 Open Access
Open Access
ARTICLE
Identification of PBL Gene Family in Tree Peonies and Its Function in Regulating Pollen Tube Growth
1 College of Agriculture, Henan University of Science and Technology, Luoyang, 471000, China
* Corresponding Authors: Lili Guo. Email: ; Xiaogai Hou. Email:
(This article belongs to the Special Issue: Ornamental Plants: Traits, Flowering, Aroma, Molecular Mechanisms, Postharvest Handling, and Application)
Phyton-International Journal of Experimental Botany 2025, 94(4), 1159-1176. https://doi.org/10.32604/phyton.2025.063737
Received 22 January 2025; Accepted 14 March 2025; Issue published 30 April 2025
Abstract
Receptor-like cytoplasmic kinases (RLCKs) play a crucial role in the physiological processes of plant growth and development and stress response. To elucidate the characteristics and functions of the PBL gene family in tree peonies, the whole genome identification of PBL family members in tree peonies was conducted using a bioinformatics approach based on the published Arabidopsis thaliana PBL protein sequence. A total of 51 PoPBL members were identified, which were distributed unevenly on five chromosomes in the tree peony. PoPBL proteins were localized in the nucleus, cytoplasm, chloroplasts, and mitochondria, with most members of the same clade exhibiting similar gene structures and conserved motif components. The promoter region of PoPBLs contained many response elements related to plant hormones and stress. The relative expression levels of PoPBL1, PoPBL4, PoPBL14, PoPBL40 and PoPBL45 were highly expressed in the pistil samples from the first hour after pollination, and all of them contained elements related to growth and development. At the same time, PoPBL40 of 2044 bp was obtained, and it was found that it had a positive regulatory effect on pollen tube growth of tree peonies, which laid a foundation for further study of PBL gene function in tree peonies.Keywords
Supplementary Material
Supplementary Material FileTree peony (Paeonia section Moutan DC.) is a member of Paeoniaceae family and Paeonia genus and is classified as a perennial deciduous subshrub. It is an endemic species in China [1]. It is mainly distributed in the central, southwestern to northwestern parts of China [2,3]. Because of its large flower size and colorful flowers, it is known as “the king of flowers”. Tree peony flowers are characterized by their large size, aesthetic appeal, vibrant colors, and pleasant fragrance. It has been loved and praised by people throughout the ages. Due to long-term natural and artificial selection, germplasm resources are very rich. Cultivated peonies in China mainly include the Central Plains cultivar group, the Northwest variety group, the Southwest variety group, the Jiangnan variety group, and the foreign variety group. In recent years, there has been a growing demand for tree peonies in various markets, including garden ornamentals, indoor floral arrangements, and medicinal applications. This trend is largely driven by the advancement and development of global economic integration [4]. The substantial economic benefits associated with the tree peony industry have stimulated the expansion of tree peony cultivation regions and areas. This expansion has increased the visibility of tree peonies within the aesthetic purview of both the emperor and the common populace, thereby promoting the rapid development of tree peony culture. Consequently, tree peony culture has emerged as a dominant force in the realm of ornamental culture [5,6].
The plant receptor-like kinases (RLKs) identified to date are related to a variety of biological processes, including development, self-incompatibility, response to pathogens, and various environmental stresses. In the RLK superfamily, there exists a class of protein kinases that lack extracellular domains and transmembrane domains, which are called receptor-like cytoplasmic kinases (RLCKs) [7]. These kinases play crucial roles in plant growth and pathogen defense [8,9]. Their structure primarily comprises an extracellular domain for the perception of extracellular signals, a transmembrane domain, and an intracellular kinase domain for signal transduction [10]. The RLCKs family contains 379 and 147 members in rice (Oryza sativa) and Arabidopsis thaliana, respectively [9]; based on sequence homology, Arabidopsis thaliana and Oryza sativa RLCKs have been categorized into 17 subgroups, namely RLCK-II and RLCK-IV to RLCK-XIX. Most RLCKs contain a single Ser/Thr kinase structural domain, while others encompass LRR, EGF, WD40, or transmembrane structural domains [11]. RLCK family genes were found to be involved in pathogen stress response in Solanum lycopersicum [12], Capsicum annuum [12], Oryza sativa [13], and Triticum aestivum [14]. Furthermore, RLCKs and RKS/RLPS act in concert to regulate plant sexual reproduction in response to abiotic stress, hormone signaling, maintenance of stem and root meristematic tissues, vascular tissue differentiation, petal abscission, and other developmental processes [15–18]. Currently, the RLCK-VII subgroup in Arabidopsis thaliana comprises 46 members, the majority of which play significant roles in plant growth and development and in response to hormonal signaling. RPM1-induced protein kinase (RIPK) acts downstream of cell membrane surface receptor FERONIA (FER) to regulate initial rooting and root hair growth [19]. MARIS (MRI) regulates pollen tube integrity by acting on two closely related polypeptide hormone receptors ANX1 (ANXUR1), ANX2 and buddha’s paper seal 1 (BUPS1) and BUPS2 downstream [20–22]. Botrytis-induced kinase 1 (BIK1) and its related RLCK member PBS1-likes (PBLs) interact with several cell membrane surface receptor kinases and participate in the regulation of immune response [15,23]. BR signaling kinases (BSKs) interact with brassinosteroid insensitive1 (BRI1) to regulate brassinosteroid signaling [24,25]. Concurrently, BSK1 interacts with the immune receptor flagellin sensing 2 (FLS2) to regulate immune signals activated by the pathogen-associated molecular pattern flg22 [26]. In addition, RIPK participated in the immune reaction by directly phosphorylating NADPH oxidase respiratory burst homolog D (RBOHD) to regulate the production of reactive oxygen species [27].
PBL (PBS1-LIKE) is a member of the RLCK-VII subfamily and exhibits high homology to PBS1. It engages in similar immune pathways due to the structural similarities of its kinase domain. In signal transduction, certain RLCKs are capable of directly phosphorylating PBL. For instance, during the immune response, PBL1 undergoes phosphorylation by RLCKs, which subsequently activates downstream defense mechanisms [28]. In Arabidopsis, mutations in pbs1 or pbl mutants within the RLCK-VII subfamily, as well as other RLCK members such as bik1, can result in immunodeficiency. This observation suggests functional complementation or overlap among these proteins [29]. RLCKs and PBL constitute a dynamic regulatory network that plays a crucial role in plant immunity. The relationship between them in terms of structural homology, functional synergy, and genetic interaction reveals the complexity and efficiency of plant immune signaling pathways.
At present, comprehensive whole-genome identification and expression analysis of the PBL gene family in tree peonies have not been reported. To investigate the characteristics and functions of the PBL gene in tree peonies, this study identified the members of the PBL gene family in the pistil of Paeonia ostii ‘Fengdan’, and conducted bioinformatics analysis on the members of the PoPBL genes. The analysis encompassed predictions of subcellular localization, gene structure, cis-regulatory elements, collinearity, and gene expression patterns, among other aspects.
The genome sequences, genome annotation files, and amino acid sequences utilized in the experiment were retrieved from the following websites: the Tree Peony Database CNSA (https://ftp.cngb.org/) (accessed on 13 March 2025) Arabidopsis thaliana TAIR website (www.arabidopsis.org) (accessed on 13 March 2025).
2.2.1 Identification and Physicochemical Properties of PBL Gene Family Members in Tree Peony
The genome data of tree peony were retrieved from the tree peony genome website CNSA (https://ftp.cngb.org/) (accessed on 13 March 2025), and 46 Arabidopsis thaliana PBL genes were obtained from the TAIR website (www.arabidopsis.org) (accessed on 13 March 2025) [22], and these genes were used as the Maker sequence. The members of PBL family of tree peonies were identified by BLAST of Tree peony gene. Download the Hidden Markov Model profile (HMM) for the PBL binding domain (PF00069) from the Pfam database (http://pfam.xfam.org/) (accessed on 13 March 2025). The Two Sequence Files in TBtools (v2.136) [30] and Simple HMM Search were employed to search for candidate PoPBL genes in the tree peony genome using PF00069 as the model. Redundant sequences were manually removed. The genes obtained by these two methods were screened using SMART (http://smart.embl-heidelberg.de/) (accessed on 13 March 2025) and Pfam CDD (http://pfam.xfam.org/) (accessed on 13 March 2025) and database (https://www.ncbi.nlm.nihgov/cdd) (accessed on 13 March 2025) for cross-validation candidate sequences in the existence of the PBL structure domain. The ExPAsy online tool (https://web.expasy.org/protparam/) (accessed on 13 March 2025) was used to analyze the physical and chemical properties of the potential PBL family members in tree peony, including molecular weight, isoelectric point, amino acid composition, atomic composition, atomic composition, and aliphatic index (relative magnitude of aliphatic amino acids). The subcellular localization of tree peony PBL protein family members was predicted using the Cell-PLoc 2.0 online tool (http://www.csbio.sjtu.edu.cn/bioinf/Cell-PLoc-2/) (accessed on 13 March 2025).
2.2.2 Phylogenetic Tree, Gene Structure and Conserved Motif Analysis of Tree Peony PBL Genes
Arabidopsis thaliana PBL protein sequences were retrieved from the TAIR website (www.arabidopsis.org) (accessed on 13 March 2025). The phylogenetic tree was constructed using the Neighbor-Joining (NJ) method in MEGA (version 11.0), and the Bootstrap repeat value was set to 1000. The phylogenetic tree was visualized using the online tool iTOLv6 (https://itol.embl.de/) (accessed on 13 March 2025). Using MEME Suite (https://meme-suite.org/meme/tools/meme) (accessed on 13 March 2025), members of the family of tree peony PBL protein sequence of the conservative motif were analyzed and set the conservative base sequence for a length of at least 6, the maximum length of 20. The conserved motifs were visualized using TBtools. Visualize Gene Structure is used to visualize the gene structure of PBL family members of tree peonies.
2.2.3 Cis-Acting ELEMENT Analysis of Tree Peony PBL Gene
To predict and elucidate the function of the PBL gene in tree peony, the 2000 bp gene sequence upstream of the initiation codon ATG in the tree peony genome was extracted using the TBtools software as the promoter region. Forecast analysis of the cis-acting elements in the promoter of the tree peony SHMT gene was conducted using the PlantCARE website (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) (accessed on 13 March 2025). The results were visually analyzed using the TBtools software.
2.2.4 Chromosome Localization and Collinearity Analysis of Tree Peony PBL Family
Using the tree peony gff file and the IDs of the PBL gene family, the gene density parameter in the Gene Location Visualization module of TBtools (v2.136) software was set to 20,000,000. This setting was employed to generate a chromosomal location diagram for the PBL gene in tree peony. For visualization purposes, TBtools software was employed. The chromosome length, gene density, and collinearity files were obtained using TBtools software, with the collinearity files being utilized as default settings. The Advanced Circos function within TBtools software was utilized to achieve collinearity visualization among the PBL family species of tree peony.
2.2.5 Analysis of Protein Interaction and Gene Expression of Tree Peony PBL Family Members
The analysis of protein interactions among members of the PBL gene family in tree peony was performed using the STRING online database (https://string-db.org/) (accessed on 13 March 2025). During the network display, disconnected nodes were hidden by selecting the appropriate option. To detect the expression of PBL family genes in tree peony at various stages post-bee pollination, qRT-PCR analysis was conducted on candidate PoPBL genes of Paeonia ostii ‘Fengdan’ at different developmental stages following pollination (T0: 0 h post-pollination, M1: 1 h post-pollination, M2: 2 h post-pollination). In all samples, genes with low expression levels were excluded. Origin 2022 was utilized to generate tables.
2.2.6 Cloning and Sequence Analysis of PoPBL40
By searching the sequence annotation information of the full-length transcriptome database of tree peony petals previously constructed, the PoPBL40 sequence was retrieved. The open reading frame (ORF) of the PoPBL40 gene was determined utilizing the NCBI ORFfinder tool (https://www.ncbi.nlm.nih.gov/orffi.nder/) (accessed on 13 March 2025). Primers were designed using Primer Premier 5.0 software. The cloning primers are presented in Table 1. Total RNA was extracted from the petals using the Tiangen polysaccharide polyphenol plant total RNA extraction kit (Tiangen) according to the manufacturer’s instructions. Following the manufacturer’s protocol, subsequent to acquiring the sequence via the cloning kit, a cloning vector was constructed and submitted for sequencing. Bacterial colonies that yielded correct sequencing results were selected for subsequent analyses.
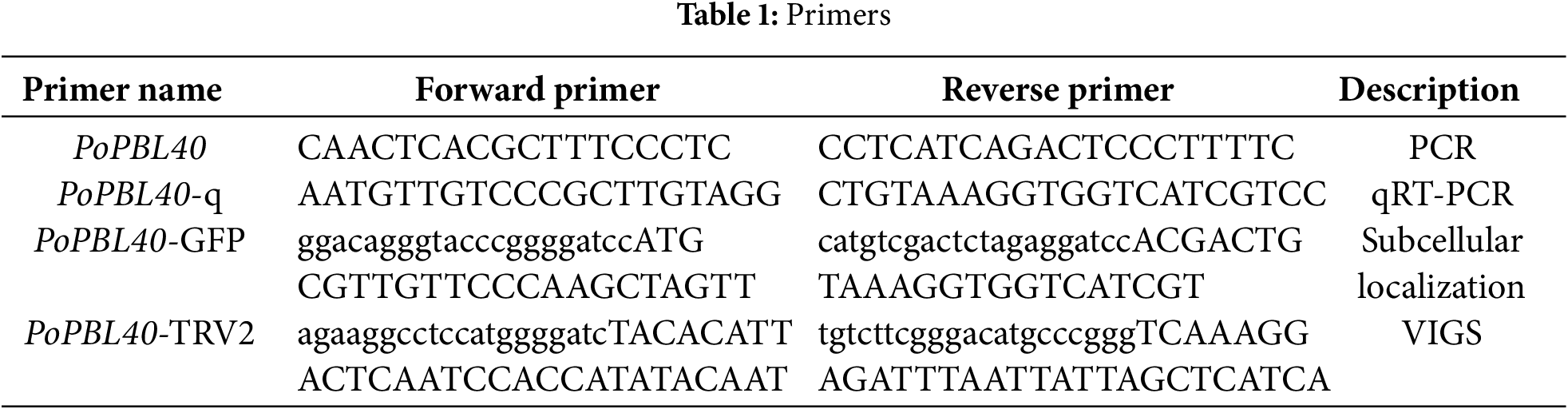
2.2.7 Subcellular Localization Analysis of PoPBL40
Using the coding region of the PoPBL40 gene, from which the stop codon had been excised, as a template, the plant expression vector pCAMBIA2300-GFP was digested with the restriction endonuclease enzyme BamHI, and the seamless cloning technology was used. The OK Clon DNA Ligation Kit II (Ecorui Biotechnology Co., Ltd., AG11807, Changsha, China) was used to obtain the recombinant vector pCAMBIA2300-PoPBL40-GFP. The primers are shown in Table 1. Both pCAMBIA2300-GFP and pCAMBIA2300-PoPBL40-GFP were transformed into the Agrobacterium GV3101 strain using the heat shock method. Colonies identified as positive were selected and grown in LB liquid medium containing 0.5 L·mL−1 kanamycin and 1 L·mL−1 rifampicin until OD600 reached 0.6 to 0.8. Tobacco leaves at the 4- to 6-leaf stage were infiltrated with a 1 mL sterile syringe. After infiltration, the leaves were incubated in the dark for 12 h, followed by 48 h of alternating 12-h light and 12-h dark cycles. The expression of GFP fluorescence in tobacco cells was observed by laser scanning confocal microscopy.
2.2.8 Transient Silencing of PoPBL40 Gene
Using the coding region of the PoPBL40 gene as a template, the VIGS silencing vector TRV-PoPBL40 was constructed using the virus-induced gene silencing (VIGS) vector tobacco-TRV2 with the restriction endonucleases BamHI and SmaI. The primers are listed Table 1. The OD600 of tobacco-TRV1, tobacco-TRV2, and TRV-PoPBL40 was adjusted to 0.6 to 0.8, and the bacterial suspensions were incubated in the dark for 3 to 6 h. Pollen from Paeonia ostii ‘Fengdan’ at full flowering stage was soaked in the infection solution for 20 min, incubated in a 15% sucrose solution at 25°C for 2 h, and pollen development was observed under an inverted fluorescence microscope.
3.1 Identification of PBL Gene Family Members and Analysis of Physicochemical Properties of Tree Peony
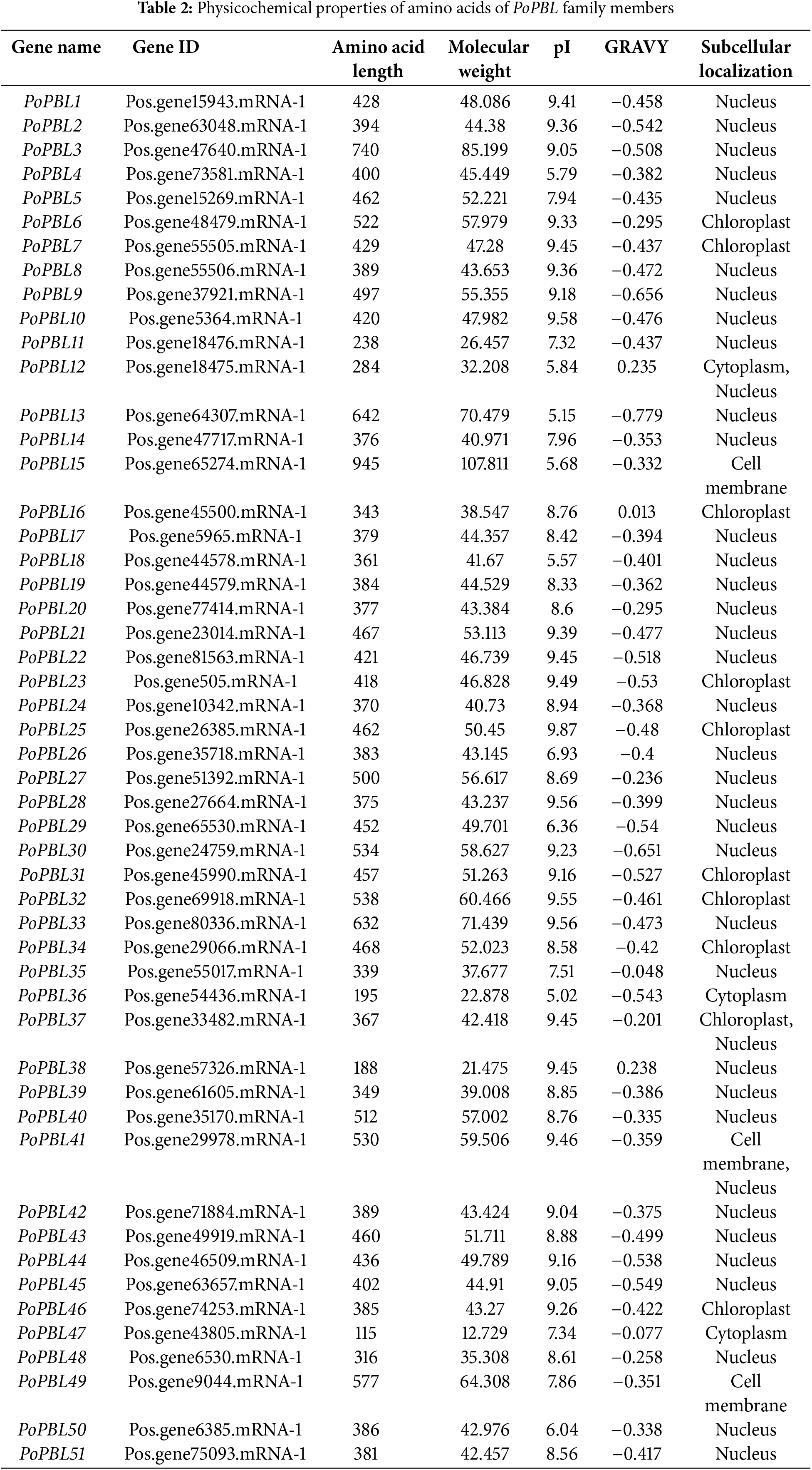
Fifty-one PBL proteins were identified from tree peony using BLAST and HMM methods and were designated according to conventional nomenclature as PoPBL1-51 based on the chromosomal positions of the genes. The Pfam and SMART databases were used to analyze the protein domains, and it was found that all members of the PoPBL family contain the conserved domain S_TKc. The number of amino acids encoded by the 51 predicted PBL proteins ranged from 115 to 945, with the predicted molecular weight ranging from 12.73 to 107.81 kDa, and the isoelectric point (pI) ranging from 5.02 to 9.87, with a mean value of 8.38 (Table 2). 9 PoPBL proteins have pI values less than 7, indicating that they are acidic proteins, whereas the remaining PoPBL proteins have pI values greater than 7, indicating that they are basic proteins (Table 2). The overall average hydrophilicity coefficient (GRAVY) of the PoPBL proteins ranges from −0.779 to 0.238 (Table 2). 94% of the PoPBL proteins had a total mean hydropathicity coefficient less than 0, indicating that most PoPBL proteins are hydrophilic (Table 2). 68% of the PoPBL family members are located in the nucleus, 9 PoPBL family members are localized in the chloroplast, PoPBL15 and PoPBL49 are localized in the plasma membrane, PoPBL36 and PoPBL47 are localized in the cytoplasm, and PoPBL37 is present in both chloroplasts and nuclei. PoPBL41 is found exclusively in the plasma membrane and the nucleus, whereas PoPBL12 is found in both the nucleus and the cytoplasm (Table 2).
3.2 Chromosome Localization and Collinearity Analysis of PBL Gene Family in Tree Peony
The 51 members were distributed across all five chromosomes, albeit unevenly. The majority of PoPBL genes were located on chromosome 1, with 20 members of the PBL family present. Chromosomes 2, 4, and 5 exhibited similar numbers of members, while chromosome 3 had the fewest members. Furthermore, it was observed that PoPBL50 and PoPBL51 were located on unassembled scaffolds (Fig. 1A). A pair of tandemly repeated genes (PoPBL6 and PoPBL23) were found on chromosomes 1 and 2, indicating that the chromosome fragment may have a duplication event in the evolutionary process and may not be fully differentiated, and there may be redundancy in function (Fig. 1B).
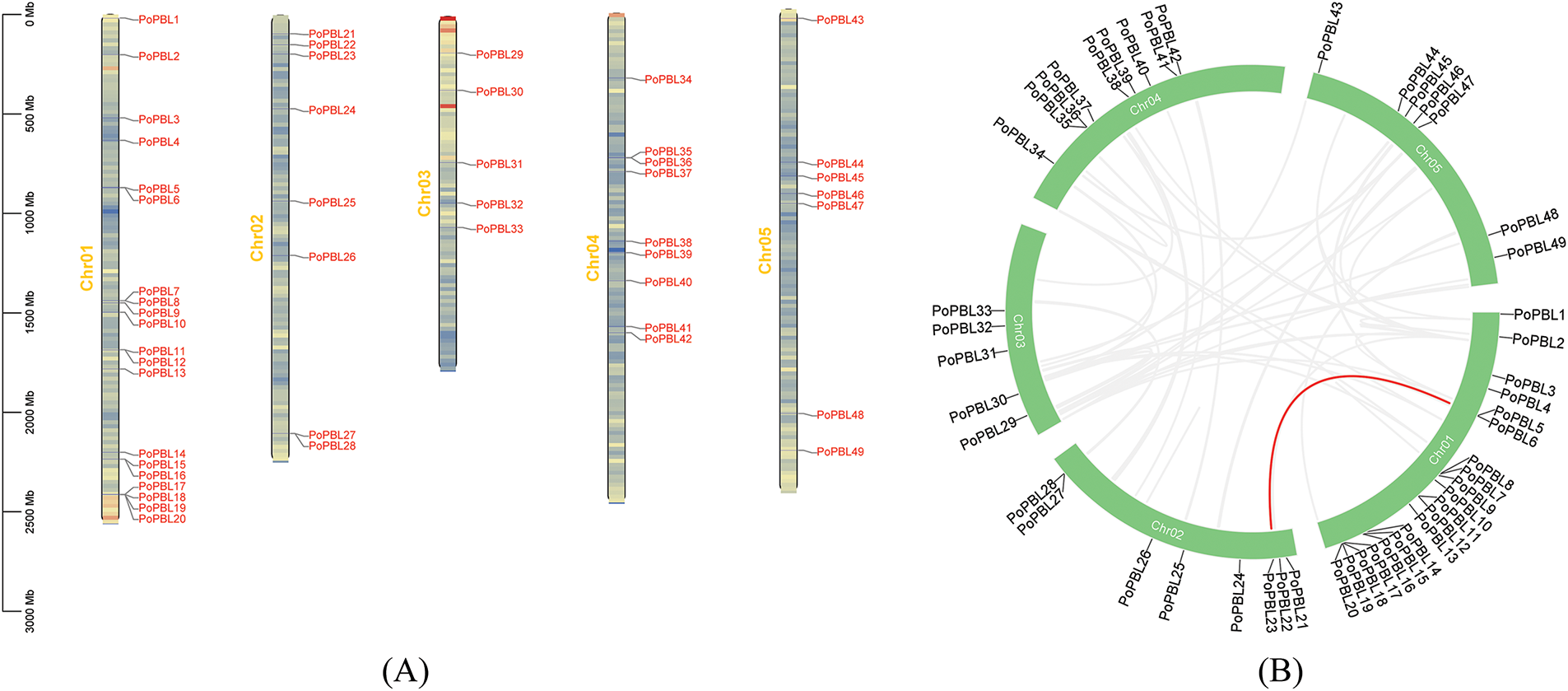
Figure 1: Chromosomal mapping of PoPBL genes in tree peony (A) collinearity analysis (B)
The interior of the chromosome is filled with gene density, and the color from blue to red indicates that the gene density goes from small to large.
3.3 Phylogenetic Tree Analysis of PBL Gene in Tree Peony
Based on the Arabidopsis thaliana PBL family members (31), 51 PBL tree peony family members were categorized into 11 groups (Fig. 2). The majority of tree peony PBL members were distributed in subgroups III, IV, VIII, X, and XI, comprising 7, 6, 5, 15, and 18 PoPBL members, respectively.
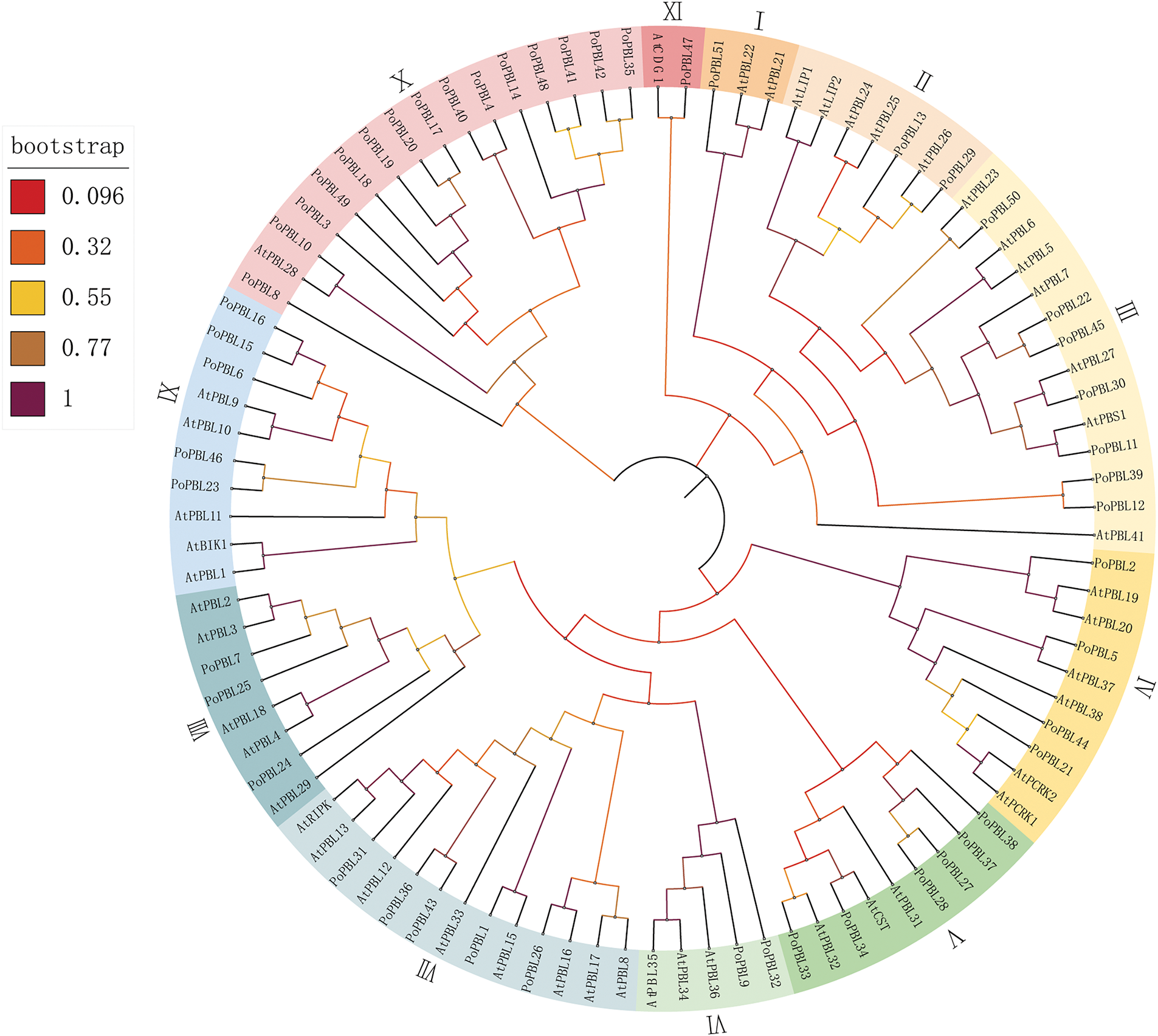
Figure 2: Phylogenetic tree of PoPBL family members
At and Po, respectively, represent members of the Arabidopsis thaliana and Paeonia section Moutan DC PBL family members, and the branching values are the self-guided values obtained from 1000 bootstrap tests using the adjacency method. Different colored areas represent different clades.
3.4 Analysis of Gene Structure and Conserved Motifs of PBL Gene in Tree Peony
A total of 20 conserved motifs were identified among the members of the tree peony PBL family (Motif 1 to Motif 20) (Fig. 3A). Motifs 1, 2, 3, 4, and 5 were found to be prevalent within the PoPBL family. The distribution of these motifs within the PoPBL family exhibits distinct patterns that can be broadly categorized into three groups: Group I comprises Motifs 13, 4, 11, 7, 2, 9, 6, 10, and 8; Group II comprises Motifs 16, 4, 7, 12, 2, 9, 3, 3, 10, and 8; Group III comprises Motifs 4, 12, 2, 5, 3, 1, and 8. All three groups contain Motifs 4, 7, 2, 9, 5, 3, 1, and 8. The three groups under consideration contain the Motifs 4, 7, 2, 9, 5, 3, 1, and 8. Despite the presence of variations in motif sequence types among the groups, members of the same group exhibited a propensity to manifest analogous motif patterns (Fig. 3A). In the PoPBL family, all PoPBL genes except PoPBL12 and PoPBL38 contain Motif 1 (Fig. 3A). The 51 PoPBL genes exhibited variation in the number of exons, with 31.4% of the members containing six exons and five introns, 25.5% containing five exons and four introns, and 15.7% containing seven exons and six introns. A small number of PBL family members contain eight or more exons (PoPBL12, PoPBL34, PoPBL41) (Fig. 3B).
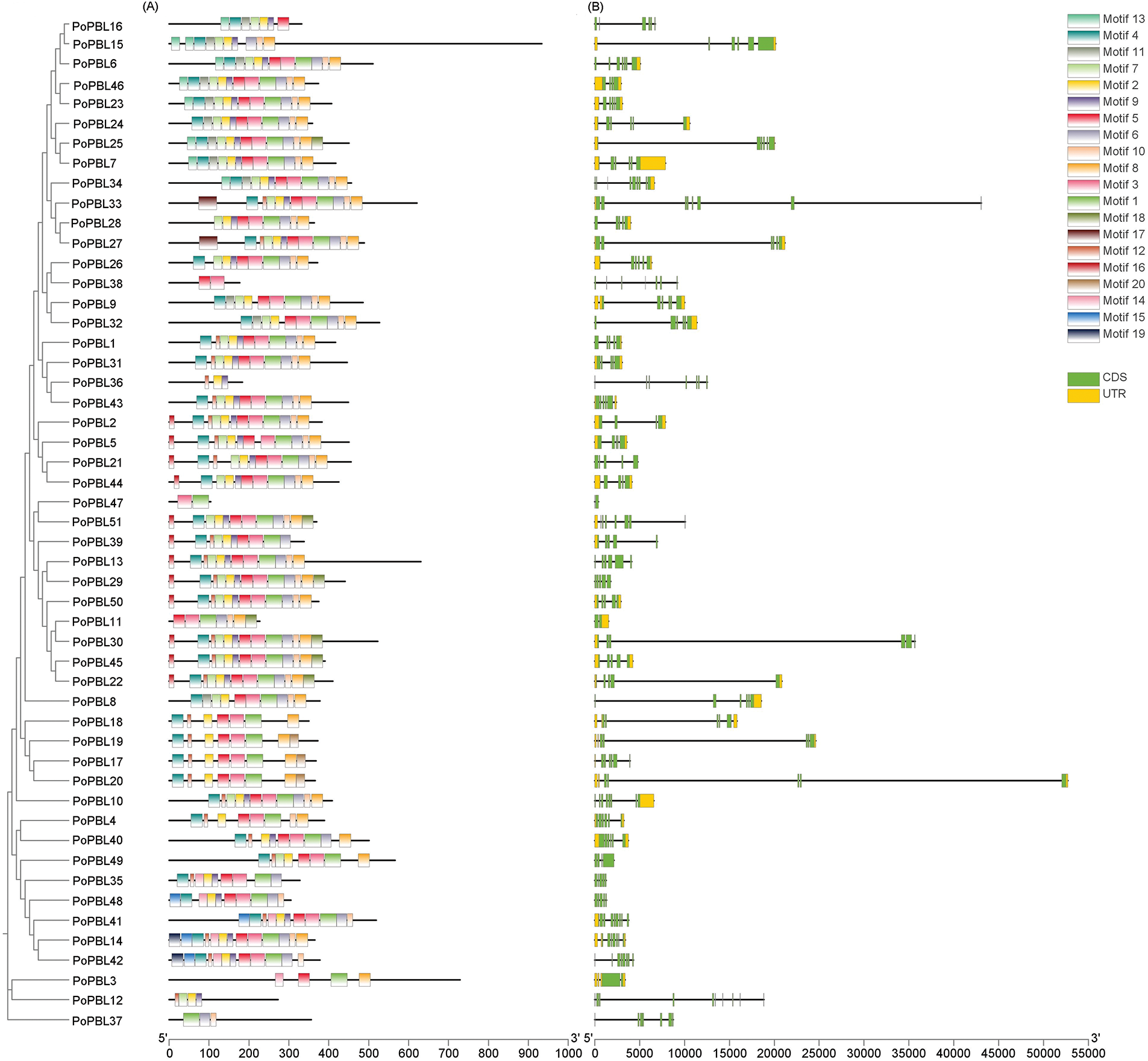
Figure 3: Conserved motif (A) and exon-intron structure (B) of PoPBL family members. (A) Distribution of conserved motifs of PoPBL family members. Motifs 1–20 are shown in boxes of different colors; (B) The gene structure of PoPBL family members. The untranslated regions, exons, and introns are represented by colored boxes and black lines, respectively, and the length of the protein is indicated by the scale at the bottom
3.5 Analysis of Cis-Acting Elements of PBL Gene in Tree Peony
The cis-acting elements of the promoter region of members of the PoPBL family are divided into three categories. The first category is the elements related to light regulation, G-Box (41 members) and Box 4 (38 members). The second category is the response hormone-related elements, including 5 types of hormone response elements, mainly ABRE response elements (ABRE, 36 members), auxin (TGA-element, 21 members), gibberellin response elements (P-box, GARE-motif, 25 members and 11 members), methyl jasmonate (MeJA, 11 members), and methyl jasmonate. 29 members) and salicylic acid (TCA-element, 13 members). It is notable that the promoter region of the majority of PoPBL family members contains between three and four hormone response elements. PoPBL30 and PoPBL47, the number of abscisic acid response elements was the largest, followed by gibberellin response elements, and MeJA response elements ranked third. The third category is the response to abiotic stress-related elements, cis-regulatory elements necessary for anaerobic induction (ARE, 42 members), cis-acting elements involved in defense and stress responses (TC-rich repeats, 25 members), cis-acting elements involved in low-temperature response (LTR, 25 members). Cis-acting elements involved in low temperature response (LTR, 22 members) and MYB binding sites involved in drought induction (MBS, 21 members) (Fig. 4). The following elements are implicated in plant growth and development in PoPBL genes: The following elements are involved in the regulation of zeatin metabolism (O2-site): the MYB binding site (CCAAT-box); the low-temperature response (LTR); the endosperm expression (GCN4_motif) element; the meristem expression (CAT-box) element; the seed-specific regulation (RY-element) element; and the circadian rhythm cis-regulatory elements (circadian).
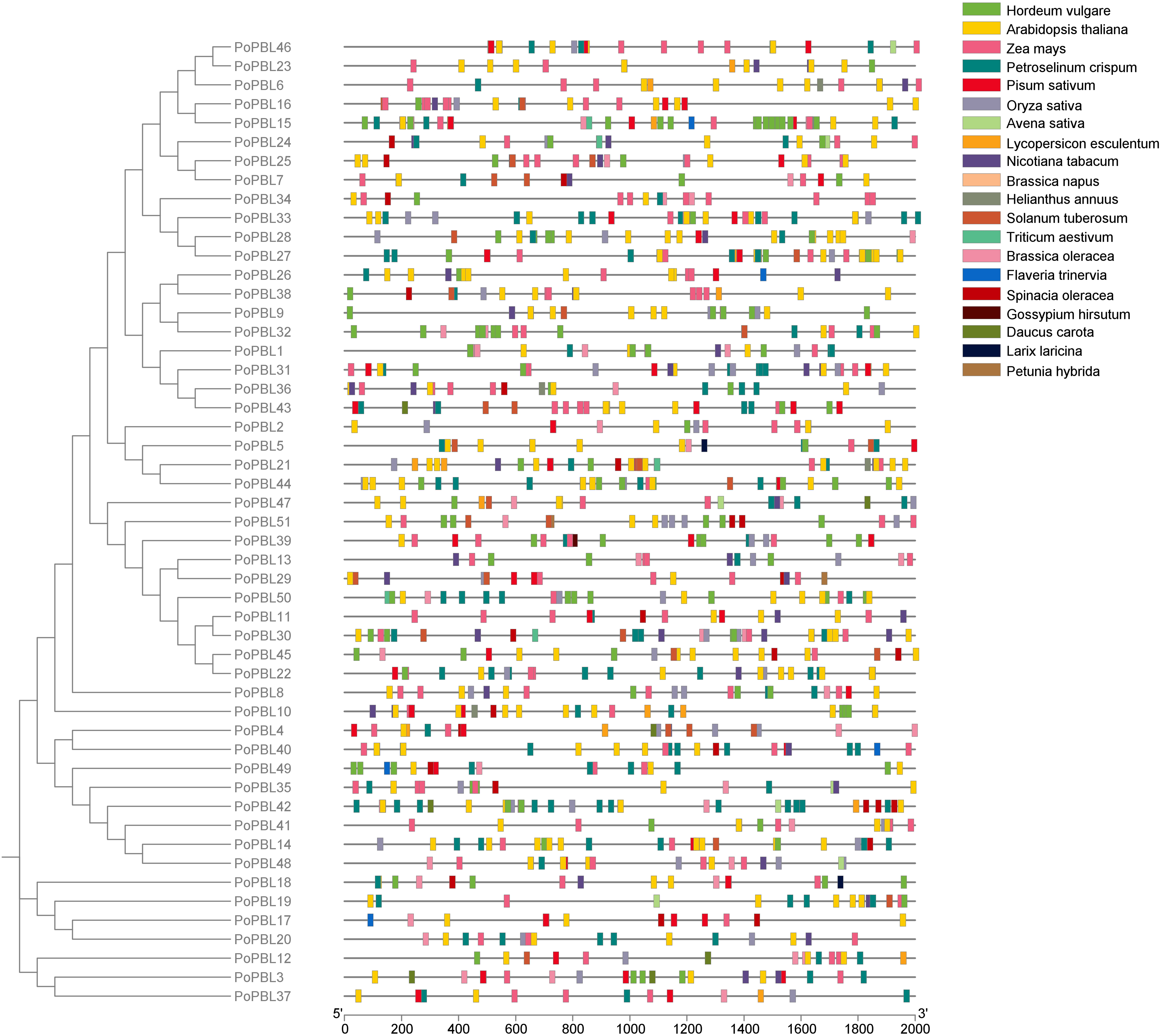
Figure 4: Analysis of cis regulatory elements of PoPBL gene members
3.6 Protein Interaction Analysis of PBL Gene in Tree Peony
There are 9 proteins (17.6%) in PoPBLs that interact with PPCK2 protein, namely PoPBL1, PoPBL2, PoPBL8, PoPBL17, PoPBL20, PoPBL28, and PoPBL46 (Fig. 5). In addition, there was an interaction between PoPBL8 and PCRK1 (Fig. 5).
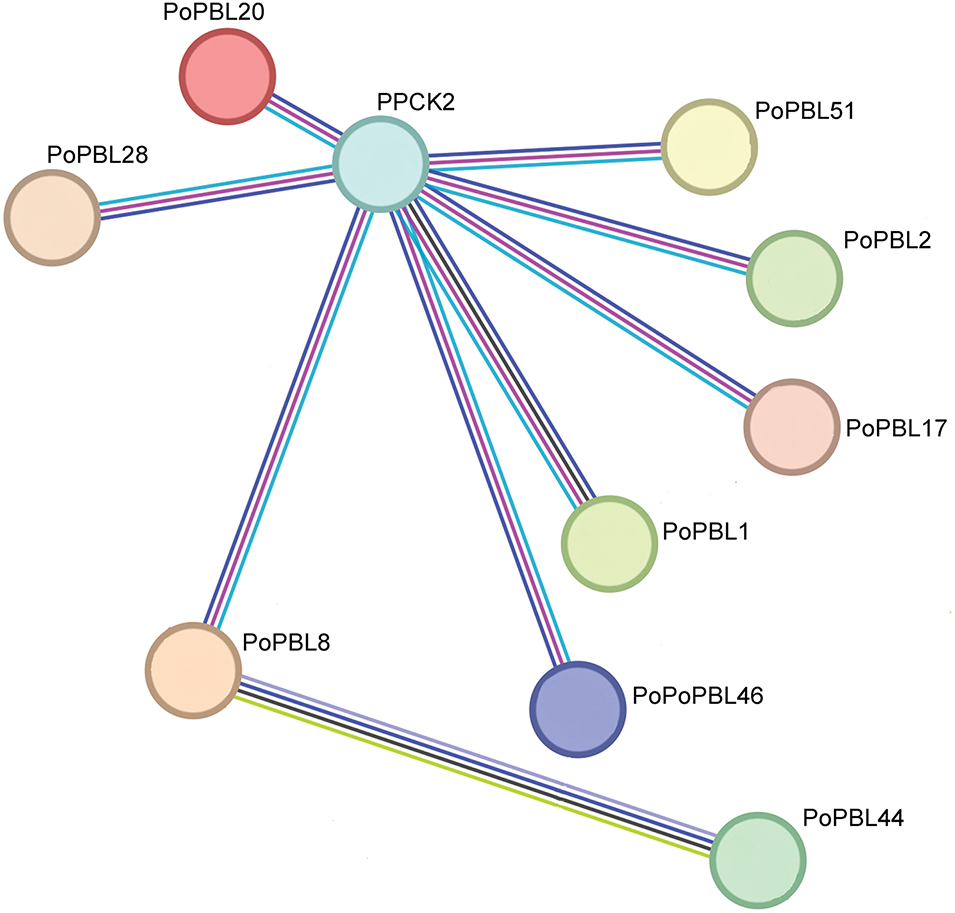
Figure 5: Protein interactions encoded by members of the PoPBL family members
3.7 Analysis on the Expression of PBL Family Members in Tree Peony
The relative expression levels of PoPBL1, PoPBL3, PoPBL4, PoPBL7, PoPBL13, PoPBL14, PoPBL25, PoPBL30, PoPBL39, PoPBL40, PoPBL45 and PoPBL48 genes in T0, M1, and M2 initially increased and then showed a downward trend with increasing time (Figs. 6, S1A–H). The relative expression levels of PoPBL5, PoPBL6, PoPBL8, PoPBL9, PoPBL11, PoPBL16, PoPBL18, PoPBL19, PoPBL20, PoPBL21, PoPBL22, PoPBL23, PoPBL27, PoPBL31, PoPBL32, PoPBL33, PoPBL34, PoPBL45, PoPBL46, PoPBL50 and PoPBL51 genes showed an increasing trend with time (Figs. 6, S2A–R). The expression levels of PoPBL10, PoPBL26, PoPBL49 and genes showed a decreasing trend with time (Fig. 6). The relative expression changes of PoPBL2, PoPBL15, and PoPBL17 genes did not fluctuate significantly with time (Figs. 6, S3A–C). Among them, the relative expression levels of PoPBL1, PoPBL4, PoPBL14, PoPBL40 and PoPBL45 were significantly higher in the M1 period (p < 0.05, Figs. 6, S4A–B). The relative expression levels of PoPBL8, PoPBL31, and PoPBL50 genes were significantly higher in the M2 period (p < 0.05, Fig. 6).
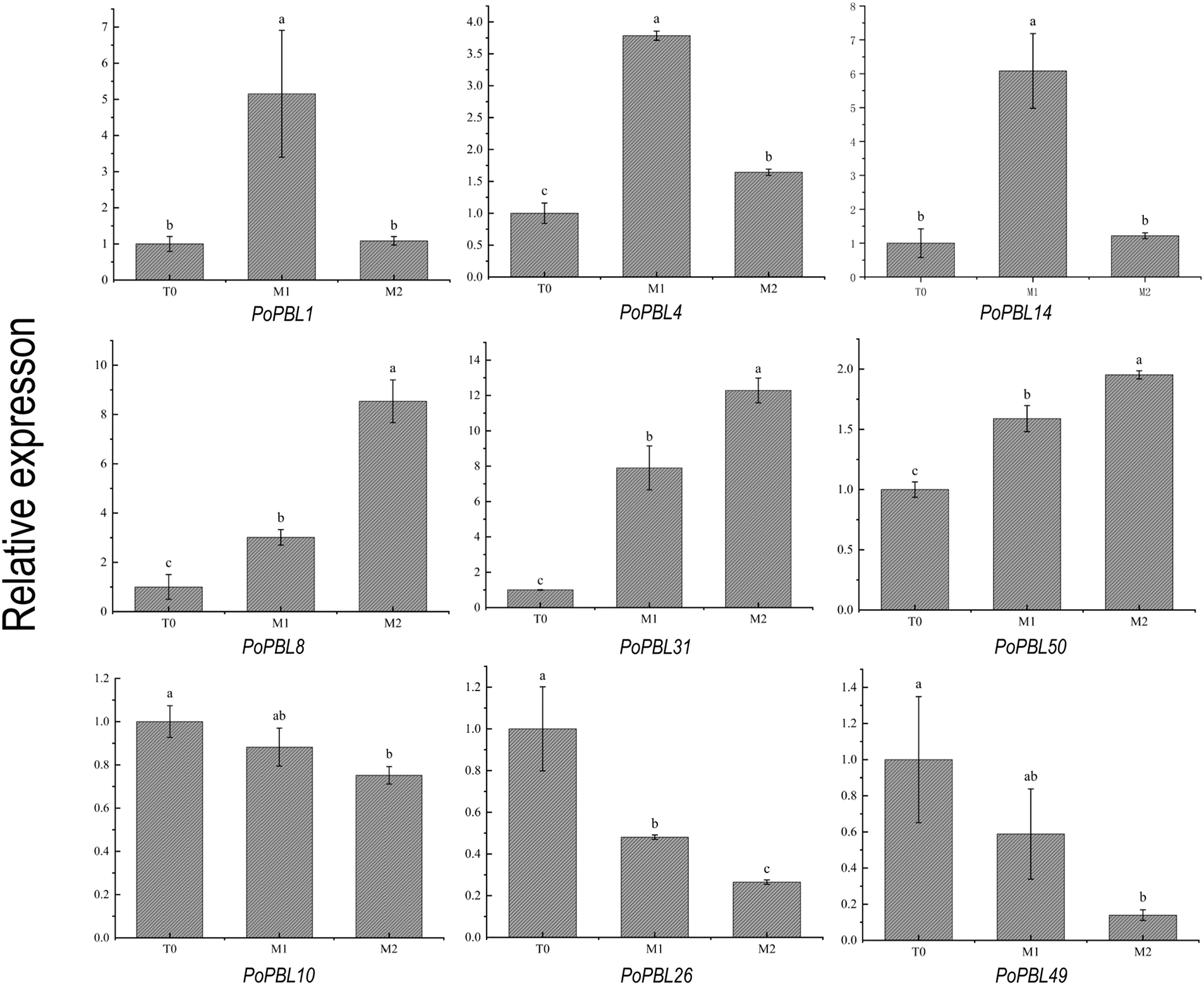
Figure 6: qRT-PCR analysis of PoPBL gene members at T0, M1 and M2 stages
3.8 Cloning and Sequence Analysis of PoPBL40
A single, bright band was cloned and obtained (Fig. 7A). After sequencing, it was the same as the expected size, and it was named PoPBL40. The sequence was 2044 bp, and the open reading frame was 1506 bp, encoding 501 amino acids (Fig. 7A). PoPBL40 protein contains 222 amino acids in random coils, accounting for 44.31%, 178 amino acids in α-helix, accounting for 35.53%, 74 amino acids in the extended strand, accounting for 14.77%, and 27 amino acids in β-sheet, accounting for 5.39% (Fig. 7B). The phylogenetic tree was constructed by comparing the protein sequences of tree peony with those of 10 species, such as Alnus glutinosa, Vitis riparia, Carpinus fangiana and Corylus avellana. The results showed that the tree peony PoPBL40 gene had the closest evolutionary relationship with Liquidambar formosana and the farthest genetic relationship with Corylus avellana (Fig. 7C).
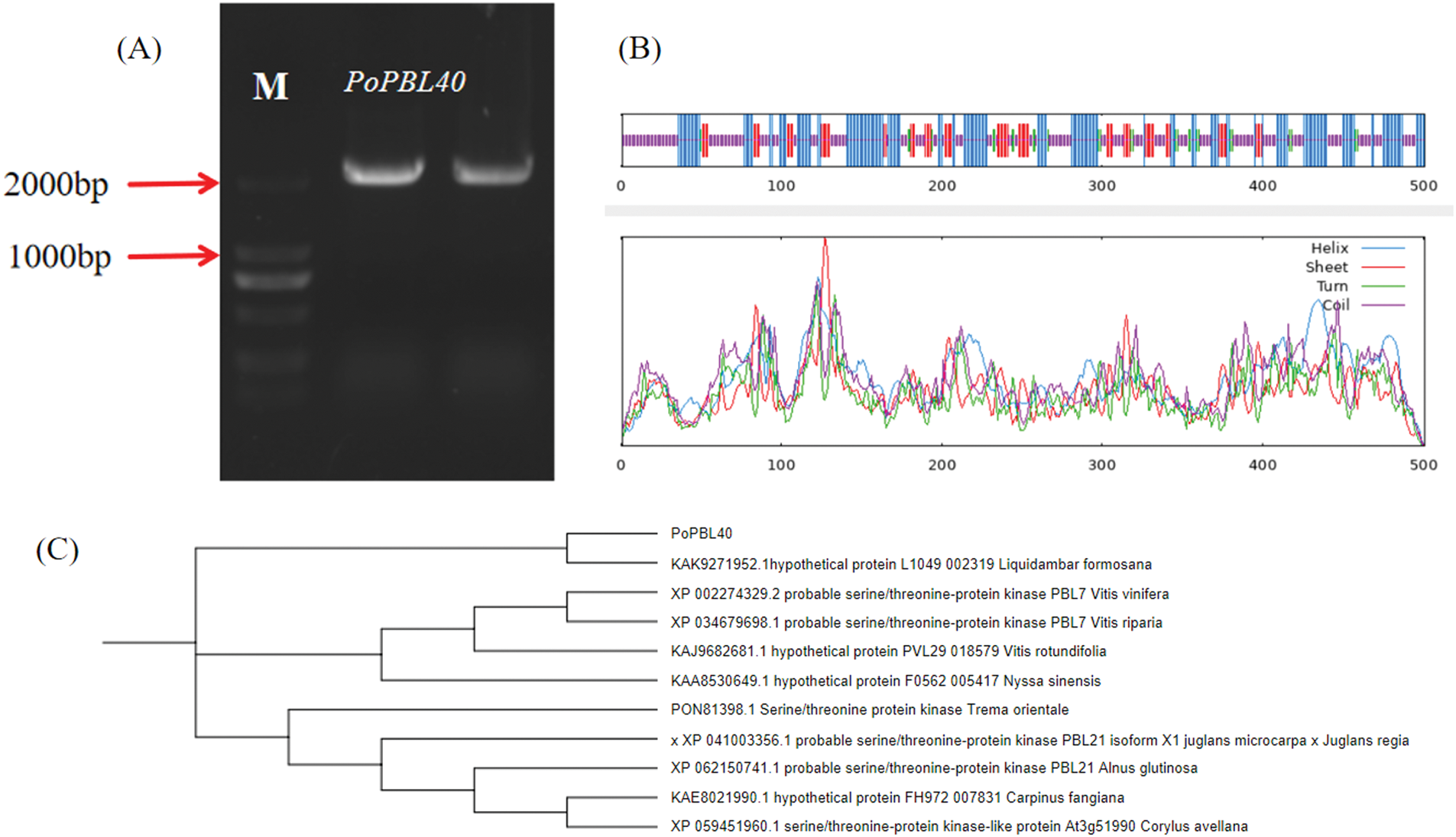
Figure 7: Cloning and sequence analysis of PoPBL40 gene in tree peony. (A): PoPBL40 gene PCR amplification product, where M is DNA maker DL2000; (B): PoPBL40 protein secondary structure prediction map; (C): Phylogenetic tree of PoPBL40 homologous proteins
3.9 Subcellular Localization of PoPBL40
The fluorescence of pCAMBIA2300-GFP was observed in the nucleus and cytoplasm by laser confocal microscopy, and the fluorescence of PoPBL40 gene was obviously expressed in the nucleus (Fig. 8).
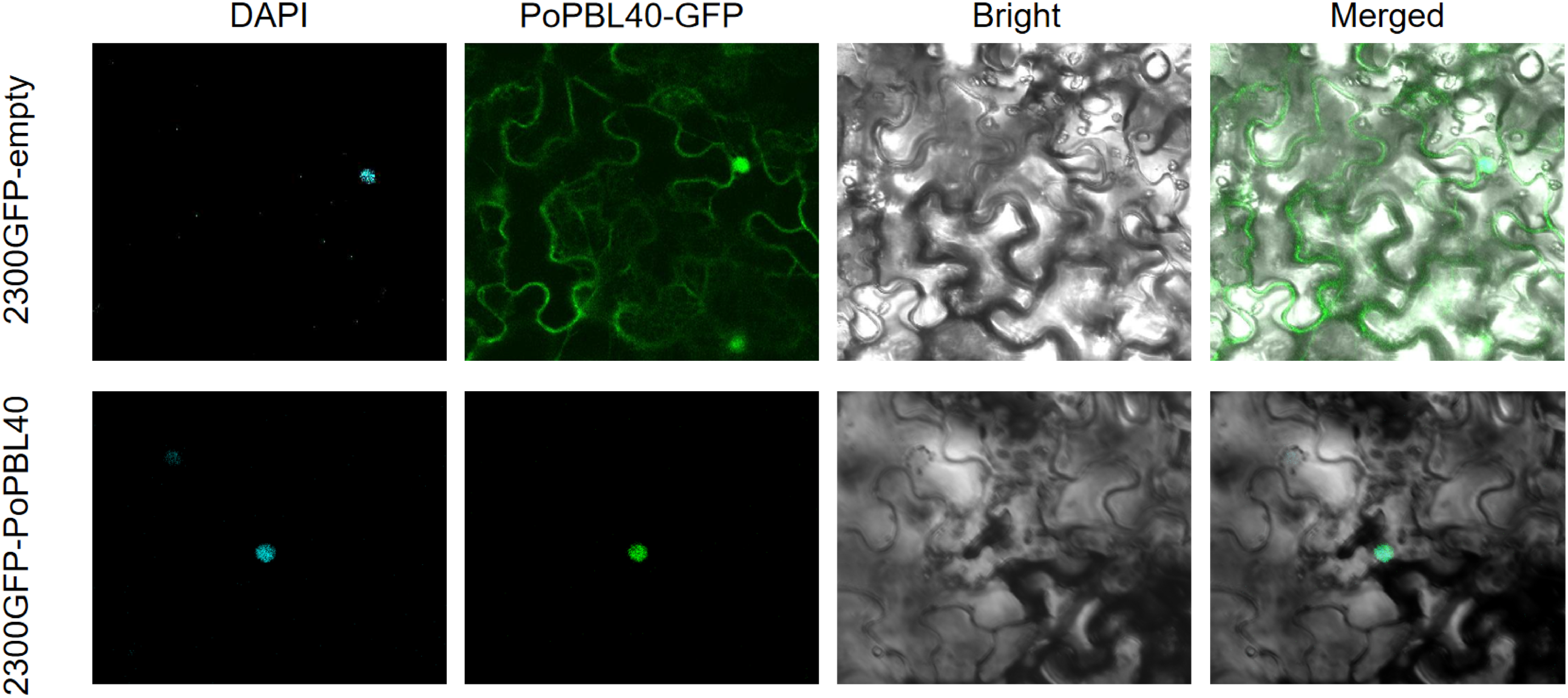
Figure 8: Subcellular localization of PoPBL40 gene
3.10 Transient Silencing of PoPBL40 Gene
VIGS technology was used to silence PoPBL40 in mature pollen of Paeonia ostii ‘Fengdan’ After 3–6 h treatment, compared with the control group (tobacco-TRV1 + tobacco-TRV2), the number of pollen germination in the gene silencing group (tobacco-TRV1 + TRV-PoPBL40) was significantly lower (6.3%) compared with the control group (tobacco-TRV1 + tobacco-TRV2), and the pollen tube grew slowly without obvious elongation. The number of pollen tube germination in the control group (tobacco-TRV1 + tobacco-TRV2) was higher (11.17%), and the growth length of the pollen tube was longer (Fig. 9).
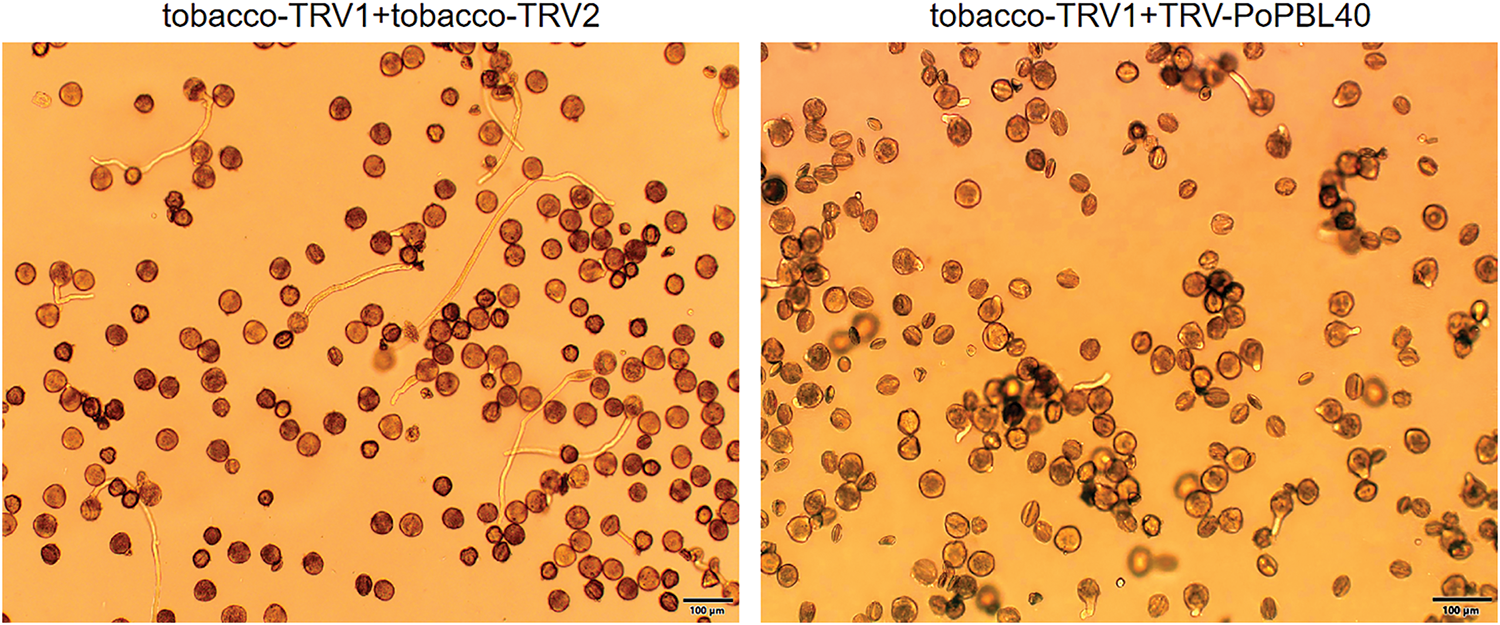
Figure 9: VIGS diagram of PoPBL40 gene
In this study, the members of the PoPBL gene family were identified through the genome of the tree peony, and the chromosome location, phylogenetic relationship, conserved motif, expression characteristics, and promoter elements of the PoPBL family members were analyzed by bioinformatics software and websites. A total of 51 PoPBL genes with typical serine/threonine protein kinase domains were identified by Pfam analysis. Subcellular localization prediction showed that PoPBL genes were distributed in the nucleus, cytoplasm, cell membrane and chloroplasts, mainly located in the nucleus. This localization feature is similar to that of the RLCK-VII subfamily members in Arabidopsis thaliana. [31]. Physicochemical property analyses revealed that the average hydrophilicity (GRAVY) of 94% of PoPBL proteins was less than zero, indicating hydrophilic protein characteristics. These results are similar to the characteristics of RLCK-VII proteins found in many plants [32]. In the phylogenetic analysis, according to the taxonomic and topological structure proposed in previous studies, the tree peony PBL genes were categorized into 11 subgroups, which differs from the number of PBL family subgroups in Arabidopsis thaliana [15]; this discrepancy may be attributed to interspecific differences. Some subgroups were only from PBL members of tree peonies, suggesting that the PBL gene family may have undergone species-specific differentiation after isolation. RLCK-VII subfamily members have a conserved role as convergent substrates of cell membrane-localized receptor-like kinases (RLKs) and regulate multiple signaling nodes, orchestrating a complex array of defense responses against phytopathogens [22]. Such as AtPCRK1 [33], AtPCRK2 [34] and AtPBL19 [35] in subgroup IV, AtPBL27 [36] in subgroup III and AtRIPK [37] in subgroup VII (Fig. 2). These findings elucidate the specific functions of PBL members in plant immune mechanisms. However, the family composition, molecular evolution, and resistance function of RLCK-VII subfamily genes in tree peonies remain unclear. Therefore, elucidating the RLCK-VII members in tree peonies is of significant importance for uncovering valuable genetic resources and innovating tree peony varieties in response to changing environments and pathogens.
After analyzing the phylogenetic tree and exon-intron structure of PoPBL family members, it was observed that the majority of PoPBL family members contain 5 to 7 exons, with varying numbers of introns. The variation in gene structure within the same subfamily suggests that PoPBLs may have undergone functional divergence or acquired novel functions during evolution.
Gene structure analysis showed that the PoPBL genes contained Motif 1 among the 51 identified PoPBL genes. Motif 1, Motif 2, Motif 3, Motif 4, and Motif 5 are widely present in the PoPBL family, which is consistent with the gene structure of PBL genes reported in other plants [38]. For example, 72 GhRLCK-VII proteins in Gossypium hirsutum contain two conserved motifs: Motif 1 and Motif 2. These motifs are located in the Pkinase_Tyr or Pkinase domain and are important features of these proteins [9]. The presence of these conserved motifs may be crucial to the function of PoPBLs proteins, particularly in plant immune responses. These motifs may facilitate interactions with various downstream proteins, thereby triggering a diverse array of responses. Therefore, further study of these conserved motifs will help to understand the role of PoPBL gene family in plant growth, development and defense responses.
The results of the cis-acting element analysis showed that the PoPBL genes contained various hormone response elements, light response elements, abiotic stress response elements, and growth and development response elements. In addition, cis-acting elements related to abiotic stress, such as ARE, ABRE, LTR, GARE, etc., are also more abundant in PoPBL genes. Studies have shown that RLCK plays an important role in plant biotic and abiotic stresses [39]. Almost all PoPBL genes have been found cis-regulatory elements necessary for anaerobic induction in the promoter region (ARE, 82%). It is suggested that PoPBL family members may be involved in the response of plants to abiotic stresses such as osmotic stress, anaerobic environment and ABA [40]. It is speculated that PoPBLs have a complex mechanism for the plant immune system to resist stresses. The existence of cis-acting elements and the number of cis-acting elements have certain reference values for analyzing whether the gene can respond to related stresses and provide a theoretical basis for further study on the molecular mechanism of tree peony PBL gene regulating the growth and development and stress response of tree peony.
Quantitative real-time PCR (qRT-PCR) analysis of 51 PoPBL family members in tree peonies at different developmental stages post-pollination revealed that the expression changes of gene family members across various developmental stages could be broadly categorized into three groups. I: With the increase in time, it showed a trend of increasing first and then decreasing. II: Expression levels showed an upward trend with increasing time. III: Expression levels demonstrated a downward trend as time increased. The expression of each gene in different periods of tree peony tissue was significantly different. It indicated that PoPBL genes may play a regulatory role in developing flower and root tissues. The expression levels of PoPBL1 and PoPBL14 were highest at 1 h post-pollination, suggesting that these genes may be involved in the regulation of flower pollination and early developmental processes. The expression levels of PoPBL8 and PoPBL31 were the highest at 2 h after pollination, suggesting that the PoPBL gene family of tree peonies may be involved in the regulation of reproductive growth and development. Protein interaction network predictions identified that the protein encoded by the PBL gene interacts with PPCK2, a calcium-independent kinase. Recent studies have demonstrated that Phosphoenolpyruvate carboxylase kinase 2 (PPCK2) is a calcium-independent kinase involved in the phosphorylation of light-dependent phosphoenolpyruvate carboxylase and belongs to the Ser/Thr protein kinase family. The protein is expressed in flowers and roots, and the expression level is low in stems and leaves [41]. Additionally, PoPBL8 was found to interact with PCRK1, a serine/threonine protein kinase involved in the activation of early immune responses. PCRK1 plays a role in pattern-triggered immunity (PTI) induced by pathogen-associated molecular patterns (PAMP) and damage-associated molecular patterns [42]. It is further proved that the PBL gene family has an important influence on plant immunity and growth and development systems.
In this study, the PBL gene family of tree peony was identified, and it was preliminarily revealed that PoPBL family members may be involved in the regulation of reproductive organ development, as well as the immune system and stress resistance regulation of tree peony. At the same time, a PBL transcription factor PoPBL40 was cloned from tree peony. The full-length cDNA of PoPBL40 was 2044 bp, encoding 502 amino acids. The protein was closely related to the KAK9271952.1 hypothetical protein L1049_002319 of Liquidambar formosana. The expression level of PoPBL40 was the highest 1 h post-pollination on the stigma of Paeonia ostii ‘Fengdan’, and then gradually decreased. Silencing PoPBL40 can reduce the growth rate of tree peony pollen tubes, and the growth rate is slow, indicating that PoPBL40 plays a positive regulatory role in the growth regulation of tree peony pollen tubes.
Acknowledgement: Thanks to all researchers for their contribution to this work.
Funding Statement: This work was supported by the National Natural Science Foundation of China [Grant Number U23A20211].
Author Contributions: Yuxin Zhao: methodology, data curation and the original draft. Yuxin Zhao and Zhanxiang Tan: investigation and data curation. Kaiyue Zhang: plant material preparation; Yuying Li: review and editing. Xiaogai Hou and Lili Guo: conceptualization, review & editing, supervision. All authors reviewed the results and approved the final version of the manuscript.
Availability of Data and Materials: The data sets generated during the current study are not publicly available due to non-disclosure of the data, but may be available from the corresponding authors upon reasonable request.
Ethics Approval: Not applicable.
Conflicts of Interest: The authors declare no conflicts of interest to report regarding the present study.
Supplementary Materials: The supplementary material is available online at 10.32604/phyton.2025.063737.
References
1. Hou XG, Duan CY, Liu SY, Lv JX, Zhang YB, Li JJ. Advances on chromosome study of Chinese tree peony. Chin Agric Sci Bull. 2006;22(2):307. (In Chinese). doi:10.11924/j.issn.1000-6850.0602307. [Google Scholar] [CrossRef]
2. Han X, Wang L, Liu ZA, Jan DR, Shu Q. Characterization of sequence-related amplified polymorphism markers analysis of tree peony bud sports. Sci Hortic. 2008;115(3):261–7. doi:10.1016/j.scienta.2007.09.003. [Google Scholar] [CrossRef]
3. Zhang J, Wang L, Shu Q, Liu Z, Li C, Zhang J, et al. Comparison of anthocyanins in non-blotches and blotches of the petals of Xibei tree peony. Sci Hortic. 2007;114(2):104–11. doi:10.1016/j.scienta.2007.05.009. [Google Scholar] [CrossRef]
4. Zhou ZQ, Pan KY, Hong DY. Advances in studies on relationships among wild tree peony species and the origin of cultivated tree peonies. Acta Hortic Sin. 2003;30(6):751–7. (In Chinese). doi:10.16420/j.issn.0513-353x.2003.06.037. [Google Scholar] [CrossRef]
5. Liu P, Fu LS, Jiang KM. Comparative analysis on economic benefit of xiangfeng peony. Sci Silvae Sin. 2019;55(3):167–74. (In Chinese). doi:10.11707/j.1001-7488.20190319. [Google Scholar] [CrossRef]
6. Zhang HK, Lu L, Huang YY, Mo MY, Wang QF, Guo CD, et al. Study on comprehensive evaluation of cortex moutan radicis quality in different harvesting periods. Mod Tradit Chin Med Mater Med World Sci Technol. 2019;21(2):240–7. (In Chinese). doi:10.11842/wst.2019.02.014. [Google Scholar] [CrossRef]
7. Dievart A, Gottin C, Périn C, Ranwez V, Chantret N. Origin and diversity of plant receptor-like kinases. Annu Rev Plant Biol. 2020;71(1):131–56. doi:10.1146/annurev-arplant-073019-025927. [Google Scholar] [PubMed] [CrossRef]
8. He HJ, Zhang DQ, Tang L, Yang D. Recent advance on biological function and signal pathway of receptor-like cytoplasmic kinase in plants. Plant Physiol J. 2014;50(7):885–90. (In Chinese). doi:10.13592/j.cnki.ppj.2014.0154. [Google Scholar] [CrossRef]
9. Vij S, Giri J, Dansana PK, Kapoor S, Tyagi AK. The receptor-like cytoplasmic kinase (OsRLCK) gene family in rice: organization, phylogenetic relationship, and expression during development and stress. Mol Plant. 2008;1(5):732–50. doi:10.1093/mp/ssn047. [Google Scholar] [PubMed] [CrossRef]
10. Liu PL, Du L, Huang Y, Gao SM, Yu M. Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol Biol. 2017;17(1):47. doi:10.1186/s12862-017-0891-5. [Google Scholar] [PubMed] [CrossRef]
11. Zhao Z, Li Y, Peng J, Zhu J, Zhang W. Cloning of GhPBL1 gene in cotton and its resistance function against Fusarium wilt. J South Agric. 2022;53(12):3307–17. doi:10.3969/j.issn.2095-1191.2022.12.002. [Google Scholar] [CrossRef]
12. Kim DS, Hwang BK. The pepper receptor-like cytoplasmic protein kinase CaPIK1 is involved in plant signaling of defense and cell-death responses. Plant J. 2011;66(4):642–55. doi:10.1111/j.1365-313X.2011.04525.x. [Google Scholar] [PubMed] [CrossRef]
13. Pei M, Cao Y, Xie X, Cao Y, Chen J, Zhang X, et al. Synergy in rice immunity: exploring strategies of coordinated disease defense through receptor-like kinases and receptor-like cytoplasmic kinases. Rice Sci. 2024;31(6):643–58. doi:10.1016/j.rsci.2024.07.002. [Google Scholar] [CrossRef]
14. Wu TC, Zhu XL, Lv LJ, Chen XY, Xu GB, Zhang ZY. The wheat receptor-like cytoplasmic kinase TaRLCK1B is required for host immune response to the necrotrophic pathogen Rhizoctonia cerealis. J Integr Agric. 2020;19(11):2616–27. doi:10.1016/S2095-3119(20)63160-4. [Google Scholar] [CrossRef]
15. Liang X, Zhou JM. Receptor-like cytoplasmic kinases: central players in plant receptor kinase-mediated signaling. Annu Rev Plant Biol. 2018;69(1):267–99. doi:10.1146/annurev-arplant-042817-040540. [Google Scholar] [PubMed] [CrossRef]
16. Lin W, Ma X, Shan L, He P. Big roles of small kinases: the complex functions of receptor-like cytoplasmic kinases in plant immunity and development. J Integr Plant Biol. 2013;55(12):1188–97. doi:10.1111/jipb.12071. [Google Scholar] [PubMed] [CrossRef]
17. Tang D, Wang G, Zhou JM. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell. 2017;29(4):618–37. doi:10.1105/tpc.16.00891. [Google Scholar] [PubMed] [CrossRef]
18. Yu X, Feng B, He P, Shan L. From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu Rev Phytopathol. 2017;55(1):109–37. doi:10.1146/annurev-phyto-080516-035649. [Google Scholar] [PubMed] [CrossRef]
19. Qiang X, Li X, Chen J, Liao H, Yu F. Preliminary analysis of functional diversity of RALF peptide family in Arabidopsis thaliana. Biotechnol Bull. 2019;35(1):2–10. doi:10.13560/j.cnki.biotech.bull.1985.2018-0921. [Google Scholar] [CrossRef]
20. Liao HZ, Zhu MM, Cui HH, Du XY, Tang Y, Chen LQ, et al. MARIS plays important roles in Arabidopsis pollen tube and root hair growth. J Integr Plant Biol. 2016;58(11):927–40. doi:10.1111/jipb.12484. [Google Scholar] [PubMed] [CrossRef]
21. Boisson-Dernier A, Franck CM, Lituiev DS, Grossniklaus U. Receptor-like cytoplasmic kinase MARIS functions downstream of CrRLK1L-dependent signaling during tip growth. Proc Natl Acad Sci U S A. 2015;112(39):12211–6. doi:10.1073/pnas.1512375112. [Google Scholar] [PubMed] [CrossRef]
22. Rao S, Zhou Z, Miao P, Bi G, Hu M, Wu Y, et al. Roles of receptor-like cytoplasmic kinase VII members in pattern-triggered immune signaling. Plant Physiol. 2018;177(4):1679–90. doi:10.1104/pp.18.00486. [Google Scholar] [PubMed] [CrossRef]
23. Veronese P, Nakagami H, Bluhm B, Abuqamar S, Chen X, Salmeron J, et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18(1):257–73. doi:10.1105/tpc.105.035576. [Google Scholar] [PubMed] [CrossRef]
24. Sreeramulu S, Mostizky Y, Sunitha S, Shani E, Nahum H, Salomon D, et al. BSKs are partially redundant positive regulators of brassinosteroid signaling in Arabidopsis. Plant J. 2013;74(6):905–19. doi:10.1111/tpj.12175. [Google Scholar] [PubMed] [CrossRef]
25. Tang W, Kim TW, Oses-Prieto JA, Sun Y, Deng Z, Zhu S, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321(5888):557–60. doi:10.1126/science.1156973. [Google Scholar] [PubMed] [CrossRef]
26. Shi H, Shen Q, Qi Y, Yan H, Nie H, Chen Y, et al. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell. 2013;25(3):1143–57. doi:10.1105/tpc.112.107904. [Google Scholar] [PubMed] [CrossRef]
27. Li P, Zhao L, Qi F, Htwe NMPS, Li Q, Zhang D, et al. The receptor-like cytoplasmic kinase RIPK regulates broad-spectrum ROS signaling in multiple layers of plant immune system. Mol Plant. 2021;14(10):1652–67. doi:10.1016/j.molp.2021.06.010. [Google Scholar] [PubMed] [CrossRef]
28. Wang C, Tang RJ, Kou S, Xu X, Lu Y, Rauscher K, et al. Mechanisms of calcium homeostasis orchestrate plant growth and immunity. Nature. 2024;627(8003):382–8. doi:10.1038/s41586-024-07100-0. [Google Scholar] [PubMed] [CrossRef]
29. Liu Z, Wu Y, Yang F, Zhang Y, Chen S, Xie Q, et al. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci U S A. 2013;110(15):6205–10. doi:10.1073/pnas.1215543110. [Google Scholar] [PubMed] [CrossRef]
30. Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020;13(8):1194–202. doi:10.1016/j.molp.2020.06.009. [Google Scholar] [PubMed] [CrossRef]
31. Cen Y, Geng S, Gao L, Wang X, Yan X, Hou Y, et al. Genome-wide identification and expression analysis of RLCK-VII subfamily genes reveal their roles in stress responses of upland cotton. Plants. 2023;12(17):3170. doi:10.3390/plants12173170. [Google Scholar] [PubMed] [CrossRef]
32. Yao K, Wang Y, Li X, Ji H. Genome-wide identification of the soybean LysM-RLK family genes and its nitrogen response. Int J Mol Sci. 2023;24(17):13621. doi:10.3390/ijms241713621. [Google Scholar] [PubMed] [CrossRef]
33. Sreekanta S, Bethke G, Hatsugai N, Tsuda K, Thao A, Wang L, et al. The receptor-like cytoplasmic kinase PCRK1 contributes to pattern-triggered immunity against Pseudomonas syringae in Arabidopsis thaliana. New Phytol. 2015;207(1):78–90. doi:10.1111/nph.13345. [Google Scholar] [PubMed] [CrossRef]
34. Kong Q, Sun T, Qu N, Ma J, Li M, Cheng YT, et al. Two redundant receptor-like cytoplasmic kinases function downstream of pattern recognition receptors to regulate activation of SA biosynthesis. Plant Physiol. 2016;171(2):1344–54. doi:10.1104/pp.15.01954. [Google Scholar] [PubMed] [CrossRef]
35. Li Y, Xue J, Wang FZ, Huang X, Gong BQ, Tao Y, et al. Plasma membrane-nucleo-cytoplasmic coordination of a receptor-like cytoplasmic kinase promotes EDS1-dependent plant immunity. Nat Plants. 2022;8(7):802–16. doi:10.1038/s41477-022-01195-x. [Google Scholar] [PubMed] [CrossRef]
36. Shinya T, Yamaguchi K, Desaki Y, Yamada K, Narisawa T, Kobayashi Y, et al. Selective regulation of the chitin-induced defense response by the Arabidopsis receptor-like cytoplasmic kinase PBL27. Plant J. 2014;79(1):56–66. doi:10.1111/tpj.12535. [Google Scholar] [PubMed] [CrossRef]
37. Du C, Li X, Chen J, Chen W, Li B, Li C, et al. Receptor kinase complex transmits RALF peptide signal to inhibit root growth in Arabidopsis. Proc Natl Acad Sci U S A. 2016;113(51):E8326–34. doi:10.1073/pnas.1609626113. [Google Scholar] [PubMed] [CrossRef]
38. Li DD, Chen QS, Yin T, Xin DW. Identification and analysis of soybean GmPBS1 gene family. Soybean Sci Technol. 2023;3:1–14, 29. (In Chinese). doi:10.20095/j.issn1674-3547.2023.03.001. [Google Scholar] [CrossRef]
39. Hailemariam S, Liao CJ, Mengiste T. Receptor-like cytoplasmic kinases: orchestrating plant cellular communication. Trends Plant Sci. 2024;29(10):1113–30. doi:10.1016/j.tplants.2024.04.006. [Google Scholar] [PubMed] [CrossRef]
40. Verma V, Ravindran P, Kumar PP. Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 2016;16(1):86. doi:10.1186/s12870-016-0771-y. [Google Scholar] [PubMed] [CrossRef]
41. Liu X, Lei Z, Yang Y, Wang Z, Ha S, Lei Z, et al. Genome-wide identification of GhRLCK-VII subfamily genes in Gossypium hirsutum and investigation of their functions in resistance to Verticillium wilt. BMC Plant Biol. 2023;23(1):421. doi:10.1186/s12870-023-04435-0. [Google Scholar] [PubMed] [CrossRef]
42. Feria AB, Bosch N, Sánchez A, Nieto-Ingelmo AI, de la Osa C, Echevarría C, et al. Phosphoenolpyruvate carboxylase (PEPC) and PEPC-kinase (PEPC-k) isoenzymes in Arabidopsis thaliana: role in control and abiotic stress conditions. Planta. 2016;244(4):901–13. doi:10.1007/s00425-016-2556-9. [Google Scholar] [PubMed] [CrossRef]
Cite This Article
 Copyright © 2025 The Author(s). Published by Tech Science Press.
Copyright © 2025 The Author(s). Published by Tech Science Press.This work is licensed under a Creative Commons Attribution 4.0 International License , which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.


 Submit a Paper
Submit a Paper Propose a Special lssue
Propose a Special lssue View Full Text
View Full Text Download PDF
Download PDF Downloads
Downloads
 Citation Tools
Citation Tools