature Science Foundation Regional Fund Project (32360455); Qingyang City Joint Research Fund Project—Major Project (QY-STK-2024A-046); Doctoral Foundation of Longdong University (XYBYZK2107); University Teachers Innovation Fund Project of Gansu Province (2025A-198).
Author Contributions: Weiliang Qi conceived and designed the study. Weiliang Qi, Cairong Yang, Wancang Sun, Li Ma, Haiqing Liu, and Ziyao Wei conducted the experiments and analyzed the data. Weiliang Qi and Xiaolong Li wrote the manuscript. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: Not applicable.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Appendix A
References
1. Wang Y, Wang J, Sarwar R, Zhang W, Geng R, Zhu KM, et al. Research progress on the physiological response and molecular mechanism of cold response in plants. Front Plant Sci. 2024;15:1334913. doi:10.3389/fpls.2024.1334913. [Google Scholar] [PubMed] [CrossRef]
2. Liu LJ, Pu YY, Fang Y, Ma L, Yang G, Niu ZX, et al. Genome-wide analysis of DNA methylation and transcriptional changes associated with overwintering memory in Brassica rapa L. grown in the field. Chem Biol Technol Agric. 2024;11(1):132. doi:10.1186/s12870-019-1670-9. [Google Scholar] [CrossRef]
3. Singh A, Kumar A, Yadav S, Singh I. Reactive oxygen species-mediated signaling during abiotic stress. Plant Gene. 2019;18:100173. doi:10.1016/j.plgene.2019.100173. [Google Scholar] [CrossRef]
4. Qi W, Wang F, Ma L, Qi Z, Liu S, Chen C, et al. Physiological and biochemical mechanisms and cytology of cold tolerance in Brassica napus. Front Plant Sci. 2020;11:1241. doi:10.3389/fpls.2020.01241. [Google Scholar] [PubMed] [CrossRef]
5. Xu Y, Zhang S, Zhang M, Jiao S, Guo Y, Jiang T. The role of reactive oxygen species in plant-virus interactions. Plant Cell Rep. 2024;43(8):197. doi:10.1007/s00299-024-03280-1. [Google Scholar] [PubMed] [CrossRef]
6. Yoshioka H, Hino Y, Iwata K, Ogawa T, Yoshioka M, Ishihama N, et al. Dynamics of plant immune MAPK activity and ROS signaling in response to invaders. Physiol Mol Plant Pathol. 2023;125:102000. doi:10.1016/j.pmpp.2023.102000. [Google Scholar] [CrossRef]
7. Mahiwal S, Pahuja S, Pandey GK. Structural-functional relationship of WRKY transcription factors: unfolding the role of WRKY in plants. Int J Biol Macromol. 2024;257(Pt 2):128769. doi:10.1016/j.ijbiomac.2023.128769. [Google Scholar] [PubMed] [CrossRef]
8. Choudhary A, Sharma S, Kaur H, Sharma N, Gadewar MM, Mehta S, et al. Plant system, abiotic stress resilience, reactive oxygen species, and coordination of engineered nanomaterials: a review. S Afr J Bot. 2024;171(2):45–59. doi:10.1016/j.sajb.2024.05.053. [Google Scholar] [CrossRef]
9. Ding C, Hao X, Wang L, Li N, Huang J, Zeng J, et al. iTRAQ-based quantitative proteomic analysis of tea plant (Camellia sinensis (L.) O. Kuntze) during cold acclimation and de-acclimation procedures. Beverage Plant Res. 2023;3(1):16. doi:10.48130/BPR-2023-0016. [Google Scholar] [CrossRef]
10. Desikan RAH, Mackerness S, Hancock JT, Neill SJ. Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 2001;127(1):159–72. doi:10.1104/pp.127.1.159. [Google Scholar] [PubMed] [CrossRef]
11. Wang XC, Zhao QY, Ma CL, Zhang ZH, Cao HL, Kong YM, et al. Global transcriptome profiles of camellia sinensis during cold acclimation. BMC Genomics. 2013;14:1–15. doi:10.1186/1471-2164-14-415. [Google Scholar] [PubMed] [CrossRef]
12. Wang M, Zhang X, Liu JH. Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata L. Raf.) in response to cold stress. BMC Genomics. 2015;16(1):1–20. doi:10.1186/1471-2164-14-415. [Google Scholar] [CrossRef]
13. Liu Y, Jiang Y, Lan J, Zou Y, Gao J. Comparative transcriptomic analysis of the response to cold acclimation in eucalyptus dunnii. PLoS One. 2014;9(11):e113091. doi:10.1371/journal.pone.0113091. [Google Scholar] [PubMed] [CrossRef]
14. Moliterni VMC, Paris R, Onofri C, Orrù L, Cattivelli L, Pacifico D, et al. Early transcriptional changes in beta vulgaris in response to low temperature. Planta. 2015;242:187–201. doi:10.1007/s00425-015-2299-z. [Google Scholar] [PubMed] [CrossRef]
15. Wu JY, Pan QW, Fahim AM, Zhang LL, Gong H, Liu LJ, et al. Effects of exogenous calcium and calcium inhibitor on physiological characteristics of winter turnip rape (Brassica rapa) under low temperature stress. BMC Plant Biol. 2024;24(1):937. doi:10.1186/s12870-024-05556-w. [Google Scholar] [PubMed] [CrossRef]
16. Zhang F. A new spectral index for monitoring leaf area index of winter oilseed rape (Brassica napus L.) under different coverage methods and nitrogen treatments. Plants. 2024;13(14):1901. doi:10.3390/plants13141901. [Google Scholar] [PubMed] [CrossRef]
17. Chen J, Luo M, Li S, Tao M, Ye X, Duan W, et al. A comparative study of distant hybridization in plants and animals. Sci China Life Sci. 2018;61:285–309. doi:10.1007/s11427-017-9094-2. [Google Scholar] [PubMed] [CrossRef]
18. Zhao Y, Mi C, Sun W, Liu Z, Wu J, Fang Y, et al. Analysis of botanical traits and self-compatibility in winter rapeseed Brassica napus and Brassica Rapa and F1 hybrid populations. Acta Prataculturae Sinica. 2016;25(3):108–19. doi:10.11686/cyxb2015103. [Google Scholar] [CrossRef]
19. Wang XF, Tian JH, Zhang YW, Zhang YF, Li DR. New early mature rapeseed germplasms created by Brassica napus × Brassica campestris. Chin Agricu Sci Bull. 2017;33(32):28–33. [Google Scholar]
20. Ma L, Coulter JA, Liu L, Zhao Y, Chang Y, Pu Y, et al. Transcriptome analysis reveals key cold stress responsive genes in winter rapeseed (Brassica rapa L.). Int J Mol Sci. 2019;20(5):1071. doi:10.3390/ijms20051071. [Google Scholar] [PubMed] [CrossRef]
21. Lai D, Mao Y, Zhou H, Li F, Wu M, Zhang J, et al. Endogenous hydrogen sulfide enhances salt tolerance by coupling the reestablishment of redox homeostasis and preventing salt-induced K+ loss in seedlings of Medicago sativa. Plant Sci. 2014;225:117–29. doi:10.1016/j.plantsci.2014.06.006. [Google Scholar] [PubMed] [CrossRef]
22. Liu X, Wang C, Xu Q, Zhao D, Liu F, Han B. Metabolic response of the lycium barbarum variety ‘Ningqi No. 7’ to drought stress. Plants. 2024;13(14):1935. doi:10.3390/plants13141935. [Google Scholar] [PubMed] [CrossRef]
23. Ehrenshaft M, Bilski P, Li MY, Chignell CF, Daub ME. A highly conserved sequence is a novel gene involved in de novo vitamin B6 biosynthesis. Proc Natl Acad Sci. 1999;96(16):9374–8. doi:10.1073/pnas.96.16.9374. [Google Scholar] [PubMed] [CrossRef]
24. Mittenhuber G. Phylogenetic analyses and comparative genomics of vitamin B6 (pyridoxine) and pyridoxal phosphate biosynthesis pathways. J Mol Microbiol Biotechnol. 2001;3(1):1–20. [Google Scholar] [PubMed]
25. Fitzpatrick TB, Moccand C, Roux C. Vitamin B6 biosynthesis: charting the mechanistic landscape. ChemBioChem. 2010;11(9):1185–93. [Google Scholar] [PubMed]
26. Mooney S, Hellmann H. Vitamin B6: killing two birds with one stone? Phytochemistry. 2010;71(5–6):495–501. doi:10.1016/j.phytochem.2009.12.015. [Google Scholar] [PubMed] [CrossRef]
27. Chen H, Xiong L. Pyridoxine is required for post-embryonic root development and tolerance to osmotic and oxidative stresses. Plant J. 2005;44(3):396–408. doi:10.1111/j.1365-313X.2005. [Google Scholar] [CrossRef]
28. Benabdellah K, Azcón-Aguilar C, Valderas A, Speziga D, Fitzpatrick TB, Ferrol N. GintPDX1 encodes a protein involved in vitamin B6 biosynthesis that is up-regulated by oxidative stress in the arbuscular mycorrhizal fungus Glomus intraradices. New Phytol. 2009;184(3):682–93. doi:10.1111/j.1469-8137.2009.02978.x. [Google Scholar] [PubMed] [CrossRef]
29. Raschke M, Boycheva S, Crèvecoeur M, Nunes-Nesi A, Witt S, Fernie AR, et al. Enhanced levels of vitamin B6 increase aerial organ size and positively affect stress tolerance in Arabidopsis. Plant J. 2011;66(3):414–32. doi:10.1111/j.1365-313X.2011.04499.x. [Google Scholar] [PubMed] [CrossRef]
30. Huang SH, Zhang JY, Wang LH, Huang LQ. Effect of abiotic stress on the abundance of different vitamin B6 vitamers in tobacco plants. Plant Physiol Biochem. 2013;66:63–7. doi:10.1016/j.plaphy.2013.02.010. [Google Scholar] [PubMed] [CrossRef]
31. Falter C, Reumann S. The essential role of fungal peroxisomes in plant infection. Mol Plant Pathol. 2022;23(6):781–794. doi:10.1111/mpp.13180. [Google Scholar] [PubMed] [CrossRef]
32. Sandalio LM, Peláez-Vico MA, Molina-Moya E, Romero-Puertas MC. Peroxisomes as redox-signaling nodes in intracellular communication and stress responses. Plant Physiol. 2021;186(1):22–35. doi:10.1093/plphys/kiab060. [Google Scholar] [PubMed] [CrossRef]
33. Knoops K, Manivannan S, Cepińska MN, Krikken AM, Kram AM, Veenhuis M, et al. Preperoxisomal vesicles can form in the absence of PEX3. J Cell Biol. 2014;204(5):659–68. doi:10.1083/jcb.201310148. [Google Scholar] [PubMed] [CrossRef]
34. Pan D, Nakatsu T, Kato H. Crystal structure of peroxisomal targeting signal-2 bound to its receptor complex Pex7p-Pex21p. Nat Struct Mol Biol. 2013;20(8):987–93. doi:10.1038/nsmb.2618. [Google Scholar] [PubMed] [CrossRef]
35. Okumoto K, Noda H, Fujiki Y. Distinct modes of ubiquitination of peroxisome-targeting signal type 1 (PTS1) receptor Pex5p regulate PTS1 protein import. J Biol Chem. 2014;289(20):14089–108. [Google Scholar] [PubMed]
36. Rodrigues TA, Alencastre IS, Francisco T, Brites P, Fransen M, Grou CP, et al. A PEX7-centered perspective on the peroxisomal targeting signal type 2-mediated protein import pathway. Mol Cell Biol. 2014;34(15):2917–28. doi:10.1128/MCB.01727-13. [Google Scholar] [PubMed] [CrossRef]
37. Sandalio LM, Collado-Arenal AM, Romero-Puertas MC. Deciphering peroxisomal reactive species interactome and redox signalling networks. Free Radic Biol Med. 2023;197(14):58–70. doi:10.1016/j.freeradbiomed.2023.01.014. [Google Scholar] [PubMed] [CrossRef]
38. Mano S, Nakamori C, Nito K, Kondo M, Nishimura M. The Arabidopsis pex12 and pex13 mutants are defective in both PTS1- and PTS2-dependent protein transport to peroxisomes. Plant J. 2006;47(4):604–18. doi:10.1111/j.1365-313X.2006.02809.x. [Google Scholar] [PubMed] [CrossRef]
39. Pedrosa AG, Francisco T, Bicho D, Dias AF, Barros-Barbosa A, Hagmann V, et al. Peroxisomal monoubiquitinated PEX5 interacts with the AAA ATPases PEX1 and PEX6 and is unfolded during its dislocation into the cytosol. J Biol Chem. 2018;293(29):11553–63. doi:10.1074/jbc.RA118.003669. [Google Scholar] [PubMed] [CrossRef]
40. Gonzalez Kim L, Ratzel SE, Burks KH, Danan CH, Wages JM, Zolman BK, et al. A pex1 missense mutation improves peroxisome function in a subset of arabidopsis pex6 mutants without restoring pex5 recycling. Proc Nat Acad Sci. 2018;115(14):E3163–72. doi:10.1073/pnas.1721279115. [Google Scholar] [PubMed] [CrossRef]
41. Ren H, Li Y, Zhao F, Pu X, Wei L, Lv X, et al. The role of autophagy in alleviating damage of aluminum stress in Arabidopsis thaliana. Plant Growth Regul. 2016;79:167–75. doi:10.1007/s10725-015-0122-2. [Google Scholar] [CrossRef]
42. Shibata M, Oikawa K, Yoshimoto K, Kondo M, Mano S, Yamada K, et al. Highly oxidized peroxisomes are selectively degraded via autophagy in Arabidopsis. Plant Cell. 2013;25(12):4967–83. doi:10.1105/tpc.113.116947. [Google Scholar] [PubMed] [CrossRef]
43. Yu H, Song L, Han J, Yu X, Wu Y, Yu Z. Exogenous hydrogen sulphide alleviates senescence of postharvest pakchoi through regulating antioxidant system, endogenous hydrogen sulphide and nitric oxide metabolism. Postharvest Biol Technol. 2024;207:112632. doi:10.1016/j.postharvbio.2023.112632. [Google Scholar] [CrossRef]
44. Hao X, Cao H, Wang Z, Jia X, Jin ZP, Pei Y. Hydrogen sulfide improves plant drought tolerance by regulating the homeostasis of reactive oxygen species. Plant Growth Regul. 2024;9(2):1–9. doi:10.1007/s10725-024-01197-z. [Google Scholar] [CrossRef]
45. Cheng J, Zhang Z, Gao Y, Dong Y, Xian X, Li C, et al. Orexpressing ATP sulfurylase improves fe-deficiency tolerance in apple calli and tobacco. Agronomy. 2024;14(3):404. doi:10.3390/agronomy14030404. [Google Scholar] [CrossRef]
46. Manne H, Kumari N, Yashveer S, Nain S, Duhan J, Avtar R, et al. Mitigation of lead toxicity in Brassica juncea L. by sulphur application-Via various biochemical and transcriptomic strategies. J King Saud Univ-Sci. 2024;36(5):103175. doi:10.1016/j.jksus.2024.103175. [Google Scholar] [CrossRef]
47. Rotte C, Leustek T. Differential subcellular localization and expression of ATP sulfurylase and 5′-adenylylsulfate reductase during ontogenesis of Arabidopsis leaves indicates that cytosolic and plastid forms of ATP sulfurylase may have specialized functions. Plant Physiol. 2000;124(2):715–24. doi:10.1104/pp.124.2.715. [Google Scholar] [CrossRef]
48. Lee S, Leustek T. APS kinase from Arabidopsis thaliana: genomic organization, expression, and kinetic analysis of the recombinant enzyme. Biochem Biophys Res Commun. 1998;247(1):171–5. doi:10.1006/bbrc.1998.8751. [Google Scholar] [CrossRef]
49. Kolupaev YE, Yemets AI, Yastreb TO, Blume YB. The role of nitric oxide and hydrogen sulfide in regulation of redox homeostasis at extreme temperatures in plants. Front Plant Sci. 2023;14:1128439. doi:10.3389/fpls.2023.1128439. [Google Scholar] [PubMed] [CrossRef]
50. Swindell J, Dos Santos PC. Interactions with sulfur acceptors modulate the reactivity of cysteine desulfurases and define their physiological functions. BBA-Mol Cell Res. 2024;1871(7):119794. doi:10.1016/j.bbamcr.2024.119794. [Google Scholar] [PubMed] [CrossRef]
51. Bhadwal SS, Verma S, Hassan S, Kaur S. Unraveling the potential of hydrogen sulfide as a signaling molecule for plant development and environmental stress responses: a state-of-the-art review. Plant Physiol Biochem. 2024;212:108730. doi:10.1016/j.plaphy.2024.108730. [Google Scholar] [PubMed] [CrossRef]
52. Capaldi FR, Gratão PL, Reis AR, Lima LW, Azevedo RA. Sulfur metabolism and stress defense responses in plants. Trop Plant Biol. 2015;8(3-4):60–73. doi:10.1007/s12042-015-9152-1. [Google Scholar] [CrossRef]
53. Zheng L, Yu P, Zhang Y, Wang P, Yan W, Guo B, et al. Evaluating the bio-application of biomacromolecule of lignin-carbohydrate complexes (LCC) from wheat straw in bone metabolism via ROS scavenging. Int J Biol Macromol. 2021;176(3):13–25. doi:10.1016/j.ijbiomac.2021.01.103. [Google Scholar] [PubMed] [CrossRef]
54. Zheng S, Li J, Ma L, Zhuang C. OsAGO2 controls ROS production and the initiation of tapetal PCD by epigenetically regulating OsHXK1 expression in rice anthers. Proc Nat Acad Sci. 2019;116(15):7549–58. doi:10.1073/pnas.1817675116. [Google Scholar] [PubMed] [CrossRef]
55. Lin S, Li Y, Zhao J, Guo W, Jiang M, Li X, et al. Transcriptome analysis of biochemistry responses to low-temperature stress in the flower organs of five pear varieties. Forests. 2023;14(3):490–14030490. doi:10.3390/f14030490. [Google Scholar] [CrossRef]
56. Zhang X, Zhang D, Zhong C, Li W, Dinesh-Kumar SP, Zhang Y. Orchestrating ROS regulation: coordinated post-translational modification switches in NADPH oxidases. New Phytol. 2025;245(2):510–22. doi:10.1111/nph.20231. [Google Scholar] [PubMed] [CrossRef]
57. Wang H, Feng M, Zhong X, Yu Q, Que Y, Xu L, et al. Identification of Saccharum CaM gene family and function characterization of ScCaM1 during cold and oxidant exposure in Pichia pastoris. Genes Genomics. 2023;45(1):103–22. doi:10.1007/s13258-022-01263-8. [Google Scholar] [PubMed] [CrossRef]
58. Wang H, Cheng X, Yin D, Chen D, Luo C, Liu H, et al. Advances in the research on plant WRKY transcription factors responsive to external stresses. Curr Issues Mol Biol. 2023;45(4):187. doi:10.3390/cimb45040187. [Google Scholar] [PubMed] [CrossRef]
59. Anthony RG, Henriques R, Helfer A, Mészáros T, Rios G, Testerink C, et al. A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J. 2004;23(3):572–81. [Google Scholar] [PubMed]
60. Son Y, Cheong YK, Kim NH, Chung HT, Kang DG, Pae HO. Mitogen-activated protein kinases and reactive oxygen species: how can ROS activate MAPK pathways? J Signal Transduct. 2011;2011(1–4):792639–6. doi:10.1155/2011/792639. [Google Scholar] [PubMed] [CrossRef]
61. Singh VP, Jaiswal S, Wang Y, Feng S, Tripathi DK, Singh S, et al. Evolution of reactive oxygen species cellular targets for plant development. Trends Plant Sci. 2024;29(8):865–77. [Google Scholar] [PubMed]
62. Liu J, Wang X, Wu H, Zhu Y, Ahmad I, Dong G, et al. Association between reactive oxygen species, transcription factors, and candidate genes in drought-resistant sorghum. Int J Mol Sci. 2024;25(12):6464. doi:10.3390/ijms25126464. [Google Scholar] [PubMed] [CrossRef]
63. Goyal P, Devi R, Verma B, Hussain S, Arora P, Tabassum R, et al. WRKY transcription factors: evolution, regulation, and functional diversity in plants. Protoplasma. 2023;260(2):331–48. doi:10.1007/s00709-022-01794-7. [Google Scholar] [PubMed] [CrossRef]
64. Fu QT, Yu DQ. Expression profiles of AtWRKY25, AtWRKY26 and AtWRKY33 under abiotic stresses. Yi Chuan Hereditas. 2010;32(8):848–56. doi:10.3724/sp.j.1005.2010.00848. [Google Scholar] [PubMed] [CrossRef]
65. Zhou LX, Cao HX. Expression characteristics of WRKY transcription factor genes in oil palm under low temperature. J South Agric. 2018;49(8):1490–7. [Google Scholar]
66. Zhang J, Wang X, Vikash V, Ye Q, Wu D, Liu Y, et al. ROS and ROS-mediated cellular signaling. Oxid Med Cell Longev. 2016;2016(1):4350965. doi:10.1155/2016/4350965. [Google Scholar] [PubMed] [CrossRef]
67. Singh R, Parihar P, Singh S, Mishra RK, Singh VP, Prasad SM. Reactive oxygen species signaling and stomatal movement: current updates and future perspectives. Redox Biol. 2017;11:213–8. doi:10.1016/j.redox.2016.11.006. [Google Scholar] [PubMed] [CrossRef]
68. Magrì A, Cubisino SA, Battiato G, Lipari CL, Conti Nibali S, Saab MW, et al. VDAC1 knockout affects mitochondrial oxygen consumption triggering a rearrangement of ETC by impacting on complex I activity. Int J Mol Sci. 2023;24(4):3687. doi:10.3390/ijms24043687. [Google Scholar] [PubMed] [CrossRef]
69. Lee SM, Hoang MHT, Han HJ, Kim HS, Lee K, Kim KE, et al. Pathogen inducible voltage-dependent anion channel (AtVDAC) isoforms are localized to mitochondria membrane in Arabidopsis. Mol Cells. 2009;27:321–7. doi:10.1007/s10059-009-0041-z. [Google Scholar] [PubMed] [CrossRef]
70. Tan W, Colombini M. VDAC closure increases calcium ion flux. BBA-Biomembr. 2007;1768(10):2510–5. doi:10.1016/j.bbamem.2007.06.002. [Google Scholar] [PubMed] [CrossRef]
71. Lai JC, Tan W, Benimetskaya L, Stein CA. A pharmacologic target of G3139 in melanoma cells may be the mitochondrial VDAC. Proc Nat Acad Sci. 2006;103(19):7494–9. doi:10.1073/pnas.0602217103. [Google Scholar] [PubMed] [CrossRef]
72. Li Y, Chen L, Mu J, Zuo J. LESION SIMULATING DISEASE1 interacts with catalases to regulate hypersensitive cell death in Arabidopsis. Plant Physiol. 2013;163(2):1059–70. doi:10.1104/pp.113.225805. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools