 Open Access
Open Access
REVIEW
The Role of Glutathione S-Transferase in the Regulation of Plant Growth, and Responses to Environmental Stresses
1 Key Laboratory of Plant Functional Genomics of the Ministry of Education, Yangzhou University, Yangzhou, 225009, China
2 Jiangsu Key Laboratory of Crop Genomics and Molecular Breeding, Yangzhou University, Yangzhou, 225009, China
* Corresponding Authors: Chen Lin. Email: ; Youping Wang. Email:
(This article belongs to the Special Issue: Plant and Environments)
Phyton-International Journal of Experimental Botany 2025, 94(3), 583-601. https://doi.org/10.32604/phyton.2025.063086
Received 04 January 2025; Accepted 26 February 2025; Issue published 31 March 2025
Abstract
Glutathione S-transferases (GSTs) represent a large and diverse enzyme family ubiquitously distributed across the plant kingdom. These proteins catalyze the conjugation of glutathione (GSH) with electrophilic substrates in response to various stress conditions. Beyond their role in stress adaptation, certain GSTs are integral regulators of plant growth and development, contributing to a range of physiological processes. Most GST proteins exhibit dual enzymatic activities, functioning as both transferases and peroxidases, which enables their involvement in diverse cellular processes, including detoxification and stress responses. Recent advancements, particularly in X-ray crystallography, have enabled detailed structural analysis of GST proteins, significantly enhancing our understanding of their biological functions. This review offers a comprehensive overview of the classification and structural characteristics of GSTs in plants. It also highlights recent findings on their roles in plant growth and development, cell signaling, catalytic transport, and stress tolerance. Furthermore, key scientific challenges related to GSTs are discussed, focusing on their potential applications in agriculture. These insights aim to facilitate the screening of functional GST genes and support molecular breeding efforts across diverse crop species.Keywords
Glutathione S-transferases (GSTs) are enzyme proteins with a wide range of biological functions in both plants and animals [1]. In the plant kingdom, GSTs were first identified in maize, where they were shown to conjugate with herbicides, thereby protecting the plant from damage [2]. Since then, various members of the GST family have been reported to participate in distinct functions in plants. Throughout their growth and development, plants must continuously adapt to fluctuations in their external environment. In response to external stimuli, plants typically activate a series of signaling cascades that trigger defense mechanisms, which, in some cases, may also negatively impact plant growth. The upregulation of GST gene expression is widely recognized as a hallmark for plant stress responses [1,3]. In recent years, a number of GST proteins have been identified in various plants including Arabidopsis thaliana [4], rice (Oryza sativa) [5], poplar (Populus tomentosa) [6], oilseed rape (Brassica napus) [7], among others. Previous studies have demonstrated that GSTs play a crucial role in plant stress responses by detoxifying both endogenous and exogenous xenobiotics as well as oxidized molecules [8,9]. In addition, certain GST members are involved in the regulation of plant growth, development and metabolism. Given their diverse roles, understanding the biological functions of GST proteins is essential. This review provides a comprehensive overview of the recent research progress on the classification, protein structure, and biological functions of GST family members in plants. These insights aim to promote further in-depth research on GSTs and facilitate their application in the molecular breeding of diverse crop species.
2 Classification and Structural Characteristic of GST Protein
The N- and C-terminal domains of GSTs are highly conserved, suggesting that these enzymes retain conserved biological functions across different species [10–12]. Based on their subcellular localization, GSTs can be classified into cytoplasmic, mitochondrial and microsomal types [13]. Microsomal GSTs are also known as Membrane Associated Proteins in Eicosanoid and Glutathione Metabolism (MAPEG proteins) [14,15]. Among these, cytoplasmic GSTs constitute the largest family and are also the most extensively studied. Given the growing number of identified GST members, it is essential to establish a systematic classification for these proteins. The widely used GST classification system was initially proposed by Droog in 1997 [16], that was further revised, supplemented, and improved by Edwards in 2000 [17]. This system divides the GST superfamily into multiple subfamilies based on gene structure (including exon and intron sequences), amino acid sequence similarity, conservation of key amino acid sites and substrate binding specificity [5]. In plants, cytoplasmic GST can be divided into at least 14 classes, namely, Tau (U), Phi (F), Zeta (Z), Theta (T),Lambda (L), Elongation Factor 1 Gamma (EF1Bγ), Metaxin, Ure2p, Hemerythrin, Lota, TetraChloro-HydroQuinone Dehalogenase (TCHQD), DeHydroAscorbate Reductase (DHAR), microsomal ProstaGlandin E-Synthase Type 2 (mPGES-2) and Glutathionyl-Hydroquinone Reductases (GHRs) [18–20]. Eight of these categories are prevalent across different plant species. Notably, GSTU, GSTF, GSTL, and DHAR are overrepresented in plants, which is commonly regarded as plant-specific features [21]. However, some studies have indicated that GSTF is also present in microorganisms, and its distribution is closely associated with the size of microbial genomes and their habitats [22].
GSTs are also classified differently across various plant species. The model plant Arabidopsis possesses 47 GST family members, which can be categorized into four main groups based on amino acid sequences (Fig. 1A), exhibiting relatively low homology among these groups. In contrast, wheat contains 330 GSTs, with the majority belonging to the GSTU and GSTF classes [4,21]. Poplar has 74 GST genes, which are classified into 11 categories [6]. Oilseed rape contains 179 GST genes, grouped into 6 categories, with these genes unevenly distributed across chromosomes [7]. Medicago (Medicago truncatula) has 73 GST genes, divided into 8 categories based on phylogenetic analysis, and these genes vary in structure, distribution and characteristics of their encoded proteins [23]. Rice (Oryza sativa) contains 59 GST genes classified into 6 categories [5] (Fig. 1B) and they are expressed in various tissues [18]. Soybean (Glycine max) and maize (Zea mays) have 42 and 25 GST genes, respectively, which can also be divided into 6 categories [5].
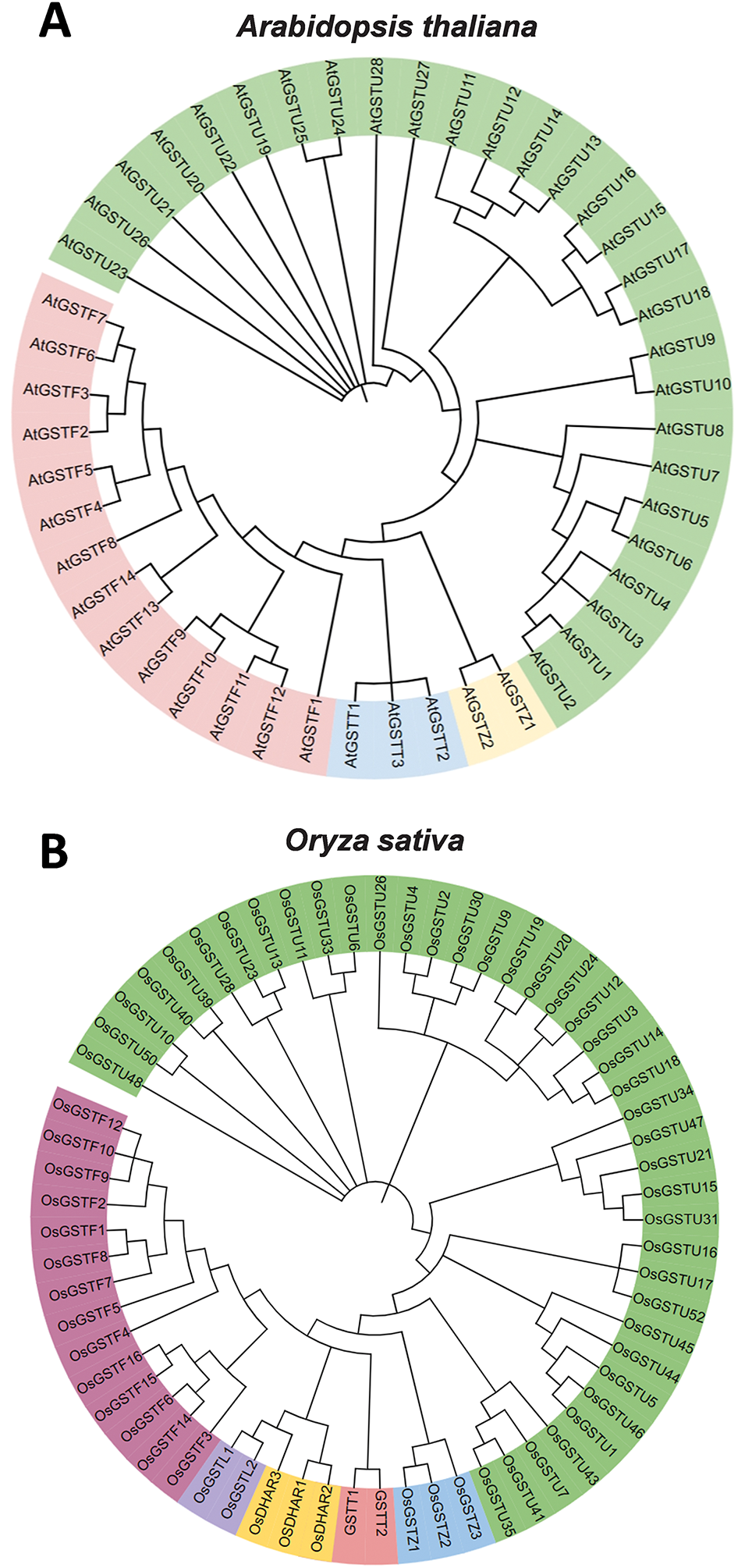
Figure 1: Alignment of GST proteins in Arabidopsis thaliana (A) and Oryza sativa (B). The Neighbor-Joining (NJ) phylogenetic tree regarding the GST proteins was constructed with MEGA7 software. These amino acid sequences of GST members in Arabidopsis thaliana (https://www.arabidopsis.org/) (accessed on 25 February 2025) and Oryza sativa (https://phytozome.jgi.doe.gov/pz/portal.html) (accessed on 25 February 2025) were obtained from online database. Different colours indicate different subgroups. This is an originally developed figure
Although the GST protein family consists of numerous members, different GSTs share conserved domains, indicative of their ancient evolutionary origins [24,25]. These conserved features facilitate the formation of stable homodimers or heterodimers, with each subunit typically ranging between 23 and 30 kilodaltons (kDa) [17]. For example, in maize, specific GST classes, such as GSTF and GSTU, form heterodimers [26–28]. Each GST monomer consists of two structural domains: the N-terminal domain (NTD) and the C-terminal domain (CTD), connected by a linker region of approximately 10 amino acid residues (Fig. 2). The NTD sequence is relatively conserved and typically includes three α-helices and four β-sheets, featuring a conserved binding site for glutathione (GSH), known as the GSH binding site (G-site). A conserved serine residue within the NTD is located in the active site, positioned to stabilize the thiolate anion of glutathione, as demonstrated by site-specific mutagenesis [29]. The NTD binds to reduced GSH through its conserved G-site, utilizing cysteine residues in GSH to provide nucleophilic thiols and exert antioxidant effects by binding with free radicals in plants [30,31]. Recent studies indicate that the G-site can also bind to other tripeptide analogues, although its affinity and catalytic efficiency for these molecules are generally low [32].
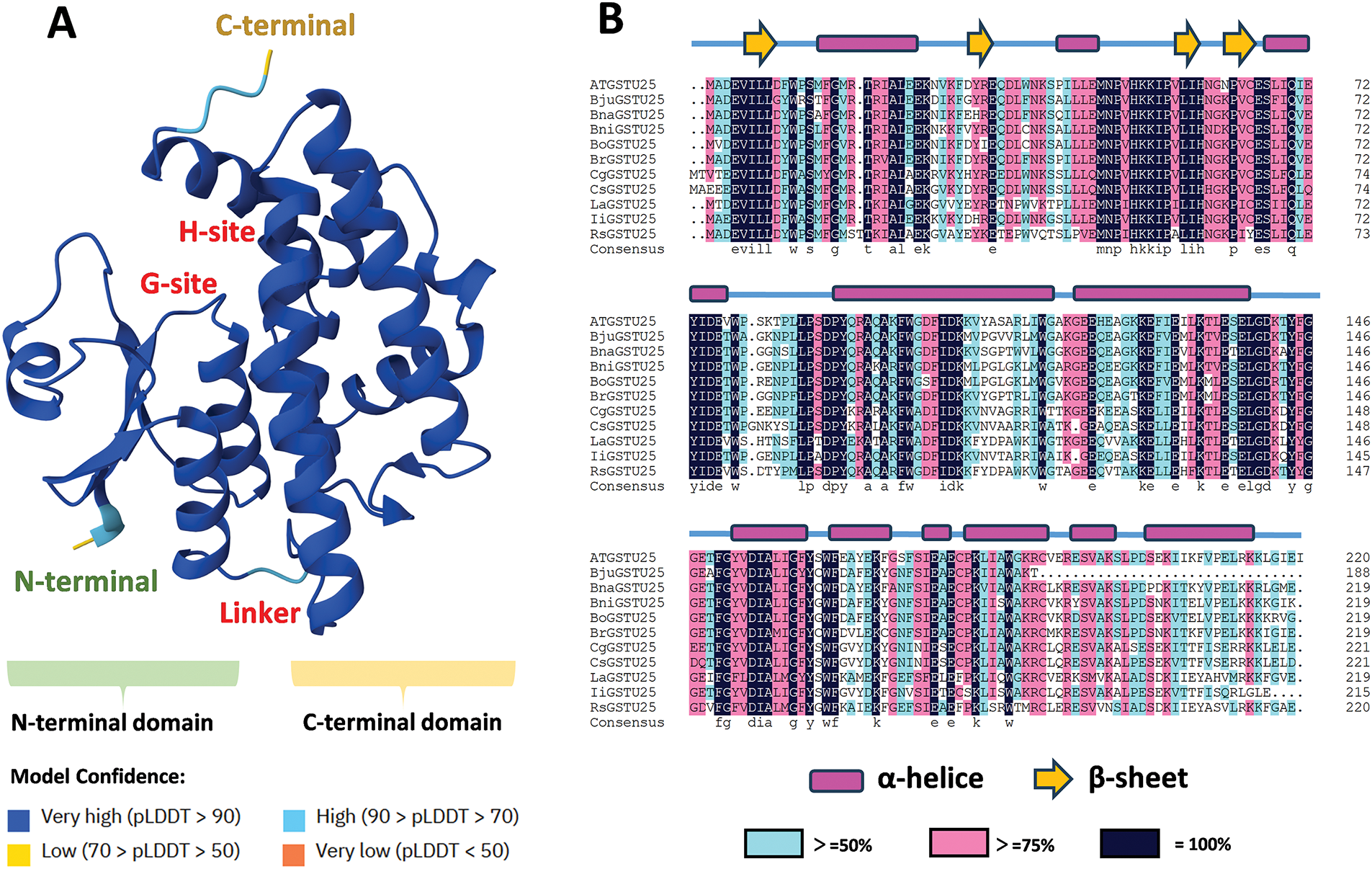
Figure 2: The predicted protein structure of AtGSTU25 and the sequence alignment of GSTU25 in different plant species. (A) The protein structure of AtGSTU25 is predicted with AlphaFold (https://alphafold.ebi.ac.uk/) (accessed on 25 February 2025). It produces a per-residue model confidence score (pLDDT) between 0 and 100. (B) The alignment of GSTU25 amino acid sequences in different plant species was generated by DNAman software. At: Arabidopsis thaliana; Bju: Brassica juncea; Bna: Brassica napus; Bni: Brassica nigra; Bo: Brassica oleracea; Br: Brassica rapa; Cg: Capsella grandiflora; Cs: Camelina sativa; La: Leavenworthia alabamica; Ii: Lsatis indigotica; Rs: Raphanus sativus. The amino acid sequences were obtained from database UniProt (https://www.uniprot.org) (accessed on 25 February 2025)
The CTD sequence exhibits considerable variation, composed of five α-helices, which form the active sites for binding hydrophobic ligands (cosubstrate-binding domain or hydrophobic binding site, H-site). These variations are important for the versatility of GST enzymes in interacting with different ligands [17]. The CTD determines substrate specificity by binding substrates at the H-site [33,34]. The H-site of GSTs generally exhibits significant structural diversity, plasticity and flexibility, enabling it to accommodate a wide range of electrophilic substrates. This versatility is crucial, as it enables GSTs to interact with a variety of compounds, thereby enhancing their role in detoxification and metabolism. A site-directed mutagenesis approach and enzyme kinetics were used to unveil the role of key amino acids at the H-sites during the conjugation of glutathione to hydrophobic compounds catalyzed by GSTs [35].
Most GSTs perform their full biological functions as homodimers or heterodimers, although individual GST subunits retain active sites. The fluidity of the G- and H-sites suggests that significant conformational changes occur in GST subunits upon substrate binding [27]. These structural adaptations are crucial for accommodating various substrates, enhancing the catalytic efficiency and versatility of these GST enzymes.
These subunits facilitate the coupling of toxic substrates with reduced GSH, resulting in the formation of GSH conjugates. The overall negative charge and increased water solubility of these conjugates enhance detoxification processes and facilitate their transfer within biological systems, thereby supporting the efficient removal of exogenous xenobiotics and intracellular oxidized molecules [31,36]. In Arabidopsis, studies have shown that serine residues at certain active sites of GSTs can be replaced by cysteine or non-proton-extracting residues, indicating the potential for different catalytic mechanisms [27,37]. The advancement of X-ray crystallography has facilitated the determination of numerous crystal structures of soluble GST proteins, significantly enhancing our understanding of their functional mechanisms and interactions with substrates.
3 The Biological Functions of GSTs
GSTs are a multifunctional enzyme family that catalyzes the nucleophilic attack of the sulfur atom in glutathione (gamma-glutamyl-cysteinylglycine) on electrophilic group of metabolic products or xenobiotic compounds. This reaction forms conjugates that yield less toxic or non-toxic derivatives, thereby protecting cells from oxidative stress [13,27]. Most GST proteins exhibit both catalase and transferase activities.
3.1 GST Involvement in Plant Response to Stress
The role of GSTs in plant stress responses has garnered considerable attention in recent decades (Fig. 3). In rice, OsGSTU17 knockout mutants exhibit markedly reduced drought tolerance compared to wild-type plants. Furthermore, the expression levels of several drought stress marker genes are significantly lower in these mutants, suggesting that OsGSTU17 positively regulates drought tolerance in rice [38]. Recently, the miR6445-NAC029-GSTU23 module has been identified as a crucial regulator of drought tolerance in poplar by modulating reactive oxygen species (ROS) homeostasis [39]. In wheat (Triticum aestivum), the TaWRKY74 transcription factor interacts with the promoter region of TaGSTU1 to regulate GSH levels, catalyzing the binding of GSH and H2O2, thereby enhancing wheat tolerance to copper stress. This finding is crucial for developing strategies to maintain food security [40]. Overexpression of OsGSTU4 from rice enhances tolerance to salt and oxidative stress in Arabidopsis [41], likely by reducing ROS levels and increasing GSH levels in plants, thereby mitigating cellular damage. Similarly, in tea (Camellia sinensis), the transcription factor CsMYC2.2 regulates the expression of CsGSTU45, leading to H2O2 accumulation in tea plants, which negatively affects resistance to tea anthracnose [42]. In tomato (Solanum lycopersicum), SlGSTU43 enhances salt stress tolerance by regulating lignin biosynthesis through its interactions with SlCOMT2, SlMYB71, and SlWRKY8 [43]. In Medicago sativa, calmodulin-like protein (CML) acts as a Ca2+ sensor that plays a crucial role in plant growth, development, and adaptation to environmental stress. Cold stress induces the activation of Ca2+ channels in alfalfa (Medicago sativa), resulting in elevated intracellular Ca2+ levels. An increase in Ca2+ concentration activates MsCML10, which subsequently triggers the downstream activation of MsGSTU8. Together, MsCML10 and MsGSTU8 regulate the cold tolerance of Medicago by maintaining Ca2+ homeostasis, a crucial factor in stabilizing cellular processes under stress conditions [44]. In wheat, the interaction between TaGSTU6 and the cysteine sulfide β-cystathionine-β-synthase (CBS) domain protein TaCBSX3 enhances resistance to powdery mildew by strengthening the plant’s defense mechanisms, likely through improved detoxification and antioxidant activities [45]. In soybean, two specific candidates, GmGSTU23 and GmGSTU24, exhibited distinct upregulation across all three cell types in response to soybean mosaic virus (SMV) infection. Transient overexpression of GmGSTU23 or GmGSTU24 in tobacco inhibited SMV infection, demonstrating the antiviral function of soybean GSTU proteins [46]. In Arabidopsis, UV-B treatment activates genes encoding the TGA2/5/6 transcription factors, which subsequently upregulate the expression of downstream genes AtGSTU7, AtGSTU8 and AtGSTU25, thereby enhancing UV-B tolerance through improved antioxidant and stress response mechanisms [47]. When trifoliate orange (Poncirus trifoliate) is subjected to cold stress, the PtrERF9 transcription factor activates the expression of the downstream gene PtrGSTU17, thereby enhancing cold tolerance in plants [48]. Recent studies have revealed that CsCIPK11 plays a central role in the cold signaling pathway and modulates antioxidant capacity by phosphorylating CsGSTU23, thereby enhancing cold tolerance in tea plants [49]. GSTs bind with toxic substances through GSH to form complexes, which are subsequently transported to vacuoles for detoxification [50]. Herbicide safeners are compounds that enhance the tolerance of cereal crops to herbicides by upregulating the expression of herbicide-detoxifying enzymes, among which GSTs represent a critical class. In Arabidopsis, the expression of AtGSTU19 is significantly induced by herbicide safeners, thereby improving the plant’s tolerance to herbicides by facilitating the detoxification process through GSH-conjugation [51]. When wheat is exposed to bromobenzyl herbicide, two safeners-cyanobacterial extract and tryptophan, activate the expression of GST genes. Consequently, the increased GST enzyme activity enhances the plant’s tolerance to herbicides by improving the detoxification of harmful herbicide compounds [52–54]. Overexpression of GST1 from maize in tobacco (Nicotiana tabacum) also enhances resistance to herbicides [55]. Similar observations of induced GST gene expression and increased herbicide tolerance have also been reported in weeds and maize, further supporting the role of safeners in enhancing plant resilience to herbicides [56–58]. In poplar (Populus trichocarpa), overexpression of PtGSTF1 positively regulates cell proliferation and salt tolerance [59]. In addition, overexpression of GmGSTL1 from Glycine max in Arabidopsis reduces ROS accumulation, thereby enhancing salt tolerance by alleviating oxidative stress in the plant [60]. In cassava (Manihot esculenta), the transcription factor MeLSD3 forms complexes with the nuclear factor (NF) MeNF-YC15, MeNF-YA2/4, and MeNF-YB18. This regulatory module activates the expression of downstream genes MeGSTU37 and MeGSTU39, which enhance tolerance to oxidative stress by facilitating the detoxification of ROS and improving overall stress response in plants [61]. In walnuts (Juglans regia), the ethylene-responsive factor JrERF2-2 interacts with the WRKY transcription factor JrWRKY7 to regulate GST activity, remarkably improving drought tolerance [62]. Similarly, overexpression of JrGSTU1 further enhances drought tolerance under osmotic stress [63]. In flax (Linum usitatissimum), GST23 and GSTU8 likely play important roles in response to aluminum (Al) stress through the regulation of ROS and modifications to the cell wall, which help mitigate oxidative damage [64]. Moreover, the overexpression of a winter rapeseed GSTF gene in Arabidopsis significantly improves cold tolerance in plants [24].
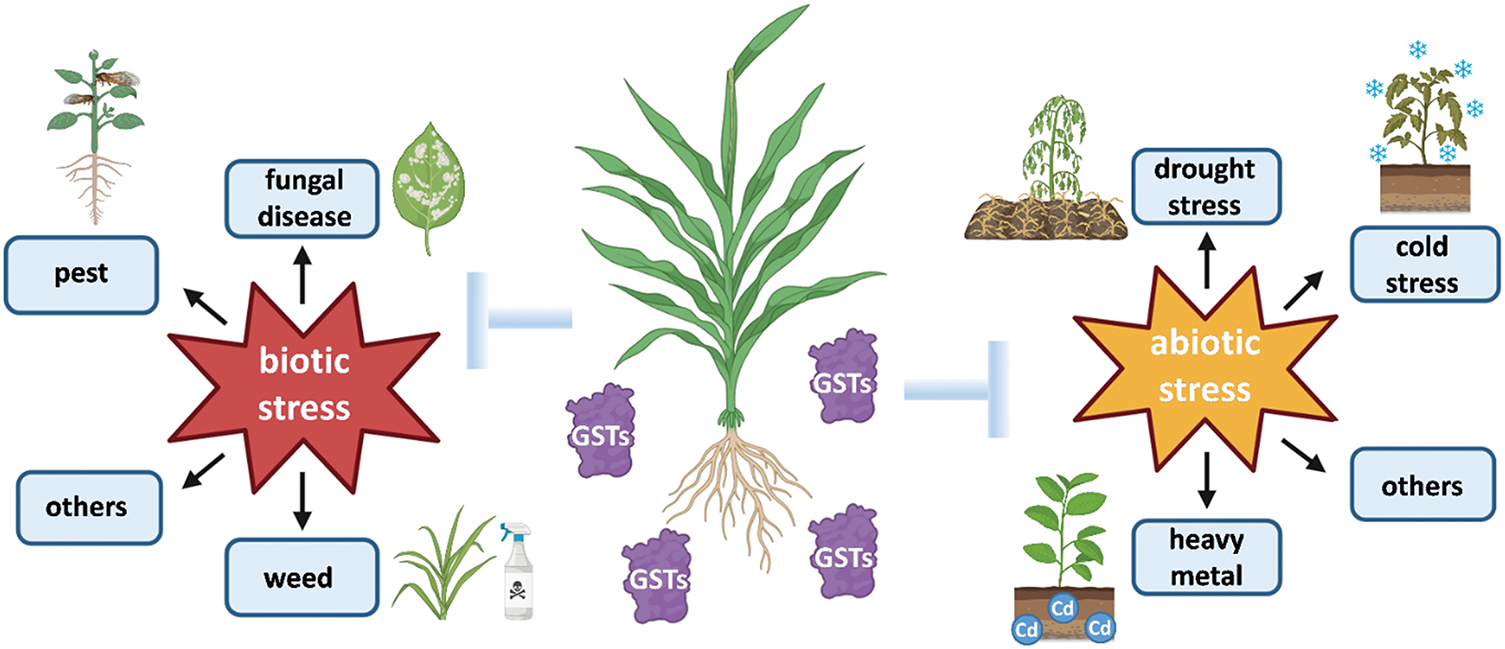
Figure 3: GST involvement in plant stress responses encompasses both biotic and abiotic stresses. Biotic stresses include fungal diseases, pests, weeds, etc., while abiotic stresses include drought, cold, heavy metal, etc. The figure illustrating these responses was originally created, and several components were prepared using the BioRender online software (https://www.biorender.com/) (accessed on 25 February 2025)
3.2 The Roles of GSTs in Catalysis and Transportation
When grapes (Vitis vinifera) are infected with pathogens, the plants release antitoxins such as trans-resveratrol. It has been reported that VvGSTU2 in grapes is involved in transporting trans-resveratrol to the intercellular space, thereby contributing to resistance against pathogen invasion [65]. Similarly, in maize, different ZmGSTs can bind to protoporphyrin IX (PPIX), mesoporphyrin, coproporphyrin, uroporphyrin, and Mg protoporphyrin. However, these GSTs do not form GSH- conjugates, indicating a distinct role in porphyrin metabolism [26]. It has been demonstrated that AtGSTF12 is involved in the transport and distribution of flavonoids [66]. In carnation (Dianthus caryophyllus), the flavonoid 3 gene encodes a GST that functions in the final step of anthocyanin biosynthesis, facilitating the storage of these synthesized anthocyanins within vacuoles, effectively isolating them from the external environment [67]. During this process, a GST-anthocyanin complex or a GST-flavonol complex can be formed [68–70]. In another study, the crucial role of GSTU1 in the transfer of anthocyanin pigments into the vacuole in blood orange was highlighted, with its expression used to discriminate non-pigmented and pigmented fruits [71]. GSTU1 is also considered a potential target for genetic modification to create novel germplasm with enhanced stress tolerance. Additionally, GSTs are involved in catalyzing the transfer of flavonoids. They also catalyze ATP hydrolysis by ABC transporters, supplying the energy necessary for flavonoid transport [69]. Several GSTs bind to the auxin indole-3-acetic acid (IAA) for its intracellular transport [72]. In tobacco and maize, auxin IAA can also induce the gene expression of some GSTs [10]. Similar findings have been observed in Arabidopsis, where the auxin analogue naphthaleneacetic acid (NAA) induces the expression of AtGSTU2 [27].
3.3 The Involvement of GSTs in the Regulation of Signal Transduction
When tobacco leaves are treated with jasmonic acid (JA), the amount of oxidized glutathione (GSSG) and cytoplasmic calcium ions increases. This feedback regulation promotes JA biosynthesis and signal transduction [73,74]. In Arabidopsis, the Far-red (FR) insensitive 219 (FIN219) interacts with FIN219-interacting protein 1 (FIP1), which exhibits GST enzyme activity, to participate in light signaling transduction and regulate plant growth and development [75]. Transcriptome analysis indicates that some signaling genes, including those for JA and abscisic acid (ABA), are activated following treatment with reduced GSH in Arabidopsis, thereby enhancing the plant’s tolerance to drought and salt stress [76]. Several studies suggest an interaction between GSTP1 and nitric oxide (NO), highlighting their potential role in cellular signaling. An elevated concentration of GSTP1 inhibits the activity of c-Jun N-terminal Kinase (JNK) in the cell apoptosis signaling pathway, thereby suppressing cell apoptosis [77].
3.4 The Roles of GSTs in Regulating Plant Growth and Development
Under salt stress conditions, overexpression of the GST gene SbGST from Salicornia Brachiate promotes seed germination and growth in tobacco [78]. The endophytic fungus Piriformospora indica, which colonizes plant roots, enhances root development in Chinese cabbage. During this process, the expression of a BcGSTU gene, highly homologous to AtGSTU24 and AtGSTU25 in Arabidopsis thaliana, is largely induced. Overexpression of this gene in Arabidopsis promotes root and stem development [79,80]. In addition, overexpression of an AtGSTF gene in Arabidopsis stimulates the regeneration of explant stems and leads to earlier flowering. In contrast, double-stranded RNA interference (RNAi) targeting AtGSTF prevents explant stem regeneration under the same cultivation conditions and delays flowering time [25]. Transcriptomic and proteomic analyses suggest that several GmGSTU genes in soybean may play a role in regulating anther development [81].
In recent years, some other functions of GSTs have been identified, including their involvement in anthocyanin accumulation and secondary metabolite biosynthesis [37]. LcGST4 and PcGST57 are involved in anthocyanin accumulation in lychee (Litchi chinensis) and pear (Pyrus communis), respectively [82,83]. In Arabidopsis, the expression of AtGSTU19 facilitates selective binding of the glutathionylated oxophytodienoic acid-GSH conjugate, with the enzyme catalyzing the conjugation reaction, highlighting its role in metabolite stabilization [84]. Furthermore, in barley (Hordeum vulgare), a senescence-induced GST (SIGST) has been linked to leaf senescence. The expression of SIGST was significantly upregulated following treatment with herbicide and low temperature, suggesting its potential involvement in secondary metabolism, antioxidant activity, and enzymatic enhancement during senescence [85]. In wheat, TaGSTU1B and TaGSTF6 are highly expressed in flag leaves during monocarpic senescence, indicating a conserved role in regulating senescence in monocot species [86]. In Arabidopsis, AtGSTU13 contributes to thioglucoside metabolism, thereby playing a role in pathogen defense [87]. In addition, studies have demonstrated that herbicide-treated Arabidopsis plants exhibit increased GST content when exposed to the safener fenclorim within a defined concentration range, reflecting the plant’s adaptive mechanism involving GST-mediated metabolism. Further research indicates that GST enzymes can also activate the metabolism of other safeners, such as glyphosate, thereby enhancing herbicide tolerance [88].
GSTs have emerged as pivotal regulators of plant stress responses, however, significant gaps persist in our understanding of their precise functions. Elucidating the functional diversity of GSTs across different plant species is essential for uncovering their specific roles and the intricate interactions within stress-response pathways. As climate change intensifies environmental stresses, including drought, salinity, and extreme temperatures [89–92], understanding the role of GSTs in stress tolerance has become increasingly critical for developing resilient crop varieties. The following issues represent key areas for further investigation.
4.1 Elucidating the Mechanisms of GST-Mediated Stress Tolerance
GSTs are well described for their essential roles in detoxification, regulation of oxidative stress, and maintenance of cellular redox balance in plants. These enzymes catalyze the conjugation of GSH to a range of toxic compounds, mitigating the effects of detrimental substances and protecting cells from oxidative stress. These detrimental substances, also referred to as xenobiotics, encompass a wide range of compounds, including heavy metals, pesticides and herbicides, pathogen-derived toxins, ROS, etc. [8,31]. These substances pose a significant threat to plant cells as they disrupt cellular functions, damage cellular structures, and interfere with metabolic processes. GSTs mitigate these stresses primarily through their two enzymatic properties. As transferases, GSTs catalyze the conjugation of the reduced tripeptide GSH with the electrophilic centers of various organic molecules [13]. As peroxidases, some GSTs exhibit GSH-dependent glutathione peroxidase (GPOX) activity, enabling the reduction of lipid peroxides to their corresponding alcohols [17,82].
Heavy metals (HM), such as cadmium, lead, and mercury, are highly toxic to plants due to their tendency to accumulate in tissues and disrupt cellular homeostasis [93,94]. These HMs impair enzyme activity, hinder protein synthesis, and induce oxidative stress by generating ROS. GSTs play a vital role in alleviating HM toxicity by catalyzing the conjugation of GSH to these metals. This process forms less reactive metal-GSH complexes, which are subsequently sequestered in vacuoles, reducing cellular damage and preserving metabolic stability [37,95]. For instance, TaGSTU1 in wheat contributes to improved tolerance to copper-induced stress [40].
4.1.2 Pesticides and Herbicides
Synthetic pesticides and herbicides, commonly used in agriculture, can accumulate in plant tissues and cause significant toxicity [96,97]. GSTs contribute to the detoxification of these chemicals by conjugating them with GSH, enhancing their water solubility and reducing their reactivity. The resulting GSH conjugates are either transported out of the cell or stored in vacuoles, minimizing their detrimental effects on cellular processes [17,98]. Overexpression of ZmGST1 in tobacco enhances resistance to herbicides [55].
During pathogen attacks, plants are exposed to various toxic compounds, including phytotoxins, which can disrupt cellular functions [99,100]. GSTs enhance plant defense mechanisms by catalyzing the conjugation of GSH to these toxins, rendering them inactive. This detoxification process prevents damage to plant tissues and strengthens resistance to pathogen infection [4,17]. CsGSTU45 regulate the plant resistance to anthracnose [48].
ROS, such as hydrogen peroxide (H2O2) and superoxide anion (O2.−), are byproducts of cellular metabolism, particularly under stress conditions like drought, high light intensity, or pathogen infection [101,102]. Excessive ROS accumulation can damage lipids, proteins, and DNA, leading to oxidative stress. GSTs, along with other antioxidant enzymes like superoxide dismutase (SOD) and catalase, neutralize ROS-derived metabolites by conjugating them with GSH. This process reduces oxidative damage and helps maintain cellular homeostasis [103,104]. For instance, GmGSTL1 plays a crucial role in soybean by mitigating the accumulation of ROS under saline conditions [60].
Despite their pivotal roles, the precise molecular mechanisms by which GSTs contribute to plant stress tolerance, particularly in the context of stress signaling and cross-talk, remain poorly understood. A key area requiring further exploration is the interaction between GSTs and plant signaling pathways, especially those involving phytohormones such as ABA, JA and salicylic acid (SA). Previous transcriptome analysis has revealed that certain GST genes play a role in the cross-talk between GSH, JA and ABA [76]. These phytohormones play crucial roles in plant responses to environmental stresses such as drought, salinity and pathogen-induced infection [105]. Although GSTs are known to play a key role in stress responses, their involvement in modulating phytohormone signaling remains largely unexplored. It is plausible that GSTs regulate phytohormone activity by balancing their biosynthesis, metabolism, and signal transduction pathways. Therefore, future research should focus on elucidating the dynamic interactions between GSTs and critical signaling molecules, aiming to uncover novel regulatory networks that link detoxification mechanisms to stress response signaling.
4.2 Functional Characterization of GST Isoforms
The increasing availability of genomic and transcriptomic data across a diverse range of plant species has opened new avenues for the identification and functional characterization of GST isoforms. While model plants such as Arabidopsis and rice have been extensively studied, many crop species lack well-characterized mutant lines, which limits the functional analysis of specific genes. The development of advanced techniques, such as CRISPR/Cas9 gene editing and RNA interference (RNAi) [106,107], have enabled the generation of knockout and overexpression lines in various crop species, such as barley, wheat, and oilseed rape, facilitating the exploration of the roles of individual GST isoforms in key physiological processes, including stress tolerance, growth regulation, and development. For example, the GSTU1 isoform in blood orange exhibits significantly higher enzymatic activity against 1-chloro-2, 4-dinitrobenzene (CDNB) as a substrate compared to that in red orange [71]. These tools allow for precise manipulation of GST gene expression, enabling us to dissect the contributions of specific isoforms to plant responses under both abiotic and biotic stress conditions.
The functional characterization of GST isoforms is crucial for understanding their distinct roles in plant stress tolerance. For instance, specific isoforms may predominantly participate in detoxification pathways, whereas others could play a pivotal role in modulating cellular redox homeostasis or hormone signaling [98,108]. In poplar, the GSTU23 isoform contributes to improved drought tolerance by regulating the homeostasis of ROS [39]. Similarly, the overexpression of OsGSTU4 isoform from rice increases tolerance to both salt and oxidative stress in Arabidopsis [41]. By clarifying the functional diversity of GST isoforms, researchers can identify the most promising candidates for integration into crop improvement programs designed to enhance stress resilience and productivity. Incorporating these insights into breeding strategies has the potential to significantly accelerate the development of crops with enhanced stress tolerance, thereby addressing the challenges imposed by climate change and contributing to global food security.
4.3 Applications of Biotechnologies for Crop Improvement
Genetic manipulation represents a promising strategy for enhancing crop performance under both biotic and abiotic stress [109–111]. GSTs play a pivotal role in detoxifying ROS and maintaining cellular redox homeostasis, which are crucial for stress tolerance. Although the upregulation of GST expression has been shown to enhance stress resistance, it is essential to consider the trade-offs between increased stress tolerance and potential effects on growth, yield, and other agronomic traits [112,113]. Overexpression of some GSTs might inadvertently interfere with metabolic pathways or compromise plant growth and development under non-stress conditions.
Future research should focus on optimizing the regulation of GST expression in crops by identifying the most effective GST isoforms for specific stress conditions and employing inducible or tissue-specific promoters to control their expression [114]. It is also possible to fine-tune the regulation of GST genes by using different CRISPR-based technologies [115–117]. Such strategies could enhance stress tolerance while minimizing negative impacts on yield, disease resistance, and plant growth, thereby ensuring a balanced improvement in crop performance.
4.4 Integrating GSTs into Multi-Omics Approaches
A comprehensive understanding of the functions of GSTs requires the integration of multiple omics technologies, including genomics, transcriptomics, proteomics, and metabolomics [118,119]. By incorporating these datasets, researchers can unravel the intricate regulatory networks that govern GST activity, stress responses, and metabolic adaptations in plants [120]. These approaches provide a more in-depth understanding of the interactions between GSTs and other key molecules, such as ROS, phytohormones, and secondary metabolites [121], which play vital roles in plant stress tolerance.
Multi-omics integration enables the identification of important biomarkers linked to GST function under various stress conditions, offering deeper insights into the mechanisms by which GSTs contribute to stress resilience. Recently, single-cell RNA sequencing of soybean has revealed significant transcriptional changes in response to soybean mosaic virus, highlighting the antiviral roles of the genes GmGSTU23 and GmGSTU24 [46]. Furthermore, these integrated datasets can reveal gene-environment interactions that regulate GST expression and activity, providing a more comprehensive understanding of how plants adapt to environmental challenges. Another promising application of these approaches lies in the development of predictive models for plant performance under diverse stress conditions. By integrating data from genomics, transcriptomics, proteomics, and metabolomics, more robust and accurate models can be constructed, facilitating the identification of candidate genes for breeding stress-resistant crops. This approach could lead to more efficient breeding strategies, ultimately accelerating the development of crops with enhanced tolerance to both biotic and abiotic stress.
4.5 Comparisons between Different Plant Species and Evolutionary Insights
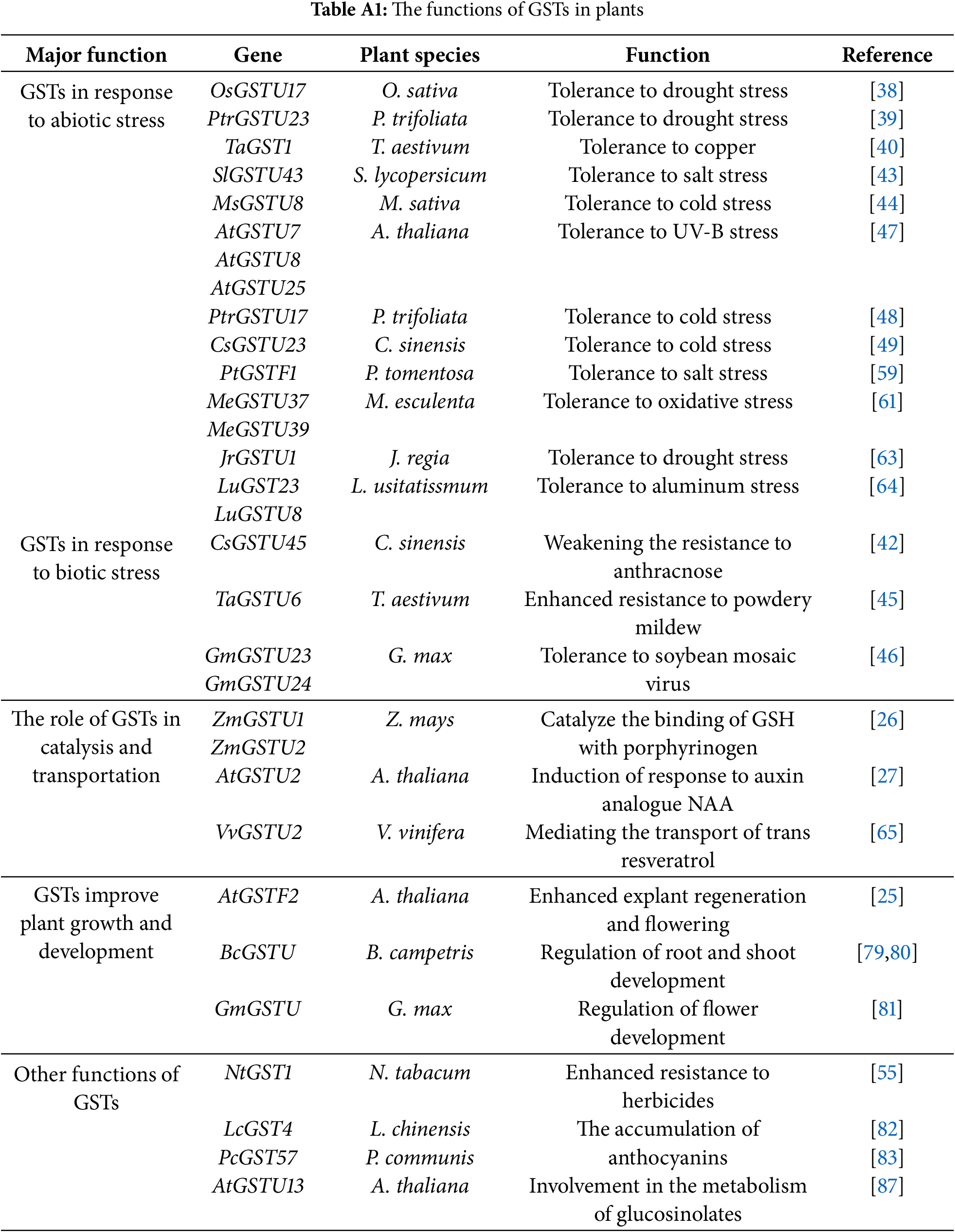
The evolutionary conservation and functional diversity of GSTs across different plant species offer valuable insights into the roles these enzymes play in mitigating environmental stresses. GSTs are highly conserved in plants [122,123] (Fig. 2), yet their functional diversity has evolved to respond to diverse environmental stresses and meet physiological demands [112,124]. Comparative studies between model plants, such as Arabidopsis, and important crops like rice, wheat, and maize, can uncover key variations in GST isoform expression, regulation, and activity (Table A1). PtGSTU23 in poplar is involved in regulating drought tolerance [45], whereas in soybean, GmGSTU23 plays a role in controlling soybean mosaic virus [52]. These investigations shed light on how different plant species have evolved distinct GST functions to adapt to environmental stresses, offering a foundation for developing targeted breeding strategies to enhance crop resilience.
The Identification of specific changes in GST functionality across diverse plant species is likely to yield a deeper understanding of the mechanisms that enable certain crops to thrive under specific environmental conditions. This knowledge could pave the way for the development of tailored crop improvement strategies by integrating the beneficial traits of various GST isoforms. Furthermore, evolutionary analyses of GSTs across species may uncover conserved functional motifs and regulatory networks, offering a broader framework for enhancing stress resilience in crops worldwide. Ultimately, comparative studies of plant species and evolutionary studies of GSTs could enable the development of more precise and effective crop breeding strategies aimed at enhancing stress tolerance and ensuring food security.
In conclusion, the current research on GSTs in plants shows great potential to substantially enhance agricultural productivity and sustainability. A deeper understanding of their molecular mechanisms and roles in crop breeding could lead to the development of plants with enhanced tolerance to environmental stresses, thereby contributing to the maintenance of global food security amidst changing environmental conditions.
Acknowledgement: None.
Funding Statement: This research is funded by National Natural Science Foundation of China (grant no. 32301870 to Chen Lin), Natural Science Foundation of Jiangsu Province (grant no. BK20230568 to Chen Lin), the Jiangsu Provincial Agricultural Science and Technology Independent Innovation Fund (grant no. CX(24)3124 to Chen Lin), Outstanding Ph.D. Program in Yangzhou (grant no. YZLYJFJH2022YXBS147 to Chen Lin), the General Project of Basic Scientific Research to colleges and universities in Jiangsu Province (grant no. 22KJB210019 to Chen Lin), and the Priority Academic Program Development of Jiangsu Higher Education Institutions is greatly acknowledged.
Author Contributions: Chen Lin and Youping Wang conceptualized the reviews, Chen Lin and Zidan Zhang wrote the manuscript with comments from all authors. Zhao Zhang, Yuxiang Long, Xuwen Shen, Jinghao Zhang and Youping Wang revised the manuscript and gave suggestions. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: All data are included in this article.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Appendix A
References
1. Aloke C, Onisuru OO, Achilonu I. Glutathione S-transferase: a versatile and dynamic enzyme. Biochem Biophys Res Commun. 2024;734(80):150774. doi:10.1016/j.bbrc.2024.150774. [Google Scholar] [PubMed] [CrossRef]
2. Shimabukuro RH, Swanson HR, Walsh WC. Glutathione conjugation. Plant Physiol. 1970;46(1):103–7. doi:10.1104/pp.46.1.103. [Google Scholar] [PubMed] [CrossRef]
3. Gullner G, Komives T, Király L, Schröder P. Glutathione S-transferase enzymes in plant-pathogen interactions. Front Plant Sci. 2018;9:1836. doi:10.3389/fpls.2018.01836. [Google Scholar] [PubMed] [CrossRef]
4. Wagner U, Edwards R, Dixon DP, Mauch F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol Biol. 2002;49(5):515–32. doi:10.1023/a:1015557300450. [Google Scholar] [PubMed] [CrossRef]
5. Ann M. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam Horm. 2005;72:155–202. doi:10.1016/S0083-6729(05)72005-7. [Google Scholar] [PubMed] [CrossRef]
6. Sylvestre-Gonon E, Morette L, Viloria M, Mathiot S, Boutilliat A, Favier F, et al. Biochemical and structural insights on the poplar tau glutathione transferase GSTU19 and 20 paralogs binding flavonoids. Front Mol Biosci. 2022;9:958586. doi:10.3389/fmolb.2022.958586. [Google Scholar] [PubMed] [CrossRef]
7. Wei L, Zhu Y, Liu R, Zhang A, Zhu M, Xu W, et al. Genome wide identification and comparative analysis of glutathione transferases (GST) family genes in Brassica napus. Sci Rep. 2019;9(1):9196. doi:10.1038/s41598-019-45744-5. [Google Scholar] [PubMed] [CrossRef]
8. Ding H, Wang B, Han Y, Li S. The pivotal function of dehydroascorbate reductase in glutathione homeostasis in plants. J Exp Bot. 2020;71(12):3405–16. doi:10.1093/jxb/eraa107. [Google Scholar] [PubMed] [CrossRef]
9. Zhuge XL, Xu H, Xiu ZJ, Yang HL. Biochemical functions of glutathione S-transferase family of Salix babylonica. Front Plant Sci. 2020;11:364. doi:10.3389/fpls.2020.00364. [Google Scholar] [PubMed] [CrossRef]
10. Droog FN, Hooykaas PJ, Libbenga KR, van der Zaal EJ. Proteins encoded by an auxin-regulated gene family of tobacco share limited but significant homology with glutathione S-transferases and one member indeed shows in vitro GST activity. Plant Mol Biol. 1993;21(6):965–72. doi:10.1007/BF00023595. [Google Scholar] [PubMed] [CrossRef]
11. Mannervik B. Five decades with glutathione and the GSTome. J Biol Chem. 2012;287(9):6072–83. doi:10.1074/jbc.X112.342675. [Google Scholar] [PubMed] [CrossRef]
12. Enayati AA, Ranson H, Hemingway J. Insect glutathione transferases and insecticide resistance. Insect Mol Biol. 2005;14(1):3–8. doi:10.1111/j.1365-2583.2004.00529.x. [Google Scholar] [PubMed] [CrossRef]
13. Cassier-Chauvat C, Marceau F, Farci S, Ouchane S, Chauvat F. The glutathione system: a journey from cyanobacteria to higher eukaryotes. Antioxidants. 2023;12(6):1199. doi:10.3390/antiox12061199. [Google Scholar] [PubMed] [CrossRef]
14. Di Micco S, Terracciano S, Pierri M, Cantone V, Liening S, König S, et al. Identification of 2,4-dinitro-biphenyl-based compounds as MAPEG inhibitors. ChemMedChem. 2022;17(22):e202200327. doi:10.1002/cmdc.202200327. [Google Scholar] [PubMed] [CrossRef]
15. Manzoor MA, Sabir IA, Shah IH, Riaz MW, Rehman S, Song C, et al. Flavonoids: a review on biosynthesis and transportation mechanism in plants. Funct Integr Genomics. 2023;23(3):212. doi:10.1007/s10142-023-01147-4. [Google Scholar] [PubMed] [CrossRef]
16. Droog F. Plant glutathione S-transferases, a tale of Theta and tau. J Plant Growth Regul. 1997;16(2):95–107. doi:10.1007/PL00006984. [Google Scholar] [CrossRef]
17. Edwards R, Dixon DP, Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000;5(5):193–8. doi:10.1016/s1360-1385(00)01601-0. [Google Scholar] [PubMed] [CrossRef]
18. Jain M, Ghanashyam C, Bhattacharjee A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genomics. 2010;11(1):73. doi:10.1186/1471-2164-11-73. [Google Scholar] [PubMed] [CrossRef]
19. Liu YJ, Han XM, Ren LL, Yang HL, Zeng QY. Functional divergence of the glutathione S-transferase supergene family in Physcomitrella patens reveals complex patterns of large gene family evolution in land plants. Plant Physiol. 2013;161(2):773–86. doi:10.1104/pp.112.205815. [Google Scholar] [PubMed] [CrossRef]
20. Yang Q, Liu YJ, Zeng QY. Biochemical functions of the glutathione transferase supergene family of Larix kaempferi. Plant Physiol Biochem. 2014;77:99–107. doi:10.1016/j.plaphy.2014.02.003. [Google Scholar] [PubMed] [CrossRef]
21. Wang R, Ma J, Zhang Q, Wu C, Zhao H, Wu Y, et al. Genome-wide identification and expression profiling of glutathione transferase gene family under multiple stresses and hormone treatments in wheat (Triticum aestivum L.). BMC Genomics. 2019;20(1):986. doi:10.1186/s12864-019-6374-x. [Google Scholar] [PubMed] [CrossRef]
22. Munyampundu JP, Xu YP, Cai XZ. Phi class of glutathione S-transferase gene superfamily widely exists in nonplant taxonomic groups. Evol Bioinform. 2016;12:59–71. doi:10.4137/EBO.S35909. [Google Scholar] [PubMed] [CrossRef]
23. Han XM, Yang ZL, Liu YJ, Yang HL, Zeng QY. Genome-wide profiling of expression and biochemical functions of the Medicago glutathione S-transferase gene family. Plant Physiol Biochem. 2018;126:126–33. doi:10.1016/j.plaphy.2018.03.004. [Google Scholar] [PubMed] [CrossRef]
24. Niu Z, Liu L, Yue J, Wu J, Wang W, Pu Y, et al. Genome-wide identification of GSTs gene family and functional analysis of BraGSTF2 of winter rapeseed (Brassica rapa L.) under cold stress. Genes. 2023;14(9):1689. doi:10.3390/genes14091689. [Google Scholar] [PubMed] [CrossRef]
25. Gong H, Jiao Y, Hu WW, Pua EC. Expression of glutathione-S-transferase and its role in plant growth and development in vivo and shoot morphogenesis in vitro. Plant Mol Biol. 2005;57(1):53–66. doi:10.1007/s11103-004-4516-1. [Google Scholar] [PubMed] [CrossRef]
26. Lederer B, Böger P. Binding and protection of porphyrins by glutathione S-transferases of Zea mays L. Biochim Biophys Acta. 2003;1621(2):226–33. doi:10.1016/s0304-4165(03)00073-4. [Google Scholar] [PubMed] [CrossRef]
27. Dixon DP, Davis BG, Edwards R. Functional divergence in the glutathione transferase superfamily in plants. Identification of two classes with putative functions in redox homeostasis in Arabidopsis thaliana. J Biol Chem. 2002;277(34):30859–69. doi:10.1074/jbc.M202919200. [Google Scholar] [PubMed] [CrossRef]
28. Dirr H, Reinemer P, Huber R. X-ray crystal structures of cytosolic glutathione S-transferases. Implications for protein architecture, substrate recognition and catalytic function. Eur J Biochem. 1994;220(3):645–61. doi:10.1111/j.1432-1033.1994.tb18666.x. [Google Scholar] [PubMed] [CrossRef]
29. Thom R, Dixon DP, Edwards R, Cole DJ, Lapthorn AJ. The structure of a Zeta class glutathione S-transferase from Arabidopsis thaliana: characterisation of a GST with novel active-site architecture and a putative role in tyrosine catabolism. J Mol Biol. 2001;308(5):949–62. doi:10.1006/jmbi.2001.4638. [Google Scholar] [PubMed] [CrossRef]
30. Sylvestre-Gonon E, Law SR, Schwartz M, Robe K, Keech O, Didierjean C, et al. Functional, structural and biochemical features of plant serinyl-glutathione transferases. Front Plant Sci. 2019;10:608. doi:10.3389/fpls.2019.00608. [Google Scholar] [PubMed] [CrossRef]
31. Zhuge XL, Xie T, Du X, Zhang XX, Hu JP, Yang HL. Non-synonymous substitution of evolutionarily conserved residue in Tau class glutathione transferases alters structural and catalytic features. Int J Biol Macromol. 2022;197(3):39–48. doi:10.1016/j.ijbiomac.2021.12.040. [Google Scholar] [PubMed] [CrossRef]
32. Lallement PA, Meux E, Gualberto JM, Dumarcay S, Favier F, Didierjean C, et al. Glutathionyl-hydroquinone reductases from poplar are plastidial proteins that deglutathionylate both reduced and oxidized glutathionylated quinones. FEBS Lett. 2015;589(1):37–44. doi:10.1016/j.febslet.2014.11.021. [Google Scholar] [PubMed] [CrossRef]
33. Sheehan D, Meade G, Foley VM, Dowd CA. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem J. 2001;360(1):1–16. doi:10.1042/bj3600001. [Google Scholar] [CrossRef]
34. Axarli IA, Rigden DJ, Labrou NE. Characterization of the ligandin site of maize glutathione S-transferase I. Biochem J. 2004;382(3):885–93. doi:10.1042/BJ20040298. [Google Scholar] [PubMed] [CrossRef]
35. Lo Piero AR, Mercurio V, Puglisi I, Petrone G. Different roles of functional residues in the hydrophobic binding site of two sweet orange tau glutathione S-transferases. FEBS J. 2010;277(1):255–62. doi:10.1111/j.1742-4658.2009.07481.x. [Google Scholar] [PubMed] [CrossRef]
36. Hilario E, De Keyser S, Fan L. Structural and biochemical characterization of a glutathione transferase from the Citrus canker pathogen Xanthomonas. Acta Crystallogr D Struct Biol. 2020;76(Pt 8):778–89. doi:10.1107/S2059798320009274. [Google Scholar] [PubMed] [CrossRef]
37. Dixon DP, Skipsey M, Edwards R. Roles for glutathione transferases in plant secondary metabolism. Phytochemistry. 2010;71(4):338–50. doi:10.1016/j.phytochem.2009.12.012. [Google Scholar] [PubMed] [CrossRef]
38. Li J, Meng L, Ren S, Jia C, Liu R, Jiang H, et al. OsGSTU17, a tau class glutathione S-transferase gene, positively regulates drought stress tolerance in Oryza sativa. Plants. 2023;12(17):3166. doi:10.3390/plants12173166. [Google Scholar] [CrossRef]
39. Niu MX, Feng CH, He F, Zhang H, Bao Y, Liu SJ, et al. The miR6445-NAC029 module regulates drought tolerance by regulating the expression of glutathione S-transferase U23 and reactive oxygen species scavenging in Populus. New Phytol. 2024;242(5):2043–58. doi:10.1111/nph.19703. [Google Scholar] [PubMed] [CrossRef]
40. Li GZ, Zheng YX, Chen SJ, Liu J, Wang PF, Wang YH, et al. TaWRKY74 participates copper tolerance through regulation of TaGST1 expression and GSH content in wheat. Ecotoxicol Environ Saf. 2021;221:112469. doi:10.1016/j.ecoenv.2021.112469. [Google Scholar] [PubMed] [CrossRef]
41. Sharma R, Sahoo A, Devendran R, Jain M. Over-expression of a rice tau class glutathione s-transferase gene improves tolerance to salinity and oxidative stresses in Arabidopsis. PLoS One. 2014;9(3):e92900. doi:10.1371/journal.pone.0092900. [Google Scholar] [PubMed] [CrossRef]
42. Lv W, Jiang H, Cao Q, Ren H, Wang X, Wang Y. A tau class glutathione S-transferase in tea plant, CsGSTU45, facilitates tea plant susceptibility to Colletotrichum camelliae infection mediated by jasmonate signaling pathway. Plant J. 2024;117(5):1356–76. doi:10.1111/tpj.16567. [Google Scholar] [PubMed] [CrossRef]
43. Yuan L, Dang J, Zhang J, Wang L, Zheng H, Li G, et al. A glutathione S-transferase regulates lignin biosynthesis and enhances salt tolerance in tomato. Plant Physiol. 2024;196(4):2989–3006. doi:10.1093/plphys/kiae504. [Google Scholar] [PubMed] [CrossRef]
44. Yu S, Wu J, Sun Y, Zhu H, Sun Q, Zhao P, et al. A calmodulin-like protein (CML10) interacts with cytosolic enzymes GSTU8 and FBA6 to regulate cold tolerance. Plant Physiol. 2022;190(2):1321–33. doi:10.1093/plphys/kiac311. [Google Scholar] [PubMed] [CrossRef]
45. Wang Q, Guo J, Jin P, Guo M, Guo J, Cheng P, et al. Glutathione S-transferase interactions enhance wheat resistance to powdery mildew but not wheat stripe rust. Plant Physiol. 2022;190(2):1418–39. doi:10.1093/plphys/kiac326. [Google Scholar] [PubMed] [CrossRef]
46. Song S, Wang J, Zhou J, Cheng X, Hu Y, Wang J, et al. Single-cell RNA-sequencing of soybean reveals transcriptional changes and antiviral functions of GmGSTU23 and GmGSTU24 in response to soybean mosaic virus. Plant Cell Environ. 2024;10:116. doi:10.1111/pce.15164. [Google Scholar] [PubMed] [CrossRef]
47. Herrera-Vásquez A, Fonseca A, Ugalde JM, Lamig L, Seguel A, Moyano TC, et al. TGA class II transcription factors are essential to restrict oxidative stress in response to UV-B stress in Arabidopsis. J Exp Bot. 2021;72(5):1891–905. doi:10.1093/jxb/eraa534. [Google Scholar] [PubMed] [CrossRef]
48. Zhang Y, Ming R, Khan M, Wang Y, Dahro B, Xiao W, et al. ERF9 of Poncirus trifoliata (L.) Raf. undergoes feedback regulation by ethylene and modulates cold tolerance via regulating a glutathione S-transferase U17 gene. Plant Biotechnol J. 2022;20(1):183–200. doi:10.1111/pbi.13705. [Google Scholar] [PubMed] [CrossRef]
49. Di T, Wu Y, Feng X, He M, Lei L, Wang J, et al. CIPK11 phosphorylates GSTU23 to promote cold tolerance in Camellia sinensis. Plant Cell Environ. 2024;47(12):4786–99. doi:10.1111/pce.15070. [Google Scholar] [PubMed] [CrossRef]
50. Délye C. Unravelling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: a major challenge for weed science in the forthcoming decade. Pest Manag Sci. 2013;69(2):176–87. doi:10.1002/ps.3318. [Google Scholar] [PubMed] [CrossRef]
51. DeRidder BP, Dixon DP, Beussman DJ, Edwards R, Goldsbrough PB. Induction of glutathione S-transferases in Arabidopsis by herbicide safeners. Plant Physiol. 2002;130(3):1497–505. doi:10.1104/pp.010066. [Google Scholar] [PubMed] [CrossRef]
52. Gaafar RM, Osman MEH, Abo-Shady AM, Almohisen IAA, Badawy GA, El-Nagar MMF, et al. Role of antioxidant enzymes and glutathione S-transferase in bromoxynil herbicide stress tolerance in wheat plants. Plants. 2022;11(20):2679. doi:10.3390/plants11202679. [Google Scholar] [PubMed] [CrossRef]
53. Taylor VL, Cummins I, Brazier-Hicks M, Edwards R. Protective responses induced by herbicide safeners in wheat. Environ Exp Bot. 2013;88:93–9. doi:10.1016/j.envexpbot.2011.12.030. [Google Scholar] [PubMed] [CrossRef]
54. Scarponi L, Quagliarini E, Del Buono D. Induction of wheat and maize glutathione S-transferase by some herbicide safeners and their effect on enzyme activity against butachlor and terbuthylazine. Pest Manag Sci. 2006;62(10):927–32. doi:10.1002/ps.1258. [Google Scholar] [PubMed] [CrossRef]
55. Karavangeli M, Labrou NE, Clonis YD, Tsaftaris A. Development of transgenic tobacco plants overexpressing maize glutathione S-transferase I for chloroacetanilide herbicides phytoremediation. Biomol Eng. 2005;22(4):121–8. doi:10.1016/j.bioeng.2005.03.001. [Google Scholar] [PubMed] [CrossRef]
56. Hu T, Fang H, Pan Q, Xu H, Lv T, Fan X, et al. Seed microbiome-mediated herbicide resistance evolution in weeds. New Phytol. 2024;242(2):333–43. doi:10.1111/nph.19459. [Google Scholar] [PubMed] [CrossRef]
57. Pang S, Duan L, Liu Z, Song X, Li X, Wang C. Co-induction of a glutathione-S-transferase, a glutathione transporter and an ABC transporter in maize by xenobiotics. PLoS One. 2012;7(7):e40712. doi:10.1371/journal.pone.0040712. [Google Scholar] [PubMed] [CrossRef]
58. Li D, Gao Q, Xu L, Pang S, Liu Z, Wang C, et al. Characterization of glutathione S-transferases in the detoxification of metolachlor in two maize cultivars of differing herbicide tolerance. Pestic Biochem Physiol. 2017;143:265–71. doi:10.1016/j.pestbp.2016.12.003. [Google Scholar] [PubMed] [CrossRef]
59. Gao H, Yu C, Liu R, Li X, Huang H, Wang X, et al. The glutathione S-transferase PtGSTF1 improves biomass production and salt tolerance through regulating xylem cell proliferation, ion homeostasis and reactive oxygen species scavenging in poplar. Int J Mol Sci. 2022;23(19):11288. doi:10.3390/ijms231911288. [Google Scholar] [PubMed] [CrossRef]
60. Chan C, Lam HM. A putative lambda class glutathione S-transferase enhances plant survival under salinity stress. Plant Cell Physiol. 2014;55(3):570–9. doi:10.1093/pcp/pct201. [Google Scholar] [PubMed] [CrossRef]
61. Zeng H, Xu H, Wang H, Chen H, Wang G, Bai Y, et al. LSD3 mediates the oxidative stress response through fine-tuning APX2 activity and the NF-YC15-GSTs module in cassava. Plant J. 2022;110(5):1447–61. doi:10.1111/tpj.15749. [Google Scholar] [PubMed] [CrossRef]
62. Yang G, Peng S, Wang T, Gao X, Li D, Li M, et al. Walnut ethylene response factor JrERF2-2 interact with JrWRKY7 to regulate the GSTs in plant drought tolerance. Ecotoxicol Environ Saf. 2021;228(5):112945. doi:10.1016/j.ecoenv.2021.112945. [Google Scholar] [PubMed] [CrossRef]
63. Yang G, Chen S, Li D, Gao X, Su L, Peng S, et al. Multiple transcriptional regulation of walnut JrGSTTau1 gene in response to osmotic stress. Physiol Plant. 2019;166(3):748–61. doi:10.1111/ppl.12833. [Google Scholar] [PubMed] [CrossRef]
64. Dmitriev AA, Krasnov GS, Rozhmina TA, Kishlyan NV, Zyablitsin AV, Sadritdinova AF, et al. Glutathione S-transferases and UDP-glycosyltransferases are involved in response to aluminum stress in flax. Front Plant Sci. 2016;7:1920. doi:10.3389/fpls.2016.01920. [Google Scholar] [PubMed] [CrossRef]
65. Martínez-Márquez A, Martínez-Esteso MJ, Vilella-Antón MT, Sellés-Marchart S, Morante-Carriel JA, Hurtado E, et al. A tau class glutathione-S-transferase is involved in Trans-resveratrol transport out of grapevine cells. Front Plant Sci. 2017;8:1457. doi:10.3389/fpls.2017.01457. [Google Scholar] [PubMed] [CrossRef]
66. Sylvestre-Gonon E, Schwartz M, Girardet JM, Hecker A, Rouhier N. Is there a role for tau glutathione transferases in tetrapyrrole metabolism and retrograde signalling in plants? Philos Trans R Soc Lond B. Biol Sci. 2020;375(1801):20190404. doi:10.1098/rstb.2019.0404. [Google Scholar] [PubMed] [CrossRef]
67. Larsen ES, Alfenito MR, Briggs WR, Walbot V. A carnation anthocyanin mutant is complemented by the glutathione S-transferases encoded by maize Bz2 and Petunia An9. Plant Cell Rep. 2003;21(9):900–4. doi:10.1007/s00299-002-0545-x. [Google Scholar] [PubMed] [CrossRef]
68. Li X, Gao P, Cui D, Wu L, Parkin I, Saberianfar R, et al. The Arabidopsis tt19-4 mutant differentially accumulates proanthocyanidin and anthocyanin through a 3’ amino acid substitution in glutathione S-transferase. Plant Cell Environ. 2011;34(3):374–88. doi:10.1111/j.1365-3040.2010.02249.x. [Google Scholar] [PubMed] [CrossRef]
69. Zhao J, Dixon RA. The ‘ins’ and ‘outs’ of flavonoid transport. Trends Plant Sci. 2010;15(2):72–80. doi:10.1016/j.tplants.2009.11.006. [Google Scholar] [PubMed] [CrossRef]
70. Petrussa E, Braidot E, Zancani M, Peresson C, Bertolini A, Patui S, et al. Plant flavonoids—biosynthesis, transport and involvement in stress responses. Int J Mol Sci. 2013;14(7):14950–73. doi:10.3390/ijms140714950. [Google Scholar] [PubMed] [CrossRef]
71. Lo Piero AR, Mercurio V, Puglisi I, Petrone G. Gene isolation and expression analysis of two distinct sweet orange [Citrus sinensis L. (Osbeck)] tau-type glutathione transferases. Gene. 2009;443(1–2):143–50. doi:10.1016/j.gene.2009.04.025. [Google Scholar] [PubMed] [CrossRef]
72. Bilang J, Sturm A. Cloning and characterization of a glutathione S-transferase that can be photolabeled with 5-azido-indole-3-acetic acid. Plant Physiol. 1995;109(1):253–60. doi:10.1104/pp.109.1.253. [Google Scholar] [PubMed] [CrossRef]
73. Wasternack C, Hause B. Jasmonates and octadecanoids: signals in plant stress responses and development. Prog Nucleic Acid Res Mol Biol. 2002;72:165–221. doi:10.1016/s0079-6603(02)72070-9. [Google Scholar] [PubMed] [CrossRef]
74. Aslam S, Gul N, Mir MA, Asgher M, Al-Sulami N, Abulfaraj AA, et al. Role of jasmonates, calcium, and glutathione in plants to combat abiotic stresses through precise signaling cascade. Front Plant Sci. 2021;12:668029. doi:10.3389/fpls.2021.668029. [Google Scholar] [PubMed] [CrossRef]
75. Chen IC, Huang IC, Liu MJ, Wang ZG, Chung SS, Hsieh HL. Glutathione S-transferase interacting with far-red insensitive 219 is involved in phytochrome A-mediated signaling in Arabidopsis. Plant Physiol. 2007;143(3):1189–202. doi:10.1104/pp.106.094185. [Google Scholar] [PubMed] [CrossRef]
76. Cheng MC, Ko K, Chang WL, Kuo WC, Chen GH, Lin TP. Increased glutathione contributes to stress tolerance and global translational changes in Arabidopsis. Plant J. 2015;83(5):926–39. doi:10.1111/tpj.12940. [Google Scholar] [PubMed] [CrossRef]
77. Russell TM, Richardson DR. The good Samaritan glutathione-S-transferase P1: an evolving relationship in nitric oxide metabolism mediated by the direct interactions between multiple effector molecules. Redox Biol. 2023;59:102568. doi:10.1016/j.redox.2022.102568. [Google Scholar] [PubMed] [CrossRef]
78. Jha B, Sharma A, Mishra A. Expression of SbGSTU (tau class glutathione S-transferase) gene isolated from Salicornia brachiata in tobacco for salt tolerance. Mol Biol Rep. 2011;38(7):4823–32. doi:10.1007/s11033-010-0625-x. [Google Scholar] [PubMed] [CrossRef]
79. Kao CW, Bakshi M, Sherameti I, Dong S, Reichelt M, Oelmüller R, et al. A Chinese cabbage (Brassica campetris subsp. Chinensis) τ-type glutathione-S-transferase stimulates Arabidopsis development and primes against abiotic and biotic stress. Plant Mol Biol. 2016;92(6):643–59. doi:10.1007/s11103-016-0531-2. [Google Scholar] [PubMed] [CrossRef]
80. Lee YC, Johnson JM, Chien CT, Sun C, Cai D, Lou B, et al. Growth promotion of Chinese cabbage and Arabidopsis by Piriformospora indica is not stimulated by Mycelium-synthesized auxin. Mol Plant Microbe Interact. 2011;24(4):421–31. doi:10.1094/MPMI-05-10-0110. [Google Scholar] [PubMed] [CrossRef]
81. Li J, Chen L, Zhi X, Wang J, Lu Y, Tian Z, et al. Integrated transcriptome and proteome analysis reveals molecular responses of soybean anther under high-temperature stress. Front Plant Sci. 2023;14:1187922. doi:10.3389/fpls.2023.1187922. [Google Scholar] [PubMed] [CrossRef]
82. Hu B, Zhao J, Lai B, Qin Y, Wang H, Hu G. LcGST4 is an anthocyanin-related glutathione S-transferase gene in Litchi chinensis Sonn. Plant Cell Rep. 2016;35(4):831–43. doi:10.1007/s00299-015-1924-4. [Google Scholar] [PubMed] [CrossRef]
83. Li B, Zhang X, Duan R, Han C, Yang J, Wang L, et al. Genomic analysis of the glutathione S-transferase family in pear (Pyrus communis) and functional identification of PcGST57 in anthocyanin accumulation. Int J Mol Sci. 2022;23(2):746. doi:10.3390/ijms23020746. [Google Scholar] [PubMed] [CrossRef]
84. Dixon DP, Edwards R. Selective binding of glutathione conjugates of fatty acid derivatives by plant glutathione transferases. J Biol Chem. 2009;284(32):21249–56. doi:10.1074/jbc.M109.020107. [Google Scholar] [PubMed] [CrossRef]
85. Kunieda T, Fujiwara T, Amano T, Shioi Y. Molecular cloning and characterization of a senescence-induced tau-class Glutathione S-transferase from barley leaves. Plant Cell Physiol. 2005;46(9):1540–8. doi:10.1093/pcp/pci167. [Google Scholar] [PubMed] [CrossRef]
86. Gallé A, Csiszár J, Secenji M, Guóth A, Cseuz L, Tari I, et al. Glutathione transferase activity and expression patterns during grain filling in flag leaves of wheat genotypes differing in drought tolerance: response to water deficit. J Plant Physiol. 2009;166(17):1878–91. doi:10.1016/j.jplph.2009.05.016. [Google Scholar] [PubMed] [CrossRef]
87. Piślewska-Bednarek M, Nakano RT, Hiruma K, Pastorczyk M, Sanchez-Vallet A, Singkaravanit-Ogawa S, et al. Glutathione transferase U13 functions in pathogen-triggered glucosinolate metabolism. Plant Physiol. 2018;176(1):538–51. doi:10.1104/pp.17.01455. [Google Scholar] [PubMed] [CrossRef]
88. Brazier-Hicks M, Evans KM, Cunningham OD, Hodgson DRW, Steel PG, Edwards R. Catabolism of glutathione conjugates in Arabidopsis thaliana. J Biol Chem. 2008;283(30):21102–12. doi:10.1074/jbc.m801998200. [Google Scholar] [PubMed] [CrossRef]
89. Campos EVR, Pereira ADES, Aleksieienko I, do Carmo GC, Gohari G, Santaella C, et al. Encapsulated plant growth regulators and associative microorganisms: nature-based solutions to mitigate the effects of climate change on plants. Plant Sci. 2023;331:111688. doi:10.1016/j.plantsci.2023.111688. [Google Scholar] [PubMed] [CrossRef]
90. Lozano-Elena F, Fàbregas N, Coleto-Alcudia V, Caño-Delgado AI. Analysis of metabolic dynamics during drought stress in Arabidopsis plants. Sci Data. 2022;9(1):90. doi:10.1038/s41597-022-01161-4. [Google Scholar] [PubMed] [CrossRef]
91. Andrew SC, Simonsen AK, Coppin CW, Arnold PA, Briceño VF, McLay TGB, et al. Expression-environment associations in transcriptomic heat stress responses for a global plant lineage. Mol Ecol. 2024;33(16):e17473. doi:10.1111/mec.17473. [Google Scholar] [PubMed] [CrossRef]
92. Horváth E, Kulman K, Tompa B, Hajnal ÁB, Pelsőczi A, Bela K, et al. Glutathione transferases are involved in the genotype-specific salt-stress response of tomato plants. Antioxidants. 2023;12(9):1682. doi:10.3390/antiox12091682. [Google Scholar] [PubMed] [CrossRef]
93. Riyazuddin R, Nisha N, Ejaz B, Khan IR, Kumar M, Ramteke M, et al. A comprehensive review on the heavy metal toxicity and sequestration in plants. Biomolecules. 2021;12(1):43. doi:10.3390/biom12010043. [Google Scholar] [PubMed] [CrossRef]
94. Goncharuk EA, Zagoskina NV. Heavy metals, their phytotoxicity, and the role of phenolic antioxidants in plant stress responses with focus on cadmium: review. Molecules. 2023;28(9):3921. doi:10.3390/molecules28093921. [Google Scholar] [PubMed] [CrossRef]
95. AbdElgawad H, Zinta G, Hamed BA, Selim S, Beemster G, Hozzein WN, et al. Maize roots and shoots show distinct profiles of oxidative stress and antioxidant defense under heavy metal toxicity. Environ Pollut. 2020;258:113705. doi:10.1016/j.envpol.2019.113705. [Google Scholar] [PubMed] [CrossRef]
96. Johnson N, Zhang G, Soble A, Johnson S, Baucom RS. The consequences of synthetic auxin herbicide on plant-herbivore interactions. Trends Plant Sci. 2023;28(7):765–75. doi:10.1016/j.tplants.2023.02.003. [Google Scholar] [PubMed] [CrossRef]
97. Sen MK, Hamouzová K, Košnarová P, Soukup J. H2O2-mediated signaling in plant stress caused by herbicides: its role in metabolism and degradation pathways. Plant Sci. 2024;346(2):112166. doi:10.1016/j.plantsci.2024.112166. [Google Scholar] [PubMed] [CrossRef]
98. Ito T, Ohkama-Ohtsu N. Degradation of glutathione and glutathione conjugates in plants. J Exp Bot. 2023;74(11):3313–27. doi:10.1093/jxb/erad018. [Google Scholar] [PubMed] [CrossRef]
99. Pirc K, Albert I, Nürnberger T, Anderluh G. Disruption of plant plasma membrane by Nep1-like proteins in pathogen-plant interactions. New Phytol. 2023;237(3):746–50. doi:10.1111/nph.18524. [Google Scholar] [PubMed] [CrossRef]
100. Nobori T, Ecker JR. Yet uninfected? Resolving cell states of plants under pathogen attack. Cell Rep Methods. 2023;3(7):100538. doi:10.1016/j.crmeth.2023.100538. [Google Scholar] [PubMed] [CrossRef]
101. Berrios L, Rentsch JD. Linking reactive oxygen species (ROS) to abiotic and biotic feedbacks in plant microbiomes: the dose makes the poison. Int J Mol Sci. 2022;23(8):4402. doi:10.3390/ijms23084402. [Google Scholar] [PubMed] [CrossRef]
102. Fedoreyeva LI. ROS as signaling molecules to initiate the process of plant acclimatization to abiotic stress. Int J Mol Sci. 2024;25(21):11820. doi:10.3390/ijms252111820. [Google Scholar] [PubMed] [CrossRef]
103. Wang P, Liu WC, Han C, Wang S, Bai MY, Song CP. Reactive oxygen species: multidimensional regulators of plant adaptation to abiotic stress and development. J Integr Plant Biol. 2024;66(3):330–67. doi:10.1111/jipb.13601. [Google Scholar] [PubMed] [CrossRef]
104. Mittler R, Zandalinas SI, Fichman Y, Van Breusegem F. Reactive oxygen species signalling in plant stress responses. Nat Rev Mol Cell Biol. 2022;23(10):663–79. doi:10.1038/s41580-022-00499-2. [Google Scholar] [PubMed] [CrossRef]
105. Shaffique S, Hussain S, Kang SM, Imran M, Injamum-Ul-Hoque M, Khan MA, et al. Phytohormonal modulation of the drought stress in soybean: outlook, research progress, and cross-talk. Front Plant Sci. 2023;14:1237295. doi:10.3389/fpls.2023.1237295. [Google Scholar] [PubMed] [CrossRef]
106. Chung SH, Feng H, Jander G. Engineering pest tolerance through plant-mediated RNA interference. Curr Opin Plant Biol. 2021;60:102029. doi:10.1016/j.pbi.2021.102029. [Google Scholar] [PubMed] [CrossRef]
107. Liu G, Lin Q, Jin S, Gao C. The CRISPR-Cas toolbox and gene editing technologies. Mol Cell. 2022;82(2):333–47. doi:10.1016/j.molcel.2021.12.002. [Google Scholar] [PubMed] [CrossRef]
108. Labrou NE, Papageorgiou AC, Pavli O, Flemetakis E. Plant GSTome: structure and functional role in xenome network and plant stress response. Curr Opin Biotechnol. 2015;32:186–94. doi:10.1016/j.copbio.2014.12.024. [Google Scholar] [PubMed] [CrossRef]
109. Nguyen HN, Lai N, Kisiala AB, Emery RJN. Isopentenyltransferases as master regulators of crop performance: their function, manipulation, and genetic potential for stress adaptation and yield improvement. Plant Biotechnol J. 2021;19(7):1297–313. doi:10.1111/pbi.13603. [Google Scholar] [PubMed] [CrossRef]
110. Mou R, Niu R, Yang R, Xu G. Engineering crop performance with upstream open reading frames. Trends Plant Sci. 2024;30(3):311–23. doi:10.1016/j.tplants.2024.10.005. [Google Scholar] [PubMed] [CrossRef]
111. Akter S, Castaneda-Méndez O, Beltrán J. Synthetic reprogramming of plant developmental and biochemical pathways. Curr Opin Biotechnol. 2024;87(1215):103139. doi:10.1016/j.copbio.2024.103139. [Google Scholar] [PubMed] [CrossRef]
112. He Z, Webster S, He SY. Growth-defense trade-offs in plants. Curr Biol. 2022;32(12):R634–9. doi:10.1016/j.cub.2022.04.070. [Google Scholar] [PubMed] [CrossRef]
113. Sparks EE, Rasmussen A. Trade-offs in plant responses to the environment. Plant Cell Environ. 2023;46(10):2943–5. doi:10.1111/pce.14689. [Google Scholar] [PubMed] [CrossRef]
114. Xun H, Zhang X, Yu J, Pang J, Wang S, Liu B, et al. Analysis of expression characteristics of soybean leaf and root tissue-specific promoters in Arabidopsis and soybean. Transgenic Res. 2021;30(6):799–810. doi:10.1007/s11248-021-00266-7. [Google Scholar] [PubMed] [CrossRef]
115. Xing S, Chen K, Zhu H, Zhang R, Zhang H, Li B, et al. Fine-tuning sugar content in strawberry. Genome Biol. 2020;21(1):230. doi:10.1186/s13059-020-02146-5. [Google Scholar] [PubMed] [CrossRef]
116. Capdeville N, Merker L, Schindele P, Puchta H. Sophisticated CRISPR/Cas tools for fine-tuning plant performance. J Plant Physiol. 2021;257(6299):153332. doi:10.1016/j.jplph.2020.153332. [Google Scholar] [PubMed] [CrossRef]
117. Pacesa M, Pelea O, Jinek M. Past, present, and future of CRISPR genome editing technologies. Cell. 2024;187(5):1076–100. doi:10.1016/j.cell.2024.01.042. [Google Scholar] [PubMed] [CrossRef]
118. Shen S, Zhan C, Yang C, Fernie AR, Luo J. Metabolomics-centered mining of plant metabolic diversity and function: past decade and future perspectives. Mol Plant. 2023;16(1):43–63. doi:10.1016/j.molp.2022.09.007. [Google Scholar] [PubMed] [CrossRef]
119. Raza A, Salehi H, Bashir S, Tabassum J, Jamla M, Charagh S, et al. Transcriptomics, proteomics, and metabolomics interventions prompt crop improvement against metal(loid) toxicity. Plant Cell Rep. 2024;43(3):80. doi:10.1007/s00299-024-03153-7. [Google Scholar] [PubMed] [CrossRef]
120. Bisht A, Saini DK, Kaur B, Batra R, Kaur S, Kaur I, et al. Multi-omics assisted breeding for biotic stress resistance in soybean. Mol Biol Rep. 2023;50(4):3787–814. doi:10.1007/s11033-023-08260-4. [Google Scholar] [PubMed] [CrossRef]
121. Jogawat A, Yadav B, Chhaya, Lakra N, Singh AK, Narayan OP. Crosstalk between phytohormones and secondary metabolites in the drought stress tolerance of crop plants: a review. Physiol Plant. 2021;172(2):1106–32. doi:10.1111/ppl.13328. [Google Scholar] [PubMed] [CrossRef]
122. Duan Q, Li GR, Qu YP, Yin DX, Zhang CL, Chen YS. Genome-wide identification, evolution and expression analysis of the glutathione S-transferase supergene family in Euphorbiaceae. Front Plant Sci. 2022;13:808279. doi:10.3389/fpls.2022.808279. [Google Scholar] [PubMed] [CrossRef]
123. Vaish S, Gupta D, Mehrotra R, Mehrotra S, Basantani MK. Glutathione S-transferase: a versatile protein family. 3 Biotech. 2020;10(7):321. doi:10.1007/s13205-020-02312-3. [Google Scholar] [PubMed] [CrossRef]
124. Micic N, Rønager AH, Sørensen M, Bjarnholt N. Overlooked and misunderstood: can glutathione conjugates be clues to understanding plant glutathione transferases? Philos Trans R Soc Lond B. Biol Sci. 2024;379(1914):20230365. doi:10.1098/rstb.2023.0365. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools